
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sens. , 16 September 2022
Sec. Lab-on-a-Chip Devices
Volume 3 - 2022 | https://doi.org/10.3389/fsens.2022.1007355
This article is part of the Research Topic Editors’ Showcase: Lab-on-a-chip Devices View all 8 articles
Multiplex detection of protein biomarkers in biological fluids facilitates high-throughput detection using small-volume samples, thereby enhancing efficacy of diagnostic assays and proteomic studies. Graphically encoded hydrogel microparticles conjugated with capture antibodies have shown great potential in multiplex immunoassays by providing superior sensitivity and specificity, a broad dynamic range, and large encoding capacity. Recently, the process of post-synthesis conjugation of reduced capture antibodies to unreacted acrylate moieties in hydrogel particles has been developed to efficiently prevent the aggregation of capture antibodies inside particles, which occurs when using conventional conjugation methods. This direct conjugation process yielded robust assay performance through homogeneous conjugation of the capture antibodies, and avoided the use of hydrolytically unstable linker additives. However, no research has been conducted to optimize the process of conjugating capture antibodies to the particles. We here present a strategy to optimize capture antibody conjugation based on the finding that excessive addition of capture antibodies during incubation can rather lower the amount of capture antibodies conjugated to the particles for some types of capture antibodies. Based on our optimized capture antibody conjugation process, a singleplex immunoassay for a selected target was conducted. Enhanced sensitivity compared with previous studies was confirmed. We also validated the increased specificity of multiplex detection through our optimization process. We believe that the optimization process presented herein for capture antibody conjugation will advance the field of encoded hydrogel microparticle-based immunoassays.
Proteins are three-dimensionally structured biomolecules that play diverse and essential roles in numerous metabolic processes, including respiration (Melin and Hellwig, 2020), immunity (Ozato et al., 2008), and digestion (Souza and Magalhães, 2010). Although the concentration of each protein involved in these metabolic processes is normally maintained within certain ranges by homeostasis, abnormal upregulation or downregulation of protein concentrations can occur due to specific diseases and disorders. Detection of abnormal protein concentrations in body systems is important for diagnosis of various diseases, such as cancer (Liang and Chan, 2007), preeclampsia (He et al., 2020), and COVID-19 (Kaur et al., 2020), and research aimed at better understanding these diseases. In particular, non-invasive liquid biopsy-based protein detection has aroused interest for its contribution to the facile monitoring and management of diseases (Lone et al., 2022) while minimizing patient discomfort.
In the field of liquid biopsy-based protein detection, multiplexing of the assay process is a powerful tool for efficient detection based on high-throughput processing of small-volume samples (Ahsan, 2021). Planar microarray and fluorescent bead technologies are representative widely used multiplex protein detection strategies (Cretich et al., 2014; Preuss et al., 2021). In these techniques, the target information can be encoded by the position or fluorescence of each system, enabling facile decoding during the multiplex detection process. Although these processes have been widely developed and commercialized, there are still inevitable limitations. For example, planar microarrays have long process durations and low sensitivities due to limited mass transport of the reactants to the planar surface (Cohen and Walt, 2018). In fluorescent bead assays, there is a vulnerability of code misreading by overlapping of the emitted wavelengths from the beads (Jun et al., 2012).
Graphically encoded hydrogel microparticles composed of polyethylene glycol (PEG) offers one means of overcoming these limitations (Lee et al., 2019a; Lee et al., 2019b). Graphical encoding of particles can provide a large encoding capacity (>106) for multiplex detection with facile distinguishability using simple imaging (Birtwell and Morgan, 2009). The anti-fouling property of the bioinert PEG components of the particles enhances specificity, which can reduce the vulnerability of false-positive bias due to signal originating from non-target molecules (Krishnan et al., 2008). These three-dimensional hydrogel networks enable incorporation of sufficient capture antibodies and targets to enhance sensitivity relative to that of the enzyme-linked immunosorbent assay (ELISA), the gold-standard immunoassay technique (Lee et al., 2019a; Lee et al., 2019b).
The role of capture antibodies is important in encoded hydrogel microparticle-based immunoassays because they directly capture the target antigens, and the amount of captured targets is quantitated by fluorescence or colorimetric signals to determine the target concentration in samples (Lee et al., 2019a; Lee et al., 2019b; Roh et al., 2020). Accordingly, the strategy for conjugating the capture antibodies into the hydrogel network is critical, and can greatly influence assay outcomes. Recently, a method for post-synthesis functionalization of reduced capture antibodies for conjugation to unreacted acrylate moieties in hydrogel particles was developed using click chemistry (Lee et al., 2019a; Roh et al., 2020). This direct and linker-free conjugation process enables robust assay performances including sensitivity and dynamic ranges based on homogeneous capture antibody conjugation into the particles and the non-requirement of hydrolytically unstable linker additives. Despite the powerful advantages of post-synthesis linker-free conjugation, no studies have addressed optimization of the post-synthesis functionalization process of the capture antibodies into particles, which can considerably influence assay performance.
Herein, we present a new optimization strategy for capture antibody conjugation based on the finding that excessive addition of capture antibodies during incubation can lower the conjugation efficiency of capture antibodies into the particles for some types of capture antibodies. First, the relationship between the assay signal and the concentration of capture antibodies in reaction buffer during incubation with the particles was characterized for two protein targets, vascular endothelial growth factor (VEGF) and choriogonadotropin subunit beta (CG beta). Unlike CG beta, there existed an optimized capture antibody concentration where the detection signal was the highest in case of VEGF assay. To clarify these dissimilar results, we scrutinized the relationship between the concentration of capture antibodies in the incubation buffer and the amount of capture antibodies attached to the particles for VEGF. The characterization identified the optimal concentration of capture antibodies during incubation to achieve maximal conjugation of VEGF. Based on the optimized capture antibody conjugation process for VEGF, a singleplex immunoassay was conducted, and the enhanced sensitivity compared with previous studies was confirmed. Finally, we validated the increased specificity of multiplex detection using the described optimization process.
Polyethylene glycol (PEG), polyethylene glycol diacrylate (PEGDA), 2-hydroxy-2-methylpropiophenone (Darocur 1,173), Tween 20, and streptavidin-phycoerythrin (SA-PE) were purchased from Sigma Aldrich (United States). Protein targets and antibodies were supplied by R&D Systems (Minneapolis, MN, United States). Bovine serum albumin (BSA) was obtained from R&D Systems. Fluorescein isothiocyanate (FITC)-labelled VEGF capture antibody was purchased from Bioss (United States). Tris (2-carboxyethyl) phosphine (TCEP) was purchased from Thermo Fisher Scientific (United States). Phosphate-buffered saline (1× PBS) and fetal bovine serum (FBS) were purchased from Welgene (Republic of Korea). PDMS reagents were obtained from Corning (United States). The SU-8 25 master mold was obtained from MicroChem (United States).
Hydrogel microparticles were produced using stop-flow lithography (SFL) on a PDMS channel. The PDMS reagents SYLGARD A and B were mixed at a 10:1 volume ratio and poured on the SU-8 25 master mold. This mixture was cured for 4 h in a 70°C incubator. After peeling the hardened PDMS channel from the master mold, inlet and outlet holes were punched. Then, the channel was placed on a PDMS-coated slide glass and cured for another 20 min in the 70°C incubator.
The particle precursor liquid comprised 40% (v/v) PEG (Mn = 600), 35% (v/v) deionized water, 20% (v/v) PEGDA (Mn = 700), and 5% (v/v) Darocur 1,173. The SFL cycle consisted of liquid flow, stop, ultraviolet light (UV) exposure, and holding for 400, 200, 75, and 200 ms, respectively. The ultraviolet light intensity was adjusted to 2,200 mW/cm2 when measured using a ×20 microscope objective. Two different photomasks were selected to encode the particles corresponding to each protein target: VEGF and CG beta. Liquid flow pressure was fixed at 10 kPa. A total of 1,000 hydrogel particles were produced and collected in microtubes containing PEG (Mn = 200). The retrieved particles were rinsed thrice with 1× PBS containing 0.05% (v/v) Tween 20 (PBST).
To optimize the concentration of capture antibodies for incubating with hydrogel microparticles, various concentrations of fluorescein isothiocyanate (FITC)-labelled antibodies were prepared. The FITC-labelled VEGF capture antibodies were diluted to 3,000, 1,200, 600, 300, 120, and 60 ng/μl in PBST. The antibodies were reduced in 0.1 mM TCEP for an hour on the benchtop prior to the downstream thiol-ene click reaction with the acrylate groups in the hydrogel particles. After TCEP reduction, each concentration of capture antibody was incubated with hydrogel particles at 25°C for 48 h with shaking at 1,500 rpm. After the reaction, the particles were rinsed five times with PBST buffer after the reaction. The immunoassay signals according to the capture antibody concentrations were acquired using the same concentrations used in the FITC-labelled antibody experiments. Varying concentrations of capture antibodies were conjugated via the method described above, followed by addition of 1,024 pg/ml of target for quantitative assessment. Bound targets were flanked by biotinylated secondary antibodies. The fluorescent substance SA-PE was labelled for signal analysis. A detailed description of the assay procedure is provided in Section 2.5. Identical procedure with that of VEGF was conducted in CG beta capture antibody conjugation to the hydrogel particles used for detection.
The degree of aggregation of VEGF and CG beta capture antibodies was observed using cryo-electron microscopy (cryo-EM). Cryo-EM enables the examination of samples in their natural state, minimizing deformation by freezing the samples in a very short period of time at cryogenic temperatures. Capture antibodies of 3,000 ng/μl were reduced in TCEP and rapidly frozen using a Thermo Vitrobot Plunge-freezing Instrument for Cryo TEM (Thermo Fisher Scientific, United States). Frozen samples were placed on a JEM-2100Plus Electron microscope (JEOL, Japan) for analysis. An acceleration voltage of 200 kV was applied, and ×150,00, ×25,000, and ×40,000 microscope objectives were used for imaging. Images were edited using ImageJ software.
Target proteins were prepared in FBS at various concentrations. The initial VEGF stock solution was made into 30,000 pg/ml. Serial dilutions were made to obtain 2048, 512, 128, 32, and 8 pg/ml working stocks. The CG beta stock was diluted to the same concentrations. Fifty capture antibody-loaded particles suspended in 40 μL PBST were mixed with 40 μl of target solution. The final target concentrations ranged from 15,000 pg/ml to 4 pg/ml for VEGF, and from 7,500 pg/ml to 4 pg/ml for CG beta. The mixture was incubated at 25°C for 2 h with agitation at 1,500 rpm. After incubation with target, the particles were rinsed thrice with PBST.
The secondary antibody was prepared in PBST containing 5% BSA at final concentrations of 25 ng/μl for VEGF, and 125 ng/μl for CG beta. 40 μl of the previous reaction solution and 10 μl of secondary antibody cocktail were combined and incubated at 25°C for 1 h at 1,500 rpm.
After washing the particles three times with PBST, the fluorescent substance SA-PE diluted in PBST containing 5% BSA and PBST was added to obtain a final dilution factor of 50. 10 μl of diluted SA-PE were added to 40 μl of the previous solution. The mixture was incubated at 25°C for 30 min at 1,500 rpm and rinsed five times with PBST.
For each 2 sets of the four combinations, 50 particles per protein target (100 particles in total) suspended in 40 μl of PBST were mixed with 40 μl of 2× target proteins in FBS. The final target concentrations of the targets in presence were 512 pg/ml and 256 pg/ml for VEGF and CG beta, respectively. A mixture of eight different combinations was placed in a 25°C incubator for 2 h at 1,500 rpm. The particles were rinsed three times using PBST, with a final volume of 30 µl for each microtube. 10 μl of secondary antibodies at concentrations of 25 ng/μl for VEGF and 125 ng/μl for CG beta were mixed in advance and added to the particle solution. The final volume in each microtube was 50 µl. The mixtures were incubated at 25°C for 1 h. After rinsing the particles with PBST three times, the secondary antibodies were incubated with 10 µl of fluorescent tag SA-PE diluted with PBST containing 5% BSA at a ratio of 1:50. The particles were rinsed five times with PBST prior to imaging.
Grayscale fluorescence images of the hydrogel particles were captured using a Prime CMOS camera (United States) connected to a microscope. The fluorescence light source was exposed with HXP 120 V (Germany). The light was filtered with fluorescent microscope filter sets of λex/λem = 546/590 nm for SA-PE and λex/λem = 450–490/515 nm for FITC. The fluorescence exposure times for imaging were 50 ms for SA-PE and 500 ms for FITC. The images were saved in TIFF format, and signal intensity was quantitated using ImageJ software. Background signals from regions outside the particle were subtracted to obtain net signal intensities.
The encoded hydrogel microparticles were synthesized using stop-flow lithography, as shown in Figure 1A. Stop-flow lithography can produce uniform graphically structured hydrogel microparticles by periodic exposure of the precursor to ultraviolet rays (UV) through a photomask in microfluidic channels. After the particles were synthesized, the unreacted acrylate groups of double-armed-PEGDA remained in the hydrogel networks owing to incomplete conversion. These remnant acrylate groups can facilitate conjugation of reduced capture antibodies via thiol-ene Michael addition reaction (Mather et al., 2006) (Figure 1A). The free thiol groups in the antibodies were supplied by reduction of disulfide bonds between the two heavy chains of the antibody, as illustrated in Figure 1A. We selected TCEP as the reducing agent because it is powerful and highly tolerant to a wide pH range in aqueous conditions (Rhee and Burke, 2004). The reaction concentration of TCEP was 0.1 mM, which was determined in previous optimization experiments (Lee et al., 2019a).
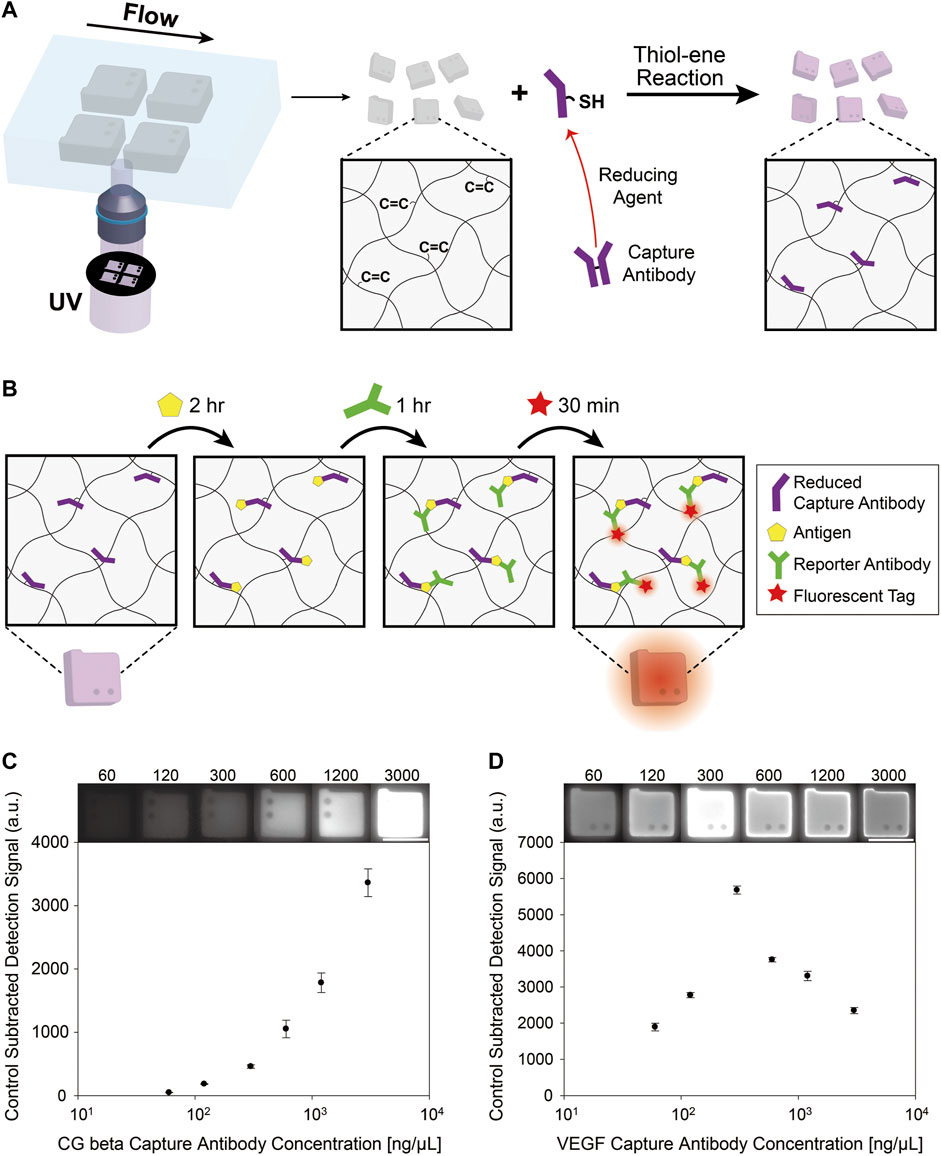
FIGURE 1. (A) Synthesis of encoded hydrogel microparticles with stop-flow lithography. Conjugation of thiol groups of the reduced antibody to the unreacted acrylate groups of the encoded hydrogel particles by thiol-ene reaction. (B) The detection process of protein antigen utilizing capture antibody conjugated hydrogel microparticles. The particles are incubated with the target protein for 2 h. Then, a biotinylated reporter antibody binds to the target. After the reporter antibody is attached, the fluorescent label, streptavidin-phycoerythrin (SA-PE) binds to the reporter antibody by streptavidin-biotin interaction. (C) Control-subtracted detection signals of hydrogel particles after immunoassay varying concentrations of CG beta capture antibodies during post-synthesis functionalization. (D) Control-subtracted detection signals of hydrogel particles after immunoassay varying concentrations of VEGF capture antibodies during post-synthesis functionalization. The scale bars are 100 μm.
To study the relationship between the assay signal and the capture antibody concentration in reaction buffer, we conducted immunoassay following the procedures presented in Figure 1B. The target antigen, biotinylated reporter antibody, and fluorescent tag linked with streptavidin were incubated consecutively with the particles, same process as the previous hydrogel microparticle based immunoassay studies. VEGF and CG beta were used for the target proteins because they are both essential biomarkers for preeclampsia (Carty et al., 2008), a pregnancy complication that induces high blood pressure and kidney damage (Tangren et al., 2018).
The characterization results of the relationship between capture antibody concentration in incubation buffer and the assay signal is presented in Figures 1C,D. Intriguingly, the signal variance aspect according to capture antibody concentration (3,000, 1,200, 600, 300, 120, and 60 ng/μl) were different between VEGF and CG beta. For VEGF samples, the capture antibody concentration at maximum signal value was 300 ng/μl, while it was 3,000 ng/μl for CG beta samples. The presence of a peak point in VEGF assay result is conflicting with the conventional notion that higher concentrations of capture antibodies in the incubation buffer increase the amount of conjugated antibodies in the particles induced by faster reaction rates.
We hypothesized that the higher concentration of VEGF capture antibodies during particle functionalization rather lowered the conjugation efficiency of VEGF capture antibodies to the particles, and thereby reduced the overall assay signals. To validate the hypothesis, we used fluorescein (FITC) labelled VEGF capture antibodies to compare the fluorescence signals after post-synthesis functionalization at various antibody concentrations (Figure 2A). It can be expected that as the amount of capture antibody conjugated to the hydrogel particles increases, fluorescence signal would also increase because more fluorescent dye molecules are incorporated into the particles. In general, when reactants are conjugated to hydrogel microparticles, a higher reactant concentration in the reaction buffer results in a higher conjugation efficiency due to reaction kinetics (Xu et al., 2018). However, in the case of VEGF capture antibodies, a reaction concentration of 300 ng/μL produced the highest signal, implying that the highest amount of antibody was conjugated (Figure 2B). The presence of peak value at 300 ng/μl of VEGF antibody concentration in Figure 2B coincide well to the result of Figure 1D, which demonstrates the hypothesis that higher concentration of VEGF capture antibodies can rather lower the conjugation efficiency in particles.
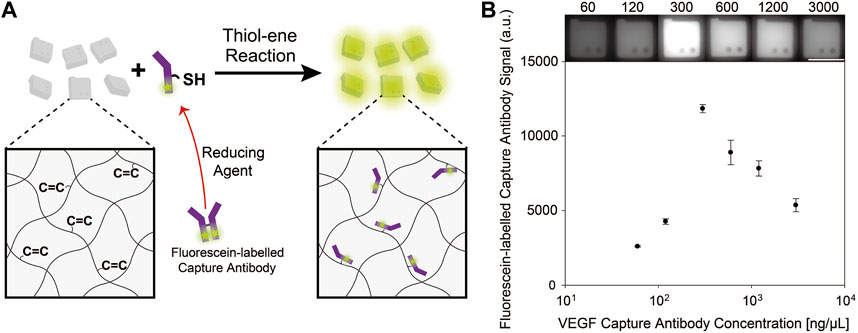
FIGURE 2. (A) Conjugation of thiol groups in reduced FITC-labelled capture antibodies to the unreacted acrylate groups in the encoded hydrogel particles. (B) Fluorescence signals of hydrogel particles incubated with varying concentrations of FITC-labelled VEGF capture antibodies. The scale bar is 100 μm.
We speculated that this inconsistent result between two protein targets is attributed to antibody aggregation in the reaction solution for some types of capture antibodies, including VEGF. To confirm this speculation, we performed cryo-EM analysis of VEGF and CG beta capture antibodies at a concentration of 3,000 ng/μl. While VEGF capture antibodies clustered, CG beta capture antibodies displayed no aggregation, as shown in Supplementary Figure S1. When capture antibodies form aggregates, their bulky forms are less likely to disperse through the hydrogel matrix, and they are impeded in capturing target antigens even when conjugated to the hydrogel particles. Accordingly, it is important to screen for optimal capture antibody concentrations in reaction solutions, considering that high concentrations can strengthen the aggregation behavior of some types of antibodies. Despite the presence of large-scale aggregates in cryo-EM image (Supplementary Figure S1), no significant aggregation was observed in the fluorescence images of the particles functionalized with VEGF capture antibodies (Figure 1D, Figure 2B). We speculate that the aggregates smaller than 200 nm, which are under the resolution of optical microscopes, cannot be observed, and the aggregates bigger than 200 nm are relatively difficult to penetrate into and diffuse through the hydrogel microparticles with nanometer-scale pores compared to optically indistinguishable less-aggregated and well-dispersed antibodies (Choi et al., 2012). To validate that the penetration of optically distinguishable large aggregates into the particles are limited, we synthesized cuboid microposts with hydrogel fabrication procedure presented in section 2.2. The microposts (side lenth∼200 μm) were synthesized in the (3-Mercaptopropyl) trimethoxysilane (MPTMS) coated channel (height∼60 μm) utilizing the process of the previous study (Jang et al., 2022). The fluorescently labelled 100 nm nanobeads were incubated in microchannels to mimic large-scale antibody aggregates, and reduced VEGF capture antibodies labelled with FITC were incubated to confirm that optically indistinguishable less-aggregated and well-dispersed antibodies can penetrate into the microgel and react with unreacted acrylate groups. The fluorescent microposts were imaged after 6 h of incubation, and fluorescent signal ratio of the post to the background signal is calculated to be 19.0% for nanobeads and 85.4% for antibody (Supplementary Figure S2A). The distinctly higher signal ratio to background of the antibody sample than that of the nanobead sample indicates that the penetration efficiency of optically indistinguishable less-aggregated and well-dispersed antibodies is higher than that of optically distinguishable large aggregates. Furthermore, a uniform and clear fluorescence signal of microposts after rinsing off the unreacted antibodies was confirmed, which indicate the reaction of penetrated antibodies and unreacted acrylate groups in microposts (Supplementary Figure S2B).
Thus, we concluded that 300 ng/μl of capture antibody is the optimal concentration to detect VEGF antigen, unlike previously used values in former studies involving hydrogel microparticles.
To confirm that optimizing the capture antibody conjugation process can enhance the assay performance and to acquire the calibration curve for multiplexing, we conducted singleplex assays of VEGF and CG beta target. For singleplex detection of CG beta, 3,000 ng/μl of capture antibody was incubated with the particles, which is the same concentration used in previous study (Lee et al., 2019a). The selected capture antibody concentrations for VEGF detection were 300 and 3,000 ng/μl, which were the optimized and previously exploited values, respectively (Lee et al., 2019a). As shown in the particle images and graph, fluorescence signals in both assays linearly increased with spiked target concentration (Figure 3). Sensitivity was determined by calculating the limit of detection (LOD), which is defined as the lowest target concentration that produces an assay signal three standard deviations larger than the control signal. The LOD of CG beta detection was 1.91 pg/ml, which is nearly identical with the previous value, 2.07 pg/ml. The LOD of the optimized VEGF assay was 1.01 pg/ml, which was approximately five times higher than that of the non-optimized case, 5.38 pg/ml. The LOD of the optimized assay was also 31.0 times more sensitive than ELISA, the gold standard for immunoassays. The dynamic range, which is the range from lowest to highest detectable target concentration, is an important feature of immunoassays. Based on the improved lower detection limit, the dynamic range was broadened in the optimized assay from 3.5 log to 4.2 log(Table 1). In conclusion, we demonstrated that the optimized capture antibody conjugation process can enhance the sensitivity and dynamic range.
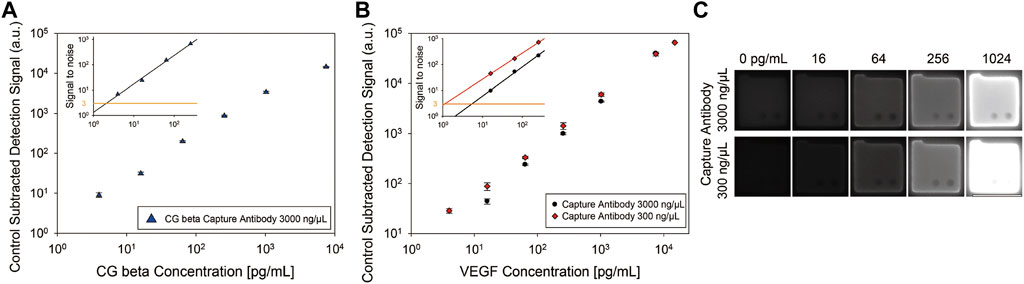
FIGURE 3. Singleplex assay results of CG beta and VEGF. (A) Control-subtracted CG beta detection signals using particles incubated with 3,000 ng/μl of capture antibody. (B) Control-subtracted VEGF detection signals using two types of particles, each incubated with 3,000 ng/μl and 300 ng/μl of capture antibody. The inset displays signal-to-noise, signals divided by standard deviation of the control for each case. LOD is the target concentration where each regression plot intersects with y = 3. (C) Fluorescence images of the hydrogel microparticles from VEGF singleplex detection. The scale bar is 100 μm.
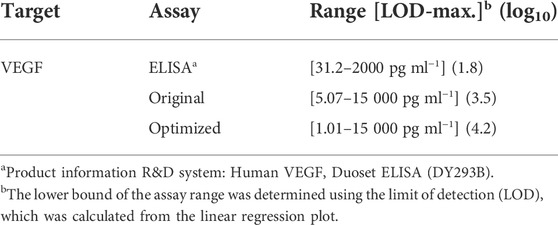
TABLE 1. The limit of detection and dynamic range of ELISA, the original particle assay, and the optimized conjugation process assay.
The ability to detect multiple protein biomarkers simultaneously (multiplexing) is a key ability in immunoassays because it can reduce the cost and time required for diagnosis. One of the most important features of multiplexing is specificity, which is the ability to accurately identify the presence and concentration of each target protein without false-positive outcomes. To characterize and compare the specificity in multiplexing between the optimized and non-optimized VEGF capture antibody conjugation processes, we conducted a duplex immunoassay on 512 pg/ml of VEGF and 256 pg/ml of CG beta antigen. When conjugating the VEGF capture antibodies to the particles, 3,000 ng/μl and 300 ng/μl of VEGF capture antibody were used during incubation, representing non-optimized and optimized conditions, respectively.
Duplex assay results are presented in Figure 4A. We calculated the recovery rate, which is the ratio of the observed target concentration to the actual target concentration, to confirm the consistency between the singleplex and multiplex assays (Table 2). The recovery rates for each capture antibody conjugation condition and target type ranged between 70% and 130%, which are acceptable values for application in industrial fields (Djoba Siawaya et al., 2008). The specificity of multiplexing was quantitatively assessed by comparing the increased non-specific signals induced by cross-reactivity for the non-optimized and optimized cases, according to the presence of the CG beta antigen and its secondary antibody in the samples. The increased non-specific signals was calculated by subtracting each control signal of the particles conjugated with VEGF capture antibodies in the singleplex assays from the control signals of the particles in the multiplex assay. As shown in Figure 4B, the increase in non-specific signals during multiplexing was much lower for the optimized case. In particular, the ratio of the increased non-specific signal between the non-optimized and optimized cases was much higher when the antigen and secondary antibody of CG beta were both included (a factor of ∼3.81), compared to the case when only secondary antibody was included (a factor of ∼2.12).
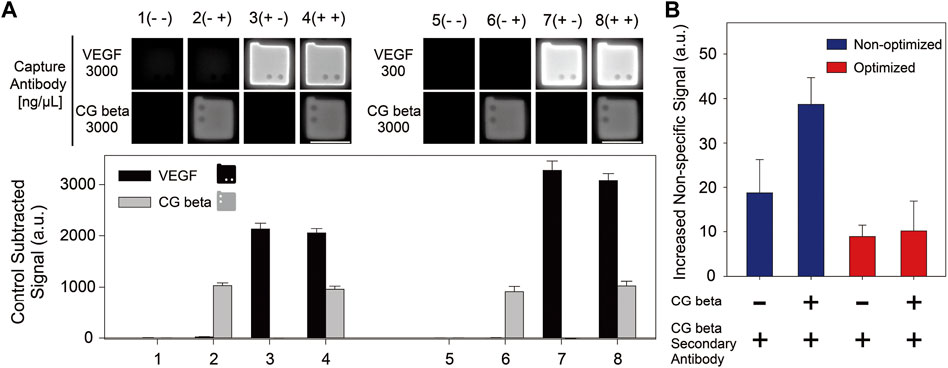
FIGURE 4. (A) Multiplex immunoassay results for VEGF and CG beta targets. Two sets of multiplex detection were conducted, using particles incubated 300 ng/μl and 3,000 ng/μL of VEGF capture antibodies, and 3,000 ng/μl of CG beta capture antibodies during post-synthesis functionalization. The spike-in concentrations for VEGF and CG beta target were 512 pg/ml and 256 pg/ml, respectively. The scale bars are 100 μm. (B) The increased non-specific signals of the particles functionalized with VEGF capture antibodies in multiplex assay induced by antigen and secondary antibody of CG beta. The increased non-specific signals was calculated by subtracting each control signal of the particles conjugated with VEGF capture antibodies in the singleplex assays from the control signals of the particles in the multiplex assay.
The smaller increase in non-specific signals in the optimized cases during multiplex immunoassay can be explained by the difference in the amount of antibody aggregates formed when VEGF capture antibodies were incubated with particles. As presented in Supplementary Figure S1, large aggregations of VEGF capture antibodies were observed at the non-optimized antibody concentration (3,000 ng/μl) during the incubation process. A considerable amount of capture antibody aggregates adsorbed to the particles in the non-optimized condition might increase cross-reactivity by inducing non-specific binding with the antigen and secondary antibodies of CG beta.
In this study, we introduced a new optimization method for conjugation of reduced capture antibodies to encoded hydrogel microparticles, that enhances immunoassay performance. We demonstrated that a lower amount of capture antibodies was conjugated when particles were reacted with excessive amounts of some capture antibodies, such as VEGF. Based on the optimization results, we conducted singleplex VEGF detection and succeeded in enhancing the sensitivity and dynamic range compared to those in a previous non-optimized study. Furthermore, duplex detection of VEGF and CG beta was conducted to show enhanced specificity in multiplex immunoassays by lowering the increase in non-specific signals when the optimized process was adopted.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
DK conceived and conducted experiments, designed figures and wrote the manuscript. JK wrote the manuscript and contributed to experiments and figure design. WJ wrote and reviewed the manuscript and figures. KB conceived of experiments and wrote, reviewed, revised, and edited the manuscript. All authors have contributed to the manuscript and approved the submitted version.
This work received financial support from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1D1A1B07046577), Engineering Research Center of Excellence Program through the NRF funded by the Ministry of Science and ICT (MSIT, South Korea) (2016R1A5A1010148), the Next-Generation Biogreen 21 Program funded by the Rural Development Administration of the Republic of Korea (PJ016004), and the Technology Innovation Program (20018111, Development of super-fast multiplex technology for the examination of diagnosis of infectious disease and in-body response test) funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsens.2022.1007355/full#supplementary-material
Ahsan, H. (2021). Monoplex and multiplex immunoassays: Approval, advancements, and alternatives. Comp. Clin. Path. 31, 333–345. doi:10.1007/s00580-021-03302-4 | |
Birtwell, S., and Morgan, H. (2009). Microparticle encoding technologies for high-throughput multiplexed suspension assays. Integr. Biol. 1 (5-6), 345–362. doi:10.1039/b905502a | |
Carty, D. M., Delles, C., and Dominiczak, A. F. (2008). Novel biomarkers for predicting preeclampsia. Trends Cardiovasc. Med. 18 (5), 186–194. doi:10.1016/j.tcm.2008.07.002 | |
Choi, N. W., Kim, J., Chapin, S. C., Duong, T., Donohue, E., Pandey, P., et al. (2012). Multiplexed detection of mRNA using porosity-tuned hydrogel microparticles. Anal. Chem. 84 (21), 9370–9378. doi:10.1021/ac302128u | |
Cohen, L., and Walt, D. R. (2018). Highly sensitive and multiplexed protein measurements. Chem. Rev. 119 (1), 293–321. doi:10.1021/acs.chemrev.8b00257 | |
Cretich, M., Damin, F., and Chiari, M. (2014). Protein microarray technology: How far off is routine diagnostics? Analyst 139 (3), 528–542. doi:10.1039/c3an01619f | |
Djoba Siawaya, J. F., Roberts, T., Babb, C., Black, G., Golakai, H. J., Stanley, K., et al. (2008). An evaluation of commercial fluorescent bead-based luminex cytokine assays. PloS one 3 (7), e2535. doi:10.1371/journal.pone.0002535 | |
He, A., Zhou, Y., Wei, Y., and Li, R. (2020). Potential protein biomarkers for preeclampsia. Cureus 12 (6), e8925. doi:10.7759/cureus.8925 | |
Jang, W., Kim, D. Y., Mun, S. J., Choi, J. H., Roh, Y. H., and Bong, K. W. (2022). Direct functionalization of cell‐adhesion promoters to hydrogel microparticles synthesized by stop‐flow lithography. J. Polym. Sci. 60, 1767–1777. doi:10.1002/pol.20210934 |
Jun, B.-H., Kang, H., Lee, Y.-S., and Jeong, D. H. (2012). Fluorescence-based multiplex protein detection using optically encoded microbeads. Molecules 17 (3), 2474–2490. doi:10.3390/molecules17032474 | |
Kaur, M., Tiwari, S., and Jain, R. (2020). Protein based biomarkers for non-invasive Covid-19 detection. Sens. Bio-Sensing Res. 29, 100362. doi:10.1016/j.sbsr.2020.100362 | |
Krishnan, S., Weinman, C. J., and Ober, C. K. (2008). Advances in polymers for anti-biofouling surfaces. J. Mat. Chem. 18 (29), 3405–3413. doi:10.1039/b801491d |
Lee, H. J., Kim, J. Y., Roh, Y. H., Kim, S. M., and Bong, K. W. (2019a). Linker-free antibody conjugation for sensitive hydrogel microparticle-based multiplex immunoassay. Analyst 144 (22), 6712–6720. doi:10.1039/c9an01243e | |
Lee, H. J., Roh, Y. H., Kim, H. U., Kim, S. M., and Bong, K. W. (2019b). Multiplexed immunoassay using post-synthesis functionalized hydrogel microparticles. Lab. Chip 19 (1), 111–119. doi:10.1039/c8lc01160e | |
Liang, S.-L., and Chan, D. W. (2007). Enzymes and related proteins as cancer biomarkers: A proteomic approach. Clin. Chim. acta 381 (1), 93–97. doi:10.1016/j.cca.2007.02.017 | |
Lone, S. N., Nisar, S., Masoodi, T., Singh, M., Rizwan, A., Hashem, S., et al. (2022). Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 21 (1), 79–22. doi:10.1186/s12943-022-01543-7 | |
Mather, B. D., Viswanathan, K., Miller, K. M., and Long, T. E. (2006). Michael addition reactions in macromolecular design for emerging technologies. Prog. Polym. Sci. 31 (5), 487–531. doi:10.1016/j.progpolymsci.2006.03.001 |
Melin, F., and Hellwig, P. (2020). Redox properties of the membrane proteins from the respiratory chain. Chem. Rev. 120 (18), 10244–10297. doi:10.1021/acs.chemrev.0c00249 | |
Ozato, K., Shin, D.-M., Chang, T.-H., and Morse, H. C. (2008). TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8 (11), 849–860. doi:10.1038/nri2413 | |
Preuss, J. M., Burret, U., and Vettorazzi, S. (2021). “Multiplex fluorescent bead-based immunoassay for the detection of cytokines, chemokines, and growth factors,” in Proteomic profiling (Springer), 247–262. |
Rhee, S. S., and Burke, D. H. (2004). Tris (2-carboxyethyl) phosphine stabilization of RNA: Comparison with dithiothreitol for use with nucleic acid and thiophosphoryl chemistry. Anal. Biochem. 325 (1), 137–143. doi:10.1016/j.ab.2003.10.019 | |
Roh, Y. H., Lee, H. J., Kim, J. Y., Kim, H. U., Kim, S. M., and Bong, K. W. (2020). Precipitation-based colorimetric multiplex immunoassay in hydrogel particles. Lab. Chip 20 (16), 2841–2850. doi:10.1039/d0lc00325e | |
Souza, P. M. d., and Magalhães, P. d. O. (2010). Application of microbial α-amylase in industry-A review. Braz. J. Microbiol. 41, 850–861. doi:10.1590/s1517-83822010000400004 | |
Tangren, J. S., Wan Md Adnan, W. A. H., Powe, C. E., Ecker, J., Bramham, K., Hladunewich, M. A., et al. (2018). Risk of preeclampsia and pregnancy complications in women with a history of acute kidney injury. Hypertension 72 (2), 451–459. doi:10.1161/hypertensionaha.118.11161 | |
Keywords: encoded hydrogel microparticles, immunoassay, capture antibody, preeclampsia, multiplex detection
Citation: Kim DY, Kim J, Jang W and Bong KW (2022) Optimizing reduced capture antibody conjugation to encoded hydrogel microparticles for enhanced multiplex immunoassays. Front. Sens. 3:1007355. doi: 10.3389/fsens.2022.1007355
Received: 30 July 2022; Accepted: 30 August 2022;
Published: 16 September 2022.
Edited by:
Chang-Soo Lee, Chungnam National University, South KoreaReviewed by:
Dong-Sik Shin, Sookmyung Women’s University, South KoreaCopyright © 2022 Kim, Kim, Jang and Bong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ki Wan Bong, Ym9uZzk4QGtvcmVhLmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.