- 1Health Informatics, Sema4, Stamford, CT, United States
- 2Division of Hematology and Medical Oncology, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 3Department of Population Health Science and Policy, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 4Molecular Oncology, Sema4, New York, NY, United States
- 5Department of Urology, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 6Medical Science, Sema4, Stamford, CT, United States
Background: Until recently there were no genome-directed therapies (GDTs) requiring next-generation sequencing (NGS) in prostate cancer. We examined whether the US approval of poly-(ADP-ribose) polymerase (PARP) inhibitors in May 2020 influenced the actionability and utilization of NGS in patients with prostate cancer.
Methods: This was a single-center, retrospective study including men with prostate cancer who received NGS testing from a single lab between 7/11/2018-7/6/2021. Clinical and testing data were derived from the electronic medical record.
Results: There were 346 patients with prostate cancer and qualifying NGS testing during the study period. Overall, 55 patients (15.9%) had qualifying homologous recombination repair (HRR) alterations for PARP inhibitor treatment. A greater proportion of alterations were actionable post-approval compared to pre-approval (22.7% vs 0%, Chi-squared p<0.001). 9 patients received olaparib during the study period. Patients receiving NGS testing after the PARP inhibitor approval were more likely to have metastatic disease than patients sequenced before the approval (74.2% vs. 41.1% Chi-squared p<0.001). Only 10.4% of patients with metastatic prostate cancer received NGS testing within 30 days of diagnosis. NGS testing was performed after a median of 1 prior line of systemic therapy. The median number of days between metastatic diagnosis and NGS testing was 196 (Q1-Q3: 54-832). The median time from NGS testing to the next treatment was 255 days (95% CI 151-300). These characteristics were not significantly different before or after the PARP inhibitor approval.
Conclusion: In this single-center cohort, the approval of PARP inhibitors for later-line treatment of metastatic prostate cancer increased the actionability of NGS findings but did not lead to earlier use of NGS testing.
Background
Next-generation sequencing (NGS) is increasingly relevant in urologic oncology, driven by the increasing number of genome-directed therapies (GDT) (1). However, the availability of GDTs – and hence the relevance of NGS – has been limited in some solid tumors, including prostate cancer. In a 2019 study of nearly 5,000 consecutive solid tumor specimens submitted for commercial NGS testing, only 67 (1.4%) were prostate cancers (2).
Prior to May 2020, there were no US Food and Drug Administration (FDA)-approved GDTs in prostate cancer. This changed with the approval of the poly (ADP-ribose) polymerase (PARP) inhibitors olaparib (May 19, 2020) and rucaparib (May 15, 2020) for selected patients with metastatic castration-resistant prostate cancer (mCRPC). Rucaparib received accelerated approval for patients with somatic or germline BRCA1 or BRCA2 alterations based on overall response (44%) in the single-arm phase 2 TRITON2 trial (3). Olaparib was granted full approval for patients with germline or somatic alterations in homologous recombination repair (HRR) genes based on improved progression-free survival in the phase 3 PROfound trial (4).
We performed this retrospective study to assess the impact of the recent PARP inhibitor approvals on NGS utility and utilization in patients with prostate cancer at a single academic cancer center.
Methods
Study design and patients
This was a retrospective single-center study including all patients with prostate cancer seen at Mount Sinai Hospital (New York, NY) between 7/11/2018-7/6/2021 who received tumor sequencing via a Sema4 Signal NGS test (See Genomic Data for details). Patients with prostate cancer were identified based on ICD codes (185 in ICD-9 and C61 in ICD-10). Among those who received tumor sequencing, the majority (81.8%) of tests were done by Sema4 (Supplemental Figure 2). Tumor sequencing for prostate cancer was confirmed based on lab requisition forms. We limited the analysis to those receiving Sema4 sequencing tests to ensure completeness of genomic data.
The objectives of the study were to describe the clinical actionability and utilization patterns of NGS testing before and after the approval of PARP inhibitors for advanced prostate cancer. Clinical actionability was defined as the proportion of patients with genomic alterations associated with an FDA-approved therapy for metastatic prostate cancer. Actionability of alterations was tiered based on therapies available: tier 1 (FDA-approved therapies for prostate cancer), tier 2a (FDA-approved therapies for a different cancer), tier 2b (clinical trial eligibility criteria). Utilization patterns included the proportion of sequenced patients with metastatic rather than localized disease and, among patients with metastatic disease, the median number of prior systemic treatments at the time of NGS testing, the time from metastatic diagnosis to NGS testing, and the time from NGS testing to the next systemic treatment (Supplemental Figure 1).
This study was reviewed and approved by the Mount Sinai Institutional Review Board (STUDY-21-01592).
Clinical data
A de-identified clinical database was generated from the electronic medical record (EMR) using a proprietary pipeline (Sema4, Stamford, CT) combining automated data extraction via natural language processing with targeted manual review for quality control. Clinically relevant features, including patient demographics and disease characteristics, were extracted from both structured tables and unstructured clinical notes. The dataset was de-identified for research use.
The date of sequencing was considered the baseline timepoint, and clinical characteristics were extracted relative to this timepoint, unless otherwise specified. Patients were assigned to the localized or metastatic sub-cohorts based on their disease status at the time of sequencing (Supplemental Figure 2). PSA values closest to NGS testing date were selected for each patient.
Genomic data
Genomic data were generated by extracting DNA and RNA from formalin-fixed paraffin-embedded tumor specimens. Sequencing was performed by Sema4 (Stamford, CT) using a 161-gene NGS assay built on the Oncomine Comprehensive Assay v3 platform (OCAv3) (Thermo Fisher Scientific, Waltham, MA) or a the Sema4 Signal whole exome and whole transcriptome sequencing test (WES/WTS). Variants of unknown significance were not included in this analysis. Clinically actionable genes and tier description are shown in Supplemental Table 1.
Statistical methods
Descriptive statistics were summarized using counts, proportions, means, and medians as appropriate. Comparisons of 2-dimensional count data were made using Pearson’s Chi-Squared Test. The Cochran-Mantel-Haenszel Chi-Squared Test was used to test for independence of 3-dimensional count data. The Wilcoxon rank-sum test was used to compare continuous values, assuming unequal variances and non-normal distribution based on the Shapiro-Wilk test for normality. Time to event outcomes with censoring were analyzed using the Kaplan-Meier estimator. We applied the Fisher’s exact test to compare alteration frequencies between localized and metastatic cohorts.
All analyses were performed in R version 4.1.0 (5).
Results
Study cohort and patient characteristics
Out of 18,752 patients with a history of prostate cancer seen within the health system during the study period, 423 (2.3%) received NGS tumor profiling of any kind (Supplemental Figure 2). Of the patients receiving NGS, we had complete genomic data on the 346 (81.8%) who underwent Sema4 testing with either a 161-gene panel or whole-exome sequencing. This was the final analysis cohort.
The cohort was predominantly White (53%), with a median age at prostate cancer diagnosis of 65 (IQR 12). At the time of NGS testing, 182 (52.6%) of patients had localized disease, while 164 (47.4%) had metastatic disease. Patients were stratified by their NGS sequencing date relative to the date of olaparib approval (May 19, 2020). The majority of patients were sequenced in the pre-approval period (N=280, 81%) compared to post-approval (N=66, 19%) (Table 1). Cohort characteristics stratified by disease status (localized vs. metastatic) are provided in Supplemental Table 2 (6–9).
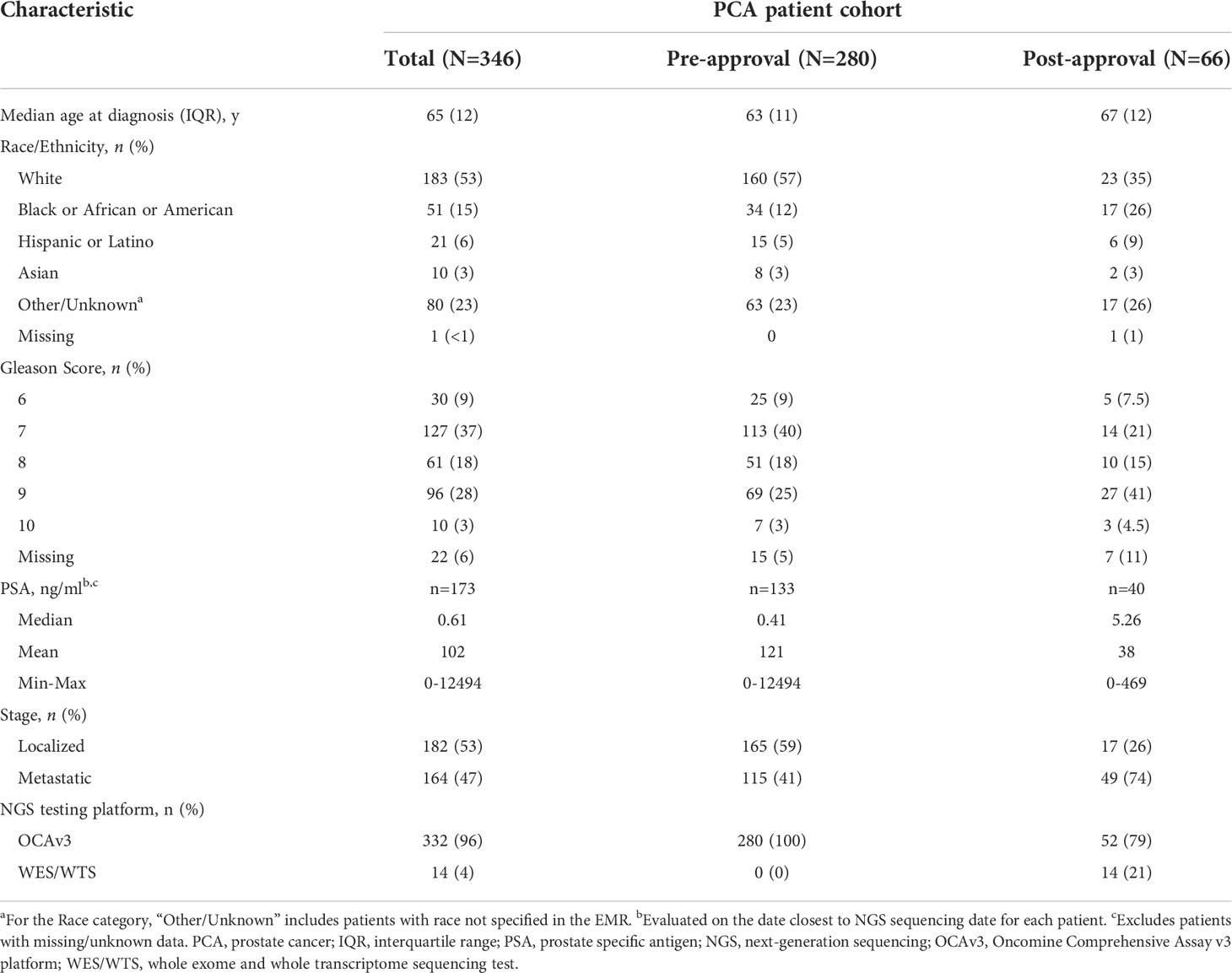
Table 1 Patient demographics and clinical characteristics, stratified by sequencing date relative to the date of olaparib approval in the United States (May 19, 2020).
NGS utilization in patients with prostate cancer
Overall, the majority of patients (N=332, 96%) were sequenced using the OCAv3 platform. Of the 14 patients who received WES/WTS, 12 were metastatic. All WES testing occurred in the post-approval cohort (Figures 1A, B).
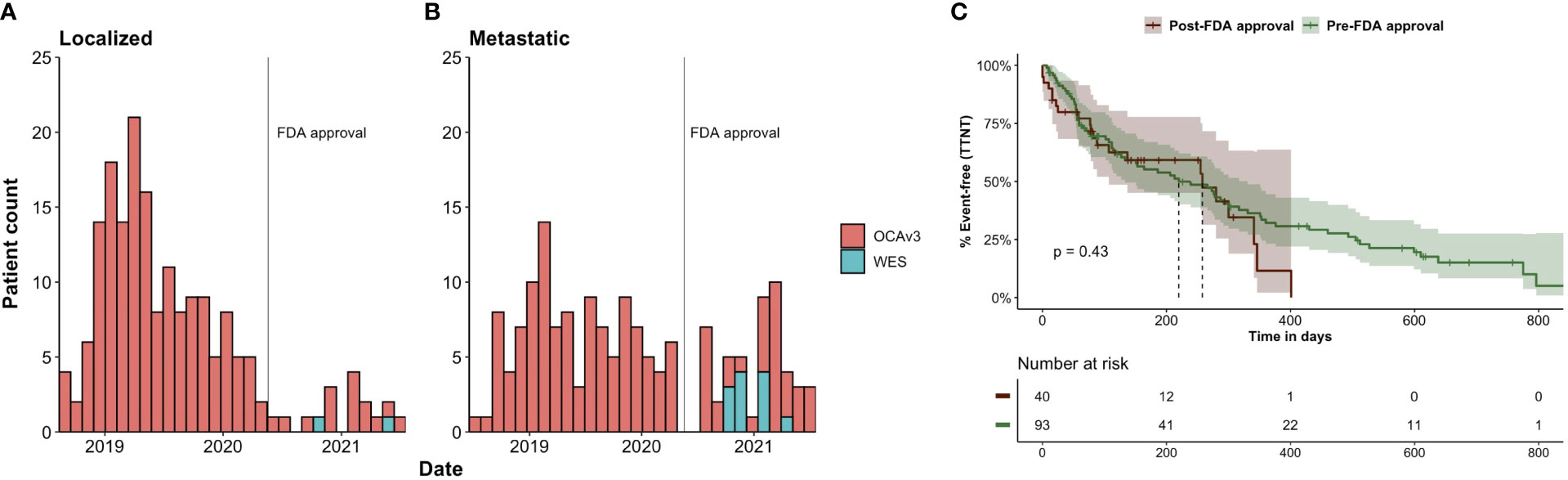
Figure 1 NGS testing date, by sequencing platform, for all patients with prostate cancer in PRODBv2 (N=346). Number of patients with (A) localized and (B) metastatic disease – at time of sequencing – who received whole exome (WES) or Oncomine (OCAv3) testing between 7/2018-7/2021. The date of FDA approval for olaparib is marked with a vertical line (May 19, 2020). (C) Kaplan-Meier plot of time from NGS testing to next treatment for patients sequenced in the pre- and post-approval periods.
Sequencing period relative to FDA approval was associated with the disease state at the time of sequencing. A greater proportion of sequencing tests were performed in patients with metastatic disease in the post-approval period than in the pre-approval period (74.2% vs. 41.1% Chi-squared p<0.001) (Table 1).
To further characterize NGS utilization in the context of metastatic disease, we examined the number of systemic treatments prior to NGS, the time from metastasis to NGS, and time from NGS to next treatment (Supplemental Figure 1). Among patients with metastatic prostate cancer (mPC) at the time of sequencing (N=164), the median number of systemic treatments prior to NGS was 1 (IQR 1 - 2) and the median time from metastasis to NGS was 196 days (Q1-Q3: 54-832). Only 10.4% of patients with mPC received NGS within 30 days of metastatic diagnosis. These quantities were not statistically different in the pre- or post-approval periods: median of 1 prior systemic therapy both pre- and post- approval (Wilcoxon rank-sum p = 0.53); median of 234 (IQR 53-809) days (pre) vs. 170 (IQR 60-1172) days (post) from metastasis to NGS testing (Wilcoxon rank-sum, p=0.8); and 11.3% (pre) vs. 8.2% (post) of patients received NGS testing within 30 days of diagnosis (Fisher’s exact p = 0.78)
To calculate time from NGS to next treatment for mPC patients, we excluded patients missing data on cancer-directed systemic treatments and patients without follow-up after NGS testing. This left 133 out of 164 mPC patients with post-NGS follow-up and known treatment status. To account for censoring (44 patients did not change treatment during follow-up), we used the Kaplan-Meier method to estimate the median time from NGS to next treatment: 255 days (95% CI 151-300) overall. Stratifying by pre/post-approval, time from NGS to next treatment was 220 days (95% CI 133-331) for the 93 patients sequenced pre-approval and 258 days (95% CI 107-NA) for the 40 patients sequenced post-approval (log-rank p = 0.43) (Figure 1C).
Genomic characteristics
Genomic alterations for selected clinically significant genes identified in >1% of localized and metastatic patients with prostate cancer are shown in Supplemental Figure 3. A complete list of alterations with patient count by stage can be found in Supplemental Table 3.
The genomic profile of our cohort was similar to previously reported cohorts: ETS family transcription factors (TMPRSS2–ERG, TP53, and PTEN) and the tumor suppressor SPOP were among the most commonly altered genes (10). We also observed an enrichment for alterations in MYC, TP53 and AR in the metastatic setting (11) and SPOP in earlier disease states (8)(Supplemental Figure 3, Fisher’s exact test p-value < 0.01).
We identified rare BRAF and IDH1 alterations with targetable potential in prostate cancer (12, 13). BRAF alterations were found in 13 patients (4% of total cohort) while IDH1 alterations were found in 7 patients (2% of total cohort). Interestingly, alterations in IDH1 were limited to the localized setting only.
The most common pathogenic genomic alterations in the localized cohort were ERG fusions (48%), followed by single nucleotide variants (SNVs) in SPOP (18%), and TP53 alterations (14%) (Supplemental Figure 3).
The most common pathogenic genomic alterations in the metastatic cohort were TP53 alterations (40%), ERG fusions (43%), and MYC gains or amplifications (16%) (Supplemental Figure 3).
Clinical actionability of NGS findings before and after PARP inhibitor approvals
Overall, 55 patients (15.9% of the whole cohort) had HRR alterations qualifying for PARP inhibitor treatment (Figure 2). The proportion of patients with HRR alterations was similar in the pre- and post-approval cohorts (14% vs. 22.7%, Chi-squared p=0.09). However, a greater proportion of alterations were actionable post-approval compared to pre-approval (22.7% vs 0%, Chi-squared p<0.001). There were an additional 138 alterations in 110 patients (31.8% of the cohort) classified as either Tier 2a (associated with an FDA-approved therapy for a different cancer) or Tier 2b (associated with clinical trial eligibility criteria) (Figure 2A).
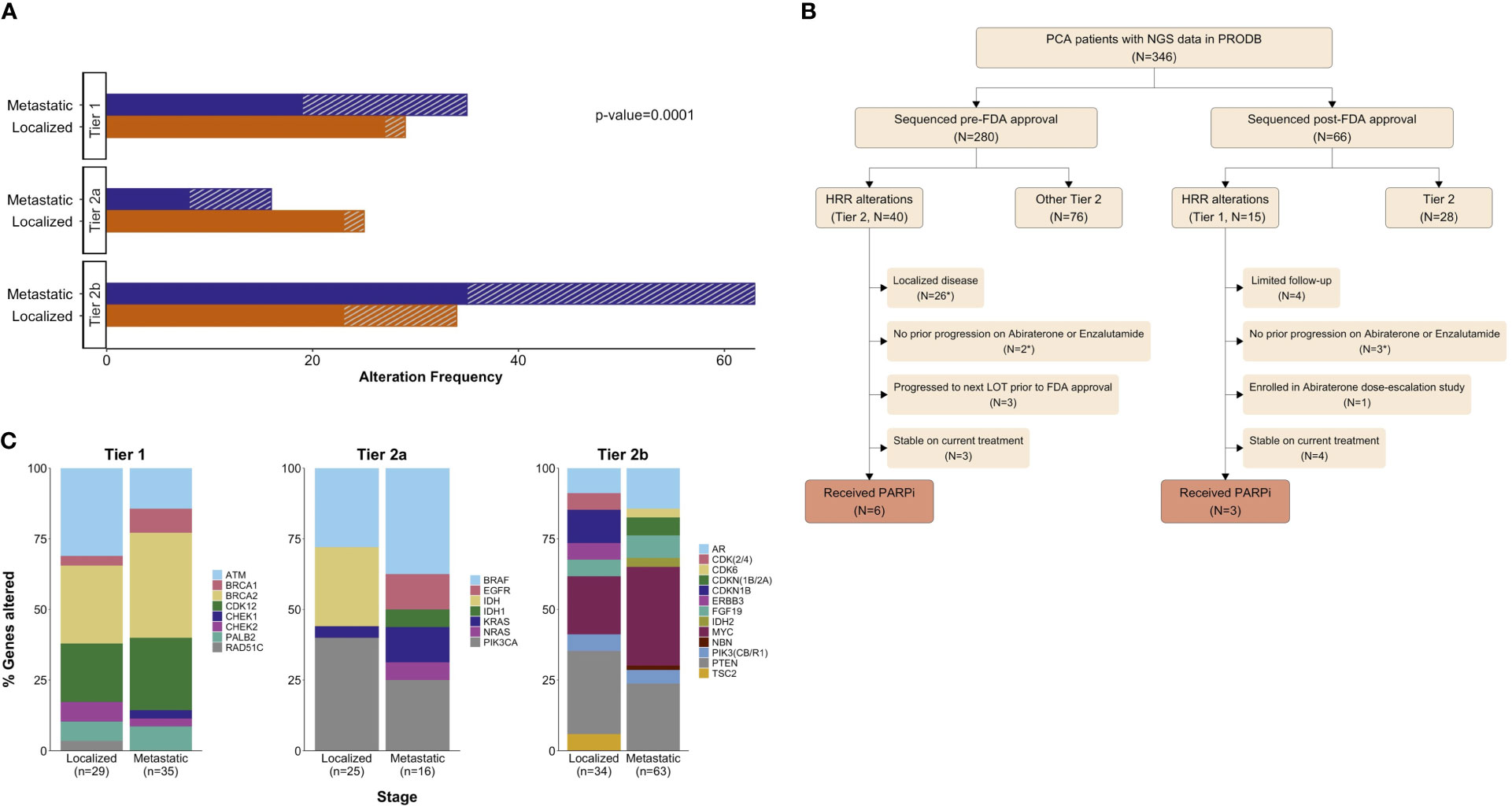
Figure 2 Clinically actionable alterations identified in 138 patients with prostate cancer (40% of total cohort). (A) Clinical actionability tier of 202 alterations (in 138 patients), stratified by stage and by sequencing period: (solid) pre- or (///) post-approval. (B) Proportions of specific genes altered within each stage and clinical actionability tier. Tier 1 = associated with an FDA-approved agent for prostate cancer, Tier 2a = associated with an off-label agent, and Tier 2b = relevant for clinical trial eligibility. For panels (A, B), clinical actionability tiers were defined based on the treatment landscape of December 2020. Patient flowchart showing clinical actionability tier (at time of sequencing) of genomic findings and patients receiving genome-directed therapy (GDT). * Indicates patients who do not meet current labelled indications for PARP inhibitor use in mPC (C).
Among the 55 patients with HRR alterations, the most common genes affected were: BRCA2 (33%), CDK12 (23%), ATM (22%), and PALB2 (8%) (Figure 2B). During the study period, 9 of these patients received GDT with Olaparib (Figure 2C). Among the remaining 46 patients with HRR alterations who did not receive GDT during the study period, 31 did not meet the labelled indication due to having localized disease (N=26) or no prior progression on abiraterone/enzalutamide (N=5). Among the 15 meeting the labelled indication: 7 were stable on current treatment, 3 progressed to the next line of treatment prior to the FDA approval, 1 enrolled on an abiraterone dose-escalation study, and 4 patients had limited follow-up (Figure 2C).
Focusing now on the 25 mPC patients with HRR alterations and adequate follow-up (Figure 3A), the median interval time from NGS to next treatment was 317 days (95% CI 239 – NA) pre-approval and 341 days (95% CI 255 – NA) post-approval (log-rank p=0.59) (Figure 3B). The most common HRR alterations were: BRCA2 (32%), CDK12 (28%), and ATM (20%). Four of these patients had biallelic alterations in BRCA2 (N=2), BRCA1 (N=1) and CDK12 (N=1) (Figure 3C; Supplemental Table 4).
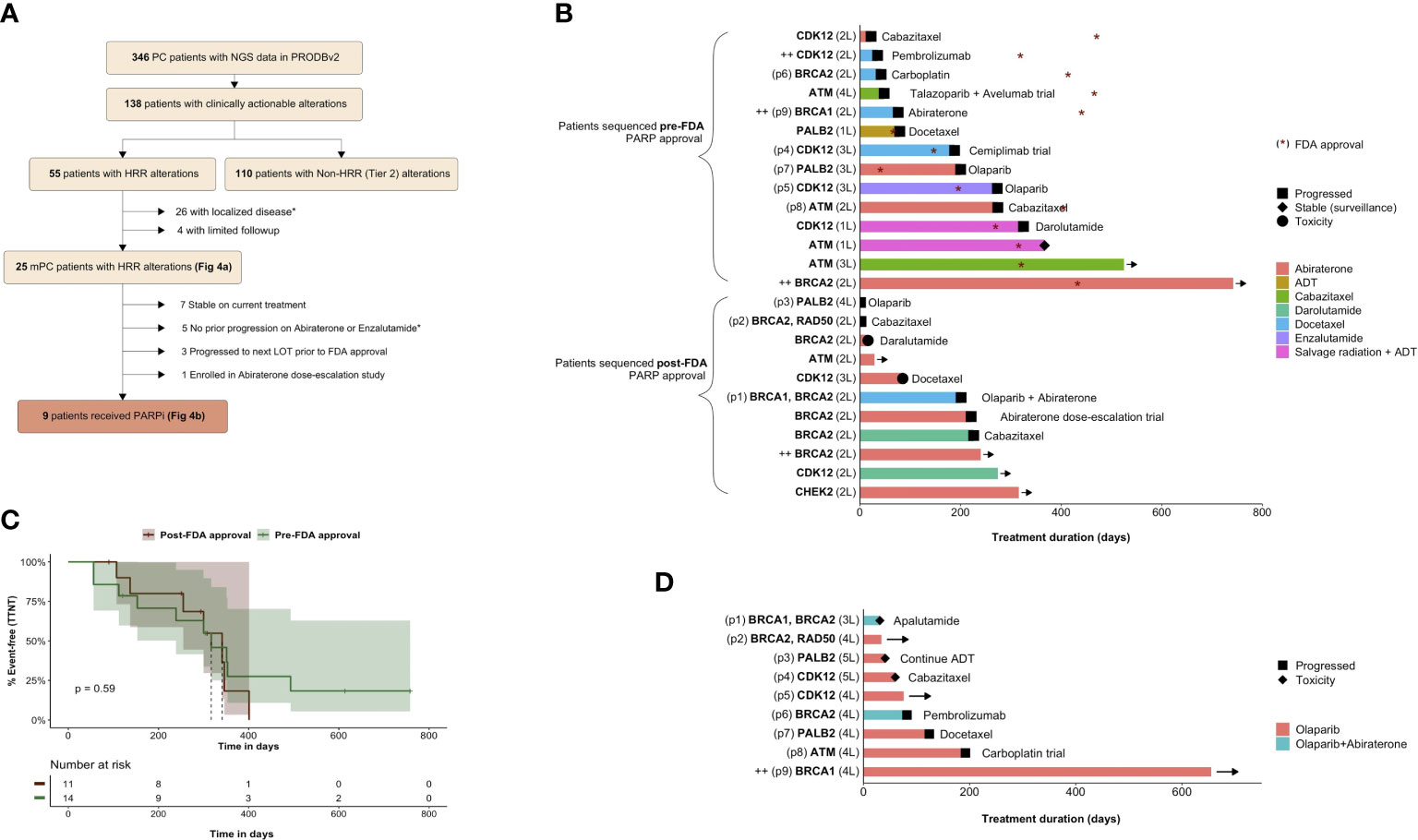
Figure 3 Treatment outcomes for patients with HRR-altered metastatic prostate cancer. Patient flow for HRR-altered patients ultimately receiving olaparib. * Indicates patients not meeting the labelled indication for Olaparib (A). Kaplan-Meier plot of time from NGS to next treatment for HRR-altered mPC patients sequenced in the pre- and post-approval periods (B). Swimmer’s plots showing treatment duration, treatment response and subsequent line of therapy (—> indicates ongoing treatment). The y-axis lists the altered gene(s) and the current line of therapy (1-5L) for individual patients. All HRR-altered mPC patients (N=25) beginning at time of NGS testing (C). Treatment response for HRR-altered mPC patients receiving genome-directed therapy (N=9; all received Olaparib) beginning at time of treatment initiation. (D). ++ indicates patients with biallelic alteration. Patients appearing in both (C) and (D) are annotated as p1-9 on the y-axis.
Olaparib usage and outcomes
Among those treated with olaparib (N=9), 3 patients harbored BRCA2 alterations (1 co-mutated with RAD50), 2 patients harbored BRCA1 alterations (1 bi-allelic and 1 co-mutated with BRCA2), 2 patients harbored PALB2 deletions, 2 patients harbored CDK12 alterations, and 1 patient harbored an ATM alteration (Figure 3D).
For these 9 patients, median lines of therapy prior to olaparib was 3 and median time to next treatment was 346 days (95% CI 239 – NA) (Figure 3D). Three patients remained on olaparib throughout the study period. Notably, one of these (p9) was an exceptional responder. This patient had biallelic BRCA1 alterations (p.V1646Sfs*12, p.H41R) with a response to off-label olaparib lasting over 600 days.
Among the 6 patients no longer on olaparib, 3 changed treatment due to toxicity while 3 changed treatment due to progression. Treatments after olaparib included chemotherapy agents (N=3; docetaxel, cabazitaxel, carboplatin), pembrolizumab (N=1), apalutamide (N=1), and ADT alone (N=1).
Discussion
“Actionability” has emerged as a key principle in debates over the clinical utility of NGS (14–20). However, the actionability of NGS is not static. Genomic variants can become actionable as novel targeted therapies are introduced (21, 22). The recent approval of PARP inhibitors for prostate cancer created an opportunity to examine changes in NGS actionability and utilization at a single academic center.
Overall, only 2.3% of patients with a history of prostate cancer had NGS testing during the study period, suggesting that NGS testing was not a routine practice for most prostate cancer patients. That said, we identified 280 cases of NGS testing prior to the approval of PARP inhibitors. This was driven by at least two factors related to the academic setting of the study. First, NGS testing may have been used to screen patients for clinical trials or to select patients for off-label PARP inhibitor therapy prior to the FDA approval (there was one notable case of durable response to off-label olaparib lasting over 600 days). Second, specific clinicians were “early adopters” of NGS testing, particularly in the localized setting. This suggests that some clinicians perceived utility in NGS testing even before the approval of PARP inhibitors, though we do not have details on the clinical decision making driving each test order.
NGS testing shifted towards metastatic patients after the approval. While we cannot attribute causation, this is consistent with clinicians focusing NGS testing on the population where NGS findings had become more actionable. However, other patterns of utilization, such as the timing of NGS testing in mPC patients, did not change significantly.
NGS testing was not a routine part of the initial evaluation for metastatic prostate cancer. Only 10.4% of mPC patients received NGS within a month of diagnosis. NGS was typically performed 6-8 months after metastatic diagnosis and usually during second-line treatment. This is unsurprising, since PARP inhibitors are currently only approved as a later-line therapy. However, recent randomized trials in mCRPC testing upfront combinations of PARP inhibitors and androgen receptor signaling inhibitors may provide the impetus for a shift towards earlier NGS testing in prostate cancer (23, 24).
Due to the effectiveness of existing mPC treatments and the reservation of PARP inhibitors for later-line treatment, the impact of NGS results on management were not immediately apparent for many patients. Among the 25 patients with mPC found to have a qualifying alteration, only 9 received a PARP inhibitor during the study period. Of the remaining 16 patients potentially eligible for a PARP inhibitor based on metastatic disease and biomarker status, 5 had yet to progress on abiraterone or enzalutamide (a prerequisite per the FDA label for olaparib), and 7 others were stable on treatment after a median of 304 days of follow-up. The approval of PARP inhibitor combinations for upfront treatment of mCRPC would make NGS results more salient earlier in the treatment course.
The limitations of this study include its single-center retrospective design, a limited post-approval sample size with limited follow-up, and a focus on somatic tumor profiling from a single vendor (though this vendor performed 81.8% of NGS testing among prostate cancer patients during this period). While some NGS testing patterns were associated with the FDA approval of PARP inhibitors, we cannot causally attribute the change in testing patterns to the approval. Other concurrent events, including the COVID-19 pandemic, may have affected NGS testing patterns (6–8, 25).
In summary, the approval of PARP inhibitors for later-line treatment of metastatic prostate cancer increased the actionability of NGS findings but did not lead to earlier use of NGS testing. Greater utilization of NGS testing in prostate cancer is expected going forward, especially as PARP inhibitors advance to earlier lines of therapy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by Mount Sinai Institutional Review Board. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
Concept: TJ; Analysis: JG; Data Acquisition and Curation: JG, C-KT, VP, BL, SG, MR, FH, SZ, AT, MG, WO, and RC; Resources: RC and WO; Writing – Original Draft: TJ and JG; Writing – Review and Editing: All authors. All authors contributed to the article and approved the submitted version.
Conflict of interest
MG reports advising and receiving research funding from AstraZeneca. JG, SG, MR, FH, XZ, WO and TJ were employed by Sema4.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fruro.2022.997396/full#supplementary-material
Supplementary Figure 1 | Schematic of NGS utilization metrics in patients with metastatic prostate cancer.
Supplementary Figure 2 | Patient cohort selection. PCA, prostate cancer; MSH, Mount Sinai Hospital; WES/WTS, whole exome and whole transcriptome sequencing test; OCAv3, Oncomine Comprehensive Assay v3 platform; hx, history; dx, diagnosis.
Supplementary Figure 3 | Clinically significant alterations identified in 303 patients with prostate cancer (88% of total cohort). Oncoprints showing alteration frequency in (A) localized and (B) metastatic patients with prostate cancer for genes altered in > 1% of patients.
Supplementary Table 1 | Clinically actionable genes. Actionability tiers as of December 2020, relevant to . Tier 1 = associated with an FDA-approved agent for prostate cancer, Tier 2a = associated with an off-label agent, and Tier 2b = relevant for clinical trial eligibility.
Supplementary Table 2 | Patient demographics and clinical characteristics for study cohort stratified by stage. aFor the Race category, “Other/Unknown” includes patients with race not specified in the EMR. bEvaluated on the date closest to NGS sequencing date for each patient. cExcludes patients with missing/unknown data. PCA, prostate cancer; IQR, interquartile range; PSA, prostate specific antigen; NGS, next-generation sequencing; WES/WTS, whole exome and whole transcriptome sequencing test.
Supplementary Table 3 | Clinically significant variants for patients with localized and metastatic prostate cancer.
Supplementary Table 4 | PARP inhibitor eligibility, variant type, and treatment response data for HRR-altered mPC patients illustrated in Figures C, D. ++, biallelic alterations; *, limited follow-up post subsequent treatment; LOT, line of therapy.
References
1. Kim JA, Ceccarelli R, Lu CY. Pharmacogenomic biomarkers in US FDA-approved drug labels (2000-2020). J Pers Med (2021) 11(3):179. doi: 10.3390/jpm11030179
2. Ikeda S, Elkin SK, Tomson BN, Carter JL, Kurzrock R. Next-generation sequencing of prostate cancer: genomic and pathway alterations, potential actionability patterns, and relative rate of use of clinical-grade testing. Cancer Biol Ther (2019) 20(2):219–26. doi: 10.1080/15384047.2018.1523849
3. Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol (2020) 38(32):3763–72. doi: 10.1200/JCO.20.01035
4. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. New Engl J Med (2020) 382(22):2091–102. doi: 10.1056/NEJMoa1911440
5. RStudio Team. RStudio: Integrated Development Environment for R. In: RStudio Boston, MA (2021). Available at: www.rstudio.com.
6. Bhalla S, Bakouny Z, Schmidt AL, Labaki C, Steinharter JA, Tremblay DA, et al. Care disruptions among patients with lung cancer: A COVID-19 and cancer outcomes study. Lung Cancer. (2021) 160:78–83. doi: 10.1016/j.lungcan.2021.07.002
7. Wu JTY, Kwon DH, Glover MJ, Henry S, Wood D, Rubin DL, et al. Changes in cancer management due to COVID-19 illness in patients with cancer in northern California. JCO Oncol Practice. (2021) 17(3):e377–85. doi: 10.1200/OP.20.00790
8. Richards M, Anderson M, Carter P, Ebert BL, Mossialos E. The impact of the COVID-19 pandemic on cancer care. Nat Cancer. (2020) 1(6):565–7. doi: 10.1038/s43018-020-0074-y
9. Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. (2019) 116(23):11428–36. doi: 10.1073/pnas.1902651116
10. Abida W, Armenia J, Gopalan A, Brennan R, Walsh M, Barron D, et al. Prospective genomic profiling of prostate cancer across disease states reveals germline and somatic alterations that may affect clinical decision making. JCO Precis Oncol (2017) 2017:1–16. doi: 10.1200/PO.17.00029
11. Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. (2015) 161(5):1215–28. doi: 10.1016/j.cell.2015.05.001
12. Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet (2020) 52(8):778–89. doi: 10.1038/s41588-020-0648-8
13. Prophet M, Xiao K, Gourdin TS, Nagy RJ, Kiedrowski LA, Ledet E, et al. Detection of actionable BRAF missense mutations by ctDNA-based genomic analysis in prostate cancer. JCO. (2018) 36:306–6. doi: 10.1200/JCO.2018.36.6_suppl.306
14. Marquart J, Chen EY, Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol (2018) 4(8):1093–8. doi: 10.1001/jamaoncol.2018.1660
15. Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open (2019) 2(5):e192535. doi: 10.1001/jamanetworkopen.2019.2535
16. Pritchard D, Goodman C, Nadauld LD. Clinical utility of genomic testing in cancer care. JCO Precis Oncol (2022) 6):e2100349. doi: 10.1200/PO.21.00349
17. Dienstmann R, Tamborero D. Genomic testing for targeted oncology drugs: hopes against hype. Ann Oncol (2021) 32(7):837–8. doi: 10.1016/j.annonc.2021.05.354
18. Beaubier N, Bontrager M, Huether R, Igartua C, Lau D, Tell R, et al. Integrated genomic profiling expands clinical options for patients with cancer. Nat Biotechnol (2019) 37(11):1351–60. doi: 10.1038/s41587-019-0259-z
19. Cobain EF, Wu YM, Vats P, Chugh R, Worden F, Smith DC, et al. Assessment of clinical benefit of integrative genomic profiling in advanced solid tumors. JAMA Oncol (2019) 7:525–33. doi: 10.1001/jamaoncol.2020.7987
20. Dunn C, Gately L, Gibbs P. Is universal next-generation sequencing testing of patients with advanced cancer ready for prime time? JAMA Oncol (2021) 7(8):1246. doi: 10.1001/jamaoncol.2021.1904
21. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J Mol Diagn. (2017) 19(1):4–23. doi: 10.1016/j.jmoldx.2016.10.002
22. Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT). Ann Oncol (2018) 29(9):1895–902. doi: 10.1093/annonc/mdy263
23. Chi KN, Rathkopf DE, Smith MR, Efstathiou E, Attard G, Olmos D, et al. Phase 3 MAGNITUDE study: First results of niraparib (NIRA) with abiraterone acetate and prednisone (AAP) as first-line therapy in patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) with and without homologous recombination repair (HRR) gene alterations. JCO. (2022) 40(6_suppl):12–2. doi: 10.1200/JCO.2022.40.6_suppl.012
24. Clarke NW, Armstrong AJ, Antoine T-V, Oya M, Shore N, Loredo E, et al. Abiraterone and olaparib for metastatic castration-resistant prostate cancer. NEJM Evidence (2022) 1:EVIDoa2200043. doi: 10.1056/EVIDoa2200043
Keywords: prostate cancer, NGS - next generation sequencing, PARP inhibitors, homologous recombination deficiency (HRD), BRCA 1/2 mutation carriers
Citation: Griffin J, Tsao C-K, Patel V, Liaw BC, Guin S, Joshi H, Rossi M, Hantash F, Zhou X, Tewari A, Galsky MD, Oh WK, Chen R and Jun T (2022) Clinical actionability and utilization of next-generation sequencing for prostate cancer in a changing treatment landscape. Front. Urol. 2:997396. doi: 10.3389/fruro.2022.997396
Received: 18 July 2022; Accepted: 13 September 2022;
Published: 06 October 2022.
Edited by:
Chia-Chang Wu, Taipei Medical University, TaiwanReviewed by:
Jure Murgic, Sisters of Charity Hospital, CroatiaSpyridon P Basourakos, NewYork Presbyterian Hospital, United States
Copyright © 2022 Griffin, Tsao, Patel, Liaw, Guin, Joshi, Rossi, Hantash, Zhou, Tewari, Galsky, Oh, Chen and Jun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomi Jun, dG9taS5qdW5Ac2VtYTQuY29t
 Jacqueline Griffin1
Jacqueline Griffin1 Che-Kai Tsao
Che-Kai Tsao Bobby C. Liaw
Bobby C. Liaw Sunny Guin
Sunny Guin Himanshu Joshi
Himanshu Joshi Feras Hantash
Feras Hantash Xiang Zhou
Xiang Zhou Matthew D. Galsky
Matthew D. Galsky William K. Oh
William K. Oh Tomi Jun
Tomi Jun