- 1Center for Genomic and Personalized Medicine, Guangxi key Laboratory for Genomic and Personalized Medicine, Guangxi Collaborative Innovation Center for Genomic and Personalized Medicine, Guangxi Medical University, Nanning, China
- 2Institute of Urology and Nephrology, The First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 3Department of Urology, The First Affiliated Hospital of Wannan Medical College (Yijishan Hospital of Wannan Medical College), Wuhu, China
- 4Department of Urology, The Second Affiliated Hospital of Guangxi Medical University, Nanning, China
Purpose: There is evidence that the Crystallin Alpha B (CRYAB) gene is involved in the regulation of the tumor microenvironment and influences tumor prognosis in some cancers. However, the role of CRYAB gene in prognosis and immunology in pan-cancer is still unclear.
Methods: In this study, we analyzed the transcriptional profiles and survival data of cancer patients from The Cancer Genome Atlas (TCGA) database. CRYAB gene and its relationships with pan-cancer were analyzed using R packages, TIMER2.0, GEPIA2, Sangerbox, UALCAN, cBioPortal, ESTIMATE algorithm, and STRING. Besides, real-time fluorescence quantitative polymerase chain reaction (RT-qPCR) was utilized to detect CRYAB expression in KIRC and a human KIRC cell line (Caki-1).
Results: We found that CRYAB expression was different in tumors and adjacent tumors in human cancers, affecting patients’ prognosis in 15 cancer types. Additionally, CRYAB expression significantly correlated with tumor microenvironment (TME), immune checkpoints (ICP), tumor mutational burden (TMB), and microsatellite instability (MSI) in human cancers. Besides, CRYAB expression was positively associated with the immune infiltration of cancer-associated fibroblasts (CAFs) and endothelial cells in most human cancers. Based on enrichment analysis, the most prevalent CRYAB gene mechanism in malignant tumors may be through anti-apoptotic activity. Moreover, some FDA-approved drugs were found to be associated with CRYAB and might be potential cancer therapeutic candidates.
Conclusions: CRYAB is a crucial component of the TME and influences immune cell infiltration, making it a promising biomarker to assess immune infiltration and prognosis in many malignancies.
Introduction
With the rapid growth and aging of the global population, cancer and related deaths are increasing (1). Therefore, there is an urgent need for cancer management across the globe. Nonetheless, it is well known that tumors are difficult to conquer due to their heterogeneity, and because of the difference of origin location, cell type, and gene mutation form, tumors can show different morphology. These factors also affect the therapeutic effect of tumors. Although it has been confirmed that many genetic changes directly lead to phenotypic changes, the complex molecular mechanisms in many tumor lineages are still not fully elucidated (2). TCGA and Gene Expression Omnibus (GEO) analyzed a large number of human tumors to find molecular alterations at the DNA, RNA, protein, and epigenetic levels. The pan-cancer analysis uses these databases to find common features from different tumors. The analysis of molecular changes and their functions in different tumors will in the future tell us how to apply effective therapies to tumors with similar genetic phenotypes (3).
CRYAB comprises three domains: N-terminal, central, and C-terminal (4). As a molecular chaperone, the CRYAB gene could prevent damages induced by heat shock, radiation, and oxidative stress, thereby escaping cell apoptosis and promoting cell survival (5). Recent studies have confirmed that the CRYAB gene regulates tumor invasion and metastasis in colorectal cancer and gastric cancer (6, 7). Studies have shown that tumors with high expression of the CRYAB gene have a poor survival rate (8–10), implying that the CRYAB gene is a potential biomarker. However, the role of the CRYAB gene in pan-cancer prognosis has not been reported.
CRYAB gene could suppress apoptosis of tumor cells and regulate the vascular endothelial growth factor (VEGF) causing tumorigenesis in breast cancer (11, 12). Besides, increasing evidence indicates that the CRYAB gene potentially modulates tumor development via the TME (13). One study (14) revealed that M2 macrophages in the TME promote lung cancer progression by upregulating CRYAB expression. Researchers (6) also discovered that the CRYAB gene increases colorectal cancer progression by inducing epithelial-mesenchymal transition (EMT). These studies implied that the CRYAB gene may have functional roles in M2 macrophages and EMT in lung cancer and colorectal cancer. With the development of tumor immunology and single-cell sequencing technology, researchers have proposed that tumor-associated macrophages play an important role in the development of cancer, and CAFs can induce EMT in human tongue squamous cell carcinoma (15–17). Tumor-associated macrophages, CAFs, and other tumor-infiltrating immune cells are important components of the tumor microenvironment. They are present in all stages of cancer and play a key role in the tumor development, progression, and metastasis (15, 18). CRYAB's effect on immune infiltration will greatly affect cancer progression and needs to be taken seriously as part of pan-cancer research. Although there has been evidence of CRYAB's effect on tumor prognosis and tumor-infiltrating lymphocytes, it has only been limited to a few tumors, and there has been no comprehensive pan-cancer analysis based on TCGA and GO. As such, we evaluated the function of the CRYAB gene in human tumors using a comprehensive pan-cancer analysis. We first evaluated the expression of CRYAB gene in pan-cancer to gain a general understanding of the gene function, and then investigated the effect of CRYAB expression on tumor prognosis and immune infiltration. In addition, we analyzed the molecular pathways of CRYAB-related genes and the drug sensitivity of CRYAB gene. Further, RT-qPCR was utilized to detect CRYAB expression in KIRC and Caki-1 to confirm the bioinformatics results. The process of this pan-cancer analysis is shown in Figure 1.
Materials and methods
Data acquisition and processing
We got the data from the TCGA via the UCSC Xena database (http://xena.ucsc.edu/), including transcriptional profiles and survival data. The R (version 4.1.3; The R Foundation for Statistical Computing, Vienna, Austria) language was used to process the original data. Online web tools were also used for data analysis.
Expression analysis of CRYAB gene
To compare the expression of CRYAB in pan-cancer and their normal tissues, the TIMER2.0 database (http://timer.cistrome.org/) was employed. In the “Gene_DE” module, we entered the gene name “CRYAB” to obtain the results. Further, CRYAB gene expression was analyzed using the GTEx database as the control by the Sangerbox website (http://sangerbox.com). The relationship between CRYAB expression and the cancer pathological stages was examined using the “Stage Plot” module of GEPIA2 (http://gepia2.Cancer-pku.cn/). On the UALCAN database (http://ualcan.path.uab.edu/analysis.html) homepage, the “Clinical Proteomic Tumor Analysis Consortium (CPTAC) analysis” module was selected, then we entered the gene name “CRYAB” to explore the total protein and phosphoprotein of CRYAB protein in different cancers.
Prognostic and survival analysis of CRYAB gene
We obtained survival data from the TCGA database. The data included complete follow-up records of patients and CRYAB expression profiles. Then, the Kaplan-Meier method and log-rank test were used for survival analysis (p < 0.05). The median expression of the CRYAB gene was selected as the cut-off value for the human cancer dichotomy, hence categorizing patients into high and low risk groups. Subsequently, the R package “survminer” and “survival” were used to plot the survival curve. Cox analysis was also performed using the R package “survival” and “forestplot”. Lastly, we obtained respective outcomes of overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI).
Correlation analysis of CRYAB expression with immune infiltration
CRYAB expression and immune infiltration in human malignancies were investigated using the “Immune-Gene” module of TIMER2.0 (http://timer.cistrome.org/). The algorithms in the above analysis process included QUANTISEQ, TIDE, XCELL, MCPCOUNTER, EPIC, TIMER, and CIBERSORT. The P-values and partial correlation (cor) values were calculated by Spearman's rank correlation test.
Correlation analysis of CRYAB expression with immune checkpoints (ICP), tumor mutational burden (TMB), and microsatellite instability (MSI)
SangerBox website (http://sangerbox.com) was used to explore the association between CRYAB expression and immune checkpoints (ICP), tumor mutational burden (TMB), and microsatellite instability (MSI) in human tumors. In the “Gene+” module of the Sangerbox website, we entered the gene name “CRYAB”, and got the results by using the Pearson correlation test.
Enrichment analysis of CRYAB-related gene
On the website of STRING (https://cn.string-db.org/), we obtained the top 50 CRYAB-binding proteins. First, the gene name “CRYAB” was entered and the organism of “Homo sapiens” was searched. Then the parameters of “the full STRING network”, “evidence”, “experiments”, “low confidence (0.150)”, and “no more than 50 interactors” were set. On the GEPIA2 website, the top 200 CRYAB-correlated targeting genes were obtained from the “Similar Gene Detection” module by setting TCGA tumor and TCGA normal. These two datasets were integrated for gene ontology (GO) enrichment and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis via R-package “org.Hs.eg.db”, “clusterProfiler”, and “enrichplot”. Lastly, The R package “ggplot2″ was used to realize the visualization of enrichment paths.
Drug sensitive analysis
RNA expression data and drug data from the NCI-60 cell line were obtained from the CellMiner database (https://discover.nci.nih.gov/cellminer/home.do). To ensure the reliability of the analysis results, 792 drugs that have passed clinical trials are and FDA approved were selected. We used Pearson correlation to explore the association between CRYAB expression and drug sensitivity, and p < 0.05 was considered statistically significant.
Clinical sample collection
We collected seven specimens of renal clear cell carcinoma and their adjacent noncancerous tissues from The Second Affiliated Hospital of Guangxi Medical University. The Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University approved the work (Approval Number: 2021-KY(0123)). The study participants signed the informed consent forms.
Cell culture
Both the human KIRC cell line (Caki-1) and the human renal cortex proximal tubule epithelial cells (HK-2) were bought from the American Type Culture Collection. Caki-1 was cultured in McCoy's 5A medium (Procell, Wuhan, China) with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, United States) and placed in an incubator set at 37°C and 5% CO2. HK-2 was cultured in Minimum Essential Medium (MEM, Procell, Wuhan, China) with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, United States) and placed in an incubator set at 37°C and 5% CO2.
RT-qPCR analysis
TRIzol (Beyotime, Shanghai, China) was used to extract total RNA from the tissues and cells. The OD260/280 values were used to detect the total RNA concentration and purity. FastKing gDNA Dispelling RT SuperMix cDNA synthesis Kit (TIANGEN, Beijing, China) was used to synthesize cDNAs, and at the same time, it was used to remove the residual genomes. Polymerase Chain Reaction was performed by using FastStart Essential DNA Green Mster (Roche Diagnostics GmbH, Mannheim, Germany) and LightCycler® 96 Instrument (Roche Diagnostics GmbH, Mannheim, Germany). All experiments were repeated three times. GAPDH was used as an internal standard, and the primer sequences of GAPDH were 5′–GGAGTCCACTGGCGTCTTCA –3′ (Forward Primer) and 5′ –GTCATGAGGCCAGAAATGAAGG– 3′ (Reverse Primer). The primer sequences of CRYAB were 5′ –AGGTGTTGGGAGATGTGATTGA– 3′ (Forward Primer) and 5′ –GGATGAAGTAATGGTGAGAGGGT– 3′ (Reverse Primer). We used the 2−ΔΔCt method to calculate the relative expression of CRYAB.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, United States). The paired T-test was used to analyze the relative expression of CRYAB in tumor tissues and adjacent non-tumor tissues. The Student's t-test was used to analyze the relative expression of CRYAB in Caki-1 and HK-2. P < 0.05 was considered statistically significant.
Results
CRYAB gene expression analysis
The results of TIMER2.0 revealed lower CRYAB gene expression in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) than that in adjacent normal tissues except for cholangiocarcinoma (CHOL), kidney renal clear cell carcinoma (KIRC), and kidney renal papillary cell carcinoma (KIRP) (Figure 2A). Several tumors in TCGA did not have paired normal tissue. The GTEx database was used as a control to further investigate CRYAB gene expression. As shown in Figure 2B, CRYAB gene expression was lower in tumors than that in normal tissues except for CHOL, glioblastoma multiforme (GBM), KIRC, KIRP, brain lower grade glioma (LGG), and pancreatic adenocarcinoma (PAAD). Besides, we analyzed the CRYAB total protein expression in cancers via CPTAC and found that, unlike normal tissues, CRYAB protein expression levels were significantly low in COAD, HNSC, PAAD, OV, UCEC, LUAD, and liver hepatocellular carcinoma (LIHC) except for KIRC (Figure 2C). Further, we examined the relationship between CRYAB gene expression and pathological stages of cancers. CRYAB gene was significantly upregulated in the advanced pathological stage of COAD, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), STAD, ovarian serous cystadenocarcinoma (OV), and BLCA (P < 0.05) (Figure 2D).
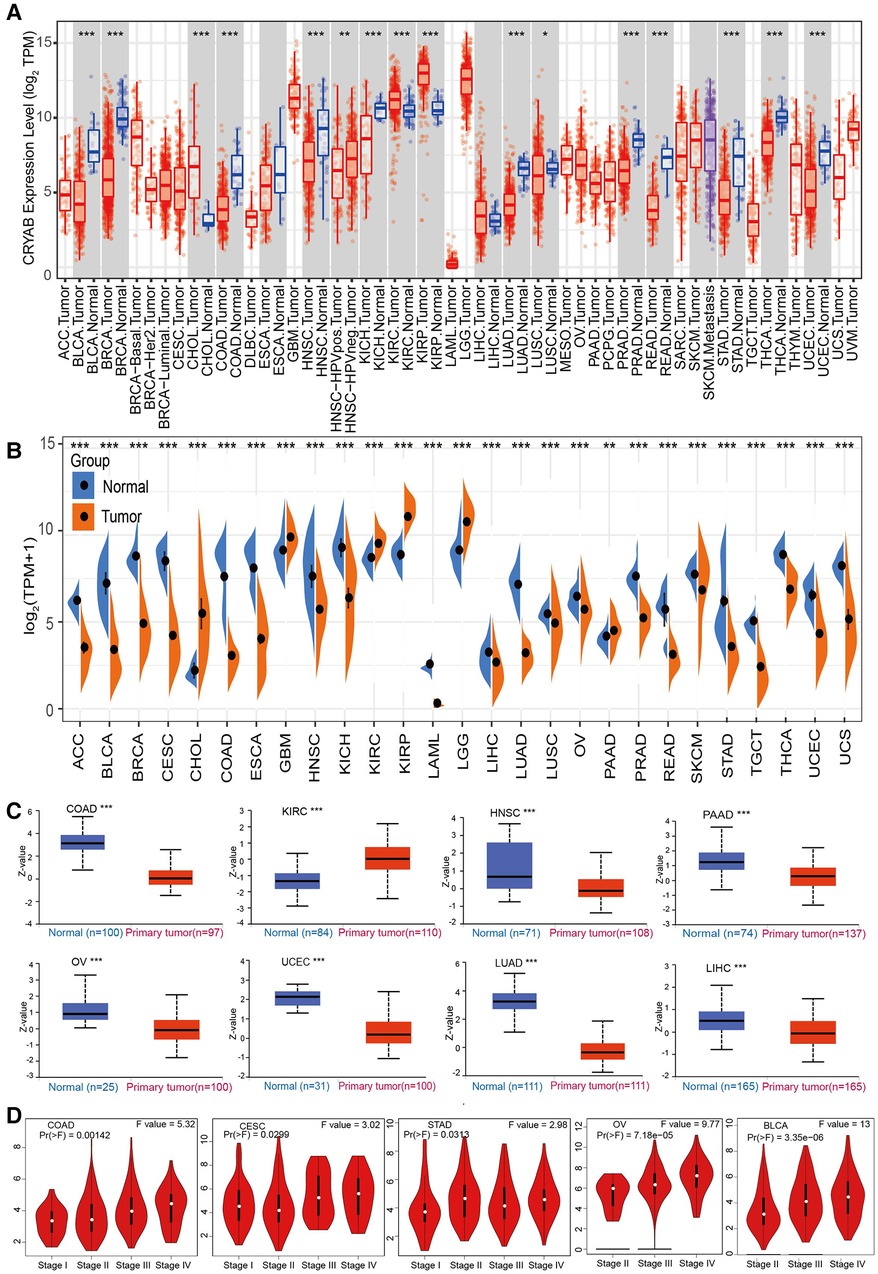
Figure 2. The expression profile of CRYAB in pan-cancer. (A) TIMER2.0 analysis of the CRYAB gene expression in pan-cancer. (B) Sangerbox analysis of CRYAB gene expression in pan-cancer in TCGA. GTEx database served as the control. (C) CRYAB protein expression in COAD, KIRC, HNSC, PAAD, OV, UCEC, LUAD, and LIHC in the CPTAC database. (D) The correlation between CRYAB and cancer pathological stages in COAD, CESC, STAD, OV, and BLCA as determined using GEPIA2 tool. *p < 0.05 **p < 0.01; ***p < 0.001.
Survival analysis of CRYAB in human tumors
To understand the prognostic significance of the CRYAB gene, we analyzed its relevance to survival outcomes in human tumors, including OS, DFI, PFI, and DSS. First, the OS of the CRYAB gene was analyzed using Cox proportional hazards model. Subsequently, we plotted Kaplan–Meier plotter survival curves for cancers with significant prognosis in the TCGA database. Figure 3A showed the results of the Cox proportional hazards model analysis, indicating that the CRYAB gene is a risk factor in BLCA, LIHC, READ, STAD, and UVM, yet a protective factor in GBM, KICH, LGG, and SARC. Cancer patients were categorized into low and high CRYAB expression groups. The Kaplan–Meier method analysis showed that high-CRYAB groups were linked to the poor OS in BLCA (Figure 3B), COAD (Figure 3C), LIHC (Figure 3D), READ (Figure 3F), and OV (Figure 3G), but with a positive prognosis in KICH (Figure 3E). Afterward, the DSS was analyzed based on the data for tumor death in the TCGA database. In the forest plot (Figure 3H), we noted that the CRYAB gene was a risk factor in BLCA, COAD, OV, STAD, UCEC, and UVM, but a protective factor in GBM, KICH, KIRP, LGG, and PRAD. Kaplan–Meier method indicated that high CRYAB expression was remarkably related to poor DSS in BLCA (Figure 3I), COAD (Figure 3J), OV (Figure 3K), and UCEC (Figure 3L), but with an optimistic prognosis in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) (Figure 3M) and KICH (Figure 3N).
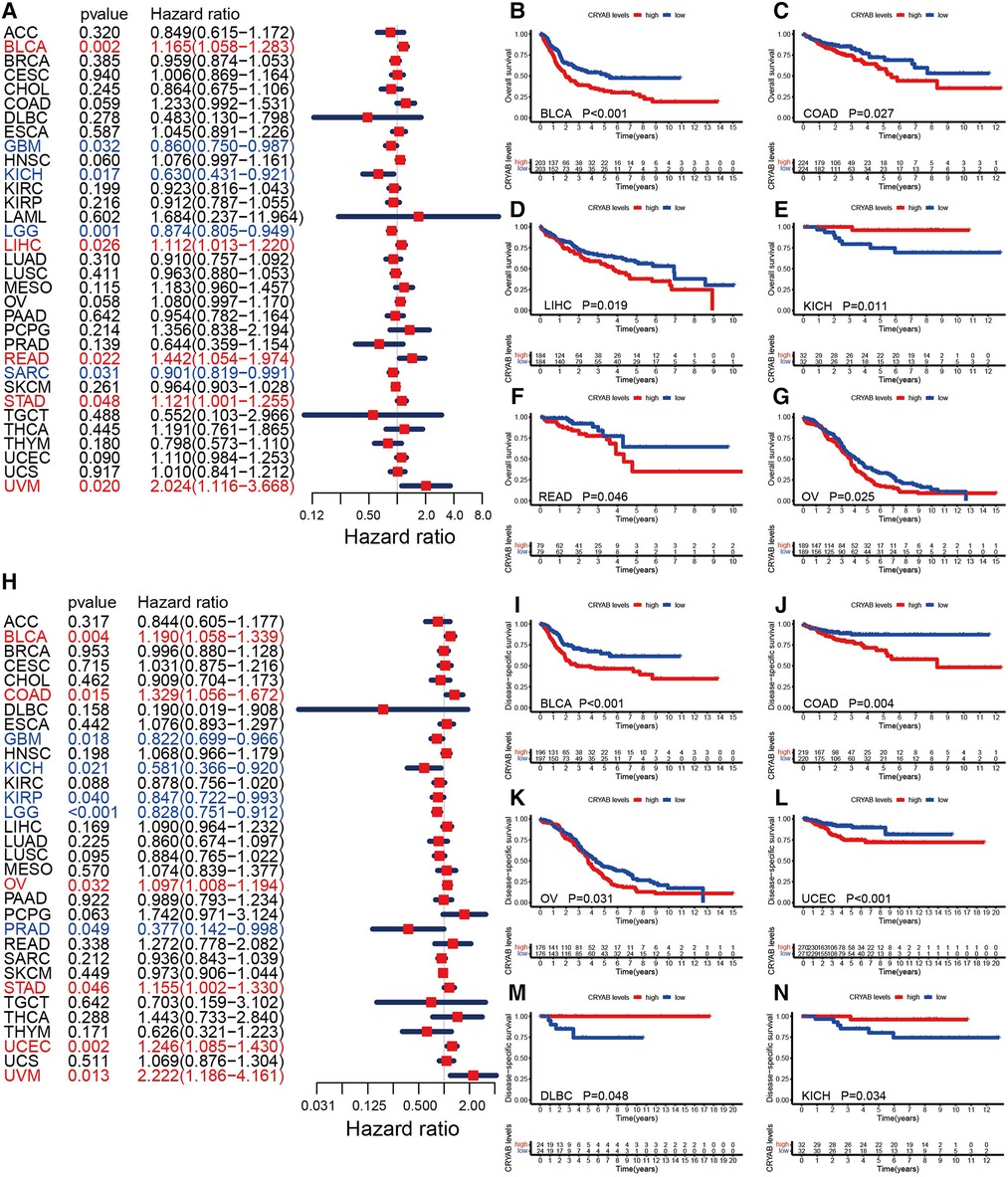
Figure 3. Relationship between CRYAB and OS and DSS in pan-cancer in TCGA database. (A) Cox model analysis of CRYAB for OS in pan-cancer. (B–G) The Kaplan–Meier plotter survival analysis of CRYAB for OS. (H) Cox model analysis of CRYAB for DSS in pan-cancer. (I–N) The Kaplan–Meier plotter survival analysis of CRYAB for DSS.
Furthermore, we investigated the association between the CRYAB gene expression and PFI. The forest plots showed that the CRYAB gene was a risk factor in BLCA, COAD, and OV, but a protective factor in KICH, LGG, PRAD, and skin cutaneous melanoma (SKCM) (Figure 4A). According to the Kaplan-Meier method, high CRYAB expression was linked to poor prognosis in BLCA (Figure 4B), COAD (Figure 4C), and OV (Figure 4D); however, high CRYAB expression was related to optimistic prognosis in KICH (Figure 4E), LGG (Figure 4F), and SKCM (Figure 4G). We also analyzed the effect of CRYAB gene on DFI of cancer patients. Based on forest plot results, the CRYAB gene was a risk factor in HNSC, but a protective factor in PRAD (Figure 4H). According to the Kaplan-Meier analysis of DFI, high CRYAB expression was related to an optimistic prognosis in KIRP (Figure 4I). The above results confirmed that CRYAB gene expression was closely linked to the clinical outcomes of various cancers.
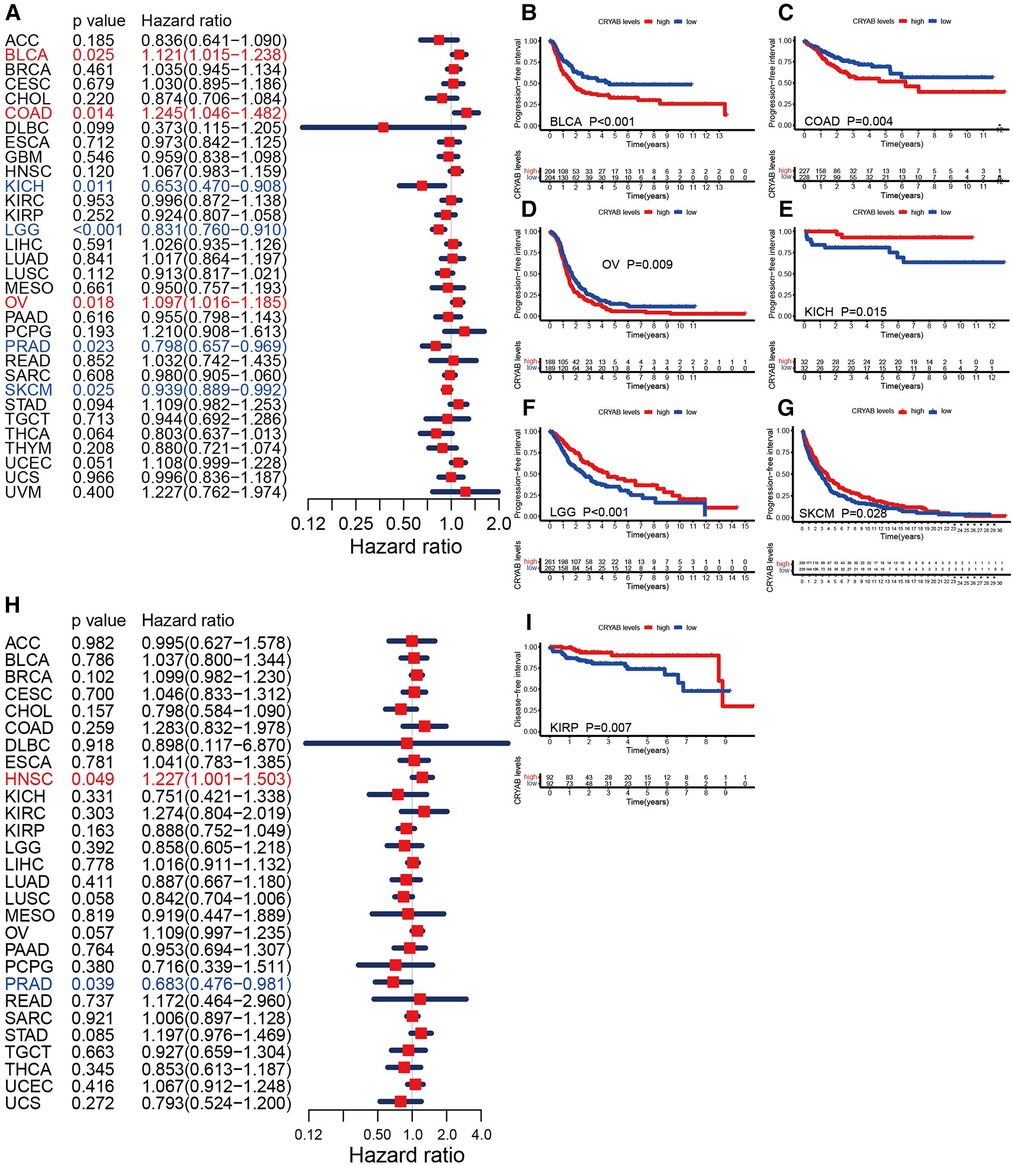
Figure 4. Relationship between CRYAB and PFI and DFI in pan-cancer in TCGA database. (A) Cox model analysis of CRYAB for PFI in pan-cancer. (B–G) The Kaplan–Meier plotter survival analysis of CRYAB for PFI. (H) Cox model analysis of CRYAB for DFI in pan-cancer. (I) The Kaplan–Meier plotter survival analysis of CRYAB for DFI in KIRP.
Association between CRYAB expression and immune infiltration in human tumors
TIMER2.0 was used to investigate the extent of immune infiltration and CRYAB gene expression in human cancers (Figure 5A, Figure 6A, and Supplementary Figure S1). Consequently, the CRYAB gene positively related to CAFs in BLCA, COAD, HNSC, HNSC-HPV-, STAD, TGCT, LUAD, LIHC, PAAD, READ, BRCA-LumA, BRCA-LumB, ESCA, CESC, PRAD, and THYM (Figure 5B–K, Supplementary Figures S1A–F), but negatively related to CAFs in KIRP, GBM, and KIRC (Figure 5L–M, Supplementary Figure S1l). Also, the CRYAB gene positively correlated with endothelial cells in BLCA, COAD, THYM, TGCT, READ, LUAD, STAD, PAAD, BRCA, BRCA-LumA, BRCA-LumB, PCPG, and PRAD (Figure 6B–I, Supplementary Figures S1G–K), but negatively associated with their counterparts in KIRP and KIRC (Figure 6J–K).
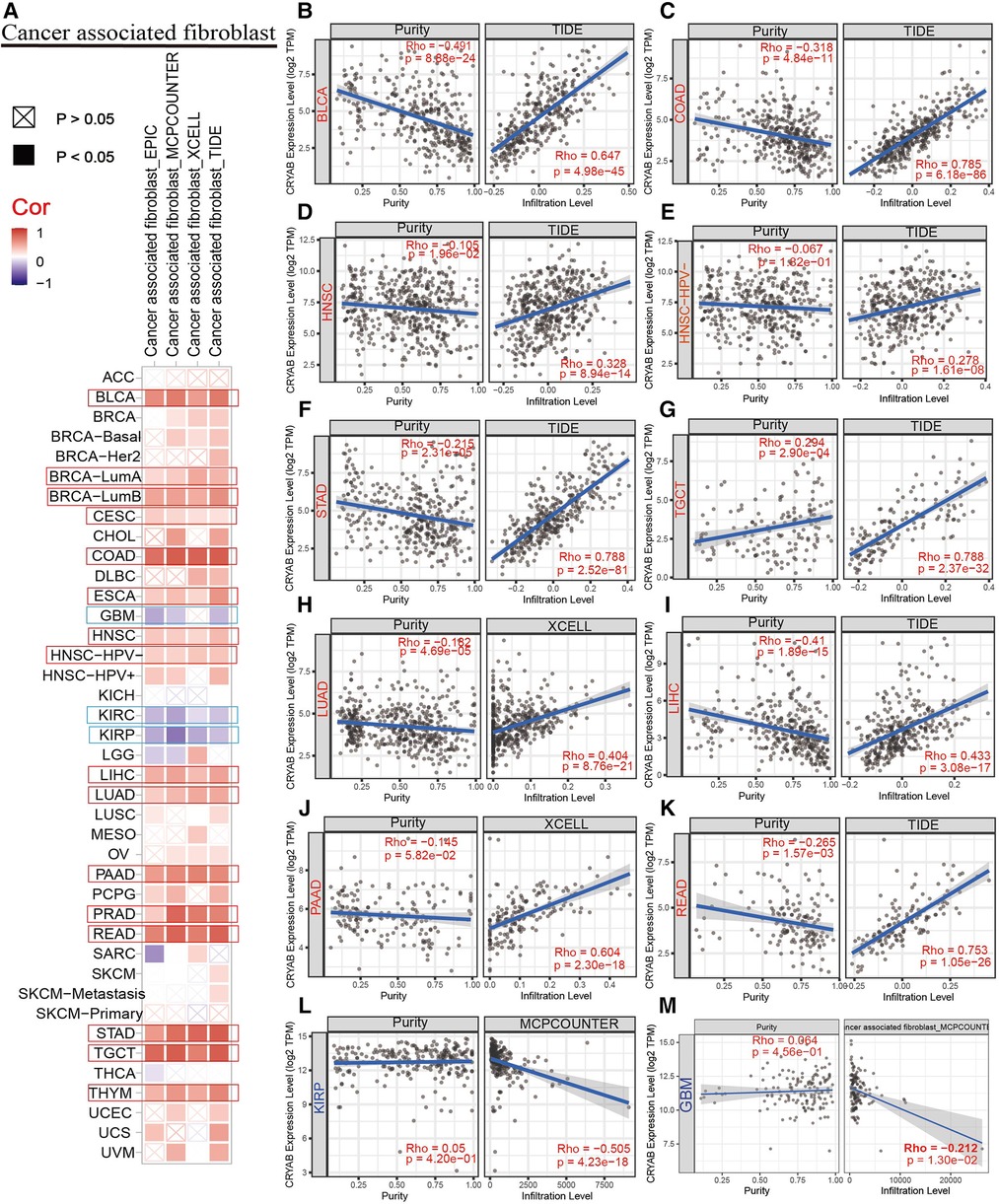
Figure 5. Association between CRYAB expression and CAFs infiltration in pan-cancer. (A) A summary of using different algorithms to explore the associations between CRYAB expression and CAFs infiltration in pan-cancer. The CRYAB gene positively related to CAFs in BLCA (B), COAD (C), HNSC (D), HNSC-HPV- (E), STAD (F), TGCT (G), LUAD (H), LIHC (I), PAAD (J), and READ (K), but negatively related to CAFs in KIRP(L) and GBM(M).
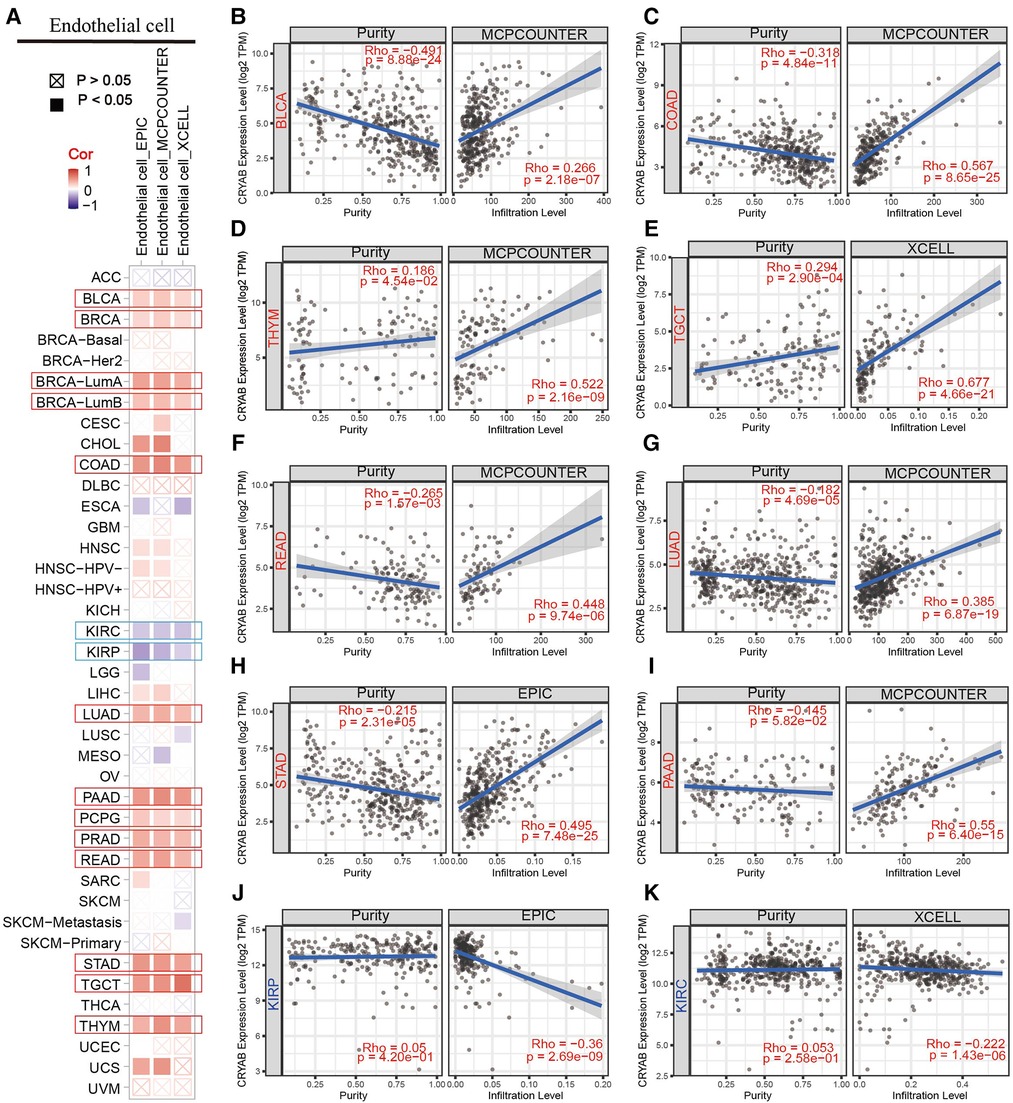
Figure 6. Association between CRYAB expression and infiltration of endothelial cells in pan-cancer. (A) A summary of using different algorithms to explore the associations between CRYAB expression and endothelial cells infiltration in pan-cancer. The CRYAB gene positively correlated with endothelial cells in BLCA (B), COAD (C), THYM (D), TGCT (E), READ (F), LUAD (G), STAD (H), and PAAD (I) but negatively correlated with endothelial cells in KIRP (J) and KIRC (K).
Association between CRYAB expression and immune checkpoints (ICP), major histocompatibility complex (MHC) molecules, tumor mutational burden (TMB), and microsatellite instability (MSI) in human tumors
Sangerbox website was used to evaluate the association between CRYAB expression and immune checkpoint (ICP) genes. As shown in Figure 7A, 47 immune checkpoint genes were evaluated, and CRYAB expression was associated with immune checkpoint genes in different cancers, including BLCA, TGCT, COAD, KIRP, KIRC, SARC, THCA, (acute myeloid leukemia) LAML, READ, and LGG. In BLCA, COAD, LAML, READ, and LGG, CRYAB expression was positively associated with immune checkpoint genes. In TGCT, KIRP, SARC, KIRC, and THCA, CRYAB expression was negatively correlated with immune checkpoints. Using the TISIDB website, the association between CRYAB expression and 21 MHCs were analyzed. CRYAB expression was positively associated with 21 MHCS in BLCA, BRCA, KICH, LIHC, OV, and UVM (Figure 7B).
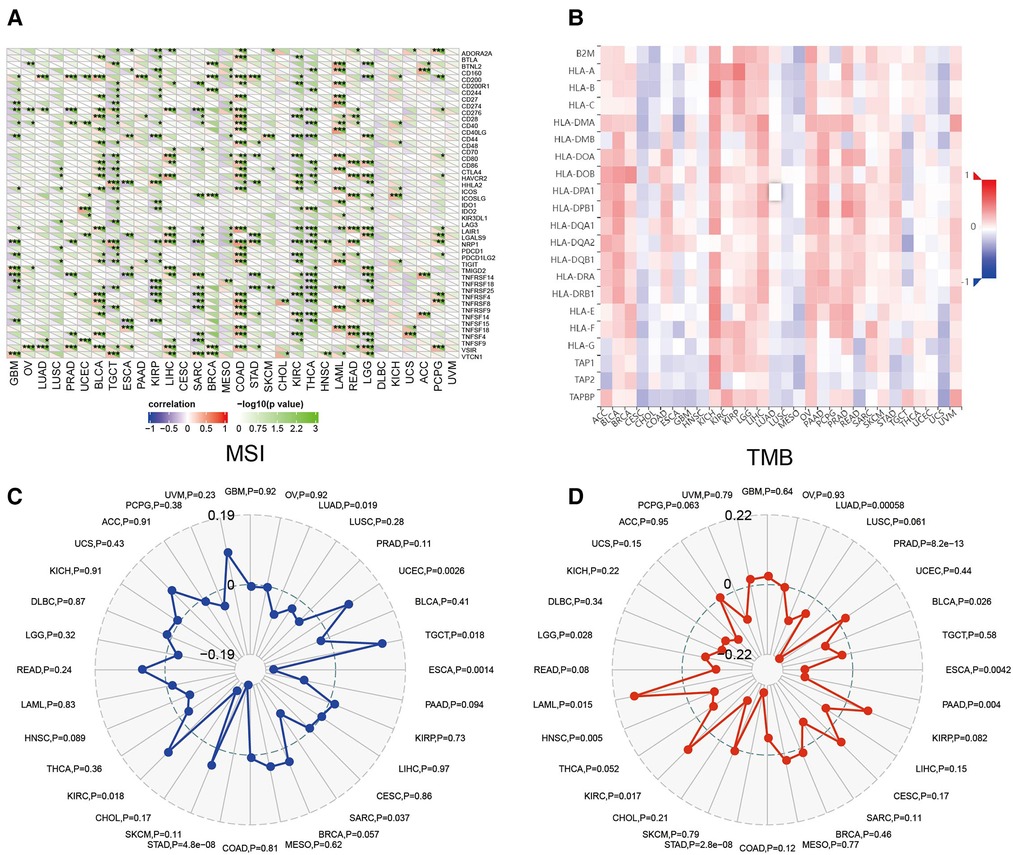
Figure 7. Association between CRYAB expression and immune checkpoints (ICP) (A), major histocompatibility complex (MHC) molecules (B), microsatellite instability (MSI) (C), and tumor mutational burden (TMB) (D) in human tumors.
In TME, MSI and TMB are predictors of tumor immunotherapy efficacy (19, 20). Sangerbox website was used to evaluate the association between CRYAB expression and MSI/TMB. CRYAB expression was positively associated with MSI in UCEC, TGCT, and KIRC, while negatively associated with MSI in LUAD, ESCA, SARC, and STAD (Figure 7C). Moreover, CRYAB was positively associated with TMB in KIRC and LAML, but negatively correlated with LUAD, PRAD, BLCA, ESCA, PAAD, STAD, and LGG (Figure 7D).
Enrichment analysis of CRYAB-related genes
Pathway enrichment analysis of CRYAB-related genes and CRYAB binding proteins was conducted to understand the mechanisms by which the CRYAB gene regulates cancer cells on a molecular level. We screened 50 CRYAB binding proteins and their protein-protein interaction (PPI) network using the STRING database (Figure 8A). The top 200 CRYAB-related genes were picked using GEPIA2 from the TCGA database. Based on these two datasets, we used the R package “clusterProfiler”, “org.Hs.eg.db”, and “enrichplot” for functional enrichment analysis. Based on the GO analysis, The major molecular functions of CRYAB included “identical protein binding”, “cysteine type endopeptidase activity involved in the execution phase of apoptosis”, and “cysteine_type endopeptidase activity involved in the apoptotic process” (Figure 8B). The major biological functions of CRYAB included “neurogenesis”, “neuron differentiation”, and “apoptotic process” (Figure 8C). Cell components related to CRYAB gene included “neuron projection”, “synapse”, and “cell_cell junctions” (Figure 8D). The KEGG results revealed that CRYAB-related genes enriched in “Alzheimer”s disease”, “Human cytomegalovirus infection”, “Hepatitis C”, “Apoptosis”, “Signaling pathways regulating pluripotency of stem cells”, “EGFR tyrosine kinase inhibitor resistance”, “Apoptosis—multiple species”, and “p53 signaling pathway” (Figure 8E). These results revealed that the CRYAB gene might be involved in regulating tumor immune microenvironment and tumor growth.
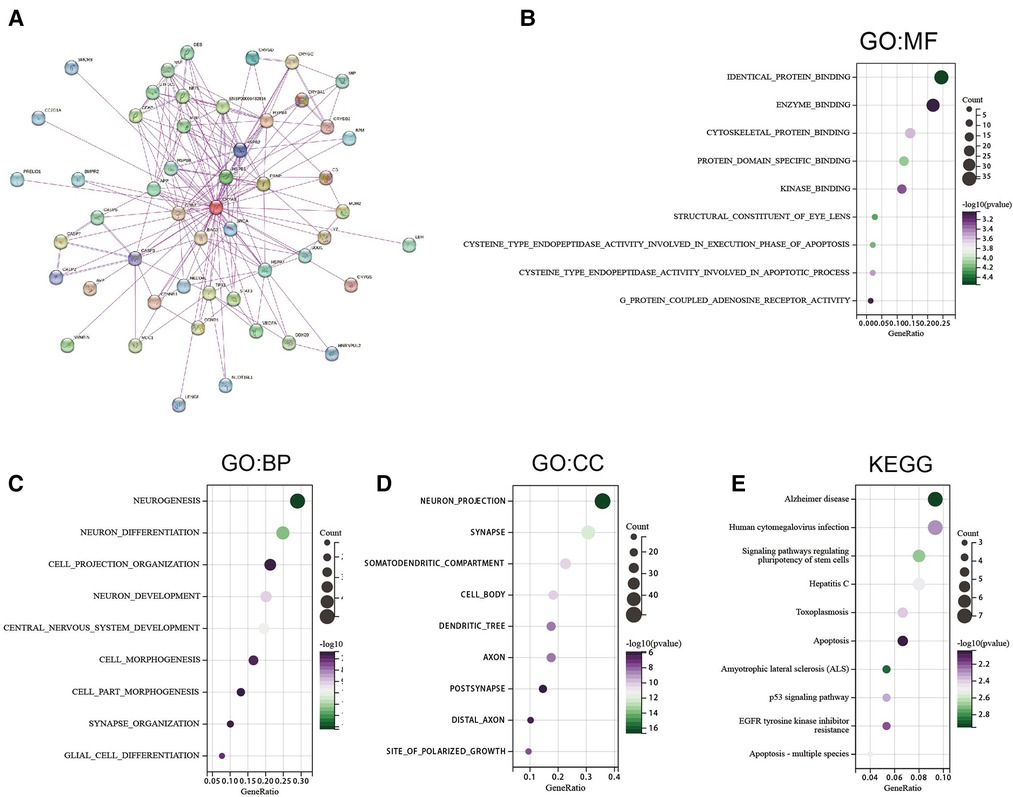
Figure 8. Gene enrichment analysis. (A) 50 CRYAB-binding proteins experimentally verified to interact with CRYAB. (B) GO molecular functions of CRYAB-binding and interacting genes (C) GO biological processes associated with CRYAB-binding and interacting genes. (D) GO cellular components associated with CRYAB-binding and interacting genes. (E) KEGG pathway analysis of CRYAB-binding and related genes.
Association between CRYAB expression and drug sensitivity
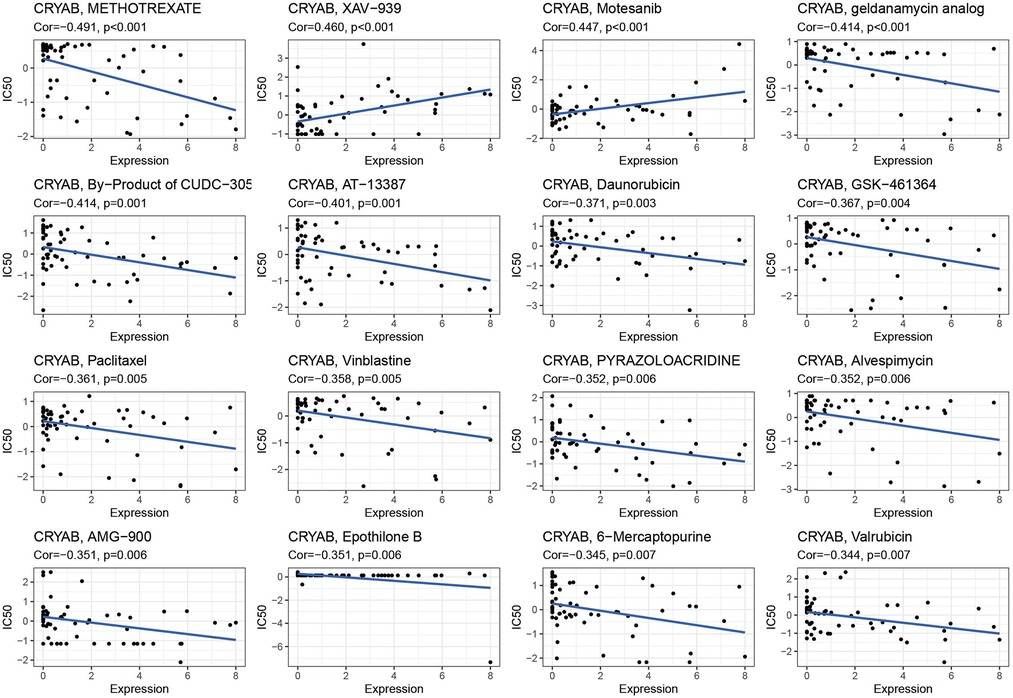
The detection of CRYAB expression in NCI-60 cell lines showed that CRYAB expression was associated with drug sensitivity (Supplementary Table S1). With the increase of CRYAB expression, the IC50 of METHOTREXATE, geldanamycin analog, By−Product of CUDC−305, AT-13387, Daunorubicin, GSK−461364, Paclitaxel Vinblastine, PYRAZOLOACRIDINE, Alvespimycin, AMG−900, Epothilone B, 6−Mercaptopurine, and Valrubicin decreased in cancer cells (Figure 9). This indicated that the CRYAB expression developed tumor cell sensitivity to these drugs. By contrast, it decreased the drug sensitivity of XAV−939 and Motesanib (Figure 9).
Validation of CRYAB expression in kidney renal clear cell carcinomas and caki-1
To detect CRYAB expression in KIRC and Caki-1, we collected the specimens of renal clear cell carcinoma and their adjacent noncancerous tissues and cultured Caki-1 and HK-2 for RT-qPCR. CRYAB mRNA expression was significantly higher in KIRC tissues than that in normal tissues (P < 0.05, Figure 10A). Besides, CRYAB mRNA expression was elevated in Caki-1 compared with HK-2 (P < 0.05, Figure 10B).
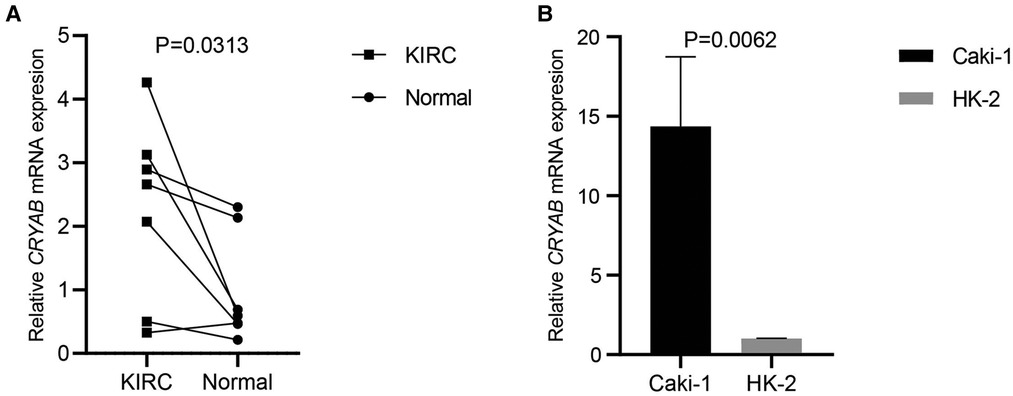
Figure 10. CRYAB expression in KIRC and caki-1. (A) Seven specimens of KIRC and their adjacent normal tissues were used to detect CRYAB expression. CRYAB mRNA expression was significantly elevated in KIRC tissues compared with normal tissues. (B) CRYAB mRNA expression was elevated in Caki-1 compared with HK-2.
Discussion
The incidence of patients with cancer is globally increasing with rapid growth and the aging population (1). A number of these cancers have an aggressive clinical presentation with a poor prognosis. Notably, it is critical to foresee the clinical outcome of tumor patients when developing a treatment plan. TNM staging is adopted to evaluate the clinical outcome of patients. Nonetheless, TNM staging is not significantly precise. For instance, although early diagnosis and advanced therapeutic interventions have shown progress in COAD, the five-year survival rate remains at approximately 45% (21). Molecular biomarkers improve the probability of precisely predicting cancer prognosis (22). Therefore, analyzing molecular biomarkers can significantly benefit most cancer patients by helping in choosing the appropriate treatment approach. Previous studies have shown that cancer patients with high levels of CRYAB demonstrated an unfavorable survival (8–10), indicating its potential as a prognostic biomarker. As a molecular chaperone, the CRYAB gene could prevent protein degradation caused by cell stress stimulation (including heat shock, oxidative stress, or radiation) (23). A previous study revealed that CRYAB blocks apoptosis by directly interacting with caspase-3, Bax, and Bcl-xS, thereby suppressing their translocation to mitochondria (24, 25). Besides, the anti-inflammatory pathway of CRYAB involves immunomodulatory responses via endosomal/phagosomal CD14 and Toll-like receptors 1 and 2 (26). However, the immunological effect of the CRYAB gene in pan-cancer background warrants additional research. Herein, we identified the role of the CRYAB gene in human cancers via a pan-cancer analysis of TCGA and CPTAC databases.
First, this work analyzed CRYAB expression in pan-cancer using the TIMER2.0. The expression of CRYAB gene in most cancers was lower than their adjacent normal tissues except for CHOL, KIRC, and KIRP. Further, we explored CRYAB gene expression in pan-cancer via the Sangerbox tool using the TCGA and GTEx databases. Consequently, the expression of CRYAB was lower in tumor tissues than in healthy tissues except for CHOL, GBM, KIRC, KIRP, LGG, and PAAD. These findings were consistent with previous studies in BLCA, OV, KIRC, COAD, HNSC, and GBM, PRAD (27–33). In this pan-cancer analysis, we found that CRYAB gene expression was elevated in KIRC through TCGA and GTEx database. Based on our previous studies on KIRC, we want to further verify whether the expression of CRYAB in our clinical samples is consistent with that in TCGA. So we selected seven clinical samples of KIRC from our hospital and a human KIRC cell line to verify CRYAB expression through RT-qPCR. CRYAB mRNA expression was significantly elevated in KIRC tissues and Caki-1. The above results were in line with the bioinformatics analysis results in KIRC.
Furthermore, the CRYAB protein expression levels were analyzed in cancers through CPTAC. As a result, CRYAB expression levels were lower in COAD, UCEC, HNSC, LIHC, LUAD, PAAD, and OV than in healthy tissues except for KIRC. The results in COAD, HNSC, LUAD, and OV were consistent with that of previous studies (29–31, 34). Based on the above findings, it appears that the protein levels matched the RNA levels.
Subsequently, we examined the relationship between CRYAB gene expression and pathological stages of cancers. The results showed that CRYAB expression was upregulated with tumor stage in COAD, CESC, STAD, OV, and BLCA; other reports are in agreement with the above results in BLCA and COAD (21, 27).
The effects of the CRYAB gene were evaluated on the survival outcome of pan-cancer. The Cox proportional hazards model analysis of OS indicated that the CRYAB gene is a risk factor in BLCA, LIHC, READ, STAD, and UVM, while a protective factor in GBM, KICH, LGG, and SARC. The Kaplan–Meier method analysis revealed that high-CRYAB groups correlated with poor OS in BLCA, COAD, LIHC, OV, and READ, but with optimistic an prognosis in KICH. The Cox proportional hazards model analysis of DSS showed that the CRYAB gene was a risk factor in BLCA, COAD, OV, STAD, UCEC, and UVM, but a protective factor in GBM, KICH, KIRP, LGG, and PRAD. The Kaplan-Meier analysis revealed that high-CRYAB groups were linked to poor DSS in BLCA, COAD, OV, and UCEC, but with a positive prognosis in DLBC and KICH. Nonetheless, the endpoint of OS potentially reduces the feasibility of clinical research. Non-cancer-related deaths do not necessarily indicate tumor biology, aggressiveness, or response to therapy. Additionally, OS or DSS warrants longer follow-up. Therefore, the use of PFI or DFI can more effectively indicate the effect of various factors on patients. We examined the PFI or DFI of the CRYAB gene in cancer patients using the Cox proportional hazards model and Kaplan-Meier analysis. The Cox proportional hazards model analysis of PFI revealed that the CRYAB gene was a risk factor in BLCA, COAD, and OV, but a protective factor in KICH, LGG, PRAD, and SKCM. According to the Kaplan-Meier method, high-CRYAB groups were associated with poor PFI in BLCA, COAD, and OV, but with optimistic prognosis in KICH, LGG, and SKCM. On the other hand, high-CRYAB groups were linked to optimistic DFI in KIRP. Based on forest plot results, the CRYAB gene was a risk factor in HNSC, but a protective factor in PRAD. Current studies show that the CRYAB gene is an oncogene, and patients with high expression of CRYAB are in the advanced tumor stage or have a poor prognosis. Juliane Volkmann et al. (30) discovered that the CRYAB gene is unexpressed or only weakly expressed in 75% of ovarian cancer samples, whereas high CRYAB expression correlates with poor outcomes (OS, PFS) in ovarian cancer patients. CRYAB expression in COAD was similar to that in OV and high expression of CRYAB positively correlated with high-grade malignancies, more advanced tumors, lymph nodes, metastatic (TNM) cancer stages, and poor survival outcomes (21, 29). In one bioinformatics analysis of BLCA, the results revealed that patients demonstrated poor OS and DFS when they expressed high levels of CRYAB (27). In head and neck cancer, Chin David et al. (31) revealed that CRYAB is an independent marker for poor survival outcomes, further emphasizing its potential role as a prognostic biomarker. So far, the CRYAB gene has been shown to suppress tumorigenesis only in prostate cancer and nasopharyngeal carcinoma (NPC) (33, 35). In the nude mice model, CRYAB activation potentially inhibits the occurrence of nasopharyngeal carcinoma. High CRYAB gene expression improves NPC progression (35). Besides, downregulated CRYAB expression was noted in patients with PRAD, serving as a protective gene for PRAD (33), which is consistent with our results. In this study, high CRYAB expression was linked to poor OS and DSS in BLCA, COAD, LIHC, READ, STAD, UCEC, and UVM patients, but with optimistic OS and DSS in GBM, KICH, KIRP, LGG, SARC, and PRAD patients. Meanwhile, high CRYAB expression was linked to poor DFI and PFI in BLCA, COAD, OV, and HNSC patients, but with optimistic DFI and PFI in KICH, LGG, PRAD, PRAD, and SKCM patients. Given that CRYAB expression has several promising interactions in eukaryotic cells, it might be beneficial to one cancer yet harmful to another (36), and with a distinct molecular mechanism of CRYAB in cancer development (25).
TME innately modulates cancer progression, allowing tumor cells to evade immune surveillance, thereby significantly influencing immunotherapy outcomes and survival prognosis of patients (13). TME comprises infiltrating immune cells, and cancer-related stromal cells, among other normal epithelial cells. Accumulating evidence suggests that stromal cells regulate the occurrence and development of many tumors (37–40). Therefore, we further investigated the relationship between CRYAB expression and the immune infiltration of cancer-related stromal cells and immune cells. In our study, the CRYAB gene positively correlated with CAFs in most cancers but negatively related to CAFs in KIRP, GBM, and KIRC. Moreover, we identified a positive correlation between the CRYAB gene and endothelial cells in the majority of cancers, but a negative correlation in KIRC and KIRP. We noted the first potential relationship between CAFs and CRYAB expression in tumors.
Of note, CAFs are a primary component of TME. In addition to their interaction with cancer cells, CAFs affect extracellular matrix (ECM), immune infiltration, and other elements of the TME (15). Histopathological analysis has shown that CAFs play a role in the prognosis of different cancers (41, 42). At present, CAFs exert a tumor-promoting effect in a majority of cancers. Patients with high levels of CAFs infiltration have a poor prognosis (43). Also, CAFs promote angiogenesis by producing abundant pro-angiogenic factors including VEGFA, PDGFC, FGF2, osteopontin, and secreting frizzled associated protein 2 (SFRP2). CAFs potentially inhibit or impair CD8+T cells' function by secreting TGFβ, increasing arginase activity, expressing abundant FAS ligand, or killing CD8+ T cells in an antigen-specific manner (15, 44–46). In many cancers, the infiltration of CD8+ T cells is associated with better clinical outcomes, but endothelial cells negatively correlate with survival (47). As such, whether CAFs promoted endothelial cells and suppressed CD8 + T cells, or regulated each other, remains to be explored.
Analysis of data on pathological stages of cancer revealed a possible connection between high CRYAB expression and advanced pathological staging in CESC, COAD, BLCA, and STAD due to the increased infiltration of CAFs and endothelial cells in the TME. Examination of prognostic data showed a potential association between high CRYAB expression and poor prognosis in BLCA, COAD, HNSC, LIHC, READ, and STAD. This was ascribed to the increased infiltration of CAFs and endothelial cells, and decreased CD8+ T cells infiltration in the TME. In contrast, high expression of the CRYAB gene in GBM and KIRP reduced infiltration of CAFs and endothelial cells, which improved the survival outcome. In conclusion, our findings demonstrated that CRYAB may influence the clinical outcomes of cancer patients by influencing CAFs and endothelial cells infiltration in tumors. CAFs may play a major role in mediating the effect of CRYAB on the clinical outcome of cancer patients. This association needs to be further studied.
In our study, the association between CRYAB expression and ICP, TMB, and MSI in human tumors also confirmed that CRYAB closely associated with TME in human cancers. Besides, we found that CRYAB was negatively related to TMB and MSI in some cancers, such as LUAD, PRAD, BLCA, ESCA, PAAD, STAD, SARC, and LGG. This meant CRYAB may influence immunotherapy response in these cancers. Moreover, we explored the association between CRYAB expression and drug sensitivity in cancer cell lines. The results indicated that CRYAB expression developed tumor cell sensitivity to a number of chemotherapeutic drugs, such as METHOTREXATE, geldanamycin analog, By−Product of CUDC−305, AT-13387, Daunorubicin, GSK−461364, Paclitaxel Vinblastine, PYRAZOLOACRIDINE,.
Alvespimycin, AMG−900, Epothilone B, 6−Mercaptopurine, and Valrubicin. By contrast, it decreased the drug sensitivity of XAV−939 and Motesanib. Among these drugs, Motesanib is often used in bladder cancer to develop the antitumor effect of cisplatin (48). METHOTREXATE can be used to treat many kinds of cancer, such as LUAD, HNSC, and BRCA (49–51). The above results indicated that CRYAB expression could influence drug response and it could serve as a potential drug target.
To explore the possible mechanism underlying the effects of CRYAB on cancer patients, we conducted GO and KEGG enrichment analyses of binding proteins and genes related to CRYAB expression. The results indicated that CRYAB-related genes enriched in “Alzheimer”s disease”, “Human cytomegalovirus infection”, “Hepatitis C”, “Apoptosis”, “Signaling pathways regulating pluripotency of stem cells”, “EGFR tyrosine kinase inhibitor resistance”, “Apoptosis—multiple species”, and “p53 signaling pathway”. The “EGFR tyrosine kinase inhibitor resistance” of KEGG results was consistent with the drug sensitivity results. In our study, we found CRYAB decreased the drug sensitivity of Motesanib which is a tyrosine kinase inhibitor. These findings expand our understanding of the role of CRYAB in human tumors. Studies have shown that CRYAB is an anti-apoptotic protein that negatively regulates the expression of pro-apoptotic proteins (24, 52). In OV, high expression of CRYAB suppressed tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and promoted tumor growth (30). CRYAB enhanced tumorigenesis by modulating VEGF and increased resistance to anti-VEGF therapy in breast cancer (11, 12). Altogether, these findings show that CRYAB has complex roles in malignant tumors. The most notable effect of CRYAB is its anti-apoptosis activity in tumors. Moreover, CRYAB may affect the prognosis of patients by influencing CAFs infiltration levels in tumors.
In this study, we performed a comprehensive analysis to evaluate the function of the CRYAB gene in human tumors, but there are still some limitations in this experiment. First, we used TCGA and GTEx datasets for pan-cancer analysis, but the data of some special tumors were still not available. Second, in this study, we mainly carried out bioinformatics analysis. The potential function of CRYAB needs to be verified in vivo/in vitro experiments. Third, even though we demonstrated that CRYAB may influence the prognosis of cancer patients, specifically targeting infiltration of CAFs and endothelial cells in TME. However, the exact pathway by which CRYAB is involved in immune regulation requires further investigation.
In conclusion, this is arguably the first pan-cancer analysis of the CRYAB gene. CRYAB gene expression varied in different cancers and their adjacent normal tissues. In addition, CRYAB is a crucial component of the TME and influences immune cell infiltration, making it a promising biomarker to assess immune infiltration and prognosis in many malignancies. Furthermore, CRYAB has potential impacts on cellular biological processes and molecular pathways in tumors. Besides, a number of FDA-approved drugs were found to be associated with CRYAB and might be potential cancer therapeutic candidates. This careful analysis provides valuable insights into the role of the CRYAB gene in prognosis and immunology in human tumors as well as new evidence for accurate and customized cancer therapy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
The studies involving human participants were reviewed and approved by The Ethics Committee of The Second Affiliated Hospital of Guangxi Medical University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization, HH and ZM; Data curation, LC; Formal analysis, LC and XZ; Funding acquisition, ZM; Methodology, LC, XZ, JW and JZ; Project administration, HH and ZM; Software, LC and XZ; Supervision, HH and ZM; Visualization, LC and XZ; Writing – original draft, LC and XZ; Writing – review & editing, Houbao Huang and Zengnan Mo. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsurg.2022.1117307/full#supplementary-material.
References
1. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the global burden of disease study 2016. Lancet (London, England). (2017) 390(10100):1151–210. doi: 10.1016/s0140-6736(17)32152-9
2. Zhang K, Wang H. [Cancer genome atlas pan-cancer analysis project]. Zhongguo Fei Ai Za Zhi. (2015) 18(4):219–23. doi: 10.3779/j.issn.1009-3419.2015.04.02
3. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. (2013) 45(10):1113–20. doi: 10.1038/ng.2764
4. Rajagopal P, Tse E, Borst AJ, Delbecq SP, Shi L, Southworth DR, et al. A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis. eLife. (2015) 4. doi: 10.7554/eLife.07304
5. Huang XY, Ke AW, Shi GM, Zhang X, Zhang C, Shi YH, et al. αB-crystallin complexes with 14-3-3ζ to induce epithelial-mesenchymal transition and resistance to sorafenib in hepatocellular carcinoma. Hepatology (Baltimore, Md). (2013) 57(6):2235–47. doi: 10.1002/hep.26255
6. Shi C, Yang X, Bu X, Hou N, Chen P. Alpha B-crystallin promotes the invasion and metastasis of colorectal cancer via epithelial-mesenchymal transition. Biochem Biophys Res Commun. (2017) 489(4):369–74. doi: 10.1016/j.bbrc.2017.05.070
7. Chen D, Cao G, Qiao C, Liu G, Zhou H, Liu Q. Alpha B-crystallin promotes the invasion and metastasis of gastric cancer via NF-kappaB-induced epithelial-mesenchymal transition. J Cell Mol Med. (2018) 22(6):3215–22. doi: 10.1111/jcmm.13602
8. Kim HS, Lee Y, Lim YA, Kang HJ, Kim LS. αB-Crystallin is a novel oncoprotein associated with poor prognosis in breast cancer. J Breast Cancer. (2011) 14(1):14–9. doi: 10.4048/jbc.2011.14.1.14
9. Mao Y, Zhang DW, Lin H, Xiong L, Liu Y, Li QD, et al. Alpha B-crystallin is a new prognostic marker for laryngeal squamous cell carcinoma. J Exp & Clin Cancer Res: CR. (2012) 31(1):101. doi: 10.1186/1756-9966-31-101
10. Annertz K, Enoksson J, Williams R, Jacobsson H, Coman WB, Wennerberg J. Alpha B-crystallin - a validated prognostic factor for poor prognosis in squamous cell carcinoma of the oral cavity. Acta Otolaryngol. (2014) 134(5):543–50. doi: 10.3109/00016489.2013.872293
11. Chen W, Lu Q, Lu L, Guan H. Increased levels of alphaB-crystallin in vitreous fluid of patients with proliferative diabetic retinopathy and correlation with vascular endothelial growth factor. Clin Experiment Ophthalmol. (2017) 45(4):379–84. doi: 10.1111/ceo.12891
12. Ruan Q, Han S, Jiang WG, Boulton ME, Chen ZJ, Law BK, et al. αB-crystallin, an effector of unfolded protein response, confers anti-VEGF resistance to breast cancer via maintenance of intracrine VEGF in endothelial cells. Mol Cancer Res: MCR. (2011) 9(12):1632–43. doi: 10.1158/1541-7786.Mcr-11-0327
13. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. (2019) 79(18):4557–66. doi: 10.1158/0008-5472.Can-18-3962
14. Guo Z, Song J, Hao J, Zhao H, Du X, Li E, et al. M2 macrophages promote NSCLC metastasis by upregulating CRYAB. Cell Death Dis. (2019) 10(6):377. doi: 10.1038/s41419-019-1618-x
15. Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discovery. (2019) 18(2):99–115. doi: 10.1038/s41573-018-0004-1
16. Zhou B, Chen WL, Wang YY, Lin ZY, Zhang DM, Fan S, et al. A role for cancer-associated fibroblasts in inducing the epithelial-to-mesenchymal transition in human tongue squamous cell carcinoma. J Oral Pathol & Med. (2014) 43(8):585–92. doi: 10.1111/jop.12172
17. Zhu C, Chrifi I, Mustafa D, van der Weiden M, Leenen PJM, Duncker DJ, et al. CECR1-mediated Cross talk between macrophages and vascular mural cells promotes neovascularization in malignant glioma. Oncogene. (2017) 36(38):5356–68. doi: 10.1038/onc.2017.145
18. Zhu C, Mustafa D, Zheng PP, van der Weiden M, Sacchetti A, Brandt M, et al. Activation of CECR1 in M2-like TAMs promotes paracrine stimulation-mediated glial tumor progression. Neuro-oncol. (2017) 19(5):648–59. doi: 10.1093/neuonc/now251
19. Lee V, Murphy A, Le DT, Diaz LA Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. (2016) 21(10):1200–11. doi: 10.1634/theoncologist.2016-0046
20. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. (2017) 377(25):2500–1. doi: 10.1056/NEJMc1713444
21. Pagano C, Navarra G, Gazzerro P, Vitale M, Notarnicola M, Caruso MG, et al. Association of alpha B-crystallin expression with tumor differentiation grade in colorectal cancer patients. Diagnostics (Basel, Switzerland). (2021) 11(5). doi: 10.3390/diagnostics11050896
22. Tamayo P, Cho YJ, Tsherniak A, Greulich H, Ambrogio L, Schouten-van Meeteren N, et al. Predicting relapse in patients with medulloblastoma by integrating evidence from clinical and genomic features. J Clin Oncol. (2011) 29(11):1415–23. doi: 10.1200/jco.2010.28.1675
23. Treweek TM, Meehan S, Ecroyd H, Carver JA. Small heat-shock proteins: important players in regulating cellular proteostasis. Cell and Mol Life Sci: CMLS. (2015) 72(3):429–51. doi: 10.1007/s00018-014-1754-5
24. Hu WF, Gong L, Cao Z, Ma H, Ji W, Deng M, et al. αA- and αB-crystallins interact with caspase-3 and bax to guard mouse lens development. Curr Mol Med. (2012) 12(2):177–87. doi: 10.2174/156652412798889036
25. Zhang J, Liu J, Wu J, Li W, Chen Z, Yang L. Progression of the role of CRYAB in signaling pathways and cancers. Onco Targets Ther. (2019) 12:4129–39. doi: 10.2147/ott.S201799
26. van Noort JM, Bsibsi M, Nacken PJ, Gerritsen WH, Amor S, Holtman IR, et al. Activation of an immune-regulatory macrophage response and inhibition of lung inflammation in a mouse model of COPD using heat-shock protein alpha B-crystallin-loaded PLGA microparticles. Biomaterials. (2013) 34(3):831–40. doi: 10.1016/j.biomaterials.2012.10.028
27. Zhang C, Berndt-Paetz M, Neuhaus J. Identification of key biomarkers in bladder cancer: evidence from a bioinformatics analysis. Diagnostics (Basel, Switzerland). (2020) 10(2). doi: 10.3390/diagnostics10020066
28. Ho P-Y, Chueh S-C, Chiou S-H, Wang S-M, Lin W-C, Lee IL, et al. αB-Crystallin in clear cell renal cell carcinoma: tumor progression and prognostic significance. Urol Oncol: Semin Orig Invest. (2013) 31(7):1367–77. doi: 10.1016/j.urolonc.2012.01.015
29. Deng J, Chen X, Zhan T, Chen M, Yan X, Huang X. CRYAB Predicts clinical prognosis and is associated with immunocyte infiltration in colorectal cancer. PeerJ. (2021) 9:e12578. doi: 10.7717/peerj.12578
30. Volkmann J, Reuning U, Rudelius M, Hafner N, Schuster T, Becker VRA, et al. High expression of crystallin alphaB represents an independent molecular marker for unfavourable ovarian cancer patient outcome and impairs TRAIL- and cisplatin-induced apoptosis in human ovarian cancer cells. Int J Cancer. (2013) 132(12):2820–32. doi: 10.1002/ijc.27975
31. Chin D, Boyle GM, Williams RM, Ferguson K, Pandeya N, Pedley J, et al. Alpha B-crystallin, a new independent marker for poor prognosis in head and neck cancer. Laryngoscope. (2005) 115(7):1239–42. doi: 10.1097/01.MLG.0000164715.86240.55
32. Kore RA, Abraham EC. Inflammatory cytokines, interleukin-1 beta and tumor necrosis factor-alpha, upregulated in glioblastoma multiforme, raise the levels of CRYAB in exosomes secreted by U373 glioma cells. Biochem Biophys Res Commun. (2014) 453(3):326–31. doi: 10.1016/j.bbrc.2014.09.068
33. Valcarcel-Jimenez L, Macchia A, Martín-Martín N, Cortazar AR, Schaub-Clerigué A, Pujana-Vaquerizo M, et al. Integrative analysis of transcriptomics and clinical data uncovers the tumor-suppressive activity of MITF in prostate cancer. Cell Death Dis. (2018) 9(10):1041. doi: 10.1038/s41419-018-1096-6
34. Qin H, Ni Y, Tong J, Zhao J, Zhou X, Cai W, et al. Elevated expression of CRYAB predicts unfavorable prognosis in non-small cell lung cancer. Med Oncol. (2014) 31(8):142. doi: 10.1007/s12032-014-0142-1
35. Huang Z, Cheng Y, Chiu PM, Cheung FM, Nicholls JM, Kwong DL, et al. Tumor suppressor alpha B-crystallin (CRYAB) associates with the cadherin/catenin adherens junction and impairs NPC progression-associated properties. Oncogene. (2012) 31(32):3709–20. doi: 10.1038/onc.2011.529
36. Caporossi D, Parisi A, Fantini C, Grazioli E, Cerulli C, Dimauro I. AlphaB-crystallin and breast cancer: role and possible therapeutic strategies. Cell Stress Chaperones. (2021) 26(1):19–28. doi: 10.1007/s12192-020-01175-0
37. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. (2022) 12(1):31–46. doi: 10.1158/2159-8290.Cd-21-1059
38. Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. (2012) 487(7408):500–4. doi: 10.1038/nature11183
39. Steenbrugge J, De Jaeghere EA, Meyer E, Denys H, De Wever O. Splenic hematopoietic and stromal cells in cancer progression. Cancer Res. (2021) 81(1):27–34. doi: 10.1158/0008-5472.Can-20-2339
40. Zhan HX, Zhou B, Cheng YG, Xu JW, Wang L, Zhang GY, et al. Crosstalk between stromal cells and cancer cells in pancreatic cancer: new insights into stromal biology. Cancer Lett. (2017) 392:83–93. doi: 10.1016/j.canlet.2017.01.041
41. Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. (2021) 20(1):131. doi: 10.1186/s12943-021-01428-1
42. Gieniec KA, Butler LM, Worthley DL, Woods SL. Cancer-associated fibroblasts-heroes or villains? Br J Cancer. (2019) 121(4):293–302. doi: 10.1038/s41416-019-0509-3
43. Hosein AN, Brekken RA, Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol & Hepatol. (2020) 17(8):487–505. doi: 10.1038/s41575-020-0300-1
44. Thomas DA, Massagué J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. (2005) 8(5):369–80. doi: 10.1016/j.ccr.2005.10.012
45. Érsek B, Silló P, Cakir U, Molnár V, Bencsik A, Mayer B, et al. Melanoma-associated fibroblasts impair CD8+ T cell function and modify expression of immune checkpoint regulators via increased arginase activity. Cell and Mol Life Sci: CMLS. (2021) 78(2):661–73. doi: 10.1007/s00018-020-03517-8
46. Lakins MA, Ghorani E, Munir H, Martins CP, Shields JD. Cancer-associated fibroblasts induce antigen-specific deletion of CD8+ T cells to protect tumour cells. Nat Commun. (2018) 9(1):948. doi: 10.1038/s41467-018-03347-0
47. Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. (2012) 12(4):298–306. doi: 10.1038/nrc3245
48. Ho JN, Byun SS, Lee SE, Youn JI, Lee S. Multikinase inhibitor motesanib enhances the antitumor effect of cisplatin in cisplatin-resistant human bladder cancer cells via apoptosis and the PI3K/akt pathway. Oncol Rep. (2019) 41(4):2482–90. doi: 10.3892/or.2019.7005
49. Zhuang J, Chen Z, Chen Z, Chen J, Liu M, Xu X, et al. Construction of an immune-related lncRNA signature pair for predicting oncologic outcomes and the sensitivity of immunosuppressor in treatment of lung adenocarcinoma. Respir Res. (2022) 23(1):123. doi: 10.1186/s12931-022-02043-4
50. Iop A, Cartei G, Isaia A. Vinorelbine, bleomycin and methotrexate as a salvage therapy for patients with head and neck squamous carcinoma in relapse after cisplatin/fluorouracil. Ann Oncolo. (1998) 9(2):225–7. doi: 10.1023/a:1008229106595
51. Abolmaali S S, Zarenejad S, Mohebi Y, Najafi H, Javanmardi S, Abedi M, et al. Biotin receptor-targeting nanogels loaded with methotrexate for enhanced antitumor efficacy in triple-negative breast cancer in vitro and in vivo models. Int J Pharm. (2022) 624:122049. doi: 10.1016/j.ijpharm.2022.122049
Keywords: CRYAB, prognosis, immune infiltration, tumor microenvironment, pan-cancer
Citation: Cheng L, Zou X, Wang J, Zhang J, Mo Z and Huang H (2023) The role of CRYAB in tumor prognosis and immune infiltration: A Pan-cancer analysis. Front. Surg. 9:1117307. doi: 10.3389/fsurg.2022.1117307
Received: 6 December 2022; Accepted: 29 December 2022;
Published: 13 January 2023.
Edited by:
Hongji Zhang, University of Virginia, United StatesReviewed by:
Amblessed Onuma, The Ohio State University, United StatesZhaoxia Zhang, Chongqing Medical University, China
© 2023 Cheng, Zou, Wang, Zhang, Mo and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zengnan Mo bW96ZW5nbmFuQGd4bXUuZWR1LmNu Houbao Huang ZHJodWFuZ2hvdWJhb0AxNjMuY29t
†These authors have contributed equally to this work
Specialty Section: This article was submitted to Surgical Oncology, a section of the journal Frontiers in Surgery
 Lang Cheng
Lang Cheng Xiong Zou1,2,†
Xiong Zou1,2,† Jiawei Wang
Jiawei Wang Houbao Huang
Houbao Huang