
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Stroke, 21 February 2024
Sec. Genetics and Omics of Stroke
Volume 3 - 2024 | https://doi.org/10.3389/fstro.2024.1338636
This article is part of the Research TopicDiversity in Stroke Omic(s) and Epidemiology Research: Opportunities and ChallengesView all 6 articles
 Chad M. Aldridge1*
Chad M. Aldridge1* Nicole D. Armstrong2
Nicole D. Armstrong2 N. Abimbola Sunmonu3
N. Abimbola Sunmonu3 Christopher Becker4
Christopher Becker4 Deepak Palakshappa5
Deepak Palakshappa5 Arne G. Lindgren6,7
Arne G. Lindgren6,7 Annie Pedersen8,9
Annie Pedersen8,9 Tara M. Stanne8,9
Tara M. Stanne8,9 Christina Jern8,9
Christina Jern8,9 Jane Maguire10
Jane Maguire10 Fang-Chi Hsu11,12
Fang-Chi Hsu11,12 Keith L. Keene13,14
Keith L. Keene13,14 Michele Sale13,14†
Michele Sale13,14† Marguerite R. Irvin2
Marguerite R. Irvin2 Bradford B. Worrall1,14
Bradford B. Worrall1,14Introduction: Stroke is a leading cause of death and disability worldwide. Recurrent strokes are seven times more lethal than initial ones, with 54% leading to long-term disability. Substantial recurrent stroke risk disparities exist among ancestral groups. Notably, Africans face double the risk and higher fatality rates compared to Europeans. Although genetic studies, particularly GWAS, hold promise for uncovering biological insights into recurrent stroke, they remain underexplored. Our study addresses this gap through meta-analyses of recurrent stroke GWAS, considering specific ancestral groups and a combined approach.
Methods: We utilized four independent study cohorts for African, European, and Combined ancestry recurrent stroke GWAS with genotyping, imputation, and strict quality control. We harmonized recurrent stroke phenotype and effect allele estimates across cohorts. The logistic regression GWAS model was adjusted for age, sex, and principal components. We assessed how well genetic risk of stroke informs recurrent stroke risk using Receiver Operating Characteristic (ROC) curve analysis with the GIGASTROKE Consortium's polygenic risk scores (PRS).
Results: Harmonization included 4,420 participants (818 African ancestry and 3,602 European ancestry) with a recurrent stroke rate of 16.8% [median age 66.9 (59.1, 73.6) years; 56.2% male]. We failed to find genome-wide significant variants (p < 5e−8). However, we found 18 distinct suggestive (p < 5e−6) genetic loci with high biological relevance consistent across African and European ancestries, including PPARGC1B, CCDC3, OPRL1, and MYH11 genes. These genes affect vascular stenosis through constriction and dilation. We also observed an association with SDK1 gene, which has been previous linked with hypertension in Nigerian and Japanese populations). ROC analysis showed poor performance of the ischemic stroke PRS in discriminating recurrent stroke status (area under the curve = 0.48).
Discussion: Our study revealed genetic associations with recurrent stroke not previously associated with incident ischemic stroke. We found suggestive associations in genes previously linked with hypertension. We also determined that knowing the genetic risk of incident stroke does currently not inform recurrent stroke risk. We urgently need more studies to understand better the overlap or lack thereof between incident and recurrent stroke biology.
Stroke is a leading cause of death and disability worldwide, with presentations and mechanisms of action that are quite heterogeneous. In the United States, the majority, nearly 87%, of the ~800,000 strokes that occur each year are ischemic, and a substantial number, roughly 185,000, are recurrent attacks (Benjamin et al., 2019).
Compared to first strokes, recurrent strokes are more deadly and more likely to cause disability. The 30-day case fatality for the first stroke and recurrent stroke is 2 and 14%, respectively (Jong et al., 2004). Likewise, 54% of individuals suffering a recurrent stroke will be disabled (Hankey et al., 2007). In addition to geographical differences in stroke incidence and prevalence, stroke disparities by race-ethnicity are alarming. Stroke has one of the starkest health disparities in the United States. African Americans (AA) endure a nearly 2-fold greater risk of stroke than European Americans (EA; Howard et al., 2016a). AA are also 2–3 times more likely to die from stroke (Howard et al., 2016b). Even among individuals who survive an initial stroke, AA have a much greater risk of stroke recurrence (Sheinart et al., 1998; Park and Ovbiagele, 2016). Considering these facts, it is imperative to investigate the role of genetics in stroke recurrence.
While genome-wide association studies (GWAS) have identified loci and pathways associated with ischemic stroke, such studies on recurrent stroke are limited. It remains unknown what impact genetic ancestry may have on an individual's risk of recurrent stroke. To address these gaps, we conducted a meta-analysis of GWAS data from four independent cohorts to explore general and ancestry-specific genetic contributors to recurrent stroke. Our findings aim to uncover novel genetic variants causally linked to recurrent stroke mechanisms, including indirect mechanisms. Ancestry-specific meta-analyses may provide insights into the genetic factors contributing to recurrent stroke risk disparities between African and European ancestral groups.
This study is a meta-analysis of recurrent stroke genome-wide association study summary statistics from four independent cohorts. Each study included adults aged 18 years or older that had an ischemic stroke that qualified them for study enrollment. The participants were followed until recurrent stroke event or length of the cohort-specific observable period. Each cohort followed participants prospectively. Below, we discuss the cohorts and further detail can be found in Supplementary material 1.
We utilized the following four independent cohorts for this study because of their prospective observation of stroke survivors and their incidence of recurrent stroke events over time: the Australian Stroke Genetics Collaboration (ASGC; Lemmens et al., 2010), the Reasons for Geographic and Racial Differences in Stroke (REGARDS; Howard et al., 2005), the Sahlgrenska Academy Study on Ischemic Stroke (SAHLSIS; Jood et al., 2005), and the Vitamin Intervention for Stroke Intervention clinical trial (VISP; Toole, 2002). Additionally, these cohorts allow the investigation of ancestry-specific genetic associations of recurrent stroke, African and European. The REGARDS and VISP cohorts have notable proportions of African ancestry, while the ASGC and SAHLSIS cohorts are European.
The Australian data comprises 1,230 ischemic stroke cases derived from four acute stroke centers in New South Wales, South Australia and Perth, Western Australia under the banner of the ASGC. These stroke cases comprised European-ancestry stroke patients admitted to clinical centers across Australia. Cases were consecutively recruited from the Neurosciences Department at Gosford Hospital, Gosford, New South Wales (NSW) and the Neurology Department at John Hunter Hospital, Newcastle, NSW between 2003 and 2008. In South Australia, cases were recruited from the Queen Elizabeth Hospital, Adelaide at the same time. For Royal Perth Hospital site, this population-based, longitudinal cohort study sought to determine the age and sex-specific incidence and case fatality of stroke and included residents from the Perth metropolitan area who had a stroke or transient ischemic attack (TIA) between 20 February 1989 and 19 August 1990 (Hankey et al., 2007; Maguire et al., 2011).
The REGARDS study is a population-based, longitudinal cohort comprising 30,239 non-Hispanic Black and White American adults 45 years or older from the 48 contiguous United States and the District of Columbia (Howard et al., 2005). By design, participants were oversampled if they were Black or if they were residents of the stroke belt. The purpose of REGARDS is to investigate the reasons why stroke mortality is higher among Black compared with White adults and residents of the Southeastern US compared with other regions. Enrollment took place in 2003–2007, and participants completed a computer-assisted telephone interview to collect demographic information, as well as an in-home visit for blood pressure measurements and collection of biologic specimens (e.g., blood, urine; Howard et al., 2005).
The SAHLSIS is a hospital-based, longitudinal cohort study, which has been described in detail elsewhere (Jood et al., 2005). In brief, patients with first-ever or recurrent acute ischemic stroke were consecutively recruited at stroke units in Western Sweden. Ischemic stroke cases were aged 18–69 years and were recruited between 1998 and 2011. Ischemic stroke was defined as an episode of focal brain dysfunction with acute onset, lasting >24 h, and of presumed vascular cause with no signs of hemorrhage on neuroimaging. Participants were excluded if further evaluation showed another etiology than stroke.
The VISP trial, a randomized double-blinded trial, has been described previously (Toole, 2002). In summary, the trial investigated the effect of vitamin supplementation dosage on the risk of recurrent stroke. The study enrolled participants with a non-disabling ischemic stroke (mRS ≤ 3) ≥ 72 h before enrollment. Participants (n = 3,680) were randomized to a high-dose or low-dose vitamin supplementation arm and reassessed every 3 months until a recurrent stroke event, but not longer than 2 years. Ten sites were not approved for genetic studies, resulting in a subset of 2,100 genotyped participants. There was no intervention effect between treatment groups, thus we considered participants from each treatment group as the same cohort.
Each cohort underwent genotyping and strict quality control procedures. Supplementary material 1 details the methods by cohort. The genotyping arrays utilized by cohort were the following: ASGC utilized the Illumina HumanHap610-Quad array. REGARDS used the Illumina Infinium Multi-Ethnic AMR/AFR Extended BeadChip arrays. The SAHLSIS performed genotyping on the Illumin Human OmniExpressExome BeadChip version 1.0 or 1.1, while VISP used the Illumina HumanOmni1-Quad-v1 array.
The cohorts had their genotype data referenced to varying genome builds. We utilized the UCSC LiftOver software (Lee et al., 2022) to bring the ASGC, SAHLSIS, and VISP cohorts to the genome assembly GRChr38. We also harmonized the beta coefficients so that the effect allele remained consistent among the cohorts.
Recurrent stroke phenotyping definitions by cohort are described here:
ASGC defined stroke by WHO criteria as a sudden focal neurologic deficit of vascular origin, lasting more than 24 h and confirmed by imaging such as computerized tomography (CT) and/or magnetic resonance imaging (MRI) brain scan. Other investigative tests such as electrocardiogram, carotid doppler and trans-esophageal echocardiogram were conducted to define ischemic stroke mechanism as clinically appropriate. Cases were excluded from participation if aged < 18 years, diagnosed with hemorrhagic stroke or transient ischemic attack rather than ischemic stroke, or were unable to undergo baseline brain imaging. A subset of stroke cases phenotypically identified as not “first ever stroke,” as recurrent and were derived from this data and are included in the current study.
REGARDS included all Black and White participants with available genotyping and informed consent. Individuals were excluded if they self-reported a history of stroke or lacked stroke follow-up information. Recurrent stroke cases are defined as participants with a primary incident ischemic stroke plus a secondary ischemic stroke during REGARDS follow-up.
For the SAHLSIS study to obtain data on non-fatal recurrent strokes, the National Hospital Discharge Registry was used. Sweden has a publicly financed healthcare system that offers healthcare to all citizens at a comparatively low cost, and all hospitals must report discharge diagnoses of all patients to this registry. The registry thus contains almost complete data (99%) on dates and codes for hospital discharge diagnoses and surgical procedures. All stroke diagnoses were confirmed by reviewing the corresponding medical record as described (Pedersen et al., 2016). When the medical record could not be found (in about 15 % of the cases), the event was still registered if it was the main diagnosis. Information on the cause of death was obtained from the Swedish Cause of Death Register, which is based on the International Classification of Diseases 10th Revision (ICD10). The medical records within 6 months before death were reviewed, both for participants who died in the hospital and for participants who died at home as described (Pedersen et al., 2016).
Recurrent stroke in VISP was defined as an incident stroke occurring after trial randomization and before the 2-year trial endpoint, which was previously described in great detail (Toole, 2002). In summary, recurrent stroke was diagnosed only with evidence of sudden onset of focal neurologic deficit lasting at least 24 h accompanied by an increased NIHSS score in a brain area that was previously normal. Determination of recurrent stroke status was done by a local neurologist and two external review committee neurologists.
For consistency, we defined recurrent stroke as an ischemic or hemorrhagic stroke that occurred after the “index” or qualifying ischemic stroke for study enrollment among the cohorts.
We performed African and European ancestry-stratified GWAS in the REGARDS and VISP cohorts due to the large proportions of these ancestries. Next, we meta-analyzed the REGARDS and VISP African ancestry GWAS summary statistics. We repeated this for European ancestry by adding the primarily European ASGC and SAHLSIS cohorts. Lastly, we performed a combined ancestry GWAS, including ASGC, REGARDS, SAHLSIS, and VISP. Each GWAS followed the same logistic regression model with Recurrent Stroke status as the response variable adjusting for age in years, sex, and the first 10 principal components. Because VISP was a clinical trial, we added the treatment arm to its respective models to account for possible confounding effects of vitamin supplementation.
We utilized the “metafor” R package (Viechtbauer, 2010) to apply an inverse-variance weighted meta-analysis regression to the summary statistics from each ancestry type. The custom R script first performed the fixed effects method and calculated the I2-value. If the I2-value exceeded 50%, the script performed the same meta-analysis regression with the “REML” method for mixed-effect and the test parameter “knha.” The “knha” test is the Knapp-Hartung method, which applies an adjustment to the standard errors to account for uncertainty due to heterogeneity in the estimates. Due to the multiple hypothesis tests, we used a Bonferroni Correction set at two thresholds, genome-wide suggestive (p < 5e−6) and significant (p < 5e−8).
We utilized the PANTHER classification system and software PANTHER18.0 (Thomas et al., 2022) to perform GO enrichment analysis for biological pathways in Homo sapiens using Fisher's Exact test and a False Discovery Rate (FDR) adjustment (Mi and Thomas, 2009). We also performed a PANTHER Pathways analysis using the same test and correction parameters (Mi et al., 2019).
Much discussion surrounds the genetic risk of first-ever ischemic risk as a continued causal vector for the recurrent stroke phenotype. To help determine if the recurrent stroke meta-analyses reflect genetically determined ischemic stroke risk, we utilized the GIGAStroke consortium's polygenetic risk scores (PRS) for European ancestry (Mishra et al., 2022). We calculated scores within the entire VISP cohort (n = 2,100) and performed a Receiver Operating Characteristic (ROC) Curve analysis.
All cohorts combined provide 4,420 stroke survivors for analysis with notable heterogeneity in recurrent stroke rates ranging from 8.6 to 29% (Table 1). Age differed similarly, with the SAHLSIS cohort having a median age of 58 compared to 76 years in the AUST cohort.
Figure 1 shows the Manhattan plots of each Ancestry meta-analysis GWAS. The results of the ancestry-specific meta-analyses did not show any genomic inflation among the GWAS meta-analyses (Figure 2).
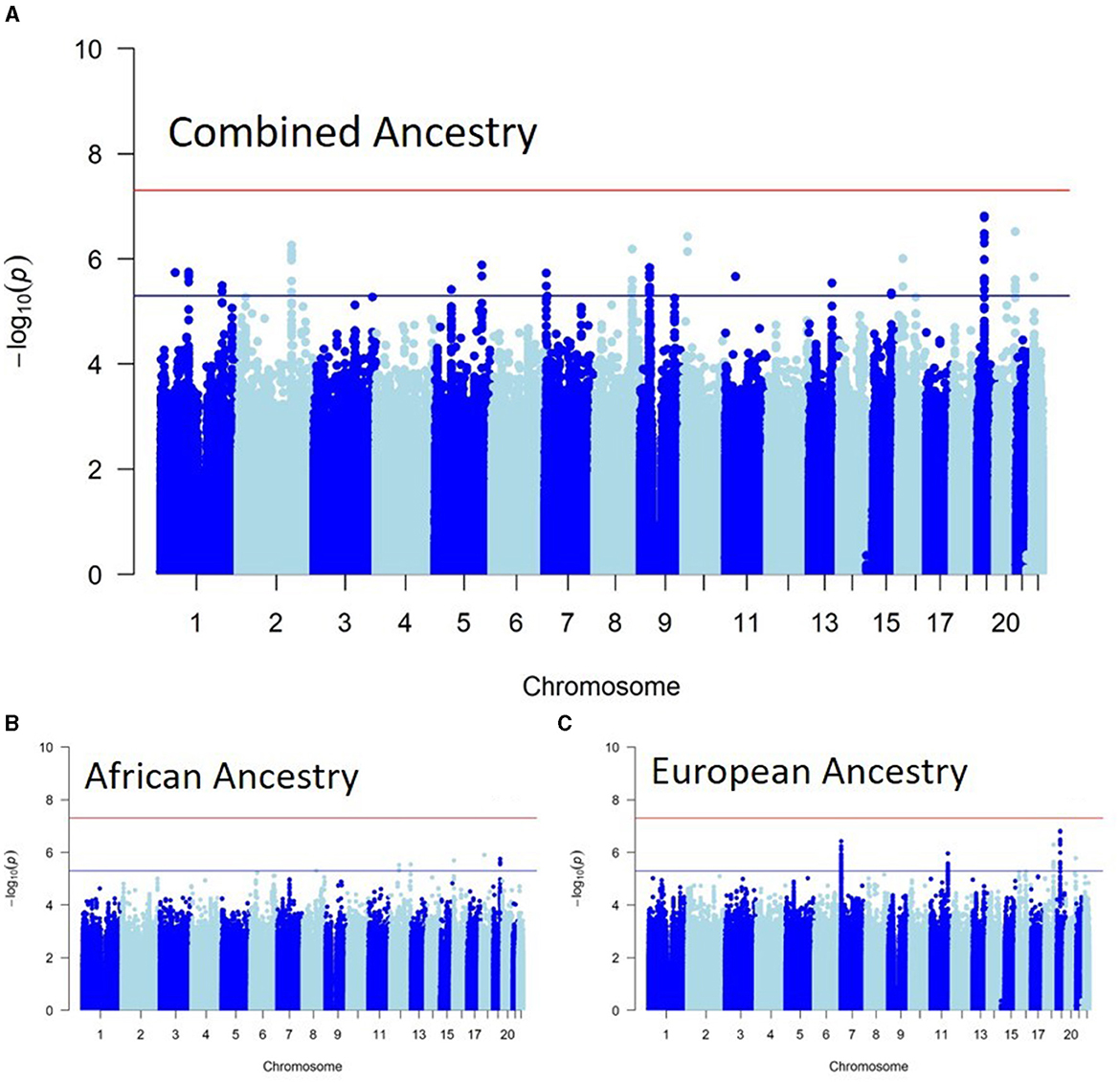
Figure 1. Ancestry-specific genome-wide association study (GWAS) meta-analyses. (A) Shows the genome-wide associations of recurrent stroke in individuals with African ancestry. (B) Shows the genome-wide associations of recurrent stroke in individuals with European ancestry, while (C) is the Combined ancestry GWAS meta-analysis.
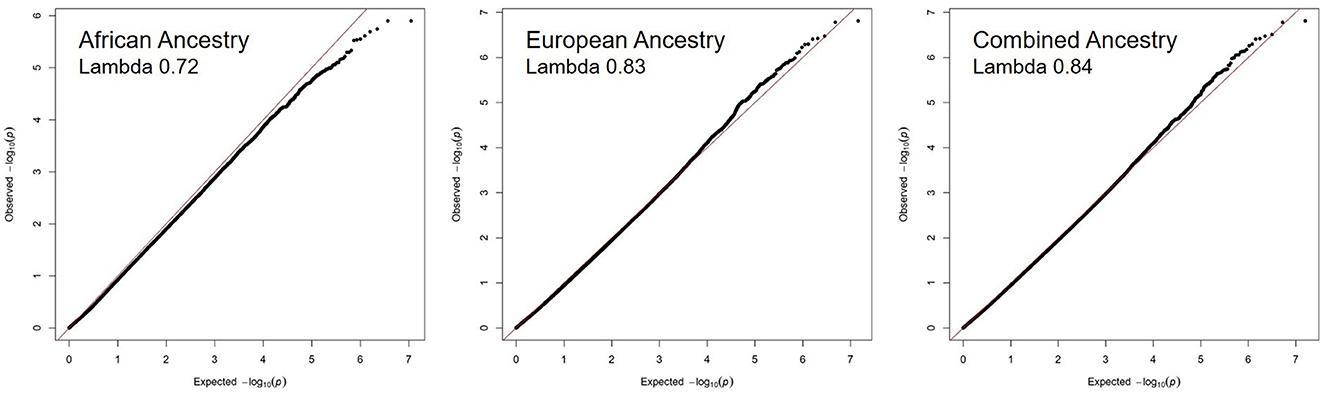
Figure 2. No evidence of genomic inflation of recurrent stroke GWAS meta-analysis across ancestries. The QQ plots for each ancestry reflect the observed –log10 (p-values) of each genetic variant vs. the –log10 (p-values) expected by chance due to multiple hypothesis testing. The lambda value quantifies the amount of inflation in the observed p-values, a value of 1.10 or less is acceptable.
The REGARDS and VISP African ancestry cohorts (n = 818) had 5,574,354 SNPs overlap. None of the genetic variants reached genome-wide significance. Nine reached suggestive significance (p < 5e−6), with some found within the ADGRD1 (protein encoding), PPL (protein encoding), and LINC01915 (long-coding RNA) genes (Supplementary material 2).
The European ancestry meta-analysis consisted of the ASGC, REGARDS, SAHLSIS, and VISP cohorts (n = 3,602), with 6,140,939 SNPs overlapping at least two cohorts. Seventy-one SNPs reached suggestive associations (p < 5e−6) with a strong locus within the SDK1 (protein encoding) gene (Supplementary material 2). There were other associations with the RDX (protein encoding) gene and GAD3P pseudogene.
The combined ancestry meta-analysis consisted of all cohorts (n = 4,420) with African and European ancestries, which resulted in 7,948,212 overlapping SNPs. None of the SNPs reached genome-wide significance (p < 5e−8). However, 70 SNPs reached suggestive (p < 5e−6) associations (Supplementary material 2). Some of the suggestive associations are within the CCDC3, CD59, CTXND1, DSCC1, GPC5, GSTT4, MAL2, MYH11, OPRL1, PELO-AS1, PPARGC1B, RN7SL396P, and SDK1 genes.
We selected all of the genes with intron genetic variants from the combined ancestry meta-analysis with P-values < 5e−6, which were CCDC3, CD59, CTXND1, DSCC1, GPC5, GSTT4, MAL2, MYH11, OPRL1, PELO-AS1, PPARGC1B, RN7SL396P, and SDK1. Both the GO Enrichment and PANTHER Pathway analyses failed to show statistically significant associations after FDR correction.
We applied an ROC analysis on the calculated ischemic stroke PRS from the GIGAStroke consortium which revealed an area under the curve (AUC) of 0.48; Figure 3, suggesting that knowing the genetic risk of first-ever ischemic risk predicts recurrent stroke no better than chance (AUC = 0.5). Because of this result, we investigated if the PRS score was significantly associated with recurrent stroke status as a continuous variable and as quartiles via a Cox proportional hazards model. We set a significant level to p < 0.05. To avoid Simpson's paradox (Wagner, 1982), we added age, sex, and principal components 1–10 as covariates. Neither the continuous form nor the quartile one showed significant associations with recurrent stroke status. To note, there was a 97% overlap between the genetic variants within the GIGAStroke PRS and the VISP genotyped data.
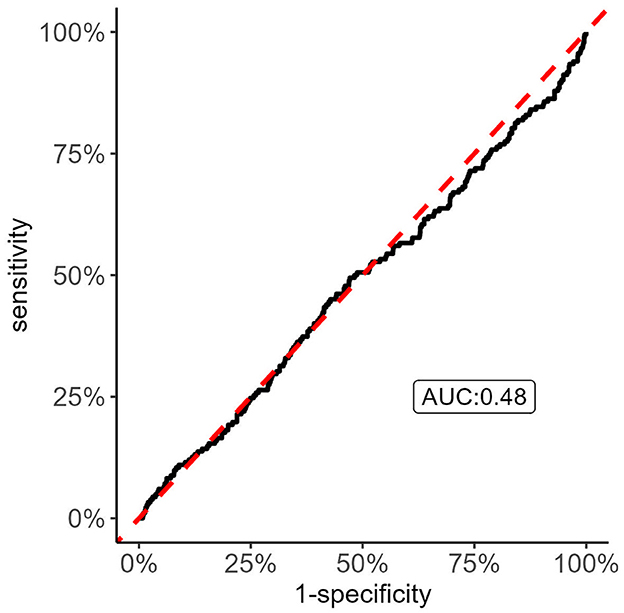
Figure 3. Receiver Operating Characteristic (ROC) curve analysis of recurrent stroke status with GIGASTROKE consortium's polygenic risk scores (PRS) of incident ischemic stroke. The figure shows that the incident ischemic stroke PRS do no better than chance in discriminating recurrent stroke status. Thus, there is the possibility that the genetic risk of recurrent stroke has distinct drivers compared to incident ischemic stroke.
Next, we investigated if any of the 60 genome-wide significant variants from GIGAStroke (Mishra et al., 2022), in their Supplementary Table 4A, were significant in our recurrent stroke meta-analysis results. None of the variants associated with stroke risk in GIGAStroke were associated with recurrent stroke in our study after Bonferroni correction (0.05/60).
Lastly, we investigated the correlation between the GIGAStroke PRS and known stroke risk factors within the VISP cohort (Figure 4). The PRS was most strongly associated with age in years (–R = 0.24; p < 0.001) followed by BMI (R = 0.12; p < 0.001).
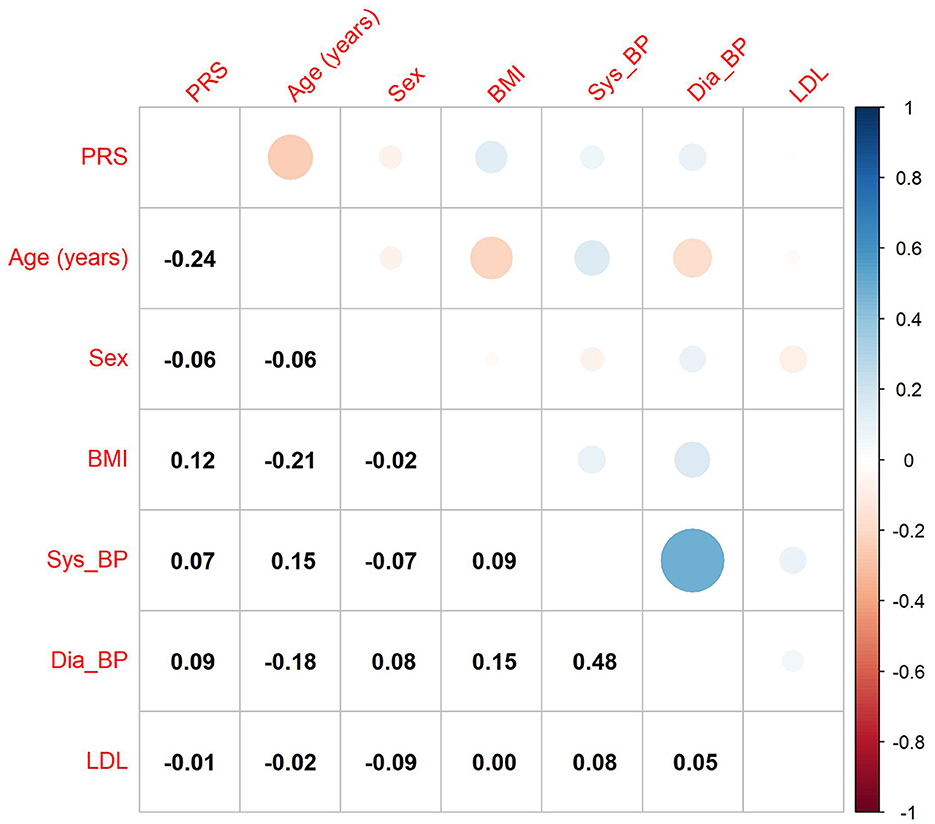
Figure 4. Correlation of GIGAStroke stroke polygenic risk score (PRS) with stroke risk factors in the VISP cohort. The black numbers show Pearson's correlation coefficient among all pairs. The size of the circles and their color reflect the direction and strength of the correlation coefficient, as seen on the upper triangle of the figure. PRS is the polygenic risk score for incident stroke, BMI is the body mass index value, and Sys_BP and Dia_BP are systolic and diastolic blood pressure.
Stroke remains a leading cause of death and long-term disability worldwide. Strikingly, recurrent strokes are seven times more fatal, with high rates of disability after survival (Jong et al., 2004). GWAS offers a strong approach to discovering biological insights associated with recurrent stroke disease. Plus, performing ancestry-specific and combined GWAS meta-analyses provides a window into the genetic determinants of the recurrent stroke risk disparities between African and European ancestral groups. This study builds on prior work that investigated candidate gene associations with recurrent stroke (Fernández-Cadenas et al., 2017).
We did not observe any genome-wide significant associations with recurrent stroke in any ancestry meta-analysis. However, we found 18 suggestive (p < 5e−6) genome-wide associated loci consistent among the African, European, and Combined ancestry meta-analyses (Supplementary material 2; Figure 5). Some appear related to stroke risk factors. For example, the PPARGC1B gene (rs61408734-T locus) is down-regulated in people with type 2 Diabetes Mellitus as well as in pre-diabetes (Patti et al., 2003; Ling et al., 2004). Additionally, the rs36097625-C locus within the CCDC3 gene suggests an indirect link with tumor necrosis factor-alpha (TNF-alpha; Azad et al., 2014), which is a potential marker for stroke and its recovery (Xue et al., 2022). CCDC3 plays an inhibitory role on TNF-alpha pro-inflammatory responses in endothelial cells, including vascular tissue (Azad et al., 2014). Similarly, OPRL1 produces a G protein receptor known as the ORL1, N/OFQ receptor. This receptor receives the endogenous neuropeptide nociceptin which directly causes vasodilation in blood vessels. There is a proposed biological mechanism for cerebral hypoxia, wherein this receptor's down-regulation leads to vasoconstriction and the slowing of cerebrovascular blood flow (Armstead, 2011). Conversely, the MYH11 gene (rs7205185-G) produces a smooth muscle myosin protein, which has been associated with aortic aneurysm/dissections (Zhu et al., 2006) with a theoretical mechanism of action being hyperplasia leading to an occlusive vascular pathology. The DSCC1 gene also has associations with ascending aortic size and dispensability (Benjamins et al., 2022). Lastly, the SDK1 gene has particular interest because of its link with hypertension in two prior GWAS in Nigerians (Tayo et al., 2009) and Japanese (Oguri et al., 2010) populations.
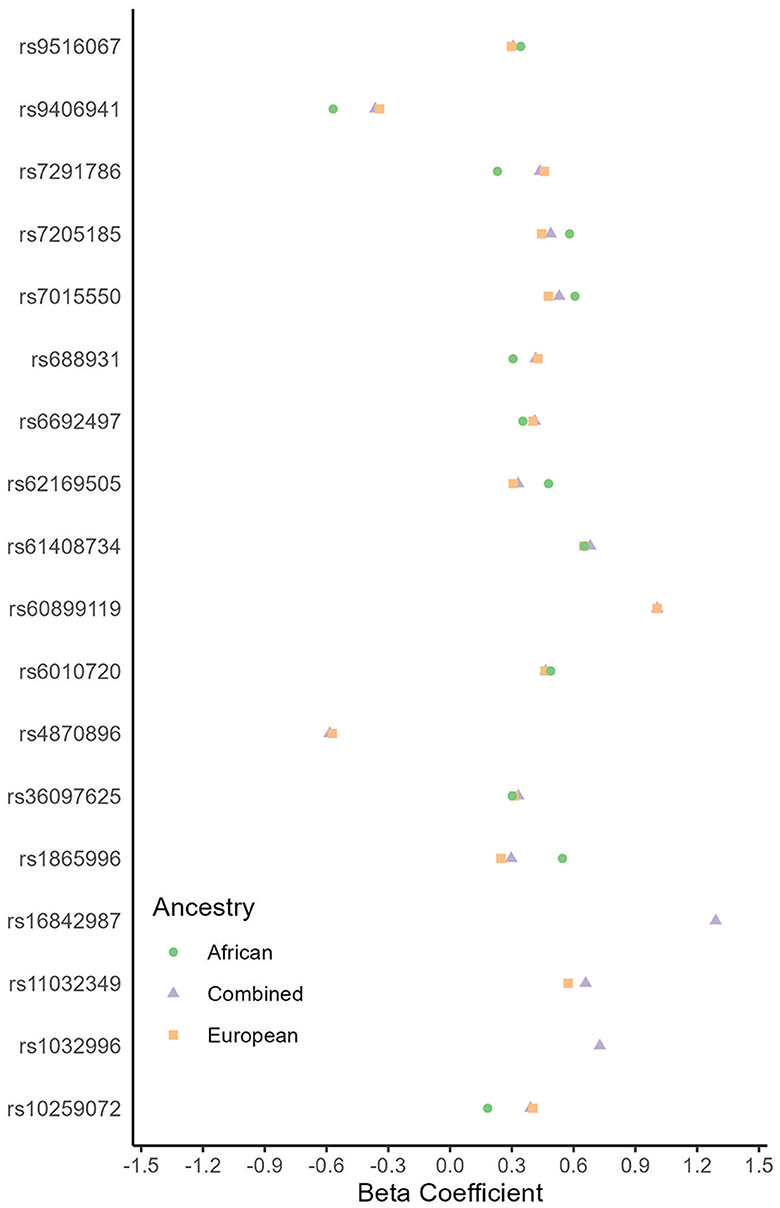
Figure 5. Genetic loci associated with recurrent stroke (p < 5e−6) by ancestry. Multiple genetic loci had estimates in the same direction (<0 or >0) between African and European ancestry genome-wide association studies. These genetic loci are consistent across ancestries.
Other loci appear to be related to the central nervous system. Interestingly, the same rs7205185-G locus above is also found in the NDE1 gene, because this gene and MYH11 overlap the same section of chromosome 16, but on opposite strands. NDE1 function is essential for the development of the cerebral cortex and may regulate production of neurons (Bakircioglu et al., 2011). The CD59 gene (rs11032349-T locus) is a potent inhibitor of the complement membrane attack complex (MAC) with known expression in human central nervous tissue (Farkas et al., 2002). The CD59 gene is slightly upregulated in neurons and glial cells in neuro-degenerative diseases associated with inflammation like Alzheimer's disease (McGeer et al., 1991).
The GO Enrichment and PANTHER Pathway analyses failed to show any biological process associations suggesting that this study has power limitations. Because of this and the desire to determine how much genetic risk of ischemic stroke can explain recurrent stroke risk, we implemented the GIGAStroke Consortium's PRS for European ancestry in the VISP cohort. The European PRS was developed from a primarily European cohort, but the cohort also had a notable proportion of individuals with African ancestry (Mishra et al., 2022). This is similar to the VISP cohort. We found that the PRS did no better than chance when discriminating recurrent stroke cases vs. control. By investigating stroke survivors, we are conditioning on the fact that they had a stroke and are already genetically “at-risk” for stroke.
Because the GIGAStroke consortium's PRS values correlated negatively with age and positively with BMI (Figure 4) in the VISP cohort, there is a measure of validity. Individuals with higher genetic risk of stroke are more likely have a stroke at a younger age, and BMI is a well-known stroke risk-factor. This raises the possibility that genetically determined recurrent stroke risk has some distinct signatures from incident stroke risk. Therefore, additional recurrent stroke GWAS must address the question: are there independent genetic drivers of recurrent stroke compared to incident stroke? This is an important question to help direct the field to new therapeutics for mechanism-specific secondary stroke prevention.
It is important to note that our study with 620/4,420 recurrent stroke cases among all ancestries is likely an underpowered GWAS for such a complex and multi-factorial disease. This may have contributed to the lack of genome-wide significant associations. Furthermore, our genome-wide suggestive (p < 5e−6) associations have higher a probability of false positive associations due to the large number of hypothesis tests. On the other hand, the majority of the suggestive genetic loci have known biological mechanisms affecting vascular contractility and stenosis as well as neuron development or response. The addition of strong biological relevance provides greater confidence in these suggestive genetic loci for further investigation.
In conclusion, we discovered several suggestive genetic loci associated with recurrent stroke from a population of African and European ancestries. Even though our recurrent stroke GWAS meta-analyses failed to find genome-wide significant associations, we observed multiple suggestive loci with high biological relevance. The application of incident ischemic stroke PRS to the VISP cohort revealed poor ability to discriminate between recurrent stroke status. We believe that this suggests that recurrent stroke genetic drivers are at least in part distinct from incident ischemic stroke.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The studies involving humans were approved by Wake Forest University School of Medicine University of North Carolina at Chapel Hill School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and institutional requirements.
CA: Formal analysis, Methodology, Writing – original draft. NA: Formal analysis, Methodology, Writing – review & editing. NS: Writing – review & editing. CB: Writing – review & editing. DP: Writing – review & editing. AL: Writing – review & editing. AP: Data curation, Writing – review & editing. TS: Data curation, Funding acquisition, Writing – review & editing. CJ: Data curation, Funding acquisition, Writing – review & editing. JM: Data curation, Funding acquisition, Writing – review & editing. F-CH: Methodology, Supervision, Writing – review & editing. KK: Methodology, Supervision, Writing – review & editing. MI: Data curation, Writing – review & editing. MS: Funding acquisition, Methodology, Writing – review & editing. BW: Data curation, Funding acquisition, Methodology, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The REGARDS genetics study was supported by the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (NHLBI) grant R01HL136666 (Leslie A. Lange, MI). This research project was supported by cooperative agreement U01 NS041588 co-funded by the NIH National Institute of Neurological Disorders and Stroke (NINDS) and the National Institute on Aging (NIA), Department of Health and Human Services. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the NIH. Representatives of the funding agency have been involved in the review of the manuscript but are not directly involved in the collection, management, analysis, or interpretation of the data. The SAHLSIS was funded by the Swedish Heart and Lung Foundation (CJ, 20220184); the Swedish Research Council (CJ, 2021-01114); the Swedish state under the agreement between the Swedish government and the county councils, the ALF agreement (CJ, ALFGBG-965328), the King Gustaf V:s and Queen Victoria's Foundation (CJ), the Agneta Prytz-Folke's, and Gösta Folke's Foundation (CJ), and Insamlingsstiftelsen for Neurological Research (TS). The GWAS component of the VISP study was supported by the United States National Human Genome Research Institute (NHGRI), Grant U01 HG005160 (PI: MS and BW), as part of the Genomics and Randomized Trials Network (GARNET). Genotyping services were provided by the Johns Hopkins University Center for Inherited Disease Research (CIDR), which was fully funded through a federal contract from the NIH to the Johns Hopkins University. Assistance with data cleaning was provided by the GARNET Coordinating Center (U01 HG005157; PI: Bruce S. Weir). Study recruitment and collection of datasets for the VISP clinical trial were supported by an Investigator-Initiated Research Grant (R01 NS34447; PI: James Toole) from the United States Public Health Service, NINDS, Bethesda, Maryland. Control data for comparison with European ancestry VISP stroke cases were obtained through the Database of Genotypes and Phenotypes (dbGAP) High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation (phs000187.v1.p1; R01CA100264, 3P50CA093459, 5P50CA097007, 5R01ES011740, 5R01CA133996, and HHSN268200782096C; PIs: Christopher Amos, Qingyi Wei, and Jeffrey E. Lee). For VISP stroke cases of African ancestry, a subset of the Healthy Aging in Neighborhoods of Diversity across the Life Span Study (HANDLS) were used as stroke free controls. HANDLS was funded by the National Institute of Aging (1Z01AG000513; PI: Michele K. Evans).
The authors thank the other investigators, the staff, and the participants of the REGARDS study for their valuable contributions. A full list of participating REGARDS investigators and institutions can be found at: https://www.uab.edu/soph/regardsstudy/. The authors acknowledge Research Computing at The University of Virginia for providing computational resources and technical support that have contributed to the results reported within this publication. URL: https://rc.virginia.edu. The authors thank the other SAHLSIS investigators, especially Katarina Jood, Petra Redfors, and Lukas Holmegaard, Institute of Neuroscience and Physiology, University of Gothenburg and Department of Neurology, Sahlgrenska University Hospital, as well as Björn Andersson, Bioinformatics and Data Center, Sahlgrenska Academy, for bioinformatic support, Riksstroke, and the SAHLSIS participants.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fstro.2024.1338636/full#supplementary-material
Armstead, W. M. (2011). Nociceptin/orphanin phenylalanine glutamine (FQ) receptor and cardiovascular disease. Cardiovasc. Ther. 29, 23–28. doi: 10.1111/j.1755-5922.2010.00241.x
Azad, A. K., Chakrabarti, S., Xu, Z., Davidge, S. T., and Fu, Y. (2014). Coiled-coil domain containing 3 (CCDC3) represses tumor necrosis factor-α/nuclear factor κB-induced endothelial inflammation. Cell Signal. 26, 2793–2800. doi: 10.1016/j.cellsig.2014.08.025
Bakircioglu, M., Carvalho, O. P., Khurshid, M., Cox, J. J., Tuysuz, B., Barak, T., et al. (2011). The essential role of centrosomal NDE1 in human cerebral cortex neurogenesis. Am. J. Hum. Genet. 88, 523–535. doi: 10.1016/j.ajhg.2011.03.019
Benjamin, E. J., Muntner, P., Alonso, A., Bittencourt, M. S., Callaway, C. W., Carson, A. P., et al. (2019). Heart disease and stroke statistics-2019 update: a report from the American Heart Association. Circulation 139, e56–528. doi: 10.1161/CIR.0000000000000659
Benjamins, J. W., Yeung, M. W., van de Vegte, Y. J., Said, M. A., van der Linden, T., Ties, D., et al. (2022). Genomic insights in ascending aortic size and distensibility. EBioMedicine 75:103783. doi: 10.1016/j.ebiom.2021.103783
Farkas, I., Baranyi, L., Ishikawa, Y., Okada, N., Bohata, C., Budai, D., et al. (2002). CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. J. Physiol. 539, 537–545. doi: 10.1113/jphysiol.2001.013381
Fernández-Cadenas, I., Mendióroz, M., Giralt, D., Nafria, C., Garcia, E., Carrera, C., et al. (2017). GRECOS project (genotyping recurrence risk of stroke): the use of genetics to predict the vascular recurrence after stroke. Stroke 48, 1147–1153. doi: 10.1161/STROKEAHA.116.014322
Hankey, G. J., Spiesser, J., Hakimi, Z., Carita, P., and Gabriel, S. (2007). Time frame and predictors of recovery from disability following recurrent ischemic stroke. Neurology 68, 202–205. doi: 10.1212/01.wnl.0000250327.73031.54
Howard, G., Kissela, B. M., Kleindorfer, D. O., McClure, L. A., Soliman, E. Z., Judd, S. E., et al. (2016a). Differences in the role of black race and stroke risk factors for first vs. recurrent stroke. Neurology 86, 637–642. doi: 10.1212/WNL.0000000000002376
Howard, G., Moy, C. S., Howard, V. J., McClure, L. A., Kleindorfer, D. O., Kissela, B. M., et al. (2016b). Where to focus efforts to reduce the Black-White disparity in stroke mortality: incidence vs. case fatality? Stroke 47, 1893–1898. doi: 10.1161/STROKEAHA.115.012631
Howard, V. J., Cushman, M., Pulley, L., Gomez, C. R., Go, R. C., Prineas, R. J., et al. (2005). The reasons for geographic and racial differences in stroke study: objectives and design. Neuroepidemiology 25, 135–143. doi: 10.1159/000086678
Jong, G., Lodder, J., Kessels, F., and Raak, L. (2004). Homogeneity of large and small vessel disease over time: arguments from a study on recurrent stroke in 998 patients with first cerebral infarct. J. Stroke Cerebrovasc. Dis. 13, 141–147. doi: 10.1016/j.jstrokecerebrovasdis.2004.06.001
Jood, K., Ladenvall, C., Rosengren, A., Blomstrand, C., and Jern, C. (2005). Family history in ischemic stroke before 70 years of age: the Sahlgrenska academy study on ischemic stroke. Stroke 36, 1383–1387. doi: 10.1161/01.STR.0000169944.46025.09
Lee, B. T., Barber, G. P., Benet-Pagès, A., Casper, J., Clawson, H., Diekhans, M., et al. (2022). The UCSC Genome Browser database: 2022 update. Nucl. Acids Res. 50, D1115–D1122. doi: 10.1093/nar/gkab959
Lemmens, R., Buysschaert, I., Geelen, V., Fernandez-Cadenas, I., Montaner, J., Schmidt, H., et al. (2010). The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke 41, 1850–1857. doi: 10.1161/STROKEAHA.110.587980
Ling, C., Poulsen, P., Carlsson, E., Ridderstråle, M., Almgren, P., Wojtaszewski, J., et al. (2004). Multiple environmental and genetic factors influence skeletal muscle PGC-1α and PGC-1β gene expression in twins. J. Clin. Invest. 114, 1518–1526. doi: 10.1172/JCI21889
Maguire, J., Thakkinstian, A., Levi, C., Lincz, L., Bisset, L., Sturm, J., et al. (2011). Impact of COX-2 rs5275 and rs20417 and GPIIIa rs5918 polymorphisms on 90-day ischemic stroke functional outcome: a novel finding. J. Stroke Cerebrovasc. Dis. Off. J. Natl. Stroke Assoc. 20, 134–144. doi: 10.1016/j.jstrokecerebrovasdis.2009.10.011
McGeer, P. L., Walker, D. G., Akiyama, H., Kawamata, T., Guan, A. L., Parker, C. J., et al. (1991). Detection of the membrane inhibitor of reactive lysis (CD59) in diseased neurons of Alzheimer brain. Brain Res. 544, 315–319. doi: 10.1016/0006-8993(91)90071-3
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, X., et al. (2019). Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721. doi: 10.1038/s41596-019-0128-8
Mi, H., and Thomas, P. (2009). PANTHER Pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol. Biol. Clifton NJ. 563, 123–140. doi: 10.1007/978-1-60761-175-2_7
Mishra, A., Malik, R., Hachiya, T., Jürgenson, T., Namba, S., Posner, D. C., et al. (2022). Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 611, 115–123. doi: 10.1038/s41586-022-05165-3
Oguri, M., Kato, K., Yokoi, K., Yoshida, T., Watanabe, S., Metoki, N., et al. (2010). Assessment of a polymorphism of SDK1 with hypertension in Japanese Individuals. Am. J. Hypertens. 23, 70–77. doi: 10.1038/ajh.2009.190
Park, J. H., and Ovbiagele, B. (2016). Association of black race with recurrent stroke risk. J. Neurol. Sci. 365, 203–206. doi: 10.1016/j.jns.2016.04.012
Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., et al. (2003). Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 100, 8466–8471. doi: 10.1073/pnas.1032913100
Pedersen, A., Redfors, P., Lundberg, L., Gils, A., Declerck, P. J., Nilsson, S., et al. (2016). Haemostatic biomarkers are associated with long-term recurrent vascular events after ischaemic stroke. Thromb. Haemost. 116, 537–543. doi: 10.1160/TH15-12-0938
Sheinart, K. F., Tuhrim, S., Horowitz, D. R., Weinberger, J., Goldman, M., Godbold, J. H., et al. (1998). Stroke recurrence is more frequent in Blacks and Hispanics. Neuroepidemiology 17, 188–198. doi: 10.1159/000026172
Tayo, B. O., Luke, A., Zhu, X., Adeyemo, A., and Cooper, R. S. (2009). Association of regions on chromosomes 6 and 7 with blood pressure in Nigerian families. Circ. Cardiovasc. Genet. 2, 38–45. doi: 10.1161/CIRCGENETICS.108.817064
Thomas, P. D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, L. P., Mi, H., et al. (2022). PANTHER making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8–22. doi: 10.1002/pro.4218
Toole, J. F. (2002). Vitamin intervention for stroke prevention. J. Neurol. Sci. 203–204, 121–124. doi: 10.1016/S0022-510X(02)00265-4
Viechtbauer, W. (2010). Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48. doi: 10.18637/jss.v036.i03
Wagner, C. H. (1982). Simpson's Paradox in real life. Am. Stat. 36, 46–48. doi: 10.1080/00031305.1982.10482778
Xue, Y., Zeng, X., Tu, W. J., and Zhao, J. (2022). Tumor necrosis factor-α: the next marker of stroke. Dis. Mark. 2022:2395269. doi: 10.1155/2022/2395269
Keywords: recurrent stroke, genetic risk, diversity, GWAS, African ancestry, meta-analysis, polygenic risk score
Citation: Aldridge CM, Armstrong ND, Sunmonu NA, Becker C, Palakshappa D, Lindgren AG, Pedersen A, Stanne TM, Jern C, Maguire J, Hsu F-C, Keene KL, Sale M, Irvin MR and Worrall BB (2024) Diversity in genetic risk of recurrent stroke: a genome-wide association study meta-analysis. Front. Stroke 3:1338636. doi: 10.3389/fstro.2024.1338636
Received: 15 November 2023; Accepted: 22 January 2024;
Published: 21 February 2024.
Edited by:
Paul Alan Nyquist, Johns Hopkins University, United StatesReviewed by:
Michelle C. Johansen, Johns Hopkins Medicine, United StatesCopyright © 2024 Aldridge, Armstrong, Sunmonu, Becker, Palakshappa, Lindgren, Pedersen, Stanne, Jern, Maguire, Hsu, Keene, Sale, Irvin and Worrall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chad M. Aldridge, Y21hN25AdXZhaGVhbHRoLm9yZw==
†Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.