- 1Key Laboratory of Buffalo Genetics, Breeding and Reproduction Technology, Ministry of Agriculture and Guangxi Buffalo Research Institute, Chinese Academy of Agricultural Sciences, Nanning, China
- 2Institute of Animal and Dairy Sciences, University of Agriculture, Faisalabad, Pakistan
- 3Dairy Science Department, National Research Centre, Giza, Egypt
This study was aimed to evaluate the effect of a mixed phytogenic (MP) on rumen bacteria and their potential association with rumen fermentation and milk yield parameters in water buffaloes. Twenty Murrah buffaloes were fed a basal diet (consisting of maize silage, brewers' grains, and concentrate mixture) for 6 weeks supplemented with 0 (control), 15 (MP15), 25 (MP25), and 35 (MP35) g of mixed phytogenic/buffalo per d. The mixed phytogenic contained fennel (seeds), ajwain (seeds), ginger (tubers), Swertia chirata (leaves), Citrullus colocynthis (fruit), turmeric, fenugreek (seeds), Terminalia chebula (fruit), licorice (roots), and Phyllanthus emblica (fruit) in equal quantities. After 2 weeks of adaptation, daily milk yield, and weekly milk composition were recorded. On the last day of the experiment (d 42), rumen contents were collected to determine rumen fermentation parameters and bacterial diversity through 16S rRNA sequencing. Results revealed no change in dry matter intake, milk yield and rumen fermentation parameters except pH, which increased (P = 0.029) in response to MP supplementation. The mixed phytogenic increased (P < 0.01) milk fatty acids (C4 to C14:0) in MP15 only. The milk C16:1 content and its unsaturation index were higher (P < 0.05) in MP35 as compared to the control and other treatments. Furthermore, C18:3n3 was higher (P < 0.05) in the control, MP15, and MP25, as compared to MP35. Supplementation of MP tended to increase (P = 0.095) the Shannon index of bacterial alpha diversity and a difference (P < 0.05) among treatment groups was observed in beta diversity. Feeding MP increased the Firmicutes, Proteobacteria, and Spirochaetes but decreased Bacteroidetes numerically. In addition, the dominant genus Prevotella decreased in all treatment groups while Pseudobutyrivibrio, Butyrivibrio, and Succinivibrioanceae increased numerically in MP25 and MP35. The mixed phytogenic promoted groups of rumen bacteria positively associated with milk and fat yield. Overall, our study revealed 14 positive correlations of rumen bacteria with milk yield and eight with rumen fermentation parameters. Our findings reveal substantial changes in the rumen bacteriome composition and milk fatty acid content in response to MP but these results should be interpreted carefully, as the sample size of our study was relatively small.
Introduction
Gut microbes perform major digestive and metabolic activities to derive energy from nutrient components of the diet and are considered one of the crucial factors affecting the feed conversion efficiency of ruminants. During rumen fermentation, fermentable dietary components are broken down into volatile fatty acids (VFA) and microbial protein (MCP) is synthesized. Volatile fatty acids and MCP satisfy a major part of the dietary energy (ca. 80%) and protein (65–85%) requirements of the host (1, 2). Since the availability of fermentation products (amount and composition) impacts milk yield, milk fat, and protein synthesis, rumen fermentation is considered to be a vital process, affecting the performance of dairy animals (3). The escape of microbial cells from the rumen is followed by their digestion and absorption in the small intestine leading to the availability of amino acids, needed to satisfy the requirements of the host animal (4). Cell membranes of rumen bacteria are composed of different fatty acids like odd and branch-chain fatty acids that also contribute to fatty acid profile of milk (5).
Some phytogenic feed additives, particularly secondary plant compounds, have shown to affect the composition of the rumen microbiome, change rumen fermentation dynamics and have impact on milk production performance (6–10). So far, the majority of in vitro and in vivo studies, aimed to evaluate the use of plant secondary metabolites in ruminants, have been conducted using one or two plants or their extract or essential oils. In contrast, we wanted to test if different plant-based compounds would act synergistically and therefore decided to supplement a relatively complex mixture of phytogenic compounds derived from 10 plants with proven antioxidant or antimicrobial activity. Combinations of phytogenic antioxidants have, for example, greater potential to scavenge free radicals than individual plant compounds (11).
The plant compounds selected for this study have previously shown to be bioactive and had beneficial effects on rumen fermentation and animal performance (12). For example, supplementation of ginger improved in vitro fermentation characteristics by reducing ammonia nitrogen (NH3-N), methane and acetate to propionate ratio along with desirable effects on fibrolytic bacteria and protozoa (13). Turmeric (Curcuma long) possesses anti-bacterial (14), anti-parasitic (15) and antioxidant properties owing to its high content of curcumin and other curcuminoids (16). In a previous study, we highlighted the potential effects of curcumin as an epigenetic modulator with potential effects on animal physiology (17). Recently, curcumin supplementation has shown to increase milk yield and unsaturated fatty acid (oleic acid) contents of milk in dairy sheep (18). In combination with other herbs, turmeric increased fat- and energy-corrected milk yields in cows, while decreasing the acetate-to-propionate ratio in the rumen fluid (19). Terminalia chebula and Phyllanthus emblica are rich sources of tannins with potential impact on rumen fermentation; particularly a reduction of methanogenesis through decreasing rumen protozoa (20). Supplementation of T. chebula at 10 g per kg diet DM in sheep improved nutrient digestibility and fiber degradability possibly through increasing numbers of fibrolytic bacteria (21, 22). Inclusion of Phyllanthus emblica has shown to increase in vitro dry and organic matter degradability and the synthesis of microbial biomass, while reducing methane production (23). A combination of ajwain, fenugreek, and fennel seeds and fruits of Terminalia chebula and Phyllanthus emblica, reduced in vitro methane production without affecting other fermentation parameters (24). Supplementation of fenugreek led to an improvement in vitro dry matter degradability and in vivo nutrient digestion and utilization in goats (25, 26). Recently, the inclusion of licorice root has shown to increase protein and saturated fatty acid contents of milk while decreasing unsaturated fatty acids and somatic cell count of goat milk (27).
In present study, we attempted to evaluate the synergistic effects of a mixture of 10 different plant-derived compounds on the rumen bacteriome, rumen fermentation, and milk yield and composition of water buffaloes.
Materials and Methods
Animals, Diet, and Experimental Design
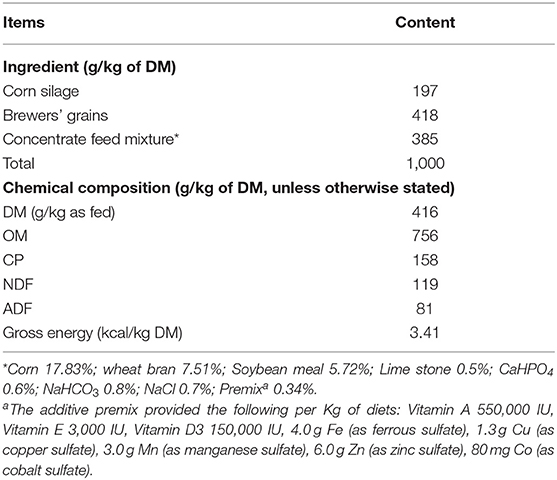
This research was carried out at Guangxi Buffalo Research Institute, Nanning, China (latitude 28° 48′N, longitude 108° 22′E). All experimental procedures used in this experiment were approved by the Ethics committee of the Chinese Academy of Agriculture Sciences, Guangxi Buffalo Research Institute, China. Twenty Murrah buffaloes of similar body weight (580 ± 25 kg), parity and stage of lactation (3–4 months) were randomly selected for this experiment and divided into four groups. The four groups of buffaloes were fed with the same basal diet supplemented with 0 (control), 15 (MP15), 25 (MP25), or 35 (MP35) g of a mixture of 10 different phytogenic substances per buffalo per d. Aside from time to exercise and swim, buffaloes were housed individually in an open-sided shed. To exercise, the buffaloes were set free in an adjacent open yard with a stocking density of 15 m2/buffalo. Free access to water was provided to all buffaloes throughout the day. Fans were installed in the buffalo barn to improve airflow. Buffaloes were allowed 30 min swimming time before milking. Buffaloes were machine milked twice a day. The same experimental diet consisting of maize silage, brewers' grains, and concentrate mixture was fed to all experimental buffaloes for 6 weeks. The buffalo were fed a total mixed ration (TMR) twice per day for ad libitum intake. The TMR was formulated to meet the dietary requirements of lactating buffalo. The respective amount of phytogenic supplement was top-dressed on TMR during morning feeding before milking and each buffalo was monitored for leftover. Details of the chemical composition of the experimental diet are given in Table 1. The first 2 weeks were considered as an adaptation period. Feed intake of individual buffaloes was measured during the last week of the experiment.
The mixed phytogenic consisted of respective parts of following plants; fennel (seeds), ajwain (seeds), ginger (tubers), Swertia chirata (leaves), Citrullus colocynthis (fruit), Turmeric, Fenugreek (seeds), Terminalia chebula (fruit), Licorice (roots), and Phyllanthus emblica (fruit). These plant parts were procured in dry, finely-ground form from Verbena Nutraceuticals Inc. (Islamabad, Pakistan). To make up the tested supplement, equal quantities of each compound were thoroughly mixed.
Chemical Composition of the Diet and Mixed Phytogenic
Dry matter (DM), crude protein (CP), and ash content of the feed samples were analyzed according to the standard procedures (28). Neutral detergent fiber (NDF) and acid detergent fiber (ADF) were determined using an ANKOM2000 Fiber Analyzer (ANKOM Technology Corp., Macedon, NY, USA) including alpha-amylase and sodium sulfite (28, 29). The total polyphenolic content of mixed phytogenic was determined using the Folin-Ciocalteau's phenol reagent as reported previously (30). Gallic acid (10–60 μg/g) was used as standard. The results were expressed as mg of gallic acid equivalent (GAE) per g of MP. Total tannins were measured as tannic acid equivalent and flavonoids were determined as catechin equivalent using UV-VIS Spectrophotometer (Labomed UVD-3500 Spectro) as described previously (31). The chemical composition of the mixed phytogenic is presented in Table S1.
Rumen Fermentation Parameters
Rumen content samples (500 ml) were collected only once, at the last day of the experiment before the morning feeding, using a stomach tube. After collection, the samples were directly transported to the laboratory. The rumen pH was measured immediately using a pH meter (HI 9024C; HANNA Instruments, Woonsocket, Rhode Island, USA). Subsequently, the rumen contents were strained through two layers of cheesecloth and subsamples were analyzed for VFA concentrations (C2, C3, C4, C5, iC4, and iC5) using a GC system (Agilent 7890A, Agilent Technologies, USA), as described by Qin (32). A sub-sample of rumen fluid (4 mL) was acidified with 4 mL of HCl (0.2 mol/L) and stored in a freezer (−20°C) for determination of NH3-N using the indophenols method (33). Microbial protein content was analyzed with a spectrophotometer at 595 nm using 1 mg/ml bovine serum albumin solution (Sigma-Aldrich Co., LLC, St. Louis, Missouri, USA) as standard equivalent (34).
Milk Yield and Composition
Milk yield in the morning (at 5:00 am) and evening (at 5:00 pm) was recorded daily for each buffalo between d 15 and 42. Milk samples for determination of milk composition were collected weekly for 4 consecutive weeks. Fresh milk samples were used to analyze milk composition (milk total solids, protein, fat, and lactose) for morning and evening separately using MilkoScanTM F120 (FOSS, Hillerød, Denmark). Energy corrected milk (ECM) was calculated according to Tyrrell and Reid (35):
Determination of Fatty Acid Profile in Milk
Samples from morning and evening milking were pooled (relative to the quantity of milk produced) for each week separately. Milk samples from each week were stored at −20°C until analysis of fatty acids. Briefly, 20 mL of milk was centrifuged in a 50 mL falcon tube at 17,800 × g for 30 min at 4°C. After centrifugation, the above fat layer (1.0 g) was transferred to a 1.5 mL Eppendorf tube and left at room temperature (~20°C) for ~20 min to allow fat to melt. After that, it was centrifuged at 19,300 × g for 20 min at room temperature in a microcentrifuge. Centrifugation of fat separated the sample into 3 layers: top layer containing lipid; middle layer containing protein, fat, and other water-insoluble solids; and bottom aqueous layer (36). Milk fatty acids were trans-esterified with sodium methoxide according to Zahran and Tawfeuk (37). Briefly, 2.0 mL of n-hexane were added to 40 ul of butterfat and vortexed for 30 s followed by the addition of 2 mL of sodium methoxide (0.4 mol). After vortexing, the mixture was allowed to settle for 15 min. The upper phase, containing the fatty acid methyl ester (FAME), was recovered and analyzed by an Agilent 7890B Gas chromatography (GC-FID) with a polar capillary column CP-Sil®-88 100 m, 0.25 mm id, 0.2 μm film thickness (Agilent Technologies, USA). Helium was used as a carrier gas at a flow rate of 20 cm s−1 and split ratio 100:1. The column temperature profile was held at 100°C for 5 min, ramp to 240°C @ 4°C min−1; hold at 240°C for 30 min. A sample volume of 1.0 μL was injected. The FAME was identified by comparing their relative and absolute retention times with FAME standards (from C4:0 to C22:0). Fatty acid contents are presented as percentage of total fat weight (wt%/wt%).
DNA Extraction and Sequencing of the 16S rRNA Gene
The DNA was extracted from 1 mL of frozen rumen content (both solid and fluid phase) using the CTAB bead-beating method (38). The quality of DNA was checked using a spectrophotometer (NanoDrop2000, Thermo Scientific, USA). High throughput (Illumina MiSeq) sequencing of the 16S rRNA gene was carried out using barcoded primers for V3–V4 region (39). DNA libraries were sequenced using a 2 × 300 paired-end sequencing module (Illumina, San Diego). Resultant paired-end sequence reads were joined together using their overlap relationship (minimum 10 bp) allowing maximum mismatch ratio of 0.2 using FLASH and Trimmomatic software. After pruning, optimized sequence reads were aligned against the SILVA database, Release128 (http://www.arb-silva.de) for identification of Operational Taxonomic Units (OTU) using cluster identity threshold of 97% (40, 41). After that taxonomy of each sequence (OTU representative) was analyzed by RDP Classifier (http://rdp.cme.msu.edu/) against the database (confidence threshold of 0.7). Taxonomic assignment of rumen bacteria was performed with bioinformatics pipeline of Qiime software (http://qiime.org/scripts/assign_taxonomy.html) as described previously (42).
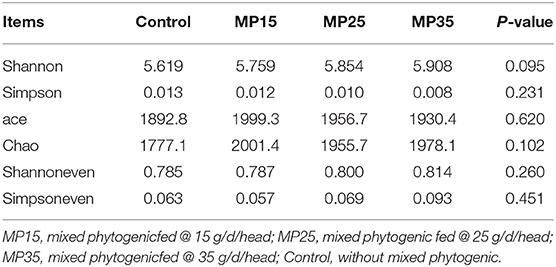
The bacterial diversity of treatment groups was determined by analyzing alpha and beta diversity indices. Population richness (Chao, ACE) and evenness (Shannoneven and Simpsoneven) of rumen bacteria were analyzed for each sample (43). Alpha diversity was estimated by determining Shannon and Simpson indices (44–47). Beta diversity index was calculated to analyze rumen bacterial diversity across different treatment groups using Bray-Curtis dissimilarities (48). Bray-Curtis dissimilarities among different treatment groups were evaluated non-parametrically by utilizing permutation analysis of variance method (PERMANOVA using 999 permutations) as previously reported (49). Redundancy analysis (RDA) was performed at the bacterial genus level using VFAs and milk yield parameters as explanatory variables in the vegan R package (version 3.2).
Statistical Analysis
Effect of MP on all parameters related to milk yield and composition; DM intake, rumen fermentation, and bacterial alpha diversity were analyzed using the general linear model in SAS (SAS Institute Inc., Cary, NC, USA) with treatment as a fixed effect and buffalo as a random effect nested in treatment group. The Duncan's multiple range test was used as a post-hoc test to identify differences among treatment groups. We also analyzed three orthogonal contrasts including all MP treatments vs. the control, linear effect of MP dose, and quadratic effect of MP dose. Treatment effects were declared significant at P < 0.05 and trends were discussed at 0.05 ≤ P < 0.1. The abundances of bacterial phyla and genera were compared using the Kruskal-Wallis H test with a false discovery rate (FDR) correction and Scheffer as a post-hoc test to elucidate differences across treatment groups.
Spearman's rank correlation (r) analyses were performed with the vegan R package (version 3.2) to analyze the association of relative abundance of bacterial genera with rumen fermentation and milk yield parameters. Correlation heatmaps were constructed using the corrplot R package. In the two-dimensional heat map, change in defined color and its depth indicates the nature and strength of the correlation, respectively. Asterisk sign was used when the r value was >0.1 and the P-values were < 0.05 (*0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001).
Results
Rumen Fermentation Parameters
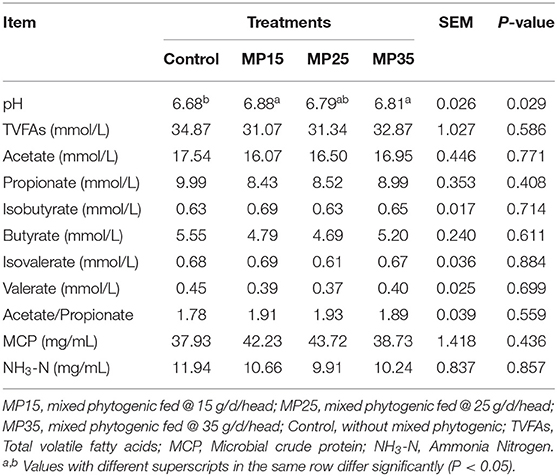
Supplementation of MP increased ruminal pH (P = 0.029) in MP15 and MP35 but no change in pH was observed in MP25 compared to the control (Table 2). There was no effect of treatment on any other rumen fermentation parameter.
Dry Matter Intake (DMI), Milk Yield, and Composition
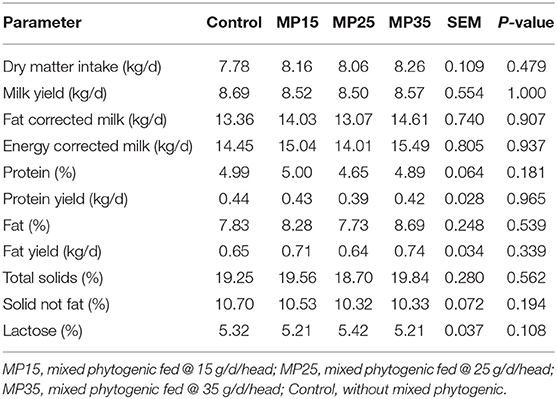
There was no treatment effect on DMI and milk production performance (Table 3).
Fatty Acids Composition of Milk
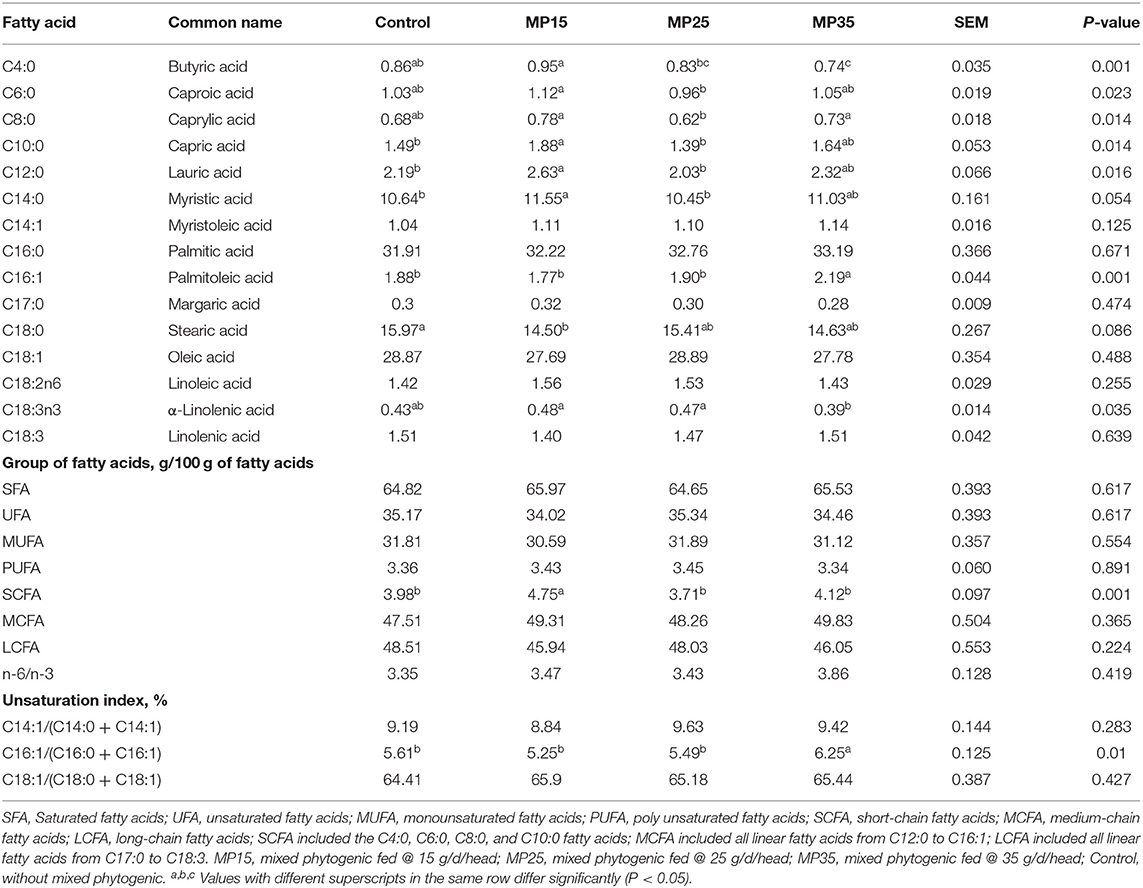
Supplementation of MP increased (P < 0.05) short-chain fatty acids in MP15 compared to other groups (Table 4). Myristic acid (C14:0) tended to increase, while C18:0 tended to decrease in MP15 as compared to other groups. The C16:1 content and its unsaturation index was higher (P < 0.05) in MP35 as compared to the control and other treatment groups. Furthermore, C18:3n3 was (P < 0.05) higher in control, MP15 and MP25, as compared to MP35. There was no treatment effect on total UFA, MUFA, and PUFA content as well as omega6 to omega3 ratio.
Rumen Bacterial Diversity
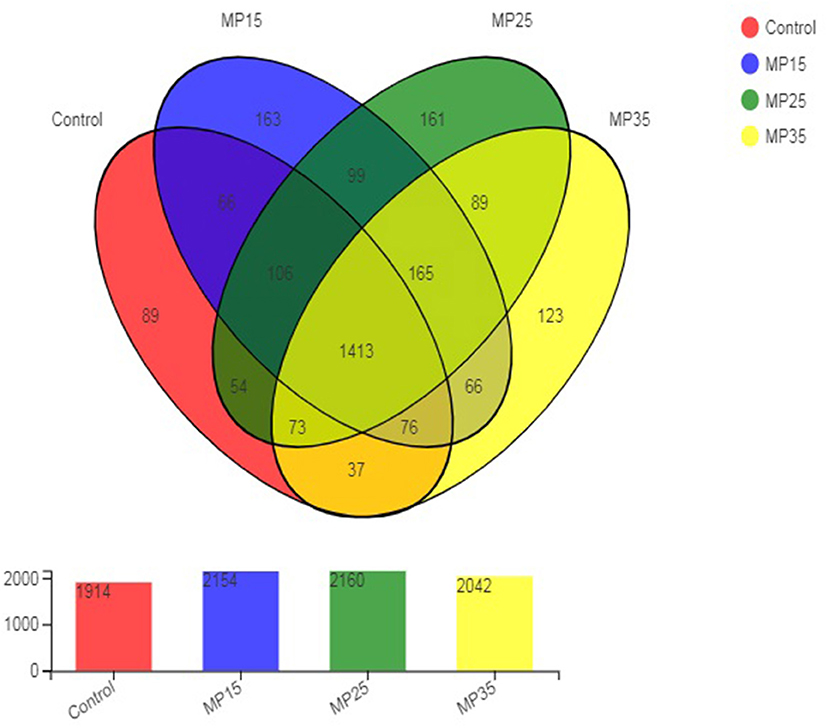
High throughput sequencing of the 16S rRNA gene revealed a total of 2,780 OTU in the rumen content samples. The distribution of shared and unique OTU of the four treatment groups is presented in Figure 1. The highest number of OTU was detected in buffaloes supplemented with MP25, compared to control and other groups. The number of OTU increased in response to MP15 and MP25 but decreased for MP35 as compared to the control. A total of 1,413 OTU were shared by all groups, while the number of unique OTU was 536. The highest number of unique OTU was found in MP15 (163) followed by MP25 (161), MP35 (123), and the control (50).
Treatment had no effects on alpha diversity parameters (Table 5). Analysis of beta diversity showed the difference between groups caused by dietary treatment. The first two dimensions from the (non-metric) multi-dimensional scaling (NDMS) of the Bray-Curtis dissimilarity matrix are presented in Figure 2. Samples were grouped by the level of MP and PERMANOVA (using 999 permutations) amongst all groups showed effect of treatment (P = 0.025).
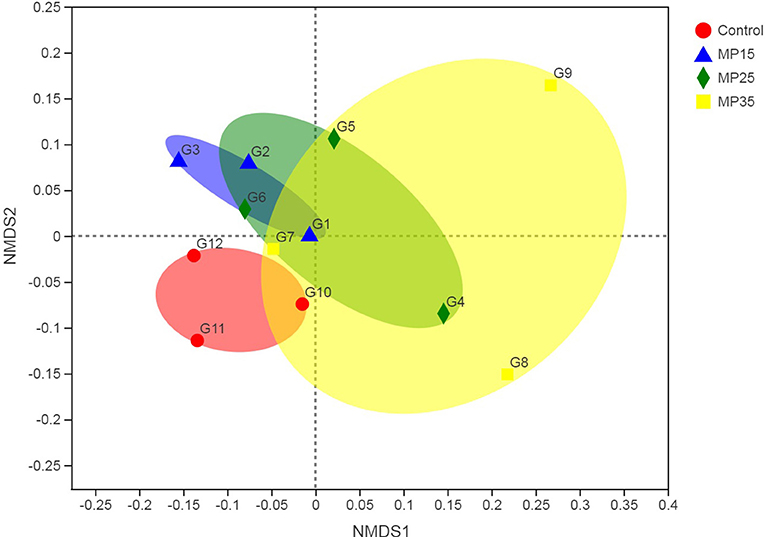
Figure 2. First two dimensions from the (non-metric) multi-dimensional scaling of the Bray-Curtis dissimilarity matrix. Samples were grouped by feed additive. PERMANOVA amongst all groups p = 0.025 (using 999 permutations).
Relative Abundance of Rumen Bacteria
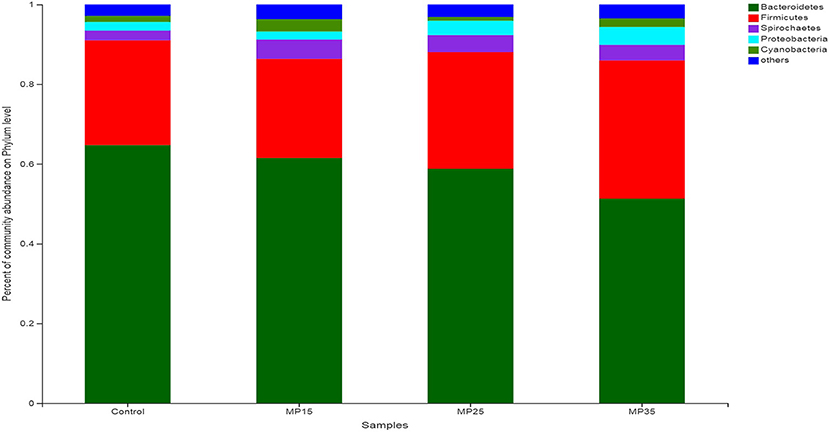
Bacteroidetes and Firmicutes were the most dominant phyla representing between 85 and 91% of total bacteria detected in the rumen of the buffaloes (Figure 3). The relative abundance of Firmicutes and Proteobacteria increased while Bacteroidetes and Spirochaetes decreased numerically in response to treatment compared to the control (Table S2). The abundance of Cyanobacteria increased in MP15 and MP35 but decreased numerically in MP25 as compared to the control group.
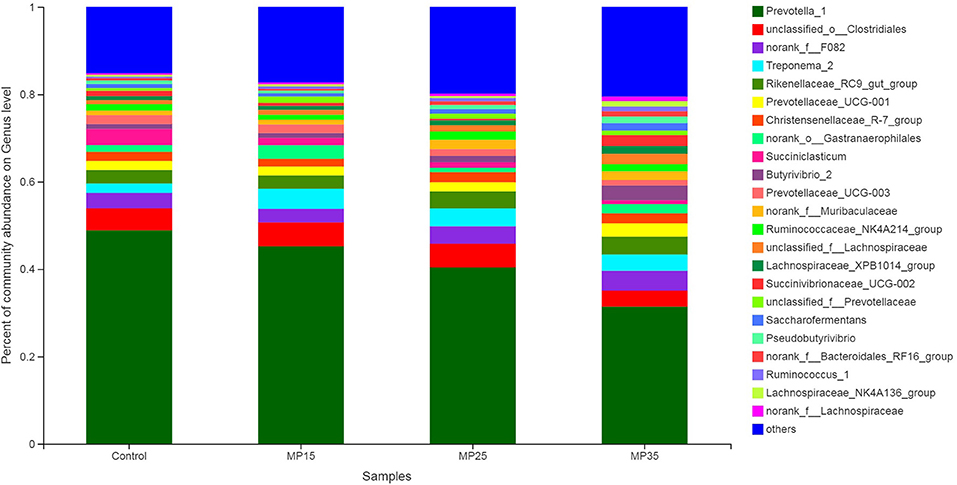
Prevotella was the dominant genus in all four treatments, representing about 31–49% of all sequences (Table S3). Relative abundance of Prevotella decreased numerically with increasing levels of MP (Figure 4). Second most abundant genus was o-Clostridales, which increased with supplementation particularly in MP15 (5.74%) and MP25 (5.53%) as compared to MP35 (3.62%) and control (4.96%). Abundance of f__F082 increased in MP25 (4.01%) and MP35 (4.40%) compared to MP15 (3.09%) and control (3.45%). Similarly, abundance of Rikenellaceae_RC9_gut_group also increased in MP25 (3.95%) and MP35 (4.01%) compared to MP15 (2.99%) and the control (3.09%). Treponema decreased linearly in response to MP supplementation. Highest relative abundance of Christensenellaceae_R-7_group was observed in MP25 (2.39%) and MP35 (2.33%) as compared to MP15 (1.80%) and control group (2.07%). Reduced abundance of Succiniclasticum and Prevotellaceae_UCG-003 was observed in MP-supplemented buffaloes compared to the control. Relative abundance of Butyrivibrio increased with supplementation of MP, particularly in MP35, compared to the control. The relative abundance of Ruminococcaceae also increased as result of MP supplementation. Pseudobutyrivibrio decreased in response to MP15 but was present in greater abundance in MP25 and MP35 compared to the control. The relative abundance of Succinibrionaceae_UCG-002 increased in MP35 (2.33%) compared to MP15 (0.66%), MP25 (0.41%), and the control (1.17%).
Association of Bacteria With Rumen Fermentation Parameters
Redundancy analysis showed acetate contributed to the bacterial community differences at genus level (contribution = 56.8%, P = 0.023) among the four treatment groups (Figure 5). Milk yield and composition parameters did not contributed to overall differences in bacterial genera. Two bacterial genera Prevotella_1 (contribution = 78.6, P = 0.04) and Treponema_2 (contribution = 13.3, P = 0.045) contributed substantially to the compositional differences in rumen bacteriome (Figure 5).
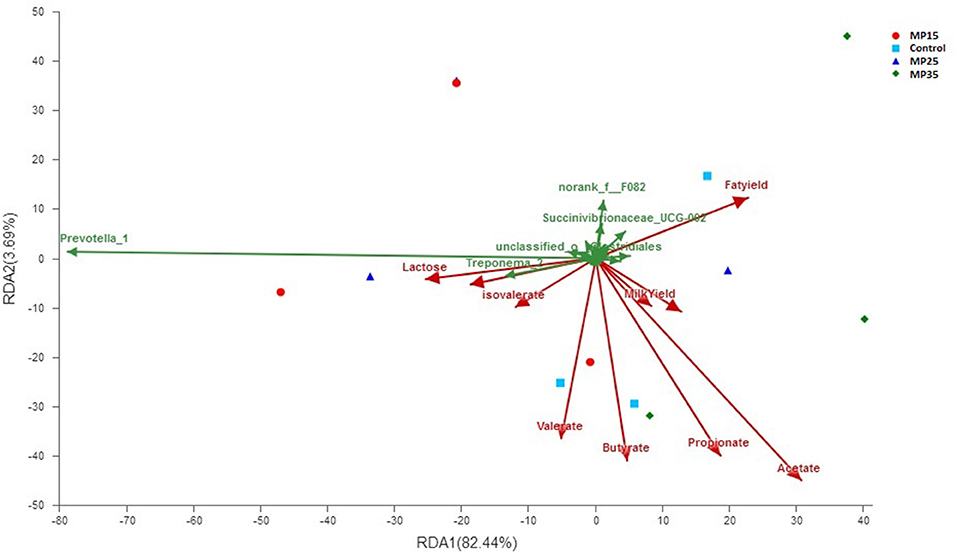
Figure 5. Biplot of RDA analysis on genus level between bacterial genera, VFA, and milk yield parameters.
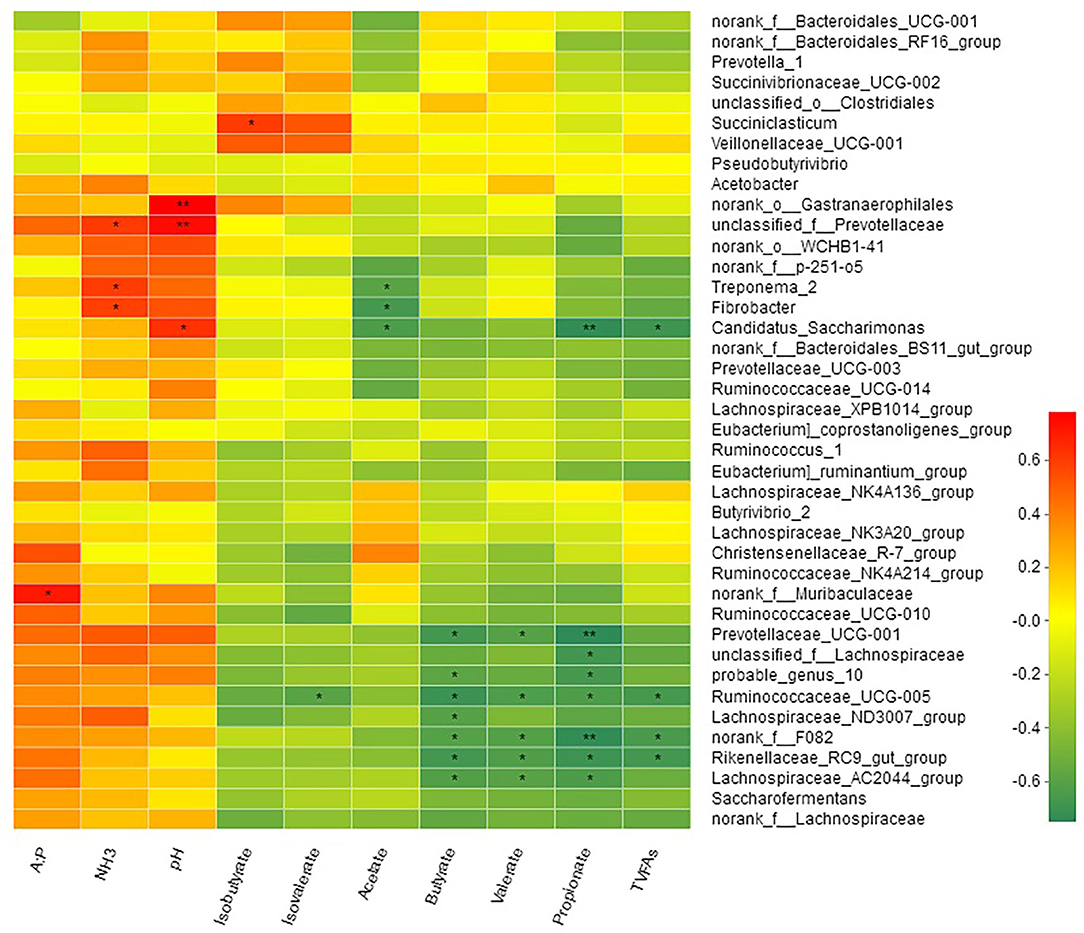
Spearman's correlation between the relative abundance of bacterial genera and rumen fermentation parameters is shown in Figure 6. Acetate concentrations were negatively correlated with Treponema_2 (R = −0.59; P < 0.05), Fibrobacter (R = −0.66; P < 0.05), and Candidatus_Saccharimonas (R = −0.64; P < 0.05). Propionate, butyrate and valerate were negatively correlated with genus f_F082, Rikenellaceae_RC9_gut_group, Prevotellaceae_UCG-001 (R = −0.75, P < 0.01, Lachnospiraceae_AC2044_group, Ruminococcaceae_UCG-005, Lachnospiraceae_ND3007_group, and probable_genus_10, while two genera Ruminococcaceae_NK4A214_group and Candidatus_Saccharimonas were negatively correlated with propionate but not with butyrate (Table S4). Isobutyrate showed a positive correlation (R = 0.60; P < 0.05) with genus Succiniclasticum, while isovalerate was negatively correlated (R = −0.59; P < 0.05) with Ruminococcaceae_UCG-005. Only one bacterial genus f_Muribaculaceae showed positive correlation (R = 0.71; P < 0.05) with acetate to propionate ratio. TVFAs showed a negative correlation with genus f_F082, Rikenellaceae_RC9_gut_group, Ruminococcaceae_UCG-005, and Candidatus_Saccharimonas. Three bacterial genera Rikenellaceae_RC9_gut_group, f_Prevotellaceae, and Fibrobacter were positively correlated with the ruminal concentration of NH3-N.
Association of Rumen Bacteria With DMI, Milk Yield, and Composition
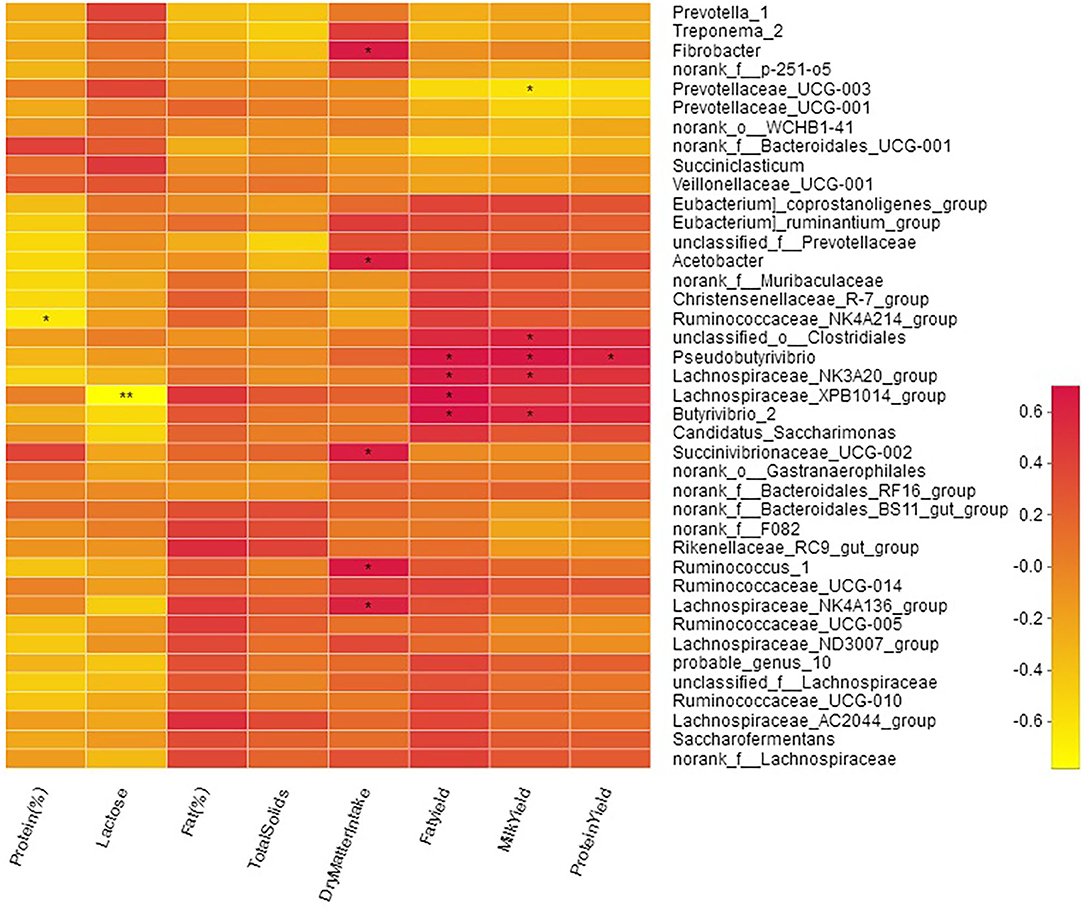
Milk yield was positively correlated with genera o_Clostridiales (R = 0.59; P < 0.05), Butyrivibrio_2 (R = 0.59; P < 0.05), Pseudobutyrivibrio (R = 0.67; P < 0.05), and Lachnospiraceae_NK3A20_group (R = 0.58; P < 0.05; Figure 7, Table S5). A moderate negative (R = −0.61; P < 0.01) correlation of milk yield was observed with Prevotellaceae_UCG-003. Milk protein contents were negatively (R = −0.64; P < 0.05) correlated with Ruminococcaceae_NK4A214_group while milk protein yield was positively correlated (R = 0.61; P < 0.05) with Pseudobutyrivibrio. Milk fat yield showed positive correlation with; o_Clostridiales (R = 0.56; P < 0.05), Butyrivibrio_2 (R = 0.69; P < 0.05), Lachnospiraceae_XPB1014_group (R = 0.70; P < 0.05), Pseudobutyrivibrio (R = 0.68; P < 0.05), and Lachnospiraceae_NK3A20_group (R = 0.64; P < 0.05). Milk lactose was negatively (R = −0.78) correlated with Lachnospiraceae_XPB1014_group. Bacteria specialized in fiber and polysaccharides degradation including Succinivibrionaceae_UCG-002 (R = 0.63; P < 0.05), Ruminococcus_1 (R = 0.65; P < 0.05), Lachnospiraceae_NK4A136_group (R = 0.60; P < 0.05), Fibrobacter (R = 0.65; P < 0.05), and Acetobacter (R = 0.62; P < 0.05) were positively correlated with DMI (Figure 7).
Discussion
Rumen Fermentation Parameters
The mixed phytogenic tested in this study had no effect on rumen fermentation parameters of buffaloes except pH. An increase in rumen pH in response to supplementation of phytochemicals (flavonoids and polyphenols) in ruminants has been reported earlier (51, 52). A stabilization of rumen pH and prevention of acidotic bouts would be particularly beneficial for ruminants fed large amount of readily fermentable carbohydrates which can decrease rumen pH rapidly and alter fermentation kinetics and the composition of rumen microbiome. Strong declines in rumen pH reduce the activity of cellulolytic bacteria (53), shift bacterial populations and promote lysis of gram-negative bacteria leading to an increase in lipopolysaccharides (LPS) in the rumen (54, 55). Plant polyphenols have shown to increase rumen pH and stimulate the diversity of rumen microbiota, which is commonly high in conditions of physiological rumen pH and regular rumen function (56).
Rumen pH plays a crucial role in fiber degradation as it directly affects bacterial adhesion to cellulosic material (57, 58). This can lead to a reduction in fiber digestion, as frequently observed in animals fed high-grain diets (59). In addition, fibrolytic bacteria like Rumiococcus and F. Succinogenes are highly sensitive to even mildly acidic pH (60). Our findings indicate that the tested combination of phytogenics might improve the performance and health of ruminants by preventing excessive drop in pH and subsequent accumulation of LPS in rumen. However, it needs to be emphasized that we measured rumen pH only once as spot sampling just before the morning feeding. It may not be reflective of pH changes over the course of the day, which are important variables to consider. Earlier studies have reported the strong diurnal variation of rumen fermentation parameters and particularly pH (61). The limited number of buffaloes enrolled in the experiment and the fact that we only sampled each buffalo once, is a limitation of our study. In addition, we also did not collect rumen samples at the beginning of the experiment but considered the control group as baseline for our experiment. A more extensive rumen sampling protocol should be followed in further experiments.
We observed an increase in the relative abundance of well-known bacterial genera like Pseudobutyrivibrio, Butyrivibrio, and Succinivibrioanceae. These bacteria form butyrate and propionate which can subsequently affect milk composition especially the milk fat content. The shift in the relative abundance of certain bacterial genera had, besides the discussed modulation of rumen pH, no other effect on rumen fermentation. This might be due to the functional redundancy of the rumen microbiota. The rumen microbiome possesses the ability to adapt to long term exposure to inhibitory substances, like some phytogenics but the effectiveness of the adaptation is dependent on the robustness and diversity of the microbiome, length of exposure, and the concentration of the inhibitor (62). No change in rumen fermentation parameters in response to phytogenic compounds like peppermint oil, garlic, and Piper sarmentosum was reported earlier (63, 64).
DMI, Milk Yield, and Composition
Earlier studies have also reported no effect of plant compounds, such as propolis polyphenols, garlic and peppermint on DMI and the apparent digestibility of nutrients in buffaloes (63, 65). As it was the case in the present study, other studies also reported that polyphenolic compounds had no negative impact on milk yield. For example, supplementation of propolis polyphenols had no effect on milk yield and concentration of milk solids in dairy cows (66). Studies using a blend of different phytochemicals like cinnamaldehyde, eugenol and capsicum also reported no effects on milk yield in dairy cattle (67–69).
Milk Fatty Acid Contents
The major milk fatty acids were C16:0 and C18:1, followed by C18:0 and C14:0, which is in agreement with earlier studies in dairy cattle (70, 71). Contents of SFA (65%) and UFA (35%) measured in our study are similar to earlier reports in cattle and buffaloes (72, 73).
Supplementation of MP15 increased the content of short-chain fatty acids (C4 to C10:0) in milk. The increase in C18:3n3 in response to MP15 and MP25 means that MP has the potential to affect de novo synthesis of fatty acids. The tendency to decrease the percentage of stearic acid (C18:0), a major saturated fatty acid, is desirable from a human health point of view. Polyphenolic compounds have shown to affect microbial biohydrogenation by inhibiting specific rumen bacteria, this can lead to a more desirable fatty acid composition of milk (74–76). Condensed tannins have shown to partially inhibit the last step of C18:3 biohydrogenation in the RUSITEC system (77). Durmic et al. (78) reported that tannins extracted from Acacia mearnsii inhibited Clostridium proteoclasticum but exhibited no effect on Butyrivibrio fibrisolven, revealing selective inhibition of rumen bacteria involved in biohydrogenation.
Earlier studies reported that polyphenolic-rich forage increased the α-linoleic acid content of milk in sheep (79, 80). Higher abundance of Butyrivibrio and Pseudobutyrivibrio was associated with an increase in the content of unsaturated fatty acids owing to their positive correlation with linoleic acid and n-3 fatty acid content of milk (81). The decrease in stearic acid (C18:0) together with the increase in n-3 fatty acid contents, is in agreement with earlier studies that reported similar findings in response to supplementation of tannins in dairy sheep (82). Based on the ratio of C14:1 to C14:0 (a proxy of desaturation), it has been suggested that tannins can enhance the activity of stearoyl Co-A desaturase enzyme (SCD), which mediates the conversion of stearic acid to oleic acid and vaccenic acid to conjugated linolenic acid (CLA). In particular, SCD has shown to contribute almost 50% of oleic acid and cis-9, trans-11 CLA secreted in sheep milk (83). This implies that tannins can increase milk unsaturated fatty acids especially n-3 fatty acids not only by mediating rumen biohydrogenation but also through enhancing SCD activity (82, 84, 85).
Since we did not determine the fatty acids content of the rumen microbial biomass, we are unable to associate microbial abundance with the fatty acid profile in milk. This should be attempted in future studies.
Rumen Bacterial Diversity
Supplementation of MP had no effect on bacterial alpha diversity. However, beta diversity was impacted by MP. Similar results regarding alpha and beta diversity have been reported earlier in response to grape-pomace which is rich in polyphenols (86).
As it was the case in this study, Bacteroidetes and Firmicutes are the major bacterial phyla in both dairy cattle and buffaloes (50, 87–90). A linear increase in Firmicutes was observed together with a decrease in Bacteroidetes. The highest increase in relative abundance in Firmicutes was observed in response to the highest dose of phytogenics (MP35) and resulted in a corresponding decrease in Bacteroidetes. An increase in Firmicutes-to-Bacteroidetes ratio in response to supplementation of plant flavonoids has been reported earlier (50). A major function of rumen Bacteroidetes is the breakdown of polysaccharide, along with various other activities (91). Firmicutes are particle-associated bacteria, which produce butyrate. Numerically higher concentration of butyrate in MP35 was associated with a greater abundance of Firmicutes. Furthermore, buffaloes in this group also had higher milk fat percentage and fat yield, likely due to the positive association of Firmicutes-to-Bacteroidetes ratio and milk fat yield, as previously reported (92).
In the present study, Prevotella was the dominant genus across all treatment groups. This is in agreement with earlier studies in buffalo (93–95). Supplementation of MP linearly decreased the abundance of Prevotella, with the greatest reduction (1.6-fold) in MP35 compared to the control. The decrease in Prevotella was correlated with numerically higher DMI, fat corrected milk (FCM), ECM, fat (%), and milk fat yield in MP35 as compared to the control, most likely due to the negative association of Prevotella with DMI and milk fat content (92, 96). We also observed a negative correlation of Prevotella with acetate, propionate, milk yield and fat (%) but these correlations were weak (R = 0.2–0.37) and not significant. In contrast, earlier studies have also reported a positive correlation of Prevotella with acetate and butyrate in dairy cows (97, 98) and butyrate in buffaloes (99). Prevotella species are more specialized in protein degradation, peptide fermentation and their uptake in the rumen (100). Similar to Prevotella, lower abundance of Succiniclasticum and Prevotellaceae_UCG-003 was observed in buffaloes supplemented with MP compared to the control. In response to MP35, we detected a 3-fold increase in the relative abundance of Butyrivibrio, together with a 1.8-fold increase in Pseudobutyrivibrio compared to the control. Bacterial taxa like Firmicutes, Butyrivibrio, and Pseudobutyrivibrio, are important degraders of polysaccharides in the rumen and produce formate, butyrate, and acetate (101).
Plant phenolic compounds like thymol; have shown to increase the relative abundance of Firmicutes in vitro (up to 82.8%) mainly by inhibiting more sensitive non-Firmicutes (Bacteroidetes) bacteria (102). In contrast, plant essentials oils extracted from Origanum vulgare, garlic and peppermint have shown to decrease the abundance of Firmicutes and methane production, while increasing Bacteroidetes (7). The increase in Proteobacteria in response to MP supplementation was interesting as a substantial increase (2-fold) in the relative abundance of Succinibrionaceae in response to MP35 was also observed. Previously, plant secondary metabolites (8-hydroxyquinoline, α-terpineol, camphor, bornyl acetate, α-pinene, thymoquinone, and thymol) have shown to increase the relative abundance of Succinibrionaceae (102). In study Succinibrionaceae was a dominant family of Proteobacteria, which is in agreement with earlier data from cattle (103). The major fermentation product of this bacterial family is succinate which is subsequently converted to propionate in the rumen, so it creates the possibility of competition between Succinivibrioanceae and methanogens to utilize hydrogen as a substrate to produce succinate and propionate instead of methane. In line with this, greater abundance of Succinivibrioanceae was negatively correlated with methane production (R = −0.72) in cattle (102). Substantially higher abundance of Succinivibrioanceae has been observed in beef cattle with low methane production compared to cattle with higher emissions (103). In addition to the fact that methane is a strong greenhouse gas, reduced losses of methane can also be associated with an improvement in feed efficiency in ruminants. Since we did not measure methane production or total methanogens, we can only speculate about the effect of MP on methane emissions. However, the detected shift in rumen bacteriome toward more beneficial bacteria like Pseudobutyrivibrio, Butyrivibrio and Succinivibrioanceae make it somewhat likely that the tested phytogenic has not only positive impact on production performance but also greenhouse gas intensity of milk.
Association of Bacteria With Rumen Fermentation and Milk Yield Parameters
The Spearman's correlation analysis revealed 28 negative and 8 positive correlations of bacterial genera with rumen fermentation parameters. Three bacterial genera Fibrobactor, Treponema_2, and f_Prevotellaceae had a modest positive correlation with ruminal NH3-N concentrations. Treponema belongs to phylum Spirochetes which mostly ferments soluble sugars to formic acid, acetic acid, lactic acid, and succinic acid (104). Succiniclasticum was positively correlated with the concentration of isobutyrate in the rumen as this genus of bacteria is associated with the formation of succinate from starch degradation leading to the subsequent production of propionate (105). Moreover, increased abundance of Succiniclasticum in high-producing dairy cows has been associated with greater propionate production (106). In our study, we observed very few positive correlations in contrary to earlier studies reporting various strong correlations of bacterial genera with VFA in the rumen of dairy cows (81, 107) and buffaloes (99). This may be attributed to the low variation observed in fermentation parameters, which was likely due to the relatively low sample size and the fact that we only sampled once instead of multiple times over the course of the day. We took rumen samples once from each buffalo using the stomach tube at the end of the experiment; consequently we had a total of five samples per treatment. The relatively low number of buffaloes per treatment and only one rumen sampling are the main limitations of the present study. To evaluate potential effects of MP on rumen fermentation and shifts in the bacterial population in more detail, further studies are required involving a larger cohort of animals and multiple rumen samplings.
Fibrobactor is one of the most active cellulolytic bacteria which ferment only cellulose, glucose, and cellobiose, its primary end products are acetic and succinic acid (104). Unsurprisingly, in this study presence of Fibrobactor was positively correlated with DMI due to its ability to breakdown fiber. However, the negative correlation between Fibrobactor and acetate is difficult to explain. All five bacterial genera (Succinivibrio, Ruminococcus, Lachnospiraceae, Fibrobacter, and Acetobacter) which were positively correlated with DMI are well-known cellulolytic and amylolytic bacteria (108). Dry matter intake has a direct association with milk production so the relationship of these bacteria with DMI, as observed in this study, indicates their potential in enhancing milk yield in buffaloes (109).
Our study showed a positive correlation of Pseudobutyrivibrio with milk, fat and protein yield. The positive impact of Pseudobutyrivibrio on milk yield parameters has been reported earlier in dairy cows (108). A positive correlation of Butyrivibrio and Lachnospiraceae with milk yield and protein has also been reported (93). Furthermore, a positive correlation of Butyrivibrio with average milk fat, milk solid, and total milk yield has been reported in buffaloes (99). Butyrivibrio and Pseudobutyrivibrio ferment structural carbohydrates (hemicellulose, xylan, and pectin) to butyrate (110). However, a negative correlation of Butyrivibrio species with milk fat yield has also been reported in dairy cattle (81, 96). The substantial increase in the relative abundance of Butyrivibrio (3-fold) and Pseudobutyrivibrio (1.8-fold) in this study was correlated with numerically higher milk fat (%), DMI, FCM, ECM, and DMI in buffaloes supplemented with MP35. An unclassified genus of family Prevotellaceae showed a negative correlation with milk yield which is also in agreement with earlier reports revealing a negative association of Prevotella with DMI and milk fat content (92, 96). One unclassified genus belonging to Clostridiales showed a positive correlation with milk yield in the present study. Previous reports have shown substantial differences in abundance of these taxa in beef steers with high and low residual feed intake (111).
Our study found that the tested mixed-phytogenic has the potential to stabilize rumen pH which may be beneficial especially for ruminants in intensive grain-based feeding systems. An increase in Firmicutes-to-Bacteroidetes ratio in response to mixed phytogenic substances reveals their synergistic potential to increase milk fat yield in buffaloes. The significant increase in omega-3 and numeric increase in PUFA in response to MP15 and MP25 may be beneficial from a human health perspective. Our study provides new information regarding the potential effect of a mixed phytogenic on the rumen microbial population, particularly rumen bacteria, and their potential association with fermentation and milk performance parameters. The use of tested mixture of different phytogenics could lead to improvements in production performance and digestive health of buffaloes. However, further studies on larger cohorts are required to solidify these first results and explore the shift in the rumen bacteriome and their impact on production related traits in depth.
Conclusions
Supplementation of MP increased rumen pH and n-3 fatty acid content of milk, while decreasing its stearic acid content. Additionally, MP promoted bacteria that are positively associated with milk and fat yield (Firmicutes-to-Bacteroidetes ratio, Pseudobutyrivibrio, Butyrivibrio, and Succinivibrioanceae). Overall, our findings provide new insight into the modulation of rumen bacteriome caused by a mixed phytogenic feed additive in water buffaloes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, no. PRJNA564158.
Ethics Statement
The animal study was reviewed and approved by Ethics committee of the Chinese Academy of Agriculture Sciences, Guangxi Buffalo Research Institute, China.
Author Contributions
FH and CY: conceptualization. HE and ZT: data curation. FH, ZT, and HE: formal analysis. CY: funding acquisition, supervision, and validation. FH: investigation and writing–original draft. FH, HE, ML, LP, KP, and XL: methodology. XL and CY: project administration. ZT, ML, LP, KP, and CY: resources. FH and HE: software. FH, HE, and CY: writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the National Key Research and Development Program of China (2016YFD0500507 and 2018YFD0501600), the National Natural Science Foundation of China (No. 31560649), and the Guangxi Natural Science Foundation (2018GXNSFAA281162).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
FH gratefully acknowledges the receipt of post-doctoral research fellowship under Talented Young Scientist Program (TYSP) of Ministry of Science and Technology, China. Authors also acknowledge help of Dr. Tariq Sarwar, Director, Research and Development, Verbena Nutraceuticals Inc. (Islamabad, Pakistan) in procurement of phytogenic compounds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.00569/full#supplementary-material
References
1. Bergman E. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. (1990) 70:567–90. doi: 10.1152/physrev.1990.70.2.567
2. Patil N, Mathur B, Patel A, Patidar M, Mathur A. Feeding and Management of Livestock During Drought and Scarcity. Jodhpur: Scientific Publishers (2012).
3. Seymour W, Campbell D, Johnson Z. Relationships between rumen volatile fatty acid concentrations and milk production in dairy cows: a literature study. Anim Feed Sci Technol. (2005) 119:155–69. doi: 10.1016/j.anifeedsci.2004.10.001
4. McDonald P, Edwards RA, Greenhalgh JFD, Morgan CA, Sinclair LA, Wilkinson RG. Animal Nutrition. 7th ed. New york, NY: Prentice Hall Publishers (2010).
5. Harfoot CG. Lipid metabolism in the rumen. Prog Lipid Res. (1978) 17:21–54. doi: 10.1016/0079-6832(78)90004-6
6. Calsamiglia S, Busquet M, Cardozo P, Castillejos L, Ferret A. Invited review: essential oils as modifiers of rumen microbial fermentation. J Dairy Sci. (2007) 90:2580–95. doi: 10.3168/jds.2006-644
7. Patra AK, Yu Z. Effects of essential oils on methane production and fermentation by, and abundance and diversity of rumen microbial populations. Appl Environ Microbiol. (2012) 78:4271–80. doi: 10.1128/AEM.00309-12
8. Oh J, Hristov AN, Lee C, Cassidy T, Heyler K, Varga G, et al. Immune and production responses of dairy cows to postruminal supplementation with phytonutrients. J Dairy Sci. (2013) 96:7830–43. doi: 10.3168/jds.2013-7089
9. Jayanegara A, Wina E, Takahashi J. Meta-analysis on methane mitigating properties of saponin-rich sources in the rumen: influence of addition levels and plant sources. Asian-Australas J Anim Sci. (2014) 27:1426. doi: 10.5713/ajas.2014.14086
10. Oh J, Giallongo F, Frederick T, Pate J, Walusimbi S, Elias R, et al. Effects of dietary Capsicum oleoresin on productivity and immune responses in lactating dairy cows. J Dairy Sci. (2015) 98:6327–39. doi: 10.3168/jds.2014-9294
11. Aftab N, Vieira A. Antioxidant activities of curcumin and combinations of this curcuminoid with other phytochemicals. Phytother Res. (2010) 24:500–2. doi: 10.1002/ptr.2960
12. Kamra DN, Pawar M, Singh B. Effect of plant secondary metabolites on rumen methanogens and methane emissions by ruminants. In: Patra A, editor. Dietary Phytochemicals and Microbes. Dordrecht: Springer (2012). p. 351–70. doi: 10.1007/978-94-007-3926-0_12
13. Kim E, Kim C-H, Min K-S, Lee S. Effects of plant extracts on microbial population, methane emission and ruminal fermentation characteristics in in vitro. Asian-Australas J Anim Sci. (2012) 25:806. doi: 10.5713/ajas.2011.11447
14. Varmuzova K, Matulova ME, Gerzova L, Cejkova D, Gardan-Salmon D, Panhéleux M, et al. Curcuma and Scutellaria plant extracts protect chickens against inflammation and Salmonella enteritidis infection. Poult Sci. (2015) 94:2049–58. doi: 10.3382/ps/pev190
15. Liu Y-M, Zhang Q-Z, Xu D-H, Fu Y-W, Lin D-J, Zhou S-Y, et al. Antiparasitic efficacy of curcumin from Curcuma longa against Ichthyophthirius multifiliis in grass carp. Vet Parasitol. (2017) 236:128–36. doi: 10.1016/j.vetpar.2017.02.011
16. Gazal M, Valente MR, Acosta BA, Kaufmann FN, Braganhol E, Lencina CL, et al. Neuroprotective and antioxidant effects of curcumin in a ketamine-induced model of mania in rats. Eur J Pharmacol. (2014) 724:132–9. doi: 10.1016/j.ejphar.2013.12.028
17. Hassan F, Rehman MS, Khan MS, Ali MA, Javed A, Nawaz A, et al. Curcumin as an alternative epigenetic modulator: mechanism of action and potential effects. Front Genet. (2019) 10:514. doi: 10.3389/fgene.2019.00514
18. Jaguezeski AM, Perin G, Bottari NB, Wagner R, Fagundes MB, Schetinger MRC, et al. Addition of curcumin to the diet of dairy sheep improves health, performance and milk quality. Anim Feed Sci Technol. (2018) 246:144–57. doi: 10.1016/j.anifeedsci.2018.10.010
19. Hashemzadeh-Cigari F, Khorvash M, Ghorbani G, Kadivar M, Riasi A, Zebeli Q. Effects of supplementation with a phytobiotics-rich herbal mixture on performance, udder health, and metabolic status of Holstein cows with various levels of milk somatic cell counts. J Dairy Sci. (2014) 97:7487–97. doi: 10.3168/jds.2014-7989
20. Patra A, Kamra D, Agarwal N. Effect of plant extracts on in vitro methanogenesis, enzyme activities and fermentation of feed in rumen liquor of buffalo. Anim Feed Sci Technol. (2006) 128:276–91. doi: 10.1016/j.anifeedsci.2005.11.001
21. Patra A, Kamra D, Agarwal N, Chatterjee P. Effect of Terminalia chebuaa and Allium sativum on rumen fermentation, enzyme activities and microbial profile in buffaloes. Indian J Anim Nutr. (2007) 24:251–5.
22. Patra A, Kamra D, Bhar R, Kumar R, Agarwal N. Plant secondary metabolites present in Terminalia chebula and Allium sativum reduce methane emission in sheep. Aust J Exp Agric. (2008) 48, lxx–lxxi.
23. Kumar D, Singh P, Chaturvedi V, Verma A. In vitro digestibility and rumen fermentation parameters of feeds as affected by supplementary amla powder and fenugreek seeds. Indian J Anim Nutr. (2016) 33:64–9. doi: 10.5958/2231-6744.2016.00011.6
24. Lakhani N, Kamra DN, Lakhani P, Kala A. Effect of Rumen modifier on methanogenesis and feed digestibility under in vitro conditions. Indian J Anim Nutr. (2019) 36:99–102. doi: 10.5958/2231-6744.2019.00018.5
25. Mir IA, Kumar R, Sharma R, Barman K. Effect of addition of herbs on in vitro digestibility of feed with rumen liquor of goat. Indian J Vet Res. (2010) 19:13–8.
26. Mir IA, Kumar R. Effect of feeding fenugreek seeds (Trigonella foenum-graecum) as feed additive on nutrient utilization in goats. Indian J Anim Nutr. (2012) 29:256–61.
27. Bennato F, Ianni A, Martino C, Di Luca A, Innosa D, Fusco AM, et al. Dietary supplementation of Saanen goats with dried licorice root modifies chemical and textural properties of dairy products. J Dairy Sci. (2020) 103:52–62. doi: 10.3168/jds.2019-16838
28. AOAC. Official Methods of Analysis. 16th ed. Washington, DC: Association of Official Analytical Chemists (1997).
29. Van Soest PV, Robertson J, Lewis B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. (1991) 74:3583–97. doi: 10.3168/jds.S0022-0302(91)78551-2
30. Singleton VL, Orthofer R, Lamuela-Raventos RM. Analysis of total phenols other oxidation substrates antioxidant by means of FolineCiocalteu reagent. Methods Enzymol. (1999) 299:153–78.
31. Chandran KC, Indira G. Quantitative estimation of total phenolic, flavonoids, tannin and chlorophyll content of leaves of Strobilanthes Kunthiana (Neelakurinji). J Med Plants. (2016) 4:282–6.
32. Qin W. Determination of rumen volatile fatty acids by means of gas chromatography. J Nanjing Agric Univ. (1982) 4:110–6.
33. Weatherburn M. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem (Wash). (1967) 39:971–4. doi: 10.1021/ac60252a045
34. Makkar H, Sharma O, Dawra R, Negi S. Simple determination of microbial protein in rumen liquor. J Dairy Sci. (1982) 65:2170–3. doi: 10.3168/jds.S0022-0302(82)82477-6
35. Tyrrell H, Reid J. Prediction of the energy value of cow's milk. J Dairy Sci. (1965) 48:1215–23. doi: 10.3168/jds.S0022-0302(65)88430-2
36. Feng S, Lock A, Garnsworthy P. A rapid lipid separation method for determining fatty acid composition of milk. J Dairy Sci. (2004) 87:3785–8. doi: 10.3168/jds.S0022-0302(04)73517-1
37. Zahran HA, Tawfeuk HZ. Physicochemical properties of new peanut (Arachis hypogaea L.) varieties. OCL. (2019) 26:19. doi: 10.1051/ocl/2019018
38. Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. (2004) 36:808–12. doi: 10.2144/04365ST04
39. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. (2013) 41:e1. doi: 10.1093/nar/gks808
40. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2012) 41:D590–6. doi: 10.1093/nar/gks1219
41. Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. (2013) 42:D643–8. doi: 10.1093/nar/gkt1209
42. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335. doi: 10.1038/nmeth.f.303
43. Smith B, Wilson JB. A consumer's guide to evenness indices. Oikos. (1996) 70–82. doi: 10.2307/3545749
44. Shannon CE. A mathematical theory of communication. Bell Syst Tech J. (1948) 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
46. Chao A. Nonparametric estimation of the number of classes in a population. Scan J Stat. (1984) 11:265–70.
47. Chao A, Lee S-M. Estimating the number of classes via sample coverage. J Am Statis Assoc. (1992) 87:210–7. doi: 10.1080/01621459.1992.10475194
48. Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. (1957) 27:325–49. doi: 10.2307/1942268
49. Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. (2001) 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x
50. Zhan J, Liu M, Wu C, Su X, Zhan K, Qi Zhao G. Effects of alfalfa flavonoids extract on the microbial flora of dairy cow rumen. Asian-Australas J Anim Sci. (2017) 30:1261. doi: 10.5713/ajas.16.0839
51. Balcells J, Aris A, Serrano A, Seradj A, Crespo J, Devant M. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J Anim Sci. (2012) 90:4975–84. doi: 10.2527/jas.2011-4955
52. De Nardi R, Marchesini G, Plaizier JC, Li S, Khafipour E, Ricci R, et al. Use of dicarboxylic acids and polyphenols to attenuate reticular pH drop and acute phase response in dairy heifers fed a high grain diet. BMC Vet Res. (2014) 10:277. doi: 10.1186/s12917-014-0277-5
53. Plaizier J, Krause D, Gozho G, Mcbride B. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. (2008) 176:21–31. doi: 10.1016/j.tvjl.2007.12.016
54. Plaizier J, Khafipour E, Li S, Gozho G, Krause D. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim Feed Sci Technol. (2012) 172:9–21. doi: 10.1016/j.anifeedsci.2011.12.004
55. Mao S, Zhang R, Wang D, Zhu W. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. (2013) 24:12–9. doi: 10.1016/j.anaerobe.2013.08.003
56. De Nardi R, Marchesini G, Li S, Khafipour E, Plaizier KJ, Gianesella M, et al. Metagenomic analysis of rumen microbial population in dairy heifers fed a high grain diet supplemented with dicarboxylic acids or polyphenols. BMC Vet Res. (2016) 12:29. doi: 10.1186/s12917-016-0653-4
57. Russell JB, Wilson DB. Why are ruminal cellulolytic bacteria unable to digest cellulose at low pH? J Dairy Sci. (1996) 79:1503–9. doi: 10.3168/jds.S0022-0302(96)76510-4
58. Miron J, Jacobovitch J, Bayer E, Lamed R, Morrison M, Ben-Ghedalia D. Subcellular distribution of glycanases and related components in Ruminococcus albus SY3 and their role in cell adhesion to cellulose. J Appl Microbiol. (2001) 91:677–85. doi: 10.1046/j.1365-2672.2001.01434.x
59. Goad D, Goad C, Nagaraja T. Ruminal microbial and fermentative changes associated with experimentally induced subacute acidosis in steers. J Anim Sci. (1998) 76:234–41. doi: 10.2527/1998.761234x
60. Russell JB, Dombrowski D. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl Environ Microbiol. (1980) 39:604–10. doi: 10.1128/AEM.39.3.604-610.1980
61. Li M, Penner G, Hernandez-Sanabria E, Oba M, Guan L. Effects of sampling location and time, and host animal on assessment of bacterial diversity and fermentation parameters in the bovine rumen. J Appl Microbiol. (2009) 107:1924–34. doi: 10.1111/j.1365-2672.2009.04376.x
62. Cobellis G, Trabalza-Marinucci M, Marcotullio MC, Yu Z. Evaluation of different essential oils in modulating methane and ammonia production, rumen fermentation, and rumen bacteria in vitro. Anim Feed Sci Technol. (2016) 215:25–36. doi: 10.1016/j.anifeedsci.2016.02.008
63. Verma V, Chaudhary L, Agarwal N, Bhar R, Kamra D. Effect of feeding mixture of garlic bulb and peppermint oil on methane emission, rumen fermentation and microbial profile in buffaloes. Anim Nutr Feed Techn. (2012) 12:157–64.
64. Cherdthong A, Khonkhaeng B, Foiklang S, Wanapat M, Gunun N, Gunun P, et al. Effects of supplementation of piper sarmentosum leaf powder on feed efficiency, rumen ecology and rumen protozoal concentration in Thai native beef cattle. Animals. (2019) 9:130. doi: 10.3390/ani9040130
65. De Paula E, Samensari R, Machado E, Pereira L, Maia F, Yoshimura E, et al. Effects of phenolic compounds on ruminal protozoa population, ruminal fermentation, and digestion in water buffaloes. Livest Sci. (2016) 185:136–41. doi: 10.1016/j.livsci.2016.01.021
66. Aguiar S, Cottica S, Boeing J, Samensari R, Santos G, Visentainer J, et al. Effect of feeding phenolic compounds from propolis extracts to dairy cows on milk production, milk fatty acid composition, and the antioxidant capacity of milk. Anim Feed Sci Technol. (2014) 193:148–54. doi: 10.1016/j.anifeedsci.2014.04.006
67. Benchaar C, Calsamiglia S, Chaves A, Fraser G, Colombatto D, Mcallister T, et al. A review of plant-derived essential oils in ruminant nutrition and production. Anim Feed Sci Technol. (2008) 145:209–28. doi: 10.1016/j.anifeedsci.2007.04.014
68. Benchaar C. Diet supplementation with cinnamon oil, cinnamaldehyde, or monensin does not reduce enteric methane production of dairy cows. Animal. (2016) 10:418–25. doi: 10.1017/S175173111500230X
69. Oh J, Harper M, Lang C, Wall E, Hristov AN. Effects of phytonutrients alone or in combination with monensin on productivity in lactating dairy cows. J Dairy Sci. (2018) 101:7190–8. doi: 10.3168/jds.2018-14439
70. Heck JM, Van Valenberg HJ, Bovenhuis H, Dijkstra J, Van Hooijdonk TC. Characterization of milk fatty acids based on genetic and herd parameters. J Dairy Res. (2012) 79:39–46. doi: 10.1017/S0022029911000641
71. Pegolo S, Cecchinato A, Casellas J, Conte G, Mele M, Schiavon S, et al. Genetic and environmental relationships of detailed milk fatty acids profile determined by gas chromatography in Brown Swiss cows. J Dairy Sci. (2016) 99:1315–30. doi: 10.3168/jds.2015-9596
72. Ebeid H, Gawad R, Mahmoud A. Influence of ration containing tomato pomace silage on performance of lactating buffaloes and milk quality. Asian J Anim Vet Adv. (2015) 10:14–24. doi: 10.3923/ajava.2015.14.24
73. Abdullah M, Akhtar M, Pasha T, Bhatti J, Ali Z, Saadullah M, et al. Comparison of oil and fat supplementation on lactation performance of Nili Ravi buffaloes. J Dairy Sci. (2019) 102:3000–9. doi: 10.3168/jds.2018-15452
74. Ellis KA, Innocent G, Grove-White D, Cripps P, Mclean W, Howard C, et al. Comparing the fatty acid composition of organic and conventional milk. J Dairy Sci. (2006) 89:1938–50. doi: 10.3168/jds.S0022-0302(06)72261-5
75. Collomb M, Bisig W, Bütikofer U, Sieber R, Bregy M, Etter L. Fatty acid composition of mountain milk from Switzerland: comparison of organic and integrated farming systems. Int Dairy J. (2008) 18:976–82. doi: 10.1016/j.idairyj.2008.05.010
76. Butler G, Stergiadis S, Seal C, Eyre M, Leifert C. Fat composition of organic and conventional retail milk in northeast England. J Dairy Sci. (2011) 94:24–36. doi: 10.3168/jds.2010-3331
77. Khiaosa-Ard R, Bryner S, Scheeder M, Wettstein H-R, Leiber F, Kreuzer M, et al. Evidence for the inhibition of the terminal step of ruminal α-linolenic acid biohydrogenation by condensed tannins. J Dairy Sci. (2009) 92:177–88. doi: 10.3168/jds.2008-1117
78. Durmic Z, Mcsweeney C, Kemp G, Hutton P, Wallace R, Vercoe P. Australian plants with potential to inhibit bacteria and processes involved in ruminal biohydrogenation of fatty acids. Anim Feed Sci Technol. (2008) 145:271–84. doi: 10.1016/j.anifeedsci.2007.05.052
79. Roy N, Knight T, Reynolds G, Deighton M, Death A, Sinclair B, et al. The effect of condensed-tannins in fresh Sulla Hedysarum coronarium on the net flux of fatty acids across the mammary gland and their secretion in the milk of lactating ewes. In: Proceedings-New Zealand Society of Animal Production: Palmerston North: New Zealand Society of Animal Production (2002). p. 231–5.
80. Cabiddu A, Molle G, Decandia M, Spada S, Fiori M, Piredda G, et al. Responses to condensed tannins of flowering sulla (Hedysarum coronarium L.) grazed by dairy sheep: part 2: effects on milk fatty acid profile. Livest Sci. (2009) 123:230–40. doi: 10.1016/j.livsci.2008.11.019
81. Bainbridge ML, Cersosimo LM, Wright ADG, Kraft J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol Ecol. (2016) 92:fiw059. doi: 10.1093/femsec/fiw059
82. Buccioni A, Pauselli M, Viti C, Minieri S, Pallara G, Roscini V, et al. Milk fatty acid composition, rumen microbial population, and animal performances in response to diets rich in linoleic acid supplemented with chestnut or quebracho tannins in dairy ewes. J Dairy Sci. (2015) 98:1145–56. doi: 10.3168/jds.2014-8651
83. Frutos P, Toral PG, Ramos-Morales E, Shingfield KJ, Belenguer A, Hervás G. Oral administration of cobalt acetate alters milk fatty acid composition, consistent with an inhibition of stearoyl-coenzyme A desaturase in lactating ewes. J Dairy Sci. (2014) 97:1036–46. doi: 10.3168/jds.2013-7327
84. Mele M, Conte G, Castiglioni B, Chessa S, Macciotta NPP, Serra A, et al. Stearoyl-coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holsteins. J Dairy Sci. (2007) 90:4458–65. doi: 10.3168/jds.2006-617
85. Vasta V, Priolo A, Scerra M, Hallett KG, Wood JD, Doran O. Δ9 desaturase protein expression and fatty acid composition of longissimus dorsi muscle in lambs fed green herbage or concentrate with or without added tannins. Meat Sci. (2009) 82:357–64. doi: 10.1016/j.meatsci.2009.02.007
86. Biscarini F, Palazzo F, Castellani F, Masetti G, Grotta L, Cichelli A, et al. Rumen microbiome in dairy calves fed copper and grape-pomace dietary supplementations: composition and predicted functional profile. PLoS ONE. (2018) 13:e0205670. doi: 10.1371/journal.pone.0205670
87. Kong Y, Teather R, Forster R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol. (2010) 74:612–22. doi: 10.1111/j.1574-6941.2010.00977.x
88. Pitta DW, Pinchak WE, Dowd SE, Osterstock J, Gontcharova V, Youn E, et al. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol. (2010) 59:511–22. doi: 10.1007/s00248-009-9609-6
89. Singh KM, Ahir VB, Tripathi AK, Ramani UV, Sajnani M, Koringa PG, et al. Metagenomic analysis of Surti buffalo (Bubalus bubalis) rumen: a preliminary study. Mol Biol Rep. (2012) 39:4841–8. doi: 10.1007/s11033-011-1278-0
90. Peng S, Yin J, Liu X, Jia B, Chang Z, Lu H, et al. First insights into the microbial diversity in the omasum and reticulum of bovine using illumina sequencing. J Appl Genet. (2015) 56:393–401. doi: 10.1007/s13353-014-0258-1
91. Thomas F, Hehemann J-H, Rebuffet E, Czjzek M, Michel G. Environmental and gut bacteroidetes: the food connection. Front Microbiol. (2011) 2:93. doi: 10.3389/fmicb.2011.00093
92. Jami E, White BA, Mizrahi I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE. (2014) 9:e85423. doi: 10.1371/journal.pone.0085423
93. Pitta D, Kumar S, Vecchiarelli B, Shirley D, Bittinger K, Baker L, et al. Temporal dynamics in the ruminal microbiome of dairy cows during the transition period. J Anim Sci. (2014) 92:4014–22. doi: 10.2527/jas.2014-7621
94. Jewell KA, Mccormick CA, Odt CL, Weimer PJ, Suen G. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol. (2015) 81:4697–710. doi: 10.1128/AEM.00720-15
95. Singh K, Jisha T, Reddy B, Parmar N, Patel A, Patel A, et al. Microbial profiles of liquid and solid fraction associated biomaterial in buffalo rumen fed green and dry roughage diets by tagged 16S rRNA gene pyrosequencing. Mol Biol Rep. (2015) 42:95–103. doi: 10.1007/s11033-014-3746-9
96. Jiang Y, Ogunade I, Arriola K, Qi M, Vyas D, Staples C, et al. Effects of the dose and viability of Saccharomyces cerevisiae. 2. Ruminal fermentation, performance of lactating dairy cows, and correlations between ruminal bacteria abundance and performance measures. J Dairy Sci. (2017) 100:8102–18. doi: 10.3168/jds.2016-12371
97. Bi Y, Zeng S, Zhang R, Diao Q, Tu Y. Effects of dietary energy levels on rumen bacterial community composition in Holstein heifers under the same forage to concentrate ratio condition. BMC Microbiol. (2018) 18:69. doi: 10.1186/s12866-018-1213-9
98. Xue D, Chen H, Luo X, Guan J, He Y, Zhao X. Microbial diversity in the rumen, reticulum, omasum, and abomasum of yak on a rapid fattening regime in an agro-pastoral transition zone. J Microbiol. (2018) 56:734–43. doi: 10.1007/s12275-018-8133-0
99. Zou C, Gu Q, Zhou X, Xia Z, Muhammad WI, Tang Q, et al. Ruminal microbiota composition associated with ruminal fermentation parameters and milk yield in lactating buffalo in Guangxi, China—a preliminary study. J Anim Physiol Anim Nutr. (2019) 103:1374–9. doi: 10.1111/jpn.13154
100. Stewart C, Flint H, Bryant M. The rumen bacteria. In: Hobson PN, Stewart CS, editors. The Rumen Microbial Ecosystem. Amsterdam: Springer (1997). p. 10–72. doi: 10.1007/978-94-009-1453-7_2
101. Palevich N, Kelly WJ, Ganesh S, Rakonjac J, Attwood GT. Butyrivibrio hungatei MB2003 competes effectively for soluble sugars released by Butyrivibrio proteoclasticus B316T during growth on xylan or pectin. Appl Environ Microbiol. (2019) 85:e02056-18. doi: 10.1101/395103
102. Joch M, Mrázek J, Skrivanová E, Cermák L, Marounek M. Effects of pure plant secondary metabolites on methane production, rumen fermentation and rumen bacteria populations in vitro. J Anim Physiol Anim Nutr. (2018) 102:869–81. doi: 10.1111/jpn.12910
103. Wallace RJ, Rooke JA, Mckain N, Duthie C-A, Hyslop JJ, Ross DW, et al. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics. (2015) 16:839. doi: 10.1186/s12864-015-2032-0
105. Hook SE, Steele MA, Northwood KS, Dijkstra J, France J, Wright A-DG, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. (2011) 78:275–84. doi: 10.1111/j.1574-6941.2011.01154.x
106. Tong J, Zhang H, Yang D, Zhang Y, Xiong B, Jiang L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS ONE. (2018) 13:e0198225. doi: 10.1371/journal.pone.0198225
107. Sandri M, Manfrin C, Pallavicini A, Stefanon B. Microbial biodiversity of the liquid fraction of rumen content from lactating cows. Animal. (2014) 8:572–9. doi: 10.1017/S1751731114000056
108. Lima FS, Oikonomou G, Lima SF, Bicalho ML, Ganda EK, De Oliveira Filho JC, et al. Prepartum and postpartum rumen fluid microbiomes: characterization and correlation with production traits in dairy cows. Appl Environ Microbiol. (2015) 81:1327–37. doi: 10.1128/AEM.03138-14
109. Dado R, Allen M. Variation in and relationships among feeding, chewing, and drinking variables for lactating dairy cows. J Dairy Sci. (1994) 77:132–44. doi: 10.3168/jds.S0022-0302(94)76936-8
110. Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. (2008) 6:121. doi: 10.1038/nrmicro1817
Keywords: mixed phytogenic, rumen bacteria, rumen fermentation, milk yield, fatty acids, buffalo
Citation: Hassan F, Ebeid HM, Tang Z, Li M, Peng L, Peng K, Liang X and Yang C (2020) A Mixed Phytogenic Modulates the Rumen Bacteria Composition and Milk Fatty Acid Profile of Water Buffaloes. Front. Vet. Sci. 7:569. doi: 10.3389/fvets.2020.00569
Received: 20 March 2020; Accepted: 16 July 2020;
Published: 26 August 2020.
Edited by:
Uchenna Anele, North Carolina Agricultural and Technical State University, United StatesReviewed by:
Martin Hünerberg, University of Göttingen, GermanyManuel Gonzalez Ronquillo, Universidad Autónoma del Estado de México, Mexico
Copyright © 2020 Hassan, Ebeid, Tang, Li, Peng, Peng, Liang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengjian Yang, eWNqMDc0NkBzaW5hLmNvbQ==
 Faiz-ul Hassan
Faiz-ul Hassan Hossam M. Ebeid
Hossam M. Ebeid Zhenhua Tang1
Zhenhua Tang1 Mengwei Li
Mengwei Li Chengjian Yang
Chengjian Yang