
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 03 November 2020
Sec. Animal Nutrition and Metabolism
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.572064
 Zhaoqing Han1†
Zhaoqing Han1† Aoyun Li2*†
Aoyun Li2*† Lulu Pei2†
Lulu Pei2† Kun Li2
Kun Li2 Taihua Jin1
Taihua Jin1 Fukuan Li1
Fukuan Li1 Zhennan Wang1
Zhennan Wang1 Shenjin Lv1*
Shenjin Lv1* Yongzhu Li1*
Yongzhu Li1*Increasing evidence has indicated the ameliorative effect of milk replacer supplementation in ruminants for regulating their early growth and rumen development. However, it is still unclear whether milk replacer supplementation has a beneficial role in the growth performance and rumen microbiota of Yimeng black goats (YBGs). Therefore, this study was performed to investigate the effects of milk replacer on growth performance and rumen microbiota of YBGs. Our results revealed that milk replacer supplementation could significantly improve the growth performance of YBGs. Additionally, the results of alpha and beta diversities indicated that there was no significant difference in richness and diversity between the control and milk replacer-treated YBGs. At the phylum level, Bacteroidetes, Firmicutes, and Proteobacteria were the most dominant phyla in all the samples at different stages. Moreover, the YBGs treated with milk replacer possessed a higher abundance of Verrucomicrobia than that in the control YBGs, while the level of Actinobacteria was obviously decreased. It is noteworthy that the abundance of Proteobacteria in the control YBGs was higher than that in the YBGs supplemented with milk replacer throughout the experiment. At the level of genus, the differences in the richness between control and milk replacer supplement YBGs were gradually observed. Compared with the control YBGs, the proportion of Akkermansia, Veillonella, Anaerovibrio, Ruminococcaceae_NK4A214_group, Ruminococcus_1, and Ruminococcus_2 was increased in the YBGs treated with milk replacer, whereas Turicibacter was decreased. In conclusion, milk replacer supplementation may serve as a good applicant for ameliorating early YBGs development and rumen microbiota.
Yimeng black goats (YBGs), mainly found in Shandong province of China, are characterized by strong resistance and excellent adaptability (1). This breed is an important source of meat, leather, and wool for local herdsmen. Previous studies have shown that the meat of YBGs have an extremely high nutritional value (2, 3). However, the population of YBGs is worryingly small due to delayed growth and low kidding rate. Therefore, it is crucial for the development of the goat industry to improve the growth performance of goats and shorten the pregnancy cycle of ewe. Currently, early-weaning and milk replacer supplementation are considered as the primary method to increase ewe breeding efficiency and goat's growth performance (4).
Early-weaning not only reduces the postpartum convalescence of the ewe but also increases the reproductive efficiency and promotes the development of digestive organs and survival of goats (5). Additionally, it may help in reducing the breeding and production costs of goats (6). However, goats may suffer from psychological, physical, and immune stress due to changes in the composition and physical form of their feed after weaning (7, 8). Importantly, the resistance of goats to various diseases will also decrease, due to the loss of maternal immunoglobulins (9, 10).
Increasing evidence has signified the beneficial effect of milk replacer supplementation in early-weaning goats for regulating their growth performance and rumen development (11). Milk replacer is artificial milk made by replacing milk protein with non-milk protein based on the nutritional standards of breast milk (12). Milk replacer supplementation not only reduces ewe consumption and the empty pregnancy rate but also improves reproductive efficiency and shortens the reproductive cycle (13). In addition, supplementing with milk replacer can also improve the growth performance of goats and decrease the morbidity and mortality caused by deficiency of early nutrient intake (14). Recent studies indicate that milk replacer supplementation is beneficial for the development and establishment of rumen microbiota (15). Rumen microbiota plays a key role in rumen function and nutrient digestion (16). However, it remains to be determined whether milk replacer supplementation has an ameliorative role in the growth performance and rumen microbiota of YBGs. Therefore, the objective of the present study was to investigate the effects of milk replacer on growth performance and rumen microbiota of YBGs.
The animal experiments were performed under the approval of the Ethics Committee of the Huazhong Agricultural University (Permit No. 4200695757).
A total of 30 1-day-old YBGs (initial weight 1.84 ± 0.86 kg) were purchased from a commercial feedlot (Shandong Province, China). All the selected YBGs had a similar genetic background and the same immune procedure. The YBGs were equally divided into two groups (n = 15), i.e., the control group (B group) and milk replacer (Table 1) supplementation group (R group). The YBGs in the B group were fed with goat's milk throughout the experimental period, i.e., from day 1 to 75, whereas the YBGs in the R group were fed with goat's milk from days 1 to 10. Afterwards, the YBGs in the R group were compulsively weaned on day 10 and were then provided with milk replacer (2% of the body weight from days 10 to 45 and 1.5% of the body weight from days 46 to 75) each day. Both groups received ad libitum regular starter feed (Table 2) and water from day 15. In addition, all animals were reared for 75 days, and body weight and average daily weight gain in each group was recorded on day 10, 15, 25, 45, and 75. During the experiment period, three YBGs from each group were randomly selected and euthanized by injecting pentobarbital (25 mg/kg) on day 10, 15, 25, 45, and 75. After euthanization, the carcasses were placed in a natural position to minimize the potential contamination among the four compartments. Afterwards, each stomach chamber was tied off using cotton rope and transferred to sterilized brown paper. The content of the rumen was homogenized and then the content which weighed approximately 200 g was stored in sterilized tubes. The collected samples were snap-frozen using liquid nitrogen and stored at −80°C for further analyses.
Bacterial genomic DNA was extracted using a QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the producer's instructions. DNA quality was determined by electrophoresis on agarose gel 0.8% (w/v) and the quantity was determined using a Nanodrop™ spectrophotometer (NanoDrop Technologies, Thermo Scientific, USA).
Specific PCR primers (338F: ACT CCT ACG GGA GGC AGC A and 806R: GGA CTA CHV GGG TWT CTA AT) based on conserved regions were synthesized to amplify the V3/V4 regions. An AxyPrep DNA Gel Extraction Kit (Axygen, CA, USA) and 2% agarose gel electrophoresis were used for target fragment recovery and PCR amplification product evaluation, respectively. The PCR amplification recovery products were quantified on a fluorescent Microplate reader (BioTek, FLx800) via a Quant-iT PicoGreen dsDNA Assay Kit based on the preliminary quantitative results of electrophoresis. For the sequence library construction, a TruSeq Nano DNA Low Throughput Library Prep Kit (Illumina, CA, USA) was used following manufacturer's specifications. Amplified products' sequence ends were repaired by End Repair Mix2. To enrich the sequencing library template, PCR amplification was carried out and the library enrichment product was purified again using BECKMAN AMPure XP Beads. The final fragment-selection and purification of the library was performed using 2% agarose gel electrophoresis.
The quality of the libraries was examined on an Agilent Bioanalyzer using the Agilent High Sensitivity DNA Kit prior to the sequencing procedure. The libraries with only one peak signal and no linker signal were considered for the procedure. Moreover, the libraries were quantified using a Quant-iT PicoGreen dsDNA Assay Kit on the Promega QuantiFluor fluorescence quantification system. The qualified library concentration must be above 2 nM. The qualified libraries were gradient diluted and mixed in proportion according to the amount of sequencing required. The MiSeq Reagent Kit V3 (600 cycles) was used to perform 2 × 300 bp paired-end sequencing on the MiSeq sequencing machine after the mixed libraries were denatured into single strands by sodium hydroxide. The original sequence data were submitted to the Sequence Read Archive (SRA) (NCBI, USA) with the accession no. PRJNA637829.
The QIIME software (Qiime1.9.1) was used for 16S rRNA original data quality preliminary screening and analysis. Interrogative and short sequences (<200 bp) were discarded using the QIIME software. The obtained sequences were clustered and OTU partitioned at ≥97% sequence similarity by using the clustering program VSEARCH (1.9.6.). A representative sequence of each OTU was classified at a confidence threshold of 0.8 according to the Ribosomal Database Program (RDP) classifier. Additionally, the obtained sequences with 97% similarity were merged to the same operational taxonomic units (OTUs). Before calculating alpha and beta diversity statistics, the sequencing depth of each sample was evaluated using sparse curves. Continuous analysis of alpha diversity and beta diversity were performed based on the output normalized date. Four metrics including Chao1, ACE, Simpson, and Shannon were used to analyze alpha diversity (18, 19). GraphPad Prism (version 6.0c) and R (v3.0.3) software were used for statistical analysis (20). The criterion of significance was performed at p < 0.05 and the values were presented as means ± SD.
To investigate the effect of milk replacer supplementation on the growth performance of YBGs, the body weight and average daily weight gain of both groups were recorded. The average initial body weight of YBGs in the B and R groups were 1.84 ± 0.073 kg and 1.85 ± 0.09 kg, respectively, with similar values. The body weight gain of the B group was higher than that of the R group from days 1 to 15, whereas the body weight of the B group was significantly higher than that of the R group on day 15 (P < 0.01) (Figure 1A). However, statistical analysis indicated that the average daily weight gain of the R group surpassed that of the B group after day 25, and a significant difference was observed on day 45 and day 75 of the experiment (P < 0.05) (Figure 1A). Additionally, the body weight differed significantly between the groups and reached 7.66 ± 0.26 kg and 8.40 ± 0.31 kg, respectively, on day 75 (P < 0.05) (Figure 1B).
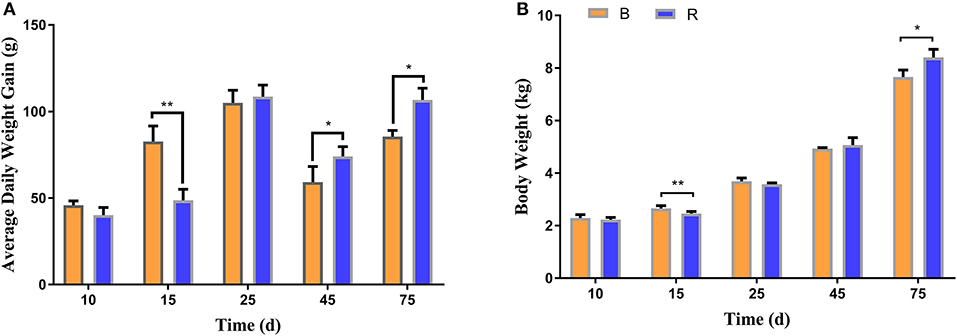
Figure 1. Milk replacer supplementation improved the growth performance of YBGs. (A,B) indicate average daily weight gain and body weight of goats on days 10, 15, 25, 45, and 75, respectively. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group. The data are expressed as the mean ± SD. *P < 0.05, **P < 0.01.
In the current study, 30 rumen samples collected from goats were subjected to high-throughput sequencing analysis. After optimizing the preliminary data, a total of 877,614 and 883,962 high-quality valid sequences were obtained from the B and R groups, respectively. The rarefaction (Chao1 and Shannon) and rank abundance curves showed a relatively flat curve and a tendency to saturate characteristic (Supplementary Figures 1A–C). The high-quality sequences were merged and OTU partitioned based on 97% nucleotide sequence similarity. The average number of OTUs in the R and B groups gradually increased from days 10 to 15, respectively, and all the samples showed the highest number of OTUs on day 75 (Figure 2). Throughout the experiment, the number of OTUs in the B group were higher than that in the R group except on day 15, other than that no significant difference was observed between the two groups (Figure 2).
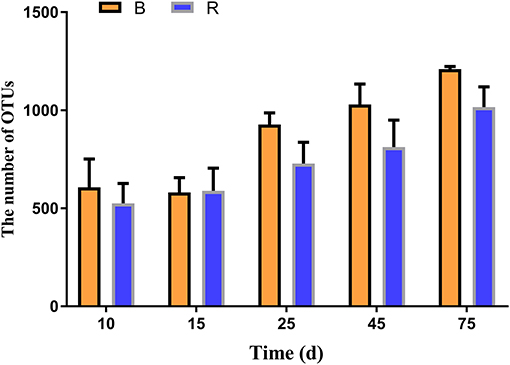
Figure 2. The number of OTUs in the B and R groups on day 10, 15, 25, 45, and 75. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group.
The alpha diversity of rumen microbiota was evaluated using Chao1, ACE, Shannon, and Simpson. Good's coverage estimates were ~100% for all samples, showing excellent coverage. The ACE and Chao1 indices in all samples showed a gradually upward trend with the increase of experiment time. Specifically, the average of the ACE index in the B group varied from 636 to 1,437, while the Chao1 index varied from 608 to 1,386. Moreover, the average value of the ACE index in the R group ranged from 627 to 1335, while the average values of the Chao1 index ranged from 601 to 1,301. The ACE and Chao1 indices of the B group were higher than that of the R group throughout the experiment period, other than that no obvious difference was observed between the two groups (P > 0.05) (Figures 3A,B). The results of the Chao1 and ACE indices suggested that there was no significant difference in the rumen microbiota abundance between the two groups. Compared with the B group, the Shannon and Simpson indexes in the R group were relatively higher on days 10 and 15, but lower on days 25, 45, and 75. Statistical analysis showed that both B and R groups exhibited the highest Shannon (7.42 and 6.83) and Simpson (0.977 and 0.973) indices on day 75. However, there was no significant difference in the average of the Shannon and Simpson indexes between the two groups during the whole experimental period (P > 0.05) (Figures 3C,D). The Shannon and Simpson indices revealed a non-significant difference in the rumen microbiota diversity among all the samples. The results of beta diversity demonstrated that the samples in the B and R groups gradually clustered with time, suggesting that rumen microbiota was not different between experimental groups (Figures 4A,B).
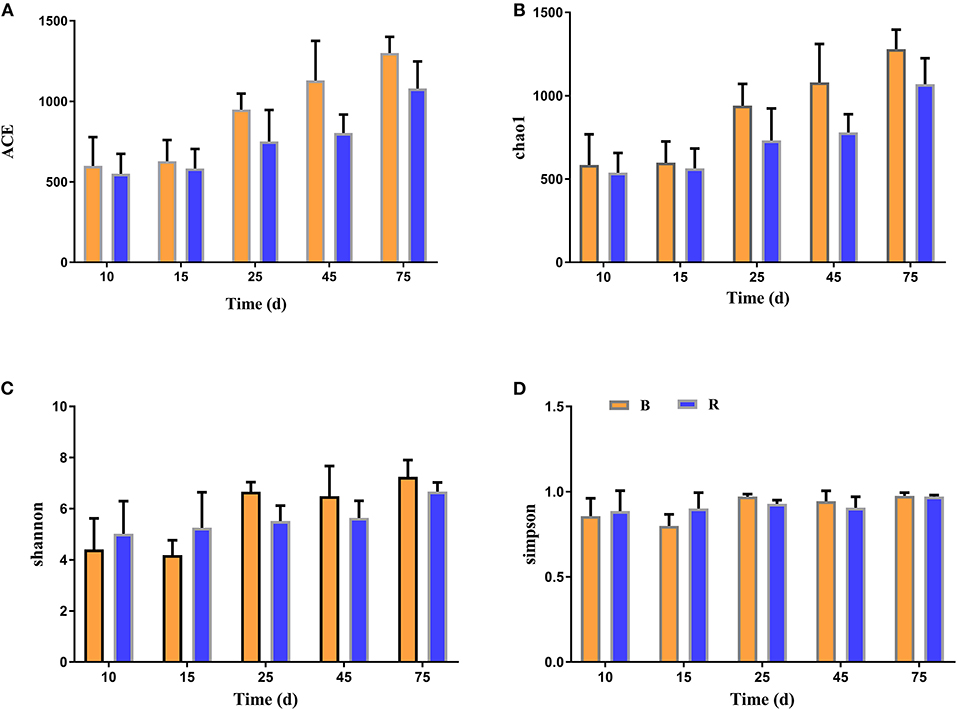
Figure 3. The diversity indices of rumen microbiota in different groups. Chao1 (A), ACE (B), Shannon (C), and Simpson (D) were used to evaluate the alpha diversity of rumen microbiota. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group.
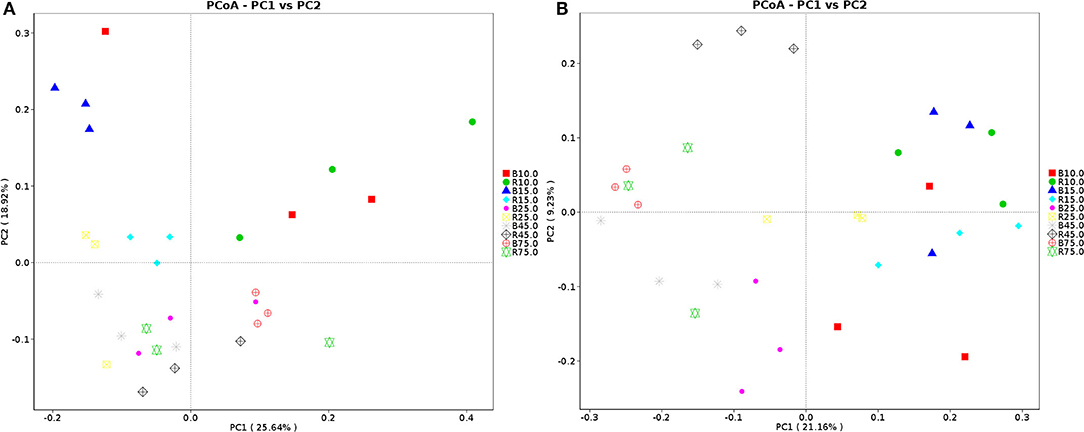
Figure 4. Principal coordinate (PCoA) analysis of rumen microbiota. (A) PCoA map based on weighted uniFrac distance; (B) PCoA map based on unweighted uniFrac distance. Each point indicates one sample. The distance of the two points indicate the difference of rumen microbiota. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group.
The rumen microbiota in the B and R groups were assessed at different taxonomical levels. The three most abundant (>90%) phyla in all samples were Firmicutes, Bacteroidetes, and Proteobacteria (Figure 5A). Other phyla including Synergistetes, Chlamydiae, Spirochaetes, Lentisphaerae, and Fusobacteria were present with a lower abundance accounting for <8% of the total 16S rRNA gene sequences in all samples. On day 10, Bacteroides (10.94%), Veillonella (7.00%), and Escherichia-Shigella (6.55%) were predominant in the B group, whereas Megasphaera (16.35%), Bacteroides (13.50%), and Lactobacillus (13.21%) were prominent in the R group at the genus level (Figure 5B). Additionally, the most abundant genera were Alloprevotella (40.84%) and Bacteroides (4.83%) in the B group, whereas Bacteroides (6.19%) and Prevotella_1 (8.85) were predominant in the R group on day 15. Interestingly, Bacteroides and Prevotella_1 were the dominant bacteria in the B and R groups on days 25, 45, and 75.
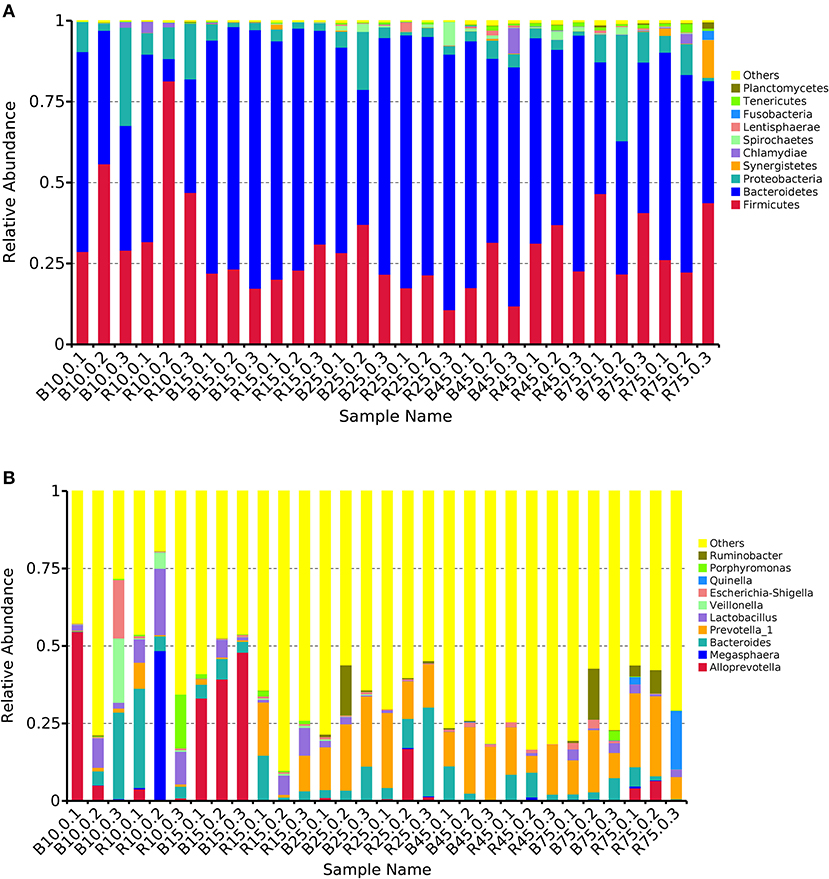
Figure 5. The relative abundance of microbial composition of different samples. (A) The top 10 dominant phylum of the rumen microbiota in the B and R groups. (B) The top 10 primary genera of the rumen microbiota in the B and R groups. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group.
The comparison results of rumen microbiota between the B and R groups using Metastats showed significant abundance of Verrucomicrobia (P < 0.05) in the R group when compared to the B group on day 15, while Actinobacteria (P < 0.01) was significantly lower on day 75 (Figures 6A,B). The abundance of Proteobacteria in the B group was found relatively higher, but it was not statistically significant (P > 0.05) when compared to the R group (data not shown). At the genus level, Akkermansia and Veillonella in the R group was significantly higher (P < 0.05) than the B group on day 15 (Figures 6C,D). Moreover, a comparison of the B and R groups showed an obvious decrease (P < 0.05) in the abundance of Anaerovibrio as well as a significant increase (P < 0.05) in the abundance of Turicibacter on day 25 (Figures 6E,F). The relative abundance of Ruminococcaceae_NK4A214_group, Ruminococcus_1, and Ruminococcus_2 were higher (P < 0.05) in the R group than in the B group on day 45, whereas the abundance of Romboutsia, Alistipes, and Anaerotruncus in the R group were lower (P < 0.05) when compared to the B group on day 75 (Figures 6G–L).
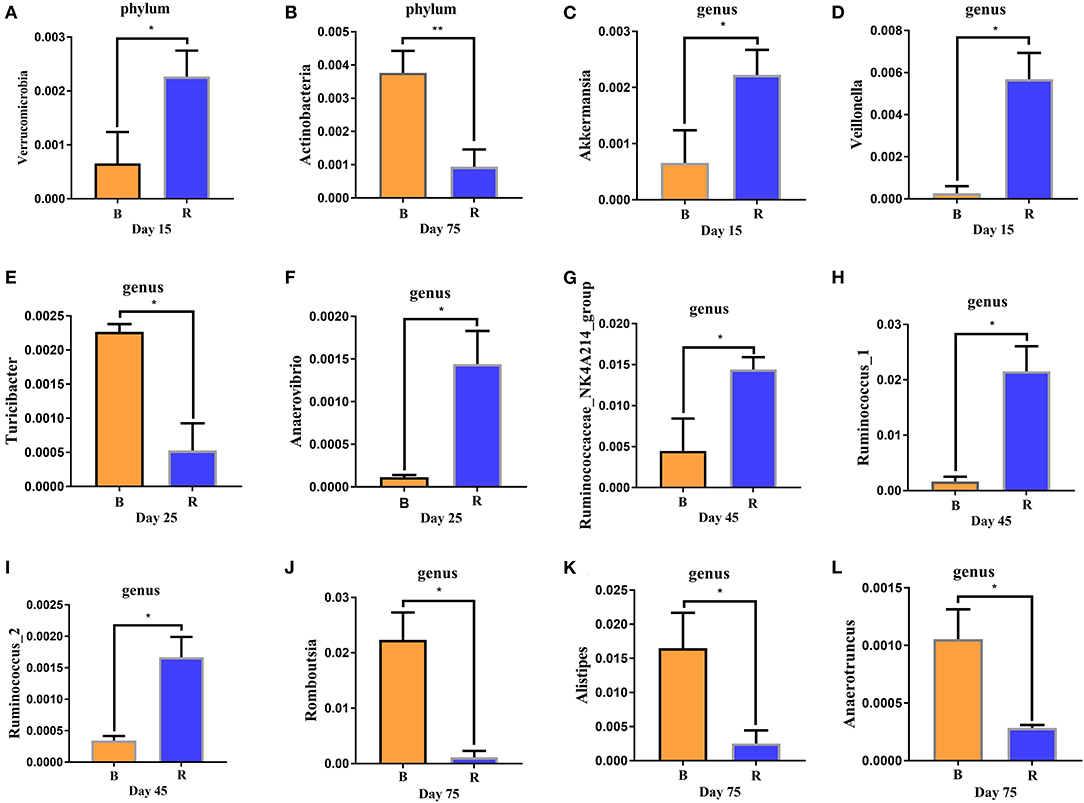
Figure 6. (A–L) Differences in rumen bacteria abundance between the B and R groups. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group. The results were evaluated through one-way ANOVA. All of the data represent means ± SD. *p < 0.05, **p < 0.01.
Currently, milk replacer has been widely used in the livestock industry due to its stable quality and high nutritional value (21, 22). However, it remains to be determined whether milk replacer supplementation has protective and ameliorative roles in the growth performance and rumen microbiota of YBGs. In this study, we investigated the effect of milk replacer supplementation on the growth performance and rumen microbiota of YBGs on different growth days. The results indicated that milk replacer supplementation improved the growth performance of YBGs, whereas the rumen microbiota abundance and diversity between experimental groups were not different.
The adaptation of animals to feed replacement is a gradual process, especially in juvenile ruminants with an immature gastrointestinal tract (23). Therefore, the abrupt change of feed from udder milk to milk replacer can result in a severe stress response, which is also considered as an important factor restricting the growth of young animals (24). In this study, the stress response may be one of the reasons for the slow growth of YBGs in the R group from day 1 to 15. The active substances and nutrients in udder milk were easily affected by multiple external factors, which may hinder the growth and development of goats (25). Conversely, high-quality milk replacer provides comprehensive nutrients to goat growth to compensate for the deficiency of udder milk (26, 27). We observed that milk replacer supplementation gradually improved the growth performance of YBGs, which was consistent with the results of numerous previous studies (23, 28). Moreover, milk replacer can reduce the stress response caused by feed alterations, which contribute to the overall health of the body and rumen development.
Generally, Proteobacteria, Bacteroidetes, and Firmicutes were the most predominant phyla in the ruminants and the content of each phylum was dynamically diverse and influenced by many factors including feeding mode, animals' species, and feed (29, 30). Our results indicated that Proteobacteria, Bacteroidetes, and Firmicutes were the three most dominant phyla in the rumen microbiota of YBGs, which were consistent with previous observations in bovine, sheep, and yak (31, 32). Remarkably, the Proteobacteria level of the R group displayed a downward trend as compared to the B group. It is well-known that Proteobacteria is one of the largest phyla, that comprises gram-negative pathogenic bacteria including Helicobacter pylori, Escherichia coli, Salmonella, and Vibrio cholerae (33, 34). The higher abundance of Proteobacteria in the rumen microbiota may induce an immune response and increase the risk of pathogen infection in the host. Previous studies have suggested that the synergy between Actinobacteria and one partner or host can easily translate into pathogenic interactions with another (35). Furthermore, (36) observed that the abundance of Actinobacteria in the rumen of dairy cattle with subacute ruminal acidosis was significantly increased. Compared with nutritionally stable milk replacer, milk is easily influenced by the feeding environment and nutritional condition, which causes the decline of the quality and safety of the milk (26). Importantly, poor quality milk may alter the rumen microbiota in a negative way (26). In the current study, the R group had lower Proteobacteria and Actinobacteria content as compared to the B group, which signified that milk replacer supplementation may decrease the risk of disease and contribute to growth and rumen microbiota balance of early-weaned goats.
At the genus level, Lactobacillus was the predominant genera in the rumen of the R group on day 10, which differs from the B group. Previous research indicated that Lactobacillus plays an important role in maintaining rumen microbiota and improving the digestive power of rumen (37–40). Additionally, Lactobacillus can also inhibit the proliferation of pathogenic bacteria in the rumen by producing organic acid and antimicrobial peptides (39, 40). All the goats remained healthy, with no sign of acidosis, in the context of Lactobacillus increase. Compared with the B group, the R group had a higher Lactobacillus level, which may be one of the reasons for the rapid growth of YBGs in the R group. Remarkably, the percentage of Akkermansia, Anaerovibrio, Veillonella, Ruminococcus, and Ruminococcaceae in the R group was significantly increased, while the ratio of Turicibacter was obviously decreased compared with the B group. Ruminococcus mostly inhabits the rumen and hindgut of cud-chewing animals and contributes to the degradation of cellulose and starch (41). Ruminococcus can produce acetic acid, formic acid, and a small amount of lactic acid (42). Previous research has indicated that short-chain fatty acid including formic acid, acetic acid play a critical role in regulating the balance of gut microbiota and maintaining morphology and function of intestinal epithelial cells (43, 44). Moreover, the short-chain fatty acid regulates energy intake by the brain-gut axis (45). Lactic acid can improve digestive enzyme activity and exert bacteriostatic effects by regulating rumen pH (39). Furthermore, lactic acid supplementation in feed not only improves the growth performance of juvenile animals but also decreases the occurrence rate of gastrointestinal bacterial diseases (46–48). Akkermansia helps to maintain digestive tract health and reduce the risk of obesity, diabetes, and inflammation (49, 50). Anaerovibrio participates in the breakdown of fats and sugars and produces propionic acid, acetic acid, and succinic acid (51). Veillonella enhances the respiratory and digestive system immunity of the host and decreases the incidence of tooth decay (52, 53). Our results conveyed important information that milk replacer supplementation improved the rumen microbiota by increasing the ratio of beneficial and pathogenic bacteria.
In conclusion, the current study investigated the effect of milk replacer on the growth performance and rumen microbiota of early-weaning YBGs. The results indicated that milk replacer supplementation can improve the growth performance and rumen microbiota of YBGs. However, this study has some limitations such as incontrollable variables including individual variability caused by eating behavior and sample collection. However, our study revealed that milk replacer supplementation may have a beneficial role in improving growth performance and rumen microbiota during dysbacteriosis and weaning stress.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA637829.
The animal study was reviewed and approved by the Ethics Committee of the Huazhong Agricultural University.
ZH, YL, and SL conceived and designed the experiments. AL, LP, KL, TJ, FL, and ZW contributed sample collection and reagents preparation. ZH, AL, and LP analyzed the data. ZH wrote the manuscript. ZH, AL, KL, and LP revised the manuscript. YL and SL provided resources. All authors reviewed the manuscript.
This work was supported by the Shandong Province Education Department Animal Behavior Welfare and Healthy Breeding Technology Skills Innovation Team Project and the Shandong Province Agriculture Major Applied Technology Innovation Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.572064/full#supplementary-material
Supplementary Figure 1. Feasibility analysis of different samples. Each curve represents a sample. The rarefaction curves (A,B) were used to evaluate the adequacy of sequencing for each sample and the Rank abundance curve (C) was used to assess the abundance and evenness of samples. B represents the YBGs in the control group, while R indicates the YBGs in the milk replacer supplementation group.
1. Li Y, Jin T, Han Z, Xia C, Chao H, Zhang N, et al. Effects of the milk replacer on the development of intestine, the flora diversity and the relative expression of glucose transporter gene of early weaned yimeng black goat lambs. Sci Agric Sin. (2018) 51:2193–205. doi: 10.3864/j.issn.0578-1752.2018.11.016
2. Zhang Y, Wang K, Liu J, Zhu H, Qu L, Chen H, et al. CSN1S1An 11-bp indel polymorphism within the gene is associated with milk performance and body measurement traits in Chinese goats. Animals. (2019) 9:114. doi: 10.3390/ani9121114
3. Zhang T, Mu Y, Zhang D, Lin X, Wang Z, Hou Q, et al. Determination of microbiological characteristics in the digestive tract of different ruminant species. Microbiol Open. (2019) 8:e00769. doi: 10.1002/mbo3.769
4. Hu W, Hill T, Dennis T, Suarez-Mena F, Aragona K, Quigley J, et al. Effects of milk replacer feeding rates on growth performance of Holstein dairy calves to 4 months of age, evaluated via a meta-analytical approach. J Dairy Sci. (2020) 103:2217–32. doi: 10.3168/jds.2019-17206
5. Li C, Wang W, Liu T, Zhang Q, Wang G, Li F, et al. Effect of early weaning on the intestinal microbiota and expression of genes related to barrier function in lambs. Front Microbiol. (2018) 9:1431. doi: 10.3389/fmicb.2018.01431
6. Kenyon P, Blair H, Jenkinson C, Morris S, Mackenzie D, Peterson S, et al. The effect of ewe size and nutritional regimen beginning in early pregnancy on ewe and lamb performance to weaning. New Zeal J Agr Res. (2009) 52:203–12. doi: 10.1080/00288230909510505
7. Greenwood P, Cafe L. Prenatal and pre-weaning growth and nutrition of cattle: long-term consequences for beef production. Animal. (2007) 1:1283–96. doi: 10.1017/S175173110700050X
8. Tang W, Chen D, Yu B, He J, Huang Z, Zheng P, et al. Capsulized faecal microbiota transplantation ameliorates post-weaning diarrhoea by modulating the gut microbiota in piglets. Vet Res. (2020) 51:55. doi: 10.1186/s13567-020-00779-9
9. Carroll J, Arthington J, Chase C. Early weaning alters the acute-phase reaction to an endotoxin challenge in beef calves. J Anim Sci. (2009) 87:4167–72. doi: 10.2527/jas.2009-2016
10. Fusani L, Day L, Canoine V, Reinemann D, Hernandez E, Schlinger B. Androgen and the elaborate courtship behavior of a tropical lekking bird. Horm Behav. (2007) 51:62–8. doi: 10.1016/j.yhbeh.2006.08.005
11. Bhatt RS, Tripathi MK, Verma DL, Karim SA. Effect of different feeding regimes on pre-weaning growth rumen fermentation and its influence on post-weaning performance of lambs. J Anim Physiol Anim Nutr. (2009) 93:568–76. doi: 10.1111/j.1439-0396.2008.00845.x
12. Chapman C, Erickson P, Quigley J, Hill T, Bateman H, Suarez-Mena F, et al. Effect of milk replacer program on calf performance and digestion of nutrients with age of the dairy calf. J Dairy Sci. (2016) 99:2740–7. doi: 10.3168/jds.2015-10372
13. Santos A, Giráldez F, Valdés C, Trevisi E, Lucini L, Frutos J, et al. Milk replacer restriction during early life impairs the live body weight and progesterone patterns of ewe lambs during the replacement period. J Dairy Sci. (2018) 101:8021–31. doi: 10.3168/jds.2018-14648
14. Emsen E, Yaprak M, Bilgin O, Ockermanb H. Growth performance of Awassi lambs fed calf milk replacer. Small Ruminant Res. (2004) 53:99–102. doi: 10.1016/j.smallrumres.2003.08.018
15. Zhang R, Zhang W, Bi Y, Tu Y, Beckers Y, Du H, et al. Early feeding regime of waste milk, milk, and milk replacer for calves has different effects on rumen fermentation and the bacterial community. Animals. (2019) 9:443. doi: 10.3390/ani9070443
16. Henderson G, Cox F, Ganesh S, Jonker A, Young W, Global R, et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. (2015) 5:14567. doi: 10.1038/srep14567
17. Wang Y, Zhang H, Zhu L, Xu Y, Liu N, Sun X, et al. Dynamic distribution of gut microbiota in goats at different ages and health states. Front Microbiol. (2018) 9:2509. doi: 10.3389/fmicb.2018.02509
19. Chao A, Yang K. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika. (1993) 80:193–201. doi: 10.1093/biomet/80.1.193
20. Liu Z, Li A, Wang Y, Iqbal M, Zheng A, Zhao M, et al. Comparative analysis of microbial community structure between healthy and Aeromonas veronii-infected Yangtze finless porpoise. Microb Cell Fact. (2020) 19:123. doi: 10.1186/s12934-020-01383-4
21. Liermann W, Schäff C, Gruse J, Derno M, Weitzel J, Kanitz E, et al. Effects of colostrum instead of formula feeding for the first 2 days postnatum on whole-body energy metabolism and its endocrine control in neonatal calves. J Dairy Sci. (2020) 103:3577–98. doi: 10.3168/jds.2019-17708
22. Monaco M, Kim DA, Gurung R, Donovan S. Evaluation of 6'-sialyllactose sodium salt supplementation to formula on growth and clinical parameters in neonatal piglets. Nutrients. (2020) 12:1030. doi: 10.3390/nu12041030
23. Sevi A, Caroprese M, Annicchiarico G, Albenzio M, Taibi L, Muscio A. The effect of a gradual separation from the mother on later behavioral.; immune and endocrine alterations in artificially reared lambs. Appl Anim Behav Sci. (2003) 83:41–53. doi: 10.1016/S0168-1591(03)00088-1
24. Zhang H, Chen Y, Chen Y, Ji S, Jia P, Li Y, et al. Comparison of the protective effects of resveratrol and pterostilbene against intestinal damage and redox imbalance in weanling piglets. J Anim Sci Biotechnol. (2020) 11:52. doi: 10.1186/s40104-020-00460-3
25. Hill T, Quigley J, Bateman H, Suarez-Mena F, Dennis T, Schlotterbeck R. Effect of milk replacer program on calf performance and digestion of nutrients in dairy calves to 4 months of age. J Dairy Sci. (2016) 99:8103–10. doi: 10.3168/jds.2016-11239
26. Toral P, Chilliard Y, Rouel J, Leskinen H, Shingfield K, Bemard L. Comparison of the nutritional regulation of milk fat secretion and composition in cows and goats. J Dairy Sci. (2015) 98:7277–97. doi: 10.3168/jds.2015-9649
27. Liu X, Liu F, Yan T, Chang S, Wanapat M, Hou F. Cistanche deserticola addition improves growth.; digestibility, and metabolism of sheep fed on fresh forage from Alfalfa/Tall fescue pasture. Animals. (2020) 10:668. doi: 10.3390/ani10040668
28. Napolitano F, Cifuni G, Pacelli C, Riviezzi AM, Girolami A. Effect of artificial rearing on lamb welfare and meat quality. Meat Sci. (2020) 60:307–15. doi: 10.1016/S0309-1740(01)00140-1
29. Dibaise J, Zhang H, Crowell M, Krajmalnik-Brown R, Decker G, Rittmann B. Gut microbiota and its possible relationship with obesity. Mayo Clin Proc. (2008) 83:460–9. doi: 10.4065/83.4.460
30. Wang W, Li C, Li F, Wang X, Zhang X, Liu T, et al. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. Sci Rep. (2016) 6:32479. doi: 10.1038/srep32479
31. Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. (2013) 7:1069–79. doi: 10.1038/ismej.2013.2
32. Xin J, Chai Z, Zhang C, Zhang Q, Zhu Y, Cao H, et al. Comparing the microbial community in four stomach of dairy cattle, yellow cattle and three yak herds in Qinghai-Tibetan Plateau. Front Microbiol. (2019) 10:1547. doi: 10.3389/fmicb.2019.01547
33. Yang H, Xiao Y, Gui G, Li J, Wang J, Li D. Microbial community and short-chain fatty acid profile in gastrointestinal tract of goose. Poult Sci. (2018) 97:1420–8. doi: 10.3382/ps/pex438
34. Huang Z, Kraus V. Does lipopolysaccharide-mediated inflammation have a role in OA?. Nat Rev Rheumatol. (2016) 12:123–9. doi: 10.1038/nrrheum.2015.158
35. Miao V, Davies J. Actinobacteria: the good.; the bad.; and the ugly. Antonie Van Leeuwenhoek. (2010) 98:143–50. doi: 10.1007/s10482-010-9440-6
36. Mao SY, Zhang RY, Wang DS, Zhu WY. (2013). Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 24, 12–9. doi: 10.1016/j.anaerobe.2013.08.003
37. Jeffrey M, MacPherson C, Mathieu O, Tompkins T, Green-Johnson J. Lactobacillus rhamnosus secretome-mediated interactions with intestinal epithelial cells: a role for secretome components from R0011 in the attenuation of serovar Typhimurium secretome and TNF-α-induced proinflammatory responses. J Immunol. (2020) 204:2523–34. doi: 10.4049/jimmunol.1901440
38. Shimazu T, Villena J, Tohno M, Fujie H, Hosoya S, Shimosato T, et al. Immunobiotic Lactobacillus jensenii elicits anti-inflammatory activity in porcine intestinal epithelial cells by modulating negative regulators of the Tolllike receptor signaling pathway. Infect Inmmunity. (2012) 80:276–88. doi: 10.1128/IAI.05729-11
39. Wang Y, Li A, Jiang X, Zhang H, Mehmood K, Zhang L, et al. Leuconostoc pseudomesenteroides probiotic potential of and strains isolated from yaks. Front Microbiol. (2018) 9:2987. doi: 10.3389/fmicb.2018.02987
40. Li N, Wang Q, Wang Y, Sun A, Lin Y, Jin Y, et al. Oral probiotics ameliorate the behavioral defcits induced by chronic mild stress in mice via the gut microbiota-infammation axis. Front Behav Neurosci. (2018) 12:266. doi: 10.3389/fnbeh.2018.00266
41. Zhao J, Yao Y, Li D, Xu H, Wu J, Wen A, et al. Characterization of the gut microbiota in six geographical populations of Chinese Rhesus macaques (Macaca mulatta); implying an adaptation to high-altitude environment. Microb Ecol. (2018) 76:565–77. doi: 10.1007/s00248-018-1146-8
42. Berg Miller M, Antonopoulos D, Rincon M, Band M, Bari A, Akraiko T, et al. Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS ONE. (2009) 4:e6650. doi: 10.1371/journal.pone.0006650
43. Tan J, Mckenzie C, Potamitis M, Thorburn A, Mackay C, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. (2014) 121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
44. Li K, Nader SM, Zhang X, Ray BC, Kim C, Das A, et al. Novel lactate dehydrogenase inhibitors with in vivo efficacy against Cryptosporidium parvum. PLoS Pathog. (2019) 15:e1007953. doi: 10.1371/journal.ppat.1007953
45. Samuel B, Shaito A, Motoike T, Rey F, Backhed F, Manchester J, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor.; Gpr41. Proc Natl Acad Sci USA. (2008) 105:16767–72. doi: 10.1073/pnas.0808567105
46. Wang Y, Li A, Zhang L, Waqas M, Mehmood K, Iqbal M, et al. Probiotic potential of Lactobacillus on the intestinal microflora against Escherichia coli induced mice model through high-throughput sequencing. Microb Pathog. (2019) 137:103760. doi: 10.1016/j.micpath.2019.103760
47. Durso L, Wells JE, Kim MS. Diversity of Microbiomes in Beef Cattle. Boston, MA: Springer (2015). p. 129–38. doi: 10.1007/978-1-4614-6418-1_730-4
48. Li A, Wang Y, Li Z, Qamar H, Mehmood K, Zhang L, et al. Probiotics isolated from yaks improves the growth performance, antioxidant activity, and cytokines related to immunity and inflammation in mice. Microb Cell Fact. (2019) 18:112. doi: 10.1186/s12934-019-1161-6
49. Liu C, Li X, Chen Y, Cheng Z, Duan Q. Age-related response of rumen microbiota to mineral salt and effects of their interactions on enteric methane emissions in cattle. Microb Ecol. (2017) 73:590–601. doi: 10.1007/s00248-016-0888-4
50. Derrien M, Belzer C, Vos W. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. (2016) 9:171–81. doi: 10.1016/j.micpath.2016.02.005
51. Ouattara A, Traore A, Garcia J. Characterization of Anaerovibrio burkinabensis sp. nov a lactate fermenting bacterium isolated from rice field soils. Int J Syst Bacteriol. (1992) 42:390–7. doi: 10.1099/00207713-42-3-390
52. Johns AT. The mechanism of propionic acid formation by Veillonella gazogenes. J Gen Microbiol. (1951) 5:326–36. doi: 10.1099/00221287-5-2-326
Keywords: Yimeng black goats, milk replacer, growth performance, rumen microbiota, early weaning
Citation: Han Z, Li A, Pei L, Li K, Jin T, Li F, Wang Z, Lv S and Li Y (2020) Milk Replacer Supplementation Ameliorates Growth Performance and Rumen Microbiota of Early-Weaning Yimeng Black Goats. Front. Vet. Sci. 7:572064. doi: 10.3389/fvets.2020.572064
Received: 12 June 2020; Accepted: 14 August 2020;
Published: 03 November 2020.
Edited by:
Vincenzo Tufarelli, University of Bari Aldo Moro, ItalyReviewed by:
Maria Giovanna Ciliberti, University of Foggia, ItalyCopyright © 2020 Han, Li, Pei, Li, Jin, Li, Wang, Lv and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenjin Lv, bHZzaGVuamluQGx5dS5lZHUuY24=; Yongzhu Li, bGl5b25nemh1QGx5dS5lZHUuY24=; Aoyun Li, YW95dW5saUBzaW5hLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.