
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Surg. , 03 March 2025
Sec. Visceral Surgery
Volume 12 - 2025 | https://doi.org/10.3389/fsurg.2025.1523684
This article is part of the Research Topic Exploring Machine Learning Applications in Visceral Surgery View all 5 articles
 Mihaela-Flavia Avram1,2*
Mihaela-Flavia Avram1,2* Nicolae Lupa3
Nicolae Lupa3 Dimitrios Koukoulas4
Dimitrios Koukoulas4 Daniela-Cornelia Lazăr5
Daniela-Cornelia Lazăr5 Mihaela-Ioana Mariș6,7
Mihaela-Ioana Mariș6,7 Marius-Sorin Murariu1,2
Marius-Sorin Murariu1,2 Sorin Olariu1,2
Sorin Olariu1,2
Background: Colorectal cancer is considered to be triggered by the malignant transformation of colorectal polyps. Early diagnosis and excision of colorectal polyps has been found to lower the mortality and morbidity associated with colorectal cancer.
Objective: The aim of this study is to offer a predictive model for the presence of colorectal polyps based on Random Forests machine learning algorithm, using basic patient information and common laboratory test results.
Materials and methods: 164 patients were included in the study. The following data was collected: sex, residence, age, diabetes mellitus, body mass index, fasting blood glucose levels, hemoglobin, platelets, total, LDL and HLD cholesterol, triglycerides, serum glutamic-oxaloacetic transaminase, chronic gastritis, presence of colonic polyps at colonoscopy. 80% of patients were included in the training set for creating a Random forests algorithm, 20% were in the test set. External validation was performed on data from 42 patients. The performance of the Random Forests was compared with the performance of a generalized linear model (GLM) and support vector machine (SVM) built and tested on the same datasets.
Results: The Random Forest prediction model gave an AUC of 0.820 on the test set. The top five variables in order of importance were: body mass index, platelets, hemoglobin, triglycerides, glutamic-oxaloacetic transaminase. For external validation, the AUC was 0.79. GLM performance in internal validation was an AUC of 0.788, while for external validation AUC-0.65. For SVN, the AUC - 0.785 for internal validation and 0.685 for the external validation dataset.
Conclusions: A random forest prediction model was developed using patient's demographic data, medical history and common blood tests results. This algorithm can foresee, with good predictive power, the presence of colonic polyps.
Colorectal cancer (CRC) is the second cause of cancer related deaths worldwide, having a 4%–5% lifetime long risk of appearing in the general population. It is estimated that, in the absence of screening strategies, 7.7%–8.5% of persons above 40 years old would develop CRC and 3.2%–3.4% would die of it (1).
CRC incidence and mortality have a decreasing trend in the majority of European Countries, USA and big part of Asia-Pacific. The incidence in USA has decreased by more than 35% since screening programs have been used in the 1990's. The detection of premalignant lesions is an important objective in CRC screening as the removal of polyps during colonoscopy is efficient in reducing the incidence of CRC (2, 3).
Studies have shown that when the progression from polyp to CRC takes places its duration is 10.6–25.8 years (4, 5). Detection and resection of these polyps reduces the incidence of CRC.
Research done on colonoscopies report an incidence of polyps of 20%–53% in adults aged >50years, with a 9.7% incidence of advanced adenomas (defined as adenomatous polyps sized >10 mm or with villous characteristics or having high grade dysplasia). Meta-analysis of these studies (for patients >50years old) determined a global prevalence rate of 24% for polyps and the prevalence of advanced adenomas – 4.5% (6–8).
The age of the screening initiation is crucial for the efficiency and rentability of screening programs. Simulation analysis in USA, which were the basis of screening recommendations for CRC made by US Preventive Service Task Force and American Cancer Society, state that 45 years old is a better age to start screening, as opposed to 50 years, providing a more efficient balance of life-years gained from screening and colonoscopy burden (5, 9). Simulation modelling analysis for CRC, taking into consideration the incidence in the younger population, have determined the American Cancer Society to recommend CRC screening to be started at 45 years for individuals with a moderate risk for CRC (10). Other countries have adjusted the starting age for CRC: Germany reduced the age from 55 to 50 years (for men only), in England, the UK National Screening Committee recommends to reduce the age from 60 to 50 years (11).
Colonoscopy is the gold standard for the diagnosis and treatment of colorectal polyps. This intervention requires the existence of adequate medical facility and dedicated personnel, so the possibility of performing colonoscopies is limited, no matter how rich the medical system is. The aim of this study is to offer a predictive model for the presence of colorectal polyps using basic patient information and laboratory test results. This model can be used for selecting patients which have a high risk of being diagnosed with colorectal polyps and to be offered a colonoscopy, even if they are not at the starting age for CRC screening, thus reducing the incidence of CRC in the general population.
Data from patients who underwent colonoscopies between January 2022 and February 2023 in one hospital, Municipal Hospital “Dr. Teodor Andrei” Lugoj, Romania, was extracted. For external validation of the algorithm data from patients who underwent colonoscopies between June 2022-June 2023 in an outpatient gastroenterology facility, “Dr.K.D.Medic” Clinic, Caransebes, Romania. The study was approved by the Local Ethics Board.
Figure 1 shows patients' selection and analysis. 200 consecutive patients with normal colonoscopies and 200 consecutive patients with polyp diagnostic colonoscopies were selected from the medical records. Exclusion criteria were: missing data (145 patients) and patients with high risk of CRC (91 patients). The dataset included 164 patients which were randomly divided 80% into a training set for the development of the model and 20% into a test set for the validation of the model. The random forests were developed on the training set. The testing set was used to perform internal validation of the model created. The dataset for external validation selected 42 patients out of 72, as exclusion was done for 30 (19 – data was missing and 11 – high risk of CRC).
The data included in this study was selected to include medical information that can be easily obtained in most adults (demographic data, medical history, common blood tests results not older than 12 months), the reason being to create a model which can be easily employed for future patients, which requires no additional costs.
Data collected from the patients' records, as seen in Table 1, included: sex (male/female), residence (urban or rural), age, diabetes mellitus (present or absent), BMI value (body mass index), fasting blood glucose levels, hemoglobin levels, platelets values, total cholesterol, LDL cholesterol, HDL cholesterol, triglycerides, serum glutamic-oxaloacetic transaminase, chronic gastritis (present or absent), the presence or absence of colonic polyps at colonoscopy.
A frequently used machine learning model, random forests is a non-parametric, supervised ensemble machine learning technique that was first put forth by Breiman as an extension to address regression and classification issues (12, 13). Fisher's discriminant is employed as a linear classifier for every branch of the random forests, which is based on techniques that train a forests of binary decision trees. To separate the observations into two homologous groups, known as branches, the algorithm in an ensemble decision tree uses a binary arithmetic technique. This splitting procedure is repeated until the “tree” has fully grown (“node purity” is reached) (14).
Data analyses was done and the random forests model was created using the statistical program R version 4.4.4. Using the random Forest package in R software, random forests of the variables were created for prediction of the variable polyp; 500 trees size was specified to be used in order to produce reliable findings. The mean loss in accuracy and Gini index values were used to assess the significance of each individual variable. In comparison to variables with lower values, those with a greater mean decline in accuracy or Gini index value were deemed more important for the algorithm. The receiver operating characteristic (ROC) curve was drawn and the area under the curve (AUC) was calculated in order to determine the diagnostic power of the variables for the prediction of colonic polyps. 80% of patients were used for the creation of the model, while 20% were included in the model validation subgroup. External validation was performed on the specific dataset.
In order to evaluate if the Random Forests was a good choice to create a prediction model for colonic polyps, two other methods were also evaluated: a generalized linear model (GLM) and Support Vector Machine (SVM). Their performance was tested on the same datasets and compared to the initial algorithm. For SVM the e1071 and pROC packages in R were used, while for GLM stats and caret packages in R were used.
The initial data set included 164 patients, 89 with normal colonoscopies, 75 with colonic polyps. 46.34% male, with a mean aged of 62.54 years, 20% had diabetes and 33.53% were previously diagnosed with gastritis. The mean BMI was 29.55, the mean fasting glucose was 120.4 mg/dl, the mean hemoglobin value was 13.49 g/dl, with a mean platelet count of 254.3/L. Mean total cholesterol levels were 201.7 mg/dl, for LDL 119.4 mg/dl and HDL 53.17 mg/dl, triglycerides had a mean value of 149.66 mg/dl, while serum glutamic-oxaloacetic transaminase was 26.64UI/L Table 2.
When comparing data of the patients without polyps and those with polyps, only 2 variables showed a statistically significant difference (p < 0.05): sex and body mass index. More male patients were in the polyp group while the BMI in the group without polyps was lower than in the polyp group (median-27 vs. 31) Table 3.
The dataset for external validation included 42 patients, 28 with normal colonoscopies, 14 with colonic polyps. 47.62% male, with a mean aged of 60.95 years, 7.14% had diabetes and 38.1% were previously diagnosed with gastritis. The mean BMI was 27.05, the mean fasting glucose was 104.6 mg/dl, the mean hemoglobin value was 13.54 g/dl, with a mean platelet count of 252.3/L. Mean total cholesterol levels were 195.1 mg/dl, for LDL 127.2 mg/dl and HDL 51.48 mg/dl, triglycerides had a mean value of 105.07 mg/dl, while serum glutamic-oxaloacetic transaminase was 23.64 UI/L Table 4.
The patients were randomly split 80–20 into a training set and a testing set. Using the training set a Random Forest model was created. The size was set at 500 trees and 3 variables were tried at each split.
While Random Forests don't require cross-validation to function (13), we used it to evaluate and tune the model. The “caret” package was used in R to specify 10-fold cross-validation. Different values for mtry (number of features considered at each split) and accuracy metric was used to evaluate the different mtry values. mtry of 2 or 3 proved to provide the highest accuracy, with minimal differences Table 5.
Using the “caret” package in R, fine tuning of mtry and number of trees (trees) was done to establish the best model and the OOBError (out of bag error) was used to select the best values. For an mtry = 1 the OOBError was 0.09923, for mtry = 2 the OOBError was 0.00763, while starting from mtry = 3 the error becomes 0. Figure 2 Mtry = 3 was selected for the algorithm.
For tunning the number of trees for the algorithm, different values were tried and 500 trees was associated with the lowest OOBError Table 6.
Analyzing the importance of the variables used while taking into consideration three measures derived from the structures of the trees (mean depth of first split of a variable, total number of nodes that split on that variable and the number of trees in which the variable splits the root) the variables with the most importance are (Figure 3):
- body mass index (IMC), mean minimum depth-2.01, number of nodes-1359, number of trees - 492
- platelets (plt), mean minimum depth-2.66, number of nodes-1266, number of trees - 470
- hemoglobin (Hb), mean minimum depth-3.06, number of nodes-1102, number of trees - 446
- triglicerides (triglic), mean minimum depth-3.08, number of nodes-1174, number of trees - 454
- glutamic-oxaloacetic transaminase (TGO), mean minimum depth-3.14, number of nodes-1172, number of trees – 455
- followed by: glycemia, HDL cholesterol, cholesterol, LDL cholesterol, and age.
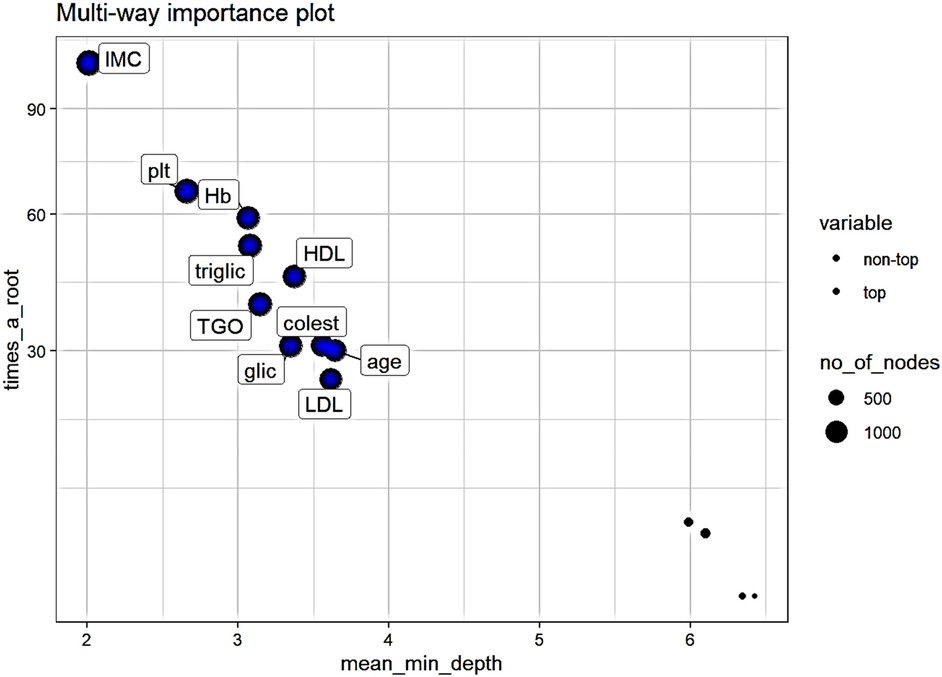
Figure 3. Multi-way importance plot of measures derived from the structures of the trees. Variables represented with bigger circles are used in more than 1000 nodes, top variables are represented with blue.
Analyzing the importance measures which take into consideration the role which the variable has in predicting (accuracy decrease, gini index decrease and p-value of a binomial distribution of the number of nodes which split on the variable assuming the variables are randomly used for splitting) (Figure 4) the top variables are (all with p < 0.01):
- body mass index (IMC), gini decrease- 13.09, accuracy decrease-0.07,
- platelets (plt), gini decrease-6.88, accuracy decrease-0.02,
- triglicerides (triglic), gini decrease- 6.36, accuracy decrease-0.02
- glutamic-oxaloacetic transaminase (TGO), gini decrease-5.77, accuracy decrease-0.01,
- hemoglobin (Hb), gini decrease-5.56, accuracy decrease- 0.01
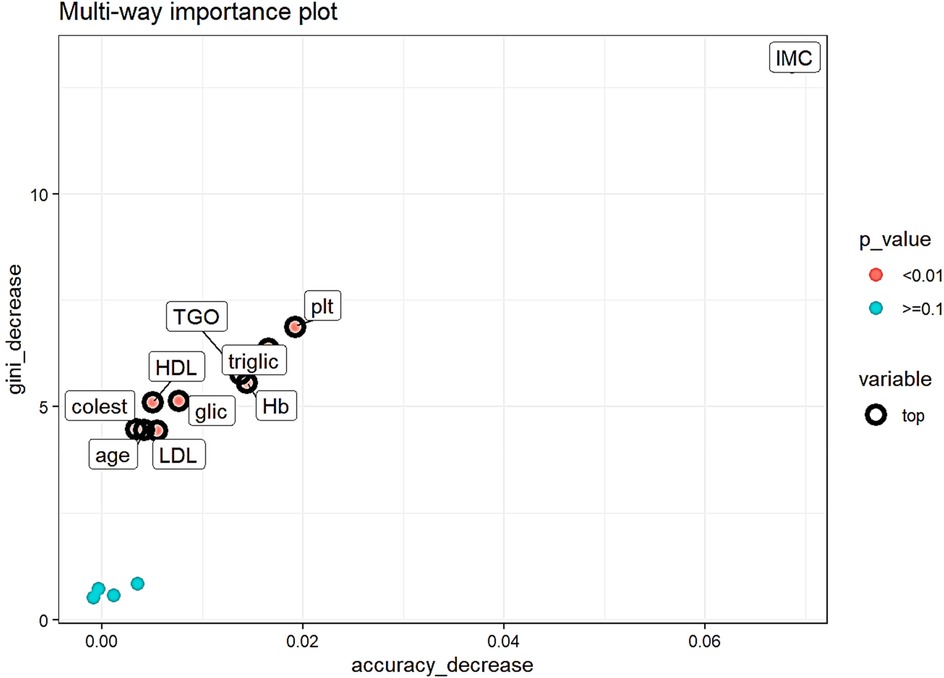
Figure 4. Multi-way importance plot of measures which take into consideration the predicting role of the variable. Top variables are represented with black circumference circles, variables with p < 0.01 are red, with p ≥ 0.1 are represented with light blue.
To evaluate the performance of the random forest model, receiver operating characteristics (ROC) curve analysis, as it takes into consideration both sensitivity as well as specificity. AUC value was 0.820 (95% CI = 0.747–0.893), having a good discriminative power Figure 5.
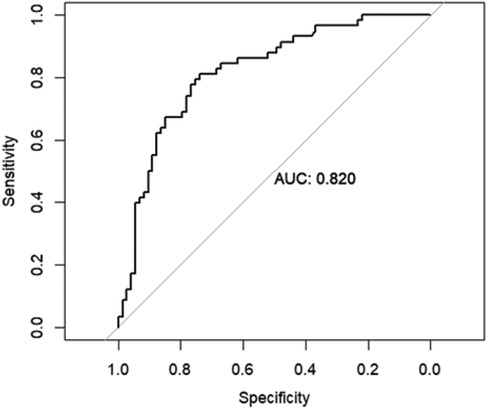
Figure 5. Area under the curve (AUC) of the receiver operating characteristics (ROC) of random forest model in the testing set.
For external validation AUC was 0.796 (95% CI = 0.718–0.851), The model's performance on the external validation dataset was slightly lower than on the internal validation dataset, which is expected. However, the drop in performance is minimal Figure 6.
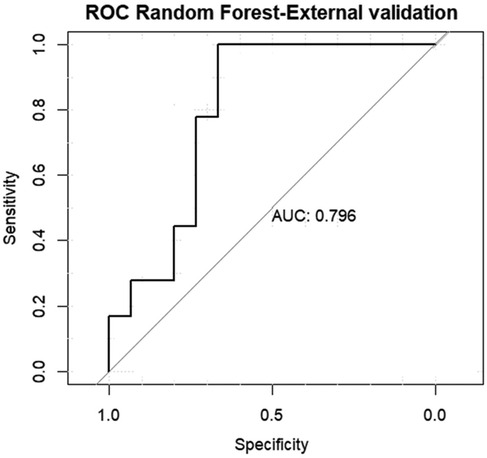
Figure 6. Area under the curve (AUC) of the receiver operating characteristics (ROC) of random forest model in the external validation data set.
The initial GLM (binomial family) created included all the variables in order to predict the presence of polyps. In order to improve its performance manual down stepping based on p-values was done, the reduced model included only the following variables: DZ, IMC, plt, HDL and triglic. This model provided an AIC-190.76.
The AUC of the ROC of this model, upon internal validation, was good: 0.788 Figure 7.
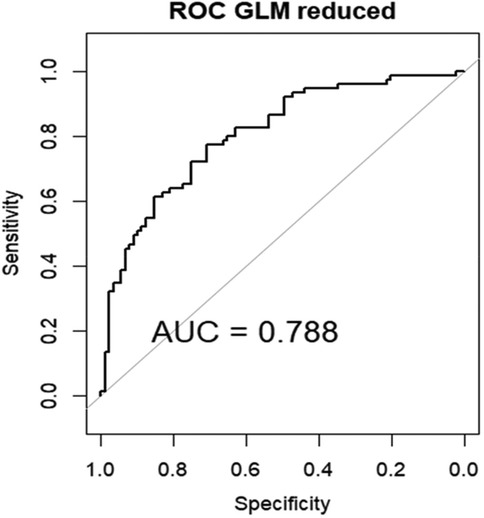
Figure 7. Area under the curve (AUC) of the receiver operating characteristics (ROC) of GLM in the internal validation data set.
Analyzing the performance of the linear model on the external validation dataset, we observe an AUC of 0.65, showing a modest performance of the model on new data Figure 8.
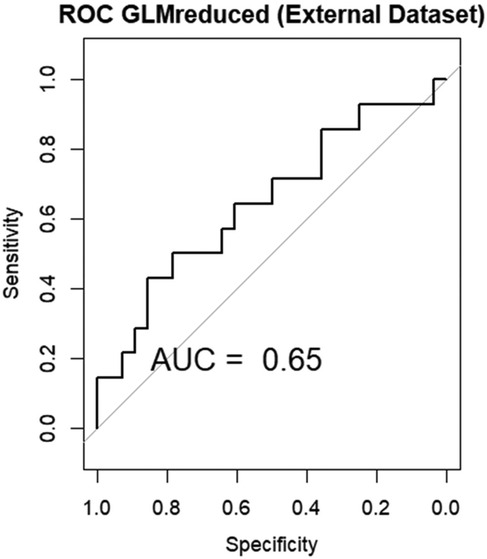
Figure 8. Area under the curve (AUC) of the receiver operating characteristics (ROC) of GLM in the external validation data set.
SVM are supervised learning models used for both classification and regression. In classification, SVM tries to find the hyperplane that divides best the data points of different classes in the feature space. The Radial Basis Function (RBF) kernel was used. Hyperparameter tuning was made using a grid search approach. The best combination found was cost = 10 and gamma = 1. Evaluation of the performance was made similar to the previous model, first on the internal validation dataset, then on the external validation dataset.
The AUC of the ROC of this model, upon internal validation, was good: 0.785 Figure 9.

Figure 9. Area under the curve (AUC) of the receiver operating characteristics (ROC) of SVM in the internal validation data set.
For external validation dataset, an AUC of 0.648 was obtained Figure 10.
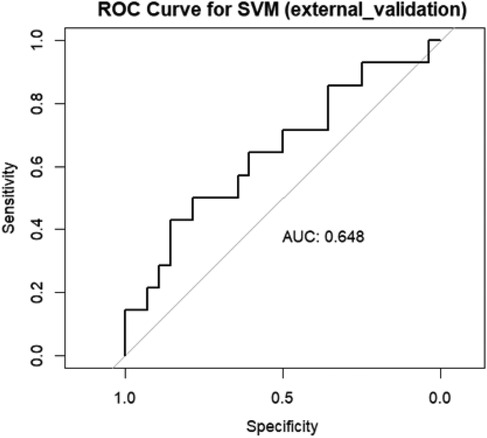
Figure 10. Area under the curve (AUC) of the receiver operating characteristics (ROC) of SVM in the external validation data set.
AI has a statistically significant positive influence on increasing the detection rate of colorectal polyps during colonoscopies (15). The application of AI algorithms is critical in reducing polyp miss rates in endoscopy. AI algorithms can analyze real-time images of the colon, highlighting alarming spots that humans may miss. This allows endoscopists to identify and remove polyps sooner, which is critical for preventing the development of colorectal cancer (16).
AI can analyse massive amounts of data from multiple sources and identify patterns in photos that indicate polyps (17). This allows AI to identify small changes in the mucosal surface that the human eye may overlook. Overall, these learning procedures have enhanced computer aided diagnostic systems (16, 18). AI algorithms rapidly scan the colon video footage and highlight suspicious areas that the endoscopist may have missed (19). This can reduce the polyp miss rate while increasing the detection rate, although it did increase the withdrawal time (20).
Computer aided diagnostic systems for colorectal polyps significantly increased adenoma detection rate or polyp detection rate with the use of different algorithms (21). The utility of AI in aiding the diagnosis of colorectal polyps during colonoscopy is questioned in certain studies (22–24). In a randomized controlled trial the computer aided diagnostic system showed a non-significant trend towards improving adenoma detection rate among patients undergoing screening or surveillance colonoscopy compared to high-definition white-light colonoscopy alone (25). The same lack of statistically significant increase in adenoma detection rate in real time endoscopies was found for using the GI Genius (Medtronic) module, an AI based computer aided diagnostic system (26). Also, no improvement in diagnosis was found when using AI in colonoscopies for patients with high risk of having colorectal lesions (27). When analyzing the clinical use of AI in colonoscopies, an improvement in lesion detection was seen for trainee endoscopists (28).
Li et al. developed a screening framework, Feature Interpretability Screening Framework, to identify patients at high risk for CRC. They used a combination of variables (sex, age, marital status), occult fecal test results, personal and family cancer history, gastrointestinal symptoms, obtained from a large patients' dataset (1,649,317) to train different artificial intelligence models in order to identify patients at high risk for CRC. The best performance was obtained by Naïve Bayes and SVM (highest sensitivity-0.779), Lasso had the highest specificity (0.868) and Logistic Regression -the highest AUC (0.859) (29). This study was done for CRC, on a large population database from a single medical center, we note that the Random Forests algorithm had an AUCs of 0.826, a value similar to our model, although a real comparison cannot be done between the studies, as they are trained for identifying patients with high risk of different lesions.
Zhang et al. constructed a ML extracellular vesicles based proteomics strategy model using a panel of 10 circulating protein markers which can predict well pre-malignant polyps and early stage CRC. The ML algorithms used, which provided excellent predictive power, were Naive Bayes, SVM, and Random Forest, having an AUC value that differentiate polyp from healthy, CRC from healthy, and CRC from polyp: 1, 0.97 and 0.94, respectively (30).
Random forests algorithms have been used to determine the relationship between gut microbiota and genetic factors in CRC. The model had good predicting potential of KRAS mutation status among CRC patients (AUC - 0.819), offering a potential new strategy for the precise treatment of CRC (31).
Artificial intelligence has also been used in differentiating adenomatous from non-adenomatous polyps on CT colonography. A Random Forest radiomics based model was developed and used for assisting radiologists in identifying polyp characteristics on CT colonography. The AI-assisted readings had higher accuracy, sensitivity, and specificity in selecting polyps eligible for polypectomy (32).
The development of clinical models of disease risk is the subject of several studies, and there are numerous relevant risk models available, such as those for colorectal cancer and coronary heart disease (33, 34). At the moment, colorectal cancer represents the basis for the majority of colorectal disease prediction models (35). Few colorectal polyp risk prediction models exist.
In this study we used a supervised learning model developed on easily obtainable and usually already available data for selecting patients with a high risk of being diagnosed with colonic polyps. Our algorithm had an AUC of 0.820. A study by Huang et al. developed a clinical predictive nomogram for the risk of a missed diagnosis of colorectal polyps in individuals based on multivariate analysis, the AUC being 0.747 (36). Their study was mainly focused on the necessity of performing a follow-up colonoscopy in certain patients at risk of having missed polyps during the initial procedure, as compared to our study which is focused on identifying which patients would benefit of a colonoscopy in order to identify and resect colorectal polyps. Ba et al. (37) developed a colorectal polyp prediction model using laboratory results, vital signs and demographic data from a big cohort of patients undergoing colonoscopies (5,426 patients). They included data similar to ours, but also more advanced lab tests (carcino embryonic antigen, hemoglobin A1c) which are not routinely done for the general population, making it harder to implement on a vast number of individuals. They tested 9 different ML methods and proved that, for their data, the adaptive boosting machine (AdaBoost) model had the best performance, providing an AUC = 0.675 on internal validation.
The incidence of colorectal polyps rises with age, according to numerous research. With every year of age gain, the risk of colorectal polyps increases by 1.03 times (38). The incidence rate of colorectal polyps rose with age and was higher in males than in females, according to the study of data of 327,785 colonoscopies performed in the US (39). In our study, the polyp group didn't have a statistically significant age difference, but it had more male participants than the no polyp group. Factors including bile acid synthesis, insulin-like growth factors, and estrogen receptor genes may be linked to females' decreased incidence of colorectal adenomatous polyps (40, 41). We noted the fact that the AI algorithm didn't include the sex variable in the top 10 most important variables, although it was statistically important, showing the completely different approach this algorithm has compared to more conventional statistical approaches regarding polyp prediction. Body mass index in the polyp group was higher than in the control group, which is consistent to other published studies (42).
Comparing the Random Forests algorithm with other two methods, generalized linear models and support vector machine, for our datasets, Random Forests provided better performance.
The model should be seen as a helpful tool for identifying unscreened individuals who are more likely to have precancerous lesions, rather than as a potential replacement for colonoscopies.
This research has a number of limitations. Data on eating habits, smoking, alcohol and drug use history, and family history, were not included, potentially excluding aspects associated with polyp formation. The medical records do not provide information on diet, while smoking and alcohol consumption information is not always realistically provided by patients. We also excluded patients with a family history of digestive tumors, as this is a separate risk factor, requiring attentive observation. This was a preliminary study, using a small number of patients. Only patients who had had a colonoscopy were included in the study population, which may not be representative of the general population. The study's retrospective design exposes it to selection bias, additionally the variables used to build the model were collected retrospectively, therefore it is uncertain how well the model performs in real time situations. The study is a single center study, the patients coming from a specific small region, which might have reduced the generalizability of our results. External validation was done on a small dataset, which contained data retrospectively obtained. Consequently, future research with bigger sample size would better evaluate our model (43, 44). Only the presence or absence of colonic polyps was assessed, without any other details. In the future, it would be useful to construct algorithms to also predict the presence of advanced adenomas or the size of the polyps as well as to create a calculator to determine the probability that asymptomatic people have colorectal polyps.
Colonic polyps have a risk of progressing into colonic cancer and their early diagnosis and removal might lead to a decrease in the incidence of colonic cancer. A random forest prediction model was developed using patient's demographic data, medical history and common blood tests results. This algorithm can foresee, with a high predictive power, the presence of colonic polyps.
The data analyzed in this study is subject to the following licenses/restrictions: data will be made available on reasonable request from the authors. Requests to access these datasets should be directed to Mihaela-Flavia Avram -YXZyYW0ubWloYWVsYUB1bWZ0LnJv.
The retrospective studies involving humans were approved by Institutional Ethics Board of Municipal Hospital “Dr. Teodor Andrei” Lugoj. The retrospective studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
M-FA: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Supervision, Writing – original draft, Writing – review & editing. NL: Formal Analysis, Software, Visualization, Writing – original draft, Writing – review & editing. DK: Data curation, Writing – original draft, Writing – review & editing. D-CL: Data curation, Formal Analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. M-IM: Investigation, Visualization, Writing – original draft, Writing – review & editing. M-SM: Supervision, Validation, Writing – original draft, Writing – review & editing. SO: Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We would like to acknowledge Victor Babes University of Medicine and Pharmacy Timisoara for their support in covering the costs of publication for this research paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sullivan BA, Noujaim M, Roper J. Cause, epidemiology, and histology of polyps and pathways to colorectal cancer. Gastrointest Endosc Clin N Am. (2022) 32(2):177–94. doi: 10.1016/j.giec.2021.12.001
2. Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. (2012) 366(8):687–96. doi: 10.1056/NEJMoa1100370
3. Kaminski MF, Wieszczy P, Rupinski M, Wojciechowska U, Didkowska J, Kraszewska E, et al. Increased rate of adenoma detection associates with reduced risk of colorectal cancer and death. Gastroenterology. (2017) 153(1):98–105. doi: 10.1053/j.gastro.2017.04.006
4. US Preventive Services Task Force, Davidson KW, Barry MJ, Mangione CM, Cabana M, Caughey AB, et al. Screening for colorectal cancer: uS preventive services task force recommendation statement. JAMA. (2021) 325(19):1965–77. Erratum in: JAMA. 2021 326(8):773. doi: 10.1001/jama.2021.6238
5. Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, et al. Estimation of benefits, burden, and Harms of colorectal cancer screening strategies: modeling study for the US preventive services task force. JAMA. (2016) 315(23):2595–609. doi: 10.1001/jama.2016.6828
6. Wong MCS, Huang J, Huang JLW, Pang TWY, Choi P, Wang J, et al. Global prevalence of colorectal neoplasia: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. (2020) 18(3):553–61.e10. doi: 10.1016/j.cgh.2019.07.016
7. Kolb JM, Hu J, DeSanto K, Gao D, Singh S, Imperiale T, et al. Early-age onset colorectal neoplasia in average-risk individuals undergoing screening colonoscopy: a systematic review and meta-analysis. Gastroenterology. (2021) 161(4):1145–55.e12. doi: 10.1053/j.gastro.2021.06.006
8. Bretthauer M, Kaminski MF, Løberg M, Zauber AG, Regula J, Kuipers EJ, et al. Population-based colonoscopy screening for colorectal cancer: a randomized clinical trial. JAMA Intern Med. (2016) 176(7):894–902. doi: 10.1001/jamainternmed.2016.0960
9. Peterse EFP, Meester RGS, Siegel RL, Chen JC, Dwyer A, Ahnen DJ, et al. The impact of the rising colorectal cancer incidence in young adults on the optimal age to start screening: microsimulation analysis I to inform the American cancer society colorectal cancer screening guideline. Cancer. (2018) 124(14):2964–73. doi: 10.1002/cncr.31543
10. Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American cancer society. CA Cancer J Clin. (2018) 68(4):250–81. doi: 10.3322/caac.21457
11. Chen C, Stock C, Hoffmeister M, Brenner H. Optimal age for screening colonoscopy: a modeling study. Gastrointest Endosc. (2019) 89(5):1017–25.e12. doi: 10.1016/j.gie.2018.12.021
12. Yuan H, Fan XS, Jin Y, He JX, Gui Y, Song LY, et al. Development of heart failure risk prediction models based on a multi-marker approach using random forest algorithms. Chin Med J (Engl). (2019) 132(7):819–26. doi: 10.1097/CM9.0000000000000149
14. Wang W, Li L, Gu H, Chen Y, Zhen Y, Dong Z. Random forest-based prediction of acute respiratory distress syndrome in patients undergoing cardiac surgery. Heart Surg Forum. (2022) 25(6):E854–9. doi: 10.1532/hsf.5113
15. Adiwinata R, Tandarto K, Arifputra J, Waleleng BJ, Gosal F, Rotty L, et al. The impact of artificial intelligence in improving polyp and adenoma detection rate during colonoscopy: systematic-review and meta-analysis. Asian Pac J Cancer Prev. (2023) 24(11):3655–63. doi: 10.31557/APJCP.2023.24.11.3655
16. Masud M, Sikder N, Nahid AA, Bairagi AK, AlZain MA. A machine learning approach to diagnosing lung and colon cancer using a deep learning-based classification framework. Sensors (Basel). (2021) 21(3):748. doi: 10.3390/s21030748
17. Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial intelligence in colorectal cancer screening, diagnosis and treatment. A new era. Curr Oncol. (2021) 28(3):1581–607. doi: 10.3390/curroncol28030149
18. Ben Hamida A, Devanne M, Weber J, Truntzer C, Derangère V, Ghiringhelli F, et al. Deep learning for colon cancer histopathological images analysis. Comput Biol Med. (2021) 136:104730. doi: 10.1016/j.compbiomed.2021.104730
19. Xu H, Tang RSY, Lam TYT, Zhao G, Lau JYW, Liu Y, et al. Artificial intelligence-assisted colonoscopy for colorectal cancer screening: a multicenter randomized controlled trial. Clin Gastroenterol Hepatol. (2023) 21(2):337–46.e3. doi: 10.1016/j.cgh.2022.07.006
20. ELKarazle K, Raman V, Then P, Chua C. Detection of colorectal polyps from colonoscopy using machine learning: a survey on modern techniques. Sensors (Basel). (2023) 23(3):1225. doi: 10.3390/s23031225
21. Kikuchi R, Okamoto K, Ozawa T, Shibata J, Ishihara S, Tada T. Endoscopic artificial intelligence for image analysis in gastrointestinal neoplasms. Digestion. (2024) 105(6):419–35. doi: 10.1159/000540251
22. Wei MT, Shankar U, Parvin R, Abbas SH, Chaudhary S, Friedlander Y, et al. Evaluation of computer-aided detection during colonoscopy in the community (AI-SEE): a multicenter randomized clinical trial. Am J Gastroenterol. (2023) 118(10):1841–7. doi: 10.14309/ajg.0000000000002239
23. Mangas-Sanjuan C, de-Castro L, Cubiella J, Díez-Redondo P, Suárez A, Pellisé M, et al. Role of artificial intelligence in colonoscopy detection of advanced neoplasias: a randomized trial. Ann Intern Med. (2023) 176(9):1145–52. doi: 10.7326/M22-2619
24. Schöler J, Alavanja M, de Lange T, Yamamoto S, Hedenström P, Varkey J. Impact of AI-aided colonoscopy in clinical practice: a prospective randomised controlled trial. BMJ Open Gastroenterol. (2024) 11(1):e001247. doi: 10.1136/bmjgast-2023-001247
25. Alali AA, Alhashmi A, Alotaibi N, Ali N, Alali M, Alfadhli A. Artificial intelligence for adenoma and polyp detection during screening and surveillance colonoscopy: a randomized-controlled trial. J Clin Med. (2025) 14(2):581. doi: 10.3390/jcm14020581
26. Rønborg SN, Ujjal S, Kroijer R, Ploug M. Assessing the potential of artificial intelligence to enhance colonoscopy adenoma detection in clinical practice: a prospective observational trial. Clin Endosc. (2024) 57(6):783–9. doi: 10.5946/ce.2024.038
27. Chow KW, Bell MT, Cumpian N, Amour M, Hsu RH, Eysselein VE, et al. Long-term impact of artificial intelligence on colorectal adenoma detection in high-risk colonoscopy. World J Gastrointest Endosc. (2024) 16(6):335–42. doi: 10.4253/wjge.v16.i6.335
28. Yamaguchi D, Shimoda R, Miyahara K, Yukimoto T, Sakata Y, Takamori A, et al. Impact of an artificial intelligence-aided endoscopic diagnosis system on improving endoscopy quality for trainees in colonoscopy: prospective, randomized, multicenter study. Dig Endosc. (2024) 36(1):40–8. doi: 10.1111/den.14573
29. Li M, Gong Y, Pang Y, Wu M, Gu K, Wang Y, et al. A novel colorectal cancer screening framework with feature interpretability to identify high-risk populations for colonoscopy. J Gastroenterol Hepatol. (2024) 39(9):1827–36. doi: 10.1111/jgh.16600
30. Zhang J, Gao Z, Xiao W, Jin N, Zeng J, Wang F, et al. A simplified and efficient extracellular vesicle-based proteomics strategy for early diagnosis of colorectal cancer. Chem Sci. (2024) 15(44):18419–30. doi: 10.1039/D4SC05518G
31. Huang Z, Huang X, Huang Y, Liang K, Chen L, Zhong C, et al. Identification of KRAS mutation-associated gut microbiota in colorectal cancer and construction of predictive machine learning model. Microbiol Spectr. (2024) 12(5):e0272023. doi: 10.1128/spectrum.02720-23
32. Grosu S, Fabritius MP, Winkelmann M, Puhr-Westerheide D, Ingenerf M, Maurus S, et al. Effect of artificial intelligence-aided differentiation of adenomatous and non-adenomatous colorectal polyps at CT colonography on radiologists’ therapy management. Eur Radiol. (2025). doi: 10.1007/s00330-025-11371-0
33. Huang X, Cai W, Yuan W, Peng S. Identification of key lncRNAs as prognostic prediction models for colorectal cancer based on LASSO. Int J Clin Exp Pathol. (2020) 13(4):675–84.32355515
34. Guan H, Dai GH, Gao WL, Zhao X, Cai ZH, Zhang JZ, et al. A 5-year survival prediction model for chronic heart failure patients induced by coronary heart disease with traditional Chinese medicine intervention. Evid Based Complement Alternat Med. (2021) 2021:4381256. doi: 10.1155/2021/4381256
35. Reidy E, Leonard NA, Treacy O, Ryan AE. A 3D view of colorectal cancer models in predicting therapeutic responses and resistance. Cancers (Basel). (2021) 13(2):227. doi: 10.3390/cancers13020227
36. Huang Y, Liu Y, Yin X, Zhang T, Hao Y, Zhang P, et al. Establishment of clinical predictive model based on the study of influence factors in patients with colorectal polyps. Front Surg. (2023) 10:1077175. doi: 10.3389/fsurg.2023.1077175
37. Ba Q, Yuan X, Wang Y, Shen N, Xie H, Lu Y. Development and validation of machine learning algorithms for prediction of colorectal polyps based on electronic health records. Biomedicines. (2024) 12(9):1955. doi: 10.3390/biomedicines12091955
38. Han X, Qian W, Liu Y, Zheng T, Su XJ, Zhang PP, et al. Effects of age, sex and pathological type on the risk of multiple polyps: a Chinese teaching hospital study. J Dig Dis. (2020) 21(9):505–11. doi: 10.1111/1751-2980.12863
39. Lieberman DA, Williams JL, Holub JL, Morris CD, Logan JR, Eisen GM, et al. Race, ethnicity, and sex affect risk for polyps >9 mm in average-risk individuals. Gastroenterology. (2014) 147(2):351–8; quiz e14–5. doi: 10.1053/j.gastro.2014.04.037
40. Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. (1994) 7(4):536–40. doi: 10.1038/ng0894-536
41. Wu J, Bai Y, Lu Y, Yu Z, Zhang S, Yu B, et al. Role of sex steroids in colorectal cancer: pathomechanisms and medical applications. Am J Cancer Res. (2024) 14(7):3200–21. doi: 10.62347/OEBS6893
42. Ben Q, An W, Jiang Y, Zhan X, Du Y, Cai QC, et al. Body mass index increases risk for colorectal adenomas based on meta-analysis. Gastroenterology. (2012) 142(4):762–72. doi: 10.1053/j.gastro.2011.12.050
43. Althubaiti A. Sample size determination: a practical guide for health researchers. J Gen Fam Med. (2022) 24(2):72–8. doi: 10.1002/jgf2.600
Keywords: colorectal polyps, random forests, machine learning, colorectal cancer prevention, risk prediction model, artificial intelligence
Citation: Avram M-F, Lupa N, Koukoulas D, Lazăr D-C, Mariș M-I, Murariu M-S and Olariu S (2025) Random forests algorithm using basic medical data for predicting the presence of colonic polyps. Front. Surg. 12:1523684. doi: 10.3389/fsurg.2025.1523684
Received: 6 November 2024; Accepted: 10 February 2025;
Published: 3 March 2025.
Edited by:
Gabriel Sandblom, Karolinska Institutet (KI), SwedenReviewed by:
Luis Capitán-Morales, University of Seville, SpainCopyright: © 2025 Avram, Lupa, Koukoulas, Lazăr, Mariș, Murariu and Olariu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mihaela-Flavia Avram, YXZyYW0ubWloYWVsYUB1bWZ0LnJv
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.