- Laboratory for Chemical Synthetic Biology and Xenobiology, Department of Chemistry, University of Manitoba, Winnipeg, MB, Canada
A Commentary on
Evolving a mitigation of the stress response pathway to change the basic chemistry of life
by Tolle I, Oehm S, Hoesl MG, Treiber-Kleinke C, Peil L, Bozukova M, Albers S, Adamu Bukari A-R, Semmler T, Rappsilber J, Ignatova Z, Gerstein AC and Budisa N (2023) Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1:1248065. doi: 10.3389/fsybi.2023.1248065
In his 1859 work “On the Origin of Species,” Darwin acknowledged evolution as a slow process not readily observable (Darwin, 1859). However, in 1878, Reverend Dallinger conducted a groundbreaking experiment (Hass, 2000). Cultivating protozoa in a controlled environment with increasing temperatures, he observed them adapt to higher heat levels. Darwin, upon learning of this, remarked on the significance of the results (Lenski, 2011). Dallinger’s experiment, the first Adaptive Laboratory Evolution study, demonstrated the adaptability of even simple organisms and provided a tangible observation of evolution within a human lifetime.
In the 1960s, non-canonical amino acids (ncAAs) were viewed as growth inhibitors and antimetabolites (Richmond, 1962). However, the pioneering works of Wong (1983) and Bacher and Ellington (2001), in which tryptophan (Trp) was replaced by fluorinated analogs, showed that microbes can adapt adeptly in synthetic microenvironments and achieve substantial replacement levels. Trp, a rare amino acid encoded by a single TGG codon, is an ideal target due to its recent addition to the genetic code (Fournier and Alm, 2015), with diverse anthropogenically produced indole side chains like fluoroindole (or fluorotryptophan) (Budisa and Paramita Pal, 2004). Despite this, reassigning over 20,000 codons in Escherichia coli to chemically modified analogs remain a formidable task (Zhang and Ellington, 2020).
Notably, strict analytical evidence for the full replacement of Trp with analogs in the proteome was elusive until 2015 when Hoesl et al. (2015) reported on the evolution of the chemical composition of the Escherichia coli. Using adaptive laboratory evolution (ALE), they were able to prove analytically that all Trp residues were completely replaced by the non-canonical amino acid analog L-beta-(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa). Subsequently, in 2019, Agostini et al. (2020) performed ALE experiments, successfully incorporating 4- and 5-fluorotryptophan into the entire E. coli proteome.
There are two reports in the current issue of Frontiers in Synthetic Biology that fundamentally address these questions. The first report by Tolle et al. (2023) provides a mechanistic understanding of the complete adaptation of E. coli to ([3,2]Tpa) as the sole replacement source for Trp. In the second report, Treiber-Kleinke et al. (2024) present a comprehensive study focusing on fermentation protocols that enable the strict, proteome-wide replacement of Trp with 6- and 7-fluorotryptophan.
Performing a “clean” proteome-wide exchange in microbial cells with a well-defined auxotrophic metabolic prototype and defined genomic background is challenging for several reasons, primarily rooted in empirical experimental design. The traditional approach involves serial dilution in minimal media with decreasing Trp concentrations over time, alongside a constant high level of analogs throughout ALE (Figure 1). However, difficulties arise due to the presence of traces of Trp in commercially available preparations, even those labeled as “ultrapure.” Cells tend to adhere to these residual traces under selection pressure for analog use.
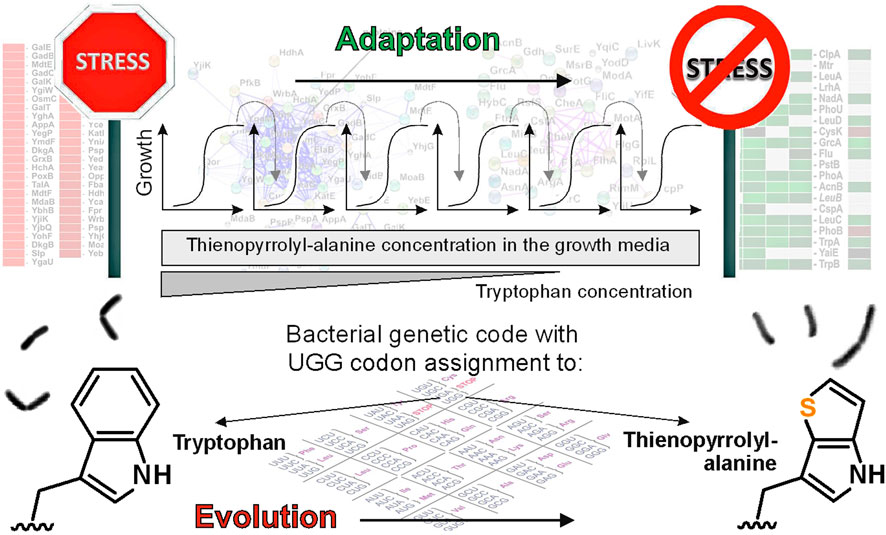
Figure 1. Escherichia coli is able to overcome the frozen state of the genetic code by mitigating its own stress response. In this context, the work of Tolle et al. (2023) is important evidence that the genetic code and the rigidity (conservation) of the protein translation machinery are also “guarded” by the regulatory network of metabolism, signal transduction and physiology in general.
In the works of Hoesl et al. (2015), Agostini et al. (2020), Tolle et al. (2023), and Treiber-Kleinke et al. (2024), overcoming challenges involved the utilization of chemically pure fluoroindole or analogous compounds like thienopyrrole. These were meticulously synthesized to exclude any presence of canonical (“natural”) Trp. In addition, state-of-the-art mass spectrometric analyses of proteomes and metabolomes were performed to ensure a thorough examination of the cellular composition and to detect any traces of Trp in the experimental setup.
Now, armed with well-established empirical protocols, we can advance to contemplate the chemical evolution of synthetic cells using various synthetic ncAAs. This involves a genomic-level approach, monitoring the emergence of key mutations in genes associated with various cellular processes, including the general stress response, amino acid metabolism, stringent response, and chemotaxis. Understanding these adaptation mechanisms to non-canonical biomass components is crucial for informing strategies in engineering synthetic metabolic pathways and cells (Lefèvre-Morand et al., 2024). With a substantial body of empirical data on ALE through ncAA proteome-wide insertions, the significance of the “oligogenic barrier” (Mat et al., 2010) becomes increasingly apparent. This barrier comprises a relatively small number of genes that must undergo mutation to facilitate the successful insertion of a new, non-canonical amino acid into the genetic code (Acevedo-Rocha and Schulze-Makuch, 2015).
The overcoming of these hurdles is illustrated by the discovery of Tolle et al. (2023): adapted bacterial strains successfully overcome the adverse effects associated with the incorporation of synthetic amino acids at Trp positions. This adaptation primarily involves the suppression of the growth-inhibitory stress response within the regulatory networks of the bacterial strains (Figure 1). Essentially, these strains have evolved to enforce a phenotype capable of utilizing [3,2] Tpa as a core building block by effectively modulating their regulatory networks, particularly by suppressing the stress response.
It is crucial to emphasize that substituting amino acids involves chemically diversifying their side chains. Efforts to modify the “alanine core” (Kubyshkin and Budisa, 2019a), as seen with proline analogs (Kubyshkin and Budisa, 2019b), are generally poorly tolerated and frequently rejected by the universally conserved protein translation machinery. Nevertheless, these experiments underscore the remarkable adaptability of the protein translation apparatus to chemical variations in amino acid side chains. The works of Hoesl, Agostini, Tolle, and Treiber-Kleinke clearly show that, among other factors, the stress response acts as both a physical and a biological constraint when attempting to alter the amino acid repertoire of the genetic code.
The findings emphasize that the universally conserved repertoire can be experimentally altered by addressing biological constraints, such as specific metabolic regulatory networks that have played a role in maintaining or “freezing” the code (Figure 1). Consequently, the laboratory-driven reassignment of codons becomes a feasible task when conservation mechanisms like the stress response or the quality of protein folding are effectively mitigated or bypassed. This illustrates the potential for further advances in our ability to “unfreeze” the genetic code through specific interventions in biological processes, using an efficient top-down approach to alter the chemical composition of living cells.
These experiments will provide a critical mass of empirical data that will enable us to use sophisticated genome-editing tools to configure chassis to adapt and thrive in man-made chemical processes. We are indeed at the very beginning of the long journey towards synthetic species away from the “old” living world (Marliere, 2009). With a built-in genetic firewall ensured by biological compounds of mostly anthropogenic origin, these chassis will allow us to create and explore strange new life forms and establish Xenobiology (Budisa et al., 2020) as the science of alien life forms.
Author contributions
NB: Conceptualization, Writing–original draft, Writing–review and editing.
Acknowledgments
Dedicated to Professor Luis Moroder on his 83rd birthday.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acevedo-Rocha, C. G., and Schulze-Makuch, D. (2015). How many biochemistries are available to build a cell? ChemBioChem 16 (15), 2137–2139. doi:10.1002/cbic.201500379
Agostini, F., Sinn, L., Petras, D., Schipp, C. J., Kubyshkin, V., Berger, A. A., et al. (2020). Multiomics analysis provides insight into the laboratory evolution of Escherichia coli toward the metabolic usage of fluorinated indoles. ACS Central Sci. 7 (1), 81–92. doi:10.1021/acscentsci.0c00679
Bacher, J. M., and Ellington, A. D. (2001). Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 183 (18), 5414–5425. doi:10.1128/JB.183.18.5414-5425.2001
Budisa, N., Kubyshkin, V., and Schmidt, M. (2020). Xenobiology: a Journey towards parallel life forms. ChemBioChem 21 (16), 2228–2231. doi:10.1002/cbic.202000141
Budisa, N., and Paramita Pal, P. (2004). Designing novel spectral classes of proteins with a tryptophan-expanded genetic code. Biol. Chem. 385 (10), 893–904. doi:10.1515/BC.2004.117
Darwin, C. (1859). On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. (London: John Murray, Albemarle Street, 1859).
Fournier, G. P., and Alm, E. (2015). Ancestral reconstruction of a pre-LUCA aminoacyl-tRNA synthetase ancestor supports the late addition of Trp to the genetic code. J. Mol. Evol. 80, 171–185. doi:10.1007/s00239-015-9672-1
Hass, J. W. (2000). The Reverend Dr William Henry Dallinger, F.R.S. (1839-1909). Notes Rec. R. Soc. Lond 54, 53–65. doi:10.1098/rsnr.2000.0096
Hoesl, M. G., Oehm, S., Durkin, P., Darmon, E., Peil, L., Aerni, H. R., et al. (2015). Chemical evolution of a bacterial proteome. Angew. Chem. Int. Ed. 54 (34), 10030–10034. doi:10.1002/anie.201502868
Kubyshkin, V., and Budisa, N. (2019a). Anticipating alien cells with alternative genetic codes: away from the alanine world. Curr. Opin. Biotechnol. 60, 242–249. doi:10.1016/j.copbio.2019.05.006
Kubyshkin, V., and Budisa, N. (2019b). The alanine world model for the development of the amino acid repertoire in protein biosynthesis. Int. J. Mol. Sci. 20 (21), 5507. doi:10.3390/ijms20215507
Lefèvre-Morand, R. Y. L., Nikel, P. I., and Acevedo-Rocha, C. G. (2024). How many mutations are needed to evolve the chemical makeup of a synthetic cell? ChemBioChem, e202300829. doi:10.1002/cbic.202300829
Lenski, R. E. (2012). Evolution in action: a 50,000-generation salute to Charles Darwin. (Editors R. Kolter, and S. Maloy). Book chapter in: Microbes and Evolution: The World That Darwin Never Saw. 9–16. doi:10.1128/9781555818470.ch1; ; ISBN: 978-1-683-67073-5 (American Society for Microbiology; 1st edition, 2012).
Marliere, P. (2009). The farther, the safer: a manifesto for securely navigating synthetic species away from the old living world. Syst. Synthetic Biol. 3 (1-4), 77–84. doi:10.1007/s11693-009-9040-9
Mat, W.-K., Xue, H., and Wong, J. T.-F. (2010). Genetic code mutations: the breaking of a three billion year invariance. PLoS One 5 (8), e12206. doi:10.1371/journal.pone.0012206
Richmond, M. H. (1962). The effect of amino acid analogues on growth and protein synthesis in microorganisms. Bacteriol. Rev. 26 (4), 398–420. doi:10.1128/mmbr.26.4.398-420.1962
Tolle, I., Oehm, S., Hoesl, M. G., Treiber-Kleinke, C., Peil, L., Bukari, A.-R. A., et al. (2023). Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1, 1248065. doi:10.3389/fsybi.2023.1248065
Treiber-Kleinke, C., Berger, A. A., Adrian, L., Budisa, N., and Koksch, B. (2024). Escherichia coli adapts metabolically to 6-and 7-fluoroindole, enabling proteome-wide fluorotryptophan substitution. Front. Synth. Biol. 1, 1345634. doi:10.3389/fsybi.2023.1345634
Wong, J. (1983). Membership mutation of the genetic code: loss of fitness by tryptophan. Proc. Natl. Acad. Sci. 80 (20), 6303–6306. doi:10.1073/pnas.80.20.6303
Keywords: expanding the genetic code, chemical evolution of cells, microbial cells, stress response, synthetic life, xenobiology
Citation: Budisa N (2024) Commentary: Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 2:1380879. doi: 10.3389/fsybi.2024.1380879
Received: 02 February 2024; Accepted: 08 February 2024;
Published: 08 March 2024.
Edited by:
Eyal Arbely, Ben-Gurion University of the Negev, IsraelReviewed by:
Chenguang Fan, University of Arkansas, United StatesCopyright © 2024 Budisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nediljko Budisa, bmVkaWxqa28uYnVkaXNhQHVtYW5pdG9iYS5jYQ==
 Nediljko Budisa
Nediljko Budisa