- 1Bioengineering for Sustainability Laboratory, Department of Biosystems Engineering, University of Manitoba, Winnipeg, MB, Canada
- 2Department of Chemistry, University of Manitoba, Winnipeg, MB, Canada
We envision the convergence of synthetic biology (SynBio) and metagenomics as a significant development for the engineering of complex biological systems. The entire biosphere with its diverse life forms can also be considered as a reservoir for evolutionary innovations and a source of modules for SynBio. Metagenomics, which is a large part of biodiversity, should be considered as an important source of modules. The abstraction hierarchy of amalgamating SynBio and metagenomics (“synthetic metagenomics”) entails the standardized integration of parts, devices, circuits, and modules into functional chassis. These principles transcend the boundaries of single cell design and apply to the engineering of biodiversity sustainability in multicellular entities, their interconnections, and their dynamics in communities and whole ecosystems. Examples include applications in environmental sustainability, such as analysis of antimicrobial resistance in waste management, bioremediation of oil spills, and degradation of plastics. Future research and experimental interventions will ultimately provide a strong link between bioengineering, metagenomics, microbial consortia, ecosystems, and biodiversity sustainability under the umbrella of synthetic biology.
1 Introduction
Synthetic biology (SynBio) aims to use a combination of natural or artificial life components and apply multidisciplinary engineering principles to biotechnology (Acevedo-Rocha and Budisa, 2016). SynBio has essentially “borrowed” the concepts from software or electrical engineering for the design of basic functional units (usually bottom-up) that could be called modules (Endy, 2005). Thus, SynBio is a technoscience based on the fundamental idea that complexity can be created through the integration, combination, or arrangement of modules as interchangeable systems (de Lorenzo, 2011). Indeed, the inherent “constructability” of living organisms enables the division of complex systems into smaller, independent subsystems that can be used for different applications (Schmidt et al., 2018). To achieve this, standardization, abstraction, and modularity are required, with modules referring to functionally independent units with minimal interactions (orthogonality) (Costello and Badran, 2021). To design complicated biological systems, a common understanding of the functionality of these modular parts is critical. In addition, these parts should be, standardized, well characterized and easily accessible (e.g., BioBrick assembly of the MIT Registry of Standard Biological Parts (Sleight et al., 2010b)). For this reason, SynBio is often assumed to be the computational analysis and modelling of complex biological systems (Chandran et al., 2008; Zomorrodi and Segrè, 2016). But it is more than mathematical modelling and tinkering by engineers of different domains.
Modularity and orthogonality are the basis for a robust and scalable circuit design inspired by natural gene circuits. These gene circuits rely on regulatory factors for cellular functions, using isolated and interacting components to achieve specific responses. Natural circuits consist of distinct functional modules, enabling adaptability to changing environments. Since the advent of recombinant DNA technology, researchers have extracted and modified elements from these circuits to create increasingly complex designs. As a result, versatile tools and components are available for the engineering of gene circuits. This journey has been accompanied by the expansion of synthetic biology tools from basic gene expression control and pathway adaptation (e.g., promoters, transcriptional activators/repressors, DNA/RNA/protein control and modification, molecular memory, etc.) to advanced genome editing control (e.g., CRISPR/Cas9, transcriptional regulation), multicellular pattern formations, genetic code expansion and engineering, and the recent innovation of synthetic cells and xenobiology [reviewed in e.g. (Bradley et al., 2016; Budisa et al., 2020)]. The latter is an excellent example of the process of assimilation of non-native substrates, also referred to in the literature as synthetic heterotrophy (Sullivan et al., 2023), which is relevant not only to the design of synthetic life but also to cell regulation, as it is a technology that diverts synthetic metabolism to optimize biomass formation.
A “device” in SynBio refers to a combination of some biological parts that work together to produce a particular useful function or behavior (Müller and Arndt, 2012). These devices can be further integrated or scaled to form more complicated structures. For example, a genetically engineered bacterium created for a specific purpose, such as the degradation of plastic polymers into monomers, will have both the ability to convert monomers into biomass through anabolic processes and catabolically “burn” the hydrolysis products of the degraded polymer. In this scenario, the polymer-degrading “device” (e.g., the catalytic enzyme cascade) and the biomass-producing “circuit” (e.g., synthetic metabolism) are functionally independent, but interact at the interface level in a controlled and predictable manner. These hierarchical levels of complexity can be described as “systems” where the combined behavior of different devices and circuits results in holistic behavior phenomena (Delgado and Porcar, 2013). Finally, the term “chassis” refers to living host that nurtures and sustains the genetic elements, systems, and processes (as described above) by providing the resources necessary for their proper functioning (Danchin, 2012).
In this context, the integration of metagenomics into SynBio promotes the pursuit of advanced modular design that leverages host cells for tailored bioactivities by tapping into the genetic and biochemical diversity in the genomes of microorganisms from environmental samples or through metagenomics (Figure 1). We anticipate that this integration will significantly boost the whole SynBio field, as its basic principles (modular integration, standardization, abstraction, and orthogonality) are applicable to the engineering of biodiversity sustainability both in single cells, and in multicellular entities, their interconnections and dynamics in communities and whole ecosystems.
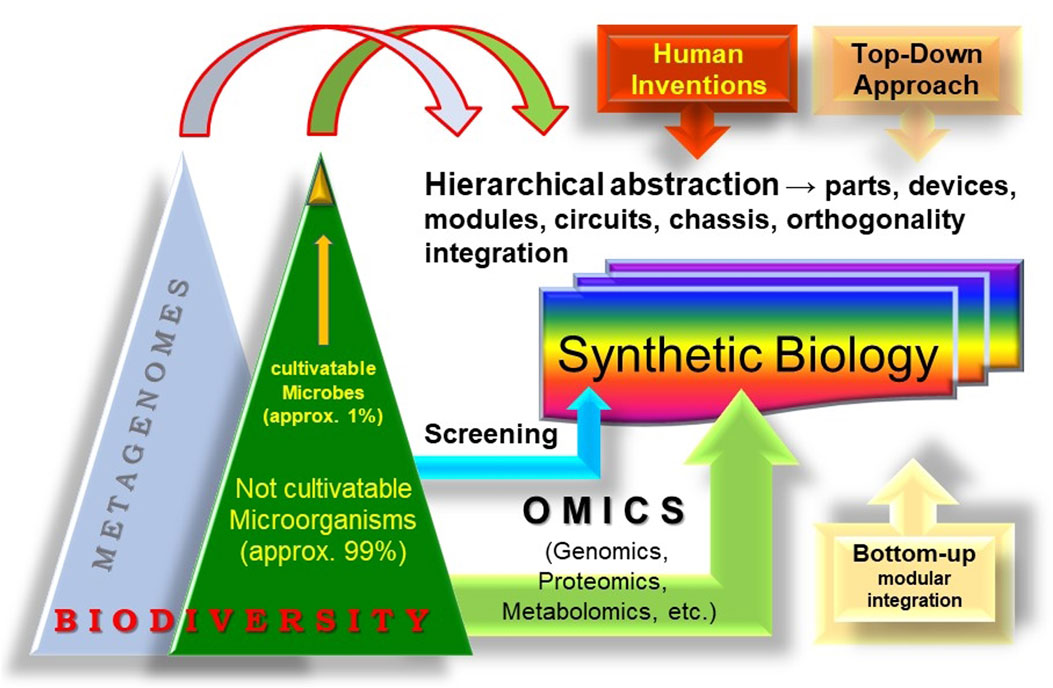
FIGURE 1. The concept of “synthetic metagenomics.” The entirety of the biosphere, rich with its diverse life forms can also be considered as a reservoir for evolutionary innovations and a source of modules for Synthetic Biology. Metagenomics, which undoubtedly constitutes a large part of biodiversity, should be considered as an important wellspring of modules. The establishment of standardized procedures can leverage the bioengineering of these modules in specific contexts to create new biobased technologies and economies and even synthetic cells and communities.
2 Topics relevant for the subject
2.1 Metagenomics
Metagenomics is a field of research that focuses on the study of genetic material obtained directly from environmental samples such as soil, water, or even the human gut (Yen and Johnson, 2021). Unlike traditional genomics, which focuses on the DNA of individual organisms, metagenomics examines the collective genetic material of all microorganisms present in a given sample (New and Brito, 2020). The core idea of metagenomics is to analyze the genetic diversity and functional potential of entire microbial communities without the need to isolate and culture individual organisms (Berini et al., 2017). This approach allows us to gain insights into the composition, structure, and functional capabilities of complex microbial ecosystems that are difficult to study using conventional molecular biological methods (Garrido-Cardenas and Manzano-Agugliaro, 2017).
Metagenomics involves sequencing and analyzing the DNA or RNA present in a sample from a particular ecological niche (Marco, 2008). Because this material is normally fragmented and piecemeal, advanced sequencing technologies are used to retrieve genetic information. By analyzing the sequences, it is possible to identify the species of microorganisms present, their relative abundance and the functional genes they carry (Kayani et al., 2021). This information can provide valuable insights into ecosystem dynamics, microbial interactions, and potential applications in biotechnology and environmental research as elaborated in our study. In principle, metagenomics allows us to explore the genetic composition of entire microbial communities in their natural habitats and thus discover the hidden biodiversity and functional potential of these ecosystems (Bouhajja et al., 2016). Here, we describe possible pathways to “synthetic metagenomics” (Figure 1) by merging SynBio with classical metagenomics.
In this context, metagenomics is evolving into a technology that complements gene and protein engineering methods and applies SynBio’s system design concepts (see above). Metagenomics exploits the wealth of genetic and biochemical diversity present in the genomes of microorganisms in environmental samples (Figure 1) and offers a range of new technologies for screening novel catalytic activities from environmental samples with potential biotechnological applications (Guazzaroni et al., 2015) or even diagnostics (Greninger, 2018). For example, metagenomic analyses of marine ecosystems have begun to reveal the extent of novel compounds encoded in the vast bacterial richness and diversity of the marine ecosystem (Tseng and Tang, 2014). A combination of unique physicochemical properties and spatial niche-specific substrates in wide-spread and extreme habitats underscores the potential of the marine environment to deliver functionally novel biocatalytic activities (Parages et al., 2016).
In recent years, metagenomics has expanded our understanding of how the collective functions of microorganisms emerge from the complex web of interactions between their genetic and metabolic potentials and the environment to enable useful applications in terms of environmental sustainability (Sharma et al., 2022). A particular focus of metagenomics is the study of the interactions of environmental pollutants (xenobiotics) of anthropogenic origin (e.g., plastics, pharmaceuticals, pesticides, cosmetics, flavorings, fragrances, food additives, industrial chemicals, and plastics) with the local environment and their global impacts (Danso et al., 2019). Metagenomic analyses have expanded our understanding of ecosystems and biodiversity in several fields such as antimicrobial resistance in waste management systems, bioremediation of oil spills, and biodegradation of plastics.
Therefore, synthetic metagenomics can be seen as an emerging field of research in which metagenomes are studied functionally following the key postulates of SynBio (see above). This approach substantially expands the set of identified biological activities and building blocks. For example, functional metagenomic of metabolic pathways in microbial communities has led to the discovery of novel bioactivities such as amidases, NF-κB modulators, naphthalene degrading enzymes, cellulases, lipases, and transporters (Robinson et al., 2021). Using these genetic devices and circuits as templates along with their further functional integration, improvements can be made by altering or redesigning metabolic pathways in selected microbial chassis to improve synthesis and yield of high-value products (van der Helm et al., 2018).
2.2 Antimicrobial resistance
The emergence and the spread of antimicrobial resistance in animal husbandry is a growing global concern (Karbalaei-Heidari and Budisa, 2020). Metagenomics has proven to be a powerful tool for analyzing the evolution of antimicrobial resistance genes (ARGs) and mobile genetic elements (MGEs) in microbiome of animals. Large quantities of veterinary pharmaceuticals and drugs are used not only to treat infectious diseases, but also as prophylactic therapies and feed enhancers, potentially leading to the selection of ARGs, MGEs, and ultimately, antimicrobial resistance bacteria (ARBs). For instance, the study of the “Resistome” (complete set of ARGs) in ruminal microbiomes has shown the presence of ARGs encoding resistance to tetracyclines, one of the most used antibiotics in farms, in the genomes of common ruminal species (Sabino et al., 2019). Not surprisingly, animal manures are significant reservoir of ARGs and MGEs and can promote the development of ARBs, which are often spread in agricultural fields when manure is applied as fertilizer (Buta-Hubeny et al., 2022).
In this context, metagenomics and microbial diversity analyses provide a valuable framework for studying the impact of different manure treatments on resistomes and mobilomes (complete set of MGEs). Through these technologies, we are gaining a better understanding of the microbial species that harbor antimicrobial resistant elements and the dynamics that drive changes in ARGs and MGEs. For example, metagenomic analyses have shown that microbial successions during the mesophilic and thermophilic stages of manure composting or the establishment of anaerobic consortia during anaerobic digestion (AD), can reduce the abundance and diversity of ARGs and MGEs in animal manure (Cheng, 2017; Flores-Orozco et al., 2023; Flores-Orozco et al., 2023). The knowledge gained from these applications is crucial for addressing the serious health problem that antimicrobial resistance poses and for developing strategies to minimize the risk of its spread in agriculture.
2.3 Bioremediation of oil spills
Advances in microbial genetics and “omics” techniques have facilitated the analysis of microbial communities beyond the limited reach of conventional culturing methods, which generally focus on the growth and activity of single organisms under optimal laboratory conditions. Metagenomics has expanded scientific knowledge of complex microbial communities, enabling analysis of interactions among microbes in the community, as well as co-metabolism, enzyme activity, and biodegradation of oil in the natural environment. “Omics” tools, such as metagenomics and meta-transcriptomics give researchers and spill responders the ability to assess the biodegradation capacity of the native microbial community at a spill site and provide them with further information on whether and to what extent further action is needed (Czaplicki and Gunsch, 2016; Ghosal et al., 2016; Techtmann and Hazen, 2016; Mukherjee et al., 2017).
Oil is a naturally occurring, non-renewable energy source that can cause severe detrimental environmental damage if released in large quantities, such as in an accidental spill. Crude oil is a mixture of thousands of compounds, of which polycyclic aromatic compounds (PACs) are of particular concern due to their acute and chronic toxicity, mutagenicity, and carcinogenicity to interacting organisms (Cao et al., 2009; Dupuis and Ucan-Marin, 2015). PACs consist of two or more fused benzene rings and vary in structure, complexity, and recalcitrance (Haritash and Kaushik, 2009; Lee, 2015). The larger the compound, the more persistent it is likely to be in the environment (Rosenberg and Ron, 1996). To limit the impact of oil on habitats, biota, and nearby communities, mechanical and chemical measures are commonly used in oil spills to facilitate removal or degradation of the oil. However, some of these conventional methods are invasive and cause damage to sensitive habitats and leave oil behind (Zhu et al., 2001). Increasing research and attention has been given to non-invasive alternatives in which native microorganisms play a role in removing oil compounds from the environment after an oil spill (Ghosal, Ghosh, Dutta and Ahn, 2016).
The potential of bioremediation to fill the sustainability gap is substantial when guided by the right insights. The refinement of bioremediation practices can be achieved through the integration of novel “OMICS” methodologies aimed at deciphering the microbiome associated with hydrocarbon plastics. To attain this objective, an exhaustive understanding of bacterial and fungal degradation pathways, their intricate interplays within microbiomes, and their interactions with the soil matrix is imperative. Leveraging “OMICS” techniques facilitates the investigation of both cultivable and non-cultivable soil microbial communities engaged in degradation processes, offering tools to pinpoint pivotal organisms responsible for degrading soil pollutants and reinstating soil vitality and resilience. To do this, we need to thoroughly investigate microbial communities, their degradation pathways, and their complex interactions. Numerous studies provided detailed explanations of genomic and metagenomic methods, as well as the tools necessary to decipher the information obtained. These methods are applicable in two ways: first, to refine bio-based strategies for effectively treating soils contaminated by petroleum hydrocarbons, and second, to facilitate the smooth up-scaling of these technologies to industrial scales (Chicca et al., 2022; Chicca et al., 2022; Becarelli et al., 2023).
2.4 Biodegradation of plastics
Petroleum-based synthetic plastics such as polyethylene (PE), polypropylene (PP), polyvinylchloride (PVC), polyethylene terephthalate (PET), and polystyrene (PS) are the indispensable components of modern industry and daily life but have caused significant environmental problems world-wide due to their extreme recalcitrance to biodegradation in the environments (Krueger et al., 2015; MacLeod et al., 2021). Since current management strategies such as landfills, incineration, and mechanical recycling, are not sufficient to cope with the increasing amount of plastic waste (Drzyzga and Prieto, 2019; Lim and San Thian, 2022; Ackerman and Levin, 2023), biodegradation of these plastics and biotechnological recycling have emerged as an alternative and become one of the research areas actively studied worldwide. Several studies also showed that certain microbial groups have been enriched in plastic films or microplastics in terrestrial environments. Chung et al. (2022) compared the microbial communities on the plastic waste films, which were mostly made of PE and had been buried in the four landfill sites for more than 20 years, with those in the nearby soils assuming that specific and plastic-degrading microorganisms were enriched on the WPFs. The Fourier-transform infrared (FTIR) spectroscopy showed that the WPFs were oxidized, though not as much as when exposed to UV-light and oxygen.
Microbial communities were primarily characterized by their geographical origins, but communities on WPFs generally had lower species richness and diversity than those in nearby soils and exhibited community structures that differed from those in nearby soils. The species taxonomically close to Bradyrhizobiaceae, Pseudarthrobacter (formerly known as Arthrobacter), and Bacillus in the bacterial communities and the one close to Mortierella in the fungal communities were enriched on the WPFs, suggesting that they might be involved in the degradation of the plastic waste films. The network analysis also indicated that the species related to Mortierella was one of the keystone species on the WPFs. Several studies reported that Arthrobacter and Mortierella were also enriched on plastics in soils (Gao et al., 2021; Wright et al., 2021; Yi et al., 2021) or had the ability to degrade plastics (Albertsson et al., 1998; Han et al., 2020). For example, metagenomic analyses have shown that specific and common microbial groups adapt to plastics in terrestrial and marine environments. While there is not yet direct evidence that these microbes are directly responsible for degrading plastics is not yet available, SynBio technologies may soon confirm this hypothesis.
3 Discussion and outlook
The combination of smaller subsystems that arise independently and are later integrated into a whole with new functions is precisely how ecosystems evolved their metagenomics and enormous biodiversity. Therefore, the fundamental principles of SynBio are entirely compatible with our perceptions of ecosystems and their sustainable development. In this context, metagenomics is the study of the structure and function of entire nucleotide sequences isolated and analyzed from all organisms (typically bacteria and fungi) in a bulk sample, based on the direct extraction and cloning of DNA from their natural environment. Metagenomics is often used to study a specific community of microorganisms, such as those residing on human skin, in the soil, or in a water sample. The difference between genomics and metagenomics is in the nature of the sample. Genomics explores examines only the complete genetic information of a single organism only, whereas metagenomics explores a mixture of DNA from multiple organisms and entities within a microbial community, and which may also include viruses, viroids, and free DNA.
Metagenomics samples undoubtedly hold great potential for the introduction of new technologies, e.g., by screening for novel catalytic activities in these samples. This should primarily lead to the development of new-to-nature (i.e., artificial) functions and systems, such as bacterial photography, tumor targeting bacteria, tunable oscillators, sensors, and counters (Sleight et al., 2010a). The integration of novel catalytic activities into controlled synthetic heterotrophic metabolism (Bayer et al., 2009) should facilitate genetic re-engineering of microorganisms e.g., combining metabolic engineering and protein engineering (Völler and Budisa, 2017) for practical applications such as degradation of plastics or remediation of xenobiotics. The production of valuable chemicals or raw materials such as detergents, antimalarials, biofuels from plant biomass, valuable components produced by adaptive evolution in the laboratory, or even a synthetic chromosome transplanted into a host are just a few examples that are well documented in the current literature (Wang et al., 2019).
Metagenomic samples often contain unique genetically encoded information derived from different spatial habitats, e.g., extreme environments (hot, cold, acidic, basic, salty), and often offer fascinating new biological activities. However, a key challenge is to find a compatible host or chassis capable of functionally expressing metagenomic DNA. Species-specific differences in metabolic profiles, regulatory networks, transcription, translation, and post-translation mechanisms complicate this endeavor (Amarelle et al., 2023). Therefore, the introduction of standardized hosts (chassis) and SynBio principles is essential to effectively exploit the biological innovations of metagenomics. These adaptable chassis, which include mesophilic, psychrophilic, thermophilic, halophilic, and other species, should be standardized platforms designed to house built-in metagenomic libraries. For example, live bacterial therapeutics could use microbial hosts (e.g., Escherichia coli-based chassis) to ensure resilient colonization and drug delivery in challenging luminal environments (Russell et al., 2022). Other approaches include finding standardized chassis for genomic clusters encoding various natural products (Beites and Mendes, 2015; Liu et al., 2022), or synthetic metagenomics in yeast—a concept has the potential to reproduce the metabolic network of an entire ecosystem in a single Saccharomyces cerevisiae cell (Belda et al., 2021).
Therefore, the integration of metagenomics, ecosystems, and biodiversity sustainability in SynBio is a compelling vision (Figure 1). The conceptual framework of SynBio, which recognizes the hierarchical structure of genes to genomes and extends to higher levels of complexity (Endy, 2005), shares many commonalities with the above topics. In addition, Systems Biology and mathematical modeling will serve as foundational elements in considering the modular redesign of individual life forms and their communities (Porcar et al., 2013). This will allow us to conceptualize and even design complex communities and environments such as the human gut, e.g., through the directed evolution of microbial species, subpopulations, and entire populations as components of a given ecosystem or microenvironment (Bacchus and Fussenegger, 2013). Not surprisingly, Synthetic Microbial Consortia have gained prominence in SynBio almost a decade ago (Shong et al., 2012). Future studies and experimental interventions, including bioremediation, will explore the intricacies of microbiomes, diverse ecological niches, and environments that are highly contaminated by pollutants such as heavy metals, plastics, oil spills, and xenobiotics.
This comprehensive approach will ultimately provide a strong link between bioengineering, metagenomics, microbial consortia, ecosystems, and biodiversity sustainability in Synthetic Biology. The ultimate goal of these efforts would be to create a portfolio of technologies for a long-term, sustainable solution to critical global problems such as marine pollution reduction, climate change mitigation, food security, and biodiversity conservation.
Author contributions
DL: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Validation, Visualization, Writing–original draft, Writing–review and editing. NB: Conceptualization, Writing–review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Natural Sciences and Engineering Research Council (NSERC) of Canada through NSERC Discovery Grants (RGPIN-04945-2017 and RGPIN-05669-2020), Agri-culture and Agri-Food Canada AgriScience BioProducts Cluster Grant (ASC-03; DL) and through the Canada Research Chairs Program (Grant No. 950-231971) to NB.
Acknowledgments
We would like to thank our students to our esteemed colleagues for the numerous discussions and great interest in our research. Specifically, we are grateful to Daniel Flores-Orozco whose research focuses on antibiotic-resistant bacteria, as well as Aidan Guttormson and Madeline Stanley, who are working on bio-remediation techniques for oil spills. We also acknowledge Dr. Hamid Reza Karbalaei-Heidari from the University of Manitoba and Dr. Jae-Hyung Ahn from the National Institute of Agricultural Sciences in the Republic of Korea for their valuable insights and fruitful discussions on topics such as synthetic biology and biodegradation of plastics. Finally, we would also like to thank the local University of Manitoba Prairie iGEM team for their efforts in promoting our synthetic biology research among undergraduates and other students and community members in our university and province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors NB and DL declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acevedo-Rocha, C. G., and Budisa, N. (2016). Xenomicrobiology: A roadmap for genetic code engineering. Microb. Biotechnol. 9, 666–676. doi:10.1111/1751-7915.12398
Ackerman, J., and Levin, D. B. (2023). Rethinking plastic recycling: A comparison between north America and europe. J. Environ. Manag. 340, 117859. doi:10.1016/j.jenvman.2023.117859
Albertsson, A-C., Erlandsson, B., Hakkarainen, M., and Karlsson, S. (1998). Molecular weight changes and polymeric matrix changes correlated with the formation of degradation products in biodegraded polyethylene. J. Environ. Polym. Degrad. 6, 187–195. doi:10.1023/a:1021873631162
Amarelle, V., Roldán, D. M., Fabiano, E., and Guazzaroni, M-E. (2023). Synthetic biology toolbox for antarctic Pseudomonas sp. strains: toward a psychrophilic nonmodel chassis for function-driven metagenomics. ACS Synth. Biol. 12, 722–734. doi:10.1021/acssynbio.2c00543
Bacchus, W., and Fussenegger, M. (2013). Engineering of synthetic intercellular communication systems. Metab. Eng. 16, 33–41. doi:10.1016/j.ymben.2012.12.001
Bayer, T. S., Hoff, K. G., Beisel, C. L., Lee, J. J., and Smolke, C. D. (2009). Synthetic control of a fitness tradeoff in yeast nitrogen metabolism. J. Biol. Eng. 3, 1–11. doi:10.1186/1754-1611-3-1
Becarelli, S., Bernabei, G., Siracusa, G., Baderna, D., Ruffini Castiglione, M., De Simone, G., et al. (2023). Bio-based decontamination and detoxification of total petroleum hydrocarbon contaminated dredged sediments: perspectives to produce constructed technosols in the frame of the circular economy. bioRxiv 2023, 545458.
Beites, T., and Mendes, M. V. (2015). Chassis optimization as a cornerstone for the application of synthetic biology based strategies in microbial secondary metabolism. Front. Microbiol. 6, 906. doi:10.3389/fmicb.2015.00906
Belda, I., Williams, T. C., de Celis, M., Paulsen, I. T., and Pretorius, I. S. (2021). Seeding the idea of encapsulating a representative synthetic metagenome in a single yeast cell. Nat. Commun. 12, 1599. doi:10.1038/s41467-021-21877-y
Berini, F., Casciello, C., Marcone, G. L., and Marinelli, F. (2017). Metagenomics: novel enzymes from non-culturable microbes. FEMS Microbiol. Lett. 364, fnx211. doi:10.1093/femsle/fnx211
Bouhajja, E., Agathos, S. N., and George, I. F. (2016). Metagenomics: probing pollutant fate in natural and engineered ecosystems. Biotechnol. Adv. 34, 1413–1426. doi:10.1016/j.biotechadv.2016.10.006
Bradley, R. W., Buck, M., and Wang, B. (2016). Tools and principles for microbial gene circuit engineering. J. Mol. Biol. 428, 862–888. doi:10.1016/j.jmb.2015.10.004
Budisa, N., Kubyshkin, V., and Schmidt, M. (2020). Xenobiology: a journey towards parallel life forms. Chembiochem 21, 2228–2231. doi:10.1002/cbic.202000141
Buta-Hubeny, M., Korzeniewska, E., Hubeny, J., Zieliński, W., Rolbiecki, D., Harnisz, M., et al. (2022). Structure of the manure resistome and the associated mobilome for assessing the risk of antimicrobial resistance transmission to crops. Sci. Total Environ. 808, 152144. doi:10.1016/j.scitotenv.2021.152144
Cao, B., Nagarajan, K., and Loh, K-C. (2009). Biodegradation of aromatic compounds: current status and opportunities for biomolecular approaches. Appl. Microbiol. Biotechnol. 85, 207–228. doi:10.1007/s00253-009-2192-4
Chandran, D., Copeland, W., Sleight, S., and Sauro, H. (2008). Mathematical modeling and synthetic biology. Drug Discov. Today Dis. Models 5, 299–309. doi:10.1016/j.ddmod.2009.07.002
Chicca, I., Becarelli, S., Bernabei, G., Siracusa, G., and Di Gregorio, S. (2022). Innovative culturomic approaches and predictive functional metagenomic analysis: the isolation of hydrocarbonoclastic bacteria with plant growth promoting capacity. Water 14, 142. doi:10.3390/w14020142
Chicca, I., Becarelli, S., and Di Gregorio, S. (2022). Microbial involvement in the bioremediation of total petroleum hydrocarbon polluted soils: challenges and perspectives. Environments 9, 52. doi:10.3390/environments9040052
Chung, J-h., Yeon, J., Seong, H. J., An, S-H., Kim, D-Y., Yoon, Y., et al. (2022). Distinct bacterial and fungal communities colonizing waste plastic films buried for more than 20 years in four landfill sites in Korea. J. Microbiol. Biotechnol. 32, 1561–1572. doi:10.4014/jmb.2206.06021
Costello, A., and Badran, A. H. (2021). Synthetic biological circuits within an orthogonal central dogma. Trends Biotechnol. 39, 59–71. doi:10.1016/j.tibtech.2020.05.013
Czaplicki, L. M., and Gunsch, C. K. (2016). Reflection on molecular approaches influencing state-of-the-art bioremediation design: culturing to microbial community fingerprinting to omics. J. Environ. Eng. 142, 03116002. doi:10.1061/(ASCE)EE.1943-7870.0001141
Danchin, A. (2012). Scaling up synthetic biology: do not forget the chassis. FEBS Lett. 586, 2129–2137. doi:10.1016/j.febslet.2011.12.024
Danso, D., Chow, J., and Streit, W. R. (2019). Plastics: environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 85, e01095-19–e01019. doi:10.1128/AEM.01095-19
de Lorenzo, V. (2011). Beware of metaphors: chasses and orthogonality in synthetic biology. Bioeng. bugs 2, 3–7. doi:10.4161/bbug.2.1.13388
Delgado, A., and Porcar, M. (2013). Designing de novo: interdisciplinary debates in synthetic biology. Syst. Synthetic Biol. 7, 41–50. doi:10.1007/s11693-013-9106-6
Drzyzga, O., and Prieto, A. (2019). Plastic waste management, a matter for the community. Microb. Biotechnol. 12, 66–68. doi:10.1111/1751-7915.13328
Dupuis, A., and Ucan-Marin, F. (2015). A literature review on the aquatic toxicology of petroleum oil: an overview of oil properties and effects to aquatic biota, Fisheries and Oceans Canada, Ottawa, dostęp. Fisheries and Oceans Canada. Science Advisory Secretariate.
Flores-Orozco, D., Patidar, R., Levin, D., Kumar, A., Sparling, R., and Cicek, N. (2023). Metagenomic analyses reveal that mesophilic anaerobic digestion substantially reduces the abundance of antibiotic resistance genes and mobile genetic elements in dairy manures. Environ. Technol. Innovation 30, 103128. doi:10.1016/j.eti.2023.103128
Gao, B., Yao, H., Li, Y., and Zhu, Y. (2021). Microplastic addition alters the microbial community structure and stimulates soil carbon dioxide emissions in vegetable-growing soil. Environ. Toxicol. Chem. 40, 352–365. doi:10.1002/etc.4916
Garrido-Cardenas, J. A., and Manzano-Agugliaro, F. (2017). The metagenomics worldwide research. Curr. Genet. 63, 819–829. doi:10.1007/s00294-017-0693-8
Ghosal, D., Ghosh, S., Dutta, T. K., and Ahn, Y. (2016). Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front. Microbiol. 7, 1369. doi:10.3389/fmicb.2016.01369
Greninger, A. L. (2018). The challenge of diagnostic metagenomics. Expert Rev. Mol. diagnostics 18, 605–615. doi:10.1080/14737159.2018.1487292
Guazzaroni, M. E., Silva-Rocha, R., and Ward, R. J. (2015). Synthetic biology approaches to improve biocatalyst identification in metagenomic library screening. Microb. Biotechnol. 8, 52–64. doi:10.1111/1751-7915.12146
Han, Y-N., Wei, M., Han, F., Fang, C., Wang, D., Zhong, Y-J., et al. (2020). Greater biofilm formation and increased biodegradation of polyethylene film by a microbial consortium of Arthrobacter sp. and Sreptomyces sp. Microorganisms 8, 1979. doi:10.3390/microorganisms8121979
Haritash, A., and Kaushik, C. (2009). Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): a review. J. Hazard. Mater. 169, 1–15. doi:10.1016/j.jhazmat.2009.03.137
Karbalaei-Heidari, H. R., and Budisa, N. (2020). Combating antimicrobial resistance with new-to-nature lanthipeptides created by genetic code expansion. Front. Microbiol. 11, 590522. doi:10.3389/fmicb.2020.590522
Kayani, MuR., Huang, W., Feng, R., and Chen, L. (2021). Genome-resolved metagenomics using environmental and clinical samples. Briefings Bioinforma. 22, bbab030. doi:10.1093/bib/bbab030
Krueger, M. C., Harms, H., and Schlosser, D. (2015). Prospects for microbiological solutions to environmental pollution with plastics. Appl. Microbiol. Biotechnol. 99, 8857–8874. doi:10.1007/s00253-015-6879-4
Lee, K. (2015). Royal society of Canada expert panel report: the behaviour and environmental impacts of crude oil released into aqueous environments. Royal Society of Canada.
Lim, B. K. H., and San Thian, E. (2022). Biodegradation of polymers in managing plastic waste—a review. Sci. Total Environ. 813, 151880. doi:10.1016/j.scitotenv.2021.151880
Liu, J., Wang, X., Dai, G., Zhang, Y., and Bian, X. (2022). Microbial chassis engineering drives heterologous production of complex secondary metabolites. Biotechnol. Adv. 59, 107966. doi:10.1016/j.biotechadv.2022.107966
MacLeod, M., Arp, H. P. H., Tekman, M. B., and Jahnke, A. (2021). The global threat from plastic pollution. Science 373, 61–65. doi:10.1126/science.abg5433
Marco, D. (2008). Metagenomics and the niche concept. Theory Biosci. 127, 241–247. doi:10.1007/s12064-008-0028-x
Mukherjee, A., Chettri, B., Langpoklakpam, J. S., Basak, P., Prasad, A., Mukherjee, A. K., et al. (2017). Bioinformatic approaches including predictive metagenomic profiling reveal characteristics of bacterial response to petroleum hydrocarbon contamination in diverse environments. Sci. Rep. 7, 1108. doi:10.1038/s41598-017-01126-3
Müller, K. M., and Arndt, K. M. (2012). Standardization in synthetic biology. Methods Mol. Biol. 813, 23–43. doi:10.1007/978-1-61779-412-4_2
New, F. N., and Brito, I. L. (2020). What is metagenomics teaching us, and what is missed? Annu. Rev. Microbiol. 74, 117–135. doi:10.1146/annurev-micro-012520-072314
Parages, M. L., Gutiérrez-Barranquero, J. A., Reen, F. J., Dobson, A. D., and O’Gara, F. (2016). Integrated (meta) genomic and synthetic biology approaches to develop new biocatalysts. Mar. drugs 14, 62. doi:10.3390/md14030062
Porcar, M., Latorre, A., and Moya, A. (2013). What symbionts teach us about modularity. Front. Bioeng. Biotechnol. 1, 14. doi:10.3389/fbioe.2013.00014
Robinson, S. L., Piel, J., and Sunagawa, S. (2021). A roadmap for metagenomic enzyme discovery. Nat. Product. Rep. 38, 1994–2023. doi:10.1039/d1np00006c
Rosenberg, E., and Ron, E. Z. (1996). Bioremediation of petroleum contamination. Biotechnol. Res. Ser. 6, 100–124.
Russell, B. J., Brown, S. D., Siguenza, N., Mai, I., Saran, A. R., Lingaraju, A., et al. (2022). Intestinal transgene delivery with native E. coli chassis allows persistent physiological changes. Cell 185, 3263–3277.e15. doi:10.1016/j.cell.2022.06.050
Sabino, Y. N. V., Santana, M. F., Oyama, L. B., Santos, F. G., Moreira, A. J. S., Huws, S. A., et al. (2019). Characterization of antibiotic resistance genes in the species of the rumen microbiota. Nat. Commun. 10, 5252. doi:10.1038/s41467-019-13118-0
Schmidt, M., Pei, L., and Budisa, N. (2018). Xenobiology: state-of-the-art, ethics, and philosophy of new-to-nature organisms. Synth. biology–metabolic Eng. 162, 301–315. doi:10.1007/10_2016_14
Sharma, P., Singh, S. P., Iqbal, H. M., and Tong, Y. W. (2022). Omics approaches in bioremediation of environmental contaminants: an integrated approach for environmental safety and sustainability. Environ. Res. 211, 113102. doi:10.1016/j.envres.2022.113102
Shong, J., Diaz, M. R. J., and Collins, C. H. (2012). Towards synthetic microbial consortia for bioprocessing. Curr. Opin. Biotechnol. 23, 798–802. doi:10.1016/j.copbio.2012.02.001
Sleight, S. C., Bartley, B. A., Lieviant, J. A., and Sauro, H. M. (2010a). Designing and engineering evolutionary robust genetic circuits. J. Biol. Eng. 4, 12–20. doi:10.1186/1754-1611-4-12
Sleight, S. C., Bartley, B. A., Lieviant, J. A., and Sauro, H. M. (2010b). In-Fusion BioBrick assembly and re-engineering. Nucleic acids Res. 38, 2624–2636. doi:10.1093/nar/gkq179
Sullivan, S. F., Shetty, A., Bharadwaj, T., Krishna, N., Trivedi, V. D., Gopinarayanan, V. E., et al. (2023). Towards universal synthetic heterotrophy using a metabolic coordinator. Metab. Eng. 79, 14–26. doi:10.1016/j.ymben.2023.07.001
Techtmann, S. M., and Hazen, T. C. (2016). Metagenomic applications in environmental monitoring and bioremediation. J. Industrial Microbiol. Biotechnol. 43, 1345–1354. doi:10.1007/s10295-016-1809-8
Tseng, C-H., and Tang, S-L. (2014). Marine microbial metagenomics: from individual to the environment. Int. J. Mol. Sci. 15, 8878–8892. doi:10.3390/ijms15058878
van der Helm, E., Genee, H. J., and Sommer, M. O. (2018). The evolving interface between synthetic biology and functional metagenomics. Nat. Chem. Biol. 14, 752–759. doi:10.1038/s41589-018-0100-x
Völler, J-S., and Budisa, N. (2017). Coupling genetic code expansion and metabolic engineering for synthetic cells. Curr. Opin. Biotechnol. 48, 1–7. doi:10.1016/j.copbio.2017.02.002
Wang, Y., Ling, C., Chen, Y., Jiang, X., and Chen, G-Q. (2019). Microbial engineering for easy downstream processing. Biotechnol. Adv. 37, 107365. doi:10.1016/j.biotechadv.2019.03.004
Wright, R. J., Langille, M. G., and Walker, T. R. (2021). Food or just a free ride? A meta-analysis reveals the global diversity of the plastisphere. ISME J. 15, 789–806. doi:10.1038/s41396-020-00814-9
Yen, S., and Johnson, J. S. (2021). Metagenomics: a path to understanding the gut microbiome. Mamm. Genome 32, 282–296. doi:10.1007/s00335-021-09889-x
Yi, M., Zhou, S., Zhang, L., and Ding, S. (2021). The effects of three different microplastics on enzyme activities and microbial communities in soil. Water Environ. Res. 93, 24–32. doi:10.1002/wer.1327
Zhu, X., Venosa, A. D., Suidan, M. T., and Lee, K. (2001). Guidelines for the bioremediation of marine shorelines and freshwater wetlands. Cincinnati, OH: U.S. Environmental Protection Agency, Office of Research and Development, National Risk Management Research Laboratory, Land Remediation and Pollution Control Division.
Keywords: circuits, chassis, devices, biodiversity, bioengineering, metagenomics, modules
Citation: Levin DB and Budisa N (2023) Synthetic biology encompasses metagenomics, ecosystems, and biodiversity sustainability within its scope. Front. Synth. Biol. 1:1255472. doi: 10.3389/fsybi.2023.1255472
Received: 08 July 2023; Accepted: 28 August 2023;
Published: 13 September 2023.
Edited by:
Carrie A. Eckert, Oak Ridge National Laboratory (DOE), United StatesReviewed by:
Alexander James Webb, Imperial College London, United KingdomCopyright © 2023 Levin and Budisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David B. Levin, ZGF2aWQubGV2aW5AdW1hbml0b2JhLmNh; Nediljko Budisa, bmVkaWxqa28uYnVkaXNhQHVtYW5pdG9iYS5jYQ==
 David B. Levin
David B. Levin Nediljko Budisa
Nediljko Budisa