
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 24 April 2019
Sec. Veterinary Infectious Diseases
Volume 6 - 2019 | https://doi.org/10.3389/fvets.2019.00128
 Dekang Zhu1,2†
Dekang Zhu1,2† Jianbang Wan1,2†
Jianbang Wan1,2† Zhishuang Yang1,2,3
Zhishuang Yang1,2,3 Jinge Xu4
Jinge Xu4 Mingshu Wang1,2,3
Mingshu Wang1,2,3 Renyong Jia1,2,3
Renyong Jia1,2,3 Shun Chen1,2,3
Shun Chen1,2,3 Mafeng Liu1,2,3
Mafeng Liu1,2,3 Xinxin Zhao1,2,3
Xinxin Zhao1,2,3 Qiao Yang1,2,3
Qiao Yang1,2,3 Ying Wu1,2,3
Ying Wu1,2,3 Shaqiu Zhang1,2,3
Shaqiu Zhang1,2,3 Yunya Liu1,3
Yunya Liu1,3 Ling Zhang1,3
Ling Zhang1,3 Yanling Yu1,3
Yanling Yu1,3 Xiaoyue Chen1,3
Xiaoyue Chen1,3 Anchun Cheng1,2,3*
Anchun Cheng1,2,3*We report for the first time the occurrence of integrative conjugative elements (ICEs) in Riemerella anatipestifer (R.anatipestifer) isolated from diseased ducks in China. For this purpose, a total of 48 genome sequences were investigated, which comprised 30 publicly available R. anatipestifer genome sequences, and 18 clinical isolates genomes sequences. Two ICEs, named ICERanRCAD0133-1 and ICERanRCAD0179-1 following the classic nomenclature system, were identified in R. anatipestifer through the use of bioinformatics tools. Comparative analysis revealed that three ICEs in Ornithobacterium rhinotracheale showed a high degree of conservation with the core genes of ICERanRCAD0133-1, while 13 ICEs with high similarity to ICERanRCAD0179-1 were found in Bacteroidetes. Based on the definition of ICE family, ICERanRCAD0179-1 was grouped in CTnDOT/ERL family; however, ICERanRCAD0133-1, which had no significant similarity with known ICEs, might be classified into a novel ICE family. The sequences of ICERanRCAD0133-1 and ICERanRCAD0179-1 were 70890 bp and 49166 bp in length, had 33.14 and 50.34% GC content, and contained 77 CDSs and 51 CDSs, respectively. Cargo genes carried by these two ICEs were predicted to encode: R-M systems, IS elements, a putative TonB-dependent receptor, a bacteriocin/lantibiotic efflux ABC transporter, a tetracycline resistance gene and more. In addition, phylogenetic analyses revealed that ICERanRCAD0179-1 and related ICEs were derived from a common ancestor, which may have undergone divergence prior to integartation into the host bacterial chromosome, and that the core genes co-evolved via a related evolutionary process or experienced only a low degree of recombination events during spread from a common CTnDOT/ERL family ancestor. Collectively, this study is the first identification and characterization of ICEs in R. anatipestifer; and provides new insights into the genetic diversity, evolution, adaptation, antimicrobial resistance, and virulence of R. anatipestifer.
Integrative conjugative elements (ICEs), a type of self-transmissible mobile genetic elements (MGEs) are widely distributed in bacterial genomes. ICEs are major mediators for horizontal gene transfer (HGT), which contribute to microbial evolution and adjustment to new niche (1, 2). ICEs have the capability to excise from their host chromosome and reintegrate into a new host's chromosome at a target site where they replicate as a part of the host chromosome. Generally, each type of ICE contains a set of core genes involved in its integration and excision, regulation and maintenance, and conjugative transfer (3). The remaining components of ICEs are accessory genes, which endow the host bacteria with multiple phenotypes that can be beneficial for the recipient bacteria, such as colonization of a eukaryotic host, nitrogen fixation or promotion of virulence and biofilm formation, or resistance to antibiotics and heavy metals (4). In particularly, homologous recombination among unrelated or distantly related ICEs probably occurs in ICE recipients, resulting in a diverse set of novel hybrid ICEs, which leads to the diversity of ICEs among bacterial genomes (5, 6).
R.anatipestifer, a member of the Flavobacteriaceae family, is a non-spore-forming and rod-shaped Gram-negative bacterium. It is a major contributor to acute and chronic septicemia associated with fibrinous pericarditis, perihepatitis, airsacculitis, and meningitis (7). R.anatipestifer can infect various types of avian species, such as ducks, geese, turkeys, and some wild birds. R.anatipestifer infection is a contagious disease, that causes enormous economic losses to the poultry industry worldwide (8).
Here, we investigated 48 R.anatipestifer genome sequences by using bioinformatics tools, comprised of 30 publicly available genomic sequences and 18 clinical isolates genomes sequenced in this study. As a result, two ICEs were identified in the genome of R.anatipestifer (RCAD0133 and RCAD0179) for the first time. Analysis of coding sequences (CDS) within the accessory regions of the two ICEs identified several putative virulence factors and an antibiotic resistance gene, which can confer hosts with selective advantages beneficial for niche adaptation. Alignment and comparative analysis of those two ICEs revealed that homologous ICE-like elements are prevalent in other bacterial genomes, and shed light on their phylogenetic relationships and on their modular evolution.
A total of 18 clinical isolates of R.anatipestifer were obtained from the livers of infected ducks and identified using PCR. Among them, 9 strains obtained from Culture Collection of the University of Gothenburg (CCUG) (Table S1). All animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals. The duck-use procedures were approved by the Animal Ethics Committee of the Sichuan Agricultural University (approval No. 2017-013). All isolates were purely cultured for genome sequencing. Genomic DNA was extracted using a TIANamp Bacteria DNA Kit (Tiangen Biotech Co, Ltd., Beijing, China), which was further quantified using a NanoDrop 2000 (Thermo-Scientific).
The sequence of strain RCAD0133 was determined by applying a single-molecule real-time sequencing platform with a PacBio RS II sequencer. The raw data were filtered, and then, a de novo assembly was executed with MHAP (9). The remaining 17 strains were sequenced using an Illumina HiSeq 2500 instrument. A de novo assembly was conducted using Velvet version 1.2.09 (10). The 18 assembled sequences were annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (http://www.ncbi.nlm.nih.gov/genome/annotation_prok). The average nucleotide identity (ANI) of all the R. anatipestifer involved in this study was calculated by pyani (a Python module, https://github.com/widdowquinn/pyani) with the ANIb algorithm.
All 48 R.anatipestifer genome sequences (18 were newly sequenced and 30 sequences were publicly available in the NCBI database) were analyzed using bioinformatics tools. Islandviewer 4 was used to predict genomic islands; also the secretion systems were identified via MacSyFinder, then the genomic islands containing a type IV secretion system (T4SS) (Table S2) and a relaxase were extracted as inputs for the ICEberg server to accurately confirm potential ICEs, and identify which ICE family they belong to (11–14). ICEs integration sites were determined by searching direct repeats (DRs) using a BLASTn alignment of the first and last 400 bp of the predicted ICEs. The ORF sets encoded on the ICERan elements were standardized using the PATRIC annotation tool (15, 16). BLASTp was used to determine the putative functions of the hypothetical proteins encoded by ORFs on ICE elements against the nr database, while conserved domains were identified using the Batch CD-search tool and the Conserved Domain Database (17, 18). Easyfig was used to visualize ICEs organization and their comparisons (19). For amino acid alignments, putative proteins with hits below 40% identity or alignment <120 base pairs were discarded (20).
Searching homologous regions (ICE-like elements) for the putative ICE were performed by NCBI BLASTn comparison analysis using the original confirmed ICE sequence as a probe against the NCBI non-redundant nucleotide sequences(nr) database and the whole-genome shotgun contigs (wgs) database limited by Riemerella taxon.
The amino acid sequences of the ICE host Chromosomal housekeeping genes infB, gyrB, and rpoB; ICE elements core genes and the analogous ICE-like elements core genes were extracted, concatenated, and aligned using MUSCLE (21). A phylogenetic tree was constructed using MEGA v 7.0 with the neighbor-joining method, by Poisson correction and a bootstrap test was performed with 1,000 replications (22).
All 18 newly sequenced nucleotide sequences of R.anatipestifer in this work were submitted to GenBank and all the accession numbers of investigated R.anatipestifer sequences are listed in the supplementary file (Table S1). For the ANI analysis of 48 R.anatipestifer, the minimum ANI was 93% (Supplementary Figure 1).
All 18 novel genome sequences obtained in this study and other publicly available genome sequences from the NCBI database (Table S1) were submitted to the Islandviewer4 server. Among these predicted genomic islands (Table S3, Table S4), two speculative integrative conjugative elements were then confirmed by the ICEberg server (Figure 1) (11, 14). Interestingly, an incomplete ICE was split into two contigs within the RA-JLLY genome sequence, which may bias the study of ICE, and therefore, was not included in this study. ICE elements identified in R.anatipestifer were named ICERan in accordance with the nomenclature system proposed by Burrus, the different ICERan elements were allocated to different strain numbers, ICERanRCAD0133-1, and ICERanRCAD0179-1 (23, 24). ICERanRCAD0133-1 was 70890 bp in size, carrying 77 protein coding sequences(CDSs) and inserted into the vicinity of hypothetical protein at position 991320-1062210, yielding 13 bp direct repeats (5'-AAGTTGAGTTACT-3') representing attL and attR sites. Similarly, ICERanRCAD0179-1 was a 49166 bp genomic region with 51 CDSs and inserted into position 54705-103871 of contig (QXQM00000004) of isolate RCAD0179 near an Arginyl-tRNA synthetase, where it yielded 7 bp direct repeats (5'-GTAACTT-3') located on either ends. The overall GC content of ICERan elements was 33.14% (ICERanRCAD0133-1) and 50.34% (ICERanRCAD0179-1). The G+C content of ICERanRCAD0133-1 (33.14%) was slightly lower than that of the rest of the chromosome (34.85% for the complete chromosomes of RCAD0133). However, the overall GC content of ICERanRCAD0179-1 was 50.34%, which was dramatically higher than the genomic GC content (34.58%) of R. anatipestifer RCAD0179. Comparison analysis of ICERanRCAD0133-1 and ICERanRCAD0179-1 was carried out through the BLASTn and BLASTp, which indicated that these two ICEs were not similar in DNA sequences or protein productions encoded by core genes and therefore were not classified into the same ICE family (11). Next, ICEberg server analysis was performed to exactly identify to which ICE family the ICERan elements belongs; consequently, ICERanRCAD0179-1 was classified into the CTnDOT/ERL family. Nevertheless, ICERanRCAD0133-1 was considered a novel ICE family because of low similarity to previously reported ICEs.
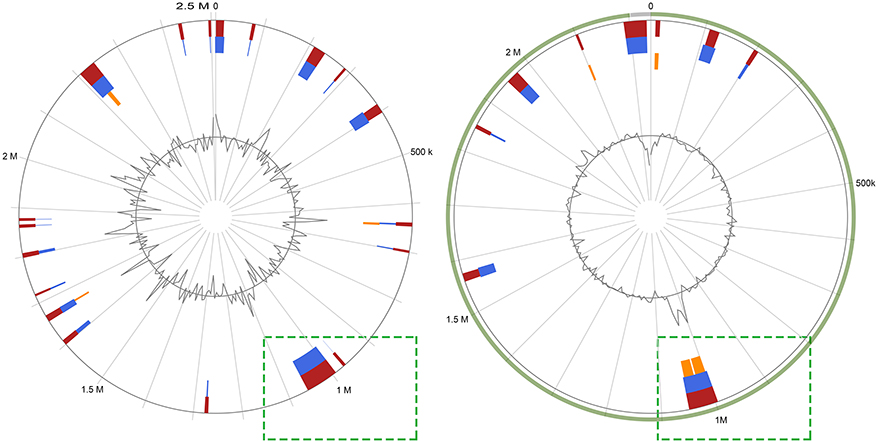
Figure 1. Identification of genomic islands (candidate integrative conjugative elements) Islandviewer 4 analyses of the R.anatipestifer RCAD0133 and R. anatipestifer RCAD0179 chromosomes, with colored regions indicating genomic islands which likely are integrative conjugative elements.
Active ICEs have the ability to excise from the host chromosome with the help of integrase and excisionase, and form an extrachromosomal circular form of the ICE (25). Therefore, to detect the circular intermediate, four pairs of primer pairs were designed that specifically targeted the DRs of ICERanRCAD0133-1 and ICERanRCAD017-1. Surprisingly, no expected product was observed (data not shown).
The cargo genes carried by ICEs often endow novel functions to the host bacteria affecting its life styles and niche adaption, such as development of virulence and acquired of antibiotic resistances (26).
For accessory gene of ICERanRCAD0133-1, it has a Toprim domain protein, which is a catalytic domain found in type IA and II topoisomerases, OLD family nucleases, and RecR proteins, proteins that are involved in DNA replication (27). This ICE had two genes that encode a bacteriocin/lantibiotic efflux ABC transporter. Bacteriocins are small ribosomally-synthesized peptides, that display a highly potent antimicrobial activity. The bacteriocin/lantibiotic efflux ABC transporter was composed of two functional domains: a peptidase C39 domain, which was involved in the processing of bacteriocin precursors at the double glycine, and an ABC transporter domain, which was involved in the export of bacteriocins (28, 29). An arylsulfatase regulator encoded by ICERanRCAD0133-1 can play a significant role in a response against sulfate-limiting conditions to fulfill its sulfate requirements in the bacterial cell (30–32). Interestingly, this ICE also had two genes encoding a putative TonB-dependent receptor, which was involved in siderophore-mediated iron acquisition and played a significant role in bacterial virulence of R.anatipestifer (33). ICERanRCAD0133-1 encoded a mycobacteriophage Barnyard protein gp56 (TIGR03299), which was found in different mycobacterium phage genomes, in Streptomyces coelicolor plasmid SCP1, and in bacterial genomes near various markers that suggest lateral gene transfer. Five insertion sequences (IS) were encoded by this ICE, which were all closely related to ISRa1, first found on the R.anatipestifer plasmid pCFC2, that belongs to the IS982 family; and is widely distributed in various R.anatipestifer strains with 2–20 copies (34).
A special region of isolate RCAD0179 was ICERanRCAD0179-1. Furthermore, comparison analysis revealed that a ribosomal protection type tetracycline resistance gene (tetQ) was carried by this ICE, which is an important contributor to the wide distribution of antibiotic resistance loci. A gene containing a lysozyme_like domain (cl00222) was found on ICERanRCAD0179-1, which is involved in the hydrolysis of β-1,4-linked polysaccharides. Bifunctional deaminase-reductase is involved in catalyzing the second and third steps in the biosynthesis of riboflavin. One such protein was present in ICERanRCAD0179-1, which contained a bifunctional deaminase-reductase domain (35). A gene encoding for prokaryotic ATPase was also identified in this ICE, and it may serve as a significant energy provider for the biological activity of ICERanRCAD0179-1 through ATP hydrolysis.
The two ICERan elements were not identical, and comparative analysis of the coding sequences (CDS) within the conserved region of ICERan elements confirmed the presence of core genes involved in ICE integration and excision, regulation, and maintenance, replication, and conjugative transfer.
In most cases, tyrosine integrase, a site-specific recombinase, frequently serves as a mediator of chromosomal integration and excision of ICEs, while in some cases, it can be replaced by a serine recombinase or DDE transposase as well. Two integrases were identified at one extremity of ICERanRCAD0133-1 and ICERanRCAD0179-1, respectively. An alignment of these two integrases with the characterized recombinases of the tyrosine integrase family suggested that they both contain a characteristic RHRH tetrad with conserved nucleophilic tyrosine at the C-terminus (5, 36). Therefore, they were grouped into the tyrosine integrase family (Supplementary Figure 2). A phylogenetic analysis was performed on the two integrases from ICERan elements with all available tyrosine integrases from the GenBank database. The phylogenetic tree showed that the integrase of ICERanRCAD0133-1 was phylogenetically distinct from other integrases and belonged to a single phylogenetic cluster but was close to Ornithobacterium rhinotracheale (Supplementary Figure 3). For another integrase of ICERanRCAD0179-1, it was closer to and grouped with integrases of the Bacteroides phylum (Supplementary Figure 4).
A set of various genes encoding for regulation and conjugative transfer was detected on ICERan elements; for ICERanRCAD0133-1, including three conjugative DNA processing and transfer clusters (mobI, traABD, traEFGGIJKMNOQ), regulators of excision and conjugative transfer (AbiEi, TetR, and RteC), A single-stranded DNA-binding protein and a DNA topoisomerase that are involved in DNA replication, recombination and repair, and often participate in single-stranded DNA recombination and replication intermediates, respectively, were also found in this ICE (5, 37). Furthermore, ICERanRCAD0133-1 also encoded a putative DNA methylase, which contains a conserved protein N6_Mtase (cl28090) domain, a member of restriction-modification (R-M) systems. These solitary methylases might provide a broad protection from the host R–M systems (38). Among the conserved core genes of ICERanRCAD0179-1, two clusters encoding for ICE conjugation processing and transfer (mobI, mobII, traABCDEGHIJKMNPQ) were identified. Moreover, this ICE had a series of two-component systems for ICE maintenance and regulation (RteA, RteB, RteC) (39, 40). A putative DNA methylase and a DNA topoisomerase III participated in ICE replication, recombination and repair (4). Therefore, all the core genes of the ICERan elements together ensured integration and excision, conjugative transfer, and regulation of ICERan elements.
Nucleotide sequence alignments were performed to identify homologs of ICERan elements among the sequences of ICE-like elements available in the GenBank database, using the nucleotide sequence of ICERan elements as a probe. BLASTn analysis of the ICE in RCAD0133 against the NCBI nr database, suggested that three ICE-like elements similar to ICERanRCAD0133-1 were identified in Ornithobacterium rhinotracheale specie (Table S5). They shared the core backbone for integration, maintenance, and dissemination, and had significant similarity (BLASTp, coverage≥99.0%, identity≥81.0%, E-value 0.0). Comparison among integrases of ICERanRCAD0133-1 and the three ICE-like elements, confirmed that they belonged to the same ICE family (Figure 2). Interestingly, bioinformatics analysis for ICERanRCAD0179-1 suggested that 13 ICE-like elements resemble ICERanRCAD0179-1 and are widely distributed in the genome sequences of several different bacteria including(Table S6):Prevotella intermedia strain KCOM 2033, Bacteroides dorei isolate HS2_L_2_B_045b, Bacteroides fragilis YCH46 DNA, Parabacteroides sp. CT06, Bacteroides caecimuris strain I48, Barnesiella viscericola DSM 18177, Ornithobacterium rhinotracheale ORT-UMN 88, Bacteroides salanitronis DSM 18170,Bacteroides fragilis strain Q1F2,Bacteroides dorei CL03T12C01,Prevotella intermedia strain KCOM 1741,and Riemerella columbina DSM 16469. Based on sequence similarity and structural comparison with the backbone of core genes in ICERanRCAD0179-1, a member of the CTnDOT/ERL family, these putative ICE elements were also classified into the CTnDOT/ERL family. Comparison between ICERan elements and their closely related ICE-like elements was performed with the genome comparison visualizer Easyfig (Figure 3).
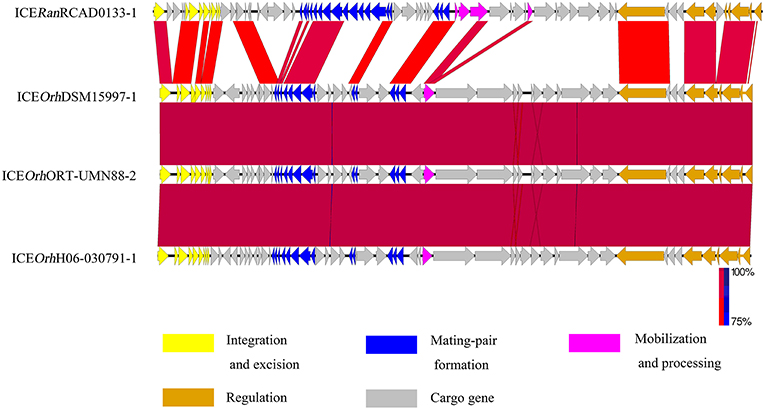
Figure 2. Comparison of the genetic organization of ICERanRCAD0133-1 with its closely related ICEs. Comparison of the genetic organization of ICERanRCAD0133-1 with its homologs in related bacteria. Its closely related ICEs were named with the nomenclature system list in Table S5. Arrows represent predicted CDSs. Highly conserved regions determined by pairwise BLASTn comparisons with E-values lower than 0.001 were plotted. Regions with forward and reverse matches are indicated by red and blue shades, respectively, with color intensity indicating nucleotide identity levels (from 75 to 100%). The absence of red and blue areas denotes no homology. ORFs with different functions are colored differently.

Figure 3. Comparison of the genetic organization of ICERanRCAD0179-1 with its closely related ICEs. Comparison of the genetic organization of ICERanRCAD0179-1 with its homologs in related bacteria. Its closely related ICEs were named with the nomenclature system list in Table S6. Arrows represent predicted CDSs. Highly conserved regions determined by pairwise BLASTn comparisons with E-values lower than 0.001 were plotted. Regions with forward and reverse matches are indicated by red and blue shades, respectively, with color intensity indicating nucleotide identity levels (from 69 to 100%). The absence of red and blue areas denotes no homology.
A high level of synteny was also indicated by alignments of ICERanRCAD0179-1 and 13 closely related ICEs. The ICE phylogenetic tree was constructed based on the concatenated amino acid sequences of 17 conserved proteins (mob1, mob2, RteC, Top, TraD, TraE, TraF, TraG, TraH, TraI, TraJ, TraK, TraL, TraM, TraO, TraP, TraQ) encoded by ICERanRCAD0179-1 and the closely related ICE-like elements to speculate on the evolution of these ICEs. Comparison of the phylogenetic tree topologies produced by neighbor-joining analysis of concatenated amino acid sequences of the conserved ICE CDS and protein products of ICE hosts chromosomal housekeeping gene (gyrA, infB, rpoB) showed that ICERanRCAD0179-1 was clustered with most ICE-like elements in Bacteroides, and the ICE-like element in Riemerella columbina DSM 16469, and the evolutionary relationship of ICEs wasn't consistent with that of the ICE host genes (Figure 4). Furthermore, to trace the evolutionary relationship among these ICEs and anticipate their ancestral root, a phylogenetic tree was produced based on four proteins (Int, TraG, TraI, RteC) corresponding to significant ICE function, such as integration, exclusion determination, transfer, and regulation, respectively. Our analysis showed that ICERanRCAD0179-1 and homologous ICE-like elements in Bacteroides were very closely related and clustered into one branch, while the ICEPinkCOM1741-1 always formed a separate branch (Figure 5).
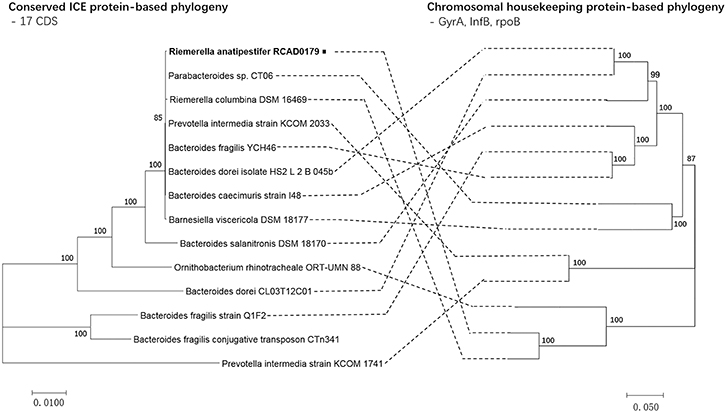
Figure 4. Phylogeny of ICERanRCAD0179-1 and closely related ICEs and comparison to chromosomal housekeeping marker phylogeny. Alignments were performed with MUSCLE using the concatenated amino acid sequences of the ICE elements (17 conserved CDSs) and housekeeping markers (gyrA, infB, rpoB). Phylogenies were constructed using MEGA v 7.0, using the neighbor-joining algorithm, Poisson correction and bootstrap analysis (n = 1000). In genbank, CTn341 only had its own sequence, and its host bacterial sequence wasn't in publicly available.
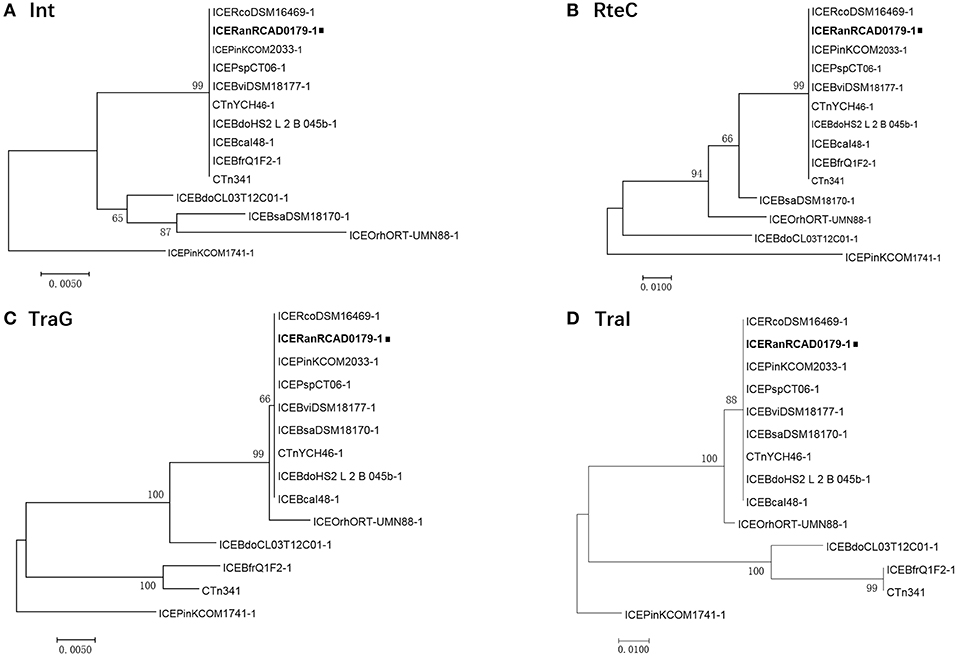
Figure 5. Phylogenetic analysis of ICERanRCAD0179-1 (A) Int, (B) RteC, (C) TraG, and (D) TraI. The trees were constructed by applying the maximum likelihood method based on the Poisson correction model using MEGA v 7.0. Bootstrap analysis with 1,000 replications was performed to test the reliability of each tree.
Genomic sequence analysis first revealed two ICEs (ICERanRCAD0133-1 and ICERanRCAD0179-1) in the R.anatipestifer genomes of RCAD0133 and RCAD0179, respectively. The ICE family is generally determined by the definition that they should have an integrase homology of ≥60% and significant structural synteny. Based on the significant similarity of ICE core modules, ICERanRCAD0133-1 and its related ICEs were grouped in one novel ICE family, due to lack of similarities to known ICE families (11). Similarly, ICERanRCAD0179-1 and its 13 related ICEs were classified into the same ICE family (CTnDOT/ERL). The high prevalence of ICE-like elements similar to ICERanRCAD0179-1 indicated that interspecies genetic exchange also occurred via ICEs, thus broadening the host range, and dissemination of combined cargo genes across different strains. In accordance with the phylogenetic tree of the integrases of these two ICEs, the integrase of ICERanRCAD0133-1 was grouped in a cluster with Ornithobacterium rhinotracheale specie, and the integrase of ICERanRCAD0179-1 had a close phylogeny relationship with Bacteroides phylum because they were in one clade of the phylogenetic tree. Interestingly, integrases of the two ICEs were both classified in the tyrosine integrase family. Therefore, it can be envisaged that the two ICEs may have a similar integration or excision behavior for mobile element DNA because integrases of the ICERan elements are in the same family.
ICEs are self-transmissible mobile genetic elements that have core modules for conjugative transfer and intricate regulatory systems to control their excision from the chromosome (1). Although integrative and conjugative genes were present in ICERanRCAD0133-1 and ICERanRCAD0179, to date we failed to detect a circular intermediate by PCR method via repeated experiments, which suggests that these two ICE elements may have lost the ability to recircularize and transfer to other bacteria.
ICERanRCAD0133-1 had a similar GC content to the host genome, and it can be envisaged that this ICE has been assimilated by the host genome at an early evolutionary timepoint or comes from the bacteria with similar GC content to RCAD0133. Cargo genes encoded on this ICE greatly expanded R.anatipestifer genome variability. R-M systems usually protect a bacterial cell against invasion of foreign DNA by endonucleolytic cleavage of DNA that is unmethylated, and also provide resistance against bacteriophages. The solitary methylase encoded on this ICE might provide a broad protection from the host R–M systems (41). IS elements often encode transposase(s) which can insert randomly into various DNA sites, utilize the replicative mechanisms to generate a copy at a new position, are frequently implicated in ICE recombination, and cause genomic rearrangements including deletion, inversion and cointegrate formation (42). Therefore, ISs shape the host bacterial genome and facilitate the diversity of microbes. Arylsulfatase has been implicated in E. coli infection of the brain microvascular endothelial cells (BMEC) of the host. The presence of arylsulfatase may contribute to the ability of the pathogen to cross the blood-brain barrier leading to meningitis (30–32). Interestingly, an arylsulfatase regulator was present on this ICE, and we hypothesize that it may be implicated in the pathogenicity of R.anatipestifer through participation in the regulation process of arylsulfatase. Based on previous research, we conferred that the putative TonB-dependent receptor encoded on this ICE may significantly influence R.anatipestifer pathogenicity via hemin and iron acquisition. Bacteriocins are synthesized ribosomally, and always display highly potent antimicrobial activity, their production relies on genes encoding a bacteriocin, a bacteriocin ABC transporter, and bacteriocin immunity proteins (43). The bacteriocin/lantibiotic efflux ABC transporter detected on this ICE may participate in the export of peptide bacteriocins across the cytoplasmic membrane. The cargo genes carried on this ICE, with potential for pathogenesis along with other genes in the RCAD0133 strain, also expanded bacterial genome diversity, which may provide further insights into the adaptation mechanisms of the pathogen to thrive under diverse ecological niches.
The ICE element identified in isolate RCAD0179 had a significantly higher GC content than the host genome, which implied that this region was not native and had been adopted by HGT. Comparative analysis confirmed that it belonged to the CTnDOT/ERL family. The spread of resistance determinants was mostly contributed by the conjugative transfer of the CTnDOT/ERL element within the Bacteroides group (44). A tetracycline resistance gene present on ICERanRCAD0179-1 will be an important vehicle for the wide distribution of antibiotic resistance loci. It provided a competitive advantage for the host to rapidly overcome harsh environment. Comparison of the phylogenetic trees of ICERanRCAD0179-1 and related ICE elements against their hosts demonstrated that the compared ICEs were derived from a common ancestor that underwent divergence prior to integration into the host bacterial chromosome. Furthermore, through analysis of the phylogenetic trees based on the single core gene of these ICEs, it was identified that they shared almost the same topology structure of phylogeny, which indicated that the individual core genes most likely have not evolved independently; rather; these ICEs underwent a related evolutionary process or experienced only a low degree of recombination events during spread from a common CTnDOT/ERL family ancestor.
In conclusion, genomic analysis revealed two ICERan elements in R.anatipestifer for the first time. In silico characterization of the ICERanRCAD0179-1 elements showed that similar ICE-like elements were found in other Bacteroides and that these ICEs are likely derived from a common ancestor. The two ICERan elements possess the core elements required for ICE DNA integration, maintenance and regulation, and dissemination. The presence of these core modules, however, can't prove that the ICERan elements can excise, circularize, conjugative transfer and then integrate within the chromosome. Genes encoding resistance to tetracycline antibiotics, DNA repair and recombination systems, virulence factors, toxin-antitoxin system, and regulation of conjugative transfer, were found to be present within cargo genes of ICERan elements. In particular, the putative antibiotic biosynthetic loci encoded on ICERan elements provide potential targets for the exploration of R.anatipestifer antibiosis phenotypes. ICERan elements could therefore play a significant role in providing beneficial phenotypes to their hosts. With the emergence of massive amounts of available genome sequences, it can be reasonably envisaged that more novel ICE elements will be identified and novel ICE families will be described. It may assist in studying the special mechanisms by which ICE impacts host metabolism and physiology.
DZ and AC conceived and designed the project. JW, JX, LZ, YL, and XC cultured bacteria and extracted genomic DNA. ZY, ML, RJ, SC, JX, and MW assembled and annotated the genomes. JW, XZ, QY, YY, and YW performed genomic islands analysis. JW, ZY, and SZ performed ICE identification and related ICE search. JW, DZ, and ZY performed the construction of the phylogenetic tree. JW and DZ drafted and revised the manuscript. All authors have read and approved the final version manuscript.
This work was supported by the National Key Research and Development Program of China under Grant No. 2017YFD050080; China Agricultural Research System under Grant No. CARS-42-17; Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System under Grant No. CARS-SVDIP; International S&T Cooperation Program of Sichuan Province under Grant No. 2017HH0026; Science and Technology Innovation Program of Guizhou Academy of Agricultural Science under Grant No. (2017)03.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2019.00128/full#supplementary-material
1. Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. (2010) 8:552. doi: 10.1038/nrmicro2382
2. Gyles C, Boerlin P. Horizontally transferred genetic elements and their role in pathogenesis of bacterial disease. Vet Pathol. (2014) 51:328–40. doi: 10.1177/0300985813511131
3. Burrus V, Marrero J, Waldor MK. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. (2006) 55:173–83. doi: 10.1016/j.plasmid.2006.01.001
4. Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. (2004) 155:376–86. doi: 10.1016/j.resmic.2004.01.012
5. Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Ann Rev Genet. (2015) 49:577–601. doi: 10.1146/annurev-genet-112414-055018
6. Burrus V. Mechanisms of stabilization of integrative and conjugative elements. Curr Opin Microbiol. (2017) 38:44–50. doi: 10.1016/j.mib.2017.03.014
7. Yu CY, Liu YW, Chou SJ, Chao MR, Weng BC, Tsay JG, et al. Genomic diversity and molecular differentiation of Riemerella anatipestifer associated with eight outbreaks in five farms. Avian Pathol. (2008) 37:273–9. doi: 10.1080/03079450802056546
8. Miao S, Xing L, Qi J, Yu H, Jiang P, Sun B, et al. Roles of the TonB1 and TonB2 proteins in haemin iron acquisition and virulence in Riemerella anatipestifer. Microbiology. (2015) 161:1592–9. doi: 10.1099/mic.0.000123
9. Berlin K, Koren S, Chin C-S, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. (2015) 33:623. doi: 10.1038/nbt.3238
10. Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. (2008) 18:821–9. doi: 10.1101/gr.074492.107
11. Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, et al. ICEberg: a web-based resource for integrative and conjugative elements found in bacteria. Nucleic Acids Res. (2011) 40:D621–6. doi: 10.1093/nar/gkr846
12. Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. MacSyFinder: a program to mine genomes for molecular systems with an application to CRISPR-cas systems. PLoS ONE. (2014) 9:e110726. doi: 10.1371/journal.pone.0110726
13. Abby SS, Rocha EPC. Identification of protein secretion systems in bacterial genomes using macsyfinder In: Journet L, Cascales E, editors Bacterial Protein Secretion Systems: Methods and Protocols (New York, NY: Springer New York) (2017), p. 1–21. doi: 10.1007/978-1-4939-7033-9_1
14. Bertelli C, Laird MR, Williams KP, Group SFURC, Lau BY, Hoad G, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. (2017) 45:W30–W35. doi: 10.1093/nar/gkx343
15. Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. (2015) 5:8365. doi: 10.1038/srep08365
16. Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. (2016) 45:D535–42. doi: 10.1093/nar/gkw1017
17. Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. (2004) 32:W327–31. doi: 10.1093/nar/gkh454
18. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. (2011) 39:D225–9. doi: 10.1093/nar/gkq1189
19. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. (2011) 27:1009–10. doi: 10.1093/bioinformatics/btr039
20. Pearson WR. An introduction to sequence similarity (“homology”) searching. Curr Prot Bioinformat. (2013) Chapter 3:Unit 3.1 3.1.1-7. doi: 10.1002/0471250953.bi0301s27
21. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7. doi: 10.1093/nar/gkh340
22. Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33, 1870. doi: 10.1093/molbev/msw054
23. Burrus V, Pavlovic G, Decaris B, Guédon G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid. (2002) 48:77–97. doi: 10.1016/S0147-619X(02)00102-6
24. Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, et al. Revised nomenclature for transposable genetic elements. Plasmid. (2008) 60:167–73. doi: 10.1016/j.plasmid.2008.08.001
25. Sultan I, Rahman S, Jan AT, Siddiqui MT, Mondal AH, Haq QM R. Antibiotics, Resistome and Resistance Mechanisms: A Bacterial Perspective. Front. Microbiol. (2018) 9:2066. doi: 10.3389/fmicb.2018.02066
26. Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. (2018) 31:e00088–e00017. doi: 10.1128/cmr.00088-17
27. Aravind L, Leipe DD, Koonin EV. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. (1998) 26:4205–13. doi: 10.1093/nar/26.18.4205
28. McAuliffe O, Ross RP, Hill C. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol Rev. (2001) 25:285–308. doi: 10.1111/j.1574-6976.2001.tb00579.x
29. Nishie M, Nagao J-I, Sonomoto K. Antibacterial peptides “bacteriocins”: an overview of their diverse characteristics and applications. Biocontrol Sci. (2012) 17:1–16. doi: 10.4265/bio.17.1
30. Hoffman JA, Badger JL, Zhang Y, Huang SH, Kim KS. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect Immun. (2000) 68:5062–7. doi: 10.1128/IAI.68.9.5062-5067.2000
31. Frank T, Gody JC, Nguyen LBL, Berthet N, Fleche-Mateos AL, Bata P, et al. First case of Elizabethkingia anophelis meningitis in the Central African Republic. Lancet. (2013) 381:1876. doi: 10.1016/S0140-6736(13)60318-9
32. Lau SKP, Wu AKL, Teng JLL, Tse H, Curreem SOT, Tsui SKW, et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg Infect Dis. (2015) 21:232–41. doi: 10.3201/eid2102.140623
33. Lu F, Miao S, Tu J, Ni X, Xing L, Yu H, et al. The role of TonB-dependent receptor TbdR1 in Riemerella anatipestifer in iron acquisition and virulence. Vet Microbiol. (2013) 167:713–8. doi: 10.1016/j.vetmic.2013.08.020
34. Weng S-C, Lin W-H, Chang C-F, Chang Y-F. Identification of a virulence-associated protein homolog gene and ISRa1 in a plasmid of Riemerella anatipestifer. FEMS Microbiol Lett. (1999) 179:11–9. doi: 10.1111/j.1574-6968.1999.tb08701.x
35. Richter G, Fischer M, Krieger C, Eberhardt S, Luttgen H, Gerstenschlager I, et al. Biosynthesis of riboflavin: characterization of the bifunctional deaminase-reductase of Escherichia coli and Bacillus subtilis. J Bacteriol. (1997) 179:2022–8. doi: 10.1128/jb.179.6.2022-2028.1997
36. Esposito D, Scocca JJ. The integrase family of tyrosine recombinases: evolution of a conserved active site domain. Nucleic Acids Res. (1997) 25:3605–14. doi: 10.1093/nar/25.18.3605
37. Cury J, Touchon M, Rocha EPC. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res. (2017) 45:8943–56. doi: 10.1093/nar/gkx607
38. Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. Restriction–modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol Microbiol. (2000) 37:1052–65. doi: 10.1046/j.1365-2958.2000.02049.x
39. Stevens AM, Sanders JM, Shoemaker NB, Salyers AA. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J Bacteriol. (1992) 174:2935. doi: 10.1128/jb.174.9.2935-2942.1992
40. Wang Y, Shoemaker NB, Salyers AA. Regulation of a bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J Bacteriol. (2004) 186:2548. doi: 10.1128/JB.186.9.2548-2557.2004
41. Jeltsch A. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene. (2003) 317:13–6. doi: 10.1016/S0378-1119(03)00652-8
42. Hickman AB, Chandler M, Dyda F. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit Rev Biochem Mol Biol. (2010) 45:50–69. doi: 10.3109/10409230903505596
43. Havarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. (1995) 16:229–40. doi: 10.1111/j.1365-2958.1995.tb02295.x
Keywords: Riemerella anatipestifer, integrative conjugative elements, cargo genes, antimicrobial resistance, virulence
Citation: Zhu D, Wan J, Yang Z, Xu J, Wang M, Jia R, Chen S, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Chen X and Cheng A (2019) First Report of Integrative Conjugative Elements in Riemerella anatipestifer Isolates From Ducks in China. Front. Vet. Sci. 6:128. doi: 10.3389/fvets.2019.00128
Received: 28 December 2018; Accepted: 05 April 2019;
Published: 24 April 2019.
Edited by:
Subhash Verma, CSK Himachal Pradesh Agricultural University, IndiaReviewed by:
Shaohui Wang, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2019 Zhu, Wan, Yang, Xu, Wang, Jia, Chen, Liu, Zhao, Yang, Wu, Zhang, Liu, Zhang, Yu, Chen and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.