- 1Department of Neurosurgery, St. Joseph’s Hospital and Medical Center, Barrow Neurological Institute, Phoenix, AZ, USA
- 2Division of Neurosurgery, College of Medicine, University of Arizona, Tucson, AZ, USA
- 3College of Medicine – Phoenix, University of Arizona, Phoenix, AZ, USA
- 4Laboratory of Neurosurgery, Irkutsk Scientific Center of Surgery and Traumatology, Irkutsk, Russia
- 5Irkutsk State Medical University, Irkutsk, Russia
Background: Intervertebral disc degeneration (IVDD) is considered a multifactorial disease that is influenced by both environmental and genetic factors. The last two decades of research strongly demonstrate that genetic factors contribute about 75% of the IVDD etiology. Recent total genome sequencing studies have shed light on the various single-nucleotide polymorphisms (SNPs) that are associated with IVDD.
Aim: This review presents comprehensive and updated information about the diversity of genetic factors in the inflammatory, degradative, homeostatic, and structural systems involved in the IVDD. An organized collection of information is provided regarding genetic polymorphisms that have been identified to influence the risk of developing IVDD. Understanding the proteins and signaling systems involved in IVDD can lead to improved understanding and targeting of therapeutics.
Materials and methods: An electronic literature search was performed using the National Library of Medicine for publications using the keywords genetics of IVDD, lumbar disc degeneration, degenerative disc disease, polymorphisms, SNPs, and disc disease. The articles were then screened based on inclusion criteria that included topics that covered the correlation of SNPs with developing IVDD. Sixty-five articles were identified as containing relevant information. Articles were excluded if they investigated lower back pain or just disc herniation without an analysis of disc degeneration. This study focuses on the chronic degeneration of IVDs.
Results: Various genes were identified to contain SNPs that influenced the risk of developing IVDD. Among these are genes contributing to structural proteins, such as COL1A1, COL9A3, COL9A3, COL11A1, and COL11A2, ACAN, and CHST3. Furthermore, various SNPs found in the vitamin-D receptor gene are also associated with IVDD. SNPs related to inflammatory cytokine imbalance are associated with IVDD, although some effects are limited by sex and certain populations. SNPs in genes that code for extracellular matrix-degrading enzymes, such as MMP-1, MMP-2, MMP-3, MMP-9, MMP-14, ADAMTS-4, and ADAMTS-5 are also associated with IVDD. Apoptosis-mediating genes, such as caspase 9 gene (CASP9), TRAIL, and death receptor 4 (DR4), as well as those for growth factors, such as growth differentiation factor 5 and VEGF, are identified to have polymorphisms that influence the risk of developing IVDD.
Conclusion: Within the last 10 years, countless new SNPs have been identified in genes previously unknown to be associated with IVDD. Furthermore, the last decade has also revealed new SNPs identified in genes already known to be involved with increased risk of developing IVDD. Improved understanding of the numerous genetic variants behind various pathophysiological elements of IVDD could help advance personalized care and pharmacotherapeutic strategies for patients suffering from IVDD in the future.
Introduction
Over 80% of all people will experience some form of lower back pain in their lifetime (1–3). Symptomatic intervertebral disc (IVD) degeneration (IVDD) is a common cause of lower back pain, yet the etiology and pathophysiology underlying IVDD remain poorly understood (4, 5). Although various environmental factors such as smoking, age, gender, and mechanical load increase the risk of IVDD, it is hypothesized that up to 74% of the etiology of IVDD is due to heritability (2, 6). With lower back pain costing over $100 billion/year in the United States, it is essential to investigate both the environmental and genetic predispositions to IVDD (5).
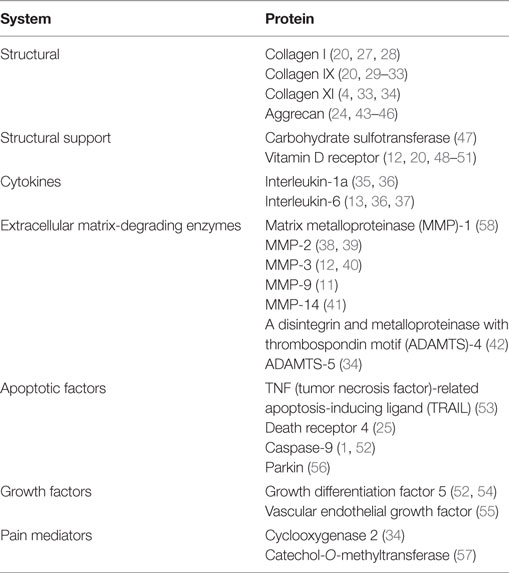
The normal IVD is composed of two parts: the outer annulus fibrosis (AF) region and the central nucleus pulposus (NP) (Figure 1). The AF consists of fibroblast-like cells with elongated nuclei placed between concentric layers of collagen fibers (3). The extracellular matrix (ECM) of the AF can be described as a fibrocartilaginous structure consisting of predominantly collagen-I fibers (60% of total dry weight), with low proteoglycan content (25%) and low water retention (5, 7). Its primary function is to provide structural integrity to the disc and hold the contents of the NP in the center (3, 5). The NP is a gelatinous structure with chondrocyte-like cells that secrete collagen II. Dispersed throughout the collagen fibers are an abundance of proteoglycans, predominantly aggrecan, which are responsible for facilitating water retention (3, 5, 8). The primary function of the NP is to create hydrostatic pressure to resist axial compression (5, 7).
Figure 1. (A) Normal intervertebral disc (IVD) from the sagittal view. (B) Normal IVD from the axial view. (C) Magnified illustration of an IVD. Used with permission from Barrow Neurological Institute, Phoenix, AZ, USA.
Intervertebral disc degeneration seems to be an irreversible process that can begin as early as the second decade (5). The first molecular change that occurs at the beginning of degeneration is a reduced ability of the NP to retain water and consequently maintain a significant hydrostatic pressure (7). These changes result in decreased disc height and reduced ability of the spine to withstand compression. Over time, the collagen fibers and other ECM components of both the NP and AF are degraded and reduced in quantity (8). Upregulation of degradative systems such as apoptosis, inflammation, and matrix metalloproteinases (MMPs) further damage the existing (9–13). The past 20 years of genomic research has revealed an astounding number of genetic polymorphisms of various genes that are correlated with increased risk of developing IVDD. Polymorphisms in the genes coding for collagen, aggrecan, interleukins (ILs), apoptosis factors, vitamin D receptor (VDR), MMPs, and other critical proteins involved in IVDD are examined in this paper. Although previous reviews have documented the various single-nucleotide polymorphisms (SNPs) that are associated with IVDD, we aim to provide an up-to-date and comprehensive review of the subject (5, 7, 8).
With an improved understanding of the genetic variants associated with IVDD, we hope to help advance personalized care and pharmaceutical therapies for patients suffering from IVDD. Across various medical specialties, genome sequencing has begun to play a significant role in improving the care provided to patients (14). Human genome analysis allows physicians to obtain a deeper understanding of the pathophysiology of diseases to provide improved risk and prognostic assessments to patients. Furthermore, information regarding genetic variants provides insight into therapeutic options as physicians are better able to target the underlying disease-causing mechanisms (15). Throughout this paper, we will explore the complexity and diversity of the molecular and genetic factors involved in IVDD. Genetic variants from various molecular pathways are investigated including inflammatory, degradative, homeostatic, and structural systems. Clinical use of genome analysis allows physicians to pinpoint which systems and particular pathways are involved with the patient’s unique case of IVDD and subsequently provide personalized and improved health care.
Methods
An electronic literature search was performed using the National Library of Medicine for publications using these keywords: genetics of IVDD, lumbar disc degeneration, degenerative disc disease, polymorphisms, SNPs, and disc disease. The articles were then screened based on inclusion criteria that included topics that covered the correlation of SNPs with developing IVDD. Furthermore, articles containing supporting information regarding the treatment and diagnosis of IVDD were included. Sixty-five total articles were identified as containing relevant information. Articles were excluded if they investigated lower back pain or disc herniation without an analysis of disc degeneration or study of correlation with SNPs. This investigation focuses on the chronic degeneration of IVDs and the genetic factors that influence its development.
Treatment for IVDD
Diagnosis of IVDD requires a careful history, physical examination, and, most importantly for the experimental studies included in this review, magnetic resonance imaging (MRI) of the spine. The majority of studies that were included in this literature review used axial and/or sagittal T2-weighted MRIs to evaluate the lumbar spine of the patients (Figure 2). Once the patient has been accurately diagnosed with disc degeneration, limited approved therapeutics are available to abate the progression of the degeneration. Therapy to combat IVDD and the associated degeneration and pain is highly complex, and it can be difficult to predict its effectiveness. Recently, researchers have found success utilizing targeted molecular and gene therapies in an attempt to mitigate degradation and even promote anabolic processes. Injection of recombinant human bone morphogenetic protein 7 (BMP-7, also known as osteogenic protein 1, OP-1) has been successful in a rabbit model (16). BMP-7 injection restored the disc height and biomechanical properties of the damaged disc. Other growth factors such as rhGDF-5 have also shown great promise (17). In that study, a single injection was shown to increase disc height. Furthermore, rhGDF-5 injection has been shown to reduce the expression of ADAMTS-4 and ADAMTS-5 proteins for which, the genes have been identified to contain SNPs associated with altered risk of developing IVDD (18). This serves as an excellent example of the intersection of providing targeted therapy and gene analysis of patients with IVDD. RhGDF-5 injections may serve as the most effective therapy in a patient who has been screened for having high-risk IVDD due to SNPs in their ADAMTS-4 and -5 genes (18). Furthermore, injection of other molecules, such as TGF-β1 and BMP-2, has been shown to inhibit MMP-1 expression and increase expression of aggrecan protein. Genes for both MMP-1 and aggrecan protein are known to contain SNPs that predispose patients to develop IVDD (19). Combining the specific effects of these anabolic therapies with an understanding of the individualized molecular profile of each patient may yield a highly effective treatment. Therefore, it is essential that research efforts continue to progress in both targeted therapies and gene analysis of IVDD.
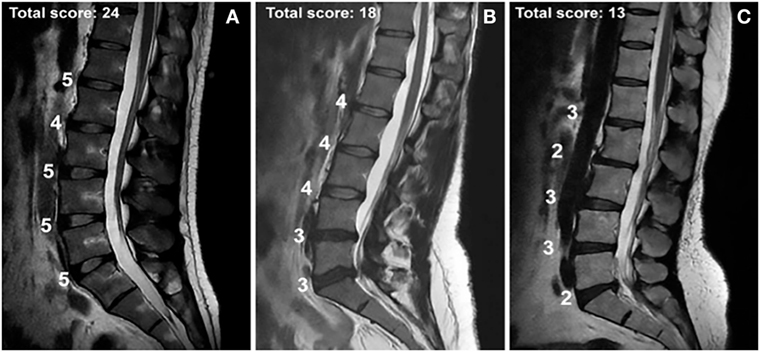
Figure 2. MRIs of intervertebral disc disease in patients with total Pfirrmann scores of (A) 24, (B) 18, and (C) 13 and the assessed score for each lumbar disc. The Pfirrmann grading scale for disc degeneration classifies discs into 5 grades according to the amount of degeneration. Grade 1 corresponds to a hyperintense healthy disc, while grade 5 corresponds to a hypointense severely degenerated disc. The figure contains a point system corresponding to each intervertebral disc from L1 to S1. Five points were given for a grade 1 score, four points for a grade 2, three for a grade 3, two for a grade 4, and one for a grade 5. The highest possible total score is 25; the lowest possible score is 5. Used with permission from Toktas et al. (20).
The most effective experimental approach in the treatment of IVDD is the use of viral vectors in gene therapy. In vitro bovine experimentation with the delivery of sex-determining region Y box 9 (SOX9) and BMP7 through an adenovirus vector revealed increased expression of type II collagen and an increase in disc height (21). Another experiment showed that cells virally transduced with Ad-BMP-4 and -14 displayed an increase in collagen deposition, whereas cells transduced with Ad-BMP-2 and -7 displayed an increase in proteoglycan accumulation (19). The consistently positive results obtained from these experiments suggest a largely uncharted frontier exists in the use of personalized medicine for IVDD.
Grading IVDD
Physicians utilize various grading systems to assist in diagnosing and measuring the severity of IVDD and to determine the most standardized and objective classification of disc degeneration. A popular and widely accepted scale is the Pfirrmann grading system. The system includes grades 1–5, where grade 1 signifies a normal disc with homogenous hyperintensity on MRI, and grade 5 signifies a collapsed disc space with a hypointense signal (Figure 2) (22). A common critique of the system is its subjectivity. It is often modified or combined with other grading systems such as Modic changes to create an objective, reproducible system (23). Some physicians and research groups opt to develop their own grading system, while others utilize classification systems such the one developed by Schneiderman et al. (13, 24–26). Once a patient’s disc degeneration is objectively graded, a standard of care can be established.
Gene Polymorphisms Associated with IVDD
Table 1 (1, 4, 11–13, 20, 24, 25, 27–57) presents comprehensive information on the research studies that have investigated genes with SNPs associated with IVDD and their protein products. Table 2 (1, 4, 11–13, 20, 24, 25, 27–58) summarizes the protein systems associated with such changes in the respective genes.
Table 1. Summary of research studies on single-nucleotide polymorphisms (SNPs) associated with intervertebral disc degeneration (IVDD).
Collagens
Collagen is the most abundant protein found in the human body, with 28 different types. Throughout the body, the various collagen types are found in the ECM and have different structural support roles. Structurally, collagen fibers are composed of three polypeptide chains, referred to as α chains, that form one or more triple-helixes along their rod-shaped structure (59). When referring to the gene that produces a specific collagen type, the gene name and subunit name are given (e.g., collagen type IX alpha 2, COL9A2). The collagen types of interest to us are the ones found within IVDs: collagen I, II, IX, and XI.
The AF consists primarily of collagen I, a fibril-forming collagen. Fibrillar collagens – I, II, and III – are essential in defining the molecular and mechanical properties of a particular tissue (59). In the AF, collagen I is responsible for maintaining the tensile strength to withstand spinal compression, hydrostatic pressure, and keeping the NP contained. Collagen II is the primary collagen of the NP and is found as a loosely connected network (3). Various minor collagens such as collagen IX play an important supporting role in forming cross-links between different types of collagen, increasing structural strength. Collagen XI, although found in small amounts, is important in structural support of collagen II as well as forming connections between proteoglycans and collagen (5, 8). Considering the integral role of collagen in maintaining the structural integrity of the IVD, genetic polymorphisms affecting the function or abundance of collagen can predispose a patient to IVDD.
Collagen I
Collagen I, although found in both the NP and AF, is much more abundant within the AF of the IVD. Collagen I is made up of a helix consisting of two α1 chains, encoded by the collagen type I alpha 1 gene, COL1A1, and one α2 chain encoded by the collagen type I alpha 2 gene COL1A2 (7). COL1A1 contains a particular polymorphism that may be involved with increased risk of IVDD. Three noteworthy studies have established an association between the COL1A1 Sp1-binding site SNP and IVDD (20, 27, 28). This particular SNP is a G to T substitution at position +1245, which is found within the first intron of the COL1A1 gene (60). The change in nucleotides reportedly increases levels of COL1A1 messenger RNA expression and subsequently COL1A1 protein expression (27). Investigators have hypothesized that the SNP leads to disequilibrium between COL1A1 and COL1A2 protein expression leading to instability of the collagen fibers (27, 28). Pluijm et al. examined 966 elderly (>65 years) Dutch individuals and reported that patients with the TT genotype had a 3.6-fold increased susceptibility to IVDD than patients with the GT or GG genotypes (27). The following year, Tilkeridis et al. examined the frequency of the Sp1-binding site polymorphism in 24 young Greek military recruits (28). The study reported that 33.3% of the patients with IVDD had the TT genotype while none of the control subjects did. Furthermore, the study indicated that 66.7% of the IVDD patients had the GT genotype while only 41.7% of the controls did. More recently, a 2015 study by Toktas et al. found that patients homozygous for the risk allele T had a significantly lower average Pfirrmann score (17.63) than patients without the allele (average score, 21.88) (20). They found a similar relationship between patients heterozygous for the allele compared with control patients. This study suggests that the COL1A1 Sp1 polymorphism may not only be associated with an increased risk of developing IVDD but also associated with more severe forms of degeneration.
Collagen IX
Collagen IX is composed of three unique polypeptides, such as α1, α2, and α3, which are encoded by genes collagen type 9 alpha 1 (COL9A1), collagen type 9 alpha 2 (COL9A2), and collagen type 9 alpha 3 (COL9A3), respectively (20). Collagen IX is thought to play a significant role in connecting various types of collagens together, particularly collagen II (8, 59). Various studies have found SNPs located on either COL9A2 or COL9A3 that may be associated with increased risk of IVDD.
Annunen et al. examined 157 unrelated Finnish subjects with IVDD-induced sciatica (29). The study characterized a COL9A2 polymorphism named Trp2, which caused a substitution of Gln or Arg for Trp in the collagen molecule. This substitution is particularly interesting because there are no naturally occurring Trp residues in collagen because the COL9 gene does not encode for the amino acid Trp. The statistical analysis showed that patients with the allele coding for Trp were at a 4.5-fold increased risk of developing IVDD than those without the allele (29). Their population analysis found that 6 of the 157 individuals with IVDD had the Trp allele while none of the 174 controls did. A few other investigators have attempted to establish a connection between the Trp2 allele and IVDD but failed. For instance, Toktas et al. (20), Kales et al. (30), and Zhang et al. (31) did not find a correlation between COL9A2 polymorphisms and IVDD.
A common SNP that has been studied in COL9A3 is Trp3. This SNP is similar to the one found in COL9A2; it is an Arg103 to Trp substitution. Paassilta et al. studied the occurrence of the Trp3 allele in 171 Finnish subjects (32). The statistical analysis showed that patients who had a copy of the Trp3 allele were at a 2.7-fold increased risk of developing IVDD compared with patients who did not have the allele. Evidence for the association between the Trp3 allele and IVDD grew with a 2006 study by Solovieva et al. (33). They examined 135 middle-aged Finnish men and found that patients who carried the Trp3 risk allele in the absence of the IL-1 βT3954 SNP allele were at a 7.0-fold increased risk of a dark NP on MRI. These men had an overall 8.0-fold increased risk of degenerative changes in the spine. Although this study qualified the association between Trp3 and IVDD as dependent on the absence of the IL-1 βT3954 SNP allele, it nonetheless established a connection between the two (33). More recently, a 2015 study by Toktas et al. established a connection between the Trp3 allele and increased severity of disc degeneration (20). The study showed that of the five cases with Trp3 alleles, the heterozygous patients with the allele had a significantly lower average Pfirrmann score (19.40) compared with the wild-type patients without the allele (average score, 21.07). This finding suggests that not only is the Trp3 allele associated with an increased risk of developing IVDD but also associated with more severe forms of degeneration.
Collagen XI
Collagen XI has a similar structure to collagen IX in that it is a heterotrimer. The three chains, such as α1, α2, and α3, are coded by collagen type XI alpha 1 (COL11A1), collagen type XI alpha 2 (COL11A2), and collagen type II alpha 1 (COL2A1), respectively (5). Collagen XI is found in both the AF and NP of IVDs and has an important role in connections between the different collagen molecules, particularly collagen II, as well as connections between proteoglycans and collagen (5, 8).
Solovieva et al. showed a relationship between a G to A substitution SNP within intron 9 of COL11A2 and disc bulging (33). Patients who were carriers of the SNP allele had a 2.1-fold increased risk of disc bulging compared with patients who did not have the allele. The study also noted a 1.6-fold increased risk of signs of disc degeneration, but the SD was too large to be statistically significant. Nonetheless, it is worth noting that the G to An SNP of COL11A2 was related to change associated with disc degeneration. A 2009 study by Videman et al. documented five different polymorphisms in collagen XI genes that were significantly associated with signs of disc degeneration such as reduced disc signal and disc bulging (4). This particular large-scale study enrolled 588 Finnish male twins ranging from 35 to 70 years of age. The rs2072915, rs9277933, and rs2076311 SNPs of COL11A2 were significantly associated with reduced disc signal on MRI, whereas the rs1337185 and rs1463035 polymorphisms of COL11A1 were significantly associated with increased risk of disc bulging. A 2015 study by Rajasekaran et al. supported these findings (34). The study revealed the rs1337185 SNP of COL11A1 was associated with a 1.55-fold increased risk of developing IVDD. Research suggests that SNPs in both COL11A2 and COL11A1 could predispose an individual to an increased risk of developing IVDD.
Cytokines
Cytokines, such as IL-6, IL-1a, IL-1b, and tumor necrosis factor (TNF)-α, are some of the key pro-inflammatory mediators that are found and released at sites of tissue injury. IL-1 is naturally found within the IVD and is responsible for indirectly degrading ECM components through the production of degradative enzymes, upregulation of other cytokines, and preventing the production of ECM components (5). IL-1 has three different subtypes: IL-1a, IL-1b, and IL-1RN. The alpha and beta subtypes are pro-inflammatory, whereas IL-1RN is anti-inflammatory (7). Within the disc, a delicate homeostasis between the pro-inflammatory and anti-inflammatory subtypes exists that is easily disturbed by trauma to the spine and genetic polymorphisms.
A common SNP of interleukin 1 alpha (IL1A) was significantly associated with IVDD in a 2007 study by Virtanen et al. who examined 150 Finnish men (35). The SNP is an -889C/T substitution where the T allele is the risk allele. Patients in the study with the TT genotype were at a 7.87-fold increased risk of developing IVDD compared with patients with the CC genotype. These findings were supported by a 2012 study by Eskola et al. of Danish adolescents (36). The study found a 2.82-fold increased risk of developing IVDD among girls who were carriers of the T allele compared with the controls. The study also described the polymorphism as increasing IL-1a expression, and thus furthering its function as a cartilage destroyer (36). These two studies, along with a few others, established the -889C/T SNP of IL1A as a genetic risk factor for IVDD (7, 35, 36).
Interleukin-6 is an important mediator of inflammation and having involvement with lumbar disc herniation (36). Despite this information, the exact role of IL-6 in disc degeneration is not fully known (5). Noponen-Hietala et al. documented an SNP in the interleukin 6 gene (IL6) that was significantly associated with IVDD (37). A 15T/A substitution was located within exon 5 of IL6. Statistical analysis showed that patients with the AA or AT genotypes were at a 4.4-fold increased risk of IVDD than patients with the TT genotype. The study documented that the 15T/A SNP results in an exon 5 amino acid substitution that replaces Asp with Glu. The researchers hypothesized that this polymorphism led to disequilibrium of the pro-inflammatory cytokines and, therefore, accelerated inflammation (37).
Another SNP associated with IL6 was described in a 2010 study by Eskola et al. (13). They identified SNP rs1800796, a 572G/C substitution, which was significantly associated with IVDD in Danish girls. The study found that female patients carrying the C allele were at a 6.71-fold increased risk of developing IVDD than those without the allele. This study did not find the same association in Danish boys (13). However, a 2012 study by Eskola et al. described two different SNPs of IL6 that were found only in adolescent boys: rs1800797 and rs1800795. The G/A genotype (risk allele, G) of SNP rs1800797 was associated with a 0.27-fold decreased risk of developing IVDD, whereas the G/C genotype (risk allele, G) of SNP rs1800795 was associated with a 0.26-fold decreased risk of IVDD. Both polymorphisms were protective and potentially reduced the inflammatory tone of IL6 (36). Overall, the research on IL6 suggests that various polymorphisms may influence a patient’s genetic risk of IVDD; however, this effect may be limited to certain genders or populations.
Matrix-Degrading Enzymes
Several types of matrix-degrading enzymes exist within the ECM of IVDs. Two of the major types of matrix-degrading enzymes that are involved in IVD degradation are MMPs and “a disintegrin and metalloproteinase with thrombospondin motif” (ADAMTS). The homeostasis of ongoing ECM turnover is managed by the balance between MMPs and tissue inhibitors of metalloproteinases (12). Various MMPs are responsible for degrading different substances. For example, collagen I, II, and III are primarily degraded by MMP-1, -8, and -13 – the collagenases, whereas denatured collagen is the target of MMP-2 and MMP-9 (59). It is important to remember that increased expression of MMPs leads to accelerated destruction of the ECM. ADAMTS are also referred to as aggrecanases because their primary function within the IVDs is to digest aggrecan (34). Similarly, an increase in expression of ADAMTS results in accelerated IVDD.
Matrix Metalloproteinase
Song et al. examined 691 southern Chinese people between the ages of 18 and 55 years and found an SNP at position -1607 in the promoter of the matrix metalloproteinase 1 gene (MMP-1) (58). The SNP was significantly associated with IVDD, and of the two alleles, D and G, the D allele was the risk allele. The statistical analysis revealed that patients carrying the D allele had a 1.41-fold increased risk of IVDD compared with those without the allele. Further analysis showed an even stronger connection in patients over the age of 40 years. In patients over the age of 40 years carrying the D allele, there was a 1.445-fold increased risk of developing IVDD. This study was particularly interesting because previous studies have shown the G allele of the -1607 SNP as increasing MMP-1 expression. The researchers hypothesized that expression of the D allele might lead to disequilibrium between the MMPs, and thus, greater degradation of the AF and NP (58).
MMP-2, one of the two gelatinases, tends to target denatured collagen as its substrate (59). Dong et al. found that the -1306C/T polymorphism of the MMP2 gene was a genetic risk factor for IVDD (38). The study examined 162 Chinese young adults with disc degeneration. The statistical analysis demonstrated that patients with the CC genotype had a 3.08-fold increased risk of developing IVDD than those with at least one T allele (CT or TT). The study also found that the CC genotype was associated with more severe forms of IVDD than the CT and TT genotypes. This study was exceptionally interesting because the SNP is a C to T substitution, where the T allele is the risk allele, and the C allele is the wild-type. The T allele reduces Sp1 transcription factor binding to the gene and thus reduces overall transcription. The polymorphism that leads to increased protein production is the most likely one associated with an increased genetic risk of IVDD; in this case, it happened to be the wild-type C allele (38). A later study in 2013 by Zhang et al. revealed a similar phenomenon in the -735C/T polymorphism of MMP2 (39). The study found that patients with the TT or CT genotypes had a 0.413-fold reduced risk of developing IVDD, whereas patients carrying the CC genotype were at nearly a 2.5-fold increased risk of developing IVDD compared with patients with the TT genotype. Similar to the -1306C/T SNP, the T allele was associated with disrupting a Sp1-binding site (CCACC box) and reducing transcription, while the C allele was considered the wild-type and was associated with increased transcription (39). These studies reveal that multiple nearby Sp1-binding sites whose polymorphisms are connected to genetic risk of IVDD exist (38, 39).
MMP-9 is also a gelatinase with variable expression that has been linked to IVDD. A 2009 study by Sun et al. revealed a -1562C/T polymorphism that affected the protein expression of MMP-9 (11). Patients with the CT/TT genotypes were at a 2.14-fold increased risk of developing IVDD compared with patients with the CC genotype. The T allele is associated with increased MMP-9 expression, and thus an imbalance between MMPs and tissue inhibitors of metalloproteinases, leading to excessive degradation of the ECM (11).
MMP-3 is one of the three MMPs that are categorized as stromelysins (59). One of the main functions of stromelysins is to degrade proteoglycans, laminas, and other components of the IVD ECM as well as indirectly degrade the disc through activating other MMPs (40). Expression of MMP-3 has also been shown to rise in response to inflammation (51).
The most commonly studied SNP of MMP3 is the 5A variant allele in the promoter region of the gene. A 2001 study by Takahashi et al. revealed that elderly patients who had the 5A/5A or 5A/6A genotype were at an increased risk of IVDD (40). However, the study did not find this association in younger patients. Yuan et al. investigated the same 5A polymorphism and found that patients who carried the shorter 5A allele were at a 1.96-fold increased risk of developing IVDD (12). More recently, Zawilla et al. found that the 5A allele was associated with a 2.5-fold greater risk of developing IVDD (51). The study also found a link between the 5A allele and increased severity of degradation. An abundance of evidence suggests that the shorter 5A polymorphism of MMP3 is linked to an increased genetic risk of IVDD (51).
MMP-14 is a membrane-anchored MMP that is found at the cell surface and is involved in degrading small fragments of collagen and activating MMP2 (41, 59). Researchers have hypothesized that overexpression of MMP-14 leads to overall disc degradation mainly through the activation of MMP2 (41). In a 2015 study by Zhang, the -378T/C SNP in MMP14 was a genetic risk factor associated with IVDD (41). Patients with the TT genotype had a 1.59-fold increased risk of IVDD compared with patients with the CC genotype.
Considering all the various SNPs associated with MMPs and their influence on patients’ risk of developing IVDD, protein expression levels are a delicate and important aspect of ECM maintenance of IVDs. It is possible that genetic manipulation of MMPs is a significant factor in the etiology behind IVDD. Furthermore, MMPs are strong candidates for therapeutic options for mitigating or reversing IVD degradation.
A Disintegrin and Metalloproteinase with Thrombospondin Motif
A disintegrin and metalloproteinase with thrombospondin motif are enzymes that play a central role in disc degeneration via aggrecan turnover (42). In particular, ADAMTS-4 and ADAMTS-5 are found at the site of disc degeneration. Various genetic polymorphisms in the ADAMTS family of genes are linked to the risk of IVDD. Liu et al. were the first to investigate a polymorphism in ADAMTS4 (42). They found that SNP rs4233367, an 1877C/T substitution, was associated with a reduced risk of IVDD. Patients with the TT genotype were at a 0.21-fold reduced risk of developing IVDD compared with those with the CC genotype. This strong connection suggests that ADAMTS4 plays an important part in proteoglycan degradation within the IVD. Rajasekaran et al. investigated SNP rs162509 in ADAMTS5 and found that the risk allele was associated with a 1.281-fold increased risk of developing IVDD (34). Although this relationship is small, it supports the notion that ADAMTS proteins are essential for the maintenance of healthy, hydrated discs.
Aggrecan
Aggrecan is the most plentiful proteoglycan found within the IVD, and its primary function is to retain water. The core protein of aggrecan contains a large number of chondroitin sulfate and keratin sulfate chains that facilitate its ability to create an osmotic gradient. Furthermore, aggrecan binds to negatively charged glycosaminoglycans to increase the hydrostatic pressure of the NP (5, 7). One of the most investigated polymorphisms of aggrecan is the variable nucleotide tandem repeat (VNTR) in the chondroitin sulfate-1 encoding domain of the aggrecan gene (ACAN) (45). The chondroitin sulfate encoding allele has VNTRs ranging from 13 to 33 nucleotides, with the most common number being 26, 27, or 28 repeats. As aggrecan water-retention abilities are heavily reliant on the number and size of chondroitin sulfate chains, it makes sense that a reduced number of repeats would impair the ability of aggrecan to retain water (24, 43–46). One of the earliest studies published on this topic was in 1999 by Kawaguchi et al. (43). The study found patients with 18 or 21 repeats in the chondroitin sulfate encoding domain were at an increased risk of multilevel disc degeneration as well as more severe forms of degeneration when compared with patients with longer alleles. A 2010 study by Eser et al. supported these results (24). They found that patients with short alleles, consisting of VNTRs of A13 to A26, were at an increased risk of severe disc degeneration compared with those with longer VNTRs in their alleles. The study also found that patients with short, A13 to A26, or normal, A27, were at an increased risk of multilevel disc degeneration. These findings were further supported by a 2012 study by Xu et al. who found that patients with less than 23 VNTRs were at a 1.95-fold increased risk of IVDD compared with those with more than 23 repeats (44). The study also found that patients with less than 25 repeats were at a 1.85-fold increased risk of IVDD compared with those with more than 25 repeats. This study helped establish the dose-dependent nature of the VNTRs of the aggrecan gene. The risk associated with VNTRs seems to follow a continuous scale, as opposed to a Boolean, or “cut-off” pattern (44). A 2013 meta-analysis by Gu et al. revealed that patients with shorter alleles, A13 to A25, were at a 1.54-fold increased risk of IVDD compared with those with either normal, A26 to A27, or longer alleles, A28 to A32. This relationship was found to be even stronger in patients of Asian descent, who were at a 1.65-fold increased risk of IVDD (45). This study helped solidify the notion that shorter VNTRs are not only associated with increased risk of IVDD but also suggest that the magnitude of the effect may be associated with race.
In 2007, Solovieva et al. investigated the VNTRs for the aggrecan gene in 132 middle-aged Finnish men (46). Their analysis found that the A26 allele was associated with an increased risk of the patient’s NP to be dark on an MRI scan, which is an indication of IVDD. The study also found that patients with A26/A26 genotype were at a 2.77-fold increased risk of a dark NP compared with patients who had longer or shorter VNTRs. This study is unique and did not follow the same trends as the previously mentioned studies. In previous studies, A26 was either considered within normal/typical range or even long (24, 43–45). This 2007 study helped support the notion that the effects of VNTRs may also be influenced by the race or ethnicity of the patient.
Various other genes that affect the aggrecan water-retention abilities or aggrecan expression have also been investigated. Carbohydrate sulfotransferase 3 (CHST-3) is an enzyme that is involved in sulfation of the aggrecan side-chains and is coded by CHST3. This function makes CHST-3 an important and indirect contributor to disc hydration. Song et al. identified the SNP rs4148941 that produced the risk allele A (47). They found that the allele A variant of CHST3 had improved binding with micro RNA sequence miR-513a-5p. Their statistical analysis showed that patients with the AA or AC genotypes were at a 1.48-fold increased risk of developing IVDD. Further analysis revealed that the A allele was associated with reduced expression of the CHST3 messenger RNA within the IVDs, suggesting reduced expression of CHST-3 protein (47). Overall, the study established the SNP of CHST3 as a genetic risk factor for IVDD.
Vitamin D Receptor
The VDR is a nuclear receptor for a vitamin D metabolite, 1a,25-dihydroxyvitamin D3 (Figure 3). Previous studies have shown that VDR polymorphisms are associated with various bone disorders including osteoarthritis, osteoporosis, and cardiovascular disease (7, 44). VDR function in IVDs is hypothesized to be through an indirect pathway for chondrocyte proliferation and the effect of chondrocytes on proteoglycans (12). Over the past two decades, various polymorphisms affecting the expression and function of VDRs in IVDD have been identified. These SNPs include FokI (rs2228570), TaqI (rs731236), and ApaI (rs7975232) (12, 20, 24, 44, 48, 49, 51, 61, 62).
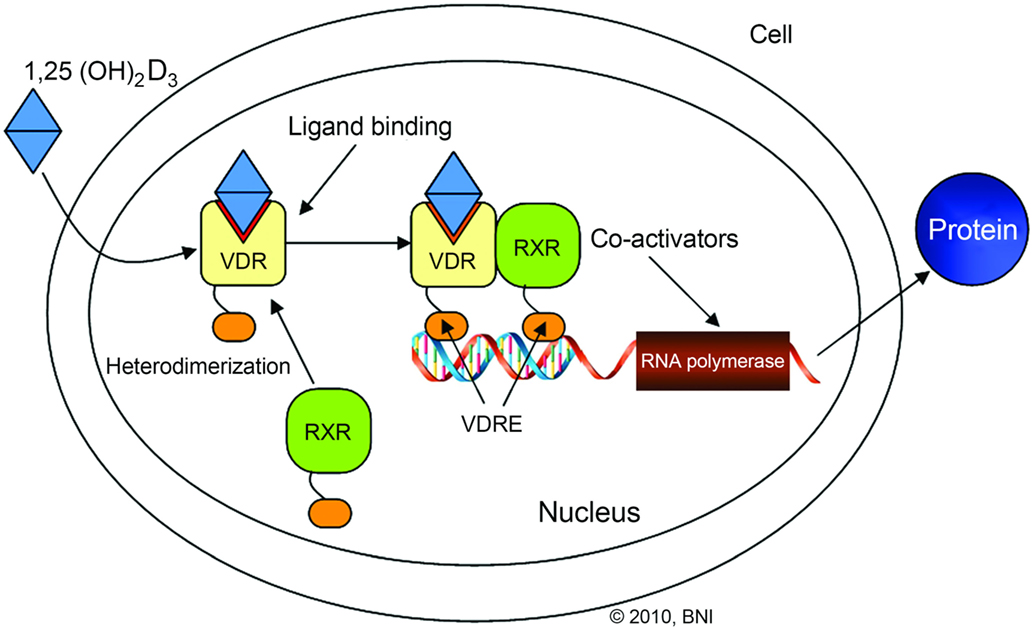
Figure 3. Illustration outlining the vitamin D receptor (VDR) signaling pathway. VDR/retinoid X receptor (RXR) interaction with vitamin D response element (VDRE) stimulates gene transcription. Used with permission from Barrow Neurological Institute, Phoenix, AZ, USA.
The FokI polymorphism of VDR is a C to T substitution found in exon 2 (49). This SNP leads to altered protein size, and subsequently, altered function. Research has shown that the shorter polypeptide of VDR is associated with the wild-type C variant. The F allele has a higher affinity for transcription factor II B. The wild-type alleles lead to normal functioning VDR, while the T substitution (risk allele f) is associated with reduced function (62). A 1998 study by Videman et al. of Finnish twins found that the ff genotype was associated with 9.3% reduced signal intensity within the T6–S1 region on an MRI compared with the FF genotype (48). They also found that the Ff genotype was associated with 4.3% reduced signal intensity within the same region. These results were supported by a 2010 study by Eser who found that the FF genotype was associated with milder grades of degradation (grades I and II), whereas the ff genotype was associated with more severe grades (grades III and IV) (24). The FokI SNP was not only associated with an increased severity of IVDD but also increased the risk of developing IVDD. Vieira et al. found that the T allele was associated with a 1.58-fold increased risk of developing IVDD compared with the C allele (49). These results were further supported by a recent 2016 study by Zhao et al. They found that Hispanic patients with the ff or Ff genotype (TT or TC alleles) were at a 1.742-fold higher risk of developing IVDD, whereas Asian patients with similar genotypes had a 1.293-fold increased risk (62). The data on the FokI SNP suggest that it is a genetic risk factor not only for IVDD but also for the severity of IVDD. Furthermore, these data suggest that the FokI polymorphism manifests differently in patients based on race or ethnicity.
Another significant SNP of VDR that has been the target of the most investigation among VDR polymorphisms is the TaqI variant (44). Interestingly, TaqI is a silent mutation in exon 9 of the VDR gene, yet it has a profound effect on a patient’s genetic risk of developing IVDD (50). One of the earliest studies of the TaqI polymorphism was the 1998 study by Videman et al. (48). They found that the patients with the tt genotype displayed 12.6% reduced signal intensities in the T6–12 range on MRI compared with patients with the TT genotype. These findings were supported by a 2002 study by Kawaguchi et al. who investigated the incidence of the TaqI SNP in Japanese young adults (50). The study found that patients with the Tt genotype were at an increased risk of multilevel IVDD and more severe forms of degeneration. The study was unable to establish the same connection for the tt genotype because none of the subjects had the tt genotype. In 2010, Eser et al. found that patients with the TT genotype displayed significantly milder forms of IVDD than patients with the tt genotype (24). A study in 2015 by Toktas et al. supported the association of the TaqI SNP with increased severity of disc degeneration. They found that patients with the homozygous tt genotype had an average Pfirrmann score of 18.45, which was significantly lower than in those with wild-type genotypes (average score, 22.15) (20). The findings from these studies suggest that the TaqI SNP of VDR is associated with both increased risk of developing IVDD and severity of IVDD.
Another common polymorphism of VDR that has received much attention is the ApaI SNP. The ApaI SNP maps to intron 8 of VDR and is associated with increased risk of IVDD (50). Yuan et al. found that the risk allele of the ApaI SNP was associated with a 1.70-fold increased risk of developing IVDD (12). These findings are supported by a 2013 study by Zawilla et al. who found that the mutant T allele of VDR was associated with a 3.1-fold increased risk of developing IVDD (51). They also found that the mutant T allele was significantly associated with increased severity of IVDD. Although the ApaI polymorphism is associated with both severity and risk of developing IVDD, the exact mechanism and its impact on the VDR protein has not been thoroughly investigated (12, 50, 51). Despite this, ApaI is a well-established genetic risk factor of IVDD.
Apoptosis
Studies regarding the molecular mechanisms of IVDD have established that degenerated discs display much higher rates of apoptosis, programed cell death (3, 5). Although the exact cascade of molecules involved in apoptosis of IVD cells remains under investigation, there are a few significant genes whose polymorphisms have been associated with increased risk of IVDD. Among these are caspase-9 (CASP9), TNF-related apoptosis-inducing ligand (TRAIL), and death receptor-4, DR4, also known as TRAIL receptor 1 (TRAILR1) (1, 10).
Caspase-9 is an important activator of the intrinsic pathway of apoptosis. Its expression levels within the IVD have been reported to increase during disc degeneration (1). The first study to report on CASP9 polymorphisms and their relationship to IVDD was a 2011 study by Guo et al. (1). The study investigated two SNPs, rs4645978 (-1263A/G) and rs4645981 (-712C/T). They analyzed data from 154 patients with IVDD and found that the mutant GG genotype was associated with a 2.760-fold increase in the risk of IVDD compared with the AA genotype (1). Mu et al. investigated the same polymorphism, -1263A/G, and found that the G allele was associated with a 2.059-fold increase in the risk of developing lower back pain compared with the A allele (52). These studies suggest that SNPs affecting the expression and function of apoptosis factors may be another way in which genetic factors influence the progression of IVDD.
DR4 and DR5 are both receptors that bind to TRAIL and induce apoptosis within the target cell. Recent studies have shown that the TRAIL/DR4/DR5 system is important in mediating apoptosis within IVDs (10). Polymorphisms that influence the function and expression of either TRAIL or DR4 can significantly impact the rate of apoptosis occurring within IVDs. Xu et al. identified two polymorphisms of TRAIL within the 3′-untranslated region, such as 1525A/G and 1595T/C, which are associated with IVDD (53). The mutant GG genotype at the 1525 locus and the mutant CC genotype of the 1595 locus were associated with increased risk of IVDD. The investigation found that both the GG1525 and CC1595 genotypes were associated with reduced TRAIL expression within the cells as well as more severe forms of IVDD (grade IV). Although reduced TRAIL expression has already been established in IVDD, the underlying pathophysiology remains under investigation (53).
The TRAIL/DR4/DR5 system is also affected by polymorphisms in DR4. Tan et al. found that degenerating IVD cells had increased expression of DR4 (25). They investigated a Chinese Han population and found that SNP rs4871857 (626C/G) in exon 4 of DR4 was associated with IVDD. Patients with the mutant G allele were at a 1.958-fold increased risk of developing IVDD. Furthermore, the GG and CG genotypes were associated with more severe grades of IVDD (25). The findings on TRAIL and DR4 revealed another aspect of IVDD that may be controlled by genetic factors.
Growth Factors
Growth differentiation factor 5 (GDF5) is part of the transforming growth factor-β superfamily involved in bone, ligament, and soft tissue development (52, 54). Increased GDF5 expression is linked to increased collagen II and aggrecan production in human IVDs (63, 64). An investigation of polymorphisms in GDF5 revealed that its variable expression and function are linked to osteoarthritis. Williams et al. investigated SNP rs143383 (a T to C substitution at position 104) located within the promoter region of the GDF5 gene. Their analysis showed that the T allele was associated with 1.72-fold increased risk of disc space narrowing and osteophyte production in women (54). These findings are supported by a 2013 study by Mu et al. who investigated the same SNP (52). They found that the T allele of GDF5 was associated with a 2.115-fold increased risk of lower back pain. Although the study revealed an association between the T allele and lower back pain, the findings still suggest the involvement of GDF5 polymorphisms in IVDD.
Similar studies have investigated the influence of vascular endothelial growth factor (VEGF) gene polymorphisms and their link to IVDD (55). IVDs are some of the largest avascular structures within the human body. Consequently, they rely on small capillaries extending from the lumbar artery to help remove metabolic waste (5). One of the main features of a severely degenerated disc is neovascularization penetrating the AF, hence, the interest in VEGF, a key mediator of angiogenesis (55). Han et al. found that when a patient possessed multiple VEGF SNPs, there was a significant association with IVDD (55). For example, a patient with the genotype of -2578CA or AA, combined with -634CC genotype, was at a 21-fold increased risk of IVDD. With limited data, it is difficult to conclude with certainty that VEGF SNPs are associated with IVDD; however, Han et al. (55) have helped establish the preliminary data to warrant further investigation into VEGF polymorphisms.
Ubiquitin-Mediated Degradation
E3 ubiquitin-protein ligase is a multiprotein complex that functions in an ubiquitin–proteasome pathway, marking proteins for degradation. A key protein in this complex named Parkin is expressed in various organs and skeletal muscles. Parkin is coded by PARK2, which was recently associated with IVDD (8, 56). In a 2013 study of 4600 Northern Europeans, Williams et al. reported that the rs926849 SNP is a T to C substitution found within an intron of PARK2 (56). Their statistical analysis revealed that the C allele was significantly associated with reduced risk of IVDD, suggesting that the C allele was protective. The underlying mechanism of how the C allele influences the expression of PARK2 and the subsequent pathology remains under investigation (56). Nonetheless, this study adds another component to the etiology of IVDD as well as highlighting the complexity and continued discoveries associated with IVDD.
Cyclooxygenase
Cyclooxygenase 2 is an essential enzyme that is involved in the production of various prostaglandins and thromboxanes. The cyclooxygenase 2 gene COX2 and its products participate in multiple pathways including inflammation and pain (8, 34, 65). In 2015, Rajasekaran et al. identified two SNPs, such as rs5277 and rs5275, in COX2 that are significantly associated with severe IVDD (34).
Catechol-O-Methyltransferase
Catechol-O-methyltransferase is an enzyme that is involved in the degradation and processing of catechol neurotransmitters such as dopamine. Previous clinical studies showed a relationship between certain polymorphisms in the catechol-O-methyltransferase (COMT) gene and pain. The IVDD researchers believed that variable catechol-O-methyltransferase expression led to increased pain in IVDD. Gruber et al. identified four COMT SNPs, such as rs4633, rs165656, rs2095019, and rs4708592, significantly associated with IVDD (57). Their findings supported results that were previously published regarding the association of rs4633 and IVDD. Although rs165656 has previously been associated with mental retardation, Gruber et al. were the first to show its significant association with IVDD (57). The rs2095019 and rs4708592 polymorphs are novel SNPs that have not been reported previously (57). The study is a strong indicator of the complexity of the acute and chronic changes that occur with IVDD as well as highlighting the ongoing research that has revealed new aspects of its etiology.
Personalized Medicine
The ultimate goal in reviewing the medical literature about the genetic polymorphisms associated with IVDD is to provide patients with personalized and targeted therapeutics. When a patient enters a clinic with lower back pain and degenerative disc disease is suspected, an MRI can provide a conclusive diagnosis. To provide targeted treatment for the specific patient, the physician must understand the patient’s unique molecular profile. Through gene sequencing and screening for SNPs, physicians can obtain a better understanding of the imbalances that led to the patient’s disc degeneration. Some patients may primarily have imbalances with ECM degrading enzymes, whereas others may have overexpression of proapoptotic factors. With this information, unique to each patient, specific therapies can be selected to provide the best long-term outcome.
Conclusion
Despite continued research, the etiology and pathophysiology underlying IVDD remain poorly understood (34). Nonetheless, a significant shift in the understanding of IVDD has occurred over the past two decades, and we now understand that roughly 75% of the etiology behind IVDD is genetic (2, 6). One of the crucial techniques that have helped researchers to realize this understanding is the advent of large-scale DNA arrays and computational analysis software to analyze polymorphisms quickly (34). These techniques have helped bring to light new proteins and associations within systems that were previously thought not to be linked to IVDD. With a better understanding of the pathophysiology of IVDD and improved technology for scanning entire genomes for SNPs than in the past, we expect to produce innovative, new therapeutic approaches.
Two important aspects of genetic polymorphisms that have come to light are variations in race and ethnicity. Some polymorphisms tend to have stronger, or even no effect, on certain races. For example, Hispanics with the FokI SNP of VDR were at a much higher risk of IVDD than their Asian counterparts. Furthermore, the same meta-analysis found that the FokI SNP was not associated with IVDD in people of Caucasian decent (62). These racial variations add a new aspect and complexity to the understanding of the genetic factors underlying IVDD.
With more than 20 unique polymorphisms associated with IVDD, the molecular changes in the associated proteins or pathology of the disc are not yet fully understood. In the coming years, research targeted toward fully understanding the protein changes due to the already identified SNPs is crucial. If we can fully understand the molecular changes involved in IVDD then creating targeted therapeutics based on genetic profiling becomes a possibility.
With improved understanding of the genetic variants associated with IVDD, and rapid genomic analysis available through next-generation genotype sequencing, the possibility of providing effective personalized medicine can become a reality (14, 15). This comprehensive literature review regarding the genetic variants associated with IVDD not only affords a better understanding of the molecular mechanisms behind IVDD but also allows physicians the possibility of providing targeted treatments. For instance, if an IVDD patient were identified to have genetic variants resulting in the overexpression of apoptotic factors, physicians would be able to refine their therapy and target the specific underlying IVDD-causing mechanism unique to that patient. Furthermore, genomic screening for the known variants associated with IVDD can help predict disease progression and severity. This knowledge can help provide more effective treatments personalized to the unique phenotypic presentation of the patient. Considering the majority of the etiology underlying IVDD is genetic, it is essential that researchers and clinicians have a keen understanding of this underlying etiology to optimize treatment (2). Consequently, DNA screening for the genetic variants explaining the pathophysiology of the patient’s IVDD should be the standard of care.
Author Contributions
All authors made substantial contributions to the conception or design of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Neuroscience Publications staff of Barrow Neurological Institute for assistance in preparing this manuscript.
Funding
Research was funded through the Russian Science Foundation grant (Project 15-15-30037).
Abbreviations
ADAMTS, a disintegrin and metalloproteinase with thrombospondin motif; AF, annulus fibrosis; ECM, extracellular matrix; GDF5, growth differentiation factor 5; IL, interleukin; IVD, intervertebral disc; IVDD, intervertebral disc degeneration; MMP, matrix metalloproteinase; MRI, magnetic resonance imaging; NP, nucleus pulposus; SNP, single-nucleotide polymorphism; VEGF, vascular endothelial growth factor; VNTR, variable nucleotide tandem repeat.
References
1. Guo TM, Liu M, Zhang YG, Guo WT, Wu SX. Association between caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res (2011) 52(2):133–8. doi:10.3109/03008207.2010.487621
2. Seki S, Kawaguchi Y, Chiba K, Mikami Y, Kizawa H, Oya T, et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat Genet (2005) 37(6):607–12. doi:10.1038/ng1557
3. Kadow T, Sowa G, Vo N, Kang JD. Molecular basis of intervertebral disc degeneration and herniations: what are the important translational questions? Clin Orthop Relat Res (2015) 473(6):1903–12. doi:10.1007/s11999-014-3774-8
4. Videman T, Saarela J, Kaprio J, Nakki A, Levalahti E, Gill K, et al. Associations of 25 structural, degradative, and inflammatory candidate genes with lumbar disc desiccation, bulging, and height narrowing. Arthritis Rheum (2009) 60(2):470–81. doi:10.1002/art.24268
5. Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG. The molecular basis of intervertebral disc degeneration. Spine J (2013) 13(3):318–30. doi:10.1016/j.spinee.2012.12.003
6. Janeczko L, Janeczko M, Chrzanowski R, Zielinski G. The role of polymorphisms of genes encoding collagen IX and XI in lumbar disc disease. Neurol Neurochir Pol (2014) 48(1):60–2. doi:10.1016/j.pjnns.2013.04.001
7. Kalb S, Martirosyan NL, Kalani MY, Broc GG, Theodore N. Genetics of the degenerated intervertebral disc. World Neurosurg (2012) 77(3–4):491–501. doi:10.1016/j.wneu.2011.07.014
8. Mayer JE, Iatridis JC, Chan D, Qureshi SA, Gottesman O, Hecht AC. Genetic polymorphisms associated with intervertebral disc degeneration. Spine J (2013) 13(3):299–317. doi:10.1016/j.spinee.2013.01.041
9. Sun Z, Yin Z, Liu C, Liang H, Jiang M, Tian J. IL-1beta promotes ADAMTS enzyme-mediated aggrecan degradation through NF-kappaB in human intervertebral disc. J Orthop Surg Res (2015) 10:159. doi:10.1186/s13018-015-0296-3
10. Bertram H, Nerlich A, Omlor G, Geiger F, Zimmermann G, Fellenberg J. Expression of TRAIL and the death receptors DR4 and DR5 correlates with progression of degeneration in human intervertebral disks. Mod Pathol (2009) 22(7):895–905. doi:10.1038/modpathol.2009.39
11. Sun ZM, Miao L, Zhang YG, Ming L. Association between the -1562 C/T polymorphism of matrix metalloproteinase-9 gene and lumbar disc disease in the young adult population in North China. Connect Tissue Res (2009) 50(3):181–5. doi:10.1080/03008200802585630
12. Yuan HY, Tang Y, Liang YX, Lei L, Xiao GB, Wang S, et al. Matrix metalloproteinase-3 and vitamin d receptor genetic polymorphisms, and their interactions with occupational exposure in lumbar disc degeneration. J Occup Health (2010) 52(1):23–30. doi:10.1539/joh.L8149
13. Eskola PJ, Kjaer P, Daavittila IM, Solovieva S, Okuloff A, Sorensen JS, et al. Genetic risk factors of disc degeneration among 12-14-year-old Danish children: a population study. Int J Mol Epidemiol Genet (2010) 1(2):158–65.
14. Cornetta K, Brown CG. Balancing personalized medicine and personalized care. Acad Med (2013) 88(3):309–13. doi:10.1097/ACM.0b013e3182806345
15. Chan IS, Ginsburg GS. Personalized medicine: progress and promise. Annu Rev Genomics Hum Genet (2011) 12:217–44. doi:10.1146/annurev-genom-082410-101446
16. Takegami K, An HS, Kumano F, Chiba K, Thonar EJ, Singh K, et al. Osteogenic protein-1 is most effective in stimulating nucleus pulposus and annulus fibrosus cells to repair their matrix after chondroitinase ABC-induced in vitro chemonucleolysis. Spine J (2005) 5(3):231–8. doi:10.1016/j.spinee.2004.11.001
17. Chujo T, An HS, Akeda K, Miyamoto K, Muehleman C, Attawia M, et al. Effects of growth differentiation factor-5 on the intervertebral disc – in vitro bovine study and in vivo rabbit disc degeneration model study. Spine (2006) 31(25):2909–17. doi:10.1097/01.brs.0000248428.22823.86
18. Bae WC, Masuda K. Emerging technologies for molecular therapy for intervertebral disk degeneration. Orthop Clin North Am (2011) 42(4):585–601,ix. doi:10.1016/j.ocl.2011.07.004
19. Fontana G, See E, Pandit A. Current trends in biologics delivery to restore intervertebral disc anabolism. Adv Drug Deliv Rev (2015) 84:146–58. doi:10.1016/j.addr.2014.08.008
20. Toktas ZO, Eksi MS, Yilmaz B, Demir MK, Ozgen S, Kilic T, et al. Association of collagen I, IX and vitamin D receptor gene polymorphisms with radiological severity of intervertebral disc degeneration in Southern European Ancestor. Eur Spine J (2015) 24(11):2432–41. doi:10.1007/s00586-015-4206-5
21. Zhang Y, An HS, Thonar EJ, Chubinskaya S, He TC, Phillips FM. Comparative effects of bone morphogenetic proteins and sox9 overexpression on extracellular matrix metabolism of bovine nucleus pulposus cells. Spine (2006) 31(19):2173–9. doi:10.1097/01.brs.0000232792.66632.d8
22. Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (2001) 26(17):1873–8. doi:10.1097/00007632-200109010-00011
23. Yu LP, Qian WW, Yin GY, Ren YX, Hu ZY. MRI assessment of lumbar intervertebral disc degeneration with lumbar degenerative disease using the Pfirrmann grading systems. PLoS One (2012) 7(12):e48074. doi:10.1371/journal.pone.0048074
24. Eser B, Cora T, Eser O, Kalkan E, Haktanir A, Erdogan MO, et al. Association of the polymorphisms of vitamin D receptor and aggrecan genes with degenerative disc disease. Genet Test Mol Biomarkers (2010) 14(3):313–7. doi:10.1089/gtmb.2009.0202
25. Tan H, Zhao J, Jiang J, Ren Y. Association of the polymorphism of DR4 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Scand J Clin Lab Invest (2012) 72(7):576–9. doi:10.3109/00365513.2012.713176
26. Schneiderman G, Flannigan B, Kingston S, Thomas J, Dillin WH, Watkins RG. Magnetic resonance imaging in the diagnosis of disc degeneration: correlation with discography. Spine (1987) 12(3):276–81. doi:10.1097/00007632-198704000-00016
27. Pluijm SM, van Essen HW, Bravenboer N, Uitterlinden AG, Smit JH, Pols HA, et al. Collagen type I alpha1 Sp1 polymorphism, osteoporosis, and intervertebral disc degeneration in older men and women. Ann Rheum Dis (2004) 63(1):71–7. doi:10.1136/ard.2002.002287
28. Tilkeridis C, Bei T, Garantziotis S, Stratakis CA. Association of a COL1A1 polymorphism with lumbar disc disease in young military recruits. J Med Genet (2005) 42(7):e44. doi:10.1136/jmg.2005.033225
29. Annunen S, Paassilta P, Lohiniva J, Perala M, Pihlajamaa T, Karppinen J, et al. An allele of COL9A2 associated with intervertebral disc disease. Science (1999) 285(5426):409–12. doi:10.1126/science.285.5426.409
30. Kales SN, Linos A, Chatzis C, Sai Y, Halla M, Nasioulas G, et al. The role of collagen IX tryptophan polymorphisms in symptomatic intervertebral disc disease in Southern European patients. Spine (2004) 29(11):1266–70. doi:10.1097/00007632-200406010-00017
31. Zhang Z, Zhang J, Ding L, Teng X. Meta-analysis of the association between COL9A2 genetic polymorphisms and lumbar disc disease susceptibility. Spine (2014) 39(20):1699–706. doi:10.1097/BRS.0000000000000497
32. Paassilta P, Lohiniva J, Goring HH, Perala M, Raina SS, Karppinen J, et al. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA (2001) 285(14):1843–9. doi:10.1001/jama.285.14.1843
33. Solovieva S, Lohiniva J, Leino-Arjas P, Raininko R, Luoma K, Ala-Kokko L, et al. Intervertebral disc degeneration in relation to the COL9A3 and the IL-1ss gene polymorphisms. Eur Spine J (2006) 15(5):613–9. doi:10.1007/s00586-005-0988-1
34. Rajasekaran S, Kanna RM, Senthil N, Raveendran M, Ranjani V, Cheung KM, et al. Genetic susceptibility of lumbar degenerative disc disease in young Indian adults. Eur Spine J (2015) 24(9):1969–75. doi:10.1007/s00586-014-3687-y
35. Virtanen IM, Karppinen J, Taimela S, Ott J, Barral S, Kaikkonen K, et al. Occupational and genetic risk factors associated with intervertebral disc disease. Spine (2007) 32(10):1129–34. doi:10.1097/01.brs.0000261473.03274.5c
36. Eskola PJ, Kjaer P, Sorensen JS, Okuloff A, Wedderkopp N, Daavittila I, et al. Gender difference in genetic association between IL1A variant and early lumbar disc degeneration: a three-year follow-up. Int J Mol Epidemiol Genet (2012) 3(3):195–204.
37. Noponen-Hietala N, Virtanen I, Karttunen R, Schwenke S, Jakkula E, Li H, et al. Genetic variations in IL6 associate with intervertebral disc disease characterized by sciatica. Pain (2005) 114(1–2):186–94. doi:10.1016/j.pain.2004.12.015
38. Dong DM, Yao M, Liu B, Sun CY, Jiang YQ, Wang YS. Association between the -1306C/T polymorphism of matrix metalloproteinase-2 gene and lumbar disc disease in Chinese young adults. Eur Spine J (2007) 16(11):1958–61. doi:10.1007/s00586-007-0454-3
39. Zhang Y, Gu Z, Qiu G. Association of the polymorphism of MMP2 with the risk and severity of lumbar disc degeneration in the Chinese Han population. Eur Rev Med Pharmacol Sci (2013) 17(13):1830–4.
40. Takahashi M, Haro H, Wakabayashi Y, Kawa-uchi T, Komori H, Shinomiya K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metalloproteinase-3 gene. J Bone Joint Surg Br (2001) 83(4):491–5. doi:10.1302/0301-620X.83B4.11617
41. Zhang J, Sun X, Liu J, Liu J, Shen B, Nie L. The role of matrix metalloproteinase 14 polymorphisms in susceptibility to intervertebral disc degeneration in the Chinese Han population. Arch Med Sci (2015) 11(4):801–6. doi:10.5114/aoms.2015.53301
42. Liu S, Wu N, Liu J, Liu H, Su X, Liu Z, et al. Association between ADAMTS-4 gene polymorphism and lumbar disc degeneration in Chinese Han population. J Orthop Res (2016) 34(5):860–4. doi:10.1002/jor.23081
43. Kawaguchi Y, Osada R, Kanamori M, Ishihara H, Ohmori K, Matsui H, et al. Association between an aggrecan gene polymorphism and lumbar disc degeneration. Spine (1999) 24(23):2456–60. doi:10.1097/00007632-199912010-00006
44. Xu G, Mei Q, Zhou D, Wu J, Han L. Vitamin D receptor gene and aggrecan gene polymorphisms and the risk of intervertebral disc degeneration – a meta-analysis. PLoS One (2012) 7(11):e50243. doi:10.1371/journal.pone.0050243
45. Gu J, Guan F, Guan G, Xu G, Wang X, Zhao W, et al. Aggrecan variable number of tandem repeat polymorphism and lumbar disc degeneration: a meta-analysis. Spine (2013) 38(25):E1600–7. doi:10.1097/BRS.0000000000000012
46. Solovieva S, Noponen N, Mannikko M, Leino-Arjas P, Luoma K, Raininko R, et al. Association between the aggrecan gene variable number of tandem repeats polymorphism and intervertebral disc degeneration. Spine (2007) 32(16):1700–5. doi:10.1097/BRS.0b013e3180b9ed51
47. Song YQ, Karasugi T, Cheung KM, Chiba K, Ho DW, Miyake A, et al. Lumbar disc degeneration is linked to a carbohydrate sulfotransferase 3 variant. J Clin Invest (2013) 123(11):4909–17. doi:10.1172/JCI69277
48. Videman T, Leppavuori J, Kaprio J, Battie MC, Gibbons LE, Peltonen L, et al. Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine (1998) 23(23):2477–85. doi:10.1097/00007632-199812010-00002
49. Vieira LA, De Marchi PL, dos Santos AA, Christofolini DM, Barbosa CP, Fonseca FL, et al. Analysis of FokI polymorphism of vitamin D receptor gene in intervertebral disc degeneration. Genet Test Mol Biomarkers (2014) 18(9):625–9. doi:10.1089/gtmb.2014.0030
50. Kawaguchi Y, Kanamori M, Ishihara H, Ohmori K, Matsui H, Kimura T. The association of lumbar disc disease with vitamin-D receptor gene polymorphism. J Bone Joint Surg Am (2002) 84-A(11):2022–8.
51. Zawilla NH, Darweesh H, Mansour N, Helal S, Taha FM, Awadallah M, et al. Matrix metalloproteinase-3, vitamin D receptor gene polymorphisms, and occupational risk factors in lumbar disc degeneration. J Occup Rehabil (2014) 24(2):370–81. doi:10.1007/s10926-013-9472-7
52. Mu J, Ge W, Zuo X, Chen Y, Huang C. Analysis of association between IL-1beta, CASP-9, and GDF5 variants and low-back pain in Chinese male soldier: clinical article. J Neurosurg Spine (2013) 19(2):243–7. doi:10.3171/2013.4.SPINE12782
53. Xu S, Liang T, Li S. Correlation between polymorphism of TRAIL gene and condition of intervertebral disc degeneration. Med Sci Monit (2015) 21:2282–7. doi:10.12659/MSM.894157
54. Williams FM, Popham M, Hart DJ, de Schepper E, Bierma-Zeinstra S, Hofman A, et al. GDF5 single-nucleotide polymorphism rs143383 is associated with lumbar disc degeneration in Northern European women. Arthritis Rheum (2011) 63(3):708–12. doi:10.1002/art.30169
55. Han IB, Ropper AE, Teng YD, Shin DA, Jeon YJ, Park HM, et al. Association between VEGF and eNOS gene polymorphisms and lumbar disc degeneration in a young Korean population. Genet Mol Res (2013) 12(3):2294–305. doi:10.4238/2013.July.8.10
56. Williams FM, Bansal AT, van Meurs JB, Bell JT, Meulenbelt I, Suri P, et al. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta-analysis of 4600 subjects. Ann Rheum Dis (2013) 72(7):1141–8. doi:10.1136/annrheumdis-2012-201551
57. Gruber HE, Sha W, Brouwer CR, Steuerwald N, Hoelscher GL, Hanley EN Jr. A novel catechol-O-methyltransferase variant associated with human disc degeneration. Int J Med Sci (2014) 11(7):748–53. doi:10.7150/ijms.8770
58. Song YQ, Ho DW, Karppinen J, Kao PY, Fan BJ, Luk KD, et al. Association between promoter -1607 polymorphism of MMP1 and lumbar disc disease in Southern Chinese. BMC Med Genet (2008) 9:38. doi:10.1186/1471-2350-9-38
59. Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol (2011) 3(1):a004978. doi:10.1101/cshperspect.a004978
60. Jin H, van’t Hof RJ, Albagha OM, Ralston SH. Promoter and intron 1 polymorphisms of COL1A1 interact to regulate transcription and susceptibility to osteoporosis. Hum Mol Genet (2009) 18(15):2729–38. doi:10.1093/hmg/ddp205
61. Cervin Serrano S, Gonzalez Villareal D, Aguilar-Medina M, Romero-Navarro JG, Romero Quintana JG, Arambula Meraz E, et al. Genetic polymorphisms of interleukin-1 alpha and the vitamin d receptor in Mexican mestizo patients with intervertebral disc degeneration. Int J Genomics (2014) 2014:302568. doi:10.1155/2014/302568
62. Zhao J, Yang M, Shao J, Bai Y, Li M. Association between VDR FokI polymorphism and intervertebral disk degeneration. Genomics Proteomics Bioinformatics (2015) 13(6):371–6. doi:10.1016/j.gpb.2015.11.003
63. Feng C, Liu H, Yang Y, Huang B, Zhou Y. Growth and differentiation factor-5 contributes to the structural and functional maintenance of the intervertebral disc. Cell Physiol Biochem (2015) 35(1):1–16. doi:10.1159/000369670
64. Belykh E, Giers M, Bardonova L, Theodore N, Preul M, Byvaltsev V. The role of bone morphogenetic proteins 2, 7, and 14 in approaches for intervertebral disk restoration. World Neurosurg (2015) 84(4):871–3. doi:10.1016/j.wneu.2015.08.011
Keywords: back pain, biomarker, degeneration, disc, gene expression, herniation, personalized care, single-nucleotide polymorphism
Citation: Martirosyan NL, Patel AA, Carotenuto A, Kalani MYS, Belykh E, Walker CT, Preul MC and Theodore N (2016) Genetic Alterations in Intervertebral Disc Disease. Front. Surg. 3:59. doi: 10.3389/fsurg.2016.00059
Received: 09 June 2016; Accepted: 13 October 2016;
Published: 21 November 2016
Edited by:
Eberval Figueiredo, Universidade de São Paulo, BrazilReviewed by:
Alberto Di Martino, Università Campus Bio-Medico, ItalyAndrei Fernandes Joaquim, Unicamp, Brazil
Copyright: © 2016 Martirosyan, Patel, Carotenuto, Kalani, Belykh, Walker, Preul and Theodore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas Theodore, bmV1cm9wdWJAZGlnbml0eWhlYWx0aC5vcmc=
 Nikolay L. Martirosyan
Nikolay L. Martirosyan Arpan A. Patel
Arpan A. Patel Alessandro Carotenuto
Alessandro Carotenuto M. Yashar S. Kalani
M. Yashar S. Kalani Evgenii Belykh
Evgenii Belykh Corey T. Walker
Corey T. Walker Mark C. Preul
Mark C. Preul Nicholas Theodore
Nicholas Theodore