
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Soil Sci. , 24 June 2022
Sec. Plant-Soil Interactions
Volume 2 - 2022 | https://doi.org/10.3389/fsoil.2022.829575
Shrub encroachment is a common phenomenon in grasslands all over the world. However, little is known about the consequences of shrub encroachment on soil microbial community structure in different layers. We investigated the effects of three common shrub encroachment (Potentilla fruticosa, Spiraea alpina, and Caragana microphylla) on grassland soil bacterial communities at the surface and deep layers in Qinghai–Tibetan Plateau. 16S rRNA gene sequencing was used to investigate the bacterial communities, and Fourier translation infrared spectroscopy (FTIR) was conducted to assess the soil organic carbon (SOC) chemical composition in surface and deep layers of shrub-encroached alpine grassland. Shrub encroachment has significantly increased SOC degradation in deep layer. After shrub invasion, the bacterial alpha-diversity in the surface and deep soil was higher than in grassland soil (except for the surface layer of C. microphylla). Factors driving bacterial community changes in soil surface and deep layer were different. Among the soil properties that were measured, SOC content was the primary factor that altered soil bacterial community composition in surface soil, while SOC chemical composition (aromatic and polysaccharides) was the main driver in the deep layer. A total of 39 and 42 biomarkers were found by linear discriminant analysis (LDA) effect size (LEfSe) in the surface and deep soil layer among the four sampling groups, respectively. Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) showed that the most abundant predicted functional genes belonged to categories of metabolism (52.83%) in the primary metabolic pathway. Redundancy analysis (RDA) results also showed that the key factors affecting bacterial metabolic function appear to be SOC, pH, and aromatics, which are largely consistent with those affecting community composition. We suggest that shrub encroachment affect the structure, diversity, and predicted functions of bacterial communities, thus affecting the C cycle in this region.
Grasslands around the world are experiencing a rapid transition from herbaceous to shrub dominance, a phenomenon known as shrub encroachment (SE) (1, 2). This process is driven by many factors, such as climate change, gas/nutrient cycling, human management, edaphic characteristics, and moisture, which vary from different regions (3). Changes in the dominance of herbaceous to woody plants alter primary production, plant distribution, and root depth, which may potentially persist to several meters belowground (4), and nutrient cycling and carbon storage (5). Soil organic carbon (SOC) is the largest reservoir of organic carbon in terrestrial ecosystems, and the effects of shrub encroachment on the soil organic carbon cycle are particularly important (6). The topsoil has more extensive organic carbon content than deep soil; hence, it plays an important role in the global C cycle (7). Previous studies showed inconsistent results of the effect of shrub encroachment on SOC, reporting an increased SOC storage (8), while some showed that it remained constant (9) or decreased (10). These discrepancies may be primarily attributed to climate, vegetation types, soil properties, historical land-use patterns, and scales (10, 11). Therefore, exploring the impact of shrub encroachment on microbial community in the Qinghai–Tibetan Plateau can help us further understand the response of soil carbon pool to shrub encroachment in alpine grassland.
Soil microorganisms are sensitive to the changes in the external environment, including dynamics of plant community composition (12). Vegetation types are the primary determinants of soil microbial community structural and functional diversity to different degrees (13). Plant community shapes bacterial communities through inputting litter and root exudates into the soil (14). Furthermore, different root exudates will largely contribute to the heterogeneity of microorganisms. Mixing of woody and herbaceous plants’ litter increased the chemical complexity of the matrix pool and further increased the microbial niche space and diversity (15). With the deeper and broader roots, the shrub can transport organic carbon from the surface layer to the deeper soil (16), potentially increasing the diversity of the bacterial community in the deeper soil. Microorganisms in deeper soils have a greater influence on soil-forming processes than topsoil due to their proximity to soil parent material (17). Soil properties are highly heterogeneous in the profile, which can be seen at different soil depths. The topsoil is rich in available carbon substrates due to the input of root exudates, litter, and root detritus. In contrast, the input ratio of C to the deep layer is generally lower (18). Soil C is generally divided into labile C (easy to mineralize, with short turnover time) and recalcitrant C (not easy to mineralize, with long turnover time) (19). Generally, according to SOM availability, the soil microbiome can be classified into oligotrophic and copiotrophic categories (20). The oligotrophics are characterized by their ability to grow at low substrate concentrations and generally have higher substrate utilization efficiency. In contrast, the copiotrophics are more sensitive to available carbon sources (21). Although the effects of shrub encroachment on aboveground and belowground communities have been extensively studied, little is known about the effects of shrub encroachment on deep soil microbial communities and the vertical spatial distribution of soil microbial community structure.
SOC mainly derives from plant litter (i.e., shoot and root litter) and root exudates (22). Plant litter comprises polysaccharides, aromatic compounds, and aliphatic compounds (23). Previous research has demonstrated that the quality and quantity of plant litters contribute to the accumulation of organic carbon at the surface (24), while root litter and root deposits are the main carbon sources in deep soils. The above- and below-ground inputs of plants are often different in chemical composition and quantity (23), which may affect C accumulation in the soil profile. Recalcitrant plant inputs lead to higher carbon stocks, while labile plant inputs lead to lower carbon stocks (25, 26). Soil carbon cycling is ultimately the result of the growth and activities of microorganisms (27), like abundance, diversity, or community composition (28). For example, dilution of soil suspension leads to a decline in microbial diversity, reducing the degradation of readily degradable and persistent decomposition of carbon sources in grasslands (29). Most soil microorganisms rely on carbon decomposition for energy, and a recent meta-analysis based on a global scale concluded that SOC content is a key driver of soil bacterial diversity (30, 31). Moreover, some microbial groups also participate in the decomposition of soil organic matter, causing changes in soil organic carbon composition and ultimately affecting soil CO2 emission (32). There are significant differences in carbon dynamics and biological processes between surface and deep soil due to vertical variations in substrate quality and microbial activity with soil depth (7). However, the changes in SOC in the soil profile after shrub encroachment in the Qinghai–Tibetan Plateau are still unclear, and there is still a lack of understanding of the relationship between SOC and soil bacterial communities.
Qinghai–Tibetan Plateau (QTP) is the highest plateau on earth, the largest single geographical unit in the world, and is known as the “Third Pole” (33). This region is also considered as the sensitive area for global climate change and an initiation zone for climate change in China. The alpine grassland is the dominant ecosystem on the QTP, accounting for more than 50% of the total area of the plateau (34). Similarly, grasslands on the QTP are encroached by shrubs. Previous studies have focused on the effects of shrub encroachment on aboveground and soil microorganisms in arid and semi-arid regions. The effects of shrub encroachment on soil microbial communities may vary with different climates, vegetation, and soil types (35). However, the relationship between shrub encroachment and the soil microbial community in different soil layers in alpine grassland remains largely unknown. Therefore, in this study, we use high-throughput techniques to investigate the diversity and composition of bacterial communities and their potential functions in the surface (0–10 cm) and deep (50–60 cm) layers of three common shrubs (Potentilla fruticosa, Spiraea alpina, and Caragana microphylla) on the Tibetan Plateau. Our major hypotheses were as follows: (1) the shrub encroachment would not significantly change the soil organic carbon content in surface and deep layers; (2) the shrub encroachment would increase the diversity of soil microbial communities and alter the soil microbial composition in the surface and deep layers; (3) the shrub encroachment would alter the carbon cycle of alpine grassland in Qinghai–Tibetan Plateau by changing the composition and predicted functions of the bacterial communities.
The study was conducted near the Qinghai–Tibetan Plateau Research Station of Southwest Minzu University (N 32°49'38", E 102°34'21"), located in Hongyuan County, Sichuan Province, China (Figure 1). The region is located on the eastern edge of the Qinghai–Tibetan Plateau, a transition zone from the mountainous region to the plateau in the northwest of Sichuan. The altitude of the study area is 3,500 m, the average annual temperature is 0.9°C, and the average annual precipitation is 690 mm (36). The soil type is alpine shrub meadow soil, and the parent material of soil is slope deposit. S. alpina, C. microphylla, and P. fruticosa are three shrubs growing in the sampling area for 20 years. The main species in the grassland matrix belong to the Gramineae family.
Soil samples were collected at 18 shrub-encroached and 6 grassland matrixes in early August 2019. In this study, we selected three typical shrubs (S. alpina, C. microphylla, and P. fruticosa) and a grassland matrix without shrub growth as a control. Each matrix is 100 m apart. The area of each plot is about 50 m×50 m. Each plot has six small quadrants of 1m×1m (Figure 1). The surface layer (0–10 cm) and deep layer (50–60 cm) soils were collected within each plot. Using the 5-point approach (four vertices and the center of plots), soil from grassland plots was collected and then mixed as one sample for control soils. Each shrub-encroached soils were collected under six shrub canopies (near the roots), which were closest to the four vertices and the center of the plot, and then mixed as a sample (Supplementary Figure S1). A total of 48 samples, 12 (6 sites × 2 layers) from control grassland and 36 (3 species ×6 sites × 2 layers) from shrub-encroached soils were analyzed in this study.
To evaluate the soil pH, the soil was sieved (2 mm mesh) to carefully remove fine roots, seeds, and plant materials after air drying. Soil pH was determined with acidity meter, and the soil to water ratio was 2.5:1. The remaining soil samples were divided into two sub-samples and sieved (0.125 and 0.15 mm) to determine the SOC content and chemical composition, respectively. The SOC content as determined using an Elementar Variomax CNS Analyzer (Elementar Corp., Hannau, Germany). Fourier translation infrared spectroscopy (FTIR) can be combined with high-throughput gene sequencing to clarify the relationship between soil microbial community structure and SOC chemical composition. The soil/97%KBr mix (1:80 w/w) was homogenized by grinding with an agate mortar and pestle. The spectral range was 4,000–400 cm−1 and an average of 16 scans with a resolution of 4 cm−1 for each sample. The relative peak area (S, %) was calculated as the ratio of the area of an obviously reflected peak (e.g., 3,435, 2,930, 1,620, or 1,034 cm−1) to the sum of all four selected peaks (37). The S provided an estimate of the relative abundance of different chemical composition in SOC: aliphatic (2,930 cm−1), aromatic (1,620 cm−1), polysaccharides (1,034 cm−1), and alcohol (3,435 cm−1) (38).
Index 1 is a relative peak area ratio of aliphatic and aromatic to polysaccharides and alcohol, an increase of which is thought to be associated with greater recalcitrance of SOC (39). S is the relative peak area; each number represents the corresponding wavelength of the characteristic peak (cm−1).
Microbial DNA was extracted using the PowerSoil® DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA) from 0.25 g of fresh soil according to the manufacturer’s protocols. After that, agarose gel and Nanodrop (Nanodrop 2000, Thermo Scientific) were used to detect the purity and concentration of DNA. The extracted DNA was diluted to 10 ng/μl with sterile water and stored at −40°C.
Using the diluted genomic DNA as a template, the V4–V5 region of 16S rRNA gene was amplified using the primers (forward primers 515F:5’-GTGCCAGCMGCCGCGGTAA-3’ and reverse primers 909R:5’-CCCCGYCAATTCMTTTRAGT-3’) (40). PCR was carried out in 25 μl reaction mixtures containing 2× Taq MasterMix, 12.5 μl; forward primer, 1 μl, reverse primer, 1 μl; ddH2O, 9.5 ul; DNA (10ng/μl), 1 μl. The PCR amplification program included initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 40 s, 56°C for 60 s, and 72°C for 60 s, and a final extension at 72°C for 10 min (41). Two PCR reactions were performed for each sample and combined after amplification. The PCR products were evaluated using 1% agarose gel electrophoresis, the target band was recovered with the DNA Gel Extraction Kit, and the concentration and quality were determined by Nanodrop. The samples were mixed according to the concentration of PCR-recovered products. Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit. After Qubit quantification, the libraries were sequenced using Illumina NovaSeq 6000 (2×250 bp).
Sequences were merged using FLASh (V1.2.7, http://ccb.jhu.edu/software/FLASH/) (42). The merged sequences with high quality (reads length >300 bp, without ambiguous base “N,” and average base quality score >30) were used for further analysis. The aligned 16S rRNA gene sequences were used for a chimera check using the Uchime algorithm (43). After removing the chimera, effective tags were obtained for the subsequent analysis. The effective tags of each sample were clustered into an operational taxonomic unit (OTU) matrix using USEARCH (http://drive5.com/usearch) at a 97% similarity threshold (44). Sequences annotated as chloroplasts or Viridiplantae were removed. Representative sequences for each OTU were selected using the command “pic_rep_set.py” in QIIME (V1.9.0) (45), and singletons were also removed. QIIME was used to annotate OTU representative sequences, and Silva V132 was used as the reference database (46). Finally, the OTU table was rarefied to a depth of 10,595 reads per sample. Alpha diversity, including Shannon and observed species, and beta diversity based on Unifrac distance were calculated by QIIME. The original sequence data are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) by accession number PRJNA801100, and additional data are available from the corresponding author on reasonable request.
For the differences in soil variables, including soil pH, SOC content, SOC chemical composition, bacterial composition, and alpha diversity, the Shapiro–Wilk normality test and Levene’s test were used to test the normality and homoscedasticity of the data, respectively. When the data could meet the normality and homoscedasticity criteria simultaneously, ANOVA and Tukey’s honestly significant difference (HSD) comparisons were used to test the significance of difference among four groups, or Wilcoxon’s test was used in SPSS version 22.0. Redundancy analysis (RDA) was employed using Canoco (version 5.0) to evaluate the relationship between soil properties and the relative abundance of bacteria and bacterial function. The differences in bacterial community composition among groups were analyzed by principal coordinate analysis (PCoA) with Unifrac distances and permutational multivariate analysis of variance (PERMANOVA; permutations = 999) using R v.3.6.1. The microbial biomarkers were obtained by the linear discriminate analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/galaxy) using the Kruskal–Wallis test to determine the significant different taxa among four groups (47). LDA was performed to evaluate the difference of each microbial taxon with a threshold value of 4.0. PICRUSt (http://huttenhower.sph.harvard.edu/galaxy) was used to predict functional characteristics that may be conveyed by bacterial communities. The functional genes were identified from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. However, there are some limitations when interpreting PICRUSt predictions. For example, the ability of PICRUSt detection patterns depends on the input data used. If the marker gene sequences used between strains are the same, the software cannot distinguish the variation at the strain level; if these genes are not included in the input genome data used, or if the path annotation is currently poor, the software cannot detect gene families (or summarize them into the path) (48).
The pH of the surface soil beneath S. alpina was significantly higher than that of the grassland soil (p =0.01), whereas the topsoil pH of P. fruticosa (p > 0.05) and C. microphylla (p > 0.05) shrubs was not significantly different from that of the grassland soil (Figure 2). The deep soil pH of the three shrubs was not significantly different from the pH of the grass matrix (Figure 2). In addition, the difference between surface and deep soil pH of the grassland was significant (p = 0.04), while the pH of the three shrub matrixes was not (p > 0.05) (Figure 2).
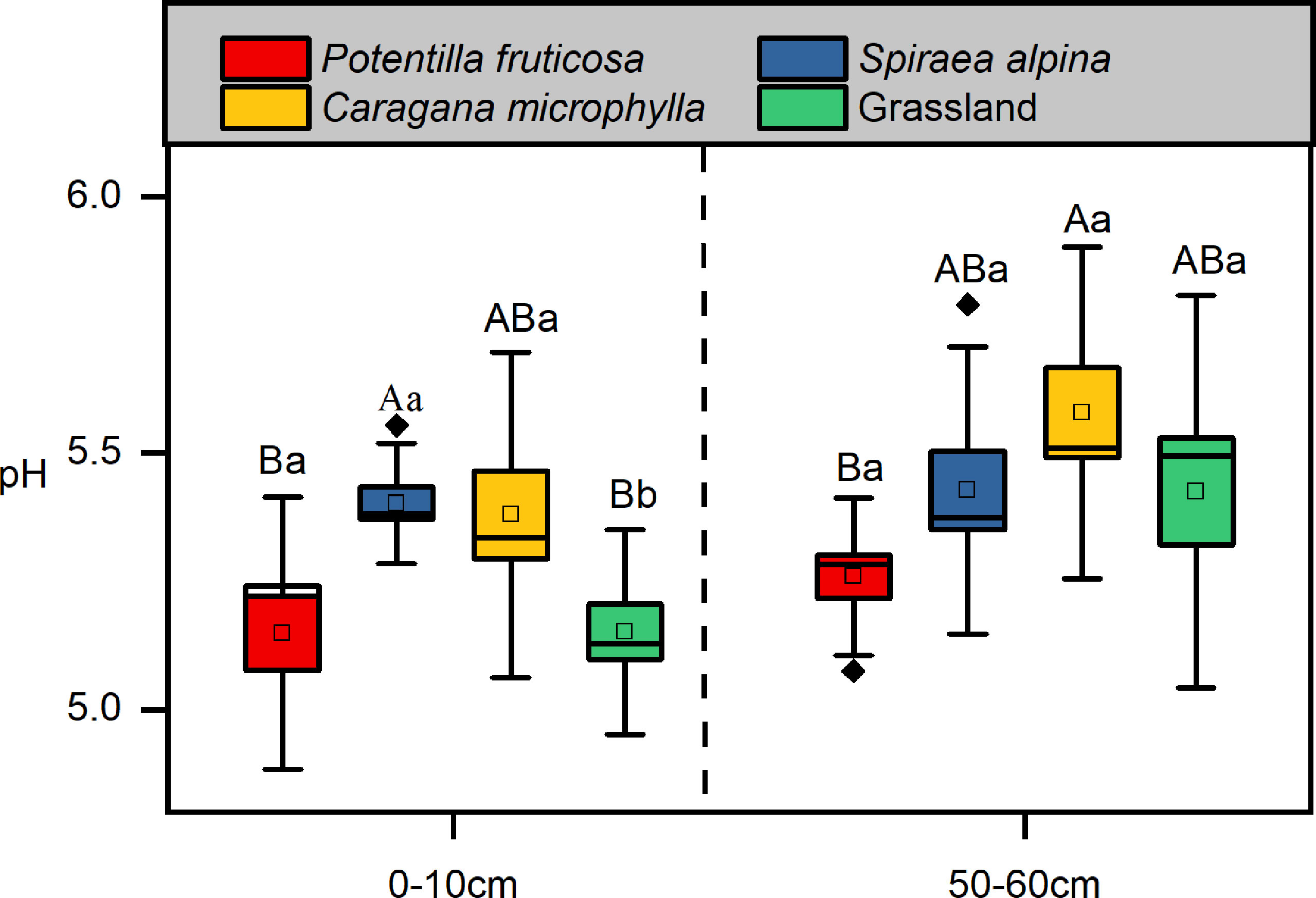
Figure 2 Soil pH. Different lowercase letters indicate significant differences between the different soil layers within the same vegetation types (p < 0.05, independent-samples t-test). Different uppercase letters indicate significant differences between the different vegetation types within the same soil layers (p < 0.05, Tukey’s HSD test).
The SOC content of three shrubs matrixes did not significantly change in surface and deep layers (Figure 3) compared with that of grassland. We found that the SOC content of the grassland was significantly higher in surface layer than deep layer (p = 0.02), and this phenomenon was not found in any of the three shrub matrixes (Figure 3).
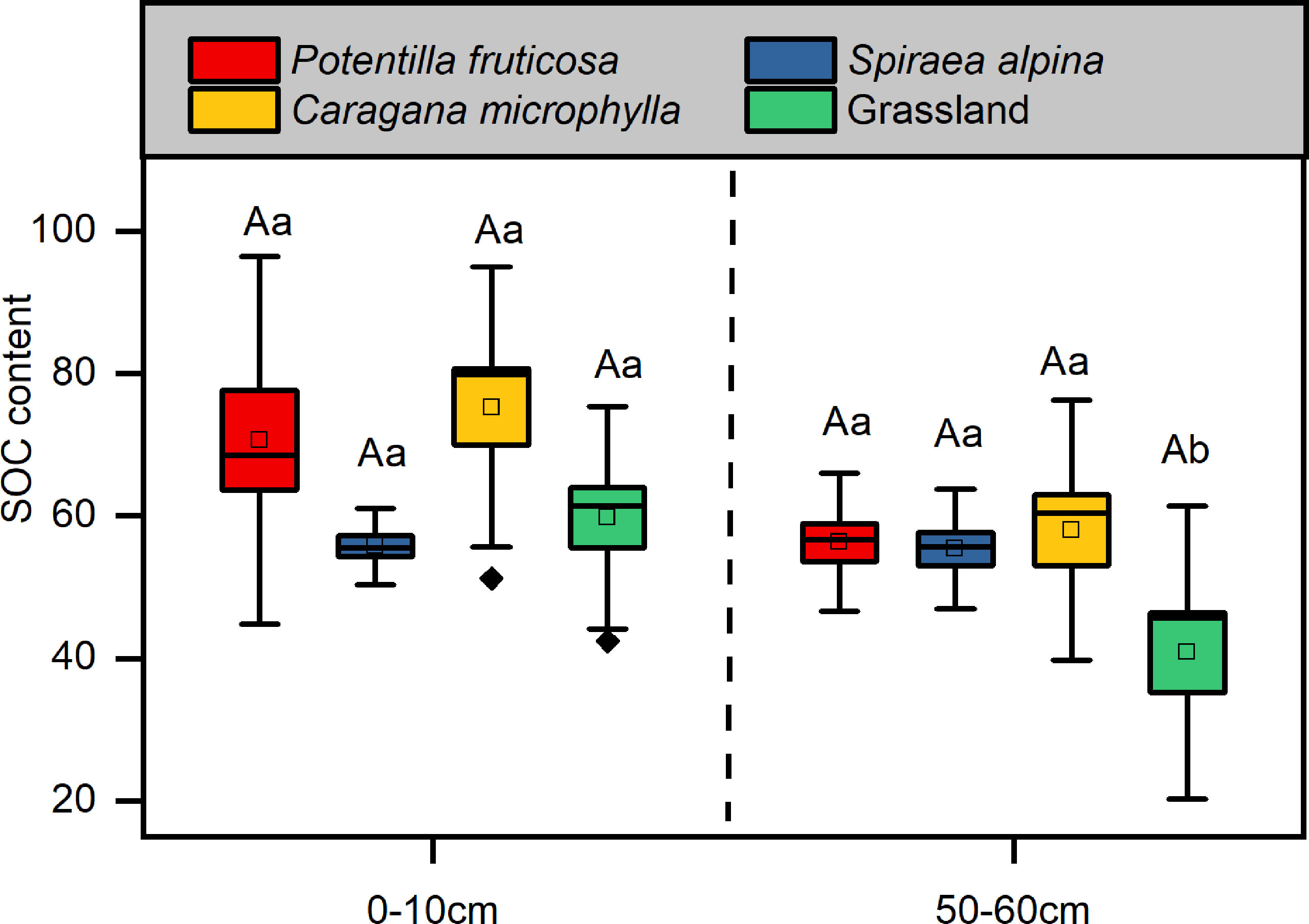
Figure 3 Soil organic content. Different lowercase letters indicate significant differences between the different soil layers within the same vegetation types (p < 0.05, independent-samples t-test). Different uppercase letters indicate significant differences between the different vegetation types within the same soil layers (p < 0.05, Tukey’s HSD test).
The infrared spectrum characteristic peaks of three shrub and grassland matrixes in alpine grassland were similar, mainly at 3,435, 2,930, 1,620, and 1,034 cm−1, and they were alcohol, aliphatic, aromatic, and polysaccharides, respectively, according to their FTIR spectral absorption peaks. There was no significant difference in SOC chemical composition between shrubs and grassland matrixes in surface layer. The invasion of C. microphylla and P. fruticosa shrubs significantly increased the content of polysaccharides (p = 0.00) and alcohol (p = 0.00), respectively, compared to the grassland deep layer. P. fruticosa also decreased the content of aliphatics in deep layer when compared with that in grassland (p = 0.03). None of the three shrubs significantly affected surface SOC recalcitrance, compared to the grassland surface layer (p > 0.05) (Table 1). For the deep layer, the invasion of S. alpina (p = 0.04), C. microphylla (p = 0.00), and P. fruticosa (p = 0.01) significantly reduced the resistance to the decomposition of SOC (Table 1).
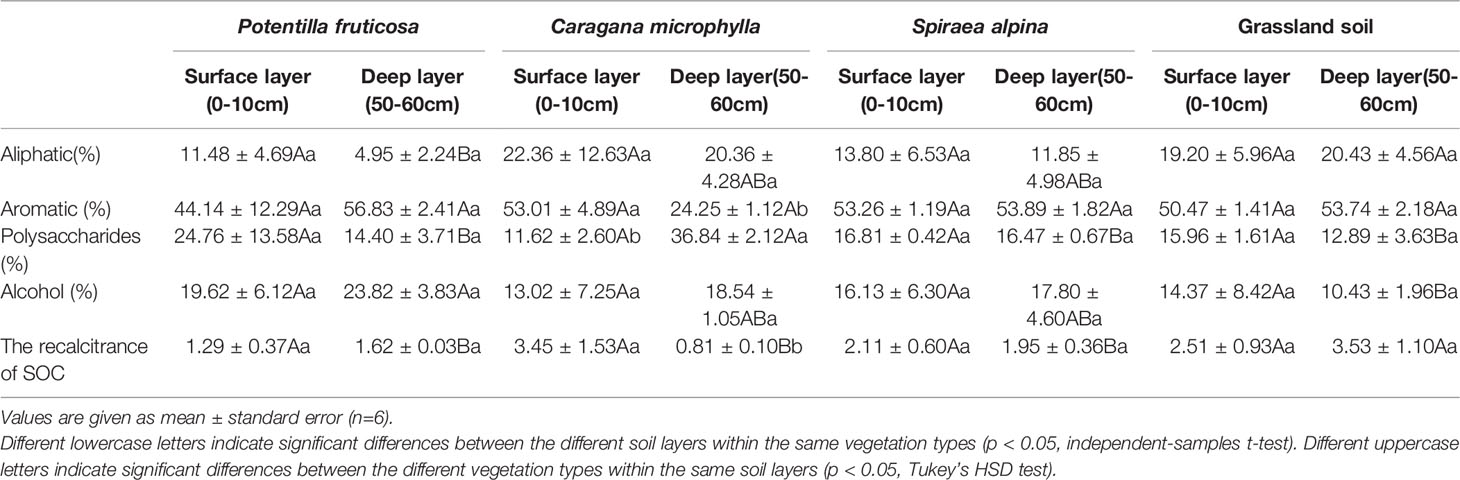
Table 1 The chemical composition and recalcitrance of SOC in surface and deep layers of shrub and grassland patches on the Qinghai–Tibet Plateau.
Soil bacterial alpha-diversity (Shannon index and observed species) was calculated at a depth of 10,595 random sequences per sample. In the 0–10 cm and 50–60 cm layer, the Shannon diversity of P. fruticosa (p = 0.002; p = 0.027) and S. alpina (p = 0.000; p = 0.001) was significantly higher than that in grassland (Figure 4A). For the observed species, P. fruticosa shrub increased the number of OTUs (p = 0.007) compared to that in grassland. In the deep layer, the observed species were higher than that in grassland (p<0.05) (Figure 4B). The Shannon diversity and observed species of soil bacteria in the surface layer of P. fruticosa (p = 0.002; p = 0.004) and grassland (p = 0.000; p = 0.000) soil were significantly higher than that in the deep layer, but not in C. microphylla (p > 0.05) (Figure 4).
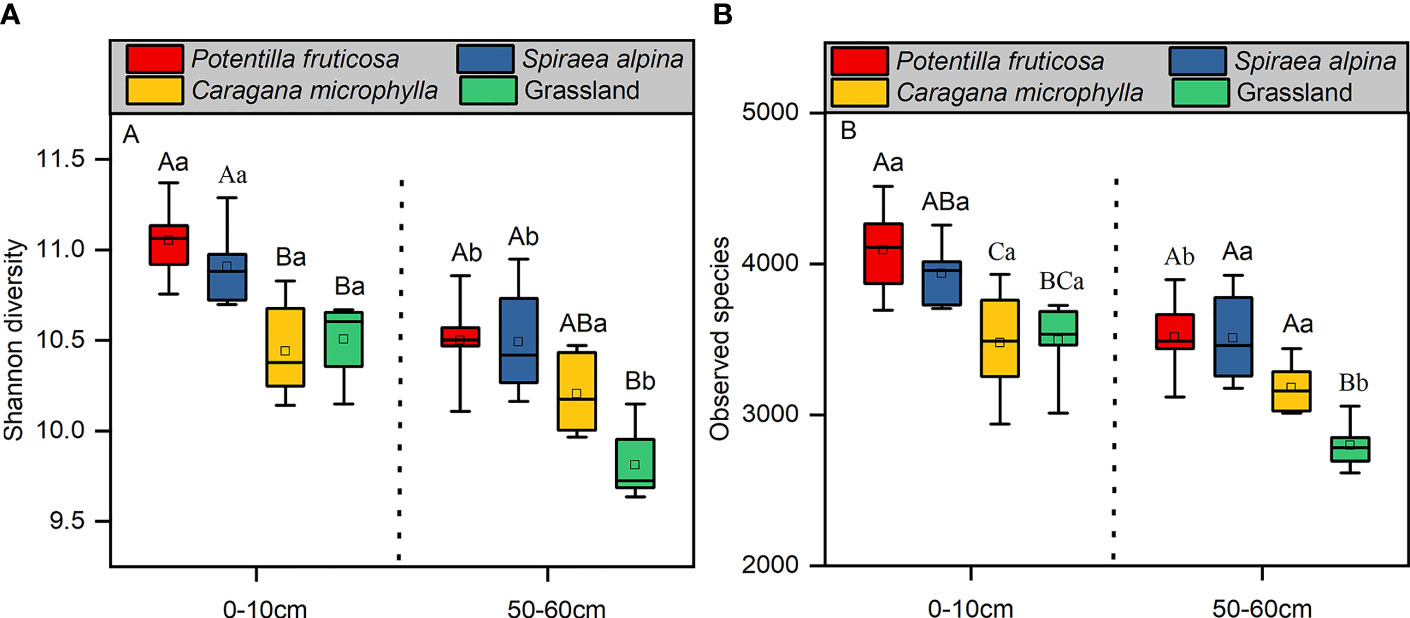
Figure 4 Soil bacterial alpha-diversity (A) Shannon diversity; (B) Observed species. Different lowercase letters indicate significant differences between the different soil layers within the same vegetation types (p < 0.05, independent-samples t-test). Different uppercase letters indicate significant differences between the different vegetation types within the same soil layers (p < 0.05, Tukey’s HSD test).
Across all soil samples, we obtained 2,798,400 high-quality bacterial sequences with 10,000–10,595 sequences per sample. Soil bacterial community composition was analyzed at a depth of 10,595 randomly selected sequences per sample. The dominant phyla (average relative abundance >1%) in the study were Acidobacteria, Proteobacteria, Actinobacteria, Chloroflexi, Gemmatimonadetes, Latescibacteria, Planctomycetes, Rokubacteria, and Bacteroidetes (Figure 5; Supplementary Figure S2). The abundance of Acidobacteria in S. alpina (p = 0.003; p = 0.032) and C. microphylla (p = 0.013; p = 0.018) matrixes was lower than in grassland in the surface and deep layer, respectively (Figure 5). However, the abundance of Actinobacteria in S. alpina (p = 0.037; p = 0.022) and C. microphylla (p = 0.000; p = 0.002) matrixes was higher than in grassland in the surface and deep layers (Figure 5).
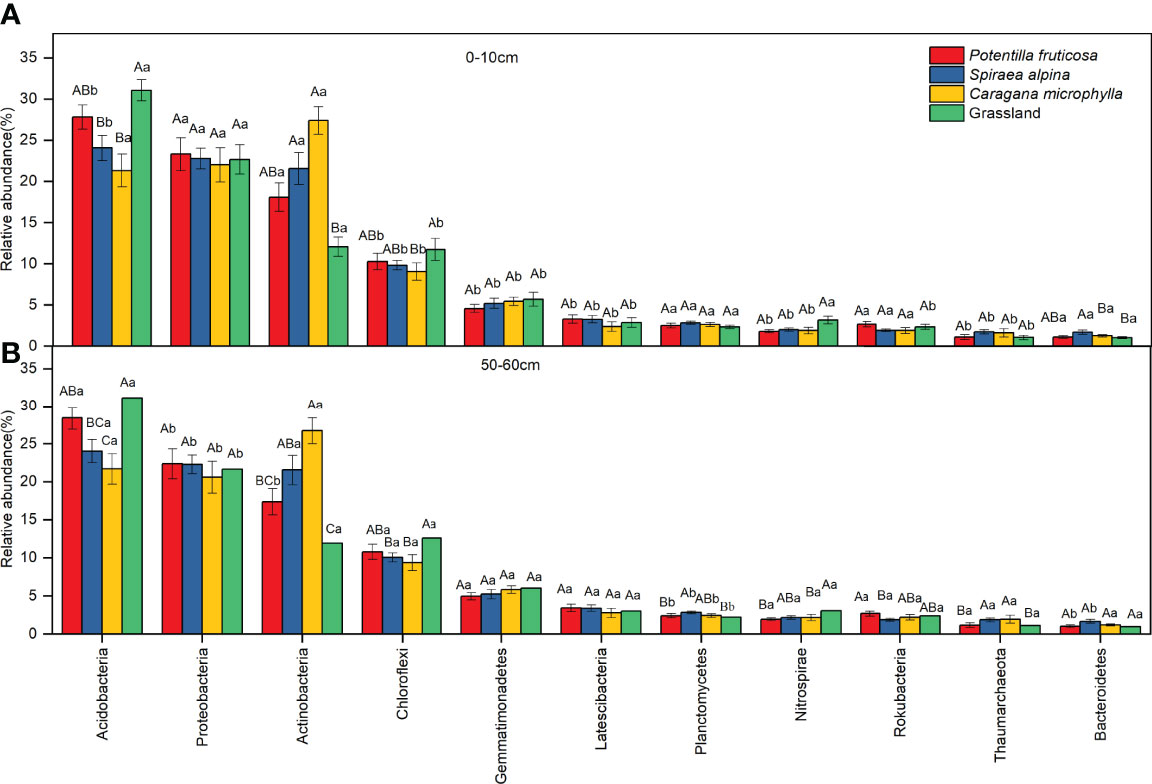
Figure 5 The relative abundance of soil microbial communities at phylum taxonomic level. (A) Surface layer; (B) Deep layer. Bars represent mean; error bars denote standard error. Different lowercase letters indicate significant differences between the different soil layers within the same vegetation types (p < 0.05, independent-samples t-test). Different uppercase letters indicate significant differences between the different vegetation types within the same soil layers (p < 0.05, Tukey’s HSD test).
We utilized PCoA to analyze differences in surface and deep soil bacterial communities between three shrub and grasslands matrixes. The two PCs explained 38.24% (PC1) and 18.12% (PC2) of the variance, respectively (Figure 6). PERMANOVA analysis confirmed that the soil microbial composition was significantly different under three typical shrub and grassland in surface and deep soil (p < 0.05), respectively (Supplementary Table S1).
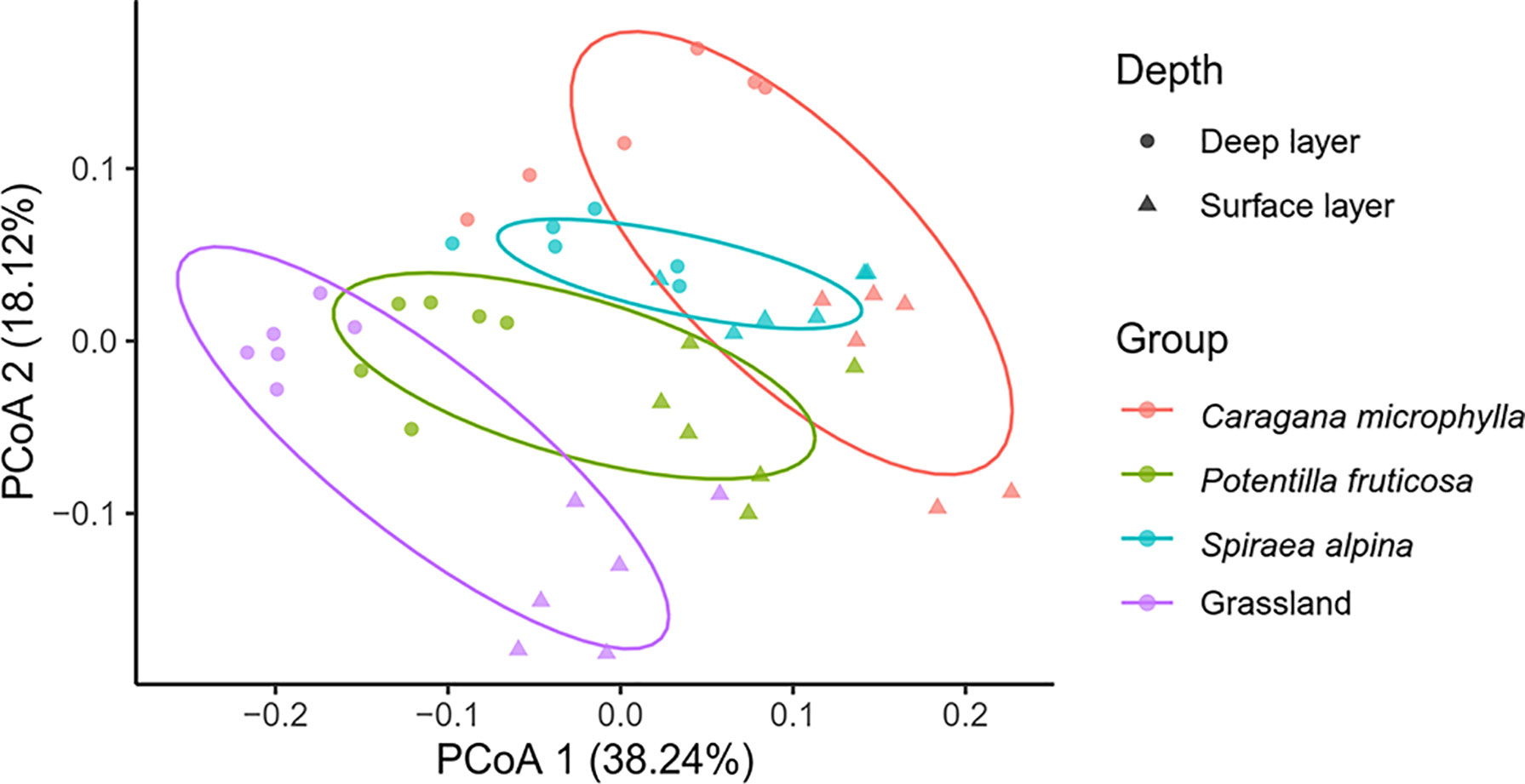
Figure 6 Principal coordinate analysis (PCoA) of soil bacterial community in the shrub and grassland patches based on weighted Unifrac distance.
LEFSe analysis further identified the microbial groups with statistical differences among the four groups of soils. The results showed 39 and 42 biomarkers in the surface and deep layer among the four groups, respectively (Figure 7). In the surface soil of grassland patches, the abundance of bacteria was significantly higher in two phyla (Acidobacteria and Chloroflexi), five genera (freshwater sediment metagenome, GOUTA6, HSB OF53 F07, Ellin6067, and Candidatus Solibacter) than in the shrub patches; C. microphylla patches had a significantly higher abundance of bacteria in one phylum (Actinobacteria) and three genera (Acidothermus, Conexibacter, and Bradyrhizobium); P. fruticosa patches had a significantly higher abundance of bacteria in two genera (Gaiella and Pseudonocardia); S. alpina patches had a significantly higher abundance of bacteria in one genus (MND1) (Figure 7A). In the deep soil of grassland patches, the abundance of bacteria was significantly higher in two phyla (Chloroflexi and Rokubacteria), three genera (freshwater sediment metagenome, GOUTA6, Candidatus Methylomirabilis, and Geobacter) than in the shrub patches; C. microphylla patches had a significantly higher abundance of bacteria in two phyla (Actinobacteria and Nitrospirae) and two genera (Gaiella, Nitrospira); P. fruticosa patches had a significantly higher abundance of bacteria in one phylum (Acidobacteria) and one genus (MND1); and S. alpina patches had a significantly higher abundance of bacteria in one genus (RB41) (Figure 7B).
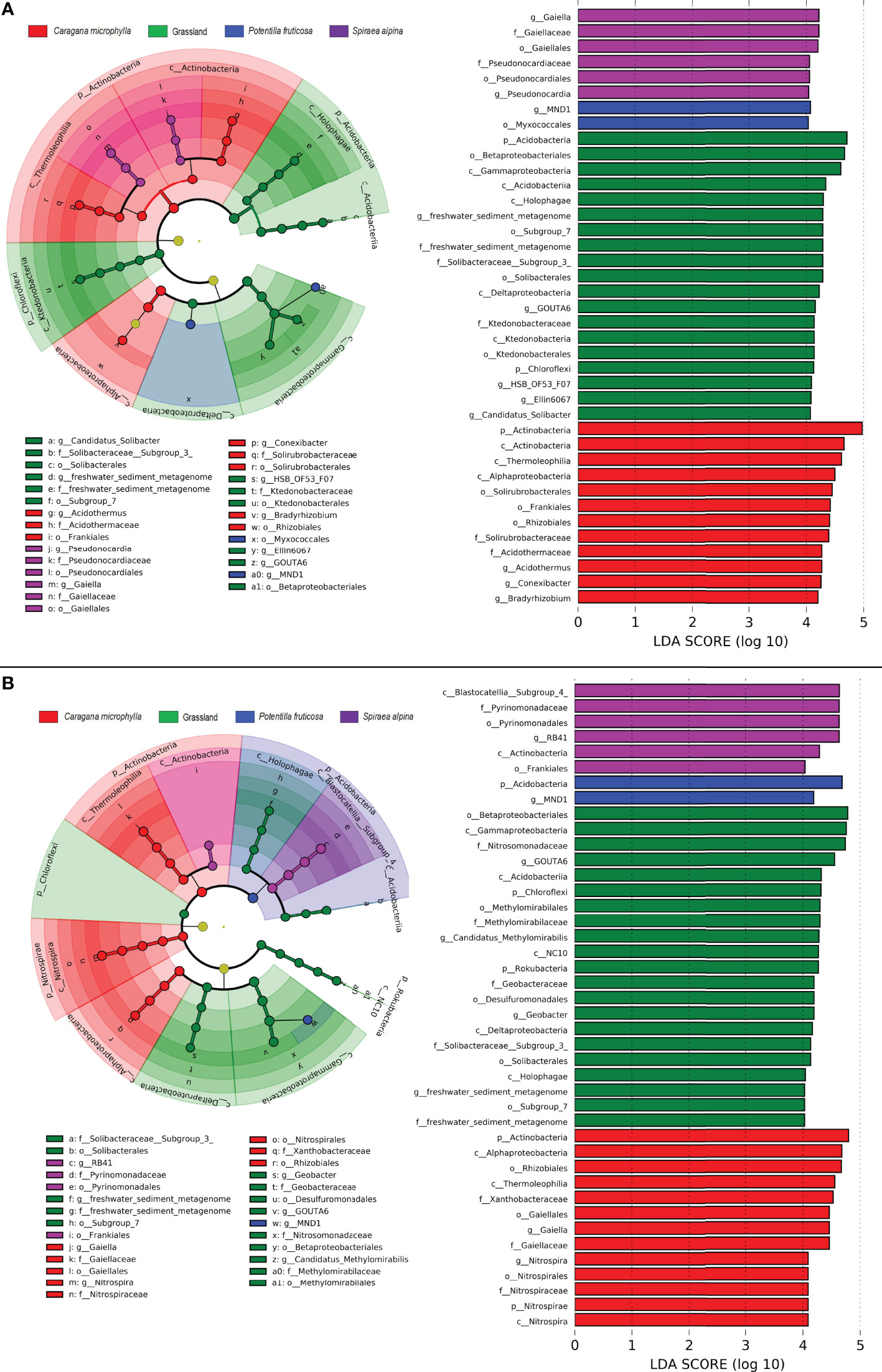
Figure 7 LEFSe analysis of biomarkers in the four groups of soil bacteria. (A) Surface layer; (B) Deep layer. Cladogram representing the taxonomic hierarchy of biomarkers for four soil types. The circles radiating from the inside to the outside represent taxonomic levels from phylum to genus. LDA score >4 indicates a biomarker with statistical differences among the groups.
Redundancy analysis showed that the explanatory rate of all physical and chemical factors was 80.62% in the surface layer and 77.78% in the deep layer (Figure 8). In the topsoil, pH, aliphatic, SOC stability, and SOC were positively correlated with the relative abundances of Actinobacteria, Bacteroidetes, Planctomycetes, and Proteobacteria (Figure 8A). Aromatic and alcohol were positively correlated with the relative abundances of Chloroflexi, Latescibacteria, Rokubacteria, and Gemmatimonadetes (Figure 8A). In the deep soil, aromatic, SOC, and SOC stability was positively associated with Latescibacteria and Acidobacteria. pH and polysaccharides were positively associated with the relative abundances of Thaumarchaeota, Actinobacteria, Bacteroidetes, Planctomycetes, and Proteobacteria (Figure 8B).
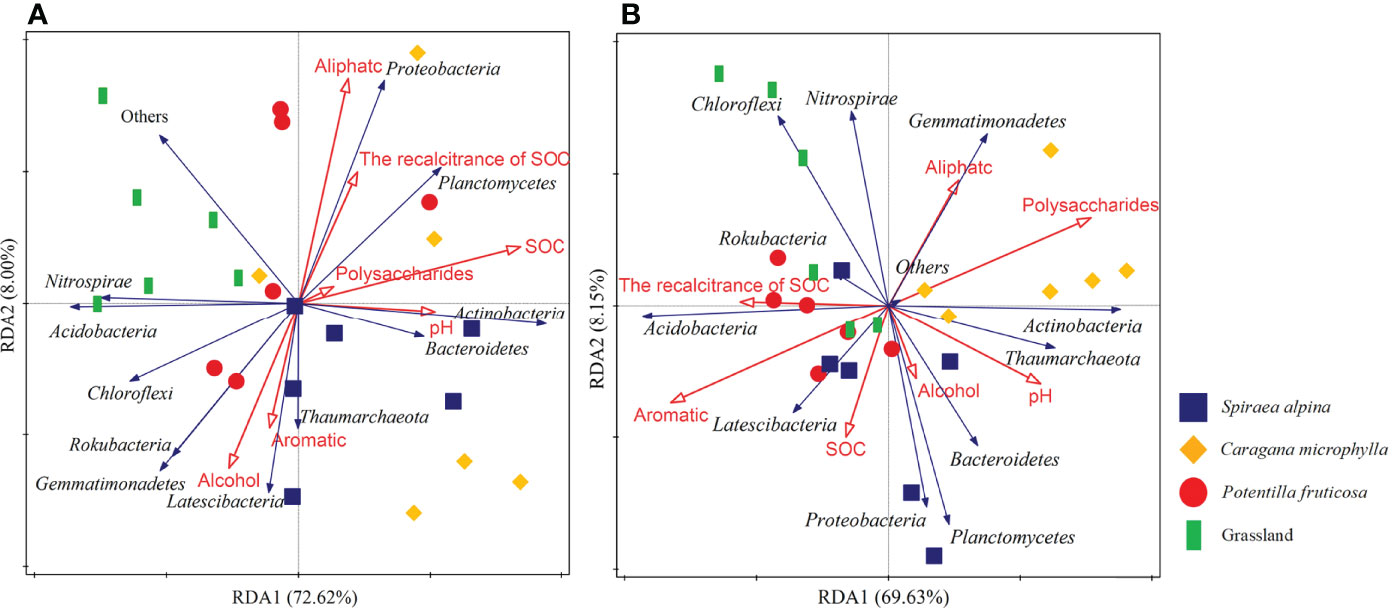
Figure 8 RDA analysis of soil physicochemical properties on microbial community composition. (A) Surface soil (0–10 cm), (B) Deep soil (50–60 cm).
Shifts in the microbial community caused changes in soil microbial predicted functions. In this study, 5 primary and 27 secondary predicted metabolic pathways were found. The most abundant predicted functional genes belonged to categories of metabolism (52.83%) in the primary metabolic pathway, followed by genetic information processing (16.20%), unclassified (13.32%), environmental information processing (13.22%), and cellular processes (4.21%) (Figure 9). In the surface layer, the richest predicted functional genes in C. microphylla, P. fruticosa, and grassland belonged to categories of cell communication, genetic information process, and metabolism, respectively (Supplementary Figure S3A), while in the deep layer, they were cell motility, genetic information process, and metabolism, respectively (Supplementary Figure S3B). No significant predicted bacterial genes were observed for S. alpina when compared to the other two shrub and grassland patches. The microbial community predicted functional genes in the surface layer were mainly influenced by SOC content (p=0.01) and pH (p=0.016), while the microbial community predicted functions in the deep layer were affected by aromatics (p=0.018) (Supplementary Table S2). In the surface layer, pH and SOC were positively associated with the relative abundances of predicted functional genes belonging to categories of transport and catabolism, metabolism of other amino acids, and xenobiotics biodegradation and metabolism (Supplementary Figure S4A). In the deep soil, pH and polysaccharides were positively associated with the relative abundances of predicted functional genes belonging to categories of membrane transport and xenobiotics biodegradation and metabolism (Supplementary Figure S4B).
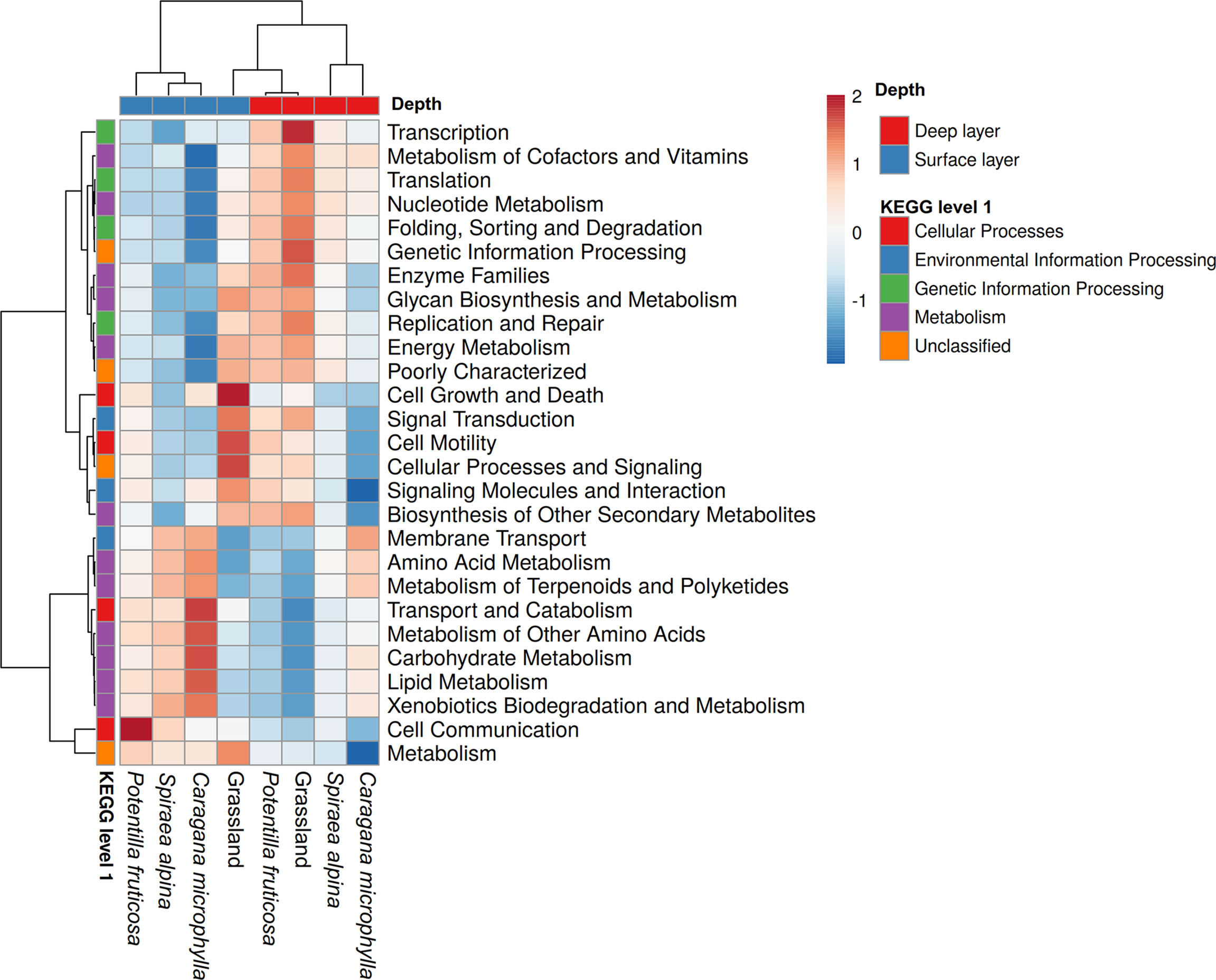
Figure 9 The metabolic function of bacterial communities in the shrub and grassland patches predicted by PICRUSt.
Soil pH is determined by the balance between acid and non-acid cations on colloid surfaces and the balance between hydrogen ions (H+) and hydroxide ions (OH−) in soil solution (49). We found that S. alpina significantly reduced soil acidity (Figure 2). Previous studies have recorded high soil pH under shrubs compared with grass areas in semi-arid systems (50). A potential mechanism explaining this phenomenon might be due to the large canopy of the shrub and the leaching intensity of surface soil being lower than that of grassland soil. Leaching may cause the loss of non-acidic cations (i.e., , ,K+, and Na+) from humus–clay colloidal exchange sites and their replacement by H+, allowing more H+ to remain in the soil and making it more acidic (51). Therefore, S. alpina may inhibit soil acidification by weakening the intensity of the leaching, probably because of the higher coverage of S. alpina compared to the other two shrubs. Furthermore, the pH of the surface layer of grassland was significantly higher than that of the deep layer, while the pH of the surface and deep layers of shrub patches did not differ significantly (Figure 2). This may be related to the fact that shrubs with deeper rooting patterns and perennial nature are more likely to trap more soil nitrate than annual pastures, thereby reducing nitrate leaching and decreasing net acid input (52).
The quantity and quality of aboveground litter and root exudates of shrubs affect carbon sequestration. Eldridge et al. (35) and Li et al. (11) observed that shrub encroachment contributed to the accumulation of SOC in arid and humid regions. However, our study showed that the invasion of C. microphylla, P. fruticosa, and S. alpina had no significant effect on the soil carbon pool of alpine grassland, supporting our first hypothesis. This is probably due to the replacement of prior carbon decompositions with the fresh input of shrub leaves and roots (24, 25). Interestingly, our results show a significant difference in SOC content between the surface and deep layers of grassland, but no such difference was found among the three shrubs (Figure 2). The C content in the surface soil of shrub patches was mainly derived from leaves, while the root and leaf C sources were more dominant in the grass matrix (53). Carbon in the deep soil layer is mainly derived from plant root secretions and fine root turnover, which, however, was not quantified in the present study and should be considered in future research. More root compounds can be produced in shrub patches than in grassland matrixes (54), which would enhance C accumulation. Therefore, compared with grassland, the amount of shrub root compounds in the deep layer weakened the difference in C content between the surface and the deep layer, resulting in no significant change in SOC content. While grasses allocate a significant amount of carbon below ground, their short, dense root systems cannot reach deeper soil layers, resulting in a heterogeneous vertical distribution of SOC content.
We found that aromatics were the highest among the four SOC chemical compositions, indicating that SOC was more stable in QTP grassland soil during chemical and biological degradation (Table 1). Previous studies have also shown that soil recalcitrant C, such as from lignin compounds, is abundant in the bark and roots of shrubs (55). Labile substances such as sugars are decomposed by microorganisms quickly, and organic carbon components such as lignins, which are difficult to be degraded by microorganisms, can accumulate in the soil and transform into recalcitrant organic carbon fractions. Moreover, pH higher or lower than 6.5 also decreases the SOC decomposition (56). In the present study, the lower soil pH in the four patches might partly account for the high resistance to decomposition. Our results showed no significant changes in the recalcitrance of SOC in the surface layer after shrub encroachment, compared to grassland soil. Prior work has shown that the harvesting energy from recalcitrant compounds does not maintain microbial activity, and soil decomposition is extremely restricted in deep soil (25), which is inconsistent with our findings that the recalcitrance of SOC in three shrubs decreased significantly with depth, compared to that in grassland soil (Table 1). The deep root distribution of shrub allows the transport of plant C (i.e., root exudates, root detritus) into deeper soil (16). As a result, deep soil microorganisms are supplied with nutrients, increasing their activity and biomass, leading to the decomposition of deep SOC (57). Therefore, shrub encroachment affected the carbon cycle in deep soil by changing the decomposition of organic carbon (Table 1).
In this study, P. fruticosa and S. alpina had significantly higher Shannon diversity relative to grassland in the surface and deep soil, while C. microphylla did not in surface soil (Figure 4A). The former conclusion is consistent with the research of shrub encroachment on soil bacterial diversity conducted by Xiang et al. (58) in Inner Mongolia grassland, supporting our second hypothesis. The latter finding is consistent with the meta-analysis performed by Custer and van Diepen (59) that plant invasion has limited and variable effects on the alpha-diversity of soil microorganisms. The differences in the Shannon diversity index for the three shrubs indicate that plant species is a significant predictor of bacterial diversity in the Qinghai–Tibetan Plateau. Even under similar environmental conditions, the chemical composition of plant root exudates may vary depending on the plant species, thus affecting the diversity of soil microorganisms (60). Our results also showed that the SOC chemical composition (polysaccharides and alcohol) was slightly higher in P. fruticosa and S. alpina than in grasslands (Table 1), which are considered to be labile substrates and can be quickly absorbed by microorganisms (39). This may also be one of the reasons for the higher microbial diversity under the two shrubs than grasslands. In deep soil, the three shrubs significantly increased the number of observed species (Figure 4B). This may be because shrubs cause an increase in nutrients in deep soil, reducing competition among bacterial communities and allowing rare species to survive, leading to an increase in bacterial alpha-diversity in soil (14). In this study, shrub encroachment triggered significant changes in soil bacterial community composition (Figure 4). This is consistent with previous studies showing that aboveground vegetation notably alters the belowground bacterial communities (61), restructuring bacterial communities of shrub encroached and grassland soils, indicating that soil bacterial community appears to be a sensitive indicator of plant cover type.
One of the aims of this study was to investigate whether the diversity and structure of the soil microbial communities were related to soil properties after shrub encroachment. We found that different factors affected the composition of microbial community in the surface and deep layers after shrub encroachment. The microbial community in the surface layer was mainly influenced by SOC content (p=0.004), while the microbial community in the deep layer was affected by aromatics (p=0.008) and polysaccharides (p=0.02) (Figures 8, 10; Supplementary Table S3). Xiang et al. (58) also showed that SOC was the most important factor for the difference in bacterial community composition after shrub encroachment in temperate grassland. Shrub encroachment may have changed the quality and composition of plant litter and root exudates, thus affecting bacterial communities (14, 62). This is consistent with the results found by Shao et al. (20) in a forest succession experiment, where changes in aliphatics, aromatics, and polysaccharides were the main drivers for the differences in microbial community composition. The statistical analysis and LDA results showed that the relative abundance of Actinobacteria was higher in surface and deep layer of the C. microphylla patches, and Acidobacteria was enriched in surface layer of grassland soils (Figures 5, 7). In addition, the relative abundance of Proteobacteria, Actinobacteria, and Acidobacteria were the highest in our study (Figure 5). Our study showed that Actinobacteria were positively correlated with SOC and polysaccharides in the surface and deep layers, respectively (Figure 8). Actinobacteria play a key role in the decomposition of organic matter and the formation of humus (63), and they play an important role in the turnover of SOC compounds and significantly influence soil C storage (64). Therefore, we suggest that shrub encroachment may have affected C cycling in alpine grassland. This is consistent with our third hypothesis that shrub encroachment alters the C cycle by changing the composition of the bacterial community. Aromatic groups and Acidobacteria were positively correlated in deep soil (Figure 8B). Bacteria belonging to Acidobacteria are considered oligotrophic, which contain genes encoding cellulose and hemicellulose decomposition and play an important role in the decomposition of recalcitrant C (65). Acidobacteria is normally enriched in soils with low nutrient availability (66). To maintain their physiological activity, they break down recalcitrant substances in the soil for energy. Proteobacteria are generally considered copiotrophic (or R-strategists) that thrive under conditions of high nutrient availability (21).
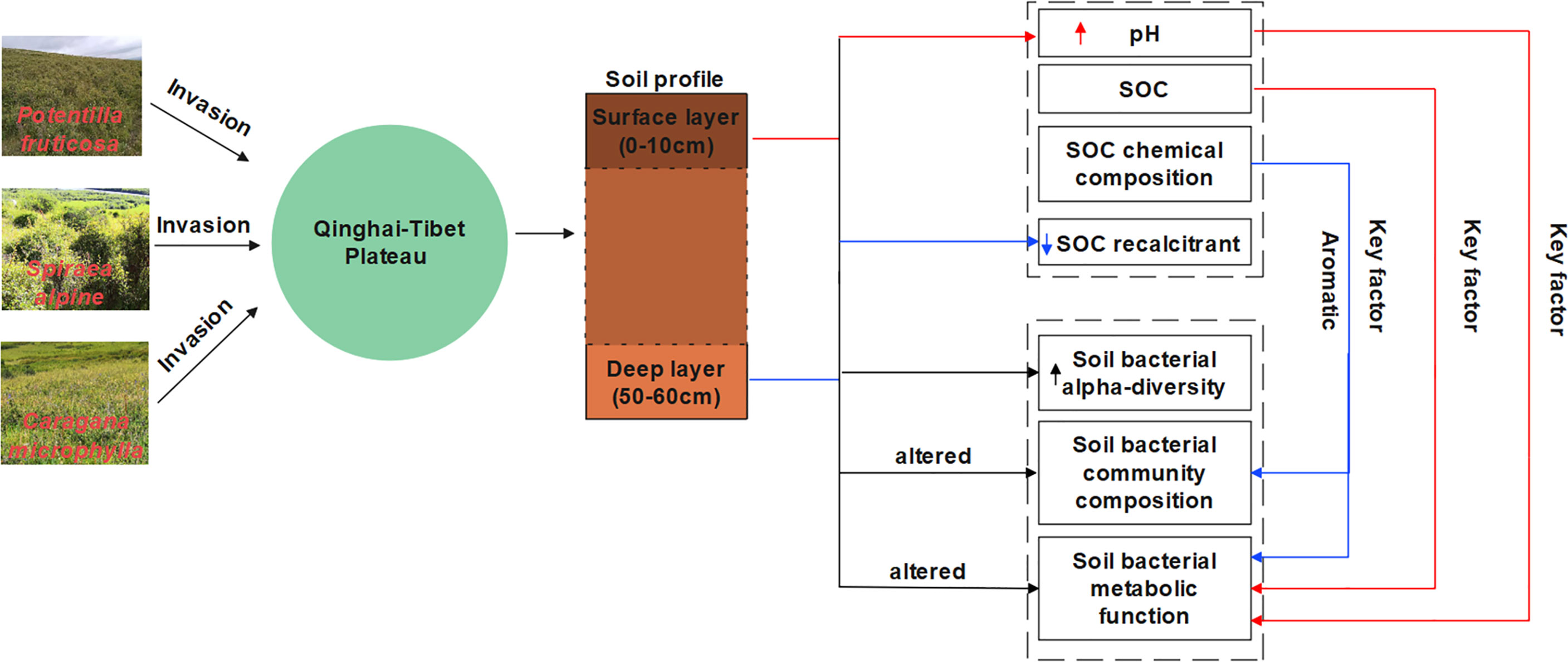
Figure 10 The summary of our findings. The red line represents the surface layer, the blue represents the deep layer, and black represents both layers.
Functional diversity of soil microbial communities is an essential indicator of structure and ecological function, which is of great importance to clarify their role in different environments (67). In the KEGG2 level of metabolic pathway, cellular motility pathway was enriched in the surface layer of grassland, compared to C. microphylla and S. alpina (Supplementary Figure S3). Our study also showed that the SOC content in the surface layer of grassland was slightly lower than that of the C. microphylla and S. alpina. This may provide a competitive advantage for bacterial communities in these two shrublands to move towards nutrients (68). In C. microphylla, lipid metabolism was enriched in the surface and deep layers (Supplementary Figure S3), which might indicate that soil bacteria preferentially use lipids as a source of carbon and energy in this shrub (69). The relative abundance of carbohydrate and amino acid metabolism were high, as carbohydrate metabolism can be closely related to the formation, breakdown, and conversion of carbohydrates. Carbohydrates are synthesized by plants from the atmosphere through photosynthesis and can be used as substrates for cellular respiration (70). Moreover, carbohydrate metabolism was enriched in surface and deep layer of C. microphylla (Supplementary Figure S3). Amino acid metabolism can help bacteria obtain amino acids, which is conducive to the survival and reproduction of bacteria (71). Some microbes (e.g., Rhizobium) can convert N2 in the air into ammonia to synthesize amino acids. The result of LEFSe also showed that amino acid metabolism and Rhizobiales were enriched in surface and deep layers of C. microphylla (Supplementary Figure S3). Therefore, we think that the invasion of C. microphylla may also affect the C cycle of alpine grassland. RDA results showed that SOC (p=0.01) and pH (p=0.016) appeared to be key factors influencing the bacterial predicted metabolic functions of shrub encroachment surface layers, as opposed to aromatics (p=0.018) in the deep layers (Supplementary Figure S4; Supplementary Table S2; Figure 10). This is similar to the key soil properties that influence the composition of bacterial communities.
This study evaluated the impact of shrub encroachment on the surface and deep SOC content and its chemical composition, microbial community composition, diversity, and predicted functions on the Qinghai–Tibetan Plateau grassland. Our results showed that different shrub-encroached grasslands had different effects on soil pH value. S. alpina significantly reduced soil acidity in the surface layer, while no similar phenomenon occurred in the other shrub species. The shrub encroachment had no significant effect on SOC content, but weakened the difference between the surface and the deep layer in the eastern edge of the Tibetan Plateau. Shrub encroachment reduced soil organic carbon recalcitrance and did not contribute to the storage of deep soil organic carbon. Shrub encroachment was associated with a significant increase in bacterial alpha diversity and changes in bacterial community composition. The key factors influencing surface and deep bacterial communities were different. Abiotic factors, such as SOC content and SOC chemical composition (aromatics and polysaccharides), regulate soil surface and deep microbial communities. By LEFSe and PICRUSt analysis, we suggest that shrub encroachment might have affected C cycling of alpine grassland by changing bacterial community composition and functioning. This work will help to further refine our knowledge of how shrub encroachment affects bacterial community structure in grassland ecosystems. However, we did not investigate the effect of encroachment on soil fungal communities, which might be more important for carbon cycling and closely related to changes in vegetation. This limitation should be addressed in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA801100.
WM and ZD were responsible for experimental design and manuscript preparation. ST was responsible for writing guidance. WM and CW were responsible for experiment performance. DZ were responsible for experimental data processing and analysis. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31600378 and U20A2008) and postgraduate innovative scientific research project of Southwest Minzu University (CX2021SZ27).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Professor Brian McGarvey for providing language help and proof reading for this article. We are grateful to the staff in our research group for their help on field sampling.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsoil.2022.829575/full#supplementary-material
1. Lunt ID, Winsemius LM, McDonald SP, Morgan JW, Dehaan RL. How Widespread is Woody Plant Encroachment in Temperate Australia? Changes in Woody Vegetation Cover in Lowland Woodland and Coastal Ecosystems in Victoria From 1989 to 2005. J Biogeogr (2010) 37:722–32. doi: 10.1111/j.1365-2699.2009.02255.x
2. Anadón JD, Sala OE, Turner BL, Bennett EM. Effect of Woody-Plant Encroachment on Livestock Production in North and South America. Proc Natl Acad Sci USA (2014) 111:12948–53. doi: 10.1073/pnas.1320585111
3. Naito AT, Cairns DM. Patterns and Processes of Global Shrub Expansion. Prog Phys Geog (2011) 35:423–42. doi: 10.1177/0309133311403538
4. Nepstad DC, de Carvalho CR, Davidson EA, Jipp PH, Lefebvre PA, Negreiros GH, et al. The Role of Deep Roots in the Hydrological and Carbon Cycles of Amazonian Forests and Pastures. Nature (1994) 372:666–9. doi: 10.1038/372666a0
5. Trumbore SE. Potential Responses of Soil Organic Carbon to Global Environmental Change. Proc Natl Acad Sci USA (1997) 94:8284–9. doi: 10.1073/pnas.94.16.8284
6. Schmidt MWI, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, et al. Persistence of Soil Organic Matter as an Ecosystem Property. Nature (2011) 478:49–56. doi: 10.1038/nature10386
7. Rumpel C, Kögel-Knabner I. Deep Soil Organic Matter—a Key But Poorly Understood Component of Terrestrial C Cycle. Plant Soil (2011) 338:143–58. doi: 10.1007/s11104-010-0391-5
8. Mckinley DC, Blair JM. Woody Plant Encroachment by Juniperus Virginiana in a Mesic Native Grassland Promotes Rapid Carbon and Nitrogen Accrual. Ecosystems (2008) 11:454–68. doi: 10.1007/s10021-008-9133-4
9. Lett MS, Knapp AK, Briggs JM, Blair JM. Influence of Shrub Encroachment on Aboveground Net Primary Productivity and Carbon and Nitrogen Pools in a Mesic Grassland. Can J Bot (2004) 82:1363–70. doi: 10.1139/b04-088
10. Jackson RB, Banner JL, Jobbagy EG, Pockman WT, Wall DH. Ecosystem Carbon Loss With Woody Plant Invasion of Grasslands. Nature (2002) 418:623–6. doi: 10.1038/nature00910
11. Li H, Shen HH, Chen LY, Liu TY, Hu HF, Zhao X, et al. Effects of Shrub Encroachment on Soil Organic Carbon in Global Grasslands. Sci Rep (2016) 6:28974. doi: 10.1038/srep28974
12. Marchante E, Kjller A, Struwe S, Freitas H. Invasive Acacia Longifolia Induce Changes in the Microbial Catabolic Diversity of Sand Dunes. Soil Biol Biochem (2008) 40:2563–8. doi: 10.1016/j.soilbio.2008.06.017
13. Wubet T, Christ S, Schöning I, Boch S, Gawlich M, Schnabel B, et al. Differences in Soil Fungal Communities Between European Beech (Fagus Sylvatica L.) Dominated Forests are Related to Soil and Understory Vegetation. PLos One (2012) 7:e47500. doi: 10.1371/journal.pone.0047500
14. Xiang XJ, Gibbons SM, Li H, Shen HH, Fang JY, Chu HY. Shrub Encroachment Is Associated With Changes in Soil Bacterial Community Composition in a Temperate Grassland Ecosystem. Plant Soil (2018) 425:539–51. doi: 10.1007/s11104-018-3605-x
15. Mcguire KL, Zak DR, Edwards IP, Blackwood CB, Upchurch R. Slowed Decomposition Is Biotically Mediated in an Ectomycorrhizal, Tropical Rain Forest. Oecologia (2010) 164:785–95. doi: 10.1007/s00442-010-1686-1
16. Biederman LA, Boutton TW. Biodiversity and Trophic Structure of Soil Nematode Communities are Altered Following Woody Plant Invasion of Grassland. Soil Biol Biochem (2009) 41:1943–50. doi: 10.1016/j.soilbio.2009.06.019
17. Buss HL, Bruns MA, Schultz MJ, Moore J, Brantley SL. The Coupling of Biological Iron Cycling and Mineral Weathering During Saprolite Formation, Luquillo Mountains, Puerto Rico. Geobiology (2010) 3:247–60. doi: 10.1111/j.1472-4669.2006.00058.x
18. Trumbore S. Age of Soil Organic Matter and Soil Respiration: Radiocarbon Constraints on Belowground C Dynamics. Ecol Appl (2000) 10:399–411. doi: 10.1890/1051-0761(2000)010[0399:AOSOMA]2.0.CO;2
19. Cheng L, Leavitt SW, Kimball BA, Pinter PJ Jr., Ottman MJ, Matthias A, et al. Dynamics of Labile and Recalcitrant Soil Carbon Pools in a Sorghum Free-Air CO2 Enrichment (FACE) Agroecosystem. Soil Biol Biochem (2007) 39:2250–63. doi: 10.1016/j.soilbio.2007.03.031
20. Shao PS, Liang C, Rubert-Nason K, Li XZ, Xie HT, Bao XL. Secondary Successional Forests Undergo Tightly-Coupled Changes in Soil Microbial Community Structure and Soil Organic Matter - ScienceDirect. Soil Biol Biochem (2019) 128:56–65. doi: 10.1016/j.soilbio.2018.10.004
21. Ho A, Di Lonardo DP, Bodelier PLE. Revisiting Life Strategy Concepts in Environmental Microbial Ecology. FEMS Microbiol Ecol (2017) 93:3. doi: 10.1093/femsec/fix006
22. Carbone MS, Trumbore SE. Contribution of New Photosynthetic Assimilates to Respiration by Perennial Grasses and Shrubs: Residence Times and Allocation Patterns. New Phytol (2007). 176:124–35. doi: 10.1111/j.1469-8137.2007.02153.x
23. Crow SE, Lajtha K, Filley TR, Swanston CW, Bowden RD, Caldwell BA. Sources of Plant-Derived Carbon and Stability of Organic Matter in Soil: Implications for Global Change. Global Change Biol (2010) 15:2003–19. doi: 10.1111/j.1365-2486.2009.01850.x
24. Filley TR, Boutton TW, Liao JD, Jastrow JD, Gamblin DE. Chemical Changes to Nonaggregated Particulate Soil Organic Matter Following Grassland-to-Woodland Transition in a Subtropical Savanna. J Geophys Res.: Atmos (2008) 113:10269. doi: 10.1029/2007JG000564
25. Fontaine S, Barot S, Barré P, Bdioui N, Mary B, Rumpel C. Stability of Organic Carbon in Deep Soil Layers Controlled by Fresh Carbon Supply. Nature (2007) 450:277–80. doi: 10.1038/nature06275
26. Kuzyakov Y. Priming Effects: Interactions Between Living and Dead Organic Matter. Soil Biol Biochem (2010) 42:1363–71. doi: 10.1016/j.soilbio.2010.04.003
27. Chao L, Balser TC. Warming and Nitrogen Deposition Lessen Microbial Residue Contribution to Soil Carbon Pool. Nat Commun (2012) 3:1–4. doi: 10.1038/ncomms2224
28. Liu YR, Delgado-Baquerizo M, Wang JT, Hu H W, Yang ZM, He JZ. New Insights Into the Role of Microbial Community Composition in Driving Soil Respiration Rates. Soil Biol Biochem (2018) 118:35–41. doi: 10.1016/j.soilbio.2017.12.003
29. Maron PA, Sarr A, Kaisermann A, Lévêque J, Mathieu O, Guigue J, et al. High Microbial Diversity Promotes Soil Ecosystem Functioning. Appl Environ Microbio (2018) 84:e02738–17. doi: 10.1128/AEM.02738-17
30. Delgado-Baquerizo M, Maestre FT, Gallardo A, Bowker MA, Wallenstein MD, Quero JL, et al. Decoupling of Soil Nutrient Cycles as a Function of Aridity in Global Drylands. Nature (2013) 502:672–6. doi: 10.1038/nature12670
31. Maestre FT, de Guevara ML, Quero JL, Lazaro R, Delgado-Baquerizo M, Ochoa V, et al. Changes in Biocrust Cover Drive Carbon Cycle Responses to Climate Change in Drylands. Global Change Biol (2013) 19:3835–47. doi: 10.1111/gcb.12306
32. Tardy V, Spor E, Mathieu O, Eque J, Maron PA. Shifts in Microbial Diversity Through Land Use Intensity as Drivers of Carbon Mineralization in Soil. Soil Biol Biochem (2015) 90:204–13. doi: 10.1016/j.soilbio.2015.08.010
33. Yao TD, Thompson LG, Mosbrugger V, Zhang F, Ma YM, Luo TX, et al. Third Pole Environment (TPE). Environ Dev (2012) 3:52–64. doi: 10.1016/j.envdev.2012.04.002
34. Gao YH, Zhou X, Wang Q, Wang CZ, Zhan ZM, Chen LF, et al. Vegetation Net Primary Productivity and its Response to Climate Change During 2001-2008 in the Tibetan Plateau. Sci Total Environ (2013) 444:356–62. doi: 10.1016/j.scitotenv.2012.12.014
35. Eldridge DJ, Bowker MA, Maestre FT, Roger E, Reynolds JF, Whitford WG. Impacts of Shrub Encroachment on Ecosystem Structure and Functioning: Towards a Global Synthesis. Ecol Lett (2011) 14:709–22. doi: 10.1111/j.1461-0248.2011.01630.x
36. Wang CT, Zhao XQ, Zi HB, Hu L, Ade LJ, Wang GX, et al. The Effect of Simulated Warming on Root Dynamics and Soil Microbial Community in an Alpine Meadow of the Qinghai-Tibet Plateau. Appl Soil Ecol (2017) 116:30–41. doi: 10.1016/j.apsoil.2017.03.005
37. Giacometti C, Demyan MS, Cavani L, Marzadori C, Kandeler E. Chemical and Microbiological Soil Quality Indicators and Their Potential to Differentiate Fertilization Regimes in Temperate Agroecosystems. Appl Soil. Ecol (2013) 64:32–48. doi: 10.1016/j.apsoil.2012.10.002
38. Calderón F, Haddix M, Conant R, Magrini-Bair K, Paul E. Diffuse-Reflectance Fourier-Transform Mid-Infrared Spectroscopy as a Method of Characterizing Changes in Soil Organic Matter. Soil Sci Soc Am J (2013) 77:1591–600. doi: 10.2136/sssaj2013.04.0131
39. Veum KS, Goyne KW, Kremer RJ, Miles RJ, Sudduth KA. Biological Indicators of Soil Quality and Soil Organic Matter Characteristics in an Agricultural Management Continuum. Biogeochemistry (2014) 117:81–99. doi: 10.1007/s10533-013-9868-7
40. Rui J, Li J, Zhang S, Yan X, Wang Y, Li X. The Core Populations and Co-Occurrence Patterns of Prokaryotic Communities in Household Biogas Digesters. Biotechnol Biofuels (2015) 8:1–15. doi: 10.1186/s13068-015-0339-3
41. Li XZ, Rui JP, Mao YJ, Yannarell A, Mackie R. Dynamics of the Bacterial Community Structure in the Rhizosphere of a Maize Cultivar. Soil Biol Biochem (2014) 68:392–401. doi: 10.1016/j.soilbio.2013.10.017
42. Magoč T, Salzberg SL. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
43. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
44. Edgar RC. UPARSE: Highly Accurate OTU Sequences From Microbial Amplicon Reads. Nat Methods (2013) 10:996. doi: 10.1038/NMETH.2604
45. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat Methods (2010) 7:335–6. doi: 10.1038/nmeth.f.303
46. Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 Years of Serving the Community With Ribosomal RNA Gene Reference Databases and Tools. J Biotechnol (2017) 261:169–76. doi: 10.1016/j.jbiotec.2017.06.1198
47. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic Biomarker Discovery and Explanation. Genome Biol (2011) 12:1–18. doi: 10.1186/gb-2011-12-6-r60
48. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat Biotechnol (2013) 31:814–21. doi: 10.1038/nbt.2676
50. Gonzalez-Polo M, Austin AT. Spatial Heterogeneity Provides Organic Matter Refuges for Soil Microbial Activity in the Patagonian Steppe, Argentina. Soil Biol Biochem (2009) 41:1348–51. doi: 10.1016/j.soilbio.2009.03.008
51. Berthrong ST, Schadt CW, Pineiro G, Jackson RB. Afforestation Alters the Composition of Functional Genes in Soil and Biogeochemical Processes in South American Grasslands. Appl Environ Microbiol (2009) 75:6240–8. doi: 10.1128/AEM.01126-09
52. Kerridge PC. “Adaptation of Shrub Legumes to Acid Soils,”. In: Wright RJ, Baligar VC, Murrmann RP, editors. Plant-Soil Interactions at Low Ph. Beckley: Kluwer Academic Publishers (1991). p. 977–87.
53. Zhou LH, Li H, Shen HH, Xu YP, Wang YH, Xing AJ, et al. Effects of Shrub Encroachment on Vertical Changes in Soil Organic Carbon in Mongolian Grasslands: Using a Multi-Biomarker Approach. Plant Soil (2018) 431:217–30. doi: 10.1007/s11104-018-3761-z
54. Zhou Y, Boutton TW, Wu XB. Soil Carbon Response to Woody Plant Encroachment: Importance of Spatial Heterogeneity and Deep Soil Storage. J Ecol (2017) 105:1738–49. doi: 10.1111/1365-2745.12770
55. Feng XJ, Simpson AJ, Schlesinger WH, Simpson MJ. Altered Microbial Community Structure and Organic Matter Composition Under Elevated CO2 and N Fertilization in the Duke Forest. Global Change Biol (2010) 16:2104–16. doi: 10.1111/j.1365-2486.2009.02080.x
56. Delaune RD, Reddy CN, Patrick WH Jr. Organic Matter Decomposition in Soil as Influenced by pH and Redox Conditions. Soil Biol Biochem (1981) 13:533–4. doi: 10.1016/0038-0717(81)90045-6
57. Song YH, Zhai JY, Zhang JY, Qiao LL, Wang GL, Ma LH, et al. Forest Management Practices of Pinus Tabulaeformis Plantations Alter Soil Organic Carbon Stability by Adjusting Microbial Characteristics on the Loess Plateau of China. Sci Total. Environ (2021) 766:144209. doi: 10.1016/j.scitotenv.2020.144209
58. Xiang XJ, Gibbons SM, Li H, Shen HH, Chu HY. Proximate Grassland and Shrub-Encroached Sites Show Dramatic Restructuring of Soil Bacterial Communities. Peer J (2019) 7:e7304. doi: 10.7717/peerj.7304
59. Custer GF, van Diepen LTA. Plant Invasion has Limited Impact on Soil Microbial α-Diversity: A Meta-Analysis. Diversity (2020) 12:112. doi: 10.3390/d12030112
60. Broeckling CD, Broz AK, Bergelson J, Manter DK, Vivanco JM. Root Exudates Regulate Soil Fungal Community Composition and Diversity. Appl Environ Microbiol (2008) 74:738–44. doi: 10.1128/AEM.02188-07
61. Gellie NJC, Mills JG, Breed MF, Lowe AJ. Revegetation Rewilds the Soil Bacterial Microbiome of an Old Field. Mol Ecol (2017) 26:2895–904. doi: 10.1111/mec.14081
62. Archer S, Schimel DS, Holland EA. Mechanisms of Shrubland Expansion: Land Use, Climate or CO2? Clim Change (1995) 29:91–9. doi: 10.1007/BF01091640
63. Wang QF, Jiang X, Guan DW, Wei D, Zhao BS, Ma MC, et al. Long-Term Fertilization Changes Bacterial Diversity and Bacterial Communities in the Maize Rhizosphere of Chinese Mollisols. Appl Soil. Ecol (2018) 125:88–96. doi: 10.1016/j.apsoil.2017.12.007
64. Lozano YM, Hortal S, Armas C, Pugnaire FI. Interactions Among Soil, Plants, and Microorganisms Drive Secondary Succession in a Dry Environment. Soil Biol Biochem (2014) 78:298–306. doi: 10.1016/j.soilbio.2014.08.007
65. Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl Environ Microbiol (2009) 75:5111–20. doi: 10.1128/AEM.00335-09
66. Belova SE, Suzina NE, Rijpstra WIC, Damsté JSS, Dedysh SN. Edaphobacter Lichenicola Sp. Nov., a Member of the Family Acidobacteriaceae From Lichen-Dominated Forested Tundra. Int J Syst Evol Microbiol (2018) 68:1265–70. doi: 10.1099/ijsem.0.002663
67. Widder S, Allen RJ, Pfeiffer T, Curtis TP, Wiuf C, Sloan WT, et al. Challenges in Microbial Ecology: Building Predictive Understanding of Community Function and Dynamics. ISME. J (2016) 10:2557–68. doi: 10.1038/ismej.2016.45
68. Veljo K, Angelica V, Armin L, Gerard T, Teresa L, Francesco F. Phylogenetic and Functional Metagenomic Profiling for Assessing Microbial Biodiversity in Environmental Monitoring. PLos One (2012) 7:e43630. doi: 10.1371/journal.pone.0043630
69. Lauro FM, Mcdougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The Genomic Basis of Trophic Strategy in Marine Bacteria. Proc Natl Acad Sci USA (2009) 106:15527–33. doi: 10.1073/pnas.0903507106
70. Gray GM. Carbohydrate Digestion and Absorption. Gastroenterology (1970) 58:96–107. doi: 10.1016/S0016-5085(70)80098-1
Keywords: shrub encroachment, bacterial community, soil organic carbon, 16S rRNA, Fourier translation infrared spectroscopy (FTIR), soil layer
Citation: Dengzeng Z, Ma W, Wang C, Tang S and Zhang D (2022) Effect of Shrub Encroachment on Alpine Grass Soil Microbial Community Assembly. Front. Soil Sci. 2:829575. doi: 10.3389/fsoil.2022.829575
Received: 06 December 2021; Accepted: 17 May 2022;
Published: 24 June 2022.
Edited by:
Vanessa Nessner Kavamura, Rothamsted Research, United KingdomReviewed by:
Bahar S. Razavi, University of Kiel, GermanyCopyright © 2022 Dengzeng, Ma, Wang, Tang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenming Ma, TWF3bXRmQHN3dW4uZWR1LmNu; Changting Wang, cXN3Z2huMjAxM0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.