
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Astron. Space Sci. , 15 June 2023
Sec. Astrobiology
Volume 10 - 2023 | https://doi.org/10.3389/fspas.2023.1183867
A correction has been applied to this article in:
Corrigendum: Co-adaptation of Streptococcus mutans to simulated microgravity and silver nitrate
 Mizpha C. Fernander
Mizpha C. Fernander Kelyah Spurgeon
Kelyah Spurgeon Jada Graves
Jada Graves Wynter Guess
Wynter Guess Jordan Miller
Jordan Miller Chanell Mangum
Chanell Mangum Joseph L. Graves Jr.
Joseph L. Graves Jr. Misty D. Thomas*
Misty D. Thomas*To sustain life on extended space missions, it is essential to maintain clean potable water. NASA currently uses iodine as the primary biocide in the potable water dispenser on the International Space Station and has recently proposed a potential switch to silver-based antimicrobials. Streptococcus mutans is the primary etiological agent of dental caries, part of the normal oral flora, and would endure direct exposure to water from the potable water dispenser. In our previous work, we examined the 100-day adaptive response of Streptococcus mutans to simulated microgravity (sMG). Here, we examined the evolutionary co-adaptation of S. mutans under sMG and silver nitrate (AgNO3) to evaluate the consequences of using silver as a primary biocide in space and the impact on the evolution of microbes from the oral microbiome. To do this, we adapted four populations of S. mutans under sMG and co-adapted four populations in simulated microgravity and silver nitrate using high-aspect ratio vessels for 100 days. Genomic analysis at multiple time points showed that S. mutans in sMG evolved variants consistent with our previous findings (SMU_1307c and SMU_399) while also acquiring novel mutations in the glutathione reductase gorA. The co-adapted populations showed mutations specific for the environment in ciaH/R, PBP1a, trkA, and trkB. We also assessed virulence phenotypes, and while simulated microgravity increased antibiotic susceptibility, sucrose-dependent adhesion, and, in some populations, acid tolerance, co-adaptation to silver nitrate reversed these effects. Overall, these data show that the use of silver as a biocide in simulated microgravity can evolve strains with novel genotypic and phenotypic traits that could alter virulence.
On the International Space Station, there are several precautions put in place to reduce microbial contamination and growth (Pierson, 2001; Mermel, 2013). One such example is the water recovery system (WRS) which recovers potable water from urine distillate, cabin air humidity condensate, and other hygiene wastewater (Carter, Tobias, & Orozco, 2013). Iodine (up to 3 mg/L) has primarily been used in the WRS to inhibit bacterial growth, but more recent reports have suggested a switch to using ionic silver (0.5 ppm or 0.5 mg/L) for future long-term missions (Slote, 2016; Li, 2018; Ley et al, 2021). Iodine has proven to be an effective antimicrobial agent on the International Space Station (ISS), although long-term exposure has been shown to have a negative effect on the thyroid gland (Slote, 2016). Therefore, iodine requires removal prior to consumption through additional hardware. This increases the complexity and required maintenance of the system (Packham et al, 1999; Li, 2018).
Silver has always been used as the primary biocide by the Russian Space Agency (RSA) as it has very few side effects on humans at bactericidal concentrations and, therefore, does not require removal prior to consumption (Mosher, 2008; Ley et al, 2021). The mechanism of antimicrobial action of silver is still not well understood. In vitro, its mechanism is multiform, and it has been shown to interact with and damage the bacteria cell wall and permeate membranes. It also binds to thiol groups in respiratory enzymes and other proteins causing inactivation and disruption of metabolism, cell signaling, DNA replication, transcription, translation, and cell division (Liau, 1997; Rai, M., Yadav, & Gade, 2009; Flores-López, Espinoza-Gómez, & Somanathan, 2019; Park et al, 2009). Despite silver having a high level of toxicity, there has been clear evidence that microbes can evolve resistance to silver-based antimicrobial agents and resistance to metals can be pleiotropic, often leading to antibiotic resistance through overlapping resistance mechanisms (Graves et al, 2015; Tajkarimi et al, 2017; Thomas et al, 2021; Engin, Engin, & Engin, 2023).
In 2018, Li et al (Li, 2018) performed a comprehensive review of the potential consequences of using silver as a biocide on the ISS. Their main conclusions were as follows: 1) silver is a powerful biocide that is relatively safe for humans, 2) we do not have a complete understanding of its biocidal activity, 3) usage should not exceed 0.400 ppm for 1000-day class missions, and 4) silver resistance is a concern, especially for long-term application. As a result, they suggested the use of complementary biocides and periodic elimination of the entire bacterial population with a secondary biocide. The use of silver-based compounds will eliminate microbes present in the PWD, but not the microbes that inhabit the astronauts themselves.
The human mouth is a very complex community made up of over 1000 different species and can reach cell densities as high as 1011 CFU/mL (Evaldson, Heimdahl, Kager, & Nord, 1982) despite having to endure a constant change in environmental pressures (Kolenbrander et al, 2006; Palmer Jr, Diaz, & Kolenbrander, 2006; Kreth, Zhang, & Herzberg, 2008). The oral microbiome plays a role in not only maintaining oral health but also in maintaining systemic health (Shroff, Meslin, & Cebra, 1995; Hooper, Littman, & Macpherson, 2012). Despite this, dental decay remains one of the highest prevailing diseases in humans (Takahashi & Nyvad, 2011). Of the oral residents, S. mutans has been actively studied for its cariogenic properties as this organism not only facilitates dental decay but also resides as a member of normal human plaque (Gross et al, 2010). As dental decay is one of the highest prevailing diseases in humans, NASA’s Space Medicine Exploration Medical Condition List lists dental health conditions as a major concern for occurrence during space flight missions (i.e., temporary fillings, crown replacements, abscesses, tooth loss, or toothaches) (Watkins, 2011). Several studies have evaluated the effects of both microgravity and simulated microgravity on astronauts’ oral health (Lloro et al, 2020) and have shown that on long-term missions (30–220 days), humans showed an increase in both IgAs and anaerobic bacteria, specifically Streptococcus mutans in both saliva and dental plaque (Brown et al, 1974; Lloro et al, 2020). Currently, it is poorly understood how these microbes, and generally microbes of the microbiome, evolve as a result of the selective pressures encountered during space missions.
Silver also has a long history of use in dentistry, and in the oral cavity specifically, silver has been shown to inhibit demineralization and exhibit effective antimicrobial properties against a wide range of both Gram-negative and Gram-positive bacteria (Fernandez et al, 2021). As the oral cavity will continuously encounter water from the PWD and, therefore, silver, if unfiltered, we explored the evolutionary consequences of silver nitrate exposure on the evolution of the oral human resident and pathogen S. mutans in simulated microgravity (sMG) as it may also alter treatment strategies. This is a follow-up study to our initial work evaluating the adaptive response of S. mutans to sMG alone (Fernander et al, 2022). Here, our objective is to better understand the consequences of using silver nitrate or silver-based compounds as the main antimicrobial mitigation strategy on future long-term space missions and provide data to support informed decisions regarding potential water treatment options and/or mitigation strategies.
Streptococcus mutans Clarke strain NCTC 10449 was purchased from ATCC (25175) and deemed our ancestral strain. All standard growth experiments were conducted using brain heart infusion (BHI) broth and incubated at 37 °C with 5% CO2 unless otherwise noted.
We first performed a range-finding study to determine the sublethal concentration of AgNO3 to use for the selection experiment. To do this, an overnight culture of the ancestral strain was grown, diluted to an O.D.600 of 0.05 and used to inoculate a 96-well plate with an increasing concentration of AgNO3 (0–50 μg/mL). After 24 h, the O.D.600 showed that 16 μg/mL AgNO3 was the highest concentration of silver nitrate that supported growth of S. mutans. Once grown in our system, we then had to reduce the concentration to 10 μg/mL to allow survival and, therefore, selection during the EE study.
High-aspect rotating vessels (HARVs) were purchased from Synthecon Inc. (Houston, TX) and used to culture S. mutans under sMG (Herranz, R. et al, 2013). Prior to inoculation, the HARVs were cleaned as described previously (Fernander et al, 2022). Using the S. mutans ancestral stock, 100 μL of an overnight culture was used to inoculate 100 mL of fresh BHI broth. This was then split into 2 × 50 mL cultures, and 10 μg/mL silver nitrate was added to one 50 mL sample. The sub-culture in BHI alone was loaded into four HARVs and deemed sMG5–8 (to distinguish these four populations from our previous study that generated sMG1–4 (Fernander et al, 2022)). The BHI supplemented with AgNO3 was split into four additional HARVs and deemed sMGAg1–4. All eight HARVs were incubated on the vertical axis to simulate microgravity and incubated at 37 °C overnight with 5% CO2 at 25 rpm. After 24 h of growth, the HARVs were then sub-cultured by adding 10 mL of fresh BHI or BHI supplemented with 10 μg/mL AgNO3, and this was repeated every 24 h for 51 days. At day 52, we increased the concentration of AgNO3 to 11 μg/mL and sub-cultured again every 24 h until 100 days. Twice a week, we performed serial dilutions and plated the cultures onto both BHI agar and mitis salivarius bacitracin agar to validate the integrity of the populations, and if deemed pure, glycerol stocks were made and stored at −80 °C. At every milestone time point (21, 42, 63, and 100 days), the remainder of the cultures were pelleted and stored at −80°C for DNA extraction and whole-genome sequencing.
The ancestral, all eight 100-day populations (sMG5–8 and sMGAg1–4) along with the normal gravity (NG) populations from our original study (Fernander et al, 2022) (NG1–4) were grown overnight in the HARVs and diluted to 0.5 based on the McFarland standard and used to swab 150 mm BHI agar plates. Disks of ampicillin, bacitracin, chloramphenicol, erythromycin, penicillin (G+), streptomycin, tetracycline, and trimethoprim (BD BBL™ Sensi-Disc™ Antimicrobial) were then added to plates. After disc placement, the plates were inverted and placed at 37 °C with 5% CO2 overnight. The following day, zones of inhibition were measured (in mm), and based on the company’s standards for each antibiotic, we evaluated changes in susceptibility. All populations were assessed in triplicate.
Acid tolerance assays were performed as described in our previous study (Fernander et al, 2022). In short, after removal of the cultures from the HARVs, a sample of each population was diluted to an O.D.600 of 0.3 using fresh BHI. These 1 mL diluted cultures were then centrifuged to pellet the cells and then washed using 0.2 M glycine pH 6.8–7.0. Samples were again pelleted and exposed to 0.2 M glycine pH 2.8–3.0 for 0, 20, 30, and 45 min. At the end of the incubation time, the samples were pelleted and re-suspended in fresh BHI twice for serial dilutions. Serial dilutions of each time point were then plated on BHI agar and CFUs were counted after 48 h incubation and compared to CFU counts of the ancestral population. Each assay was performed in triplicate.
All eight 100-day populations from this study were assessed for changes in their adhesion abilities as previously described (Fernander et al, 2022). Populations from the HARVs were diluted to an O.D.600 of 0.05 and 10 mL were used to inoculate 190 mL of fresh BHI broth supplemented with 0.1% sucrose to measure sucrose-dependent adhesion (SDA) or 0.1% glucose to measure sucrose-independent adhesion (SIA) in a 96-well plate. Both SDA and SIA were also measured with the addition of 11 μg/mL AgNO3. All populations were assessed in triplicate. The plates were then incubated without shaking at 37 °C with 5% CO2 for 24 and 48 h. After 24 and 48 h, the media were removed and the plates were washed with distilled water two times. Also, 125 mL of 1% crystal violet solution was then added to the washed plate and left to incubate for 15 min at room temperature. The plates were then washed with distilled water four times and left to dry overnight upside down. The following day, 125 mL of 30% acetic acid was used to re-suspend any adhered cells and then the plate was read at 595 nm.
Chromosomal DNA was extracted from the pellets originating from the 21-, 42-, 63-, and 100-day populations using the E. Z.N.A. bacterial DNA extraction kit from Omega Biotek® as per the manufacturers’ protocol. Eluted DNA was then quantified using the QuantiFluor® dsDNA system (Promega) and shipped to the Microbial Sequencing Center at the University of Pittsburgh. Sample libraries were prepared using the Illumina DNA Prep kit and IDT 10 bp UDI indices and sequenced on an Illumina NextSeq 2000, producing 2 × 151 bp reads. Demultiplexing, quality control, and adapter trimming were performed with BCL-convert (v 3.9.3) (Illumina, 2021). Sequence alignment and variant calling from the samples were achieved by using the breseq 0.33.2 pipeline (Deatherage & Barrick, 2014).
All phenotypic data were plotted in GraphPad Prism ® Version 9.2.0. Statistics for pairwise comparisons between ancestral populations and each of the individual treatment populations were calculated using an ordinary one-way ANOVA. Significance is defined by the two-tailed p-value using an *. * <0.05, ** <0.001, *** <0.0001, and **** <0.0001.
After 24 h, the O.D.600 showed that 16 μg/mL AgNO3 was the highest concentration of silver nitrate that supported growth of S. mutans. After two attempts at carrying out an EE study (in the following section) at this concentration, populations continuously went extinct; therefore, we reduced the silver concentration to 10 μg/mL to allow survival and, therefore, selection during the EE study.
After the 100-day EE study, we assessed the genetic changes that resulted from both adaptation to sMG and co-adaptation of sMG and AgNO3 (sMGAg). To do this, we performed whole-genome DNA resequencing at multiple time points (21, 42, 63, and 100 days). The S. mutans NTCC 10449 genome was used as the alignment sequence in this study (S.mutans_NCTC10449. gbk), and variant calling was conducted using breseq 33.2 (Deatherage & Barrick, 2014). The ancestral population was sequenced in our previous work (Fernander et al, 2022) and used to negate variation from the sequence database. Coverage distribution plots of mapped reads output from breseq showed a depth of coverage for unique positions ranging from ∼150 x to over 200 x with most showing ∼200 x coverage. All sequencing data with frequencies of mutation (f) above 0.1 are summarized in Supplementary Table S1 and compared to the data generated in our initial study. f represents the fraction of the total population that carries the indicated mutation. The datasets generated in this study can be found via the NCBI Bioproject database under Bioproject number PRJNA923356.
Four new independent populations of S. mutans were adapted to sMG in this study and deemed sMG5–8. We, therefore, began by comparing the mutations obtained in this study with those obtained in our previous work to further validate the importance of these mutations during adaptation to sMG (Supplementary Table S2).
Our initial study (Fernander et al, 2022) showed that sMG1–4 treatments displayed unique variants in three genes: SMU_1307c, which encodes for a DUF1003 domain containing protein (MG1, 3, and 4), pknB, a serine/threonine kinase (MG1 and 4), and SMU_399, the C3-glycoprotein degrading protease (C3-GDP) (sMG1, 2 and 4). We also detected mutations in three genes earlier than when they were detected in our normal gravity (NG) NG1–4 populations, in SMU_399, ptsH, and rex. The sMG5–8 population did not acquire any mutations in pknB, ptsH, or rex. They did, however, acquire mutations in the SMU_399, albeit not until 42 days (sMG5 and 6) and all at low frequencies (f > 0.01). Although, by 100 days, sMG5–8 all show mutations in SMU_399, sMG5 and 6 to fixation (f = 1.000), and sMG7 and 8 with multiple variants at high frequency. A single non-synonymous mutation in SMU_1307c was also detected in sMG5 and 6 both reaching fixation (f = 1.00) by day 100, again reiterating the importance of both SMU_1307c and SMU_399 in the adaptation of S. mutans to sMG.
In addition to the examples of parallel evolution mentioned above, we also detected novel variants in our new sMG5–8 populations (Supplementary Table S3). This includes variants in gorA, the glutathione-disulfide reductase which acquired mutations at day 63, and by day 100, sMG5, 7, and 8 all held gorA mutations all with at least one variant with f > 0.2. In our initial study, sMG2 also held a gorA mutation (E251*) at 100 day with f of 0.114, and as it was the only mutation to be acquired across the four populations at all time points, we did not originally note this gene important for sMG1–4 adaptation.
Finally, as with the initial study, each of the four new sMG5–8 populations has a variety of mutations that are unique to the individual population (Supplementary Table S3), reiterating again the uniqueness in each of the evolutionary trajectories for each biological replicate due to the selective nature of sMG.
There is also significant evidence of parallel evolution between the populations that were co-adapted to sMG and silver nitrate (sMGAg1–4) with those adapted in sMG alone (1–8) (Supplementary Table S1). This includes variants in SMU_399 in two populations and variants in SMU_1307c, which, at 100 day, showed strong evidence of positive selection in all four sMGAg populations.
The sMGAg populations also acquired a variety of novel mutations indicative of selection specific for sMGAg (Supplementary Table S4). First, at 100 days, each population has a unique set of mutations to fixation. sMGAg1 carries an intergenic mutation between ahpC ←/← DQM59_RS06790 and an intergenic mutation between DQM59_RS03470 →/→ lacA. sMGAg2 carries an A327S mutation in DQM59_RS04610, a phosphopyruvate hydratase, and an E18* in DQM59_RS06795 a U32 family peptidase. sMGAg3 does not carry any unique mutations to fixation but does have three high-frequency mutations in DQM59_RS01815, a LacI family transcriptional regulator (S15I, f = 0.839), in DQM59_RS10135, a hypothetical protein (P117S, f = 0.833), and in DQM59_RS10150, an NAD-dependent succinate-semi-aldehyde dehydrogenase (Q261*, f = 0.888). Finally, sMGAg4 carries three high-frequency variants in DQM59_RS04410, a TVP38/TMEM64 family protein (+T, f = 0.943; +G, f = 1.00 and Y34F, f = 0.943), and a +T to fixation in DQM59_RS10040, an APC family permease.
We also observed parallel evolution across the sMGAg populations themselves. At 21 days, sMGAg1 acquired an S67I (f = 0.170) and a G351V (f = 0.257) in the PBP1A family penicillin-binding protein and by day 42, all four populations carried high-frequency mutations in this gene. By day 63, sMGAg2–4 all lost their mutations, and by day 100, we were no longer able to detect any variants in these populations for this gene. At day 21, all four populations selected the same L253F mutation in the DQM59_RS04495 gene which encodes the regulatory subunit of an ATP phosphoribosyl transferase (f = 0.356, 0.261, 0.158, and 0.310, respectively). These mutations all increase in frequency by day 42, and by day 100, all lost this variant.
At day 21, sMGAg2 and 3 acquired mutations (f = 0.060 and 0.713, respectively) in DQM59_RS05155, the histidine kinase CiaH. By day 42, sMGAg4 acquired a mutation in DQM59_RS05155, which encodes for the response regulator CiaR, part of a two-component response system with CiaH. At 100 days, all three mutations went to fixation supporting positive selection on this gene under these experimental conditions. Finally, at day 100, three of our four populations (sMGAg1–3) each acquired four to five different variants in both trkA and trkB. All have at least one variant with an f > 0.2, but the number of total variants across each population indicates that selection is still taking place on these two genes.
As SDA is important for establishment of S. mutans within the oral biofilm (Signoretto, C., et al, 2009), we measured this trait for all eight populations and compared to the ancestral in a pairwise fashion. As with sMG2–4, sMG5–8 showed a 1.3-fold decrease in O.D.595 values from 3.25 in the ancestral to an average of 2.5 across sMG populations (p ˂ 0.0001) at 24 h (Figure 1A). Again, as with sMG1–4, this decrease was restored to ancestral levels when analyzed after 48 h (Figure 1B). On the contrary, the co-adapted populations (sMGAg1–4) showed no significant changes from the ancestral population at 24 or 48 h (Figures 1A, B). Interestingly, when the BHI media were supplemented with 11 μg/mL AgNO3, adherence of the sMG populations at 24 h was restored to ancestral levels and the sMGAg populations again showed no change from the ancestral (Figures 1C, D). Therefore, adaptation in sMG appears to decrease SDA in S. mutans, albeit both the short-term presence of silver nitrate in the environment (Figures 1C, D) and adaptation to silver nitrate can reverse this effect (Figure 1B).
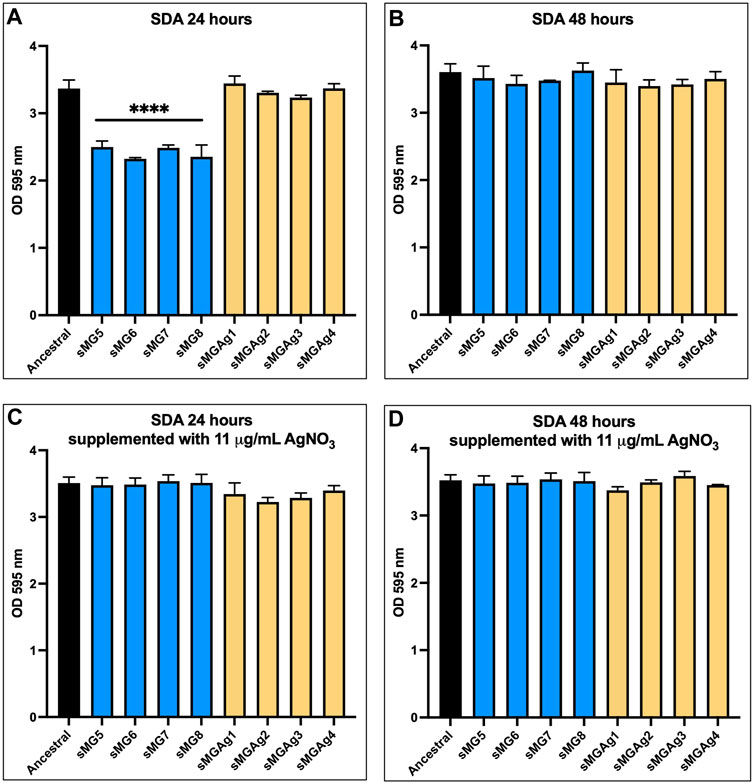
FIGURE 1. Silver nitrate reverses the reduction in SDA observed in sMG adapted populations. Sucrose-dependent adhesion was assessed after 24 and 48 h of stationary growth in BHI supplemented with 1% sucrose alone (A and B) and with the addition of 11 μg/mL silver nitrate (C and D) using the 100-day populations.
SIA allows S. mutans to achieve early colonization on the surface of the tooth enamel (Signoretto, C., et al, 2009). In our original study, all populations showed a significant reduction in SIA when compared to the ancestral population except for NG4 and sMG3. In this study, after 24 h and 48 h (Figure 2) sMG6 showed a five-fold (p ˂ 0.0001) and a three-fold (p < 0.05) increase compared to the ancestral, while the remainder, including the co-adapted populations, showed no change, respectively. The results with the addition of 11 μg/mL AgNO3 were very similar with sMG6 having a six-fold increase (p < 0.0001) over the ancestral at 24 h, and at 48 h, sMG8 also showed a four-fold increase (p < 0.05) at 24 h and 48 h (p < 0.05) over the ancestral.
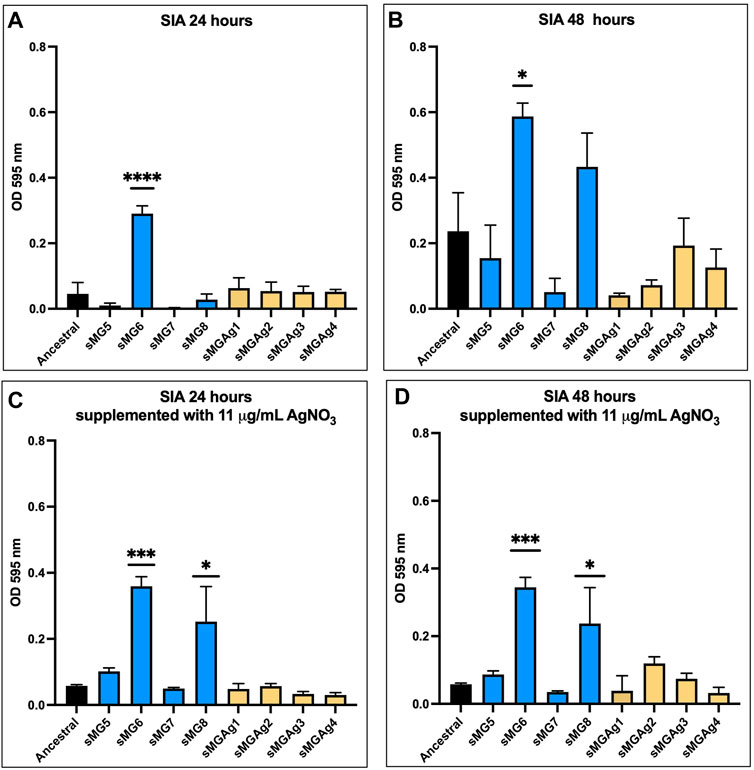
FIGURE 2. SIA shows variability between biological replicates in sMG. Sucrose-independent adhesion was assessed after 24 and 48 h of stationary growth in BHI supplemented with 1% glucose alone (A and B) and with the addition of 11 μg/mL silver nitrate (C and D) using the 100-day populations.
In our initial study (sMG1–4), we detected significant variance in acid tolerance between biological replicates at 100 days with most changes indicating that the populations became less tolerant (Fernander et al, 2022). In this study, we found slightly different results (Figure 3). After 20 min of exposure to low pH, all populations showed a decrease (1.6–3.8-fold) in acid tolerance as compared to the ancestral except for sMG6 and sMGAg2 which showed no change. By 30 min, all populations except sMG7 and sMGAg2 showed an increase in acid tolerance as compared to the ancestral. The sMG population showed an average of a 2–3-fold increase, whereas the sMGAg populations showed an average of a 3–7-fold increase. All sMG populations after 45 min of acid exposure showed an increase in acid resistance as compared to the ancestral population; both sMG5 and 6 displayed a 9-fold increase over the ancestral, sMG7 showed a 36-fold increase, and sMG8 showed a 4-fold increase. The co-adapted sMGAg showed more variance, sMGAg1 showed no change over the ancestral, sMGAg2 and 3 showed an increase in acid resistance (6- and 7-fold, respectively), and sMGAg4 showed a three-fold decrease in acid resistance over the ancestral population (sMGAg1 showed no change). The increase in acid resistance observed in these populations was the first time we observed this phenotype and reiterates the importance of biological replicates while using a weak selection environment such as sMG.
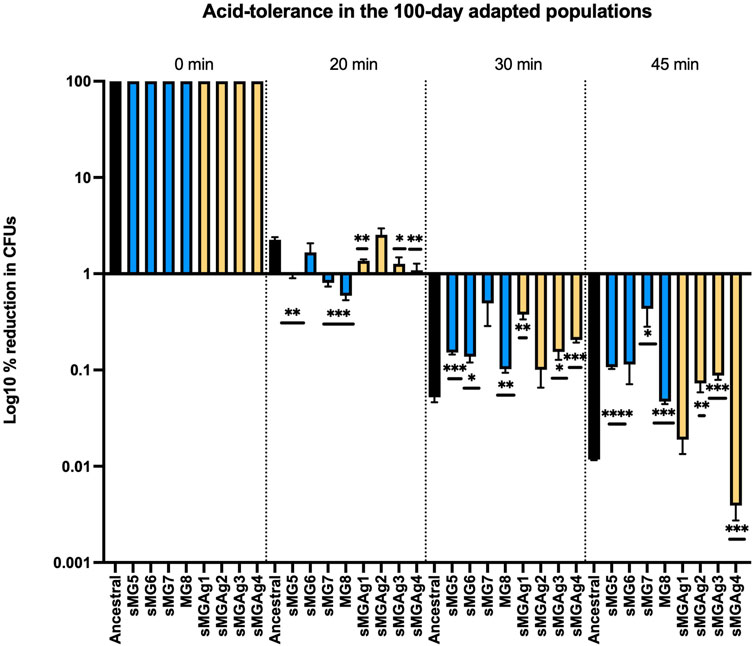
FIGURE 3. Changes in acid tolerance were identified as a result of microgravity and co-adaptation to microgravity and silver. Changes in acid tolerance were assessed after 100-day adaptation to sMG (blue) and simulated microgravity and co-adaptation in silver nitrate (yellow). Populations were compared to the ancestral population (black). The 100-day populations were exposed to an acidic environment (glycine pH 2.8) for 0, 20, 30, and 45 min, rescued, and serial diluted, and CFUs were counted after 48 h of growth on BHI agar plates. The CFU counts at the 0-time point were then normalized to 100%, and the percentage reduction was then calculated by dividing the CFU count at the indicated time points by the CFU count at 0. The values were then plotted on a log10 scale to visualize the data. For this dataset, prior to the one-way ANOVA, we performed a normality check (GraphPad Prism ® Version 9.2.0), and the Shapiro–Wilk test showed normality in all three individual time points (20, 30, and 45 min).
We used the disc diffusion method to assess changes in antibiotic susceptibility to eight common antibiotics (ampicillin, bacitracin, chloramphenicol, erythromycin, penicillin (G +), streptomycin, tetracycline, and trimethoprim) (Figure 4). For comparison, we analyzed both the ancestral and the normal gravity populations (NG1–4) adapted in our original experiment (Fernander et al, 2022). First, all the normal gravity adapted populations showed no change in susceptibility in comparison to the ancestral control for any of the eight tested antibiotics. The most significant change was observed in the sMG5–8 adapted populations. All four populations showed a 1.2–1.3-fold increase in susceptibility to erythromycin, penicillin G+, and tetracycline. sMG6 was also 1.3-fold more susceptible to bacitracin and streptomycin. sMG7 was 1.3-fold more susceptible to ampicillin, while sMG8 showed a 1.3-fold increase in susceptibility to ampicillin, bacitracin, and chloramphenicol. We saw very few changes in the sMGAg populations with only an increase in susceptibility of sMGAg1 by 1.4-fold to bacitracin, sMGAg4 by 1.3-fold to chloramphenicol, sMGAg3 and 4 by 1.2-fold to erythromycin, and sMGAg4 by 1.2-fold to tetracycline. In both this study and our previous work, we found increased susceptibility to antibiotics as a result of adaptation to sMG, albeit this positive effect is reversed for many of our tested antibiotics when co-adapted in the presence of silver nitrate.
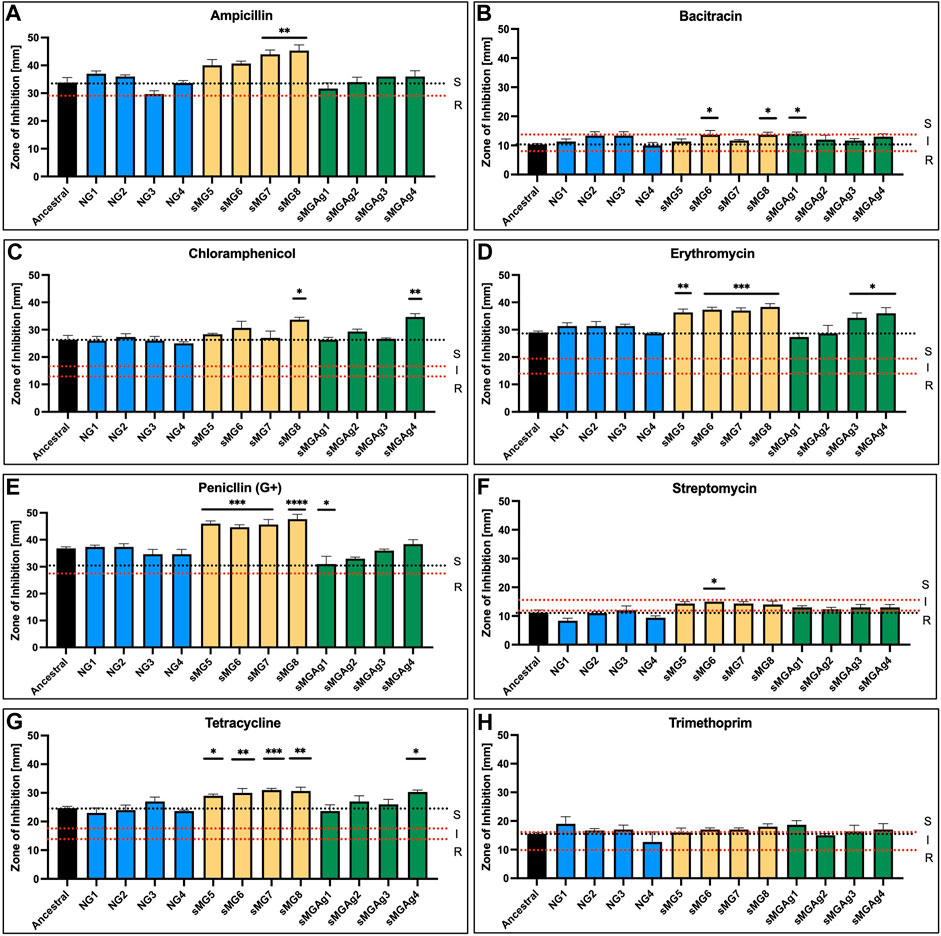
FIGURE 4. sMG adaptation increases susceptibility to antibiotics while co-adaptation reverses the effect. Antibiotic susceptibility was tested via disc diffusion assays using six antibiotics of varying mechanisms. (A) Ampicillin, (B) bacitracin, (C) chloramphenicol, (D) erythromycin, (E) penicillin (G+), (F) streptomycin, (G) tetracycline, and (H) trimethoprim. Zones of inhibition in (mm) were plotted in triplicate for 100-day adapted (yellow) and co-adapted populations (green). Normal gravity populations were also assessed (Fernander et al, 2022) as a second control (blue). The black dotted line represents the average ancestral level for easier comparison; the dotted red lines represent the “S” susceptibility, “I” intermediate, and “R” resistant zones as outlined by the manufacturer.
The goal of this study was two-fold, first, to add four additional long-term sMG adapted genomic replicates to the online public databases and, second, to begin to understand the impact of using silver as a biocide on long-term space missions.
We observed mutations in two of the five genes we previously deemed important for sMG adaptation (Fernander et al, 2022), and this included mutations in the DUF1003 domain containing protein SMU_1307c and SMU_399 the C3-GDP. We also found novel gorA mutations in sMG5, 7, and 8. We did identify a gorA mutation in sMG2 albeit not until 100 days in the original study. gorA is a glutathione-disulfide reductase that has not actually been characterized in Streptococcus species and is inferred from homology (uniprot.org). In Escherichia coli, gorA has been shown to be induced by reactive oxygen species, specifically H2O2 stress (Zheng, M., Åslund, & Storz, 1998). In our initial study, we identified mutations in rex. Rex is a redox sensor that specifically senses NADH/NAD + levels in the cell (Bitoun, Liao, Yao, Xie, & Wen, 2012; Zheng, Y. et al, 2014). GorA is predicted to have two NAD + binding sites, indicating the potential for redundant mutations and parallel evolution. This may also be why we never observed rex mutations in these new populations. Together, these data reiterate the importance of both SMU_1307c and SMU_399 in adaptation to the sMG alone. In addition, multiple experiments, along with our data, point to redox stress as a source of selection during sMG exposure (Fernander et al, 2022; Sharma & Curtis, 2022).
Despite the potent antimicrobial effects of ionic silver, there are still some major issues with its use as a sole antimicrobial agent in space. First, deposition of silver on the container and flow line surfaces reduces the concentration of silver ions to an unsatisfactory disinfection level (Slote, 2016). A few studies have tried to address this point by constructing silver-based filters and by using silver fluoride as opposed to the typical silver nitrate (Birmele, Michele N., L. E. McCoy, and Michael S.Roberts., 2011; Slote, 2016). These methods seem to prevent silver deposits from acting as seeds to precipitate the silver ions from solution. Second, as water is continuously recycled, any microorganism that survives a round of filtration and antimicrobial exposure will be exposed to silver ions. In addition, if this exposure occurs at sublethal levels, there should not only be a concern for infection, but more importantly, there should be a concern for the potential evolution of silver resistance strains which will survive all subsequent rounds of filtration. Therefore, to better understand the consequences of using silver or silver-based compounds as the primary biocide in the PWD, it is important to preemptively investigate the potential evolutionary consequences it may have on microbes of the human microbiome which will continuously be exposed through consumption. After 100 days of co-adaptation of S. mutans to sMG and silver nitrate, evaluation of whole-genome resequencing data showed several clear genomic mutations specific for silver nitrate. Despite several unique genomic sweeps in each of the individual sMGAg populations, there were three systems that acquired mutations within multiple populations which were maintained into day 100. This includes the two-component response system (TCRS) ciaH/R, the potassium transporters trkA and trkB, and the penicillin-binding protein pbp1a. sMGAg1 acquired mutations in PBP1a, trkA, and trkB and then unique mutations in the upstream region of a U32 peptidase and an intergenic insertion in front of lacA. sMGAg2 acquired the ciaH and trkB mutations and unique mutations in a phosphopyruvate hydratase and the U32 peptidase. sMGAg3 acquired ciaH, trkA, and trkB along with unique mutations in a hypothetical protein and an NAD-dependent succinate-semi-aldehyde dehydrogenase. sMGAg4 acquired mutations in ciaR and pbp1a and then unique mutations in a TMEM64 family protein, an APC family protease, and an energy-coupling factor transporter ATPase.
ciaH/R is a TCRS and a global regulator for multiple stress responses including biofilm formation, acid tolerance, bacteriocin production, and genetic competence (Qi, Merritt, Lux, & Shi, 2004; Ahn, Wen, & Burne, 2006; Biswas, Drake, Erkina, & Biswas, 2008). Ultimately, CiaH is important for global stress tolerance and, interestingly, does so through not only regulating its cognate response regulator CiaR but also phosphorylation of other response regulators (Ahn et al, 2006). We have mapped both the CiaH and CiaR mutations onto predicted structures generated through AlphaFold (Jumper et al, 2021; Varadi et al, 2022) and available through uniprot.org (Figure 5A; B). It has been shown previously that deletions and mutations (Figure 5A, black) in ciaH leads to the overexpression of ciaR. The overexpression of ciaR confers a pleiotropic phenotype, increasing antibiotic resistance, decreasing competence, and protection during lysis-inducing conditions (Halfmann, Kovács, Hakenbeck, & Brückner, 2007; Mascher, Heintz, Zä;hner, Merai, & Hakenbeck, 2006; Muller, 2018). ciaR mutants are typically lysis prone (Mascher et al, 2006). This pleiotropic response is why we hypothesize that the adaptive mutations are deleterious to CiaH function. When our adapted populations were tested for antibiotic susceptibility, many showed increased resistance over the sMG populations adapted alone for restoration of ancestral resistance profiles. CiaH is a histidine kinase that binds ATP and autophosphorylates at H226. One of the observed mutations, V364F, resides on the back surface of the ATP-binding pocket. This valine to phenylalanine mutation could occlude ATP from binding, potentially inactivating the protein. The second mutation, I136S resides in the sensor domain. It is possible that this mutation prevents interactions with either its dimeric partner which is required for activation or potentially with other molecules/proteins required for activation but require further testing to confirm. ciaR mutation (A80V) lies in the response regulator domain (Figure 5B), and this residue is extremely close to the phosphorylated aspartate (D51). Therefore, this may be inducing a conformational change that mimics the transphosphorylation state which is required to decrease inhibition by CiaH. Both predicted phenotypes could lead to increased cellular CiaR which has been shown to increase resistance to cell lysis and oxidative stress (Ibrahim, Kerr, McCluskey, & Mitchell, 2004; Mascher et al, 2006), both consequences of environmental silver. Additionally, ciaH/R-deficient strains have demonstrated severe growth defects (Ahn et al, 2006), and we did not observe this while monitoring daily O.D.600s (data not shown). In addition, it has also been shown that ciaH-defective mutants increase glutathione reductase expression (Wu et al, 2010) and the two populations (sMGAg2 and 3) that carry ciaH mutations are also our only silver adaptation populations to also carry mutations in the glutathione-disulfide reductase gorA. We believe that these results further support likely the activation of this system and, specifically, the activation of ciaR (Mascher et al, 2006). Albeit as stated, these effects require functional validation.
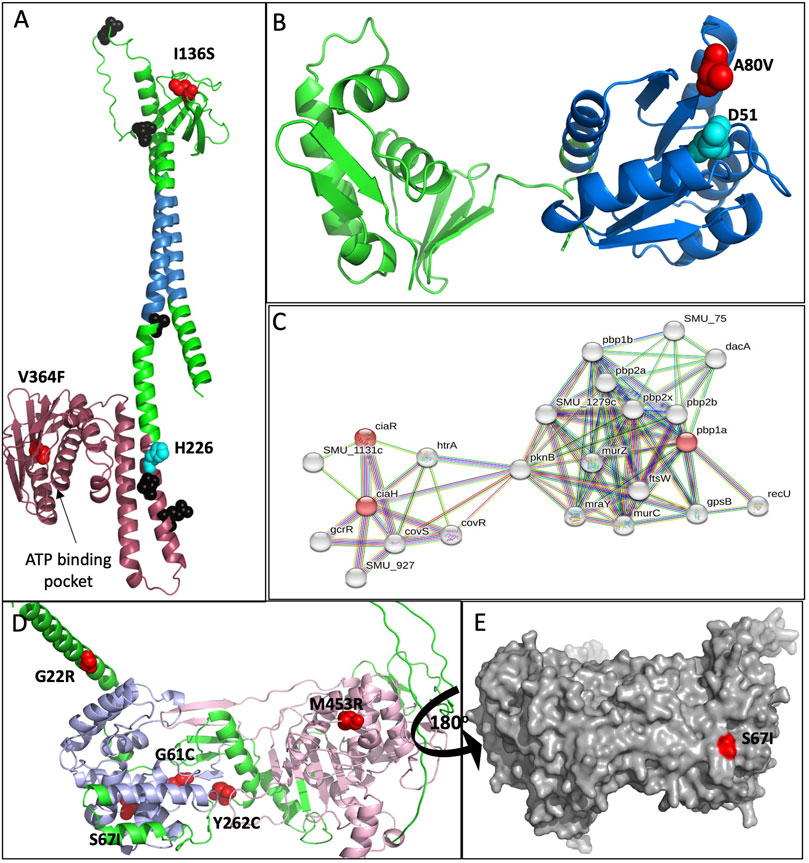
FIGURE 5. Molecular modeling of adaptive mutations. None of the affected proteins had structures present in the protein data bank; therefore, predicted structures generated by AlphaFold (Jumper et al, 2021; Varadi et al, 2022) were downloaded from Uniprot.org. (A) Adaptive mutations mapped onto the predicted CiaH cartoon structure. Mutations resulting from this study are mapped in red, and the phosphorylated histidine (H266) is mapped in cyan. Mutations generated in other studies shown to inactivate or reduce CiaH function are mapped in black (Uniprot.org). The pink region represents the C-terminal ATPase domain, blue depicts the two transmembrane domains, and the green region at the top of the structure is the extracellular sensor domain. (B) An AlphaFold cartoon model of CiaR with the adaptive mutation mapped in red. The phosphorylated aspartic acid (D51) is highlighted in cyan. (C) String. db was used to generate an interaction map between CiaH and PBP1A. Important nodes are highlighted in red. The edges in the map indicate both physical and functional associations that originate from text mining, experiments, databases, co-expression, neighborhood, gene fusions, and co-occurrence. For this map, we used CiaH, CiaR, and PBP1a to generate the map with no more than 20 nodes in the second shell. (D) Cartoon and (E) surface AlphaFold model of PBP1a with adaptive mutations mapped in red. All structures were generated using PyMOL.
Genes regulated by cia have also been shown to respond to PBP1a-mediated changes in cell wall regulation. Penicillin-binding proteins (PBPs) are a major player in the synthesis of peptidoglycan in the bacterial cell wall (Schweizer et al, 2017). The interplay between these two systems may explain why we detected PBP1a mutations during early adaptation (21 days) in sMGAg1 which was the only population at this time point not to acquire mutations in the ciaH/R TRCS. Then, after 42 days, the frequency of ciaH/R mutations decreases in sMGAg2–4 while all four populations acquired PBP1a mutations to high f (0.5–0.8). At day 63, sMGAg2–4 populations lost PBP1a mutations while increasing frequency for ciaH/R mutations, while sMGAg1 was again the only to further increase the frequency of PBP1a mutations while never acquiring ciaH/R variants. These two independent evolutionary trajectories support the idea that silver may be damaging the cell wall and inducing lysis. While PBP1a and the ciaH/R regulon have not been shown to interact directly, (Figure 5C), they have both been shown to be affected by the heat shock HtrA protease. HtrA protects the cell from oxidative stress and lysis-inducing conditions while also playing a role in biofilm formation and genetic competence (Lemos & Burne, 2008). We predict that the PBP1a mutations would again not have a deleterious effect on the function of the protein, and like ciaR defective mutants, PBP1a-defective mutans exhibit major defects in growth, cell division, and biofilm capabilities (Wen, Bitoun, & Liao, 2015) again, none of which we observed during this adaptation study. Adaptive mutations in PBP1a (Figure 5D) occurred throughout the protein in both the glycosyl transfer domain (blue) and in the transpeptidase domain (pink). The highest frequency mutation that remains at day 100 in sMGAg1 is the S67I mutation, and this mutation is harder to characterize in terms of predicted function as it sits on the rear surface far from enzymatic binding sites and could potentially be involved in protein–protein interactions (Figure 5E).
Finally, at 100 days, three of our four sMGAg populations held mutations in the tkr genes at a total f of ∼0.4–0.6. sMGAg1 carries four mutations in tkr genes, two in tkrA and two in tkrB, sMGAg2 carries five in tkrB, and sMGAg3 carries one in tkrA and three in tkrB. One NG and three sMG populations did carry mutations in both genes as well but not to the extent observed in sMGAg populations (12 of the 20 mutations in these genes). The Tkr system plays a major role in potassium (K+) uptake important for homeostasis, regulation of membrane potential, pathogenesis, antimicrobial resistance, and biofilm formation (Do, E. A. & Gries, 2021; Zhang et al, 2020); (Do, T. T. et al, 2022; Girgis, Hottes, & Tavazoie, 2009; Lee et al, 2009). Specifically in S. mutans, Trks have been shown to be important for acid stress, hyperosmotic tolerance, and biofilm formation (Binepal et al, 2016; Peng, Zhang, Bai, Zhou, & Wu, 2016). Streptococcus mutans encodes two tkr potassium transporters, Trk1 (trkB, trk, and pacL), which is critical for growth under low potassium, membrane potential, acidic/osmotic stress, and biofilm formation, and Trk2 (trkA and trkH), which plays a minor/redundant role to Trk1 (Binepal et al, 2016). Many of the detected variants are premature stop codons (four), two are deletions, one is an insertion, three are intergenic mutations, and the remainders are SNPs. Of those SNPs, one changes the stop codon to a leucine and is the only SNP in trkA. All four trkB SNPs are in the N-terminal NAD+ binding domain. The nature of these mutations suggests that they are preventing the production of a functional protein product. Specifically, sMGAg1 carries a premature stop codon in both trkA and trkB, and sMGAg3 carries a premature stop codon in trkB and a 1 bp deletion in trkA indicating that neither system is functional in these two populations at 100 days. sMGAg2 not only carries a 3 bp insertion and 3 bp deletions which would not disrupt the reading frame but also carries a variety of SNPs. It is important to note that these variants only appeared at 100 days and have f = 0.4–0.6 of the total population while the remainder carry the ancestral sequence. This balance of variation in the population may explain why we do not see any major differences in biofilm formation nor a reduction in acid tolerance in these populations as the ancestral phenotype may be fitter in the assay conditions. Interestingly, sMGAg4 is the only population to not carry any trk variants and is the only to become more susceptible to acid stress. Currently, there is no evidence in the literature showing that the Trk system can import silver, albeit there is evidence that the bacterial Trk systems can uptake other monovalent cations such as cesium (Cs+) and rubidium (Ru+) (Bakker, 1983; Avery, 1995; Perkins & Gadd, 1995). The parallel evolution exhibited across three of our sMGAg populations may suggest a novel mechanism of silver transport in bacteria that remains to be elucidated.
We co-adapted planktonic S. mutans to simulated microgravity and silver nitrate to better understand the consequences of using silver as a biocide in space. Control sMG adapted populations showed similar results to our previous adaptation study (Fernander et al, 2022) in the acquisition of adaptive mutations in the DUF1003 domain containing protein SMU_1307c and SMU_399 in the C3-glycoprotein degrading protease. We also detected novel mutations consistent with selection due to redox stress. This demonstrates consistent parallel evolution and supports our previous results demonstrating unique evolutionary trajectories between biological replicates and the importance a biological replicate number in sMG studies. Our co-adaptation study demonstrated the evolution of populations that acquired unique mutations in the TCRS ciaH/R and pbp1a which could potentially hold overlapping adaptive mechanisms protecting the cell from the oxidative stress of silver and deleterious mutations in the trkA and trkB genes which could limit silver entry. Silver nitrate also specifically contributes to the evolution of unique genetic variants that display phenotypes that are more tolerant to acid than ancestral strains and have increased SDA and antibiotic resistance over populations adapted in sMG alone. Altogether, our data show that the addition of silver nitrate into the sMG environment influences the evolutionary trajectory of the dental pathogens S. mutans and could influence treatment of resultant infections in space. In addition, our data show the importance for additional studies predicting and evaluating the evolutionary outcome of not only potential PWD mitigation strategies in space but also on the general evolutionary impact of the selective pressures of space on the microbes of the human microbiome.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA923356.
MF was responsible for overseeing and conducting all the experiments outlined in the manuscript. KS and JG helped in carrying out the evolution experiment, and KS also performed acid tolerance assays. WG, JM, and CM performed the antibiotic susceptibility assays. MT was responsible for overseeing the work of MF, KS, JG, WG, and CM, writing the manuscript, and acquiring funding. JG contributed to the acquisition of funding, conception and design of the study, analysis, and interpretation of all data and performed the statistical analysis. All authors contributed to the article and approved the submitted version.
This work was funded via support by start-up funds provided to MT by the North Carolina Agricultural and Technical State University, by Biocomputational Evolution in Action (BEACON): An NSF Center for the Study of Evolution in Action (National Science Foundation Cooperative Agreement No. DBI-0939454) and MCF held a Triangle Center for Evolutionary Medicine (TriCEM) Graduate Student Award. KS was funded by the North Carolina A&T State University Bridges-to-Doctorate Program (R25 GM119987) funded by the National Institute of General Medical Sciences (NIGMS) and the National Institutes of Health (NIH). WG, JM, and CM were funded by a National Institutes of Health NIGMS MARC Undergraduate NRSA Institutional Grant (Award number 5T34GM083980).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2023.1183867/full#supplementary-material
Ahn, Sug-Joon, Ahn, Sang-Joon, Browngardt, Christopher M., and Burne, Robert A. (2009). Changes in biochemical and phenotypic properties of Streptococcus mutans during growth with aeration. Appl. Environ. Microbiology75 8, 2517–2527. doi:10.1128/aem.02367-08
Ahn, S., Wen, Z. T., and Burne, R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74 (3), 1631–1642. doi:10.1128/iai.74.3.1631-1642.2006
Avery, S. V. (1995). Caesium accumulation by microorganisms: uptake mechanisms, cation competition, compartmentalization and toxicity. J. Industrial Microbiol. 14, 76–84. doi:10.1007/bf01569888
Bakker, E. P. (1983). pH-dependent transport of rubidium by the constitutive potassium uptake system TrkA of escherichia coli K-12. FEMS Microbiol. Lett. 16 (2-3), 229–233. doi:10.1111/j.1574-6968.1983.tb00293.x
Binepal, G., Gill, K., Crowley, P., Cordova, M., Brady, L. J., Senadheera, D. B., et al. (2016). Trk2 potassium transport system in Streptococcus mutans and its role in potassium homeostasis, biofilm formation, and stress tolerance. J. Bacteriol. 198 (7), 1087–1100. doi:10.1128/jb.00813-15
Birmele, Michele N., McCoy, L. E., and Roberts, Michael S. (2011). Disinfection of spacecraft potable water systems by passivation with ionic silver. Washingdon, United States: NASA. NASA Technical Report., Retrieved from internal-pdf://3211441679/20110014435-2.pdf.
Biswas, I., Drake, L., Erkina, D., and Biswas, S. (2008). Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. Streptococcus mutans J. Bacteriol. 190 (1), 68–77. doi:10.1128/jb.00990-07
Bitoun, J. P., Liao, S., Yao, X., Xie, G. G., and Wen, Z. T. (2012). The redox-sensing regulator rex modulates central carbon metabolism,stress tolerance response and biofilm formation by Streptococcus mutans. Peer-Reviewed. doi:10.1371/journal.pone.0044766
Brown, Lee R., Merrill, G., Wheatcroft, W. J. F., and Rider, L. J. (1974). Effects of a simulated skylab mission on the oral health of astronauts. J. Dent. Research53 5, 1268–1275. doi:10.1177/00220345740530053201
Carter, D. L., Tobias, B., and Orozco, N. Y. (2013). “Status of ISS water management and recovery,” in Paper presented at the 43rd International Conference on Environmental Systems, Vail, Colorado, July 14-18, 2013, 3509.
Deatherage, D. E., and Barrick, J. E. (2014). Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Eng. Anal. Multicell. Syst. Methods Protoc. 1151, 165–188. doi:10.1007/978-1-4939-0554-6_12
Do, E. A., and Gries, C. M. (2021). Beyond homeostasis: potassium and pathogenesis during bacterial infections. Infect. Immun. 89 (7), e0076620. doi:10.1128/iai.00766-20
Do, T. T., Rodríguez-Beltran, J., Cebrián-Sastre, E., Rodríguez-Rojas, A., Castañeda-García, A., and Blázquez, J. (2022). Inactivation of a new potassium channel increases rifampicin resistance and induces collateral sensitivity to hydrophilic antibiotics in mycobacterium smegmatis. Antibiotics 11 (4), 509. doi:10.3390/antibiotics11040509
Engin, A. B., Engin, E. D., and Engin, A. (2023). Effects of co-selection of antibiotic-resistance and metal-resistance genes on antibiotic-resistance potency of environmental bacteria and related ecological risk factors. Environ. Toxicol. Pharmacol. 98, 104081. doi:10.1016/j.etap.2023.104081
Evaldson, G., Heimdahl, A., Kager, L., and Nord, C. E. (1982). The normal human anaerobic microflora. Scand. J. Infect. Dis. 35, 9–15.
Fernander, M. C., Parsons, P. K., Khaled, B., Bradley, A., Graves, J., Joseph, L., et al. (2022). Adaptation to simulated microgravity in Streptococcus mutans. Streptococcus mutans NPJ Microgravity 8 (1), 17. doi:10.1038/s41526-022-00205-8
Fernandez, C. C., Sokolonski, A. R., Fonseca, M. S., Stanisic, D., Araújo, D. B., Azevedo, V., et al. (2021). Applications of silver nanoparticles in dentistry: advances and technological innovation. Int. J. Mol. Sci. 22 (5), 2485. doi:10.3390/ijms22052485
Flores-López, L. Z., Espinoza-Gómez, H., and Somanathan, R. (2019). Silver nanoparticles: electron transfer, reactive oxygen species, oxidative stress, beneficial and toxicological effects. mini review. J. Appl. Toxicol. 39 (1), 16–26. doi:10.1002/jat.3654
Girgis, H. S., Hottes, A. K., and Tavazoie, S. (2009). Genetic architecture of intrinsic antibiotic susceptibility. PloS One 4 (5), e5629. doi:10.1371/journal.pone.0005629
Graves, J. L., Tajkarimi, M., Cunningham, Q., Campbell, A., Nonga, H., Harrison, S. H., et al. (2015). Rapid evolution of silver nanoparticle resistance in escherichia coli. Front. Genet. 6, 42. doi:10.3389/fgene.2015.00042
Gross, E. L., Leys, E. J., Gasparovich, S. R., Firestone, N. D., Schwartzbaum, J. A., Janies, D. A., et al. (2010). Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48, 4121–4128. doi:10.1128/jcm.01232-10
Halfmann, A., Kovács, M., Hakenbeck, R., and Brückner, R. (2007). Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66 (1), 110–126. doi:10.1111/j.1365-2958.2007.05900.x
Herranz, R., Anken, R., Boonstra, J., Braun, M., Christianen, P. C., de Geest, M., et al. (2013). Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiology 13, 1–17. doi:10.1089/ast.2012.0876
Hooper, D. R., Littman, A. J., and Macpherson, L. V. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. doi:10.1126/science.1223490
Ibrahim, Y. M., Kerr, A. R., McCluskey, J., and Mitchell, T. J. (2004). Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. Streptococcus pneumoniae J. Bacteriol. 186 (16), 5258–5266. doi:10.1128/jb.186.16.5258-5266.2004
Illumina, (2021). BCL convert: A proprietary illumina software for the conversion of bcl files to basecalls. California, United States: Illumina.
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kolenbrander, P. E., Palmer, R. J., Rickard, A. H., Jakubovics, N. S., Chalmers, N. I., and Diaz, P. I. (2006). Bacterial interactions and successions during plaque development. Periodontol 42, 47–79. doi:10.1111/j.1600-0757.2006.00187.x
Kreth, J., Zhang, Y., and Herzberg, M. C. (2008). Streptococcal antagonism in oral biofilms: streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190 (13), 4632–4640. doi:10.1128/JB.00276-08
Lee, S., Hinz, A., Bauerle, E., Angermeyer, A., Juhaszova, K., Kaneko, Y., et al. (2009). Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. 106 (34), 14570–14575. doi:10.1073/pnas.0903619106
Lemos, J. A., and Burne, R. A. (2008). A model of efficiency: stress tolerance by Streptococcus mutans. Streptococcus mutans Microbiol. 154 (11), 3247–3255. doi:10.1099/mic.0.2008/023770-0
Ley, S., Li, W., Rodell, A., Meyer, M., Calle, L., Lersch, T., et al. (2021). “Fate of silver biocide on the international space station living environment,” in 50th International Conference on Environmental Systems, Lisbon, Portugal, July, 2021.
Li, W., C. (2018). “Investigation of silver biocide as a disinfection tehcnology for Spacecraft–An early literature review,” in 48th International Conference on Environmental Systems, United States, July 08-12, 2018.
Liau, S. Y. R., Read, D., Pugh, W., Furr, J., and Russell, A. (1997). Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterialaction of silver ions. Lett. Appl. Microbiol. 25, 279–283. Retrieved from internal-pdf://2306743118/j.1472-765X.1997.00219.x.pdf. doi:10.1046/j.1472-765x.1997.00219.x
Lloro, V., Giovannoni, L. M., Lozano-de Luaces, V., Lloro, I., and Manzanares, M. C. (2020). Is oral health affected in long period space missions only by microgravity? A systematic review. Acta Astronautica167 167, 343–350. doi:10.1016/j.actaastro.2019.11.015
Mascher, T., Heintz, M., Zähner, D., Merai, M., and Hakenbeck, R. (2006). The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in β-lactam resistance. J. Bacteriol. 188 (5), 1959–1968. doi:10.1128/jb.188.5.1959-1968.2006
Menon, A., B. (2012). Dental working group meeting. summary report. Hanover, MD, United States: Nasa Casi. NASA/TM-2012-217367.
Mermel, L. A. (2013). Infection prevention and control during prolonged human space travel. Clin. Infect. Dis. 56 (1), 123–130. doi:10.1093/cid/cis861
Muller, M., and Tajkhorshid, E. (2018). Molecular simulations provide insite on lysine snorkeling modulation of the integrin transmembrane domain. Biophysical J. 114 (3), 21a–22a. doi:10.1016/j.bpj.2017.11.161
Packham, N., Brasseaux, H., Rotter, H., Chhipwadia, K., Sauer, R., and Veselka, D. (1999). SAE Technical Paper No. 1999-01-2118. SAE international, Warrendale, PA. Removal of iodine for spacecraft applications.
Palmer, R. J., Diaz, P. I., and Kolenbrander, P. E. (2006). Rapid succession within the veillonella population of a developing human oral biofilm in situ. J. Bacteriol. 188, 4117–4124. doi:10.1128/jb.01958-05
Park, H., Kim, J. Y., Kim, J., Lee, J., Hahn, J., Gu, M. B., et al. (2009). Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity. Water Res. 43 (4), 1027–1032. doi:10.1016/j.watres.2008.12.002
Peng, X., Zhang, Y., Bai, G., Zhou, X., and Wu, H. (2016). Cyclic di-AMP mediates biofilm formation. Mol. Microbiol. 99 (5), 945–959. doi:10.1111/mmi.13277
Perkins, J., and Gadd, G. M. (1995). The influence of pH and external H+ concentration on caesium toxicity and accumulation inEscherichia coli andBacillus subtilis. J. Industrial Microbiol. Biotechnol. 14 (3-4), 218–225. doi:10.1007/bf01569931
Pierson, D. L. (2001). Microbial contamination of spacecraft. Gravitational Space Res. 14 (2), 1–6. Retrieved from internal-pdf://3006968069/261-780-1-PB.pdf.
Qi, F., Merritt, J., Lux, R., and Shi, W. (2004). Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect. Immun. 72 (8), 4895–4899. doi:10.1128/iai.72.8.4895-4899.2004
Rai, B., and Kaur, J. (2011). The history and importance of aeronautic dentistry. J. Oral Sci. 53 (2), 143–146. doi:10.2334/josnusd.53.143
Rai, M., Yadav, A., and Gade, A. (2009). Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 27 (1), 76–83. doi:10.1016/j.biotechadv.2008.09.002
Randall, C. P., Gupta, A., Jackson, N., Busse, D., and O'Neill, A. J. (2015). Silver resistance in gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 70 (4), 1037–1046. doi:10.1093/jac/dku523
Schweizer, I., Blättner, S., Maurer, P., Peters, K., Vollmer, D., Vollmer, W., et al. (2017). New aspects of the interplay between penicillin binding proteins, murM, and the two-component system CiaRH of penicillin-resistant Streptococcus pneumoniae serotype 19A isolates from Hungary. Antimicrob. Agents Chemother. 61 (7), e00414-e00417. doi:10.1128/aac.00414-17
Sharma, G., and Curtis, P. D. (2022). The impacts of microgravity on bacterial metabolism. Life 12 (6), 774. doi:10.3390/life12060774
Shroff, K. E., Meslin, K., and Cebra, J. J. (1995). Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect. Immun. 63, 3904–3913. doi:10.1128/iai.63.10.3904-3913.1995
Signoretto, C., Canepari, P., Pruzzo, C., and Gazzani, G. (2009). Anticaries and antiadhesive properties of food constituents and plant extracts and implications for oral health. InFood Const. oral health, 240–262. Woodhead Publishing. doi:10.1533/9781845696290.2.240
Slote, B. M. (2016). “Silver ion biocide delivery system for water disinfection,” in 46th International Conference on Environmental Systems, Vienna, Austria, July 2016.
Tajkarimi, M., Campbell, A., Rhinehardt, K., Thomas, M., Akamu Ewunkem, J., Boyd, S., et al. (2017). Selection for ionic-silver confers silver nanoparticle resistance in escherichia coli. JSM Nanotechnol. Nanomedicine 5 (1), 1047. Retrieved from internal-pdf://2474738234/nanotechnology-5-1047-3.pdf.
Takahashi, B., and Nyvad, N. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303. doi:10.1177/0022034510379602
Thomas, M. D., Ewunkem, A. J., Boyd, S., Williams, D. K., Moore, A., Rhinehardt, K. L., et al. (2021). Too much of a good thing: adaption to iron (II) intoxication in escherichia coli. Evol. Med. Public Health 9 (1), 53–67. doi:10.1093/emph/eoaa051
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50 (D1), D439–D444. doi:10.1093/nar/gkab1061
Watkins, S., B. (2011). The space medicine exploration medical condition list. Washington, United States: NASA.
Wen, Z. T., Bitoun, J. P., and Liao, S. (2015). PBP1a-Deficiency causes major defects in cell division, growth and biofilm formation by Streptococcus mutans. Streptococcus mutans PLoS One 10 (4), e0124319. doi:10.1371/journal.pone.0124319
Wu, C., Ayala, E. A., Downey, J. S., Merritt, J., Goodman, S. D., and Qi, F. (2010). Regulation of ciaXRH operon expression and identification of the CiaR regulon in Streptococcus mutans. Streptococcus mutans J. Bacteriol. 192 (18), 4669–4679. doi:10.1128/jb.00556-10
Zhang, H., Pan, Y., Hu, L., Hudson, M. A., Hofstetter, K. S., Xu, Z., et al. (2020). TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential. Nat. Commun. 11 (1), 547. doi:10.1038/s41467-019-14240-9
Zheng, M., Åslund, F., and Storz, G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279 (5357), 1718–1722. doi:10.1126/science.279.5357.1718
Keywords: adaptation, simulated microgravity, Streptococcus mutans, experimental evolution, silver nitrate (AgNO3)
Citation: Fernander MC, Spurgeon K, Graves J, Guess W, Miller J, Mangum C, Graves JL and Thomas MD (2023) Co-adaptation of Streptococcus mutans to simulated microgravity and silver nitrate. Front. Astron. Space Sci. 10:1183867. doi: 10.3389/fspas.2023.1183867
Received: 10 March 2023; Accepted: 30 May 2023;
Published: 15 June 2023.
Edited by:
Dayong Wang, Southeast University, ChinaReviewed by:
Ricardo Amils, Autonomous University of Madrid, SpainCopyright © 2023 Fernander, Spurgeon, Graves, Guess, Miller, Mangum, Graves and Thomas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Misty D. Thomas, bXRob21hczFAbmNhdC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.