
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Water , 20 August 2021
Sec. Environmental Water Quality
Volume 3 - 2021 | https://doi.org/10.3389/frwa.2021.710853
This article is part of the Research Topic Functional Diversity of Aquatic Microorganisms and Their Roles in Water Quality View all 10 articles
 Yannan Huang1
Yannan Huang1 Colin M. Ragush1
Colin M. Ragush1 Lindsay H. Johnston1
Lindsay H. Johnston1 Michael W. Hall2
Michael W. Hall2 Robert G. Beiko2
Robert G. Beiko2 Rob C. Jamieson1
Rob C. Jamieson1 Lisbeth Truelstrup Hansen1,3*
Lisbeth Truelstrup Hansen1,3*Wastewater stabilization ponds (WSPs) are commonly used to treat municipal wastewater in the Canadian Arctic. Bacterial community structure and functionality remain mostly uncharacterized for arctic WSPs, yet are presumed important for treatment outcomes during the 3-month summer treatment season with open water in the WSPs. The objective of this study was to investigate treatment performance and related temporal and spatial changes in the structure and putative function of bacterial communities during treatment of municipal wastewater in the WSPs of Pond Inlet and Clyde River, Nunavut over two consecutive summer treatment seasons. Influent raw wastewater contained a high organic load and large bacterial communities (~9 log 16S rRNA copies/mL) belonging mainly to Proteobacteria. Although designed to be facultative ponds, both WSPs remained anaerobic with neutral pH values (7.5–7.8) throughout the summer treatment season. Water quality data showed that nutrients [measured as carbonaceous biological oxygen demand (CBOD5)], total suspended solids, and total ammonia nitrogen were progressively reduced during treatment in the ponds as the summer progressed. The pond bacterial population size and species richness depended on the pond temperature (2–18°C), with 8.5 log 16S rRNA copies/mL and the largest alpha diversities (Shannon-Wiener index of 4-4.5) observed mid-season (late July). While the phylogenetic beta diversity in raw wastewater from the two locations remained similar, pond bacterial communities underwent significant (p < 0.05) changes to dominance of Comamonadaceae, Geobacteracea, and Porphyromonadaceae. Multivariate distance based redundancy analysis and predicted gene functionalities in the microbiota agreed with water quality results that microbial removal of nutrients (e.g., CBOD5) peaked in the middle of the summer coinciding with the treatment period with the highest pond temperatures. Information from this study will be useful for further development of models to predict biological treatment outcomes, which could be used to size and assess the feasibility of WSPs in extreme climates. Higher pond temperatures resulted in optimal biological processes and nutrient removal in the middle of the summer. While it is challenging to control environmental factors in a passive wastewater treatment system there are some design considerations that could be used to optimize temperature regimes, such as the depth of the pond.
Wastewater stabilization ponds (WSPs) are commonly used to treat municipal wastewater in remote arctic communities in Canada. The WSPs consist of either naturally occurring lakes with uncontrolled release or engineered structures with a lining and berm for controlled release of the water. Engineered WSPs have typically been designed to be facultative (i.e., aerobic) and to hold the annual volume of wastewater generated within the community (Ragush et al., 2015).
These WSPs are passive (i.e., non-mechanical) treatment systems that depend on physical settling and biological processes and are strongly affected by the climatic conditions including ambient temperature (Heinke et al., 1991). Operations of arctic WSPs are unique in that the content will remain frozen for 9 months of the year limiting the treatment season to the three summer months with open water and temperatures that support biological activity (Balch et al., 2018). Also, due to the low water consumption per capita (~100 L/person × day) in arctic regions such as Nunavut compared to Southern Canada (average of 274 L/ person × day), the deposited sewage tends to be more concentrated (Daley et al., 2014; Ragush et al., 2015). This can lead to high organic loading rates (OLRs) and anaerobic conditions in the facultative ponds which may affect the microbiological processes negatively (Ragush et al., 2017). Past studies have shown that primary treatment can be attained in arctic WSPs with removal of around 25–40% of nutrients, oxygen demanding material, suspended solids, and 99% of fecal indicator Escherichia coli (Marais, 1974; Heinke et al., 1991), even in WSPs with high OLRs and anaerobic conditions (Ragush et al., 2015; Huang et al., 2018).
It is well-known that microbial communities play an important role in biological wastewater treatment (Wagner et al., 2002). Insights in the dynamics of the wastewater microbiome during treatment can therefore be useful in determining which organisms are present and how they relate to the treatment of the wastewater for the benefit of future process optimization.
Studies of full scale mechanical wastewater treatment facilities show that low temperature generally reduces the bacterial diversity and slows down the growth and metabolic activity of functional bacteria to the extent where the treatment is affected. Rodriguez-Caballero et al. (2012) observed that low wastewater temperatures during the winter (10–15°C) caused difficulties in maintaining the required rates of nitrogen removal in biological wastewater treatment plants (WWTPs) operated in Northern Sweden. Another cold-climate study (Ju et al., 2014) found that the relative abundances of functional groups in activated sludge communities were shaped by seasonal temperature changes, i.e., when the temperature dropped during winter season, the abundances of nitrite-oxidizing bacteria (NOB) and hydrolysers decreased, leading to the incomplete nitrification and hydrolysis of organic material. The relative temperature-driven reduction in microbial diversity was also observed in a Finnish study conducted in the seven Northern communities, where wastewater was treated in bioreactors operated at low temperatures of 3–7°C resulting in good nutrient removal but variable performance for nitrogen removal (Gonzalez-Martinez et al., 2018).
In terms of microbial communities related to treatment in arctic WSPs, Gromala et al. (2021) recently examined water samples from WSPs located in Baker Lake (Qamani'tuaq), Cambridge Bay (Iqaluktuuttiaq), and Kugluktuk (Qurluktuk), all located in Western Nunavut, and reported temporal and spatial differences in the abundance of major bacterial phyla that were relatable to specific WSPs and the time after the Spring thaw. However, studies of the structure and function of microbial communities in relation to the treatment performance of arctic WSP systems throughout the summer treatment season have to the best of our knowledge not been performed.
The objective of this study was to investigate the temporal and spatial changes in the structure and putative function of bacterial communities and their link to treatment performance during treatment of municipal wastewater in arctic WSP systems located in the communities of Pond Inlet (Mittimatalik) and Clyde River (Kangiqtugaapik) in Nunavut, Canada. This study was conducted over two summer treatment seasons (June to early-September) and involved monitoring microbial populations in influent raw wastewater and the progressively treated water in the WSPs as well as selected environmental and water quality parameters. Information about the microbiology of these systems can help in understanding passive, biological wastewater treatment systems operated in cold climates.
From June, 2013 to September, 2014, six and five sampling trips were made to Pond Inlet and Clyde River, Nunavut, Canada. Both communities are located on Qikitaaluk/Baffin Island. The area's polar climate is characterized by long cold winters (~9 months from mid-September to May) and short cool summers (June to mid-September).
Domestic wastewater is collected daily or several times a week from holding tanks located inside buildings (homes, library, schools, health clinic, municipal offices, etc.) and trucked to the WSPs for treatment. Wastewater is loaded into the WSPs throughout the year, however, treatment mainly occurs after the Spring thaw and during summer months, where the content of the WSPs is liquid. None of the communities have any industries or agricultural activities that would contribute to the wastewater.
Pond Inlet (72° 41′ 57″ N, 77° 57′ 33″ W) has a population of 1612 (Statistics Canada, 2012). The average daily temperature is −34°C in February (coldest month) and 6°C in July (warmest month, Environment Canada, 2014a). The community's WSP system consists of one engineered pond with a surface area of ~40,000 m2 and an average depth of ~1.9 m during the summer. The treated wastewater (effluent) is discharged once annually in September by pumping the water over the berm after which it goes through a 500 m channel and into the ocean (Eclipse Sound). The annual estimated wastewater generation for the community is 8.0 × 107 L (140 L/person × day).
Clyde River (70° 28′ 26″ N, 68° 35′ 10″ W) has a population of 1004 (Statistics Canada, 2012). The community experiences average daily temperatures of −30°C in February (coldest month) and 5°C in July (warmest month, Environment Canada, 2014b). The WSP system in Clyde River consists of a small primary pond (6,000 m2) and a larger secondary pond (15,000 m2). The estimated annual wastewater volume is 3.1 × 107 L (88 L/person × day). This two-cell WSP system is designed to be operated in a staged manner, where the raw wastewater is deposited in the primary pond. The pre-processed wastewater would then be transferred into the secondary pond for further treatment before the annual decant in September, where the water is pumped into a wetland area that leads into the ocean (Patricia Bay). In September 2013, operations deviated as untreated wastewater was deposited in both ponds.
On each visit, grab samples of wastewater were obtained from the wastewater trucks (i.e., raw wastewater, n ≥ 3), and from different locations (n = 2–5) and at varying depths (0.1, 1.1, and 1.9 m) in the ponds to examine spatial variability. Effluent samples were also obtained during the decant (n = 3) in Pond Inlet or from the secondary pond (n = 3) in Clyde River just prior to the annual decant.
Samples from the Pond Inlet WSP were obtained in the beginning (late June/early July, just after the Spring thaw,), middle (late July/early August), and end (early/middle September, time of the annual decant which is just before to the freeze-up) of the summer treatment season in 2013 and 2014. Sludge samples (one composite sample per visit) were also obtained from the Pond Inlet WSP in 2014.
Samples from Clyde River were collected three times in 2013, while sampling in 2014 took place at the beginning and end of the summer just prior to the decant event.
Wastewater samples were collected in 1-L or 500-mL sterile containers (Nalgene, Fisher Scientific, Nepean, ON, Canada) from an inflatable boat or from the shore of the pond using a subsurface pole sampler and then stored in a cooler (4°C). Within 24 h of the sampling event, samples were flown to the Northern Water Quality Laboratory (NWQL) at the Nunavut Research Institute in Iqaluit, NU, for further processing.
Deployment of multi-parameter sondes (YSI Inc., Yellow Spring, OH) allowed for in-situ hourly measurements of wastewater temperature, pH, and dissolved oxygen (DO). Sondes were installed in the WSPs (0.1 m below the surface) during the first sampling trip and retrieved at the end of each treatment season.
Grab samples were analyzed for the content of carbonaceous biological oxygen demand (CBOD5), total ammonia (TA), total phosphorus (TP), total nitrogen (TN), and total suspended solids (TSS) using standard methods. Briefly, CBOD5 was analyzed in duplicate following the APHA standard method 5210 B (American Public Health Association, 1999). Dissolved oxygen was measured using a probe (Orion 83005MD, Fisher Scientific, Ottawa, ON, Canada) and an Orion StarTM series meters (Fisher Scientific). The APHA standard methods 2540 D (American Public Health Association, 1999) with Whatman TM934-AH 47 mm glass fiber filters (Fisher Scientific) were used for measurements of TSS. Measurements of total ammonia (TA) was done using a Thermo Scientific OrionTM High-Performance Ammonia Electrode with the addition of Ionic Strength Adjuster (Fisher Scientific). The ascorbic acid method via Hach® TNT™ or TNT plus™ test kits was used to test TP. TN was analyzed using Hach® TN Test ‘N Tubes™ (0.5 to 25.0 mg/L N), according to the manufacturer's procedure.
Genomic DNA was extracted from samples immediately upon arrival to the NWQL. DNA was extracted from three biological replicates (separate samples) per sampling site whenever available, with two technical replicates per sample. Ten mL of each sample was centrifuged at 3,200 × g for 10 min (min) to obtain a pellet, which was subjected to genomic DNA extraction using the MO BIO PowerSoil DNA isolation kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer's instructions. Within 24–48 h, all DNA samples were transported to the laboratory at Dalhousie University in Halifax, NS in a cooler (4°C) and stored at −20°C until further analysis.
Bacterial 16S rRNA gene copy numbers were quantified in a quantitative PCR (qPCR) assay with the BACT 2 primer set (1369F: 5'-CGGTGAATACGTTCYCGG-3'; 1492R: 5'-GGWTACCTTGTTACGACTT-3') (Suzuki et al., 2000). The qPCR amplification was performed on a Bio-Rad CFX96 TouchTM Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). Each reaction consisted of: 20-μL total reaction volumes consisting of 4.0 μL of template DNA, 4.4 μL of sterile and nuclease-free water, 0.8 μL of each primer (10 μM), and 10 μL of 2× Power SYBR Green PCR master mix (ThermoFisher Scientific). The thermocycler program consisted of 10 min of initial denaturation at 95°C, followed by 40 cycles of 15 s denaturation at 94°C, 30 s annealing at 55°C, and 30 s extension at 72°C. Melt curve analysis (30 s at 55°C and then increasing by 0.5°C each cycle until reaching 95°C) showed that the melting temperature of the amplicon was 84.5 ± 0.5°C.
A standard curve was constructed using 10-fold dilutions of a plasmid DNA extracted from an Escherichia coli DH5α culture containing a positive control plasmid (pCR2.1, TOPO TA PCR 2.1 with an insertion of the 16S rRNA fragment, gift from Dr. C. Yost, University of Regina, Canada), with triplicate measurements of diluted standards. Assay efficiency was calculated to be 102%. The coefficient of determination values (R2) of the standard curve was 0.999, the limits of detection (LOD) and quantification (LOQ) were 68.6 and 686 copies/reaction, respectively. Absence of PCR inhibitors was confirmed by comparison of the threshold cycle (Ct) values for 10-fold diluted DNA extracts.
The V6-V8 regions of the bacterial 16S rRNA gene amplicon library preparation and Illumina MiSeq Paired-End (PE) sequencing were performed following the established protocol (Comeau et al., 2017) at the Dalhousie University Integrated Microbiome Resource (IMR; http://cgeb-imr.ca). The demultiplexed PE reads were obtained in the fastq format from IMR and subjected to the bioinformatics analysis workflow briefly described in the following section.
The raw PE sequencing reads were preprocessed by stitching PE reads and removing low quality reads in PEAR (Zhang et al., 2014), and removing chimeras in VSEARCH (version 1.11.1, 28) in the Microbiome Helper pipeline at IMR (Comeau et al., 2017). As a result, an average of 73.1% of the reads (range 49.6–94.6% for individual samples) was generated as final high-quality sequences. The final high-quality sequences were then clustered into Operational Taxonomic Units (OTUs) at an identity level of ≥97% using the de novo UPARSE-OTU algorithm in the USEARCH pipeline (Edgar, 2013). OTUs from UPARSE were deposited in the Microbiome Helper pipeline to filter out spurious OTUs (Comeau et al., 2017), resulting in a total of 2,093 OTUs (from 139 samples). Subsequent analyses were carried out using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline version 1.9.1 (Caporaso et al., 2010a), unless otherwise noted. The taxonomic assignment was performed in the Ribosome Database Project (RDP) classifier version 2.2 (Wang et al., 2007) based on the Greengenes database (version 13_8) at the cut-off value of 60% (Claesson et al., 2009). The rarefied OTU table was generated by subsampling a minimal number of reads (10,539 sequences per sample), resulting in a total of 1,924 OTUs. The sequence alignment was built using the Python Nearest Alignment Space Termination (PyNAST) tool (Caporaso et al., 2010b). This alignment was used to build a phylogenetic tree in FastTree (version 2.1.10, http://www.microbesonline.org/fasttree/) which infers approximately-maximum-likelihood phylogenetic trees (Price et al., 2010). The rarefied OTU table and the phylogenetic tree were used in the downstream analysis, which included determining the core bacterial community that requires the core OTUs to be present in 100% of the samples (Huse et al., 2012), alpha-diversity measures (number of the observed OTUs, Chao1, the Shannon-Wiener (SWI) and the Simpson evenness indexes), and phylogenetic beta-diversity measures, respectively. The phylogenetic beta-diversity was calculated using both the weighted and unweighted UniFrac metrics (Lozupone and Knight, 2005). Principal Coordinate Analysis (PCoA) and an Unweighted Pair Group Method with Arithmetic Mean (UPGMA) tree were used to visualize the diversity trends based on the weighted UniFrac beta-diversity distance matrix. The trends revealed by the unweighted UniFrac analyses resembled the weighted UniFrac results and are therefore not presented. GenGIS (Parks et al., 2013) was applied to visualize information about bacterial communities representing different locations, times and years in a sampling map.
A distance-based redundancy analysis (dbRDA) was performed on the weighted UniFrac beta-diversity matrix and selected environmental variables for pond samples obtained from each of the two communities over the course of the 2-year study period. The models were assessed using the ANOVA function (R Core Team, 2021) to determine the significance of the environmental predictors (temperature, CBOD5, TSS, TP, TN, and TA) on the beta-diversity among communities. The dbRDA was run using the capscale function in the R package vegan (Oksanen et al., 2020) and ordination plots were generated using the ggord package (Beck, 2020).
The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt, version 1.1.0, Langille et al., 2013) in the Microbiome Helper pipeline (http://github.com/mlangill/microbiome_helper/wiki/PICRUSt-workflow) was used to predict gene contents based on 16S rRNA gene surveys. The PICRUSt analysis works from the observation that there is a good correlation between the phylogenetic information inferred from 16S rRNA marker gene sequences and genomic content when related reference genomes are available. Prior to the analysis, the accuracy of predictions is measured by the Nearest Sequenced Taxon Index (NSTI), with lower values indicating a closer relationship and availability of closely related reference genomes (Langille et al., 2013). Low NSTI values of 0.07 ± 0.01 and 0.08 ± 0.02 were calculated for wastewater samples from Pond Inlet and Clyde River, respectively. For comparison, it has been reported (Langille et al., 2013) that human-associated samples had the lowest (best) NSTI values (0.03 ± 0.02), while higher NSTI values (>0.14) were reported for other mammalian gut and soil samples. Thus, arctic municipal wastewater samples appeared to constitute a suitable data set to derive functional predictions by using PICRUSt and the recommended workflow.
Significant differences (p < 0.05) in the log 16S rRNA copy numbers/mL and alpha diversity among sample groups were tested using the non-parametric Kruskal-Wallis test performed in Prism 7 (version 7.0b, Graph Pad Software, Inc., La Jolla, CA, USA). In the study of phylogenetic beta-diversity measures, the nonparametric statistical test through permutations analysis of similarities (ANOSIM) approach was performed in the QIIME pipeline (Caporaso et al., 2010a) to test whether there were statistically significant differences among the trends that were observed in PCoA plots. In the study of inferring functional content in the bacterial communities, significant differences at a 5% level between two groups were tested using Welch's t-test with the Benjamini-Hochberg false-discovery rate (FDR) correction, while significant differences at a 5% level among three or more groups were tested using the Kruskal-Wallis H-test with FDR correction. These two tests were conducted in the statistical analysis of taxonomic and functional profiles (STAMP) software version 2.1.3 (Parks et al., 2014).
Raw wastewater samples from both communities consistently contained an average of 9.0 log 16S rRNA copies/mL, a level which showed no change (p > 0.05) over the duration of the study (data not shown). The water quality varied but tended be contain higher levels of CBOD5 and other nutrients in Pond Inlet compared to Clyde River (Table 1).
Surface pond temperatures in Pond Inlet gradually increased from 2 to 8°C to mid-season peak temperatures of 18°C followed by a decrease to below 5°C in the month of September (Figures 1A,B). The bacterial population size followed the changes in pond temperatures with midseason peaks of 8–8.9 log 16S rRNA copies/mL followed by significant (p < 0.05) decrease to 6.5–7 log 16S rRNA copies/mL at the end. The population size in 2014 was significantly (p < 0.05) higher than in 2013, which coincided with the higher pond temperatures recorded in 2014. Results from the deployed DO and pH sensors revealed negligible DO levels (<0.2 mg/L), except for two one-week mid-season spikes to measurable levels (see Supplementary Figure 1), and stable pH-values from 7.5 to 7.8 (Supplementary Figure 2). Absence of marked increases in pond pH and DO indicated that algal blooms were absent in 2013 and 2014.
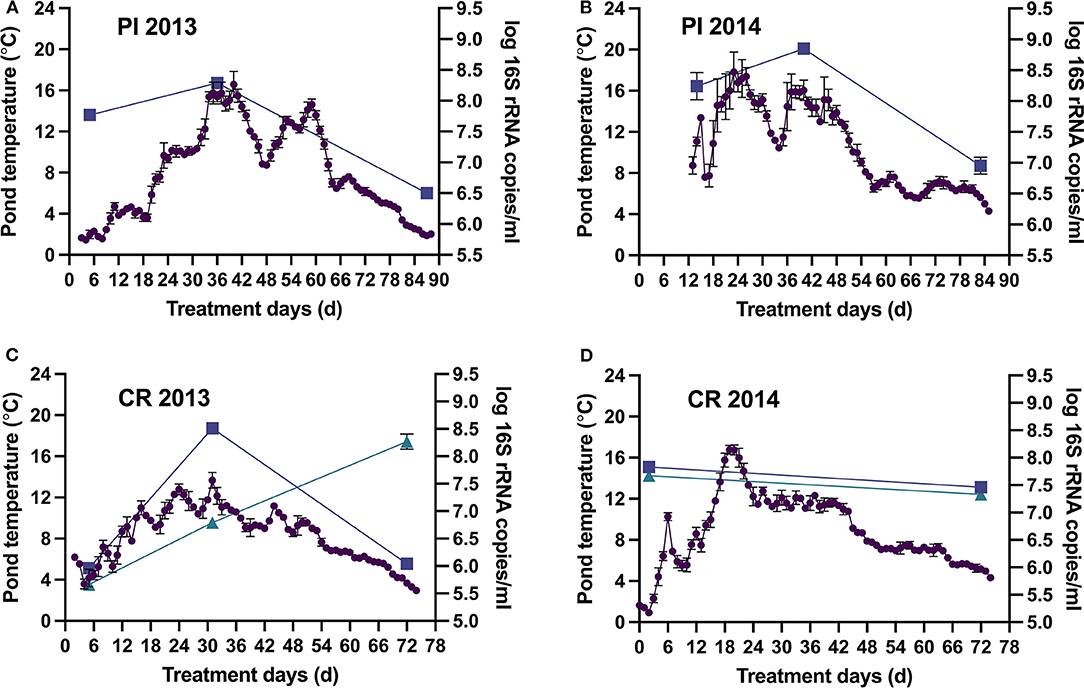
Figure 1. Pond Inlet (PI) and the Clyde River (CR) WSP system surface pond temperatures (•) and bacterial populations [log 16S rRNA copies/mL, ■ primary pond, ▴ secondary pond (only CR), mean ± standard deviation] during the 2013 (A,C) and 2014 (B,D) treatment seasons. Bacterial populations were determined at the beginning, middle, and end of treatment seasons. Temperatures are represented by daily averages of hourly measurements (n = 24, mean ± standard deviation) from the deployed sensors.
In Clyde River, the highest and lowest bacterial levels in the primary pond (~8.5 and ~6.0 log 16S rRNA copies/mL, respectively) also coincided with the highest (13.7°C) and lowest (3.7°C) pond temperatures in 2013 (Figure 1C). Normal operation of the Clyde River WSP systems in 2014 reduced populations in the secondary pond to 7.4 log 16S rRNA copies/mL at the end of the treatment season (Figure 1D). However, in September 2013, the secondary pond contained 8.3 log 16S rRNA copies/mL (Figure 1C), which was likely caused by direct discharge of raw wastewater into that pond. DO levels remained negligible (<0.2 mg/L) throughout both treatment seasons, and pH ranged from 7.5 to 7.8, indicating absence of algal blooms in both years (Supplementary Figures 1, 2).
Overall, arctic WSP treatment effected a >2 log reduction in bacterial numbers leading to an effluent with a content of 6–7.0 log 16S rRNA copies/mL ready to be released into the receiving environment (Figure 1). Water quality data showed that treatment in the WSPs removed 67–83, 84–92, 9–43, 22–59% and none to 36% of the CBOD5, TSS, TN, TP, and TA, respectively, found in the raw wastewater (Table 1). The content of CBOD5 in the ponds decreased over the summer treatment season, with the lowest values 70–90 mg/L observed at the end of the summer in the secondary pond in Clyde River as opposed to 103–115 mg/L in Pond Inlet.
The alpha diversity decreased during the treatment of the wastewater as shown by the significant (p < 0.05) decreases in the SWI from values of ~5 for the raw wastewater to values ranging between 2.5 and 4.5 in the ponds of both communities (Figure 2). Comparison of pond samples across the treatment seasons showed that the alpha diversity was lower at the start and end of the treatment season (2.5–4) while SWI values peaked at 4.3–4.5 during the warmer mid-season. The alpha diversity tended to be lower in Clyde River samples, which coincided with pond temperatures overall being colder in Clyde River compared to Pond Inlet (Figure 1). Other alpha-diversity indices (observed_OTUs and chao1) revealed the same trends (Supplementary Tables 1, 2). However, the community evenness (Simpson_e), ranged from 0.02 to 0.06 regardless of the sample type or season or year (Supplementary Tables 1, 2).
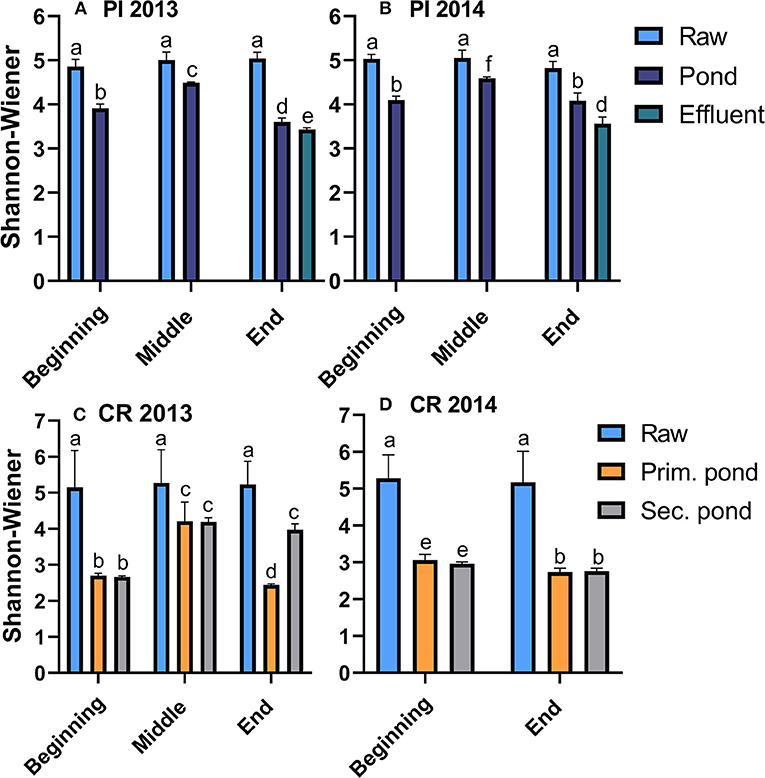
Figure 2. Alpha diversity (Shannon-Wiener Index) in raw and treated wastewater samples from Pond Inlet [PI, (A,B)] and Clyde River [CR, (C,D)] in 2013 and 2014. Bars with different letters within the same community are significantly (p < 0.05) different from each other.
The alpha diversity of bacterial community in Pond Inlet sludge samples, which were only obtained in 2014, increased over the treatment season, a change which occurred simultaneously with growing bacterial populations with final levels of 9.9 log 16S rRNA copies/mL (Supplementary Table 1).
The weighted and unweighted UniFrac measures showed that the bacterial beta diversity was not significantly (p > 0.05) different in raw wastewater samples obtained from the two communities during the study period. The raw wastewater core microbial community was comprised of 18 bacterial families mainly belonging to the Proteobacteria phylum. Dominant Proteobacteria families were Aeromonadaceae (25.2–41.8%), followed by Rhodospirillaceae (8.5–20.9%), Comamonadaceae (2.7–18.6%), Enterobacteriaceae (3.3–11.5%), Pseudomonadaceae (5.4–10.5%), and Campylobacteraceae (1.1–6.2%) (Figure 3). Within the Bacteroidetes phylum and Bacteroidia class, two families were present; Bacteroidaceae with a prevalence of 4.2–12.3% and Porphyromonadaceae with abundances ranging from 2.4 to 8.5%. The genus level composition of the core microbiomes in the raw wastewater samples is shown in Supplementary Figure 3.
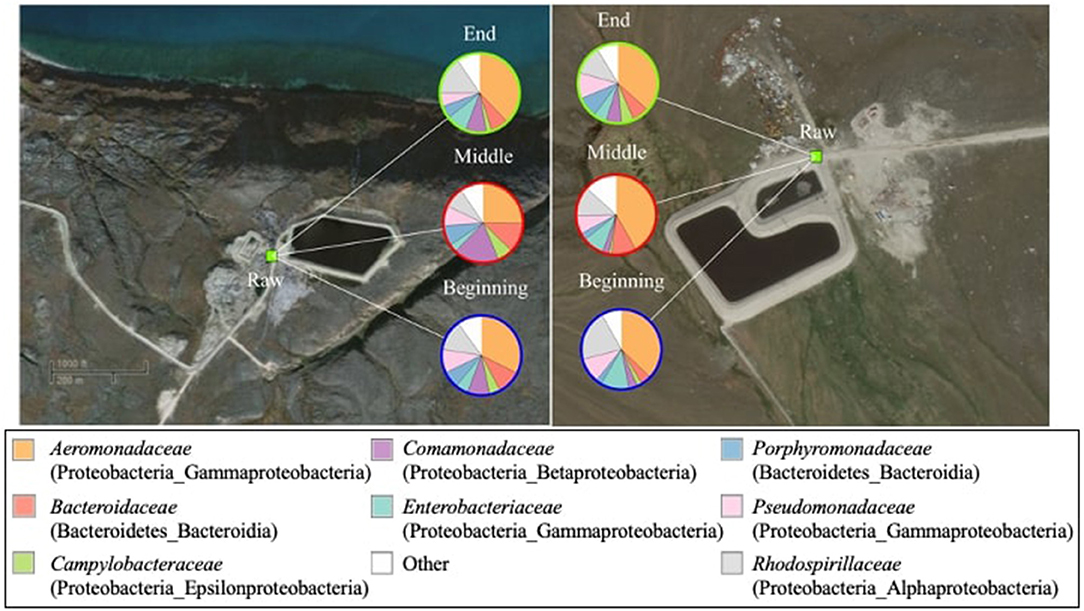
Figure 3. Composition of the core bacterial community (family level) in raw wastewater samples (n = 34 in total) from Pond Inlet (left) and Clyde River (right) obtained in 2013–2014. The relative abundances of bacterial families (top eight of 18 families) are shown in the pie charts. Legends on top of the pie charts (Beginning, Middle, and End) indicate the sampling time. Bacterial families and their associated phylum and class names are listed in brackets. Other: sum of bacterial families with relative abundances below 10%.
Treatment in the WSP systems in Pond Inlet and Clyde River significantly (p < 0.05) altered the phylogenetic beta diversity of bacterial communities. For Pond Inlet, this observation is exemplified by a comparison of raw, pond, sludge and effluent samples from September 2014, which showed different distributions of bacterial families along the treatment process (Figure 4A). Here, the Aeromonadaceae family (40.0%) dominated in the raw wastewater samples but became a minor group (1.8%) in the pond samples. Sludge samples were dominated by the Pseudomonadaceae family (29.7%). Pond and effluent samples resembled each other with the most abundant group being Comamonadaceae (~41%), Campylobacteraceae (~15%), and Geobacteraceae (~13%). The above mentioned families were represented at the genus level by Rhodoferax, Arcobacter, and Geobacter in the pond and effluent samples (Supplementary Figure 4). The resulting PCoA plot (Figure 4B) from the weighted UniFrac analysis shows that 53.3% of the variation in microbial beta diversity depended on the sample type while another 24.0% of the variation was due to the difference between raw wastewater vs. samples from the WSP. Results from Clyde River showed similar changes in the composition of bacterial communities along the WSP treatment train with Geobacteraceae, Campylobacteraceae, and Comamonadaceae making up 50% of the bacterial community in the secondary pond (Supplementary Figure 5).
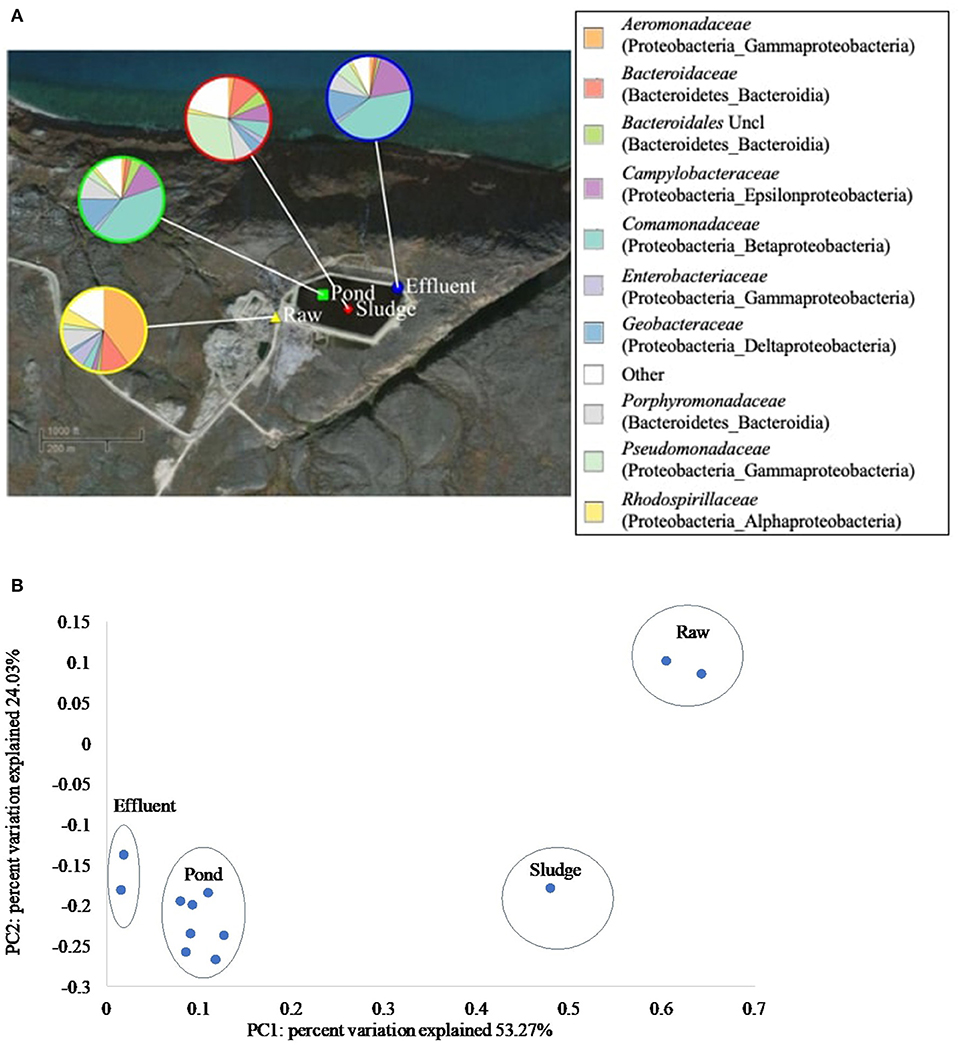
Figure 4. Composition and phylogenetic beta-diversity of bacterial communities along the Pond Inlet WSP treatment train in September 2014. (A) Relative abundance of bacterial families (top 10 of 269 families) in wastewater samples collected along the treatment process. The legend shows the bacterial families with the phylum and class names listed in brackets. Uncl: unclassified. Other: sum of bacterial families with relative abundances below 10%. (B) PCoA plot presented the phylogenetic beta-diversity of bacterial communities measured by the weighted UniFrac metric.
The time of sampling within the treatment season significantly (p < 0.05) influenced the phylogenetic beta diversity of bacterial communities in the arctic WSPs as did the sampling year. These seasonal and annual differences in pond bacterial communities are shown for Pond Inlet in Figure 5. Results from Clyde River showed similar trends and the results from 2014 are presented in Figure 6.
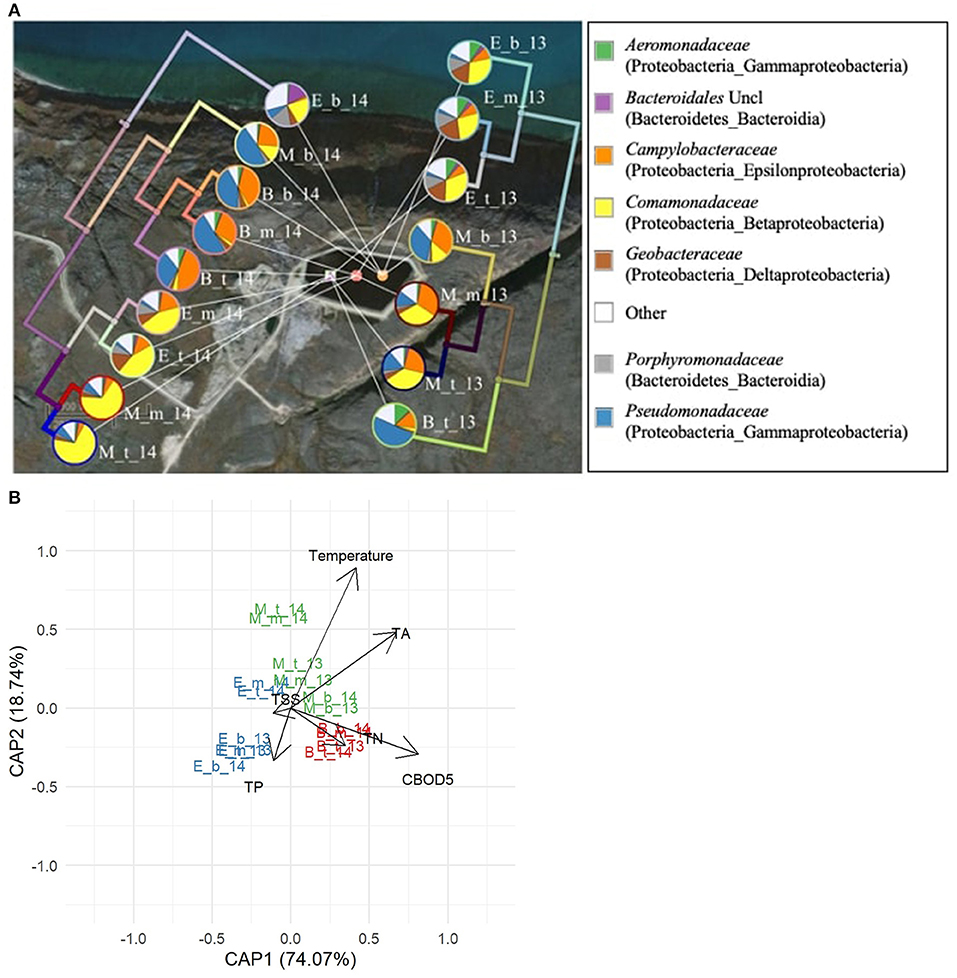
Figure 5. Bacterial communities in the Pond Inlet WSP during the 2013 (right side) and 2014 (left side) treatment seasons. (A) The seven most abundant families (relative abundances ≥ 10% among 269 bacterial families) are presented in the pie charts. The UPGMA tree shows the bacterial phylogenetic beta-diversity (weighted UniFrac metric) in each sampling year. Legends beside the pie charts: B – Beginning, M – Middle, E – End of summer treatment season; t: top, m: middle, and b: bottom sampling depths in the WSP, respectively. The legend (right) denotes the bacterial families with the phylum and class names in brackets. Uncl: unclassified. Other: sum of bacterial families with relative abundances below 10%. (B) Distance-based RDA plot showing the relationship between temperature, water quality measures and the seasonal and annual phylogenetic bacterial beta-diversity in pond samples. Significant factors (p < 0.01) were temperature, CBOD5 and TP.
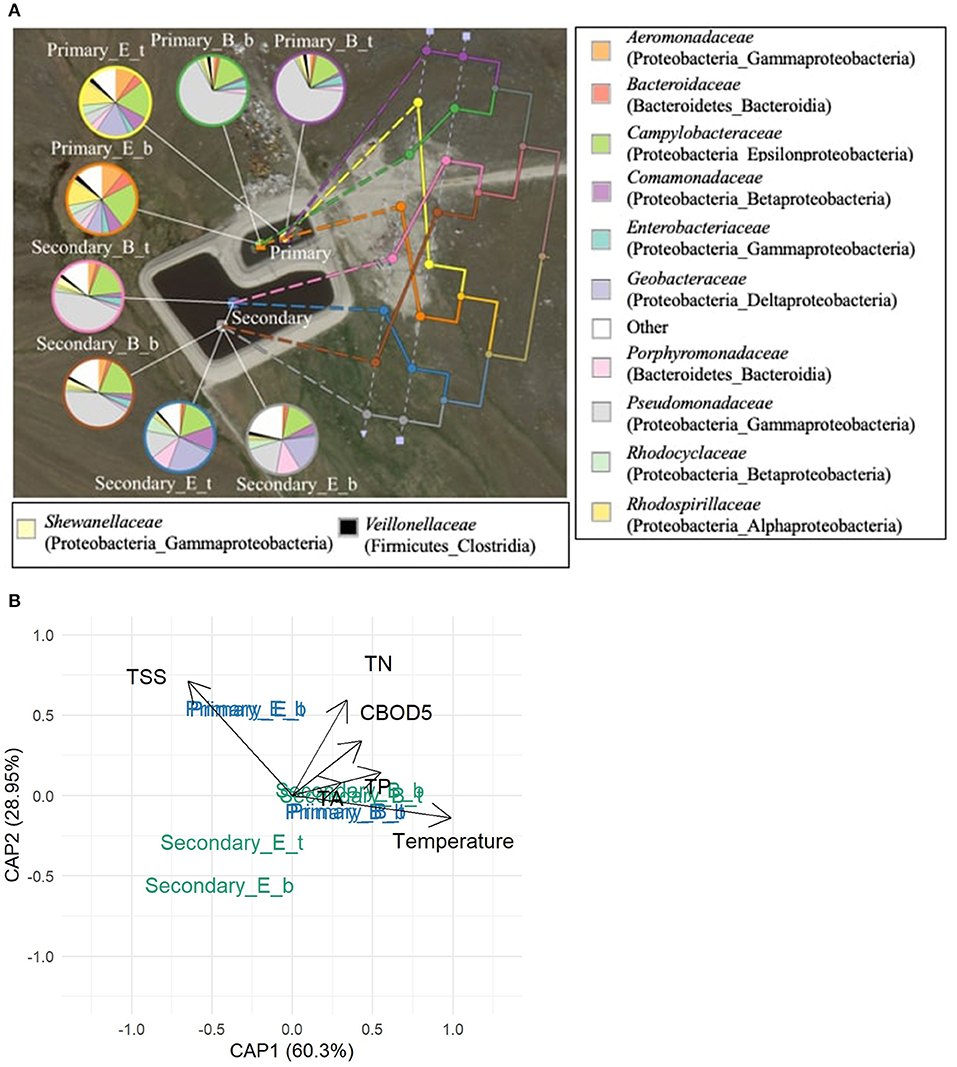
Figure 6. Bacterial communities in Clyde River WSP system during the 2014 treatment season. (A) The UPGMA tree shows the phylogenetic beta-diversity of bacterial communities measured by the weighted UniFrac metric. The 12 most abundant bacterial families (relative abundances ≥ 10%, total of 269 bacterial families) are presented in the pie charts. Legends beside the pie charts: Primary – Primary pond, Secondary – Secondary pond; B – Beginning, E – End of summer treatment season; t – top and b – bottom indicates the sampling sites in the WSP, respectively. The legend beside the map shows bacterial families: the phylum and class names are listed in the bracket of each family name. Other: sum of bacterial families with relative abundances <10%. (B) Distance-based RDA plot showing the relationship between temperature, water quality measures (TSS, TN, CBOD5, TA, TP, all significant, p < 0.05) and the seasonal phylogenetic bacterial beta-diversity in the pond samples.
In Pond Inlet in 2013, early season samples were dominated by Pseudomonadaceae (49.5%) and Campylobacteraceae (13.6%) (Figure 5A). Communities then shifted to a mid-season mix dominated by Comamonadaceae (36.8%) and Campylobacteraceae (24.5%) and finally to an end-season blend with Comamonadaceae (31.7%), Geobacteraceae (16.8%), and Porphyromonadaceae (10%) dominating. For the annual differences, the mid-season dominating family Comamonadaceae varied between 36.8 and 70.0% in 2013 and 2014, respectively. When switching to the end-season, the relative abundance of Comamonadaceae decreased to 31.7% in 2013 and 44.3% in 2014 (Figure 5A).
The dbRDA plot relating the measured environmental and water quality factors to the weighted UniFrac beta diversity among pond communities in Pond Inlet revealed that temperature, CBOD5 and TA were significant factors (p < 0.05) driving differences in the bacterial communities (Figure 5B). The CBOD5 content and Pseudomonadaceae and Campylobacteraceae correlated positively with the beginning of the treatment season, while the negative correlation at the middle and end of the treatment season showed the role of the microbiota (i.e., Comamonadaceae) in removing CBOD5, a development that correlated with the higher temperatures in the middle of the season and its effect on the bacterial communities. The decrease in TP correlated with increases in Geobacteraceae, and members of the anaerobic Bacteroidales (incl. Porphyromonadaceae) at the end of the treatment season. The dbRDA plot also revealed differences between 2013 and 2014 microbial communities obtained in the middle and end of the season supporting the marked effect of the ambient temperature on the microbiota and wastewater treatment in the Pond Inlet WSP system.
For Clyde River, Pseudomonadaceae dominated in the primary pond at the beginning of the 2014 season, while late season primary pond samples contained a diverse community including Campylobacteraceae, Geobacteraceae, Rhodopirillaceae, and Comamonadaceae (Figure 6A). A similar change from a Pseudomonadaceae dominated (about 50%) community to a highly diverse community at the end of the season was seen in the secondary pond samples, where the proportion of Porphyromonadaceae and Comamonadaceae increased together with Geobacteraceae. These seasonal trends were reflected in the phylogenic UPGMA tree with the first branch splitting at the beginning/end of season followed by a second split into the primary/secondary pond. The dbRDA plot relating the measured environmental and water quality factors to the beta diversity among pond communities in Clyde River revealed that temperature, CBOD5, TA, TSS, TN, and TP were significantly (p < 0.05) correlated to the observed differences (Figure 6B).
The UPGMA tree for Pond Inlet revealed that bacterial communities in the bottom layer differed significantly (p < 0.05) from the top or middle layers during the 2014 treatment season, but not in 2013 (Figure 5A). Interestingly, the different sampling depths in Clyde River two-cell WSP did not affect (p > 0.05) microbial community diversity in 2014 (Figure 6A).
The two most significant (p < 0.05) KEGG pathways, which were predicted to be present in the bacterial communities in WSP samples, are associated with carbohydrate and energy metabolism. As shown in Figure 7A, the predicted proportion of sequences associated with carbohydrate metabolism was significantly higher (p < 0.05) during the middle of the season compared to the beginning and end of the 2014 treatment season in Pond Inlet. The carbohydrate metabolism was predicted to be significantly (p < 0.05) higher in the bacterial community of the secondary pond in Clyde River compared to the primary pond (Figure 7B). The energy metabolism in Pond Inlet and Clyde River bacterial WSP communities followed the same trend (data not shown). The predicted abundance of genes responsible for the carbohydrate and energy metabolism in mid-season pond samples were significantly (p < 0.05) higher in 2014 compared to 2013 (data not shown).
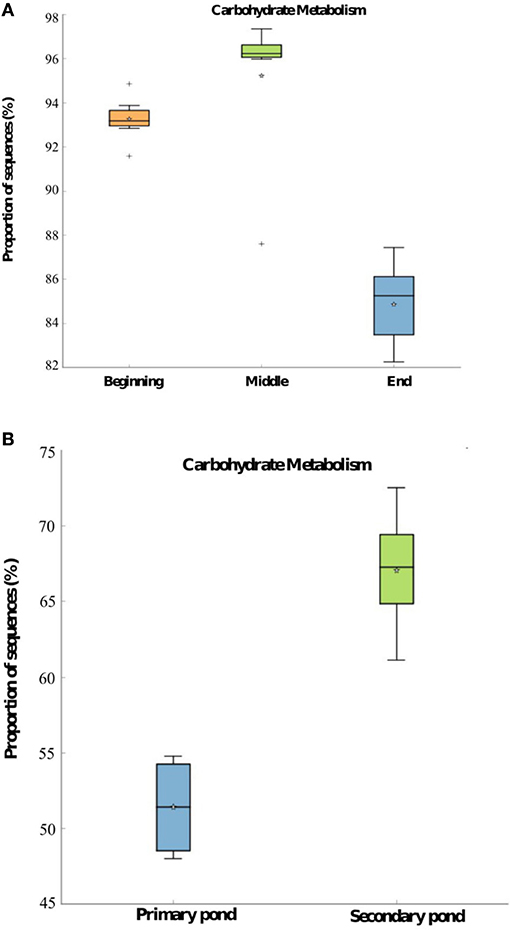
Figure 7. PICRUSt predicted abundance of genes belonging to carbohydrate metabolism with significant differences (p < 0.05) based on the mean proportions and identities of 16S rRNA genes in pond samples obtained (A) from the beginning to the end of the 2014 treatment season in Pond Inlet, and (B) from the primary and secondary pond in Clyde River at the end of the 2014 treatment season. Significant differences (p < 0.05) were detected by the Kruskal-Wallis H-test with a multiple test correction Benjamini-Hochberg FDR for the Pond Inlet analysis and the Welch's t-test with a multiple test correction Benjamini-Hochberg FDR for the Clyde River analysis. *: mean. +: Outliers.
In both Pond Inlet and Clyde River WSPs, PICRUSt identified low proportions (0.002–0.006%) of 16S rRNA genes associated with the three KEGG pathways that encode ammonia monooxygenase, an enzyme which is involved in microbial removal of ammonia. This agrees with the relative low abundance (0.01%) of both the Nitrosomonadaceae and Crenotrichaceae families, which are associated with the three pathways, in the WSPs bacterial communities.
The treatment of the wastewater resulted in removal of 70–90% of the content of TSS and CBOD5 and reduction of E. coli levels to 104−6 MPN/100 mL (Huang et al., 2018) leading to treated water qualities (effluent in Pond Inlet and water in the secondary pond at the end of the season in Clyde River), which complied with the criteria set by the licensing Nunavut Water Board of there being <104-106 E. coli MPN/100 mL, 180 mg/L TSS, and 120 mg/L CBOD5 in water released into the recipient (Nunavut Water Board, 2014). These criteria have been set in consideration of the climate, nature of the treatment systems, receiving environment and population density in Nunavut and are different from the Canadian Wastewater Systems Effluent Regulations (WSERs), which require maximum contents of 25 mg/L CBOD5, 25 mg/L TSS, and 1.25 mg/L un-ionized ammonia for wastewater effluents in Southern Canada (Environment Canada, 2015). The observed removal of nutrients is likely due to a combination of biological processes, physical settling processes, dilution (due to snow and rainfalls) and evaporation events following the disposal of the wastewater into the WSPs (Heinke et al., 1991; Ragush et al., 2015).
The bacterial communities loaded into the WSPs were similar as the influent raw wastewater contained a core set of bacteria in spite of coming from the two geographically separated arctic communities. It may be that low per capita consumption of drinking water and absence of industrial or agricultural activities in both communities led to the presence of this core bacterial community dominated by Proteobacteria (Figure 3). Treatment subsequently led to significant changes to the composition of bacterial communities and a 2-log reduction in 16S rRNA gene copies/ml in the treated water at the end of the treatment season. This reduction was similar to the 2-3 log removal of total coliform bacteria and Escherichia coli previously reported for these WSPs (Huang et al., 2018). This suggests a preferential reduction in enteric bacteria, i.e., a disinfection effect, which was supported by a shift from dominance of fecally related Aeromonadaceae, Bacteroidaceae, Campylobacteraceae, and Enterobacteriaceae in influent wastewater toward Comamonadaceae, Porphyromonadaceae (Paludibacter), and Geobacteraceae in the treated water (Figure 4 and Supplementary Figure 5). A similar reduction in the content of enteric bacteria and shift in bacterial communities was observed in a comparative study of the influent and effluent of a German WWTP (Numberger et al., 2019). The disinfection effect may be due to pond environmental factors (temperature, DO, pH, etc.), microbial completion and/or antagonism and solar irradiation although the underlying mechanisms remain unknown (Ho and Goethals, 2020).
Monitoring of the WSP environment throughout the 2013 and 2014 summer treatment season showed that the intended facultative ponds stayed anaerobic with very low levels of oxygen, neutral pH-values and therefore no signs of algae growth (Supplementary Figures 1, 2). This was different from 2011 where a reduced operational depth in the pond, yielding a lower OLR (kg/m2/day), and an unusual warm summer led to an algal bloom in the Pond Inlet WSP (Ragush et al., 2015). The lack of algae in the WSPs could affect the wastewater treatment as normal WSP treatment is based on growth of algae to supply oxygen to the aerobic decomposing bacteria, sequester phosphorous and disinfection due to the increased alkalinity of the water during algal blooms (Amengual-Morro et al., 2012; Ho and Goethals, 2020). The absence of algae growth may be related to the high OLRs and low temperatures in the ponds as demonstrated in model experiments (Ragush et al., 2017). Since the WSPs remained anaerobic and with neutral pH throughout the treatment season, temperature became an important driver of changes in the bacterial communities over the treatment season. When the pond temperatures peaked mid-season, the pond bacterial community became more abundant and diverse. This coincided with improved removal of fecal indicator and pathogenic bacteria as well as nutrients (Table 1; Huang et al., 2018), suggesting that disinfection and nutrient removal are correlated to bacterial richness and diversity in WSPs. The correlation between temperature, nutrient removal and bacterial richness and alpha-diversity was similarly observed in seasonal studies of sewage batch reactors (Ebrahimi et al., 2010) and WWTPs operated in colder climates (Rodriguez-Caballero et al., 2012; Ju et al., 2014; Kang et al., 2018). The multivariate dbRDA supported that largely anaerobic biological treatment of the wastewater was closely related to bacterial community shifts and temperature in the WSPs (Figures 5B, 6B). This potentially makes the treatment success unpredictable due to annual differences in the ambient summer temperatures. However, when comparing a cold (2013) vs. a warm (2014) year, treatment outcomes in terms of effluent CBOD5, TSS, and TA were still comparable (Table 1), indicating some robustness of the biological processes as well as concurrent impact of the physical processes.
Decomposing microorganisms with a wide range of metabolisms are important for removal of CBOD5 and other nutrients in WSP systems (Wagner et al., 2002; Ferrera and Sánchez, 2016; Ho and Goethals, 2020). Bacterial community changes along the treatment process (raw to pond, and season) led to reductions in alpha diversity and increases in Comamonadaceae, Geobacteraceae, and Porphyromonadaceae. Similar impacts of the treatment process were observed in the study of several full-scale municipal WWTPs (Hu et al., 2012; Gao et al., 2016; Numberger et al., 2019; Xue et al., 2019). The members of the bacterial communities were not evenly composed in the arctic WSP systems as indicated by low Simpson evenness measures (0.02–0.06) in all samples. However, the unevenness of bacterial community composition in arctic WSPs was not unexpected as Proteobacteria was by far the most abundant bacterial phylum with a relative abundance of 70–90% in all samples. The same dominance of Proteobacteria was also reported by Gromala et al. (2021) for three arctic WSPs located in Western Nunavut and in seven Finnish WWTPs operated above the Arctic Circle (Gonzalez-Martinez et al., 2018). Members of this bacterial phylum are known to be responsible for removal of the organic waste in municipal wastewater (Wagner et al., 2002). Comamonadaceae (member of the Burkholderiales order in Betaproteobacteria) was the most dominant group in pond and treated effluent samples from Pond Inlet and was in the dbRDA related to removal of nutrients (CBOD5). This family of bacteria has previously been reported to be involved in the biological nutrient removal processes, especially carbon and nitrogen containing compounds when the oxygen supply was limited (Sato et al., 2016; Usharani, 2019) as was the case in the present study. At the genus-level, Rhodoferax spp. (Comamonadaceae) became the dominant group in the pond and effluent samples (Supplementary Figure 4). This genus was recently reported to be present in the WSP in Baker Lake (Gromala et al., 2021), indicating its presence in the arctic sewage system, and is known to decompose organic material (Zhang et al., 2012). The Geobacter genus [Geobacteraceae, Deltaproteobacteria or the proposed Desulfobacterota (Waite et al., 2020)], which started as a minor group in the raw wastewater and rose in abundance in the WSP samples, is associated with cold anaerobic soil and aquatic environments and an ability to reduce iron and sulfate, and can complete oxidize organic compounds (Lovley et al., 1987), and was observed in the anaerobic tank of industrial wastewater treatment systems operating at low temperatures (8–16°C) in China (Zhang et al., 2017). Porphyromonadaceae and other members of the anaerobic Bacteroidetes are known to degrade complex organic substances in WWTPs (Gao et al., 2016; Zhang et al., 2017).
In 2014 samples from the Pond Inlet WSP, the bacterial diversity depended on the sampling depth, which may be due to the solids settling processes, temperature shifts between the top and bottom layer of the pond, DO gradients, and other unmonitored wastewater and operational factors. However, this depth-related difference was not observed in samples from 2013 or from Clyde River, which may indicate variable mixing of the wastewater in the ponds depending on the year and WSP design. Sludge is known to accumulate in arctic WSPs (Miyamoto and Heinke, 1979). An examination of the sludge in Pond Inlet during the 2014 treatment season revealed increases in sludge community size and diversity (Figure 4 and Supplementary Table 1), indicating the role of the sludge community in treatment of the wastewater in arctic WSPs should be the subject of further investigations.
The predicted functional gene content of wastewater bacterial communities pointed to higher levels of carbohydrate and energy metabolism in the middle of the treatment season in Pond Inlet WSP, concurring with the CBOD5 removal observed between the beginning and middle of the season in 2014 (Table 1). In the two-cell WSP system in Clyde River, the predicted higher carbohydrate and energy metabolism of the secondary pond bacterial communities similarly coincided with the further reduction of CBOD5 levels following initial treatment in the primary pond (Table 1). Gromala et al. (2021) also reported presence of KEGG pathways associated with carbohydrate and energy metabolisms in the Western Nunavut WSPs. Taken together, it seems likely that optimal CBOD5 removal can be related to higher pond temperatures, larger bacterial populations, greater bacterial diversity, and enhanced carbohydrate and energy metabolic activity levels in mid-season WSPs.
The low proportion of genes associated with ammonium oxidizing pathways matched the low relative abundance of ammonia oxidizing bacteria in both WSPs. This finding was supported by the water quality data (Table 1), which showed a modest ammonia (TA) removal in both WSPs during the study period. Interestingly, Gromala et al. (2021) reported absence of ammonia oxidation pathways in WSPs in Baker Lake, and Cambridge Bay, while the genes in the pathway were detected in Kugluktuk. These findings overall suggest that the current design of arctic WSPs does not promote growth of microbial communities, which substantially contribute to the removal of ammonia.
The study explored the composition and putative function of bacterial communities in arctic WSPs treating municipal wastewater during two consecutive summer treatment seasons. The WSPs received raw wastewater which had a high nutrient content and similar bacterial communities despite the geographical separation. Although designed to be facultative, the ponds stayed anaerobic with neutral pH-values, and temperature in the WSPs became a major determinant of the size, composition and diversity of the bacterial communities. The multivariate dbRDA showed correlations between temperature, nutrient removal and increased levels of Comamonadaceae, Geobacteraceae, and Porphyromonadaceae. The predicted gene functions (KEGG pathway study) supported that increased microbial removal of nutrients occurred mid-season and in the secondary pond of the two-cell Clyde River WSP system.
While it is challenging to control environmental factors in a passive wastewater treatment system there are some design considerations that could be used to optimize temperature regimes, such as the depth of the pond. Also, use of ponds in sequence would improve treatment. Information from this study would also be useful for further development of models that predict biological treatment, which could be used to size and assess the feasibility of WSPs in extreme climates.
The permit for this work is registered with the Nunavut Research Institute under the following title:
“Northern Municipal Wastewater Effluent Discharge Quality Objectives in the Context of Canadian Council of Ministers of the Environment Strategy and Environment Canada's Wastewater Systems Effluent.”
The sequencing dataset presented in this study has been deposited at NCBI as a Bioproject PRJNA630784. The remaining data can be found in the Supplemental Material.
YH: conception, execution of field work and experiments, data analyses, bioinformatics analysis, and writing of manuscript. CR: execution of field work, analysis of wastewater quality, and review of manuscript. LJ: statistical analyses, graphing, and review of manuscript. MH: bioinformatics analysis and review of manuscript. RB: conception, supervision, bioinformatics analysis, and review of manuscript. RJ: funding acquisition, conception, supervision, and writing and review of manuscript. LT: funding acquisition, conception, supervision, and writing and review of manuscript. All authors contributed to the article and approved the submitted version.
The Dalhousie University Northern Wastewater Research team would like to thank the Government of Nunavut, Natural Science and Engineering Research Council of Canada (NSERC): Systems Training and Education in Water Assets Research and Development (STEWARD) and the Canadian Water Network for providing funding in support of this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Our team would like to extend our gratitude to the communities of Pond Inlet and Clyde River for their support of this study. Our gratitude also goes to the Nunavut Research Institute for providing laboratory space in Iqaluit. Finally, our team members (Amy Jackson, Emmalina Corriveau, Joanna Poltarowicz, Justine Lywood, Mark Greenwood, and Meggie Letman) are sincerely thanked for their help in both laboratory and field work. The funding agencies were not involved in conducting the research or preparing this paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frwa.2021.710853/full#supplementary-material
Amengual-Morro, C., Niell, G. M., and Martínez-Taberner, A. (2012). Phytoplankton as bioindicator for waste stabilization ponds. J. Env. Man. 95, S71–S76. doi: 10.1016/j.jenvman.2011.07.008
American Public Health Association (1999). Standard Methods for the Examination of Water and Wastewater. Baltimore: United Book Press.
Balch, G., Hayward, J., Jamieson, R., Wooton, B., and Yates, C. N. (2018). “Recommendations for the use of tundra wetlands for treatment of municipal wastewater in Canada's far North,” in Multifunctional Wetlands, eds N. Nagabhatla and C. Metcalfe (Cham: Springer Nature), 83–120. doi: 10.1007/978-3-319-67416-2_3
Beck, M. W. (2020). ggord: Ordination PlotsWith ggplot2. R Package Version 1.1.5. Available online at: https://zenodo.org/record/3828862#.YQqirS271pQ (accessed July 18, 2021).
Caporaso, J. G., Bittinger, K., Bushman, F. D., DeSantis, T. Z., Andersen, G. L., and Knight, R. (2010b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinform 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010a). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Claesson, M. J., O'Sullivan, O., Wang, Q., Nikkilä, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 4:e6669. doi: 10.1371/journal.pone.0006669
Comeau, A. M., Douglas, G. M., and Langille, M. G. I. (2017). Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems 2, e00127–e00116. doi: 10.1128/mSystems.00127-16
Daley, K. D., Castleden, H., Jamieson, R., Furgal, C., and Ell, L. (2014). Municipal water qualities and health in Nunavut households: an exploratory case study in Coral Harbour, Nunavut, Canada. Int. J. Circumpolar Hlth. 73:23843. doi: 10.3402/ijch.v73.23843
Ebrahimi, S., Gabus, S., Rohrbach-Brandt, E., Hosseini, M., Rossi, P., Maillard, J., et al. (2010). Performance and microbial community composition dynamics of aerobic granular sludge from sequencing batch bubble column reactors operated at 20°C, 30°C, and 35°C. Appl. Microbiol. Biotechnol. 87, 1555–1568. doi: 10.1007/s00253-010-2621-4
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Environment Canada (2014a). Canadian Climate Normals 1981-2010 Station Data. Pond Inlet A Station, Nunavut. Available online at: http://climate.weather.gc.ca/climate_normals/results_1981_2010_e.html?stnID=1774&autofwd=1 (accessed May 3, 2014).
Environment Canada (2014b). Canadian Climate Normals 1981-2010 Station Data. Clyde River A Station, Nunavut. Available online at: http://climate.weather.gc.ca/climate_normals/results_1981_2010_e.html?stnID=1743&lang=e&dCode=0&province=NU&provBut=Go&month1=0&month2=12 (accessed July 18, 2021).
Environment Canada (2015). Wastewater Systems Effluent Regulations. Available online at: http://www.ec.gc.ca/eu-ww/ (accessed December 15, 2016).
Ferrera, I., and Sánchez, O. (2016). Insights in microbial diversity in wastewater treatment systems: How far have we come? Biotechnol. Adv. 34, 790–802. doi: 10.1016/j.biotechadv.2016.04.003
Gao, P., Xu, W., Sontag, P., Li, X., Xue, G., Liu, T., et al. (2016). Correlating microbial community compositions with environmental factors in activated sludge from four full-scale municipal wastewater treatment plants in Shanghai, China. Appl. Microbiol. Biotechnol. 100, 4663–4673. doi: 10.1007/s00253-016-7307-0
Gonzalez-Martinez, A., Sihvonen, M., Muñoz-Palazon, B., Rodriguez-Sanchez, A., Mikola, A., and Vahala, R. (2018). Microbial ecology of full-scale wastewater treatment systems in the Polar Arctic Circle: archae, bacteria and fungi. Sci. Rep. 8:2208. doi: 10.1038/s41598-018-20633-5
Gromala, M., Neufeld, J. D., and McConkey, B. J. (2021). Monitoring microbial population and antibiotic reistance gene enrichment associated with arctic waste stabilization ponds. Appl. Environ. Microbiol. 87, e02914–e02920. doi: 10.1128/AEM.02914-20
Heinke, G. W., Smith, D. W., and Finch, G. R. (1991). Guidelines for the planning and design of wastewater lagoon systems in cold climates. Can. J. Civ. Eng. 18, 556–567. doi: 10.1139/l91-068
Ho, L., and Goethals, P. L. M. (2020). Municipal wastewater treatment with pond technology: Historical review and future outlook. Eco. Eng. 148:105791. doi: 10.1016/j.ecoleng.2020.105791
Hu, M., Wang, X., Wen, X., and Xia, Y. (2012). Microbial community structures in different wastewater treatment plants as revealed by 454-Pyrosequencing analysis. Bioresour. Technol. 117, 72–79. doi: 10.1016/j.biortech.2012.04.061
Huang, Y., Truelstrup Hansen, L., Ragush, C. M., and Jamieson, R. C. (2018). Disinfection and removal of human pathogenic bacteria in arctic waste stabilization ponds. Environ. Sci. Pollut. Res. 25, 32881–32893. doi: 10.1007/s11356-017-8816-9
Huse, S. M., Ye, Y., Zhou, Y., and Fodor, A. A. (2012). A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE 7:e34242. doi: 10.1371/journal.pone.0034242
Ju, F., Guo, F., Ye, L., Xia, Y., and Zhang, T. (2014). Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 6, 80–89. doi: 10.1111/1758-2229.12110
Kang, X., Wu, X., Wang, H., Zeng, X., Leng, Y., and Li, S. (2018). Illumina sequencing reveals bacterial community shift and its role in a full-scale A2O sewage treatment process at low temperatures. Desalin. Water Treat. 109, 193–204. doi: 10.5004/dwt.2018.22142
Langille, M. G., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Lovley, D. R., Stolz, J. F., Nord, G. L., and Phillips, E. J. P. (1987). Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature 330:252–254. doi: 10.1038/330252a0
Lozupone, C., and Knight, R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005
Marais, G. V. R. (1974). Fecal bacterial kinetics in stabilization ponds. J. Environ. Eng. 100, 119–139. doi: 10.1061/JEEGAV.0000138
Miyamoto, H. K., and Heinke, G. W. (1979). Performance evaluation of an arctic sewage lagoon. Can. J. Civ. Eng. 6, 324–328. doi: 10.1139/l79-033
Numberger, D., Ganzert, L., Zoccarato, L., Mülhdorfer, K., Sauer, S., Grossart, H.-P., et al. (2019). Characterization of bacterial communities in wastewater with enhance taxonomic resolution by full-length 16S rRNA sequencing. Sci. Rep. 9:9673. doi: 10.1038/s41598-019-46015-z
Nunavut Water Board (2014). Public Registry FTP. Nunavut Water Board. Available online at: https://www.nwb-oen.ca/content/public-registry (accessed January 5, 2014).
Oksanen, J., Guillaume Blanchet, F., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al (2020). vegan: Community Ecology Package. R package version 2.5-7. Available online at: https://CRAN.R-project.org/package=vegan (accessed July 18, 2021).
Parks, D. H., Mankowski, T., Zangooei, S., Porter, M. S., Armanini, D. G., Baird, D. J., et al. (2013). GenGIS 2: geospatial analysis of traditional and genetic biodiversity, with new gradient algorithms and an extensible plugin framework. PLoS ONE 8:e69885. doi: 10.1371/journal.pone.0069885
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. J. Bioinform. 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
R Core Team (2021). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/ (accessed July 18, 2021).
Ragush, C. M., Poltarowicz, J. M., Lywood, J., Gagnon, G. A., Truelstrup Hansen, L., and Jamieson, R. C. (2017). Environmental and operational factors affecting carbon removal in model arctic waste stabilization ponds. Ecol. Eng. 98, 91–97. doi: 10.1016/j.ecoleng.2016.10.031
Ragush, C. M., Schmidt, J. J., Krkosek, W. H., Gagnon, G. A., Truelstrup Hansen, L., and Jamieson, R. C. (2015). Performance of municipal waste stabilization ponds in the Canadian Arctic. Ecol. Eng. 83, 413–421. doi: 10.1016/j.ecoleng.2015.07.008
Rodriguez-Caballero, A., Hallin, S., Påhlson, C., Odlare, M., and Dahlquist, E. (2012). Ammonia oxidizing bacterial community composition and process performance in wastewater treatment plants under low temperature conditions. Water Sci. Technol. 65, 197–204. doi: 10.2166/wst.2012.643
Sato, Y., Hori, T., Navarro, R. R., Habe, H., Yanagishita, H., and Ogata, A. (2016). Fine-scale monitoring of shifts in microbial community composition after high organic loading in a pilot-scale membrane bioreactor. J. Biosci. Bioeng. 121, 550–556. doi: 10.1016/j.jbiosc.2015.10.003
Statistics Canada (2012). Census Profile. 2011 Census. Statistics Canada Catalogue no. 98-316-XWE. Ottawa. Available online at: http://www12.statcan.gc.ca/census-recensement/2011/dp-pd/prof/index.cfm?Lang=E (accessed December 30, 2016).
Suzuki, M. T., Taylor, L. T., and DeLong, E. F. (2000). Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66, 4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000
Usharani, B. (2019). Metagenomics study of the microbes in constructed wetland system treating sewage. Int. Lett. Nat. Sci. 74, 26–48. doi: 10.18052/www.scipress.com/ILNS.74.26
Wagner, M., Loy, A., Nogueira, R., Purkhold, U., Lee, N., and Daims, H. (2002). Microbial community composition and function in wastewater treatment plants. Ant. Leeuwenhoek 81, 665–680. doi: 10.1023/A:1020586312170
Waite, D. W., Chuvochina, M., Pelikan, C., Parks, D. H., Yilmaz, P., Wagner, M., et al. (2020). Proposal to reclassify the proteobacterial class Deltaproteobacteria and Oligoflexia and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 70, 5972–6016. doi: 10.1099/ijsem.0.004213
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Xue, J., Schmitz, B. W., Caton, K., Zhang, B., Zabaleta, J., Garai, J., et al. (2019). Assessing the spatial and temporal variability of bacterial communities in two Bardenpho wastewater treatment systems via Illumina MiSeq sequencing. Sci. Tot. Environ. 657, 1543–1552. doi: 10.1016/j.scitotenv.2018.12.141
Zhang, B., Xu, X., and Zhu, L. (2017). Structure and function of the microbial consortia of activated sludge in typical municipal wastewater treatment plants in winter. Sci. Rep. 7:17930. doi: 10.1038/s41598-017-17743-x
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. J. Bioinform. 30, 614–620. doi: 10.1093/bioinformatics/btt593
Keywords: pond temperature, bacterial diversity, nutrient removal, cold climate, sewage microbiology, treatment season, remote communities
Citation: Huang Y, Ragush CM, Johnston LH, Hall MW, Beiko RG, Jamieson RC and Truelstrup Hansen L (2021) Changes in Bacterial Communities During Treatment of Municipal Wastewater in Arctic Wastewater Stabilization Ponds. Front. Water 3:710853. doi: 10.3389/frwa.2021.710853
Received: 17 May 2021; Accepted: 26 July 2021;
Published: 20 August 2021.
Edited by:
Jasna Kovac, The Pennsylvania State University (PSU), United StatesReviewed by:
Yun Zhou, Huazhong Agricultural University, ChinaCopyright © 2021 Huang, Ragush, Johnston, Hall, Beiko, Jamieson and Truelstrup Hansen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisbeth Truelstrup Hansen, bGl0ckBmb29kLmR0dS5kaw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.