
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 18 December 2019
Sec. Veterinary Infectious Diseases
Volume 6 - 2019 | https://doi.org/10.3389/fvets.2019.00447
Salmonella Enteritidis is a major cause of salmonellosis worldwide and more than 80% of outbreaks investigated in Europe have been associated with the consumption of poorly cooked eggs or foods containing raw eggs. Vaccination has been proven to be one of the most important measures to control Salmonella Enteritidis infections in poultry farms as it can decrease colonization of the reproductive organs and intestinal tract of laying hens, thereby reducing egg contamination. Differentiation of live vaccine from field or wild type S. Enteritidis isolates in poultry is essential for monitoring of veterinary isolates and targetting control actions. Due to decreasing costs, whole genome sequencing (WGS) is becoming a key tool for characterization of Salmonella isolates, including vaccine strains. Using WGS we described the genetic changes in the live attenuated Salmovac 440 and AviPro SALMONELLA VAC E vaccine strains and developed a method for differentiation from the wildtype S. Enteritidis strains. SNP analysis confirmed that streptomycin resistance was associated with a Lys43Arg missense mutation in the rpsL gene whilst 3 missense mutations in acrB and 1 missense mutation in acrA confer erythromycin sensitivity in AviPro SALMONELLA VAC E. Further mutations Arg242His in purK and Gly236Arg in the hisB gene were related to adenine and histidine dependencies in Salmovac 440. Unique SNPs were used to construct a database of variants for differentiation of vaccine from the wildtype isolates. Two fragments from each vaccine were represented in the database to ensure high accuracy. Each of the two selected Salmovac 440 fragments differed by 6 SNPs from the wildtype and the AviPro SALMONELLA VAC E fragments differed by 4 and 6 SNPs, respectively. CD-hit software was applied to cluster similar fragments that produced the best fit output when searched with SRST2. The developed vaccine differentiation method was tested with 1,253 genome samples including field isolates of Salmovac 440 (n = 51), field isolates of AviPro SALMONELLA VAC E (n = 13), S. Gallinarum (n = 19), S. Pullorum (n = 116), S. Enteritidis (n = 244), S. Typhimurium (n = 810) and achieved 100% sensitivity and specificity.
Salmonella Enteritidis is a leading cause of salmonellosis worldwide (1) and more than 80% of investigated outbreaks in Europe have been associated with the consumption of inadequately cooked eggs or foods containing uncooked eggs (https://doi.org/10.2903/j.efsa.2019.5596) (2). Over the last few decades the Colindale phage typing (PT) scheme has played a central role in epidemiological studies of S. Enteritidis while there are limitations regarding Pulsed-field Gel Electrophoresis (PFGE) and Multilocus Variable-Number Tandem Repeat Analysis (MLVA) methods and “neither PFGE nor MLVA could distinguish all of the S. Enteritidis PT30 from various sources,” for example (3–5). Recent phylogenetic studies based on whole genome sequencing (WGS) have revealed the presence of two separate clonal lineages of S. Enteritidis (6, 7). Phage types that dominated in western Europe and Asia, including PT1, PT4, and PT21 occurred in clonal lineage I, while PTs that were most common in North America (i.e., PT8, PT13a, and PT13) comprised the majority of clonal lineage II (6).
Vaccination has been proven to be one of the most successful measures to reduce Salmonella Enteritidis infections in poultry farms (8) as it can decrease colonization of the reproductive organs and intestinal tract of laying hens by Salmonella, thus reducing egg contamination (9). The prevalence of S. Enteritidis in large-scale laying hen holdings may be reduced by 88 percent by vaccination (www.efsa.europa.eu/EFSA/efsa_locale-1178620753812_1178620761896.htm). Live attenuated vaccines have been proposed to provide better protection and are better suited for mass vaccination than inactivated (killed) vaccines (10), although there can be practical problems with effective administration in the field (11). There are two commercially available S. Enteritidis live-vaccines in UK; Salmovac 440 (Gallivac SE) (Merial Animal Health Ltd, Lyon, France) and AviPro SALMONELLA VAC E (Lohmann Animal Health GmbH Heinz, Germany). Salmovac 440 vaccine strain has been derived through chemical mutagenesis from S. Enteritidis PT 4 that lacks the serovar specific plasmid (https://www.efsa.europa.eu/en/efsajournal/pub/114) and has no antimicrobial resistance genes but contains point mutations resulting in auxotrophism for histidine and adenine. S. Enteritidis field isolates are differentiated from the vaccine strain by growing on minimal media with and without histidine and adenine (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/551476/pub-salm15-intro.pdf). AviPro SALMONELLA VAC E is a metabolic drift mutant strain derived by chemical mutagenesis from S. Enteritidis PT4 (http://www.baltivet.com/en/products/veterinary-products/avipro-salmonella-vac-e/). It has been selected according to attenuation criteria such as prolonged generation time, super-sensitivity to quinolones and increased permeability of the bacterial membrane. It is capable of surviving long enough inside the bird to stimulate immunity, if administered properly, but incapable of surviving in the environment. Sensitivity to erythromycin and resistance to rifampicin are tested to distinguish the vaccine from Salmonella field isolates. It is also highly resistant to streptomycin (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/551476/pub-salm15-intro.pdf).
Whole genome sequencing (WGS) has been applied as an epidemiological tool for outbreak investigations as it provides high resolution for comparing genomes (12). In the United Kingdom, where human isolates of Salmonella are routinely sequenced, WGS has been used successfully to identify and investigate Salmonella outbreaks (13–15). Typing of all Salmonella isolates with WGS is planned at Animal and Plant Health Agency (APHA) in a near future and will include differentiation of the vaccine from the field isolates, removing the need for phenotypic testing of vaccine types. Also, phenotypic methods can be time- consuming and subject to some variability, requiring confirmation of colony purity and repeat testing of a proportion of isolates. The national control programs for Salmonella in chickens introduced in UK in 2007, 2008, and 2009 for breeders, layers, and broilers, respectively, seek to reduce or maintain low Salmonella levels of specified serotypes to targets set out in EU regulations (https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/183065/salmonella-breeders.pdf). Differentiation of vaccine from field S. Enteritidis isolates is essential for monitoring of S. Enteritidis in poultry and for targeted disease control measures. As a component of the proposed APHA Salmonella WGS typing scheme, we characterized the 2 live S. Enteritidis vaccines: Salmovac 440 and AviPro SALMONELLA VAC E used in UK poultry by comparing them with wildtype S. Enteritidis using WGS-based approaches. Here we describe the method developed to differentiate the vaccine from field isolates based on SNP differences.
The vaccine (Salmovac 440, n = 5; AviPro SALMONELLA VAC E, n = 5) and the S. Enteritidis wildtype (n = 6) strains used for method development were from the APHA Salmonella Archives (Addlestone, UK) (Table 1). All strains were kept at −80°C in 1% (w/v) proteose peptone water containing 10% (v/v) glycerol. The methods supplied by the manufacturers for differentiation with S. Enteritidis wildtype field isolates, based on growth in minimal media without adding histidine and adenine for Salmovac 440 or in media containing rifampicin, streptomycin and erythromycin to distinguish AviPro SALMONELLA VAC E, were used to confirm the vaccinal identity of isolates (16). A further 1,237 isolates, including field isolates of Salmovac 440 (n = 46), field isolates of AviPro SALMONELLA VAC E (n = 8), S. Gallinarum (n = 19), S. Pullorum (n = 116), S. Enteritidis (n = 238) and S. Typhimurium (n = 810) were used to test the developed SNP (single nucleotide polymorphism) differentiation method. All the samples in the study were collected from the environment, such as chicken feces. Therefore, there was no need for the APHA ethics committee to approve the study.
Overnight bacterial isolates were collected by centrifugation and resuspended in 0.5 mL 0.1 M PBS (pH 7.2) solution. Genomic DNA was purified with the ArchivePure DNA Cell/Tissue (1 g) kit (5 Prime, Gaithersburg, USA). Purified genomic DNA was fragmented, tagged using the Nextera XT DNA Sample Preparation Kit (Illumina UK) and sequenced at the APHA on the Illumina MiSeq platform based on the manufacturer's instructions. Phylogenetic analysis was carried out with Snippy (https://github.com/tseemann/snippy) to identify all SNPs. Tree of life (iTol) was used to produce midpoint rooted trees (17). To identify differential features that could separate the S. Enteritidis wildtype from either one of the two vaccine strains, the draft genomes of S. Enteritidis field isolates, Salmovac 440 and AviPro SALMONELLA VacE live vaccine strains as well as 4 field isolates of each of the vaccine strains (Table 1) were analyzed to identify plasmids, plasmid replicons, virulence genes, antimicrobial resistant genes as well as point mutations. SRST2 (18) database searches were carried out to identify plasmids with PlasmidFinder, replicons with PlasmidReplicon database (https://github.com/katholt/srst2), virulence factors with Virulence Factor database (http://www.mgc.ac.cn/VFs/main.htm) and a vaccine database (this study). Antimicrobial resistance genes were identified by Ariba (19) with the Card database (20). The gene presence and absence tables were generated through genome annotation by Prokka (21) after the fastq data were assembled to fasta files with Spades (22) and summarized by Roary (23). Student's t-test was carried out compare data between the wildtype and vaccine strains (two tailed distribution for two-sample populations with unequal variance) with Excel spreadsheet function (Microsoft). Blastx was performed with isolate fasta files from Spades assemblies after a protein sequence database was made (24).
Comparative genomic analyses of S. Enteritidis field isolates, Salmovac 440 and AviPro SALMONELLA VacE live vaccine strains as well as four field isolates of each vaccine strain idendified by standard laboratory methods were carried out in order to identify genomic features that could differentiate the strains. The searches and identified features are summarized in Table 2. SRST2 plasmid searches identified presence of a Salmonella Paratyphi C strain RKS4594 plasmid pSPCV (plasmid CP000858) in the wild type S. Enteritidis genomes, the vaccine strain SALMONELLA Vac E and field isolates. However, the 262 bp fragment in PlasmidFinder that identifies the 55,414 base pairs plasmid CP000858 is identical with the 262 bp fragment that identifies the 59,372 bp of S. Enteritidis strain-specific plasmid pSEN (HG970000) or pSENV (JN885080) which is present in most S. Enteritidis wild type isolates. As expected, the presence of the pSENV plasmid and associated plasmid replicon was confirmed in all wild type and the SALMONELLA Vac E vaccine strain and field isolates but not in the genome of the vaccine strain Salmovac 440, one of the features of this vaccine strain (Rows 1–2, Table 2). The analysis of the AMR genes or mutations identified rifampicin resistance related to the rpoB gene in Avipro SALMONELLA Vac E strains (Row 3, Table 2).
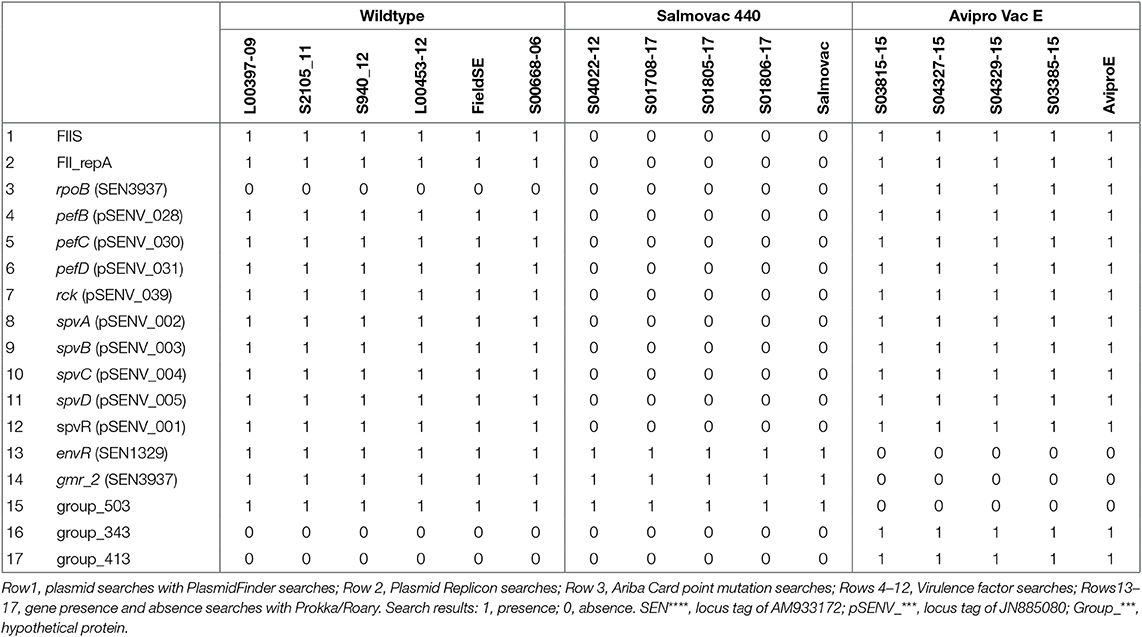
Table 2. Identification of differential features between groups of the wildtype, Salmovac 440, and Avipro SALMONELLA Vac E from the search results of PlasmidFinder.
Furthermore, the 9 pSENV plasmid-associated virulence genes identified in the wildtype and Avipro SALMONELLA Vac E vaccine and field isolates were not present in the Salmovac 440 strains (Rows 4–12, Table 2), confirming the absence of the plasmid in these strains.
The comparison of the annotated wildtype and vaccine genomes revealed the absence of 3 genes: envR, a regulator in the Salmonella pathogenicity island 2 (SPI2) gmr_2, cyclic di-GMP phosphodiesterase and group_503, and a hypothetical protein (Rows 13–15, Table 2) and presence of two hypothetical genes: group_343 and group_413 (Rows 16–17) in the vaccine strain Avipro SALMONELLA Vac E strains. A further 62 pSENV plasmid-related genes were absent in Salmovac 440 but present in the wildtype and Avipro SALMONELLA Vac E (Table S1).
Phylogenetic analysis of the core SNPs indicated a close relatedness of both vaccine strains with the reference S. Enteritidis strain P125109 (AM933172). The distance was 115 SNPs between AviPro SALMONELLA VAC E and the reference and 213 SNPs between Salmovac 440 and the reference (Figure 1). The Salmovac 440 vaccine reference strain and the vaccine strains recovered from farms differed by up to 3 SNPs; while the AviPro SALMONELLA VAC E reference strain differed from the vaccine strains recovered from farms by 0–2 SNPs. All SNPs are listed between the reference and AviPro Salmonella VAC E or Salmonvac 440 (Table S2).
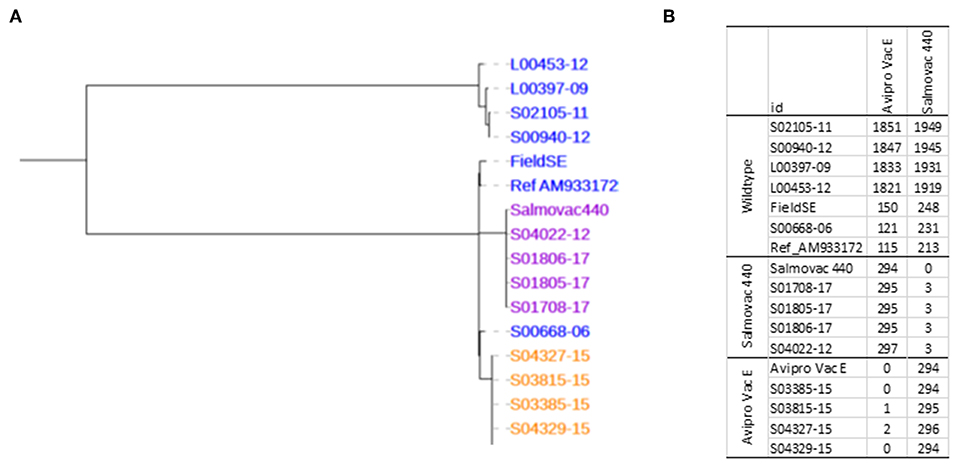
Figure 1. (A), Maximum likelihood tree of S. Enteritidis vaccines. Salmovac 440 (purple), AviproVacE= AviPro SALMONELLA VAC E (orange) and the wildtype (blue). (B), SNP distance from AviPro SALMONELLA VAC E and Salmovac 440 to all the samples including the reference (S. Enteritidis strain P125109) in the tree.
A point mutation in the rpoB gene was identified by Ariba Card relating to AviPro SALMONELLA VAC E's rifampicin resistance (Table 2). Ariba Card does not identify the position in the gene, however, we identified a missense mutation, Ser531Phe in the rpoB gene (Row 1, Table 3) that could explain the rifampicin resistance of the AviPro SALMONELLA VAC E vaccine strain and a missence mutation Lys43Arg in the rpsL gene that could explain the resistance to streptomycin (Row 2, Table 3).
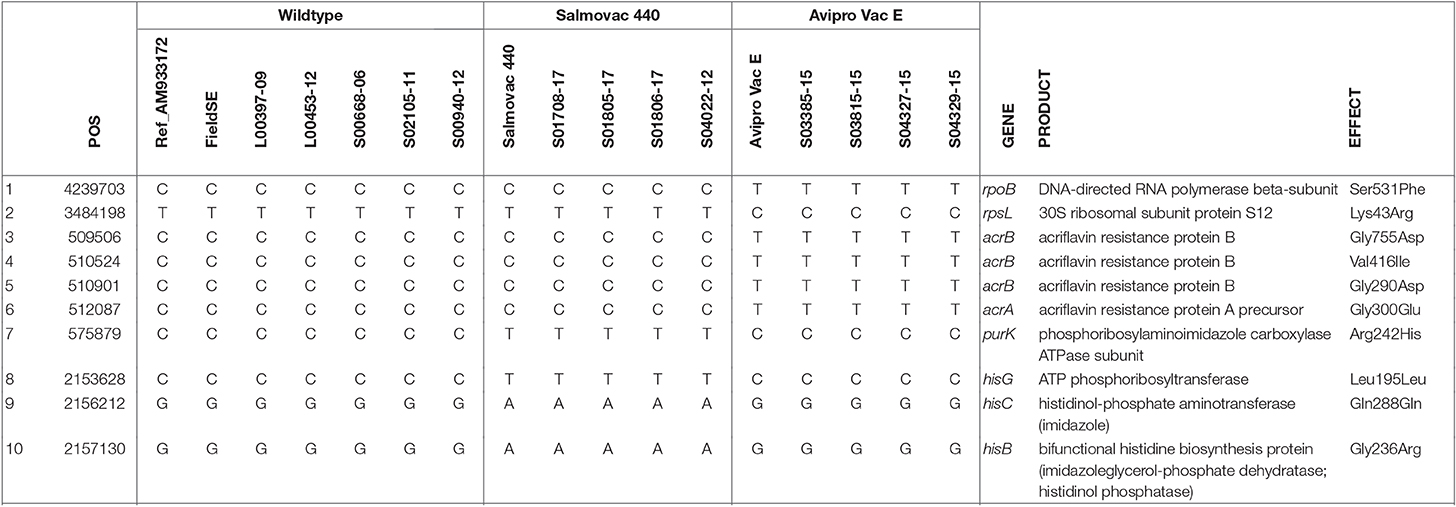
Table 3. Identification of unique SNPs linked to vaccine strain properties of antimicrobial resistance and histidine and adenine dependencies.
Furthermore, 3 missense mutations in acrB and 1 missense mutation in acrA genes found in AviPro SALMONELLA VAC E are likely to be associated with erythromycin sensitivity (Rows 3–6, Table 3).
The adenine dependency of Salmovac 440 could be explained by a missense mutation Arg242His in purK, the N5-Carboxyaminoimidazole ribonucleotide (N5-CAIR) synthetase gene of the purine biosynthesis pathway (Row 7, Table 3). Three mutations in the histidine biosynthesis pathway were identified when Salmovac 440 was compared with the wildtype (Rows 8–10, Table 3); the mutation in the hisB gene was a missense mutation, Gly236Arg, suggesting that the mutation might give Salmovac 440 the property of histidine dependence.
Phylogenetic analysis considers only single nucleotide changes. To rule out any possibility of insertions or deletions in the pathways of efflux pump, histidine biosynthesis, and purine biosynthesis, Blastx was carried out to find out any amino acid sequence changes. The analyses showed no evidence of insertions or deletions and the mis-sense mutations 1 in acrA, 3 in acrB unique to AviPro SALMONELLA VAC E and 1 in hisB and 1 in purK unique to Salmovac 440 identified by phylogenetic analysis, were confirmed (Table 3 and Table S3).
Salmovac 440 has lost the pathogenic plasmid that encodes a number of virulence factors (Table 2). This may partially explain the attenuation of Salmovac 440. However, for AviPro SALMONELLA VAC E, Prokka/Roary/TTest only identified envR as a potential candidate for attenuation (Row 13, Table 2) a regulator in the Salmonella pathogenicity island 2 (SPI2). Other possible sources for attenuation were sought, such as point mutations in known genes associated with virulence. Phylogenetic analysis identified a total of 96 SNPs unique to AviPro SALMONELLA VAC E of which 56 were missense SNPs. Among these 56 missense SNPs, 20 were in genes reported to be associated with virulence (Table 4). This group of genes were from diverse functions: 1 in iron uptake (Row 6), 3 in potassium transport (Rows 9–12) and 7 genes related to antimicrobial resistance through point mutations (Rows 1–4 and 15–17).
To construct vaccine differentiation database, total SNPs were identified from the phylogenetic analysis of the 16 sequenced genomes (Table 1) using the S. Enteritidis strain P125109 (accession AM933172) as reference. Unique SNPs to Salmovac 440 were identified after comparison with the wildtype S. Enteritidis and AviPro SALMONELLA VAC E genomes. Two regions were selected to represent each vaccine strain, Salmovac 440 and AviPro SALMONELLA VAC E, and pair wise fragments were created so that one represented Salmovac 440 and the other the wildtype. The paired fragments were identical except for 6 SNPs in each region (Figure S1). CD-hit (45) was performed to cluster the paired fragments together so that when the fragments in the database are used as references for SRST2 (18) only one fragment from a pair gets reported: either a vaccine fragment or a wildtype fragment but not both.
The SNPs unique to Salmovac 440 were located in two genomic regions, region 831G from position 2145373 to 2157130 of the reference AM933172 with 6 different nucleotides in positions: 2145373 (WT, G; 831G, A), 2148431 (WT, G; 831G, A), 2150004 (WT, G; 831G, A), 2153628 (WT, C; 831G, T), 2156212 (WT, G; 831G, A), and 2157130 (WT, G; 831G, A) and region 832G from position 854486 to 863710 with 6 different nucleotides in positions: 854486 (WT, C; 832G, T), 855295 (WT, C; 832G, T), 856153 (WT, C; 832G, T), 858894 (WT, C; 832G, T), 863334 (WT, C; 832G, T), 863710 (WT, C; 832G, T) (Figure S1). The two regions for the AviPro SALMONELLA VAC E strain were 509256 to 512337 of the reference AM933172 with 4 SNPs differences in positions: 509506 (WT, C; 8321, T), 510524 (WT, C; 8321, T), 510901 (WT, C; 8321, T), 512087 (WT, C; 8321, T), and Avipro Vac E 8322 from position 802121 to 806926 with 6 SNPs differences in positions: 802371 (WT, C; 8322, T), 802460 (WT, C; 8322, T), 803918 (WT, C; 8322, T), 805115 (WT, C; 8322, T), 805353 (WT, C; 8322, T), 806676 (WT, C; 8322, T) (Figure S1). The identified genomic regions were used to create pair wise fragments, one fragment representing the vaccine strain and the second representing the wild type, differing only by the identified SNPs.
The sensitivity and specificity of the vaccine database of variants in distinguishing field vaccine strains from wild type strains was 100%. The database was tested with a total of 1,253 sequenced genomes including Salmovac 440 (n = 51) and AviPro SALMONELLA VAC E (n = 13) field isolates, S. Gallinarum (n = 19), S. Pullorum (n = 116), wildtype S. Enteritidis (n = 244), and S. Typhimurium (n = 810) (Table 5).
In this study, using whole sequencing approaches, we characterized some of the genomic properties of the vaccine strains Salmovac 440 and AviPro SALMONELLA VAC E. Deletion of the tolC, acrB or acrAB genes is linked to strains with increased susceptibility to antimicrobials, including erythromycin (46). In AviPro SALMONELLA VAC E, phylogenetic analysis identified 3 missense mutations in acrB and 1 missense mutation in acrA (Rows 3–6, Table 3). In E. coli, multiple mutations in the acrB gene increase the susceptibility to erythromycin while individually they do not result in any changes of sensitivity, and the mutation in strain T37W even increases resistance to antimicrobials (47). Therefore, the sensitivity in AviPro SALMONELLA VAC E may be due to the possibility that these 4 mutations in acrAB have changed some properties of AcrAB efflux pump. The strA-strB genes are most likely associated with high levels of streptomycin resistance, whereas the aadA gene confers low-level resistance (48). Streptomycin resistance in M. tuberculosis isolates is frequently linked to missense mutations in the rpsL gene for ribosomal protein S12 or in the rrs gene for nucleotide substitutions in the 16S rRNA gene (49). In this study, we detected a missense mutation, Lys43Arg, in the rpsL gene (Table 3). The same amino acid replacement Lys43Arg of rpsL has been described in Mycobacterium tuberculosis and Helicobacter pylori (50, 51); while in E. coli the change is Lys42Arg (52). A missence point mutation Ser531Phe in the rpoB gene we detected in this study is most likely linked to the rifampicin resistance of SALMONELLA VAC E strain. Several mutations in the rpoB gene have been described to reduce the susceptibility to rifampicin in clinical Mycobacterium tuberculosis isolates that contribute to various degree of fitness cost to the strain (53).
HisB is bifunctional since the C-terminal domain catalyzes as IGP dehydratase (the sixth step) while the N-terminal domain as Hol-P phosphatase (the eighth step) in the histidine biosynthesis pathway (54). A total of 1,020 independent histidine-requiring mutations were isolated in the histidine operon after strain LT2 of S. Typhimurium was treated with N-methy1-N'-nitro-N-nitrosoguanidine, the same agent used for Salmovac 440 mutagenesis; many of these mutations were found in the hisB gene (55).
Salmovac 440 is lacking the pathogenic plasmid pSENV that encodes a number of virulence factors (Table 2). The plasmid could be associated with much of the virulence as the spv (Salmonella plasmid virulence) is considered crucial for the phenotype of S. Enteritidis (56) and it has been shown that spvB mutants are avirulent in mice (57). Histidine-requiring mutations have also been shown to lead to attenuation. In Aspergillus fumigatus, mutation in hisB causes histidine auxotrophy and attenuation of virulence in 3 murine models: pulmonary infection, systemic infection, corneal infection, and in the wax moth larvae model (58). In Xanthomonas oryzae pv. Oryzicola which triggers bacterial leaf streak in rice, two genes in histidine biosynthesis operon, trpR and hisB, were identified to be essential for virulence and bacterial growth in plants (59). Adenine is one of the products of purine biosynthesis. Disrupted de novo purine biosynthesis has been revealed to attenuate the virulence of several pathogens, such as Salmonella, Burkholderia, Brucella, and Francisella (60–64). Therefore, the attenuation of Salmovac 440 is most likely the result of the combination of absence of the virulence plasmid and histidine and adenine dependencies.
As for AviPro SALMONELLA VAC E, the attenuation may also be the result of multiple factors: e.g., the missing envR gene (Table 2) and point mutations in 20 virulence-associated genes (Table 4). EnvR as a potential candidate for attenuation (Row 13, Table 2) is a regulator in the Salmonella pathogenicity island 2 (SPI2) that encodes type III secretion system (T3SS) that changes the host cell functions and facilitate intracellular replication (65, 66). The RpoB H526D mutant (Rif) displayed reduced survival compared with control strains in Mycobacterium tuberculosis (36). Mutations in rpsL that result in streptomycin resistance indicated that the K43N and K43T mutations were pleiotropic, showing reduced virulence in Erwinia carotovora (39). Attenuation of an avian pathogenic Escherichia coli strain resulted from a point mutation in rpsL (40). Direct evidence of point mutations leading to attenuation is also observed in Salmonella Typhimurium mutants resistant to streptomycin or rifampicin that become avirulent in mice (41).
With the wide use of S. Enteritidis live vaccines on chicken farms, a reliable and rapid differentiation method is essential. Currently, there are two methods in use. One is phenotypic typing based on manufacturers' instructions. For AviPro SALMONELLA VAC E, the vaccine strain is rifampicin resistant and erythromycin sensitive; while for Salmovac 440, the vaccine strain requires histidine and adenine supplements in order to grow on minimal media. The second method uses TaqMan-qPCR to differentiate Salmovac 440 and AviPro SALMONELLA VAC E vaccine strains from the wildtype Enteritidis (67). To produce this test, the authors sequenced the whole genomes of both vaccine strains and identified SNPs that were used to design PCR probes based on 2 SNPs for AviPro SALMONELLA VAC E and 1 SNP for Salmovac 440. The real time PCR method identified all 30 Salmovac 440 and 7 AviPro SALMONELLA VAC E vaccine strains (100% sensitivity) and eliminated all of the 97 wild type S. Enteritidis as well as other S. enterica strains (100% specificity). The method we developed in this study was also based on SNP differences between the vaccine strains and the wildtype however, the short regions described in Maurischat et al. (67) used in real tim PCR differentiation, 146 bp in nhaA for Salmovac SE and 88 bp in kdpA for AviPro SALMONELLA VAC E, were not sufficient for alignment with Illumina short reads with high coverage. To ensure high sensitivity and specificity we selected two regions each to represent the vaccine strains and more SNPs than the PCR method (67) (Figure S1). In the real time PCR differentiation study, the authors tested the specificity with non-Salmonella species as well as Salmonella serovars (67); while in this study, we aimed to differentiate vaccine strains from genomes of isolates typed as Salmonella Enteritidis, Typhimurium, Gallinarum or Pullorum by our WGS serotyping pipeline. Further database development will include an additional 3 live vaccines: AviPro SALMONELLA VAC T, Salmoporc STM and Nobilis SG 9R (Tang et al. unpublished data). Although the differentiation using the developed vaccine database showed 100% sensitivity and 100% specificity (Table 5), we have also developed a scheme to ensure typing accuracy in case of mixed results. If a sample is typed as wildtype in one region and vaccine in another, this sample will be analyzed using phylogeny based on a panel of isolates shown in Figure 1, including wildtype S. Enteritidis isolates and both Salmovac 440 and AviPro SALMONELLA VAC E vaccines so that all SNPs will be considered. With the mean mutation rate across all S. Enteritidis lineages being 2.2 × 10–7 substitutions per site per year or 1.01 SNPs per genome per year (68) both attenuation of the vaccine strains and SNPs used for differentiation should be stable.
In conclusion, we characterized Salmovac 440 and AviPro SALMONELLA VAC E vaccine strains and identified genomic features that could have resulted in attenuation, resistance to rifampicin and streptomycin in AviPro SALMONELLA VAC E and histidine and adenine dependencies in Salmovac 440. We developed a database of highly specific SNP variants that could differentiate vaccine from wild type strains with 100% sensitivity and 100% specificity. The knowledge and methods from this study could be applied for characterization and differentiation of other Salmonella vaccine strains that are in use outside UK.
The whole genome sequencing fastq files for this study can be found in PRJEB33366 of the European Nucleotide Archive (https://www.ebi.ac.uk/ena).
LP, YT, and RD designed the study. YT and LP analyzed the data and drafted the manuscript. All authors read through and approved the manuscript.
This work was supported by funding from Defra (CR2000F).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Drs. Shaun Cawthraw and Carmen Garcia-Pelayo for sample preparations.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2019.00447/full#supplementary-material
1. Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. (2011) 8:887–900. doi: 10.1089/fpd.2010.0787
2. Tirado C, Schmidt K. WHO surveillance programme for control of foodborne infections and intoxications: preliminary results and trends across greater Europe. World Health Organization. J Infect. (2001) 43:80–4. doi: 10.1016/S0163-4453(01)90861-8
3. Threlfall EJ, Chart H. Interrelationships between strains of Salmonella Enteritidis. Epidemiol Infect. (1993) 111:1–8. doi: 10.1017/S0950268800056612
4. De Lappe N, Doran G, O'Connor J, O'Hare C, Cormican M. Characterization of bacteriophages used in the Salmonella enterica serovar Enteritidis phage-typing scheme. J Med Microbiol. (2009) 58:86–93. doi: 10.1099/jmm.0.000034-0
5. Parker CT, Huynh S, Quinones B, Harris LJ, Mandrell RE. Comparison of genotypes of Salmonella enterica serovar Enteritidis phage type 30 and 9c strains isolated during three outbreaks associated with raw almonds. Appl Environ Microbiol. (2010) 76:3723–31. doi: 10.1128/AEM.03053-09
6. Zheng J, Pettengill J, Strain E, Allard MW, Ahmed R, Zhao S, et al. Genetic diversity and evolution of Salmonella enterica serovar Enteritidis strains with different phage types. J Clin Microbiol. (2014) 52:1490–500. doi: 10.1128/JCM.00051-14
7. Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci USA. (2015) 112:863–8. doi: 10.1073/pnas.1416707112
8. O'Brien SJ. The “decline and fall” of nontyphoidal Salmonella in the United Kingdom. Clin Infect Dis. (2013) 56:705–10. doi: 10.1093/cid/cis967
9. Gantois I, Ducatelle R, Timbermont L, Boyen F, Bohez L, Haesebrouck F, et al. Oral immunisation of laying hens with the live vaccine strains of TAD Salmonella vac E and TAD Salmonella vac T reduces internal egg contamination with Salmonella Enteritidis. Vaccine. (2006) 24:6250–5. doi: 10.1016/j.vaccine.2006.05.070
10. Harrison JA, Villarreal-Ramos B, Mastroeni P, Demarco de Hormaeche R, Hormaeche CE. Correlates of protection induced by live Aro- Salmonella typhimurium vaccines in the murine typhoid model. Immunology. (1997) 90:618–25. doi: 10.1046/j.1365-2567.1997.00158.x
11. EFSA-BIOHAZ-Panel. Salmonella control in poultry flocks and its public health impact. EFSA J. (2019) 17:e05596. doi: 10.2903/j.efsa.2019.5596
12. Dallman T, Inns T, Jombart T, Ashton P, Loman N, Chatt C, et al. Phylogenetic structure of European Salmonella Enteritidis outbreak correlates with national and international egg distribution network. Microb Genom. (2016) 2:e000070. doi: 10.1099/mgen.0.000070
13. Ashton PM, Peters T, Ameh L, McAleer R, Petrie S, Nair S, et al. Whole genome sequencing for the retrospective investigation of an outbreak of Salmonella Typhimurium DT 8. PLoS Curr. (2015) 7:ecurrents.outbreaks.2c05a47d292f376afc5a6fcdd8a7a3b6. doi: 10.1371/currents.outbreaks.2c05a47d292f376afc5a6fcdd8a7a3b6
14. Hormansdorfer S, Messelhausser U, Rampp A, Schonberger K, Dallman T, Allerberger F, et al. Re-evaluation of a 2014 multi-country European outbreak of Salmonella Enteritidis phage type 14b using recent epidemiological and molecular data. Euro Surveill. (2017) 22. doi: 10.2807/1560-7917.ES.2017.22.50.17-00196
15. Kanagarajah S, Waldram A, Dolan G, Jenkins C, Ashton PM, Carrion Martin AI, et al. Whole genome sequencing reveals an outbreak of Salmonella Enteritidis associated with reptile feeder mice in the United Kingdom, 2012-2015. Food Microbiol. (2018) 71:32–8. doi: 10.1016/j.fm.2017.04.005
16. Rabsch W, Liesegang A, Tschape H. Laboratory-based surveillance of salmonellosis of humans in Germany–safety of Salmonella typhimurium and Salmonella Enteritidis live vaccines. Berl Munch Tierarztl Wochenschr. (2001) 114:433–7.
17. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. (2016) 44:W242–5. doi: 10.1093/nar/gkw290
18. Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. (2014) 6:90. doi: 10.1186/s13073-014-0090-6
19. Hunt M, Mather AE, Sanchez-Buso L, Page AJ, Parkhill J, Keane JA, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. (2017) 3:e000131. doi: 10.1099/mgen.0.000131
20. Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2017) 45:D566–73. doi: 10.1093/nar/gkw1004
21. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
22. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
23. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. (2015) 31:3691–3. doi: 10.1093/bioinformatics/btv421
24. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. (2009) 10:421. doi: 10.1186/1471-2105-10-421
25. Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. (2006) 59:126–41. doi: 10.1111/j.1365-2958.2005.04940.x
26. Wang-Kan X, Blair JMA, Chirullo B, Betts J, La Ragione RM, Ivens A, et al. Lack of AcrB efflux function confers loss of virulence on Salmonella enterica Serovar Typhimurium. MBio. (2017) 8:e00968–17. doi: 10.1128/mBio.00968-17
27. Endley S, McMurray D, Ficht TA. Interruption of the cydB locus in Brucella abortus attenuates intracellular survival and virulence in the mouse model of infection. J Bacteriol. (2001) 183:2454–62. doi: 10.1128/JB.183.8.2454-2462.2001
28. Gotz F, Mayer S. Both terminal oxidases contribute to fitness and virulence during organ-specific Staphylococcus aureus colonization. MBio. (2013) 4:e00976–13. doi: 10.1128/mBio.00976-13
29. Nagy TA, Moreland SM, Andrews-Polymenis H, Detweiler CS. The ferric enterobactin transporter Fep is required for persistent Salmonella enterica serovar typhimurium infection. Infect Immun. (2013) 81:4063–70. doi: 10.1128/IAI.00412-13
30. Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, et al. Ferrioxamine-mediated iron(III) utilization by Salmonella enterica. Appl Environ Microbiol. (1999) 65:1610–8.
31. Bucarey SA, Villagra NA, Martinic MP, Trombert AN, Santiviago CA, Maulen NP, et al. The Salmonella enterica serovar Typhi tsx gene, encoding a nucleoside-specific porin, is essential for prototrophic growth in the absence of nucleosides. Infect Immun. (2005) 73:6210–9. doi: 10.1128/IAI.73.10.6210-6219.2005
32. Mac Siomoin RA, Nakata N, Murai T, Yoshikawa M, Tsuji H, Sasakawa C. Identification and characterization of ispA, a Shigella flexneri chromosomal gene essential for normal in vivo cell division and intracellular spreading. Mol Microbiol. (1996) 19:599–609. doi: 10.1046/j.1365-2958.1996.405941.x
33. Moule MG, Monack DM, Schneider DS. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. (2010) 6:e1001065. doi: 10.1371/journal.ppat.1001065
34. Alegado RA, Chin CY, Monack DM, Tan MW. The two-component sensor kinase KdpD is required for Salmonella typhimurium colonization of Caenorhabditis elegans and survival in macrophages. Cell Microbiol. (2011) 13:1618–37. doi: 10.1111/j.1462-5822.2011.01645.x
35. Perinet S, Jeukens J, Kukavica-Ibrulj I, Ouellet MM, Charette SJ, Levesque RC. Molybdate transporter ModABC is important for Pseudomonas aeruginosa chronic lung infection. BMC Res Notes. (2016) 9:23. doi: 10.1186/s13104-016-1840-x
36. Rifat D, Campodonico VL, Tao J, Miller JA, Alp A, Yao Y, et al. In vitro and in vivo fitness costs associated with Mycobacterium tuberculosis RpoB mutation H526D. Future Microbiol. (2017) 12:753–65. doi: 10.2217/fmb-2017-0022
37. Dong H, Zhang X, Pan Z, Peng D, Liu X. Identification of genes for biofilm formation in a Salmonella Enteritidis strain by transposon mutagenesis. Wei Sheng Wu Xue Bao. (2008) 48:869–73. doi: 10.1637/10008-980611-DIGEST.1
38. Latasa C, Echeverz M, Garcia B, Gil C, Garcia-Ona E, Burgui S, et al. Evaluation of a Salmonella strain lacking the secondary messenger C-di-GMP and RpoS as a live oral vaccine. PLoS ONE. (2016) 11:e0161216. doi: 10.1371/journal.pone.0161216
39. Barnard AM, Simpson NJ, Lilley KS, Salmond GP. Mutations in rpsL that confer streptomycin resistance show pleiotropic effects on virulence and the production of a carbapenem antibiotic in Erwinia carotovora. Microbiology. (2010) 156:1030–9. doi: 10.1099/mic.0.034595-0
40. Amoako KK, Prysliak T, Potter AA, Collinson SK, Kay WW, Allan BJ. Attenuation of an avian pathogenic Escherichia coli strain due to a mutation in the rpsL gene. Avian Dis. (2004) 48:19–25. doi: 10.1637/6025
41. Bjorkman J, Hughes D, Andersson DI. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA. (1998) 95:3949–53. doi: 10.1073/pnas.95.7.3949
42. Jelsbak L, Hartman H, Schroll C, Rosenkrantz JT, Lemire S, Wallrodt I, et al. Identification of metabolic pathways essential for fitness of Salmonella Typhimurium in vivo. PLoS ONE. (2014) 9:e101869. doi: 10.1371/journal.pone.0101869
43. Llama-Palacios A, Lopez-Solanilla E, Rodriguez-Palenzuela P. The ybiT gene of Erwinia chrysanthemi codes for a putative ABC transporter and is involved in competitiveness against endophytic bacteria during infection. Appl Environ Microbiol. (2002) 68:1624–30. doi: 10.1128/AEM.68.4.1624-1630.2002
44. Aussel L, Loiseau L, Hajj Chehade M, Pocachard B, Fontecave M, Pierrel F, et al. ubiJ, a new gene required for aerobic growth and proliferation in macrophage, is involved in coenzyme Q biosynthesis in Escherichia coli and Salmonella enterica serovar Typhimurium. J Bacteriol. (2014) 196:70–9. doi: 10.1128/JB.01065-13
45. Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. (2006) 22:1658–9. doi: 10.1093/bioinformatics/btl158
46. Yamasaki S, Fujioka T, Hayashi K, Yamasaki S, Hayashi-Nishino M, Nishino K. Phenotype microarray analysis of the drug efflux systems in Salmonella enterica serovar Typhimurium. J Infect Chemother. (2016) 22:780–4. doi: 10.1016/j.jiac.2016.03.015
47. Zwama M, Yamasaki S, Nakashima R, Sakurai K, Nishino K, Yamaguchi A. Multiple entry pathways within the efflux transporter AcrB contribute to multidrug recognition. Nat Commun. (2018) 9:124. doi: 10.1038/s41467-017-02493-1
48. Sunde M, Norstrom M. The genetic background for streptomycin resistance in Escherichia coli influences the distribution of MICs. J Antimicrob Chemother. (2005) 56:87–90. doi: 10.1093/jac/dki150
49. Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. (1995) 8:496–514. doi: 10.1128/CMR.8.4.496
50. Dailidiene D, Dailide G, Kersulyte D, Berg DE. Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori. Appl Environ Microbiol. (2006) 72:5908–14. doi: 10.1128/AEM.01135-06
51. Khosravi AD, Etemad N, Hashemzadeh M, Khandan Dezfuli S, Goodarzi H. Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J Glob Antimicrob Resist. (2017) 9:51–6. doi: 10.1016/j.jgar.2017.01.005
52. Taylor DE, Trieber CA, Trescher G, Bekkering M. Host mutations (miaA and rpsL) reduce tetracycline resistance mediated by Tet(O) and Tet(M). Antimicrob Agents Chemother. (1998) 42:59–64. doi: 10.1128/AAC.42.1.59
53. Brandis G, Pietsch F, Alemayehu R, Hughes D. Comprehensive phenotypic characterization of rifampicin resistance mutations in Salmonella provides insight into the evolution of resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. (2015) 70:680–5. doi: 10.1093/jac/dku434
54. Winkler ME, Ramos-Montanez S. Biosynthesis of Histidine. EcoSal Plus. (2009) 3. doi: 10.1128/ecosal.3.6.1.9
55. Hartman PE, Hartman Z, Stahl RC. Classification and mapping of spontaneous and induced mutations in the histidine operon of Salmonella. Adv Genet. (1971) 16:1–34. doi: 10.1016/S0065-2660(08)60352-1
57. Roudier C, Fierer J, Guiney DG. Characterization of translation termination mutations in the spv operon of the Salmonella virulence plasmid pSDL2. J Bacteriol. (1992) 174:6418–23. doi: 10.1128/jb.174.20.6418-6423.1992
58. Dietl AM, Amich J, Leal S, Beckmann N, Binder U, Beilhack A, et al. Histidine biosynthesis plays a crucial role in metal homeostasis and virulence of Aspergillus fumigatus. Virulence. (2016) 7:465–76. doi: 10.1080/21505594.2016.1146848
59. Su P, Song Z, Wu G, Zhao Y, Zhang Y, Wang B, et al. Insights into the roles of two genes of the histidine biosynthesis operon in pathogenicity of Xanthomonas oryzae pv. oryzicola. Phytopathology. (2018) 108:542–51. doi: 10.1094/PHYTO-09-17-0332-R
60. Alcantara RB, Read RD, Valderas MW, Brown TD, Roop RM II. Intact purine biosynthesis pathways are required for wild-type virulence of Brucella abortus 2308 in the BALB/c mouse model. Infect Immun. (2004) 72:4911–7. doi: 10.1128/IAI.72.8.4911-4917.2004
61. Pilatz S, Breitbach K, Hein N, Fehlhaber B, Schulze J, Brenneke B, et al. Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect Immun. (2006) 74:3576–86. doi: 10.1128/IAI.01262-05
62. Mantena RK, Wijburg OL, Vindurampulle C, Bennett-Wood VR, Walduck A, Drummond GR, et al. Reactive oxygen species are the major antibacterials against Salmonella Typhimurium purine auxotrophs in the phagosome of RAW 264.7 cells. Cell Microbiol. (2008) 10:1058–73. doi: 10.1111/j.1462-5822.2007.01105.x
63. Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, et al. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy. (2012) 8:1342–56. doi: 10.4161/auto.20808
64. Cummins J, Gahan CG. Signature tagged mutagenesis in the functional genetic analysis of gastrointestinal pathogens. Gut Microbes. (2012) 3:93–103. doi: 10.4161/gmic.19578
65. Xu X, Hensel M. Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect Immun. (2010) 78:49–58. doi: 10.1128/IAI.00931-09
66. Figueiredo R, Card R, Nunes C, AbuOun M, Bagnall MC, Nunez J, et al. Virulence Characterization of Salmonella enterica by a new microarray: detection and evaluation of the cytolethal distending toxin gene activity in the unusual host S. Typhimurium. PLoS ONE. (2015) 10:e0135010. doi: 10.1371/journal.pone.0135010
67. Maurischat S, Szabo I, Baumann B, Malorny B. Rapid real-time PCR methods to distinguish Salmonella Enteritidis wildtype field isolates from vaccine strains Salmovac SE/Gallivac SE and AviPro SALMONELLA VAC E. J Microbiol Methods. (2015) 112:92–8. doi: 10.1016/j.mimet.2015.03.015
Keywords: Salmonella Enteritidis, vaccine, whole genome sequencing, differentiation, characterization
Citation: Tang Y, Davies R and Petrovska L (2019) Identification of Genetic Features for Attenuation of Two Salmonella Enteritidis Vaccine Strains and Differentiation of These From Wildtype Isolates Using Whole Genome Sequencing. Front. Vet. Sci. 6:447. doi: 10.3389/fvets.2019.00447
Received: 26 June 2019; Accepted: 26 November 2019;
Published: 18 December 2019.
Edited by:
Timothy J. Johnson, University of Minnesota Twin Cities, United StatesReviewed by:
Jean Guard, US National Poultry Research Centre, United StatesCopyright © 2019 Tang, Davies and Petrovska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yue Tang, eXVlLnRhbmdAYXBoYS5nb3YudWs=; Liljana Petrovska, bGlsamFuYS5wZXRyb3Zza2FAYXBoYS5nb3YudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.