- 1The Shmunis School of Biomedicine and Cancer Research, Tel Aviv University, Tel Aviv-Yafo, Israel
- 2Desert Mars Analog Ramon Station, Mitzpe Ramon, Israel
- 3Faculty of Biology, Technion-Israel Institute of Technology, Haifa, Israel
- 4Eco Encounter Studies Institute, Hod Hasharon, Israel
- 5Department of Earth and Planetary Sciences, Weizmann Institute of Science, Rehovot, Israel
- 6Dead Sea and Arava Science Center, Masada, Israel
As human exploration missions to Mars are on the horizon, microbial cross-contamination remains a key issue to address. These issues can be approached today using advances in molecular metagenomics methods, which include rapid and sensitive sequencing platforms for characterizing microbial populations. Combined with analog missions, these methods provide powerful tools for assessing the challenges associated with planetary exploration. Here, we designed a protocol to monitor forward and backward contamination events and progression in an 11-days Mars analog mission in the Ramon crater in Israel. Forward contamination soil samples were collected daily from three sites–two sites in close proximity to the habitat and one isolated site. Backward contamination was determined in samples from nitrile gloves of six analog astronauts before and after extravehicular activities Temperature, relative humidity and soil composition data were also collected for all sites. Environmental DNA samples were extracted in the main habitat and 16S (bacterial) and 18S (eukaryotic, fungal) rRNA gene amplicons were sequenced and analyzed to study microbial population diversity and composition. Shannon Diversity index analysis and Principal Coordinates analysis (PCoA) of rRNA genes indicated that differences in the diversity and population composition were significant in sites closer to the habitat when compared to a reference site. These samples also demonstrated the introduction of human-associated taxa to the environment. Backward contamination consisted of bacterial taxa found on gloves upon return from EVA and also detected in soil, altogether 44 genera, indicating backward contamination events. To our knowledge, this is the first protocol to utilize advanced molecular technologies to investigate forward and backward contamination in a Mars analog mission.
Introduction
Even before the Viking landers successfully landed on Mars in 1976, NASA’s principles of planetary-protection policies were conceived and embedded in mission design. The Committee on Space Research (COSPAR, est. 1958) provides recommendations which are then taken by space agencies to create specific mission requirements. COSPAR policies have undergone many iterations, until reaching their current and present configuration (Fisk et al., 2020; NASA, 2021). Prevention of forward and backward contamination is the main goal of planetary-protection policies, to eliminate and reduce the potential of cross-transferring life forms from one planet to another. Such contamination is possible either by carrying terrestrial microorganisms via a spacecraft (forward contamination) or by bringing extraterrestrial microorganisms from other planets or celestial bodies back to Earth (backward contamination), via various mechanisms such as sample return missions (Cockell 2005; Pratt and Smith, 2020). Protection against contamination has significant ethical, scientific and safety related aspects (Rummel, 2001; Schwartz, 2019). Forward and backward contamination is becoming a pronounced risk due to an increasing number of planned spaceflight missions, as highlighted by the National Research Council 2002 Safe on Mars report (National Research Council, 2002), and the 2015 COSPAR workshop (Race et al., 2015).
A recent example of such risk is the case of the Israeli SpaceIL Beresheet lunar lander. Beresheet’s hard landing on the Moon in April 2020, while carrying a small payload of tardigrades, raised a vigorous global debate on both ethical and legal implications of a possible forward contamination event (Caplin, 2019; Shahar and Greenbaum, 2020). Although the question whether life arose on Mars is still under investigation (Cabrol, 2018), the potential of Mars-sourced biological cross-contamination via robotic, and in the future, human missions to Mars, stresses the need for a better understanding of the forward and backward contamination potential of such missions (Goh and Kazeminejad, 2004). Several in-depth interdisciplinary analyses of contamination potential were previously done in Mars analog missions, such as the AustroMars simulation, performed in the Mars Desert Research Station (MDRS), located in the Moab desert in Utah, United States (Grömer et al., 2007). An additional approach to study forward contamination was performed under simulated Martian conditions such as desiccation, freezing, Martian-like atmosphere, pressure and UV radiation climate, either in a Mars Simulation Chamber (MSC) (Schuerger et al., 2003) or by using wind tunnels (Van Heereveld et al., 2017) to mimic eolian Martian transport conditions. Most of the current and past studies focus on a single organism, such as Bacillus subtilis (Schuerger et al., 2003; Nicholson et al., 2012), or Staphylococcus xylosus (Van Heereveld et al., 2017), Desulfotalea psychrophila and other sulfate-reducing bacteria (Silver, 2018).
Recent advances in high-throughput molecular biology methods, such as amplicon sequencing, have significantly expanded humanity knowledge on the evolutionary relations and taxonomic structure of life forms and life niches around the world (Liu et al., 2007; Huse et al., 2008), and have enabled deeper understanding of such communities. Characterization of bacterial communities by DNA extraction has already been successfully performed in extreme and pristine environments, such as acidic hypersaline lakes in Australia (Mormile, 2009), the Antarctic Dry Valleys (Cary et al., 2010), the hyper arid Atacama Desert (Piubeli et al., 2015) and the high altitude Andean lakes (Ordoñez et al., 2009). This technology can be efficiently adapted to characterize low-biomass environments and to study delicate shifts in microbial and fungal population dynamics.
Extreme analog environments on earth possess geological or environmental characteristics that are similar to those that exist on an extra-terrestrial planetary body. Therefore, they can serve as a tool to study specific aspects of a space mission to such an extra-terrestrial body (Martins et al., 2017).
In this study, we aimed to investigate the backward and forward contamination potential in a Mars analog. We chose the Ramon crater for an analog mission due to its Mars-like geomorphology and geology, isolation and hyper arid climate conditions (Rubinstein et al., 2019). The Ramon crater is the largest of the three erosional valleys formed along anticlinal axes in the Negev desert in Israel. The crater is 40 km long and 12 km wide, with an area of 241 km2 (Notesco et al., 2016). The climate of the area is arid to hyper arid and the mean multiannual rainfall is 56 mm in the central part of the crater (Schmidt and Karnieli, 2001). The Ramon crater is well-studied and rich in its geological history, with some unique features such as the largest anticline in the region, exposure of layers from the Triassic period, volcanoes, dikes, sills and basalt flows created from extensive Lower Cretaceous volcanic activity (Schmidt and Karnieli, 2001). It contains marine fossils from the Triassic, Lower Cretaceous, and Upper Cretaceous periods and has additional unique features which have promoted its inclusion in the Negev Craterland Geopark initiative (Finzi et al., 2019).
Our working assumption was as follows: We hypothesized that during the analog mission, soil population composition will shift between sites and between sampling points, and that differences in its diversity and composition will be significant in the sites which are closer to the habitat, compared to the isolated site. We assumed that it will take several days until a forward or backward contamination event will occur and the microbial composition will shift, in all sites. If forward contamination occurs, population diversity and composition will be affected by the artificial introduction of new taxa, potentially human-associated species. New species will be introduced to the habitat, if they remain on the astronaut gloves upon returning to the mock pressure chamber for spacesuit doffing.
We utilized, for the first time to our knowledge, advanced molecular microbiology tools to better understand cross contamination potential. We offer a field demonstration of this methodology, in the longest analog mission performed in Israel to date.
Materials and Methods
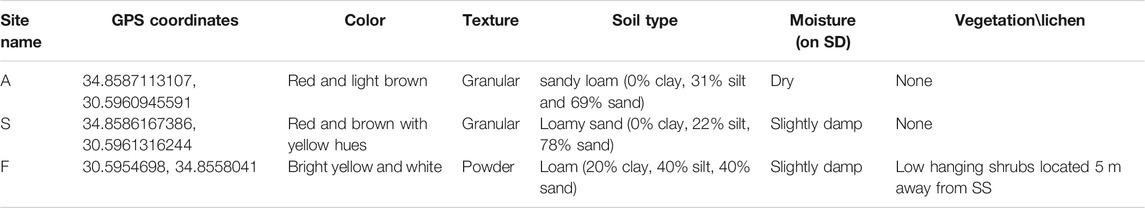
Site Selection and Soil Sampling
D-MARS03 analog mission took place between the 3rd and 14th of March 2019 (Rubinstein et al., 2019) and was situated in a 500 m radius area within Ramon crater, at elevation of 520 m above sea level. During the 11-days mission, six isolated analog astronauts (“Ramonauts,” “AAs”) performed daily EVAs, fully suited with a simulated space suit (∼13 kg) and a personal life support system. During EVAs, the AAs collected soil samples for forward analysis and soil characterization. For the initial soil analyses, samples (three samples per site ∼0.5 kg each) were transferred to sterile zip lock bags from three different sites: “A”—∼0.5 m from the habitat’s airlock entrance, “F”—an undisturbed, isolated site, approximately 280 m west of site A, and site “S”—located at the back of the habitat, ∼0.3 m from the sanitary disposal tank, 10.7 m north of site A (see Figure 1). The GPS coordinates for each site are detailed in Table 1. Plastic jars (non-sterile, 450 ml volume) were used for soil type analysis. Jars were filled with soil (∼250 g), and DDW was added to each jar (approx. 350 ml). Jars were shaken vigorously and left to stabilize overnight, at room temperature. Soil texture and type were diagnosed using the available paradigm and texture calculator from the United States Agriculture Department (USDA), Natural Resources Conservation Service Soils (NRCS) calculator (Natural Resource Conservation Service, 1999). For forward contamination analysis, three soil samples were collected at each site from the top soil layer down to a 3 cm depth, with approximately 10 cm distance between individual samples. The daily sampling time was at 08:00–09:00 AM.

FIGURE 1. An orthophoto and elevation model of the habitat and the sampling sites. Sites are highlighted with white circles. Site A is denoted “A,” site F is denoted “F” and site S is denoted “S.” The white arrow points toward the habitat. Small inset shows the general location of the experiment in Israel, Ramon Crater.
Nitrile Gloves Sampling
For backward contamination analysis, XXL nitrile gloves (SemperGuard® nitrile powder-free disposable gloves, Sigma-Aldrich) were placed over the AAs’ suit gloves, in the airlock before exiting to an EVA. In brief, a 4 × 4 cm2 section of the palm of the hand, and then five fingers, were gently swabbed with a sterile cotton swab, which was then stored in RT, inside a sealed tube. A second sampling took place immediately upon the AA’s return to the habitat from EVA, in the simulated airlock area. Then, both samples were extracted as detailed in Sample Processing and DNA Sequencing. Control samples were obtained by either using a new pair of disposable nitrile gloves, or by extracting DNA from a sterile swab.
Sample Processing and DNA Sequencing
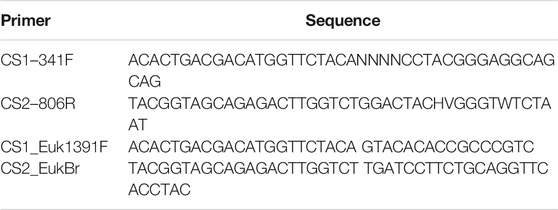
In total, 71 soil samples were collected, of which 40 were further used for DNA extraction. All sample manipulations and DNA extractions were carried out in a laminar flow hood (BSC-8, MRC Ltd.). Genomic DNA was extracted from swabs or soil samples using DNeasy PowerSoil Pro Kit (QIAGEN), as recommended by the manufacturer. Controls for DNA extraction reagents and procedure were either sterile swabs or sterile water to verify the integrity of the extraction in the given conditions. Long-term storage of the samples prior to DNA extraction was avoided, and DNA was immediately isolated from the soil on the same sampling day. No positive control was done for DNA extraction. Extracted DNA samples were stored at −20°C until further processing. Directly after the mission, PCR amplifications of 16S and 18S rRNA genes were carried out with universal primers containing 5′-end common sequences. Primers used in this study are detailed in Table 2. A total of 28–31 PCR cycles (95°C for 15°s, 53°C for 15°s, and 72°C for 15°s) were conducted using the PCR master mix KAPA2G Fast (Kapa Biosystems), and successful amplification was verified by agarose gel electrophoresis. A control reaction without a template but containing all other reagents was included in each PCR mix. DNA extraction from a human fecal sample was used as a positive control for PCR reactions.
Data Analysis
Paired-end deep sequencing of the PCR products was performed on an Illumina MiSeq platform at the University of Illinois at Chicago Sequencing Core (UICSQC). Sequencing depth ranged from 5,797 to 145,126 sequences per sample for 16S, and 870 to 95,418 sequences per sample for 18S; Raw data processing was done by the University of Illinois at Chicago Core for Research Informatics (UICCRI). Forward and reverse reads were merged using PEAR (Zhang et al., 2014). Quality trimming based on quality threshold and length parameters was done with p = 0.01 and min length = 100. Adaptor and ambiguous nucleotide trimming was done prior to further applications. Chimeric sequences were identified using the USEARCH (Edgar 2010) algorithm as compared with a reference database (silva_132_16S.97 and silva_132_18S.97) (Oliver et al., 2017). Sequences were de-replicated and clustered at a threshold of 0.97; sequences appearing fewer than 10 times were excluded. Taxonomic assignments were identified using the USEARCH algorithm against the silva_132_16S.97 and silva_132_18S.97 reference databases. A table of relative abundance data and associated taxonomic annotation was created after read counts were normalized as a fraction of total sequence counts in each sample. Raw sequences were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/), and can be accessed under BioProject accession number PRJNA662496 (release date 31-10-2020).
Statistical Analyses
R vegan (Oksanen et al., 2013) package was used to calculate Shannon diversity index and ANOSIM probability tests. PCoAs were constructed using the base R function “cmdscale” and plotted with ggplot2 package (Wickham 2016). Data rearrangement was done using the Reshape package (Wickham, 2007). Kruskal-Wallis test was conducted using base R functions. The contribution of different variables to the microbiome composition was assayed by PERMANOVA, using the function adonis (vegan) with default parameters and setting temperature, relative humidity and sampling site as independent variables.
Temperature and Relative Humidity Monitoring
In order to monitor the environmental temperature and relative humidity, three HOBO data loggers (HOBO® MX2300 Series Data Logger, HOBO® MX2301A Data Logger and HOBOware 3.7.18 by Onset Computer Corporation, Bourne, MA, United States) were placed, one in each site, 3 cm depth, throughout the mission. The loggers were shielded from direct sunlight by a white plastic cover and retrieved at the last sampling day. The data loggers monitored the temperature and relative humidity at all sites continuously.
Results
Soil Analysis and Environmental Data Collection
Characterization and analysis of soil composition were performed as described above and the results are detailed in Table 1. An orthophoto and elevation model (DEM) of the mission area and sampling sites can be viewed in Figure 1. Soil composition was found to be sandy loam in site A, loamy sand in site S and loam in site F. Sites A and S had no vegetation or lichen present, while site F contained low hanging shrubs located 5 m away from the sampling point. Soil type results correspond to previously documented layers of Mishor and Ardon formations, Lower Jurassic, which include mainly sandstone, limestone, siltstone, clay and gypsum (Sneh et al., 1998; Notesco et al., 2015). Environmental data were collected using HOBO data loggers, a well-established tool for environmental temperature and moisture monitoring (Da Cunha, 2015) (results are summarized in Table 3). Data collected throughout the mission indicated that the sites differ from each other in terms of average temperature, day and night temperature fluctuations, and relative humidity. While the maximal temperature was equal across all sites, both minimal and mean temperatures were higher in site A (Table 3). In terms of relative humidity, site A was drier than sites F and S (52.89 vs. 66.95% and 67.43%, respectively). This observation is expected as site A, located at the entrance of the habitat, was exposed to direct sunlight most of the day, unlike site S, which was located at the backend side of the habitat, and shaded half of the day. Since it was previously shown that microbial activity and biomass are affected by temperature and moisture in arid environments (Vishnevetsky and Steinberger, 1997), we first examined the correlation of temperature and humidity between all sites. While environmental conditions differed slightly between sampling sites, the changes of both temperature and RH across time were positively intercorrelated between all three sites (Pearsons correlation coefficient 0.8 to 0.87 when correlating RH across sites; 0.61 to 0.87 when correlating temperature). Permutational multivariate analysis of variance (PERMANOVA) indicated that the only 12% of the microbial variance can be explained by RH (p = 0.002), and only 9% by temperature (p = 0.001), while 37% of the variance is explained by the site (p = 0.001).
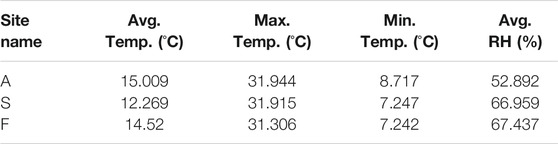
TABLE 3. Summary of environmental temperatures and relative humidity in the different sites throughout the mission.
Population Diversity Comparison Among Sites Throughout the Mission
To detect forward contamination events, temporal changes in microbial communities were monitored using 16S and 18S rRNA genes analysis.
16S rRNA sequences were clustered at identity threshold of 97%, yielding a total of 1,485 distinct genera in soil samples (Supplementary Table S1). 419 genera were found in all sites (but not in all samples).197 genera were unique to site F, while 387 and 78 were present in sites A and S only, respectively. A distance matrix, based on pairwise distances between a pair of samples from each site, was calculated and visualized using Principal Coordinates Analysis (PCoA). Comparison of the PCR control and kit control to the soil samples from all sites showed strong separation (ANOSIM R-value = 0.5 for S site, R-value = 0.85 for A site and R-value = 1 for F site, Supplementary Figures S1–S3) between the controls and the samples.
A strong separation (analysis of similarity [ANOSIM] R-value = 0.83, p-value = 0.001) of soil microbial populations composition between sites was evident (Figure 2A). Notably, the population composition within sites near the habitat exhibited temporal fluctuations; this is reflected in the highly dispersed A and S clusters in Figure 2A. The mean pairwise distances for samples in A and S sites was significantly lower than for the F site (means of 0.52 and 0.72 vs. 0.25 receptively, Kruskal-Wallis p = 1.19e−10). Shannon diversity index was calculated based on genera (Figure 2B) and significantly differs among sites F, S, and A (p = 0.004, Kruskal-Wallis test).
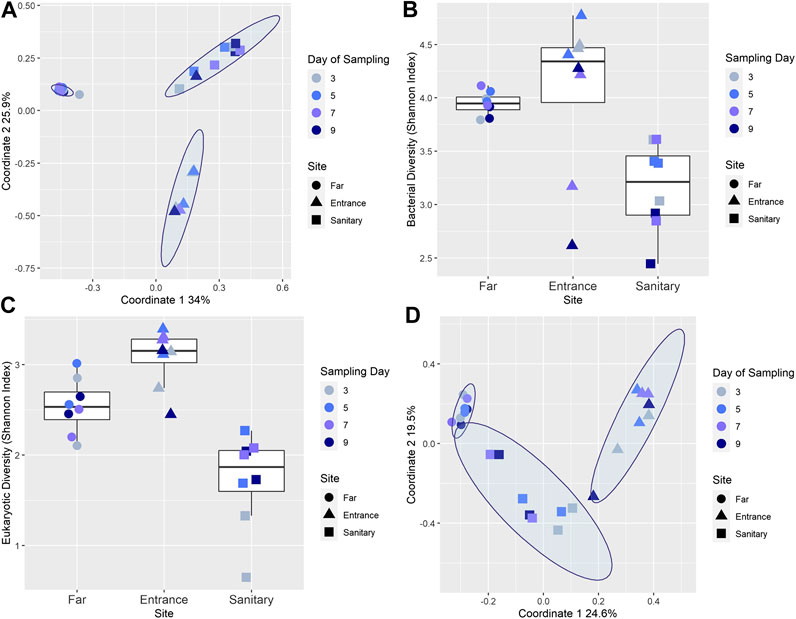
FIGURE 2. (A) Similarity in microbial composition across different sites and days. PCoA of all soil samples throughout the mission, visualizing the spatial relationships between samples. A strong separation is observed between the three sites, with ANOSIM statistic R = 0.8307, Significance: 0.001. (B) Microbial Shannon diversity index of soil samples. Colors denote the sampling day, and shape denotes the origin site. (C) Similarity in eukaryotic composition across different sites and days. PCoA of all soil samples throughout the mission, visualizing the spatial relationships between samples. A separation is observed between the three sites, with ANOSIM statistic R = 0.6, Significance: 0.001. (D) Eukaryotic Shannon diversity index of soil samples. Colors denote the sampling day, and shape denotes the origin site.
While the Shannon diversity index for bacterial populations (16S) of site F is relatively high and stable across all time points (min = 3.79, max = 4.11, and mean = 3.94), the Shannon diversity index for sites S and especially A fluctuated considerably (min = 2.61, max = 4.772, and mean = 4.05 for site A; min = 2.44, max = 3.61, and mean = 3.15 for site S), with particularly low diversity observed for some samples taken on days 7 and 9. Similar trends were observed in the 18S rRNA diversity analysis; Clustering of sequences at an identity level cutoff of 97% detected a total of 664 unique genera in the soil samples (Supplementary Table S2). Shannon diversity index was calculated based on genera (Figure 2C) with samples from the third days presenting relative lower diversity in site S, and samples from the third and ninth day for site A. PCoA (Figure 2D) indicated a separation (ANOSIM R-value = 0.6, p-value = 0.001), similarly to the results acquired with 16 S sequences.
Population Dynamics Throughout the Mission Suggest Forward Contamination
To better examine the population composition of each site and each sample, dominant taxa across sites were compared. Bar plots showing detailed taxonomic composition across each site, as a factor of time, are presented in Figures 3A–C. Rare taxa that had a relative abundance (RA) of less than 1% were eliminated from the bar plots to simplify the visualization.
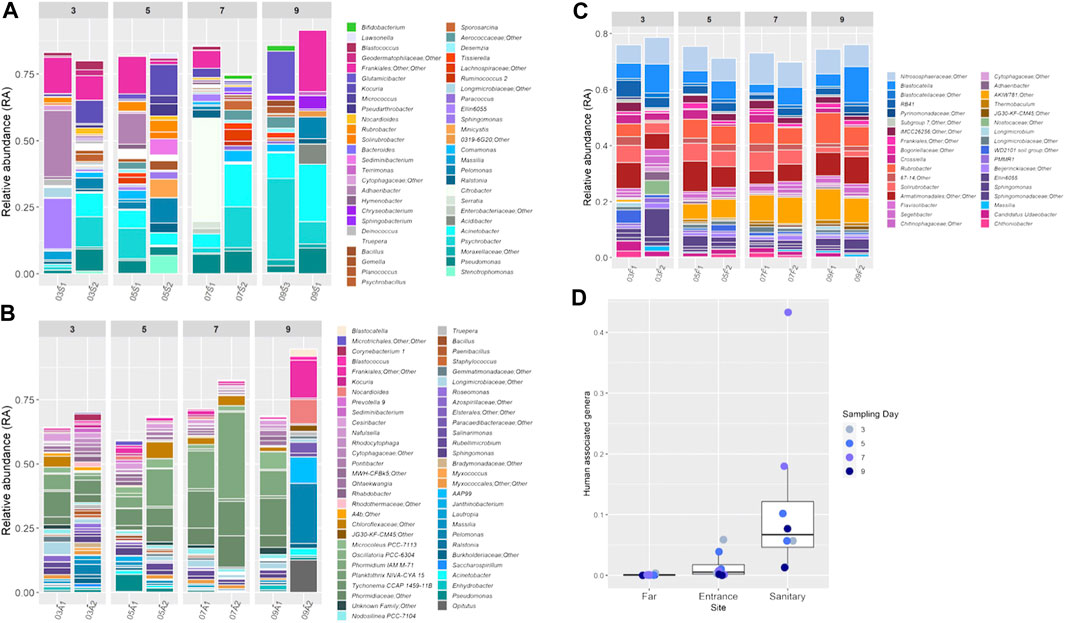
FIGURE 3. (A–C) Relative abundance of bacterial genera in the soil samples during the mission. Each color on the graph represents a distinct family level OTU. Only taxa that constituted >1% of an individual sample are presented, unless stated otherwise. Bar plots were generated using R software. (D) RA of human associated genera in different sites during the mission. Colors denote the sampling day.
Most genera were present at RA of less than 5% (Supplementary Figure S4).
A certain degree of variation between paired samples (taken from the same location on the same day) is apparent when examining the bar graphs (Figures 3A–C). To further investigate and quantify this variation, we extracted from the Bray-Curtis distance matrix all pairwise distances for paired samples and compared those to all other pairwise distances (random). Overall, smaller pairwise distances are observed for paired samples, with samples taken from the far site having the lowest within-pair variation (p Kruskal-Wallis = 0.0002; p Permutations test = 0.15).
In site S (sanitary), which was the least diverse, Moraxellaceae was the dominant bacterial family with the genus Psychrobacter most prevalent (mean RA: 17.8 ± 9%). Actinobacter was the second most dominant genus with average RA of 16.4 ± 6.2% (Figure 3A).
In site A (habitat entrance), Phormidiaceae was the most prevalent family with average RA of 12% (±9%) (Figure 3B). Site F showed the most consistent bacterial and archaeal population composition (Figure 3C). The most dominant families were Nitrososphaeraceae (average RA 8.7 ± 1.2%), Blastocatellaceae (6.6 ± 3%) and an unidentified family from the class Armatimonadia (7.7 ± 2%). In addition, site F was the only site to show a time affected microbial population shift (R = 0.4, p = 0.007, Supplementary Figure S5–S7).
To detect forward contamination, we first compiled a list of taxa found to be human-associated in previous studies, utilizing the dbBact database (http://dbbact.org/enrichment; Supplementary Table S3). These taxa can serve as markers for such contamination. The sum RA of all human associated genera in each site is presented in Figure 3D. Site S presented the highest RA of human associated genera, with samples collected on the seventh day being the most contaminated. As an example, the genus Citrobacter had an average RA of 0.09 ± 0.015% on the third and fifth days, and on the seventh day its RA in sample 07S1 increased to 38%. Site A was less contaminated, but still had higher RA of human associated genera as compared to site F, which presented very low levels of contamination (Figure 3D).
Introduction of Soil Microorganisms Into the Habitat
The study of backward contamination was designed to detect microorganisms carried on the AA’s gloves, upon return from EVAs. Baseline for the nitrile gloves was conducted so each “after EVA” sample was compared and paired to its “before EVA” sample (new glove). In addition, three samples of sterile swabs, DDW, and never worn nitrile gloves were included in the analysis. PCoA indicated that the “before” samples clustered together, along with the controls, suggesting a similar population composition. In contrast, the “after” samples were more dispersed and did not cluster with the controls (Figure 4A). The dominant and shared family in both “after” and “before” samples was Moraxellaceae, specifically the genus Acinetobacter, with average RA 9.6 ± 16% in the “before” samples, and average RA of 8 ± 14.9% in the “after” samples. Analysis of the controls with “after” samples showed a strong separation (ANOSIM R-value = 0.7, p-value = 0.003, Supplementary Figure S8), while clustering the control samples with the “before” samples, showed a much lower degree of separation (ANOSIM R-values = 0.26, p-value = 0.035, Supplementary Figure S9).
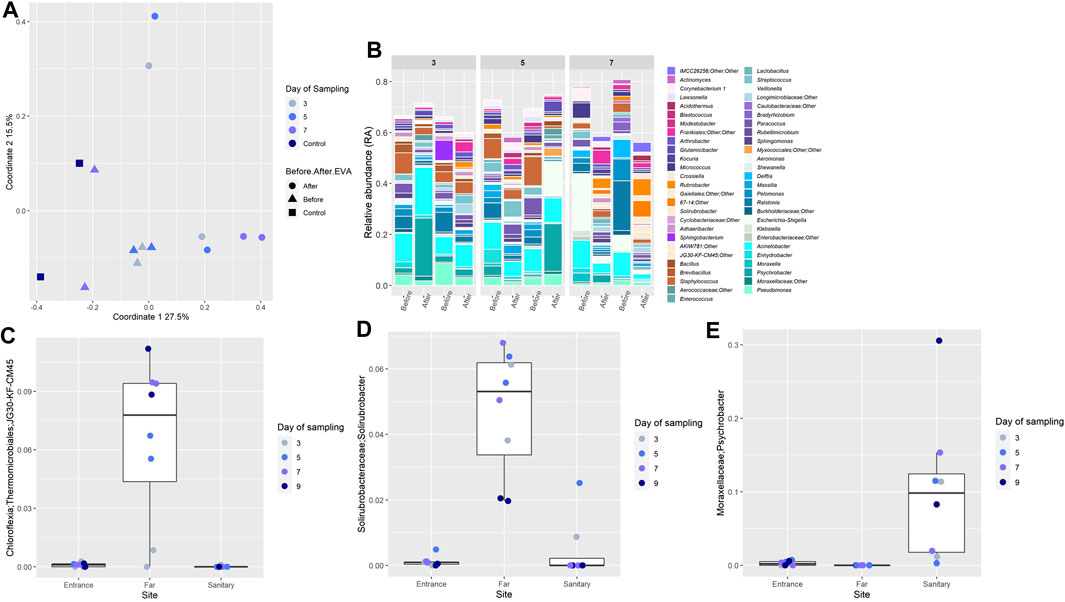
FIGURE 4. (A) Similarity in microbial composition on nitrile gloves before and after EVAs. PCoA of all glove samples throughout the mission, visualizing the spatial relationships between samples. (B) Relative abundance of bacterial OTUs comprising samples of gloves samples. Each sample is paired, e.g. before and after an EVA. Each color on the graph represents a distinct family level OTU. Only taxa that constituted RA of >1% in individual samples are presented, unless stated otherwise. Bar plots were generated using R. (C–E) Relative abundance of selected bacterial OTUs shared between soil and nitrile gloves samples. In the boxplots, lines indicate median relative abundances, boxes denote third and first quantile, whiskers denote ± 1.5 * IQR, and outliers are shown as dots.
To detect the most probable candidates for backward contamination, genera detected from “after-EVA” gloves were crossed reference with those detected in soil samples, yielding 44 genera that were shared between both sample types (very rare taxa, not reaching a threshold of 0.005 in any sample, were excluded before intersection). We then examined the distribution patterns of these shared genera across three sites. Ten shared genera appeared mainly in the Far site, and belong to commonly arid soil taxa, such as Chloroflexia (green non-sulfur bacteria, Figure 4C) and Solirubrobacterales (Figure 4D). These may thus be viewed as classic examples of backward contamination.
A member of the Moraxellaceae family, genus Psychrobacter (Figure 4E) presented a possible case of a cross backward contamination event (taken from inside the habitat, to the outside, and then back again into the habitat). This genus was found with low RA (0.2%) in the “before” sample taken on the third day, and upon return to the habitat from an EVA, it was found in much higher average RA (10 ± 9% of genera in the sample). A similar pattern was observed in a sample from the fifth day (3.7% before, 18% after) and the seventh day (0.006% before, 3% after). However, Psychrobacter was present in sites F and A with extremely low average RA (0.003 ± 0.006% and 0.28 ± 0.29%, respectively), and showed high RA in site S (17.8 ± 9%), suggesting its source is not in the local environment. Although several Psychrobacter species are environmental psychrotrophs (Bowman et al., 1996), some are also known to be human associated (Deschaght et al., 2012; Bonwitt et al., 2018).
Discussion
Analog missions are powerful platforms to gain broader and deeper understanding of the challenges accompanying human spaceflight and space missions. Forward and backward contamination potential in space exploration missions possess various aspects and hazards that, in order to be handled properly, should be further investigated. As discussed in the 2015 COSPAR workshop, there is a need for research on sampling methodology in low biomass environments, with special attention to determining spatial and temporal detection limits (Race et al., 2015). NASA’s updated policies on biological planetary protection for human missions to Mars (NASA, 2021) focus on the control of forward contamination of Mars, and possible backward contamination in the Earth-Moon system, associated with human presence in space vehicles. NASA, alongside other key players in this field, continuously advocate for the development of internationally accepted protocols to prevent backward and forward contamination.
In this study, deep amplicon sequencing of the bacterial 16S rRNA and the eukaryotic 18S rRNA genes were used to assess forward and backward contamination potential during a Mars analog mission. Our forward contamination study included two sites with high and almost daily human interruptions: A location very close to the sanitary disposal tanks (S), checked daily by the crew, and a location near the airlock exit (A), which the AAs stepped on, upon exit and return to the habitat. As a reference point, a third isolated site was selected 280 m west of the habitat, and AAs were not allowed to enter its perimeter, except for the purpose of sampling. We hypothesized that differences in the diversity and population composition will be significant in the sites closer to the habitat when compared to the “Far” site, and that presence of human associated microorganisms will be observed. Indeed, sites S and A presented major differences, as compared to site F. Site F, the least visited site, was the only site to present a time dependent population composition change (R = 0.4, Supplementary Figure S6) and maintained a high and consistent diversity. The changes in microbial composition were not correlated to the changes in relative humidity and temperature, according to our statistical analysis. We believe this to be a natural manifestation of the temporal dynamics in the soil microbial communities’ composition, as has been previously shown for bacterial groups in Namib Desert soil (Gunnigle et al., 2017). Site A was located outside the airlock, where the AAs frequently visited, soil and microorganisms carried on the soles of the boots could have been transported to the site from other sites, and from inside the habitat itself. The relatively high diversity score in site A supports our hypothesis and can be explained by the site’s location. As for site S, which presented a lower average diversity score, it is important to note that DMARS03 was not the first analog mission to take place in the exact same locale. Thus, pollution may have affected the natural population and its diversity in the soil close to the habitat. The population composition in site S, indicated high RA of several families likely to be human associated, such as Enterobacteriaceae, Moraxellaceae, and Pseudomonadaceae. The fact that some of the samples taken on the same day and from the same site exhibited differences in their population composition highlights the spatial effect on soil sampling, as previously reported (Raynaud and Nunan, 2014).
Cross contamination was also possible between sites, and its level remains unclear. AAs sometimes carried equipment that might have transported microorganisms from one site to the next. Another possible vector for cross contamination transport is the AAs’ boot soles, which were not sterilized between sites. This might explain the presence (even at low RA level) of human-associated genera in site F.
We were interested to see if these changes in microbial population will occur within days of establishing a human presence in an isolated environment, based on D-MARS03 mission settings.
Our results indicate that forward contamination occurs within days in the area most adjacent to the habitat and does not manifest itself within days in an isolated location even 280 m away from the habitat. It will take a longer mission to see if changes occur in target areas 200 m or more, from the habitat or activity area of a human crew. Current mission included analog astronaut walking on foot with mock spacesuits on, it did not include any kind of vehicles or mobile robotic components that could’ve potentially caused forward contamination as well.
Presently, these results show that backward contamination occurred, within a few days, with one dominant genus–Acinetobacter (family Moraxellaceae). Also, several genera from the “F” site-such as Chloroflexia (green non-sulfur bacteria, and Solirubrobacterales) were detected.
The case of Psychrobacter is intriguing. This genus was present in high RA in site S, in contrast to sites A and F, in which its presence was rare to negligible. This genus was also found on the gloves of almost all AA’s upon return from EVAs. This genus might be human associated, since it was specifically enriched in the soil around the sanitary disposal tanks, and exhibited some changes in RA during the mission. Therefore, it is possible that the presence of Psychrobacter in site S, is due to forward contamination, and its return to the habitat is an evidence of backward contamination, hence cross contamination. In order to better handle and reduce cross contamination, an in-depth analysis of possible vectors, that are prone to facilitate the transfer of microorganisms, must be performed.
Investigation of backward contamination is complex; in the current study, we cannot predict which species will colonize the indoors of the habitat. Microbial succession, that should also be considered when examining forward contamination events, is affected by environmental factors, such as pH, salinity, and organic carbon and by stochastic processes (Dini-Andreote et al., 2015). This should be further tested with additional monitoring experiments which will follow the establishment of bacterial species inside the habitat, during a longer period, perhaps in a similar bacterial monitoring method to the one done in Kibō on the ISS (Ichijo et al., 2016). This proof of concept demonstration of cross contamination monitoring can be modified to be used on Mars, with several considerations. While the use of deep sequencing of marker genes is a powerful and sensitive technique to detect and identify bacteria in low biomass settings, it nevertheless possesses inherent technical limitations, which may distort taxonomic information (Salter et al., 2014; Silverman et al., 2019). In particular, the impact of a contaminated extraction kit on low-biomass environmental investigation, was shown to be a possible hazard. Our controls (sterile DDW, sterile cotton swab, sterile nitrile glove) indicated low presence of background DNA, which for most analyses was negligible. However, when examining sensitive questions of contamination, a better and more credible control is needed. Additionally, our suggested method cannot quantify the exact level of contamination. This can be easily solved with addition of a quantitative or semi-quantitative method, such as RT-PCR (Kim et al., 2014). Also, the presented approach is tailored to nucleic acids based genetic material and ribosomal rRNA, yet life that originated elsewhere could be unrecognizable to our technology and methods, rendering it undetectable. Generally, nucleic acids-based detection might not necessarily be the most suitable technique to detect foreign life forms, and different biomarkers should be examined (Davila and McKay, 2014).
Future studies will look for the presence of all the reported genera on the habitat surfaces, and mobile robotic elements or vehicles if those will be used. Also, the impact on habitat systems health and on astronaut health should be investigated in depth regarding these genera.
To conclude, the ability to control and reduce forward and backward contamination is crucial for the future of humankind in space. As our knowledge of the extent and range of possible contamination grows, future space related missions can be utilized to establish the proper protocols to reduce cross contamination events and protect both Earth and extra-terrestrial environments.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject [Accession: PRJNA662496].
Author Contributions
YY conceived and designed the experiment, collected the data, performed the analysis and wrote most of the paper. LR performed data analysis. RA conceived and designed the experiment, wrote small part of the paper and performed other contributions. CG collected the data. GA contributed to data analysis. GY collected the data and performed data analysis and other contributions. OA supported the design and execution of the experiment. All authors reviewed the article and approved submission.
Funding
This research was funded by The Minerva Center, Origin of Life Under Extreme Planetary Conditions, The Weizmann Institute of Science, and by The Dead Sea and Arava Science Center. D-MARS03 mission was supported by the Israeli Space Agency, the Ministry of Science and Technology, Israel.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge the support of Desert Mars Analog Ramon Station volunteers and support crew in this experiment. We would like to thank the analog astronauts of D-MARS03 science mission who carried out the experiment: YY, Carmit Avidan Shpalter, Noa Breuer, Camelia Gochev, Yiftah Curiel and Salman Abdalla. We would like to thank the Minerva Center for Life under Extreme Planetary Conditions at the Weizmann Institute of Science for supporting the science mission. D-MARS03 mission was supported by ICA foundation and Shika design collective. The authors would also like to thank Prof. Eliora Z. Ron, Prof. Uri Gophna and Dr Dvora Biran from Tel Aviv University for their useful comments and insights. Lastly, the authors thank Jonathan Shvartzberg, Bar-Ilan University, for his generous support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspas.2021.589147/full#supplementary-material.
Abbreviations
AA, analog astronaut; EVA, extravehicular activity; RA, relative abundance
References
Bonwitt, J., Tran, M., Droz, A., Gonzalez, A., and Glover, W. A. (2018). Psychrobacter sanguinis wound infection associated with marine environment exposure, Washington, United States. Emerg. Infect. Dis. 24 (10), 1942–1944. doi:10.3201/eid2410.171821
Bowman, J. P., Cavanagh, J., Austin, J. J., and Sanderson, K. (1996). Novel psychrobacter species from antarctic ornithogenic soils. Int. J. Syst. Bacteriol. 46 (4), 841–848. doi:10.1099/00207713-46-4-841
Cabrol, N. A. (2018). The coevolution of life and environment on Mars: an ecosystem perspective on the robotic exploration of biosignatures. Astrobiology 18 (1), 1–27. doi:10.1089/ast.2017.1756
Cary, S. C., Mcdonald, I. R., Barrett, J. E., and Cowan, D. A. (2010). On the rocks: the microbiology of antarctic dry valley soils. Nat. Rev. Microbiol. 8, 129–138. doi:10.1038/nrmicro2281
Cockell, C. S. (2005). Planetary protection-A microbial ethics approach. Space Policy 21, 287–292. doi:10.1016/j.spacepol.2005.08.003
Davila, A. F., and McKay, C. P. (2014). Chance and necessity in biochemistry: implications for the search for extraterrestrial biomarkers in earth-like environments. Astrobiology 14 (6), 534–540. doi:10.1089/ast.2014.1150
Rubinstein, H., Abramovich, R. S., Shikar, A., Zagai, M., Nevenzal, H., Finzi, Y., et al. (2019). “The 2019 analog Mars mission season at the Desert Mars analog ramon station”, in Conference: 70th International Astronautical Congress (IAC-19), Washington, DC, October 21–25, 2019.
Deschaght, P., Janssens, M., Vaneechoutte, M., and Wauters, G. (2012). Psychrobacter isolates of human origin, other than Psychrobacter phenylpyruvicus, are predominantly Psychrobacter faecalis and Psychrobacter pulmonis, with emended description of P. faecalis. Int. J. Syst. Evol. Microbiol. 62 (3), 671–674. doi:10.1099/ijs.0.032631-0
Dini-Andreote, F., Stegen, J. C., Van Elsas, J. D., and Salles, J. F. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 112 (11), E1326–E1332. doi:10.1073/pnas.1414261112
Finzi, Y., Avni, S., Maroz, A., Avriel-Avni, N., Ashckenazi-Polivoda, S., and Ryvkin, I. (2019). Extraordinary geodiversity and geoheritage value of erosional craters of the Negev Craterland. Geoheritage 11 (3), 875–896. doi:10.1007/s12371-018-0335-7
Fisk, L., Worms, J. C., Coustenis, A., Hedman, N., and Kminek, G. (2020). Introduction to the new COSPAR policy on planetary protection. Space Res. 208, 9. doi:10.1016/j.srt.2020.07.008
Grömer, G., Frischauf, N., Soucek, A., and Sattler, B. (2007). “AustroMars-a simulated high-fidelity human Mars analogue mission,” in Mars2030-AustroMars Science Workshop, Salzburg, Austria, December, 2007, 4–12.
Gunnigle, E., Frossard, A., Ramond, J.-B., Guerrero, L., Seely, M., and Cowan, D. A. (2017). Diel-scale temporal dynamics recorded for bacterial groups in Namib Desert soil. Sci. Rep. 7, 40189. doi:10.1038/srep40189
Huse, S. M., Dethlefsen, L., Huber, J. A., Welch, D. M., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. Plos Genet. 4 (11), e1000255. doi:10.1371/journal.pgen.1000255
Ichijo, T., Yamaguchi, N., Tanigaki, F., Shirakawa, M., and Nasu, M. (2016). Four-year bacterial monitoring in the international space station-Japanese experiment module “kibo” with culture-independent approach. NPJ Microgravity 2, 1–6. doi:10.1038/npjmgrav.2016.7
Goh, G. M., and Kazeminejad, B. (2004). Mars through the looking glass : an interdisciplinary analysis of forward and backward contamination. Space Policy 20, 217–225. doi:10.1016/j.spacepol.2004.06.008
Liu, Z., Lozupone, C., Hamady, M., Bushman, F. D., and Knight, R. (2007). Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35 (18), e120. doi:10.1093/nar/gkm541
Martins, Z., Cottin, H., Kotler, J. M., Carrasco, N., Cockell, C. S., de la Torre Noetzel, R., et al. (2017). Earth as a tool for astrobiology-a European perspective. Space Sci. Rev. 209 (1–4), 43–81. doi:10.1007/s11214-017-0369-1
Mormile, M. R. (2009). Molecular analysis of the microbial communities of Mars analog lakes in western Australia. Astrobiology 9 (10), 919. doi:10.1089/ast.2008.0293
NASA (2021). Nid 8715.129: biological planetary protection for human missions to Mars. Washington, DC: NASA, 1–6.
National Research Council (2002). Safe on Mars: precursor measurements necessary to support human operations on the martian surface. Washington, DC: The National Academies Press.
Nicholson, W. L., McCoy, L. E., Kerney, K. R., Ming, D. W., Golden, D. C., and Schuerger, A. C. (2012). Aqueous extracts of a Mars analogue regolith that mimics the Phoenix landing site do not inhibit spore germination or growth of model spacecraft contaminants Bacillus subtilis 168 and Bacillus pumilus SAFR-032. Icarus 220 (2), 904–910. doi:10.1016/j.icarus.2012.06.033
Notesco, G., Ogen, Y., and Ben-Dor, E. (2016). Integration of hyperspectral shortwave and longwave infrared remote-sensing data for mineral mapping of Makhtesh Ramon in Israel. Remote Sens. 8 (4), 318. doi:10.3390/rs8040318
Notesco, G., Ogen, Y., and Ben-Dor, E. (2015). Mineral classification of makhtesh ramon in Israel using hyperspectral longwave infrared (LWIR) remote-sensing data. Remote Sens. 7 (9), 12282–12296. doi:10.3390/rs70912282
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O’Hara, R., et al. (2013). Package “vegan”. Community ecology package, version 2. Available at: http://CRAN.R-project.org/package=vegan.
Oliver, F., Yilmaz, P., Quast, C., Greken, J., Beccati, A., Ciuprina, A., et al. (2017). 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176. doi:10.1016/j.jbiotec.2017.06.1198
Ordoñez, O. F., Flores, M. R., Dib, J. R., Paz, A., and Farías, M. E. (2009). Extremophile culture collection from andean lakes: extreme pristine environments that host a wide diversity of microorganisms with tolerance to UV radiation. Microb. Ecol. 58, 461–473. doi:10.1007/s00248-009-9527-7
Piubeli, F., de Lourdes Moreno, M., Kishi, L. T., Henrique-Silva, F., García, M. T., and Mellado, E. (2015). Phylogenetic profiling and diversity of bacterial communities in the death valley, an extreme habitat in the atacama desert. Indian J. Microbiol. 55 (4), 392–399. doi:10.1007/s12088-015-0539-3
Pratt, L. M., and Smith, A. L. (2020). “Expectations for backward planetary protection planning during Mars sample return planning,” in 2020 IEEE Aerospace Conference, Big Sky, Montana, March, 2020, 1–7. doi:10.1109/aero47225.2020.9172722
Race, M. S., Johnson, J. E., Spry, J. A., Siegel, B., and Conley, C. A. (2015). “Planetary protection knowledge gaps for human extraterrestrial missions: COSPAR workshop report,” in Workshop on Planetary Protection Knowledge Gaps for Human Extraterrestrial Missions, Moffett Field, CA, March 24–26, 2015. Available at: https://ntrs.nasa.gov/archive/nasa/casi.ntrs.nas.
Raynaud, X., and Nunan, N. (2014). Spatial ecology of bacteria at the microscale in soil. PLoS One 9 (1), e87217. doi:10.1371/journal.pone.0087217
Da Cunha, A. R. (2015). Evaluation of measurement errors of temperature and relative humidity from HOBO data logger under different conditions of exposure to solar radiation. Environ. Monit. Assess. 187, 4458. doi:10.1007/s10661-015-4458-x
Rummel, J. D. (2001). Planetary exploration in the time of astrobiology: protecting against biological contamination. Proc. Natl. Acad. Sci. 98 (5), 2128–2131. doi:10.1073/pnas.061021398
Salter, S. J., Cox, M. J., Turek, E. M., Calus, S. T., Cookson, W. O., Moffatt, M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12 (1), 87. doi:10.1186/s12915-014-0087-z
Schmidt, H., and Karnieli, A. (2001). Sensitivity of vegetation indices to substrate brightness in hyper-arid environment: the Makhtesh Ramon Crater (Israel) case study. Int. J. Remote Sens. 22 (17), 3503–3520. doi:10.1080/01431160110063779
Schuerger, A. C., Mancinelli, R. L., Kern, R. G., Rothschild, L. J., and Mckay, C. P. (2003). Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated martian environments:implications for the forward contamination of Mars. Icarus 165, 253–276. doi:10.1016/S0019-1035(03)00200-8
Schwartz, J. S. J. (2019). Where no planetary protection policy has gone before. Int. J. Astrobiology 18 (4), 353–361. doi:10.1017/S1473550418000228
Shahar, K., and Greenbaum, D. (2020). Lessons in space regulations from the lunar tardigrades of the Beresheet hard landing. Nat. Astron. 4, 208–209. doi:10.1038/s41550-020-1034-2
Silver, M. M. W. (2018). An investigation into the suitability of sulfate-reducing bacteria as models for Martian forward contamination. Thesis and Dissertations. Fayetteville (AR): University of Arkansas.
Silverman, J. D., Bloom, R. J., Jiang, S., Durand, H. K., Mukherjee, S., and David, L. A. (2019). Measuring and mitigating PCR bias in microbiome data. bioRxiv, 1604025. doi:10.1101/604025
Sneh, A., Bartov, Y., Weissbrodt, T., and Rosensaft, M. (1998). Geology map of Israel, 1: 200000, 4 sheets. Jerusalem, Israel: Geological Survey of Israel.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 (19), 2460–2461. doi:10.1093/bioinformatics/btq461
Kim, T. G., Jeong, S. Y., and Cho, K. S. (2014). Comparison of droplet digital PCR and quantitative real-time PCR for examining population dynamics of bacteria in soil. Appl. Microbiol. Biotechnol. 98 (13), 6105–6113. doi:10.1007/s00253-014-5794-4
Van Heereveld, L., Merrison, J., Nørnberg, P., and Finster, K. (2017). Assessment of the forward contamination risk of Mars by clean room isolates from space-craft assembly Facilities through aeolian transport-a model study. Orig Life Evol. Biosph. 47 (2), 203–214. doi:10.1007/s11084-016-9515-0
Vishnevetsky, S., and Steinberger, Y. (1997). Bacterial and fungal dynamics and their contribution to microbial biomass in desert soil. J. Arid Environ. 37 (1), 83–90. doi:10.1006/jare.1996.0250
Wickham, H. (2007). Reshaping data with the reshape Package. J. Stat. Soft. 21 (12), 1–20. doi:10.18637/jss.v021.i12
Keywords: planetary protection, contamination, mars, analog research, ramon crater, 16S, 18S
Citation: Yair Y, Reshef L, Shopen-Gochev C, Yoffe G, Azulay G, Aharonson O and Sorek-Abramovich R (2021) Temporal and Spatial Analysis of Forward and Backward Microbial Contamination in a Mars Analog Mission. Front. Astron. Space Sci. 8:589147. doi: 10.3389/fspas.2021.589147
Received: 30 July 2020; Accepted: 11 January 2021;
Published: 18 March 2021.
Edited by:
Dov Greenbaum, Yale University, United StatesReviewed by:
Mihaela Glamoclija, Rutgers University, United StatesJames Benardini, NASA Jet Propulsion Laboratory (JPL), United States
Copyright © 2021 Yair, Reshef, Shopen-Gochev, Yoffe, Azulay, Aharonson and Sorek-Abramovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reut Sorek-Abramovich, UmV1dC5zb3Jla0BnbWFpbC5jb20=
†ORCID: Oded Aharonson orcid.org/0000-0001-9930-2495
 Yael Yair
Yael Yair Leah Reshef1
Leah Reshef1 Camelia Shopen-Gochev
Camelia Shopen-Gochev Gal Yoffe
Gal Yoffe Gil Azulay
Gil Azulay Reut Sorek-Abramovich
Reut Sorek-Abramovich