- 1Department of Surgery, University of Pittsburgh, Pittsburgh, PA, United States
- 2Center for Inflammation and Regeneration Modeling, McGowan Institute for Regenerative Medicine, University of Pittsburgh, Pittsburgh, PA, United States
- 3Center for Systems Immunology, University of Pittsburgh, Pittsburgh, PA, United States
Introduction: My Personal Journey to the Frontier of Systems Biology
This manuscript is intended as a general discussion on the role and potential impact of systems biology in the broader context of biomedical research, as a brief meditation on my own journey in systems biology, and as a statement of purpose for our new journal. To me, these elements are all intertwined, since I fell into systems biology somewhat coincidentally during a time (the turn of the 21st century) when this discipline was truly new [at least in the way it was defined then (Ideker et al., 2001) and largely still now, as opposed to the fairly long history of mathematics applied to the study of biological systems (Edelstein-Keshet, 1988)].
As an introduction into the mission of Frontiers in Systems Biology, I think it is useful to recapitulate my own path into systems biology, since the concepts espoused in this journal’s mission statement derive fairly directly from my own experiences; I believe that the same holds true for my Section Chief Editor colleagues. I obtained my Ph.D. in Immunology in 1993, but my life in biological research was not a foregone conclusion since I had an early interest in the application of mathematics to biology (the setting in which I understood the power of mathematics for the first time, in high school) as well as a concurrent interest in the application of computers to medical practice (having generated a medical practice software program in the mid-1980s to help my father run his surgical practice). My undergraduate, graduate, and postdoctoral research focused on biochemistry and immunology in diverse contexts (sepsis, cancer, neurodegenerative diseases), leading to the realization that diverse diseases seemed to be more alike than different with regard to the involvement of inflammation in their etiology or pathogenesis. This realization was influenced strongly by the generalist view of biology—and especially of inflammation—of my mentors (Nathan and Sporn, 1991; Nathan, 2002). Generalism and synthesis, however, were out of favor at that time, a culmination of multiple efforts at gaining insights into the molecular biology of disease at the cellular level that, unfortunately, resulted in a plethora of data from micro-focused studies but a dearth of true insight into how biology was wired (Mesarovic et al., 2004; Kirschner, 2005), with the consequence being insufficient clinical translation and other practical applications of this vast body of reductionist science (Ideker et al., 2001; An, 2014).
As luck would have it, I came to this realization after arriving at the University of Pittsburgh in 1999, shortly after which I was invited to collaborate on a quixotic project aimed at mathematical modeling of critical illness (Clermont et al., 2001; Kumar et al., 2001). Via this interdisciplinary collaboration, I learned the power of being able to express biology in the language of intertwined differential equations or programming rules, and the challenges inherent in interdisciplinary interactions to make this happen on a larger scale (Vodovotz et al., 2007). We (and others) helped define the theoretical dynamics of acute inflammation (An, 2001; Kumar et al., 2004), to apply these models for the design of in silico clinical trials (An, 2004; Clermont et al., 2004), to suggest the presence of a single set of mechanisms that drive inflammation induced by diverse stimuli (Chow et al., 2005), and to create the first individual-specific, mechanistic models of inflammation in humans (Li et al., 2008; Brown et al., 2015). In the process, I came to know true interdisciplinary research, and have lived through ∼20 of the most fascinating, productive, engaging, fulfilling, frustrating, and at times terrifying years of my life.
This has culminated in my now taking on the role of Field Chief Editor of this journal. Frontiers in Systems Biology represents the unification, under one title, of multiple sections on systems biology previously listed under separate Frontiers journals. As I noted, this is a deeply personal journey for me. I am both tremendously grateful to have this opportunity and acutely aware of the need to push beyond the status quo. This is because despite major conceptual, methodological, and practical advances that have brought systems biology and related fields (e.g., bioinformatics) to the fore, many challenges remain if systems biology is to achieve the lofty goals envisioned by researchers (Ideker et al., 2001; Kitano, 2002; An, 2014; Levine, 2017), policymakers (Brüninghaus et al., 2015), and the public at large (Burke and Trinidad, 2011; Schleidgen et al., 2017). When viewed individually, these challenges may seem as disparate and specific to particular domains and/or constituencies. In contrast, I see these challenges as stemming from a fundamental lack of agreement on underlying principles, conceptual approaches, and ultimate use that have led to a state of fragmentation. As I outline below, the goal of this journal is to address this core challenge not to drive conflict among constituents or to pick favorites, but as an opportunity to improve systems biology as a discipline. After all, systems biology is about integrating the parts to discern the whole. You may concur or disagree; the editorial staff and I welcome your feedback.
Integration, A Grand Challenge for Systems Biology
Research in biology and biomedicine is tackling more complex questions than ever before, due to the seemingly intractable nature of the urgent questions affecting us all. The ongoing COVID-19 pandemic has brought this into sharp relief for scientists, policymakers, and the population at large. Addressing these issues requires an integrated, interdisciplinary approach, and yet much of research in the field of biology—systems biology included—remains entrenched in silos based on distinct disciplines and methods. Leading journals have taken steps to drive integration across fields by the very nature of the way they present their content (Nature_Biotechnology, 2016).
Unfortunately, systems biology (much like the reductionist research that this discipline is rapidly supplanting) suffers from fragmentation. While the challenge in the early 2000s was to demonstrate the value of systems biology relative to reductionist approaches (Neugebauer et al., 2001; Ahn et al., 2006), the current challenge lies more at the interface of pure data-driven machine learning approaches vs. principles-based, mechanistic approaches (Azhar et al., 2013; Alber et al., 2019). Another area of fragmentation in systems biology, reflecting that seen in the broader context of biomedical research, involves the tension between gleaning basic insights vs. driving clinical translation or other practical applications to better human health and wellbeing (Vodovotz et al., 2008). The advent of “multi-omics” (Haas et al., 2017; Hasin et al., 2017) has highlighted the paradoxically siloed nature of individual omics approaches. Perhaps less obvious is the fragmentation inherent in systems biology research focused on a specific biological goal vs. methodologically driven work aimed at driving advances in modeling theory. Finally, there is currently no meaningful interaction between the systems biology community and the systems engineering community and its rich tradition and toolset of systems thinking (Mesarovic et al., 2004).
The New Frontier in Systems Biology
Frontiers in Systems Biology is therefore dedicated to the concept of integration across disciplines, across modeling scales, across datasets, and across computational methodologies. Though this journal is still in its formative stages, we have structured Frontiers in Systems Biology into nine sections that should be viewed as integral parts of a whole (Figure 1), wherein discrete fields (evolution, genetics/genomics, neuroscience, immunology, and hopefully more to come) leverage a common set of methods (e.g., multiscale mechanistic modeling, integrative omics) in a manner that is, undergirded by a clear grounding in systems concepts and theory and with the ultimate goal of driving clinical and other practical advances to better the human condition in this increasingly complex and intertwined world. We hope you will join us in integrating mindsets and toolsets at Frontiers in Systems Biology, as we do our small part in helping this discipline to live up to the promise of integrating theory, experimentation, and practical application in an ethical and sustainable context.
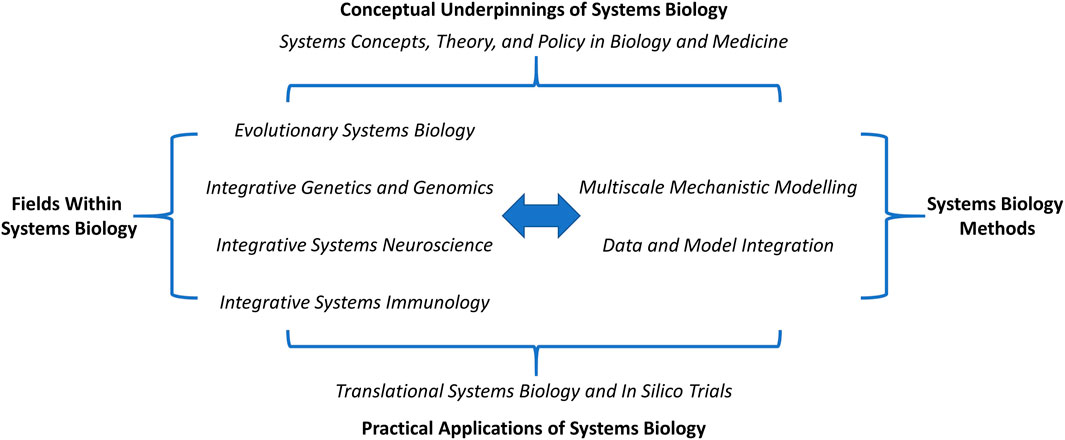
FIGURE 1. Frontiers in Systems Biology, a platform for integration across disciplines, methods, and applications. This journal has been structured into nine sections to highlight common conceptual underpinnings and methodological approaches as a means for driving practical outcomes in core systems biology content areas.
Author Contributions
YV is the sole author, having conceived, written, and edited this manuscript.
Conflict of Interest
YV is also a co-founder of, and stakeholder in, Immunetrics, Inc. He holds several patents related to commercial applications of systems and computational biology.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
My colleagues who are Section Chief Editors share in this vision and have contributed to it; I am especially thankful to my collaborators and now journal colleagues Gary An (Section Chief Editor, Translational Systems Biology and In Silico Trials) and Harinder Singh (Section Chief Editor, Integrative Systems Immunology) in this regard.
References
Ahn, A. C., Tewari, M., Poon, C.-S., and Phillips, R. S. (2006). The Limits of Reductionism in Medicine: Could Systems Biology Offer an Alternative? Plos Med. 3 (6), e208. doi:10.1371/journal.pmed.0030208
Alber, M., Buganza Tepole, A., Cannon, W. R., De, S., Dura-Bernal, S., Garikipati, K., et al. (2019). Integrating Machine Learning and Multiscale Modeling-Perspectives, Challenges, and Opportunities in the Biological, Biomedical, and Behavioral Sciences. Npj Digit. Med. 2, 115. doi:10.1038/s41746-019-0193-y
An, G. (2001). Agent-based Computer Simulation and SIRS: Building a Bridge Between Basic Science and Clinical Trials. Shock 16 (4), 266–273. doi:10.1097/00024382-200116040-00006
An, G. (2004). In Silico Experiments of Existing and Hypothetical Cytokine-Directed Clinical Trials Using Agent-Based Modeling. Crit. Care Med. 32, 2050–2060. doi:10.1097/01.ccm.0000139707.13729.7d
An, G. V. (2014). Translational Systems Biology: Concepts and Practice for the Future of Biomedical Research. New York, NY: Elsevier.
Azhar, N., Mi, Q., Ziraldo, C., Buliga, M., Constantine, G., and Vodovotz, Y. (2013). “Integrating Data Driven and Mechanistic Models of the Inflammatory Response in Sepsis and Trauma,” in Complex Systems and Computational Biology Approaches to Acute Inflammation. Editors Y. Vodovotz, and G. An (New York: Springer). doi:10.1007/978-1-4614-8008-2_8
Brown, D., Namas, R. A., Almahmoud, K., Zaaqoq, A., Sarkar, J., Barclay, D. A., et al. (2015). Trauma In Silico: Individual-specific Mathematical Models and Virtual Clinical Populations. Sci. Transl. Med. 7, 285ra61. doi:10.1126/scitranslmed.aaa3636
Brüninghaus, A., Petersen, I., Kollek, R., and Döring, M. (2015). Science Policy of Systems Biology. Contextualizing Systems Biology., 213–260. doi:10.1007/978-3-319-17106-7_5
Burke, W., and Trinidad, S. (2011). Systems Medicine and the Public's Health. Genome Med. 3 (7), 47. doi:10.1186/gm263
Chow, C. C., Clermont, G., Kumar, R., Lagoa, C., Tawadrous, Z., Gallo, D., et al. (2005). The Acute Inflammatory Response in Diverse Shock States. Shock 24, 74–84. doi:10.1097/01.shk.0000168526.97716.f3
Clermont, G., Bartels, J., Kumar, R., Constantine, G., Vodovotz, Y., and Chow, C. (2004). In Silico Design of Clinical Trials: a Method Coming of Age. Crit. Care Med. 32, 2061–2070. doi:10.1097/01.ccm.0000142394.28791.c3
Clermont, G., Chow, C., Kumar, R., and Vodovotz, Y. (2001). Mathematical Simulation of the Innate Immune Response. Crit. Care Med. 29 (Suppl. l), A111.
Haas, R., Zelezniak, A., Iacovacci, J., Kamrad, S., Townsend, S., and Ralser, M. (2017). Designing and Interpreting 'Multi-Omic' Experiments that May Change Our Understanding of Biology. Curr. Opin. Syst. Biol. 6, 37–45. doi:10.1016/j.coisb.2017.08.009
Hasin, Y., Seldin, M., and Lusis, A. (2017). Multi-Omics Approaches to Disease. Genome Biol. 18 (1), 83. doi:10.1186/s13059-017-1215-1
Ideker, T., Galitski, T., and Hood, L. (2001). A NEWAPPROACH TODECODINGLIFE: Systems Biology. Annu. Rev. Genom. Hum. Genet. 2, 343–372. doi:10.1146/annurev.genom.2.1.343
Kirschner, M. W. (2005). The Meaning of Systems Biology. Cell 121 (4), 503–504. doi:10.1016/j.cell.2005.05.005
Kitano, H. (2002). Systems Biology: a Brief Overview. Science 295 (5560), 1662–1664. doi:10.1126/science.1069492
Kumar, R., Chow, C. C., Clermont, G., and Vodovotz, Y. (2001). A Theoretical Model for the Simulation of Sepsis. J. Leukoc. Biol. Supplement, 90.
Kumar, R., Clermont, G., Vodovotz, Y., and Chow, C. C. (2004). The Dynamics of Acute Inflammation. J. Theor. Biol. 230, 145–155. doi:10.1016/j.jtbi.2004.04.044
Levine, A. J. (2017). The Future of Systems Biology. Curr. Opin. Syst. Biol. 1, v. doi:10.1016/j.coisb.2017.02.007
Li, N. Y. K., Verdolini, K., Clermont, G., Mi, Q., Rubinstein, E. N., Hebda, P. A., et al. (2008). A Patient-specific In Silico Model of Inflammation and Healing Tested in Acute Vocal Fold Injury. Plos One 3, e2789. doi:10.1371/journal.pone.0002789
Mesarovic, M. D., Keene, J. D., and Sreenath, S. N. (2004). Search for Organising Principles: Understanding in Systems Biology. Syst. Biol. (Stevenage) 1 (1), 19–27. doi:10.1049/sb:20045010
Nathan, C. (2002). Points of Control in Inflammation. Nature 420 (6917), 846–852. doi:10.1038/nature01320
Nathan, C., and Sporn, M. (1991). Cytokines in Context. J. Cell Biol. 113, 981–986. doi:10.1083/jcb.113.5.981
Nature_Biotechnology, (2016). So Long to the Silos. Nat. Biotechnol. 34 (4), 357. doi:10.1038/nbt.3544
Neugebauer, E. A. M., Willy, C., and Sauerland, S. (2001). Complexity and Non-Linearity in Shock Research: Reductionism or Synthesis? Shock 16 (4), 252–258. doi:10.1097/00024382-200116040-00003
Schleidgen, S., Fernau, S., Fleischer, H., Schickhardt, C., Oßa, A.-K., and Winkler, E. C. (2017). Applying Systems Biology to Biomedical Research and Health Care: a Précising Definition of Systems Medicine. BMC Health Serv. Res. 17 (1), 761. doi:10.1186/s12913-017-2688-z
Vodovotz, Y., Csete, M., Bartels, J., Chang, S., and An, G. (2008). Translational Systems Biology of Inflammation. Plos Comput. Biol. 4, e1000014–6. doi:10.1371/journal.pcbi.1000014
Keywords: systems biology, computational biology, immunology, neuroscience, genomics, microbiome
Citation: Vodovotz Y (2021) Integrating Mindsets and Toolsets at the Frontier of Systems Biology. Front. Syst. Biol. 1:745692. doi: 10.3389/fsysb.2021.745692
Received: 22 July 2021; Accepted: 10 August 2021;
Published: 30 August 2021.
Edited and reviewed by:
Rongling Wu, The Pennsylvania State University (PSU), United StatesCopyright © 2021 Vodovotz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoram Vodovotz, dm9kb3ZvdHp5QHVwbWMuZWR1
 Yoram Vodovotz
Yoram Vodovotz