- 1Department of Cell and Systems Biology, University of Toronto, Toronto, ON, Canada
- 2Department of Chemical and Physical Sciences, University of Toronto Mississauga, Mississauga, ON, Canada
- 3Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON, Canada
- 4Department of Chemistry, University of Toronto, Toronto, ON, Canada
- 5Department of Immunology, University of Toronto, Toronto, ON, Canada
The procedures involved in diagnoses of intestinal diseases are often costly, invasive, expensive and dependent on specific technical expertise; these factors can hinder clinicians’ efforts to monitor and treat inflammatory bowel disease and other conditions. Simpler, less expensive and more widely accessible methods could help evaluate the efficacy of treatments and assess the risk of relapse. Yeast, including the probiotic strain Saccharomyces boulardii, can access the gastrointestinal tract, coming into contact with otherwise difficult-to-access disease biomarkers. These yeast cells can be recovered from patient stool samples after ingestion and used to report on prevailing conditions within the gastrointestinal tract during their passage. Here, we report a yeast-based assay offering a simple output based on colour change of colonies plated from recovered stool samples and demonstrate that the reporter can respond to an externally-supplied small-molecule signal while passing through the gastrointestinal system of mice.
Introduction
The diagnosis of intestinal health issues often requires procedures which are costly, invasive, expensive and rely on high levels of technical expertise (Schiavone and Romano, 2015; Rasmussen et al. 2022). These barriers can hamper clinicians in their efforts to monitor the progress of inflammatory bowel disease (IBD) and other conditions (Seyedian et al. 2019; Jayasooriya et al. 2023). Methods that lowered these barriers (by being simpler, less expensive and invoking more widely available technical expertise) could broaden access to accurate evaluations of the efficacy of treatments or assessments of the risk of relapse (Baumgart and Sandborn, 2007; Alatab et al., 2020). The probiotic yeast Saccharomyces boulardii (also designated Saccharomyces cerevisiae var boulardii) can travel intact through the gastrointestinal tract (Edwards-Ingram et al. 2007; Scott et al. 2021), offering the cells access to disease biomarkers in the intestinal lumen. The yeast cells can be recovered from patient stool samples after ingestion and used to report on the conditions they encountered during their passage (Scott et al. 2021).
S. boulardii, initially identified by H. Boulard as having a protective effect against chlorella (Palma et al., 2015), offers attractive features as a microbial chassis for intestinal biosensing. It is an established commercial probiotic, with several strains having been shown in animal models to protect against bacterial infections (Palma et al., 2015) and inflammation (Zhou et al., 2018) in the intestine. While being highly similar to the widely-employed S. cerevisiae S288C laboratory strain (>95% homology; Khatri et al., 2017), it differs in key ways, including having fewer transposable elements (Douradinha et al., 2014), showing improved growth at 37°C and under anaerobic conditions (Fietto et al., 2004; Hudson et al., 2014), and possessing higher resistance to stresses associated with the gastrointestinal tract, including low pH levels and exposure to bile salts (Fietto et al., 2004; Hudson et al., 2014). S. boulardii has been successfully modified to express IL-10, GFP, mRuby, lacZ lysozyme, the vitamin precursor β-carotene, and the yeast surface markers AGA1 and AGA2 (Michael et al., 2013; Pöhlmann et al., 2013; Hudson et al., 2014; Wang et al., 2014; Liu et al., 2016; Durmusoglu et al., 2021).
Here, we describe a novel assay implemented in S. boulardii that produces a simple output in the form of a colour change of colonies plated from recovered stool samples. Our focus is on the yeast-based output method itself, using an externally-applied small molecule as a proxy for disease biomarkers, and demonstrating a proof of principle for future studies that might couple the reporter output to intestinal biomarker detection. In designing the sensor, we had two main goals: we wanted the system to be stable in the yeast for long enough for stool samples to be collected without losing the reported signal; and we sought an approach that could be implemented with a minimum of specialized equipment and specialized training (lowering the cost and potentially enabling deployment in under-resourced settings).
A number of systems have been developed as stable intestinal reporters, most often in bacterial species. A genetic switch controlled by a serine-recombinase in bacteria was shown to hold the expression of a fluorescent reporter for over 100 generations after induction with an arabinose-inducible system (Bonnet et al., 2012). Another genetic switch, the cl/cro system from bacteriophage lambda, encoded the response to a stimulus into the stable expression of β-galactosidase for over 150 generations (Kotula et al., 2014). Both these systems were subsequently implemented in bacteria to monitor specific stimuli within the intestine of mice (Mimee et al., 2015; Riglar et al., 2017). Similar strategies could be employed for a yeast-based assay. This would allow the sensor’s signal to remain active throughout the time the stool sample would be recovered from the patient and delivered to the technician.
A range of reporter genes could be used to generate visual outputs, including through fluorescence (such as the green fluorescent protein, GFP, among many others) or chemiluminescence (such as the luciferase family of enzymes). However, the detection of potentially low-abundance fluorescent or luminescent signals often requires sophisticated instruments capable of highly sensitive photon detection, placing these outputs in conflict with our goal of simplicity and broad accessibility. We chose instead to pursue a colorimetric approach that generated macroscopically visible colour changes.
Colorimetric reporting is a well-established approach in microbiology. A common example is blue/white colony colour screening in E. coli based on β-galactosidase activity. The β-galactosidase enzyme, encoded by the lacZ gene, breaks down β-glycosidic bonds of lactose to release sugar monomers (Juers et al., 2012) (and can also cleave other molecules with a β-glycosidic bond). In blue/white colony colour screening, a bacterial colony that expresses β-galactosidase will be able to hydrolyze a substrate molecule added to the media (such as 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside, generally abbreviated as X-gal) to release a blue-coloured compound that can be readily distinguished by eye from the white bacterial colonies not expressing β-galactosidase. This is frequently used in cloning techniques to confirm the insertion of heterologous DNA into a plasmid backbone. Importantly, this assay does not require special equipment to measure the signal from the output gene: the bacterial cells simply need to be grown into colony-forming units on solid bacterial growth media. We aimed to develop a similar colorimetric assay with S. boulardii.
The output we selected for our sensor is based on the ade2-dependent white-to-red phenotypic conversion of yeast colony colour (Weng and Nickoloff, 1997). Upon detection of its stimuli, the genetically engineered probiotic yeast colonies convert from their typical white colour to either a red or partially-red (sectored) colour phenotype, which can be observed with the naked eye. This colorimetric output has previously been implemented by immobilizing yeast on alginate beads, using a copper-sensitive promoter to trigger the white/red shift in response to copper concentrations in water samples (Vopálenská et al., 2015). In our application, the engineered yeast are passed through the intestinal tract, collected in stool samples, and plated to obtain colonies. Calculating the percentage of white vs. red/sectored colonies then offers a semi-quantitative report of a stimulus level encountered by the yeast while in transit through the gastrointestinal tract.
Methods
Media, reagents, and chemicals
Yeast cultures were prepared for transformation and oral gavage using standard YPD broth (1% yeast extract, 2% peptone, 2% dextrose). CRISPR-Cas9 mediated integration was carried out using YED solid media plates (1% yeast extract, 2% dextrose, 2% agar) with 200 μg/mL G-418 (Wisent). Yeast-based colorimetric assays employed YPD solid media plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) with 40 mg/L of adenine and 50 μg/mL ampicillin or 10 μg/mL chloramphenicol. E. coli strains with cloned plasmids were cultured in standard LB Miller broth (1% yeast extract, 0.5% Tryptone, 1% NaCl) with 100 μg/mL of ampicillin. Doxycycline (Thermo Fisher) was resuspended in sterile water at a concentration of 10 mg/mL, filter sterilized through a 0.22 µM filter, and stored at −20°C until use.
Yeast strains, plasmids, and oligos
A complete list of yeast strains and plasmids used in this study is provided in Supplementary Table S1. We purchased the commercially-available Fluorastor® product and isolated the probiotic strain Saccharomyces boulardii CNCM I-745. Plasmids were assembled by NEBuilder® HiFi DNA Assembly (New England Biolabs) or Golden Gate assembly. To design primers for amplifying the donor DNA for yeast genome integration, oligos 5′-TCGAGGTCGACGGTATCG-3′ and 5′-CCGGTAGAGGTGTGGTCAATAAG-3′ or 5′-ATTAACCCTCACTAAAGGGA-3′ were used to amplify the GFP expression cassettes from pLD024. Oligos 5′-CCTGATGCGGTATTTTCTCC -3′ and 5′-CCTCTGACACATGCAGCTC-3′ were used to amplify the lox-flanked ade2 cassette from pLD141, pLD201, pLD205, and pLD206. Appended to those primers were 50 bp overhangs on the 5′end of the primer appropriate for the integration site targeted. For insertion into the ade2 locus, these 50 bp overhangs were used instead:
Forward primer = 5′-GACATTACTATTTTGCATTTTAATTTAATTAGAACTTGACTAGCGCACTA-3'; reverse primer = 5′- TCTTATGTATGAAATTCTTAAAAAAGGACACCTGTAAGCGTTGATTTCTA -3'. For genotyping the ade2 locus, the forward and reverse primers used were 5′- TTAGAACTTGACTAGCGCAC -3′ and 5′- CACCTGTAAGCGTTGATTTC-3'.
Yeast transformation
Plasmid transformation into S. boulardii followed the standard LiAc/PEG protocol (Gietz and Schiestl, 2007). Transformed cells were left to recover in 2 mL of YPD at 30°C for 24 h with shaking (200–250 rpm) before plating on selective media (YED media plus G-418 (200 μg/mL).
CRISPR-Cas9 mediated integration into yeast chromosome
Site specific guide RNA were designed using the online tool available from Benchling (https://benchling.com). Guides were designed to target intergenic sites previously determined to be suitable for high stable expression in yeast (Flagfeldt et al., 2009). Up to three guide RNA were tested for each integration site, selection being based on integration efficiency of a GFP reporter. CRISPR-Cas9 guide RNA expression vectors were expanded and purified by plasmid DNA Miniprep (New England Biolabs). The donor DNA was amplified by PCR using primers with 50 bp overhangs added to the 5′end of the primer appropriate for the integration site targeted. For transformation, 4 µg of Cas9-gRNA plasmid with 3–6 µg of the purified PCR product was added to the reaction mix before proceeding with the transformation protocol described above. After recovery in YPD media, cells were plated on YED with G-418 agar plates. After PCR confirmation, if needed, positive colonies were cured of plasmid by successive passaging in YPD media for 1–2 days and selected for loss of drug resistance(s). For multiplex genome editing in S. boulardii, donor DNA was created by PCR with 50 bp overhangs homologous for each integration site. For transformation, the cells were prepared as above before adding 4 µg of the CRISPR-Cas9 plasmid with the GFP-specific gRNA and roughly 3–6 µg of each purified PCR product before proceeding with the transformation protocol. Colonies were screened for loss of fluorescence using a blue-light illuminator (Safe Imager, Invitrogen).
Colony PCR
DNA was extracted from the colonies with sodium hydroxide to genotype colonies and confirm genomic integration. Briefly, a small portion of the colony was mixed into 20 µL of a 20 mM solution of NaOH inside a 200 µL PCR tube. Tubes were inserted into PCR thermocycler and incubated twice at 95°C for 5 min with 10-s mixing after every cycle, then the cells were pelleted by centrifugation and 0.5 µL of the supernatant used in a 10 µL PCR reaction with appropriate primers for genotyping.
In vitro characterization of the yeast-based colorimetric assay
Strains were inoculated in a 96-deep well plate (VWR Canada) in 1 mL of YPD media and grown at 30°C overnight on a plate shaker (600–700 rpm) (VWR Canada). In the morning, cultures were diluted 1/100 in 1 mL of fresh media and grown for 4 h under the same conditions. After initial culturing, doxycycline stock (10 mg/mL) was diluted in sterile water at concentration 100X the final concentration desired. Then 10 µL of diluted doxycycline were added into the appropriate well making final concentrations ranging from 10 ng/mL to 10 μg/mL. Cultures were incubated for another 4 h under the same growth conditions. Then, cultures were diluted 1/100,000 in water, and 67 µL were plated on YPD solid media plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) with 40 mg/L of adenine and 50 μg/mL ampicillin or 10 μg/mL chloramphenicol. Each sample was plated on six YPD solid media plates that were incubated for 3 days at 30°C and then stored in the fridge. Across the six plates per sample, we obtained approximately 330–660 colony-forming units; colonies were counted by eye and characterized as either white, sectored, or red, yielding a percentage of red + sectored colonies. A complete set of plate images and counts is available in the Supplementary Material.
Testing the yeast-based colorimetric assay in mice
Conventionally raised C57BL/6 J mice were maintained in the Division of Comparative Medicine (DCM) at the University of Toronto. All mice were provided with an antibiotic treatment, with or without doxycycline, 3 days prior to the oral gavage of yeast. This antibiotic cocktail consisted of 0.3 g/L ampicillin sodium salt, 0.3 g/L kanamycin sulfate, 0.3 g/L metronidazole and 0.15 g/L vancomycin hydrochloride mixed with 10 g/L sucrose in autoclaved drinking water. Doxycycline stock (10 mg/mL) was then added to the drinking water at a final concentration of 1 μg/mL, 5 μg/mL, 10 μg/mL, 50 μg/mL, or 100 μg/mL. Equivalent amounts of sterile water were added to the control drinking water (which did not contain doxycycline, but still contained the antibiotics cocktail). Water was filter sterilized through a 0.22 µM membrane and transferred into the drinking canteen of the mice cage. Water was changed every 2 days throughout the period of the experiment, up to 1 week in total.
The yeast sensor strain was prepared by inoculating the yeast into 5 mL of YPD media and grown at 30°C overnight with shaking (200–250 rpm). The next day, about 1 hour before administering the yeast strain to the mice, the cultures were harvested by centrifugation, washed in 1 mL of ice-cold PBS, and resuspended in 600 µL of ice-cold PBS and kept on ice. This was enough resuspended yeast cells for three mice, or roughly 108 yeast cells per 200 µL. If more cells were needed, culture sizes were increased accordingly. 150 μL of the diluted yeast was added to a 3 mL sterile syringe and attached with a sterile metal oral gavage needle. The yeast sensor strain was then administered to the mice by oral gavage. After 6 h (unless otherwise indicated), two pellets of stool samples were collected from each mouse and added to pre-weighed 1.7 mL centrifuge tubes. Each tube was weighed, and the stool samples were homogenized in PBS at 0.1 g/mL concentration. Stool samples were kept on ice or in the fridge (never frozen) until needed. Each sample was serially diluted 1/10 in PBS 5 times for the initial determination of yeast colony counts. Then 67 µL of each dilution were plated on YPD solid media plates with 40 mg/L of adenine and 50 μg/mL ampicillin or 10 μg/mL chloramphenicol. The plates were incubated at 30°C for at least 24 h while the remaining samples were kept in the fridge (never frozen). The next day, the concentration of colony-forming units was estimated for each sample. The appropriate dilution used for full plating was the one with roughly 60–80 colony-forming units per plate (10 × 15 mm). After choosing the appropriate dilution, each sample was plated on five YPD solid media plates with 40 mg/L of adenine and 50 μg/mL ampicillin or 10 μg/mL chloramphenicol. Plates were incubated for 3 days at 30°C and then stored in the fridge. Across the five plates per sample, we obtained approximately 220–450 colony-forming units; colonies were counted by eye and characterized as either white, sectored, or red, yielding a percentage of red + sectored colonies. A complete set of plate counts is available in the Supplementary Material, and the original plate images will be provided upon request.
Results and discussion
Designing a colorimetric yeast-based assay to detect small molecules in its environment
Yeast colonies can be converted from a white colour to a red colour by disrupting the expression of either the ade2 or ade1 genes in the yeast purine biosynthesis pathway (Jones and Fink, 1982; Weng and Nickoloff, 1997). Deleting either of these genes (leading to the ade1d or ade2d genotype) causes the cells to accumulate a red pigment within their vacuole (Weisman et al., 1987). When the colonies are fully grown, the red-coloured colonies are distinguishable from the white colonies. This convenient phenotype has long been used in yeast research as a genetic tool to study cellular processes, including recombination events (Weng and Nickoloff, 1997), chromosomal loss (Hieter, 1985), and genome evolution (Wightman et al., 2020). Notably, the phenotypic conversion is stable as long as the gene (ade2 or ade1) remains disrupted. One unfortunate consequence of the accumulation of the red pigment is its cytotoxic effect on the cells, which reduces the growth rate of the colonies; these slower-growing colonies may be outcompeted by wild type yeast. Supplementing adenine to the media suppresses the ade2/ade1 -dependent pathway, preventing excessive accumulation of the cytotoxic red pigment. We demonstrated that by adding a non-saturating concentration of adenine to the yeast in either liquid or solid growth media, the growth defects of ade2 deficient S. boulardii colonies could be partially reversed. As shown in Supplementary Figure S1, the doubling time in ade2d yeast in liquid culture can be restored to within approximately 10% of the wild type doubling time with adenine supplementation. On solid media plates, increasing the adenine concentration too much results in a loss of the distinctively red colony phenotype, but with an intermediate concentration of adenine, both cell types can grow sufficiently quickly to be seen on the same plate, while maintaining the visually distinct red colour of the ade2d colonies (Supplementary Figure S2).
We took advantage of this phenotype to create a probiotic yeast-based colorimetric (red/white) colony assay to detect small molecules in the intestine. This was accomplished by positioning the ade2 locus between two Cre recognition sites (lox sites) in the genome of S. boulardii, so that Cre-specific recombination would lead to the excision of the ade2 gene and cause the accumulation of the red pigment (Figure 1A). The expression of the Cre recombinase was then coupled with the detection of a small molecule that could be sensed in the gastrointestinal environment.
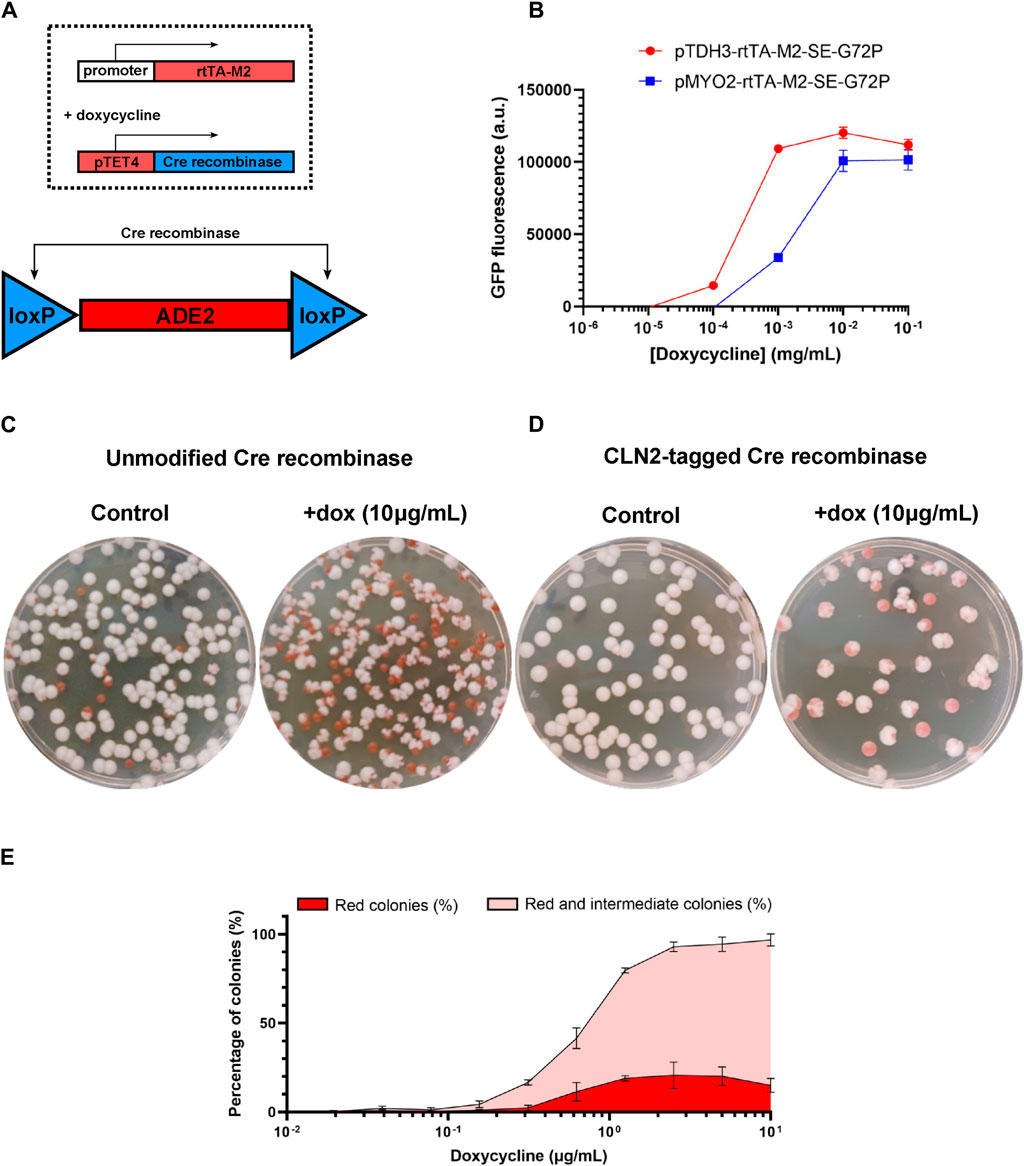
Figure 1. Design and in vitro tests of the colony-colour yeast sensor. (A) Constitutive expression of rtTA leads, in the presence of doxycycline (4 hour exposure), to expression of the Cre recombinase, which in turn excises the loxP-flanked ade2 region of the genome, converting yeast colonies from a white to a red phenotype. (B) Expressing GFP from promoter pTET4 shows that varying the constitutive promoter expressing rtTA shifts the doxycycline response curve. (C) Expression of unmodified Cre recombinase results in a large number of red colony phenotypes in the absence of doxycycline, but (D) tagging the Cre recombinase with the degradation tag CLN2 substantially reduces the rate of red colony formation in the absence of the dox signal. (E) In the strain in which Cre is CLN2-tagged, the combined percentage of fully-red and sectored-red colonies exhibits a smooth dose response to doxycycline concentration in bacterial culture. (Throughout: error bars represent 1 s.d. over three biological replicates (independently-grown yeast cultures).
As a proof of concept, we used a doxycycline-inducible yeast expression system that would allow the yeast cells to sense doxycycline (Roney et al., 2016). In this system, the reverse tetracycline/doxycycline transactivator (rtTA) binds to its target promoter containing tetO operator sequences upon detection of tetracycline or doxycycline, leading to the expression of the downstream gene. We tested the activity of this transcriptional activation system in S. boulardii by genomically integrating the rtTA gene under two different constitutive promoters (pTDH3 or pMYO2) as well as a GFP reporter under a promoter with four tetO operator sequences (pTET4-GFP). The generated yeast strains were exposed to different amounts of doxycycline in their growth media and fluorescence was measured in the cultures. As expected, increasing concentrations of doxycycline led to increased expression of GFP (Figure 1B). As was previously reported (Roney et al., 2016), using different strength constitutive promoters to drive the expression of rtTA affected the dose response curves, with the lower strength promoter (pMYO2) shifting the response curve to higher concentrations of doxycycline compared with the higher strength promoter (pTDH3), an example of tuning response curves by horizontal scaling (Ang et al., 2013). The lower strength promoter (pMYO2) was selected for subsequent experiments because it produced a lower rate of false positives (red or red-sectored colonies appearing in the absence of doxycycline) while still offering a good level of true positives in response to the presence of the target molecule.
We placed the Cre recombinase under the control of the tetO-containing promoter (pTET4) and tested its ability to excise the loxP-flanked ade2 gene cassette and convert S. boulardii colonies from white to red. Using the unmodified Cre recombinase, a substantial portion of the yeast colonies (∼10%) would spontaneously convert to a red colour without the addition of doxycycline (Figure 1C). Despite that, when the same yeast strain was exposed to saturating concentration of doxycycline (10 μg/mL), virtually all the colonies converted to a red phenotype, confirming doxycycline-inducible Cre-specific recombination (Figure 1C). To reduce basal levels of Cre activity under uninduced conditions, a degradation signal peptide (CLN2) was fused to the recombinase. The degradation signal peptide, Cln2, is a G1 cyclin in yeast whose C-terminal domain results in the rapid and constitutive degradation of the attached protein (Salama et al., 1994; Mateus and Avery, 2000), thus reducing overall protein levels and activity. The addition of CLN2 reduced basal levels of Cre activity to near zero, avoiding the spontaneous formation of red colonies, while also maintaining relatively high Cre activation levels with the presence of saturating concentrations of doxycycline (10 μg/mL; Figure 1D).
However, not all colonies converted into a strictly red colour and instead adopted a sectored red phenotype where sections of the colonies would be white whereas others would be red, leading to the formation of diverse types of cauliflower-shaped colonies (Supplementary Figure S3). The sectoring events are not simply caused by doxycycline-induced expression of the Cre recombinase, since placing the Cre recombinase under a oxidative stress sensing expression system (pSynOS_4 (alt1); Dacquay and McMillen 2021) still results in the formation of sectored red colonies (Supplementary Figure S4). The sectored phenotype appears to be dependent on Cre-mediated ade2 excision: Cre-specific excision of a fluorescent reporter between two loxP sites in S. boulardii does not cause the formation of sectored GFP-expressing colonies (Supplementary Figure S5). Expressing the Cre recombinase under constitutive promoters of varying strength still led to the formation of sectored red colonies; use of a higher strength constitutive promoter (Lee et al., 2015) did increase the ratio of strictly red colonies to red-sectored colonies, but did not completely avoid its formation (Supplementary Figure S6). ade2 deletion and accumulation of the red pigment causes a growth defect for yeast cells, creating a selective pressure in favour of phenotypes that avoid red pigment accumulation. Thus, the formation of red-sectored intermediate S. boulardii colonies from Cre-specific recombination of loxP-flanked ade2 does not seem to be entirely preventable. However, sectored-red colonies are still visually distinct from the unmodified white colonies, allowing us to use the total amount of red and red-sectored colony formation as an indicator of sensor activation.
When the doxycycline-induced yeast colorimetric sensor strain was incubated with increasing concentrations of its inducer, the proportion of red and red-sectored colony forming units counted on the plates increased, eventually saturating at nearly complete conversion of the colonies from white to red or red-sectored (Figure 1E). The colorimetric sensor response curve is roughly equivalent to its fluorescent counterparts, allowing us to reliably distinguish between concentrations in the range of 0.156–2.5 μg/mL of doxycycline (the sensor saturates its output for higher concentrations). Thus the colorimetric yeast-based assay can not only be used to report on the presence of specific molecules within its environment, but also to gauge the relative abundance of its inducer.
Testing colorimetric yeast-based assay functional capabilities in an intestinal environment
The colorimetric yeast-based assay could be applied to monitor the levels of a disease-specific biomarker within the gastrointestinal tract of a patient. To validate the assay’s functional capabilities in a real gastrointestinal environment, we designed a protocol to test the activity of the colorimetric yeast sensor strain in mice. The protocol involved colonizing the mouse intestine with the doxycycline-inducible yeast sensor strain through oral administration, recovering the yeast cells from fecal pellets, and plating diluted samples on yeast nutrient agar plates to produce yeast colonies. To be able to distinguish between the yeast colonies and other potentially contaminating bacterial colonies from the native gut microbiota, we integrated bacterial antibiotics (ampicillin and/or chloramphenicol) into the yeast nutrient agar plates to prevent bacteria colony growth while allowing yeast colony growth. Using either 50 μg/mL of ampicillin or 10 μg/mL of chloramphenicol was sufficient to prevent outgrowth of any bacteria from stool collected from specific pathogen free (SPF) raised mice. Administration of the engineered S. boulardii strain by oral gavage led to the presence of yeast colonies within recovered stool samples at dilutions between 10−3 to 10−6 CFUs per Gram of stool. The antibiotics added to the media would not prevent the growth of other yeast or fungal colonies from the mice native gut microbiota, but none of the plated samples included visually distinct non-yeast colonies, leading us to conclude that any non-S. boulardii colonies were vastly outnumbered by those resulting from the orally administered strain.
Saccharomyces boulardii cannot stably colonize the mouse intestine when in competition with the full microbiota of SPF mice, with yeast colonies becoming absent from recovered stools samples only 24 h after initial oral gavage (Pais et al., 2020). Therefore, to maintain the yeast within the intestine for a longer period, the mice were provided with an antibiotic cocktail previously shown to increase residency time (Hedin et al., 2022). Two groups of mice (n = 3) were provided with the antibiotic cocktail and either with or without doxycycline (10 μg/mL) in their drinking water (Figure 2A). After a few days of acclimation, the mice were administered with the doxycycline-inducible colorimetric yeast reporter strain by oral gavage (∼108 CFUs). Stool samples were collected every subsequent day for four total days and plated unto yeast agar media plates supplemented with antibiotics (ampicillin and chloramphenicol) as well as additional adenine (40 mg/L). Yeast colonies were successfully recovered from all mice between 106 to 109 CFUs per Gram of stool, and remained roughly constant throughout the 4 day collection period (Figure 2B). Clear phenotypic differences were observed from yeast colonies collected between control and doxycycline-fed mice (Figure 2C). Yeast colonies from control mice remained mostly white, whereas yeast colonies from doxycycline-fed mice showed formation of either red or red-sectored colonies starting from the first day. The proportion of red or intermediate colonies on the plates of doxycycline-fed mice gradually increased as the days progressed, starting from ∼25% on the first day and ending with ∼75% on the fourth day (Figure 2D). The slower response to the doxycycline in mice compared to in vitro conditions (where yeast colonies in liquid culture converged to close to 100% red/red-sectored phenotypes after only a 4-h incubation), is not entirely surprising given that the doxycycline was only passively administered to the mice, and was likely being absorbed within the intestine, reducing its effective dose. Administering the doxycycline through drinking water also implies that we cannot currently distinguish between the yeast being exposed to doxycycline in the stomach from exposure in the intestine itself. (We note that previous measurements suggest that an ingested sample passes into the mouse intestine within 1–3 h (Padmanabhan et al., 2013), implying that the majority of the 48 h before sample collection were likely spent in the intestine.) It is clear that the yeast is able to respond to doxycycline in the murine gastrointestinal environment, and that it correctly transduces doxycycline into red/red-sectored colony formation after passing through the intestines and emerging in stool samples.
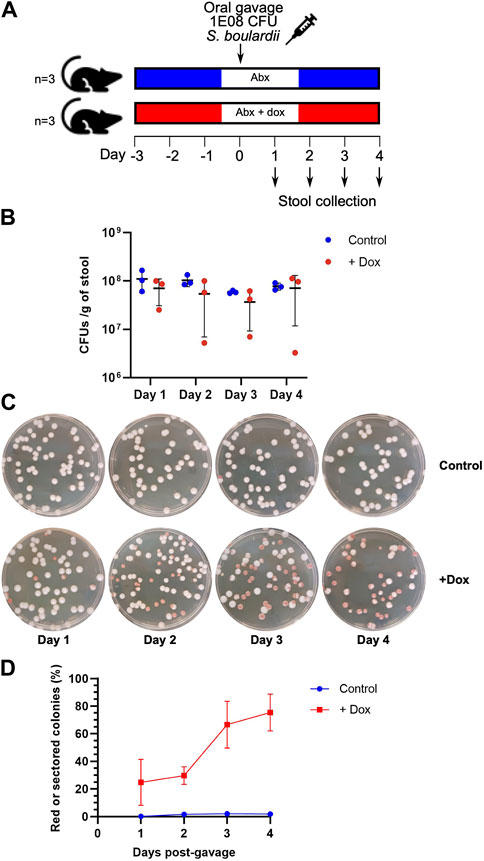
Figure 2. Characterization of the in vivo time course of the colony-colour sensor’s response to exposure to doxycycline in mice. (A) Mouse protocol: after 3 days of exposure to an antibiotic cocktail (Abx), with or without doxycycline (dox, at 10 μg/mL) provided in the mouse’s drinking water, the yeast sensor strain is introduced by oral gavage. Stool samples are then collected for 4 days post-gavage. (B) Total colony-forming unit (CFU) counts per Gram of stool. (C) Representative images of plated stool samples from the control mice (Abx only) and dox-exposed mice. (D) Percentages of red + red-sectored colonies obtained for 4 days after introduction of the yeast sensor strain. (Throughout: error bars represent 1 s.d. over three biological replicates (samples from three mice).
To determine if the sensor strain could distinguish between different concentrations of doxycycline within the intestine, the activity of the sensor strain was assessed between five groups of mice that were provided with concentrations of doxycycline in their drinking water that varied from 1.0 to 100 μg/mL. In general, the proportion of red + red-sectored colonies increases with doxycycline concentration (Figure 3), though there are some deviations from this trend at the lowest and highest concentrations. The inconsistency in the trend may relate to our use of a passive administration method of doxycycline: the inducer was provided in the animals’ drinking water, meaning that different doses may have been ingested by individual mice. A more direct route such as rectal administration could result in more consistent dose-response measurements.
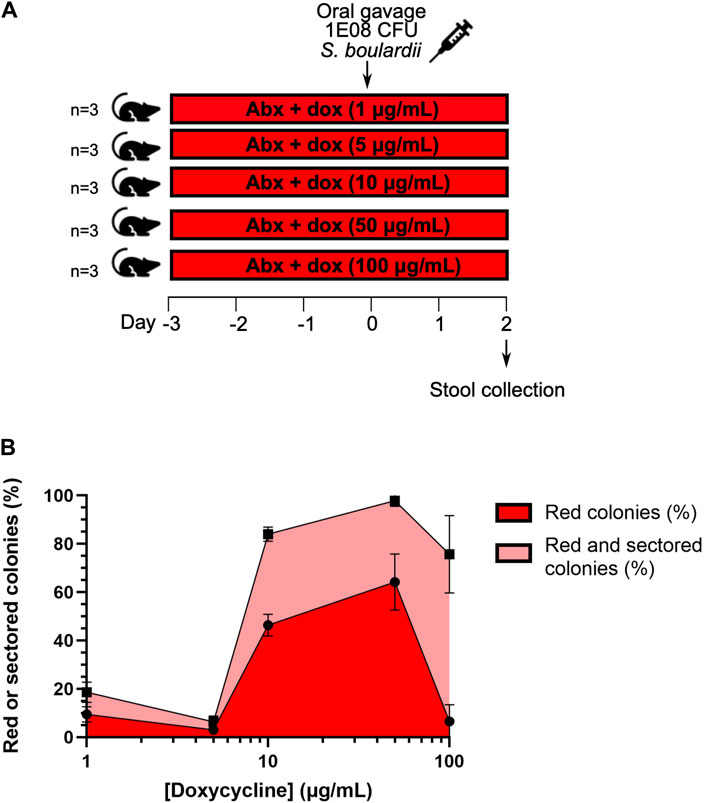
Figure 3. Characterization of the in vivo dose response of the colony-colour sensor to exposure to doxycycline in mice. (A) Mouse protocol for doxycycline concentration-dependence tests. Drinking water was supplemented with an antibiotic cocktail (Abx) and varying concentrations of doxycycline (dox) for 3 days prior to administration of the yeast sensor strain by oral gavage. Stool samples were collected 2 days post-gavage. (B) The percentages of red and red + red-sectored colonies as a function of doxycycline concentration. (Throughout: error bars represent 1 s.d. over three biological replicates (samples from three mice).
We have established that the colorimetric yeast-based sensor can be used to detect doxycycline in a whole-animal intestinal environment, which suggests that it has promise for further development into a diagnostic tool. The individual system elements (ade2-based colony colour shifting; Cre-based genomic modification; and plating of samples for visual counting) have been shown to work together, and to exhibit the desired ability to respond to a sensed signal while the yeast pass through the gastrointestinal tract.
Future work will involve connecting the colorimetric output to input from a disease biomarker; several methods of detecting intestinal disease states have been explored previously, detecting tetrathionate (Riglar et al., 2017), thiosulfate (as well as tetrathionate) (Daeffler et al., 2017), nitrate (Woo et al., 2020), and extracellular ATP (Scott et al., 2021). Efforts are underway to use an existing sensing mode to trigger the excision event in our sensor; the yeast-based detection of extracellular ATP employed by Scott et al. (2021) is a particularly promising possibility.
We envision an eventual application in which the yeast would provide a short-term “snapshot” of the current intestinal environment: after swallowing a sensor, a patient’s stool sample would be collected in one or 2 days, then plated and scored for colony-colour percentages. Over that short duration, we would not anticipate that the fitness defect seen in the ade2d (red) yeast would have the opportunity to shift the population percentages significantly in favour of the wild-type ade2 (white) yeast, but further mouse experiments will be required to test this assumption. For longer-duration intestinal residence times (or other sensing contexts), two factors could act to shift the reported colony-colony percentages: 1) since the ade2 excision process is irreversible, any basal level of uninduced excisions could lead to a gradual accumulation of false positives (the in vitro plating tests in the Supplementary Material, along with the control mice results in Figure 2D, indicate that 4 days is not enough time for such a shift, but the limited intestinal residence times of our yeast leads to a lack of in vivo data on longer time scales); and 2) in contexts where the yeast are able to grow during their application as a sensor, the slower-growing ade2d (red) yeast could be out-competed, artificially lowering their representation in the population. Further testing of these and other issues (including the in vivo dynamics of the sensor response after a sensed signal has been removed) will be required to establish the real-world utility of sensors using this output mode, but these initial results indicate that the sensor has the key properties we sought to achieve. Comparing this method with standard-of-care monitoring by endoscopy or colonoscopy, this approach provides an instrument-free visual output with the potential to monitor gastrointestinal conditions, relying only on relatively simple technical skills and inexpensive materials.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Ethics committee, Department of Immunology, University of Toronto. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LD: Conceptualization, Writing–review and editing, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft. BS: Investigation, Methodology, Resources, Writing–review and editing. MPN: Investigation, Methodology, Formal analysis, Writing-review and editing. DP: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–review and editing. DM: Conceptualization, Writing–review and editing, Funding acquisition, Project administration, Resources, Supervision.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Support for operating costs (including personnel, consumables, and animal facility fees) was provided by the Canada Research Coordinating Committee, through the New Frontiers in Research Program (grant number NFRFE-2019-00742).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsybi.2024.1265343/full#supplementary-material
References
Alatab, S., Sepanlou, S. G., Ikuta, K., Vahedi, H., Bisignano, C., Safiri, S., et al. (2020). The global, regional, and national Burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol. Hepatol. 5 (1), 17–30. doi:10.1016/S2468-1253(19)30333-4
Ang, J., Harris, E., Hussey, B. J., Kil, R., and McMillen, D. R. (2013). Tuning response curves for synthetic biology. ACS Synth. Biol. 2 (10), 547–567. doi:10.1021/sb4000564
Baumgart, D. C., and Sandborn, W. J. (2007). Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 369 (9573), 1641–1657. doi:10.1016/S0140-6736(07)60751-X
Bonnet, J., Subsoontorn, P., and Endy, D. (2012). Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. 109 (23), 8884–8889. doi:10.1073/pnas.1202344109
Dacquay, L. C., and McMillen, D. R. (2021). Improving the design of an oxidative stress sensing biosensor in yeast. FEMS Yeast Res. 21 (4), foab025. doi:10.1093/femsyr/foab025
Daeffler, K. N., Galley, J. D., Sheth, R. U., Ortiz-Velez, L. C., Bibb, C. O., Shroyer, N. F., et al. (2017). Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13 (4), 923. doi:10.15252/msb.20167416
Douradinha, B., Reis, V. C. B., Rogers, M. B., Torres, F. A. G., Evans, J. D., and Marques, E. T. A. (2014). Novel insights in genetic transformation of the probiotic yeast Saccharomyces boulardii. Bioengineered 5 (1), 21–29. doi:10.4161/bioe.26271
Durmusoglu, D., Al’Abri, I. S., Collins, S. P., Cheng, J., Eroglu, A., Beisel, C. L., et al. (2021). In situ biomanufacturing of small molecules in the mammalian gut by probiotic Saccharomyces boulardii. ACS Synth. Biol. 10 (5), 1039–1052. doi:10.1021/acssynbio.0c00562
Edwards-Ingram, L., Gitsham, P., Burton, N., Warhurst, G., Clarke, I., Hoyle, D., et al. (2007). Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 73 (8), 2458–2467. doi:10.1128/AEM.02201-06
Fietto, J. L. R., Araújo, R. S., Valadão, F. N., Fietto, L. G., Brandão, R. L., Neves, M. J., et al. (2004). Molecular and physiological comparisons between Saccharomyces cerevisiae and Saccharomyces boulardii. Can. J. Microbiol. 50 (8), 615–621. doi:10.1139/w04-050
Flagfeldt, D. B., Siewers, V., Huang, L., and Nielsen, J. (2009). Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26, 545–551. doi:10.1002/yea.1705
Gietz, R., and Schiestl, R. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi:10.1038/nprot.2007.13
Hedin, K. A., Rees, V. E., Zhang, H., Kruse, V., Vazquez-Uribe, R., and Sommer, M. O. A. (2022). Effects of broad-spectrum antibiotics on the colonisation of probiotic yeast Saccharomyces boulardii in the murine gastrointestinal tract. Sci. Rep. 12 (1), 8862. doi:10.1038/s41598-022-12806-0
Hieter, P., Mann, C., Snyder, M., and Davis, R. W. (1985). Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 40 (2), 381–392. doi:10.1016/0092-8674(85)90152-7
Hudson, L. E., Fasken, M. B., McDermott, C. D., McBride, S. M., Kuiper, E. G., Guiliano, D. B., et al. (2014). Functional heterologous protein expression by genetically engineered probiotic yeast Saccharomyces boulardii. PLoS One 9 (11), e112660. doi:10.1371/journal.pone.0112660
Jayasooriya, N., Baillie, S., Blackwell, J., Bottle, A., Petersen, I., Creese, H., et al. (2023). Systematic review with meta-analysis: time to diagnosis and the impact of delayed diagnosis on clinical outcomes in inflammatory bowel disease. Aliment. Pharmacol. Ther. 57, 635–652. doi:10.1111/apt.17370
Jones, E. W., and Fink, G. R. (1982). Regulation of amino acid and nucleotide biosynthesis in yeast. Cold Spring Harb. Monogr. Arch., 181–299.
Juers, D. H., Matthews, B. W., and Huber, R. E. (2012). LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 21 (12), 1792–1807. doi:10.1002/pro.2165
Khatri, I., Tomar, R., Ganesan, K., Prasad, G. S., and Subramanian, S. (2017). Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci. Rep. 7 (1), 371–413. doi:10.1038/s41598-017-00414-2
Kotula, J. W., Kerns, S. J., Shaket, L. A., Siraj, L., Collins, J. J., Way, J. C., et al. (2014). Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. 111 (13), 4838–4843. doi:10.1073/pnas.1321321111
Lee, M. E., DeLoache, W. C., Cervantes, B., and Dueber, J. E. (2015). A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 4 (9), 975–986. doi:10.1021/sb500366v
Liu, J., Kong, I. I., Zhang, G., Jayakody, L. N., Kim, H., Xia, P., et al. (2016). Metabolic engineering of probiotic Saccharomyces boulardii. Appl. Environ. Microbiol. 82 (8), 2280–2287. doi:10.1128/AEM.00057-16
Mateus, C., and Avery, S. V. (2000). Destabilized Green Fluorescent Protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast 16 (14), 1313–1323. doi:10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O
Michael, S., Keubler, L. M., Smoczek, A., Meier, M., Gunzer, F., Pöhlmann, C., et al. (2013). Quantitative phenotyping of inflammatory bowel disease in the IL-10-deficient mouse by use of noninvasive magnetic resonance imaging. Inflamm. Bowel Dis. 19 (1), 185–193. doi:10.1002/ibd.23006
Mimee, M., Tucker, A. C., Voigt, C. A., and Lu, T. K. (2015). Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell. Syst. 1 (1), 62–71. doi:10.1016/j.cels.2015.06.001
Padmanabhan, P., Grosse, J., Asad, A., Radda, G., and Golay, X. (2013). Gastrointestinal transit measurements in mice with 99mTc-DTPA-labeled activated charcoal using NanoSPECT-CT. EJNMMI Res. 3 (1), 60. doi:10.1186/2191-219X-3-60
Pais, P., Almeida, V., Yılmaz, M., and Teixeira, M. C. (2020). Saccharomyces boulardii: what makes it tick as successful probiotic? J. Fungi (Basel) 6 (2), 78. doi:10.3390/jof6020078
Palma, M. L., Zamith-Miranda, D., Martins, F. S., Bozza, F. A., Nimrichter, L., Montero-Lomeli, M., et al. (2015). Probiotic Saccharomyces cerevisiae strains as biotherapeutic tools: is there room for improvement? Appl. Microbiol. Biotechnol. 99 (16), 6563–6570. doi:10.1007/s00253-015-6776-x
Pöhlmann, C., Thomas, M., Förster, S., Brandt, M., Hartmann, M., Bleich, A., et al. (2013). Improving health from the inside: use of engineered intestinal microorganisms as in situ cytokine delivery system. Bioengineered 4 (3), 172–179. doi:10.4161/bioe.22646
Rasmussen, N. F., Green, A., Allin, K. H., Højgaard Allin, K., Iversen, A. T., Madsen, G. I., et al. (2022). Clinical procedures used to diagnose inflammatory bowel disease: real-world evidence from a Danish nationwide population-based study. BMJ Open Gastroenterol. 9, e000958. doi:10.1136/bmjgast-2022-000958
Riglar, D. T., Giessen, T. W., Baym, M., Kerns, S. J., Niederhuber, M. J., Bronson, R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35 (7), 653–658. doi:10.1038/nbt.3879
Roney, I. J., Rudner, A. D., Couture, J.-F., and Kærn, M. (2016). Improvement of the reverse tetracycline transactivator by single amino acid substitutions that reduce leaky target gene expression to undetectable levels. Sci. Rep. 6 (1), 27697. doi:10.1038/srep27697
Salama, S. R., Hendricks, K. B., and Thorner, J. (1994). G1 cyclin degradation: the PEST motif of yeast Cln2 is necessary, but not sufficient, for rapid protein turnover. Mol. Cell. Biol. 14 (12), 7953–7966. doi:10.1128/mcb.14.12.7953
Schiavone, C., and Romano, M. (2015). Diagnosis and management of Crohn's disease. J. Ultrasound 18 (1), 1–2. doi:10.1007/s40477-015-0159-0
Scott, B. M., Gutiérrez-Vázquez, C., Sanmarco, L. M., da Silva Pereira, J. A., Li, Z., Plasencia, A., et al. (2021). Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nat. Med. 27, 1212–1222. doi:10.1038/s41591-021-01390-x
Seyedian, S. S., Nokhostin, F., and Malamir, M. D. (2019). A review of the diagnosis, prevention, and treatment methods of inflammatory bowel disease. J. Med. Life 12 (2), 113–122. doi:10.25122/jml-2018-0075
Vopálenská, I., Váchová, L., and Palková, Z. (2015). New biosensor for detection of copper ions in water based on immobilized genetically modified yeast cells. Biosens. Bioelectron. 72, 160–167. doi:10.1016/j.bios.2015.05.006
Wang, T., Sun, H., Zhang, J., Liu, Q., Wang, L., Chen, P., et al. (2014). The establishment of Saccharomyces boulardii surface display system using a single expression vector. Fungal Genet. Biol. 64, 1–10. doi:10.1016/j.fgb.2013.11.006
Weisman, L. S., Bacallao, R., and Wickner, W. (1987). Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J. Cell. Biol. 105 (4), 1539–1547. doi:10.1083/jcb.105.4.1539
Weng, Y., and Nickoloff, J. A. (1997). Nonselective URA3 colony-color assay in yeast Ade1 or Ade2 mutants. Biotechniques 23 (2), 237–241. doi:10.2144/97232bm13
Wightman, E. L. I., Kroukamp, H., Pretorius, I. S., Paulsen, I. T., and Nevalainen, H. K. M. (2020). Rapid colorimetric detection of genome evolution in SCRaMbLEd synthetic Saccharomyces cerevisiae strains. Microorganisms 8 (12), 1914. doi:10.3390/microorganisms8121914
Woo, S.-G., Moon, S.-J., Kim, S. K., Kim, T. H., Lim, H. S., Yeon, G.-H., et al. (2020). A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron. 168, 112523. doi:10.1016/j.bios.2020.112523
Keywords: synthetic biology, colorimetric sensor, yeast, mouse model, gastrointestinal sensing
Citation: Dacquay LC, Samman B, Newman MP, Philpott DJ and McMillen DR (2024) Engineering a colorimetric yeast-based assay to detect small molecules in the gastrointestinal system. Front. Synth. Biol. 2:1265343. doi: 10.3389/fsybi.2024.1265343
Received: 22 July 2023; Accepted: 31 May 2024;
Published: 27 June 2024.
Edited by:
Tae Seok Moon, EBRC, United StatesReviewed by:
Finn Edward Stirling, University of Cambridge, United KingdomXinyi Wan, Hainan University, China
Alexander James Webb, Imperial College London, United Kingdom
Copyright © 2024 Dacquay, Samman, Newman, Philpott and McMillen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dana J. Philpott, ZGFuYS5waGlscG90dEB1dG9yb250by5jYQ==; David R. McMillen, ZGF2aWQubWNtaWxsZW5AdXRvcm9udG8uY2E=
 Louis C. Dacquay1,2
Louis C. Dacquay1,2 Bana Samman
Bana Samman Dana J. Philpott
Dana J. Philpott David R. McMillen
David R. McMillen