
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Urol. , 15 September 2022
Sec. Urologic Oncology
Volume 2 - 2022 | https://doi.org/10.3389/fruro.2022.967480
 Yuki Kohada1,2
Yuki Kohada1,2 Akito Kuromoto1
Akito Kuromoto1 Kazuya Takeda3
Kazuya Takeda3 Hiromichi Iwamura1,4
Hiromichi Iwamura1,4 Yuri Atobe5
Yuri Atobe5 Jun Ito1
Jun Ito1 Tomonori Kaifu3
Tomonori Kaifu3 Yasuhiro Kaiho1
Yasuhiro Kaiho1 Ichiro Nakashima5
Ichiro Nakashima5 Nobuyuki Hinata2
Nobuyuki Hinata2 Akira Nakamura3*
Akira Nakamura3* Makoto Sato1
Makoto Sato1Introduction: This study aims to investigated for the first time the role of myeloid-derived suppressor cells (MDSCs) in metastatic-hormone sensitive prostate cancer (mHSPC), which has not been investigated previously.
Materials and methods: This was a prospective observational cohort study. MDSC subsets in peripheral blood samples were classified and evaluated by flow cytometry as early-stage MDSCs (e-MDSCs), polymorphonuclear MDSCs (PMN-MDSCs), and monocytic MDSCs (M-MDSCs). The prostate-specific antigen progression free survival (PSA–PFS) and overall survival (OS) were evaluated to assess the prognostic value of each of the MDSC subsets. The immune cell dynamics and gene expression alteration were analyzed by single-cell RNA-sequencing (scRNA-seq) in a representative case.
Results: Thirty-one mHSPC patients and 11 healthy controls (HCs) were included in this study. There were significantly more PMN/M-MDSCs in mHSPC patients than in HCs (p <0.05) before treatment, but the numbers became similar to those in HCs after treatment. Although there were no marked differences in the high and low ratios of e-MDSCs and M-MDSCs, patients with a high ratio of PMN-MDSCs (≥0.30%) had a poorer PSA–PFS and OS than those with a low ratio (<0.30%) (p <0.05). scRNA-seq showed that the expression of genes implicated in tumor progression was upregulated in a representative mHSPC case.
Conclusions: A high frequency of PMN-MDSCs correlated with poor prognosis in mHSPC patients. PMN-MDSCs and their highly expressed genes are potential novel therapeutic targets for mHSPC.
Along with lung and colorectal cancer, prostate cancer (PC) is one of the three most common cancers among men. Metastatic PC accounts for only 6% of all newly diagnosed PC patients but has a poor prognosis, with a 5-year relative survival rate of only 30% (1). Furthermore, some cases of metastatic PC develop after radical cure treatment for localized PC. Indeed, in localized PC patients underwent radical prostatectomy, 6% had recurrence and developed metastatic PC at 5 years after surgery (2).
Androgen deprivation therapy (ADT), which is the primary treatment of metastatic PC, provides temporary disease control in the majority of patients (3). Within two years, however, more than half of patients with metastatic PC progress to castration-resistant prostate cancer (CRPC), which is refractory to ADT (4). The prognosis of CRPC is still poor even with other treatments, including taxane chemotherapy. Various immunotherapies widely used in other cancers have been attempted to improve the prognosis of metastatic PC patients, such as immune checkpoint inhibitors (5). However, there are still no approved therapies for PC except for the autologous dendritic cell vaccine called sipuleucel-T (6).
To establish a novel effective treatment, it is necessary to investigate how immunotherapy-resistance metastatic PC affects the host immune system. Myeloid-derived suppressor cells (MDSCs) are immunosuppressive immature myeloid cells that expanded under tumor-bearing conditions (7). MDSCs are heterogenous cells that can be divided into three representative subsets based on their phenotypic and morphological characteristics (8). Polymorphonuclear MDSCs (PMN-MDSCs) and monocytic MDSCs (M-MDSCs) are phenotypically and morphologically similar to neutrophils and monocytes, respectively. The immature subsets of MDSCs that lack monocytic and granulocytic markers are called early-stage MDSCs (e-MDSCs).
Each of these MDSC subsets are not only phenotypically and morphologically distinct, but also have unique functional characteristics and biochemical traits that suppress anti-tumor immunity in the tumor-associated immune system. For example, PMN-MDSCs have been reported to enhance angiogenesis through the production of matrix metallopeptidase (MMP)-9 and vascular endothelial growth factor (VEGF) and also to have a suppressive effect against T cells (9). M-MDSCs have been shown to differentiate into tumor associated macrophages (TAMs) after migration to the tumor site and play an immunosuppressive role (10). In head and neck canner and ovarian cancer, e-MDSCs reportedly have an immunosuppressive property, but their specific roles in other tumors are still unclear (11).
The high frequency of MDSCs in peripheral blood is reportedly associated with disease progression and a poor prognosis in many malignant neoplasms, including lung cancer and gastric cancer (12, 13). Several studies that have described the roles of MDSCs in localized PC or CRPC have mainly focused on either PMN-MDSCs or M-MDSCs, however, with no reports published concerning the involvement of the three MDSC subsets in untreated metastatic PC patients (14–17).
To investigate which of the MDSC subtypes is associated with tumor progression and a poor prognosis in pre-treated metastatic PC, we analyzed the presence of the three types of MDSCs in the periphery before and after treatment with a GnRH antagonist and clarified the correlation between each of the MDSC subsets ratios and the progression-free survival (PFS) and overall survival (OS) in metastatic-hormone sensitive prostate cancer (mHSPC). We also analyzed the immune cell dynamics and gene expression alteration by single-cell RNA-sequencing (scRNA-seq) in a representative case. We found that the PMN-MDSC fractions were correlated with a poor prognosis in mHSPC patients, and the presence of different immune cell subsets and immunosuppressive gene expression of PMN-MDSCs according to scRNA-seq suggested that PMN-MDSCs are involves in the response to ADT for mHSPC.
The present study was a prospective analysis of data acquired from patients with newly diagnosed mHSPC between 2017 and 2021. This study was approved by the clinical research ethics board of Tohoku Medical and Pharmaceutical Hospital. All patients were diagnosed with metastatic prostate cancer because metastatic findings were detected in the lymph node outside the pelvis, bone, and viscera, and the following were determined by pathological and clinical tests: biopsy Gleason score; neutrophil, lymphocyte and monocyte counts; and the levels of prostate-specific antigen (PSA), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), hemoglobin (Hb) and albumin (Alb). Patients who had not been diagnosed pathologically PC by prostatic biopsy or did not consent to this study were excluded. PSA progression was defined as a first increase in PSA greater than 25% and > 2 ng/mL above nadir based on the Prostate Cancer Clinical Trials Working Group 3 (18). PSA progression-free survival (PSA-PFS) was defined as the duration between the start of ADT to PSA progression. Patients showing normalized PSA levels after 3 months of ADT with GnRH antagonist (degarelix) were diagnosed with mHSPC and were included in this study. The patients’ characteristics are detailed in Table 1 and Supplementary (Supplementary Table 1). A total of 31 patients with untreated metastatic PC patients (median age; 73, range; 63–84, gender; 31 males) were included in this study, and 11 healthy volunteers (median age; 41, range; 24–52, gender; males, four females) served as normal controls.
Blood samples (4 mL) from the patients were drawn into heparin blood collection tubes and processed within 2 hours. After the removal of red blood cells (RBCs) using HetaSep (STEMCELL Technologies, Vancouver, Canada), the peripheral white blood cells (pWBCs; peripheral blood cells without RBCs) were enriched by Ficoll density gradient centrifugation (GE Healthcare Life Sciences, TOMY, Tokyo, Japan) at 90 x g for 4 minutes at room temperature. pWBCs were isolated from the interface and washed in 35 ml of phosphate-buffered saline (PBS). The solution was centrifuged at 120 x g for 11 minutes by density centrifugation.
The fractions of MDSCs in the peripheral blood was tested by using Flow cytometry (Attune NxT Flow Cytometer, Thermo Fisher Scientific, Waltham, MA). Flow cytometry was conducted using the following antibodies (8): anti-CD3-FITC (UCHT1, eBioscience, San Diego, CA.), anti-CD11b-Super Bright436 (ICRF44, eBioscience), anti-CD14-PE-Cy5 (61D3, eBioscience), anti-CD15-Brilliant Violet711 (W6D3, Biolegend, Sandiego, CA.), anti-CD19-FITC (HIB19, Tonbo Biosciences, Sandiego, CA.), anti-CD33-PE (WM53, eBioscience), anti-CD45-PerCP/Cy5.5 (HI30, Biolegend), anti-CD56-FITC (TULY56, Thermo, Tokyo, Japan), anti-HLA-DR-APC-eFluor780 (LN3, eBioscience). The data of 10,000–100,000 events were analyzed using the FlowJo software programs (BD bioscience, San Jose, CA). The MDSC subsets were defined as following surface markers (7): M-MDSCs; CD11b+CD15−CD14+HLA-DR−, PMN-MDSCs; CD11b+CD14−CD15+, e-MDSCs; CD11b+CD33+CD15−CD14−Lineage (CD3/19/56)− HLA-DR−. The proportions of each MDSC subsets calculated using CD45-positive cell fraction.
An scRNA-seq of gene expression and immune cell profiling analysis were performed using the 10X Genomics Chromium Single Cell platform according to the manufacturer’s instructions. Briefly, RBC removal pWBCs were capsulized using microfluidics technology, and were then barcoded by a unique molecular identifier (UMI). Complementary DNA was sequenced on an Illumina 3000 HiSeq system. Raw reads were aligned to the GRCh38 human transcriptome using STAR (Spliced Transcripts Alignment to a Reference) and gene expression was quantified by the Cell Ranger software package (version 5.0.0) with default parameters. The total number of cells passing the filters was 3,885 cells for the patient and 4,800 cells for the healthy subject, with 21, 697 and 21, 280 genes, respectively, which were passed the quality control check. We compressed the dimensions using T-distributed stochastic neighbor embedding (t-SNE) and plotted the single cells in two dimensions. Data were log-normalized for expression levels and scaled around zero to calculate the relative expression of the genes. Data have been archived in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE207006.
Differences in variables with a continuous distribution across dichotomous categories were assessed using the Wilcoxon signed-rank test. Spearman’s rank correlation coefficient (rs) was used to assess the respective correlations between each MDSC sub-domains and clinicopathological factors. PSA-PFS and overall survival (OS) were estimated using the Kaplan–Meier method. All statistical analyses were conducted using JMP pro 16.0.0 (SAS Institute Inc., Cary, NC, USA), with p < 0.05 indicating statistical significance.
We investigated the three MDSC subsets of e-MDSCs, PMN-MDSCs, and M-MDSCs from pWBCs in patients with newly diagnosed mHSPC. From 74 total metastatic PC patients, 43 were excluded for the following reasons: CRPC patients, no prostatic biopsy, or did not consent to this study. A total of 31 patients with untreated metastatic PC patients were ultimately included in this study (Figure 1).
We first evaluated the therapeutic effect of ADT in patients with untreated metastatic PC and confirmed that all patients were sensitive to hormonal therapy. As shown in Supplementary Figure 1, the PSA levels in all 31 patients declined from pre-ADT to 3 months post-treatment, indicating that all subjects had mHSPC. Figures 2A shows the representative dot plots for one of the patients in the study to illustrate the gating strategy. Since an expansion of MDSCs has been reported in patients with diabetes as well as in smokers (19, 20), the possibility of this was considered in the population of MDSCs in this study. However, no effect on MDSC frequencies from either diabetes or smoking was found in the subjects in our study (Supplementary Figure 2).
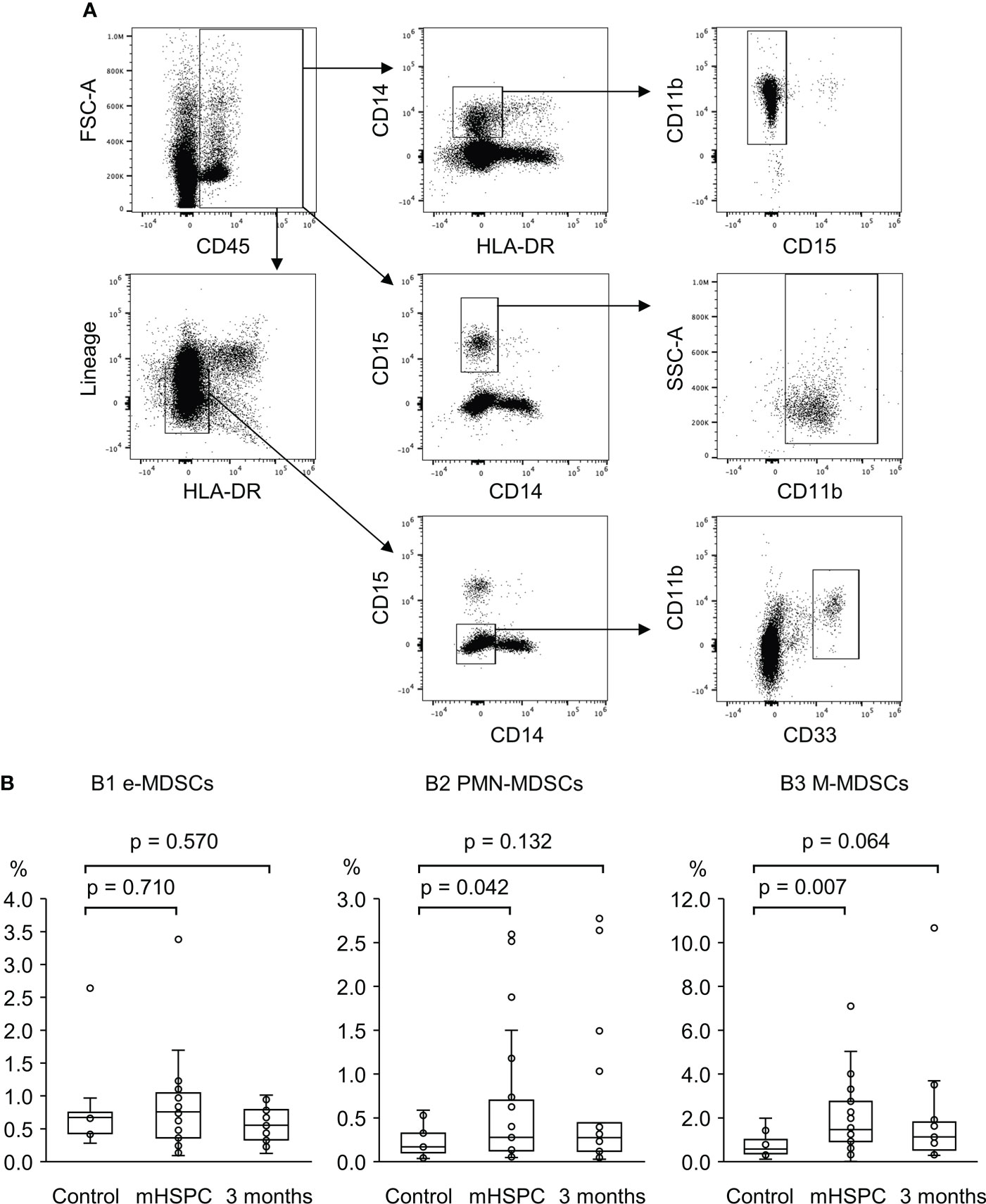
Figure 2 (A) Gating strategy for the identification of peripheral MDSC subsets of the mHSPC patient by Flow cytometry. Each of the MDSC subsets listed below was detected in the cell fraction excluding CD45-negative cells (RBCs and endothelial cells). M-MDSCs; CD11b+CD15−CD14+HLA-DR− cells, PMN-MDSCs; CD11b+CD14−CD15+ cells, e-MDSCs; CD11b+CD33+CD15−CD14−Lineage (CD3/19/56)−HLA-DR− cells. (B) A comparing of the percentage of each of the MDSC subsets of the healthy control, pretreated mHSPC patients, and those treated by ADT for 3 months. B1. e-MDSCs, healthy control; median 0.67%, range 0.28–2.64%, mHSPC; median 0.76%, range 0.09–3.38%, 3 months; median 0.57%, range 0.13–1.01%, B2. PMN-MDSCs, healthy control; median 0.11%, range 0.04–0.53%, mHSPC; median 0.28%, range 0.05–2.60%, 3 months; median 0.25%, range 0.03–2.76%, B3. M-MDSCs, healthy control; median 0.58%, range 0.11–1.98%, mHSPC; median 1.46%, range 0.01–7.10%, 3 months; median 1.20%, range 0.29–10.67%. MDSCs, myeloid-derived suppressor cells; mHSPC, metastatic-hormone sensitive prostate cancer, PMN-MDSCs; polymorphonuclear MDSCs; M-MDSCs, monocytic MDSCs; e-MDSCs, early-stage MDSCs.
We next investigated the cell fractions from hormone-sensitive patients before and three months after treatment. As shown in Figures 2B, while there was no significant difference in the proportion of the e-MDSC subset between mHSPC patients (median value: 0.76%) and healthy controls (0.67%), the PMN/M-MDSC fraction was significantly elevated in patients relative to healthy controls before the treatment (PMN-MDSCs: 0.28% vs. 0.11%, M-MDSCs: 1.46% vs. 0.58%) (Figures 2B). The percentage of PMN/M-MDSCs in patients declined to almost the same level as in the healthy controls after treatment, suggesting that these MDSC subsets fluctuated in conjunction with the therapeutic effects of the GnRH antagonist.
While PSA levels are reportedly associated with the frequencies of M-MDSCs (15, 17), their correlation with e-MDSCs and/or PMN-MDSCs has not been investigated in PC patients. Furthermore, there are no reports describing the association between each MDSC subset and other serological/hematological tests in PC patients. Therefore, we investigated the correlation between each MDSC subset and the results of serological and hematological examinations (PSA, ALP, LDH, peripheral blood factions, biopsy Gleason score) in mHSPC patients at the start of the treatment (Figure 3, and Supplementary Figures 2, 3). The frequency of e-MDSCs did not correlate with serum PSA or ALP levels, but the population of PMN-MDSCs was significantly positively correlated with ALP values (rs = 0.369, p = 0.041), and M-MDSC fractions were positively correlated with both PSA (rs = 0.413, p = 0.021) and ALP levels (rs = 0.464, p = 0.008). M-MDSC fractions were positively correlated with PSA levels as previously reported (15, 17), in addition to ALP levels. MDSCs have been reported to differentiate into osteoclast-like cells and cause bone destruction in mice with bone metastatic carcinoma (21), which may be reflected in the positive correlation with increased ALP levels. In contrast, no marked correlation was found between the percentages of neutrophils, monocytes, and lymphocytes and the ratios of each of the MDSC subsets.
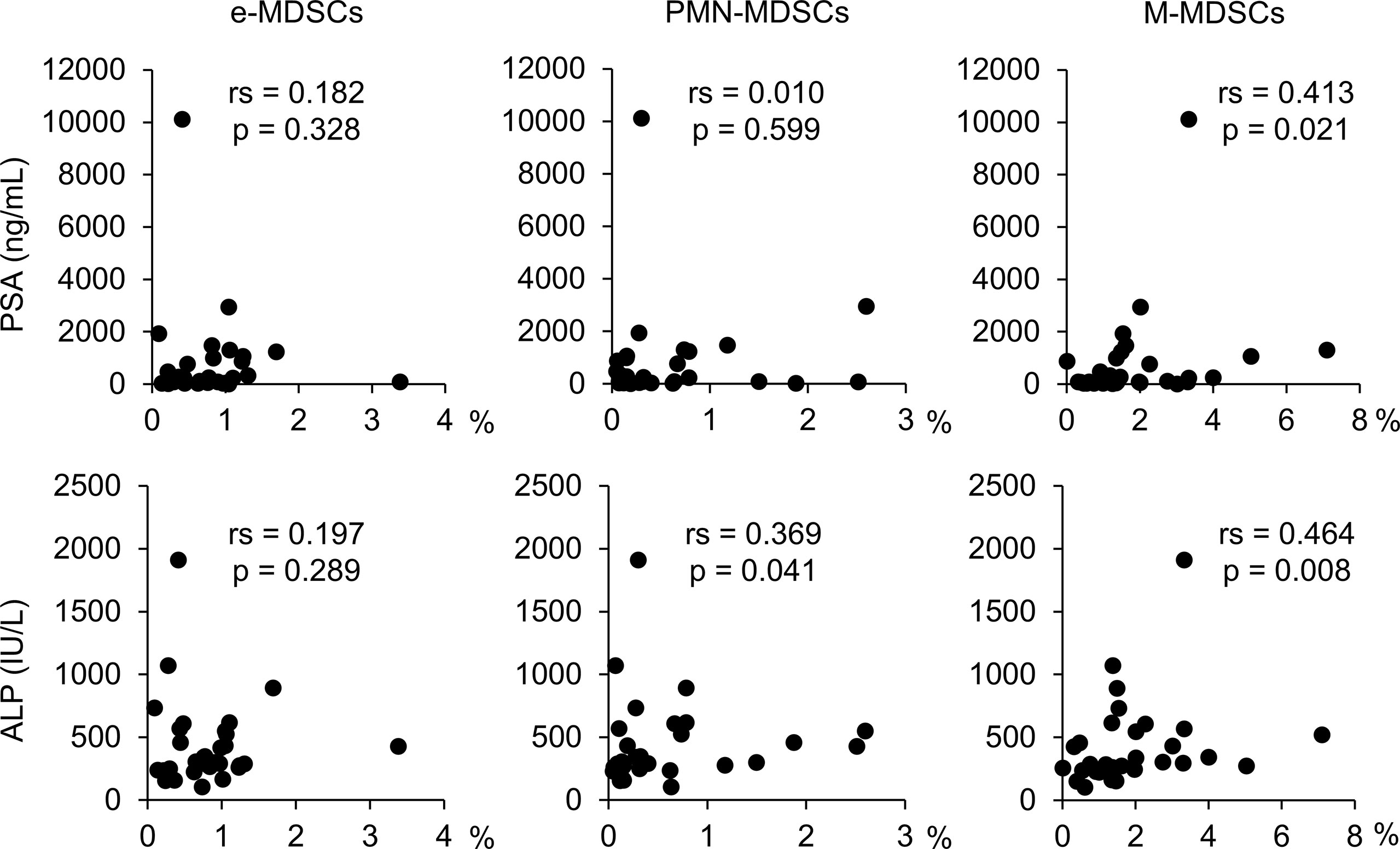
Figure 3 Relevance between each MDSC subset and PSA and ALP. rs, Spearman’s rank correlation coefficient; MDSCs, myeloid-derived suppressor cells; PMN-MDSCs, polymorphonuclear MDSCs; M-MDSCs, monocytic MDSCs; e-MDSCs, early-stage MDSCs; PSA, prostate-specific antigen; ALP, alkaline phosphatase.
According to these results, there is a clear positive correlation between PMN-MDSC and M-MDSC frequencies and the PSA and/or ALP levels in patients with newly diagnosed mHSPC.
The high frequency of MDSCs was shown to be associated with a poor prognosis with various malignant neoplasms via the immunosuppressive effect in the tumor-associated immune system (12, 13). While the association between total MDSC subsets and the OS in PC patients has been reported (16), no studies have examined the impact of each MDSC subset on the prognosis of mHSPC patients. Therefore, we assessed the prognostic value of pretreatment MDSC counts in mHSPC patients (The mean follow-up time: 567 days). During the follow-up period, 19 mHSPC patients had PSA progression, and three patients expired due to tumor aggravation.
To evaluate the prognostic value of MDSCs, we compared mHSPC patients with high and low percentages of each of the MDSC subsets. To investigate the association between clinicopathological factors/each MDSC subset and the PSA-PFS, we performed univariate/multivariate analyses. While cut-off values from retrospective studies have been used in previous studies (22, 23), it is necessary to set a more sensitive cut-off value for a prospective therapeutic intervention study in untreated subjects. We therefore set the cut-off value for the high and well-assessed percentage of PMN-MDSCs and M-MDSCs, except for e-MDSCs, at three times the median value of normal subjects, and a value well above the standard deviation (SD) of normal subjects. The cut-off value for the high percentage of e-MDSCs was set to be just above the median values, as there were no significant differences between patients and healthy controls. Therefore, we set the following values as cut-off values for each MDSC subset: e-MDSCs, 0.75%; PMN-MDSCs, 0.30%; and M-MDSCs, 1.70%.
Table 2 shows the results of univariate and multivariate analyses concerning the PSA-PFS in mHSPC patients. A univariate analysis showed that a high ratio of PMN-MDSCs was significantly associated with a worse PSA-PFS in mHSPC patients. A multivariate analysis of PSA, which is the established biomarker, and PMN-MDSCs showed that PMN-MDSCs were significantly associated with the prognosis of mHSPCs. We then performed a Kaplan–Meier analysis for the PSA-PFS and OS using the defined cut-off values for e-MDSCs, PMN-MDSCs, and M-MDSCs (Figure 4). As shown in Figure 4A, patients with a high ratio of PMN-MDSCs had a worse PSA–PFS than those with a low ratio. In addition, a high frequency of PMN-MDSCs was associated with a worse OS in subjects (Figure 4B). However, high percentages of e-MDSCs and/or M-MDSCs did not relate to the PSA–PFS or OS (Figure 4). These results suggested that PMN-MDSC subsets have prognostic value for metastatic PC patients receiving first-line treatment.
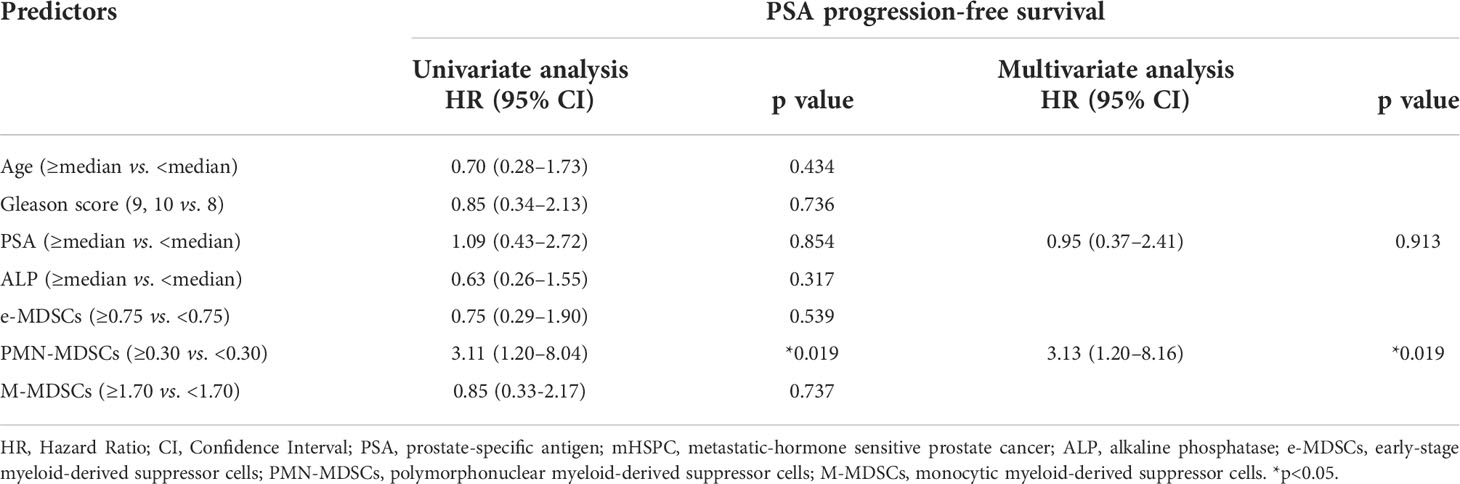
Table 2 Univariate and multivariate analyses regard of the PSA progression-free survival in patients with mHSPC.
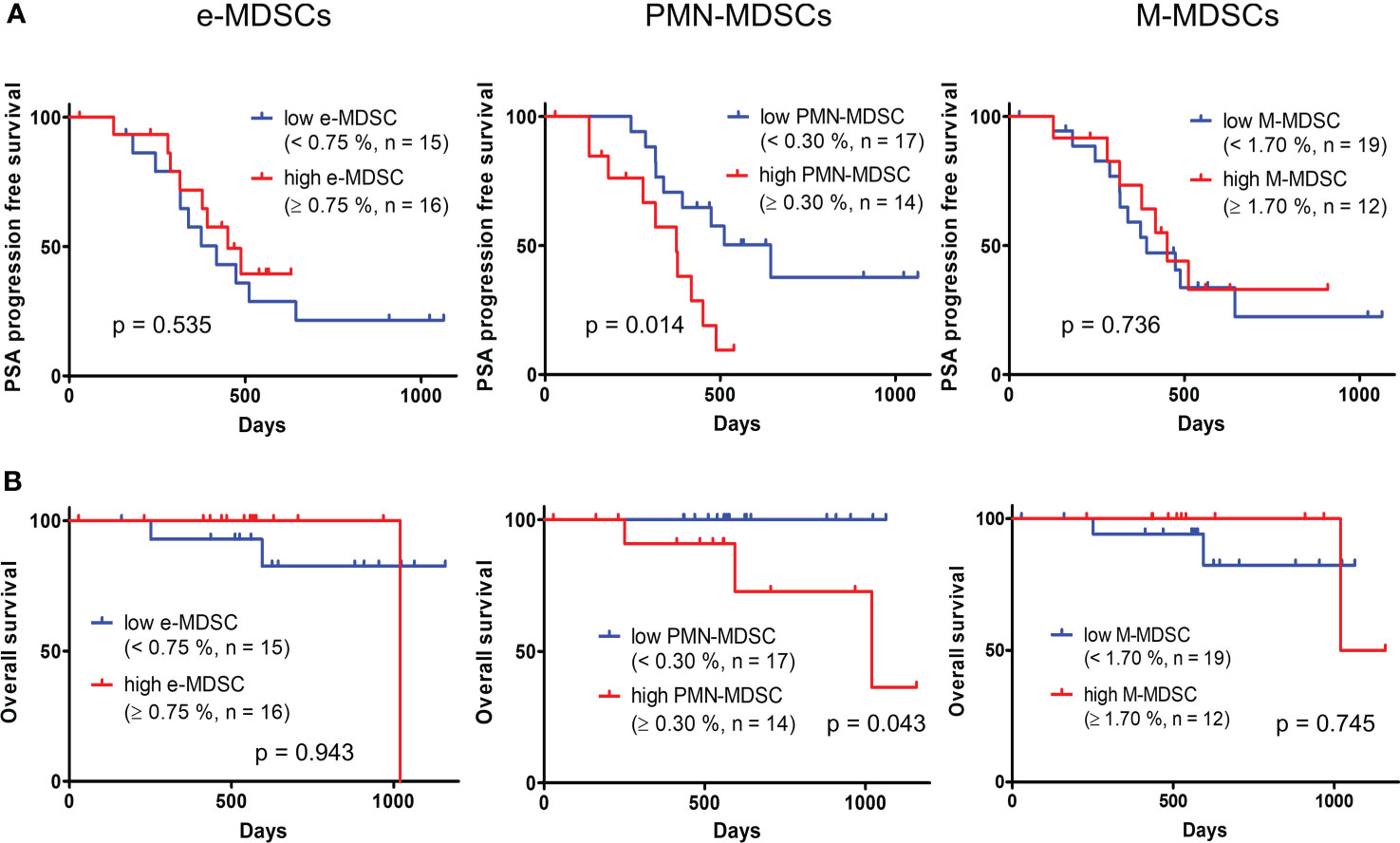
Figure 4 Kaplan–Meier curve of PSA–progression free survival (A) and OS (B) in patients with high and low percentages of each MDSC fraction. PSA, prostate-specific antigen; OS, overall survival; MDSCs, myeloid-derived suppressor cells; PMN-MDSCs, polymorphonuclear MDSCs; M-MDSCs, monocytic MDSCs; e-MDSCs, early-stage MDSCs.
Since the fraction of MDSCs was significantly increased in mHSPC patients, we investigated the immune cell dynamics and gene expressions of MDSCs using an scRNA-seq analysis of pWBCs from a patient with an average percentage increase in the fraction of PMN-MDSCs (neutrophils: 64.4%, lymphocytes: 26.3%, monocytes: 8.4%, e-MDSCs: 1.10%, PMN-MDSCs: 0.79%, M-MDSCs: 3.24%) and a healthy volunteer (neutrophils; 51.6%, lymphocytes: 43.3%, monocytes: 3.4%, e-MDSCs: 0.74%, PMN-MDSCs: 0.04%, M-MDSCs: 2.76%). The transcripts of cells were obtained by the 10X Genomics platform, and we acquired 21,280 and 21,697 genes of single cells from the subject and control, respectively.
Figure 5A shows the t-SNE plots displaying clusters of all immune cells of the mHSPC patient (3885 cells) and the healthy control (4639 cells). We obtained 14 clusters by unbiased graph-based clustering and identified cell groups consisting of T cells, B cells, innate immune cells (γδT cells, NK cells, and NKT cells), and myeloid cells. A greater increase in the frequencies of innate immune cells and myeloid cells was shown in the patient than in the control (Figure 5A), and the gene expressions of these immune cells differed among subjects (Supplementary Table 2). For example, the expression of S100 proteins, which relates to the development and progression of various cancers, was increased in the innate immune cells and myeloid cells of the PC patient. We also confirm that the gene expressions differed even in the same cell subset due to the reclustering of T cells, innate lymphoid cells, and myeloid cells (Supplementary Figure 4). While the population of effecter memory CD4/8 T cells was almost the same between the patient and control, the ratio of central memory CD4/8 T cells was markedly decreased in the patient, implying that the differentiation of T cells into effector cells was enhanced in the tumor-bearing state.
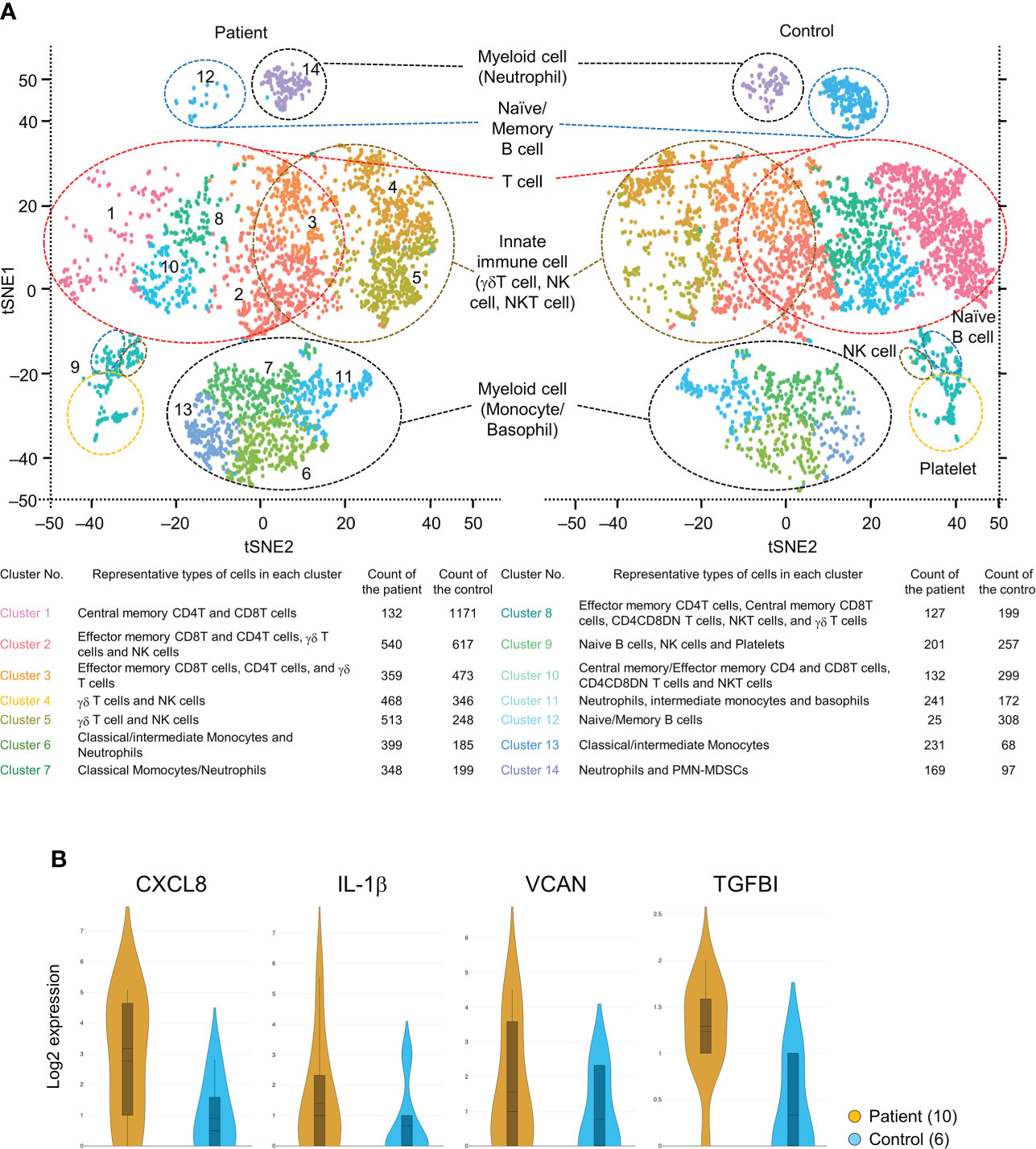
Figure 5 (A) t-SNE plot showing clusters of all immune and non-immune cells of both the mHSPC patient and the healthy control generated from 10X Genomics platform. Cell fractions identified for each cluster and their counts are shown. (B) Violin plots showing the elevated genes of secreted proteins in the patient PMN-MDSCs compared with that of the healthy control. t-SNE, T-distributed stochastic neighbor embedding; mHSPC, metastatic-hormone sensitive prostate cancer; PMN-MDSCs, poly-morphonuclear MDSCs.
Immune checkpoint molecules were not detected as differentially expressed genes in T cell subsets (Supplementary Table 2), but the S100 gene-expressing γδT cells and monocytes were increased in the mHSPC sample (Supplementary Figure 4). In addition, we were able to detect small cell groups of PMN-MDSCs and found the enhanced expression of genes involved in tumor progression [CXCL8, IL-1β, proteoglycan versican (VCAN), and transforming growth factor β-induced protein (TGFBI)] (Figure 5B). While we acknowledge that further data accumulation is needed, our present RNA-seq data were largely consistent with those from previous reports on other cancers (24, 25), suggesting that these genes may also be potential therapeutic targets in mHSPC.
This is the first study implying the prognostic value of MDSC subset for mHSPC patients. While the expansion of PMN-MDSC and M-MDSC subsets has been reported to occur in the peripheral blood and tumor tissue of CRPC (14–17, 26), whether or not PMN-MDSCs have predictive value in the progress of mHSPC has been unclear. In this study, we noted the association of elevated PMN-MDSC fractions with a poor prognosis. In addition, the scRNA-seq analysis in the mHSPC patient showed differing immune cell subsets and immunosuppressive gene expression in PMN-MDSCs.
A unique feature of this study is the finding that the PSA-PFS and OS were poorer when the percentage of PMN-MDSCs was above a certain ratio (≥0.30%) than when it was below that ratio (<0.30%). High percentages of MDSCs have been reported to be associated with a poor prognosis in various malignant neoplasms (22, 23, 27). However, this correlation was found between the cancer prognosis and total MDSC fractions rather than the three subsets of MDSCs (22, 23). In addition, while further measures are required to confirm the contribution of MDSCs to the cancer prognosis on how many times the percentage of MDSCs in the target population increases compared to those of responders or healthy individuals, the high ratio of MDSC fractions in earlier studies was defined as a greater than the mean + 2SD of healthy controls (28), with no proper assessment of MDSC subsets or ratios. In the present study, we defined a high percentage of PMN-MDSCs as above 0.30%, which is close to the median value (0.28%) of patients and about three times that of healthy subjects (0.11%), and we found a significant correlation with the prognosis of mHSPC. This method provides useful insight into the importance of determining the distinct ratio of the specific MDSC subset.
It has been shown that high M-MDSC ratios are associated with a shorter OS in CRPC, indicating a relationship between M-MDSCs and a poor clinical outcome (14–17, 26). In the present study, while the proportion of M-MDSCs was increased remarkably in mHSPC patients and higher than that of PMN-MDSCs, there was no significant difference in either the PSA-PFS or OS of patients and the healthy controls. While PD-L1 was reported to be highly expressed on M-MDSCs (29), no expression of PD-L1 on M-MDSCs patients was found in our flow cytometric analysis (data not shown). One limitation of this study is that the present scRNA-seq analysis also detected only a small number of M-MDSCs in both the patient (10 cells) and the control (2 cells), and the data were not conclusive with regard to the presence or absence of suppressive gene expressions. It is difficult to determine why the percentage of M-MDSCs did not correlate with the prognosis of mHSPC patients based on the scRNA-seq analysis findings. However, the administration of an androgen receptor antagonist was shown to increase the immunosuppressive effect of M-MDSCs via glycolysis enhancement in a murine model (30). Although comparative studies between mHSPC and CRPC patients are needed to clarify the effect of ADT on the inhibitory function of M-MDSCs, it is possible that M-MDSCs grown under mHSPC conditions are less immuno-suppressive before treatment than other malignant neoplasms.
We also investigated e-MDSCs in this study, but the proportion of e-MDSCs did not increase in mHSPC patients, and no correlation with either the PSA-PFS or OS was noted. Because of the difficulty of detecting e-MDSCs, there are few previous studies that investigated the correlation with malignant neoplasms, and their association with diseases is reported to be low (11). For example, it has been demonstrated that survival rates of head and neck cancer and gastric cancer patients correlate with the fraction of peripheral PMN-MDSCs but not with the percentage of e-MDSCs (31, 32). e-MDSCs did not affect T cell proliferation or cytokine production, suggesting that they are less suppressive than PMN-/M-MDSCs because of their immature state (31). To assess the role of e-MDSCs in malignant neoplasms, further studies that accurately detect the e-MDSCs are needed.
Our scRNA-seq results showed different proportions of immune cells in the mHSPC patient. Despite the obvious limitation of the present data in that they were derived from the analysis of a representative case, a greater increase in the ratio of γδT cells and monocytes was found in the PC subject than in the control (Figure 5A and Supplementary Figure 4). γδT cells are reportedly involved in the growth of MDSCs and their recruitment to tumor sites (33). In the present study, the patient’s γδT cells displayed high levels of S100B, PTGDS, ALOX5P, and SPON2 genes, which have been shown to be involved in the MDSC differentiation or tumor progression, suggesting that γδT cells affect MDSC characteristics, including its differentiation in PC with metastasis (34–36). Classical monocytes have been reported to show an elevated gene expression on fibrin-binding F13A1 (37) and to function as a scaffold for lung cancer metastasis, while intermediate monocytes have been reported to express the scavenger receptor (38), MARCO, which is associated with a poor prognosis in various cancers (39). Since peripheral blood monocyte counts are reportedly correlated with TAM infiltration and the prognosis in PC patients after prostatectomy (40), further studies are required to clarify whether or not the above molecules (S100B, PTGDS, ALOX5P, SPON2, and F13A1) are involved in the progress of mHSPC.
In this scRNA-seq analysis, we were able to detect a small number of PMN-MDSCs that showed an augmented expressions of several genes whose presence in PMN-MDSCs has already been reported (Figure 5B) (41). For example, CXCL-8 (IL-8) and IL-1β have been shown to promote cancer progression as well as traffic MDSCs to the tumor microenvironment (25, 42). VCAN and TGFBI enhance tumor metastasis (43, 44). Because PMN-MDSCs aid in circulating tumor cell extravasation and contribute to the pre-metastatic niche formation (45), it is possible that these highly expressed genes are involved in the metastatic status of PC. Although we measured these molecules in cases with high percentages of PMN-MDSCs by an enzyme-linked immunosorbent assay, all were below the detection limit (data not shown). In addition to the accumulation of further cases, their expressions of these molecules in MDSCs in tumor tissue rather than in peripheral blood will need to be investigated in the future. In contrast, our scRNA-seq did not detect a significant difference in the expression of immune checkpoint molecules expressed on T cells and myeloid cells between the mHSPC patient and control (Supplementary Table 2). As with cytokine measurements, it may be that detection is only possible when data from a number of subjects are accumulated, although differences in the expression of immune check point molecules may also have contributed to the finding that those inhibitors are less effective in PCs than in other carcinomas.
Several limitations associated with the present study warrant mention. First, the study was conducted at a single institution and involved a relatively small number of patients. Second, it lacked long-term follow-up. However, the study had uniform patient characteristics in that all mHSPC patients received the same treatment and suggested an association between PMN-MDSCs despite the small number of cases. Further studies with larger cohorts and a longer follow-up are needed to confirm the results of this study.
Our present study showed that PMN-MDSCs are correlated with a poor prognosis in mHSPC patients. An scRNA-seq analysis detected the high expression of genes involved in cancer progress in PMN-MDSCs. According to these results, PMN-MDSCs, including their highly expressed genes, have potential utility as novel therapeutic targets against mHSPC.
The data presented in the study are deposited in the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE207006.
The studies involving human participants were reviewed and approved by clinical research ethics boards of Tohoku Medical and Pharmaceutical Hospital (approval number: 2018-2-062). The patients/participants provided their written informed consent to participate in this study.
All authors contributed to the article and approved the submitted version. Conceptualization was performed by YUK, AK, AN, and MS. Material preparation, data collection and analysis were performed by YUK, AK, KT, HI, YA, JI, TK, and AN. The first draft of the manuscript was written by YUK. The review and editing of the manuscript were performed by AN. Supervision was provided by YUK, IN, NH, AN, and MS.
This work was supported in part by JSPS KAKENHI Grant Number 19K07511 to KT, 19K06471 to TK, and 20K09719 to AN.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fruro.2022.967480/full#supplementary-material
Supplementary Figure 1 | Parallel fluctuation of the PSA levels in mHSPC patients before treatment to 3 months for ADT. The PSA levels declined in all 31 patients after 3 months (median; 186.0, range 2.1–10115.1). PSA, prostate-specific antigen; mHSPC, metastatic-hormone sensitive prostate cancer; ADT, androgen deprivation therapy.
Supplementary Figure 2 | Relevance between each of the MDSC subsets and diabetes, smoking status, and GS. MDSCs, myeloid-derived suppressor cells; PMN-MDSCs, polymorphonuclear MDSCs; M-MDSCs, monocytic MDSCs; e-MDSCs, early-stage MDSCs; DM, diabetes; GS, gleason score.
Supplementary Figure 3 | Relevance between each of the MDSC subsets and LDH, Neutrophil, Lymphocyte, and Monocyte. Spearman’s rank correlation coefficient, MDSCs, myeloid-derived suppressor cells; PMN-MDSCs, polymorphonuclear MDSCs; M-MDSCs, monocytic MDSCs; e-MDSCs, early-stage MDSCs; LDH, lactate dehydrogenase.
Supplementary Figure 4 | Reclustered t-SNE plots comparing T cell cluster, innate immune cell cluster, and myeloid cell cluster between mHSPC patient and healthy control. Parentheses indicate the representative highly expressed gene in each cell fraction. mHSPC, metastatic-hormone sensitive prostate cancer. t-SNE, T-distributed stochastic neighbor embedding.
Supplementary Table 1 | Characteristics of the patients in this study
Supplementary Table 2 | All gene expressions that are upregulated in patient compared to the control in immune and non-immune cells identified by 10x single cell RNA-sequencing.
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71:7–33. doi: 10.3322/caac.21654
2. Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol (2013) 190:2047–53. doi: 10.1016/j.juro.2013.06.017
3. Mohler JL, Armstrong AJ, Bahnson RR, D'Amico AV, Davis BJ, Eastham JA, et al. Prostate cancer, version 1.2016. J Natl Compr Canc Netw (2016) 14:19–30. doi: 10.6004/jnccn.2016.0004
4. Armstrong AJ, Szmulewitz RZ, Petrylak DP, Holzbeierlein J, Villers A, Azad A, et al. ARCHES: A randomized, phase III study of androgen deprivation therapy with enzalutamide or placebo in men with metastatic hormone-sensitive prostate cancer. J Clin Oncol (2019) 37:2974–86. doi: 10.1200/JCO.19.00799
5. Kim TJ, Koo KC. Current status and future perspectives of checkpoint inhibitor immunotherapy for prostate cancer: A comprehensive review. Int J Mol Sci (2020) 21:5484. doi: 10.3390/ijms21155484
6. Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med (2010) 363:411–22. doi: 10.1056/NEJMoa1001294
7. Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, et al. The terminology issue for myeloid-derived suppressor cells. Cancer Res (2007) 67:425; author reply 6. doi: 10.1158/0008-5472.CAN-06-3037
8. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun (2016) 7:12150. doi: 10.1038/ncomms12150
9. Zhou J, Nefedova Y, Lei A, Gabrilovich D. Neutrophils and PMN-MDSC: Their biological role and interaction with stromal cells. Semin Immunol (2018) 35:19–28. doi: 10.1016/j.smim.2017.12.004
10. Tcyganov E, Mastio J, Chen E, Gabrilovich DI. Plasticity of myeloid-derived suppressor cells in cancer. Curr Opin Immunol (2018) 51:76–82. doi: 10.1016/j.coi.2018.03.009
11. Khan ANH, Emmons TR, Wong JT, Alqassim E, Singel KL, Mark J, et al. Quantification of early-stage myeloid-derived suppressor cells in cancer requires excluding basophils. Cancer Immunol Res (2020) 8:819–28. doi: 10.1158/2326-6066.CIR-19-0556
12. Cui C, Lan P, Fu L. The role of myeloid-derived suppressor cells in gastrointestinal cancer. Cancer Commun (Lond) (2021) 41:442–71. doi: 10.1002/cac2.12156
13. Yang Z, Guo J, Weng L, Tang W, Jin S, Ma W. Myeloid-derived suppressor cells-new and exciting players in lung cancer. J Hematol Oncol (2020) 13:10. doi: 10.1186/s13045-020-0843-1
14. Hossain DM, Pal SK, Moreira D, Duttagupta P, Zhang Q, Won H, et al. TLR9-targeted STAT3 silencing abrogates immunosuppressive activity of myeloid-derived suppressor cells from prostate cancer patients. Clin Cancer Res (2015) 21:3771–82. doi: 10.1158/1078-0432.CCR-14-3145
15. Idorn M, Kollgaard T, Kongsted P, Sengelov L, Thor Straten P. Correlation between frequencies of blood monocytic myeloid-derived suppressor cells, regulatory T cells and negative prognostic markers in patients with castration-resistant metastatic prostate cancer. Cancer Immunol Immunother (2014) 63:1177–87. doi: 10.1007/s00262-014-1591-2
16. Chi N, Tan Z, Ma K, Bao L, Yun Z. Increased circulating myeloid-derived suppressor cells correlate with cancer stages, interleukin-8 and -6 in prostate cancer. Int J Clin Exp Med (2014) 7:3181–92.
17. Vuk-Pavlovic S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, et al. Immunosuppressive CD14+HLA-DRlow/- monocytes in prostate cancer. Prostate (2010) 70:443–55. doi: 10.1002/pros.21078
18. Scher HI, Morris MJ, Stadler WM, Higano C, Basch E, Fizazi k., et al. Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the prostate cancer clinical trials working group 3. J Clin Oncol (2016) 34:1402–18. doi: 10.1200/JCO.2015.64.2702
19. Wang S, Tan Q, Hou Y, Dou H. Emerging roles of myeloid-derived suppressor cells in diabetes. Front Pharmacol (2021) 12:798320. doi: 10.3389/fphar.2021.798320
20. Ortiz ML, Lu L, Ramachandran I, Gabrilovich DI. Myeloid-derived suppressor cells in the development of lung cancer. Cancer Immunol Res (2014) 2:50–8. doi: 10.1158/2326-6066.CIR-13-0129
21. Sawant A, Deshane J, Jules J, Lee CM, Harris BA, Feng X, et al. Myeloid-derived suppressor cells function as novel osteoclast progenitors enhancing bone loss in breast cancer. Cancer Res (2013) 73:672–82. doi: 10.1158/0008-5472
22. Yang G, Shen W, Zhang Y, Liu M, Zhang L, Liu Q, et al. Accumulation of myeloid-derived suppressor cells (MDSCs) induced by low levels of IL-6 correlates with poor prognosis in bladder cancer. Oncotarget (2017) 8:38378–88. doi: 10.18632/oncotarget.16386
23. Wang L, Chang EW, Wong SC, Ong SM, Chong DQ, Ling KL. Increased myeloid-derived suppressor cells in gastric cancer correlate with cancer stage and plasma S100A8/A9 proinflammatory proteins. J Immunol (2013) 190:794–804. doi: 10.4049/jimmunol.1202088
24. Cedervall J, Zhang Y, Olsson AK. Tumor-induced NETosis as a risk factor for metastasis and organ failure. Cancer Res (2016) 76:4311–5. doi: 10.1158/0008-5472.CAN-15-3051
25. Groth C, Weber R, Lasser S, Ozbay FG, Kurzay A, Petrova V, et al. Tumor promoting capacity of polymorphonuclear myeloid-derived suppressor cells and their neutralization. Int J Cancer (2021) 149:1628–38. doi: 10.1002/ijc.33731
26. Wen J, Huang G, Liu S, Wan J, Wang X, Zhu Y, et al. Polymorphonuclear MDSCs are enriched in the stroma and expanded in metastases of prostate cancer. J Pathol Clin Res (2020) 6:171–7. doi: 10.1002/cjp2.160
27. Gao XH, Tian L, Wu J, Ma XL, Zhang CY, Zhou Y, et al. Circulating CD14(+) HLA-DR(-/low) myeloid-derived suppressor cells predicted early recurrence of hepatocellular carcinoma after surgery. Hepatol Res (2017) 47:1061–71. doi: 10.1111/hepr.12831
28. de Goeje PL, Bezemer K, Heuvers ME, Dingemans AC, Groen HJ, Smit EF, et al. Immunoglobulin-like transcript 3 is expressed by myeloid-derived suppressor cells and correlates with survival in patients with non-small cell lung cancer. Oncoimmunology (2015) 4:e1014242. doi: 10.1080/2162402X.2015.1014242
29. Okla K, Rajtak A, Czerwonka A, Bobinski M, Wawruszak A, Tarkowski R, et al. Accumulation of blood-circulating PD-L1-expressing m-MDSCs and monocytes/macrophages in pretreatment ovarian cancer patients is associated with soluble PD-L1. J Transl Med (2020) 18:220. doi: 10.1186/s12967-020-02389-7
30. Zhao Y, Peng X, Baldwin H, Zhang C, Liu Z, Lu X. Anti-androgen therapy induces transcriptomic reprogramming in metastatic castration-resistant prostate cancer in a murine model. Biochim Biophys Acta Mol Basis Dis (2021) 1867:166151. doi: 10.1016/j.bbadis.2021.166151
31. Lang S, Bruderek K, Kaspar C, Höing B, Kanaan O, Dominas , et al. Clinical relevance and suppressive capacity of human myeloid-derived suppressor cell subsets. Clin Cancer Res (2018) 24:4834–44. doi: 10.1158/1078-0432.CCR-17-3726
32. Tavukcuoglu E, Horzum U, Yanik H, Uner A, Yoyen-Ermis D, Nural SK, et al. Human splenic polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) are strategically located immune regulatory cells in cancer. Eur J Immunol (2020) 50:2067–74. doi: 10.1002/eji.202048666
33. Park JH, Lee HK. Function of gammadelta T cells in tumor immunology and their application to cancer therapy. Exp Mol Med (2021) 53:318–27. doi: 10.1038/s12276-021-00576-0
34. Charmsaz S, Hughes E, Bane FT, Tibbitts P, McIlroy M, Byrne C, et al. S100beta as a serum marker in endocrine resistant breast cancer. BMC Med (2017) 15:79. doi: 10.1186/s12916-017-0836-2
35. Trabanelli S, Chevalier MF, Martinez-Usatorre A, Usatorre A, Gomez-Cadena A, Salome B, et al. Tumour-derived PGD2 and NKp30-B7H6 engagement drives an immunosuppressive ILC2-MDSC axis. Nat Commun (2017) 8:593. doi: 10.1038/s41467-017-00678-2
36. Schmid F, Wang Q, Huska MR, Andrade-Navarro MA, Lemm M, Fichtner I, et al. SPON2, a newly identified target gene of MACC1, drives colorectal cancer metastasis in mice and is prognostic for colorectal cancer patient survival. Oncogene (2016) 35:5942–52. doi: 10.1038/onc.2015.451
37. Porrello A, Leslie PL, Harrison EB, Gorentla BK, Kattula S, Ghosh SK, et al. Factor XIIIA-expressing inflammatory monocytes promote lung squamous cancer through fibrin cross-linking. Nat Commun (2018) 9:1988. doi: 10.1038/s41467-018-04355-w
38. Kiss M, Caro AA, Raes G, Laoui D. Systemic reprogramming of monocytes in cancer. Front Oncol (2020) 10:1399. doi: 10.3389/fonc.2020.01399
39. Shi B, Chu J, Huang T, Wang X, Li Q, Gao Q, et al. The scavenger receptor MARCO expressed by tumor-associated macrophages are highly associated with poor pancreatic cancer prognosis. Front Oncol (2021) 11:771488. doi: 10.3389/fonc.2021.771488
40. Hayashi T, Fujita K, Nojima Hayashi S Y, Nakano K, Ishizuya Y. Peripheral blood monocyte count reflecting tumor-infiltrating macrophages is a predictive factor of adverse pathology in radical prostatectomy specimens. Prostate (2017) 77:1383–8. doi: 10.1002/pros.23398
41. Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol (2018) 19:108–19. doi: 10.1038/s41590-017-0022-x
42. Yang Y, Li C, Liu T, Dai X, Bazhin AV. Myeloid-derived suppressor cells in tumors: From mechanisms to antigen specificity and microenvironmental regulation. Front Immunol (2020) 11:1371. doi: 10.3389/fimmu.2020.01371
43. Wang Z, Li Z, Wang Y, Cao D, Wang X, Jiang M, et al. Versican silencing improves the antitumor efficacy of endostatin by alleviating its induced inflammatory and immunosuppressive changes in the tumor microenvironment. Oncol Rep (2015) 33:2981–91. doi: 10.3892/or.2015.3903
44. Lakshmanachetty S, Cruz-Cruz J, Hoffmeyer E, Cole AP, Mitra SS. New insights into the multifaceted role of myeloid-derived suppressor cells (MDSCs) in high-grade gliomas: From metabolic reprograming, immunosuppression, and therapeutic resistance to current strategies for targeting MDSCs. Cells (2021) 10:893. doi: 10.3390/cells10040893
Keywords: myeloid-derived suppressor cells, prostatic neoplasms, androgen receptor antagonists, flow cytometry, single-cell analysis, sequence analysis, RNA
Citation: Kohada Y, Kuromoto A, Takeda K, Iwamura H, Atobe Y, Ito J, Kaifu T, Kaiho Y, Nakashima I, Hinata N, Nakamura A and Sato M (2022) Circulating PMN-MDSC level positively correlates with a poor prognosis in patients with metastatic hormone-sensitive prostate cancer. Front. Urol. 2:967480. doi: 10.3389/fruro.2022.967480
Received: 12 June 2022; Accepted: 22 August 2022;
Published: 15 September 2022.
Edited by:
Oluwaseun Adebayo Bamodu, Taipei Medical University, TaiwanReviewed by:
Aihua Lei, University of South China, ChinaCopyright © 2022 Kohada, Kuromoto, Takeda, Iwamura, Atobe, Ito, Kaifu, Kaiho, Nakashima, Hinata, Nakamura and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Nakamura, YWtpLW5AdG9ob2t1LW1wdS5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.