- 1Nanomedicine Group, UCIBIO, Dept, Ciências da Vida, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, Caparica, Portugal
- 2i4HB, Associate Laboratory—Institute for Health and Bioeconomy, Faculdade de Ciências e Tecnologia, Universidade NOVA de Lisboa, Caparica, Portugal
- 3ALMASCIENCE, Caparica, Portugal
Nucleic acid amplification technologies (NAATs) have become fundamental tools in molecular diagnostics, due to their ability to detect small amounts of target molecules. Since its development, Polymerase Chain Reaction (PCR) has been the most exploited method, being stablished as the “gold standard” technique for DNA amplification. However, the requirement for different working temperatures leads to the need of a thermocycler machine or complex thermal apparatus, which have been preventing its application in novel integrated devices for single workflow and high throughput analysis. Conversely, isothermal amplification methods have been gaining attention, especially for point-of-care diagnosis and applications. These non-PCR based methods have been developed by mimicking the in vivo amplification mechanisms, while performing the amplification with high sensitivity, selectivity and allowing for high-throughput analysis. These favorable capabilities have pushed forward the implementation and commercialization of several platforms that exploit isothermal amplification methods, mostly against virus, bacteria and other pathogens in water, food, environmental and clinical samples. Nevertheless, the future of isothermal amplification methods is still dependent on achieving technical maturity and broader commercialization of enzymes and reagents.
Introduction
Innovations in biotechnology and molecular biology have push forward novel nucleic acid amplification technologies (NAATs) to meet the demands for more efficient, sensitive, specific protocols capable to be integrated into portable systems and for high-throughput analyses. Polymerase Chain Reaction (PCR), introduced by Mullis in 1985 (Mullis et al., 1986) has become the leading method for DNA amplification with application in all types of molecular detection strategies, such as recognition and identification of infectious pathogens, characterization of genetic disorders, identification of disease biomarkers, gene expression studies, sample preparation for downstream applications and techniques (e.g., sequencing, labelling, etc.), among many others (Klein 2002). In fact, PCR is considered the “gold standard” in molecular analysis of nucleic acids, owing to its capability to amplify from as few as 1 to 10 molecules of target, ultimately leading to an increase to the sensitivity of molecular assays. As a result of the hype regarding PCR, many variants have been developed to address the ever-growing need for additional features, such as multiplex (Edwards and Gibbs 1994), nested (Porter-Jordan et al., 1990), quantitative (Porter-Jordan et al., 1990), reverse (Porter-Jordan et al., 1990) and digital forms (Kopp et al., 1998), which will be further addressed. This is indeed the technique against all others compare.
Still, inherent features to PCR require dedicated instrumentation capable of thermal cycling with appropriate temperature control, which is usually found in laboratory settings. This small but critical bottleneck has spurred the development of non-PCR based techniques, preferably suitable to be performed at a constant temperature—isothermal amplification (IA) methods, which aim at simplifying protocols for point-of-care (PoC) use, allowing to move the molecular diagnosis from centralized labs.
This review shall focus on the most representative and applied isothermal amplification methods, such as Nucleic Acid Sequence-based Amplification (NASBA), Loop-mediated Isothermal Amplification (LAMP), Strand Displacement Amplification (SDA), Recombinase Polymerase Amplification (RPA) and Rolling Circle Amplification (RCA), as well as their characteristics, applications and prospects to become the next gold standard amplification technique (Figure 1).
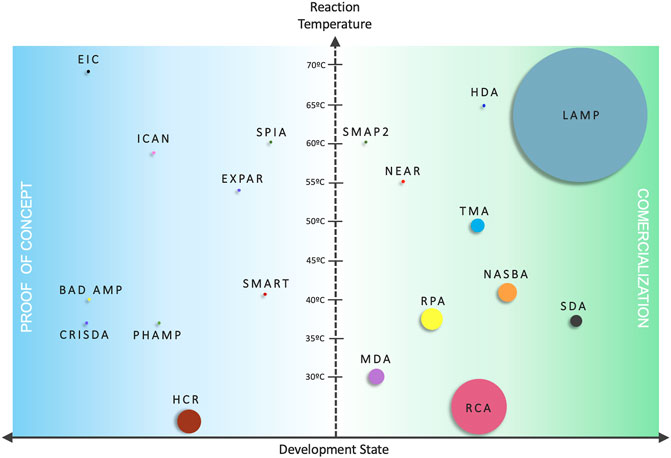
FIGURE 1. Comparison between isothermal amplification mechanisms. Vertical axis represents the reaction temperature and the horizontal axis the development status—(Blue) IA methods not developed much beyond proof of concept/academic research only; (Green) IA methods commercialized under kit formats/exploited by biotech companies. The size of the circles is proportional to the number of scientific items in literature.
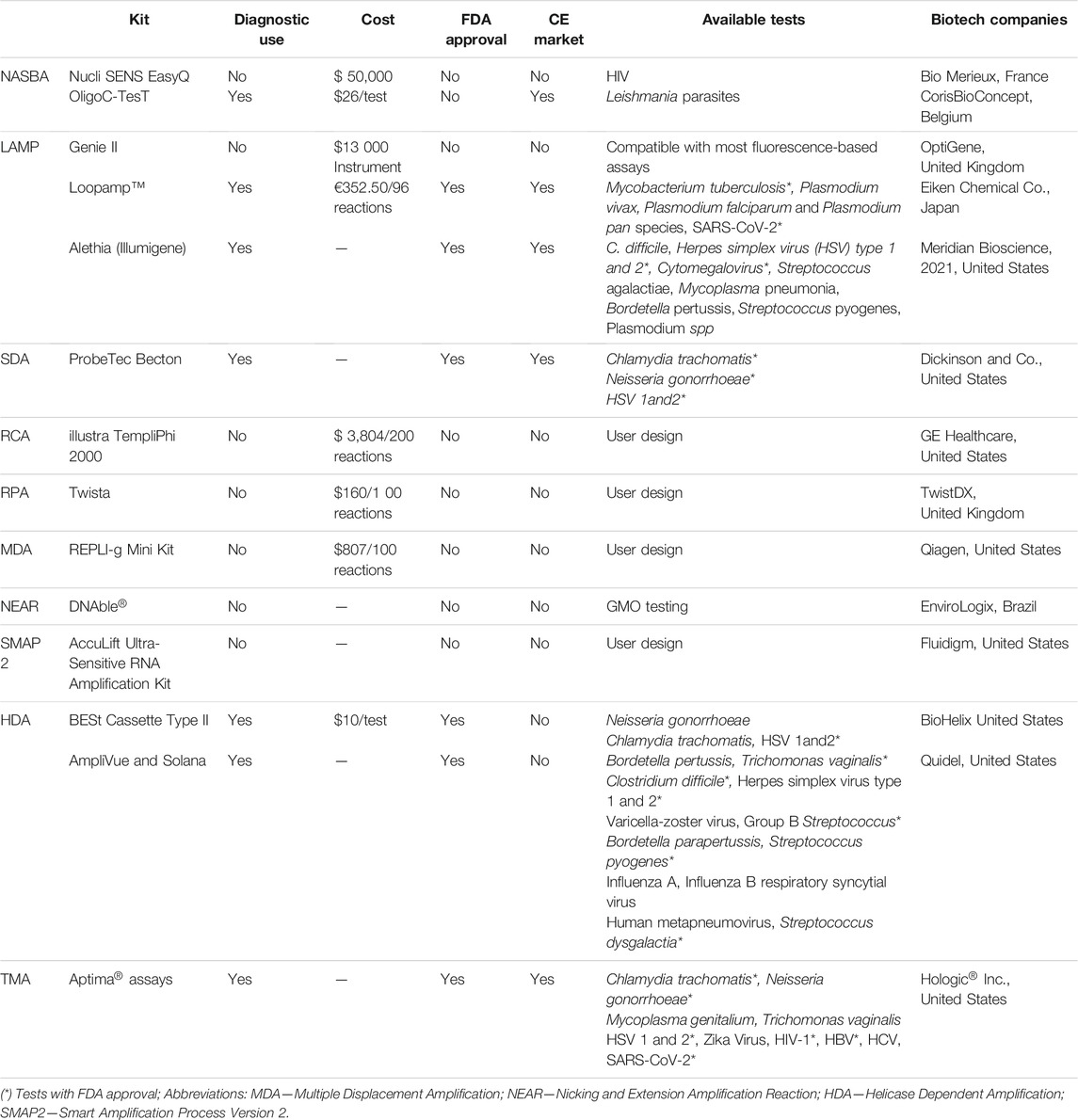
Despite the plethora of different IA concepts, they converge in the need to remove temperature cycling requirements for specific amplification (Li and Macdonald, 2015). While PCR requires the lowering and rising of two or three working temperatures, to allow annealing and denaturation of DNA strands and primers, and to facilitate strand displacement, IA methods proceed at a single working temperature, which removes the need for a thermocycler. Some of these isothermal amplification mechanisms also allow direct amplification from non-DNA targets, such as RNA (without the need for an additional retro transcription step) or protein-nucleic acids conjugates. Another interesting feature of these IA reactions is that they show increased tolerance to (bio-)chemical inhibitors often present in clinical samples (Bachmann et al., 2009; Craw and Balachandran 2012; Kaneko et al., 2007; Mori and Notomi 2009; Vincent et al., 2004). A summary of the main IA mechanisms proposed in the literature are presented in Table 1, together with key advantages and disadvantages.
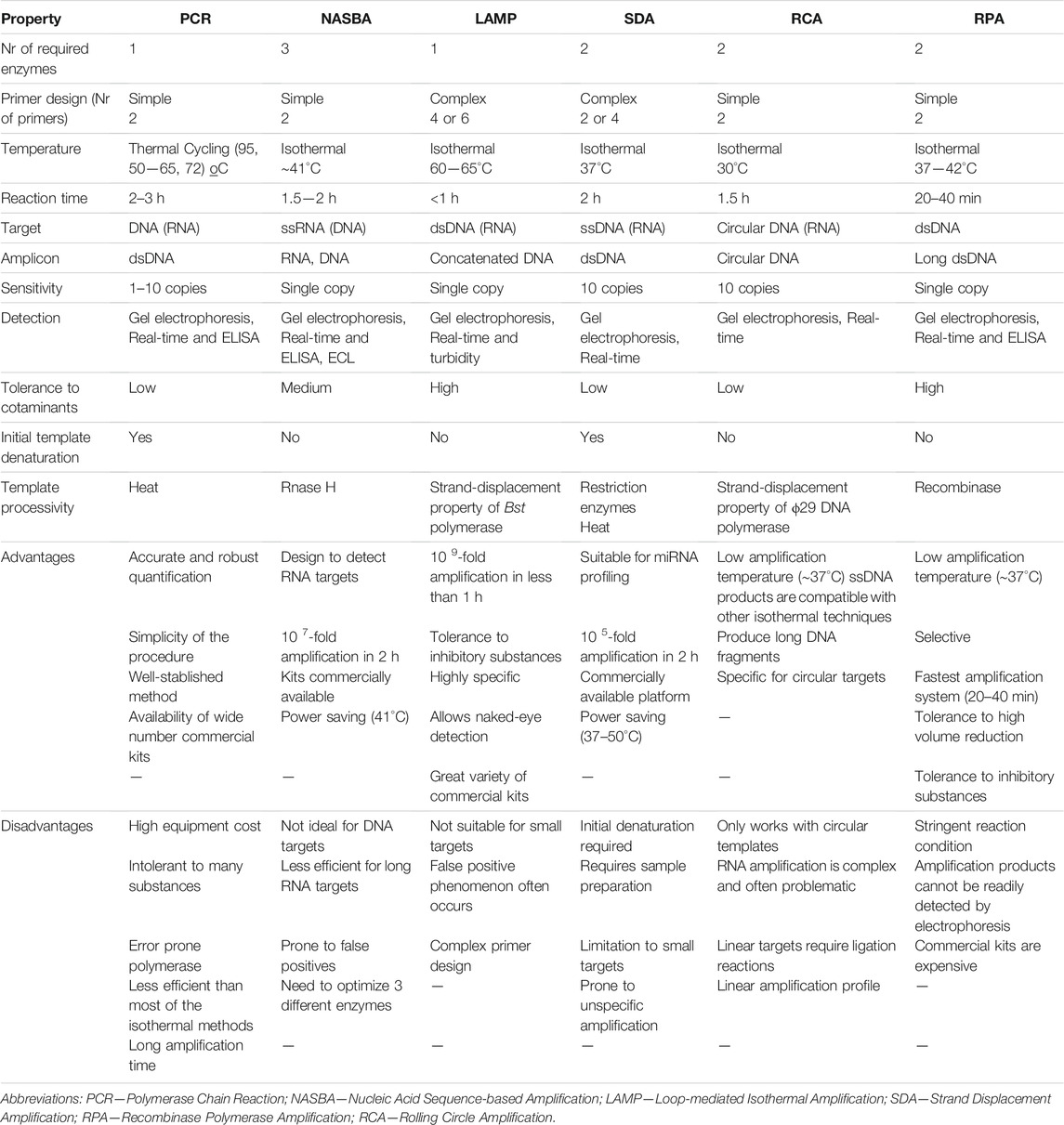
TABLE 1. Summary of the characteristics of PCR and the main isothermal amplification systems, highlighting key advantages and disadvantages.
Despite the exciting advantages of the IA schemes, these approaches also face some challenges, such as complex primer design, unspecific amplification, high background signal, confuse reaction mechanisms and the requirement for more enzymes and denaturing agents (Karami et al., 2011), These methods differ from each other in terms of primers, enzymes, reaction conditions, attainable sensitivity, specificity and ideal target molecules (miRNA, long DNA fragments, circular DNA and RNA targets) (Chang et al., 2012). Overall, isothermal amplification processes produce longer amplicons, have greater amplification efficiency, produce higher amplification yields and require less sample preparation steps than PCR (Klein 2002). In fact, it has been reported that non-PCR based clinical tests often outmatch the PCR results in clinical diagnosis (Aryan et al., 2010; Bachmann et al., 2009). From these, a risen trend concerning IA systems has been observable, over the last decades (Figure 2).
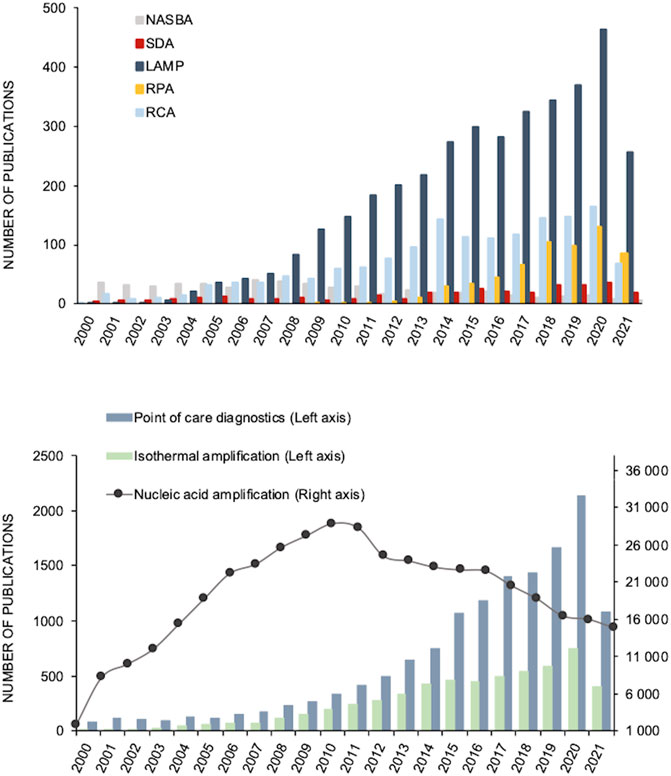
FIGURE 2. Number of scientific publications per year concerning amplification systems from 2000 until 2021. (A) Number of publications with the key words “Nucleic acid sequence-based amplification (NASBA)”, “Strand displacement amplification (SDA)”, “Loop mediated isothermal amplification (LAMP)”, “Rolling circle amplification (RCA)” and “Recombinase polymerase amplification (RPA)”. (B) Number of with the key words “DNA amplification”, “Point of care diagnosis” and “Isothermal amplification”. All the data was obtained from PubMed (accessed May 21, 2021).
In fact, scientific literature has been reporting the application of isothermal amplification systems in a wide number of fields, among them: pathogen detection in clinical, environmental and food samples (Fang et al., 2010; Chow et al., 2008; Zhao et al., 2012; Mahmoudian et al., 2008a; Krõlov et al., 2014), diagnosis of various infectious diseases (Schopf et al., 2011; Hellyer and Nadeau 2004; B. A. Rohrman and Richards-Kortum 2012; Torres-Chavolla and Alocilja 2011; S. Y. Lee et al., 2007) and Single-Nucleotide Polymorphisms (SNP) detection (Li et al., 2010). Furthermore, some clinical trials based on IA systems for diagnosis and detection of infectious diseases have been performed: LAMP—(ClinicalTrials.gov Identifier:NCT02371395 2018a; ClinicalTrials.gov Identifier: NCT04579549, 2021; ClinicalTrials.gov Identifier:NCT03829735, 2020a; NASBA—ClinicalTrials.gov Identifier: NCT01838902 2018a; HDA—ClinicalTrials.gov Identifier: NCT01838902 2018b; RPA—ClinicalTrials.gov Identifier: NCT04500873 2020b). Among the publications concerning these amplification techniques, most are focused on the analysis of water and food, with LAMP covering 67 and 59% of the publications, respectively. Then RPA covers for circa 20%, and the remainders, RPA, RCA, HDA and NASBA add up to 5%.
Despite the continuous efforts to make IA strategies more robust, there are still some hurdles before these approaches might eventually counteract current PCR limitations. Some constraints relate to complex primer design, different kinetics for template denaturation, the high proficiency of the amplification might indivertibly lead to unspecific amplification, and the need for multiple enzymes that are still not in the mainstream production (Karami et al., 2011). Other bottlenecks are shared between PCR and IA, such as requirements for apparatus towards test portability. Still, some IA schemes have been endorsed for particular applications, some already being approved by Regulatory Agencies, such as the Food and Drug Administration (FDA), European Medicines Agency (EMA), and even the World Health Organization (WHO) through the Foundation for Innovative New Diagnostics (FIND), for molecular diagnostics of critical pathogens (Guichón et al., 2004; Eiken 2021; Lucigen 2021; New England BioLabs, 2021), which has attracted further attention by diagnostics-oriented biotech companies (Karami et al., 2011). Some of these advances are depicted in Table 2.
Mature Isothermal Amplification Systems—How Close to Become the Next “Gold Standard”
Nucleic Acid Sequence-Based Amplification
Nucleic acid sequence-based amplification (NASBA) is an isothermal transcription-based technique, that mimics the retroviral RNA replication (Compton 1991). NASBA mechanism entails two phases: 1) non-cycling, where the target RNA is converted to dsDNA by reverse transcription; and 2) cycling, where the dsDNA molecules are actively transcribed into RNA products, leading to a yield of 10–100 copies of RNA from each template molecule (Hønsvall and Robertson 2017) (Figure 3A).
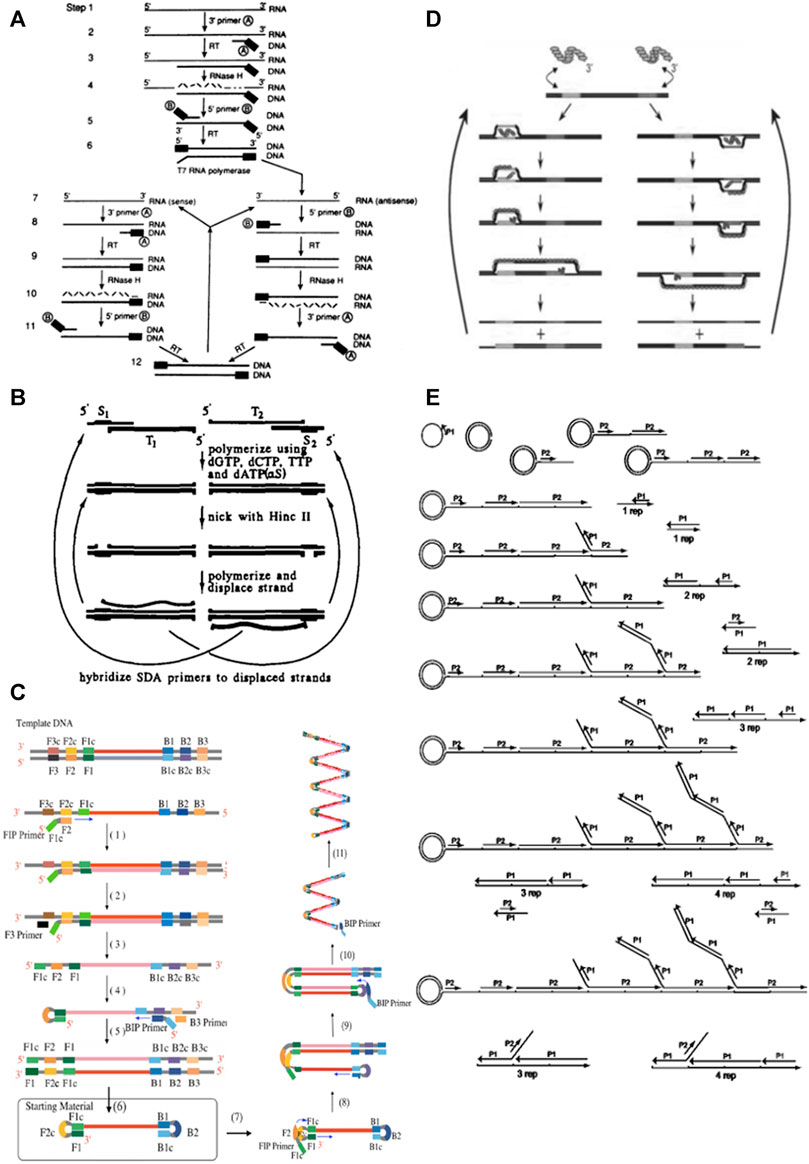
FIGURE 3. Schematic representations of the main isothermal amplification techniques. (A) Schematic representation of NASBA. Initially, the forward primer hybridizes to the target RNA molecule, leading to the formation of complementary DNA (cDNA) intermediate by reverse transcriptase and RNase H activity. Secondly, the reverse primer hybridizes in the cDNA intermediate forming a double stranded (ds) cDNA intermediate with a promoter region, which is recognized by T7 DNA-dependent RNA polymerase to produce more initial RNA template targets by transcription of the cDNA intermediate. The newly formed antisense RNA targets and cDNA serve as templates for the continuous cycling of reverse transcription (rt) and transcription reactions, resulting on the exponential accumulation of antisense RNA molecules complementary to the initial RNA target. (B) Schematic representation of SDA. The SDA mechanism starts with heat denaturation in the presence of two primers. After this step, primers hybridize forming two primer-template duplexes with 5’ overhangs, each containing one restriction site for the endonuclease (NEase) to cleave. Following, DNA polymerase extends the 3’ ends of the duplexes to produce dsDNA with complete recognition sites, that will be cleaved after by NEase. These nicks create new 3’ ends that promote new extension reaction with the displacement of the downstream fragment by DNA polymerase. The cycles of these sequenced events of cleavage and polymerization/displacement continuously produce a ssDNA molecule complementary to each of the primer-template duplexes, resulting in exponential accumulation of target sequences. (C) Schematic representation of LAMP. Steps (1) to (6) correspond to the first stage of LAMP—starting material producing stage. Steps (7) to (10) correspond to the second stage—Cycling amplification. Step (11) represent the third and final step of LAMP—Elongation and rolling cycle step. Adapted from reference (Notomi, 2000). (D) Schematic representation of RPA. The recombinase-primer filaments scan the dsDNA target molecule for the homologous site. Recombinase catalyzes the primer hybridization with the homologous template sequence. Strand exchange is promoted by recombinase in the cognate sites. The resulting structures are stabilized by ssDNA-binding proteins to prevent primers displacement by branch migration. The DNA polymerase recognize the 3’ end of the primer and starts primer extension reaction. The binding/extension events of two opposing primers generate one complete copy amplicon together with the original template. (E) Schematic representation of RCA of a circularized probe using two primers. The first primer (P1) initiates an RCA reaction, and the reverse primer (P2) binds to each tandem repeat generated by the rolling circle. Multiple priming events are initiated by P2 as the original RCA strand elongates. As these priming events elongate and generate displaced DNA strands, new priming sites for the first primer (P1) are generated. To follow the sequence of strand displacement events, note that as the reverse primer P2 binds to the fifth repeat, the primer at the third repeat begins to displace a branch; subsequently, as P2 binds to the seventh repeat, the elongating strand at the fifth repeat begins to displace a branch, and so forth. By the time a reverse primer binds to the tenth repeat, the DNA product already contains three growing branches. New primer extension events initiated in released DNA molecules also generate branches, as shown at the bottom of figure. As the displaced DNA becomes completely double-stranded, it accumulates in fragments of unit length containing one, two, three or four repeats (shown as: 1 rep, 2 rep, 3 rep, 4 rep). Thus, in the presence of a circular template, the two primers generate a self-propagating, ever-increasing pattern of alternating strand-displacement, branching and DNA fragment release events, which we call hyperbranching.
Even though this mechanism requires three enzymes - Reverse transcriptase, RNase H and T7 RNA polymerase, there are some advantages when compared to Reverse-transcription-PCR (RT-PCR), such as isothermal working temperature (41°C), which allows amplification on a simple heating block, shorter reaction time and less prone to inhibitory components in clinical and environmental samples (Dyer et al., 1996; Rutjes et al., 2006). In addition, NASBA uses RNA as the main target molecule, removing the need to previously prepare cDNA, thus saving time, labor and reducing the risk of contamination (Hønsvall and Robertson 2017). Also, contamination with DNA does not interfere with the reaction, because the temperature is kept bellow the DNA melting temperature, so strand displacement does not occur (Simpkins et al., 2000) (Table 3).
Nevertheless, there are limitations concerning NASBA implementation, such as the challenge to optimize three different enzymes that leads to higher costs, and the susceptibility to false positives that require additional probe-specific detection methods, which are generally more expensive than non-specific dyes (Cordray and Richards-Kortum 2012). Once amplification has been achieved, there are several ways to detect and monitor products, such as: molecular beacons, which is the most commonly used (Leone et al., 1998), fluorescence markers that bind and/or intercalate into the produced amplicons (Hønsvall and Robertson 2017) and gel electrophoresis for end-point detection (Morabito et al., 2013). Additionally, other colorimetric methods have been proposed that rely on nanoparticles, e.g., gold nanorods (Niazi et al., 2013) and spherical gold nanoparticles functionalized with specific probes (Mollasalehi and Yazdanparast, 2012).
Despite its promising performance, NASBA has yet to be widely acknowledge by users in mainstream labs and thus it remains in the “shadow” of other amplification techniques. NASBA has already shown it usefulness for the detection of common waterborne pathogens, such as Escherichia coli (Heijnen and Medema, 2009), Pseudo-nitschia multiseries (Delaney et al., 2011), norovirus (Rutjes et al., 2006) and notoriously it has been adopted by international space agencies for monitorization of recycled water in the International Space Station (Bechy-Loizeau et al., 2015). Another example suitable for the standard user is the NucliSENS EasyQ, an automatized commercially available NASBA system directed at the real-time amplification detection by fluorescence (Yao et al., 2005), (Biomerieux, 2021).
Depending on the detection method used for monitorization of the isothermal amplification profile, there is the possibility of NASBA integration into Lab-on-chip (LOC) devices, suitable for point-of-need, low-cost and portable platforms. In fact, there are some examples of LOC devices using NASBA for the assessment of environmental pathogens (Table 4), among which the first integrated chip for RNA isolation, NASBA amplification and real-time detection proposed (Dimov et al., 2008).
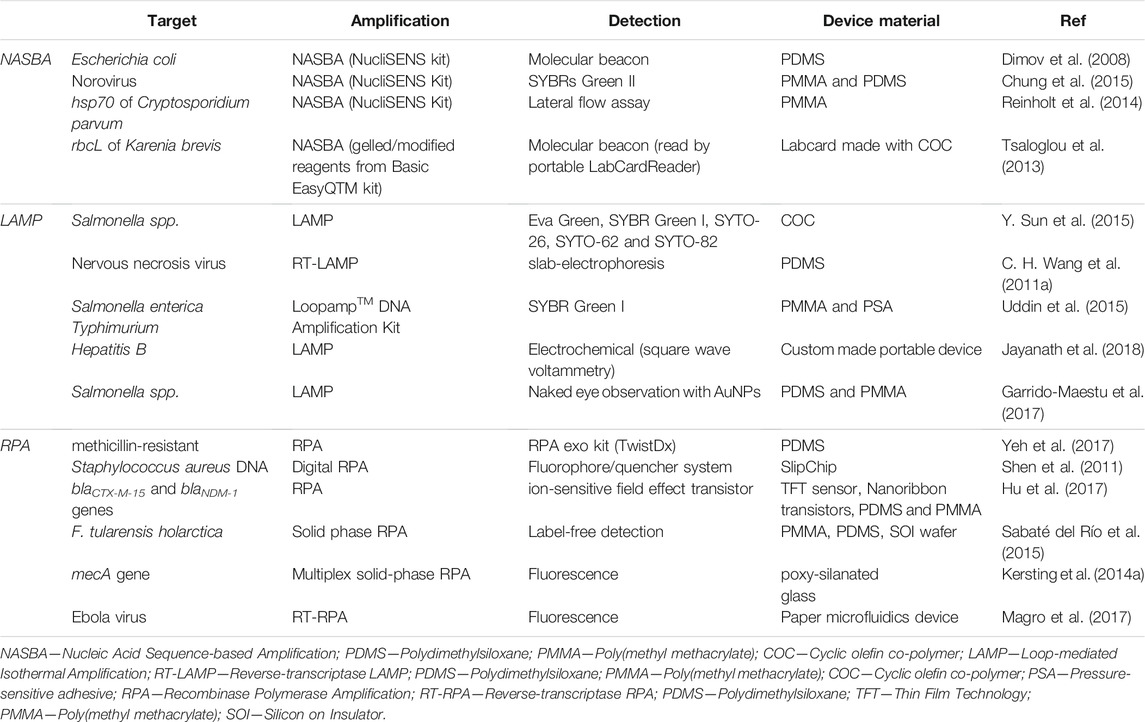
TABLE 4. Top. Examples of LOC devices using NASBA for amplification and detection of targets with environmental interest. Middle. Examples of LOC devices using LAMP for the amplification and detection of targets relevant in infectious diseases and food quality control. Bottom. Examples of LOC devices using RPA for the amplification and detection of targets relevant in infectious diseases and antibiotic resistance.
In summary, NASBA has been reported as more sensitive and less prone to inhibitory substances than PCR, which in combination with the isothermal profile makes it a useful point-of-need (PoN) tool. Still, the high cost of the reaction kits, the need to optimize multiple enzymes and the lack of specificity are crucial factors that prevent its widespread uptake by the general community.
Strand Displacement Amplification
In 1992, Walker et al. proposed to use the strand displacement capability of primers and enzymes for amplification of a given target (Walker et al., 1992). The strand displacement amplification (SDA) combines the action of an endonuclease, an exonuclease deficient DNA polymerase and two sets of primers, one with single-stranded restriction site overhangs, and a second set called “bumper primers” to support the displacement of the amplification product from the first set of primers. Briefly, dsDNA is initially heat denatured, allowing for the hybridization of the first set of primers with 5′ overhangs with specific recognition site for HincII restriction enzyme. Because of thiol-modified dATP, only the original primer is cleaved by HincII but not the newly synthetized strand, resulting in 3’ overhangs, which are extended by the exonuclease deficient klenow (exo-klenow) polymerase and further displace the downstream DNA strand, at temperatures ranging from 30—50°C. The exponential amplification is achieved though the coupling of sense and antisense reactions, in which the displaced sense strand serve as template for the antisense reaction and vice versa—(Deng and Gao 2015) for deeper insights into the molecular mechanism (Figure 3B).
Despite the advantages of isothermal reaction, the low temperature (between 30 and 55°C) makes it prone to unspecific primer-binding that might cause unspecific amplifications (Karami et al., 2011), similarly to what happens with NASBA. This is a critical aspect that prevent expansive implementation for diagnostics, since DNA/RNA retrieved from clinical samples usually contains several potential similar target sequences (e.g., gene families, homologous loci, etc.) that might miss-prime in relation to the desired target (de Pazet al., 2014). Generally, SDA presents high amplification efficiency for short target sequences that require initial template denaturation.
The high amplification efficiency of SDA associated to the capability to amplify small fragments (ranging from 50 to 120 bp) has highlighted its potential for miRNA expression profiling (Ye et al., 2019; Deng and Gao 2015). Still, SDA takes a long-time to detection (2.5 h), which makes SDA inadequate for on-site amplifications and detection. As such, SDA has been mainly applied to clinical diagnosis of Chlamydia trachomatis, Neisseria gonorrhoeae and herpes simplex virus from urogenital samples, on a high throughput platform commercialized since 1999 (Little et al., 1999; Akduman et al., 2002). This semiautomated system offers several advantages over the traditional methods for STDs detection, such as the exploit of an alternative NAAT while maintaining high sensibility and specificity, offers high-throughput and removes the need for separate work areas or unidirectional workflow. Additionally, offers medium-free transport for swabs and room temperature storage and the total time of the assays ranges from 3 to 4 h (Van Der Pol et al., 2001). Another example refers SDA inclusion into an automated device that combines sample preparation from whole cells and detection (J. M. Yang et al., 2002).
Loop-Mediated Isothermal Amplification
Loop-Mediated Isothermal Amplification (LAMP) (Notomi et al., 2000) has been the most widely used IA method, referenced in approximately 3,700 scientific publications, 8 clinical trials (ClinicalTrials.gov, 2021) and approved by WHO as an alternative molecular diagnostics method for pulmonary tuberculosis (World Health Organization 2016) and SARS-CoV-2 (Kashir and Ahmed, 2020).
LAMP relies on the strand-displacement activity of a DNA polymerase combined with a set of four unique primers, that may be extended to six for additional acceleration of amplification (Notomi et al., 2000; Mori and Notomi 2009). Compared to other NAATs, LAMP is 10 to 100-fold more sensitive than PCR (X. Wang X et al., 2014), with a detection limit as low as 1 copy per µL of template (H. Zhang et al., 2019), showing a higher specificity (Fujino et al., 2005) with an amplification time usually less than 1 h at 60–65°C, yielding 109 copies (Notomi et al., 2000). Additional advantages include not requiring an initial template denaturation step at 95°C and being less prone to inhibitory substances often present in biological samples (Enomoto et al., 2005; Kaneko et al., 2007). Perhaps one of the most interesting benefits is that LAMP output may be visualized by the naked eye (e.g., turbidimetry), making it ideal for poor resource settings and PoC applications (Besuschio et al., 2017).
In brief, LAMP takes advantage of a polymerase with strand displacement activity and four pairs of primers, that recognize a total of six distinct sequences on the target DNA. The forward inner primer (FIP) can hybridize on the dsDNA target an initiate LAMP. The DNA synthesis is initiated by the DNA polymerase upon the annealing of the outer primer (F3) to its complementary region and its subsequent extension causes strand displacement, releasing a ssDNA molecule. This molecule serves as template for the annealing on the other end of the second inner (BIP) and outer (B3) primers, producing a stem-loop DNA structure. After this step, LAMP enters in the cycling phase in which one inner primer hybridizes to the stem-loop and initiates displacement DNA synthesis, yielding the original stem–loop molecule and a new stem–loop DNA with a stem twice as long. The cycling reaction continues leading to an accumulation of around 109 copies of target under the form of concatemers in less than 1 h (Notomi et al., 2000) (Figure 3C).
The main features of LAMP, and in particular its outstanding specificity and ability to amplify without highly sophisticated equipment, makes it a valuable alternative to PCR (H. Zhang et al., 2019). In fact, LAMP has been used for detection of pathogens, e.g., detection of RNA from severe acute respiratory syndrome virus (M. M. Parida et al., 2008), identification of Methicillin Resistant Staphylococcus aureus (MRSA) (C. H. Wang et al., 2011a), tuberculosis (M. F. Lee et al., 2009), influenza A (Jung et al., 2015), Zika (J. Song et al., 2016), nervous necrosis (C. H. Wang et al., 2011b), biomarkers of disease such as Adenoviral keratoconjunctivitis (Wakabayashi et al., 2004), Varicella-zoster virus (Okamoto et al., 2004) and SNP genotyping (Iwasaki et al., 2003) and cancer diagnostics like detecting metastasis of gastric cancer (Horibe et al., 2007), KRas gene mutation (Itonaga et al., 2016) and screening of other cancer-related mutations (Srividya et al., 2019). More recently, LAMP as also been applied to the molecular detection of SARS-CoV-2 (Srividya et al., 2019), due to the rapidness and specificity of its amplification mechanisms, being exploited by many biotech-oriented companies.
Additionally, LAMP has also played an important role in quality control of food and dietary products, such as rapid detection of Salmonella (Q. Yang et al., 2018), Staphylococcus aureus (H. Yang et al., 2011), E. coli O157 (Zhao et al., 2010), Vibrio parahaemolyticus (Oh et al., 2016) and food allergens (Yuan et al., 2018).
Continuous evolution of LAMP led to improvements to reaction conditions and accuracy, which spun it into a mature and reliable assay, suitable for stand-alone molecular diagnostics (Eiken 2021; Meridian Bioscience, 2021) (Table 5). Some of the available commercially and certified detections assays take between 15 and 30 min to result. Additionally, for real-time monitorization of LAMP reaction, low complexity apparatus are also available, such as Instrument Genie II from Optigene (Horsham, United Kingdom) for fluorescence-based measurements and Ilumipro-10 from Meridian Bioscience that detects amplification products based on turbidimetry (de Paz et al., 2014).
Due to its robust and fast isothermal amplification profile, LAMP has become a suitable solution for on field molecular assays (M. Parida et al., 2004), especially for integration in lab-on-chip platforms based on microfluidics. Since the development of the first microfluidic device for the detection of hepatitis B virus, that accomplished a sensitivity of about 50 copies per reaction within 60 min (S. Y. Lee et al., 2008), other PoC devices based on LAMP have been developed targeting relevant biomarkers in infectious diseases and food quality control (Table 4).
Over the years, several modifications have been made to LAMP, such as digital LAMP where reactions occur in individualized “reactors” (micelles) yielding a 0/1 output, ideal for applications requiring quantification of target (Rane et al., 2015; Gansen et al., 2012), reverse transcription LAMP for assessing gene expression (C. H. Wang et al., 2011a), multiplex LAMP for simultaneous detection of pathogens using dedicated platforms (Abe et al., 2011) and LAMP-on-a-chip (Xuzhi Zhang et al., 2014). These developments allow for high throughput, up to 1,200 samples simultaneously, while remaining extremely sensitive (less than 10 fg of target DNA) in small sample volumes (∼10 pL) (Q. Zhu et al., 2012; C. H. Wang et al., 2011b; Rane et al., 2015). However, the complex primer design and the susceptibility to false positives still need to be addressed before LAMP is routinely applied globally (Tanner et al., 2012).
Perhaps one of the most relevant acknowledgements to LAMP has been its recognition by the WHO to fulfil all the criteria for an ideal NAAT for diagnostics (Wong et al., 2018). In fact, considering the continuous evolution of LAMP will allow expansion to other targets of relevance, which in turn will push costs of reagents down and make them more accessible, it is expected that LAMP will become a universal tool (H. Zhang et al., 2019).
Recombinase Polymerase Amplification
Recombinase Polymerase Amplification (RPA) is an isothermal amplification mechanism that operates at low temperature, usually between 37°C and 42°C, which exploits the activity of two different enzymes: a recombinase and a DNA polymerase and single stranded DNA-binding proteins. (SSBs)to ensure ssDNA stabilization (Piepenburg et al., 2006). RPA allows amplification to occur in 20—40 min, exhibiting a detection limit of 1 copy of target, being considered as one of the fastest NAATs available (de Paz et al., 2014). Briefly, RPA amplification begins with the binding of a recombinase protein (RecA from E. coli or as usually used uvsX from T4-like bacteriophages) to primers in the presence of ATP and a crowding agent (high molecular weight PEG or Carbowax20M), leading to the formation of a recombinase-primer complex. Then the complex scans the dsDNA target seeking for a homologous sequence, once homology is found the complex promotes strand displacement, forming a D-loop structure, which is stabilized by SSBs and further promotes primer-target hybridization. Finally, recombinase disassembly allows for a DNA polymerase with strand displacement activity (Bacillus subtilis Pol 1 or Sau Recombinase polymerase) to bind to the 3’ end of the primer and elongate it in the presence of dNTPs. An exponential amplification is achieved by cyclic repetition of this process (Lobato et al., 2018) (Figure 3D).
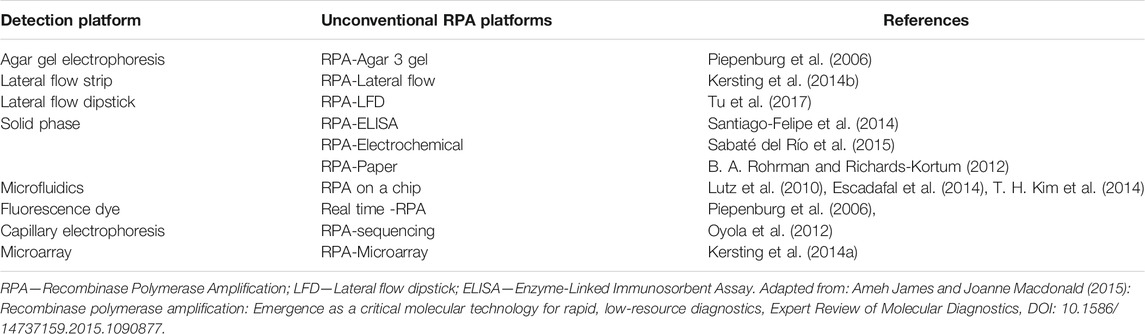
Besides the established RPA reaction setup, several improvements have been made allowing RPA to evolve to different formats, such as RT-RPA (Magro et al., 2017), digital RPA (Yeh et al., 2017) and multiplex RPA, an example is the study conducted by Kersting et al. for multiplex detection of Neisseria gonorrhoeae, Salmonella enterica and Staphylococcus aureus (Kersting et al., 2014a). Additionally, several proof of concepts on RPA incorporation with varied detection technologies have been described for improved sensitivity, reduced manual handling, improved cost effectiveness and are suitable for rapid molecular diagnosis (James and MacDonald 2015) (Table 6).
These RPA-based assays attain comparable sensitivities to those obtained with PCR, but with a significant reduction in the reaction time—time to full reaction between 10 and 20 min (Kunze et al., 2016). Moreover, RPA assays show a detection limit around 10–20 copies of target, reduced sample and reagent volumes, and the capability to amplify under high concentrations of known PCR inhibitors (Polymerase and Rpa 2017; Kersting et al., 2014b; Crannell et al., 2014a). Also, RPA does not require an initial template denaturation step and complex thermal instrumentation (Deng and Gao 2015).
RPA has been mainly applied to the detection of pathogens (Shen et al., 2011; Crannell et al., 2014b; B. Rohrman and Richards-Kortum, 2015). For example, Rohrman and Richards-Kortum accomplished the detection of 10 copies of HIV DNA in under 15 min through RPA incorporated with a lateral flow assay allowing to perform HIV diagnostics compatible with resource poor settings (B. A. Rohrman and Richards-Kortum, 2012). Additionally, an RPA assay was also developed for the detection of Tuberculosis under 20 min with a limit of detection in the femtograms of template (Boyle et al., 2014). RPA has long been translated to the commercial and laboratory stetting - TwistDX (Cambridge, United Kingdom) for both end-point detection (TwistAmpTM LF probe) and real-time monitorization (TwistAmpTM exo probe) (TwistDx 2021).
Apart from the traditional detection methods usually applied for PCR, a unique FRET-based fluorescence probe and a lateral-flow strip system have also been employed in RPA detection of SARS-CoV-2 N gene, which decrease background noise and enable an instrumentation-free readout (Xia and Chen 2020). In fact, the RPA’s low temperature reaction profile coupled to the high efficiency and specificity have been supporting the growing application of RPA in LOC devices for the amplification and detection of targets relevant in infectious diseases and antibiotic resistance (Table 4).
These examples demonstrate the advantages of RPA, especially as the quest for decentralized NAATs for diagnostics at PoC (Lobato et al., 2018; Daher et al., 2016). Additional reports show the possibility to use RPA for biomedical applications in surveillance of disease biomarkers (Euler et al., 2012), in the food industry (T. H. Kim et al., 2014) and in the agriculture sector (Mekuria et al., 2014). Although, no RPA assays have been approved by FDA or CE market thus far, it is anticipated that its operational advantages will prompt for the uptake of this technology for the detection of human pathogens in low resource settings, which would have a tremendous impact in reducing the high burden of infectious diseases and associated morbidity.
Rolling Circle Amplification
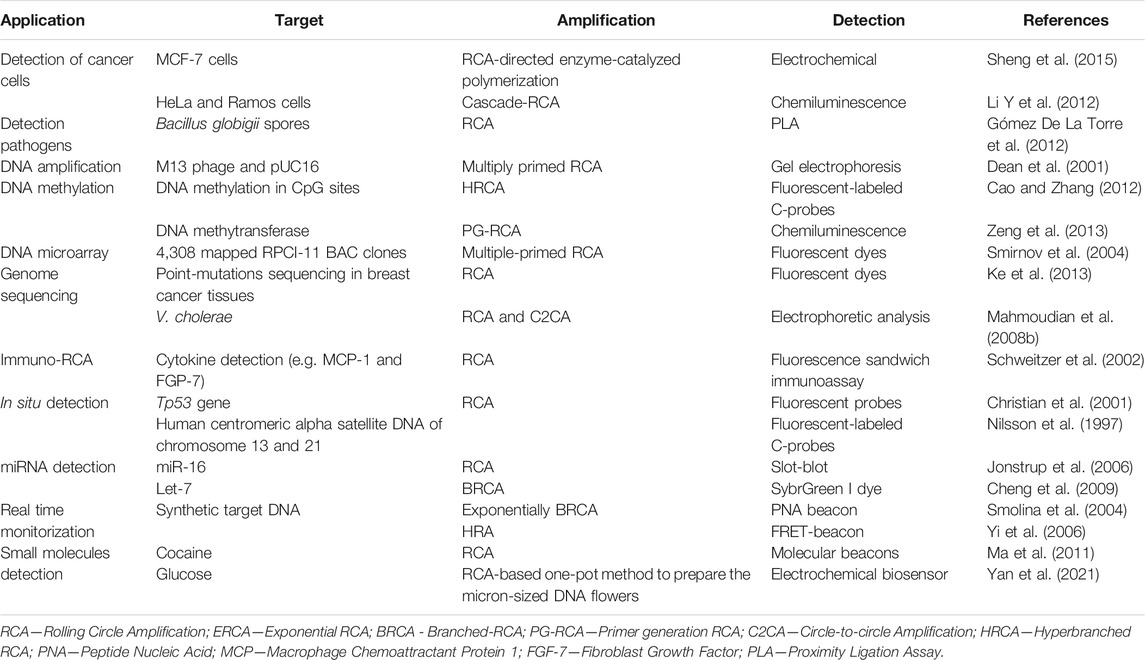
Rolling Circle Amplification (RCA) is an isothermal amplification protocol that operates at low temperature (23°C—60°C), which has been adapted from the in vivo RCA mechanism (Fire and Xu, 1995). This system requires a DNA polymerase with strand displacement activity (such as Phi29 DNA polymerase), that in presence of a circular template (e.g., plasmid, bacteriophages, DNA/RNA from virus or bacteria, etc.) and a specific primer generates a long DNA molecule with tandem repeats (D. Liu et al., 1996) (Figure 3E). The RCA mechanism has circular DNA as the predominant type of template, but in presence of linear DNA, a surrogate circular intermediate template is formed, that is then used for the subsequent amplification step. The RCA architecture offers two main benefits: 1) the low temperature isothermal scheme, with incubation temperatures as low as 23°C; and 2) the simplicity of its mechanism requirements (single primer and a productive DNA polymerase) (Li and Macdonald 2015), which yields high amplification efficiencies (∼103 copies in under 1 h), that could be enhanced though the addition of ssDNA-binding proteins (Y. Zhao et al., 2015).
Interestingly, the early RCA mechanism results in a linear amplification profile over time. In order to overcome this hurdle, several techniques have been developed to allow for an exponential mechanism, such as ramification amplification (RAM) (D. Y. Zhang et al., 2001), hyper-branched RCA (HRCA) (Lizardi et al., 1998), cascade RCA (Thomas et al., 1999) and multiply primed RCA (Dean et al., 2001).
One key feature of RCA, i.e., the capability to produce long fragments has been proven of great value to produce material for whole genome amplification, especially in the analysis of viral DNA genomes (Rector et al., 2004; Inoue-Nagata et al., 2004). RCA can also be performed on a solid support or complex biological matrixes (e.g., inside the cell or on cell surfaces) allowing for molecular level detection (Konry et al., 2011).
These advantageous characteristics have spurred the development of highly sensitive RCA-based detection methods (Yue et al., 2021) (Table 7).
The low temperature isothermal profile of RCA also makes it an attractive candidate for integration in point-of-care devices (Giuffrida and Giuseppe Spoto, 2017), for example for the detection of bacteria and virus (Sato et al., 2010; Schopf et al., 2008). Other schemes have evolved from the original RCA, such as C2CA, digital RCA, microbead-based RCA, particle-based RCA, performed in a great diversity of device formats, e.g., centrifugal valve-less (Heo et al., 2016) and droplet-based devices (Giuffrida and Giuseppe Spoto, 2017), DMF chips and paper-strips (Ali et al., 2009).
Despite RCA use has been evolving over 2 decades, no RCA-based diagnostic kits have yet reached the market (de Paz et al., 2014). So far, there are only RCA research-type kits being commercialized, such as the Illustra TempliPhi DNA amplification kit from GE HealthCare (Buckinghamshire, United Kingdom) (Reagin et al., 2003).
Presently, the large number of RCA developments have focused on biotechnology applications and molecular detection schemes, several innovative concepts have been merging RCA and nanotechnology that promise extension of RCA potential to new a totally different scale and scope.
Blooming Contenders to the “Gold Standard” Title
Despite the great number of isothermal amplification systems already showing technological maturity and consolidated applications, such as the reviewed before, the quest for improved systems and specific application still encourages the development of more isothermal amplification methods. The ones with most expression have been the Multiple Displacement Amplification (MDA) (Spits et al., 2006), Exponential Amplification Reaction (EXPAR) and Nicking and Extension Amplification Reaction (NEAR) (Van Ness et al., 2003), Helicase Dependent Amplification (HDA) (Vincent et al., 2004) and Hybridization Chain Reaction (HCR) (Evanko 2004; Dirks and Pierce 2004) (Figure 4). Next, these methods are briefly overviewed.
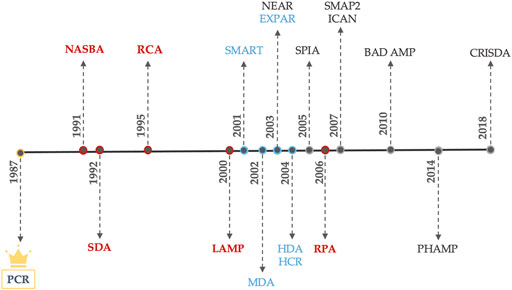
FIGURE 4. Development year of each amplification scheme. (Red) Main nucleic acids amplification techniques. (Blue) Blooming NA amplification techniques. (Black) Poorly applied NA amplification schemes.
The ability to amplify really short fragments (∼10 bp), perform whole genome amplification, use miRNA as targets, generate amplification products under linear ss-DNA forms, error prone reduction and the pursue for more efficient and specific reactions were the driving forces behind the development of these methods.
⁃ Signal-Mediated Amplification of RNA Technology (SMART)—Used for detecting both DNA and RNA targets by a tree-way junction mechanism (Wharam et al., 2001).
⁃ Multiple Displacement Amplification (MDA)—Ability to amplify very long fragments (around 50 kilobases) (Paul M. Lizardi 1997; Maragh et al., 2008; Y. Q. Sun et al., 2007).
⁃Nicking and Extension Amplification Reaction (NEAR)—Most efficient with short fragments (21–28 bp). Amplifies directly from RNA. Generates ssDNA products (Maples et al., 2007).
⁃ Exponential Amplification Reaction (EXPAR)—Most applied to miRNA detection, DNA, proteins, enzyme activity and metal ions. Exponential version of NEAR (R. D. Li et al., 2016; Jia et al., 2010; Van Ness et al., 2003; Wang K et al., 2014; Zhang et al., 2015; J. Chen et al., 2016; H. Liu et al., 2017; Q. Xue et al., 2015; Jia et al., 2014).
⁃ Helicase Dependent Amplification (HDA)—Diagnosis of dsDNA pathogens such as Neisseria gonorrhoeae, Clostridium difficile, Staphylococcus aureus and HIV-1 (Goldmeyer et al., 2008; Chow et al., 2008; Pandori and Branson 2010; Tang et al., 2010).
⁃ Hybridization Chain Reaction (HCR)—Non-enzymatic mechanism based on successive hairpin hybridization. Applied to mRNA imaging, signal amplification and photoelectrochemical detection (Wu et al., 2015; Sha, Zhang, and Wang 2016; Chu et al., 2019).
⁃ Single Chimeric Primer Isothermal Amplification (SPIA)—Specifically used to amplify linear mRNA. It has been applied to Whole genome amplification and Whole transcriptome amplification (Peng et al., 2018; Myrmel et al., 2017).
⁃ Smart Amplification Process Version 2 (SMAP 2)—SPN identification and genotyping (Watanabe et al., 2007; Tatsumi et al., 2008; Araki et al., 2010).
⁃ Isothermal and Chimeric primer-initiated Amplification of Nucleic acids (ICAN)—Specifically amplify short DNA sequences from complex DNA samples, (Mukai et al., 2007; Shimada et al., 2002).
⁃ Beacon-assisted molecular detection (BAD AMP)—Emerging method to detect and amplify short DNA fragments (<22 bp) (Connolly and Matt, 2010; Connolly and Matt, 2011).
⁃ Hairpin Florescence Probe Assisted Isothermal Amplification (PHAMP)—Detect and amplify microRNA (<22 bb). Mechanism similar to BADAMP (Ma, Liu, and Shi 2014)
⁃ Exonuclease III-induced cascade two-stage isothermal amplification-mediated zinc (II) protoporphyrin IX/G-quadruplex supramolecular fluorescent nanotags (EIC)—Emerging method for early diagnosis of gene-related diseases (Q. Xue et al., 2014).
⁃ CRISPR-Cas9-triggered nicking endonuclease-mediated strand displacement amplification mediated strand displacement amplification (CRISDA)—Emerging method with single nucleotide specificity and attomolar sensitivity (Zhou et al., 2018).
Multiple Displacement Amplification (2002)
MDA is an alternative DNA amplification based on random hexamer primers and the activity of a high-fidelity enzyme, such as Phi29 DNA polymerase (Spits et al., 2006). Compared with PCR and other amplification techniques, MDA do not require sequence specific primers, once its mechanism exploits the random hybridization to generate large sized DNA fragments. Beside the isothermal profile (30°C), Phi 29 polymerase has proofreading activity, consequently MDA products contain less errors and higher sizes than the ones obtain via Taq amplification for the same time interval (2–3 h) (Blanco et al., 1989; Paez et al., 2004; Esteban, Salas, and Blanco 1993).
These beneficial features have spurred the application of MDA for Single Cell Genome Sequencing (Dean et al., 2002; Yoon et al., 2011; Rinke et al., 2013). Since it yields roughly 1–2 µg of DNA (enough for sequencing studies) with a genome coverage around 99% (Paez et al., 2004; Hellani et al., 2005), which allowed for the screening of genetic health condition in early-staged embryos before implantation (Hellani et al., 2005). MDA is also valuable for SPN allele detection (Ling et al., 2012) and for identifying the size of allelic polymorphic repeats (Ballantyne et al., 2006).
Exponential Amplification Reaction and Nicking and Extension Amplification Reaction (2003)
EXPAR is the exponential version of NEAR system, both simulate the SDA mechanism, where the difference in the melting temperature is exploited for target release and regeneration (Van Ness et al., 2003). A target template comprising a nicking site is designed to hybridize with the desire target. The polymerase replicates the template target, which is then nicked, producing a short fragment (8–16 bp) that is released by duplex instability, rather than by strand displacement activity (Van Ness et al., 2003). Conversely, the exponential version of NEAR uses a target template with two target complementary regions, instead of one, which after being nicked, releases a synthetized target, that can also pair up with the template target, creating a cycle of target regeneration and empowering NEAR to the desired exponential scale (Jia et al., 2010).
NEAR has a linear amplification profile, nevertheless it allows the amplification of any small targets by adding restriction sequences in the flanking regions (Maples et al., 2007). On the other hand, EXPAR is strongly limited by the requirement of targets natively flanked by restriction sites (Jia et al., 2010). Both NEAR and EXPAR (Tan et al., 2005) are particularly well suited for the detection of small fragments (8–16 bp) with high analytical sensitivity. Applications of NEAR include the detection of DNA (Zhou et al., 2014; Y. Song et al., 2014), proteins (A. X. Zheng et al., 2012; L. Xue, Zhou, and Xing 2010; Y. Huang et al., 2013; L. Xue, Zhou, and Xing 2012) and enzymes (Y. Zhao et al., 2013; X. Liu et al., 2014). Additionally, NEAR’s ability to amplify at room temperature, without initial template denaturation and higher tolerance to reaction inhibitors are two beneficial criteria for POC integration. In this regard, Wang et al., (L. Wang et al., 2017) reported on a highly specific detection assay for transgenic soya based on NEAR that is promising for PoC analysis. NEAR system is also being commercialized under kit formats. DNAble® by EnviroLogix (Portland, United States) test kit detects Salmonella and Clavibacter michiganesis (a plant pathogen) in 15 min with a sensitivity of 103 cfu per reaction (Envirologix, 2021). Alere-i POC allows the detection of influenza A and B virus in 15 min (Alere, Waltham, United States) (Nie et al., 2014; AlereTM now Abbott, 2021). Even though, not much is known about NEAR and EXPAR technology, their simplicity, specificity, and high adaptability are attractive features pushing for wider application.
Helicase Dependent Amplification (2004)
HDA is regarded as one of the simplest isothermal amplification schemes since it is based on the in vivo human DNA replication process (Vincent, Xu, and Kong 2004). HDA retains most of the PCR advantages, while improving the efficiency and selectivity, reducing the reaction and analysis complexity and requires less sample preparation steps, denoting the potential for integration in decentralized settings (An et al., 2005).
In the same fashion as PCR, HDA is compatible with different detection mechanisms such as gel electrophoresis for end-point measurement, fluorescent detection [with fluorescent DNA intercalator (Barbieri et al., 2014) and specific probes (Tong et al., 2008)] and electrochemical detection [with electroactive intercalator (Kivlehan et al., 2011)]], for real-time monitorization. However, HDA can also be monitored though Lateral Flow Devices (LFDs), which are much more suitable for low-cost and point-of-need scenario, than real-time assays (Du et al., 2017; Mahalanabis et al., 2010; Kolm et al., 2019; Tomlinson, Dickinson, and Boonham 2010).
Regarding HDA applications, the majority are devoted to the detection of infectious diseases. In fact, HDA has found a market niche in point-of-need testing (Craw et al., 2015), driven forward the development of commercial test kits (Gaydos et al., 2017) for pathogens such as, Herpes simplex [approved by FDA in 2011 (Lemieux et al., 2012); H. J. Kim et al., 2011)], Bordetella pertussis, Clostridium difficile Toxin A and Streptococcus from vaginal/rectal swabs (Faron et al., 2015). Quidel (proprietary for HDA technology) commercializes the AmpliVue and Solana test kits (Quidel 2021), based on HDA-Lateral Flow Devices, allowing the DNA amplification to occur in a CLIA-waiver, meaning without any sample preparation step (Weber et al., 2016). Additionally, BioHelix developed qualitative amplification detection kits (New England BioLabs, 2021) that recently receive FDA 501 (k) approval (Craw and Balachandran 2012).
Despite the usefulness of HDA in pathogen detection, it can be easily misinterpreted, since HDA presents issues often posed by the isothermal profile, such as the high risk of primer-dimer artifacts and off-target effects, ultimately leading to false positive phenomena, high background signal, inability to multiplex, low sensibility and selectivity (Barreda-García et al., 2018). Together, single-propriety commercialization, the false positive phenomena and the lack of validated assays have been hampered HDA widespread. However, if developers pushed forward the integration with LOC systems, HDA will likely show a significant increase in point-of-need pathogen detection, ultimately fueling the development of more isothermal amplification schemes.
Hybridization Chain Reaction (HCR) (2004)
HCR Evanko (2004), Dirks and Pierce (2004) differs from all the previously reported methods, since isothermal amplification is achieved via signal amplification, without requiring the generation of DNA or RNA products. Comparing to the polymerase-based methods, the signal amplification schemes are not subject to product or enzyme inhibition. Depending on the strategy for signal amplification mechanism, there are three categories: nuclease-assisted, DNAzyme-assisted and enzyme-free reactions. Regarding the last type, HCR was the first being proposed in 2004 (Dirks and Pierce, 2004) being based in the differential hybridization of two partially complementary probes (hairpin form), that can only attain the energy required for conformational change (at room temperature) when in the presence of target ssDNA or RNA, generating a long-nicked copolymer. Trough the introduction of a fluorescent label on a specific hairpin moiety, real-time tracking of HCR can be achieved (Chemeris, Nikonorov, and Vakhitov 2008). This method pioneered the development of nanostructures from DNA self-assembly (Xuan and Hsing 2014; G. Zhu et al., 2013; Ding et al., 2015; J. Zheng et al., 2013; Xuan, Fan, and Hsing 2015) and introduced DNA as an amplifying transducer for biosensing (J. Huang et al., 2011; Y. Chen et al., 2012; L. Yang et al., 2012; B. Zhang et al., 2012; P. Liu et al., 2013) and bioimaging (Choi et al., 2010; Choi, Beck, and Pierce 2014; G. Zhu et al., 2013). HCR have shown applicability in in situ analysis (Choi et al., 2010; Choi, Beck, and Pierce 2014), miRNA (L. Zhou et al., 2019) and mRNA (Choi et al., 2010) detection. Additionally, the combination of HCR with other methods such as Catalyze Hairpin Assembly for detection single nucleotide polymorphisms (Li B et al., 2012), immuno-HCR (B. Zhang et al., 2012; J. Zhou et al., 2012) and AuNPs-supporting HCR (W. J. Wang et al., 2015) as electrochemical immunoassays and RCA incorporating HCR (Bi et al., 2013).
HCR’s attractive features such as enzyme-free, isothermal profile and excellent efficiency, have contributed for its rapid gain of popularity (Deng and Gao 2015). Despite all the hype around non-enzymatic methods, they have been put aside due to leakage problems (Bi, Yue, and Zhang 2017).
Other Isothermal Amplification Schemes
Beside the up-mentioned methods, some other “niche” methods have also been developed, among them are SMART, SPIA, ICAN, SMAP 2 and BAD AMP (Figure 5).
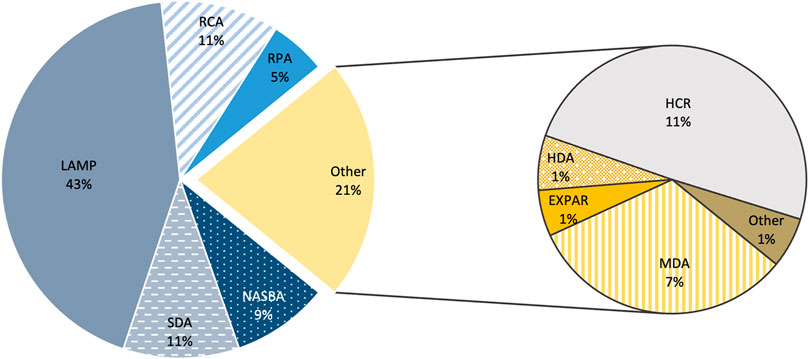
FIGURE 5. Expression of each amplification scheme. Percentage of the Number of publications until 28 of October of 2020 in PUBMED. On the second plot, “Other” comprise to the following schemes: SMART, NEAR, SPIA, ICAN, BAD-AMP, SMAP 2, Exonuclease III-induced cascade two-stage isothermal amplification-mediated zinc (II) protoporphyrin IX/G-quadruplex supramolecular fluorescent nanotags, Hairpin Florescence Probe Assisted Isothermal Amplification and CRISDA.
These new isothermal amplification schemes cover broad applications, from nucleic acids detection (DNA, RNA, long RNA, SNPs, miRNA, DNA methylation) to the detection of proteins, enzymes, cancer cells, pathogens, small molecules and metal-ions (Y. Zhao et al., 2015; S. Kumar J. et al., 2018). Despite their range of applications, not all of these “blooming” amplification schemes have been applied much beyond their initial “proof-of-concept” purpose, since many of these methods still lack technical maturity (e.g., PHAMP, EIC, SPIA, ICAN and BAD AMP). Conversely, others have been endorsed over the years (e.g., MDA, SMART, HDA, EXPAR and HCR), nevertheless the strictness of conditions and reagents and the lack of accessible amplification kits and literature has been preventing wider use, forcing them to set as “niche” application schemes. As so, all schemes must undergo through improvements before they can emerge as competitors to the previously stablished methods (Li and Macdonald 2015).
Conclusions and Future Perspectives
PCR is the most widespread method and the current standard for nucleic acids amplification, its inherent features, such as the requirement for a thermocycler and the strict reaction conditions, increase the complexity of device integration and operating settings, overall rising the cost of PCR-based devices and assays. Conversely, isothermal schemes have constant and low temperature profiles, ability to use different types of molecules as starting material, low sensibility to inhibitors and offer a wide range of detection methods, which are crucial features rendering for a significantly easier LOC integration, ultimately pushing forward their implementation and rising consideration. Furthermore, the integration under chip formats contributes to enhance the sensibility, reproducibility, rapidness, cost-effectiveness and achieve more accurate results, reducing the amplification bias. All together, these features are true “game changers”, defying PCR technique for the “gold standard” title.
Generally, isothermal amplification methods offer a great potential for lower-cost, higher speed, smaller consumption of sample and reagents and the opportunity to automation of all processes from sample preparation to signal detection under lab-on-chip formats. In this regard, the isothermal amplification schemes were grouped in two main categories: the mature and consolidated ones, such as SDA, NASBA, LAMP, RPA and RCA; and the blooming schemes, that despite their promising features, still lack technical maturity and widespread application or commercialization under kit formats. This last group comprises SMART, MDA, EXPAR, HDA, HCR, SPIA and others.
Besides the nucleic acid detection purposes, isothermal amplification schemes have also been applied much further than this initial concept, such as: whole genome sequencing, single cell sequencing, electrochemical immunoassays, single-nucleotide polymorphism detection, virus and bacterial detection from biological swabs, environmental point-of-need niche applications, amplification of non-DNA templates and amplification of long, short and micro-RNA fragments. Nevertheless, the ultimate success of the isothermal schemes is still dependent on wider demonstration of specificity and cost-effectiveness. In fact, most of these methods show unspecific products formation and high background noise, due to the low amplification temperature, issues that need to be addressed before translation to the clinical scenario. Additionally, many of the schemes discussed are still protected by patents or licensed by a single proprietary company, which may be limiting the access to the necessary enzymes and reagents, ultimately leading to high market prices, thus preventing broader application. Once these rights have expired, IA-based methods should meet the requirements to become a reference to nucleic acid diagnosis and POC applications, with a profitable commercialization worldwide.
Author Contributions
BO, BV, and PB planned and conceptualized the work, drafted the manuscript, and edited the final version. BO designed the schematics and figures.
Funding
This work is financed by national funds from FCT—Fundação para a Ciência e a Tecnologia, I.P., in the scope of the project UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences—UCIBIO and the project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy—i4HB; and for 2020.07660. BD to BO.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, T., Segawa, Y., Watanabe, H., Yotoriyama, T., Kai, S., Yasuda, A., et al. (2011). Point-of-Care Testing System Enabling 30 Min Detection of Influenza Genes. Lab. Chip 11 (6), 1166–1167. doi:10.1039/c0lc00519c
Akduman, D., Ehret, J. M., Messina, K., Ragsdale, S., and Judson, F. N. (2002). Evaluation of a Strand Displacement Amplification Assay (BD ProbeTec-SDA) for Detection of Neisseria Gonorrhoeae in Urine Specimens. J. Clin. Microbiol. 40 (1), 281–283. doi:10.1128/JCM.40.1.281-283.2002
AlereTM now Abbott. 2021. “ID NOWTM INFLUENZA A & B 2.” 2021. Available at: https://www.globalpointofcare.abbott/en/product-details/id-now-influenza-ab-2.html.
Ali, M. M., Aguirre, S. D., Xu, Y., Filipe, C. D. M., Pelton, R., and Li, Y. (2009). Detection of DNA Using Bioactive Paper Strips. Chem. Commun. 43, 6640–6642. doi:10.1039/b911559e
An, L., Tang, W., Ranalli, T. A., Kim, H.-J., Wytiaz, J., and Kong, H. (2005). Characterization of a Thermostable UvrD Helicase and its Participation in Helicase-dependent Amplification. J. Biol. Chem. 280 (32), 28952–28958. doi:10.1074/jbc.M503096200
Araki, T., Shimizu, K., Nakamura, K., Nakamura, T., Mitani, Y., Obayashi, K., et al. (2010). Usefulness of Peptide Nucleic Acid (PNA)-Clamp Smart Amplification Process Version 2 (SmartAmp2) for Clinical Diagnosis of KRAS Codon12 Mutations in Lung Adenocarcinoma. J. Mol. Diagn. 12 (1), 118–124. doi:10.2353/jmoldx.2010.090081
Aryan, E., Makvandi, M., Farajzadeh, A., Huygen, K., Bifani, P., Mousavi, S.-L., et al. (2010). A Novel and More Sensitive Loop-Mediated Isothermal Amplification Assay Targeting IS6110 for Detection of Mycobacterium Tuberculosis Complex. Microbiol. Res. 165 (3), 211–220. doi:10.1016/j.micres.2009.05.001
Biomerieux. 2021. “NUCLISENS EASYQ®.” 2021. Available at: https://www.biomerieux-diagnostics.com/nuclisens-easyqr.
Bachmann, L. H., Johnson, R. E., Cheng, H. John. R. Papp., Markowitz, L. E., Papp, J. R., and Hook, E. W. (2009). Nucleic Acid Amplification Tests for Diagnosis of Neisseria Gonorrhoeae Oropharyngeal Infections. J. Clin. Microbiol. 47 (4), 902–907. doi:10.1128/JCM.01581-08
Ballantyne, K. N., van Oorschot, R. A. H., John Mitchell, R., and Koukoulas, I. (2006). Molecular Crowding Increases the Amplification Success of Multiple Displacement Amplification and Short Tandem Repeat Genotyping. Anal. Biochem. 355 (2), 298–303. doi:10.1016/j.ab.2006.04.039
Barbieri, D., Venturoli, S., Rösl, F., and Rincon-Orozco, B. (2014). Detection of High-Risk Human Papillomavirus Type 16 and 18 Using Isothermal Helicase-dependent Amplification. Diagn. Microbiol. Infect. Dis. 79 (2), 178–182. doi:10.1016/j.diagmicrobio.2014.02.012
Barreda-García, S., Miranda-CastroMiranda-Castro, R., de-los-Santos-Álvarez, N., Miranda-Ordieres, A. J., and Lobo-Castañón, M. J. (2018). Helicase-Dependent Isothermal Amplification: A Novel Tool in the Development of Molecular-Based Analytical Systems for Rapid Pathogen Detection. Anal. Bioanal. Chem. 410 (3), 679–693. doi:10.1007/s00216-017-0620-3
Bechy-Loizeau, A.-L., Flandrois, J.-P., Abaibou, H., and Abaibou, Hafid. (2015). Assessment of Polycarbonate Filter in a Molecular Analytical System for the Microbiological Quality Monitoring of Recycled Waters Onboard ISS. Life Sci. Space Res. 6, 29–35. doi:10.1016/j.lssr.2015.06.002
Besuschio, S. A., Llano Murcia, M., Benatar, A. F., Monnerat, S., Cruz, I., Picadode Puig, A., et al. (2017). Analytical Sensitivity and Specificity of a Loop-Mediated Isothermal Amplification (LAMP) Kit Prototype for Detection of Trypanosoma Cruzi DNA in Human Blood Samples. Plos Negl. Trop. Dis. 11 (7), e0005779–18. doi:10.1371/journal.pntd.0005779
Bi, S., Yue, S., and Zhang, S. (2017). Hybridization Chain Reaction: A Versatile Molecular Tool for Biosensing, Bioimaging, and Biomedicine. Chem. Soc. Rev. 46 (14), 4281–4298. doi:10.1039/c7cs00055c
Bi, S., Zhao, T., Luo, B., and Zhu, J.-J. (2013). Hybridization Chain Reaction-Based Branched Rolling Circle Amplification for Chemiluminescence Detection of DNA Methylation. Chem. Commun. 49 (61), 6906–6908. doi:10.1039/C3CC43353F
Blanco, L., Bernad, A., Lázaro, J. M., Martín, G., Garmendia, C., and Salas, M. (1989). Highly Efficient DNA Synthesis by the Phage ϕ 29 DNA Polymerase. J. Biol. Chem. 264 (15), 8935–8940. doi:10.1016/s0021-9258(18)81883-x
Boyle, D. S., McNerney, R., Teng Low, H., Leader, B. T., Meyer, A. C., Meyer, J. C., et al. (2014). Rapid Detection of Mycobacterium Tuberculosis by Recombinase Polymerase Amplification. PLoS ONE 9 (8), e103091–9. doi:10.1371/journal.pone.0103091
Cao, A., and Zhang, C.-y. (2012). Sensitive and Label-free DNA Methylation Detection by Ligation-Mediated Hyperbranched Rolling Circle Amplification. Anal. Chem. 84 (14), 6199–6205. doi:10.1021/ac301186j
Chang, C.-C., Chen, C.-C., Lu, S.-C. H.-H., Hui, Y.-H., Liang, C.-W., and Lin, C. W. (2012). Diagnostic Devices for Isothermal Nucleic Acid Amplification. Sensors 12 (6), 8319–8337. doi:10.3390/s120608319
Chemeris, D. A., Nikonorov, Y. M., and Vakhitov, V. A. (2008). Real-Time Hybridization Chain Reaction. Dokl Biochem. Biophys. 419, 53–55. doi:10.1134/s1607672908020014
Chen, J., Zhou, X., Ma, Y., Lin, X., Dai, Z., and Zou, X. (2016). Asymmetric Exponential Amplification Reaction on a Toehold/Biotin Featured Template: An Ultrasensitive and Specific Strategy for Isothermal MicroRNAs Analysis. Nucleic Acids Res. 44 (15), gkw504. doi:10.1093/nar/gkw504
Chen, Y., Xu, J., Su, J., Xiang, Y., Yuan, R., and Chai, Y. (2012). In Situ Hybridization Chain Reaction Amplification for Universal and Highly Sensitive Electrochemiluminescent Detection of DNA. Anal. Chem. 84 (18), 7750–7755. doi:10.1021/ac3012285
Cheng, Y., Zhang, X., Li, Z., Jiao, X., Wang, Y., and Zhang, Y. (2009). Highly Sensitive Determination of Microrna Using Target-Primed and Branched Rolling-Circle Amplification. Angew. Chem. Int. Ed. 48 (18), 3268–3272. doi:10.1002/anie.200805665
Choi, H. M. T., Beck, V. A., and Pierce, N. A. (2014). Next-Generation In Situ Hybridization Chain Reaction: Higher Gain, Lower Cost, Greater Durability. ACS Nano 8 (5), 4284–4294. doi:10.1021/nn405717p
Choi, H. M. T., Chang, J. Y., Padilla, J. E., Fraser, S. E., and Pierce, N. A. (2010). Programmable In Situ Amplification for Multiplexed Imaging of MRNA Expression. Nat. Biotechnol. 28 (11), 1208–1212. doi:10.1038/nbt.1692
Chow, W. H. A., McCloskey, C., Tong, Y., Hu, L., You, Q., Kelly, C. P., et al. (2008). Application of Isothermal Helicase-dependent Amplification with a Disposable Detection Device in a Simple Sensitive Stool Test for Toxigenic Clostridium Difficile. J. Mol. Diagn. 10 (5), 452–458. doi:10.2353/jmoldx.2008.080008
Christian, A. T., Pattee, M. S., Attix, C. M., Reed, B. E., Sorensen, K. J., and Tucker, J. D. (2001). Detection of DNA Point Mutations and MRNA Expression Levels by Rolling Circle Amplification in Individual Cells. Proc. Natl. Acad. Sci. 98 (25), 14238–14243. doi:10.1073/pnas.251383598
Chu, Y., Deng, A.-P., Wang, W., and Zhu, J.-J. (2019). Concatenated Catalytic Hairpin Assembly/Hyperbranched Hybridization Chain Reaction Based Enzyme-free Signal Amplification for the Sensitive Photoelectrochemical Detection of Human Telomerase RNA. Anal. Chem. 91 (5), 3619–3627. doi:10.1021/acs.analchem.8b05610
Chung, S. H., Baek, C., Cong, V. T. J., and Min, J. (2015). The Microfluidic Chip Module for the Detection of Murine Norovirus in Oysters Using Charge Switchable Micro-bead Beating. Biosens. Bioelectron. 67, 625–633. doi:10.1016/j.bios.2014.09.083
ClinicalTrials.gov 2021Clinical Trials with LAMP Assay. Available at: https://www.clinicaltrials.gov/ct2/results?cond=&term=Loop+mediated+isothermal+amplification&cntry=&state=&city=&dist= (Accessed July 27, 2021).
ClinicalTrials.gov Identifier: NCT04500873. 2020a. “Performance of RPA-LF for Cutaneous Leishmaniasis (RPA-LF).” 2020. Available at: https://clinicaltrials.gov/ct2/show/NCT04500873?term=recombinase+polymerase+amplification&draw=2&rank=1.
ClinicalTrials.gov Identifier: NCT04579549. 2021. “Repeat Testing for SARS-CoV-2.” 2021. Available at: https://clinicaltrials.gov/ct2/show/NCT04579549?term=Loop+mediated+isothermal+amplification&draw=2&rank=5.
ClinicalTrials.gov Identifier:NCT02371395. 2018b. “Validation of RealAmp Method for the Diagnosis of Malaria in Endemic Areas of Brazil.” 2018. Available at: https://clinicaltrials.gov/ct2/show/NCT02371395.
ClinicalTrials.gov Identifier: NCT01838902 (2018a). “Primaquine’s Gametocytocidal Efficacy in Malaria Asymptomatic Carriers (PRINOGAM).” 2018.
ClinicalTrials.gov Identifier:NCT03829735 (2020b). “Neonatal Vaccination against Hepatitis B in Africa - Sero-Survey in Senegal (NeoVac2S).” 2020.
Compton, J. (1991). Nucleic Acid Sequence-Based Amplification. Nature 350 (6313), 91–92. doi:10.1038/350091a0
Connolly, A. R., and Trau, M. (2010). Isothermal Detection of DNA by Beacon-Assisted Detection Amplification. Angew. Chem. Int. Edition 49 (15), 2720–2723. doi:10.1002/anie.200906992
Connolly, A. R., and Trau, M. (2011). Rapid DNA Detection by Beacon-Assisted Detection Amplification. Nat. Protoc. 6 (6), 772–778. doi:10.1038/nprot.2011.326
Cordray, M. S., and Richards-Kortum, R. R. (2012). Emerging Nucleic Acid-Based Tests for Point-of-Care Detection of Malaria. Am. J. Trop. Med. Hyg. 87 (2), 223–230. doi:10.4269/ajtmh.2012.11-0685
Crannell, Z. A., Rohrman, B., and Richards-Kortum, R. (2014b). Equipment-Free Incubation of Recombinase Polymerase Amplification Reactions Using Body Heat. PLoS ONE 9 (11), e112146–7. doi:10.1371/journal.pone.0112146
Crannell, Z. A., Rohrman, B., and Richards-kortum, R. (2014a). Quantification of HIV-1 DNA Using Real-Time Recombinase Polymerase Amplification. Anal. Chem. 86 (12), 5615–5619. doi:10.1021/ac5011298
Craw, P., and Balachandran, W. (2012). Isothermal Nucleic Acid Amplification Technologies for Point-of-Care Diagnostics: A Critical Review. Lab. Chip 12 (14), 2469–2486. doi:10.1039/c2lc40100b
Craw, P., Mackay, R., Naveenathayalan, A., Hudson, C., Branavan, M., Sadiq, S., et al. (2015). A Simple, Low-Cost Platform for Real-Time Isothermal Nucleic Acid Amplification. Sensors 15 (9), 23418–23430. doi:10.3390/s150923418
Cuadros, J., Martin Ramírez, A., González, I. J., Ding, X. C., Perez Tanoira, R., Rojo-Marcos, G., et al. (2017). LAMP Kit for Diagnosis of Non-falciparum Malaria in Plasmodium Ovale Infected Patients. Malar. J. 16 (1), 1–5. doi:10.1186/s12936-016-1669-8
Daher, R. K., Stewart, G., Boissinot, M., and Bergeron, M. G. (2016). Gale Stewart, Maurice Boissinot, and Michel G. Bergeron.Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 62 (7), 947–958. doi:10.1373/clinchem.2015.245829
de Paz, H. D., Brotons, P., and Muñoz-Almagro, C. (2014). Molecular Isothermal Techniques for Combating Infectious Diseases: Towards Low-Cost Point-of-Care Diagnostics. Expert Rev. Mol. Diagn. 14 (7), 827–843. doi:10.1586/14737159.2014.940319
Dean, F. B., Hosono, S., Fang, L., Wu, X., Faruqi, A. F., Bray-Ward, P., et al. (2002). Comprehensive Human Genome Amplification Using Multiple Displacement Amplification. Proc. Natl. Acad. Sci. 99 (8), 5261–5266. doi:10.1073/pnas.082089499
Dean, F. B., Nelson, J. R., Giesler, T. L., and Lasken, R. S. (2001). Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 11 (6), 1095–1099. doi:10.1101/gr.180501
Delaney, J. A., Ulrich, R. M., and Paul, J. H. (2011). Detection of the Toxic Marine Diatom Pseudo-nitzschia Multiseries Using the RuBisCO Small Subunit (RbcS) Gene in Two Real-Time RNA Amplification Formats. Harmful Algae 11, 54–64. doi:10.1016/j.hal.2011.07.005
Deng, H., and Gao, Z. (2015). Bioanalytical Applications of Isothermal Nucleic Acid Amplification Techniques. Analytica Chim. Acta 853 (1), 30–45. doi:10.1016/j.aca.2014.09.037
Dimov, I. K., Garcia-Cordero, J. L., O'Grady, J., Poulsen, C. R., Viguier, C., Kent, L., et al. (2008). Integrated Microfluidic TmRNA Purification and Real-Time NASBA Device for Molecular Diagnostics. Lab. Chip 8 (12), 2071–2078. doi:10.1039/b812515e
Ding, J., Gu, Y., Li, F., Zhang, H., and Qin, W. (2015). DNA Nanostructure-Based Magnetic Beads for Potentiometric Aptasensing. Anal. Chem. 87 (13), 6465–6469. doi:10.1021/acs.analchem.5b01576
Dirks, R. M., and Pierce, N. A. (2004). From the Cover: Triggered Amplification by Hybridization Chain Reaction. Proc. Natl. Acad. Sci. 101 (43), 15275–15278. doi:10.1073/pnas.0407024101
Du, X.-j., Zhou, T.-j., Li, P., and Wang, S. (2017). A Rapid Salmonella Detection Method Involving Thermophilic Helicase-dependent Amplification and a Lateral Flow Assay. Mol. Cell Probes 34, 37–44. doi:10.1016/j.mcp.2017.05.004
Dyer, J. R., Gilliam, B. L., Gilliam, J. J., Grosso, L., Cohen, M. S., and Fiscus, S. A. (1996). Quantitation of Human Immunodeficiency Virus Type 1 RNA in Cell Free Seminal Plasma: Comparison of NASBA with Amplicor Reverse Transcription-PCR Amplification and Correlation with Quantitative Culture. J. Virol. Methods 60 (2), 161–170. doi:10.1016/0166-0934(96)02063-0
Edwards, M. C., and Gibbs, R. A. (1994). Multiplex PCR: Advantages, Development, and Applications. Genome Res. 3 (4), S65–S75. doi:10.1101/gr.3.4.S65
Eiken. 2021. “Products, Eiken Chemical Co., LTD.,” 2021. Available at: http://www.eiken.co.jp/en/product/index.html.
Enomoto, Y., Yoshikawa, T., Ihira, M., Akimoto, S., Miyake, F., Usui, C., et al. (2005). Rapid Diagnosis of Herpes Simplex Virus Infection by a Loop-Mediated Isothermal Amplification Method. J. Clin. Microbiol. 43 (2), 951–955. doi:10.1128/JCM.43.2.951-955.2005
Envirologix. 2021 DNAble. Available at: https://www.envirologix.com/technology/molecular-detection/ (Accessed July 27, 2021).
Escadafal, C., Faye, O., Sall, A. A., Faye, O., Weidmann, M., Strohmeier, O., et al. (2014). Rapid Molecular Assays for the Detection of Yellow Fever Virus in Low-Resource Settings. Plos Negl. Trop. Dis. 8 (3), e2730. doi:10.1371/journal.pntd.0002730
Esteban, J. A., Salas, M., and Blanco, L. (1993). Fidelity of Phi 29 DNA Polymerase. Comparison between Protein-Primed Initiation and DNA Polymerization. J. Biol. Chem. 268 (4), 2719–2726. doi:10.1016/s0021-9258(18)53833-3
Euler, M., Wang, Y., Otto, P., Tomaso, H., Escudero, R., Anda, P., et al. (2012). Recombinase Polymerase Amplification Assay for Rapid Detection of Francisella Tularensis. J. Clin. Microbiol. 50 (7), 2234–2238. doi:10.1128/JCM.06504-11
Evanko, D. (2004). Hybridization Chain Reaction. Nat. Methods 1 (3), 186. doi:10.1038/nmeth1204-186a
Fang, X., Liu, Y., Kong, J., and Jiang, X. (2010). Loop-Mediated Isothermal Amplification Integrated on Microfluidic Chips for Point-of-Care Quantitative Detection of Pathogens. Anal. Chem. 82 (7), 3002–3006. doi:10.1021/ac1000652
Faron, M. L., Ledeboer, N. A., Granato, P., Daly, J. A., Pierce, K., Pancholi, P., et al. (2015). Detection of Group A Streptococcus in Pharyngeal Swab Specimens by Use of the AmpliVue Gas Isothermal Helicase-dependent Amplification Assay. J. Clin. Microbiol. 53 (7), 2365–2367. doi:10.1128/JCM.01085-15
Fire, A., and Xu, S. Q. (1995). Rolling Replication of Short DNA Circles. Proc. Natl. Acad. Sci. 92 (10), 4641–4645. doi:10.1073/pnas.92.10.4641
Fujino, M., Yoshida, N., Yamaguchi, S., Hosaka, N., Ota, Y., Notomi, T., et al. (2005). A Simple Method for the Detection of Measles Virus Genome by Loop-Mediated Isothermal Amplification (LAMP). J. Med. Virol. 76 (3), 406–413. doi:10.1002/jmv.20371
Gansen, A., Herrick, A. M., Dimov, I. K., Lee, L. P., and Chiu, D. T. (2012). Digital LAMP in a Sample Self-Digitization (SD) Chip. Lab. Chip 12 (12), 2247–2254. doi:10.1039/c2lc21247a
García-Fernández, S., Morosini, M.-I., Marco, F., Gijon, D., Vergara, A., Vila, J., et al. (2014). Evaluation of the Eazyplex(R) SuperBug CRE System for Rapid Detection of Carbapenemases and ESBLs in Clinical Enterobacteriaceae Isolates Recovered at Two Spanish Hospitals. J. Antimicrob. Chemother. 70 (4), 1047–1050. doi:10.1093/jac/dku476
Garrido-Maestu, A., Azinheiro, S., Carvalho, J., Abalde-Cela, S., Carbó-Argibay, E., Diéguez, L., et al. (2017). Combination of Microfluidic Loop-Mediated Isothermal Amplification with Gold Nanoparticles for Rapid Detection of Salmonella Spp. In Food Samples. Front. Microbiol. 8, 2159. doi:10.3389/fmicb.2017.02159
Gaydos, C., Schwebke, J., Dombrowski, J., Marrazzo, J., Coleman, J., Silver, B., et al. (2017). Clinical Performance of the Solana Point-of-Care Trichomonas Assay from Clinician-Collected Vaginal Swabs and Urine Specimens from Symptomatic and Asymptomatic Women. Expert Rev. Mol. Diagn. 17 (3), 303–306. doi:10.1080/14737159.2017.1282823
Giuffrida, M. C., and Spoto, G. (2017). Integration of Isothermal Amplification Methods in Microfluidic Devices: Recent Advances. Biosens. Bioelectron. 90, 174–186. doi:10.1016/j.bios.2016.11.045
Goldmeyer, J., Li, H., McCormac, M., Cook, S., Stratton, C., Lemieux, B., et al. (2008). Identification of Staphylococcus Aureus and Determination of Methicillin Resistance Directly from Positive Blood Cultures by Isothermal Amplification and a Disposable Detection Device. J. Clin. Microbiol. 46 (4), 1534–1536. doi:10.1128/JCM.02234-07
Gómez de la Torre, T. Z., Ke, R., Mezger, A., Svedlindh, P., Strømme, M., Nilsson, M., et al. (2012). Sensitive Detection of Spores Using Volume-Amplified Magnetic Nanobeads. Small 8 (14), 2174–2177. doi:10.1002/smll.201102632
Guichón, A., Chiparelli, H., Martínez, A., Rodríguez, C., Trento, A., Russi, J. C., et al. (2004). Evaluation of a New NASBA Assay for the Qualitative Detection of Hepatitis C Virus Based on the NucliSens Basic Kit Reagents. J. Clin. Virol. 29 (2), 84–91. doi:10.1016/S1386-6532(03)00091-X
Heijnen, L., and Medema, G. (2009). Method for Rapid Detection of Viable Escherichia Coli in Water Using Real-Time NASBA. Water Res. 43 (12), 3124–3132. doi:10.1016/j.watres.2009.04.025
Hellani, A., Coskun, S., Tbakhi, A., and Al-Hassan, S. (2005). Clinical Application of Multiple Displacement Amplification in Preimplantation Genetic Diagnosis. Reprod. BioMedicine Online 10 (3), 376–380. doi:10.1016/S1472-6483(10)61799-3
Hellyer, T. J., and Nadeau, J. G. (2004). Strand Displacement Amplification: A Versatile Tool for Molecular Diagnostics. Expert Rev. Mol. Diagn. 4 (2), 251–261. doi:10.1586/14737159.4.2.251
Heo, H. Y., Chung, S., Kim, Y. T., Kim, D. H., and Seo, T. S. (2016). A Valveless Rotary Microfluidic Device for Multiplex Point Mutation Identification Based on Ligation-Rolling Circle Amplification. Biosens. Bioelectron. 78, 140–146. doi:10.1016/j.bios.2015.11.039
Hønsvall, B. K., and Robertson., L. J. (2017). From Research Lab to Standard Environmental Analysis Tool: Will NASBA Make the Leap. Water Res. 109, 389–397. doi:10.1016/j.watres.2016.11.052
Horibe, D., Ochiai, T., Shimada, H., Tomonaga, T., Nomura, F., Gun, M., et al. (2007). Rapid Detection of Metastasis of Gastric Cancer Using Reverse Transcription Loop-Mediated Isothermal Amplification. Int. J. Cancer 120 (5), 1063–1069. doi:10.1002/ijc.22397
Hu, C., Kalsi, S., Zeimpekis, I., Sun, K., Ashburn, P., Turner, C., et al. (2017). Ultra-Fast Electronic Detection of Antimicrobial Resistance Genes Using Isothermal Amplification and Thin Film Transistor Sensors. Biosens. Bioelectron. 96 (January), 281–287. doi:10.1016/j.bios.2017.05.016
Huang, J., Wu, Y., Chen, Y., Zhu, Z., Yang, X., Yang, C. J., et al. (2011). Pyrene-Excimer Probes Based on the Hybridization Chain Reaction for the Detection of Nucleic Acids in Complex Biological Fluids. Angew. Chem. Int. Ed. 50 (2), 401–404. doi:10.1002/anie.201005375
Huang, Y., Chen, J., Zhao, S., Shi, M., Chen, Z.-F., and Liang, H. (2013). Label-Free Colorimetric Aptasensor Based on Nicking Enzyme Assisted Signal Amplification and DNAzyme Amplification for Highly Sensitive Detection of Protein. Anal. Chem. 85 (9), 4423–4430. doi:10.1021/ac3037443
Ibarra-Meneses, A. V., Cruz, I., Chicharro, C., Sánchez, C., Biéler, S., Broger, T., et al. (2018). Evaluation of Fluorimetry and Direct Visualization to Interpret Results of a Loop-Mediated Isothermal Amplification Kit to Detect Leishmania DNA. Parasites Vectors 11 (1), 1–9. doi:10.1186/s13071-018-2836-2
Inoue-Nagata, A. K., Albuquerque, L. C., Rocha, W. B., and Nagata, N. (2004). A Simple Method for Cloning the Complete Begomovirus Genome Using the Bacteriophage φ29 DNA Polymerase. J. Virol. Methods 116 (2), 209–211. doi:10.1016/j.jviromet.2003.11.015
Itonaga, M., Matsuzaki, I., Warigaya, K., Tamura, T., Shimizu, Y., Fujimoto, M., et al. (2016). Novel Methodology for Rapid Detection of KRAS Mutation Using PNA-LNA Mediated Loop-Mediated Isothermal Amplification. PLoS ONE 11 (3), e0151654–12. doi:10.1371/journal.pone.0151654
Iwasaki, M., Yonekawa, T., Otsuka, K., Suzuki, W., Nagamine, K., Hase, T., et al. 2003. “Validation of the Loop-Mediated Isothermal Amplification Method for Single Nucleotide Polymorphism Genotyping with Whole Blood.” Genome Lett. 2 (8): 119–126. doi:10.1166/gl.2003.028
James, A., and MacDonald, J. (2015). Recombinase Polymerase Amplification: Emergence as a Critical Molecular Technology for Rapid, Low-Resource Diagnostics. Expert Rev. Mol. Diagn. 15 (11), 1475–1489. doi:10.1586/14737159.2015.1090877
Jayanath, N. Y., Nguyen, L. T., Vu, T. T., and Tran, L. D. (2018). Development of a Portable Electrochemical Loop Mediated Isothermal Amplification (LAMP) Device for Detection of Hepatitis B Virus. RSC Adv. 8 (61), 34954–34959. doi:10.1039/c8ra07235c
Jia, H., Li, Z., Liu, C., and Cheng, Y. (2010). Ultrasensitive Detection of MicroRNAs by Exponential Isothermal Amplification. Angew. Chem. Int. Edition 49 (32), 5498–5501. doi:10.1002/anie.201001375
Jia, H., Wang, Z., Wang, C., Chang, L., and Li, Z. (2014). Real-Time Fluorescence Detection of Hg2+ Ions with High Sensitivity by Exponentially Isothermal Oligonucleotide Amplification. RSC Adv. 4 (19), 9439–9444. doi:10.1039/c3ra45986a
Jonstrup, S. P., Koch, J., and Kjems, J. (2006). A MicroRNA Detection System Based on Padlock Probes and Rolling Circle Amplification. Rna 12 (9), 1747–1752. doi:10.1261/rna.110706
Jung, J. H., Park, B. H., Oh, S. J., Choi, G., and Seo, T. S. (2015). Integration of Reverse Transcriptase Loop-Mediated Isothermal Amplification with an Immunochromatographic Strip on a Centrifugal Microdevice for Influenza A Virus Identification. Lab. Chip 15 (3), 718–725. doi:10.1039/c4lc01033g
Kaneko, H., Kawana, T., Fukushima, E., and Suzutani, T. (2007). Tolerance of Loop-Mediated Isothermal Amplification to a Culture Medium and Biological Substances. J. Biochem. Biophysical Methods 70 (3), 499–501. doi:10.1016/j.jbbm.2006.08.008
Kashir, J., and Yaqinuddin, A. (2020). Loop Mediated Isothermal Amplification (LAMP) Assays as a Rapid Diagnostic for COVID-19. Med. Hypotheses 141 (March), 109786. doi:10.1016/j.mehy.2020.109786
Ke, R., Mignardi, M., Pacureanu, A., Svedlund, J., Botling, J., Wählby, C., et al. (2013). In Situ Sequencing for RNA Analysis in Preserved Tissue and Cells. Nat. Methods 10 (9), 857–860. doi:10.1038/nmeth.2563
Kersting, S., Rausch, V., Bier, F. F., and von Nickisch-Rosenegk, M. (2014a). Multiplex Isothermal Solid-phase Recombinase Polymerase Amplification for the Specific and Fast DNA-Based Detection of Three Bacterial Pathogens. Microchim Acta 181 (13–14), 1715–1723. doi:10.1007/s00604-014-1198-5
Kersting, S., Rausch, V., Bier, F. F., and Von Nickisch-Rosenegk, M. (2014b). Rapid Detection of Plasmodium Falciparum with Isothermal Recombinase Polymerase Amplification and Lateral Flow Analysis. Malar. J. 13 (1): 1–9. doi:10.1186/1475-2875-13-99
Kim, H.-J., Tong, Y., Tang, W., Quimson, L., Cope, V. A., Pan, X., et al. (2011). A Rapid and Simple Isothermal Nucleic Acid Amplification Test for Detection of Herpes Simplex Virus Types 1 and 2. J. Clin. Virol. 50 (1), 26–30. doi:10.1016/j.jcv.2010.09.006
Kim, T.-H., Park, J., Kim, C.-J., and Cho, Y.-K. (2014). Fully Integrated Lab-On-A-Disc for Nucleic Acid Analysis of Food-Borne Pathogens. Anal. Chem. 86 (8), 3841–3848. doi:10.1021/ac403971h
Kivlehan, F., Mavré, F., Talini, L., Limoges, B., and Marchal, D. (2011). Real-Time Electrochemical Monitoring of Isothermal Helicase-dependent Amplification of Nucleic Acids. Analyst 136 (18), 3635–3642. doi:10.1039/c1an15289k
Klein, D. (2002). Quantification Using Real-Time PCR Technology: Applications and Limitations. Trends Mol. Med. 8 (6), 257–260. doi:10.1016/S1471-4914(02)02355-9
Kolm, C., Martzy, R., Führer, M., Mach, R. L., Krska, R., Baumgartner, S., et al. (2019). Mach, Rudolf Krska, Sabine Baumgartner, Andreas H. Farnleitner, and Georg H. Reischer. Detection of a Microbial Source Tracking Marker by Isothermal Helicase-dependent Amplification and a Nucleic Acid Lateral-Flow Strip Test. Sci. Rep. 9 (1), 1–9. doi:10.1038/s41598-018-36749-7
Konry, T., Smolina, I., Yarmush, J. M., Irimia, D., and Yarmush, M. L. (2011). Ultrasensitive Detection of Low-Abundance Surface-Marker Protein Using Isothermal Rolling Circle Amplification in a Microfluidic Nanoliter Platform. Small 7 (3), 395–400. doi:10.1002/smll.201001620
Kopp, M. U., De Mello, A. J., and Manz, A. (1998). Continuous-Flow PCR on a Chip Concept of a Chemical Amplifier for DNA. Rep. 1046 Sci. 280 (May), 1046–1048. doi:10.1126/science.280.5366.1046
Krõlov, K., Frolova, J., Tudoran, O., Suhorutsenko, J., Lehto, T., Sibul, H., et al. (2014). Sensitive and Rapid Detection of Chlamydia Trachomatis by Recombinase Polymerase Amplification Directly from Urine Samples. J. Mol. Diagn. 16 (1), 127–135. doi:10.1016/j.jmoldx.2013.08.003
Kumar, J., Saxena, D., Parida, M., and Rathinam, S. (2018a). Evaluation of Real-Time Reverse-Transcription Loop-Mediated Isothermal Amplification Assay for Clinical Diagnosis of West Nile Virus in Patients. Indian J. Med. Res. 147 (March), 293–298. doi:10.4103/0971-5916.234607
Kumar, S., Amit, K., and Venkatesan, G. (2018b). Isothermal Nucleic Acid Amplification System: An Update on Methods and Applications. J. Genet. Genomens 2 (1), 1–5. doi:10.1128/9781555819156.ch12 https://www.omicsonline.org/open-access/isothermal-nucleic-acid-amplification-system-an-update-on-methods-andapplications.pdf
Kunze, A., Dilcher, M., Abd El Wahed, A., Hufert, F., Niessner, R., and Seidel, M. (2016). On-Chip Isothermal Nucleic Acid Amplification on Flow-Based Chemiluminescence Microarray Analysis Platform for the Detection of Viruses and Bacteria. Anal. Chem. 88 (1), 898–905. doi:10.1021/acs.analchem.5b03540
Lee, M.-F., Chen, Y.-H., and Peng, C.-F. (2009). Evaluation of Reverse Transcription Loop-Mediated Isothermal Amplification in Conjunction with ELISA-Hybridization Assay for Molecular Detection of Mycobacterium tuberculosis. J. Microbiol. Methods 76 (2), 174–180. doi:10.1016/j.mimet.2008.10.005
Lee, S.-Y., Huang, J.-G., Chuang, T.-L., Sheu, J.-C., Chuang, Y.-K., Holl, M., et al. (2008). Compact Optical Diagnostic Device for Isothermal Nucleic Acids Amplification. Sensors Actuators B: Chem. 133 (2), 493–501. doi:10.1016/j.snb.2008.03.008
Lee, S.-Y., Lee, C.-N., Mark, H., Meldrum, D. R., and Lin, C.-W. (2007). Efficient, Specific, Compact Hepatitis B Diagnostic Device: Optical Detection of the Hepatitis B Virus by Isothermal Amplification. Sensors Actuators B: Chem. 127 (2), 598–605. doi:10.1016/j.snb.2007.05.015
Lemieux, B., Li, Y., Kong, H., and Tang, Y.-W. (2012). Near Instrument-free, Simple Molecular Device for Rapid Detection of Herpes Simplex Viruses. Expert Rev. Mol. Diagn. 12 (5), 437–443. doi:10.1586/erm.12.34
Leone, G., Van Schijndel, H., Van Gemen, B., Kramer, F. R., and Schoen, C. D. (1998). Molecular Beacon Probes Combined with Amplification by NASBA Enable Homogeneous, Real-Time Detection of RNA. Nucleic Acids Res. 26 (9), 2150–2155. doi:10.1093/nar/26.9.2150
Li, B., Jiang, Y., Chen, X., and Ellington, A. D. (2012). Probing Spatial Organization of DNA Strands Using Enzyme-free Hairpin Assembly Circuits. J. Am. Chem. Soc. 134 (34), 13918–13921. doi:10.1021/ja300984b
Li, J., Deng, T., Chu, X., Yang, R., Jiang, J., Shen, G., et al. (2010). Rolling Circle Amplification Combined with Gold Nanoparticle Aggregates for Highly Sensitive Identification of Single-Nucleotide Polymorphisms. Anal. Chem. 82 (7), 2811–2816. doi:10.1021/ac100336n
Li, J., and Macdonald, J. (2015). Advances in Isothermal Amplification: Novel Strategies Inspired by Biological Processes. Biosens. Bioelectron. 64, 196–211. doi:10.1016/j.bios.2014.08.069
Li, R.-D., Yin, B.-C., and Ye, B.-C. (2016). Ultrasensitive, Colorimetric Detection of MicroRNAs Based on Isothermal Exponential Amplification Reaction-Assisted Gold Nanoparticle Amplification. Biosens. Bioelectron. 86, 1011–1016. doi:10.1016/j.bios.2016.07.042
Li, Y., Zeng, Y., Ji, X., Li, X., and Ren, R. (2012). Cascade Signal Amplification for Sensitive Detection of Cancer Cell Based on Self-Assembly of DNA Scaffold and Rolling Circle Amplification. Sensors Actuators B: Chem. 171-172, 361–366. doi:10.1016/j.snb.2012.04.060
Ling, J., Deng, Y., Long, X., Liu, J., Du, H., Cao, B., et al. (2012). Single-Nucleotide Polymorphism Array Coupled with Multiple Displacement Amplification: Accuracy and Spatial Resolution for Analysis of Chromosome Copy Numbers in Few Cells. Biotechnol. Appl. Biochem. 59 (1), 35–44. doi:10.1002/bab.64
Little, M. C., Andrews, J., Moore, R., Bustos, S., Jones, L., Embres, C., et al. (1999). Strand Displacement Amplification and Homogeneous Real-Time Detection Incorporated in a Second-Generation DNA Probe System, BDProbeTecET. Clin. Chem. 45 (6 I), 777–784. doi:10.1093/clinchem/45.6.777
Liu, D., Daubendiek, S. L., Zillman, M. A., Ryan, K., and Kool, E. T. (1996). Rolling Circle DNA Synthesis: Small Circular Oligonucleotides as Efficient Templates for DNA Polymerases. J. Am. Chem. Soc. 118 (7), 1587–1594. doi:10.1021/ja952786k
Liu, H., Tian, T., Zhang, Y., Ding, L., Yu, J., and Yan, M. (2017). Sensitive and Rapid Detection of MicroRNAs Using Hairpin Probes-Mediated Exponential Isothermal Amplification. Biosens. Bioelectron. 89 (November 2016), 710–714. doi:10.1016/j.bios.2016.10.099
Liu, P., Yang, X., Sun, S., Wang, Q., Wang, K., Huang, J., et al. (2013). Enzyme-Free Colorimetric Detection of DNA by Using Gold Nanoparticles and Hybridization Chain Reaction Amplification. Anal. Chem. 85 (16), 7689–7695. doi:10.1021/ac4001157
Liu, X., Chen, M., Hou, T., Wang, X., Liu, S., and Li, F. (2014). Label-Free Colorimetric Assay for Base Excision Repair Enzyme Activity Based on Nicking Enzyme Assisted Signal Amplification. Biosens. Bioelectron. 54: 598–602. doi:10.1016/j.bios.2013.11.062
Lizardi, P. M., Huang, X., Zhu, Z., Bray-Ward, P., Thomas, D. C., and Ward, D. C. (1998). Mutation Detection and Single-Molecule Counting Using Isothermal Rolling-Circle Amplification. Nat. Genet. 19 (3), 225–232. doi:10.1038/898
Lobato, I. M., O'Sullivan, C. K., Ciara, K., and O’Sullivan, (2018). Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. Trac Trends Anal. Chem. 98, 19–35. doi:10.1016/j.trac.2017.10.015
Lucigen, . 2021. “LavaLAMPTM DNA Master Mix for Amplification.” 2021. Available at: https://www.lucigen.com/LavaLAMP-DNA-Master-Mix-loop-mediated-isothermal-amplification/.
Lutz, S., Weber, P., Focke, M., Faltin, B., Hoffmann, J., Müller, C., et al. (2010). Microfluidic Lab-On-A-Foil for Nucleic Acid Analysis Based on Isothermal Recombinase Polymerase Amplification (RPA). Lab. Chip 10 (7), 887–893. doi:10.1039/b921140c
Ma, C., Liu, S., and Shi, C. (2014). Ultrasensitive Detection of MicroRNAs Based on Hairpin Fluorescence Probe Assisted Isothermal Amplification. Biosens. Bioelectron. 58, 57–60. doi:10.1016/j.bios.2014.02.034
Ma, C., Wang, W., Yang, Q., Shi, C., and Cao, L. (2011). Cocaine Detection via Rolling Circle Amplification of Short DNA Strand Separated by Magnetic Beads. Biosens. Bioelectron. 26 (7), 3309–3312. doi:10.1016/j.bios.2011.01.003
Magro, L., Jacquelin, B., Escadafal, C., Garneret, P., Kwasiborski, A., Manuguerra, J.-C., et al. (2017). Paper-Based RNA Detection and Multiplexed Analysis for Ebola Virus Diagnostics. Sci. Rep. 7 (1), 1–9. doi:10.1038/s41598-017-00758-9
Mahalanabis, M., Do, J., Almuayad, H., Zhang, J. Y., and Klapperich, C. M. (2010). An Integrated Disposable Device for DNA Extraction and Helicase Dependent Amplification. Biomed. Microdevices 12 (2), 353–359. doi:10.1007/s10544-009-9391-8
Mahmoudian, L., Kaji, N., Tokeshi, M., Nilsson, M., and Baba, Y. (2008a). Rolling Circle Amplification and Circle-to-Circle Amplification of a Specific Gene Integrated with Electrophoretic Analysis on a Single Chip. Anal. Chem. 80 (7), 2483–2490. doi:10.1021/ac702289j
Mahmoudian, L., Melin, J., Mohamadi, M. R., Yamada, K., Ohta, M., Kaji, N., et al. (2008b). Microchip Electrophoresis for Specific Gene Detection of the Pathogenic Bacteria V. Cholerae by Circle-to-Circle Amplification. Anal. Sci. 24 (3), 327–332. doi:10.2116/analsci.24.327
Maples, K., Brian, , Holmberg, R. C., Miller, A. P., Provins, J., and Roth, R. B. (2007). Nicking and Extension Amplification Reaction for the Exponential Amplification of Nucleic Acids. US20090081670A1.
Maragh, S., Jakupciak, J. P., Wagner, P. D., Rom, W. N., Sidransky, D., Srivastava, S., et al. 2008. “Multiple Strand Displacement Amplification of Mitochondrial DNA from Clinical Samples.” BMC Med. Genet. 9: 1–9. doi:10.1186/1471-2350-9-7
Mekuria, T. A., Zhang, S., and Eastwell, K. C. (2014). Rapid and Sensitive Detection of Little Cherry Virus 2 Using Isothermal Reverse Transcription-Recombinase Polymerase Amplification. J. Virol. Methods 205, 24–30. doi:10.1016/j.jviromet.2014.04.015
Meridian Bioscience. 2021. Alethia® Molecular Platform. Available at: https://www.meridianbioscience.com/platform/molecular/alethia/. (Accessed July 29, 2021).
Mollasalehi, H., and Yazdanparast, R. (2012). Non-Crosslinking Gold Nanoprobes for Detection of Nucleic Acid Sequence-Based Amplification Products. Anal. Biochem. 425 (2), 91–95. doi:10.1016/j.ab.2012.03.008
Morabito, K., Wiske, C., and Tripathi, A. (2013). Engineering Insights for Multiplexed Real-Time Nucleic Acid Sequence-Based Amplification (NASBA): Implications for Design of Point-of-Care Diagnostics. Mol. Diagn. Ther. 17 (3), 185–192. doi:10.1007/s40291-013-0029-4
Mori, Y., and Notomi, T. (2009). Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 15 (2), 62–69. doi:10.1007/s10156-009-0669-9
Motamedi, M. K., Saghafinia, M., Karami, A., and Gill, P. (2011). A Review of the Current Isothermal Amplification Techniques: Applications, Advantages and Disadvantages. J. Glob. Infect Dis 3 (3), 293–302. doi:10.4103/0974-777X.83538
Mukai, H., Uemori, T., Takeda, O., Kobayashi, E., Yamamoto, J., Nishiwaki, K., et al. (2007). Highly Efficient Isothermal DNA Amplification System Using Three Elements of 5′-DNA-RNA-3′ Chimeric Primers, RNaseH and Strand-Displacing DNA Polymerase. J. Biochem. 142 (2), 273–281. doi:10.1093/jb/mvm138
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., and Erlich, H. (1986). Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harbor Symposia Quantitative Biol. 51 (0): 263–273. doi:10.1101/sqb.1986.051.01.032
Myrmel, M., Oma, V., Khatri, M., Hansen, H. H., Stokstad, M., Berg, M., et al. (2017). Single Primer Isothermal Amplification (SPIA) Combined with Next Generation Sequencing Provides Complete Bovine Coronavirus Genome Coverage and Higher Sequence Depth Compared to Sequence-independent Single Primer Amplification (SISPA). PLoS ONE 12 (11), e0187780–9. doi:10.1371/journal.pone.0187780
New England BioLabs 2021. IsoAmp® II Universal THDA Kit. Available at: https://international.neb.com/products/h0110-isoamp-ii-universal-thda-kit#Product Information (Accessed June 26, 2021).
Nguyen, V. A. T., Nguyen, H. V., Dinh, T. V., Du, H. H., Do, C. N., Marks, G. B., et al. (2018). Evaluation of LoopampMTBC Detection Kit for Diagnosis of Pulmonary Tuberculosis at a Peripheral Laboratory in a High burden Setting. Diagn. Microbiol. Infect. Dis. 90 (3), 190–195. doi:10.1016/j.diagmicrobio.2017.11.009
Niazi, A., Jorjani, O.-N., Nikbakht, H., and Gill, P. (2013). A Nanodiagnostic Colorimetric Assay for 18S RRNA of Leishmania Pathogens Using Nucleic Acid Sequence-Based Amplification and Gold Nanorods. Mol. Diagn. Ther. 17 (6), 363–370. doi:10.1007/s40291-013-0044-5
Nie, S., Roth, R. B., Stiles, J., Mikhlina, A., Lu, X., Tang, Y.-W., et al. (2014). Evaluation of Alere I Influenza A&B for Rapid Detection of Influenza Viruses A and B. J. Clin. Microbiol. 52 (9), 3339–3344. doi:10.1128/JCM.01132-14
Nilsson, M., Krejci, K., Koch, J., Kwiatkowski, M., Gustavsson, P., and Landegren, U. (1997). Padlock Probes Reveal Single-Nucleotide Differences, Parent of Origin and In Situ Distribution of Centromeric Sequences in Human Chromosomes 13 and 21. Nat. Genet. 16 (3), 252–255. doi:10.1038/ng0797-252
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. 2000. “Loop-Mediated Isothermal Amplification of DNA.” Nucleic Acids Res. 28 (12): 63e – 63. doi:10.1093/nar/28.12.e63
Oh, S. J., Jung, J. H., Choi, G., Lee, D. C., Kim, D. H., and Seo, T. S. (2016). Centrifugal Loop-Mediated Isothermal Amplification Microdevice for Rapid, Multiplex and Colorimetric Foodborne Pathogen Detection. Biosens. Bioelectron. 75, 293–300. doi:10.1016/j.bios.2015.08.052
Okamoto, S., Yoshikawa, T., Ihira, M., Suzuki, K., Shimokata, K., Nishiyama, Y., et al. (2004). Rapid Detection of Varicella-Zoster Virus Infection by a Loop-Mediated Isothermal Amplification Method. J. Med. Virol. 74 (4), 677–682. doi:10.1002/jmv.20223
Oyola, S. O., Otto, T. D., Gu, Y., Maslen, G., Manske, M., Campino, S., et al. (2012). Optimizing Illumina Next-Generation Sequencing Library Preparation for Extremely At-Biased Genomes. BMC Genomics 13 (1), 1–12. doi:10.1186/1471-2164-13-1
Paez, J. G., Lin, M., Beroukhim, R., Lee, J. C., Zhao, X., Richter, D. J., et al. (2004). Genome Coverage and Sequence Fidelity of 29 Polymerase-Based Multiple Strand Displacement Whole Genome Amplification. Nucleic Acids Res. 32 (9), e71. doi:10.1093/nar/gnh069
Pancholi, P., Kelly, C. M., Raczkowski, M., and Balada-Llasat, J. M. (2012). Detection of Toxigenic Clostridium Difficile: Comparison of the Cell Culture Neutralization, Xpert C. Difficile, Xpert C. Difficile/Epi, and Illumigene C. Difficile Assays. J. Clin. Microbiol. 50 (4), 1331–1335. doi:10.1128/JCM.06597-11
Pandori, M. W., and Branson, B. M. (2010). 2010 HIV Diagnostics Conference. Expert Rev. Anti-Infective Ther. 8 (6), 631–633. doi:10.1586/eri.10.48
Parida, M., Posadas, G., Inoue, S., Hasebe, F., and Morita, K. (2004). Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification for Rapid Detection of West Nile Virus. J. Clin. Microbiol. 42 (1), 257–263. doi:10.1128/JCM.42.1.257-263.2004
Parida, M., Sannarangaiah, S., DashRao, P. K., Rao, P. V. L., and Morita, K. (2008). Loop Mediated Isothermal Amplification (LAMP): A New Generation of Innovative Gene Amplification Technique; Perspectives in Clinical Diagnosis of Infectious Diseases. Rev. Med. Virol. 18 (6), 407–421. doi:10.1002/rmv.593
Peng, Y., Li, D., Yuan, R., and Xiang, Y. (2018). A Catalytic and Dual Recycling Amplification ATP Sensor Based on Target-Driven Allosteric Structure Switching of Aptamer Beacons. Biosens. Bioelectron. 105 (October), 1–5. doi:10.1016/j.bios.2018.01.017
Piepenburg, O., Williams, C. H., Stemple, D. L., and Armes, N. A. (2006). DNA Detection Using Recombination Proteins. Plos Biol. 4 (7), e204–21. doi:10.1371/journal.pbio.0040204
Polymerase, R., and Rpa, A. (2017). Recombinase Polymerase Amplification (RPA) an Isothermal Technique Suitable for a Range of Applications- Evaluation of RPA Volume Tolerance. BioTechniques 62 (6), 295. doi:10.2144/000114561
Porter-Jordan, K., Keiser, J. F., Allan, M., Gross, A. M., Nasim, S., and Garrett, C. T. (1990). Nested Polymerase Chain Reaction Assay for the Detection of Cytomegalovirus Overcomes False Positives Caused by Contamination with Fragmented DNA. J. Med. Virol. 30 (2): 85–91. doi:10.1002/jmv.1890300202
Rane, T. D., Chen, L. H., Zec, H. C., and Wang, T.-H. (2015). Microfluidic Continuous Flow Digital Loop-Mediated Isothermal Amplification (LAMP). Lab. Chip 15 (3), 776–782. doi:10.1039/c4lc01158a
Ratliff, A. E., Duffy, L. B., and Waites, K. B. (2014). Comparison of the Illumigene Mycoplasma DNA Amplification Assay and Culture for Detection of Mycoplasma Pneumoniae. J. Clin. Microbiol. 52 (4), 1060–1063. doi:10.1128/JCM.02913-13
Reagin, M. J., GieslerGieslerAlia L. Merla, T. L., Merla, A. L., Resetar-Gerke, J. M., Kapolka, K. M., and Mamone, J. A. (2003). Templiphi: A Sequencing Template Preparation Procedure that Eliminates Overnight Cultures and DNA Purification. J. Biomol. Tech. 14 (2), 143–148. doi:10.1080/02713680490905817
Rector, A., Tachezy, R., and Van Ranst, M. (2004). A Sequence-independent Strategy for Detection and Cloning of Circular DNA Virus Genomes by Using Multiply Primed Rolling-Circle Amplification. J. Virol. 78 (10), 4993–4998. doi:10.1128/jvi.78.10.4993-4998.2004
Reinholt, S. J., Behrent, A., Greene, C., Kalfe, A., and Baeumner, A. J. (2014). Isolation and Amplification of MRNA within a Simple Microfluidic Lab on a Chip. Anal. Chem. 86 (1), 849–856. doi:10.1021/ac403417z
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J.-F., et al. (2013). Insights into the Phylogeny and Coding Potential of Microbial Dark Matter. Nature 499 (7459), 431–437. doi:10.1038/nature12352
Rohrman, B. A., and Richards-Kortum, R. R. (2012). A Paper and Plastic Device for Performing Recombinase Polymerase Amplification of HIV DNA. Lab. Chip 12 (17), 3082–3088. doi:10.1039/c2lc40423k
Rohrman, B., and Richards-Kortum, R. (2015). Inhibition of Recombinase Polymerase Amplification by Background Dna: A Lateral Flow-Based Method for Enriching Target DNA. Anal. Chem. 87 (3), 1963–1967. doi:10.1021/ac504365v
Rutjes, S. A., van den BergVan Den Berg, H. H. J. L., Lodder, W. J., and de Roda Husman, A. M. (2006). Real-Time Detection of Noroviruses in Surface Water by Use of a Broadly Reactive Nucleic Acid Sequence-Based Amplification Assay. Appl. Environ. Microbiol. 72 (8), 5349–5358. doi:10.1128/AEM.00751-06
Sabaté del Río, J., Steylaerts, T., Henry, O. Y. F., Bienstman, P., Stakenborg, T., Van Roy, W., et al. (2015). Real-Time and Label-free Ring-Resonator Monitoring of Solid-phase Recombinase Polymerase Amplification. Biosens. Bioelectron. 73, 130–137. doi:10.1016/j.bios.2015.05.063
Santiago-Felipe, S., Tortajada-Genaro, L. A., Puchades, R., and Maquieira, A. (2014). Recombinase Polymerase and Enzyme-Linked Immunosorbent Assay as a DNA Amplification-Detection Strategy for Food Analysis. Analytica Chim. Acta 811, 81–87. doi:10.1016/j.aca.2013.12.017
Sato, K., Tachihara, A., Renberg, B., Mawatari, K., Sato, K., Tanaka, Y., et al. (2010). Microbead-Based Rolling Circle Amplification in a Microchip for Sensitive DNA Detection. Lab. Chip 10 (10), 1262–1266. doi:10.1039/b927460j
Schopf, E., Fischer, N. O., Chen, Y., and Tok, J. B.-H. (2008). Sensitive and Selective Viral DNA Detection Assay via Microbead-Based Rolling Circle Amplification. Bioorg. Med. Chem. Lett. 18 (22), 5871–5874. doi:10.1016/j.bmcl.2008.07.064
Schopf, E., Liu, Y., Deng, J. C., Yang, S., Cheng, G., and Chen, Y. (2011). Mycobacterium Tuberculosis Detection via Rolling Circle Amplification. Anal. Methods 3 (2), 267–273. doi:10.1039/c0ay00529k
Schweitzer, B., Roberts, S., Grimwade, B., Shao, W., Wang, M., Fu, Q., et al. (2002). Multiplexed Protein Profiling on Microarrays by Rolling-Circle Amplification. Nat. Biotechnol. 20 (4), 359–365. doi:10.1038/nbt0402-359
Sha, L., Zhang, X., and Wang, G. (2016). A Label-free and Enzyme-free Ultra-sensitive Transcription Factors Biosensor Using DNA-Templated Copper Nanoparticles as Fluorescent Indicator and Hairpin DNA Cascade Reaction as Signal Amplifier. Biosens. Bioelectron. 82, 85–92. doi:10.1016/j.bios.2016.03.066
Shen, F., Davydova, E. K., Du, W., Kreutz, J. E., Piepenburg, O., and Ismagilov, R. F. (2011). Digital Isothermal Quantification of Nucleic Acids via Simultaneous Chemical Initiation of Recombinase Polymerase Amplification Reactions on SlipChip. Anal. Chem. 83 (9), 3533–3540. doi:10.1021/ac200247e
Sheng, Q., Cheng, N., Bai, W., and Zheng, J. (2015). Ultrasensitive Electrochemical Detection of Breast Cancer Cells Based on DNA-Rolling-Circle-Amplification-Directed Enzyme-Catalyzed Polymerization. Chem. Commun. 51 (11), 2114–2117. doi:10.1039/c4cc08954e
Shimada, M., Hino, F., Sagawa, H., Mukai, H., Asada, K., and Kato, I. (2002). Development of the Detection System for Mycobacterium tuberculosis DNA by Using the Isothermal DNA Amplification Method ICAN. Rinsho Byori 50 (5), 528–532.
Simpkins, S. A., Chan, A. B., Hays, J., opping, B. P., and Cook, N. (2000). An RNA Transcription-Based Amplification Technique (NASBA) for the Detection of Viable Salmonella Enterica. Lett. Appl. Microbiol. 30 (1), 75–79. doi:10.1046/j.1472-765x.2000.00670.x
Smirnov, D. A., Burdick, J. T., Morley, M., and Cheung, V. G. (2004). Method for Manufacturing Whole-Genome Microarrays by Rolling Circle Amplification. Genes Chromosom. Cancer 40 (1), 72–77. doi:10.1002/gcc.20015
Smolina, I. V., Demidov, V. V., Cantor, C. R., and Broude, N. E. (2004). Real-Time Monitoring of Branched Rolling-Circle DNA Amplification with Peptide Nucleic Acid Beacon. Anal. Biochem. 335 (2), 326–329. doi:10.1016/j.ab.2004.07.022
Song, J., Mauk, M. G., Hackett, B. A., Cherry, S., Bau, H. H., and Liu, C. (2016). Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 88 (14), 7289–7294. doi:10.1021/acs.analchem.6b01632
Song, Y., Li, W., Duan, Y., Li, Z., and Deng, L. (2014). Nicking Enzyme-Assisted Biosensor for Salmonella Enteritidis Detection Based on Fluorescence Resonance Energy Transfer. Biosens. Bioelectron. 55, 400–404. doi:10.1016/j.bios.2013.12.053
Spits, C., Le Caignec, C., De Rycke, M., Van Haute, L., Van Steirteghem, A., Liebaers, I., et al. (2006). Whole-Genome Multiple Displacement Amplification from Single Cells. Nat. Protoc. 1 (4), 1965–1970. doi:10.1038/nprot.2006.326
Srividya, A., Maiti, B., Chakraborty, A., and Chakraborty, G. (2019). Loop Mediated Isothermal Amplification: A Promising Tool for Screening Genetic Mutations. Mol. Diagn. Ther. 23 (6), 723–733. doi:10.1007/s40291-019-00422-0
Sun, Y.-Q., Monstein, H.-J., Ryberg, A., and Borch, K. (2007). Multiple Strand Displacement Amplification of DNA Isolated from Human Archival Plasma/Serum: Identification of Cytokine Polymorphism by Pyrosequencing Analysis. Clinica Chim. Acta 377 (1–2), 108–113. doi:10.1016/j.cca.2006.09.003
Sun, Y., Quyen, T. L., Hung, T. Q., Chin, W. H., Wolff, A., and Bang, D. D. (2015). A Lab-On-A-Chip System with Integrated Sample Preparation and Loop-Mediated Isothermal Amplification for Rapid and Quantitative Detection of Salmonella Spp. In Food Samples. Lab. Chip 15 (8), 1898–1904. doi:10.1039/c4lc01459f
Tan, E., Wong, J., Nguyen, D., Zhang, Y., Erwin, B., Van Ness, L. K., et al. (2005). Isothermal DNA Amplification Coupled with DNA Nanosphere-Based Colorimetric Detection. Anal. Chem. 77 (24), 7984–7992. doi:10.1021/ac051364i
Tang, W., Chow, W. H. A., Li, Y., Kong, H., Tang, Y. W., and Lemieux, B. (2010). Nucleic Acid Assay System for Tier II Laboratories and Moderately Complex Clinics to Detect HIV in Low‐Resource Settings. J. Infect. Dis. 201 (Suppl. 1), S46–S51. doi:10.1086/650388
Tanner, N. A., Zhang, Y., and Evans, T. C. (2012). Simultaneous Multiple Target Detection in Real-Time Loop-Mediated Isothermal Amplification. BioTechniques 53 (2), 81–89. doi:10.2144/0000113902
Tatsumi, K., Mitani, Y., Watanabe, J., Takakura, H., Hoshi, K., Kawai, Y., et al. (2008). Rapid Screening Assay for KRAS Mutations by the Modified Smart Amplification Process. J. Mol. Diagn. 10 (6), 520–526. doi:10.2353/jmoldx.2008.080024
Thomas, D. C., Nardone, G. A., and Randall, S. K. (1999). Amplification of Padlock Probes for DNA Diagnostics by Cascade Rolling Circle Amplification or the Polymerase Chain Reaction. Arch. Pathol. Lab. Med. 123 (12), 1170–1176. doi:10.1043/1543-2165-123.20.1170
Tomlinson, J. A., Dickinson, M. J., and Boonham, N. (2010). Rapid Detection of Phytophthora Ramorum and P. Kernoviae by Two-Minute DNA Extraction Followed by Isothermal Amplification and Amplicon Detection by Generic Lateral Flow Device. Phytopathology 100 (2), 143–149. doi:10.1094/PHYTO-100-2-0143
Tong, Y., Tang, W., Kim, H.-J., Pan, X., Ranalli, T. A., and Kong, H. (2008). Development of Isothermal TaqMan Assays for Detection of Biothreat Organisms. BioTechniques 45 (5), 543–557. doi:10.2144/000112959
Torres-Chavolla, E., and Alocilja, E. C. (2011). Nanoparticle Based DNA Biosensor for Tuberculosis Detection Using Thermophilic Helicase-dependent Isothermal Amplification. Biosens. Bioelectron. 26 (11), 4614–4618. doi:10.1016/j.bios.2011.04.055
Tsaloglou, M.-N., Laouenan, F., Loukas, C.-M., Monsalve, L. G., Thanner, C., Morgan, H., et al. (2013). Real-Time Isothermal RNA Amplification of Toxic Marine Microalgae Using Preserved Reagents on an Integrated Microfluidic Platform. Analyst 138 (2), 593–602. doi:10.1039/c2an36464f
Tu, P.-A., Shiu, J.-S., Lee, S.-H., Pang, V. F., Wang, D.-C., Wang, P.-H., et al. (2017). Development of a Recombinase Polymerase Amplification Lateral Flow Dipstick (RPA-LFD) for the Field Diagnosis of Caprine Arthritis-Encephalitis Virus (CAEV) Infection. J. Virol. Methods 243: 98–104. doi:10.1016/j.jviromet.2017.01.023
TwistDx. 2021. “TwistAmp® Liquid Exo.” 2021. Available at: https://www.twistdx.co.uk/en/products/product/twistamp-liquid-exo.
Uddin, S., Ibrahim, F., Sayad, A., Thiha, A., Pei, K., Mohktar, M., et al. (2015). A Portable Automatic Endpoint Detection System for Amplicons of Loop Mediated Isothermal Amplification on Microfluidic Compact Disk Platform. Sensors 15 (3), 5376–5389. doi:10.3390/s150305376
Van Der Pol, B., Ferrero, D. V., Buck-Barrington, L., Hook, E., Lenderman, C., Quinn, T., et al. (2001). Multicenter Evaluation of the BDProbeTec ET System for Detection of Chlamydia Trachomatis and Neisseria Gonorrhoeae in Urine Specimens, Female Endocervical Swabs, and Male Urethral Swabs. J. Clin. Microbiol. 39 (3), 1008–1016. doi:10.1128/JCM.39.3.1008-1016.2001
Van Ness, J., Van Ness, L. K., Galas, D. J., and Galas, David. J. (2003). Isothermal Reactions for the Amplification of Oligonucleotides. Proc. Natl. Acad. Sci. 100 (8), 4504–4509. doi:10.1073/pnas.0730811100
Vincent, M., Xu, Y., and Kong, H. (2004). Helicase‐dependent Isothermal DNA Amplification. EMBO Rep. 5 (8), 795–800. doi:10.1038/sj.embor.7400200
Wakabayashi, T., Yamashita, R., Kakita, T., Kakita, M., and Oshika, T. (2004). Rapid and Sensitive Diagnosis of Adenoviral Keratoconjunctivitis by Loop-Mediated Isothermal Amplification (LAMP) Method. Curr. Eye Res. 28 (6), 445–450. doi:10.1080/02713680490905817doi:10.1080/02713680490503796
Walker, G. T., Little, M. C., Nadeau, J. G., and Shank, D. D. (1992). Isothermal In Vitro Amplification of DNA by a Restriction Enzyme/DNA Polymerase System. Proc. Natl. Acad. Sci. 89 (1), 392–396. doi:10.1073/pnas.89.1.392
Wang, C.-H., Lien, K.-Y., Wang, T.-Y., Chen, T.-Y., and Lee, G.-B. (2011a). An Integrated Microfluidic Loop-Mediated-Isothermal-Amplification System for Rapid Sample Pre-treatment and Detection of Viruses. Biosens. Bioelectron. 26 (5), 2045–2052. doi:10.1016/j.bios.2010.08.083
Wang, C.-H., Lien, K.-Y., Wu, J.-J., and Lee, G.-B. (2011b). A Magnetic Bead-Based Assay for the Rapid Detection of Methicillin-Resistant Staphylococcus aureus by Using a Microfluidic System with Integrated Loop-Mediated Isothermal Amplification. Lab. Chip 11 (8), 1521–1531. doi:10.1039/c0lc00430h
Wang, K., Zhang, K., Lv, Z., Zhu, X., Zhu, L., and Zhou, F. (2014). Ultrasensitive Detection of MicroRNA with Isothermal Amplification and a Time-Resolved Fluorescence Sensor. Biosens. Bioelectron. 57, 91–95. doi:10.1016/j.bios.2014.01.058
Wang, L., Qian, C., Qian, W., Wang, R., Wu, J., Ying, Y., et al. (2017). A Highly Specific Strategy for in Suit Detection of DNA with Nicking Enzyme Assisted Amplification and Lateral Flow. Sensors Actuators B: Chem. 253: 258–265. doi:10.1016/j.snb.2017.06.124
Wang, W.-J., Li, J.-J., Rui, K., Gai, P.-P., Zhang, J.-R., and Zhu, J.-J. (2015). Sensitive Electrochemical Detection of Telomerase Activity Using Spherical Nucleic Acids Gold Nanoparticles Triggered Mimic-Hybridization Chain Reaction Enzyme-free Dual Signal Amplification. Anal. Chem. 87 (5), 3019–3026. doi:10.1021/ac504652e
Wang, X., Seo, D. J., Lee, M. H., Choi, C., and Onderdonk, A. B. (2014). Comparison of Conventional PCR, Multiplex PCR, and Loop-Mediated Isothermal Amplification Assays for Rapid Detection of Arcobacter Species. J. Clin. Microbiol. 52 (2), 557–563. doi:10.1128/JCM.02883-13
Watanabe, J., Mitani, Y., Kawai, Y., Kikuchi, T., Kogo, Y., Oguchi-Katayama, A., et al. (2007). Use of a Competitive Probe in Assay Design for Genotyping of the UGT1A1*28 Microsatellite Polymorphism by the Smart Amplification Process. BioTechniques 43 (4), 479–484. doi:10.2144/000112563
Weber, N. C., Klepser, M. E., Akers, J. M., Klepser, D. G., and Adams, A. J. (2016). Use of CLIA-Waived Point-of-Care Tests for Infectious Diseases in Community Pharmacies in the United States. Expert Rev. Mol. Diagn. 16 (2), 253–264. doi:10.1586/14737159.2015.1116388
Wharam, S. D., Peter Marsh, J. S., McPhee, J. E., Weston, A., Donald, L., and Cardy, N. (2001). Specific Detection of DNA and RNA Targets Using a Novel Isothermal Nucleic Acid Amplification Assay Based on the Formation of a Three-Way Junction Structure. Nucleic Acids Res. 29 (11), 54e–54. doi:10.1093/nar/29.11.e54
Wong, Y.-P., Othman, S., Lau, Y.-L., Radu, S., and Chee, H.-Y. (2018). Loop-Mediated Isothermal Amplification (LAMP): A Versatile Technique for Detection of Micro-organisms. J. Appl. Microbiol. 124 (3), 626–643. doi:10.1111/jam.13647
World Health Organization (2016). The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. Geneva, Switzerland: WHO Library Cataloguing-in-Publication Data.
Wu, C., Cansiz, S., Zhang, L., Teng, I.-T., Qiu, L., Li, J., et al. (2015). A Nonenzymatic Hairpin DNA Cascade Reaction Provides High Signal Gain of MRNA Imaging inside Live Cells. J. Am. Chem. Soc. 137 (15), 4900–4903. doi:10.1021/jacs.5b00542
Xia, S., and Chen, X. (2020). Single-copy Sensitive, Field-Deployable, and Simultaneous Dual-Gene Detection of SARS-CoV-2 RNA via Modified RT-RPA. Cell Discov. 6 (1), 4–7. doi:10.1038/s41421-020-0175-x
Xuan, F., Fan, T. W., and Hsing, I.-M. (2015). Electrochemical Interrogation of Kinetically-Controlled Dendritic DNA/PNA Assembly for Immobilization-free and Enzyme-free Nucleic Acids Sensing. ACS Nano 9 (5), 5027–5033. doi:10.1021/nn507282f
Xuan, F., and Hsing, I.-M. (2014). Triggering Hairpin-free Chain-Branching Growth of Fluorescent DNA Dendrimers for Nonlinear Hybridization Chain Reaction. J. Am. Chem. Soc. 136 (28), 9810–9813. doi:10.1021/ja502904s
Xue, L., Zhou, X., and Xing, D. (2010). Highly Sensitive Protein Detection Based on Aptamer Probe and Isothermal Nicking Enzyme Assisted Fluorescence Signal Amplification. Chem. Commun. 46 (39), 7373–7375. doi:10.1039/C0CC02038A
Xue, Q., Lv, Y., Xu, S., Zhang, Y., Wang, L., Li, R., et al. (2015). Highly Sensitive Fluorescence Assay of DNA Methyltransferase Activity by Methylation-Sensitive Cleavage-Based Primer Generation Exponential Isothermal Amplification-Induced G-Quadruplex Formation. Biosens. Bioelectron. 66, 547–553. doi:10.1016/j.bios.2014.12.017
Xue, Q., Lv, Y., Zhang, Y., Xu, S., Li, R., Yue, Q., et al. (2014). Ultrasensitive Fluorescence Detection of Nucleic Acids Using Exonuclease III-Induced Cascade Two-Stage Isothermal Amplification-Mediated Zinc (II)-Protoporphyrin IX/G-Quadruplex Supramolecular Fluorescent Nanotags. Biosens. Bioelectron. 61, 351–356. doi:10.1016/j.bios.2014.05.047
Xue, L., Zhou, X., and Xing., D. (2012). Sensitive and Homogeneous Protein Detection Based on Target-Triggered Aptamer Hairpin Switch and Nicking Enzyme Assisted Fluorescence Signal Amplification. Anal. Chem. 84 (8), 3507–3513. doi:10.1021/ac2026783
Yan, Y., Qiao, Z., Hai, X., Song, W., and Bi, S. (2021). Versatile Electrochemical Biosensor Based on Bi-enzyme Cascade Biocatalysis Spatially Regulated by DNA Architecture. Biosens. Bioelectron. 174: 112827. doi:10.1016/j.bios.2020.112827
Yang, H., Ma, X., Zhang, X., Wang, Y., and Zhang, W. (2011). Development and Evaluation of a Loop-Mediated Isothermal Amplification Assay for the Rapid Detection of Staphylococcus Aureus in Food. Eur. Food Res. Technol. 232 (5), 769–776. doi:10.1007/s00217-011-1442-8
Yang, J. M., Bell, J., Huang, Y., Tirado, M., Thomas, D., Forster, A. H., et al. (2002). An Integrated, Stacked Microlaboratory for Biological Agent Detection with DNA and Immunoassays. Biosens. Bioelectron. 17 (6–7), 605–618. doi:10.1016/S0956-5663(02)00023-4
Yang, L., Liu, C., Ren, W., and Li, Z. (2012). Graphene Surface-Anchored Fluorescence Sensor for Sensitive Detection of MicroRNA Coupled with Enzyme-free Signal Amplification of Hybridization Chain Reaction. ACS Appl. Mater. Inter. 4 (12), 6450–6453. doi:10.1021/am302268t
Yang, Q., Domesle, K. J., and Ge, B. (2018). Loop-Mediated Isothermal Amplification forSalmonellaDetection in Food and Feed: Current Applications and Future Directions. Foodborne Pathog. Dis. 15 (6), 309–331. doi:10.1089/fpd.2018.2445
Yao, J., Liu, Z., Ko, L.-S., Pan, G., and Jiang, Y. (2005). Quantitative Detection of HIV-1 RNA Using NucliSens EasyQ HIV-1 Assay. J. Virol. Methods 129 (1), 40–46. doi:10.1016/j.jviromet.2005.04.017
Ye, J., Xu, M., Tian, X., Cai, S., and Zeng, S. (2019). Research Advances in the Detection of MiRNA. J. Pharm. Anal. 9 (4), 217–226. doi:10.1016/j.jpha.2019.05.004
Yeh, E.-C., Fu, C.-C., Hu, L., Thakur, R., Feng, J., and Lee, L. P. (2017). Self-powered Integrated Microfluidic point-of-care Low-Cost Enabling (SIMPLE) Chip. Sci. Adv. 3, e1501645–12. doi:10.1126/sciadv.1501645
Yi, J., Zhang, W., and Zhang, D. Y. (2006). Molecular Zipper: A Fluorescent Probe for Real-Time Isothermal DNA Amplification. Nucleic Acids Res. 34 (11), e81. doi:10.1093/nar/gkl261
Yoon, H. S., Price, D. C., Stepanauskas, R., Rajah, V. D., Sieracki, M. E., Wilson, W. H., et al. (2011). Single-Cell Genomics Reveals Organismal Interactions in Uncultivated Marine Protists. Science 332 (6030), 714–717. doi:10.1126/science.1203163
Yoshino, M., Watari, H., Kojima, T., Ikedo, M., and Kurita, J. (2009). Rapid, Sensitive and Simple Detection Method Forkoi Herpesvirususing Loop-Mediated Isothermal Amplification. Microbiol. Immunol. 53 (7), 375–383. doi:10.1111/j.1348-0421.2009.00145.x
Yuan, D., Kong, J., Li, X., Fang, X., and Chen, Q. (2018). Colorimetric LAMP Microfluidic Chip for Detecting Three Allergens: Peanut, Sesame and Soybean. Sci. Rep. 8 (1), 1–8. doi:10.1038/s41598-018-26982-5
Yue, S., Li, Y., Qiao, Z., Song, W., and Bi, S. (2021). Rolling Circle Replication for Biosensing, Bioimaging, and Biomedicine. Trends Biotechnol. S0167-7799 (21), 00039–41. doi:10.1016/j.tibtech.2021.02.007
Zeng, Y.-p., Hu, J., Long, Y., and Zhang, C.-y. (2013). Sensitive Detection of DNA Methyltransferase Using Hairpin Probe-Based Primer Generation Rolling Circle Amplification-Induced Chemiluminescence. Anal. Chem. 85 (12), 6143–6150. doi:10.1021/ac4011292
Zhang, B., Liu, B., Tang, D., Niessner, R., Chen, G., and Knopp, D. (2012). DNA-based Hybridization Chain Reaction for Amplified Bioelectronic Signal and Ultrasensitive Detection of Proteins. Anal. Chem. 84 (12), 5392–5399. doi:10.1021/ac3009065
Zhang, D. Y., Brandwein, M., Hsuih, T., and Li, H. B. (2001). Ramification Amplification: A Novel Isothermal DNA Amplification Method. Mol. Diagn. 6 (2), 141–150. doi:10.1054/modi.2001.2532310.2165/00066982-200106020-00010
Zhang, H., Xu, Y., Fohlerova, Z., Chang, H., Iliescu, C., and Neuzil, P. (2019). LAMP-on-a-Chip: Revising Microfluidic Platforms for Loop-Mediated DNA Amplification. Trac Trends Anal. Chem. 113, 44–53. doi:10.1016/j.trac.2019.01.015
Zhang, X., Liu, C., Sun, L., Duan, X., and Li, Z. (2015). Lab on a Single Microbead: An Ultrasensitive Detection Strategy Enabling MicroRNA Analysis at the Single-Molecule Level. Chem. Sci. 6 (11), 6213–6218. doi:10.1039/c5sc02641e
Zhang, X., Lowe, S. B., and Gooding, J. J. (2014). Brief Review of Monitoring Methods for Loop-Mediated Isothermal Amplification (LAMP). Biosens. Bioelectron. 61, 491–499. doi:10.1016/j.bios.2014.05.039
Zhao, X., Dong, T., Yang, Z., Pires, N., and Høivik, N. (2012). Compatible Immuno-NASBA LOC Device for Quantitative Detection of Waterborne Pathogens: Design and Validation. Lab. Chip 12 (3), 602–612. doi:10.1039/c1lc20836e
Zhao, X., Li, Y., Wang, L., You, L., Xu, Z., Li, L., et al. (2010). Development and Application of a Loop-Mediated Isothermal Amplification Method on Rapid Detection Escherichia Coli O157 Strains from Food Samples. Mol. Biol. Rep. 37 (5), 2183–2188. doi:10.1007/s11033-009-9700-6
Zhao, Y., Chen, F., Li, Q., Wang, L., and Fan, C. (2015). Isothermal Amplification of Nucleic Acids. Chem. Rev. 115 (22), 12491–12545. doi:10.1021/acs.chemrev.5b00428
Zhao, Y., Chen, F., Wu, Y., Dong, Y., and Fan, C. (2013). Highly Sensitive Fluorescence Assay of DNA Methyltransferase Activity via Methylation-Sensitive Cleavage Coupled with Nicking Enzyme-Assisted Signalamplification. Biosens. Bioelectron. 42. 56–61. doi:10.1016/j.bios.2012.10.022
Zheng, A.-X., Wang, J.-R., Li, J., Song, X.-R., Chen, G.-N., and Yang, H.-H. (2012). Nicking Enzyme Based Homogeneous Aptasensors for Amplification Detection of Protein. Chem. Commun. 48 (3), 374–376. doi:10.1039/c1cc15853h
Zheng, J., Zhu, G., Li, Y., Li, C., You, M., Chen, T., et al. (2013). A Spherical Nucleic Acid Platform Based on Self-Assembled DNA Biopolymer for High-Performance Cancer Therapy. ACS Nano 7 (8), 6545–6554. doi:10.1021/nn402344v
Zhou, J., Xu, M., Tang, D., Gao, Z., Tang, J., and Chen, G. (2012). Nanogold-Based Bio-Bar Codes for Label-free Immunosensing of Proteins Coupling with an In Situ DNA-Based Hybridization Chain Reaction. Chem. Commun. 48 (100), 12207–12209. doi:10.1039/c2cc36820j
Zhou, L., Wang, Y., Yang, C., Xu, H., Luo, J., Zhang, W., et al. (2019). A Label-free Electrochemical Biosensor for MicroRNAs Detection Based on DNA Nanomaterial by Coupling with Y-Shaped DNA Structure and Non-linear Hybridization Chain Reaction. Biosens. Bioelectron. 126 (February), 657–663. doi:10.1016/j.bios.2018.11.028
Zhou, W., Hu, L., Ying, L., Zhao, Z., Paul, C., Yu, X.-F., et al. (2018). A CRISPR-Cas9-Triggered Strand Displacement Amplification Method for Ultrasensitive DNA Detection. Nat. Commun. 9 (1), 5012. doi:10.1038/s41467-018-07324-5
Zhou, W., Su, J., Chai, Y., Yuan, R., and Xiang, Y. (2014). Naked Eye Detection of Trace Cancer Biomarkers Based on Biobarcode and Enzyme-Assisted DNA Recycling Hybrid Amplifications. Biosens. Bioelectron. 53, 494–498. doi:10.1016/j.bios.2013.10.020
Zhu, G., Zhang, S., Song, E., Zheng, J., Hu, R., Fang, X., et al. (2013). Building Fluorescent DNA Nanodevices on Target Living Cell Surfaces. Angew. Chem. Int. Ed. 52 (21), 5490–5496. doi:10.1002/anie.201301439
Keywords: isothermal amplification, non-pcr amplification, point—of—care, nucleic acids amplification, NAAT
Citation: Oliveira BB, Veigas B and Baptista PV (2021) Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2:752600. doi: 10.3389/fsens.2021.752600
Received: 03 August 2021; Accepted: 13 September 2021;
Published: 28 September 2021.
Edited by:
Gorachand Dutta, Indian Institute of Technology Kharagpur, IndiaCopyright © 2021 Oliveira, Veigas and Baptista. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pedro Viana Baptista, cG12YkBmY3QudW5sLnB0
 Beatriz B. Oliveira
Beatriz B. Oliveira Bruno Veigas3
Bruno Veigas3 Pedro Viana Baptista
Pedro Viana Baptista