- 1Department of General Pediatrics, Medical Faculty, Heinrich Heine University, Düsseldorf, Germany
- 2Roche Diagnostics GmbH, Nonnenwald, Germany
- 3AMSilk GmbH, Neuried, Germany
- 4Institute of Chemistry and Biochemistry, Freie Universität Berlin, Berlin, Germany
- 5Icosagen Cell Factory OÜ, Tartumaa, Estonia
- 6European Science Communication Institute (ESCI) GmbH, Oldenburg, Germany
- 7Institute of Biochemistry and Molecular Biology, University of Hamburg, Hamburg, Germany
- 8Departement of Microbiology and Statistics, University of Manitoba, Winnipeg, MB, Canada
- 9Robert Koch Institute, Berlin, Germany
- 10Institute of Biotechnology, TU Berlin, Berlin, Germany
- 11Department of Chemistry, University of Manitoba, Winnipeg, MB, Canada
- 12Biocatalysis Group, Institute for Chemistry, Technische Universität Berlin, Berlin, Germany
Despite billions of years of evolution, there have been only minor changes in the number and types of proteinogenic amino acids and the standard genetic code with codon assignments across the three domains of life. The rigidity of the genetic code sets it apart from other aspects of organismal evolution, giving rise to key questions about its origins and the constraints it places on innovation in translation. Through adaptive laboratory evolution (ALE) in Escherichia coli, we aimed to replace tryptophan (Trp) in the genetic code with an analogue L-β-(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa). This required Escherichia coli to recruit thienopyrrole instead of indole and allowed reassignment of UGG codons. Crossing the stress response system emerged as a major obstacle for ancestral growth in the presence of [3,2]Tp and Trp limitation. During ALE, a pivotal innovation was the deactivation of the master regulon RpoS, which allowed growth solely in the presence of [3,2]Tp in minimal medium. Notably, knocking out the rpoS gene in the ancestral strain also facilitated growth on [3,2]Tp. Our findings suggest that regulatory constraints, not just a rigid translation mechanism, guard Life’s canonical amino acid repertoire. This knowledge will not only facilitate the design of more effective synthetic amino acid incorporation systems but may also shed light on a general biological mechanism trapping organismal configurations in a status quo.
1 Introduction
“Evolution is, as an engineer, an opportunist, not a perfectionist.” ― Stanisław Lem, His Master’s Voice
Evolution is an ongoing and pervasive process for all life on the planet and has yielded extant organisms with an immense diversity of body shapes, structures, and functions. The genetic code, which emerged >3 billion years ago, relates each of the 20 canonical amino acids to specific nucleotide triplets in genes and mRNAs (Wong, 1988). Given the adaptative potential that allows organisms to live in virtually all known ecosystems, one would expect to find the appropriate variation in the building blocks of life; yet it has remained an almost invariant (“frozen”) feature of life on Earth (Crick, 1968). The genetic code likely started with a few small, simple amino acids that can be produced via a proto-metabolism, and gradually evolved over time to add more complex amino acids with diverse functionalities to the repertoire. The discovery of selenocysteine and pyrrolysine, the 21st and 22nd proteinogenic amino acids implies some flexibility to the code, yet they are incorporated at very specific positions in only a small set of proteins. This prompts a fundamental inquiry into why evolution appears to be constrained to these 20 canonical amino acids (Hartman and Smith, 2014; Koonin and Novozhilov, 2017; Kun and Radványi, 2018) (cAAs) and whether it is feasible for a species to naturally or artificially evolve from an existing organism with a modified set of cAAs (canonical amino acids). The emergence of a species using a non-standard genetic code with synthetic amino acids would pave the way for the creation of artificial diversity and synthetic life.
The amino acid tryptophan (Trp), which has the highest metabolic cost for biosynthetic production, is thought to be the latest addition to the genetic code (Trifonov, 2009; Fournier and Alm, 2015). Trp and methionine are the only two cAAs encoded by a single codon (Lajoie et al., 2013). Trp is the least abundant amino acid in eukaryotic proteomes (1.24%) and viruses (1.19%), and the second least abundant in both Archaeal (1.03%) and bacterial proteomes (1.27%) (Kozlowski, 2017). In contrast, the most prevalent amino acid in all life forms, leucine, has an abundance between 8.84% (in viruses) and 10.09% (in bacteria) (Kozlowski, 2017). For a long time, Trp has been a focal point in showcasing the genetic code’s chemical mutability by substituting it with noncanonical synthetic analogues (Wong, 1983; Bacher and Ellington, 2001).
The use of Adaptive Laboratory Evolution (ALE) experiments has been instrumental in advancing our comprehension of the genetic modifications that enable bacteria to grow in the presence of and incorporate noncanonical amino acids (ncAA). In long-term serial dilution experiments (i.e., ALE), bacterial metabolic prototypes are exposed to high concentrations of ncAAs over extended periods, creating significant selection pressure and leading to increased incorporation. Understanding the intermediate stages in this crucial process would offer valuable insights into the number of steps necessary for the transition from the parent strain to the adapted bacteria, providing a molecular blueprint for constructing new strains.
Recently, we successfully achieved complete substitution of Trp with its surrogate L-β-(thieno[3,2-b]pyrrolyl)-alanine ([3,2]Tpa, (Hoesl et al., 2015, Figure 1) in the E. coli proteome. However, the key knowledge gap that still needed to be elucidated was the underlying mechanism of adaptation.
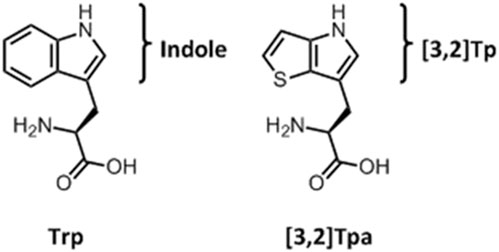
FIGURE 1. Chemical structures of Tryptophan and L-β-(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa). The brackets indicate the structures of the precursor’s indole and β-thieno[3,2-b]pyrrole ([3,2]Tp).
Selective pressure was exerted to a Trp-auxotrophic strain, compelling it to convert ([3,2]Tp) and L-Ser to [3,2]Tpa through the catalytic reaction of the endogenous enzyme Trp synthase (TrpS). However, upon analyzing the genetic sequencing and mass spectrometry data, we discovered that the genomic configuration of the ancestral strain made it impractical to determine the genetic factors responsible for adaptation to [3,2]Tpa alone, as it was intertwined with other off-target selective pressures (manuscript in prep). In 2014, Yu et al. (Yu et al., 2014) sequenced and analyzed a Trp-auxotrophic Bacillus subtilis strain adapted towards the ncAA 4-fluorotryptophan. (Wong, 1983). After undergoing just a few rounds of selection, the strain become tolerant to 4-F-Trp by acquiring mutations in rpoC and in the sigma factors sigI (Zuber et al., 2001) and sigB (Hecker et al., 2007), all general stress response factors in B. subtilis. A similar experiment with Trp-auxotrophic Escherichia coli revealed that point mutations arose in genes encoding the non-essential vegetative sigma factors rpoA and rpoC. These sigma factors play a crucial role in reconfiguring transcription under unfavorable environmental conditions (Agostini et al., 2021). Combined, the present work points to the potential significance of the stress response pathway in promoting the growth and incorporation of ncAAs.
The stress response is triggered in bacteria when they enter the stationary phase, providing protection against various environmental stresses in their natural habitats, such as nutrient limitation, resource competition, temperature fluctuations, pH changes, and osmolarity variations. These mechanisms are crucial for E. coli to thrive in their natural habitats (Jenkins et al., 1990; Lange and Hengge-Aronis, 1991; McCann et al., 1991; Hengge, 2014). The general stress response is mediated by the alternative sigma factor RpoS (σS), which interacts with the core RNA polymerase (RNAP) (Gottesman, 2019). RpoS plays a key role in regulating the expression of a large set of genes (Weber et al., 2005) and controls various promoters shared with the vegetative sigma factor RpoD (σ70). The response mediated by RpoS is subject to regulation at multiple levels, including transcription, translation, degradation, and activity (Battesti et al., 2010).
In this study, we devised a novel ALE experiment using an E. coli strain with an optimized genomic configuration (Blattner et al., 1997). Through this new experimental approach, we demonstrate that E. coli can overcome the frozen state of the genetic code to completely replace tryptophan with the [3,2]Tpa analogue. Furthermore, we identified that the primary adaptive mechanism involved in this process is an alternation of the RpoS-mediated general stress response.
2 Materials and methods
2.1 Inoculation method of the adaptive evolution experiment
In a 100 mL Erlenmeyer flask covered with aluminium foil, 10 mL of the respective NMM-medium (Budisa et al., 1995; Minks et al., 2000) was inoculated with the previous culture at a ratio of 1:100. The culture grew under shaking for 1 or 2 days until the OD600 was constant. Medium supplements containing indole and amino acids were removed as soon as possible while maintaining the population density above 0.1 OD600 to avoid bottleneck effects.
2.2 Doubling times of adapted strains
Isolates were re-grown from a cryostock in 5 mL LB at 37°C overnight. To remove the LB medium, cells were washed twice with NMM0. 10 mL of the respective NMM was inoculated with 100 μL of these washed NMM0 cells and grown at 30°C or 37°C until stationary phase was reached (generally between 1–2 days). This step was repeated twice and then these cultures were used to inoculate a 96-well plate with 200 μL of cultures at a ratio of 1:100. OD600 values were determined using the Infinite®M200 plate reader (Tecan Group AG, Männedorf, Switzerland). The maximum growth rate was calculated using an R script performing spline-fitting. During the adaptation experiment, a concentration of 25 µM [3,2]Tp was utilized, whereas for the characterization experiments, a slightly higher concentration of 30 µM indole analogue was employed. This adjustment was made to enhance culturing stability during the latter experiments.
2.3 Next-generation sequencing and analysis
Genomic DNA was sent to Beijing Genomics Institute in Hong Kong for re-sequencing. An Illumina HiSeq 2000 sequencing platform was used for the adapted strains TUB40, TUB85, and TUB145, and an Illumina HiSeq 2,500 sequencing platform was used for TUB170. Sequencing data were aligned to E. coli MG1655 (NC_000913.3) and analyzed by the company.
To assess evidence of strain evolution, we utilized breseq v0.34.0 (Deatherage and Barrick, 2014) in consensus mode, running the analysis on 15 processors with a limit-fold-coverage (-l) of 100. Consensus mode is the suitable approach for re-sequencing a clonal haploid genome. E. coli MG1655 (NC_000913.3) was used as reference in all analyses. The number of mutations in each genome was counted using the gdtools utility from breseq. The gdtools utility was also used to combine all mutations from each strain into a gd file, which was then converted into a vcf file, and the effects of the mutations were evaluated using SnpEff (Cingolani et al., 2012).
To construct the phylogeny of the clones, MG1655 was used as an outgroup. A phylip file for multiple sequence alignment was generated from the gd files of the strains using gdtools COMPARE as part of the Breseq analyses. The phylip file, containing the alignment data, was imported into Seaview (Gouy et al., 2010) to obtain alignment statistics. Subsequently, a phylogenetic tree was constructed using PhyML with a GTR model, 100 bootstraps, and the best of NNI and SPR as tree searching operations. Finally, the resulting tree was visualized using iTO (Letunic and Bork, 2007).
2.4 Cluster plots using string.db
Cluster plots were generated using the STRING v10 database (http://string-db.org/). This bioinformatics tool allows the detection of interactions between two proteins based on data mining. A detailed description of the software can be found in Szklarczyk et al. (Szklarczyk et al., 2015).
2.5 Chromosomal gene deletions: phage P1 transduction and CRISPR/Cas9
A 1:100 dilution of the overnight culture of the donor strain was inoculated into 5 mL of LB medium. The cells were then grown under agitation at 37°C reaching an OD600 of approximately 0.6. Following the addition of 5 mM CaCl2, the cell suspension was incubated at 37°C for 30 min. Subsequently, 100 μL of the treated cell suspension was mixed with 100 μL of a phage P1 solution. After incubation at 37°C for 20 min without shaking, the mixture was added to 4 mL 0.6% soft agar containing 5 mM CaCl2. After infecting the donor cells, the soft agar-containing mixture was poured onto LB agar plates and left to incubate overnight. Following incubation, the soft agar-containing plaques were removed, and 800 μL of CHCl3 were added to the plates. The suspension was vigorously mixed, and then subjected to centrifugation at 15,000 x g for 10 min. After centrifugation, the supernatant was preserved by adding a few drops of CHCl3 and stored at 4°C. The recipient strain was cultured with agitation at 37°C until reaching an OD600 of approximately 0.6. After adding 5 mM CaCl2, the cell suspension was incubated at 37°C for 30 min. Subsequently, 1 mL of the cell suspension was infected with 30 μL of the donor phage solution. The cells were incubated at 37°C for 15 min without shaking, then pelleted and resuspended in 1 mL of LB supplemented with 0.1 M sodium citrate. After shaking for 45 min at 37°C, the cells were plated on LB/Kan agar plates and incubated overnight at 37°C. The knockout was verified through colony PCR.
CRISPR/Cas9 deletions were performed using the CAGO technique, as previously described (Zhao et al., 2017).
2.6 Proteomic analysis - Sample preparation and measurement
Isolates were revived from a cryostock by growing them in 5 mL of LB medium at 37°C overnight. To remove any remaining LB medium, the cells were washed twice with NMM0. Subsequently, 10 mL of the appropriate NMM were inoculated with 100 μL of the NMM0 washed cells and incubated at either 30°C or 37°C until they reached the stationary phase, which typically occurred within 1 to 2 days. Afterward, these cultures were used to inoculate triplicate 50 mL cultures. The triplicate cultures were allowed to grow until reaching the early stationary phase, after which the cells were harvested by centrifugation at 4°C, 5,000 g for 15 min. Cell pellets were lysed using a solution containing 100 mM TRIS (pH 7.5), 4% sodium dodecyl sulfate (SDS), and 100 mM DTT. The lysis process was performed at 95°C for 10 min, followed by sonication in a Bioruptor Standard (Diagenode, Belgium) for 30 s on and 30 s off, totaling 10 cycles, with a power setting at high (H); throughout this process, samples were constantly kept on ice. After lysing, the samples were clarified by centrifugation at 4°C, 17,000 g for 5 min.
The ‘heavy’-labeled (H) TUB00dKO lysate was prepared using NMM(17/30/1) supplemented with 13C615N4-arginine and 13C615N2-lysine from Silantes, Munich, Germany. This lysate was mixed at a 1:1 ratio with various ‘light’-labeled (L) lysates, namely, TUB00 (NMM(19/0/30)), TUB85 (NMM(19/30/1)), TUB85 (NMM(19/30/0)), TUB170 (NMM(19/30/1)), and TUB170 (NMM(19/30/0)). Afterward, the proteins were digested following previously described methods (Wiśniewski et al., 2009).
For LC-MS/MS analysis, peptides were desalted using C18-StageTips and then separated through reversed-phase liquid chromatography using a Dionex UltiMate RSLCnano 3000 system, which was coupled to a Q Exactive mass-spectrometer from Thermo Fisher Scientific, United States. Peptides were loaded onto a 75 μm × 300 mm fused silica emitter (New Objective, United States) packed in-house with Reprosil-Pur C18-AQ 3 μm particles (Dr. Maisch, Germany) at a flow rate of 500 nL/min for 10 min. The separation was achieved using a 230-min gradient of 2%–40% B (A 0.1% formic acid, B 0.1% formic acid/80% acetonitrile) at a flow rate of 200 nL/min. This was followed by an 11-min ramp to 95% B, a 5-min wash at 95% B, and a return to the initial conditions for the next run.
The eluted peptides were directly sprayed into a Q Exactive mass spectrometer operating in data-dependent mode. For each precursor ion scan, up to ten MS/MS scans were recorded with a normalized collision energy (NCE) of 25. Precursor ion spectra were recorded in profile mode with a mass range of m/z 350–1,400 and a resolution (R) of 70,000. The data-dependent MS/MS spectra were acquired in profile mode with an NCE of 25 and a resolution (R) of 17,500. Mono-isotopic precursor selection was enabled, and ions with a single charge or an unassigned charge state were rejected. Additionally, each fragmented ion was dynamically excluded for 90 s to avoid repetitive fragmentation.
The MS data were analyzed using MaxQuant 1.3.0.5, and the Escherichia coli MG1655 UniProtKB reference proteome sequence database (downloaded on 20110909) was utilized. The analysis was conducted with the following parameters: enzyme - Trypsin/P, precursor mass accuracy - 10 ppm, fragment mass accuracy - 20 ppm, fixed modification—Carbamidomethyl (C), variable modifications - Oxidation (M) and Trp->[3,2]Tpa (W + 5.956 Da). The False Discovery Rate (FDR) threshold was set at 0.01.
3 Results
3.1 Set-up of the ALE on [3,2]Tp
We constructed an E. coli MG1655 strain by deleting the tryptophanase gene and most of the genes in the Trp operon (trpLEDC). However, we left the tryptophan synthase genes (trpBA) intact, and their regulation is controlled by trpR (MG1655 ΔtnaA ΔtrpLEDC). This modification enables us to supplement the growth medium with indole or [3,2]Tp, which the cells can take up and convert directly into Trp or [3,2]Tpa within the cytoplasm. We named the modified starting strain TUB00 (refer to Supplementary Figure S1A). The Adaptive Laboratory Evolution (ALE) experiment was carried out using a new minimal medium (NMM) with varying compositions denoted as NMM(a/b/c) where a represents the number of canonical amino acids (cAAs), b represents the concentration of [3,2]Tp in µM, and c represents the concentration of indole in µM. The ALE was initiated with NMM(19/30/1) and concluded with NMM(0/25/0) (see Figure 2A).
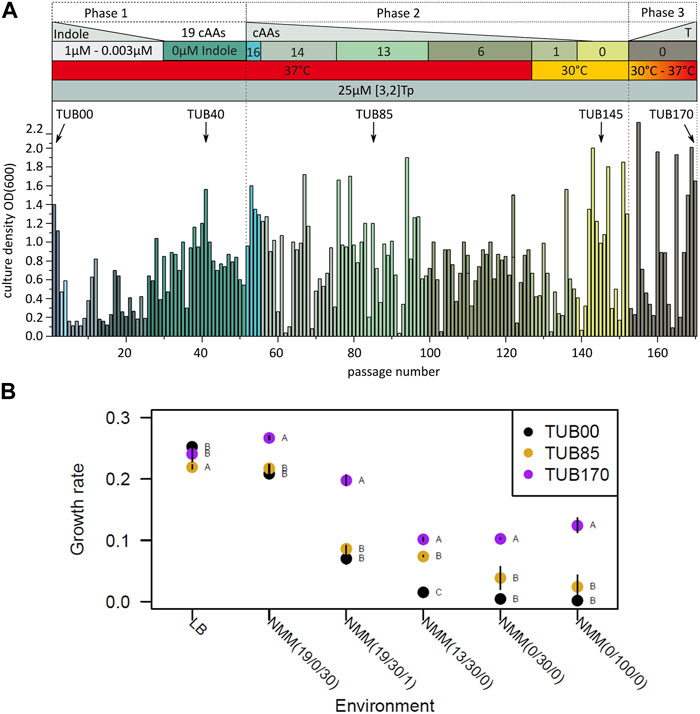
FIGURE 2. ALE on [3,2]Tp. (A) Optical densities achieved during the adaptation experiment. Phase 1: the indole concentration was reduced from 1 µM to 0 µM in several steps. Phase 2 all other 19 cAAs were removed following the metabolic blocks pattern, and growth temperature was reduced to 30°C in order to remove the last amino acid, Met. The numbers in the bars above the graph denote the total number of the remaining cAAs. Phase 3 The temperature was gradually increased back to 37°C. (B) Doubling times of TUB00, TUB85, and TUB170 at 30°C, each dot is the average of three replicates with the standard error bar. The letters A, B, C indicate Tukey test results (statistical significance, p < 0.05), when different strains have different letters, they are statistically different (see also Supplementary Table S2).
During the first phase of ALE, the indole concentration was systematically decreased over 29 passages, equivalent to approximately 190 generations. Surprisingly, at this point, the bacterial population demonstrated the ability to grow in a medium without the need for any supplemental indole (refer to Figure 2A). To prevent any potential contamination with Trp traces from commercial amino acid preparations, similar to what has been observed in other experiments (Bacher and Ellington, 2001), the second phase involved removing the remaining 19 canonical amino acids (cAAs) from the medium. This removal was performed in metabolically related amino acid groups, known as “metabolic blocks” (Agostini et al., 2021).
After 97 additional passages, which corresponds to about 630 more generations, and with a concurrent decrease in cultivation temperature from 37°C to 30°C, all canonical amino acids (cAAs) were removed from the medium, except for methionine. In the subsequent 10 passages (∼66 generations), the methionine was removed from the medium without compromising the growth of the bacterial population. During this process, the cultivation temperature was gradually raised back to 37°C in what we refer to as phase 3 (see Figure 2A). The adaptation experiment was concluded after a total of 170 passages (approximately 1,100 generations), leading to the isolation of the final evolved strain denoted as TUB170.
The rationale of switching cultivation temperatures from 37°C to 30°C is as follows. In phase 2 of the evolution experiment, we gradually removed the additional amino acids from the medium and at the same time performed side experiments to find out which amino acids could be removed without completely stopping growth. Interestingly, in one of these experiments we reduced the temperature and found that at 30°C we could already remove all but one amino acid from the medium. The lower cellular stress at 30°C probably contributed to this result and led to a more relaxed selection. However, since we were not satisfied with cells growing only at 30°C, we adjusted them in phase 3 to grow again at 37°C. In this way, we finally achieved cells that could grow at 37°C in pure mineral medium.
We observed considerable variation in the growth rates among the three strains TUB00, TUB85 (isolated after 85 passages in phase 2), and TUB170, when tested in six distinct environments that represent different experimental phases (refer to Figure 2B and ANOVA results in Supplementary Table S3). Across all NMM environments, including the starting condition medium (NMM (19/30/1)), TUB170 consistently displayed a significantly higher growth rate compared to TUB00 and TUB85. The growth rate of TUB170 was notably higher when using 100 µM [3,2]Tp (NMM(0/100/0)) in comparison to 30 µM [3,2]Tp (NMM(0/30/0)). This difference is likely attributed to either the enhanced efficiency of tRNATrp to be aminoacylated with [3,2]Tpa or the more efficient conversion of [3,2]Tp to [3,2]Tpa by the endogenous TrpBA in TUB170. On the other hand, TUB170 exhibited a significantly lower growth rate in the standard rich LB medium. Notably, TUB85 and TUB00 showed very similar growth rates, except in NMM (13/30/0), indicating that TUB85 might carry a beneficial mutation (or mutations) that specifically benefits growth in this particular environment.
3.2 Mutations occurred in a wide range of genes
During the ALE experiment, we isolated single colonies from different experimental phases, specifically TUB40 (phase 1), TUB85, and TUB145 (phase 2), as well as TUB170 (phase 3). The whole genomes of these four isolates were sequenced, and a total of 42 mutations were identified, offering insights into the evolutionary dynamics of the ALE (see Figure 3A and Supplementary Table S4). An analysis of the mutations revealed both shared mutations, which were present in at least two isolates, and mutations that were not shared with other lineages, which were unique to a single isolate. This analysis indicated that the populations likely consisted of multiple lineages throughout the 170 transfers of the ALE (Figure 3B). Among the identified mutations, nine persisted in all sequenced isolates, suggesting that they likely emerged during the initial 40 transfers in the same genetic background (lineage) from which all sequenced isolates originated. These nine mutations were found in genes related to amino acid biosynthesis and processing (aroG, astB, and leuS), cell morphogenesis (fliD, cvrA), membrane proteins (ylcJ, yejM), oxidoreduction (ydfI), and in the promoter region of yobF, a small protein with an unknown function.
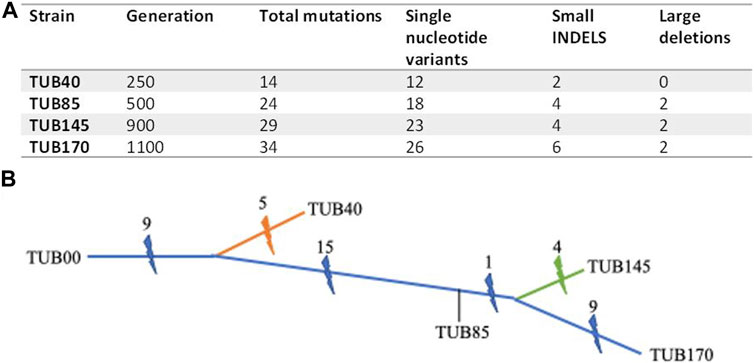
FIGURE 3. Genomic analysis of the ALE. (A) Number of mutations identified in each sequenced isolate. Generations are estimated as described elsewhere27. (B) A cartoon dendrogram depiction of the evolutionary history of the sequenced strains from the ALE experiment as they accumulate mutations (lightning bolt). Mutations that arose in the lineage leading towards the final sequenced isolate, TUB170, are shown in blue. Orange and green lines, and corresponding mutations that were not shared with other lineages, leading to TUB40 and TUB145 are shown along the orange and green trajectories respectively. The precise timing of mutations is unknown, by example, the one mutation shared by TUB145 and TUB170 could have arisen at any point between transfer 85 and the branching point (at an unknown transfer) where the lineages leading to TUB145 and TUB170 arose. Branch lengths are intended to be proportional to the number of accumulated mutations.
Before reaching transfer 40, a divergence occurred, leading to the formation of at least two distinct lineages. One of these lineages gave rise to TUB40, which has five mutations that were not shared with other lineages (i.e., unique to its genetic profile). The second lineage served as the ancestor to the other three sequenced isolates (TUB85, TUB145, TUB170). The second lineage contains 13 shared single-nucleotide variants in kinases (glcK, pyrH, phoR), phosphatases (gpp), reductases (nfeF), proteases (lon, ftsH), stress response factors (rpoS, lrhA, gpp, sanA, sgrR), and a mutation in a pseudogene (ybfl). Additionally, this lineage showed two large deletions, with the larger one spanning 23 kb and involving 35 genes, including the Rac prophage. The smaller is a 6 kb deletion encompassing 10 genes. This deletion commenced within the putative defective integrase of the Qin prophage (intQ) and terminated within the putative selenite reductase (ynfE). Importantly, no mutations that were not shared with other lineages were found in TUB85, suggesting that this isolate serves as a common ancestor to both TUB145 and TUB170.
The presence of only one shared mutation, specifically in ppc (a phosphoenolpyruvate carboxylase), in both TUB145 and TUB170, suggests that these two lineages likely diverged shortly after transfer 85. Furthermore, TUB145 had an additional four mutations that were not shared with other lineages, while TUB170 had nine of them (i.e., mutations that were not shared with other lineages), indicating unique genetic changes in each lineage. Notably, two genes, cysN (sulfate adenylyltransferase) and gltA (citrate synthetase), exhibited distinct mutations in the terminal isolates (TUB145 and TUB170). This observation suggests a robust selection pressure for variants of these genes, making them of particular interest in the context of the evolutionary dynamics of the experiment.
3.3 Mutations involved in stress response
The mutation in the stress response gene rpoS, found in TUB85, TUB145 and TUB170, caught immediate attention. This mutation specifically affected residue D118 (shown in Figure 4A), leading to the disruption of the salt bridge interaction between D118 in σS and R275 in the RpoC-subunit of the core-RNAP (Figure 4B). The aspartate is conserved in most sigma factors (σS, σD, σH) (Gruber and Gross, 2003). To verify whether an altered stress response is beneficial for adapting to [3,2]Tp, we used CRISPR/Cas9 to delete the rpoS in TUB00. Our rationale was that by decreasing σS-mediated stress response, we might enhance the growth on the indole-analogue compound. We conducted a growth comparison among TUB00, TUB170, and the ΔrpoS strain in media containing different indole concentrations. The optical density of the ΔrpoS strain showed an intermediate level of growth between the ancestral (TUB00) and the evolved strain (TUB170) (Figure 4C, Supplementary Table S5). This indicates that the ΔrpoS strain exhibits increased tolerance towards the adverse effects of [3,2]Tp supplementation, suggesting that the absence of the rpoS gene provides some benefits. It is noteworthy that improved growth of the ΔrpoS strain relative to the ancestor was evident even in media lacking a source of Trp such as NMM (19/30/0) and NMM(0/30/0)).
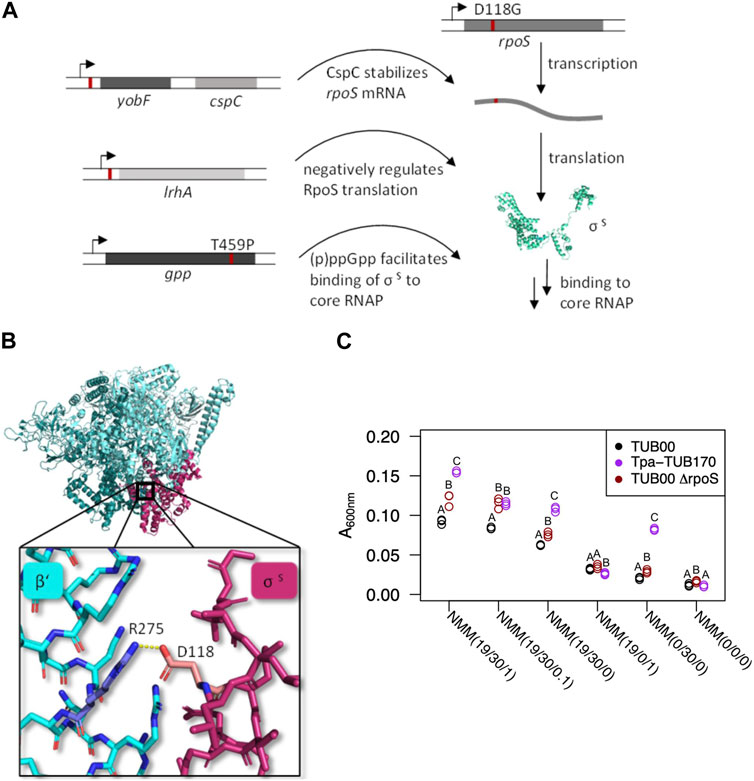
FIGURE 4. Mutations involved in the RpoS-mediated stress response. (A) Schematic representation of the mutations directly involved in the general stress response and their impact on the stress response. (B) The interaction between Asp118 (rose) of the sigma factor S and Arg275 (blue) of the RpoC subunit of the core RNAP (PDB 6OMF). The D118G mutation abolishes the salt bridge. (C) Optical densities of the ΔrpoS mutant compared to the ancestral strain and the adapted strain TUB170 in the presence of varying indole concentrations after 24 h. NMM(19/30/1) starting conditions, NMM(19/30/0) end of phase 3, NMM(19/0/1) without [3,2]Tp, NMM(0/30/0) end conditions, NMM(0/0/0) without amino acids or precursors. Each point is an individual colony (n = 3 biological replicates) and indicates the mean of two experiments. The letters A, B, C indicate Tukey test results (statistical significance, p < 0.05), when different strains have different letters, they are statistically different (see also Supplementary Table S5 and Supplementary Figure S5).
Other general stress response proteins were also identified from ALE strains. Notably, two mutations that emerged before transfer 40 and were present in all sequenced isolates were located in the promoter regions of the transcription units yobF-cspC and lrhA. The stress protein CspC plays a role in stabilizing rpoS mRNA (Cohen-Or et al., 2010), whereas the DNA-binding transcriptional dual regulator LrhA is involved in repressing rpoS translation (Peterson et al., 2004). In TUB85, TUB145, and TUB170, a threonine-to-proline mutation was identified in the gpp gene. This mutation is predicted to have a deleterious effect according to the Protein Variation Effect Analyzer (PROVEAN) (Choi, 2012; Choi et al., 2012). The enzyme GppA modulates the balance between pppGpp and ppGpp nucleotides by hydrolyzing the 5′-phosphate of pppGpp, leading to the generation of ppGpp (Mechold et al., 2013). As a part of the stringent response, the alarmone ppGpp accumulates in response to amino acid starvation or other forms of stress. Additionally, ppGpp is also associated with the general stress response mediated by σS (Grace et al., 2017). (pp)Gpp plays a crucial role in enhancing the synthesis and stability of RpoS, which, in turn, leads to increased expression of genes involved in stress protection (Gentry et al., 1993; Spira and Ospino, 2020). However, it is worth noting that an excess accumulation of (p)ppGpp or RpoS can lead to reduced fitness under nutrient-limited conditions (King et al., 2004). Moreover, (p)ppGpp has been shown to increase the availability of core RNAP (Barker et al., 2001), while indirectly decreasing cellular RpoD, thereby promoting binding of RpoS to RNAP (Durfee et al., 2008; Traxler et al., 2008).
3.4 Mutations deactivating proteases accompany the adaptation
Three mutations that arose in protease genes (lon and ftsH, identified in TUB85, TUB145, and TUB170; clpP, found in TUB170) were predicted to have deleterious effects according to PROVEAN. Based on the physicochemical differences between [3,2]Tpa and Trp, we propose that the insertion of [3,2]Tpa into proteins could lead to local or global protein misfolding. This hypothesis is supported by previous observations showing a reduction in protein stability upon [3,2]Tpa incorporation (Budisa et al., 2001). In typical circumstances, misfolded entities are cleared by proteases. However, in our evolved strain, each Trp position is replaced by [3,2]Tpa, potentially leading to an increase in partially misfolded or sub-optimally folded proteins. During the adaptation experiment, reducing proteolytic activity could have been beneficial for the host bacteria, as it likely increased the half-life of certain proteins affected by these changes. The occurrence of mutations in proteases, particularly during the early stages of the ALE, indicates that protein folding might be a potential concern when substituting Trp with [3,2]Tpa. However, it seems that the misfolding and impairment of protein structure may not have a significant impact, as a slight reduction in proteolytic activity appears to counterbalance this effect and promote adaptation to the use of non-canonical amino acids.
3.5 Proteomic analysis reveals major upregulation of stress proteins
Proteomic analysis was performed using stable isotope labeling by amino acids in cell culture (SILAC). In this method, proteins are labeled with isotope-labeled amino acids, specifically 13C615N4-arginine and 13C615N2-lysine. To achieve this, the biosynthesis of arginine and lysine in the ancestral strain TUB00 was disabled through phage P1 transduction, using strains from the Keio collection as donors (Datsenko and Wanner, 2000). The resulting strain was designated TUB00dKO and served as the reference in all SILAC experiments. Digested proteins from cells grown with and without the isotope-labeled amino acids were mixed and quantified using high-resolution mass spectrometry.
To assess the impact of [3,2]Tp addition on the proteome of the ancestral strain, TUB00, two conditions were compared. The first condition involved cultivating in indole-rich medium without [3,2]Tp (NMM(19/0/30)). The second condition utilized the reference strain (TUB00dKO) cultured in the presence of [3,2]Tp (NMM(17/30/1), supplemented with the two isotope-labeled amino acids (+13C615N4-arginine and 13C615N2-lysine), which resulted in NMM(19/30/1) with the labeled amino acids. In the absence of [3,2]Tp, a total of eighty-seven proteins displayed significant up- or downregulation (log2-value > ± 2.5) (Supplementary Figure S2). Among the 36 proteins that were downregulated in the ancestor upon [3,2]Tp supplementation, eight of them are associated with chemotaxis (e.g., CheW, CheA) and motility (e.g., FliC, FliN, FliA, FliL). These changes in protein expression are a common response observed during the transition to stationary phase and also a part of the general stress response (Barembruch and Hengge, 2007; Pesavento et al., 2007; Dong et al., 2011). In the ancestral strain with [3,2]Tp supplementation, the top 51 upregulated proteins were primarily associated with the general stress response, known to be regulated by RpoS, or controlled by the transcriptional regulator H-NS (Supplementary Figure S2, cluster plot). H-NS plays a crucial role in regulating RpoS expression during stress conditions by facilitating RpoS-dependent expression (Arnqvist et al., 1994; Barth et al., 1995; Colland et al., 2000; Grainger et al., 2008) through the housekeeping sigma factor RpoD. (Arnqvist et al., 1994; Barth et al., 1995; Colland et al., 2000; Waterman and Small, 2003; Shin et al., 2005; Weber et al., 2006; Grainger et al., 2008). Additionally, H-NS also destabilizes the RpoS protein (Barth et al., 1995; Yamashino et al., 1995; Battesti et al., 2012).
Proteins participating in the RpoS-mediated general stress response, such as GadC, GadB, and GadA from the glutamate-dependent acid resistance system, exhibited downregulation (Table 1). GadAB serves as a glutamate decarboxylase that removes one proton during the conversion of glutamate to γ-aminobutyrate (GABA) (Foster, 2004; Tramonti et al., 2008). Subsequently, the produced GABA is secreted by the glutamate/GABA antiporter GadC (Capitani et al., 2003; Foster, 2004). Another downregulated protein associated with the general stress response is the multidrug efflux transporter MdtEF (Table 1), which plays a role in detoxification and has been reported to be induced by indole (Hirakawa et al., 2005). Taken together, these findings demonstrate that the ancestral strain employs multiple facets of the stress response pathway to cope with the presence of [3,2]Tp supplementation in the medium.
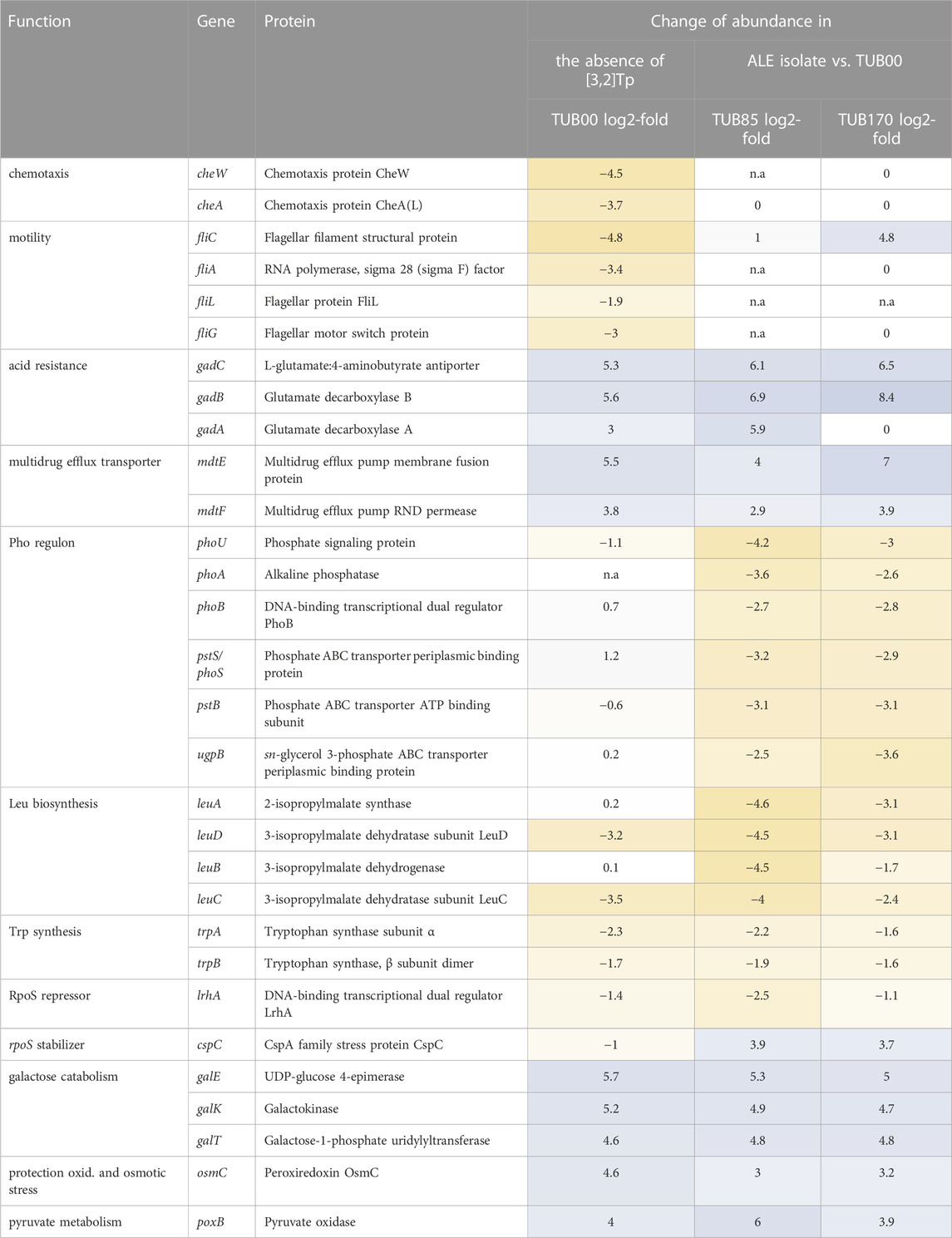
TABLE 1. Overview of up- and downregulated proteins in response to [3,2]Tp-supplementation. Isolates from the beginning of the ALE (TUB00, cultivated in NMM(19/0/30)) are compared to isolates from the middle (TUB85, cultivated in NMM(13/30/0)) and the end of the experiment (TUB170, cultivated in NMM(0/30/0)). The reference (TUB00dKO) was cultivated in a medium supplied with [3,2]Tp mirroring the starting conditions (NMM(19/30/1)). The change in abundance is reported as log2-fold (>2.5) and color coded in yellow for upregulated, blue for downregulated, white for no change in abundance. n.a. not available.
Subsequently, TUB85 and TUB170 were cultivated in different media to examine the evolution of their expression profiles during the adaptation process. They were cultured in media resembling the initial conditions (NMM(19/30/1)) as well as in their respective adapted media (NMM(13/30/0) and NMM(0/30/0)). The aim was to distinguish between the impact of cAA depletion and the adaptive response to [3,2]Tp. Interestingly, TUB85 and TUB170 exhibited similar expression profiles under all three culturing conditions, with the upregulation of amino acid biosynthesis enzymes in media lacking cAA as the only major difference (as expected, Supplementary Figure S3, S4). Therefore, it appears that in response to the presence of [3,2]Tp, the ancestral strain activates the RpoS-mediated general stress response. However, both adapted strains seem to readjust this response and revert to an unstressed state similar to the ancestral strain observed in the medium without [3,2]Tp (Table 1). When comparing the adapted strains to the ancestral strain cultivated in [3,2]Tp-supplemented media, 105 proteins are significantly up- or downregulated in TUB85 and TUB170 in the presence of [3,2]Tp.
The most prevalent functional feature observed among the downregulated proteins was related to the σS-mediated general stress response (Supplementary Figure S4), indicating a potential compensatory mechanism to counteract the negative impact induced by [3,2]Tp in TUB00. An example of this downregulation is observed in the rpoS mRNA-stabilizing protein CspC (Figure 4A), where a mutation in the promoter region was detected.
In both TUB85 and TUB170, proteins from the leucine biosynthesis pathway were found to be upregulated. This pathway is activated by the transcriptional regulator LeuO, (Chen et al., 2001) which indirectly inhibits rpoS translation by repressing the transcription of the sRNA DsrA (Klauck et al., 1997). Consequently, DsrA stabilizes rpoS mRNA (McCullen et al., 2010), leading to the activation of its translation (Sledjeski et al., 1996; Lease et al., 1998; Majdalani et al., 1998). Notably, in the adapted strains (TUB85 and TUB170), the tryptophan synthase TrpBA was prominently upregulated. TrpBA is responsible for the synthesis of tryptophan from indole and serine (Lane and Kirschner, 1991) (or in this case [3,2]Tpa synthesis from the precursor [3,2]Tp and serine) (Bae et al., 2001; Budisa and Pal, 2004). Additionaly, LrhA, for which a mutation in the promoter region was detected, was also upregulated. LrhA is a repressor of rpoS translation (Peterson et al., 2004).
A significant portion of the upregulated proteins (Supplementary Figure S3) are associated with the regulation of the Pho-regulon (PhoU, PhoA, PstS, PhoB, PhoS, PstB, UgpB). Typically, these proteins are responsible for Pi uptake in phosphate-limiting environments (Santos-Beneit, 2015). However, it remains unclear why these proteins were upregulated in our study, considering that the cultivation media used contained phosphate buffer in non-limiting concentrations. We hypothesize that the upregulation of the Pho regulon proteins might be a response to the mutation in sgrR, which leads to the activation of the small RNA gene sgrS under glucose-phosphate stress conditions. Previous studies have demonstrated that induction of the Pho regulon can alleviate the growth defect observed in an Escherichia coli sgrS mutant (Richards and Vanderpool, 2012). Others have established a connection between the Pho regulon and the synthesis of polyphosphate and guanosine tetraphosphate (ppGpp), a key phosphate-containing molecule involved in the stringent response (Barker et al., 2001). The stringent response plays a vital role in inhibiting RNA synthesis under conditions of amino acid scarcity (Traxler et al., 2008). E. coli mutants that are unable to accumulate ppGpp exhibit repression of the Pho regulon, although the precise mechanism linking these two processes remains unclear (Loewen et al., 1998).
4 Discussion
Our aim was to investigate the possibility of evolving an existing species to have an altered set of canonical amino acids. This process would represent a significant step towards creating artificial diversity and synthetic life. We performed ALE using a modified version of the tryptophan-auxotrophic E. coli MG1655. The goal was to generate strains that underwent chemical evolution leading to proteome-wide replacement of a canonical amino acid. We selected tryptophan (Trp) as our target for substitution and replaced it with a non-canonical analogue known as L-β-(thieno[3,2-b]pyrrolyl)alanine ([3,2]Tpa). This analogue possesses a physicochemical structure similar to Trp, making it a suitable candidate for our experiments, particularly due to Trp’s rare role as an essential catalytic moiety in E. coli enzymes. We initiated the experiments in a medium containing NMM supplemented with indole and amino acids. Through a series of 170 transfers, our goal was achieved, resulting in the evolution of a strain capable of growing in NMM with 25 µM [3,2]Tp without any additional supplements.
In our study, we replaced more than 20,000 Trp indole side chains in the proteome with thienopyrroles, prompting the question of whether these alterations should be deemed “radical,” “moderate,” or “negligible” changes in the chemistry of life (Budisa et al., 2020). Our experiments fall under the top-down approach of Synthetic Biology, in which the chemical composition of existing life forms is gradually changed. We demonstrated that the genetic code of Escherichia coli was altered compared to ancestor cells, resulting in changes in the chemistry of life. The classification of this change may vary depending on individual scientific perspectives (Kubyshkin et al., 2018). However, it is undeniable that our experiments resulted in significant, or “deep,” changes in the chemical composition of the cells. Thienopyrrole and indole are distinct chemical entities, even though they share certain chemical and steric similarities (Budisa et al., 2004).
In the initial phase of the experiment, prior to transfer 40 (generation 250), the indole concentration was gradually reduced from 1 µM to zero. This gradual reduction enabled E. coli to adapt to both the absence of Trp and the presence of the non-canonical amino acid precursor [3,2]Tp simultaneously. We identified three mutations in genes directly associated with amino acid metabolism (astB, aroG, and leuS). The first mutation introduced a stop codon, resulting in a truncation in N-succinylarginine dihydrolase (astB). This enzyme is part of the arginine succinyltransferase (astCADBE) operon, which is responsible for producing ammonia during arginine degradation under nitrogen-limited growth conditions. Moreover, this operon also contributes to the breakdown of various other amino acids. Given that the growth medium used in our study contains a non-limiting ammonia source (ammonium sulfate), it is essential to decrease the ammonia producing capacity of this operon while ensuring its retention of functionality for other essential processes. By disrupting astB, it has been demonstrated that succinylarginine dihydrolase activity is eliminated, preventing arginine utilization, while ornithine catabolism remains unaffected (Schneider et al., 1998). This reduced reliance on arginine as a nitrogen source allows arginine to fulfill other important roles, such as contributing to the biosynthesis of essential polyamines, putrescine, and spermidine. These polyamines play a critical role in various physiological processes (Charlier and Glansdorff, 2004), thereby supporting optimal growth.
During the second phase of the experiment, we systematically reduced the number of canonical amino acids from 19 to 0. Over the course of transfers 40 to 85 (generation 500), the evolving population acquired many mutations in genes associated with chemotaxis and flagella synthesis. E. coli typically exhibits a reduction in the synthesis of chemotaxis and flagella when entering stationary phase or facing various stresses (Barembruch and Hengge, 2007; Pesavento et al., 2007; Dong et al., 2011). Simultaneously, it enhances the production of stress-related proteins and drug exporters. During this state, E. coli undergoes a metabolic shift, prioritizing the allocation of limited resources towards survival and maintenance purposes rather than the production of growth-related (macro)molecules (Hengge, 2014). During this second phase, the population of E. coli acquired a mutation in rpoS, leading to the inactivation of RpoS. This mutation enhanced the adaptability of the population. Particularly, the mutation of the Asp118 residue of RpoS diminishes RpoS binding to the core-RNAP, thereby reducing the overall stress response.
The careful examination of rpoS mutations observed in laboratory settings is crucial due to the potential influence of storage and shipping methods, such as stab cultures or glycerolized cultures on filter discs, which can inadvertently select for rpoS mutants (Sutton et al., 2000; Spira et al., 2011). Despite the possibility of storage-related artifacts with rpoS mutations in other contexts, it is unlikely that the rpoS mutation and the subsequent downregulation of RpoS protein observed in this study were influenced by storage. For this experiment, samples collected during the ALE experiment were promptly frozen at −80°C, and all isolates used for subsequent characterization and analysis were directly derived from these frozen fossils, ensuring the integrity of the findings.
Indeed, the deletion of rpoS in the ancestral strain alone is sufficient to support growth on [3,2]Tp. Moreover, previous ALE experiments conducted under nutrient-limited conditions, such as in glucose-limited chemostats (Notley-Mcrobb et al., 2002) have also identified rpoS mutations. Another experiment was performed using a modified version of MG1655 with a reduced genome, and this particular strain exhibited growth defects in minimal medium. Deletion of rpoS resulted in the restoration of the unevolved strain’s growth rate to 80% of the growth rate observed in the evolved strain (Choe et al., 2019). In stationary phase cells or cells experiencing stress, the abundance of RpoS protein leads to competition between RpoS and the vegetative sigma factor RpoD for a limited number of RNA polymerase core subunits (Stoebel et al., 2009). Mutations in rpoS thus likely alleviate this competition, favoring nutrient scavenging through increased expression of RpoD-dependent genes.
In a natural habitat characterized by fluctuations in environmental conditions, such as temperature, osmolarity, or pH, loss-of-function of RpoS is likely to be disadvantageous. Furthermore, under specific stress conditions, the loss of RpoS function can also have detrimental effects (Ferenci, 2008). As an example, rpoS mutants that evolved under osmotic stress have been found to insert IS10 in the promoter region of the otsBA operon. This insertion partially restores the wildtype response to osmotic stress, transforming the expression from being RpoS-dependent to RpoS-independent (Stoebel et al., 2009). Additionally, in certain cases, attenuated rpoS mutations have been observed in long-term stationary phase cultures (Zambrano et al., 1993), and glucose-limited chemostats under acidic pH (Notley-Mcrobb et al., 2002) implying that less drastic mutations may offer benefits. The rarity of rpoS deletions in natural E. coli populations (Snyder et al., 2012), coupled with the findings from experiments, indicates that the trade-off between reduced stress resistance and increased nutrient depletion is delicately balanced. In isolates TUB85 and TUB170, we observed an increase in the production of proteins involved in the leucine biosynthesis pathway, specifically from the leuLABCD operon. This operon’s transcription is regulated not only by LeuO but also by guanosine tetraphosphate (ppGpp). When leucine levels are scarce (i.e., leucine starvation), ppGpp facilitates the binding of RNA polymerase to the leuLABCD promoter (leuLp), leading to the activation of transcription. Intriguingly, the leucine supplement was ceased for the TUB85 and TUB170 strains. During our ALE experiment, ppGpp production was induced by guanosine-5′-triphosphate,3′-diphosphate phosphatase (GppA). Notably, we observed a T459P mutation in the GppA gene during the course of the experiment. Consequently, the observed upregulation of proteins from the leucine biosynthesis pathway might result from the hyperactivation of leuLABCD operon transcription, facilitated by both LeuO and ppGpp. Furthermore, besides its role in activating leucine biosynthesis, LeuO indirectly hinders rpoS translation, leading to the deactivation of the general stress response.
During the process of adaptation to a gradual depletion of a crucial canonical substrate, E. coli mitigated its stress response to prioritize cell growth. Notably, the proteomic data revealed that the levels of stress-related proteins gradually decreased over time, eventually reaching levels similar to those observed in the ancestral strain when grown in relaxing media (i.e., in non-stressful conditions). Genomic data provided further evidence for this discovery, as mutations related to four stress proteins were identified, including both upregulated and downregulated proteins, along with alterations in the master regulator RpoS. The adapted strain appears to counteract or mitigate the negative effects of [3,2]Tpa incorporation by reducing its protein quality management, specifically by decreasing proteolytic activity. This is supported by the presence of mutations in three proteases.
Our study provides compelling evidence that adapting to proteome-wide ncAA-insertions leads to extensive effects throughout the cell. These effects are likely to be the outcome of a complex interplay of various interactions in which pleiotropy is also significantly involved. Indeed, these interactions result from changes in metabolic pathways and intracellular networking and are crucial for the adaptive mechanisms triggered in response to the large-scale incorporation of ncAAs into the proteome. They enable the modulation of folding pathways, stress responses, and various other functions, contributing significantly to the overall adaptive process. Considering the multifaceted roles of canonical amino acids beyond translational activity, it is reasonable to expect that the incorporation of ncAAs will have far-reaching effects beyond mere proteome alterations. Indeed, the incorporation ncAAs can exert influences on a wide range of cellular aspects, encompassing cell physiology, signaling pathways, and various metabolic and structural processes. These diverse effects underscore the significance and complexity of ncAA utilization in biological systems.
It is essential to recognize that the evolution of bacteria and other organisms over billions of years cannot be directly equated to proteome-wide ncAA insertion experiments conducted over the last 50 years (Cowie and Cohen, 1957). This distinction arises from the differences in the experimental methods used. In the laboratory setting, selection pressures can be significantly intensified and controlled, and the availability of diverse ncAAs permits an unparalleled exploration and diversification of life’s chemistry. As a result, the experimental approach allows for unique insights into the potential impacts of ncAA incorporation on cellular processes and evolution, while acknowledging that it may not fully mirror the complexities and dynamics of natural evolutionary processes (Kubyshkin and Budisa, 2017). In essence, our efforts are advancing us towards the creation of artificial life, where human influence amplifies the process of evolution. This approach frees us from the constraints of the slow and random course of natural evolution and enables more deliberate and rapid changes to the basic building blocks of life (Wiltschi and Budisa, 2007). By incorporating non-canonical amino acids and other innovative techniques, we are breaking new ground and exploring possibilities that were previously beyond the reach of traditional evolutionary processes.
In Synthetic Biology, our experimental approach is a top-down approach to create life by evolving synthetic bacterial cells with different genetic codes (Diwo and Budisa, 2019). The universal genetic code governs the use of 20 amino acids for protein synthesis in all organisms. Yet, the reason for the selection of these 20 amino acids remains unknown (Weber and Miller, 1981). There are several hypotheses, such as the “frozen accident” (Crick, 1968) the RNA world hypothesis, (Joyce and Orgel, 1993), the Hartman-Smith model, (Hartman and Smith, 2014) coevolution theory, (Wong, 1988) and the Alanine-World Model, (Kubyshkin and Budisa, 2019a; Kubyshkin and Budisa, 2019b) which try to explain or justify the selection of these amino acids based on peptide and protein chemistry, metabolic constraints, and physicochemical logic, to name just a few.
While various models and theories offer plausible explanations for the selection of amino acids in the genetic code, experimental verification is still needed. Our research findings contribute to this effort by shedding light on how environmental factors can induce genetic changes that drive evolution and regulate cellular metabolism. This knowledge could prove useful in designing synthetic cells.
Evolutionary alienation of the basic chemical makeup of living cells may be the best approach to experimentally create parallel biological worlds (Budisa, 2014), that exhibit variations in their morphology, chemistry, metabolism, energy, and information transfer from contemporary life forms (Schmidt et al., 2018). Furthermore, these cells ought to possess genetic codes that are entirely distinct from those found in life as we know it.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here https://www.ncbi.nlm.nih.gov/bioproject/991156.
Author contributions
IT performed crucial steps in the final phase of the project, writing the final version of the manuscript and providing samples for additional and control experiments. MH, NB and SO designed the experimental setup, and SO performed the serial dilution experiments and associated analytical/microbiological procedures. ZI, MB, SA, and AG were involved in data analysis and discussion. AG contributed to writing/editing the final version of the manuscript. AG, TS, and A-RA performed genetic analyses and constructed pedigrees. LP and JR performed proteomic analyses, and together with SO interpreted the data. CT-K. created the ∆rpoS strain and performed the corresponding growth assay. NB conceived the project, wrote a first draft, directed the experimental work, and supervised the entire preparation and revision of the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
IT and SO were supported by GRK 1582 “Fluorine as a Key Element”, funded by Deutsche Forschungsgemeinschaft (DFG). AG and A-RA were funded by an NSERC Discovery Grant. LP was supported by a Marie Curie Intra European Fellowship within the seventh European Community Framework Program and by an Estonian Research Council grant (PUT626). The Wellcome Centre for Cell Biology supported JR by core funding from the Wellcome Trust (03149). This work was supported by European Union’s Horizon 2020 research and innovation program as part of the SynCrop ETN under the Marie Skłodowska-Curie grant 764591 (to ZI and NB).
Acknowledgments
NB gratefully acknowledges support from the Natural Sciences and Engineering Research Council (NSERC) of Canada (Discovery Grant RGPIN-05669-2020) and the Canada Research Chairs Program (Grant No. 950-231971).
Conflict of interest
SO is employed by Roche Diagnostics GmbH. MH is employed by AMSilk GmbH. LP is employed by Icosagen Cell Factory OÜ. MB is employed by European Science Communication Institute (ESCI) GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors TS, AG, and NB declared that they were editorial board members of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fsybi.2023.1248065/full#supplementary-material
References
Agostini, F., Sinn, L., Petras, D., Schipp, C. J., Kubyshkin, V., Berger, A. A., et al. (2021). Multiomics analysis provides insight into the laboratory evolution of Escherichia coli toward the metabolic usage of fluorinated indoles. ACS Central Sci. 7, 81–92. doi:10.1021/acscentsci.0c00679
Arnqvist, A., Olsen, A., and Normark, S. (1994). Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13, 1021–1032. doi:10.1111/j.1365-2958.1994.tb00493.x
Bacher, J. M., and Ellington, A. D. (2001). Selection and characterization of escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 183. doi:10.1128/JB.183.18.5414-5425.2001
Bae, J. H., Alefelder, S., Kaiser, J. T., Friedrich, R., Moroder, L., Huber, R., et al. (2001). Incorporation of β-selenolo [3, 2-b] pyrrolyl-alanine into proteins for phase determination in protein X-ray crystallography. J. Mol. Biol. 309 (4), 925–936. doi:10.1006/jmbi.2001.4699
Barembruch, C., and Hengge, R. (2007). Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 65, 76–89. doi:10.1111/j.1365-2958.2007.05770.x
Barker, M. M., Gaal, T., Josaitis, C. A., and Gourse, R. L. (2001). Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688. doi:10.1006/jmbi.2000.4327
Barth, M., Marschall, C., Muffler, A., Fischer, D., and Hengge-Aronis, R. (1995). Role for the histone-like protein H-NS in growth phase-dependent and osmotic regulation of σ(S) and many σ(S)-dependent genes in Escherichia coli. J. Bacteriol. 177, 3455–3464. doi:10.1128/jb.177.12.3455-3464.1995
Battesti, A., Majdalani, N., and Gottesman, S. (2010). The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65, 189–213. doi:10.1146/annurev-micro-090110-102946
Battesti, A., Tsegaye, Y. M., Packer, D. G., Majdalani, N., and Gottesman, S. (2012). H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J. Bacteriol. 194, 2470–2478. doi:10.1128/JB.00132-12
Blattner, F. R., Plunkett, G. r., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., et al. (1997). The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462. doi:10.1126/science.277.5331.1453
Budisa, N., Alefelder, S., Bae, J. H., Golbik, R., Minks, C., Huber, R., et al. (2001). Proteins with β-(thienopyrrolyl)alanines as alternative chromophores and pharmaceutically active amino acids. Protein Sci. 10, 1281–1292. doi:10.1110/ps.51601
Budisa, N., Kubyshkin, V., and Schmidt, M. (2020). Xenobiology A journey towards parallel life forms. ChemBioChem 21 (16), 2228–2231. doi:10.1002/cbic.202000141
Budisa, N., Pal, P. P., Alefelder, S., Birle, P., Krywcun, T., Rubini, M., et al. (2004). Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code. Biol. Chem. 385. doi:10.1515/bc.2004.038
Budisa, N., and Pal, P. (2004). Designing novel spectral classes of proteins with a tryptophan-expanded genetic code. Biol. Chem. 385, 893–904. doi:10.1515/BC.2004.117
Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J., and Huber, R. (1995). High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem., 230 (2), 788–796. doi:10.1111/j.1432-1033.1995.0788h.x
Budisa, N. (2014). Xenobiology, new-to-nature synthetic cells and genetic firewall. Curr. Org. Chem. 18 (8), 936–943. doi:10.2174/138527281808140616154301
Capitani, G., Gut, H., Grütter, M. G., De Biase, D., Aurizi, C., and Bossa, F. (2003). Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J. 22, 4027–4037. doi:10.1093/emboj/cdg403
Charlier, D., and Glansdorff, N. (2004). Biosynthesis of arginine and polyamines. EcoSal Plus 1. doi:10.1128/ecosalplus.3.6.1.10
Chen, C. C., Fang, M., Majumder, A., and Wu, H. Y. (2001). A 72-base pair AT-rich DNA sequence element functions as a bacterial gene silencer. J. Biol. Chem. 276, 9478–9485. doi:10.1074/jbc.M010501200
Choe, D., Lee, J. H., Yoo, M., Hwang, S., Sung, B. H., Cho, S., et al. (2019). Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 10, 935. doi:10.1038/s41467-019-08888-6
Choi, Y. “A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein,” in Proceedings of the BCB' 12 ACM International Conference on Bioinformatics, Computational Biology and Biomedicine, Orlando Florida, October 2012. doi:10.1145/2382936.2382989
Choi, Y., Sims, G. E., Murphy, S., Miller, J. R., and Chan, A. P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7, e46688. doi:10.1371/journal.pone.0046688
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. fly 6 (2), 80–92. doi:10.4161/fly.19695
Cohen-Or, I., Shenhar, Y., Biran, D., and Ron, E. Z. (2010). CspC regulates rpoS transcript levels and complements hfq deletions. Res. Microbiol. 161 (8), 694–700. doi:10.1016/j.resmic.2010.06.009
Colland, F., Barth, M., Hengge-Aronis, R., and Kolb, A. (2000). Sigma factor selectivity of Escherichia coli RNA polymerase role for CRP, IHF and Lrp transcription factors. EMBO J. 19, 3028–3037. doi:10.1093/emboj/19.12.3028
Cowie, D. B., and Cohen, G. N. (1957). Biosynthesis by Escherichia coli of active altered proteins containing selenium instead of sulfur. BBA - Biochimica Biophysica Acta 26, 252–261. doi:10.1016/0006-3002(57)90003-3
Crick, F. H. C. (1968). The origin of the genetic code. J. Mol. Biol. 38, 367–379. doi:10.1016/0022-2836(68)90392-6
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 6, 6640–6645. doi:10.1073/pnas.120163297
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Eng. Anal. Multicell. Syst. Methods Protoc. 1151, 165–188. doi:10.1007/978-1-4939-0554-6_12
Diwo, C., and Budisa, N. (2019). Alternative biochemistries for alien life: basic concepts and requirements for the design of a robust biocontainment system in genetic isolation. Genes 10, 17. doi:10.3390/genes10010017
Dong, T., Yu, R., and Schellhorn, H. (2011). Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79, 375–386. doi:10.1111/j.1365-2958.2010.07449.x
Durfee, T., Hansen, A.-M., Zhi, H., Blattner, F. R., and Jin, D. J. (2008). Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190, 1084–1096. doi:10.1128/JB.01092-07
Ferenci, T. (2008). The spread of a beneficial mutation in experimental bacterial populations The influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100, 446–452. doi:10.1038/sj.hdy.6801077
Foster, J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2, 898–907. doi:10.1038/nrmicro1021
Fournier, G. P., and Alm, E. J. (2015). Ancestral reconstruction of a pre-LUCA aminoacyl-tRNA synthetase ancestor supports the late addition of Trp to the genetic code. J. Mol. Evol. 80, 171–185. doi:10.1007/s00239-015-9672-1
Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B., and Cashel, M. (1993). Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 175, 7982–7989. doi:10.1128/jb.175.24.7982-7989.1993
Gottesman, S. (2019). Trouble is coming signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 294, 11685–11700. doi:10.1074/jbc.rev119.005593
Gouy, M., Guindon, S., and Gascuel, O. (2010). SeaView version 4 A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27 (2), 221–224. doi:10.1093/molbev/msp259
Grace, E. D., Halliday, J. A., Gourse, R. L., Girard, M. E., Herman, C., and Gopalkrishnan, S. (2017). DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J. Bacteriol. 200, e00463-17. doi:10.1128/jb.00463-17
Grainger, D. C., Goldberg, M. D., Lee, D. J., and Busby, S. J. W. (2008). Selective repression by Fis and H-NS at the Escherichia coli dps promoter. Mol. Microbiol. 68, 1366–1377. doi:10.1111/j.1365-2958.2008.06253.x
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi:10.1146/annurev.micro.57.030502.090913
Hartman, H., and Smith, T. F. (2014). The evolution of the ribosome and the genetic code. Life 4, 227–249. doi:10.3390/life4020227
Hecker, M., Pané-Farré, J., and Uwe, V. (2007). SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61, 215–236. doi:10.1146/annurev.micro.61.080706.093445
Hengge, R. (2014). Stationary-phase gene regulation in Escherichia coli §. EcoSal Plus 4. doi:10.1128/ecosalplus.5.6.3
Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T., and Yamaguchi, A. (2005). Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55, 1113–1126. doi:10.1111/j.1365-2958.2004.04449.x
Hoesl, M. G., Oehm, S., Durkin, P., Darmon, E., Peil, L., Aerni, H. R., et al. (2015). Chemical evolution of a bacterial proteome. Angew. Chem. Int. Ed. Engl. 54, 10030–10034. doi:10.1002/anie.201502868
Jenkins, D. E., Chaisson, S. A., and Matin, A. (1990). Starvation-induced cross protection against osmotic challenge in Escherichia coli. J. Bacteriol. 172, 2779–2781. doi:10.1128/jb.172.5.2779-2781.1990
Joyce, G. F., and Orgel, L. E. (1993). Prospects for understanding the origin of the RNA world. Cold Spring Harb. Monogr. Ser. 24, 1.
King, T., Ishihama, A., Kori, A., and Ferenci, T. (2004). A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186, 5614–5620. doi:10.1128/JB.186.17.5614-5620.2004
Klauck, E., Böhringer, J., and Hengge-Aronis, R. (1997). The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25, 559–569. doi:10.1046/j.1365-2958.1997.4911852.x
Koonin, E. V., and Novozhilov, A. S. (2017). Origin and evolution of the genetic code: the universal enigma. Annu. Rev. Genet. 61, 99–111. doi:10.1002/iub.146
Kozlowski, L. P. (2017). Proteome-pI Proteome isoelectric point database. Nucleic acids Res. 45, D1112–D1116. doi:10.1093/nar/gkw978
Kubyshkin, V., Acevedo-Rocha, C. G., and Budisa, N. (2018). On universal coding events in protein biogenesis. Biosystems 164, 16–25. doi:10.1016/j.biosystems.2017.10.004
Kubyshkin, V., and Budisa, N. (2019b). Anticipating alien cells with alternative genetic codes: away from the alanine world. Curr. Opin. Biotechnol. 60, 242–249. doi:10.1016/j.copbio.2019.05.006
Kubyshkin, V., and Budisa, N. (2017). Synthetic alienation of microbial organisms by using genetic code engineering: why and how? Biotechnol. J. 12 (8), 1600097. doi:10.1002/biot.201600097
Kubyshkin, V., and Budisa, N. (2019a). The alanine world model for the development of the amino acid repertoire in protein biosynthesis. Int. J. Mol. Sci. 20, 5507. doi:10.3390/ijms20215507
Kun, Á., and Radványi, Á. (2018). The evolution of the genetic code: impasses and challenges. Biosystems 164, 217–225. doi:10.1016/j.biosystems.2017.10.006
Lajoie, M. J., Rovner, A. J., Goodman, D. B., Aerni, H.-R., Haimovich, A. D., Kuznetsov, G., et al. (2013). Genomically recoded organisms expand biological functions. Science 342, 357–360. doi:10.1126/science.1241459
Lane, A. N., and Kirschner, K. (1991). Mechanism of the physiological reaction catalyzed by tryptophan synthase from Escherichia coli. Biochemistry 30, 479–484. doi:10.1021/bi00216a025
Lange, R., and Hengge-Aronis, R. (1991). Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 5, 49–59. doi:10.1111/j.1365-2958.1991.tb01825.x
Lease, R. A., Cusick, M. E., and Belfort, M. (1998). Riboregulation in Escherichia coli DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. 95, 12456–12461. doi:10.1073/pnas.95.21.12456
Letunic, I., and Bork, P. (2007). Interactive tree of life (iTOL) An online tool for phylogenetic tree display and annotation. Bioinformatics 23 (1), 127–128. doi:10.1093/bioinformatics/btl529
Loewen, P. C., Hu, B., Strutinsky, J., and Sparling, R. (1998). Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44, 707–717. doi:10.1139/cjm-44-8-707
Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. (1998). DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 95, 12462–12467. doi:10.1073/pnas.95.21.12462
McCann, M. P., Kidwell, J. P., and Matin, A. (1991). The Putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J. Bacteriol. 173, 4188–4194. doi:10.1128/jb.173.13.4188-4194.1991
McCullen, C. A., Benhammou, J. N., Majdalani, N., and Gottesman, S. (2010). Mechanism of positive regulation by DsrA and RprA small noncoding RNAs Pairing increases translation and protects rpoS mRNA from degradation. J. Bacteriol. 192, 5559–5571. doi:10.1128/JB.00464-10
Mechold, U., Potrykus, K., Murphy, H., Murakami, K. S., and Cashel, M. (2013). Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41, 6175–6189. doi:10.1093/nar/gkt302
Minks, C., Alefelder, S., Moroder, L., Huber, R., and Budisa, N. (2000). Towards new protein engineering In vivo building and folding of protein shuttles for drug delivery and targeting by the selective pressure incorporation (SPI) method. Tetrahedron 56 (48), 9431–9442. doi:10.1016/s0040-4020(00)00827-9
Notley-Mcrobb, L., King, T., and Ferenci, T. (2002). rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. Am. Soc. Microbiol. 184, 806–811. doi:10.1128/jb.184.3.806-811.2002
Pesavento, C., Becker, G., Sommerfeldt, N., Possling, A., Tschowri, N., Mehlis, A., et al. (2007). Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22, 2434–2446. doi:10.1101/gad.475808
Peterson, C. N., Ruiz, N., and Silhavy, T. J. (2004). RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J. Bacteriol. 186, 7403–7410. doi:10.1128/JB.186.21.7403-7410.2004
Richards, G. R., and Vanderpool, C. K. (2012). Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J. Bacteriol. 194, 2520–2530. doi:10.1128/JB.00009-12
Santos-Beneit, F. (2015). The Pho regulon: a huge regulatory network in bacteria. Front. Microbiol. 6, 402. doi:10.3389/fmicb.2015.00402
Schmidt, M., Pei, L., and Budisa, N. (2018). Xenobiology: state-of-the-art, ethics, and philosophy of new-to-nature organisms. Synth. biology–metabolic Eng. 162, 301–315. doi:10.1007/10_2016_14
Schneider, B. L., Kiupakis, A. K., and Reitzer, L. J. (1998). Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J. Bacteriol. 180, 4278–4286. doi:10.1128/JB.180.16.4278-4286.1998
Shin, M., Song, M., Rhee, J., Hong, Y., Kim, Y., Seok, Y., et al. (2005). DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev., doi:10.1101/gad.1316305.of
Sledjeski, D. D., Gupta, A., and Gottesman, S. (1996). The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15, 3993–4000. doi:10.1002/j.1460-2075.1996.tb00773.x
Snyder, E., Gordon, D. M., and Stoebel, D. M. (2012). Escherichia coli lacking RpoS are rare in natural populations of non-pathogens. G3 Genes, Genomes, Genet. 2, 1341–1344. doi:10.1534/g3.112.003855
Spira, B., De Almeida Toledo, R., Maharjan, R. P., and Ferenci, T. (2011). The uncertain consequences of transferring bacterial strains between laboratories - RpoS instability as an example. BMC Microbiol. 11, 248. doi:10.1186/1471-2180-11-248
Spira, B., and Ospino, K. (2020). Diversity in E. coli (p)ppGpp levels and its consequences. Front. Microbiol. 11, 1759. doi:10.3389/fmicb.2020.01759
Stoebel, D. M., Hokamp, K., Last, M. S., and Dorman, C. J. (2009). Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 5, e1000671. doi:10.1371/journal.pgen.1000671
Sutton, A., Buencamino, R., and Eisenstark, A. (2000). rpoS mutants in archival cultures of Salmonella enterica serovar typhimurium. J. Bacteriol. 182, 4375–4379. doi:10.1128/JB.182.16.4375-4379.2000
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids Res. 43 (D1), D447–D452. doi:10.1093/nar/gku1003
Tramonti, A., De Canio, M., and De Biase, D. (2008). GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70, 965–982. doi:10.1111/j.1365-2958.2008.06458.x
Traxler, M. F., Summers, S. M., Nguyen, H.-T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi:10.1111/j.1365-2958.2008.06229.x
Trifonov, E. N. (2009). The origin of the genetic code and of the earliest oligopeptides. Res. Microbiol. 160 (7), 481–486. doi:10.1016/j.resmic.2009.05.004
Waterman, S. R., and Small, P. L. C. (2003). Transcriptional expression of Escherichia coli glutamate-dependent acid resistance genes gadA and gadBC in an hns rpoS mutant. J. Bacteriol. 185, 4644–4647. doi:10.1128/JB.185.15.4644-4647.2003
Weber, A. L., and Miller, S. L. (1981). Reasons for the occurrence of the twenty coded protein amino acids. J. Mol. Evol. 17, 273–284. doi:10.1007/BF01795749
Weber, H., Pesavento, C., Possling, A., Tischendorf, G., and Hengge, R. (2006). Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62, 1014–1034. doi:10.1111/j.1365-2958.2006.05440.x
Weber, H., Polen, T., Heuveling, J., Wendisch, V. F., Hengge, R., Ju, F., et al. (2005). Genome-wide analysis of the general stress response network in Escherichia coli σ S -dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187, 1591–1603. doi:10.1128/jb.187.5.1591-1603.2005
Wiltschi, B., and Budisa, N. (2007). Natural history and experimental evolution of the genetic code. Appl. Microbiol. Biotechnol. 74, 739–753. doi:10.1007/s00253-006-0823-6
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. methods 6 (5), 359–362. doi:10.1038/nmeth.1322
Wong, J. T. (1983). Membership mutation of the genetic code: loss of fitness by tryptophan. Proc. Nati. Acad. Sci. U. S. A. 80, 6303–6306. doi:10.1073/pnas.80.20.6303
Yamashino, T., Ueguchi, C., and Mizuno, T. (1995). Quantitative control of the stationary phase-specific sigma factor, sigma S, in Escherichia coli involvement of the nucleoid protein H-ns. EMBO J. 14, 594–602. doi:10.1002/j.1460-2075.1995.tb07035.x
Yu, A. C. S., Yim, A. K. Y., Mat, W. K., Tong, A. H. Y., Lok, S., Xue, H., et al. (2014). Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol. Evol. 6, 629–641. doi:10.1093/gbe/evu044
Zambrano, M. M., Siegele, D. A., Almirón, M., Tormo, A., and Kolter, R. (1993). Microbial competition Escherichia coli mutants that take over stationary phase cultures. Science 259, 1757–1760. doi:10.1126/science.7681219
Zhao, D., Feng, X., Zhu, X., Wu, T., Zhang, X., and Bi, C. (2017). CRISPR/Cas9-assisted gRNA-free one-step genome editing with no sequence limitations and improved targeting efficiency. Sci. Rep. 7 (1), 16624. doi:10.1038/s41598-017-16998-8
Keywords: adaptive laboratory evolution, canonical/noncanonical amino acids, evolution, genetic code, genomics, proteomics, stress response, synthetic life
Citation: Tolle I, Oehm S, Hoesl MG, Treiber-Kleinke C, Peil L, Bozukova M, Albers S, Adamu Bukari A-R, Semmler T, Rappsilber J, Ignatova Z, Gerstein AC and Budisa N (2023) Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1:1248065. doi: 10.3389/fsybi.2023.1248065
Received: 26 June 2023; Accepted: 31 July 2023;
Published: 24 August 2023.
Edited by:
Patrick O’Donoghue, Western University, CanadaReviewed by:
Jeffery M. Tharp, Indiana University Bloomington, United StatesNoah Reynolds, University of Illinois at Springfield, United States
Copyright © 2023 Tolle, Oehm, Hoesl, Treiber-Kleinke, Peil, Bozukova, Albers, Adamu Bukari, Semmler, Rappsilber, Ignatova, Gerstein and Budisa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aleeza C. Gerstein, YWxlZXphLmdlcnN0ZWluQHVtYW5pdG9iYS5jYQ==; Nediljko Budisa, bmVkaWxqa28uYnVkaXNhQHR1LWJlcmxpbi5kZQ==, bmVkaWxqa28uYnVkaXNhQHVtYW5pdG9iYS5jYQ==
†These authors have contributed equally to this work
 Isabella Tolle
Isabella Tolle Stefan Oehm2†
Stefan Oehm2† Lauri Peil
Lauri Peil Abdul-Rahman Adamu Bukari
Abdul-Rahman Adamu Bukari Torsten Semmler
Torsten Semmler Zoya Ignatova
Zoya Ignatova Aleeza C. Gerstein
Aleeza C. Gerstein Nediljko Budisa
Nediljko Budisa