- Center for Biomedical Engineering, School of Engineering, Brown University, Providence, RI, United States
Decentralized Point-of-Care (PoC) diagnostics hold momentous potential for rapid and accessible viral infection disease detection. Presented is a unique design application of an easy-to-use (plug-and-play) platform for viral detection. The platform leverages a simplified multiplex Reverse-Transcription Loop-mediated Isothermal Amplification (RT-LAMP) Lateral Flow Biosensor (LFB) assay with a lyophilized master mix, eliminating the need for RNA isolation or special reporting equipment. A user-friendly Saliva Measuring Tube (SMT) ensures accurate saliva volume self-collection, and a Syringe-based PoC (SPoC) platform automates sample treatment, reagent mixing, and temperature control using readily available components and consumables. The platform’s performance was evaluated by multiplexed detection of the SARS-CoV-2 N2 target gene and human ACTB gene from saliva samples. The SPoC platform achieved a detection limit of spiked 500 copies/mL for SARS-CoV-2 and consistent internal control readout. The presented PoC system offers a promising initial step for further development toward a decentralized solution for viral infection testing.
1 Introduction
Infectious diseases remain a significant challenge in global public health, causing morbidity and mortality worldwide (Bloom and Cadarette, 2019). Viral infections, in particular, pose a substantial threat due to their rapid transmission and diverse clinical presentations, exemplified by the 2019 global COVID-19 pandemic (Kaur et al., 2021). Accurate and timely diagnosis of viral pathogens is crucial for effective patient management, infection control, viral genotyping, and the development of targeted therapies (Fraser et al., 2004). Traditional diagnostic methods, while definitive, often require specialized equipment and laboratory expertise, leading to delays in diagnosis and hindering patient care, particularly in resource-limited settings (Matsumura et al., 2023; Yalley et al., 2022). This highlights the critical need for a rapid, sensitive, cost-effective, and user-friendly point-of-care (PoC) diagnostic platform for viral infections at the early stages of infection.
Most global diagnostics efforts to fight against viral infection have been site-centralized: sample collection and laboratory detection of viral load are at a centralized site where patient-provided samples are collected and transported to a specialized laboratory for diagnostics. On the other hand, a site-decentralized diagnostics platform tests at the location of patients’ convenience on a low-throughput scale and poses a great alternative to centralized diagnostic efforts. Decentralized diagnostics could help decrease community spread drastically if patients are often screened at early asymptomatic infection stages (Nikolai et al., 2020).
Presented is an assay workflow for detecting viral infections, specifically SARS-CoV-2, using an automated duplex single-throughput PoC platform. The platform’s performance was evaluated by multiplexed detection of the SARS-CoV-2 N2 target gene and human ACTB gene from saliva samples. The N2 gene is a region of the nucleocapsid (N) gene of SARS-CoV-2 responsible for developing the N protein with essential structural functions (Wu et al., 2023). The N2 gene is highly conserved and specific to the virus, making it ideal for specific diagnostics. The ACTB gene, on the other hand, encodes the beta-actin protein, a common housekeeping gene in humans. It is often included as an internal control to ensure the integrity of the sample and the amplification process in many nucleic acid amplification tests (Ostheim et al., 2022).
Nasopharyngeal (NP) swab, a sample collection method that inserts a long shaft into the nasal cavity in the upper respiratory tract of patients, is a reliable and standard process for collecting specimens from patients with a respiratory viral infection (Kinloch et al., 2020). However, the collection process can be mildly painful and often requires contact with medical personnel. Saliva is a comparable alternative specimen, as it is minimally invasive, easily self-administered, and has a higher viral load compared to NP swabs in early detection of SARS-CoV-2 (Wyllie et al., 2020; McPhillips and MacSharry, 2022). For example, Beyene et al. (2021) found that saliva samples had higher viral load than NP swabs (proxy measure of viral load indicated by cycle threshold (Ct) value for NP swabs was 32.66 while saliva was 24.21) suggesting saliva as an effective in detecting the virus early in the infection (Beyene et al., 2021). Positive viral infection testing from saliva has also been reported in cases of human papillomavirus (HPV), positive oropharyngeal cancer (OPC), hepatitis C virus, human herpes simplex virus, human cytomegalovirus, and influenza A and B viruses (Tang et al., 2019; Sosa-Jurado et al., 2014; Waters et al., 2019; Vohra et al., 2020; Galar et al., 2021). Hence, the proposed PoC platform was developed to work directly with saliva samples, further simplifying testing efforts.
The reverse transcription-quantitative PCR (RT-qPCR) amplification testing method has been the gold standard for the sensitive and specific detection of viral RNA (Hasan et al., 2020). However, typical RT-qPCR methods often require complex processes such as RNA isolation from collected specimens, reverse transcription, amplification via thermal cycling, and fluorescence signal detection. These processes, therefore, require complex instruments with thermal drive and imaging capacity, sufficient technical expertise for prepping samples, and relatively long turnaround times. Such challenges can be addressed with high throughput automation systems but at a high cost (Cassedy et al., 2021; Dhibika et al., 2023). Additionally, the manual nature of traditional diagnostic methods often suffers from human error and is a significant factor in cross-contamination. Automated systems can help mitigate this risk by reducing the number of manual steps or reagent interactions and the required direct handling (Chen et al., 2021a).
Here, we sought to streamline the testing process using an alternative assay, reverse transcription-loop mediated isothermal amplification (RT-LAMP), a one-pot nucleic acid amplification and detection method. LAMP has proven to be a reliable PoC diagnostic technique because of its specificity and ability to amplify target genes using a single constant temperature (Notomi et al., 2000). The unique properties of the assay complemented the simple design of the platform through two designs: 1) LAMP requires a single temperature during amplification instead of a temperature cycle, and 2) lateral flow biosensors (LFBs) to detect amplicons easily without special detection modules. Compared to PCR workflows, where amplifying viral loads requires expensive thermocyclers to cycle through different temperatures, the LAMP reaction can sufficiently happen, allowing for a simple heating instrument with a constant single temperature to amplify the target gene, making it ideal for PoC workflows. While LAMP has been used in multiple PoC testing efforts, detecting amplified amplicon products has become a significant difficulty due to the lack of simple analytical tools. For example, different detection techniques, such as fluorescent agents, colorimetric agents, and gel electrophoresis, have been used to analyze LAMP products but require sophisticated instrumentation to collect and interpret the results (Alhamid et al., 2022; Saxena et al., 2022).
In this study, the LAMP reaction was coupled with LFBs as an amplicon detector due to their low cost, ease of use, and fast visualization capabilities, bypassing any need for advanced detection tools. LFBs are common point-of-care detection paper-based lateral-flow tool that employs a sandwich detection system and uses tracers with conjugates to generate detectable signals that are easily observable with the naked eye. They are commonly used in antigen tests and are ideal for rapid diagnostics. They are ubiquitous in infectious disease detection due to their robustness, low cost, and quick assay performance ability (Vealan et al., 2023). Another bottleneck of PoC systems is the difficulty of supplying reagents in cold-chain transportation; hence, lyophilization of master mixes has also been explored to bypass the cold-chain transportation difficulty in SARS-CoV-2 (Song et al., 2022; Prado et al., 2024). Multiple researchers have reported different iterations of the RT-LAMP’s primer mix to detect SARS-CoV-2 on LFB (Zhu et al., 2020; Zasada et al., 2022; Thompson and Lei, 2020; Li et al., 2022; Chen et al., 2021b; Choi et al., 2023; Agarwal et al., 2022; Zhang et al., 2021; Tang et al., 2024). However, a unique combination of LFB with LAMP utilizing multiplexed Forward inner primer and Loop forward-tagged primers for detecting spiked saliva in a PoC instrument has yet to be explored. This provides an easily accessible platform for patients to use at their convenience at a high sensitivity level and with low consumables. The proposed design of our platforms uses a simple consumable tube for collecting self-administrative saliva samples, a control system for handling saliva volumes, and techniques for resuspending lyophilized master mix with primers and reagents specific to SARS-CoV-2 detection by RT-LAMP LFB assay (Figure 1).
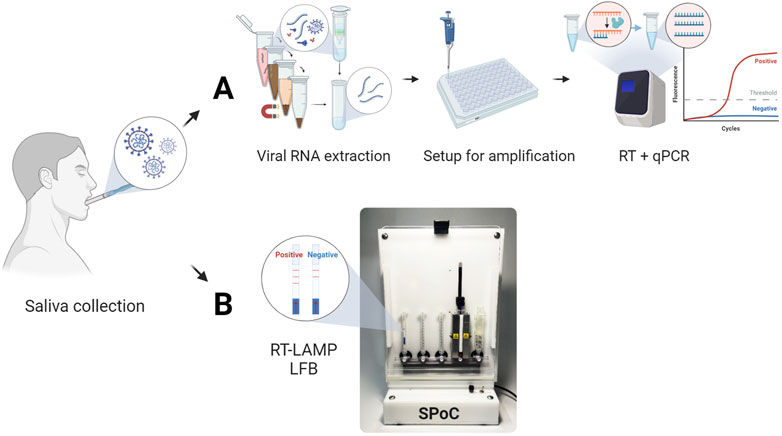
Figure 1. Comparison of conventional and proposed PoC platform workflows for detecting SARS-CoV-2 from saliva samples. (A) Conventional workflow consisting of viral RNA extraction, amplification setup, and RT-qPCR amplification for typical viral infection detection via fluorescence signal. (B) Proposed PoC detection systems utilizing RT-LAMP reaction and LFB strips.
2 Results and discussion
2.1 RT-LAMP LFB assay
RT-LAMP with LFB assay was designed to detect the highly conserved N2 target region of the SARS-CoV-2 genome and human ACTB target housekeeping gene as an internal control (Table 1). (Lee et al., 2021; Valente et al., 2009) Similar RT-LAMP LFB assays have been verified and reported for detecting various infectious diseases but have yet to integrate the whole workflow for a PoC using saliva samples (Saxena et al., 2022; Zhang et al., 2021; Tang et al., 2024; Mytzka et al., 2023). The assay required no RNA isolation but utilized lysis treatment to expose the ssRNA from the virus. A custom lyophilized reverse-transcriptase and Bst warm start polymerase were used to transcribe and amplify the target RNA (Figure 2). Similar to Notomi et al., 2000, six LAMP primers, including the loop forward and reverse primers, were utilized for amplification in this study (Notomi et al., 2000). However, the Forward inner primer (FIP) was modified to have Biotin conjugate at the 5′-end. Similarly, the Loop forward (LF) primer was modified to have Fluorescein Amidite (FAM)or Digoxigenin (DIG) at the 5′-end of the primer specific to the target sequence. These conjugates allow the detection of any amplified products on the LFB. The LFB detectable amplicons in the sixth generation of amplification had target sequences of interest, the N2 gene and ACTB gene, sandwiched between the FAM-Biotin conjugation and FAM-DIG conjugation, respectively (Figure 2). The amplicons were then added to the sample pad section of the LFB strip, wherein the presence of SARS-CoV-2 target RNA or housekeeping ACTB gene, the FAM conjugates bind with the goat anti-FAM antibody gold nanoparticle complex. The products immobilize when streptavidin or anti-DIG antibodies bind to their respective conjugate from N2 or ACTB gene amplicons. The concentrated gold nanoparticles then make the bands observable with visual inspection when immobilized. Any leftover FAM-gold nanoparticle complexes bind to the anti-goat antibody in the control band. Amplicons with biotin or DIG but without FAM end are common and can occupy the binding sites on the LFB and go undetected (Figure 2VI). These undetected products are inherent byproducts of the amplification method and can compete with the detectable products, potentially compromising the detection limit on the LFB. Such products may be mitigated by modifying different primers with the conjugates, but doing so will change the detectable products and may interfere with the sensitivity, which will need further investigation.
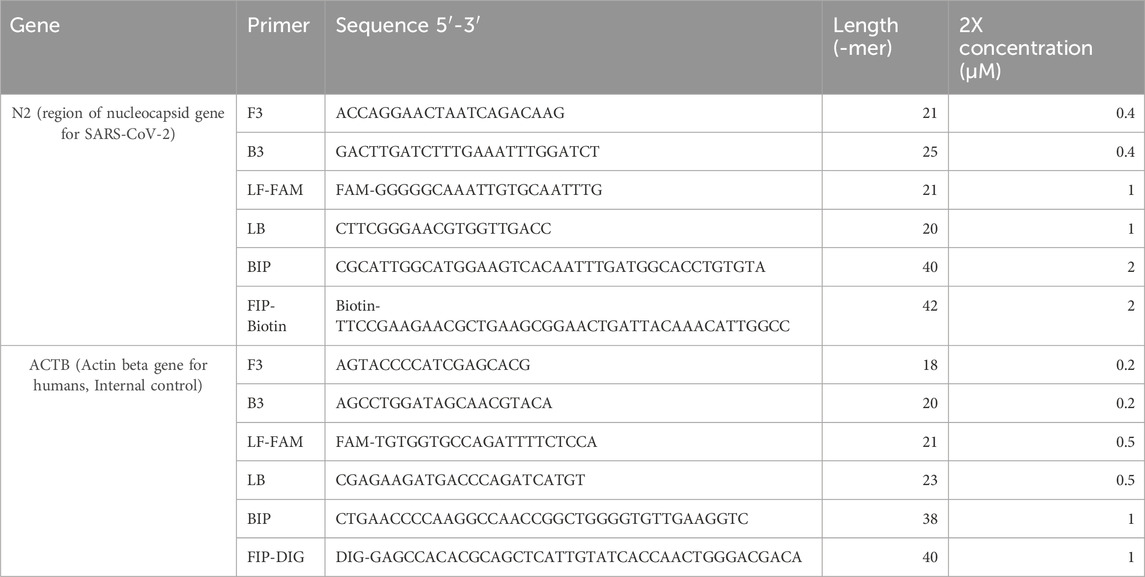
Table 1. Custom primer sequences targeting N2 and ACTB genes with respective Biotin and DIG conjugate tags for detecting amplicons on LFB.
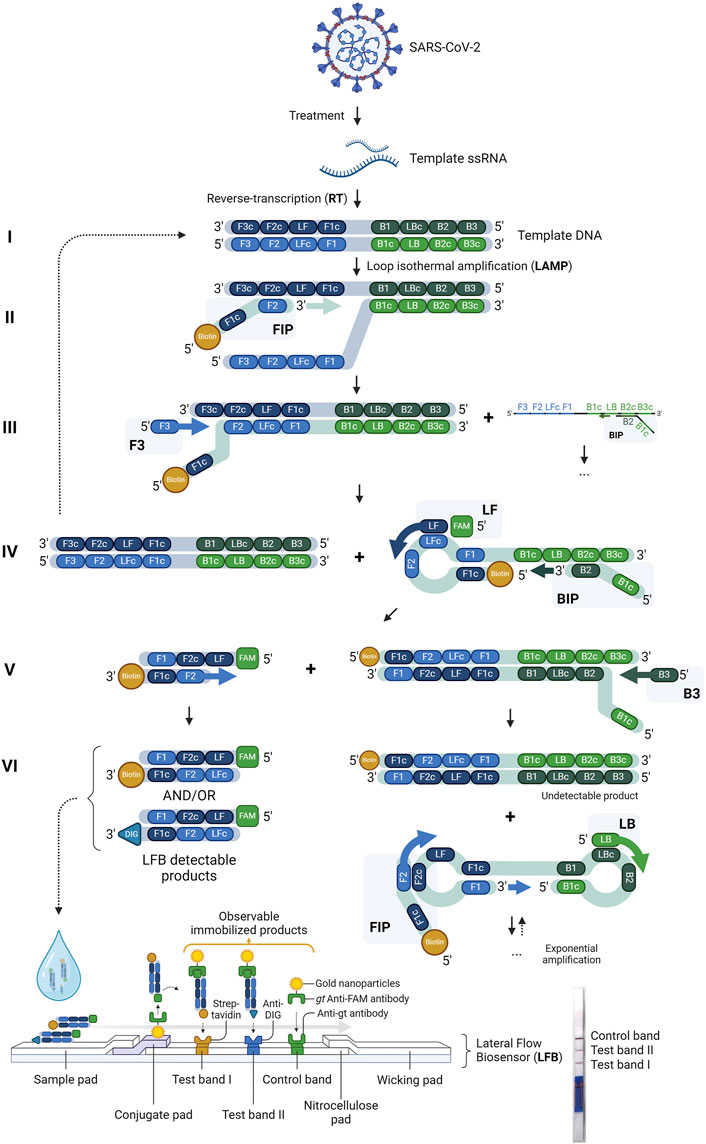
Figure 2. Schematic illustration and principles of RT-LAMP LFB assay. The viral RNA from SARS-CoV-2 is isolated and used as a template for converting the RNA to complementary DNA (cDNA) through reverse transcription (RT). Six primers (FIP, BIP, LF, LB, F3, and B3) amplify the cDNA using Loop-mediated isothermal amplification (LAMP), where various amplicon byproducts are made. The LFB-detectable products are formed with FAM-Biotin or FAM-DIG conjugates after six generations (VI) of strand displacement and hybridization from LF and FIP primers. The sandwiched products are added to the sample pad of the LFB strips, where two test bands and an LFB control band with corresponding streptavidin, anti-DIG antibodies, and anti-goat (gt) antibodies—the goat anti-FAM gold nanoparticle—where the amplicons complex exhibit visible red bands when immobilized. Any excess or unimmobilized products are absorbed in the wicking pad. The presence of visible lines on the test bands indicates a positive result for SARS-CoV-2 and the presence of the ACTB housekeeping gene (human control). The control band is always visible for a valid test.
The RT-LAMP assay is designed to work with at least 50 μL input saliva sample. Cases of direct SARS-CoV-2 detection from patient saliva samples have been explored, and this is often done by pretreating the saliva samples before the amplification step (Naranbat et al., 2022; Castillo-Bravo et al., 2022; Fathi Karkan et al., 2022). The addition of thermolabile Proteinase K directly to saliva has been a common way to digest proteins, inactivate nucleases, and lyse cells and viruses (Kim et al., 2024; Vogels et al., 2021). Often, Proteinase K is stored at −20°C or 4°C depending on the composition of the stock mix (Sweeney et al., 1993). Hence, to eliminate cold chain transportation struggles in PoC platforms, a room-temperature stable thermolabile Proteinase K was used directly in this assay. A final concentration of 2.22 mg/mL of Proteinase K was used to inactivate the sample. Proteinase K retains most of its activity at nearly 60°C but will degrade irreversibly at 95°C (Sweeney et al., 1993; Schokker and van Boekel, 1997). Any residual effect of Proteinase K impeded the LAMP reaction by denaturing the polymerase and invalidating any LFB reading. Hence, it was crucial to deactivate the Proteinase K-saliva sample mix by heat-inactivation before any downstream processing.
A 1:1 ratio of 2X RT-LAMP primer mix and deactivated Proteinase K-saliva sample mix was homogenized before the lyophilized 2X RT-LAMP enzyme master mix pellet was dissolved (1:1, primer-Proteinase K-saliva mix: RT-LAMP enzyme master mix). The entire mix was incubated at 63°C for 30 min for a complete LAMP reaction and diluted with chase buffer before being loaded onto the LFB. The LAMP primers mix had a total of 12 primers for the duplex reaction - a set of six primers for each target gene. Benchtop experiments showed LFB valid results for all tested conditions - NTC, positive readouts for ACTB gene amplification from saliva, positive readouts for N2 gene amplification from SARS-CoV-2 spiked waters, and positive readouts for ACTB + N2 gene amplification from spiked saliva (Figure 3).
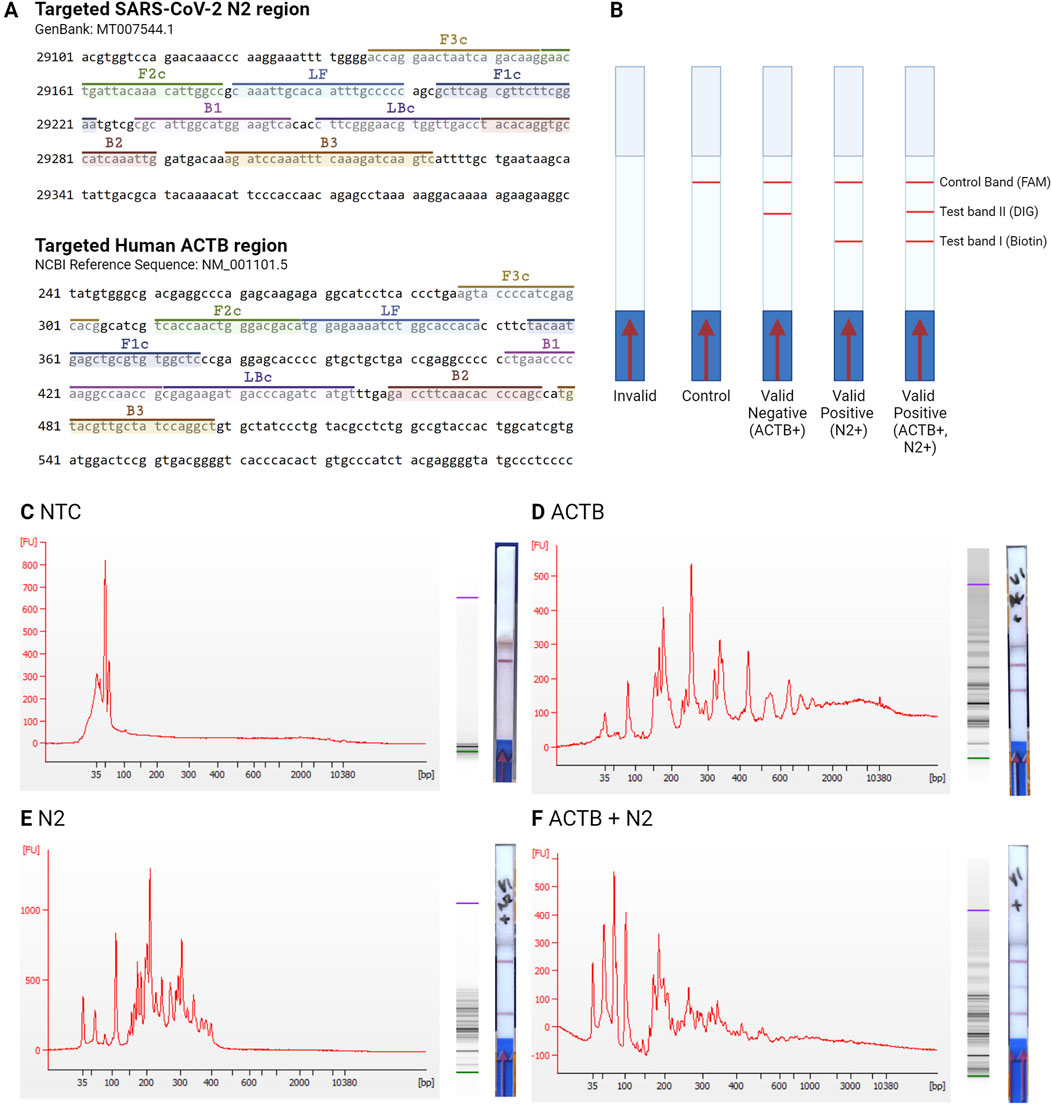
Figure 3. Interpretation of LFB RT-LAMP results. (A) Targeted regions of the N2 and the ACTB genes. (B) The different versions of LFB readout. No band is an invalid result; a single line at the control band is a negative LFB readout; a red line at the test band II is a positive result for internal control ACTB gene amplification and a valid negative result for N2 gene; a red line at the test band I is a positive result for N2 target gene amplification; and a red line at both of the test band is a positive result for ACTB and N2 gene amplification. Comparison of electropherogram, gel electrophoresis, and LFB results in (C) NTC sample with no target genes, (D) saliva sample with ACTB gene, (E) sample with spiked N2 gene, and (F) saliva sample with ACTB gene and spiked N2 gene.
The corresponding electropherogram from the LFB readouts showed the different amplicons from the LAMP reaction in size distribution depending on the location of the targeted region (Figure 3A). In the negative control condition, without any amplification, the primers, ranging from 20–42 nucleotide lengths, are shown in the NTC condition electrophoresis below 100 bp size (Figure 3C). Positive results, however, produce longer strands of LAMP products, as seen in both the ACTB and N2 targeted duplex amplification (Figures 3D–F). The predicted dumbbell shapes of the amplicon sizes were analyzed using a mathematical model to predict false positive results, as mentioned by Schneider et al. (2019), but with the addition of LF and BF primers (Schneider et al., 2019). The first dumbbell amplicon size from the LAMP reaction for the N2 target gene was 134 mers, while the ACTB target gene was 165 mers (Supplementary Table S1), as reflected on the electropherogram with correct amplicons produced (Figures 3C–F). The capacity to multiplex targeted genes in a single reaction and detection was advantageous compared to other PoC RT-LAMP assays detection, such as colorimetric assay limited to a single targeted gene amplification (Rivas-Macho et al., 2023). However, the limitation of the assay is that to interpret the amplification results, the lateral strips need to be submerged into the final amplicon, which can lead to false-positive results due to aerosol contamination when the reaction tube is opened (Tang et al., 2024). Hence, this study focused on a closed PoC system capable of containing RT-LAMP reaction and adapting sample input.
2.2 Saliva measuring tube (SMT)
Ideal PoC platforms are self-administrable, requiring minimal user interaction with the device and no need for expert laboratory skills. The proposed platform in this study requires only a single point of contact for the user to run self-diagnostic testing: the saliva input step. During this step, it is crucial to input a consistent volume of saliva, which is often done by a pipette with laboratory-proficient personnel in a laboratory setting. The ratio of lysis reagent volume for the rupture of the viral lipid membrane and the input saliva sample volume is critical as this dictates the success of downstream amplification procedures (Bustos-Garcia et al., 2022). Hence, part of this study’s goal was to design an easy-to-use, self-saliva-collection tube capable of accurately and precisely measuring and displacing 200 μL saliva directly from the mouth by passive drooling. The saliva measuring tube (SMT) design consists of three parts: 1) the mouthpiece, 1) the metering tube, and 3) the piercer (Figure 4B).
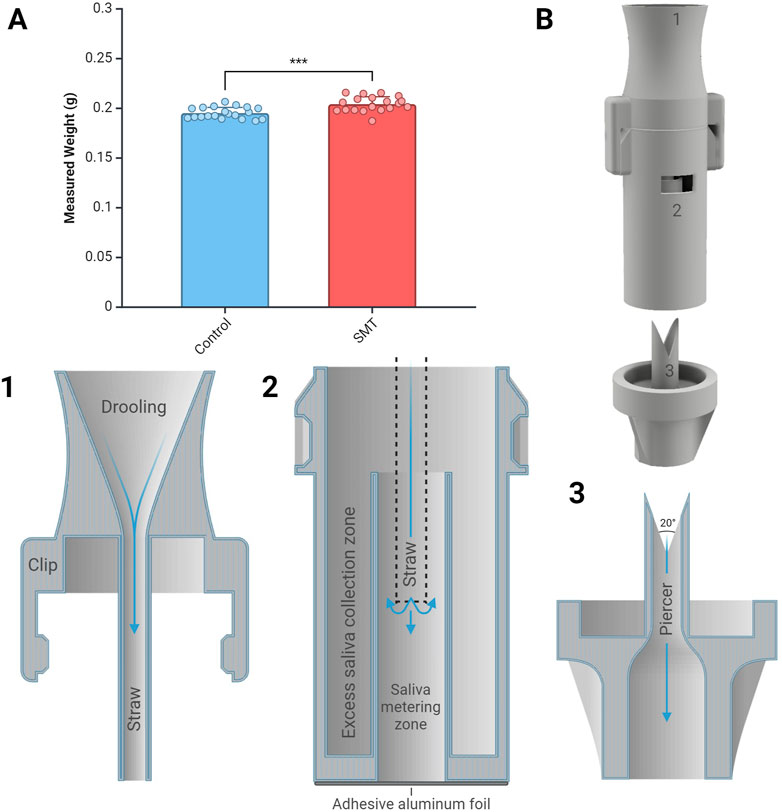
Figure 4. Saliva measuring tube volume displacement capacity and design. (A) Comparison of saliva measuring tube (SMT) and bench-top pipette (control) set at 200 μL. Statistical significance level ***p < 0.001 observed between two groups (n=20 per condition). (B) SMT assembly and the cross-sectional view of SMT: 1) The mouthpiece, 2) the metering tube, and 3) the piercer. The mouthpiece and the metering tube are assembled. Users passively drool into the mouthpiece, and the saliva is collected in the metering tube. Samples are accessed through the double piercer by transporting the saliva to the next chamber by puncturing the aluminum foil.
The custom-designed SMT was 3D-printed using a medical-grade resin to minimize any carryover or compromise integrity of the sample. The SMT was designed in three parts to simplify assembly. The clip pre-attaches the mouthpiece and the metering tube, as shown in Figure 4 B1 and B2. The user places the mouthpiece into their mouth and drools passively, without spitting, and the targeted volume of the saliva is collected in the saliva metering zone (Figure 4 B2). The junction between the mouthpiece and the metering tube is a straw to consolidate the saliva at the bottom of the metering zone without forming any bubbles. Saliva creates air pockets without the straw and does not fill the saliva metering zone due to its viscous nature. Any excess saliva is collected in the excess saliva collection zone, positioned radially around the saliva metering zone. The measured saliva volume is directly correlated with the height of the saliva collection zone wall, and the height was adjusted to compensate for the loss of saliva by the piercing mechanism and the walling in between. The collected saliva was kept intact using an adhesive aluminum foil. The foil was punctured using a 20॰ double-sided piercer that collects any leftover aluminum foil between the piercers, allowing all liquid to dispense (Figure 4 B3). Multiple different piercing mechanisms were tested (data not shown) to maximize the accuracy and precision of the collection, where the double piercer showed the best volume transport capacity and least failure rate. The piercer has an adaptative end to transport the saliva to the PoC device.
The SMT collected saliva from an individual, and the volume displacement was measured to assess transport accuracy and precision (Supplementary Table S2). The individual’s saliva viscosity was measured at 1.22 ± 0.12 cPa (n=3). The saliva metering tube section had a height of 12.67 mm with a radius of 2.5 mm to collect ∼250 μL. During displacement, saliva volumes were inherently adhered to the wall and left in the junction between the metering tube and the piercer. The larger volume capacity of the metering tube compensated for this volume loss. Spraying the tubing with surfactant improved the consistency of displaced volume. After collection, the mouthpiece is removed, and the piercer punctures the metering tube to transport the remaining measured saliva sample. A p200 pipettor set at 200 μL displaced 0.1953 ± 0.0056 g of saliva while the SMT transported 0.2043 ± 0.0074 g of saliva (n=20; Figure 4A). The higher viscosity of saliva or the timestamp of when the saliva samples were collected (e.g., after waking up or after eating) may interfere with the precision of the saliva volume transportation. Additionally, the onset of symptoms or physiological stress response could also affect the viscosity of saliva in patients, potentially affecting the precision and accuracy of sample collection. For example, Govindaraj et al. (2019) found that saliva viscosity increased under stress and may be attributed to increased sympathetic activity and altered salivary gland function (Gholami et al., 2017; Govindaraj et al., 2019). While Katayama et al. (2022) found that there was no significant difference in viral loads based on the timing of saliva collection within 9 days of symptom onset, Serrato et al. (2023) highlighted the number and diversity of other microbiomes presence due to the initial infection which may affect the physical properties of the saliva (Katayama et al., 2022; Larios Serrato et al., 2023; Rereddy et al., 2023).
The set volume of the input saliva can be altered depending on the specific assay or required input volume by changing the volume of the saliva metering zone and the height of the piercer. The angle between the two piercer edges must be 20° to keep the sharp edge. To handle low volumes of saliva (<100 μL), the diameter of the piercer can be decreased, and at the same time, the angles between piercers may need to be increased to shorten the height of the metering tube. It is anticipated that an increased piercer angle will not be as precise at transporting the saliva volume. The SMT was designed as a plug-in to the PoC platform for the RT-LAMP LFB assay.
2.3 Syringe-based PoC (SPoC) platform
The duplex RT-LAMP LFB assay was adopted onto a stand-alone single-pot SPoC platform (Figure 5A). Based on the assay procedure, the platform was designed to displace volume, mix reagents, and control temperature at a specified sequence order. Users load their measured saliva sample onto the SPoC platform using the SMT and start the installed script. The script displaces the liquid using off-the-shelf leur lock syringes and mixes the saliva samples with a pre-loaded primer and Proteinase K before heat treatment (Figure 5B I). The heat-treated samples are displaced and mixed with lyophilized RT-LAMP before running a 30-min LAMP reaction (Figure 5B II and III). The amplicons were displaced to mix with the chase buffer (1X TE buffer) before being loaded onto the LFB (Figure 5B IV and V). At each step, the 3-way valves are positioned for 2-way openings to connect the syringe with the desired reagent to the syringe pump during volume displacement.
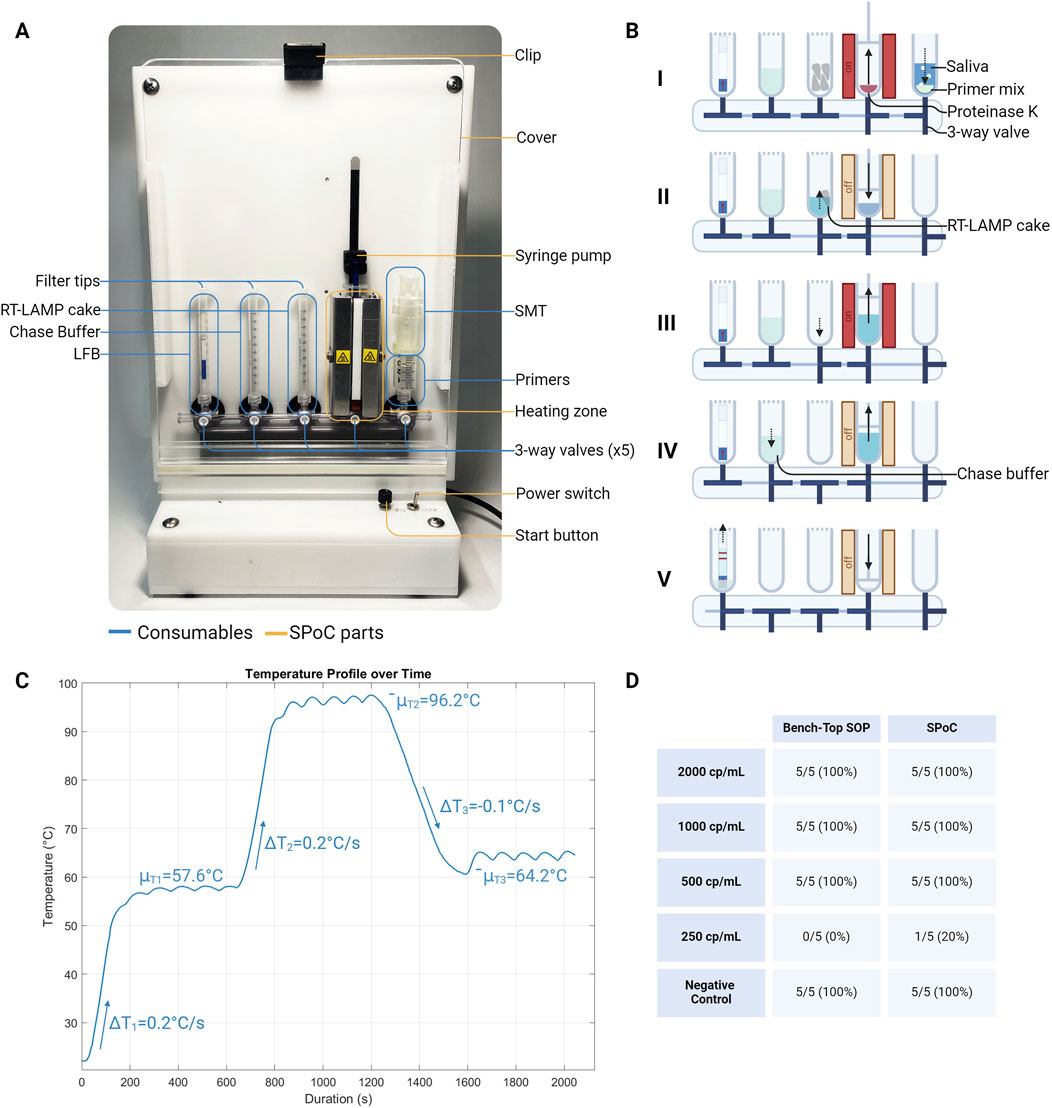
Figure 5. A syringe-based PoC (SPoC) platform for running the RT-LAMP LFB assay. (A) Overview of SPoC assembly with specified device parts (yellow) and the consumables (blue). The device uses 3-way valves to connect 5x leur lock syringe ports, displace liquid, and heat at different temperatures. (B) Schematic illustration of operation order: I) Mix saliva and primer mix with Proteinase K and heat treat sample, II) dissolve lyophilized RT-LAMP master mix pellets after heat treatment, III) isothermal amplification, IV) mix the chase buffer (1X TE buffer) with the amplicon, V) add diluted amplicon with chase buffer to LFB. (C) Temperature profile of the heating station set at three different temperatures (55°C, 95°C, 63°C). (D) Sensitivity of the control assay (RT-LAMP LFB bench-top SOP) and the SPoC platform (see Supplementary Figure S1 for LFB readouts).
The platform aimed to introduce the least number of custom cartridge consumables to run the RT-LAMP LFB assay. Hence, off-the-shelf valving and reagent containers (leur-lock syringes) were utilized. The off-the-shelf parts were then adapted to the platform using custom 3D-printed parts and servo motors to drive the syringe plunger and the valving positions. A series of simple resistor heaters drove the heat conduction for the sample treatment and amplification steps. The system’s capacity to displace the required volume was a crucial function in the RT-LAMP LFB procedure; given the larger volume of saliva sample input (200 μL) added to the system, scaled-up volumes of subsequent downstream reagents were required. A volume of ∼20 μL was consistently lost in each junction between the valving and the syringes. Therefore, air was pumped into each syringe container before changing valving positions.
To compensate for any volume loss and to converse, the reagents were displaced at 8 mm/s during operation. Bubble formation between each volume displacement step was typical; hence, variations of syringe pumping speeds were enforced to ensure the mixed samples’ complete homogeneity and bubble removal. A lower speed of 8 mm/s was first implemented to aspirate the reagent from its containing syringe to the heating zone, and then a faster speed of 16 mm/s was used to mix reagents.
To maximize the activity of and to deactivate Proteinase K, 55°C and 95°C temperatures were driven by a heat resistor, respectively. A simple bang-bang controller feedback system was used to control the desired temperatures, where the local minimum and maximum temperatures fluctuated from 57.02°C ± 0.39°C to 58.08°C ± 0.18°C at an average temperature of μT1=57.6°C and from 95.61°C ± 0.38°C to 97.31°C ± 0.20°C at an average temperature of μT2=96.2°C for set 55°C and 95°C, respectively. From lab room temperature to 55°C and 95°C, the average ramp-up rate was ΔT1,2=0.2°C/s, while the average cool-down rate from 95°C to 65°C was ΔT3=−0.1°C/s. Conduction of the heat was distributed from the heat resistors by an aluminum block around the 1 mL syringe, while brushless fans next to the heat resistors dissipated the heat. The LAMP reaction rate was most optimum between 60°C and 65°C (Saxena et al., 2022); hence, the SPoC platform was set at 63°C for 30 min for amplification to yield consistent detectable products. Here, local minimum and maximum temperatures fluctuated from 63.50°C ± 0.05°C to 65.11°C ± 0.06°C at an average temperature of μT3=64.2°C for set 63°C (Figure 5C).
In LFB detection, a high concentration of label-specific analytes (the label-conjugated amplicons from the LAMP reaction) can cause a high dose hook effect on the lateral flow’s finite quantity of binding (anti-conjugate) sites (Ross et al., 2020; Ma et al., 2019). When the concentration of the labeled products surpassed the capacity of available binding sites without dilution, a reduction in the formation of signal sandwiches occurred, leading to false-negative results. Hence, the high dose hook effect is often compensated by dilution of the amplicons. Therefore, in the SPoC platform, the amplicons are diluted with chase buffer after the LAMP reaction before loaded onto the LFB to avoid false-negative results. On the other hand, false-positive results are avoided by minimizing cross-contamination issues and ensuring the heating unit syringe path is never open to the atmosphere. The contamination is prevented by never positioning the syringe pump’s valve to the open atmosphere and using a polyethylene frit filter at the syringe opening to block any aerosol contamination while not creating vacuum conditions during aspirate and dispense (Figure 5A). The single-use cartridge consumable (the syringe and the syringe valve connector) is completely disposed of in a biohazard bag after the run is complete. The cartridge consumable is replaced run-to-run.
SARS-CoV-2 spiked in saliva samples at 250, 500, 1000, and 2000 copies/mL was used to determine the limit of detection (LoD) for the two-target gene RT-LAMP LFB assay in the SPoC platform and the bench-top SOP control procedures (Figure 5D). 500 copies/mL was the established LoD for both procedures at which the target gene N2 (with the presence of ACTB) was amplified and detected on the LFB simultaneously. The LoD values are important for comparing different tests, where tests with high analytical sensitivity methods (such as RT-qPCR test) have LoD of ∼102–103 copies/mL compared to tests with low analytical sensitivity methods (such as antigen test) that have LoD of ∼105–107 copies/mL (Caruana et al., 2021; Cubas-Atienzar et al., 2021; Corman et al., 2021; Olearo et al., 2021). The viral load in patients with COVID-19 may vary significantly depending on several factors, including the stage of the infection, symptom severity, and the presence of different SARS-CoV-2 variants (Zheng et al., 2020; Han et al., 2020; Stankiewicz Karita et al., 2022). Multiple studies have found that in the early stage of the infection, patients suffering from COVID-19 may have a SARS-CoV-2 viral load as low as 103–105 copies/mL of saliva; therefore, a highly sensitive detection method is important in early detection of SARS-CoV-2 (Savela et al., 2022; Winnett et al., 2020; Kissler et al., 2021a; Kissler et al., 2021b). Testing the device with real patient saliva samples would be a critical milestone in determining the full capacity of the instrument.
The RT-LAMP method simplifies the workflow by bypassing the RNA isolation protocol and combining lysis, reverse transcription, and amplification steps. The reported results in LFB from amplification are easily read by visual inspection without requiring specialized analytical instruments such as a quantitative thermo-cycler. The SPoC platform utilized simple off-the-shelf syringes and valving consumables to automate the amplification and detection workflow. This module signified the possibility of adopting the lyophilized duplex RT-LAMP master mix with added proteinase K and primers of interest with LFB assay into an automated workflow using off-the-shelf parts without expensive detection equipment. Further simplification and miniaturization of the instrument and the assay can make the instrument truly PoC-friendly, promoting the decentralization of infectious disease diagnostics. The system is limited by its inability to quantify the amplified products in the LAMP reaction in real time or estimate the viral load from the sample. While the RT-LAMP master mix has been lyophilized and used in this study, the Proteinase K and the specific target primers were not lyophilized. However, cases of lyophilized mixes of amplification enzymes and primers are a common practice that can be utilized (Promega, 2024).
3 Materials and methods
3.1 RT-LAMP LFB assay design and workflow
Saliva samples were collected with the individual’s informed consent and were verified for SARS-CoV-2 negative through a routine Verily COVID-19 RT-PCR Test (Verily, San Francisco, CA). The viscosity of the collected saliva samples was measured using an AR2000 Rheometer (TA Instruments, New Castle, DE). The COVID-19 negative saliva samples were spiked at different 2-fold serial dilutions (250, 500, 1000, and 2000 copies per mL of saliva), concentrations with NATtrol SARS-CoV-2 Stock External Quality Control (ZeptoMatrix, Franklin, MA). Water-based samples were spiked with the SARS-CoV-2 control to detect the N2 target gene separately. At least 50 μL of saliva samples were treated with room temperature stable 20 mg/mL Proteinase K solution (Promega, Madison, WI) to a final concentration of 2.22 mg/mL (50 μL saliva +6.25 μL of 20 mg/mL Proteinase K). The saliva-Proteinase K sample mix was heat-treated using SimpliAmp Thermal Cycler (Applied Biosystems, Waltham, MA) set at 55°C and 95°C, 10 min each. The treated samples were mixed with a custom 2X 100 μL per pellet lyophilized WarmStart RT-LAMP Kit (New England Biolabs, Ipswich, MA), where a single pellet was used per 50 μL of saliva sample. The primers for the targeted genes N2 (region of nucleocapsid gene for SARS-CoV-2) and ACTB (Actin beta gene for humans) were previously designed and described by Zhang et al. (2020). (Zhang et al., 2020). The N2 gene primers set targeted the SARS-CoV-2 genome region between 29,136 and 29323 (GeneBank: MT007544.1), while the ACTB gene primers set targeted Homo sapiens actin beta sequence between 287 and 498 (NCBI Reference Sequence: NM_001101.5) (NCBI, 2020; NCBI, 2024). The primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA), as shown in Table 1. The stock RT-LAMP mix and the primers (2X) were mixed at a final concentration of 1X and were amplified at 63°C for 30 min in the SimpliAmp Thermal Cycler. The amplicons were diluted with 1X TE buffer (Promega, Madison, WI) as the chase buffer at 2 parts to 1 part of the amplicons. Finally, an LFB HybriDetect 2T (Milenia-Biotec, Gießen, Germany) was dipped into the final diluted amplicons.
3.2 Saliva measuring tube (SMT) fabrication and measurement
Custom SMT parts were designed using Fusion 360 (Autodesk, San Francisco, CA). The piercer had a thickness of 0.5 mm, an inner radius of 1.9mm, a height of 10.46 mm, and a cut angle of 20° between edges (Figure 4 B3). The outer lip of the piercer where the metering tube is placed was designed with a 7.35 mm radius at a thickness of 2 mm. The metering tube had a base diameter of 14.50, while the saliva metering zone had a height of 12.67 mm and an inner radius of 2.5 mm. The total height of the metering tube was 30.79 mm (Figure 4 B2). The straw for the mouthpiece part had a height of 21.88 mm with a thickness of 0.5 mm. The distance between the mouthpiece’s base and the straw’s end was 16.88 mm long. The mouthpiece opening had a diameter of 6.25 mm and a thickness of 1 mm (Figure 4 B1). The SMT was printed using a Form 3 3D printer (Formlabs, Somerville, MA) with BioMed Clear Resin (Formlabs, Somerville, MA). The SMT was assembled, and an Adhesive PCR Plate Foil (Thermo Scientific, Waltham, MA) was applied at the bottom of the metering tube. Gravimetric measurement by an LB-200-124e Analytical Balance (Cole-Palmer, Vernon Hills, IL) was used to measure the displaced saliva into a 5 mL tube. When used with the SPoC platform, the collected saliva samples were spiked in with SARS-CoV-2 Verification Panel Full Genome before the aluminum seal was pierced with the piercer on the SPoC platform.
3.3 Syringe-based PoC (SPoC) platform fabrication
The SPoC platforms’ consumables were assembled from 4x sterile 1 mL Luer Lock Syringe (Filtrous, Poway, CA), cut 1x sterile 10 mL Luer Lock Syringe (Filtrous, Poway, CA) where the SMT piercer sits, 1x 5-Gang Stopcock Manifold Female Luer Lock (QOSINA, Ronkonkoma, NY), and 3x 5.33 mm Standard Polyethylene Cone Filter (Transcat Pipettes, Milford, MA). From the left to the right end of the 5-gang stopcock valve, the syringes were leur locked in and encapsulated the 1 strip LFB, 850 μL chase buffer, 4 pellets of RT-LAMP, 25 μL of Proteinase K, and 200 μL primers mix, as shown in Figure 5A. The polyethylene frit filters were fitted into the three of the 1 mL syringes in place of the plungers, while the remaining 1 mL syringe was used with the plunger to drive the displacement of reagents, as shown in Figure 5B.
The heating unit of the SPoC instrument used 4x 10W 20R Metal Aluminum Chassis Mount Resistors (Riedon, Alhambra, CA) mounted to a 1 mL syringe-fit custom aluminum block, 2x at each side. The temperature was controlled using a thermocouple and amplifier (Sparkfun, Boulder, CO). 2x brushless fans were positioned behind the heat resistors (MA2506L12C; Mechatronics Fan Group, Preston, WA) to dissipate heat during cooling. The syringe valves and plungers were aligned and fitted to custom PLA 3D-printed parts (Creality, Shenzhen, China). The valves and syringe plunger were moved using a simple metal gear servo motor (MG995 and DS3218MG; Amazon, Seattle, WA). The sequence of liquid displacement and heating was controlled by a Mini Maestro 12-Channel USB Servo Controller (Pololu Robotics and Electronics, Las Vegas, NV). The script for the procedure was written by Pololu Control Center software. The temperature inside the syringe was measured with the thermocouple submerged in water during the heating and cooling steps. The measured temperatures were plotted using a custom Matlab script (MathWorks, Natick, MA). Comparison of statistical differences was analyzed using a two-way ANOVA test (Turkey comparison analysis) with R scripts (R Foundation for Statistical Computing, Vienna, Austria). Illustrations and comparison graphs were created with BioRender.com (BioRender, Toronto, Ontario, Canada).
4 Conclusion
This study successfully developed and validated a platform for an automated and user-friendly Point-of-Care (PoC) diagnostic system for detecting SARS-CoV-2 from saliva samples. The platform utilized a simplified lyophilized duplex RT-LAMP assay with an LFB reporter, providing several advantages over conventional RT-qPCR detection methods, such as the elimination of RNA isolation, rapid result visualization without specialized reporter equipment, easy self-administration with minimal user interaction, and using readily available consumables and components for building the instrument. A custom self-saliva collection tube Saliva Measuring Tube (SMT) ensured accurate and precise measurement of 200 μL saliva volume displacement to the Syringe-based PoC (SPoC) automated system. The SPoC system used off-the-shelf components, single-use syringes, and valves to handle sample processing, reagent mixing, and temperature controlling for the RT-LAMP LFB assay. The platform demonstrated a Limit of Detection (LoD) of 500 copies/mL SARS-CoV-2 detection sensitivity. The presented system provides the potential for a PoC testing system to detect viral infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies involving humans because Saliva samples were self-collected by the authors. Saliva samples were anonymized, and verbal consent was provided. Throughout the study, confidentiality was maintained by omitting any personally identifiable information, and no personal information was collected. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DN: Conceptualization, Data curation, Investigation, Methodology, Software, Visualization, Writing–original draft, Writing–review and editing. JM: Conceptualization, Investigation, Resources, Writing–review and editing. AT: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partly supported by Revvity’s research grant to Brown University and research funds at Brown University.
Acknowledgments
Special thanks to Dr. Richard Joseph, Dr. Morey Krause, Ifelola Olabode, and Amy Oh for their valuable contribution and guidance to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frlct.2024.1450891/full#supplementary-material
References
Agarwal, S., Warmt, C., Henkel, J., Schrick, L., Nitsche, A., and Bier, F. F. (2022). Lateral flow–based nucleic acid detection of SARS-CoV-2 using enzymatic incorporation of biotin-labeled dUTP for POCT use. Anal. Bioanal. Chem. 414 (10), 3177–3186. doi:10.1007/s00216-022-03880-4
Alhamid, G., Tombuloglu, H., Motabagani, D., Motabagani, D., Rabaan, A. A., Unver, K., et al. (2022). Colorimetric and fluorometric reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for diagnosis of SARS-CoV-2. Funct. Integr. Genomics 22 (6), 1391–1401. doi:10.1007/s10142-022-00900-5
Beyene, G. T., Alemu, F., Kebede, E. S., Alemayehu, D. H., Seyoum, T., Tefera, D. A., et al. (2021). Saliva is superior over nasopharyngeal swab for detecting SARS-CoV2 in COVID-19 patients. Sci. Rep. 11 (1), 22640. doi:10.1038/s41598-021-02097-2
Bloom, D. E., and Cadarette, D. (2019). Infectious disease threats in the twenty-first century: strengthening the global response. Front. Immunol. 10, 549. doi:10.3389/fimmu.2019.00549
Bustos-Garcia, B., Garza-Manero, S., Cano-Dominguez, N., Lopez-Sanchez, D. M., Salgado-Montes de Oca, G., Salgado-Aguayo, A., et al. (2022). Development and testing of a low-cost inactivation buffer that allows for direct SARS-CoV-2 detection in saliva. Vaccines (Basel) 10 (5), 730. doi:10.3390/vaccines10050730
Caruana, G., Croxatto, A., Kampouri, E., Kritikos, A., Opota, O., Foerster, M., et al. (2021). Implementing SARS-CoV-2 rapid antigen testing in the emergency ward of a Swiss university hospital: the INCREASE study. Microorganisms 9 (4), 798. doi:10.3390/microorganisms9040798
Cassedy, A., Parle-McDermott, A., and O’Kennedy, R. (2021). Virus detection: a review of the current and emerging molecular and immunological methods. Front. Mol. Biosci. 8, 637559. doi:10.3389/fmolb.2021.637559
Castillo-Bravo, R., Lucca, N., Lai, L., Marlborough, K., Brychkova, G., Sakhteh, M. S., et al. (2022). Clinical performance of direct RT-PCR testing of raw saliva for detection of SARS-CoV-2 in symptomatic and asymptomatic individuals. Microbiol. Spectr. 10 (6), e0222922–22. doi:10.1128/spectrum.02229-22
Chen, X., Cheng, K., Sun, X., Zhang, Y., Cao, Z., Li, J., et al. (2021a). Comparison of traditional methods and high-throughput genetic sequencing in the detection of pathogens in pulmonary infectious diseases. Ann. Transl. Med. 9 (8), 702. doi:10.21037/atm-21-1322
Chen, X., Zhou, Q., Li, S., Yan, H., Chang, B., Wang, Y., et al. (2021b). Rapid and visual detection of SARS-CoV-2 using multiplex reverse transcription loop-mediated isothermal amplification linked with gold nanoparticle-based lateral flow biosensor. Front. Cell Infect. Microbiol. 11, 581239. doi:10.3389/fcimb.2021.581239
Choi, G., Moehling, T. J., and Meagher, R. J. (2023). Advances in RT-LAMP for COVID-19 testing and diagnosis. Expert Rev. Mol. Diagnostics 23 (1), 9–28. doi:10.1080/14737159.2023.2169071
Corman, V. M., Haage, V. C., Bleicker, T., Schmidt, M. L., Mühlemann, B., Zuchowski, M., et al. (2021). Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: a single-centre laboratory evaluation study. Lancet Microbe 2 (7), e311–e319. doi:10.1016/S2666-5247(21)00056-2
Cubas-Atienzar, A. I., Kontogianni, K., Edwards, T., Wooding, D., Buist, K., Thompson, C. R., et al. (2021). Limit of detection in different matrices of 19 commercially available rapid antigen tests for the detection of SARS-CoV-2. Sci. Rep. 11, 18313. doi:10.1038/s41598-021-97489-9
Dhibika, M., Madhusudhan, N. S., Malini, A., and Natarajan, M. (2023). Comparison of manual and automated nucleic acid (RNA) extraction methods for the detection of SARS-CoV-2 by qRT-PCR. Cureus 15 (3), e36773. doi:10.7759/cureus.36773
Fathi Karkan, S., Maleki Baladi, R., Shahgolzari, M., Gholizadeh, M., Shayegh, F., and Arashkia, A. (2022). The evolving direct and indirect platforms for the detection of SARS-CoV-2. J. Virol. Methods 300, 114381. doi:10.1016/j.jviromet.2021.114381
Fraser, C., Riley, S., Anderson, R. M., and Ferguson, N. M. (2004). Factors that make an infectious disease outbreak controllable. Proc. Natl. Acad. Sci. 101 (16), 6146–6151. doi:10.1073/pnas.0307506101
Galar, A., Catalán, P., Vesperinas, L., Miguens, I., Muñoz, I., García-Espona, A., et al. (2021). Use of saliva swab for detection of influenza virus in patients admitted to an emergency department. Microbiol. Spectr.;9, e0033621(1). doi:10.1128/spectrum.00336-21
Gholami, N., Hosseini Sabzvari, B., Razzaghi, A., and Salah, S. (2017). Effect of stress, anxiety and depression on unstimulated salivary flow rate and xerostomia. J. Dent. Res. Dent. Clin. Dent. Prospects 11 (4), 247–252. doi:10.15171/joddd.2017.043
Govindaraj, S., Daniel, M. J., Vasudevan, S. S., and Kumaran, J. V. (2019). Changes in salivary flow rate, pH, and viscosity among working men and women. Dent. Med. Res. 7 (2), 56. doi:10.4103/dmr.dmr_20_19
Han, M. S., Seong, M. W., Kim, N., Shin, S., Cho, S. I., Park, H., et al. (2020). Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, seoul, South Korea. Emerg. Infect. Dis. 26 (10), 2497–2499. doi:10.3201/eid2610.202449
Hasan, M. R., Mirza, F., Al-Hail, H., Sundararaju, S., Xaba, T., Iqbal, M., et al. (2020). Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLOS ONE 15 (7), e0236564. doi:10.1371/journal.pone.0236564
Katayama, Y., Murai, R., Moriai, M., Nirasawa, S., Saeki, M., Yakuwa, Y., et al. (2022). Does the timing of saliva collection affect the diagnosis of SARS-CoV-2 infection? J. Infect. Chemother. 28 (7), 1012–1014. doi:10.1016/j.jiac.2022.03.009
Kaur, S., Bherwani, H., Gulia, S., Vijay, R., and Kumar, R. (2021). Understanding COVID-19 transmission, health impacts and mitigation: timely social distancing is the key. Environ. Dev. Sustain 23 (5), 6681–6697. doi:10.1007/s10668-020-00884-x
Kim, M. J., Park, P. G., Hwang, S. J., Bang, S. J., Jung, J. H., Kown, E., et al. (2024). Saliva-based Proteinase K method: a rapid and reliable diagnostic tool for the detection of SARS-COV-2 in children. J. Med. Virology 96 (1), e29361. doi:10.1002/jmv.29361
Kinloch, N. N., Shahid, A., Ritchie, G., Dong, W., Lawson, T., Montaner, J. S. G., et al. (2020). Evaluation of nasopharyngeal swab collection techniques for nucleic acid recovery and participant experience: recommendations for COVID-19 diagnostics. Open Forum Infect. Dis. 7 (11), ofaa488. doi:10.1093/ofid/ofaa488
Kissler, S. M., Fauver, J. R., Mack, C., Olesen, S. W., Tai, C., Shiue, K. Y., et al. (2021a). Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLOS Biol. 19 (7), e3001333. doi:10.1371/journal.pbio.3001333
Kissler, S. M., Fauver, J. R., Mack, C., Tai, C. G., Breban, M. I., Watkins, A. E., et al. (2021b). Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N. Engl. J. Med. 385 (26), 2489–2491. doi:10.1056/NEJMc2102507
Larios Serrato, V., Meza, B., Gonzalez-Torres, C., Gaytan-Cervantes, J., González Ibarra, J., Santacruz Tinoco, C. E., et al. (2023). Diversity, composition, and networking of saliva microbiota distinguish the severity of COVID-19 episodes as revealed by an analysis of 16S rRNA variable V1-V3 region sequences. mSystems 8 (4), e0106222–22. doi:10.1128/msystems.01062-22
Lee, S. H., McGrath, J., Connolly, S. P., and Lambert, J. (2021). Partial N gene sequencing for SARS-CoV-2 verification and pathway tracing. Int. Med. Case Rep. J. 14, 1–10. doi:10.2147/IMCRJ.S291166
Li, M., Ge, H., Sun, Z., Fu, J., Cao, L., Feng, X., et al. (2022). A loop-mediated isothermal amplification-enabled analytical assay for the detection of SARS-CoV-2: a review. Front. Cell Infect. Microbiol. 12, 1068015. doi:10.3389/fcimb.2022.1068015
Ma, Q., Yao, J., Yuan, S., Liu, H., Wei, N., Zhang, J., et al. (2019). Development of a lateral flow recombinase polymerase amplification assay for rapid and visual detection of Cryptococcus neoformans/C. gattii in cerebral spinal fluid. BMC Infect. Dis. 19, 108. doi:10.1186/s12879-019-3744-6
Matsumura, Y., Nakazaki, T., Kitamori, K., Kure, E., Shinohara, K., Tsuchido, Y., et al. (2023). Development and evaluation of the automated multipurpose molecular testing system PCRpack for high-throughput SARS-CoV-2 testing. Microbiol. Spectr. 11 (6), e0271623–23. doi:10.1128/spectrum.02716-23
McPhillips, L., and MacSharry, J. (2022). Saliva as an alternative specimen to nasopharyngeal swabs for COVID-19 diagnosis: review. Access Microbiol. 4 (5), acmi000366. doi:10.1099/acmi.0.000366
Mytzka, N., Arbaciauskaite, S., Sandetskaya, N., Mattern, K., and Kuhlmeier, D. (2023). A fully integrated duplex RT-LAMP device for the detection of viral infections. Biomed. Microdevices 25 (4), 36. doi:10.1007/s10544-023-00676-w
Naranbat, D., Schneider, L., Kantor, R., Beckwith, C. G., Bazerman, L., Gillani, F., et al. (2022). DirectDetect SARS-CoV-2 direct real-time RT-PCR study using patient samples. ACS Omega 7 (6), 4945–4955. doi:10.1021/acsomega.1c05595
NCBI (2020). Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/human/AUS/VIC01/2020, complete genome. Available at: http://www.ncbi.nlm.nih.gov/nuccore/MT007544.1 Accessed June 5, 2024.
NCBI (2024). Homo sapiens actin beta (ACTB), mRNA. Accessed June 5, 2024. Available at: http://www.ncbi.nlm.nih.gov/nuccore/NM_001101.5
Nikolai, L. A., Meyer, C. G., Kremsner, P. G., and Velavan, T. P. (2020). Asymptomatic SARS coronavirus 2 infection: invisible yet invincible. Int. J. Infect. Dis. 100, 112–116. doi:10.1016/j.ijid.2020.08.076
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28 (12), e63. doi:10.1093/nar/28.12.e63
Olearo, F., Nörz, D., Heinrich, F., Sutter, J. P., Roedl, K., Schultze, A., et al. (2021). Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. J. Clin. Virology 137, 104782. doi:10.1016/j.jcv.2021.104782
Ostheim, P., Alemu, S. W., Tichý, A., Sirak, I., Davidkova, M., Stastna, M. M., et al. (2022). Examining potential confounding factors in gene expression analysis of human saliva and identifying potential housekeeping genes. Sci. Rep. 12 (1), 2312. doi:10.1038/s41598-022-05670-5
Prado, N. O., Marin, A. M., Lalli, L. A., Sanchuki, H. B. S., Wosniaki, D. K., Nardin, J. M., et al. (2024). Development and evaluation of a lyophilization protocol for colorimetric RT-LAMP diagnostic assay for COVID-19. Sci. Rep. 14 (1), 10612. doi:10.1038/s41598-024-61163-7
Promega (2024). Lyophilized master mixes. Available at: https://www.promega.com/custom-solutions/custom-manufacturing/lyophilized-master-mixes/ Accessed June 17, 2024.
Rereddy, S. K., Cao, A. C., Blackwell, B., Poling-Skutvik, R., Arratia, P. E., and Mirza, N. (2023). Rheology of saliva in health and disease. Biorheology 59 (1-2), 19–27. doi:10.3233/BIR-210014
Rivas-Macho, A., Sorarrain, A., Marimón, J. M., Goñi-de-Cerio, F., and Olabarria, G. (2023). Extraction-free colorimetric RT-LAMP detection of SARS-CoV-2 in saliva. Diagn. (Basel) 13 (14), 2344. doi:10.3390/diagnostics13142344
Ross, G. M. S., Filippini, D., Nielen, M. W. F., and Salentijn, G. I. J. (2020). Unraveling the hook effect: a comprehensive study of high antigen concentration effects in sandwich lateral flow immunoassays. Anal. Chem. 92 (23), 15587–15595. doi:10.1021/acs.analchem.0c03740
Savela, E. S., Viloria Winnett, A., Romano, A. E., Porter, M. K., Shelby, N., Akana, R., et al. (2022). Quantitative SARS-CoV-2 viral-load curves in paired saliva samples and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J. Clin. Microbiol. 60 (2), e0178521–21. doi:10.1128/jcm.01785-21
Saxena, A., Rai, P., Mehrotra, S., Baby, S., Singh, S., Srivastava, V., et al. (2022). Development and clinical validation of RT-LAMP-based lateral-flow devices and electrochemical sensor for detecting multigene targets in SARS-CoV-2. Int. J. Mol. Sci. 23 (21), 13105. doi:10.3390/ijms232113105
Schneider, L., Blakely, H., and Tripathi, A. (2019). Mathematical model to reduce loop mediated isothermal amplification (LAMP) false-positive diagnosis. ELECTROPHORESIS 40 (20), 2706–2717. doi:10.1002/elps.201900167
Schokker, E. P., and van Boekel, A. J. S. (1997). Kinetic modeling of enzyme Inactivation: kinetics of heat inactivation at 90−110 °C of extracellular proteinase from Pseudomonas fluorescens 22F. J. Agric. Food Chem. 45 (12), 4740–4747. doi:10.1021/jf970429i
Song, X., Coulter, F. J., Yang, M., Smith, J. L., Tafesse, F. G., Messer, W. B., et al. (2022). A lyophilized colorimetric RT-LAMP test kit for rapid, low-cost, at-home molecular testing of SARS-CoV-2 and other pathogens. Sci. Rep. 12 (1), 7043. doi:10.1038/s41598-022-11144-5
Sosa-Jurado, F., Hernández-Galindo, V. L., Meléndez-Mena, D., Mendoza-Torres, M. A., Martínez-Arroniz, F. J., Vallejo-Ruiz, V., et al. (2014). Detection of hepatitis C virus RNA in saliva of patients with active infection not associated with periodontal or liver disease severity. BMC Infect. Dis. 14 (1), 72. doi:10.1186/1471-2334-14-72
Stankiewicz Karita, H. C., Dong, T. Q., Johnston, C., Neuzil, K. M., Paasche-Orlow, M. K., Kissinger, P. J., et al. (2022). Trajectory of viral RNA load among persons with incident SARS-CoV-2 G614 infection (wuhan strain) in association with COVID-19 symptom onset and severity. JAMA Netw. Open 5 (1), e2142796. doi:10.1001/jamanetworkopen.2021.42796
Sweeney, P. J., and Walker, J. M. (1993). “Proteinase K (EC 3.4.21.14),” in Enzymes of molecular biology. Editor M. M. Burrell (Advanced Technologies Limited, Cambridge, United Kingdom: Humana Press), 305–311. doi:10.1385/0-89603-234-5:305
Tang, K. D., Baeten, K., Kenny, L., Frazer, I. H., Scheper, G., and Punyadeera, C. (2019). Unlocking the potential of saliva-based test to detect HPV-16-Driven oropharyngeal cancer. Cancers (Basel) 11 (4), 473. doi:10.3390/cancers11040473
Tang, J., Zhu, J., Wang, J., Qian, H., Liu, Z., Wang, R., et al. (2024). Development and clinical application of loop-mediated isothermal amplification combined with lateral flow assay for rapid diagnosis of SARS-CoV-2. BMC Infect. Dis. 24 (1), 81. doi:10.1186/s12879-023-08924-3
Thompson, D., and Lei, Y. (2020). Mini review: recent progress in rt-lamp enabled covid-19 detection. Sensors Actuators Rep. 2 (1), 100017. doi:10.1016/j.snr.2020.100017
Valente, V., Teixeira, S. A., Neder, L., Okamoto, O. K., Oba-Shinjo, S. M., Marie, S. K., et al. (2009). Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative RT-PCR. BMC Mol. Biol. 10, 17. doi:10.1186/1471-2199-10-17
Vealan, K., Joseph, N., Alimat, S., Karumbati, A. S., and Thilakavathy, K. (2023). Lateral flow assay: a promising rapid point-of-care testing tool for infections and non-communicable diseases. Asian Biomed. 17 (6), 250–266. doi:10.2478/abm-2023-0068
Vogels, C. B. F., Watkins, A. E., Harden, C. A., Brackney, D. E., Shafer, J., Wang, J., et al. (2021). SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. (N Y) 2 (3), 263–280.e6. doi:10.1016/j.medj.2020.12.010
Vohra, P., Vohra, P., Belkhode, V., Nimonkar, S., Potdar, S., Bhanot, R., et al. (2020). Evaluation and diagnostic usefulness of saliva for detection of HIV antibodies: a cross-sectional study. J. Fam. Med. Prim. Care 9 (5), 2437–2441. doi:10.4103/jfmpc.jfmpc_138_20
Waters, S., Lee, S., Lloyd, M., Irish, A., and Price, P. (2019). The detection of CMV in saliva can mark a systemic infection with CMV in renal transplant recipients. Int. J. Mol. Sci. 20 (20), 5230. doi:10.3390/ijms20205230
Winnett, A., Cooper, M. M., Shelby, N., Romano, A. E., Reyes, J. A., Ji, J., et al. (2020). SARS-CoV-2 viral load in saliva rises gradually and to moderate levels in some humans. medRxiv Prepr. Serv. health Sci. doi:10.1101/2020.12.09.20239467
Wu, W., Cheng, Y., Zhou, H., Sun, C., and Zhang, S. (2023). The SARS-CoV-2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostics. Virology J. 20 (1), 6. doi:10.1186/s12985-023-01968-6
Wyllie, A. L., Fournier, J., Casanovas-Massana, A., Campbell, M., Tokuyama, M., Vijayakumar, P., et al. (2020). Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. Published online April 22, 2020:2020.04.16.20067835. doi:10.1101/2020.04.16.20067835
Yalley, A. K., Ahiatrogah, S., Kafintu-Kwashie, A. A., Amegatcher, G., Prah, D., Botwe, A. K., et al. (2022). A systematic review on suitability of molecular techniques for diagnosis and research into infectious diseases of concern in resource-limited settings. Curr. Issues Mol. Biol. 44 (10), 4367–4385. doi:10.3390/cimb44100300
Zasada, A. A., Mosiej, E., Prygiel, M., Polak, M., Wdowiak, K., Formińska, K., et al. (2022). Detection of SARS-CoV-2 using reverse transcription helicase dependent amplification and reverse transcription loop-mediated amplification combined with lateral flow assay. Biomedicines 10 (9), 2329. doi:10.3390/biomedicines10092329
Zhang, Y., Ren, G., Buss, J., Barry, A. J., Patton, G. C., and Tanner, N. A. (2020). Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 69 (3), 178–185. doi:10.2144/btn-2020-0078
Zhang, C., Zheng, T., Wang, H., Chen, W., Huang, X., Liang, J., et al. (2021). Rapid one-pot detection of SARS-CoV-2 based on a lateral flow assay in clinical samples. Anal. Chem. 93 (7), 3325–3330. doi:10.1021/acs.analchem.0c05059
Zheng, S., Fan, J., Yu, F., Feng, B., Lou, B., Zou, Q., et al. (2020). Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ 369, m1443. doi:10.1136/bmj.m1443
Keywords: loop mediated isothermal amplication (LAMP), PoC (point of care), lateral flow biosensor (LFB), saliva test, infectious disease diagnostics, liquid handling automation
Citation: Naranbat D, Murphy J and Tripathi A (2024) An automated syringe-based PoC RT-LAMP LFB platform for infectious disease detection from saliva. Front. Lab. Chip. Technol. 3:1450891. doi: 10.3389/frlct.2024.1450891
Received: 18 June 2024; Accepted: 15 August 2024;
Published: 27 August 2024.
Edited by:
Xiaohua Huang, University of Memphis, United StatesReviewed by:
Zhengpeng Wan, Massachusetts Institute of Technology, United StatesGeorgette B. Salieb-Beugelaar, Münster University of Applied Sciences, Germany
Copyright © 2024 Naranbat, Murphy and Tripathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anubhav Tripathi, YW51Ymhhdl90cmlwYXRoaUBicm93bi5lZHU=
 Dulguunnaran Naranbat
Dulguunnaran Naranbat John Murphy
John Murphy Anubhav Tripathi
Anubhav Tripathi