- 1Department of Dermatology, China–Japan Friendship Hospital, Beijing, China
- 2Graduate School of Peking Union Medical College, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
The skin is the outermost barrier of the body. It has developed a sophisticated system against the ever-changing environment. The application of single-cell technologies has revolutionized dermatology research and unraveled the changes and interactions across skin resident cells in the healthy and inflamed skin. Single-cell technologies have revealed the critical roles of stromal cells in an inflammatory response and explained a series of plausible previous findings concerning skin immunity. Here, we summarized the functional diversity of skin stromal cells defined by single-cell analyses and how these cells orchestrated events leading to inflammatory diseases, including atopic dermatitis, psoriasis, vitiligo, and systemic lupus erythematosus.
Introduction
The skin is the body’s outermost barrier and is composed of the epidermis and dermis. It is defined by a multilayered architecture based on diverse cell populations of keratinocytes, melanocytes, fibroblasts, and endothelial cells, as well as various immune cells. Facing the ever-changing environment, it has developed a complex system that responds to stimuli such as pathogen infection, UV exposure, and toxic compounds (1). In this system, skin stromal cells have long been considered passive participants and bystanders in immunoregulation. However, recent studies have revealed that they actively participate in skin inflammatory responses.
RNA sequencing (RNA-seq) technology has long been used to dissect the molecular differences or commonalities of different inflammatory skin diseases (2). Previous bulk RNA sequencing and microarray studies on skin inflammatory diseases have revealed dysregulation of inflammatory and barrier genes, but many cell-specific transcripts often remain masked in whole-skin transcriptomic analyses. In addition, it has been difficult to systematically understand the sophisticated interplay among cells in the skin (3). Sing-cell RNA-seq (scRNA-seq) can capture tissue transcripts at the single-cell resolution and effectively solve this problem (3). The experimental procedure of scRNA-seq mainly includes four steps: sample preparation, cell enrichment, library preparation, and data analysis (4). It has been leveraged in various fields, contributing to novel discoveries and improving our understanding of embryonic medicine, developmental biology, tumor ecosystems, immunology, and inflammatory diseases (5). scRNA-seq studies have elucidated how skin resident cells are involved in skin homeostasis regulation and inflammatory responses and revealed their immunoregulatory role (6–8). However, scRNA-seq transcriptome analysis remains challenging to define cell subpopulations due to the heterogeneity of skin cells during inflammation.
In this review, we recapitulated the main cell populations and their subpopulations of skin-resident stromal cells observed by scRNA-seq technologies in healthy and inflamed human skin and discussed their pathological alterations and “feedforward” role in recruiting immune cells in skin inflammatory diseases, including atopic dermatitis (AD), psoriasis, vitiligo, and systemic lupus erythematosus (SLE). We attempted to highlight current disagreements and consensus on skin stromal cell subsets and functions in inflammatory diseases.
Skin Resident Cells Identified by scRNA-seq
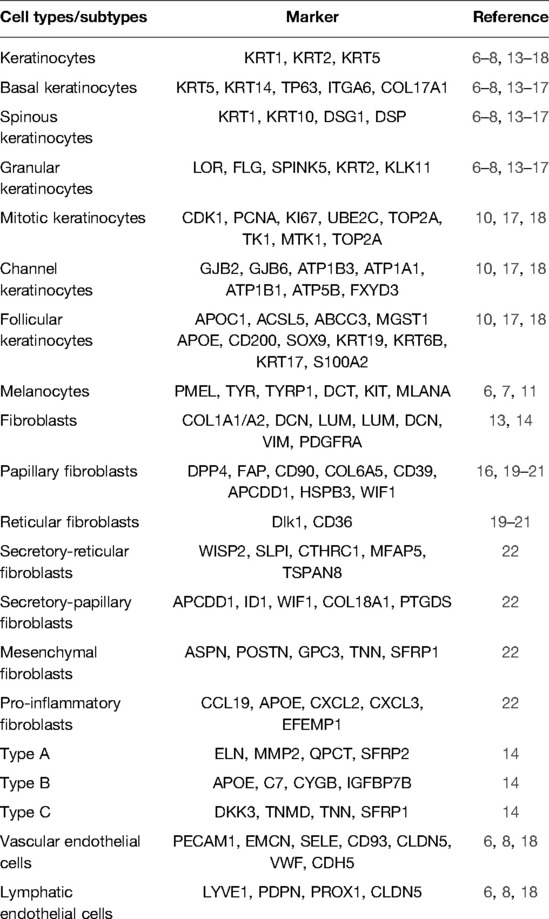
Defining subsets of cells is an important step in scRNA-seq bioinformatics analysis. The type of obtained cells varied with different collection methods. Full-thickness skin samples contained complete skin cells, while suction blisters would lose information on fibroblasts, endothelial cells, and immune cells in the dermis (9). However, suction blisters were more accessible and often used in the study of AD (7, 10, 11). Most studies on the skin referred to the Human Cell Atlas (12) for cell identification, which could accurately distinguish skin stromal cells. However, when it comes to specific cell subtypes, especially fibroblasts with obvious heterogeneity, it was difficult for researchers to reach a consensus on grouping and naming. We would describe the disagreements and consensus on each cell subset classification in the following. The biological markers used in scRNA-seq studies are listed in Table 1.
Keratinocytes Actively Contributed to Immunopathology in Inflamed Skin
Keratinocytes are the major cells in the epidermis, accounting for 66% of the epidermic cells from suction blisters and 37.96% of the full-thickness skin from skin biopsies in scRNA-seq (13). Keratinocytes were distinguished from other skin cells by cluster-specific expression of keratin1 (KRT1), KRT2, and KRT5 (6–8, 13–18).
Cell subtype identification of keratinocytes was consistent across various papers (6, 7, 13). Three subpopulations were separated corresponding to their stereotyped differentiation process in the normal epidermis: undifferentiated basal keratinocytes had high expression of basal epidermal proteins (KRT5 and KRT14), hemidesmosome molecules [integrin alpha-6, ITGA6; and collagen alpha-1 (XVII) chain, COL17A1] and stemness factor (tumor protein 63, TP63); upper spinous keratinocytes expressed suprabasal cell transcripts (KRT1 and KRT10); and granular keratinocytes expressed KRT2, KLK11, and late differentiation markers (Loricrin, LOR; Filaggrin, FLG; and serine protease inhibitor Kazal-type 5, SPINK5) (6–8, 13–18).
In addition, three subpopulations were unable to be distinguished by the classical differentiation model. The first undergoing proliferation was deemed as mitotic keratinocytes, as characterized by DNA synthesis and cell division transcripts, including cyclin-dependent kinase 1 (CDK1), proliferating cell nuclear antigen (PCNA), KI67, ubiquitin-conjugating enzyme E2 C (UBE2C), DNA topoisomerase 2-alpha (TOP2A), thymidine kinase (TK1), and mitogen-activated protein kinase kinase kinase 4 (MAP3K4). The second was termed “channel” keratinocytes by coordinate elevation of ion channel and cell-cell communication transcripts [gap junction beta (GJB2/6), potassium-transporting ATPase (ATP1B3/1A1/1B1/5B), and FXYD domain-containing ion transport regulator 3 (FXYD3)]. Finally, the third was separated individually as follicular keratinocytes that expressed genes associated with the sebaceous gland [apolipoprotein (APOC1/E), long-chain-fatty-acid–CoA ligase 5 (ACSL5), ATP-binding cassette sub-family C member 3 (ABCC3), and microsomal glutathione S-transferase 1 (MGST1)], follicles (TRK19, CD200, SOX9), and (or) outer root sheath (KRT6B, KRT17, and S100A2) (10, 17, 18).
Interestingly, force-directed graph (FDG) and PAGA analyses on scRNA-seq data defined two differentiation pathways of basal keratinocytes to suprabasal cells, with or without a high level of the lamellar body (LB)-related transcripts (glucosylceramide transporter ABCA12; cytoskeleton-associated protein 4, CKAP4; and CAP-Gly domain-containing linker protein 1, CLIP1), that characterized late epidermal differentiation in healthy skin (6). When it comes to inflammation, keratinocytes showed a more complicated picture of gene expression. Gene differential expression analysis revealed that keratinocytes, especially those in spinous and granular layers, overexpressed a panel of interferon response genes [interferon alpha-inducible protein 27 (IFI27) and interferon-induced transmembrane protein 1 (IFITM1)] and damage-associated transcripts, including antimicrobial peptides, also known as alarmis (S100A7/A8/A9), wound healing states (KRT6A/16), and serpins (SERPINB4/13), consistent with previous study results (4, 6, 13). It was reported that evoked S100A8/A9 expression was a molecular event preceding any histological alteration or deregulation of cytokines and played a crucial role in enhancing keratinocyte differentiation while inhibiting proliferation (23). In addition to damage-associated molecules, keratinocytes also expressed chemokine ligands and cytokines to recruit immune cells. Reynolds et al. defined a new subpopulation of celled inflammatory differentiated keratinocytes, as suggested by coexpressing lower levels of undifferentiated and differentiated markers but additionally expressing intercellular adhesion molecule1 (ICAM1), tumor necrosis factor (TNF), and CCL20 (6). CCL20 is the only known high-affinity ligand of CCR6 and attracts IL17A-producing CCR6+ immune cells, such as dendritic cells (DCs) and T cells (24). It has been reported that the CCL20/CCR6 axis contributes to the formation of an IL17A-rich milieu in psoriasis (25). Pruritus-induced mechanical scratching also upregulated CCL20 production from keratinocytes, which could partially explain high IL17A levels in AD (24).
Several disease-specific alterations were described in keratinocytes of AD patients. Abnormally elevated genes included aquaporin-3 (AQP3, a water channel component associated with keratinocyte proliferation and transepidermal water loss) (26), tumor necrosis factor receptor superfamily member 12A (TNFRSF12A), cornifin-B (SPRR1B, promoting the cornification of keratinocytes) (10, 27), CCL2 (binding to CCR2 to polarize Th2 response) (28), CCL27, and proinflammatory cytokine IL32 (modulating keratinocyte apoptosis) (17, 29). Downregulated genes involved DEFB1, an antimicrobial against Staphylococcus aureus (10, 30). After dupilumab treatment, most of the activated transcripts were normalized, including S100A7/A8/A9, KRT6A, KRT16, SERPINB4, CCL2, and CCL27, whereas IFI27, TNFRSF12A, and SPRR1B remained dysregulated 1 year later (31). The relationship between these genes and recurrence is worth exploring.
Psoriatic epidermis is remarkable for the expansion of mitotic and channel subfractions with increased GJB6, which is described as a psoriasis risk gene (18). In addition, keratinocytes significantly overexpress MHC molecules (HLA-DRA, HLA-DMA, HLA-C), WNT5A, CD58, and pro-inflammatory cytokines IL36G and NFKBIZ (transcriptional regulator of IL36-driven gene expression) (13, 32). It has been described that keratinocytes are capable of directly presenting antigen to T cells and initiating an immune response (33). IL36γ could induce keratinocytes to produce CXCL1, CXCL10, and CCL20 and function as a linker between keratinocytes and DCs (34). Unexpectedly, Hughes et al. found that FOS-related antigen 1 (FOSL1) was positively correlated with diffusion pseudo-time in psoriatic KCs compared with healthy KCs by differential pseudo-time correlation analysis and further validated the distribution at the protein level by immunofluorescence staining (8). FOSL1 belongs to the FOS gene family that has been implicated as regulators of cell differentiation, proliferation, and transformation. FOSL1 was identified as the TF encoding gene of psoriasis (35), and the enhanced FOSL1 expression was significantly correlated with high psoriasis area and severity index (36). FOSL1 knockdown inhibited IL22-induced proliferation and enhanced apoptosis of keratinocytes (37). These results suggest that FOSL1 may serve as a new therapeutic target for psoriasis.
It is an intriguing finding that CCL27 expression of keratinocytes was increased in AD (17) but decreased in psoriasis (32). CCL27 is a chemotactic factor binding to CCR10 that plays an important role in the homeostatic establishment of resident lymphocytes (38). CCL27/CCR10-derived regulation would promote activated T cell product IL17A/IL22 (38). Interactions between CCL2 and CCL27 (keratinocytes) and CCR1/2 and CCR10 (T cells) might cooperatively induce the Th2 milieu in AD (17). Nevertheless, it was confusing that impaired CCL27–CCR10 interaction was present in IL17-driven psoriasis (32). This contradictory result suggested that CCL27 might serve as a molecular marker for differentiating psoriasis and AD.
Keratinocytes respond to inflammation even without visible skin damage. It was reported that keratinocytes upregulated IFN-responsive genes and fibrotic pathways in non-lesional, non-sun-exposed skin of SLE (39, 40). Keratinocytes can even recruit CXCR3+ T cells to kill surrounding melanocytes by expressing high levels of chemokines CXCL9/10/11 (16). These results emphasized that keratinocytes collaborate with immune cells and actively contribute to immunopathology.
Melanocytes Not Bystanders of Skin Inflammation
Melanocytes account for 10% of the basal cells of the epidermis, and they can be captured at a relatively large proportion by suction blistering, defined by PMEL, tyrosinase (TYR), tyrosinase-related protein 1/2 (TYRP1/2), KIT, and melanoma antigen (MLANA) in scRNA-seq. Melanocytes express many molecules necessary for the maintenance of the extracellular matrix and cell–cell adhesion, including fibronectins, collagens, and laminins, and adhesion to the basal keratinocyte layer by expressed cognate ligands (FN1, LAMBA4, and LAMB1) (6, 7, 11).
Pigmentation changes are very common in inflammatory skin diseases. Consistently, scRNA-seq studies indicated that melanocytes display considerable numbers of dysregulated genes, including S100 transcripts and pigmentation-associated genes (TYR and MFSD12) in inflamed skin. Despite clinical clearance of the skin, these genes are still overexpressed in spontaneously healed or dupilumab-treated AD (7, 11).
Melanocytes were previously seen as bystanders of skin inflammation, but scRNA-seq demonstrated that they are involved in a variety of inflammatory responses. They increase MHC-I signaling transcript (HLA-A, HLA-B, and HLA-E) in inflamed skin, targeting T cells (HLA-CD3D) and natural killer (NK) cells (HLA-E-KLRC1) (16). It was reported that HLA-C-presented melanocyte was attacked by CD8± T cells and further induced T cells to secrete IL17A in psoriasis lesions (41). Notably, HLA-C might also play a crucial role in transformation from acute inflammation to a chronic disease (42). Furthermore, some melanocytes significantly evoked IL4Rα, CCL18, and CXCL1 expression responses to type 2 inflammation in AD (7, 43). In vitiligo, a disease characterized by melanocyte loss, CCL18 was also overexpressed in melanocytes, which attracted CCR8+ immune cells (T and NK cells) to kill themselves (16, 44). These pieces of evidence imply that melanocytes are part of a “feedforward” inflammatory response in the epidermis, resembling keratinocytes.
Despite their pro-inflammatory role, melanocytes might be involved in inflammatory diminishing, as suggested by overexpressing matricellular protein CCN family member 3 (CCN3, limiting proinflammatory activation) (45), SERPINF1 (antiangiogenic factor) (46), and Annexin A1 (ANXA1, an anti-inflammatory factor functioning as a major mediator of glucocorticoid responses) in AD (7, 47). Consistent with this study, Rindler et al. found that melanocytes upregulated T cell reactivity inhibitor dickkopf-related protein 3 (DKK3) (48), TGF-β signaling promotor CD81 (49), and platelet-activating factor acetylhydrolase (PLA2G7), which catalyzed the degradation of the strongly pro-inflammatory phospholipid mediator platelet-activating factor (PAF) (11, 50) in spontaneously healed AD (11). Similarly, anti-inflammatory genes (CCN3 and ANXA1) remained among the top upregulated genes after 1-year dupilumab treatment, whereas inflammatory factor expression was normalized (CCL18 and CXCL1) (31).
Heterogeneity, Dysregulated Extracellular Matrix, and Immune Regulation of Fibroblasts in Diseases
Fibroblasts make up the majority of dermal cells, marked by collagen alpha-1 (I) chain (COL1A1/A2), decorin (DCN), lumican (LUM), vimentin (VIM), and platelet-derived growth factor receptor alpha (PDGFRA). Fibroblasts play an important role in epidermal stem and progenitor cell (EpSC) maintenance (51, 52) and immune surveillance of the dermis (53). Fibroblasts account for 31.53% of the full-thickness skin cells from skin biopsies (13, 14) but are lost in suction blisters for scRNA-seq.
Fibroblasts can be spatially divided into upper papillary fibroblasts and lower reticular fibroblasts. Driskell et al. first defined dipeptidyl peptidase 4 (DPP4, also termed CD26) as a papillary fibroblast marker and delta-like non-canonical notch ligand 1 (Dlk1)+ cell as a fibroblast within the reticular layers (19). It is interesting that DPP4± fibroblasts deposit the matrix during wound healing in mice (54). In line with this study, Guerrero-Juarez. et al. identified CRABP1± papillary fibroblasts in large wounds by conducting scRNA-seq (55). These studies implied that upper papillary dermal compartments seem to be more actively involved in wound regeneration. Subsequently, fibroblast activation protein (FAP), CD90, α5 chain of collagen VI (COL6A5), CD39, APCDD1, heat shock protein beta-3 (HSPB3), and WNT inhibitory factor 1 (WIF1) were identified as papillary fibroblast markers, whereas FAP− CD90+ CD36+ fibroblasts were classified as reticular fibroblasts (20, 21). Philippeos et Al. revealed distinct properties of the upper and lower dermis using RNA-seq on the dermis separated by microdissection (21). The study demonstrated that the papillary dermis exhibited upregulation of the Wnt signaling pathway (WIF1, APCDD1, RSP01, axin-2, AXIN2) and presented an anti-inflammatory phenotype, as suggested by a significant reduction in the upregulation of PDL-1 and CD40 compared with lower fibroblasts in response to IFNγ stimulation. Reticular layers highly expressed several extracellular matrix (ECM) components (ADAM12, ADAMTS9, COL1A2, COL1A1, ELN, and FN1) and inflammatory mediators (CCL19, CCL7, CXCR4, and IL6) (21).
However, the classical markers rarely distinguish the two populations in scRNA-seq studies. Ascensión et al. conducted an integrated analysis of fibroblasts in four scRNA-seq studies and found that papillary and reticular fibroblast categories were composed of a mixture of cell subtypes (14). The classical papillary marker CD26 transcript was present in virtually all cells in the dermis without regional restriction (56). It might result from the functional heterogeneity of fibroblasts (57) or the progressive loss of clear genetic distinctions during the aging process (22). In addition, the heterogeneity of fibroblastic cells also existed across developmental time, between different anatomic skin locations (58). Therefore, fibroblast subsets identified varied among studies. How to unify the nomenclature of fibroblast subtypes is an urgent problem.
A possible strategy is to combine spatial location and function. Solé-Boldo et al. classified fibroblasts into four subpopulations: secretory-reticular fibroblasts [CCN5, antileukoproteinase, SLPI, collagen triple helix repeat-containing protein 1 (CTHRC1), microfibrillar-associated protein 5 (MFAP5), and tetraspanin-8 (TSPAN8)], secretory-papillary fibroblasts [(APCDD1, ID1, WIF1, COL18A1, prostaglandin-H2 d-isomerase (PTGDS)], mesenchymal fibroblasts [asporin (ASPN), periostin (POSTN), glypican-3 (GPC3), tenascin-N (TNN), and secreted frizzled-related protein 1 (SFRP1)], and pro-inflammatory fibroblasts [CCL19, APOE, CXCL2, CXCL3, and EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1)] that are widely spread within the dermis (22). Another study re-analyzed the four published datasets and classified fibroblasts into three major subsets: type A accounted for about 49.5% of the fibroblasts (defined by ELN, MMP2, QPCT, and SFRP2) and was responsible for dermal cell and ECM homeostasis; type B represented 30.5% of the fibroblasts (defined by APOE, C7, CYGB, and IGFBP7B) and might play a clear role in immune surveillance and inflammation promotion; and type C represented 49.5% of the fibroblasts (defined by DKK3, TNMD, TNN, and SFRP1) and included more specialized subpopulations, such as dermal papilla cells and dermo-hypodermal junction fibroblasts (14). There were still significant differences between the two methods. To clearly describe the expression changes of fibroblasts, it is necessary to identify function-based cellular markers and arrive at a consensus for nomenclature.
Due to the different classification of fibroblast subsets among various studies, fibroblasts will be considered as a group in discussing their changes in inflammation. As expected, fibroblasts significantly increased the expression of MHC-I and MHC-II molecules (HLA-A, HLA-B, HLA-C, and HLA-DRA) in inflamed conditions (13), driving T and NK cell activation via targeting CD4, CD8, and CD94 (59, 60). In addition to antigen presentation, Reynolds et al. found that COL1A1+ COL1A2+ COL6A1+ fibroblasts were significantly enriched in AD and psoriatic conditions, which specialized toward ECM remodeling and maintenance, and overexpressed chemokines CXCL12 and CCL19 (6).
Consistent with the abovementioned study, Gao et al. also demonstrated that ECM-related proteins (SDK1, FGF7, COMP, COL5A3, and COL1A1) were elevated in psoriatic fibroblasts, indicating ECM remodeling in the inflamed condition (13). As major contributors to and regulators of inflammation, psoriatic fibroblasts also significantly increased cytokines CCL26, IL6, LIF, IL17B, CCL19, CXCL12, TNFRSF11B, and TNFSF13B (BAFF) (8, 13). CCL26 was a classical type 2 chemokine, while IL6 was considered a hallmark inflammatory cytokine formed by fibroblasts, and the autocrine LIF-LIFR positive feedback loop was critical for maintaining sustained I-6 transcription (61). Increased IL17B was enriched in perivascular fibroblasts (PDGFRβ+), which might be involved in the surface expression of CD80 and CD86 proteins in DCs (13, 62). CCL19 was one of the ligands of CCR7 that was widely expressed by DCs and T cells (63), suggesting that fibroblasts might communicate with these cells frequently in psoriasis. Elevated BAFF expression was strongly correlated with male psoriatic arthritis (PsA) activity and the progression of rheumatoid arthritis (64, 65), implying a consistent response of fibroblasts to inflammation throughout the body. All these results underlined dysregulated ECM and immune regulation of fibroblasts in psoriasis.
Of note, Rojahn et al. reported a new inflammatory fibroblast subpopulation only present in the upper dermis of AD lesions, marked by COL6A5 and COL18A1. These fibroblasts expressed inflammatory cytokines, including CCL2, CCL19, and IL32, similar to inflammatory keratinocytes. The study further confirmed that fibroblasts were juxtaposed to CD3+ T cells, indicating that these inflammatory fibroblasts were engaged in T cell recruitment and organization (10). The special role of this group of cells is worth studying. In addition, like upper keratinocytes and melanocytes, fibroblasts also secret CXCL9/10 in vitiligo. Xu et al. found that IFNγ-responsive fibroblasts were responsible for melanocyte loss via recruiting CD8± T cells through single-cell analysis, along with cell-type-specific genetic knockouts and engraftment experiments (66). These findings suggested that skin stromal cells induced a specific immune milieu via secreting the same regulatory factors in an inflammatory response.
Endothelial Cells in Regulating Leukocyte Adhesion and Migration
There are few endothelial cells in the dermis that are not even detected in studies. Endothelial cells in the healthy adult dermis constitute the vascular endothelium (PECAM1, EMCN, SELE, CD93, CLDN5, VWF, and CDH5) and lymphatic endothelium (LYVE1, PDPN, PROX1, and CLDN5) (6, 8, 18). Reynolds et al. reported that some vascular endothelial cells, which expressed γ-synuclein (SNCG) and high level of ACKR1 (the venular capillary marker), might function as postcapillary venular cells. They found that ACKR1+ endothelial cells and CXCL8 (IL8)+ macrophages interacted with each other by cell communication analysis and further confirmed this finding with immunofluorescence staining in situ (6). Besides, ACKR1+ endothelial cells coexpressed inflammatory cytokines, chemokines, and leukocyte adhesion molecules including IL6, IL33, SELE, and ICAM1, implying that they might regulate leukocyte adhesion and migration (6).
Conclusion
Although many technical issues remain to be resolved, scRNA-seq has demonstrated alterations in skin-intrinsic stromal cells in many inflammatory diseases (9). Thanks to scRNA-seq, we can precisely and systematically understand the heterogeneity of skin resident stromal cells, their pathological responses, and their interactions with immune cells in inflamed skin (Table 2). Skin stromal cells played a feedforward role by expressing chemokines, (pro)inflammatory factors, presenting antigens to immune cells, or limiting inflammation by expression regulators (Figure 1). These findings will greatly accelerate the development of personalized diagnostics and precision treatment.
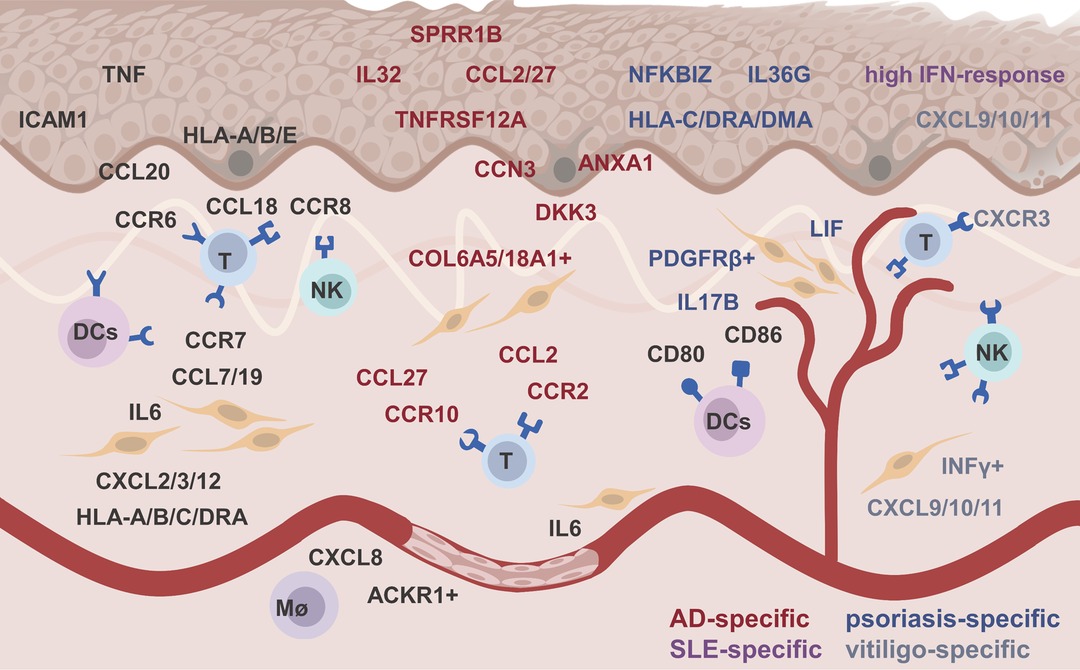
Figure 1. The feedforward role of skin stromal cell in inflamed skin. Black, red, blue, purple, and gray words represented ubiquitous, atopic dermatitis -specific, psoriasis-specific, systemic lupus erythematosus-specific, and vitiligo-specific altered genes, respectively.
Author Contributions
BL drafted and wrote the initial manuscript. AL, JX, and YC reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81872516, 82173418, and 82103100).
Conflict of Interest
The reviewer YZ declared a shared parent affiliation with the authors BL, AL, and YC to the handling editor at the time of review.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wong R, Geyer S, Weninger W, Guimberteau JC, Wong JK. The dynamic anatomy and patterning of skin. Exp Dermatol. (2016) 25(2):92–8. doi: 10.1111/exd.12832
2. Schwingen J, Kaplan M, Kurschus FC. Review-current concepts in inflammatory skin diseases evolved by transcriptome analysis: in-depth analysis of atopic dermatitis and psoriasis. Int J Mol Sci. (2020) 21(3):699. doi: 10.3390/ijms21030699
3. Theocharidis G, Tekkela S, Veves A, McGrath JA, Onoufriadis A. Single-cell transcriptomics in human skin research: available technologies, technical considerations and disease applications. Exp Dermatol. (2022) 31(5):655–73. doi: 10.1111/exd.14547
4. Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. (2017) 14(5):637–50. doi: 10.1080/15476286.2016.1201618
5. Dubois A, Gopee N, Olabi B, Haniffa M. Defining the skin cellular community using single-cell genomics to advance precision medicine. J Invest Dermatol. (2021) 141(2):255–64. doi: 10.1016/j.jid.2020.05.104
6. Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, et al. Developmental cell programs are co-opted in inflammatory skin disease. Science. (2021) 371(6527):eaba6500. doi: 10.1126/science.aba6500
7. Bangert C, Rindler K, Krausgruber T, Alkon N, Thaler FM, Kurz H, et al. Persistence of mature dendritic cells, T(H)2A, and Tc2 cells characterize clinically resolved atopic dermatitis under IL-4Rα blockade. Sci Immunol. (2021) 6(55):eabe2749. doi: 10.1126/sciimmunol.abe2749
8. Hughes TK, Wadsworth MH 2nd, Gierahn TM, Do T, Weiss D, Andrade PR, et al. Second-strand synthesis-based massively parallel scRNA-seq reveals cellular states and molecular features of human inflammatory skin pathologies. Immunity. (2020) 53(4):878–94.e7. doi: 10.1016/j.immuni.2020.09.015
9. Kim D, Chung KB, Kim TG. Application of single-cell RNA sequencing on human skin: technical evolution and challenges. J Dermatol Sci. (2020) 99(2):74–81. doi: 10.1016/j.jdermsci.2020.06.002
10. Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J Allergy Clin Immunol. (2020) 146(5):1056–69. doi: 10.1016/j.jaci.2020.03.041
11. Rindler K, Krausgruber T, Thaler FM, Alkon N, Bangert C, Kurz H, et al. Spontaneously resolved atopic dermatitis shows melanocyte and immune cell activation distinct from healthy control skin. Front Immunol. (2021) 12:630892. doi: 10.3389/fimmu.2021.630892
12. Rozenblatt-Rosen O, Shin JW, Rood JE, Hupalowska A, Regev A, Heyn H. Building a high-quality human cell atlas. Nat Biotechnol. (2021) 39(2):149–53. doi: 10.1038/s41587-020-00812-4
13. Gao Y, Yao X, Zhai Y, Li L, Li H, Sun X, et al. Single cell transcriptional zonation of human psoriasis skin identifies an alternative immunoregulatory axis conducted by skin resident cells. Cell Death Dis. (2021) 12(5):450. doi: 10.1038/s41419-021-03724-6
14. Ascensión AM, Fuertes-Álvarez S, Ibañez-Solé O, Izeta A, Araúzo-Bravo MJ. Human dermal fibroblast subpopulations are conserved across single-cell RNA sequencing studies. J Invest Dermatol. (2021) 141(7):1735–44.e35. doi: 10.1016/j.jid.2020.11.028
15. Thompson SM, Phan QM, Winuthayanon S, Driskell IM, Driskell RR. Parallel single cell multi-omics analysis of neonatal skin reveals transitional fibroblast states that restricts differentiation into distinct fates. J Invest Dermatol. (2021) 17:S0022-202X(21)02608-7. doi: 10.1016/j.jid.2021.11.032
16. Gellatly KJ, Strassner JP, Essien K, Refat MA, Murphy RL, Coffin-Schmitt A, et al. scRNA-seq of human vitiligo reveals complex networks of subclinical immune activation and a role for CCR5 in T(reg) function. Sci Transl Med. (2021) 13(610):eabd8995. doi: 10.1126/scitranslmed.abd8995
17. He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol. (2020) 145(6):1615–28. doi: 10.1016/j.jaci.2020.01.042
18. Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, et al. Transcriptional programming of normal and inflamed human epidermis at single-cell resolution. Cell Rep. (2018) 25(4):871–83. doi: 10.1016/j.celrep.2018.09.006
19. Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. (2013) 504(7479):277–81. doi: 10.1038/nature12783
20. Korosec A, Frech S, Gesslbauer B, Vierhapper M, Radtke C, Petzelbauer P, et al. Lineage identity and location within the dermis determine the function of papillary and reticular fibroblasts in human skin. J Invest Dermatol. (2019) 139(2):342–51. doi: 10.1016/j.jid.2018.07.033
21. Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, et al. Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations. J Invest Dermatol. (2018) 138(4):811–25. doi: 10.1016/j.jid.2018.01.016
22. Solé-Boldo L, Raddatz G, Schütz S, Mallm JP, Rippe K, Lonsdorf AS, et al. Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol. (2020) 3(1):188. doi: 10.1038/s42003-020-0922-4
23. Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol. (2018) 9:1298. doi: 10.3389/fimmu.2018.01298
24. Furue K, Ulzii D, Tanaka Y, Ito T, Tsuji G, Kido-Nakahara M, et al. Pathogenic implication of epidermal scratch injury in psoriasis and atopic dermatitis. J Dermatol. (2020) 47(9):979–88. doi: 10.1111/1346-8138.15507
25. Furue K, Ito T, Tsuji G, Nakahara T, Furue M. The CCL20 and CCR6 axis in psoriasis. Scand J Immunol. (2020) 91(3):e12846. doi: 10.1111/sji.12846
26. Bollag WB, Aitkens L, White J, Hyndman KA. Aquaporin-3 in the epidermis: more than skin deep. Am J Physiol Cell Physiol. (2020) 318(6):C1144–53. doi: 10.1152/ajpcell.00075.2020
27. Scholz GM, Sulaiman NS, Al Baiiaty S, Kwa MQ, Reynolds EC. A novel regulatory relationship between RIPK4 and ELF3 in keratinocytes. Cell Signal. (2016) 28(12):1916–22. doi: 10.1016/j.cellsig.2016.09.006
28. Kim Y, Park JY, Kim H, Chung DK. Differential role of lipoteichoic acids isolated from Staphylococcus aureus and Lactobacillus plantarum on the aggravation and alleviation of atopic dermatitis. Microb Pathog. (2020) 147:104360. doi: 10.1016/j.micpath.2020.104360
29. Meyer N, Zimmermann M, Bürgler S, Bassin C, Woehrl S, Moritz K, et al. IL-32 is expressed by human primary keratinocytes and modulates keratinocyte apoptosis in atopic dermatitis. J Allergy Clin Immunol. (2010) 125(4):858–65.e10. doi: 10.1016/j.jaci.2010.01.016
30. Morrison G, Kilanowski F, Davidson D, Dorin J. Characterization of the mouse beta defensin 1, Defb1, mutant mouse model. Infect Immun. (2002) 70(6):3053–60. doi: 10.1128/iai.70.6.3053-3060.2002
31. Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Persistence of mature dendritic cells, TH2A, and Tc2 cells characterize clinically resolved atopic dermatitis under IL-4Rα blockade. J Allergy Clin Immunol. (2020) 146(5):1056–69. doi: 10.1016/j.jaci.2020.03.041
32. Kim J, Lee J, Kim HJ, Kameyama N, Nazarian R, Der E, et al. Single-cell transcriptomics applied to emigrating cells from psoriasis elucidate pathogenic versus regulatory immune cell subsets. J Allergy Clin Immunol. (2021) 148(5):1281–92. doi: 10.1016/j.jaci.2021.04.021
33. Black AP, Ardern-Jones MR, Kasprowicz V, Bowness P, Jones L, Bailey AS, et al. Human keratinocyte induction of rapid effector function in antigen-specific memory CD4 + and CD8+ T cells. Eur J Immunol. (2007) 37(6):1485–93. doi: 10.1002/eji.200636915
34. Furue K, Yamamura K, Tsuji G, Mitoma C, Uchi H, Nakahara T, et al. Highlighting interleukin-36 signalling in plaque psoriasis and pustular psoriasis. Acta Derm Venereol. (2018) 98(1):5–13. doi: 10.2340/00015555-2808
35. Zeng F, Liu H, Lu D, Liu Q, Chen H, Zheng F. Integrated analysis of gene expression profiles identifies transcription factors potentially involved in psoriasis pathogenesis. J Cell Biochem. (2019) 120(8):12582–94. doi: 10.1002/jcb.28525
36. Sobolev VV, Zolotorenko AD, Soboleva AG, Elkin AM, Il’ina SA, Serov DN, et al. Effects of expression of transcriptional factor AP-1 FOSL1 gene on psoriatic process. Bull Exp Biol Med. (2011) 150(5):632–4. doi: 10.1007/s10517-011-1208-0
37. Meng J, Chen FR, Yan WJ, Lin YK. MiR-15a-5p targets FOSL1 to inhibit proliferation and promote apoptosis of keratinocytes via MAPK/ERK pathway. J Tissue Viability. (2021) 30(4):544–51. doi: 10.1016/j.jtv.2021.08.006
38. Li C, Xu M, Coyne J, Wang WB, Davila ML, Wang Y, et al. Psoriasis-associated impairment of CCL27/CCR10-derived regulation leads to IL-17A/IL-22-producing skin T-cell overactivation. J Allergy Clin Immunol. (2021) 147(2):759–63.e9. doi: 10.1016/j.jaci.2020.05.044
39. Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. (2019) 20(7):915–27. doi: 10.1038/s41590-019-0386-1
40. Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight. (2017) 22(8):3879. doi: 10.1172/jci.insight.93009
41. Arakawa A, Siewert K, Stöhr J, Besgen P, Kim SM, Rühl G, et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J Exp Med. (2015) 212(13):2203–12. doi: 10.1084/jem.20151093
42. van den Bogaard EH, Tijssen HJ, Rodijk-Olthuis D, van Houwelingen KP, Coenen MJ, Marget M, et al. Cell surface expression of HLA-Cw6 by human epidermal keratinocytes: positive regulation by cytokines, lack of correlation to a variant upstream of HLA-C. J Invest Dermatol. (2016) 136(9):1903–6. doi: 10.1016/j.jid.2016.05.112
43. Bitton A, Avlas S, Reichman H, Itan M, Karo-Atar D, Azouz NP, et al. A key role for IL-13 signaling via the type 2 IL-4 receptor in experimental atopic dermatitis. Sci Immunol. (2020) 228(2):476–84. doi: 10.1126/sciimmunol.aaw2938
44. Amniai L, Ple C, Barrier M, de Nadai P, Marquillies P, Vorng H, et al. Natural killer cells from allergic donors are defective in their response to CCL18 chemokine. Int J Mol Sci. (2021) 135(8):1996–2004. doi: 10.3390/ijms22083879
45. Lin Z, Natesan V, Shi H, Hamik A, Kawanami D, Hao C, et al. A novel role of CCN3 in regulating endothelial inflammation. J Cell Commun Signal. (2010) 4(3):141–53. doi: 10.1007/s12079-010-0095-x
46. Eslani M, Putra I, Shen X, Hamouie J, Afsharkhamseh N, Besharat S, et al. Corneal mesenchymal stromal cells are directly antiangiogenic via PEDF and sFLT-1. Invest Ophthalmol Vis Sci. (2017) 58(12):5507–17. doi: 10.1167/iovs.17-22680
47. Jia Y, Morand EF, Song W, Cheng Q, Stewart A, Yang YH. Regulation of lung fibroblast activation by annexin A1. J Cell Physiol. (2013) 228(2):476–84. doi: 10.1002/jcp.24156
48. Meister M, Tounsi A, Gaffal E, Bald T, Papatriantafyllou M, Ludwig J, et al. Self-antigen presentation by keratinocytes in the inflamed adult skin modulates T-Cell autor-reactivity. J Invest Dermatol. (2015) 135(8):1996–2004. doi: 10.1038/jid.2015.130
49. Wang HX, Sharma C, Knoblich K, Granter SR, Hemler ME. EWI-2 negatively regulates TGF-β signaling leading to altered melanoma growth and metastasis. Cell Res. (2015) 25(3):370–85. doi: 10.1038/cr.2015.17
50. Schauberger E, Peinhaupt M, Cazares T, Lindsley AW. Lipid mediators of allergic disease: pathways, treatments, and emerging therapeutic targets. Curr Allergy Asthma Rep. (2016) 16(7):48. doi: 10.1007/s11882-016-0628-3
51. Schumacher M, Schuster C, Rogon ZM, Bauer T, Caushaj N, Baars S, et al. Efficient keratinocyte differentiation strictly depends on JNK-induced soluble factors in fibroblasts. J Invest Dermatol. (2014) 134(5):1332–41. doi: 10.1038/jid.2013.535
52. Taniguchi K, Arima K, Masuoka M, Ohta S, Shiraishi H, Ontsuka K, et al. Periostin controls keratinocyte proliferation and differentiation by interacting with the paracrine IL-1α/IL-6 loop. J Invest Dermatol. (2014) 134(5):1295–304. doi: 10.1038/jid.2013.500
53. Chambers ES, Vukmanovic-Stejic M. Skin barrier immunity and ageing. Immunology. (2020) 160(2):116–25. doi: 10.1111/imm.13152
54. Tabib T, Morse C, Wang T, Chen W, Lafyatis R. SFRP2/DPP4 and FMO1/LSP1 define major fibroblast populations in human skin. J Invest Dermatol. (2018) 138(4):802–10. doi: 10.1016/j.jid.2017.09.045
55. Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun. (2019) 10(1):650. doi: 10.1038/s41467-018-08247-x
56. Vorstandlechner V, Laggner M, Kalinina P, Haslik W, Radtke C, Shaw L, et al. Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. Faseb j. (2020) 34(3):3677–92. doi: 10.1096/fj.201902001RR
57. Wang T, Zhou Z, Luo E, Zhong J, Zhao D, Dong H, et al. Comprehensive RNA sequencing in primary murine keratinocytes and fibroblasts identifies novel biomarkers and provides potential therapeutic targets for skin-related diseases. Cell Mol Biol Lett. (2021) 26(1):42. doi: 10.1186/s11658-021-00285-6
58. Jiang D, Rinkevich Y. Distinct fibroblasts in scars and regeneration. Curr Opin Genet Dev. (2021) 70:7–14. doi: 10.1016/j.gde.2021.04.005
59. Fleury S, Thibodeau J, Croteau G, Labrecque N, Aronson HE, Cantin C, et al. Sékaly: HLA-DR polymorphism affects the interaction with CD4. J Exp Med. (1995) 182(3):733–41. doi: 10.1084/jem.182.3.733
60. Sun J, Leahy DJ, Kavathas PB. Interaction between CD8 and major histocompatibility complex (MHC) class I mediated by multiple contact surfaces that include the alpha 2 and alpha 3 domains of MHC class I. J Exp Med. (1995) 182(5):1275–80. doi: 10.1084/jem.182.5.1275
61. Nguyen HN, Noss EH, Mizoguchi F, Huppertz C, Wei KS, Watts GFM, et al. Autocrine loop involving IL-6 family member LIF, LIF receptor, and STAT4 drives sustained fibroblast production of inflammatory mediators. Immunity. (2017) 46(2):220–32. doi: 10.1016/j.immuni.2017.01.004
62. Shan B, Shao M, Zhang Q, Hepler C, Paschoal VA, Barnes SD, et al. Perivascular mesenchymal cells control adipose-tissue macrophage accrual in obesity. Nat Metab. (2020) 2(11):1332–49. doi: 10.1038/s42255-020-00301-7
63. Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. (2008) 8(5):362–71. doi: 10.1038/nri2297
64. Pongratz G, Straub RH, Schölmerich J, Fleck M, Härle P. Serum BAFF strongly correlates with PsA activity in male patients only – is there a role for sex hormones? Clin Exp Rheumatol. (2010) 28(6):813–9.20863443
65. Reyes LI, León F, González P, Rozas MF, Labarca C, Segovia A, et al. Dexamethasone inhibits BAFF expression in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Cytokine. (2008) 42(2):170–8. doi: 10.1016/j.cyto.2007.12.005
66. Xu Z, Chen D, Hu Y, Jiang K, Huang H, Du Y, et al. Anatomically distinct fibroblast subsets determine skin autoimmune patterns. Nature. (2022) 601(7891):118–24. doi: 10.1038/s41586-021-04221-8
Keywords: scRNA-seq, skin resident cells, skin stromal cells, immunoregulatory axis, cellular communication
Citation: Liu B, Li A, Xu J and Cui Y (2022) Single-Cell Transcriptional Analysis Deciphers the Inflammatory Response of Skin-Resident Stromal Cells. Front. Surg. 9:935107. doi: 10.3389/fsurg.2022.935107
Received: 3 May 2022; Accepted: 23 May 2022;
Published: 14 June 2022.
Edited by:
Ronghua Yang, First People’s Hospital of Foshan, ChinaReviewed by:
Jinhua Xu, Huashan Hospital, Fudan University, ChinaYa-gang Zuo, Peking Union Medical College Hospital (CAMS), China
Aie Xu, Third People’s Hospital of Hangzhou, China
Copyright © 2022 Liu, Li, Xu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Cui d3VodWN1aXlvbmdAdmlwLjE2My5jb20=
Specialty section: This article was submitted to Reconstructive and Plastic Surgery, a section of the journal Frontiers in Surgery
 Baoyi Liu1,2
Baoyi Liu1,2 Ang Li
Ang Li Jingkai Xu
Jingkai Xu Yong Cui
Yong Cui