- 1Department of Food Science, University of Copenhagen, Frederiksberg, Denmark
- 2Division of Physical Chemistry, Department of Chemistry, Lund University, Lund, Sweden
- 3NanoLund, Lund University, Lund, Sweden
- 4LINXS Institute of Advanced Neutron and X-Ray Science, Lund, Sweden
- 5School of Chemical Engineering and Translational Nanobioscience Research Center, Sungkyunkwan University, Suwon, Republic of Korea
- 6Physical Chemistry of Biosystems, Institute of Physical Chemistry, Heidelberg University, Heidelberg, Germany
- 7Center for Integrative Medicine and Physics, Institute for Advanced Study, Kyoto University, Kyoto, Japan
Food is a complex soft matter, because various components, such as proteins, lipids, and carbohydrates, are self-assembled via non-covalent, colloidal interactions and form hierarchical structures at multiple length scales. Soft matter scientists have shown an increasing interest in understanding the general principles governing the food structure formation. During the last several decades, an increasing number of studies have shown that the maintenance of healthy gastrointestinal tract and its microbiome is essential for human health and wellbeing. The realization of the importance of the gastrointestinal microbiome has led to the development of probiotics, which are defined as living bacteria that confer a health benefit on the host. Probiotic bacteria and enzymes can be delivered to the intestinal system by formulating appropriate carriers and including these into food ingested by humans. Despite this simple statement, it involves many challenges in the field of soft matter science. This review aims to highlight how the key concepts in soft matter science can be used to design, characterize, and evaluate self-assembled formulations of probiotics and enzymes based on lipids and biopolymers. The topics covered in this review includes the emulsification of oil-water mixtures, the self-assembly of lipids and polymers at interfaces, the electrostatics and viscoelasticity of interfaces, and the wetting/adhesion of colloidal particles.
1 Introduction: food science and soft matter
Food is a complex material, consisting of macromolecules, such as proteins, lipids, and carbohydrates, together with water, minerals, and many other minor but nutritionally significant compounds, such as vitamins and polyphenols. These components are self-assembled via colloidal forces and form hierarchical structures at multiple length scales. The understanding of such forces is fundamental to not only understand the structures naturally present in plant and animal tissues, but also how they can be disrupted and reassembled into new structures with higher nutritional value by employing appropriate processing. It is noteworthy that food structures are not constant but are prone to changes during harvesting, storage, distribution, and digestion. Soft matter scientists have shown an increasing interest in understanding the general principles governing the food structure formation. As Assenza and Mezzenga pointed out, the application of theoretical and experimental tools from soft condensed matter physics to describe foods and macronutrients is challenging because the length scales involved is ranging from angstroms to tens of micrometers (Assenza and Mezzenga, 2019). The use of techniques, such as neutron and x-ray scattering, has provided new insights (Gilbert, 2019).
An important aspect that controls the nutritional value of food is that the structure is broken down with the help of enzymes in the gastro-intestinal tract. The structural changes are directly linked to physiological events, as reported for digestion of lipids by Patton and Carrey in the 1970s (Patton and Carey, 1979). Modern analytical tools, such as synchrotron x-ray scattering, have provided new insights on the changes in the lipid assembly structures that occur due to lipolysis (Barauskas and Nylander, 2008; Salentinig et al., 2011; Wadsäter et al., 2018), as well as for more complex substrates, such as milk, under simulated in vivo gastrointestinal tract conditions (Salentinig et al., 2013; Salentinig et al., 2015). The structure formed during digestion depends on the lipid composition of the food, as shown by Pham et al. for different infant formulas (Pham et al., 2020). Such studies offer the possibility to design human milk-like systems that control nutrient transport and the uptake of bioactive substances. Because lipolytic processes occur under physiological conditions involving a water soluble enzyme acting on water insoluble lipid substrates, digestion always occurs at the interfaces and is strongly influenced by pH and other solution conditions (Humphreys et al., 2022). X-ray fluorescence microscopy, X-ray and neutron scattering were applied to study the digestion of protein gels formed from canola seed storage proteins in vitro, which enabled to follow the process on the individual protein and the structure connectivity (Pasquier et al., 2019).
Another key factor is the effect of diet on physiological processes and health. Food supplements like anthocyanins from blueberries have been demonstrated to hamper weakening of neuronal functions but are challenging to use due to their limited bioavailability (Tran and Tran, 2021). It should be noted Porphyromonas gingivalis, which is a pathogen associated chronic periodontitis also has been identified in the brain of Alzheimer’s disease patients (Dominy et al., 2019). The use of brain-penetrant inhibitors can reduce P. gingivalis brain infection and hamper neurodegeneration in e.g. AD patients (Dominy et al., 2019). Again, formulation is a challenge and might possible through food supplement. The brain contains a rich variety of lipids, assembled to form intriguing structures for which the links to the brain functions are not yet fully understood (Siegel et al., 1999). These structures are dependent on the cholesterol content, the composition of salts, and temperature (Alfredsson et al., 2021). Lipidomics studies have demonstrated that a certain lipid composition pattern in the blood can be linked to the clinical diagnosis of Alzheimer Disease (Proitsi et al., 2017). It is tempting to assume that this in turn is related to differences in the lipid self-assembly structure. Some of the work has been concerned with the Alzheimer disease and the possibility to use food components, such as extracts from mulberry, as neuroprotective agents in this respect as it is has been considered to inhibit neuroinflammation (Liu and Du, 2020). Our understanding of the uptake of nutrients and drugs in the intestine is complicated by the fact that many food components, such as fat, starch, and fibers, are insoluble. The content of the lumen should be considered as a dispersed colloidal system rather than a simple solution. The maintenance of healthy gastrointestinal tract and its microbiome is essential for human health and wellbeing. The total number of cells in the microbiome in the gastrointestinal tract is larger than that of the human/mammalian cells and it has therefore been suggested that the human body should be considered a “superorganism” (Woolfson, 2016). The realization of the importance of the gastrointestinal microbiome has led to the development of so-called probiotics. Probiotics are defined as living bacteria that, when administered in adequate amounts, confer a health benefit on the host (FAO/WHO, 2001). A mechanistic understanding of the uptake of bio-actives (comprising nutrients and other functional components such as drugs) in the intestine also requires knowledge of the interaction within in the mucus layer, which is partly lacking. In particular, the link between the interactions on the molecular, colloidal, and macroscopic levels are often missing partly due to the lack of suitable model systems. Some studies on mucus expressing cell cultures have provided some new insights (Gagnon et al., 2013; Kleiveland, 2015; Ferraretto et al., 2018; Eshrati et al., 2019). The digestive system is a challenging environment where the pH varies from pH = 1 in the stomach to pH = 8 in the intestine. Additionally, lipases and bile salts challenge any self-assembled structure used to orally deliver any functional ingredients (Assenza and Mezzenga, 2019). Bile salts have been shown to interact not only with lipids but also with a range of biopolymers (Maldonado-Valderrama et al., 2011). Numerous strategies have been proposed to overcome these challenges based on the understanding of colloidal aspects (McClements, 2015).
This review article is structured as follows. In Section 2, we provide an overview of how one can characterize and modify the surface and colloidal properties of probiotic bacteria and how probiotic bacteria can be formulated in colloidal food materials. In Section 3, some representative methodological strategies are introduced to show how to formulate probiotics and enzymes in lipid self-assembliess, silica-based particles, and polymer-based microcapsules. Section 4 highlights recent results on the physical characterization and functional delivery of probiotics and enzymes using the above-mentioned formulations. Section 5 covers how to assess the functions of probiotics and enzymes after delivery using the simulator of the human gastrointestinal tract and the in vitro model of intestinal epithelium integrated into microfludic devices. After this, review is summarized with some future perspectives and challenges for soft matter sciences.
2 Surface and colloidal properties of probiotic bacteria
Probiotic bacteria belonging to either the Gram-negative or Gram-positive groups are micron-sized, living colloidal particles (Behnsen et al., 2013). This review primarily focuses on Gram-positive bacteria, which are the largest group claiming to have probiotic properties. The bacterial surface properties, formation of biofilms on surfaces, aggregation of bacteria (in microbiology termed auto-aggregation) are important, but also constitute the foundation for various approaches for the formulation and the colloidal behavior of bacteria in multiphase systems, such as foams and emulsions.
2.1 Characterization of Gram-negative bacteria cell walls
The cell wall of Gram-positive bacteria (Figure 1A) is characterized by its main entities, namely, an inner cytoplasmic membrane and an outer peptidoglycan layer facing the external environment. In contrast, Gram-negative bacteria have a significantly thinner peptidoglycan layer that is surrounded by a second outer phospholipid membrane, which is decorated with poly-saccharides facing the extracellular environment (Figure 1B) (Rohde, 2019). The peptidoglycan layer is a water-containing cross-linked system of di-saccharides formed by N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), which are cross-linked by flexible pentapeptide amino acid chains forming a mesh-like system (Vollmer et al., 2008), and has embedded proteins and teichoic acid. The main component of teichoic acid is a co-polymer (poly-di-ester) of glycerol and phosphate groups, which is negatively charged at neutral pH. Therefore, the surface of Gram-positive bacteria is considered to be hydrophilic and negatively charged due to the nature of the peptidoglycan layer and the teichoic acid content. However, as discussed in the later section, the natural variation of the cell wall composition also manifests itself by varying its surface charge (from almost neutral to negatively charged) and hydrophobicity. The joint electrostatic effect of the charged groups on the bacterial surface are routinely characterized using zeta-potential measurements, either performed at neutral (physiological) pH or as function of pH to reveal the effect of the protonation and deprotonation of bacterial surface groups (Jiang et al., 2023a).
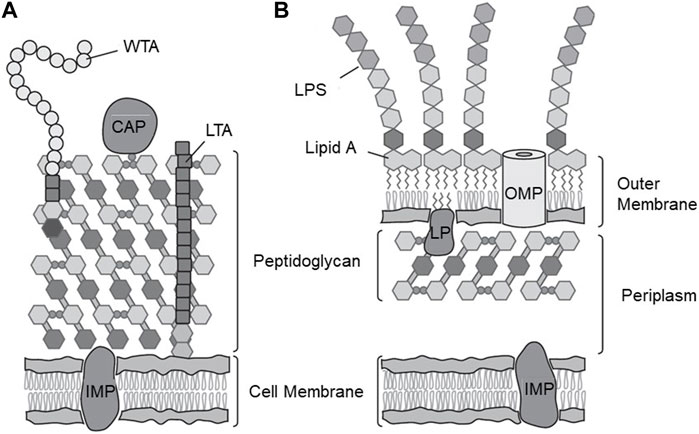
FIGURE 1. Cell wall structures of Gram-positive and Gram-Negative bacteria. (A) Gram positive-bacteria highlighting the plasma membrane, membrane proteins, thick peptidoglycan layer, S-layer proteins, teichoic acid, and polysaccharides (B) Gram-negative cell wall highlighting the plasma membrane, membrane proteins, thin peptidoglycan layer, outer membrane with trans-membrane pores, and LPSs facing the external environment. CAP, covalently attached protein; IMP, integral membrane protein; LP, lipoprotein; LPS, lipopolysaccharide; LTA, lipoteichoic acid; OMP, outer membrane protein; WTA, wall teichoic acid. Adapted from (Silhavy et al., 2010), with permission from Cold Spring Harbor Publications.
Revealing the structure and properties of bacterial surfaces are non-trivial both experimentally and theoretically. Small- and wide-angle X-ray scattering measurements have been widely used to determine the supramolecular self-assembly of purified lipopolysaccharides (LPSs) in suspension experimentally (Brandenburg et al., 1992; Garidel et al., 2005). Specular neutron and X-ray reflectivity at the solid/water and air/water interface have been used to further investigate the molecular-level structures in the z-direction perpendicular to the membrane xy-plane. The monolayer of LPSs at the air/water interface can be used to study the structural integrity of the outermost LPSs (Oliveira et al., 2010; Thoma et al., 2020), whereas asymmetric bilayers formed at the solid/aqueous interface can be used as a more realistic model of the outer membrane, demonstrating the ion-specific collapse of the saccharide head groups (Schneck et al., 2009b; Clifton et al., 2013). The neutron-based techniques provide structural information with Å resolution by optimizing the contrast in the scattering length densities, by selective deuteration and using appropriate mixtures of H2O and D2O. However, these methods are less sensitive to specific ions/elements. Grazing-incidence X-ray fluorescence (GIXF) was proposed as a promising strategy to analyze the electrostatics of LPS surfaces covered by charged saccharide head groups because the X-ray fluorescence signals measured near the critical angle of total reflection can be used to calculate the density profiles of ions in the head group of LPS (Schneck et al., 2010; Abuillan et al., 2013). It is notable that the charge density near the LPS surface is neither uniform nor static, and charged LPS moieties can be crosslinked using multivalent ions. These considerations render the classical Derjaguin-Landau-Verwey-Overbeek theory inadequate. Moreover, atomistic molecular dynamics simulations are not suited to handle the typical length scale (>103 nm3) and characteristic time window of diffusion-limited reactions (∼10–3 s). Pink et al. employed a coarse-grained, minimal model of LPS membranes and calculated the ion density profiles and conformational changes within the framework of a linearized Poisson-Boltzmann theorem derived by Netz (Netz, 1999). They demonstrated that the divalent cations bound to core saccharides form electrostatic potential barrier (Pink et al., 2003; Oliveira et al., 2009), which explains the data obtained by GIXF and reflectivity experiments.
Microbial adhesion to hexadecane (MATH) or microbial adhesion to solvents (MATS) are microbiological standard techniques that provide indirect information about the bacterial surface properties (Rosenberg et al., 1980; Geertsema-Doornbusch et al., 1993). Upon vigorous shaking of a two-phase system, a fraction of the bacteria is trapped in the organic phase and a MATH value is estimated from the optical density (OD) measurements. A larger MATH value means that the surface is predominantly hydrophobic. It has been observed that a coarse and creaming emulsion that retains a fraction of bacteria through a Pickering mechanism is formed (Rosenberg, 1984; Rosenberg, 2006). Unlike real distribution coefficients, no simple equilibrium thermodynamic interpretation can me made using MATH values.
The contact angle at the three-phase contact is related to interfacial tensions of wetting liquid, between the liquid and the bacterial film, and the bacterial film and air through the Young’s equation, that is, in principle the thermodynamic quantities that can be extracted from contact angle measurements. It should be noted that the contact angle of the “dry” bacterial layer in contact to air does not represent the hydrated surfaces of bacteria under water; these measurements represent the contact angles with the “collapsed and dry constituents” of the bacterial cell wall (Van Oss et al., 1988; Jiang et al., 2023a). The Lewis base character (A−) is defined as the difference between the MATS values for chloroform and hexadecane and the Lewis acid character (A+) is derived from the differences of the solvent pair, acetylacetate, and decane. Using contact angle measurements in combination with reference solvents, e.g. water, formamide, and 1-bromo-naphtalene, and the van Oss−Good model, the Lewis acid and base components of the bacterial interfacial tensions can be separated and the bacterial-water interfacial tensions can be estimated. In this approach, the bacterial cohesive free energy density,
Atomic force microscopy (AFM) can be used to characterize the bacterial cell wall and isolated cell wall components such as the peptidoglycan layers. For a review, see (Dufrêne et al., 2021). AFM can be used to detect the surface morphology of the bacterial cell wall, revealing that Gram-positive bacteria exhibit a regular architecture of a peptidoglycan layer and adhesins (Dupres et al., 2005). Moreover, the force-distance curves can be obtained with the µm-scale spatial resolution. A small deformation can be used to detect the elasticity of the cell wall, whereas a larger deformation provides information on the cell stiffness, reflecting the turgor pressure of the cell.
In addition to AFM, defined model systems can be used to study the mechanical properties of bacterial surfaces more quantitatively. Schneck et al. fabricated planar stacks of purified LPS membranes with various strains and preformed off-specular neutron scattering at defined disjoining pressures. Although the model consisted of symmetric bilayer stacks of LPSs, this approach enabled quantitative insights into how saccharide chain modifications modulate the mechanical parameters of outer membranes, such as the bending rigidity and vertical compression modulus (Schneck et al., 2009a). Measuring the response function under the application of a shear stress, which is called interfacial stress rheology, is another technique that is employed to measure the complex viscoelastic modulus of model membranes (Brooks et al., 1999; Schneider et al., 2002). Compared to the commonly used rotating disk rheometry, a small probe-membrane contact enables sensitivity for determining the loss and storage moduli of “soft” membranes, such as the membranes of LPSs of Gram-negative bacteria and lipids mimicking lipoteic acid of Gram-positive bacteria (Tanaka et al., 2004; Herrmann et al., 2015). Experiments using model systems, as compared to experiments using living (or fixed) bacteria, enable the understanding of the contributions of the outermost surfaces, such as the viscoelasticity of the peptidoglycans and inner membranes, as well as the turgor pressure pushing the cell wall from the inside.
Yet, despite the major progress made over the past decades as described above, there is a clear demand for experimental tools that bridge the well-defined model systems and more complex, real bacterial surfaces. On one hand, well-defined two-dimensional model systems, such as LPS membranes, enable the precise measurement of detailed structural features, ion density profiles, and interfacial viscoelasticity. On the other hand, most of the model systems cannot address the effect of the physico-chemical properties of food on biological functions. Moreover, from the theoretical viewpoint, the development of multiscale modeling that connects atomistic simulations and coarse-scale models is required to cover several orders of magnitude in space and time.
2.2 Modification of bacterial surfaces via chemical and physical approaches
Different approaches have recently been used to change the native surface properties of bacteria, with a particular focus on increasing hydrophobicity and thereby the adhesion behavior of Gram-positive bacteria to promote the ability of bacteria to be active in building structure and texture of food by for example be active Pickering stabilizers at surfaces and interphases. Three approaches can be implemented, namely i) adsorption of macro molecules, ii) chemical grafting of molecules to the bacterial wall components, and iii) selective removal of chemical constituents from the bacterial cell wall using enzymes. The first category contains studies by means of the deposition of cationic and anionic polymers (Priya et al., 2011; Yucel Falco et al., 2017b). This procedure changes the sign of the surface potential of the bacterial surface, depending on the effective charge of the outer polyelectrolyte layer. As the polymers used in these studies were mostly hydrophilic, the bacterial surface remained hydrophilic.
2.2.1 Modifications of group i
Falco et al. adsorbed milk proteins (sodium caseinate or β-casein) on Lactobacillus acidophilus (La-5) at a low pH, where the proteins and bacteria are oppositely charged (Yucel Falco et al., 2017a). The modified bacteria showed higher surface activities at the air/water interface, and the change in the bacterial surface potential was assessed by the zeta potential measurements, and Shekarforoush et al. used the adsorption of hydrophobic corn protein zein to enhance the hydrophobicity (Shekarforoush et al., 2022). The MATH values were moderately enhanced without lowering the colony forming unit (CFU) of the strains. The MATH values for Lactobacillus rhamnosus (LGG) and Lactobacillus delbruekii subs. lactis ATCC 4797 (LBD) increased from approximately 7%–20% and from 12% to 45%, respectively.
2.2.2 Modifications of group ii
Fatty acid anhydrides and fatty acid chlorides can be used as hydrophobizing agents by forming either fatty acid amides or esters with the amino or hydroxyl groups of the macro molecules under investigation. The component octanyl succinic anhydride (OSA) is used in the food industry to convert hydrolyzed starch to components with emulsifying properties. OSA is only mildly reactive with water and the reaction can occur under aqueous conditions. Jiang et al. noted that Gram-positive cell walls contain both amino and hydroxy groups capable of reacting with OSA, and used this principle to enhance the hydrophobicity of LA-5 (Jiang et al., 2019). The dosing was determined according to the percentage of OSA per dry bio-mass of the bacteria. An OSA dose between 0% and 10% increased the MATH hydrophobicity from approximately 40%–60%. CFU measurements at a dose of 6% resulted in a substantial increase in hydrophobicity without compromising culturability, whereas higher dosing did not increase the MATH values significantly but was accompanied by massive loss of culturability.
Fatty acid chlorides, such as lauryl chloride, are very reactive with water, and bacterial modifications must be performed in water-free but non-harmful environments, such as MCT oil, and involve prior water removal, such as, lyophilization. Using such a procedure can increase the MATH values of LGG from approximately 15%–75%, depending on the concentration of lauryl chloride (Jiang et al., 2021). At a 20% dose, the water contact angle continuously increases to approximately 100°, whereas the MATH values remained unchanged at the dose of ≥3%. These data suggest that the MATH may not be sensitive enough to discriminate the difference between hydrophobic bacteria. The most hydrophobic bacteria could not be fully dispersed in water and appeared as “bacterial chunks” in the microscopy images. CFU measurements revealed that the chemical treatment was not detrimental beyond the effect of the initial lyophilization step, which caused a marked decrease in CFU.
A recent study involved bacterial modification of LGG and Lactobacillus crispatus (LBC) in both the exponential and stationary phase (Jiang et al., 2023a). The surface of LGG and LBC were chemically modified using either octanal or hexanoic anhydride. The octanal selectively reacts with amino groups, whereas hexanoic anhydride is reactive with both the amino and hydroxy groups. X-ray photoelectron spectroscopy showed that LGG has a higher O/C ratio and a lower N/C ratio, suggesting that the LGG surface was more enriched with polysaccharides than that of LBC. In contrast, the LBC surface contained higher amount of proteins than the LGG surface. Figure 2A shows the zeta potentials of unmodified (UM), octanal-treated (OCT), and hexanoic anhydride-treated (HA) LGG and LBC measured under different pH conditions. LGG exhibited no remarkable difference between UM, OCT, and HA under all pH conditions. In contrast, LBC exhibited a clear decrease in the zeta potential with an increasing pH, and the difference between UM, OCT, and HA became more distinct under higher pH conditions. As shown in Figure 2B, UM, LGG, and LBC exhibited a large difference in
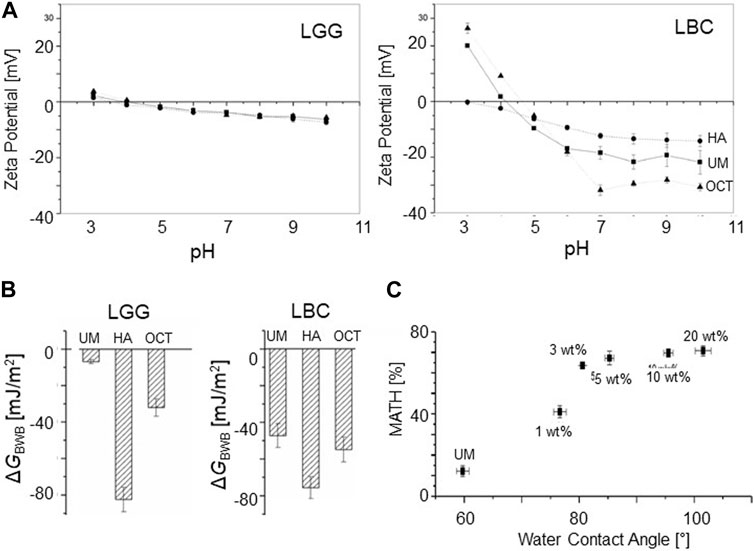
FIGURE 2. Properties of modified bacterial surfaces. (A) Zeta potential as a function of pH for UM, HA-, and OCT-modified LGG and LBC measured at various pHs (B)
Enzymatic degradation of the poly-saccharide and protein components of Gram-positive cell walls is a standard procedure for making protoplasts, which are cell wall-free bacteria only having a bacterial membrane and are spherical despite their original shape (Martin, 1963). Protoplasts are used for extracting or introducing DNA to bacteria as the cell wall constitutes a major barrier against DNA extraction and the protoplast can regrow their cell wall again after mild manipulation (such as DNA insertion). Shakarfourosh et al. used an even milder enzymatic treatment employing polysaccharide degrading enzymes (such as lysozyme) and proteolytic enzymes (such as protease K) to degrade the components of the LGG and LBD bacterial cell walls without forming protoplasts (Shekarforoush et al., 2022). Despite the fact that polysaccharides are predominantly hydrophilic and proteins are more hydrophobic, both enzyme treatments increased the hydrophobicity, as quantified using MATH measurements. This surprising effect was attributed to the fact that the MATH method is based on a Pickering effect, and thus changes the repulsive steric effect of the peptidoglycan layer (Shekarforoush et al., 2022).
2.3 Colloidal food materials containing bacteria
Probiotic lactic acid bacteria can be incorporated into food structures and have the dual role of being a functional part of the food structure as well as delivering the probiotic function of the bacteria. Several studies have focused on Pickering emulsions stabilized by bacteria adhering to the oil/water interface using natural UM bacteria (Firoozmand and Rousseau, 2016; Muhammed et al., 2023) and chemically modified bacteria (Jiang et al., 2019; Jiang et al., 2021). The emulsions exhibited large emulsions droplets of 10—100 μm, which was one to two orders of magnitude larger than the size of bacteria (∼1 µm). For food emulsions, the size limitation mainy comes from the size of bacteria that are used as the stabilizers. Mayonnaise-like emulsions were produced using food grade, Streptococcus thermophilus with a high internal oil phase load exceeding 40% (Firoozmand and Rousseau, 2016). MCT oil in water emulsions were produced using a selection of 31 naturally hydrophobic lactobacillus bacteria (Muhammed et al., 2023), as well as enzymatically treated LGG and LBD bacteria (Jiang et al., 2021). The highly hydrophobic LGG bacteria modified by laurylchloride exhibited the surprising ability to promote the formation of W/O/W double emulsions (Figures 3A–C using a simple one-step procedure (Jiang et al., 2021). The outer surface of the oil droplets (diameter ≈60 µm) was stabilized by a densely packed layer of adsorbed bacteria, whereas the inner water droplets (diameter ≈5 µm) were jointly stabilized by the bacteria and unknown low molecular weight surfactants (Figure 3D). Foams can also be made using natural and modified lactic acid bacteria as Pickering stabilizers. Falco et al. used β-casein- and sodium caseinate-modified La-5 to produce dry foams (foams showing fast drainage of the serum phase) that exhibited stability for more than 180 min (Yucel Falco et al., 2017a). Jiang et al. modified the same bacteria using OSA to produce dry foams that were stable for more than 180 min with superior stability, as compared to those using UM bacteria (Jiang et al., 2019). Because the air bubbles in food foam are typically significantly larger than the typical oil droplets of emulsions and due to the micron size of bacteria, bacterial stabilization may be more suited for foams than for emulsions. The new field of bacteria as Pickering stabilizers for emulsions and foams lacks systematic studies of the interrelationships between bacterial surface properties of natural or modified bacteria and the properties of emulsions.
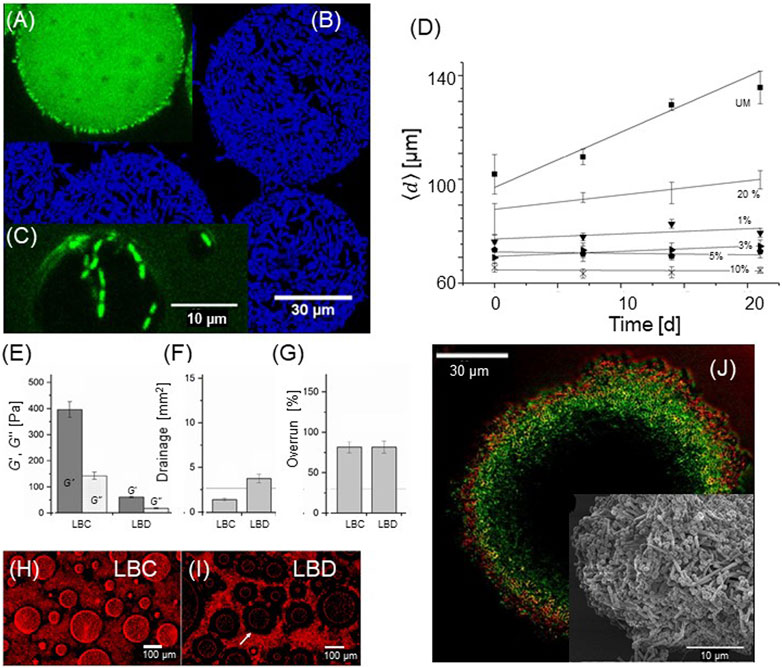
FIGURE 3. Colloidal food materials containing bacteria. Emulsions and double emulsions stabilized using 10% laurylchloride-treated LGG bacteria. (A) Primary oil droplet with inner water droplets. The oil and lipid phases are labeled with BODIPY (green). (B) Maximum intensity projections of a z-stack series of 30 images (DAPI, blue). (C) Inner droplet partially stabilized by bacteria (D) Average droplet diameter plotted as a function of time, measured in the presence of different laurylchloride concentrations. The stability of emulsions depends on the bacterial surface properties. (A-D): Adapted from (Jiang et al., 2021), with permission from Elsevier. (E) Rheological properties of whipping foam based on hydrophobic LBC and hydrophilic LBD (F) Drainage of the two types of bacterial whipping foam. The reference line indicates drainage of ordinary dairy-based whipping foam. (G) Overrun (degree of air incorporation) of bacterial whipping foam, as compared to standard dairy whipping foam. (H) Microstructure of the whipping foam showing the adsorption of hydrophobic LBC to air bubbles (I) Structure of hydrophilic LBD-based foam with almost no surface adsorption of bacteria. (E-I): Adapted from (Jiang et al., 2023b), licensed CC-BY-4.0. (J) Colloidosome based on the self-assembled hydrophobic LBD (green). The outer most surface is coated with a highly anionic PCP layer (red). Insert: scanning electron microscopy image of the surface, showing both cylindrical LBD and spherical PCP. Adapted from (Jiang et al., 2022), licensed CC-BY-4.0.
A more complex structure was fabricated using bacteria as colloidal building blocks (Jiang et al., 2022; Jiang et al., 2023b). Jiang et al. noted that fat globules and lactic acid have the same approximate size and share functionality with respect to aggregation and the ability to adhere at the water/air interphase, and proposed that the fat globules of a whippeable emulsion could be substituted with lactic acid bacteria (Jiang et al., 2023b). A whipping cream was formulated using bacteria (35% (V/V), either the hydrophobic LBC or more hydrophilic LBD, as well as sodium caseinate, and a thickening agent, both in low concentrations. The naturally hydrophobic LBC imparted the foam structure with bacteria densely covering the air phase with a network of bacterial aggregates, producing foam with a high modulus and good drainage stability (Figures 3E–I). In contrast, the LBD-based foam was softer with poorer drainage resistance and exhibited a structure where the internal air/water interphase was depleted of bacteria. The bacteria culturability for both formulations was intact upon whipping, although the aggregation of LBC lowered the apparent CFU quantification.
“Colloidosomes” are hollow, water-filled structures analogue to liposomes, but formed by colloidal particles rather than lipid molecules (Thompson et al., 2015). Simple stable colloidosomes were generated by spontaneous water ingression into micro-bubbles Pickering stabilized by OSA-modified LBC bacteria (Jiang et al., 2022). The fact that the structure does not disintegrate after removal of the air/water interphase indicate that a stabilizing principle beyond Pickering phenomena is in action, that is, inter-bacterial attraction of the modified hydrophobic bacteria. The basic anionic bacterial colloidosome was modified using the layer-by-layer method involving cationic and anionic polymers or hybrid combinations of polymers and a second layer of the strongly anionic bacterial strain Pediococcus pentosaceus (PCP), thus forming a bacterial double layer of two strains (Figure 3J). The permeability of the anionic fluorescein permeant could be rationalized by electrostatic repulsions (capsule zeta potential) for anionic colloidosomes and porosity for cationic capsules (Jiang et al., 2022).
Finally is should be mentioned that the health effect of ingestion of modified bacteria is currently understudied. Physical modification based on food components such as proteins will not require approval and is unlikely to induce any toxic effects. Chemical modification by amide and ester bonded fatty acids will most like require approval from authorities, however it can be argued that upon hydrolysis natural fatty acids will be released and a toxic response is unlikely. An interesting future research topic is that the relationship between bacterial surface properties, their binding to the intestinal interphase and the ability of bacteria to colonize the intestine.
3 Commonly used formulation strategies
3.1 Formulations of probiotic bacteria and enzymes in lipid self-assemblies
Polar lipids are key components of cell membranes that on their own form liquid crystals that possess the long-range orientational order of crystals and the high molecular mobility of liquids. The interplay between the hydrocarbon chain and polar head group interactions controls the assembly in aqueous solvents. This can be understood by the packing parameter of the amphiphilic molecule (Israelachvili et al., 1976; Mitchell and Ninham, 1981). The packing parameter (v/al) is defined as the ratio between the volume of the hydrophobic chain (v) and the product of the head group area (a) and the chain length (l). It is important to note that the packing parameter is not a fixed molecular parameter but depends on the environment. Therefore, different lipid liquid crystalline (LLC) structures illustrated in Figure 4 can be formed by both changing the lipid composition and environmental factors, such as temperature, pH, and ionic strength (Larsson, 1989; Larsson et al., 2006; Barriga et al., 2019). The so-called inverse LLC, where the lipid aqueous interface is curved towards the aqueous solvent, forming aqueous cavities, is of particular interest. This is because the size of the cavity often matches or can be formulated to match the size of functional protein or an enzyme. Varying the lipid composition results in an LLC with different structures and curvatures of the aqueous lipid interface, including inverse bicontinuous cubic (Q2), sponge phase (L3), or inverse hexagonal (H2) structure (Larsson, 1989; Seddon and Templer, 1995; Mulet et al., 2013; Gilbert et al., 2019). The phase behavior of polar lipids with decreasing water content and increasing temperature often follows the sequence (left to right in Figure 4): hexagonal phase (HI) → lamellar phase (Lα) for water soluble lipids and lamellar phase (Lα) → reversed hexagonal phase (HII) → inverse micellar phase for lipids (L2) for lipids with low water solubility. Different types of phases with cubic symmetry (micellar cubic I1 and I2, bicontinuous cubic V1 and V2) often occur in between these phases. When it comes to bicontinuous cubic phases the two systems of water channels allow the substrates to be transported in and out of the structure. The self-assembled character of the structure also offers advantages when it comes to formulation. The flexibility and softness of the structure makes it easier to include enzyme that are as large or even larger than the water channel dimensions.
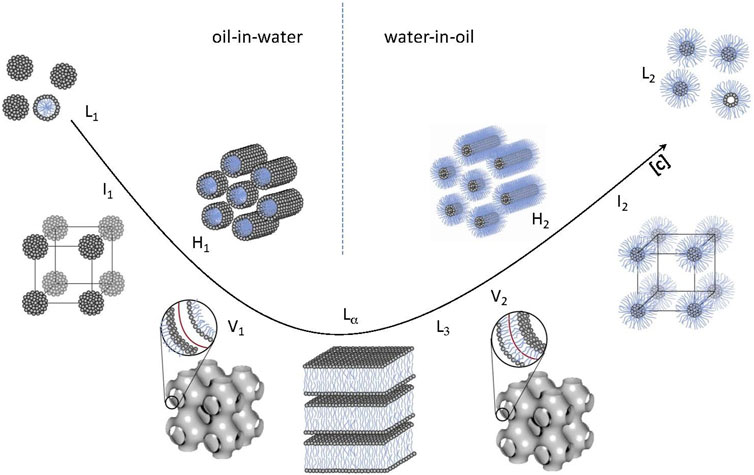
FIGURE 4. Various lipid liquid crystalline phases (LLC). From left to right: micellar cubic (I1), hexagonal (H1), bicontinuous cubic (V1), lamellar (Lα), inversed bicontinuous cubic (V2), inversed hexagonal (H2), and inversed micellar cubic (I2). The micellar (L1), inversed micellar (L2), and typical location of the “sponge” or L3 phase are also indicated. Adapted from (Barauskas and Nylander, 2008), with permission from Elsevier.
The LLC phases can be dispersed into colloidally stable nanoparticles, such as cubosomes, spongosomes/sponge-like nanoparticles, and hexosomes, which are more convenient for many scientific and technological applications that require continuous processing, such as dispensing or injection (Larsson, 2000; Mulet et al., 2013; Barriga et al., 2019; Valldeperas et al., 2019b). Inverse bicontinuous phases (V2) are nanostructures where the lipid bilayer curves to form water channels in a three-dimensional network (Larsson, 1989). The curvature of such a bilayer can be described as a minimal surface, which is as concave as it is convex, that is, it has a zero mean curvature. The bicontinuous lipid sponge phase can be regarded as a “melted” bicontinuous cubic phase, and as such, it does not present the long-range order of cubic phases (Anderson et al., 1989). The scattering pattern only features two broad diffraction peaks, indicating the mobility of the system (Porcar et al., 2003). It has a more flexible bilayer structure than that of the V2 phases, and consequently, it is more macroscopically fluid and swollen than the V2 phases. A dispersing agent is usually added to form LLC nanoparticles in a large excess of water (>90%), but the inner structure is closely related to the corresponding fully hydrated bulk phase (Valldeperas et al., 2016).
Entrapping proteins, including enzymes, with retained bioactivity is challenging due to their large size, complexity, and limited stability outside their natural biological environment. A lipid self-assembly with large water pores is required for the encapsulation of such large biomolecules. Inverse bicontinuous cubic (V2) and sponge (L3) phases have been successfully applied for this purpose (Razumas et al., 1994; Angelova et al., 2011; Sun et al., 2015; Valldeperas et al., 2019c; Gilbert et al., 2019). A recent summary shows that cubosomes can be loaded with a wide range of compounds from small drug molecules (<1 kDa) to large proteins with a molecular weight up to 174 kDa (Barriga et al., 2019). However, the activity of entrapped enzymes depends on the size of the water channels in relation to size of the enzyme molecules (Razumas et al., 1994; Sun et al., 2015). It has been shown that the long-term stability from days to a month in terms of retained activity of biosensors based on electrodes coated with monoolein and entrapped enzyme increase in the urease (molecular weight, Mw ≈ 590 kDa) < creatinine deiminase (Mw ≈ 200 kDa) ≈ glucose oxidase (Mw ≈ 160 kDa) < lactate oxidase (Mw ≈ 80 kDa) (Razumas et al., 1994). The enzyme stability can be improved when the water channels are larger than the protein size (Sun et al., 2015). It should be noted that the bicontinuous cubic phase used here are liquid crystalline and can adopt to size of the water channel to the enzyme size. This is even easier for the more flexible sponge (L3) phase. Here the inclusion of enzymes was found to increase the bending rigidity of the lipid bilayer of the sponge phase as determined by neutron spin echo measurements (Gilbert et al., 2022). The size of the water channels in the LLC structure could also be tuned by changing the environmental conditions, which also opens up the possibility to control the encapsulation and release of enzymes using external stimuli (Fong et al., 2016). Oral delivery of lipid-based carriers, such as liquid crystalline nanoparticles, vesicles, or nano-emulsions, also need to survive the harsh environment through the gastrointestinal tract, where lipolysis is a particular challenge as pointed out by Dully et al. (2020). The explored a strategy using lipase inhibitors, pointed out that there are other strategies like use of non-digestible lipids or use of capsules. If the lipid particle survives the harsh conditions in the stomach all the way down the small intestine the cargo need to traverse the mucus layer prior to absorption of the drug. In some cases, such as in inflammatory bowel disease, local delivery to the intestine is desirable. In this case, absorption is not needed, but the drug or nanoparticle still must penetrate the mucus (Date et al., 2018). In other cases, mucus adhesion is needed, where the nanoparticle adheres to the mucus for a prolonged time, releasing the drug and resulting in a prolonged effect (Gamboa and Leong, 2013). Therefore, the colloidal aspect is clearly crucial for the optimization of the formulation. This is a very wide field of research beyond the scope of this review. A comprehensive review of the topic and associated challenges has been presented by Zhai et al. (2019).
3.2 Formulation of probiotic bacteria and enzymes in silica-based particles
Amorphous forms of silica and silicates are recognized by the US Food and Drug Administration (FDA) and the European Food Safety Authority (EFSA) as the safe ingredients for oral delivery up to the dose of 1.5 g per day (Diab et al., 2017). In biological systems, silica can be found in a variety of nano- and micro-structures, such as silica diatoms (Sumper and Brunner, 2006) and spicules of sponges (Uriz et al., 2003), which inspired the synthesis of various hybrid materials (Nassif and Livage, 2011). Mesoporous silica nanoparticles are extensively used for the encapsulation of drugs with small molecular weights (Manzano and Vallet-Regí, 2020) and for the imaging of biological specimens (Tasciotti et al., 2008). The sol-gel process is a promising and effective strategy to synthesize mesoporous silica for food applications because of the compatibility and non-cytotoxicity to bio-macromolecules, bioactive enzymes, and probiotic bacteria (Diab et al., 2017).
Mesoscopic pores can be used as cavities that facilitate the stability of the encapsulated bioactive enzymes (or proteins in general) under broader pH and temperature conditions (Hudson et al., 2008; Carlsson et al., 2014). As the pore size, typically in the order of several nm to 10 nm, is large enough to allow large molecules to enter and diffuse inside the pores. Therefore, enzymes or their aggregates bind not only to the outer surface of particles, but many of them can also bind to the significantly larger, inner pore surfaces either by physisorption or by chemisorption. It should be noted that the immobilization of the bioactive enzymes into small pores or into cavities with only one opening can block the diffusion of substrates and products, which could reduce the catalytic activity of the confined enzymes (Lee et al., 2009). In contrast, the use of “macropores” facilitates the diffusion of substrates and improves the enzymatic activity, but the enzymes can also exit confinement and diffuse away from the formulation. Therefore, the chemical coupling of enzymes to the inner pore surface or the use of crosslinked or aggregated enzymes is desirable to keep the enzymes inside the mesopores (Wang et al., 2010).
3.3 Formulation of probiotic bacteria in polymer-based microcapsules
A variety of formulations have been developed and examined for the delivery of probiotics, such as spray-drying, freeze-drying, extrusion, and emulsification (Gouin, 2004; Champagne and Fustier, 2007; Gasperini et al., 2014). Among these methods, emulsification with the aid of surfactants is a gentle technique that causes minimal damage to microorganisms (Krasaekoopt et al., 2003). When an aqueous polymer solution is mixed with oil under stirring, the presence of surfactants (such as polyglycerol polyricinoleate (PGPR)) further contributes to stabilizing the water droplets by reducing the surface tension at the oil/water interface. If the polymers in aqueous droplets can be crosslinked, the microparticles of crosslinked hydrogels can easily be extracted from the oil-in-water emulsions using centrifugation.
The use of crosslinked hydrogels based on biopolymers approved by the US FDA is a straightforward strategy to deliver such formulations to the gastrointestinal tract (Figure 5A). For this purpose, probiotic bacteria can be encapsulated in particles of hydrogels based on anionic polysaccharides, such as alginate, pectin, and xanthan gum, to form hydrogels crosslinked by divalent cations, such as Ca2+ (Figure 5B) (Kwiecień and Kwiecień, 2018). Polyalginate (PA−), which is crosslinked by divalent cations such as Mg2+ and Ca2+ by the formation of “egg-box” junctions hosting divalent cations between the stacks of guluronic acid blocks (Katchalsky et al., 1961), is one of the most common encapsulation matrices for probiotic bacteria and enzymes (Orive et al., 2002). PA− is an FDA-approved food ingredient and is a linear polysaccharide extracted from the cell wall of marine brown algae, consisting of disaccharide units based on L-guluronic and D-mannuronic acids (Lee and Mooney, 2012). It is expected that formulations based on anionic polysaccharides, such as PA−, are protonated and hence are less soluble at low pH. The formulations are meant to protect probiotic bacteria under gastric conditions, whereas the bacteria are released in the intestines because the de-protonated polyelectrolytes are more soluble at higher pHs.
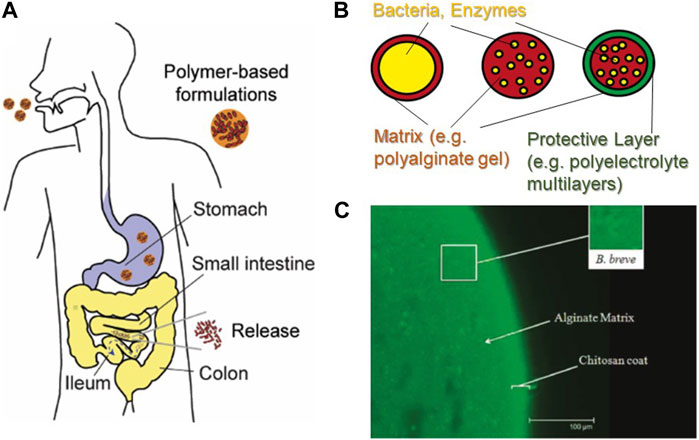
FIGURE 5. Formulation of probiotic bacteria in polymer-based microcapsules. (A) Schematic illustration of the delivery of probiotic bacteria using polymer-based formulations (B) Examples of polymer-based formulations. (C) Encapsulation of Bifidobacterium breve in PA− particles coated with one chitosan layer. Scale bar: 100 µm. Note that the size is significantly larger than the sensory limit of the human tongue (80 µm). Adapted from (Cook et al., 2011), with permission from the American Chemical Society.
As PA− microcapsules are not sufficiently stable to protect probiotic bacteria under gastric conditions, the deposition of additional protective layers is desired (Matricardi et al., 2008). For example, the layer-by-layer deposition of cationic and anionic polyelectrolytes (Lvov et al., 1993; Decher, 1997) has been used for the stable encapsulation of enzymes (Caruso et al., 2000), antigens (De Koker et al., 2009), and living cells (Diaspro et al., 2002). 1H NMR data showed that polyelectrolyte multilayers act as a diffusion barrier by modulating the mobility of water molecules, especially when are stacked (Schwarz and Schönhoff, 2002). Notably, only one additional layer of chitosan on PA− capsules increases the survival rate of Bifidobacterium and Lactobacillus in gastric fluids by two to three orders of magnitude (Figure 5C) (Krasaekoopt et al., 2004; Cook et al., 2011). However, it should also be noted that the size of the particles used in these studies was one order of magnitude larger than the sensory limit of the human tongue (Krasaekoopt et al., 2004), or were not even controlled (Cook et al., 2011). Consequently, several key factors remain unclear, such as (a) the minimum requirements for stable formulations, (b) the status of the probiotic bacteria release from the polymer capsules after reaching the target colon, and (c) whether the functionality of the probiotics is sustained after delivery. Systematic investigations addressing these points are still largely missing.
4 Physical properties of formulations for functional delivery
4.1 Formulations of enzymes in lipid self-assemblies
As examples of the delivery of bioactive molecules using lipid-based formulations, the encapsulation of two key types of enzymes of different sizes, namely aspartic protease (34 kDa or a diameter of about 5.7 nm) and β-galactosidase (460 kDa or a diameter of about 14 nm), are discussed (Valldeperas et al., 2019c; Gilbert et al., 2019). Here we note that the typical water channel diameter for fully swollen, bicontinuous cubic phase is about 5 nm while the corresponding value for a sponge phase is about 13 nm, bearing mind that both phases are flexible structures. Both enzymes are important for food processing, but the storage and delivery of the bioactivity is challenging. Aspartic protease from C. parasitica is a milk-clotting enzyme that is highly specific for κ-casein and promotes curd formation as well as cheese ripening. β-galactosidase from K. lactis (465 kDa, size = 14 nm) hydrolyses lactose to a mixture of glucose and galactose; therefore, it is used in food formulations intended for lactose-intolerant consumers. The ability to deliver iron is becoming increasingly important as the shift to more plant-based foods can lead to iron deficiency. The delivery of iron-carrying heme proteins has become important for food iron supply (Gilbert et al., 2023). Although the curvature of the lipid aqueous interfaces in the non-lamellar phases determines the size of the aqueous cavities, and hence the space given to the enzyme, the interaction between the enzyme and the lipid layer is an important factor that controls the efficiency of the encapsulation.
Mixtures of food-grade acylglycerides and acyldiglycerides can form highly swollen sponge phases (L3) with aqueous pores up to 13 nm in diameter, and can form well defined nanoparticles in excess water with the help of the dispersing agent polysorbate 80 (P80) (Figure 6) (Valldeperas et al., 2016). Aspartic protease (Mw = 34 kDa) assayed by milk clotting capability (78) and β-galactosidase (Mw = 476 kDa, tetramer) assayed by monitoring the Ortho-Nitrophenyl-β-galactoside - > O-nitrophenol (yellow) transition was found to be still active after inclusion into the sponge phase lipid matrix even after 3 months at 25°C (Gilbert et al., 2019). This is not possible if the enzyme is delivered from a solution where the stability at 25°C is a matter of days. It should here be noted that the lipid matrix used only consisted by approved food grade lipids. Interestingly, the two proteins affect the phase behavior of the lipid system differently (Figure 7). The smaller enzyme has a larger effect on the phase structure. The enzyme “likes” the lipid bilayer to be wrapped around itself, indicating that the protein is inside the water channels and that the change in curvature effect is induced by protein-lipid interaction.
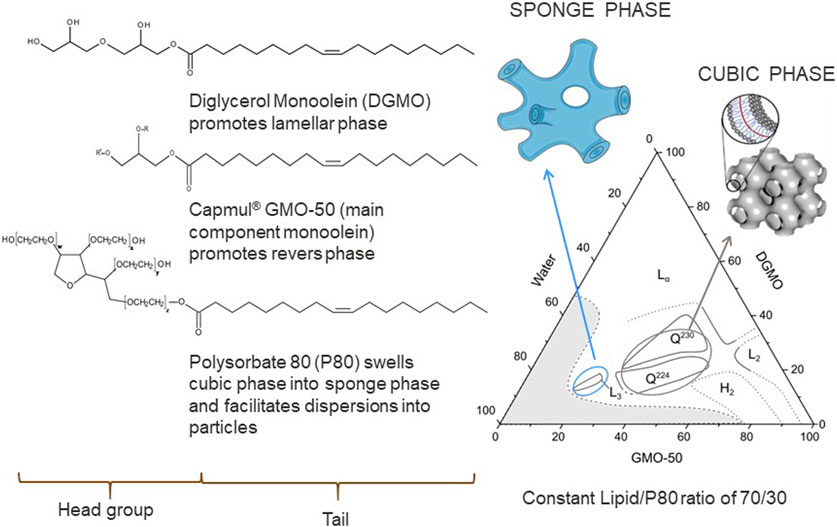
FIGURE 6. Phase diagram of the diglycerol monoolein (DGMO), monoolein, and polysorbate 80 system and the molecular structure of the components. Adapted from (Valldeperas et al., 2016), with permission from the American Chemical Society.
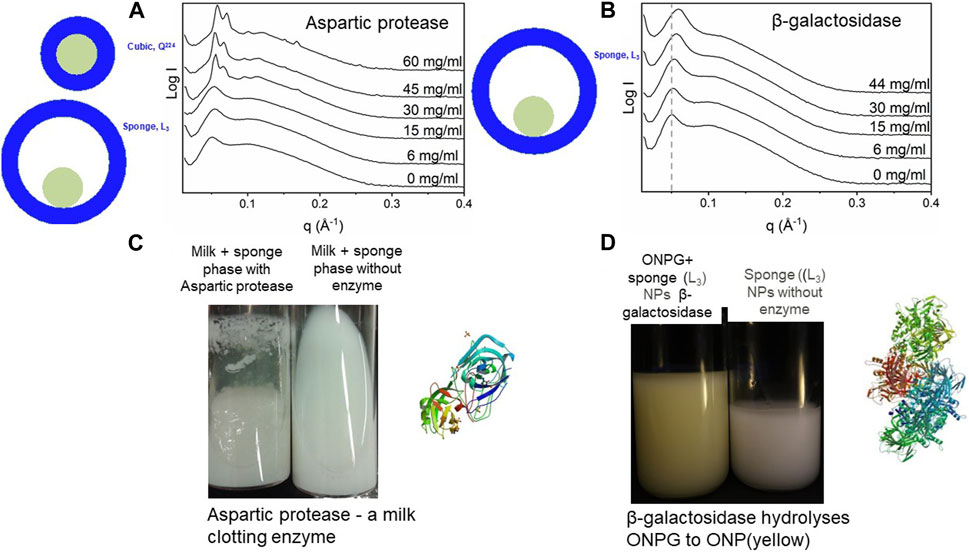
FIGURE 7. Effect of enzymes on LLC structures. (A) Small-angle x-ray scattering data of lipid LLC structures measured at different aspartic protease concentrations, indicating the transition from a sponge phase to a cubic phase at higher enzyme concentrations (B) Addition of β-galactosidase leads to a shift of main peak to a larger q with an increasing enzyme concentration, suggesting a decrease in the degree of hydration. (C) Aspartic protease is still active after inclusion into the lipid matrix, even after 3 months at 25°C as judged by its ability to clot milk (D) β-galactosidase maintained its function, that is, hydrolyses of galactose analogue in the lipid system, even after 3 months, that is o-nitrophenyl-β-galactoside (ONPG) is hydrolyzed, in the presence of β-galactosidase to o-nitrophenol (D). (A,C): Adapted from (Valldeperas et al., 2019c), with permission from Elsevier. (B,D): Adapted from (Gilbert et al., 2019), with permission from the Royal Society of Chemistry.
The particles adsorb at the interface to form a lipid bilayer, as shown using quartz crystal microbalance with dissipation (QCM-D) and neutron reflectometry (Valldeperas et al., 2019a). Raman spectroscopy results for the sponge phases show large similarities in the lipid chain confirmation and head group interactions as in the lamellar and reverse bicontinuous cubic phase in the same lipid system (Talaikis et al., 2019). Size exclusion chromatography demonstrated efficient encapsulation of both enzymes, yet they retained their enzymatic activity over months, surpassing the storage stability of pure enzymes in solution (Valldeperas et al., 2019c; Gilbert et al., 2019). This may be owing to the penetration of the enzymes into the formed lipid bilayer, as shown using Raman spectroscopy (Valldeperas et al., 2019c) and neutron reflectometry (Valldeperas et al., 2019a). Neutron spin echo and molecular dynamics simulations have confirmed this (Figure 8) (Gilbert et al., 2022).
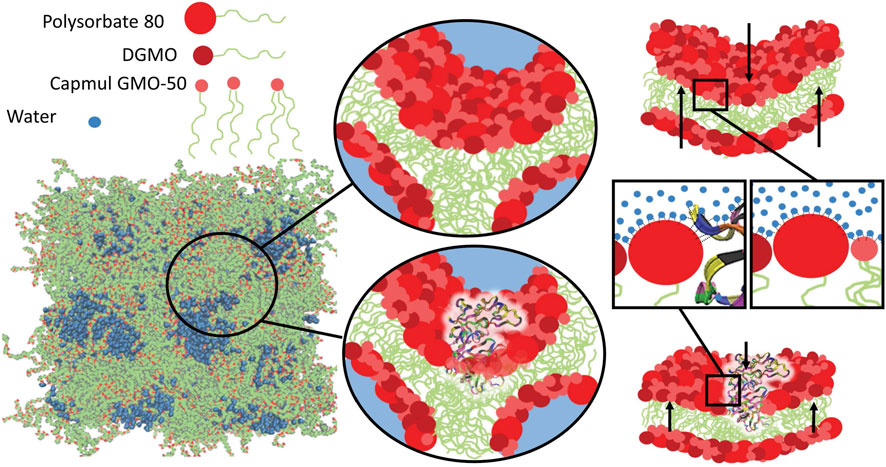
FIGURE 8. Molecular dynamics simulations of aspartic protease encapsulated in LLC suggests the penetration of enzymes into the lipid bilayers. This was confirmed using neutron spin echo measurements, which that showed an increase in the membrane bending rigidity upon the encapsulation of enzymes in the lipid sponge phase (data not shown). Reproduced from (Gilbert et al., 2022), licensed CC-BY-3.0.
4.2 Formulations of probiotic bacteria and enzymes in silica-based particles
The formulation of bioactive enzymes in porous silica particles is a highly promising approach (Lee et al., 2009). To address the trade-off between “mesopores” vs. “macropores” (Section 3.2), the use of materials displaying both types of pores has been proposed. Yang et al. reported the “fish-in-net” encapsulation of enzymes by the condensation of pre-formed mesoporous silica particles around the enzyme aggregates in the presence of glycerol (Yang et al., 2006). Bernal et al. synthesized meso-macroporous silica materials (mesopore diameter: 10–40 nm, macropore diameter: 0.1–20 µm) by the polycondensation of sodium silicate using the micelles of cationic surfactants (cetyltriammoniumbromide (CTAB)) in the presence of ethylacetate as the template and encapsulated bioactive enzymes, such as β-galactosidase and lipase (Bernal et al., 2011; Bernal et al., 2014). Blin et al. employed double emulsions based on water/oil/water (W/O/W) or water/solid lipid nanoparticles/water (W/SLPs/W) as templates and synthesized meso-macroporous silica particles, resulting in hexagonally aligned mesopores and scalable macropores (Blin et al., 2014). Pavel et al. applied this formulation (mesopore diameter: 9 nm, macropore diameter: 0.2 µm) to encapsulate β-galactosidase (Pavel et al., 2017). The adsorption of enzymes to meso- or macropores show a dependence on concentration and thus the aggregation state of enzymes. Notably, the oligomers of β-galactosidase adsorbed on the surface of macropores exhibit a high catalytic activity.
The encapsulation of probiotic bacteria requires another strategy because the pore size is too small to accommodate µm-sized cells. For this purpose, a protective silicate layer can be mineralized on polymer-based microparticles using electro-spraying or emulsification. Callone et al. (Callone et al., 2008) reported the encapsulation of Saccharomyces cerevisiae and Oenococcus oeni in PA− particles by injecting cell/PA− droplets into an M2+-loaded bath, resulting in the formation of 400–500 µm-sized particles. Subsequently, the surface was coated with amorphous silica synthesized using three methods, namely (i) dipping the particles in silica sol, (ii) consolidating the particles with tetraethoxysilane in an apolar solvent, and (iii) coating the particle surface with methyltriethoxysilane in the gas phase. The particles prepared using (ii) exhibited cytotoxicity, but those prepared using (i) and (iii) exhibited an increase in the function of the cells, such as the fermentation of glucose by Saccharomyces and maleic acid by Oenococcus. Haffner et al. (Haffner et al., 2016) implemented a similar strategy and fabricated Lactobacillus/PA− particles by electrospraying into a bath containing Ca2+ ions, yielding 200 µm-sized hydrogel particles encapsulating Lactobacillus. The protective amorphous silica layer was deposited by the electrostatic adsorption of aminotrimethoxysilane, followed by the condensation of tetramethyl orthosilicate. The bacterial viability was assessed by counting the colony forming units (CFUs) before and after incubation in solutions mimicking gastric and intestinal fluids (Minekus et al., 2014). The Lactobacillus encapsulated in silica-PA− particles exhibited only very minor leakages and maintained the viability of the bacteria, whereas all the naked bacteria with no protection died in both fluids. The metabolic function of bacteria can be further evaluated by using an in vitro gastrointestinal simulator (see Section 5.1 for more details) (Haffner et al., 2017). The coating of organic particles with “porous” silica layers further allows for the diffusion of molecules, which helps the bacteria maintain their functions and stabilizes the encapsulated enzymes (Blin et al., 2014; Haffner et al., 2016).
4.3 Formulations of probiotic bacteria in composite polymer microcapsules
As summarized in Section 3.3, probiotic bacteria can be encapsulated in formulations based on biopolymer-based hydrogel droplets using emulsification (Krasaekoopt et al., 2003). Maintaining the size of the formulation particles to smaller than the sensory limit of the human tongue (80 μm, according to (Tyle, 1993)) is an important requirement for food applications, otherwise consumers would feel the formulations as “additional, foreign ingredients”. For this purpose, it is necessary to optimize the emulsification process. For example, the combination of shear-thinning agents and surfactants enables the formation of small and stable droplets even at a significantly lower emulsification speed. However, this has been either difficult to control or even overlooked in previous studies (Krasaekoopt et al., 2004; Cook et al., 2011). Moreover, hydrogel-based microcapsules prepared by emulsification, such as those based on PA− hydrogels crosslinked by Ca2+, cannot maintain their structural integrity in gastrointestinal environments or sustain the functions of the encapsulated probiotic bacteria (Eshrati et al., 2018). The deposition of additional polymer layers is a possible strategy to increase the stability of PA− microparticles and hence the protection of the probiotic bacteria (Matricardi et al., 2008). For example, the layer-by-layer deposition of cationic and anionic polyelectrolytes (Lvov et al., 1993; Decher, 1997) has been used for the stable encapsulation of enzymes (Caruso et al., 2000), antigens (De Koker et al., 2009), and living cells (Diaspro et al., 2002). The deposition of one chitosan layer increases the survival rate of Bifidobacterium and Lactobacillus encapsulated in PA particles by a factor of 100–1,000 under gastric conditions, suggesting that even one additional polymer layer could increase the stability of the formulations (Krasaekoopt et al., 2004; Cook et al., 2011).
Eshrati et al. demonstrated the encapsulation of LGG into small, highly monodisperse PA− particles by the systematic optimization of the preparatory parameters. To avoid mechanical damage to probiotic bacteria during emulsification, it is necessary to stir the oil/water mixture at a low speed (such as 300 rpm) in the presence of surfactants (such as PGPR) and shear-thinning agents (such as xanthan gum). Although LGG encapsulated in crosslinked PA–particles exhibits an increase in viability, which was assessed by the number of CFUs, by four orders of magnitude, as compared to “naked” LGG, the fraction of viable LGG after 2 h in gastric fluid (3 g/L pepsin and 85 mM NaCl, pH 2.0) is still 0.1%. The fraction of viable LGG in crosslinked PA–particles is 0.01% after incubating LGG/PA− particles in intestinal fluid (1 g/L pancreatin, 1 g/L bile salt, and 85 mM NaCl, pH 6.5) for 3 h. These results clearly indicate that PA–particle cores alone do not have a sufficiently high protective function. In contrast, the layer-by-layer deposition of positively charged chitosan (Ch+) and negatively charged PA– (Figure 9A) significantly increases the viability of LGG (Caruso et al., 2000; Cook et al., 2011). Remarkably, the deposition of only three polyelectrolyte layers increases the CFU values by a factor of more than 106 in stomach fluid and 105 in small intestine fluid (Figure 9B). It is notable that a further increase in the layer number did not improve the viability, suggesting that the deposition of three polyelectrolyte layers (chitosan + PA + chitosan), corresponding to a total thickness of only 20 nm, is the minimum requirement to achieve the highest protective function (Eshrati et al., 2018).
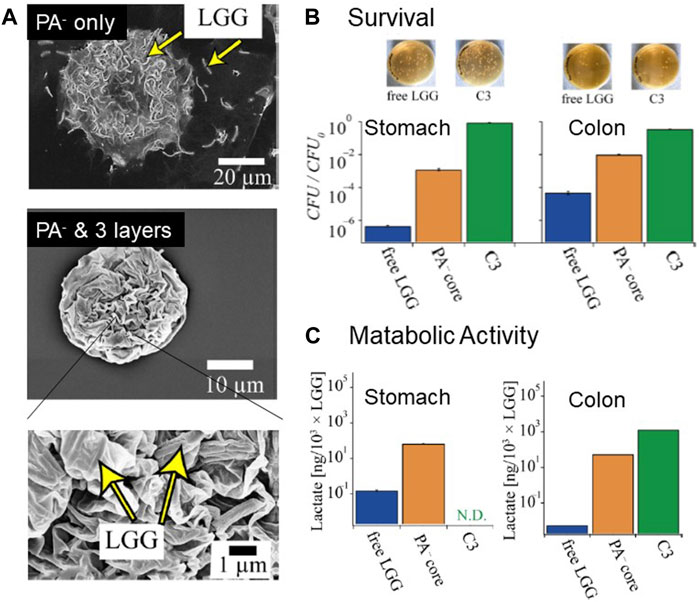
FIGURE 9. Encapsulation of Probiotic Bacteria in Composite Polymer Formulations. (A) Scanning electron micrographs of LGG encapsulated in a PA− microcapsule and PA–-coated with three additional polyelectrolyte layers. The LGGs were well-protected by the 20 nm-thick polyelectrolyte layers. The particle size was <40 μm, which is not sensed by the human tongue (B) Photographs of Petri dishes and normalized CFUs after treatment with fluids simulating gastric (left) and small intestine environments. The PA–core with three additional layers increased the survival rates by 107 and 104 in the stomach and small intestine environments, respectively. (C) Metabolic activity of the “released” LGG assessed by the lactate synthesis determined using a gastrointestinal simulator. The composite polymer formulations increased the lactate production by 105. Adapted from (Eshrati et al., 2018), with permission from the American Chemical Society.
Unfortunately, chitosan is not an ingredient approved by the FDA or EFSA, and it is claimed that chitosan is antimicrobial (No et al., 2002; Tsai and Hwang, 2004). The use of food-derived milk proteins is a promising strategy for the stabilization of PA–particles by the layer-by-layer deposition of charged macromolecules because milk proteins possess both positive and negative net charges near neutral pH, such as lactoferrin (+), caseinate (−), and whey protein isolates (−). Falco et al. used several milk proteins for the layer-by-layer coating of 150 µm-diameter PA− particles encapsulating LGG (Yucel Falco et al., 2019). The layer-by-layer deposition was monitored using the change in the zeta potential and QCM-D. The coating with only milk proteins did not lead to sufficiently high stability. However, the deposition of Ch+ as the first layer improved the stability. The treatment of PA–particles coated with Ch+ and additional milk protein layers in gastric fluid for 30 min resulted in a decrease in the CFU number by three orders of magnitude. Unfortunately, a longer exposure to gastric fluids resulted in the death of all bacteria, indicating insufficient stability. The use of materials approved by the FDA and EFSA is a necessary step to apply such (bio)polymer-based formulations as food ingredients.
Moreover, layer-by-layer coating with polyelectrolytes can be applied on the surfaces of the negatively charged cell wall of bacteria, enabling the use of a single bacteria as the smallest delivery vehicle (Priya et al., 2011; Yucel Falco et al., 2017b). Falco et al. coated individual bacteria with sulfated β-glucan and Ch+, because β-glucan can resist the human enzymatic system but not the bacterial glycanases in the small intestine (Yucel Falco et al., 2017b). The layer-by-layer deposition was confirmed on planar model surfaces using QCM-D and spectroscopic ellipsometry, whereas the deposition on the bacteria was confirmed using zeta-potential measurements. Although the formulations could resist the acidic gastric conditions, they were partially degraded during exposure to a combination of simulated intestinal fluid containing glucanase, indicating the release mechanism of bacteria in the intestine.
4.4 Formulations of probiotic bacteria in cellulose-based particles
Cellulose derivatives, such as carboxymethyl cellulose (CMC) and hydroxyethyl cellulose (HEC), have been used for the development of probiotic delivery systems based on crosslinked edible films (Singh et al., 2019), water in water (W/W) emulsions with gelatin (Singh et al., 2018a), and coacervates with Ch+ with genepin (Singh et al., 2018b) or without (Singh et al., 2017). Food grade cellulose derivatives are water soluble, typically due to the presence of carboxylic acid groups, and serve as thickeners in the food industry. Therefore, these components require additional gelling agents or crosslinkers to form stable particles or films that do not readily dissolve or disintegrate. CMC- and HEC-based edible films were cast and crosslinked under mild conditions by drying at 50°C after the addition of citric acid (Singh et al., 2019). The lactic acid bacteria in LGG could be absorbed into the films and the pH dependency of film swelling could be tuned to provide a low degree of swelling at gastric pH and high degree of swelling at intestinal pH by adjusting the CMC/HEC ratio and citric acid content. Overall, the films could be loaded with a high quantity of bacteria and provided good protection.
Water in water emulsion is a phenomenon driven by thermodynamic incompatibility between pairs of water-soluble polymers. The interfacial tensions are extremely small, the interphase has a large width, and both inhibit the stabilization with small (molecular) emulsifiers; therefore, Pickering stabilization using nano- or micron-scaled particles must rather be employed. Singh et al. (2018a) used CMS and gelatin at temperatures above 35°C to avoid the gelation of gelatin and to introduce LGG bacteria in dispersed CMC droplets. The bacteria exhibited a specific affinity for the CMC phase and was intensively present at the interphase of the particles, which was attributed to Pickering stabilization phenomena and the high desorption energy of the large size of the bacteria. The formulation strategy resulted in the enhanced survival of entrapped LGG when exposed to simulated gastric and intestinal conditions. Sub-millimeter CMC particles were produced following a polymer coacervation route by the dropwise addition of a CMC solution into a chitosan solution (Singh et al., 2018b) in combination with a mild crosslinker, namely genipin. The addition of sodium caseinate prevented particle aggregation due to electrostatic repulsion of the otherwise nearly neutral particles. The formulation significantly improved the survival rate of the bacteria when exposed to simulated gastric and intestinal conditions, as compared with the unformulated LGG bacteria. Since probiotics can be administered through food, the long-term culturability was tested over 30 days, showing a significantly improved stability, as compared with the unformulated samples. However still up to five log units of loss was detected even for the improved CMC-chitosan formulations which still limits the practical applications.
5 Functional assessment using in vitro model of gastrointestinal environments
5.1 In vitro gastrointestinal simulator
As the physical access to the digestive product after each compartment in the gastrointestinal tract in vivo is not realistic, several types of in vitro systems have been developed to simulate the entire gastrointestinal tract (Van de Wiele et al., 2015). In vitro gastrointestinal simulators are powerful tools to not only assess the protective capacity of formulations but also to quantify the metabolic function of LGG during its passage through the entire gastrointestinal tract in the presence of nutrients (starch and other carbohydrates) and other bacteria. This assay is distinct from the previous CFU counting assay because only the probiotic bacteria that are released from the formulation can be detected using flow cytometry. Namely, the bacteria that are encapsulated and protected by the formulation are not detected. Once the LGG/PA− particles were coated with three polyelectrolyte layers, no released bacteria could be detected throughout the entire gastric process (1 h at pH 4.8, 0.5 h at pH 3.0, and 0.5 h at pH 2.0). Notably, the residence in the small intestine chamber showed an increase in the released LGG by a factor of five, as compared with the initial level, suggesting the release of LGG that proliferated in the formulations by the deprotonation of chitosan (pKb = 6.5) (Guarino et al., 2015). Most significantly, the amount of lactate (metabolic product) produced increased in the ileum, small intestine, and colon chambers by five orders of magnitude (105 ×), as compared with the free LGG (Eshrati et al., 2018), demonstrating that the 40 µm composite polymer formulations significantly improved the delivery of functional probiotic bacteria without being detected by the tongue of the consumer (Figure 9C).
5.2 Adhesion of released probiotics on in vitro model of intestinal mucosa
After their release from the formulations, probiotic bacteria adhere on the surface of the colon epithelium and create colonies (Sonnenburg et al., 2004). The surface of the gastrointestinal tract is covered with a mucosal layer consisting of mucin, which protects against the invasion of pathological bacteria (Pelaseyed et al., 2014). An increasing number of studies have demonstrated that the protective function of mucus is supported by the presence of phospholipids, especially, phosphatidylcholine and lysophosphatidylcholine (Ehehalt et al., 2004).
AFM is a commonly used method to reveal the mechanism of the initial step, which is the adhesion of LGG on the mucosal surface of intestinal epithelial cells. Earlier studies have shown that bacteria can be immobilized on a cantilever surface by polycations, such as poly (lysine) and poly (ethyleneimine), and the force-distance curves can be measured on various substrates (Cao et al., 2006; Boks et al., 2008; Rowan-Nash et al., 2019). Dague et al. coated plastic dish surfaces with porcine gastric mucin using physisorption as a model of the gastrointestinal tract surfaces, and measured the force-distance curves with a Lactococcus lactis-coated cantilever (Dague et al., 2010). The force-distance curves measured using bacteria-coated cantilevers are distinct from those measured using intact Si3N4 cantilevers. As the surface density of mucin is hardly controllable by physisorption, Amadei et al. reported the immobilization of biotin-functionalized mucin onto planar, supported lipid membranes incorporating biotinylated lipids via neutravidin crosslinkers (Amadei et al., 2018). A high-affinity binding of biotin and neutravidin (KD ∼ 10–15 M) enables the grafting of biopolymers by controlling the self-assembly of biotinylated lipids (Kaindl et al., 2012; Rieger et al., 2015). Instead of measuring interfacial forces using AFM, the microinterferometry technique has been used to calculate the mean force potential from the height fluctuation of cell-sized beads (Higaki et al., 2017). Although mucin is highly glycosylated, carrying negative net charges, the mucin-functionalized surface exhibits a markedly stronger interaction with silica particles coated with phosphatidylcholine than those coated with positively charged lipids, suggesting that the mucin-phosphatidylcholine interaction is not driven by electrostatic interactions, as suggested by a previous isothermal calorimetry study (Korytowski et al., 2017).
As a more realistic model for the interaction of the released probiotic bacteria with the intestinal tract surfaces, Eshrati et al. fabricated a polarized monolayer of enterocyte-like cells and goblet cell-like cells differentiated from CaCo2 and HT29 MTX cells (Figure 10A). After the establishment of a polarized epithelial monolayer with tight junctions, the sample was transferred into a parallel plate flow chamber. The adhesion of LGG on the surface was monitored under the shear stress that covers the bacteria in vivo, 0.1–0.5 Pa (Eshrati et al., 2019). It should be noted that the cells need to be cultured in Transwell® culture until they establish the apico-basal polarity because the commonly used cell culture on plastic or glass substrates is not able to establish the polarized epithelial cell monolayer connected with tight junctions to express the layer of mucin proteins. Therefore, the differentiated cell monolayer with a sufficiently high transepithelial electric resistance (≥400 Ω cm2) was transferred into a microfluidic chamber (Fröhlich et al., 2021). In addition to the establishment of tight junctions (Figure 10B), the expression of mucin 2, which is a major constituent of mucus layers that build up a protective barrier together with phospholipids on the epithelial surfaces against pathogens (Pelaseyed et al., 2014; Korytowski et al., 2017), was confirmed by immunofluorescence imaging (Figure 10B). Subsequently, the Young’s modulus of the mucin layer was measured using AFM nano-indentation, which indicated that the goblet cell-like cells derived from HT29 MTX cells produced thicker and softer mucin layers, EHT29 = 0.2 kPa and ECaCo2 = 10 kPa. Interestingly, the surface density of adhered LGG (Figure 10C) exhibited a clear maximum at approximately 0.3 Pa, suggesting that the adhesion of LGG to the mucosal surface in the colon is shear-dependent. Moreover, the persistence of LGG adhesion was further examined by a stepwise increase in the shear stress exerted on the adherent LGG, demonstrating that some LGG cells can withstand high shear stress even beyond the physiological levels. The presence of such “strong binders” might contribute to the maintenance of the chronic homeostasis of microbiomes in the gastrointestinal tract.
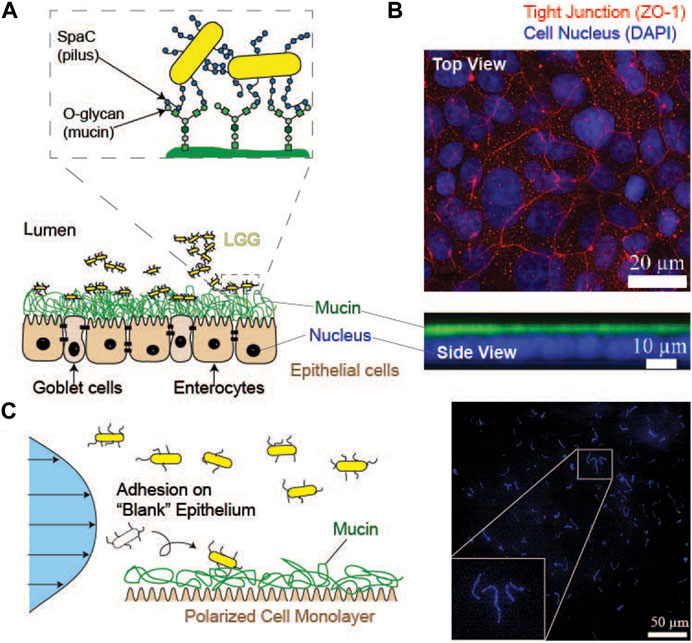
FIGURE 10. Adhesion of Probiotic Bacteria (LGG) to the in vitro Intestinal Epithelium Model. (A) Schematic representation of the adhesion of “released” probiotic bacteria (LGG) to the surface of the cultured epithelium model expressing heavily glycosylated mucin proteins. LGG binds to the glycan chains of mucin using SpaC expressed in pili. (B) Immunofluorescence top and side view images of the cultured colon epithelium model. Red: ZO-1 (tight junction), blue: DAPI (cell nucleus), and green: mucin (C) Left: monitoring of LGG adhesion to the in vitro model of the intestinal epithelium. The polarized epithelial monolayer was transferred from the Transwell® culture to the flow chamber after confirming the establishment of tight junctions. Right: a representative image of LGG adhered to the surface of a model epithelium. Adapted from (Eshrati et al., 2019), with permission from the American Chemical Society.
Finally, adhered bacteria start forming new microbiomes by forming colonies. To characterize the interactions between bacteria, the timescale of bacterial auto-aggregation is commonly used in microbiology. The assay involves monitoring the time evolution of the loss of OD due to sedimentation of the aggregated bacteria (Thomas et al., 2018). Freely suspended single bacteria remain in suspension for a significantly longer time because the sedimentation velocity (v) of a particle with a radius (r) increases with
6 Final remarks and perspectives
Probiotic bacteria and enzymes can be delivered to the intestinal system by formulating appropriate carriers and including these into food ingested by humans. Despite this simple statement, it involves many challenges in the field of soft matter science. This review aimed to highlight how the key concepts in soft matter science can be used to design, characterize, and evaluate self-assembled formulations based on lipids and biopolymers. The topics covered in this review included the emulsification of oil-water mixtures, the self-assembly of lipids and polymers at interfaces, the electrostatics and viscoelasticity of interfaces, and the wetting/adhesion of colloidal particles.
One of the specific challenges in food applications is the series of diverse chemical conditions and timescales that the carriers experience throughout their journeys from storage to the colon through the entire gastrointestinal tract. This calls for the design of responsive, food-grade materials that ensure the protection of the probiotic bacteria and enzymes under highly acidic gastric conditions (pH 2, pepsin) and the intestinal conditions exposed to pancreatic juice (pH 6.5, pancreatin and bile acid). Many approaches introduced here have demonstrated that exploiting the self-assembly of lipids and biopolymers, such as the sponge phase of lipids and layer-by-layer coating with nm-thick polyelectrolyte layers, opens the possibility to tune the encabsulation capability and stability of the carriers. This offers the possibility not only to protect the bacteria and enzymes under the harsh gastrointestinal environments, but also to release the active component at the right place.
The protection of the functions of probiotic bacteria and enzymes throughout delivery can be evaluated using in vitro systems that simulate the microenvironments of the human gastrointestinal tract. For example, the survival of bacteria can be monitored by counting the CFUs after their passage through chambers mimicking the stomach, small intestine, ileum, and colon environments, whereas the function of bacteria and enzymes can be assessed by measuring the amount of bacteria or enzyme products. Upon reaching their destination, the delivered probiotic bacteria must adhere to the surface of the colon epithelium coated with mucin and form a new microbiome. The initial phase, that is, the adhesion of bacteria onto the mucin surface, can be monitored either by coating the surface of dishes or microfluidic channels with isolated mucin proteins or by transferring the polarized monolayers of enterocyte-like cells and goblet cell-like cells expressing mucin. This enables the determination of the binding/unbinding force and critical shear stress for adhesion/detachment.
The increasing knowledge on the key roles of compositions of healthy and less healthy microbiomes in sustaining health and wellbeing has drawn increasing attention in improving microbiomes by the delivery of probiotic bacteria and enzymes. Changing the composition of the human diet (such as a more plant-based diet) is one direction; however, the delivery of missing gut bacteria and enzymes could be a more direct approach. The development of efficient carriers for the delivery of gut bacteria opens a new challenge because the natural gut bacteria are strictly anaerobic, unlike Lactobacillus in yoghurt; therefore, they need to be protected against oxygen before ingestion.
Author contributions
JR: Conceptualization, Writing–original draft, Writing–review and editing. TN: Conceptualization, Writing–original draft, Writing–review and editing. MT: Conceptualization, Writing–original draft, Writing–review and editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. Much of the work presented in this review were performed with the support by EU FP7 under and REA grant agreement no 606713 BIBAFOODS. JR is thankful to the Independent Research Fund Denmark for support (grant 8022-00139B). TN acknowledges the financial support from the Swedish Research Council under contract 2017-06716 and 2020-05421 and NanoLund. MT thanks JSPS (JP19H05719) and Nakatani Foundation for their support.
Acknowledgments
The authors didicate this manuscript to the memory of Maria Miguel, who passed away too early. The authors thank the following people for fruitful collaboration and stimulating discussions: Björn Lindman, Maria Miguel, Andreea Pasc, Marité Cardenas, Stefan Kaufmann, Tom Van Wiele, Bruno Medronho, Stefan Ulvenlund, Gemma Montalvo, Sam Possemiers, Jordi Esquena, Hans van den Brink. Dennis S. Nielsen, Nils Arneborg, and Kathryn A. Whitehead. The authors are especially thankful to the graduate students and junior and senior researchers who contributed to the work presented here, including M. Valldeperas Badell, J. Gilbert, I. Ermilova, J. Swenson, M. Nagao, J. Barauskas, C. Yucel Falco, I. Pavel, F. Haffner, M. Eshrati, F. Amadei, P. Singh, M. Veschgini, W. Abuillan, X. Jiang, E. Shekarforoush, M.K. Muhammed. We thank Editage (www.editage.jp) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors MT and TN declared that they were editorial board members of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abuillan, W., Schneck, E., Körner, A., Brandenburg, K., Gutsmann, T., Gill, T., et al. (2013). Physical interactions of fish protamine and antisepsis peptide drugs with bacterial membranes revealed by combination of specular x-ray reflectivity and grazing-incidence x-ray fluorescence. Phys. Rev. E 88 (1), 012705. doi:10.1103/PhysRevE.88.012705
Alfredsson, V., Lo Nostro, P., Ninham, B., and Nylander, T. (2021). Morphologies and structure of brain lipid membrane dispersions. Front. Cell Dev. Biol. 9, 675140. doi:10.3389/fcell.2021.675140
Amadei, F., Fröhlich, B., Stremmel, W., and Tanaka, M. (2018). Nonclassical interactions of phosphatidylcholine with mucin protect intestinal surfaces: a microinterferometry study. Langmuir 34 (46), 14046–14057. doi:10.1021/acs.langmuir.8b03035
Anderson, D., Wennerstroem, H., and Olsson, U. (1989). Isotropic bicontinuous solutions in surfactant-Solvent systems: the L3 phase. J. Phys. Chem. 93 (10), 4243–4253. doi:10.1021/j100347a067
Angelova, A., Angelov, B., Mutafchieva, R., Lesieur, S., and Couvreur, P. (2011). Self-assembled multicompartment liquid crystalline lipid carriers for protein, peptide, and nucleic acid drug delivery. Accounts Chem. Res. 44 (2), 147–156. doi:10.1021/ar100120v
Assenza, S., and Mezzenga, R. (2019). Soft condensed matter physics of foods and macronutrients. Nat. Rev. Phys. 1 (9), 551–566. doi:10.1038/s42254-019-0077-8
Barauskas, J., and Nylander, T. (2008). “Lyotropic liquid crystals as delivery vehicles for food ingredients,” in Delivery and controlled release of bioactives in foods and nutraceuticals. Editor N. Garti (Cambridge: Woodhead Publishing), 107–131.
Barriga, H. M. G., Holme, M. N., and Stevens, M. M. (2019). Cubosomes: the next generation of smart lipid nanoparticles? Angew. Chem. Int. Ed. 58 (10), 2958–2978. doi:10.1002/anie.201804067
Behnsen, J., Deriu, E., Sassone-Corsi, M., and Raffatellu, M. (2013). Probiotics: properties, examples, and specific applications. Cold Spring Harb. Perspect. Med. 3 (3), a010074. doi:10.1101/cshperspect.a010074
Bernal, C., Illanes, A., and Wilson, L. (2014). Heterofunctional hydrophilic–hydrophobic porous silica as support for multipoint covalent immobilization of lipases: application to lactulose palmitate synthesis. Langmuir 30 (12), 3557–3566. doi:10.1021/la4047512
Bernal, C., Sierra, L., and Mesa, M. (2011). Application of hierarchical porous silica with a stable large porosity for β-galactosidase immobilization. ChemCatChem 3 (12), 1948–1954. doi:10.1002/cctc.201100174
Bezryadina, A., C Preece, D., Chen, J. C., and Chen, Z. (2016). Optical disassembly of cellular clusters by tunable ‘tug-of-war’ tweezers. Light Sci. Appl. 5 (10), e16158. doi:10.1038/lsa.2016.158
Blin, J.-L., Jacoby, J., Kim, S., Stébé, M.-J., Canilho, N., and Pasc, A. (2014). A meso-macro compartmentalized bioreactor obtained through silicalization of “green” double emulsions: W/O/W and W/SLNs/W. . Chem. Commun. 50 (80), 11871–11874. doi:10.1039/C4CC06007E
Boks, N. P., Busscher, H. J., van der Mei, H. C., and Norde, W. (2008). Bond-strengthening in staphylococcal adhesion to hydrophilic and hydrophobic surfaces using atomic force microscopy. Langmuir 24 (22), 12990–12994. doi:10.1021/la801824c
Brandenburg, K., Koch, M. H. J., and Seydel, U. (1992). Phase diagram of deep rough mutant lipopolysaccharide from Salmonella Minnesota R595. J. Struct. Biol. 108 (2), 93–106. doi:10.1016/1047-8477(92)90010-8
Brooks, C. F., Fuller, G. G., Frank, C. W., and Robertson, C. R. (1999). An interfacial stress rheometer to study rheological transitions in monolayers at the Air−Water interface. Langmuir 15 (7), 2450–2459. doi:10.1021/la980465r
Callone, E., Campostrini, R., Carturan, G., Cavazza, A., and Guzzon, R. (2008). Immobilization of yeast and bacteria cells in alginate microbeads coated with silica membranes: procedures, physico-chemical features and bioactivity. J. Mater. Chem. 18 (40), 4839–4848. doi:10.1039/B807301E
Cao, T., Tang, H., Liang, X., Wang, A., Auner, G. W., Salley, S. O., et al. (2006). Nanoscale investigation on adhesion of E. coli to surface modified silicone using atomic force microscopy. Biotechnol. Bioeng. 94 (1), 167–176. doi:10.1002/bit.20841
Carlsson, N., Gustafsson, H., Thörn, C., Olsson, L., Holmberg, K., and Åkerman, B. (2014). Enzymes immobilized in mesoporous silica: a physical–chemical perspective. Adv. Colloid Interface Sci. 205, 339–360. doi:10.1016/j.cis.2013.08.010
Caruso, F., Trau, D., Möhwald, H., and Renneberg, R. (2000). Enzyme encapsulation in layer-by-layer engineered polymer multilayer capsules. Langmuir 16, 1485–1488. doi:10.1021/la991161n
Champagne, C. P., and Fustier, P. (2007). Microencapsulation for the improved delivery of bioactive compounds into foods. Curr. Opin. Biotechnol. 18, 184–190. doi:10.1016/j.copbio.2007.03.001
Clifton, L. A., Skoda, M. W. A., Daulton, E. L., Hughes, A. V., Le Brun, A. P., Lakey, J. H., et al. (2013). Asymmetric phospholipid: lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J. R. Soc. Interface 10 (89), 20130810. doi:10.1098/rsif.2013.0810
Cook, M. T., Tzortzis, G., Charalampopoulos, D., and Khutoryanskiy, V. V. (2011). Production and evaluation of dry alginate-chitosan microcapsules as an enteric delivery vehicle for probiotic bacteria. Biomacromolecules 12, 2834–2840. doi:10.1021/bm200576h
Dague, E., Le, D. T. L., Zanna, S., Marcus, P., Loubière, P., and Mercier-Bonin, M. (2010). Probing in vitro interactions between Lactococcus lactis and mucins using AFM. Langmuir 26 (13), 11010–11017. doi:10.1021/la101862n
Date, A. A., Halpert, G., Babu, T., Ortiz, J., Kanvinde, P., Dimitrion, P., et al. (2018). Mucus-penetrating budesonide nanosuspension enema for local treatment of inflammatory bowel disease. Biomaterials 185, 97–105. doi:10.1016/j.biomaterials.2018.09.005
De Koker, S., De Geest, B. G., Singh, S. K., De Rycke, R., Naessens, T., Van Kooyk, Y., et al. (2009). Polyelectrolyte microcapsules as antigen delivery vehicles to dendritic cells: uptake, processing, and cross-presentation of encapsulated antigens. Angew. Chem. 121, 8637–8641. doi:10.1002/ange.200903769
Decher, G. (1997). Fuzzy nanoassemblies: toward layered polymeric multicomposites. Science 277, 1232–1237. doi:10.1126/science.277.5330.1232
Diab, R., Canilho, N., Pavel, I. A., Haffner, F. B., Girardon, M., and Pasc, A. (2017). Silica-based systems for oral delivery of drugs, macromolecules and cells. Adv. Colloid Interface Sci. 249, 346–362. doi:10.1016/j.cis.2017.04.005
Diaspro, A., Silvano, D., Krol, S., Cavalleri, O., and Gliozzi, A. (2002). Single living cell encapsulation in nano-organized polyelectrolyte shells. Langmuir 18, 5047–5050. doi:10.1021/la025646e
Dominy, S. S., Lynch, C., Ermini, F., Benedyk, M., Marczyk, A., Konradi, A., et al. (2019). Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5, eaau3333. doi:10.1126/sciadv.aau3333
Dufrêne, Y. F., Viljoen, A., Mignolet, J., and Mathelié-Guinlet, M. (2021). AFM in cellular and molecular microbiology. Cell. Microbiol. 23 (7), e13324. doi:10.1111/cmi.13324
Dully, M., Brasnett, C., Djeghader, A., Seddon, A., Neilan, J., Murray, D., et al. (2020). Modulating the release of pharmaceuticals from lipid cubic phases using a lipase inhibitor. J. colloid interface Sci. 573, 176–192. doi:10.1016/j.jcis.2020.04.015
Dupres, V., Menozzi, F. D., Locht, C., Clare, B. H., Abbott, N. L., Cuenot, S., et al. (2005). Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat. Methods 2 (7), 515–520. doi:10.1038/nmeth769
Ehehalt, R., Wagenblast, J., Erben, G., Lehmann, W. D., Hinz, U., Merle, U., et al. (2004). Phosphatidylcholine and lysophosphatidylcholine in intestinal mucus of ulcerative colitis patients. A quantitative approach by nanoelectrospray-tandem mass spectrometry. Scand. J. gastroenterology 39 (8), 737–742. doi:10.1080/00365520410006233
Eshrati, M., Amadei, F., Staffer, S., Stremmel, W., and Tanaka, M. (2019). Shear-enhanced dynamic adhesion of lactobacillus rhamnosus GG on intestinal epithelia: correlative effect of protein expression and interface mechanics. Langmuir 35 (2), 529–537. doi:10.1021/acs.langmuir.8b02931
Eshrati, M., Amadei, F., Van de Wiele, T., Veschgini, M., Kaufmann, S., and Tanaka, M. (2018). Biopolymer-based minimal formulations boost viability and metabolic functionality of probiotics lactobacillus rhamnosus GG through gastrointestinal passage. Langmuir 34 (37), 11167–11175. doi:10.1021/acs.langmuir.8b01915
FAO/WHO (2001). Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Acid Bacteria. report: report of a Joint FAO/WHO Expert Consultation, Córdoba, Argentina. Available at: http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf?ua=1.
Ferraretto, A., Bottani, M., De Luca, P., Cornaghi, L., Arnaboldi, F., Maggioni, M., et al. (2018). Morphofunctional properties of a differentiated Caco2/HT-29 co-culture as an in vitro model of human intestinal epithelium. Biosci. Rep. 38 (2). doi:10.1042/bsr20171497
Firoozmand, H., and Rousseau, D. (2016). Microbial cells as colloidal particles: pickering oil-in-water emulsions stabilized by bacteria and yeast. Food Res. Int. 81, 66–73. doi:10.1016/j.foodres.2015.10.018
Fong, W.-K., Negrini, R., Vallooran, J. J., Mezzenga, R., and Boyd, B. J. (2016). Responsive self-assembled nanostructured lipid systems for drug delivery and diagnostics. J. Colloid Interface Sci. 484, 320–339. doi:10.1016/j.jcis.2016.08.077
Fröhlich, B., Dasanna, A. K., Lansche, C., Czajor, J., Sanchez, C. P., Cyrklaff, M., et al. (2021). Functionalized supported membranes for quantifying adhesion of P. falciparum-infected erythrocytes. Biophysical J. 120 (16), 3315–3328. doi:10.1016/j.bpj.2021.07.003
Gagnon, M., Zihler Berner, A., Chervet, N., Chassard, C., and Lacroix, C. (2013). Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 94 (3), 274–279. doi:10.1016/j.mimet.2013.06.027
Gamboa, J. M., and Leong, K. W. (2013). In vitro and in vivo models for the study of oral delivery of nanoparticles. Adv. drug Deliv. Rev. 65 (6), 800–810. doi:10.1016/j.addr.2013.01.003
Garidel, P., Rappolt, M., Schromm, A. B., Howe, J., Lohner, K., Andrä, J., et al. (2005). Divalent cations affect chain mobility and aggregate structure of lipopolysaccharide from Salmonella Minnesota reflected in a decrease of its biological activity. Biochimica Biophysica Acta (BBA) - Biomembr. 1715 (2), 122–131. doi:10.1016/j.bbamem.2005.07.013
Gasperini, L., Mano, J. F., and Reis, R. L. (2014). Natural polymers for the microencapsulation of cells. J. R. Soc. Interface 11, 20140817. doi:10.1098/rsif.2014.0817
Geertsema-Doornbusch, G. I., van der Mei, H. C., and Busscher, H. J. (1993). Microbial cell surface hydrophobicity the involvement of electrostatic interactions in microbial adhesion to hydrocarbons (MATH). J. Microbiol. Methods 18 (1), 61–68. doi:10.1016/0167-7012(93)90072-P
Gilbert, E. P. (2019). Small-angle X-Ray and neutron scattering in food colloids. Curr. Opin. Colloid & Interface Sci. 42, 55–72. doi:10.1016/j.cocis.2019.03.005
Gilbert, J., Christensen, S., Nylander, T., and Bülow, L. (2023). Encapsulation of sugar beet phytoglobin BvPgb 1.2 and myoglobin in a lipid sponge phase system. Front. Soft Matter 3. doi:10.3389/frsfm.2023.1201561
Gilbert, J., Ermilova, I., Nagao, M., Swenson, J., and Nylander, T. (2022). Effect of encapsulated protein on the dynamics of lipid sponge phase: a neutron spin echo and molecular dynamics simulation study. Nanoscale 14 (18), 6990–7002. doi:10.1039/D2NR00882C
Gilbert, J., Valldeperas, M., Dhayal, S. K., Barauskas, J., Dicko, C., and Nylander, T. (2019). Immobilisation of β-galactosidase within a lipid sponge phase: structure, stability and kinetics characterisation. Nanoscale 11 (44), 21291–21301. doi:10.1039/C9NR06675F
G. J. Siegel, B. W. Agranoff, R. Wayne Albers, S. K. Fisher, and M. D. Uhler (Editors) (1999). Basic neurochemistry (Philadelphia: Lippincott-Raven).
Gouin, S. (2004). Microencapsulation: industrial appraisal of existing technologies and trends. Trends Food Sci. Technol. 15, 330–347. doi:10.1016/j.tifs.2003.10.005
Guarino, V., Caputo, T., Altobelli, R., and Ambrosio, L. (2015). Degradation properties and metabolic activity of alginate and chitosan polyelectrolytes for drug delivery and tissue engineering applications. AIMS Mater. Sci. 2 (4), 497–502. doi:10.3934/matersci.2015.4.497
Haffner, F. B., Girardon, M., Fontanay, S., Canilho, N., Duval, R. E., Mierzwa, M., et al. (2016). Core–shell alginate@silica microparticles encapsulating probiotics. J. Mater. Chem. B 4 (48), 7929–7935. doi:10.1039/C6TB02802K
Haffner, F. B., van de Wiele, T., and Pasc, A. (2017). Original behavior of L. rhamnosus GG encapsulated in freeze-dried alginate–silica microparticles revealed under simulated gastrointestinal conditions. J. Mater. Chem. B 5 (38), 7839–7847. doi:10.1039/C7TB02190A
Herrmann, M., Schneck, E., Gutsmann, T., Brandenburg, K., and Tanaka, M. (2015). Bacterial lipopolysaccharides form physically cross-linked, two-dimensional gels in the presence of divalent cations. Soft Matter 11 (30), 6037–6044. doi:10.1039/C5SM01002K
Higaki, Y., Frohlich, B., Yamamoto, A., Murakami, R., Kaneko, M., Takahara, A., et al. (2017). Ion-specific modulation of interfacial interaction potentials between solid substrates and cell-sized particles mediated via zwitterionic, super-hydrophilic poly (sulfobetaine) brushes. J. Phys. Chem. B 121 (6), 1396–1404. doi:10.1021/acs.jpcb.6b11540
Hudson, S., Cooney, J., and Magner, E. (2008). Proteins in mesoporous silicates. Angew. Chem. Int. Ed. 47 (45), 8582–8594. doi:10.1002/anie.200705238
Humphreys, B. A., Campos-Terán, J., Arnold, T., Baunsgaard, L., Vind, J., Dicko, C., et al. (2022). The influence of pH on the lipase digestion of nanosized triolein, diolein and monoolein films. Front. Soft Matter 2. doi:10.3389/frsfm.2022.929104
Israelachvili, J. N., Mitchell, D. J., and Ninham, B. W. (1976). Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 2 Mol. Chem. Phys. 72, 1525–1568. doi:10.1039/f29767201525
Jiang, X., Martens, H. J., Shekarforoush, E., Muhammed, M. K., Whitehead, K. A., Arneborg, N., et al. (2022). Multi-species colloidosomes by surface-modified lactic acid bacteria with enhanced aggregation properties. J. Colloid Interface Sci. 622, 503–514. doi:10.1016/j.jcis.2022.04.136
Jiang, X., Shekarforoush, E., Muhammed, M. K., Bovet, N., Ceccato, M., Whitehead, K. A., et al. (2023a). Probing surface properties of lactic acid bacteria - comparative modification by anhydride and aldehyde grafting. Surfaces Interfaces 38, 102848. doi:10.1016/j.surfin.2023.102848
Jiang, X., Shekarforoush, E., Muhammed, M. K., Whitehead, K. A., Arneborg, N., and Risbo, J. (2023b). Lactic acid bacteria as structural building blocks in non-fat whipping cream analogues. Food Hydrocoll. 135, 108137. doi:10.1016/j.foodhyd.2022.108137
Jiang, X., Shekarforoush, E., Muhammed, M. K., Whitehead, K., Simonsen, A. C., Arneborg, N., et al. (2021). Efficient chemical hydrophobization of lactic acid bacteria – one-step formation of double emulsion. Food Res. Int. 147, 110460. doi:10.1016/j.foodres.2021.110460
Jiang, X., Yucel Falco, C., Dalby, K. N., Siegumfeldt, H., Arneborg, N., and Risbo, J. (2019). Surface engineered bacteria as Pickering stabilizers for foams and emulsions. Food Hydrocoll. 89, 224–233. doi:10.1016/j.foodhyd.2018.10.044
Kaindl, T., Rieger, H., Kaschel, L.-M., Engel, U., Schmaus, A., Sleeman, J., et al. (2012). Spatio-temporal patterns of pancreatic cancer cells expressing CD44 isoforms on supported membranes displaying hyaluronic acid oligomers arrays. PLoS One 7, e42991. doi:10.1371/journal.pone.0042991
Katchalsky, A., Cooper, R. E., Upadhyay, J., and Wassermann, A. (1961). 1028. Counter-ion fixation in alginates. J. Chem. Soc. (Resumed) (0), 5198–5204. doi:10.1039/JR9610005198
Kleiveland, C. R. (2015). “Co-Cultivation of caco-2 and HT-29mtx,” in The Impact of Food Bioactives on Health: in vitro and ex vivo models. Editors K. Verhoeckx, P. Cotter, I. López-Expósito, C. Kleiveland, T. Lea, A. Mackieet al. (Cham: Springer International Publishing), 135–140.
Korytowski, A., Abuillan, W., Amadei, F., Makky, A., Gumiero, A., Sinning, I., et al. (2017). Accumulation of phosphatidylcholine on gut mucosal surface is not dominated by electrostatic interactions. Biochimica Biophysica Acta (BBA) - Biomembr. 1859 (5), 959–965. doi:10.1016/j.bbamem.2017.02.008
Krasaekoopt, W., Bhandari, B., and Deeth, H. (2003). Evaluation of encapsulation techniques of probiotics for yoghurt. Int. Dairy J. 13, 3–13. doi:10.1016/s0958-6946(02)00155-3
Krasaekoopt, W., Bhandari, B., and Deeth, H. (2004). The influence of coating materials on some properties of alginate beads and survivability of microencapsulated probiotic bacteria. Int. Dairy J. 14, 737–743. doi:10.1016/j.idairyj.2004.01.004
Kwiecień, I., and Kwiecień, M. (2018). Application of polysaccharide-based hydrogels as probiotic delivery systems. Gels 4 (2), 47. doi:10.3390/gels4020047
Larsson, K. (2000). Aqueous dispersions of cubic lipid–water phases. Curr. Opin. Colloid & Interface Sci. 5 (1), 64–69. doi:10.1016/S1359-0294(00)00040-6
Larsson, K. (1989). Cubic lipid-water phases: structures and biomembrane aspects. J. Phys. Chem. 93 (21), 7304–7314. doi:10.1021/j100358a010
Larsson, K., Quinn, P., Sato, K., and Tyberg, F. (2006). Lipids: structure, physical properties and functionality. Bridgewater: The Oily Press.
Lee, C.-H., Lin, T.-S., and Mou, C.-Y. (2009). Mesoporous materials for encapsulating enzymes. Nano today 4 (2), 165–179. doi:10.1016/j.nantod.2009.02.001
Lee, K. Y., and Mooney, D. J. (2012). Alginate: properties and biomedical applications. Prog. Polym. Sci. 37, 106–126. doi:10.1016/j.progpolymsci.2011.06.003
Liu, D., and Du, D. (2020). Mulberry fruit extract alleviates cognitive impairment by promoting the clearance of amyloid-β and inhibiting neuroinflammation in Alzheimer’s disease mice. Neurochem. Res. 45 (9), 2009–2019. doi:10.1007/s11064-020-03062-7
Lvov, Y., Decher, G., and Moehwald, H. (1993). Assembly, structural characterization, and thermal behavior of layer-by-layer deposited ultrathin films of poly (vinyl sulfate) and poly (allylamine). Langmuir 9, 481–486. doi:10.1021/la00026a020
Maldonado-Valderrama, J., Wilde, P., Macierzanka, A., and Mackie, A. (2011). The role of bile salts in digestion. Adv. Colloid Interface Sci. 165 (1), 36–46. doi:10.1016/j.cis.2010.12.002
Manzano, M., and Vallet-Regí, M. (2020). Mesoporous silica nanoparticles for drug delivery. Adv. Funct. Mater. 30 (2), 1902634. doi:10.1002/adfm.201902634
Martin, H. H. (1963). Bacterial protoplasts—a review. J. Theor. Biol. 5 (1), 1–34. doi:10.1016/0022-5193(63)90034-1
Matricardi, P., Meo, C. D., Coviello, T., and Alhaique, F. (2008). Recent advances and perspectives on coated alginate microspheres for modified drug delivery. Expert Opin. Drug Deliv. 5, 417–425. doi:10.1517/17425247.5.4.417
McClements, D. J. (2015). Encapsulation, protection, and release of hydrophilic active components: potential and limitations of colloidal delivery systems. Adv. Colloid Interface Sci. 219, 27–53. doi:10.1016/j.cis.2015.02.002
Minekus, M., Alminger, M., Alvito, P., Ballance, S., Bohn, T., Bourlieu, C., et al. (2014). A standardised static in vitro digestion method suitable for food – an international consensus. Food & Funct. 5 (6), 1113–1124. doi:10.1039/C3FO60702J
Mitchell, D. J., and Ninham, B. W. (1981). Micelles, vesicles and microemulsions. J. Chem. Soc. Faraday Trans. 2 Mol. Chem. Phys. 77 (4), 601–629. doi:10.1039/f29817700601
Muhammed, M. K., Jiang, X., Shekarforoush, E., Whitehead, K., Vogensen, F. K., Risbo, J., et al. (2023). Ex-<em>Lactobacillus</em> strains with intrinsic propensity to stabilize pickering oil-in-water emulsions. bioRxiv, 540633. doi:10.1101/2023.05.13.540633
Mulet, X., Boyd, B. J., and Drummond, C. J. (2013). Advances in drug delivery and medical imaging using colloidal lyotropic liquid crystalline dispersions. J. Colloid Interface Sci. 393, 1–20. doi:10.1016/j.jcis.2012.10.014
Nassif, N., and Livage, J. (2011). From diatoms to silica-based biohybrids. Chem. Soc. Rev. 40 (2), 849–859. doi:10.1039/C0CS00122H
Netz, R. R. (1999). Debye-Hückel theory for interfacial geometries. Phys. Rev. E 60 (3), 3174–3182. doi:10.1103/PhysRevE.60.3174
No, H. K., Park, N. Y., Lee, S. H., and Meyers, S. P. (2002). Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. food Microbiol. 74 (1-2), 65–72. doi:10.1016/s0168-1605(01)00717-6
Oliveira, R. G., Schneck, E., Quinn, B. E., Konovalov, O. V., Brandenburg, K., Gutsmann, T., et al. (2010). Crucial roles of charged saccharide moieties in survival of gram negative bacteria against protamine revealed by combination of grazing incidence x-ray structural characterizations and Monte Carlo simulations. Phys. Rev. E 81 (4), 041901. doi:10.1103/PhysRevE.81.041901
Oliveira, R. G., Schneck, E., Quinn, B. E., Konovalov, O. V., Brandenburg, K., Seydel, U., et al. (2009). Physical mechanisms of bacterial survival revealed by combined grazing-incidence X-ray scattering and Monte Carlo simulation. Comptes Rendus Chim. 12 (1), 209–217. doi:10.1016/j.crci.2008.06.020
Orive, G., Ponce, S., Hernández, R. M., Gascón, A. R., Igartua, M., and Pedraz, J. L. (2002). Biocompatibility of microcapsules for cell immobilization elaborated with different type of alginates. Biomaterials 23, 3825–3831. doi:10.1016/s0142-9612(02)00118-7
Pasquier, J., Brûlet, A., Boire, A., Jamme, F., Perez, J., Bizien, T., et al. (2019). Monitoring food structure during digestion using small-angle scattering and imaging techniques. Colloids Surfaces A Physicochem. Eng. Aspects 570, 96–106. doi:10.1016/j.colsurfa.2019.02.059
Patton, J. S., and Carey, M. C. (1979). Watching fat digestion. Science 204 (4389), 145–148. doi:10.1126/science.432636
Pavel, I.-A., Prazeres, S. F., Montalvo, G., Garcia Ruiz, C., Nicolas, V., Celzard, A., et al. (2017). Effect of meso vs macro size of hierarchical porous silica on the adsorption and activity of immobilized β-galactosidase. Langmuir 33 (13), 3333–3340. doi:10.1021/acs.langmuir.7b00134
Pelaseyed, T., Bergström, J. H., Gustafsson, J. K., Ermund, A., Birchenough, G. M., Schütte, A., et al. (2014). The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 260 (1), 8–20. doi:10.1111/imr.12182
Pham, A. C., Peng, K.-Y., Salim, M., Ramirez, G., Hawley, A., Clulow, A. J., et al. (2020). Correlating digestion-driven self-assembly in milk and infant formulas with changes in lipid composition. ACS Appl. Bio Mater. 3 (5), 3087–3098. doi:10.1021/acsabm.0c00131
Pink, D. A., Truelstrup Hansen, L., Gill, T. A., Quinn, B. E., Jericho, M. H., and Beveridge, T. J. (2003). Divalent calcium ions inhibit the penetration of protamine through the polysaccharide brush of the outer membrane of gram-negative bacteria. Langmuir 19 (21), 8852–8858. doi:10.1021/la030193e
Porcar, L., Hamilton, W. A., Butler, P. D., and Warr, G. (2003). Scaling of structural and rheological response of L3 sponge phases in the “sweetened” cetylpyridinium/hexanol/dextrose/brine system. Langmuir 19 (26), 10779–10794. doi:10.1021/la034661w
Priya, A. J., Vijayalakshmi, S. P., and Raichur, A. M. (2011). Enhanced survival of probiotic lactobacillus acidophilus by encapsulation with nanostructured polyelectrolyte layers through layer-by-layer approach. J. Agric. Food Chem. 59 (21), 11838–11845. doi:10.1021/jf203378s
Proitsi, P., Kim, M., Whiley, L., Simmons, A., Sattlecker, M., Velayudhan, L., et al. (2017). Association of blood lipids with Alzheimer's disease: a comprehensive lipidomics analysis. Alzheimer's Dementia 13 (2), 140–151. doi:10.1016/j.jalz.2016.08.003
Razumas, V., Kanapieniené, J., Nylander, T., Engström, S., and Larsson, K. (1994). Electrochemical biosensors for glucose, lactate, urea, and creatinine based on enzymes entrapped in a cubic liquid crystalline phase. Anal. Chim. acta 289 (2), 155–162. doi:10.1016/0003-2670(94)80098-7
Rieger, H., Yoshikawa, H. Y., Quadt, K., Nielsen, M. A., Sanchez, C. P., Salanti, A., et al. (2015). Cytoadhesion of Plasmodium falciparum–infected erythrocytes to chondroitin-4-sulfate is cooperative and shear enhanced. Blood, J. Am. Soc. Hematol. 125 (2), 383–391. doi:10.1182/blood-2014-03-561019
Rohde, M. (2019). The gram-positive bacterial cell wall. Microbiol. Spectr. 7(3), doi:10.1128/microbiolspec.gpp1123-0044-2018
Rosenberg, M. (1984). Bacterial adherence to hydrocarbons: a useful technique for studying cell surface hydrophobicity. FEMS Microbiol. Lett. 22 (3), 289–295. doi:10.1111/j.1574-6968.1984.tb00743.x
Rosenberg, M., Gutnick, D., and Rosenberg, E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9 (1), 29–33. doi:10.1111/j.1574-6968.1980.tb05599.x
Rosenberg, M. (2006). Microbial adhesion to hydrocarbons: twenty-five years of doing MATH. FEMS Microbiol. Lett. 262 (2), 129–134. doi:10.1111/j.1574-6968.2006.00291.x
Rowan-Nash, A. D., Korry, B. J., Mylonakis, E., and Belenky, P. (2019). Cross-domain and viral interactions in the microbiome. Microbiol. Mol. Biol. Rev. 83 (1), e00044–e00018. doi:10.1128/MMBR.00044-18
Salentinig, S., Phan, S., Hawley, A., and Boyd, B. J. (2015). Self-assembly structure formation during the digestion of human breast milk. Angew. Chem. Int. Ed. 54 (5), 1600–1603. doi:10.1002/anie.201408320
Salentinig, S., Phan, S., Khan, J., Hawley, A., and Boyd, B. J. (2013). Formation of highly organized nanostructures during the digestion of milk. ACS Nano 7 (12), 10904–10911. doi:10.1021/nn405123j
Salentinig, S., Sagalowicz, L., Leser, M. E., Tedeschi, C., and Glatter, O. (2011). Transitions in the internal structure of lipid droplets during fat digestion. Soft Matter 7 (2), 650–661. doi:10.1039/C0SM00491J
Schneck, E., Oliveira, R. G., Rehfeldt, F., Demé, B., Brandenburg, K., Seydel, U., et al. (2009a). Mechanical properties of interacting lipopolysaccharide membranes from bacteria mutants studied by specular and off-specular neutron scattering. Phys. Rev. E 80 (4), 041929. doi:10.1103/PhysRevE.80.041929
Schneck, E., Papp-Szabo, E., Quinn, B. E., Konovalov, O. V., Beveridge, T. J., Pink, D. A., et al. (2009b). Calcium ions induce collapse of charged O-side chains of lipopolysaccharides from Pseudomonas aeruginosa. J. R. Soc. Interface 6 (5), S671–S678. doi:10.1098/rsif.2009.0190.focus
Schneck, E., Schubert, T., Konovalov, O. V., Quinn, B. E., Gutsmann, T., Brandenburg, K., et al. (2010). Quantitative determination of ion distributions in bacterial lipopolysaccharide membranes by grazing-incidence X-ray fluorescence. Proc. Natl. Acad. Sci. 107 (20), 9147–9151. doi:10.1073/pnas.0913737107
Schneider, M. F., Lim, K., Fuller, G. G., and Tanaka, M. (2002). Rheology of glycocalix model at air/water interface. Phys. Chem. Chem. Phys. 4 (10), 1949–1952. doi:10.1039/B110631G
Schwarz, B., and Schönhoff, M. (2002). A 1H NMR relaxation study of hydration water in polyelectrolyte mono and multilayers adsorbed to colloidal particles. Colloids Surfaces A Physicochem. Eng. Aspects 198, 293–304. doi:10.1016/s0927-7757(01)00945-1
Seddon, J., and Templer, R. (1995). “Polymorphism of lipid-water systems,” in Handbook of biological physics (Elsevier), 97–160.
Shekarforoush, E., Jiang, X., Kedir Muhammed, M., Whitehead, K. A., Arneborg, N., and Risbo, J. (2022). Enzymatic modification and adsorption of hydrophobic zein proteins on lactic acid bacteria stabilize Pickering emulsions. Food Res. Int. 161, 111783. doi:10.1016/j.foodres.2022.111783
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2 (5), a000414. doi:10.1101/cshperspect.a000414
Singh, P., Magalhães, S., Alves, L., Antunes, F., Miguel, M., Lindman, B., et al. (2019). Cellulose-based edible films for probiotic entrapment. Food Hydrocoll. 88, 68–74. doi:10.1016/j.foodhyd.2018.08.057
Singh, P., Medronho, B., Alves, L., da Silva, G. J., Miguel, M. G., and Lindman, B. (2017). Development of carboxymethyl cellulose-chitosan hybrid micro- and macroparticles for encapsulation of probiotic bacteria. Carbohydr. Polym. 175, 87–95. doi:10.1016/j.carbpol.2017.06.119
Singh, P., Medronho, B., Miguel, M. G., and Esquena, J. (2018a). On the encapsulation and viability of probiotic bacteria in edible carboxymethyl cellulose-gelatin water-in-water emulsions. Food Hydrocoll. 75, 41–50. doi:10.1016/j.foodhyd.2017.09.014
Singh, P., Medronho, B., Santos, T. d., Nunes-Correia, I., Granja, P., Miguel, M. G., et al. (2018b). On the viability, cytotoxicity and stability of probiotic bacteria entrapped in cellulose-based particles. Food Hydrocoll. 82, 457–465. doi:10.1016/j.foodhyd.2018.04.027
Sonnenburg, J. L., Angenent, L. T., and Gordon, J. I. (2004). Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5 (6), 569–573. doi:10.1038/ni1079
Sumper, M., and Brunner, E. (2006). Learning from diatoms: nature's tools for the production of nanostructured silica. Adv. Funct. Mater. 16 (1), 17–26. doi:10.1002/adfm.200500616
Sun, W., Vallooran, J. J., and Mezzenga, R. (2015). Enzyme kinetics in liquid crystalline mesophases: size matters, but also topology. Langmuir 31 (15), 4558–4565. doi:10.1021/acs.langmuir.5b00579
Talaikis, M., Valldeperas, M., Matulaitienė, I., Borzova, J. L., Barauskas, J., Niaura, G., et al. (2019). On the molecular interactions in lipid bilayer–water assemblies of different curvatures. J. Phys. Chem. B 123 (12), 2662–2672. doi:10.1021/acs.jpcb.8b11387
Tanaka, M., Schiefer, S., Gege, C., Schmidt, R. R., and Fuller, G. G. (2004). Influence of subphase conditions on interfacial viscoelastic properties of synthetic lipids with gentiobiose head groups. J. Phys. Chem. B 108 (10), 3211–3214. doi:10.1021/jp0367934
Tasciotti, E., Liu, X., Bhavane, R., Plant, K., Leonard, A. D., Price, B. K., et al. (2008). Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic applications. Nat. Nanotechnol. 3 (3), 151–157. doi:10.1038/nnano.2008.34
Thoma, J., Abuillan, W., Furikado, I., Habe, T., Yamamoto, A., Gierlich, S., et al. (2020). Specific localisation of ions in bacterial membranes unravels physical mechanism of effective bacteria killing by sanitiser. Sci. Rep. 10 (1), 12302. doi:10.1038/s41598-020-69064-1
Thomas, T., Hawzeen, S. K., and Jack, C. L. (2018). Bacterial autoaggregation. AIMS Microbiol. 4 (1), 140–164. doi:10.3934/microbiol.2018.1.140
Thompson, K. L., Williams, M., and Armes, S. P. (2015). Colloidosomes: synthesis, properties and applications. J. Colloid Interface Sci. 447, 217–228. doi:10.1016/j.jcis.2014.11.058
Tran, P. H. L., and Tran, T. T. D. (2021). Blueberry supplementation in neuronal health and protective technologies for efficient delivery of blueberry anthocyanins. Biomolecules 11 (1), 102. doi:10.3390/biom11010102
Tsai, G.-J., and Hwang, S.-P. (2004). In vitro and in vivo antibacterial activity of shrimp chitosan against some intestinal bacteria. Fish. Sci. 70 (4), 675–681. doi:10.1111/j.1444-2906.2004.00856.x
Tyle, P. (1993). Effect of size, shape and hardness of particles in suspension on oral texture and palatability. Acta Psychol. 84, 111–118. doi:10.1016/0001-6918(93)90077-5
Uriz, M. J., Turon, X., Becerro, M. A., and Agell, G. (2003). Siliceous spicules and skeleton frameworks in sponges: origin, diversity, ultrastructural patterns, and biological functions. Microsc. Res. Tech. 62 (4), 279–299. doi:10.1002/jemt.10395
Valldeperas, M., Dabkowska, A. P., Pálsson, G. K., Rogers, S., Mahmoudi, N., Carnerup, A., et al. (2019a). Interfacial properties of lipid sponge-like nanoparticles and the role of stabilizer on particle structure and surface interactions. Soft Matter 15 (10), 2178–2189. doi:10.1039/C8SM02634C
Valldeperas, M., Salis, A., Barauskas, J., Tiberg, F., Arnebrant, T., Razumas, V., et al. (2019b). Enzyme encapsulation in nanostructured self-assembled structures: toward biofunctional supramolecular assemblies. Curr. Opin. Colloid & Interface Sci. 44, 130–142. doi:10.1016/j.cocis.2019.09.007
Valldeperas, M., Talaikis, M., Dhayal, S. K., Velička, M., Barauskas, J., Niaura, G., et al. (2019c). Encapsulation of aspartic protease in nonlamellar lipid liquid crystalline phases. Biophysical J. 117 (5), 829–843. doi:10.1016/j.bpj.2019.07.031
Valldeperas, M., Wiśniewska, M., Ram-On, M., Kesselman, E., Danino, D., Nylander, T., et al. (2016). Sponge phases and nanoparticle dispersions in aqueous mixtures of mono- and diglycerides. Langmuir 32 (34), 8650–8659. doi:10.1021/acs.langmuir.6b01356
Van de Wiele, T., Van den Abbeele, P., Ossieur, W., Possemiers, S., and Marzorati, M. (2015). The simulator of the human intestinal microbial ecosystem (SHIME®). Impact Food Bioact. Health vitro ex vivo models 2015, 305–317.
Van Oss, C. J., Chaudhury, M. K., and Good, R. J. (1988). Interfacial Lifshitz-van der Waals and polar interactions in macroscopic systems. Chem. Rev. 88 (6), 927–941. doi:10.1021/cr00088a006
Verbelen, C., Raze, D., Dewitte, F., Locht, C., and Dufrene, Y. F. (2007). Single-molecule force spectroscopy of mycobacterial adhesin-adhesin interactions. J. Bacteriol. 189 (24), 8801–8806. doi:10.1128/jb.01299-07
Vollmer, W., Blanot, D., and De Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32 (2), 149–167. doi:10.1111/j.1574-6976.2007.00094.x
Wadsäter, M., Barauskas, J., Tiberg, F., and Nylander, T. (2018). The lipolytic degradation of highly structured cubic micellar nanoparticles of soy phosphatidylcholine and glycerol dioleate by phospholipase A2 and triacylglycerol lipase. Chem. Phys. Lipids 211, 86–92. doi:10.1016/j.chemphyslip.2017.11.011
Wang, M., Qi, W., Yu, Q., Su, R., and He, Z. (2010). Cross-linking enzyme aggregates in the macropores of silica gel: a practical and efficient method for enzyme stabilization. Biochem. Eng. J. 52 (2-3), 168–174. doi:10.1016/j.bej.2010.08.003
Yang, X.-Y., Li, Z.-Q., Liu, B., Klein-Hofmann, A., Tian, G., Feng, Y.-F., et al. (2006). Fish-in-Net encapsulation of enzymes in macroporous cages for stable, reusable, and active heterogeneous biocatalysts. Adv. Mater. 18 (4), 410–414. doi:10.1002/adma.200502003
Yucel Falco, C., Amadei, F., Dhayal, S. K., Cárdenas, M., Tanaka, M., and Risbo, J. (2019). Hybrid coating of alginate microbeads based on protein-biopolymer multilayers for encapsulation of probiotics. Biotechnol. Prog. 35 (3), e2806. doi:10.1002/btpr.2806
Yucel Falco, C., Geng, X., Cárdenas, M., and Risbo, J. (2017a). Edible foam based on Pickering effect of probiotic bacteria and milk proteins. Food Hydrocoll. 70, 211–218. doi:10.1016/j.foodhyd.2017.04.003
Yucel Falco, C., Sotres, J., Rascón, A., Risbo, J., and Cárdenas, M. (2017b). Design of a potentially prebiotic and responsive encapsulation material for probiotic bacteria based on chitosan and sulfated β-glucan. J. Colloid Interface Sci. 487, 97–106. doi:10.1016/j.jcis.2016.10.019
Keywords: self-assembly, lipids, polymers, colloids, probiotics, enzymes, foods
Citation: Risbo J, Nylander T and Tanaka M (2023) Delivery of probiotics and enzymes in self-assemblies of lipids and biopolymers based on colloidal principles. Front. Soft Matter 3:1257688. doi: 10.3389/frsfm.2023.1257688
Received: 12 July 2023; Accepted: 11 October 2023;
Published: 01 November 2023.
Edited by:
Kai Yang, Soochow University, ChinaCopyright © 2023 Risbo, Nylander and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Risbo, anJpQGZvb2Qua3UuZGs=; Tommy Nylander, VG9tbXkuTnlsYW5kZXJAZmtlbTEubHUuc2U=; Motomu Tanaka, dGFuYWthQHVuaS1oZWlkZWxiZXJnLmRl
 Jens Risbo1*
Jens Risbo1* Tommy Nylander
Tommy Nylander Motomu Tanaka
Motomu Tanaka