
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 July 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.00376
Canine superficial pyoderma (CSP) is a bacterial infection secondary to several skin diseases of the dog. Staphylococcus pseudintermedius, which is a commensal bacterium of the dog's skin, is the leading agent found in dogs affected by CSP, which can progress to deep pyoderma. It is also of clinical significance because S. pseudintermedius strains carry antimicrobial resistance genes, mainly the mecA gene. In this descriptive longitudinal study, molecular characterization of bacterial isolates from dogs affected by CSP was performed in addition to phenotyping, antimicrobial profiling, and assessment of resistance carriage status. Fifty dogs (24 females and 26 males) attending the CES University Veterinary Teaching Hospital were included in the study. CSP was confirmed according to clinical signs and cytological examination. Swabs were taken from active skin lesions for bacterial culture, and phenotyping and antimicrobial resistance profiles were assessed using API-Staph phenotyping and the Kirby–Bauer method, respectively. We also performed molecular detection and characterization of the mecA and nuc encoding gene of coagulase-positive Staphylococci. The mecA gene frequency was established by qPCR amplification of a 131bp gene fragment. Data were evaluated by descriptive statistics. Erythema, peeling, pruritus, and alopecia were the predominant symptoms (72, 56, and 46%, respectively). We isolated bacteria compatible with Staphylococcus species from all samples tested. API phenotyping showed 83.1 to 97.8% compatibility with S. pseudintermedius. PCR-genotyping resulted in 15, 3, and 1 isolates positive for S. pseudintermedius, S. aureus, and S. schleiferi, respectively. Isolated strains showed high susceptibility to Imipenem, Ampicillin/Sulbactam, and Rifampicin (100, 94, and 92%, respectively). The highest resistance was against Vancomycin and Trimethoprim/Sulfamethoxazole (98 and 74%, respectively). S. pseudintermedius, S. aureus, and S. schleiferi isolates were cloned and shared 96% sequence homology. Finally, we found 62% carriage status of the mecA gene in isolates of CSP patients, although only 36% of the isolates were methicillin-resistant. Identification of three Staphylococcus species causing CSP, high-level resistance against conventional antimicrobials, and carriage of the mecA gene highlight the importance of performing molecular characterization of bacteria causing dermatological conditions in dogs.
In dogs affected by several dermatological pathologies, including atopic dermatitis, endocrine-related dermatitis, and seborrheic dermatitis, superficial pyoderma is a frequent secondary complication (1) known as canine superficial pyoderma (CSP). Almost 90% of CSP symptoms are caused by gram-positive bacteria Staphylococcus pseudintermedius (2). Lesions of CSP are circumscribed to the stratum corneum of the skin and hair follicle. Most clinical symptoms arise from complications with ectoparasite infestation, hypersensitivity reactions, endocrinopathies, and impaired keratinization. Common lesions of CSP patients include papules, pustules, scabs, epidermal collarettes, and focal and multifocal alopecia (1), affecting the nasal, pharyngeal, and anal regions of the skin, which are frequently colonized by S. pseudintermedius (3). This commensal bacterium of the dog's skin is the most frequent within the Staphylococcus intermedius group, although it is implicated in bacterial infections of the skin (4, 5). Other bacterial species found in healthy skin of dogs are S. aureus, S. schleiferi (6), and Pseudomonas aeruginosa (7). Because S. pseudintermedius occasionally colonizes the human skin causing bacterial dermatitis, it has been proposed and evidenced as a zoonotic bacterium transmitted from the dog to their owner (8–12). One critical problem in clinical settings requiring confirmatory phenotyping of staphylococci causing CSP is the inability to differentiate between S. aureus and S. pseudintermedius because of the lack of differentiation tests between these two species (13), which can result in incorrect diagnoses. Diagnostic laboratories in our clinical settings are more accustomed to phenotyping S. aureus isolates; although S. pseudintermedius is the most frequent agent causing CSP, they report S. aureus (14). Although the Staph-API kit can diagnose 92.49% of Staphylococci species, the Staphylococcus species of several clinical samples cannot be diagnosed. For these reasons, it is necessary to perform molecular diagnosis for differentiating between S. aureus and S. pseudintermedius in samples from CSP patients. The clinical implications of the wrong diagnosis are that several S. pseudintermedius strains carry resistance to methicillin (methicillin-resistant Staphylococcus pseudintermedius) or multidrug resistance (MDR) genes (15), resulting in lack of response to antimicrobial treatments, impairment of clinical signs, and progression of CSP toward deep pyoderma (11, 12, 16).
Several authors reported the carriage status of antimicrobial resistance-encoding genes by the genus Staphylococcus, the most critical being the methicillin-resistance encoding mec gene (11, 12, 17, 18). The mec gene encodes an alternative penicillin-binding protein (PBP2a) with a high affinity to β-lactamic antibiotics. This gene is encoded by the staphylococcal chromosomal cassette mec (SCCmec), which is a motile genetic element composed of three components: the mec complex, consisting of five classes (A, B, C, D, and E); the ccr complex, consisting of eight allotypes (1 to 8); and the joining (J) region (19). The variable SCCmec types: I [1B], II [2A], III [3A], IV [2B], V [5C2], VI [4B], VII [5C1], and VIII, are the result of several combinations between the five classes of the mec complex and eight ccr allotypes (20, 21). The thermonuclease encoding nuc gene sequence allows the design of specific primers (22, 23) for the molecular identification of S. pseudintermedius by PCR methods.
Based on the discrepancy regarding the etiology of CSP in our clinical setting, our working hypothesis was that S. pseudintermedius instead of the commonly diagnosed S. aureus was the staphylococcal species causing CSP in our patients. The present study aimed to identify the main microorganism present in dogs with CSP, their antimicrobial resistance profile, if there was a correlation between phenotyping, API-identification, and their molecular characterization. Finally, we evaluate the carriage status for the mecA gene in clinical isolates from dogs with CSP.
This study consisted of a descriptive prospective longitudinal study, including a sample of dogs with clinical symptoms typical of CSP (n = 50) attending the Veterinary Teaching Hospital of University CES at Envigado, Colombia, between June 2016 and March 2017. The inclusion criteria for the dogs of the study were as follows: (1) clinical findings compatible with pyoderma; (2) presence of bacterial cocci by cytological exam and cocci growth under bacteriological culture; and (3) growth of colonies compatible with Staphylococcus species in blood agar and McConkey agar. The exclusion criteria were as follows: (1) cytology results indicating the presence of Bacilli; and (2) growth of cocci different from Staphylococcus species. All dogs were evaluated by the same researcher who performed the clinical exam, cytological smears from the most reliable skin lesions, and swabs sampling for bacterial culture. Samples were sent to the Veterinary Hospital Clinical Laboratory for evaluation and interpretation. Results of cytological smears and bacterial culture were available within the first hour and at 48 h after sampling, respectively. Dogs received the antimicrobial treatment according to the results of the antibiogram.
Fifty cytological samples positive for cocci were stained with Hemacolor® for confirmation of cocci. Swab samples from these 50 dogs were cultured at 37°C/18–24 h on Blood agar and McConkey agar. Diagnosis of Staphylococcus species was performed by the API Staph® kit (Reference # 20500, API Systems S.A., Montalieu-Vercieu, France) (24). Briefly, isolates were subjected to 20 biochemical tests added with a homogenous bacterial suspension at 0.5 McFarland turbidity and cultured at 37°C/24 h. The VP1, VP2, NIT1, NIT2, ZYM-A, and ZYM-B reagents were added, and microorganisms were identified using the analytical catalog provided by the manufacturer. The identification was performed based on the API numerical system (24). We only selected isolates with more than 80% Staphylococcal compatibility (Table 1).
Antimicrobial profiles of each isolated Staphylococcus were established by the Kirby–Bauer method on colonies cultured in a Mueller–Hinton agar (Becton Dickinson, Heidelberg, Germany), according to the CLSI protocol (25). The antimicrobials tested were selected based on the frequency of their use for the treatment of dermatological conditions in our setting (Table 2). The minimum inhibitory concentrations were as follows: (1) Cephalexin, 30 μg; (2) Oxacillin, 5 μg; (3) Amoxicillin Clavulanate 30 μg; (4) Ampicillin sulbactam, 20 μg; (5) Enrofloxacin, 5 μg; (6) Ciprofloxacin, 5 μg; (7) Doxycycline, 30 μg; (8) Rifampicin, 5 μg; (9) Trimethoprim sulfa, 25 μg; (10) Imipenem, 10 μg; (11) Vancomycin, 30 μg.
A sample of each isolate compatible with Staphylococcus species was harvested with a sterile loop and shed in sterile DNase-free 1.5-mL tubes containing 200-μL sterile 1 × PBS. The tube was centrifuged at 20,000 × g, and the pellet was resuspended in a 100-μL TE buffer (10 mM Tris, 1 mM EDTA), with 20-μL lysozyme. Samples were incubated at 37°C/30 min, followed by 55°C/10 min, and then centrifuged at 6,000 × g. After centrifugation, the supernatant was transferred to a new DNase-free 1.5-mL tube and was stored at −20°C until further use as a template for PCR. DNA quality was evaluated by determining the concentration through spectrophotometry using the Nanodrop 2000® its purity was analyzed by the relationship between absorbance at 260 and 280 nm.
Specific primers were designed for amplifying a segment of the nuc gene (a gene encoding the thermostable endonuclease of coagulase-positive Staphylococcus) by real-time PCR sybr green I. The gene sequence for each of the three Staphylococcus species considered in this study was aligned using the GenBank/NCBI (26) software for identification of sequence homologies shared among the nuc gene (27). We used the forward and reverse primers described by Sasaki et al. (28) for the in silico search. Sequences were aligned in Bioedit 7.0 software (Ibis Therapeutics, Carlsbad, CA, USA) to generate a consensus sequence that was used to obtain oligonucleotide sequences for different Staphylococcus species using the primer3plus software. The annealing temperature was set to 60°C, to obtain PCR products of 127, 99, and 115 bp for S. aureus, S. pseudintermedius, and S. schleiferi subspecies coagulans, respectively (Table 3).
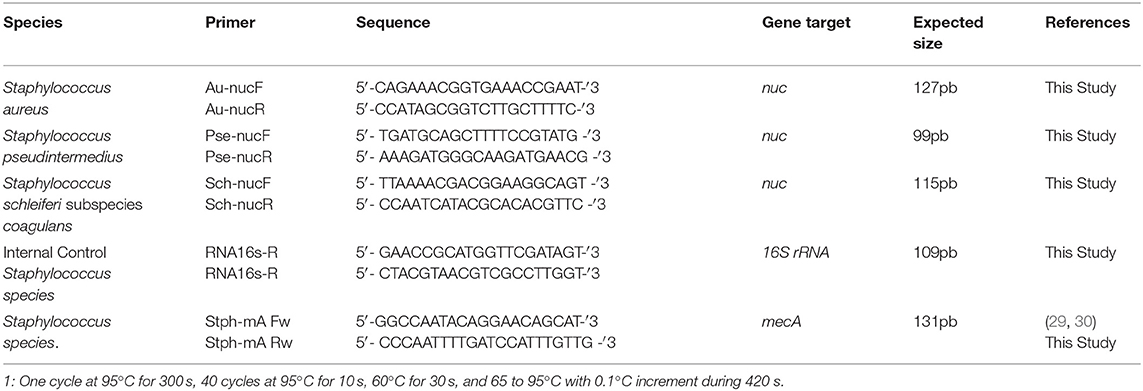
Table 3. Oligonucleotide sequence and conditions used for Staphylococci amplification of the nuc gene of bacteria isolated from dogs with superficial pyoderma.
In order to evaluate the presence or absence of the mecA gene in our samples, a 131-bp fragment of the gene was amplified using real-time PCR. The primers were designed based on an alignment of three Staphylococcus sequences available in GenBank/NCBI (S. aureus KP336394.1, S. epidermidis KP336396.1, and S. saprophyticus KP336397.1), following the same procedure indicated for species-specific primers. In order to confirm the identity of amplified fragments, products were sequenced and edited using the BioEdit software v7.1.316.
Amplification was achieved using the commercial kit QuantiNova® SYBR® Green PCR Kit (Qiagen®, Germantown, USA) in a total volume of 12 μL, containing 0.5 μM of each primer and 2-μL template DNA (average DNA amount was 66.6 ng/μL). PCR was run as follows: denaturation at 95°C/5min, 40 cycles at 95°C/10 s, and 60°C/30 s. Bound SYBR Green I fluorescence was measured after each amplification cycle. Finally, a Melting Curve Analysis was performed in a temperature gradient between 65 and 95°C. Melting temperature (MT) of each amplified segment was determined and compared to the predicted MT with uMelt® (melting curve predictions software—University of UTAH®).
Amplicons were visualized in a 2.5% agarose gel electrophoresis and run at 100 V for 45 min. Fragments corresponding to the expected size (Table 3) were cut and put into 1.5-mL tubes for purification using the QIAquick Gel Extraction Kit®, according to the manufacturer's instructions. Briefly, each gel fragment was added with 6 vol QG buffer and incubated at 50°C until gel dissolution. One volume of isopropanol was added, and the resulting solution was poured into the affinity column, washed, and eluted twice with a 50-μL EB buffer. The obtained DNA was stored at −20°C until further processing.
Amplicons obtained after real-time PCR were diluted at 12 μM and sequenced by Macrogen, Seoul, Korea, using the forward primer. The resulting sequences were analyzed by BLAST against the nr (GenBank/NCBI) database. For sequence analysis and phonogram construction, sequences in FASTA format were recorded in BioEdit v7.1.316 software (Ibis Therapeutics, Carlsbad, CA, USA) and aligned using the ClustalW program. Nucleotide Blast searches against the GenBank database in NCBI confirmed that each of the sequences generated has sequence identity scores of 96% or higher to each of the three Staphylococcal species of interest.
Data were consigned in Microsoft Excel (Microsoft Office); quantitative variables were analyzed by descriptive statistics, and the qualitative variables were represented in absolute and relative frequencies. To explore the possible association between clinical variables and the presence of S. pseudintermedius, crossed tables were performed using the IBM SPSS 22 statistical package to obtain the Pearson chi-square value. Significant P-value was 0.05, with 95% confidence interval.
The Ethics Committee of CES University approved this study. All procedures were performed according to the Code of Ethics for Animal Subject Experimentation for Veterinary Medicine (Ley 576, 2000 and Ley 84, 1989, Republic of Colombia).
Demographic data comprised 24 females and 26 males; age ranged from 7 to 168 months (average 58, and 3 and 61 months for females and males, respectively) (Figure 1A). The dog breeds are presented in Figure 1A, with most of the dogs belonging to a single breed (Figure 1B). All dogs exhibited at least one clinical sign compatible with CSP. The most frequent lesions were as follows: erythema, 36%; peeling, 28%; pruritus 23%; alopecia, 23%, epidermal collarettes, 18%; scabs, 15%; and pustules, 14% (Table 4). Thirty-three patients presented previous lesions mostly dermatological; 14 patients did not present systemic pathologies, and no data related to the presence or absence of associated skin diseases was obtained from three patients.
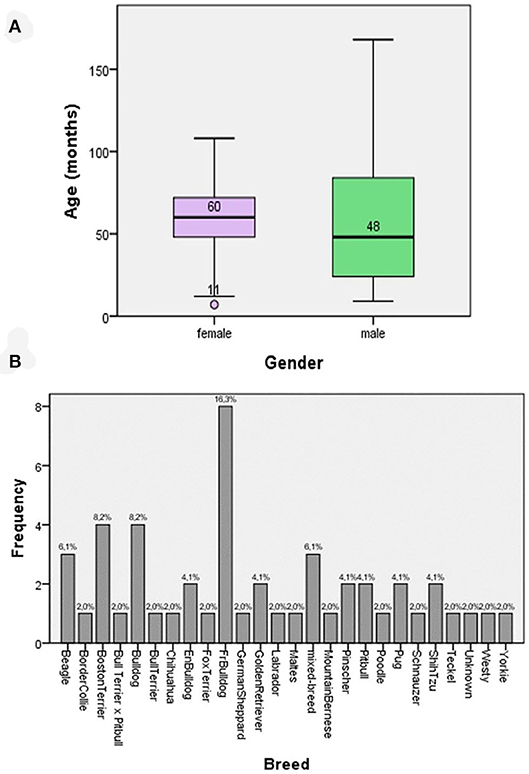
Figure 1. (A) Box and whisker plots for the age of the patients according to gender. (B) The frequency of breeds of patients suffering from superficial pyoderma included in the study.
All 50 dogs tested positive for Staphylococcus species and were evaluated for the antimicrobial profile. Within these, 88% were positive for colonies compatible with Staphylococcus species within the inhibition halo, indicating the presence of at least two species of the genus Staphylococcus in these isolates. These samples were further processed by real-time PCR resulting in the growth of S. pseudintermedius (Figure 2) and S. schleiferi (Figure 3). The most common antimicrobial effective against the isolated strains were Imipenem, Ampicillin/Sulbactam, and Rifampicin, exhibiting 100, 94, and 92% of sensitivity, respectively (Table 3). The higher level of resistance was 98 and 74% for Vancomycin and Trimethoprim/Sulfamethoxazole, respectively (Table 3). We found 36% isolates resistant to Oxacillin (Figure 4). The S. pseudintermedius PCR results were significantly associated with females (P < 0.01) (Table 5). API phenotyping showed 83.1 to 97.8% of isolates compatible with S. pseudintermedius. PCR-genotyping showed 15, 3, and 1 isolates positive for S. pseudintermedius, the three Staphylococcal species, and S. schleiferi, respectively.
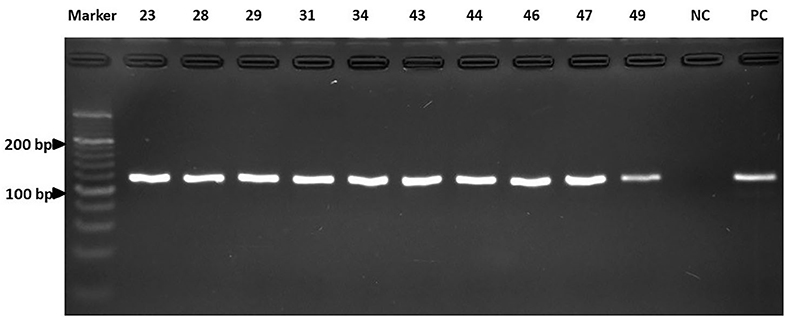
Figure 2. The results of the real-time PCR assay. Detection of the nuc gene of S. pseudintermedius by electrophoresis. Line A: Marker. Lines B to K: Isolates. NC, Negative control; PC, Positive control. Electrophoresis was run as indicated in Materials and Methods.
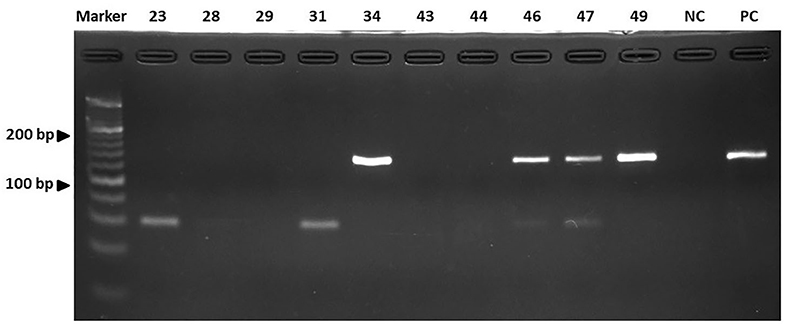
Figure 3. The results of the real-time PCR assay. Detection of the nuc gene of S. schleiferi by electrophoresis. Line A: Marker. Lines B to K: Isolates. NC, Negative control; PC, Positive control. Electrophoresis was run as indicated in Materials and Methods.
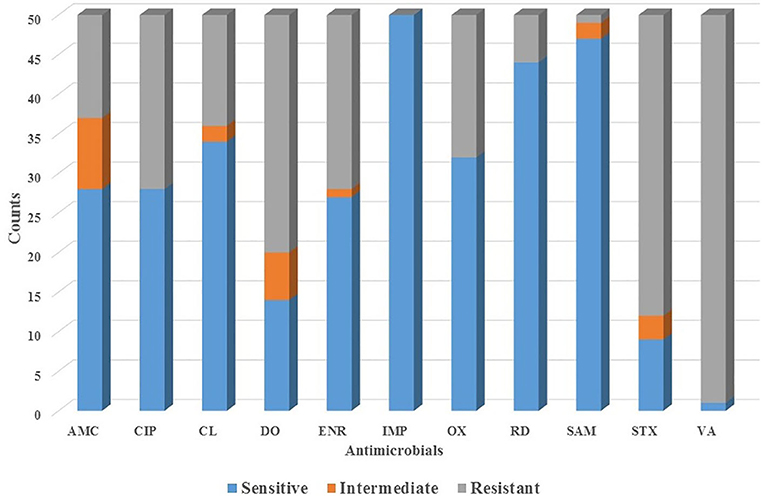
Figure 4. The profile of antimicrobial sensibility of Staphylococcal isolates obtained from canine patients suffering from CSP. AMC, Amoxicillin/Clavulanic acid; CIP, Ciprofloxacin; CL, Cephalexin; DO, Doxycycline; ENR, Enrofloxacin; IPM, Imipenem; OX, Oxacillin; SAM, Ampicillin/Sulbactam; VA, Vancomycin; RM, Rifampicin; TMX, Trimethoprim/Sulfonamide.
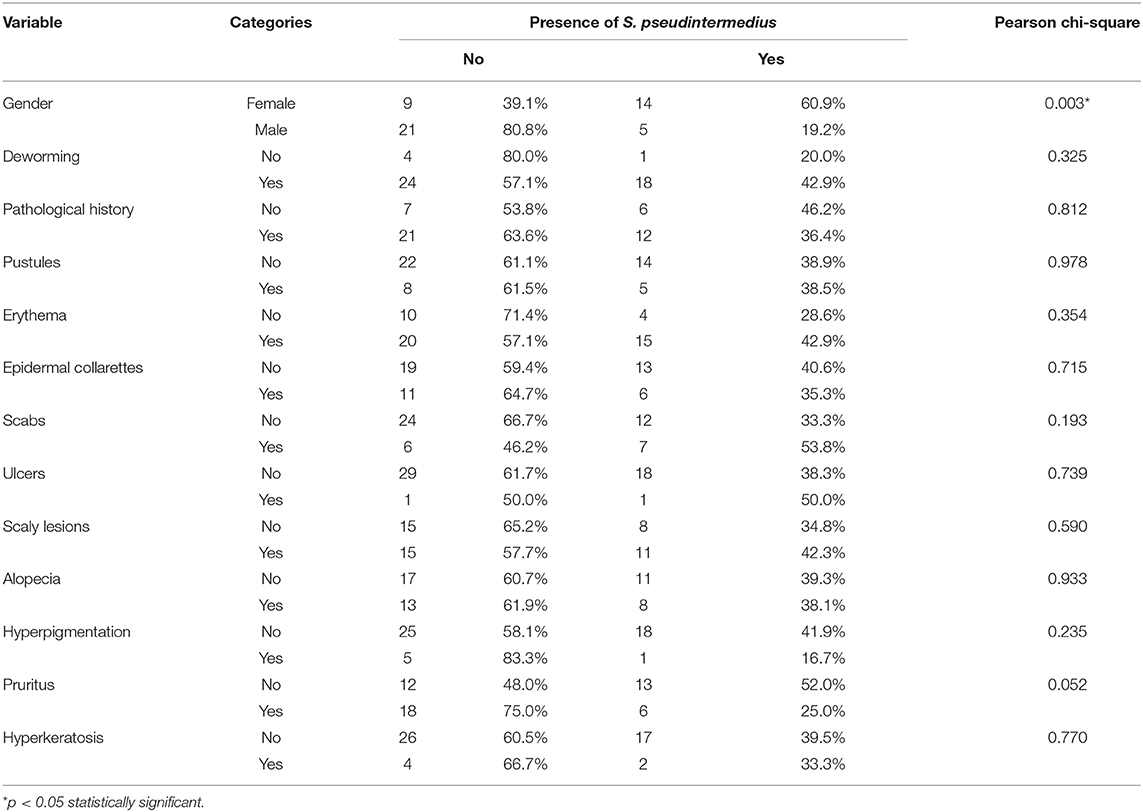
Table 5. Relationship between qualitative variables and the confirmed presence of Staphylococcus pseudintermedius.
The fragment corresponding to the mec gene was amplified in 62% of all isolates in this study. From the total number of isolates compatible with S. pseudintermedius by PCR, 73.6% harbored the mec gene, although without statistically significant differences (P = 0.237, chi-squared test) (Figures 5A,B). Ten amplification fragments of the mec gene were sequenced, exhibiting 98.8% homology to Staphylococcus sequences, particularly to a region of the mecA gene. The sequences were aligned and compared using the ClustalW method, resulting in a complementary sequence to the mecA gene of the S. aureus strain IQH (accession number MK214488.1).
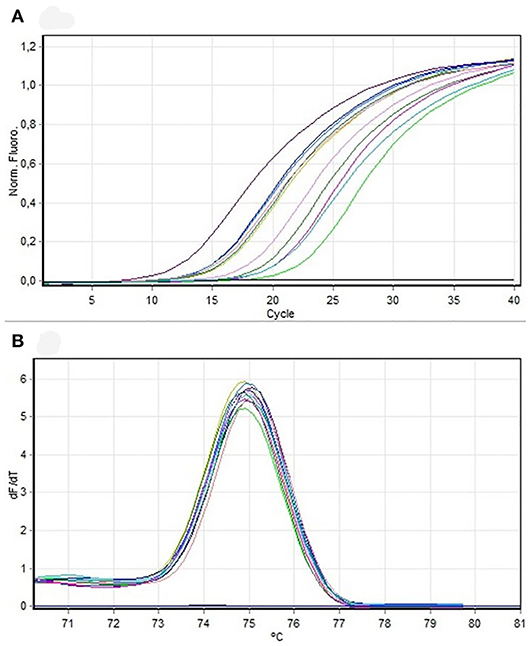
Figure 5. (A) Amplification curve. PCR mecA samples to be sequenced. (B) Dissociation curve. PCR mecA samples to be sequenced.
In this study for the first time in Latin America, the concomitant infection with two or three Staphylococcal species in dogs with CSP was reported, whereas the diagnosis of infection with one species (S. pseudintermedius) was reported in the remaining cases. Most previous studies reported infection with S. aureus, mainly. The finding of S. pseudintermedius in dogs with CSP provides the criteria for a more precise definition of the etiology of CSP and its corresponding treatment. S. aureus is the most frequent commensal found in humans, particularly in the nasal cavity of 30% healthy individuals. Within coagulase-negative staphylococci, S. epidermidis is most commonly isolated from nostrils, perineum, inguinal, axillary, and interdigital skin. In dogs, S. pseudintermedius is considered the most common coagulase-positive commensal isolated from 37 to 92% of healthy dogs, whereas S. aureus is only isolated in 4.3 to 12% of healthy dogs (31). Superficial pyoderma and superficial folliculitis are current entities in canine dermatology, and S. pseudintermedius is frequently isolated in these clinical entities. CSP is not a life-threatening condition, although it contributes to impair health and well-being because of pruritus and inflammation; it is often recurrent, resulting in repeated treatments (32). Our clinical findings are similar to the report by Khoshnegah et al. (33). The lack of association with the breed is similar to this previous report (33). Our methods do not support a possible explanation for the association between sex and pruritus.
Bacterial culture and antimicrobial sensitivity tests must be established on a routine basis for the diagnosis of CSP. In scientific literature, there is growing evidence on the resistance of S. pseudintermedius to methicillin [methicillin-resistant S. pseudintermedius (MRSP)] and other antimicrobials of beta-lactamase group, which is related to the mecA (methicillin-resistance) gene (32, 34, 35), which complicate the treatment of insidious infections such as those occurring in dogs suffering from deep pyoderma. In our study, 100% of dogs with CSP tested positive for Staphylococcus species, suggesting a high incidence of this genus in dogs with dermatologic pathologies.
The high percentage of resistance of Staphylococcus species isolates to Trimethoprim-Sulfamethoxazole, Vancomycin, Doxycycline, Ciprofloxacin, Enrofloxacin, and Amoxicillin/Clavulanate agrees with previous studies reporting the resistance of Staphylococcus species to amikacin (36), lincosamides (37), clarithromycin (38), cephalexin (39), and quinolones (40, 41). We also found a 36% resistance to Oxacillin, suggesting that these isolates probably have the mecA gene. Other studies had reported resistance to methicillin (17, 35, 42, 43). On the contrary, we found a high percentage of isolates exhibiting susceptibility to Imipenem, Ampicillin/Sulbactam, Rifampicin, and Cephalexin (Table 2). Other studies reported variable degrees of susceptibility of this genus to synthetic thiazole compounds (44), cationic peptides (45), naturals antimicrobial peptides and cathelicidin (46), orbifloxacin (47), marbofloxacin (48), pradofloxacin (49), and phosphomicin (50).
In most patients in our clinical setting, we have found Staphylococcus species isolates resistant to methicillin and other antimicrobials commonly used as therapies in dermatitis caused by Staphylococcus species. The persistence of resistant isolates limits the effectiveness of antimicrobials frequently used, as indicated by Cain (51). Accordingly, performing bacterial culture and susceptibility tests becomes mandatory for patients with recurrent clinical sings. Therefore, Cain (51) proposed to perform a bacterial culture and antimicrobial resistance test for all patients meeting the following criteria:
1. They are affected by recurrent pyoderma.
2. Suffering from skin infections that have not responded to appropriate empirical therapy.
3. Present clinical lesions typical of deep pyoderma, including nodules, hemorrhagic blisters, fistulas, and boils.
4. Cytological exam evidencing mixed infection by bacilli and cocci.
5. Repeated antimicrobial therapy predisposing to colonization and subsequent infection by methicillin-resistant strains.
6. Previous infection with methicillin-resistant Staphylococci, because colonization can persist for prolonged periods.
We propose that all those patients that meet the criteria mentioned above must undergo bacterial culture and antibiogram, which will help establish appropriate therapies, shorten diagnosis times, and shorten patients' recovery time. This procedure will be of critical importance for the outcome of the patients.
One and three of our dogs were concomitantly infected with three and two Staphylococcal species, S. aureus, S. pseudintermedius, and S. schleiferi, respectively. Larsen et al. (52) found that samples of dogs from several lesions resulted in the isolation of genetically different strains of Staphylococcus species taken from different regions of the skin, and up to four strains isolated of the same dog showed variable resistance profiles, requiring treatment with several topical, and systemic antimicrobials (52).
For achieving more accurate diagnoses of staphylococcal infections of the skin of dogs, it is essential to combine more advanced protocols such as API and PCR (17, 35). In our study, we diagnosed S. pseudintermedius in 44 out of 50 isolates by API, whereas PCR resulted in only 15 sample positives for S. pseudintermedius. However, this may be attributable to recent changes in Staphylococcus nomenclature that could account for erroneous identifications: most isolates identified as S. intermedius before 2005 now are probably S. pseudintermedius or S. delphini (1).
Molecular diagnosis identifying the 5S, 16S, and 23S ribosomal subunits, and their intergenic spaces, is one of the most useful tools in bacterial taxonomy (53, 54). PCR-based diagnosis and genome sequencing have become necessary for the study of bacterial epidemiology. Accordingly, it has been shown that S. pseudintermedius exhibits high genetic variability and an epidemiologic structure, suggesting the occurrence of recombination events rather than mutations. Most of the MRSP strains that arose during the last 10 years are genetically distinct regarding their antimicrobial resistance profiles, SCCmec content, and geographic distribution (55).
In a study performed on Staphylococcus isolated from dogs in New Zealand, the mecA gene was isolated in 38% out of 176 presumptive S. pseudintermedius isolates (15). On the contrary, in our study, 73.6% (19 cases) of isolates corresponding to S. pseudintermedius were presumptive to carry the mec gene. Interestingly, it has been found that S. pseudintermedius from companion animals was the causative agent of infection in humans (9, 11, 12, 16, 56), highlighting the importance of considering those isolates found in the present study as potential zoonotic agents. On the other hand, the SCCmec is a complex element. Although in the present study only the mec fragment was evaluated, its presence in our S. pseudintermedius isolates suggests the circulation of Staphylococcus strains causing infection in our population of dermatologic patients, which represents the first report in our country.
In conclusion, it is common to observe increasing cases of antimicrobial resistance of staphylococcal species to the most common antimicrobials used in the current veterinary practice, as evidenced in our study for vancomycin trimethoprim/sulfonamide and methicillin. The use of API and real-time PCR for the identification of S. pseudintermedius, S. aureus, and S. schleiferi is a valuable tool for achieving better results regarding antibiotic choice and clinical outcome. The presence of nuc and mecA genes in Staphylococcal isolates from dogs with dermatological pathologies warns about their potential zoonotic behavior and must be considered for preventive strategies as potential causative agents of antimicrobial resistance-gene transmission to the human population. Finally, we provide evidence that S. pseudintermedius was the most prevalent Staphylococcus species in our setting, contrary to the typical report of S. aureus. From the clinical point of view, to achieve a precise diagnosis of the current disease is critical for the establishment of the most appropriate therapeutic protocol. In the context of the present study, performing accurate bacterial culture and antimicrobial profiles and the molecular identification of the presence of Methicillin resistance genes in Staphylococcal isolates provide a comprehensive approach that benefits clinical diagnosis, treatment, and outcome of patients suffering CSP.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
The animal study was reviewed and approved by Institutional Board on Animal Subject Experimentation of CES University (CICUA) Minute # 19 from April 26, 2016. Written informed consent was obtained from the owners for the participation of their animals in this study.
MG-D designed the study, performed patients' selection and their clinical exam for verifying inclusion and exclusion criteria, sampled the dogs, analyzed the data, wrote the manuscript, and prepared its final version. HC performed bacterial culture, cytological evaluation, and API identification, and contributed to data analysis and manuscript preparation. DC-E performed DNA extraction and molecular protocols, and contributed to data analysis and manuscript preparation. DC-C performed molecular experiments, sequencing of the genes, and data analysis and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by CES University (Research and Innovation Office).
DC-C was employed by Laboratorio de Genética Animal Mascolab S.A, Colombia.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Special thanks to Maria C. Arango, Alejandra Correa-Gomez, Carolina Restrepo-Llanos, and Natalia A. Cadavid-Hincapie for technical support.
2. Riley MC, Perreten V, Bemis DA, Kania SA. Complete genome sequences of three important methicillin-resistant clinical isolates of Staphylococcus pseudintermedius. Genome Announc. (2016) 4:e01194–16. doi: 10.1128/genomeA.01194-16
3. Rubin JE, Chirino-Trejo M. Prevalence, sites of colonization, and antimicrobial resistance among Staphylococcus pseudintermedius isolated from healthy dogs in Saskatoon, Canada. J Vet Diagn Invest. (2011) 23:351–4. doi: 10.1177/104063871102300227
4. Bannoehr J, Guardabassi L. Staphylococcus pseudintermedius in the dog: taxonomy, diagnostics, ecology, epidemiology and pathogenicity. Vet Dermatol. (2012) 23:e51–2. doi: 10.1111/j.1365-3164.2012.01046.x
5. Devriese LA, Hermans K, Baele M, Haesebrouck F. Staphylococcus pseudintermedius versus Staphylococcus intermedius. Vet Microbiol. (2009) 133:206–7. doi: 10.1016/j.vetmic.2008.06.002
6. Ravens P, Vogelnest L, Ewen E, Bosward K, Norris J. Canine superficial bacterial pyoderma: evaluation of skin surface sampling methods and antimicrobial susceptibility of causal Staphylococcus isolates. Aust Vet J. (2014) 92:149–55. doi: 10.1111/avj.12176
7. Hillier A, Alcorn JR, Cole LK, Kowalski JJ. Pyoderma caused by Pseudomonas aeruginosa infection in dogs: 20 cases. Vet Dermatol. (2006) 17:432–9. doi: 10.1111/j.1365-3164.2006.00550.x
8. Novick RP, Morse SI. In vivo transmission of drug resistance factors between strains of Staphylococcus aureus. J Exp Med. (1967) 125:45–59. doi: 10.1084/jem.125.1.45
9. Guardabassi L, Loeber ME, Jacobson A. Transmission of multiple antimicrobial-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet Microbiol. (2004) 98:23–7. doi: 10.1016/j.vetmic.2003.09.021
10. Loeffler A, Pfeiffer DU, Lloyd DH, Smith H, Soares-Magalhaes R, Lindsay JA. Meticillin-resistant Staphylococcus aureus carriage in UK veterinary staff and owners of infected pets: new risk groups. J Hosp Infect. (2010) 74:282–8. doi: 10.1016/j.jhin.2009.09.020
11. Gagetti P, Wattam AR, Giacoboni G, De Paulis A, Bertona E, Corso A, et al. Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina. BMC Vet Res. (2019) 15:264. doi: 10.1186/s12917-019-1990-x
12. Meroni G, Soares Filipe JF, Drago L, Martino PA. Investigation on antibiotic-resistance, biofilm formation and virulence factors in multi drug resistant and non multi drug resistant Staphylococcus pseudintermedius. Microorganisms. (2019) 7:702. doi: 10.3390/microorganisms7120702
13. Feng Y, Tian W, Lin D, Luo Q, Zhou Y, Yang T, et al. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius in pets from South China. Vet Microbiol. (2012) 160:517–24. doi: 10.1016/j.vetmic.2012.06.015
14. Paul NC, Moodley A, Ghibaudo G, Guardabassi L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission: methicilln-resistant Staphylococcus pseudintermedius in veterinarians. Zoonoses Public Health. (2011) 58:533–9. doi: 10.1111/j.1863-2378.2011.01398.x
15. Nisa S, Bercker C, Midwinter AC, Bruce I, Graham CF, Venter P, et al. Combining MALDI-TOF and genomics in the study of methicillin resistant and multidrug resistant Staphylococcus pseudintermedius in New Zealand. Sci Rep. (2019) 9:1271. doi: 10.1038/s41598-018-37503-9
16. Brooks MR, Padilla-Vélez L, Khan TA, Qureshi AA, Pieper JB, Maddox CW, et al. Prophage-mediated disruption of genetic competence in Staphylococcus pseudintermedius. mSystems. (2020) 5:e00684–19. doi: 10.1128/mSystems.00684-19
17. Duim B, Verstappen KMHW, Kalupahana RS, Ranathunga L, Fluit AC, Wagenaar JA. Methicillin-resistant Staphylococcus pseudintermedius among dogs in the description of novel SCCmec variants. Vet Microbiol. (2018) 213:136–41. doi: 10.1016/j.vetmic.2017.11.022
18. Krapf M, Müller E, Reissig A, Slickers P, Braun SD, Müller E, et al. Molecular characterisation of methicillin-resistant Staphylococcus pseudintermedius from dogs and the description of their SCCmec elements. Vet Microbiol. (2019) 233:196–203. doi: 10.1016/j.vetmic.2019.04.002
19. Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, et al. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. (2016) 101:56–67. doi: 10.1016/j.micpath.2016.10.028
20. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. (2001) 45:1323–36. doi: 10.1128/AAC.45.5.1323-1336.2001
21. Zhang X, Luo T, Keysary A, Baneth G, Miyashiro S, Strenger C, et al. Genetic and antigenic diversities of major immunoreactive proteins in globally distributed Ehrlichia canis strains. Clin Vaccine Immunol. (2008) 15:1080–8. doi: 10.1128/CVI.00482-07
22. Baron F, Cochet M-F, Pellerin J, Ben Zakour N, Lebon A, Navarro A, et al. Development of a PCR test to differentiate between Staphylococcus aureus and Staphylococcus intermedius. J Food Prot. (2004) 67:2302–5. doi: 10.4315/0362-028X-67.10.2302
23. Soedarmanto I, Kanbar T, Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, et al. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res Vet Sci. (2011) 91:e25–7. doi: 10.1016/j.rvsc.2011.01.027
24. bioMérieux Inc. API Staph. Rf 20 500. 07468K - es - 2009/11. IVD. (2009) Available online at: https://amyd.quimica.unam.mx/pluginfile.php/9914/mod_resource/content/1/API.STAPH.instructivo%20REF_20500.pdf (accessed May 26, 2020).
25. Cockerill FR, Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute (2012).
26. Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. (1997) 25:3389–402. doi: 10.1093/nar/25.17.3389
27. Gandra EA, Fernandez MA, Silva JA, da Silva WP. Standardization of a multiplex PCR for the identification of coagulase-positive Staphylococcus. Ciência Tecnologia Alimentos. (2011) 31:946–9. doi: 10.1590/S0101-20612011000400019
28. Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. (2010) 48:765–9. doi: 10.1128/JCM.01232-09
29. Giles T, Yon L, Hannant D, Barrow P, Abu-Median AB. Corrigendum to “Development of a DNA-based microarray for the detection of zoonotic pathogens in rodent species”. Mol Cell Probes. (2016) 30:277. doi: 10.1016/j.mcp.2016.07.007
30. Giles T, Yon L, de Bree F, Bossers A, Hannant D, Barrow P, et al. Development of a DNA-based microarray for the detection of zoonotic pathogens in rodent species. Mol Cell Probes. (2015) 29:427–37. doi: 10.1016/j.mcp.2015.07.005
31. Schmidt VM, Williams NJ, Pinchbeck G, Corless CE, Shaw S, McEwan N, et al. Antimicrobial resistance and characterisation of staphylococci isolated from healthy labrador retrievers in the United Kingdom. BMC Vet Res. (2014) 10:17. doi: 10.1186/1746-6148-10-17
32. Loeffler A, Lloyd DH. What has changed in canine pyoderma? A narrative review. Vet J. (2018) 235:73–82. doi: 10.1016/j.tvjl.2018.04.002
33. Khoshnegah J, Movassaghi AR, Rad M. Survey of dermatological conditions in a population of domestic dogs in Mashhad, northeast of Iran (2007-2011). Vet Res Forum. (2013) 4:99–103.
34. Grönthal T, Eklund M, Thomson K, Piiparinen H, Sironen T, Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J Antimicrob Chemother. (2017) 72:1021–30. doi: 10.1093/jac/dkx086
35. Kang JH, Chung TH, Hwang CY. Clonal distribution of methicillin-resistant Staphylococcus pseudintermedius isolates from skin infection of dogs in Korea. Vet Microbiol. (2017) 210:32–7. doi: 10.1016/j.vetmic.2017.08.017
36. Gold RM, Cohen ND, Lawhon SD. Amikacin resistance in Staphylococcus pseudintermedius isolated from dogs. J Clin Microbiol. (2014) 52:3641–6. doi: 10.1128/JCM.01253-14
37. Larsen R, Boysen L, Berg J, Guardabassi L, Damborg P. Lincosamide resistance is less frequent in Denmark in Staphylococcus pseudintermedius from first-time canine superficial pyoderma compared with skin isolates from clinical samples with unknown clinical background. Vet Dermatol. (2015) 26:e43–4. doi: 10.1111/vde.12209
38. Dicicco M, Neethirajan S, Singh A, Weese JS. Efficacy of clarithromycin on biofilm formation of methicillin-resistant Staphylococcus pseudintermedius. BMC Vet Res. (2012) 8:225. doi: 10.1186/1746-6148-8-225
39. Papich MG, Lindeman C. Cephalexin susceptibility breakpoint for veterinary isolates: clinical laboratory standards institute revision. J Vet Diagn Invest. (2018) 30:113–20. doi: 10.1177/1040638717742434
40. Kang MH, Chae MJ, Yoon JW, Lee SY, Yoo JH, Park HM. Resistance to fluoroquinolones and methicillin in ophthalmic isolates of Staphylococcus pseudintermedius from companion animals. Can Vet J. (2014) 55:678–82.
41. Schink A-K, Kadlec K, Hauschild T, Brenner Michael G, Dörner JC, Ludwig C, et al. Susceptibility of canine and feline bacterial pathogens to pradofloxacin and comparison with other fluoroquinolones approved for companion animals. Vet Microbiol. (2013) 162:119–26. doi: 10.1016/j.vetmic.2012.08.001
42. Davis JA, Jackson CR, Fedorka-Cray PJ, Barrett JB, Brousse JH, Gustafson J, et al. Carriage of methicillin-resistant staphylococci by healthy companion animals in the US. Lett Appl Microbiol. (2014) 59:1–8. doi: 10.1111/lam.12254
43. Lehner G, Linek M, Bond R, Lloyd DH, Prenger-Berninghoff E, Thom N, et al. Case–control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet Microbiol. (2014) 168:154–60. doi: 10.1016/j.vetmic.2013.10.023
44. Mohammad H, Reddy PVN, Monteleone D, Mayhoub AS, Cushman M, Hammac GK, et al. Antibacterial characterization of novel synthetic thiazole compounds against methicillin-resistant Staphylococcus pseudintermedius. PLoS ONE. (2015) 10:e0130385. doi: 10.1371/journal.pone.0130385
45. Mohamed MF, Hammac GK, Guptill L, Seleem MN. Antibacterial activity of novel cationic peptides against clinical isolates of multi-drug resistant Staphylococcus pseudintermedius from infected dogs. PLoS ONE. (2014) 9:e116259. doi: 10.1371/journal.pone.0116259
46. Santoro D, Maddox CW. Canine antimicrobial peptides are effective against resistant bacteria and yeasts. Vet Dermatol. (2014) 25:35–e12. doi: 10.1111/vde.12091
47. Scott DW, Peters J, Miller WH. Efficacy of orbifloxacin tablets for the treatment of superficial and deep pyoderma due to Staphylococcus intermedius infection in dogs. Can Vet J. (2006) 47:999–1002.
48. Paradis M, Abbey L, Baker B, Coyne M, Hannigan M, Joffe D, et al. Evaluation of the clinical efficacy of marbofloxacin (Zeniquin) tablets for the treatment of canine pyoderma: an open clinical trial. Vet Dermatol. (2001) 12:163–9. doi: 10.1046/j.1365-3164.2001.00195.x
49. Restrepo C, Ihrke PJ, White SD, Spiegel IB, Affolter VK. Evaluation of the clinical efficacy of pradofloxacin tablets for the treatment of canine pyoderma. J Am Anim Hosp Assoc. (2010) 46:301–11. doi: 10.5326/0460301
50. DiCicco M, Weese S, Neethirajan S, Rousseau J, Singh A. Fosfomycin susceptibility of canine methicillin-resistant Staphylococcus pseudintermedius isolates. Res Vet Sci. (2014) 96:251–3. doi: 10.1016/j.rvsc.2014.02.004
51. Cain CL. Antimicrobial resistance in staphylococci in small animals. Vet Clin North Am Small Anim Pract. (2013) 43:19–40. doi: 10.1016/j.cvsm.2012.09.003
52. Larsen RF, Boysen L, Jessen LR, Guardabassi L, Damborg P. Diversity of Staphylococcus pseudintermedius in carriage sites and skin lesions of dogs with superficial bacterial folliculitis: potential implications for diagnostic testing and therapy. Vet Dermatol. (2018) 29:291–e100. doi: 10.1111/vde.12549
53. Mühlhauser M, Rivas L. Laboratorio de microbiología: conocimientos básicos para un clínico. Revista Médica Clínica Las Condes. (2014) 25:569–79. doi: 10.1016/S0716-8640(14)70072-0
54. Koneman EW, Procop GW. Koneman Diagnóstico Microbiológico: Texto y Atlas en Color. Barcelona. Wolters Kluwer. (2018).
55. Pires dos Santos T, Damborg P, Moodley A, Guardabassi L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: inference of population structure from multilocus sequence typing data. Front Microbiol. (2016) 7:1599. doi: 10.3389/fmicb.2016.01599
Keywords: bacterial phenotyping, dogs, mecA gene, Staphylococcus aureus, Staphylococcus pseudintermedius, Staphylococcus schleiferi
Citation: González-Domínguez MS, Carvajal HD, Calle-Echeverri DA and Chinchilla-Cárdenas D (2020) Molecular Detection and Characterization of the mecA and nuc Genes From Staphylococcus Species (S. aureus, S. pseudintermedius, and S. schleiferi) Isolated From Dogs Suffering Superficial Pyoderma and Their Antimicrobial Resistance Profiles. Front. Vet. Sci. 7:376. doi: 10.3389/fvets.2020.00376
Received: 17 January 2020; Accepted: 28 May 2020;
Published: 23 July 2020.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Md.Tanvir Rahman, Bangladesh Agricultural University, BangladeshCopyright © 2020 González-Domínguez, Carvajal, Calle-Echeverri and Chinchilla-Cárdenas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: María S. González-Domínguez, bWdvbnphbGV6QGNlcy5lZHUuY28=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.