
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Surg. , 06 January 2023
Sec. Surgical Oncology
Volume 9 - 2022 | https://doi.org/10.3389/fsurg.2022.1050242
This article is part of the Research Topic Intercellular Information Transmission and Communication During Tumor Development View all 5 articles
Background: Pleural effusion (PE) caused by lung cancer is prevalent, and it is difficult to differentiate it from PE caused by tuberculosis. Exosome-based liquid biopsy offers a non-invasive technique to diagnose benign and malignant PE. Exosomal miRNAs are potential diagnostic markers and play an essential role in signal transduction and biological processes in tumor development. We hypothesized that exosomal miRNA expression profiles in PE would contribute to identifying its diagnostic markers and elucidating the molecular basis of PE formation in lung cancer.
Methods: The exosomes from PE caused by lung adenocarcinoma (LUAD) and pulmonary tuberculosis were isolated and verified by transmission electron microscopy. The exosomal miRNA profiles were identified using deep sequencing and validated with quantitative real-time PCR (qRT-PCR). We performed bioinformatic analysis for differentially expressed miRNAs to explore how exosomal miRNAs regulate pleural effusion.
Results: We identified 99 upregulated and 91 downregulated miRNAs in malignant pleural effusion (MPE) compared to tuberculous pleural effusion (TPE). Seven differentially expressed miRNAs (DEmiRNAs) were validated by qRT-PCR, out of which 5 (71.4%) were confirmed through sequencing. Gene Ontology (GO) analysis revealed that most exosomal miRNAs target genes were involved in regulating cellular processes and nitrogen compound metabolism. According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, the exosomal miRNAs target genes were mainly involved in Fc gamma R-mediated phagocytosis, Rap1 signaling pathway, and breast cancer. The hub genes, including ITGAM, FOXO1, MAPK14, YWHAB, GRIN1, and PRF1, were screened through plug-in cytoHubba. The PFR1 was identified as a critical gene in MPE formation using single-cell sequencing analysis. Additionally, we hypothesized that tumor cells affected natural killer cells and promoted the generation of PE in LUAD via the exosomal hsa-miR-3120-5p-PRF1 axis.
Conclusions: We identified exosomal miRNA profiles in LUAD-MPE and TPE, which may help in the differential diagnosis of MPE and TPE. Bioinformatic analysis revealed that these miRNAs might affect PE generation through tumor immune response in LUAD. Our results provided a new theoretical basis for understanding the function of exosomal miRNAs in LUAD-MPE.
Pleural effusion (PE), the accumulation of fluid in the pleural space, is the presenting symptom of numerous diseases. Lung cancer and pulmonary tuberculosis are the leading causes of PE. However, a clinical challenge is a differential diagnosis of tuberculosis pleural effusion (TPE) and lung cancer-related malignant pleural effusion (MPE). The diagnosis of MPE is always based on PE cytological analysis. However, the sensitivity of cytology is low (1), and finding effective prognostic biomarkers is a research hotspot. Pleural biopsy is an invasive procedure with a potentially high risk that may not provide a comprehensive biological profile of malignant tissue due to tumor heterogeneity. Liquid biopsy is a non-invasive, rapid, and safe alternative to tissue biopsy for cancer diagnosis, and it is likely to provide a thorough understanding of tumor heterogeneity (2–4).
Exosome-based liquid biopsies have been proposed for use in lung cancer diagnosis, prognosis, and surveillance in numerous studies in the past (5–7). Exosomes are small extracellular vesicles released by various cells into their microenvironment and are present in most body fluids (8, 9). They contain ncRNA, DNA, proteins, and lipids that derive from the parental cells, reflecting the characteristics of the donor cell (10). Exosomes are enriched in PE and may be used as biomarkers for malignant pleural mesothelioma (11), lung cancer (12), and tuberculosis (13).
Exosomes can also serve as a critical mediator of intercellular communication, transmitting biological signals like proteins, mRNAs, miRNAs, and DNAs to recipient cells, resulting in morphological and functional changes in the corresponding cells. The miRNAs are a family of small, non-coding, endogenous molecules involved in the post-transcriptional regulation of gene expression (14). Recent studies suggested that miRNAs are abundant in exosomes and are resistant to RNase degradation, making them useful for diagnosis, prognosis, treatment monitoring, and exploration of disease mechanisms (15–17). PE contains many immune cells, such as dendritic cells, macrophages, neutrophils, and T cells (18, 19). It is well known that angiogenesis and increased vascular permeability are essential for forming PE. Recently, numerous studies have suggested that exosomal miRNAs can affect mRNA expression/translation in the recipient cells, including various immune cells and endothelial cells, indicating that miRNAs may play potential roles in mediating intercellular communication, immunoregulation, and angiogenesis in tumor progression (20–22). Tumor-derived exosomes may play important roles in PE formation. However, the underlying mechanisms are unclear.
In this study, exosomes derived from PE of patients with lung cancer and pulmonary tuberculosis were isolated, and the miRNA profiles were analyzed to identify those which are differentially expressed. We unveiled the most important miRNAs for diagnosing lung cancer and characterized the biological progression of LUAD-induced PE. The study established the viability of exosome-associated miRNA signatures in PE as tumor biomarkers and explored the potential mechanisms underlying PE formation in LUAD.
PE samples from six LUAD and six tuberculosis patients were collected between July 2019 and December 2020 at the Beilun Branch Hospital of the First Affiliated Hospital of the Medical School of Zhejiang University. The PE fluid was collected and stored at −80 °C. LUAD was diagnosed by pathology or cytology. The patients with tuberculous PE were confirmed by bacteriology or clinical diagnosis, along with observing several distinguishing characteristics, including a high lymphocyte ratio and adenosine deaminase levels > 40 U/L. This study was approved by the regional ethical committee, approval No. 2019-53 (K), date of approval: 2019-04-17. Informed consent was provided by all participants.
Exosomes were isolated from PEs using the ultracentrifugation technique described previously (23). Briefly, PE samples were centrifuged at 500 × g for 5 min to eliminate cells. The supernatant was centrifuged at 2000 × g for 10 min to remove the component from the samples. The PE supernatants were then centrifuged sequentially at 10,000 × g for 30 min and were filtered through a 0.22 µm membrane filter (Merck Millipore). It was then centrifuged at 100,000 × g for 2 h. The exosome pellet was washed once with PBS and centrifuged again. Finally, the pellets were resuspended in PBS and stored at 80 °C until further use.
The exosomes were resuspended in 50–100 µl of 2% PFA, and 5 µl of the exosomal suspension was added to a formvar-carbon-coated copper grid and washed with PBS. The formvar membranes were kept moist at every step, but the other side remained dry. The copper mesh was placed on 50 µl of 1% glutaraldehyde droplets for 5 min before being washed eight times in 100 µl of ultrapure water for 5 min. The copper mesh-bearing exosomes were incubated for 5 min with 50 µl of uranyl oxalate droplets (pH 7) and 10 min with 50 µl of methylcellulose droplets. The excess samples were blotted off the filter paper, and the grid was air-dried for 5–10 min. Exosome morphologies were observed using the FEI Tecnai G2 Spirit transmission electron microscope.
Total RNA, including miRNA, was extracted from exosome samples using the miRNeasy MiniKit (Qiagen), according to the manufacturer's instructions. Total RNA was eluted with 30 µl of nuclease-free water (Ambion) and stored at −80 °C until further processing. RNA samples were qualified and quantified by agarose gel electrophoresis and NanoDrop ND-1000 (NanoDrop, USA) before deep sequencing analysis or qRT-PCR validation.
RNA samples were qualified and quantified using agarose gel electrophoresis and Nanodrop ND-1000 (NanoDrop, USA). Small RNA libraries were constructed using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, Ipswich, MA, USA). Their quality was evaluated using Agilent Bioanalyzer 2100 (Agilent Technologies, USA). The libraries were denatured with 0.1 M NaOH to generate single-stranded DNA. Sequencing was performed for 50 cycles on the Illumina NextSeq 500 system following the manufacturer's instructions. FastQC examined sequencing quality.
To confirm miRNA sequencing results, qRT-PCR was used to compare expression levels of randomly selected seven miRNAs between the two groups. For miRNA detection, the RNA was reverse-transcribed into cDNA before PCR. The amplification conditions were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. The miRNA level was quantitatively determined using a real-time RT-PCR kit. has-miR935p was used as a reference as its expression level remains constant across different samples.
miRDeep2 was used to obtain the miRNA counts, which were normalized using counts per million (CPM). Significant differences among groups were assessed using edgeR analysis. The differential genes were identified using the criteria |log2FoldChange| ≥ 1.5 and P value ≤ 0.05. The target genes of the top ten upregulated and downregulated differentially expressed miRNAs (DEmiRNAs) were predicted by the intersection of miRDB (24) and Targetscan (25) databases. Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed for the target genes of DEmiRNAs. The Search Tool for the Retrieval of Interacting Genes (STRING; http://string.embl.de/) database was used to construct a protein-protein interaction (PPI) of target genes for DEmiRNAs with an interaction score > 0.7 (26). The PPI network was visualized using Cytoscape, and the cytoHubba plug-in was used to identify hub genes (27). Based on the degree of the genes in the PPI network, the top 20 ranked genes were identified as hub genes. Finally, single-cell RNA profiles of MPE samples of GSE185058 were obtained from GEO (http://www.ncbi.nlm.nih.gov/geo), and differential genes were screened by iDEP.96 (http://bioinformatics.sdstate.edu/idep/) (28). The intersection of miRNA target genes and differential genes was expected to play a vital role in forming PE.
Exosomes were successfully enriched from each pleural fluid sample using the ultracentrifugation technique. Morphology of the exosome was confirmed by TEM. Representative TEM images are presented in Figure 1. TEM demonstrated that purified exosomes were round or oval with a diameter approaching 100 nm.
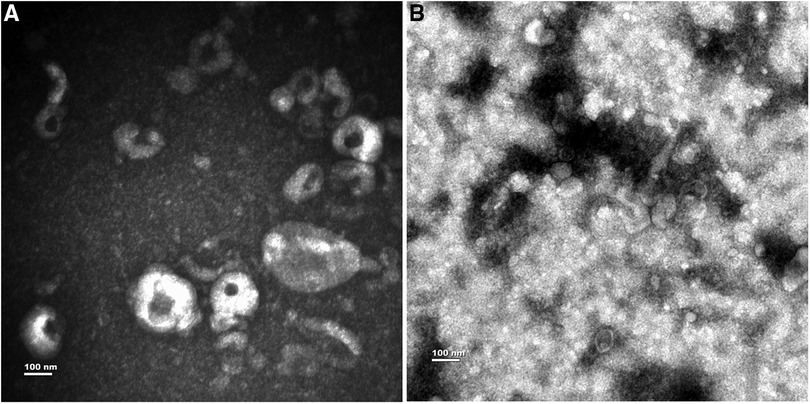
Figure 1. Characterization of exosomes from PE. (A) The exosomes from MPE. (B) The exosomes from TPE.
This study identified 974 miRNAs in clinical samples. The DEmiRNAs were filtered based on P ≤ 0.05 and |log2FC| ≥ 1.5. We identified 99 upregulated and 91 downregulated miRNAs in MPE compared to TPE. The top 10 upregulated and downregulated miRNAs are listed in Table 1. The distribution and heatmap of cluster analysis and the volcano plot of DEmiRNAs are displayed in Figure 2.
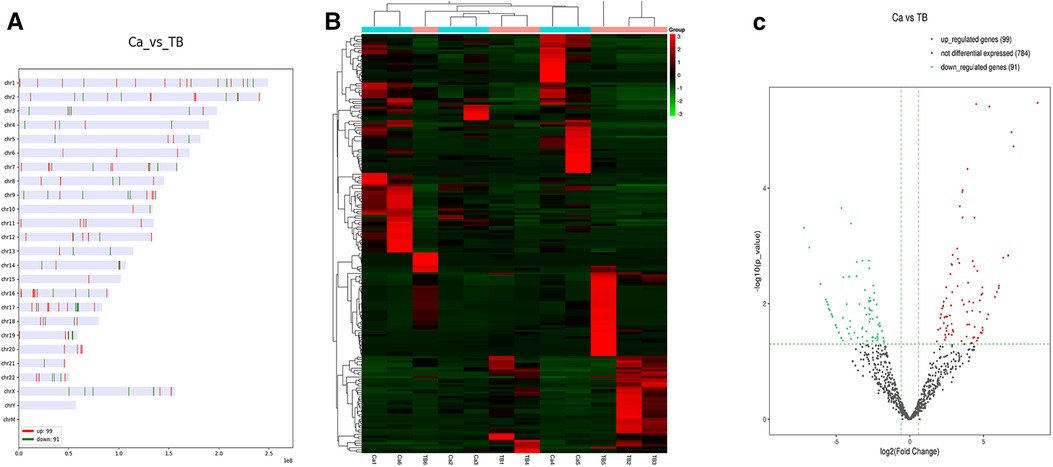
Figure 2. The distribution, heatmap, and volcano plot of DEmiRNAs. (A) The distribution of DEmiRNAs. (B) Heatmap of DEmiRNAs. (C) Volcano plot of DEmiRNAs.
We randomly chose seven DEmiRNAs for qRT-PCR analysis (Figure 3). The hsa-miR-93-5p was selected as the internal reference. The results from real-time PCR analysis revealed that five of seven (71.4%) DEmiRNAs were consistent with sequencing results. Two miRNAs (hsa-miR-150-5p and hsa-miR-3614-5p) were downregulated in MPE compared to TPE. According to sequencing and PCR, hsa-miR-150-5p was down-regulated 0.18- and 0.35-fold in MPE than TPE. Three miRNAs (hsa-miR-200b-3p, hsa-miR-182-5p, and hsa-miR-629-5p) were upregulated in MPE compared to TPE. For instance, hsa-miR-629-5p was upregulated 2.61- and 3.78-fold in MPE compared to TPE, as confirmed by sequencing and PCR. However, the PCR result of hsa-miR-517a-3p and hsa-miR-185-5p were inconsistent with sequencing.
To elucidate the exo-miRNAs regulatory mechanism in MPE, we predicted the target genes for DEmiRNAs using miRDB and TargetScan databases. The genes at the intersection of two gene sets were considered miRNA target genes. As a result, 305 and 739 target genes were predicted for upregulated and downregulated DEmiRNAs, respectively (Supplementary Material S1). Finally, the overlapped genes were included for further study.
GO and KEGG pathway enrichment analyses were performed for the overlapped target genes. The top three GO analysis terms for the target genes of upregulated and downregulated DEmiRNAs are listed in Table 2. For the target genes of upregulated DEmiRNAs, the most enriched GO terms were regulation of the cellular process, nitrogen compound metabolic process, and the developmental process in the biological process (BP) group. The most meaningful GO terms were cytoplasm, membrane-bounded organelle, and membrane in the cellular components (CC) group. The most important GO terms were protein binding, catalytic activity, and RNA polymerase II cis-regulatory region sequence-specific DNA binding in the molecular function (MF) group. For the target genes of downregulated DEmiRNAs, the most significant GO terms were regulation of the cellular process, nitrogen compound metabolic process, and RNA metabolic process in the BP group. The most enriched GO terms were cytoplasm, nucleoplasm, and membrane-bounded organelle in the CC group. The most significantly enriched GO terms were protein binding, transcription regulator activity, and DNA binding. KEGG pathway enrichment analysis revealed that the target genes of upregulated DEmiRNAs were significantly enriched in Fc gamma R-mediated phagocytosis, Rap1 signaling pathway, and Leishmaniasis pathways. While the majority of the target genes of downregulated DEmiRNAs were associated with breast cancer, gastric cancer, and autophagy-animal pathways. The top ten GO and KEGG terms of the overlapped target genes are displayed in Figure 4.
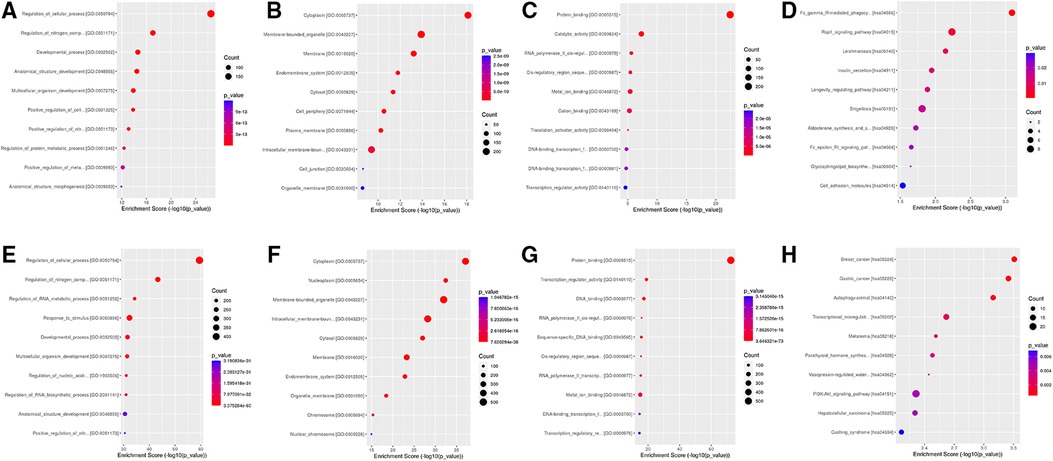
Figure 4. Go and KEGG analysis for target genes of DEmiRNAs. (A) BP group for upregulated DEmiRNAs. (B) CC group for upregulated DEmiRNAs. (C) MF group for upregulated DEmiRNAs. (D) KEGG for upregulated DEmiRNAs. (E) BP group downregulated DEmiRNAs. (F) CC group for downregulated DEmiRNAs. (G) MF group for downregulated DEmiRNAs. (H) KEGG for downregulated DEmiRNAs.
The PPI network of target genes for DEmiRNAs was constructed by the STRING online database. Subsequently, the PPI network was visualized by cytoscape software, and hub genes were screened by degree methods through plug-in cytoHubba. The top 20 ranked genes are listed in Table 3. Three significant modules for target genes of upregulated and downregulated miRNAs were obtained using MCODE (Figure 5).
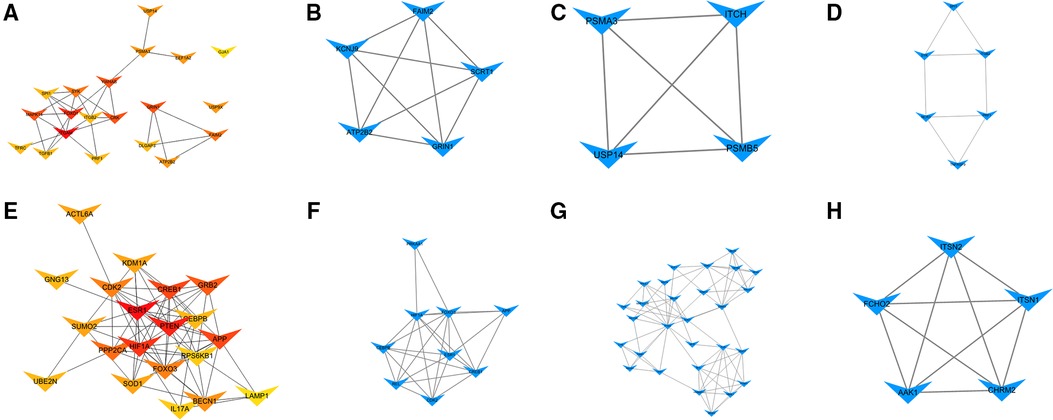
Figure 5. Hub genes and modules analysis. (A) PPI for the top 20 ranked hub genes for upregulated DEmiRNAs. (B) Module 1, (C) Module 2, and (D) Module 3 for target genes of upregulated DEmiRNAs. (E) PPI for the top 20 ranked hub genes for downregulated DEmiRNAs. (F) Module 1, (G) Module 2, and (H) Module 3 for target genes of downregulated DEmiRNAs.
Immune cells play an essential role in the pathogenesis of PE (29). Single-cell RNA sequencing was performed to explore the immunological microenvironment of MPE in GSE185058. We screened 12 overexpressed and 26 underexpressed genes (Supplementary Material S2). We crossed between 26 underexpressed genes and target genes of upregulated miRNAs and acquired a key gene, PRF1. The KEGG analysis indicated that PRF1 was involved in the natural killer cell-mediated cytotoxicity pathway (Figure 6).
PE is a common and devastating complication of advanced lung cancer and pulmonary tuberculosis. It is difficult to differentiate clinically between lung cancer and tuberculosis based on the pleural fluid. However, there is inconsistency in the pathogenesis of PEs due to lung cancer and pulmonary tuberculosis. In this study, we identified exosome-derived miRNA profiles and explored the underlying mechanism of PE in LUAD.
Our study demonstrated that a substantial number of miRNAs differ between MPE and TPE, especially some miRNAs that were extraordinarily highly expressed in MPE. The PCR and sequencing results confirmed that hsa-miR-200b-3p, hsa-miR-182-5p, and hsa-miR-629-5p were upregulated in MPE compared to TPE. Several studies indicated that hsa-miR-200b-3p is upregulated in renal cell carcinoma (30), gastric adenocarcinoma (31), and LUAD (32, 33). Our findings showed that hsa-miR-200b-3p was overexpressed in MPE, suggesting that it has the potential to serve as a biomarker to distinguish between benign and malignant PE. It was reported that hsa-miR-182-5p was upregulated in small cell lung cancer (34), ovarian cancer (35), and LUAD (36). Moreover, hsa-miR-629-5p was highly expressed in bladder urothelial carcinoma (37). These investigations validated the accuracy of our analysis, which implied that these miRNAs were associated with the malignant phenotypes and may aid in distinguishing between malignant and benign PE.
According to KEGG analysis, the target genes of upregulated DEmiRNAs were significantly enriched in Fc gamma R-mediated phagocytosis and Rap1 signaling pathway. Fc gamma receptors are involved in several immune system functions, such as phagocytosis and the release of inflammatory mediators. Several immune cells, including macrophages, neutrophils, and monocytes, participate in Fc gamma R-mediated phagocytosis. Many studies have demonstrated that tumor-associated macrophages and neutrophils have a pro-tumor effect, enhancing tumor cell invasion, metastasis, angiogenesis, and extracellular matrix remodeling (38). A previous study found that HUVEC cultured with malignant-associated PE promoted cell proliferation, migration, and angiogenesis (39); however, the molecular mechanism was not discussed. We speculated that the exosomes in MPE interacted with HUVEC to increase the formation of pleural capillaries and PE.
In the present study, we identified hub genes by PPI network construction. The hub gene with the highest degree for elevated miRNAs was ITGAM, which encodes for CD11b. As an immune-related gene, it regulates macrophage and monocyte activities and predicts prognosis and outcome in malignant tumors (40). It also plays an essential role in the adhesion of macrophages, neutrophils, and monocytes (41, 42). A previous study indicated that macrophages were associated with MPE and pleural metastasis of cancer triggered immune responses (43). We hypothesized that ITGAM was implicated in the development of MPE by tumor immune response, even though its significance in the PE of lung cancer is yet unknown.
Different categories of immune cells play distinct roles in the development of MPE. The wide geography of immune cells in human MPE was revealed by single-cell RNA sequencing, which also improved knowledge of the pathophysiology of MPE. In this study, we explored the DEGs of immune cells in the MPE microenvironment by analyzing single-cell sequencing data, which revealed that PRF1 was downregulated in MPE and was also identified as a hub gene. The KEGG analysis indicated that PRF1 is involved in the natural killer cell-mediated cytotoxicity pathway. The role of PRF1 in PE remains unclear. According to reports, T-lymphocyte cytotoxic function depends on the presence of PRF1, and its deficiency may impair antitumor immunity (44, 45). In this study, PRF1 was identified as the target gene of hsa-miR-3120. Hence, we proposed that the overexpression of hsa-miR-3120-5p in MPE decreases PRF1 expression and inhibits the immune response against cancer cells, leading to the formation of MPE. But there is still a lot to learn about this theory.Bioinformatic analysis depicted that the target genes of exosomal miRNAs were mainly immune-related, indicating that the tumor immune response might play a pivotal role in the generation of MPE in LUAD. Additionally, we speculated that tumor cells affected NK cells and promoted the generation of MPE in LUAD via the exosomal hsa-miR-3120-5p-PRF1 axis. Our results provided a new theoretical basis regarding the function of exosomal miRNAs in LUAD-MPE. However, further validation and investigation of the underlying mechanisms are required to validate our findings.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by This study received ethical approval from Beilun District People's Hospital, Ningbo, Zhejiang, China, and the current study conformed to the principles of the Declaration of Helsinki. The patients/participants provided their written informed consent to participate in this study.
XZ and LB: were responsible for identifying the exosomal miRNA profile and RT-PCR. GY and HW: performed bioinformatics. XZ and HW: were responsible for the subject design, experimental quality control, and paper writing. All authors contributed to the article and approved the submitted version.
The present study was supported by Natural Science Foundation of Ningbo City (2019A610235).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsurg.2022.1050242/full#supplementary-material.
BP, biological process; CC, cellular components; CPM, counts per million; DEmiRNAs, differentially expressed miRNAs; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; LUAD, lung adenocarcinoma MF, molecular function; MPE, malignant pleural effusion; PE, pleural effusion; PPI, protein-protein interaction; qRT-PCR, quantitative real-time PCR; TEM, transmission electron microscopy; TPE, tuberculous pleural effusion.
1. Krishnan VG, Kunoor A, Keechilath P, Mehta AA. Diagnostic utility of pleural fluid carcinoembryonic antigen in patients with exudative pleural effusion. Lung India. (2021) 38(2):139–43. doi: 10.4103/lungindia.lungindia_196_20
2. Buscail E, Alix-Panabières C, Quincy P, Cauvin T, Chauvet A, Degrandi O, et al. High clinical value of liquid biopsy to detect circulating tumor cells and tumor exosomes in pancreatic ductal adenocarcinoma patients eligible for up-front surgery. Cancers (Basel). (2019) 11(11):1656. doi: 10.3390/cancers11111656
3. Vitiello PP, De Falco V, Giunta EF, Ciardiello D, Cardone C, Vitale P, et al. Clinical practice use of liquid biopsy to identify RAS/BRAF mutations in patients with metastatic colorectal cancer (mCRC): a single institution experience. Cancers. (2019) 11(10):1504. doi: 10.3390/cancers11101504
4. Miller BF, Petrykowska HM, Elnitski L. Assessing ZNF154 methylation in patient plasma as a multicancer marker in liquid biopsies from colon, liver, ovarian and pancreatic cancer patients. Sci Rep. (2021) 11(1):221. doi: 10.1038/s41598-020-80345-7
5. De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. (2019) 40(3):172–86. doi: 10.1016/j.tips.2019.01.006
6. Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. (2020) 13(1):152. doi: 10.1186/s13045-020-00987-y
7. Wang C, Li YM, Fang XD. Research progress on liquid biopsy in oncology and its clinical applications. Yi Chuan. (2017) 39(3):220–31. doi: 10.16288/j.yczz.16-386
8. Emami A, Talaei-Khozani T, Tavanafar S, Zareifard N, Azarpira N, Vojdani Z. Synergic effects of decellularized bone matrix, hydroxyapatite, and extracellular vesicles on repairing of the rabbit mandibular bone defect model. J Transl Med. (2020) 18(1):361. doi: 10.1186/s12967-020-02525-3
9. Mohankumar S, Patel T. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Funct Genomics. (2016) 15(3):249–56. doi: 10.1093/bfgp/elv058
10. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18(1):75. doi: 10.1186/s12943-019-0991-5
11. Ahmadzada T, Kao S, Reid G, Clarke S, Grau GE, Hosseini-Beheshti E. Extracellular vesicles as biomarkers in malignant pleural mesothelioma: a review. Crit Rev Oncol Hematol. (2020) 150:102949. doi: 10.1016/j.critrevonc.2020.102949
12. Kim Y, Shin S, Lee KA. Exosome-based detection of EGFR T790M in plasma and pleural fluid of prospectively enrolled non-small cell lung cancer patients after first-line tyrosine kinase inhibitor therapy. Cancer Cell Int. (2021) 21(1):50. doi: 10.1186/s12935-021-01761-x
13. Lin J, Wang Y, Zou YQ, Chen X, Huang B, Liu J, et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Bio. (2016) 12(15):15835–45. doi: 10.1007/s13277-016-5410-6
14. Louloupi A, Ntini E, Liz J, Ørom UA. Microprocessor dynamics shows co- and post-transcriptional processing of pri-miRNAs. RNA. (2017) 23(6):892–8. doi: 10.1261/rna.060715.117
15. Xu D, Wang Y, Zhou K, Wu J, Zhang Z, Zhang J, et al. Identification of an extracellular vesicle-related gene signature in the prediction of pancreatic cancer clinical prognosis. Biosci Rep. (2020) 40(12):BSR20201087. doi: 10.1042/BSR20201087
16. Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z, et al. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. (2013) 41(22):e203. doi: 10.1093/nar/gkt1054
17. Kang HJ, Yoon S, Lee S, Choi K, Seol S, Park S, et al. FKBP5-associated miRNA signature as a putative biomarker for PTSD in recently traumatized individuals. Sci Rep. (2020) 10(1):3353. doi: 10.1038/s41598-020-60334-6
18. Kim CH, Lee SY, Lee YD, Yoo SS, Lee SY, Cha SI, et al. Atypical pleural fluid profiles in tuberculous pleural effusion: sequential changes compared with parapneumonic and malignant pleural effusions. Inter Med. (2016) 55(13):1713–9. doi: 10.2169/internalmedicine.55.5803
19. Yi FS, Zhai K, Shi HZ. Helper T cells in malignant pleural effusion. Cancer Lett. (2021) 500:21–8. doi: 10.1016/j.canlet.2020.12.016
20. Wang L, Chen X, Zhou X, Roizman B, Zhou GG. miRNAs targeting ICP4 and delivered to susceptible cells in exosomes block HSV-1 replication in a dose-dependent manner. MolTher. (2018) 26(4):1032–9. doi: 10.1016/j.ymthe.2018.02.016
21. Wang J, Ni J, Beretov J, Thompson J, Graham P, Li Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit Rev Oncol Hematol. (2020) 145:102860. doi: 10.1016/j.critrevonc.2019.102860
22. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. (2015) 8:83. doi: 10.1186/s13045-015-0181-x
23. Marqués-García F, Isidoro-García M. Protocols for exosome isolation and RNA profiling. Methods Mol Biol. (2016) 1434:153–67. doi: 10.1007/978-1-4939-3652-6_11
24. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. (2019) 48(D1):D127–31. doi: 10.1093/nar/gkz757
25. McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, et al. The biochemical basis of microRNA targeting efficacy. Science. (2019) 366(6472):eaav1741. doi: 10.1126/science.aav1741
26. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47(D1):D607–d613. doi: 10.1093/nar/gky1131
27. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. Cytohubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. (2014) 8 Suppl 4(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11
28. Ishino T, Takeno S, Takemoto K, Yamato K, Oda T, Nishida M, et al. Distinct gene set enrichment profiles in eosinophilic and non-eosinophilic chronic rhinosinusitis with nasal polyps by bulk RNA barcoding and sequencing. Int J Mol Sci. (2022) 23(10):5653. doi: 10.3390/ijms23105653
29. Jin D, Chen Y, Wang Z, Wang S, Bunjhoo H, Zhu J, et al. Diagnostic value of interleukin 22 and carcinoembryonic antigen in tuberculous and malignant pleural effusions. Exp Ther Med. (2011) 2(6):1205–9. doi: 10.3892/etm.2011.344
30. Sequeira JP, Constâncio V, Salta S, Lobo J, Barros-Silva D, Carvalho-Maia C, et al. Likidmirs: a ddPCR-based panel of 4 circulating miRNAs for detection of renal cell carcinoma. Cancers. (2022) 14(4):858. doi: 10.3390/cancers14040858
31. Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J, Yang SH, et al. MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int J Oncol. (2014) 45(6):2403–10. doi: 10.3892/ijo.2014.2680
32. Chi Y, Zheng W, Bao G, Wu L, He X, Gan R, et al. Circular RNA circ_103820 suppresses lung cancer tumorigenesis by sponging miR-200b-3p to release LATS2 and SOCS6. Cell Death Dis. (2021) 12(2):185. doi: 10.1038/s41419-021-03472-7
33. Liu K, Zhang W, Tan J, Ma J, Zhao J. MiR-200b-3p functions as an oncogene by targeting ABCA1 in lung adenocarcinoma. Technol Cancer Res Treat. (2019) 18:1533033819892590. doi: 10.1177/1533033819892590
34. Han D, Li L, Ge X, Li D, Zhang X. MicroRNA expression integrated analysis and identification of novel biomarkers in small cell lung cancer: a meta-analysis. Transl Cancer Res. (2020) 9(5):3339–53. doi: 10.21037/tcr.2020.04.12
35. Li Y, Li L. Prognostic values and prospective pathway signaling of MicroRNA-182 in ovarian cancer: a study based on gene expression omnibus (GEO) and bioinformatics analysis. J Ovarian Res. (2019) 12(1):106. doi: 10.1186/s13048-019-0580-7
36. Hsu YL, Hung JY, Lee YL, Chen FW, Chang KF, Chang WA, et al. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget. (2017) 8(62):104831–54. doi: 10.18632/oncotarget.21022
37. Sathipati S Y, Tsai MJ, Shukla SK, Ho SY, Liu Y, Beheshti A. MicroRNA signature for estimating the survival time in patients with bladder urothelial carcinoma. Sci Rep. (2022) 12(1):4141. doi: 10.1038/s41598-022-08082-7
38. Kim J, Bae JS. Tumor-Associated macrophages and neutrophils in tumor microenvironment. Mediat Inflamm. (2016) 2016:6058147. doi: 10.1155/2016/6058147
39. Changchien CY, Chen Y, Chang HH, Chang SY, Tsai WC, Tsai HC, et al. Effect of malignant-associated pleural effusion on endothelial viability, motility and angiogenesis in lung cancer. Cancer Sci. (2020) 111(10):3747–58. doi: 10.1111/cas.14584
40. Liang T, Chen J, Xu G, Zhang Z, Xue J, Zeng H, et al. TYROBP, TLR4 and ITGAM regulated macrophages polarization and immune checkpoints expression in osteosarcoma. Sci Rep. (2021) 11(1):19315. doi: 10.1038/s41598-021-98637-x
41. Tian S, Huang P, Gu Y, Yang J, Wu R, Zhao J, et al. Systems biology analysis of the effect and mechanism of qi-jing-sheng-bai granule on leucopenia in mice. Front Pharmacol. (2019) 10:408. doi: 10.3389/fphar.2019.00408
42. Jaime-Lara RB, Roy A, Wang Y, Stanfill A, Cashion AK, Joseph PV. Gene co-expression networks are associated with obesity-related traits in kidney transplant recipients. BMC Med Genomics. (2020) 13(1):37. doi: 10.1186/s12920-020-0702-5
43. Chae G, Jun JB, Jung HS, Park CY, Kim JH, Kang BJ, et al. Histiocytic pleural effusion: the strong clue to malignancy. World J Surg Oncol. (2021) 19(1):180. doi: 10.1186/s12957-021-02296-1
44. Hechinger AK, Smith BA, Flynn R, Hanke K, McDonald-Hyman C, Taylor PA, et al. Therapeutic activity of multiple common γ-chain cytokine inhibition in acute and chronic GVHD. Blood. (2015) 125(3):570–80. doi: 10.1182/blood-2014-06-581793
Keywords: exosomes, miRNAs, pleural effusions, bioinformatic analysis, lung adenocarcinoma
Citation: Zhang X, Bao L, Yu G and Wang H (2023) Exosomal miRNA-profiling of pleural effusion in lung adenocarcinoma and tuberculosis. Front. Surg. 9:1050242. doi: 10.3389/fsurg.2022.1050242
Received: 21 September 2022; Accepted: 21 November 2022;
Published: 6 January 2023.
Edited by:
Wenjie Zheng, Affiliated Hospital of Nantong University, ChinaReviewed by:
Rocco Cappellesso, University Hospital of Padua, Italy© 2023 Zhang, Bao, Yv and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haifeng Wang d2FuZ2gwODMxQDEyNi5jb20=
Specialty Section: This article was submitted to Surgical Oncology, a section of the journal Frontiers in Surgery
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.