
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 12 November 2020
Sec. Animal Nutrition and Metabolism
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.575801
This article is part of the Research Topic The Use of Phytogenic Feed Additives to Enhance Productivity and Health in Ruminants View all 14 articles
 Faiz-ul Hassan1,2†
Faiz-ul Hassan1,2† Muhammad Adeel Arshad2†
Muhammad Adeel Arshad2† Hossam M. Ebeid3
Hossam M. Ebeid3 Muhammad Saif-ur Rehman2
Muhammad Saif-ur Rehman2 Muhammad Sajjad Khan2
Muhammad Sajjad Khan2 Shehryaar Shahid2
Shehryaar Shahid2 Chengjian Yang1*
Chengjian Yang1*Ruminants inhabit the consortia of gut microbes that play a critical functional role in their maintenance and nourishment by enabling them to use cellulosic and non-cellulosic feed material. These gut microbes perform major physiological activities, including digestion and metabolism of dietary components, to derive energy to meet major protein (65–85%) and energy (ca 80%) requirements of the host. Owing to their contribution to digestive physiology, rumen microbes are considered one of the crucial factors affecting feed conversion efficiency in ruminants. Any change in the rumen microbiome has an imperative effect on animal physiology. Ruminal microbes are fundamentally anaerobic and produce various compounds during rumen fermentation, which are directly used by the host or other microbes. Methane (CH4) is produced by methanogens through utilizing metabolic hydrogen during rumen fermentation. Maximizing the flow of metabolic hydrogen in the rumen away from CH4 and toward volatile fatty acids (VFA) would increase the efficiency of ruminant production and decrease its environmental impact. Understanding of microbial diversity and rumen dynamics is not only crucial for the optimization of host efficiency but also required to mediate emission of greenhouse gases (GHGs) from ruminants. There are various strategies to modulate the rumen microbiome, mainly including dietary interventions and the use of different feed additives. Phytogenic feed additives, mainly plant secondary compounds, have been shown to modulate rumen microflora and change rumen fermentation dynamics leading to enhanced animal performance. Many in vitro and in vivo studies aimed to evaluate the use of plant secondary metabolites in ruminants have been conducted using different plants or their extract or essential oils. This review specifically aims to provide insights into dietary interactions of rumen microbes and their subsequent consequences on rumen fermentation. Moreover, a comprehensive overview of the modulation of rumen microbiome by using phytogenic compounds (essential oils, saponins, and tannins) for manipulating rumen dynamics to mediate CH4 emanation from livestock is presented. We have also discussed the pros and cons of each strategy along with future prospective of dietary modulation of rumen microbiome to improve the performance of ruminants while decreasing GHG emissions.
Improving feed efficiency and livestock production is a more coveted goal in animal agriculture being sought through selective breeding, scientific management, and improvement of feed composition. Feed efficiency in ruminants mainly depends upon the quality of feed, rumen fermentation, and dynamics mediated by rumen microbiomes. The rumen in animals is inhabited by the diverse microbiome, including bacteria, protozoa, fungi, and archaea. Different factors like temperature (38–42°C), pH (5.5–7), and redox potential (250–450 mV) regulated by saliva buffering provide a specific environment for degradation of cellulolytic plant material by microbes (1).
Degradation of various feed components is being accomplished by mutual interaction of microbiota to yield mainly acetate, propionate, butyrate, hydrogen (H2), carbon dioxide (CO2), and ammonia (NH3). Total VFA (75% of total amount) are the primary source of energy for the animal (2). Besides, microbial cell biomass is also utilized as the primary origin of protein and amino acids by host animals (3). The microbial ecosystem likewise produces vitamins B and K and utilizes the products of phytotoxin and mycotoxin detoxification processes (4). The ingested fiber is mainly degraded by bacteria and fungi into soluble nutrients (5). These soluble nutrients are subsequently used for the maintenance, growth, production, and reproduction of animals. During rumen fermentation of feed, some by-products are additionally produced, such as CO2 and H2, which are further converted into CH4 by some methanogens like Methanopyrales, Methanomicrobiales, Methanobacteriales, Methanococcales, Methanocellales, and Methanosarcinales). Some archaea (Methanoplasmatales or Thermoplasmatales) can also form CH4 through other substrates, such as methanol and mono-, di-, and tri-methylamine (6–8). Major greenhouse gases (CH4 and CO2) are released during enteric fermentation from ruminants. Production of CH4 also deprives the host animal of carbon resources and results in loss of energy (13.3 Mcal/kg CH4), leading to poor feed efficiency (9). Maximizing the flow of metabolic hydrogen ([H]) in the rumen away from CH4 and toward VFA would increase the efficiency of ruminant production and decrease its environmental impact. Czerkawski (10) proposed that inhibiting methanogenesis could favor microbial biomass production as an alternative [H] sink. Chalupa (11) suggested that metabolic hydrogen incorporated into excess NADH was redirected to fatty acid synthesis and fermentation end products such as lactate and ethanol, although the latter sinks were not quantitatively important (12).
Owing to its diverse physiological and metabolic functions, the rumen microbiome is considered the ultimate target to improve the energetic efficiency of animals while reducing environmental hazards like CH4 emissions. Highly efficient animals produce less CH4 and produce more milk, consuming less feed owing to their unique set of rumen microbiome (13). Specific physiological processes in lactating animals are correlated with specific rumen microbes owing to their unique fermentation and metabolic activities (14). The association of VFA composition with rumen bacteria has been reported in dairy cows possessing different efficiencies of production (15). Moreover, the rumen microbiome varies significantly among different animals, but intra-animal variation in microflora is quite less (16). These facts indicate the crucial role of the rumen microbiome in shaping the physiology of digestion and production in lactating animals and its potential utility for manipulation of performance and health.
Greenhouse gases (GHG) produced from ruminants have been an area of environmental concern (17, 18). Improving animal production systems must understand societal concerns and should realize the effect of such systems on the environment (19). Increasing feed efficiency to enhance animal production should also focus on CH4 mitigation strategies to reduce GHG emissions. In this regard, identification and manipulation of the microbes associated with methanogenesis are considered a significant and most crucial step (19, 20). Methanogenesis occurs both in the rumen and hindgut, but 90% of the total CH4 production originates from the rumen (21). A better understanding of digestive physiology and feed fermentation in rumen is necessary to ensure further improvement of production efficiency in ruminants to overcome the increasing demand for food by growing the human population. To achieve this daring task, manipulation of rumen fermentation is required to increase feed conversion efficiency while decreasing energy losses in the form of CH4 emanations through dietary interventions (22).
Manipulation of rumen fermentation is considered as an optimization process to seek suitable conditions for maximization and/or minimization of the specific rumen fermentation pathways, depending on factors such as type and level of feeding and animal production. The basic target behind such manipulation is the alteration in ruminal microflora that can be achieved by dietary intervention and the use of additives that selectively affect rumen communities. Improvement in ruminant production is possible with the manipulation of rumen fermentation to increase total VFA and propionate production while decreasing CH4 emission through reducing rumen methanogenesis (23). Many feed additives such as antibiotics, ionophores, and defaunating agents have been utilized to mediate rumen fermentation to improve the productivity of ruminants and reduce methanogenesis. However, most chemical additives either are noxious to host animals or present a temporary impact on methanogenesis (24, 25). Therefore, nutritionists and microbiologists are continuously trying to explore some natural substances with anti-methanogenic activity for eco-friendly animal production by reducing CH4 emission and its greenhouse effects (26).
We need to strengthen our understanding of diet–microbe interactions to devise dietary interventions to modulate the rumen microbiome to improve production efficiency and reduce energy losses in the form of GHG emissions. Moreover, we need to explore natural feed additives with limited or no adverse effects to manipulate rumen fermentation to improve feed digestibility and utilization. Plant secondary metabolites are natural substances with the potential ability to alter rumen fermentation without causing microbial resistance, and their residual effects can positively affect the animal end products (27, 28). Owing to the excellent antimicrobial activities of phytochemicals, they are considered as a potential modulator of the rumen microbiome to alter rumen physiology (29). Many experiments, including both in vitro and in vivo studies, have been conducted to explore the potential of phytochemicals on rumen fermentation to increase feed digestibility and reduce methanogenesis (30–32). Many of them have shown promising results, but applicability in terms of efficient animal production is questionable. Therefore, efforts are still underway to find an appropriate feed additive to mitigate rumen CH4 production, simultaneously improving livestock production while reducing greenhouse effects on the environment. To accomplish this challenging task, an in-depth understanding of rumen development, microbial colonization, the interaction of rumen microbiome with the host, and diet is indispensable. Therefore, this review aims to provide insights into the effect of different phytogenic and dietary interventions on ruminal microbes to mediate rumen fermentation and methanogenesis to increase overall feed efficiency to make livestock production sustainable and more profitable.
The ruminant digestive system switches from monogastric to become fully active post-weaning rumen with the ability to digest fibrous feed. During the suckling period of the calf, milk bypasses the rumen due to the esophageal groove. Developed rumen comprises 60–80% of the total digestive system as compared to the monogastric stomach in early life. Besides this, rumen villi are not yet developed, which are necessary for the absorption of nutrients (33, 34). Rumen microbial populations exhibit an incredible impact on rumen structure and physiological development. Initial inoculation of rumen microbes in calves constitutes both aerobic and facultative anaerobic microbial taxa following birth, which later on mostly are replaced by anaerobic taxa (35). That is why 1-day-old calves have a massively different bacterial population as compared to 3-days-old calves (14).
The oxidative condition of the rumen is a primary regulator of shifts in the newborn rumen ecosystem, and redox has an inert impact on the colonization of methanogenic species (36). Ruminal bacteria such as cellulolytic species, Ruminococcus flavefaciens, and Ruminococcus albus and members of the Prevotella genus can already be detected on day one after birth. These microbes are involved in various rumen functions, such as cellulose and hemicellulose degradation (14). One of the primary changes observed throughout the rumen development includes modification in configuration within the Bacteroidetes phylum. In the developed rumen, this phylum is dominated by the genus Prevotella across several ruminant species (37). Nevertheless, during the primary stages of development, Bacteroides is the main genus within Bacteroidetes and is subsequently replaced by the Prevotella during the first 2 months (38).
Quick fluctuations in community configuration also affect methanogenic archaeal communities along with bacteria. Rumen methanogenic communities in calves and lambs have been detected as early as 20 min after birth. Like bacterial populations, the primary methanogenic population varies significantly between young and adult animals (39–41). Both preweaning calves and mature animals have a Methanobacteriales order, but rumen of preweaning calves contains two additional orders, Methanosarcinales and Methanomicrobiales (41). The compositional variations in rumen archaea lead to shifts in substrate utilization, methanogenic pathway, and extent of CH4 production (42).
The establishment and colonization of microbiota play a key role in the development and function of the gastrointestinal tract (GIT), which is subsequently associated with higher body weight and feed efficiency of growing ruminants (43). Developing a rumen ecosystem during weaning age is key to getting improved growth rates and better health at a later stage of life (44, 45). The main objective of such strategies is to overcome the risk of undesirable health consequences associated with an altered gut microbiome in neonatal animals and restoration of the gut microbial community following dysbiosis. A complete understanding of early gut colonization is necessary to designing different effective strategies to manipulate the GIT microbiome. Although a wealth of literature is available on different aspects of rumen microbiome in adult animals and early colonization of gut microbiota, information regarding the role of host genetics and microbial interactions in the early development of the gut microbiota is limited.
Ruminants can transform fibrous and non-fibrous plant material into valuable products like meat and milk with the help of rumen microbes (46). Rumen inhabits various microbes like bacteria, protozoa, fungi, archaea, and bacteriophages (47). A symbiotic relationship exists between rumen microbes and the host animal in which both provide coveted substance to each other mainly in three ways: (1) mastication and rumination expand the surface area of feed particles for microbial attachment and digestion, and consequently, microbes secrete fibrolytic enzymes for degradation of cellulose, and hemicelluloses; (2) ruminal movements (peristalsis and antiperistalsis) bring microbes in contact with the fresh substrate by mixing of digesta and consequently yield fermentation products, especially VFA; and (3) elimination of fermentation products by belching and absorption is essential for keeping ideal conditions (pH) for microbial development and utilizing non-protein nitrogen (48).
Ruminal bacteria are the most prevailing microbiome, and their population measured by direct counts is usually 1011 cells per gram of rumen contents (4) comprising more than 200 species (49). Bacteria colonize inside rumen and have a major role in the metabolism of dietary carbohydrates and nitrogen and utilize fiber, starch, protein, and sugars. Generally, ruminal bacteria used homoserine lactone-based quorum sensing to communicate with each other (50). The most important genera of ruminal bacteria are Butyrivibrio, Prevotella, Ruminococcus, and Pseudobutyrivibrio. Mainly CH4 emissions depend upon the abundance of H2-producing bacteria in the rumen (51).
Ruminal fungi comprise 5–20% of the total microbiota in the rumen (52, 53). Anaerobic fungi are known as key players for the breakdown of lignocellulosic fiber (54). Anaerobic fungi are considered one of the most potent fiber-degrading agents, because of their active and extensive set of enzymes for the breakdown of plant polymers (55). Fungi produce enzymes vital for the digestion of plant materials, including cellulases, xylanases, mannanases, esterases, glucosidases, and glucanases (56). Rumen fungi also possess amylolytic (57) and proteolytic activities (58). The action of anaerobic fungi is promoted by the methanogenic archaea (59). However, the present understanding of rumen eukaryote function is far less than that of rumen bacteria, primarily due to the restricted annotation of the transcriptome and multiple-genome sequence availability (60). A recent in vitro study reported that a combination of anaerobic fungi (Caecomyces) and methanogens (Methanobrevibacter) have a greater ability to degrade lignocellulose and to produce CH4 as compared to the combination of bacteria and methanogens, and whole rumen content enrichment (61).
Ruminal protozoa represent about 20–50% of total microbial biomass and are commonly grouped into flagellates and ciliates. The flagellate proportion to overall ruminal fermentation is negligible (62). However, ciliate protozoa have a fundamental function in rumen fermentation as they engulf fermentable carbohydrates (63) and prevent alternative bacterial fermentation that would otherwise decrease pH and increase the onset of lactic acid acidosis (64). There is a positive correlation between ruminal protozoa and volatile fatty acid and CH4 production. Ciliate protozoa can enhance the metabolic output of the rumen microbiome; for instance, acetate, butyrate, iso-butyrate, and iso-valerate concentrations were improved in microcosms incubated with the protozoa population (65). The hydrogenosomes of rumen protozoa are involved in the production of H2, which is subsequently converted to CH4 by the methanogens through the hydrogenotrophic pathway (66, 67). Approximately 11% reduction in the CH4 output has been observed due to the defaunation of protozoa (64, 68).
Archaea represent the third major domain of rumen microbes that constitute about 21% of the rumen microbiome (69). Methanogenic archaea belong to the phylum Euryarchaeota and are ubiquitously involved in methanogenesis (7, 69). Rumen methanogens have a synergistic association with bacteria and a symbiotic association with protozoa as <1% of the total microbial population (70). Different substrates are utilized during methanogenesis including formate, or acetate, methanol, H2, methylamines, and CO2 (71). Methane is produced mainly through three pathways: (i) primarily by reduction of CO2 through the hydrogenotrophic pathway, (ii) less from the use of methyl groups (methylotrophic pathway), and (iii) even less through acetate (acetoclastic pathway) production (Figure 1). Methanogenic paths comprise three stages: exchange of the methyl set to coenzyme M (CoM-SH), reduction of methyl-coenzyme M with coenzyme B (CoB-SH), and reuse of heterodisulfide CoM-S-S-CoB (51, 68, 73).
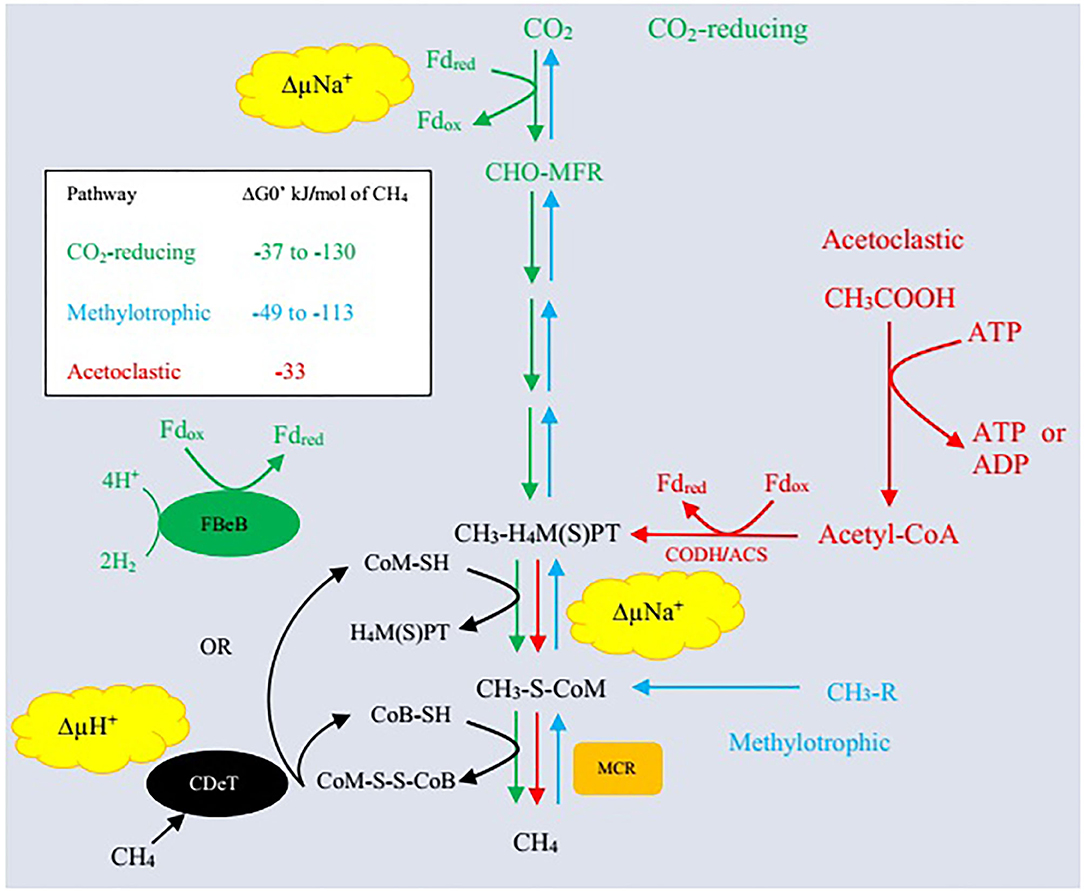
Figure 1. Three enzymatic pathways of methanogenesis. Δ, CO2-reducing pathway (hydrogenotrophic pathway); Δ, acetoclastic pathway; Δ, methylotrophic pathway; MFR, methanofuran; H4MPT, tetrahydromethanopterin; H4SPT, tetrahydrosarcinapterin; ΔG0′, standard free energy change; CH3-R, methyl-containing compounds such as methanol, methanethiol, dimethylsulfide, monomethylamine, dimethylamine, trimethylamine, and tetramethylammonium; Fdred, reduced form of ferredoxin; Fdox, oxidized form of ferredoxin; ΔμNa+, electrochemical sodium ion potential; ΔμH+, electrochemical proton potential; FBeB, flavin-based electron bifurcation; CDeT, cytochrome-dependent electron transfer; MCR, methyl-coenzyme M reductase; CODH/ACS, carbon monoxide dehydrogenase/acetylCoA synthase/decarbonlyase complex. Adapted from Lyu et al. (72).
The bacteriophage community is also an important component of the rumen microbial ecosystem. Studies have reported inconsistent findings of bacteriophage counts ranging from >109 particles of phages (74) to between 3 × 109 and 1.6 × 1010 particles per ml of rumen content (75). Bacteriophages possess a specific lysogenic ability against different bacteria that helps in bacterial mass turnover in the rumen. Due to a lack of information regarding the mechanisms of rumen phage–host interactions and the environmental factors affecting the relative proportions and dynamics of the phage population in the rumen, it is not possible to definitively determine whether the presence of phage in the rumen is disadvantageous or advantageous. However, possible functional consequences of rumen phages have been proposed as (1) the negative nutritional consequences of phage-induced bacterial lysis resulting in the recycle of nutrients within the rumen, (2) the positive effects of maintaining bacterial population diversity and facilitating gene transfer, and (3) the negative consequences of phage-mediated gene transfer.
Keeping in view their critical role in digestive physiology and nutrient metabolism, modulation of the rumen microbiome is envisioned as a practical strategy to mediate fermentation kinetics and methanogenesis. Modulation of the rumen microbiome can be possible through different dietary interventions; however, in this regard plant secondary metabolites possess a greater potential as compared with antibiotics to modulate the ruminal microbiome and mitigate CH4 emission through diverse antimicrobial mechanisms such as perturbation of cell membrane, modulation of signal transduction or gene expression pathways, enzyme inhibition, and inhibition of bacterial colonization (76, 77). Plant secondary metabolites usually enhance the permeability and fluidity of the cellular membranes, further causing an efflux of metabolites and ions and ultimately leading to cell leakage and microbial death. Besides, they can also desirably manipulate the rumen metabolism by increasing the cell membrane's permeability of few specific rumen bacteria (78, 79). Putative mechanisms of actions mainly include disturbance of the cytoplasmic membrane, disruption of the proton motive force, electron flow, active transport mechanisms, and coagulation of cell composition (80).
The host diet has a major influence on the relative abundance and diversity of the rumen microbiome. However, genetic manipulation of rumen microbiota is also possible through different techniques as host genetics influences some heritable microbial traits (81–83). In this regard, recent biological techniques such as transgenesis are getting attention for improving the efficiency of animal production and reducing environmental impacts (84). New genome editing tools provide an efficient way to produce gene-edited ruminants, having resistance against certain diseases and specific product quality (85). In transgenic animals usually, a foreign gene of interest is inserted into its genome to express a desirable trait (86). In a study of pig transgenesis, neomycin phosphotransferase transgene has been evaluated using high-throughput sequencing. Neo-transgenic expression in transgenic pigs showed a significant increase in the relative abundance of some bacteria (Firmicutes, Bacteroidetes, and Proteobacteria) with a reduction of potentially harmful bacteria such as Escherichia–Shigella–Hafnia (87). A recent study by Yang et al. (88) showed inactivation of the ABO acetyl-galactosaminyl-transferase gene through a deletion of 2.3 Kb, potentially affecting the microbiota composition and its relative abundance (particularly Christensenellaceae and Erysipelotrichaceae families).
Presently, different studies are being focused to identify heritable microbes and microbial features in humans and animals, considering gut microbiomes as heritable phenotypes, (89). Elucidation of the association between host genetics and rumen microbiome composition will help to identify persistent microbial taxa and functions that are characteristic of efficient animals, thereby directly facilitating efforts to genetically select or permanently alter the rumen microbiome. Within the next 5–10 years, significant progress is expected regarding the relationship between gut microbiomes and the genetics of their ruminant hosts, unraveling an intricate network that paves the way for the genetic selection of heritable microbes and keystone microbial species. These efforts will rapidly advance microbial ecology research and animal production to efficiently and sustainably produce high-quality protein for human consumption to considerably contribute to global food security (90).
Emerging feeding techniques to limit CH4 emissions are necessarily required both for preserving the environment and for increasing the efficacy of energy utilization. Different microbes are involved in the production of VFA (Figure 2). The rumen of dairy cow possesses diverse consortia of microbes that produce significant amounts of GHG gases (mainly CH4) during feed digestion (48). Methodologies to divert rumen carbon and nitrogen metabolism away from these products offer opportunities for improving the efficiency of ruminant production by enhancing nutrient utilization while reducing GHG emissions.
Many studies have been conducted regarding the development of nutritional interventions to cut down CH4 emissions from ruminants (91–93). With increasing food safety concerns, different natural compounds of plants are being considered ideal for moderating/mitigating CH4 emissions (94). The diet fed to ruminants is the primary determinant of bacterial community structure (95–97). Co-oscillations of microbiota are very important to maintaining homeostasis in gastrointestinal ecology after dietary perturbations (98). It is important to adjust the animal diet according to their age and physiological condition and to provide proper time to adapt to different dietary changes. Diet is also considered as an important factor to ensure proper animal health and performance because digestion and utilization of nutrients mainly depend upon collaboration and competition of the microbiome. Important diet–microbe interactions are discussed as below.
Rumen microbiology involves the characterization of microbes and processes associated with fiber digestion (17, 99, 100). Rumen bacteria play a dominant role in fiber digestion, although anaerobic fungi and protozoa have been reported to contribute to lignocellulose breakdown by attacking lignocellulosic material differently (60, 64) and discharging different enzyme complexes (101). Microbial communities found in the rumen of cows fed total mix ration (TMR) vary as compared to pasture-fed cows, owing to the variable dietary composition (102, 103). Different rumen bacteria have shown associations with specific diets like Fibrobacteraceae with TMR and Veillonellaceae with pasture-based diets (104).
Feeding concentrate diets have shown to lower the ruminal pH, increase VFA concentrations and osmolality, and induce metabolic disorders (105). Moreover, high starch feeding substantially increases the activity of lactic acid consuming and producing bacteria in the rumen because these microbes are not susceptible to lower pH and hence opportunistically utilize higher substrate availability (106). In contrast, feeding a considerable amount of roughages may limit feed consumption, energy efficiency, and microbial protein synthesis in ruminants (107). Therefore, an increasing quantity of starch in the diet is considered as a promising strategy to decrease methanogenesis per unit of dry matter intake through shifting ruminal fermentation toward propionogenesis (108). It is mainly attributed to the fact that the supply of substrates and microbial growth depends upon the dietary fiber or starch contents (109). Increased dietary starch levels have shown a relatively lower proportion of cellulolytic bacteria R. albus and R. flavefaciens, owing to their higher sensitivity toward low pH. However, in an acidic ruminal environment F. succinogenes have shown to remain stable due to their gram-negative nature and different cell membranes than R. albus and R. flavefaciens (110). Feeding grain-based diets to cattle could reduce the bacterial diversity compared to forage-based diet (111). This reduction might be attributed to less availability of substrates for bacteria that ferment structural carbohydrates and the subsequent lower pH.
Feeding of high starch and low fiber diets has also been shown to enhance the growth of amylolytic bacteria in the rumen. Propionate is produced by S. ruminantium through decarboxylation of succinate (6). This bacterium is capable of using starch and sugar for its growth. Higher dietary starch contents have been shown to increase the concentrations of propionic acid in the rumen substantially. Furthermore S. ruminantium is also capable of using lactic acid to stabilize the rumen pH (112). Thus, an increase in the population of these bacteria can enhance the utilization of fermentable substrates generated in the rumen following a high concentrate diet. A facultative anaerobe (S. bovis) predominated in ruminants fed an increased amount of concentrate during lactic acidosis (109). The activity of S. bovis only increases as a result of low pH (<5.75) conditions in the bovine rumen (113). According to McCaughern et al. (114), feeding a high-starch (220 g/kg of DM) diet to dairy cows could reduce rumen pH (0.15 units lower than normal pH) and increase milk yield (0.09 kg/d) and milk protein content (2.8 g/kg). Feeding a high-starch diet can directly affect the colonic lumen environment, which in turn alters the lumen-specific functional taxonomic groups (Akkermansia, unclassified Christensenellaceae, and vadinBB60). Consequently, the colonic epithelium makes a new niche that triggers cell apoptosis to achieve a functional transformation (98). These studies suggested that microbe–host interaction is vital for remodeling of hindgut homeostasis to allow adaptation to dietary perturbations.
Silage from various sources harbors different rumen microbial communities. Feeding of alfalfa silage increased the relative abundance of F. succinogenes and R. flavefaciens while reducing CH4 production in the cow rumen as compared to sweet sorghum silage. However, populations of Ruminococcus albus and Ruminobacter amylophilus showed no change (115). Contrarily, sheep fed an alfalfa hay diet had higher ruminal Fibrobacter succinogenes compared to Ruminococcus (116). According to Guo et al. (117), fermented corn stover showed a positive effect on ruminal bacterial diversity favoring four bacterial phyla; Bacteroidetes, Lentisphaerae, Firmicutes, and Fibrobacteres, which constituted 77% of total bacterial abundance. Additionally, feeding of fermented corn stover shifted the rumen fermentation kinetics in cows through increasing the relative abundance of Prevotella and stabilizing the rumen microbial ecosystem. Recently, it has been reported that feeding temperate grasses produce less enteric CH4 than tropical grasses in ruminants (118) as feeding low-quality tropical grasses emitted 17 g CH4/kg DM intake. However, a 10–25% decrease in CH4 production has been observed, when foliage and pods of trees and shrubs are included in the cattle diet (118).
A recent meta-analysis showed that feeding high-forage diets (>40% DM) reduced milk production (0.087 L/d) and milk lactose content (0.065 g/100 g) compared to high-concentrate diets (>40% DM) in sheep. However, fat content and conjugated linoleic acid concentrations were higher in the high-forage group (119). These findings convincingly reveal that the appropriate ratio of roughage and concentrate is required to optimize the rumen microbial ecosystem for better digestion and utilization of the dietary components while minimizing CH4 emission. Chemostatic feedback regulation (energy feedback), physical fill and feed passage rate of concentrate, and forage-type diets are involved in affecting the DM intake of ruminants. Moreover, inoculation of silage with specific groups of beneficial microbes can positively influence rumen fermentation kinetics and performance of animals on a sustainable basis.
Generally, supplementation of fatty acids (FA) is not required for microbial proliferation in the rumen because microbes can synthesize their own FA. Basal feed ingredients, including forages and grains, provide about 3–3.5% fat on a dry matter (DM) basis. However, for high-producing dairy cows, additional fat supplementation up to 2% of rumen-active fat (vegetable blends, oilseeds) and rumen-inert fat is usually recommended to make total dietary lipids up to 6–7% of DM (120). These dietary lipids are usually enriched in polyunsaturated fatty acids (PUFA) (121), which can make complexes with bacterial cell walls and are considered toxic to gram-positive bacteria in the rumen (122). In most ruminant diets, fat is below 5% of total DM. Higher dietary fat contents, primarily unsaturated FA, are discouraged owing to their adverse impacts on ruminal bacteria and feed degradation (123). However, rumen microbiota can detoxify unsaturated FA through the biohydrogenation process to reduce/eliminate the adverse effects on rumen fermentation (121, 124).
The addition of fats from plant or animal sources is an accepted approach for mitigation of CH4. However, consideration of fat supplementation to mitigate enteric CH4 emission depends on the cost and expected adverse effect on feed intake and digestibility (19). Improving the nutritive quality (high fat and fiber digestibility) of the offered diets has shown to reduce the DM intake in lambs but showed no effect on total tract digestibility of DM, organic matter (OM), crude protein, acid detergent fiber (ADF), and neutral detergent fiber (NDF) contents (125). Adding fat up to 6–7% of DM has shown no adverse effects on total tract digestibility (126). Recent studies have reported no effects of rumen-protected fats on NH3-N concentrations, total VFA, and overall bacterial population in sheep (127). Changes in rumen microbiota (Acetitomaculum, Lachnospira, and Prevotella) caused by an increased proportion of concentrates were considerably more significant than fat (128). Fibrobacter and Ruminococcus were most adversely affected among different bacterial genera, but such effects were highly variable for Butyrivibrio and Prevotella. These two genera (Butyrivibrio and Prevotella) include many species with diverse functions in metabolic pathways (129).
A decrease or no change was observed in major protozoa genera in response to the addition of linseed oil, particularly in high-concentrate diets (130). Moreover, increasing the degree of unsaturation reduces the protozoal count, but due to high random and animal variations, this change can be challenging to assess, which may explain inconsistent experimental data (131). Dietary supplementation of camelina oil has not shown any effect on ruminal protozoa (132), but a decrease in bacterial N and the number of cellulolytic bacteria was observed in diets supplemented with 8% dietary lipids (133). However, Bayat et al. (134) reported that the inclusion of camelina oil in the diet exhibited no effects on the relative abundance of protozoa, total bacteria, methanogens, fungi, and fiber-degrading bacteria.
Lipids from oilseeds, vegetable oils, and rumen-protected fat of vegetable oils are usually used as energy sources for dairy cattle (135). Oilseeds can be one of the efficient ways to reduce enteric CH4 production to mitigate CH4 emission from ruminants. Plant oils can mitigate CH4 by directly inhibiting rumen protozoa and methanogens and increasing the biohydrogenation of PUFA to act as a sink for hydrogen produced by rumen microbes (136). The utility of lipids to reduce enteric CH4 production is a better strategy as compared to antibiotics and ionophores like monensin. Several studies have reported adverse effects of FA, especially PUFA, on methanogenesis in the rumen (136). The anti-methanogenic effects of PUFA generally get intensified with the increase in double bond number per FA, as suggested by Czerkawski and Clapperton (137).
Supplementation of fat has been shown to reduce CH4 emission in ruminants consistently. However, various factors such as fat source, FA profile, basal diet, and fat type can affect the anti-methanogenic efficiency of dietary fats (93). This reduction in CH4 emission by dietary fat is mainly through the depressed fiber digestion in the rumen (138). However, according to McGeough et al. (125), a highly digestible fiber diet with higher fat content tended to increase CH4 emissions per kg of DMI and OMI, while the same amount of fat showed no effect on these parameters in a diet with low fiber digestibility. The inclusion of fat in a high-concentrate diet of sheep improved fat and conjugated linoleic acid contents of milk (119). Bayat et al. (139) reported a decrease in daily CH4 emission in lactating cows fed a low concentrate diet supplemented with sunflower oil.
Recently, moringa and camelina oils have shown to effectively reduce enteric in vitro CH4 production in different TMR through modulation of rumen microbes and shifting rumen kinetics (140, 140). Variable effects of vegetable oils and unsaturated FA on CH4 emission might be associated with the double bond number per FA, type of oil (free oil or whole seed), and composition (roughage-to-concentrate ratio) of the rations (134). An in vitro study of Vargas et al. (141) shows that supplementation of vegetable oil (sunflower and linseed) at 6% in high-concentrate TMR has the potential to reduce CH4 emission (up to 21–28%), butyrate concentration, and A:P, while increasing propionate concentration. Recently, a meta-analysis showed that addition of nitrates and vegetable oils in cattle diet has the ability to reduce CH4 emission up to 6–20% (118). Considering different studies regarding the reduction of CH4 emission, the addition of plant oils to ruminant rations is suggested as a feasible nutritional strategy with a cleaner repercussion on the environment.
Dietary changes have been reported as a major factor that influences the dynamics of rumen microbial populations and resultant metabolic shifts leading to significant changes in ruminant production (142, 143). Many dietary interventions have been used in ruminants for the manipulation of the rumen microbiome to improve overall feed efficiency while reducing methanogenesis. A few dietary methodologies have been assessed for enhancing rumen fermentation, mainly to reduce CH4 emission. These strategies were focused to (i) improve feed efficiency using the quality feed, (ii) shift rumen fermentation pathways using assorted feed additives, and (iii) genetically manipulate host animals using selective breeding. Each strategy has some potential advantages and limitations. Recent issues of drug residues and antibiotic resistance have shifted the interest toward natural feed additives with potential abilities to modulate performance in ruminants. Many encouraging results have been observed by the application of different feed additives, including organic acids, probiotics, enzymes, and phytochemicals. Ideally, feed additives should diminish CH4 emission, enhance animals' energetic efficiency by increasing propionate concentration, improve N2 utilization efficiency by decreasing its excretion, optimize rumen pH, and improve fiber digestion (144).
Most important natural feed additives are phytochemicals produced by plants as secondary metabolites with diverse biological activities. Some potential effects of these feed additives and their mechanism of action as rumen modulators are described as below.
Despite the sustainability of functional redundancy in the rumen microbiome, plant secondary metabolites have shown significant manipulation of the rumen microflora leading to a shift in fermentation dynamics and milk production in lactating animals (29, 31, 145–147). They have also shown to reduce the methanogenesis both in vitro and in vivo (29, 31, 76, 148, 149). Recently, numerous plant extracts have been investigated for their capacity to manipulate gut physiology and antimicrobial activity. Some plant metabolites, for example, saponins, tannins, and essential oils (EO), have shown promising potential for decreasing CH4 emission from animals. They have shown significant impacts on methanogens as well as protozoa, feed degradation/absorption, and fermentation parameters.
Saponins are a class of plant secondary compounds with diverse chemical compositions and biological activities (31). Saponins comprise mainly sapogenins and glycosides found mostly in angiosperms. Steroidal and triterpenoid saponins are two significant groups of saponins, which protect plants from bacterial and fungal invasions (150). Saponin-rich plants such as lucerne and soybeans are broadly utilized for ruminant feeding. Additionally, Quillaja saponaria (soapbark), Yucca shidigera (yucca), and Sapindus sp. (soap berries) are considered as well-known sources of saponins (151). Saponins have a fat-soluble nucleus, and they have shown antibacterial, antitumor, and anti-inflammatory properties in animals (152, 153). Saponins mediate rumen fermentation mainly by decreasing protein degradation and concentrations of urea and NH3 in the rumen, leading to an increased flow of amino acids to the small intestine. Potential effects of saponins are associated with N2 metabolism, mainly through their lethal effect on protozoa, which are primarily responsible for proteolytic activity in the rumen (154). Saponins reduce the protozoal population and some methanogens associated with protozoa, although their effect on methanogens does not always correlate with the effect on protozoa (31). The interaction of sterol moiety with saponin, present in the protozoa membrane, has an association with the antiprotozoal effect of saponins (154).
It is generally expected that a reduction in the population of methanogens can decrease CH4 emissions. Extracts of S. sesban have shown to decrease protozoal and methanogen populations but surprisingly did not decrease CH4 production (155). There was a frail relationship between methanogenesis and methanogens in the rumen. This is mainly because the enhanced expression of some methanogenic genes may lead to enhanced methanogenesis that ultimately compensated a decrease in the overall number of methanogens (156). These findings provide explicit insight regarding, managing gene interactions from the microbiome to enhance nutrient utilization efficiency.
Tea saponins have been extensively used in many studies in vitro and in vivo to evaluate the effects on rumen fermentation and methanogenesis. Variable results regarding microbial population and rumen fermentation parameters have been observed in response to the supplementation of different sources of saponins (Tables 1, 2). Despite the decline observed in methanogenesis through a direct decrease in protozoa populations under in vitro conditions, the same plant extract (0.52% on DM basis) failed to reduce daily CH4 production in lactating dairy cows (162). This suggests that the impacts of tea saponins under in vitro conditions must be confirmed in vivo to develop effective CH4-mitigating strategies.
Inactivation of saponins has been observed through deglycosylation into sapogenins by the rumen microorganisms that lead to the transitory antiprotozoal property of saponins. There are two approaches to improve the effectiveness of saponins and reduce their degradation by rumen microbes. One possible method is to use a combination of saponins with glycosidase-inhibiting iminosugars (167). The second option is altering the saponin structure, such as by combining ivy saponins with stevia extract. Hederagenin bis-succinate (HBS) obtained by hydrolysis of ivy fruit extract has shown to shift fermentation toward propionate, attributed to its structural modifications that mediated the diversity of bacterial communities (167).
Recently, tea saponins have been supplemented in alfalfa hay and soybean hull-based fiber diets and exhibited their ability to alter ruminal lipid metabolism in cattle through reducing the relative abundance of Lachnospiraceae (168). Furthermore, tea saponins have also been shown to effectively decrease N2 emission in sheep (157). However, studies have revealed that the activity of saponins fluctuates and even reduces during long-term studies (158), probably because of microbial adaptation (169). Moreover, saponins can increase the propionate ratio at the expense of both acetate and butyrate (158). Studies have shown that a combination of garlic oil, nitrate, and saponins can additively lower CH4 emission with similar rumen fermentation and degradability (163). Archaeal growth was inhibited by all treatments, but the abundance of F. succinogenes R. albus, and R. flavefaciens varied. According to Liu et al. (157), tea saponin did not affect methanogens and the total bacterial population including R. flavefaciens R. albus, and Butyrivibrio. However, protozoa were effectively reduced in response to tea saponins.
All saponins have no inherent antiprotozoal activity; that is why their biological activity can be affected by even small changes in their structure. For instance, sapogenins like asiatic acid and madecassic acid have more ability for the inhibition of protozoa than their corresponding saponins (Re and Rh1 and madecassoside). Therefore, further research is warranted to understand the deglycosylation of saponins and the nature of their antiprotozoal activity to devise effective ways to use saponins for CH4 mitigation in ruminants (170).
Tannins are polyphenolic compounds with molecular weights ranging from 500 to 5,000 Da with two major groups [i.e., condensed tannins (CT) and hydrolyzable tannins (HT)]. Tannins can bind with dietary proteins, starch, and sugar by making strong complexes at pH 3.5–7 (150). Tannins are widely distributed in different plant species, particularly in cereals, legumes, and fruits. They can limit the digestibility and nutritional value of plants considerably when their concentration reaches more than 5% (171). The action of tannins in the rumen is not entirely well-conceived yet (172). Although they possess bacteriostatic effects, the association of tannins with the rumen microbes is different, as hydrolyzable tannin is more susceptible to microbial hydrolysis than condensed tannin (173). They can limit the degree of microbial hydrolysis along with direct inhibition of methanogens. Additionally, they can also lower methanogenesis indirectly by decreasing H2 availability by reducing fiber digestion (Figure 3). Tannins can modify the ruminal microbiome, reduce protein degradation, decrease methanogenesis, and inhibit FA biohydrogenation (174, 175).
Variable results regarding the shifting of microbial population and rumen fermentation parameters have been observed in response to the supplementation of different sources of tannins (Tables 3, 4). Studies have reported quite different effects of tannin supplementations regarding CH4 mitigation. Some studies have also shown that tannins indirectly impede the degradation of fiber (76). A recent study showed that supplementation of acacia tannin (15 g/d/animal) reduced short-chain fatty acids (SCFA) and acetate (molar percentage) in lambs affected by gastrointestinal nematode infection. Furthermore, supplementation also increased the diversity and abundance of butyrate-producing and other beneficial bacteria (including probiotic species like Bifidobacterium and Lactobacillusamino), while enhancing the amino acid metabolic pathways and purine, pyrimidine, and sphingolipid metabolism (176). However, the precise mechanism of action and extent of contributory effects of acacia tannin on the ruminal microbiome are still not clear. Supplementation of tannic acid has shown to reduce the CP digestibility and CH4 production in beef cattle (198). Dietary supplementation of HT (chestnut) and CT (quebracho) increased the relative abundance of Butyrivibrio fibrisolvens by 3 and 5-fold, respectively, in the rumen of dairy sheep. On the other hand, they decreased the B. proteoclasticus population by 5 and 15-fold, respectively (199). Inhibition of rumen bacteria by CT is probably due to interactions between CT present in the tannin structure and the specific substrate (e.g., protein, bacterial cell walls, etc.) to which it binds (200). The addition of CT extracts to the diet reduced populations of methanogenic archaea and some cellulolytic bacteria (R. flavefaciens) (201). These reports suggest that dietary sources of HT and CT can affect the rumen microbiome quite differently owing to their structural variations.
Different sources of tannin have been used to mitigate CH4 emission while increasing animal performance. Gallic acid is a phenolic monomer and one of the ruminal decomposed metabolites of tannic acid (173), and it serves as an essential bioactive component in modifying the rumen fermentation. As a subunit of HT, gallic acid has the potential to decrease the environmental impact of ruminants (by lowering CH4 and NH3 emissions) without decreasing animal performance (180). Gallic acid (0.015%) decreased the urine nitrogen emissions by 28.5% (CP 0.11% DM) and 30.9% (CP 0.15% DM) when applied to the soil (202). A recent study showed that gallic acid can inhibit undesirable microorganisms such as Clostridium, Listeria, and Escherichia coli during ensiling and can improve fermentation quality and protein preservation (203). Extracts of HT (tara) and CT (mimosa and gambier) inhibited the activity of methanogens and protozoa without affecting ruminal fermentation and animal production (185). Condensed tannins have shown better protein efficiency and growth rate of lambs as it can protect dietary proteins (e.g., soybean meal) from ruminal degradation, leading to reduction in digestive losses (204). Recently an in vitro study of Saminathan et al. (205) showed that tropical legumes having CT with different molecular weights can serve as potential feed additives to mitigate CH4 production with no adverse effects on rumen fungal microflora and fiber digestion. Contrarily, Rira et al. (196) reported that in vitro HT (A. nilotica) are more promising for suppressing methanogenesis than CT (from C. calothyrsus and L. leucocephala). Chestnut tannin possesses sufficient potential to reduce methanogenesis, without compromising feed efficiency and animal performance due to its neutral effect on NDF digestibility (177). According to Witzig et al. (184), CH4 emission was reduced in response to monensin and chestnut tannin supplementation, owing to the lower abundances of M. ruminantium and M. stadtmanae.
Tannic acid (0–1.25 mg/mL) can alter microbial activities and improve feed efficiency in ruminants. However, the increased tannic acid concentration may lead to the complete inhibition of ruminal bacteria in sheep (206). Mimosa CT could reduce the abundance of specialized fibrolytic bacteria and inhibit the biohydrogenation process as compared to chestnut HT. Further investigations are required to evaluate the impact of different sources of tannins on ruminal biohydrogenation (186). Hydrolyzable tannins are considered more suitable for CH4 mitigation than CT. A. nilotica (HT) showed a more potent inhibitory effect on CH4 production compared to C. calothyrsus and L. leucocephala (CT). It may be attributed to the fact that HT (e.g., gallic acid subunits) directly inhibit methanogens, but the action of CT on rumen CH4 production is variable (172, 207). However, long-term trials are required to assess the possible adaptation of rumen microbes toward the optimal level of HT and its subunit, gallic acid, to avoid their adverse effects on animal performance (196).
Tannin containing hay has been shown to reduce the CH4 emission (5.4 DM vs. 3.5 ml/g) and urea N excretion in beef cattle (187). To reduce the adverse effect of tannin on DM intake, the encapsulation of tannin extract could be considered as a better strategy as the slow release of tannin also improves its utilization (208). Recently, Adejoro et al. (189) determined the effect of crude (40 g/kg feed) and lipid encapsulated-acacia tannin (50 g/kg feed) extracts on sheep fed TMR. They reported a 30% and 19% reduction in CH4 production (g/kg DM) with crude and encapsulated-acacia tannin, respectively. However, crude tannin also imparted an adverse effect on NDF digestibility compared to encapsulated-acacia tannin. Supplementation of tannin could reduce the NH3 toxification which is usually produced in response to NPN addition in ruminant diets (208). More recently, Adejoro et al. (188) reported that supplementation of 42 g acacia tannin /kg feed DM did not reduce CH4 production in lamb fed nitrate or urea as an NPN source. A possible reason is the comparatively higher affinity of acacia tannin for feed protein than microbial protein or microbial enzymes (209). Tannin has the potential to reduce the excretion of a more volatile form of N into the environment by decreasing rumen degradability of CP and shifting N excretion from urine to feces (210). A meta-analysis showed reduction in ruminal ammonia N (16%), milk urea (9%), and urinary N excretion (11%) in response to supplementation of tannin in lactating dairy cows. However, tannin exhibited no effect on fat- and protein-corrected milk yield (211). A short-term effect of A. mearnsii (30 g/kg) showed a negative effect on CH4 production in dairy cows.
Dietary supplementation of oak tannin has also been shown to reduce the urinary N excretion by 12% while increasing α-linolenic acid content in milk by 17.7% without affecting CH4 production (190). Such divergent findings may possibly be due to different dietary concentrations of tannin and a variable number of hydroxyl groups in their structure (207). Studies have suggested an association of milk FA profile with CH4 emission, which can assist in determining the impact of tanniferous supplement against enteric CH4 production (212). The medium-chain FA (lauric and myristic acids) have shown a positive correlation with enteric CH4 production as these FA are synthesized (de novo synthesis) in the mammary gland from ruminal acetate and butyrate (213). In this regard, it has been reported that supplementation of quebracho tannins (30 g/kg DM) could reduce myristic acid content in dairy cows, which reveals the negative effects of tannin on fiber digestibility (214). However, long-chain FA (pentadecanoic and heptadecanoic acid) exhibited a negative correlation with CH4 emission as their de novo synthesis is mediated from ruminal propionate (215). However, further careful investigations are warranted to corroborate this relationship.
Tannins also possess antioxidant properties as they can scavenge free radicals due to hydroxyl groups, degree of polymerization, and redox activities (216–218). Supplementation of tannin has been shown to improve the antioxidant status of cattle and sheep (219–221). Hydrolyzable tannins are considered the most potent antioxidants, which can prevent cellular damage and neutralize free radicals (222), while condensed tannins (catechin) also possess antioxidant activities (223). Pomegranate as hydrolyzable tannin has shown better antioxidant activity as tested on cultured bovine aortic endothelial cells; it has no adverse effects on cell viability and apoptosis. Pomegranate has also exhibited protective effects against membrane lipid peroxidation, owing to its potent ability to reduce the production of intracellular reactive oxygen species (224). However, the optimum level of tannin for antioxidant capacity and its putative mechanism of action in animal tissues require further elucidation. A recent study has revealed that rumen microbial taxa (Bifidobacterium, Lactobacillus, and Schwartzia) exhibited a strong association with host antioxidant capacity and immunomodulatory functions (225).
Prolonged use of the purified form of secondary compounds can lead to antimicrobial resistance; however, tannin supplementation as a crude extract of mixtures (having different molecular sizes) offers a major advantage to control antimicrobial resistance (226). Moreover, studies have shown that CT-rich diets can effectively decrease CH4 emissions per unit of DMI over a range of dietary CP from 15 to 25%. For example, a decrease up to 25 to 50% was observed in in vitro CH4 production in steers grazing on winter wheat forage (15 to 18% CP) supplemented with quebracho CT extract at 10–20 g/kg DMI (227, 228). This shows the effective inhibitory effects of tannins on ruminal protein degradation and CH4 emission but requires careful selection of diets and nutrient composition to avoid adverse effects on feed digestibility and efficiency (229). However, further studies are required to fully understand the mechanism of action of tannins regarding modulation of the rumen microbiome, potential inhibitory effects on methanogens and protozoa, and their optimum inclusion levels to elucidate their potential for CH4 mitigation. Furthermore, focused investigations are required to explore the optimum levels and types of tannins and feeding conditions to reduce GHG emission in commercial ruminant production systems.
Essential oils are terpenoids (monoterpenoids and sesquiterpenoids) and phenylpropanoid compounds with characteristic flavors and odors, formed by different plants (herbs and spices). They contain numerous chemical substances, for example, alcohols, hydrocarbons, ketones, aldehydes, ethers, and esters, and mostly EO are lipophilic complexes (230, 231). Various studies have been performed to evaluate the effect of EO on rumen fermentation and feed degradability. Many in vitro and in vivo trials have proved the favorable effect of EO in reducing CH4 production and altering microbial populations (Tables 5, 6). The potential effect of EO on rumen fermentation and methanogenesis is mainly mediated by their antimicrobial activities owing to their interaction with cell membranes of microbes (by disrupting membrane stability of lipid bilayer). They are most effective against gram +ve bacteria and possess almost no activity against gram –ve (because of their hydrophilic bilayer) except thymol and carvacrol (232). Garlic oil has shown inhibition of HMG-CoA reductase, leading to membrane instability and, eventually, cell death in methanogenic archaea. Recently, metagenomic analysis of goat rumen revealed that EO cobalt complexes significantly manipulated the structural and functional profile of rumen microbiota. It was revealed that Bacteroides sp. and Succinivibrio sp. showed a positive correlation with enhanced VFA production in supplemented groups. Moreover, functional prediction pathway analysis exhibited upregulation of lipid and carbohydrate pathways by EO (233).
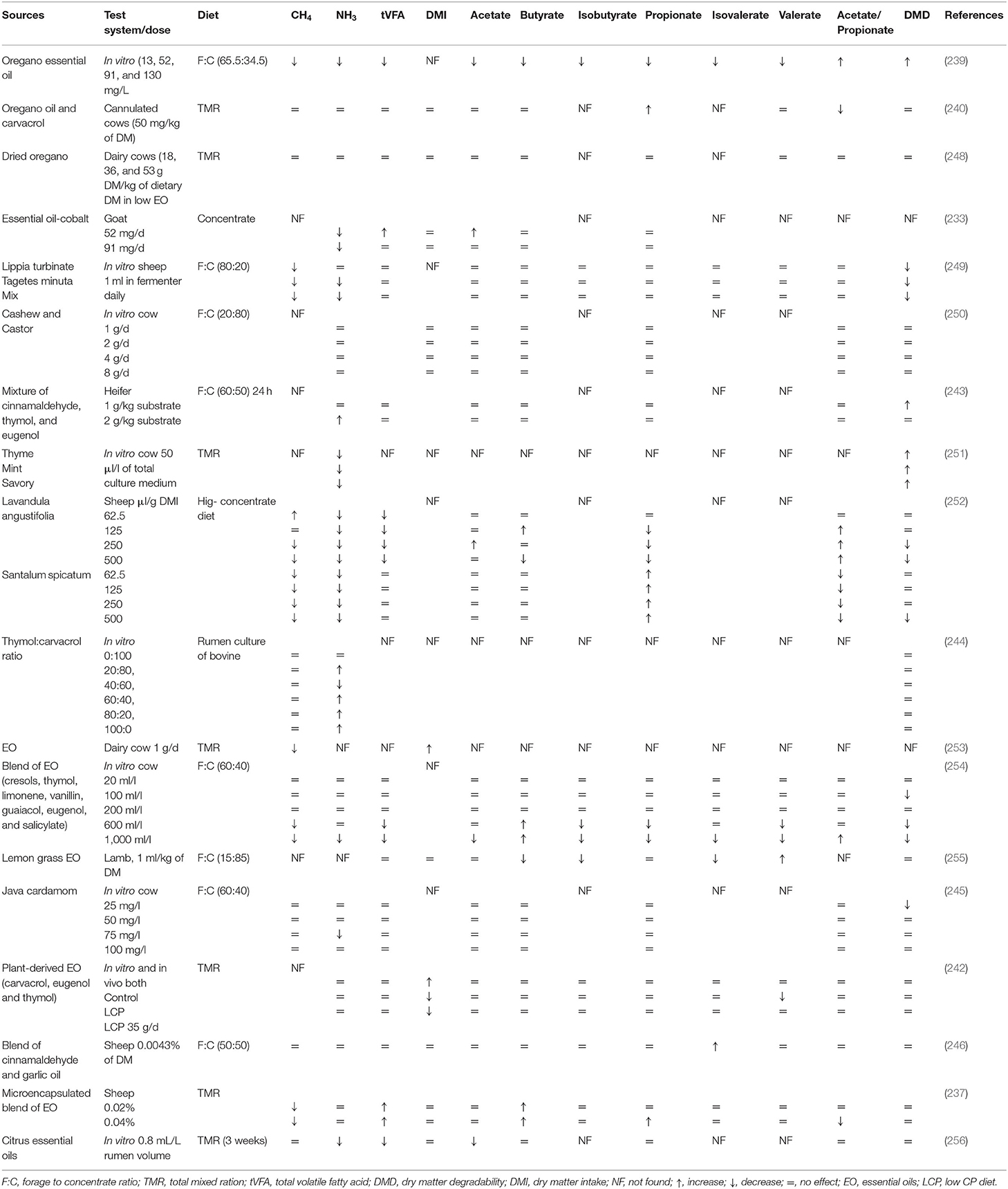
Table 6. Effects of various EO and their compounds on methanogenesis, rumen fermentation, and feed degradability.
Some studies have also reported a few unfavorable effects of using EO as feed additives as they depressed synthesis of VFA by reducing feed degradability (234). These harmful effects might be due to their extensive and non-specific antimicrobial properties in the rumen. In a study, no effect on the rumen microbiome has been observed by supplementation of a blend of EO having thymol, guaiacol, eugenol, vanillin, salicylaldehyde, and limonene (235). The desirable effects of EO on the rumen physiology are mainly attributed to their phenolic compounds, which possess the potent ability to affect the activity of both gram-positive and gram-negative bacteria (236). Inhibition of gram-positive bacteria in the rumen can potentially increase the propionate concentration (235). Supplementing a blend of EO (cinnamaldehyde, eugenol, carvacrol, and capsicum oleoresin) firstly increased rumen acetate concentration, which was replaced by propionate concentration afterward. This shift indicated a combined effect of low pH and antimicrobial activity of EO. Furthermore, this blend of EO showed the ability to improve microbial protein synthesis in sheep (237). Higher microbial protein might be attributed to the enhanced post-ruminal protein supply and absorption. Moreover, it might possibly due to the reduction of protozoal counts as ruminal protozoa devour many bacteria and their protein flow toward the small intestine (237). Recently, Garcia et al. (238) revealed that the chemical composition of EO, especially proportion of oxygenated compounds, has a positive interaction with fermentation pattern and indicate promising potential regarding CH4 mitigation. However, EO have shown inconsistent effects on rumen microbes and feed degradability in different studies, owing to different types of EO used, their chemical composition, and their variable dietary and host responses (Tables 5, 6).
Supplementation of oregano EO at 4 and 7 g/d promoted the population of primary cellulolytic bacteria and ruminal fungi, respectively, in sheep (241). An in vitro study revealed that the inclusion of EO of S. spicatum in a high-concentrate diet significantly improved the rumen fermentation characteristics by reducing CH4 and NH3-N while promoting propionate concentration (252). Dietary supplementation of EO (coriander, geranyl acetate, and eugenol) reduced CH4 production up to 6% per cow/day and 20% less CH4 per kg of milk. It can be speculated that energy saved through this reduced CH4 production may be diverted toward milk production (253). A blend of EO (carvacrol, caryophyllene, p-cymene, cineole, terpinene, and thymol) altered the rumen functions by selectively promoting the growth of rumen bacteria (by decreasing Firmicutes while increasing Bacteroidetes) in calves (257). Essential oils of Lippia turbinata and Tagetes minuta have shown a 10-fold decrease of in vitro CH4 production coupled with modification of N metabolism in the rumen (249). Recently, a meta-analysis showed that dietary supplementation of a blend of EO (coriander, eugenol, and geraniol) increased the milk yield (3.6%), milk fat and protein (4.1%), and feed efficiency (4.4%), while decreasing DM intake (12.9%) and CH4 production (8.8%) during long term trial in dairy cattle (258).
Essential oil–cobalt complexes have shown positive effects in ruminants by enhancing productive performance while decreasing NH3 emissions (233). Likewise, synergetic effects of EO (thyme, mint, and savory) in a high-concentrate diet have been observed regarding desirable shifts in microbial fermentation and higher microbial protein yield in dairy cows (251). Increased feed efficiency and calcium homeostasis have been observed with supplementation of a plant bioactive EO blend (>80% menthol, eugenol, and anethol). Increased uptake of calcium and ammonium was also observed as a result of specific cation-transporting proteins expressed by the rumen. However, further investigations are needed to evaluate the fate of the absorbed nutrients, especially calcium and N (259). Recently, Zhou et al. (239) suggested that oregano EO (52 mg/L) in mature ruminants can modify ruminal fermentation and mitigate in vitro CH4 production through mediating ruminal bacteria (Prevotella and Dialister). Some studies involving supplementation of EO in ruminants have shown contrary findings as the feeding of oregano EO did not reduce CH4 yield together with no effect on animal performance and rumen fermentation (240, 248). These divergent findings may be partially explained by variable experimental conditions of studies including the type of diets, plant species, dose and type of EO, pH of rumen fluid, and host animal (260, 261).
Studies have suggested the use of a combination or blend of different EO as a better strategy to modulate rumen microbiome to manipulate rumen fermentation than using individual EO. This is mainly because each EO possesses a complex mixture of phytochemicals and their synergistic effects can lead to the synthesis of new compounds with a quite different bioactivity that could not be harvested with individual compounds (29, 262). Additionally, using a combination of phytochemicals is also advantageous for the host regarding the provision of various phytonutrients from different plant combinations. Moreover, the benefits of such a combination are its ultimate utility for using on large scale in the animal industry as a commercial feed additive to have an overall impact on the improvement of global animal production while mitigating GHG emissions.
Rumen microbiome plays a critical functional role in N2 utilization, rumen feed fermentation, and CH4 production, ultimately influencing the production, health, and welfare in ruminants. Rumen microbes are highly active and can adapt to an extensive range of dietary fluctuations or host physiological conditions. Extensive literature supports the supplementation of phytogenic feed additives like saponin, tannins, and EO for the manipulation of rumen microbiome to modulate ruminal fermentation to increase VFA and decrease NH3 and CH4 production. Decreasing methanogenesis using dietary interventions at the expense of decreased VFA production is nutritionally adverse and unadvisable. Inhibition of enteric CH4 emission in ruminants is possible through the use of plant bioactive compounds; however, studies on the long-term effects of these compounds to reduce methanogenesis are essentially required. Studies summarized above clearly demonstrate that although phytochemicals possess a potent ability to modulate rumen microbiome and reduce methanogenesis in vitro, the observed in vivo effects varied greatly. Many factors, including variations of the chemical compositions of the compounds due to the differences in plant origin, growing conditions, and processing methods as well as different application methods, feeding conditions, and progressive adaptation of microbes for specific phytochemicals, contribute to this vast variability. Because of the complexity of these issues, it is difficult to conduct systematic and comprehensive evaluations of the efficacy and safety of these compounds for commercial applications in the animal industry. Therefore, controlling this variability is key to developing phytogenic substances as natural feed additives. This ideally should include all procedures from production, extraction, processing, and application. Optimization of different conditions during these steps can definitely help to address problems like inconsistency and transient and adverse effects of phytogenic feed additives in ruminants. Recent developments in molecular docking analysis and three-dimensional structure databases of phytochemicals have opened a new horizon for the discovery of putative functions (particularly antimicrobial and antimethanogenic) of different compounds by evaluating the structure affinity relationships with different microbes and their substrates. It will be of interest to identify potent phytochemicals based on their structural homology and binding affinity with functional proteins of rumen microbes particularly methanogenic archaea through molecular docking analysis first and then testing their biological activity in vitro and in vivo. This approach will not only help to find out new phytochemicals with potent activities but also help to understand their mechanism of action and exploit their synergistic effects and interactions with other compounds. Moreover, there is a dire need to exploit advances in molecular chemistry like encapsulation techniques to avoid ruminal degradation of phytochemicals and use their nanostructures to enhance their bioactivity and bioavailability, which seems to be an exciting area to explore their promising effects on the rumen microbiome.
It has been proposed that higher molecular weight compounds (such as polyphenols) were not dissolved well in water (263). Nanoparticles are formulated with hydrophobic groups inside and polar groups on the surface of particles and have shown to significantly enhance the solubility and bioavailability of less-water-soluble phenolic phytochemicals (264). Moreover, nanoparticles of plant extracts and EO have shown higher antioxidant and antimicrobial activities as compared with their crude extracts or EO (265). The higher antimicrobial activity of the nanoparticles or nanoemulsion of phytochemicals is related to the size of the nanoparticles or nanoemulsion droplets, which is in the subcellular size range. This allows the penetration of the nanoparticles or nanoemulsion droplets to the microbial cells leading to enhanced activity (265, 266). It is anticipated that the use of nanoparticles and nanoemulsions can potentially enhance the modulatory effects of phytochemicals on the rumen microbiome, subsequently leading to better health and performance of ruminants.
FH and CY: conceptualization. MAA, SS, and HME: data curation. CY: funding acquisition, project administration, supervision, and validation. FH, MSK, and MSR: investigation. MSR and CY: resources. FH and MSR: software. MAA, MSR, MSK, and FH: writing—original draft and formal analysis. MAA, FH, HME, and CY: writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was funded by the National Key Research and Development Program of China, grant numbers 2016YFD0500507 and 2018YFD0501600.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Valente TNP, da Silva Lima E, dos Santos WBR, Cesario A, Santose E. Ruminal microorganism consideration and protein used in the metabolism of the ruminants: a review. Afr J Microbiol Res. (2016) 10:456–64. doi: 10.5897/AJMR2016.7627
2. Bergman E. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. (1990) 70:567–90. doi: 10.1152/physrev.1990.70.2.567
3. Storm E, Orskov E. The nutritive value of rumen micro-organisms in ruminants: 1. Large-scale isolation and chemical composition of rumen micro-organisms. Br J Nutr. (1983) 50:463–70. doi: 10.1079/BJN19830114
4. Mackie R, Aminov R, White B, McSweeney C. Molecular ecology and diversity in gut microbial ecosystems. In: PB Cronje, editor, Ruminant Physiology: Digestion, Metabolism, Growth and Reproduction. Wallingford: CABI Publishing (2000). p. 61–77. doi: 10.1079/9780851994635.0061
5. Kittelmann S, Pinares-Patino CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, et al. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS ONE. (2014) 9:e103171. doi: 10.1371/journal.pone.0103171
6. Stewart C, Flint H, Bryant M, Stewart CS, Flint HJ, Bryant MP. The rumen bacteria. In: Hobson PN, Stewart CS, editors. The Rumen Microbial Ecosystem II. London: Chapman and Hall (1997). p. 10–72. doi: 10.1007/978-94-009-1453-7_2
7. Paul K, Nonoh JO, Mikulski L, Brune A. “Methanoplasmatales:” thermoplasmatales-related archaea in termite guts and other environments are the seventh order of methanogens. Appl Environ Microbiol. (2012) 78:8245–53. doi: 10.1128/AEM.02193-12
8. Poulsen M, Schwab C, Jensen BB, Engberg RM, Spang A, Canibe N, et al. Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat Commun. (2013) 4:1–9. doi: 10.1038/ncomms2432
9. Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. (1995) 73:2483–92. doi: 10.2527/1995.7382483x
10. Czerkawski JW. An Introduction to Rumen Studies. Exter: Pergamon Press (1986). doi: 10.1016/B978-0-08-025486-9.50007-4
11. Chalupa W. Manipulating rumen fermentation. J Anim Sci. (1977) 46:585–99. doi: 10.2527/jas1977.453585x
12. Ungerfeld EM. Shifts in metabolic hydrogen sinks in the methanogenesis-inhibited ruminal fermentation: a meta-analysis. Front Microbiol. (2015) 6:1–17. doi: 10.3389/fmicb.2015.00037
13. Huws SA, Creevey CJ, Oyama LB, Mizrahi I, Denman SE, Popova M, et al. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: past, present, and future. Front Microbiol. (2018) 9:2161. doi: 10.3389/fmicb.2018.02161
14. Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. (2013) 7:1069–79. doi: 10.1038/ismej.2013.2
15. Hernandez-Sanabria E, Goonewardene LA, Li M, Mujibi DF, Stothard P, Moore SS, et al. Correlation of particular bacterial PCR-denaturing gradient gel electrophoresis patterns with bovine ruminal fermentation parameters and feed efficiency traits. Appl Environ Microbiol. (2010) 76:6338–50. doi: 10.1128/AEM.01052-10
16. Jami E, Shabtay A, Nikbachat M, Yosef E, Miron J, Mizrahi I. Effects of adding a concentrated pomegranate-residue extract to the ration of lactating cows on in vivo digestibility and profile of rumen bacterial population. J Dairy Sci. (2012) 95:5996–6005. doi: 10.3168/jds.2012-5537
17. Morgavi DP, Kelly W, Janssen P, Attwood G. Rumen microbial (meta) genomics and its application to ruminant production. Animal. (2013) 7:184–201. doi: 10.1017/S1751731112000419
18. Knapp J, Laur G, Vadas P, Weiss W, Tricarico J. Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J Dairy Sci. (2014) 97:3231–61. doi: 10.3168/jds.2013-7234
19. Hristov A, Oh J, Lee C, Meinen R, Montes F, Ott T, et al. Mitigation of greenhouse gas emissions in livestock production: A review of technical options for non-CO2 emissions. Rome: Food and Agriculture Organization of the United Nations (2013).
20. Connor E. Invited review: improving feed efficiency in dairy production: challenges and possibilities. Animal. (2015) 9:395–408. doi: 10.1017/S1751731114002997
21. Kumar S, Choudhury PK, Carro MD, Griffith GW, Dagar SS, Puniya M, et al. New aspects and strategies for methane mitigation from ruminants. Appl Microbiol Biotechnol. (2014) 98:31–44. doi: 10.1007/s00253-013-5365-0
22. Jassim SA, Limoges RG. Reducing greenhouse gas emissions from livestock farms. In: SA Jassim and RG Limoges, editors. Bacteriophages: Practical Applications for Nature's Biocontrol. Cham: Springer (2017). p. 165–77. doi: 10.1007/978-3-319-54051-1_5
23. Hart KJ, Yáñez-Ruiz DR, Duval SM, McEwan NR, Newbold CJ. Plant extracts to manipulate rumen fermentation. Anim Feed Sci Technol. (2008) 147:8–35. doi: 10.1016/j.anifeedsci.2007.09.007
24. Moss AR, Jouany J-P, Newbold J. Methane production by ruminants: its contribution to global warming. Annal Zootechnie. (2000) 49:231–53. doi: 10.1051/animres:2000119
25. Khattab MSA, El-Zaiat HM, Abd El Tawab AM, Matloup OH, Morsy AS, Abdou MM, et al. Impact of lemongrass and galangal as feed additives on performance of lactating Barki goats. Int J Dairy Sci. (2017) 12:184–9. doi: 10.3923/ijds.2017.184.189
26. Ugbogu EA, Elghandour MM, Ikpeazu VO, Buendía GR, Molina OM, Arunsi UO, et al. The potential impacts of dietary plant natural products on the sustainable mitigation of methane emission from livestock farming. J Clean Prod. (2019) 213:915–25. doi: 10.1016/j.jclepro.2018.12.233
27. Wallace RJ, McEwan NR, McIntosh FM, Teferedegne B, Newbold CJ. Natural products as manipulators of rumen fermentation. Asian-Australas J Anim Sci. (2002) 15:1458–68. doi: 10.5713/ajas.2002.1458
28. Kholif AE, Hassan AA, El Ashry GM, Bakr MH, El-Zaiat HM, Olafadehan OA, et al. Phytogenic feed additives mixture enhances the lactational performance, feed utilization and ruminal fermentation of Friesian cows. Animal Biotechnol. (2020) 4:1–11. doi: 10.1080/10495398.2020.1746322
29. Calsamiglia S, Busquet M, Cardozo P, Castillejos L, Ferret A. Invited review: essential oils as modifiers of rumen microbial fermentation. J Dairy Sci. (2007) 90:2580–95. doi: 10.3168/jds.2006-644
30. Patra A, Stiverson J, Yu Z. Effects of quillaja and yucca saponins on communities and select populations of rumen bacteria and archaea, and fermentation in vitro. J Appl Microbiol. (2012) 113:1329–40. doi: 10.1111/j.1365-2672.2012.05440.x
31. Jayanegara A, Wina E, Takahashi J. Meta-analysis on methane mitigating properties of saponin-rich sources in the rumen: influence of addition levels and plant sources. Asian-Australas J Anim Sci. (2014) 27:1426–35. doi: 10.5713/ajas.2014.14086
32. Marrez DA, Cieślak A, Gawad R, Ebeid HM, Chrenková M, Gao M, et al. Effect of freshwater microalgae Nannochloropsis limnetica on the rumen fermentation in vitro. J Anim Feed Sci. (2017) 26:359–64. doi: 10.22358/jafs/81275/2017
33. Krishnamoorthy U, Moran J. Rearing young ruminants on milk replacers and starter feeds. Rome, Italy: Food and Agriculture Organization of the United Nations (FAO). (2012).
34. Van Soest PJ. Nutritional Ecology of the Ruminant. Ithaca, New York, NY: Cornell University Press (2018).
35. Minato H, Otsuka M, Shirasaka S, Itabashi H, Mitsumori MJ. Colonization of microorganisms in the rumen of young calves. J General Appl Microbiol. (1992) 38:447–56. doi: 10.2323/jgam.38.447
36. Friedman N, Shriker E, Gold B, Durman T, Zarecki R, Ruppin E, et al. Diet-induced changes of redox potential underlie compositional shifts in the rumen archaeal community. Environ Microbiol. (2017) 19:174–84. doi: 10.1111/1462-2920.13551
37. Henderson G, Cox F, Ganesh S, Jonker A, Young W, Collaborators GRC, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. (2015) 5:14567–79. doi: 10.1038/srep14567
38. Rey M, Enjalbert F, Combes S, Cauquil L, Bouchez O, Monteils V. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. J Appl Microbiol. (2014) 116:245–57. doi: 10.1111/jam.12405
39. Skillman LC, Evans PN, Naylor GE, Morvan B, Jarvis GN, Joblin KN. 16S ribosomal DNA-directed PCR primers for ruminal methanogens and identification of methanogens colonising young lambs. Anaerobe. (2004) 10:277–85. doi: 10.1016/j.anaerobe.2004.05.003
40. Guzman CE, Bereza-Malcolm LT, De Groef B, Franks AE. Presence of selected methanogens, fibrolytic bacteria, and proteobacteria in the gastrointestinal tract of neonatal dairy calves from birth to 72 hours. PLoS ONE. (2015) 10:e0133048. doi: 10.1371/journal.pone.0133048
41. Friedman N, Jami E, Mizrahi I. Compositional and functional dynamics of the bovine rumen methanogenic community across different developmental stages. Environ Microbiol. (2017) 19:3365–73. doi: 10.1111/1462-2920.13846
42. Mizrahi I, Jami E. The compositional variation of the rumen microbiome and its effect on host performance and methane emission. Animal. (2018) 12:s220–32. doi: 10.1017/S1751731118001957
43. Wickramasinghe HKJP, Anast JM, Schmitz-Esser S, Serão NVL, Appuhamy JADRN. Beginning to offer drinking water at birth increases the species richness and the abundance of Faecalibacterium and Bifidobacterium in the gut of preweaned dairy calves. J Dairy Sci. (2020) 103:4262–74. doi: 10.3168/jds.2019-17258
44. De Barbieri I, Hegarty RS, Silveira C, Gulino LM, Oddy VH, Gilbert RA, et al. Programming rumen bacterial communities in newborn Merino lambs. Small Ruminant Res. (2015) 129:48–59. doi: 10.1016/j.smallrumres.2015.05.015
45. O'Hara E. Investigating Early Life Microbial and Host Transcriptomic Dynamics in the 1028 Bovine Gastrointestinal Tract. Edmonton: Doctor of Philosophy, University of Alberta. (2019).
46. Ross EM, Moate PJ, Bath CR, Davidson SE, Sawbridge TI, Guthridge KM, et al. High throughput whole rumen metagenome profiling using untargeted massively parallel sequencing. BMC Genetics. (2012) 13:53–66. doi: 10.1186/1471-2156-13-53
47. Gilbert RA, Klieve AV. Ruminal viruses (Bacteriophages, Archaeaphages). In: Puniya AK, Singh R and Kamra DN, editors. Rumen Microbiology: From Evolution to Revolution. New Delhi: Springer (2015). p. 121–41. doi: 10.1007/978-81-322-2401-3_9
48. Millen DD, Arrigoni MDB, Pacheco RDL. Rumenology. Switzerland: Springer International Publishing. (2016). doi: 10.1007/978-3-319-30533-2
49. McSweeney CS, Denman SE, Mackie RI. Rumen bacteria. In: HPS Makkar and CS McSweeney, editors. Methods in Gut Microbial Ecology for Ruminants. Dordrecht: Springer (2005). p. 23–37. doi: 10.1007/1-4020-3791-0_2
50. Won MY, Oyama LB, Courtney SJ, Creevey CJ, Huws SA. Can rumen bacteria communicate to each other? Microbiome. (2020) 8:1–8. doi: 10.1186/s40168-020-00796-y
51. Tapio I, Snelling TJ, Strozzi F, Wallace RJ. The ruminal microbiome associated with methane emissions from ruminant livestock. J Anim Sci Biotechnol. (2017) 8:7–17. doi: 10.1186/s40104-017-0141-0
52. Rezaeian M, Beakes GW, Parker DS. Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol Res. (2004) 108:1227–33. doi: 10.1017/S0953756204000929
53. Elekwachi CO, Wang Z, Wu X, Rabee A, Forster RJ. Total rRNA-Seq analysis gives insight into bacterial, fungal, protozoal and archaeal communities in the rumen using an optimized RNA isolation method. Front Microbiol. (2017) 8:1814–27. doi: 10.3389/fmicb.2017.01814
54. Lee S, Ha J, Cheng K-J. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl Environ Microbiol. (2000) 66:3807–13. doi: 10.1128/AEM.66.9.3807-3813.2000
55. Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, et al. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science. (2016) 351:1192–5. doi: 10.1126/science.aad1431
56. Haitjema CH, Solomon KV, Henske JK, Theodorou MK, O'Malley MA. Anaerobic gut fungi: advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol Bioeng. (2014) 111:1471–82. doi: 10.1002/bit.25264
57. Gordon GL, Phillips MW. The role of anaerobic gut fungi in ruminants. Nutr Res Rev. (1998) 11:133–68. doi: 10.1079/NRR19980009
58. Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, et al. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol. (2014) 90:1–17. doi: 10.1111/1574-6941.12383
59. Cheng YF, Edwards JE, Allison GG, Zhu W-Y, Theodorou MK. Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresource Technol. (2009) 100:4821–8. doi: 10.1016/j.biortech.2009.04.031
60. Edwards JE, Forster RJ, Callaghan TM, Dollhofer V, Dagar SS, Cheng Y, et al. PCR and omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities. Front Microbiol. (2017) 8:1657–83. doi: 10.3389/fmicb.2017.01657
61. Ma Y, Li Y, Li Y, Cheng Y, Zhu W. The enrichment of anaerobic fungi and methanogens showed higher lignocellulose degrading and methane producing ability than that of bacteria and methanogens. World J Microbiol Biotechnol. (2020) 36:1–9. doi: 10.1007/s11274-020-02894-3
62. Williams AG, Coleman GS. The rumen protozoa. In: PN Hobson, CS Stewart, editors. The Rumen Microbial Ecosystem. Dordrecht: Springer (1997). p. 73–139. doi: 10.1007/978-94-009-1453-7_3
63. Kamra D. Rumen microbial ecosystem. Curr Sci. (2005) 89:124–35. Available online at: https://www.jstor.org/stable/24110438?seq=1
64. Newbold CJ, de la Fuente G, Belanche A, Ramos-Morales E, McEwan NR. The role of ciliate protozoa in the rumen. Front Microbiol. (2015) 6:1313–26. doi: 10.3389/fmicb.2015.01313
65. Solomon R, Wein T, Levy B, Reiss V, Zehavi T, Furman O, et al. Rumen protozoa shape microbiome composition and metabolic output of fermentation. bioRxiv. (2020). doi: 10.1101/2020.05.15.080218
66. Vogels GD, Hoppe WF, Stumm CK. Association of methanogenic bacteria with rumen ciliates. Appl Environ Microbiol. (1980) 40:608–12. doi: 10.1128/AEM.40.3.608-612.1980
67. Belanche A, de la Fuente G, Newbold CJ. Study of methanogen communities associated with different rumen protozoal populations. FEMS Microbiol Ecol. (2014) 90:663–77. doi: 10.1111/1574-6941.12423
68. Morgavi D, Forano E, Martin C, Newbold C. Microbial ecosystem and methanogenesis in ruminants. Animal. (2010) 4:1024–36. doi: 10.1017/S1751731110000546
69. Kim M, Morrison M, Yu Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol Ecol. (2011) 76:49–63. doi: 10.1111/j.1574-6941.2010.01029.x
70. Wright A-DG, Klieve AV. Does the complexity of the rumen microbial ecology preclude methane mitigation? Anim Feed SciTechnol. (2011) 166:248–53. doi: 10.1016/j.anifeedsci.2011.04.015
71. Ferry JG, Kastead KA. Methanogenesis. In: R Cavicchioli, editor. Archaea. Washington, DC: American Society of Microbiology (2007). p. 288–314. doi: 10.1128/9781555815516.ch13
72. Lyu Z, Shao N, Akinyemi T, Whitman WB. Methanogenesis. Curr Biol. (2018) 28:R727–32. doi: 10.1016/j.cub.2018.05.021
73. Lu C, Li K, Wang H, Zhu X, Wei Y, Zheng M, et al. Chemical looping reforming of methane using magnetite as oxygen carrier: structure evolution and reduction kinetics. Appl Energy. (2018) 211:1–14. doi: 10.1016/j.apenergy.2017.11.049
74. Ritchie A, Robinson I, Allison M. Rumen bacteriophage: survey of morphological types. Microscopie Electronique. (1970) 3:333–4.
75. Klieve AV, Swain RA. Estimation of ruminal bacteriophage numbers by pulsed-field gel electrophoresis and laser densitometry. Appl Environ Microbiol. (1993) 59:2299–303. doi: 10.1128/AEM.59.7.2299-2303.1993
76. Bodas R, Prieto N, Garcia-Gonzalez R, Andres S, Giraldez FJ, Lopez S. Manipulation of rumen fermentation and methane production with plant secondary metabolites. Anim Feed Sci Technol. (2012) 176:78–93. doi: 10.1016/j.anifeedsci.2012.07.010
77. Reddy PRK, Elghandour MMMY, Salem AZM, Yasaswini D, Reddy PPR, Reddy AN, et al. Plant secondary metabolites as feed additives in calves for antimicrobial stewardship. Anim Feed Sci Technol. (2020) 264:114469. doi: 10.1016/j.anifeedsci.2020.114469
78. Patra AK, Amasheh S, Aschenbach JR. Modulation of gastrointestinal barrier and nutrient transport function in farm animals by natural plant bioactive compounds–a comprehensive review. Crit Rev Food Sci Nutr. (2019) 59:3237–66. doi: 10.1080/10408398.2018.1486284
79. Omojate Godstime C, Enwa Felix O, Jewo Augustina O, Eze Christopher O. Mechanisms of antimicrobial actions of phytochemicals against enteric pathogens–a review. J Pharm Chem Biol Sci. (2014) 2:77–85. Available online at: http://www.jpcbs.info/2014_2_2_3_%20Enwa.pdf
80. Lillehoj H, Liu Y, Calsamiglia S, Fernandez-Miyakawa ME, Chi F, Cravens RL, et al. Phytochemicals as antibiotic alternatives to promote growth and enhance host health. Vet Res. (2018) 49:76. doi: 10.1186/s13567-018-0562-6
81. Li F, Li C, Chen Y, Liu J, Zhang C, Irving B, et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome. (2019) 7:92. doi: 10.1186/s40168-019-0699-1
82. Brito L, Rojas de Oliveira H, Houlahan K, Fonseca PA, Lam S, Butty AM, et al. Genetic mechanisms underlying feed utilization and implementation of genomic selection for improved feed efficiency in dairy cattle. Can J Anim Sci. (2020) 1–18. doi: 10.1139/cjas-2019-0193
83. Terry SA, Basarab JA, Guan L, McAllister TA. Invited review: strategies to improve the efficiency of beef cattle production. Can J Anim Sci. (2020). doi: 10.1139/CJAS-2020-0022
84. Wu G, Bazer FW. Application of new biotechnologies for improvements in swine nutrition and pork production. J Anim Sci Biotechnol. (2019) 10:28. doi: 10.1186/s40104-019-0337-6
85. Yuan M, Gao Y, Han J, Wu T, Zhang J, Wei Y, et al. The development and application of genome editing technology in ruminants: a review. Front Agri Sci Eng. (2020) 7:171–80. doi: 10.15302/J-FASE-2019302
86. Laible G. Production of transgenic livestock: overview of transgenic technologies. In: Niemann H, Wrenzycki C, editors. Animal Biotechnology 2. Cham: Springer (2018). p. 95–121. doi: 10.1007/978-3-319-92348-2_6
87. Wang Q, Qian L, Jiang S, Cai C, Ma D, Gao P, et al. Safety evaluation of neo transgenic pigs by studying changes in gut microbiota using high-throughput sequencing technology. PLoS ONE. (2016) 11:e0150937. doi: 10.1371/journal.pone.0150937
88. Yang H, Wu J, Huang X, Zhou Y, Zhang Y, Liu M, et al. An ancient deletion in the ABO gene affects the composition of the porcine microbiome by altering intestinal N-acetyl-galactosamine concentrations. bioRxiv. (2020). doi: 10.1101/2020.07.16.206219
89. Sasson G, Ben-Shabat SK, Seroussi E, Doron-Faigenboim A, Shterzer N, Yaacoby S, et al. Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow's capacity to harvest energy from its feed. MBio. (2017) 8:17. doi: 10.1128/mBio.00703-17
90. Myer PR. Bovine genome-microbiome interactions: metagenomic frontier for the selection of efficient productivity in cattle systems. Msystems. (2019) 4:19. doi: 10.1128/mSystems.00103-19
91. Takahashi J, Mwenya B, Santoso B, Sar C, Umetsu K, Kishimoto T, et al. Mitigation of methane emission and energy recycling in animal agricultural systems. Asian-Australas J Anim Sci. (2005) 18:1199–208. doi: 10.5713/ajas.2005.1199
92. Hristov A, Oh J, Firkins J, Dijkstra J, Kebreab E, Waghorn G, et al. SPECIAL TOPICS—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options 1. J Anim Sci. (2013) 91:5045–69. doi: 10.2527/jas.2013-6583
93. Beauchemin K, Kreuzer M, O'mara F, McAllister T. Nutritional management for enteric methane abatement: a review. Aust J Exp Agric. (2008) 48:21–7. doi: 10.1071/EA07199
94. Makkar H, Francis G, Becker K. Bioactivity of phytochemicals in some lesser-known plants and their effects and potential applications in livestock and aquaculture production systems. Animal. (2007) 1:1371–91. doi: 10.1017/S1751731107000298
95. Henderson G, Cox F, Kittelmann S, Miri VH, Zethof M, Noel SJ, et al. Effect of DNA extraction methods and sampling techniques on the apparent structure of cow and sheep rumen microbial communities. PLoS ONE. (2013) 8:e74787. doi: 10.1371/journal.pone.0074787
96. Pitta D, Pinchak W, Indugu N, Vecchiarelli B, Sinha R, Fulford J. Metagenomic analysis of the rumen microbiome of steers with wheat-induced frothy bloat. Front Microbiol. (2016) 7:689–99. doi: 10.3389/fmicb.2016.00689
97. Pitta D, Indugu N, Vecchiarelli B, Rico D, Harvatine K. Alterations in ruminal bacterial populations at induction and recovery from diet-induced milk fat depression in dairy cows. J Dairy Sci. (2018) 101:295–309. doi: 10.3168/jds.2016-12514
98. Lin L, Wang Y, Xu L, Liu J, Zhu W, Mao S. Microbiome–host co-oscillation patterns in remodeling of colonic homeostasis during adaptation to a high-grain diet in a sheep model. Animal Microbiome. (2020) 2:1–12. doi: 10.1186/s42523-020-00041-9
99. McSweeney C, Mackie R. Micro-organisms and ruminant digestion: state of knowledge, trends and future prospects. Rome: Commission of Genetic Resources for Food and Agriculture. Background study paper No. 61. (2012). Available online at: http://www.fao.org/docrep/016/me992e/me992e (accessed December 4, 2012).
100. Khattab MS, Ebeid HM, Abd El Tawab AM, El-Nor SA, Aboamer AA. Effect of supplementing diet with herbal plants on ruminal fiber digestibility and gas production. Res J Pharma Biol Chem Sci. (2016) 7:1093–7. Available online at: https://www.rjpbcs.com/pdf/2016_7(6)/%5B140%5D.pdf
101. Puniya AK, Salem AZ, Kumar S, Dagar SS, Griffith GW, Puniya M, et al. Role of live microbial feed supplements with reference to anaerobic fungi in ruminant productivity: a review. J Integr Agric. (2015) 14:550–60. doi: 10.1016/S2095-311960837-6
102. Eastridge M. Major advances in applied dairy cattle nutrition. J Dairy Sci. (2006) 89:1311–23. doi: 10.3168/jds.S0022-030272199-3
103. Hills J, Wales W, Dunshea F, Garcia S, Roche J. Invited review: an evaluation of the likely effects of individualized feeding of concentrate supplements to pasture-based dairy cows. J Dairy Sci. (2015) 98:1363–401. doi: 10.3168/jds.2014-8475
104. De Menezes AB, Lewis E, O'Donovan M, O'Neill BF, Clipson N, Doyle EM. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol Ecol. (2011) 78:256–65. doi: 10.1111/j.1574-6941.2011.01151.x
105. DeVries T, Schwaiger T, Beauchemin K, Penner G. Impact of severity of ruminal acidosis on feed-sorting behaviour of beef cattle. Anim Prod Sci. (2014) 54:1238–42. doi: 10.1071/AN14227
106. Nagaraja T, Titgemeyer E. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci. (2007) 90:E17–38. doi: 10.3168/jds.2006-478
107. Yang W, Beauchemin K. Physically effective fiber: method of determination and effects on chewing, ruminal acidosis, and digestion by dairy cows. J Dairy Sci. (2006) 89:2618–33. doi: 10.3168/jds.S0022-0302(06)72339-6
108. Doreau M, Van Der Werf H, Micol D, Dubroeucq H, Agabriel J, Rochette Y, et al. Enteric methane production and greenhouse gases balance of diets differing in concentrate in the fattening phase of a beef production system. J Anim Sci. (2011) 89:2518–28. doi: 10.2527/jas.2010-3140
109. González L, Manteca X, Calsamiglia S, Schwartzkopf-Genswein K, Ferret A. Ruminal acidosis in feedlot cattle: Interplay between feed ingredients, rumen function and feeding behavior (a review). Anim Feed SciTechnol. (2012) 172:66–79. doi: 10.1016/j.anifeedsci.2011.12.009
110. Granja-Salcedo YT, Ribeiro Júnior CS, de Jesus RB, Gomez-Insuasti AS, Rivera AR, Messana JD, et al. Effect of different levels of concentrate on ruminal microorganisms and rumen fermentation in Nellore steers. Arch Anim Nutr. (2016) 70:17–32. doi: 10.1080/1745039X.2015.1117562
111. Carvalho PH, Pinto AC, Millen DD, Felix TL. Effect of cattle breed and basal diet on digestibility, rumen bacterial communities, and eating and rumination activity. J Anim Sci. (2020) 98:114. doi: 10.1093/jas/skaa114
112. Russell JB, Baldwin R. Substrate preferences in rumen bacteria: evidence of catabolite regulatory mechanisms. Appl Environ Microbiol. (1978) 36:319–29. doi: 10.1128/AEM.36.2.319-329.1978
113. Russell JB, Dombrowski D. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl Environ Microbiol. (1980) 39:604–10. doi: 10.1128/AEM.39.3.604-610.1980
114. McCaughern JH, Mackenzie AM, Sinclair LA. Dietary starch concentration alters reticular pH, hepatic copper concentration, and performance in lactating Holstein-Friesian dairy cows receiving added dietary sulfur and molybdenum. J Dairy Sci. (2020) 103:9024–36. doi: 10.3168/jds.2020-18453
115. Chen L, Dong Z, Li J, Shao T. Ensiling characteristics, in vitro rumen fermentation, microbial communities and aerobic stability of low-dry matter silages produced with sweet sorghum and alfalfa mixtures. J Sci Food Agric. (2019) 99:2140–51. doi: 10.1002/jsfa.9406
116. Michalet-Doreau B, Fernandez I, Peyron C, Millet L, Fonty G. Fibrolytic activities and cellulolytic bacterial community structure in the solid and liquid phases of rumen contents. Reprod Nutr Dev. (2001) 41:187–94. doi: 10.1051/rnd:2001122
117. Guo W, Guo X, Zhu B, Guo Y, Zhou X. In situ degradation, ruminal fermentation, and the rumen bacterial community of cattle fed corn stover fermented by lignocellulolytic microorganisms. Anim Feed Sci Technol. (2019) 248:10–9. doi: 10.1016/j.anifeedsci.2018.07.007
118. Ku-Vera JC, Castelán-Ortega OA, Galindo-Maldonado FA, Arango J, Chirinda N, Jiménez-Ocampo R, et al. Strategies for enteric methane mitigation in cattle fed tropical forages. Animal. (2020) 1–11. doi: 10.1017/S1751731120001780
119. Hernández JCÁ, Alberto RV, Kebreab E, Appuhamy JADRN, Dougherty HC, Ortega OC, et al. Effect of forage to concentrate ratio and fat supplementation on milk composition in dairy sheep: a meta-analysis. Livestock Sci. (2020) 238: 104069. doi: 10.1016/j.livsci.2020.104069
120. Palmquist D, Jenkins T. Fat in lactation rations. J Dairy Sci. (1980) 63:1–14. doi: 10.3168/jds.S0022-030282881-5
121. Lourenco M, Ramos-Morales E, Wallace R. The role of microbes in rumen lipolysis and biohydrogenation and their manipulation. Animal. (2010) 4:1008–23. doi: 10.1017/S175173111000042X
122. Kim Y, Liu R, Rychlik J, Russell J. The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J Appl Microbiol. (2002) 92:976–82. doi: 10.1046/j.1365-2672.2002.01610.x
123. Brooks C, Garner G, Gehrke C, Muhrer M, Pfander W. The effect of added fat on the digestion of cellulose and protein by ovine rumen microorganisms. J Anim Sci. (1954) 13:758–64. doi: 10.2527/jas1954.134758x
124. Maia MR, Chaudhary LC, Bestwick CS, Richardson AJ, McKain N, Larson TR, et al. Toxicity of unsaturated fatty acids to the biohydrogenating ruminal bacterium, Butyrivibrio fibrisolvens. BMC Microbiol. (2010) 10:52–61. doi: 10.1186/1471-2180-10-52
125. McGeough E, Passetti L, Chung Y-H, Beauchemin K, McGinn S, Harstad OM, et al. Methane emissions, feed intake and total tract digestibility in lambs fed diets differing in fat content and fibre digestibility. Can J Anim Sci. (2019) 99:4. doi: 10.1139/cjas-2018-0185
126. Grainger C, Beauchemin K. Can enteric methane emissions from ruminants be lowered without lowering their production? Anim Feed SciTechnol. (2011) 166:308–20. doi: 10.1016/j.anifeedsci.2011.04.021
127. Behan AA, Loh TC, Fakurazi S, Kaka U, Kaka A, Samsudin AA. Effects of supplementation of rumen protected fats on rumen ecology and digestibility of nutrients in sheep. Anim. (2019) 9:400–17. doi: 10.3390/ani9070400
128. Asma Z, Sylvie C, Laurent C, Jérôme M, Christophe K, Olivier B, et al. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol Ecol. (2013) 83:504–14. doi: 10.1111/1574-6941.12011
129. Hackmann TJ, Firkins JL. Electron transport phosphorylation in rumen butyrivibrios: unprecedented ATP yield for glucose fermentation to butyrate. Front Microbiol. (2015) 6:622–33. doi: 10.3389/fmicb.2015.00622
130. Enjalbert F, Combes S, Zened A, Meynadier A. Rumen microbiota and dietary fat: a mutual shaping. J Appl Microbiol. (2017) 123:782–97. doi: 10.1111/jam.13501
131. Oldick B, Firkins J. Effects of degree of fat saturation on fiber digestion and microbial protein synthesis when diets are fed twelve times daily. J Anim Sci. (2000) 78:2412–20. doi: 10.2527/2000.7892412x
132. Halmemies-Beauchet-Filleau A, Shingfield KJ, Simpura I, Kokkonen T, Jaakkola S, Toivonen V, et al. Effect of incremental amounts of camelina oil on milk fatty acid composition in lactating cows fed diets based on a mixture of grass and red clover silage and concentrates containing camelina expeller. J Dairy Sci. (2017) 100:305–24. doi: 10.3168/jds.2016-11438
133. Brandao V, Dai X, Paula E, Silva L, Marcondes M, Shenkoru T, et al. Effect of replacing calcium salts of palm oil with camelina seed at 2 dietary ether extract levels on digestion, ruminal fermentation, and nutrient flow in a dual-flow continuous culture system. J Dairy Sci. (2018) 101:5046–59. doi: 10.3168/jds.2017-13558
134. Bayat A, Kairenius P, Stefański T, Leskinen H, Comtet-Marre S, Forano E, et al. Effect of camelina oil or live yeasts (Saccharomyces cerevisiae) on ruminal methane production, rumen fermentation, and milk fatty acid composition in lactating cows fed grass silage diets. J Dairy Sci. (2015) 98:3166–81. doi: 10.3168/jds.2014-7976
135. Chilliard Y, Ferlay A, Doreau M. Contrôle de la qualité nutritionnelle des matières grasses du lait par l'alimentation des vaches laitières: acides gras trans, polyinsaturés, acide linoléique conjugué. Productions Animal. (2001) 14:323–35. doi: 10.20870/productions-animales.2001.14.5.3758
136. Wang S, Kreuzer M, Braun U, Schwarm A. Effect of unconventional oilseeds (safflower, poppy, hemp, camelina) on in vitro ruminal methane production and fermentation. J Sci Food Agric. (2017) 97:3864–70. doi: 10.1002/jsfa.8260
137. Czerkawski J, Clapperton J. Fats as energy-yielding compounds in the ruminant diet. In: Proceedings-Easter School in Agricultural Science. Nottingham: University of Nottingham (1984). doi: 10.1016/B978-0-408-10864-5.50018-8
138. Mathison G. Effect of canola oil on methane production in steers. Can J Anim Sci. (1997) 77:545–6.
139. Bayat A, Ventto L, Kairenius P, Stefański T, Leskinen H, Tapio I, et al. Dietary forage to concentrate ratio and sunflower oil supplement alter rumen fermentation, ruminal methane emissions, and nutrient utilization in lactating cows. Transl Anim Sci. (2017) 1:277–86. doi: 10.2527/tas2017.0032
140. Ebeid HM, Mengwei L, Kholif AE, Hassan FU, Lijuan P, Xin L, et al. Moringa oleifera oil modulates rumen microflora to mediate in vitro fermentation kinetics and methanogenesis in total mix rations. Curr Microbiol. (2020) 77:1271–82. doi: 10.1007/s00284-020-01935-2
141. Vargas JE, Andrés S, López-Ferreras L, Snelling TJ, Yáñez-Ruíz DR, García-Estrada C, et al. Dietary supplemental plant oils reduce methanogenesis from anaerobic microbial fermentation in the rumen. Sci Rep. (2020) 10:1–9. doi: 10.1038/s41598-020-58401-z
142. Ornaghi M, Prado RM, Ramos TR, Catalano FR, Mottin C, Creevey CJ, et al. Natural plant-based additives can improve ruminant performance by influencing the rumen microbiome. Researchsquare (2020). doi: 10.21203/rs.3.rs-29748/v1
143. Kocherginskaya SA, Aminov RI, White BA. Analysis of the rumen bacterial diversity under two different diet conditions using denaturing gradient gel electrophoresis, random sequencing, and statistical ecology approaches. Anaerobe. (2001) 7:119–34. doi: 10.1006/anae.2001.0378
144. Jouany JP, Morgavi DP. Use of'natural'products as alternatives to antibiotic feed additives in ruminant production. Anim Int J Anim Biosci. (2007) 1:1443. doi: 10.1017/S1751731107000742
145. Patra AK, Yu Z. Effects of essential oils on methane production and fermentation by, and abundance and diversity of, rumen microbial populations. Appl Environ Microbiol. (2012) 78:4271–80. doi: 10.1128/AEM.00309-12
146. Oh J, Hristov AN, Lee C, Cassidy T, Heyler K, Varga G, et al. Immune and production responses of dairy cows to postruminal supplementation with phytonutrients. J Dairy Sci. (2013) 96:7830–43. doi: 10.3168/jds.2013-7089
147. Oh J, Giallongo F, Frederick T, Pate J, Walusimbi S, Elias R, et al. Effects of dietary Capsicum oleoresin on productivity and immune responses in lactating dairy cows. J Dairy Sci. (2015) 98:6327–39. doi: 10.3168/jds.2014-9294
148. Patra AK, Saxena J. Dietary phytochemicals as rumen modifiers: a review of the effects on microbial populations. Antonie Leeuwenhoek. (2009) 96:363–75. doi: 10.1007/s10482-009-9364-1
149. Patra A, Park T, Kim M, Yu Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J Anim Sci Biotechnol. (2017) 8:13. doi: 10.1186/s40104-017-0145-9
150. Szumacher-Strabel M, Cieslak A. Potential of phytofactors to mitigate rumen ammonia and methane production. J Anim Feed Sci. (2010) 19:319–37. doi: 10.22358/jafs/66296/2010
151. Majinda RR. Extraction and isolation of saponins. In: SD Sarker, L Nahar, editors. Natural Products Isolation. United Kingdom: Springer (2012). p. 415–26. doi: 10.1007/978-1-61779-624-1_16
152. Cheok CY, Salman HAK, Sulaiman R. Extraction and quantification of saponins: a review. Food Res Int. (2014) 59:16–40. doi: 10.1016/j.foodres.2014.01.057
153. Wojciechowski K, Orczyk M, Gutberlet T, Geue T. Complexation of phospholipids and cholesterol by triterpenic saponins in bulk and in monolayers. Biochim Biophys Acta. (2016) 1858:363–73. doi: 10.1016/j.bbamem.2015.12.001
154. Patra A, Saxena J. The effect and mode of action of saponins on the microbial populations and fermentation in the rumen and ruminant production. Nutr Res Rev. (2009) 22:204–19. doi: 10.1017/S0954422409990163
155. Goel G, Makkar HP, Becker K. Effects of Sesbania sesban and Carduus pycnocephalus leaves and Fenugreek (Trigonella foenum-graecum L.) seeds and their extracts on partitioning of nutrients from roughage-and concentrate-based feeds to methane. Anim Feed Sci Technol. (2008) 147:72–89. doi: 10.1016/j.anifeedsci.2007.09.010
156. Henderson G, Cook GM, Ronimus RS. Enzyme-and gene-based approaches for developing methanogen-specific compounds to control ruminant methane emissions: a review. Anim Prod Sci. (2018) 58:1017–26. doi: 10.1071/AN15757
157. Liu Y, Ma T, Chen D, Zhang N, Si B, Deng K, et al. Effects of tea saponin supplementation on nutrient digestibility, methanogenesis, and ruminal microbial flora in dorper crossbred ewe. Animals. (2019) 9:29–39. doi: 10.3390/ani9010029
158. Yuliana P, Laconi E, Jayanegara A, Achmadi S, Samsudin A. Extracted saponin from sapindus rarak and hibiscus sp. As an additive in cassava leaf silage: effects on chemical composition, rumen fermentation and microbial population. Adv Anim Vet Sci. (2019) 7:530–6. doi: 10.17582/journal.aavs/2019/7.7.530.536
159. Wu H, Meng Q, Zhou Z, Yu Z. Ferric citrate, nitrate, saponin and their combinations affect in vitro ruminal fermentation, production of sulphide and methane and abundance of select microbial populations. J Appl Microbiol. (2019) 127:150–8. doi: 10.1111/jam.14286
160. Molina-Botero IC, Arroyave-Jaramillo J, Valencia-Salazar S, Barahona-Rosales R, Aguilar-Pérez CF, Burgos AA, et al. Effects of tannins and saponins contained in foliage of Gliricidia sepium and pods of Enterolobium cyclocarpum on fermentation, methane emissions and rumen microbial population in crossbred heifers. Anim Feed SciTechnol. (2019) 251:1–11. doi: 10.1016/j.anifeedsci.2019.01.011
161. Popova M, Guyader J, Silberberg M, Seradj AR, Saro C, Bernard A, et al. Changes in the rumen microbiota of cows in response to dietary supplementation with nitrate, linseed, and saponin alone or in combination. Appl Environ Microbiol. (2019) 85:e02657–18. doi: 10.1128/AEM.02657-18
162. Guyader J, Eugène M, Doreau M, Morgavi D, Gérard C, Martin C. Tea saponin reduced methanogenesis in vitro but increased methane yield in lactating dairy cows. J Dairy Sci. (2017) 100:1845–55. doi: 10.3168/jds.2016-11644
163. Patra A, Yu Z. Effects of garlic oil, nitrate, saponin and their combinations supplemented to different substrates on in vitro fermentation, ruminal methanogenesis, and abundance and diversity of microbial populations. J Appl Microbiol. (2015) 119:127–38. doi: 10.1111/jam.12819
164. Sidhu A, Wadhwa M. Effect of supplementation of saponin containing herbs on in vitro methane production under different feeding systems. Indian J Anim Scis. (2019) 89:82–9.
165. Singh RK, Dey A, Paul SS, Singh M, Dahiya SS, Punia BS. Associative effects of plant secondary metabolites in modulating in vitro methanogenesis, volatile fatty acids production and fermentation of feed in buffalo (Bubalus bubalis). Agroforestry Syst. (2019) 94:1555–66. doi: 10.1007/s10457-019-00395-3
166. Liu C, Qu Yh, Guo Pt, Xu Cc, Ma Y, Luo Hl. Effects of dietary supplementation with alfalfa (Medicago sativa L.) saponins on lamb growth performance, nutrient digestibility, and plasma parameters. Anim Feed Sci Technol. (2018) 236:98–106. doi: 10.1016/j.anifeedsci.2017.12.006
167. Ramos-Morales E, De La Fuente G, Duval S, Wehrli C, Bouillon M, Lahmann M, et al. Antiprotozoal effect of saponins in the rumen can be enhanced by chemical modifications in their structure. Front Microbiol. (2017) 8:399. doi: 10.3389/fmicb.2017.00399
168. Wang B, Ma M, Diao Q, Tu Y. Saponin-induced shifts in the rumen microbiome and metabolome of young cattle. Front Microbiol. (2019) 10:356–69. doi: 10.3389/fmicb.2019.00356
169. Patra AK. Enteric methane mitigation technologies for ruminant livestock: a synthesis of current research and future directions. Environ Monitor Assess. (2012) 184:1929–52. doi: 10.1007/s10661-011-2090-y
170. Ramos-Morales E, Lyons L, de la Fuente G, Braganca R, Newbold C. Not all saponins have a greater antiprotozoal activity than their related sapogenins. FEMS Microbiol Lett. (2019) 366:fnz144. doi: 10.1093/femsle/fnz144
171. Atiku A, Oladipo OO, Forcados GE, Usman AS, Mancha MD. Anti-nutritional and phytochemical profile of some plants grazed upon by ruminants in north central nigeria during the dry season (January to April). Int J Livest Prod. (2016) 7:19–23. doi: 10.5897/IJLP2015.0268
172. Aboagye IA, Beauchemin KA. Potential of molecular weight and structure of tannins to reduce methane emissions from ruminants: a review. Animals. (2019) 9:856. doi: 10.3390/ani9110856
173. McSweeney C, Palmer B, McNeill D, Krause D. Microbial interactions with tannins: nutritional consequences for ruminants. Anim Feed SciTechnol. (2001) 91:83–93. doi: 10.1016/S0377-8401(01)00232-2
174. Patra AK, Min B-R, Saxena J. Dietary tannins on microbial ecology of the gastrointestinal tract in ruminants. In: AK Patra, editor. Dietary Phytochemicals and Microbes. Dordrecht: Springer (2012). p. 237–62. doi: 10.1007/978-94-007-3926-0_8
175. Canul-Solis J, Campos-Navarrete M, Piñeiro-Vázquez A, Casanova-Lugo F, Barros-Rodríguez M, Chay-Canul A, et al. Mitigation of rumen methane emissions with foliage and pods of tropical trees. Animals. (2020) 10:843. doi: 10.3390/ani10050843
176. Corrêa PS, Mendes LW, Lemos LN, Crouzoulon P, Niderkorn V, Hoste H, et al. Tannin supplementation modulates the composition and function of ruminal microbiome in lambs infected with gastrointestinal nematodes. FEMS Microbiol Ecol. (2020) 96:fiaa024. doi: 10.1093/femsec/fiaa024
177. Mannelli F, Daghio M, Alves SP, Bessa RJ, Minieri S, Giovannetti L, et al. Effects of chestnut tannin extract, vescalagin and gallic acid on the dimethyl acetals profile and microbial community composition in rumen liquor: an in vitro study. Microorganisms. (2019) 7:202–17. doi: 10.3390/microorganisms7070202
178. Zhou K, Bao Y, Zhao G. Effects of dietary crude protein and tannic acid on rumen fermentation, rumen microbiota and nutrient digestion in beef cattle. Arch Anim Nutr. (2019) 73:30–43. doi: 10.1080/1745039X.2018.1545502
179. Matra M, Wanapat M, Cherdthong A, Foiklang S, Mapato C. Dietary dragon fruit (Hylocereus undatus) peel powder improved in vitro rumen fermentation and gas production kinetics. Trop Anim Health Prod. (2019) 51:1531–8. doi: 10.1007/s11250-019-01844-y
180. Aboagye IA, Oba M, Koenig KM, Zhao GY, Beauchemin KA. Use of gallic acid and hydrolyzable tannins to reduce methane emission and nitrogen excretion in beef cattle fed a diet containing alfalfa silage. J Anim Sci. (2019) 97:2230–44. doi: 10.1093/jas/skz101
181. Sharifi A, Chaji M, Vakili A. Effect of treating recycled poultry bedding with tannin extracted from pomegranate peel on rumen fermentation parameters and cellulolytic bacterial population in Arabian fattening lambs. In: Veterinary Research Forum: Faculty of Veterinary Medicine. Urmia: Urmia University (2019). p. 145–52.
182. Wei C, Guyader J, Collazos L, Beauchemin K, Zhao G. Effects of gallic acid on in vitro rumen fermentation and methane production using rumen simulation (Rusitec) and batch-culture techniques. Anim Prod Sci. (2019) 59:277–87. doi: 10.1071/AN17365
183. Baruah L, Malik PK, Kolte AP, Goyal P, Dhali A, Bhatta R. Rumen methane amelioration in sheep using two selected tanniferous phyto-leaves. Carbon Manag. (2019) 10:299–308. doi: 10.1080/17583004.2019.1605480
184. Witzig M, Zeder M, Rodehutscord M. Effect of the ionophore monensin and tannin extracts supplemented to grass silage on populations of ruminal cellulolytics and methanogens in vitro. Anaerobe. (2018) 50:44–54. doi: 10.1016/j.anaerobe.2018.01.012
185. Salami SA, Valenti B, Bella M, O'Grady MN, Luciano G, Kerry JP, et al. Characterisation of the ruminal fermentation and microbiome in lambs supplemented with hydrolysable and condensed tannins. FEMS Microbiol Ecol. (2018) 94:fiy061. doi: 10.1093/femsec/fiy061
186. Costa Mn, Alves SP, Cappucci A, Cook SR, Duarte A, Caldeira RM, et al. Effects of condensed and hydrolyzable tannins on rumen metabolism with emphasis on the biohydrogenation of unsaturated fatty acids. J Agri Food Chem. (2018) 66:3367–77. doi: 10.1021/acs.jafc.7b04770
187. Stewart EK, Beauchemin KA, Dai X, MacAdam JW, Christensen RG, Villalba JJ. Effect of tannin-containing hays on enteric methane emissions and nitrogen partitioning in beef cattle. J Anim Sci. (2019) 97:3286–99. doi: 10.1093/jas/skz206
188. Adejoro FA, Hassen A, Akanmu AM, Morgavi DP. Replacing urea with nitrate as a non-protein nitrogen source increases lambs' growth and reduces methane production, whereas acacia tannin has no effect. Anim Feed Sci Technol. (2020) 259:114360. doi: 10.1016/j.anifeedsci.2019.114360
189. Adejoro FA, Hassen A, Akanmu AM. Effect of lipid-encapsulated acacia tannin extract on feed intake, nutrient digestibility and methane emission in sheep. Animals. (2019) 9:863. doi: 10.3390/ani9110863
190. Focant M, Froidmont E, Archambeau Q, Van QD, Larondelle Y. The effect of oak tannin (Quercus robur) and hops (Humulus lupulus) on dietary nitrogen efficiency, methane emission, and milk fatty acid composition of dairy cows fed a low-protein diet including linseed. J Dairy Sci. (2019) 102:1144–59. doi: 10.3168/jds.2018-15479
191. Pineiro-Vazquez A, Jimenez-Ferrer G, Alayon-Gamboa J, Chay-Canul A, Ayala-Burgos A, Aguilar-Pérez C, et al. Effects of quebracho tannin extract on intake, digestibility, rumen fermentation, and methane production in crossbred heifers fed low-quality tropical grass. Trop Anim Health Prod. (2018) 50:29–36. doi: 10.1007/s11250-017-1396-3
192. Koenig KM, Beauchemin KA. Effect of feeding condensed tannins in high protein finishing diets containing corn distillers grains on ruminal fermentation, nutrient digestibility, and route of nitrogen excretion in beef cattle. J Anim Sci. (2018) 96:4398–413. doi: 10.1093/jas/sky273
193. Dentinho MTP, Paulos K, Portugal PV, Moreira OC, Santos-Silva J, Bessa RJ. Proteolysis and in situ ruminal degradation of lucerne ensiled with Cistus ladanifer tannins. Grass Forage Sci. (2019) 74:78–85. doi: 10.1111/gfs.12394
194. Lee S-J, Eom J-S, Kim H-S, Kim H-S, Lee S-S. Effects of dietary Allium fistulosum L. and tannic acid on in vitro ruminal fermentation characteristics and methane emission. Kor J Org Agri. (2018) 26:775–87. doi: 10.11625/KJOA.2018.26.4.775
195. Nascimento T, Bezerra L, Menezes D, Lucena A, Queiroz MÁ, Trajano J, et al. Condensed tannin-amended cassava silage: fermentation characteristics, degradation kinetics and in-vitro gas production with rumen liquor. J Agri Sci. (2018) 156:83–91. doi: 10.1017/S0021859617000867
196. Rira M, Morgavi DP, Genestoux L, Djibiri S, Sekhri I, Doreau M. Methanogenic potential of tropical feeds rich in hydrolysable tannins. J Anim Sci. (2019) 97:2700–10. doi: 10.1093/jas/skz199
197. Syahniar TM, Ridla M, Jayanegara A, Samsudin AA. Effects of glycerol and chestnut tannin addition in cassava leaves (Manihot esculenta Crantz) on silage quality and in vitro rumen fermentation profiles. J Appl Anim Res. (2018) 46:1207–13. doi: 10.1080/09712119.2018.1485568
198. Yang K, Wei C, Zhao G, Xu Z, Lin S. Effects of dietary supplementing tannic acid in the ration of beef cattle on rumen fermentation, methane emission, microbial flora and nutrient digestibility. J Anim Physiol Anim Nutr. (2017) 101:302–10. doi: 10.1111/jpn.12531
199. Buccioni A, Pauselli M, Viti C, Minieri SARA, Pallara G, Roscini V, et al. Milk fatty acid composition, rumen microbial population, and animal performances in response to diets rich in linoleic acid supplemented with chestnut or quebracho tannins in dairy ewes. J Dairy Sci. (2015) 98:1145–56. doi: 10.3168/jds.2014-8651
200. Bae HD, McAllister TA, Muir AD, Yanke LJ, Bassendowski KA, Cheng KJ. Selection of a method of condensed tannin analysis for studies with rumen bacteria. J Agric Food Chem. (1993) 41:1256–60. doi: 10.1021/jf00032a018
201. Bhatta R, Uyeno Y, Tajima K, Takenaka A, Yabumoto Y, Nonaka I, et al. Difference in the nature of tannins on in vitro ruminal methane and volatile fatty acid production and on methanogenic archaea and protozoal populations. J Dairy Sci. (2009) 92:5512–22. doi: 10.3168/jds.2008-1441
202. Bao Y, Zhou K, Zhao G. Nitrous oxide emissions from the urine of beef cattle as regulated by dietary crude protein and gallic acid. J Anim Sci. (2018) 96:3699–711. doi: 10.1093/jas/sky252
203. He L, Chen N, Lv H, Wang C, Zhou W, Chen X, Zhang Q. Gallic acid influencing fermentation quality, nitrogen distribution and bacterial community of high-moisture mulberry leaves and stylo silage. Bioresource Technol. (2020) 295:122255. doi: 10.1016/j.biortech.2019.122255
204. Dentinho MTP, Paulos K, Francisco A, Belo AT, Jerónimo E, Almeida J, et al. Effect of soybean meal treatment with Cistus ladanifer condensed tannins in growth performance, carcass and meat quality of lambs. Livestock Sci. (2020) 236:104021. doi: 10.1016/j.livsci.2020.104021
205. Saminathan M, Kumari Ramiah S, Gan HM, Abdullah N, Wong CM, Ho YW, et al. In vitro study on the effects of condensed tannins of different molecular weights on bovine rumen fungal population and diversity. Ital J Anim Sci. (2019) 18:1451–62. doi: 10.1080/1828051X.2019.1681304
206. Cipriano-Salazar M, Rojas-Hernández S, Olivares-Perez J, Jimenez-Guillen R, Cruz-Lagunas B, Camacho-Diaz LM, et al. Antibacterial activities of tannic acid against isolated ruminal bacteria from sheep. Microb Pathogenesis. (2018) 117:255–8. doi: 10.1016/j.micpath.2018.01.045
207. Goel G, Makkar HP. Methane mitigation from ruminants using tannins and saponins. Trop Anim Health Prod. (2012) 44:729–39. doi: 10.1007/s11250-011-9966-2
208. Adejoro FA, Hassen A, Thantsha MS. Characterization of starch and gum arabic-maltodextrin microparticles encapsulating acacia tannin extract and evaluation of their potential use in ruminant nutrition. Asian-Australas J Anim Sci. (2019) 32:977–87. doi: 10.5713/ajas.18.0632
209. Beauchemin KA, McGinn SM, Martinez TF, McAllister TA. Use of condensed tannin extract from quebracho trees to reduce methane emissions from cattle. J Anim Sci. (2007) 85:1990–6. doi: 10.2527/jas.2006-686
210. Grainger C, Clarke T, Auldist MJ, Beauchemin KA, McGinn SM, Waghorn GC, et al. Potential use of Acacia mearnsii condensed tannins to reduce methane emissions and nitrogen excretion from grazing dairy cows. Can J Anim Sci. (2009) 89:241–51. doi: 10.4141/CJAS08110
211. Herremans S, Vanwindekens F, Decruyenaere V, Beckers Y, Froidmont E. Effect of dietary tannins on milk yield and composition, nitrogen partitioning and nitrogen use efficiency of lactating dairy cows: a meta-analysis. J Anim Physiol Anim Nutr. (2020) 104:1029–18. doi: 10.1111/jpn.13341
212. Denninger TM, Schwarm A, Birkinshaw A, Terranova M, Dohme-Meier F, Münger A, et al. Immediate effect of Acacia mearnsii tannins on methane emissions and milk fatty acid profiles of dairy cows. Anim Feed Sci Technol. (2020) 261:114388. doi: 10.1016/j.anifeedsci.2019.114388
213. Bernard L, Leroux C, Chilliard Y. Expression and nutritional regulation of lipogenic genes in the ruminant lactating mammary gland. In: Bösze Z, editor. Bioactive components of milk. New York, NY: Springer (2008). p. 67–108. doi: 10.1007/978-0-387-74087-4_2
214. Henke A, Dickhoefer U, Westreicher-Kristen E, Knappstein K, Molkentin J, Hasler M, et al. Effect of dietary Quebracho tannin extract on feed intake, digestibility, excretion of urinary purine derivatives and milk production in dairy cows. Archiv Anim Nutr. (2017) 71:37–53. doi: 10.1080/1745039X.2016.1250541
215. French EA, Bertics SJ, Armentano LE. Rumen and milk odd-and branched-chain fatty acid proportions are minimally influenced by ruminal volatile fatty acid infusions. J Dairy Sci. (2012) 95:2015–26. doi: 10.3168/jds.2011-4827
216. Ariga T, Hamano M. Radical scavenging action and its mode in procyanidins B-1 and B-3 from azuki beans to peroxyl radicals. Agri Biol Chem. (1990) 54:2499–504. doi: 10.1080/00021369.1990.10870369
217. Ricci A, Olejar KJ, Parpinello GP, Mattioli AU, Teslić N, Kilmartin PA, et al. Antioxidant activity of commercial food grade tannins exemplified in a wine model. Food Additiv Contaminants A. (2016) 33:1761–74. doi: 10.1080/19440049.2016.1241901
218. Nawab A, Li G, An L, Nawab Y, Zhao Y, Xiao M, et al. The potential effect of dietary tannins on enteric methane emission and ruminant production; as an alternative to antibiotic feed additives–a review. Annal Anim Sci. (2020) 1:5. doi: 10.2478/aoas-2020-0005
219. Barreira JC, Ferreira IC, Oliveira MBP, Pereira JA. Antioxidant activities of the extracts from chestnut flower, leaf, skins and fruit. Food Chem. (2008) 107:1106–13. doi: 10.1016/j.foodchem.2007.09.030
220. Huang QQ, Jin L, Xu Z, Barbieri LR, Acharya S, Hu TM, et al. Effects of purple prairie clover (Dalea purpurea Vent.) on feed intake, nutrient digestibility and faecal shedding of Escherichia coli O157: H7 in lambs. Anim Feed Sci Technol. (2015) 207:51–61. doi: 10.1016/j.anifeedsci.2015.06.009
221. Peng K, Shirley DC, Xu Z, Huang Q, McAllister TA, Chaves AV, et al. Effect of purple prairie clover (Dalea purpurea Vent.) hay and its condensed tannins on growth performance, wool growth, nutrient digestibility, blood metabolites and ruminal fermentation in lambs fed total mixed rations. Anim Feed Sci Technol. (2016) 222:100–10. doi: 10.1016/j.anifeedsci.2016.10.012
222. Chambi F, Chirinos R, Pedreschi R, Betalleluz-Pallardel I, Debaste F, Campos D. Antioxidant potential of hydrolyzed polyphenolic extracts from tara (Caesalpinia spinosa) pods. Indus Crops Product. (2013) 47:168–75. doi: 10.1016/j.indcrop.2013.03.009
223. Anggraini T, Tai A, Yoshino T, Itani T. Antioxidative activity and catechin content of four kinds of Uncaria gambir extracts from West Sumatra, Indonesia. African J Biochem Res. (2011) 5:33–8.
224. Ciampi F, Sordillo LM, Gandy JC, Caroprese M, Sevi A, Albenzio M, et al. Evaluation of natural plant extracts as antioxidants in a bovine in vitro model of oxidative stress. J Dairy Sci. (2020) 103:8938–47. doi: 10.3168/jds.2020-18182
225. Wang B, Luo H. Mining novel specific rumen bacteria to enhance the bioactive function of mulberry leaf in promoting host antioxidant activity and immunomodulatory. Researchsquare (2020). doi: 10.21203/rs.3.rs-36704/v1
226. Huang Q, Liu X, Zhao G, Hu T, Wang Y. Potential and challenges of tannins as an alternative to in-feed antibiotics for farm animal production. Anim Nutr. (2018) 4:137–50. doi: 10.1016/j.aninu.2017.09.004
227. Min BR, Pinchak WE, Fulford JD, Puchala R. Effect of feed additives on in vitro and in vivo rumen characteristics and frothy bloat dynamics in steers grazing wheat pasture. Anim Feed Sci Technol. (2005) 123:615–29. doi: 10.1016/j.anifeedsci.2005.04.050
228. Min BR, Pinchak WE, Anderson RC, Fulford JD, Puchala R. Effects of condensed tannins supplementation level on weight gain and in vitro and in vivo bloat precursors in steers grazing winter wheat. J Anim Sci. (2006) 84:2546–54. doi: 10.2527/jas.2005-590
229. Min BR, Solaiman S, Waldrip HM, Parker D, Todd RW, Brauer D. Dietary mitigation of enteric methane emissions from ruminants: a review of plant tannins mitigation options. Anim Nutr. (2020) 6:231–46. doi: 10.1016/j.aninu.2020.05.002
230. Benchaar C, Chaves A, Fraser G, Beauchemin K, McAllister T. Effects of essential oils and their components on in vitro rumen microbial fermentation. Can J Anim Sci. (2007) 87:413–9. doi: 10.4141/CJAS07012
231. Bakkali F, Averbeck S, Averbeck D, Idaomar M. Biological effects of essential oils–a review. Food Chem Toxicol. (2008) 46:446–75. doi: 10.1016/j.fct.2007.09.106
232. Zengin H, Baysal A. Antibacterial and antioxidant activity of essential oil terpenes against pathogenic and spoilage-forming bacteria and cell structure-activity relationships evaluated by SEM microscopy. Molecules. (2014) 19:17773–98. doi: 10.3390/molecules191117773
233. Lei Z, Zhang K, Li C, Jiao T, Wu J, Wei Y, et al. Ruminal metagenomic analyses of goat data reveals potential functional microbiota by supplementation with essential oil-cobalt complexes. BMC Microbiol. (2019) 19:30–9. doi: 10.1186/s12866-019-1400-3
234. Busquet M, Calsamiglia S, Ferret A, Carro M, Kamel C. Effect of garlic oil and four of its compounds on rumen microbial fermentation. J Dairy Sci. (2005) 88:4393–404. doi: 10.3168/jds.S0022-0302(05)73126-X
235. Schären M, Drong C, Kiri K, Riede S, Gardener M, Meyer U, et al. Differential effects of monensin and a blend of essential oils on rumen microbiota composition of transition dairy cows. J Dairy Sci. (2017) 100:2765–83. doi: 10.3168/jds.2016-11994
236. Cobellis G, Trabalza-Marinucci M, Yu Z. Critical evaluation of essential oils as rumen modifiers in ruminant nutrition: a review. Sci Total Environ. (2016) 545:556–68. doi: 10.1016/j.scitotenv.2015.12.103
237. Soltan YA, Natel AS, Araujo RC, Morsy AS, Abdalla AL. Progressive adaptation of sheep to a microencapsulated blend of essential oils: ruminal fermentation, methane emission, nutrient digestibility, and microbial protein synthesis. Anim Feed SciTechnol. (2018) 237:8–18. doi: 10.1016/j.anifeedsci.2018.01.004
238. Garcia F, Colombatto D, Brunetti MA, Martínez MJ, Moreno MV, Scorcione Turcato M, et al. The reduction of methane production in the in vitro ruminal fermentation of different substrates is linked with the chemical composition of the essential oil. Animals. (2020) 10:786. doi: 10.3390/ani10050786
239. Zhou R, Wu J, Lang X, Liu L, Casper DP, Wang C, et al. Effects of oregano essential oil on in vitro ruminal fermentation, methane production, and ruminal microbial community. J Dairy Sci. (2020) 103:2303–14. doi: 10.3168/jds.2019-16611
240. Benchaar C. Feeding oregano oil and its main component carvacrol does not affect ruminal fermentation, nutrient utilization, methane emissions, milk production, or milk fatty acid composition of dairy cows. J Dairy Sci. (2020) 103:1516–27. doi: 10.3168/jds.2019-17230
241. Zhou R, Wu J, Zhang L, Liu L, Casper DP, Jiao T, et al. Effects of oregano essential oil on the ruminal pH and microbial population of sheep. PLoS ONE. (2019) 14:e0217054. doi: 10.1371/journal.pone.0217054
242. Oh J, Harper M, Hristov AN. Effects of lowering crude protein supply alone or in a combination with essential oils on productivity, rumen function and nutrient utilization in dairy cows. Animal. (2019) 13:2510–18. doi: 10.1017/S1751731119001083
243. Kim H, Jung E, Lee HG, Kim B, Cho S, Lee S, et al. Essential oil mixture on rumen fermentation and microbial community–an in vitro study. Asian-Australas J Anim Sci. (2019) 32:808–14. doi: 10.5713/ajas.18.0652
244. Castañeda-Correa A, Corral-Luna A, Hume ME, Anderson RC, Ruiz-Barrera O, Castillo-Castillo Y, et al. Effects of thymol and carvacrol, alone or in combination, on fermentation and microbial diversity during in vitro culture of bovine rumen microbes. J Environ Sci Health B. (2019) 54:170–5. doi: 10.1080/03601234.2018.1536580
245. Kurniawati A, Widodo W, Artama WT, Yusiati LM. Amomum compactum soland ex maton addition as essential oil source and its effect on ruminal feed fermentation by in vitro analysis. BIOTROPIA. (2019) 26:1089–102. doi: 10.11598/btb.2019.26.3.1089
246. Ye D, Karnati S, Wagner B, Firkins J, Eastridge M, Aldrich J. Essential oil and monensin affect ruminal fermentation and the protozoal population in continuous culture. J Dairy Sci. (2018) 101:5069–81. doi: 10.3168/jds.2017-13646
247. Chahaardoli A, Soroor M, Foroughi A. The effects of Anise (Pimpinella anisum) essential oil and extract on in vitro rumen fermentation parameters and protozoa population of sheep. Int J Vet Sci. (2018) 7:21–7.
248. Olijhoek DW, Hellwing ALF, Grevsen K, Haveman LS, Chowdhury MR, Løvendahl P, et al. Effect of dried oregano (Origanum vulgare L.) plant material in feed on methane production, rumen fermentation, nutrient digestibility, and milk fatty acid composition in dairy cows. J Dairy Sci. (2019) 102:9902–18. doi: 10.3168/jds.2019-16329
249. Garcia F, Vercoe P, Martínez M, Durmic Z, Brunetti M, Moreno M, et al. Essential oils from Lippia turbinata and Tagetes minuta persistently reduce in vitro ruminal methane production in a continuous-culture system. Anim Prod Sci. (2019) 59:709–20. doi: 10.1071/AN17469
250. Coneglian SM, Serrano RDC, Cruz OTB, Branco AF. Effects of essential oils of Cashew and Castor on intake, digestibility, ruminal fermentation and purine derivatives in beef cattle fed high grain diets. Semina. (2019) 40:2057–70. doi: 10.5433/1679-0359.2019v40n5p2057
251. Davoodi SM, Mesgaran MD, Vakili AR, Valizadeh R, Pirbalouti AG. In vitro effect of essential oils on rumen fermentation and microbial nitrogen yield of high concentrate dairy cow diet. Biosci Biotechnol Res Asia. (2019) 16:333–41. doi: 10.13005/bbra/2749
252. Jahani-Azizabadi H, Durmic Z, Vadhanabhuti J, Vercoe P. Effect of some australian native shrubs essential oils on in vitro rumen microbial fermentation of a high-concentrate diet. J Anim Plant Sci. (2019) 29:8–15. Available online at: http://www.thejaps.org.pk/docs/V-29-01/02.pdf
253. Hart KJ, Jones HG, Waddams KE, Worgan HJ, Zweifel B, Newbold CJ. An essential oil blend decreases methane emissions and increases milk yield in dairy cows. Open J Anim Sci. (2019) 9:259–67. doi: 10.4236/ojas.2019.93022
254. Joch M, Kudrna V, Hakl J, BoŽik M, Homolka P, Illek J, et al. In vitro and in vivo potential of a blend of essential oil compounds to improve rumen fermentation and performance of dairy cows. Anim Feed Sci Technol. (2019) 251:176–86. doi: 10.1016/j.anifeedsci.2019.03.009
255. Lodi F, Ferreira EM, Biava JS, Martins MC, de Assis RG, Moletta JL, et al. Lemon grass essential oil (Cymbopogum flexuosus) in high-concentrate diets for lambs. Arch Vet Sci. (2019) 24:33–47. doi: 10.5380/avs.v24i2.61062
256. Wu P, Liu Z, He W, Yu S, Gao G, Wang J. Intermittent feeding of citrus essential oils as a potential strategy to decrease methane production by reducing microbial adaptation. J Clean Prod. (2018) 194:704–13. doi: 10.1016/j.jclepro.2018.05.167
257. Poudel P, Froehlich K, Casper DP, St-Pierre B. Feeding essential oils to neonatal holstein dairy calves results in increased ruminal prevotellaceae abundance and propionate concentrations. Microorganisms. (2019) 7:120–34. doi: 10.3390/microorganisms7050120
258. Belanche A, Newbold CJ, Morgavi DP, Bach A, Zweifel B, Yáñez-Ruiz DR. A meta-analysis describing the effects of the essential oils blend agolin ruminant on performance, rumen fermentation and methane emissions in dairy cows. Animals. (2020) 10:620. doi: 10.3390/ani10040620
259. Braun H-S, Schrapers KT, Mahlkow-Nerge K, Stumpff F, Rosendahl J. Dietary supplementation of essential oils in dairy cows: evidence for stimulatory effects on nutrient absorption. Animal. (2019) 13:518–23. doi: 10.1017/S1751731118001696
260. Christaki E, Bonos E, Giannenas I, Florou-Paneri P. Aromatic plants as a source of bioactive compounds. Agriculture. (2012) 2:228–43. doi: 10.3390/agriculture2030228
261. Tajodini M, Moghbeli P, Saeedi HR, Effati M. The effect of medicinal plants as a feed additive in ruminant nutrition. Iran J Appl Anim Sci. (2014) 4:681–6. Available online at: http://ijas.iaurasht.ac.ir/article_513470.html
262. McIntosh FM, Williams P, Losa R, Wallace RJ, Beever DA, Newbold CJ. Effects of essential oils on ruminal microorganisms and their protein metabolism. Appl Environ Microbiol. (2003) 69:5011–4. doi: 10.1128/AEM.69.8.5011-5014.2003
263. Gao S, Hu M. Bioavailability challenges associated with development of anti-cancer phenolics. Mini Rev Med Chem. (2010) 10:550–67. doi: 10.2174/138955710791384081
264. Li Z, Gu L. Fabrication of self-assembled ()-epigallocatechin gallate (EGCG) ovalbuminedextran conjugate nanoparticles and their transport across monolayers of human intestinal epithelial Caco-2 cells. J Agri Food Chem. (2014) 62:1301–9. doi: 10.1021/jf404621f
265. Gopal J, Muthu M, Paul D, Kim D-H, Chun S. Bactericidal activity of green tea extracts: the importance of catechin containing nano particles. Sci Rep. (2016) 6:19710. doi: 10.1038/srep19710
Keywords: rumen, microbiome, methane, fermentation, VFA, plant secondary metabolites
Citation: Hassan F, Arshad MA, Ebeid HM, Rehman MS, Khan MS, Shahid S and Yang C (2020) Phytogenic Additives Can Modulate Rumen Microbiome to Mediate Fermentation Kinetics and Methanogenesis Through Exploiting Diet–Microbe Interaction. Front. Vet. Sci. 7:575801. doi: 10.3389/fvets.2020.575801
Received: 24 June 2020; Accepted: 06 October 2020;
Published: 12 November 2020.
Edited by:
Zora Varadyova, Slovak Academy of Sciences, SlovakiaReviewed by:
Jorge Avila Stagno, University of Concepcion, ChileCopyright © 2020 Hassan, Arshad, Ebeid, Rehman, Khan, Shahid and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengjian Yang, eWNqMDc0NkBzaW5hLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.