
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Water , 22 May 2023
Sec. Water and Human Health
Volume 5 - 2023 | https://doi.org/10.3389/frwa.2023.1136066
This article is part of the Research Topic Wastewater-Based Epidemiology as a Tool for Monitoring Public Health View all 5 articles
 Karin Yaniv1†
Karin Yaniv1† Hillary A. Craddock2†
Hillary A. Craddock2† Fareed Mahameed3
Fareed Mahameed3 Marilou Shagan1
Marilou Shagan1 Ikram Salah2
Ikram Salah2 Satish Lakkakula1
Satish Lakkakula1 Keren Resnick2
Keren Resnick2 Corinne Haber2
Corinne Haber2 Nadav Davidovitch2
Nadav Davidovitch2 Jacob Moran-Gilad2
Jacob Moran-Gilad2 Ariel Kushmaro1,4,5*
Ariel Kushmaro1,4,5* Clive Lipchin3*
Clive Lipchin3*Background: Wastewater based epidemiology (WBE) has become an important tool in SARS-CoV-2 surveillance and epidemiology. While WBE measurements generally correlate with observed case numbers in large municipal areas on sewer grids, there are few studies on its utility in communities that are off-grid (non-sewered).
Methods and materials: To explore the applicability of wastewater surveillance in our region, five Bedouin communities along the Hebron Stream in Southern Israel (Negev desert) were sampled. One point (El-Sayed) represents a community with partial connection to the sewer grid system and another point (Um Batin) represents a community with no access to the sewer grid system. The towns of Hura, Lakia, and Tel Al-Sabi/Tel Sheva were on-grid. A total of 87 samples were collected between August 2020 to January 2021 using both grab and composite sampling. RNA was extracted from the raw sewage and concentrated sewage. RT-qPCR was carried out with N1, N2, and N3 gene targets, and findings were compared to human case data from the Israeli Ministry of Health.
Results: SARS-CoV-2 was detected consistently over time in on-grid Bedouin towns (Lakia, Tel Sheva/Tel as-Sabi, and Hura) and inconsistently in smaller, off-grid communities (El-Sayed and Um Batin). The trend in maximum copy number/L appears to be driven by population size. When comparing case numbers normalized to population size, the amount of gene copies/L was inconsistently related to reported case numbers. SARS-CoV-2 was also detected from sewage-impacted environmental waters representing communities with no access to the wastewater grid. When grab sampling and composite sampling data were compared, results were generally comparable however composite sampling produced superior results.
Conclusions: The mismatch observed between detected virus and reported cases could indicate asymptomatic or “silent” community transmission, under-testing within these communities (due to factors like mistrust in government, stigma, misinformation) or a combination therein. While the exact reason for the mismatch between environmental SARS-CoV-2 signals and case numbers remains unresolved, these findings suggest that sewage surveillance, including grab sampling methodologies, can be a critical aspect of outbreak surveillance and control in areas with insufficient human testing and off-grid communities.
While the primary mode of surveillance for SARS-CoV-2 has been individual testing of clinical samples, wastewater sampling to assess community-level COVID-19 activity is gaining in popularity (Ahmed et al., 2020, 2021; Nemudryi et al., 2020; Scott et al., 2021; Yaniv et al., 2021a). Although vaccinations have curtailed disease levels (Fontanet et al., 2021), the emergence of variants of concern as well as surges of cases in unvaccinated populations and reduction in vaccine effectiveness call for continued surveillance. Using a relatively passive, community-level surveillance methodology such as wastewater-based epidemiology (WBE) has the potential to provide beneficial data under such circumstances. WBE is useful for monitoring variants, as surveillance can continue with standard gene targets (which often focus on well-conserved areas of the genome) while variants can be observed via specific PCR screens design (based on specific mutations) or through sequencing efforts.
Although the main SARS-CoV-2 transmission route is through the respiratory system, the virus can be found in other bodily fluids including stool. A study by Zheng et al. (2020), shows that the viral shedding period is longer in stool than in respiratory samples and for the most part can last for up to 30 days after infection. The clinical surveillance for SARS-CoV-2 is limited by personal willingness to be tested, access to testing and overall healthcare system testing capacity. WBE has been previously employed for monitoring other microbial and viral diseases, including the rapid development of assays for emerging diseases such as monkeypox (Tiwari et al., 2022a) and established surveillance protocols for ancient threats like polio, distribution of antibiotic resistance or even chemical water pollution (Asghar et al., 2014; Elkayam et al., 2018; Lorenzo and Picó, 2019; Pruden et al., 2021; Tiwari et al., 2022b). In Israel, virus surveillance in wastewater has been utilized for several decades and played a central role in the response to the silent poliovirus outbreak of 2013/14 (Manor et al., 2014; Shulman et al., 2016; Brouwer et al., 2018). Since the beginning of the COVID-19 pandemic, many studies have been conducted on optimizing and utilizing WBE for estimations of viral burden (Lorenzo and Picó, 2019; Hart and Halden, 2020; Polo et al., 2020; Hemalatha et al., 2021; Wurtzer et al., 2021; Yaniv et al., 2021a,b, 2022), however there is a lack of studies investigating WBE to monitor SARS-CoV-2 in under-resourced and under-serviced communities (Street et al., 2020). Specifically, there have been few studies investigating WBE in communities not connected or partially-connected to a wastewater grid and research has called for more effort in this regard (Aguiar-Oliveira et al., 2020; Street et al., 2020; Iglesias et al., 2021; Driver et al., 2022; Islam et al., 2023; Naughton et al., 2023).
The Bedouin communities are an indigenous Arab ethnic group residing within Israel that have been documented as facing greater social and health disparities when compared to the majority of the Israeli population (Razon, 2016; Abu-Rabia-Queder, 2017; Daoud et al., 2018) as well as disparities in terms of infrastructure access (Porob et al., 2020). This represents a situation where there are difficulties in acquiring accurate data via clinical surveillance or wastewater-based surveillance, as individuals may not be willing or able to access healthcare services (Voeten et al., 2009; Hermesh et al., 2020). The COVID-19 response in the region has shown geopolitical discrepancy, potentially fostering similar trends of mistrust (Negev et al., 2021). Even within Israel, a recent study among the various Arab ethnic groups found that there is a discrepancy and mistrust in the healthcare system and increased psychological stress regarding the COVID-19 pandemic was observed in certain groups (Lavie et al., 2020; Satran et al., 2021). Therefore, the Bedouin in Israel represent a population where testing for coronavirus could be impacted negatively by numerous factors, including but not limited to healthcare access disparities, information access, and trust in service providers and the government. Simultaneously, WBE is limited in these communities as they are frequently denied access to the wastewater grid, therefore they will not be adequately represented in national surveillance efforts (Porob et al., 2020; Bar-Or et al., 2021a). In this study WBE was used to assess SARS-CoV-2 in wastewater in on-grid, partially off-grid, and fully off-grid Bedouin communities and compare it to clinical surveillance data in order to assess differences in clinical and sewage data, which has utility both in Israel as well as globally in communities with disparities in healthcare access and infrastructure.
Samples were collected from five Bedouin communities (Figure 1), all of which are recognized by the Israeli government but represent different levels of connectivity to the wastewater grid. Hura, Tel Sheva/Tel al-Sabi, and Lakia are fully connected to the wastewater grid, El-Sayed village is partially on the wastewater grid and partially off-grid, and Um Batin is a recognized village with no access to the grid. Sample collection methods that this was undertaken with permission and involvement of the Al-Kaysom, Hura, and Lakia regional councils. Off-grid villages are completely without connection to the wastewater grid (i.e., non-sewered); wastewater (including solids and supernatant) is stored in unlined cesspits. Wastewater leaches into the ground, and cesspits are sealed when full.
Samples for partially-off grid and on-grid municipalities were collected from the Shoket WWTP. This WWTP treats ~15,000 m3 per day, and it uses activated sludge for tertiary water treatment. Shoket services municipalities including Meitar, Carmit, Hura, Lakiya, and the wastewater from the southern West Bank that is first received at the West Bank catchment (WBCF) facility located on the Green Line. The WBCF was constructed by Israel to handle the treated sewage, untreated sewage, and industrial effluent discharged from Hebron and surrounding Palestinian areas into the Hebron stream which crosses the Green Line into Israel. The primary purpose of the catchment facility is to lower the turbidity of the sewage which is extremely high due to the large influx of stone cutting slurry that is very high in calcium carbonate. The facility uses a coagulant polymer and mechanical press to lower the turbidity of the sewage. Once this is done the sewage is sent by pipeline to the Shoket WWTP where it is mixed and treated with local municipal sewage.
Estimated populations and spelling of municipality names in Arabic and Hebrew are in Supplementary Table 1 (Israeli Central Bureau of Statistics, 2019). Samples from Um Batin were collected from an environmental water source (Hebron Stream) via large-volume (100 L) grab sampling methodology. Grab and composite samples were also collected from two upstream sampling points, Meitar, which is upstream of Um Batin, and Hebron Catchment, which is upstream of Meitar) along the Hebron Stream as comparison sites (Figure 1).
Samples from the West Bank Sewage Catchment facility, which represents surface water as well as untreated sewage from off-grid communities gathered at the point where the Hebron Stream crosses the Green Line into Israel, was collected via grab sample and autosampler methodologies and used for protocol development and comparison. Stormwater drains to natural valleys and is only integrated into the wastewater system in the situation of the WBCF. Samples were collected via grab samplers and 18-h composite autosampler methodologies (as described in Yaniv et al. (2021a). The automated sampler pumped 250 mL every half hour between 5 a.m. to 11 p.m. on a Saturday. Upon arrival at the lab, samples were kept at 4°C for up to 2 days until processing them for virus concentration and RNA extraction. Full details on sampling dates and locations are in Supplementary Table 2.
For the grab samples, the following protocol was used. Prior to centrifugation of 50 mL to remove solids, 200 μL of raw sewage was aliquoted for direct extraction and stored at 4°C. The 50 mL aliquot of sewage was spun for 30 min at 4,700 RCF at 4°C over centrifugal devices with a 10 kDa filter size (Pall Corporation, Port Washington, NY, USA and Merck, Darmstadt, Germany) (Ahmed et al., 2020; Gonçalves et al., 2021).
For the composite and surface water samples, the following protocol was used. The samples were concentrated using dialysis filters with 3 to 30 nanometer pore size (recycled dialysis filter, NUFiltration©, Caesarea, Israel) by “dead end” flow. In the case of “dead-end” flow, the applied pressure drives the entire water flow through a membrane filter (similar to the procedure described in Obayomi et al., 2019). The filter was then back-washed with 0.07–0.1 Liter of distilled water. For composite samples, all initial and final volumes are reported in Supplementary Table 3.
For extraction of all samples, 200 μL of filtered samples (raw or concentrated) underwent RNA extraction using the Machery-Nagel NucleoSpin RNA isolation kit (catalog number 740955, Düren, Germany). Five microliter of 20 mg per mL Proteinase K (cas number 70663-4, Sigma Aldrich, St. Louis, Missouri, US) and five μL of MS2 phage at a 106 copies per μL concentration were added to the lysis step. The RNA was eluted with 50 μL of nuclease-free water.
Grab samples were subject to RT-qPCR on the MIC thermocycler (BioMolecular Systems, Upper Coomera, Australia) and composite samples were subject to RT-qPCR on the Applied Biosystems (Thermo Scientific, Waltham, MA, USA) qPCR instrument. N1 and N2 gene targets were quantified on both devices using United States Centers for Disease Control and Prevention primer probe mixtures (catalog number 10006713, IDT, Coralville, IA, USA) and recommended cycling conditions (US Centers for Disease Control Prevention, 2020), and the N3 gene target was quantified with the N3R primer-probe set (Yaniv et al., 2021a). The initial filtration volumes and the final concentrated fraction for each sample were used to calculate the final RNA gene copies/Liter of wastewater. Calibration curves for Thermo Scientific and MIC devices are included in the Supplementary Figure 1. As a part of quality control, we calculated MS2 recovery (Yaniv et al., 2021b) percentage. If the recovery percentage was lower than 5%, a second RNA extraction or sample concentration was performed. Low recovery percentage can imply inhibition presence.
Israeli cases and test data were extracted from publicly available Israeli Ministry of Health databases. Data extracted included daily total case numbers by municipality and daily number of tests administered by municipality, and these data were used to calculate cases per day as well as percent positive tests per day by municipality. Data were extracted from relevant date ranges to provide concordant data with sewage sampling dates by municipality. Due to the discrepancy in size among the studied municipalities, case and test data were normalized to population size. All parameters use in this study can be found in the Appendix (cases and tests per 10,000 people). Statistics including frequency comparison (via chi squared and Fisher's exact tests) and means comparisons (via Mann Whitney and Kruskal Wallis tests) regarding the comparison of composite to grab samples) were carried out in R (Version 4.0.2), specifically the psych package. Maps were generated using ArcGIS Pro (Version: 2.9.5, ESRI, Redlands CA, USA).
Figure 2 presents copy number overlaid with case data for all four sites sampled in the study period (August 2020 to January 2021). Regarding epidemiology parameters as measured by the Ministry of Health (active cases, number of tests, and percentage of tests found to be positive), some fluctuations and patterns can be observed. In Hura, a major virus activity peak was evident from mid-November to mid-December 2020. In Lakia, the main peak was observed from late August throughout September 2020 and a minor peak at the end of 2020 until the beginning of 2021. In Tel Sheva/Tel al-Sabi, more modest case rates were reported, with higher numbers of cases during December 2020 and January 2021. In El-Sayed, no large peaks were observed as the total test number per day remained very low (under 10 tests per day). All peaks correspond to peaks in the total number of tests, suggesting that asymptomatic or subclinical cases may have gone undiagnosed outside of these waves of infection. SARS-CoV-2 was detected in Hura and Lakia during the first sampling efforts in August of 2020, however at this timepoint only Lakia had a wave of testing and subsequent reported cases, indicating the possibility of silent, or unreported, transmission. In Hura, Lakia, and Tel Sheva/Tel al-Sabi, spikes in observed SARS-CoV-2 N gene were observed 2–3 weeks before spikes in observed cases. In El-Sayed, spikes in observed SARS-CoV-2 N gene were inconsistently associated with observed cases. Given that this community is only partially on-grid, it is possible that at some points positive sewage was not part of the sampled catchment area. Overall, the highest observed copy numbers per sample were observed in the larger communities (Supplementary Figure 2). When observing trends by, the number of N2 gene copies/L was generally higher in the larger communities. This trend was not as strong in regards to N1 gene copies/L (Supplementary Figure 2).
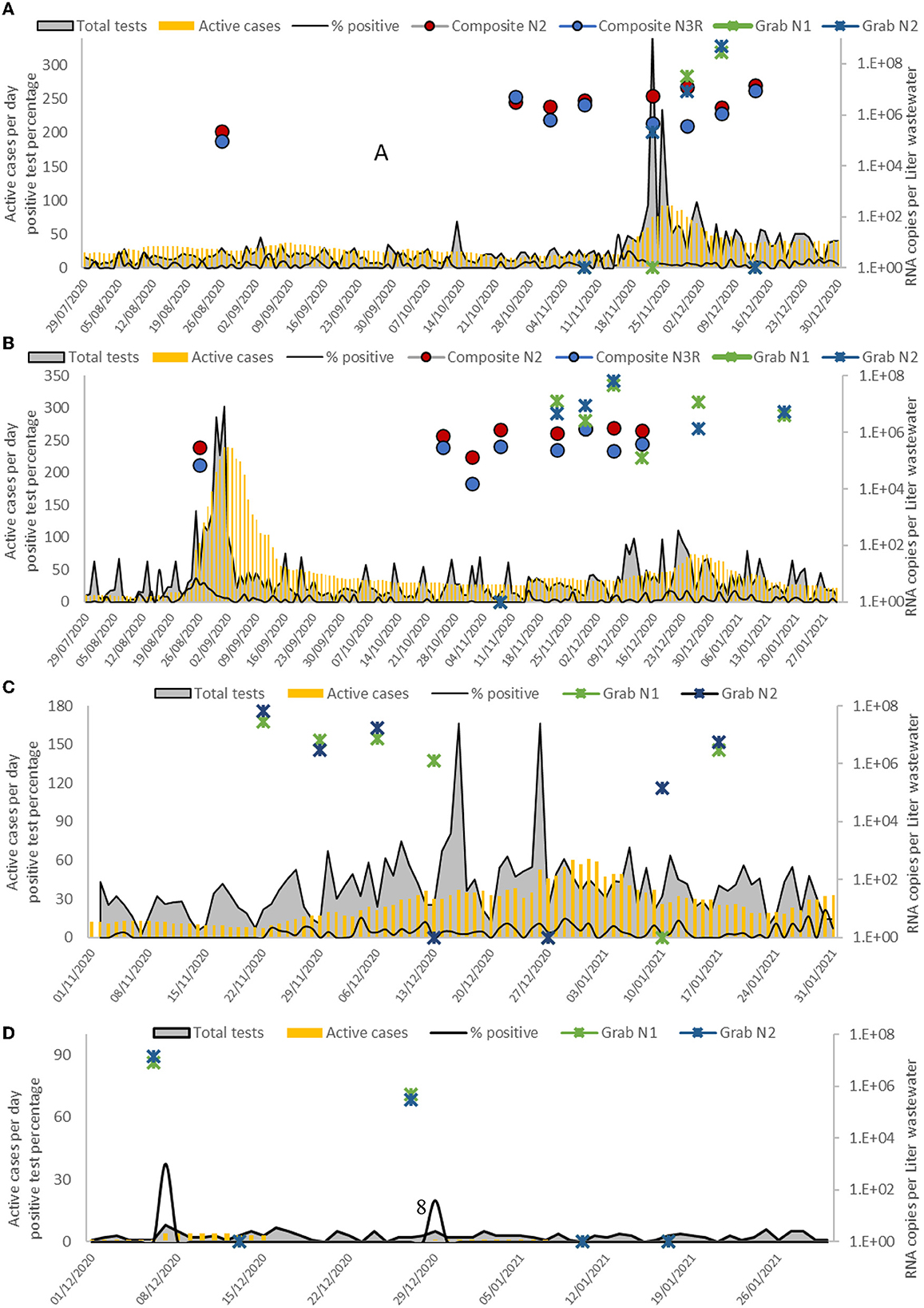
Figure 2. SARS-CoV-2 detection in four sampling locations- Hura (A), Lakia (B), Tel-Sheva/Tel al-Sabi (C), and El-Sayed (D) during the study time period (X-axis). Right Y axis presents RNA copy numbers per Liter of wastewater per RT-qPCR results. A Circle icon present composite sampling, blue for N3R detection set and red for N2 detection set. Asterisk icons indicate grab sampling, green for N1 detection set and blue for N2 detection set. In a case of Not Detected result, a value of one appears on the graph. The left Y-axis presents current confirmed cases per 10,000 people (yellow bars), positive test percentage (black line) and total COVID-19 tests performed per 10,000 people (in gray).
A total of 14 pairs of grab and composite samples were available for analysis. Statistically the differences were not significant for N1 (p = 0.6) and N3 (p = 0.3). N2 (p = 0.06) was approaching significance. Copy numbers were comparable for N1 (Composite: Mean: 5.1 × 106 Range: 1.4 × 106–9.3 × 106, Grab: Mean: 5.1 × 106, Range: 5.8 × 105–1.7 × 107) and N3 (Composite: Mean: 6.8 × 105 Range: 2.3 × 105–6.5 × 105, Grab: Mean: 2.7 × 106 Range: 1.9 × 105–1.3 × 107), and slightly higher for grab sample data for N2 (Composite: Mean: 3.8 × 106 Range: 4.0 × 106–9.6 × 106, Grab: Mean: 6.5 × 106, Range: 2.2 × 106–1.3 × 107) (Figure 3, Supplementary Table 4). Of the composite samples, 100% were detected with at least one gene target. Of the grab samples, four samples (28.6%) were negative for all gene targets. When frequencies were compared there was not a statistically significant difference in overall detection frequency for the N1 and N2 gene targets (p-value = 0.3 and 0.2, respectively), however the difference was statistically significant for N3 (p = 0.004).
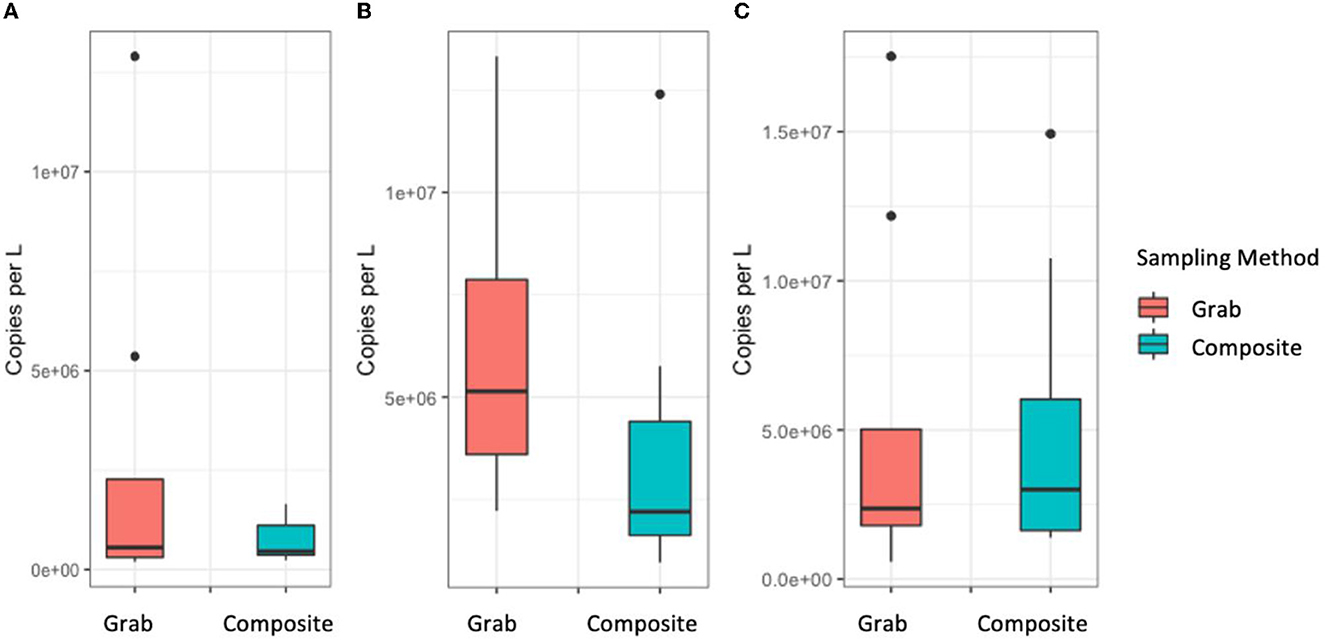
Figure 3. Comparison of copy number per L recovered from grab samples and 18-h composite samples from an autosampler for the N1 (A), N2 (B), and N3 (C) gene targets.
Environmental water samples (Grab samples) were collected from three points along the Hebron River, a transboundary waterway originating near Hebron, West Bank, Palestinian Territories which passes through numerous Israeli towns and villages after crossing the Green Line (Figure 1). Um Batin sewage discharges into the Hebron River, and it was observed that sampling points near Um Batin were positive for SARS-CoV-2 RNA at one timepoint in October 2020 and one in January 2021, but not at an additional timepoint in January 2021 (Supplementary Table 5). During and in the week preceding the October timepoint there were no recorded active cases in Um Batin, and during the entire January sampling timeframe there were 1–2 recorded active cases in Um Batin (Data not shown). It should be noted that in timepoints where upstream (Meitar) points are positive for SARS-CoV-2, Um Batin is negative, suggesting that dilution effect and/or surface water treatment at the Shoket WWTP (Figure 1) reduced the virus levels below detection limit.
In this study we investigated SARS-CoV-2 in sewage-impacted environmental waters as well as utilizing WBE in on-grid and partially off-grid communities. It is known that the communities sampled experience disparities regarding healthcare access and utilization, ergo it should be noted that peaks in SARS-CoV-2 quantification are not completely aligned with known cases in these communities. However, it should be also noted that testing rates were overall low and case rates were closely associated with testing rates. While large, on-grid municipalities (i.e., Hura) had more robust data, grab samples and composite samples from smaller and partially-off-grid municipalities were able to detect SARS-CoV-2. Copy numbers/L were in line with findings from other municipalities in Israel in other studies (Yaniv et al., 2021a,b). Furthermore, large-volume grab samples of surface water impacted by sewage from fully off-grid communities were able to detect SARS-CoV-2.
Throughout the COVID-19 outbreak and including the study period described here, Yaniv et al. (2021b) monitored wastewater samples from Beer-Sheva and several other non-Bedouin locations in Israel (August 2020–October 2021) for the presence of SARS-CoV-2 virus using RT-qPCR including CDC's standard detection set. They documented the SARS-CoV-2 RNA levels in wastewater and monitored these values together with the corresponding reported active cases and percent vaccination in the city populations. It was evident that an increase in detected SARS-CoV-2 RNA levels in the city wastewater appeared just prior to every one of the three peaks in reported active cases. Thus, the appearance in the city's wastewater foresaw the increase in reported clinical cases of COVID-19 (Yaniv et al., 2021b). Given the findings of Yaniv et al. (2021a,b) as well as other studies that have noted strong correlations between epidemiological data and sewage data, the lack of alignment of these data in communities with access disparities is noteworthy (Gonzalez et al., 2020; Ahmed et al., 2021; Bar-Or et al., 2021b). The findings of this study support the hypothesis regarding “under the radar” morbidity and emphasize the importance of using WBE to learn about morbidity levels within the community (Hamouda et al., 2021; Karthikeyan et al., 2021; Lee et al., 2022).
Composite samples identified SARS-CoV-2 RNA with greater frequency than grab samples, however when compared statistically the detection frequency was not significantly different. While composite sampling is undeniably the preferred standard for wastewater surveillance, it can be logistically and economically difficult to set up in low-income communities, and even if setup is accomplished it may not be sustainable (D'Aoust et al., 2021). Composite sampling raises questions of sustainability in terms of equipment functionality over time, maintenance, access and affordability of replacement parts or units, and costs and access to electricity for running the systems. One question of debate is the relevance of grab vs. composite samples when surveying for SARS-CoV-2 at various catchment scales (George et al., 2022). George et al. (2022) demonstrated that at the high-flow site, grab samples were reasonably comparable to composite samples. However, as the flow rates decreased, the percentage of false-negative grab samples increased and the SARS-CoV-2 concentrations of grab samples varied by up to 1–2 orders of magnitude compared to their respective composite sample concentrations (George et al., 2022). They further conclude that composite sampling is superior to grab sampling, especially as flow decreases. Therefore, for municipalities seeking to augment surveillance for SARS-CoV-2 but without the resources to install and maintain autosamplers, grab sampling is an acceptable method for WBE however flow rates must be considered and areas of higher-flow should be prioritized.
When considering these issues of flow as well as grid access, it is important to consider the implications of these findings for populations not fully connected to the wastewater grid. In this study, we were able to access a WWTP that served a community partially connected to the grid as well as a surface water site known to receive untreated sewage from an off-grid community. This is in line with a recent strategy employed by researchers in Bangladesh, who employed a methodology for off-grid settings that focused on “hotspot” or “nodal” sampling via surface water sites or community gathering places. Using this modality, they were able to effectively track COVID-19 dynamics in connected and unconnected populations (Jakariya et al., 2021). A study from Ecuador investigating rivers impacted by the sewage of cities noted a viral copy load similar to that of raw WWTP influent in other countries and a study of rivers in Brazil also reported observing SARS-CoV-2 loads tracked with population-level case data (Aguiar-Oliveira et al., 2020; Guerrero-Latorre et al., 2020). The level and detection of SARS-CoV-2 genetic material in our surface water sample was low, however considering that our sampling points along the Hebron Stream represented the sewage of a population of under 5,000, the low level of viral material is reasonable. A study in Japan noted that river water receiving fully treated wastewater did not test positive for SARS-CoV-2 RNA (Haramoto et al., 2020), whereas an Italian study investigating river water receiving combined sewage overflow and improperly treated wastewater did identify SARS-CoV-2 genetic material in surface water (Rimoldi et al., 2020). As such, future studies investigating regions with combined on-grid and off-grid populations should consider the presence of improperly treated wastewater in the entire surface water catchment area.
This study was limited in terms of numbers of sampling sites as well as in being able to sample in all locations at all timepoints. It represents a proof of concept of what may be accomplished in a pandemic scenario in under-serviced communities. Such studies may be difficult, in particular during periods of country-wide lockdowns requiring researchers to obtain essential worker permits and due to delays in obtaining reagents due to supply chain disruptions. It is recommended that future WBE research be carried out in non-pandemic scenarios (i.e., with other diseases such as Polio, Brucellosis, or seasonal influenza) with more comprehensive sampling to fully understand how sewer grid access, healthcare access, and health inequality interface in regards to the utility of WBE (Heijnen and Medema, 2011). This study also focused on Israel's southern Bedouin population, and further research even within Israel itself is needed among various rural off-grid populations experiencing infrastructure access and healthcare inequality as well as healthcare system mistrust to establish broad applicability.
Numerous studies have emerged detailing the utility of WBE regarding SARS-CoV-2 surveillance as well as surveillance for other pathogenic threats such as antimicrobial resistance (Asghar et al., 2014; Thakali et al., 2022; Tiwari et al., 2022a,b). These studies have primarily been carried out in communities with greater resources primarily representing on-grid communities, however, given the resource limitations and supply chain disruptions observed during the COVID-19 pandemic, it is critical to ensure that all communities can utilize surveillance methods like WBE. The findings of this study suggest that sampling, with grab sample or composite sample methods, can identify SARS-CoV-2 virus in wastewater from communities on the grid, partially on the grid, and off-grid communities. Furthermore, given that the communities of focus in this study experience health inequalities so case data may not represent the full picture of infection in the community. These findings are also useful for SARS-CoV-2 surveillance for communities without infrastructure or equitable access to healthcare resources.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
KY and HC: data curation, formal analysis, investigation, methodology, supervision, visualization, writing—original draft, and writing—review and editing. FM: data curation, investigation, project administration, and writing—review and editing. MS, SL, KR, and CH: data curation, investigation, and writing—review and editing. IS: data curation, formal analysis, methodology, investigation, and writing—review and editing. ND: conceptualization, data curation, and writing—review and editing. JM-G and AK: conceptualization, funding acquisition, methodology, project administration, resources, supervision, writing—original draft, and writing—review and editing. CL: conceptualization, funding acquisition, project administration, supervision, and writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was funded in part by the US Agency for International Development Middle East Regional Cooperation project: Greywater Reuse for Smallholder Agricultural Development in Off-Grid Communities in Israel, the Palestinian Authority and Jordan (project number: M35-009, award number: SIS70018GR35009). We would like to acknowledge additional funding from Ben Gurion University, The Corona Challenge COVID-19, the Israeli Ministry of Health, and the Jewish National Fund USA. HC received postdoctoral funding from Fulbright Israel and the Zuckerman Institute.
We would like to thank Kando for helping with sewage composite sampling. We gratefully acknowledge Esti Kramarsky-Winter's assistance for comments and scientific editing of the manuscript and Eyas ElAtawneh for preparing the maps. Thank you to the Neve-Midbar water corporation, Al-Kaysom regional council, Hura regional council, Lakia regional council, Shoket WWTP personnel, and the Hebron catchment facility operators, who all facilitated access to sampling sites.
AK, ND, and JM-G serve on the Kando Scientific Advisory Board. HC received funding from the Zuckerman STEM Foundation and Fulbright Israel. This research was funded by the US Agency for International Development Middle East Regional Cooperation, Ben Gurion University, the Israeli Ministry of Health, and the Jewish National Fund USA.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2023.1136066/full#supplementary-material
Abu-Rabia-Queder, S. (2017). The paradox of professional marginality among Arab-Bedouin women. Sociology 51, 1084–1100. doi: 10.1177/0038038516641621
Aguiar-Oliveira, M. D. L., Campos, A., R. Matos, A., Rigotto, C., Sotero-Martins, A., Teixeira, P. F., et al. (2020). Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: a valuable tool for COVID-19 surveillance—a brief review. Int. J. Environ. Res. Public Health 17, 9251. doi: 10.3390/ijerph17249251
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J. W., et al. (2020). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728, 138764. doi: 10.1016/j.scitotenv.2020.138764
Ahmed, W., Tscharke, B., Bertsch, P. M., Bibby, K., Bivins, A., Choi, P., et al. (2021). SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 761, 144216. doi: 10.1016/j.scitotenv.2020.144216
Asghar, H., Diop, O. M., Weldegebriel, G., Malik, F., Shetty, S., El Bassioni, L., et al. (2014). Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 210 (Suppl_1), S294–S303. doi: 10.1093/infdis/jiu384
Bar-Or, I., Weil, M., Indenbaum, V., Bucris, E., Bar-Ilan, D., Elul, M., et al. (2021a). Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 789, 148002. doi: 10.1016/j.scitotenv.2021.148002
Bar-Or, I., Yaniv, K., Shagan, M., Ozer, E., Weil, M., Indenbaum, V., et al. (2021b). Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. Front. Public Health. 9, 561710. doi: 10.3389/fpubh.2021.561710
Brouwer, A. F., Eisenberg, J. N., Pomeroy, C. D., Shulman, L. M., Hindiyeh, M., Manor, Y., et al. (2018). Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Nat. Acad. Sci. U. S. A. 115, E10625–E10633. doi: 10.1073/pnas.1808798115
Daoud, N., Gao, M., Osman, A., and Muntaner, C. (2018). Interpersonal and institutional ethnic discrimination, and mental health in a random sample of Palestinian minority men smokers in Israel. Soc. Psychiatry Psychiatr. Epidemiol. 53, 1111–1122. doi: 10.1007/s00127-018-1531-0
D'Aoust, P. M., Towhid, S. T., Mercier, É., Hegazy, N., Tian, X., Bhatnagar, K., et al. (2021). COVID-19 wastewater surveillance in rural communities: comparison of lagoon and pumping station samples. Sci. Total Environ. 801, 149618. doi: 10.1016/j.scitotenv.2021.149618
Driver, E. M., Bowes, D. A., Halden, R. U., and Conroy-Ben, O. (2022). Implementing wastewater monitoring on American Indian reservations to assess community health indicators. Sci. Total Environ. 823, 153882. doi: 10.1016/j.scitotenv.2022.153882
Elkayam, R., Aharoni, A., Vaizel-Ohayon, D., Sued, O., Katz, Y., Negev, I., et al. (2018). Viral and microbial pathogens, indicator microorganisms, microbial source tracking indicators, and antibiotic resistance genes in a confined managed effluent recharge system. J. Environ. Eng. 144, 05017011. doi: 10.1061/(ASCE)EE.1943-7870.0001334
Fontanet, A., Autran, B., Lina, B., Kieny, M. P., Karim, S. S. A., and Sridhar, D. (2021). SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 397, 952–954. doi: 10.1016/S0140-6736(21)00370-6
George, A. D., Kaya, D., Layton, B. A., Bailey, K., Mansell, S., Kelly, C., et al. (2022). Impact of sampling type, frequency, and scale of the collection system on SARS-CoV-2 quantification fidelity. Environ. Sci. Technol. Lett. 9, 160–165. doi: 10.1101/2021.07.07.21260158
Gonçalves, J., Koritnik, T., Mioč, V., Trkov, M., Bolješič, M., Berginc, N., et al. (2021). Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 755, 143226. doi: 10.1016/j.scitotenv.2020.143226
Gonzalez, R., Curtis, K., Bivins, A., Bibby, K., Weir, M. H., Yetka, K., et al. (2020). COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 186, 116296. doi: 10.1016/j.watres.2020.116296
Guerrero-Latorre, L., Ballesteros, I., Villacrés-Granda, I., Granda, M. G., Freire-Paspuel, B., and Ríos-Touma, B. (2020). SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 743, 140832. doi: 10.1016/j.scitotenv.2020.140832
Hamouda, M., Mustafa, F., Maraqa, M., Rizvi, T., and Hassan, A. A. (2021). Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 759, 143493. doi: 10.1016/j.scitotenv.2020.143493
Haramoto, E., Malla, B., Thakali, O., and Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 737, 140405. doi: 10.1016/j.scitotenv.2020.140405
Hart, O. E., and Halden, R. U. (2020). Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 730, 138875. doi: 10.1016/j.scitotenv.2020.138875
Heijnen, L., and Medema, G. (2011). Surveillance of Influenza A and the pandemic influenza A (H1N1) 2009 in sewage and surface water in the Netherlands. J. Water Health 9, 434–442. doi: 10.2166/wh.2011.019
Hemalatha, M., Kiran, U., Kuncha, S. K., Kopperi, H., Gokulan, C. G., Mohan, S. V., et al. (2021). Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: comprehensive study. Sci. Total Environ. 768, 144704. doi: 10.1016/j.scitotenv.2020.144704
Hermesh, B., Rosenthal, A., and Davidovitch, N. (2020). The cycle of distrust in health policy and behavior: lessons learned from the Negev Bedouin. PLoS ONE 15, e0237734. doi: 10.1371/journal.pone.0237734
Iglesias, N. G., Gebhard, L. G., Carballeda, J. M., Aiello, I., Recalde, E., Terny, G., et al. (2021). SARS-CoV-2 surveillance in untreated wastewater: detection of viral RNA in a low-resource community in Buenos Aires, Argentina. Rev Panam Salud Pública 45, e137. doi: 10.26633/RPSP.2021.137
Islam, M. A., Rahman, M. A., Jakariya, M., Bahadur, N. M., Hossen, F., Mukharjee, S. K., et al. (2023). A 30-day follow-up study on the prevalence of SARS-CoV-2 genetic markers in wastewater from the residence of COVID-19 patient and comparison with clinical positivity. Sci. Total Environ. 858, 159350. doi: 10.1016/j.scitotenv.2022.159350
Israeli Central Bureau of Statistics. (2019). Localities in Israel—Files of Localities. Settlements in Israel—Files of Settlements. Available online at: https://www.cbs.gov.il/he/publications/Pages/2019/ .aspx
.aspx
Jakariya, M., Ahmed, F., Islam, M. A., Ahmed, T., Marzan, A. A., Hossain, M., et al. (2021). Wastewater based surveillance system to detect SARS-CoV-2 genetic material for countries with on-site sanitation facilities: an experience from Bangladesh. MedRxiv. doi: 10.1101/2021.07.30.21261347
Karthikeyan, S., Nguyen, A., McDonald, D., Zong, Y., Ronquillo, N., Ren, J., et al. (2021). Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. Msystems 6, e00793-21. doi: 10.1128/mSystems.00793-21
Lavie, E., Elran, M., Sawaed, K., Abu Mokh, M., and Dallashi, M. (2020). Israel's Arab Society and the Coronavirus Challenge (No. 1288; INSS Insight). Institute for National Security Studies (INSS). Available online at: https://www.inss.org.il/publication/coronavirus-and-the-israeli-arabs/ (accessed March 01, 2021).
Lee, W. L., Gu, X., Armas, F., Leifels, M., Wu, F., Chandra, F., et al. (2022). Monitoring human arboviral diseases through wastewater surveillance: challenges, progress and future opportunities. Water Res. 223, 118904. doi: 10.1016/j.watres.2022.118904
Lorenzo, M., and Picó, Y. (2019). Wastewater-based epidemiology: current status and future prospects. Curr. Opin. Environ. Sci. Health 9, 77–84. doi: 10.1016/j.coesh.2019.05.007
Manor, Y., Shulman, L. M., Kaliner, E., Hindiyeh, M., Ram, D., Sofer, D., et al. (2014). Intensified environmental surveillance supporting the response to wild poliovirus type 1 silent circulation in Israel, 2013. Eurosurveillance 19, 20708. doi: 10.2807/1560-7917.ES2014.19.7.20708
Naughton, C. C., Roman, F. A. Jr, Alvarado, A. G. F., Tariqi, A. Q., Deeming, M. A., Kadonsky, K. F., et al. (2023). Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. FEMS Microbes 4, xtad003. doi: 10.1093/femsmc/xtad003
Negev, M., Dahdal, Y., Khreis, H., Hochman, A., Shaheen, M., Jaghbir, M. T. A., et al. (2021). Regional lessons from the COVID-19 outbreak in the Middle East: from infectious diseases to climate change adaptation. Sci. Total Environ. 768, 144434. doi: 10.1016/j.scitotenv.2020.144434
Nemudryi, A., Nemudraia, A., Wiegand, T., Surya, K., Buyukyoruk, M., Cicha, C., et al. (2020). Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 1, 100098. doi: 10.1016/j.xcrm.2020.100098
Obayomi, O., Bernstein, N., Edelstein, M., Vonshak, A., Ghazayarn, L., Ben-Hur, M., et al. (2019). Importance of soil texture to the fate of pathogens introduced by irrigation with treated wastewater. Sci. Total Environ. 653, 886–896. doi: 10.1016/j.scitotenv.2018.10.378
Polo, D., Quintela-Baluja, M., Corbishley, A., Jones, D. L., Singer, A. C., Graham, D. W., et al. (2020). Making waves: wastewater-based epidemiology for SARS-CoV-2–Developing robust approaches for surveillance and prediction is harder than it looks. Water Res. 186, 116404. doi: 10.1016/j.watres.2020.116404
Porob, S., Craddock, H. A., Motro, Y., Sagi, O., Gdalevich, M., Ezery, Z., et al. (2020). Quantification and characterization of antimicrobial resistance in greywater discharged to the environment. Water 12, 1460. doi: 10.3390/w12051460
Pruden, A., Vikesland, P. J., Davis, B. C., and de Roda Husman, A. M. (2021). Seizing the moment: now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 64, 91–99. doi: 10.1016/j.mib.2021.09.013
Razon, N. (2016). Entangled bodies: jews, bedouins, and the making of the secular Israeli. Med. Anthropol. 35, 291–304. doi: 10.1080/01459740.2016.1138950
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., et al. (2020). Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 744, 140911. doi: 10.1016/j.scitotenv.2020.140911
Satran, C., Ali-Saleh, O., Mashiach-Eizenberg, M., and Bord, S. (2021). Stress and perceived discrimination among the Arab population in Israel: the mediation role of the perceived COVID-19 threat and trust in the healthcare system. Ethn. Health 27, 1377–94. doi: 10.1080/13557858.2021.1899139
Scott, L. C., Aubee, A., Babahaji, L., Vigil, K., Tims, S., and Aw, T. G. (2021). Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 200, 111374. doi: 10.1016/j.envres.2021.111374
Shulman, L. M., Manor, Y., Hindiyeh, M., Sofer, D., and Mendelson, E. (2016). “Molecular characterization of polio from environmental samples: ISSP, The Israeli Sewage Surveillance Protocol,” in Poliovirus (New York, NY: Humana Press), 55–107.
Street, R., Malema, S., Mahlangeni, N., and Mathee, A. (2020). Wastewater surveillance for Covid-19: an African perspective. Sci. Total Environ. 743, 140719. doi: 10.1016/j.scitotenv.2020.140719
Thakali, O., Raya, S., Malla, B., Tandukar, S., Tiwari, A., Sherchan, S. P., et al. (2022). Pilot study on wastewater surveillance of dengue virus RNA: lessons, challenges, and implications for future research. Environ. Challenges 9, 100614. doi: 10.1016/j.envc.2022.100614
Tiwari, A., Adhikari, S., Kaya, D., Islam, M. A., Malla, B., Sherchan, S. P., et al. (2022a). Monkeypox outbreak: Wastewater and environmental surveillance perspective. Sci. Total Environ 856(Pt 2), 159166. doi: 10.1016/j.scitotenv.2022.159166
Tiwari, A., Kurittu, P., Al-Mustapha, A. I., Heljanko, V., Johansson, V., Thakali, O., et al. (2022b). Wastewater surveillance of antibiotic-resistant bacterial pathogens: a systematic review. Front. Microbiol. 13, 977106. doi: 10.3389/fmicb.2022.977106
US Centers for Disease Control Prevention (2020). CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel, For Emergency Use Only, Instructions for Use. Available online at: https://stacks.cdc.gov/view/cdc/85028/cdc_85028_DS1.pdf
Voeten, H. A. C. M., de Zwart, O., Veldhuijzen, I. K., Yuen, C., Jiang, X., Elam, G., et al. (2009). Sources of Information and Health Beliefs Related to SARS and Avian Influenza among Chinese Communities in the United Kingdom and The Netherlands, compared to the general population in these countries. Int. J. Behav. Med. 16, 49–57. doi: 10.1007/s12529-008-9006-4
Wurtzer, S., Waldman, P., Ferrier-Rembert, A., Frenois-Veyrat, G., Mouchel, J. M., Boni, M., et al. (2021). Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 198, 117183. doi: 10.1016/j.watres.2021.117183
Yaniv, K., Ozer, E., Lewis, Y., and Kushmaro, A. (2021a). RT-qPCR assays for SARS-CoV-2 variants of concern in wastewater reveals compromised vaccination-induced immunity. Water Res. 207, 117808. doi: 10.1016/j.watres.2021.117808
Yaniv, K., Ozer, E., Shagan, M., Paitan, Y., Granek, R., and Kushmaro, A. (2022). Managing an evolving pandemic: Cryptic circulation of the Delta variant during the Omicron rise. Sci. Total Environ. 836, 155599. doi: 10.1016/j.scitotenv.2022.155599
Yaniv, K., Shagan, M., Lewis, Y. E., Kramarsky-Winter, E., Weil, M., Indenbaum, V., et al. (2021b). City-level SARS-CoV-2 sewage surveillance. Chemosphere 283, 131194. doi: 10.1016/j.chemosphere.2021.131194
Keywords: wastewater monitoring, off-grid, WBE, Bedouin communities, SARS-CoV-2
Citation: Yaniv K, Craddock HA, Mahameed F, Shagan M, Salah I, Lakkakula S, Resnick K, Haber C, Davidovitch N, Moran-Gilad J, Kushmaro A and Lipchin C (2023) Wastewater monitoring of SARS-CoV-2 in on-grid, partially and fully off-grid Bedouin communities in Southern Israel. Front. Water 5:1136066. doi: 10.3389/frwa.2023.1136066
Received: 02 January 2023; Accepted: 21 April 2023;
Published: 22 May 2023.
Edited by:
Mohammad Hoque, University of Portsmouth, United KingdomReviewed by:
Jakariya Md., North South University, BangladeshCopyright © 2023 Yaniv, Craddock, Mahameed, Shagan, Salah, Lakkakula, Resnick, Haber, Davidovitch, Moran-Gilad, Kushmaro and Lipchin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clive Lipchin, Y2xpdmVhcmF2YUBnbWFpbC5jb20=; Ariel Kushmaro, YXJpZWxrdXNAYmd1LmFjLmls
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.