- 1Department of Community Health, Faculty of Science and Medicine, University of Fribourg, Fribourg, Switzerland
- 2Department of Paediatrics, Fribourg Hospital, Fribourg, Switzerland
- 3Infectious Diseases Research Group, Murdoch Children’s Research Institute, Melbourne, VIC, Australia
- 4Department of Paediatrics, The University of Melbourne, Melbourne, VIC, Australia
Introduction: The nasopharynx harbours a diverse and dynamic microbial community, which plays an important role in maintaining the health and homeostasis of the respiratory tract, as well as in immune system development. Understanding factors that influence the composition of the nasopharyngeal microbiome in children and its association with diseases is of particular importance, as children are at a heightened risk for respiratory infections and other adverse health outcomes.
Objectives: This review systematically summarises studies which investigated the nasopharyngeal microbiome in children, including its dynamics, stability over time, and the influence of intrinsic and extrinsic factors on its composition.
Methods: MEDLINE was searched using the OVID interface. Original studies which investigated the nasopharyngeal microbiome using next generation sequencing in children were summarised.
Results: The search identified 736 studies, of which 77 were included. The studies show that the nasopharyngeal microbiome in children is dynamic and influenced by many external factors. A high abundance of Haemophilus, Moraxella, and Streptococcus and a low abundance of Corynebacterium and Dolosigranlum are associated with adverse health outcomes such as respiratory tract infections, wheezing and asthma exacerbations. Factors which have been identified as risk factors for these adverse health outcomes, such as being born by Caesarean section, not being breast-fed, having siblings, day-care attendance, and antibiotic exposure have been shown to be associated with the aforementioned features in the nasopharyngeal microbiome.
Conclusion: The association between specific nasopharyngeal microbial profiles and adverse health outcomes highlights the potential of the nasopharyngeal microbiome as a marker for identifying children at risk for disease and even more importantly, as an avenue for targeted interventions and preventive strategies.
Introduction
The nasopharynx is the gateway to the respiratory tract and a site of continuous contact with the environment. It harbours a diverse and dynamic microbial community, collectively known as the nasopharyngeal microbiome, which plays an important role in maintaining the health and homeostasis of the respiratory tract (Cleary and Clarke, 2017). The colonising bacteria not only influence the development of the immune system but are also important for metabolism (Enoksson et al., 2020; Chun et al., 2021; Hou et al., 2022). Commensals and pathogens of the microbiome have agonistic and antagonistic interactions and disturbance of homeostasis can lead to overgrowth, diminished resilience to pathogen invasion and infections. From the nasopharynx pathogens can spread to cause acute otitis media (AOM), pneumonia or invade the bloodstream to cause sepsis and meningitis. Some children are more susceptible to infections, especially respiratory tract infections.
Recent advances in next-generation sequencing technologies have revolutionised our ability to comprehensively characterise the nasopharyngeal microbiome and its dynamics in health and disease. Understanding the nasopharyngeal microbiome in children is of particular importance, as children are at a heightened risk for respiratory infections and other diseases, such as recurrent wheezing or asthma.
While it is well known that the nasopharynx is rapidly colonised by Streptococcus pneumoniae, non-typeable Haemophilus influenzae and Moraxella catarrhalis in early infancy, less is known about the colonisation dynamics of commensal bacteria (van Meel et al., 2020). Various environmental factors, such as temperature, humidity, nutrient and oxygen availability, can influence the colonisation of the nasopharynx (Camarinha-Silva et al., 2012). In addition, the composition of the nasopharyngeal microbiome is also shaped by age, genetics, and microbial interactions (Enoksson et al., 2020; Reyman et al., 2021).
This systematic review summarises studies that have investigated the nasopharyngeal microbiome in children using next generation sequencing, including its dynamics, stability over time, and the influence of intrinsic and extrinsic factors on its composition. Studying the nasopharyngeal microbiome in children is crucial for advancing our knowledge of respiratory health and disease and for developing effective interventions to improve paediatric health outcomes.
Methods
Data sources
In May 2023, MEDLINE (1946 to present) was searched using the Ovid interface with the following search terms: ‘bacteria or microbiome or DNA or sequencing’ AND ‘nasopharynx’ AND ‘neonates or infant or children’ (see Supplementary Data for detailed search terms). No language or geographical limitations were used. References of retrieved articles were searched for additional publications.
Study selection
Original studies which investigated the bacterial composition of the nasopharyngeal microbiome in neonates and (less than 28 days of age), infants (29 days to 12 months of age), and children and adolescents (1 to 18 years of age) with next-generation sequencing were included. Exclusion criteria were studies which: (i) investigated the microbiome of the anterior nares; (ii) investigated only certain bacteria and not the overall composition of the nasopharyngeal microbiome (iii) did not report results from children separately from these of adults; (iv) investigated children with cystic fibrosis; (v) investigated immunocompromised children; and (vi) investigated the effect probiotics on the nasopharyngeal microbiome.
Data extraction
The following variables were extracted from the included studies: author, publication year, country, study type, number and characteristics of participants, age of participants, number of samples, timing of testing, collection method and storage conditions, analysis technique, important findings, strengths, limitations, and potential bias.
The studies were identified, selected, appraised, and synthesised following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for systematic reviews, ensuring a comprehensive and rigorous approach to the synthesis of evidence (Page et al., 2021). Findings are presented as described in the original publications, as data to re-calculate the associations was not available in many publications.
Primary aim
The aim of the study was to describe the composition of the nasopharyngeal microbiome in children, including its dynamics, stability over time, and the influence of intrinsic and extrinsic factors on its composition.
Level of evidence
The level of evidence for each study was graded using the classification from the Oxford Centre for evidence-based medicine (Phillips et al., 2009).
Results
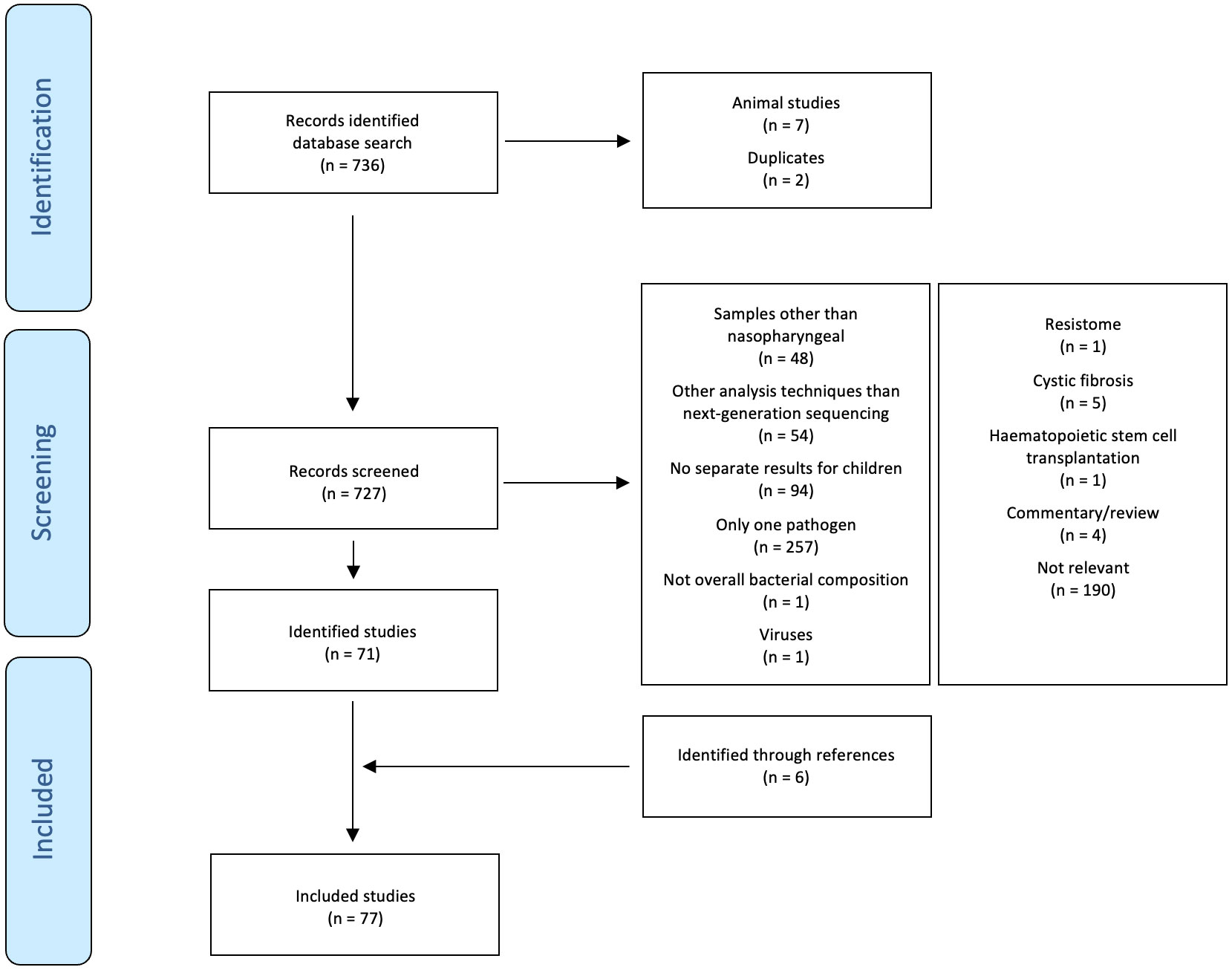
The search identified 736 studies. Of these, 71 fulfilled the inclusion criteria (Bogaert et al., 2011; Hilty et al., 2012; Biesbroek et al., 2014b; Biesbroek et al., 2014c; Sakwinska et al., 2014; Feazel et al., 2015; Jervis-Bardy et al., 2015; Stearns et al., 2015; Teo et al., 2015; Bosch et al., 2016; Hasegawa et al., 2016; Pérez-Losada et al., 2016; Rosas-Salazar et al., 2016a; Rosas-Salazar et al., 2016b; Shilts et al., 2016; Bosch et al., 2017; Chonmaitree et al., 2017; Hasegawa et al., 2017; Kelly et al., 2017; Pérez-Losada et al., 2017; Perez et al., 2017; Salter et al., 2017; Stewart et al., 2017; Dai et al., 2018; Ederveen et al., 2018; Kelly et al., 2018; Lappan et al., 2018; Luna et al., 2018; Pérez-Losada et al., 2018; Rosas-Salazar et al., 2018; Teo et al., 2018; Toivonen et al., 2018; Wen et al., 2018; Boelsen et al., 2019; Man et al., 2019c; Man et al., 2019b; Man et al., 2019a; Mansbach et al., 2019; McCauley et al., 2019; Stewart et al., 2019; Toivonen et al., 2019; Walker et al., 2019; Yau et al., 2019; Accorsi et al., 2020; Chapman et al., 2020; Enoksson et al., 2020; Haro et al., 2020; Liu et al., 2020; Mansbach et al., 2020; Salgado et al., 2020; Shilts et al., 2020; Thapa et al., 2020; Zhou et al., 2020; Aydin et al., 2021; Binia et al., 2021; Chun et al., 2021; Coleman et al., 2021; Elling et al., 2021; Folino et al., 2021; Fujiogi et al., 2021; Henares et al., 2021; McCauley et al., 2021; Reyman et al., 2021; Tang et al., 2021; Tozzi et al., 2021; Verhagen et al., 2021; Xu et al., 2021b; Hou et al., 2022; Kelly et al., 2022; McCauley et al., 2022; Tan et al., 2023). An additional six relevant studies were identified by hand-searching of references (Biesbroek et al., 2014a; Zhou et al., 2016; Lu et al., 2017; Man et al., 2020; Raita et al., 2021; Xu et al., 2021a). The selection of included studies is summarised in Figure 1. Of the 77 included studies, 36 reported findings from overlapping participants (Bogaert et al., 2011; Biesbroek et al., 2014b; Biesbroek et al., 2014c; Biesbroek et al., 2014a; Teo et al., 2015; Bosch et al., 2016; Hasegawa et al., 2016; Pérez-Losada et al., 2016; Rosas-Salazar et al., 2016a; Rosas-Salazar et al., 2016b; Bosch et al., 2017; Hasegawa et al., 2017; Kelly et al., 2017; Lu et al., 2017; Pérez-Losada et al., 2017; Stewart et al., 2017; Dai et al., 2018; Kelly et al., 2018; Luna et al., 2018; Pérez-Losada et al., 2018; Rosas-Salazar et al., 2018; Teo et al., 2018; Toivonen et al., 2018; Wen et al., 2018; Man et al., 2019a; Mansbach et al., 2019; Stewart et al., 2019; Toivonen et al., 2019; Mansbach et al., 2020; Zhou et al., 2020; Fujiogi et al., 2021; Raita et al., 2021; Reyman et al., 2021; Xu et al., 2021b; Xu et al., 2021a; Tan et al., 2023). Counting these studies only once, the following study designs were used: randomised placebo-controlled trial (2), prospective birth cohort study (6), prospective cohort study (17), retrospective cohort study (4), prospective case-control study (5), and cross-sectional study (17) (one study had two sub-studies with different study designs). The studies were done in Australia (4), Bangladesh (1), Botswana (2), Brazil (1), Canada (1), China (2), Fiji (1), Germany (1), Hong Kong (2), Israel (1), Italy (2), Japan (1), Kenya (1), the Netherlands (5), Venezuela (1), New Zealand (1), Spain (1), Switzerland (2), Sweden (1), Thailand (1), United Kingdom (1), and United States of Amerika (USA) (17). Excluding overlapping participants, the studies investigated 7,780 children (mean 151 children/study, range 11 to 1,005). The children included were healthy children (2,619), preterm-born children (487), children with an acute respiratory infection (ARTI) (200), an upper respiratory tract infection (URTI) (341), acute or chronic otitis media or middle ear effusion (777), a lower respiratory tract infection (LRTI) (1,987), allergic rhinoconjunctivitis (23), chronic wheezing (23), asthma (963), atopy, allergic rhinitis or recurrent wheezing (36), gastrointestinal infection (28), invasive pneumococcal disease (27), a chronic illness not further specified (11), and children prone to infection and allergy (492). Excluding overlapping samples, 20,571 samples (mean 375 samples/study, range 11 to 3,122) were analysed. Of these, 12,169 were nasopharyngeal swabs (30 taken with brushes), 2,115 nasopharyngeal washes, 3,332 nasopharyngeal aspirates, 2,922 swabs or aspirates, and 33 nasal filters. The analysis techniques used for the evaluation of the nasopharyngeal microbiome were shotgun metagenomic sequencing (2), 16S rRNA sequencing (50) (V1-V3 (8), V3 (3), V1-V4 (1), V3-V4 (13), V3-V5 (2), V4 (23), V5-V6 (1), V5-V7 (1), V4+ITS2 (1)), shotgun metagenomic plus 16S rRNA sequencing (V4) (3) and metatranscriptomics (1). The following platforms were used for sequencing: NovaSeq 6000 (1), NextSeq 500 (3), HiSeq (4), MiSeq (37), MiSeq plus NovaSeq 6000 (1), GS FLX Titanium (9), PacBio RS II (1), and 3130xl Genetic Analyzer (1). The following databases were used for taxonomic identification of 16S rRNA sequencing: SILVA (25), GreenGenes (9), Expanded Human Oral Microbiome (1), Human Oral Microbiome plus National Center for Biotechnology Information (NCBI) (1), Live Tree Project (1), MiSeq standard operating procedure plus SILVA (1), Ribosomal Database Project (RDP) (8), RDP plus SILVA (3), RDP, SILVA plus NCBI (1). The studies which used shotgun metagenomics for taxonomic identification used the NCBI RefSeq (1), MetaPhlAn2 (1), the GOTTCHA (1) or custom databases (2).
The main results of the studies are summarised in Table 1, Figures 2, 3 and Supplementary Table 1.

Table 1 Summary of findings of studies investigating the association between intrinsic and extrinsic factors and the composition of the nasopharyngeal microbiome in children using next generation sequencing.
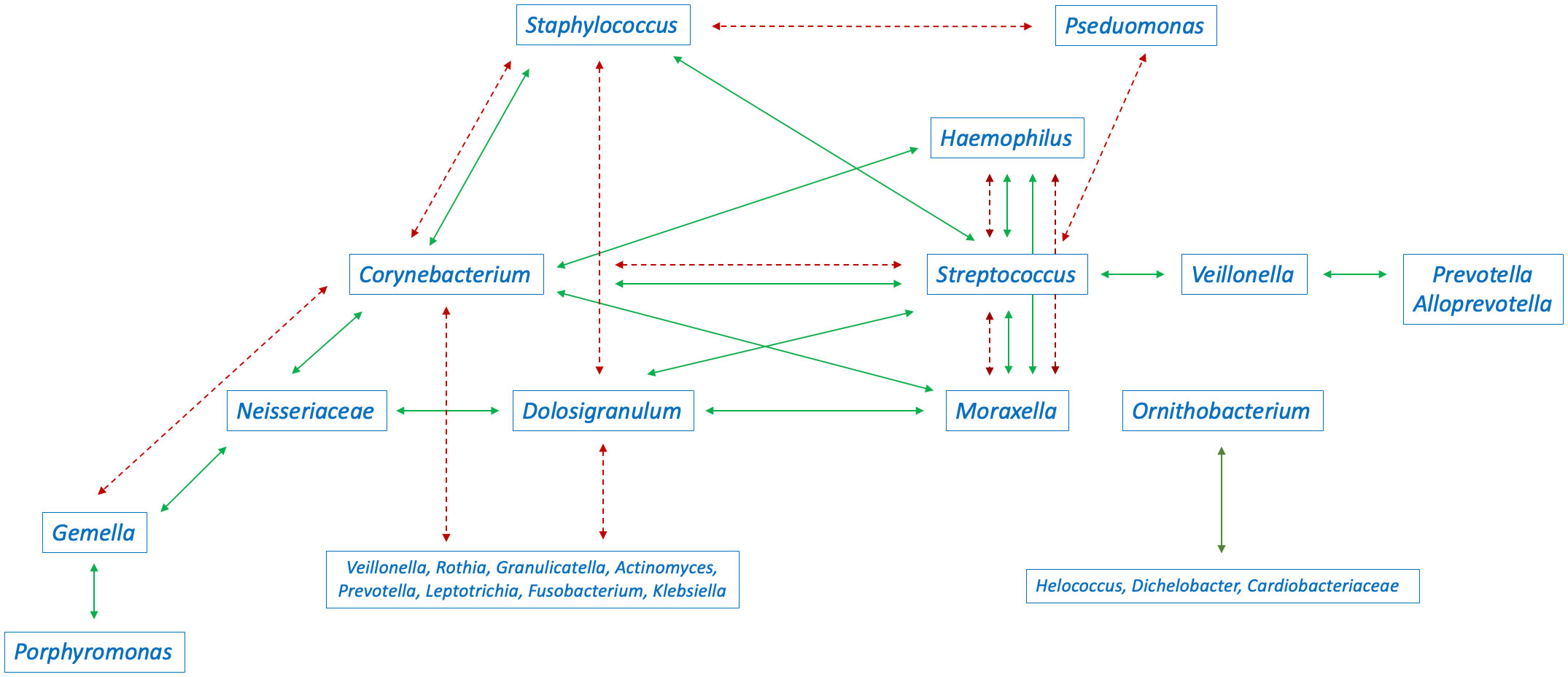
Figure 2 Interaction between the abundance of bacteria in the nasopharyngeal microbiome in children (green continuous arrows indicate positive correlations, red dashed arrows indicate negative correlations). Conflicting arrows result from studies reporting conflicting results.
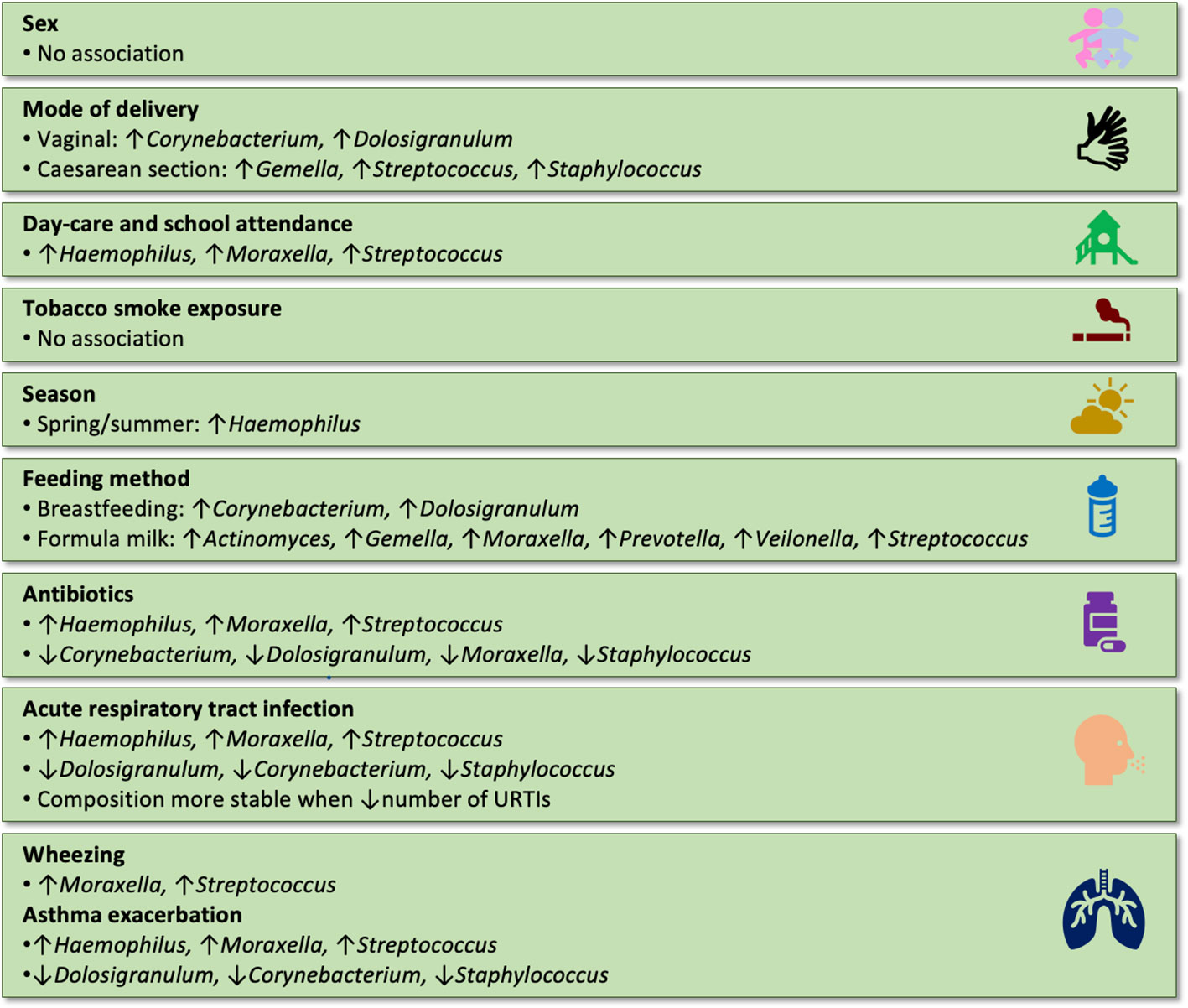
Figure 3 Summary of the associations between extrinsic factors and the composition of the nasopharyngeal microbiome in children (results were included when reported in at least two independent studies).
Host factors
Age
The composition of microbial communities in the nasopharynx in different age groups of children has been investigated in numerous studies, revealing varying levels of diversity and taxonomic identification. In healthy infants up to six months of age, 13 bacterial phyla (Biesbroek et al., 2014a), 29 to 328 genera (Shilts et al., 2016; Chapman et al., 2020), 60 species (Shilts et al., 2020), and 895 to 1,354 operational taxonomic units (OTUs) (Biesbroek et al., 2014a; Bosch et al., 2016; Reyman et al., 2021) were identified. In studies including healthy children up to 24 months of age, ten phyla (Biesbroek et al., 2014b; Biesbroek et al., 2014c), and 297 to 314 OTUs (Biesbroek et al., 2014b; Biesbroek et al., 2014c; Salter et al., 2017) were identified and in studies in toddlers up to 73 genera (Feazel et al., 2015; Coleman et al., 2021) and 121 OTUs (Verhagen et al., 2021). Studies including both healthy children and children with ARTI identified up to 15 phyla (Bosch et al., 2017; Tozzi et al., 2021), 95 families (Tozzi et al., 2021), 21 genera (Chonmaitree et al., 2017), and 245 (Tozzi et al., 2021) to 13,982 OTUs (Bosch et al., 2016; Bosch et al., 2017; Chonmaitree et al., 2017; Boelsen et al., 2019; McCauley et al., 2021). Studies including children with asthma identified 30 phyla (Hou et al., 2022), 396 to 436 genera (Pérez-Losada et al., 2017; Hou et al., 2022), and 21 to 8,034 OTUs (Shilts et al., 2016; Pérez-Losada et al., 2017; Pérez-Losada et al., 2018).
Several prospective birth cohort studies have investigated the dynamics of the nasopharyngeal microbiome during early life, revealing diverse and sometimes contrasting findings regarding the relationship between age and microbial diversity, richness, and density. Two prospective birth cohort studies found a higher diversity of the nasopharyngeal microbiome at birth compared with later periods during the first year of life (Biesbroek et al., 2014a; Kelly et al., 2022). The first study also found a positive association between increasing age and an increase in bacterial richness (Kelly et al., 2022). In contrast to these findings, a decrease in richness between the age of two to four months, without any difference in diversity has been reported in another study (Binia et al., 2021). An increase in the stability of species composition with increasing age has been described during the first year of life (Accorsi et al., 2020). Another prospective birth cohort study reported an increase in diversity with age, especially in children older than two years of age (Teo et al., 2018), while another study reported a positive individual correlation between diversity at one month of age and during the following eight months, but no overall association between age and diversity (Chonmaitree et al., 2017). Yet another prospective birth cohort study reported an increase in diversity during the first three months of life with the most changes in the microbiome composition during the first two months of life (Bosch et al., 2016), while another study reported a decrease in diversity and evenness during the first six months of life (Biesbroek et al., 2014a). One study found no differences in diversity or density during the first two years of life (Biesbroek et al., 2014b). An increase in bacterial density during the first month of life has been reported in a prospective birth cohort study (Bosch et al., 2017; Man et al., 2019a), while a retrospective cohort study found no change in density during the first two years of life (Biesbroek et al., 2014b).
The composition of the nasopharyngeal microbiome in infants and young children exhibits distinct patterns, with specific phyla and genera dominating at different stages. In infants, the most abundant phyla identified were Proteobacteria (39%), Actinobacteria (26%), Firmicutes (26%), and Bacteroidetes (6%) (Chonmaitree et al., 2017). The most abundant genera present at birth were Staphylococcus (22%), Corynebacterium (5%), Lactobacillus (4%), Gardnerella (3%), Prevotella (3%), and Gemella (3%) (Kelly et al., 2022) and in infants Moraxella (4-31%), Corynebacterium (3-28%), Streptococcus (6-22%), Staphylococcus (10-22%), Prevotella (20%), Dolosigranulum (4-14%), Veillonella (12%), Haemophilus (4-11%), and Neisseria (3%) (Teo et al., 2015; Bosch et al., 2016; Shilts et al., 2016; Bosch et al., 2017; Man et al., 2019a; Chapman et al., 2020; Shilts et al., 2020; Kelly et al., 2022). One prospective birth cohort study found higher abundances of Acinetobacter, Gardnerella, Lactobacillus, and Sneathia at birth compared with later periods during the first year of life (Kelly et al., 2022). Other prospective birth cohort studies reported that the most abundant species and genera at birth were Streptococcus (S. viridans), Gemella, Staphylococcus, and Dolosigranulum (Bosch et al., 2016; Bosch et al., 2017; Man et al., 2019a). On species level, S. pneumoniae, H. influenzae, M. catarrhalis, Moraxella nonliquefaciens, Moraxella lincolnii, Dolosigranulum pigrum, Corynebacterium pseudodiphteriticum/propinquum, Streptococcus viridans and Staphylococcus aureus were commonly identified in infants (Bosch et al., 2016; Bosch et al., 2017; Binia et al., 2021). Three studies reported a decrease in the abundance of S. aureus and an increase in the abundance of Moraxella (M. catarrhalis/nonliquefaciens), Corynebacterium (C. propinquum), Dolosigranulum (D. pigrum), H. influenzae, and S. pneumoniae during the first six months of life (Biesbroek et al., 2014a; Bosch et al., 2016; Reyman et al., 2021). Another study reported a decrease in the abundance Staphylococcus and Corynebacterium and an increase in the abundance of Dolosigranulum and Moraxella during the first year of life (Teo et al., 2015). Yet another study a decrease in the abundance of Corynebacterium, Dolosigranulum and Staphylococcus and an increase of Haemophilus (Biesbroek et al., 2014b). At six weeks of age five different microbiome profiles were found dominated by either Streptococcus, Moraxella, Staphylococcus, Corynebacterium or Corynebacterium/Dolosigranulum. Afterwards the Staphylococcus-dominated profile disappeared, and a Haemophilus-dominated profile emerged, the Corynebacterium/Dolosigranulum-dominated profile was replaced by Moraxella/Dolosigranulum-dominated profile (Biesbroek et al., 2014b).
In studies including children less than 24 months of age, the most commonly identified phyla were Proteobacteria (64%), Firmicutes (21%), Bacteroidetes (11%), Actinobacteria (3%), and Fusobacteria (1.4%) (Bogaert et al., 2011) and the most commonly identified genera Moraxella, Streptococcus, Haemophilus, Dolosigranulum, Corynebacterium, Staphylococcus, Flavobacteria, Prevotella, Pasteurella, Neisseria, Pseudomonas, Acinetobacter, and Helococcus (Bogaert et al., 2011; Kinabo et al., 2013; Biesbroek et al., 2014b; Feazel et al., 2015; Mika et al., 2015; Chonmaitree et al., 2017; Salter et al., 2017; Boelsen et al., 2019; Tang et al., 2021). A prospective cohort study reported a higher abundance of Staphylococcus and Corynebacterium in the first 3 months of life compared with later timepoints during the first two years of life. The study also reported a decrease in the abundance of Streptococcus and an increase of unclassified Flavobacteriaceae during the first year of life and an increase in abundance of Moraxella in the first 21 months (Salter et al., 2017). A further prospective birth cohort study identified four different microbiome profiles in the nasopharyngeal microbiome during the first six months of life: a Dolosigranulum/Corynebacterium-, a Moraxella-, a Staphylococcus-, and a Streptococcus-dominated microbiome profile. At the age of two years, all children had mixed composition profiles, but many were dominated by Moraxella (Tang et al., 2021).
In toddlers, Moraxella, Streptococcus, Haemophilus, Dolosigranulum and Corynebacterium were identified on a genera level (Feazel et al., 2015; Verhagen et al., 2021), and M. catarrhalis, S. pneumoniae, H. influenzae, D. pigrum, C. pseudodiphteriticum, S. aureus, M. nonliquefaciens, S. epidermidis, and S. mitis on species or OTU level (Feazel et al., 2015; Tang et al., 2021). A prospective cohort study including children aged 6 to 17 years, reported that younger children had a higher abundance of Dolosigranulum, Haemophilus, and Moraxella; as well as Ascochyta, Cladosporium, and Verticillium in their nasopharyngeal microbiome, while older children had a higher abundance of Corynebacterium and Staphylococcus (McCauley et al., 2022).
Sex
The impact of sex on the composition of the nasopharyngeal microbiome in children has been investigated in several studies. One prospective birth cohort study reported a higher abundance of Moraxella in males in the first year of life during healthy periods but not during ARTI (Teo et al., 2015). Ten other studies did not find an association between sex and the composition of the nasopharyngeal microbiome in children (Hilty et al., 2012; Feazel et al., 2015; Shilts et al., 2016; Thapa et al., 2020; Boelsen et al., 2019; Man et al., 2019c; Liu et al., 2020; Zhou et al., 2020; Kelly et al., 2022).
Ethnicity
Studies exploring the influence of ethnicity on the nasopharyngeal microbiome in children reported varied findings. A prospective cohort study from the USA reported that non-Hispanic White children more frequently had a Haemophilus-dominated microbiome profile compared with children from other ethnicities (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021). Another prospective cohort study from the USA found an association between ethnicity and the nasopharyngeal microbiome composition without specifying further details (McCauley et al., 2021). A retrospective cohort study from Fiji reported that infants of Indian descent had a higher diversity compared with Indigenous infants, while ethnicity was not associated with differences in richness (Boelsen et al., 2019). Furthermore, Indigenous infants had a higher abundance of Moraxella, Haemophilus, and Helcococcus and lower abundance of Staphylococcus, Dolosigranulum, and Corynebacterium compared with infants of Indian descent (Boelsen et al., 2019). Two other studies from the USA did not find an association between ethnicity and diversity or the composition of the nasopharyngeal microbiome (Shilts et al., 2016; Chonmaitree et al., 2017).
Perinatal factors
Delivery mode
Multiple studies investigating the potential influence of delivery mode on the composition of the nasopharyngeal microbiome in children have reported a range of associations. A prospective birth cohort study reported that in the first year of life infants born by Caesarean section (CS) had a prolonged predominance of Actinomyces, Granulicatella, Neisseria, and Prevotella and a higher abundance of Gemella and Streptococcus in their nasopharyngeal microbiome, while infants born vaginally had a prolonged predominance of Corynebacterium and Dolosigranulum, and a late enrichment of Moraxella (Bosch et al., 2017). Another prospective birth cohort study reported that during the first six months of life infants born by CS had a higher abundance of Gemella, S. aureus, S. viridans, and S. salivarius, while infants born vaginally had a higher abundance of C. pseudodiphteriticum/propinquum and D. pigrum. Furthermore, infants born by CS stayed longer in a S. aureus-dominated microbiome profile, while infants born vaginally switched earlier to a Moraxella- or Corynebacterium/Dolosigranulum-dominated microbiome profile (Bosch et al., 2016). A third prospective birth cohort study reported an association between mode of delivery and the composition of the nasopharyngeal microbiome in the first six months of life without specifying further details (Reyman et al., 2021). A cross-sectional study reported a higher richness and diversity of the nasopharyngeal bacterial microbiome with a lower abundance of Corynebacterium and a higher abundance of Staphylococcus in infants born by CS compared with these born vaginally (Shilts et al., 2016).
Six other studies did not find an association between mode of delivery and the composition of the nasopharyngeal microbiome in the first year of life (Teo et al., 2015; Bosch et al., 2016; Binia et al., 2021) or at an older age (Thapa et al., 2020; Chonmaitree et al., 2017; Zhou et al., 2020).
Feeding method
The influence of breastfeeding on the composition of the nasopharyngeal microbiome in infants has been investigated in several studies, yielding diverse results and highlighting potential associations between breastfeeding and specific microbial taxa. A prospective birth cohort study observed an early abundance of Dolosigranulum and a prolonged predominance of Corynebacterium and Dolosigranulum, as well as a late enrichment of Moraxella in breastfed compared to formula-fed infants in the first year of life. In contrast, formula-fed infants were reported to have a higher abundance of Gemella and Streptococcus from birth onwards; and a higher abundance of Streptococcus, Porphyromonas, Prevotella, and Veillonella and a prolonged predominance of Actinomyces, Granulicatella, Neisseria, and Prevotella after one month of age (Bosch et al., 2017). Another prospective birth cohort study reported that being breastfed was associated with a higher abundance of Corynebacterium and lower abundances of Haemophilus, Moraxella, and Streptococcus during the first year of life (Kelly et al., 2022). A third prospective birth cohort study reported an association between being breastfed and the composition of the nasopharyngeal microbiome in the first six months of life without further specifying details (Reyman et al., 2021). A prospective cohort study reported a higher richness with a higher abundance of Moraxella in the nasopharyngeal microbiome in infants who were fed formula milk (Shilts et al., 2016). A retrospective cohort study reported that breastfed infants had a higher evenness, and more stable composition of their nasopharyngeal microbiome in the first two years of life (Biesbroek et al., 2014b). Furthermore, breastfeeding was associated more frequently with a Corynebacterium/Dolosigranulum-dominated microbiome profile and a higher abundance of Corynebacterium (C. pseudodiphteriticum, C. propinquum, C. accolens, C. fastidiosum, C. segmentosum) and Dolosigranulum (D. pigrum), while formula-fed infants were reported to have a higher abundance of Actinomyces, Gemella, Granulicatella, Prevotella, Rothia, Staphylococcus, and Veillonella at the age of six weeks, and a higher abundance of Granulicatella at the age of six months (Biesbroek et al., 2014b).
Eight other studies did not find an association between breastfeeding and richness, diversity or the composition of the nasopharyngeal microbiome in infants or older children (Teo et al., 2015; Thapa et al., 2020; Chonmaitree et al., 2017; Boelsen et al., 2019; Man et al., 2019c; Zhou et al., 2020; Binia et al., 2021; Henares et al., 2021). One prospective cohort study reported that there was no association between the presence of fucosylated oligosaccharides in breast milk and the composition of the nasopharyngeal microbiome in infants between two to four months of age (Binia et al., 2021).
Environmental factors
Siblings and household size
The presence of siblings, particularly those under the age of five, has been investigated in relation to the nasopharyngeal microbiome composition in infants, revealing potential associations and accelerated microbiome maturation in the first year of life. A prospective birth cohort study reported that infants with siblings less than five years of age had an accelerated microbiome maturation and a higher abundance of Pasteurellaceae in their nasopharynx in the first year of life (Bosch et al., 2017). Another prospective birth cohort study found a higher abundance of Haemophilus, Moraxella, and Streptococcus and a lower abundance of Staphylococcus in infants who had older siblings at 12 months of age (Teo et al., 2015). A third prospective birth cohort study found an association between having siblings less than five years of age and the composition of the nasopharyngeal microbiome in the first six months of life without further specifying details (Reyman et al., 2021). A retrospective cohort study reported that in children under the age of 18 months, the abundance of S. pneumoniae was positively associated with increasing number of children in the household (Bogaert et al., 2011). A cross-sectional study found no association between household size and the composition of the nasopharyngeal microbiome in healthy children and children with otitis media (Coleman et al., 2021).
Day-care attendance
Day-care attendance during the first year of life has been examined in relation to the nasopharyngeal microbiome composition, revealing potential associations with specific microbial profiles, including increased abundance of certain bacteria such as M. catarrhalis and H. influenzae, as reported in prospective birth cohort studies. A prospective birth cohort study reported that in the first year of life day-care attendance was associated with an increase in the abundance of M. catarrhalis and H. influenzae in the nasopharynx (Accorsi et al., 2020). Another prospective birth cohort study observed that day-care attendance was associated with a higher abundance of Haemophilus, Streptococcus, and Moraxella and a lower abundance of Staphylococcus in the first year of life (Teo et al., 2015). A third prospective birth cohort study reported that day-care attendance was associated with an accelerated microbiome maturation and a higher abundance of Moraxella and a lower abundance of Staphylococcus (Bosch et al., 2017). A further prospective birth cohort study reported an association between day-care attendance and the composition of the nasopharyngeal microbiome in the first six months of life without further specifying details (Reyman et al., 2021). A prospective cohort study reported that infants in the first year of life who attended day-care attendance were more frequently colonised with S. pneumoniae (Xu et al., 2021b; Xu et al., 2021a). Three other studies did not find an association between day-care attendance and the composition of the nasopharyngeal microbiome (Bogaert et al., 2011; Hilty et al., 2012; Man et al., 2019c).
Pets
The influence of pets, particularly furry animals, on the nasopharyngeal microbiome during early life has been investigated in several studies, with varying results. One prospective birth cohort study reported that having furry pets was associated with a lower abundance of Streptococcus in the nasopharynx in the first year of life (Teo et al., 2015). Another prospective birth cohort study reported an association between having pets and the composition of the nasopharyngeal microbiome in the first six months of life without specifying further details (Reyman et al., 2021). A small prospective cohort study did not find an association between having pets and the composition of the nasopharyngeal microbiome (Shilts et al., 2016).
Tobacco smoke exposure The impact of tobacco smoke exposure on the composition of the nasopharyngeal microbiome has been a subject of limited research. There were only two studies which investigated the effect of tobacco smoke exposure on the composition of the nasopharyngeal microbiome. Both studies did not find an association (up to five months of age (Boelsen et al., 2019) or at 18 months of age (Bogaert et al., 2011)).
Season
The influence of seasonal variations on the composition of the nasopharyngeal microbiome has been investigated through several studies. These studies have provided insights into the association between different microbial profiles and specific seasons in healthy children and those with upper respiratory tract infections. Findings from these studies have been diverse, with some reporting significant associations between season and microbiome composition while others did not find any such relationship. A prospective cohort study from the USA reported that in either healthy children or children with an URTI an Dolosigranulum/Corynebacterium-dominated microbiome profile was more frequent during summer and autumn, while a Haemophilus-dominated microbiome profile was more frequent during winter and spring (McCauley et al., 2021). A prospective birth cohort study from Australia reported that during the first year of life in healthy infants or infants with an ARTI a Haemophilus-dominated microbiome profile was more frequent during spring/summer and a Moraxella-dominated microbiome profile more frequent during autumn/winter (Teo et al., 2015). A retrospective cohort study from the Netherlands observed a higher abundance of Proteobacteria, Fusobacteria, and Cyanobacteria during autumn/winter and a higher abundance of Bacteroides during spring. Furthermore, the study reported a higher abundance of Bacillus, Brevibacillus, Flavobacterium, and Lactobacillus during spring (Bogaert et al., 2011). On the species level a higher abundance of B. fragilis was found during spring (Bogaert et al., 2011). A prospective cohort study from the USA reported that children with asthma had a higher abundance of Moraxella in their nasopharynx during spring and a higher abundance of Staphylococcus during autumn (McCauley et al., 2022). In the same cohort, season had no influence on the fungal composition of the nasopharyngeal microbiome in children with asthma, expect during ARTI, when a higher abundance of Malassezia was found during spring and of Candida and Cladosporium during autumn (McCauley et al., 2022). Another prospective cohort study from the USA reported that in children with asthma, season did not influence the diversity of the nasopharyngeal microbiome but that a higher abundance of Haemophilus was found during summer (Pérez-Losada et al., 2017). Another reported an association between season and the overall microbiome composition without specifying further details (Reyman et al., 2021). Three other studies did not find an association between season and the composition of the nasopharyngeal microbiome (Boelsen et al., 2019; Man et al., 2019c; Coleman et al., 2021).
Health-care associated factors
Vaccination
The impact of vaccination, particularly with pneumococcal conjugate vaccines (PCVs), on the composition of the nasopharyngeal microbiome has been investigated in several studies. These studies have explored the association between vaccination status and the presence or abundance of specific bacterial taxa in the nasopharynx during early childhood. However, findings from these studies have been inconsistent, with some reporting differences in colonization patterns or microbial diversity between vaccinated and non-vaccinated children, while others did not observe significant changes. A prospective cohort study found that children who were vaccinated with 7-valent pneumococcal conjugate vaccine (PCV7) were less frequently colonised with Streptococcaceae and Corynebacteriaceae in the nasopharynx during the first two years of life (Hilty et al., 2012). A prospective birth cohort study reported that infants who were vaccinated with PCV13 had a lower abundance of S. pneumoniae in the first year of life (Kelly et al., 2022). A cross-sectional study reported that children who were vaccinated with H. influenzae type b (Hib)/PCV10 had a higher richness but similar diversity of the nasopharyngeal microbiome compared with these who were not vaccinated (Salgado et al., 2020). The study did not find a difference in the abundance of bacteria between vaccinated and non-vaccinated children (Salgado et al., 2020). A prospective case-control study reported that children who were vaccinated with PCV had a higher diversity in their nasopharyngeal microbiome compared with these who were not vaccinated (Henares et al., 2021). A retrospective cohort study did not find differences in diversity, richness or overall composition of the microbiome between children who were vaccinated with PCV7 and these who were not. However, vaccinated children had a lower abundance of S. pneumoniae (Boelsen et al., 2019). Another retrospective cohort study reported that infants who were vaccinated with 3 doses of PCV7 had a higher diversity in their nasopharyngeal microbiome with a higher abundance of Haemophilus, Staphylococcus, Veillonella, Prevotella, Bacteroidetes, Leptotrichia, and Streptococcus at one year of age. At two years of age, no differences in the microbiome composition were found between vaccinated and non-vaccinated children (Biesbroek et al., 2014c). Further retrospective cohort studies did not find that Hib/PCV10 (Feazel et al., 2015) or PCV7 (Hilty et al., 2012; Biesbroek et al., 2014b) were associated with changes in diversity or composition of the nasopharyngeal microbiome.
Antibiotic exposure
The impact of antibiotic exposure on the nasopharyngeal microbiome in infants has been a subject of investigation in several prospective and retrospective cohort studies. These studies have explored the association between antibiotic use and the abundance and diversity of specific bacterial taxa in the nasopharynx during early life. However, findings from these studies have been varied, with some reporting changes in microbial composition following antibiotic exposure, while others did not observe significant associations. A prospective birth cohort study reported that infants who had been exposed to antibiotics (drug not specified) in the previous four months had a higher abundance of Haemophilus, Streptococcus, and Moraxella and lower abundance of Dolosigranulum and Corynebacterium in their nasopharynx (Teo et al., 2015). Another prospective birth cohort study reported that infants who had been exposed to antibiotics (mostly amoxicillin for seven to ten days) in the previous six months had a higher diversity and lower abundance of Corynebacterium and Dolosigranulum within seven days, Enterobacter within seven to 14 days, and Staphylococcus within 14 to 30 days after exposure, and a higher abundance of Bifidobacterium and Firmicutes incertae sedis within seven days after exposure (Chonmaitree et al., 2017). A third prospective birth cohort study, which did not specify the drug or time interval which was investigated, reported that antibiotic exposure was associated with a lower abundance of Corynebacterium and Dolosigranulum (Bosch et al., 2017). A further prospective birth cohort study reported that infants who were exposed to antibiotics during the first year of life (amoxicillin, metronidazole or trimethoprim/sulfamethoxazole) had a lower abundance of Corynebacterium and Lactobacillus and a higher abundance of Haemophilus, Moraxella, and Streptococcus in their nasopharynx (Kelly et al., 2022).
A prospective cohort study reported a lower abundance of Moraxella and a higher abundance of Brachybacterium, Dolosigranulum, and Streptococcus during antibiotic treatment (mostly oral amoxicillin given for LRTI) (Salter et al., 2017). Another prospective cohort study reported antibiotic exposure in the previous two months was associated with a lower abundance of Moraxellaceae, Staphylococcaceae, and Streptococcaceae, and a higher abundance Pasteurellaceae (Hilty et al., 2012). A prospective birth cohort study reported an association between exposure to antibiotics (drug not specified) in the 30 days prior to sampling and the composition of the nasopharyngeal microbiome in the first six months of life without further specifying details (Reyman et al., 2021). A randomised placebo-controlled trial reported a lower abundance of Moraxella after exposure to azithromycin for 14 days. A higher abundance of Dolsigranulum and Corynebacterium and a lower abundance of Streptococcus was observed after exposure to placebo for 14 days (Zhou et al., 2016).
A cross-sectional study reported that exposure to antibiotics in the three previous months was associated with a higher abundance of Haemophilus (Zhou et al., 2016). Several prospective cohort studies with overlapping participants reported that infants who were hospitalised in the first few months of life with bronchiolitis and exposed to antibiotics (drug not specified) more frequently had a Haemophilus-dominated microbiome profile and also had a higher a higher abundance of H. influenzae compared with infants who were hospitalised with bronchiolitis and did not receive antibiotics (Hasegawa et al., 2016; Hasegawa et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021; Raita et al., 2021).
A retrospective cohort study found no association between antibiotic exposure two weeks before sampling (drug not specified) and the composition of the nasopharyngeal microbiome (Boelsen et al., 2019), and a cross-sectional study between antibiotic exposure six months before sampling (drug not specified) (Man et al., 2019c). Two retrospective studies, which did not specify the time interval or drugs that were investigated, also did not find an association between antibiotic exposure and the composition of the nasopharyngeal microbiome (Bogaert et al., 2011; Feazel et al., 2015).
Disease-associated factors
Acute respiratory tract infection
The nasopharyngeal microbiome composition in infants and children during ARTIs has been the focus of several prospective cohort and cross-sectional studies. A prospective cohort study reported that in infants during the first few months of life the most abundant genera in the nasopharynx during ARTI were Moraxella, Streptococcus, Corynebacterium, Haemophilus, and Dolosigranulum (Rosas-Salazar et al., 2016a). A cross-sectional study reported that the genera with the highest abundance in children under the age of two years with ARTI were Streptococcus, Haemophilus, and Moraxella (Salgado et al., 2020). Another prospective birth cohort study reported that children less than two years of age with an ARTI more often had a H. influenzae-, M. catarrhalis-, or S. pneumoniae-dominated microbiome profile, and less frequently a D. pigrum-, C. pseudodiphtheriticum-, S. mitis- or Staphylococcus-dominated microbiome profile than healthy children (Tang et al., 2021). Another prospective birth cohort study observed a lower abundance of Corynebacterium (in children less than three years of age) and of Dolosigranulum and Staphylococcus (in children less than four years of age) in children with ARTI compared with healthy children. Furthermore, a higher frequency of Haemophilus-, Streptococcus-, and Moraxella-dominated microbiome profiles were observed during ARTIs and a high abundance of one of these three bacteria was associated with a decrease in the diversity of the nasopharyngeal microbiome and an increase in severity of the ARTI (Teo et al., 2018). A third prospective birth cohort study reported that Haemophilus-, Moraxella- and Streptococcus-dominated microbiome profiles were more frequent during ARTI, while Dolosigranulum-, Staphylococcus-, and Corynebacterium-dominated microbiome profiles were less frequent during ARTI. Furthermore, Neisseria was more commonly found in nasopharyngeal samples taken during ARTI (Teo et al., 2015). A further prospective birth cohort study reported a decrease in presence and abundance of Corynebacterium, Dolosigranulum, and Moraxella and an increase in Fusobacterium and Streptococcus before and during an ARTI (Man et al., 2019a). On species level, the study found an increase in abundance of Janthinobacterium lividum, Neisseria lactamica, and Prevotella nanceiensis before and during ARTIs in infants (Man et al., 2019a).
A prospective birth cohort study reported that infants with frequent ARTIs during the first year of life had a less stable composition of the nasopharyngeal microbiome with a higher abundance of Moraxella and Haemophilus early in life, and a higher abundance of Neisseria, Prevotella, and Alloprevotella from the age of two months onwards. Infants with frequent ARTIs also had an absence or lower abundance of Corynebacterium, Dolosigranulum, and Streptococcus (Bosch et al., 2017). Another prospective birth cohort study observed an increase in the abundance of Moraxella one to two months before the occurrence of an ARTI. The study also reported a negative corelation between the abundance of Streptococcus closely matching to S. gordonni or S. thermophilus/salivarius/vestibularis and the risk for an ARTI (Teo et al., 2018). A third prospective birth cohort study reported that frequent ARTIs were associated with a higher abundance of Moraxella and lower abundance of Dolosigranulum and Corynebacterium (Teo et al., 2015). In another prospective birth cohort study a H. influenzae-dominated microbiome profile was associated with frequent ARTIs (Bosch et al., 2016). A cross-sectional study reported that ARTIs are more frequent in children colonised with S. pneumoniae (Kelly et al., 2018). while a prospective cohort study did not find an association between the composition of the nasopharyngeal microbiome and the risk for ARTI (Binia et al., 2021).
In a prospective cohort study of children with asthma a higher abundance of Cladosporium was associated with longer duration from baseline to the occurrence of an ARTI (McCauley et al., 2022). During a ARTI, a higher abundance of Moraxella and Haemophilus was associated with the presence of a virus in the nasopharynx (McCauley et al., 2022).
Two studies did not find an association between a prior ARTI and the composition of the nasopharyngeal microbiome (Man et al., 2019c; Binia et al., 2021).
Upper respiratory tract infection
The nasopharyngeal microbiome composition during URTIs in infants and children has been investigated in various prospective and retrospective cohort studies, as well as cross-sectional studies. A prospective birth cohort study found a higher abundance of Haemophilus, Moraxella, and Streptococcus and a lower abundance of Myroides, Pseudomonas, Sphingomonas, and Yersinia in infants less than nine months of age with URTI compared with healthy infants (Chonmaitree et al., 2017). A higher abundance of Moraxella and Streptococcus during URTI was associated with the presence of a virus (Chonmaitree et al., 2017). A retrospective cohort study found a higher abundance of Haemophilus, Moraxella, and Streptococcus and lower abundance of Dolosigranulum and Corynebacterium during URTI in infants at one year of age (Boelsen et al., 2019). A cross-sectional study reported that children with URTI had a lower abundance of Staphylococcus (S. aureus) and Neisseriaceae compared with healthy children (Coleman et al., 2021). Another cross-sectional study observed that in infants Moraxella- and Streptococcus-dominant microbiome profiles were more frequent during URTI than during healthy periods (Kelly et al., 2017). A further cross-sectional study reported a higher abundance of Haemophilus, Moraxella, and Streptococcus and a lower abundance of Staphylococcus during an URTI (Kelly et al., 2017). A prospective birth cohort study did not find an influence of an URTI on the diversity of the nasopharyngeal microbiome (Chonmaitree et al., 2017). A prospective cohort study reported that during an URTI the most abundant genera were Haemophilus, Moraxella, Streptococcus Pseudomonas, Novosphingobium, Corynebacterium, and Dolosigranulum (Haro et al., 2020).
A prospective birth cohort study reported that a higher diversity at one month of age was associated with frequent URTIs within the first six months of life without increasing the risk for acute otitis media (AOM) (Chonmaitree et al., 2017). A retrospective cohort study found that children with Corynebacterium/Dolosigranulum-dominated microbiome profile at six weeks and six months of age and Moraxella-dominated microbiome profiles at six weeks, six and 12 months had less frequent URTIs. The composition of the nasopharyngeal microbiome was more stable over time in children with less frequent URTIs (Biesbroek et al., 2014b).
A prospective birth cohort study reported that early colonisation with Moraxella was associated with an earlier occurrence of an URTI (Teo et al., 2015). Another prospective cohort study reported that in pre-school children a Haemophilus- and Moraxella-dominated microbiome profile was associated with an increased risk of URTIs and sinusitis, while an Dolosigranulum/Corynebacterium-dominated microbiome profile (which was also richer and more diverse) was associated with less frequent URTIs and sinusitis. Furthermore, children who developed URTI or sinusitis had a higher abundance of Moraxella and a lower abundance of Prevotella, Acetobacteraceae, and Chryseobacterium (McCauley et al., 2021).
A further prospective cohort study reported that children with URTI who were colonised with S. pneumoniae had a higher abundance of Haemophilus and Streptococcus, while children without URTI colonised with S. pneumoniae had a higher abundance of Streptococcus and a lower abundance of Corynebacterium_1 at one year of age (Xu et al., 2021b; Xu et al., 2021a). A retrospective cohort study reported that a higher abundance of Dolosigranulum was associated with less frequent URTIs, while a higher abundance of Gemella was associated with more frequent URTI (Biesbroek et al., 2014a).
Acute otitis media
The nasopharyngeal microbiome composition has been extensively investigated in relation to acute otitis media (AOM) and chronic otitis media, utilizing various prospective cohort studies, cross-sectional studies, and case-control studies. A prospective birth cohort study found that infants who develop AOM in the first year of life had a higher abundance of Bifidobacterium, Enterobacter, Haemophilus, and Yersinia, and a lower abundance of Corynebacterium, Myroides, and Pseudomonas in their nasopharynx. A higher abundance of Staphylococcus and Sphingobium was associated with a reduced risk of developing AOM after an URTI. Compared with healthy infants, no difference in diversity, but a higher abundance of Haemophilus, Moraxella, and Streptococcus was observed in the nasopharynx of infants with AOM. Furthermore, in infants with recurrent AOM a lower abundance of Micrococcus was found (Chonmaitree et al., 2017). Another prospective cohort study reported that compared with healthy children, children with AOM had a lower richness and diversity, but a higher density in their nasopharyngeal microbiome with a lower abundance of Acidaminococcaceae, Comamonadaceae, Corynebacteriaceae, and Staphylococcaceae. This study did not find an association between recurrent AOM and the composition of the nasopharyngeal microbiome (Hilty et al., 2012). A small prospective cohort study reported that in children with otitis media with effusion the most abundant genera in the nasopharynx were Moraxella, Corynebacterium, Dolosigranulum, Haemophilus, and Streptococcus. The presence of Haemophilus and C. propinquum was associated with anti-inflammatory mediators, while the presence of Turicella and Dolosigranulum was associated with pro-inflammatory mediators (Enoksson et al., 2020). Another prospective cohort study reported that in children with otitis media with effusion the most prevalent OTUs were M. catarrhalis, H. influenzae, Streptococcus, Ornithobacterium, D. pigrum, and C. pseudodiphtheriticum (Jervis-Bardy et al., 2015). A cross-sectional study reported that the abundances of Acinetobacter, Klebsiella, Neisseria, and Haemophilus were associated with longer duration of otorrhoea, while the abundances of Corynebacterium, Dolosigranulum, and Haemophilus were associated with shorter duration (Man et al., 2019b).
Another cross-sectional study reported that in children with recurrent AOM the most abundant genera were Moraxella (42%), Streptococcus (20%), Haemophilus (11%), Dolosigranulum (17%), and Corynebacterium (9%) (Folino et al., 2021). A further cross-sectional study reported that there was no difference in diversity in children who had recurrent AOM compared with healthy children, but that the former had a higher abundance of Moraxella. Furthermore, children with middle ear effusion had a higher abundance of Ornithobacterium compared with children who never had AOM (Coleman et al., 2021). A cross-sectional study reported that children who were carrier of a variant of the fucosyltransferase genes, which encodes an enzyme involved in the production of the H antigen in body fluids and associated with an increased risk for AOM, had a higher abundance of Cutibacterium and a lower abundance of Actinobacillus, Selenomonas, and Saccharibacteria. Furthermore, children who were carrier of a Ras interacting protein 1 variant which is also associated with an increased risk for AOM, had a higher abundance of Cutibacterium, Escherichia-Shigella, and Staphylococcus and a lower abundance of Acintobacillus (Elling et al., 2021). Another cross-sectional study reported that there was no difference in diversity between children with recurrent AOM and children with atopy. However, the former had a lower abundance of Dolosigranulum and Corynebacterium and a higher abundance of Haemophilus, especially these children with recurrent AOM with tympanic membrane perforation (Folino et al., 2021). A prospective cohort study reported that compared with infants without AOM, these with recurrent AOM had a lower diversity of their nasopharyngeal microbiome at six but not 12 months of age. Infants with recurrent AOM had a lower abundance Bacillus, Gemella, Fusobacterium, Prevotella, and Veillonella and a higher abundance of Moraxella and Dolosigranulum (Xu et al., 2021b; Xu et al., 2021a). A cross-sectional study reported that compared with healthy children, children with recurrent AOM had a higher diversity in their nasopharyngeal microbiome with a lower abundance of Corynebacterium and Dolosigranulum and a higher abundance of Neisseria, Gemella, Porphyromonas, Alloprevotella, and Fusobacterium (Lappan et al., 2018).
A prospective case-control study reported that compared with healthy children, children with chronic otitis media had a lower diversity of their nasopharyngeal microbiome and more frequently had Corynebacterium-, Moraxella-, and Streptococcus-dominated microbiome profiles. Furthermore, a higher abundance of H. influenzae, M. catarrhalis, Moraxella caprae, and S. pneumoniae, and a lower abundance of C. acnes, Capnocytophaga, Lactococcus, Lautropia, Neisseria, two Oxalobacteraceae OTUs, and Salmonella infantis was found in children with chronic otitis media (Walker et al., 2019).
Lower respiratory tract infection
The nasopharyngeal microbiome has been extensively studied in infants and children with LRTIs, including bronchiolitis and pneumonia, providing valuable insights into the microbial profiles associated with these conditions. A prospective case-control study reported that in infants who were admitted to an intensive care unit with a LRTI the most abundant species in the nasopharynx were M. catarrhalis/nonliquefaciens, H. influenzae/haemolyticus, and S. pneumoniae. Compared to healthy children, children with LRTI more frequently had H. influenzae/haemolyticus- and S. pneumoniae-dominated microbiome profiles and less frequently M. catarrhalis/nonliquefaciens- and C. propinquum/D. pigrum-dominated microbiome profiles. Children with LRTI also had a higher abundance of H. influenzae/haemolyticus, S. pneumoniae, Actinomyces, Prevotella and a lower abundance of Moraxella, C. propinquum, D. pigrum, and Helococcus in the nasopharynx (Man et al., 2019c). A prospective birth cohort reported that early colonisation with Streptococcus was associated with earlier LRTI (Teo et al., 2015), while a prospective cohort study found that the acquisition of a new S. pneumoniae serotype was not associated with LRTI (Salter et al., 2017).
Several studies reported that during bronchiolitis the most abundant genera in the nasopharynx were Streptococcus, Moraxella, Haemophilus (Hasegawa et al., 2016; Hasegawa et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Stewart et al., 2019; Toivonen et al., 2019; Fujiogi et al., 2021), and Prevotella and Staphylococcus (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021). The studies identified between four to six different microbiome profiles: Haemophilus-, Moraxella-, Streptococcus-, Staphylococcus-, Corynebacterium-, Enterobacter-dominant profiles and a mixed profile (Hasegawa et al., 2016; Hasegawa et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Mansbach et al., 2019; Toivonen et al., 2019; Fujiogi et al., 2021). A prospective cohort study reported Haemophilus-dominant microbiome profile associated with increased severity of bronchiolitis and length of hospital stay, while a Moraxella-dominant microbiome profile was associated with less frequent admission to an intensive care unit (Hasegawa et al., 2016; Hasegawa et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021).
A cross-sectional study reported that compared with healthy children, children with pneumonia more frequently had Haemophilus-, Staphylococcus-, and Streptococcus-dominant microbiome profiles, a higher abundance of Haemophilus, Streptococcus, Escherichia, and Klebsiella and a lower abundance of Corynebacterium and Dolosigranulum (Kelly et al., 2017). A prospective cohort study, reported that compared with healthy children, children with pneumonia had a lower diversity of their nasopharyngeal microbiome with a higher abundance of Firmicutes, Mycoplasma, Streptococcus, Staphylococcus, Lactobacillus, Ralstonia, Acinetobacter, and Actinomyces, and a lower abundance of Bacteroidetes, Prevotella, Moraxella, and Dolosigranulum (Lu et al., 2017; Dai et al., 2018). A prospective case-control study reported that children with pneumonia had a lower richness and diversity with a higher abundance of Moraxella, Haemophilus, and Streptococcus during non-viral pneumonia and a higher abundance of Moraxella lacunata during viral pneumonia (Sakwinska et al., 2014).
Viral infections
The nasopharyngeal microbiome plays a crucial role in respiratory viral infections, and several prospective cohort and cross-sectional studies have shed light on the microbial profiles associated with specific pathogens. Several prospective cohort studies reported that during RSV infection the most abundant genera in the nasopharynx were Moraxella (38-39%), Streptococcus (20-27%), Staphylococcus (27%), Haemophilus (11-14%), Corynebacterium (5-19%), and Dolosigranulum (4-5%) (Rosas-Salazar et al., 2016b; Rosas-Salazar et al., 2018; Tan et al., 2023). Compared to healthy children, a lower richness and diversity at OTU level was reported. Children infected with RSV also had a lower abundance of Staphylococcus, and Corynebacterium and higher abundance of Haemophilus, Moraxella, and Streptococcus (Rosas-Salazar et al., 2016a; Rosas-Salazar et al., 2016b).
A higher abundance of Moraxella was found in children with RSV-A compared with children with RSV-B infection (Tan et al., 2023). A cross-sectional study reported that RSV infection was associated with a S. pneumoniae-dominated microbiome profile (Toivonen et al., 2019; Raita et al., 2021). Another cross-sectional study reported a delayed clearance of RSV after bronchiolitis in infants with a Haemophilus-dominated microbiome profile (Mansbach et al., 2019). A further cross-sectional study found an association between RSV viral load and the overall composition of the nasopharyngeal microbiome. A lower abundance of Veillonella, and a higher abundance of Achromobacter and Haemophilus was found in children infected with RSV compared with healthy children. The study found a positive correlation between the abundance of Haemophilus and C-X-C motif chemokine ligand 8 levels, which are indicative for a higher disease severity (Ederveen et al., 2018). A prospective birth cohort study reported that children who are colonised with Dolosigranulum had fewer RSV infections, especially RSV LRTIs (Teo et al., 2015).
A cross-sectional study reported that during rhinovirus infection the most abundant genera in the nasopharynx were Streptococcus (34%), Moraxella (19%), Staphylococcus (10%), Burkholderia (9%), Neisseria (6%), Haemophilus (6%), and Janthinobacterium (5%) (Perez et al., 2017). Another cross-sectional study reported that infants with rhinovirus infection had a lower abundance of Streptococcus compared with infants with other viral infections (Toivonen et al., 2019). Rhinovirus A infection was associated with a Haemophilus-dominant microbiome profile and rhinovirus C infection with a Moraxella-dominant microbiome profile (Toivonen et al., 2019; Raita et al., 2021). Children with a Haemophilus-dominated microbiome profile were more often infected with rhinovirus only (compared with co-infection with other or several viruses) (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021). A further cross-sectional study reported that rhinovirus infection was associated with a higher abundance of Moraxella (Tozzi et al., 2021).
A prospective cohort study reported that compared with healthy children, children with an influenza infection had a higher diversity with a lower abundance of Moraxella, Staphylococcus, Corynebacterium, and Dolosigranulum, and a higher abundance of Phyllobacterium, Acinetobacter, unclassified Acidobacteria, Ralstonia, Pseudomonas, Lachnoclostridium, and Halomonas (Wen et al., 2018; Zhou et al., 2020). Five microbiome profiles were identified: Moraxella-,Streptococcus-, Staphylococcus-, Corynebacterium-, and Dolosigranulum-dominant microbiome profiles (Zhou et al., 2020).
Another prospective cohort study reported that children infected with alpha coronaviruses more often had a Haemophilus-dominated microbiome profile compared with children infected with beta coronaviruses (Fujiogi et al., 2021).
A prospective cohort study reported that compared to healthy children, children with Mycoplasma pneumoniae pneumonia had a lower diversity, more often a Staphylococcus-dominated microbiome profile and a higher abundance of Ralstonia and Acidobacteria (Zhou et al., 2020).
A cross-sectional study reported that pertussis infection was associated with a higher abundance of Alcaligenaceae and Achromobacter (Tozzi et al., 2021).
Invasive pneumococcal disease
Invasive pneumococcal disease poses a significant health threat, and understanding the nasopharyngeal microbiome composition in affected children can provide valuable insights into the disease. A prospective case-control study reported that compared with healthy children, children who suffered from an invasive pneumococcal disease were more frequently colonised with S. pneumoniae and had a lower abundance of D. pigrum and M. lincolnii in their nasopharynx (Henares et al., 2021).
Atopy
The relationship between the nasopharyngeal microbiome and the development of allergic conditions has attracted significant attention, and several studies have explored this association. A prospective cohort study reported that children with atopy had a higher abundance of Burkholderiaceae, Enterobacteriaceae, Sphingomonadaceae, Staphylococcaceae, and Xanthobacteraceae in their nasopharynx compared with healthy children (Aydin et al., 2021). A prospective case-control study found that children with allergic rhinoconjunctivitis had a higher diversity of their nasopharyngeal microbiome and that there was an association between increasing diversity and disease severity (Yau et al., 2019). A cross-sectional study observed a lower diversity and lower abundance of Corynebacterium and S. epidermidis in children with cat allergy and a lower abundance of Corynebacterium in children with dog allergy. The study did not find association between bacterial composition and pollen allergy (Chun et al., 2021).
In a retrospective cohort study, healthy children with an increased risk for developing allergies were reported to have a higher abundance of M. catarrhalis in their nasopharynx (Chapman et al., 2020).
Wheezing
The relationship between the nasopharyngeal microbiome and wheezing, a common respiratory symptom in children, has been a subject of extensive investigation. During acute wheezing, a prospective cohort study more frequently observed a S. pneumoniae- and less frequently a D. pigrum-dominated microbiome profile in the nasopharynx of children (Tang et al., 2021).
In a prospective cohort study, a lower abundance of Lactobacillus and Staphylococcus during RSV infection was associated with an increased rate of recurrent wheezing at the age of two years (Rosas-Salazar et al., 2018). In a randomised double-blinded, placebo-controlled trial, which evaluated the effect of azithromycin in infants hospitalised with RSV bronchiolitis, a higher abundance of Moraxella in the nasopharynx was associated with higher rates of recurrent wheezing in the future (Zhou et al., 2016).
A prospective cohort study reported a higher abundance of Proteobacteria, Burkholderiaceae, Enterobacteriaceae, Sphingomonadaceae, Staphylococcaceae, and Xanthobacteraceae, as well as a higher abundance of Haemophilus (H. influenzae), Moraxella (M. catarrhalis), S. aureus, and S. pneumoniae in the nasopharynx of children with chronic wheezing (Aydin et al., 2021).
A prospective birth cohort found an association between a high abundance of Streptococcus before first the first ARTI and the development of chronic wheezing (Teo et al., 2015). Another prospective cohort study reported that children with a higher abundance of Moraxella or Streptococcus three weeks after hospitalisation for bronchiolitis and a higher abundance of Streptococcus in summer had a higher risk for recurrent wheezing at three years of age (Mansbach et al., 2020).
Asthma
The nasopharyngeal microbiome has emerged as a significant factor in the development and progression of asthma in children. A prospective cohort study reported that compared with healthy children, children with asthma had a higher abundance of Firmicutes and Staphylococcaceae in their nasopharynx (Aydin et al., 2021). Another prospective cohort study reported that the most abundant genera in the nasopharynx of children with asthma were Moraxella (35%), Staphylococcus (14%), Dolosigranulum (9%), Corynebacterium (9%), Prevotella (6%), Streptococcus (5%), Haemophilus (4%), and Fusobacterium (3%) (Pérez-Losada et al., 2017). A third prospective cohort study reported that children with asthma had a stable composition of their nasopharyngeal microbiome over time despite viral infections or exacerbations. Six microbiome profiles were identified: Moraxella-, Staphylococcus-, Corynebacterium-, Streptococcus-, Dolosigranulum-, and Haemophilus-dominated profiles. Moraxella-dominated microbiome profiles were associated with an increased risk for exacerbation, while Staphylococcus- or Corynebacterium-dominated microbiome profiles were associated with reduced risk for respiratory illness and exacerbations (McCauley et al., 2019). A further prospective cohort study reported that the most abundant genera in the nasopharynx of children with asthma were Moraxella (28%), Staphylococcus (18%), Corynebacterium (10%), Dolosigranulum (8%), Prevotella (6%), Streptococcus (6%), Fusobacterium (3%), and Haemophilus (3%). The study found an association between the microbiome composition of the nasopharynx and the asthmatic phenotype. Children who were older when they were diagnosed with asthma, had persisting symptoms despite treatment or a higher body mass index (BMI) had a higher abundance of Corynebacterium and Prevotella and a lower abundance of Moraxella and Dolosigranulum. Children who were youngest when they were diagnosed, had a high rate of positive allergen tests, high blood eosinophil and immunoglobulin E levels and a high rate of needing inhaled corticosteroids had a lower diversity and a higher abundance of Moraxella and a lower abundance of Corynebacterium, Staphylococcus, and Prevotella. Children with a lower BMI, low rate of positive skin prick test and a better response to treatment with bronchodilators had an intermediate abundance of the main genera (Pérez-Losada et al., 2018). Another prospective cohort study, reported that a higher abundance of Moraxella and Haemophilus was associated with asthma exacerbations (McCauley et al., 2022). Furthermore, a further prospective cohort study a lower diversity was observed during an asthma exacerbation with a higher abundance of Moraxella. Additionally, metabolic pathways associated with Moraxella (methane, ketone bodies, and vitamin B3 metabolism) were enhanced during an exacerbation (Hou et al., 2022). In the same study, Dolosigranulum- and Corynebacterium 1-dominated microbiome profiles were more frequent at baseline and in healthy controls compared with children during an asthma exacerbation (Hou et al., 2022).
A high abundance of H. influenzae and S. pneumoniae during hospitalisation with severe bronchiolitis in infancy was associated with a higher risk of developing asthma at the age of five years, while a high abundance of M. nonliquefaciens was associated with lower risk (Raita et al., 2021). Children with reversible airway obstruction had a higher abundance of Haemophilus and S. pneumoniae and lower abundance of Moraxella, Corynebacterium, Dolosigranulum, and Staphylococcus at the age of six years (Man et al., 2020). A cross-sectional study reported a negative correlation between the abundance of Corynebacterium and S. epidermidis and the expression of genes involved in inflammatory processes (Chun et al., 2021).
Positive pressure ventilation
The composition of the nasopharyngeal microbiome in infants during hospitalization for bronchiolitis has been linked to the severity of the respiratory condition. In a cross-sectional study, infants requiring positive pressure ventilation exhibited distinct microbial profiles in their nasopharynx compared to those who did not require such intervention. A cross-sectional study reported that infants needing positive pressure ventilation during hospitalisation for bronchiolitis had a higher abundance of Haemophilus, Klebsiella, Rothia, and Streptococcus, and a lower abundance of M. catarrhalis in their nasopharyngeal microbiome. Sphingolipid metabolites were enriched in infants needing positive pressure ventilation and correlated to the abundance of S. pneumoniae (Stewart et al., 2017). The abundance of Streptococcus positively correlated with metabolites (glucuronate and 1-palmitoyl-2-palyitoleoyl-GPC 16:0/16:1) associated with a higher risk of needing positive pressure ventilation and negatively correlated with metabolites (plasmalogen sub-pathway) associated with a lower risk of needing positive pressure ventilation, for the abundance Moraxella an opposite correlation was found (Stewart et al., 2019).
Other factors
The composition of the nasopharyngeal microbiome in infants has been found to be influenced by various factors and has implications for their respiratory health. A prospective cohort study reported that infants with low vitamin D levels had a lower richness and diversity and a higher abundance of Staphylococcus in their nasopharyngeal microbiome. In infants with low vitamin D levels a Haemophilus-dominant microbiome profile was associated with a higher risk of intensive care admission during hospitalisation with bronchiolitis (Toivonen et al., 2018). A cross-sectional study reported that infants who were perinatally exposed to HIV had a higher abundance of Klebsiella in their nasopharynx (Kelly et al., 2017). A prospective birth cohort study reported an association between pacifier use and composition of the nasopharyngeal microbiome in the first 6 months of life without further specifying any details (Reyman et al., 2021). A randomised, placebo-controlled trial reported that children who were born preterm and had received palivizumab had a lower frequency of Staphylococcus-dominated microbiome profiles and a higher abundance of biomarker species, such as Klebsiella, as well as a more diverse set of oral taxa, including Streptococcus in their nasopharynx at one year of age. Furthermore, they had a higher abundance of Haemophilus and a lower abundance of Moraxella and Neisseriaceae at six years of age (Man et al., 2020).
Collection method
The collection method used to assess the nasopharyngeal microbiome in children can have a impact on the observed microbial composition. In children with asthma a higher diversity and a higher mean number of OTUs was found in samples taken by nasal brushes compared to nasal washes. Furthermore, a higher abundance of Bacteroides and Pseudomonas and a lower abundance of Haemophilus, Fusobacterium, Moraxella, Prevotella, Staphylococcus, Streptococcus, and Treponema was found in samples taken by nasal brushes (Pérez-Losada et al., 2016). A study which compared results from nasal filters with nasal washed found very similar results. The only genus that was differently abundant between the two collection methods was Sphingobium (Shilts et al., 2020).
Interaction between bacteria
An early presence and high abundance of Moraxella, Dolosigranulum, and Corynebacterium, as well as Moraxella-, Dolosigranulum-, Haemophilus-, and Streptococcus-dominated microbiome profiles have been reported to be associated with a more stable composition of the nasopharyngeal microbiome over time (Biesbroek et al., 2014b; Hou et al., 2022; Kelly et al., 2022). In contrast, a high abundance of Streptococcus, Haemophilus, and Bacteroidetes, and a H. influenzae-dominated microbiome profile have been reported to be associated with a less stable microbial composition over time (Biesbroek et al., 2014b; Bosch et al., 2016; Kelly et al., 2022). Colonisation with M. catarrhalis, S. pneumoniae, and H. influenzae have been associated with a lower bacterial diversity (Chonmaitree et al., 2017; Chapman et al., 2020).
A positive correlation has been found between the abundance of Dolosigranulum (D. pigrum) and Corynebacterium (C. pseudodiphteriticum) (Biesbroek et al., 2014a; Lappan et al., 2018; Coleman et al., 2021), Moraxella (Coleman et al., 2021), and Neisseriaceae (Coleman et al., 2021). A positive correlation has also been observed between the abundance of Streptococcus (S. pneumoniae) and Moraxella (Kelly et al., 2018; Boelsen et al., 2019), Haemophilus (Boelsen et al., 2019; Coleman et al., 2021), Corynebacterium (including C. accolens) (Salter et al., 2017), and Staphylococcus (Salter et al., 2017). Furthermore, a positive correlation between the abundance of Corynebacterium (including C. accolens) and Staphylococcus (Salter et al., 2017), and between the abundance of Ornithobacterium and Helococcus, Dichelobacter, and Cardiobacteriaceae has been reported (Coleman et al., 2021). Furthermore, a positive correlation has also been observed between the abundance of Haemophilus and Moraxella, and Gemella and Porphyromonas and Neisseria (Lappan et al., 2018), as well as between the abundance of Veillonella and Streptococcus, Prevotella and Alloprevotella (Figure 2) (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021). On species level, positive correlations between the abundance of M. nonliquefaciens and S. pneumoniae, H. influenzae and M. catarrhalis (Binia et al., 2021), and between Acinetobacter, and Streptococcus parasanguinis, Streptococcus salivarius, and Veillonella have been reported (Accorsi et al., 2020).
Negative correlations have been reported between the abundance of Corynebacterium (C. pseudodiphtheriticum/propinquum, C. accolens/macginleyi, and C. tuberculostearicum), Dolosigranulum, and Streptococcus (including S. pneumoniae) (Biesbroek et al., 2014b; Biesbroek et al., 2014a; Salter et al., 2017; Kelly et al., 2018; Kelly et al., 2022), Staphylococcus (Biesbroek et al., 2014b; Biesbroek et al., 2014a; Salter et al., 2017), Moraxella (Biesbroek et al., 2014a), Veillonella (Biesbroek et al., 2014a), Rothia (Biesbroek et al., 2014a), Granulicatella (Biesbroek et al., 2014a), Actinomyces (Biesbroek et al., 2014a) Prevotella (Biesbroek et al., 2014a), Gemella (Biesbroek et al., 2014a), Leptotrichia (Biesbroek et al., 2014a), Fusobacterium (Biesbroek et al., 2014a), and Klebsiella (Biesbroek et al., 2014a). A negative correlation has also been reported between the abundance of Streptococcus (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021) and Haemophilus (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021), Moraxella (Hasegawa et al., 2016; Hasegawa et al., 2017; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021), Staphylococcus (Salter et al., 2017; Kelly et al., 2018), and Pseudomonas (Figure 2) (Boelsen et al., 2019). Children colonised with S. pneumoniae have been reported to have a lower abundance of Actinomyces, Prevotella, Dolosigranulum, Veillonella, Corynebacterium_1, Gemella, and Anoxybacillus (Xu et al., 2021b; Xu et al., 2021a). On species level, a negative correlation has been observed between the abundance D. pigrum and the acquisition of S. aureus (Accorsi et al., 2020). Furthermore, children not colonised with S. pneumoniae, H. influenzae or M. catarrhalis have been reported to have a higher abundance of Lactococcus lactis subsp. cremoris, Cutibacterium acnes, Moraxella osloensis, Acinetobacter pittii, C. accolens, Staphylococcus hominis, Staphylococcus epidermidis, S. aureus, S. viridans, Staphylococcus haemolyticus, D. pigrum, and Staphylococcus arlettae (Binia et al., 2021).
Discussion
The findings of this comprehensive review show that the nasopharyngeal microbiome in children is dynamic and influenced by many extrinsic factors. Colonisation of the nasopharynx with certain bacteria is a risk factor for developing infections and other diseases, while other bacteria are protective (Salter et al., 2017). A high abundance of Haemophilus, Moraxella, and Streptococcus and a low abundance of Corynebacterium and Dolosigranlum are associated with respiratory tract infections (including AOM, bronchiolitis and pneumonia), wheezing and asthma exacerbations (Teo et al., 2015; Hasegawa et al., 2016; Rosas-Salazar et al., 2016a; Zhou et al., 2016; Chonmaitree et al., 2017; Hasegawa et al., 2017; Kelly et al., 2017; Pérez-Losada et al., 2017; Luna et al., 2018; Pérez-Losada et al., 2018; Teo et al., 2018; Toivonen et al., 2018; Boelsen et al., 2019; Man et al., 2019a; Stewart et al., 2019; Toivonen et al., 2019; Mansbach et al., 2020; Salgado et al., 2020; Aydin et al., 2021; Fujiogi et al., 2021; Tang et al., 2021). Importantly, this review also shows that the extrinsic factors identified as risk factors for these adverse health outcomes (Schuez-Havupalo et al., 2017; Sandall et al., 2018; Branger et al., 2022; Duong et al., 2022), are associated with the aforementioned changes in the nasopharyngeal microbiome. This includes being born by CS (Bosch et al., 2016; Shilts et al., 2016), not being breastfed (Biesbroek et al., 2014b; Shilts et al., 2016; Kelly et al., 2022), antibiotic exposure (Teo et al., 2015; Hasegawa et al., 2016; Thapa et al., 2020; Chonmaitree et al., 2017; Hasegawa et al., 2017; Salter et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021; Raita et al., 2021; Kelly et al., 2022), having siblings (Teo et al., 2015), and day-care attendance (Teo et al., 2015; Xu et al., 2021b; Xu et al., 2021a).
Understanding the dynamics, composition, and function of the nasopharyngeal microbiome in healthy children is a prerequisite to investigate the role of the microbiome in children with diseases. The presence or abundance of an individual bacteria affects those of others due to ecologic interactions. Research on the pathogenesis and prevention of diseases should, therefore, not only focus on pathogens but also on commensals, and even more importantly on interactions between bacterial communities, as well as interactions with the immune system and metabolism. In addition to identifying microbial composition, identification of microbial community function is likely even more important.
The interaction between bacteria within the microbiome can be direct, such as competition for nutrients or receptors (for example iron or epithelial bindings sites) or metabolites (antimicrobial products such as bacteriocins), or indirect trough other bacteria or the immune system (Hertli and Zimmermann, 2022). Several studies have found a negative association between the abundance of Corynebacterium and Streptococcus (S. pneumoniae) (Laufer et al., 2011; Biesbroek et al., 2014b; Biesbroek et al., 2014a; Teo et al., 2015; Bomar et al., 2016; Salter et al., 2017; Kelly et al., 2018; Kelly et al., 2022). C. accolens hydrolyses human triacylglycerols into free fatty acids which have antimicrobial activity and inhibit growth of S. pneumoniae (Bomar et al., 2016; Kelly et al., 2022). In vitro studies have also shown that C. propinquum and C. pseudodiphtheriticum inhibit growth of S. pneumoniae (Xu et al., 2021b). Corynebacterium is often found together with Dolosigranulum. Dolosigranulum is thought to lower the pH trough the production of lactic acids, creating a more favourable condition for Corynebacterium to grow (de Steenhuijsen Piters et al., 2015). A negative relationship has also been described between the abundance of Streptococcus and Staphylococcus (Salter et al., 2017; Kelly et al., 2018), more specifically between S. pneumoniae and S. aureus (Veenhoven et al., 2003; Bogaert et al., 2004; Regev-Yochay et al., 2004; McNally et al., 2006; Watson et al., 2006; Zemlicková et al., 2006; Madhi et al., 2007; Pettigrew et al., 2008). It has been hypothesised that S. pneumoniae inhibits the growth of S. aureus trough the production of hydrogen peroixide (Pericone et al., 2000; Regev-Yochay et al., 2006; Park et al., 2008). In vitro, hydrogen peroxide produced by S. pneumoniae has been shown to be bactericidal against H. influenzae, N. meningitidis, and M. catarrhalis (Pericone et al., 2000). However, in clinical studies, both negative (Madhi et al., 2007; Pettigrew et al., 2008) and positive (Jacoby et al., 2007; Pettigrew et al., 2008; Shiri et al., 2013; Feazel et al., 2015; Boelsen et al., 2019) correlations between the abundance of S. pneumoniae and H. influenzae have been described.
Most respiratory tract infections are polymicrobial and the bacteria involved in the infection influence each other. In AOM, for example, co-infection with H. influenzae and M. catarrhalis leads to an increased resistance of biofilms to antibiotics and to a decreased host clearance (Armbruster et al., 2010). Vaccination against one species (e.g. H. influenzae), might therefore also impact colonisation or infection with another species (e.g. M. catarrhalis), as the bacteria might be easier cleared by the host if not in a biofilm maintained by the other bacteria.
In addition to biofilm production, bacteria might also influence each other through interaction with the immune system. In infants, a Haemophilus-dominant microbiome profile, has been associated with upregulated T helper (TH)2 and TH17-type inflammatory response in the airways (Hasegawa et al., 2016). In a murine study, colonisation of the nasopharynx by either H. influenzae or S. pneumoniae results in persistent colonisation. However, co-administration of these bacteria leads to rapid clearance of S. pneumoniae from the nasopharynx. This clearance is likely attributed to enhanced opsonophagocytic killing, as components of H. influenzae stimulate complement-dependent phagocytic elimination of S. pneumoniae (Lysenko et al., 2005). Consequently, this raises the question of whether underlying host factors, such as immunity parameters, independently drive both outcomes, thereby compromising microbiome stability and increasing susceptibility to infections. Changes in the nasopharyngeal microbiome can be observed before ARTIs with a decrease of beneficial microbes and increased abundance of potential pathogenic bacteria with an influx of oral species into the nasopharyngeal (Man et al., 2019a).
Furthermore, there are also interactions between viruses and bacteria. Respiratory viruses disrupt the nasopharyngeal microbiome and lead to a higher abundance of Haemophilus, Moraxella, and Streptococcus (Rosas-Salazar et al., 2016a; Rosas-Salazar et al., 2016b; Ederveen et al., 2018; Toivonen et al., 2019; Raita et al., 2021; Tozzi et al., 2021). On the other hand, children colonised with Dolosigranulum have fewer RSV infections, especially RSV LRTIs (Teo et al., 2015). RSV has been reported to increase the virulence of S. pneumoniae (Smith et al., 2014) and influenza adhesion of S. pneumoniae to host cells (Avadhanula et al., 2006).
In addition to URTIs, the nasopharyngeal microbiome also influences the risk for LRTIs, including pneumonia (Sakwinska et al., 2014; Kelly et al., 2017; Lu et al., 2017; Dai et al., 2018). Pneumonia is the leading cause of childhood death globally and a leading cause of hospitalization in children in developed countries (World Health Association, [[NoYear]]; Jain et al., 2015). Children with pneumonia have lower diversity of their nasopharyngeal microbiome with a higher abundance of Moraxella, Haemophilus, Staphylococcus, and Streptococcus (Sakwinska et al., 2014; Kelly et al., 2017; Lu et al., 2017; Dai et al., 2018). Microbiome manipulations that enhance stability of the upper airway microbiome in infants could conceivably diminish childhood pneumonia.
A Cochrane meta-analysis shows that oral administration of probiotics (mainly Lactobacillus and Streptococcus-containing products) decreases the number of AOM and other respiratory tract infection in healthy children, but not in children with recurrent AOM (Scott et al., 2019). However, the range of species that have been tested is limited and includes mostly bacteria deriving from the intestine. Furthermore, the studies included in the review did not investigate local administration of probiotics to the nasopharynx. Probiotics are able to modulate the epithelial barrier function. For Lactobacillus, for example, it has been shown that it promotes the expression and regulation of tight junctions and adherence junctions, resulting in the restoration of a defective epithelial barrier. Furthermore, probiotic bacteria also interact with the immune system through pattern recognition receptors, such as Toll-like receptors, which upon activation can stimulate or suppress various immune responses (Martens et al., 2018). Further studies are needed to investigate, nasopharyngeal microbiome-manipulation in prevention and treatment of respiratory disease.
In the studies included in this review, antibiotic exposure has been associated with a lower abundance of bacteria in the nasopharynx which have been associated with respiratory health such as Dolosigranulum (Teo et al., 2015; Bosch et al., 2017; Chonmaitree et al., 2017), and Corynebacterium (Teo et al., 2015; Bosch et al., 2017; Chonmaitree et al., 2017; Kelly et al., 2022), but an increase in genera associated with ARTI such as Haemophilus (Teo et al., 2015; Hasegawa et al., 2016; Thapa et al., 2020; Hasegawa et al., 2017; Luna et al., 2018; Toivonen et al., 2018; Toivonen et al., 2019; Fujiogi et al., 2021; Raita et al., 2021; Kelly et al., 2022), and Moraxella (Teo et al., 2015; Kelly et al., 2022). However, there are other studies which show that children treated with erythromycin are less commonly colonised by S. pneumoniae and M. catarrhalis, while colonisation by H. influenzae remains unchanged (Sundberg et al., 1984). Furthermore, even a single dose of ceftriaxone has been shown to significantly decrease the abundance of M. catarrhalis, H. influenzae, and S. pneumoniae in the nasopharynx (Heikkinen et al., 2000), and ten days of amoxicillin/clavulanate leads to an even greater decrease in the abundance of M. catarrhalis and H. influenzae (Cohen et al., 1999). As with the intestinal microbiome (Zimmermann and Curtis, 2019).
the effect of antibiotics on the nasopharyngeal microbiome likely depends on the spectrum of activity, route of administration, as well as dose and duration of administration.
Conclusions and future directions
The association between the composition of the nasopharyngeal microbiome and health outcomes in children is intriguing and highlights the potential of the nasopharyngeal microbiome as a marker for identifying children at risk for disease and even more importantly, as an avenue for targeted interventions and preventive strategies.
Future studies should include larger numbers of participants and longitudinal follow-up. Samples need to be sequenced sufficiently deep and/or combine short- with long-read sequencing approaches. Standardising methods and reporting will help comparison and pooling of results from different studies. This will enable stronger quantitative analysis of the contribution of each risk factor.
Limitations
One of the main limitations of current studies that have investigating the nasopharyngeal microbiome in children is the large overlap of participants (Bogaert et al., 2011; Biesbroek et al., 2014b; Biesbroek et al., 2014c; Biesbroek et al., 2014a; Teo et al., 2015; Bosch et al., 2016; Hasegawa et al., 2016; Pérez-Losada et al., 2016; Rosas-Salazar et al., 2016a; Rosas-Salazar et al., 2016b; Bosch et al., 2017; Hasegawa et al., 2017; Kelly et al., 2017; Lu et al., 2017; Pérez-Losada et al., 2017; Stewart et al., 2017; Dai et al., 2018; Kelly et al., 2018; Luna et al., 2018; Pérez-Losada et al., 2018; Rosas-Salazar et al., 2018; Teo et al., 2018; Toivonen et al., 2018; Wen et al., 2018; Man et al., 2019a; Mansbach et al., 2019; Stewart et al., 2019; Toivonen et al., 2019; Mansbach et al., 2020; Zhou et al., 2020; Fujiogi et al., 2021; Raita et al., 2021; Reyman et al., 2021; Xu et al., 2021b; Xu et al., 2021a; Tan et al., 2023). Furthermore, many of the studies were underpowered and might therefore not have identified factors that influence the composition of the nasopharyngeal microbiome. Furthermore, as with all microbiome studies, it is difficult to account for effect modifiers. For example, differences in the nasopharyngeal microbiome between breastfed and formula-fed infants might contribute to the protective effect of breastfeeding from respiratory tract infections (Biesbroek et al., 2014a). However, formula-fed infants more often have older siblings and are more often exposed to tobacco smoke (Biesbroek et al., 2014a). Similarly, infants born by CS, are more often exposed to antibiotics. Moreover, geographic location will likely also influence the composition of the nasopharyngeal microbiome.
Other limitations include those associated with diagnostics. Results obtained by metagenomic sequencing are influenced by sampling methods, DNA extraction, library preparation, as well as by the choice of primers, sequencing method and platform, bioinformatic pipelines and tools. In 16S rRNA sequencing, usually only a short segment of the gene is sequenced, which can lead to incomplete taxonomic identification. The choice of variable (V) region can significantly impact results and interpretation of microbial community analysis. Different V regions have varying levels of sequence conservation and discriminatory power, leading to differences in taxonomic resolution and the ability to detect specific microbial taxa. Identification by the use of references databases relieson previously sequenced bacteria, hence the use of different databases also influences results It is known that Dolosigranulum is misclassified as Alloiococcus in the GreenGenes database and therefore Dolosigranulum was used throughout the manuscript.
These limitations partly explain why some studies report conflicting results (as summarised in Table 1 and Figure 2) and that further research to clarify the association between different components of the nasopharyngeal microbiome and external factors are needed.
Author contributions
PZ did the literature research, wrote the manuscript, and designed the figures.
Funding
PZ is supported by grants from the Swiss National Foundation (PZPGP3_193140), the University of Fribourg and the Fribourg Hospital, Switzerland. None of the funders had a role in designing the study or in the study conduct.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author PZ declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frmbi.2023.1231271/full#supplementary-material
Supplementary Table 1 | Summary of findings of studies investigating the nasopharyngeal microbiome in children using next generation sequencing.
References
Accorsi E. K., Franzosa E. A., Hsu T., Joice Cordy R., Maayan-Metzger A., Jaber H., et al. (2020). Determinants of Staphylococcus aureus carriage in the developing infant nasal microbiome. Genome Biol. 21 (1), 301. doi: 10.1186/s13059-020-02209-7
Armbruster C. E., Hong W., Pang B., Weimer K. E., Juneau R. A., Turner J., et al. (2010). Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1 (3). doi: 10.1128/mBio.00102-10
Avadhanula V., Rodriguez C. A., Devincenzo J. P., Wang Y., Webby R. J., Ulett G. C., et al. (2006). Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J. Virol. 80 (4), 1629–1636. doi: 10.1128/jvi.80.4.1629-1636.2006
Aydin M., Weisser C., Rué O., Mariadassou M., Maaß S., Behrendt A. K., et al. (2021). The rhinobiome of exacerbated wheezers and asthmatics: insights from a German pediatric exacerbation network. Front. Allergy 2, 667562. doi: 10.3389/falgy.2021.667562
Biesbroek G., Bosch A. A., Wang X., Keijser B. J., Veenhoven R. H., Sanders E. A., et al. (2014a). The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 190 (3), 298–308. doi: 10.1164/rccm.201401-0073OC
Biesbroek G., Tsivtsivadze E., Sanders E. A., Montijn R., Veenhoven R. H., Keijser B. J., et al. (2014b). Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 190 (11), 1283–1292. doi: 10.1164/rccm.201407-1240OC
Biesbroek G., Wang X., Keijser B. J., Eijkemans R. M., Trzciński K., Rots N. Y., et al. (2014c). Seven-valent pneumococcal conjugate vaccine and nasopharyngeal microbiota in healthy children. Emerging Infect. Dis. 20 (2), 201–210. doi: 10.3201/eid2002.131220
Binia A., Siegwald L., Sultana S., Shevlyakova M., Lefebvre G., Foata F., et al. (2021). The influence of FUT2 and FUT3 polymorphisms and nasopharyngeal microbiome on respiratory infections in breastfed Bangladeshi infants from the microbiota and health study. mSphere 6 (6), e0068621. doi: 10.1128/mSphere.00686-21
Boelsen L. K., Dunne E. M., Mika M., Eggers S., Nguyen C. D., Ratu F. T., et al. (2019). The association between pneumococcal vaccination, ethnicity, and the nasopharyngeal microbiota of children in Fiji. Microbiome 7 (1), 106. doi: 10.1186/s40168-019-0716-4
Bogaert D., Keijser B., Huse S., Rossen J., Veenhoven R., van Gils E., et al. (2011). Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One 6 (2), e17035. doi: 10.1371/journal.pone.0017035
Bogaert D., van Belkum A., Sluijter M., Luijendijk A., de Groot R., Rümke H. C., et al. (2004). Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363 (9424), 1871–1872. doi: 10.1016/s0140-6736(04)16357-5
Bomar L., Brugger S. D., Yost B. H., Davies S. S., Lemon K. P. (2016). Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7 (1), e01725–e01715. doi: 10.1128/mBio.01725-15
Bosch A., de Steenhuijsen Piters W. A. A., van Houten M. A., Chu M., Biesbroek G., Kool J., et al. (2017). Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am. J. Respir. Crit. Care Med. 196 (12), 1582–1590. doi: 10.1164/rccm.201703-0554OC
Bosch A., Levin E., van Houten M. A., Hasrat R., Kalkman G., Biesbroek G., et al. (2016). Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9, 336–345. doi: 10.1016/j.ebiom.2016.05.031
Branger B., Bainier A., Martin L., Darviot E., Forgeron A., Sarthou L., et al. (2022). Breastfeeding and respiratory, ear and gastro-intestinal infections, in children, under the age of one year, admitted through the paediatric emergency departments of five hospitals. Front. Pediatr. 10, 1053473. doi: 10.3389/fped.2022.1053473
Camarinha-Silva A., Jáuregui R., Pieper D. H., Wos-Oxley M. L. (2012). The temporal dynamics of bacterial communities across human anterior nares. Environ. Microbiol. Rep. 4 (1), 126–132. doi: 10.1111/j.1758-2229.2011.00313.x
Chapman T. J., Morris M. C., Xu L., Pichichero M. E. (2020). Nasopharyngeal colonization with pathobionts is associated with susceptibility to respiratory illnesses in young children. PLoS One 15 (12), e0243942. doi: 10.1371/journal.pone.0243942
Chonmaitree T., Jennings K., Golovko G., Khanipov K., Pimenova M., Patel J. A., et al. (2017). Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One 12 (7), e0180630. doi: 10.1371/journal.pone.0180630
Chun Y., Do A., Grishina G., Arditi Z., Ribeiro V., Grishin A., et al. (2021). The nasal microbiome, nasal transcriptome, and pet sensitization. J. Allergy Clin. Immunol. 148 (1), 244–249.e4. doi: 10.1016/j.jaci.2021.01.031
Cleary D. W., Clarke S. C. (2017). The nasopharyngeal microbiome. Emerg. Top. Life Sci. 1 (4), 297–312. doi: 10.1042/etls20170041
Cohen R., Navel M., Grunberg J., Boucherat M., Geslin P., Derriennic M., et al. (1999). One dose ceftriaxone vs. ten days of amoxicillin/clavulanate therapy for acute otitis media: clinical efficacy and change in nasopharyngeal flora. Pediatr. Infect. Dis. J. 18 (5), 403–409. doi: 10.1097/00006454-199905000-00002
Coleman A., Zaugg J., Wood A., Cottrell K., Håkansson E. G., Adams J., et al. (2021). Upper respiratory tract microbiome of Australian aboriginal and torres strait islander children in ear and nose health and disease. Microbiol. Spectr. 9 (2), e0036721. doi: 10.1128/Spectrum.00367-21
Dai W., Wang H., Zhou Q., Feng X., Lu Z., Li D., et al. (2018). The concordance between upper and lower respiratory microbiota in children with Mycoplasma pneumoniae pneumonia. Emerg. Microbes Infect. 7 (1), 92. doi: 10.1038/s41426-018-0097-y
de Steenhuijsen Piters W. A., Sanders E. A., Bogaert D. (2015). The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 370 (1675), 20140294. doi: 10.1098/rstb.2014.0294
Duong Q. A., Pittet L. F., Curtis N., Zimmermann P. (2022). Antibiotic exposure and adverse long-term health outcomes in children: a systematic review and meta-analysis. J. Infect. 117 (6), 1297–307. doi: 10.1016/j.jinf.2022.01.005
Ederveen T. H. A., Ferwerda G., Ahout I. M., Vissers M., de Groot R., Boekhorst J., et al. (2018). Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome 6 (1), 10. doi: 10.1186/s40168-017-0395-y
Elling C. L., Scholes M. A., Streubel S. O., Larson E. D., Wine T. M., Bootpetch T. C., et al. (2021). The FUT2 variant c.461G>A (p.Trp154*) is associated with differentially expressed genes and nasopharyngeal microbiota shifts in patients with otitis media. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.798246
Enoksson F., Ruiz Rodriguez A., Peno C., Balcazar Lopez C., Tjernström F., Bogaert D., et al. (2020). Niche- and gender-dependent immune reactions in relation to the microbiota profile in pediatric patients with otitis media with effusion. Infection Immun. 88 (10), e00147-20. doi: 10.1128/iai.00147-20
Feazel L. M., Santorico S. A., Robertson C. E., Bashraheil M., Scott J. A., Frank D. N., et al. (2015). Effects of vaccination with 10-valent pneumococcal non-typeable haemophilus influenza protein D conjugate vaccine (PHiD-CV) on the nasopharyngeal microbiome of Kenyan toddlers. PLoS One 10 (6), e0128064. doi: 10.1371/journal.pone.0128064
Folino F., Fattizzo M., Ruggiero L., Oriano M., Aliberti S., Blasi F., et al. (2021). Nasopharyngeal microbiota analysis in healthy and otitis-prone children: focus on history of spontaneous tympanic membrane perforation. Pediatr. Infect. Dis. J. 40 (1), 16–21. doi: 10.1097/inf.0000000000002895
Fujiogi M., Camargo C. A. Jr., Raita Y., Toivonen L., Freishtat R. J., Mansbach J. M., et al. (2021). Association of endemic coronaviruses with nasopharyngeal metabolome and microbiota among infants with severe bronchiolitis: a prospective multicenter study. Pediatr. Res. 89 (7), 1594–1597. doi: 10.1038/s41390-020-01154-2
Haro K., Ogawa M., Saito M., Kusuhara K., Fukuda K. (2020). Bacterial composition of nasal discharge in children based on highly accurate 16S rRNA gene sequencing analysis. Sci. Rep. 10 (1), 20193. doi: 10.1038/s41598-020-77271-z
Hasegawa K., Mansbach J. M., Ajami N. J., Espinola J. A., Henke D. M., Petrosino J. F., et al. (2016). Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur. Respir. J. 48 (5), 1329–1339. doi: 10.1183/13993003.00152-2016
Hasegawa K., Mansbach J. M., Ajami N. J., Petrosino J. F., Freishtat R. J., Teach S. J., et al. (2017). The relationship between nasopharyngeal CCL5 and microbiota on disease severity among infants with bronchiolitis. Allergy 72 (11), 1796–1800. doi: 10.1111/all.13160
Heikkinen T., Saeed K. A., McCormick D. P., Baldwin C., Reisner B. S., Chonmaitree T. (2000). A single intramuscular dose of ceftriaxone changes nasopharyngeal bacterial flora in children with acute otitis media. Acta paediatrica (Oslo Norway 1992) 89 (11), 1316–1321. doi: 10.1080/080352500300002499
Henares D., Brotons P., de Sevilla M. F., Fernandez-Lopez A., Hernandez-Bou S., Perez-Argüello A., et al. (2021). Differential nasopharyngeal microbiota composition in children according to respiratory health status. Microb. Genom 7 (10), 000661. doi: 10.1099/mgen.0.000661
Hertli S., Zimmermann P. (2022). Molecular interactions between the intestinal microbiota and the host. Mol. Microbiol. 117 (6), 1297–1307. doi: 10.1111/mmi.14905
Hilty M., Qi W., Brugger S. D., Frei L., Agyeman P., Frey P. M., et al. (2012). Nasopharyngeal microbiota in infants with acute otitis media. J. Infect. Dis. 205 (7), 1048–1055. doi: 10.1093/infdis/jis024
Hou J., Song Y., Leung A. S. Y., Tang M. F., Shi M., Wang E. Y., et al. (2022). Temporal dynamics of the nasopharyngeal microbiome and its relationship with childhood asthma exacerbation. Microbiol. Spectr. 10 (3), e0012922. doi: 10.1128/spectrum.00129-22
Jacoby P., Watson K., Bowman J., Taylor A., Riley T. V., Smith D. W., et al. (2007). Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine 25 (13), 2458–2464. doi: 10.1016/j.vaccine.2006.09.020
Jain S., Williams D. J., Arnold S. R., Ampofo K., Bramley A. M., Reed C., et al. (2015). Community-acquired pneumonia requiring hospitalization among U. S. children. N Engl. J. Med. 372 (9), 835–845. doi: 10.1056/NEJMoa1405870
Jervis-Bardy J., Rogers G. B., Morris P. S., Smith-Vaughan H. C., Nosworthy E., Leong L. E., et al. (2015). The microbiome of otitis media with effusion in Indigenous Australian children. Int. J. Pediatr. otorhinolaryngology 79 (9), 1548–1555. doi: 10.1016/j.ijporl.2015.07.013
Kelly M. S., Plunkett C., Yu Y., Aquino J. N., Patel S. M., Hurst J. H., et al. (2022). Non-diphtheriae Corynebacterium species are associated with decreased risk of pneumococcal colonization during infancy. ISME J. 16 (3), 655–665. doi: 10.1038/s41396-021-01108-4
Kelly M. S., Surette M. G., Smieja M., Pernica J. M., Rossi L., Luinstra K., et al. (2017). The nasopharyngeal microbiota of children with respiratory infections in Botswana. Pediatr. Infect. Dis. J. 36 (9), e211–e218. doi: 10.1097/inf.0000000000001607
Kelly M. S., Surette M. G., Smieja M., Rossi L., Luinstra K., Steenhoff A. P., et al. (2018). Pneumococcal colonization and the nasopharyngeal microbiota of children in Botswana. Pediatr. Infect. Dis. J. 37 (11), 1176–1183. doi: 10.1097/inf.0000000000002174
Kinabo G. D., van der Ven A., Msuya L. J., Shayo A. M., Schimana W., Ndaro A., et al. (2013). Dynamics of nasopharyngeal bacterial colonisation in HIV-exposed young infants in Tanzania. Trop. Med. Int. Health 18 (3), 286–295. doi: 10.1111/tmi.12057
Lappan R., Imbrogno K., Sikazwe C., Anderson D., Mok D., Coates H., et al. (2018). A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 18 (1), 13. doi: 10.1186/s12866-018-1154-3
Laufer A. S., Metlay J. P., Gent J. F., Fennie K. P., Kong Y., Pettigrew M. M. (2011). Microbial communities of the upper respiratory tract and otitis media in children. mBio 2 (1), e00245–e00210. doi: 10.1128/mBio.00245-10
Liu Q., Liu Q., Meng H., Lv H., Liu Y., Liu J., et al. (2020). Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27 (1), 68–78.e5. doi: 10.1016/j.chom.2019.11.003
Lu Z., Dai W., Liu Y., Zhou Q., Wang H., Li D., et al. (2017). The alteration of nasopharyngeal and oropharyngeal microbiota in children with MPP and non-MPP. Genes (Basel) 8 (12), 380. doi: 10.3390/genes8120380
Luna P. N., Hasegawa K., Ajami N. J., Espinola J. A., Henke D. M., Petrosino J. F., et al. (2018). The association between anterior nares and nasopharyngeal microbiota in infants hospitalized for bronchiolitis. Microbiome 6 (1), 2. doi: 10.1186/s40168-017-0385-0
Lysenko E. S., Ratner A. J., Nelson A. L., Weiser J. N. (2005). The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PloS Pathog. 1 (1), e1. doi: 10.1371/journal.ppat.0010001
Madhi S. A., Adrian P., Kuwanda L., Cutland C., Albrich W. C., Klugman K. P. (2007). Long-term effect of pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae–and associated interactions with Staphylococcus aureus and Haemophilus influenzae colonization–in HIV-Infected and HIV-uninfected children. J. Infect. Dis. 196 (11), 1662–1666. doi: 10.1086/522164
Man W. H., Clerc M., de Steenhuijsen Piters W. A. A., van Houten M. A., Chu M., Kool J., et al. (2019a). Loss of microbial topography between oral and nasopharyngeal microbiota and development of respiratory infections early in life. Am. J. Respir. Crit. Care Med. 200 (6), 760–770. doi: 10.1164/rccm.201810-1993OC
Man W. H., Scheltema N. M., Clerc M., van Houten M. A., Nibbelke E. E., Achten N. B., et al. (2020). Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: a subanalysis of the MAKI randomised controlled trial. Lancet Respir. Med. 8 (10), 1022–1031. doi: 10.1016/s2213-2600(19)30470-9
Man W. H., van Dongen T. M. A., Venekamp R. P., Pluimakers V. G., Chu M., van Houten M. A., et al. (2019b). Respiratory microbiota predicts clinical disease course of acute otorrhea in children with tympanostomy tubes. Pediatr. Infect. Dis. J. 38 (6), e116–e125. doi: 10.1097/inf.0000000000002215
Man W. H., van Houten M. A., Mérelle M. E., Vlieger A. M., Chu M., Jansen N. J. G., et al. (2019c). Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. Lancet Respir. Med. 7 (5), 417–426. doi: 10.1016/s2213-2600(18)30449-1
Mansbach J. M., Hasegawa K., Piedra P. A., Avadhanula V., Petrosino J. F., Sullivan A. F., et al. (2019). Haemophilus-dominant nasopharyngeal microbiota is associated with delayed clearance of respiratory syncytial virus in infants hospitalized for bronchiolitis. J. Infect. Dis. 219 (11), 1804–1808. doi: 10.1093/infdis/jiy741
Mansbach J. M., Luna P. N., Shaw C. A., Hasegawa K., Petrosino J. F., Piedra P. A., et al. (2020). Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J. Allergy Clin. Immunol. 145 (2), 518–527.e8. doi: 10.1016/j.jaci.2019.10.034
Martens K., Pugin B., De Boeck I., Spacova I., Steelant B., Seys S. F., et al. (2018). Probiotics for the airways: Potential to improve epithelial and immune homeostasis. Allergy 73 (10), 1954–1963. doi: 10.1111/all.13495
McCauley K. E., DeMuri G., Lynch K., Fadrosh D. W., Santee C., Nagalingam N. N., et al. (2021). Moraxella-dominated pediatric nasopharyngeal microbiota associate with upper respiratory infection and sinusitis. PloS One 16 (12), e0261179. doi: 10.1371/journal.pone.0261179
McCauley K., Durack J., Valladares R., Fadrosh D. W., Lin D. L., Calatroni A., et al. (2019). Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J. Allergy Clin. Immunol. 144 (5), 1187–1197. doi: 10.1016/j.jaci.2019.05.035
McCauley K. E., Flynn K., Calatroni A., DiMassa V., LaMere B., Fadrosh D. W., et al. (2022). Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J. Allergy Clin. Immunol. 150 (1), 204–13. doi: 10.1016/j.jaci.2022.01.020
McNally L. M., Jeena P. M., Gajee K., Sturm A. W., Tomkins A. M., Coovadia H. M., et al. (2006). Lack of association between the nasopharyngeal carriage of Streptococcus pneumoniae and Staphylococcus aureus in HIV-1-infected South African children. J. Infect. Dis. 194 (3), 385–390. doi: 10.1086/505076
Mika M., Mack I., Korten I., Qi W., Aebi S., Frey U., et al. (2015). Dynamics of the nasal microbiota in infancy: a prospective cohort study. J. Allergy Clin. Immunol. 135 (4), 905–912.e11. doi: 10.1016/j.jaci.2014.12.1909
Page M. J., McKenzie J. E., Bossuyt P. M., Boutron I., Hoffmann T. C., Mulrow C. D., et al. (2021). The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ (Clinical Res. ed) 372, n71. doi: 10.1136/bmj.n71
Park B., Nizet V., Liu G. Y. (2008). Role of Staphylococcus aureus catalase in niche competition against Streptococcus pneumoniae. J. bacteriology 190 (7), 2275–2278. doi: 10.1128/jb.00006-08
Perez G. F., Pérez-Losada M., Isaza N., Rose M. C., Colberg-Poley A. M., Nino G. (2017). Nasopharyngeal microbiome in premature infants and stability during rhinovirus infection. J. Investig. Med. 65 (6), 984–990. doi: 10.1136/jim-2017-000414
Pérez-Losada M., Alamri L., Crandall K. A., Freishtat R. J. (2017). Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS One 12 (1), e0170543. doi: 10.1371/journal.pone.0170543
Pérez-Losada M., Authelet K. J., Hoptay C. E., Kwak C., Crandall K. A., Freishtat R. J. (2018). Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome 6 (1), 179. doi: 10.1186/s40168-018-0564-7
Pérez-Losada M., Crandall K. A., Freishtat R. J. (2016). Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome 4 (1), 25. doi: 10.1186/s40168-016-0170-5
Pericone C. D., Overweg K., Hermans P. W., Weiser J. N. (2000). Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infection Immun. 68 (7), 3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000
Pettigrew M. M., Gent J. F., Revai K., Patel J. A., Chonmaitree T. (2008). Microbial interactions during upper respiratory tract infections. Emerging Infect. Dis. 14 (10), 1584–1591. doi: 10.3201/eid1410.080119
Phillips B., Ball C., Sackett D., Badenoch D., Straus S., Haynes B., et al. Oxford Centre for Evidence-Based Medicine: Levels of Evidence 2009. Available at: https://www.cebm.ox.ac.uk/resources/levels-of-evidence/oxford-centre-for-evidence-based-medicine-levels-of-evidence-march-2009.
Raita Y., Pérez-Losada M., Freishtat R. J., Hahn A., Castro-Nallar E., Ramos-Tapia I., et al. (2021). Nasopharyngeal metatranscriptome profiles of infants with bronchiolitis and risk of childhood asthma: a multicentre prospective study. Eur. Respir. J. 60 (1), 2102293. doi: 10.1183/13993003.02293-2021
Regev-Yochay G., Dagan R., Raz M., Carmeli Y., Shainberg B., Derazne E., et al. (2004). Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. Jama 292 (6), 716–720. doi: 10.1001/jama.292.6.716
Regev-Yochay G., Trzcinski K., Thompson C. M., Malley R., Lipsitch M. (2006). Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. bacteriology 188 (13), 4996–5001. doi: 10.1128/jb.00317-06
Reyman M., Clerc M., van Houten M. A., Arp K., Chu M., Hasrat R., et al. (2021). Microbial community networks across body sites are associated with susceptibility to respiratory infections in infants. Commun. Biol. 4 (1), 1233. doi: 10.1038/s42003-021-02755-1
Rosas-Salazar C., Shilts M. H., Tovchigrechko A., Schobel S., Chappell J. D., Larkin E. K., et al. (2016a). Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J. Infect. Dis. 214 (12), 1924–1928. doi: 10.1093/infdis/jiw456
Rosas-Salazar C., Shilts M. H., Tovchigrechko A., Chappell J. D., Larkin E. K., Nelson K. E., et al. (2016b). Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am. J. Respir. Crit. Care Med. 193 (10), 1180–1183. doi: 10.1164/rccm.201512-2350LE
Rosas-Salazar C., Shilts M. H., Tovchigrechko A., Schobel S., Chappell J. D., Larkin E. K., et al. (2018). Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J. Allergy Clin. Immunol. 142 (5), 1447–1456.e9. doi: 10.1016/j.jaci.2017.10.049
Sakwinska O., Bastic Schmid V., Berger B., Bruttin A., Keitel K., Lepage M., et al. (2014). Nasopharyngeal microbiota in healthy children and pneumonia patients. J. Clin. Microbiol. 52 (5), 1590–1594. doi: 10.1128/jcm.03280-13
Salgado V. R., Fukutani K. F., Fukutani E., Lima J. V., Rossi E. A., Barral A., et al. (2020). Effects of 10-valent pneumococcal conjugate (PCV10) vaccination on the nasopharyngeal microbiome. Vaccine 38 (6), 1436–1443. doi: 10.1016/j.vaccine.2019.11.079
Salter S. J., Turner C., Watthanaworawit W., de Goffau M. C., Wagner J., Parkhill J., et al. (2017). A longitudinal study of the infant nasopharyngeal microbiota: The effects of age, illness and antibiotic use in a cohort of South East Asian children. PloS Negl. Trop. Dis. 11 (10), e0005975. doi: 10.1371/journal.pntd.0005975
Sandall J., Tribe R. M., Avery L., Mola G., Visser G. H., Homer C. S., et al. (2018). Short-term and long-term effects of caesarean section on the health of women and children. Lancet 392 (10155), 1349–1357. doi: 10.1016/s0140-6736(18)31930-5
Schuez-Havupalo L., Toivonen L., Karppinen S., Kaljonen A., Peltola V. (2017). Daycare attendance and respiratory tract infections: a prospective birth cohort study. BMJ Open 7 (9), e014635. doi: 10.1136/bmjopen-2016-014635
Scott A. M., Clark J., Julien B., Islam F., Roos K., Grimwood K., et al. (2019). Probiotics for preventing acute otitis media in children. Cochrane Database systematic Rev. 6 (6), Cd012941. doi: 10.1002/14651858.CD012941.pub2
Shilts M. H., Rosas-Salazar C., Lynch C. E., Tovchigrechko A., Boone H. H., Russell P. B., et al. (2020). Evaluation of the upper airway microbiome and immune response with nasal epithelial lining fluid absorption and nasal washes. Sci. Rep. 10 (1), 20618. doi: 10.1038/s41598-020-77289-3
Shilts M. H., Rosas-Salazar C., Tovchigrechko A., Larkin E. K., Torralba M., Akopov A., et al. (2016). Minimally invasive sampling method identifies differences in taxonomic richness of nasal microbiomes in young infants associated with mode of delivery. Microb. Ecol. 71 (1), 233–242. doi: 10.1007/s00248-015-0663-y
Shiri T., Nunes M. C., Adrian P. V., Van Niekerk N., Klugman K. P., Madhi S. A. (2013). Interrelationship of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus colonization within and between pneumococcal-vaccine naïve mother-child dyads. BMC Infect. Dis. 13, 483. doi: 10.1186/1471-2334-13-483
Smith C. M., Sandrini S., Datta S., Freestone P., Shafeeq S., Radhakrishnan P., et al. (2014). Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am. J. Respir. Crit. Care Med. 190 (2), 196–207. doi: 10.1164/rccm.201311-2110OC
Stearns J. C., Davidson C. J., McKeon S., Whelan F. J., Fontes M. E., Schryvers A. B., et al. (2015). Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 9 (5), 1268. doi: 10.1038/ismej.2015.49
Stewart C. J., Mansbach J. M., Ajami N. J., Petrosino J. F., Zhu Z., Liang L., et al. (2019). Serum metabolome is associated with the nasopharyngeal microbiota and disease severity among infants with bronchiolitis. J. Infect. Dis. 219 (12), 2005–2014. doi: 10.1093/infdis/jiz021
Stewart C. J., Mansbach J. M., Wong M. C., Ajami N. J., Petrosino J. F., Camargo C. A. Jr, et al. (2017). Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A multiomic analysis. Am. J. Respir. Crit. Care Med. 196 (7), 882–891. doi: 10.1164/rccm.201701-0071OC
Sundberg L., Cederberg A., Edén T., Ernstson S. (1984). The effect of erythromycin on the nasopharyngeal pathogens in children with secretory otitis media. Acta Otolaryngol 97 (3-4), 379–383. doi: 10.3109/00016488409131004
Tan Y., Shilts M. H., Rosas-Salazar C., Puri V., Fedorova N., Halpin R. A., et al. (2023). Influence of sex on respiratory syncytial virus genotype infection frequency and nasopharyngeal microbiome. J. Virol. 97 (3), e0147222. doi: 10.1128/jvi.01472-22
Tang H. H. F., Lang A., Teo S. M., Judd L. M., Gangnon R., Evans M. D., et al. (2021). Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J. Allergy Clin. Immunol. 147 (5), 1683–1691. doi: 10.1016/j.jaci.2020.10.009
Teo S. M., Mok D., Pham K., Kusel M., Serralha M., Troy N., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17 (5), 704–715. doi: 10.1016/j.chom.2015.03.008
Teo S. M., Tang H. H. F., Mok D., Judd L. M., Watts S. C., Pham K., et al. (2018). Airway microbiota dynamics uncover a critical window for interplay of pathogenic bacteria and allergy in childhood respiratory disease. Cell Host Microbe 24 (3), 341–352.e5. doi: 10.1016/j.chom.2018.08.005
Thapa S., Runge J. K., Venkatachalam A., Denne C., Luna R. A., Anon J. B.. (2020). The Nasopharyngeal and Gut Microbiota in Children in a Pediatric Otolaryngology Practice. Pediatr Infect Dis J. 39 (9), e226–e33.
Toivonen L., Camargo C. A. Jr., Gern J. E., Bochkov Y. A., Mansbach J. M., Piedra P. A., et al. (2019). Association between rhinovirus species and nasopharyngeal microbiota in infants with severe bronchiolitis. J. Allergy Clin. Immunol. 143 (5), 1925–1928.e7. doi: 10.1016/j.jaci.2018.12.1004
Toivonen L., Hasegawa K., Ajami N. J., Celedón J. C., Mansbach J. M., Petrosino J. F., et al. (2018). Circulating 25-hydroxyvitamin D, nasopharyngeal microbiota, and bronchiolitis severity. Pediatr. Allergy Immunol. 29 (8), 877–880. doi: 10.1111/pai.12977
Tozzi A. E., Del Chierico F., Pandolfi E., Reddel S., Gesualdo F., Gardini S., et al. (2021). Nasopharyngeal microbiota in hospitalized children with Bordetella pertussis and Rhinovirus infection. Sci. Rep. 11 (1), 22858. doi: 10.1038/s41598-021-02322-y
van Meel E. R., Jaddoe V. W. V., Looman K. I. M., de Jongste J. C., Moll H. A., Duijts L. (2020). Airway bacterial carriage and childhood respiratory health: A population-based prospective cohort study. Pediatr. Allergy Immunol. 31 (7), 774–782. doi: 10.1111/pai.13310
Veenhoven R., Bogaert D., Uiterwaal C., Brouwer C., Kiezebrink H., Bruin J., et al. (2003). Effect of conjugate pneumococcal vaccine followed by polysaccharide pneumococcal vaccine on recurrent acute otitis media: a randomised study. Lancet 361 (9376), 2189–2195. doi: 10.1016/s0140-6736(03)13772-5
Verhagen L. M., Rivera-Olivero I. A., Clerc M., Chu M., van Engelsdorp Gastelaars J., Kristensen M. I., et al. (2021). Nasopharyngeal Microbiota Profiles in Rural Venezuelan Children Are Associated With Respiratory and Gastrointestinal Infections. Clin Infect Dis. 72 (2), 212–21.
Walker R. E., Walker C. G., Camargo C. A. Jr., Bartley J., Flint D., Thompson J. M. D., et al. (2019). Nasal microbial composition and chronic otitis media with effusion: A case-control study. PLoS One 14 (2), e0212473. doi: 10.1371/journal.pone.0212473
Watson K., Carville K., Bowman J., Jacoby P., Riley T. V., Leach A. J., et al. (2006). Upper respiratory tract bacterial carriage in Aboriginal and non-Aboriginal children in a semi-arid area of Western Australia. Pediatr. Infect. Dis. J. 25 (9), 782–790. doi: 10.1097/01.inf.0000232705.49634.68
Wen Z., Xie G., Zhou Q., Qiu C., Li J., Hu Q., et al. (2018). Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. BioMed. Res. Int. 2018, 6362716. doi: 10.1155/2018/6362716
World Health Association Revised WHO classification and treatment of childhood pneumonia at health facilities. Available at: https://www.who.int/publications/i/item/9789241507813.
Xu L., Earl J., Bajorski P., Gonzalez E., Pichichero M. E. (2021a). Nasopharyngeal microbiome analyses in otitis-prone and otitis-free children. Int. J. Pediatr. otorhinolaryngology 143, 110629. doi: 10.1016/j.ijporl.2021.110629
Xu L., Earl J., Pichichero M. E. (2021b). Nasopharyngeal microbiome composition associated with Streptococcus pneumoniae colonization suggests a protective role of Corynebacterium in young children. PLoS One 16 (9), e0257207. doi: 10.1371/journal.pone.0257207
Yau J. W., Hou J., Tsui S. K. W., Leung T. F., Cheng N. S., Yam J. C., et al. (2019). Characterization of ocular and nasopharyngeal microbiome in allergic rhinoconjunctivitis. Pediatr. Allergy Immunol. 30 (6), 624–631. doi: 10.1111/pai.13088
Zemlicková H., Urbásková P., Adámková V., Motlová J., Lebedová V., Procházka B. (2006). Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, Moraxella catarrhalis and Staphylococcus aureus isolated from the nasopharynx of healthy children attending day-care centres in the Czech Republic. Epidemiol. Infect. 134 (6), 1179–1187. doi: 10.1017/s0950268806006157
Zhou Y., Bacharier L. B., Isaacson-Schmid M., Baty J., Schechtman K. B., Sajol G., et al. (2016). Azithromycin therapy during respiratory syncytial virus bronchiolitis: Upper airway microbiome alterations and subsequent recurrent wheeze. J. Allergy Clin. Immunol. 138 (4), 1215–1219.e5. doi: 10.1016/j.jaci.2016.03.054
Zhou Q., Xie G., Liu Y., Wang H., Yang Y., Shen K., et al. (2020). Different nasopharynx and oropharynx microbiota imbalance in children with Mycoplasma pneumoniae or influenza virus infection. Microbial pathogenesis 144, 104189. doi: 10.1016/j.micpath.2020.104189
Keywords: 16S rRNA, shotgun analysis, nasal, respiratory tract infection, microbiome, asthma, bronchiolitis, otitis media
Citation: Zimmermann P (2023) Exploring the microbial landscape of the nasopharynx in children: a systematic review of studies using next generation sequencing. Front. Microbiomes 2:1231271. doi: 10.3389/frmbi.2023.1231271
Received: 30 May 2023; Accepted: 14 September 2023;
Published: 19 October 2023.
Edited by:
Jin Xu, Moffitt Cancer Center, United StatesReviewed by:
Yu-Wei Wu, Taipei Medical University, TaiwanPengfan Zhang, Innovative Genomics Institute (IGI), United States
Copyright © 2023 Zimmermann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Zimmermann, cGV0cmEuemltbWVybWFubkB1bmlmci5jaA==
 Petra Zimmermann
Petra Zimmermann