- 1Clinical Research Institute, The First People’s Hospital of Foshan, Foshan, China
- 2R/D Center, Visionwise Medical Technology, Foshan, China
- 3Department of Dermatology, The First People’s Hospital of Foshan, Foshan, China
- 4Department of Burn and Plastic Surgery, Guangzhou First People's Hospital, South China University of Technology, Guangzhou, China
- 5Guangdong Medical University, Zhanjiang, China
Introduction: Skin cancer is one of the most common types of cancer. An accessible tool to the public can help screening for malign lesion. We aimed to develop a deep learning model to classify skin lesion using clinical images and meta information collected from smartphones.
Methods: A deep neural network was developed with two encoders for extracting information from image data and metadata. A multimodal fusion module with intra-modality self-attention and inter-modality cross-attention was proposed to effectively combine image features and meta features. The model was trained on tested on a public dataset and compared with other state-of-the-art methods using five-fold cross-validation.
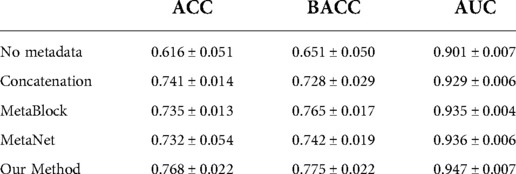
Results: Including metadata is shown to significantly improve a model's performance. Our model outperformed other metadata fusion methods in terms of accuracy, balanced accuracy and area under the receiver-operating characteristic curve, with an averaged value of 0.768±0.022, 0.775±0.022 and 0.947±0.007.
Conclusion: A deep learning model using smartphone collected images and metadata for skin lesion diagnosis was successfully developed. The proposed model showed promising performance and could be a potential tool for skin cancer screening.
Introduction
Skin cancer is one of the most common types of cancer. The number of skin cancer diagnosed all over the world reached 1.2 million in 2020 (1). Skin cancer is caused by damaged skin cells or abnormal growth of skin cells, which is closely related to excessive exposure to ultraviolet radiation, chemical carcinogens, and radioactive radiation (2). Early detection and treatment can improve the survival of patients. Dermoscope is a powerful tool used to observe pigmented skin disorders (2). By examining dermoscopic images, dermatologists can diagnose and grade a patient's lesion condition. However, reading dermoscopic images mainly relies on the experience and subjective judgments of dermatologists, which may introduce errors in the diagnosis process. Moreover, the cost of dermoscopy is relatively high, which imposes a considerable burden on patients in some underdeveloped country. Therefore, it is necessary to develop an automated tool to help to diagnose skin cancer and disease using affordable devices such as smartphone. Such a tool can reduce the cost, shorten the waiting time, and improve the accuracy of diagnosis.
Deep learning has emerged as a promising technology of artificial intelligence in recent years. With the continuous progress in the field of deep learning, artificial intelligence has made great breakthroughs in the field of medicine. Researchers have also shown positive outcomes in the smart diagnosis of skin cancer based on medical images. Various methods are proposed to diagnose skin cancers based on images (3). A mole classification system for early diagnosis of melanoma skin was proposed (4). The features were extracted according to the ABCD (5) rules of the lesions and classified them into common moles, rare moles and melanoma moles using a back-propagation feed-forward neural network. Aswin et al. (6) proposed a method for skin cancer detection based on Genetic Algorithm (GA) and Neural Network Algorithm, which divided lesion images into two categories: cancerous and non-cancerous. A skin cancer detection system based on convolutional neural network (CNN) was proposed by Mahbod et al. (7) who extracted deep features from pretrained deep CNNs for classification of skin diseases. Kalouche (8) proposed to finetune a pretrained deep CNN architecture VGG-16 and achieved an accuracy of 78%. A method combining a self-organizing neural network and a radial basis function (RBF) neural network was proposed to diagnose three different types of skin cancers with an accuracy of 93%, a significant improvement over traditional classifiers (9). Bisla et al. (10) proposed a deep learning method for data cleaning and a GAN method for data augmentation, which achieved 86.01% classification accuracy. Ali et al. also proposed a skin damage data enhancement method based on self-attention progressive GAN (11).
In real-world clinical setting, dermatologists make medical decision by considering a variety of information, including different types of imaging result, laboratory result, demographic information and patient feedback on their own feeling. To better utilize information from multiple sources, multi-modal fusion classification is introduced into the detection and classification of skin cancer. Cai et al. (12) proposed a multimodal transformer to fuse multimodal information. Chen et al. (13) proposed a skin cancer Multimodal Data Fusion Diagnosis Network (MDFNet) framework based on a data fusion strategy to effectively fuse clinical skin images with patient clinical data. Yap et al. (15) propose a method that combines multiple imaging modalities with patient metadata to improve the performance of automated skin lesion diagnosis. Li et al. proposed the MetaNet, which uses a sequence of 1D convolution on metadata to extract coefficients to assist visual features extracted from images in the classification task (16). Pacheco et al. developed a metadata processing block (MetaBlock) to fuse metadata with image data, which outperformed MetaNet and conventional feature concatenation method (17).
However, previous studies mainly focus the combination of metadata and image feature without exploring the underlying relationship between the two modalities. We argued that metadata provides extra information which can guide the interpretation of image. Similarly, image features also contain unique information which can guide the understanding of metadata. The two data modalities can facilitate each other to better unveil features that are most relevant to disease classification. We therefore proposed an attention-guided multimodal fusion network to better integrate imaging data and metadata for skin lesion diagnosis.
Methods
Model development
In medical diagnosis, clinicians are usually faced with multiple sources of information. Such information usually includes medical images and metadata (clinical or demographic supporting information that are not in the form of images). Medical decision is made after aggregating various aspects of information. In deep learning, the simplest method to combine information from different sources is channel concatenation. However, since imaging data is usually in higher dimensional than metadata, simply concatenating or adding them may not be the optimal solution for information fusion. We therefore proposed our method called multimodal fusion network (MMF-Net) to better solve the problem of image and metadata fusion.
Experiment description
In this study, we used the images and meta information provided in PAD-UPES-20 as our experimental data (17). Data were obtained from the Dermatological and Surgical Assistance Program at the Federal University of Espírito Santo (UFES), and all samples were representative of skin lesions in patients and consisted of images and meta data. The dataset includes 2,298 images collected from smartphones including 6 different types of skin lesions. Each image also contains up to 21 clinical characteristics, including but not limited to age, lesion location, Fitzpatrick skin type, and lesion diameter. All data can be accessed on the following website https://data.mendeley.com/datasets/zr7vgbcyr2/1.1. In the dataset, among the 2,298 cases, 730 of them are actinic keratosis (ACK), 845 are Basal Cell Carcinoma (BCC), 52 are malignant melanoma (MEL), 244 are Melanocytic Nevus of Skin (NEV), 192 are Squamous Cell Carcinoma (SCC) and 235 are Seborrheic Keratosis (SEK).
Model construction
To classify skin lesion types, each sample is given a smartphone photo and accompanying meta information, shown in Table 1. A convolutional neural network such as ResNet-50 was used to extract features from the smartphone image, denoted as ximg. Metadata is preprocessed as follow: numerical features were kept the same; Boolean features were converted to 0 and 1. Categorical features were one-hot encoded. For example, for the gender attribute, the one-hot encoding is [1,0] for male, [0,1] for female and [0,0] for missing information. A multi-layer perceptron was applied to extract features from meta information, denoted as xmeta. Our goal is to estimate the probability y of a skin lesion belonging to a certain class c in one of the six categories: Basal Cell Carcinoma (BCC), Squamous Cell Carcinoma (SCC), Actinic Keratosis (ACK), Seborrheic Keratosis (SEK), Melanoma (MEL), and Nevus (NEV), given the input image and meta information.
The overall design of the network is shown in Figure 1. The image encoder and meta encoder extract features from image and meta information, respectively. The extracted features are then fused together by the multimodal fusion module and passed into the classifier module composed of a fully connected layer. The final output is a six-channel vector representing the probability of six lesion types.
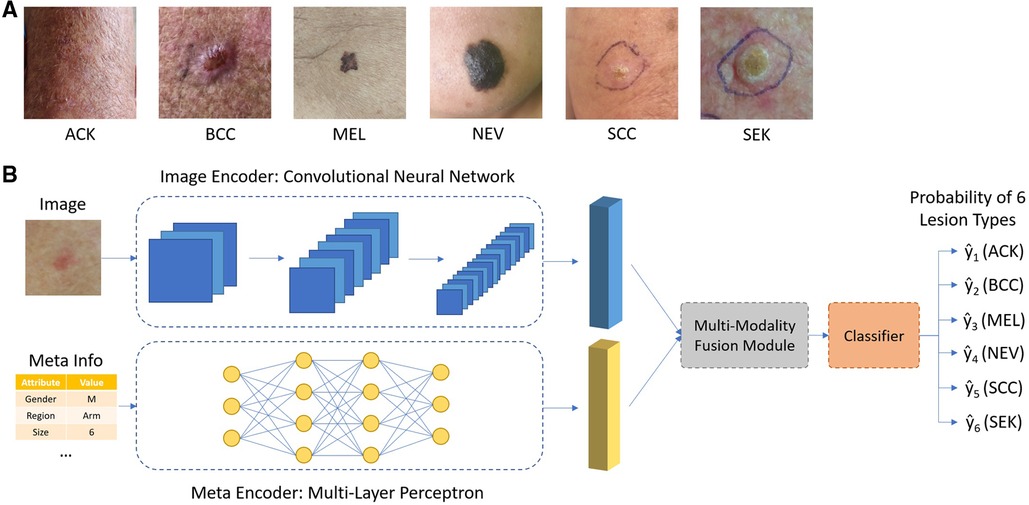
Figure 1. (A) Typical images of different types of skin lesions; (B) overall network architecture of the proposed network.
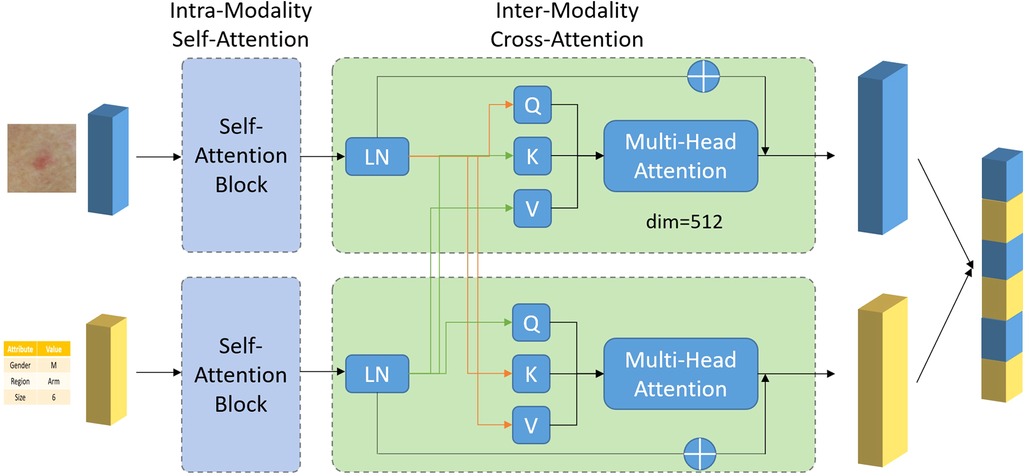
The simplest method of combining image and meta information, is by concatenating their corresponding features ximg and xmeta in the channel dimension or by summation. Yet these approaches essentially treat image and meta information as the same, ignoring the underlying differences between them. Moreover, there are inter-correlation between image and meta information, complementing of each other, which may be overlooked by simple combination. To exploit such intrinsic connection between two modalities, an attention-based multi-modality fusion module is proposed. Basically, what the attention mechanism does is to assign different weights to different features. Features with larger weights are considered to be more important in the diagnosis process and vice versa. One typical example for the use of attention mechanism is, if a specific body location is related to just one or few types of skin diseases, when such information is available as meta data, it could be used to directly suppress the prediction probability of other irrelevant diseases, thus guiding the network to focus on the rest possible types. The overall design of the fusion module, composed of intra-modality self-attention and inter-modality cross-attention, is shown in Figure 2.
Intra-modality self-attention
There may exist irrelevant information in each modality (e.g., image background in image). To avoid irrelevant information to confuse the neural network and to better guide the network to focus on key features, a multi-head self-attention module is applied to the two features, respectively. A typical attention module is consisting of two steps, linearly projecting the input feature x into Query (Q), Key (K) and Value (V) vector, followed by multiplying V by the attention weight obtained from dot-product of Q and K to get the weighted feature x’, shown as below:
After passing through the self-attention module, we obtained the weighted ximg and xmeta, which gives high weighting to cancer-relevant information and low weighting to irrelevant information.
Inter-modality cross-attention
This module contains two paths. One path is designed to use meta information to guide the selection of most relevant information from image feature. The other path is designed the other way around, to use image feature to guide the selection of most relevant information from meta data. For the first path, a cross-attention module is designed with input vector Query and Value from ximg projection and Key from xmeta, shown as below:
For the second part, a cross-attention module is designed with input vector Query and Value from xmeta projection and Key from ximg, shown as below:
We then concatenated xmeta’’ and ximg’’ to obtained the final feature vector xfinal that is passed into a fully connected layer and a softmax layer to output the probability of six lesion classes.
Training and evaluation procedures
To ensure a fair comparison with other methods, we followed the experimental setup as that in Andre's work (14). We measured different methods’ performance by calculating the following metrics including, accuracy (ACC), balanced accuracy (BACC) and aggregated area under the curve (AUC). The balanced accuracy is calculated by the arithmetic mean of sensitivity and specificity. As we can see that the portion of different diseases in the dataset is imbalanced, balanced accuracy is therefore more favored in such case as accuracy may be biased. The aggregated area under the curve is obtained by computing the average AUC of all possible pairwise combinations of the six classes, which will not be biased by imbalanced class distribution in our data. Five-fold cross-validation stratified by the label's frequency was applied for each competing method, namely baseline feature concatenation, MetaNet, MetaBlock and our proposed method. In each split, the whole dataset is split into five folds. Four folds were used as training set and the remaining fold was used as test set. The process was repeated five times until each fold has been used as test set at least once. The averaged performance of the five folds were reported.
During training, we used the SGD optimizer with an initial learning rate of 0.001. The learning rate is reduced to 0.1 of its current value if training loss stop to decrease within 10 epochs. A total of 100 epochs were trained for each method. Data augmentations such as horizontal and vertical flipping, color jittering, gaussian noise and random contrast were applied on-the-fly during training. All codes were written in Python 3.6.10. PyTorch framework (v1.7.0) was used by constructed the neural network.
Statistical analysis
Metadata attributes were examined by univariate analysis. Continuous variables were first examined by Shapiro–Wilk test to determine if they were normally distributed. Student t test (for normally distributed parameters) or Mann–Whitney U test (for non-normally distributed parameters) were used to identify statistically significant variable between different skin lesion types. Categorical variables were compared by chi-square test. To compare different methods’ performance, non-parametric Friedman test followed by the Wilcoxon test were used. P < 0.05 is consider to be statistically significant. The statistical analysis was performed with SPSS (IBM, version 26).
Results
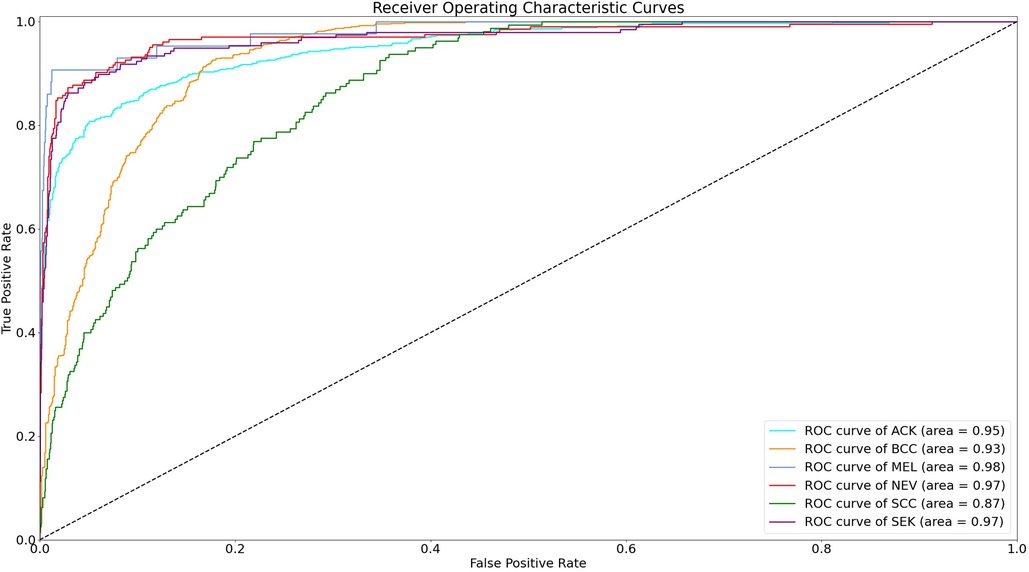
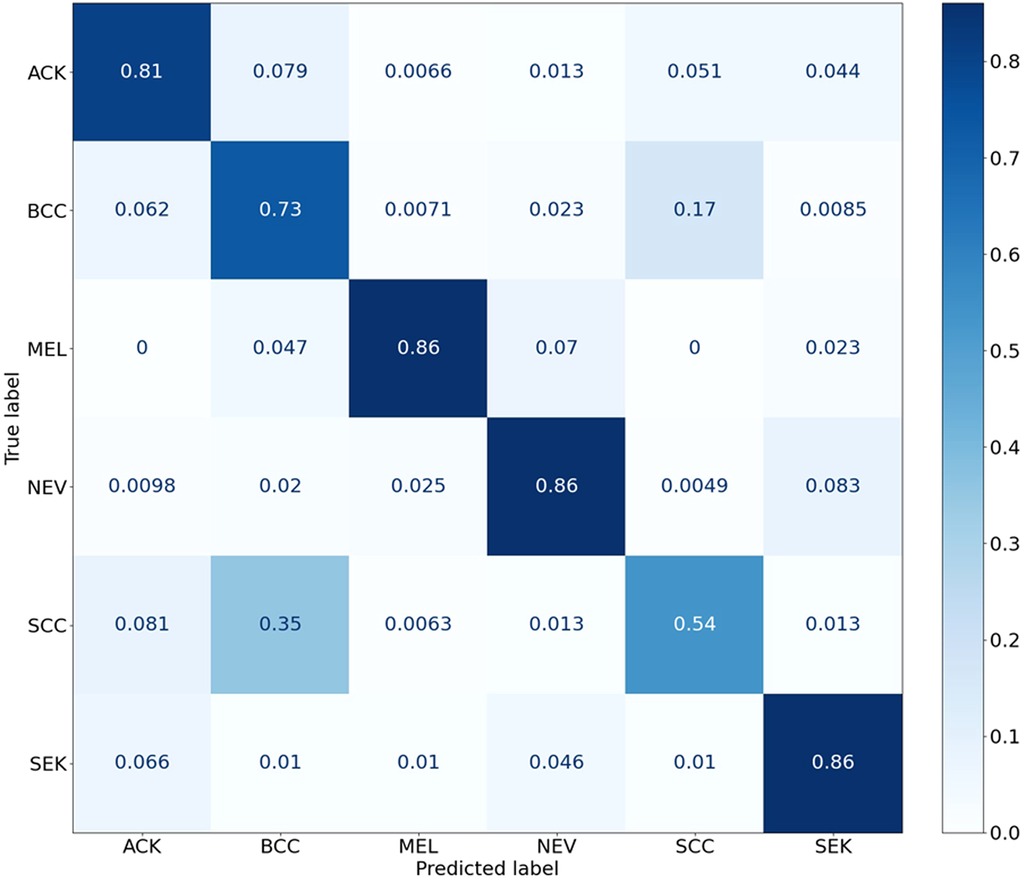
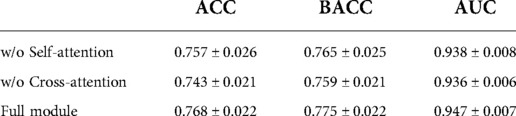
The statistical significance of the difference of metadata attributes between different lesion types is shown in Table 1. Most metadata attributes are significantly correlated with different types of skin lesion. The averaged five-fold cross-validation performance of each method is presented in Table 2. Our proposed method achieved the best performance in ACC (0.768 ± 0.022), BACC (0.775 ± 0.022) and AUC (s) metrics. We can observe that including meta information significantly improves the performance in terms of AUC (0.947 ± 0.007 vs. 0.901 ± 0.007, P < 0.001), ACC (0.768 ± 0.022 vs. 0.616 ± 0.051, P < 0.001) and BACC (0.775 ± 0.022 vs. 0.651 ± 0.050, P < 0.001), as shown in Table 3. Our proposed method utilized meta information in a more effective way, which significantly outperformed the other two methods MetaNet (P = 0.035) and MetaBlock (P = 0.028). The ROC curves for six lesion types are plotted in Figure 3. The proposed method achieved the best performance for MEL (AUC = 0.98), followed by NEV (AUC = 0.97), SEK (AUC = 0.97), ACK (AUC = 0.95) and BCC (AUC = 0.93). SCC is the most difficult one to identify, with AUC = 0.87. The averaged AUC of six disease types is 0.947. We further examined the failed cases. Figure 4 shows the averaged confusion matrix, from which we can see that SCC has the lowest accuracy because it is often mistaken as BCC. This is understandable as our images are collected from smartphone. Clinically, there is a tendency of SCC misdiagnosed as BCC. Using dermoscopy can improve the diagnostic accuracy (22). We further performed an ablation study to analyze different components’ effect on the final result. Table 4 shows the performance of the model without the inter-modality self-attention and model without the intra-modality cross-attention. This indicates that the two modules act synergistically to improve the fusion of image and metadata.
Discussion
We have developed a deep learning model to diagnose skin lesion by using clinical images and meta information obtained from smartphones. We proposed a new module which uses the combination of intra-modality self-attention and inter-modality cross-attention to better fuse image and metadata. The proposed model achieved promising performance on the public dataset, outperforming other state-of-the-art methods.
Compared with previous work (MetaNet, MetaFuse) on metadata fusion, our method has made two improvements. First, we used a multi-head self-attention module which calculates the feature correlation matrix to assign attention weights to different features within the same modality, removing irrelevant information in each modality. Second, unlike previous works where the attention is one-way (metadata feature ≥ image feature), our attention mechanism works two-way. Metadata features are used to guide the attention on image features and vice versa, image features are also used to guide the attention on metadata features. This has enabled our model to better exploit the information in each modality and outperform the other methods, as demonstrated in our experiments.
Many previous studies applying artificial intelligence to skin disease diagnosis focused on dermoscopy images only. However, dermoscope is not always accessible to many people in rural area or in underdeveloped country. In contrast, smartphone is a more accessible tool to many patients. Moreover, many patients tend to seek advices online instead of scheduling a visit to the clinic at the early stage of skin disease, which can delay their treatment and result in poor prognosis. The model developed in the current study can be integrated into a smartphone-based automatic screening app, which can help to identify high risk lesion and urge the patient for immediate treatment.
In real world clinical settings, dermatologists seldom make decision solely based on images. They usually make their judgement based on patient demographics (age, gender) and lesion characteristics (anatomical region) (18) which cannot be simply obtained from images. They also consider other risk factors such as cancer history, exposure to chemical and types of skin (19). Including meta information and multiple sources of data has been proved to be helpful for deep learning model in some previous studies (20, 21). In this study, we incorporated meta information into neural network and have showed that it can significantly improve the skin lesion diagnostic performance. Such information can be easily obtained in the form of online questionnaire which costs only a few clicks on the app from the user.
Though we have developed a promising model for skin lesion diagnosis, there are several limitations in our study. The number of included skin lesion types is relatively few compared to real world scenario. This can be improved by collecting data from more diseases in the future. External validation using data collected from different sites is needed before the model is put into clinical practice.
Conclusion
We have developed a deep learning model to diagnose skin lesion using clinical images and meta information obtained from smartphones. A new module consisting of intra-modality self-attention and inter-modality cross-attention is proposed to better fuse image data and metadata and is shown to outperformed other state-of-the-art methods. Our proposed model could be integrated into smartphone as a potential and handy tool to screen for skin disease and skin cancer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
CO, SZ, and RY contributed to conception and design of the study. CO, HH and WJ completed the code and analysis. SZ and RY performed the statistical analysis. CO and HH wrote the first draft of the manuscript. SZ, RY, WG, WC and XQ wrote sections of the manuscript. WL, XP and JL supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (82002913), Guangdong Basic and Applied Basic Research Foundation (2021A1515011453, 2022A1515012160, 2021B1515120036, and 2022A1515012245) and the Medical Scientific Research Foundation of Guangdong Province (A2022293).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. (2021) 149(4):778–89. doi: 10.1002/ijc.33588
2. Khan IU, Aslam N, Anwar T, Aljameel SS, Ullah M, Khan R, et al. Remote diagnosis and triaging model for skin cancer using efcientnet and extreme gradient boosting. Complexity. (2021) 2021:5591614. doi: 10.1155/2021/5591614
3. Dildar M, Akram S, Irfan M, Khan HU, Ramzan M, Mahmood AR, et al. Skin cancer detection: a review using deep learning techniques. Int J Environ Res Public Health. (2021) 18(10):5479. doi: 10.3390/ijerph18105479
4. Cueva WF, Munoz F, Vasquez G, Delgado G. Detection of skin cancer “melanoma” through computer vision. In: Proceedings of the 2017 IEEE XXIV international conference on electronics, electrical engineering and computing (INTERCON), Cusco, Peru; 15–18 August 2017. p. 1–4.
5. Nachbar F, Stolz W, Merkle T, Cognetta AB, Vogt T, Landthaler M, et al. The ABCD rule of dermatoscopy: high prospective value in the diagnosis of doubtful melanocytic skin lesions. J Am Acad Dermatol. (1994) 30(4):551–9. doi: 10.1016/S0190-9622(94)70061-3
6. Aswin RB, Jaleel JA, Salim S. Hybrid genetic algorithm: artificial neural network classifier for skin cancer detection. In: Proceedings of the 2014 international conference on control, instrumentation, communication and computational technologies (ICCICCT), kanyakumari, India; 10–11 July 2014. p. 1304–9.
7. Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I. Skin lesion classification using hybrid deep neural networks. In: Proceedings of the ICASSP 2019–2019 IEEE international conference on acoustics, speech and signal processing (ICASSP), brighton, UK; 12–17 May 2019. p. 1229–33.
8. Kalouche S. Vision-based classification of skin cancer using deep learning (2016). Available at: https://www.semanticscholar.org/paper/Vision-Based-Classification-of-Skin-Cancer-using-Kalouche/b57ba909756462d812dc20fca157b397 2bc1f533 (Accessed January 10, 2021).
9. Mengistu AD, Alemayehu DM. Computer vision for skin cancer diagnosis and recognition using RBF and SOM. Int J Image Process. (2015) 9:311–9. doi: 10.1007/s00432-022-04180-1
10. Bisla D, Choromanska A, Stein JA, Polsky D, Berman R. Towards automated melanoma detection with deep learning: data purification and augmentation. arXiv. (2019) 2720–28. doi: 10.48550/arXiv.1902.06061
11. Abdelhalim ISA, Mohamed MF, Mahdy YB. Data augmentation for skin lesion using self-attention based progressive generative adversarial network. Expert Systems with Applications. (2021) 165:113922. doi: 10.48550/arXiv.1910.11960
12. Cai G, Zhu Y, Wu Y, Jiang X, Ye J, Yang D. A multimodal transformer to fuse images and metadata for skin disease classification. Vis Comput. (2022):1–13. doi: 10.1007/s00371-022-02492-4
13. Chen Q, Li M, Chen C, Zhou P, Lv X, Chen C. MDFNet: application of multimodal fusion method based on skin image and clinical data to skin cancer classification. J Cancer Res Clin Oncol. (2022) 1–13. doi: 10.1007/s00432-022-04180-1
14. Pacheco AG, Krohling RA. An attention-based mechanism to combine images and metadata in deep learning models applied to skin cancer classification. IEEE J Biomed Health Inform. (2021) 25(9):3554–63. doi: 10.1109/JBHI.2021.3062002
15. Yap J, Yolland W, Tschandl P. Multimodal skin lesion classification using deep learning. Exp Dermatol. (2018) 27(11):1261–7. doi: 10.1111/exd.13777
16. Li W, Zhuang J, Wang R, Zhang J, Zheng W-S. Fusing metadata and dermoscopy images for skin disease diagnosis. In: 2020 IEEE 17th international symposium on biomedical imaging (ISBI). IEEE (2020). pp. 1996–2000.
17. Pacheco AG, Lima GR, Salomão AS, Krohling B, Biral IP, de Angelo GG, et al. PAD-UFES-20: a skin lesion dataset composed of patient data and clinical images collected from smartphones. Data Brief. (2020) 32:106221. doi: 10.1016/j.dib.2020.106221
18. Wolff K, Johnson RA, Saavedra AP, Roh EK. Fitzpatrick’s color atlas and synopsis of clinical dermatology. 8th edn. New York, USA: McGraw-Hill Education (2017).
19. Duarte AF, Sousa-Pinto B, Haneke E, Correia O. Risk factors for development of new skin neoplasms in patients with past history of skin cancer: a survival analysis. Sci Rep. (2018) 8(1):1–6. doi: 10.1038/s41598-018-33763-7
20. Kharazmi P, Kalia S, Lui H, Wang Z, Lee T. A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile. Skin Res Technol. (2018) 24(2):256–64. doi: 10.1111/srt.12422
21. Ou C, Li C, Qian Y, Duan CZ, Si W, Zhang X, et al. Morphology-aware multi-source fusion–based intracranial aneurysms rupture prediction. Eur Radiol. (2022) 32:5633–41. doi: 10.1007/s00330-022-08608-7
Keywords: deep learning - artificial neural network, multimodal fusion, metadata, skin cancer, attention
Citation: Ou C, Zhou S, Yang R, Jiang W, He H, Gan W, Chen W, Qin X, Luo W, Pi X and Li J (2022) A deep learning based multimodal fusion model for skin lesion diagnosis using smartphone collected clinical images and metadata. Front. Surg. 9:1029991. doi: 10.3389/fsurg.2022.1029991
Received: 28 August 2022; Accepted: 15 September 2022;
Published: 4 October 2022.
Edited by:
Kun Xiong, Central South University, China© 2022 Ou, Zhou, Yang, Jiang, He, Gan, Chen, Qin, Luo, Pi and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiehua Li amllaHVhbGlfZnNAMTYzLmNvbQ== Xiaobing Pi cHhiaW5naWNlQDE2My5jb20= Wei Luo bHVvd2VpXzQyMUAxNjMuY29t
†These authors have contributed equally to this work
Specialty Section: This article was submitted to Reconstructive and Plastic Surgery, a section of the journal Frontiers in Surgery
 Chubin Ou
Chubin Ou Sitong Zhou
Sitong Zhou Ronghua Yang
Ronghua Yang Weili Jiang
Weili Jiang Haoyang He2
Haoyang He2 Wei Luo
Wei Luo