
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 04 August 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00407
This article is part of the Research Topic The Belt and Road of Animal Diseases View all 12 articles
 Hongchao Gou1,2,3,4
Hongchao Gou1,2,3,4 Zhibiao Bian1,2,3,4
Zhibiao Bian1,2,3,4 Rujian Cai1,2,3,4
Rujian Cai1,2,3,4 Zhiyong Jiang1,2,3,4
Zhiyong Jiang1,2,3,4 Shuai Song1,2,3,4
Shuai Song1,2,3,4 Yan Li1,2,3,4
Yan Li1,2,3,4 Pinpin Chu1,2,3,4
Pinpin Chu1,2,3,4 Dongxia Yang1,2,3,4
Dongxia Yang1,2,3,4 Ying-An Zang5*
Ying-An Zang5* Chunling Li1,2,3,4*
Chunling Li1,2,3,4*In 2016, a novel porcine circovirus (PCV), PCV3, was identified in USA. Subsequently, it was proved to be also epidemic in China, Poland, and Korea. To analyze and control the epidemic situation of PCV3, it is necessary to establish accurate and high-throughput detection methods. In this study, the colorimetric isothermal multiple-self-matching-initiated amplification (IMSA) using cresol red was developed to detect PCV3 for the first time. The reaction can be easily performed by incubating the tube at 63°C for 60 min. By the addition of pH-sensitive indicator dye cresol red, the initial color of the reaction mixture is red. When PCV3 capsid gene DNA was positive in the sample, the color of the reaction mixture changed from red to yellow after the isothermal incubation at 63°C, while the negative control maintained the red color. The colorimetric IMSA displayed good specificity in detecting PCV3, PCV2, and PCV1 and 4 porcine DNA pathogens. Moreover, it has a low and repeatable detection limit of 10 copies, which is consistent with TaqMan-based qPCR, but 10 times more sensitive than PCR. In diagnosing 128 clinical specimens, it not only showed 100% agreement with qPCR but also detected 15 positive results more than PCR. The colorimetric IMSA we offered might be a good choice for PCV3 epidemiological investigation and point-of-care testing.
Porcine circovirus (PCV) is a non-enveloped, circular single-stranded DNA virus, belonging to the genus Circovirus within the family Circoviridae. PCV1 and PCV2 were reported to be the only two members of PCV for a long time (1). PCV1 was considered non-pathogenic to pigs. However, PCV2 was known to be connected with clinical features including post-weaning multisystemic wasting syndrome (PMWS), porcine dermatitis and nephropathy syndrome (PDNS), porcine respiratory disease complex (PRDC), proliferative and necrotizing pneumonia (PNP), and reproductive disorders (2, 3). PCV2 has caused severe economic losses to the swine industry because of its circulation worldwide (4).
In 2016, a novel porcine circovirus, PCV3, was identified in USA. Similar symptoms with pigs infected by PCV2, such as PDNS and reproductive disorders, also appear in pigs infected by PCV3 (5, 6). Successive studies proved it was epidemic in China, Poland, and Korea (7–10). Considering this, it is necessary to develop a rapid and simple assay for PCV3 detection. Although PCR-based methods have been utilized to detect PCV3, it is not easy to be performed onsite, or laboratories lack expensive thermal cycling equipment and skilled operators (7, 11). Therefore, isothermal amplification methods are suggested to be a suitable choice for point-of-need diagnostics. To meet this demand, recombinase polymerase amplification (RPA) assay for rapid detection of PCV3 has been described (12). However, at least two kinds of enzymes are needed to start the RPA reaction. The expensive reaction reagent may limit its wide application in rural areas.
Compared with RPA, loop-mediated isothermal amplification (LAMP) or isothermal multiple-self-matching-initiated amplification (IMSA) only utilizes strand-displacing DNA polymerase to accomplish robust DNA synthesis (13, 14). This makes them to be good choice for low-cost isothermal methods. What is more, IMSA proved to be more sensitive than LAMP in the previous study (13, 15). Recently, rapid, sensitive, and visual detection assay was achieved by the addition of pH-sensitive indicator dyes. To develop a rapid, convenient, low-cost method suitable for rural areas, the colorimetric IMSA using cresol red was utilized to detect PCV3 in our study. This method showed good potential on rapid examination of clinical samples suspected to be infected by PCV3.
All animal experiments were reviewed and approved by the ethical and ethics commission (Institute of Animal Health, Guangdong Academy of Agricultural Sciences, China). The license number was SYXK (Yue) 2011–0116. Moreover, samples collecting treatment in this study were performed in accordance with national and local laws and guidelines.
PCV2 isolate HN6 (GenBank no: KM035762.1), PCV1, pseudorabies virus (PRV) GD-WH strain (GenBank no: KT936468.1), Haemophilus parasuis (HPS) serotype 5, Streptococcus suis (SS) serotype 2, and Actinobacillus pleuropneumoniae (APP) Serovar 1 were preserved in our laboratory. They were used to evaluate the specificity of the colorimetric IMSA.
In Guangdong province, a total number of 128 clinical samples were collected from pigs suspected to be infected with PCV3. The tissues include blood, tonsil, lymph gland, lung, kidney, and brain. In addition, 15 blood samples of specific-pathogen-free (SPF) pigs (5 months old), from the Laboratory Animal Center of Southern Medical University, China, were collected. All samples were stored at −80°C until DNA extraction.
According to the instructions of the manufacturer, viral DNA in supernatant of cell culture or clinical samples was purified by using the HiPure Viral RNA/DNA Kit (Magen, China). Finally, viral DNA was dissolved in 50 μL nuclease-free water and stored at −80°C. Bacterial DNA was extracted by using the HiPure Bacterial DNA Kit (Magen, China) according to the manufacturer's protocol.
According to the previously reported methods, PCR was performed in a 25-μL reaction mixture (7). The mixture contained 0.3 μM of each primer, 1 × Premix Tag (Takara Biotechnology, China) and 2 μL template DNA. The PCR program was as follows: 94°C for 5 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 10 min. The products were analyzed on 2% agarose gel.
TaqMan-based qPCR for PCV3 genome identification was carried out as is previously reported (6). The 25-μL reaction mixture contained 0.4 μM of each primer and probe, 1 × qPCR Probe Master Mix (Vazyme, China), and 2 μL template DNA. The reaction program was set as follows: 95°C for 3 min, followed by 40 cycles at 95°C for 10 s and 60°C for 60 s. FAM fluorescence signals were obtained at the end of each annealing step by the real-time PCR detection system (Roche Light Cycler 480 II, Switzerland). Results with a cycle threshold (Ct) value of <40 were considered positive, while results with no Ct value within 40 cycles were considered negative.
As in the report previously described, partial sequences of capsid gene of PCV3 were amplified by the PCR method from the clinical samples (7). The PCR product was purified by using the Cycle Pure Kit according to the instructions of the manufacturer (Omega, USA). Then, the purified fragment was cloned into pMDTM19-T Vector (TaKaRa Biotechnology, China) and the pMD19T-capsid plasmid was constructed. After the plasmid was transformed into DH5α competent cells, the plasmid DNA was extracted by using the Plasmid Mini Kit I (Omega, USA).
The conserved region of the capsid gene was determined by alignment of PCV3 strains indexed in the GenBank (accession no: MF589105.1, MF589107.1, MF769811.1, MF769807.1, MF084994.1, KX778720.1, KX898030.1, MG310152.1, MF079254.1, and MG250187.1). According to the principle of IMSA assay, capsid gene sequences of PCV3-US/MO2015 strain (accession no: KX778720.1) were input for IMSA primer design by using the software Primer Premier 5.0 (15). Among multiple sets of primers, the primers targeting to the conserved regions of the capsid gene were selected for subsequent analysis. IMSA primers are listed in Table 1.
As the previous study described, Tris–HCl and betaine was not included in the colorimetric assay (16). To establish this method, the reaction mixture containing 10 mM (NH4)2SO4, 10 mM KCl, 8 mM MgSO4, 0.1% v/v Tween-20, 16 mM cresol red, 0.8 mM dNTPs, and 8 U Bst WarmStart DNA Polymerase (New England Bio-labs, USA) was prepared, and the pH of the mixture was adjusted to 9.0 by using 100 mM KOH. To perform the IMSA reaction, primers' concentration was optimized and determined as 1.6 μM SteF/SteR, 0.8 μM FIT/RIT, and 0.2 μM DsF/DsR. Then, the reaction tube was incubated in a thermostatic device (HB-202, BIOER TECHNOLOGY, China) at 63°C for 60 min. The results can be directly judged with the naked eyes. The yellow color indicated the positive results, while the red color indicated the negative results. In each colorimetric IMSA test, 3 observers were invited to identify colors and read out the results. For each result, every observer read it out 3 times.
DNA extracted from PCV2, PCV1, PRV GD-WH strain, HPS, SS, and APP were used to evaluate the specificity of the colorimetric IMSA.
Ten-fold serial dilutions of pMD19T-capsid plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies) were used to determine the detection limit of the colorimetric IMSA. The negative control was conducted at the same time. The detection limit of the colorimetric IMSA was compared with qPCR and PCR.
To analyze the reproducibility of the colorimetric IMSA, ten-fold serial dilutions of pMD19T-capsid plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies) and the negative control were detected for 3 different times.
A total number of 128 suspected clinical samples and 15 blood samples from SPF pigs were used for DNA extraction and colorimetric IMSA detection. To evaluate the test accuracy of the colorimetric IMSA, its results were identified by TaqMan-based qPCR and PCR.
To establish the colorimetric IMSA assay, Tris–HCl was removed from the reaction mixture but the cresol red was added. After 60 min of incubation at 63°C, the color of the reaction mixture changed to yellow when the plasmid containing partial sequences of the PCV3 capsid gene was used as DNA template, while the negative control keeps the red color (Figure 1). So, we judged the positive result of the colorimetric IMSA by observing the yellow color of the reaction tube.
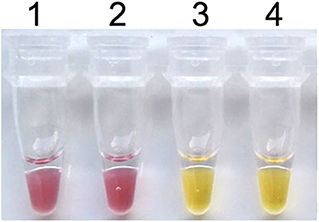
Figure 1. Establishment of the colorimetric IMSA assay for PCV3. Tubes 1–2, negative control. Tubes 3–4, 105 copies of PMD19T-capsid plasmid DNA.
To evaluate the specificity of the colorimetric IMSA, pathogens inducing similar clinical syndromes with PCV3, such as PCV2, PCV1, PRV GD-WH strain, HPS, SS, and APP, were used for DNA extraction. For multiple viral or bacterial DNAs, only the plasmid containing partial sequences of the PCV3 capsid gene could start the reaction and the reaction mixture displayed the yellow color (Figure 2). This manifested the good specificity of the colorimetric IMSA.
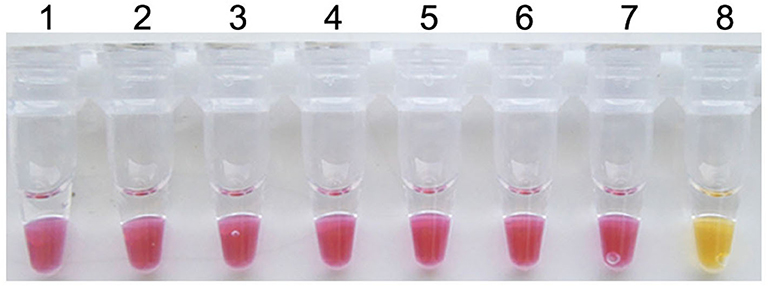
Figure 2. Specificity of the colorimetric IMSA assay for PCV3. Tubes 1–6, DNA of PCV2, PCV1, PRV GD-WH strain, HPS, SS, and APP. Tube 7, negative control. Tube 8, 105 copies of PMD19T-capsid plasmid DNA.
Ten-fold serial dilutions of plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies) were, respectively, used as DNA template to determine the detection limit of the colorimetric IMSA. Moreover, the results were compared with qPCR and PCR. In this study, as low as 10 copies of plasmid DNA can be detected by the colorimetric IMSA and qPCR, while the detection limit of PCR was 100 copies, which was 10 times lower than the colorimetric IMSA and qPCR (Figure 3).
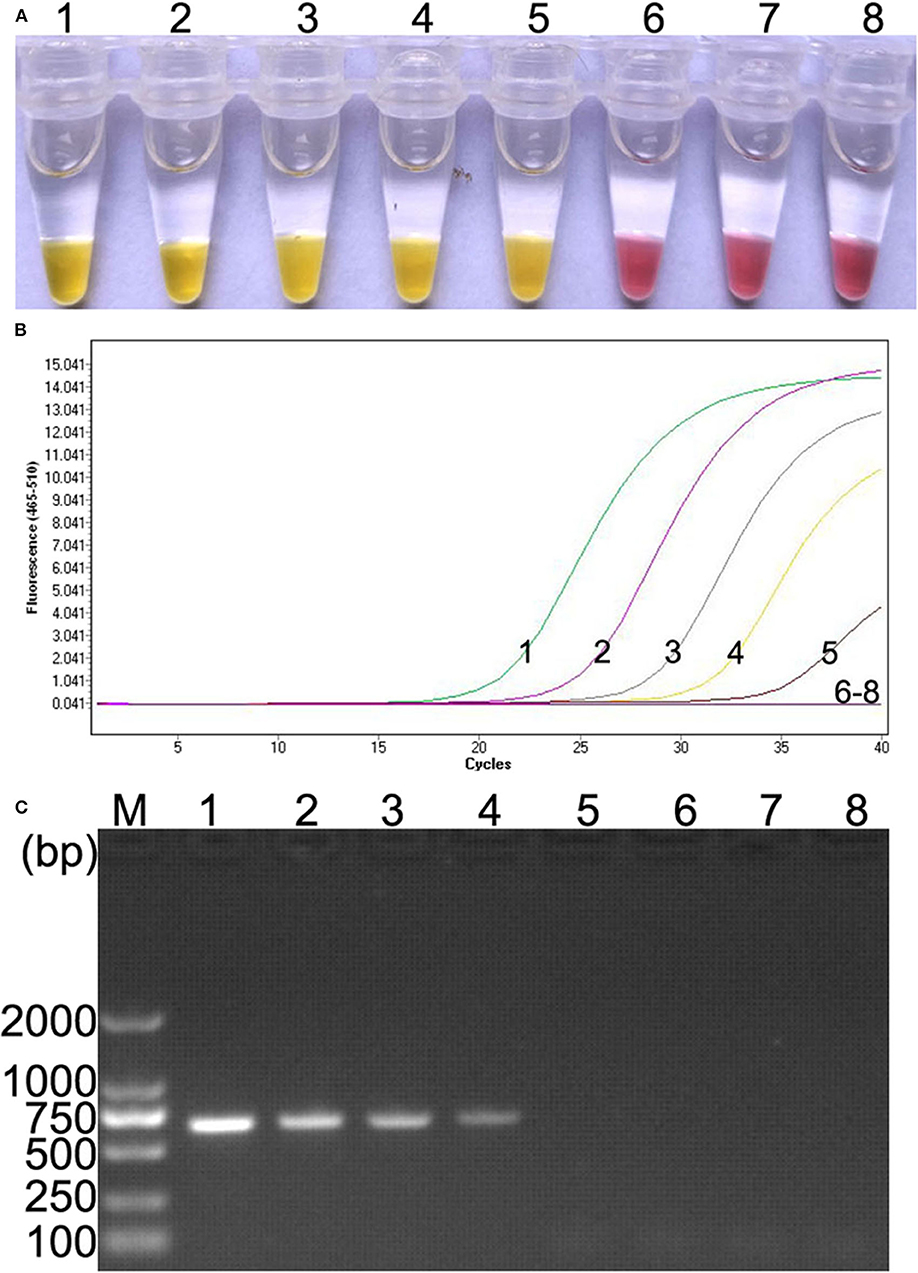
Figure 3. Comparison of sensitivity of the colorimetric IMSA with qPCR and PCR. (A) Sensitivity of the colorimetric IMSA. (B) Sensitivity of qPCR. (C) Sensitivity of PCR analyzed by agarose gel electrophoresis. Tubes or lanes 1–7, ten-fold serially diluted PMD19T-capsid plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies). Tube or lane 8: negative control. Lane M, 2,000-bp DNA marker.
When the colorimetric IMSA was evaluated by detecting ten-fold serial dilutions of pMD19T-capsid plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies) and the negative control for 3 different times, the same results were obtained for each dilution sample every time (Figure 4). Hence, the colorimetric IMSA showed stable detection reproducibility for samples of different concentrations.

Figure 4. Reproducibility analysis of the colorimetric IMSA. Tubes 1–7, ten-fold serially diluted PMD19T-capsid plasmid DNA (105, 104, 103, 102, 101, 100, and 10−1 copies). Tube 8: negative control.
Among 128 clinical samples, 35 positive results were detected by the PCR method. The positive rate was 27.3% (35/128). In addition to these 35 positive results, the colorimetric IMSA showed 15 positive results more than PCR, which were also positive for TaqMan-based qPCR (Table 2). The positive rate was 39.1% (50/128). To analyze the accuracy of IMSA in testing negative animals, 15 blood tissues sampled from SPF pigs were detected by using the colorimetric IMSA, qPCR, and PCR. Fifteen negative results were obtained for these 3 methods (data not shown).
PCV2, the main inducer of porcine circovirus-associated diseases (PCAD), severely damaged efficiency of swine production worldwide (1). PCV3, which was firstly identified in PCV2-negative pigs in 2016, causes cardiac pathology and multisystemic inflammation. In addition, PCV2-like syndromes including PDNS and reproductive failure are also associated with PCV3 infection (6). To analyze and control the epidemic situation of PCV3, it is necessary to establish accurate and high-throughput detection methods. Herein, a simple and rapid colorimetric IMSA using cresol red was developed to detect PCV3 for the first time.
In our study, the IMSA primers were designed according to the conserved region of the PCV3 capsid gene, which was the usual detective marker of PCR-based methods (7, 11). The primer design work was not as complicated as those of LAMP, because the IMSA primers consisted of seven basic primers specifically recognizing distinct regions of the target, which can be easily designed by using the software Primer Premier 5.0 (15). The results of colorimetric IMSA can be visually judged within 60 min, which was at least 60 min shorter than qPCR or PCR methods. However, the detection limit of the colorimetric IMSA is as low as 10 copies, which is consistent with qPCR and even 10 times more sensitive than PCR previously reported (7, 11). Moreover, the reproducibility analysis shows that the detection accuracy of the colorimetric IMSA is very stable for samples of different concentrations. In the whole detection process, the colorimetric IMSA only needs simple isothermal equipment to perform the reaction, but results of PCR or qPCR need to be analyzed by agarose gel electrophoresis or sophisticated thermal cycling apparatus. This makes it very suitable for point-of-care tests using the colorimetric IMSA assay. Compared with the LAMP assay using hydroxynaphthol blue for PCV3 detection, the color change from red to yellow of the reaction tube in this study is more easily to be distinguished with the naked eye than that of the color change from purple to sky blue (17). Hence, it is very convenient for the observers to read out and judge the results of the colorimetric IMSA.
When the colorimetric IMSA are used to detect clinical samples, all IMSA-positive samples can be exactly determined by TaqMan-based qPCR, which was proved to be an accurate method for PCV3 DNA detection. Moreover, the colorimetric IMSA displayed no false-positive results for detection of blood samples from SPF pigs. This further manifests its precise test ability. In our results, some IMSA-positive samples were negative for PCR detection. This may be attributed to the more sensitive detection limit of the colorimetric IMSA and qPCR than PCR.
In recent years, PCV3 was continuously identified in China and further surveillance studies showed that PCV3 are epidemic in a number of pig farms in many provinces (7, 10). In our study, the positive detection rate of the suspected clinical samples by colorimetric IMSA and qPCR is 39.1% (50/128), which was higher than the data in the previous study (11, 12). Our data offers new evidence that PCV3 was epidemic in sick pigs in China. As an emerging virus in 2016, some reports have manifested its connection with PDNS. Pathological lesions and PCV3-specific antigens are detected in various tissues and organs, including the blood, lung, heart, kidney, lymph nodes, and spleen (6, 18). Our results also showed that PCV3 is widely distributed in multiple tissues of the suspected infection pigs. What is more, we find that PCV3 DNA can be detected in the brain tissues of some sick pigs. This is consistent with the findings in some recent reports (19, 20).
In summary, the IMSA assay using cresol red was used for a rapid and sensitive detection of PCV3 for the first time. Not only was this method easy to be performed, but also its results were clearly distinguished with naked eyes. It is likely that the simple method was widely applied in the PCV3 detection in laboratories or rural areas.
The datasets generated for this study are available on request to the corresponding author.
This animal study was reviewed and approved by the ethical and ethics commission (Institute of Animal Health, Guangdong Academy of Agricultural Sciences, China). The license number was SYXK (Yue) 2011–0116. Moreover, sample collecting treatment in this study were performed in accordance with national and local laws and guidelines.
HG carried out the experiment design and drafted the manuscript. PC and DY prepared materials for the experiments. ZB participated in the experiments. SS, ZJ, and YL participated in the analysis of the data. CL, Y-AZ, and RC conceived the study. All authors read and approved the final manuscript.
This work was supported by grants from the Key Projects in the National Research and Development Program during the thirteenth Five-year Plan Period (No. 2018YFD0500804), the Natural Science Foundation of Guangdong, China (Nos. 2019A1515010757, 2017A030310074, and 2020A1515010475), the Science and Technology plan Program of Guangdong, China (Nos. 2017B020202002 and 2014A010107020), the 2018 Rural Revitalization Strategy Project (No. Guangdong Agriculture Planning 2018-54), the Science and Technology Program of Guangzhou, China (Nos. 201804010071 and 201607010380), the Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science (Nos. R2017YJ-YB2005 and R2018QD-094), and the Social Public Welfare Science and Technology Research Project of Zhongshan, China (No. 2018B1012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Allan G, Krakowka S, Ellis J, Charreyre C. Discovery and evolving history of two genetically related but phenotypically different viruses, porcine circoviruses 1 and 2. Virus Res. (2012) 164:4–9. doi: 10.1016/j.virusres.2011.09.013
2. Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. (1982) 295:64–6. doi: 10.1038/295064a0
3. Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. (1986) 91:271–6. doi: 10.1007/BF01314286
4. Segales J. Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. (2012) 164:10–9. doi: 10.1016/j.virusres.2011.10.007
5. Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. (2016) 13:184. doi: 10.1186/s12985-016-0642-z
6. Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2017) 91:e01879-16. doi: 10.1128/JVI.01879-16
7. Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. (2017) 64:703–8. doi: 10.1111/tbed.12638
8. Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. (2017) 207:178–80. doi: 10.1016/j.vetmic.2017.06.013
9. Stadejek T, Wozniak A, Milek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. (2017) 64:1350–3. doi: 10.1111/tbed.12672
10. Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, et al. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg Dis. (2018) 65:264–6. doi: 10.1111/tbed.12639
11. Wang J, Zhang Y, Liu L, Pang X, Yuan W. Development of a TaqMan-based real-time PCR assay for the specific detection of porcine circovirus 3. J Virol Methods. (2017) 248:177–80. doi: 10.1016/j.jviromet.2017.07.007
12. Wang J, Zhang Y, Zhang R, Han Q, Liu L, Li R, et al. Recombinase polymerase amplification assay for rapid detection of porcine circovirus 3. Mol Cell Probes. (2017) 36:58–61. doi: 10.1016/j.mcp.2017.09.001
13. Ding X, Nie K, Shi L, Zhang Y, Guan L, Zhang D, et al. Improved detection limit in rapid detection of human enterovirus 71 and coxsackievirus A16 by a novel reverse transcription-isothermal multiple-self-matching-initiated amplification assay. J Clin Microbiol. (2014) 52:1862–70. doi: 10.1128/JCM.03298-13
14. Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. (2015) 53:1–5. doi: 10.1007/s12275-015-4656-9
15. Yang Z, Liu W, Liang H, Wen R, Zhang Y. Development and evaluation of LAMP, CPA and IMSA methods for rapid detection of the AML1/ETO fusion gene in acute myeloid leukemia. Exp Ther Med. (2018) 16:3353–62. doi: 10.3892/etm.2018.6617
16. Tanner NA, Zhang Y, Evans TC Jr. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. (2015) 58:59–68. doi: 10.2144/000114253
17. Park YR, Kim HR, Kim SH, Lee KK, Lyoo YS, Yeo SG, et al. Loop-mediated isothermal amplification assay for the rapid and visual detection of novel porcine circovirus 3. J Virol Methods. (2018) 253:26–30. doi: 10.1016/j.jviromet.2017.12.006
18. Jiang H, Wang D, Wang J, Zhu S, She R, Ren X, et al. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol. (2019) 93:e02045-18. doi: 10.1128/JVI.02045-18
19. Chen GH, Mai KJ, Zhou L, Wu RT, Tang XY, Wu JL, et al. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg Dis. (2017) 64:1650–4. doi: 10.1111/tbed.12702
Keywords: PCV3, isothermal multiple-self-matching-initiated amplification (IMSA), detection, colorimetric assay, cresol red
Citation: Gou H, Bian Z, Cai R, Jiang Z, Song S, Li Y, Chu P, Yang D, Zang Y-A and Li C (2020) The Colorimetric Isothermal Multiple-Self-Matching-Initiated Amplification Using Cresol Red for Rapid and Sensitive Detection of Porcine Circovirus 3. Front. Vet. Sci. 7:407. doi: 10.3389/fvets.2020.00407
Received: 20 January 2020; Accepted: 08 June 2020;
Published: 04 August 2020.
Edited by:
Feng Li, South Dakota State University, United StatesReviewed by:
Francisco Rivera-Benítez, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), MexicoCopyright © 2020 Gou, Bian, Cai, Jiang, Song, Li, Chu, Yang, Zang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-An Zang, MTEzNDU4MDkwMEBxcS5jb20=; Chunling Li, bGNsY2FyZUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.