- 1Council for Agricultural Research and Economics (CREA), Research Center for Animal Production and Aquaculture, Monterotondo, Italy
- 2Laboratory of Microbiology, Wageningen University & Research, Wageningen, Netherlands
New approaches are needed to improve the sustainability of feed production and utilization by ruminants. Promising approaches include increased use of buffaloes for more sustainable milk production, and arbuscular mycorrhizal fungi (AMF) to reduce crop production input needs. However, studies assessing the effect of crops grown in the presence of AMF on rumen microbial utilization are limited. Based on current knowledge, we hypothesized that maize grain grown on AMF-inoculated soil affected ruminal fermentation and microbiota, and that this effect differed between buffalo and cattle. A dietary cross-over study (four weeks per diet) was conducted using rumen-cannulated cattle (n = 5) and buffalo (n = 6) to assess the effect of maize grain (3.9% (w/v) of diet) grown on soil with or without AMF (15 kg/ha) on ruminal fermentation and microbiota. Production of maize on AMF-treated soil did not affect any of the assessed ruminal fermentation parameters, microbial concentrations, or prokaryotic community composition (using prokaryotic 16S rRNA gene sequence analysis). In contrast, host type had numerous effects. Protozoal counts, lactate, total VFA and isobutyrate, were significantly higher in buffaloes compared to cattle. Conversely, butyrate was significantly lower in buffaloes than in cattle. Host type explained 9.3% of the total variation in prokaryotic community composition, and relative abundance of nine amplicon sequence variants significantly differed between host types. These findings indicate that AMF treatment of maize crops has no detrimental impact on the value of the resulting maize grains as a ruminant feed, and provides additional insight into rumen-based differences between cattle and buffalo.
Introduction
Increasing world population, urbanization, and the growing concern over the environmental impact of animal farming means a long-term global strategy for more intensive and sustainable ruminant production is needed (1). Ruminant livestock are important not only for the production of high quantity and quality animal protein (milk and meat) in human diets, but also their ability to produce this using fibrous feeds that cannot be used by humans (2). Furthermore, low-quality feedstuffs, which contain high levels of fiber and low levels of fermentable carbohydrates, are more efficiently utilized by buffaloes than cows. As such, there is increasing interest in this more sustainable ruminant species for livestock production. This is evidenced by the 15.7% increase in buffalo milk production worldwide compared to 4.09% in dairy cows during 2014–2018 (FAOstat data). In Italy, the increase in the same period was up to 21%. This is because in Italy, buffalo milk is used to produce mozzarella cheese which is the third largest Italian DOP (i.e., protected designation of origin) cheese in terms of market value.
Buffalo are more efficient at utilizing low-quality feedstuffs, compared to cattle, due to differences in their digestive system anatomy and physiology (3). Compared to cattle, buffalo have higher rumen retention time (4) and higher feed digestibility (5–7). Associated with this, rumen microbial concentrations and community composition have also been shown to differ between cattle and buffalo (7–9). In order to decrease the impact of both buffalo and cattle production on the environment, new approaches are also needed to improve the sustainability of feed production, for example by decreasing the need for crop fertilization, pest and weed control, and water input. Mycorrhizal fungi are one means of doing this, as they can decrease crop production costs and associated environmental pollution risk.
Mycorrhizal associations of fungi with plants are present in almost all ecosystems, from deserts to arable land (10). Arbuscular mycorrhizal fungi (AMF) are the most common type of fungi that form a symbiotic relationship with terrestrial plants (11). The mycorrhiza formed essentially result in an extension of the plant's root system (12). AMF obtain carbohydrates from their plant host, and in return they provide the plant with mineral nutrients including phosphorus, nitrogen, and potassium (11, 13). AMF positively influence the accumulation of iron and zinc in the plant, which are among the numerous minerals that are relevant to ruminant livestock nutrition (14–16).
As well as exchanging minerals for nutrients, AMF relieve plants from abiotic stress such as drought, salinity and heavy metals (17–19). Protection of host plants from biotic stresses, such as microbial pathogens, has also been reported (20, 21). Furthermore, AMF can contribute to mycotoxin control (22) and induction of the expression of defense-related genes (23). In broader terms, AMF also improve soil structure (24) and can even play an important role in maintaining plant biodiversity and ecosystem stability (25).
Some commercially available AMF preparations include other biological components, such as plant growth promoting rhizobacteria (PGPR) (26–28). PGPR are very effective in promoting plant growth by releasing growth stimulating hormones, and can also give some protection against soil-borne pathogens (29, 30). With maize, PGPR have been shown to improve the mineral status of the plant, promote mycorrhiza colonization of maize roots and increase maize growth (30, 31). As maize is a basic feed for ruminants, differences in its quality can have a significant impact on milk yield and composition. It has been reported that silages from AMF treated forage crops have a higher protein and dry matter content compared to non-AMF treated forage crops (26, 28). Inoculation with both the PGPR strain Pseudomonas fluorescens Pf4 and AMF has also been shown to promote maize growth under field conditions, and can differentially affect grain nutritional content (16). However, only limited studies to date have looked at the effect of AMF applied during the production of feed on livestock.
A study with dairy cows and buffalo fed silages of sorghum and maize produced in the presence of AMF looked at milk production parameters and in vitro feed digestibility (28). Another study assessed the effect of AMF on the in vivo digestibility of fresh barley and berseem clover in goats (27). Both studies found a positive effect of AMF on forage digestibility, however, neither of these studies directly assessed ruminal fermentation parameters and the associated microbiota. In a preliminary communication, we have reported on a feeding trial with lactating Holstein cows where we evaluated the effect of maize grain grown on mycorrhized soil (32). Differences were found in term of maize grain degradation characteristics and, compared to the control maize grain fed animals, the grain grown on soil inoculated with AMF increased rumen microbial concentrations, dry matter intake and milk protein content.
To date, no other information is available regarding the effect of maize grains grown on soil inoculated with AMF on the rumen microbiota and associated fermentation in cattle, and nothing is known in buffalo. Based on the available knowledge outlined above, we hypothesized that maize grain grown on soil inoculated with AMF affects ruminal fermentation and associated microbiota, and that this effect will differ between buffalo and cattle. In order to test this hypothesis, a dietary cross-over study was conducted using rumen-cannulated Holstein Friesian dairy cows and Mediterranean buffaloes. Ruminal fermentation parameters and rumen microbial concentrations were measured, and barcoded amplicon sequencing of the 16S rRNA gene was used to assess rumen prokaryotic community composition.
Materials and Methods
Animals and Experimental Design
Rumen cannulated, non-lactating Mediterranean buffalo (Bubalus bubalis) cows (n = 6) and Holstein-Friesian (Bos taurus) dairy cows (n = 6) were used for the trial. Animals were kept in paddocks, with three animals of the same host type in each paddock. Animals were fed the same basal diet for 4 weeks prior to the start of the trial, which had a crossover design comprised of two experimental periods. Within the first experimental period, half of the animals for each host type were assigned to the diet containing maize grain produced with AMF, and the other half were assigned to the diet containing maize grain produced without AMF. These experimental diets were then fed for 4 weeks. After this, the basal diet was fed again for 4 weeks as a “washout” period before start of the second experimental period. In the second experimental period, diets were then switched over within the same host type, and were fed for 4 weeks.
Both host type groups started the study with six animals, however, due to one Holstein-Friesian cow having to be euthanized during the first experimental period the number of cattle was subsequently decreased to n = 5. Rumen sampling was performed during the last 3 consecutive days of each experimental period. Statistical analysis was performed using animal replication, i.e., for cattle n = 5, and for buffalo n = 6.
The animal study research protocol was approved by the National Ethics Committee (Ministry of Health Decree 26/2014, authorization n°399/2018, Italy) in accordance with the guidelines established by the EU Council/Directives 86/609/EEC.
Maize Grains and Diet
Mycorrhized soil maize grains (M) as well as control maize grains (C), were both produced on two farms in a maize cropping area in the northern Italy near Cremona. For only the mycorrhized soil produced maize grains, Micosat ® Cereal, was applied (15 kg/ha) as a granular preparation when the maize crop was sowed according to the manufacturer's recommendations. The Micosat® Cereal product is a biostimulant microbial consortium consisting mainly of 40% AMF species (Glomus caledonium GM 24, G. coronatum GU 5, and Rhizophagus irregularis RI 31) in the form of ground spores, hyphae, and root fragments of donor plants, along with 9.3% (2.0 × 107 C.F.U./g) of both PGPR (Bacillus subtilis BR 62, Streptomyces sp. ST 60, Paenibacillus durus PD 76) and saprophytic fungi (Trichoderma harzianum TH 01, Trichoderma atroviride TA 28). The composition of the remainder of the product is abiotic (C.C.S. Aosta s.r.l., Quart, AO, Italy). The rhizobacteria and the fungi are added by the producer, as they are thought to have additional value alongside AFM.
Diets containing the maize grains (Diet M or Diet C) were fed once per day (11 am) as a total mixed ration (TMR) with the same proportion of all ingredients for both species, and each group receiving 60 kg feed/day (i.e. 20kg/head/day on average). Feed refusals were not collected (it was observed during the study that feed refusals were all generally <1kg for both host types), and consequently dry matter intake (DMI) was not assessed or predicted in this study. After the loss of the Holstein-Friesian cow from the trial, the corresponding group then received 40 kg feed/day. Details of the composition of the TMR and basal diet are provided in the Supplementary Methods. Dry matter and chemical analysis of the basal diet, maize grains and the associated TMR diets (Table 1) were performed according to the methods described in the Supplementary Methods. Buffalo and cattle nutritional requirements are provided in Supplementary Tables 1 and 2, respectively.
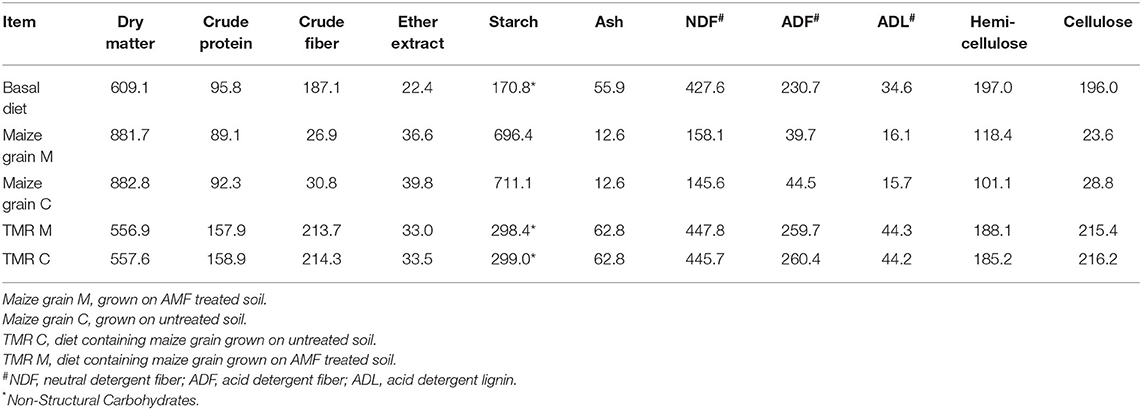
Table 1. Dry matter (g/kg) and chemical analysis (g/kg DM) of the basal diet, maize grains and the associated total mixed ration (TMR) diets.
Rumen Sampling
Rumen digesta samples (1 L) were collected by hand from each animal 1 h before morning feeding on 3 consecutive days. Sampling was done from the dorsal and ventral sac of the rumen via the rumen cannula, and the samples pooled. Two aliquots from each rumen sample were made. The first aliquot was strained through three layers of cheesecloth and used for direct determination of pH and protozoa counts. The second aliquot was bag filtered (BagPage. Interscience, France) and treated with a homogenizer for 5 min (Stomacher, VWR International, Pennsylvania, USA). Aliquots of the homogenate were then processed as follows for later analysis: frozen at −80°C for microbial analysis, preserved with sulfuric acid and stored at −20°C for ammonia, volatile fatty acids (VFA) and lactate analysis.
Fermentation Analysis
Fermentation analysis was performed on all the rumen samples, i.e., 2 experimental periods × 11 animals (5 cows + 6 buffalo) × 3 days. Metabolic fermentation products (i.e. VFA and lactate) were analyzed using high-performance liquid chromatography (HPLC). The procedure is described in detail in the Supplementary Methods. Ammonia samples were analyzed according to the 4030-method reported in the “Analytical Methods for Water” (33).
DNA Extraction
DNA was extracted from the rumen homogenate samples using a protocol involving a combination of bead beating, Stool Transport and Recovery (STAR) buffer (Roche Diagnostics Nederland BV, Almere, The Netherlands) and the Maxwell ® 16 Instrument (Promega, Leiden, The Netherlands). The procedure is described in detail in the Supplementary Methods.
Microbial Concentrations
Analysis of microbial concentrations was also performed on all the rumen samples as reported for fermentation analysis. Protozoa were counted using a Fuchs-Rosenthal chamber according to the Warner procedure (34). For absolute quantification of bacteria and archaea, SYBR green based qPCR assays were performed on DNA extracts using a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories BV, Veenendaal, Netherlands) as previously described (35). All qPCR analyses were carried out in triplicate with a reaction volume of 10 μL, which contained 2 μL of 1:50 dilutions of sample DNA extracts. Standard curves (102-108 copies/μL) were generated using serial dilutions of custom synthesized DNA prepared from known 16S rRNA gene sequences of Ruminococcus albus and Methanobrevibacter olleyae (accessible under ENA accession numbers: CP002403.1 and CP014265.1, respectively).
For absolute quantification of anaerobic fungi, a 5.8S rRNA gene Taqman probe based method was used as previously described (36) with the exception that a CFX384 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories BV) was used. All qPCR analyses were carried out in triplicate in a reaction volume of 10 μL, which contained 2 μL of 1:50 dilutions of sample DNA extracts. The standard curve (101-108 copies/μL) for the assay was generated using serially diluted custom synthesized DNA prepared from a known partial sequence of the Neocallimastix sp. CF17 rrn operon that contains the full 5.8S rRNA gene (accessible under ENA accession number: GU055516.1).
Prokaryotic Community Composition Analysis
Samples from the first day of rumen sampling, for each animal and period, were used for prokaryotic community composition profiling by amplification and barcoding (37) of the V4 variable region of the 16S rRNA gene using the barcoded primers 515F (Parada): 5′-GTGYCAGCMGCCGCGGTAA-3′ (38) and 806R (Apprill): 5′ GGACTACNVGGGTWTCTAAT-3′ (39).
For each sample, triplicate PCRs were performed in a volume of 50 μL containing 1 × HF buffer (Finnzymes, Vantaa, Finland), 1 μL dNTP Mix (10 mM; Promega Benelux, Leiden, Netherlands), 1 U of Phusion® Hot Start II High-Fidelity DNA polymerase (Finnzymes), 500 nM of barcoded primers and 2 ng of template DNA. Cycling conditions consisted of an initial denaturation at 98°C for 30 s followed by 25 cycles of 98°C for 10 s, 56°C for 10 s, and 72°C for 10 s, and a final extension at 72°C for 7 min. PCR success and product size was assessed by agarose gel [2% (w/v)] electrophoresis with 1 × SYBR® Safe (Thermo Scientific).
Triplicate reactions were pooled, purified using HighPrep™ (MagBio Europe Ltd, Kent, United Kingdom), quantified using a Qubit dsDNA BR Assay Kit (Thermo Scientific) and subsequently mixed in equimolar amounts. In order to assess the resulting data quality and potential effects of technical biases, a non-template control (NTC) reaction and a defined synthetic mock community of known composition (37) were included in the equimolar pool. The resulting library was sent for adaptor ligation and 150 nt paired-end sequencing on an Illumina HiSeq4000 instrument (GATC-Biotech, Konstanz, Germany, now part of Eurofins Genomics Germany GmbH).
The resulting raw data was processed with NG-Tax 2.0 using default settings (40). The processing was performed as follows. Paired-end libraries were demultiplexed using read pairs with valid and perfectly matching barcodes. Amplicon sequence variants (ASVs) were then picked. ASVs are defined as individual sequence variants, rather than a cluster of sequence variants with a shared similarity above a user specified threshold as was traditionally the case for operational taxonomic units. ASV selection used an open reference approach as follows. Sequences were ordered by relative abundance for each sample independently, and sequences were considered valid when their relative abundance was >0.1%. Initially discarded low-abundance sequences were then added to ASVs in the dataset, allowing for a single nucleotide mismatch. Taxonomy was assigned using the SILVA 16S rRNA gene reference database version 132 (41). The raw sequence data generated in this study is deposited in ENA under study accession number PRJEB37928, with individual sample accession numbers ranging from ERS4525212-ERS4525234.
Statistical Analysis
The microbial concentration and fermentation data were analyzed using the PROC GLM procedure of SAS 9.4 software (42) using the following univariate linear effects models: Yijkml = μ + Ai + Bj + Ck + (AB)ij + am + eijkml where: Y is the observation vector for the all traits; μ is total average for the trait; Ai is the fixed effect of species: buffalo or cattle; Bj is the fixed effect of diet: M or C; Ck is the fixed experimental period (AB)ij is the interaction between species and diet; am is the animal modeled as random effect; eijkl is the random residual. The statistical significance of all traits and least-squares means were determined using Student's t-test in the GLM procedure. A probability value <0.05 was considered significant.
Prokaryotic community composition data was analyzed in R version 3.4.0 (43). Stacked bar graphs of individual microbiota composition were created by summarizing the relative abundance of all ASVs to either the phylum level, or to the top 15 most abundant prokaryotic genus level groupings. A stacked bar graph of the average microbiota composition of the treatment groups was also created using the summarized relative abundance of all ASVs to the top 20 most abundant prokaryotic genus level groupings. In terms of both genus level graphs, for clarity all other genus level groupings were summarized as “Other.” Pairwise distance between microbiota samples (beta-diversity) was visualized with Non-metric Multidimensional Scaling (NMDS). The NMDS algorithm places each sample into a two-dimensional space such that the between-sample distances are preserved as well as possible. The weighted UniFrac and unweighted UniFrac metrics, which are based on the phylogenetic relatedness of the ASVs, were used as estimator of microbial beta diversity between the animals. While weighted UniFrac considers the abundance of each ASV, unweighted UniFrac provides equal weight to all ASVs, thereby giving equal importance to presence or absence of low and high abundance ASVs. Redundancy Analysis (RDA) was performed to determine the multivariate effects of the environmental variables on the microbiota composition using the rda function from the vegan package (44). RDA is a gradient technique summarizing the linear relationships between a set of variables i.e., prokaryotic community composition and a set of explanatory variables such as host type, dietary intervention, and experimental period. ASV abundance was normalized by centered log ratio scaling to account for compositionality of the data (45). To determine the effect sizes and overlap of the significant environmental variables, variation partitioning was performed using the varpart function from vegan.
Results
Ruminal Fermentation and Microbial Concentrations
For all of the analyzed ruminal fermentation parameters there was no significant effect of diet (Table 2). However, some of the ruminal fermentation parameters were affected by host type (Table 2). Lactate concentrations were significantly (P = 0.04) higher in buffaloes compared to cattle. Total VFA concentrations showed the same trend, being significantly (P < 0.001) higher in buffaloes compared to cattle. The molar proportion of isobutyrate was also significantly (P = 0.02) higher in buffaloes compared to cattle. In contrast, the molar proportion of butyrate was significantly (P = 0.02) lower in buffalo compared to cattle. No other ruminal fermentation parameters were significantly affected by host type (Table 2).
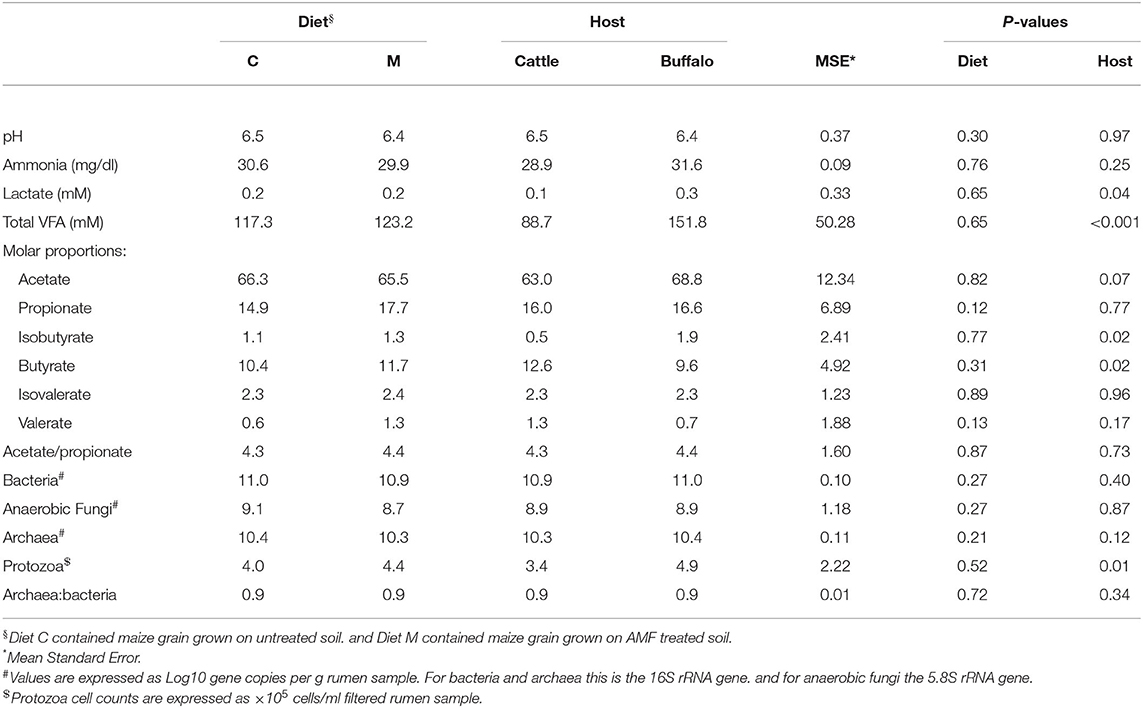
Table 2. Effect of diet and host type on ruminal fermentation parameters and microbial concentrations.
As with the ruminal fermentation parameters, diet had no significant effect on any of the analyzed rumen microbial concentrations or the archaea/bacteria ratio (Table 2). Host type had a significant (P = 0.01) effect on protozoal counts, being higher in buffaloes compared to cattle (Table 2). No other microbial concentrations or ratios were affected by host type (Table 2).
Prokaryotic Community Composition
Of the 960 bacterial and archaeal ASVs detected in the dataset, only three could not be annotated by version 132 of the SILVA database. The number of ASVs annotated at the different taxonomic ranks were as follows: kingdom, 957; phylum 957; class 956; order, 955; family 945; genus 840. The two bacterial phyla Bacteroides and Firmicutes accounted for the majority of the prokaryotic community composition at the phylum level (Supplementary Figure 1). The 20 most predominant (>1.5% mean abundance) genus level groupings of the ASVs in the dataset belonged to the phyla Firmicutes, Bacteroidetes, Fibrobacteres, and Euryarchaeota (Figure 1). Among these, genus level groups within the Firmicutes were annotated as belonging to Succiniclasticum, Acetitomaculum, Butyrivibrio 2, Christensenellaceae R−7 group, Lachnospiraceae NK3A20 group, Ruminococcaceae NK4A214 group, Ruminococcaceae UCG−005 and Ruminococcus 2. Those within the Bacteroidetes were annotated as belonging to Prevotella 1, Prevotellaceae UCG−001, Prevotellaceae UCG−003, Rikenellaceae RC9 gut group and to families F082, Muribaculaceae and p−251–o5. Within the phyla Fibrobacteres and Euryarchaeota, only the genera Fibrobacter and Methanobrevibacter, respectively, were represented within the top 20 genus level groupings. Within each treatment group, variation between individual animals was observed at both the phylum and genus levels (Supplementary Figures 1, 2).
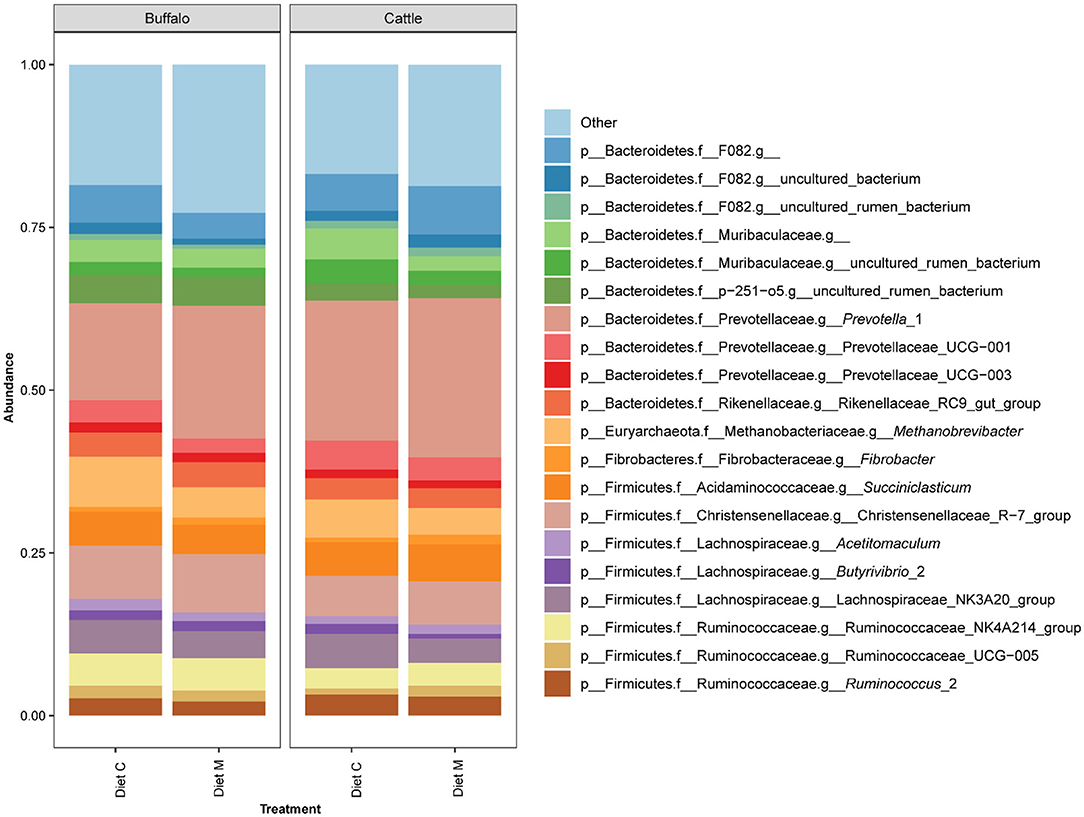
Figure 1. Bar graph showing the average of the 20 most abundant genus level groupings of ASVs from the rumen microbiota of Holstein-Friesian cattle (Cattle) and Mediterranean buffalo (Buffalo) fed diets that differed in terms of containing maize grain which was grown on soil that was treated (Diet M) or not (Diet C) with a commercial AMF preparation.
Weighted and unweighted analysis of beta diversity using unsupervised NMDS showed no separation of samples by diet, and only separation of host type in the unweighted analysis (Supplementary Figure 3). This indicated that differences between host types was largely due to low abundance taxa unique to one host type.
Redundancy analysis (RDA) was performed with ASV level data (i.e., not grouped at the genus level) (Figure 2) to determine the multivariate effects of the experiment on rumen prokaryotic community composition. In contrast to diet (P = 0.9), host type significantly affected rumen prokaryotic community composition and explained 9.3% of the total microbiota variation (P < 0.001). The difference in host type can be clearly observed in the RDA biplot (Figure 2). Of the 11 plotted ASVs, of which the variation in relative abundance was most accurately fitted with the model, two (numbered 8 and 11 in Figure 2) were positively associated with buffalo. Four of the 11 plotted ASVs were positively associated with cattle (numbered 5, 6, 7, and 9 in Figure 2). Subsequently, an unpaired Wilcoxon test was used to identify ASVs the relative abundance of which was significantly different between host types. Nine of the 960 ASVs (i.e., 0.94%) were significantly affected by host type (Table 3). Four of the nine had a higher mean relative abundance in buffalo compared to cattle, and these belonged to the genera Butyrivibrio, Prevotella 1, Methanobrevibacter and the Bacteroidetes unclassified family F082. The other five had a lower mean relative abundance in buffalo compared to cattle, and these all belonged to the Bacteroidetes phylum: two ASVs belonged to Prevotella 1 and the others belonged to the families Muribaculaceae, p-251-o5 and F082.
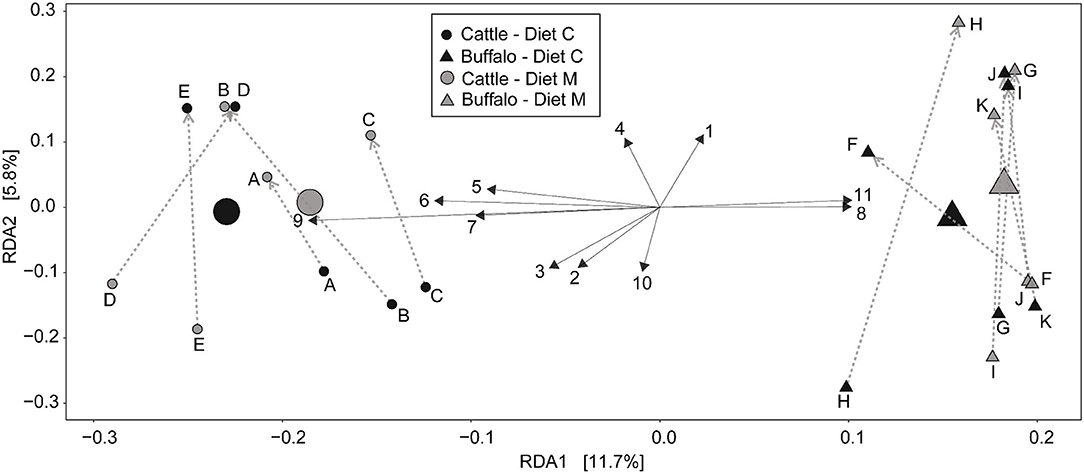
Figure 2. Redundancy analysis biplot of the rumen prokaryotic communities at the level of individual ASVs. constrained by host type (cattle or buffalo) and experimental period. Small symbols are data points represent of rumen microbial communities of individual samples and are labeled with the respective animal codes for the Holstein-Friesian cattle (A–E) and Mediterranean buffalo (animals F–K). Large symbols indicate the weighted center of all samples from a group. The diets containing maize grains grown on soil that was treated (Diet M) or not (Diet C) with a commercial AMF preparation are also indicated. Gray dotted arrows indicate the direction of the cross-over from the first to the second experimental period. Black solid arrows depict the 11 ASV's whose abundance most accurately fits the shown variation. The longer the arrow. the stronger the association with the samples in that direction. Due to space restrictions on the plot these arrows are numbered and the corresponding ASV identification number and taxonomic annotation is stated in this legend. Annotation is only given to the taxonomic level [i.e., phylum (p), class (c), order (o), family (f), or genus (g)] to which it can be reliably annotated. The ASV details are: [1] ASV 350279275 = f_Prevotellaceae.g_; [2] ASV 35027951 = g_Prevotellaceae_UCG-001; [3] ASV 35027962 = g_Prevotellaceae_NK3B31_group; [4] ASV 350279171 = g_Prevotella_1; [5] ASV 3502796 = g_Prevotella_1; [6] ASV 35027983 =p_Bacteroidetes.f_F082.g_; [7] ASV 3502793 = p_Bacteroidetes.f_F082.g_; [8] ASV 35027920 = p_Bacteroidetes.f_p-251-o5. g_uncultured _bacterium; [10] ASV 35027993 = g_Christensenellaceae_R-7_group; [11] ASV 350279543 = g_Lachnospiraceae_XPB1014_group.
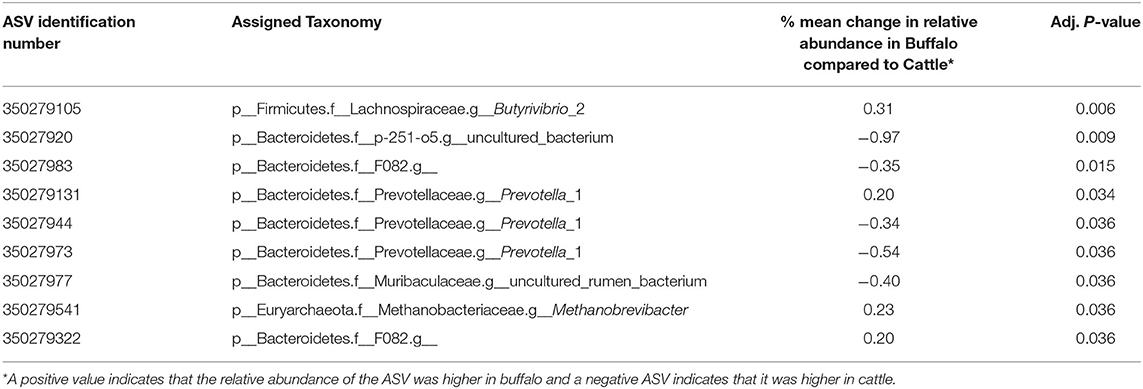
Table 3. The effect of host type on ASVs was compared using an unpaired Wilcoxon test with data normalized using relative abundance and P values adjusted using the Benjamini–Hochberg procedure to decrease the false discovery rate.
As well as host type, experimental period also had a significant (P = 0.012) but small affect on the prokaryotic community compostion, explaining 3.1% of the total microbiota variation. The effect of the experimental period is evident from the uniform direction of the arrows which depict time (Figure 2). Independent of the dietary intervention, samples from the first and second experimental period are located toward the bottom and top half of the plot, respectively.
Discussion
Based on current knowledge, it was hypothesized that maize grain grown on soil inoculated with AMF would affect ruminal fermentation and associated microbiota, and that this effect would differ between buffalo and cattle. This study clearly showed that presence of AMF during production of maize grains had no effect on either host type in terms of ruminal fermentation parameters, microbial concentrations or prokaryotic community composition. This is in contrast to findings of our previous study, with lactating dairy cows, where rumen bacterial and protozoal concentrations were found to increase (32). However, the findings of the current study is perhaps not surprising considering the limited differences that were observed in the chemical composition of diets M and C (Table 1). The lack of effect of the commercial AMF soil treatment on the resulting ruminal utilization of the maize grains indicates there are no issues associated with the use of these grains in ruminant feed.
The maize grain, however, is only one part of the maize crop. Maize silage for ruminants is normally prepared using the whole plant. However, AMF inoculation has been reported not to affect subsequent maize silage composition, and when fed to dairy cows also had no impact on milk yield (28). Similarly, no impact of grain produced on soil inoculated with AMF on milk yield was found in our previous study, although a beneficial increase in milk protein yield did occur (32). This would suggest that irrespective of which part of the plant is used as a ruminant feedstuff, AMF inoculation of maize crops has no subsequent deterimental impact on ruminal processes or ruminant productivity. However, as in this study the commercial AMF preparation used also contained some other biostimulating microbes, comparisons between studies that use commercial products containing AMF and those that use only AMF should be made cautiously.
Host type affected protozoal concentration as well as prokaryotic community composition. Differences between cattle and buffalo have been previously reported in terms of ruminal fermentation, microbial counts, rumen degradability, and digestibility of CP and hemicellulose (3, 5, 7, 8, 46). Longer retention time of feed particles in the rumen has also been reported for buffalo bulls compared to Friesian bulls (4). As ruminal feed retention time of feed influences its ruminal digestion (due to having more time to be degraded by the rumen microbiota), buffaloes are likely to degrade nutrients more extensively than cattle (5). Whilst DMI was not measured in this study, it is likely that differences in DMI reported previously (32) contributed to the higher VFA concentration of buffaloes. However, differences in feed intake pattern between the host types over the day may also be a contributing factor.
It is also important to consider that in this trial the cattle's diet was used for the buffalo, which is richer in nutrients relative to the buffalo's dietary requirements (see Supplementary Table 1). The significantly higher ruminal total VFA concentration in buffalo, compared to cattle, is similar to other data reported by Parmar et al. (47) where buffalo were also given a high concentrate diet. However, despite the higher VFA concentration in buffalo, we did not observe an associated decrease in pH. Buffalo fed the same basal diet on the research farm where this study was conducted usually have a ruminal pH between 6.7 and 7. This pH is higher than was observed in this study. As such, there may well have been a reduction in the buffalo rumen pH during the study due to the high concentration of VFA.
Buffaloes were found in this study to have a higher lactate concentration and molar proportion of isobutyrate. The 3-fold higher concentration of lactate in buffalo compared to cattle in this study is likely to be due to the more efficient ruminal feed degradation in buffalo. Wanapat and Pimpa (48) reported that lactate in the rumen liquid increased significantly with the increase in the dietary proportion of concentrate. According to Nikolov (49), high lactate concentration in the rumen can occur when it is not completely metabolized to propionate. The animal attempts to neutralize it by increasing buffering capability by enhancing salivation. This is normally also associated with increasing amounts of ammonia in the rumen, as was also observed in this study. The increased lactate concentration was not associated with a decreased pH relative to the cattle fed the same diet, and there was no evidence of the buffalo suffering from subacute ruminal acidosis (49).
Branched-chain VFAs such as isobutyrate are required by many rumen bacteria for protein synthesis, particularly cellulolytic bacteria (50). Isobutyrate supplementation of cattle diets has been reported to improve nutrient utilization and ruminal fermentation characteristics (51). As such, it is speculated that the higher molar proportion of isobutyrate in buffalo, compared to cattle, positively affects their ruminal degradation of feed.
The finding of a significantly lower butyrate molar proportion in buffalo, compared to cattle, was consistent with a previous study that compared swamp buffalo with beef cattle (7). Furthermore, it has also been reported that water buffalo have a lower ruminal butyrate concentration relative to Jersey cows (9). The reason for the increased butyrate molar proportion in cattle in this study may be due to it being produced by one or more of the bacteria represented by the five ASVs that were significantly increased in cattle compared to buffalo. However, in this study this cannot be confirmed as the Bacteroidetes F082 and p-251-o5 families are uncultured, fermentation products of members of the Muribaculaceae family have not yet been described (52) and ruminal Prevotella spp. do not produce butyrate (53).
It is interesting to note that one of the ASVs annotated as the butyrate producing genus Butyrivibrio 2 was found to be significantly decreased in cattle compared to buffalo. This is the opposite from what would be expected based on the butyrate molar proportion data. However, due to the higher total VFA in buffalo, there is actually more butyrate production in buffalo compared to cattle.
Three other ASVs belonging to the genera Prevotella 1, Methanobrevibacter, and the Bacteroidetes unclassified family F082 were also significantly increased in buffalo compared to cattle. ASVs from Prevotella 1 and the Bacteroidetes F082 family were also significantly increased in cattle. This indicates that within this genus and uncultured family significant ecological diversity exists with respect to bovine host type. Prevotella spp. are known to be involved in protein degradation and polysaccharide digestion (54), however, the ruminal role of the Bacteroidetes unclassified family F082 is not yet known. In contrast, the role of hydrogenotrophic Methanobrevibacter in the rumen is well known due to it's central role in ruminal methane production. An increase in Methanobrevibacter in buffalo compared to cattle is consistent with the higher VFA production in these animals, which will also result in more hydrogen being available for methane production. Predictive equations have been used to estimate the enteric methane production in buffalo, however, this needs to be directly assessed in vivo (55, 56).
In this study, a higher protozoal concentration was found in buffalo, compared to cattle. This result is consistent with findings of other studies (9, 57). As methanogenic archaea are extra- and intra-cellulary associated with protozoa, this may also explain the higher relative abundance of Methanobrevibacter in buffalo compared to cattle. Although the protozoal community composition was not analyzed in the present study, it has been previously shown to differ between water buffalo and Jersey cows (9). In sheep, increased protozoal concentrations have been associated with a decreased rumen bacterial concentration (due to their bacteriolytic activity) and increased ammonia and VFA concentrations (58). However, in our study no differences between buffalo and cattle were found in terms of ammonia or bacterial concentrations. It is now realized though that the role of protozoa in the rumen is more complex than previously thought, as indicated by a review which included a meta-analysis of over 20 different defaunation studies (59).
As well as host type effects on the rumen prokaryotic composition, an effect of experimental period was evident. The reason for this is not clear, and may be related to environmental factors. Temperature could be an indirect trigger to shift rumen community composition, via changes in the physiology of animals in response to heat stress (60, 61). For example, the animal housing area was not temperature controlled and the ambient temperature in the first experimental period (23.9 + 1.7°C) was lower than that of the second experimental period (27.6 + 1.0°C). Regardless though, this finding confirms the importance of using a cross-over design when comparing different diets within the same animals.
Conclusion
Based on current knowledge, it was hypothesized that maize grain produced in the presence of AMF would affect ruminal fermentation and associated microbiota, and that this effect would differ between buffalo and cattle. This was proven not to be the case as no effect on ruminal fermentation or the associated ruminal microbiota in either cattle or buffaloes was observed when feeding maize grains produced from crops grown on soil treated with a commercial AMF preparation. Considering the beneficial effect of AMF on maize cultivation, in terms of environmental and economic sustainability of forage production, this is clearly a positive outcome. Prokaryotic community composition, protozoal concentrations, and several ruminal fermentation parameters significantly differed between buffalo and cattle, even though they were fed and managed under the same conditions. The different nutrional requirements of these non-lactating animals may have been a contributing factor to this, along with previously reported differences in digestive physiology and rumen retention time between these two bovine species.
Data Availability Statement
The raw sequence data generated in this study is deposited in ENA under study accession number PRJEB37928, with individual sample accession numbers ranging from ERS4525212-ERS4525234. Supplementary Material includes supplementary methods (diet composition, feed chemical analysis, rumen fermentation analysis, and DNA extraction), host nutritional requirements (Supplementary Tables 1, 2), phylum level prokaryotic community composition data for individual animals (Supplementary Figure 1), genus level prokaryotic community composition data for individual animals (Supplementary Figure 2), and weighted and unweighted UniFrac NMDS plots (Supplementary Figure 3).
Ethics Statement
The animal study research protocol was approved by the National Ethics Committee (Ministry of Health Decree 26/2014, authorization n°399/2018, Italy) in accordance with the guidelines established by the EU Council/Directives 86/609/EEC.
Author Contributions
AC designed and performed the experiment. DM and SD performed feed analyses and processing. GC performed the statistical analysis and interpretation of univariate data. AC together with JE performed the rumen prokaryotic community composition and microbial concentration analysis. GH performed the statistical analysis of the rumen prokaryotic community composition data. AC, JE, and GH wrote the manuscript with input from all authors. All authors approved the final manuscript.
Funding
This study was part of a larger project (Use of non-toxigenic strains of Aspergillus flavus and mycorrhized microbial consortia for the reduction of the mycotoxin content of maize), which was funded by the FEASR, Rural Development Program 2014–2020, Lombardy Region, Italy (N° 9571 30/9/2016). AC also gratefully acknowledges CREA for funding her short stage placement at WUR, during which the rumen prokaryotic community composition and microbial concentration analysis was performed.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Assistance provided during sampling and protozoa counts by Emanuela Rossi was greatly appreciated. We are also grateful to farm workers for animal care.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.556764/full#supplementary-material
References
1. Pulina G, Francesconi AHD, Stefanon B, Sevi A, Calamari L, Lacetera N, et al. Sustainable ruminant production to help feed the planet. Italian J Anim Sci. (2017) 16:140–71. doi: 10.1080/1828051X.2016.1260500
2. Eisler MC, Lee MRF, Tarlton JF, Martin GB, Beddington J, Dungait JAJ, et al. Agriculture: steps to sustainable livestock. Nature. (2014) 507:32–4. doi: 10.1038/507032a
3. Gultepe EE, Uyarlar C, Cetingul IS, Iqbal A, Ozcinar U, Bayram I. Comparison of ruminal digestibility of Origanum onites L. leaves in dairy buffalo and cows. Trop Anim Health Prod. (2020) 52:2063–71. doi: 10.1007/s11250-020-02233-6
4. Bartocci S, Amici A, Verna M, Terramoccia S, Martillotti F. Solid and fluid passage rate in buffalo, cattle and sheep fed diets with different forage to concentrate ratios. Livest Prod Sci. (1997) 52:201–8. doi: 10.1016/S0301-6226(97)00132-2
5. Wanapat M, Nontaso N, Yuangklang C, Wora-Anu S, Ngarmsang A, Wachirapakorn C, et al. Comparative study between swamp buffalo and native cattle in feed digestibility and potential transfer of buffalo rumen digesta into cattle. Asian Austr J Anim Sci. (2003) 16:504–10. doi: 10.5713/ajas.2003.504
6. Calabrò S, Moniello G, Piccolo V, Bovera F, Infascelli F, Tudisco R, et al. Rumen fermentation and degradability in buffalo and cattle using the in vitro gas production technique. J Anim Physiol Anim Nutr. (2008) 92:356–62. doi: 10.1111/j.1439-0396.2007.00799.x
7. Chanthakhoun V, Wanapat M, Kongmun P, Cherdthong A. Comparison of ruminal fermentation characteristics and microbial population in swamp buffalo and cattle. Livest Sci. (2012) 143:172–6. doi: 10.1016/j.livsci.2011.09.009
8. Puppo S, Bartocci S, Terramoccia S, Grandoni F, Amici A. Rumen microbial counts and in vivo digestibility in buffaloes and cattle given different diets. Anim Sci. (2002) 75:323–9. doi: 10.1017/S135772980005308X
9. Iqbal MW, Zhang Q, Yang Y, Li L, Zou C, Huang C, et al. Comparative study of rumen fermentation and microbial community differences between water buffalo and Jersey cows under similar feeding conditions. J Appl Anim Res. (2018) 46:740–8. doi: 10.1080/09712119.2017.1394859
10. Brundrett MC. Mycorrhizal associations and other means of nutrition of vascular plants: understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil. (2009) 320:37–77. doi: 10.1007/s11104-008-9877-9
12. Ryan MH, Graham JH. Is there a role for arbuscular mycorrhizal fungi in production agriculture? Plant Soil. (2002) 244:263–71. doi: 10.1007/978-94-017-1284-2_26
13. Liu A, Hamel C, Hamilton RI, Ma BL, Smith DL. Acquisition of Cu, Zn, Mn and Fe by mycorrhizal maize (Zea mays L.) grown in soil at different P and micronutrient levels. Mycorrhiza. (2000) 9:331–6. doi: 10.1007/s005720050277
14. Subramanian KS, Bharathi C, Jegan A. Response of maize to mycorrhizal colonization at varying levels of zinc and phosphorus. Biol Fertil Soils. (2008) 45:133–44. doi: 10.1007/s00374-008-0317-z
15. Balakrishnan N, Subramanian KS. Mycorrhizal symbiosis and bioavailability of micronutrients in maize grain. Maydica. (2012) 57:129–38.
16. Berta G, Copetta A, Gamalero E, Bona E, Cesaro P, Scarafoni A, et al. Maize development and grain quality are differentially affected by mycorrhizal fungi and a growth-promoting pseudomonad in the field. Mycorrhiza. (2014) 24:161–70. doi: 10.1007/s00572-013-0523-x
17. Li T, Lin G, Zhang X, Chen Y, Zhang S, Chen B. Relative importance of an arbuscular mycorrhizal fungus (Rhizophagus intraradices) and root hairs in plant drought tolerance. Mycorrhiza. (2014) 24:595–602. doi: 10.1007/s00572-014-0578-3
18. Wu QS. Arbuscular mycorrhizas and stress tolerance of plants. Groundwater Sustain Dev. (2017) 7:490–4. doi: 10.1007/978-981-10-4115-0
19. Ranganathswamy M, Kadam GL, Jhala YK. An insight into Mycorrhiza involved in building soil and plant health. In: Panpatte DG, Jhala YK, editors, Soil Fertility Management for Sustainable Development. Singapore: Springer (2019). p. 211–29.
20. Gianinazzi S, Gollotte A, Binet MN, van Tuinen D, Redecker D, Wipf D. Agroecology: the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza. (2010) 20:519–30. doi: 10.1007/s00572-010-0333-3
21. Singh R, Soni SK, Kalra A. Synergy between Glomus fasciculatum and a beneficial Pseudomonas in reducing root diseases and improving yield and forskolin content in Coleus forskohlii Briq. under organic field conditions. Mycorrhiza. (2013) 23:35–44. doi: 10.1007/s00572-012-0447-x
22. Siddiqui ZA, Pichtel J. Mycorrhizae: an overview. In: Siddiqui ZA, Akhtar, MS, Kazuyoshi F, editors, Mycorrhizae: Sustainable Agriculture and Forestry. Dordrecht: Springer Netherlands (2008). p. 1–35.
23. Ismail Y, Mccormick S, Hijri M. The arbuscular mycorrhizal fungus, glomus irregulare, controls the mycotoxin production of fusarium sambucinum in the pathogenesis of potato. FEMS Microbiol Lett. (2013) 348:46–51. doi: 10.1111/1574-6968.12236
24. Miller RM, Jastrow JD. Mycorrhizal fungi influence soil structure. In: Kapulnick Y, Douds DD, editors, Arbuscular Mycorrhizas: Physiology and Function. Dordrecht: Kluwer Academic Publisher (2000). p. 3–18.
25. van der Heijden MGA, Martin FM, Selosse MA, Sanders IR. Mycorrhizal ecology and evolution: the past, the present, the future. New Phytol. (2015) 205:1406–23. doi: 10.1111/nph.13288
26. Sabia E, Claps S, Morone G, Bruno A, Sepe L, Aleandri R. Field inoculation of arbuscular mycorrhiza on maize (Zea mays L.) under low inputs: preliminary study on quantitative and qualitative aspects. Ital J Agron. (2015) 10:30–3. doi: 10.4081/ija.2015.607
27. Sabia E, Claps S, Napolitano F, Annicchiarico G, Bruno A, Francaviglia R, et al. In vivo digestibility of two different forage species inoculated with arbuscular mycorrhiza in Mediterranean red goats. Small Rumin Res. (2015) 123:83–7. doi: 10.1016/j.smallrumres.2014.10.008
28. Uzun P, Masucci F, Serrapica F, Varricchio ML, Pacelli C, Claps S, et al. Use of mycorrhizal inoculum under low fertilizer application: effects on forage yield, milk production, and energetic and economic efficiency. J Agric Sci. (2018) 156:127–35. doi: 10.1017/S0021859618000072
29. Vassilev N, Vassileva M, Nikolaeva I. Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl Microbiol Biotechnol. (2006) 71:137–44. doi: 10.1007/s00253-006-0380-z
30. Krey T, Vassilev N, Baum C, Eichler-Löbermann B. Effects of long-term phosphorus application and plant-growth promoting rhizobacteria on maize phosphorus nutrition under field conditions. Eur J Soil Biol. (2013) 55:124–30. doi: 10.1016/j.ejsobi.2012.12.007
31. Tripaldi C, Novero M, Di Giovanni S, Chiarabaglio PM, Lorenzoni P, Meo Zilio D, et al. Impact of Mycorrhizal fungi and rhizosphere microorganisms on maize grain yield and chemical composition. Pak J Agri Sci. (2017) 54:857–65. doi: 10.21162/PAKJAS/17.4840
32. Chiariotti A, Meo Zilio D, Contò G, Di Giovanni S, Tripaldi C. Mycorrhized maize grain: effect on rumen environment and degradability. In: Book of Abstracts, EAAP 67th Aunnual Meeting. Belfast (2016). p. 553.
34. Warner AC. Enumeration of rumen micro-organisms. J Gen Microbiol. (1962) 28:119–28. doi: 10.1099/00221287-28-1-119
35. van Lingen HJ, Edwards JE, Vaidya JD, van Gastelen S, Saccenti E, van den Boger B, et al. Diurnal dynamics of gaseous and dissolved metabolites and microbiota composition in the bovine rumen. Front Microbiol. (2017) 8:425. doi: 10.3389/fmicb.2017.00425
36. Edwards JE, Kingston-Smith AH, Jimenez HR, Huws SA, Skøt KP, Griffith GW, et al. Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. FEMS Microbiol Ecol. (2008) 66:537–45. doi: 10.1111/j.1574-6941.2008.00563.x
37. Ramiro-Garcia J, Hermes GDA, Giatsis C, Sipkema D, Zoetendal EG, Schaap PJ, et al. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research. (2018) 5:1791. doi: 10.12688/f1000research.9227.2
38. Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. (2016) 18:1403–14. doi: 10.1111/1462-2920.13023
39. Apprill A, Mcnally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. (2015) 75:129–37. doi: 10.3354/ame01753
40. Poncheewin W, Hermes GDA, van Dam JCJ, Koehorst JJ, Smidt H, Schaap PJ. NG-Tax 2.0: a semantic framework for high-throughput amplicon analysis. Front. Genet. (2020) 10:1366. doi: 10.3389/fgene.2019.01366
41. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. (2013) 41:590–6. doi: 10.1093/nar/gks1219
43. R Core Team. R: A Language and Environment for Statistical Computing. Available online at: https://www.r-project.org/ (2018).
44. Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. Vegan: Community Ecology Package. R Package Version 23-0 2015. (2015). Available online at: https://cran.r-project.org/web/packages/vegan
45. Aitchison J. The statistical analysis of compositional data. Stat Anal Compos Data. (1986) 44:139–77. doi: 10.1007/978-94-009-4109-0
46. Bartocci S, Terramoccia S, Puppo S. New acquisition on the digestive physiology of the Mediterranean Buffalo. In: Buffalo Production and Research. Rome: Food and Agriculture Organization of the United Nations (FAO) (2005). p. 145–60. Available online at: https://www.cabdirect.org/cabdirect/abstract/20063074777
47. Parmar NR, Solanki JV, Patel AB, Shah TM, Patel AK, Parnerkar S, et al. Metagenome of Mehsani buffalo rumen microbiota: an assessment of variation in feed-dependent phylogenetic and functional classification. J Mol Microb Biotech. (2014) 24:249–61. doi: 10.1159/000365054
48. Wanapat M, Pimpa O. Effect of ruminal NH3-N levels on ruminal fermentation, purine derivatives, digestibility and rice straw intake in swamp buffaloes. Asian Aust J Anim Sci. (1999) 12:904–7. doi: 10.5713/ajas.1999.904
49. Nikolov Y. Clinical experimental studies on acute rumen acidosis in buffaloes (Bubalus bubalis L.). V. Influence on several blood and rumen biochemical parameters. Vet Arhiv. (1998) 68:205–12
50. Zeoula LM, Machado E, Carraro J, Aguiar SC, Yoshimura EH, Agustinho BC, et al. Antioxidant action in diets with ground soybeans on ruminal microbial production, digestion, and fermentation in buffaloes. Rev. Brasil Zoot. (2019) 48:1–11. doi: 10.1590/rbz4820180167
51. Misra AK, Thakur SS. Effect of dietary supplementation of sodium salt of isobutyric acid on ruminal fermentation and nutrient utilization in a wheat straw based low protein diet fed to crossbred cattle. Asian Austr J Anim Sci. (2001) 14:479–84. doi: 10.5713/ajas.2001.479
52. Lagkouvardos I, Lesker TR, Hitch TC, Gálvez EJ, Smit N, Neuhaus K, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. (2019) 7:28. doi: 10.1186/s40168-019-0637-2
53. Bryant MP, Small N, Bouma C, Chu H. Bacteroides ruminicola n. sp. Succinimonas amylolytica the new genus species: species of succinic acid-producing anaerobic bacteria of the bovine rumen. J Bacteriol. (1957) 76:15–23. doi: 10.1128/JB.76.1.15-23.1958
54. Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. (2012) 3:289–306. doi: 10.4161/gmic.19897
55. Sarubbi F, Baculo R, Palomba R, Auriemma G. Estimation of the methane emission factor for buffalo cattle bulls. Iran J Appl Anim Sci. (2013) 3:223–7.
56. Sarubbi F, Chiariotti A, Baculo R, Contò G, Huws SA. Nutritive value of maize and sorghum silages: fibre fraction degradation and rumen microbial density in buffalo cows. Czech J Anim Sci. (2014) 59:278–87. doi: 10.17221/7498-CJAS
57. Franzolin R, Dehority BA. Comparison of protozoal populations and digestion rates between water buffalo and cattle fed an all forage diet1. J Appl Anim Res. (1999) 16:33–46. doi: 10.1080/09712119.1999.9706260
58. Kurihara Y, Eadie JM, Hobson PN, Mann SO. Relationship between bacteria and ciliate protozoa in the sheep rumen. J Gen Microbiol. (1968) 51:267–88. doi: 10.1099/00221287-51-2-267
59. Newbold CJ, de La Fuente G, Belanche A, Ramos-Morales E, McEwan NR. The role of ciliate protozoa in the rumen. Front Microbiol. (2015) 6:1313. doi: 10.3389/fmicb.2015.01313
60. Tajima K, Nonaka I, Higuchi K, Takusari N, Kurihara M, Takenaka A, et al. Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe. (2007) 13:57–64. doi: 10.1016/j.anaerobe.2006.12.001
Keywords: rumen microbiome, bacteria, archaea, protozoa, anaerobic fungi, fermentation
Citation: Chiariotti A, Edwards JE, Hermes GDA, Catillo G, Meo Zilio D, Di Giovanni S, Smidt H and Buttazzoni L (2020) Increasing the Sustainability of Maize Grain Production by Using Arbuscular Mycorrhizal Fungi Does Not Affect the Rumen of Dairy Cattle (Bos taurus) and Buffalo (Bubalus bubalis). Front. Vet. Sci. 7:556764. doi: 10.3389/fvets.2020.556764
Received: 28 April 2020; Accepted: 31 August 2020;
Published: 15 October 2020.
Edited by:
Domenico Vecchio, Istituto Zooprofilattico Sperimentale del Mezzogiorno, ItalyReviewed by:
Malgorzata Szumacher-Strabel, Poznan University of Life Sciences, PolandEduardo Marostegan Paula, Instituto de Zootecnia (IZ), Brazil
Xiaoxia Dai, U.S. Dairy Forage Research Center, United States
Copyright © 2020 Chiariotti, Edwards, Hermes, Catillo, Meo Zilio, Di Giovanni, Smidt and Buttazzoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Chiariotti, YW50b25lbGxhLmNoaWFyaW90dGlAY3JlYS5nb3YuaXQ=
 Antonella Chiariotti
Antonella Chiariotti Joan E. Edwards
Joan E. Edwards Gerben D. A. Hermes
Gerben D. A. Hermes Gennaro Catillo
Gennaro Catillo David Meo Zilio
David Meo Zilio Sabrina Di Giovanni1
Sabrina Di Giovanni1 Hauke Smidt
Hauke Smidt Luca Buttazzoni
Luca Buttazzoni