- 1Department of Civil and Environmental Engineering, University of New Hampshire, Durham, NH, United States
- 2Application and Development Department, New England Biolabs, Ipswich, MA, United States
Municipal sewage carries SARS-CoV-2 viruses shed in the human stool by infected individuals to wastewater treatment plants (WWTPs). It is well-established that increasing prevalence of COVID-19 in a community increases the viral load in its WWTPs. Despite the fact that wastewater treatment facilities serve a critical role in protecting downstream human and environmental health through removal or inactivation of the virus, little is known about the fate of the virus along the treatment train. To assess the efficacy of differing WWTP size and treatment processes in viral RNA removal we quantified two SARS-CoV-2 nucleocapsid (N) biomarkers (N1 and N2) in both liquid and solids phases for multiple treatment train locations from seven coastal New England WWTPs. SARS-CoV-2 biomarkers were commonly detected in the influent, primary treated, and sludge samples (returned activated sludge, waste activated sludge, and digested sludge), and not detected after secondary clarification processes or disinfection. Solid fractions had 470 to 3,700-fold higher concentrations of viral biomarkers than liquid fractions, suggesting considerably higher affinity of the virus for the solid phase. Our findings indicate that a variety of wastewater treatment designs are efficient at achieving high removal of SARS CoV-2 from effluent; however, quantifiable viral RNA was commonly detected in wastewater solids at various points in the facility. This study supports the important role municipal wastewater treatment facilities serve in reducing the discharge of SARS-CoV-2 viral fragments to the environment and highlights the need to better understand the fate of this virus in wastewater solids.
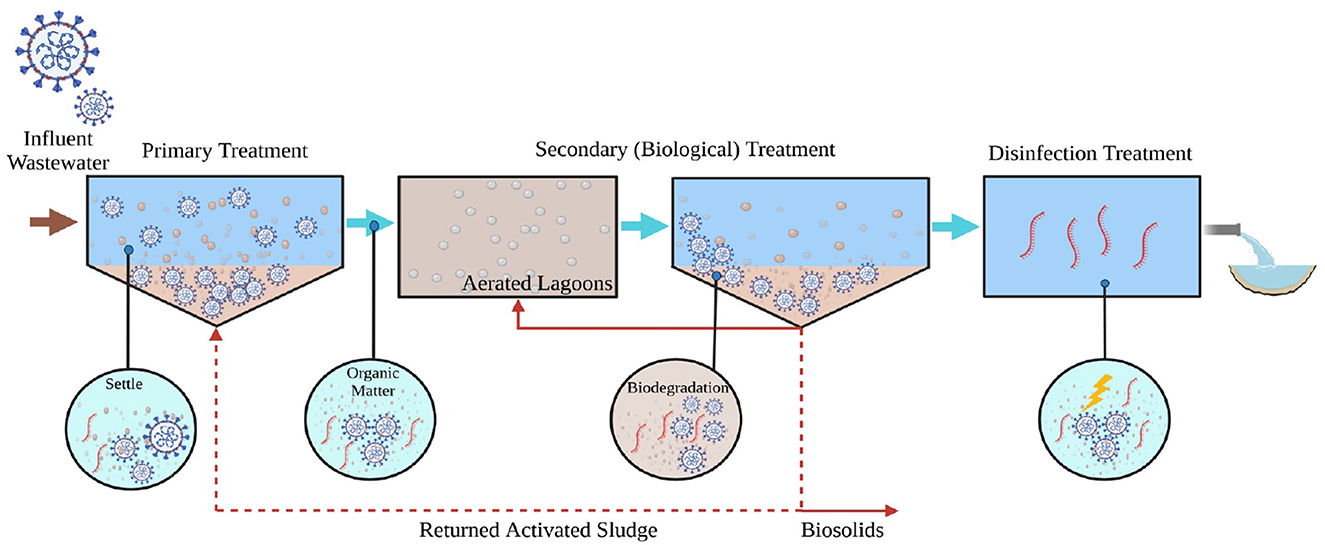
Graphical Abstract. SARS-CoV-2 viral RNA that enters municipal wastewater treatment facilities is efficiently removed across a range of facility sizes and treatment trains.
1. Introduction
For more than a decade, wastewater surveillance has been used to track chemical markers of human activity, such as illicit drugs, pharmaceuticals, tobacco and alcohol (Causanilles et al., 2017; Choi et al., 2018; Mercan et al., 2019; Estévez-Danta et al., 2022), as well as pathogens including enteric viruses (Bisseux et al., 2018), Poliovirus (Hovi et al., 2012), Hepatitis A Virus and Norovirus (Hellmér et al., 2014). Between January and March 2020, the early detection of the new human coronavirus (SARS-CoV-2) in the stool of confirmed COVID-19 patients (Wu Y. et al., 2020) and in sewage samples (Medema et al., 2020) suggested wastewater surveillance could also be useful in monitoring the spread of COVID-19. Since that time, SARS-CoV-2 has been found in stool samples of both symptomatic and asymptomatic individuals (Amirian, 2020; Mizumoto et al., 2020; Nishiura et al., 2020) and in community wastewater systems across the globe (Ahmed et al., 2020a). It is clear that increasing prevalence of COVID-19 in the population increases the viral load in community wastewater treatment plants (WWTPs) (Amoah et al., 2020; Bogler et al., 2020; Paul et al., 2021), making monitoring of SARS-CoV-2 biomarkers in wastewater systems a useful public health tool. Therefore, wastewater surveillance is considered a promising approach for the early detection of outbreaks and to monitor the spread of SARS-CoV-2 within the catchment areas of wastewater treatment plants (Bisseux et al., 2018; Gonzalez et al., 2020; Graham et al., 2020; Xagoraraki and O'Brien, 2020; Gonçalves et al., 2021).
Wastewater treatment personnel may come in contact with wastewater media (liquids, solids, aerosols) during sampling or system maintenance; therefore, there was early concern of SARS-CoV-2 exposure to personnel at treatment facilities (Ghernaout and Elboughdiri, 2020; Oliver et al., 2020; Shutler et al., 2020). However, several studies have shown the risk of COVID-19 transmission during wastewater treatment to be limited (Arora et al., 2020; Kitajima et al., 2020). Nevertheless, only a few studies have assessed the fate of SARS-CoV-2 in wastewater systems including their discharge to freshwater bodies (Kumar et al., 2021; Mohan et al., 2021; Westhaus et al., 2021; Wurtzer et al., 2021). Bivins et al., (Bivins et al., 2020) documented 90% reduction of SARS-CoV-2 viability after only 1.5 days in wastewater, which is longer than the travel time in a sewage gathering network, and considerably shorter than the typical hydraulic residence time during treatment (Xing et al., 2021). Recent studies have shown that SARS-CoV-2 genetic material was detected in hospital wastewater (Gonçalves et al., 2021; Pourakbar et al., 2021), WWTP influent (Sherchan et al., 2020; Hata et al., 2021) and effluent (Nasseri et al., 2021; Westhaus et al., 2021), as well as in water bodies receiving treated wastewater (Naddeo and Liu, 2020). This indicates a potential downstream human and environmental health risk via the fecal-oral and fecal- aerosol infection routes (Lewis, 2020; Gholipour et al., 2021). For instance, Pourakbar and coauthors (Pourakbar et al., 2021) reported the presence of SARS-CoV-2 RNA in aerosols released during aeration within secondary treatment reactors. Altogether, this suggests SARS-CoV-2 may be variably removed at wastewater treatment facilities.
Like other enteric pathogens present in human sewage, the fate of SARS-CoV-2 in wastewater treatment facilities is likely to be influenced by facility design and operational factors. Recent studies have suggested that the efficiency of engineered facilities in eliminating enteric virus such as SARS-CoV-2 or neutralizing its infectivity depends on population infection rate, the treatment facility design (Wigginton et al., 2015), the disinfection approach (Arslan et al., 2020; Bogler et al., 2020) and environmental factors (inc. temperature, pH, organic matter, oxidizing agents and presence of antagonistic bacteria) (Gundy et al., 2009). To date, only a few studies have assessed SARS-CoV-2 biomarker removal during the key stages of a wastewater treatment process (Haramoto et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). Furthermore, there is limited data available regarding the removal or decay of SARS-CoV-2 present in sludge after treatment (Kocamemi et al., 2020; Abu Ali et al., 2021; Balboa et al., 2021; Bhattarai et al., 2021; Serra-Compte et al., 2021; Westhaus et al., 2021). These studies suggest that viral RNA is mostly diverted to the sludge, and that RNA degradation may also contribute to their absence after secondary treatment. The majority of existing studies are focused on the comparison of SARS-CoV-2 genetic material loads in the liquid phase, and very little attention has been paid to their fate in the solid phase of wastewater (Kocamemi et al., 2020; Peccia et al., 2020). It is known that enteric viruses such as SARS-CoV-2 can adsorb to solid and/or colloidal particles due to the presence of a lipid bilayer surrounding the protein capsid, via electrostatic and hydrophobic interactions (Arraj et al., 2005; Verbyla and Mihelcic, 2015). Balboa et al. (2021) also found SARS-CoV-2 genetic material to have high affinity to primary sludge and sludge thickened solids.
Here, we set out to document how SARS-CoV-2 genetic material moves through seven coastal New England wastewater treatment facilities. Our three objectives were: (1) to track changes in SARS-CoV-2 RNA biomarkers along the treatment train and assess viral RNA removal before discharge; (2) to evaluate the fate of SARS-CoV-2 within the facility, specifically its partitioning between the liquid and solid phases; and (3) to compare the log removal and adsorption–desorption distribution coefficient (KD) of SARS-CoV-2 to other well-studied viruses in wastewater systems. This study demonstrates the important role played by municipal wastewater treatment facilities in collecting, concentrating, and removing SARS-CoV-2 in sewage systems before discharge to the environment, and highlights the need to better understand the fate of this virus in wastewater solids.
2. Materials and methods
2.1. Collection of wastewater samples
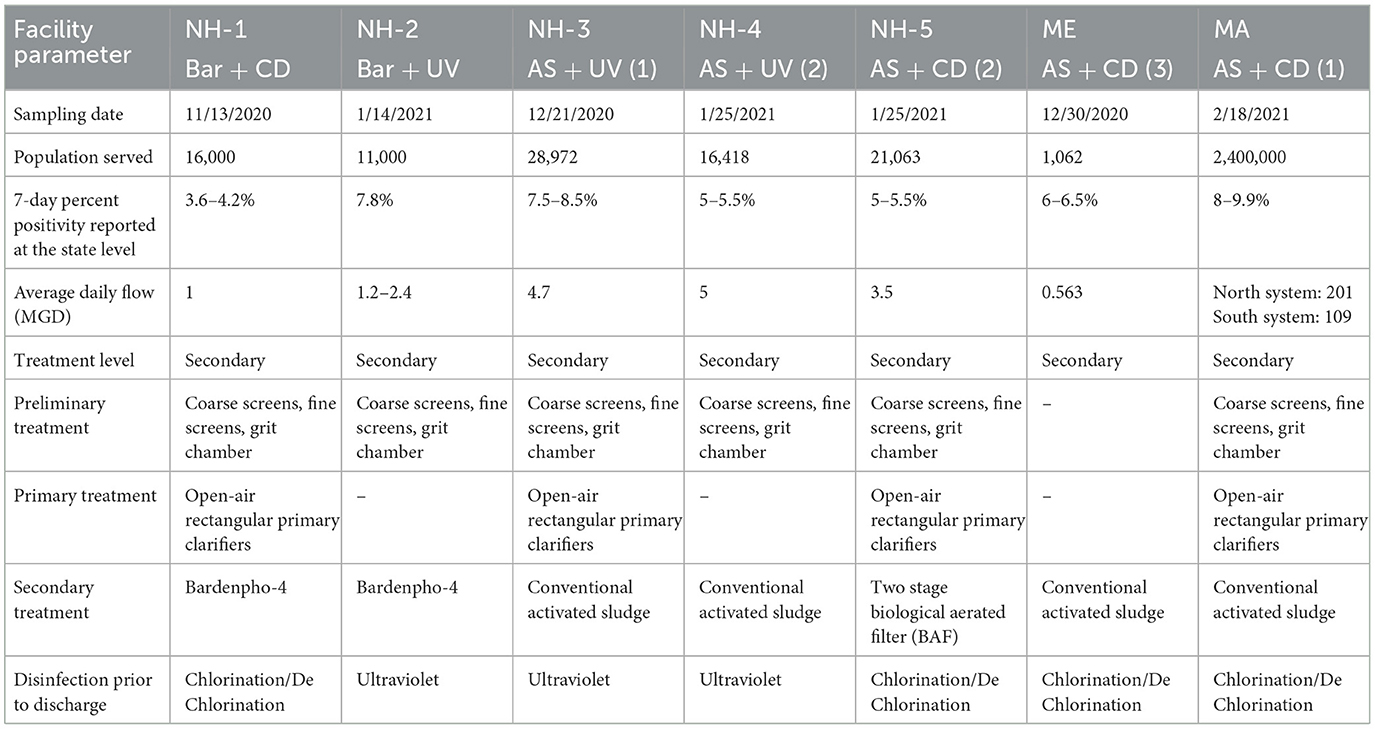
Samples were collected between October 2020 and February 2021 from seven WWTPs located within 70 miles of each other in three coastal New England states: New Hampshire, Maine, and Massachusetts. The WWTPs differ considerably by flow, population served, treatment train, and estimated positivity (based on 7-day average) at the time of sampling (Table 1). WWTPs are referenced in this manuscript based on their location (e.g., NH-1), secondary treatment process (Bar = multi-stage Bardenpho or AS = activated sludge) and their disinfection system (CD = chlorination/dechlorination or UV = UV disinfection). A total of 45 samples were collected in duplicate from the seven WWTPs, including untreated wastewater (n = 9), primary treated (n = 4), secondary treated (n = 13), solids/sludge including return activated sludge, waste activated sludge, and digested sludge (n = 12), and final effluent (after disinfection, n = 7). Samples were collected from locations from the beginning to the end of the facility, designated as BP = before primary, AP = after primary, AL = aerated lagoon, AS = after secondary, ASC = after secondary clarifier, RAS = returned activated sludge, WAS = waste activated sludge, CD = after chlorination/dechlorination, and UV = after UV disinfection (Supplementary Figure S1). Samples were collected in sterile 1-L RNase/DNase free polystyrene containers in the morning (between 10:00 am to 12:00 noon) and immediately transported on ice to the University of New Hampshire, where they were processed within 24 h.
2.2. Extraction and quantification of SARS-CoV-2 viral RNA
Viral RNA was extracted separately from solid and liquid fractions and quantified (Supplementary Figure S2). Of the 45 samples, 33 were fractionated into solid and liquid fractions, with the remaining 12 solely analyzed for liquids because of insufficient solids generated during centrifugation. The solid fraction was collected from the pre-centrifuge step, where subsamples were centrifuged at 5,000 × g for 30 min in order to pellet suspended solids including microbial cells and associated viral particles (Ahmed et al., 2020b). Nucleic acids were extracted from 0.25 g of the pelleted solids using the Allprep PowerViral DNA/RNA Kit (Qiagen, Hilden, Germany) and a QIAcube Connect (Qiagen, Hilden, Germany). The supernatant generated from the pre-centrifuge step, representing the liquid fraction, was subsequently transferred into a sterile 50 ml conical tube containing 10% (w/v) Polyethylene Glycol 8000 (PEG; Millipore Sigma) and 2.25% (w/v) NaCl (0.3 M, Millipore Sigma) (Bibby and Peccia, 2013; Ahmed et al., 2020b). The liquid fraction viral genetic material was concentrated with PEG/NaCl by centrifugation at 12,000 × g for 2 h. After the supernatant was removed, the resulting liquid fraction pellet was resuspended in 400 μl of RNAse free dH2O. Nucleic acids were extracted from the liquid fraction pellet suspension using the Allprep PowerViral DNA/RNA Kit (Qiagen, Hilden, Germany) and a QIAcube Connect (Qiagen, Hilden, Germany). Viral RNA for both fractions (solids and liquids) were each eluted into a 50 uL final volume RNase free dH2O for quantification.
Two SARS-CoV-2 viral nucleocapsid biomarkers (N1 and N2) and a biomarker for the human RNase P gene (RP) (CDC, 2020) were quantified in RNA extracts using reverse transcriptase droplet digital PCR (RT-ddPCR). See Supporting Information for thermocycling parameters and digital droplet quantification details, and Supplementary Table S1 for primer and probes used in this study. Although quantified RNA was not adjusted based on percent recovered, we determined the recovery of SARS-CoV-2 RNA from both solid and liquid fractions by spiking synthetic SARS-CoV-2 RNA quantified control (catalog no. COV019; Exact Diagnostics, Fort Worth, TX) into sterilized wastewater at two concentrations. Recovered concentrations were converted to percent recovery by dividing by the total spiked concentration. An average 58 to 54% recovery of N1 and N2 biomarkers in liquid fractions and average of 37 to 31% recovery of N1 and N2 biomarkers in solid fractions was observed. These recoveries are comparable with those obtained by similar studies using the electronegative filtration method (Gonzalez et al., 2020; Sherchan et al., 2020) but lower than studies using aluminum hydroxide adsorption-precipitation concentration method (Randazzo et al., 2020). Note we did not use the aluminum hydroxide approach because we wanted to maintain a consistent method for processing liquid and solid fractions in this work. Differences in RNA recovery between data here and other studies is likely related to different matrices, RNA extraction approach, and differences in RT-ddPCR workflow.
2.3. Field and wastewater physicochemical parameters analyses
In addition to collecting samples for SARS-CoV-2 RNA quantification, we collected parallel samples into 1L HDPE sterile bottles for analyses of total suspended solids (TSS), total dissolved solids (TDS), volatile suspended solids (VSS), and chemical oxygen demand (COD). TDS, TSS, and VSS were determined within 24 h of sampling following EPA methods 2540-C, 2540-D and 2540-E, respectively. COD analysis was conducted on unfiltered samples within 2 days of sample collection using a spectrophotometer (DR6000, Hach, USA), following Method 8000 TNT Plus 821/822. Field measurements were collected at each sample location for electrical conductivity (EC), pH, dissolved oxygen (DO), and redox potential using a Thermo Scientific Orion Star A329 meter (ThermoFisher Scientific, Waltham, MA) calibrated before each sampling event.
2.4. Data normalization and statistical analyses
Differences between SARS-CoV-2 RNA concentrations before and after each treatment stage were evaluated using a paired t-test (p ≤ 0.05), while associations between viral RNA concentrations and physicochemical parameters were evaluated with Pearson's correlation coefficient in SigmaPlot version 14.5 (Systat Software Inc., San Jose, CA, USA). Viral RNA concentrations that were below method Limits of Detection (LODs) were treated as half of the LOD value in our statistical comparisons.
To elucidate sorption behavior of SARS-CoV-2 viral fragments in wastewater, we calculated adsorption–desorption distribution coefficient (KD in L/kg) at different stages of treatment based on viral RNA biomarker concentrations measured in the solid and liquid fractions. The KD was obtained by dividing viral biomarker concentration in the solids fraction (copies/Kg of wet weight solid) by the biomarker concentration in the liquid fraction (copies/L).
Log removal of SARS-CoV-2 biomarkers between influent and effluent samples for each treatment stage was calculated as follows: Log removal = .
Influent concentration refers to the SARS-CoV-2 RNA concentration of the influent (both liquid and solid phase) of each WWTP, while effluent concentration refers to SARS-CoV-2 RNA concentration (liquid and solid phase) of the effluent from the corresponding treatment stage.
Liquid-solid partitioning (%abundance) was calculated using the fraction of SARS-CoV-2 biomarkers mean loadings discharged in liquid or sludge, where the loadings were calculated from SARS-CoV-2 biomarker concentrations per respective volume (for liquid samples) or wet weighted mass load (for sludge samples).
3. Results
3.1. SARS-CoV-2 viral RNA is removed from effluent during wastewater treatment
To investigate the efficiency of SARS-CoV-2 viral RNA removal in WWTPs differing in size and treatment processes, we quantified two biomarkers (N1 and N2), and the human RNase P gene (RP), in both liquid and solid phases of wastewater samples from seven coastal New England WWTPs. The average daily flow of the WWTPs studied ranged from <1 million gallons per day (MGD) to >1,000 MGD, representing small towns (1,062 persons) to a major metropolitan area (>3 million persons). The estimated positivity (based on 7-day average) ranged from 3.6 to 9.9 at the time of sampling, respectively (Table 1). Despite significant differences in population size and percent positivity during sampling, SARS-CoV-2 viral biomarkers N1 and N2 were consistently quantified at concentrations <2,200 copies/100 ml in the liquid phase and <16,000 copies/g in the solid phase of the influent wastewater samples. Specifically, viral biomarker concentrations ranged from 1,070 to 2,200 copies/100 ml for N1, and 910 to 2,100 copies/100 ml for N2 in raw (untreated) wastewater (Figure 1 and Supplementary Table S3). Levels of viral biomarkers N1 and N2 were below detection after disinfection for all facilities sampled, highlighting the efficiency of these WWTPs in removing viral RNA during treatment (Figures 1A–C and Supplementary Table S3). In all WWTPs sampled, viral biomarkers N1 and N2 decreased in the liquid phase from the influent until after the secondary clarifier. However, N1 and N2 biomarker concentrations in the liquid phase of return activated sludge (RAS) and/or waste activated sludge (WAS) were on par with and sometimes larger than liquid phase biomarker concentrations before primary treatment. Similarly, viral biomarkers in the solids fraction also decreased from the influent until after the secondary treatment, with N1 and N2 biomarkers concentrated in the RAS and/or WAS at levels similar to before primary treatment (Figure 1 and Supplementary Table S3). These data are in line with recent studies conducted in Spain, France, Iran, rural Canada, and Utah (USA) documenting the removal of SARS-CoV-2 RNA during wastewater treatment (Balboa et al., 2021; Bhattarai et al., 2021; D'Aoust et al., 2021; Pourakbar et al., 2021; Serra-Compte et al., 2021).
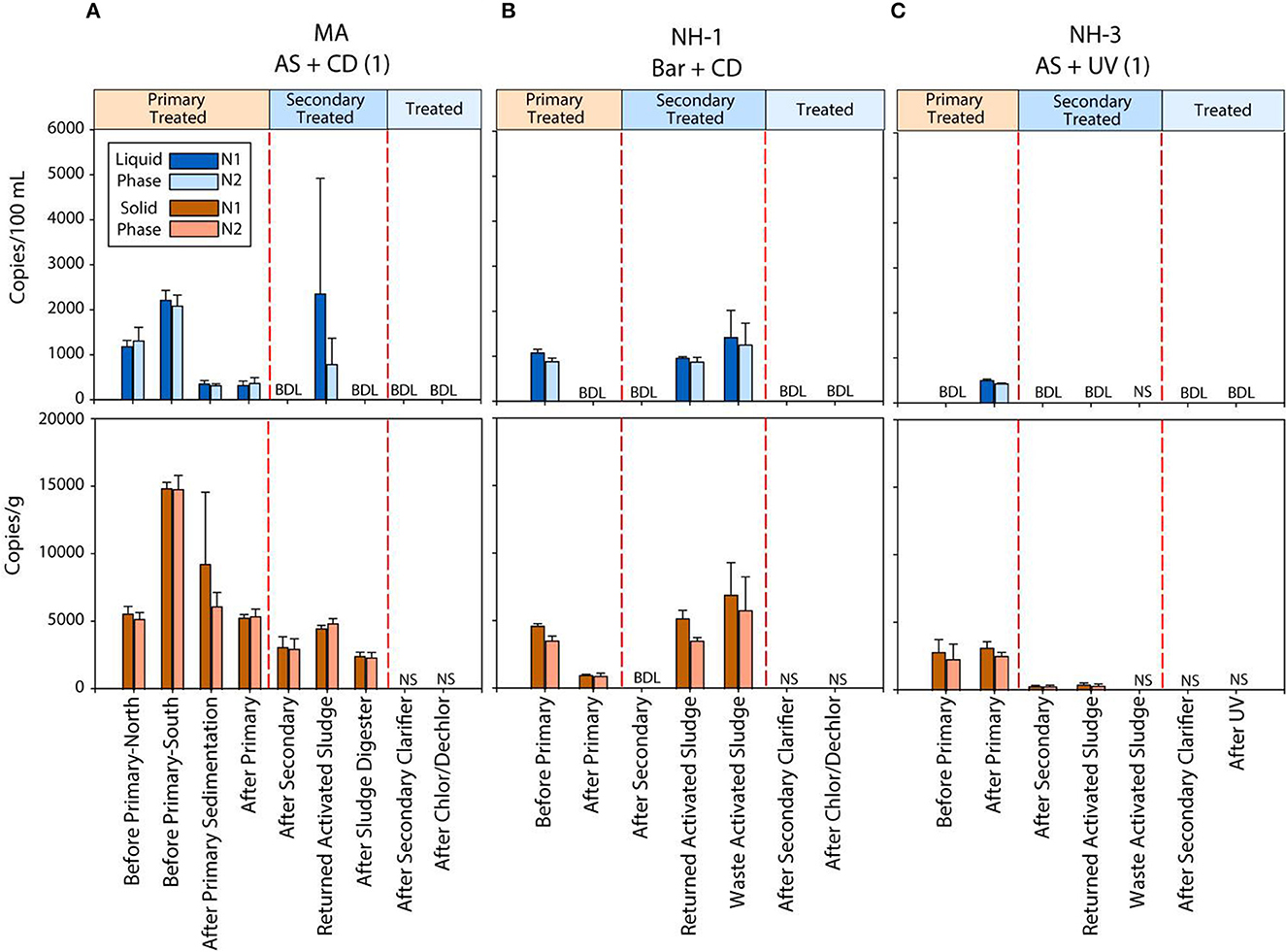
Figure 1. Concentration of two SARS-CoV-2 RNA biomarkers (N1 and N2) measured using RT ddPCR in the liquid and solid phases of untreated and treated wastewater samples from three coastal New England WWTPs (MA, NH-1 and NH-3) (A–C). Liquid phase concentrations are shown as blue bars, while solid phase concentrations are shown in brown. N.S. indicates no sample was analyzed due to limited solids recovery. BDL represents concentrations below the limit of detection of the instrument.
We observed that primary treatment (n = 5) resulted in an average log removal of 0.25 to 0.27 for N1 and N2 biomarkers, respectively, from the liquid phase of the effluents collected from WWTPs after this stage (Supplementary Table S4). Viral particle removal during secondary treatment processes showed a similar range. However, we observed differences in removal depending on the type of secondary treatment process. In WWTPs with AS systems (NH-3, NH-4, NH-5, MA, and ME), which used aeration to enhance biological degradation of suspended and dissolved solids, an average 0.17 to 0.19 log reduction of N1 and N2 biomarkers was observed (Supplementary Table S4). In contrast, wastewater systems designed with Bar (NH-1 and NH-2) had 0.5 to 0.53 log reduction in SARS-CoV-2 biomarkers. Enhanced nutrient and suspended solids removal in the Bar system than in other secondary treatment processes may improve viral particle reduction (Schmitz et al., 2016). Finally, no viral RNA was detected after the secondary clarifier stage, resulting in 1.39 log reduction or higher (Supplementary Table S4). These results provide evidence that secondary treatment and the subsequent clarification processes effectively remove the majority of detectable SARS- CoV-2 genetic material from the liquid phase during wastewater treatment.
The presence of SARS-CoV-2 RNA in the solid phase of these investigated WWTPs was also evaluated. We detected N1 and N2 biomarkers in sample solids from influent wastewater through secondary treatment (at concentrations up to 3,560 copies/g). Primary treatment (n = 5) resulted in an average log removal of 0.25–0.26 for N1 and N2 biomarkers from the solid phase of the effluents (Supplementary Table S4). Viral particle removal during secondary treatment processes showed a similar range. In WWTPs with AS systems (NH-3, NH-4, NH-5, MA, and ME), an average of 0.2–0.23 log reduction was observed (Supplementary Table S4). Wastewater systems designed with advanced Bardenpho process (NH-1 and NH-2) had 0.5–0.45 log reduction in SARS-CoV-2 biomarkers (N1 and N2) which is higher than facilities using conventional processes. We were unable to quantify log removal for solids after secondary clarification because insufficient mass was recovered for viral extraction. Results of SARS-CoV-2 removal in both solid and liquid phases reported here (Supplementary Table S4 and Figure 1) demonstrate that only a portion of viral genetic material present in the influent is removed during the first stage of wastewater treatment, through physical procedures like gravitational precipitation of suspended colloids or settling of organic matter (Saawarn and Hait, 2021; Sangkham, 2021). Consequently, secondary treatment and/or disinfection is necessary for complete removal of SARS-CoV-2.
Interestingly, we found relatively high SARS-CoV-2 RNA concentrations in waste activated sludge samples (up to 5,400 copies/g) from the three smallest WWTPs (NH-1, NH-5, and ME) where the sludge is only treated by volume reduction methods. Similarly, SARS-CoV-2 biomarkers were detected in the solid phase of digested sludge samples (up to 2,600 copies/g) where waste activated sludge discharge into an anaerobic sludge digester (MA WWTP). This data indicates that SARS-CoV-2 genetic material partitions into the solids fraction of samples to various degrees (Zhang et al., 2017).
3.2. Solids carry a sizeable portion of SARS-CoV-2 viral RNA in wastewater
Balboa et al. (2021) showed SARS-CoV-2 was preferentially associated with wastewater sludge. To further assess the affinity of SARS-CoV-2 genetic material for wastewater solids, we estimated adsorption-desorption distribution coefficients (KD in L/kg) for different stages of treatment based on N1 and N2 concentrations measured in liquid and solid phases (Supplementary Table S5). In general, log KD increased with higher solids content of wastewater samples along the treatment train. For instance, the mean log KD for the N1 biomarker increased from 2.7 L/kg in the influent, to 3.2 L/kg after-primary, and 3.6 L/kg after-secondary (MA) (Supplementary Table S5). These estimates indicated SARS-CoV-2 is ~470 to 3,700 × more concentrated in the solids as compared the liquid phase. Moreover, despite solids making up only a small fraction of the sample (solids content ~0.02%), the majority of SARS-CoV-2 genetic material is associated with the solids fraction.
Based on these differences in the adsorption-desorption distribution constant, we further explored sample partitioning at three facilities (MA, NH-1, and NH-2) by considering the percent solids in each sample. We assumed in our calculation that the majority of solids were separated and extracted in our wet weighted solids fraction, with the remaining sample volume extracted as liquid phase. Using a straight water density conversion (1 g = 1 ml), we calculated the portion of biomarkers associated with each phase (Supplementary Table S6). SARS-CoV-2 N1 biomarkers in the solids therefore comprised from 51 to 81% in raw wastewater, 60 to 89% after primary treatment, then 66 to 90% after secondary treatment (Figure 2 and Supplementary Table S6). The portion of N1 viral biomarkers in the solids for RAS and WAS samples showed a similar range (54 to 92%). These values indicated that the majority of viral RNA in wastewater samples prior to the secondary clarifier are associated with the solids, which in turn represents a very small mass fraction of the sample. This is supported by limited data reported by others (Balboa et al., 2021; Serra-Compte et al., 2021).
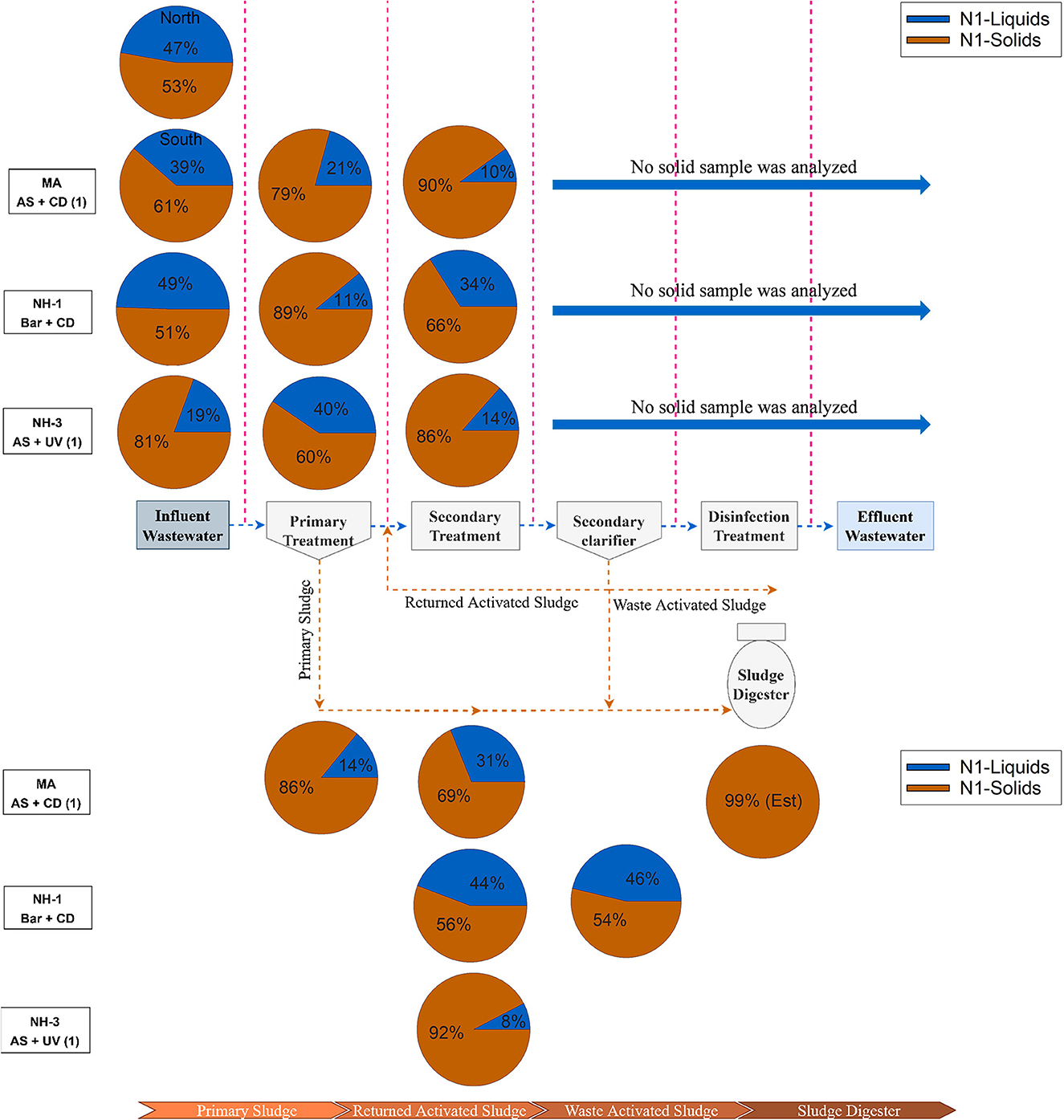
Figure 2. Percentage abundance of SARS-CoV-2 biomarkers across treatment stages in three different wastewater treatment plants in coastal New England area. See Supplementary Table S6 for calculations.
The WWTPs sampled in this work vary in several aspects that may influence the liquid-solid partitioning of SARS-CoV-2 RNA. Influent total suspended solids (TSS) concentrations differed 20-fold between the seven WWTPs (104 to 3,050 mg/L), which may have affected SARS-CoV-2 RNA partitioning (Supplementary Table S7). Similarly, influent organic matter percentage (VSS/TSS) varied from 11 to 98%, indicating that in some cases the TSS was comprised primarily of inorganic content (MA, NH-2) as compared to organics (NH-1, NH-3, NH-5, ME). Furthermore, the average removal rates of TSS measured after secondary treatment were higher in MA and NH-1 (93 and 99.5% respectively) as compared with those measured in NH-2, NH-3, NH-4, NH-5 and ME (60%, 81.9 %, 16%, 86% and 81.6% respectively) WWTPs. This potentially affected the removal rate of viral RNA attached to the suspended solids during this treatment step.
In order to determine whether EC, TSS, TDS, VSS, or COD concentrations were predictive of SARS-CoV-2 viral biomarker concentrations, we next assessed the relationship between these parameters using Pearson's correlation coefficient. No significant correlation (p > 0.05) was found between TSS, TDS, VSS, pH, redox, or COD and SARS-CoV-2 biomarkers N1 and N2 in either the liquid or solid wastewater fractions (Figure 3). Interestingly, SARS-CoV-2 biomarkers were strongly associated with EC values in both liquid (ρ = 0.36; p < 0.05) and solid phases (ρ = 0.48; p < 0.05; Supplementary Table S2). Electrical conductivity, which is a measure of the ability of water to conduct an electrical current, depends on the concentration of conductive ions (cations and ions) in the water (Liu et al., 2020). One possible explanation for this correlation is that high cation content disrupts floc structures, reducing its settleability (Kara et al., 2008) and enhancing adsorption sites for viruses (Wong et al., 2013). The presence of cations can also reduce the electrostatic double layer, altering charge structure on solids (Nasser et al., 1993; Wong et al., 2012; Yang et al., 2022).
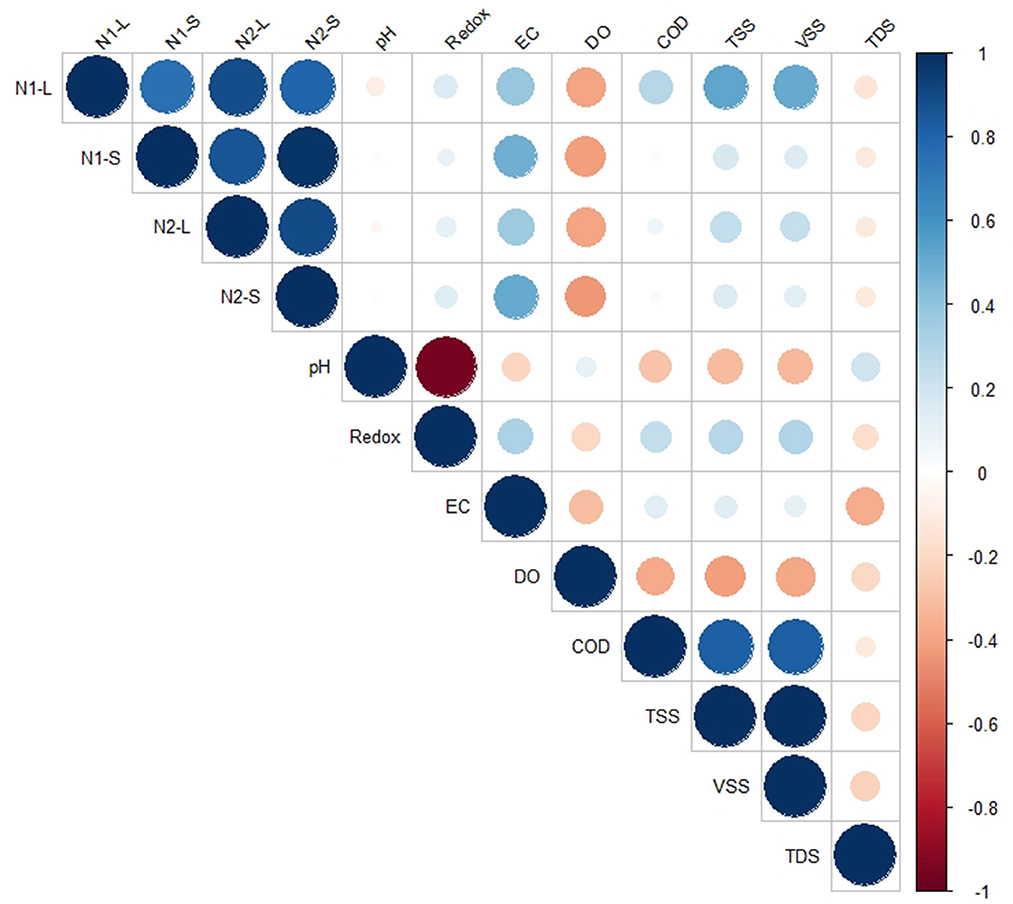
Figure 3. Correlation between SARS-CoV-2 biomarker concentrations in the liquid and solid phase, and physicochemical parameters measured across coastal New England WWTPs.
4. Discussion
In this work, we sought to investigate changes in concentrations of SARS-CoV-2 RNA biomarkers along the wastewater treatment train and assess viral RNA removal before discharge. We also aimed to evaluate the fate of SARS-CoV-2 within the facility, specifically its partitioning between the liquid and solid phases. Our work indicates that primary and secondary treatment achieves variable but consistent removal of SARS-CoV-2 RNA biomarkers from wastewater liquids across all investigated WWTPs. This finding is consistent with related studies assessing viral RNA removal during the key stages of a wastewater treatment process (Haramoto et al., 2020; Randazzo et al., 2020; Sherchan et al., 2020). In contrast to previous work that reported detectable viral RNA in both the influent and treated sewage (Westhaus et al., 2021), here we found no quantifiable viral RNA after the secondary clarification process. Although viral RNA detection is influenced by the sensitivity of our extraction and quantification approach (Dimitrakopoulos et al., 2022), our results support the effectiveness of several different wastewater treatment sizes and designs in removing SARS-CoV-2 RNA before discharge to receiving water bodies at the reported detection limits. Nevertheless, molecular methods for concentrating viral RNA in wastewater continue to evolve with research knowledge and improved biotechnology tools. For example, our current approach uses magnetic nanopolymers rather than PEG/NaCl to concentrate viral particles in wastewater. Magnetic nanoparticles or electronegative filtration (LaTurner et al., 2021) may have improved recovery of SARS-CoV-2 in this study. Additionally, a second limitation comes from our use of grab samples at these facilities. Recent studies have indicated that 24-h composite samples or swabs deployed over a specific temporal interval may better account for fluctuations in SARS-CoV-2 than grab samples, particularly in low flow catchments (Acer et al., 2022; George et al., 2022). However, given the samples in our study derive from the treatment facility, some degree of community mixing would have occured in the sewershed before reaching the facility influent. Therefore, community variations should be dampened to some degree.
Removal for different types of indicator viruses including human adenovirus, human polyomavirus and pepper mild mottle virus during secondary and disinfection processes has been reported as being between 0.6 and 3.4 log (Kitajima et al., 2014; Schmitz et al., 2016; Tandukar et al., 2020; Wu Z. et al., 2020). These results of other indicator virus elimination rates are in agreement with the log removal of SARS-CoV-2 RNA described in the present work. This suggests secondary treatment followed by disinfection is effective in elimination of various groups of indicator viruses and pathogens. Although the number of facilities investigated here was small, we did observe that Bar systems had greater removal of SARS-CoV-2 genetic material over activated sludge systems (Schmitz et al., 2016). Differing microbial communities, redox conditions, and/or longer solids retention time associated with the Bar systems may all play a role in these observed differences. Moreover, it was clear from this work that higher SARS-CoV-2 RNA removal occurred following the primary clarification stage, suggesting that the interaction between viral particles and primary solids is critically important for viral particle removal (Graham et al., 2020; Balboa et al., 2021).
It is known that enteric viruses can adsorb to solid and/or colloidal particles due to the presence of a lipid bilayer surrounding the protein capsid, via electrostatic and hydrophobic interactions (Arraj et al., 2005; Verbyla and Mihelcic, 2015). Here, our analyses showed measurable levels of SARS-CoV-2 biomarkers in primary, secondary, waste activated sludge and anaerobic digested sludge samples. This data indicates this enveloped virus may have high affinity toward solids, similar to non-enveloped enteric viruses (e.g., adenovirus, rotavirus, enteroviruses and norovirus) (Berg, 1973; Ward, 1981; Rao et al., 1987; Yin et al., 2018; Gholipour et al., 2022). A previous study suggested viral particle sorption to solids was positively correlated with the organic matter content (Gundy et al., 2009), which may play a role in protecting viral particles from physical, chemical, or biological degradation (Gundy et al., 2009; Paul et al., 2021). The strong association with organic matter may also partially explain the efficient removal of SARS-CoV-2 material in the primary treatment phase, as well as its higher removal observed in the Bar systems, which typically have longer solids retention time (SRT). A higher SRT allows greater interaction time between SARS-CoV-2 and activated sludge solids, increasing its potential for sorption.
Interestingly, almost 70% of measured SARS-CoV-2 biomarkers left the treatment system in the solids, demonstrating the importance of this wastewater media in concentrating viral RNA during treatment. Our results are in agreement with recent studies reporting that solids are responsible for sorptive protection of viral RNA, suggesting that the SARS-CoV-2 RNA biomarkers are mostly diverted to the sludge (Kocamemi et al., 2020; Balboa et al., 2021; Serra-Compte et al., 2021). Moreover, our work is consistent with those of Serra-Compte et al. (2021) who reported the presence of SARS-CoV-2 genetic material in anaerobic digester sludge. These results indicate that viral biomarkers may continue to partition into the solids fraction of samples and are not inactivated by the anaerobic condition of the digester (Zhang et al., 2017). Similarly, previous studies with other human coronaviruses reported the occurrence of viral RNA in sludge samples after anaerobic digestion (Bibby et al., 2011; Bibby and Peccia, 2013; Bhattarai et al., 2021). Although PCR-based molecular tools do not indicate infectious SARS-CoV-2 virus is present, biomarkers are useful for monitoring the prevalence and fate of viruses in wastewater treatment media, especially when treated sludge is reused for beneficial purposes such as soil amendment (Wigginton et al., 2015). Recent studies indicated that biosolids containing detectable SARS-CoV-2 genetic material may leach into the surrounding soils and even contaminate groundwater (Li et al., 2020; Yang et al., 2020). Moreover, greater adsorption capacity and longer survivability for enveloped (e.g., SARS-CoV-2 and Phi6) compared with non-enveloped viral surrogates (e.g., MS2, T4, and Phix174) has been observed in wastewater solids, which is of interest for sludge discharge or reuse strategies (Katz et al., 2018; Yang et al., 2020, 2022). Therefore, further studies assessing the fate of SARS-CoV-2 biomarkers in wastewater sludge, as well as studies determining the viability of viral fragments in biosolids would further inform our understanding of any public health risks associated with their handling.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
PM, MA, and FC conceived and designed the study. PM, FC, and JM supervised and reviewed the design of laboratory experiments. FC and MA processed the raw ddPCR data. MA performed the statistical analyses, conducted the laboratory experiments, and wrote the first draft of the manuscript. MA and PM developed the manuscript figures. All authors revised and approved the submitted version of the manuscript.
Funding
This work was funded by the New Hampshire Sea Grant (NHSG) Development funds and a UNH STAF fellowship.
Acknowledgments
We are grateful to Dr. Jenna L. Luek for her constructive feedback which greatly improved the manuscript. We also thank Kellen Sawyer and wastewater treatment personnel for assisting with facility sampling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2023.1130114/full#supplementary-material
References
Abu Ali, H., Yaniv, K., Bar-Zeev, E., Chaudhury, S., Shagan, M., Lakkakula, S., et al. (2021). Tracking SARS-CoV-2 RNA through the wastewater treatment process. ACS ESandT Water 1, 1161–1167. doi: 10.1021/acsestwater.0c00216
Acer, P. T., Kelly, L. M., Lover, A. A., and Butler, C. S. (2022). Quantifying the relationship between SARS-CoV-2 wastewater concentrations and building-level COVID-19 prevalence at an isolation residence: a passive sampling approach. Int. J. Environ. Res. Public Health 19, 11245. doi: 10.3390/ijerph191811245
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O'Brien, J. W., et al. (2020a). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 728, 138764. doi: 10.1016/j.scitotenv.2020.138764
Ahmed, W., Bertsch, P. M., Bivins, A., Bibby, K., Farkas, K., Gathercole, A., et al. (2020b). Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 739, 139960. doi: 10.1016/j.scitotenv.2020.139960
Amirian, E. S. (2020). Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 95, 363–370. doi: 10.1016/j.ijid.2020.04.057
Amoah, I. D., Kumari, S., and Bux, F. (2020). Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 143, 105962. doi: 10.1016/j.envint.2020.105962
Arora, S., Nag, A., Sethi, J., Rajvanshi, J., Saxena, S., Shrivastava, S. K., et al. (2020). Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. Water Sci. Technol. 82, 2823–2836. doi: 10.2166/wst.2020.540
Arraj, A., Bohatier, J., Laveran, H., and Traore, O. (2005). Comparison of bacteriophage and enteric virus removal in pilot scale activated sludge plants. J. Appl. Microbiol. 98, 516–524. doi: 10.1111/j.1365-2672.2004.02485.x
Arslan, M., Xu, B., and El-Din, M. G. (2020). Transmission of SARS-CoV-2 via fecal-oral and aerosols–borne routes: environmental dynamics and implications for wastewater management in underprivileged societies. Sci. Total Environ. 743, 140709. doi: 10.1016/j.scitotenv.2020.140709
Balboa, S., Mauricio-Iglesias, M., Rodriguez, S., Martínez-Lamas, L., Vasallo, F. J., Regueiro, B., et al. (2021). The fate of SARS-CoV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 772, 145268. doi: 10.1016/j.scitotenv.2021.145268
Berg, G. (1973). Removal of viruses from sewage, effluents, and waters: 1. A review. Bull. World Health Organ. 49, 451.
Bhattarai, B., Sahulka, S. Q., Podder, A., Hong, S., Li, H., Gilcrease, E., et al. (2021). Prevalence of SARS-CoV-2 genes in water reclamation facilities: from influent to anaerobic digester. Sci. Total Environ. 796, 148905. doi: 10.1016/j.scitotenv.2021.148905
Bibby, K., and Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 47, 1945–1951. doi: 10.1021/es305181x
Bibby, K., Viau, E., and Peccia, J. (2011). Viral metagenome analysis to guide human pathogen monitoring in environmental samples. Lett. Appl. Microbiol. 52, 386–392. doi: 10.1111/j.1472-765X.2011.03014.x
Bisseux, M., Colombet, J., Mirand, A., Roque-Afonso, A-. M., Abravanel, F., Izopet, J., et al. (2018). Monitoring human enteric viruses in wastewater and relevance to infections encountered in the clinical setting: a one-year experiment in central France, 2014 to 2015. Eurosurveillance 23, 17–00237. doi: 10.2807/1560-7917.ES.2018.23.7.17-00237
Bivins, A., Greaves, J., Fischer, R., Yinda, K. C., Ahmed, W., Kitajima, M., et al. (2020). Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol Lett. 7, 937–942. doi: 10.1021/acs.estlett.0c00730
Bogler, A., Packman, A., Furman, A., Gross, A., Kushmaro, A., Ronen, A., et al. (2020). Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 3, 981–990. doi: 10.1038/s41893-020-00605-2
Causanilles, A., Ruepert, C., Ibáñez, M., Emke, E., Hernández, F., de Voogt, P., et al. (2017). Occurrence and fate of illicit drugs and pharmaceuticals in wastewater from two wastewater treatment plants in Costa Rica. Sci. Total Environ. 599, 98–107. doi: 10.1016/j.scitotenv.2017.04.202
CDC (2020). CDC2019.-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primer and Probe information. Atlanta GA: CDC.
Choi, P. M., Tscharke, B. J., Donner, E., O'Brien, J. W., Grant, S. C., Kaserzon, S. L., et al. (2018). Wastewater-based epidemiology biomarkers: past, present and future. Trends Analyt. Chem. 105, 453–469. doi: 10.1016/j.trac.2018.06.004
D'Aoust, P. M., Towhid, S. T., Mercier, É., Hegazy, N., Tian, X., Bhatnagar, K., et al. (2021). COVID-19 wastewater surveillance in rural communities: comparison of lagoon and pumping station samples. Sci. Total Environ. 801, 149618. doi: 10.1016/j.scitotenv.2021.149618
Dimitrakopoulos, L., Kontou, A., Strati, A., Galani, A., Kostakis, M., Kapes, V., et al. (2022). Evaluation of viral concentration and extraction methods for SARS-CoV-2 recovery from wastewater using droplet digital and quantitative RT-PCR. Case Stud. Chem. Environ. Eng. 6, 100224. doi: 10.1016/j.cscee.2022.100224
Estévez-Danta, A., Bijlsma, L., Capela, R., Cela, R., Celma, A., Hernández, F., et al. (2022). Use of illicit drugs, alcohol and tobacco in Spain and Portugal during the COVID-19 crisis in 2020 as measured by wastewater-based epidemiology. Sci. Total Environ. 836, 155697. doi: 10.1016/j.scitotenv.2022.155697
George, A. D., Kaya, D., Layton, B. A., Bailey, K., Mansell, S., Kelly, C., et al. (2022). Impact of sampling type, frequency, and scale of the collection system on SARS-CoV-2 quantification fidelity. Environ. Sci. Technol Lett. 9, 160–165. doi: 10.1021/acs.estlett.1c00882
Ghernaout, D., and Elboughdiri, N. (2020). Antibiotics resistance in water mediums: background, facts, and trends. Appl. Eng. 4, 1–6. doi: 10.4236/oalib.1106374
Gholipour, S., Ghalhari, M. R., Nikaeen, M., Rabbani, D., Pakzad, P., Miranzadeh, M. B., et al. (2022). Occurrence of viruses in sewage sludge:/a systematic review. Sci. Total Environ. 153886. doi: 10.1016/j.scitotenv.2022.153886
Gholipour, S., Mohammadi, F., Nikaeen, M., Shamsizadeh, Z., Khazeni, A., Sahbaei, Z., et al. (2021). COVID-19 infection risk from exposure to aerosols of wastewater treatment plants. Chemosphere 273, 129701. doi: 10.1016/j.chemosphere.2021.129701
Gonçalves, J., Koritnik, T., Mioč, V., Trkov, M., Bolješič, M., Berginc, N., et al. (2021). Detection of SARS-CoV-2 RNA in hospital wastewater from a low COVID-19 disease prevalence area. Sci. Total Environ. 755, 143226. doi: 10.1016/j.scitotenv.2020.143226
Gonzalez, R., Curtis, K., Bivins, A., Bibby, K., Weir, M. H., Yetka, K., et al. (2020). COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 186, 116296. doi: 10.1016/j.watres.2020.116296
Graham, K. E., Loeb, S. K., Wolfe, M. K., Catoe, D., Sinnott-Armstrong, N., Kim, S., et al. (2020). SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 55, 488–498. doi: 10.1021/acs.est.0c06191
Gundy, P. M., Gerba, C. P., and Pepper, I. L. (2009). Survival of coronaviruses in water and wastewater. Food Environ. Virol. 1, 10–14. doi: 10.1007/s12560-008-9001-6
Haramoto, E., Malla, B., Thakali, O., and Kitajima, M. (2020). First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 737, 140405. doi: 10.1016/j.scitotenv.2020.140405
Hata, A., Hara-Yamamura, H., Meuchi, Y., Imai, S., and Honda, R. (2021). Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 758, 143578. doi: 10.1016/j.scitotenv.2020.143578
Hellmér, M., Paxéus, N., Magnius, L., Enache, L., Arnholm, B., Johansson, A., et al. (2014). Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 80, 6771–6781. doi: 10.1128/AEM.01981-14
Hovi, T., Shulman, L., Van Der Avoort, H., Deshpande, J., Roivainen, M., Gourville, D. e., et al. (2012). of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 140, 1–13. doi: 10.1017/S095026881000316X
Kara, F., Gurakan, G., and Sanin, F. D. (2008). Monovalent cations and their influence on activated sludge floc chemistry, structure, and physical characteristics. Biotechnol. Bioeng. 100, 231–239. doi: 10.1002/bit.21755
Katz, A., Peña, S., Alimova, A., Gottlieb, P., Xu, M., Block, K. A., et al. (2018). Heteroaggregation of an enveloped bacteriophage with colloidal sediments and effect on virus viability. Sci. Total Environ. 637, 104–111. doi: 10.1016/j.scitotenv.2018.04.425
Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., Hamilton, K. A., et al. (2020). SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 739, 139076. doi: 10.1016/j.scitotenv.2020.139076
Kitajima, M., Iker, B. C., Pepper, I. L., and Gerba, C. P. (2014). Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci. Total Environ. 488, 290–296. doi: 10.1016/j.scitotenv.2014.04.087
Kocamemi, B., Kurt, H., Sait, A., Sarac, F., Saatci, A., Pakdemirli, B., et al. (2020). SARS-CoV-2 detection in istanbul wastewater treatment plant sludges. medRxiv [Preprint]. doi: 10.1101/2020.05.12.20099358
Kumar, M., Kuroda, K., Patel, A. K., Patel, N., Bhattacharya, P., Joshi, M., et al. (2021). Decay of SARS-CoV-2 RNA along the wastewater treatment outfitted with upflow Anaerobic Sludge Blanket (UASB) system evaluated through two sample concentration techniques. Sci. Total Environ. 754, 142329. doi: 10.1016/j.scitotenv.2020.142329
LaTurner, Z. W., Zong, D. M., Kalvapalle, P., Gamas, K. R., Terwilliger, A., Crosby, T., Ali, P., Avadhanula, V., Santos, H. H., and Weesner, K. (2021). Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 197, 117043. doi: 10.1016/j.watres.2021.117043
Lewis, D. (2020). Mounting evidence suggests coronavirus is airborne—but health advice has not caught up. Nature 583, 510–513. doi: 10.1038/d41586-020-02058-1
Li, M., Yang, Y., Lu, Y., Zhang, D., Liu, Y., Cui, X., et al. (2020). Natural host–environmental media–human: a new potential pathway of COVID-19 outbreak. Engineering 6, 1085–1098. doi: 10.1016/j.eng.2020.08.010
Liu, J., Yang, P., and Yang, Z. J. (2020). Electrical properties of frozen saline clay and their relationship with unfrozen water content. Cold Reg. Sci. Technol. 178, 103127. doi: 10.1016/j.coldregions.2020.103127
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., and Brouwer, A. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 7, 511–516. doi: 10.1021/acs.estlett.0c00357
Mercan, S., Kuloglu, M., and Asicioglu, F. (2019). Monitoring of illicit drug consumption via wastewater: development, challenges, and future aspects. Curr. Opin. Environ. Sci. Health 9, 64–72. doi: 10.1016/j.coesh.2019.05.002
Mizumoto, K., Kagaya, K., Zarebski, A., and Chowell, G. (2020). Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the diamond princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance 25, 2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180
Mohan, S. V., Hemalatha, M., Kopperi, H., Ranjith, I., and Kumar, A. K. (2021). SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 405, 126893. doi: 10.1016/j.cej.2020.126893
Naddeo, V., and Liu, H. (2020). Editorial Perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environ. Sci. Water Res. Technol. 6, 1213–1216. doi: 10.1039/D0EW90015J
Nasser, A., Adin, A., and Fattal, B. (1993). Adsorption of poliovirus 1 and F+ bacteriophages onto sand. Water Sci. Technol. 27, 331–338. doi: 10.2166/wst.1993.0567
Nasseri, S., Yavarian, J., Baghani, A. N., Azad, T. M., Nejati, A., Nabizadeh, R., et al. (2021). The presence of SARS-CoV-2 in raw and treated wastewater in 3 cities of Iran: Tehran, Qom and Anzali during coronavirus disease 2019 (COVID-19) outbreak. J. Environ. Health Sci. Eng. 19, 573–584. doi: 10.1007/s40201-021-00629-6
Nishiura, H., Kobayashi, T., Miyama, T., Suzuki, A. Jung, S.-M., Hayashi, K., et al. (2020). Estimation of the asymptomatic ratio of novel coronavirus infections (COVID-19). Int. J. Infect. Dis. 94, 154. doi: 10.1016/j.ijid.2020.03.020
Oliver, M. M. H., Hewa, G. A., Pezzaniti, D., Haque, M. A., Haque, S., Haque, M. M., et al. (2020). COVID-19 and recycled wastewater irrigation: a review of implications. Preprints. doi: 10.20944/preprints202006.0105.v1
Paul, D., Kolar, P., and Hall, S. G. A. (2021). review of the impact of environmental factors on the fate and transport of coronaviruses in aqueous environments. Npj Clean Water 4, 1–13. doi: 10.1038/s41545-020-00096-w
Peccia, J., Zulli, A., Brackney, D. E., Grubaugh, N. D., Kaplan, E. H., Casanovas-Massana, A., et al. (2020). Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 38, 1164–1167. doi: 10.1038/s41587-020-0684-z
Pourakbar, M., Abdolahnejad, A., Raeghi, S., Ghayourdoost, F., Yousefi, R., Behnami, A., et al. (2021). Comprehensive investigation of SARS-CoV-2 fate in wastewater and finding the virus transfer and destruction route through conventional activated sludge and sequencing batch reactor. Sci. Total Environ. (2021) 806:151391. doi: 10.1016/j.scitotenv.2021.151391
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., Sánchez, G., et al. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 181, 115942. doi: 10.1016/j.watres.2020.115942
Rao, V. C., Metcalf, T. G., and Melnick, J. L. (1987). Removal of indigenous rotaviruses during primary settling and activated-sludge treatment of raw sewage. Water Res. 21, 171–177. doi: 10.1016/0043-1354(87)90046-7
Saawarn, B., and Hait, S. (2021). Occurrence, fate and removal of SARS-CoV-2 in wastewater: current knowledge and future perspectives. J. Environ. Chem. Eng. 9, 104870. doi: 10.1016/j.jece.2020.104870
Sangkham, S. (2021). A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral particles. J. Environ. Manag. 299:113563. doi: 10.1016/j.jenvman.2021.113563
Schmitz, B. W., Kitajima, M., Campillo, M. E., Gerba, C. P., and Pepper, I. L. (2016). Virus reduction during advanced bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 50, 9524–9532. doi: 10.1021/acs.est.6b01384
Serra-Compte, A., González, S., Arnaldos, M., Berlendis, S., Courtois, S., Loret, J. F., et al. (2021). Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 202, 117435. doi: 10.1016/j.watres.2021.117435
Sherchan, S. P., Shahin, S., Ward, L. M., Tandukar, S., Aw, T. G., Schmitz, B., et al. (2020). First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Total Environ. 743, 140621. doi: 10.1016/j.scitotenv.2020.140621
Shutler, J., Zaraska, K., Holding, T., Machnik, M., Uppuluri, K., Ashton, I., et al. (2020). Risk of SARS-CoV-2 infection from contaminated water systems. medRxiv [Preprint]. doi: 10.1101/2020.06.17.20133504
Tandukar, S., Sherchan, S. P., and Haramoto, E. (2020). Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 10, 1–8. doi: 10.1038/s41598-020-60547-9
Verbyla, M. E., and Mihelcic, J. R. (2015). A review of virus removal in wastewater treatment pond systems. Water Res. 71, 107–124. doi: 10.1016/j.watres.2014.12.031
Ward, R. (1981). “Virus survival during sludge treatment,” in Viruses and Wastewater Treatment, ed M. Goddard (Amsterdam: Elsevier), 65–77. doi: 10.1016/B978-0-08-026401-1.50015-4
Westhaus, S., Weber, F-. A., Schiwy, S., Linnemann, V., Brinkmann, M., Widera, M., et al. (2021). Detection of SARS-CoV-2 in raw and treated wastewater in Germany–suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 751, 141750. doi: 10.1016/j.scitotenv.2020.141750
Wigginton, K., Ye, Y., and Ellenberg, R. (2015). Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ. Sci. Water Res. Technol. 1, 735–746. doi: 10.1039/C5EW00125K
Wong, K., Mukherjee, B., Kahler, A. M., Zepp, R., and Molina, M. (2012). Influence of inorganic ions on aggregation and adsorption behaviors of human adenovirus. Environ. Sci. Technol. 46, 11145–11153. doi: 10.1021/es3028764
Wong, K., Voice, T. C., and Xagoraraki, I. (2013). Effect of organic carbon on sorption of human adenovirus to soil particles and laboratory containers. Water Res. 47, 3339–3346. doi: 10.1016/j.watres.2013.03.029
Wu, Y., Guo, C., Tang, L., Hong, Z., Zhou, J., Dong, X., et al. (2020). Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 5, 434–435. doi: 10.1016/S2468-1253(20)30083-2
Wu, Z., Greaves, J., Arp, L., Stone, D., and Bibby, K. (2020). Comparative fate of CrAssphage with culturable and molecular fecal pollution indicators during activated sludge wastewater treatment. Environ. Int. 136, 105452. doi: 10.1016/j.envint.2019.105452
Wurtzer, S., Waldman, P., Ferrier-Rembert, A., Frenois-Veyrat, G., Mouchel, J-. M., Boni, M., et al. (2021). Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 198, 117183. doi: 10.1016/j.watres.2021.117183
Xagoraraki, I., and O'Brien, E. (2020). “Wastewater-based epidemiology for early detection of viral outbreaks,” in Women in Water Quality, ed D. J. O'Bannon (New York, NY: Springer), 75–97. doi: 10.1007/978-3-030-17819-2_5
Xing, Y., Wang, S.-p., Zhang, Z.-q., Liu, X., and Lu, J.-S. (2021). Effect of minocycline on the changes in the sewage chemical index and microbial communities in sewage pipes. J. Hazard. Mater. 402, 123792. doi: 10.1016/j.jhazmat.2020.123792
Yang, W., Cai, C., and Dai, X. (2020). The potential exposure and transmission risk of SARS-CoV-2 through sludge treatment and disposal. Resour. Conserv. Recycl. 162, 105043. doi: 10.1016/j.resconrec.2020.105043
Yang, W., Cai, C., and Dai, X. (2022). Interactions between virus surrogates and sewage sludge vary by viral analyte: recovery, persistence, and sorption. Water Res. 210, 117995. doi: 10.1016/j.watres.2021.117995
Yin, Z., Voice, T. C., Tarabara, V. V., and Xagoraraki, I. (2018). Sorption of human adenovirus to wastewater solids. J. Environ. Eng. 144, 06018008. doi: 10.1061/(ASCE)EE.1943-7870.0001463
Keywords: COVID-19, wastewater-based epidemiology, SARS-CoV-2 in wastewater, viral RNA removal, wastewater solids, ddPCR
Citation: Aghababaei M, Colosimo F, Malley JP Jr and Mouser PJ (2023) The fate of SARS-CoV-2 viral RNA in coastal New England wastewater treatment plants. Front. Water 5:1130114. doi: 10.3389/frwa.2023.1130114
Received: 22 December 2022; Accepted: 22 March 2023;
Published: 20 April 2023.
Edited by:
Karin Yaniv, University of California, Berkeley, United StatesReviewed by:
Mohamed Azab El-Liethy, National Research Centre, EgyptSihem JEBRI, National Center for Nuclear Science and Technology, Tunisia
Copyright © 2023 Aghababaei, Colosimo, Malley and Mouser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula J. Mouser, cGF1bGEubW91c2VyQHVuaC5lZHU=
 Mina Aghababaei
Mina Aghababaei Fabrizio Colosimo
Fabrizio Colosimo James P. Malley Jr.1
James P. Malley Jr.1 Paula J. Mouser
Paula J. Mouser