
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Signal Process. , 30 November 2022
Sec. Biomedical Signal Processing
Volume 2 - 2022 | https://doi.org/10.3389/frsip.2022.1019253
This article is part of the Research Topic Adversarial Machine Learning and Domain Generalization in Neurophysiological Signal Analysis View all 4 articles
 Shiva Salsabilian
Shiva Salsabilian Laleh Najafizadeh*
Laleh Najafizadeh*Developing models for identifying mild traumatic brain injury (mTBI) has often been challenging due to large variations in data from subjects, resulting in difficulties for the mTBI-identification models to generalize to data from unseen subjects. To tackle this problem, we present a long short-term memory-based adversarial variational autoencoder (LSTM-AVAE) framework for subject-invariant mTBI feature extraction. In the proposed model, first, an LSTM variational autoencoder (LSTM-VAE) combines the representation learning ability of the variational autoencoder (VAE) with the temporal modeling characteristics of the LSTM to learn the latent space representations from neural activity. Then, to detach the subject’s individuality from neural feature representations, and make the model proper for cross-subject transfer learning, an adversary network is attached to the encoder in a discriminative setting. The model is trained using the 1 held-out approach. The trained encoder is then used to extract the representations from the held-out subject’s data. The extracted representations are then classified into normal and mTBI groups using different classifiers. The proposed model is evaluated on cortical recordings of Thy1-GCaMP6s transgenic mice obtained via widefield calcium imaging, prior to and after inducing injury. In cross-subject transfer learning experiment, the proposed LSTM-AVAE framework achieves classification accuracy results of 95.8% and 97.79%, without and with utilizing conditional VAE (cVAE), respectively, demonstrating that the proposed model is capable of learning invariant representations from mTBI data.
Mild traumatic brain injury (mTBI) is a common form of brain injury and a growing public health problem. mTBI can have long-lasting effects on patients’ cognitive abilities and social functioning. The diagnosis of mTBI, specially at its early stages, has remained challenging despite the negative effects it has on patient’s quality of life (Iverson et al., 2000; Kou and Iraji, 2014; Schmid et al., 2021). The main reasons for this include rapid recovery of symptoms (e.g., loss of consciousness, confusion, disorientation), and incapability of imaging methods (e.g., computerized tomography (CT) or magnetic resonance imaging (MRI)) in detecting injury at the mild level (Ruff et al., 2009). Hence, developing accurate mTBI diagnostic methods is essential for an early diagnosis of mTBI and providing proper and timely treatments to patients (Eierud et al., 2014; Levin and Diaz-Arrastia, 2015). One major challenge in mTBI identification is the undesirable variability in the data obtained from subjects. Such variability poses difficulties for accurate mTBI diagnosis by preventing mTBI-identification models from being generalizable and transferable to data from new subjects. Therefore, a robust feature extractor for learning accurate subject-invariant injury-related features is desired. In addition, limited data from mTBI subjects are often collected or are available, posing the problem of having inadequate data for training reliable models.
Recently, transfer learning has been utilized to extract features by leveraging knowledge across domains, tasks, or subjects. Cross-subject transfer learning aims at discovering and exploiting invariant and generalizable features across subjects. For example, transfer learning approaches such as learning population level common spatial base dictionaries (Morioka et al., 2015), spectral transfer using information geometry (Waytowich et al., 2016), and regularizing classifiers (Fazli et al., 2009) or feature extractors (Lotte and Guan, 2010), have been proposed for brain computer interfaces (BCIs). In (Bethge et al., 2022a), a multi-source learning framework based on maximum mean discrepancy (MMD) alignment of electroencephalography (EEG) data for emotion classification was presented. In (Peterson et al., 2021), to classify arm movements, a decoder model based on Hilbert transform was trained using pooled electrocorticography (ECOG) data, and tested on ECOG or EEG data from unseen subjects.
Invariant representation learning in neural networks, suggesting the idea of using a latent space shared across subjects, was presented in (Louppe et al., 2017; Xie et al., 2017). In (Angjelichinoski et al., 2020) linear classifiers are trained with multiple transfer functions to transfer data from one subject to another. The idea of generative models, algorithms that learn the posterior distribution of data via Bayesian rules, has been recently proposed. Using deep generative models, the original data are refined and converted into features that increase the intra-class variations and minimize the inter-class variations in the dataset. The most extensively used deep generative learning models include the variational autoencoder (VAE) and generative adversarial networks (GAN) (Goodfellow et al., 2014). The advantage of VAE is the smooth latent representation learning of data and bringing the ability to control the distribution of the latent space, which can be combined with feature learning methods. The principle of GAN has also been applied to transfer learning to address the data variability problem for domain- and subject-invariant feature learning (Ming et al., 2019; Wu et al., 2020; Özdenizci et al., 2020; Salsabilian and Najafizadeh, 2021a). For example, in (Li et al., 2019), to generalize the models for EEG emotion recognition across subjects and sessions, the marginal distribution is adapted in the early layers of the neural networks using adversarial training. In another work (Özdenizci et al., 2020), using an adversarial inference approach, subject variability for motor imagery decoding is decreased. A subject adaptation network inspired by GAN was proposed in (Ming et al., 2019) to align the distribution of data from different subjects. Autoencoder-based neural representation learning models have also recently adapted adversarial regularization for feature disentanglement. For example, subject-invariant representations were learned via a conditional variational autoencoder (cVAE) and an adversarial network from unseen users’ EEG data in motor imagery BCIs (Özdenizci et al., 2019). In (Han et al., 2020, 2021) disentangled adversarial autoencoder (AE) and rateless AE (RAE) feature extractors were proposed to extract nuisance-robust universal features from physiological signals for stress level assessment, demonstrating improvements in cross-subject transfer analysis.
In this paper, we present an mTBI-identification model using cross-subject transfer learning and adversarial networks. The proposed method consists of a long short-term memory-based variational autoencoder (LSTM-VAE) representation learning model with an attached adversarial network. In the proposed model, the adversary network is utilized as a constraint for latent representations to learn representations that are invariant to cross-subject variability. The model can therefore, learn the common structure of the data shared among subjects, making it suitable for cross-subject feature learning and mTBI identification. The LSTM-VAE model combines the representation learning abilities of the VAE with the temporal modeling capabilities of the LSTM. The adversarial network is attached to the encoder in an attempt to ensure the latent representation contains minimum subject-specific information. After training, the trained encoder is used as a feature extractor, and a separate classifier learns to predict mTBI or normal class labels, given the latent representation obtained from the trained encoder. We evaluate the proposed model using cortical activity recordings of Thy1-GCaMP6s transgenic mice that were obtained via widefield calcium imaging, before and after inducing injury.
The rest of the paper is organized as follows. Methods, including the description of the dataset as well as the proposed framework, are presented in Section 2. Results are discussed in Section 3, and the paper is concluded in Section 4.
In this section, we first describe the details of the experimental procedure, data collection, and preprocessing steps. Then, we present the proposed LSTM-based adversarial variational autoencoder (AVAE) and other feature extraction models that we used for comparison.
Cortical recordings from Thy1-GCaMP6 transgenic mice were acquired in the Department of Cell Biology and Neuroscience of Rutgers University using widefield optical imaging. All procedures were approved by the Rutgers University Institutional Animal Care and Use Committee. Animal models of mTBI, in comparison to studying mTBI in human patients offer the opportunity of having control over the experimental parameters and conditions, such as maintaining the same site of injury and similar injury severity levels across subjects. Among the animal models, mice have been widely used (Morganti-Kossmann et al., 2010; Marshall and Mason, 2019). The mouse model mimics many features of TBI seen in humans, including cell death, neuroinflammation, and changes in behavior (Morganti-Kossmann et al., 2010; Wiltschko et al., 2015; Ellenbroek and Youn, 2016; Marshall and Mason, 2019). An important similarity between mice and humans is in their genetic makeup, suggesting that findings from mouse studies can often be related to human (Breschi et al., 2017; Beauchamp et al., 2022). Nevertheless, while mice and human brains exhibit similarities, there are differences between mice and humans that must be considered when using them as models for brain injuries. Among them is their quick healing process from injuries, which should be taken into consideration when studying brain injury’s long-term effects and recovery in mice vs. human TBI models (You et al., 2007; Cortes and Pera, 2021). Widefield calcium imaging in animals enables recording of the neural activity with high temporal and spatial resolutions. This imaging technique has been used to study the relationship between cortical activity and behavior (Zhu et al., 2017; Salsabilian et al., 2018; 2020b; Lee et al., 2020; Salsabilian and Najafizadeh, 2021b), as well as investigating brain’s functional changes in response to injury (Cramer et al., 2019; Salsabilian et al., 2020a; Koochaki et al., 2020; Salsabilian and Najafizadeh, 2020; Salsabilian and Najafizadeh, 2021a; Koochaki and Najafizadeh, 2021; Cramer et al., 2022).
Data acquisition process, the experimental setting and the injury procedure were described previously in (Zhu et al., 2018; Salsabilian et al., 2019; Salsabilian and Najafizadeh, 2020). In summary, Thy1-GCaMP6 transgenic mice were prepared with a transparent skull and a fixation post to record cortical Ca2+ transient activity (Lee and Margolis, 2016; Salsabilian et al., 2020b). The left hemisphere and a portion of the right hemisphere were visualized using a custom-designed microscope. The excitation light was filtered (479/40 nm; Chroma) and reflected by a 50 dichroic mirror (Q470/lp, Chroma). Through a 100 × 100 pixel sensor, filtered fluorescence emission was captured at 100 frames per second with a MiCam Ultima CMOS camera (Brain vision). On the day of injury, a small craniotomy (
Spontaneous cortical activity from 12 animals were acquired in two sessions, one prior to and one after inducing the injury. Data obtained from the sessions prior to and after inducing injury are referred here to as normal and mTBI data, respectively. Each recording session included 8 trials of duration of 20.47 s.
Relative GCaMP6s fluorescence calcium signal changes (ΔF/F%) were computed in every pixel value by subtracting and then dividing each pixel by the baseline. The baseline for each pixel was defined as the average of the fluorescence intensities of that pixel in the first 49 frames. We selected twenty-five 5 × 5-pixel regions of interest (ROIs) or channels (i.e., C = 25) distributed over the cortex based on their location according to S1 (Salsabilian et al., 2019). We obtained timeseries from each ROI by calculating the average pixel intensities within the ROI.
A sliding window with the duration of T = 400 and step size of w = 20 time points is moved over the timeseries in each trial (Figure 1). This duration was found to be optimal for capturing the necessary information required for classification. The data within each window is formed as a sampled data matrix
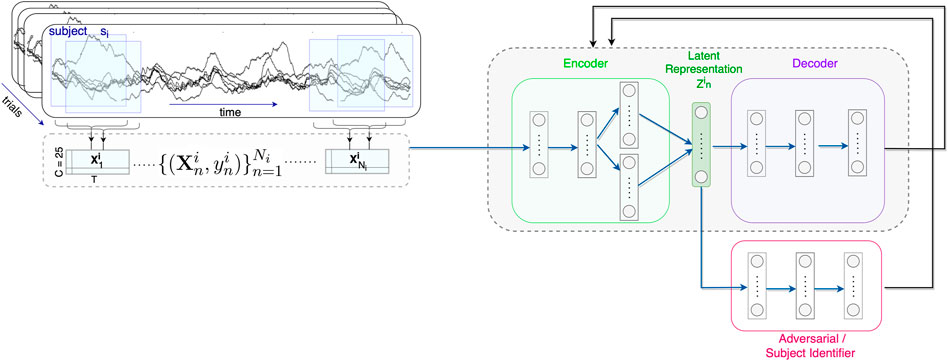
FIGURE 1. Schematic representation of the proposed model architecture for mTBI subject-invariant feature extraction. Cortical activity from C =25 channels are collected. For each subject si, a sliding window with the duration of T and step size of w time points is used, and the sampled data matrices
To evaluate the subject-invariant feature learning performance of the models in a cross-subject transfer learning experiment, the subject attribute si is defined as the subject one-hot encoded label, i.e., a zero vector of size 1 × 12 with 1 at the ith index. The goal is to learn subject-invariant features and an mTBI discriminative model that predicts class label
We now present the proposed subject-invariant feature extraction approach and the mTBI-identification model. The proposed model aims to achieve discriminative properties that are robust to subject variability. The proposed method consists of two components: an LSTM-based variational autoencoder and an adversarial network. The autoencoder is first trained to minimize the reconstruction error of the decoder to ensure that the latent representation contains enough information to allow for the reconstruction of the input. Next, the latent representation is refined to include minimum subject-dependent information by preventing it from predicting the correct subject attribute using adversarial regularization. With this approach, the model is capable of achieving latent representations that contain discriminative properties that correspond to the structure of the data that is common among subjects and therefore, becomes robust to subject variability. Finally, a separate classifier is trained to predict mTBI or normal class labels, given the latent representation obtained from the trained encoder as the feature extractor. In addition, we incorporate conditional VAE (cVAE) in the decoder architecture of the proposed model, to further explore the benefits of removing subject-dependent information from learned representations.
We compare the performance of the proposed model with that of different variants of autoencoder models for feature extraction. The considered models are a simple autoencoder (AE), a variational autoencoder (VAE), a supervised VAE (SVAE), and an adversarial variational autoencoder (AVAE). The schematic illustration of the proposed model and other considered autoencoder feature extraction models are shown in Figure 2. Except SVAE, all the autoencoder models are trained based on the mean squared error (MSE).
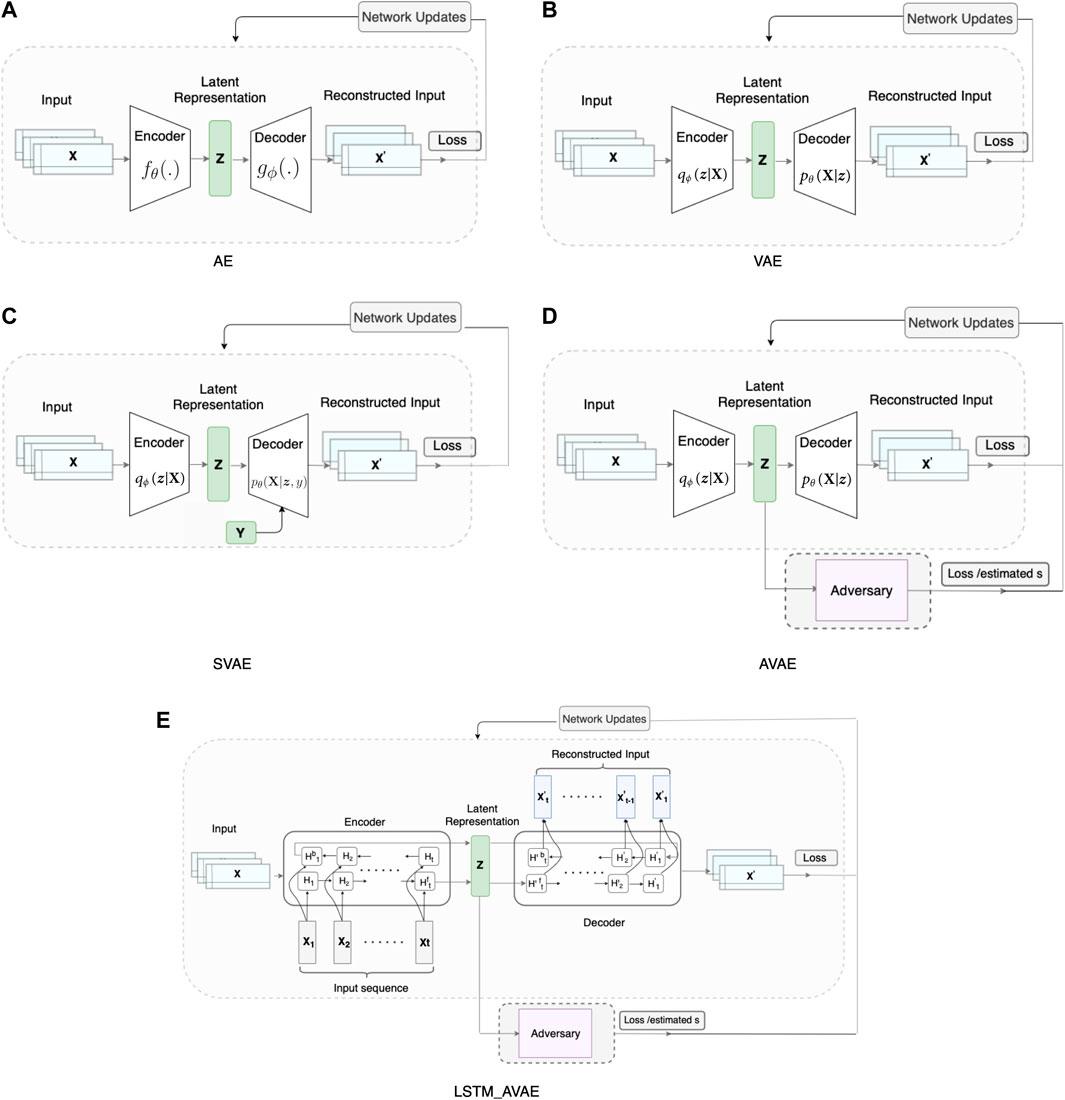
FIGURE 2. Autoencoder-based data representation learning models: (A) simple autoencoder (AE), (B) variational autoencoder (VAE), (C) supervised VAE (SVAE), (D) adversarial VAE (AVAE), (E) proposed LSTM-AVAE.
AE (Figures 2A) reconstructs the input data X as
VAE (Figure 2B) is a probabilistic variant of AE, that uses variational lower bound of the marginal likelihood, based on Bayesian inference, to identify multivariate patterns in data. The latent variable z is a stochastic variable given the input data X. The probabilistic encoder approximates the posterior qϕ(z|X), and the generative decoder represents the likelihood of data X generation by the conditional probability pθ(X|z). To make the model trainable, the reparameterization approach introduced in (Doersch, 2016) was used. The loss function of the VAE,
where ϕ, and θ are the encoder and decoder parameters, respectively. The first term in (Eq. 2) represents the autoencoder reconstruction loss, and the second term is the Kullback-Leibler (KL) (Kullback and Leibler, 1951) divergence. The KL divergence is a measure of the similarity between the prior distribution p(z) and the posterior distribution qϕ(z|X) of the latent variable. Minimizing the KL divergence regularizes the latent space.
SVAE (Figure 2C) method differs from VAE only in that the data label
In order for the model to be generalizable across subjects, representations should be invariant to the subject attribute si. To achieve this goal, we utilize an adversary network (Makhzani et al., 2015) parameterized by qψ(.). The adversary network is attached to the encoder to enforce the latent representations to include minimum subject-dependent information (Figure 2D). The adversary network is trained to maximize the likelihood qψ(si|z), which maximizes its ability to predict subject attribute si. The VAE is simultaneously trained based on two objectives: the decoder’s reconstruction loss is minimized to ensure that the latent representations include sufficient information for minimizing the input reconstruction error; and the latent representation is enforced to include minimum subject-dependent information by preventing the adversary network from predicting the correct subject attribute. This leads to a model, capable of extracting discriminative features that are common across subjects. The AVAE network is trained simultaneously by these objectives with the loss function,
where λ ≥ 0 denotes the weight parameter that adjusts the impact of the adversary network. Note that AVAE is equivalent to VAE when λ = 0. At each iteration, first, the log-likelihood (max objective) is maximized and the parameters of the adversary network are updated. Then, the parameters of the autoencoder in the min Objective are updated towards the overall loss back-propagation.
In the proposed LSTM-AVAE model (Figure 2E), we combine the timeseries feature representation learning advantages of the VAE with the temporal modeling performance of LSTM to extract features from data. Bidirectional recurrent neural networks (RNN) have shown to be advantageous over unidirectional RNN (Yu Y. et al., 2019), and hence, here, we use a variant of VAE with its encoder and decoder implemented by bidirectional LSTM networks.
In the proposed model, a bidirectional LSTM layer has two sets of LSTM cells. For each input data sample
The details of the architectures for the LSTM, the encoder, the decoder, and the adversary network are summarized in Table 1. The number of hidden layers is set to 1. As a result of the bidirectional structure, the total number of hidden layers is doubled. The number of LSTM hidden units is searched and the results with the highest accuracy are reported. The learning rate of 0.001, the batch size of 32, and the regularization drop-out ratio of 0.2 were selected. For the encoder architecture, 20 temporal and 10 spatial convolutional units are used, embedding the temporal and spatial filtering. The last fully connected layer at the output of the encoder generates the dz-dimensional latent parameter vector. Our experiments with different activation functions in the architectures showed that, on average, adding an activation function to the last layer and using ReLU activation function offered slightly better results. The classifier utilizes representation z as an input to a fully connected layer with a softmax unit for class label discrimination. The adversary network is realized as a fully connected layer with 8 softmax units for subject discrimination, to obtain normalized log-probabilities that are used to calculate the losses. We used temporal convolution kernel size of 20, and spatial convolution kernel size of 30.
In the proposed model, after training the autoencoder model, the trained encoder with frozen network weights is utilized as an static feature extractor. The feature representations are sampled from the learned encoder μx and σx. An independent classifier is attached to the encoder and is trained to estimate the class label
The classifier is optimized to minimize the cross-entropy loss,
After training, new input data
We considered, multilayer perceptron (MLP) (realized as a single layer with 15 neurons), nearest neighbors (NN), linear discriminant analysis (LDA), linear regression (LR), and support vector machine (SVM), as classifiers.
7The goal of this work is to utilize subject-invariant feature extraction to develop models for accurate mTBI identification. The performance of the models were evaluated in a cross-subject transfer learning task with a 1 held-out subject training and testing approach. That is, data from one subject was held out for testing. Data from the remaining subjects were used for training. Data for training and validation sets was formed by randomly selecting 80% and 20% of the data from each of the remaining subjects, respectively. This procedure was repeated for each held-out subject, and the accuracy result averaged over all cross-subject runs was computed. The autoencoder models were first trained using the training data that were normalized to have zero mean. To prevent overfitting, the validation set was used to stop the training process early. The training process was terminated if the performance over the validation set was decreased compared to the previous training epoch. In our models, following the training of the autoencoder models, the trained encoders with frozen weights were used as feature extractors. Next, utilizing the feature representations from the training set and their corresponding class labels y, a separate classifier was trained as shown in Figure 3. To train the classifiers, the binary cross-entropy loss was minimized with the class labels y. In the last step, the held-out subject was used to assess the models’ cross-subject transfer learning performance. We repeated the described training and testing procedures for all subjects, and averaged the accuracy results over all cross-subject runs. The average accuracy results for different feature extraction models using the considered classifiers are reported in Figure 4. To be consistent, these results are based on setting the dimension of the latent representation feature, dz, to 10 for all models. This value was selected based on further investigation about the effects of dz on the models’ performance, as will be discussed in Section 3.3.
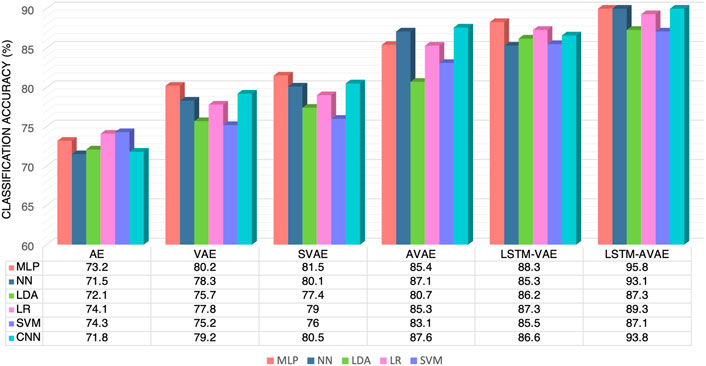
FIGURE 4. Average accuracy results of different models and classifiers in subject-invariant feature extraction for mTBI identification.
Overall, comparing the performance of all the classifiers, we observe that on average, the MLP classifier offers the highest accuracy with various models. The highest accuracy (= 95.8%) is achieved by the proposed LSTM-AVAE using this classifier.
To assess the impact of variational inference in learning features for mTBI identification, we compare the results of AE and VAE. From Figure 4, one can observe that VAE achieves higher accuracy than AE for all the considered classifiers, suggesting that variational representation learning allows for a better feature representation extraction, ultimately resulting in a more accurate mTBI identification model. The reason is due to the additional tunable parameters in the VAE model, in comparison to the AE model, that provide more control over the latent distribution learning ability (Higgins et al., 2016b). It has also been shown that VAE models are capable of learning representations with disentangled factors (Higgins et al., 2016a) due to the isotropic Gaussian priors on the latent variable, the known power of the Bayesian models. The better performance of VAE compared to AE models has also been shown previously in other applications such as anomaly detection, object identification, and BCIs (Dai et al., 2019; Tahir et al., 2021; Zhou et al., 2021). As an additional point, comparing the results of VAE with SVAE, suggests the added value of supervised learning in training better models.
The positive impact of adversarial networks in learning generalizable representations that are domain-, task-, subjects- and source-invariant has been shown recently in many applications such as in drug molecular analysis (Hong et al., 2019), decoding brain states (Du et al., 2019), brain lesion segmentation (Kamnitsas et al., 2017), and evaluating subjects’ mental states (Bethge et al., 2022b). These methods learn representations that are independent of some nuisance variables such as subject-specific or task-specific variations. In this case, there will be a trade-off between enforcing representations that are independent of the nuisance variable via adversary and retaining enough information for the successful data reconstruction of the decoder. In our model architecture, this trade-off can be balanced via the weight parameter λ.
To investigate the impact of adversary networks on subject-invariant feature learning in cross-subject mTBI identification, we varied the value of the weight parameter λ, which adjusts the impact of adversary network in feature learning, for λ ∈ {0, 0.01, 0.05, 0.1, 0.2, 0.5}. The AVAE and the proposed LSTM-AVAE models with the MLP and LR classifiers were considered for performance comparison. Note that for λ = 0 the models are equivalent to simple VAE and LSTM-VAE models, respectively. We computed the accuracy and F1 score of the classifiers for each λ value, and the results are summarized in Table 2.
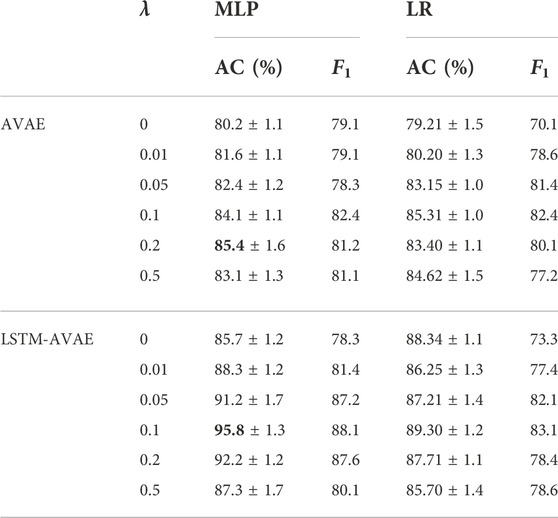
TABLE 2. Comparing the impact of the adversary network on the model accuracy (MLP: multilayer perceptron, LR: linear regression).
From Table 2, one can observe that for both methods and considered classifiers, adding the adversary network to the models (i.e., λ ≠ 0) increases the accuracy, further emphasizing the positive impact of the inclusion of the adversary networks in subject-invariant feature learning. Moreover, it can be seen that the LSTM-AVAE and the MLP classifier with λ = 0.1 achieves the highest classification accuracy of 95.8%, which signifies the robustness of the proposed method in cross-subject transfer learning for mTBI identification.
Moreover, we performed a repeated measures analysis of variance (ANOVA) statistical test on the results of LSTM-VAE and LSTM-AVAE, to statistically compare the performances of the adversary and non-adversary LSTM models, i.e., LSTM-AVAE and LSTM-VAE. We compared the accuracy results for each held-out subject (test data) and across different repeated runs using the MLP classifier. Results, as shown in Figure 5, indicate a significant increase in accuracy with the adversarial training (p = 0.02), which rejects the hypothesis that results are equal across subjects and runs.
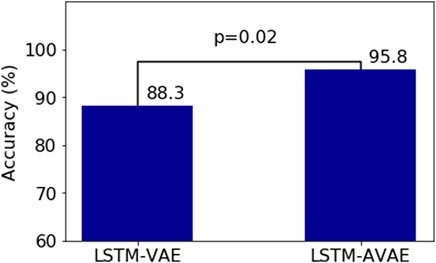
FIGURE 5. Statistical analysis using ANOVA test comparing adversary (LSTM-AVAE) and non-adversary (LSTM-VAE) models (p =0.02).
To investigate the effect of the feature dimension dz on the models’ cross-subject classification performance and determine the proper feature dimension dz, we considered different feature dimensions dz ∈ {3, 5, 7, 8, 9, 10, 11, 12, 13, 15, 18, 20}. The VAE, AVAE, and the proposed LSTM-AVAE models were trained with the extracted latent feature of different dimensions and their optimized parameters, and the corresponding accuracy for the 1 held-out subject averaged over all subjects using the MLP classifier was computed. The results are shown in Figure 6. As can be seen, the highest accuracy is obtained when dz = 10. Increasing the size of features further than dz = 10, does not improve the accuracy results of the models, suggesting that higher feature dimension does not provide additional information for the models. Moreover, the result illustrates that LSTM-AVAE provides higher accuracy than VAE and AVAE for all feature dimensions, indicating the importance of including temporal information for mTBI identification, as will be discussed further in the next section.
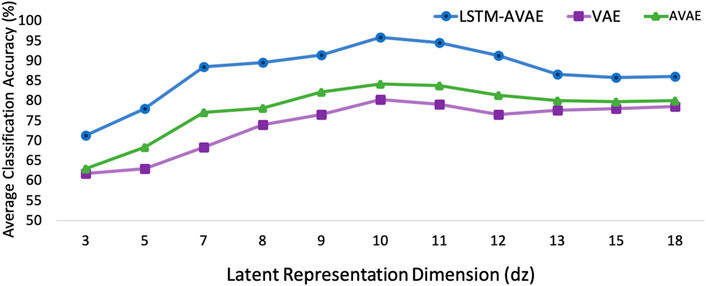
FIGURE 6. Classification accuracy results of the LSTM-AVAE and the VAE models (λ =0.1) for different dimensions of the latent feature, dz, using MLP classifier.
The significance of considering temporal dependency in the analysis of brain activity has been previously noted (Linkenkaer-Hansen et al., 2001; Cornblath et al., 2020; Gu et al., 2021). For example, the added value of considering temporal dependencies has been shown in applications such as emotion recognition (Alhagry et al., 2017) or gait decoding from EEG data (Tortora et al., 2020), or using LSTM recurrent neural networks in discriminating motor tasks from EEG data (Shamsi et al., 2021). Recently, it has been shown that considering the temporal dependency in the GCaMP brain dynamics also improves the performance of the analysis. For example, considering temporal data in studies such as (Salsabilian and Najafizadeh, 2021b; Perich et al., 2021) has improved the behavior decoding and modeling performance using GCaMP data.
Moreover, the improved performance of the VAE models along with LSTM models has been shown in some studies. For example, in (Niu et al., 2020), the added value of considering temporal information is shown using an LSTM-based VAE-GAN network for timeseries anomaly detection. In (Park et al., 2018), using an LSTM-based VAE detector has improved the performance of the robot-assistive model.
In the case of brain injury, brain functional connections may have been disrupted, and considering temporal information may indeed help to find these disruptions and alterations of the brain communication flow. To investigate whether including the temporal dependencies can lead to more precise predictions of mTBI, we used an LSTM network to introduce the temporal dependency to the VAE model. In the presented structure for the LSTM-VAE, the model projects the multivariate timeseries into the latent space representations at each time step, and the decoder uses the latent space representations to reconstruct the input. In this approach, the temporal dependency between the points in each data sample (i.e. X) is processed by the LSTM in the VAE model.
To assess the impact of capturing the spatio-temporal features of the data for mTBI identification, we compare the results of VAE to LSTM-VAE and the results of AVAE to LSTM-AVAE in Figure 4. It can be observed that although we considered temporal and spatial convolution layers in the structure of the autoencoders (see Table 2), all classifiers achieve higher accuracy results with the LSTM-based models (LSTM-VAE and LSTM-AVAE) compared to their non-LSTM counterparts (VAE and AVAE). This result demonstrates that the LSTM-based models are effective in extracting and learning the temporal dependencies in the neural data that are informative for mTBI identification. The highest accuracy of 95.8% is achieved by the proposed LSTM-AVAE using the MLP classifier.
The accuracy result of the LSTM-AVAE model with the MLP classifier, dz = 10, and λ = 0.1 obtained for each subject is reported in Table 3. We observe that the proposed model is capable of achieving high accuracy among all subjects with a minimum and maximum mean accuracy of 91.96% and 98.79%, respectively.
Inspired by recent studies that suggest the experimental results benefit from utilizing conditional VAE (cVAE) (Sohn et al., 2015) in removing the impact of a nuisance variable from the learning representations (e.g., (Özdenizci et al., 2019)), here, we adapt cVAE in the proposed architecture to explore its impact in further removing subject-dependency from mTBI latent representations during training of the encoder. In cVAE, the decoder is conditioned on a nuisance variable as an additional input besides the latent representations. In this way, as the nuisance variable is already given to the decoder, the encoder is expected to only learn representations that are invariant of the nuisance variable. The loss function of the cVAE,
Considering that only the encoder is used for classification, conditioning the decoder on subject variable s, will not affect the rest of the modeling and classification chain. Here, we consider the LSTM-A-cVAE model, by conditioning the decoder in the LSTM-AVAE model on the subject label si. We compare the performance of LSTM-A-cVAE with LSTM-AVAE considering the LR and MLP classifiers with λ = {0.1, 0.2}. The training and testing procedures were kept similar to as described earlier. Results are presented in Table 4. We observe that conditioning the VAE on the subject variable has increased the accuracy by 2% on average in cross-subject transfer learning, suggesting the added value of cVAE in removing subject-dependent information from representations for cross-subject mTBI identification.
In this paper, by taking advantage of adversary networks and proposing an LSTM-AVAE model, we addressed the problem of subject variability, which imposes challenges in extracting injury-related features for accurate mTBI diagnosis. The proposed model considers the temporal dependency of neural data and learns representations from neural data, while the attached adversary network disentangles the subject-related information from learned representations, making the model proper for cross-subject feature extraction. The experimental results demonstrated the benefits of the proposed LSTM-AVAE model for accurate mTBI identification, proving the ability of the model in extracting robust subject-invariant features. The proposed approach can be generalized to other domains to learn subject-invariant features.
The data analyzed in this study is subject to the following licenses/restrictions: The dataset was collected in another research laboratory. Requests to access these datasets should be directed to laleh.najafizadeh@rutgers.edu.
The animal study was reviewed and approved by the Rutgers University Institutional Animal Care and Use Committee.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the National Science Foundation (NSF) under award 1605646, and the New Jersey Commission on Brain Injury Research (NJCBIR) under award CBIR16IRG032.
The authors thank Prof. David J. Margolis and Dr. Christian R. Lee, with the Department of Cell Biology and Neuroscience at Rutgers University, for providing the experimental data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alhagry, S., Fahmy, A. A., and El-Khoribi, R. A. (2017). Emotion recognition based on EEG using LSTM recurrent neural network. ijacsa. 8. doi:10.14569/ijacsa.2017.081046
Angjelichinoski, M., Choi, J., Banerjee, T., Pesaran, B., and Tarokh, V. (2020). Cross-subject decoding of eye movement goals from local field potentials. J. Neural Eng. 17, 016067. doi:10.1088/1741-2552/ab6df3
Beauchamp, A., Yee, Y., Darwin, B., Raznahan, A., Mars, R. B., and Lerch, J. P. (2022). Whole-brain comparison of rodent and human brains using spatial transcriptomics. bioRxiv.
Bethge, D., Hallgarten, P., Grosse-Puppendahl, T., Kari, M., Mikut, R., Schmidt, A., et al. (2022a). “Domain-invariant representation learning from EEG with private encoders,” in ICASSP 2022 IEEE international conference on acoustics, speech and signal processing (IEEE), 1236–1240.
Bethge, D., Hallgarten, P., Özdenizci, O., Mikut, R., Schmidt, A., and Grosse-Puppendahl, T. (2022b). Exploiting multiple EEG data domains with adversarial learning. arXiv preprint arXiv:2204.07777.
Breschi, A., Gingeras, T. R., and Guigó, R. (2017). Comparative transcriptomics in human and mouse. Nat. Rev. Genet. 18, 425–440. doi:10.1038/nrg.2017.19
Cornblath, E. J., Ashourvan, A., Kim, J. Z., Betzel, R. F., Ciric, R., Adebimpe, A., et al. (2020). Temporal sequences of brain activity at rest are constrained by white matter structure and modulated by cognitive demands. Commun. Biol. 3, 1–12. doi:10.1038/s42003-020-0961-x
Cortes, D., and Pera, M. F. (2021). The genetic basis of inter-individual variation in recovery from traumatic brain injury. npj Regen. Med. 6, 5–9. doi:10.1038/s41536-020-00114-y
Cramer, J. V., Gesierich, B., Roth, S., Dichgans, M., Duering, M., and Liesz, A. (2019). In vivo widefield calcium imaging of the mouse cortex for analysis of network connectivity in health and brain disease. Neuroimage 199, 570–584.
Cramer, S. W., Haley, S. P., Popa, L. S., Carter, R. E, Scott, E., Flaherty, E. B., et al. (2022). Wide-field calcium imaging reveals widespread changes in cortical connectivity following repetitive, mild traumatic brain injury in the mouse. bioRxiv 11, 1. doi:10.1038/s41598-021-02371-3
Dai, M., Zheng, D., Na, R., Wang, S., and Zhang, S. (2019). EEG classification of motor imagery using a novel deep learning framework. Sensors 19, 551. doi:10.3390/s19030551
Du, C., Jinpeng, L., Lijie, H., and Huiguang, H. (2019). Brain encoding and decoding in fMRI with bidirectional deep generative models. Engineering 5, 948–953.
Eierud, C., Craddock, R. C., Fletcher, S., Aulakh, M., King-Casas, B., Kuehl, D., et al. (2014). Neuroimaging after mild traumatic brain injury: Review and meta-analysis. NeuroImage Clin. 4, 283–294. doi:10.1016/j.nicl.2013.12.009
Ellenbroek, B., and Youn, J. (2016). Rodent models in neuroscience research: Is it a rat race? Dis. models Mech. 9, 1079–1087. doi:10.1242/dmm.026120
Fazli, S., Popescu, F., Danóczy, M., Blankertz, B., Müller, K.-R., and Grozea, C. (2009). Subject-independent mental state classification in single trials. Neural Netw. 22, 1305–1312. doi:10.1016/j.neunet.2009.06.003
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). Generative adversarial nets. Adv. Neural Inf. Process. Syst. 27.
Gu, H., Schulz, K. P., Fan, J., and Yang, Y. (2021). Temporal dynamics of functional brain states underlie cognitive performance. Cereb. Cortex 31, 2125–2138. doi:10.1093/cercor/bhaa350
Han, M., Özdenizci, O., Koike-Akino, T., Wang, Y., and Erdoğmuş, D. (2021). Universal physiological representation learning with soft-disentangled rateless autoencoders. IEEE J. Biomed. Health Inf. 25, 2928–2937. doi:10.1109/jbhi.2021.3062335
Han, M., Özdenizci, O., Wang, Y., Koike-Akino, T., and Erdoğmuş, D. (2020). Disentangled adversarial autoencoder for subject-invariant physiological feature extraction. IEEE Signal Process. Lett. 27, 1565–1569. doi:10.1109/lsp.2020.3020215
Higgins, I., Matthey, L., Glorot, X., Pal, A., Uria, B., Blundell, C., et al. (2016a). Early visual concept learning with unsupervised deep learning. arXiv preprint arXiv:1606.05579.
Higgins, I., Matthey, L., Pal, A., Burgess, C., Glorot, X., Botvinick, M., et al. (2016b). beta-vae: Learning basic visual concepts with a constrained variational framework
Hong, S. H., Ryu, S., Lim, J., and Kim, W. Y. (2019). Molecular generative model based on an adversarially regularized autoencoder. J. Chem. Inf. Model. 60, 29–36. doi:10.1021/acs.jcim.9b00694
Iverson, G. L., Lovell, M. R., Smith, S., and Franzen, M. D. (2000). Prevalence of abnormal CT-scans following mild head injury. Brain Inj. 14, 1057–1061. doi:10.1080/02699050050203559
Kamnitsas, K., Baumgartner, C., Ledig, C., Newcombe, V., Simpson, J., Kane, A., et al. (2017). “Unsupervised domain adaptation in brain lesion segmentation with adversarial networks,” in International conference on information processing in medical imaging (Springer), 597–609.
Koochaki, F., and Najafizadeh, L. (2021). “A convolutional autoencoder for identification of mild traumatic brain injury,” in 10th international IEEE/EMBS conference on neural engineering (NER), 412–415.
Koochaki, F., Shamsi, F., and Najafizadeh, L. (2020). “Detecting mTBI by learning spatio-temporal characteristics of widefield calcium imaging data using deep learning,” in 42nd annual international conference of the IEEE engineering in medicine & Biology society (EMBC), 2917–2920.
Kou, Z., and Iraji, A. (2014). Imaging brain plasticity after trauma. Neural Regen. Res. 9, 693. doi:10.4103/1673-5374.131568
Kullback, S., and Leibler, R. A. (1951). On information and sufficiency. Ann. Math. Stat. 22, 79–86. doi:10.1214/aoms/1177729694
Lee, C. R., and Margolis, D. J. (2016). Pupil dynamics reflect behavioral choice and learning in a go/nogo tactile decision-making task in mice. Front. Behav. Neurosci. 10, 200. doi:10.3389/fnbeh.2016.00200
Lee, C. R., Najafizadeh, L., and Margolis, D. J. (2020)., 225. Springer, 467–480.Investigating learning-related neural circuitry with chronic in vivo optical imagingBrain Struct. Funct.
Levin, H. S., and Diaz-Arrastia, R. R. (2015). Diagnosis, prognosis, and clinical management of mild traumatic brain injury. Lancet Neurology 14, 506–517. doi:10.1016/s1474-4422(15)00002-2
Li, J., Qiu, S., Du, C., Wang, Y., and He, H. (2019). Domain adaptation for EEG emotion recognition based on latent representation similarity. IEEE Trans. Cogn. Dev. Syst. 12, 344–353. doi:10.1109/tcds.2019.2949306
Linkenkaer-Hansen, K., Nikouline, V. V., Palva, J. M., and Ilmoniemi, R. J. (2001). Long-range temporal correlations and scaling behavior in human brain oscillations. J. Neurosci. 21, 1370–1377. doi:10.1523/jneurosci.21-04-01370.2001
Lotte, F., and Guan, C. (2010). Regularizing common spatial patterns to improve BCI designs: Unified theory and new algorithms. IEEE Trans. Biomed. Eng. 58, 355–362. doi:10.1109/tbme.2010.2082539
Louppe, G., Kagan, M., and Cranmer, K. (2017). Learning to pivot with adversarial networks. Adv. Neural Inf. Process. Syst. 30.
Makhzani, A., Shlens, J., Jaitly, N., Goodfellow, I., and Frey, B. (2015). Adversarial autoencoders. ArXiv Preprint ArXiv:1511.05644.
Marshall, J. J., and Mason, J. O. (2019). Mouse vs man: Organoid models of brain development & disease. Brain Res. 1724, 146427. doi:10.1016/j.brainres.2019.146427
Ming, Y., Ding, W., Pelusi, D., Wu, D., Wang, Y.-K., Prasad, M., et al. (2019). Subject adaptation network for EEG data analysis. Appl. Soft Comput. 84, 105689. doi:10.1016/j.asoc.2019.105689
Morganti-Kossmann, M. C., Yan, E., and Bye, N. (2010). Animal models of traumatic brain injury: Is there an optimal model to reproduce human brain injury in the laboratory? Injury 41, S10–S13. doi:10.1016/j.injury.2010.03.032
Morioka, H., Kanemura, A., Hirayama, J.-i., Shikauchi, M., Ogawa, T., Ikeda, S., et al. (2015). Learning a common dictionary for subject-transfer decoding with resting calibration. NeuroImage 111, 167–178. doi:10.1016/j.neuroimage.2015.02.015
Niu, Z., Yu, K., and Wu, X. (2020). LSTM-based VAE-GAN for time-series anomaly detection. Sensors 20, 3738. doi:10.3390/s20133738
Özdenizci, O., Wang, Y., Koike-Akino, T., and Erdoğmuş, D. (2020). Learning invariant representations from EEG via adversarial inference. IEEE access 8, 27074–27085. doi:10.1109/access.2020.2971600
Özdenizci, O., Wang, Y., Koike-Akino, T., and Erdoğmuş, D. (2019). “Transfer learning in brain-computer interfaces with adversarial variational autoencoders,” in 2019 9th international IEEE/EMBS conference on neural engineering (NER) (IEEE)–210.
Park, D., Hoshi, Y., and Kemp, C. C. (2018). A multimodal anomaly detector for robot-assisted feeding using an lstm-based variational autoencoder. IEEE Robot. Autom. Lett. 3, 1544–1551. doi:10.1109/lra.2018.2801475
Perich, M. G., Arlt, C., Soares, S., Young, M. E., Mosher, C. P., Minxha, J., et al. (2021). Inferring brain-wide interactions using data-constrained recurrent neural network models. BioRxiv, 1–37.
Peterson, S. M., Steine-Hanson, Z., Davis, N., Rao, R. P., and Brunton, B. W. (2021). Generalized neural decoders for transfer learning across participants and recording modalities. J. Neural Eng. 18, 026014. doi:10.1088/1741-2552/abda0b
Ruff, R. M., Iverson, G. L., Barth, J. T., Bush, S. S., Broshek, D. K., Policy, N., et al. (2009). Recommendations for diagnosing a mild traumatic brain injury: A national academy of neuropsychology education paper. Archives Clin. neuropsychology 24, 3–10. doi:10.1093/arclin/acp006
Salsabilian, S., Bibineyshvili, E., Margolis, D. J., and Najafizadeh, L. (2019). “Quantifying changes in brain function following injury via network measures,” in 41st annual international conference of the IEEE engineering in medicine and Biology society (Berlin, Germany: EMBC), 5217–5220.
Salsabilian, S., Bibineyshvili, E., Margolis, D. J., and Najafizadeh, L. (2020a). “Study of functional network topology alterations after injury via embedding methods,” in Optics and the brain (Washington, D.C: Optical Society of America). BW4C–3.
Salsabilian, S., Lee, C. R., Margolis, D. J., and Najafizadeh, L. (2018). “Using connectivity to infer behavior from cortical activity recorded through widefield transcranial imaging,” in Optics and the brain (Hollywood, FL: Optical Society of America). BTu2C–4.
Salsabilian, S., and Najafizadeh, L. (2021b). “A variational encoder framework for decoding behavior choices from neural data,” in 43rd annual international Conference of the IEEE Engineering in medicine & Biology society (EMBC).
Salsabilian, S., and Najafizadeh, L. (2021a). “An adversarial variational autoencoder approach toward transfer learning for mTBI identification,” in 10th international IEEE/EMBS conference on neural engineering (NER), 408–411.
Salsabilian, S., and Najafizadeh, L. (2020). “Detection of mild traumatic brain injury via topological graph embedding and 2D convolutional neural networks,” in 42st annual international conference of the IEEE engineering in medicine and Biology society (EMBC).
Salsabilian, S., Zhu, L., Lee, C. R., Margolis, D. J., and Najafizadeh, L. (2020b). “Identifying task-related brain functional states via cortical networks,” in IEEE international symposium on circuits and systems (ISCAS), 1–4.
Schmid, W., Fan, Y., Chi, T., Golanov, E., Regnier-Golanov, A. S., Austerman, R. J., et al. (2021). Review of wearable technologies and machine learning methodologies for systematic detection of mild traumatic brain injuries. J. Neural Eng. 18, 041006. doi:10.1088/1741-2552/ac1982
Shamsi, F., Haddad, A., and Najafizadeh, L. (2021). Early classification of motor tasks using dynamic functional connectivity graphs from EEG. J. Neural Eng. 18, 016015. doi:10.1088/1741-2552/abce70
Sohn, K., Lee, H., and Yan, X. (2015). Learning structured output representation using deep conditional generative models. Adv. neural Inf. Process. Syst. 28.
Srivastava, N., Mansimov, E., and Salakhudinov, R. (2015). “Unsupervised learning of video representations using LSTMs,” in International conference on machine learning (Lille, France: PMLR), 843–852.
Tahir, R., Sargano, A. B., and Habib, Z. (2021). Voxel-based 3D object reconstruction from single 2D image using variational autoencoders. Mathematics 9, 2288. doi:10.3390/math9182288
Tortora, S., Ghidoni, S., Chisari, C., Micera, S., and Artoni, F. (2020). Deep learning-based BCI for gait decoding from EEG with LSTM recurrent neural network. J. Neural Eng. 17, 046011. doi:10.1088/1741-2552/ab9842
Waytowich, N. R., Lawhern, V. J., Bohannon, A. W., Ball, K. R., and Lance, B. J. (2016). Spectral transfer learning using information geometry for a user-independent brain-computer interface. Front. Neurosci. 10, 430. doi:10.3389/fnins.2016.00430
Wiltschko, A. B., Johnson, M. J., Iurilli, G., Peterson, R. E., Katon, J. M., Pashkovski, S. L., et al. (2015). Mapping sub-second structure in mouse behavior. Neuron 88, 1121–1135. doi:10.1016/j.neuron.2015.11.031
Wu, F., Jing, X.-Y., Wu, Z., Ji, Y., Dong, X., Luo, X., et al. (2020). Modality-specific and shared generative adversarial network for cross-modal retrieval. Pattern Recognition 104.
Xie, Q., Dai, Z., Du, Y., Hovy, E., and Neubig, G. (2017). Controllable invariance through adversarial feature learning. Adv. Neural Inf. Process. Syst. 30.
You, Z., Yang, J., Takahashi, K., Yager, P. H., Kim, H.-H., Qin, T., et al. (2007). Reduced tissue damage and improved recovery of motor function after traumatic brain injury in mice deficient in complement component c4. J. Cereb. Blood Flow. Metab. 27, 1954–1964. doi:10.1038/sj.jcbfm.9600497
Yu, W., Kim, I. Y., and Mechefske, C. (2019a). Remaining useful life estimation using a bidirectional recurrent neural network based autoencoder scheme. Mech. Syst. Signal Process. 129, 764–780. doi:10.1016/j.ymssp.2019.05.005
Yu, Y., Si, X., Hu, C., and Zhang, J. (2019b). A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 31, 1235–1270. doi:10.1162/neco_a_01199
Zhou, Y., Liang, X., Zhang, W., Zhang, L., and Song, X. (2021). VAE-based deep SVDD for anomaly detection. Neurocomputing 453, 131–140. doi:10.1016/j.neucom.2021.04.089
Zhu, L., Lee, C. R., Margolis, D. J., and Najafizadeh, L. (2018). Decoding cortical brain states from widefield calcium imaging data using visibility graph. Biomed. Opt. Express 9, 3017–3036. doi:10.1364/boe.9.003017
Keywords: variational autoencoder (VAE), adversarial regularization, cross-subject transfer learning, mild TBI (mTBI), LSTM-long short-term memory
Citation: Salsabilian S and Najafizadeh L (2022) Subject-invariant feature learning for mTBI identification using LSTM-based variational autoencoder with adversarial regularization. Front. Sig. Proc. 2:1019253. doi: 10.3389/frsip.2022.1019253
Received: 14 August 2022; Accepted: 14 November 2022;
Published: 30 November 2022.
Edited by:
Ozan Özdenizci, Graz University of Technology, AustriaReviewed by:
Ye Wang, Mitsubishi Electric Research Laboratories (MERL), United StatesCopyright © 2022 Salsabilian and Najafizadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laleh Najafizadeh, bGFsZWgubmFqYWZpemFkZWhAcnV0Z2Vycy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.