
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Astron. Space Sci., 07 October 2021
Sec. Astrobiology
Volume 8 - 2021 | https://doi.org/10.3389/fspas.2021.755100
This article is part of the Research TopicCurrent and Future Instrumentation for the Detection and Identification of Signatures of Life on Mars and BeyondView all 8 articles
 Luoth Chou1,2*
Luoth Chou1,2* Paul Mahaffy1
Paul Mahaffy1 Melissa Trainer1
Melissa Trainer1 Jennifer Eigenbrode1
Jennifer Eigenbrode1 Ricardo Arevalo3
Ricardo Arevalo3 William Brinckerhoff1
William Brinckerhoff1 Stephanie Getty1
Stephanie Getty1 Natalie Grefenstette4,5
Natalie Grefenstette4,5 Victoria Da Poian1
Victoria Da Poian1 G. Matthew Fricke6
G. Matthew Fricke6 Christopher P. Kempes4
Christopher P. Kempes4 Jeffrey Marlow7
Jeffrey Marlow7 Barbara Sherwood Lollar8
Barbara Sherwood Lollar8 Heather Graham1
Heather Graham1 Sarah Stewart Johnson2
Sarah Stewart Johnson2For the past fifty years of space exploration, mass spectrometry has provided unique chemical and physical insights on the characteristics of other planetary bodies in the Solar System. A variety of mass spectrometer types, including magnetic sector, quadrupole, time-of-flight, and ion trap, have and will continue to deepen our understanding of the formation and evolution of exploration targets like the surfaces and atmospheres of planets and their moons. An important impetus for the continuing exploration of Mars, Europa, Enceladus, Titan, and Venus involves assessing the habitability of solar system bodies and, ultimately, the search for life—a monumental effort that can be advanced by mass spectrometry. Modern flight-capable mass spectrometers, in combination with various sample processing, separation, and ionization techniques enable sensitive detection of chemical biosignatures. While our canonical knowledge of biosignatures is rooted in Terran-based examples, agnostic approaches in astrobiology can cast a wider net, to search for signs of life that may not be based on Terran-like biochemistry. Here, we delve into the search for extraterrestrial chemical and morphological biosignatures and examine several possible approaches to agnostic life detection using mass spectrometry. We discuss how future missions can help ensure that our search strategies are inclusive of unfamiliar life forms.
Biosignatures are the tantalizing chemical and physical imprints associated with life, and the possibility that life exists elsewhere beyond Earth drives us to search for these biosignatures on other planets and moons. The enterprise of space exploration, galvanized by the question of “Are we alone in the Universe?”, demands a stronger understanding of the diversity of biosignatures that life could express, thereby driving payload instruments on board astrobiology missions to offer broader and more advanced detection capabilities. In tandem with cutting-edge instrument platforms, research in data processing and data analysis on Earth-based (Terran) astrobiology analogs and on extraterrestrial materials also serves to increase the breadth of interpretations possible with mission data. Thus, a robust search for present or past life on other planetary bodies will be facilitated by the development of analytical tools that enable the search for multiple types of biosignatures, whether they are based on familiar biochemistry or not.
There have been many attempts to describe life for scientific, philosophical, or cultural purposes, though no formal definition has been universally accepted or agreed upon. Despite this lack of consensus, NASA has formulated an operational working definition of life—“a self-sustaining chemical system capable of Darwinian evolution”—to aid in the development of life detection strategies and encourage the establishment of standards needed to qualify a measurement as an unequivocal sign of life (Cleland and Chyba, 2002). This definition broadly captures all biological systems on Earth (Terran life) as well as life forms with potentially differing underlying molecular frameworks. While search strategies for extraterrestrial life are traditionally rooted in our knowledge of Terran life, widely-accepted properties of life, such as being primarily composed of carbon, nitrogen, hydrogen, oxygen, phosphorous, and sulfur (CNHOPS) elements or being based on informational storage or catalytic biopolymers, such as DNA or proteins, respectively, are all attributes that directly reflect the historical contingency of life on Earth. This contingency is due to the fact that the evolution and selection of biochemical regimes that pervade Terran life are inherently tied to Earth’s geochemical, hydrological, and (eventually) biological history, as well as the astronomical and geophysical circumstances that led to the Earth’s formation and planetary evolution. Yet, other life forms that originated on Earth or elsewhere may be based on substantially different chemistries, and it may be possible for “a self-sustaining system capable of Darwinian evolution” to operate in an unfamiliar, exotic biochemical system. This possibility motivates the need to broaden our search criteria beyond biosignatures that are fundamentally based on Terran life, and focus on those that are universally inclusive of all life forms—thus the emerging focus on “agnostic biosignatures.”
Claims of life detection require robust proof that meets a series of principles that have been previously proposed by the scientific community (Des Marais et al., 2008). More recently, Neveu et al. (2018) formulated the Ladder of Life Detection, a tool designed to guide life detection investigations during robotic astrobiology space missions. This tool draws from examples of previous life detection experiments to provide current and future efforts with decision rules that allow for the rejection of all abiotic explanations, pointing to a life detection claim as a “last resort hypothesis” (Sagan et al., 1993). Briefly, for a life detection claim to be convincing, the measurement or sets of measurements must be sensitive, contamination-free, and repeatable; the features being measured must be detectable, preservable (survivable), reliable (distinctly different from abiotic background), and compatible with our understanding of life; ultimately, all other abiotic hypotheses must be rejected (Neveu et al., 2018). These criteria are often met not only with one particular analytical instrument, but by using multiple sets of measurements deriving from a variety of technologies that are able to provide the contextual background information needed to assess the biosignature’s presence and provenance as well as the unlikelihood of its production through abiotic means alone. This necessitates a checks-and-balances approach between scientific measurements and instrumentation that enhances the certainty of a potential biosignature while decreasing the likelihood of false positive and false negative interpretations (NAS, 2019; Neveu et al., 2018; Chou et al., 2021). In some cases, statistical frameworks can also help establish the qualitative criteria necessary to confirm a series of measurements as life, measurements which can be evaluated using tools such as Bayesian hypothesis testing (Johnson et al., 2018; Lorenz, 2019; Pohorille and Sokolowska, 2020).
On Earth, the detection and characterization of chemical and morphological biosignatures have benefited from advances in technology, including the development of analytical tools such as mass spectrometers. Biosignatures observable through mass spectrometry span physical scales and chemical characteristics, including elemental distributions, molecular structures, isotopic signature, and spatially-resolvable information via imaging. Such a diverse toolkit has led to a number of revolutionary advances in chemistry (and astrobiology), including the characterization of molecular fossils to provide organic geochemists with a window into the history of Terran life in deep time (Peters et al., 2005; Eigenbrode, 2008). These advances have also enabled a detailed investigation into the chemistry of biopolymers, such as proteins (using proteomics), which are the molecular machineries that permit life as we know it to function with enormous flexibility and efficiency.
However, mass spectrometry investigations, especially of novel or unknown molecules, are often limited by our capacity to manually interpret never-before-seen data. This is because de novo mass spectrometry interpretation (identifying chemical structures from patterns of mass fragments) is often an exceptionally arduous task. This process necessitates not only contextual information (e.g., an a priori hypothesis of what compounds to expect) and an experienced user, but also the ultimate authentication of the identified molecular structures with synthesized standards. While these challenges have been somewhat alleviated by the development of high-throughput automated pipelines for de novo metabolite identification (Neumann and Böcker, 2010; Peironcely et al., 2013) and peptide sequencing (Dančík et al., 1999; Frank and Pevzner, 2005; Jeong et al., 2013), these methods are often designed for use with high resolution mass spectrometers (such as Orbitrap analyzers) with specific types of mass fragmentation protocol (e.g., collision induced dissociation), and for classes of biological molecules that are based on Terran biochemistry, such as proteins. This can limit our ability to use Terran-based spectral databases for life detection efforts that are focused on agnostic biochemistry. Despite this, ongoing advances in understanding fragmentation reactions, including in tandem mass spectrometry (Wolf et al., 2010; Zapadinsky et al., 2019), as well as computational methods designed to make predictions of fragmentation patterns (Rasche et al., 2011; Hufsky and Bocker, 2017; Dührkop et al., 2021) continue to inform our future in situ investigations of organic molecules, and potentially molecular biosignatures, on other planetary bodies.
While a number of existing reviews provide a thorough description of planetary mass spectrometry in terms of historical instrumentation in spaceflight (Nier, 1960; Palmer and Limero, 2001) and advanced space-capable mass analyzer technologies (Arevalo et al., 2020), this review focuses on the applications of planetary mass spectrometry instruments for astrobiology investigations, and how they can enable in situ life detection missions on other worlds. We discuss types of Terran-based chemical biosignatures that can be analyzed by mass spectrometry, including their associated caveats and false positive (abiotic) potentials. We then draw on contemporary examples of agnostic biosignatures and highlight the utility of mass spectrometry in detecting them. Finally, we consider future planetary astrobiology missions that may carry mass spectrometers, and the potential infusion of agnostic approaches to life detection in these missions.
To date, the Viking mission remains the only successfully-operated previous mission with a prime focus on in situ life detection on another planet. The mission boasted a life detection payload composed of three biological experiments: 1) the Gas Exchange experiment, which aimed to identify gases released from the Martian soil upon adding a nutrient solution (Oyama et al., 1976), 2) the Labeled Release experiment, which monitored radioactive gases for biological activity after infusing the soil with 14C-labeled nutrients (Levin and Straat, 1977), and 3) the Pyrolytic Release experiment, which sought evidence of biological carbon assimilation into radio-labeled organic matter after exposing potential Martian life to 14CO2 and 14CO gases (Horowitz et al., 1972). Two gas chromatography-mass spectrometers (GC-MS), then state-of-the-art, were also included on board to study Mars organic matter and search for complex organic compounds that could be indicative of life, which could help to independently verify a positive life detection for one or more of the biological experiments. However, despite a positive observation of the release of radioactive gas in the Labeled Release experiments (Levin and Straat, 1977), the fully-functional and highly capable mass spectrometers failed to detect diagnostic organic compounds, even with a mass spectrometer detection limit on the order of ppm to ppb (Biemann et al., 1977).
The initial results of the Viking GC-MS experiments were revisited after the 2008 discovery of native oxychlorines during the Mars Phoenix mission (Hecht et al., 2009), and later confirmed by the 2013 detection of perchlorates and chlorates with the Sample Analysis at Mars (SAM) instrument suite on the Mars Science Laboratory (MSL) mission’s Curiosity rover (Ming et al., 2014). Oxychlorines are a type of chlorinated oxidant that can substantially alter or destroy organic matter in the presence of thermal or ionizing radiation, ultimately changing its chemistry. Thus, oxidized Martian organic matter may be unrecognizable or undetectable with the use of thermal volatilization extraction or pyrolysis (Navarro-González et al., 2006; Biemann, 2007). Since then, the more advanced pyrolysis-GC-MS of the SAM instrument suite (Mahaffy et al., 2012) has shown that some chlorinated compounds (Freissinet et al., 2015) and more molecularly diverse organic matter (Eigenbrode et al., 2018) are detectable by this approach even when oxychlorines are present, potentially due to a higher carbon to oxychlorine ratio (Kenig et al., 2016). In addition, the search for organic matter on Mars has evolved to include chemical derivatization and thermochemolysis techniques that can enhance the amenability of non-volatile organic compounds to GC-MS (Mahaffy et al., 2012; Mißbach et al., 2019), and laser desorption ionization (LDI) technique of the Mars Organic Molecular Analyzer (MOMA) instrument on board the 2022 ExoMars’ Rosalind Franklin rover that circumvents the problem of oxidation of organic compounds during sampling and ionization (Li et al., 2015; Goesmann et al., 2017). These technologies continue to employ mass spectrometers for their high sensitivity in detecting compounds that establish chemical context and enable the search for potential biosignatures. Despite the challenges that were faced, the Viking experiments’ first in situ attempt at life detection beyond Earth greatly inspired the SAM and MOMA investigations, as well as next generation of planetary missions that will visit Mars and beyond, further driving the community’s interest in exploring our Solar System to seek out potential evidence of life on another planet.
Mass spectrometers (MS) have been part of space exploration for a number of decades, and MS technology continues to advance rapidly. Earlier generations of MS comprise miniaturized magnetic sector mass analyzers, which are used to filter ions and separate them through a magnetic field. These instruments were flown to examine the atmospheric composition of the Earth by way of high altitude rockets (Nier, 1960), to study the Moon via Apollo 15, 16, and 17 missions (Hoffman and Hodges, 1972), to explore Venus through the Pioneer Venus Large Probe (Hoffman et al., 1980b,a), to interrogate comet 67P/Churyumov-Gerasimenko with the Rosetta orbiter (Balsiger et al., 2007), and to investigate the surface of Mars with the Viking landers and the Mars Phoenix lander (Biemann et al., 1977; Hoffman et al., 2008) (Figure 1A). Magnetic sector MS have contributed to numerous valuable science discoveries on remote bodies, such as conducting one of the first in situ characterizations of Venus’ atmosphere with the Pioneer Venus Bus Probe (Hoffman et al., 1980a), and investigating carbonate mineral phases on Mars during the Viking mission (Sutter et al., 2012). In some cases, magnetic sector analyzers are coupled with electrostatic analyzers, which are used to separate ions in flight using an electric field: when combined, the instrument suite is called a double focusing mass spectrometer (DFMS). Electrostatic analyzers have been used on board the VeGa-1 and VeGa-2 spacecrafts to measure the composition of gaseous materials near comet Halley (Gringauz et al., 1985) whereas the DFMS helped enabled the characterization of organic species on comet 67P/Churyumov-Gerasimenko (Le Roy et al., 2015). While magnetic sector MS represents some of the earlier prototypes of planetary MSs, they are generally predecessors to the more ubiquitous quadrupole mass analyzers.
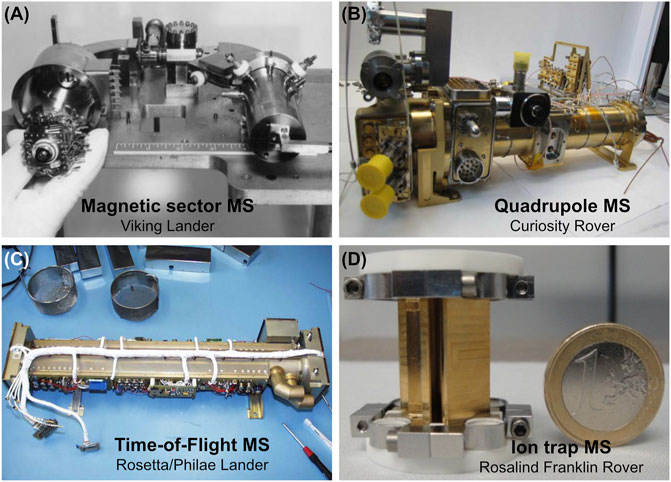
FIGURE 1. A variety of miniaturized mass analyzers that have been flown and will fly on planetary missions. (A) A magnetic sector MS was carried on the Viking landers. (B) A quadrupole MS was carried on board the Curiosity rover. (C) A time-of-flight MS was flown on the Rosetta/Philae lander (as part of the COSAC instrument suite). (D) An ion trap MS as part of the MOMA instrument suite will be flown on board the Rosalind Franklin rover. Reprinted with permission from Levin et al. (2000) and from Dr. Fred Goesmann.
Quadrupole mass spectrometer (QMS) consists of four cylindrical or hyperbolic rods with applied direct-current (DC) and radio-frequency (RF) electric fields that are used for ion separation. In planetary flight implementations, QMS analyzers typically offer unit mass resolution and mass ranges from two to hundreds of Daltons (depending on rod configuration and radio frequency range). QMSs have visited numerous worlds including Venus on the Pioneer Venus Orbiter (Niemann et al., 1979), Jupiter aboard the Galileo probe (Niemann et al., 1979; Mahaffy et al., 2000), Saturn and its moons, Titan and Enceladus, on the Cassini spacecraft (Kasprzak et al., 1996; Waite et al., 2004) and its accompanying Huygens probe (Niemann et al., 2010), our Moon on the Lunar Atmosphere and Dust Environment Explorer (LADEE) mission (Mahaffy et al., 2014), the upper atmospheres of Mars on board the Mars Atmosphere and Volatile EvolutioN (MAVEN) orbiter (Mahaffy et al., 2015), and most notably, the surface of Mars via the SAM instrument suite on the Curiosity rover (Mahaffy et al., 2012) (Figure 1B). Enigmas across our Solar System are being slowly unraveled by these planetary missions that leverage QMSs. For example, studies of elemental ratios of noble gases and the light elements H, C, N, O, and S in various molecules in the deep atmosphere of Jupiter with the Galileo probe mass spectrometer helped resolve models of the formation of the gas giant (Mahaffy et al., 2000; Atreya et al., 2003). More recently, the Curiosity rover and its analytical payload helped uncover evidence pointing to ancient habitable lake deposits (Grotzinger et al., 2014) in addition to potentially refractory organic matter preserved by sedimentary sulfurization at Gale Crater on Mars (Eigenbrode et al., 2018), which were discovered using evolved gas analysis and GC that both relied on a QMS.
Time-of-flight mass spectrometer (TOFMS) has been miniaturized for spaceflight since the 1980s but has been used less commonly in planetary missions. TOFMS analyzes charged species based on the flight time of ions that have been accelerated to a uniform kinetic energy and separated in a drift region. In order to reduce the physical space needed to resolve spectral peaks with a TOFMS, a reflectron, which is a device that acts as an ion mirror, can be used to effectively increase the drift tube distance without substantially increasing the overall instrument dimensions, leading to considerable improvements in the mass resolution of TOF mass analyzers (Scherer et al., 2006). TOFMSs were first flown on the VeGa-1 and Vega-2 spacecrafts to Venus, and then on the Giotto spacecraft that investigated the composition of cometary dust of comet Halley (Kissel and Krueger, 1987). In these experiments, the impact of dust at the source region of the MS produces a plasma that was extracted and focused into the TOFMS for analysis. In 2004, a TOFMS was also used to successfully measure the first negative ion mass spectra of cometary particles during the Stardust mission to comet 91P/Wild2 (Tsou et al., 2004). In addition, several TOFMSs were also used aboard the Rosetta spacecraft and Philae lander that explored comet 67P/Churyumov-Gerasimenko: the COmetary Secondary Ion Mass Analyzer (COSIMA) which was used as an analyzer for secondary ion mass spectrometry (SIMS) (Kissel et al., 2007), a reflectron type TOFMS called RTOF that is part of the Rosetta Orbiter Spectrometer for Ion and Neutral Analysis (ROSINA) instrument suite (Balsiger et al., 2007), and the Cometary Sampling and Composition (COSAC) experiment on Philae, which was implemented in the form of a high resolution and wide mass range TOFMS instrument suite that enabled the in situ analysis of organic molecules on the comet’s surface (Goesmann et al., 2007) (Figure 1C). Owing to their successes in chemical investigations in space environments, TOFMS remains a critical payload in future selected and proposed missions. For example, the Jupiter Icy Moon Explorer (JUICE) mission will utilize a compact TOFMS capable of measuring neutral molecules and ionospheric ions in the atmospheres of the icy moons of Jupiter, Europa, Ganymede, and Callisto (Föhn et al., 2021). High resolution TOFMSs are also currently being developed for flight applications, including the MAss Spectrometer for Planetary EXploration (MASPEX) instrument that is capable of 25,000 m/Δm resolution at 10% peak height, allowing for the elucidation of the isotopic composition of volatiles, such as CH4, H2O, NH4, CO, N2, and CO2, as well as low molecular weight (C2-C4) organic compounds in a complex mixtures (Brockwell et al., 2016).
Finally, ion trap mass spectrometer (ITMS) leverages mass analyzers that can be used to capture and subsequently analyze ions across a tailored mass range through a combination of electric or magnetic fields. They have a variety of configurations including 2D (linear ion trap), 3D (Paul ion trap), electrostatic (Orbitrap™), and magnetically-induced with electric trapping plates [ion cyclotron resonance (ICR) trap]. While 2D and 3D traps can operate at higher pressures, potentially reducing resource requirements, others such as ICRMS can offer remarkably high mass resolving power but have yet to be matured to flight readiness. The Philae lander onboard the Rosetta spacecraft housed the first ITMS that was utilized on a planetary mission, and it was coupled to a GC that targeted a measurement of the elemental and stable isotope abundances of the cometary materials (Todd et al., 2007; Morse et al., 2015; Wright et al., 2015). In 2022, the ExoMars mission will launch with MOMA, the most comprehensive ITMS instrument suite to date, comprised of a dual source 2D ion trap capable of analyzing ions generated via two different ionization mechanism: laser desorption ionization, and electron impact ionization (Brinckerhoff et al., 2013; Goesmann et al., 2017; Li et al., 2017) (Figure 1D). In addition, MOMA is able to analyze ions via two different operational modes, as a standard ITMS, and via tandem mass spectrometry (MSn) using the Stored Waveform Inverse Fourier Transform (SWIFT) mode (Goesmann et al., 2017; Li et al., 2017). Importantly, MSn enables the selection and accumulation of ions within a particular mass range, allowing for subsequent fragmentation of ions of interest, leading to a greater discernibility of the mass spectrum, and thus improved structural characterization of molecules. Ultrahigh mass resolution systems, such as Orbitraps mass analyzers, are presently being developed by a number of researchers due to the value of separating isobaric interferences with additional sample processing (Arevalo et al., 2018; Selliez et al., 2019; Willhite et al., 2021).
Whether ions are filtered or spatially separated by a mass analyzer, a device is needed to measure the intensity of the ions and record it as an electrical signal that can be digitized and read by a data acquisition system. Measuring ions can be achieved via a collector that amplifies and measures the ion current directly (i.e., Faraday cup) or via a series of dynodes that can convert individual ions into a cascade of electrons that are then transformed to an electrical signal (i.e., electron multiplier). Similarly, microchannel plate detectors can amplify ion signals by secondary emission of electrons in the detector material; additionally, the nature of the array of wells in a microchannel plate also allows for spatial resolution in some applications. Ion detection systems have varying sensitivities, response times, and noise and pressure tolerances. The electron multiplier, the most common type of ion detector in space, has been used with a number of planetary mass spectrometers, including those on board Cassini/Huygens probe, Viking landers, and Curiosity rover, to name a few (Biemann et al., 1977; Kasprzak et al., 1996; Mahaffy et al., 2012). Microchannel plate detector, especially suitable for high speed current digitization of ion signals, has also been implemented in flight, such as on the RTOF mass analyzer onboard the ROSINA instrument suite (Balsiger et al., 2007). This detector serves as a heritage for the MASPEX instrument suite (Brockwell et al., 2016).
Mass spectrometers fundamentally measure the mass-to-charge ratios (m/z) of particles that have been ionized. There is a wide range of ionization techniques, some of which have high heritage in planetary exploration. The most common, used in laboratory instruments and on mass spectrometers flown to space, is electron impact (EI) ionization, where energetic electrons (emitted from a filament) bombard molecules in gas phase causing them to become ionized and often fragmented. These sources and other MS components have been miniaturized and ruggedized for numerous missions, such as Viking, Venera, VeGa, Cassini, Rosetta, and Curiosity, among others. EI ionization is considered a “hard ionization” source, where the high degree of fragmentation can yield a detailed product spectrum that can be used for structural elucidation. In some missions (e.g., Galileo), a variable electron energy was utilized to provide more information on the molecular composition of the targets (Niemann et al., 1992). However, EI ionization may not be particularly well suited for studies of some compounds where observing the preserved molecular ion is desirable and would provide the most definitive identification. In addition, EI ionization is not amenable to certain molecular separation techniques, such as liquid chromatography (LC), because ion source pressure may cause filament degradation, decreasing the longevity of the source.
Other “soft ionization” techniques, such as chemical ionization (CI), atmospheric-pressure chemical ionization (APCI), and electrospray ionization (ESI), transfer residual energy onto the analytes, resulting in ionization while preserving the molecular ions and minimizing fragmentation. These techniques require liquid or gaseous reagents and pressurized systems which can add to the complexity of their deployment in space environments. Several studies have tested the utility of CI and ESI for the detection of potential biosignatures on ocean worlds (Sokol et al., 2011; Waller et al., 2019). In comparison, LDI sources are also considered “soft ionization” techniques; here, a pulsed laser is used to irradiate samples, causing the desorption and ionization of surface materials (Ligterink et al., 2020). Sometimes aided with matrix reagents (i.e., compounds that have the propensity to efficiently absorb the laser energy, donate protons, or act as electron acceptors for analytes), matrix-assisted laser desorption ionization (MALDI) has been used extensively on Earth to study compounds of relatively high molecular mass like long-chain peptides and other biomolecules with masses up to ∼25,000 Da (Rappsilber et al., 2003; Remoortere et al., 2010). LDI will be employed for the first time in planetary exploration on the MOMA instrument suite aboard the Rosalind Franklin rover (Goesmann et al., 2017; Li et al., 2017). Other laser ionization techniques, such as a two-step laser induced post-ionization, have also been considered for flight (Getty et al., 2012).
While there are many different types and configurations of mass analyzers and ionization sources, one requirement is consistent across all MS: the need for the target analytes to be in gas phase, and in some cases, in plasma phase. Molecular separation techniques such as gas chromatography (GC) is naturally compatible with compounds that are able to be readily volatilized via thermal extraction and have relatively low molecular weights. GCs have been high heritage instruments since the 1970s (Biemann et al., 1977), and continue to serve as a critical payload in recent missions like Rosetta/Philae and MSL (Goesmann et al., 2007; Mahaffy et al., 2012) and future missions like ExoMars (Brinckerhoff et al., 2013; Goesmann et al., 2017). Miniaturized GC using micro-electro-mechanical system (MEMS) technology is also being pursued for future missions, which encompasses a 10 m analytical column (relative to the 30 m GC columns on SAM) that has been compacted into a 150 µm by 240 µm rectangular space (Blase et al., 2020). Likewise, LC can be combined with ESI sources that help simultaneously evaporate the solvents carrying the target analytes and ionizing them as solvent evaporation occurs prior to the interface with a mass analyzer. This technique has also been miniaturized and considered for flight applications (Getty et al., 2013). Capillary electrophoresis (CE) is another liquid-based method used to separate ions based on their electrokinetic mobility under application of a high voltage through a capillary tube. Ions that are separated based on their charge and ionic radius can then be detected using fluorescence techniques (Cable et al., 2013; Willis et al., 2015; Creamer et al., 2017), or MS if coupled to an ionization source like ESI. Significant advances have been made in maturing CE techniques for space applications (Zamuruyev et al., 2021).
Inductively coupled plasma (ICP) techniques atomize and ionize target analytes using a plasma source that is inductively heated with an electromagnetic coil, and have already been miniaturized for potential spaceflight applications (Yin et al., 1999). Plasmas can also be generated by laser desorption, which can then be coupled to MS for mass separation and detection (Rohner et al., 2003). This technique has been used in combination with machine learning to provide exceptional molecular details of Terran-based microfossils (Lukmanov et al., 2021). Because ICP-MS techniques generate electrons at high densities, causing very efficient ionization of target analytes, they are very sensitive to low concentrations and can be used for trace analysis of elements in geological materials (Jenner et al., 1990).
Despite the high resolution and sensitivity of many mass spectrometry platforms, chemical biosignatures of astrobiological interest are likely to occur in complicated mixtures with other organic matter that have been processed (altered) or those that came from abiotic sources, as well as minerals, salts, and metals, some of which may have detrimental effects on MS analysis without proper sample pre-treatment. This can present a tremendous challenge in the sample processing requirements for in situ chemical measurements on other planets, especially for samples that contain oxidants like perchlorates on the Martian surface (Hecht et al., 2009), or high levels of salts like the surface and/or subsurface of Europa (Schmidt et al., 2011; Schmidt, 2020). For these reasons, select sample processing techniques can be employed to overcome the problems associated with sample matrix and media. Reagents such as N-methyl-N-tert-butyldimethylsilyl-trifluoroacetamide (MBSTFA) can react with non-volatile polar molecules (e.g., amino acids) to create volatile derivatives amenable to gas analyses, whereas thermochemolysis with reagents such as tetramethylammonium hydroxide (TMAH) in methanol causes hydrolysis followed by methylation, allowing larger analytes to be broken into components with lower molecular weight, enhancing thermal stability and volatility of the reaction products (He et al., 2020). Both of these approaches are currently being employed by SAM onboard the Curiosity rover as part of the MSL mission (Mahaffy et al., 2012; Williams et al., 2019; Millan et al., 2021, accepted) and by the MOMA instrument (Goesmann et al., 2017; Mißbach et al., 2019; Reinhardt et al., 2020). Some methods (e.g., derivatization approaches) may be sensitive to some salts and water (Freissinet et al., 2015; Mißbach et al., 2019) while others (e.g., TMAH thermochemolysis) are more tolerable of both (Mißbach et al., 2019; He et al., 2021). Thus, sample matrix composition for Martian and other astrobiology targets, particularly the briny, ice-covered oceans harbored by moons of the outer planets, may require instruments to adapt new hardware, methods, and operations for salt-laden samples (e.g., addition of desalting or dilution steps). Advances in sample processing technologies can critically improve the detection capabilities of organic molecules, including those that may be indicative of life, on other worlds.
For Solar System exploration, the search for life greatly benefits from our ability to robotically explore high-priority astrobiology destinations. The goal for many in situ methods for chemical analysis such as using mass spectrometers is to directly sample the materials present on a planetary surface (or in the atmospheres or plumes of terrestrial or ocean worlds), and then determine whether those materials could harbor chemical biosignatures. Chemical biosignatures include specific organic or inorganic molecules, molecules with preferential configurations, as well as structural and compositional patterns among families of molecules.
It is important to note, however, that we have no concrete theory for predicting what molecules and macromolecules life would use in the Universe. If we use Terran life as an example and consider what could be analyzed by mass spectrometers, then it is interesting to note that Terran macromolecules would have some unique features which we review below. However, we strongly emphasize that these particular properties are not guaranteed to be shared by life anywhere else (e.g., Kempes and Krakauer, 2021). The below review should be seen as an exercise of the types of measurements that mass spectrometers could make on Terran life, but this does not address the challenges of finding more universal properties of life that would transcend specific evolutionary history, which will be discussed in the next section.
All life on Earth employs the same essential set of building blocks: nucleotides, amino acids, carbohydrates, and lipids. All these compounds and their derivatives and polymeric forms (i.e., repeated subunits) can, in theory, be indicative of life. However, some compounds, such as amino acids, are also produced abiotically (Miller and Urey, 1959; McDermott et al., 2015; LaRowe and Regnier, 2008; Summons et al., 2008; Hörst et al., 2012; Ménez et al., 2018; Sherwood Lollar et al., 2006) and have been found in extraterrestrial materials (Pizzarello and Cronin, 2000; Glavin et al., 2006, 2018, 2020; Burton et al., 2012), suggesting that their presence may be ubiquitous in the Solar System and, thus, their detection alone is not diagnostic of life. However, certain organic compounds can provide a particularly robust indication of life because they require suites of biological machineries (i.e., enzymes) to be synthesized and have a very low probability of being produced abiotically. For example, sterols, a pervasive group of compounds found in eukaryotic life require numerous enzymes as well as molecular oxygen to be produced (Summons et al., 2008). These compounds, often referred to as biomarkers, have served as not only biosignatures of life, but also taxonomic indicators of specific types of organisms (Peters et al., 2005). While this specificity provides the diagnostic ability to interpret the data of Terran life, it is unknown what similarly definitive biomolecules may look like on another planetary target (Figure 2).
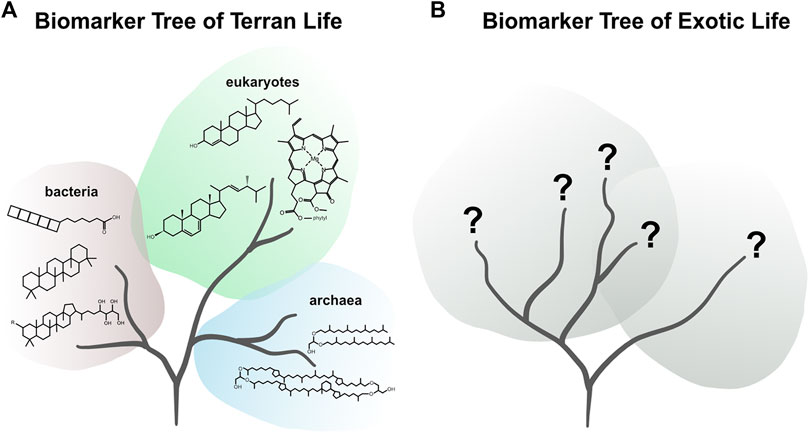
FIGURE 2. (A) Figurative biomarker tree of Terran life representing the diversity of chemical biosignatures detectable via mass spectrometry. (B) Figurative biomarker tree of exotic life representing our lack of knowledge on the biochemical ancestry and thus potential chemical biosignatures of exotic life.
Metabolic processes of Terran life also contribute to the presence of gas-phase biosignatures observed in the Earth’s atmosphere, which are often invoked when considering potential evidence for life on other exoplanets and can be detected in situ via mass spectrometry within our own Solar System. Gases such as molecular oxygen (O2) have been attributed to the advent of specific metabolic capability—oxygenic photosynthesis—that explicitly evolved on Earth, but may not have developed elsewhere (Blankenship and Hartman, 1998; Lehmer et al., 2018). In addition, despite the ample production of O2 by photosynthetic organisms on Earth, several potential planetary mechanisms can generate abundant O2 without life being present (Meadows, 2017) and can confound interpretations of such a detection. Methane (CH4) is another prospective biosignature gas, although care must be taken when interpreting a detection, as CH4 can also be produced abiotically via both high temperature geological processes (Judd, 2000; Guzmán-Marmolejo et al., 2013; Reeves and Fiebig, 2020) and low temperature water-rock reactions (Etiope and Sherwood Lollar, 2013, and references therin). Due to significant overlap in the isotopic signature of C and H in CH4, additional lines of evidence are required to discriminate between biogenic CH4 from those that form geologically (Warr et al., 2021). One avenue for distinguishing between methane sources is measuring the clumped isotopologues of methane, such as the abundances of multiply substituted isotopes (e.g., 13CH3D and 12CH2D2, which can be used to infer the formation temperature, and by proxy, the formation environment of the methane in question (Stolper et al., 2014). Variability of CH4 and O2 in the atmosphere of Mars is thus of great interest and has been measured over a time period of several years by the SAM instrument suite on MSL (Trainer et al., 2019; Webster et al., 2021). Additionally, combinations of gases can also serve as Terran biosignatures, or used to differentiate biogenic CH4 from abiogenic CH4. For example, the low ratios of CH4/C2H6, and high ratios of H/CH4 have been shown to identify abiogenic CH4 sources on Earth analog systems, due to the low trace concentrations of C2H6 and H associated with biogenic sources (Sherwood Lollar et al., 2006; Warr et al., 2021). The co-occurrence of some atmospheric species at particular concentrations (e.g., CH4 with O2, or CH4 with CO2, N2, and H2O) could indicate a chemical disequilibrium that may only persist when there is a consistent source of one or more of those species (Krissansen-Totton et al., 2018), thus pointing to the potential presence of life, especially on exoplanets.
Terran molecular biosignatures also exist as patterns of structural regularities and abundances. Because biological molecules are constructed from a limited set of simpler building blocks, they can contain consistent and repeating units associated with those building blocks. For example, lipids on Earth are produced from two different biochemical pathways: the acetogenic pathway (2-carbon building block) that form fatty acids, the basis for bacterial and eukaryotic membrane lipids, and the isoprenoid pathway (5-carbon building block) that form terpenoid compounds, a large class of molecules that constitute archeael lipids and other metabolites (Caforio and Driessen, 2017; Magnuson et al., 1993). Because life draws from the same pool of molecules, the resulting biological products often display a carbon-number preference. In particular, for fatty acid lipids, carbon chains tend to have an even number of carbons (i.e., even-over-odd patterning) (Volkman, 2006) and are limited in chain length distribution to meet physiological needs (C12-C20 being the most common). Both features (carbon number preference and chain lengths) are key signatures of Terran life and may be more broadly common to life elsewhere (Summons et al., 2008; Georgiou and Deamer, 2014). This is in contrast to abiotically synthesized fatty acids that exhibit no carbon-number preference and have either 1) shorter carbon chains (<C12), as observed in carbonaceous chondrites (Yuenk and Kvenvolden, 1973; Naraoka et al., 1999), or 2) shorter-chain dominance within a broad distribution of up to C30, as is expected of Fischer-Tropsch-type reactions (McCollom et al., 1999). Furthermore, molecules with specific structures like C20, C30, and C40 hydrocarbons (diterpenoid, triterpenoid, and tetraterpenoid, respectively) derive from the isoprenoid pathway, and their specificity, including cyclicity and stereoisomeric configuration, is determined by key biological enzymes, lending to their reliability as biosignatures for Terran life (Summons et al., 2008).
Another intriguing attribute of Terran life is enantiomeric excess (ee). Enantiomers are pairs of a chiral molecule that are mirror images of each other. Many biological organic molecules are “homochiral,” with peptides and proteins exclusively made of L-amino acids, and nucleic acids exclusively composed of D-sugars (Barron, 2008). Though knowledge of the onset of homochirality in Terran life remains an unsolved mystery in the origin of life and astrobiology community, it has been postulated that physiological properties of proteins (e.g., exclusively right-handed helical structures, etc.) are what lead most biological compounds to be synthesized in one configuration over the other (Schwartz, 1994; Tverdislov and Malyshko, 2020). While this predominance is actively maintained when an organism is alive, free homochiral molecules can racemize after the organism ceases to maintain metabolic functions, leading to the ratio between the two enantiomers to move towards 50:50 after death. The racemization rate of amino acids is largely influenced by a number of environmental factors, including temperature (Bada and Schroeder, 1975; Smith et al., 1978), which is an important factor to consider when searching for life on icy worlds. Enantiomeric excesses as high as 18.5% have been observed in meteoritic amino acids (Glavin and Dworkin, 2009). Though an ee threshold of >20% for multiple proteinogenic amino acids has been proposed as a chemical biosignature (Neveu et al., 2018), Glavin et al. (2020) instead suggested a constraint based on a specific ee may not be necessary, and rather that the distributions of the enantiomeric and isotopic composition of amino acids and other organic molecules (e.g., polyols) in combination with structural composition can be used as a set of criteria for life detection.
Other molecular patterns in Terran life manifest in isotopic abundances and elemental distributions. For example, the contrasting variability in isotopic values for C, H, and N in biomolecules compared to meteoritic organic matter is a hallmark of isotopic biosignatures (Engel et al., 1990; Hayes, 2018). In the case of isotopic biosignatures, equilibrium isotopic fractionations can overlap with the kinetic fractionations induced by biological processes (Bottinga, 1969; Galimov, 2006). Further, kinetic fractionations associated with abiotic processes have been shown to produce fractionations of comparable scale to those produced by kinetically-controlled biological processes (Etiope and Sherwood Lollar, 2013; Ojeda et al., 2020). The isotopic discrimination between certain elements in organic molecules and the surrounding local environment (e.g., rock matrix, aqueous media, etc.) is an important strategy to infer the presence of life. Furthermore, position-specific isotopic analysis is an emerging powerful technique for understanding molecular formation pathway that allow for the discrimination between biotic and abiotic compounds (Tenailleau et al., 2004) and to provide contextual information about mineral-mediated reactions relevant to molecular formation in space environments (Fox et al., 2021). However, isotopic interpretations require knowledge of both the isotopic compositions of source materials and the process by which isotopes are fractionated (Hayes, 2001). Thus, isotopic measurements have great potential in their interpretive power, but they are dependent on our understanding of the system from which they are derived. Similarly, because life on Earth is based on a set of common building blocks (CNHOPS), the composition and distribution of elements in biomass is often reflective of the types of atoms that are found in those building blocks. When looking for life on other planets, it could be possible to use elemental ratios to infer the presence of a biological system based on a particular stoichiometric proportion (Elser, 2003; Young et al., 2014; Kempes et al., 2019, 2021).
The list of all the possible chemical derivatives and patterns of Terran life provided above is non-exhaustive, and novel biological compounds are still being discovered every day (Sleiman et al., 2021). On Earth, these biosignatures are routinely analyzed using commercial mass spectrometers. In the case of specific organic molecules, mass fragmentation by hard ionization sources such as EI, or by MSn methods (e.g., collision induced dissociation), is one of the key tools needed to elucidate structural details of the molecules or for full molecular identification. The field of organic geochemistry has tremendously benefited from the standardization of EI methods in MS, with the identification of many important biomarkers of ancient life (e.g., steranes, hopanes, pigments, etc.) whose spectra are now available in comprehensive databases [e.g., the National Institute of Standards and Technology (NIST) Chemistry WebBook] (Wallace, 2021). These tools are often complementary to manual spectral interpretation by trained human investigators (Peters et al., 2005; Simoneit, 2005). For example, the molecular patterns of glycerol dialkyl glycerol tetraether (GDGT) membrane lipids found in most archaea and some bacteria are interpreted to contain a number of cyclic moieties based on their fragmentation patterns using quadrupole and tandem MS (Schouten et al., 2007, 2013; Liu et al., 2012). While fragmentation processes are generally well understood for certain biomolecular classes (e.g., aliphatic compounds and peptides, etc.) (Paizs and Suhai, 2005), it is difficult to fully elucidate molecular structures without a strong knowledge of the chemistry of the analyte and the associated fragmentation pathway. Soft ionization methods can help reduce the propensity for fragmentation, sometimes preserving the molecular ion in high abundance which could allow for the stoichiometric formula confirmation, especially when used with high resolution mass spectrometers (e.g., Orbitrap™). Fragmentation of molecular ions require a delicate balance among the ionization technique, the capability of the mass analyzer, and the sensitivity of the detection method.
While MS offers a detailed look into the molecular composition of life, the complex nature of organic chemistry can complicate in situ analyses that rely solely on MS. This is why it is often critical to choose the appropriate separation or MSn techniques to help resolve mixtures and allow for different analytes to be observed independently by the mass analyzer. For example, without a separation dimension, MS alone does not allow discrimination between isomers (Awad and El-Aneed, 2013). Separating the compounds via chromatography after derivatization could aid in the measurement of enantiomeric excess or homochirality. On the other hand, larger compounds such as peptides that may be analyzed via LDI techniques can experience fragmentation at fluences above the desorption threshold of the material, which can compromise the ability to distinguish those fragments from the molecular ions of other distinct molecules without an ultrahigh resolution mass analyzer (e.g., Orbitrap™). Tandem MS can circumvent this problem by selecting and accumulating ions of interest, fragmenting them in a secondary (or n) MS dimension, leading to “pure” mass spectra that belong only to that particular compound. Tandem MS technology will achieve flight heritage with the ExoMars mission launching in 2022.
Some molecular biosignatures require in situ contextual spatial information that can be aided by MS imaging. Elemental distribution of biogenic elements or molecules at micron scales can be detected using techniques such as LDI-MS or SIMS. For example, Riedo et al. (2020) was able to distinguish biologically relevant elements, such as H, C, S, N, P, and Fe, between microbial life and its mineral matrix in a biologically-spiked Mars analog sample via a high spatial resolution LDI-MS. Similarly, Tulej et al. (2015) was able to study the elemental composition of Terran-based natural samples using LDI-MS, and used the information to distinguish the differences between microfossils and mineral matrix background. Larger molecules have also been investigated using MS imaging, such as the characterization of lipid biomarkers in modern microbial mats and fluid inclusions using a TOF-SIMS (Siljeström et al., 2010; Leefmann et al., 2013). The spatial resolution of MS imaging approaches such as MALDI have typically been on the order of 10 s of microns (Cornett et al., 2007; Riedo et al., 2020), implying the requirement of multicellular structures to ensure biological detection, but advances in matrix application has allowed for 1.4 µm resolution in a study of metabolite, lipid, and peptide arrangements (Kompauer et al., 2017). As MS imaging resolution continues to improve toward that of a single microbial cell, the approach will gain relevance in an astrobiological context, where single-cell detection is a priority.
Contextual spatial information obtained from TOF-SIMS analysis also aided in the elucidation of abiological processes driving the alteration of organic matter in Martian meteorites, suggesting that MS imaging techniques can be critical in understanding the provenance of materials of astrobiological interests (Steele et al., 2018). While LDI techniques will achieve flight heritage with MOMA onboard the ExoMars mission, SIMS methods have already enabled the collection of spectra from space for the first time on the COSIMA instrument suite on board the Rosetta spacecraft (Kissel et al., 2007). MS imaging combined with other analytical techniques represent some of the most promising tools in future life detection missions as it allows for chemical elucidation in a spatial context.
In addition to traditional laboratory techniques, MS investigations have also benefited from automated data-driven pre-processing and analyses using machine learning (ML). A variety of ML techniques such as multi-layer perceptrons, decision trees, and genetic algorithms have been applied to MS data to aid in noise reduction, peak detection, and normalization (Liebal et al., 2020, and references therein). For discovery-driven analysis, classification of novel metabolites, biomarkers related to pathology such as peptides, have also been achieved using combined MS and ML approaches (Bạczek et al., 2004; Hilario et al., 2006; Azari et al., 2018; Liebal et al., 2020). Spatial techniques such as LDI-MS have also significantly advanced with the use of ML, which can help discriminate biotic materials from abiotic background (Lukmanov et al., 2021). Given the low computational cost, ML may become critical in planetary exploration where the classification of instrument output can aid in the reduction of data size that are produced from advanced MS instruments. As deep-space network bandwidth is a critical limiting factor for transmitting high resolution MS data back to Earth from outer Solar System destinations (Theiling et al., 2021), preprocessing techniques could remove unwanted noise, filter out corrupt mass spectra, and preliminarily classify data output so that only relevant data is transmitted. Onboard processing of raw MS data by ML algorithms would greatly enhance the science return of future missions, especially for distant worlds in the outer Solar System.
Many MS techniques are capable of analyzing and characterizing molecules relevant to life, affirming the need of MS in future life detection missions. From a different point of view however, some MS techniques are, in fact, used to investigate the underlying principles that drive the chemistry of life to be different from non-life. It is these principles that could be applied to the search for non-Terran biochemical systems using MS approaches. Mass spectrometers that reveal molecular complexity (“molecular assembly” index) (Marshall et al., 2021), chemical structures, and patterns will play an important role in the future of agnostic biosignatures detection on other worlds.
While search strategies that exclusively focus on Terran biosignatures are biased towards life as we know it, it is critical to note that any attempts to develop novel agnostic biosignature detection methods are still, inescapably, informed by Earth-based life. This is because life on Earth provides a single (N = 1) datum that serves as the reference point for searching for extraterrestrial life in the Universe. Even so, we can take inspiration and interpretive techniques from MS applications to the detection of Terran life, and in turn, design novel in situ detection techniques that are not only agnostic to the underlying biochemistry of the life forms in question, but also take advantage of existing high heritage instrumentation like MS.
On other worlds, life may be based on a different and potentially unfamiliar biochemistry. In order to capture the expressions of those life forms, we should relax our constraints on the criteria that we think a system must exhibit to qualify as life (e.g., even-over-odd fatty acids preference, homochiral amino acids, etc.), and broaden the scope of biosignatures to include the possible attributes of life that we think could be more common in the Universe. While it is not yet known what those attributes are, we can leverage our understanding of general principles of life, such as theories relating to biological processes or behaviors, to inform our intuition about how life elsewhere could have evolved (e.g., Kempes and Krakauer, 2021). Such principles allow us to extract properties that may be unique to life (compared to non-life), and determine what their potential expressions or imprints in the environment could be. The field of agnostic biosignatures is growing in astrobiology, and leveraging agnostic techniques in biosignatures investigations on other worlds may greatly enhance the science return of planetary exploration missions. Importantly, the general principles behind agnostic biosignatures also dictate the features of Terran life, suggesting that techniques that are designed to be agnostic to the life forms in question are also capable of identifying Terran or Terran-like biosignatures (Chou et al., 2021). Newly proposed ideas to detect life more generally are still being developed and formalized. The following section aims to give an overview of this new area of research as it stands today, including some ongoing work that provides a theoretical foundation for possible applications to non-Terran life.
One of the hallmarks of life on Earth is its ability to produce organic compounds through multitudes of enzymatic reactions that act as the machinery of life. However, rather than looking for molecular biosignatures (e.g., sterols) that require known Terran enzymes to form, we can instead search for complex organic molecules that would require enzymatic-type synthesis and are otherwise unlikely to form through abiotic means alone. Such enzymatic-type reactions are a result of evolutionary pressures that funneled life toward preferred molecular pathways that resulted in the suite of complex compounds observed in life. This type of “chemical complexity” has been explored by a number of researchers seeking to determine if there is a relationship between a chemical structure and the quantitative efforts (i.e., number of steps) necessary to produce that particular structure (Wiener, 1947; Rashevsky, 1955; Corey and Wipke, 1969; Bertz, 1981). The idea of chemical complexity (also referred to as “molecular complexity”) has been widely applied to a number of fields such as synthetic chemistry and natural products discovery.
The concept of intrinsic chemical complexity has led to the possibility that a complexity metric could be a way to compare abiotic and biotic compounds, and that a certain complexity threshold for compounds could be used to infer a biogenic origin (Marshall et al., 2017). “Molecular assembly (MA)” index which is computationally derived to express the number of types of operations needed to generate a compound, is a novel mathematical method that can agnostically determine whether a certain compound requires information processing systems (and hence life) to produce. In contrast to previous approaches, the MA index has provided the first intrinsic experimentally tractable measure of chemical complexity using MS (Marshall et al., 2021) (Figure 3A).
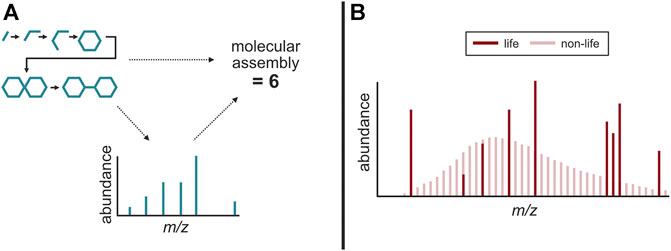
FIGURE 3. Possible agnostic biosignatures detectable using mass spectrometry. (A) Molecular assembly measurements of a molecule, correlated to the number of fragments in its mass spectra. (B) The uniform distribution of compounds found in non-life versus the discrete distribution of compounds found in life, potentially the result of the Lego principle. Adapted from McKay (2008).
MA indices deriving from molecular-level information obtained from MS2 spectra (i.e., the number of fragment ions above certain detection criteria) were shown to predict the potential biogenicity of molecules in a variety of environmental mixtures, including inorganic mineral samples, biological cultures, extracts of ∼14 million-year-old Antarctic lake sediments, Miller–Urey spark discharge mixtures, and meteoritic extracts (Marshall et al., 2021). While the utility of this agnostic method was established on an ESI-Orbitrap MS platform, MA index can be applied to other types of MS as well; in particular for flight instrumentation with lower mass resolution when using other separation techniques and ionization sources to optimize its use for agnostic life detection.
Drawing on other general biological principles, the ability for life to replicate on a molecular scale, by transcribing DNA to mRNA, and translating mRNA to proteins, may be indeed tied to Terran life, but the use of polymers to perform functional chemistry and store information may be a common trait of life in the Universe. This means that rather than looking for DNA or proteins specifically, we can develop methods that can detect polymers in general, and design analytical protocols that can help distinguish between biological polymers and abiotically-formed polymers. Benner (2017) suggested that genetic polymers, especially those that are composed of small, homochiral, repeating monomers (i.e., a molecular alphabet) and a repeating backbone charge can serve as a signature of non-Terran life. While no known device currently exists that is able to observe all of those phenomena, such a task can be aided by the use of MS. The detection and characterization of peptides, for example, have been credited to the advent of MS, including the use of MSn technologies coupled with LC and MALDI techniques in proteomic and synthetic polymer analyses (Rappsilber et al., 2003; Montaudo et al., 2006). Agnostic studies of the composition of polymers using MS, without a prior knowledge of the monomer building blocks, is currently in development for flight mass spectrometers, including the MOMA instrument suite.
With higher resolution MS, it is also possible to use mass defects (the difference between the exact mass of a molecule or atom and its nominal unit mass or atomic mass, respectively) to uncover the underlying pattern of the polymer in question and precisely obtain its molecular formula (Fouquet, 2019, and references therein). This method, known as Kendrick mass analysis, can only be achieved with highly resolved mass peaks that allow for the discrimination between repeating mass defects, and thus the presence of repeating subunits in a sample. Fortunately, high and ultrahigh resolution MS is the next frontier of planetary MS exploration; for example, the MASPEX instrument is capable of mass resolution (m/Δm) of 25,000 at 10% peak height (up to m/Δm 50,000 FWHM) (Waite et al., 2019), and Orbitrap analyzers capable of up to m/Δm > 100,000 (FWHM at m/z 100) (Arevalo et al., 2018) are currently in development for spaceflight applications (Brockwell et al., 2016; Willhite et al., 2021).
Despite a number of avenues that can be explored with polymer detection using MS, it is critical to note that the detection of a polymer does not directly indicate the presence of life. Rather, once a polymer is detected, further analysis is needed to determine if it can feasibly form abiotically or if it requires molecular information (i.e., genetic instructions provided by life-like systems) to form. More research is needed to establish the compositional differences between biotic versus abiotic polymers, such as the polymer sequences or distribution of monomers in those polymers.
An underlying mechanism that seemingly drives many patterns in the molecular composition of life is the usage and recycling of a relatively small set of chemical building blocks (e.g., acetate as the building block for the acetogenic lipid biosynthesis, etc.) (Simon, 1962). While life elsewhere may not harbor the exact biological machineries that have been selected and proliferated by Terran life, one potential shared property with Terran life is the use of smaller building blocks to create larger, more abundant ensembles of macromolecules. This is in stark contrast to the random and diverse set of compounds found in abiotic environments (e.g., meteoritic organic matter, etc.) constrained by rate of formations that are thermodynamically and kinetically directed (Sephton, 2005; Schmitt-Kopplin et al., 2010). These contrasting distributions have been observed in several Terran examples, such as in amino acid and fatty acid distributions in biological samples (e.g., terrestrial sediments, soils, and waters, etc.) compared to abiotic examples (e.g., laboratory synthesis mixtures, meteorite extracts, etc.) (McCollom et al., 1999; Dorn et al., 2011; Salter et al., 2021). Because the discrete distribution of compounds in a biological sample could indicate disequilibrium processes driving the formation and destruction of molecular constituents, it is possible such distributions are a signature of life. This phenomenon, known as the “the Lego principle,” surmises that biology uses an irregular subset of compounds whereas abiotic systems generate a continuous spectrum of compounds (McKay, 2008; Davila and McKay, 2014) (Figure 3B).
The abundances of compounds in biotic versus abiotic samples could be obtained from quantitative MS measures. While studies of amino acid or fatty acid abundance biosignatures rely on specific extraction, separation, and ionization techniques, agnostic methods that leverage the use of the Lego principle would greatly benefit from non-targeted analysis, or suites of MS instruments and coupled separation techniques that can target as many compounds as possible. Indeed, LDI techniques, such as pioneered by MOMA, and EI combined with adequate separation techniques, are particularly well-suited for non-targeted analysis and can directly and indiscriminately ablate homogeneous samples. Chromatography methods that require the compounds to be extracted, concentrated, and sometimes chemically derivatized may also be effective in capturing a diverse set of possible compounds. Combined with statistical analysis, multi-platform approaches could allow to differentiate between abiotic and biotic samples without immediate knowledge of the underlying biochemistry.
Spatial patterns may also serve as agnostic biosignatures. Specifically, life is a metabolic entity that distinguishes itself from the background, a property that is likely true for both Terran life and exotic life. The notion that living organisms are universally dissimilar from the local environment can lead to a partition in the elemental composition of biological units (cells). The Redfield ratio is a classic example of a Terran biogeochemical regularity that might serve as a target biosignature. However, it is important to note that how different the elemental ratio in cells is from the environment depends on a variety of coupled dynamics associated with environmental inputs and the ecological assortment of cells (see Kempes et al., 2021, for a discussion and review). Given that life on other planetary bodies may arise from different starting constituents, it is possible that exotic biochemistry expresses different elemental ratios based on different dynamics. In Terran microbial communities, a given organism’s “sphere of influence” is often on the order of a few microns (Dal Co et al., 2020; Steinberg et al., 2021), leading to patchiness and localized enrichment or depletion of biogenic structures. Extending these observations to an agnostic framework will require better developed models of how energy dissipation associated with metabolism creates and degrades spatial heterogeneities over time. In this context, the use instruments such as LDI-MS and/or SIMS to resolve spatial distribution of elemental or isotopic compositions of surfaces at cellular scales (µm to nm resolution) will become increasingly important.
The use of SIMS has enabled exceptionally detailed investigation of paleobiological records of microfossils (House et al., 2000; Oehler et al., 2010; Oehler and Cady, 2014) as well as detailed physiological studies of single microbial cells in modern environments (Musat et al., 2008). The enrichment or co-localization of certain elements can provide evidence for cells that are actively scavenging materials to sustain its biomass. The sequestration of those elements (into biomass or biominerals) in a nonrandom manner compared to the expected abiotic background processes (e.g., geochemical precipitation of minerals) is a consequence of life that is agnostic to the underlying biochemistry. Thus, it is possible to use techniques like LDI-MS and SIMS to search for enrichment or co-localization of elemental materials. For example, elemental enrichments are readily observable in marine phytoplankton (Slaveykova et al., 2009). Furthermore, some rare elements may also be concentrated and at disequilibrium due to the presence of biological systems that scavenge and concentrate them for purposeful use, much like the acquisition of P, Fe, or Ni from the ocean in some marine organisms (Williams and Da Silva, 2000). The use of SIMS techniques, potentially combined with particle sorting technologies (e.g., flow cytometry or microfluidics), could allow for an in-depth investigation of putative cells on an elemental or molecular level (Feng et al., 2020).
While life is delineated from its environment in a manner that is detectable using spatially resolved MS techniques, it is also important to consider non-biological processes that can also concentrate certain elements or chemicals, leading to potential false positive interpretations. For example, in the famous case of the Allan Hills 84001 (ALH84001) Martian meteorite, scanning electron microscopy images of carbonate globules revealed the presence of “microfossil”-like structures containing ordered magnetic crystals, which were thought to be of biogenic origin (McKay et al., 1996; Thomas-Keprta et al., 2002). However, upon investigating the carbonate-rich phases in ALH84001 within a geologic and petrographic context, it was determined that the magnetic crystals could form via non-biological processes (Golden et al., 2001). Furthermore, TOF-SIMS of aromatic hydrocarbons in the meteorite also provided contextual evidence necessary to refute some of the earlier interpretations about the potential biogenic origin of the ALH84001 structures (Stephan et al., 2003). Therefore, prior to studying elemental and chemical fractionation processes, it is critical to understand the potential formation pathways of specific elemental or mineral phases that is relevant to the sample of interest, to rule out potential abiotic explanations (Steele et al., 2007, 2016, 2018).
Exciting new advances in agnostic biosignatures research greatly complement future frontiers in life detection missions. Since the Viking era, the astrobiology community has gained an appreciable sense of conscientiousness regarding how to define experimental protocols in searching for life on other worlds and the guiding principles needed to interpret possible return data (Green et al., 2021, accepted). Current and planned planetary missions aim to investigate the physical and chemical characteristics of other extraterrestrial environments to better understand planetary formation, evolution, and propensity to support life. Mission concepts that aim to initiate the search for familiar and agnostic biosignatures on planetary bodies (i.e., Mars, Europa, Enceladus) are under development and have been proposed. For example, one of ExoMars’ mission objectives is to search for evidence of morphological and chemical biosignatures on the surface of Mars, using a confidence scoring methodology that is based on the geological context and the quality of the measurements (e.g., relative to in situ blank analyses and knowledge of potential spacecraft contamination) (Vago et al., 2017). Other mission concepts to Europa have also begun to integrate agnostic search strategies to account for the presence of potentially unfamiliar life forms (Mahaffy et al., 2021). In addition, sample return missions have been proposed for Mars and Enceladus to leverage the use of multiple instrument platforms to look for signs of life once the samples are back on Earth (Muirhead et al., 2020; Neveu et al., 2020; MacKenzie et al., 2021), especially those that have yet to be miniaturized for flight (e.g., nano-SIMS). Because agnostic life detection strategies can be implemented in near future missions, emerging concepts in agnostic biosignatures are concurrently being optimized in the laboratory in order to enhance the science return and increase our confidence in any future potential life detection measurements.
Selected and proposed planetary missions that will employ mass spectrometers include those that will visit Mars, Titan, Europa, Enceladus, and Venus. For example, the Rosalind Franklin rover, part of ESA’s ExoMars mission, is slated to collect samples from the subsurface of Mars (down to 2 m), where protection from damaging radiation and oxidation offers a potential oasis for the preservation of past or present life (Vago et al., 2017). Onboard this rover is the MOMA instrument suite (Goesmann et al., 2017), equipped with dual EI and LDI sources coupled to an ITMS, with the advanced capabilities to accumulate ions and fragment them using MSn. MOMA can target high molecular weight compounds (50–1,000 Da) relative to other high heritage instruments (e.g., the QMS on the SAM instrument suite is 2–535 Da). Agnostic approaches, such as searching for chemical complexity, are naturally applicable to MOMA data as it is capable of MS2 measurements (Marshall et al., 2021). Along with this promising new direction for MS flight data analysis, it is imperative to formulate mass ion selection processes for elucidating large compound structures and determine MA index measurement that can help inform ground-in-the-loop activities for MSn experiments on MOMA.
A similar instrument to MOMA, the Dragonfly Mass Spectrometer (DraMS) will explore an even more distant and stranger world, Titan. This moon of Saturn boasts a captivating chemosphere, where historical and geological circumstances have led to a surface rich in (presumably abiotic) organic synthesis, resulting in organic haze that blankets the entire moon (Gupta et al., 1981). With its mildly reducing atmosphere (primarily composed of N2 and CH4) and constant flux of UV light, cosmic rays, and energetic electrons from Saturn’s magnetosphere (Raulin and Owen, 2003), Titan is one of the prime locations in the Solar System to search for Terran-like prebiotic chemistry as well as evidence of exotic biochemistry. For example, given the existence of a non-water solvent on Titan’s surface (liquid methane and ethane), it is seemingly possible for life with a completely alternate biochemistry to have arisen (McKay, 2016). Such life would need to withstand far different physical regimes than Terran life (e.g., extremely cold surface temperatures approaching 90 K). Titan has a methanological cycle that drives dynamic surface processes, leading to the formation of lakes, rivers, rain, and clouds of liquid methane and ethane that may contribute to the distribution of organic material across the surface and atmosphere (Hörst, 2017). If undertaken, the search for “hydrocarbon-based” life on Titan would inevitably necessitate a less Terran-centric approach.
Given the organically abundant environment, one avenue would be to search for potential biopolymers on Titan. Several studies have found repeating patterns in Δm within the mass spectra of Titan’s atmophsere using in situ observations (Waite et al., 2007; Crary et al., 2009) as well as in Titan haze and aerosol analogs that could be attributed to polymers (Pernot and Schmitz-afonso, 2010; Cable et al., 2012), although the composition and distribution of the monomers as well as the polymer architecture (e.g., linear or cross-linked) have yet to be determined (Gautier et al., 2017). Titan haze analogs present an important laboratory simulant reference sample that can be used to study the abiotic endmembers of polymers that could be produced in the Titan environment, and in similar extraterrestrial environments. Studying the patterns associated with polymers in MS data will be essential to support the science of the Dragonfly mission and ongoing scientific work in the field of agnostic bioisgnatures will enhance the science return upon data downlink and interpretation.
Closer to home, our neighboring planet Venus, will also be a near future destination. The Deep Atmosphere Venus Investigation of Noble gases, Chemistry, and Imaging (DAVINCI) mission was selected as part of the NASA Discovery Program, with a major objective to study the chemical composition of Venus’s atmosphere (Arney et al., 2020) from above the clouds during flybys and the descent of its probe. Onboard the DAVINCI probe element is a mass spectrometer, related to the SAM-QMS-based instrument on the Curiosity rover, capable of in situ measurements of noble gas species, atmospheric composition, and isotopic analysis. Venus has recently fueled significant interests from the astrobiology community, with the putative discovery of phosphine (PH3) by remote spectroscopy; PH3 is a type of biosignature gas which has been attributed to Terran anaerobic life (Greaves et al., 2020), although admittedly few investigations have been undertaken to explore potential abiotic processes that could also produce this compound. This surprising detection has subsequently given rise to an ongoing debate centered around the detection and interpretation of potential PH3 signatures in the atmosphere of Venus (Encrenaz et al., 2020; Snellen et al., 2020; Lincowski et al., 2021). The origin, evolution, and composition of the Venusian atmosphere is an enigma that will soon be revealed by DAVINCI. Understanding atmospheric and planetary processes on Venus could provide important insight into its history as a potentially habitable inner rocky planet, and will provide inputs to atmospheric models that will allow the community to better interpret agnostic biosignatures in our Solar System and planetary systems beyond our own.
Icy moons of Jupiter, Europa, Ganymede, and Callisto, will welcome a new mass spectrometer aboard an ESA orbiter mission scheduled for launch in 2022. The Jupiter Icy Moon Explorer (JUICE) will harbor the Neutral gas and Ion MS (NIM), which will make the first direct measurement of the exospheres of Jupiter’s three icy moons (Grasset et al., 2013; Föhn et al., 2021). The NIM is a TOFMS that can capture neutral molecules as well as ionospheric ions with a mass range of m/z 1–650, and a resolution m/Δm of ∼750 (FWHM) (Föhn et al., 2021). Detailed investigation of the chemical composition of the exospheres of these moons not only would shed light on the potential formation of life or ingredients for life, but may also reveal how different these moons are from each other and from other Solar System bodies.
Lastly, several proposed missions are currently under development for one of the most exciting astrobiology targets in the Solar System: Europa. The Europa Clipper mission will set out to investigate the habitability of Europa using the MAss SPectrometer for Planetary Exploration/Europa (MASPEX) instrument, a high resolution TOF-MS intending to measure trace organic compounds at parts per billion levels (Brockwell et al., 2016) as well as search for evidence of life on Europa (Salter et al., 2021). As the community looks ahead to surface exploration at Europa, instrumental capabilities such as those discussed throughout this review will be critical to meet the mission’s scientific goals (Hand et al., 2018). The Characterization of Ocean Residues And Life Signatures (CORALS) instrument, a candidate instrument for a Europa Lander mission, comprise of a solid-state UV laser source capable of active beam scanning and an ultrahigh resolution Orbitrap mass analyzer, to derive 2D chemical images of Europa samples (Arevalo et al., 2018; Willhite et al., 2021) (Figure 4). Such ultrahigh resolution MS instruments can be used to distinguish polymeric patterns, or chemical complexity, helping assess the potential biogenicity of samples indigenous to Europa’s surface or subsurface (via plumes) (Quick et al., 2013; Jia et al., 2018). In addition, the Europan Molecular Indicators of Life Investigation (EMILI) instrument suite combines both liquid processing via capillary electrophoresis and gas chromatography techniques with an ITMS to provide the broadest range of chemical targets and concentrations of any instruments to date (Brinckerhoff et al., 2018, in this special issue).
FIGURE 4. The miniaturized Orbitrap mass analyzer of the Characterization of Ocean Residues And Life Signatures (CORALS) instrument, one of the next generation mass spectrometers currently being developed for spaceflight. Adapted from Willhite et al. (2021).
Since the dawn of the space age, mass spectrometers have transformed our understanding of our Solar System, and the coming decade will usher in yet another revolution in planetary mass spectrometry. Space agencies around the world are developing missions to explore enticing planetary targets with mass spectrometry investigations that could not only improve our understanding of planetary processes, but also seek evidence of life beyond Earth. Because life elsewhere may not necessarily share the same heritage as life on Earth, it is imperative to infuse agnostic approaches into these future missions. Fortunately, high heritage mass spectrometers are already capable of conducting agnostic searches for life on planetary missions. Beyond the continual drive to improve flight-capable mass spectrometry technologies, it may also be fruitful to define criteria that broadly encompass the detection of the universal traits of life, such as developing statistical frameworks for assessing biogenicity of putative agnostic biosignatures and benchmarking novel methods with Earth-based planetary analogs or extraterrestrial materials in the laboratory. Mission designs having multiple, independent biosignatures measurements that encompass both familiar and agnostic biosignatures offer a robust science strategy for in situ life detection.
LC wrote the first draft of the manuscript. All authors contributed to the writing and editing of the manuscript.
LC was supported by an appointment to the NASA Postdoctoral Program at the NASA Goddard Space Flight Center, administered by USRA through a contract with NASA and by the NASA-funded Laboratory for Agnostic Biosignatures Project (NASA Grant 80NSSC18K1140 to SJ). MT was supported by the Dragonfly Mission, part of the NASA New Frontiers Program. HG and BSL are CIFAR Fellows in the Earth 4D: Subsurface Science and Exploration Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to acknowledge the two independent reviewers, the associate editor, and the Laboratory for Agnostic Biosignatures team for providing feedback which helped improve the quality of the manuscript.
APCI, atmospheric-pressure chemical ionization; CE, capillary electrophoresis; CI, chemical ionization; CNHOPS, carbon, nitrogen, hydrogen, oxygen, phosphorous, and sulfur; DFMS, double focusing mass spectrometer; EI, electron impact; ESI, electrospray ionization; GC, gas chromatography; GC-MS, gas chromatography-mass spectrometers; ICP, inductively coupled plasma; ICR, ion cyclotron resonance; ITMS, ion trap mass spectrometer; LC, liquid chromatography; LDI, laser desorption ionization; MA, molecular assembly; MALDI, matrix-assisted laser desorption ionization; ML, machine learning; MOMA, Mars organic molecular analyzer; MS, mass spectrometers; MSL, Mars science laboratory; NIST, national institute of standards and technology; QMS, quadrupole mass spectrometer; SAM, sample analysis at Mars; SIMS, secondary ion mass spectrometry; TOFMS, time-of-flight mass spectrometer.
Arevalo, R., Ni, Z., and Danell, R. M. (2020). Mass Spectrometry and Planetary Exploration: A Brief Review and Future Projection. J. Mass. Spectrom. 55, 1–17. doi:10.1002/jms.4454
Arevalo, R., Selliez, L., Briois, C., Carrasco, N., Thirkell, L., Cherville, B., et al. (2018). An Orbitrap-Based Laser Desorption/ablation Mass Spectrometer Designed for Spaceflight. Rapid Commun. Mass. Spectrom. 32, 1875–1886. doi:10.1002/rcm.8244
Arney, G., Getty, S., Johnson, N., Kiefer, W., Lorenz, R., Ravine, M., et al. (2005). Venus as an Analog for Hot Earths. Lunar Planet. Sci. Conf. 2599, 1. doi:10.2458/azu_uapress_9780816540068-ch014
Atreya, S. K., Mahaffy, P. R., Niemann, H. B., Wong, M. H., and Owen, T. C. (2003). Composition and Origin of the Atmosphere of Jupiter-an Update, and Implications for the Extrasolar Giant Planets. Planet. Space Sci. 51, 105–112. doi:10.1016/S0032-0633(02)00144-7
Awad, H., and El-Aneed, A. (2013). Enantioselectivity of Mass Spectrometry: Challenges and Promises. Mass. Spec. Rev. 32, 466–483. doi:10.1002/mas10.1002/mas.21379
Azari, S., Zhang, M., Xue, B., and Peng, L. (2018). “Genetic Programming for Preprocessing Tandem Mass Spectra to Improve the Reliability of Peptide Identification,” in 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, July 8–13, 2018 (IEEE), 1–8. doi:10.1109/cec.2018.8477810
Bada, J. L., and Schroeder, R. A. (1975). Amino Acid Racemization Reactions and Their Geochemical Implications. Naturwissenschaften 62, 71–79. doi:10.1007/bf00592179
Balsiger, H., Altwegg, K., Bochsler, P., Eberhardt, P., Fischer, J., Graf, S., et al. (2007). Rosina - Rosetta Orbiter Spectrometer for Ion and Neutral Analysis. Space Sci. Rev. 128, 745–801. doi:10.1007/s11214-006-8335-3
Barron, L. D. (2008). “Chirality and Life,” in Strategies of Life Detection (Boston, MA: Springer US), 187–201. doi:10.1007/978-0-387-77516-6_13
Bạczek, T., Buciński, A., Ivanov, A. R., and Kaliszan, R. (2004). Artificial Neural Network Analysis for Evaluation of Peptide Ms/ms Spectra in Proteomics. Anal. Chem. 76, 1726–1732. doi:10.1021/ac041516u
Benner, S. A. (2017). Detecting Darwinism from Molecules in the Enceladus Plumes, Jupiter's Moons, and Other Planetary Water Lagoons. Astrobiology 17, 840–851. doi:10.1089/ast.2016.1611
Bertz, S. H. (1981). The First General Index of Molecular Complexity. J. Am. Chem. Soc. 103, 3599–3601. doi:10.1021/ja00402a071
Biemann, K. (2007). On the Ability of the Viking Gas Chromatograph-Mass Spectrometer to Detect Organic Matter. Proc. Natl. Acad. Sci. 104, 10310–10313. doi:10.1073/pnas.0703732104
Biemann, K., Oro, J., Toulmin, P., Orgel, L. E., Nier, A. O., Anderson, D. M., et al. (1977). The Search for Organic Substances and Inorganic Volatile Compounds in the Surface of Mars. J. Geophys. Res. 82, 4641–4658. doi:10.1029/js082i028p04641
Blankenship, R. E., and Hartman, H. (1998). The Origin and Evolution of Oxygenic Photosynthesis. Trends Biochemical Sciences 23, 94–97. doi:10.1016/s0968-0004(98)01186-4
Blase, R. C., Libardoni, M. J., Miller, G. P., Miller, K. E., Phillips-Lander, C. M., Waite, J. H., et al. (2020). Experimental Coupling of a MEMS Gas Chromatograph and a Mass Spectrometer for Organic Analysis in Space Environments. ACS Earth Space Chem. 4, 1718–1729. doi:10.1021/acsearthspacechem.0c00131
Bottinga, Y. (1969). Calculated Fractionation Factors for Carbon and Hydrogen Isotope Exchange in the System Calcite-Carbon Dioxide-Graphite-Methane-Hydrogen-Water Vapor. Geochimica et Cosmochimica Acta. 33, 49–64. doi:10.1016/0016-7037(69)90092-1
Brinckerhoff, W. B., Grubisic, A., Getty, S. A., Danell, R. M., Arevalo Jr, R. D., and Li, X (2018). EMILI: Europan Molecular Indicators of Life Investigation. In Earth and Space 2018: Engineering for Extreme Environments (Reston, VA: American Society of Civil Engineers), 524–532.
Brinckerhoff, W. B., Pinnick, V. T., Van Amerom, F. H. W., Danell, R. M., Arevalo, R. D., Atanassova, M. S., et al. (2013). “Mars Organic Molecule Analyzer (MOMA) Mass Spectrometer for ExoMars 2018 and beyond,” in 2013 IEEE Aerospace Conference, Big Sky, MT, March 2–9, 2013. doi:10.1109/AERO.2013.6496942
Brockwell, T. G., Meech, K. J., Pickens, K., Waite, J. H., Miller, G., Roberts, J., et al. (2016). “The Mass Spectrometer for Planetary Exploration (MASPEX),” in 2016 IEEE Aerospace Conference, Big Sky, MT, March 5–12, 2016, 1–17. doi:10.1109/aero.2016.7500777
Burton, A. S., Stern, J. C., Elsila, J. E., Glavin, D. P., and Dworkin, J. P. (2012). Understanding Prebiotic Chemistry through the Analysis of Extraterrestrial Amino Acids and Nucleobases in Meteorites. Chem. Soc. Rev. 41, 5459–5472. doi:10.1039/c2cs35109a
Cable, M. L., Hörst, S. M., Hodyss, R., Beauchamp, P. M., Smith, M. A., and Willis, P. A. (2012). Titan Tholins: Simulating Titan Organic Chemistry in the Cassini-Huygens Era. Chem. Rev. 112, 1882–1909. doi:10.1021/cr200221x
Cable, M. L., Stockton, A. M., Mora, M. F., and Willis, P. A. (2013). Low-Temperature Microchip Nonaqueous Capillary Electrophoresis of Aliphatic Primary Amines: Applications to Titan Chemistry. Anal. Chem. 85, 1124–1131. doi:10.1021/ac3030202
Caforio, A., and Driessen, A. J. M. (2017). Archaeal Phospholipids: Structural Properties and Biosynthesis. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids. 1862, 1325–1339. doi:10.1016/j.bbalip.2016.12.006
Chou, L., Grefenstette, N., Johnson, S. S., Graham, H., Mahaffy, P., Kempes, C., et al. (2021). Towards a More Universal Life Detection Strategy. Bull. AAS. 53, 1–7. doi:10.3847/25c2cfeb.53a24171
Cleland, C. E., and Chyba, C. F. (2002). Defining ‘Life’. Orig. Life. Evol. Biosph. 32, 387–393. doi:10.1023/A:1020503324273
Corey, E. J., and Wipke, W. T. (1969). Computer-Assisted Design of Complex Organic Syntheses. Science 166, 178–192. doi:10.1126/science.166.3902.178
Cornett, D. S., Reyzer, M. L., Chaurand, P., and Caprioli, R. M. (2007). MALDI Imaging Mass Spectrometry: Molecular Snapshots of Biochemical Systems. Nat. Methods. 4, 828–833. doi:10.1038/nmeth1094
Crary, F. J., Magee, B. A., Mandt, K., Waite, J. H., Westlake, J., and Young, D. T. (2009). Heavy Ions, Temperatures and Winds in Titan's Ionosphere: Combined Cassini CAPS and INMS Observations. Planet. Space Sci. 57, 1847–1856. doi:10.1016/j.pss.2009.09.006
Creamer, J. S., Mora, M. F., and Willis, P. A. (2017). Enhanced Resolution of Chiral Amino Acids with Capillary Electrophoresis for Biosignature Detection in Extraterrestrial Samples. Anal. Chem. 89, 1329–1337. doi:10.1021/acs.analchem.6b04338
Dal Co, A., van Vliet, S., Kiviet, D. J., Schlegel, S., and Ackermann, M. (2020). Short-range Interactions Govern the Dynamics and Functions of Microbial Communities. Nat. Ecol. Evol. 4, 366–375. doi:10.1038/s41559-019-1080-2
Dančík, V., Addona, T. A., Clauser, K. R., Vath, J. E., and Pevzner, P. A. (1999). De NovoPeptide Sequencing via Tandem Mass Spectrometry. J. Comput. Biol. 6, 327–342. doi:10.1089/106652799318300
Davila, A. F., and McKay, C. P. (2014). Chance and Necessity in Biochemistry: Implications for the Search for Extraterrestrial Biomarkers in Earth-like Environments. Astrobiology 14, 534–540. doi:10.1089/ast.2014.1150
Des Marais, D. J., Nuth, J. A., Allamandola, L. J., Boss, A. P., Farmer, J. D., Hoehler, T. M., et al. (2008). The NASA Astrobiology Roadmap. Astrobiology 8, 715–730. doi:10.1089/ast.2008.0819
Dorn, E. D., Nealson, K. H., and Adami, C. (2011). Monomer Abundance Distribution Patterns as a Universal Biosignature: Examples from Terrestrial and Digital Life. J. Mol. Evol. 72, 283–. doi:10.1007/s00239-011-9429-4
Dührkop, K., Nothias, L. F., Fleischauer, M., Reher, R., Ludwig, M., Hoffmann, M. A., et al. (2021). Systematic Classification of Unknown Metabolites Using High-Resolution Fragmentation Mass Spectra. Nat. Biotechnol. 39, 462–471. doi:10.1038/s41587-020-0740-8
Eigenbrode, J. L. (2008). Fossil Lipids for Life-Detection: A Case Study from the Early Earth Record. Space Sci. Rev. 135, 161–185. doi:10.1007/s11214-007-9252-9
Eigenbrode, J. L., Summons, R. E., Steele, A., Freissinet, C., Millan, M., Navarro-González, R., et al. (2018). Organic Matter Preserved in 3-Billion-Year-Old Mudstones at Gale Crater, Mars. Science 360, 1096–1101. doi:10.1126/science.aas9185
Elser, J. J. (2003). Biological Stoichiometry: A Theoretical Framework Connecting Ecosystem Ecology, Evolution, and Biochemistry for Application in Astrobiology. Int. J. Astrobiology. 2, 185–193. doi:10.1017/S1473550403001563
Encrenaz, T., Greathouse, T. K., Marcq, E., Widemann, T., Bézard, B., Fouchet, T., et al. (2020). A Stringent Upper Limit of the PH3 Abundance at the Cloud Top of Venus. Astron. Astrophysics. 643, 1–4. doi:10.1051/0004-6361/202039559
Engel, M. H., Macko, S. A., and Silfer, J. A. (1990). Carbon Isotope Composition of Individual Amino Acids in the Murchison Meteorite. Nature 348, 47–49. doi:10.1038/348047a0
Etiope, G., and Sherwood Lollar, B. (2013). Abiotic Methane on Earth. Rev. Geophys. 51, 276–299. doi:10.1002/rog.20011
Feng, D., Xu, T., Li, H., Shi, X., and Xu, G. (2020). Single-cell Metabolomics Analysis by Microfluidics and Mass Spectrometry: Recent New Advances. J. Anal. Test. 4, 198–209. doi:10.1007/s41664-020-00138-9
Föhn, M., Munz, H. P., Wurz, P., and Gerber, M. (2021). “Description of the Mass Spectrometer for the Jupiter Icy Moons Explorer Mission,” in IEEE Aerospace Conference, Big Sky, MT, March 6–13, 2021, 1–14. doi:10.1109/aero50100.2021.9438344
Fouquet, T. N. J. (2019). The Kendrick Analysis for Polymer Mass Spectrometry. J. Mass. Spectrom. 54, 933–947. doi:10.1002/jms.4480
Fox, A. C., Martineau, E., Remaud, G. S., and Freeman, K. H. (2021). Position-specific Isotope Fractionation in Amino Acids Sorbed to Ice: Implications for the Preservation of Isotopologue Biosignatures. Geochimica et Cosmochimica Acta. 309, 45–56. doi:10.1016/j.gca.2021.06.023
Frank, A., and Pevzner, P. (2005). PepNovo: De Novo Peptide Sequencing via Probabilistic Network Modeling. Anal. Chem. 77, 964–973. doi:10.1021/ac048788h
Freissinet, C., Glavin, D. P., Mahaffy, P. R., Miller, K. E., Eigenbrode, J. L., Summons, R. E., et al. (2015). Organic Molecules in the Sheepbed Mudstone, Gale Crater, Mars. J. Geophys. Res. Planets 120, 495–514. doi:10.1002/2014je004737
Galimov, E. M. (2006). Isotope Organic Geochemistry. Org. Geochem. 37, 1200–1262. doi:10.1016/j.orggeochem.2006.04.009
Gautier, T., Sebree, J. A., Li, X., Pinnick, V. T., Grubisic, A., Loeffler, M. J., et al. (2017). Influence of Trace Aromatics on the Chemical Growth Mechanisms of Titan Aerosol Analogues. Planet. Space Sci. 140, 27–34. doi:10.1016/j.pss.2017.03.012
Georgiou, C. D., and Deamer, D. W. (2014). Lipids as Universal Biomarkers of Extraterrestrial Life. Astrobiology 14, 541–549. doi:10.1089/ast.2013.1134
Getty, S. A., Brinckerhoff, W. B., Cornish, T., Ecelberger, S., and Floyd, M. (2012). Compact Two-step Laser Time-Of-Flight Mass Spectrometer for In Situ Analyses of Aromatic Organics on Planetary Missions. Rapid Commun. Mass. Spectrom. 26, 2786–2790. doi:10.1002/rcm.6393
Getty, S. A., Dworkin, J. P., Glavin, D. P., Martin, M., Zheng, Y., Balvin, M., et al. (2013). “Organics Analyzer for Sampling Icy Surfaces: A Liquid Chromatograph-Mass Spectrometer for Future In Situ Small Body Missions,” in IEEE Aerospace Conference, Big Sky, MT, March 2–9, 2013. doi:10.1109/AERO.2013.6497391
Glavin, D. P., Alexander, C. M. O. D., Aponte, J. C., Dworkin, J. P., Elsila, J. E., and Yabuta, H. (2018). “The Origin and Evolution of Organic Matter in Carbonaceous Chondrites and Links to Their Parent Bodies,” in Primitive Meteorites and Asteroids. Editors N. B. T. P. M. Abreu, and Asteroids (Elsevier), 205–271. doi:10.1016/B978-0-12-813325-5.00003-3
Glavin, D. P., Burton, A. S., Elsila, J. E., Aponte, J. C., and Dworkin, J. P. (2020). The Search for Chiral Asymmetry as a Potential Biosignature in Our Solar System. Chem. Rev. 120, 4660–4689. doi:10.1021/acs.chemrev.9b00474
Glavin, D. P., Dworkin, J. P., Aubrey, A., Botta, O., Doty, J. H., Martins, Z., et al. (2006). Amino Acid Analyses of Antarctic CM2 Meteorites Using Liquid Chromatography-Time of Flight-Mass Spectrometry. Meteoritics Planet. Sci. 41, 889–902. doi:10.1111/j.1945-5100.2006.tb00493.x
Glavin, D. P., and Dworkin, J. P. (2009). Enrichment of the Amino Acid L-Isovaline by Aqueous Alteration on CI and CM Meteorite Parent Bodies. Proc. Natl. Acad. Sci. 106, 5487–5492. doi:10.1073/pnas.0811618106
Goesmann, F., Brinckerhoff, W. B., Raulin, F., Goetz, W., Danell, R. M., Getty, S. A., et al. (2017). The Mars Organic Molecule Analyzer (MOMA) Instrument: Characterization of Organic Material in Martian Sediments. Astrobiology 17, 655–685. doi:10.1089/ast.2016.1551
Goesmann, F., Rosenbauer, H., Roll, R., Szopa, C., Raulin, F., Sternberg, R., et al. (2007). COSAC, the Cometary Sampling and Composition experiment on Philae. Space Sci. Rev. 128, 257–280. doi:10.1007/s11214-006-9000-6
Golden, D. C., Ming, D. W., Schwandt, C. S., Lauer, H. V., Socki, R. A., Morris, R. V., et al. (2001). A Simple Inorganic Process for Formation of Carbonates, Magnetite, and Sulfides in Martian Meteorite ALH84001. Am. Mineral. 86, 370–375. doi:10.2138/am-2001-2-321
Grasset, O., Dougherty, M. K., Coustenis, A., Bunce, E. J., Erd, C., Titov, D., et al. (2013). JUpiter ICy Moons Explorer (JUICE): An ESA mission to Orbit Ganymede and to Characterise the Jupiter System. Planet. Space Sci. 78, 1–21. doi:10.1016/j.pss.2012.12.002
Greaves, J. S., Richards, A. M. S., Bains, W., Rimmer, P. B., Sagawa, H., Clements, D. L., et al. (2020). Phosphine Gas in the Cloud Decks of Venus. Nat. Astron. 5, 655–664. doi:10.1038/s41550-020-1174-4
Green, J., Hoehler, T., Neveu, M., Domagal-Goldman, S., Scalice, D., and Voytek, M. (2021). Call for a Framework for Reporting Evidence for Life beyond Earth. arXiv. preprint arXiv:2107.10975.
Gringauz, K. I., Remizov, A. P., Verigin, M. I., Denchikova, L. I., Dyachkov, A. V., Klimenko, I. N., et al. (1985). First Results of Plasma and Neutral Gas Measurements from Near Comet Halley. Adv. Space Res. 5, 165–174. doi:10.1016/0273-1177(85)90082-1
Grotzinger, J. P., Sumner, D. Y., Kah, L. C., Stack, K., Gupta, S., Edgar, L., et al. (2014). A Habitable Fluvio-Lacustrine Environment at Yellowknife Bay, Gale Crater, Mars. Science 343, 1242777. doi:10.1126/science.1242777
Gupta, S., Ochiai, E., and Ponnamperuma, C. (1981). Organic Synthesis in the Atmosphere of Titan. Nature 293, 725–727. doi:10.1038/293725a0
Guzmán-Marmolejo, A., Segura, A., and Escobar-Briones, E. (2013). Abiotic Production of Methane in Terrestrial Planets. Astrobiology 13, 550–559. doi:10.1089/ast.2012.0817
Hand, K. P., Phillips, C., Cable, M., Craft, K. L., Hofmann, A., Dooley, J., et al. (2018). The Europa Lander Mission Concept. American Astronomical Society. Tech. Rep.
Hayes, J. M. (2001). Fractionation of Carbon and Hydrogen Isotopes in Biosynthetic Processes. Stable Isotope Geochemi. Woods Hole, MA: De Gruyter Mouton, 43, 225–278. doi:10.1515/9781501508745-006
Hayes, J. M. (2001). Fractionation of the Isotopes of the Isotopes of the Isotopes of Carbon and Hy Carbon and Hy Carbon and Hydrogen in Biosynthetic Pr in Biosynthetic Processes. Mineralogical Society of America.
He, Y., Buch, A., Szopa, C., Williams, A. J., Millan, M., Guzman, M., et al. (2020). The Search for Organic Compounds with Tmah Thermochemolysis: From Earth Analyses to Space Exploration Experiments. Trac Trends Anal. Chem. 127, 115896. doi:10.1016/j.trac.2020.115896
He, Y., Buch, A., Szopa, C., Williams, A. J., Millan, M., Malespin, C. A., et al. (2021). Influence of Calcium Perchlorate on the Search for Organics on mars with Tetramethylammonium Hydroxide Thermochemolysis. Astrobiology 21, 279–297. doi:10.1089/ast.2020.2252
Hecht, M. H., Kounaves, S. P., Quinn, R. C., West, S. J., Young, S. M. M., Ming, D. W., et al. (2009). Detection of Perchlorate and the Soluble Chemistry of Martian Soil at the phoenix lander Site. Science 325, 64–67. doi:10.1126/science.1172466
Hilario, M., Kalousis, A., Pellegrini, C., and Müller, M. (2006). Processing and Classification of Protein Mass Spectra. Mass. Spectrom. Rev. 25, 409–449. doi:10.1002/mas.20072
Hoffman, H., and Hodges, R. (1972). Lunar Orbital Mass Spectrometer experiment. Proc. Third Lunar Sci. Conf. 3, 2205–2216.
Hoffman, J. H., Chaney, R. C., and Hammack, H. (2008). Phoenix Mars Mission-The Thermal Evolved Gas Analyzer. J. Am. Soc. Mass. Spectrom. 19, 1377–1383. doi:10.1016/j.jasms.2008.07.015
Hoffman, J. H., Hodges, R. R., Donahue, T. M., and McElroy, M. B. (1980a). Composition of the Venus Lower Atmosphere from the Pioneer Venus Mass Spectrometer. J. Geophys. Res. 85, 7882–7890. doi:10.1029/JA085iA13p07882
Hoffman, J. H., Oyama, V. I., and von Zahn, U. (1980b). Measurements of the Venus Lower Atmosphere Composition: A Comparison of Results. J. Geophys. Res. 85, 7871–7881. doi:10.1029/ja085ia13p07871
Horowitz, N. H., Hubbard, J. S., and Hobby, G. L. (1972). The Carbon-Assimilation experiment: The Viking Mars Lander. Icarus 16, 147–152. doi:10.1016/0019-1035(72)90142-x
Hörst, S. M. (2017). Titan's Atmosphere and Climate. J. Geophys. Res. Planets. 122, 432–482. doi:10.1002/2016JE005240
Hörst, S. M., Yelle, R. V., Buch, A., Carrasco, N., Cernogora, G., Dutuit, O., et al. (2012). Formation of Amino Acids and Nucleotide Bases in a Titan Atmosphere Simulation experiment. Astrobiology 12, 809–817. doi:10.1089/ast.2011.0623
House, C. H., William Schopf, J., McKeegan, K. D., Coath, C. D., Mark Harrison, T., and Stetter, K. O. (2000). Carbon Isotopic Composition of Individual Precambrian Microfossils. Geology 28, 707–710. doi:10.1130/0091-7613(2000)028<0707:cicoip>2.3.co;2
Hufsky, F., and Böcker, S. (2017). Mining Molecular Structure Databases: Identification of Small Molecules Based on Fragmentation Mass Spectrometry Data. Mass. Spec. Rev. 36, 624–633. doi:10.1002/mas10.1002/mas.21489
Jenner, G. A., Longerich, H. P., Jackson, S. E., and Fryer, B. J. (1990). ICP-MS - A Powerful Tool for High-Precision Trace-Element Analysis in Earth Sciences: Evidence from Analysis of Selected U.S.G.S. Reference Samples. Chem. Geology. 83, 133–148. doi:10.1016/0009-2541(90)90145-W
Jeong, K., Kim, S., and Pevzner, P. A. (2013). UniNovo: A Universal Tool for De Novo Peptide Sequencing. Bioinformatics 29, 1953–1962. doi:10.1093/bioinformatics/btt338
Jia, X., Kivelson, M. G., Khurana, K. K., and Kurth, W. S. (2018). Evidence of a Plume on Europa from Galileo Magnetic and Plasma Wave Signatures. Nat. Astron. 2, 459–464. doi:10.1038/s41550-018-0450-z
Johnson, S. S., Anslyn, E. V., Graham, H. V., Mahaffy, P. R., and Ellington, A. D. (2018). Fingerprinting Non-terran Biosignatures. Astrobiology 18, 915–922. doi:10.1089/ast.2017.1712
Judd, A. G. (2000). “Geological Sources of Methane,” in Atmospheric Methane. Heidelberg, Germany: Springer Berlin Heidelberg, 280–303. doi:10.1007/978-3-662-04145-1_15
Kasprzak, W., Nieman, H., Harpold, D., Richards, J., Manning, H., Patrick, E., et al. (1996). Cassini Orbiter Ion and Neutral Mass Spectrometer Instrument. SPIE 2803, 129–140.
Kempes, C. P., Follows, M. J., Smith, H., Graham, H., House, C. H., and Levin, S. A. (2021). Generalized Stoichiometry and Biogeochemistry for Astrobiological Applications. Bull. Math. Biol. 83, 1–22. doi:10.1007/s11538-021-00877-5
Kempes, C. P., Koehl, M. A. R., and West, G. B. (2019). The Scales that Limit: The Physical Boundaries of Evolution. Front. Ecol. Evol. 7, 1–20. doi:10.3389/fevo.2019.00242
Kempes, C. P., and Krakauer, D. C. (2021). The Multiple Paths to Multiple Life. J. Mol. Evol. 89, 415–426. doi:10.1007/s00239-021-10016-2
Kenig, F., Chou, L., Mckay, C. P., Jackson, W. A., Doran, P. T., Murray, A. E., et al. (2016). Perchlorate and Volatiles of the Brine of Lake Vida (Antarctica): Implication for the In Situ Analysis of Mars Sediments. J. Geophys. Res. Planets. 121, 1190–1203. doi:10.1002/2015JE004964
Kissel, J., Altwegg, K., Clark, B. C., Colangeli, L., Cottin, H., Czempiel, S., et al. (2007). COSIMA - High Resolution Time-Of-Flight Secondary Ion Mass Spectrometer for the Analysis of Cometary Dust Particles Onboard Rosetta. Space Sci. Rev. 128, 823–867. doi:10.1007/s11214-006-9083-0
Kissel, J., and Krueger, F. R. (1987). The Organic Component in Dust from Comet Halley as Measured by the PUMA Mass Spectrometer on Board Vega 1. Nature 326, 755–760. doi:10.1038/326755a0
Kompauer, M., Heiles, S., and Spengler, B. (2017). Atmospheric Pressure MALDI Mass Spectrometry Imaging of Tissues and Cells at 1.4-μm Lateral Resolution. Nat. Methods. 14, 90–96. doi:10.1038/nmeth.4071
Krissansen-Totton, J., Olson, S., and Catling, D. C. (2018). Disequilibrium Biosignatures over Earth History and Implications for Detecting Exoplanet Life. Sci. Adv. 4, eaao5747. doi:10.1126/sciadv.aao5747
LaRowe, D. E., and Regnier, P. (2008). Thermodynamic Potential for the Abiotic Synthesis of Adenine, Cytosine, Guanine, Thymine, Uracil, Ribose, and Deoxyribose in Hydrothermal Systems. Orig Life Evol. Biosph. 38, 383–397. doi:10.1007/s11084-008-9137-2
Le Roy, L., Altwegg, K., Balsiger, H., Berthelier, J. J., Bieler, A., Briois, C., et al. (2015). Inventory of the Volatiles on Comet 67P/Churyumov-Gerasimenko from Rosetta/ROSINA. Astron. Astrophysics. 583, 1–12. doi:10.1051/0004-6361/201526450
Leefmann, T., Heim, C., Kryvenda, A., Siljeström, S., Sjövall, P., and Thiel, V. (2013). Biomarker Imaging of Single Diatom Cells in a Microbial Mat Using Time-Of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). Org. Geochem. 57, 23–33. doi:10.1016/j.orggeochem.2013.01.005
Lehmer, O. R., Catling, D. C., Parenteau, M. N., and Hoehler, T. M. (2018). The Productivity of Oxygenic Photosynthesis Around Cool, M Dwarf Stars. ApJ 859, 171. doi:10.3847/1538-4357/aac104
Levin, G., Kuznetz, L., and Lafleur, A. (2000). Approaches to Resolving the Question of Life on Mars. Proc. SPIE: Instr. Methods Missions Astrobiology 4137, 48–62. doi:10.1117/12.411620
Levin, G. V., and Straat, P. A. (1977). Life on Mars? the Viking Labeled Release experiment. BioSystems 9, 165–174. doi:10.1016/0303-2647(77)90026-0
Li, X., Danell, R. M., Brinckerhoff, W. B., Pinnick, V. T., van Amerom, F., Arevalo, R. D., et al. (2015). Detection of Trace Organics in mars Analog Samples Containing Perchlorate by Laser Desorption/ionization Mass Spectrometry. Astrobiology 15, 104–110. doi:10.1089/ast.2014.1203
Li, X., Danell, R. M., Pinnick, V. T., Grubisic, A., van Amerom, F., Arevalo, R. D., et al. (2017). Mars Organic Molecule Analyzer (MOMA) Laser Desorption/ionization Source Design and Performance Characterization. Int. J. Mass Spectrom. 422, 177–187. doi:10.1016/j.ijms.2017.03.010
Liebal, U. W., Phan, A. N. T., Sudhakar, M., Raman, K., and Blank, L. M. (2020). Machine Learning Applications for Mass Spectrometry-Based Metabolomics. Metabolites 10, 243. doi:10.3390/metabo10060243
Ligterink, N. F. W., Grimaudo, V., Moreno-García, P., Lukmanov, R., Tulej, M., Leya, I., et al. (2020). ORIGIN: a Novel and Compact Laser Desorption - Mass Spectrometry System for Sensitive In Situ Detection of Amino Acids on Extraterrestrial Surfaces. Sci. Rep. 10, 1–10. doi:10.1038/s41598-020-66240-1
Lincowski, A. P., Meadows, V. S., Crisp, D., Akins, A. B., Schwieterman, E. W., Arney, G. N., et al. (2021). Claimed Detection of PH3 in the Clouds of Venus Is Consistent with Mesospheric SO2. ApJL 908, L44. doi:10.3847/2041-8213/abde47
Liu, X.-L., Summons, R. E., and Hinrichs, K.-U. (2012). Extending the Known Range of Glycerol Ether Lipids in the Environment: Structural Assignments Based on Tandem Mass Spectral Fragmentation Patterns. Rapid Commun. Mass. Spectrom. 26, 2295–2302. doi:10.1002/rcm.6355
Lorenz, R. D. (2019). A Bayesian Approach to Biosignature Detection on Ocean Worlds. Nat. Astron. 3, 466–467. doi:10.1038/s41550-019-0810-3
Lukmanov, R. A., Tulej, M., Ligterink, N. F. W., De Koning, C., Riedo, A., Grimaudo, V., et al. (2021). Chemical Identification of Microfossils from the 1.88-Ga Gunflint Chert: Towards Empirical Biosignatures Using Laser Ablation Ionization Mass Spectrometer. J. Chemometrics e3370, 1. doi:10.1002/cem.3370
MacKenzie, S. M., Neveu, M., Davila, A. F., Lunine, J. I., Craft, K. L., Cable, M. L., et al. (2021). The Enceladus Orbilander mission Concept: Balancing Return and Resources in the Search for Life. Planet. Sci. J. 2, 77. doi:10.3847/psj/abe4da
Magnuson, K., Jackowski, S., Rock, C. O., and Cronan, J. E. (1993). Regulation of Fatty Acid Biosynthesis in escherichia Coli. Microbiol. Rev. 57, 522–542. doi:10.1128/mr.57.3.522-542.1993
Mahaffy, P. R., Benna, M., King, T., Harpold, D. N., Arvey, R., Barciniak, M., et al. (2015). The Neutral Gas and Ion Mass Spectrometer on the Mars Atmosphere and Volatile Evolution Mission. Space Sci. Rev. 195, 49–73. doi:10.1007/s11214-014-0091-1
Mahaffy, P. R., Niemann, H. B., Alpert, A., Atreya, S. K., Demick, J., Donahue, T. M., et al. (2000). Noble Gas Abundance and Isotope Ratios in the Atmosphere of Jupiter from the Galileo Probe Mass Spectrometer. J. Geophys. Res. 105, 15061–15071. doi:10.1029/1999JE001224
Mahaffy, P. R., Richard Hodges, R., Benna, M., King, T., Arvey, R., Barciniak, M., et al. (2014). The Neutral Mass Spectrometer on the Lunar Atmosphere and Dust Environment Explorer Mission. Space Sci. Rev. 185, 27–61. doi:10.1007/s11214-014-0043-9
Mahaffy, P. R., Webster, C. R., Cabane, M., Conrad, P. G., Coll, P., Atreya, S. K., et al. (2012). The Sample Analysis at Mars Investigation and Instrument Suite. Space Sci. Rev. 170, 401–478. doi:10.1007/s11214-012-9879-z
Mahaffy, P., Weng, M., Arevalo, R., Benna, M., Summons, R., Brinckerhoff, W., et al. (2021). Agnostic Biosignature Exploration at Europa through Plume Sampling. Bull. AAS 53, 1–7. Available at: Https://baas.aas.org/pub/2021n4i059. doi:10.3847/25c2cfeb.4be98902
Marshall, S. M., Mathis, C., Carrick, E., Keenan, G., Cooper, G. J. T., Graham, H., et al. (2021). Identifying Molecules as Biosignatures with Assembly Theory and Mass Spectrometry. Nat. Commun. 12, 1–9. doi:10.1038/s41467-021-23258-x
Marshall, S. M., Murray, A. R. G., and Cronin, L. (2017). A Probabilistic Framework for Identifying Biosignatures Using Pathway Complexity. Phil. Trans. R. Soc. A. 375, 20160342. doi:10.1098/rsta.2016.0342
McCollom, T. M., Ritter, G., and Simoneit, B. R. T. (1999). Lipid Synthesis under Hydrothermal Conditions by Fischer-tropsch-type Reactions. Origins Life Evol. Biosph. 29, 153–166. doi:10.1023/A:1006592502746
McDermott, J. M., Seewald, J. S., German, C. R., and Sylva, S. P. (2015). Pathways for Abiotic Organic Synthesis at Submarine Hydrothermal fields. Proc. Natl. Acad. Sci. USA. 112, 7668–7672. doi:10.1073/pnas.1506295112
McKay, C. P. (2008). An Approach to Searching for Life on Mars, Europa, and Enceladus. Space Sci. Rev. 135, 49–54. doi:10.1007/s11214-007-9229-8
McKay, D. S., Gibson, E. K., Thomas-Keprta, K. L., Vali, H., Romanek, C. S., Clemett, S. J., et al. (1996). Search for Past Life on mars: Possible Relic Biogenic Activity in Martian Meteorite Alh84001. Science 273, 924–930. doi:10.1126/science.273.5277.924
Meadows, V. S. (2017). Reflections on O2as a Biosignature in Exoplanetary Atmospheres. Astrobiology 17, 1022–1052. doi:10.1089/ast.2016.1578
Ménez, B., Pisapia, C., Andreani, M., Jamme, F., Vanbellingen, Q. P., Brunelle, A., et al. (2018). Abiotic Synthesis of Amino Acids in the Recesses of the Oceanic Lithosphere. Nature 564, 59–63. doi:10.1038/s41586-018-0684-z
Millan, M., Teinturier, C. A., Malespin, J. Y., Bonnet, A., Buch, J. P., and Dworkin, J. L. (2021). Organic molecules revealed in Mars’s Bagnold Dunes by Curiosity’s derivatization experiment. Nat. Astron.. doi:10.1038/s41550-021-01507-9
Miller, S. L., and Urey, H. C. (1959). Organic Compound Synthes on the Primitive Eart: Several Questions about the Origin of Life Have Been Answered, but Much Remains to Be Studied. Science 130, 245–251. doi:10.1126/science.130.3370.245
Ming, D. W., Archer, P. D., Glavin, D. P., Eigenbrode, J. L., Franz, H. B., Sutter, B., et al. (2014). Volatile and Organic Compositions of Sedimentary Rocks in Yellowknife Bay, Gale Crater, Mars. Science 343, 1245267. doi:10.1126/science.1245267
Mißbach, H., Steininger, H., Thiel, V., and Goetz, W. (2019). Investigating the Effect of Perchlorate on Flight-like Gas Chromatography-Mass Spectrometry as Performed by MOMA on Board the ExoMars 2020 Rover. Astrobiology 19, 1339–1352. doi:10.1089/ast.2018.1997
Montaudo, G., Samperi, F., and Montaudo, M. S. (2006). Characterization of Synthetic Polymers by MALDI-MS. Prog. Polym. Sci. 31, 277–357. doi:10.1016/j.progpolymsci.2005.12.001
Morse, A., Mousis, O., Sheridan, S., Morgan, G., Andrews, D., Barber, S., et al. (2015). Low CO/CO2ratios of Comet 67P Measured at the Abydos landing Site by thePtolemymass Spectrometer. A&A 583, A42–A44. doi:10.1051/0004-6361/201526624
Muirhead, B. K., Nicholas, A. K., Umland, J., Sutherland, O., and Vijendran, S. (2020). Mars Sample Return Campaign Concept Status. Acta Astronautica. 176, 131–138. doi:10.1016/j.actaastro.2020.06.026
Musat, N., Halm, H., Winterholler, B., Hoppe, P., Peduzzi, S., Hillion, F., et al. (2008). A Single-Cell View on the Ecophysiology of Anaerobic Phototrophic Bacteria. Pnas 105, 17861–17866. doi:10.1073/pnas.0809329105
Naraoka, H., Shimoyama, A., and Harada, K. (1999). Molecular Distribution of Monocarboxylic Acids in Asuka Carbonaceous Chondrites from Antarctica. Origins Life Evol. Biosph. 29, 187–201. doi:10.1023/A:1006547127028
NAS (2019). An Astrobiology Strategy for the Search for Life in the Universe. Washington, DC: National Academies of Sciences, Engineering, and Medicine. doi:10.17226/25252
Navarro-González, R., Navarro, K. F., Rosa, J. d. l., Iñiguez, E., Molina, P., Miranda, L. D., et al. (2006). The Limitations on Organic Detection in Mars-like Soils by thermal Volatilization-Gas Chromatography-MS and Their Implications for the Viking Results. Proc. Natl. Acad. Sci. 103, 16089–16094. doi:10.1073/pnas.0604210103
Neumann, S., and Böcker, S. (2010). Computational Mass Spectrometry for Metabolomics: Identification of Metabolites and Small Molecules. Anal. Bioanal. Chem. 398, 2779–2788. doi:10.1007/s00216-010-4142-5
Neveu, M., Anbar, A. D., Davila, A. F., Glavin, D. P., MacKenzie, S. M., Phillips-Lander, C. M., et al. (2020). Returning Samples from Enceladus for Life Detection. Front. Astron. Space Sci. 7, 26. doi:10.3389/fspas.2020.00026
Neveu, M., Hays, L. E., Voytek, M. A., New, M. H., and Schulte, M. D. (2018). The Ladder of Life Detection. Astrobiology 18, 1375–1402. doi:10.1089/ast.2017.1773
Niemann, H. B., Atreya, S. K., Demick, J. E., Gautier, D., Haberman, J. A., Harpold, D. N., et al. (2010). Composition of Titan's Lower Atmosphere and Simple Surface Volatiles as Measured by the Cassini-Huygens Probe Gas Chromatograph Mass Spectrometer experiment. J. Geophys. Res. 115, 1–22. doi:10.1029/2010JE003659
Niemann, H. B., Harpold, D. N., Atreya, S. K., Carignan, G. R., Hunten, D. M., and Owen, T. C. (1992). “Galileo Probe Mass Spectrometer Experiment,” in Space Science Reviews. Editor C. T. Russell (Dordrecht: Springer Netherlands), 111–142. doi:10.1007/978-94-011-2512-3_5
Niemann, H. B., Hartle, R. E., Kasprzak, W. T., Spencer, N. W., Hunten, D. M., and Carignan, G. R. (1979). Venus Upper Atmosphere Neutral Composition: Preliminary Results from the Pioneer Venus Orbiter. Science 203, 770–772. doi:10.1126/science.203.4382.770
Nier, A. O. (1960). Small General Purpose Double Focusing Mass Spectrometer. Rev. Scientific Instr. 31, 1127–1132. doi:10.1063/1.1716824
Oehler, D., and Cady, S. (2014). Biogenicity and Syngeneity of Organic Matter in Ancient Sedimentary Rocks: Recent Advances in the Search for Evidence of Past Life. Challenges 5, 260–283. doi:10.3390/challe5020260
Oehler, D. Z., Robert, F., Walter, M. R., Sugitani, K., Meibom, A., Mostefaoui, S., et al. (2010). Diversity in the Archean Biosphere: New Insights from nanoSIMS. Astrobiology 10, 413–424. doi:10.1089/ast.2009.0426
Ojeda, A. S., Phillips, E., and Sherwood Lollar, B. (2020). Multi-element (C, H, Cl, Br) Stable Isotope Fractionation as a Tool to Investigate Transformation Processes for Halogenated Hydrocarbons. Environ. Sci. Process. Impacts. 22, 567–582. doi:10.1039/C9EM00498J
Oyama, V. I., Berdahl, B. J., Carle, G. C., Lehwalt, M. E., and Ginoza, H. S. (1976). The Search for Life on Mars: Viking 1976 Gas Changes as Indicators of Biological Activity. Origins Life Evol. Biosph. 7, 313–333. doi:10.1007/bf00926949
Paizs, B., and Suhai, S. (2005). Fragmentation Pathways of Protonated Peptides. Mass. Spectrom. Rev. 24, 508–548. doi:10.1002/mas.20024
Palmer, P. T., and Limero, T. F. (2001). Mass Spectrometry in the U.S. Space Program: Past, Present, and Future. J. Am. Soc. Mass. Spectrom. 12, 656–675. doi:10.1016/S1044-0305(01)00249-5
Peironcely, J. E., Rojas-Chertó, M., Tas, A., Vreeken, R., Reijmers, T., Coulier, L., et al. (2013). Automated Pipeline for De Novo Metabolite Identification Using Mass-Spectrometry-Based Metabolomics. Anal. Chem. 85, 3576–3583. doi:10.1021/ac303218u
Pernot, P., Carrasco, N., Thissen, R., and Schmitz-Afonso, I. (2010). Tholinomics-Chemical Analysis of Nitrogen-Rich Polymers. Anal. Chem. 82, 1371–1380. doi:10.1021/ac902458q
Peters, K., Walters, C., and Moldowan, J. (2005). The Biomarker Guide: Biomarkers and Isotopes in the Environment and Earth History. Cambridge, MA: Cambridge University Press.
Pizzarello, S., and Cronin, J. R. (2000). Non-racemic Amino Acids in the Murray and Murchison Meteorites. Geochimica et Cosmochimica Acta. 64, 329–338. doi:10.1016/S0016-7037(99)00280-X
Pohorille, A., and Sokolowska, J. (2020). Evaluating Biosignatures for Life Detection. Astrobiology 20, 1236–1250. doi:10.1089/ast.2019.2151
Quick, L. C., Barnouin, O. S., Prockter, L. M., and Patterson, G. W. (2013). Constraints on the Detection of Cryovolcanic Plumes on Europa. Planet. Space Sci. 86, 1–9. doi:10.1016/j.pss.2013.06.028
Rappsilber, J., Moniatte, M., Nielsen, M. L., Podtelejnikov, A. V., and Mann, M. (2003). Experiences and Perspectives of MALDI MS and MS/MS in Proteomic Research. Int. J. Mass Spectrom. 226, 223–237. doi:10.1016/S1387-3806(02)00976-4
Rasche, F., Svatoš, A., Maddula, R. K., Böttcher, C., and Böcker, S. (2011). Correction to Computing Fragmentation Trees from Tandem Mass Spectrometry Data. Anal. Chem. 83, 6911. doi:10.1021/ac201785d
Rashevsky, N. (1955). Life, Information Theory, and Topology. Bull. Math. Biophys. 17, 229–235. doi:10.1007/bf02477860
Raulin, F., and Owen, T. (2003). Organic Chemistry and Exobiology on Titan. The Cassini-Huygens Mission 1. doi:10.1007/978-94-017-3251-2_10
Reeves, E. P., and Fiebig, J. (2020). Abiotic Synthesis of Methane and Organic Compounds in Earth's Lithosphere. Elements 16, 25–31. doi:10.2138/gselements.16.1.25
Reinhardt, M., Goetz, W., and Thiel, V. (2020). Testing Flight-like Pyrolysis Gas Chromatography-Mass Spectrometry as Performed by the Mars Organic Molecule Analyzer Onboard the ExoMars 2020 Rover on Oxia Planum Analog Samples. Astrobiology 20, 415–428. doi:10.1089/ast.2019.2143
Remoortere, A. V., van Zeijl, R. J. M., van den Oever, N., Franck, J., Longuespée, R., Wisztorski, M., et al. (2010). MALDI Imaging and Profiling MS of Higher Mass Proteins from Tissue. J. Am. Soc. Mass. Spectrom. 21, 1922–1929. doi:10.1016/j.jasms.2010.07.011
Riedo, A., De Koning, C., Stevens, A. H., Cockell, C. S., McDonald, A., López, A. C., et al. (2020). The Detection of Elemental Signatures of Microbes in Martian Mudstone Analogs Using High Spatial Resolution Laser Ablation Ionization Mass Spectrometry. Astrobiology 20, 1224–1235. doi:10.1089/ast.2019.2087
Rohner, U., Whitby, J. A., Wurz, P., Whitby, J. A., and Wurz, P. (2003). A Miniature Laser Ablation Time-Of-Flight Mass Spectrometer Forin Situplanetary Exploration. Meas. Sci. Technol. 14, 2159–2164. doi:10.1088/0957-0233/14/12/017
Sagan, C., Thompson, W. R., Carlson, R., Gurnett, D., and Hord, C. (1993). A Search for Life on Earth from the Galileo Spacecraft. Nature 365, 715–721. doi:10.1038/365715a0
Salter, T., Magee, B., Waite, J., and Sephton, M. (2021). Mass Spectrometric Fingerprints of Bacteria and Archaea for Life Detection on Icy Moons. Astrobiology 52 (6).
Scherer, S., Altwegg, K., Balsiger, H., Fischer, J., Jäckel, A., Korth, A., et al. (2006). A Novel Principle for an Ion Mirror Design in Time-Of-Flight Mass Spectrometry. Int. J. Mass Spectrom. 251, 73–81. doi:10.1016/j.ijms.2006.01.025
Schmidt, B. E., Blankenship, D. D., Patterson, G. W., and Schenk, P. M. (2011). Active Formation of 'chaos Terrain' over Shallow Subsurface Water on Europa. Nature 479, 502–505. doi:10.1038/nature10608
Schmidt, B. E. (2020). The Astrobiology of Europa and the Jovian System. Tucson, AZ: The University of Arizona Press. doi:10.2458/azu_uapress/_/9780816540068-ch008
Schmitt-Kopplin, P., Gabelica, Z., Gougeon, R. D., Fekete, A., Kanawati, B., Harir, M., et al. (2010). High Molecular Diversity of Extraterrestrial Organic Matter in Murchison Meteorite Revealed 40 Years after its Fall. Proc. Natl. Acad. Sci. 107, 2763–2768. doi:10.1073/pnas.0912157107
Schouten, S., Hopmans, E. C., and Sinninghe Damsté, J. S. (2013). The Organic Geochemistry of Glycerol Dialkyl Glycerol Tetraether Lipids: A Review. Org. Geochem. 54, 19–61. doi:10.1016/j.orggeochem.2012.09.006
Schouten, S., Van Der Meer, M. T. J., Hopmans, E. C., Rijpstra, W. I. C., Reysenbach, A.-L., Ward, D. M., et al. (2007). Archaeal and Bacterial Glycerol Dialkyl Glycerol Tetraether Lipids in hot springs of Yellowstone National Park. Appl. Environ. Microbiol. 73, 6181–6191. doi:10.1128/AEM.00630-07
Schwartz, A. W. (1994). Origin of Life: The Origin of Macromolecular Chirality. Curr. Biol. 4, 758–760. doi:10.1016/S0960-9822(00)00171-8
Selliez, L., Briois, C., Carrasco, N., Thirkell, L., Thissen, R., Ito, M., et al. (2019). Identification of Organic Molecules with a Laboratory Prototype Based on the Laser Ablation-CosmOrbitrap. Planet. Space Sci. 170, 42–51. doi:10.1016/j.pss.2019.03.003
Sephton, M. A. (2005). Organic Matter in Carbonaceous Meteorites: Past, Present and Future Research. Phil. Trans. R. Soc. A. 363, 2729–2742. doi:10.1098/rsta.2005.1670
Sherwood Lollar, B., Lacrampe-Couloume, G., Slater, G. F., Ward, J., Moser, D. P., Gihring, T. M., et al. (2006). Unravelling Abiogenic and Biogenic Sources of Methane in the Earth's Deep Subsurface. Chem. Geology. 226, 328–339. doi:10.1016/j.chemgeo.2005.09.027
Siljestrã–M, S., Lausmaa, J., Sjã–Vall, P., Broman, C., Thiel, V., and Hode, T. (2010). Analysis of Hopanes and Steranes in Single Oil-Bearing Fluid Inclusions Using Time-Of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). Geobiology 8, 37–44. doi:10.1111/j.1472-4669.2009.00223.x
Simoneit, B. R. T. (2005). A Review of Current Applications of Mass Spectrometry for Biomarker/molecular Tracer Elucidations. Mass. Spectrom. Rev. 24, 719–765. doi:10.1002/mas.20036
Slaveykova, V. I., Guignard, C., Eybe, T., Migeon, H.-N., and Hoffmann, L. (2009). Dynamic NanoSIMS Ion Imaging of Unicellular Freshwater Algae Exposed to Copper. Anal. Bioanal. Chem. 393, 583–589. doi:10.1007/s00216-008-2486-x
Sleiman, D., Garcia, P. S., Lagune, M., Loc’h, J., Haouz, A., Taib, N., et al. (2021). A Third Purine Biosynthetic Pathway Encoded by Aminoadenine-Based Viral DNA Genomes. Science 372, 516–520. doi:10.1126/science.abe6494
Smith, G. G., Williams, K. M., and Wonnacott, D. M. (1978). Factors Affecting the Rate of Racemization of Amino Acids and Their Significance to Geochronology. J. Org. Chem. 43, 1–5. doi:10.1021/jo00395a001
Snellen, I. A. G., Guzman-Ramirez, L., Hogerheijde, M. R., Hygate, A. P. S., and van der Tak, F. F. S. (2020). Re-analysis of the 267 GHz ALMA Observations of Venus. A&A 644, L2. doi:10.1051/0004-6361/202039717
Sokol, E., Noll, R. J., Cooks, R. G., Beegle, L. W., Kim, H. I., and Kanik, I. (2011). Miniature Mass Spectrometer Equipped with Electrospray and Desorption Electrospray Ionization for Direct Analysis of Organics from Solids and Solutions. Int. J. Mass Spectrom. 306, 187–195. doi:10.1016/j.ijms.2010.10.019
Steele, A., Benning, L. G., Wirth, R., Siljeström, S., Fries, M. D., Hauri, E., et al. (2018). Organic Synthesis on Mars by Electrochemical Reduction of CO2. Sci. Adv. 4, eaat5118. doi:10.1126/sciadv.aat5118
Steele, A., Fries, M. D., Amundsen, H. E. F., Mysen, B. O., Fogel, M. L., Schweizer, M., et al. (2007). Comprehensive Imaging and Raman Spectroscopy of Carbonate Globules from Martian Meteorite ALH 84001 and a Terrestrial Analogue from Svalbard. Meteoritics Planet. Sci. 42, 1549–1566. doi:10.1111/j.1945-5100.2007.tb00590.x
Steele, A., McCubbin, F. M., and Fries, M. D. (2016). The Provenance, Formation, and Implications of Reduced Carbon Phases in Martian Meteorites. Meteorit Planet. Sci. 51, 2203–2225. doi:10.1111/maps.12670
Steinberg, S., Grinberg, M., Beitelman, M., Peixoto, J., Orevi, T., and Kashtan, N. (2021). Two-way Microscale Interactions between Immigrant Bacteria and Plant Leaf Microbiota as Revealed by Live Imaging. Isme J. 15, 409–420. doi:10.1038/s41396-020-00767-z
Stephan, T., Jessberger, E. K., Heiss, C. H., and Rost, D. (2003). Tof-sims Analysis of Polycyclic Aromatic Hydrocarbons in Allan hills 84001. Meteoritics Planet. Sci. 38, 109–116. doi:10.1111/j.1945-5100.2003.tb01049.x
Stolper, D. A., Sessions, A. L., Ferreira, A. A., Santos Neto, E. V., Schimmelmann, A., Shusta, S. S., et al. (2014). Combined 13C-D and D-D Clumping in Methane: Methods and Preliminary Results. Geochimica et Cosmochimica Acta. 126, 169–191. doi:10.1016/j.gca.2013.10.045
Summons, R. E., Albrecht, P., McDonald, G., and Moldowan, J. M. (2008). Molecular Biosignatures. Space Sci. Rev. 135, 133–159. doi:10.1007/s11214-007-9256-5
Sutter, B., Boynton, W. V., Ming, D. W., Niles, P. B., Morris, R. V., Golden, D. C., et al. (2012). The Detection of Carbonate in the Martian Soil at the Phoenix Landing Site: A Laboratory Investigation and Comparison with the Thermal and Evolved Gas Analyzer (TEGA) Data. Icarus 218, 290–296. doi:10.1016/j.icarus.2011.12.002
Tenailleau, E. J., Lancelin, P., Robins, R. J., and Akoka, S. (2004). Authentication of the Origin of Vanillin Using Quantitative Natural Abundance 13C NMR. J. Agric. Food Chem. 52, 7782–7787. doi:10.1021/jf048847s
Theiling, B., Brinckerhoff, W., Castillo-Rogez, J., Chou, L., Da Poian, V., Graham, H., et al. (2021). Non-robotic Science Autonomy Development. Bull. Am. Astronomical Soc. 53, 048. doi:10.3847/25c2cfeb.ee4e6b64
Thomas-Keprta, K. L., Clemett, S. J., Bazylinski, D. A., Kirschvink, J. L., McKay, D. S., Wentworth, S. J., et al. (2002). Magnetofossils from Ancient mars: a Robust Biosignature in the Martian Meteorite Alh84001. Appl. Environ. Microbiol. 68, 3663–3672. doi:10.1128/aem.68.8.3663-3672.2002
Todd, J. F. J., Barber, S. J., Wright, I. P., Morgan, G. H., Morse, A. D., Sheridan, S., et al. (2007). Ion Trap Mass Spectrometry on a Comet Nucleus: the Ptolemy Instrument and the Rosetta Space mission. J. Mass. Spectrom. 42, 1–10. doi:10.1002/jms.1147
Trainer, M. G., Wong, M. H., McConnochie, T. H., Franz, H. B., Atreya, S. K., Conrad, P. G., et al. (2019). Seasonal Variations in Atmospheric Composition as Measured in Gale Crater, Mars. J. Geophys. Res. Planets. 124, 3000–3024. doi:10.1029/2019JE006175
Tsou, P., Brownlee, D. E., Anderson, J. D., Bhaskaran, S., Cheuvront, A. R., Clark, B. C., et al. (2004). Stardust Encounters Comet 81P/Wild 2. J. Geophys. Res. 109, 1–8. doi:10.1029/2004JE002317
Tulej, M., Neubeck, A., Ivarsson, M., Riedo, A., Neuland, M. B., Meyer, S., et al. (2015). Chemical Composition of Micrometer-Sized Filaments in an Aragonite Host by a Miniature Laser Ablation/Ionization Mass Spectrometer. Astrobiology 15, 669–682. doi:10.1089/ast.2015.1304
Tverdislov, V. A., and Malyshko, E. V. (2020). Chiral Dualism as an Instrument of Hierarchical Structure Formation in Molecular Biology. Symmetry 12, 1–9. doi:10.3390/sym12040587
Vago, J. L., Westall, F., Pasteur Instrument Teams, Landing S, A. J., Coates, A. J., Jaumann, R., Korablev, O., et al. (2017). Habitability on Early Mars and the Search for Biosignatures with the ExoMars Rover. Astrobiology 17, 471–510. doi:10.1089/ast.2016.1533
Volkman, J. (2006). Marine Organic Matter: Biomarkers, Isotopes and DNA. Heidelberg, Germany: Springer-Verlag. doi:10.1007/b11682Marine Organic Matter: Biomarkers, Isotopes and DNA
Waite, J. H., Brockwell, T., Glein, C., Perry, R., Bolton, S., and Mcgrath, M. (2019). MASPEX-europa Aboard Clipper : A Mass Spectrometer for Investigating the Habitability of Europa 13, 1–2. ( Geneva, Switzerland: EPSC-DPS Joint Meeting 2019).
Waite, J. H., Lewis, W. S., Kasprzak, W. T., Anicich, V. G., Block, B. P., Cravens, T. E., et al. (2004). The Cassini Ion and Neutral Mass Spectrometer (INMS) Investigation. Space Sci. Rev. 114, 113–231. doi:10.1007/s11214-004-1408-2
Waite, J. H., Young, D. T., Cravens, T. E., Coates, A. J., Crary, F. J., Magee, B., et al. (2007). The Process of Tholin Formation in Titan's Upper Atmosphere. Science 316, 870–875. doi:10.1126/science.1139727
Wallace, W. E. (2021). “Mass Spectra,” in NIST Chemistry WebBook. Editors P. Linstrom, and W. Mallard (Gaithersburg, MD 20899: National Institute of Standards and Technology).
Waller, S. E., Belousov, A., Kidd, R. D., Nikolić, D., Madzunkov, S. M., Wiley, J. S., et al. (2019). Chemical Ionization Mass Spectrometry: Applications for the In Situ Measurement of Nonvolatile Organics at Ocean Worlds. Astrobiology 19, 1196–1210. doi:10.1089/ast.2018.1961
Warr, O., Young, E. D., Giunta, T., Kohl, I. E., Ash, J. L., and Sherwood Lollar, B. (2021). High-resolution, Long-Term Isotopic and Isotopologue Variation Identifies the Sources and Sinks of Methane in a Deep Subsurface Carbon Cycle. Geochimica et Cosmochimica Acta. 294, 315–334. doi:10.1016/j.gca.2020.12.002
Webster, C. R., Mahaffy, P. R., Pla-Garcia, J., Rafkin, S. C. R., Moores, J. E., Atreya, S. K., et al. (2021). Day-night Differences in Mars Methane Suggest Nighttime Containment at Gale Crater. Astron. Astrophysics 650, 1–14. doi:10.1051/0004-6361/202040030
Wiener, H. (1947). Structural Determination of Paraffin Boiling Points. J. Am. Chem. Soc. 69, 17–20. doi:10.1021/ja01193a005
Willhite, L., Ni, Z., Arevalo, R., Bardyn, A., Gundersen, C., Minasola, N., et al. (2021). “CORALS: A Laser Desorption/Ablation Orbitrap Mass Spectrometer for In Situ Exploration of Europa,” in IEEE Aerospace Conference, Big Sky, MT, March 6–13, 2021, 1–13. doi:10.1109/aero50100.2021.9438221
Williams, A. J., Eigenbrode, J., Floyd, M., Wilhelm, M. B., O'Reilly, S., Johnson, S. S., et al. (2019). Recovery of Fatty Acids from Mineralogic Mars Analogs by TMAH Thermochemolysis for the Sample Analysis at Mars Wet Chemistry Experiment on the Curiosity Rover. Astrobiology 19, 522–546. doi:10.1089/ast.2018.1819
Williams, R. J. P., and Fraústo da Silva, J. J. R. (2000). The Distribution of Elements in Cells. Coord. Chem. Rev. 200-202, 247–348. doi:10.1016/s0010-8545(00)00324-6
Willis, P. A., Creamer, J. S., and Mora, M. F. (2015). Implementation of Microchip Electrophoresis Instrumentation for Future Spaceflight Missions. Anal. Bioanal. Chem. 407, 6939–6963. doi:10.1007/s00216-015-8903-z
Wolf, S., Schmidt, S., Müller-Hannemann, M., and Neumann, S. (2010). In Silico fragmentation for Computer Assisted Identification of Metabolite Mass Spectra. BMC Bioinformatics 11, 1–12. doi:10.1186/1471-2105-11-148
Wright, I. P., Sheridan, S., Barber, S. J., Morgan, G. H., Andrews, D. J., and Morse, A. D. (2015). CHO-bearing Organic Compounds at the Surface of 67P/Churyumov-Gerasimenko Revealed by Ptolemy. Science 349, aab0673. doi:10.1126/science.aab0673
Yin, Y., Messier, J., and Hopwood, J. A. (1999). Miniaturization of Inductively Coupled Plasma Sources. IEEE Trans. Plasma Sci. 27, 1516–1524. doi:10.1109/27.799834
Young, P. A., Desch, S. J., Anbar, A. D., Barnes, R., Hinkel, N. R., Kopparapu, R., et al. (2014). Astrobiological Stoichiometry. Astrobiology 14, 603–626. doi:10.1089/ast.2014.1143
Yuen, G. U., and Kvenvolden, K. A. (1973). Monocarboxylic Acids in Murray and Murchison Carbonaceous Meteorites. Nature 246, 301–303. doi:10.1038/246301a0
Zamuruyev, K., Ferreira Santos, M. S., Mora, M. F., Kurfman, E. A., Noell, A. C., and Willis, P. A. (2021). Automated Capillary Electrophoresis System Compatible with Multiple Detectors for Potential In Situ Spaceflight Missions. Anal. Chem. 93, 9647–9655. doi:10.1021/acs.analchem.1c02119
Keywords: mass spectrometry, life detection, agnostic biosignatures, planetary exploration, astrobiology
Citation: Chou L, Mahaffy P, Trainer M, Eigenbrode J, Arevalo R, Brinckerhoff W, Getty S, Grefenstette N, Da Poian V, Fricke GM, Kempes CP, Marlow J, Sherwood Lollar B, Graham H and Johnson SS (2021) Planetary Mass Spectrometry for Agnostic Life Detection in the Solar System. Front. Astron. Space Sci. 8:755100. doi: 10.3389/fspas.2021.755100
Received: 07 August 2021; Accepted: 17 September 2021;
Published: 07 October 2021.
Edited by:
Andreas Riedo, University of Bern, SwitzerlandReviewed by:
Jack Waite, Southwest Research Institute (SwRI), United StatesCopyright © 2021 Chou, Mahaffy, Trainer, Eigenbrode, Arevalo, Brinckerhoff, Getty, Grefenstette, Da Poian, Fricke, Kempes, Marlow, Sherwood Lollar, Graham and Johnson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luoth Chou, bHVvdGguY2hvdUBuYXNhLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.