
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. RNA Res. , 18 September 2024
Sec. RNP Biogenesis and Function
Volume 2 - 2024 | https://doi.org/10.3389/frnar.2024.1448194
This article is part of the Research Topic Ribonucleoprotein formation and regulation View all 5 articles
 A. Gregory Matera1,2,3*
A. Gregory Matera1,2,3* Rebecca E. Steiner1†‡
Rebecca E. Steiner1†‡ C. Allie Mills4
C. Allie Mills4 Benjamin D. McMichael1
Benjamin D. McMichael1 Laura E. Herring4
Laura E. Herring4 Eric L. Garcia1,5‡
Eric L. Garcia1,5‡Introduction: Molecular chaperones and co-chaperones are highly conserved cellular components that perform a variety of duties related to the proper three-dimensional folding of the proteome. The web of factors that carries out this essential task is called the proteostasis network (PN). Ribonucleoproteins (RNPs) represent an underexplored area in terms of the connections they make with the PN. The Survival Motor Neuron (SMN) complex is an assembly chaperone and serves as a paradigm for studying how specific RNAs are identified and paired with their client substrate proteins to form RNPs. SMN is the eponymous component of a large complex, required for the biogenesis of uridine-rich small nuclear ribonucleoproteins (U-snRNPs), that localizes to distinct membraneless organelles in both the nucleus and cytoplasm of animal cells. SMN protein forms the oligomeric core of this complex, and missense mutations in the human SMN1 gene are known to cause Spinal Muscular Atrophy (SMA). The basic framework for understanding how snRNAs are assembled into U-snRNPs is known. However, the pathways and mechanisms used by cells to regulate their biogenesis are poorly understood.
Methods: Given the importance of these processes to normal development as well as neurodegenerative disease, we set out to identify and characterize novel SMN binding partners. We carried out affinity purification mass spectrometry (AP-MS) of Drosophila SMN complexes using fly lines exclusively expressing either wildtype or SMA-causing missense alleles.
Results: Bioinformatic analyses of the pulldown data, along with comparisons to proximity labeling studies carried out in human cells, revealed conserved connections to at least two other major chaperone systems including heat shock folding chaperones (HSPs) and histone/nucleosome assembly chaperones. Notably, we found that heat shock cognate protein Hsc70-4 and other HspA family members preferentially associated with SMA-causing alleles of SMN.
Discussion: Hsc70-4 is particularly interesting because its mRNA is aberrantly sequestered by a mutant form of TDP-43 in mouse and Drosophila ALS (Amyotrophic Lateral Sclerosis) disease models. Most important, a missense allele of Hsc70-4 (HspA8 in mammals) was recently identified as a bypass suppressor of the SMA phenotype in mice. Collectively, these findings suggest that chaperone-related dysfunction lies at the etiological root of both ALS and SMA.
Cellular stressors, both extrinsic and intrinsic, come in a countless variety of shapes and flavors but they all lead to the same end: macromolecular chaos. In order to survive the pull of these entropic forces, at least for a lifetime, organisms have evolved defense mechanisms that help maintain homeostasis in a dynamic environment. These defense mechanisms orchestrate the activation of stress response pathways across tissues and organs to promote cellular and, ultimately, organismal health (Miles et al., 2019). Heat stress is unsurprisingly among the most significant barriers to life. All organisms respond to the presence of too much heat by inducing the synthesis of heat shock proteins, or HSPs (Lindquist and Craig, 1988). Indeed HSPs, are highly conserved in all three kingdoms of life and participate in protein quality control, reviewed in (Calderwood and Murshid, 2017).
Proteostasis (protein homeostasis) is a term used to describe the overall process of maintaining a functional proteome. The web of cellular components that carries out this essential task is called the proteostasis network, or PN (Miles et al., 2019). A natural aspect of proteostasis involves the degradation and/or recycling of proteins and complexes that are misfolded beyond repair, aggregated or have simply reached the end of their normal lifetimes. Thus the PN includes the ubiquitin proteosome system (UPS) along with several membrane-associated processes that we collectively term autophagy (Kwon and Ciechanover, 2017). Additionally, the PN also includes the HSPs that serve as molecular chaperones, many of which are constitutively expressed along with their close partners (co-chaperones), reviewed in (Kampinga et al., 2009; Hartl et al., 2011).
Molecular chaperones and co-chaperones not only prevent co-translational misfolding, but also carry out re-folding of stress-denatured proteins, re-directing non-native intermediates to their native states (Hartl et al., 2011; Richter et al., 2010). Thus, a molecular chaperone is often defined as a protein that helps another protein to acquire its active conformation, without actually being present in the final product (Hartl and Hayer-Hartl, 2009). Distinct classes of structurally unrelated chaperones exist in cells, forming cooperative pathways and sub-networks (Brehme et al., 2014). HSPs belong to the broad category of “folding” chaperones, that also includes the ring-shaped chaperonins (Richter et al., 2010). The so-called ‘assembly’ chaperones represent another class of molecular chaperones that help carry out the biogenesis of large macromolecular complexes such as nucleosomes, proteasomes, ribosomes and spliceosomes (Gill et al., 2022; Johal et al., 2023; Kato and Satoh, 2018; Matera and Wang, 2014; Woodson, 2008; Baßler and Hurt, 2019).
The Survival Motor Neuron (SMN) complex directs assembly of the uridine-rich small nuclear ribonucleoproteins (U-snRNPs) that make up the spliceosome (Paushkin et al., 2002; Matera et al., 2007). Because the SMN complex is essential for proper formation of the RNP, but is not part of the final particle (Chari et al., 2008), it can be viewed as an assembly chaperone (see (Matera and Wang, 2014; Gruss et al., 2017; Raimer et al., 2017) for details). In human cells, the complex is composed of SMN and eight main partner proteins (Matera and Wang, 2014; Gruss et al., 2017; Battle et al., 2006), collectively called Gemins (Gem2-8 plus STRAP/UNRIP). In Drosophilids and other Dipteran genomes, the Gem6·7·8 subunit has been lost (Matera et al., 2019). The best-studied client substrates of the SMN complex are the Sm-proteins and U-rich snRNAs (Matera and Wang, 2014; Matera et al., 2007), although it likely plays a role in facilitating assembly of other RNP classes as well (Lu et al., 2014; Singh et al., 2017; Massenet, 2019). The core of the complex is composed of SMN·Gem2 dimers which, in vitro, are fully sufficient for catalyzing assembly of the heteroheptameric Sm-ring onto human or fruitfly snRNAs (Kroiss et al., 2008). Higher-order multimerization of SMN·Gem2 dimers is required for metazoan viability (Gupta et al., 2021). Note that budding yeast genomes have lost SMN and all of the other Gemins, except Gem2 (Matera and Wang, 2014). Given the fact that these organisms have introns and express spliceosomal snRNPs, it seems likely that the rest of the Gemins are involved in regulatory or other aspects of SMN function.
Although the basic framework for understanding how RNPs traffic through the cell during their maturation cycle is known and is conserved among metazoans (Matera and Shpargel, 2006; Stanek and Neugebauer, 2006), the pathways and mechanisms used by cells to regulate snRNP biogenesis are poorly understood (Matera and Wang, 2014). How are the activities of other macromolecular assembly machineries connected to snRNP biogenesis and how do they change in disease states? What signaling pathways are used to coordinate these essential cellular functions and how are the signals received? To begin to address these questions, we decided to search beyond the Gemins for additional SMN binding partners. Over the past decade, we have developed Drosophila as model system to understand SMN function using an allelic series of transgenic flies expressing SMA-causing missense mutations that recapitulates the full range of phenotypic severity seen in human patients (Matera et al., 2019; Gupta et al., 2021; Praveen et al., 2012; Praveen et al., 2014; Garcia et al., 2013; Garcia et al., 2016; Gray et al., 2018; Spring et al., 2019; Raimer et al., 2020; Garcia et al., 2024).
Here, we carried out affinity purification, coupled with mass spectrometry (AP-MS), to identify novel SMN binding partners present in embryonic lysates of wildtype and hypomorphic SMA animal models. Bioinformatic analyses of the hits, along with comparisons to proximity labeling data from human cells revealed connections to at least two other major chaperone systems. Links to the proteostasis network of folding chaperones were even more apparent in the lysates of SMA-causing missense alleles. In particular, we identified the heat shock cognate protein Hsc70-4 and other Hsc70/Hsp70 family members. Hsc70-4 is notable because the mRNA encoding this protein is targeted by a dominant negative, ALS (Amyotrophic Lateral Sclerosis) causing form of TDP-43 in mouse and Drosophila disease models (Coyne et al., 2017). Furthermore, a missense allele of Hsc70-4 (HspA8 in mammals) was recently identified as a suppressor of the SMA phenotype in mice (Kim et al., 2023). Taken together, these findings strongly suggest that the underlying neuromuscular phenotypes of SMA and ALS stem from defects in a common pathway served by heat shock folding chaperones.
Oregon-R was used as the wild-type control. The creation of transgenic Smn fly lines was previously described (Praveen et al., 2014). Generation of ‘proteomic stocks’ (i.e., stable fly lines expressing only transgenic Flag-SMN protein in the absence of endogenous SMN) was also described previously (Gray et al., 2018; Spring et al., 2019). Briefly, virgin females from the SmnX7/TM6B-GFP (null/balancer) line were crossed to SmnX7,Flag-SmnTG/TM6B-GFP males at 25°C, where TG represents the WT, G73R, I93F or G210C allele. To reduce stress from overpopulation and/or competition from heterozygous siblings, crosses were performed on molasses plates with yeast paste and GFP negative (SmnX7,Flag-SmnTG/SmnX7) larvae were sorted into vials containing standard molasses fly food during the second instar larval stage. Large numbers of these progeny were allowed to intercross in order to establish stable stocks. Following expansion, a stable population was formed and then maintained for 1–5 years (35–150 generations). Although Flag-Smn was initially hemizygous, this process allowed for meiotic recombination and selection to generate populations that are essentially homozygous (spot-checked by PCR) at the Flag-Smn locus at band 86F, but remain null at the endogenous Smn locus at band 73A.
0–24 h embryos were collected from Oregon-R control and the various Flag-SMN stocks, dechorionated, flash frozen, and stored at −80°C prior to use. Roughly 75–100 ul of the packed dechorionized embryos were used for each replicate, which were resuspended on ice into 300 ul of Lysis Buffer containing 100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2 mM magnesium acetate, 0.5 mM dithiothreitol (DTT) plus ½ tablet of protease inhibitor cocktail. Lysates were generated inside a 1.5 mL centrifuge tube by crushing with a pestle and then centrifuged for 10 min at 4°C at 13,000 rpm in a microfuge. The soluble (middle) layer of the lysate was transferred to a new tube, and assayed (Bradford) for protein content.
30ul of anti-FLAG M2 agarose beads (Sigma) were resuspended and then pre-washed in 1 mL lysis buffer (by inverting 5 times and then centrifuging at 100 × g for 1 min) a total of four times. After pre-washes, beads were resuspended in 100 ul lysis buffer. For each replicate, 5 mg of embryonic lysate were then added to the prepared beads and incubated for 2 h at 4°C with end-over-end rotation. After incubation, beads were spun down (4°C, 500 × g, 1 min) resuspended in 1 mL of Wash Buffer 1 (WB1: 20 mM Tris·HCl, pH 8.0, 100 mM KCl, 2 mM MgCl2, 10% glycerol, 0.05% Tween-20, 0.5 mM DTT and 0.2 mM PMSF) and washed 2x with (4°C, 100 × g, 1 min) centrifugation. This process was repeated 3 × in Wash Buffer 2 (same as WB1 except 250 mM KCl was used) and then a final time in WB1 for a total of six washing steps.
Samples were eluted 3 × into 50 ul of Elution Buffer (20 mM Tris·HCl, pH 8.0, 100 mM KCl, 10% glycerol, 0.5 mM DTT, 0.2 mM PMSF) containing 200 ug/mL of 3xFLAG peptide (Sigma). The first two elutions were combined and small portions were used for subsequent quality control steps (10% each for silver staining and Western blotting) prior to sending for mass spectrometic analysis. Once checked, the immunoprecipitated samples were subjected to SDS-PAGE and stained with coomassie. Lanes (∼1 cm) for each sample were excised and the proteins were reduced, alkylated, and in-gel digested with trypsin overnight at 37°C. Peptides were extracted, desalted with C18 spin columns (Pierce) and dried via vacuum centrifugation. Peptide samples were stored at −80°C until further analysis.
Immunoprecipitated samples were analyzed by LC-MS/MS using an Easy nLC 1200 coupled to a QExactive HF mass spectrometer (Thermo). Samples were injected onto an Easy Spray PepMap C18 column (75 μm id × 25 cm, 2 μm particle size; Thermo) and separated over a 90 min time period. The gradient for separation consisted of 5%–45% mobile phase B at a 250 nL/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in 80% ACN. The QExactive HF was operated in data-dependent mode where the 15 most intense precursors were selected for subsequent fragmentation. Resolution for the precursor scan (m/z 350–1700) was set to 60,000, while MS/MS scans resolution was set to 15,000. The normalized collision energy was set to 27% for HCD. Peptide match was set to preferred, and precursors with unknown charge or a charge state of 1 and ≥7 were excluded.
Raw data files were processed using Proteome Discoverer version 2.4 (Thermo Scientific). Peak lists were searched against a reviewed Uniprot Drosophila database (downloaded February 2020, containing 21,973 sequences), appended with a common contaminants database, using Sequest HT within Proteome Discoverer. The following parameters were used to identify tryptic peptides for protein identification: 10 ppm precursor ion mass tolerance; 0.02 Da product ion mass tolerance; up to two missed trypsin cleavage sites; carbamidomethylation of Cys was set as a fixed modification; oxidation of Met and acetylation of N-terminus were set as variable modifications. Scaffold (version 4.7.3, Proteome Software) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95% probability to achieve an FDR less than 0.1% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Relative quantitation was performed using the calculated quantitative values (spectral counts) within Scaffold. The data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez-Riverol et al., 2022) and can be accessed using the dataset identifier PXD053506.
Once identified by mass spectrometry, proteins were assigned to specific UniProt IDs. Official gene names, FBgn IDs, annotations, and a variety of other pertinent information used in this study were obtained from FlyBase (Gramates et al., 2022). Gene ontology (GO) analyses were performed with FBgn IDs using g:Profiler (Reimand et al., 2007; Consortium et al., 2023)
Until recently, SMA was untreatable and recognized as the most prevalent genetic cause of early childhood mortality (Pearn, 1980; Talbot and Tizzano, 2017; Wirth et al., 2020). Humans have two paralogous SMN genes, named SMN1 and SMN2. SMA is typically caused by deletion of both copies of SMN1; left on its own, SMN2 does not provide enough full-length SMN protein to fully compensate for loss of SMN1 (Monani et al., 2000; Hsieh-Li et al., 2000). Notably, a small fraction (<5%) of SMA patients present with a deletion in one copy of SMN1 and a missense mutation in the other copy (Wirth et al., 2020; Wirth, 2000). Patients bearing only one copy of SMN2 and a homozygous missense mutation in SMN1 are even more exceptional (Li et al., 2023). Thus, disease presentation in humans varies dramatically, depending on the SMN1 allele, and the number of SMN2 gene copies present in the background (which can vary from 0 to 6).
Our use of Drosophila melanogaster as a model system to study SMN function solves a number of the problems that confound phenotypic interpretation in humans and mammals. First, human SMN2 copy number variation can mask the phenotype of SMN1 point mutations (Gupta et al., 2021; Calucho et al., 2018; Yamamoto et al., 2014). Second, alternative splicing of SMN2 creates a feedback loop (Jodelka et al., 2010; Ruggiu et al., 2012) that can negatively regulate SMN expression. Third, purification of native or epitope-tagged SMN from cell lines or tissues may limit the number of binding partners to those that are expressed in a given cell lineage. In flies, there is only one Smn gene and its protein coding region is located within a single, constitutively expressed exon. Thus, one can eliminate alternative splicing as a variable when the goal is to understand SMN protein function.
We have developed a set of fly stocks whose survival depends entirely on transgenic expression of Flag-SMN protein (Gray et al., 2018; Spring et al., 2019). That is, these animals do not express any endogenous SMN (maternally or zygotically). The stocks have the overall genotype: SmnX7/X7,Flag-SmnTg/Tg, where SmnX7 is a null allele and Tg denotes a: WT, D20V, G73R, I93F or G210C transgene (Figure 1A). Thus, these models arguably provide a more accurate readout of SMN protein function because the mutants can be expressed and analyzed in the absence of wild-type SMN. Furthermore, the stocks are ideal for carrying out proteomics because we do not have to perform crosses to obtain animals of the desired genotype and we are able to collect large quantities of material. We chose to use embryos because they contain a wide-variety of cell types and they naturally express ∼100x the amount of SMN protein present in larval or adult stages (Raimer et al., 2020).
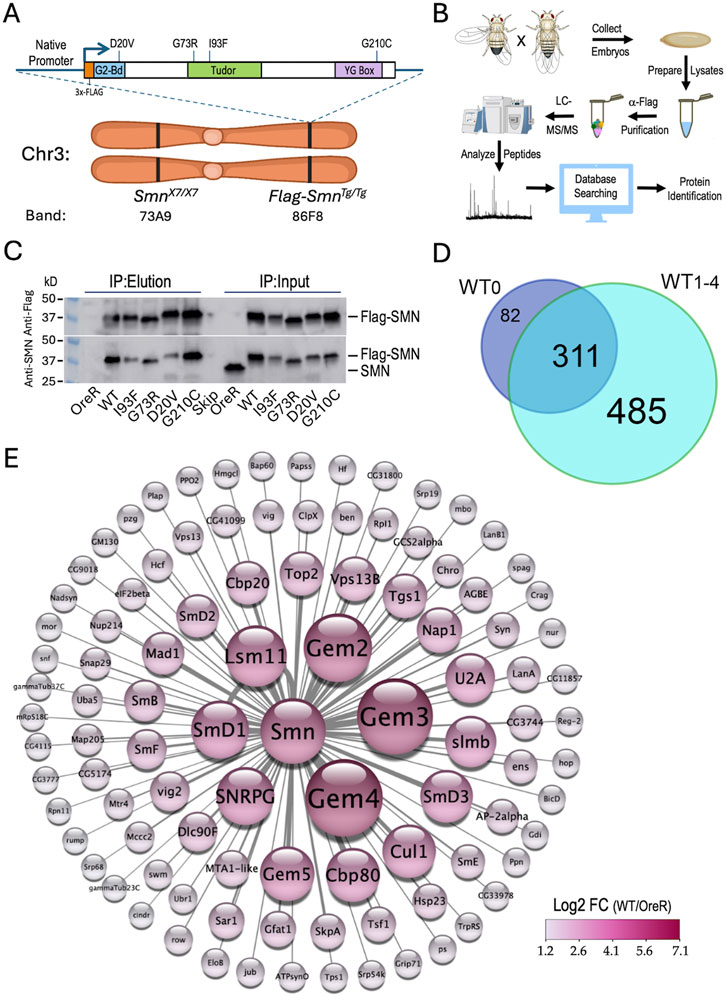
Figure 1. Experimental setup, workflow and overview of results. (A) Ideogram of fruitfly chromosome 3 (Chr3) showing cytogenetic band locations of the endogenous Smn gene at 73A9 and the Flag-Smn rescue transgene (Tg) at a PhiC31 landing site located at 86F8. Cartoon above shows the features of the rescue transgene driven by the native Smn promoter and control region. The SMN coding region was tagged with a 3x-FLAG epitope at the N-terminus. In addition to the WT Smn line, stocks expressing four different missense alleles (D20V, G73R, I93F and G210C) were also generated. (B) Diagram of experimental workflow, beginning with embryo collection and ending with identification of peptides and protein predictions. Panels created using Biorender.com. (C) Western blot of immunoprecipitation (IP) experiment for Oregon-R (OreR) control or the various stable transgenic stocks described above. The upper blot was probed with anti-Flag and the lower blot was probed with anti-SMN, verifying the presence of the untagged endogenous protein in the OreR input lane, but not in any of the other lanes. (D) Venn diagram of the total number of proteins identified in Flag-IPs from SmnWT transgenic animals, comparing the four biological replicates (WT1-4) generated in this study with a fifth one from a previous dataset, WT0 (Gray et al., 2018). (E) Graphical heat representation of the top 100 protein hits as determined by Log2 fold-change (LFC) from the WT samples vs. the OreR controls. Heatscale is at bottom right. Diameters of the circles are proportional to calculated LFC values for each protein. Panel was created using Cytoscape.com.
As outlined in Figure 1B, we used population cages of these “proteomic stocks” to set up embryo collections (0–24 h) from which we prepared lysates. The embryonic lysates were then subjected to anti-Flag purification followed by LC-MS/MS, peptide mapping and protein identification (see Methods for details). Western blot analysis of the five Flag-Smn lines, along with a wildtype negative control (OreR) is shown in Figure 1C, demonstrating the presence of endogenous (untagged) SMN only in the OreR input lane, and its absence from the immunoprecipitated material in all of the lanes.
Altogether, we performed three different mass spectrometry runs that included a total of 23 samples and identified a total of 893 different Drosophila proteins (see Supplementary Table S1; data are also available via ProteomeXchange with identifier PXD053506). Among the four WT biological replicates (WT1-4), there were 796 proteins that copurified with Flag-SMN (Figure 1D), 79% of which (311/393) overlapped with those we identified previously (Gray et al., 2018) using a single replicate (WT0; Supplementary Table S2). A visual summary of the top 100 hits, as ranked by Log2 fold-change differences between WT and OreR controls, is shown in Figure 1E. Clearly identified among the highest ranked partner proteins are the core members of the SMN complex (SMN, Gem2, Gem3, Gem4/Glos and Gem5/rig) and its best known RNP assembly clients, the Sm proteins (SmB, SmD1, SmD2, SmD3, SmE, SmF, SNRPG/SmG and Lsm11). Other RNP biogenesis factors include Cbp80·Cbp20, Snf, U2A and Tgs1. Also prominent atop this list are components of the SCFslmb E3 ligase (Slmb, Cul1 and SkpA) and the E2 ubiquitin conjugase, ben (Figure 1E) both of which have been previously reported to interact physically and genetically with SMN (Gray et al., 2018; Garcia et al., 2024). These data show that many orthologs of the well known mammalian SMN binding partners co-purified with wildtype Drosophila SMN.
To assess the reproducibility of these interactions and to help establish appropriate cutoffs for downstream comparisons, we performed a Student’s T-test (two tailed, homoscedastic) between the WT and OreR replicates (four each) and generated a volcano plot versus the fold-change data (Log2 transformed), see Supplementary Table S1. As shown in Figure 2A, many of the aforementioned SMN partner proteins are well above the traditional threshold of p < 0.05 (-Log10 > 1.25). Conspicuously below that line are other known SMN binding partners like SkpA, ben, Tgs1, Cbp20, SmD3 and Gem2, most of which are relatively small proteins (15–25 kDa). Gem2 and SMN form the heterodimeric core of the SMN complex (Gupta et al., 2021; Gupta et al., 2015); whereas Gem4 and Gem3 are not part of the core, but are much larger in size (100–120 kDa). Small and/or hydrophobic proteins have fewer peptides that can be detected by MS and thus are at a disadvantage compared to larger ones.
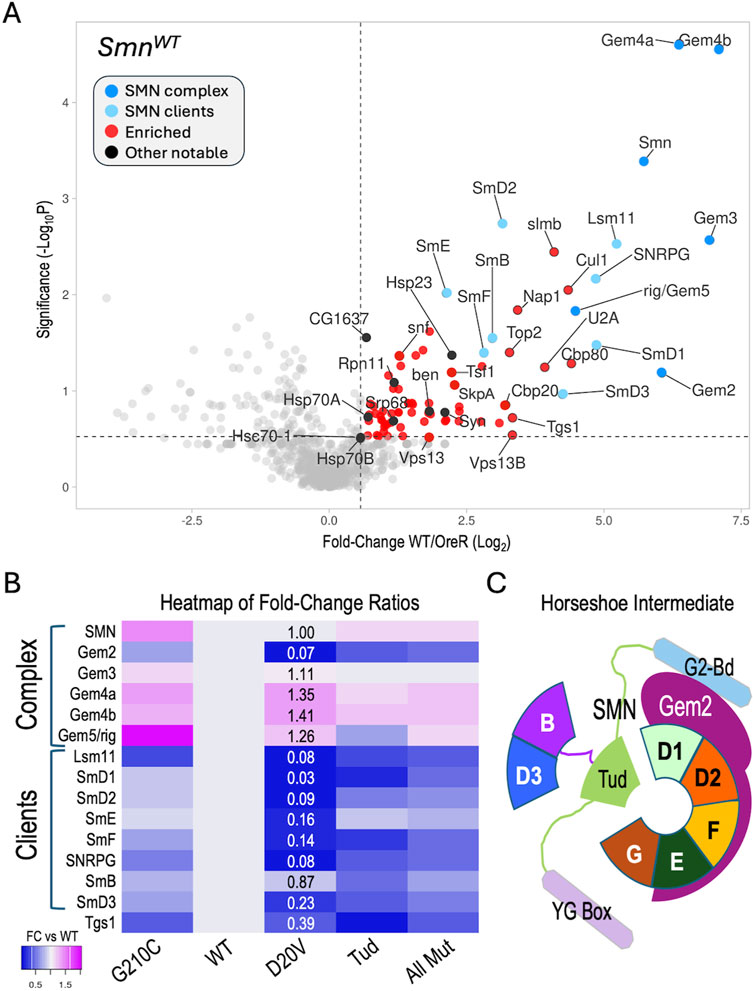
Figure 2. Analysis of AP-MS data. (A) Volcano plot of the full SmnWT dataset. Dots represent individual proteins. The SMN complex is shown in dark blue, known SMN clients (Sm proteins) are shaded light blue. Other notable proteins are shaded black. Vertical dotted line represents a fold-change cutoff of 1.5x (LFC ≥0.58) for enriched proteins (shaded in red). Horizontal dotted line is shown for display purposes, see text for details regarding significance cutoffs. (B) Heatmap of fold-change ratios for well-known SMN binding partners, comparing the data from the WT pulldowns to those of the G210C, D20V and Tud mutants. Tud = combined results for G73R and I93F. AllMut = combined results for all of the mutants. (C) Cartoon of known intermediate in spliceosomal snRNP assembly pathway, showing the seven canonical Sm proteins (B, D3, D1, D2, F, E and G), Gemin2 (Gem2), and SMN (with its three differentially shaded domains corresponding to those in Figure 1A). See text for details.
To determine if our MS data could be used to identify changes in the association of known binding partners among the SMA-causing missense alleles, we next focused on the members of the SMN complex and their well-known clients, the Sm proteins. For example, the human G279C allele (G210C in fly) is reported to be slightly hyper-oligomeric (Gupta et al., 2021), whereas D44V (D20V in fly) is reported to reduce binding affinity to Gem2 (Zhang et al., 2011). The two Tudor domain (Tud) mutations (G73R and I93F in the fly; G95R and I116F in human) are known to cause temperature-sensitive misfolding (Raimer et al., 2020; Tripsianes et al., 2011) and are plausibly expected to interfere with binding to Sm client proteins. Therefore, we generated a heatmap of fold-change ratios for various SMN alleles versus that of the WT (Figure 2B). As shown, the G210C mutant pulled down slightly higher levels of SMN and Gemins 3-5, consistent with its reported hyper-oligomeric nature. Concordantly, the D20V mutant co-purified considerably less Gem2 than did the WT construct, despite the fact that there were nearly identical levels of SMN detected in the two pulldowns (Figure 2B).
During U-snRNP assembly, Gem2 is known to directly bind to five of the seven Sm proteins, forming a key horseshoe-shaped intermediate (Figure 2C). Interestingly, those same five proteins (SmD1, D2, E, F, G) were the most reduced in the D20V mutant pulldowns (Figure 2B), whereas SmB and D3 were the two least affected clients. As predicted, the G73R and I93F mutants (Tud) also pulled down fewer Sm client proteins, but the contrast among the five Gem2-binders was less apparent (Figure 2B). Moreover, association of the Sm clients was relatively unaffected in the G210C mutants (Figure 2B). Note that Lsm11 (which is also reduced in D20V) dimerizes with Lsm10 to replace the SmD1·D2 dimer in the context of the U7 snRNP (Matera et al., 2007). Fold-change ratios for the combined set of mutants (AllMut) vs. WT are provided for general comparison, as are data for the snRNP biogenesis factor, Tgs1. In summary, these data show that the binding profiles of the fruitfly SMN missense mutants are consistent with the previously reported activities of their human counterparts (Gupta et al., 2021; Zhang et al., 2011; Raimer et al., 2020; Tripsianes et al., 2011).
The overall profiles of polypeptides identified by AP-MS in the wildtype and mutant Flag-SMN pulldowns were relatively similar (Supplementary Figure S1A), particularly among the top hits. Given the comparatively mild phenotypes of these SMA models, this was perhaps unsurprising. Among the co-purifying proteins with fold-change values ≥1.5, the WT, G210C and D20V constructs pulled down 210, 239 and 223 partners, respectively (Supplementary Figure S1A). By contrast, the Tud mutants (G73R and I93F) copurified only 144 such proteins. This general trend of reduced binding to targets shared between the WT and Tud samples is illustrated in Supplementary Figure S1B, by a difference plot. A similar comparison of shared hits between WT and G210C, for example, showed a more or less equal distribution of increases and decreases (Supplementary Figure S1B). To gain insight into biological processes that might be most affected by these mutations, we carried out gene ontology/functional enrichment analysis using gProfiler (Kolberg et al., 2023) with the top 100 hits (measured by fold-change) from each of the SMNWT and SMNTud experiments as queries. As shown in Figure 3A, spliceosomal RNP assembly and mRNA processing categories (along with many overlapping ones not illustrated) were clearly the top hits in both samples.
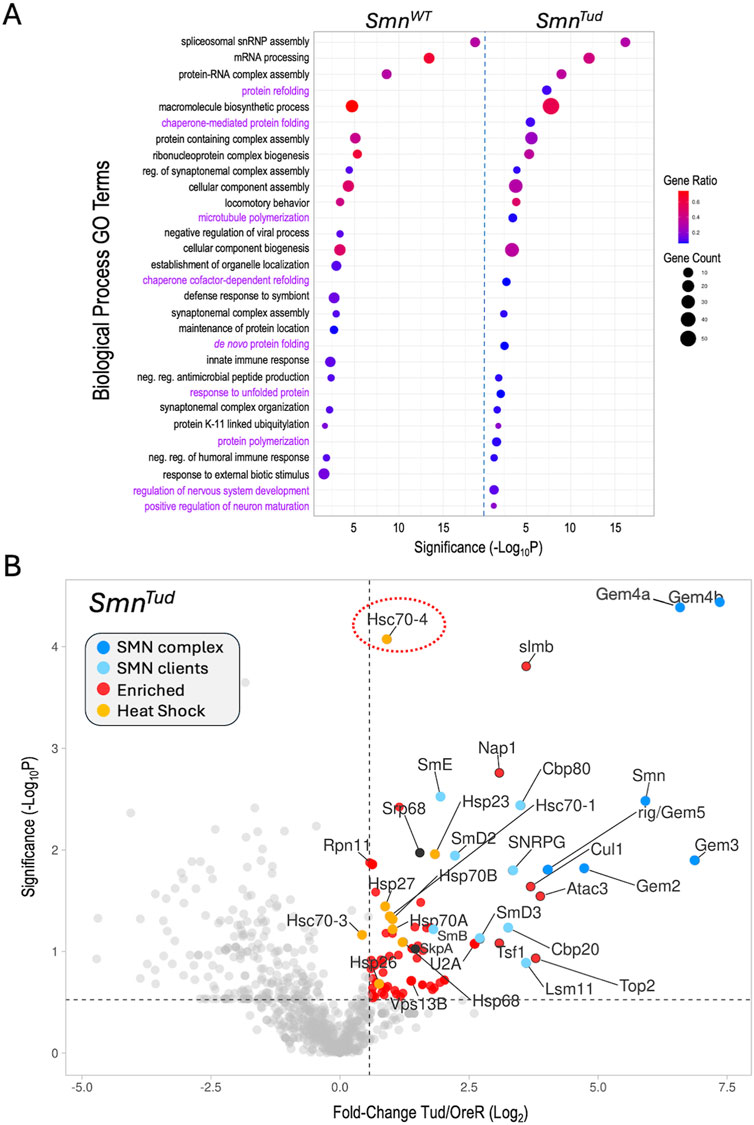
Figure 3. Functional enrichment analysis. (A) Comparative gene ontology (GO) terms associated with the top 100 proteins identified in the SmnWT and SmnTud AP-MS datasets. For each GO term listed, the size of the dot is proportional to the number of genes contained within that term (gene count), and the fraction of those genes scoring significantly (gene ratio) is represented using a heatmap (legend at right). Adjusted p-values (−log10 transformed) for each GO term were calculated and plotted separately for the WT and Tud results. (B) A volcano plot of the full SmnTud dataset. Dots represent individual proteins, shaded as per Figure 2A and shown in the color key (inset). Heat shock proteins are highlighted in orange. Hsc70-4 is circled. See text for details.
Other shared categories include the regulation of synaptonemal complex formation, cellular component assembly and biogenesis, antimicrobial peptide synthesis, locomotory behavior, and the humoral immune response. Given that we and others have identified connections between SMN and many of these pathways including innate immune signaling (Raimer et al., 2017; Gray et al., 2018; Garcia et al., 2024; Maccallini et al., 2020; Deguise et al., 2017), these categories were not unexpected. Prominent among the significant GO terms not shared by the two lists were those centered on aspects of protein folding and re-folding (Figure 3A). Because the GO terms were chosen on the basis of fold-change data alone, we next wanted to assess the significance of the full set of proteins identified in the SMNTud pulldowns. We therefore generated a volcano plot of the Tud mutants compared to OreR controls. Many of the SMNWT binding partners, including members of the SMN complex, the Sm clients, the Cbp80·20 cap-binding complex, the histone chaperone Nap1, and the SCFslmb E3 ligase complex (Cul1, SkpA, slmb) were significantly enriched in the SMNTud pulldowns as well (Figure 3B). The small HspB family member, Hsp23, was also significantly enriched in both the WT and Tud samples (Figures 2A, 3B).
Conspicuous among the factors identified in the Tud pulldowns (Figure 3B), are members of the large HspA family (Kampinga et al., 2009) that includes Hsp70 and Hsc70-type proteins. Although many members of this family were also identified in the WT samples, they were not as highly enriched. Most prominent among the HspA family members that were significantly enriched in the SMNTud pulldown is Hsc70-4 (Figure 3B, circled). This protein is particularly noteworthy because it has been recently linked to neuromuscular disease phenotypes in mouse and fruitfly models of both SMA and ALS, as discussed below (Coyne et al., 2017; Kim et al., 2023). Furthermore, Hsc70 (subtype not specified) was originally shown to co-purify with SMN in HeLa cells (Meister et al., 2001), though its importance to SMA was unrecognized at the time (discussed below). Because heat shock proteins constitute a major class of molecular (folding) chaperones, they interact with a large fraction of the proteome (Hartl et al., 2011). Thus HSPs are often identified as “contaminants” in many AP-MS experiments. Nevertheless, the data in Figure 3 identify clear signatures of a proteotoxic stress response (e.g., to unfolded proteins).
In human cells, the SMN complex is known to localize to stress granules (SGs), non-membrane bound cytoplasmic structures that form in response to a variety of cellular stressors (Alberti et al., 2017; Wolozin and Ivanov, 2019). Low levels of SMN are thought to impair the cell’s ability to form SGs (Zou et al., 2011). Previous studies have used proximity labeling approaches (e.g., BioID, APEX) to analyze the composition of RNP granules, finding that many of the interactions that take place during SG assembly are pre-existing in unstressed cells (Youn et al., 2018; Padron et al., 2019; Markmiller et al., 2021). One such study generated more than a hundred different stable cell lines expressing biotin ligase (BirA) tagged versions of RNP granule associated proteins, including two members of the human SMN complex, GEMIN3/DDX20 and STRAP/UNRIP (Youn et al., 2018). As expected, these two proteins were among the top five BirA-tagged “baits” to identify SMN as one of its biotinylated “prey” (Figure 4, table inset).
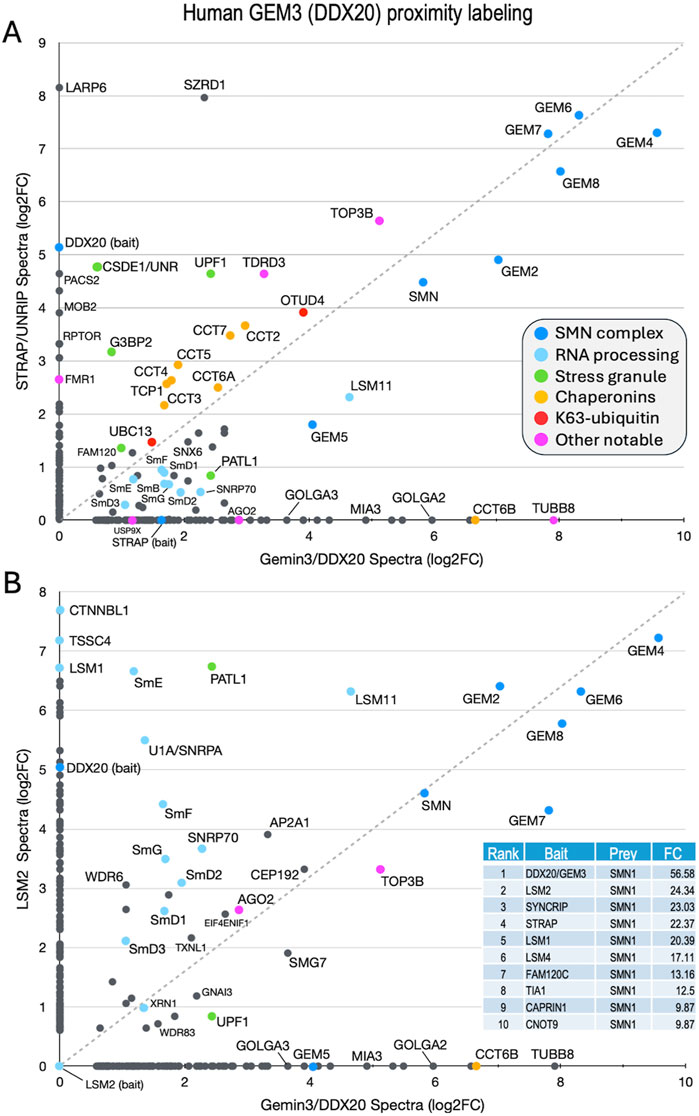
Figure 4. Comparison of previously published (Youn et al., 2018) BioID proximity labeling data for human stress granule proteins. Fold-change scatter plots of GEMIN3/DDX20 versus (A) STRAP/UNRIP and (B) LSM2, are shown. Color key is inset in panel (A). The inset in panel B provides a table listing the top ten BirA-tagged baits that included SMN as a prey. Fold-change (FC) values shown for comparison. See text.
Both STRAP (serine/threonine kinase receptor associated protein) and GEMIN3 (GEM3) are known to form complexes outside of their interaction with SMN (Reiner and Datta, 2011; Kamenska et al., 2016; Grundhoff et al., 1999; Gillian and Svaren, 2004; Lee et al., 2005). Originally identified as a component of the TGF-beta signaling machinery, STRAP is also known to interact with UNR/CSDE1 and LARP6 (Kamenska et al., 2016), serving as a translational regulatory factor (Vukmirovic et al., 2013). STRAP is a peripheral member of the SMN complex, tethered via its direct interaction with GEMIN7 as part of the GEMIN6·7·8 subunit (Otter et al., 2007). GEM3/DDX20 is a putative RNA helicase that heterodimerizes with GEMIN4 (GEM4) and has many reported activities (Gillian and Svaren, 2004; Lee et al., 2005; He et al., 2023; Miralles et al., 2022; Panek et al., 2023). To identify additional factors and complexes that associate with the human SMN complex and compare them to those identified in the fruitfly, we utilized the mass spectrometry data for proximity labeling of STRAP and GEM3 in unstressed cells (Youn et al., 2018) to generate a scatter plot (Figure 4A). We reasoned that proteins forming associations with the SMN complex would tend to plot along the diagonal, whereas those that are uniquely associated with either GEM3 or STRAP would localize along the X or Y axes, respectively. Consistent with that interpretation, and with what is known about the interactions among and between members of the SMN complex (Otter et al., 2007), GEM4 and the GEM6·7·8 subunit are very strongly labeled, localizing to the upper right hand corner of the plot (Figure 4A).
Other members of the complex, including GEM2, SMN and GEM5 align along the diagonal with the Sm client proteins. Of particular note, LARP6 and CSDE1 were well labeled only by STRAP, whereas the Golgin proteins GOLGA2 and GOLGA3 and the microtubule protein TUBB8 were detected only in the GEM3/DDX20 experiment (Figure 4A). Proteins that are known to be important for stress granule formation (PATL1, FAM120, UPF1 and G3BP2) were identified by both baits but are not well aligned along the diagonal. In contrast, TDRD3 and TOP3B form a complex with FMRP that is implicated in synapse formation and neurological development (Xu et al., 2013; Stoll et al., 2013) and are labeled by both GEM3 and STRAP. Two factors involved in aspects of K63-linked ubiquitylation, UBC13 (bendless in flies) and OTUD4, and all eight members of the chaperonin ring complex (TCP1/CCT1 thru CCT8) were also labeled by both baits (Figure 4A).
As a control, we plotted the proximity labeling data for one of the other top bait proteins for which SMN was identified as a prey, LSM2 (Figure 4B, inset). LSM2 is an Sm-like protein with manifold connections to the mRNA surveillance machinery and is also part of the U6 snRNP particle, though it is not a known SMN client. As shown in Figure 4B, factors involved in RNA processing and quality control (e.g., XRN1, UPF1, LSM1, TSSC4 and CTNNBL1) are well labeled by LSM2 but neither UBC13 nor the chaperonins were identified. These latter two categories are important because they point to a role for the SMN complex specifically in innate immune signaling and chaperone-mediated protein folding, both of which processes are deeply integrated with the proteostasis network (discussed below).
The fruitfly ortholog of STRAP is called wing morphogenesis defect (Wmd) and its sequence is well conserved in metazoa. Whether or not it forms part of the SMN complex in the fly is an open question. The Gem6·7·8 subunit is entirely missing from Drosophilid genomes, although these proteins are conserved in other insects (Matera et al., 2019). Consistent with the absence of its Gem7 tether (Otter et al., 2007) in the fly, neither Wmd nor any putative Gem6-8 paralogs were detected in our Flag-SMN pulldowns (Supplementary Table S1). We therefore conclude that Wmd is not part of (i.e., does not stably associate with) the Drosophila SMN complex (Figure 5).
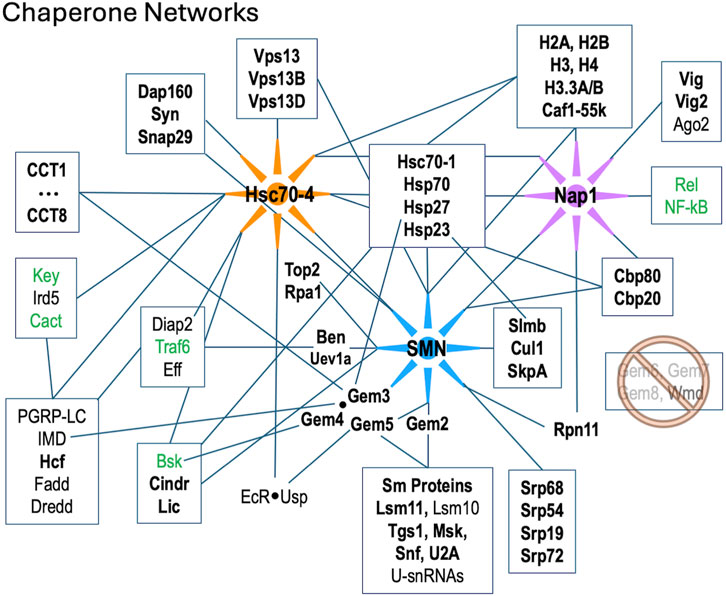
Figure 5. Diagram of protein-protein interactions between three key molecular chaperone systems: SMN (RNP assembly), Hsc70-4 (protein folding) and Nap1 (nucleosome assembly). Solid lines indicated known interactions. Factors listed in bold text are were identified in this work; those in green are innate immune signaling factors that were shown to interact genetically with SMN (Garcia et al., 2024).
In addition to the many novel findings reported above, the AP-MS data confirm and extend our knowledge of cellular components and pathways that are connected to the SMN complex in diverse organisms. Beyond the obvious core members of the SMN complex and its Sm protein clients, these additional partners include, but are not limited to: the nuclear cap biding complex, the signal recognition particle, autophagic lumenal markers of the endoplasmic reticulum and Golgi, various kinds of ATPases, innate immune signaling factors such as kinases and ubiquitylases, presynaptic cystosolic components, spindle proteins, synaptonemal complex factors, and chromatin remodelers (Table 1; Figure 5).
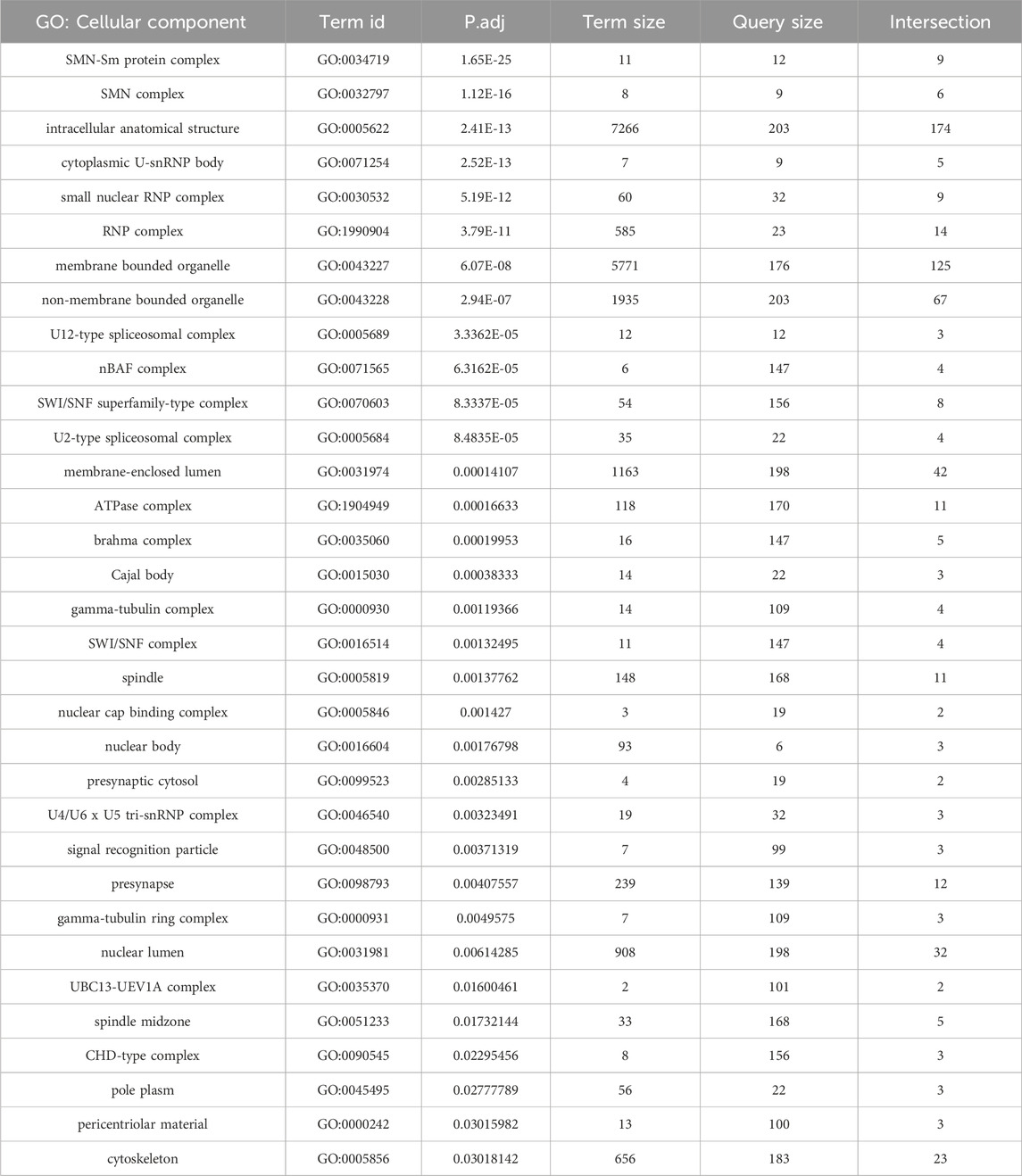
Table 1. Functional enrichment analysis of AP-MS data from Figure 2A. GO terms focusing on cellular components are shown.
A few of these conserved connections deserve particular note. Perhaps unsurprisingly we identified numerous proteins with strong connections to RNP biogenesis and transport. U-snRNP components like U2A, Snf (U2B“and U1A”) and Lsm11·10 (U7 snRNP), the nuclear import factor Msk (Importin-7), the m7G-cap binding complex (Cbp80·20) and the trimethylguanosine synthase protein, Tgs1 (Table 1; Figure 5). Tgs1 was recently shown to play a role in snRNA 3′-end processing (Chen et al., 2022) with loss-of-function impacts in eye development but no obvious neuromuscular defects (Maccallini et al., 2020).
SMN has also been implicated in the biogenesis and/or regulation of other RNPs besides the U-snRNPs. For example, assembly of the signal recognition particle takes place in the nucleolus and cytoplasm, and this process is thought to involve the activity of the SMN complex (Massenet, 2019). In total, our AP-MS data identified Srp-19, −54, −68, and −72, suggesting that this reported function of the SMN complex is conserved in metazoa. Other connections to the nucleolus are evident; SMN and other Cajal body components are known to relocalize to the nucleolus during genotoxic stress (Tapia et al., 2012; Musawi et al., 2023). In support of these findings, we identified the large subunit of RNA pol I (Rpa1), topoisomerase 2 (Top2) and fibrillarin (Fib) in our dataset (Figure 5; Supplementary Table S1). SMN, via its interaction with symmetric dimethylarginine (sDMA) residues on the pol II large subunit, is reportedly involved in R-loop resolution in the nucleoplasm (Yanling Zhao et al., 2016; Cuartas and Gangwani, 2022). We hypothesize that this same type of activity could be important for RNA pol I transcripts in the nucleolus upon cell stress.
We recently showed that hypomorphic mutation or depletion of SMN induces a systemic hyperactivation of the Toll and IMD signaling pathways leading to formation of larval melanotic nodules (Garcia et al., 2024). Importantly, depletion of U-snRNPs due to mutation of Phax (an snRNA export factor) does not elicit such an immune response (Garcia et al., 2016). Thus, immune dysfunction in these SMA models is a direct consequence of reduced SMN levels and not a downstream or indirect consquence of snRNP loss (Garcia et al., 2024). As outlined in Figure 5, there are numerous physical associations ((Gray et al., 2018; Guruharsha et al., 2011; Oughtred et al., 2021); this work) and genetic interactions (Garcia et al., 2024) between members of the SMN complex and innate immune signaling factors like Ben, Traf6 and Imd. Not to be overlooked are conserved interactions between members of the SMN complex and the small (B-type) heat shock ‘holdases’ like Hsp23 and Hsp27 (Figure 5). In human cells, GEM3 directly interacts with HspB5/αB-Crystallin (den Engelsman et al., 2013) and the SMN complex is thought to participate in its nuclear import and localization to nucleoplasmic speckles (Van Den Ijssel et al., 2003). Overexpression of GEM4, the direct binding parter of GEM3, causes the SMN complex to localize to the nucleoplasm (Meier et al., 2018). Interestingly, missense mutations in the human orthologues of two closely related small heat shock proteins, HspB8 (Hsp22) and HspB1 (Hsp27), are known to form abnormally strong interactions with GEM3 that are associated with motor neuron diseases (Sun et al., 2010). Hence, our work points to the heterodimeric Gem3·Gem4 subunit of the SMN complex as a conduit between the RNP assembly chaperones, the folding chaperones/co-chaperones (heat shock proteins and chaperonins), and the innate immune system (Figure 5 and (Garcia et al., 2024)).
As mentioned above, Wmd/STRAP forms a connection to the TGF-beta signaling pathway in mammals, but this link appears to be severed in flies due to loss of the Gem7 bridge. Perhaps a new signaling link was established in flies via the activity of Gem5/rig. This protein was originally thought to serve as a nuclear hormone receptor/co-factor (Gates et al., 2004), as it interacts physically and functionally with the ecdysone receptor, EcR (Figure 5). Subsequently, rig was identified as the orthologue of human GEM5 (Matera et al., 2019; Kroiss et al., 2008). Together with its binding partner Usp, EcR forms functional complexes with Hsc70-4 and numerous other shared interactors (Oughtred et al., 2021; Ozturk-Colak et al., 2024). Ecdysone is the central driver of developmental decision making in arthropods (Ecdysozoa); indeed, pulses of similar steroid hormones are known to control the timing of developmental transitions in all types of animals (Yamanaka et al., 2013).
Viewed in that light, it is perhaps unsurprising that there would be a link between the snRNP assembly machinery and regulators of cellular proliferation (Bruns et al., 2009). In all animals, high levels of SMN protein are really only required when building an organism. In flies, SMN is maternally provided and its levels remain high throughout embryogenesis, dropping to basal levels during the three larval stages (Raimer et al., 2020). It rises again during pupariation (metamorphosis is tantamount to building a new organism), only to fall back again to basal levels upon eclosion as adults (Raimer et al., 2020; Casas-Vila et al., 2017). In mice, depletion of SMN protein later in development has relatively little consequence compared to early depletion (Kariya et al., 2014). Thus, high levels of SMN are required in rapidly proliferating cells. How do cells communicate these needs and coordinate them with other biosynthetic pathways across and between cellular compartments?
Given SMN’s primary cellular location in the cytoplasm, it was somewhat surprising to see so many highly significant GO terms (Table 1) for nucleosome remodeling complexes like SWI/SNF, CHD, Brahma and nBAF. As mentioned above, connections between SMN and Cajal bodies or nucleoli are to be expected, but direct links to bulk chromatin are hard to fathom. It is important to remember that, like any other protein, nucleosomal subunits are born in the cytoplasm. Similar to Sm protein subcomplexes, histones are assembled into heterodimers prior to their nuclear import and incorporation into chromatin. The key finding in our AP-MS datasets is nucleosome assembly protein 1, Nap1 (Figure 5). This novel, high-confidence SMN partner was reproducibly detected in both the wildtype and mutant Flag-pulldowns (Figures 1E, 2A, 3B; Supplementary Figure S1B). Nap1 was also highly enriched in our previous study (WT0, Figure 1D (Gray et al., 2018)) and its co-purification with Flag-SMN was confirmed by Western blotting (Supplementary Figure S2).
Nap1 has well-established functions in chromatin remodeling and gene expression regulation (Walfridsson et al., 2007; Okuwaki et al., 2010), but it is also thought to play a role in the innate immune response, particularly to viral infection (Tanaka et al., 2017; Kayesh et al., 2022). In humans, there are five Nap1-like genes (NAP1L1-L5), three of which are retrogenes that are expressed exclusively in the nervous system (Attia et al., 2011; Attia et al., 2013). NAP1L1 and NAP1L4 are the most ancestral family members, and depletion of NAP1L1 interferes with the nuclear translocation of RelA, the 65kD subunit of NF-kB, leading to a weakened TLR3 (Toll-like receptor 3) response (Çevik et al., 2017). Nap1L1 also interacts with A- and C-type (Hsc70 and Hsp90, respectively) heat shock proteins, which are known to interact with H3·H4 heterodimers in the cytoplasm (Seebart et al., 2010; Keck and Pemberton, 2012). As illustrated in Figure 5, Nap1 and SMN are central members of two different assembly chaperone systems that share conserved connections to the folding chaperones and the innate immune signaling system. From a conceptual standpoint, the findings reported here expand our understanding and appreciation for the extent to which macromolecular assembly chaperones intersect with innate immune signaling proteins that function within the larger context of the proteostasis and ribostasis networks.
As protectors of the proteome (and the RNPome), molecular chaperones of the HspA (70 kD) family are important players in intracellular signaling pathways because they regulate the folding and activity of signaling proteins (Mayer and Bukau, 2005). Importantly, expression of a misfolded protein is known to change the profile of HSP binding partners (Ryu et al., 2020). We recently identified a subset of SMA-causing missense mutations in the Tudor domain of SMN (G73R, I93F, V72G and F70S) that are temperature-sensitive (Raimer et al., 2020). In response to relatively small increases in culturing temperature (e.g., from 25°C to 27° or 29°C), these Tud mutants display reduced SMN protein levels and fairly dramatic changes in organismal viability (Raimer et al., 2020). As detailed in Figure 3, the SMNTud mutants exhibit striking changes in binding-partner profiles even at the ‘sub-clinical’ temperature of 25°C. Most notable among the many HSPs that co-purify with the Tud mutants is Hsc70-4. Reciprocally, a high throughput screen for Hsc70-4 (HspA8 in mammals) client substrates identified GEM3 and SMN as potential targets (Ryu et al., 2020).
Most important, a G470R missense mutation in Hsc70-4/HspA8 was recently identified as a potent suppressor of the SMA phenotype in a murine disease model (Kim et al., 2023). Monani and colleagues identified a spontaneous point mutation in the background of their mouse colony that suppresses SMA-like phenotypes by bypassing the need for high levels of full-length SMN (Kim et al., 2023). They mapped the mutation to the substrate recognition domain of Hspa8 and found that this allele binds less efficiently to SMN (Kim et al., 2023). Although the precise molecular mechanism behind the genetic suppression is unclear, it requires the presence of a transgene that expresses the truncated SMN∆7 isoform. The authors posit that the reduced affinity (partial loss-of-function) of HspA8G470R for SMN allows the cell to ‘repurpose’ the folding chaperone away from SMN and onto other clients (e.g., SNAREs) that are important for neurotransmission (Kim et al., 2023). Consistent with this view, malformed complexes containing mutant or suboptimal SMN isoforms (e.g., SMN∆7) could change the profile of Hsc70/HspA binding partners (Kim et al., 2023; Ryu et al., 2020; this work).
If changing the overall balance of well-vs. poorly-folded clients can redirect HSPs to different pools of substrates, then what happens to the clients upon changes in the pools of HSPs? Perhaps equally important in this regard are the findings of Zarnescu and colleagues (Coyne et al., 2017). These authors found that a dominant-negative (gain-of-function), ALS-causing allele of TDP-43 aberrantly sequesters Hsc70-4/Hspa8 mRNA away from translating ribosomes in mouse and Drosophila disease models (Coyne et al., 2017). Recent evidence also suggests that, in addition to proteins, HSPs may also assist in the folding of certain non-coding RNAs (Dos Santos et al., 2019; Lakhotia, 2012; Place and Noonan, 2014). Ancestrally, Sm and Sm-like proteins are known to function as RNA chaperones. Could SMN be involved in regulating the localization and/or translation of HspA8 mRNA? Interestingly, we previously showed that SmD3, an SMN client, is involved in the transport and localization of oskar mRNA in the fruitfly ovary (Gonsalvez et al., 2010). Moreover, we also identified TDP-43 mRNA as a target of Sm proteins in human cells via RIP-seq (Lu et al., 2014). Additional studies will be needed in this area to precisely determine the degree of etiological overlap between SMA and ALS.
However, given the well-known roles of HspA8 and its close relatives in synaptic vesicle recycling and micro-autophagy, the idea that chaperone-related dysfunction lies at the root of two of the most prominent motor neuron diseases is very appealing. Taken together with our findings, these two studies (Coyne et al., 2017; Kim et al., 2023) provide clear examples of how the lines between loss-of-function and gain-of-function muations can become blurry when viewed in the context of a large network. In 2006, Csermely and colleagues posited that chaperone-related immune dysfunction is an emergent property of a distorted proteostasis network (Nardai et al., 2006). They defined an emergent property as one that “can not be elucidated from the properties of any single network element” rather, it emerges as a consequence of interactions within the entire network (Nardai et al., 2006). Indeed, this notion that chaperone deficiencies or polymorphisms, might distort signaling networks in unpredictable ways to induce (or suppress) disease states has turned out to be rather prescient.
Despite the fact that the primary therapeutic agents used to treat SMA are splice altering drugs (Antonaci et al., 2023), defects in pre-mRNA splicing do not cause SMA. The drugs alter the normal splicing pattern of SMN2 to increase levels of full-length SMN. Previous work in our laboratory and others strongly suggests that the underlying cause of the disease is related to functions of SMN that are independent of its role in spliceosomal snRNP biogenesis. Animals bearing hypomorphic SMN point mutations that cause milder forms of SMA complete development and display normal snRNP levels, but still exhibit neuromuscular defects (Garcia et al., 2016; Spring et al., 2019). As outlined here, manifold connections among and between molecular chaperones like HspA/Hsc70, SMN, and Nap1 with innate immune signaling pathways are conserved between insects and mammals. We therefore hold that the neuromuscular dysfunction in SMA and ALS is a direct consequence of perturbations within the proteostasis network.
The datasets presented in this study have been deposited to the ProteomeXchange Consortium and can be accessed via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) using the identifier PXD053506.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements. The manuscript presents research on animals that do not require ethical approval for their study.
AM: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing–original draft, Writing–review and editing. RS: Formal Analysis, Investigation, Methodology, Validation, Writing–review and editing. CM: Data curation, Formal Analysis, Methodology, Software, Validation, Writing–review and editing. BM: Software, Visualization, Writing–review and editing. LH: Data curation, Methodology, Project administration, Resources, Supervision, Writing–review and editing. EG: Formal Analysis, Investigation, Methodology, Software, Visualization, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH/NIGMS grant R35-GM136435 (to AM). This research was also supported by work conducted by the UNC Proteomics Core Facility, which is funded in part by an NCI Center Core Support Grant (P30-CA016086) to the UNC Lineberger Comprehensive Cancer Center. The funders had no role in the design and conduct of the study.
We thank C. Hamilton for assistance with the embryo collections, V. Vandadi for help with fly husbandry, K. Gray for Western blot analysis, and N. Barker-Krantz for sample preparation and MS data collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frnar.2024.1448194/full#supplementary-material
Alberti, S., Mateju, D., Mediani, L., and Carra, S. (2017). Granulostasis: protein quality control of RNP granules. Front. Mol. Neurosci. 10, 84. doi:10.3389/fnmol.2017.00084
Antonaci, L., Pera, M. C., and Mercuri, E. (2023). New therapies for spinal muscular atrophy: where we stand and what is next. Eur. J. Pediatr. 182, 2935–2942. doi:10.1007/s00431-023-04883-8
Attia, M., Förster, A., Rachez, C., Freemont, P., Avner, P., and Rogner, U. C. (2011). Interaction between nucleosome assembly protein 1-like family members. J. Mol. Biol. 407, 647–660. doi:10.1016/j.jmb.2011.02.016
Attia, M., Rachez, C., Avner, P., and Rogner, U. C. (2013). Nucleosome assembly proteins and their interacting proteins in neuronal differentiation. Archives Biochem. Biophysics 534, 20–26. doi:10.1016/j.abb.2012.09.011
Battle, D. J., Kasim, M., Yong, J., Lotti, F., Lau, C. K., Mouaikel, J., et al. (2006). The SMN complex: an assembly machine for RNPs. Cold Spring Harb. Symposia Quantitative Biol. 71, 313–320. doi:10.1101/sqb.2006.71.001
Baßler, J., and Hurt, E. (2019). Eukaryotic ribosome assembly. Annu. Rev. Biochem. 88, 281–306. doi:10.1146/annurev-biochem-013118-110817
Brehme, M., Voisine, C., Rolland, T., Wachi, S., Soper, J. H., Zhu, Y., et al. (2014). A chaperone subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell. Rep. 9, 1135–1150. doi:10.1016/j.celrep.2014.09.042
Bruns, A. F., van Bergeijk, J., Lorbeer, C., Nolle, A., Jungnickel, J., Grothe, C., et al. (2009). Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 106, 12747–12752. doi:10.1073/pnas.0900122106
Calderwood, S. K., and Murshid, A. (2017). Molecular chaperone accumulation in cancer and decrease in alzheimer's disease: the potential roles of HSF1. Front. Neurosci. 11, 192. doi:10.3389/fnins.2017.00192
Calucho, M., Bernal, S., Alías, L., March, F., Venceslá, A., Rodríguez-Álvarez, F. J., et al. (2018). Correlation between SMA type and SMN2 copy number revisited: an analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord. 28, 208–215. doi:10.1016/j.nmd.2018.01.003
Casas-Vila, N., Bluhm, A., Sayols, S., Dinges, N., Dejung, M., Altenhein, T., et al. (2017). The developmental proteome of Drosophila melanogaster. Genome Res. 27, 1273–1285. doi:10.1101/gr.213694.116
Çevik, R. E., Cesarec, M., Da Silva Filipe, A., Licastro, D., McLauchlan, J., and Marcello, A. (2017). Hepatitis C virus NS5A targets nucleosome assembly protein NAP1L1 to control the innate cellular response. J. virology 91, e00880. doi:10.1128/jvi.00880-17
Chari, A., Golas, M. M., Klingenhager, M., Neuenkirchen, N., Sander, B., Englbrecht, C., et al. (2008). An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 135, 497–509. doi:10.1016/j.cell.2008.09.020
Chen, L., Roake, C. M., Maccallini, P., Bavasso, F., Dehghannasiri, R., Santonicola, P., et al. (2022). TGS1 impacts snRNA 3'-end processing, ameliorates survival motor neuron-dependent neurological phenotypes in vivo and prevents neurodegeneration. Nucleic Acids Res. 50, 12400–12424. doi:10.1093/nar/gkac659
Consortium, T. G. O., Aleksander, S. A., Balhoff, J., Carbon, S., Cherry, J. M., Drabkin, H. J., et al. (2023). The gene ontology knowledgeable in 2023. Genetics 224, iyad031. doi:10.1093/genetics/iyad031
Coyne, A. N., Lorenzini, I., Chou, C. C., Torvund, M., Rogers, R. S., Starr, A., et al. (2017). Post-transcriptional inhibition of hsc70-4/HSPA8 expression leads to synaptic vesicle cycling defects in multiple models of ALS. Cell. Rep. 21, 110–125. doi:10.1016/j.celrep.2017.09.028
Cuartas, J., and Gangwani, L. (2022). R-Loop mediated DNA damage and impaired DNA repair in spinal muscular atrophy. Front. Cell. Neurosci. 16, 826608. doi:10.3389/fncel.2022.826608
Deguise, M. O., De Repentigny, Y., McFall, E., Auclair, N., Sad, S., and Kothary, R. (2017). Immune dysregulation may contribute to disease pathogenesis in spinal muscular atrophy mice. Hum. Mol. Genet. 26, 801–819. doi:10.1093/hmg/ddw434
den Engelsman, J., van de Schootbrugge, C., Yong, J., Pruijn, G. J., and Boelens, W. C. (2013). Pseudophosphorylated αB-crystallin is a nuclear chaperone imported into the nucleus with help of the SMN complex. PloS one 8, e73489. doi:10.1371/journal.pone.0073489
Dos Santos, R. F., Arraiano, C. M., and Andrade, J. M. (2019). New molecular interactions broaden the functions of the RNA chaperone Hfq. Curr. Genet. 65, 1313–1319. doi:10.1007/s00294-019-00990-y
Garcia, E. L., Lu, Z., Meers, M. P., Praveen, K., and Matera, A. G. (2013). Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. RNA 19, 1510–1516. doi:10.1261/rna.038919.113
Garcia, E. L., Steiner, R. E., Raimer, A. C., Herring, L. E., Matera, A. G., and Spring, A. M. (2024). Dysregulation of innate immune signaling in animal models of Spinal Muscular Atrophy. BMC Biol. 22, 94. doi:10.1186/s12915-024-01888-z
Garcia, E. L., Wen, Y., Praveen, K., and Matera, A. G. (2016). Transcriptomic comparison of Drosophila snRNP biogenesis mutants reveals mutant-specific changes in pre-mRNA processing: implications for spinal muscular atrophy. RNA 22, 1215–1227. doi:10.1261/rna.057208.116
Gates, J., Lam, G., Ortiz, J. A., Losson, R., and Thummel, C. S. (2004). Rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development 131, 25–36. doi:10.1242/dev.00920
Gill, J., Kumar, A., and Sharma, A. (2022). Structural comparisons reveal diverse binding modes between nucleosome assembly proteins and histones. Epigenetics Chromatin 15, 20. doi:10.1186/s13072-022-00452-9
Gillian, A. L., and Svaren, J. (2004). The Ddx20/DP103 dead box protein represses transcriptional activation by Egr2/Krox-20. J. Biol. Chem. 279, 9056–9063. doi:10.1074/jbc.m309308200
Gonsalvez, G. B., Rajendra, T. K., Wen, Y., Praveen, K., and Matera, A. G. (2010). Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development 137, 2341–2351. doi:10.1242/dev.042721
Gramates, L. S., Agapite, J., Attrill, H., Calvi, B. R., Crosby, M. A., Dos Santos, G., et al. (2022). FlyBase: a guided tour of highlighted features. Genetics 220, iyac035. doi:10.1093/genetics/iyac035
Gray, K. M., Kaifer, K. A., Baillat, D., Wen, Y., Bonacci, T. R., Ebert, A. D., et al. (2018). Self-oligomerization regulates stability of survival motor neuron protein isoforms by sequestering an SCF(Slmb) degron. Mol. Biol. Cell. 29, 96–110. doi:10.1091/mbc.e17-11-0627
Grundhoff, A. T., Kremmer, E., Tureci, O., Glieden, A., Gindorf, C., Atz, J., et al. (1999). Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274, 19136–19144. doi:10.1074/jbc.274.27.19136
Gruss, O. J., Meduri, R., Schilling, M., and Fischer, U. (2017). UsnRNP biogenesis: mechanisms and regulation. Chromosoma 126, 577–593. doi:10.1007/s00412-017-0637-6
Gupta, K., Martin, R., Sharp, R., Sarachan, K. L., Ninan, N. S., and Van Duyne, G. D. (2015). Oligomeric properties of survival motor Neuron·Gemin2 complexes. J. Biol. Chem. 290, 20185–20199. doi:10.1074/jbc.m115.667279
Gupta, K., Wen, Y., Ninan, N. S., Raimer, A. C., Sharp, R., Spring, A. M., et al. (2021). Assembly of higher-order SMN oligomers is essential for metazoan viability and requires an exposed structural motif present in the YG zipper dimer. Nucleic Acids Res. 49, 7644–7664. doi:10.1093/nar/gkab508
Guruharsha, K. G., Rual, J. F., Zhai, B., Mintseris, J., Vaidya, P., Vaidya, N., et al. (2011). A protein complex network of Drosophila melanogaster. Cell. 147, 690–703. doi:10.1016/j.cell.2011.08.047
Hartl, F. U., Bracher, A., and Hayer-Hartl, M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. doi:10.1038/nature10317
Hartl, F. U., and Hayer-Hartl, M. (2009). Converging concepts of protein folding in vitro and in vivo. Nat. Struct. and Mol. Biol. 16, 574–581. doi:10.1038/nsmb.1591
He, L., Yang, J., Hao, Y., Yang, X., Shi, X., Zhang, D., et al. (2023). DDX20: a multifunctional complex protein. Molecules 28, 7198. doi:10.3390/molecules28207198
Hsieh-Li, H. M., Chang, J. G., Jong, Y. J., Wu, M. H., Wang, N. M., Tsai, C. H., et al. (2000). A mouse model for spinal muscular atrophy. Nat. Genet. 24, 66–70. doi:10.1038/71709
Jodelka, F. M., Ebert, A. D., Duelli, D. M., and Hastings, M. L. (2010). A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum. Mol. Genet. 19, 4906–4917. doi:10.1093/hmg/ddq425
Johal, K. S., Cheema, M. S., and Stefanelli, G. (2023). Histone variants and their chaperones: an emerging epigenetic mechanism in neurodevelopment and neurodevelopmental disorders. J. Integr. Neurosci. 22, 108. doi:10.31083/j.jin2205108
Kamenska, A., Simpson, C., Vindry, C., Broomhead, H., Bénard, M., Ernoult-Lange, M., et al. (2016). The DDX6–4E-T interaction mediates translational repression and P-body assembly. Nucleic acids Res. 44, 6318–6334. doi:10.1093/nar/gkw565
Kampinga, H. H., Hageman, J., Vos, M. J., Kubota, H., Tanguay, R. M., Bruford, E. A., et al. (2009). Guidelines for the nomenclature of the human heat shock proteins. Cell. Stress Chaperones 14, 105–111. doi:10.1007/s12192-008-0068-7
Kariya, S., Obis, T., Garone, C., Akay, T., Sera, F., Iwata, S., et al. (2014). Requirement of enhanced Survival Motoneuron protein imposed during neuromuscular junction maturation. J. Clin. Investig. 124, 785–800. doi:10.1172/jci72017
Kato, K., and Satoh, T. (2018). Structural insights on the dynamics of proteasome formation. Biophys. Rev. 10, 597–604. doi:10.1007/s12551-017-0381-4
Kayesh, M. E. H., Kohara, M., and Tsukiyama-Kohara, K. (2022). Toll-like receptor response to hepatitis C virus infection: a recent overview. Int. J. Mol. Sci. 23, 5475. doi:10.3390/ijms23105475
Keck, K. M., and Pemberton, L. F. (2012). Histone chaperones link histone nuclear import and chromatin assembly. Biochimica Biophysica Acta (BBA)-Gene Regul. Mech. 1819, 277–289. doi:10.1016/j.bbagrm.2011.09.007
Kim, J. K., Jha, N. N., Awano, T., Caine, C., Gollapalli, K., Welby, E., et al. (2023). A spinal muscular atrophy modifier implicates the SMN protein in SNARE complex assembly at neuromuscular synapses. Neuron 111, 1423–1439.e4. doi:10.1016/j.neuron.2023.02.004
Kolberg, L., Raudvere, U., Kuzmin, I., Adler, P., Vilo, J., and Peterson, H. (2023). g: profiler—interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic acids Res. 51, W207–W212. doi:10.1093/nar/gkad347
Kroiss, M., Schultz, J., Wiesner, J., Chari, A., Sickmann, A., and Fischer, U. (2008). Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 105, 10045–10050. doi:10.1073/pnas.0802287105
Kwon, Y. T., and Ciechanover, A. (2017). The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886. doi:10.1016/j.tibs.2017.09.002
Lakhotia, S. C. (2012). Long non-coding RNAs coordinate cellular responses to stress. WIREs RNA 3, 779–796. doi:10.1002/wrna.1135
Lee, M. B., Lebedeva, L. A., Suzawa, M., Wadekar, S. A., Desclozeaux, M., and Ingraham, H. A. (2005). The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 25, 1879–1890. doi:10.1128/mcb.25.5.1879-1890.2005
Li, L., Perera, L., Varghese, S. A., Shiloh-Malawsky, Y., Hunter, S. E., Sneddon, T. P., et al. (2023). A homozygous missense variant in the YG box domain in an individual with severe spinal muscular atrophy: a case report and variant characterization. Front. Cell. Neurosci. 17, 1259380. doi:10.3389/fncel.2023.1259380
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi:10.1146/annurev.ge.22.120188.003215
Lu, Z., Guan, X., Schmidt, C. A., and Matera, A. G. (2014). RIP-seq analysis of eukaryotic Sm proteins identifies three major categories of Sm-containing ribonucleoproteins. Genome Biol. 15, R7. doi:10.1186/gb-2014-15-1-r7
Maccallini, P., Bavasso, F., Scatolini, L., Bucciarelli, E., Noviello, G., Lisi, V., et al. (2020). Intimate functional interactions between TGS1 and the Smn complex revealed by an analysis of the Drosophila eye development. PLoS Genet. 16, e1008815. doi:10.1371/journal.pgen.1008815
Markmiller, S., Sathe, S., Server, K. L., Nguyen, T. B., Fulzele, A., Cody, N., et al. (2021). Persistent mRNA localization defects and cell death in ALS neurons caused by transient cellular stress. Cell. Rep. 36, 109685. doi:10.1016/j.celrep.2021.109685
Massenet, S. (2019). In vivo assembly of eukaryotic signal recognition particle: a still enigmatic process involving the SMN complex. Biochimie 164, 99–104. doi:10.1016/j.biochi.2019.04.007
Matera, A. G., Raimer, A. C., Schmidt, C. A., Kelly, J. A., Droby, G. N., Baillat, D., et al. (2019). Composition of the survival motor neuron (SMN) complex in Drosophila melanogaster. G3 Genes.|Genomes|Genetics 9, 491–503. doi:10.1534/g3.118.200874
Matera, A. G., and Shpargel, K. B. (2006). Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell. Biol. 18, 317–324. doi:10.1016/j.ceb.2006.03.005
Matera, A. G., Terns, R. M., and Terns, M. P. (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell. Biol. 8, 209–220. doi:10.1038/nrm2124
Matera, A. G., and Wang, Z. (2014). A day in the life of the spliceosome. Nat. Rev. Mol. Cell. Biol. 15, 108–121. doi:10.1038/nrm3742
Mayer, M. P., and Bukau, B. (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi:10.1007/s00018-004-4464-6
Meier, I. D., Walker, M. P., and Matera, A. G. (2018). Gemin4 is an essential gene in mice, and its overexpression in human cells causes relocalization of the SMN complex to the nucleoplasm. Biol. Open 7, bio032409. doi:10.1242/bio.032409
Meister, G., Buhler, D., Pillai, R., Lottspeich, F., and Fischer, U. (2001). A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell. Biol. 3, 945–949. doi:10.1038/ncb1101-945
Miles, J., Scherz-Shouval, R., and van Oosten-Hawle, P. (2019). Expanding the organismal proteostasis network: linking systemic stress signaling with the innate immune response. Trends Biochem. Sci. 44, 927–942. doi:10.1016/j.tibs.2019.06.009
Miralles, M. P., Sansa, A., Beltran, M., Soler, R. M., and Garcera, A. (2022). Survival motor neuron protein and neurite degeneration are regulated by Gemin3 in spinal muscular atrophy motoneurons. Front. Cell. Neurosci. 16, 1054270. doi:10.3389/fncel.2022.1054270
Monani, U. R., Sendtner, M., Coovert, D. D., Parsons, D. W., Andreassi, C., Le, T. T., et al. (2000). The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 9, 333–339. doi:10.1093/hmg/9.3.333
Musawi, S., Donnio, L. M., Zhao, Z., Magnani, C., Rassinoux, P., Binda, O., et al. (2023). Nucleolar reorganization after cellular stress is orchestrated by SMN shuttling between nuclear compartments. Nat. Commun. 14, 7384. doi:10.1038/s41467-023-42390-4
Nardai, G., Vegh, E. M., Prohaszka, Z., and Csermely, P. (2006). Chaperone-related immune dysfunction: an emergent property of distorted chaperone networks. Trends Immunol. 27, 74–79. doi:10.1016/j.it.2005.11.009
Okuwaki, M., Kato, K., and Nagata, K. (2010). Functional characterization of human nucleosome assembly protein 1-like proteins as histone chaperones. Genes. Cells 15, 13–27. doi:10.1111/j.1365-2443.2009.01361.x
Otter, S., Grimmler, M., Neuenkirchen, N., Chari, A., Sickmann, A., and Fischer, U. (2007). A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 282, 5825–5833. doi:10.1074/jbc.m608528200
Oughtred, R., Rust, J., Chang, C., Breitkreutz, B. J., Stark, C., Willems, A., et al. (2021). The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200. doi:10.1002/pro.3978
Ozturk-Colak, A., Marygold, S. J., Antonazzo, G., Attrill, H., Goutte-Gattat, D., Jenkins, V. K., et al. (2024). FlyBase: updates to the Drosophila genes and genomes database. Genetics 227, iyad211. doi:10.1093/genetics/iyad211
Padron, A., Iwasaki, S., and Ingolia, N. T. (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell. 75, 875–887.e5. doi:10.1016/j.molcel.2019.07.030
Panek, J., Roithova, A., Radivojevic, N., Sykora, M., Prusty, A. B., Huston, N., et al. (2023). The SMN complex drives structural changes in human snRNAs to enable snRNP assembly. Nat. Commun. 14, 6580. doi:10.1038/s41467-023-42324-0
Paushkin, S., Gubitz, A. K., Massenet, S., and Dreyfuss, G. (2002). The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell. Biol. 14, 305–312. doi:10.1016/s0955-0674(02)00332-0
Pearn, J. (1980). Classification of spinal muscular atrophies. Lancet 315, 919–922. doi:10.1016/s0140-6736(80)90847-8
Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic acids Res. 50, D543–D552. doi:10.1093/nar/gkab1038
Place, R. F., and Noonan, E. J. (2014). Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell. Stress Chaperones 19, 159–172. doi:10.1007/s12192-013-0456-5
Praveen, K., Wen, Y., Gray, K. M., Noto, J. J., Patlolla, A. R., Van Duyne, G. D., et al. (2014). SMA-causing missense mutations in survival motor neuron (Smn) display a wide range of phenotypes when modeled in Drosophila. PLoS Genet. 10, e1004489. doi:10.1371/journal.pgen.1004489
Praveen, K., Wen, Y., and Matera, A. G. (2012). A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell. Rep. 1, 624–631. doi:10.1016/j.celrep.2012.05.014
Raimer, A. C., Gray, K. M., and Matera, A. G. (2017). SMN - a chaperone for nuclear RNP social occasions? RNA Biol. 14, 701–711. doi:10.1080/15476286.2016.1236168
Raimer, A. C., Singh, S. S., Edula, M. R., Paris-Davila, T., Vandadi, V., Spring, A. M., et al. (2020). Temperature-sensitive spinal muscular atrophy-causing point mutations lead to SMN instability, locomotor defects and premature lethality in Drosophila. Dis. Model. Mech. 13, dmm043307. doi:10.1242/dmm.043307
Reimand, J., Kull, M., Peterson, H., Hansen, J., and Vilo, J. (2007). g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35, W193–W200. doi:10.1093/nar/gkm226
Reiner, J. E., and Datta, P. K. (2011). TGF-beta-dependent and -independent roles of STRAP in cancer. Front. Biosci. Landmark Ed. 16, 105–115. doi:10.2741/3678
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell. 40, 253–266. doi:10.1016/j.molcel.2010.10.006
Ruggiu, M., McGovern, V. L., Lotti, F., Saieva, L., Li, D. K., Kariya, S., et al. (2012). A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell. Biol. 32, 126–138. doi:10.1128/mcb.06077-11
Ryu, S. W., Stewart, R., Pectol, D. C., Ender, N. A., Wimalarathne, O., Lee, J. H., et al. (2020). Proteome-wide identification of HSP70/HSC70 chaperone clients in human cells. PLoS Biol. 18, e3000606. doi:10.1371/journal.pbio.3000606
Seebart, C., Prenni, J., Tomschik, M., and Zlatanova, J. (2010). New nuclear partners for nucleosome assembly protein 1: unexpected associations. Biochem. Cell. Biol. 88, 927–936. doi:10.1139/o10-115
Singh, R. N., Howell, M. D., Ottesen, E. W., and Singh, N. N. (2017). Diverse role of survival motor neuron protein. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1860, 299–315. doi:10.1016/j.bbagrm.2016.12.008
Spring, A. M., Raimer, A. C., Hamilton, C. D., Schillinger, M. J., and Matera, A. G. (2019). Comprehensive modeling of spinal muscular atrophy in Drosophila melanogaster. Front. Mol. Neurosci. 12, 113. doi:10.3389/fnmol.2019.00113
Stanek, D., and Neugebauer, K. M. (2006). The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 115, 343–354. doi:10.1007/s00412-006-0056-6
Stoll, G., Pietilainen, O. P. H., Linder, B., Suvisaari, J., Brosi, C., Hennah, W., et al. (2013). Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 16, 1228–1237. doi:10.1038/nn.3484
Sun, X., Fontaine, J. M., Hoppe, A. D., Carra, S., DeGuzman, C., Martin, J. L., et al. (2010). Abnormal interaction of motor neuropathy-associated mutant HspB8 (Hsp22) forms with the RNA helicase Ddx20 (gemin3). Cell. Stress Chaperones 15, 567–582. doi:10.1007/s12192-010-0169-y
Talbot, K., and Tizzano, E. F. (2017). The clinical landscape for SMA in a new therapeutic era. Gene Ther. 24, 529–533. doi:10.1038/gt.2017.52
Tanaka, T., Hozumi, Y., Iino, M., and Goto, K. (2017). NAP1L1 regulates NF-κB signaling pathway acting on anti-apoptotic Mcl-1 gene expression. Biochimica Biophysica Acta (BBA) - Mol. Cell. Res. 1864, 1759–1768. doi:10.1016/j.bbamcr.2017.06.021
Tapia, O., Bengoechea, R., Palanca, A., Arteaga, R., Val-Bernal, J. F., Tizzano, E. F., et al. (2012). Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell. Biol. 137, 657–667. doi:10.1007/s00418-012-0921-8
Tripsianes, K., Madl, T., Machyna, M., Fessas, D., Englbrecht, C., Fischer, U., et al. (2011). Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18, 1414–1420. doi:10.1038/nsmb.2185
Van Den Ijssel, P., Wheelock, R., Prescott, A., Russell, P., and Quinlan, R. A. (2003). Nuclear speckle localisation of the small heat shock protein alpha B-crystallin and its inhibition by the R120G cardiomyopathy-linked mutation. Exp. Cell. Res. 287, 249–261. doi:10.1016/s0014-4827(03)00092-2
Vukmirovic, M., Manojlovic, Z., and Stefanovic, B. (2013). Serine-threonine kinase receptor-associated protein (STRAP) regulates translation of type I collagen mRNAs. Mol. Cell. Biol. 33, 3893–3906. doi:10.1128/mcb.00195-13
Walfridsson, J., Khorosjutina, O., Matikainen, P., Gustafsson, C. M., and Ekwall, K. (2007). A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 26, 2868–2879. doi:10.1038/sj.emboj.7601728
Wirth, B. (2000). An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum. Mutat. 15, 228–237. doi:10.1002/(sici)1098-1004(200003)15:3<228::aid-humu3>3.0.co;2-9
Wirth, B., Karakaya, M., Kye, M. J., and Mendoza-Ferreira, N. (2020). Twenty-five years of spinal muscular atrophy research: from phenotype to genotype to therapy, and what comes next. Annu. Rev. Genomics Hum. Genet. 21, 231–261. doi:10.1146/annurev-genom-102319-103602
Wolozin, B., and Ivanov, P. (2019). Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20, 649–666. doi:10.1038/s41583-019-0222-5
Woodson, S. A. (2008). RNA folding and ribosome assembly. Curr. Opin. Chem. Biol. 12, 667–673. doi:10.1016/j.cbpa.2008.09.024
Xu, D., Shen, W., Guo, R., Xue, Y., Peng, W., Sima, J., et al. (2013). Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 16, 1238–1247. doi:10.1038/nn.3479
Yamamoto, T., Sato, H., Lai, P. S., Nurputra, D. K., Harahap, N. I., Morikawa, S., et al. (2014). Intragenic mutations in SMN1 may contribute more significantly to clinical severity than SMN2 copy numbers in some spinal muscular atrophy (SMA) patients. Brain Dev. 36, 914–920. doi:10.1016/j.braindev.2013.11.009
Yamanaka, N., Rewitz, K. F., and O'Connor, M. B. (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58, 497–516. doi:10.1146/annurev-ento-120811-153608
Yanling Zhao, D., Gish, G., Braunschweig, U., Li, Y., Ni, Z., Schmitges, F. W., et al. (2016). SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 529, 48–53. doi:10.1038/nature16469
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 69, 517–532.e11. doi:10.1016/j.molcel.2017.12.020
Zhang, R., So, B. R., Li, P., Yong, J., Glisovic, T., Wan, L., et al. (2011). Structure of a key intermediate of the SMN complex reveals Gemin2's crucial function in snRNP assembly. Cell. 146, 384–395. doi:10.1016/j.cell.2011.06.043
Keywords: spinal muscular atrophy (SMA), Amyotrophic lateral sclerosis (ALS), AP-MS, affinity purification coupled with mass spectrometry, ribonucleoprotein (RNP) biogenesis, proteostasis networks, chaperone mediated autophagy
Citation: Matera AG, Steiner RE, Mills CA, McMichael BD, Herring LE and Garcia EL (2024) Proteomic analysis of the SMN complex reveals conserved and etiologic connections to the proteostasis network. Front. RNA Res. 2:1448194. doi: 10.3389/frnar.2024.1448194
Received: 12 June 2024; Accepted: 19 August 2024;
Published: 18 September 2024.
Edited by:
Graydon Gonsalvez, Augusta University, United StatesReviewed by:
Daniela Zarnescu, The Pennsylvania State University, United StatesCopyright © 2024 Matera, Steiner, Mills, McMichael, Herring and Garcia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Gregory Matera, bWF0ZXJhQHVuYy5lZHU=
†Present address: Rebecca E. Steiner, Lake Erie College of Osteopathic Medicine, Bradenton FL, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.