- 1Jet Propulsion Laboratory, California Institute of Technology, Pasadena, CA, United States
- 2NASA Astrobiology Institute—Icy Worlds, Pasadena, CA, United States
The search for life in Solar System bodies such as Mars and Ocean Worlds (e.g., Europa and Enceladus) is an ongoing and high-priority endeavor in space science, even ∼ five decades after the first life detection mission at Mars performed by the twin Viking landers. However, the in situ detection of biosignatures remains highly challenging, both scientifically and technically. New instruments are being developed for detecting extinct or extant life on Mars and Ocean Worlds due to new technology and fabrication techniques. These instruments are becoming increasingly capable of both detecting and identifying in situ organic biosignatures that are indicative of life and will play a pivotal role in the search for evidence of life through robotic lander missions. This review article gives an overview of techniques used for space missions (gas chromatography, mass spectrometry, and spectroscopy), the further ongoing developments of these techniques, and ion mobility spectrometry. In addition, current developments of techniques used in the next-generation instruments for organic biosignature detection are reviewed; these include capillary electrophoresis, liquid chromatography, biosensors (primarily immunoassays), and nanopore sensing; whereas microscopy, biological assays, and isotope analysis are beyond the scope of this paper and are not covered.
1 Background
Is there life elsewhere in our Solar System? This inquiry, grandiose in scope but modest in words, gives rise to an array of much-debated follow-up questions: What is life, and if it is there, how do we detect it? There is no firm consensus regarding the definition of life (Cleland and Chyba, 2002; Tsokolov, 2009); however, NASA’s current working definition is a “self-sustaining chemical system capable of Darwinian evolution” (Benner, 2010). In the absence of a precise classification, searching for signs of life with Earth as a reference is a pragmatic approach, such as visual (e.g., cells and fossils) or chemical (e.g., biological macromolecules, molecular evidence of metabolism, and molecular fossils) biosignatures (Domagal-Goldman et al., 2016). Organic biosignatures can be represented by complex organic molecules at concentrations requiring biotic synthesis or relative abundance of organic molecules not found in abiotic chemical systems (Barge et al., 2021). Consequently, key objectives are to identify and quantify the abundance and distribution of organic molecules of planetary bodies such as Mars, Europa, and Enceladus, where signatures of past or present life could exist. In situ robotic missions accompanied by chemical analysis instruments will play a pivotal role in this endeavor (Seaton et al., 2021)—because selecting samples with the highest probability of containing biosignatures with limited prior knowledge, maintaining pristineness, and returning them to Earth is currently not feasible. However, in situ detection of organic biosignatures in the space environment remains a scientific and technical challenge. One of the major technical difficulties is that space-bound scientific instruments must be of small size, low mass, and low power.
Candidate biosignatures are assessed by their reliability (differentiation between abiotic and biotic), survivability (no degradation in harsh environments), and detectability (having features that could be observed or detected; National Academies of Sciences and Medicine, 2019). Biosignatures can range from patterns observed by visual inspection to chemical patterns. The Ladder of Life Detection is currently perhaps the most notable attempt at summarizing potential biosignatures (Neveu et al., 2018). On the basis of life as we know it, the paper provides a framework of biosignatures, how specific they are to life, and their likelihood of indicating life. However, it is important to underline that biosignatures are contextual—for example, it is essential to consider the environment in which they are evaluated.
The technical difficulties of performing reliable analysis for the identification of chemical biosignatures beyond Earth are numerous and not easily addressed. Instruments and analysis methods need to be fit for the purpose—namely, they must be within the requirements regarding detection limits, trueness, precision, reproducibility, robustness, and ruggedness (for standardized terminology, see Thompson et al., 2002; Joint Committee for Guides in Metrology, 2008; Magnusson and Örnemark, 2014). Robustness is defined here as the capacity to remain unaffected by instrument or method parameter (internal factors) fluctuation, and ruggedness refers to external factors, such as different instruments, operational environments, reagents from different batches, or sample composition variations; however, the two terms are sometimes used interchangeably in the literature.
Instruments must be able to survive and operate in extreme extra-terrestrial environments (pressure, temperature, radiation, and mechanical stress). Furthermore, they must satisfy stringent mass, volume, and power requirements. The scientific payloads typically include various instruments such as cameras, spectrometers, radiation sensors, and meteorological sensors. As a reference, a commercial benchtop gas chromatograph (GC; e.g., Agilent 7890A GC System) weighs more than 50 kg, a typical ultra-high-performance liquid chromatograph weighs 140 kg (e.g., Waters ACQUITY UPLC system), and a typical mass spectrometer (MS) weighs about 140 kg (e.g., Thermo Fisher LTQ XL). These chemical analysis systems take up entire workbenches and cannot be used for space applications without extensive reengineering.
It is challenging to develop in situ chemical analysis instruments with detection limits in the low-ppb (parts-per-billion, µg/kg) range, capable of quantitative analysis, and that provide sufficient chemical information for identification. Specifications are mission-dependent. However, lower limits of detection (LOD) of 1 ppb (Mahaffy et al., 2012; Pappalardo et al., 2013) or even lower (Hand et al., 2017; MacKenzie et al., 2022b) are often required.
A broad goal of chemical analysis instruments is to identify and quantify relevant analytes that can provide insight into prebiotic chemistry and analytes that are biosignatures for extant or extinct life. The strategies of agnostic life detection are discussed elsewhere (e.g., Botta et al., 2008; McKay, 2008; Summons et al., 2008; Domagal-Goldman et al., 2016; Neveu et al., 2018; Barge et al., 2021). High-priority analyte classes often include, but are not limited to, lipids (e.g., fatty acids and hopanes), amino acids, informational polymers (e.g., DNA and RNA), and biopolymers (e.g., proteins and saccharides; Neveu et al., 2018).
Previous review papers have primarily covered chemical analysis instruments previously used in spaceflight applications. Seaton et al. (2021), Seaton et al. (2022) have summarized analytical techniques and instruments used in previous and planned spaceflight missions; including those used for organic biosignature detection, inorganic analysis, and remote sensing; but also covered instruments used in space probes in addition to lander missions. Poinot and Geffroy-Rodier (2015) have provided a detailed overview of previous spaceflight-GC instruments used for organic detection. The authors also briefly discussed ongoing developments of chip-based instruments capable of detecting organic biosignatures.
We present a detailed examination of in situ organic biosignature detection instruments that have been developed, are under development, or are suitable for adaptation for use in upcoming in situ planetary applications (Figure 1). For example, a detailed and up-to-date survey is provided of recent fluorescence spectroscopy, Raman spectroscopy, and GC-MS spaceflight instruments previously used in lander missions, along with ongoing developments of the techniques. However, we also assess the next-generation novel approaches for the future (i.e., beyond planned missions) detection and analysis of biosignatures; as low-mass, low-power alternatives to commonly used conventional analytical techniques in the context of future astrobiology-focused space missions to those bodies. The next-generation techniques have not been used at planetary bodies; some are still in the method development and validation phase and have only taken some steps towards miniaturization and spaceflight compatibility.
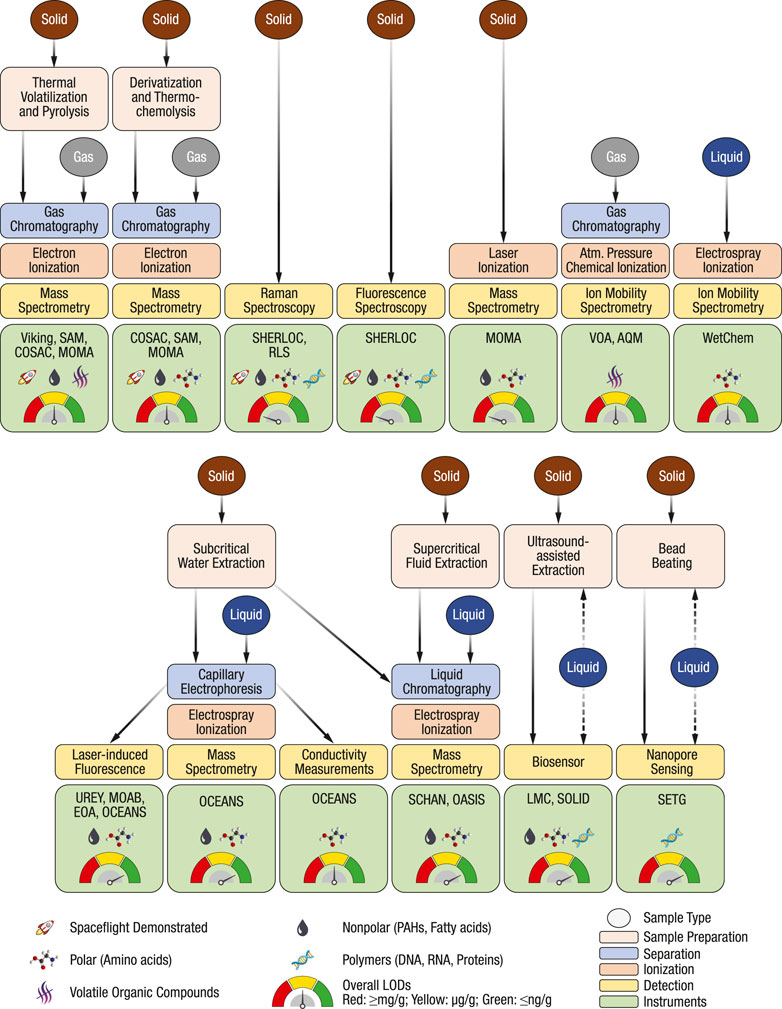
FIGURE 1. Overview of chemical analysis techniques discussed in this review. Examples of combined techniques, relevant instruments, frequently targeted analyte classes, and overall LODs are denoted with symbols. The evaluation is based on spaceflight instruments and adaptions of techniques for in situ space applications. The overview comes with the caveat that the target analyte classes can be extended through method development, all analytes in one class are often not detectable with a specific technique, some specific analytes may have substantially higher or lower response factors, and LODs are highly sample-dependent.
Unlike previous review papers, we focus on chemical analysis techniques for organic biosignatures rather than individual instruments that may use a combination of different techniques. More techniques are covered than previously, including the often-overlooked sample preparation step (e.g., extraction techniques). Additionally, this review assesses next-generation techniques: capillary electrophoresis, liquid chromatography, biosensors (primarily immunoassays), and nanopore sensing. We have mainly focused on chemical analysis techniques for detecting biotic organic compounds relevant to life detection in space exploration. Other avenues, although exciting but outside the scope of this review, include microscopy (e.g., Wallace et al., 2019), biological experiments (Touchette et al., 2022), and isotope analysis (Rodin et al., 2020), which provide orthogonal approaches to life detection. This manuscript can also be seen as a stepping stone, outlining influential reviews covering each technique.
The techniques and instruments have been grouped into sections based on their generalized technology readiness level (TRL; for more details, see Smoker and Smith, 2007; Mankins, 2009). Those included in spaceflight missions are presented in Section 2. The continued developments of previously flown techniques (including ion mobility spectrometry due to long operational presence onboard the International Space Station, ISS) are presented in Section 3. Techniques without spaceflight heritage at planetary bodies are assessed in Section 4 and Section 5. Sample preparation is discussed separately to highlight its importance, which is often overlooked, because these techniques may be combined with each other and in tandem with multiple downstream analysis techniques.
All instrument developments (based on either existing or new technology) start in the laboratory environment, and some have a higher potential to evolve into spaceflight instruments than the rest. This review also reflects those intricacies by including examples of instrumentation on a clear path towards spaceflight readiness and studies demonstrating progress towards fitness for purpose, miniaturization, or spaceflight compatibility.
The path for a typical technology development from an early concept to a demonstrated spaceflight qualification is long (typically >10 years) and expensive. Costs and time can be saved by utilizing instruments and parts with spaceflight heritage. However, each newly developed technique and instrument typically start with studies in laboratory environments. Once a technique has been established as fit for purpose, it is followed by miniaturization, adaptation to the relevant environment (e.g., vacuum, low temperature, and high radiation), and then the final instrument is designed and built. The fundamental knowledge of the techniques is often fragmented; laboratory studies are essential for continuously evaluating what science can be conducted with in situ measurements. Consequently, many studies and reported instruments that are in development are, in fact, not flight-like—as is the case in this review. Nevertheless, they still help us understand each chemical analysis technique’s performance, limitations, and opportunities.
Instruments determined as fit for purpose, miniaturized, and demonstrated in relevant environments may undergo substantial redesign after being selected for a planned mission. These redesigns are driven by, for instance, spacecraft restrictions or reduced budgets. For example, the GC-based instrument onboard the Curiosity rover was first under development with liquid extraction of solid samples prior to derivatization (Buch et al., 2003); however, a one-pot derivatization approach was finally used (Mahaffy et al., 2012; Stalport et al., 2012), due to budget constraints. Hence, it is not always possible to accurately predict the performance of the final spaceflight instrument while it is still in development (i.e., Sections 3–5 in this review). Finally, many in-development instruments have been given an acronym—this is not an indicator that it is close to spaceflight-ready, unless stated otherwise.
2 Overview of instruments used for planetary missions
Space instrumentation has advanced substantially due to research over the last few decades in engineering and manufacturing techniques. As a result, a vast and diverse number of instruments utilize a wide variety of techniques and methods that are relevant for the in situ detection of biosignatures in planetary missions. Comparable performance characteristics in space instrumentation with their laboratory counterparts have been progressively achieved since the 70s’ in terms of high measurement sensitivity, selectivity, high precision, and resolving power while providing small instrument size, low mass, and low power required by space missions. Among these instruments, traditionally, MS-based instruments have been the most recurrent for in situ planetary exploration mission applications. GC, often accompanied by MS, has also been commonly used for organic detection. In this section, we will review applied techniques in planetary exploration missions (including those onboard the Rosalind Franklin rover, although they have not been flown at the time this paper was written): gas chromatography, mass spectroscopy, and optical spectroscopic techniques.
2.1 Gas chromatography
One of the most commonly used techniques for organic detection is GC (for a summary of instruments and validation data, see Poinot and Geffroy-Rodier, 2015). Table 1 summarizes GC-onboard space missions (Akapo et al., 1999; Johnson et al., 2012; Szopa et al., 2017). The first successful planetary lander mission equipped with GC involved the twin Viking landers that landed on Mars in 1976 (Rushneck et al., 1978). At the time, GC was the only chemical separation technique considered mature; alternatives such as capillary electrophoresis (Lunte and Radzik, 1996), and high-performance liquid chromatography (Snyder, 1997), did not gain mainstream popularity until the mid-80s and 90s’. Still today, GC provides unmatched peak capacity, which is the number representing how many analytes theoretically can be separated in one experiment.
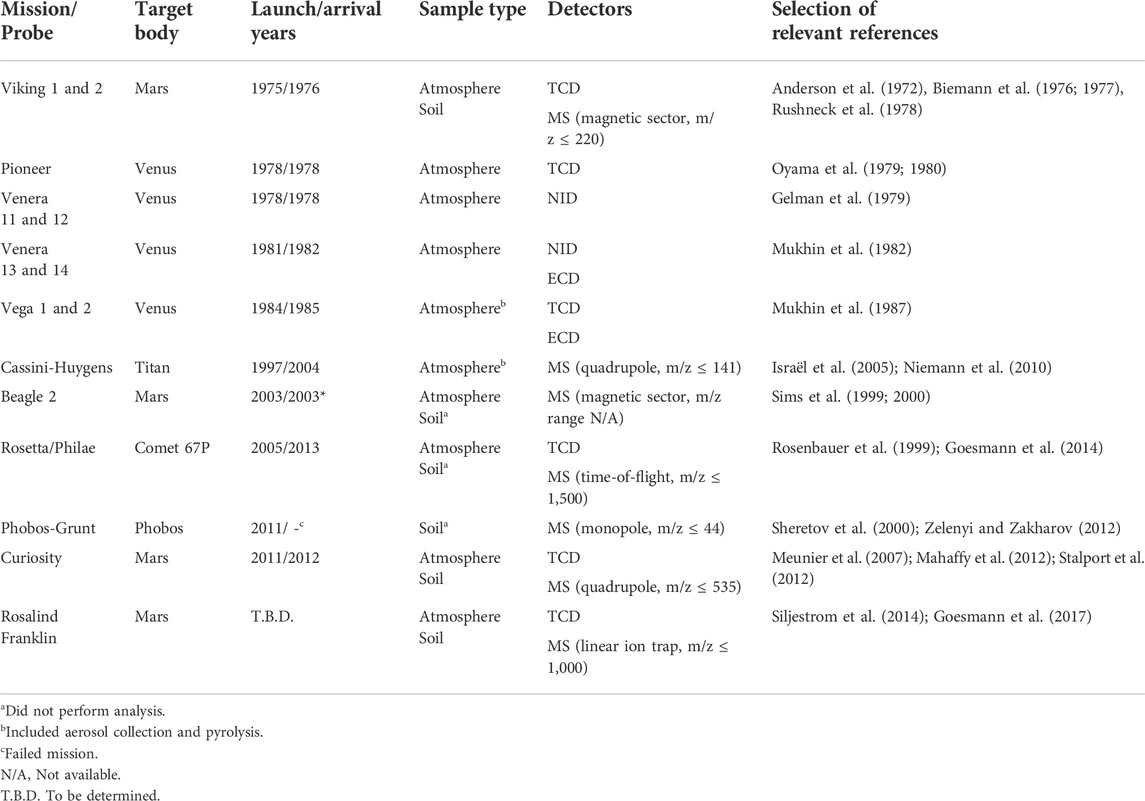
TABLE 1. GC onboard space missions (Akapo et al., 1999; Johnson et al., 2012; Szopa et al., 2017). The references are not a complete compilation and should be considered a starting point. The following abbreviations are used: thermal conductivity detector (TCD), neon ionization detector (NID), and electron-capture detector (ECD).
The basic principle of chromatography (here we discuss GC, and liquid chromatography is discussed in Section 5.2) is based on the distribution of analytes between a mobile phase (called carrier gas in GC) and a stationary phase (an immobile material inside of a column). The sample is introduced into a stream of carrier gas that passes through the GC column; analytes that interact more with the stationary phase (e.g., a thin film coated on the walls of a GC column) are retained more strongly and exit (elutes) the column later than less-retained analytes. The exiting carrier gas (effluent), along with eluted analytes, are continuously transported to a detector. The retention can be shortened by increasing the temperature because it increases the analyte’s vapor pressure, hence, increasing the relative time spent in the carrier gas. Analytes in a mixture, like in a natural sample, are separated based on their interaction with the stationary phase—providing different retention times that can be used for identifying unknown analytes, often together with mass spectra.
Two main requirements are imposed on analytes that are analyzed by GC. Firstly, they have to be sufficiently volatile to at least partially partition into the gas phase at the separation temperature (typically <400°C), and secondly, they need to be sufficiently thermally stable not to decompose during their migration through the capillary chromatography column. The upper temperature is limited by the stability of the stationary phase, which has improved over the decades. For example, the maximum temperature of the Viking GC was 200°C (Biemann et al., 1976). Utilizing a GC column close to its upper-temperature limit will cause so-called column bleed, where the stationary phase hydrolyses and contributes to the background signal and consequently diminishes the lower detection limits of less volatile analytes. Finally, essentially all GC methods use temperature programming by increasing the temperature over time (increases analyte vapor pressure) to allow separation of both volatile (starting at low temperatures for volatile analytes to partition into the stationary phase) and less volatile analytes (high enough temperature for them to partition into the carrier gas). Temperature-programmed GC methods require cooling down to the initial conditions before analyzing the subsequent sample, which is not trivial considering the ample thermal insulation of payload instruments.
Chemical analysis of the atmospheric composition has been rather straightforward because it only requires an atmosphere inlet port and a dust filter. Analysis of the atmosphere is also readily done with GC, as has been demonstrated by several missions: multiple Venus probes (Venera 11–14 and the multiprobe Pioneer Venus 2), Huygens (Titan), and Philae (Comet 67P). Huygens detected evaporated methane, ethane, acetylene, cyanogen, and carbon dioxide at the surface of Titan (Niemann et al., 2010). Philae detected several alcohols, carbonyls, amines, nitriles, amides, and isocyanates in the dust and vapor stirred up by the lander (Goesmann et al., 2015).
The history of analysis of solid samples with GC has been more intriguing. Viking is the only landed mission so far that has carried dedicated life-detection instruments. The twin landers included three experiments that added substrate to the Martian regolith, which studied gas exchange (GEX), labeled release (LR), and pyrolytic release (PR; Klein et al., 1976; klein, 1978). The GEX experiment was the only analysis that involved GC.
In the GEX experiment, the gas release was determined by GC after exposure first to humidity only, followed by water, and then to immersion in a nutrient solution (Oyama, 1972; oyama et al., 1976; oyama and berdahl, 1977). The GEX experiment was primarily designed to detect heterotrophic activity in an atmosphere of mainly CO2. The LR experiment added 14C-labeled nutrients to the soil, then determined evolved radioactive gases released by potential organisms by a radiation counter (Levin, 1972; Levin and Straat, 1977). The PR experiment (also known as the carbon assimilation experiment) incubated soil with 14CO and 14CO2, followed by pyrolysis (635°C), trap and release (to separate CO and CO2 from organic compounds), and determination of released radioactive analytes from the soil by a radiation counter (Horowitz et al., 1972; Horowitz et al., 1977).
The results and methods of these biological experiments have been put into a broader life-detection context by Neveu et al. (2018), and references therein. In summary, no conclusive results were obtained from the experiments. Surface chemistry most likely explained the positive results; however, biological activity could not be entirely rejected (Levin and Straat, 2016; Neveu et al., 2018).
Although the GC analysis of untreated soil was not classified as a life detection experiment, detected organic compounds could have served as a confirmation of life. In these experiments, the soil was stepwise heated up to 50, 200, 350, or 500°C to either volatilize analytes or produce pyrolysates of non-volatile compounds that subsequently could be separated by GC (Biemann et al., 1977).
No native organic compounds were detected in the soil with an overall detection limit on the order of ppb at either of the two landing sites (Biemann et al., 1977; Biemann and lavoie, 1979). The absence of detected native organic molecules by the Viking landers has been a subject of much debate (Benner et al., 2000; Glavin et al., 2001; Navarro-González et al., 2006; Biemann, 2007). Due to the rigorous validation and verification experiments performed with the Viking engineering breadboard and the detection of cleaning solvents during its flight to the red planet, it is without a doubt that the instrument performed as intended (Biemann et al., 1976; biemann et al., 1977; Biemann and lavoie, 1979). However, one of the major findings of the Phoenix lander (launched in 2007) was that the surface of Mars is rich in perchlorates (ClO4; Hecht et al., 2009). The impact of the discovered perchlorates on the past interpretations have indeed also been a subject of great debate (Navarro-González et al., 2010; Biemann and Bada, 2011; Navarro-González and McKay, 2011). Later findings derived from data generated by the Sample Analysis at Mars (SAM, see Figure 2) instrument on the Mars Science Laboratory (MSL) of the Curiosity rover, laboratory experiments, and further re-analysis of the Viking data have provided sound evidence that Viking indirectly did detect organic molecules. The native organic molecules likely reacted during the thermal volatilization (TV)/pyrolysis (Py) process to form chlorinated organic molecules, some of which were previously and incorrectly identified as cleaning contaminants (Miller et al., 2016; Guzman et al., 2018; Szopa et al., 2020). Over the last few years, the SAM GC-MS has detected chlorobenzene (Freissinet et al., 2015), isomers of dichlorobenzene (Szopa et al., 2020), and derivatized (derivatization methods are described later) benzoic acid, ammonia, and potentially phosphoric acid and phenol (Millan et al., 2021). All chlorinated compounds detected at Mars so far have been summarized elsewhere (Table 3 in He et al., 2021a).
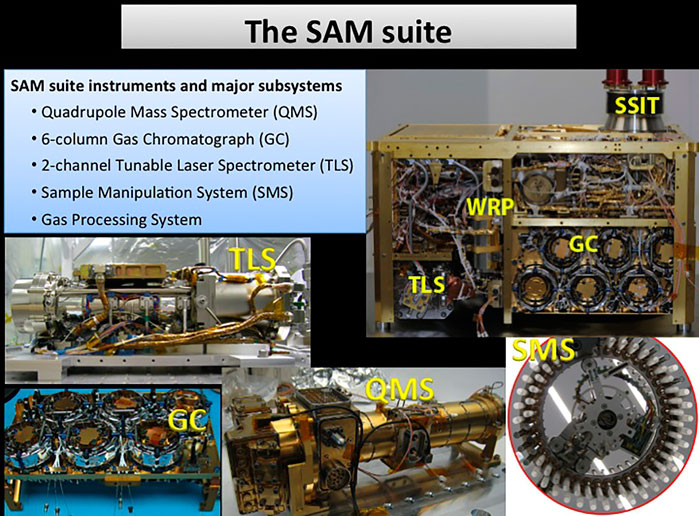
FIGURE 2. Overview of the Sample Analysis at Mars suite onboard the Curiosity rover. The suite includes a tunable laser spectrometer (TLS), a gas chromatograph (GC) with six different columns, a quadrupole mass spectrometer (QMS), a solid sample inlet tube (SSIT), sample manipulation system (SMS), and wide-range pumps (WRP; Mahaffy et al., 2012). Images courtesy of NASA/JPL-Caltech.
The Rosetta mission to Comet 67P, carrying the lander Philae equipped with the Cometary Sampling and Composition (COSAC) system, was the second mission that attempted to analyze solid samples with GC on a planetary body. Unfortunately, the Philae lander never acquired any solid sample due to issues during the landing sequence because the lander finally settled on its side. Although no GC analysis was performed, several important developments were made that were later adopted by both SAM onboard MSL and the Mars Organic Molecule Analyzer (MOMA) onboard the European Space Agency’s (ESA) Rosalind Franklin rover (previously known as the ExoMars rover).
GC is only viable for volatile and heat-stable analytes, which do not include, for example, amino acids, nucleobases, and fatty acids. These important organic biosignatures can be analyzed with GC by first performing derivatization, which converts them into volatile and more stable products; or using alternative analysis techniques described later.
All three instruments (COSAC, SAM, and MOMA) feature a so-called one-pot wet chemistry processing in addition to conventional pyrolysis, where the solid sample and the solution containing the derivatization agents are mixed without any further addition of solutions. Three derivatization reagents have been employed, namely N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA), N,N-dimethylformamide dimethyl acetal (DMF-DMA), and tetramethylammonium hydroxide (TMAH). COSAC carried DMF-DMA (Goesmann et al., 2014), SAM had cups containing either MTBSTFA or TMAH (Mahaffy et al., 2012), and MOMA was designed to have the option of using any of the three derivatization reagents (Goesmann et al., 2017).
MTBSTFA is a very reactive silyl reagent, usually facilitated by DMF that acts as a proton acceptor, which enables low LODs; for example, pure amino acids can be detected between 0.3 µM and 50 µM after derivatization, followed by direct injected into a GC (Rodier et al., 2001). The derivatization reagent is suitable for carboxylic acids, amino acids, nucleobases, amines, and alcohols. However, MTBSTFA is prone to react with water, which can reduce the product yield and create a strong interfering background. The derivatization methodology is not rugged towards samples containing hydrated minerals, inorganic anions, or iron oxides; these inhibitors may render analytes completely undetectable (Stalport et al., 2012). Even so, He et al. (2021a) have extensively studied the products of MTBSTFA/DMF formed during flash pyrolysis and concluded that the derivatization reagents remained stable in the presence of calcium perchlorate during slow SAM-like temperature ramping up to 850°C. Furthermore, MTBSTFA does not allow for chiral analysis of amino acids. Derivatized benzoic acid and ammonia were detected at Mars with this methodology (Millan et al., 2021).
DMF-DMA is a methylation reagent less reactive than MTBSTFA and targets fatty acids, primary amines, and amino acids. Higher LODs are generally obtained with DMF-DMA than MTBSTFA in matrix-free samples. Most importantly, enantiomeric amino acids derivatized with DMF-DMA can be separated on chiral stationary phases, unlike MTBSTFA. It has been demonstrated that 9 out of 19 proteogenic and enantiomeric amino acids can be separated using this methodology (Freissinet et al., 2010).
TMAH is not only a methylation agent, but also facilitates hydrolysis of macromolecules at elevated temperatures, also known as thermochemolysis. The reagent is usually dissolved in methanol. TMAH is more rugged regarding sample matrix composition, and its applications have been demonstrated on a wide variety of sample types (Geffroy-Rodier et al., 2009; Williams et al., 2019). Nucleobases can also be detected with a TMAH-based methodology (He et al., 2019). The applications and reaction mechanisms of TMAH thermochemolysis have been extensively reviewed by He et al. (2020). Perhaps most importantly, TMAH appears to be better suited for samples containing perchlorates. He et al. (2021b) demonstrated that up to 5 wt% of calcium perchlorate did not significantly impact the recovery of free fatty acids at a concentration of 300 ppb. In situ analysis with TMAH at Mars has been performed; however, data analysis is, at this moment, ongoing (Roach et al., 2021).
Limited validation of the one-pot methodologies for COSAC, SAM, or MOMA has been published. Nonetheless, a multitude of demonstration and qualitative studies have been performed with flight-like sample chambers coupled to commercial GC-MS systems where pyrolysis, derivatization, or thermochemolysis were applied (e.g., Lewis et al., 2018; Reinhardt et al., 2020; He et al., 2021b). Whereas the one-pot derivatization methodology is versatile, it also adds risk. SAM demonstrated this, when one of the cups leaked, and the derivatization reagent formed several products, which adversely affected data interpretation and detection limits (Glavin et al., 2013; Leshin et al., 2013).
Poinot and Geffroy-Rodier (2015) summarized publicly available validation data and pointed out that quantitative comparison of these instruments is impossible due to the lack of validation data. It has been claimed that MOMA is capable of detecting one or more amino acids with LODs of 80 ppb and 1.3 ppm with MTBSTFA or DMF-DMA, respectively (Goesmann et al., 2017). Two validation studies have been conducted using a laboratory pilot resembling SAM and MOMA instrumentation (Meunier et al., 2007; David et al., 2016). The earlier study showed an instrument mean LOD of 10 pmol of 28 organic acids derivatized with MTBSTFA, which were injected into the GC-MS system. However, the reactor’s transfer yield in on-line experiments was generally low (<5.5%), and the method LODs were not determined. The second study demonstrated LODs of ∼1 nmol for amino acids with off-line derivatization with MTBSTFA and where the derivatized amino acids were subsequently spiked onto 50 mg of solid sample (David et al., 2016). This corresponds to approximately 1.5–3.6 ppm LODs; however, the experiments did not consider matrix effects in the derivatization step. Comparisons between laboratory instruments and engineering test units (nearly equivalent spaceflight instruments) are rare. In the case of MOMA, the separation efficiency is substantially lower (broader peaks) due to a relatively slow heating (10–15 s) of the injection trap, compared to the conventional pre-heated injection port used on commercial instruments (Guzman et al., 2020). The same authors also extensively characterized the GC columns used in MOMA and demonstrated the vast number of analytes (e.g., noble gases, alkanes, alcohols, carboxylic acid, and amino acids) that could be separated with either of the four columns.
Despite the mentioned shortcomings of the current implementation of thermal volatilization and pyrolysis GC-MS with or without derivatization agents, the technique provides the ability to detect a wide range of relevant organic biosignatures from a variety of samples (Bishop et al., 2013; Reinhardt et al., 2020). It provides these capabilities with relatively low instrument complexity. On the basis of the successful deployments of GC-MS on Mars, it is safe to say that GC columns do not significantly degrade in the high radiation environment. Common capillary columns and consumables have been tested before and after radiation, corresponding to the levels expected for missions to the Jovian moons, without any substantial reduction in performance (Freissinet et al., 2016; Freissinet et al., 2019). Nonetheless, future missions will likely require lower LODs of biosignatures and better ruggedness regarding sample composition than what current GC-MS implementation can provide.
2.2 Mass spectroscopy
With MS-based techniques, it is possible to obtain information such as composition, atomic or molecular weight, and molecular structure of the constituents in a sample being analyzed, which is difficult to obtain by other means. An excellent general introduction to MS is given by de Hoffmann and Stroobant (2013). Mass spectrometers have a wide variety of configurations; however, they all have four essential components. These four components are the sample inlet, the ionization source, the mass analyzer, and the detector. Some instruments combine the sample inlet and the ionization source, whereas others combine the mass analyzer and the detector. The development for space applications over the last few decades has focused on minimizing power and mass while improving the mass resolution (m/Δm, Δm being at full width at half maximum, FWHM) of these instruments. These developments with emphasis on space applications have been extensively reviewed and covered by others (Palmer and Limero, 2001; Johnson et al., 2012; Snyder et al., 2016; Ren et al., 2018; Arevalo et al., 2020; Chou et al., 2021). The mass analyzer and the detector (often referred to as the back-end) function relatively independently of the front-end, which may involve a GC or a laser desorption and ionization (LDI) instrument. However, the optimum choice of ionization source depends greatly on the type of incoming sample and the environment in which it operates. The basics of various ionization techniques have been covered by Siuzdak (2005).
To date, mass spectrometry on landed robotic missions to other Solar System bodies has exclusively been used to analyze incoming gas streams such as direct atmospheric measurements, evolved gases from pyrolysis, and effluent from GC (Johnson et al., 2012). Electron ionization (EI) has therefore been the natural choice of ionization; because it provides high sensitivity, robustness, and capability of ionizing most analytes. EI is classified as a hard ionization technique that typically fragments the analyte. Hence, EI mass spectra tend to be rather complicated, but they also provide a unique opportunity for identification, as opposed to soft ionization techniques. EI provides reproducible spectra which can be matched with databases for identification, which is not always the case for other ionization techniques. Figure 3 illustrates all MS instruments utilized for spaceflight missions. Table 1 provides the mass ranges of MS instruments on the Vikings, COSAC, SAM, and MOMA.
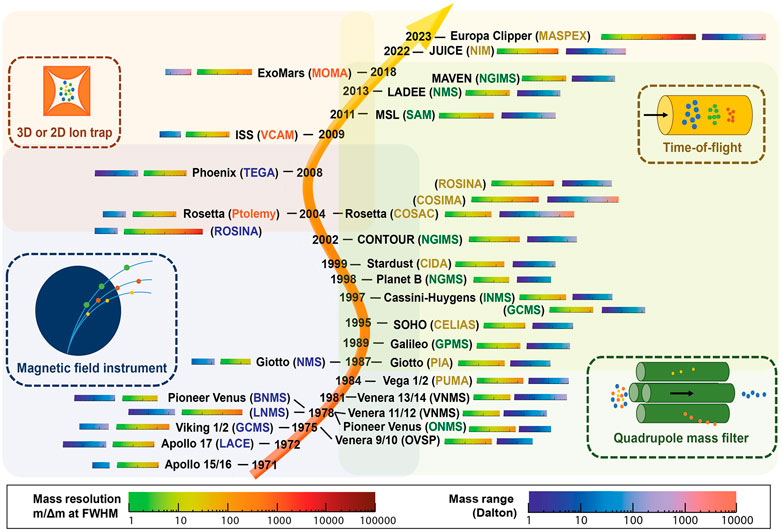
FIGURE 3. An illustrated summary of MS instruments utilized for space flight missions and their performance metrics. Reprinted by permission from Arevalo et al. (2020). Copyright 2020 Wiley.
The earliest MS instruments used for spaceflight were based on magnetic sector mass spectrometry. This was the preferred technique at the time as it was less dependent on electronic circuits (simple design) and did not require high power levels (Anderson et al., 1972); for example, for a radiofrequency generator used in the quadrupole mass analyzer. However, the magnetic sector MS is not ideal for scanning at a high m/z (mass to charge ratio) at a rate compatible with modern-day separation techniques, and its performance is greatly affected by miniaturization (Ren et al., 2018). Over the last few decades, single quadrupoles, time-of-flight (ToF), and ion traps (IT) have been the main techniques utilized. ToF and IT are the focus of ongoing developments, which are discussed below. The main reason for the change in focus is that the quadrupole mass analyzer suffers from reduced ion transmission and resolution when it is further miniaturized. In miniaturized versions, the assembly is critical, with positioning tolerances in the micrometer scale of the four rods that act as the ion filter (Ren et al., 2018).
The sensitivity and signal-to-noise ratio of an MS instrument cannot be determined accurately without considering the ionization and ion transmission efficiencies. Therefore, it is difficult to compare these instruments’ detection limits. Validation data are, unfortunately, very scarce. Nevertheless, tremendous advancements have been made on several fronts, and development is still ongoing (see Section 3.2). The GC-MS system onboard the Viking landers weighed 25 kg and only had a scan rate of one spectrum per 10.24 s. In comparison, the MS onboard MOMA weighs 7.5 kg (11.5 kg including GC and LDI) and has a scan rate of several Hz (Goesmann et al., 2017).
2.3 Optical spectroscopic techniques
Non-invasive remote or standoff spectroscopic techniques have gained importance over the last few decades. Their role has mostly been focused on elemental composition and mineralogy analysis. Although these instruments’ primary purpose was not detecting organic biosignatures, the development of the optical spectroscopic techniques has paved the way for more capable detection instruments. Standoff sensing provides unique possibilities for identifying samples or areas of interest for further chemical analysis with other techniques (e.g., GC-MS) where the total number of analyses is limited. A brief overview of existing space flight instruments is given here, and ongoing developments are discussed in detail later (see Section 3.1).
Virtually all Mars landers have employed optical spectroscopic techniques for analyzing regolith, with X-ray spectroscopy for elemental analysis being the most common (Inza and Lopez-Reyes, 2020). In fact, the most current and future planned landers have an increased number of spectroscopic instruments in their payloads, compared to past missions. The reader is referred to payload summaries of each mission: Curiosity rover (MSL; Grotzinger et al., 2012), Perseverance rover (Mars 2020; Farley et al., 2020), Rosalind Franklin rover (Vago et al., 2017), and the Tianwen-1 lander (Zou et al., 2021).
The combination of spectroscopic techniques employed by instruments onboard the Perseverance rover can be used to characterize and identify samples of interest, which has been demonstrated at a Mars analog field site in the Mojave Desert (see Figure 4; Martin et al., 2020), and in the laboratory with microbial mats from the ∼3.42 Ga Buck Reef Chert in South Africa (Hickman-Lewis et al., 2022). Two remote sensing instruments, Mastcam-Z (Bell et al., 2021) and SuperCam (Wiens et al., 2021), and two microscopic proximity instruments, Scanning for Habitable Environments with Raman and Luminescence for Organics and Chemicals (SHERLOC; Beegle et al., 2015) and Planetary Instrument for X-ray Lithochemistry (PIXL; Allwood et al., 2020), are a part of the Mars 2020 rover mission (Perseverance) instrument suite (Farley et al., 2020). The instrument suite will be used to identify samples of interest for caching for potential future return to Earth. Similarly, the Raman Laser Spectrometer (RLS, 532 nm) onboard the Rosalind Franklin rover will characterize the mineralogy of collected powdered samples, with the possibility of detecting organic compounds (Lopez-Reyes et al., 2013; Rull et al., 2017; Veneranda et al., 2020).
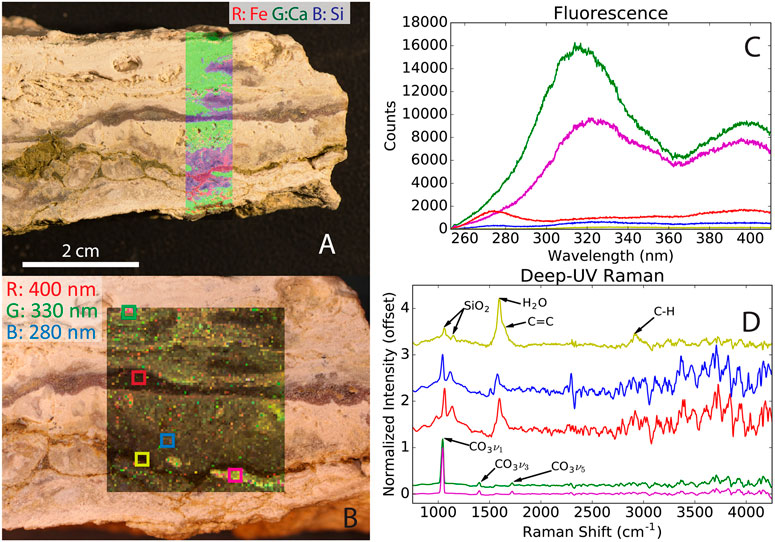
FIGURE 4. Stand-off measurements of a calcite-bearing sample at a Mars analog field site in the Mojave Desert with the techniques used by PIXL [X-ray spectroscopy, (A)] and SHERLOC [UV fluorescence and Raman spectroscopy, (B–D)]. (A) Overlay of the elemental abundance of Fe, Ca, and Si. (B) Fluorescence scan map, (C) averaged fluorescence spectra of the selected areas, and (D) averaged Raman spectra of the selected areas. The figure highlights that stand-off spectroscopy can identify interesting samples and determine mineralogy. Reprinted from Martin et al. (2020).
Fluorescence and Raman spectroscopy are the most relevant techniques for direct in situ measurements of organic biosignatures. In the case of fluorescence, analytes are excited (timescale of femtoseconds) by the photons from a light source (e.g., a laser) through absorption. The excited analyte then undergoes internal conversion and vibrational relaxation (picoseconds), followed by the emission of a photon (fluorescence, nanoseconds) at a lower energy (longer wavelength) than the initial excitation energy. Fluorescence competes with other processes to reach the ground state, which is molecule dependent; hence, providing different quantum yields. On the other hand, Raman spectroscopy does not rely on absorbed light—it is based on the scattering of monochromatic light. Most scattered light is at the same wavelength as the source (called Rayleigh scatter); however, a small amount (10−8 to 10−12) of the incident light will be scattered at lower or higher energies (called Raman scattering). The transfer of energy from the photon to the analyte is called the Stokes-Raman effect, and the transfer of energy from the analyte is called the anti-Stokes effect, which is what is typically measured. The peaks in the Raman spectrum correspond to specific molecular bond vibrations, such as C-C, C=C, and O-H.
Among the spectroscopy instruments, SHERLOC is specifically geared to provide information about organic biosignatures. SHERLOC combines deep-UV (DUV, <250 nm excitation) native fluorescence (250–360 nm emission) and resonance Raman spectroscopy (810 to >3,600 cm−1) with a high-resolution imager (Beegle et al., 2015). Combining the two techniques comes naturally because aromatic compounds excited by UV generate both fluorescence and resonantly enhanced Raman scattering. DUV fluorescence spectroscopy provides general LODs of sub-ppm levels for aromatic compounds and LODs of <100 ppm for aliphatic compounds with a spatial resolution of 30 μm (Farley et al., 2020). Fluorescence typically offers minimal data to facilitate the identification of organic compounds. Raman spectroscopy is better suited for deconvolution and identification; however, it has much poorer LODs (Abbey et al., 2017). For example, 5 wt% adenosine 5′-monophosphate spiked in clay samples containing 1 wt% magnesium perchlorate was near the LOD when analyzed by Raman spectroscopy (Razzell Hollis et al., 2021).
An essential benefit of using DUV excitation for fluorescence and Raman measurements is that it reduces background autofluorescence from minerals (Bhartia et al., 2010; Abbey et al., 2017). The Raman region is also free of fluorescence from organics by DUV (Asher and Johnson, 1984). However, there is a limited capability of differentiating carbonaceous material from abiotic and biotic sources with Raman spectroscopy, particularly through the commonly used D (∼1,350 cm−1) and G (∼1,580 cm−1) band parameters (Bower et al., 2013). Even though the resolving power and the ability of analyte identification are limited, Raman and fluorescence spectroscopy (preferably combined) remain capable of identifying samples containing preserved fossilized, complex, macromolecular compounds embedded in minerals (Shkolyar et al., 2018, and references therein).
Although these spectroscopic techniques may not be ideally suited for detecting and identifying organic biosignatures, they are useful standoff sensing tools that facilitate sample selection; either for instruments limited by the number of samples they can analyze, or for potential sample return and analysis in terrestrial laboratories.
3 Current developments in organic biosignature analysis techniques
In organic-detection space missions, the main driving forces are minimizing measurement error, LODs, power, mass, volume, and risk. Lessons learned from the previous Mars missions also suggest that techniques and methods must be less affected by the sample composition. This is demonstrated by the chemical degradation of organics at elevated temperatures in the presence of perchlorates and the inhibitory matrix effects of minerals during derivatization (Navarro-González et al., 2006; Stalport et al., 2012). Furthermore, current instrumentation is not fit for the purpose in regards to targeted LODs (ca. 1 ppb) of key organic biosignatures such as fatty acids, amino acids, and informational polymers (i.e., DNA and RNA; Mahaffy et al., 2012; Pappalardo et al., 2013; Hand et al., 2017; Neveu et al., 2018). On the basis of the summary of previously used in situ organic detection techniques, further development is needed in order to detect a greater span of organic biosignatures at lower concentrations than what is currently possible.
Currently, no technique can universally detect all high-priority target analyte classes at desired LODs and with satisfactory ruggedness, trueness, and precision. Each mission has its scientific scope and engineering constraints; the sample type can range from dry solids to completely liquid. It is essential to underline that current developments for in situ organic biosignature analysis are primarily infusions of techniques utilized in larger sectors such as the biomedical industry. The main challenge is adopting these techniques for space flight applications.
3.1 Recent advances in gas chromatography
Although GC has been a part of instrument suites on several missions, the development of primarily system miniaturization is still ongoing. More recent attention has focused on scaling down the size by utilizing chip-based platforms as opposed to traditional capillary columns. The Spacecraft Atmosphere Monitor (S.A.M.) onboard the ISS is perhaps currently the most notable instrument, which consists of a very compact chip-based GC and a compact quadrupole ion trap MS (Schowalter et al., 2019). Of note here, S.A.M. should not be confused with the SAM instrument suite onboard the Curiosity rover on Mars. The instrument onboard the ISS weighs only 10 kg and is capable of measuring major air constituents and trace concentrations of VOCs. Other developments in chip-based GC instruments are in progress (Figure 5; Blase et al., 2020b; Blase et al., 2022).
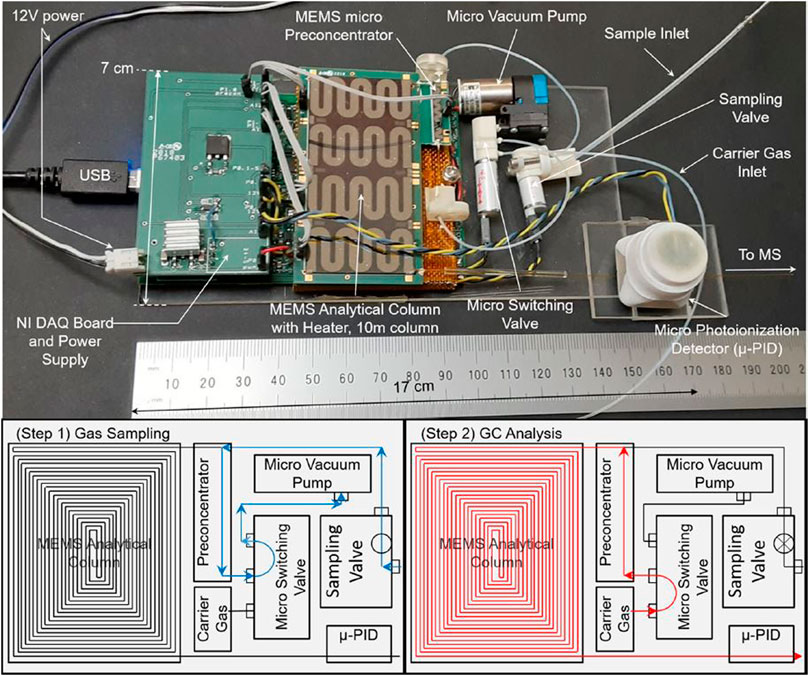
FIGURE 5. Top image shows a micro-electromechanical system (MEMS) GC and its major components. The bottom figures illustrate the operation, with first the loading step followed by the analysis step. Adapted with permission from Blase et al. (2020b). Copyright 2020 American Chemical Society.
So far, applications of chip-GC have mainly been restricted to the analysis of VOCs due to the limited upper-temperature range (∼200°C). For example, silicon, glass, and adhesives that are frequently used for fabrication have mismatching thermal expansion coefficients, which leads to cracking and leaking after thermal cycling (Ghosh et al., 2018). GC methods for analyzing methylated fatty acids and polycyclic aromatic hydrocarbons (PAHs) almost always employ temperatures above 200°C, or even 300°C. There is a vast literature covering fundamental research to improve the upper-temperature limit of microchip GC (Ghosh et al., 2018). Among various approaches, stainless steel microchip GC columns appear particularly promising due to their high thermal conductivity and compatibility with high operating temperatures up to 350°C (Ghosh et al., 2019). Nonetheless, a chip-GC manufactured out of etched silicon substrate and bonded with anodized Pyrex glass was able to operate up to 300°C; with the ability to separate alkanes and derivatized fatty acids up to C22 (Blase et al., 2022). The authors demonstrated that the chip-GC hyphenated with an MS provided LODs between 4 μM and 43 µM (4–43 pmol), a peak capacity of 124, and retention variations of <1% RSD.
Shrinking GC from traditional capillary columns to chip-based separation also comes at the expense of chromatographic efficiency. Chip-based GC suffers from a less homogeneous stationary phase coating due to the square geometry of the flow channels as compared to the cylindrical geometry of conventional capillary columns. The excessive accumulation of stationary phase in the corners, also known as pooling, creates an uneven film thickness which contributes to peak broadening (Wang et al., 2014). There is currently a multitude of ongoing research efforts to improve the separation efficiency by optimizing the stationary phase coating technique, column architecture, and layout. For example, serpentine, circular-spiral, and square-spiral column layouts are the most frequently utilized designs (Regmi and Agah, 2018). The channel width and the coating thickness play an important role in the trade-off between separation efficiency and sample capacity. Narrow channels with a thin coating provide higher separation capacity but limited sample capacity. Multi-capillary columns and semi-packed columns (pillar array columns) have been developed to increase the sample capacity by increasing the relative volume of the stationary phase; yet, they still have a thin film of the stationary phase to maintain the separation efficiency, which consequently keeps the mass transfer resistance low (Zareian-Jahromi et al., 2008; Jespers et al., 2017). Furthermore, micro-columns often exhibit tailing of polar analytes compared to conventional capillary columns due to surface adsorption (Regmi et al., 2018).
Comprehensive two-dimensional microscale GC (µGC × µGC) is currently being developed for future planetary missions (Blase et al., 2020a; Blase et al., 2022). Under ideal conditions, the peak capacity of GC × GC is almost an order of magnitude higher than conventional GC for the equivalent analysis time (Klee et al., 2015). In brief, GC × GC is a multidimensional separation technique achieved by coupling two columns with different stationary phases and where the effluent of the first column is cyclically trapped, refocused, and injected onto the second column. The technique has gained immense acceptance in petroleum analysis, where the first dimension typically separates based on volatility and the second dimension primarily separates based on polarity, facilitating analysis of samples even when all analytes are not entirely resolved (Abrahamsson et al., 2017; Prebihalo et al., 2018). The interface between the first column and the second column is called modulation. Conventional modulation is typically achieved by either thermal modulation or flow modulation. Thermal modulation is accomplished by cryogenic cooling through a jet of liquid nitrogen to trap analytes in the column and then rapid heating to elute the analytes into the second column. Flow modulation typically uses sample loops and valves to sample the effluent from the first column and reintroduce the analytes to the second column. Whereas the former approach is usually preferred for sharp peaks and lower detection limits, the latter is preferred for very volatile analytes which are not readily trapped even under cryogenic conditions. Conventional thermal modulation uses large volumes of liquid N2 or liquid CO2 and is unsuitable for space applications.
A microfabricated thermal modulator (μTM) has been developed for µGC × µGC, which completely retires the need for cryogenic fluids (Figure 6; Kim et al., 2010; Kim et al., 2011). The device measures 13 mm × 6 mm × 0.5 mm and is thermally controlled by a thermoelectric cooler (−35°C to −25°C at a rate as high as 168°C/s) and thin-film resistors (up to 210°C at a rate as high as 2,400°C/s; Kim et al., 2011). The cooling requires 21 W, and the heating requires 10 W. The authors reported FWHM of second dimension peaks as narrow as 90 ms. This is an impressive feat; however, it could potentially pose an interface issue with the limited scan speed of current state-of-the-art mass spectrometers.
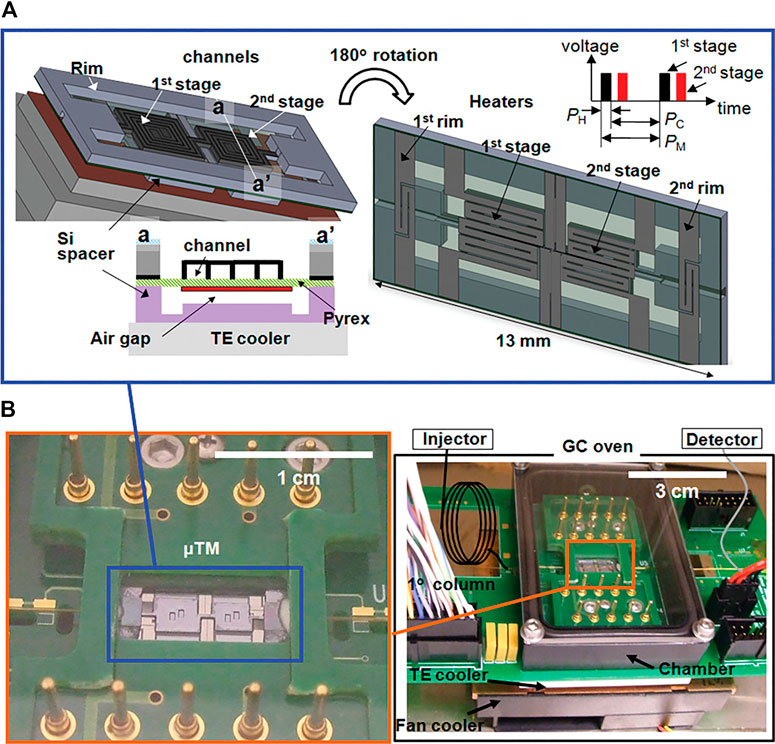
FIGURE 6. Illustration of a two-stage μTM that is both miniaturized and essential for comprehensive µGC × µGC. The two stages are cooled in order to trap analytes from the first dimension, followed by rapid heating so that the analytes are introduced in a narrow band into the second dimension. The upper figure (A) shows the diagram and the duration of the heating (PH) and cooling (PC) of one modulation period (PM). The lower photographs (B) show the assembly mounted on a printed circuit board. Reprinted with permission from Kim et al. (2011). Copyright 2011 American Chemical Society.
Despite all the recent development in the miniaturization of GC, the engineering of connectors and interfaces are still lagging behind the individual components, such as, those between chip columns, sample introduction sub-systems, and detectors (Ghosh et al., 2018). These connections must be compatible with the high temperature of GC for biosignature analysis and maintain the separation’s peak fidelity. Connections that introduce additional dead volume increase the dispersion in the system and consequently give rise to broader peaks and poorer separation. Currently, the performance of chip-based GC does not match conventional capillary column GC; however, rapid progress is being made. For example, Blase et al. (2022) demonstrated that capillaries between chip-GC and an MS could simply be sealed with polyimide/Hysol 1C epoxy, which provided separation efficiencies close to commercial capillary columns.
Miniaturization of GC will enable a substantial reduction of size, weight, and power compared to previous instruments applied in spaceflight missions. CE or LC cannot match the peak capacity of GC when performing conventional injections, and this may be further improved by using GC×GC at the cost of additional power requirements. However, current implementations with a trap column to interface the sample processing unit and the GC give rise to broad peaks (Guzman et al., 2020). Additionally, GC is limited to the analysis of thermally stable and volatile analytes. Most high-priority organic biosignatures are incompatible with GC and must be derivatized first. Herein lies the primary drawback of current implementations; the sample composition considerably impacts the derivatization methods. Overall LODs are often magnitudes lower than for the next-generation instruments (see Section 5), primarily due to analyte transfer efficiencies and limited sample load (∼50 mg). Wet-chemistry sample preparation techniques (see Section 4) could mitigate some of these issues; however, it has not been extensively pursued. Furthermore, several techniques discussed later (e.g., LC and biosensors) do not require derivatization, which reduces risks associated with matrix effects and contamination. Finally, GC has an extensive flight heritage and may be the preferred choice for missions in the near future, with limited budgets, and when LOD requirements can be relaxed.
3.2 Recent advances in mass spectrometry
Mass spectrometry has been an essential part of space exploration, as described in Section 2.2. In the context of organic biosignature detection, in addition to mass spectrometers for landed missions, developments are ongoing for fly-by missions to attempt to capture the ejecta from the plumes of Enceladus and Europa. Recent research has studied the ionization through impact at 3–10 km/s of organic biosignatures such as amino acids and fatty acids (Klenner et al., 2020a; 2020b). The LODs achieved in these studies were generally in the low-ppm level. The upcoming Europa Clipper (Howell and Pappalardo, 2020) will carry two mass spectrometry instruments, MAss SPectrometer for Planetary Exploration (MASPEX; Brockwell et al., 2016) and SUrface Dust Analyser (SUDA; Kempf et al., 2014). Although these developments contribute to the development of MS instruments, they are beyond the scope of this review because we focus on in situ organic biosignature analysis primarily for landed missions.
3.2.1 Mass analyzers
The development of miniaturized mass spectrometers has experienced a surge over the last decade, and we refer the interested reader to a few excellent general review papers (Snyder et al., 2016; Mielczarek et al., 2020) and a few related to planetary exploration applications (Arevalo et al., 2020; Chou et al., 2021). A few ongoing developments are worth highlighting here, such as the quadrupole ion trap (QIT) MS onboard ISS (Darrach et al., 2015; Madzunkov et al., 2016; Schowalter et al., 2019), the linear ion trap (LIT) MS onboard the Rosalind Franklin rover (Arevalo et al., 2015; Goesmann et al., 2017), the multi-bounce time-of-flight MS developed for the Europa Clipper mission (Brockwell et al., 2016), and the adaption and miniaturization of the Orbitrap (Arevalo et al., 2018). Validation data are scarce; consequently, a quantitative performance comparison is not currently possible without such data.
Current progress toward non-spaceflight laboratory instruments mainly focuses on hybridization and combining various mass analyzers, as well as increasing the ion transmission from ambient pressure ionization sources by increased volumetric flow, which is enabled by more efficient vacuum pumps (Hager, 2004). However, an increase in complexity, mass, and power does not align with current general strategies of miniaturizing instruments (including mass and power) for spaceflight applications. Particularly, miniaturizing vacuum pumps and simpler designs of electronics are vital efforts. Heritage plays an important role in the selection and development of MS for in situ missions. For example, Arevalo et al. (2020) illustrated that the neutral mass spectrometer (NMS) onboard the LADEE spacecraft, with heritage components from Cassini-Huygens, took less than 3 years to deliver, as opposed to the LIT developed for MOMA, with fewer heritage components, that took more than 10 years to deliver.
Ion traps are typically easier to miniaturize because they can often operate at higher pressures (∼10−3 Torr), and miniaturization can even improve mass resolution (Johnson et al., 2019), contrary to other types of mass analyzers. Compared to ToF-MS, the ion trap offers simpler electrical design and MS/MS analysis, but provides lower resolution (Ren et al., 2018). A handheld rectilinear ion trap MS (∼10 kg, 50 W) equipped with electrospray ionization was able to detect amino acids, nucleobases, nucleosides, and peptides with LODs of approximately 1 mg/L (1 ppm) or lower (Sokol et al., 2011). The instrument was also demonstrated with multiple-stage MS up to MS5, which greatly facilitates the identification of larger analytes. Discontinuous atmospheric pressure ionization (DAPI) was utilized to reduce the load on the vacuum pumps. The principle of the technique is that the flow path between the ionization source and the mass analyzer is only open for a short duration, allowing a short pulse of ions (Gao et al., 2008). Two newer versions of the rectilinear ion trap MS system have since then been developed, with a range up to m/z 900 and a resolution of 500 (Gao et al., 2008; Li et al., 2014).
The LIT (e.g., used in MOMA) is principally a miniaturized version of the Thermo LTQ XL (Thermo Fisher Scientific), with lower performance characteristics than its commercial counterpart due to miniaturization (Brinckerhoff et al., 2013). On one side is an EI source to interface with GC and on the opposing side is an inlet for ions generated by the laser ionization sub-system, which utilizes the DAPI technique to reduce the requirement of the vacuum pumps. The valve is opened between 10 and 100’s milliseconds while the sample is subjected to laser ionization, and the ions are pulled in by a potential applied to the inlet (Li et al., 2015a). The instrument, including GC and laser ionization sub-system, weighs approximately 12 kg and provides unit resolution up to 500 Da (Goesmann et al., 2017). Recent developments of the same MS instrument have improved the performance to an m/z range of 20–800 with an FWHM of 0.5 Da over the entire range (Brinckerhoff et al., 2022).
Although the MOMA-MS only supports positive mode MS, the LIT can operate in both negative and positive modes with minor modifications (Li et al., 2019). The instrument is being repurposed for various missions concepts, such as the Dragonfly Mass Spectrometer (DraMS) that is planned for an aerial lander mission to Titan (Lorenz et al., 2018), the EMILI instrument suite comprising capillary electrophoresis (CE) and GC that is being developed as a contender for a potential Europa lander mission (Brinckerhoff et al., 2019; Brinckerhoff et al., 2022), integration with an Orbitrap MS in the Advanced Resolution Organic Molecule Analyzer (AROMA; Arevalo et al., 2016), and the Molecular Analyzer for Complex Refractory Organic-rich Surfaces (MACROS; Getty et al., 2017).
A miniaturized Paul trap mass spectrometer, also known as a quadrupole ion trap mass spectrometer (QITMS), has been developed and is currently operating onboard the ISS for cabin atmosphere monitoring (Shortt et al., 2005; Madzunkov et al., 2016; Schowalter et al., 2019). The QITMS weighs about 7.5 kg and is hyphenated with a GC system (described in Section 3.1). The ideal operating pressure is < 10−6 Torr, which is lower than the typical LIT (Madzunkov et al., 2016). The current configuration enables detection of m/z 30 to 300; however, it is extendable up to m/z 600, with a mass resolution of 18,000 at m/z 40, and a resolution of 600 throughout the entire mass range at a scanning frequency of 50 Hz (Simcic et al., 2021).
Several miniaturized ToF systems have been conceived; for example, a conventional but miniaturized ToF-MS has been interfaced with a miniaturized pyrolysis unit (Getty et al., 2010; Glavin et al., 2012b), or with laser-ionization (Getty et al., 2012). Recent trends and development have mostly focused on multibounce, or multiturn, ToF instruments. Multibounce ToF systems enable smaller instruments, even though they maintain a long travel distance in order to retain the high resolution. Instruments based on this technology have mainly been used with laser-ablation (Rohner et al., 2003; Wiesendanger et al., 2018, 2019; Riedo et al., 2020; Ligterink et al., 2021). The ToF instrument used in these studies resulted from developments based on the Rosetta TOF-MS (Hohl et al., 1999). It measured 160 mm in length and 60 mm in diameter and enabled two turns of the injected ions through a set of reflectrons ion optics (Riedo et al., 2017). A resolution of <900 was acquired when interfaced with laser ionization (Riedo et al., 2013).
MASPEX is another ToF instrument (Figure 7), with an adjustable number of turns or bounces, which will be a part of the Europa Clipper payload (Brockwell et al., 2016) and is also being developed for GC-MS applications as a candidate for a potential Europa lander mission (Blase et al., 2020a; Blase et al., 2020b). An average MS resolution of 380 was achieved when coupled with chip-based GC and scanning m/z 40–140 (Blase et al., 2020b). However, with slower measurement frequency, the resolution can be as high as 46,000 at the expense of mass range and sensitivity (Brockwell et al., 2016).
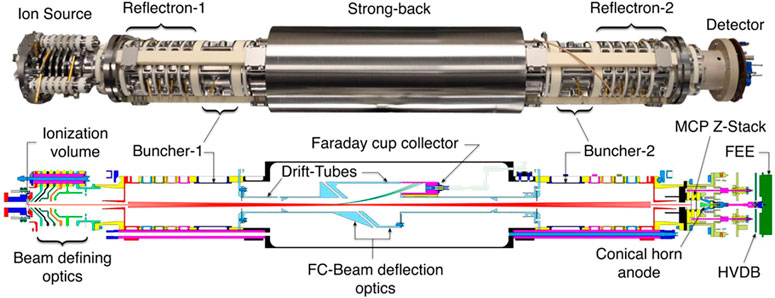
FIGURE 7. Image and illustration of the Mass Spectrometer for Organic Analysis in Space Environments (MASPEX) instrument. Multibounce is utilized to increase the flight path to achieve high-resolution measurements despite miniaturization. Adapted with permission from Blase et al. (2020b). Copyright 2020 American Chemical Society.
Orbitrap is the latest addition to MS techniques currently under investigation and development for spaceflight applications. It was introduced first in 2000 (Makarov, 2000). The Orbitrap traps the ions in an orbital motion around the inner electrode rather than in a space confined by outer electrodes like the LIT or QITMS. GC-Orbitrap, with either EI or chemical ionization, has been used to detect volatile organics from interstellar ice analogs (Javelle et al., 2021). A benchtop laser ablation (LAb)-Orbitrap has been developed (Briois et al., 2016). The MS operates at a pressure of <10−6 Torr (typically <10−10 Torr), which may require powerful vacuum pumps depending on the mission and the front-end instrumentation. The mass resolution increases with lower pressure. The benchtop Orbitrap instrument presents an impressive mass resolution of >90,000 up to m/z 209 even at 1 Hz scan rate (Briois et al., 2016). Multiple studies have highlighted how the benchtop instrument provides exceptional capabilities for identifying unknown organics by high-resolution MS (Arevalo et al., 2018; Selliez et al., 2019; Selliez at al., 2020). The Characterization of Ocean Residues And Life Signatures (CORALS) instrument, which is also based on LAb-Orbitrap, is estimated to weigh 8 kg and consume 41 W at peak power; and provides an m/z range of 20–600 with a mass resolution of 120,000 (at m/z 133; Willhite et al., 2021). The LIT-MS mentioned earlier, hyphenated (also known as hybridization) with a miniaturized Orbitrap, is also being investigated (Arevalo et al., 2016). The capabilities of the hybrid MS under investigation remain unknown; however, the configuration is popular in commercial systems used in analytical chemistry laboratories and provides improved LODs due to the accumulation of ions in the LIT, which are then delivered as coherent ion packs with high ion transmission to the Orbitrap (Makarov et al., 2006).
It is important to consider the pressure at which each mass analyzer typically operates when comparing them. The typical ITMS operates at ∼10−3 Torr, the ToF at ∼10−8 Torr, and the Orbitrap at ∼10−10 Torr. Vacuum pumps and electronics are the main contributors to the overall mass and volume. Appropriate pumping capacity must be selected depending on the mission environment (ambient pressure) and the type of front-end selected. For example, the most common ionization sources for liquid streams operate at atmospheric pressure (760 Torr).
3.2.2 Laser-based ionization
Laser ionization MS (LIMS), including laser desorption ionization (LDI) and laser ablation (LAb), enables the detection of biosignatures through a single laser pulse (typically a few nanoseconds) that desorbs and ionizes material that is subsequently MS-detected. It can be used at various ambient pressures, including high vacuum. LIMS requires no consumables such as liquids for extraction or carrier gases and further reduces instrument size and complexity. Consequently, the number of possible analyses is not limited by consumables, and countless samples can be analyzed throughout an in situ mission. LIMS enables spatial resolution (micrometer scale or lower) that cannot be achieved by conventional sample acquisition techniques for downstream chemical analysis. The downside of the technique is that the ionization efficiency is relatively low, which results in low sensitivity (Feider et al., 2019).
LIMS has historically mainly been used for the elemental and inorganic analysis of solid samples in the laboratory (Azov et al., 2022). The technique has proven powerful for the elemental analysis of signatures of microbes in solid samples (Stevens et al., 2019; Riedo et al., 2020). So far, limited validation data related to planetary analogs has been reported. The MOMA engineering test unit was used for analyzing microbial-containing silica sinter and previously extracted lipids (Siljeström et al., 2021). Only chlorophyll A was detected in the untreated sample. Spiked coronene (10 ppm) and Rhodamine 6G (10 ppm) in Mars analog samples, with added ∼1 wt% perchlorate salt, have been successfully analyzed with a breadboard setup of MOMA (Li et al., 2015a). Spiked tryptophan was detected at 0.1 wt% in various mineral substrates (Uckert et al., 2018). However, LIMS spectra of solid samples are often difficult to interpret and may be dominated by mineral fragments (Bishop et al., 2013). Two-stage LIMS can reduce mineral interference by tuning the desorption and ionization independently for aromatic analytes; however, the LODs remain high (Getty et al., 2012; Uckert et al., 2018).
It has been shown that most amino acids mixed with NaCl on a stainless steel surface can be detected, of which tryptophan had an LOD of 1.2 fmol mm−2 (Ligterink et al., 2020). Another study that used an Orbitrap adapted for spaceflight (CosmOrbitrap) showed that twelve amino acids could be detected at a concentration of 1 pmol/mm2, which was achieved by drying 10 µL of water with 10 µM of each amino acid onto a plate of 80 mm2 (Arevalo et al., 2018). The same authors demonstrated that pure uracil could also be detected. LIMS has also been applied to analyze isolated single cells with sub-micron resolution, of which mainly phospholipids were detected (Wang et al., 2018).
For Ocean Worlds applications, the ice or aqueous samples could be sublimated or evaporated to further increase the concentration of analytes in order to improve method LODs. However, further validation and comparison with alternative analysis techniques are still needed to assess its utility. Nonetheless, a LIMS instrument is included in the payload of the Rosalind Franklin rover (Li et al., 2017), and was included onboard the unsuccessful Phobos-Grunt mission (Managadze et al., 2010).
LIMS is a versatile technique that requires minimal or no sample preparation. However, compared to some of the more complicated techniques that also utilize MS (e.g., GC, CE, and LC), it has several orders of magnitude higher LODs. Additionally, LIMS offers similar performance to stand-off spectroscopic techniques; however, when combined with high-resolution MS, it may offer much higher ability to differentiate analytes in a complex sample.
3.2.3 Mass spectrometer interfaces for separation techniques
Electron ionization (EI) has been used extensively for ionization in the analysis of atmospheres and the coupling of GC-MS, and has extensive spaceflight heritage. Other ionization sources that are routinely used in laboratories are electrospray ionization (ESI), atmospheric pressure chemical ionization (APCI), atmospheric photoionization ionization (APPI), matrix-assisted laser desorption/ionization (MALDI), fast atom/ion bombardment (FAB), and chemical ionization (CI). The choice of ionization technique depends primarily on the sample; for example, solid sample, gas from a GC, or liquid from a liquid chromatography (LC) system; and secondarily on the analytes of interest.
EI is easily implemented and generates reproducible mass spectra that can be matched with databases. However, current the usage of EI is not suitable for the emerging instruments that utilize wet chemistry for separation or direct analysis of aqueous samples on Ocean Worlds. EI requires a high vacuum (<10−6 Torr) for ideal operation and to not burn out the filament. EI interfaces for liquid streams have been developed; however, their suitability for thermo-labile biosignatures has not yet been assessed (Termopoli et al., 2017; Termopoli et al., 2019; Rigano et al., 2019).
Conventional ionization techniques for liquid streams include ESI, APCI, and APPI, or variants of these (Covey et al., 2009; Feider et al., 2019). They all have in common that the ionization process occurs at atmospheric pressure (760 Torr). Consequently, additional instrument design considerations are added due to the lower atmospheric pressure at various target planetary bodies. In the case of atmospheric pressure ionization sources, the ions need to be transferred into the mass analyzer, which operates at vacuum <10−3 Torr, depending on the MS technique. Among these atmospheric ionization techniques, ESI is the most commonly used. ESI, and other atmospheric pressure ionization techniques, are adversely affected by matrix effects such as salts through ion suppression, and result in diminished sensitivity (Furey et al., 2013). During the last decade, low flow (nL/min) nano-ESI, also known as nanospray ionization, has gained widespread popularity. Among the benefits mentioned are low-solvent consumption, increased sensitivity, and reduced matrix effects from components such as non-volatile salts (Juraschek et al., 1999). However, to obtain these benefits, the flow rate needs to be a few tens of nL/min or less through the emitter (Schmidt et al., 2003; Jarvas et al., 2017). The total flow rate can be increased using multiple emitters (Kelly et al., 2008; Mao et al., 2011; Jender et al., 2021).
Although ionization sources, compatible with liquid samples, are essential for many techniques in development (see Section 5), no hardware compatible with operation in a vacuum environment (while maintaining ∼760 Torr in an ionization chamber) has been demonstrated. Additionally, ion optic interfaces with gradually increasing vacuum, from atmospheric pressure to vacuum levels required by MS analyzers, have just recently been interfaced with a flight-like MS (Brinckerhoff et al., 2022). Although these are standard techniques in the laboratory and portable instruments, they have not yet been fully adapted for spaceflight applications.
3.3 Ion mobility spectrometry
In this section, we discuss ion mobility spectrometry, which includes ion mobility spectrometry (IMS) and a sub-technique named differential mobility spectrometry (DMS). IMS resembles MS in many practical aspects, but the two techniques are not interchangeable. Ionized analytes in the gas phase are separated based on their mobility in a carrier buffer gas, often at atmospheric pressures. Hence, the same atmospheric pressure ionization techniques used in MS, such as ESI and APCI, are also combined with IMS. IMS has, over the last decade, established itself as a routine analysis technique hyphenated with MS in order to increase the overall resolving power by adding a dimension of separation. IMS has not been used for any planetary application; however, it has been routinely used on the ISS for air quality monitoring.
Commercial instruments are readily available and fully integrated. However, conventional IMS is not orthogonal to MS; the drift time often correlates with the m/z separation, and frequently only adds a marginal improvement in tandem instruments (Valentine et al., 2005; Shvartsburg et al., 2011). It is important to note that IMS can be used as a stand-alone technique, without MS; though some IMS techniques are only used in combination with MS to increase selectivity (Cumeras et al., 2015). In fact, for many smaller analytes, the correlation can be used to approximate the molecular weight based on their IMS drift times (Kim et al., 2005, 2009). IMS is readily miniaturized, and most IMS techniques are ideally operated at ∼760 Torr, which renders coupling with atmospheric pressure ionization techniques straightforward (Cumeras et al., 2015). Consequently, the technique may be seen as a more straightforward but less powerful substitute for MS. This section focuses on describing IMS as a stand-alone analyzer, with or without chromatography, but without MS. Johnson et al. (2007) have given a brief overview of IMS and its potential in space exploration applications.
Although many variations of IMS have been reported, they all include a drift tube in which the analytes are separated under a constant or variable electric field. The various commonly used types are covered elsewhere (Cumeras et al., 2015; Dodds and Baker, 2019). The most uncomplicated design involves insulated stacked rings that act as electrodes to form a uniform electric field inside the drift tube. Ions are injected as packages using an ion gate or a shutter. As the ions move through the electric field (e.g., towards a Faraday plate), their mobility depends on their surface area and charge. The ions travel through a gas typically held at ambient pressures, and molecules with a larger collision cross-section (CCS) reach the detector later than smaller molecules due to their interactions with the buffer gas. The separation is carried out on the order of micro-to milliseconds, making it ideal for coupling with fast chromatographic techniques with very narrow peaks (Valentine et al., 2005; Piendl et al., 2019). Isomers that MS does not resolve can be separated using IMS because CCS influences the migration time as opposed to the molecular mass (Zhu et al., 2012).
Current state-of-the-art IMS instruments have resolving powers above 300, sufficient to discriminate analytes with <0.5% CCS differences (Dodds et al., 2017). The resolving power (Rp) is here defined as the quotient of the drift time td and the full width at half maximum wFWHM. Some IMS techniques have also been demonstrated with on-chip separation (Costanzo et al., 2017; Deng et al., 2017).
The drift tube IMS has a simple design and operation. It consists of an ion gate, a drift region, and a shielded Faraday plate as an ion detector. The ions travel through a constant electric field that usually requires a few kV to maintain. The ions always travel through an environment of the counter-current flow of a neutral gas (mostly nitrogen, helium, argon, or carbon dioxide), called the drift gas. The separation can be optimized; for example, by the selection of buffer gas, pressure, and temperature (Matz et al., 2002; Wyttenbach and Bowers, 2003). Neutral analytes are flushed away by the buffer gas in the opposite direction of the detector. Ahrens et al. (2019) presented a simple and portable drift tube IMS (Figure 8), manufactured from PEEK and stainless-steel foils and snapped into place on a printed circuit board. The instrument provided a Rp of 63, with a drift length of 40 mm and a cross-section of 15 mm × 15 mm. Although this effort was not toward spaceflight applications, it demonstrates the ability to miniaturize IMS instruments with a simple design.
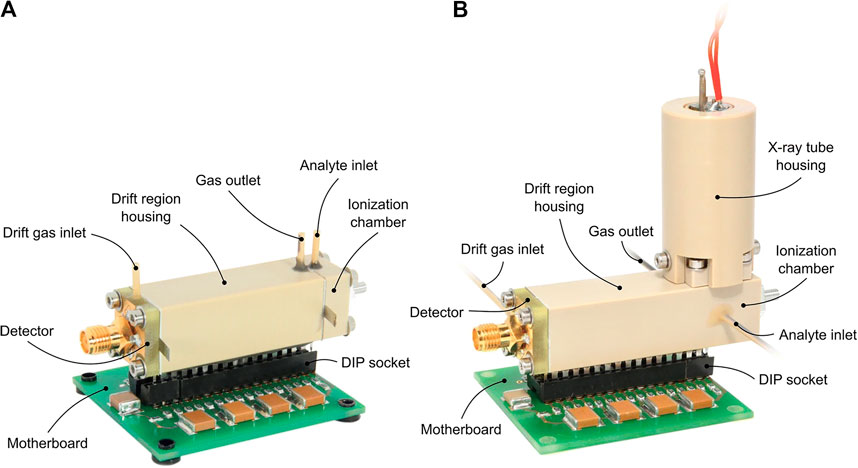
FIGURE 8. Photo of a miniaturized drift tube IMS with either a 3H ionization source (A) or an X-ray ionization source (B). This IMS is not being developed for space applications; however, it demonstrates that manufacturing a low-mass and small-volume instrument is possible. Reprinted from Ahrens et al. (2019).
DMS has proven to be a valuable addition to mobility spectrometry applications and analyses. DMS utilizes planar electrodes, and similar high-field asymmetric waveform ion mobility (FAIMS) utilizes cylindrical electrodes (Kolakowski and Mester, 2007). An electric field, perpendicular to the analytes’ direction, oscillates between low and high. Depending on the properties of the asymmetric electric field, also known as the dispersion field, stable trajectories towards the detector will be achieved for some analytes. With some similarity to a quadrupole MS, FAIMS can be used as a filter to selectively single out analytes for detection, or it can be used in a sweeping mode to produce a spectrum. Spectra are frequently plotted as intensity against the sweeping compensation voltage or compensation field, rather than a time for a drift tube IMS. Because FAIMS is not a time-based separation technique, it can accept a continuous input of ions and does, therefore, not require an ion gate (Cumeras et al., 2015). Although DMS/FAIMS can offer orthogonal separation to MS and offers more flexibility in method optimization (Shvartsburg et al., 2013), it requires more demanding electronics due to changing potentials.
In most chemical analysis laboratories, IMS is typically integrated with MS systems. However, stand-alone IMS instruments have found a niche application area where simplicity, reliability, and portability are highly valued; such as, in airport security or for military purposes. Examples of such applications are: field-analysis of explosives (Eiceman et al., 2004), chemical warfare agents (Eiceman and Stone, 2004), food and air monitoring (Eiceman, 2002), and illicit drug screening (Verkouteren and Staymates, 2011). A field-portable Py/GC-IMS instrument for aerosol analysis demonstrated that 1 µg of B. subtilis spores could be detected (Snyder et al., 1996; Dworzanski et al., 1997). More recent developments have included tandem IMS/IMS to provide similar structural information and improved resolving power as in tandem MS/MS (Merenbloom et al., 2006; Chiluwal et al., 2019).
Although, no IMS (including DMS) instrument has been used at any planetary body, it has been extensively used for air quality monitoring onboard ISS. A GC-IMS instrument was deployed to the ISS in 2001 for air quality measurements. It was the first time a real-time trace volatile organic compounds (VOCs) air quality instrument was deployed in a spacecraft. The system used radioactive nickel-63 foil as an ionization source to detect VOCs (Limero et al., 1992, 2011). Although its operation was successful, it suffered several technical issues. This sparked the development of a smaller and modularized GC-DMS instrument, the Volatile Organic Analyzer (VOA), which was installed onboard the ISS in 2009 (Limero et al., 2008). The VOA instrument was calibrated and validated for 15 VOCs. The operation of VOA lasted for about 7 years and was replaced by two units of a new GC-DMS system named the Air Quality Monitor (AQM). The AQM was able to quantify 22 VOCs (Limero et al., 2014; Wallace et al., 2017).
The Wet Chemistry Experiment at Mars (WetChem) instrument, an early-stage project that has not been used in spaceflight, included IMS in tandem with MS to analyze samples previously characterized by electrochemical techniques (Kanik et al., 2006). In the context of astrobiology, IMS has been proven suitable for detecting and identifying of many analytes; for example, organic acids (Kim et al., 2005), amino acids (Beegle et al., 2001; Johnson et al., 2004), phosphatidylcholines (Kim et al., 2009). Although no LODs were determined, the technique was able to separate the majority of 19 tested amino acids at 10 mg/L using ESI-IMS (Beegle et al., 2001).
In summary, IMS cannot operate in a vacuum environment because atmospheric pressure (∼760 Torr) is often required to achieve separation, which is based on gas-analyte interactions. An enclosure would be needed to maintain a suitable gas pressure when operating in space environments and would also require bringing additional carrier gas. However, for Mars (∼5 Torr) applications, MS instruments require capable vacuum pumps (see Figure 2), making IMS instruments a potential contender. Furthermore, the electrical subsystems for MS are substantially larger than for IMS instruments. IMS could be a smaller, but less capable regarding resolving power and identification compared to MS for in situ organic biosignature analysis. Only a handful of studies have investigated IMS for spaceflight applications at planetary bodies; however, recent progress in the state-of-the-art has proven it to be a readily miniaturized technique.
3.4 Standoff spectroscopic techniques
Remote and standoff spectrometers (e.g., X-ray spectrometers) have been an essential part of previous space missions and will continue to play an important role in future missions. However, fluorescence and Raman spectrometers for detecting organic material were first deployed on the Perseverance rover. These two techniques are among the most convenient and suitable techniques for detecting organic content in samples. No sample collection or sample preparation is needed, and the number of analyses is only limited by the laser lifetime. Nowadays, most space instrument suites include either or both Raman spectroscopy and fluorescence spectroscopy techniques in their arsenal. The same excitation wavelengths can be used in both techniques. The miniaturization of electronics and optics enables the continuous downsizing for the suitability of these instruments for space applications. Hence, a vast literature covers field demonstrations of fluorescence and Raman spectrometers onboard various platforms, including a rock-climbing robot (Uckert et al., 2020), and an ice-sheet-penetrating platform (Eshelman et al., 2019).
3.4.1 Raman spectroscopy
Recent developments in Raman spectroscopy have improved both the spatial resolution as well as the detection limits. High spatial resolution (∼1 µm) Raman imaging was used to study the hydrogen to carbon ratio in organic matter from fossilized cells (Ferralis et al., 2016). However, this study was conducted with a laboratory-based instrument, and these resolutions have not been achieved with miniaturized components. The in situ Spectroscopic Europa Explorer (iSEE) concept uses a digital micromirror device/photomultiplier assembly (DMD/PMT) and a microchip diode laser into a miniature Raman spectrometer which improves the signal-to-noise ratio by 3-orders of magnitude compared to conventional CCD arrays (Sobron et al., 2019). The authors also claim that the instrument has an LOD of <1 ppb for organic content. The Raman spectrometer designed for the Martian Moons eXploration (MMX) mission measures only 81 mm × 125 mm × 98 mm and weighs approximately 1.4 kg (Cho et al., 2021). There are other examples in the literature of other miniaturized standoff Raman instruments (Wang et al., 2003; Misra et al., 2016; Abedin et al., 2018; Sandford et al., 2021).
Background fluorescence, however, remains an important issue, which is often several orders of magnitude stronger than the Raman scattering signal, and inhibits the detection—for example, of cells in Martian simulant samples (Stevens et al., 2019). Two main strategies are used to reduce the background fluorescence signal: UV-range excitation wavelengths and time-gated (TG) Raman spectroscopy (Frosch et al., 2007; Shkolyar et al., 2018; Hanke et al., 2019). Fluorescence signals from minerals have a decay lifetime of >1 ms compared to organic fluorophores that typically have decay lifetimes of ∼<100 ns (Misra et al., 2016; Shkolyar et al., 2018). Blacksberg et al. (2020) demonstrated improved signal-to-noise ratios through fluorescence rejection by TG Raman spectroscopy with a 100 ps laser pulse, and a 1 ns detector gate duration with 550 ps rise time and 250 ps fall time. Consequently, the authors could detect kerogen (i.e., the insoluble sedimentary organic fraction formed through diagenesis of biological material; see Vandenbroucke, 2003 for a detailed definition) content down to 1% in various samples.
Research is still ongoing regarding how the sample composition influences the response factors of organic compounds in mineral matrices. Samples synthesized in the laboratory do not capture all matrix effects that may be present in a natural sample. In fact, most of the available literature only covers the study of pure analyte or single analyte spiking on solid samples. The latter does not consider the distribution of analytes or adsorption processes that only occur through geological diagenesis. Therefore, it is difficult to evaluate the performance of the techniques and the instruments. For example, Montagnac et al. (2021) found that 0.4 mM deoxyguanosine-5′-monophosphate (dGMP), spiked onto clay samples, could be detected; however, the response factor depended on the sample matrix.
3.4.2 Fluorescence spectroscopy
Fluorescence spectroscopy has long been a standard tool for marine sample analysis, in the laboratory or in situ, and is now being adapted for the Ocean Worlds applications. Of note here, the Ocean Worlds in the Solar System of particular interest, other than the Earth, are Callisto, Enceladus, Europa, Ganymede, and Titan. Many of the optical sensor technologies and marine platforms that may be applicable for Ocean Worlds have been reviewed by Aguzzi et al. (2020). The typical instrument involves absorption and fluorescence spectroscopy applied to a flow cell for the detection of organic compounds such as pigments, polycyclic aromatic hydrocarbons (PAHs), proteins (indirectly detected through tryptophan), and organic polymers such as humic acids (Wollschläger et al., 2013; Wollschläger et al., 2016). Naturally, fluorescence spectroscopy is only suitable for detecting the fluorescent fraction of the dissolved organic matter. Cells and molecules can be labeled with a fluorescent dye; however, inefficient staining, non-specific binding, and autofluorescence of mineral particles (especially those including rare-earth elements) may result in both false positives and false negatives of potential biosignatures (Gaft et al., 2001; Li et al., 2004; Shkolyar et al., 2021).
Single-wavelength excitation with full spectrum emission scanning provides limited chemical information for identification and deconvolution. Hence, development for Earth applications is moving towards using excitation and emission scanning fluorescence, or excitation-emission matrices (EEMs), which provide a 2D dataset for each analyzed sample (Bhartia et al., 2008; Ferdinand et al., 2017; Carstea et al., 2020). Such data provide more rugged deconvolution for identification and quantification through multi-way (tensorial) chemometric techniques such as parallel factor analysis (PARAFAC; Smilde et al., 2005; Toraman et al., 2018). For example, excitation and emission scanning fluorescence followed by PARAFAC enabled analysis of terrestrial humic-like species, marine humic-like species, and protein-like tryptophan in natural waters (Zielinski et al., 2018). Although no instrument is specifically being developed for space applications, EEMs have proven valuable for analyzing biological (Bhartia et al., 2008) and Ocean World (Malaska et al., 2020) analog samples, and may be a future spaceflight instrument avenue.
Ice can be directly analyzed with fluorescence spectroscopy, thereby retaining the spatial distribution. The Wireline Analysis Tool for the Subsurface Observation of Northern ice sheets (WATSON) leverages the previously developed SHERLOC instrument (currently onboard the Mars 2020 Perseverance rover) for this specific purpose (Eshelman et al., 2019). WATSON uses DUV excitation (248.6 nm) and acquires spectra between 265 nm and 440 nm. The LOD for tryptophan, tyrosine, and E. coli were 2 μM, 20 μM, and 5 × 106 cells/mL, respectively (Eshelman et al., 2019). The absolute cell LOD was estimated to be < 300 cells, achieved through the spatial resolution (100 µm) and low interrogation volume, where each scan covers a 75 mm by 25 mm area. The instrument demonstrated that it could identify hot spots in the Greenland ice sheet containing organic matter from the surface down to 105 m depth (Malaska et al., 2020).
TG-fluorescence spectroscopy provides valuable utility in improving detection limits and identification. Its applications and recent developments have been reviewed by Yang and Chen (2020). The detection limits are improved with TG-detection because organic materials display fluorescence decay lifetimes, which are orders of magnitude shorter than mineral (inorganic) decay lifetimes. This can allow organics to be detected as biosignatures in the presence of a mineral matrix (i.e., fossil organics in a rock; Shkolyar et al., 2018). Identification can be facilitated by time-resolved measurements, in which a sequence of TG measurements is taken with incremental gate delay. The decay profiles and spectra together provide information for analyte identification. For example, time-resolved fluorescence with a gate width of 1.5 ns was used to discriminate PAHs over the course of 20 ns following the laser pulse (Eshelman et al., 2018). Lymer et al. (2021) used UV time-resolved laser-induced fluorescence to detect and identify amino acids, intending to later be used on returned meteorite samples. Other similar instruments, such as the OrganiCam (TG-fluorescence and Raman spectroscopy) are currently being developed for potential Mars and Ocean Worlds applications (Wiens et al., 2019, 2020; Gasda et al., 2021).
Mineralogy greatly affects response factors, and more research is needed (Laurent et al., 2019). For example, matrices rich in transition metals (e.g., Fe and Cr) may have strong UV absorbance, greatly reducing the sensitivity (Chua et al., 2001; Shkolyar et al., 2018; Carrier et al., 2019). Most studies have been conducted with pure analytes or spiked samples; therefore, they do not reflect the true LOD of the reported methods. Hence, fluorescence spectroscopy can currently be viewed as a qualitative, or possibly semi-quantitative, tool for standoff measurements that facilitate sample selection for analysis by more capable techniques.
4 Sample preparation
Most chemical analysis techniques used in the laboratory are performed with the analyte in solution. In the context of methods developed for space applications, hybrid approaches have been investigated, such as sublimation followed by derivatization and gas chromatography analysis (Glavin and Bada, 1998; Glavin et al., 2006). However, sublimation is several orders of magnitude less efficient than solvent extraction of amino acids (Skelley et al., 2005). Direct analysis of liquid samples, including liquid extracts from solids, typically provides lower LODs. Furthermore, liquid samples are easier to prepare for downstream analysis instruments; nevertheless, preconcentration or desalting may still be necessary. Extraction techniques applicable to solid samples, primarily for Mars applications, have been reported but are relatively unexplored compared to separation and detection instruments. Although sample preparation techniques, including extraction, are generally perceived as being of secondary importance to detection technologies, it is perhaps the most crucial step to achieve required LODs and measurements with acceptable errors (Traks et al., 2005).
Analytes strongly adsorbed to minerals may be more resistant to degradation (Aerts et al., 2014 and references therein), and microbes may have higher an affinity toward certain types of minerals (Röling et al., 2015 and references therein). For example, strong adsorption to solid matrices such as clay and other silicate-rich samples aids the preservation of nucleic acids, but may result in lower recoveries from the extraction process (Direito et al., 2012). Similarly, Mars-relevant minerals have proven to be difficult to extract DNA from due to adsorption (Carr et al., 2017; Mojarro et al., 2017). Competitive binders such as phosphate, pyrophosphate, ATP, random hexamers, or skim milk are often used to increase the recoveries (Takada-Hoshino and Matsumoto, 2004; Direito et al., 2012; Mojarro et al., 2017). Furthermore, although DNA may be locally well-preserved, when extracted it may degrade due to the presence of metals or extreme pH. The impact of Mars-relevant components on nucleic acid degradation has been covered by Mojarro et al. (2021). There is no universal extraction protocol that is effective for all types of soil samples, and repeatability and reproducibility varies greatly between commercial extraction kits, whose components are often trade secrets (Zielińska et al., 2017). Consequently, sample matrices such as clay-rich minerals become a double-edged sword—organic biosignatures may survive for extended periods of time, but they are at the same time challenging to extract.
The overall instrument complexity varies for sample preparation subsystems. For example, multiple solvents and reagents, that need to be stored separately, require additional pumping units and valves. A typical DNA extraction and purification protocol illustrates such an instance: the sample is desalted; the DNA is extracted with a binding buffer with or without competitive binders, and sometimes in combination with a cell lysing step; the DNA is purified on an adsorbent and washed; lastly, the DNA is eluted from the adsorbent and can be analyzed downstream (often with additional preparation steps). On the other side, sample preparation protocols may be as simple as only using water as an extraction solvent. The storage of reagents and solvents must be carefully considered. Solutions or reagents stored dry, need to be stable over several years (or even longer), potentially also in high-radiation environments. Even solvents may degrade; for example, acetonitrile degrades into cyanide when exposed to 300 krad (Creamer et al., 2019), which could be expected during a mission to Europa (Hand et al., 2017). Storage experiments of solutions and reagents are discussed later, whenever data are available, together with their accompanying technique or instrument.
Miniaturized liquid sample handling devices are currently being developed for a holistic approach to sample distribution, filtering, the addition of reagents, and concentration by evaporation, such as the Sample Processor for Life on Icy Worlds (SPLIce; Chin et al., 2017), or the similar Extractor for Chemical Analysis of Lipid Biomarkers in Regolith (ExCALiBR) for lipid analysis (Wilhelm et al., 2021).
4.1 Derivatization for gas chromatography
Pyrolysis, derivatization, and thermochemolysis have all previously been used in combination with GC for in situ organic analysis (see Section 2.1). Research is still ongoing to further understand the impact of oxidation agents such as perchlorates on the various methods. In the presence of perchlorates, pyrolysis forms chlorinated molecules and in situ derivatization with MTBSTFA or DMA/DMF is also adversely affected by perchlorates. It has been shown that fatty acids detected through thermochemolysis with TMAH remain unaffected up to 10 wt% perchlorate (Helge Mißbach et al., 2019; He et al., 2021b). However, TMAH has been found to cause isomerization of unsaturated fatty acids, resulting in additional GC peaks (Challinor, 1991). Strategies are still being formed for selecting Mars samples that are compatible with current methods; for example, samples that do not release excessive amounts of oxygen during thermal decomposition (Lewis et al., 2018). It has been shown that the commonly used MTBSTFA, DMA-DMF, and TMAH are not adversely affected by radiation corresponding to levels expected at Europa during an in situ mission (<300 krad; Freissinet et al., 2019).
Alternatives to TMAH for in situ organic detection are also being studied, such as trimethylsulfonium hydroxide (TMSH; Kivrak et al., 2021). The processing temperatures are lower, which may reduce unwanted reactions. TMSH is commonly used for the derivatization of free fatty acids prior to GC analysis (Ishida et al., 1999; Chiu and Kuo, 2020; Gries et al., 2021). However, it has been reported that TMSH is not efficient at derivatizing some of the unsaturated fatty acids (Ostermann et al., 2014). Similarly to TMAH, TMSH can also be used to derivatize other analytes, such as nucleobases (Kivrak et al., 2021).
4.2 Subcritical water extraction
Subcritical water extraction (SCWE) and conventional liquid extraction are currently the most studied extraction techniques for extracting analytes from solid planetary analog samples. The technique is also referred to as pressurized hot water extraction, superheated water extraction, pressurized liquid extraction, or accelerated solvent extraction. At elevated temperatures (>100°C), the water is maintained in its liquid state by applying pressure (see Figure 9). Increased desorption and increased solubility are the two main benefits of SCWE. The solubility of medium-polar analytes is increased at higher temperatures due to a decreased relative static permittivity (ε, also known as the dielectric constant) of water. Liquid water at 275°C has similar solubility parameters to methanol in terms of polarity, and hydrogen-bond donating and accepting abilities; hence, can dissolve even rather non-polar analytes.
FIGURE 9. Phase diagrams of carbon dioxide (top) and water (bottom). The triple points and critical points are labeled. Extraction conditions are typically maintained at 50–150°C and 15–30 MPa for supercritical fluid extraction with CO2 and 150–250°C and 2–10 MPa for subcritical water extraction. The pressure does not affect water as long as the liquid state is sustained; however, the density of supercritical CO2 depends on the pressure, which positively affects analyte solubility.
SCWE has gained particular popularity as an extraction technique for amino acids bound to their sample matrix (e.g., Amashukeli et al., 2008; Kehl et al., 2019; Brinckerhoff et al., 2022). The recovery of amino acids is influenced by desorption, hydrolysis, and degradation, which are all positively associated with increased temperatures. For example, amino acids in solid samples from the Atacama Desert could not be extracted at 30°C; however, higher recoveries were achieved at elevated temperatures up to 250°C (Amashukeli et al., 2007). The same study showed that no amino acids were detected in extracts obtained at 325°C, likely due to degradation of amino acids.
SCWE can also hydrolyze biopolymers such as proteins into a mixture of peptides and amino acids at temperatures above 200°C (Marcet et al., 2016). Complete hydrolysis is typically achieved at very acidic conditions (i.e., 6 M HCl) for approximately 24 h at ambient temperatures, or 1 h at temperatures above 160°C (Csapá et al., 1997). Noell et al. (2018) demonstrated that hydrolysis at 185°C over 20 min was improved approximately 5-fold with 10 mM of HCl (3.2% versus 15.7% recoveries of total amino acids, respectively). It is noteworthy that the recoveries of most amino acids did improve with higher temperature and longer extraction times; however, some amino acids, such as aspartic acid and glycine, degraded. SCWE is also suitable for lysing cells and spores, while simultaneously hydrolyzing proteins that can subsequently be analyzed by, for example, CE (Cieslarova et al., 2022).
Urey, an instrument based on SCWE and microchip electrophoresis (Mars Organic Analyzer, MOA, see Section 5.1), was initially under development for the Rosalind Franklin rover; however, it was descoped before the first prototype was manufactured (Amashukeli et al., 2008; Aubrey et al., 2008; Bada et al., 2008). However, a portable breadboard version of Urey was field-tested in the Atacama Desert (Skelley et al., 2007). Several follow-up studies were later published in which capillary electrophoresis—laser induced fluorescence (a separation and detection technique discussed in Section 5.1) was used to analyze amino acids extracted by SCWE with subsequent derivatization (Skelley et al., 2007; Stockton et al., 2009a; Chiesl et al., 2009). The instrument later continued developing at the Jet Propulsion Laboratory (Beegle et al., 2011; Kehl et al., 2019). The first fully automated prototype was demonstrated onboard a remotely controlled rover, as a Mars rover simulation mission, where it performed SCWE on solid samples in the Atacama Desert (Kehl et al., 2019; Mora et al., 2020). A microfluidic device with a slurry generated from a solid sample to facilitate sample transport has also been evaluated for SCWE applications (Sherrit et al., 2017). SCWE with carbonated water (5 wt% CO2, pH ∼3, 150°C) was used to extract amino acids from solid samples acquired from a geyser in Yellowstone National Park, followed by online chiral LC-MS (Abrahamsson et al., 2021).
Although (subcritical) water is efficient at extracting polar analytes, it can be used at elevated temperatures to dissolve non-polar analytes such as PAHs with limited solubility. For example, the solubility of pyrene (logP = 4.9) in water is 3 mg/L and 78 mg/L at 200°C and 300°C, respectively (Andersson et al., 2005). Consequently, large volumes of water (>8× solvent to sample ratio) are needed even for the extraction of light PAHs (Stockton et al., 2009b), and recoveries of analytes such as stigmasterol (logP = 7.0, no solubility data reported for subcritical water) are practically zero (Luong et al., 2014). It has been shown that 300°C is required to extract hydrophobic organic matter from sedimentary rocks (Luong et al., 2015). Performing extraction above 300°C is typically not desirable because of degradation issues of most analytes, particularly beyond the critical point of water (Tc = 374°C, Pc = 22 MPa). Other techniques, such as supercritical fluid extraction or liquid extraction with organic solvents, may be preferred and are discussed later.
4.3 Supercritical fluid extraction
Supercritical CO2 (scCO2) is a complementary solvent to water with similar solubility properties as n-hexane. Hence, scCO2 is well-suited for wet chemistry analysis that targets fatty acids, sterols, steranes, polycyclic aromatic compounds, and other lipids. Faster extractions are possible compared to conventional liquid extraction techniques due to higher diffusivity and zero surface tension of scCO2. The extraction temperature can also be ramped up (150°C) to desorb analytes bound to the solid sample matrix (Björklund et al., 1999), much like SCWE, except that scCO2 maintains its polarity. The solubility is directly related to the density of the CO2, which enables efficient preconcentration of analytes (e.g., on trap columns) at ambient pressures prior to downstream analysis techniques such as chromatography (Abrahamsson et al., 2019). Field-portable supercritical fluid extraction (SFE) instruments have previously been used to analyze polychlorinated biphenyls and PAHs in soils (Bøwadt et al., 1997).
One particular benefit is that salts are insoluble in neat scCO2, rendering analysis of organic biosignatures in brines and other salt-rich samples compatible with downstream techniques, such as on-line mass spectrometry. For example, coronene was extracted from Martian regolith simulant by SFE with no measurable salts in the extract (McCaig et al., 2016). Furthermore, the Martian atmosphere contains 95% CO2, which could potentially be harvested for SFE applications that use neat scCO2. It has also been proposed that dry ice in the polar regions of Mars could be utilized as a natural extraction solvent for in situ SFE (Lang et al., 2002). SFE, in parallel with SCWE, followed by supercritical fluid chromatography (SFC) and LC with mass spectrometry detection, are utilized in the Supercritical CO2 and Subcritical H2O Analysis instrument (SCHAN; Lin et al., 2021).
4.4 Ultrasound-assisted extraction
Analysis techniques targeting biopolymers benefit or require that these targeted species remain intact. High-temperature extraction may not be feasible in these situations due to decomposition (Parro et al., 2011b). Instead, liquid extraction has been evaluated with various solvent compositions and surfactants at lower temperatures. Ultrasound-assisted extraction (UAE) has been applied to increase the extraction kinetics and to lyse cells (Fykse et al., 2003; Albero et al., 2019). The same mechanics also facilitate rupturing of solid particles to increase the accessibility of analytes. The sound waves originating from a probe form short-lived microscopic vapor bubbles that implode (cavitation process) and cause shockwaves in the sample (Albero et al., 2019). Relatively short processing times, less than a few minutes, are typically applied to avoid analyte degradation. It has been shown that UAE can be as efficient as the traditional Soxhlet extraction technique for PAHs and hopanes (Blanco-Zubiaguirre et al., 2015), and that it is more efficient than microwave-assisted extraction for multiple organic biosignatures (Timoumi et al., 2022). UAE is a part of most current biosensor instruments designed for solid samples (see Section 5.3). Although UAE improves the extraction kinetics (the extraction rate), it has been suggested that it does not improve the desorption of bound analytes. For example, nucleobases adsorbed to magnesium oxide were not extracted with higher recoveries when UAE or surfactants were applied, as opposed to traditional liquid extraction (Fornaro et al., 2013).
Non-polar organic solvents such as dichloromethane and hexane are not compatible with most affinity assays (Court et al., 2010). In order to extract hydrophobic analytes, surfactants are used in aqueous solutions with or without a small fraction of organic modifier such as methanol. Common buffers for molecular biology assays are often used, such as tris-buffered saline (TBS) and Polysorbate 20, also known as Tween 20 (Parro et al., 2011b). By adding Polysorbate 80 (1.5 g/L) to a water-methanol mixture (80:20 v/v), hydrophobic analytes such as squalene, stigmasterol, and PAHs could be extracted from pre-cleaned and spiked JSC Mars-1 sample analogs with recoveries of 17%, 8%, and >49%, respectively (Court et al., 2010, 2012). Liquid extraction with surfactants generally outperforms SCWE at ≤200°C for the very hydrophobic analytes mentioned earlier (Luong et al., 2014).
One of the main issues with surfactants is that they are not stable in solution for extended periods of time (Court et al., 2014). The same authors showed that the degradation rate greatly diminished at -20°C; however, recoveries were still lower than with fresh solutions. The degradation products can form interferents that may complicate downstream analysis. Finally, it should be noted that surfactants are generally not compatible with ESI-MS due to ion suppression and the fact that they give rise to large interfering signals.
4.5 Bead beating for cell lysis
For most chemical analysis instruments to detect metabolites inside cells or components of cell membranes, these must first be made accessible and extracted. Microorganisms, including bacteria, fungi, and algae, require lysing prior to extraction. Gram-positive bacteria and spores can be difficult to lyse due to their protective cell walls and coating structure (Harrison, 1991). Bead beating is a mechanical lysing technique often used in the laboratory and is currently investigated as a sample preparation step for DNA and RNA sequencing (see Section 5.4). It has been shown that bead beating is more efficient than UAE for extracting DNA from permafrost soil samples; however, its efficiency is sample-dependent (Raymond-Bouchard et al., 2022). A complete overview of nucleic acid sample preparation, including extraction, has been given by Emaus et al. (2020).
Miniature commercial bead beating kits with a large selection of microbeads are readily available (see Figure 10). Examples of such are the OmniLyse from ClaremontBio and SuperFastPrep from MP Biomedicals. Automated small systems that combine bead beating and extraction are also available, such as SimplePrep from ClaremontBio.
FIGURE 10. A miniaturized bead beater (OmniLyse) from Claremont BioSolutions. Some bead materials can act as an adsorbent from which the analytes can be subsequently eluted with appropriate buffer. Consequently, the bead beater can be used for lysis with or without solid-phase extraction of nucleic acids. Reprinted with permission from Vandeventer et al. (2011). Copyright 2011 American Society for Microbiology.
Combined pretreatment and extraction with zirconia/silica microbeads (10 µm) in a miniaturized system that lyses cells and spores, and adsorbs DNA and RNA to the beads been has shown to be a fit-for-purpose technique (Vandeventer et al., 2011). NASA’s Wetlab-2 platform onboard the ISS used the microbead-beating technique without extraction buffers (OmniLyse from ClaremontBio) during the cell lysing step to extract and purify RNA (Parra et al., 2017). The Search for Extra-Terrestrial Genomes (SETG) instrument uses a modified Claremont BioSolutions solid-phase Purelyse bacterial genomic DNA extraction kit to isolate DNA and RNA from solid and liquid samples that are mixed with a binding buffer (low pH with ethylenediaminetetraacetic acid, EDTA; Mojarro et al., 2019). The adsorbed polynucleotides are then eluted with a high pH buffer containing EDTA. Extraction recoveries of 5%–14% from various Mars soil analogs spiked with 2.2 × 108 Bacillus subtilis spores (∼1 µg DNA) per 50 mg of sample have been achieved by a method that included desalting, competitive binders (random hexamers), microbead beating, adsorption, and elution (Carr et al., 2017; Mojarro et al., 2017). Similar performance was later demonstrated with 50 mg Mars soil analogs containing 1.6 × 104 Escherichia coli cells or 1.6 × 104 Bacillus subtilis spores (∼70 pg DNA) with recoveries ranging from 2%–15% and 5%–11%, respectively (Mojarro et al., 2019). The extraction recovery and the optimum methodology are sample-dependent.
4.6 Solid-phase extraction
Solid-phase extraction (SPE) is a commonly used technique for extraction from fluidic streams, mainly liquids, but also gas. It can also be used for sample clean-up by selectively adsorbing analytes of interest; for example, from a sample with high salt content or for preconcentrating dilute samples upstream from analytical instruments (Andrade-Eiroa et al., 2016b; Andrade-Eiroa et al., 2016a). In short, the SPE involves a small packed bed of material into which analytes adsorb or partition favorably. Subsequently, the captured analytes are most commonly released using a strong elution solvent.
Examples of applications for planetary science applications include desalting of samples containing amino acids on a micro-chip (Craft et al., 2020; Volkenburg et al., 2022), on-line preconcentration of amino acids before chromatographic analysis (Abrahamsson et al., 2021), and refocusing of extracted analytes for on-line extraction and chromatography (Abrahamsson et al., 2019).
SPE can also be used to adsorb analytes in the headspace of a sample, where small fibers are used, namely, solid-phase microextraction (SPME). The SPME-fiber can then be directly inserted into the desorption chamber of a GC-MS system. This technique has been used to identify and quantify PAHs in a Mars analog sample (Orzechowska et al., 2011).
5 Next-generation biosignature detection techniques for planetary applications
Certain trends have greatly influenced the viability of implementing various techniques, as is later shown for chip-based GC, CE, and LC. The concept of a completely integrated microfluidic system, also known as micro total analysis systems (µTAS), for sampling, sample pretreatment, separation, detection, and so on, was introduced in 1990 (Manz et al., 1990). Since then, major advances have been made in traditional manufacturing processes of microfluidic devices, such as wet etching, micromachining, micromilling, casting, hot embossing, and injection molding. Although many various materials, such as silicon, glass, polydimethylsiloxane (PDMS), poly (methyl methacrylate) (PMMA), paper, and 3D printed materials, are used for fabricating microfluidic devices, PDMS and glass remain the most commonly used (Yamamoto et al., 2021). The material must be carefully chosen depending on the application based on, for example, biocompatibility (adsorption of analytes), chemical resistance, thermal properties, physical strength, and employed detection technique. Developments are further facilitated by recent breakthroughs in 3D printing (Weisgrab et al., 2019; Balakrishnan et al., 2021). This emerging technology enables the fabrication of truly 3D devices rather than layered 2D build-up and avoids chemical bonding to form the enclosure around fluidic channels. Currently, 100 μm-sized microchannels can be manufactured by 3D printing; however, smaller sizes remain challenging (Balakrishnan et al., 2021). The recent developments and applications have been extensively reviewed (Berlanda et al., 2021; Yamamoto et al., 2021). Most of these technologies have not yet been adopted in instrument development for space flight applications but will likely do so in the upcoming decade. The advances in materials and engineering benefit many emerging technologies, such as chip-based gas chromatography, liquid chromatography, electrophoresis, nanopore sensing, and immunoassays.
The recent surge in nanotechnology has given rise to nanopore sensing, among other techniques. This has led to the development of the third-generation sequencing techniques and has enabled single-molecule detection (see Section 5.4). Advancements in electronics and specifically integrated circuits have been instrumental to nanopore sensing but also the general miniaturization of most chemical analysis instruments.
The techniques discussed in this section have never been used at any planetary body outside of Earth, and do not have spaceflight heritage in relevant environments. Consequently, the development cost of these techniques will likely be higher than for those techniques discussed earlier. However, on the basis of demonstrated performances of the next-generation techniques, these are also more capable in terms LODs and the number of organic biosignatures that can be detected. The order in which the techniques are discussed does not entail any judgment on their performance, TRL, or whether they are fit for purpose.
5.1 Capillary electrophoresis
5.1.1 Background
The first modern capillary electrophoresis (CE; inner diameters of ≤75 µm instead of ≥3 mm) instrument was developed in 1981 (Jorgenson and Lukacs, 1981), and gained tremendous popularity during the following decades (Lunte and Radzik, 1996). In essence, CE is a separation technique in which the analytes migrate in an established electric field within a channel as the primary mechanism, where the velocity depends on their electrophoretic mobility and the strength of the electric field. The secondary migration mechanism is governed by the electroosmotic flow (EOF), typically established by deprotonating the silanol groups in a fused-silica capillary, thus forming a double diffuse layer of cations that becomes the driving force of the bulk fluid within the capillary. The EOF velocity depends on the strength of the electric field and the potential between the two layers of cations (zeta potential) that is dependent on the capillary surface charge and altered by regulating the pH of the buffer. The flat flow profile in CE reduces band broadening effects, which results in very narrow peaks, unlike in pressure-induced flow like in chromatography, where the flow profile is parabolic due to friction at the walls. CE is a well-established chemical analysis technique that is particularly useful for, but not limited to, the separation of inorganic ions, nucleic acids, amino acids, peptides, proteins, and metabolites. The interested reader is referred to the following sources regarding the fundamentals and applications of CE (Cech and Enke, 2001; Harstad et al., 2016; Voeten et al., 2018; Bernardo-Bermejo et al., 2020).
CE is a widely used separation technique in many disciplines, and its development is moving rapidly, where the most exciting topics over the last 2 years have been covered by Kristoff et al. (2020). Over the last few years, a plethora of advances have been made in preconcentration and sample introduction techniques, pseudo-stationary phases such as micellar electrokinetic chromatography (MEKC), chiral selectors and two-dimensional CE (Voeten et al., 2018). Nevertheless, CE has been criticized for its lack of repeatability, reproducibility, robustness, and ruggedness (Faller and Engelhardt, 1999; Boone et al., 2000; Schaeper and Sepaniak, 2000). However, in recent years, it has matured into a highly reliable technique. In an inter-laboratory trial, the relative migration time and peak area reproducibility were below 1.4% RSD and 30% RSD, respectively (Wenz et al., 2015). It should still be noted that the absolute migration times still vary substantially due to variations in the EOF and for in situ applications, and typically, an internal standard will be needed in order to establish the relative migration time. Kehl et al. (2022) have developed an alternative approach to continuously measure the flow, to normalize migration times based on the flow rate. The authors adapted a thermal flowmeter for CE and provided flow-compensated electropherograms with 25× improved RSD of migration times.
Various modes of separation are used and combined to achieve acceptable resolution of the analytes. Often capillary zone electrophoresis (CZE) and MEKC are used in combination or in a sequence of runs to analyze various analytes such as amino acids (Creamer et al., 2017). In brief, MEKC is used to separate analytes with a neutral net charge, which is performed by adding surfactants that form micelles that act as a pseudo-stationary phase. Often a strong EOF is utilized, and the micelles have an electrophoretic mobility in the opposing direction and, therefore, migrate at a reduced speed through the capillary. The migration times of analytes will be influenced by their partitioning between the micelles and the background electrolyte—similar to conventional chromatography. Furthermore, cyclodextrin is used to obtain chiral separation of enantiomers, which is further enhanced by coupling it with MEKC (Lu and Chen, 2002).
The ionic strength and the pH of samples have a major impact on the EOF in CE. To avoid or reduce the need for separate desalting steps, a high concentration (30–80 mM) of borate buffer is typically used and further improved by adding EDTA (Stockton et al., 2009a; Creamer et al., 2017). Duca et al. (2022) used a 35 mM borate buffer as the background electrolyte, which allowed for a maximum of 1 mM H2SO4, 10 mM Na2CO2, and 10 mM MgSO4 (with 5 mM EDTA) in the injected sample before separation failed when analyzing amino acids. Alternatively, high concentrations of acetic acid (1–5 M) can be used to enable analysis of samples with high ionic strength (Santos et al., 2018; Mora et al., 2022). Samples containing up to 3 M NaCl or 1.5 M MgSO4 were successfully analyzed using 5 M of acetic acid as BGE (Mora et al., 2022). Therefore, high concentrations of acetic acid may be a way forward; however, uncertainties remain in terms of compatibility with chiral separation and other methods that rely on a strong EOF, which is heavily suppressed by the low pH.
Miniaturized and portable CE systems have frequently been reported for purposes outside of space exploration. In brief, there are six main considerations in the design and manufacturing of an in situ CE system: channel dimensions, device material, the shape of the channel path, sample injection, detection technique, and placement, and generation of the electric field (Lewis et al., 2013). The systems typically fall into two categories, microchip electrophoresis (ME) systems and conventional capillary-based electrophoresis. An overview and the trade-off between the two platforms in miniaturized and portable systems are extensively discussed by Lewis et al. (2013). The benefits of CE are that capillaries are well-characterized and provide the maximum volume-to-surface ratio due to the cylindrical geometry. However, repeatable injections can be challenging with CE through the commonly used pneumatic injection technique. Alternatively, an injector valve can improve repeatability (Zamuruyev et al., 2021). ME, on the other hand, struggles with injection bias that consequently affects the measurement trueness (Duca et al., 2022). Furthermore, the channels often consist of two or more materials that may or may not be fully characterized, ultimately leading to unknown EOF characteristics. Furthermore, the fabrication process of creating the channels also influences the properties of the surface (Pugmire et al., 2002). Consequently, method development becomes more cumbersome for ME than for CE. In both cases of CE and ME, small channel dimensions are crucial to ensure low current to reduce Joule heating and lower power consumption. However, more narrow channels reduce the sensitivity because the optical pathway is diminished when optical detection techniques are used.
5.1.2 In situ planetary applications and state-of-the-art
CE was first proposed as a potential in situ instrument for Mars (Bada and McDonald, 1996; Bada et al., 1997) and the ISS (Orta et al., 1997) already in the late 90s’, but has so far not been tested in a space environment. Whereas CE does not operate at high pressures like its chromatographic cousins, the lab-on-a-chip concept was easily conceived. The first chiral separation of derivatized amino acids (fluorescein isothiocyanate, FITC) with cyclodextrin on a ME system with 190 mm long × 150 µm wide × 20 µm deep channels on a 100 mm diameter glass wafer was demonstrated by Hutt et al. (1999). However, only a handful number of enantiomers were entirely resolved.
Since the first microchip electrophoresis (ME) instruments were developed, the research has focused on method development instead. ME for in situ analysis in space applications has almost exclusively been developed with laser-induced fluorescence (LIF) detection, and consequently, much attention has been given to optimizing labeling protocols for various target analytes. Examples of CE-LIF and ME-LIF include chiral and achiral separation of amino acids (Skelley and Mathies, 2003; Stockton et al., 2009a; Chiesl et al., 2009; Creamer et al., 2017; Fujishima et al., 2019), peptides (Fujishima et al., 2019), amines (Skelley et al., 2006; Stockton et al., 2009a; Cable et al., 2013; Cable et al., 2014a), aldehydes and ketones (Stockton et al., 2010), nucleobases (Skelley et al., 2006; Fujishima et al., 2019), carboxylic acids (Stockton et al., 2011; Cable et al., 2014b), thiols (Mora et al., 2015), and PAHs (Stockton et al., 2009b). The instrument LODs are typically in the µg/L range (see Table 2), where the most high-performance methods targeting amino acids have LODs in the range from 5 nM to 750 nM (∼0.4 μg/L up to 100 μg/L; Creamer et al., 2017), except for valine that could be detected as low as 75 pM (9 ng/L; Chiesl et al., 2009).
TABLE 2. Summary of a selection of separated analytes and their LODs (given in parentheses as nM) using electrophoresis-based instruments in development for space applications. A slash (/) indicates co-eluting peaks without separation. Derivatization occasionally produces multiple compounds (byproducts and analyte derivatives) resulting in several peaks, which is not indicated below.
LIF has almost exclusively been the detector of choice for detecting organic biosignatures. However, very few analytes of interest contain a natural chromophore; therefore, fluorescent labeling is necessary. Several issues arise when performing ultra-trace analysis (<1 ppm) in combination with derivatization. Derivatization reagents form byproducts during the reaction step that need to be separated from the labeled analyte, which in many applications is not completely viable due to the vast number and relatively high concentration of byproducts interfering with low-concentration analysis. An excess of reagent is, however, needed to ensure efficient labeling of analytes at very low concentrations. Derivatization efficiency also depends on the sample matrix (Fernández-Fı́gares et al., 2004); however, this has not been studied in regard to environmental samples. It is noteworthy that a substantial number of publications report LODs based on derivatizations performed at high concentrations that are subsequently diluted and hence only represent instrumental LODs rather than the relevant method LODs. In this vein, a considerable amount of work has been dedicated to developing derivatization protocols that provide efficient labeling at low concentrations with minimal byproduct formation.
Further development of the microchip platform has enabled on-chip derivatization, dilution, and addition of internal standard, as well as carrying a large number of solutions that are needed for each unique and selective CE method (Mora et al., 2011; Mora et al., 2012; Kim et al., 2013). These instruments, which use LIF detection, only weigh approximately 11 kg, are approximately 13 cm × 31 cm × 33 cm, and require only <15 W during operation (see Figure 11). Channel redundancy is readily implemented on a microchip, which reduces the overall risk of, for example, clogging by having several CE channels available for separation (Benhabib et al., 2010). On-line SCWE coupled with ME-LIF onboard a remotely controlled rover was used to detect five amino acids from soil in the Atacama Desert, Chile (Mora et al., 2020).
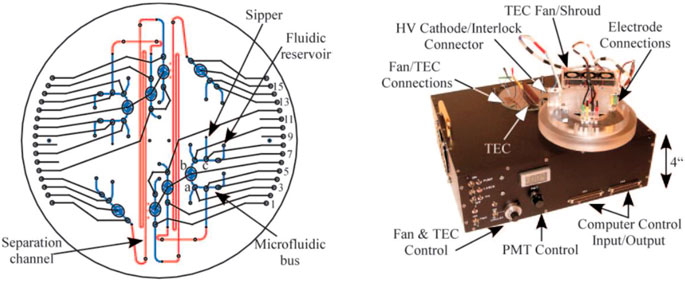
FIGURE 11. Illustration of the Mars Organic Analyzer (MOA) instrument that uses ME-LIF. In the left diagram the CE channel and other fluidic channels are highlighted. The packaged instrument is shown on the right. Reprinted from Skelley et al. (2005). Copyright 2005 National Academy of Sciences.
Capacitively coupled contactless conductivity detection (C4D) is an electrochemical detection technique that has seen tremendous growth and quick adaption in the general field of CE over the two decades (Kubáň and Hauser, 2011, 2018). C4D is well-suited for many analytes, particularly those that do not contain a chromophore or are not suitable for derivatization, and it is especially suitable for inorganic analysis. Two of the key benefits are that the electrodes are not in direct contact with the fluid hence avoiding dispersion, corrosion of the electrodes, and decoupling from the high voltages of the CE; and secondly, due to the simplicity, the detector is much smaller in size than optical-based detectors, allowing for further miniaturization. C4D in tandem with LIF (Santos et al., 2018), or MS (Beutner et al., 2018), provides additional information that is useful for identifying or resolving co-migrating species. Santos et al. (2018) demonstrated how CE-C4D could be utilized to simultaneously analyze inorganic species and amino acids. The C4D is easily incorporated onto ME platforms (Kubáň and Hauser, 2018), and has been proposed for an integrated system based on CE-C4D-LIF-MS (Creamer et al., 2020; Zamuruyev et al., 2021).
Over the last decade, CE hyphenated with MS (CE-MS), has gained mainstream popularity and has recently been conceptualized as an in situ instrument for the future exploration of the Ocean Worlds (Creamer et al., 2020; Brinckerhoff et al., 2022). A miniaturized and automated microfluidic sample delivery platform compatible with CE-MS has been developed (Figure 12; Zamuruyev et al., 2021). The multifaceted implications and considerations of the required instrumentation for CE-MS have been thoroughly described (Bonvin et al., 2012; Lindenburg et al., 2015). Applying and decoupling the voltages on the CE (>15 kV) and the ESI (<4 kV) are the primary obstacles. Typically, the outlet, which also functions as the spray tip, is grounded, and the potentials are applied at the inlet of the CE and MS, respectively. However, because capillaries are made of fused-silica (electrically insulating), the electrical connection must be made elsewhere. The connection can be achieved through a conductive liquid pumped co-axially with the CE eluent (sheath liquid), and the two liquids come in contact at the end of the CE capillary (the spray capillary tip). Alternatively, the end of the CE capillary can be made porous (e.g., by hydrofluoric acid etching), allowing contact between the two liquids (sheathless interface). In the sheathless approach, the capillary extends beyond the outer liquid, and only the solution from the CE separation is partaking in the ESI process.
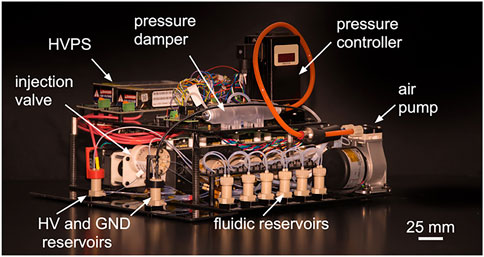
FIGURE 12. A portable and automated CE system with the main components denoted. Reprinted with permission from Zamuruyev et al. (2021). Copyright 2021 American Chemical Society.
The co-axial sheath liquid interface (2–10 μL/min) is the most commonly used approach for hyphenating CE with MS (Stolz et al., 2019). However, nano-ESI benefits are only obtained at flow rates of a few tens of nL/min or lower (Schmidt et al., 2003; Jarvas et al., 2017). The commercially available sheathless CE-ESI-MS interface (CESI, Sciex) based on the work of Moini (2007) has, over the years, gained much attention. The sheathless porous interface for CE-MS can provide these flow rates so that true nano-ESI with high sensitivity (and less influence by matrix effects) is obtained (Busnel et al., 2010).
Narrow separation capillaries (≤30 µm inner diameter) and a reduced EOF are essential to maintain flow rates within the effective nano-ESI regime. However, the EOF plays a significant role in many separation methods, especially for analytes without a net charge, and it might therefore be difficult to adapt these methods for CE-MS with true nano-ESI. As mentioned earlier, methods robust against the ionic strength utilize high background electrolyte (BGE) concentrations that either require operation in the low nanoliter/min regime or substantial dilution by an added sheath liquid. Furthermore, the ionic strength of the BGE is important as excessive current may cause the electrospray voltage to deviate from its set point depending on the interface design, and potentially cause arcing in the ionization source (Flaherty et al., 2017). Even with these challenges, CE-MS is a routine analysis method in many laboratories despite not operating in the nano-ESI regime; however, there is much room for improvement. For example, sheathless CE-MS provides LODs of cationic species such as (underivatized) amino acids and biogenic amines (e.g., spermine and spermidine) in the range of 30–1,000 nM (Hirayama et al., 2018), which are approximately 100× higher LODs than for CE-LIF. Mora et al. (2022) demonstrated that CE-MS (sheathless CESI) could detect several amino acids, nucleobases, and nucleosides with LODs of 1–2 μM, in the presence of 1 M NaCl, when using 5 M acetic acid as BGE and low injection volumes (7 nL). Nevertheless, MS provides additional resolving power for co-migrating analytes and identification potential of unknown analytes.
Long-term stability tests have shown that the fluorescent derivatization reagent carboxyfluorescein succinimidyl ester (CFSE) is stable for up to 2 years if stored at 4°C (Creamer et al., 2019). If the necessary reagents and solutions for MEKC for chiral separation of amino acids are stored dry and separated, they can withstand 300 krad TID radiation (Creamer et al., 2018). A C4D detector has been developed for high-radiation environments, and the BGE for a C4D method was shown to be stable for 2 years and survive radiation (Ferreira Santos et al., 2022).
CE is already suited for scenarios where limited sample is available, because, CE inherently uses little sample volume (low nL range) for each analysis due to the narrow capillaries or channels used for separation. Hence, ME is being developed as an instrument with the possibility of sampling plumes and detecting organic biosignatures; for applications such as a fly-by mission at Enceladus (Mathies et al., 2017). Research and development are also being made to enable an integrated ME system to withstand high-speed impacts (Stockton et al., 2016), which would be well-suited for potential future missions that rely on kinetic penetrators for in situ subsurface investigations of Ocean Worlds (Gowen et al., 2011).
Due to the early miniaturization of CE, there is a vast amount of literature regarding method development and evaluation of the fitness for purpose in space applications. Although many analyte classes can be analyzed (see Table 2), they require different derivatization and separation methods. Furthermore, the peak capacity is often limited, and relatively few analytes are separated in each run. Coupling with MS adds a mode of separation, does not require derivatization, and enables a wider range of analytes analyzed with one method. However, CE-MS is not as sensitive as ME-LIF and would be a much larger instrument when combined. Compared to the previous generation of instruments (i.e., GC-MS, fluorescence spectroscopy, and Raman spectroscopy), CE provides orders of magnitude lower LODs, especially for amino acids and other polar analytes. Engineering concerns for spaceflight applications remain rather unknown at the moment. However, several integrated and miniaturized ME-LIF systems have been reported (Kim et al., 2013; Mora et al., 2020; Golozar et al., 2022). Method demonstrations and validation have mostly been conducted on benchtop instruments with off-line sample preparation, and some caution is advised when interpreting the fitness of purpose. More research is needed with integrated, end-to-end, analysis of environmental sample analogs of Mars or Ocean Worlds.
5.2 Liquid chromatography
5.2.1 Background
It is estimated that only 20% of known organic compounds can be analyzed by GC in their native form; as in, without decomposition or derivatization to render the target analytes volatile (Snyder et al., 2011). Although GC analysis is simpler than other chromatographic and electrophoretic techniques, and offers unmatched resolution and peak capacity, LC is often the preferred method in the laboratory for less volatile analytes. Modern ultra-performance liquid chromatography (UPLC)/SFC—MS platforms offers high reproducibility; making it possible for research groups to verify, compare, and scrutinize results from in situ missions (Chocholoušková et al., 2021). This is currently not easily achievable with conventional TV/Py-GC-MS methods. There have been a limited number of reported developments and adaptions of LC for astrobiology applications. Consequently, this section covers said developments and the general status of miniaturized LC instruments primarily targeting point-of-care applications. The interested reader is encouraged to read the review series Miniaturization of Liquid Chromatography Coupled to Mass Spectrometry for a more in-depth coverage of the field (Medina et al., 2020; Mejía-Carmona et al., 2020; Vargas Medina et al., 2020).
The typical LC system consists of one or more pumps, an injector, a chromatography column, and a detector. A small volume of sample is injected into a stream of mobile phase, where the analytes are separated on the stationary phase packed inside the column based on molecular interactions. Columns packed with functionalized small particles (≤5 µm) are generally used to achieve comparable resolving power; consequently, high pressures are needed to achieve acceptable flow rates due to the flow impedance. Hence, high-performance liquid chromatography (HPLC) is often incorrectly referred to as high-pressure liquid chromatography. The most common LC technique is called reversed-phase LC, where the stationary phase is generally non-polar, and the mobile phase is mainly aqueous with an organic modifier such as methanol or acetonitrile. This mode of LC is generally used along with gradient elution—that is, when the fraction of organic solvent is increased over time to increase the elution strength. Consequently, sufficient separation of a wide range of polarities of the analytes can be achieved along with peak compression (sharper peaks).
Miniaturized separation is a relatively mature technology that mimics many of the trends seen in capillary electrophoresis. On-chip separation channels and capillary columns are readily available. However, traditional columns of 1 mm–2.1 mm inner diameter are rarely the main contributors to an LC instrument’s overall mass and volume. Nonetheless, columns with narrow IDs substantially reduce the required flow rate and solvent consumption. Miniaturized detectors based on MS, spectroscopy, and electrochemistry have been covered earlier in this review. This section mainly focuses on fluid delivery technology because it remains the most challenging component to miniaturize to render LC truly portable and suitable for in situ biosignature detection.
5.2.2 Fluid delivery systems
Mechanical pumps in commercial benchtop instruments, such as syringe and dual piston reciprocating pumps, capable of delivering 40 MPa or above, are bulky and limited to the laboratory environment. Most commercial UPLC systems are operated at flow rates of a few hundred µL/min; however, miniaturized systems are generally designed for a few µL/min down to tens of nL/min. Achieving accurate and pulse-free flow at these low flow rates is challenging. For example, in a dual piston pump, pulses arise when one piston engages in the delivery, while the other piston is retracted to refill the chamber. In order to achieve stable flow rates, modern systems use feedback control based on flow rate measured downstream (Šesták et al., 2015). The development of commercial nanoLC has not reduced the instruments’ size, but has substantially reduced the sample volume and mobile phase consumption rate (Sharma et al., 2015b).
Multiple portable LC instruments with mechanical fluid delivery sub-systems have been demonstrated; for example, using a single stepper motor-driven syringe pump that operated up to 110 MPa and weighed 1.4 kg (Sharma et al., 2014). A second pump was added to provide the option of gradient elution, with a volume of 32.5 μL in each syringe. This fluid delivery system weighed 4 kg and consumed 29 W (Sharma et al., 2015a). Subsequent developments of the same instrument resulted in a 5.9 kg LC system that utilized larger stepper motors to increase the maximum pressure and pumping accuracy (Zhao et al., 2017). A series of studies utilized syringe pumps and small breadboard valves from LabSmith that together formed portable instruments with a pressure limit of approximately <20 MPa (Li et al., 2015b; Coates et al., 2020; Lam et al., 2020; Hemida et al., 2021a; Hemida et al., 2021b). For example, Lam et al. (2020) demonstrated a portable medium-pressure capillary LC instrument (2 kg, 245 mm × 185 mm × 160 mm) that included UV/Vis absorbance detection with LODs of <10 μg/L for a few pharmaceutical compounds. The developed instrument and method provided retention time shifts of <0.1% RSD and peak areas <0.3% RSD (Lam et al., 2020). A similar instrumental setup was packaged into a briefcase (7.2 kg, 300 mm × 450 mm × 120 mm) and tested in the field along with a portable (not miniaturized) mass spectrometer for the analysis of polyfluorinated compounds in environmental samples (Hemida et al., 2021a).
A conceptually different approach uses a high-pressure gas source to drive the mobile phase in a reservoir. This approach enabled the detection of PAHs with high flowrate stability (0.29% RSD at 148.8 μL/min, limited to isocratic elution) with a portable instrument weighing 4.2 kg, including 150 ml of mobile phase and UV/Vis absorbance detection (Chatzimichail et al., 2021). Constant-pressure fluid delivery systems operating at lower pressures have been demonstrated multiple times (Wang et al., 2009, 2010; Kiplagat et al., 2010).
Electrochemical pumps without moving parts have been under development for over 20 years (Lynch et al., 2018; Rahimi et al., 2020). They provide an impressive footprint that can fit onto a chip, and are free of moving parts. Electroosmotic pumps (EOPs) offer pulse-free flow where the direction and the flow rate are easily manipulated by varying the applied potential and its polarity. The technique has been demonstrated up to 120 MPa (Gu et al., 2012), and has generally been used to deliver a few hundreds of nL/min (Haghighi et al., 2018). Lynch et al. (2017) demonstrated a typical implementation of EOP in a miniaturized instrument of about 3 kg (excluding detector) capable of gradient elution. Despite the potential of EOPs, it has not yet reached commercial and mainstream success, partly due to the need for high voltage (typically several kV), incompatibility with high organic content mobile phases, flowrate fluctuation, and chemical breakdown within the pumping elements (Haghighi et al., 2018; Lynch et al., 2018). Electrolysis-based fluid delivery for low-pressure chip-based LC has also been demonstrated (Xie et al., 2005). Far too many on-chip fluidic delivery system concepts have been developed to be accurately covered here, and the reader is referred to the comprehensive review by Haghighi et al. (2018). However, to our knowledge, none of these are currently in development for space applications.
Open-tubular columns, as opposed to conventional packed columns, do not require as high pressures to function due to lower flow impedance. This provides an opportunity to use smaller mechanical pumps or pneumatic systems based on constant pressure (Kiplagat et al., 2010; Shelor et al., 2014). This approach has been adopted for an ion-exchange LC instrument intended for in situ astrobiology applications (e.g., Shelor et al., 2014).
5.2.3 Astrobiology applications and state-of-the-art
Compared to the other techniques discussed in this manuscript, there have been relatively few reported studies on the viability of LC for in situ organic biosignature detection; likely due to the difficulties in manufacturing small pump devices that are mechanically rugged for space application. The reported literature with an astrobiology focus can be classified into four categories: analysis of analog samples using benchtop instrumentation, on-chip reversed-phase LC and ESI, supercritical fluid chromatography, and miniaturized ion chromatography (see Table 3).

TABLE 3. Summary of a selection of separated analytes and their LODs (given in parentheses as µg/L, unless stated otherwise) using liquid and supercritical-based chromatography instruments in development for space applications.
For example, benchtop LC-MS was used to detect amino acids, dipeptides, and sugars from a cometary ice analog (Eddhif et al., 2018). A similar study that also incorporated a trap-column prior to separation, also known as column-switching methodology, showed that amino acids, oligopeptides, and nucleobases could be separated and detected at approximately 1 μg/L (Ribette et al., 2019). Liu et al. (2008) used ion-pair reversed-phase LC coupled with MS to detect underivatized amino acids with instrument LODs ranging from 3 μg/L to 800 μg/L. The authors used the developed methodology to analyze amino acids from Mono Lake, Death Valley, and Haughton Crater. LC has been extensively used to characterize meteorites, including chiral amino acids, nucleobases, and other biosignature precursors (e.g., Glavin et al., 2012a; Burton et al., 2012; Ruf et al., 2019; Oba et al., 2020). Most methods require gradient elution and frequently use derivatization or ion-pairing to analyze amino acids. Abrahamsson et al. (2021) developed a column-switching method with strong cation exchange as SPE followed by enantiomeric separation on a chiral stationary phase using only carbonated water as a mobile phase in an isocratic method. LODs determined with MS detection were in the range of 0.5–500 μg/L (median LOD was 2.5 μg/L), and 12 of the 18 chiral pairs tested were separated. The same study also demonstrated on-line SCWE applied to a spicular sinter sample from Lemon Spring in Yellowstone National Park (Abrahamsson et al., 2021).
On-chip columns with integrated ESI capabilities are readily available commercially, such as the ionKey/MS (Murphy et al., 2014), Trizaic nanoTile (Hughes et al., 2011), HPLC-Chip (Yin and Killeen, 2007), and cHiPLC-nanoflex (Hebert et al., 2011). Some of these commercial options include on-chip trapping (column switching), and on-chip flow channels that are interfaced with an external rotor to form valves; however, none incorporate on-chip injection or fluid delivery. Similar devices have also been developed for in situ organic analysis applications (Getty et al., 2013; Southard et al., 2014; Southard et al., 2016a; Southard et al., 2016b). As with any nano-fluidic device; clogging, detecting tiny leaks, and extra-column dispersion remain practical challenges due to miniaturization (Desmet and Eeltink, 2013; Sharma et al., 2015b; Desmet and Broeckhoven, 2019).
SFC is another avenue currently being pursued. Although it is not strictly classified as an LC technique, modern SFC with packed columns is very similar to conventional LC (Tarafder, 2016). The main difference is that CO2, with or without co-solvents, is used as the mobile phase in its subcritical or supercritical state. Due to the non-polar nature of scCO2 (similar to hexane), it is ideal for extracting and separating lipids. Abrahamsson et al. (2019) developed a novel SFE-SFC-MS method for analyzing free fatty acids and polycyclic aromatic compounds from aqueous samples. These techniques, along with conventional LC, are used in the Supercritical CO2 and Subcritical H2O Analysis instrument (SCHAN; Abrahamsson et al., 2022), an organic biosignature analysis instrument, which is a part of the payload on an astrobiology-focused mission concept to the Mars north polar region, Mars Astrobiology Science Exploration (MASEX; Lin et al., 2021).
Another area of research currently geared towards in situ chemical analysis is ion chromatography. Although it is not strictly a technique for organic biosignature detection, it does enable the analysis of small ionic organic compounds in addition to inorganic compounds. Analysis of both inorganic and small organic cations (e.g., alkali metals, amines, and alkanolamines) and anions (e.g., chloride, oxychlorides, and organic acids) are readily analyzed by ion chromatography coupled with conductivity measurements (Shelor et al., 2014; Shelor and Dasgupta, 2017). Separation is achieved with a purely aqueous mobile phase that is modified; for example, with carbon dioxide or an inorganic base (Shelor and Dasgupta, 2017; Shelor et al., 2017; Sricharoen et al., 2019). Detection limits of <100 μg/L are obtained for many small ionic analytes (Shelor et al., 2014; Shelor and Dasgupta, 2017; Sricharoen et al., 2019). Ion chromatography also allows for the analysis of perchlorates (Shelor et al., 2014), which has been of particular interest on Mars (e.g., Hecht et al., 2009; Glavin et al., 2013).
Ion chromatography differs from other LC techniques, such as reversed-phase LC, by the mode of separation and by the required hardware. In addition to a conventional HPLC system, adding a mobile phase generator and an ion suppressor is often desirable. The mobile phase generator, as the name suggests, increasingly adds the modifier to the aqueous mobile phase to increase the elution strength over time, providing gradient elution. The ion suppressor lowers the ionic strength of the mobile phase prior to conductivity measurements by neutralizing the acid or base that was added by the mobile phase generator. Ion exchange membranes are instrumental to this end, and their role in analytical chemistry has been extensively reviewed (Dasgupta and Maleki, 2019). These components have been demonstrated at a miniaturized scale. For example, Chouhan et al. (2020) demonstrated a small hydroxide eluent generator suitable for flowrates in the nL regime and capable of gradient elution.
In the pursuit of a portable or in situ small ion analysis instrument, open tubular columns have been the preferred choice. The commercially available capillaries of various materials with narrow inner diameter (<50 µm) have increased tremendously over the last decade, which offers the analytical chemist more options in method development. Open tubular columns’ benefits are extremely high theoretical separation efficiency, low flow impedance, and simplicity. However, the loading capacity is very limited compared to packed columns due to the much lower surface area. For example, 4 nL of injection volume is slightly too large for 25 µm inner diameter capillary columns (Chouhan et al., 2020). Lower injection volumes are currently difficult to achieve with acceptable repeatability. Naturally, the optimum injection volume also depends on the ionic strength of the sample. Due to the low flow impedance, less than 1 MPa is needed to achieve flowrates <1 μL/min using a <1 m capillary column with 25 µm inner diameter (Huang et al., 2021).
Bare open tubular capillary columns, such as those described for ion chromatography applications, circumvent many challenges relating to high-pressure fluid delivery systems. The approach is also applicable to conventional LC techniques, and other capillary column variants such as wall-coated open tubular and porous layer open tubular columns, and these may in the future offer a compromise between chromatographic efficiency, loading capacity, solvent usage, and backpressure (Xiang et al., 2021).
In summary, LC offers many of the same capabilities as CE regarding targeted analyte classes and LODs. LC achieves orders of magnitude lower LODs compared to techniques previously used in spaceflight applications. Derivatization may be used but is often not required for LC analysis, which reduces some risks and issues with sample matrix effects. However, the technique has been relatively unexplored for spaceflight applications due to issues with miniaturization in the previous decade, particularly regarding fluid delivery systems. Adaptation for spaceflight is currently ongoing, and a miniaturized integrated system was recently reported (Abrahamsson et al., 2022). More validation and demonstrations would increase the confidence in the suitability for spaceflight adaption.
5.3 Biosensors
5.3.1 Background
Bioreceptors in analytical chemistry have extremely high affinity and specificity for analytes of interest and form biosensors when combined with a detection scheme. Antibodies, nucleic acids, cellular structures, biomimetic materials, and molecular imprinted polymers are the most commonly used receptors in the laboratory. Methods and instruments in development for space applications have primarily adopted antibodies and nucleic acids, also known as aptamers, for sensitive organic biosignature detection—these are covered in this section. However, there is a vast number of alternative bioreceptors or affinity receptors that are not covered here. A few noteworthy instruments that have been developed, or are under development, are the Life Marker Chip (LMC; Sims et al., 2005; sims et al., 2012; Martins, 2011), the Signs Of Life Detector (SOLID; Parro et al., 2011b; Moreno-Paz et al., 2018), the Biochip for Organic Matter Analysis in Space (BiOMAS; Baqué et al., 2011b), and the Planetary Life Explorer with Integrated Analytical Detection and Embedded Sensors (PLEIADES) chip (Nascetti et al., 2019). The LMC mainly targeted small organic molecules (ca. <1,000 Da), and the SOLID instrument has focused on larger biomolecules (ca. >1,000 Da). The LMC was initially intended to be a part of the Rosalind Franklin rover instrument suite, but was later descoped.
Antibodies are the conventional subset of protein-based receptors. The variable region known as the complementarity-determining region (CDR), which consists of an amino acid sequence, has a high affinity towards a target antigen (analyte) through non-covalent interactions (Lipman et al., 2005). The specific site to which an antibody binds is called an epitope. An antigen may have multiple epitopes to which different antibodies bind. Furthermore, epitopes may also be very similar, causing the same antibody to bind several different antigens and consequently producing false positives, also known as cross-reactivity.
Antibodies can be obtained as monoclonal antibodies (mAbs) that are produced in vitro using tissue-culture techniques or as polyclonal antibodies that are easily obtained through the production in animals, typically rabbit. Monoclonal antibodies ideally only bind to one antigen and therefore have very high specificity. The process of identifying and producing monoclonal antibodies is, unfortunately, very laborious. On the other hand, polyclonal antibodies contain multiple epitope specificities. Whether to use monoclonal or polyclonal antibodies is usually decided by their commercial availability or availability from other researchers. If an antibody is not available off-the-shelf, then polyclonal antibodies are rather easily produced within a few months, contrary to monoclonal antibodies that take years and require more expertise to produce (Lipman et al., 2005).
Aptamers are an alternative to antibodies. These are typically single-stranded oligonucleotides (single-stranded DNA or RNA) that may have high specificity for a range of target molecules such as nucleic acids, proteins, metal ions, and other small molecules.
The antibodies and aptamers can be used in various configurations for detecting analytes of interest, with either mobile (in solution) or immobilized antigens or bioreceptors, with or without fluorescent labeling. The various configurations relevant for life detection have been reviewed elsewhere (Parro et al., 2008b). The following is a short overview of the two most commonly used methodologies: the sandwich and competitive assays.
Instruments that target large molecules (e.g., proteins and other biopolymers) primarily utilize a sandwich immunoassay where the immobilized bioreceptors bind to an antigen that is in solution, followed by the addition of fluorescently-labeled antibodies that also bind to the antigen to form a sandwich (Parro et al., 2008b). Sample and fluorescent probes are washed off in multiple steps. Consequently, bright spots will arise under UV excitation where an antigen has been sandwiched. The immobilized antibodies are sometimes referred to as capturing antibodies, and the fluorescently labeled antibodies are detector antibodies. This methodology requires that an antigen carry two similar epitopes (i.e., larger molecules) to generate a positive result. The assay is useful for detecting whole cells, high molecular weight complexes (organelles, membranes, cellular debris, and humic substances), biological polymers (proteins, polysaccharides, and nucleic acids), and organo-mineral complexes (Blanco et al., 2015). An example of a complete protocol for multiplex fluorescent antibody microarray that includes analysis of cross-reaction and unspecific binding has been reported by Blanco et al. (2015).
Smaller molecules with only one epitope require a fluorescent competitive inhibition assay. In the competitive assay, a conjugate of the analyte of interest and a carrier protein (e.g., bovine serum albumin) is immobilized on a solid support. The immobilized conjugates act as a capturing probe for fluorescent antibodies. When the sample is added, the analyte present will bind to the fluorescent-labeled antibodies that will be washed out in the following step; because, they no longer bind to the immobilized conjugate. Contrarily to the sandwich array, the competitive assay will produce lower fluorescent signal with higher analyte concentration as the fluorescent probes are washed off. Competitive assays do not provide as low LODs as sandwich assays (Parro et al., 2011b).
Multiplex detection (i.e., detection of multiple analytes) is performed by applying a cocktail solution to all spots simultaneously, where each spot contains affinity receptors for each analyte (see Figure 13). Such operation generally comprises injection of the sample to a flow cell containing the microarray, incubation, washing, dissolution, and addition of a fluorescent antibody cocktail from accessory chambers, incubation, washing, fluorescent excitation with a laser beam, and image capture with a CCD camera. Such methodology has, for example, been described in detail by Parro et al. (2008a). As the number of target analytes increases, the sensitivity decreases and the risk of cross-reactivity needs to be more carefully monitored (Rivas et al., 2008). Thus, it is essential to factor in that LODs deteriorates as the number of target analytes increases, and the LODs reported for a single analyte assay are not necessarily representative.
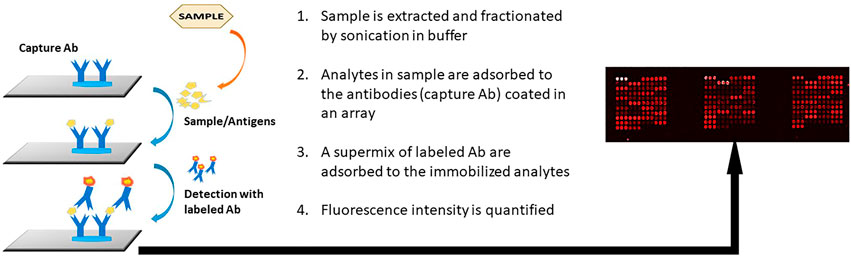
FIGURE 13. The principles of a sandwich-based immunoassay (left) and an illustration of the acquired data from a microarray where each antibody is printed in triplicate. Adapted from García-Descalzo et al. (2019).
More recent developments include utilizing autonomous capillary force-driven microfluidic networks and chemiluminescence bioassays instead of fluorescent probes (Roda et al., 2018; Nascetti et al., 2019; Zangheri et al., 2019). Chemiluminescence detection, where light is generated through a chemical reaction, provides a few distinct advantages, such as not needing an excitation light source, and consequently, there is no light scattering or autofluorescence from interfering sample components (Roda et al., 2016).
5.3.2 Applications
Antibodies can be produced against specific molecules, environmental extracts, or whole cells (see Table 4). For example, Parro et al. (2008b) have focused on producing antibodies against analytes in extracts from the acidic, iron-, and sulfur-rich Rio Tinto area. This area serves as a Mars analog, and chemolithoautotrophic bacteria such as Leptospirillum ferrooxidans and Acidithiobacillus ferrooxidans are dominant in these samples. These bacteria can subsequently rapidly be detected and identified in unknown samples (Parro et al., 2005). The multiplex sandwich array has mainly been used to pursue detecting microbial communities, which has been demonstrated multiple times with extracts of samples from extreme environments such as the Atacama Desert (Parro et al., 2011a; Blanco et al., 2013; Crits-Christoph et al., 2013; Fernández-Martínez et al., 2019), Antarctica (Lezcano et al., 2019), and other sites (Martin-Cuadrado et al., 2019). Unlike other techniques discussed in this review, immunoassays with receptors against cells do not require a cell lysing step prior to detection (Rivas et al., 2008).
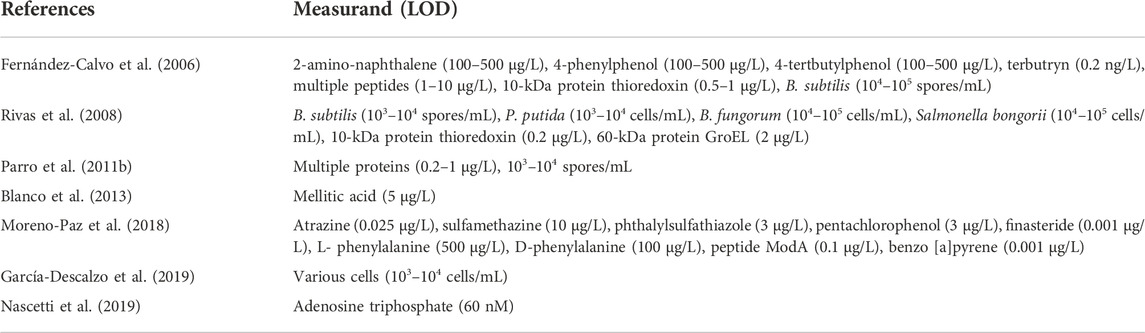
TABLE 4. A brief selection of detected measurands and their LODs (given in parentheses) using biosensors being developed for space applications.
The first stereoselective antibody against phenylalanine was demonstrated by Kassa et al. (2011). Later chiral discrimination of amino acids was adopted in the development plan of the LMC (Sims et al., 2012). A multiplex inhibitor immunoassay with specific antibodies towards D- or L-aromatic amino acids showed LODs of 100 μg/L and 500 μg/L, respectively (Moreno-Paz et al., 2018). The reported method did not discriminate between aromatic amino acids; specifically, it could not differentiate between phenylalanine and tryptophan. Additionally, other aromatic compounds were detected, such as finasteride and benzo [a]pyrene. The authors applied the method to extracts from Atacama Desert and kerogen-containing samples.
It was demonstrated with an early version of the SOLID instrument that some proteins (GroEL, thioredoxin, and glutathione-S-transferase) in solution could be detected at 10 μg/L (Parro et al., 2005). Another study using the same technique could detect 5 μg/L of mellitic acid, which was then successfully applied to extracts from Atacama Desert samples (Blanco et al., 2013). Smaller analytes such as naphthalene, 4-phenylphenol, and 4-tertbutylphenol were detected with LODs between 100 μg/L and 500 μg/L, and approximately 104–105 spores/mL with an inhibition methodology (Fernández-Calvo et al., 2006). Subsequent improvements enabled the detection of 103–104 spores/mL, approximately 1 μg/L of peptides, and 0.2–1 μg/L of thioredoxin with an instrument (SOLID3) that could perform both competitive and sandwich assays (Parro et al., 2011b).
The current state-of-the-art instrumentation designed for astrobiology applications contains <300 specific affinity receptors that include both antibodies and aptamers (Rivas et al., 2008; Parro et al., 2011b; Fairén et al., 2020). As mentioned earlier, the risks of unwanted interfering background, false-positive signals, and a reduction of LODs increase as the number of biosensors increase along with the increased complexity of the cocktail solution containing detector antibodies. This is exemplified by an order of magnitude increased LODs (from 103–104 to 104–105 spores/mL) when using a 200 antibody multiplex array compared to an assay with individual antibodies (Rivas et al., 2008). The SOLID3 instrument currently weighs 7 kg, including optics and sample preparation (ultrasonic-assisted extraction), whereas the sample analysis unit only weighs 1 kg (Parro et al., 2011b). The instrument is capable of performing both competitive and sandwich assays.
An example of a chemiluminescence bioassay includes a capillary force-driven device based on chemiluminescence that was used onboard the ISS to measure the health of an astronaut by quantifying the level of cortisol in saliva with an LOD of 0.2 μg/L (Zangheri et al., 2019). However, its potential for in situ space applications has remained relatively undocumented.
Compared to CE and LC, biosensors provide a very different approach to organic biosignature detection. The specificity is much higher than for separation-based chemical analysis techniques; however, the targeted analytes also have to be specified; an intrinsic difficulty unless it is clear precisely which biosignatures should be targeted. LODs are comparable with the other techniques discussed in this section; however, one tremendous benefit is that biopolymers (including those of whole cells) can easily be detected. Complete end-to-end instruments, including sample preparation, have been manufactured and demonstrated. Besides the limited number of analytes (≤300) that can be analyzed at once, the stability of the biological receptors has presented some minor drawbacks. However, the stability of the biosensors in space environments has been scrutinized to a much larger degree than the other techniques (CE, LC, and nanopores) and is discussed separately below.
5.3.3 Environmental testing for spaceflight applications
Multiple studies have investigated the impact of radiation, thermal cycling, and long-time storage on affinity sensors (Le Postollec et al., 2009a; Le Postollec et al., 2009b; Baqué et al., 2011a; Baqué et al., 2011b; de Diego-Castilla et al., 2011; Derveni et al., 2012; Derveni et al., 2013; Vigier et al., 2013; Coussot et al., 2019b). In summary, these studies showed that antibodies and aptamers were resistant to radiation and maintained their performance. However, it was observed that grafted (immobilized) antibodies were more sensitive to general storage parameters (Baqué et al., 2011a). Antibodies are stable for extended periods of time when they are freeze-dried (Chang et al., 2005; Wang et al., 2007). Generally, the storage conditions such as temperature and moisture appear to be more critical than levels of radiation exposure.
Although ground-based radiation testing is valuable, it does not capture the reality of varying radiation fluxes, energies, and types during long-term storage and thermal variations. Two studies evaluated affinity sensors (i.e., antibodies and aptamers) that were stored for 566 days outside of the ISS (Vigier et al., 2013; Coussot et al., 2019a; Coussot et al., 2019b). Extensive validation and functionality testing were performed with a previously developed methodology (Coussot et al., 2018a; Coussot et al., 2018b; Coussot et al., 2018c). In summary, antibodies (anti-horseradish peroxidase Ab) or aptamers were evaluated either as covalently immobilized (grafted) or in the free form. The samples stored outside the ISS were exposed to 220 mGy. It was shown that both immobilized and free antibodies retained at least 40% of the binding efficiency after being exposed to freeze-drying, long-term storage, temperature fluctuations, and radiation (Coussot et al., 2019a). Similar experiments with aptamers showed that at least 50% retained their functionality after the entire study (Coussot et al., 2019b). The same study, complemented with ground-based experiments, suggested that radiation has minimal influence on assay performance; however, long-term storage with thermal cycles has a more considerable impact on the sensitivity. The chemiluminescence approach will be tested onboard a CubeSat mission, where the effect of radiation will be monitored (Brucato et al., 2021).
5.4 Nanopore sensing
5.4.1 Background
Earlier research and instrument designs have focused on detecting the presence of specific DNA or RNA by polymerase chain reaction (PCR), followed by staining and fluorescent detection (Thiel et al., 2011; Carr et al., 2013). Primarily, the well-conserved 16S and 23S ribosomal RNA (rRNA) has been of great interest. For example, within the ca. 1,500 nucleotides of the 16S rRNA, there are several 15–20-nucleotide segments that are almost identical in all known organisms on Earth (Pace et al., 1999). This fact has sparked interest in investigating the possibility that life first evolved on Mars while it was potentially habitable and organisms were transferred to Earth through meteorites (Gladman et al., 1996; Mileikowsky et al., 2000; Benner and Kim, 2015; Carr, 2022). The oldest DNA that has been recovered was over one million years old (van der Valk et al., 2021), which corresponds to the theoretical limit based on current lower detection limits of current techniques and the degradation rate of DNA in the most favorable conditions on Earth (Willerslev et al., 2004). Due to the lower temperatures on Mars and Ocean Worlds, it is possible that nucleic acids might survive even longer.
The development of third-generation sequencing (e.g., single-molecule real-time sequencing and nanopore sequencing) has dramatically improved portability and speed. The third-generation sequencing performs longer nucleotide sequence reads at a single molecule level as opposed to the second-generation, which involves fragmentation of long strands followed by amplification and synthesis from which the sequence is derived (Bleidorn, 2016). Hence, DNA and RNA can be detected and sequenced without amplification by PCR.
Nanopore sequencers, in general, use biological nanopores such as α-Hemolysin, MspA, CsgG, and Aerolysin nanopore proteins (Zhou et al., 2020). The most widely utilized and tested platform is the MinION from Oxford Nanopore Technologies (ONT), which is currently pocket-sized (105 mm × 23 mm × 33 mm). In nanopore sequencing, single-stranded DNA or RNA is driven through a small orifice (10−9 m in diameter), located in a membrane, by electrophoresis (Rezzonico, 2014). The electric field is held constant, and the ionic current is measured. Any polymer occupying the pore’s space will partially block the flow of ions across the membrane. The ionic current depends on the nucleic acid composition that occupies the space within the nanopore from which the sequence can be derived as the DNA or RNA passes through.
Currently, ONT recommends a minimum input of 400 ng of nucleic acids without PCR and 1 ng of nucleic acids with PCR for sequencing with the MinION. However, low-input sequencing of 2 pg of DNA has been demonstrated without PCR (Mojarro et al., 2019). The long reads from single molecules are particularly useful for taxonomic identification at, or below, the species level (Benítez-Páez et al., 2016). Although nanopore sequencing at high concentrations of DNA or RNA can be achieved within minutes (Wang et al., 2017), lower amounts (on the order of pg) may require longer runs up to multiple days (e.g., Mojarro et al., 2019); depending on throughput goals. In addition, the stability of protein-based nanopores and data processing workflow have improved, almost removing any spurious signals that could cause false-positive results; however, with adequately set noise thresholds, this risk is eliminated (Mojarro et al., 2018; Pontefract et al., 2018).
Due to favorable LODs, low power, and low mass, nanopores have quickly been adopted for instrument development that targets metagenomics (Rezzonico, 2014; Carr et al., 2016). Two key features of nanopore sequencing without PCR amplification include that it avoids PCR bias (Sabina and Leamon, 2015), and the ability to sequence non-standard bases, also known as xeno nucleic acids (XNA; Carr et al., 2017).
Nanopores can also be used to partially sequence proteins. Currently, neither nanopore nor tunneling techniques (see below) can distinguish all of the 20 different amino acids in terrestrial proteins (Restrepo-Pérez et al., 2018). However, in one study, 13 out of 20 proteinogenic amino acids have been successfully differentiated in a protein sequence of a single molecule in an aerolysin nanopore with the help of a polycationic carrier—namely, arginine heptapeptide (Ouldali et al., 2020).
Biological pores often require tight control of the feed solution, including pH, ionic strength, and temperature. Any deviation may adversely affect its performance. Solid-state nanopores, on the contrary, are more chemically, thermally, and mechanically stable (Haque et al., 2013). The holes can be manufactured with an electron beam from a transmission electron microscope (Danda and Drndić, 2019), among other methods. Currently, solid-state nanopores have limited spatial and temporal resolution, where the translation of DNA is too fast to generate more than a few data points (Yuan et al., 2020). A tradeoff exists between temporal resolution and signal-to-noise ratio, based on the bandwidth of the current measurement. At this moment, 10 MHz bandwidth (100 ns measurements) is the fastest demonstrated, with a dwell time of ∼2−21 ns per nucleotide (Chien et al., 2019). Therefore, solid-state nanopores can determine the approximate diameter (based on the current) and the approximate polymer length (duration of signal) of, for example, DNA (Xia et al., 2022). Another demonstration detected ∼1-nm-size charged particulates (including DNA) in pre-filtered Antarctic soil extracts using silicon nitride pores (Niedzwiecki et al., 2020).
Nanogaps are closely related to nanopore-based techniques (see Figure 14), where the tunneling current is measured between nanoelectrodes, located in nanopores or on a surface, rather than the ionic current across the membrane. The technique is used in an early-stage instrument called Electronic Life-detection Instrument for Enceladus/Europa (ELIE; Carr et al., 2020b; Lee, 2021). A thorough technical introduction and description of applications of detection by quantum tunneling and nanogaps have been given by Di Ventra and Taniguchi (2016), and a review covering the fundamentals has been given by (Lindsay et al., 2010). Whereas biological nanopores and solid-state nanopores are efficient for detecting single polymers such as DNA, RNA, or proteins (Restrepo-Pérez et al., 2018), their capabilities are limited regarding small molecules. However, sensing over unmodified nanogaps enables the detection of single small molecules with low selectivity (Taniguchi, 2015). Additionally, the analyte interaction with a nanogap is on the order of milliseconds (equivalent to the translation speed of biological nanopores), compared to nanoseconds for typical solid-state nanopore applications, reducing the bandwidth requirement.
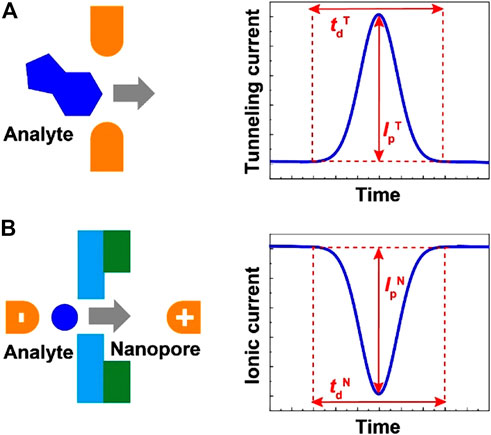
FIGURE 14. (A) The nanogap technique. A signal is acquired when a molecule passes the gap, allowing tunneling. The nanogap can be located in a nanopore or on a surface, and the analyte does not necessarily need to pass through a membrane. Analyte-nanogap interactions are on the order of milliseconds, which gives rise to a square-looking signal. (B) The nanopore technique. The current is decreased when an analyte occupies the pore’s space. In the case of solid-state nanopores (illustrated here), the event occurs on the order of nanoseconds and the order of milliseconds for biological nanopores. Nanopores and nanogaps give chemical information about the analyte that may facilitate identification, derived from the signal’s duration and intensity. Reprinted from Taniguchi (2020).
Even though nanogap sensing with functionalized electrodes has demonstrated that multiple amino acids can be distinguished, data complexity remains a challenge because each analyte can orient in various ways giving rise to several unique current signals (Zhao et al., 2014). Hence, chemometric techniques such as machine learning are needed to correctly classify each signal (Taniguchi, 2020). Finally, the emergence of nanodevices for single molecule detection has opened up multiple alternatives that have not yet been pursued for spaceflight applications; these have been reviewed elsewhere (Ohshiro and Taniguchi, 2022).
5.4.2 Applications
Many demonstrations with ONT’s MinION have shown its capability of sequencing DNA and RNA in remote locations such as Atacama Desert (Bywaters et al., 2016), Antarctic Dry Valleys (Johnson et al., 2017), paleochannels (Maggiori et al., 2020), and Arctic Permafrost Ice (Goordial et al., 2017). The MinION was also tested under Mars-like conditions at −60°C, 100% CO2, and pressures between 400 and 500 Pa (Carr et al., 2019). Successful demonstrations have been performed during and after vibration exposure under microgravity (Castro-Wallace et al., 2017; Carr et al., 2020a). In its current format, it is not hampered by radiation it would experience during travel to Mars; however, additional shielding and transitioning from protein-based pores to solid-state pores is likely required for missions to Ocean Worlds (Sutton et al., 2019). Nanopore sequencing has been used onboard the ISS (Castro-Wallace et al., 2017; Burton et al., 2020). However, it is still unknown whether biological nanopores could survive the multiple years of a space mission.
The third-generation sequencing has certainly overcome previous issues for in situ detection of informational polymers in terms of power and mass. However, the main remaining challenges relate to the extraction and perseverance of native nucleic acids from complex and oxidative samples. In terrestrial applications, workup of environmental samples includes filtering aqueous samples to remove debris and removing enzyme inhibitors such as humic acid prior to amplification or sequencing (Tringe and Rubin, 2005). Additionally, cells and spores need to be lysed to render the DNA or RNA accessible (see Section 4.5). Purification of nucleic acid also tends to co-purify other organic material that may interfere with downstream analysis (Tringe and Rubin, 2005).
A capable in situ instrument will need on-line sample pretreatment, extraction, purification, library preparation, and analysis (sequencing)—end-to-end or “sample to sequence.” The many steps with various solutions make developing a fully automated and on-line instrument challenging. A concept for an end-to-end system and its challenges is described by Bhattaru. (2018), Bhattaru et al. (2019). Besides desalting, extraction, and purification, the methodology must also include library preparation prior to sequencing. Library preparation is a multistep procedure that includes, for example: enzymatic DNA repair, the addition of adenine to both ends of the ssDNA, multiple cleanup steps that use magnetic beads, and the addition of a motor protein at the end of the DNA or RNA fragments (Quick, 2018). The overall fluid management system is also prone to bubble formation, which needs to be specifically addressed (Parra et al., 2017; Bhattaru, 2018; Bhattaru et al., 2019). Nucleic acids are also prone to adsorption throughout the fluid management system, where such losses impact the overall LOD of the system (Bhattaru, 2018).
So far, only manual sample handling has been demonstrated, and end-to-end analysis is still left as work for the future. Through decoupled experiments, it has been established that the LOD could be as low as 104 spores per 50 mg of sample without amplification (Mojarro et al., 2019).
Nanopore and nanogap sensing is the latest addition to the techniques with spaceflight potential. Biological nanopores fill a void in required capabilities that other techniques cannot provide, namely, analysis of informational biopolymers down to 2 pg (e.g., DNA and RNA). Due to the fundamental difference, nanopores cannot be directly compared to conventional spectroscopy and separation-based analysis techniques, and should be considered an orthogonal technique. Although the small biological nanopore sequencing unit has been tested in relevant environments to a limited degree, it requires extensive sample preparation, which has not yet been sufficiently demonstrated on-line. Considering that the concept is relatively young, this may be resolved in the near future. Due to extensive interest and development in other research fields, solid-state nanopores and nanogaps may take a more prominent role; however, this remains to be seen.
6 Concluding remarks
The search for signatures of extinct or extant life in Solar System bodies continues to be a challenging endeavor, even five decades after the Viking missions to Mars. Adapting instruments that fully automate rather complicated workflow, otherwise conducted in multiple steps by scientists in the laboratory, while fulfilling highly stringent size, weight, and power requirements, is not an easy task. This is an exciting new era for life detection in planetary science where detecting organic biosignatures such as informational polymers (e.g., DNA), proteins, amino acids, and various lipids at the 1 ppb level or even lower is now within our grasp, which is far beyond any capabilities that existed one or two decades ago.
Generally speaking, there has been a paradigm shift from the 70s’ Viking missions where Py/GC-MS constituted the main payload, to more recent Mars lander missions where TV-GC/MS equipped with derivatization capability has played a prominent role. On the basis of current trends in the literature, we are now in the middle of a new paradigm shift towards the next-generation instruments for organic biosignatures that are liquid-based analytical chemistry techniques such as CE, LC, biosensors, and nanopores. These techniques and instruments provide lower detection limits and a more comprehensive range of biosignatures to be targeted for interrogation than their predecessors. However, the realization of this shift remains to be seen depending on payload selection for future missions to Mars, Enceladus, Europa, Ceres, Titan, and other worlds.
Standoff spectroscopy techniques (e.g., fluorescence and Raman spectroscopy) that can identify fine-scale features will likely continue to gain importance in future missions. These instruments do not currently provide required detection limits or sufficient chemical information for the identification of organic biosignatures. However, their simplicity and ability to interrogate a vast number of samples will render them invaluable for sample selection for further analysis by instruments that are limited by the number of samples that can be analyzed.
On the basis of the reviewed literature, the next-generation biosignature detection techniques for planetary applications (CE, LC, biosensors, and nanopore sensing) are mostly fit for science requirements in mission concepts that include life detection (Pappalardo et al., 2013; Hand et al., 2017; MacKenzie et al., 2022a; MacKenzie et al., 2022b; Williams and Muirhead, 2022). Current GC-MS, fluorescence, and Raman spectroscopy instruments cannot achieve the required LODs. However, it is possible that additional sample preparation (extraction and preconcentration) and sample introduction that provides sharper peaks could remedy some of the current weaknesses (e.g., matrix effects and insufficient LODs); such efforts have not yet been reported. Nevertheless, future space missions to Ocean Worlds (e.g., Europa and Enceladus) will have different requirements than those for Mars, such as the ability to analyze aqueous samples, operate at higher vacuum, lower temperatures, high radiation environments, and with reduced data bandwidth. Within that context, the discussed techniques and instruments would require substantial development, despite having flight heritage.
We conclude that CE (including ME), LC, biosensors, and nanopore sensing have potential as payload instruments in future space missions. They have similar orders of magnitude LODs; however, this varies from one analyte to another. CE and LC are more suited for small molecules (<1,000 Da), but larger molecules can be hydrolyzed, which has been demonstrated with subcritical water. Both techniques can separate multiple enantiomeric amino acids, unlike biosensors. CE-C4D and ion chromatography can analyze inorganic and small organic ions. CE requires multiple solutions for derivatization, rinsing, and BGE; on the other hand, only low-pressure pumping is required. LC does typically not rely on derivatization but, in most cases, requires high-pressure fluid delivery. Furthermore, LC and CE can both be coupled with MS for improved separation and identification; however, most applications have utilized ME-LIF analysis, which provides a smaller format, and has predominantly focused on amino acid analysis.
Nanopore sensing is the most suitable technique for detecting and sequencing informational biopolymers (e.g., DNA and RNA), which is impossible with CE and LC. Currently, biological nanopores are required for sequencing, but these are not as stable as solid-state nanopores. However, the sample preparation is more complicated than the other techniques and has only partially been demonstrated in an on-line setup. Biosensors are capable of detecting both small (<1,000 Da), large (>1,000 Da) molecules, and intact microbes. Most receptors have high specificity, and deciding on specific target biosignatures is problematic under the assumption that extraterrestrial life may be different from Earth. Finally, we cannot accurately assess the readiness for flight because environmental testing is not always reported in the literature. Nonetheless, CE, LC, biosensors, and nanopore sensing have been miniaturized without substantial loss in performance (e.g., separation efficiency and LODs) or even with increased performance. However, complete adaptation to spaceflight followed by extensive validation is required to determine the ultimate fitness of purpose.
It is also apparent that the next generation of life detection instruments is being scrutinized to a much higher degree than their predecessors. There is a lack of publicly available validation data for the previously flown GC-based instruments (Poinot and Geffroy-Rodier, 2015). Nonetheless, it is important to note that current instruments in development are rarely validated in an end-to-end fashion; namely, automated analysis from the point that the primary sample is loaded, including sample preparation. The task of validation is becoming an increasingly cumbersome effort as the targeted compounds and sample types (i.e., ice, water, liquid methane, etc.) have increased with the number of relatively recently targeted planetary bodies (e.g., Europa, Enceladus, and Titan) in the Solar System. Historically, it has become apparent that sample matrices may have unanticipated effects in the analysis of organic molecules, such as strong oxidizing effects of perchlorates on organic molecules generated during pyrolysis and thermal volatilization at Mars. Many other disciplines where analytical chemistry has a prominent role, such as pharmaceutical, food, and forensic sciences, have precise guidelines on conducting method validation to ensure reliable results (Raposo and Ibelli-Bianco, 2020). This kind of framework does not currently exist in the domain of astrobiology. We recommend that such a framework be established so instruments can be appropriately compared and avoid representing misleading performance metrics.
The emergence of high-resolution mass spectrometry and more capable separation techniques opens new possibilities for so-called non-targeted analysis (Milman and Zhurkovich, 2017). There is a wide range of potential chemical biosignatures of interest, and they may include several thousands of chemical compounds. This calls for chemical analysis techniques that are non-targeted that enable the detection and identification of compounds that have not been considered a priori (Ruf et al., 2018). A combined approach of comprehensive non-targeted analysis and suspect screening may be the most beneficial, where the latter includes a list of high-priority targets as outlined by The Ladder of Life Detection (Neveu et al., 2018).
No analysis technique is truly universal, and each has its strengths and weaknesses. Therefore, combining different instruments, each utilizing different but complementary detection techniques, into a single instrument suite capable of uniquely identifying biosignatures is necessary. These efforts have already been pursued in the last few years. For example, EMILI, which includes CE, GC, MS, pH, and conductivity measurements (Brinckerhoff et al., 2022), the Complex Molecules Detector (CMOLD) that includes microscopy, Raman spectroscopy, and a biosensor array (Fairén et al., 2020), and the Mine Analogue Research (MINAR 5) project that is not an instrument but a wide array of sample acquisition instruments, spectroscopic instruments, biosensors, nanopore sequencing, and so on (Cockell et al., 2019).
Four techniques being developed for in situ organic biosignature detection have been identified. More may follow with possible breakthroughs in analytical chemistry in the future. However, additional development is still needed before they are spaceflight-ready. Additional end-to-end validation with miniaturized assemblies is, in many cases, still needed to prove the fitness of purpose, which is also the case for current spaceflight instruments. Nevertheless, proposed mission concepts are becoming more demanding in their measurement requirements, and the continued development of the next-generation biosignature detection techniques may be the favored path to provide an answer if there is extraterrestrial life in the Solar System.
Author contributions
VA and IK both contributed to writing, revising, and editing this manuscript.
Funding
The synthesis of this article was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration (80NM0018D0004). Funding from the NASA Astrobiology Institute—Icy Worlds Program and the JPL Senior Research Scientist Funding Program from the JPL Office of Chief Scientist is gratefully acknowledged.
Acknowledgments
Bryana Henderson, Jessica Weber, and Robert Treuhaft are thanked for providing valuable input during the writing of this manuscript. We are grateful to the two reviewers for their careful reading of our manuscript, and their many insightful comments and suggestions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbey, W. J., Bhartia, R., Beegle, L. W., DeFlores, L., Paez, V., Sijapati, K., et al. (2017). Deep UV Raman spectroscopy for planetary exploration: The search for in situ organics. Icarus 290, 201–214. doi:10.1016/j.icarus.2017.01.039
Abedin, M. N., Bradley, A. T., Misra, A. K., Bai, Y., Hines, G. D., and Sharma, S. K. (2018). Standoff ultracompact micro-Raman sensor for planetary surface explorations. Appl. Opt. 57 (1), 62–68. doi:10.1364/AO.57.000062
Abrahamsson, V., Henderson, B. L., Herman, J., Zhong, F., Lin, Y., Kanik, I., et al. (2021). Extraction and separation of chiral amino acids for life detection on Ocean Worlds without using organic solvents or derivatization. Astrobiology 21 (5), 575–586. doi:10.1089/ast.2020.2298
Abrahamsson, V., Henderson, B. L., Zhong, F., Lin, Y., and Kanik, I. (2019). Online supercritical fluid extraction and chromatography of biomarkers analysis in aqueous samples for in situ planetary applications. Anal. Bioanal. Chem. 411 (30), 8091–8101. doi:10.1007/s00216-019-02189-z
Abrahamsson, V., Henderson, B. L., Zhong, F., Prothmann, J., Lin, Y., and Kanik, I. (2022). “Schan: Supercritical CO2 and subcritical H2O analysis instrument for in-situ detection of organics and life,” in 2022 astrobiology science conference (Washington, D.C.: AGU).
Abrahamsson, V., Ristic, N., Franz, K., and Van Geem, K. (2017). Comprehensive two-dimensional gas chromatography in combination with pixel-based analysis for fouling tendency prediction. J. Chromatogr. A 1501, 89–98. doi:10.1016/j.chroma.2017.04.021
Aerts, J. W., Röling, W. F. M., Elsaesser, A., and Ehrenfreund, P. (2014). Biota and biomolecules in extreme environments on Earth: Implications for life detection on Mars. Life 4 (4), 535–565. doi:10.3390/life4040535
Aguzzi, J., Flexas, M. d. M., Flögel, S., Lo Iacono, C., Tangherlini, M., Costa, C., et al. (2020). Exo-ocean exploration with deep-sea sensor and platform technologies. Astrobiology 20 (7), 897–915. doi:10.1089/ast.2019.2129
Ahrens, A., Hitzemann, M., and Zimmermann, S. (2019). Miniaturized high-performance drift tube ion mobility spectrometer. Int. J. Ion. Mobil. Spectrom. 22 (2), 77–83. doi:10.1007/s12127-019-00248-w
Akapo, S. O., Dimandja, J. M., Kojiro, D. R., Valentin, J. R., and Carle, G. C. (1999). Gas chromatography in space. J. Chromatogr. A 843 (1-2), 147–162. doi:10.1016/s0021-9673(98)00947-9
Albero, B., Tadeo, J. L., and Perez, R. A. (2019). Ultrasound-assisted extraction of organic contaminants. TrAC Trends Anal. Chem. 118, 739–750. doi:10.1016/j.trac.2019.07.007
Allwood, A. C., Wade, L. A., Foote, M. C., Elam, W. T., Hurowitz, J. A., Battel, S., et al. (2020). Pixl: Planetary instrument for X-ray Lithochemistry. Space Sci. Rev. 216 (8), 134. doi:10.1007/s11214-020-00767-7
Amashukeli, X., Grunthaner, F. J., Patrick, S. B., and Yung, P. T. (2008). Subcritical water extractor for Mars analog soil analysis. Astrobiology 8 (3), 597–604. doi:10.1089/ast.2007.0154
Amashukeli, X., Pelletier, C. C., Kirby, J. P., and Grunthaner, F. J. (2007). Subcritical water extraction of amino acids from Atacama Desert soils. J. Geophys. Res. 112 (4), 308. doi:10.1029/2006JG000308
Anderson, D. M., Biemann, K., Orgel, L. E., Oro, J., Owen, T., Shulman, G. P., et al. (1972). Mass spectrometric analysis of organic compounds, water and volatile constituents in the atmosphere and surface of Mars: The Viking Mars Lander. Icarus 16 (1), 111–138. doi:10.1016/0019-1035(72)90140-6
Andersson, T. A., Hartonen, K. M., and Riekkola, M.-L. (2005). Solubility of acenaphthene, anthracene, and pyrene in water at 50 °C to 300 °C. J. Chem. Eng. Data 50 (4), 1177–1183. doi:10.1021/je0495886
Andrade-Eiroa, A., Canle, M., Leroy-Cancellieri, V., and Cerdà, V. (2016a). Solid-phase extraction of organic compounds: A critical review (Part I). TrAC Trends Anal. Chem. 80, 641–654. doi:10.1016/j.trac.2015.08.015
Andrade-Eiroa, A., Canle, M., Leroy-Cancellieri, V., and Cerdà, V. (2016b). Solid-phase extraction of organic compounds: A critical review. Part ii. TrAC Trends Anal. Chem. 80, 655–667. doi:10.1016/j.trac.2015.08.014
Arevalo, R., Brinckerhoff, W., van Amerom, F., Danell, R., Pinnick, V., Li, X., et al. (2015). “Design and demonstration of the mars organic molecule analyzer (MOMA) on the ExoMars 2018 rover,” in 2015 IEEE aerospace conference (Piscataway, NJ: IEEE), 1–11.
Arevalo, R., Danell, R., Gundersen, C., Hovmand, L., Southard, A., Tan, F., et al. (2016). “Advanced resolution organic molecule analyzer (AROMA): Simulations, development and initial testing of a linear ion trap-Orbitrap instrument for space,” in 3rd international workshop on instrumentation for planetary mission (Houston, TX: Lunar and Planetary Institute).
Arevalo, R., Ni, Z., and Danell, R. M. (2020). Mass spectrometry and planetary exploration: A brief review and future projection. J. Mass Spectrom. 55 (1), e4454. doi:10.1002/jms.4454
Arevalo, R., Selliez, L., Briois, C., Carrasco, N., Thirkell, L., Cherville, B., et al. (2018). An Orbitrap-based laser desorption/ablation mass spectrometer designed for spaceflight. Rapid Commun. Mass Spectrom. 32 (21), 1875–1886. doi:10.1002/rcm.8244
Asher, S. A., and Johnson, C. R. (1984). Raman spectroscopy of a coal liquid shows that fluorescence interference is minimized with ultraviolet excitation. Science 225 (4659), 311–313. doi:10.1126/science.6740313
Aubrey, A. D., Chalmers, J. H., Bada, J. L., Grunthaner, F. J., Amashukeli, X., Willis, P., et al. (2008). The Urey instrument: An advanced in situ organic and oxidant detector for Mars exploration. Astrobiology 8 (3), 583–595. doi:10.1089/ast.2007.0169
Azov, V. A., Mueller, L., and Makarov, A. A. (2022). Laser ionization mass spectrometry at 55: Quo Vadis? Mass Spectrom. Rev. 41 (1), 100–151. doi:10.1002/mas.21669
Bada, J., Becker, L., and McDonald, G. (1997). “For which compounds do we search in extraterrestrial samples for evidence of abiotic and/or biotic chemistry?,” in Optical science, engineering and instrumentation (Bellingham, WA: SPIE), 97.
Bada, J. L., Ehrenfreund, P., Grunthaner, F., Blaney, D., Coleman, M., Farrington, A., et al. (2008). Urey: Mars organic and oxidant detector. Space Sci. Rev. 135 (1-4), 269–279. doi:10.1007/s11214-007-9213-3
Bada, J. L., and McDonald, G. D. (1996). Peer reviewed: Detecting amino acids on Mars. Anal. Chem. 68 (21), 668A–673A. doi:10.1021/ac9621231
Balakrishnan, H. K., Badar, F., Doeven, E. H., Novak, J. I., Merenda, A., Dumée, L. F., et al. (2021). 3D printing: An alternative microfabrication approach with unprecedented opportunities in design. Anal. Chem. 93 (1), 350–366. doi:10.1021/acs.analchem.0c04672
Baqué, M., Le Postollec, A., Coussot, G., Moreau, T., Desvignes, I., Incerti, S., et al. (2011a). Biochip for astrobiological applications: Investigation of low energy protons effects on antibody performances. Planet. Space Sci. 59 (13), 1490–1497. doi:10.1016/j.pss.2011.06.009
Baqué, M., Le Postollec, A., Ravelet, C., Peyrin, E., Coussot, G., Desvignes, I., et al. (2011b). Investigation of low-energy proton effects on aptamer performance for astrobiological applications. Astrobiology 11 (3), 207–211. doi:10.1089/ast.2010.0520
Barge, L. M., Rodriguez, L. E., Weber, J. M., and Theiling, B. P. (2021). Determining the “biosignature threshold” for life detection on biotic, abiotic, or prebiotic worlds. Astrobiology 22 (4), 481–493. doi:10.1089/ast.2021.0079
Beegle, L., Bhartia, R., White, M., DeFlores, L., Abbey, W., Yen-Hung, W., et al. (2015). “Sherloc: Scanning habitable environments with Raman & luminescence for organics & chemicals,” in 2015 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Beegle, L., Kirby, J. P., Fisher, A., Hodyss, R., Saltzman, A., Soto, J., et al. (2011). “Sample handling and processing on Mars for future astrobiology missions,” in 2011 aerospace conference (New York, NY: IEEE), 1.
Beegle, L. W., Kanik, I., Matz, L., and Hill, H. H. (2001). Electrospray ionization high-resolution ion mobility spectrometry for the detection of organic compounds, 1. Amino acids. Anal. Chem. 73 (13), 3028–3034. doi:10.1021/ac001519g
Bell, J. F., Maki, J. N., Mehall, G. L., Ravine, M. A., Caplinger, M. A., Bailey, Z. J., et al. (2021). The mars 2020 perseverance rover mast camera zoom (Mastcam-Z) multispectral, stereoscopic imaging investigation. Space Sci. Rev. 217 (1), 24. doi:10.1007/s11214-020-00755-x
Benhabib, M., Chiesl, T. N., Stockton, A. M., Scherer, J. R., and Mathies, R. A. (2010). Multichannel capillary electrophoresis microdevice and instrumentation for in situ planetary analysis of organic molecules and biomarkers. Anal. Chem. 82 (6), 2372–2379. doi:10.1021/ac9025994
Benítez-Páez, A., Portune, K. J., and Sanz, Y. (2016). Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION™ portable nanopore sequencer. GigaScience 5 (1), 4–9. doi:10.1186/s13742-016-0111-z
Benner, S. A., Devine, K. G., Matveeva, L. N., and Powell, D. H. (2000). The missing organic molecules on Mars. Proc. Natl. Acad. Sci. U. S. A. 97 (6), 2425–2430. doi:10.1073/pnas.040539497
Benner, S., and Kim, H.-J. (2015). “The case for a Martian origin for Earth life,” in SPIE optical engineering + applications (Bellingham, WA: SPIE).
Berlanda, S. F., Breitfeld, M., Dietsche, C. L., and Dittrich, P. S. (2021). Recent advances in microfluidic technology for bioanalysis and diagnostics. Anal. Chem. 93 (1), 311–331. doi:10.1021/acs.analchem.0c04366
Bernardo-Bermejo, S., Sánchez-López, E., Castro-Puyana, M., and Marina, M. L. (2020). Chiral capillary electrophoresis. TrAC Trends Anal. Chem. 124, 115807. doi:10.1016/j.trac.2020.115807
Beutner, A., Piendl, S. K., Wert, S., and Matysik, F.-M. (2018). Methodical studies of the simultaneous determination of anions and cations by IC× CE–MS using arsenic species as model analytes. Anal. Bioanal. Chem. 410 (24), 6321–6330. doi:10.1007/s00216-018-1241-1
Bhartia, R., Hug, W. F., Salas, E. C., Reid, R. D., Sijapati, K. K., Tsapin, A., et al. (2008). Classification of organic and biological materials with deep ultraviolet excitation. Appl. Spectrosc. 62 (10), 1070–1077. doi:10.1366/000370208786049123
Bhartia, R., Salas, E. C., Hug, W. F., Reid, R. D., Lane, A. L., Edwards, K. J., et al. (2010). Label-free bacterial imaging with deep-UV-laser-induced native fluorescence. Appl. Environ. Microbiol. 76 (21), 7231–7237. doi:10.1128/AEM.00943-10
Bhattaru, S. A. (2018). Design, testing, and validation of the search for Extra-Terrestrial Genomes instrument. Master’s thesis. Cambridge (MA): Massachusetts Institute of Technology.
Bhattaru, S. A., Tani, J., Saboda, K., Borowsky, J., Ruvkun, G., Zuber, M. T., et al. (2019). “Development of a nucleic acid-based life detection instrument Testbed,” in 2019 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Biemann, K., and Bada, J. L. (2011). Comment on “Reanalysis of the Viking results suggests perchlorate and organics at midlatitudes on Mars” by Rafael Navarro-González et al. J. Geophys. Res. 116 (E12), E12001. doi:10.1029/2011JE003869
Biemann, K., and Lavoie, J. M. (1979). Some final conclusions and supporting experiments related to the search for organic compounds on the surface of Mars. J. Geophys. Res. 84 (B14), 8385–8390. doi:10.1029/JB084iB14p08385
Biemann, K. (2007). On the ability of the Viking gas chromatograph–mass spectrometer to detect organic matter. Proc. Natl. Acad. Sci. U. S. A. 104 (25), 10310–10313. doi:10.1073/pnas.0703732104
Biemann, K., Oro, J., Toulmin, P., Orgel, L. E., Nier, A. O., Anderson, D. M., et al. (1976). Search for organic and volatile inorganic compounds in two surface samples from the chryse planitia region of Mars. Science 194 (4260), 72–76. doi:10.1126/science.194.4260.72
Biemann, K., Oro, P., Toulmin, P., Orgel, L. E., Nier, A. O., Anderson, D. M., et al. (1977). The search for organic substances and inorganic volatile compounds in the surface of Mars. J. Geophys. Res. 82 (28), 4641–4658. doi:10.1029/JS082i028p04641
Bishop, J. L., Franz, H. B., Goetz, W., Blake, D. F., Freissinet, C., Steininger, H., et al. (2013). Coordinated analyses of Antarctic sediments as Mars analog materials using reflectance spectroscopy and current flight-like instruments for CheMin, SAM and MOMA. Icarus 224 (2), 309–325. doi:10.1016/j.icarus.2012.05.014
Björklund, E., Bøwadt, S., Mathiasson, L., and Hawthorne, S. B. (1999). Determining PCB sorption/desorption behavior on sediments using selective supercritical fluid extraction. 1. Desorption from historically contaminated samples. Environ. Sci. Technol. 33 (13), 2193–2203. doi:10.1021/es981071p
Blacksberg, J., Alerstam, E., Cochrane, C. J., Maruyama, Y., and Farmer, J. D. (2020). Miniature high-speed, low-pulse-energy picosecond Raman spectrometer for identification of minerals and organics in planetary science. Appl. Opt. 59 (2), 433–444. doi:10.1364/AO.59.000433
Blanco, Y., Moreno-Paz, M., Aguirre, J., and Parro, V. (2015). “Multiplex fluorescent antibody microarrays and antibody graphs for microbial and biomarker detection in the environment,” in Hydrocarbon and lipid Microbiology protocols. Editors T. J. McGenity, K. N. Timmis, and B. Nogales (Heidelberg: Springer Berlin), 207–224.
Blanco, Y., Rivas, L. A., Ruiz-Bermejo, M., and Parro, V. (2013). Immunological detection of mellitic acid in the Atacama desert: Implication for organics detection on Mars. Icarus 224 (2), 326–333. doi:10.1016/j.icarus.2012.06.006
Blanco-Zubiaguirre, L., Arrieta, N., Iturregui, A., Martinez-Arkarazo, I., Olivares, M., Castro, K., et al. (2015). Focused ultrasound solid–liquid extraction for the determination of organic biomarkers in beachrocks. Ultrason. Sonochem. 27, 430–439. doi:10.1016/j.ultsonch.2015.06.012
Blase, R. C., Libardoni, M. J., Miller, G. P., Miller, K. E., Phillips-Lander, C. M., Glein, C. R., et al. (2022). MEMS GC column performance for analyzing organics and biological molecules for future landed planetary missions. Front. Astron. Space Sci. 9, 1–19. doi:10.3389/fspas.2022.828103
Blase, R. C., Libardoni, M. J., Miller, G. P., Miller, K. E., Phillips-Lander, C. M., Waite, J. H., et al. (2020b). Experimental coupling of a MEMS gas chromatograph and a mass spectrometer for organic analysis in space environments. ACS Earth Space Chem. 4 (10), 1718–1729. doi:10.1021/acsearthspacechem.0c00131
Blase, R. C., Libardoni, M., Miller, K., Waite, J. H., Glein, C., Miller, G., et al. (2020a6009). “MAss spectrometer for planetary EXploration-ORganic composition analyzer (MASPEX-ORCA) for europa lander,” in Outer planets assessment group (OPAG) 2020 (Houston, TX: Lunar and Planetary Institute).
Bleidorn, C. (2016). Third generation sequencing: Technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers. 14 (1), 1–8. doi:10.1080/14772000.2015.1099575
Bonvin, G., Schappler, J., and Rudaz, S. (2012). Capillary electrophoresis–electrospray ionization-mass spectrometry interfaces: Fundamental concepts and technical developments. J. Chromatogr. A 1267, 17–31. doi:10.1016/j.chroma.2012.07.019
Boone, C. M., Franke, J. P., de Zeeuw, R. A., and Ensing, K. (2000). Intra- and interinstrument reproducibility of migration parameters in capillary electrophoresis for substance identification in systematic toxicological analysis. Electrophoresis, 21, 15452–15516. doi:10.1002/(SICI)1522-2683(20000501)21:8<1545::AID-ELPS1545>3.0
Botta, O., Bada, J. L., Gomez-Elvira, J., Javaux, E., Selsis, F., and Summons, R. (2008). Strategies of life detection": Summary and outlook. Space Sci. Rev. 135 (1-4), 371–380. doi:10.1007/s11214-008-9357-9
Bøwadt, S., Mazeas, L., Miller, D. J., and Hawthorne, S. B. (1997). Field-portable determination of polychlorinated biphenyls and polynuclear aromatic hydrocarbons in soil using supercritical fluid extraction. J. Chromatogr. A 785 (1-2), 205–217. doi:10.1016/S0021-9673(97)00571-2
Bower, D. M., Steele, A., Fries, M. D., and Kater, L. (2013). Micro Raman spectroscopy of carbonaceous material in microfossils and meteorites: Improving a method for life detection. Astrobiology 13 (1), 103–113. doi:10.1089/ast.2012.0865
Brinckerhoff, W. B., Pinnick, V. T., Amerom, F. H. W. v., Danell, R. M., Arevalo, R. D., Atanassova, M. S., et al. (2013). “Mars organic molecule analyzer (MOMA) mass spectrometer for ExoMars 2018 and beyond,” in 2013 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Brinckerhoff, W. B., Willis, P. A., Ricco, A. J., Kaplan, D. A., Danell, R. M., Grubisic, A., et al. (2022). Europan molecular indicators of life investigation (EMILI) for a future europa lander mission. Front. Space Technol. 2. doi:10.3389/frspt.2021.760927
Brinckerhoff, W. B., Willis, P. A., Stern, J. C., Grubisic, A., Mora, M. F., Creamer, J. S., et al. (2019). “Europan molecular indicators of life investigation (EMILI) technology development,” in 2019 astrobiology science conference (Washington, D.C.: American Geophysical Union).
Briois, C., Thissen, R., Thirkell, L., Aradj, K., Bouabdellah, A., Boukrara, A., et al. (2016). Orbitrap mass analyser for in situ characterisation of planetary environments: Performance evaluation of a laboratory prototype. Planet. Space Sci. 131, 33–45. doi:10.1016/j.pss.2016.06.012
Brockwell, T. G., Meech, K. J., Pickens, K., Waite, J. H., Miller, G., Roberts, J., et al. (2016). “The mass spectrometer for planetary exploration (MASPEX),” in 2016 IEEE aerospace conference (Piscataway, NJ: IEEE), 1–17.
Brucato, J., Nascetti, A., Iannascoli, L., Meneghin, A., Paglialunga, D., Poggiali, G., et al. (2021). “Astrobio-cubesat: A highly integrated laboratory to test in space immunoassay techniques to detect biomolecules,” in Lunar and planetary science conference (Houston, TX: Lunar and Planetary Science Institute), 2494.
Buch, A., Sternberg, R., Meunier, D., Rodier, C., Laurent, C., Raulin, F., et al. (2003). Solvent extraction of organic molecules of exobiological interest for in situ analysis of the Martian soil. J. Chromatogr. A 999 (1), 165–174. doi:10.1016/S0021-9673(03)00494-1
Burton, A. S., Stahl, S. E., John, K. K., Jain, M., Juul, S., Turner, D. J., et al. (2020). Off Earth identification of bacterial populations using 16S rDNA nanopore sequencing. Genes. 11 (1), 76. doi:10.3390/genes11010076
Burton, A. S., Stern, J. C., Elsila, J. E., Glavin, D. P., and Dworkin, J. P. (2012). Understanding prebiotic chemistry through the analysis of extraterrestrial amino acids and nucleobases in meteorites. Chem. Soc. Rev. 41 (16), 5459–5472. doi:10.1039/c2cs35109a
Busnel, J.-M., Schoenmaker, B., Ramautar, R., Carrasco-Pancorbo, A., Ratnayake, C., Feitelson, J. S., et al. (2010). High capacity capillary electrophoresis-electrospray ionization mass spectrometry: Coupling a porous sheathless interface with transient-isotachophoresis. Anal. Chem. 82 (22), 9476–9483. doi:10.1021/ac102159d
Bywaters, K., McKay, C., Davila, A., and Quinn, R. (2016). “In situ life and biosignature detection at Mars analog sites using the Oxford Nanopore Minion sequencer,” in Biosignature preservation and detection in Mars analog environments (Houston, TX: Lunar and Planetary Science Institute).
Cable, M. L., Hörst, S. M., He, C., Stockton, A. M., Mora, M. F., Tolbert, M. A., et al. (2014a). Identification of primary amines in Titan tholins using microchip nonaqueous capillary electrophoresis. Earth Planet. Sci. Lett. 403, 99–107. doi:10.1016/j.epsl.2014.06.028
Cable, M. L., Stockton, A. M., Mora, M. F., Hand, K. P., and Willis, P. A. (2014b). Microchip nonaqueous capillary electrophoresis of saturated fatty acids using a new fluorescent dye. Anal. Methods 6 (24), 9532–9535. doi:10.1039/c4ay01243g
Cable, M. L., Stockton, A. M., Mora, M. F., and Willis, P. A. (2013). Low-temperature microchip nonaqueous capillary electrophoresis of aliphatic primary amines: Applications to Titan chemistry. Anal. Chem. 85 (2), 1124–1131. doi:10.1021/ac3030202
Carr, C. E. (2022). Resolving the history of life on Earth by seeking life as we know it on Mars. Astrobiology 22 (7), 880–888. doi:10.1089/ast.2021.0043
Carr, C. E., Bryan, N. C., Saboda, K. N., Bhattaru, S. A., Ruvkun, G., and Zuber, M. T. (2020a). Nanopore sequencing at Mars, Europa, and microgravity conditions. NPJ Microgravity 6, 24. doi:10.1038/s41526-020-00113-9
Carr, C. E., Duzdevich, D., Szostak, J. W., Lee, S., Taniguchi, M., Ohshiro, T., et al. (2020b). “Electronic life-detection instrument for Enceladus/Europa (ELIE),” in AGU fall meeting 2020 (Washington, D.C.: American Geophysical Union), P044.
Carr, C. E., Mojarro, A., Hachey, J., Saboda, K., Tani, J., Bhattaru, S. A., et al. (2017). “Towards in situ sequencing for life detection,” in 2017 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Carr, C. E., Mojarro, A., Tani, J., Bhattaru, S. A., Zuber, M. T., Doebler, R., et al. (2016). “Advancing the search for extra-terrestrial genomes,” in 2016 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Carr, C. E., Rowedder, H., Vafadari, C., Lui, C. S., Cascio, E., Zuber, M. T., et al. (2013). Radiation resistance of biological reagents for in situ life detection. Astrobiology 13 (1), 68–78. doi:10.1089/ast.2012.0869
Carr, C. E., Saboda, K., Mojarro, A., Bhattaru, S. A., Hachey, J., Ruvkun, G., et al. (2019). “Nucleic acid sequencing under Mars-like conditions,” in 2019 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Carrier, B. L., Abbey, W. J., Beegle, L. W., Bhartia, R., and Liu, Y. (2019). Attenuation of ultraviolet radiation in rocks and minerals: Implications for Mars science. J. Geophys. Res. Planets 124 (10), 2599–2612. doi:10.1029/2018JE005758
Carstea, E. M., Popa, C. L., Baker, A., and Bridgeman, J. (2020). In situ fluorescence measurements of dissolved organic matter: A review. Sci. Total Environ. 699, 134361. doi:10.1016/j.scitotenv.2019.134361
Castro-Wallace, S. L., Chiu, C. Y., John, K. K., Stahl, S. E., Rubins, K. H., McIntyre, A. B., et al. (2017). Nanopore DNA sequencing and genome assembly on the international space station. Sci. Rep. 7 (1), 1–12. doi:10.1038/s41598-017-18364-0
Cech, N. B., and Enke, C. G. (2001). Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 20 (6), 362–387. doi:10.1002/mas.10008
Challinor, J. (1991). The scope of pyrolysis methylation reactions. J. Anal. Appl. Pyrolysis 20, 15–24. doi:10.1016/0165-2370(91)80059-H
Chang, L. L., Shepherd, D., Sun, J., Tang, X. C., and Pikal, M. J. (2005). Effect of sorbitol and residual moisture on the stability of lyophilized antibodies: Implications for the mechanism of protein stabilization in the solid state. J. Pharm. Sci. 94 (7), 1445–1455. doi:10.1002/jps.20363
Chatzimichail, S., Rahimi, F., Saifuddin, A., Surman, A. J., Taylor-Robinson, S. D., and Salehi-Reyhani, A. (2021). Hand-portable HPLC with broadband spectral detection enables analysis of complex polycyclic aromatic hydrocarbon mixtures. Commun. Chem. 4 (1), 17. doi:10.1038/s42004-021-00457-7
Chien, C.-C., Shekar, S., Niedzwiecki, D. J., Shepard, K. L., and Drndic, M. (2019). Single-stranded DNA translocation recordings through solid-state nanopores on glass chips at 10 MHz measurement bandwidth. ACS Nano 13 (9), 10545–10554. doi:10.1021/acsnano.9b04626
Chiesl, T. N., Chu, W. K., Stockton, A. M., Amashukeli, X., Grunthaner, F., and Mathies, R. A. (2009). Enhanced amine and amino acid analysis using Pacific Blue and the Mars Organic Analyzer microchip capillary electrophoresis system. Anal. Chem. 81 (7), 2537–2544. doi:10.1021/ac8023334
Chiluwal, U., Lee, G., Rajapakse, M. Y., Willy, T., Lukow, S., Schmidt, H., et al. (2019). Tandem ion mobility spectrometry at ambient pressure and field decomposition of mobility selected ions of explosives and interferences. Analyst 144 (6), 2052–2061. doi:10.1039/C8AN02041H
Chin, T. N., Lee, A. K., Boone, T. D., Tan, M. X., Chin, M. M., McCutcheon, G. C., et al. (2017). “Sample processor for life on icy worlds (SPLIce): Design and test results,” in International conference on miniaturized systems for chemistry and life sciences (MicroTAS 2017) (San Diego, CA: MicroTAS 2017 Conference), 22–26.
Chiu, H.-H., and Kuo, C.-H. (2020). Gas chromatography-mass spectrometry-based analytical strategies for fatty acid analysis in biological samples. J. Food Drug Anal. 28 (1), 60–73. doi:10.1016/j.jfda.2019.10.003
Cho, Y., Böttger, U., Rull, F., Hübers, H.-W., Belenguer, T., Börner, A., et al. (2021). In-situ science on Phobos with the Raman spectrometer for MMX (RAX): Preliminary design and feasibility of Raman measurements. Earth Planets Space 73 (232), 232. doi:10.1186/s40623-021-01496-z
Chocholoušková, M., Wolrab, D., Jirásko, R., Študentová, H., Melichar, B., and Holčapek, M. (2021). Intra-laboratory comparison of four analytical platforms for lipidomic quantitation using hydrophilic interaction liquid chromatography or supercritical fluid chromatography coupled to quadrupole - time-of-flight mass spectrometry. Talanta 231, 122367. doi:10.1016/j.talanta.2021.122367
Chou, L., Mahaffy, P., Trainer, M., Eigenbrode, J., Arevalo, R., Brinckerhoff, W., et al. (2021). Planetary mass spectrometry for agnostic life detection in the Solar System. Front. Astron. Space Sci. 173. doi:10.3389/fspas.2021.755100
Chouhan, B., Shelor, C. P., Huang, W., Chen, Y., and Dasgupta, P. K. (2020). Nanovolume gas-free hydroxide eluent generator for open tubular ion chromatography. Anal. Chem. 92 (7), 5561–5568. doi:10.1021/acs.analchem.0c00505
Chua, Y. T., Stair, P. C., and Wachs, I. E. (2001). A comparison of ultraviolet and visible Raman spectra of supported metal oxide catalysts. J. Phys. Chem. B 105 (36), 8600–8606. doi:10.1021/jp011366g
Cieslarova, Z., Noell, A. C., Willis, P. A., and Mora, M. F. (2022). From microorganisms to biosignatures: Subcritical water extraction as a sample preparation technique for future life detection missions. Geophys. Res. Lett. 49 (12), e2022GL098082. doi:10.1029/2022gl098082
Cleland, C. E., and Chyba, C. F. (2002). Defining ‘life. Orig. Life Evol. Biosph. 32 (4), 387–393. doi:10.1023/A:1020503324273
Coates, L. J., Lam, S. C., Gooley, A. A., Haddad, P. R., Paull, B., and Wirth, H.-J. (2020). Modular, cost-effective, and portable capillary gradient liquid chromatography system for on-site analysis. J. Chromatogr. A 1626, 461374. doi:10.1016/j.chroma.2020.461374
Cockell, C. S., Holt, J., Campbell, J., Groseman, H., Josset, J.-L., Bontognali, T. R., et al. (2019). Subsurface scientific exploration of extraterrestrial environments (MINAR 5): Analogue science, technology and education in the boulby mine, UK. Int. J. Astrobiol. 18 (2), 157–182. doi:10.1017/S1473550418000186
Costanzo, M. T., Boock, J. J., Kemperman, R. H., Wei, M. S., Beekman, C. R., and Yost, R. A. (2017). Portable FAIMS: Applications and future perspectives. Int. J. Mass Spectrom. 422, 188–196. doi:10.1016/j.ijms.2016.12.007
Court, R. W., Baki, A. O., Sims, M. R., Cullen, D., and Sephton, M. A. (2010). Novel solvent systems for in situ extraterrestrial sample analysis. Planet. Space Sci. 58 (11), 1470–1474. doi:10.1016/j.pss.2010.07.004
Court, R. W., Rix, C. S., Sims, M. R., Cullen, D. C., and Sephton, M. A. (2012). Extraction of polar and nonpolar biomarkers from the martian soil using aqueous surfactant solutions. Planet. Space Sci. 67 (1), 109–118. doi:10.1016/j.pss.2012.03.006
Court, R. W., Sims, M. R., Cullen, D. C., and Sephton, M. A. (2014). Searching for life on mars: Degradation of surfactant solutions used in organic extraction experiments. Astrobiology 14 (9), 733–752. doi:10.1089/ast.2013.1105
Coussot, G., Faye, C., Le Postollec, A., and Dobrijevic, M. (2018a). A methodological approach for the thermal stability and stress exposure studies of a model antibody. Anal. Biochem. 548, 23–31. doi:10.1016/j.ab.2018.02.019
Coussot, G., Faye, C., Le Postollec, A., and Dobrijevic, M. (2018b). One-step direct immunoassay with horseradish peroxidase as antigen for studying the functionality of antibody surfaces. Talanta 178, 922–927. doi:10.1016/j.talanta.2017.10.032
Coussot, G., Le Postollec, A., Faye, C., Baqué, M., Vandenabeele-Trambouze, O., Incerti, S., et al. (2019a). Photochemistry on the space station — Antibody resistance to space conditions after exposure outside the international space station. Astrobiology 19 (8), 1053–1062. doi:10.1089/ast.2018.1907
Coussot, G., Le Postollec, A., Faye, C., and Dobrijevic, M. (2018c). A gold standard method for the evaluation of antibody-based materials functionality: Approach to forced degradation studies. J. Pharm. Biomed. Anal. 152, 17–24. doi:10.1016/j.jpba.2018.01.041
Coussot, G., Le Postollec, A., Incerti, S., Baque, M., Faye, C., Vandenabeele-Trambouze, O., et al. (2019b). Photochemistry on the space station-aptamer resistance to space conditions: Particles exposure from irradiation facilities and real exposure outside the international space station. Astrobiology 19 (8), 1063–1074. doi:10.1089/ast.2018.1896
Covey, T. R., Thomson, B. A., and Schneider, B. B. (2009). Atmospheric pressure ion sources. Mass Spectrom. Rev. 28 (6), 870–897. doi:10.1002/mas.20246
Craft, K. L., Van Volkenburg, T. B., Ohiri, K. A., Skerritt, J. K., Irons, K. M., Kilhefner, A. Y., et al. (2020). “On-chip purification of amino acids for Ocean world in situ biosignature analyses,” in 51st lunar and planetary science conference (Houston, TX: Lunar and Planetary Institute), 2560.
Creamer, J., Noell, A., Zamuruyev, K., Kehl, F., Cretu, V., Ferreira Santos, M., et al. (2020). “The organic capillary electrophoresis analyzer (OCEAN) instrument concept,” in Outer planets assessment group (OPAG) (Houston, TX), 6005.
Creamer, J. S., Mora, M. F., Noell, A. C., and Willis, P. A. (2019). Long-term thermal stability of fluorescent dye used for chiral amino acid analysis on future spaceflight missions. Electrophoresis 40 (23-24), 3117–3122. doi:10.1002/elps.201900268
Creamer, J. S., Mora, M. F., and Willis, P. A. (2017). Enhanced resolution of chiral amino acids with capillary electrophoresis for biosignature detection in extraterrestrial samples. Anal. Chem. 89 (2), 1329–1337. doi:10.1021/acs.analchem.6b04338
Creamer, J. S., Mora, M. F., and Willis, P. A. (2018). Stability of reagents used for chiral amino acid analysis during spaceflight missions in high-radiation environments. Electrophoresis 39 (22), 2864–2871. doi:10.1002/elps.201800274
Crits-Christoph, A., Robinson, C. K., Barnum, T., Fricke, W., Davila, A. F., Jedynak, B., et al. (2013). Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 1 (1), 28. doi:10.1186/2049-2618-1-28
Csapá, J., Csapó-Kiss, Z., Wágner, L., Tálos, T., Martin, T. G., Folestad, S., et al. (1997). Hydrolysis of proteins performed at high temperatures and for short times with reduced racemization, in order to determine the enantiomers of d- and l-amino acids. Anal. Chim. Acta X. 339 (1), 99–107. doi:10.1016/S0003-2670(96)00452-7
Cumeras, R., Figueras, E., Davis, C. E., Baumbach, J. I., and Gràcia, I. (2015). Review on ion mobility spectrometry. Part 1: Current instrumentation. Analyst 140 (5), 1376–1390. doi:10.1039/c4an01100g
Danda, G., and Drndić, M. (2019). Two-dimensional nanopores and nanoporous membranes for ion and molecule transport. Curr. Opin. Biotechnol. 55, 124–133. doi:10.1016/j.copbio.2018.09.002
Darrach, M., Madzunkov, S., Schaefer, R., Nikolic, D., Simcic, J., Kidd, R., et al. (2015). “The mass analyzer for real-time investigation of neutrals at europa (MARINE),” in 2015 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Dasgupta, P. K., and Maleki, F. (2019). Ion exchange membranes in ion chromatography and related applications. Talanta 204, 89–137. doi:10.1016/j.talanta.2019.05.077
David, M., Musadji, N. Y., Labanowski, J., Sternberg, R., and Geffroy-Rodier, C. (2016). Pilot for validation of online pretreatments for analyses of organics by gas chromatography–mass spectrometry: Application to space research. Anal. Chem. 88 (10), 5137–5144. doi:10.1021/acs.analchem.6b00052
de Diego-Castilla, G., Cruz-Gil, P., Mateo-Martí, E., Fernández-Calvo, P., Rivas, L. A., and Parro, V. (2011). Assessing antibody microarrays for space missions: Effect of long-term storage, gamma radiation, and temperature shifts on printed and fluorescently labeled antibodies. Astrobiology 11 (8), 759–773. doi:10.1089/ast.2011.0647
de Hoffmann, E., and Stroobant, V. (2013). Mass spectrometry: Principles and applications. Hoboken, NJ: Wiley.
Deng, L., Webb, I. K., Garimella, S. V. B., Hamid, A. M., Zheng, X., Norheim, R. V., et al. (2017). Serpentine ultralong path with extended routing (SUPER) high resolution traveling wave ion mobility-MS using structures for lossless ion manipulations. Anal. Chem. 89 (8), 4628–4634. doi:10.1021/acs.analchem.7b00185
Derveni, M., Allen, M., Sawakuchi, G. O., Yukihara, E. G., Richter, L., Sims, M. R., et al. (2013). Survivability of immunoassay reagents exposed to the space radiation environment on board the ESA BIOPAN-6 platform as a prelude to performing immunoassays on Mars. Astrobiology 13 (1), 92–102. doi:10.1089/ast.2012.0871
Derveni, M., Hands, A., Allen, M., Sims, M. R., and Cullen, D. C. (2012). Effects of simulated space radiation on immunoassay components for life-detection experiments in planetary exploration missions. Astrobiology 12 (8), 718–729. doi:10.1089/ast.2011.0727
Desmet, G., and Broeckhoven, K. (2019). Extra-column band broadening effects in contemporary liquid chromatography: Causes and solutions. TrAC Trends Anal. Chem. 115619, 115619. doi:10.1016/j.trac.2019.115619
Desmet, G., and Eeltink, S. (2013). Fundamentals for LC miniaturization. Anal. Chem. 85 (2), 543–556. doi:10.1021/ac303317c
Di Ventra, M., and Taniguchi, M. (2016). Decoding DNA, RNA and peptides with quantum tunnelling. Nat. Nanotechnol. 11 (2), 117–126. doi:10.1038/nnano.2015.320
Direito, S. O., Marees, A., and Röling, W. F. (2012). Sensitive life detection strategies for low-biomass environments: Optimizing extraction of nucleic acids adsorbing to terrestrial and Mars analogue minerals. FEMS Microbiol. Ecol. 81 (1), 111–123. doi:10.1111/j.1574-6941.2012.01325.x
Dodds, J. N., and Baker, E. S. (2019). Ion mobility spectrometry: Fundamental concepts, instrumentation, applications, and the road ahead. J. Am. Soc. Mass Spectrom. 30 (11), 2185–2195. doi:10.1007/s13361-019-02288-2
Dodds, J. N., May, J. C., and McLean, J. A. (2017). Correlating resolving power, resolution, and collision cross section: Unifying cross-platform assessment of separation efficiency in ion mobility spectrometry. Anal. Chem. 89 (22), 12176–12184. doi:10.1021/acs.analchem.7b02827
Domagal-Goldman, S. D., Wright, K. E., Adamala, K., De La Rubia, L. A., Bond, J., Dartnell, L. R., et al. (2016). The astrobiology primer v2. 0. Astrobiology 16 (8), 561–653. doi:10.1089/ast.2015.1460
Duca, Z. A., Craft, K. L., Cable, M. L., and Stockton, A. M. (2022). Quantitative and compositional analysis of trace amino acids in icy moon analogues using a microcapillary electrophoresis laser-induced fluorescence detection system. ACS Earth Space Chem. 6 (2), 333–345. doi:10.1021/acsearthspacechem.1c00297
Dworzanski, J. P., McClennen, W. H., Cole, P. A., Thornton, S. N., Meuzelaar, H. L. C., Arnold, N. S., et al. (1997). Field-portable, automated pyrolysis-GC/IMS system for rapid biomarker detection in aerosols: A feasibility study. Field Anal. Chem. Technol. 1, 295–305. doi:10.1002/(SICI)1520-6521
Eddhif, B., Allavena, A., Liu, S., Ribette, T., Abou Mrad, N., Chiavassa, T., et al. (2018). Development of liquid chromatography high resolution mass spectrometry strategies for the screening of complex organic matter: Application to astrophysical simulated materials. Talanta 179, 238–245. doi:10.1016/j.talanta.2017.11.008
Eiceman, G. A., and Stone, J. A. (2004). Peer reviewed: Ion mobility spectrometers in national defense. Anal. Chem. 76 (21), 390A–397A. doi:10.1021/ac041665c
Eiceman, G. (2002). Ion-mobility spectrometry as a fast monitor of chemical composition. TrAC Trends Anal. Chem. 21 (4), 259–275. doi:10.1016/S0165-9936(02)00406-5
Eiceman, G., Krylov, E., Krylova, N., Nazarov, E., and Miller, R. (2004). Separation of ions from explosives in differential mobility spectrometry by vapor-modified drift gas. Anal. Chem. 76 (17), 4937–4944. doi:10.1021/ac035502k
Emaus, M. N., Varona, M., Eitzmann, D. R., Hsieh, S.-A., Zeger, V. R., and Anderson, J. L. (2020). Nucleic acid extraction: Fundamentals of sample preparation methodologies, current advancements, and future endeavors. TrAC Trends Anal. Chem. 130, 115985. doi:10.1016/j.trac.2020.115985
Eshelman, E., Daly, M., Slater, G., and Cloutis, E. (2018). Detecting aromatic compounds on planetary surfaces using ultraviolet time-resolved fluorescence spectroscopy. Planet. Space Sci. 151, 1–10. doi:10.1016/j.pss.2017.09.003
Eshelman, E. J., Malaska, M. J., Manatt, K. S., Doloboff, I. J., Wanger, G., Willis, M. C., et al. (2019). Watson: In situ organic detection in subsurface ice using deep-UV fluorescence spectroscopy. Astrobiology 19 (6), 771–784. doi:10.1089/ast.2018.1925
Fairén, A. G., Gomez-Elvira, J., Briones, C., Prieto-Ballesteros, O., Rodriguez-Manfredi, J. A., Lopez Heredero, R., et al. (2020). The complex molecules detector (CMOLD): A fluidic-based instrument suite to search for (bio)chemical complexity on mars and icy moons. Astrobiology 20 (9), 1076–1096. doi:10.1089/ast.2019.2167
Faller, T., and Engelhardt, H. (1999). How to achieve higher repeatability and reproducibility in capillary electrophoresis. J. Chromatogr. A 853 (1-2), 83–94. doi:10.1016/S0021-9673(99)00382-9
Farley, K. A., Williford, K. H., Stack, K. M., Bhartia, R., Chen, A., de la Torre, M., et al. (2020). Mars 2020 mission overview. Space Sci. Rev. 216 (8), 142. doi:10.1007/s11214-020-00762-y
Feider, C. L., Krieger, A., DeHoog, R. J., and Eberlin, L. S. (2019). Ambient ionization mass spectrometry: Recent developments and applications. Anal. Chem. 91 (7), 4266–4290. doi:10.1021/acs.analchem.9b00807
Ferdinand, O. D., Friedrichs, A., Miranda, M. L., Voß, D., and Zielinski, O. (2017). “Next generation fluorescence sensor with multiple excitation and emission wavelengths—NeXOS MatrixFlu-UV,” in Oceans 2017 (Piscataway, NJ: IEEE), 1.
Fernández-Calvo, P., Näke, C., Rivas, L. A., García-Villadangos, M., Gómez-Elvira, J., and Parro, V. (2006). A multi-array competitive immunoassay for the detection of broad-range molecular size organic compounds relevant for astrobiology. Planet. Space Sci. 54 (15), 1612–1621. doi:10.1016/j.pss.2006.02.007
Fernández-Fı́gares, I., Rodrı́guez, L. C., and González-Casado, A. (2004). Effect of different matrices on physiological amino acids analysis by liquid chromatography: Evaluation and correction of the matrix effect. J. Chromatogr. B 799 (1), 73–79. doi:10.1016/j.jchromb.2003.10.012
Fernández-Martínez, M. Á., dos Santos Severino, R., Moreno-Paz, M., Gallardo-Carreño, I., Blanco, Y., Warren-Rhodes, K., et al. (2019). Prokaryotic community structure and metabolisms in shallow subsurface of Atacama Desert playas and alluvial fans after heavy rains: Repairing and preparing for next dry period. Front. Microbiol. 10, 1641. doi:10.3389/fmicb.2019.01641
Ferralis, N., Matys, E. D., Knoll, A. H., Hallmann, C., and Summons, R. E. (2016). Rapid, direct and non-destructive assessment of fossil organic matter via microRaman spectroscopy. Carbon 108, 440–449. doi:10.1016/j.carbon.2016.07.039
Ferreira Santos, M. S., Metz, B. C., da Costa, E. T., do Lago, C. L., Willis, P. A., Mora, M. F., et al. (2022). Towards a radiation-tolerant contactless conductivity detector for use with capillary electrophoresis systems in spaceflight applications. Acta Astronaut. 190, 299–307. doi:10.1016/j.actaastro.2021.10.023
Flaherty, R. J., Sarver, S. A., Sun, L., Brownell, G. A., Go, D. B., and Dovichi, N. J. (2017). A high voltage power supply that mitigates current reversals in capillary zone electrophoresis-electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 28 (2), 247–252. doi:10.1007/s13361-016-1529-3
Fornaro, T., Brucato, J. R., Pucci, A., and Branciamore, S. (2013). Development of extraction protocols for life detection biosensor-based instruments. Planet. Space Sci. 86, 75–79. doi:10.1016/j.pss.2013.08.013
Freissinet, C., Buch, A., Sternberg, R., Szopa, C., Geffroy-Rodier, C., Jelinek, C., et al. (2010). Search for evidence of life in space: Analysis of enantiomeric organic molecules by N, N-dimethylformamide dimethylacetal derivative dependant gas chromatography-mass spectrometry. J. Chromatogr. A 1217 (5), 731–740. doi:10.1016/j.chroma.2009.11.009
Freissinet, C., Getty, S. A., Trainer, M. G., Glavin, D. P., Mahaffy, P. R., McLain, H. L., et al. (2016). Evaluation of the robustness of chromatographic columns in a simulated highly radiative Jovian environment. Planet. Space Sci. 122, 38–45. doi:10.1016/j.pss.2016.01.004
Freissinet, C., Glavin, D. P., Mahaffy, P. R., Miller, K. E., Eigenbrode, J. L., Summons, R. E., et al. (2015). Organic molecules in the sheepbed mudstone, gale crater, mars. JGR. Planets 120 (3), 495–514. doi:10.1002/2014JE004737
Freissinet, C., Millan, M., Glavin, D. P., Li, X., Grubisic, A., Eigenbrode, J. E., et al. (2019). Investigating the effects of gamma radiation on selected chemicals for use in biosignature detection instruments on the surface of Jupiter's moon Europa. Planet. Space Sci. 175, 1–12. doi:10.1016/j.pss.2019.05.009
Frosch, T., Tarcea, N., Schmitt, M., Thiele, H., Langenhorst, F., and Popp, J. (2007). UV Raman imaging a promising tool for astrobiology: Comparative Raman studies with different excitation wavelengths on SNC martian meteorites. Anal. Chem. 79 (3), 1101–1108. doi:10.1021/ac0618977
Fujishima, K., Dziomba, S., Yano, H., Kebe, S. I., Guerrouache, M., Carbonnier, B., et al. (2019). The non-destructive separation of diverse astrobiologically relevant organic molecules by customizable capillary zone electrophoresis and monolithic capillary electrochromatography. Int. J. Astrobiol. 18 (6), 562–574. doi:10.1017/S1473550419000065
Furey, A., Moriarty, M., Bane, V., Kinsella, B., and Lehane, M. (2013). Ion suppression; A critical review on causes, evaluation, prevention and applications. Talanta 115, 104–122. doi:10.1016/j.talanta.2013.03.048
Fykse, E. M., Olsen, J. S., and Skogan, G. (2003). Application of sonication to release DNA from Bacillus cereus for quantitative detection by real-time PCR. J. Microbiol. Methods 55 (1), 1–10. doi:10.1016/S0167-7012(03)00091-5
Gaft, M., Panczer, G., Reisfeld, R., and Uspensky, E. (2001). Laser-induced time-resolved luminescence as a tool for rare-Earth element identification in minerals. Phys. Chem. Min. 28 (5), 347–363. doi:10.1007/s002690100163
Gao, L., Cooks, R. G., and Ouyang, Z. (2008). Breaking the pumping speed barrier in mass spectrometry: Discontinuous atmospheric pressure interface. Anal. Chem. 80 (11), 4026–4032. doi:10.1021/ac800014v
García-Descalzo, L., Parro, V., García-Villadangos, M., Cockell, C. S., Moissl-Eichinger, C., Perras, A., et al. (2019). Microbial markers profile in anaerobic Mars analogue environments using the LDChip (Life Detector Chip) antibody microarray core of the SOLID (Signs of Life Detector) platform. Microorganisms 7 (9), 365. doi:10.3390/microorganisms7090365
Gasda, P. J., Wiens, R. C., Reyes-Newell, A., Ganguly, K., Newell, R. T., Peterson, C., et al. (2021). OrganiCam: A lightweight time-resolved laser-induced luminescenceimager and Raman spectrometer for planetary organic materialcharacterization. Appl. Opt. 60 (13), 3753–3763. doi:10.1364/AO.421291
Geffroy-Rodier, C., Grasset, L., Sternberg, R., Buch, A., and Amblès, A. (2009). Thermochemolysis in search for organics in extraterrestrial environments. J. Anal. Appl. Pyrolysis 85 (1), 454–459. doi:10.1016/j.jaap.2008.10.005
Gelman, B., Zolotukhin, V., Lamonov, N., Levchuk, B., Mukhin, L., Nenarokov, D., et al. (1979). An analysis of the chemical composition of the atmosphere of Venus on an AMS of the Venera-12 using a gas chromatograph. Kosmicheskie Issled. 17 (5), 708
Getty, S. A., Brinckerhoff, W. B., Cornish, T., Ecelberger, S., and Floyd, M. (2012). Compact two-step laser time-of-flight mass spectrometer for in situ analyses of aromatic organics on planetary missions. Rapid Commun. Mass Spectrom. 26 (23), 2786–2790. doi:10.1002/rcm.6393
Getty, S. A., Dworkin, J. P., Glavin, D. P., Martin, M., Zheng, Y., Balvin, M., et al. (2013). “Organics analyzer for sampling icy surfaces: A liquid chromatograph-mass spectrometer for future in situ small body missions,” in 2013 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Getty, S. A., Elsila, J., Balvin, M., Brinckerhoff, W. B., Cornish, T., Li, X., et al. (2017). “Molecular analyzer for complex refractory organic-rich surfaces (MACROS),” in 2017 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Getty, S. A., ten Kate, I. L., Feng, S. H., Brinckerhoff, W. B., Cardiff, E. H., Holmes, V. E., et al. (2010). Development of an evolved gas-time-of-flight mass spectrometer for the Volatile Analysis by Pyrolysis of Regolith (VAPoR) instrument. Int. J. Mass Spectrom. 295 (3), 124–132. doi:10.1016/j.ijms.2010.06.020
Ghosh, A., Foster, A. R., Johnson, J. C., Vilorio, C. R., Tolley, L. T., Iverson, B. D., et al. (2019). Stainless-steel column for robust, high-temperature microchip gas chromatography. Anal. Chem. 91 (1), 792–796. doi:10.1021/acs.analchem.8b04174
Ghosh, A., Vilorio, C. R., Hawkins, A. R., and Lee, M. L. (2018). Microchip gas chromatography columns, interfacing and performance. Talanta 188, 463–492. doi:10.1016/j.talanta.2018.04.088
Gladman, B. J., Burns, J. A., Duncan, M., Lee, P., and Levison, H. F. (1996). The exchange of impact ejecta between terrestrial planets. Science 271 (5254), 1387–1392. doi:10.1126/science.271.5254.1387
Glavin, D., Elsila, J., Burton, A., Callahan, M., Dworkin, J., Hilts, R., et al. (2012a). Unusual nonterrestrial L-proteinogenic amino acid excesses in the Tagish Lake meteorite. Meteorit. Planet. Sci. 47, 1347–1364. doi:10.1111/j.1945-5100.2012.01400.x
Glavin, D. P., and Bada, J. L. (1998). Isolation of amino acids from natural samples using sublimation. Anal. Chem. 70 (15), 3119–3122. doi:10.1021/ac9803784
Glavin, D. P., Cleaves, H. J., Buch, A., Schubert, M., Aubrey, A., Bada, J. L., et al. (2006). Sublimation extraction coupled with gas chromatography-mass spectrometry: A new technique for future in situ analyses of purines and pyrimidines on mars. Planet. Space Sci. 54 (15), 1584–1591. doi:10.1016/j.pss.2005.12.023
Glavin, D. P., Freissinet, C., Miller, K. E., Eigenbrode, J. L., Brunner, A. E., Buch, A., et al. (2013). Evidence for perchlorates and the origin of chlorinated hydrocarbons detected by SAM at the Rocknest aeolian deposit in Gale Crater. J. Geophys. Res. Planets 118 (10), 1955–1973. doi:10.1002/jgre.20144
Glavin, D. P., Malespin, C., Kate, I. L. t., Getty, S. A., Holmes, V. E., Mumm, E., et al. (2012b). “Volatile analysis by pyrolysis of regolith for planetary resource exploration,” in 2012 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Glavin, D. P., Schubert, M., Botta, O., Kminek, G., and Bada, J. L. (2001). Detecting pyrolysis products from bacteria on Mars. Earth Planet. Sci. Lett. 185 (1), 1–5. doi:10.1016/S0012-821X(00)00370-8
Goesmann, F., Brinckerhoff, W. B., Raulin, F., Goetz, W., Danell, R. M., Getty, S. A., et al. (2017). The mars organic molecule analyzer (MOMA) instrument: Characterization of organic material in martian sediments. Astrobiology 17 (6-7), 655–685. doi:10.1089/ast.2016.1551
Goesmann, F., Raulin, F., Bredehöft, J. H., Cabane, M., Ehrenfreund, P., MacDermott, A. J., et al. (2014). COSAC prepares for sampling and in situ analysis of cometary matter from comet 67P/Churyumov–Gerasimenko. . Planet. Space Sci. 103, 318–330. doi:10.1016/j.pss.2014.08.006
Goesmann, F., Rosenbauer, H., Bredehöft, J. H., Cabane, M., Ehrenfreund, P., Gautier, T., et al. (2015). COMETARY SCIENCE. Organic compounds on comet 67P/Churyumov-Gerasimenko revealed by COSAC mass spectrometry. Science 349 (6247), aab0689. doi:10.1126/science.aab0689
Golozar, M., Casto, L., Estlack, Z., Kim, J., Butterworth, A. L., and Mathies, R. A. (2022). “Microfluidic organic analyzer for biosignatures (moab): A flight-design instrument with the capabilities required to detect trace biosignatures on icy moons,” in 2022 astrobiology science conference (Washington, D.C.: AGU).
Goordial, J., Altshuler, I., Hindson, K., Chan-Yam, K., Marcolefas, E., and Whyte, L. G. (2017). In situ field sequencing and life detection in remote (79 26′ N) Canadian high arctic permafrost ice wedge microbial communities. Front. Microbiol. 8, 2594. doi:10.3389/fmicb.2017.02594
Gowen, R. A., Smith, A., Fortes, A. D., Barber, S., Brown, P., Church, P., et al. (2011). Penetrators for in situ subsurface investigations of Europa. Adv. Space Res. 48 (4), 725–742. doi:10.1016/j.asr.2010.06.026
Gries, P., Rathore, A. S., Lu, X., Chiou, J., Huynh, Y. B., Lodi, A., et al. (2021). Automated trimethyl sulfonium hydroxide derivatization method for high-throughput fatty acid profiling by gas chromatography–mass spectrometry. Molecules 26 (20), 6246. doi:10.3390/molecules26206246
Grotzinger, J. P., Crisp, J., Vasavada, A. R., Anderson, R. C., Baker, C. J., Barry, R., et al. (2012). Mars Science Laboratory mission and science investigation. Space Sci. Rev. 170 (1), 5–56. doi:10.1007/s11214-012-9892-2
Gu, C., Jia, Z., Zhu, Z., He, C., Wang, W., Morgan, A., et al. (2012). Miniaturized electroosmotic pump capable of generating pressures of more than 1200 bar. Anal. Chem. 84 (21), 9609–9614. doi:10.1021/ac3025703
Guzman, M., McKay, C. P., Quinn, R. C., Szopa, C., Davila, A. F., Navarro-González, R., et al. (2018). Identification of chlorobenzene in the viking gas chromatograph-mass spectrometer data sets: Reanalysis of viking mission data consistent with aromatic organic compounds on mars. J. Geophys. Res. Planets 123 (7), 1674–1683. doi:10.1029/2018JE005544
Guzman, M., Szopa, C., Freissinet, C., Buch, A., Stalport, F., Kaplan, D., et al. (2020). Testing the capabilities of the Mars Organic Molecule Analyser (MOMA) chromatographic columns for the separation of organic compounds on Mars. Planet. Space Sci. 186, 104903. doi:10.1016/j.pss.2020.104903
Hager, J. W. (2004). Recent trends in mass spectrometer development. Anal. Bioanal. Chem. 378 (4), 845–850. doi:10.1007/s00216-003-2287-1
Haghighi, F., Talebpour, Z., and Nezhad, A. S. (2018). Towards fully integrated liquid chromatography on a chip: Evolution and evaluation. TrAC Trends Anal. Chem. 105, 302–337. doi:10.1016/j.trac.2018.05.002
Hand, K., Murray, A., Garvin, J., Brinckerhoff, W., Christner, B., Edgett, K., et al. (2017). Europa lander study 2016 report: Europa lander mission. Washington, DC: The National Aeronautics and Space Administration.
Hanke, F., Mooij, B. J., Ariese, F., and Böttger, U. (2019). The evaluation of time-resolved Raman spectroscopy for the suppression of background fluorescence from space-relevant samples. J. Raman Spectrosc. 50 (7), 969–982. doi:10.1002/jrs.5586
Haque, F., Li, J., Wu, H.-C., Liang, X.-J., and Guo, P. (2013). Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today 8 (1), 56–74. doi:10.1016/j.nantod.2012.12.008
Harrison, S. T. L. (1991). Bacterial cell disruption: A key unit operation in the recovery of intracellular products. Biotechnol. Adv. 9 (2), 217–240. doi:10.1016/0734-9750(91)90005-G
Harstad, R. K., Johnson, A. C., Weisenberger, M. M., and Bowser, M. T. (2016). Capillary electrophoresis. Anal. Chem. 88 (1), 299–319. doi:10.1021/acs.analchem.5b04125
He, Y., Buch, A., Morisson, M., Szopa, C., Freissinet, C., Williams, A., et al. (2019). Application of TMAH thermochemolysis to the detection of nucleobases: Application to the MOMA and SAM space experiment. Talanta 204, 802–811. doi:10.1016/j.talanta.2019.06.076
He, Y., Buch, A., Szopa, C., Millan, M., Freissinet, C., Navarro-Gonzalez, R., et al. (2021a). Influence of calcium perchlorate on the search for Martian organic compounds with MTBSTFA/DMF derivatization. Astrobiology 21 (9), 1137–1156. doi:10.1089/ast.2020.2393
He, Y., Buch, A., Szopa, C., Williams, A. J., Millan, M., Guzman, M., et al. (2020). The search for organic compounds with TMAH thermochemolysis: From Earth analyses to space exploration experiments. TrAC Trends Anal. Chem. 127, 115896. doi:10.1016/j.trac.2020.115896
He, Y., Buch, A., Szopa, C., Williams, A. J., Millan, M., Malespin, C. A., et al. (2021b). Influence of calcium perchlorate on the search for organics on mars with tetramethylammonium hydroxide thermochemolysis. Astrobiology 21 (3), 279–297. doi:10.1089/ast.2020.2252
Hebert, N., Young, J., van Soest, R., Neyer, D., and Hunter, C. (2011). “Increasing protein identification using coupled chip based nanoLC columns,” in Technologies to enable personalized medicine (Lexington, KY: Association of Biomolecular Resource Facilities), 210.
Hecht, M., Kounaves, S., Quinn, R., West, S., Young, S., Ming, D., et al. (2009). Detection of perchlorate and the soluble chemistry of martian soil at the Phoenix lander site. Science 325 (5936), 64–67. doi:10.1126/science.1172466
Hemida, M., Ghiasvand, A., Gupta, V., Coates, L. J., Gooley, A. A., Wirth, H.-J., et al. (2021a). Small-footprint, field-deployable LC/MS System for on-site analysis of per- and polyfluoroalkyl substances in soil. Anal. Chem. 93, 12032–12040. doi:10.1021/acs.analchem.1c02193
Hemida, M., Haddad, P. R., Lam, S. C., Coates, L. J., Riley, F., Diaz, A., et al. (2021b). Small footprint liquid chromatography-mass spectrometry for pharmaceutical reaction monitoring and automated process analysis. J. Chromatogr. A 1656, 462545. doi:10.1016/j.chroma.2021.462545
Hickman-Lewis, K., Moore, K. R., Hollis, J. J. R., Tuite, M. L., Beegle, L. W., Bhartia, R., et al. (2022). In situ identification of Paleoarchean biosignatures using colocated Perseverance Rover analyses: Perspectives for in situ Mars science and sample return. Astrobiology 22, 1143–1163. In press. doi:10.1089/ast.2022.0018
Hirayama, A., Abe, H., Yamaguchi, N., Tabata, S., Tomita, M., and Soga, T. (2018). Development of a sheathless CE-ESI-MS interface. Electrophoresis 39 (11), 1382–1389. doi:10.1002/elps.201800017
Hohl, M., Wurz, P., Scherer, S., Altwegg, K., and Balsiger, H. (1999). Mass selective blanking in a compact multiple reflection time-of-flight mass spectrometer. Int. J. Mass Spectrom. 188 (3), 189–197. doi:10.1016/S1387-3806(99)00040-8
Horowitz, N. H., Hubbard, J. S., and Hobby, G. L. (1972). The carbon-assimilation experiment: The viking mars lander. Icarus 16 (1), 147–152. doi:10.1016/0019-1035(72)90142-X
Horowitz, N., Hobby, G., and Hubbard, J. S. (1977). Viking on mars: The carbon assimilation experiments. J. Geophys. Res. 82 (28), 4659–4662. doi:10.1029/JS082i028p04659
Howell, S. M., and Pappalardo, R. T. (2020). NASA’s europa clipper—A mission to a potentially habitable Ocean World. Nat. Commun. 11 (1), 1311. doi:10.1038/s41467-020-15160-9
Huang, W., Plistil, A., Stearns, S. D., and Dasgupta, P. K. (2021). Gradient nanopump based suppressed ion chromatography using PEEK open tubular columns. Talanta Open 3, 100029. doi:10.1016/j.talo.2020.100029
Hughes, C., Langridge, J., and McKenna, T. (2011). “Investigating the robustness of a new microfluidic device,” in Technologies to enable personalized medicine (Lexington, KY: Association of Biomolecular Resource Facilities), 218.
Hutt, L. D., Glavin, D. P., Bada, J. L., and Mathies, R. A. (1999). Microfabricated capillary electrophoresis amino acid chirality analyzer for extraterrestrial exploration. Anal. Chem. 71 (18), 4000–4006. doi:10.1021/ac9903959
Inza, A. G. M., and Lopez-Reyes, G. (2020). “Evolution of the scientific instrumentation for in situ Mars exploration,” in Mars exploration: A step forward. Editors G. Pezzella, and A. Viviani (London: IntechOpen).
Ishida, Y., Wakamatsu, S., Yokoi, H., Ohtani, H., and Tsuge, S. (1999). Compositional analysis of polyunsaturated fatty acid oil by one-step thermally assisted hydrolysis and methylation in the presence of trimethylsulfonium hydroxide. J. Anal. Appl. Pyrolysis 49 (1-2), 267–276. doi:10.1016/S0165-2370(98)00095-3
Israël, G., Szopa, C., Raulin, F., Cabane, M., Niemann, H. B., Atreya, S. K., et al. (2005). Complex organic matter in Titan's atmospheric aerosols from in situ pyrolysis and analysis. Nature 438 (7069), 796–799. doi:10.1038/nature04349
Jarvas, G., Fonslow, B., Yates, J. R., Foret, F., and Guttman, A. (2017). Characterization of a porous nano-electrospray capillary emitter at ultra-low flow rates. J. Chromatogr. Sci. 55 (1), 47–51. doi:10.1093/chromsci/bmw148
Javelle, T., Righezza, M., and Danger, G. (2021). Identify low mass volatile organic compounds from cometary ice analogs using gas chromatography coupled to an Orbitrap mass spectrometer associated to electron and chemical Ionizations. J. Chromatogr. A 1652, 462343. doi:10.1016/j.chroma.2021.462343
Jender, M., Höving, S., Novo, P., Freier, E., and Janasek, D. (2021). Coupling miniaturized free-flow electrophoresis to mass spectrometry via a multi-emitter ESI interface. Anal. Chem. 93 (19), 7204–7209. doi:10.1021/acs.analchem.1c00200
Jespers, S., Schlautmann, S., Gardeniers, H., De Malsche, W., Lynen, F., and Desmet, G. (2017). Chip-based multicapillary column with maximal interconnectivity to combine maximum efficiency and maximum loadability. Anal. Chem. 89 (21), 11605–11613. doi:10.1021/acs.analchem.7b03036
Johnson, J. T., Lee, K. W., Bhanot, J. S., and McLuckey, S. A. (2019). A miniaturized Fourier transform electrostatic linear ion trap mass spectrometer: Mass range and resolution. J. Am. Soc. Mass Spectrom. 30 (4), 588–594. doi:10.1007/s13361-018-02126-x
Johnson, P. V., Beegle, L. W., and Kanik, I. (2012). in Mass spectrometry in solar system explorationMass spectrometry handbook. Editor M. S. Lee (Hoboken, NJ: Wiley), 389.
Johnson, P. V., Beegle, L. W., Kim, H. I., Eiceman, G. A., and Kanik, I. (2007). Ion mobility spectrometry in space exploration. Int. J. Mass Spectrom. 262 (1), 1–15. doi:10.1016/j.ijms.2006.11.001
Johnson, P. V., Kim, H. I., Beegle, L. W., and Kanik, I. (2004). Electrospray ionization ion mobility spectrometry of amino acids: Ion mobilities and a mass−mobility correlation. J. Phys. Chem. A 108 (27), 5785–5792. doi:10.1021/jp0492117
Johnson, S. S., Zaikova, E., Goerlitz, D. S., Bai, Y., and Tighe, S. W. (2017). Real-time DNA sequencing in the Antarctic dry valleys using the Oxford Nanopore sequencer. J. Biomol. Tech. 28 (1), 2–7. doi:10.7171/jbt.17-2801-009
Joint Committee for Guides in Metrology (2008). Evaluation of measurement data — guide to the expression of uncertainty in measurement. Sèvres, France: JCGM.
Jorgenson, J. W., and Lukacs, K. D. (1981). Zone electrophoresis in open-tubular glass capillaries. Anal. Chem. 53 (8), 1298–1302. doi:10.1021/ac00231a037
Juraschek, R., Dülcks, T., and Karas, M. (1999). Nanoelectrospray — More than just a minimized-flow electrospray ionization source. J. Am. Soc. Mass Spectrom. 10 (4), 300–308. doi:10.1016/S1044-0305(98)00157-3
Kanik, I., Beegle, L. W., Kounaves, S., Cooks, R. G., Hecht, M., and Johnson, P. V. (2006). “Wet chemistry experiment at mars (WETCHEM),” in 37th annual lunar and planetary science conference (Houston, TX: Lunar and Planetary Institute).
Kassa, T., Undesser, L. P., Hofstetter, H., and Hofstetter, O. (2011). Antibody-based multiplex analysis of structurally closely related chiral molecules. Analyst 136 (6), 1113–1115. doi:10.1039/c0an00814a
Kehl, F., Drevinskas, T., Creamer, J. S., DeMartino, A. J., and Willis, P. A. (2022). Providing enhanced migration time reproducibility with a high-voltage-compatible flow sensor for capillary electrophoresis. Anal. Chem. 94, 5734–5740. doi:10.1021/acs.analchem.2c00038
Kehl, F., Kovarik, N. A., Creamer, J. S., Costa, E. T., and Willis, P. A. (2019). A subcritical water extractor prototype for potential astrobiology spaceflight missions. Earth Space Sci. 6 (12), 2443–2460. doi:10.1029/2019ea000803
Kelly, R. T., Page, J. S., Zhao, R., Qian, W.-J., Mottaz, H. M., Tang, K., et al. (2008). Capillary-based multi nanoelectrospray emitters: Improvements in ion transmission efficiency and implementation with capillary reversed-phase LC-ESI-MS. Anal. Chem. 80 (1), 143–149. doi:10.1021/ac701647s
Kempf, S., Altobelli, N., Briois, C., Grün, E., Horanyi, M., Postberg, F., et al. (2014). “Suda: A dust mass spectrometer for compositional surface mapping for a mission to europa,” in European planetary science congress (Cascais, Portugal: Europlanet Society),
Kim, H. I., Johnson, P. V., Beegle, L. W., Beauchamp, J. L., and Kanik, I. (2005). Electrospray ionization ion mobility spectrometry of carboxylate anions: Ion mobilities and a mass−mobility correlation. J. Phys. Chem. A 109 (35), 7888–7895. doi:10.1021/jp051274h
Kim, H. I., Kim, H., Pang, E. S., Ryu, E. K., Beegle, L. W., Loo, J. A., et al. (2009). Structural characterization of unsaturated phosphatidylcholines using traveling wave ion mobility spectrometry. Anal. Chem. 81 (20), 8289–8297. doi:10.1021/ac900672a
Kim, J., Jensen, E. C., Stockton, A. M., and Mathies, R. A. (2013). Universal microfluidic automaton for autonomous sample processing: Application to the Mars Organic Analyzer. Anal. Chem. 85 (16), 7682–7688. doi:10.1021/ac303767m
Kim, S.-J., Reidy, S. M., Block, B. P., Wise, K. D., Zellers, E. T., and Kurabayashi, K. (2010). Microfabricated thermal modulator for comprehensive two-dimensional micro gas chromatography: Design, thermal modeling, and preliminary testing. Lab. Chip 10 (13), 1647–1654. doi:10.1039/c001390k
Kim, S.-J., Serrano, G., Wise, K. D., Kurabayashi, K., and Zellers, E. T. (2011). Evaluation of a microfabricated thermal modulator for comprehensive two-dimensional microscale gas chromatography. Anal. Chem. 83 (14), 5556–5562. doi:10.1021/ac200336e
Kiplagat, I. K., Kubáň, P., Pelcová, P., and Kubáň, V. (2010). Portable, lightweight, low power, ion chromatographic system with open tubular capillary columns. J. Chromatogr. A 1217 (31), 5116–5123. doi:10.1016/j.chroma.2010.06.017
Kivrak, L., Williams, A., Buch, A., and He, Y. (2021). “Trimethylsulfonium hydroxide (TMSH) thermochemolysis with Py-GC-MS as a method of organic biosignature detection: Optimization for nucleobase detection,” in Lunar and planetary science conference (Houston, TX: Lunar and Planetary Institute), 2468.
Klee, M. S., Cochran, J., Merrick, M., and Blumberg, L. M. (2015). Evaluation of conditions of comprehensive two-dimensional gas chromatography that yield a near-theoretical maximum in peak capacity gain. J. Chromatogr. A 1383, 151–159. doi:10.1016/j.chroma.2015.01.031
Klein, H. P., Horowitz, N. H., Levin, G. V., Oyama, V. I., Lederberg, J., Rich, A., et al. (1976). The viking biological investigation: Preliminary results. Science 194 (4260), 99–105. doi:10.1126/science.194.4260.99
Klein, H. P. (1978). The Viking biological experiments on Mars. Icarus 34 (3), 666–674. doi:10.1016/0019-1035(78)90053-2
Klenner, F., Postberg, F., Hillier, J., Khawaja, N., Cable, M. L., Abel, B., et al. (2020a). Discriminating abiotic and biotic fingerprints of amino acids and fatty acids in ice grains relevant to Ocean Worlds. Astrobiology 20 (10), 1168–1184. doi:10.1089/ast.2019.2188
Klenner, F., Postberg, F., Hillier, J., Khawaja, N., Reviol, R., Stolz, F., et al. (2020b). Analog experiments for the identification of trace biosignatures in ice grains from extraterrestrial ocean worlds. Astrobiology 20 (2), 179–189. doi:10.1089/ast.2019.2065
Kolakowski, B. M., and Mester, Z. (2007). Review of applications of high-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential mobility spectrometry (DMS). Analyst 132 (9), 842–864. doi:10.1039/b706039d
Kristoff, C. J., Bwanali, L., Veltri, L. M., Gautam, G. P., Rutto, P. K., Newton, E. O., et al. (2020). Challenging bioanalyses with capillary electrophoresis. Anal. Chem. 92 (1), 49–66. doi:10.1021/acs.analchem.9b04718
Kubáň, P., and Hauser, P. C. (2018). 20th anniversary of axial capacitively coupled contactless conductivity detection in capillary electrophoresis. TrAC Trends Anal. Chem. 102, 311–321. doi:10.1016/j.trac.2018.03.007
Kubáň, P., and Hauser, P. C. (2011). Capacitively coupled contactless conductivity detection for microseparation techniques – recent developments. Electrophoresis 32 (1), 30–42. doi:10.1002/elps.201000354
Lam, S. C., Coates, L. J., Hemida, M., Gupta, V., Haddad, P. R., Macka, M., et al. (2020). Miniature and fully portable gradient capillary liquid chromatograph. Anal. Chim. Acta X. 1101, 199–210. doi:10.1016/j.aca.2019.12.014
Lang, Q., Cheng, I. F., Wai, C. M., Paszczynski, A., Crawford, R. L., Barnes, B., et al. (2002). Supercritical fluid extraction and high-performance liquid chromatography-diode array-electrochemical detection of signature redox compounds from sand and soil samples. Anal. Biochem. 301 (2), 225–234. doi:10.1006/abio.2001.5502
Laurent, B., Cousins, C. R., Gunn, M., Huntly, C., Cross, R., and Allender, E. (2019). UV luminescence characterisation of organics in Mars-analogue substrates. Icarus 321, 929–937. doi:10.1016/j.icarus.2018.12.031
Le Postollec, A., Coussot, G., Baque, M., Incerti, S., Desvignes, I., Moretto, P., et al. (2009a). Investigation of neutron radiation effects on polyclonal antibodies (IgG) and fluorescein dye for astrobiological applications. Astrobiology 9 (7), 637–645. doi:10.1089/ast.2008.0303
Le Postollec, A., Incerti, S., Dobrijevic, M., Desorgher, L., Santin, G., Moretto, P., et al. (2009b). Monte Carlo simulation of the radiation environment encountered by a biochip during a space mission to Mars. Astrobiology 9 (3), 311–323. doi:10.1089/ast.2008.0255
Lee, S. S. (2021). Single molecule detection and classification using nanogaps.” in Bachelor's degree. Cambridge (MA): Massachusetts Institute of Technology.
Leshin, L. A., Mahaffy, P. R., Webster, C. R., Cabane, M., Coll, P., Conrad, P. G., et al. (2013). Volatile, isotope, and organic analysis of martian fines with the mars curiosity rover. Science 341 (6153), 1238937. doi:10.1126/science.1238937
Levin, G. V. (1972). Detection of metabolically produced labeled gas: The Viking Mars Lander. Icarus 16 (1), 153–166. doi:10.1016/0019-1035(72)90143-1
Levin, G. V., and Straat, P. A. (1977). Recent results from the Viking labeled release experiment on Mars. J. Geophys. Res. 82 (28), 4663–4667. doi:10.1029/JS082i028p04663
Levin, G. V., and Straat, P. A. (2016). The case for extant life on Mars and its possible detection by the Viking labeled release experiment. Astrobiology 16 (10), 798–810. doi:10.1089/ast.2015.1464
Lewis, A. P., Cranny, A., Harris, N. R., Green, N. G., Wharton, J. A., Wood, R. J. K., et al. (2013). Review on the development of truly portable and in-situ capillary electrophoresis systems. Meas. Sci. Technol. 24 (4), 042001. doi:10.1088/0957-0233/24/4/042001
Lewis, J. M. T., Najorka, J., Watson, J. S., and Sephton, M. A. (2018). The search for hesperian organic matter on Mars: Pyrolysis studies of sediments rich in sulfur and iron. Astrobiology 18 (4), 454–464. doi:10.1089/ast.2017.1717
Lezcano, M. Á., Moreno-Paz, M., Carrizo, D., Prieto-Ballesteros, O., Fernández-Martínez, M. Á., Sánchez-García, L., et al. (2019). Biomarker profiling of microbial mats in the geothermal band of cerro caliente, deception island (Antarctica): Life at the edge of heat and cold. Astrobiology 19 (12), 1490–1504. doi:10.1089/ast.2018.2004
Li, L., Chen, T.-C., Ren, Y., Hendricks, P. I., Cooks, R. G., and Ouyang, Z. (2014). Mini 12, miniature mass spectrometer for clinical and other applications — Introduction and characterization. Anal. Chem. 86 (6), 2909–2916. doi:10.1021/ac403766c
Li, X., Danell, R. M., Brinckerhoff, W. B., Pinnick, V. T., van Amerom, F., Arevalo, R. D., et al. (2015a). Detection of trace organics in Mars analog samples containing perchlorate by laser desorption/ionization mass spectrometry. Astrobiology 15 (2), 104–110. doi:10.1089/ast.2014.1203
Li, X., Danell, R. M., Pinnick, V. T., Grubisic, A., van Amerom, F., Arevalo, R. D., et al. (2017). Mars Organic Molecule Analyzer (MOMA) laser desorption/ionization source design and performance characterization. Int. J. Mass Spectrom. 422, 177–187. doi:10.1016/j.ijms.2017.03.010
Li, X., Grubisic, A., Brinckerhoff, W. B., Castillo, M., Arevalo, R., Amerom, F. v., et al. (2019). “Linear ion trap mass spectrometer (LITMS) for in situ astrobiology,” in 2019 IEEE aerospace conference (Piscataway, NJ: IEEE), 1–11.
Li, Y., Dick, W. A., and Tuovinen, O. H. (2004). Fluorescence microscopy for visualization of soil microorganisms—A review. Biol. Fertil. Soils 39 (5), 301–311. doi:10.1007/s00374-004-0722-x
Li, Y., Dvořák, M., Nesterenko, P. N., Stanley, R., Nuchtavorn, N., Krčmová, L. K., et al. (2015b). Miniaturised medium pressure capillary liquid chromatography system with flexible open platform design using off-the-shelf microfluidic components. Anal. Chim. Acta X. 896, 166–176. doi:10.1016/j.aca.2015.09.015
Ligterink, N. F. W., Grimaudo, V., Moreno-García, P., Lukmanov, R., Tulej, M., Leya, I., et al. (2020). Origin: A novel and compact laser desorption – mass spectrometry system for sensitive in situ detection of amino acids on extraterrestrial surfaces. Sci. Rep. 10 (1), 9641. doi:10.1038/s41598-020-66240-1
Ligterink, N., Riedo, A., Tulej, M., Lukmanov, R., Grimaudo, V., Koning, C. d., et al. (2021). Detecting the elemental and molecular signatures of life: Laser-based mass spectrometry technologies. Bull. Am. Astron. Soc. 53 (4). doi:10.3847/25c2cfeb.782ec4d0
Limero, T., Brokenshire, J., Cumming, C., Overton, E., Carney, K., Cross, J., et al. (1992). A volatile organic analyzer for space station: Description and evaluation of a gas chromatography/ion mobility spectrometer. SAE Trans., 921385–921424. doi:10.4271/921385
Limero, T., Reese, E., and Cheng, P. (2008). “Demonstration of the microanalyzer's measurement of common trace volatile organic compounds in spacecraft atmospheres,” in International conference on environmental systems (Warrendale, PA: SAE International).
Limero, T., Reese, E., Cheng, P., and Trowbridge, J. (2011). Preparation of a gas chromatograph-differential mobility spectrometer to measure target volatile organic compounds on the international space station. Int. J. Ion. Mobil. Spectrom. 14 (2), 81–91. doi:10.1007/s12127-011-0071-z
Limero, T., Wallace, W., and James, J. T. (2014). “Operational validation of the air quality monitor on the International Space Station,” in 44th international Conference on environmental systems, (emmaus, PA: The international conference on environmental systems).
Lin, Y., Zhong, F., Henderson, B. L., Abrahamsson, V., Kanik, I., Gross, J., et al. (2021). Masex — a dedicated life detection mission on Mars. Bull. Am. Astron. Soc. 53 (4), 333. doi:10.3847/25c2cfeb.33226b42
Lindenburg, P. W., Haselberg, R., Rozing, G., and Ramautar, R. (2015). Developments in interfacing designs for CE–MS: Towards enabling tools for proteomics and metabolomics. Chromatographia 78 (5-6), 367–377. doi:10.1007/s10337-014-2795-5
Lindsay, S., He, J., Sankey, O., Hapala, P., Jelinek, P., Zhang, P., et al. (2010). Recognition tunneling. Nanotechnology 21 (26), 262001. doi:10.1088/0957-4484/21/26/262001
Lipman, N. S., Jackson, L. R., Trudel, L. J., and Weis-Garcia, F. (2005). Monoclonal versus polyclonal antibodies: Distinguishing characteristics, applications, and information resources. ILAR J. 46 (3), 258–268. doi:10.1093/ilar.46.3.258
Liu, D.-L., Beegle, L. W., and Kanik, I. (2008). Analysis of underivatized amino acids in geological samples using ion-pairing liquid chromatography and electrospray tandem mass spectrometry. Astrobiology 8 (2), 229–241. doi:10.1089/ast.2007.0176
Lopez-Reyes, G., Rull, F., Venegas, G., Westall, F., Foucher, F., Bost, N., et al. (2013). Analysis of the scientific capabilities of the ExoMars Raman laser spectrometer instrument. Eur. J. Mineral. 25 (5), 721–733. doi:10.1127/0935-1221/2013/0025-2317
Lorenz, R. D., Turtle, E. P., Barnes, J. W., Trainer, M. G., Adams, D. S., Hibbard, K. E., et al. (2018). “Dragonfly: A rotorcraft lander concept for scientific exploration at titan,” in Johns hopkins APL technical digest. Laurel, MD: Johns Hopkins APL.
Lu, X., and Chen, Y. (2002). Chiral separation of amino acids derivatized with fluoresceine-5-isothiocyanate by capillary electrophoresis and laser-induced fluorescence detection using mixed selectors of β-cyclodextrin and sodium taurocholate. J. Chromatogr. A 955 (1), 133–140. doi:10.1016/s0021-9673(02)00186-3
Lunte, S. M., and Radzik, D. M. (1996). “Chapter 1 principles of capillary electrophoresis,” in Progress in pharmaceutical and biomedical analysis. Editors S. M. Lunte, and D. M. Radzik (Elsevier), 3.
Luong, D., Court, R. W., Sims, M. R., Cullen, D. C., and Sephton, M. A. (2014). Extracting organic matter on mars: A comparison of methods involving subcritical water, surfactant solutions and organic solvents. Planet. Space Sci. 99, 19–27. doi:10.1016/j.pss.2014.05.001
Luong, D., Sephton, M. A., and Watson, J. S. (2015). Subcritical water extraction of organic matter from sedimentary rocks. Anal. Chim. Acta X. 879, 48–57. doi:10.1016/j.aca.2015.04.027
Lymer, E. A., Konstantinidis, M., Lalla, E. A., Daly, M. G., and Tait, K. T. (2021). UV time-resolved laser-induced fluorescence spectroscopy of amino acids found in meteorites: Implications for space science and exploration. Astrobiology 21 (11), 1350–1362. doi:10.1089/ast.2021.0006
Lynch, K. B., Chen, A., and Liu, S. (2018). Miniaturized high-performance liquid chromatography instrumentation. Talanta 177, 94–103. doi:10.1016/j.talanta.2017.09.016
Lynch, K. B., Chen, A., Yang, Y., Lu, J. J., and Liu, S. (2017). High-performance liquid chromatographic cartridge with gradient elution capability coupled with UV absorbance detector and mass spectrometer for peptide and protein analysis. J. Sep. Sci. 40 (13), 2752–2758. doi:10.1002/jssc.201700185
MacKenzie, S. M., Kirby, K. W., and Greenauer, P. J. (2022a). “Enceladus orbilander: A flagship mission concept for astrobiology,” in Planetary mission concept study for the 2023-2032 decadal survey. National Academies of Sciences, Engineering, and Medicine. Available at: https://science.nasa.gov/science-red/s3fs-public/atoms/files/Enceladus%20Orbilander.pdf.
MacKenzie, S. M., Neveu, M., Davila, A. F., Lunine, J. I., Cable, M. L., Phillips-Lander, C. M., et al. (2022b). Science objectives for flagship-class mission concepts for the search for evidence of life at Enceladus. Astrobiology 22 (7), 685–712. doi:10.1089/ast.2020.2425
Madzunkov, S., Bae, B., Simcic, J., Rellergert, W., Gill, J., Schaefer, R., et al. (2016). “Progress report on the spacecraft atmosphere monitor,” in International conference on environmental systems (Warrendale, PA: SAE International).
Maggiori, C., Stromberg, J., Blanco, Y., Goordial, J., Cloutis, E., García-Villadangos, M., et al. (2020). The limits, capabilities, and potential for life detection with MinION sequencing in a Paleochannel Mars analog. Astrobiology 20 (3), 375–393. doi:10.1089/ast.2018.1964
Magnusson B., and Örnemark U. (Editors) (2014). Eurachem guide: The fitness for purpose of analytical methods – a laboratory guide to method validation and related topics. Teddington, United Kingdom: Eurachem. Available at: http://www.eurachem.org.
Mahaffy, P. R., Webster, C. R., Cabane, M., Conrad, P. G., Coll, P., Atreya, S. K., et al. (2012). The sample analysis at Mars investigation and instrument suite. Space Sci. Rev. 170 (1), 401–478. doi:10.1007/s11214-012-9879-z
Makarov, A., Denisov, E., Kholomeev, A., Balschun, W., Lange, O., Strupat, K., et al. (2006). Performance evaluation of a hybrid linear ion trap/Orbitrap mass spectrometer. Anal. Chem. 78 (7), 2113–2120. doi:10.1021/ac0518811
Makarov, A. (2000). Electrostatic axially harmonic orbital trapping: A high-performance technique of mass analysis. Anal. Chem. 72 (6), 1156–1162. doi:10.1021/ac991131p
Malaska, M. J., Bhartia, R., Manatt, K. S., Priscu, J. C., Abbey, W. J., Mellerowicz, B., et al. (2020). Subsurface in situ detection of microbes and diverse organic matter hotspots in the Greenland ice sheet. Astrobiology 20 (10), 1185–1211. doi:10.1089/ast.2020.2241
Managadze, G., Wurz, P., Sagdeev, R., Chumikov, A., Tuley, M., Yakovleva, M., et al. (2010). Study of the main geochemical characteristics of Phobos’ regolith using laser time-of-flight mass spectrometry. Sol. Syst. Res. 44 (5), 376–384. doi:10.1134/S0038094610050047
Mankins, J. C. (2009). Technology readiness assessments: A retrospective. Acta Astronaut. 65 (9-10), 1216–1223. doi:10.1016/j.actaastro.2009.03.058
Manz, A., Graber, N., and Widmer, H. á. (1990). Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sensors actuators B Chem. 1 (1-6), 244–248. doi:10.1016/0925-4005(90)80209-I
Mao, P., Wang, H.-T., Yang, P., and Wang, D. (2011). Multinozzle emitter arrays for nanoelectrospray mass spectrometry. Anal. Chem. 83 (15), 6082–6089. doi:10.1021/ac2011813
Marcet, I., Alvarez, C., Paredes, B., and Diaz, M. (2016). The use of sub-critical water hydrolysis for the recovery of peptides and free amino acids from food processing wastes. Review of sources and main parameters. Waste Manag. 49, 364–371. doi:10.1016/j.wasman.2016.01.009
Martin, P. E., Ehlmann, B. L., Thomas, N. H., Wiens, R. C., Hollis, J. J., Beegle, L. W., et al. (2020). Studies of a Lacustrine-volcanic Mars analog field site with Mars-2020-like instruments. Earth Space Sci. 7 (2), e2019EA000720. doi:10.1029/2019EA000720
Martin-Cuadrado, A. B., Senel, E., Martinez-Garcia, M., Cifuentes, A., Santos, F., Almansa, C., et al. (2019). Prokaryotic and viral community of the sulfate-rich crust from Penahueca ephemeral lake, an astrobiology analogue. Environ. Microbiol. 21 (10), 3577–3600. doi:10.1111/1462-2920.14680
Martins, Z. (2011). In situ biomarkers and the life marker chip. Astron. Geophys. 52 (1), 1.34–1.35. doi:10.1111/j.1468-4004.2011.52134.x
Mathies, R. A., Razu, M. E., Kim, J., Stockton, A. M., Turin, P., and Butterworth, A. (2017). Feasibility of detecting bioorganic compounds in Enceladus plumes with the Enceladus organic analyzer. Astrobiology 17 (9), 902–912. doi:10.1089/ast.2017.1660
Matz, L. M., Hill, H. H., Beegle, L. W., and Kanik, I. (2002). Investigation of drift gas selectivity in high resolution ion mobility spectrometry with mass spectrometry detection. J. Am. Soc. Mass Spectrom. 13 (4), 300–307. doi:10.1016/S1044-0305(01)00366-X
McCaig, H. C., Stockton, A., Crilly, C., Chung, S., Kanik, I., Lin, Y., et al. (2016). Supercritical carbon dioxide extraction of coronene in the presence of perchlorate for in situ chemical analysis of Martian regolith. Astrobiology 16 (9), 703–714. doi:10.1089/ast.2015.1443
McKay, C. P. (2008). An approach to searching for life on Mars, Europa, and Enceladus. Space Sci. Rev. 135 (1), 49–54. doi:10.1007/s11214-007-9229-8
Medina, D. A. V., Maciel, E. V. S., de Toffoli, A. L., and Lancas, F. M. (2020). Miniaturization of liquid chromatography coupled to mass spectrometry.: 2. Achievements on modern instrumentation for miniaturized liquid chromatography coupled to mass spectrometry. Trac. Trends Anal. Chem. 128, 115910. doi:10.1016/j.trac.2020.115910
Mejía-Carmona, K., Soares da Silva Burato, J., Borsatto, J. V. B., de Toffoli, A. L., and Lanças, F. M. (2020). Miniaturization of liquid chromatography coupled to mass spectrometry: 1. Current trends on miniaturized LC columns. Trac. Trends Anal. Chem. 122, 115735. doi:10.1016/j.trac.2019.115735
Merenbloom, S. I., Koeniger, S. L., Valentine, S. J., Plasencia, M. D., and Clemmer, D. E. (2006). IMS-IMS and IMS-IMS-IMS/MS for separating peptide and protein fragment ions. Anal. Chem. 78 (8), 2802–2809. doi:10.1021/ac052208e
Meunier, D., Sternberg, R., Mettetal, F., Buch, A., Coscia, D., Szopa, C., et al. (2007). A laboratory pilot for in situ analysis of refractory organic matter in Martian soil by gas chromatography–mass spectrometry. Adv. Space Res. 39 (3), 337–344. doi:10.1016/j.asr.2005.05.008
Mielczarek, P., Silberring, J., and Smoluch, M. (2020). Miniaturization in mass spectrometry. Mass Spectrom. Rev. 39 (5-6), 453–470. doi:10.1002/mas.21614
Mileikowsky, C., Cucinotta, F. A., Wilson, J. W., Gladman, B., Horneck, G., Lindegren, L., et al. (2000). Natural transfer of viable microbes in space: 1. From mars to earth and earth to mars. Icarus 145 (2), 391–427. doi:10.1006/icar.1999.6317
Millan, M., Teinturier, S., Malespin, C. A., Bonnet, J. Y., Buch, A., Dworkin, J. P., et al. (2021). Organic molecules revealed in Mars’s Bagnold Dunes by Curiosity’s derivatization experiment. Nat. Astron. 6, 129–140. doi:10.1038/s41550-021-01507-9
Miller, K. E., Eigenbrode, J. L., Freissinet, C., Glavin, D. P., Kotrc, B., Francois, P., et al. (2016). Potential precursor compounds for chlorohydrocarbons detected in Gale Crater, Mars, by the SAM instrument suite on the Curiosity Rover. J. Geophys. Res. Planets 121 (3), 296–308. doi:10.1002/2015je004939
Milman, B. L., and Zhurkovich, I. K. (2017). The chemical space for non-target analysis. TrAC Trends Anal. Chem. 97, 179–187. doi:10.1016/j.trac.2017.09.013
Misra, A. K., Acosta-Maeda, T. E., Sharma, S. K., McKay, C. P., Gasda, P. J., Taylor, G. J., et al. (2016). Standoff Biofinder" for fast, noncontact, nondestructive, large-area detection of biological materials for planetary exploration. Astrobiology 16 (9), 715–729. doi:10.1089/ast.2015.1400
Mißbach, H., Steininger, H., Thiel, V., and Goetz, W. (2019). Investigating the effect of perchlorate on flight-like gas chromatography–mass spectrometry as performed by MOMA on board the ExoMars 2020 Rover. Astrobiology 19 (11), 1339–1352. doi:10.1089/ast.2018.1997
Moini, M. (2007). Simplifying CE−MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Anal. Chem. 79 (11), 4241–4246. doi:10.1021/ac0704560
Mojarro, A., Hachey, J., Bailey, R., Brown, M., Doebler, R., Ruvkun, G., et al. (2019). Nucleic acid extraction and sequencing from low-biomass synthetic Mars analog soils for in situ life detection. Astrobiology 19 (9), 1139–1152. doi:10.1089/ast.2018.1929
Mojarro, A., Hachey, J., Ruvkun, G., Zuber, M. T., and Carr, C. E. (2018). CarrierSeq: A sequence analysis workflow for low-input nanopore sequencing. BMC Bioinforma. 19 (1), 108. doi:10.1186/s12859-018-2124-3
Mojarro, A., Jin, L., Szostak, J. W., Head, J. W., and Zuber, M. T. (2021). In search of the RNA world on Mars. Geobiology 19, 307–321. doi:10.1111/gbi.12433
Mojarro, A., Ruvkun, G., Zuber, M. T., and Carr, C. E. (2017). Nucleic acid extraction from synthetic Mars analog soils for in situ life detection. Astrobiology 17 (8), 747–760. doi:10.1089/ast.2016.1535
Montagnac, G., Hao, J., Pedreira-Segade, U., and Daniel, I. (2021). Detection of nucleotides adsorbed onto clay by UV resonant Raman spectroscopy: A step towards the search for biosignatures on mars. Appl. Clay Sci. 200, 105824. doi:10.1016/j.clay.2020.105824
Mora, M. F., Greer, F., Stockton, A. M., Bryant, S., and Willis, P. A. (2011). Toward total automation of microfluidics for extraterrestial in situ analysis. Anal. Chem. 83 (22), 8636–8641. doi:10.1021/ac202095k
Mora, M. F., Kehl, F., Tavares da Costa, E., Bramall, N., and Willis, P. A. (2020). Fully automated microchip electrophoresis analyzer for potential life detection missions. Anal. Chem. 92 (19), 12959–12966. doi:10.1021/acs.analchem.0c01628
Mora, M. F., Kok, M. G. M., Noell, A., and Willis, P. A. (2022). Detection of biosignatures by capillary electrophoresis mass spectrometry in the presence of salts relevant to Ocean Worlds missions. Astrobiology 22 (8), 914–925. doi:10.1089/ast.2021.0091
Mora, M. F., Stockton, A. M., and Willis, P. A. (2013). Analysis of thiols by microchip capillary electrophoresis for in situ planetary investigations. Electrophoresis 34 (2), 309–316. doi:10.1002/elps.201200379
Mora, M. F., Stockton, A. M., and Willis, P. A. (2015). “Analysis of thiols by microchip capillary electrophoresis for in situ planetary investigations,” in Microchip capillary electrophoresis protocols (New York, NY: Humana Press), 43.
Mora, M. F., Stockton, A. M., and Willis, P. A. (2012). Microchip capillary electrophoresis instrumentation for in situ analysis in the search for extraterrestrial life. Electrophoresis 33 (17), 2624–2638. doi:10.1002/elps.201200102
Moreno-Paz, M., Gómez-Cifuentes, A., Ruiz-Bermejo, M., Hofstetter, O., Maquieira, A., Manchado, J. M., et al. (2018). Detecting nonvolatile life-and nonlife-derived organics in a carbonaceous chondrite analogue with a new multiplex immunoassay and its relevance for planetary exploration. Astrobiology 18 (8), 1041–1056. doi:10.1089/ast.2017.1747
Mukhin, L., Gelman, B., Lamonov, N., Melnikov, V., Nenarokov, D., Okhotnikov, B., et al. (1982). VENERA-13 and VENERA-14 gas chromatography analysis of the Venus atmosphere composition. Sov. Astron. Lett. 8, 216–218.
Mukhin, L., Nenarokov, D., Porschnev, N., Bondarev, V., Gelman, B., Israe, G., et al. (1987). Preliminary calibration results of Vega 1 and 2 SIGMA-3 gas chromatograph. Adv. Space Res. 7 (12), 329–335. doi:10.1016/0273-1177(87)90240-7
Murphy, J. P., Johnson, J., and Rainville, P. D. (2014). Enhancing mass spectrometry sensitivity by reducing chromatographic flow rates with ionKey. Milford, MA: Watersw Corp.
Nascetti, A., Mirasoli, M., Marchegiani, E., Zangheri, M., Costantini, F., Porchetta, A., et al. (2019). Integrated chemiluminescence-based lab-on-chip for detection of life markers in extraterrestrial environments. Biosens. Bioelectron. X. 123, 195–203. doi:10.1016/j.bios.2018.08.056
National Academies of Sciences and Medicine (2019). An astrobiology strategy for the search for life in the universe. Washington, DC: The National Academies Press.
Navarro-González, R., and McKay, C. P. (2011). Reply to comment by Biemann and Bada on “Reanalysis of the Viking results suggests perchlorate and organics at midlatitudes on Mars”. J. Geophys. Res. 116 (E12), E12002. doi:10.1029/2011je003880
Navarro-González, R., Navarro, K. F., De La Rosa, J., Iñiguez, E., Molina, P., Miranda, L. D., et al. (2006). The limitations on organic detection in Mars-like soils by thermal volatilization–gas chromatography–MS and their implications for the Viking results. Proc. Natl. Acad. Sci. U. S. A. 103 (44), 16089–16094. doi:10.1073/pnas.0604210103
Navarro-González, R., Vargas, E., de la Rosa, J., Raga, A. C., and McKay, C. P. (2010). Reanalysis of the Viking results suggests perchlorate and organics at midlatitudes on Mars. J. Geophys. Res. 115, E12010. doi:10.1029/2010JE003599
Neveu, M., Hays, L. E., Voytek, M. A., New, M. H., and Schulte, M. D. (2018). The ladder of Life detection. Astrobiology 18 (11), 1375–1402. doi:10.1089/ast.2017.1773
Niedzwiecki, D. J., Chou, Y.-C., Xia, Z., Thei, F., and Drndić, M. (2020). Detection of single analyte and environmental samples with silicon nitride nanopores: Antarctic dirt particulates and DNA in artificial seawater. Rev. Sci. Instrum. 91 (3), 031301. doi:10.1063/1.5138210
Niemann, H. B., Atreya, S. K., Demick, J. E., Gautier, D., Haberman, J. A., Harpold, D. N., et al. (2010). Composition of Titan's lower atmosphere and simple surface volatiles as measured by the Cassini-Huygens probe gas chromatograph mass spectrometer experiment. J. Geophys. Res. 115 (E12), E12006. doi:10.1029/2010je003659
Noell, A. C., Fisher, A. M., Fors-Francis, K., and Sherrit, S. (2018). Subcritical water extraction of amino acids from Mars analog soils. Electrophoresis 39 (22), 2854–2863. doi:10.1002/elps.201700459
Oba, Y., Takano, Y., Naraoka, H., Furukawa, Y., Glavin, D. P., Dworkin, J. P., et al. (2020). Extraterrestrial hexamethylenetetramine in meteorites—A precursor of prebiotic chemistry in the inner solar system. Nat. Commun. 11 (1), 6243–6248. doi:10.1038/s41467-020-20038-x
Ohshiro, T., and Taniguchi, M. (2022). Review of the use of nanodevices to detect single molecules. Anal. Biochem. 654, 114645. doi:10.1016/j.ab.2022.114645
Orta, D. R., Levy, R. J., Mudgett, P. D., Homan, M. E., Schultz, J. R., and Sauer, R. L. (1997). “Capillary electrophoresis for spacecraft drinking water analysis: Methods and breadboard development,” in International conference on environmental systems (Warrendale, PA: SAE International).
Orzechowska, G. E., Kidd, R. D., Foing, B. H., Kanik, I., Stoker, C., and Ehrenfreund, P. (2011). Analysis of Mars analogue soil samples using solid-phase microextraction, organic solvent extraction and gas chromatography/mass spectrometry. Int. J. Astrobiol. 10 (3), 209–219. doi:10.1017/S1473550410000443
Ostermann, A. I., Müller, M., Willenberg, I., and Schebb, N. H. (2014). Determining the fatty acid composition in plasma and tissues as fatty acid methyl esters using gas chromatography–a comparison of different derivatization and extraction procedures. Prostagl. Leukot. Essent. Fat. Acids 91 (6), 235–241. doi:10.1016/j.plefa.2014.10.002
Ouldali, H., Sarthak, K., Ensslen, T., Piguet, F., Manivet, P., Pelta, J., et al. (2020). Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 38 (2), 176–181. doi:10.1038/s41587-019-0345-2
Oyama, V., Berdahl, B., Carle, G., Lehwalt, M., and Ginoza, H. (1976). The search for life on Mars: Viking 1976 gas changes as indicators of biological activity. Orig. Life 7 (3), 313–333. doi:10.1007/BF00926949
Oyama, V. I., and Berdahl, B. J. (1977). The Viking gas exchange experiment results from Chryse and Utopia surface samples. J. Geophys. Res. 82 (28), 4669–4676. doi:10.1029/JS082i028p04669
Oyama, V. I., Carle, G. C., Woeller, F., and Pollack, J. B. (1979). Laboratory corroboration of the pioneer venus gas chromatograph analyses. Science 205 (4401), 52–54. doi:10.1126/science.205.4401.52
Oyama, V. I., Carle, G. C., Woeller, F., Pollack, J. B., Reynolds, R. T., and Craig, R. A. (1980). Pioneer Venus gas chromatography of the lower atmosphere of Venus. J. Geophys. Res. 85 (A13), 7891–7902. doi:10.1029/JA085iA13p07891
Oyama, V. I. (1972). The gas exchange experiment for life detection: The Viking Mars Lander. Icarus 16 (1), 167–184. doi:10.1016/0019-1035(72)90144-3
Pace, N. R., Thomas, B. C., and Woese, C. R. (1999). “Probing RNA structure, function, and history by comparative analysis,” in The RNA world. Editors R. F. Gesteland, T. R. Cech, J. F. Atkins, and Cold Spring Harbor. Second Edition (NY: Cold Spring Harbor Laboratory Press), 113.
Palmer, P. T., and Limero, T. F. (2001). Mass spectrometry in the US space program: Past, present, and future. J. Am. Soc. Mass Spectrom. 12 (6), 656–675. doi:10.1016/S1044-0305(01)00249-5
Pappalardo, R. T., Vance, S., Bagenal, F., Bills, B. G., Blaney, D. L., Blankenship, D. D., et al. (2013). Science potential from a europa lander. Astrobiology 13 (8), 740–773. doi:10.1089/ast.2013.1003
Parra, M., Jung, J., Boone, T. D., Tran, L., Blaber, E. A., Brown, M., et al. (2017). Microgravity validation of a novel system for RNA isolation and multiplex quantitative real time PCR analysis of gene expression on the International Space Station. PLOS ONE 12 (9), e0183480. doi:10.1371/journal.pone.0183480
Parro, V., de Diego-Castilla, G., Moreno-Paz, M., Blanco, Y., Cruz-Gil, P., Rodríguez-Manfredi, J. A., et al. (2011a). A microbial oasis in the hypersaline Atacama subsurface discovered by a life detector chip: Implications for the search for life on mars. Astrobiology 11 (10), 969–996. doi:10.1089/ast.2011.0654
Parro, V., de Diego-Castilla, G., Rodríguez-Manfredi, J. A., Rivas, L. A., Blanco-López, Y., Sebastián, E., et al. (2011b). SOLID3: A multiplex antibody microarray-based optical sensor instrument for in situ life detection in planetary exploration. Astrobiology 11 (1), 15–28. doi:10.1089/ast.2010.0501
Parro, V., Fernández-Calvo, P., Rodriguez Manfredi, J. A., Moreno-Paz, M., Rivas, L. A., García-Villadangos, M., et al. (2008a). SOLID2: An antibody array-based life-detector instrument in a mars drilling simulation experiment (MARTE). Astrobiology 8 (5), 987–999. doi:10.1089/ast.2007.0126
Parro, V., Rivas, L. A., and Gómez-Elvira, J. (2008b). “Protein microarrays-based strategies for life detection in astrobiology,” in Strategies of life detection. Space sciences series of ISSI. Editors O. Botta, J. L. Bada, J. Gomez-Elvira, E. Javaux, F. Selsis, and R. Summons (Boston, MA: Springer), 293.
Parro, V., Rodríguez-Manfredi, J., Briones, C., Compostizo, C., Herrero, P., Vez, E., et al. (2005). Instrument development to search for biomarkers on mars: Terrestrial acidophile, iron-powered chemolithoautotrophic communities as model systems. Planet. Space Sci. 53 (7), 729–737. doi:10.1016/j.pss.2005.02.003
Piendl, S. K., Raddatz, C.-R., Hartner, N. T., Thoben, C., Warias, R., Zimmermann, S., et al. (2019). 2D in seconds: Coupling of chip-HPLC with Ion mobility spectrometry. Anal. Chem. 91 (12), 7613–7620. doi:10.1021/acs.analchem.9b00302
Poinot, P., and Geffroy-Rodier, C. (2015). Searching for organic compounds in the Universe. TrAC Trends Anal. Chem. 65, 1–12. doi:10.1016/j.trac.2014.09.009
Pontefract, A., Hachey, J., Zuber, M. T., Ruvkun, G., and Carr, C. E. (2018). Sequencing nothing: Exploring failure modes of nanopore sensing and implications for life detection. Life Sci. space Res. 18, 80–86. doi:10.1016/j.lssr.2018.05.004
Prebihalo, S. E., Berrier, K. L., Freye, C. E., Bahaghighat, H. D., Moore, N. R., Pinkerton, D. K., et al. (2018). Multidimensional gas chromatography: Advances in instrumentation, chemometrics, and applications. Anal. Chem. 90 (1), 505–532. doi:10.1021/acs.analchem.7b04226
Pugmire, D. L., Waddell, E. A., Haasch, R., Tarlov, M. J., and Locascio, L. E. (2002). Surface characterization of laser-ablated polymers used for microfluidics. Anal. Chem. 74 (4), 871–878. doi:10.1021/ac011026r
Quick, J. (2018). One-pot ligation protocol for Oxford Nanopore libraries. Berkeley, CA: Protocols.io. Available at: https://www.protocols.io/view/one-pot-ligation-protocol-for-oxford-nanopore-libr-36wgqpr5vk57.
Rahimi, F., Chatzimichail, S., Saifuddin, A., Surman, A. J., Taylor-Robinson, S. D., and Salehi-Reyhani, A. (2020). A review of portable high-performance liquid chromatography: The future of the field? Chromatographia 83 (10), 1165–1195. doi:10.1007/s10337-020-03944-6
Raposo, F., and Ibelli-Bianco, C. (2020). Performance parameters for analytical method validation: Controversies and discrepancies among numerous guidelines. TrAC Trends Anal. Chem. 129, 115913. doi:10.1016/j.trac.2020.115913
Raymond-Bouchard, I., Maggiori, C., Brennan, L., Altshuler, I., Manchado, J. M., Parro, V., et al. (2022). Assessment of automated nucleic acid extraction systems in combination with MinION sequencing as potential tools for the detection of microbial biosignatures. Astrobiology 22 (1), 87–103. doi:10.1089/ast.2020.2349
Razzell Hollis, J., Fornaro, T., Rapin, W., Wade, J., Vicente-Retortillo, Á., Steele, A., et al. (2021). Detection and degradation of adenosine monophosphate in perchlorate-spiked martian regolith analog, by deep-ultraviolet spectroscopy. Astrobiology 21 (5), 511–525. doi:10.1089/ast.2020.2362
Regmi, B. P., and Agah, M. (2018). Micro gas chromatography: An overview of critical components and their integration. Anal. Chem. 90 (22), 13133–13150. doi:10.1021/acs.analchem.8b01461
Regmi, B. P., Chan, R., Atta, A., and Agah, M. (2018). Ionic liquid-coated alumina-pretreated micro gas chromatography columns for high-efficient separations. J. Chromatogr. A 1566, 124–134. doi:10.1016/j.chroma.2018.06.058
Reinhardt, M., Goetz, W., and Thiel, V. (2020). Testing flight-like pyrolysis gas chromatography-mass spectrometry as performed by the Mars Organic Molecule Analyzer onboard the ExoMars 2020 Rover on Oxia Planum analog samples. Astrobiology 20 (3), 415–428. doi:10.1089/ast.2019.2143
Ren, Z., Guo, M., Cheng, Y., Wang, Y., Sun, W., Zhang, H., et al. (2018). A review of the development and application of space miniature mass spectrometers. Vacuum 155, 108–117. doi:10.1016/j.vacuum.2018.05.048
Restrepo-Pérez, L., Joo, C., and Dekker, C. (2018). Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 13 (9), 786–796. doi:10.1038/s41565-018-0236-6
Rezzonico, F. (2014). Nanopore-based instruments as biosensors for future planetary missions. Astrobiology 14 (4), 344–351. doi:10.1089/ast.2013.1120
Ribette, T., Leroux, B., Eddhif, B., Allavena, A., David, M., Sternberg, R., et al. (2019). Primary step towards in situ detection of chemical biomarkers in the UNIVERSE via liquid-based analytical system: Development of an automated online trapping/liquid chromatography system. Molecules 24 (7), 1429. doi:10.3390/molecules24071429
Riedo, A., Bieler, A., Neuland, M., Tulej, M., and Wurz, P. (2013). Performance evaluation of a miniature laser ablation time-of-flight mass spectrometer designed for in situ investigations in planetary space research. J. Mass Spectrom. 48 (1), 1–15. doi:10.1002/jms.3104
Riedo, A., de Koning, C., Stevens, A. H., Cockell, C. S., McDonald, A., Lopez, A. C., et al. (2020). The detection of elemental signatures of microbes in martian mudstone analogs using high spatial resolution laser ablation ionization mass spectrometry. Astrobiology 20 (10), 1224–1235. doi:10.1089/ast.2019.2087
Riedo, A., Tulej, M., Rohner, U., and Wurz, P. (2017). High-speed microstrip multi-anode multichannel plate detector system. Rev. Sci. Instrum. 88 (4), 045114. doi:10.1063/1.4981813
Rigano, F., Tranchida, P. Q., Dugo, P., and Mondello, L. (2019). High-performance liquid chromatography combined with electron ionization mass spectrometry: A review. TrAC Trends Anal. Chem. 118, 112–122. doi:10.1016/j.trac.2019.05.032
Rivas, L. A., García-Villadangos, M., Moreno-Paz, M., Cruz-Gil, P., Gómez-Elvira, J., and Parro, V. (2008). A 200-antibody microarray biochip for environmental monitoring: Searching for universal microbial biomarkers through immunoprofiling. Anal. Chem. 80 (21), 7970–7979. doi:10.1021/ac8008093
Roach, J., Williams, A. J., Eigenbrode, J. L., Millan, M., Williams, R. H., Buch, A., et al. (2021). “Organic molecules on mars: Results from the first in situ TMAH thermochemolysis experiment at gale crater, mars,” in AGU fall meeting 2021 (Washington, D.C.: American Geophysical Union).
Roda, A., Mirasoli, M., Guardigli, M., Zangheri, M., Caliceti, C., Calabria, D., et al. (2018). Advanced biosensors for monitoring astronauts’ health during long-duration space missions. Biosens. Bioelectron. X. 111, 18–26. doi:10.1016/j.bios.2018.03.062
Roda, A., Mirasoli, M., Michelini, E., Di Fusco, M., Zangheri, M., Cevenini, L., et al. (2016). Progress in chemical luminescence-based biosensors: A critical review. Biosens. Bioelectron. X. 76, 164–179. doi:10.1016/j.bios.2015.06.017
Rodier, C., Sternberg, R., Raulin, F., and Vidal-Madjar, C. (2001). Chemical derivatization of amino acids for in situ analysis of Martian samples by gas chromatography. J. Chromatogr. A 915 (1), 199–207. doi:10.1016/S0021-9673(01)00625-2
Rodin, A., Vinogradov, I., Zenevich, S., Spiridonov, M., Gazizov, I., Kazakov, V., et al. (2020). Martian multichannel diode laser spectrometer (M-DLS) for in-situ atmospheric composition measurements on Mars onboard ExoMars-2022 landing platform. Appl. Sci. (Basel). 10 (24), 8805. doi:10.3390/app10248805
Rohner, U., Whitby, J. A., and Wurz, P. (2003). A miniature laser ablation time-of-flight mass spectrometer for in situ planetary exploration. Meas. Sci. Technol. 14 (12), 2159–2164. doi:10.1088/0957-0233/14/12/017
Röling, W. F. M., Aerts, J. W., Patty, C. H. L., Ten Kate, I. L., Ehrenfreund, P., and Direito, S. O. L. (2015). The significance of microbe-mineral-biomarker interactions in the detection of life on Mars and beyond. Astrobiology 15 (6), 492–507. doi:10.1089/ast.2014.1276
Rosenbauer, H., Fuselier, S. A., Ghielmetti, A., Greenberg, J. M., Goesmann, F., Ulamec, S., et al. (1999). The COSAC experiment on the Lander of the ROSETTA mission. Adv. Space Res. 23 (2), 333–340. doi:10.1016/S0273-1177(99)00054-X
Ruf, A., d'Hendecourt, L. L. S., and Schmitt-Kopplin, P. (2018). Data-driven astrochemistry: One step further within the origin of life puzzle. Life 8 (2), 18. doi:10.3390/life8020018
Ruf, A., Poinot, P., Geffroy, C., Le Sergeant d’Hendecourt, L., and Danger, G. (2019). Data-driven UPLC-Orbitrap ms analysis in astrochemistry. Life 9 (2), 35. doi:10.3390/life9020035
Rull, F., Maurice, S., Hutchinson, I., Moral, A., Perez, C., Diaz, C., et al. (2017). The Raman laser spectrometer for the ExoMars rover mission to mars. Astrobiology 17 (6-7), 627–654. doi:10.1089/ast.2016.1567
Rushneck, D. R., Diaz, A. V., Howarth, D. W., Rampacek, J., Olson, K. W., Dencker, W. D., et al. (1978). Viking gas chromatograph-mass spectrometer. Rev. Sci. Instrum. 49 (6), 817–834. doi:10.1063/1.1135623
Sabina, J., and Leamon, J. H. (2015). Bias in whole genome amplification: Causes and considerationsWhole genome amplification. Methods in molecular biology. Editor T. Kroneis (New York, NY: Humana Press), 15.
Sandford, M. W., Misra, A. K., Acosta-Maeda, T. E., Sharma, S. K., Porter, J. N., Egan, M. J., et al. (2021). Detecting minerals and organics relevant to planetary exploration using a compact portable remote Raman system at 122 Meters. Appl. Spectrosc. 75 (3), 299–306. doi:10.1177/0003702820943669
Santos, M. S. F., Cordeiro, T. G., Noell, A. C., Garcia, C. D., and Mora, M. F. (2018). Analysis of inorganic cations and amino acids in high salinity samples by capillary electrophoresis and conductivity detection: Implications for in-situ exploration of ocean worlds. Electrophoresis 39 (22), 2890–2897. doi:10.1002/elps.201800266
Schaeper, J. P., and Sepaniak, M. J. (2000). Parameters affecting reproducibility in capillary electrophoresis. Electrophoresis 21, 14212–14297. doi:10.1002/(SICI)1522-2683(20000401)21:7<1421::AID-ELPS1421>3.0
Schmidt, A., Karas, M., and Dülcks, T. (2003). Effect of different solution flow rates on analyte ion signals in nano-ESI MS, or: When does ESI turn into nano-ESI? J. Am. Soc. Mass Spectrom. 14 (5), 492–500. doi:10.1016/S1044-0305(03)00128-4
Schowalter, S., Madzunkov, S., Darrach, M., Diaz, E., Moore, B., Simcic, J., et al. (2019). “The technology demonstration of the spacecraft atmosphere monitor,” in International conference on environmental systems (Warrendale, PA: SAE International).
Seaton, K. M., Cable, M. L., and Stockton, A. M. (2021). Analytical chemistry in astrobiology. Anal. Chem. 93, 5981–5997. doi:10.1021/acs.analchem.0c04271
Seaton, K. M., Cable, M. L., and Stockton, A. M. (2022). Analytical chemistry throughout this solar system. Annu. Rev. Anal. Chem. Palo. Alto. Calif. 15 (1), 197–219. doi:10.1146/annurev-anchem-061020-125416
Selliez, L., Briois, C., Carrasco, N., Thirkell, L., Thissen, R., Ito, M., et al. (2019). Identification of organic molecules with a laboratory prototype based on the Laser Ablation-CosmOrbitrap. Planet. Space Sci. 170, 42–51. doi:10.1016/j.pss.2019.03.003
Selliez, L., Maillard, J., Cherville, B., Gautier, T., Thirkell, L., Gaubicher, B., et al. (2020). High-resolution mass spectrometry for future space missions: Comparative analysis of complex organic matter with LAb-CosmOrbitrap and laser desorption/ionization Fourier transform ion cyclotron resonance. Rapid Commun. Mass Spectrom. 34 (10), e8645. doi:10.1002/rcm.8645
Šesták, J., Moravcová, D., and Kahle, V. (2015). Instrument platforms for nano liquid chromatography. J. Chromatogr. A 1421, 2–17. doi:10.1016/j.chroma.2015.07.090
Sharma, S., Plistil, A., Barnett, H. E., Tolley, H. D., Farnsworth, P. B., Stearns, S. D., et al. (2015a). Hand-portable gradient capillary liquid chromatography pumping system. Anal. Chem. 87 (20), 10457–10461. doi:10.1021/acs.analchem.5b02583
Sharma, S., Plistil, A., Simpson, R. S., Liu, K., Farnsworth, P. B., Stearns, S. D., et al. (2014). Instrumentation for hand-portable liquid chromatography. J. Chromatogr. A 1327, 80–89. doi:10.1016/j.chroma.2013.12.059
Sharma, S., Tolley, L. T., Tolley, H. D., Plistil, A., Stearns, S. D., and Lee, M. L. (2015b). Hand-portable liquid chromatographic instrumentation. J. Chromatogr. A 1421, 38–47. doi:10.1016/j.chroma.2015.07.119
Shelor, C. P., Dasgupta, P. K., Aubrey, A., Davila, A. F., Lee, M. C., McKay, C. P., et al. (2014). What can in situ ion chromatography offer for Mars exploration? Astrobiology 14 (7), 577–588. doi:10.1089/ast.2013.1131
Shelor, C. P., and Dasgupta, P. K. (2017). Automated programmable pressurized carbonic acid eluent ion exclusion chromatography of organic acids. J. Chromatogr. A 1523, 300–308. doi:10.1016/j.chroma.2017.05.036
Shelor, C. P., Yoshikawa, K., and Dasgupta, P. K. (2017). Automated programmable preparation of carbonate-bicarbonate eluents for ion chromatography with pressurized carbon dioxide. Anal. Chem. 89 (18), 10063–10070. doi:10.1021/acs.analchem.7b02808
Sheretov, É., Gurov, V., Dubkov, M., and Korneeva, O. (2000). A new monopole mass-analyzer based on a hyperboloid electrode system. Tech. Phys. Lett. 26 (1), 21–22. doi:10.1134/1.1262726
Sherrit, S., Noell, A. C., Fisher, A., Lee, M. C., Takano, N., Bao, X. Q., et al. (2017). A microfluidic sub-critical water extraction instrument. Rev. Sci. Instrum. 88 (11), 114101. doi:10.1063/1.4999932
Shkolyar, S., Eshelman, E. J., Farmer, J. D., Hamilton, D., Daly, M. G., and Youngbull, C. (2018). Detecting kerogen as a biosignature using colocated UV time-gated Raman and fluorescence spectroscopy. Astrobiology 18 (4), 431–453. doi:10.1089/ast.2017.1716
Shkolyar, S., Lalla, E., Konstantindis, M., Cote, K., Daly, M. G., and Steele, A. (2021). Detecting Ce3+ as a biosignature mimicker using UV time-resolved laser-induced fluorescence and Raman spectroscopy: Implications for planetary missions. Icarus 354, 114093. doi:10.1016/j.icarus.2020.114093
Shortt, B. J., Darrach, M. R., Holland, P. M., and Chutjian, A. (2005). Miniaturized system of a gas chromatograph coupled with a Paul ion trap mass spectrometer. J. Mass Spectrom. 40 (1), 36–42. doi:10.1002/jms.768
Shvartsburg, A. A., Anderson, G. A., and Smith, R. D. (2013). Pushing the frontier of high-definition ion mobility spectrometry using FAIMS. Mass Spectrom. (Tokyo, Jpn. 2, S0011. doi:10.5702/massspectrometry.S0011
Shvartsburg, A. A., Isaac, G., Leveque, N., Smith, R. D., and Metz, T. O. (2011). Separation and classification of lipids using differential ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 22 (7), 1146–1155. doi:10.1007/s13361-011-0114-z
Siljestrom, S., Freissinet, C., Goesmann, F., Steininger, H., Goetz, W., Steele, A., et al. (2014). Comparison of prototype and laboratory experiments on MOMA GCMS: Results from the AMASE11 campaign. Astrobiology 14 (9), 780–797. doi:10.1089/ast.2014.1197
Siljeström, S., Li, X., Brinckerhoff, W., van Amerom, F., and Cady, S. L. (2021). ExoMars Mars Organic Molecule Analyzer (MOMA) laser desorption/ionization mass spectrometry (LDI-MS) analysis of phototrophic communities from a silica-depositing hot spring in Yellowstone National Park, USA. Astrobiology 21 (12), 1515–1525. doi:10.1089/ast.2020.2368
Simcic, J., Nikolić, D., Belousov, A., Atkinson, D., Lee, C., Madzunkov, S., et al. (2021). Quadrupole ion trap mass spectrometer for ice giant atmospheres exploration. Space Sci. Rev. 217 (1), 13–27. doi:10.1007/s11214-020-00785-5
Sims, M., Cullen, D., Bannister, N., Grant, W., Henry, O., Jones, R., et al. (2005). The specific molecular identification of life experiment (SMILE). Planet. Space Sci. 53 (8), 781–791. doi:10.1016/j.pss.2005.03.006
Sims, M. R., Cullen, D. C., Rix, C. S., Buckley, A., Derveni, M., Evans, D., et al. (2012). Development status of the life marker chip instrument for ExoMars. Planet. Space Sci. 72 (1), 129–137. doi:10.1016/j.pss.2012.04.007
Sims, M. R., Pillinger, C. T., Wright, I. P., Dowson, J., Whitehead, S., Wells, A., et al. (1999). Beagle 2: A proposed exobiology lander for ESA's 2003 mars express mission. Adv. Space Res. 23 (11), 1925–1928. doi:10.1016/S0273-1177(99)00280-X
Sims, M. R., Pillinger, C., Wright, I., Morgan, G., Praine, I., Fraser, G. W., et al. (2000). “Instrumentation on beagle 2: The astrobiology lander on ESA's 2003 mars express mission,” in Instruments, methods, and missions for astrobiology III (Bellingham, WA: International Society for Optics and Photonics), 36.
Siuzdak, G. (2005). Expanding role of mass spectrometry in biotechnology. 2nd ed. San Diego, CA: MCC Press.
Skelley, A. M., Aubrey, A. D., Willis, P. A., Amashukeli, X., Ehrenfreund, P., Bada, J. L., et al. (2007). Organic amine biomarker detection in the Yungay region of the Atacama Desert with the Urey instrument. J. Geophys. Res. 112 (G4), 329. doi:10.1029/2006jg000329
Skelley, A. M., Cleaves, H. J., Jayarajah, C. N., Bada, J. L., and Mathies, R. A. (2006). Application of the Mars Organic Analyzer to nucleobase and amine biomarker detection. Astrobiology 6 (6), 824–837. doi:10.1089/ast.2006.6.824
Skelley, A. M., and Mathies, R. A. (2003). Chiral separation of fluorescamine-labeled amino acids using microfabricated capillary electrophoresis devices for extraterrestrial exploration. J. Chromatogr. A 1021 (1), 191–199. doi:10.1016/j.chroma.2003.08.096
Skelley, A. M., Scherer, J. R., Aubrey, A. D., Grover, W. H., Ivester, R. H. C., Ehrenfreund, P., et al. (2005). Development and evaluation of a microdevice for amino acid biomarker detection and analysis on Mars. Proc. Natl. Acad. Sci. U. S. A. 102 (4), 1041–1046. doi:10.1073/pnas.0406798102
Smilde, A., Bro, R., and Geladi, P. (2005). Multi-way analysis: Applications in the chemical sciences. Hoboken, NJ: John Wiley & Sons.
Smoker, R. E., and Smith, S. (2007). System cost growth associated with technology-readiness level. J. Parametrics 26 (1), 8–38. doi:10.1080/10157891.2007.10462276
Snyder, A. P., Thornton, S. N., Dworzanski, J. P., and Meuzelaar, H. L. C. (1996). Detection of the picolinic acid biomarker in Bacillus spores using a potentially field-portable pyrolysis—Gas chromatography—Ion mobility spectrometry system. Field Anal. Chem. Technol. 1 (1), 492–599. doi:10.1002/(sici)1520-6521(1996)1:1<49:aid-fact7>3.0.co;2-9
Snyder, D. T., Pulliam, C. J., Ouyang, Z., and Cooks, R. G. (2016). Miniature and fieldable mass spectrometers: Recent advances. Anal. Chem. 88 (1), 2–29. doi:10.1021/acs.analchem.5b03070
Snyder, L. R., Kirkland, J. J., and Dolan, J. W. (2011). Introduction to modern liquid chromatography. Hoboken, NJ: John Wiley & Sons.
Snyder, L. R. (1997). Modern practice of liquid chromatography: Before and after 1971. J. Chem. Educ. 74 (1), 37. doi:10.1021/ed074p37
Sobron, P., Barge, L., Davila, A., Fahey, M., Krainak, M., Rehnmark, F., et al. (2019). “Programmable Raman sensing for in-situ planetary exploration,” in Lunar and planetary science conference (Houston, TX: Lunar and Planetary Institute).
Sokol, E., Noll, R. J., Cooks, R. G., Beegle, L. W., Kim, H. I., and Kanik, I. (2011). Miniature mass spectrometer equipped with electrospray and desorption electrospray ionization for direct analysis of organics from solids and solutions. Int. J. Mass Spectrom. 306 (2), 187–195. doi:10.1016/j.ijms.2010.10.019
Southard, A. E., Getty, S. A., Balvin, M., Elsila, J. E., Melina, A. E., Kotecki, C., et al. (2014). “Liquid chromatography-mass spectrometry interface for detection of extraterrestrial organics,” in 2014 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Southard, A., Getty, S., Ferrance, J., Balvin, M., Elsila, J., Stewart, D., et al. (2016b). “Design of an integrated LC-MS prototype for an in situ mission to an icy body in the Solar System,” in 3rd international workshop on instrumentation for planetary mission (Houston, TX: Lunar and Planetary Science Institute), 4080.
Southard, A., Getty, S., Ferrance, J., Balvin, M., Elsila, J., and Stewart, D. (2016a). “Oasis: A liquid chromatograph-mass spectrometer for detection of organics on icy surfaces,” in 47th lunar and planetary science conference (Houston, TX: Lunar and Planetary Science Institute), 2606.
Sricharoen, P., Limchoowong, N., Shelor, C. P., and Dasgupta, P. K. (2019). Carbonic acid eluent ion chromatography. Anal. Chem. 91 (5), 3636–3644. doi:10.1021/acs.analchem.8b05627
Stalport, F., Glavin, D. P., Eigenbrode, J. L., Bish, D., Blake, D., Coll, P., et al. (2012). The influence of mineralogy on recovering organic acids from Mars analogue materials using the “one-pot” derivatization experiment on the Sample Analysis at Mars (SAM) instrument suite. Planet. Space Sci. 67 (1), 1–13. doi:10.1016/j.pss.2012.02.010
Stevens, A. H., McDonald, A., de Koning, C., Riedo, A., Preston, L. J., Ehrenfreund, P., et al. (2019). Detectability of biosignatures in a low-biomass simulation of martian sediments. Sci. Rep. 9 (1), 9706. doi:10.1038/s41598-019-46239-z
Stockton, A., Duca, Z., Cato, M., Cantrell, T., Foreman, S., Kim, J., et al. (2016). “An organic analyzer instrument for highly sensitive in situ organic detection on an ice shell impact penetrator descent probe,” in 3rd international workshop on instrumentation for planetary mission (Houston, TX: Lunar and Planetary Science Institute), 4093.
Stockton, A. M., Chiesl, T. N., Lowenstein, T. K., Amashukeli, X., Grunthaner, F., and Mathies, R. A. (2009a). Capillary electrophoresis analysis of organic amines and amino acids in saline and acidic samples using the Mars Organic Analyzer. Astrobiology 9 (9), 823–831. doi:10.1089/ast.2009.0357
Stockton, A. M., Chiesl, T. N., Scherer, J. R., and Mathies, R. A. (2009b). Polycyclic aromatic hydrocarbon analysis with the Mars Organic Analyzer microchip capillary electrophoresis system. Anal. Chem. 81 (2), 790–796. doi:10.1021/ac802033u
Stockton, A. M., Tjin, C. C., Chiesl, T. N., and Mathies, R. A. (2011). Analysis of carbonaceous biomarkers with the Mars Organic Analyzer microchip capillary electrophoresis system: Carboxylic acids. Astrobiology 11 (6), 519–528. doi:10.1089/ast.2011.0634
Stockton, A. M., Tjin, C. C., Huang, G. L., Benhabib, M., Chiesl, T. N., and Mathies, R. A. (2010). Analysis of carbonaceous biomarkers with the Mars Organic Analyzer microchip capillary electrophoresis system: Aldehydes and ketones. Electrophoresis 31 (22), 3642–3649. doi:10.1002/elps.201000424
Stolz, A., Jooß, K., Höcker, O., Römer, J., Schlecht, J., and Neusüß, C. (2019). Recent advances in capillary electrophoresis-mass spectrometry: Instrumentation, methodology and applications. Electrophoresis 40 (1), 79–112. doi:10.1002/elps.201800331
Summons, R. E., Albrecht, P., McDonald, G., and Moldowan, J. M. (2008). Molecular biosignatures. Space Sci. Rev. 135 (1-4), 133–159. doi:10.1007/s11214-007-9256-5
Sutton, M. A., Burton, A. S., Zaikova, E., Sutton, R. E., Brinckerhoff, W. B., Bevilacqua, J. G., et al. (2019). Radiation tolerance of nanopore sequencing technology for life detection on mars and europa. Sci. Rep. 9 (1), 5370–5410. doi:10.1038/s41598-019-41488-4
Szopa, C., Coscia, D., Cabane, M., and Buch, A. (2017). “Miniaturized gas chromatography for space exploration: A 50 years history,” in 2017 symposium on design, test, integration and packaging of MEMS/MOEMS (DTIP) (Piscataway, NJ: IEEE), 1.
Szopa, C., Freissinet, C., Glavin, D. P., Millan, M., Buch, A., Franz, H. B., et al. (2020). First detections of dichlorobenzene isomers and trichloromethylpropane from organic matter indigenous to mars mudstone in gale crater, mars: Results from the sample analysis at mars instrument onboard the curiosity rover. Astrobiology 20 (2), 292–306. doi:10.1089/ast.2018.1908
Takada-Hoshino, Y., and Matsumoto, N. (2004). An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ. 19 (1), 13–19. doi:10.1264/jsme2.19.13
Taniguchi, M. (2020). Combination of single-molecule electrical measurements and machine learning for the identification of single biomolecules. ACS omega 5 (2), 959–964. doi:10.1021/acsomega.9b03660
Taniguchi, M. (2015). Selective multidetection using nanopores. Anal. Chem. 87 (1), 188–199. doi:10.1021/ac504186m
Tarafder, A. (2016). Metamorphosis of supercritical fluid chromatography to SFC: An Overview. TrAC Trends Anal. Chem. 81, 3–10. doi:10.1016/j.trac.2016.01.002
Termopoli, V., Famiglini, G., Palma, P., Piergiovanni, M., and Cappiello, A. (2017). Atmospheric pressure vaporization mechanism for coupling a liquid phase with electron ionization mass spectrometry. Anal. Chem. 89 (3), 2049–2056. doi:10.1021/acs.analchem.6b04646
Termopoli, V., Famiglini, G., Palma, P., Piergiovanni, M., Rocio-Bautista, P., Ottaviani, M. F., et al. (2019). Evaluation of a liquid electron ionization liquid chromatography–mass spectrometry interface. J. Chromatogr. A 1591, 120–130. doi:10.1016/j.chroma.2019.01.034
Thiel, C. S., Ehrenfreund, P., Foing, B., Pletser, V., and Ullrich, O. (2011). PCR-Based analysis of microbial communities during the EuroGeoMars campaign at mars desert research station, Utah. Int. J. Astrobiol. 10 (3), 177–190. doi:10.1017/S1473550411000073
Thompson, M., Ellison, S. L., and Wood, R. (2002). Harmonized guidelines for single-laboratory validation of methods of analysis (IUPAC Technical Report). Pure Appl. Chem. 74 (5), 835–855. doi:10.1351/pac200274050835
Timoumi, R., François, P., Le Postollec, A., Dobrijevic, M., Grégoire, B., Poinot, P., et al. (2022). Focused ultrasound extraction versus microwave-assisted extraction for extraterrestrial objects analysis. Anal. Bioanal. Chem. 414, 3643–3651. doi:10.1007/s00216-022-04004-8
Toraman, H. E., Abrahamsson, V., Vanholme, R., Van Acker, R., Ronsse, F., Pilate, G., et al. (2018). Application of Py-GC/MS coupled with PARAFAC2 and PLS-DA to study fast pyrolysis of genetically engineered poplars. J. Anal. Appl. Pyrolysis 129, 101–111. doi:10.1016/j.jaap.2017.11.022
Touchette, D., Altshuler, I., Raymond-Bouchard, I., Fernandez-Martinez, M. A., Bourdages, L. J., O'Connor, B., et al. (2022). Microfluidics microbial activity microassay: An automated in situ microbial metabolic detection system. Astrobiology 22 (2), 158–170. doi:10.1089/ast.2021.0072
Traks, J., Sooväli, L., and Leito, I. (2005). Uncertainty in photometric analysis: A case study. Accredit. Qual. Assur. 10 (5), 197–207. doi:10.1007/s00769-005-0909-3
Tringe, S. G., and Rubin, E. M. (2005). Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6 (11), 805–814. doi:10.1038/nrg1709
Tsokolov, S. A. (2009). Why is the definition of life so elusive? Epistemological considerations. Astrobiology 9 (4), 401–412. doi:10.1089/ast.2007.0201
Uckert, K., Grubisic, A., Li, X., Brinckerhoff, W., Cornish, T., Farcy, B., et al. (2018). IR resonance-enhanced organic detection with two-step laser desorption time-of-flight mass spectrometry. Icarus 299, 15–21. doi:10.1016/j.icarus.2017.07.006
Uckert, K., Parness, A., Chanover, N., Eshelman, E. J., Abcouwer, N., Nash, J., et al. (2020). Investigating habitability with an integrated rock-climbing robot and astrobiology instrument suite. Astrobiology 20 (12), 1427–1449. doi:10.1089/ast.2019.2177
Vago, J. L., Westall, F., Coates, A. J., Jaumann, R., Korablev, O., Ciarletti, V., et al. (2017). Habitability on early mars and the search for biosignatures with the ExoMars rover. Astrobiology 17 (6-7), 471–510. doi:10.1089/ast.2016.1533
Valentine, S. J., Liu, X., Plasencia, M. D., Hilderbrand, A. E., Kurulugama, R. T., Koeniger, S. L., et al. (2005). Developing liquid chromatography ion mobility mass spectometry techniques. Expert Rev. Proteomics 2 (4), 553–565. doi:10.1586/14789450.2.4.553
van der Valk, T., Pečnerová, P., Díez-del-Molino, D., Bergström, A., Oppenheimer, J., Hartmann, S., et al. (2021). Million-year-old DNA sheds light on the genomic history of mammoths. Nature 591 (7849), 265–269. doi:10.1038/s41586-021-03224-9
Vandenbroucke, M. (2003). Kerogen: From types to models of chemical structure. Oil Gas Sci. Technol. -. Rev. IFP. 58 (2), 243–269. doi:10.2516/ogst:2003016
Vandeventer, P. E., Weigel, K. M., Salazar, J., Erwin, B., Irvine, B., Doebler, R., et al. (2011). Mechanical disruption of lysis-resistant bacterial cells by use of a miniature, low-power, disposable device. J. Clin. Microbiol. 49 (7), 2533–2539. doi:10.1128/jcm.02171-10
Vargas Medina, D. A., Maciel, E. V. S., and Lanças, F. M. (2020). Miniaturization of liquid chromatography coupled to mass spectrometry. 3. Achievements on chip-based LC–MS devices. TrAC Trends Anal. Chem. 131, 116003. doi:10.1016/j.trac.2020.116003
Veneranda, M., Lopez-Reyes, G., Manrique-Martinez, J. A., Sanz-Arranz, A., Lalla, E., Konstantinidis, M., et al. (2020). ExoMars Raman laser spectrometer (RLS): Development of chemometric tools to classify ultramafic igneous rocks on mars. Sci. Rep. 10 (1), 16954. doi:10.1038/s41598-020-73846-y
Verkouteren, J. R., and Staymates, J. L. (2011). Reliability of ion mobility spectrometry for qualitative analysis of complex, multicomponent illicit drug samples. Forensic Sci. Int. 206 (1-3), 190–196. doi:10.1016/j.forsciint.2010.08.005
Vigier, F., Le Postollec, A., Coussot, G., Chaput, D., Cottin, H., Berger, T., et al. (2013). Preparation of the biochip experiment on the EXPOSE-R2 mission outside the international space station. Adv. Space Res. 52 (12), 2168–2179. doi:10.1016/j.asr.2013.09.026
Voeten, R. L. C., Ventouri, I. K., Haselberg, R., and Somsen, G. W. (2018). Capillary electrophoresis: Trends and recent advances. Anal. Chem. 90 (3), 1464–1481. doi:10.1021/acs.analchem.8b00015
Volkenburg, T. V., Benzing, J. S., Craft, K. L., Ohiri, K., Kilhefner, A., Irons, K., et al. (2022). Microfluidic chromatography for enhanced amino acid detection at Ocean Worlds. Astrobiology 22, 1116–1128. doi:10.1089/ast.2021.0182
Wallace, J. K., Serabyn, E., Lindensmith, C., Nadeau, J., Rider, S., and Bedrossian, M. (2019). “A multiwavelength digital holographic microscope architecture for enhancing life detection,” in 2019 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Wallace, W. T., Limero, T. F., Loh, L. J., Mudgett, P. D., and Gazda, D. B. (2017). “Monitoring of the atmosphere on the international space station with the air quality monitor,” in 47th international Conference on environmental systems). Emmaus, PA: ICES—International Conference on Environmental Systems.
Wang, A., Haskin, L. A., Lane, A. L., Wdowiak, T. J., Squyres, S. W., Wilson, R. J., et al. (2003). Development of the Mars microbeam Raman spectrometer (MMRS). J. Geophys. Res. 108 (E1), 5005. doi:10.1029/2002JE001902
Wang, A., Hynynen, S., Hawkins, A. R., Tolley, S. E., Tolley, H. D., and Lee, M. L. (2014). Axial thermal gradients in microchip gas chromatography. J. Chromatogr. A 1374, 216–223. doi:10.1016/j.chroma.2014.11.035
Wang, J., Ke, Y. H., Zhang, Y., Huang, K. Q., Wang, L., Shen, X. X., et al. (2017). Rapid and accurate sequencing of enterovirus genomes using MinION nanopore sequencer. Biomed. Environ. Sci. 30 (10), 718–726. doi:10.3967/bes2017.097
Wang, J., Wang, Z., Liu, F., Cai, L., Pan, J.-b., Li, Z., et al. (2018). Vacuum ultraviolet laser desorption/ionization mass spectrometry imaging of single cells with submicron craters. Anal. Chem. 90 (16), 10009–10015. doi:10.1021/acs.analchem.8b02478
Wang, W., Singh, S., Zeng, D. L., King, K., and Nema, S. (2007). Antibody structure, instability, and formulation. J. Pharm. Sci. 96 (1), 1–26. doi:10.1002/jps.20727
Wang, X., Cheng, C., Wang, S., Zhao, M., Dasgupta, P. K., and Liu, S. (2009). Nanocapillaries for open tubular chromatographic separations of proteins in femtoliter to picoliter samples. Anal. Chem. 81 (17), 7428–7435. doi:10.1021/ac901265t
Wang, X., Veerappan, V., Cheng, C., Jiang, X., Allen, R. D., Dasgupta, P. K., et al. (2010). Free solution hydrodynamic separation of DNA fragments from 75 to 106 000 base pairs in a single run. J. Am. Chem. Soc. 132 (1), 40–41. doi:10.1021/ja909233n
Weisgrab, G., Ovsianikov, A., and Costa, P. F. (2019). Functional 3D printing for microfluidic chips. Adv. Mat. Technol. 4 (10), 1900275. doi:10.1002/admt.201900275
Wenz, C., Barbas, C., López-Gonzálvez, Á., Garcia, A., Benavente, F., Sanz-Nebot, V., et al. (2015). Interlaboratory study to evaluate the robustness of capillary electrophoresis-mass spectrometry for peptide mapping. J. Sep. Sci. 38 (18), 3262–3270. doi:10.1002/jssc.201500551
Wiens, R. C., Gasda, P. J., Misra, A. K., Sharma, S. K., Quinn, H. M., Ganguly, K., et al. (2019). “OrganiCam: A lightweight time-resolved fluorescence imager and Raman spectrometer for icy world organic detection and characterization,” in AGU fall meeting 2019 (Washington, D.C.: American Geophysical Union), P53E.
Wiens, R. C., Maurice, S., Robinson, S. H., Nelson, A. E., Cais, P., Bernardi, P., et al. (2021). The SuperCam instrument suite on the NASA Mars 2020 rover: Body unit and combined system tests. Space Sci. Rev. 217 (1), 4–87. doi:10.1007/s11214-020-00777-5
Wiens, R., Gasda, P., Misra, A., Acosta-Maeda, T., Sharma, S., Quinn, H., et al. (2020). “OrganiCam: A lightweight time-resolved fluorescence imager and Raman spectrometer for mars cave or icy-world surface organic characterization,” in 51st lunar and planetary science conference (Houston, TX: LPI).
Wiesendanger, R., Tulej, M., Grimaudo, V., Cedeño-López, A., Lukmanov, R., Riedo, A., et al. (2019). A method for improvement of mass resolution and isotope accuracy for laser ablation time-of-flight mass spectrometers. J. Chemom. 33 (1), e3081. doi:10.1002/cem.3081
Wiesendanger, R., Wacey, D., Tulej, M., Neubeck, A., Ivarsson, M., Grimaudo, V., et al. (2018). Chemical and optical identification of micrometer-sized 1.9 billion-year-old fossils by combining a miniature laser ablation ionization mass spectrometry system with an optical microscope. Astrobiology 18 (8), 1071–1080. doi:10.1089/ast.2017.1780
Wilhelm, M., Ricco, A., Buckner, D., Boone, T., Chin, M., Eigenbrode, J., et al. (2021). “Extraction instruments to enable detection of origin-diagnostic lipids for life detection,” in Lunar and planetary science conference (Houston, TX: Lunar and Planetary Institute), 2634.
Willerslev, E., Hansen, A. J., Rønn, R., Brand, T. B., Barnes, I., Wiuf, C., et al. (2004). Long-term persistence of bacterial DNA. Curr. Biol. 14 (1), R9–R10. doi:10.1016/j.cub.2003.12.012
Willhite, L., Ni, Z., Arevalo, R., Bardyn, A., Gundersen, C., Minasola, N., et al. (2021). “Corals: A laser desorption/ablation Orbitrap mass spectrometer for in situ exploration of europa,” in 2021 IEEE aerospace conference (Piscataway, NJ: IEEE), 1.
Williams, A. J., Eigenbrode, J., Floyd, M., Wilhelm, M. B., O'Reilly, S., Johnson, S. S., et al. (2019). Recovery of fatty acids from mineralogic Mars analogs by TMAH thermochemolysis for the sample analysis at Mars wet chemistry experiment on the Curiosity Rover. Astrobiology 19 (4), 522–546. doi:10.1089/ast.2018.1819
Williams, A., and Muirhead, B. (2022). “Mars Life Explorer, mission concept study,” in Planetary mission concept study for the 2023-2032 decadal survey. National Academies of Sciences, Engineering, and Medicine. Available at: https://tinyurl.com/2p88fx4f.
Wollschläger, J., Grunwald, M., Röttgers, R., and Petersen, W. (2013). Flow-through PSICAM: A new approach for determining water constituents absorption continuously. Ocean. Dyn. 63 (7), 761–775. doi:10.1007/s10236-013-0629-x
Wollschläger, J., Voß, D., Zielinski, O., and Petersen, W. (2016). In situ observations of biological and environmental parameters by means of optics — Development of next-generation ocean sensors with special focus on an integrating cavity approach. IEEE J. Ocean. Eng. 41 (4), 753–762. doi:10.1109/JOE.2016.2557466
Wyttenbach, T., and Bowers, M. T. (2003). “Gas-phase conformations: The ion mobility/ion chromatography method,” in Modern mass spectrometry. Topics in current chemistry. Editor C. A. Schalley (Heidelberg: Springer Berlin), 207
Xia, Z., Patchin, M., McKay, C. P., and Drndic, M. (2022). Deoxyribonucleic acid extraction from Mars analog soils and their characterization with solid-state nanopores. Astrobiology 22 (8), 992–1008. doi:10.1089/ast.2021.0051
Xiang, P., Yang, Y., Chen, H., Chen, A., and Liu, S. (2021). Liquid chromatography using ≤5 μm open tubular columns. TrAC Trends Anal. Chem. 142, 116321–116329. doi:10.1016/j.trac.2021.116321
Xie, J., Miao, Y., Shih, J., Tai, Y.-C., and Lee, T. D. (2005). Microfluidic platform for liquid chromatography− tandem mass spectrometry analyses of complex peptide mixtures. Anal. Chem. 77 (21), 6947–6953. doi:10.1021/ac0510888
Yamamoto, K., Ota, N., and Tanaka, Y. (2021). Nanofluidic devices and applications for biological analyses. Anal. Chem. 93 (1), 332–349. doi:10.1021/acs.analchem.0c03868
Yang, W., and Chen, S.-L. (2020). Time-gated fluorescence imaging: Advances in technology and biological applications. J. Innov. Opt. Health Sci. 13 (03), 2030006. doi:10.1142/S1793545820300062
Yin, H., and Killeen, K. (2007). The fundamental aspects and applications of Agilent HPLC-Chip. J. Sep. Sci. 30 (10), 1427–1434. doi:10.1002/jssc.200600454
Yuan, Z., Liu, Y., Dai, M., Yi, X., and Wang, C. (2020). Controlling DNA translocation through solid-state nanopores. Nanoscale Res. Lett. 15 (1), 80–89. doi:10.1186/s11671-020-03308-x
Zamuruyev, K., Ferreira Santos, M. S., Mora, M. F., Kurfman, E. A., Noell, A. C., and Willis, P. A. (2021). Automated capillary electrophoresis system compatible with multiple detectors for potential in situ spaceflight missions. Anal. Chem. 93 (27), 9647–9655. doi:10.1021/acs.analchem.1c02119
Zangheri, M., Mirasoli, M., Guardigli, M., Di Nardo, F., Anfossi, L., Baggiani, C., et al. (2019). Chemiluminescence-based biosensor for monitoring astronauts’ health status during space missions: Results from the International Space Station. Biosens. Bioelectron. X. 129, 260–268. doi:10.1016/j.bios.2018.09.059
Zareian-Jahromi, M. A., Ashraf-Khorassani, M., Taylor, L. T., and Agah, M. (2008). Design, modeling, and fabrication of MEMS-based multicapillary gas chromatographic columns. J. Microelectromech. Syst. 18 (1), 28–37. doi:10.1109/JMEMS.2008.2007267
Zelenyi, L., and Zakharov, A. (2012). Project “phobos-grunt”: Instruments for scientific research. Sol. Syst. Res. 46 (7), 489–492. doi:10.1134/S0038094612070210
Zhao, X., Xie, X., Sharma, S., Tolley, L. T., Plistil, A., Barnett, H. E., et al. (2017). Compact ultrahigh-pressure nanoflow capillary liquid chromatograph. Anal. Chem. 89 (1), 807–812. doi:10.1021/acs.analchem.6b03575
Zhao, Y., Ashcroft, B., Zhang, P., Liu, H., Sen, S., Song, W., et al. (2014). Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol. 9 (6), 466–473. doi:10.1038/nnano.2014.54
Zhou, W., Qiu, H., Guo, Y., and Guo, W. (2020). Molecular insights into distinct detection properties of α-hemolysin, MspA, CsgG, and aerolysin nanopore sensors. J. Phys. Chem. B 124 (9), 1611–1618. doi:10.1021/acs.jpcb.9b10702
Zhu, F., Lee, S., Valentine, S. J., Reilly, J. P., and Clemmer, D. E. (2012). Mannose7 glycan isomer characterization by IMS-MS/MS analysis. J. Am. Soc. Mass Spectrom. 23 (12), 2158–2166. doi:10.1007/s13361-012-0491-y
Zielińska, S., Radkowski, P., Blendowska, A., Ludwig-Gałęzowska, A., Łoś, J. M., and Łoś, M. (2017). The choice of the DNA extraction method may influence the outcome of the soil microbial community structure analysis. MicrobiologyOpen 6 (4), e00453. doi:10.1002/mbo3.453
Zielinski, O., Rüssmeier, N., Ferdinand, O. D., Miranda, M. L., and Wollschläger, J. (2018). Assessing fluorescent organic matter in natural waters: Towards in situ excitation–emission matrix spectroscopy. Appl. Sci. (Basel). 8 (12), 2685–2710. doi:10.3390/app8122685
Keywords: astrobiology, life detection, analytical chemistry, biomarker, instruments
Citation: Abrahamsson V and Kanik I (2022) In situ organic biosignature detection techniques for space applications. Front. Astron. Space Sci. 9:959670. doi: 10.3389/fspas.2022.959670
Received: 02 June 2022; Accepted: 02 November 2022;
Published: 28 November 2022.
Edited by:
Assimo Maris, University of Bologna, ItalyReviewed by:
Svetlana Shkolyar, University of Maryland, College Park, United StatesChristopher E. Carr, Massachusetts Institute of Technology, United States
Copyright © 2022 Abrahamsson and Kanik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor Abrahamsson, VmljdG9yLlMuQWJyYWhhbXNzb25AanBsLm5hc2EuZ292
 Victor Abrahamsson
Victor Abrahamsson Isik Kanik
Isik Kanik