- Microbial Ecology Laboratory, Department of Natural Resources and Environmental Sciences, University of Illinois at Urbana-Champaign, Urbana, IL, United States
The agricultural imprints on soil microbial processes manifest at various timescales, leaving many temporal patterns to present slowly. Unfortunately, the lack of long-term continuous agricultural field sites in North America has left gaps in our understanding of agricultural management on biogeochemical processes and their controlling microbiota. Nitrification, ammonium oxidation by bacteria and archaea, is a critical control point in terrestrial nitrogen fluxes by oxidizing cationic ammonium to anionic nitrate, promoting nitrate leaching. Moreover, nitrous oxide is produced during nitrification, contributing to massive nitrous oxide emissions from fertilized agroecosystems. Nitrification is sensitive to many macro and micro-ecological filters, as nitrifiers are obligate aerobes and are sensitive to numerous non-growth substrates and metal ions. This study sought to understand the long-term implications of various rotation and fertilizer regimes on nitrification potential and nitrifying bacterial communities in the Morrow Plots (Urbana, IL). The Morrow Plots was established in 1876 and are the longest continuous field experiments in North America, making it the only site in America capable of assessing the impact of over 140 years of agricultural management on nitrification. The Morrow Plots contrasts fertilizer (manure, inorganic, unfertilized) and rotation (continuous corn, corn-soy, corn-oat alfalfa), allowing us to explore how conventional vs. regenerative agriculture practices impact nitrifier communities. The results of this study suggest that fertilizer and rotation interact to promote distinct bacterial nitrifier communities. Nitrification potential is highest in manure corn-oat-alfalfa plots, suggesting ammonium availability is not solely responsible for active nitrifier communities. Various soil chemical variables, like CEC, Mg, and Ca, significantly influenced nitrifier community beta-diversity, using 16S rRNA amplicon sequencing, suggesting long-term accumulation of specific cations diverge microbial community assembly. While this study only uses nitrification potential enzyme activity instead of isotope analyses, it sheds light on the importance of various physiochemical drivers on nitrification potential and communities. The results support the need for a more precise exploration of the mechanisms controlling field-scale nitrification rates over large temporal scales. Put together, this study supports the importance of long-term field sites for understanding agricultural manipulations of microbial biogeochemical cycling and sheds light on the micronutrients influencing nitrifier communities and potential activity.
Introduction
Soil nitrification, the aerobic oxidation of to and , is a critical control point in terrestrial nitrogen (N) cycling by modulating N-loss capacity from fertilized soils. Agronomic management practices may directly impact nitrifiers, through manipulation of nitrifier growth, or indirectly through the alteration in their soil physiochemical habitat. However, soil matrix integrity shifts temporally as the impacts of management on soil structure manifest slowly over decades (1). The lack of long-term agricultural experiments within North America which contrast regenerative (organic fertilizers and tri-rotational regimes) and intensive (synthetic fertilizers and monocultures) practices, limits the ability to investigate long-term management-driven shifts in nitrifying communities and activities. The Morrow Plots long-term agricultural experiment, established in 1867 at the University of Illinois Urbana-Champaign, contrasts conventional with regenerative practices presenting the opportunity to study the relationship between soil nitrifying potential, agronomic management strategies, and soil physiochemical factors. Long-term factorial experiments can be used to evaluate the impacts of fertilizer and rotational management strategies on nitrogen biogeochemistry in order to understand if nitrogen losses in industrial systems can be mitigated with regenerative practices. Understanding anthropogenic disruption in nitrification will assist in the optimization of agronomic management strategies, ensuring the sustainability and protection of soil resources.
Nitrifying microorganisms consists of ammonium oxidizing bacteria (AOB) and archaea (AOA), nitrite-oxidizing bacteria (NOB), and comammox bacteria (2). Nitrification involves the stepwise oxidation of ammonium to nitrite by AOB and AOA, with subsequent oxidation of to by NOB. The primary step is catalyzed by ammonium monooxygenase (AMO), a Cu-containing membrane-bound monooxygenase (3), and is coupled to the reduction of oxygen to water, producing hydroxylamine. Hydroxylamine is further oxidized by hydroxylamine oxidoreductase (HAO), producing . NOB catalyze the second step of nitrification, the oxidation of to via nitrite oxidoreductase (NXR). In addition to variations in the enzymatic and coordination chemistry of nitrification redox enzymes, nitrifier genera utilize several different carbon fixation strategies. Nitrifiers use the Calvin-Benson cycle (AOB: Nitrosomonas; NOB: Nitrospirae), 3-hydroxypropionate−4-hydroxybutyrate cycle (AOA: aerobic Crenarcheota), reductive tricarboxylic acid pathway (NOB: Nitrospirae), and the dicarboxylate 4-hydroxybutyrate pathway (AOA: anaerobic Crenarcheota) (4–6). The variation in environmental sensitivity between enzymes within the carbon fixation pathways, as well as energy-generating ammonium oxidations, influence the ecophysiology and species distribution of nitrifying organisms.
Agricultural systems modify nitrogen pools through N fertilizers, and legume regimes (7). However, due to variations in biochemical characteristics of AOB, AOA, NOB, and comammox genera, predicting the response of these microorganisms to agronomic practices is complicated. Shifts in the abundance, diversity, composition, and functional potential of nitrifying microbes have been shown to correlate with soil quality parameters, like temperature (8), soil organic carbon (SOC)/ organic matter (OM) (9), pH (10), cation exchange capacity [CEC; (11)], / (12, 13). Many of these parameters, like SOC/OM, accumulate gradually, as soil forming processes are slow and depend complex geophysical and biotic processes. When management strategies, like monocultures or fertilization, manipulate carbon and nitrogen inputs, soil structure and abiotic characteristics of soil which govern biotic interactions change (14). Therefore, assessing not only nitrifier response to major selective factors, like OM and pH, but also micronutrients, may provide insight regarding nitrifier species responses to long-term disturbances.
Nitrifiers are a particularly sensitive to both large-scale ecological filters and subtle deviations in substrates and micronutrients. Bacterial AMO has the ability to oxidize numerous non-growth substrates such as methane, methanol, benzene, and phenols (15–17). Moreover, ammonium oxidation is sensitive to numerous metal cations, like Zn, Cu, Mg, and Cd. Metal cations also drive soil formation, promoting aggregation through cationic bridging (18). Fertilizer, particularly manure, can be a dominant source of soil metal fluxes, which through time, promote soil stabilization (19, 20). However, they are easily over-applied and toxicity of cationic metals perturbed microbial communities (21, 22). Monitoring the long-term shifts in nitrification and its chemical and physical drivers can illuminate human-driven changes in N-dynamics over a deep temporal scale.
While previous research has identified management-driven changes to the soil microbial community as a whole within the Morrow Plots (23), that study did not evaluate biogeochemical transformations or the microbial functional groups responsible for them. Given the importance of nitrification for environmental quality, the sensitivity of nitrifiers to edaphic factors, and the limited opportunities to study this process in long-term agricultural experiments (particularly one so historic), the goal of this study was to assess how long-term fertilizer and rotational management impact nitrification potential activity (NP), as well as community structure in the Morrow Plots. The Morrow Plots experiment is an agricultural mesocosm for observing management-driven effects on nitrification and functionally important soil microbiota over very long time scales. The Morrow Plots are a particularly advantageous study site as Illinois is situated in a region with a high density of industrial agriculture and is a major contributor to nitrogen loading into the Mississippi River. This experiment allows a glimpse into the future of industrial agriculture on nitrogen loss processes, and the potential for regenerative management practices to mitigate nitrogen losses and transition modern American agricultural to minimally detrimental and environmentally sustainable systems.
Methods
Study Site—The Morrow Plots
The Morrow Plots, located on the campus of the University of Illinois at Urbana-Champaign, are the oldest continuously maintained agricultural research plots in the United States. The plots were established in 1876 to demonstrate the long-term effects of crop rotation, soil nutrient depletion, and the effects of synthetic and natural fertilizers (24, 25). Briefly, this long-term experiment (Supplementary Figure 1) consists of three blocks of crop rotation treatments: continuous corn (Zea mays) (C), a 2-year corn and soybean rotation (CS), and a 3-year corn-oats-alfalfa rotation (COA). Each crop rotation block is split into eight plots comprising replicated fertilizer treatments: unfertilized (UF); inorganic fertilizers (IN) with nitrogen (as urea), phosphorus (as P2O5), potassium (as K2O), and limestone; and organic fertilization (OR) with dairy manure, limestone, and phosphorus (Supplementary Figure 1). Refer to Aref and Wander (25) or Odell et al. (24) for a more detailed description of the Morrow Plots site (24, 25).
Soil Sampling
Soil samples were collected on June 4 and August 25 in 2015—a year when all the plots were planted in corn. Sampling during an all-corn year controls enables the investigation of long-term effects of the crop-rotation treatments on soil microbial communities, by avoiding the short-term effects of plant-microbe interactions from the annual rotations. Each sample consisted of five cores (1.9 cm dia × 12 cm deep). Bulk soil cores from each plot were placed in sealed plastic bags on ice while in the field and transported back to the lab and processed within 2 h of collection. The cores from each plot were composited and homogenized with a 2 mm sieve. Subsamples from each the composited, homogenized soil sample were processed as appropriate for chemical analyses, nitrification assays, and DNA extraction for soil microbiome analysis.
Soil Chemical Analyses
Soil chemical analyses were conducted by Waypoint Analytical (Champaign, IL). Soil NH4-N and NO3-N were quantified using Lachat QuickChem methods 12-107-06-2-F and 12-107-04-1-J, respectively [Soil, Plant and Water Reference Methods for the Western Region (Mod), 2013]. Soil P, K, S, Fe, K, Mg, Ca, Mn, B, and CEC, as well as percent cation saturations, were measured using Mehlich 3 extraction protocols (Handbook on Reference Methods for Soil Analysis-1999, Soil and Plant Analysis Council, Inc.). Percent organic matter (OM) was quantified using the LOI method, the results of which were used to calculate the estimated-N-released in pounds per acre. Buffer pH (SMB buffer pH) and 1:1 soil pH (pH) were quantified for each sampled (Soil, Plant and Water Reference Methods for the Western Region 2013, 4th Edition).
Potential Nitrification Assay
Soil nitrification potential (NP) was quantified colorimetrically using the Griess-Ilosvay's method (26, 27), originally adapted from Berg and Rosswall (28). Briefly, 5 g of homogenized field soil in 50 mL Falcon tubes was shaken for 5 h at room temperatures after adding 1 mM (NH4)2SO4, and 1.5 M sodium chlorate. Each sample had a corresponding control sample which was treated identically, but frozen at −20°C for the 5-h incubation. 2M KCl was added after the incubation, and the tubes were manually shaken, then centrifuged for 2 min at 2,000 RPM. The supernatant was filtered using Whatman 42 filter papers. NO2-N was measured using a Genesys 20 spectrophometer (Thermo Scientific, Rochester, NY) after adding Griess-Ilosavay reagent (sulfanilamide and N-napthylethyldiamine) at 520 nm. NO2-N concentration was quantified against a NaNO2-N standard curve. Potential nitrification rates were measured as the change in NO2-N concentration between the aerated and frozen samples, by the soil gram dry weight (% dry matter) per hour.
DNA Extraction and qPCR
DNA Extraction
Total genomic DNA was extracted from soils using the FastDNA SPIN Kit for Soil (MP Biomedicals, Solon, OH), and further purified using cetyl trimethyl ammonium bromide (CTAB) extraction to remove contaminating humic acids (29). DNA concentration was adjusted to 30 ng/μl and subjected 16S rRNA V4 region amplicon sequencing and amoA qPCR analyses at the University of Illinois Biotechnology Center (Urbana, IL).
Fluidigm qPCR
Bacterial amoA (BamoA) and archaeal amoA (AmoA) genes were quantified with fluidigm qPCR, using the amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′), amoA-2R (5′ -CCCCTCKGSAAAGCCTTCTTC−3′) for BamoA, and the CrenamoA23f (5′-ATGGTCTGGCTWAGACG-3′) and CrenamoA616r (5′-GCCATCCATCTGTATGTCCA-3′) primers for AamoA (30, 31). To increase the amount of template DNA prior to Fluidigm qPCR, a preamplification (specific target amplification; STA) reaction was performed in 5 μl reaction mixtures containing 2 × Taqman PreAmp Master Mix (Applied Biosysterms), 0.5 μM of each primer, and 1.25 μl of the DNA template. The STA reaction was performed on an MJ Research Tetrad thermal cycler with the following cycling program: 95°C for 10 min followed by 14 cycles of 95°C for 15 s and 58°C for 4 min. Standards for each gene were mixed and 5-fold diluted from 1 × 105 to 3.2 × 101 copies/μl, and amplified by the STA reaction together with the soil genomic DNA to provide standard curves for Fluidigm qPCR. The STA products were treated by exonuclease to remove excessive primers. For Fluidigm qPCR, 5 μl of sample premix was prepared containing 2 × SsoFast Evagreen Supermix with Low Rox (BioRad), 20 × DNA Binding Dye Sample Loading Reagent (Fluidigm), and 2.25 μl exonuclease treated products. Five μl of assay mix was prepared containing 2 × Assay Loading Reagent (Fluidigm), 1 × DNA Suspension Buffer (Teknova), and 50 μM each mixed forward and reverse primer. The sample premix and assay mix were loaded on a 96.96 chip (Fluidigm), and the target genes were amplified on the Fluidigm Biomark HD Real Time PCR system using the following cycling program: 70°C for 40 min, 58°C for 30 s, 95°C for 1 min followed by 30 cycles of 96°C for 5 s, 58°C for 20 s, and followed by dissociation curve. All the samples and standards were analyzed in 12 replicates with molecular grade water as no template control. The CT values (cycle threshold) were determined using Fluidigm Real-Time PCR Analysis software version 4.1.3. The copy number of genes per μl was determined for each soil sample by comparison to the standard curve in the assay, and then normalized to ng of DNA.
16S rRNA Gene Amplicon Sequencing
Illumina sequencing was used to target the prokayotic 16S rRNA V4 region for nitrifier community analyses (Illumina, San Diego, CA). Sequencing amplicons were prepared by PCR using a Fluidigm Access Array IFC chip, which allowed simultaneous amplification of each target gene (Fluidigm, San Francisco, CA). Initial reactions were carried out according to a 2-step protocol using Fluidigm-recommended reagent concentrations, and an annealing temperature of 55°C. The first PCR was performed in a 100-μL reaction volume using 2 ng DNA template, and this PCR amplified the target DNA region using the 16S rRNA V4 primers 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) augmented with Fluidigm-specific amplification primer pads CS1 (5′-ACACTGACGACATGGTTCTACA-3′) and CS2 (5′-TACGGTAGCAGAGACTTGGTCT-3′), producing amplicons that consisted of (2) CS1 Fluidigm primer pad, (3) 16S rRNA forward primer 515F, (4) 16S rRNA V4 amplicon (5) 16S rRNA reverse PCR primer 806R, and (6) CS2 Fluidigm primer pad. A secondary 30-μL PCR used 1 μL of 1:100 diluted product from the first PCR as template, and PCR primers with CS1 and CS2 sequences and Illumina-specific sequencing linkers P5 (5′-AATGATACGGCGACCACCGAGATCT-3′) and P7 (5′-CAAGCAGAAGACGGCATACGAGAT-3′), along with a 10-bp sample-specific barcode sequence, so the final construct consisted of (1) Illumina linker P5, (2) CS1, (3) 515F primer, (4) 16S rRNA V4 amplicon, (5) 806R primer, (6) CS2, (7) sample-specific 10-bp barcode, and (8) the Illumina linker P7. Final amplicons were gel-purified, quantified (Qubit; Invitrogen, Carlsbad CA, USA), combined to the same concentration, and then sequenced from both directions on an Illumina HiSeq 2,500 2 ×250 bp Rapid Run. Fluidigm amplification and Illumina sequencing was conducted at the Roy J. Carver Biotechnology Center (Urbana, IL, USA).
Barcodes were used to assign each sequence to its original sample. After de-multiplexing, paired-end sequences generated for 16S rRNA were merged using software FLASH (Fast Length Adjustment of SHort reads) (32). Quality filtering of fastq files was performed using software in the FASTX-Toolkit (33), which removed sequences with more than 10% bases with quality score lower than 30 and sequences containing ambiguous bases “N” from downstream processing. Filtered sequences were clustered into operational taxonomic units (OTUs) using USEARCH64 and a 97% similarity threshold (33). USEARCH was used to (1) de-replicate sequences and remove singletons; (2) remove chimeras contained in the sequences using GOLD (34) as a reference database; (3) form OTU clusters from sequences that were 97% similar and represent each OTU by representative sequences. The cluster file was converted into an OTU table using functions available in MacQIIME (35). Representative sequences for 16S rRNA OTUs were assigned taxonomic attribution in QIIME with the uclust algorithm (36) using the August 2013 Greengenes database (37) as a reference. Amplicon sequence data for 16S rRNA genes is available for download on the NCBI SRA database at accession number: PRJNA789310 (https://www.ncbi.nlm.nih.gov/sra/PRJNA789310). Nitrifier community diverse was then assessed by subsetting the OTU table based on nitrifier Order: specifically, Nitrosomonadales (AOB), Nitrososphaerales (AOA), Nitrospirales (NOB).
Statistical Analyses
All statistical analyses were performed in R Studio statistical software (Version 4.1.2, 2021) (38). All figures were produced using ggplot2 v. 3.3.5 (39). Two-way ANOVA's, with interaction effects, were conducted to understand the impact of fertility and rotation on potential nitrification rates, qPCR abundances, alpha-diversity metrics (Observed Richness, Chao1, Shannon Diversity index). Potential rates and qPCR abundances were natural log transformed to ensure normality. Shapiro-Wilk test was used to identify deviations in residual variances of all models and calculated with the shapiro.test() function of the stats package v. 4.1.1.; W > 0.9 was used to indicate normally distributed residual variances. Levene's Test for homogeneity of variances across fertility and rotation groups was calculated using LeveneTest() function of the car package v. 3.0-11. Means were separated using Tukey's honestly significant difference test using the HSD.test() function from the agricolae package v. 1.3-5. One-way regressions were used to identify significant chemical drivers influencing potential nitrification rates. Shapiro-Wilk's test was used to ensure no violations of regression assumptions, and the regression P-values were adjusted using a false-discovery rate test. Outliers of ANOVA and regression models were only removed to assume normality, due to low sample size.
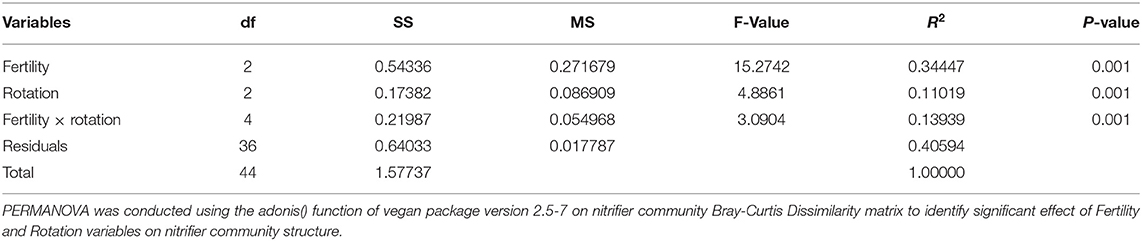
Microbial community data was analyzed using phyloseq (40) and vegan package v. 2.57 (41). Richness parameters were calculated using phyloseq package v. 1.36 (40). The complete OTU table was subset by Nitrifier taxa (Order: Nitrosomonadales, Nitrososphaerales, Nitrospirales). Nitrifier beta-diversity was calculated using a Non-metric Multiscale Dimensional Analysis (NMDS) on the Bray-Curtis dissimilarity matrix using the metaMDS() function of vegan package v. 2.57. Chemical variables were fit onto the NMDS matrix to identify significant drivers of matrix structure using the envfit() function from the vegan package v. 2.57. PERMANOVA analysis was conducted on the dissimilatory matrix to identify the influence of fertility and rotation on matrix structure using the adonis() function of the vegan package v. 2.57.
Results
Long-Term Fertilization and Rotation Impact on Potential Nitrification
The influence of fertility and rotational management strategies on potential nitrification rates and nitrifying microbial communities was assessed. Nitrification data is reported in Supplementary Table 1. Two-way ANOVA analyses (Table 1) concluded that potential nitrification was significantly influenced by fertility [2-Way ANOVA: F(2, 37) = 29.8442, P < 0.0001], rotation [F(2, 37) = 3.3028, P = 0.0478], and the interaction of both [2-Way ANOVA: F(4, 37) = 5.624, P = 0.001221]. The highest nitrification potential was in the MLP-COA plots, followed by IN-CC and IN-COA (Figure 1). Regression analyses identified the chemical and physical drivers significantly influencing the transformed nitrification rates (Figure 2). (R2 = 0.1347, P = 0.0218), (R2 = 0.2453, P = 0.0023), OM (R2 = 0.2155, P = 0.0034), Est-N-Released (R2 = 0.2155, P = 0.0034), Ca+2 (R2 = 0.1106, P = 0.03801), Mg+2 (R2 = 0.1575, P = 0.0131), Na+ (R2 = 0.1574, P = 0.0131), B (R2 = 0.2445, P = 0.0023), and CEC (R2 = 0.3084, P = 0.0009) positively influenced potential nitrification rates, individually (Figure 2; Table 2). Copper (Cu+2) content negatively impacted potential nitrification rates (R2 =0.0957, P = 0.0516).
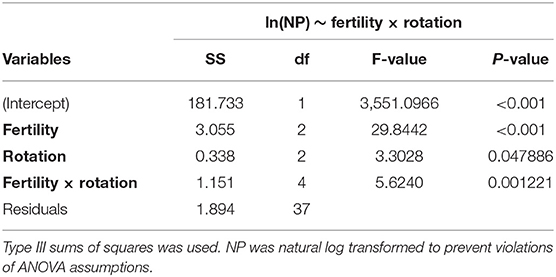
Table 1. Two-way analysis of variance (ANOVA) table for model assessing the influence of long-term fertility (MLP, IN, UF) and rotation (CC, CS, COA) on natural log transformed nitrification potential (NP) activity in the Morrow Plots.
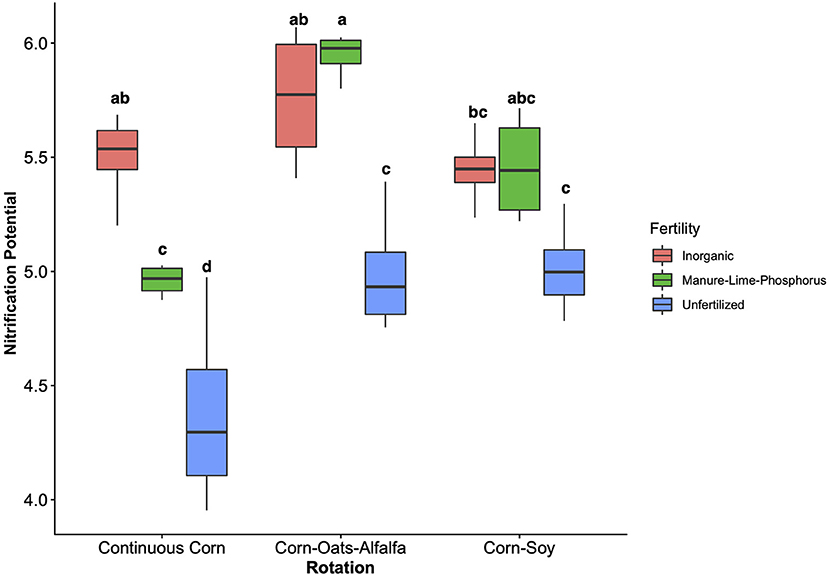
Figure 1. Nitrification potential by management treatment (fertility and rotation). Nitrification potential is measured in μg NO2-N g−1 DM−1 hr−1 and natural log transformed to assume normality. Tukey's HSD was used for separations of group means, and group membership is indicated via lettering on top of each bar. Model coefficients and P-values are reported in Table 1. Fertilization and rotational treatments are as follows: manure-lime-phosphorus (MLP), inorganic urea fertilizer (IN), unfertilized (UF); corn-oat-alfalfa (COA), corn-soy (CS), continuous corn (CC).
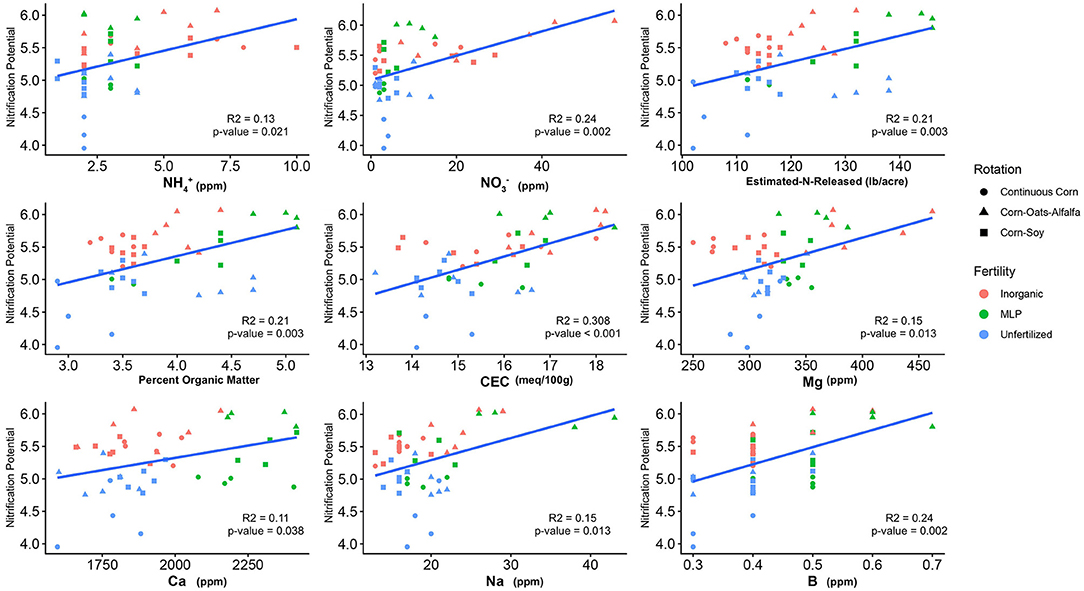
Figure 2. Relationship between nitrification potential and soil edaphic variables. Only edaphic factors which significantly influenced nitrification potential are reported. Model coefficients and P-values are reported in Table 2.
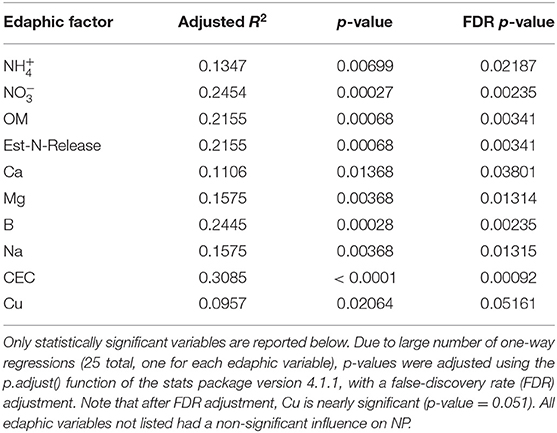
Table 2. Results of one-way linear regressions assessing the influence of individual edaphic variables on natural-log transformed nitrification potential (NP) activity.
The influence of fertility and rotation on nitrifier microbial communities' abundance was assessed using qPCR of BamoA and AamoA genes (Figure 3; Table 3). Similar to the potential nitrification rates, BamoA gene copy number was significantly influence by fertilizer [2-Way ANOVA: F(2, 36) = 56.4657, P <0.0001], and the interaction of fertility and rotation [2-Way ANOVA: F(4, 35) = 9.1299, P <0.0001], but not by rotation [F(2, 36) = 0.3767, P = 0.6888]. The highest BamoA gene copy numbers were within the IN-CC, IN-COA, and MLP-COA treatments (Figure 3A). BamoA abundances significantly influenced potential rates (Supplementary Figure 7), based on a one-way linear regression between natural log-transformed BamoA copy number/ng DNA and nitrification potential activity. AamoA did not vary by fertility or rotation (Figure 3B), and did not influence potential nitrification rates (P > 0.05).
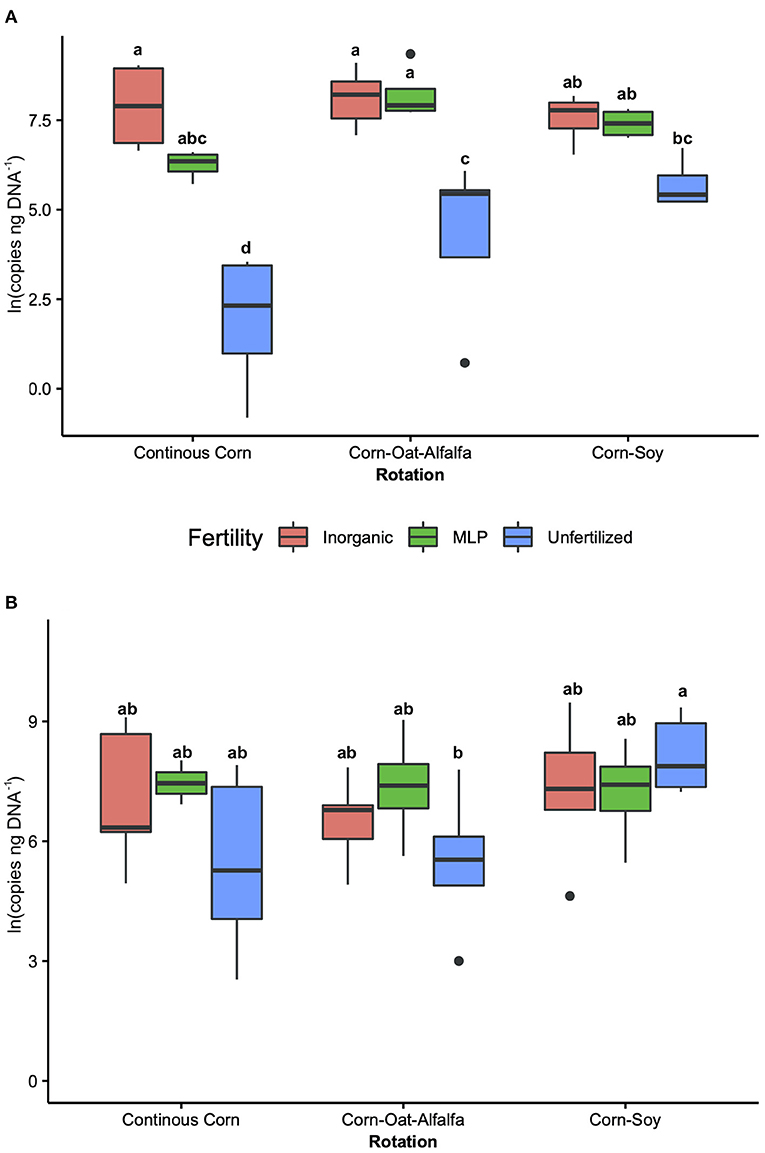
Figure 3. Bacterial (BamoA) and archaeal (AamoA) shown in A and B, respectively, compared among long-term management treatments (fertility and rotation). Abundance, determined by amoA-specific qPCR, is reported as gene copy number/ng DNA. Tukey's HSD was used for separations of group means, and group membership is indicated via lettering on top of each bar. 2-way ANOVA results are reported in Table 3. Fertilization and rotational treatments are as follows: manure-lime-phosphorus (MLP), inorganic urea fertilizer (IN), unfertilized (UF); corn-oat-alfalfa (COA), corn-soy (CS), continuous corn (CC).
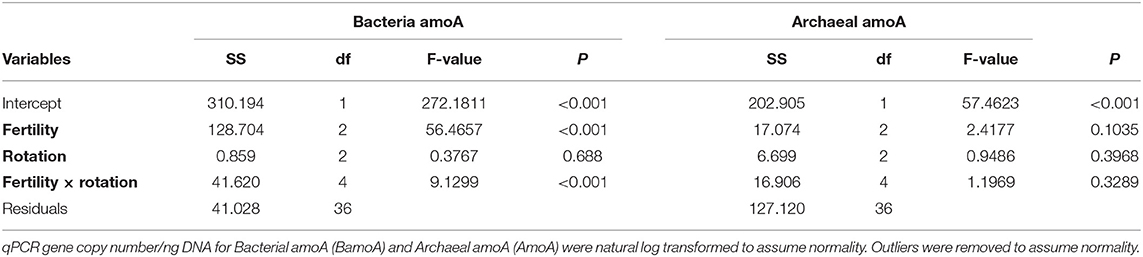
Table 3. Two-way Analysis of Variance (ANOVA) for model assessing the influence of fertility and rotation on quantitative polymerase chain reaction (qPCR) ammonium oxidation gene abundances.
Long-Term Fertility and Rotation Impact on Nitrifying Microbial Communities
16S rRNA V4 amplicon sequencing revealed shifts in the nitrifier community alpha and beta diversity. Observed nitrifier richness was significantly influenced by fertilizer [F(2, 36) = 3.6893, P = 0.03487], and the interaction of fertilizer and rotation [F(2, 36) = 3.2706, P = 0.02188]. Chao1 richness was not influenced by fertility, rotation, or the interaction of the two. Shannon Diversity Index was significantly impacted by fertility [2-Way ANOVA: F(2, 36) =13.97, P < 0.0001]. A pairwise comparison revealed no significant differences between inorganic and manure treatment Shannon values, regardless of rotation. The primary differences in Shannon index values were between inorganic and unfertilized rotations, as well as manure and unfertilized rotations (Supplementary Figure 6). While Chao1 index was the only richness index not influenced by fertility and rotation, all three indices influenced potential nitrification rates. Observed richness was nearly significant in positively influencing potential nitrification rates [F(1, 43) = 3.957, P = 0.05206]. Chao1 [F(1, 43) = 6.806, P = 0.01245], and Shannon [F(1, 43) = 14.7883, P = 0.0003926] positively influenced nitrification potential.
Effects of management on beta-diversity was analyzed using non-metric multidimensional scaling (NMDS) of the Bray-Curtis dissimilatory matrix (Figure 4), paired with a PERMANOVA analyses (Table 4). The NMDS ordination (Stress = 0.1319) was significantly influenced by fertility [PERMANOVA: R2 (2, 36) = 0.344, P = 0.001], rotation [R2 (2, 36) = 0.11019, P = 0.001] and the interaction of both [R2 (4, 36) = 0.13939, P = 0.001]. Chemical and physical variables were fit onto the ordination to identify the variables significantly influencing the ordination structure. In agreement with the multiple regressions against the potential nitrification rates, (R2 = 0.1418, P = 0.038), (R2 = 0.1474, P = 0.034), OM (R2 = 0.3037, P = 0.001), Est-N-Release (R2 = 0.3037, P = 0.001), Ca+2 (R2 = 0.4483, P = 0.001), Mg+2 (R2 = 0.5245, P = 0.001), B (R2 = 0.6175, P = 0.001), Na+ (R2 = 0.3164, P = 0.001), and CEC (R2 = 0.3725, P = 0.001) influenced the NMDS ordination (Table 5). However, community composition was additionally influenced by soil pH (R2 = 0.2110, P = 0.007), S (R2 = 0.1729, P = 0.022), Mg-Saturation (R2 = 0.1677, P = 0.021), Na-Saturation (R2 = 0.2411, P = 0.004), and H-saturation (R2 = 0.1743, P = 0.015). Within the bulk soil nitrifier community, AOA within the phylum Crenarcheota had the largest abundances (Figure 5), yet regression analysis identified no relationship between total abundance of Order Nitrososphaerales and NP (Supplementary Figure 3; P > 0.05). Of the bacterial nitrifiers, the genus Nitrospira (NOB) had highest relative abundances within all manure plots, compared to the other fertility treatments (Supplementary Figure 5). Linear regression identified Order Nitrospirales as positively influencing NP (Supplementary Figure 4). Nitrosovibrio (AOB) had the highest relative abundances within the inorganic fertilizer treatments, with remarkably lower abundances in the manure and unfertilized plots (Supplementary Figure 5). Order Nitrosomonadales also significantly influenced NP, but non-linearly, following a non-linear regression (Supplementary Figure 2).
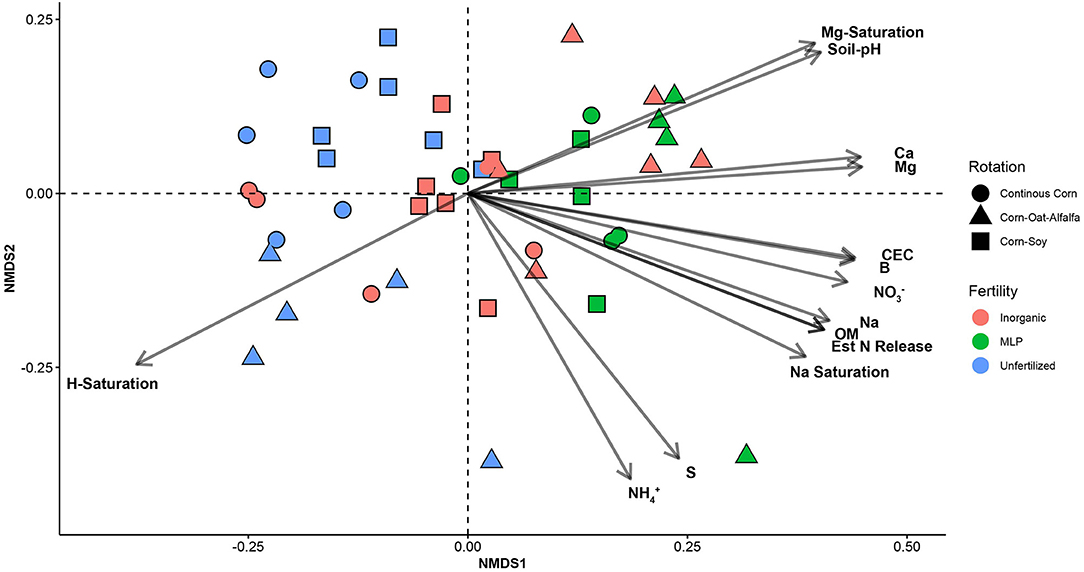
Figure 4. Non-metric multidimensional scaling (NMDS) ordination constructed from a Bray-Curtis dissimilarity matrix of the soil nitrifier community, based on 16S rRNA sequence reads assigned to nitrifier taxa. Edaphic factors were correlated to NMDS1 and NMDS2 using the envfit() function of the vegan package in R. Only significant factors are displayed for visual clarity.
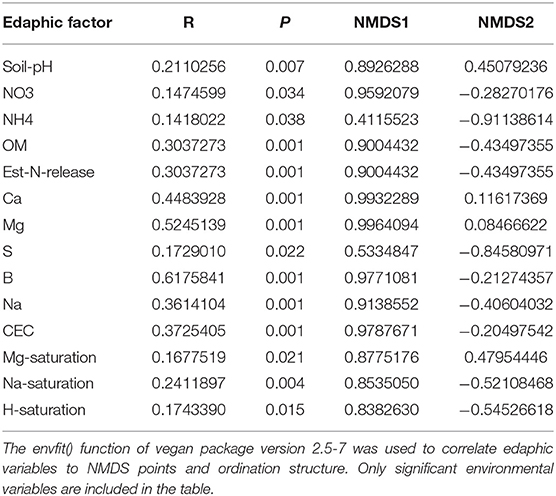
Table 5. Influence of edaphic factors on non-metric multidimensional scaling (NMDS) ordination constructed using the 16S rRNA-based nitrifier Bray-Curtis dissimilarity matrix.
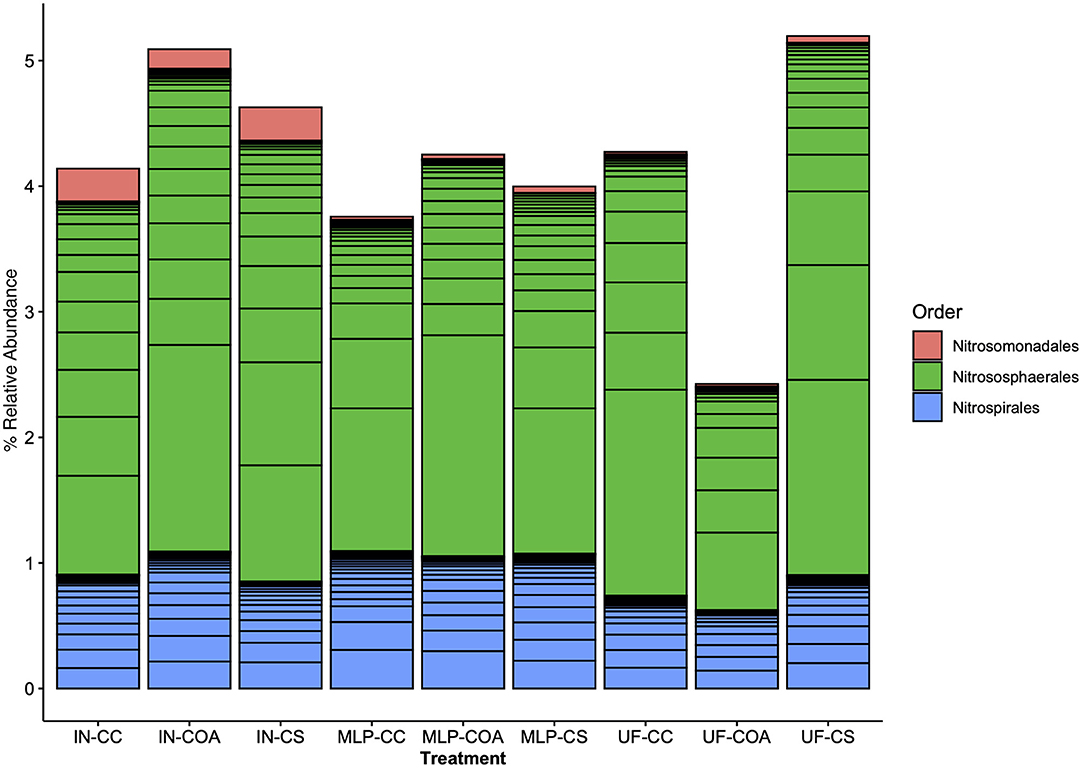
Figure 5. Nitrifier composition compared among long-term agronomic management treatments, represented as percent relative abundance of the total bulk soil 16S rRNA-based community. The bulk soil bacterial 16S rRNA community was subset by nitrifier taxa (AOA, AOB, NOB). Results are represented here with color representing order-level taxonomic classifications: Nitrosomonadales (AOB), Nitrososphaerales (AOA), Nitrospirales (NOB). Fertilization and rotational treatments are as follows: manure-lime-phosphorus (MLP), inorganic urea fertilizer (IN), unfertilized (UF); corn-oat-alfalfa (COA), corn-soy (CS), continuous corn (CC).
Discussion
This study explored the edaphic and management drivers of nitrification potential (NP) and nitrifier community structure in the Morrow Plots long-term agricultural experiment. Fertilizer and rotation significantly influence NP, as well as nitrifier community structure and evenness. The most striking observation was the enriched NP in the MLP-COA treatment, when compared to IN-CS and IN-COA treatments. The IN fertilizer in the Morrow Plots is urea based; urea is hydrolyzed to NH3 and CO2, acting as a source of both energy (ammonium oxidation) and biomass (carbon fixation) yielding substrates (42). However, these results suggest the complexity and diversity of soil physiochemistry under regenerative management promotes nitrifier growth and activity in dynamic, and potentially stimulatory, ways.
Nitrogen losses through soil nitrification have been a major focus in agronomy and microbial ecology for over a century (43–45). Yet, many of the long-term drivers of nitrification remain a major topic of exploration, due to the spatiotemporal variation in NP and nitrifier communities, and the influence of soil type, but also due to the difficulty studying this fastidious functional group. Many studies report AOB as more responsive to anthropogenic perturbations (46, 47). AOB are also reported as disproportionately contributing to soil nitrification activity (12, 47). Identifying the selective agents that determine the tradeoffs between AOB and AOA communities is of major importance for understanding controls on nitrification (48). AOA have higher substrate affinity for (49), and it is hypothesized that this higher substrate affinity allows AOA to persist at lower concentrations in oligotrophic environments, resisting the aggressive agriculture-induced variations in content (50, 51). This may be species-dependent, however as a novel AOA taxon has recently been discovered to withstand -rich environments (52). Additionally, nitrifiers have a wide variety of substrate affinities (53). In this study, Crenarcheota was the largest phylum of nitrifiers, yet AamoA gene abundance and AOA (Order Nitrosophaerales) total abundance did not influence NP. Additionally, archaeal amoA copy number was not significantly influenced by long-term fertilizer or crop rotation treatments. Future research should prioritize identifying and validating the contributions of AOA to agricultural nitrogen biogeochemistry. Due to their lack of response to the long-term agricultural treatments in this study, the remainder of the discussion will focus on AOB and NOB.
Exploring the Impact of Manure on Nitrification
Numerous studies have explored the long-term influence of agronomic management on nitrification and nitrifier communities (12, 54, 55). Large-scale ecological filters, such as organic matter (%OM), pH, and are among the most well-documented drivers of nitrification rates (56, 57). Our study unsurprisingly identified as a significant factor influencing NP and nitrifier community composition. The primary step of nitrification is the oxidation of ammonia by AMO, yielding two electrons and reducing O2 to H2O, and is the rate-limiting step of nitrification. Many studies have identified content in soils as a dominant driver of nitrification and nitrifier niche differentiation (2, 46, 48, 58, 59). As stated above, variations in enzymatic affinity to among AOB, NOB, and comammox microorganisms influence their success in oligotrophic or copiotrophic environments (2, 53). However, availability is strongly influenced by agricultural management, human-driven manipulations of soil physiochemistry, such as pH and CEC, as well as microbial resource competition. These interconnected processes require more precise methods to analyze and model predictable relationships that can inform soil management, but it is clear that is a strong predictor of NP in agricultural soils.
Carbon quantity and quality are a fundamental difference induced by management practices (e.g., fertilizer regime and crop rotation) between industrial and regenerative agriculture. Carbon inputs from manure and crop residues promote soil matrix stabilization through increased soil surface area and water holding capacity. This, in turn, maximizes the buffering capacity of the soil matrix, stabilizing pH fluctuations which may alter abiotic and biotic components. pH fluctuations dictate the ratio of NH3/ (60), which is the primary reason that nitrifiers are consistently reported as sensitive to pH fluctuations. Soil matrix pH levels, in turn, influence AOB community distributions (pH: R2 = 0.2110, P = 0.007; H-saturation: R2 = 0.1743, P = 0.015). Interestingly, neither soil-pH, nor buffer-pH, significantly influenced NP, suggesting pH drives nitrifier species distributions more than function.
Organic matter incorporation through regenerative practices would also promote heterotrophy and microbial biomass accumulation (61). Labile carbon from manure amendments stimulates microbial biomass and enzyme activity (62). Microbial biomass may promote N-immobilization and could reduce NP (63). Yet, N-mineralization may be stimulated due to the high urea content and lower C:N ratio in manures when compared to the unfertilized treatments (64, 65). However, as the IN treatments are directly fertilized with urea, it is unlikely urease activity, alone, contributed to the high NP in the MLP treatments. Certain Nitrospira (NOB/Comammox) possess genes encoding cyanase and urease enzymes (66). The dynamics of cyanate availability have only recently been explored (67), and research suggests that soil microorganisms rapidly consume cyanate, when compared to urea hydrolysis (67). AOB can utilize liberated CO2 and NH3, a term called “reciprocal feeding” between AOB and Nitrospira (66). Nitrospira can form symbioses with AOB within biofilms, occupying microsites called nitrification aggregates (68). While reciprocal feeding was not measured in this study, it may contribute to the comparable NP between the MLP-COA and MLP-CS, and the IN-COA and IN-CS treatment, as well as the higher relative abundance of Nitrospira in the MLP-COA treatment (69). This study also identified a direct relationship between Nitrospira abundance and NP (Supplementary Figure 4), determined using 16S rRNA gene sequencing, implicating Nitrospira in ammonium oxidation potential.
Ammonium oxidizers require CO2 for carbon fixation (4, 70). Promotion of not only ammonium oxidation, but also carbon fixation, would benefit nitrifiers, maintaining nitrifier biomass and overall soil nitrification potential (71). Heterotrophic respiration and the release of CO2 supply nitrifiers with carbon for growth. Additionally, NOB can utilize both the Calvin-Benson cycle and the reductive TCA cycle for carbon fixation (5, 72); the enzymes of the reductive TCA cycle are more sensitive to oxygen, forcing certain NOB to occupy microaerophilic sites in soil (72). This may further promote NOB occupation of biofilms within microsites; shifts in water retention and labile carbon inputs stimulate biofilm formation through exopolysaccharide production (73). Nitrification potential has been reported to be greatest in the clay fraction of soil, suggesting the physiochemical properties of microaggregates (74) compared to macroaggregates, benefit nitrification (75). In biofilm reactors, the nitrification rate was particularly high even at low pH, suggesting that biofilms are ideal environments for autotrophic nitrification (76). Moreover, biomass aggregation was associated with stress avoidance in Nitrosomonas mobilis Ms1 and in late stages of aggregation was associated with an upregulation in biosynthesis genes (77). Nitrosomonadaceae and Nitrospiraceae have recently been characterized as important exopolysaccharide producers under alfalfa regimes cultivated on reclaimed soils (78). As Vuko et al. (78) did not compare alfalfa cultivation to other legumes, it is uncertain if alfalfa has a unique capacity for supporting nitrifier biofilm production. Since biofilms contribute to soil structure, as well as microbial microhabitats, future research should expand on the findings of Vuko et al. (78) to investigate long-term management shifts in biofilms and importance for AOB and NOB in agricultural soils.
Micronutrients Influence on Nitrification and Nitrifiers
Long-term field experiments offer unique insight and opportunities to evaluate drivers of soil microorganisms and their activities. Over time, soil physiochemical properties which seem irrelevant to nitrification may emerge as important drivers of nitrifier distribution and function. Nitrifiers are particularly fastidious and are sensitive to micronutrients and non-growth substrates (79), making them a particularly difficult functional guild to study. Organic matter-induced changes in CEC would promote the accumulation of cations, which over time, could influence nitrification (80). In this study, (R2 = 0.1347, P = 0.0218), Ca+2 (R2 = 0.1106, P = 0.03801), Mg+2 (R2 = 0.1575, P = 0.0131), and Na+ (R2 = 0.1574, P = 0.0131) positively influenced NP. Prior research identified as a primary driver of nitrification in soil (8); this is unsurprising, considering ammonium oxidation is the primary and rate-limiting step of nitrification (8). However, few studies assess the impact of additional micronutrients within soils, although engineered systems, such as wastewater treatment plants (WWTP), have explored micronutrient drivers of NP for decades. Therefore, this section will discuss the role of micronutrients on nitrification. It is important to note that the mode of action is not identified during this study, particularly because the biological activity of metal cations is dependent on the form in which these cations exist in the soil matrix, i.e., occluded, exchangeable, organic bound, etc. (81), which is heavily influenced by physiochemical controls such as pH (82). Moreover, many of these cations influence soil aggregation (18), and biofilm formation (83), so it is not clear if they directly act on nitrifier cellular machinery or indirectly through altering their microhabitats. Ultimately, this section serves to explore literature that supports the findings of this study, and to pose avenues of exploration for future research.
Calcium positively influenced NP in this study. The manure treatments were neutralized with the addition of lime (CaCO3/limestone). Nitrifiers can use CaCO3 as a biomass substrate for adherence, as well as a buffer for pH during reactor cultivation (84). The interaction between nitrifiers and calcium promotes high ammonia-removal rates (85). It is also reported that liming promotes CH4-oxidation, the effect of which is dependent on soil type and acidification (86). Methanotrophs are phylogenetically (87) and enzymatically (15, 88) related to ammonium oxidizers, causing substrate infidelity between ammonium monooxygenase and particulate-methane monooxygenase (88). It is therefore possible that liming also promotes ammonium oxidation, due to similar mechanisms as the liming-induced methane-oxidation stimulation (86), but the mechanism remains inconclusive.
Nitrosovibrio is a genus of AOB isolated from oligotrophic environments, such as building sandstone, and is associated with biodegradation of natural building materials (89). They represent a very small percent of the bulk soil microbial community (ranging from 0.021 to 0.265% relative abundance), but are enriched within all the inorganically fertilized treatments—most particularly the IN-CC (0.262%) and IN-CS (0.265%). The soil within the Morrow Plots is a Flanagan Silt loam formed over calcareous glacial till (25). Presence of a small percentage of Nitrosovibrio may indicate soil acidification and promotion of soil erosion, as their presence in building material is associated with acidification and salt stress of calcareous material. Shi et al. (90) identify soil salt content as a major driver of Nitrosovibrio abundance (90). In addition to calcium, Na+ was a significant driver of nitrifier community structure (R2 = 0.3164, P = 0.001), in agreement with Shi et al. (90), but did not influence NP, suggesting that salt stress enriches specific nitrifying genera.
Magnesium (Mg2+) is present in soils in an exchangeable and mobile form (91). Magnesium influences the coordination chemistry of nucleoside triphosphates and is obligatory for maximum activity of succinyl-CoA synthetase (SCS) activity in Nitrosomonas europeae pure culture (92). Succinyl-CoA synthetase (SCS) produces one ATP via substrate-level phosphorylation during the TCA cycle (92). In the same study, Cu2+ had a strong capacity for SCS inhibition (92). The results of this study agree with Kondo et al. (92). After P-values were corrected with a false-discovery test, Cu2+ was nearly significant in negatively influencing nitrification potential (P = 0.051). While research regarding the inhibitory effect of copper on nitrification has yielded variable results (93), Mertens et al. (94) saw a positive correlation between nitrification inhibition and Cu2+concentrations in soils (94). Cu2+ has also been shown to decrease soil urease activity (95, 96), and influence AOB community structure (97). More recently, manure application was shown to increase heavy metal concentrations in soils, with Cu and Cd negatively correlating to net nitrification rates (20). Therefore, it is possible that the high cation exchange capacity in the MLP-COA promotes Cu2+accumulation and negatively influenced nitrifiers, whereas Mg2+ positively influence nitrifier growth. It is also important to note that Cu restores the specific growth rate of AOA inhibited by organic carbon substrates in WWTP (98). The differential effect of Cu on AOB and AOA is potentially due to AOA possessing Cu-dominant catalytic centers within electron transport enzymes (99). This implies the negative relationship between Cu and NP in our study is specific to AOB. However, the mode of action cannot be determined with this study.
Sulfur contributes to the activity of numerous redox metalloproteins by complexing with metal ions in the catalytic site (100). For example, numerous nitrogen-cycling metalloenzymes, like assimilatory nitrate reductase, periplasmic nitrate reductase, and the nitrogenase enzyme, have molybdenum catalytic sites complexed to cysteine ligands (101, 102). While sulfur would be required for the replication of N-cycling redox enzymes, sulfur within the Morrow Plots could contribute to niche differentiation between AOB and AOA. In agreement, sulfur influenced beta-diversity, but not NP (R2 = 0.1729, P = 0.022). A recent analysis identified a correlation between sulfatase activity and AOA community abundances, potentially due to their ability to adapt to hypoxic ecosystems, like those seen in marine ecosystems (103). Moreover, in reactors, AOB ammonium oxidization is particularly sensitive to hydrogen sulfide (103, 104). Sulfur was also identified as a significant factor influencing BamoA terminal restriction fragment analyses in tropical soils, along with Cu, Na, and B (97). Therefore, agricultural manipulation of sulfur may drive the differentiation of nitrifier community structures.
Finally, our results demonstrate that boron (B) significantly influenced both NP and nitrifier beta-diversity. Boron is an essential micronutrient that is required for cell wall synthesis and proper cellular replication in plants (105), and is important for cyanobacteria heterocyst stability. In fact, B plays a crucial role in legume nodulation, and is important for nodule membrane and cell wall structure, nodule infection, and the development of the symbiosome during legume-rhizobia symbioses (106, 107). In alfalfa, it is particularly important for reproductive phenology and seed quality and yield (108). Regenerative fertilization management approaches often accumulate B through time (109), but B availability can interact with calcium from liming to reduce B assimilation into plant biomass (110). Adsorption of B increases with soil pH (111), reducing B availability with liming due to neutralization of soil pH and complexation with calcium ions (105). The comparatively larger pool of B in the MLP-COA (Table 2) could be due to the combination of liming and pH buffering, promoting B accumulation in this treatment. Boron accumulation may influence microbial respiration and N-liberating activity, as B has been shown to increase urease and dehydrogenase activity in soil (112), as well as nitrate reductase activity (113). Boron also has been shown to influence nitrification by increasing nitrifying bacterial populations when applied with molybdenum (Mo), and had a strong effect on nitrification activity when applied without Mo (113). Additional research is required to dissect the synergistic effects of alfalfa rotations, liming, and B accumulation on nitrification and nitrifying microbial communities.
Study Limitations
The most significant limitation of this study is the lack of spatiotemporal resolution. The samples during this preliminary study were collected in June and August, but due to the low number of samples (and lack of in-field replication that reflects modern statistical methods), we could not assess the intra-annual variation in nitrification potential, nitrifier communities, or the edaphic drivers. Biotic factors, such as microbial enzyme potential and community structure, vary significantly spatiotemporally (8, 114, 115). Particularly, N-mineralization (116) and urease enzyme activity (117) increase with temperature, resulting in late-season pulses in these enzyme activities. These two processes contribute to ammonium availability, potentially impacting nitrification potential through time. Moreover, nitrification potential differs among soil particle fractions and depths (118) as well as temperature (119). This highlights the importance of assessing the long-term effect of abiotic variables, in addition to single-season effects, as fertilization and rotational practices significantly alter the physical structure of soil (18). Future sample collection should include a finer scale temporal resolution to understand the interaction of various N-cycling enzyme activity and nitrifier communities.
It is important to note that nitrification potential is not the same metric as field nitrification (120). This is important to distinguish because it is unclear if sustainable practices such as manure fertilization or tri-rotational regimes promote field nitrifier-induced N loss (12). The manipulation of carbon, nitrogen, and the promotion of aggregation (121) influences anaerobic microsites and anaerobic respiration strategies, e.g., denitrification. The presence of crop varieties such as legume species (122) also influences the factors driving field N-loss (56). However, due to the oxygen requirement of nitrification, it is uncertain if the high nitrification potential in the MLP-COA corresponds to increased nitrogen loss from these treatments (13, 63, 123, 124). Both denitrification and nitrifier denitrification (125, 126) contribute to global N2O emissions (127). Recent studies have shed light on dissimilatory nitrate reduction to ammonium (DNRA) as a competitor for available , particularly under rewetted soils high in labile carbon inputs and water-filled pore space (128, 129). It is therefore uncertain whether the high nitrification potential in the MLP-COA treatment increases N2O emissions through nitrifier denitrification and respiration by denitrifiers, or stimulates DNRA due to an increase in heterotrophic respiration and lowering of soil redox potential (128). These questions require precise analytical methods, such as 15N pool dilutions or soil transcriptomics analyses.
Conclusion
This study identified a significant influence of long-term rotation and fertilization on nitrification potential in the Morrow Plots. Surprisingly, the most regenerative management treatment (MLP-COA) possessed the greatest capacity for nitrification. While this study did not employ precise methods like 15N-isotope tracer analyses to pinpoint the N-cycling processes supporting the high nitrification potential, it does point to the influence of numerous abiotic macro- and micronutrients on both nitrification potential and nitrifier community structure. As agronomic management practices greatly alter soil matrix structure through time, the resulting variation in physiochemical parameters may slowly shift nitrifier communities. Moreover, chemical constituents which impact both ammonium oxidation and carbon fixation enzymes could partially explain the distinct nitrifier communities and their resulting activities. Understanding these slow-acting distal drivers of soil nitrification and how they vary through time is critical for predicting the long-term outcome of agronomic practices on soil health and sustainability.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA789310.
Author Contributions
SR: writing and data analysis. AK: writing—review and editing. CS: study design, sampling, and laboratory assays. YM: qPCR and amplicon sequencing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Cooperative State Research, Education, and Extension Service, US Department of Agriculture, under Project Number ILLU 875-374. This work was supported by a UIUC Graduate College Fellowship to SR. CS was supported by the National Science Foundation through the Science, Engineering, and Education for Sustainability Post-Doctoral Fellowship, Award # 1314064. YM was supported by funding from the National Institute of Food and Agriculture, U.S. Department of Agriculture, under Award Number 2015-67019-23584.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Robert Dunker and previous managers and the University of Illinois Agricultural Experiment Station for long-term support and maintenance of the Morrow Plots. We are very grateful to Alonso Favela, Dora Cohen, Maya Scott, Natalie Stevenson, and Dongfang Li for technical assistance with sample collection and laboratory assays.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsoil.2022.838497/full#supplementary-material
References
1. Bienes R, Marques MJ, Sastre B, García-Día AZ, Esparza I, Antón O, et al. Tracking changes on soil structure and organic carbon sequestration after 30 years of different tillage and management practices. Agronomy. (2021) 11. doi: 10.3390/agronomy11020291
2. Lehtovirta-Morley LE. Ammonia oxidation: ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol Lett. (2018) 365:1–9. doi: 10.1093/femsle/fny058
3. Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. (2002) 178:250–5. doi: 10.1007/s00203-002-0452-0
4. Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. (2011) 108:4170–5. doi: 10.1073/pnas.1010981108
5. Alfreider A, Grimus V, Luger M, Ekblad A, Salcher MM, Summerer M. Autotrophic carbon fixation strategies used by nitrifying prokaryotes in freshwater lakes. FEMS Microbiol Ecol. (2018) 94:1–12. doi: 10.1093/femsec/fiy163
6. Ren M, Zhang Z, Wang X, Zhou Z, Chen D, Zeng H, et al. Diversity and contributions to nitrogen cycling and carbon fixation of soil salinity shaped microbial communities in Tarim Basin. Front Microbiol. (2018) 9:431. doi: 10.3389/fmicb.2018.00431
7. Gelfand I, Robertson GP. Mitigation of greenhouse gases in agricultural ecosystems. In: Hamilton SK, Doll JE, Robertson GP, editors. The Ecology of Agricultural Landscapes: Long-Term Research on the Path to Sustainability. New York, NY: Oxford University Press (2015). p. 310–39. Available online at: https://books.google.com/books?hl=en&lr=&id=gPFxBgAAQBAJ&oi=fnd&pg=PA310&dq=info:b60_93ehv_UJ:scholar.google.com&ots=MNm385kwgW&sig=ZVnYvVBc8ngcHNz-T5QN-E1FlTs#v=onepage&q&f=false
8. Ouyang Y, Norton JM, Stark JM. Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol Biochem. (2017) 113:161–72. doi: 10.1016/j.soilbio.2017.06.010
9. Chen Z, Luo X, Hu R, Wu M, Wu J, Wei W. (2010). Impact of long-term fertilization on the composition of denitrifier communities based on nitrite reductase analyses in a paddy soil. Microb Ecol. (2010) 60:850–61. doi: 10.1007/s00248-010-9700-z
10. Barton L, Gleeson DB, Maccarone LD, Zúñiga LP, Murphy DV. Is liming soil a strategy for mitigating nitrous oxide emissions from semi-arid soils? Soil Biol Biochem. (2013) 62:28–35. doi: 10.1016/j.soilbio.2013.02.014
11. Robertson GP, Groffman PM. Nitrogen transformations. Soil Microbiol Ecol Biochem. (2015) 421–46. doi: 10.1016/B978-0-12-415955-6.00014-1
12. Wang F, Chen S, Wang Y, Zhang Y, Hu C, Liu B. Long-term nitrogen fertilization elevates the activity and abundance of nitrifying and denitrifying microbial communities in an upland soil: implications for nitrogen loss from intensive agricultural systems. Front Microbiol. (2018) 9:2424. doi: 10.3389/fmicb.2018.02424
13. Meng L, Ding W, Cai Z. Long-term application of organic manure and nitrogen fertilizer on N2O emissions, soil quality and crop production in a sandy loam soil. Soil Biol Biochem. (2005) 37:2037–45. doi: 10.1016/j.soilbio.2005.03.007
14. Gross A, Glaser B. Meta-analysis on how manure application changes soil organic carbon storage. Sci Rep. (2021) 11:1–13. doi: 10.1038/s41598-021-82739-7
15. Bedard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4, and CO oxidation by methanotrophs and nitrifiers. Microbiology. (1989) 53:68–84. doi: 10.1128/mr.53.1.68-84.1989
16. Lauchnor EG, Semprini L. Inhibition of phenol on the rates of ammonia oxidation by Nitrosomonas europaea grown under batch, continuous fed, and biofilm conditions. Water Res. (2013) 47:4692–700. doi: 10.1016/j.watres.2013.04.052
17. Hyman MR, Page CL, Arp DJ. Oxidation of methyl fluoride and dimethyl ether by ammonia monooxygenase in Nitrosomonas europaea. Appl Environ Microbiol. (1994) 60:3033–5. doi: 10.1128/aem.60.8.3033-3035.1994
18. Bronick CJ, Lal R. Soil structure and management: a review. Geoderma. (2005) 124:3–22. doi: 10.1016/j.geoderma.2004.03.005
19. Nath M, Bhatt D, Bhatt MD, Prasad R, Tuteja N. Microbe-mediated enhancement of nitrogen and phosphorus content for crop improvement. In: Prasad R, Gill SS, Tuteja N, editors. Crop Improvement Through Microbial Biotechnology. Elsevier (2018). p. 293–304. doi: 10.1016/B978-0-444-63987-5.00014-1
20. Ali MM, Khanom A, Nahar K, Ali MY, Azad MAK, Rahman MM. Effect of manure application on net nitrification rates, heavy metal concentrations and nitrifying archaea/bacteria in soils. Bull Environ Contam Toxicol. (2021) 106:707–13. doi: 10.1007/s00128-021-03112-y
21. Gupta AK, Singh RP, Singh A, Ibrahim MH. Effects of heavy metal and metalloid contamination on the soil microbial response: an overview. Microb Ecol Trop Soils. (2011) 44:303–18.
22. Roane TM, Pepper IL, Gentry TJ. Microorganisms and Metal Pollutants. 2nd ed. London; California; Massachusetts: Elsevier Inc. (2014).
23. Soman C, Li D, Wander MM, Kent AD. Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil. (2017) 413:145-59. doi: 10.1007/s11104-016-3083-y
24. Odell RT, Walker W, Boone L, Oldham M. The Morrow Plots: a century of learning. Bulletin-Agricultural Experiment Station, College of Agriculture, University of Illinois at Urbana-Champaign. Agric Exp Station Coll Agric Univ Illinois Urbana Champaign Urbana. (1982) 775:775. doi: 10.5962/bhl.title.16691
25. Aref S, Wander MM. Long-term trends of corn yield and soil organic matter in different crop sequences and soil fertility treatments on the morrow plots. Adv Agron. (1997) 62:153–97. doi: 10.1016/S0065-2113(08)60568-4
26. Kandeler E. Potential nitrification. In: Methods in Soil Biology, editors Schinner F, Kandeler E, Ohlinger R, dans Margesin R. Berlin: Spinger-Verlag Berlin Heidelberg (1995). p. 146–9.
27. Schinner F, Öhlinger R, Kandeler E, Margesin RTATT. Methods in Soil Biology. NV-1 onl. Berlin, Heidelberg: Springer Berlin Heidelberg (1996).
28. Berg P, Rosswall T. Ammonium oxidizer numbers, potential and actual oxidation rates in two swedish arable soils. Biol Fertil Soils. (1985) 1:131–40. doi: 10.1007/BF00301780
29. Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press (2001).
30. Rotthauwe JH, Witzel KP, Liesack W. The ammonia monooxygenase structural gene amoa as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. (1997) 63:4704–12. doi: 10.1128/aem.63.12.4704-4712.1997
31. Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. (2008) 10:2966–78. doi: 10.1111/j.1462-2920.2008.01701.x
32. Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
33. Gordon A, Hannon G. FASTX-Toolkit (2010). Available online at: http://hannonlab.cshl.edu/fastx_toolkit/index.html
34. Reddy TBK, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, et al. (2015). The Genomes OnLine Database (GOLD) v.5: A metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. (2015) 43:D1099–106. doi: 10.1093/nar/gku950
35. Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRna gene sequences from microbial communities. Curr Protoc Bioinforma. (2011) 1–20. doi: 10.1002/0471250953.bi1007s36
36. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
37. McDonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. (2012) 6:610–8. doi: 10.1038/ismej.2011.139
38. R Core Team. R: A Language Environment for Statistical Computing. R Found. Stat. Comput. Vienna, Austria 3 (2021). Available online at: https://www.r-project.org/
40. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
41. Oksanen AJ, Blanchet FG, Friendly M, Kindt R, Legendre P, Mcglinn D, et al. Vegan: Community Ecology Package. R Package Version 2.5-7. CRAN (2020). p. 2395–2396.
42. Antonious GF, Turley ET, Dawood MH. Monitoring soil enzymes activity before and after animal manure application. Agric. (2020) 10:1–12. doi: 10.3390/agriculture10050166
43. Warington R. IV. - On nitrification. J Chem Soc Trans. (1878) 33:44–51. doi: 10.1039/CT8783300044
44. Lees H. Effect of copper-enzyme poisons on soil nitrification. Nature. (1946) 4003:97. doi: 10.1038/158097a0
45. Meiklejohn J. Minimum phosphate and magnesium requirements of nitrifying bacteria. Nature. (1952) 170:1131. doi: 10.1038/1701131a0
46. Carey CJ, Dove NC, Beman JM, Hart SC, Aronson EL. Meta-analysis reveals ammonia-oxidizing bacteria respond more strongly to nitrogen addition than ammonia-oxidizing archaea. Soil Biol Biochem. (2016) 99:158–66. doi: 10.1016/j.soilbio.2016.05.014
47. Wang J, Wang J, Rhodes G, He JZ, Ge Y. Adaptive responses of comammox Nitrospira and canonical ammonia oxidizers to long-term fertilizations: implications for the relative contributions of different ammonia oxidizers to soil nitrogen cycling. Sci Total Environ. (2019) 668:224–33. doi: 10.1016/j.scitotenv.2019.02.427
48. Hatzenpichler R. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl Environ Microbiol. (2012) 78:7501–10. doi: 10.1128/AEM.01960-12
49. Martens-Habbena W, Berube PM, Urakawa H, De La Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. (2009) 461:976–9. doi: 10.1038/nature08465
50. Di HJ, Cameron KC, Shen JP, Winefield CS, O'Callaghan M, Bowatte S, et al. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. (2010) 72:386–94. doi: 10.1111/j.1574-6941.2010.00861.x
51. Straka LL, Meinhardt KA, Bollmann A, Stahl DA, Winkler MKH. Affinity informs environmental cooperation between ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (Anammox) bacteria. ISME J. (2019) 13:1997–2004. doi: 10.1038/s41396-019-0408-x
52. Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C, et al. Isolation of “Candidatus Nitrosocosmicus franklandus”, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol. (2016) 92:1–10. doi: 10.1093/femsec/fiw057
53. Jung MY, Sedlacek CJ, Kits KD, Mueller AJ, Rhee SK, Hink L, et al. Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. (2021) 16:272–83. doi: 10.1038/s41396-021-01064-z
54. Enwall K, Nyberg K, Bertilsson S, Cederlund H, Stenström J, Hallin S. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biol Biochem. (2007) 39:106–15. doi: 10.1016/j.soilbio.2006.06.015
55. Chu H, Fujii T, Morimoto S, Lin X, Yagi K. Population size and specific nitrification potential of soil ammonia-oxidizing bacteria under long-term fertilizer management. Soil Biol Biochem. (2008) 40:1960–3. doi: 10.1016/j.soilbio.2008.01.006
56. Roberston G. Nitrification and denitrification in humid tropical ecosystems: potential controls on nitrogen retention. Miner Nutr Trop For Savanna Ecosyst. (1989) 473:55–69.
57. Tao R, Wakelin SA, Liang Y, Chu G. Response of ammonia-oxidizing archaea and bacteria in calcareous soil to mineral and organic fertilizer application and their relative contribution to nitrification. Soil Biol Biochem. (2017) 114:20–30. doi: 10.1016/j.soilbio.2017.06.027
58. Ke X, Angel R, Lu Y, Conrad R. Niche differentiation of ammonia oxidizers and nitrite oxidizers in rice paddy soil. Environ Microbiol. (2013) 15:2275–92. doi: 10.1111/1462-2920.12098
59. Guo J, Ling N, Chen H, Zhu C, Kong Y, Wang M, et al. Distinct drivers of activity, abundance, diversity and composition of ammonia-oxidizers: evidence from a long-term field experiment. Soil Biol Biochem. (2017) 115:403–14. doi: 10.1016/j.soilbio.2017.09.007
60. Suzuki I, Dular U, Kwok SC. Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol. (1974) 120:556–8. doi: 10.1128/jb.120.1.556-558.1974
61. Chu H, Lin X, Fujii T, Morimoto S, Yagi K, Hu J, et al. Soil microbial biomass, dehydrogenase activity, bacterial community structure in response to long-term fertilizer management. Soil Biol Biochem. (2007) 39:2971–6. doi: 10.1016/j.soilbio.2007.05.031
62. Stark C, Condron LM, Stewart A, Di HJ, O'Callaghan M. Influence of organic and mineral amendments on microbial soil properties and processes. Appl Soil Ecol. (2007) 35:79–93. doi: 10.1016/j.apsoil.2006.05.001
63. Gao S, Zhou G, Liao Y, Lu Y, Nie J, Cao W. Contributions of ammonia-oxidising bacteria and archaea to nitrification under long-term application of green manure in alkaline paddy soil. Geoderma. (2020) 374:114419. doi: 10.1016/j.geoderma.2020.114419
64. Luxhøi J, Elsgaard L, Thomsen IK, Jensen LS. Effects of long-term annual inputs of straw and organic manure on plant N uptake and soil N fluxes. Soil Use Manag. (2007) 23:368–73. doi: 10.1111/j.1475-2743.2007.00126.x
65. Azeez JO, Van Averbeke W. Nitrogen mineralization potential of three animal manures applied on a sandy clay loam soil. Bioresour Technol. (2010) 101:5645–51. doi: 10.1016/j.biortech.2010.01.119
66. Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci USA. (2015) 112:11371–6. doi: 10.1073/pnas.1506533112
67. Mooshammer M, Wanek W, Jones SH, Richter A, Wagner M. Cyanate is a low abundance but actively cycled nitrogen compound in soil. Commun Earth Environ. (2021) 2:1–10. doi: 10.1038/s43247-021-00235-2
68. Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. (2016) 24:699–712. doi: 10.1016/j.tim.2016.05.004
69. Li X, Han S, Wan W, Zheng L, Chen W, Huang Q. Manure fertilizes alter the nitrite oxidizer and comammox community composition and increase nitrification rates. Soil Tillage Res. (2020) 204:104701. doi: 10.1016/j.still.2020.104701
70. Wendeborn S. The chemistry, biology, and modulation of ammonium nitrification in soil. Angew Chemie Int Ed. (2020) 59:2182–202. doi: 10.1002/anie.201903014
71. Smith JM, Mosier AC, Francis CA. Spatiotemporal relationships between the abundance, distribution, and potential activities of ammonia-oxidizing and denitrifying microorganisms in intertidal sediments. Microb Ecol. (2015) 69:13–24. doi: 10.1007/s00248-014-0450-1
72. Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. (2010) 107:13479–84. doi: 10.1073/pnas.1003860107
73. Davey ME, O'toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. (2000) 64:847–67. doi: 10.1128/MMBR.64.4.847-867.2000
74. Li P-P, Han Y-L, He J-Z, Zhang S-Q, Zhang L-M. Soil aggregate size and long-term fertilization effects on the function and community of ammonia oxidizers. Geoderma. (2019) 338:107–17. doi: 10.1016/j.geoderma.2018.11.033
75. Zhang Q, Liang G, Myrold DD, Zhou W. Variable responses of ammonia oxidizers across soil particle-size fractions affect nitrification in a long-term fertilizer experiment. Soil Biol Biochem. (2017) 105:25–36. doi: 10.1016/j.soilbio.2016.11.005
76. Tarre S, Green M. 2004). High-rate nitrification at low pH in suspended- and attached-biomass reactors. Appl Environ Microbiol. (2004) 70:6481–7. doi: 10.1128/AEM.70.11.6481-6487.2004
77. Isshiki R, Fujitani H, Tsuneda S. Transcriptome analysis of the ammonia-oxidizing bacterium Nitrosomonas mobilis Ms1 reveals division of labor between aggregates and free-living cells. Microbes Environ. (2020) 35:1–9. doi: 10.1264/jsme2.ME19148
78. Vuko M, Cania B, Vogel C, Kublik S, Schloter M, Schulz S. Shifts in reclamation management strategies shape the role of exopolysaccharide and lipopolysaccharide-producing bacteria during soil formation. Microb Biotechnol. (2020) 13:584–98. doi: 10.1111/1751-7915.13532
79. Fujitani H, Nomachi M, Takahashi Y, Hasebe Y, Eguchi M, Tsuneda S. Successful enrichment of low-abundant comammox Nitrospira from nitrifying granules under ammonia-limited conditions. FEMS Microbiol Lett. (2020) 367:1–8. doi: 10.1093/femsle/fnaa025
80. Smith JH. (1964). Relationships between soil cation-exchange capacity and the toxicity of ammonia to the nitrification process. Soil Sci Soc Am J. (1964) 28:640-4. doi: 10.2136/sssaj1964.03615995002800050019x
81. Gadd GM, Griffiths AJ. Microorganisms and heavy metal toxicity. Microb Ecol. (1977) 4:303–17. doi: 10.1007/BF02013274
83. Ansari FA, Jafri H, Ahmad I, Abulreesh HH. Factors affecting biofilm formation in in vitro and in the rhizosphere. Biofilms Plant Soil Heal. (2017) 275–90. doi: 10.1002/9781119246329.ch15
84. Green M, Ruskol Y, Tarre S, Loewenthal RE. Nitrification utilizing CaCO3 as the buffering agent. Environ Technol. (2002) 23:303–8. doi: 10.1080/09593332508618410
85. Sun M, Wang H, Zhang H. Effect of Ca2+ and Fe3+ addition on nitrification-denitrification process in a membrane bioreactor. Adv Mater Res. (2013) 610–3:422–7. doi: 10.4028/www.scientific.net/AMR.610-613.422
86. Kunhikrishnan A, Thangarajan R, Bolan NS, Xu Y, Mandal S, Gleeson DB, et al. Functional relationships of soil acidification, liming, and greenhouse gas flux. In: Advances in Agronomy. London; California, CA; Massachussets, MA; Oxford: Academic Press Inc. (2016). p. 1–71.
87. Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. (1995) 132:203–8. doi: 10.1111/j.1574-6968.1995.tb07834.x
88. Suzuki I, Kwok SC, Dular U. Competitive inhibition of ammonia oxidation in Nitrosomonas europaea by methane, carbon monoxide or methanol. FEBS Lett. (1976) 72:117–20. doi: 10.1016/0014-5793(76)80825-3
89. Meincke M, Krieg E, Bock E. (1989). Nitrosovibrio spp., the dominant ammonia-oxidizing bacteria in building sandstone. Appl Environ Microbiol. (1989) 55:2108–10. doi: 10.1128/aem.55.8.2108-2110.1989
90. Shi Y, Liu X, Zhang Q, Gao P, Ren J. Biochar and organic fertilizer changed the ammonia-oxidizing bacteria and archaea community structure of saline–alkali soil in the North China Plain. J Soils Sediments. (2019) 20:12–23. doi: 10.1007/s11368-019-02364-w
91. Gransee A, Führs H. Magnesium mobility in soils as a challenge for soil and plant analysis, magnesium fertilization and root uptake under adverse growth conditions. Plant Soil. (2013) 368:5–21. doi: 10.1007/s11104-012-1567-y
92. Kondo H, Ohmori T, Shibata H, Sase K, Takahashi R, Tokuyama T. Thermostable succinyl-Coenzyme A synthetase from Nitrosomonas europaea ATCC 25978: purification and properties. J Ferment Bioeng. (1995) 79:499–502. doi: 10.1016/0922-338X(95)91270-F
93. Premi PR, Cornfield AH. Effects of addition of copper, manganese, zinc and chromium compounds on ammonification and nitrification during incubation of soil. Plant Soil. (1969) 31:345–52. doi: 10.1007/BF01373578
94. Mertens J, Wakelin SA, Broos K, Mclaughlin MJ, Smolders E. Extent of copper tolerance and consequences for functional stability of the ammonia-oxidizing community in long-term copper-contaminated soils. Environ Toxicol Chem. (2010) 29:27–37. doi: 10.1002/etc.16
95. Doelman P, Haanstra L. Short- and long-term effects of heavy metals on urease activity in soils. Biol Fertil Soils. (1986) 2:213–8. doi: 10.1007/BF00260846
96. Hemida SK, Omar SA, Abdel-Mallek AY. Microbial populations and enzyme activity in soil treated with heavy metals. Water Air Soil Pollut. (1997) 95:13–22. doi: 10.1007/BF02406152
97. De Gannes V, Eudoxie G, Hickey WJ. Impacts of edaphic factors on communities of ammonia-oxidizing archaea, ammonia-oxidizing bacteria and nitrification in tropical soils. PLoS ONE. (2014) 9:e89568. doi: 10.1371/journal.pone.0089568
98. Gwak JH, Jung MY, Hong H, Kim JG, Quan ZX, Reinfelder JR, et al. Archaeal nitrification is constrained by copper complexation with organic matter in municipal wastewater treatment plants. ISME J. (2020) 14:335–46. doi: 10.1038/s41396-019-0538-1
99. Reyes C, Hodgskiss LH, Kerou M, Pribasnig T, Abby SS, Bayer B, et al. Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J. (2020) 14:2659–74. doi: 10.1038/s41396-020-0715-2
100. Sparacino-Watkins C, Stolz JF, Basu P. Nitrate and periplasmic nitrate reductases. Chem Soc Rev. (2014) 43:676–706. doi: 10.1039/C3CS60249D
101. Gani B. Enzyme chemistry. Annu Rep Prog Chem Sect B Org Chem. (1986) 83:303–30. doi: 10.1039/oc9868300303
102. Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, et al. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem Rev. (2014) 114:4366–9. doi: 10.1021/cr400479b
103. Zhang Q, Zhou W, Liang G, Sun J, Wang X, He P. Distribution of soil nutrients, extracellular enzyme activities and microbial communities across particle-size fractions in a long-term fertilizer experiment. Appl Soil Ecol. (2015) 94:59–71. doi: 10.1016/j.apsoil.2015.05.005
104. Sears K, Alleman JE, Barnard JL, Oleszkiewicz JA. Impacts of reduced sulfur components on active and resting ammonia oxidizers. J Ind Microbiol Biotechnol. (2004) 31:369–78. doi: 10.1007/s10295-004-0157-2
105. Arunkumar B, Thippeshappa G, Anjali M, Prashanth K. Boron : a critical micronutrient for crop growth and productivity. J Pharmacogn Phytochem. (2018) 7:2738–41.
106. Redondo-Nieto M, Rivilla R, El-Hamdaoui A, Bonilla I, Bolaños L. Boron deficiency affects early infection events in the pea-Rhizobium symbiotic interaction. Aust J Plant Physiol. (2001) 28:819–23. doi: 10.1071/PP01020
107. Bolaños L, Lukaszewski K, Bonilla I, Blevins D. Why boron? Plant Physiol Biochem. (2004) 42:907–12. doi: 10.1016/j.plaphy.2004.11.002
108. Chen L, Xia F, Wang M, Wang W, Mao P. Metabolomic analyses of alfalfa (Medicago sativa L. cv ‘Aohan') reproductive organs under boron deficiency and surplus conditions. Ecotoxicol Environ Saf. (2020) 202:111011. doi: 10.1016/j.ecoenv.2020.111011
109. Bulluck LR, Brosius M, Evanylo GK, Ristaino JB. Organic and synthetic fertility amendments influence soil microbial, physical and chemical properties on organic and conventional farms. Appl Soil Ecol. (2002) 19:147–60. doi: 10.1016/S0929-1393(01)00187-1
110. Tsadilas CD, Kassioti T, Mitsios IK. Influence of liming and nitrogen forms on boron uptake by tobacco. Commun Soil Sci Plant Anal. (2005) 701–8. doi: 10.1081/CSS-200043348
111. Dhaliwal SS, Naresh RK, Mandal A, Singh R, Dhaliwal MK. Dynamics and transformations of micronutrients in agricultural soils as influenced by organic matter build-up: a review. Environ Sustain Indic. (2019) 1–2:100007. doi: 10.1016/j.indic.2019.100007
112. Bilen S, Bilen M, Bardhan S. The effects of boron management on soil microbial population and enzyme activities. African J Biotechnol. (2011) 10:5311–9. doi: 10.4314/AJB.V10I27
113. Sun T, Wang YP, Wang ZY, Liu P, Xu GD. The effects of molybdenum and boron on the rhizosphere microorganisms and soil enzyme activities of soybean. Acta Physiol Plant. (2013) 35:763–70. doi: 10.1007/s11738-012-1116-6
114. Lauber CL, Ramirez KS, Aanderud Z, Lennon J, Fierer N. Temporal variability in soil microbial communities across land-use types. ISME J. (2013) 7:1641–50. doi: 10.1038/ismej.2013.50
115. Jeong J, Bolan N, Kim C. Heterotrophic soil respiration affected by compound fertilizer types in red pine (Pinus densiflora S. et Z) stands of Korea. Forests. (2016) 7:309. doi: 10.3390/f7120309
116. Cabrera ML, Kissel DE, Vigil M. Nitrogen mineralization from organic residues: researchh opportunities. J Environ Qual. (2005) 34:75–9. doi: 10.2134/jeq2005.0075
117. Xu JG, Heeraman DA, Wang Y. Fertilizer and temperature effects on urea hydrolysis in undisturbed soil. Biol Fertil Soils. (1993) 16:63–5. doi: 10.1007/BF00336517
118. Lu X, Nicol GW, Neufeld JD. Differential responses of soil ammonia-oxidizing archaea and bacteria to temperature and depth under two different land uses. Soil Biol Biochem. (2018) 120:272–82. doi: 10.1016/j.soilbio.2018.02.017
119. Taylor AE, Myrold DD, Bottomley PJ. Temperature affects the kinetics of nitrite oxidation and nitrification coupling in four agricultural soils. Soil Biol Biochem. (2019) 136:107523. doi: 10.1016/j.soilbio.2019.107523
120. Dick RP. A review: long-term effects of agricultural systems on soil biochemical and microbial parameters. Agric Ecosyst Environ. (1992) 40:25–36. doi: 10.1016/0167-8809(92)90081-L
121. Kiani M, Hernandez-Ramirez G, Quideau S, Smith E, Janzen H, Larney FJ, et al. Quantifying sensitive soil quality indicators across contrasting long-term land management systems: crop rotations and nutrient regimes. Agric Ecosyst Environ. (2017) 248:123–35. doi: 10.1016/j.agee.2017.07.018
122. Rochette P, Angers DA, Bélanger G, Chantigny MH, Prévost D, Lévesque G. Emissions of N2O from Alfalfa and Soybean Crops in Eastern Canada. Soil Sci Soc Am J. (2004) 68:493–506. doi: 10.2136/sssaj2004.4930
123. Menéndez S, López-Bellido RJ, Benítez-Vega J, González-Murua C, López-Bellido L, Estavillo JM. Long-term effect of tillage, crop rotation and N fertilization to wheat on gaseous emissions under rainfed Mediterranean conditions. Eur J Agron. (2008) 28:559–69. doi: 10.1016/j.eja.2007.12.005
124. Duan Y, Xu M, Gao S, Liu H, Huang S, Wang B. Long-term incorporation of manure with chemical fertilizers reduced total nitrogen loss in rain-fed cropping systems. Sci Rep. (2016) 6:33611. doi: 10.1038/srep33611
125. Wrage N, Velthof GL, Van Beusichem ML, Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem. (2001) 33:1723–32. doi: 10.1016/S0038-0717(01)00096-7
126. Wrage-Mönnig N, Horn MA, Well R, Müller C, Velthof G, Oenema O. The role of nitrifier denitrification in the production of nitrous oxide revisited. Soil Biol Biochem. (2018) 123:A3–A16. doi: 10.1016/j.soilbio.2018.03.020
127. Prosser JI, Hink L, Gubry-Rangin C, Nicol GW, Gubry-Rangin C, Nicol GW. Nitrous oxide production by ammonia oxidizers: Physiological diversity, niche differentiation and potential mitigation strategies. Glob Chang Biol. (2020) 26:103–18. doi: 10.1111/gcb.14877
128. Friedl J, De Rosa D, Rowlings DW, Grace PR, Müller C, Scheer C. Dissimilatory nitrate reduction to ammonium (DNRA), not denitrification dominates nitrate reduction in subtropical pasture soils upon rewetting. Soil Biol Biochem. (2018) 125:340–9. doi: 10.1016/j.soilbio.2018.07.024
Keywords: crop rotation, microbial community composition, nitrification, fertilizer treatment, ammonium oxidizing bacteria (AOB), ammonium oxidizing archaea (AOA), nitrite-oxidizing bacteria (NOB)
Citation: Raglin SS, Soman C, Ma Y and Kent AD (2022) Long Term Influence of Fertility and Rotation on Soil Nitrification Potential and Nitrifier Communities. Front. Soil Sci. 2:838497. doi: 10.3389/fsoil.2022.838497
Received: 17 December 2021; Accepted: 07 February 2022;
Published: 14 March 2022.
Edited by:
Patricia Dorr De Quadros, University of Waterloo, CanadaReviewed by:
Bharati Kollah, Indian Institute of Soil Science (ICAR), IndiaYang Ouyang, University of Oklahoma, United States
Shigeto Otsuka, The University of Tokyo, Japan
Copyright © 2022 Raglin, Soman, Ma and Kent. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela D. Kent, YWtlbnRAaWxsaW5vaXMuZWR1
†Present addresses: Chinmay Soman, EarthSense, Inc., Urbana, IL, United States
Yanjun Ma, DeWitt, LLP, Madison, WI, United States
 Sierra S. Raglin
Sierra S. Raglin Chinmay Soman†
Chinmay Soman† Angela D. Kent
Angela D. Kent