- 1College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 2Jiangsu Co-Innovation Center for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
- 3Jiangsu Research Centre of Engineering and Technology for Prevention and Control of Poultry Disease, Yangzhou, China
- 4Joint Laboratory Safety of International Cooperation of Agriculture and Agricultural-Products, Yangzhou University, Yangzhou, China
The role of alternative sigma factor RpoS in regulating biofilm formation may differ in various Salmonella Pullorum strains. In this study, the biofilm-forming ability of two Salmonella Pullorum strains S6702 and S11923-3 were compared. The biofilm forming ability of S11923-3 was much stronger than that of S6702. After knocking out the rpoS gene, S11923-3ΔrpoS had significantly reduced biofilm while S6702ΔrpoS demonstrated similar biofilm compared with each parent strain. The analysis of RpoS sequences indicated two amino acid substitutions (L193P and R293C) between S6702 and S11923-3 RpoS. A complementation study confirmed that the expression of S11923-3 RpoS rather than S6702 RpoS could restore the biofilm-forming ability of ΔrpoS strains and the L193P mutation contributed to the restoration of the biofilm-forming ability. Further study indicated that RpoS with the L193P mutant had significantly improved expression level and binding activity to RNAP and csgD gene promoter, which increased the efficacy of the csgD gene promoter and biofilm-forming ability. Therefore, the L193P mutation of RpoS is critical for stronger biofilm formation of Salmonella Pullorum.
Introduction
Bacteria have evolved various cellular stress responses to survive in different environments. Using alternative sigma (σ) subunits of RNA polymerase (RNAP) that consists of five principal subunits (β, β', ω, and two α subunits) to directly initiate transcription of different classes of promoters is a major strategy employed by bacteria to modify the expression of genes (1, 2). The σS/38 (RpoS), as an alternative sigma factor, is a central regulator enabling many Gram-negative bacteria to adapt to stress conditions and specialized environments (3, 4). RpoS can also play important roles in biofilm formation by regulating the central regulator CsgD (5).
Curli fimbriae and cellulose are two important components of biofilms. As a central regulator in the Salmonella biofilm-forming pathway, CsgD controlled by RpoS regulates the expression of the curli fimbriae csgBAC operon and putative transmembrane protein AdrA. AdrA is involved in regulating the production of cellulose by activating basABZC-bcsEFG operons (5, 6).
Pullorum disease caused by Salmonella Pullorum is a vertical or horizontal transmission disease that causes serious losses in the poultry industry (7). S. Pullorum can form biofilms, and different strains' biofilm-forming abilities vary (8, 9). Bacteria form biofilms to resist antimicrobials (10), host defense (11), desiccation, and disinfectants (12). Many studies showed that biofilms cause the majority of chronic infections and make them difficult to eradicate (13). In a previous study, we found that S. Pullorum strain S6702 could form biofilm, but deletion of the rpoS gene in S6702 had no effects on its biofilm formation (14). In the present study, we identified a stronger rpoS-dependent biofilm producer S. Pullorum strain S11923-3 and compared the role of RpoS in biofilm formation between the two S. Pullorum strains.
Materials and Methods
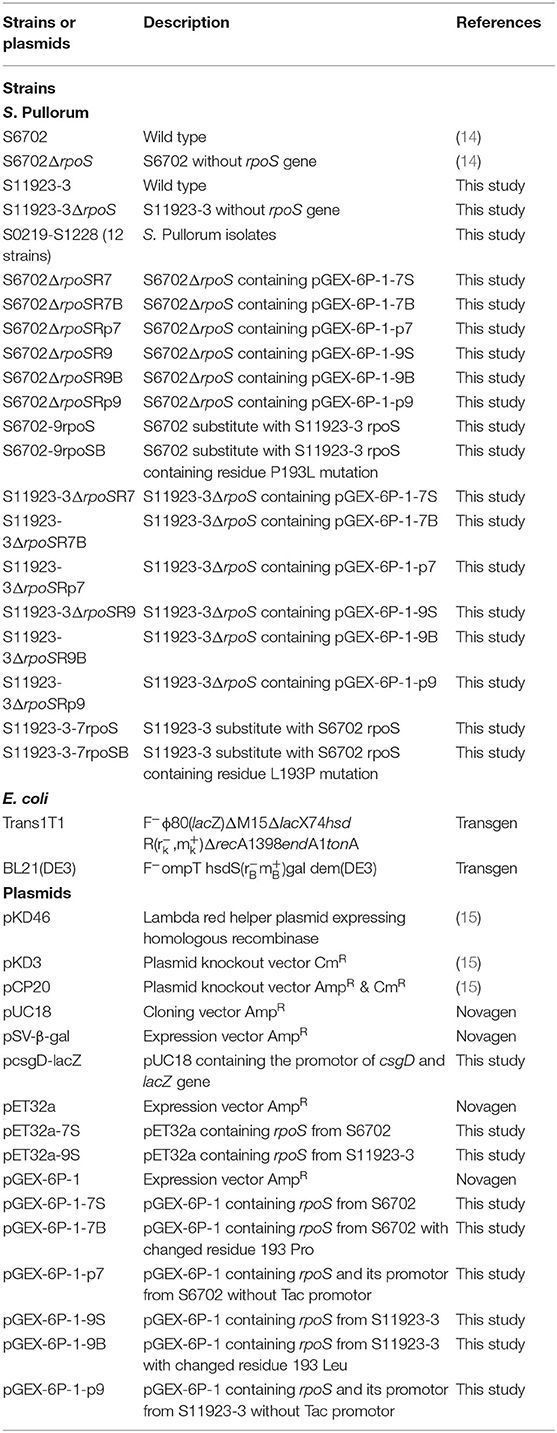
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. All of the mutants were derived from S. Pullorum S6702 (14) and S11923-3 (this study). The strains were routinely cultured in Luria-Bertani (LB, Oxoid) broth and LB agar medium containing 1.5% (w/v) agar with appropriate antibiotics at the following concentrations: 20 μg/mL−1 of chloramphenicol and 100 μg/mL−1 of ampicillin. Tryptic soy broth diluted 1:10 (1/10 TSB, BD) in distilled water was used for the biofilm assays.
Construction of Mutant and Complementary Strains
The rpoS genes of S. Pullorum strains S6702 and S11923-3 were deleted using lambda red-mediated mutagenesis procedures (16). All of the mutants were verified by PCR amplification using the primer pair rpoS-F/R. The PCR products were confirmed by DNA sequencing (TsingKe Biological Technology Company, China).
To construct the complementary strains, the rpoS genes were amplified using primer pair rpoS-HF/HR and chromosomal DNA from the S6702 and S11923-3 strains as templates. The PCR products were digested by restriction endonuclease BamHI and XhoI and cloned into pGEX-6P-1 to obtain pGEX-6P-1-7S and pGEX-6P-1-9S plasmids. The pGEX-6P-1-7B and pGEX-6P-1-9B plasmids with residue 193 mutants in RpoS were constructed using a Mut Express MultiS Fast Mutagenesis Kit V2 (Vazyme) based on the pGEX-6P-1-7S and pGEX-6P-1-9S plasmids. The pGEX-6P-1-p7 and pGEX-6P-1-p9 plasmids containing RpoS promotor without Tac promotor were constructed using a ClonExpress II One Step Cloning Kit (Vazyme) with primers listed in Supplementary Table 1. All of the plasmids (pGEX-6P-1-7S, pGEX-6P-1-7B, pGEX-6P-1-p7, pGEX-6P-1-9S, pGEX-6P-1-9B, and pGEX-6P-1-p9) were transformed into S. Pullorum S6702ΔrpoS and S11923-3ΔrpoS by electroporation to produce complementary strains S6702ΔrpoSR7, S6702 ΔrpoSR7B, S6702ΔrpoSR9, S6702ΔrpoSR9B, S6702ΔrpoSRp7, S6702ΔrpoSRp9, S11923-3ΔrpoSR7, S11923-3ΔrpoSR7B, S11923-3ΔrpoSR9, S11923-3ΔrpoSR9B, S11923-3ΔrpoSRp7, and S11923-3ΔrpoSRp9.
To construct the rpoS substitution strains, the rpoS genes with residues 193/293 substitution were amplified using primer pair rpoS-F1/R1 (Supplementary Table 1), the cat gene was amplified from plasmid pKD3 using primer pair cat-F/R, the rpoS-cat genes were amplified using overlapped rpoS and cat gene PCR products as template and rpoS-cat-F/R as primer pair. The rpoS-cat PCR products with DNA homology to the DNA regions flanking rpoS gene were transformed into S6702 or S11923-3 competent cells carrying lambda red helper plasmid pKD46. Transformants were selected on LB plates containing 20 μg/mL−1 of chloramphenicol. The mutant strains were verified by PCR amplification and DNA sequencing (TsingKe Biological Technology Company, China), and named as S6702-9rpoS, S6702-9rpoSB, S11923-3-7rpoS and S11923-3-7rpoSB (Table 2).
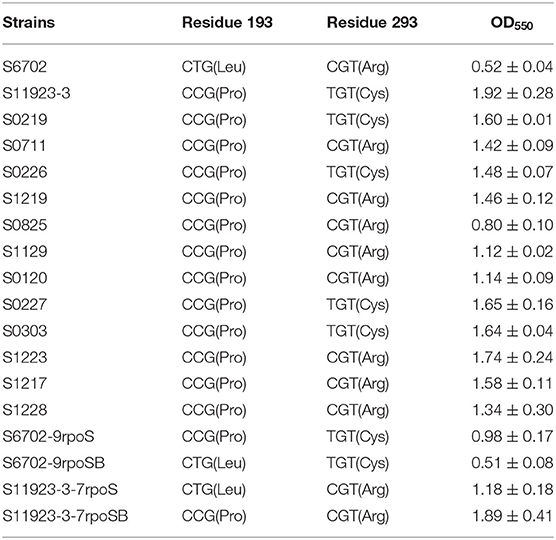
Table 2. Amino acid substitutions of RpoS and crystal violet staining quantification in S. Pullorum strains.
Biofilm Assays
Biofilm formation ability was measured as previously described (14, 17). Overnight broth cultures of each strain were diluted at 1:100 in 1/10 TSB and 100 μL of each bacterial suspension was added to 96-well U-bottomed plates (Corning). The plates were incubated at 28°C for 24 h without shaking. The supernatant was discarded and the wells were gently washed three times with 200 μL of distilled water to remove non-adherent bacteria. A total of 100 μL of 0.4% crystal violet was then added for 20 min. The wells were washed three times with distilled water. The remaining crystal violet bound to the adherent cells was solubilized with 100 μL of 25% acetone with anhydrous ethanol and the optical density (OD550) was measured in Microplate Reader (Bio-Rad). The assays were conducted three times using duplicate wells in each independent assay.
Curli fimbriae and cellulose production was evaluated by colony morphology in Congo red plates with 40 μg/mL of Congo red (Sangon Biotech) and 20 μg/mL of Coomassie brilliant blue (Sangon Biotech) and LB agar plates with 200 μg/mL of calcofluor (Sigma-Aldrich) after the inoculation plates were incubated at 28°C for 4 days (18). The colonies' fluorescence was observed under UV light.
Field Emission Scanning Electron Microscopy
The biofilm morphology was also determined using scanning electron microscopy as previously described (14). Polystyrene coverslips (d = 14 mm) were inoculated with diluted overnight broth cultures and incubated at 28°C for 24 h so biofilm would form on the coverslips. Then the coverslips were washed three times with 0.1 M of phosphate-buffered saline (PBS, pH 7.0) and fixed in 3% glutaraldehyde in PBS for 2 h at 4°C. Afterward, the coverslips were washed three times with PBS and dehydrated with increasing concentrations of ethanol. The samples were dried using critical point-drying for 5 h, coated with gold palladium alloy, and observed with a Gemini 300 SEM (Carl Zeiss).
Quantitative Real-Time PCR (qRT-PCR) Analysis
Bacteria were grown in 1/10 TSB medium at 28°C for 24 h in 60 mm dishes (Corning). The supernatant was discarded and the bacteria accumulated in the biofilms under the dishes were scraped. The total RNA was extracted using a Bacterial RNA Kit (Omega). The cDNA was synthesized using a PrimeScript RT reagent Kit with gDNA Eraser (Takara) and quantified via TB Green Premix Ex Taq (Takara). The gene transcript levels were tested in triplicate for real-time PCR in a Linegene 9600 Plus machine (Bioer). Primer pairs of Q-gyrB-F/R, Q-csgD-F/R, Q-csgA-F/R, and Q-bcsA-F/R (Supplementary Table 1) were used for the mRNA detection of gyrB, csgD, csgA, and bcsA, respectively. The target genes' mRNA levels were normalized to the gyrB mRNA levels (2−ΔΔCt) (19–21).
Immunoblotting Analyses
Bacteria were grown in 1/10 TSB in 60 mm dishes at 28°C for 8 and 24 h to test the protein expression. After the dishes were washed with PBS, the bacterial cells were collected and quantified to OD600 = 1.0, then suspended in SDS sample buffer (Beyotime Biotechnology). The samples were resolved in 12% SDS polyacrylamide gels, transferred to PVDF membranes (Millipore), and analyzed by immunoblotting using anti-RpoS (NeoClone) and anti-RpoA (NeoClone) antibodies. Bands were developed using anti-mouse-HRP (Abcam) and an ECL detection system (Tanon).
Protein Expression and Purification
The rpoS genes were amplified using rpoS-yF/yR as primers and chromosomal DNA from the S6702 and S11923-3 as templates. The PCR products were inserted between the BamHI and XhoI sites of the pET32a, which were called pET32a-7S and pET32a-9S, respectively. The plasmids were transformed to Escherichia coli BL21 (DE3) for the overexpression and purification of recombinant RpoS. Bacterial cells were grown at 37°C until OD600 of 0.4–0.6 and 1 mM of IPTG was added. The cells were then grown at 16°C for 12 h and harvested by centrifugation at 5,000 g for 10 min. His-tagged proteins were purified by incubation with Ni-NTA resin (Genscript). The resin was washed with at least 8 column volumes (CV) of wash buffer (300 mM of NaCl, 50 mM of Na2HPO4 with a pH of 8, and 10 mM of imidazole). The protein was eluted with elution buffer (300 mM of NaCl, 250 mM of Na2HPO4 with a pH of 8, and 250 mM of imidazole) in 1 CV fractions. The fractions were analyzed by SDS-PAGE and the purified RpoS was prepared for binding assays and electrophoretic mobility shift assays (EMSAs).
Binding Assay
RNAP-clarified lysates were prepared via the following steps: 50 mL of each bacterial culture incubated in TSB at 37°C overnight was pelleted at 5,000 g for 10 min. The pellets were resuspended in 8 mL pull-down buffer (140 mM of NaCl, 6.5 mM of sodium-phosphate with a pH of 7.4, and 0.02% Tween-20) and then sonicated on ice for up to 15 min at 1 min intervals. Cell debris was cleared by centrifugation at 14,000 rpm for 30 min at 4°C, and the supernatant was passed through a 0.45 μm filter. The clarified lysate protein concentration was measured by Bradford assays (Thermo Fisher Scientific).
His-tag Dynabeads (50 μL, Invitrogen) were incubated with 700 μL of purified RpoS protein (2.0 mg/mL) and rotated at 25 rpm for at least 20 min at room temperature. The beads were washed according to the manufacturer's protocols with 300 μL wash buffer (600 mM of NaCl, 100 mM of sodium-phosphate with a pH of 8, and 0.02% Tween-20). Then the clarified lysates were incubated with the beads for 15 min and washed four times. The samples were then eluted from the beads by incubation in 100 μL of elution buffer (300 mM of NaCl, 50 mM of sodium-phosphate with a pH of 8, 300 mM of imidazole, and 0.01% Tween-20). The samples were resolved in SDS-PAGE, transferred onto PVDF membranes (Millipore), and probed with primary anti-RpoS, anti-RpoA, and anti-RpoB antibodies (22, 23) and secondary antibody goat anti-mouse-HRP (Abcam). Images were captured using the Tanon Imaging System (Tanon).
Electrophoretic Mobility Shift Assays (EMSAs)
EMSAs were conducted as previously described (24). Briefly, a 646-bp csgD gene promoter including 237 nucleotides in the open reading frame was amplified from S11923-3 and purified with an AxyPrep DNA Gel Extraction Kit (Axygen), and the purified products were PCR labeled using FAM-modified primer. The binding reaction was conducted with non-specific competitor DNA (poly dI-dC, Sigma-Aldrich) in buffer (pH 7.4) containing 750 mM of NaCl, 0.5 mM of DTT, 0.5 mM of EDTA, and 50 mM of Tris at 25°C for 30 min. A total of 20 μL of each reaction system contained 4 μL 5 × binding buffer (Beyotime Biotechnology) and 200 ng of poly dI-dC (Sigma-Aldrich). The final mixtures including DNA fragments and RpoS protein were run on a 6% SDS-PAGE. Then the images were scanned and observed using a fluorescence imaging system (Typhoon FLA 9500, GE Healthcare).
β-Galactosidase Activity Assays
The promoters of csgD genes from strains S6702 and S11923-3 were amplified using primer pair csgD-F/R and the PCR products were inserted into pUC18 with KpnI and BamHI digestion to form pcsgD. The lacZ gene from plasmid pSV-β-gal was amplified using primer pair lacZ-F/R and the PCR products were inserted into pcsgD with BamHI and PstI digestion to form pcsgD-lacZ, in which the lacZ gene was downstream from the csgD gene promoter. The pcsgD-lacZ was transformed into S6702, S11923-3, and their rpoS deletion mutants. The bacteria harboring the plasmids were cultured in dishes at 28°C for 24 and 48 h without shaking. After collecting and measuring the absorbance at 600 nm, the samples were mixed with reaction buffer (60 mM of Na2HPO4, 40 mM of NaH2PO4, 10 mM of KCl, 1 mM of MgCl2, and 0.4 mg/mL of ONPG) and stop buffer (1 M of Na2CO3). The absorbance was measured at 420 nm. The β-galactosidase activity (Miller units) was calculated as previously described (25).
Statistical Analysis
GraphPad Prism 6 was used for graph plotting and statistical analysis. All of the data are expressed as mean and standard deviations (SD). All of the statistical analyses were assessed using the two-tailed t-test. P < 0.05 were considered significant.
Results
RpoS Had Different Effects on Biofilm-Forming Ability in Salmonella Pullorum S6702 and S11923-3
To explore the effects of RpoS on the biofilm formation of S. Pullorum, we constructed rpoS gene deletion mutants S6702ΔrpoS and S11923-3ΔrpoS. The biofilm-forming ability of strains S6702 and S11923-3 and their rpoS deletion mutants were determined. Crystal violet staining of bacterial strains on polystyrene plates showed that wild-type (WT) strain S6702 and its mutant strain S6702ΔrpoS had similar circle staining in the plate well walls (Figure 1A). The WT strain S11923-3 formed spot staining covering the wall and bottom of the plate well, while its mutant strain S11923-3ΔrpoS formed circle staining. After quantifying the crystal violet staining, the OD550 value of S11923-3 was significantly higher than that of S6702. The OD550 value of S11923-3ΔrpoS was significantly lower than that of S11923-3 and similar to that of S6702ΔrpoS, while S6702ΔrpoS had similar OD550 values as parent strain S6702 (Figure 1B).
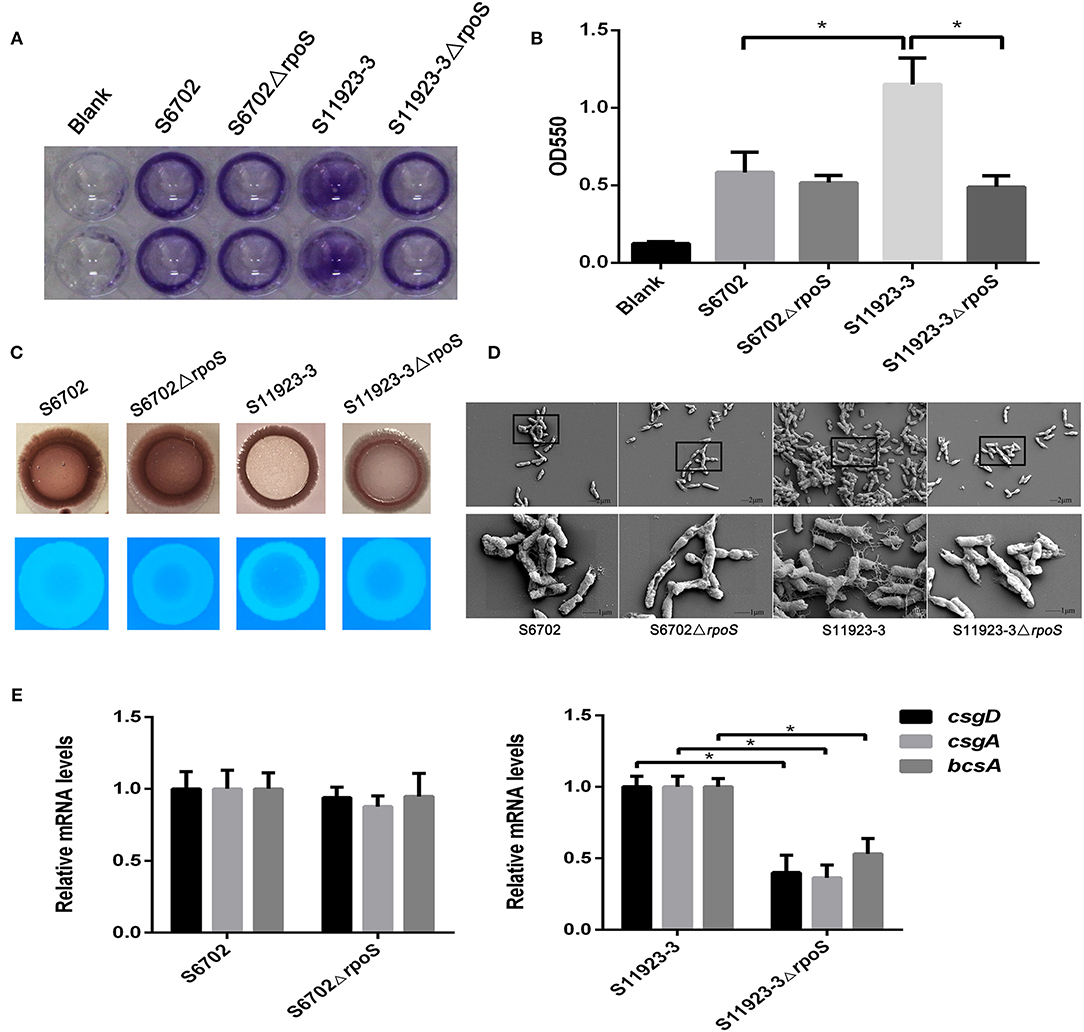
Figure 1. Effect of RpoS on biofilm formation in Salmonella Pullorum S6702 and S11923-3. (A) Crystal violet staining of bacteria grown in 96-well plates. Duplicate wells were used in each independent assay. (B) Quantification of crystal violet staining by measuring the optical density (OD550). Means and standard deviations from three independent experiments are shown. (C) Morphology of colonies after growth on Congo red and calcofluor agar plates. (D) Field emission scanning electron microscope observation of bacteria. The bottom figure (bars = 1 μm) originated from the black boxes in the top figure (bar = 2 μm). (E) The mRNA levels of csgD, csgA, and bcsA genes determined by qRT-PCR. The mRNA level of each gene was normalized by the mRNA level of the gyrB gene. The bars represent the means of three independent assays. *P < 0.05.
Colony morphologies on the Congo red plates showed that WT strain S11923-3 appeared as a red, dry, and rough colony (rdar) after incubation at 28°C for 4 days, while the S11923-3ΔrpoS produced a pink and smooth colony (Figure 1C). However, S6702 and S6702ΔrpoS produced an rdar colony. In calcofluor staining assays, the S11923-3ΔrpoS strain exhibited reduced fluorescence compared with the parent strain, while S6702 and S6702ΔrpoS showed similar fluorescence (Figure 1C).
Field emission scanning electron microscopy analysis of the biofilm formation showed that the WT strain S11923-3 had clusters of bacterial cells and meshwork-like structures surrounding the bacterial surface, while the WT strain S6702 and mutant strains S6702ΔrpoS and S11923-3ΔrpoS had a small number of bacterial cells and tiny fiber structures surrounding the bacterial surface (Figure 1D).
The transcriptional levels of csgD, csgA, and bcsA genes related to biofilm formation were determined by relative qRT-PCR. The results showed that the three genes' transcriptional levels were significantly reduced in S11923-3ΔrpoS compared with the parent strain (Figure 1E). The three genes' transcriptional levels were almost the same in strains S6702 and S6702ΔrpoS.
Two Amino Acid Substitutions (L193P and R293C) Were Identified in the S6702 and S11923-3 RpoS Sequences
To investigate the differences in RpoS in the two strains, we compared the amino acid sequences of RpoS in strains S6702 and S11923-3. Only two amino acid substitutions of L193P and R293C were found in the S6702 and S11923-3 RpoS (Supplementary Figure 1). To detect the distribution of the two residues in S. Pullorum, another 12 S. Pullorum strains isolated from 2015 to 2017 in Jiangsu were randomly selected and their RpoS sequences were analyzed (Table 2). All of the S. Pullorum strains had Pro in residue 193 of the RpoS except for S6702, while the S. Pullorum strains had Arg (9/14) or Cys (5/14) in RpoS residue 293.
To verify effects of residues 193 and 293 of RpoS on biofilm formation, all S. Pullorum strains were subjected to polystyrene plate culture and crystal violet staining (Table 2). The OD550 value of S6702 with 193L and 293R pattern was significantly lower than that of other strains with 193P and 293R/C pattern, while the OD550 values of S. Pullorum strains with 193P and 293R pattern varied from 0.80 to 1.74 and the OD550 values of S. Pullorum strains with 193P and 293C pattern varied from 1.48 to 1.92, indicating that residue 193 of RpoS might make more contribution to biofilm formation.
RpoS Residue 193 Was Critical for Biofilm Formation in S. Pullorum
To further ensure the residue effects of RpoS on biofilm formation in S6702 and S11923-3, rpoS complementary strains and substitution strains were constructed and determined by crystal violet staining test using the wild-type strains and rpoS mutant strains as contrast. Crystal violet staining of the biofilm formation in these strains showed that S6702ΔrpoSR7 complemented with S6702 RpoS demonstrated similar staining or OD550 values than S6702ΔrpoS, while S6702ΔrpoSR9 complemented with S11923-3 RpoS had stronger staining or higher OD550 values than S6702ΔrpoS and S6702 (Figure 2A). S11923-3ΔrpoSR7 complemented with S6702 RpoS showed similar staining or OD550 values as S11923-3ΔrpoS, while S11923-3ΔrpoSR9 complemented with S11923-3 RpoS recovered the staining and had higher OD550 values (Figure 2B). These data indicated the efficacy of S11923-3 RpoS in biofilm formation is higher than that of S6702 RpoS.
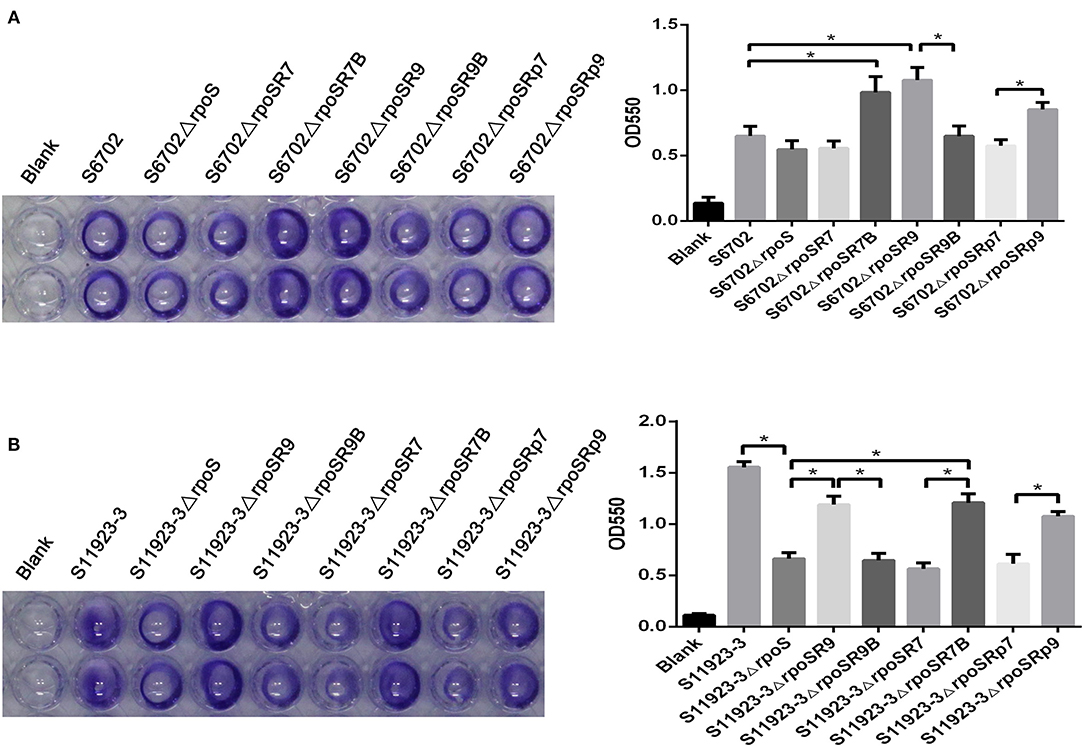
Figure 2. Determination of biofilm formation of S6702ΔrpoS (A) and S11923-3ΔrpoS (B) complemented with two point mutants in residues 193 and 293 of RpoS. All of the strains were cultured in TSB medium first. After overnight cultivation, cultures were diluted in 1/10 TSB and grown in 96-well plates (100 μL/ well) at 28°C for 24 h without shaking. Discarding the supernatant and washing the wells gently with distilled water to remove non-adherent bacteria. After staining with 0.4% crystal violet for 20 min, washed the wells and the remaining crystal violet was solubilized with 100 μL of 25% acetone with anhydrous ethanol. Crystal violet staining quantification was tested by measuring the optical density (OD550). Means and standard deviations from three independent experiments are shown. *P < 0.05.
After exchanging residue 193 between S6702 RpoS and S11923-3 RpoS, the complementary strain S6702ΔrpoSR7B demonstrated significantly stronger staining or higher OD550 values than the mutant S6702ΔrpoS strain; in contrast, the complementary S6702ΔrpoSR9B strain had similar staining or OD550 values as the mutant strain S6702ΔrpoS (Figure 2A). The same result was also observed in S11923-3 and its related strains (Figure 2B).
Biofilm formation of the substitution strains were also determined by crystal violet staining test, the OD550 values of S6702-9rpoS (193P, 293C) were significantly increased when compared with that of S6702, while OD550 values of S6702-9rpoSB (193L, 293C) were significantly decreased when compared with that of S6702-9rpoS (Table 2). In contrast, the OD550 values of S11923-3-7rpoS (193L, 293R) were significantly decreased when compared with that of S11923-3, while OD550 values of S11923-3-7rpoSB (193P, 293R) were significantly increased when compared with that of S11923-3-7rpoS. These data indicated that RpoS residue 193 rather than residue 293 facilitated the biofilm-forming ability in these two strains.
Residue L193P Contributed to the RpoS Expression During Bacterial Biofilm Formation
To determine the RpoS expression among different S. Pullorum strains, six strains were selected for determination of RpoS expression. Samples were collected after 8 h and 24 h of incubation and subjected to immunoblotting analysis. As shown in Figure 3A, the expression level of RpoS in strains S6702 with residues 193L and 293R pattern was the lowest at each time point, the expression levels of RpoS in strains S1129 and S0120 with residues 193P and 293R pattern were moderate and that of RpoS in strains S11923-3, S0227, and S0303 with residues 193P and 293C pattern were the highest, indicating that both residues 193 and 293 contribute to RpoS expression in S. Pullorum.
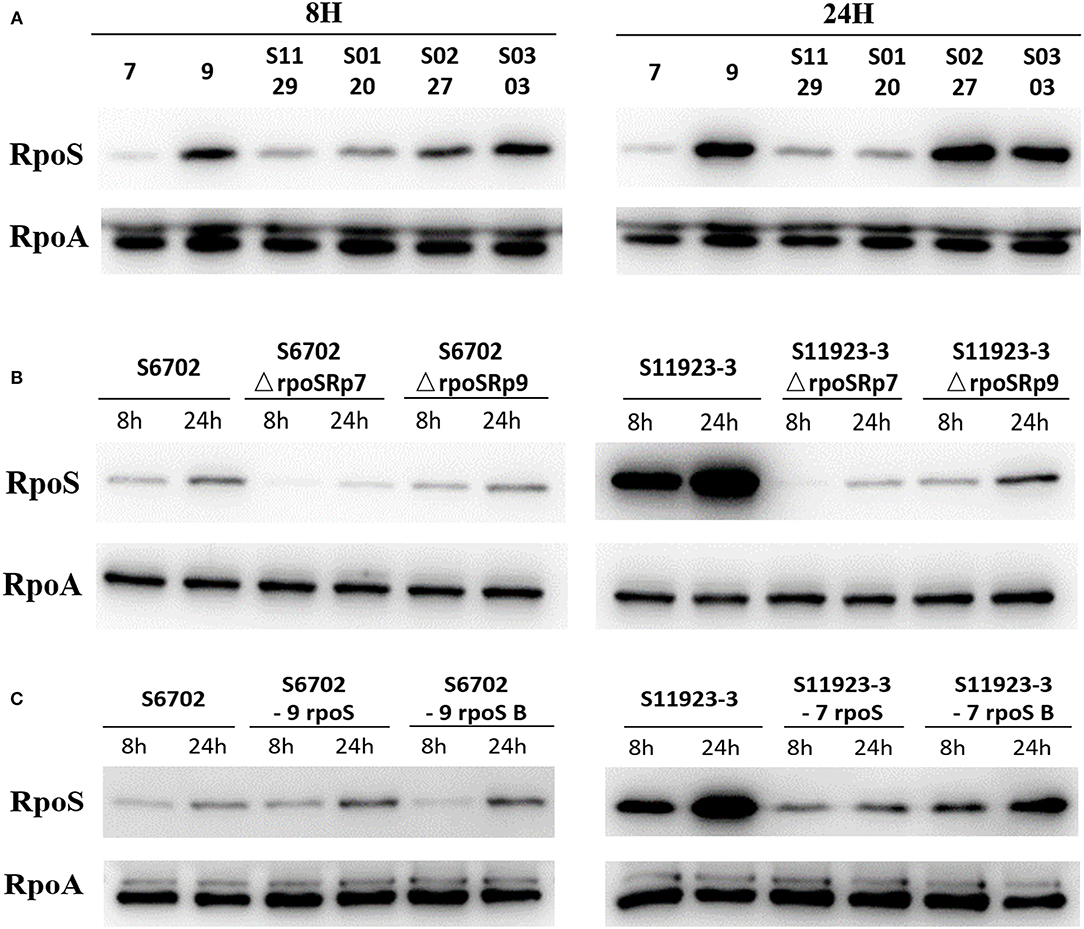
Figure 3. Determination of RpoS expression by immunoblotting analysis in different S. Pullorum isolates (A), rpoS gene deletion mutants and complementary strains (B), and ropS gene substitution strains (C) during biofilm formation. All of the strains were cultured in TSB medium first. Overnight cultures were diluted in 1/10 TSB medium in small dishes at 28°C without shaking for 8 and 24 h. Supernatant was discarded and the scraped samples were collected. The samples were quantified to OD600 = 1.0, then suspended in SDS sample buffer (Beyotime Biotechnology). The samples were resolved in 12% SDS polyacrylamide gels, transferred to PVDF membranes (Millipore), and analyzed by immunoblotting using anti-RpoS (NeoClone) and anti-RpoA (NeoClone) antibodies.
To test the RpoS expression level under the same conditions and eliminate the effects of plasmid promoter on the rpoS gene expression, the pGEX-6p-1-p7 and pGEX-6p-1-p9 plasmids with the substitution of Tac promoter by RpoS promoter of S6702 and S11923-3 were constructed and then transformed into S6702ΔrpoS and S11923-3ΔrpoS. The immunoblotting results showed that complement of S11923-3 RpoS had higher expression level than that of S6702 RpoS either in the mutant strains S6702ΔrpoS or S11923-3ΔrpoS (Figure 3B), indicating that residue L193P and R293C mutations could increase the RpoS expression significantly.
To verify the role of residues 193 and 293 on RpoS expression level, four rpoS gene substitution strains S6702-9rpoS, S6702-9rpoSB, S11923-3-7rpoS, and S11923-3-7rpoSB were constructed, and their RpoS expression levels were determined. As shown in Figure 3C, the RpoS expression level in S6702-9rpoS (193P, 293C) was higher than that in S6702 at each time point, and the RpoS expression level in S6702-9rpoSB (193L, 293C) was lower than that in S6702-9rpoS at 8 h incubation. In contract, the RpoS expression level in S11923-3-7rpoS (193L, 293R) was significantly lower than that in S11923-3 at each time point, and the RpoS expression level in S11923-3-7rpoSB (193P, 293R) was significantly higher than that in S11923-3-7rpoS. These data indicated that residue L193P increased the RpoS expression level.
RpoS From the S11923-3 Strain Had Higher Activity to Initiate Biofilm Formation
To determine affinity of RpoS to RNAP, a pull-down assay with purified His-tagged RpoS (His-RpoS) and clarified lysates from mutant strains lacking RpoS was conducted. The RNAP pulled down by His-RpoS from the lysate was confirmed by immunoblotting analyses with anti-RpoS, anti-RpoA, and anti-RpoB antibodies (Figure 4A). When the RpoS was adjusted at the same concentration, the band intensities of RpoA and RpoB bound to S11923-3 RpoS were stronger than that bound to S6702 RpoS, indicating that S11923-3 RpoS had stronger binding capacity to RNAP.
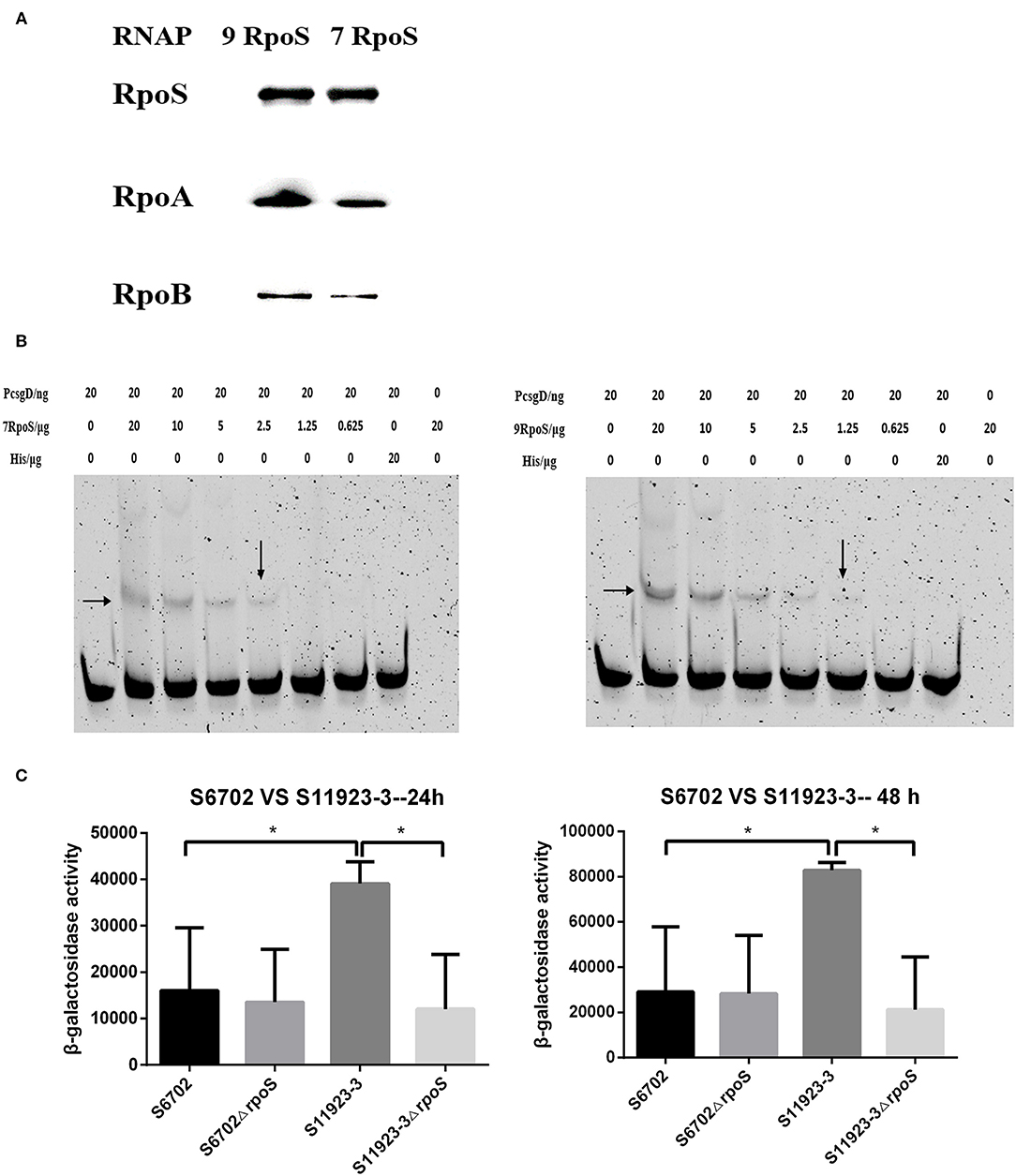
Figure 4. Determination of the binding ability of RpoS to RNAP and csgD promoter. (A) Immunoblotting analyses for binding activity of RpoS to RNAP. The purified RpoS protein (2.0 mg/mL) were incubated with his-tag Dynabeads (50 μL, Invitrogen). The beads were incubated with the clarified lysates including RNAP for 15 min. After being washed, the samples were resolved in SDS-PAGE, transferred onto PVDF membranes (Millipore), and probed with primary anti-RpoS, anti-RpoA, and anti-RpoB antibodies. (B) EMSA assays. The csgD promoter genes were PCR labeled using FAM-modified primer. The labeled DNA fragments and purified RpoS protein were incubated in the reaction system contained 4 μL 5 × binding buffer (Beyotime Biotechnology) and 200 ng of poly dI-dC (Sigma-Aldrich). The final mixtures were run on a 6% SDS-PAGE. The images were scanned and observed using a fluorescence imaging system (Typhoon FLA 9500, GE Healthcare). The data showed one representative experiment of three independent assays. (C) Determination of β-galactosidase activity. Plasmid pcsgD-lacZ was transformed into S6702, S11923-3, and their rpoS deletion mutants. The transformants were cultured in dishes at 28°C for 24 h and 48 h without shaking. After measuring the absorbance at 600 nm, the samples were mixed with reaction buffer and stop buffer. The absorbance was measured at 420 nm and the β-galactosidase activity was calculated. The bars represent the means of three independent assays. *P < 0.05.
CsgD plays an important role in regulating biofilm formation of Salmonella and its expression is controlled by RpoS (5). To test the recognition of RpoS to downstream csgD gene promoter, EMSA assays were conducted. As shown in Figure 4B, RpoS bound and shifted with DNA fragments in a dose-dependent manner, but S11923-3 RpoS bound a lower concentration (1.25 μg) of DNA probes than S6702 RpoS (2.5 μg), indicating that S11923-3 RpoS had stronger binding activity to csgD promoter.
To test the role of RpoS in regulating CsgD expression, a report plasmid was constructed. The results showed that β-galactosidase activity in the WT S6702 strain was lower than that in the WT S11923-3 strain after 24 h and 48 h of incubation (Figure 4C). β-galactosidase activity in the mutant S6702ΔrpoS strain was similar as that in S6702. However, β-galactosidase activity in S11923-3ΔrpoS was significantly lower than that in S11923-3 (Figure 4C). These data indicated that S11923-3 RpoS enhanced the expression activity of csgD promoter, and loss of the RpoS resulted in reduced expression activity of csgD promoter.
Discussion
Gram-negative bacteria have many σ factors including RpoD, RpoS, RpoE, RpoH, and RpoN that bind with core RNAP (E) to form holoenzyme Eσ. Eσ enables specific binding to gene promoters and is required for transcription initiation (26). S. Typhimurium can form biofilms that are coordinated by a sophisticated network of signaling pathways through the expression of central biofilm regulator CsgD (5). However, the dependence of CsgD expression on RpoS may be variety. Römling proved that biofilm formation and the expression of central regulator CsgD is dependent on the alternative sigma factor RpoS (6). Further study found that the regulation of CsgD is partially independent of RpoS in S. Enteritidis (27). In a previous study, we found that S. Pullorum strain S6702 could form biofilm independent of sigma factor RpoS (14). In this study, we compared the role of RpoS in biofilm formation between S. Pullorum strains S6702 and S11923-3. And three methods were applied to determine the biofilm-forming ability of S6702, S11923-3, and their rpoS deletion mutants. All data confirmed that the biofilm-forming ability of S11923-3ΔrpoS was significantly reduced compared to that of parent strain S11923-3, while there were no changes in the biofilm formation in S6702ΔrpoS. The qRT-PCR results showed that the transcriptional levels of csgD, csgA, and bcsA genes related to biofilm formation also decreased significantly in strain S11923-3ΔrpoS. These data indicated that the biofilm-forming ability of S11923-3 is dependent on RpoS.
By comparing the RpoS sequences of S6702 and S11923-3, only two nucleotides (T578C and C877T) or two amino acid substitutions (L193P and R293C) were found. Then complementary strains and substitution strains with combinational substitution of residues 193 and 293 were constructed and their biofilm formation abilities were determined by crystal violet staining (Table 2 and Figure 2). The results indicated that residue L193P of RpoS could enhance the biofilm formation in S. Pullorum. It has been shown that accumulation of RpoS coincided with the expression of curli fimbriae and other morphological and physiological changes (28). In this study, S11923-3 RpoS (193P, 293C) had higher expression level than S6702 RpoS (193L, 293R) and residue L193P substitution increased the RpoS expression significantly while residue P193L substitution decreased the RpoS expression significantly (Figure 3C). According to Table 2 and Supplementary Figure 2, the biofilm formation ability was consistent with the RpoS expression in Figure 3C indicating that residue L193P of RpoS could increase the RpoS expression level further to enhance the biofilm formation in S. Pullorum.
Regulation of RpoS synthesis in E. coli is well-studied (29). Multiple levels including rpoS transcription, translation, and protein stability of regulation affect synthesis of RpoS. Transcription regulation is primarily from the rpoS promoter, embedded within the upstream nlpD gene contributing to basal level expression of RpoS in exponential phase (30, 31). 5′-UTR part of rpoS mRNA with relevant secondary structures and sRNA binding regions folds back to occlude ribosome entry and translation. sRNAs (for example, DsrA, ArcZ, RprA, etc.) can open the secondary structure, promoting translation (32–34). Synthesis of anti-adaptors like IraD enhances RssB to deliver RpoS to the clpPX protease for RpoS degradation (35, 36). Our study showed that there was no difference in rpoS gene promoter sequences between strains S6702 and S11923-3 (data not shown) and the transcript level of S11923-3 was higher than that of S6702 (Supplementary Figure 3). The mechanism for enhanced RpoS expression by L193P substitution need to be further studied.
RpoS combines with RNA polymerase (RNAP) to regulate many genes by binding to gene promotors and initiating transcription that enables bacteria to survive adverse conditions (37, 38). Research also showed that missense mutants in RpoS can affect its function including interacting with RNAP in E. coli (STEC) isolates. Substitution of Ile128 with Pro128 in E. coli abolishes RpoS activity including response to oxidative stresses and ability to bind RNAP (3, 39). Our study identified two amino acid substitutions in RpoS (L193P and R293C) and the binding assay results demonstrated that S11923-3 RpoS had stronger affinity for RNAP (Figure 4A). Amino acid substitutions of R141S and A157T in RpoS impair its ability to bind downstream promoters but do not affect its affinity to RNAP (40). Thus, we conducted EMSA assays to verify the promotor-binding ability of RpoS. The EMSA assays showed that RpoS could bind the csgD promoter DNA and S11923-3 RpoS had stronger ability to bind promoter DNA than S6702 RpoS (Figure 4B). In the reporter assay, S11923-3 had higher β-galactosidase activity than S6702, which demonstrated that S11923-3 RpoS induced more CsgD expression than S6702 RpoS (Figure 4C). In addition, when the csgD promoter sequences in these two strains were compared, no difference was found in the two promoters (data not shown), which removed the impact of the csgD promoter. Overall, the results of RpoS activities indicated that S11923-3 RpoS (193P) had stronger effects on biofilm formation in S. Pullorum by enhancing the affinity to RNAP and activating csgD promotor.
The cellular concentration of RpoD molecules exceeds that of core RNAP, suggesting that σ factors compete for binding to a limited number of core RNAP (40, 41). Both RpoD and RpoS can interact with RNA polymerase and regulate csgBA promoter expression in Salmonella (5). RpoE is another sigma factor that can affect biofilm formation in S. Pullorum (42). In other bacteria such as Pseudomonas putida, RpoD can mediate biofilm formation at a low level when RpoS is absent (43). In our study, we found that the biofilm-forming ability of S. Pullorum S6702 was RpoS-independent, which might be because other σ factors maintain a low level of biofilm formation after deleting RpoS. In strain S11923-3, other σ factors cannot initiate high levels of biofilm formation after deleting RpoS, showing a RpoS-dependent biofilm formation characteristic.
In conclusion, we found a new potential amino acid site (residue 193P) in RpoS that can enhance the RpoS expression level, binding activity to RNAP and expression activity of csgD gene promoter, resulting in enhanced biofilm formation in S. Pullorum.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
ZF and DP conceived this study. ZF and MH conducted the research. ZF, TQ, and DP analyzed the data. ZF, YD, SC, and DP wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31572530), the Key R&D Plan of Jiangsu Province (BE2018315), and a project funded by the Priority Academic Development Program of Jiangsu Higher Education Institutions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the testing center at Yangzhou University for assistance with the emission scanning electron microscopy experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.571361/full#supplementary-material
References
1. Cavaliere P, Norel FO. Recent advances in the characterization of Crl, the unconventional activator of the stress sigma factor σS/RpoS. Biomol Concepts. (2016) 7:197–204. doi: 10.1515/bmc-2016-0006
2. Erickson AFW, Deighan P, Garcia CP, Weinzierl RO J, Hochschild A, et al. An amino acid substitution in RNA polymerase that inhibits the utilization of an alternative sigma factor. J Bacteriol.(2017) 199:e00277–17. doi: 10.1128/JB.00277-17
3. Landini P, Egli T, Wolf J, Lacour S. sigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ Microbiol Rep. (2014) 6:1–13. doi: 10.1111/1758-2229.12112
4. Schellhorn, Herb E. Elucidating the function of the RpoS regulon. Future Microbiology. (2014) 9:497–507. doi: 10.2217/fmb.14.9
5. Simm R, Ahmad I, Rhen M, Guyon SL, Römling U. Regulation of biofilm formation in Salmonella Typhimurium. Future Microbiology. (2014) 9:1261–82. doi: 10.2217/fmb.14.88
6. Römling U, Bian Z, Hammar, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. (1998) 180:722–31. doi: 10.1128/JB.180.3.722-731.1998
7. Li Q, Hu Y, Chen J, Liu Z, Han J, Sun L, et al. Identification of Salmonella enterica Serovar Pullorum antigenic determinants expressed in vivo. Infect Immunity. (2013) 81:3119–27. doi: 10.1128/IAI.00145-13
8. Thi NN, Niwat C, Carrique-Mas JJ. Antimicrobial resistance in bacterial poultry pathogens: a review. Fron Vet Sci. (2017) 4:1–17. doi: 10.3389/fvets.2017.00126
9. Guo X, Wang H, Cheng Y, Zhang W, Luo Q, Wen G, et al. Quinolone resistance phenotype and genetic characterization of Salmonella enterica serovar Pullorum isolates in China, during 2011 to 2016. BMC Microbiol. (2018) 18:225. doi: 10.1186/s12866-018-1368-4
10. Hoiby N, Bjarnsholt T, Givskov M, S. r. Molin, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrobial Agents. (2010) 35:322–32. doi: 10.1016/j.ijantimicag.2009.12.011
11. Jensen PØ, Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. Pathogens Dis. (2010) 59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x
12. Römling U, Balsalobre C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J Internal Med. (2012) 272:541–61. doi: 10.1111/joim.12004
13. Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. (2003) 2:114–22. doi: 10.1038/nrd1008
14. Lu Y, Chen S, Dong H, Sun H, Peng D, Liu X. Identification of genes responsible for biofilm formation or virulence in salmonella enterica serovar pullorum. Avian Dis. (2012) 7:e48–e51. doi: 10.1637/10008-980611-DIGEST.1
15. He S, Yuan Z, Zhu [Construction G and characterization of type I fimbriae fimH deletion mutant from avian pathogenic Escherichia coli]. Wei Sheng Wu Xue Bao. (2008) 48:252–6. doi: 10.13343/j.cnki.wsxb.2008.02.020
16. El Hag M, Zheng F, Su Y, Xiao W, Liu X. Contribution of the csgA and bcsA genes to Salmonella enterica Serovar Pullorum Biofilm Formation and Virulence. Avian Pathol. (2017) 46:1–31. doi: 10.1080/03079457.2017.1324198
17. Crawford RW, Gibson DL, Kay WW, Gunn JS. Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect Immun. (2008) 76:5341. doi: 10.1128/IAI.00786-08
18. Anriany Y, Sahu SN, Wessels KR, McCann LM, Joseph SW. Alteration of the Rugose Phenotype in waaG and ddhC Mutants of Salmonella enterica Serovar Typhimurium DT104 Is Associated with Inverse Production of Curli and Cellulose. Appl Environ Microbiol. (2006) 72:5002–12. doi: 10.1128/AEM.02868-05
19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR. Method. (2002) 25:402–8. doi: 10.1006/meth.2001.1262
20. García B, Latasa C, Solano C, PF, García-del, Gamazo C, et al. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol Microbiol. (2004) 54:264–77. doi: 10.1111/j.1365-2958.2004.04269.x
21. Liu H, Xiao Y, Nie H, Huang Q, Chen W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol Res. (2017) 204:1–8. doi: 10.1016/j.micres.2017.07.003
22. Traag BA, Ramirez-Peralta A, Wang Erickson AF, Setlow P, Losick R. A novel RNA polymerase-binding protein controlling genes involved in spore germination in Bacillus subtilis. Mol Microbiol. (2013) 89:113–22. doi: 10.1111/mmi.12262
23. Erickson AFW, Deighan P, Chen S, Barrasso K, Garcia CP, Martínez-Lumbreras S, et al. A Novel RNA polymerase-binding protein that interacts with a sigma-factor docking site. Mol Microbiol. (2017). 105:652–62. doi: 10.1111/mmi.13724
24. Guo Y, Li Y, Zhan W, Wood TK, Wang X. Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS. Microbial Biotechnology. (2018) 12:392–404. doi: 10.1111/1751-7915.13369
25. Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. (2005) 187:2233–43. doi: 10.1128/JB.187.7.2233-2243.2005
26. Kazmierczak MJ, Wiedmann M, Boor KJ. Alternative sigma factors and their roles in bacterial virulence. Microbiol Mol Biol Rev Mmbr. (2005) 69:527–43. doi: 10.1128/MMBR.69.4.527-543.2005
27. Römling U, Bokranz W, Rabsch W, Zogaj X, Nimtz M, Tschäpe Occurrence H, et al. Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. IJMM. (2003) 293:273–85. doi: 10.1078/1438-4221-00268
28. Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Roy C. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol. (2001) 41:349–63. doi: 10.1046/j.1365-2958.2001.02529.x
29. Gottesman S. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem. (2019) 294:11685–700. doi: 10.1074/jbc.REV119.005593
30. Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, protein stability. Genes Dev. (1994) 8:1600–12. doi: 10.1101/gad.8.13.1600
31. Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of therpoSgene of Escherichia coli. (1994) 243:525–31. doi: 10.1007/BF00284200
32. Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. (1996) 15:3993–4000. doi: 10.1002/j.1460-2075.1996.tb00773.x
33. Majdalani N, Chen S, Murrow J, John KS, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol. (2001) 39:1382–94. doi: 10.1111/j.1365-2958.2001.02329.x
34. Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. Embo J. (2010) 29:3094–107. doi: 10.1038/emboj.2010.179
35. Zhou YN, Gottesman S, Hoskins JR, Maurizi MR, Wickner S. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. (2001) 15:627–37. doi: 10.1101/gad.864401
36. Merrikh H, Ferrazzoli AE, Lovett ST. Growth Phase and (p)ppGpp Control of IraD, a Regulator of RpoS Stability, in Escherichia coli. J Bacteriol. (2009) 191:7436–46. doi: 10.1128/JB.00412-09
37. Battesti A, Majdalani N, Gottesman S. The RpoS-Mediated General Stress Response in Escherichia coli *. Ann Rev Microbiol. (2011) 65:189–213. doi: 10.1146/annurev-micro-090110-102946
38. Park M, Nam DD, Kweon H, Shin D. ATP reduction by MgtC and Mg 2+ homeostasis by MgtA and MgtB enables Salmonella to accumulate RpoS upon low cytoplasmic Mg 2+ stress. Mol Microbiol. (2018) 110:283–95. doi: 10.1111/mmi.14105
39. Tadayuki I, Takashi M, Saiko N, Akiko T, Yoshimitsu M. Hydrophobicity of residue 128 of the stress-inducible sigma factor rpos is critical for its activity. Front Microbiol. (2017) 8:656. doi: 10.3389/fmicb.2017.00656
40. Levi-Meyrueis C, Monteil V, Sismeiro O, M.-,Dillies A, Kolb A., Monot M, et al. Repressor activity of the RpoS/σS-dependent RNA polymerase requires DNA binding. Nucleic Acids Res. (2015) 3:3. doi: 10.1093/nar/gku1379
41. Feklístov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, genomic perspective. Ann Rev Microbiol. (2014) 68:357–76. doi: 10.1146/annurev-micro-092412-155737
42. Huang J, Chen S, Huang K, Yang L, Wu B, Peng D. [Identification of rpoE gene associated with biofilm formation of Salmonella pullorum]. Wei sheng wu xue bao = Acta microbiologica Sinica. (2015) 55:156–63. doi: 10.13343/j.cnki.wsxb.20140230
Keywords: Salmonella pullorum, rpoS, biofilm, promoter, mutation
Citation: Feng Z, El Hag M, Qin T, Du Y, Chen S and Peng D (2020) Residue L193P Mutant of RpoS Affects Its Activity During Biofilm Formation in Salmonella Pullorum. Front. Vet. Sci. 7:571361. doi: 10.3389/fvets.2020.571361
Received: 24 June 2020; Accepted: 16 October 2020;
Published: 05 November 2020.
Edited by:
Shengqing Yu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Min Yue, Zhejiang University, ChinaDavid Smith, Heriot-Watt University, United Kingdom
Copyright © 2020 Feng, El Hag, Qin, Du, Chen and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daxin Peng, cGVuZ2R4QHl6dS5lZHUuY24=; Sujuan Chen, Y2hlbnNqQHl6dS5lZHUuY24=
 Zheng Feng1,2,3,4
Zheng Feng1,2,3,4 Yinping Du
Yinping Du Daxin Peng
Daxin Peng