
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 08 October 2020
Sec. Parasitology
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.575881
This article is part of the Research Topic Neglected and Under-Researched Parasitic Diseases of Veterinary and Zoonotic Interest View all 18 articles
 Lucy J. Robertson1*
Lucy J. Robertson1* Øystein Haarklau Johansen2,3
Øystein Haarklau Johansen2,3 Tsegabirhan Kifleyohannes1,4
Tsegabirhan Kifleyohannes1,4 Akinwale Michael Efunshile5,6
Akinwale Michael Efunshile5,6 Getachew Terefe7
Getachew Terefe7Cryptosporidium, a protozoan parasite in the phylum Apicomplexa, is the etiological agent of cryptosporidiosis, an intestinal infection characterized by profuse watery diarrhea. Over 30 species of Cryptosporidium are recognized, some host specific whereas others infect a broader host range. Cryptosporidium hominis and Cryptosporidium parvum are the species most commonly associated with human infection; C. hominis is largely associated only with human infections, but C. parvum is also associated with infection in animals, especially young ruminants. In some regions, cryptosporidiosis is a serious veterinary problem, particularly for calves, and lambs. Many outbreaks of human cryptosporidiosis have been associated with zoonotic transmission following contact with infected animals. In Africa, where cryptosporidiosis is a major contributor to pediatric morbidity and mortality, evidence suggests transmission is principally anthroponotic. Given the frequent close contact between humans and animals in Africa, the apparent predominance of human-to-human transmission is both interesting and puzzling. In this article, after a brief “text book” introduction to the parasite, we consider in separate sections the different aspects of relevance to Cryptosporidium transmission in African countries, describing different aspects of the various species and subtypes in human and animal infections, considering livestock management practices in different African countries, and looking for any characteristic “hot spots” where zoonotic transmission has apparently occurred. Studies where transmission networks have been investigated are particularly relevant. Finally, in a separate section, we try to gather these different strands of evidence together in order to assess the reasons behind the apparent predominance of anthroponotic transmission in Africa. Reviewing the available evidence provides an opportunity to re-think transmission pathways, not only in Africa but also elsewhere, and also to pose questions. Does the predominance of human-to-human transmission in Africa reflect a relative absence of zoonotic C. parvum in African livestock? Are Africans less susceptible to zoonotic Cryptosporidium infection, perhaps resulting from early immunostimulation by C. hominis or due to inherent genetic traits? Is the African environment—in all its variety—simply more detrimental to oocyst survival? Will the so-called hypertransmissible subtypes, currently relatively rare in Africa, be introduced from Europe or elsewhere, and, if so, will they fade out or establish and spread? Our intention with this manuscript is not only to summarize and consolidate diverse data, thereby providing an overview of data gaps, but also to provide food for thought regarding transmission of a parasite that continues to have a considerable impact on both human and animal health.
When Cryptosporidium was first discovered it was considered primarily as a parasite of animals, with the first human cases not identified until some 70 years later. The importance of Cryptosporidium as a pathogen was first really understood in the subsequent decade, and, at this time, transmission was considered to be largely zoonotic. Anthroponotic transmission was soon recognized, and, with the advent of more precise molecular tools, it became clear that there was a multiplicity of species and genotypes with different host-specificities. However, despite frequently being described as “ubiquitous,” there are clearly geographical differences in the distribution of species, genotypes, and transmission routes. We are now more acutely aware of the global disease burden due to cryptosporidiosis, with the brunt of that burden borne by young children in African countries. At the same time, we have the enigma that, despite the closer relationship between people and animals in African countries compared with more industrialized countries, zoonotic transmission seems to occur less frequently in Africa (and some other regions) than in more developed regions, such as Europe.
In this article we explore this further, first giving a general introduction to the parasite itself, then providing a background on the parasite as a zoonosis and some background information on the burden from cryptosporidiosis in Africa, concerning both human and animal health. An overview of the species and subtypes of Cryptosporidium identified in infections in African countries is provided based on published papers, and also current perspectives on the potential for waterborne transmission. It should be noted, however, that identification of species and subtypes of Cryptosporidium is reliant on molecular techniques, which, in turn, require a relatively sophisticated laboratory with steady electricity supply and reagents that must be transported, and stored, frozen. In many parts of Africa, the infrastructure for molecular characterization is not yet developed and this means that our insights are, likewise, patchy.
We then consider animal husbandry in African countries, with emphasis on cattle, the species most associated with zoonotic transmission elsewhere. We tie this to an overview of those places and situations in Africa where zoonotic transmission has apparently occurred and try to identify defining characteristics. Finally, we extract from the previous sections those issues that are relevant regarding possible reasons why zoonotic transmission may occur less frequently in African countries than elsewhere, and compare and discuss their likely effects on transmission routes. In addition, we discuss whether, on both a global basis and from the African perspective, there is likely to be a shift toward an increase or decrease in zoonotic and anthroponotic transmission.
Cryptosporidium is a unicellular parasitic protozoan in the phylum Apicomplexa. Although considered a member of coccidia, evidence indicates that it has a closer affinity with gregarines, a large group of Apicomplexa considered particularly primitive (1). This classification has implications for the survival and spread of this parasite. To date, over 30 species of Cryptosporidium have been identified, some of which are host specific, whereas others are more promiscuous regarding host infectivity. Furthermore, whereas infection with some species of Cryptosporidium tend to be associated with little or no illness, others are particularly pathogenic with severe symptoms, which may even result in mortality. However, whether infection manifests as disease (cryptosporidiosis), and the severity of that disease, also depends on host factors, particularly those associated with host immunity and other health challenges.
Cryptosporidiosis usually manifests as a gastrointestinal disease, with diarrhea the most common clinical presentation. The lifecycle is predominantly fecal-oral, although often indirect with transmission by a vehicle such as water or food (see Figure 1). Although this article focuses on gastrointestinal infection, it should be mentioned that, for some Cryptosporidium species and hosts, respiratory cryptosporidiosis is also relevant. The oocyst transmission stage, which is infectious upon excretion without any requirement for maturation in the environment, is very robust and can be shed in high quantities, both characteristics that facilitate transmission via environmental contamination. When an infective oocyst is ingested it excysts in the small intestine and the released sporozoites invade the epithelial cells, where, in an epicellular location (intracellular but extracytoplasmic), asexual multiplication occurs. The resulting merozoites invade neighboring cells, and sexual multiplication occurs with the production of microgamonts and macrogamonts; following fertilization of the macrogamonts, oocysts are produced that sporulate within the host before being shed in host feces.
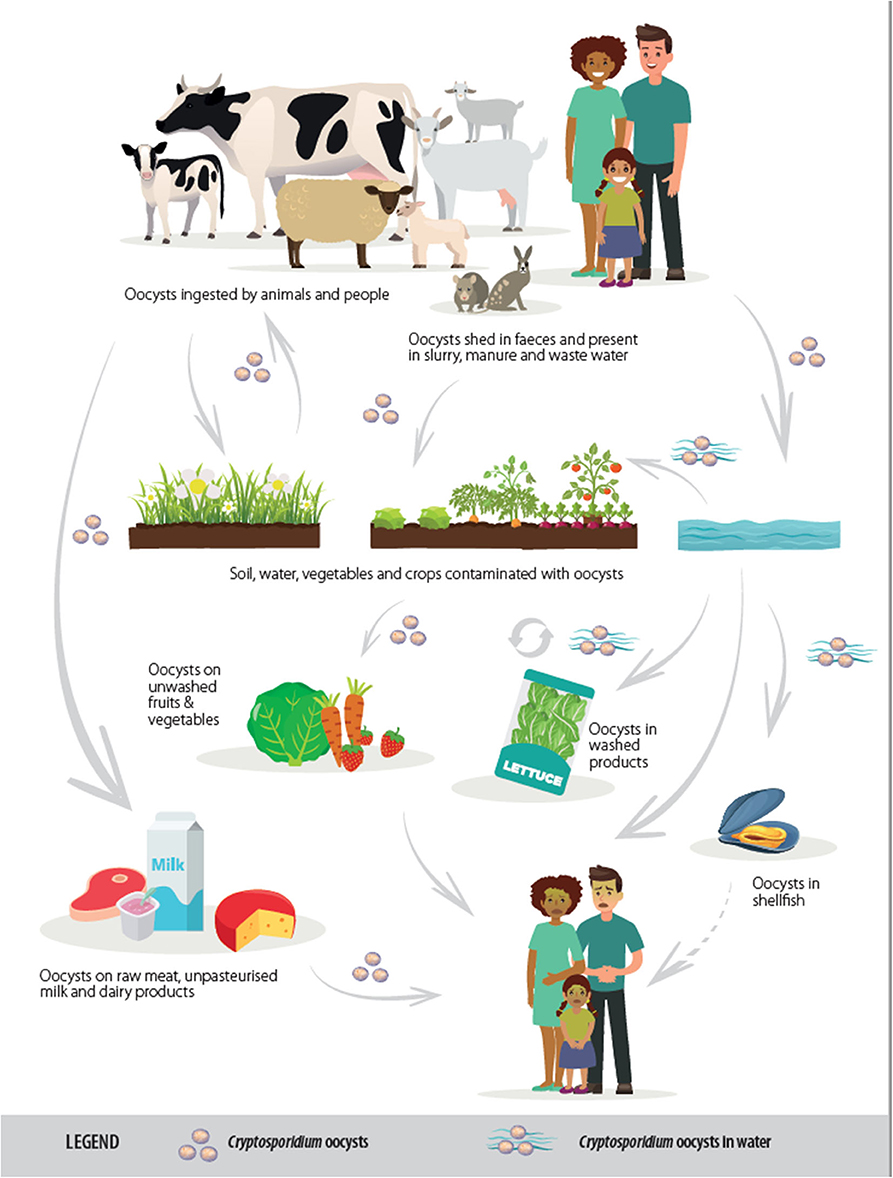
Figure 1. Transmission routes of Cryptosporidium spp. [from (2), copyright European Food Safety Authority].
The pathogenesis of cryptosporidiosis is associated with damage and changes to the cells of the intestinal wall; the interaction of Cryptosporidium sporozoites and merozoites with host cells results in signaling cascades with molecules (such as proteases and hemolysins) damaging cells, increasing fluid secretion, and causing malabsorption. Although usually self-limiting in the immunologically robust host, post-infection sequelae have been reported in human cases, although may reflect host immune responses or gut dysbiosis, rather than infection per se.
Cryptosporidium was first identified as an infection of animals, with human infections not reported until the mid-1970s. Even by 1980 only a handful of human cases had been reported, and, as a primary cause of acute diarrheal disease, it was largely unrecognized until the global HIV pandemic emerged, when Cryptosporidium became one of the first defining entities of AIDS. At this point, the potential for large communitywide outbreaks of waterborne cryptosporidiosis was also recognized, with various sizeable waterborne outbreaks documented.
For many years, only a single species, C. parvum, was really noted as the cause of human cryptosporidiosis, with C. hominis not recognized as a separate species until 2002 (3). Among the 30 or so Cryptosporidium species now identified, C. parvum is considered of substantial veterinary relevance to young livestock (calves and lambs), being considered as one of the most important causes of neonatal enteritis in young ruminants globally (4), and is also considered of major importance as a zoonotic species. Other zoonotic species include C. meleagridis (also commonly found in poultry), C. cuniculus (found in rabbits), and C. ubiquitum (commonly found in sheep); C. hominis largely infects humans. Other species are also generally host specific.
In people, where most infections are caused by C. hominis and C. parvum, the disease is generally acute and self-limiting, with symptom onset within about a week, and causes prolonged or persistent diarrheal episodes more often than other enteric pathogens. However, as treatment options are limited (the only FDA licensed treatment, nitazoxanide, licensed for treating patients aged 1-year and older, is only considered effective in those with healthy immune systems), it can be a serious illness in the very young, malnourished, and immunocompromised. In low-income countries, cryptosporidiosis is a major cause of infectious-disease mortality in children below 2 years (5).
We are now well aware of the multiplicity of species of Cryptosporidium and, in particular, the two main species infecting humans and the important differences in their epidemiology and transmission routes. As oocysts of C. hominis and C. parvum (and of most other zoonotic species) are morphologically identical, determining the infecting species relies on use of molecular tools. Such techniques are now used in many studies to determine species, and, often, subtypes, in Cryptosporidium infections, both in humans and animals, and have provided some insights into distributions and risk factors. However, many publications, even recent, refer to human Cryptosporidium infection as being with C. parvum, even when the species has not actually been determined. In many African countries, the infrastructure and trained personnel for such molecular analyses to determine species are not yet in place and this is reflected in the information available.
As noted, the first infections with Cryptosporidium were identified in animals, and many subsequent reports in the early 1980s concentrated on infections in various animals, with particular emphasis on livestock and rodents. The two first recorded human cases of cryptosporidiosis were from people living on farms (6, 7), and although the likelihood that cattle may have been the infection source was not raised, these cases strengthened the supposition that cryptosporidiosis is primarily an animal infection, with zoonotic potential.
Although cross-infection studies from Cryptosporidium isolates from guinea pigs failed to infect other animal species (8), further cross-infection studies with isolates from calves resulted in infections being established in lambs, calves, pigs, rats, mice, guinea pigs, and chicks (9). The latter authors suggested that their success with cross-infection studies, compared with the lack of success of Vetterling et al. (8), reflected that in their own experiments the challenged hosts were less than 1-day old and specific-pathogen free. Based on their results, they went so far as to propose that Cryptosporidium could be a single-species genus, much like Toxoplasma gondii (9). In retrospect, it is easy for us to see that this confusion arose due to the latter cross-infection studies using C. parvum, and the first cross-infection studies using a host-specific Cryptosporidium species (presumably C. wrairi). However, further experimental infection studies also supported the hypothesis that Cryptosporidium lacks host specificity and should therefore be regarded as a potential zoonosis (10). The first description of cryptosporidiosis in a veterinary student was published in the same year (11) and has been followed by at least a dozen more such reports since then.
Although a review of the taxonomy of Cryptosporidium in 1984, did not support the view of a single species, it concluded that although 19 species had been named at that time, only 4 should be considered valid: C. muris in mammals, C. meleagridis in birds, C. crotali in reptiles, and C. nasorum in fish (12). One year later, C. parvum was proposed to be the Cryptosporidium species infecting most mammals, including humans, distinct from C. muris for which the reported oocyst size was larger (which Levine had apparently overlooked) (13).
Although cryptosporidiosis as a zoonosis was rapidly accepted, at around the same time various studies reported that not all human infections were associated with animal contact. A study in a British children's hospital noted that most patients infected with Cryptosporidium did not have a history of close animal contact, suggesting that person-to-person transmission was as important as zoonotic transmission (14). Similarly, a UK survey of patients with gastrointestinal symptoms found that of the 5% with cryptosporidiosis, contact with animals was not a feature (15). Indeed, an extensive long-term study from Wales (16), culminated with the authors concluding that although animals may be a source of Cryptosporidium infection in people, human-to-human infection probably occurs more commonly, and cryptosporidiosis should not be regarded primarily as a zoonosis.
Most of these first epidemiological studies exploring Cryptosporidium infections, uncovering evidence of both zoonotic and anthroponotic transmission, are from industrialized countries, particularly Europe and North America. However, among these earlier reports are two from Liberia that describe associations of Cryptosporidium infections in children under 5-years of age with a range of different factors (17, 18). As with the reports from UK (15, 16), the authors of the Liberian study concluded by questioning the general belief of that time, that cryptosporidiosis is primarily a zoonosis. They grounded their suspicion on their findings that cryptosporidiosis in Liberian children seemed to occur in regions where domestic animals were uncommon, was associated with household crowding and bottle feeding, and that peak prevalence was among infants still carried on their mothers' backs and thus not in particularly close contact with animals or the wider environment (17, 18). Thus, although no suggestion was made that more than one species of Cryptosporidium may be involved in gastrointestinal cryptosporidiosis in humans, it was recognized from studies, including in Africa, that different epidemiologies could be important, and that there could be a role for both animal-to-human and human-to-human transmission.
It was not until the late 1980s and early 1990s that evidence began to mount, based initially on isoenzyme analysis and thereafter molecular tools, such as PCR-RFLP and sequence investigation, that, as well as there being an animal-to-human or a human-to-human cycle of C. parvum infections, there was also another type of Cryptosporidium that essentially infected solely humans. These two groups were initially designated as the zoonotic “cattle” genotype (usually designated genotype II or sometimes genotype C) and the anthroponotic “human” genotype (genotype I or genotype H); this latter type of Cryptosporidium received a formal species designation, C. hominis, in 2002 (3). It is now well established that not only are there two species of Cryptosporidium causing most cases of human cryptosporidiosis, C. parvum with its two potential cycles of animal-to-human or human-to-human and C. hominis being almost exclusively human-to-human, but that there are also subtypes within these species that also seem to have virulence and host infectivity differences. Indeed, some C. parvum subtypes (e.g., IIc, IIe, and IIm) are apparently almost exclusively limited to human infections, despite the species being generally considered zoonotic. Indeed, C. parvum subtype IIc has recently been proposed as being classified as an anthroponotic subspecies - C. parvum anthroponosum (19). Whereas PCR and sequencing at the SSU rRNA gene is now the most common method for determining Cryptosporidium species, for determining subtype within species most reports use sequence variations in part of the hypervariable 60 kDa glycoprotein (gp60) gene; use of these markers has been described in several publications [e.g., (20–22)]. These molecular tools provide not only a means of exploring transmission pathways in greater detail, but are also useful in outbreak investigations; an outbreak of waterborne cryptosporidiosis in which C. hominis is identified in those infected will point investigators toward considering sewage contamination, rather than runoff from agricultural land.
There is no doubt that cryptosporidiosis has a substantial health impact globally, particularly in lower-income countries. Most African countries are classified using World Bank definitions (23), as having low-income or lower-middle income economies, with the exception of Algeria, Botswana, Equatorial Guinea, Gabon, Libya, Mauritius, Namibia, and South Africa, which are classified as upper-middle income, and Seychelles being high income. Of the 31 countries globally classified as being in the lowest income group, 24 (77%) are in Africa.
One of the earliest studies investigating the impact of Cryptosporidium in an African country was from Guinea Bissau, and demonstrated that Cryptosporidium was associated with excess mortality in children younger than 12 months, with this excess mortality persisting into the second year of life (24). Although this impact from cryptosporidiosis in particular countries has long been assumed, the first comprehensive data demonstrating this were produced relatively recently, from the Global Burden of Disease (GBD) and the Global Enteric Multicenter Study (GEMS) outputs [e.g., (5, 25, 26) etc.]. These studies provided the first global estimates on impacts of cryptosporidiosis (among other diseases) in different age groups and different countries, in terms of mortality, morbidity, and disability-adjusted life-years (DALYs). A meta-analysis published in 2018 showed that earlier reports probably under-estimated the true burden by not taking into account impacts occurring after the acute phase of infection, such as decreased growth, particularly weight gain, and a greater risk of subsequent episodes of infection (27). As Cryptosporidium diarrhea damages gut endothelial cells and microvilli, absorption of macronutrients, and micronutrients are impaired (28, 29). In addition, Cryptosporidium-related malnutrition results in secondary impairment of cell-mediated immunity, which is associated with increased susceptibility to other infectious diseases. Other long-term sequalae include reduced cognitive development, poor school performance, and elevated risk of cardiovascular and metabolic diseases later in life (30, 31), all likely to have a disproportionate effect on the global poor.
Cryptosporidium infection in children under 5 years was estimated to be associated with 44.8 million diarrheal episodes and 48,300 deaths globally (27). Of these, the vast majority were from Africa, accounting for 75% of the diarrheal episodes and 88% of the deaths (27). In particular, the burden of Cryptosporidium-associated diarrhea is greatest in Sub-Saharan Africa, especially Nigeria and the Democratic Republic of the Congo (DRC) where about 48% of the under-5 associated deaths occur (27). When including downstream effects of growth shortfalls associated with cryptosporidiosis, it was estimated that the burden of this parasite could be 2.5 times higher than previous estimates (27), and recognized that accounting for the direct or indirect burden of asymptomatic infections could elevate these estimates even further.
It is well known that whereas infection with some species of Cryptosporidium has apparently marginal impact on host health, ruminants, particularly young animals, infected with C. parvum may suffer from profuse watery diarrhea, inappetence, lethargy, and dehydration; it is not unusual for death to occur, particularly in neonates. With an infectious dose for neonatal calves as low as 17 oocysts (32), ensuring that young stock are not exposed to an infectious dose on farms where other stock are already infected can be challenging. As with humans (see previous subsection), it has also been shown that severe cryptosporidiosis in calves and lambs may have long-term consequences regarding growth, weight gain, and productivity (33–35), as well as the more immediate effects from the acute infection.
Cryptosporidiosis outbreaks on farms are not commonly investigated and reported, but have been described among cattle and goats from farming enterprises in Europe and Asia [e.g., (36–38)]. Although there are no published reports of cryptosporidiosis outbreaks among livestock in Africa, several studies from different African regions have reported on calf diarrhea without any clear attribution to a specific etiological agent (39–41). Given that a variety of etiological agents, as well as Cryptosporidium, can cause calf diarrhea (e.g., rotavirus, coronavirus, bovine viral diarrhea virus, E. coli, Clostridium perfringens, Salmonella spp., and coccidia such as Eimeria zuernii and E. bovis), these data do not necessarily indicate cryptosporidiosis. A similar situation applies to lambs and goat kids.
A systematic review of Cryptosporidium infections in livestock (42) noted the prevalence being highest in the Americas and Europe—and commented that under-investigation in particular regions was not the reason for this skewed distribution. However, publication bias and insufficient information may have excluded some relevant studies (42). Nevertheless, some researchers report that Cryptosporidium oocysts are frequently detected in diarrheic calves in different African countries (43–46), but usually without determining whether cryptosporidiosis is the cause of the symptoms. Many of these studies use modified Ziehl-Neelsen (mZn) for identification; this has low sensitivity and specificity, and does not enable identification of the Cryptosporidium species. Studies using more accurate tests have revealed contrasting results; for example, a study from Tanzania used, in addition to mZn, immunofluorescent antibody testing (IFAT), auramine phenol staining, and molecular methods to investigate calves for Cryptosporidium infection and, using the latter methods, did not detect Cryptosporidium shedding in 943 calf samples, of which over 6% were diarrheic, despite some positive results with mZn (47). The authors suggest that data obtained using mZn should be treated with caution. In contrast, a study in Egypt using molecular tools reported a 32% Cryptosporidium prevalence in cattle (48). Two studies from Ethiopia, both of which used molecular methods for determining infecting species, provide contrasting data: a study from two large dairy farms in central Ethiopia showed that Cryptosporidium infection was common (40% cumulative incidence), with C. parvum most common in pre-weaned calves and C. andersoni in post-weaned calves (49). In contrast, an earlier study in an overlapping area, included 449 calves from both smallholder farmers and dairy farms and detected less than 10% infection, with C. andersoni, C. bovis, and C. ryanae identified, but not C. parvum (50). It is noteworthy that whereas the first two studies (48, 49) associated Cryptosporidium infection with a calf-health impact, the latter (50) reported that the calves were generally healthy, with only a few cases of watery diarrhea.
Two reviews of Cryptosporidium in Africa were published relatively recently (51, 52), the latter of which also considers Giardia. Although these articles have slightly different overall perspectives, both contain information on molecular epidemiology and have assimilated data from the literature describing the occurrence of different Cryptosporidium species/genotypes in various host species in different regions. Aldeyarbi et al. (51) used a defined literature search to gather data, but the authors excluded studies considered to be weakly designed or biased (although how these criteria were determined is unclear). The conclusion from this review is that both anthroponotic and zoonotic transmission cycles have potential for infecting people in Africa, that infections in wild animals are “essential contributors” to environmental contamination that threatens human health, but that C. hominis was the predominant species infecting people in many studies, regardless of host immune status (51). In addition, the authors noted that among human C. parvum infections in various sub-Saharan African countries, the GP60-subtype family IIc [previously Ic, proposed now as C. parvum anthroponosum; (19)] often predominates. As noted, this is a human-adapted subtype occurring almost exclusively in human infections. A recent systematic review and meta-analysis investigating geographical distribution of this subtype in human infections (53) found its occurrence was associated with countries with low GDP per capita and poor sanitation; of 81 relevant single-country articles included in their study (20 from Asia, 20 from Africa, 21 from Europe, 7 from North America, 3 from South America, 10 from Australia/Oceania), 35 reported the presence of C. parvum IIc subtype (C. p. anthroponosum), of which 14 were from Africa (53). Thus, we can extrapolate a significant association between this particular subtype and Africa, as compared with the rest of the world combined (p < 0.0086). Furthermore, the proportion of this subtype among C. parvum isolates in those studies reporting its presence was also higher in Africa (2–100%; mean 75%, median 76%) compared with the rest of the world (3–100%; mean 36%, median 22%).
In a study based on the GEMS data, but looking specifically children younger than 2 years, in some regions of sub-Saharan Africa and south Asia, of 28 C. parvum infections from Mali (n = 13), Kenya (n = 9), Mozambique (n = 5), and Gambia (n = 1), all were anthroponotic IIc or IIe (54). In industrialized countries, subtype IIaA15G2R1 predominates among both dairy cattle and human C. parvum infections, and is sometimes described as hypertransmissible (55, 56). However, in Africa, this subtype has been reported from both cattle and humans in only two countries, Egypt and Tunisia, and also in people in Nigeria—indicating that it does not (yet) predominate here. Thus, despite presently predominating in some circumstances and regions, whether it always or intrinsically transmits more successfully has not been clearly demonstrated.
A relatively high frequency of C. meleagridis infections has been reported in African studies, as mentioned by Aldeyarbi et al. (51) (of the studies considered, the authors report C. meleagridis among 21% of the immunocompromised and 10% of non-immunocompromised people, compared with below 1% in the developed world). Although C. meleagridis is also a zoonotic Cryptosporidium species, many of the papers from Africa do not indicate an association with infected animals or birds [although an association with chicken C. meleagridis infections has been suggested in Côte d'Ivoire (57) and Nigeria (58)] and some actually indicate a lack of association with chicken infections [e.g., (59)], perhaps suggesting transmission from a human source.
The more-recent review paper (52) tabulates Cryptosporidium infections in papers from African countries published from 2010 to 2016 according to prevalence (occurrence) in human cohorts (along with information on diagnostic technique), and by Cryptosporidium species and genotype according to patient group. Similar data for animal hosts, both wildlife and domestic animals, are tabulated separately. This is a comprehensive undertaking (although data published separately from the same research studies are sometimes listed twice), and, similarly to Aldeyarbi et al. (51), the authors note the predominance of anthroponotic transmission, with C. hominis and anthroponotically transmitted C. parvum being reported principally in human infections.
Using a literature survey of PubMed (using the search terms of each African country in turn along with Boolean AND and cryptosporidi*) we identified a further 45 papers not included in the Squire and Ryan (52) review, 34 of which had been published from 2017 and onwards (Figures 2, 3). Of these additional articles, 17 involved Cryptosporidium in human hosts only, 19 involved animal hosts only, and 9 considered both human and animal hosts. One article was from the Gambia (60), a country not featuring in the Squire and Ryan (52) review. This article reported more closely on Cryptosporidium infections in children enrolled in the GEMS study, and, although most (>80%) were C. hominis, a significant association with animals (cats or cows) living in the compound was also reported (60). However, given the host specificity of C. hominis, and that these animals were not themselves tested for infection, the presence of animals in the compound could be an indicator of another risk factor, rather than being the infection source.
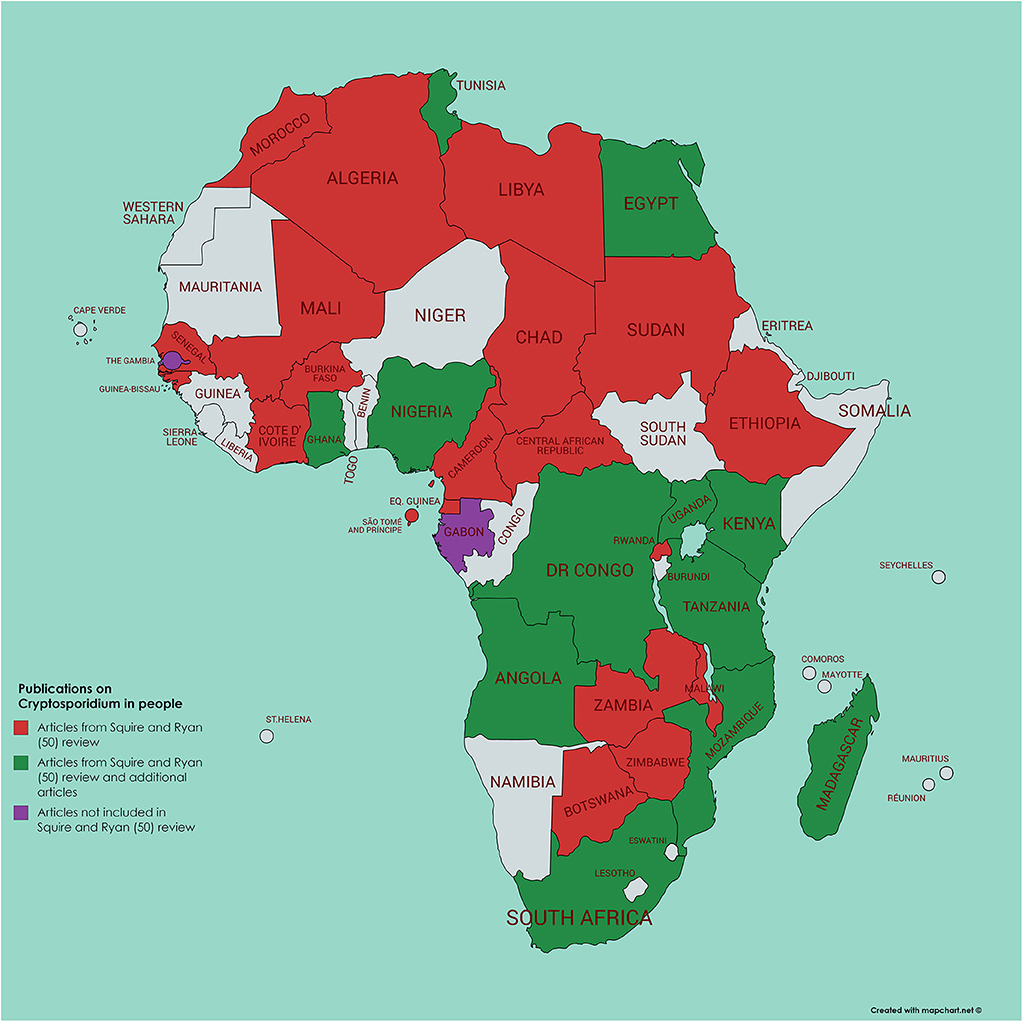
Figure 2. Map of Africa indicating those countries where human infection with Cryptosporidium has been investigated; articles either referenced in Squire and Ryan (52) or identified by current literature search.
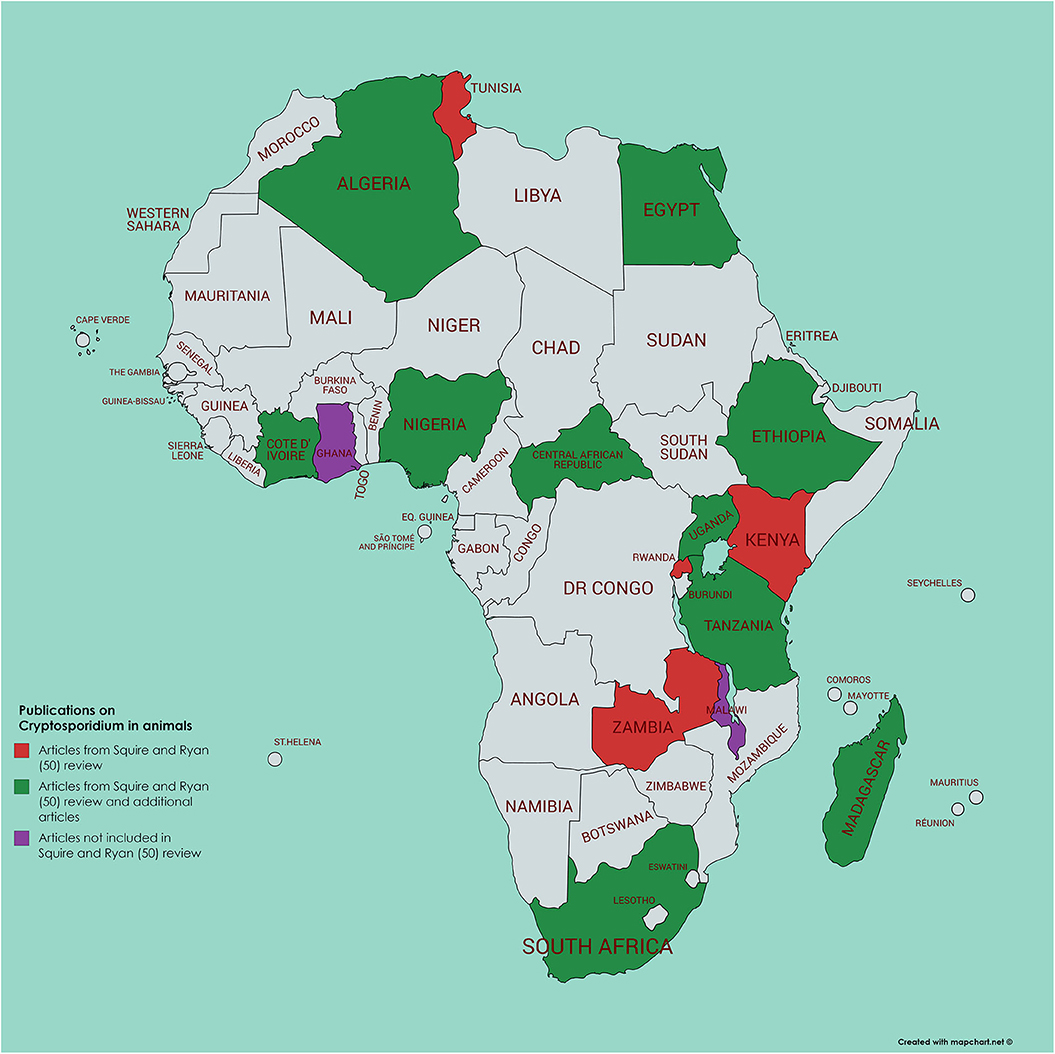
Figure 3. Map of Africa indicating those countries where animal infection with Cryptosporidium has been investigated; articles either referenced in Squire and Ryan (52) or identified by current literature search.
Of relevance regarding C. hominis infections in Africa, is that subtype IbA10G2, which is associated with most outbreaks in industrialized countries (22, 61–63), and has been described as being hyper-virulent (55, 61, 62), despite evidence for differences in clinical symptoms or advanced transmission within gp60 allele families being weak, seems to occur rarely in Africa. It was not found in a subtyping performed with the GEMS study samples (54), and has been reported only sporadically in surveys [from 3 children (of 28 with subtyped C. hominis) in Nigeria (64); from 1 HIV/AIDS patient (of 19 with subtyped C. hominis) in Ethiopia (65); a maximum of 2 children (of 19 with subtyped C. hominis) in South Africa (66); and 1 HIV and TB patient (of 2 typed with C. hominis) in Mozambique (67)].
Among the 32 articles investigating Cryptosporidium in animals in Africa included by Squire and Ryan (52), 16 (50%) reported on cattle and 4 reported on sheep or goats; in the additional articles that we identified, 8 reported on cattle and 7 on sheep and/or goats. These articles do not indicate any clear patterns regarding infectious species or subtypes, although one with data from cattle reported the presence of the “hyper-transmissible” IIaA15G2R1 subtype (68); however, as reported from other global regions, the data indicate that younger animals (both calves, lambs, and goat kids) seem more likely to be infected with C. parvum than older animals (69). Nevertheless, age does not seem to be the only determinant regarding infection with zoonotic Cryptosporidium species in cattle husbandry. For example, a Zambian study investigating the species of Cryptosporidium in calves demonstrated that whereas calves from intensive dairy farms and extensive commercial beef farms (mean calf age 15 and 26 days, respectively) were largely infected with C. parvum, among calves infected with Cryptosporidium on small traditional farms (mean calf age 22 days), only C. bovis was identified (70).
Among small ruminants, the occurrence of C. parvum infection seems to be rather low in studies from Africa; among the 6 studies compiled by Squire and Ryan (52), of those including small ruminants (sheep, goats), only 3 (50%) reported the presence of C. parvum, with the other studies reporting only C. xiaoi. Of the more recent papers, a couple with very low numbers of samples (between 1 and 8 samples) report C. parvum in sheep and goats [e.g., from Nigeria (58); from Sudan, (71); from Uganda, (72)]. Larger studies, however, indicate that C. parvum occurs relatively infrequently in African small ruminants, with C. xiaoi predominating in sheep and goats in Ghana (73) and a study in Ethiopia including 389 lambs under 5 months of age found only C. ubiquitum (74). Although C. ubiquitum has zoonotic potential (75), human infections in Africa have been reported only extremely rarely [one study from Nigeria, with two publications noted by Squire and Ryan, (52)]. Of interest is that the single C. parvum isolate from a goat in the study from Ghana was typed as being the anthroponotic IIc subtype (C. p. anthroponosum); with molecular methods being the only analytical tool used, it is possible that this single C. parvum infection among 285 goat samples of which 95 were positive, represents carriage rather than infection. Furthermore, a comprehensive contact-network analysis study conducted in 4 African countries [Gabon, Ghana, Madagascar, and Tanzania; (76)], not only reported that C. hominis predominated among human isolates (from children below 5 years), but also that C. hominis occurred not infrequently in their animal contacts (goats, sheep, cows, dogs). However, given that these animals may well have ingested feces of infected children, it is unclear whether we can infer zoonotic transmission here rather than carriage in those animals in which C. hominis DNA was detected (12 cows, 5 goats, 1 sheep, and 3 dogs), given how rarely this species has previously been identified in animal infections. Indeed, the authors themselves emphasize that human-to-human transmission appears to be the predominant route in their 4 study sites, with zoonotic transmission contributing only marginally (76). It is unfortunate that environmental samples were not analyzed in this study, as this could, potentially, have added even greater weight to the study findings.
In addition to the two review papers concerned with Cryptosporidium in Africa (51, 52), a slightly older review considers Cryptosporidium in the Arab world (77), which also includes some countries from Africa (specifically Algeria, Comoros, Djibouti, Egypt, Libya, Morocco, Mauritania, Somalia, Sudan, and Tunisia). Although the authors concluded that zoonotic transmission is important, little supportive molecular evidence was presented.
The potential for water to be a transmission vehicle for Cryptosporidium is accepted globally, with communitywide outbreaks and smaller outbreaks reported from multiple countries. The dearth of such outbreaks being reported from Africa probably represents limitations in technological capabilities and surveillance systems (78). It must also be considered that with a high background prevalence of diarrheal disease (of whatever etiology)—albeit varying regionally (79)—it is probably more difficult for an outbreak to be identified in Africa unless extremely dramatic. Even in countries with a relatively low incidence of diarrheal diseases and well-developed reporting systems, identifying a cryptosporidiosis outbreak may not be straightforward; not all cases seek medical attention, doctors may not request stool samples, and stool samples may not be analyzed appropriately (80, 81). Thus, lack of reporting of cryptosporidiosis outbreaks does not necessarily mean they do not happen. In order to improve our understanding of whether such outbreaks occur, ensuring the etiology of diarrhea in African countries is diagnosed and reported is probably the best place to start, rather than analyzing water samples. Although such analyses provide clues, the procedures are expensive and result interpretation may be difficult.
A review (82) identified 60 papers addressing Cryptosporidium in water in Africa. However, from the information presented it is difficult to extrapolate how many were concerned with surveillance of drinking water, the analytical methodology used, and the results obtained. By means of a literature survey of PubMed (using the search terms of each African country in turn along with Boolean AND and cryptosporidi* AND water) we identified just 21 papers from Africa (originating from 8 countries) for which drinking water (of different types) had been analyzed for contamination with Cryptosporidium (see Figure 4 and Supplementary Table 1). Of these, 8 had been published after the Ahmed et al. (82) review, so would not have been included there; of the 13 remaining, 5 were cited by Ahmed et al. (82). Of relevance is that many of these 21 papers used methods that seem unlikely to provide convincing results (e.g., sample volumes as low as 10 ml, with minimal processing steps, and mZn for detection of oocysts). Nevertheless, it is these articles that often provide data indicating the highest proportion of samples considered positive (over 40%), while papers using standardized methods for water analysis (those of ISO or US Environmental Protection Agency) tend to report lower proportions of positive samples [e.g., the work of Morris et al. from Kenya (83), Kifleyohannes and Robertson from Ethiopia (84), or Potgjeter et al. from South Africa (85)]. This suggests that the lack of specific detection techniques used in other studies may have resulted in false positive results. In addition, some of the studies using non-specific methods report extremely high concentrations of oocysts (tens or hundreds per liter), which may also indicate false positives (or excessively high contamination). None of the studies report subtypes of Cryptosporidium in the water samples, but 10 reported species, all of which mention C. parvum, with one of these from Egypt also noting the predominance of C. hominis (86) and one from Kenya reporting 6 samples with C. parvum and 3 containing C. andersoni (87). In this Kenyan study, the samples in which Cryptosporidium was detected were from water with likely animal contamination, as cattle were watered there and elephants were known to use it (87). However, the evidence of animals being the source of the contamination is weak. Among the other 8 articles in which C. parvum was reported from the water samples, one [from the same region of Kenya as that of Muchiri et al. (87)], reports Cryptosporidium in one of 14 samples, with C. parvum detected (88). However, the other 6 articles (published between 1997 and 2019), although reporting C. parvum, did not apparently undertake any molecular analyses, and the species definition appears to be based upon supposition rather than results (Supplementary Table 1).
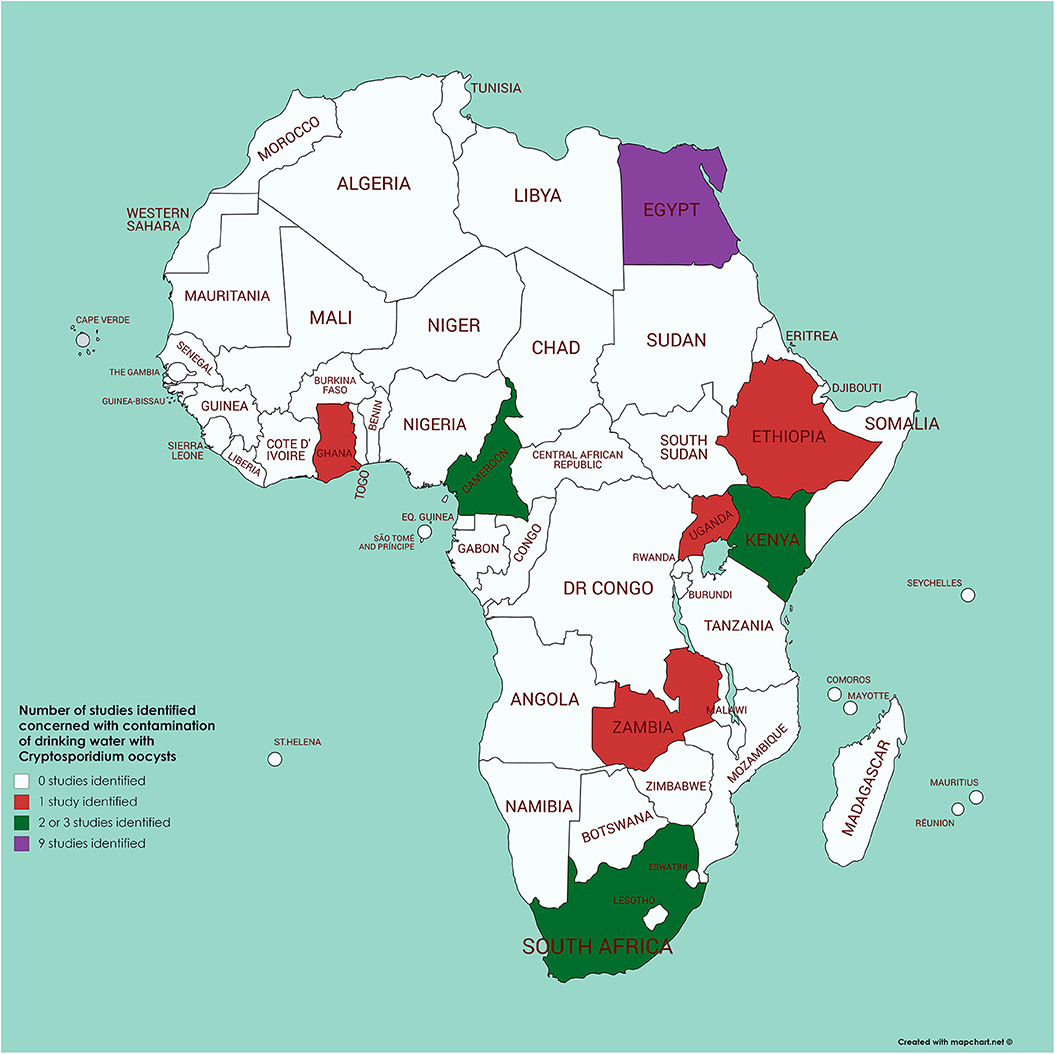
Figure 4. Map of Africa indicating those countries where contamination of drinking water with Cryptosporidium has been investigated (see Supplementary Table 1 for further details).
Two recent articles from the same research project used a modeling approach to consider Cryptosporidium contamination of rivers globally (89) and disease burden due to Cryptosporidium in surface water in sub-Saharan Africa explicitly (90). Worth noting is that hot spots for river contamination were identified in Nigeria, Algeria, and South Africa, with human contamination (point sources) considered to dominate over contamination from animals (diffuse sources). However, in some African countries (e.g., DRC, South Sudan, Chad, Ethiopia) more diffuse sources may predominate, although contamination of river water with oocysts in these countries was also considered to be amongst the lowest globally (89). Using not only the modeled contamination data, but also information on the proportion of population using surface water as a drinking water supply, along with drinking water treatments, Limaheluw et al. (90) estimated the cryptosporidiosis burden due to oocysts in the surface-water drinking water supply in sub-Saharan Africa to be 1.6 × 106 DALYs. The highest number of DALYs per 100,000 of population were in Eswatini (1022.8), Mozambique (828.5), and Kenya (715.2), and the lowest in Senegal (1.3). The extremely high DALYs in south and south-east Sub-Saharan Africa were partly explained by higher estimates of life-years lost in people with HIV/AIDS (90).
Zoonotic transmission of Cryptosporidium is most likely to originate from domestic livestock (cattle, sheep, goats), although other animals, notably rabbits (C. cuniculus) and poultry (C. meleagridis), may also be important sources. It is therefore relevant to consider ruminant livestock production systems in Africa, particularly those in countries with large cattle populations (91), when considering the potential for zoonotic transmission of Cryptosporidium. Livestock production systems vary between and within continents and countries and are classified on the basis of different criteria and metrics (92). In Africa, classification of livestock production systems is complicated by a plethora of factors such as sociocultural, agro-climate, land use, livestock densities and levels of intensification. On the basis of agro-climate or how feed for the animals is produced, Africa is dominated by land-based production systems, predominantly “mixed, rain-fed” and “grazing” (93). The pastoral grazing system occurs mainly in arid and semi-arid areas and the mixed rain-fed system is common in the humid, semi-humid, and tropical highland areas, but also occurs in the arid-semiarid climatic zones (94). Grassland utilization ranges from total nomadism (no permanent place of residence, no regular cultivation) via semi-nomadism, transhumance, and partial nomadism, through to stationary animal husbandry (92). Ruminant livestock production can also be divided by intensity of production, from extensive to intensive. Extensive production ranges from small-scale, subsistence production (smallholder farms) dependent on mixed crop-livestock systems to large pastoral holdings that rely mainly on rangeland grazing. In contrast, intensive production system involves geographically-concentrated and commercially-oriented specialized production that may develop into industrialization, possibly involving multinational firms and contract farming (92). Although both intensive and extensive production of ruminant livestock occurs in Africa, extensive and pastoral systems tend to predominate. Of relevance is that calving rate is relatively low (overall about 60%) and calf mortality relatively high (around 20%) in the majority of management systems in Africa (94, 95) compared with rates reported from other countries. Similarly, for small ruminants, lamb and kid mortality risk is high, although the production rate is high and prolificacy between 1 and 1.5, lamb and kid mortality risks are equally high (94). These parameters, in association with year-round breeding, mean that neonate density tends to be relatively low in most production systems.
The largest sub-Saharan African livestock populations are in East Africa with 55.3% of the total livestock units, followed by West Africa, Southern Africa, and Central Africa, with 27.1, 9.4, and 8.2%, respectively (94). In arid and semi-arid zones, the dominant species are goats and sheep followed by cattle, whereas in sub-humid zones, cattle predominate followed by goats and sheep with the highest densities of livestock in the highlands (94). Two countries in East Africa, Ethiopia and Sudan, have far higher cattle populations than other countries in the region. An overview table of some relevant key figures regarding cattle production in some African countries (Burkina Faso, Egypt, Ethiopia, Kenya, Nigeria, Uganda) is provided (Table 1), in which the data have been extracted from the series of FAO publications concerned with livestock production systems (96).
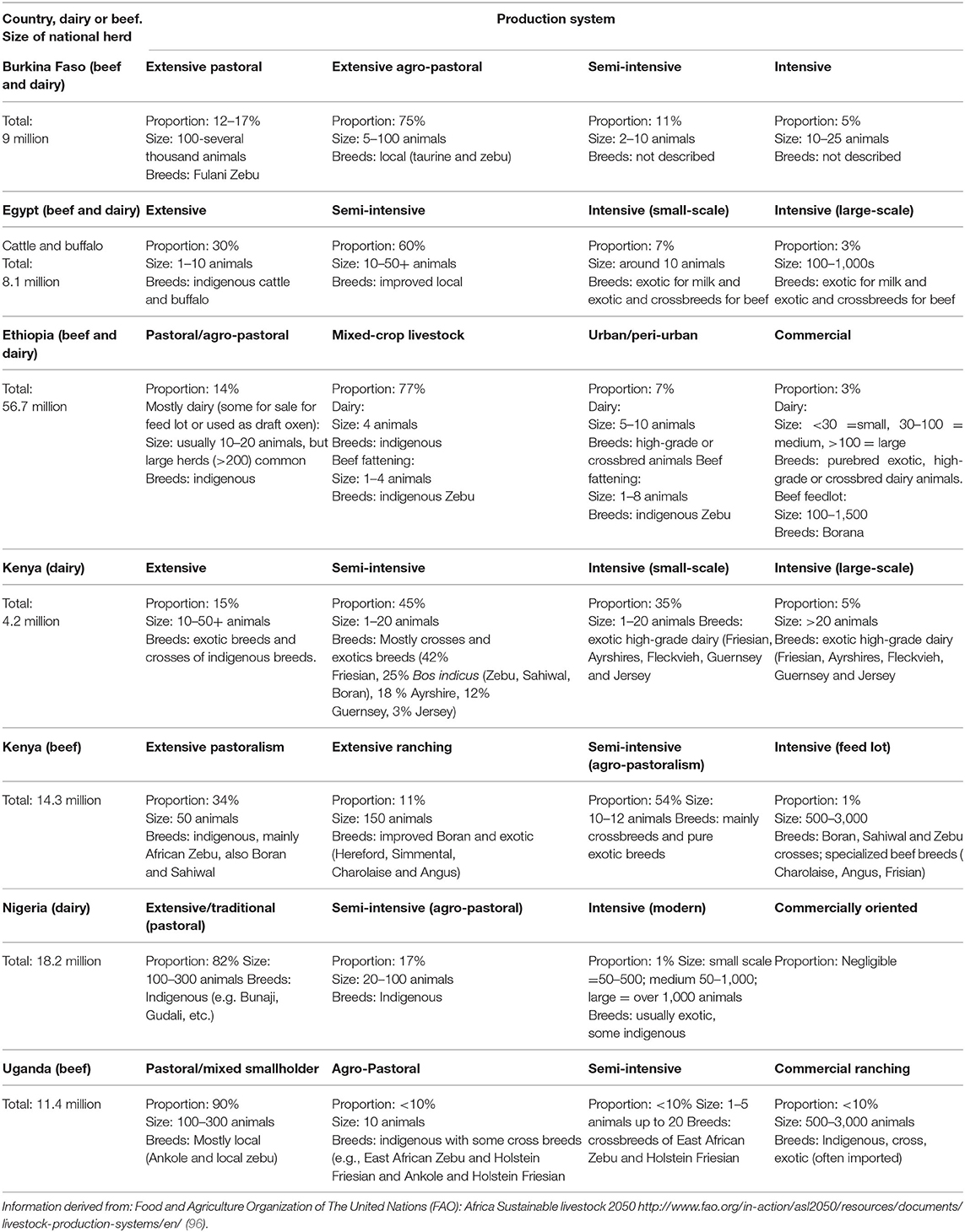
Table 1. Relevant comparative data of different cattle-production systems in some African countries.
Given the heterogeneous cattle-production systems, the varying lifestyles of human populations, and the climatic variability in Africa, the environmental load of Cryptosporidium oocysts and the risk of human infection from them probably vary significantly throughout the continent. In arid and semi-arid environments, where cattle are kept under extensive management systems in pastoral and agro-pastoral settings, large numbers of cattle graze together. Although this can potentially lead to high pasture contamination, it tends to be seasonal as pastoralists move their animals in search of pasture and water, and Cryptosporidium oocyst loads are expected only to be higher where these resources are available (97). In the pastoral cattle management system, cows with younger calves remain at home under the management of women and children on the limited food reserved for them, while other animals may travel long distances. Whether such division of labor and animal care among family members increases the risk of transmission of Cryptosporidium to young children requires further investigation.
By numbers of livestock, Ethiopia ranks first in Africa. The cattle production sector here is highly heterogeneous, comprising both traditional pastoral/agro-pastoral and mixed crop–livestock production systems and the market-oriented, intensive and specialized producers. Pastoral/agro-pastoral production dominates in the Ethiopian lowlands, where livestock are managed under pasture-based extensive systems. Cattle dominate the livestock population here, accounting for 25% of the national herd (Table 1). The mixed crop-livestock system of Ethiopia carries more than 70% of the cattle population, with extensive management systems and supplementation from crop residues [Table 1, (98)]. This system occurs often in densely populated regions where animals are frequently kept close to residential areas. Cattle manure here is used as fertilizer and for fuel as dried dung cakes, mainly prepared by women and children, potentially resulting in a high risk of exposure to Cryptosporidium oocysts (and other pathogens) (99).
Specialized commercial dairy systems (exotic and cross breeds) and feedlot (local zebu) operations in Ethiopia are concentrated in densely populated urban and peri-urban settings and constitute a very small fraction of the livestock population (98). However, in terms of environmental load of Cryptosporidium oocysts, these operations probably represent major hot spots in comparison with traditional or extensive livestock management. Interestingly, Aldeyarbi et al. (51) mention that most C. parvum infections seem to occur in urban settings in Africa, where, according to these authors, animals are not found close to residences. However, as described above, even in residential areas, people and animals often live in close proximity and Aldeyarbi et al. (51) may have under-estimated the extent of such urban-based farming in some areas of Africa.
Dairy cattle production in Kenya is the second largest contributor to the agricultural GDP and is classified into three production systems: intensive, semi-intensive, and extensive. Unlike in Ethiopia, the intensive and semi-intensive dairy farms predominate in Kenya [Table 1; (100)], whereas all beef production is by the extensive system. The intensive system is predominant in the Mount Kenya and Central Rift Valley regions, where crop production is also practiced. It is also common in many urban and peri-urban centers in humid and sub-humid areas of the country. In the extensive dairy production system, 3% of farms hold 35% of the dairy cattle population, and 70% of the national livestock herd is found in Kenya's arid and semi-arid lands (100).
In Nigeria there are three dairy cattle production systems: extensive or traditional, semi-intensive (agro-pastoral), and intensive (modern) system [Table 1; (101)]. Whereas commercial farms raise imported exotic breeds and their crosses, local breeds predominate. These are mainly managed by semi-sedentary and transhumant pastoralists in large herds. However, compared with herd sizes in North America, these may be considered relatively small; for example, concentrated animal feeding operations (CAFOs) predominate in some countries, with more than 70% of beef produced in the USA in 2002 being derived from CAFOs holding more than 5000 head of cattle (102). Commercially oriented, urban cattle farming has started to emerge in Nigeria, but is still relatively marginal.
As Egypt has limited natural pastures, cattle (and buffalo) production here is well integrated with cropland. There are three main production systems: intensive, semi-intensive, and extensive [Table 1; (103)]. The semi-intensive system is dominated by improved local breeds, producing both beef and milk and comprise almost 60% of the total bovine population.
With the majority of articles from Africa indicating the predominance of C. hominis in human cases of Cryptosporidium infection, and as many livestock infections in Africa are with non-zoonotic species, such as C. xiaoi, we tried to identify African studies providing convincing evidence of zoonotic transmission (“black swan” events). The intention was that such studies may indicate risk factors for transmission not occurring in other settings in Africa—given that the overwhelming number of studies suggest that non-zoonotic transmission predominates.
A review of the literature identified some studies in which the titles or abstracts indicated that the authors were considering zoonotic transmission of Cryptosporidium in specific settings. Most of these studies were based upon investigating prevalence, sometimes species, and less frequently subtype, of Cryptosporidium infections in people and animals residing in similar areas and inferring from these data any potential for zoonotic transmission [e.g., (104, 105)]. Less frequently case-control type studies were reported in which the prevalence and species of Cryptosporidium in people reporting close contact with animals were compared with prevalence and species in people reporting little or no contact with animals [e.g., (106)]. However, the large number of possible confounders and difficulties in appropriate matching of those with and without animal contact means the results of such studies are difficult to interpret. Finally, contact-network analysis, in which samples are analyzed from both human and animal contacts of people infected with Cryptosporidium, was reported in two papers (48, 76). Those studies that have collected the most information (species, subtype, matched case-control, or contact-network analysis) are likely to provide the strongest indications regarding the likelihood of zoonotic transmission. It is therefore interesting that the two papers providing the most detailed analysis, with molecular investigation of Cryptosporidium isolates from humans and animals along with cluster analysis of the results (48, 76), both suggest that animal-to-human transmission is a minor, and probably separate, transmission component, although does occur.
Nevertheless, some studies indicate some likelihood of zoonotic Cryptosporidium transmission, and it is thus relevant to consider these more closely. For C. parvum, some articles from Egypt, Ethiopia, Kenya, Nigeria, São Tomé and Príncipe, and Tunisia indicate the potential for zoonotic transmission. These are summarized in Table 2. We considered each article in turn, and, based on the information reported, gave a subjective analysis of the strength of evidence (strong, moderate, or weak) of zoonotic transmission provided in the situations described. Thus, articles were only considered where subtype families IIa or IId were identified. The evidence was considered strong if similar subtypes were reported from animals and humans with a plausible connection in time and space, or with other strong epidemiological and statistical evidence of animal contact being a risk factor. The evidence was scored as moderate if the study reported zoonotic species or subtypes from ≥ 3 humans, plus at least one of the following: detection by at least one other testing modality (e.g., microscopy), immunocompromised person, and gastrointestinal symptoms. The additional criteria were used to increase the probability that detection reflected infection—either because the symptoms indicated infection or because of host susceptibility. The additional testing modalities were also an attempt to exclude molecular detection reflecting transient passage rather than infection. For articles that presented neither strong nor moderate evidence, the evidence was classified as “weak.”
Of the 10 studies identified, only three, two from Egypt, and one from Tunisia, provided strong evidence that zoonotic transmission might have occurred. Thus, on the whole, the majority of articles do not provide convincing evidence of any “hot spots” of zoonotic transmission, and the overall picture is that human infections of Cryptosporidium are predominantly C. hominis and non-IIa/IId C. parvum. Regarding the two countries (Tunisia and Egypt) where the evidence for zoonotic transmission was relatively strong, we speculate cautiously that there are some relevant factors. Both countries are classified as lower-middle income economies and considered to have a relatively strong commercial sector and drive for nationally produced meat and dairy products (113). In addition, due to geographic and climate factors in both countries, cattle-raising land is restricted, being basically only available in the northern areas (specifically the districts of Beja and Bizerte) in Tunisia and along the Nile and in the Nile delta in Egypt. Thus, by necessity, there is close integration between cattle raising (both dairy and beef) and human settlements in both countries. However, with just a few papers from both countries, it is incorrect to label them as transmission hot spots, and many other regions also have close associations between people and cattle raising.
Other articles that indicate zoonotic transmission are concerned with species other than C. parvum, with some circumstantial evidence of C. meleagridis, C. muris, C. canis, C. suis, C. ubiquitum, and C. xiaoi infections in people and associated animals (see Table 3). As with the C. parvum articles, these too have been evaluated for the strength of evidence indicating zoonotic transmission. Interestingly, we were unable to find any that provided strong evidence of zoonotic transmission, and probably the strongest indicator that these are examples of zoonotic transmission are reflected in that these are typically animal-associated species and that in several of the studies the infected people are immunocompromised, and therefore probably likely to be susceptible to pathogens that do not tend to infect humans. As pointed out in an article that also discusses the public health threat from zoonotic enteric protozoa in wildlife (130), the terminology may be loaded and it is questionable whether a pathogen should be considered zoonotic that usually infects only animals and is reported rarely in low numbers from a highly immunocompromised human patient. The example given in that article is C. suis, and such pathogens are described therein as “potentially zoonotic” (130).
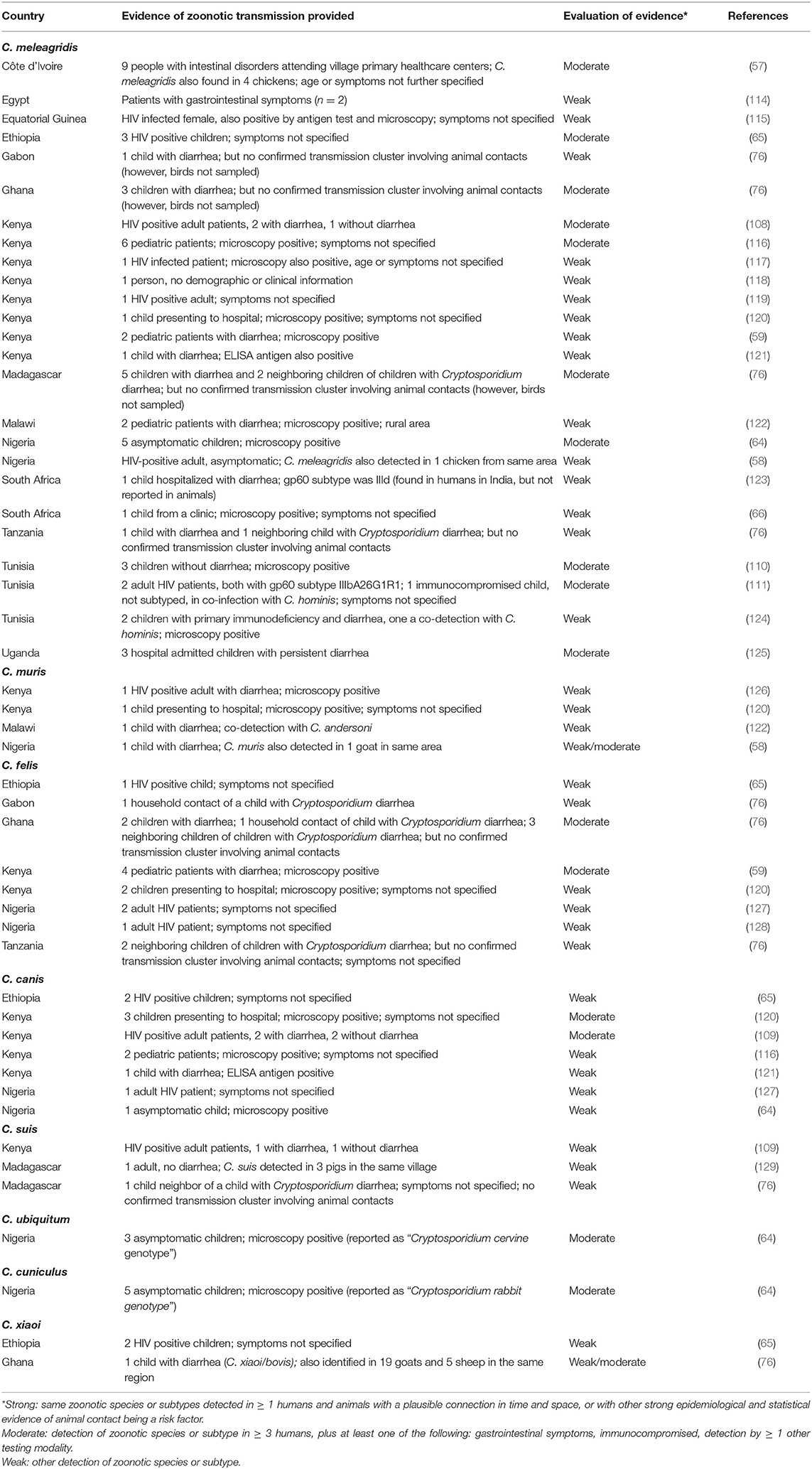
Table 3. Overview of articles for which zoonotic transmission of Cryptosporidium species other than C. parvum in Africa is suggested.
Based on the preceding sections of this article, as well as on previous reviews on Cryptosporidium infections in Africa (51, 52), it is clear that transmission of Cryptosporidium infection of people in Africa is currently largely anthroponotic (human to human), being mostly C. hominis or non-zoonotic C. parvum. However, from a superficial perspective, there are several factors that would suggest that zoonotic transmission in Africa would be at least as likely, if not more likely, to occur than in some other parts of the world. First, the relationships between animals and their owners in Africa are often much closer than in, for example, European countries; in Africa, people and their livestock may literally share the same sleeping quarters. Secondly, there is probably a greater likelihood of contamination of drinking water supplies by livestock in Africa than in many parts of the world, with water supplies often limited, and a general absence of catchment control for protection of water supplies, with surface waters used both as drinking water supplies and also for watering animals (82, 87, 131, 132). Thirdly, animals are often closely associated with the growth of fruit and vegetables that are consumed raw—with animals being involved in the plowing, harvesting, and transport of such crops, and often standing close by in marketplaces where such crops are sold (132). Fourthly, in many parts of Africa, there is close contact between people and animal manure, which is used as a resource for fertilizer, fuel, and building materials, with the pats often prepared by hand, frequently by women (also frequently involved in food preparation) and young children (99, 133). Indeed, animals in the domestic environment have been cited as being a contributor to the substantial burden of zoonotic disease, including cryptosporidiosis (134), either directly or indirectly (132). Although Aldeyarbi et al. (51) comment that animals are not found in close proximity to residences in urban settings in Africa, this seems not to be the case. For example, a survey from Burkina Faso indicated that more 25% of households in Ouagadougou kept livestock (135) and in 2019, FAO noted the potential zoonotic dangers associated with livestock in rapidly expanding African cities (136).
Thus, with this apparently great potential for zoonotic transmission in African countries, the question arises about why it does not seem to occur to a greater extent. We put forward the following possible reasons, and suggest that all or some of these may play a role. We also suggest that the large number of articles from Africa apparently suggesting the importance of zoonotic transmission in these settings, may be perpetuating a misleading myth.
Our first suggestion is that, C. parvum infection is not well established among the ruminant livestock populations in many regions in Africa. Although larger-scale herds do occur in some parts of Africa, as detailed in the section of this article on livestock production, livestock rearing is usually extensive, pastoral, or semi-pastoral. Although there are notable exceptions, and some large cattle enterprises may be found, in general the average number of cattle per farm is around 50. A multivariable analysis of risk factors for pre-weaned calves acquiring C. parvum infection and C. bovis infection has demonstrated an increased risk of C. parvum infection with greater herd size, with calves in herds of over 200 animals being at significantly greater risk of infection than calves in herds of below 100 animals (137). Furthermore, with high mortality of neonates and year-round, relatively low production rates, the neonatal density in African herds tends to remain low and constant; peaks in zoonotic transmission at the same time as seasonal lambing and calving are well recognized in some non-African countries (22, 138).
In addition, other relevant factors that significantly increased the risk of C. parvum infection in calves was mean monthly precipitation of 100–150 mm (compared with below 100 mm), being housed inside, and the use of hay bedding (137). These factors are thus those that favor close contact between animals (herd size and housing) and oocyst survival (hay bedding and precipitation); the association with hay bedding has also been reported from a study in Mexico (139). These risk factors for C. parvum infection in calves are therefore often lacking in the cattle-husbandry systems predominant in many African countries, with most herds being below 100 animals. Furthermore, even in places where large herd sizes may occur (e.g., in Burkina Faso), these are often being managed in pastoral systems where other risk factors (e.g., housing and hay bedding) are lacking (140). Furthermore, in such animal management systems, exposure to the climate is also likely to be detrimental to transmission, with desiccation and UV exposure also playing a part (see the later section on oocyst survival in the African envirnoment). Indeed, a study from Tanzania (47) in which a notable lack of Cryptosporidium infection was identified among 601 dairy calves from different management systems, small herd size and climatic factors were considered to be important factors that could have reduced the potential for establishment of infection and/or contributed to disease fade out (141). Similar arguments may also be proposed for why C. parvum may be less established in small ruminants in various African countries (142).
Another potential factor of importance for the lack of infection with C. parvum in African cattle is cattle breed (and, correspondingly, could also be relevant for small ruminant livestock). As noted by Chang'a et al. (47), most studies on Cryptosporidium in cattle involve Bos taurus breeds, but B. indicus breeds (which often predominate in African farms) may be more resistant. A study from Nigeria (143) involving 195 calves of the White Fulani and Sokoto Gudali breeds (both B. indicus breeds) reported 16% prevalence of Cryptosporidium, but none were C. parvum. Although systematic investigation of Cryptosporidium infections in general, and C. parvum in particular, are lacking, different management routines may be associated with different breeds [e.g., some breeds of cattle, particularly European taurine breeds, tend to need to be housed indoors due to their susceptibility to African trypanosomiasis; (144)]. A study from Malaysia (145) also supports the suggestion that particular breeds may be more susceptible to Cryptosporidium infection, with significantly higher prevalences of infection reported from Mafriwal cattle (Sahiwal × Friesian crosses) and from Jersey × Friesian crosses.
In Africa, a study from Zambia (70) considered that the significantly higher prevalence of Cryptosporidium infection in dairy-farm cattle compared with beef calves or “traditionally reared” calves was due to the management factors. These include higher density of dairy calves favoring propagation of infection in confined housing, whereas calves in extensively reared beef and traditional husbandry systems were not only fewer in number but outside, where any oocysts would be exposed to environmental pressures such as desiccation and direct sunlight, resulting in a reduced infection pressure (70). Cattle breed may also have played a role, as the calves on the dairy farms were Jersey, Friesian, or crossbreeds, but a mixture of cattle breeds predominated in the other management systems, including Brahman and Boran, both B. indicus breeds. Similarly, based on the results of their study in two relatively large dairy farms in the central highlands of Ethiopia (49), exotic breeds (Holstein-Friesian; B. taurus) were suggested as being more vulnerable to Cryptosporidium infection than the local Zebu breed (B. indicus), as crossbred calves with a greater proportion of Holstein-Friesian “blood” had a higher prevalence of Cryptosporidium infection than calves with a lower proportion. Considering the three articles that we considered showed relatively strong evidence of zoonotic transmission of C. parvum, only the one from Egypt (105) mentions breed; however, although the authors state that, in general, most livestock in the region of the study were native crossbreeds, the animal breeds in the study, or in the two specific farms where zoonotic transmission was suggested, were not stated.
Thus, although disentangling the potential risk factors from each other is clearly difficult, it is also apparent that for various reasons, under current conditions, African cattle may be generally less likely to be shedding C. parvum oocysts than cattle elsewhere. However, this does not exclude this parasite establishing and spreading in African livestock populations as circumstances change. Such a scenario could be devastating for African livestock production, as well as having potential for disseminating further to people.
This leads us to explore whether the “other part” of a potential zoonotic transmission cycle may also exhibit some factors that contribute to the relative lack of this transmission route; namely the potential human hosts. Although animal-human contact is probably more extensive in Africa than in other areas of the world, there may be some aspects of people in Africa that result in them having a different probability of acquiring zoonotic Cryptosporidium infection than elsewhere. Of particular relevance in this respect is the relatively high prevalence of infection with C. hominis in young children in many African countries (5, 25–27); babies and young infants are generally more likely to be exposed to Cryptosporidium oocysts from human infections than from animal infections. Thus, it seems likely that C. hominis infections early in life may provide some protection against infection with zoonotic Cryptosporidium species later. Although immune responses to cryptosporidiosis are currently not completely understood, it is clear that both innate and adaptive immune responses have a relevant role in both protection from, and resolution of, Cryptosporidium infections and cryptosporidiosis. However, the level of immunity has not been determined, nor the extent to which there is cross-protection between different species of Cryptosporidium. An early human experimental study with C. parvum, in which primary infection of seronegative adults with a challenge dose was followed by another challenge with the same C. parvum oocyst isolate approximately 1 year later, showed that initial exposure may be insufficient to protect against clinical illness 1 year later (146). However, it is likely that young children receive several low-level exposures, and this may have a different outcome to that described in the human-challenge study. In a later study, adults with pre-existing anti-C. parvum serum IgG only became infected when challenged with higher Cryptosporidium oocyst doses, and did not excrete as many oocysts, indicating that prior exposure to C. parvum does provide protection from infection and disease at lower oocyst doses (147). Using another approach to investigate exposure protection, serological investigations in two UK cities with high and low incidences of reported cryptosporidiosis indicated that exposure to non-pathogenic strains of Cryptosporidium or repeated low-level exposure to pathogenic strains could provide a protective effect (148). Indeed, it has been suggested that by eliminating a source of low-level Cryptosporidium oocyst exposure may, paradoxically, increase the risk of symptomatic infection from other exposure sources (149).
Regarding evidence of whether exposure to C. hominis provides cross-immunity against subsequent C. parvum challenge, analysis of antibody responses in children in Peru (150) and Bangladesh (151) suggests that C. hominis infection results in development of notable antibody responses against C. parvum antigens, indicating that these responses are directed toward epitopes conserved across species and subtypes. However, the importance of this is unclear as the roles of B-cells and antibody responses in cryptosporidiosis remain controversial, although they do seem to contribute to protection (152). T-cell mediated responses, particularly with CD4+ type-1-cells, are, recognized as a more crucial component. An experimental study using gnotobiotic piglets found that a substantial (one million oocysts) C. hominis challenge conferred full immunity against further challenge with the same C. hominis isolate, and partial immunity (i.e., infected but with significantly lower oocyst excretion than non-challenged controls) when further challenged by a substantial (10 million oocysts) C. parvum oocyst dose (153). Again, these large dose experiments beg the question regarding whether trickle exposures may be similarly (or more) likely to provide protection against future challenge, including with C. parvum or other species.
However, although exposure to Cryptosporidium early in life may be relevant for limiting future infection (as older children with responsibility, for example, for herding small ruminants), including with zoonotic species, other risk factors may occur in Africa that may increase infection likelihood. These could include concurrent infections or conditions that may limit the robustness or effect of an immune response against future challenge. It is well known that most people living with HIV reside in Africa, with 25.7 million estimated by World Health Organization in 2018, compared with 3.8 million in southeast Asia and 3.5 million in the Americas (154). Cryptosporidiosis is known as one of the major causes of diarrhea in patients with HIV, and is associated with significant morbidity and mortality in the AIDS population; a recent systematic review and meta-analysis indicated that the pooled prevalence of Cryptosporidium in HIV-positive patients in Africa was around 11.9% (CI: 8.8%−16.0%) (155), only marginally below that of SE Asia, which topped the list at 12.7% (CI: 9.7%−16.4%). It is not surprising that reports on zoonotic Cryptosporidium infections in Africa with species other than C. parvum are often in people with immunodeficiencies, particularly from HIV. Of the 51 articles listed in Table 2, 17 refer to infections in people with immunodeficiencies. It is also clear that other insults to human health, including malnutrition and other infections that may occur more commonly in some African countries, may not only exacerbate symptoms but also contribute toward individuals being more susceptible to infection due, among other reasons, to suppressed immunity.
In addition to immune effects of infections and other factors, genetic variations in the population itself may make individuals, or populations, more or less susceptible to specific infections. For cryptosporidiosis, candidate gene studies indicating an increased risk of cryptosporidiosis include HLA class I and II genes, SNPs in the mannose binding lectin (MBL) gene, and variation within the protein kinase C alpha (PRKCA) gene (156). Some of these variations may be of particular relevance to Africans; for example, it has been noted that the median MBL protein concentration in blood is considerably lower in Africans than in other racial groups (157), whereas, in contrast, the “risk” T allele in the PRKCA gene is reported to occur at relatively low frequencies in Africa, and least frequently in West Africa (156). Whether these genetic variations may affect zoonotic transmission (zoonotic C. parvum infection) has not yet been explored.
Finally, the third player in zoonotic transmission of Cryptosporidium, is the environment. Cryptosporidium oocysts have long been recognized as being robust to many environmental pressures (158), and is one reason why waterborne and foodborne transmission occurs. Some African environments, with low humidity and high UV index, may have a negative impact on oocyst survival. Although not all of Africa is continuously dry and sunny, and there are extremes of weather and temperature, in general, in several places in Africa where livestock are grazed, environmental conditions may not be optimal for prolonged oocyst survival. This reduces the likelihood of animal-to-animal transmission (see previous section), and also animal-to-human transmission. Whether different species or subtypes of Cryptosporidium oocysts may have greater environmental robustness has been scarcely investigated, but a tentatively forwarded hypothesis (63) is that mutation in the COWP9 gene, which as other genes in the COWP family are associated with oocyst wall formation (159) may affect robustness, and thus transmission possibilities.
Where water is contaminated with feces of an infected individual, the potential for oocyst survival, and thus onward transmission, increases. As animals may have greater opportunities to contaminate drinking water in Africa than in other parts of the world, this may argue for an increased likelihood of zoonotic transmission of Cryptosporidium in Africa rather than in places where catchment protection measures are the norm. Indeed, Vermeulen et al. (160) note that the Cryptosporidium load from manure could be reduced substantially in several African countries by manure treatment with elevated temperatures, such as composting. Nevertheless, data modeling indicates that human, rather than animal, feces are the more predominant source of oocyst contamination, with most contamination around growing urban centers, and with the potential of these urban hot spots to grow and multiply as sewer connections are installed without corresponding and appropriate sewage treatment (89).
Contamination of water sources with Cryptosporidium oocysts varies over shorter timesteps than say, monthly averages, with water contamination likely to respond strongly to major weather events, such as prolonged and heavy rain or flooding events. It should be noted that oocysts of animal origin in runoff from grazing land are more likely to have been already inactivated, than oocysts from overloaded human sewerage systems that have spent less time exposed to desiccation and UV radiation. Of potential relevance in this context, is that even in some countries where C. parvum tends to be the predominant species associated with sporadic human infections, C. hominis seems to be the most usual species associated with waterborne outbreaks. One example is Sweden, where C. parvum causes most sporadic cases of cryptosporidiosis [between 2006 and 2008, there was just under double the number of C. parvum cases compared with C. hominis cases, with most C. hominis cases infected abroad; (161)], but major waterborne outbreaks in this country have been associated with C. hominis [e.g., (162)]. The subtype here was IbA10G2, which is discussed in greater detail in the section on species and genotypes. That this subtype occurs rarely in Africa but is associated with large-scale outbreaks elsewhere, may suggest that African populations are at risk should it be introduced; alternatively, should infection with this subtype be particularly associated with the T allele in the PRKCA gene (156), then African populations may be partially protected.
That both zoonotic and anthroponotic transmission of Cryptosporidium occur has long been accepted, and it is also well established that these routes are associated with different Cryptosporidium species and subtypes. However, in-depth exploration of transmission patterns and what they mean for public and veterinary health and interventions is scanty. In our opinion, it is all too common for publications to quote the serious toll that cryptosporidiosis takes on pediatric health in countries in Africa (and other low-income areas) and tie those figures to the potential for zoonotic transmission of this parasite and the necessity of a One Health perspective [e.g., (163)]. While we applaud the One Health approach, such juxtapositions can be misleading for some readers. Establishment of the facilities for more identification of species and subtyping of Cryptosporidium in different African countries in the coming years will provide further data regarding relative occurrences in different countries, hosts, and situations, and will be an essential tool for implementing appropriate control measures. This calls for not only more sophisticated laboratory infrastructure, but also scientists trained in the various techniques and with the appropriate skillsets and knowledge for such investigations.
It is essential that we are aware that in much of Africa, and probably for a variety of reasons, as discussed in preceding sections, anthroponotic transmission predominates at present. We add the words “at present” with emphasis; we want to stress that the current situation can change and is probably changing. Globalization may result in introduction of new species/subtypes of Cryptosporidium; it is common to think that visitors to Africa from Europe, for example, may return home with diarrheal pathogens. But it is of equal or greater importance to think that they may also export specific currently non-established Cryptosporidium subtypes to Africa, including C. hominis IbA10G2 and C. parvum IIaA15G2R1. Both of these are hypothesized to be hypertransmissible (55), but apparently occur only rarely in Africa currently; if these specific subtypes are globally hypertransmissible, rather than merely well suited and established in their current niches, then their introduction to areas of Africa could be disastrous for both human and animal health. In addition, changes in farm management (e.g., less extensive farming, more intensive farming, more urban farms) may result in animal feces being less exposed to environmental pressures that inactivate Cryptosporidium oocysts and also increase the possibilities of between-cow transmission, and hence environmental contamination and infection pressure.
It may be of relevance that the two countries that seemed to have clearest evidence of zoonotic transmission of C. parvum (Tunisia and Egypt), and from where subtype IIaA15G2R1 has been reported, have relatively limited regions suitable for livestock rearing and thus the potential for direct or indirect (water contamination) transmission may be exacerbated by the requirement for high animal densities in a restricted area.
Although reducing pediatric cryptosporidiosis in Africa, with its substantial mortality and morbidity burden, should clearly be a goal, it should also be borne in mind that reduced childhood immunity may, at population level, result in an epidemiological shift from an “endemic and predominantly anthroponotic” toward an “epidemic and predominantly zoonotic” pattern. Obviously, the negative impact of diarrheal disease is more damaging in young children, but slightly older children with acute malnutrition, or children or adults with untreated HIV, are also vulnerable groups. Although the extent of waterborne transmission of cryptosporidiosis in Africa is almost impossible to determine, a water, sanitation, and hygiene (WASH) perspective is a fundamental concept to limit the transmission of any diarrheal pathogen, zoonotic or not; however, WASH initiatives should be “transformative” in order to have a lasting and substantial impact (164). At the other end of the technology scale, as genome sequencing studies uncover relevant mutations, we will gradually gain greater information that may provide the basis for implementation of different approaches to limit, or prevent, both anthroponotic and zoonotic transmission.
The theme of this article was proposed by LR and derived from discussion with all co-authors. AE had main responsibility for section Human Health Impacts, TK and GT for sections Veterinary Health Impacts and Ruminant Livestock Production in Africa and Potential Effects on Cryptosporidium Transmission, ØJ for sections Cryptosporidium Infections in Africa: Distributions of Species/Genotypes and Are There Hot Spots for Zoonotic Transmission in Africa? and Tables 2, 3. LR had main responsibility for all other sections, and for overall structure. All authors commented on all sections of the drafts and approved the final submission.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.575881/full#supplementary-material
1. Ryan U, Paparini A, Monis P, Hijjawi N. It's official—Cryptosporidium is a gregarine: what are the implications for the water industry? Water Res. (2016) 105:305–13. doi: 10.1016/j.watres.2016.09.013
2. EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards). Scientific Opinion on the public health risks associated with food-borne parasites. EFSA J. (2018) 16:5495. doi: 10.2903/j.efsa.2018.5495
3. Morgan-Ryan UM, Fall A, Ward LA, Hijjawi N, Sulaiman I, Fayer R, et al. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J Eukaryot Microbiol. (2002) 49:433–40. doi: 10.1111/j.1550-7408.2002.tb00224.x
4. Thomson S, Hamilton CA, Hope JC, Katzer F, Mabbott NA, Morrison LJ, et al. Bovine cryptosporidiosis: impact, host-parasite interaction and control strategies. Vet Res. (2017) 48:42. doi: 10.1186/s13567-017-0447-0
5. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. (2013) 382:209–22. doi: 10.1016/S0140-6736(13)60844-2
6. Meisel JL, Perera DR, Meligro C, Rubin CE. Overwhelming watery diarrhea associated with a Cryptosporidium in an immunosuppressed patient. Gastroenterology. (1976) 70:1156–60. doi: 10.1016/S0016-5085(76)80331-9
7. Nime FA, Burek JD, Page DL, Holscher MA, Yardley JH. Acute enterocolitis in a human being infected with the protozoan Cryptosporidium. Gastroenterology. (1976) 70:592–8. doi: 10.1016/S0016-5085(76)80503-3
8. Vetterling JM, Jervis HR, Merrill TG, Sprinz H. Cryptosporidium wrairi sp. n. from the guinea pig Cavia porcellus, with an emendation of the genus. J Protozool. (1971) 18:243–7. doi: 10.1111/j.1550-7408.1971.tb03315.x
9. Tzipori S, Angus KW, Campbell I, Gray EW. Cryptosporidium: evidence for a single-species genus. Infect Immun. (1980) 30:884–6.
10. Tzipori S, Angus KW, Campbell I, Gray EW. Experimental infection of lambs with Cryptosporidium isolated from a human patient with diarrhoea. Gut. (1982) 23:71–4. doi: 10.1136/gut.23.1.71
11. Anderson BC, Donndelinger T, Wilkins RM, Smith J. Cryptosporidiosis in a veterinary student. J Am Vet Med Assoc. (1982) 180:408–9.
12. Levine ND. Taxonomy and review of the coccidian genus Cryptosporidium (protozoa, Apicomplexa). J Protozool. (1984) 31:94–8. doi: 10.1111/j.1550-7408.1984.tb04296.x
13. Upton SJ, Current WL. The species of Cryptosporidium (Apicomplexa: Cryptosporidiidae) infecting mammals. J Parasitol. (1985) 71:625–9. doi: 10.2307/3281435
14. Hart CA, Baxby D, Blundell N. Gastro-enteritis due to Cryptosporidium: a prospective survey in a children's hospital. J Infect. (1984) 9:264–70. doi: 10.1016/S0163-4453(84)91117-4
15. Hunt DA, Shannon R, Palmer SR, Jephcott AE. Cryptosporidiosis in an urban community. Br Med J (Clin Res Ed). (1984) 289:814–816. doi: 10.1136/bmj.289.6448.814
16. Casemore DP, Jackson FB. Hypothesis: cryptosporidiosis in human beings is not primarily a zoonosis. J Infect. (1984) 9:153–6.
17. Højlyng N, Mølbak K, Jepsen S, Hansson AP. Cryptosporidiosis in Liberian children. Lancet. (1984) 1:734. doi: 10.1016/S0140-6736(84)92244-X
18. Højlyng N, Mølbak K, Jepsen S. Cryptosporidium spp., a frequent cause of diarrhea in Liberian children. J Clin Microbiol. (1986) 23:1109–13. doi: 10.1128/JCM.23.6.1109-1113.1986
19. Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, et al. Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol. (2019) 4:826–36. doi: 10.1038/s41564-019-0377-x
20. Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. (2010) 124:80–9. doi: 10.1016/j.exppara.2009.03.018
21. Xiao L, Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. (2017) 8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002
22. Chalmers RM, Robinson G, Elwin K, Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors. (2019) 12:95. doi: 10.1186/s13071-019-3354-6
23. The World Bank. Country Classification: World Bank Country and Lending Groups. Available online at: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups [accessed 22 June 2020].
24. Mølbak K, Højlyng N, Gottschau A, Sa JC, Ingholt L, Da Silva AP, et al. Cryptosporidiosis in infancy and childhood mortality in Guinea Bissau, West Africa. BMJ. (1993) 307:417–20. doi: 10.1136/bmj.307.6901.417
25. GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. (2017) 390:1151–10. doi: 10.1016/S0140-6736(17)32152-9
26. GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. (2017). 17:909–48. doi: 10.1016/S1473-3099(17)30276-1
27. Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Global Health. (2018) 6:e758–68. doi: 10.1016/S2214-109X(18)30283-3
28. Colombara DV, Khalil IA, Rao PC, Troeger C, Forouzanfar MH, Riddle MS, et al. Chronic health consequences of acute enteric infections in the developing world. Am J Gastroenterol Suppl. (2016) 3:4. doi: 10.1038/ajgsup.2016.9
29. Weisz AJ, Manary MJ, Stephenson K, Agapova S, Manary FG, Thakwalakwa C, et al. Abnormal gut integrity is associated with reduced linear growth in rural Malawian children. J Pediatr Gastroenterol Nutr. (2012) 55:747–50. doi: 10.1097/MPG.0b013e3182650a4d
30. Sudfeld CR, McCoy DC, Danaei G, Fink G, Ezzati M, Andrews KG, et al. Linear growth and child development in low-and middle-income countries: a meta-analysis. Pediatrics. (2015) 135:e1266–75. doi: 10.1542/peds.2014-3111
31. DeBoer MD, Lima AA, Oría RB, Scharf RJ, Moore SR, Luna MA. Early childhood growth failure and the developmental origins of adult disease: do enteric infections and malnutrition increase risk for the metabolic syndrome? Nutr Rev. (2012) 70:642–53. doi: 10.1111/j.1753-4887.2012.00543.x
32. Zambriski JA, Nydam DV, Bowman DD, Bellosa ML, Burton AJ, Linden TC, et al. Description of fecal shedding of Cryptosporidium parvum oocysts in experimentally challenged dairy calves. Parasitol Res. (2013) 112:1247–54. doi: 10.1007/s00436-012-3258-2
33. Sweeny JP, Ryan UM, Robertson ID, Jacobson C. Cryptosporidium and Giardia associated with reduced lamb carcase productivity. Vet Parasitol. (2011) 182:127–39. doi: 10.1016/j.vetpar.2011.05.050
34. Jacobson C, Williams A, Yang R, Ryan U, Carmichael I, Campbell AJ, et al. Greater intensity and frequency of Cryptosporidium and Giardia oocyst shedding beyond the neonatal period is associated with reductions in growth, carcass weight and dressing efficiency in sheep. Vet Parasitol. (2016) 228:42–51. doi: 10.1016/j.vetpar.2016.08.003
35. Shaw HJ, Innes EA, Morrison LJ, Katzer F, Wells B. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int J Parasitol. (2020) 50:371–6. doi: 10.1016/j.ijpara.2020.03.002
36. Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol. (2019) 49:569–77. doi: 10.1016/j.ijpara.2019.02.006
37. Niine T, Dorbek-Kolin E, Lassen B, Orro T. Cryptosporidium outbreak in calves on a large dairy farm: effect of treatment and the association with the inflammatory response and short-term weight gain. Res Vet Sci. (2018) 117:200–8. doi: 10.1016/j.rvsc.2017.12.015
38. Johnson EH, Muirhead DE, Windsor JJ, King GJ, Al-Busaidy R, Cornelius R. Atypical outbreak of caprine cryptosporidiosis in the Sultanate of Oman. Vet Rec. (1999) 145:521–4. doi: 10.1136/vr.145.18.521
39. Wudu T, Kelay B, Mekonnen HM, Tesfu K. Calf morbidity and mortality in smallholder dairy farms in Ada'a Liben district of Oromia, Ethiopia. Trop Anim Health Prod. (2008) 40:369–76. doi: 10.1007/s11250-007-9104-3
40. Gitau GK, McDermott JJ, Waltner-Toews D, Lissemore KD, Osumo JM, Muriuki D. Factors influencing calf morbidity and mortality in smallholder dairy farms in Kiambu District of Kenya. Prev Vet Med. (1994) 21:167–77. doi: 10.1016/0167-5877(94)90005-1
41. Wymann MN, Bonfoh B, Schelling E, Bengaly S, Tembely S, Tanner M, et al. Calf mortality rate and causes of death under different herd management systems in peri-urban Bamako, Mali. Livestock Sci. (2005) 100:169–78. doi: 10.1016/j.livprodsci.2005.08.010
42. Hatam-Nahavandi K, Ahmadpour E, Carmena D, Spotin A, Bangoura B, Xiao L. Cryptosporidium infections in terrestrial ungulates with focus on livestock: a systematic review and meta-analysis. Parasites Vectors. (2019) 12:453. doi: 10.1186/s13071-019-3704-4
43. Abebe R, Wossene A, Kumsa B. An epidemiological study of Cryptosporidium infection in dairy calves on selected dairy farms of central Ethiopia. Rev Méd Vét. (2008) 159:107.
44. Regassa A, Gizaw O, Abunna F, Abebe R, Beyene D, Megersa B, et al. Cryptosporidium in calves, lambs and kids in Haramaya, eastern Ethiopia. Ethiop Vet J. (2013) 17:81–94. doi: 10.4314/evj.v17i1.7
45. Ayele A, Seyoum Z, Leta S. Cryptosporidium infection in bovine calves: prevalence and potential risk factors in northwest Ethiopia. BMC Res Notes. (2018) 11:15. doi: 10.1186/s13104-018-3219-7
46. Peter SG, Gitau GK, Mulei CM, Vanleeuwen J, Richards S, Wichtel J, et al. Prevalence of Cryptosporidia, Eimeria, Giardia, and Strongyloides in pre-weaned calves on smallholder dairy farms in Mukurwe-ini district, Kenya. Vet World. (2015) 8:1118–25. doi: 10.14202/vetworld.2015.1118-1125
47. Chang'a JS, Robertson LJ, Mtambo MM, Mdegela RH, Løken T, Reksen O. Unexpected results from large-scale cryptosporidiosis screening study in calves in Tanzania. Ann Trop Med Parasitol. (2011) 105:513–9. doi: 10.1179/2047773211Y.0000000007
48. Helmy YA, von Samson-Himmelstjerna G, Nöckler K, Zessin KH. Frequencies and spatial distributions of Cryptosporidium in livestock animals and children in the Ismailia province of Egypt. Epidemiol. Infect. (2015) 143:1208–18. doi: 10.1017/S0950268814001824
49. Zeleke MA, Francesca S, Gezahegn M, Getachew T. Cryptosporidium infection in dairy cattle calves and its public health significance in Central Ethiopia. J Adv Vet Res. (2017) 7:59–65. Available online at: http://advetresearch.com/index.php/avr/index
50. Wegayehu T, Karim R, Anberber M, Adamu H, Erko B, Zhang L, et al. Prevalence and genetic characterization of Cryptosporidium species in dairy calves in Central Ethiopia. PLoS ONE. (2016) 11:e0154647. doi: 10.1371/journal.pone.0154647
51. Aldeyarbi HM, El-Ezz NM, Karanis P. Cryptosporidium and cryptosporidiosis: the African perspective. Environ Sci Pollut Res. (2016) 23:13811–21. doi: 10.1007/s11356-016-6746-6
52. Squire SA, Ryan U. Cryptosporidium and Giardia in Africa: current and future challenges. Parasit Vectors. (2017) 10:195. doi: 10.1186/s13071-017-2111-y
53. King P, Tyler KM, Hunter PR. Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: a systematic review and meta-analysis. Parasit Vectors. (2019) 12:16. doi: 10.1186/s13071-018-3263-0
54. Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, et al. The burden of Cryptosporidium diarrheal disease among children <24 months of age in moderate/high mortality regions of sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis. (2016) 10:e0004729. doi: 10.1371/journal.pntd.0004729
55. Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. (2018) 34:997–1011. doi: 10.1016/j.pt.2018.07.009
56. Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int J Parasitol. (2013) 43:1141–7. doi: 10.1016/j.ijpara.2013.09.002
57. Berrilli F, D'Alfonso R, Giangaspero A, Marangi M, Brandonisio O, Kaboré Y, et al. Giardia duodenalis genotypes and Cryptosporidium species in humans and domestic animals in Côte d'Ivoire: occurrence and evidence for environmental contamination. Trans R Soc Trop Med Hyg. (2012) 106:191–5. doi: 10.1016/j.trstmh.2011.12.005
58. Ayinmode AB, Oliveira BCM, Obebe OO, Dada-Adgebola HO, Ayede AI, Widmer G. Genotypic characterization of Cryptosporidium species in humans and peri-domestic animals in Ekiti and Oyo States, Nigeria. J Parasitol. (2018) 104:639–44. doi: 10.1645/17-74
59. Mbae C, Mulinge E, Waruru A, Ngugi B, Wainaina J, Kariuki S. Genetic diversity of Cryptosporidium in children in an urban informal settlement of Nairobi, Kenya. PLoS ONE. (2015) 10:e0142055. doi: 10.1371/journal.pone.0142055
60. Hossain MJ, Saha D, Antonio M, Nasrin D, Blackwelder WC, Ikumapayi UN, et al. Cryptosporidium infection in rural Gambian children: epidemiology and risk factors. PLoS Negl Trop Dis. (2019) 13:e0007607. doi: 10.1371/journal.pntd.0007607
61. Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, et al. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis. (2013) 19:1573–82. doi: 10.3201/eid1910.121361
62. Segura R, Prim N, Montemayor M, Valls ME, Muñoz C. Predominant virulent IbA10G2 subtype of Cryptosporidium hominis in human isolates in Barcelona: a five-year study. PLoS ONE. (2015) 10:e0121753. doi: 10.1371/journal.pone.0121753
63. Sikora P, Andersson S, Winiecka-Krusnell J, Hallström B, Alsmark C, Troell K, et al. Genomic variation in IbA10G2 and other patient-derived Cryptosporidium hominis subtypes. J Clin Microbiol. (2017) 55:844–58. doi: 10.1128/JCM.01798-16
64. Molloy SF, Smith HV, Kirwan P, Nichols RA, Asaolu SO, Connelly L, et al. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg. (2010) 82:608–13. doi: 10.4269/ajtmh.2010.09-0624
65. Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, et al. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis. (2014) 8:e2831. doi: 10.1371/journal.pntd.0002831
66. Abu Samra N, Jori F, Cacciò SM, Frean J, Poonsamy B, Thompson PN. Cryptosporidium genotypes in children and calves living at the wildlife or livestock interface of the Kruger National Park, South Africa. Onderstepoort J Vet Res. (2016) 83:1–7. doi: 10.4102/ojvr.v83i1.1024
67. Irisarri-Gutiérrez MJ, Mingo MH, de Lucio A, Gil H, Morales L, Seguí R, et al. Association between enteric protozoan parasites and gastrointestinal illness among HIV- and tuberculosis-infected individuals in the Chowke district, southern Mozambique. Acta Trop. (2017) 170:197–203. doi: 10.1016/j.actatropica.2017.03.010
68. Naguib D, El-Gohary AH, Mohamed AA, Roellig DM, Arafat N, Xiao L. Age patterns of Cryptosporidium species and Giardia duodenalis in dairy calves in Egypt. Parasitol Int. (2018) 67:736–41. doi: 10.1016/j.parint.2018.07.012
69. Robertson LJ, Björkman C, Axén C, Fayer R. Cryptosporidiosis in farmed animals. In: Caccio, S, Widmer, G, editors. Cryptosporidium: Parasite and Disease. Vienna: Springer (2014). p. 149–235.
70. Geurden T, Goma FY, Siwila J, Phiri IG, Mwanza AM, Gabriel S, et al. Prevalence and genotyping of Cryptosporidium in three cattle husbandry systems in Zambia. Vet Parasitol. (2006) 138:217–22. doi: 10.1016/j.vetpar.2006.02.009
71. Adam KY, Ismail AA, Masri MA, Gameel AA. First report and molecular characterization of Cryptosporidium spp. in humans and animals in Khartoum state, Sudan. Vet. World. (2019) 12:183. doi: 10.14202/vetworld.2019.183-189
72. Nolan MJ, Unger M, Yeap YT, Rogers E, Millet I, Harman K, et al. Molecular characterisation of protist parasites in human-habituated mountain gorillas (Gorilla beringei beringei), humans and livestock, from Bwindi impenetrable National Park, Uganda. Parasit Vectors. (2017) 10:340. doi: 10.1186/s13071-017-2283-5
73. Squire SA, Yang R, Robertson I, Ayi I, Ryan U. Molecular characterization of Cryptosporidium and Giardia in farmers and their ruminant livestock from the Coastal Savannah zone of Ghana. Infect Genet Evol. (2017) 55:236–43. doi: 10.1016/j.meegid.2017.09.025
74. Wegayehu T, Karim MR, Li J, Adamu H, Erko B, Zhang L, et al. Prevalence and genetic characterization of Cryptosporidium species and Giardia duodenalis in lambs in Oromia Special Zone, Central Ethiopia. BMC Vet Res. (2017) 13:22. doi: 10.1186/s12917-016-0916-0
75. Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. (2014) 20:217–24. doi: 10.3201/eid2002.121797
76. Krumkamp R, Aldrich C, Maiga-Ascofare O, Mbwana J, Rakotozandrindrainy N, Borrmann S, et al. Transmission of Cryptosporidium spp. among human and animal local contact networks in sub-Saharan Africa: a multi-country study. Clin Infect Dis. (2020) ciaa223. doi: 10.1093/cid/ciaa223. [Epub ahead of print].
77. Ghenghesh KS, Ghanghish K, El-Mohammady H, Franka E. Cryptosporidium in countries of the Arab world: the past decade (2002–2011). Libyan J Med. (2012) 7:19852. doi: 10.3402/ljm.v7i0.19852
78. Schaefer LM, le Roux WJ, Abia ALK. Reservoirs of Cryptosporidium and Giardia in Africa. In: Abia A, Lanza G, editors. Current Microbiological Research in Africa. Cham: Springer (2020).
79. Reiner RC Jr, Graetz N, Casey DC, Troeger C, Garcia GM, Mosser JF, et al. Variation in childhood diarrheal morbidity and mortality in Africa, 2000–2015. N Engl J Med. (2018) 379:1128–38. doi: 10.1056/NEJMoa1716766
80. Hajdu A, Vold L, Østmo TA, Helleve A, Helgebostad SR, Krogh T, et al. Investigation of Swedish cases reveals an outbreak of cryptosporidiosis at a Norwegian hotel with possible links to in-house water systems. BMC Infect Dis. (2008) 8:152. doi: 10.1186/1471-2334-8-152
81. Chalmers RM. Waterborne outbreaks of cryptosporidiosis. Ann Ist Super Sanita. (2012) 48:429–46. doi: 10.4415/ANN_12_04_10
82. Ahmed SA, Guerrero Flórez M, Karanis P. The impact of water crises and climate changes on the transmission of protozoan parasites in Africa. Pathog Glob Health. (2018) 112:281–93. doi: 10.1080/20477724.2018.1523778
83. Morris JF, Murphy J, Fagerli K, Schneeberger C, Jaron P, Moke F, et al. A randomized controlled trial to assess the impact of ceramic water filters on prevention of diarrhea and cryptosporidiosis in infants and young children—Western Kenya, 2013. Am J Trop Med Hyg. (2018) 98:1260–8. doi: 10.4269/ajtmh.17-0731
84. Kifleyohannes T, Robertson LJ. Preliminary insights regarding water as a transmission vehicle for Cryptosporidium and Giardia in Tigray, Ethiopia. Food Waterborne Parasitol. (2020) 19:e00073. doi: 10.1016/j.fawpar.2020.e00073
85. Potgieter N, Karambwe S, Mudau LS, Barnard T, Traore A. Human enteric pathogens in eight rivers used as rural household drinking water sources in the northern region of South Africa. Int J Environ Res Public Health. (2020) 17:E2079. doi: 10.3390/ijerph17062079
86. Hamdy D, El-Badry A, Abd El Wahab W. Assessment of Giardia and Cryptosporidium assemblages/species and their viability in potable tap water in Beni-Suef, Egypt using nested PCR/RFLP and staining. Iran J Parasitol. (2019) 14:368–78. doi: 10.18502/ijpa.v14i3.1475
87. Muchiri JM, Ascolillo L, Mugambi M, Mutwiri T, Ward HD, Naumova EN, et al. Seasonality of Cryptosporidium oocyst detection in surface waters of Meru, Kenya as determined by two isolation methods followed by PCR. J Water Health. (2009) 7:67–75. doi: 10.2166/wh.2009.109
88. Kato S, Ascolillo L, Egas J, Elson L, Gostyla K, Naples L, et al. Waterborne Cryptosporidium oocyst identification and genotyping: use of GIS for ecosystem studies in Kenya and Ecuador. J Eukaryot Microbiol. (2003) 50(Suppl.):548–9. doi: 10.1111/j.1550-7408.2003.tb00624.x
89. Vermeulen LC, van Hengel M, Kroeze C, Medema G, Spanier JE, van Vliet MTH, et al. Cryptosporidium concentrations in rivers worldwide. Water Res. (2019) 149:202–14. doi: 10.1016/j.watres.2018.10.069
90. Limaheluw J, Medema G, Hofstra N. An exploration of the disease burden due to Cryptosporidium in consumed surface water for sub-Saharan Africa. Int J Hyg Environ Health. (2019) 222:856–63. doi: 10.1016/j.ijheh.2019.04.004
91. Cook R. World Cattle Inventory: Ranking of Countries (FAO). (2015). Available online at: https://www.drovers.com/article/world-cattle-inventory-ranking-countries-fao [accessed 19 June 2020].
92. Robinson TP, Thornton PK, Franceschini G, Kruska RL, Chiozza F, Notenbaert AM, et al. Global Livestock Production Systems. Rome: Food and Agriculture Organization of the United Nations (FAO) and International Livestock Research Institute (ILRI) (2011).
93. Steinfeld H, Wassenaar T, Jutzi S. Livestock production systems in developing countries: status, drivers, trends. Rev Sci Tech. (2006) 25:505–16. doi: 10.20506/rst.25.2.1677
94. Otte MJ, Chilonda P. Cattle and Small Ruminant Production Systems in Sub-Saharan Africa. A Systematic Review. Livestock Information Sector Analysis and Policy Branch. Rome: Food and Agriculture Organization of the United Nations (2002).
95. Moran JB. Factors affecting high mortality rates of dairy replacement calves and heifers in the tropics and strategies for their reduction. Asian–Australasian J Anim Sci. (2011) 24:1318–28. doi: 10.5713/ajas.2011.11099
96. Food and Agriculture Organization of The United Nations (FAO). Africa Sustainable Livestock 2050. (2018). Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020].
97. Tate KW, Atwill ER, Mcdougald NK, George MR. Spatial and temporal patterns of cattle feces deposition on rangeland. J Range Manage Arch. (2003) 56:432–8. doi: 10.2307/4003833
98. Food and Agriculture Organization of The United Nations (FAO). Africa Sustainable Livestock 2050: Livestock Production Systems Spotlight, Ethiopia. Cattle Sectors. (2018). Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020].
99. Tegegne A. Urban livestock production and gender in Addis Ababa, Ethiopia. Urb Agric Mag. (2004) 12:31–2. Available online at: https://hdl.handle.net/10568/29819
100. Food and Agriculture Organization of The United Nations (FAO). Africa Sustainable Livestock 2050: Livestock Production Systems Spotlight, Kenya. Cattle and Poultry Sectors. Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020].
101. Food and Agriculture Organization of The United Nations (FAO). Africa Sustainable Livestock 2050: Livestock Production Systems Spotlight, Nigeria. Dairy Cattle and Poultry (chicken) sectors. Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020].
102. Hribar A. Understanding Concentrated Animal Feeding Operations and Their Impact on Communities. National Association of Local Boards of Health. (2010). Available online at: https://www.cdc.gov/nceh/ehs/docs/understanding_cafos_nalboh.pdf?fbclid=IwAR3DK7qlkmPsTg6D8CG [accessed 19 June 2020].
103. Food and Agriculture Organization of The United Nations (FAO). Africa Sustainable Livestock 2050: Livestock Production Systems Spotlight, Egypt. Cattle, Buffaloes and Poultry Sectors. Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020].
104. El-Sherbini GT, Mohammad KA. Zoonotic cryptosporidiosis in man and animal in farms, Giza Governorate, Egypt. J Egypt Soc Parasitol. (2006) 36(2 Suppl.):49–58.
105. Helmy YA, Krücken J, Nöckler K, von Samson-Himmelstjerna G, Zessin KH. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet Parasitol. (2013) 193:15–24. doi: 10.1016/j.vetpar.2012.12.015
106. Shoukry NM, Dawoud HA, Haridy FM. Studies on zoonotic cryptosporidiosis parvum in Ismailia Governorate, Egypt. J Egypt Soc Parasitol. (2009) 39:479–88.
107. Ibrahim MA, Abdel-Ghany AE, Abdel-Latef GK, Abdel-Aziz SA, Aboelhadid SM. Epidemiology and public health significance of Cryptosporidium isolated from cattle, buffaloes, and humans in Egypt. Parasitol Res. (2016) 115:2439–48. doi: 10.1007/s00436-016-4996-3
108. Adamu H, Petros B, Hailu A, Petry F. Molecular characterization of Cryptosporidium isolates from humans in Ethiopia. Acta Trop. (2010) 115:77–83. doi: 10.1016/j.actatropica.2010.02.003
109. Wanyiri JW, Kanyi H, Maina S, Wang DE, Steen A, Ngugi P, et al. Cryptosporidiosis in HIV/AIDS patients in Kenya: clinical features, epidemiology, molecular characterization and antibody responses. Am J Trop Med Hyg. (2014) 91:319–28. doi: 10.4269/ajtmh.13-0254
110. Lobo ML, Augusto J, Antunes F, Ceita J, Xiao L, Codices V, et al. Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi and other intestinal parasites in young children in Lobata province, Democratic Republic of São Tomé and Principe. PLoS ONE. (2014) 9:e97708. doi: 10.1371/journal.pone.0097708
111. Rahmouni I, Essid R, Aoun K, Bouratbine A. Glycoprotein 60 diversity in Cryptosporidium parvum causing human and cattle cryptosporidiosis in the rural region of Northern Tunisia. Am J Trop Med Hyg. (2014) 90:346–50. doi: 10.4269/ajtmh.13-0522
112. Essid R, Menotti J, Hanen C, Aoun K, Bouratbine A. Genetic diversity of Cryptosporidium isolates from human populations in an urban area of Northern Tunisia. Infect Genet Evol. (2018) 58:237–42. doi: 10.1016/j.meegid.2018.01.004
113. Delgado C, Rosegrant M, Steinfeld H, Ehui S, Courbois C. Livestock to 2020: the next food revolution. Outlook Agric. (2001) 30:27–9. doi: 10.5367/000000001101293427
114. Eida AM, Eida MM, El-Desoky A. Pathological studies of different genotypes of human Cryptosporidium Egyptian isolates in experimentally mice. J Egypt Soc Parasitol. (2009) 39:975–90.
115. Blanco MA, Iborra A, Vargas A, Nsie E, Mbá L, Fuentes I. Molecular characterization of Cryptosporidium isolates from humans in Equatorial Guinea. Trans R Soc Trop Med Hyg. (2009) 103:1282–4. doi: 10.1016/j.trstmh.2009.02.016
116. Wasike WE, Kutima HL, Muya MS, Wamachi A. Diagnostic procedures, epidemiology and genetic diversity of Cryptosporidium species in Bungoma County, Kenya. Int J Sci Res. (2015) 4:1135–40.
117. Morgan U, Weber R, Xiao L, Sulaiman I, Thompson RA, Ndiritu W, et al. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. (2000) 38:1180–3. doi: 10.1128/JCM.38.3.1180-1183.2000
118. Peng MM, Matos O, Gatei W, Das P, Stantic-Pavlinic M, Bern C, et al. A comparison of Cryptosporidium subgenotypes from several geographic regions. J Eukaryot Microbiol. (2001) 48:28–31s. doi: 10.1111/j.1550-7408.2001.tb00442.x
119. Gatei W, Greensill J, Ashford RW, Cuevas LE, Parry CM, Cunliffe NA, et al. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J Clin Microbiol. (2003) 41:1458–62. doi: 10.1128/JCM.41.4.1458-1462.2003
120. Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, et al. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg. (2006) 75:78–82. doi: 10.4269/ajtmh.2006.75.78
121. Delahoy MJ, Omore R, Ayers TL, Schilling KA, Blackstock AJ, Ochieng JB, et al. Clinical, environmental, and behavioral characteristics associated with Cryptosporidium infection among children with moderate-to-severe diarrhea in rural western Kenya, 2008–2012: The Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis. (2018). 12:e0006640. doi: 10.1371/journal.pntd.0006640
122. Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect. (2007) 135:1307–15. doi: 10.1017/S0950268806007758
123. Abu Samra N, Thompson PN, Jori F, Frean J, Poonsamy B, Du Plessis D, et al. Genetic characterization of Cryptosporidium spp. in diarrhoeic children from four provinces in South Africa. Zoonoses Public Health. (2013) 60:154–9. doi: 10.1111/j.1863-2378.2012.01507.x
124. Ben IA, Essid R, Mellouli F, Aoun K, Bejaoui M, Bouratbine A. Cryptosporidium infection in patients with major histocompatibility complex class II deficiency syndrome in Tunisia: description of five cases. Arch Pediatr. (2011) 18:939–44. doi: 10.1016/j.arcped.2011.06.015
125. Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, et al. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. (2005) 73:921–5. doi: 10.4269/ajtmh.2005.73.921
126. Gatei W, Ashford RW, Beeching NJ, Kamwati SK, Greensill J, Hart CA. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg Infect Dis. (2002) 8:204–6. doi: 10.3201/eid0802.010256
127. Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, Xiao L. Molecular characterization of Cryptosporidium spp. in HIV-infected persons in Benin City, Edo State, Nigeria. Fooyin J Health Sci. (2010) 2:85–9. doi: 10.1016/S1877-8607(11)60003-9
128. Akinbo FO, Okaka CE, Omoregie R, Adamu H, Xiao L. Unusual Enterocytozoon bieneusi genotypes and Cryptosporidium hominis subtypes in HIV-infected patients on highly active antiretroviral therapy. Am J Trop Med Hyg. (2013) 89:157–61. doi: 10.4269/ajtmh.12-0635
129. Bodager JR, Parsons MB, Wright PC, Rasambainarivo F, Roellig D, Xiao L, et al. Complex epidemiology and zoonotic potential for Cryptosporidium suis in rural Madagascar. Vet Parasitol. (2015) 207:140–3. doi: 10.1016/j.vetpar.2014.11.013
130. Robertson LJ, Clark CG, Debenham JJ, Dubey JP, Kváč M, Li J, et al. Are molecular tools clarifying or confusing our understanding of the public health threat from zoonotic enteric protozoa in wildlife? Int J Parasitol Parasites Wildl. (2019) 9:323–41. doi: 10.1016/j.ijppaw.2019.01.010
131. Marshak A, Young H, Bontrager EN, Boyd EM. The relationship between acute malnutrition, hygiene practices, water and livestock, and their program implications in eastern Chad. Food Nutr Bull. (2017) 38:115–27. doi: 10.1177/0379572116681682
132. Penakalapati G, Swarthout J, Delahoy MJ, McAliley L, Wodnik B, Levy K, et al. Exposure to animal feces and human health: a systematic review and proposed research priorities. Environ Sci Technol. (2017) 51:11537–52. doi: 10.1021/acs.est.7b02811
133. Lupindu AM, Ngowi HA, Dalsgaard A, Olsen JE, Msoffe PL. Current manure management practices and hygiene aspects of urban and peri-urban livestock farming in Tanzania. Livest Res Rural Dev. (2012) 24:167. Available online at: http://www.lrrd.org/lrrd24/9/lupi24167.htm
134. Delahoy MJ, Wodnik B, McAliley L, Penakalapati G, Swarthout J, Freeman MC, et al. Pathogens transmitted in animal feces in low- and middle-income countries. Int J Hyg Environ Health. (2018) 221:661–76. doi: 10.1016/j.ijheh.2018.03.005
135. Thys E, Oueadraogo M, Speybroeck N, Geerts S. Socio-economic determinants of urban household livestock keeping in semi-arid Western Africa. J Arid Environ. (2005) 63:475–96. doi: 10.1016/j.jaridenv.2005.03.019
136. Food and Agriculture Organization of The United Nations (FAO). Regional Office for Africa. FAO Urges African Governments to Regulate Urban Livestock Sector. (2019). Available online at: http://www.fao.org/africa/news/detail-news/en/c/1204899/ [accessed 19 June 2020].
137. Szonyi B, Chang YF, Wade SE, Mohammed HO. Evaluation of factors associated with the risk of infection with Cryptosporidium parvum in dairy calves. Am J Vet Res. (2012) 73:76–85. doi: 10.2460/ajvr.73.1.76
138. Garcia-R JC, Pita AB, Velathanthiri N, French NP, Hayman DTS. Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitol Res. (2020) 119:2317–26. doi: 10.1007/s00436-020-06729-w
139. Maldonado-Camargo S, Atwill ER, Saltijeral-Oaxaca JA, Herrera-Alonso LC. Prevalence of and risk factors for shedding of Cryptosporidium parvum in Holstein Freisian dairy calves in central México. Prev Vet Med. (1998) 36:95–107. doi: 10.1016/S0167-5877(98)00084-1
140. Food and Agriculture Organization of The United Nations (FAO). (2018). Africa Sustainable Livestock 2050: Livestock Production Systems Spotlight, Burkina Faso. Cattle and Poultry Sectors. Available online at: http://www.fao.org/in-action/asl2050/resources/documents/livestock-production-systems/en/ [accessed 19 June 2020] .
141. Anderson RM, May RM. Infectious Diseases of Humans: Dynamics and Control. Oxford: Oxford University Press (1992).
142. Goma FY, Geurden T, Siwila J, Phiri IGK, Mwanza AM, Gabriel S, et al. The prevalence and molecular characterisation of Cryptosporidium spp. in small ruminants in Zambia. Small Ruminant Res. (2007) 72:77–80. doi: 10.1016/j.smallrumres.2006.08.010
143. Maikai BV, Umoh JU, Kwaga JK, Lawal IA, Maikai VA, Cama V, Xiao L. Molecular characterization of Cryptosporidium spp. in native breeds of cattle in Kaduna State, Nigeria. Vet Parasitol. (2011) 178:241–5. doi: 10.1016/j.vetpar.2010.12.048
144. Paling RW, Dwinger RH. Potential of trypanotolerance as a contribution to sustainable livestock production in tsetse affected Africa. Vet Q. (1993) 15:60–7. doi: 10.1080/01652176.1993.9694374
145. Abdullah DA, Ola-Fadunsin SD, Ruviniyia K, Gimba FI, Chandrawathani P, Lim YAL, et al. Molecular detection and epidemiological risk factors associated with Cryptosporidium infection among cattle in Peninsular Malaysia. Food Waterborne Parasitol. (2019) 14:e00035. doi: 10.1016/j.fawpar.2019.e00035
146. Okhuysen PC, Chappell CL, Sterling CR, Jakubowski W, DuPont HL. Susceptibility and serologic response of healthy adults to reinfection with Cryptosporidium parvum. Infect Immun. (1998) 66:441–3. doi: 10.1128/IAI.66.2.441-443.1998
147. Chappell CL, Okhuysen PC, Sterling CR, Wang C, Jakubowski W, Dupont HL. Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg. (1999) 60:157–64. doi: 10.4269/ajtmh.1999.60.157
148. Elwin K, Chalmers RM, Hadfield SJ, Hughes S, Hesketh LM, Rothburn MM, et al. Serological responses to Cryptosporidium in human populations living in areas reporting high and low incidences of symptomatic cryptosporidiosis. Clin Microbiol Infect. (2007) 13:1179–85. doi: 10.1111/j.1469-0691.2007.01823.x
149. Ramsay CN, Wagner AP, Robertson C, Smith HV, Pollock KG. Effects of drinking-water filtration on Cryptosporidium seroepidemiology, Scotland. Emerg Infect Dis. (2014) 20:70–6. doi: 10.3201/eid2001.120386
150. Priest JW, Bern C, Xiao L, Roberts JM, Kwon JP, Lescano AG, et al. Longitudinal analysis of Cryptosporidium species-specific immunoglobulin G antibody responses in Peruvian children. Clin Vaccine Immunol. (2006) 13:123–31. doi: 10.1128/CVI.13.1.123-131.2006
151. Borad AJ, Allison GM, Wang D, Ahmed S, Karim MM, Kane AV, et al. Systemic antibody responses to the immunodominant p23 antigen and p23 polymorphisms in children with cryptosporidiosis in Bangladesh. Am J Trop Med Hyg. (2012) 86:214–22. doi: 10.4269/ajtmh.2012.11-0273
152. Riggs MW. Recent advances in cryptosporidiosis: the immune response. Microbes Infect. (2002) 4:1067–80. doi: 10.1016/S1286-4579(02)01631-3
153. Sheoran A, Wiffin A, Widmer G, Singh P, Tzipori S. Infection with Cryptosporidium hominis provides incomplete protection of the host against Cryptosporidium parvum. J Infect Dis. (2012) 205:1019–23. doi: 10.1093/infdis/jir874
154. World Health Organization. HIV/AIDS. Data and Statistics. Available online at: https://www.who.int/hiv/data/en/. [accessed 22 June 2020].
155. Ahmadpour E, Safarpour H, Xiao L, Zarean M, Hatam-Nahavandi K, Barac A, et al. Cryptosporidiosis in HIV-positive patients and related risk factors: a systematic review and meta-analysis. Parasite. (2020) 27:27. doi: 10.1051/parasite/2020025
156. Wojcik GL, Korpe P, Marie C, Mentzer AJ, Carstensen T, Mychaleckyj J, et al. Genome-wide association study of cryptosporidiosis in infants implicates PRKCA. mBio. (2020) 11:e03343–1. doi: 10.1128/mBio.03343-19
157. Mombo LE, Lu CY, Ossari S, Bedjabaga I, Sica L, Krishnamoorthy R, et al. Mannose-binding lectin alleles in sub-Saharan Africans and relation with susceptibility to infections. Genes Immun. (2003) 4:362–7. doi: 10.1038/sj.gene.6363979
158. Robertson LJ, Campbell AT, Smith HV. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl Environ Microbiol. (1992) 58:3494–500. doi: 10.1128/AEM.58.11.3494-3500.1992
159. Templeton TJ, Lancto CA, Vigdorovich V, Liu C, London NR, Hadsall KZ, et al. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect Immun. (2004) 72:980–7. doi: 10.1128/IAI.72.2.980-987.2004
160. Vermeulen LC, Benders J, Medema G, Hofstra N. Global Cryptosporidium loads from livestock manure. Environ Sci Technol. (2017) 51:8663–71. doi: 10.1021/acs.est.7b00452
161. Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. (2013) 141:1009–20. doi: 10.1017/S0950268812001665
162. Widerström M, Schönning C, Lilja M, Lebbad M, Ljung T, Allestam G, et al. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg Infect Dis. (2014) 20:581–9. doi: 10.3201/eid2004.121415
163. Innes EA, Chalmers RM, Wells B, Pawlowic MC. A One Health approach to tackle cryptosporidiosis. Trends Parasitol. (2020) 36:290–303. doi: 10.1016/j.pt.2019.12.016
Keywords: Africa, anthroponosis, Cryptosporidium, epidemiology, subtype, transmission, water, zoonosis
Citation: Robertson LJ, Johansen ØH, Kifleyohannes T, Efunshile AM and Terefe G (2020) Cryptosporidium Infections in Africa—How Important Is Zoonotic Transmission? A Review of the Evidence. Front. Vet. Sci. 7:575881. doi: 10.3389/fvets.2020.575881
Received: 24 June 2020; Accepted: 27 August 2020;
Published: 08 October 2020.
Edited by:
Olivier Andre Sparagano, City University of Hong Kong, Hong KongReviewed by:
Rafael Gutiérrez López, University of Porto, PortugalCopyright © 2020 Robertson, Johansen, Kifleyohannes, Efunshile and Terefe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucy J. Robertson, bHVjeS5yb2JlcnRzb25Abm1idS5ubw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.