- Robert F. Smith School of Chemical and Biomolecular Engineering, Cornell University, Ithaca, NY, United States
Carbohydrates, also known glycans, are ubiquitous in nature and exhibit a wide array of biological functions essential to life. Glycans often exist as conjugates of proteins or lipids and reside predominantly at the surface of cells, where their structure and composition are known to vary in a disease-dependent fashion. This observation has encouraged the development of tools for monitoring glycan patterns on individual molecules, cells, and tissues, to elucidate the links between glycosylation and disease for therapeutic and diagnostic applications. Over the past 2 decades, microfluidic technology has emerged as an advantageous tool for profiling the glycan content of biological systems. Miniaturizing carbohydrate analysis can circumvent several challenges commonly encountered with conventional-scale analytical techniques such as low throughput and poor detection sensitivity. The latter is often complicated by the low abundance of glycans in biological specimens and the complexity of carbohydrate structures, which often necessitates extensive concentration and purification of glycans to discern their structural features. We previously examined the application of microfluidics in the synthesis of carbohydrates in a recent paper (Pinnock et al., Anal. Bioanal. Chem., 2022, 414 (18), 5139–63). This review builds upon that discussion by delving into the application of microfluidics in the complementary field of carbohydrate analysis. Special attention is given to applications related to glycomics and the ways that microfluidics have enhanced the sensitivity, reproducibility, and throughput of carbohydrate identification and structural characterization.
1 Introduction
Carbohydrates, also known as glycans, are everywhere in nature. Since the early 19th century, it has been known that cells across all domains of life produce glycans essential to cell survival (Marth, 2008; Varki, 2011). Indeed, the most abundant biopolymers on earth are polysaccharides expressed on the cell walls of plants (e.g., cellulose) and fungi (e.g., chitin) (Gooday, 1990). These biopolymers provide structural rigidity and protection to the cells that they coat, which has made them attractive as biodegradable raw materials for the construction, textiles, and paper industries (Klemm et al., 2005). The abundance of polymeric carbohydrates reinforced early notions that glycans functioned solely as structural materials and energy depots in nature. In the early 20th century, advances in analytical technologies led to major breakthroughs in the knowledge of carbohydrate structure that suggested otherwise (Cabezas, 1994). Notably, the isolation of lipid-linked glycans, or glycolipids, from animal brains provided some of the earliest evidence that glycans not only existed as free monomers or polymers but were also commonly found covalently bound to other macromolecules. These so-called glycoconjugates included protein-linked carbohydrates, or glycoproteins, the first of which was isolated from human serum towards the middle of the century (Schmid, 1950). The half-life of serum glycoproteins was shown to depend on the identity of the terminal sugar residue, suggesting that the structure of the carbohydrate moiety is a key determinant of the activity and transport of its non-carbohydrate conjugate (Ashwell and Morell, 2006). Several additional key discoveries soon followed, including an observation that carbohydrate-binding proteins (CBP), termed lectins, agglutinated red blood cells in a blood-type dependent fashion (Sharon and Lis, 1987). Not only did this reveal sugars to be the determinants of ABO blood type but also provided some of the earliest proof of the presence of sugars on the surface of animal cells (Sharon and Lis, 1987). Thus, by the 1980s the diversity of glycan structure and glycans’ important cellular functions were finally considered a critical dimension of biology and culminated in the birth of the field of Glycobiology.
It is now known that glycoconjugates exist in high abundance at the cell surface, where they form a dense network of carbohydrates called the glycocalyx (Möckl, 2020). It is here where the varied and vital roles that glycans play in biology are put on full display. This sugary coat provides the first point of contact between a cell and its extracellular environment. Many host-pathogen encounters begin with interactions between cell surface glycans and the CBPs displayed on or secreted by the attacking pathogen. Several enveloped viruses express CBPs called hemagglutinins which bind to specific sugar residues on host cell membranes to initiate viral entry (Y. Suzuki et al., 2000; Matrosovich, Herrler, and Klenk, 2015; Walther et al., 2013). The glycocalyx also serves as a protective layer. Heavily glycosylated proteins, such as mucins and polysaccharide-covered proteoglycans, form a physical barrier to foreign objects attempting to enter cells. Mucin-bound glycans further behave as a viscous, gel-like material capable of trapping and clearing infectious particles before they reach the plasma membrane (Lillehoj et al., 2013). Alterations in the composition and length of glycocalyx glycans is a hallmark of many oncological diseases and has inspired interest in these molecules as robust diagnostic markers of tumor growth and metastasis.
Elucidating the relationship between the observed functions of glycans and the structure of these molecules is glycobiology’s central yet formidable challenge. As such, much work in the field has centered on developing tools for the identification and sequencing of glycans. To characterize carbohydrate structures, researchers must contend with the heterogeneity of natural glycan pools and their low abundance in complex biological mixtures. Conventional analytical techniques, such as Mass Spectrometry (MS), have dynamic range limitations that call for extensive purification and concentration of glycans before analysis. For these reasons, glycoscience continues to demand advances in measurement sensitivity and the creation of streamlined approaches for glycan capture and clean-up. These demands coincide with a growing interest in “omics” style investigations that seek to define the glycome of whole cells, which includes the full set of glycan structures expressed by a cell under a given set of environmental and physiological conditions. Glycomics requires the ability to analyze larger numbers of glycan structures and has necessitated the development of analytical technologies with high-throughput potential.
A parallel focus of glycobiology has been to define the functions of glycans. The goal of many functional studies is to measure interactions of discrete glycan structures with select CBPs or carbohydrate motifs. These analyses require carbohydrate pools that are both well-defined and homogenous, and these criteria are rarely met by glycans derived from natural sources. It is no surprise that many of the earliest studies of glycan function centered on the most abundant classes of glycoconjugates. A reliance on synthetic carbohydrates to fill this gap has catalyzed advances in carbohydrate chemistry that have made the synthesis of these molecules increasingly more tractable. These breakthroughs along with the prevailing challenges in the field of carbohydrate synthesis have been extensively reviewed elsewhere by our lab (Pinnock and Daniel, 2022) and others (Koeller and Wong, 2000; Sears and Wong, 2001; Seeberger, 2008; Hsu et al., 2011; Nielsen and Pedersen, 2018; Panza et al., 2018), and will not be discussed further in this review.
Over the last 20 years, microfluidic technology has advanced analytical processes in several different sectors including chemical manufacturing, pharmaceutical synthesis, and nucleic acid isolation. Several features of these micron-sized devices make them attractive tools for glycomics-related investigations. The small dimensions provide users with tight control of fluid flow, which can be exploited to separate analytes with higher resolution. The sub-microliter volumes can reduce the time and size of samples required to isolate glycans in the quantities and purity required for structural analysis. Micro-structured platforms are also highly modular, allowing easy integration of multiple processing units in series and in parallel. Carbohydrates often must undergo several preparatory steps before analysis, which can be streamlined by integrating these unit operations on-chip. This last feature makes microfluidics particularly well-suited for process automation. It is therefore not surprising that microfluidics have often improved the efficiency and sensitivity of glycan and glycoconjugate analyses.
This review explores the ways in which microfluidics have helped researchers overcome bottlenecks in the structural analysis of glycans particularly in the context of glycomics. We begin with an overview of the current challenges in carbohydrate analysis and delve deeper into the features of microfluidics that make them highly attractive for glycomics research (Section 2). We then explore applications of microfluidics at different stages of the glycan analysis workflow, including the capture of glycoproteins and cleavage of glycan moieties (Section 3), separation of released glycans (Section 4), and the evaluation of glycan structure and content (Section 5). Lastly, we highlight a subset of platforms designed as micro total analysis systems to automate carbohydrate analysis and the potential of such systems to accelerate the characterization of complex glycans (Section 6). While the miniaturization of carbohydrate analysis is still in its infancy, micro-analytical systems have already helped researchers uncover new relationships between glycosylation and human health and disease. We have selected here interesting studies concerned with the structural analysis of glycans comprising diverse biological samples, with a focus on applications that have aided the glycomic analysis of clinical samples or biotherapeutic molecules.
2 The current state of carbohydrate analysis
2.1 The challenges of glycan structural analysis
A major bottleneck in the field of glycobiology is the analysis of glycan structure. Several qualities of glycans make their structural determination challenging. Foremost is the heterogeneity of glycans structure which is reflected in the exquisite structural diversity of these molecules. Glycans exist as non-conjugated molecules, such as sugar monomers and oligomers (i.e., monosaccharides and oligosaccharides), and as conjugates of other biomolecules, namely, proteins and lipids (e.g., glycoproteins and glycolipids). Glycoproteins are one of the most studied classes of glycosylated molecules. Protein-linked glycans are sub-classified by the linkage between the amino acid and the carbohydrate moiety, with glycans that are N-linked to asparagine (N-glycans) and O-linked to serine or threonine (O-glycans) being the predominant types.
Glycans display rich compositional diversity which stems from the vast number of monosaccharide building blocks found in nature. Human glycans are generally constructed from ten types of monosaccharides, including galactose (Gal), glucose (Glc), N-acetylgalactosamine (GalNAc), N-acetylglucosamine (GlcNAc), mannose (Man), xylose (xyl), fucose (Fuc), sialic acid (Nue5Ac), glucuronic acid (GlcA), and iduronic acid (IdoA). These monomers are linked together by glycosidic bonds that form between the anomeric carbon of one monosaccharide and the hydroxyl group of another. Because monosaccharides carry several hydroxyl groups, the conjugation of two monomers can yield multiple disaccharide molecules with distinct regiochemistry, stereochemistry, and biological activities. The number of possible configurations increases exponentially with each additional monomer. For instance, constructing a tetrasaccharide using one type of monosaccharide could yield 1792 distinct structures (Lebrilla et al., 2022).
The structural diversity of glycans also arises from the heterogeneous nature of glycosylation, which is the process of glycan biosynthesis. Glycosylation is mediated by glycosyltransferases, a class of biosynthetic enzymes that reside in sub-cellular membrane compartments such as the Endoplasmic Reticulum and the Golgi apparatus in eukaryotes. Unlike protein biosynthesis, glycosylation is not template driven. Rather, a myriad of factors dictates glycan structure, including the availability of sugar donor substrates, the expression of specific biosynthetic enzymes, and the competition between biosynthetic pathways. Consequently, a single glycoprotein can have multiple glycoforms, differing in the number and sites of glycosylation on the peptide backbone (macro-heterogeneity) as well as the monosaccharide composition, sequence, and branching of the carbohydrate chain at each site (micro-heterogeneity) (Thaysen-Andersen, Packer, and Schulz, 2016) (Figure 1, left-hand side). Erythropoietin (EPO), a biotherapeutic glycoprotein used to treat anemia, offers a prime example of glycoprotein heterogeneity. EPO contains four sites of glycosylation, and variations in the carbohydrate moiety occupying each site have been detected in recombinant versions of the protein (Y. Yang et al., 2016; Čaval et al., 2018). Furthermore, sialylation and branching of its N-linked glycans have been shown to strongly influence the half-life and hematopoietic activity of the molecule (Fukuda et al., 1989; Walsh and Jefferis, 2006).
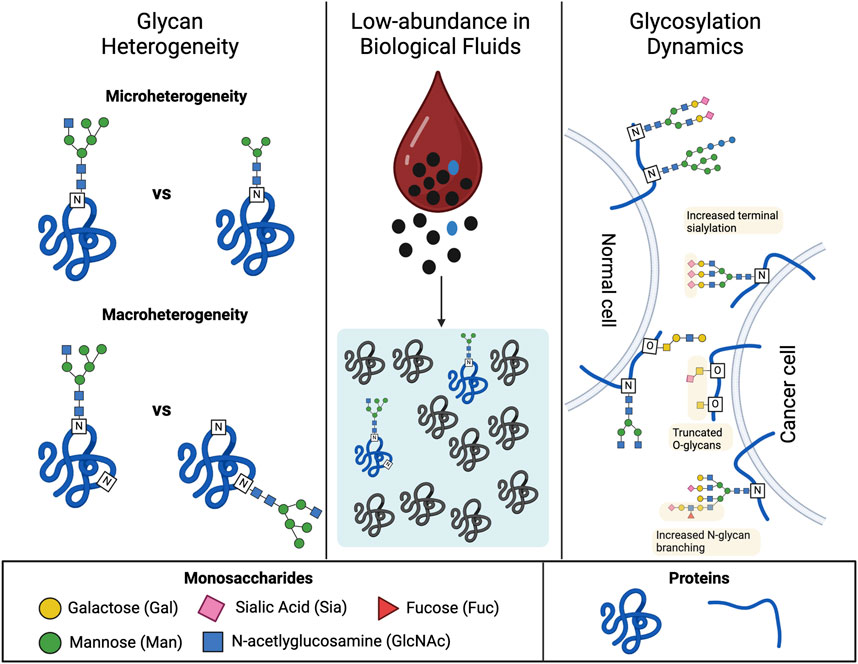
FIGURE 1. Major sources of complexity in the structural analysis of glycans. The left panel illustrates elements of glycan heterogeneity observed at the molecular level for glycoproteins. The top section depicts how the structure of the carbohydrate moiety at each glycosylation site can also vary (microheterogeneity). The bottom section depicts how a single glycoprotein can exist as multiple glycoforms, which differ in the number and/or location of glycosylation sites (macroheterogeneity). The middle panel depicts the characteristic low abundance of glycoproteins in biological fluids, such as blood, which hinders their detection. Many glycoproteins exist at or below the pg/mL level which is nine orders of magnitude below the mg/mL levels observed for the most abundant serum proteins (i.e., albumin). The right panel portrays the dynamics of glycosylation in disease progression. The diagram highlights glycosylation patterns that correlate with healthy versus cancerous cells. Signatures of cell-surface glycosylation observed on tumor cells include increased N-glycan branching, truncated O-glycans, and increased terminal sialylation. Created with BioRender.com.
Another factor complicating the structural analysis of glycans is that they generally exist in trace amounts within complex biological mixtures such as cells, tissues, and bodily fluids (Figure 1, middle). Recovery of carbohydrates from these biological soups often requires multiple rounds of purification and concentration to enable sensitive detection of the glycan components. Strategies that reduce the time, cost, and labor currently required to isolate glycans with high enough concentrations and purity are highly desired.
A final source of difficulty in the study of glycans stems from the desire to examine glycosylation at the level of whole cells, tissues, and even organisms. Systemic changes in glycosylation are known to accompany different cellular events, particularly disease progression. For instance, a common signature of malignant tumor cells is the truncation, branching, and increased sialylation of cell surface glycans, which has inspired interest in glycans as biomarkers for cancer (Guo et al., 2022) (Figure 1, right-hand side). Profiling the glycome of individual cells and tissue, thus, serves as a powerful strategy for identifying new disease biomarkers and novel molecular targets for therapeutic intervention. However, these studies present a unique challenge given the magnitude of glycans that must be examined to define a glycome, many of which are conjugated to membrane-bound molecules that require additional processing steps to recover glycans for analysis. Analytical technologies with high throughput potential have received increasing attention due to the breadth of glycan structures that must be characterized.
2.2 Conventional approaches to carbohydrate analysis
For readers who may be new to the field of glycobiology, we provide a brief overview of traditional approaches to carbohydrate structural analysis. For an in-depth primer on these state-of-the-art techniques, we refer readers to several reviews (Gaunitz et al., 2017; Gray et al., 2019; Kinoshita and Yamada, 2022) and book chapters (Haslam et al., 2022; Rudd et al., 2022) that have recently been published on the topic.
Structural analysis of carbohydrates includes both the methods used to prepare glycan samples and the techniques used to characterize their structural features. The general experimental workflow for carbohydrate analysis is depicted in Figure 2. Procedures begin with the collection of a biological sample, such as an aliquot of cultured cells, tissue, or bodily fluid, which is then processed to extract and prepare constituent carbohydrate molecules for analysis. The type and sequence of preparatory steps undertaken depend on the sample under investigation as well as the intended analytical approach(s). Figure 2 provides a breakdown of multiple possible analytical pathways with an exemplary protocol outlined by bold arrows and lines. In this procedure, once crude liquid or cellular extracts are obtained, samples are subjected to an enrichment step to isolate glycoproteins from other biomolecular contaminants present in the crude mixture. Enzymatic or chemical methods are then used to release glycans from the protein scaffold to enable the evaluation of the glycan separate from the protein molecules. Enzymatic cleavage is the gold standard for the release of N-linked glycans, which we discuss in more detail in Section 3. After their release, glycans are derivatized with chemical tags to facilitate their separation and/or enhance their detection downstream. Glycans are often subjected to a separation step ahead of structural evaluation. As discussed below, some separation methods can also provide qualitative information about the structure of the glycans in the sample. The final step in the workflow is the actual structural evaluation.
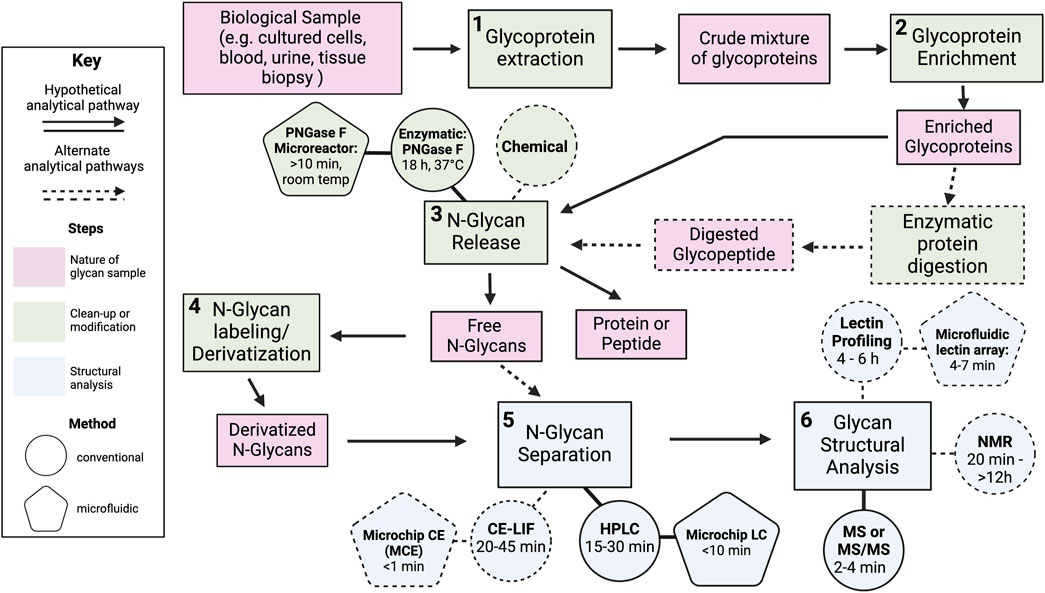
FIGURE 2. Overview of the workflow for the structural analysis of N-linked glycans. Several possible analytical pathways are indicated by dashed arrows. A hypothetical pathway is highlighted by bold arrows and lines. In this pathway, analysis begins with (1) the extraction of glycoproteins from a biological sample such as cultured cells, blood, urine, or a tissue biopsy. The crude mixture is then serially subjected to (2) a glycoprotein enrichment step to concentrate and/or purify the glycoproteins, (3) enzymatic glycan cleavage with PNGase F to release N-linked glycans, (4) a labeling step to tag released N-glycans with molecules to improve their separation and/or detection downstream, (5) separation of N-glycans by high-performance liquid chromatography, and (6) analysis by mass spectrometry (MS) to evaluate glycan structural features. Pink boxes describe the nature of the carbohydrate sample before and after each procedural step listed in the green boxes. Green boxes represent steps involving the modification or clean-up carbohydrates prior to their structural analysis. Blue boxes represent steps related to the characterization of carbohydrate structure. Several boxes are linked to circles which denote methods used to carry out the task described in the box. For instance, the release of N-linked glycans (green box) from “enriched proteins” (pink box) or “digested glycopeptides” (pink box) can be performed with chemical (green circle) or enzymatic (green circle) methods. Several preparatory and analytical method have been adapted into microfluidic platforms to improve processing speed. These microfluidic techniques are depicted as pentagons and linked to the circles representing the conventional scale technique. The average processing time for a single sample is noted in parentheses for both the macro-scale technique and its microfluidic analogue. Created with BioRender.com.
The primary structure of a glycan is defined by a wide range of features. Among them are monosaccharide composition and sequence, connectivity (position and type of glycosidic linkage), branching, as well as the number and length of antennae. Several of these structural features are depicted in Figure 3. Following glycan release, parent proteins may also be examined to gather information about the site(s) of glycan attachment. No one analytical method can account for all these structural features. As such, most analyses leverage multiple analytical techniques to increase the confidence of the findings. The choice of method(s) is often dictated by the type of glycan under investigation and/or the proposed biological question.
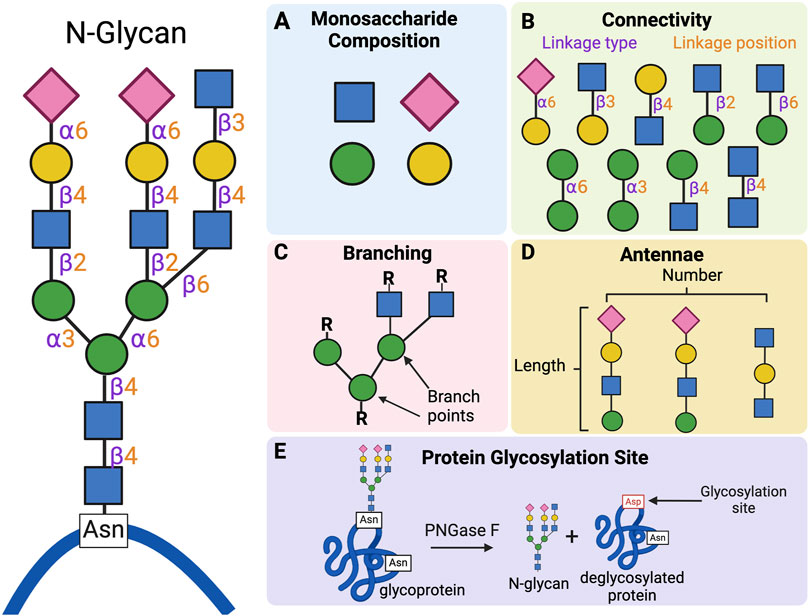
FIGURE 3. Examples of glycan structural features evaluated during structural studies of carbohydrates. Glycans possess many structural features that can be probed during structural analysis. Some of the most analyzed features are depicted here for a prototypical N-linked glycan (left panel). (A) The first feature is monosaccharide composition. The N-glycan shown here contains sialic acid (pink rhombus), mannose (green circle), galactose (yellow circle), and N-acetylglucosamine (blue square). (B) Another element is connectivity information, which includes the type of linkages (α or β) and the positions (e.g., α3, α6, β2, β4, β6) between the two sugars. (C) The location of branches in the structure, which occurs anytime a single sugar is linked to three or more other sugars, may also be elucidated. Branch points are found at two mannose residues within the core of the glycan structure. (D) Each branch extends into a linear oligosaccharide chain called an antennae, the number and length of which may also be determined. The depicted structure possesses three antennae containing three to four monosaccharides each. (E) Information about the location of glycosylation site may also be obtained by analyzing the deglycosylated protein. After treatment with PNGase F, asparagine (Asn) residues that were previously conjugated to a glycan moiety are converted to aspartic acid (Asp). Analytical techniques can be used to identify the location of aspartic acids generated after glycan removal to predict the location of glycosylation sites. Created with BioRender.com.
Conventional carbohydrate analytical techniques can be classified based on the level of structural detail they provide. The simplest techniques detect the presence or absence of a monosaccharide or oligosaccharide motif. This is commonly done using lectin profiling assays, wherein lectins, a class of proteins that bind to specific monosaccharide or oligosaccharides motifs, are used to monitor the presence of the corresponding carbohydrate binding partner. In this way, general information about glycan composition can be discerned. Fluorescently tagged lectins are typically used to visualize and quantify the abundance of the corresponding monosaccharide(s). Over 60 lectins are commercially available (Bojar et al., 2022), making it possible to assay a wide range of structural motifs in a high-throughput manner. Nevertheless, many lectins exhibit promiscuous binding, making these assays susceptible to false structural assignments. Carbohydrate-specific antibodies can be applied in a similar manner to mitigate non-specific binding, but the high cost and higher specificity of antibodies makes antibody-based profiling a less affordable and versatile alternative.
Many separation techniques employed in carbohydrate analysis can also be used to indirectly identify glycan structures. Two popular techniques include liquid chromatography and capillary electrophoresis. High-performance liquid chromatography (HPLC) is the most robust technique for carbohydrate separation. During HPLC, mixtures of glycans suspended in a mobile phase are passed through a column packed with a stationary phase by pressure-driven flow. Components of the mobile phase interact differently with the stationary phase, resulting in their elution from the column at different speeds. The resultant retention data can be compared with annotated chromatographic data obtained under similar conditions to make inferences about glycan structure. Various types of HPLC methods are employed in carbohydrate analysis, with the most popular being hydrophilic interaction liquid chromatography (HILIC) and reverse-phase LC (RPLC). Major hindrances in the use of HPLC are its low throughput, with run times ranging from 20 to 45 min per experiment, and the high cost to execute separation experiments. Capillary electrophoresis (CE) is an electrophoretic separation method commonly used in carbohydrate analysis. During CE, glycans are subjected to an electric field resulting in their differential migration through the separation medium based on the charge of the molecule. CE is attractive because separations proceed quickly relative to liquid chromatography. The main limitation of CE is its poor coupling to MS, as CE operates at negligible flow rates (<20 nL/min) that are incompatible with the flow rates used with MS.
Mass spectrometry (MS) and nuclear magnetic resonance spectroscopy (NMR) are analytical techniques used to probe fine structural details of glycans. MS is an analytical tool used to determine the mass-to-charge ratio (m/z) of one or more molecules present in a sample. During MS experiments, carbohydrates in the sample of interest are converted to ions by an ionization source and transmitted to a detector (i.e., mass analyzer), that measures ions according to their m/z. MS-based methods are the dominant analytical tool in carbohydrate analysis, as they can provide information about carbohydrate primary structure and quantity. Advances in the MS instrumentation now permit sensitive detection down to femtomole concentrations of glycans (Gray et al., 2019; Haslam et al., 2022). However, given that glycans have poor ionization efficiency, sensitive detection often requires derivatizing or labeling glycans with more ionizable groups. A major limitation of MS is its inability to distinguish structural isomers or stereochemical information. For this reason, MS is often coupled to separation techniques, like LC and to a lesser extent CE, that can resolve isomeric structures prior to MS analysis. Aside from liquid-phase separation, gas phase separation techniques, like ion-mobility spectrometry (IMS) (Gray et al., 2019), have recently begun to be interfaced with MS instrumentation and leveraged in structural studies of carbohydrates. Despite the many details that MS provides, this method cannot deduce the complete primary structure of a glycan on its own. NMR is the only method that allows for de novo structural analysis of glycans. Given that all glycans contain hydrogen atoms, H1 NMR is a broadly applicable in structural glycobiology. The method is both non-destructive and quantitative, and can provide information about primary, secondary, and tertiary structure of glycans. However, NMR is a highly specialized technique that requires a high level of expertise for data acquisition and interpretation, much of which comes at high cost for non-expert users. The biggest hurdle in NMR is its low sensitivity. Resolving the simplest structure can require up to several milligrams of sample. Moreover, structure assignment can be a Herculean task when no complementary structural information is provided. Thus, most structural studies employ one or more of the aforementioned methods in combination with MS.
To address some of the above-mentioned limitations in glycan preparation and analysis, several of these methods have been adapted into microfluidic platforms (Figure 2). The benefits that microfluidics afford are expounded upon below.
2.3 Why microfluidics for carbohydrate analysis?
For readers new to and interested in the field of microfluidics, below we provide a brief introduction to general attributes and merits of microfluidic systems. We also highlight key advantages that microfluidic technology can provide to studies of carbohydrate structure and composition. Many of these points will be explored in greater detail using illustrative examples in the subsequent sections of the review.
Since their inception, microfluidic platforms have found broad applications in chemical and bioanalytical assays. The term “microfluidics” encompasses a broad range of technologies designed to manipulate fluid flow at length scales ranging from a few micrometers up to a millimeter (Whitesides, 2006). Several qualities of microfluidics make them robust tools for analyzing complex carbohydrates. An obvious merit is that micro-technologies require small sample volumes, allowing the consumption of precious samples and associated costs to be kept to a minimum. A typical micro-flow device accommodates from picoliter up to microliter volumes (Figeys and Pinto, 2000). Microfluidic systems also afford a high degree of control over the movement of fluid. Through the design of channel geometries (e.g., inertial microfluidics) and the selection of the fluid driving force, such as pressure (e.g., continuous-flow microfluidics) or an electric field (e.g., digital microfluidics), various flow modes can be achieved to effectively mix, sort, or isolate analytes from complex fluids. Miniaturization is, therefore, an appealing approach for executing challenging separations involving highly heterogeneous sample mixtures. Indeed, proteins and other analytes have been concentrated by over a million-fold using microflow platforms (Wang, Stevens, and Han, 2005).
Microfluidics are also appealing because of their modularity. Through segmentation, a single device can incorporate multiple processing units to allow several, if not all, steps of a workflow to be integrated on one chip. Indeed, this is the concept behind micro-total analysis systems (μTAS) (Manz, Graber, and Widmer, 1990), or the more commonly known lab-on-chip technologies (Figeys and Pinto, 2000). These microfluidic systems are designed to perform many laboratory operations such as sample preparation, reaction, detection, and analysis in a single device. Such systems have already been realized for PCR analysis (Burns et al., 1998), pathogen detection (Zhu et al., 2020), and ELISA (Sun et al., 2010). Advances in lab-on-chip technologies have been aided by the development and integration of micro-sized components like valves (Weibel et al., 2005), mixers (Nguyen and Wu, 2004), pumps (Laser and Santiago, 2004), and connectors (Temiz et al., 2015) that enable fine control over liquid transport as well as sensors that allow experimental parameters (e.g., temperature, pressure, flow rate, etc.) to be monitored or tuned in real-time (Berg and Lammerink, 1998; Burns et al., 1998). Nevertheless, combining all of the steps required for proper sample preparation and analysis onto one chip is rarely straightforward. In these cases, microfluidics have been readily coupled to analytical instruments to detect or quantify analytes processed on-chip (X. Wang et al., 2015). The ease with which microfluidics can be multiplexed with other platforms also makes them ideal for automation and robotic handling.
The main strength of microfluidics for glycan analysis is their high-throughput potential. Many copies of a microchannel can be fabricated into a small area of a device to run multiple assays in parallel. Processing volumes and speed can further be scaled by operating devices for longer periods of time, otherwise known as “scaling out”, or by increasing the number of devices operating in parallel, otherwise known as “numbering up”. Accordingly, vast numbers of sample types and conditions can potentially be tested in minutes or hours instead of the typical days or weeks required at the macro-scale.
Based on the features outlined above, employing microfluidics in carbohydrate analysis presents many benefits to the field of glycobiology. In the remainder of this article, we review applications of microfluidics in the analysis of glycan structures. Table 1 summarizes the major classes of microfluidic technologies that will be discussed in the remaining sections. To date, microfluidics devices have been incorporated at several major steps of the analytical workflow for glycans. Most applications have centered on the analysis of N-linked glycans derived from glycoproteins isolated from clinical samples or commercial sources. As such, our discussion will focus on the analytical steps used to characterize N-glycans. Overall, these studies reveal the potential of microfluidics to reduce the processing times and costs that have complicated the analogous macro-scale procedures.
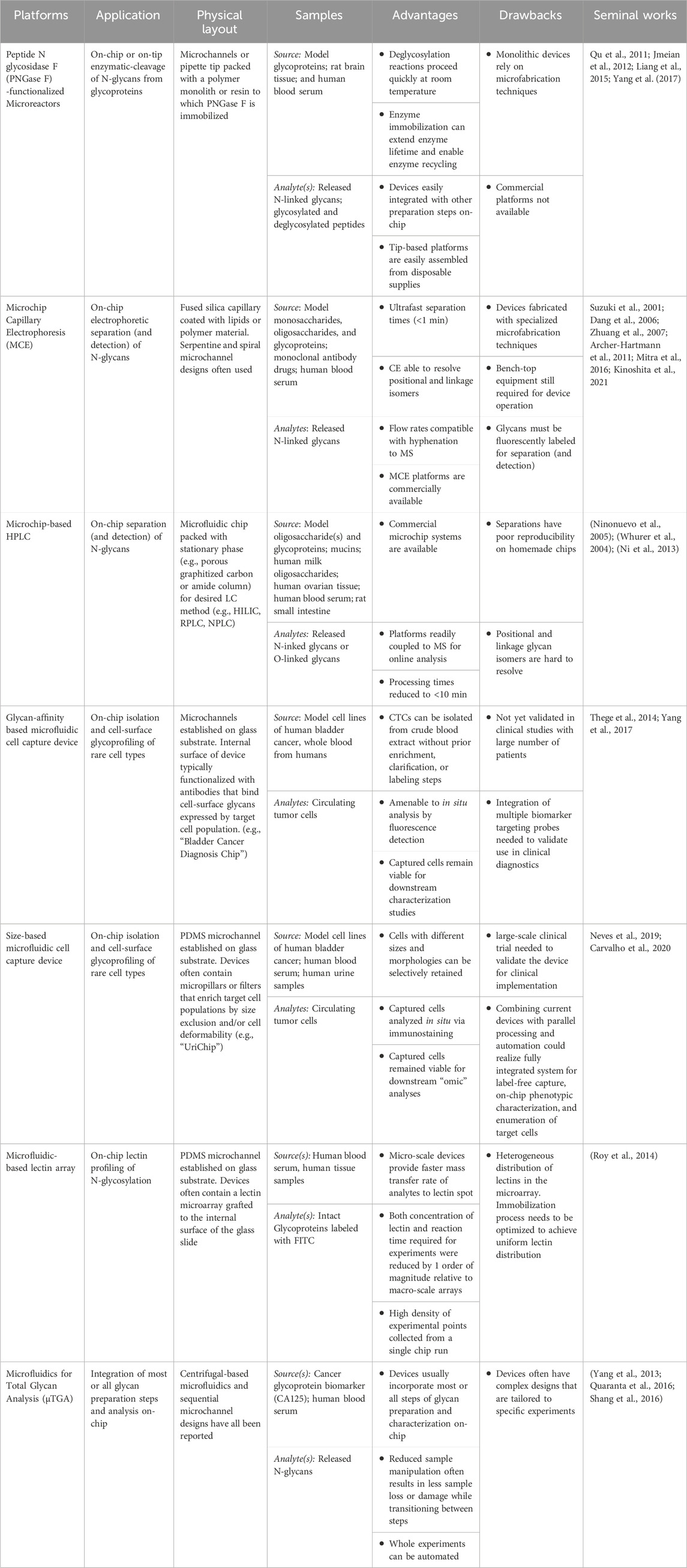
TABLE 1. Features of the current state-of-the-art microfluidics developed for carbohydrate structural analysis. Several major categories of “platforms” are defined and general descriptions of their intended “application” and “physical layout” are provided. A list of the types of biological samples and analytes processed by each device class thus far is described in the “sample” column. Lastly, “advantages” and “drawbacks” of each technology are highlighted along with a list of seminal works for each platform category. Abbreviations: PNGase F (Peptide N glycosidase F), HPLC (high-performance liquid chromatography), MS (mass spectrometry).
3 Microfluidic platforms for glycan enrichment and cleavage
Glycoproteins and oligosaccharides typically require several clean-up and/or modification steps prior to their separation and structural characterization. Most analytical workflows begin with the isolation of glycoproteins from complex biological mixtures. If the goal is to analyze the oligosaccharide separate from the non-carbohydrate moiety, as in glycomics, then the recovered glycoprotein must be enzymatically or chemically treated to release the carbohydrate moieties. Following cleavage, free glycans are subjected to one or more additional processing steps to derivatize, purify, concentrate, or separate glycan analytes prior to their evaluation with analytical methods like MS. Adapting these preparatory procedures to microfluidics can reduce the time, steps, and manual intervention required to prepare samples for the intended analytical technique. This section explores the use of microdevices in early steps of sample preparation, including the enrichment of glycoproteins from complex mixtures and the cleavage of glycans from glycoprotein isolates.
3.1 Microfluidics for deglycosylation of glycoproteins
The enzymatic release of glycans from glycoproteins is an essential step of glycan structural analysis. Cleavage is carried out enzymatically or chemically depending on the type of glycan. For the well-studied N-linked glycoproteins, enzymatic cleavage is preferred due to the commercial availability and broad substrate specificity of peptide-N-glycosidase F (PNGase F), an enzyme that cleaves the amide bond between the asparagine residues of proteins and the reducing end of N-linked glycans. Cleavage protocols employing PNGase F are often performed in solution, where they suffer from long incubation times, low deglycosylation yields, and reaction conditions that risk protein integrity (Vilaj et al., 2021). An attractive alternative has been to immobilize these biocatalysts in microfluidics to increase analytical throughput and to reduce consumption of the costly PNGase F enzyme.
The use of microfluidics functionalized with PNGase F to perform glycan cleavage on-chip was first reported in 2005. The device in question consisted of a capillary containing a polymer monolith onto which PNGase F was covalently anchored (Palm and Novotny, 2005). The final column volume was 1.6 μL. Early platforms were often designed in this fashion–as monolithic microreactors comprised of polymer supports functionalized with PNGase F using various chemistries. Popular immobilization techniques included covalent conjugation and the use of affinity interactions. In particular, glutathione-S-transferase (GST) tagged PNGaseF was used to tether enzymes to glutathione-coated supports with improved control over enzyme orientation (Krenkova et al., 2013; Szigeti et al., 2016). Devices were typically assembled inside glass capillaries, and featured monoliths synthesized with diverse polymers such as methyacrylate (Krenkova et al., 2013), poly (glycidyl methacrylate co-oly (ethylene glycol) diacrylate) (Liang et al., 2015), and 3-(trimethoxysilyl)propyl methacryltate (Krenkova, Lacher, and Svec, 2009). These early devices served as prototypes and allowed researchers to test the performance of on-chip deglycosylation relative to solution-based reactions using mixtures of standard glycoproteins. Reaction speeds and yield significantly improved in these microcolumns. Cleavage efficiencies that previously required overnight incubation steps at 37°C were achieved in minutes at room temperature using microfluidic platforms (Krenkova, Lacher, and Svec, 2009; Jmeian, Hammad, and Mechref, 2012).
3.2 Combinatorial microfluidics for enrichment and deglycosylation of glycoproteins
After microfluidics were shown to improve the performance of PNGaseF cleavage reactions, investigators began to explore integrating other preparatory steps, such as glycoprotein enrichment, on chip with glycan cleavage. Several popular strategies for glycoprotein enrichment have been adapted to microfluidics and coupled to PNGaseF devices to further streamline the preparation of glycan samples. The most notable example is hydrophilic interaction liquid chromatography (HILIC). In HILIC, a hydrophilic stationary phase is used to selectively enrich or separate glycoproteins via interactions with the hydrophilic glycan moieties. Qu and colleagues were one of the first groups to integrate HILIC on-chip with PNGaseF microfluidics (Qu et al., 2011). In this integrated system, preparation took between ∼1/28th to ∼1/17th of the time observed with conventional offline procedures. Notably, the detection limit was also reduced to 5 fmol for offline MS analysis of the deglycosylated peptides. Liang and colleagues also combined HILIC on-chip with PNGaseF microfluidics (Liang et al., 2015). A monolith was coated with gold nanoparticles, which were variably functionalized with cysteine residues or PNGaseF for the protein enrichment step or enzymatic deglycosylation steps, respectively. The coupled platform enabled the identification of 196 N-linked glycopeptides from 5 µg of human plasma.
PNGaseF microfluidics have also been paired with downstream analytical steps, such as liquid chromatography (LC) and MS. Combinatorial systems reduce procedural complexity and minimize the potential for sample loss from repeated manipulation. To this end, PNGaseF microreactors have been directly coupled to analytical scale LC/MS systems to run deglycosylation in line with glycan separation and detection (Krenkova et al., 2013). In many cases, the LC step is also conducted in a microfluidic (Jmeian, Hammad, and Mechref, 2012), which is a topic discussed in more detail in section 4. To prevent interference from the bare proteins, hydrophobic trap columns are often placed between the PNGaseF microreactor and the LC column to catch the deglycosylated proteins. This in-line technique has been shown to enhance the sensitivity of LC/MS, allowing the detection of glycans at concentrations as low as 100 fmol (Jmeian, Hammad, and Mechref, 2012). Additionally, these integrated systems were shown to be highly reusable. In one case, PNGaseF activity was preserved on-chip after 1 month of continuous use (Jmeian, Hammad, and Mechref, 2012).
3.3 Tip-based microfluidics for high-throughput capture and deglycosylation of glycoproteins
Several groups have realized high-throughput platforms for both glycoprotein enrichment and glycan cleavage by fashioning microfluidics out of disposable pipette tips. In these devices, plastic pipette tips are packed with resin or gel matrices modified with functional groups for glycoprotein capture or with digestive enzymes for glycan cleavage. Tip-based platforms are attractive because they enable facile transport of samples by manual pipetting, and, in turn, can be readily automated using liquid handlers. Yamamoto and colleagues increased the efficiency of glycoprotein digestion and deglycosylation by performing these enzymatic reactions in a tip format (Yamamoto et al., 2020). Pipette tips were filled with enzyme-impregnated gel containing trypsin or PNGaseF to digest proteins or to release N-glycans respectively via manual and iterative pipetting of glycoprotein solutions. Yang and colleagues designed an “AutoTip” system to automate N-glycomic profiles of urine samples collected from prostate cancer patients. A schematic of the platform is shown in Figure 4. The platform consisted of 200 µL pipette tips loaded with aldehyde-coated resin, on which glycoproteins were immobilized via conjugation of aldehyde groups with the N-termini or lysine residues on the glycoproteins. The immobilized glycoproteins were subsequently derivatized at sialic acid residues and deglycosylated by aspirating solutions containing the derivatization agent or PNGaseF, respectively. All three processing steps were automated with a liquid handler, and the tip format allowed parallel processing of 58 urine samples (Yang et al., 2017).
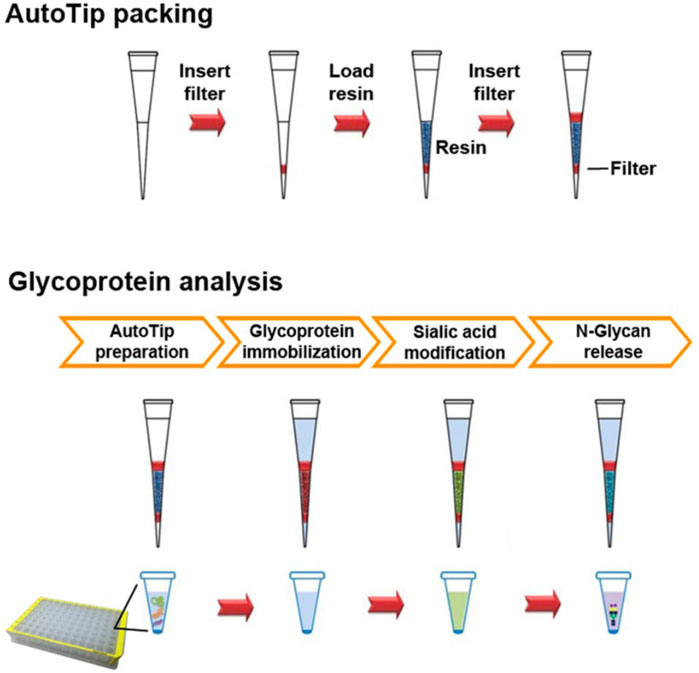
FIGURE 4. Schematic of the AutoTip packing process and operation. The top panel illustrates the steps taken to load plastic pipette tips with an aldehyde coated resin. A polyethylene sheet (red bar) was first inserted into the tip, followed by the addition of the aldehyde resin (dark blue), and then the insertion of another polyethylene sheet to secure the resin. The bottom panel depicts the steps for glycan analysis on the assembled AutoTip. After preparation, glycoproteins are adhered to the resin via iterative pipetting of glycoprotein solutions (red). The sialic acids on the resin-bound glycoproteins were derivatized by iterative pipetting of a solution containing carbodiimide (green). Finally, derivatized N-glycans were released by successive pipetting of a PNGaseF solution. Reproduced with permission from (Yang et al., 2017).
4 Microfluidics platforms for glycan separation
Following enzymatic cleavage, released glycans are often captured in complex mixtures with molecular contaminants. Non-glycan molecules present in medium or high abundance can prevent detection of less abundant oligosaccharides, making the removal of contaminants a critical step in the structural analysis of carbohydrates. Decontamination is typically achieved by separating glycans using one or more chromatographic techniques. Two prominent separation methods employed in glycan analysis include capillary electrophoresis (CE) and LC. Since these techniques are limited by low sample throughput at the macroscale, several groups have explored miniaturizing LC and CE to expedite the purification of oligosaccharides prior to their structural characterization.
4.1 Microchip capillary electrophoresis
CE is an analytical technique that separates charged species based on their electrophoretic mobility in the presence of an applied electric field. CE systems typically employ fused silica capillaries and a negatively charged background electrolyte which induces electroosmotic flow (EOF) from a positive to a negative electrode. Separated analytes are detected by laser-induced fluorescence (LIF) via an online detector module. Because carbohydrates lack intrinsic fluorescence, glycans are labeled with fluorescent tags, such as the aromatic amine 8-aminopyrene-1,3,6-trisulfonic acid (APTS), prior to injection into the CE system. CE has also been coupled with MS for offline analysis of separated glycans.
Microchip CE (MCE) emerged as a tool for oligosaccharide separation in the early 2000s following its widespread adoption in protein analysis and DNA separation (Henry, 2006). These systems were fabricated as single or multi-compartment channels and used to separate both free glycans and intact glycoconjugates. One of the first MCE systems developed for glycan separation utilized glass capillaries and was used to separate a mixture of sucrose, GlcNAc, and raffinose (Burggraf et al., n. d.). However, the initial detection limits were poor relative to other techniques. Not long after, Suzuki and colleagues achieved both rapid separation and sensitive detection of hexosamine, their reduced analogs, hexosamintols, and amino sugar derivatives of mucin O-glycans with MCE (Suzuki et al., 2001). Their home-built system featured a long separation channel totaling 33 mm in length and integrated both LIF and a charge-coupled device camera for single and multipoint detection, respectively. The system showed remarkable efficiency, separating samples in less than 60 s and detecting N-glycans at concentrations as low as 0.5 fmol. Callewaert et al. later showed MCE platforms to be suitable for the separation of complex glycan samples (Callewaert et al., 2004). A MCE device made of alumina-silicate glass was used to assay the N-glycome of human sera derived from healthy volunteers and patients with cirrhosis or chronic hepatitis. Electropherograms with resolutions matching that of commercial CE analyzers were obtained within 12 min and exceeded the performance of commercial systems at longer separation times. Ratio analysis of select peak intensities were used to diagnose patients with cirrhosis and chronic hepatitis, highlighting MCE as a potentially powerful tool for clinical assays.
Subsequent work with MCE aimed to address several shortcomings of these early devices. One point of concern was the reliance on glass-based devices, which required hazardous and specialized microfabrication techniques. Polymer-based platforms emerged as a safer and user-friendly alternative. However, the performance of the polymer systems initially lagged behind glass-based analogues because APTS-labeled glycans were more prone to non-specific adsorption on the hydrophobic, polymer surface. Dynamic coatings, wherein polymer scaffolds are doped with additive materials that help regulate EOF and suppress analyte-surface interactions, were found to mitigate this issue (Dang et al., 2003; 2006). Neutral polymers containing polyhydroxyl groups, such as methyl cellulose (MC), were shown to be highly efficient in preventing oligosaccharide absorption on poly (methyl methacrylate) (PMMA) microchips. Under optimal conditions, such coatings enabled the separation of 15 oligosaccharides in 45 s, the fastest separation on record at the time. However, the presence of cellulose polymers was found to increase the viscosity of the loading buffer which could have deleterious effects on separation efficiency. Incorporating a hybrid dynamic coating consisting of n-dodecyl β-D-maltoside (DDM) and MC was later shown to suppress glycan adsorption and EOF while keeping the viscosity low (Dang et al., 2006).
Another drawback of early MCE designs was their inability to resolve positional and linkage isomers of glycans. Jacobson and colleagues addressed this issue by altering the dimensions and geometry of the separation channel (Zhuang et al., 2007; 2011). They hypothesized that the poor resolution of linkage and positional isomers stemmed from the short separation lengths of MCE, ranging from 3.3 to 7 cm. Implementing a spiral channel design allowed the authors to extend the separation length to 22 cm while limiting the issue of sample dispersion (Figure 5A). The inner channel walls were also coated with polyacrylamide to mitigate EOF. With this setup, positional isomers of N-glycans derived from ribonuclease B were resolved in under 3 minutes, showing separation efficiencies eight to 50 times higher than prior reports (Figures 5B, C). Excitingly, similar resolution and efficiency were obtained for clinical samples, including the blood serum of stage IV breast cancer patients (Figure 5D). Structural isomers of glycans have similarly been resolved by MCE devices employing serpentine microchannels for both model and clinical glycan samples (Zhuang et al., 2011).
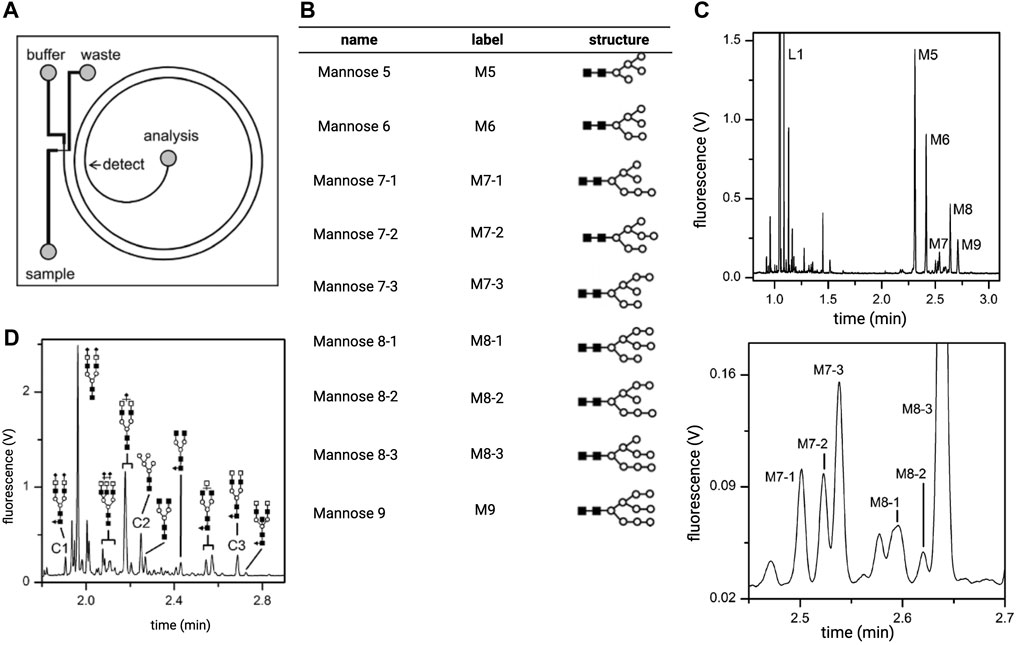
FIGURE 5. Analysis of ribonuclease B derived N-glycans on a spiral microchip capillary electrophoresis (MCE) platform. (A) A diagram of the spiral channel MCE platform. (B) The structures of the N-glycans found on model protein ribonuclease (B). These oligosaccharides were separated and detected using the MCE platform depicted in panel (A). (C) The top panel shows an electropherogram obtained from separation of mixtures of APTS-labeled N-glycans shown in (B). The bottom panel is an enlarged section of the electropherogram where peaks corresponding to positional isomers of mannose 7 (M7) and mannose 8 (M8) are resolved. (D) Electropherogram of N-glycans isolated from the blood serum of patients with stage IV breast cancer. Reproduced with permission from (Zhuang et al., 2007).
Later efforts also sought to resolve isobaric glycans using MCE analysis. Isobaric glycans vary in composition but share the same mass. They are common in biological samples and often require complex derivatization methods to be distinguished by MS. A simpler approach has been to pair MCE with lectin and exoglycosidase treatment. Lectins are proteins that bind to specific carbohydrate motifs, making them excellent tools for probing oligosaccharide composition. Exoglycosidase enzymes cleave glycosidic bonds at the extremities of glycans and release terminal monosaccharides in the process. Exoglycosidases recognize specific monosaccharides and linkages and are defined as such; β1-4 galactosidase describes exoglycosidases that cleave terminal β1-4-linked galactose. As such, exoglycosidase activity can reveal structural features such as the identity of terminal sugar residues, the stereochemistry of glycosidic bonds (e.g., 𝛂-vs β-configuration), and the regiochemistry of the linkage (e.g., 𝛂1-2 versus 𝛂1-3). Thus, when applied upstream of MCE, lectin and exoglycosidase treatment can provide compositional and structural information that may further distinguish isobaric glycans subjected to separation. Archer-Hartmann et al. leveraged this concept to differentiate isobaric glycans and to evaluate structural features of N-glycans isolated from MCF7 breast cancer cells (Archer-Hartmann et al., 2011). The MCE devices consisted of capillaries coated with phospholipid additives in which the lectin concanavalin A (conA) or exoglycosidases, 𝛂1-2,3 mannosidase and β1-4 galactosidase, were incorporated upstream of a separation module. Glycan standards treated with conA, which binds high-mannose type glycans, before separation produced electropherograms lacking glycan peaks previously observed when samples were analyzed without con A treatment (Figure 6A). This revealed the location and relative abundance peaks corresponding to high mannose species. An analogous experiment performed with β1-4 galactosidase, lowered the abundance of peaks corresponding to structures with terminal Gal (β1-4) residues and caused the appearance of peaks corresponding to cleaved products. In this way, the authors were able to distinguish an isobaric isomer pair, which differed only by the type of linkage between a terminal galactose and the underlying GalNAc residues (Gal (β1-4)-GalNAc versus Gal (β1-3)-GalNAc) (Figure 6B). Applying conA, 𝛂1-2,3 mannosidase, and β1-4 galactosidase treatments to N-glycans derived from MCF7 breast cancer cells demonstrated that similar compositional and structural insights could be acquired from highly complex, low abundance biological samples (Figure 6C). The integration of exoglycosidase sequencing and MCE was subsequently used to profile N-glycans derived from the commercial cancer drug, Trastuzumab, illustrating how this technique might aid carbohydrate analyses critical to the discovery, manufacturing, and quality control of glycoprotein therapeutics (Archer et al., 2011).
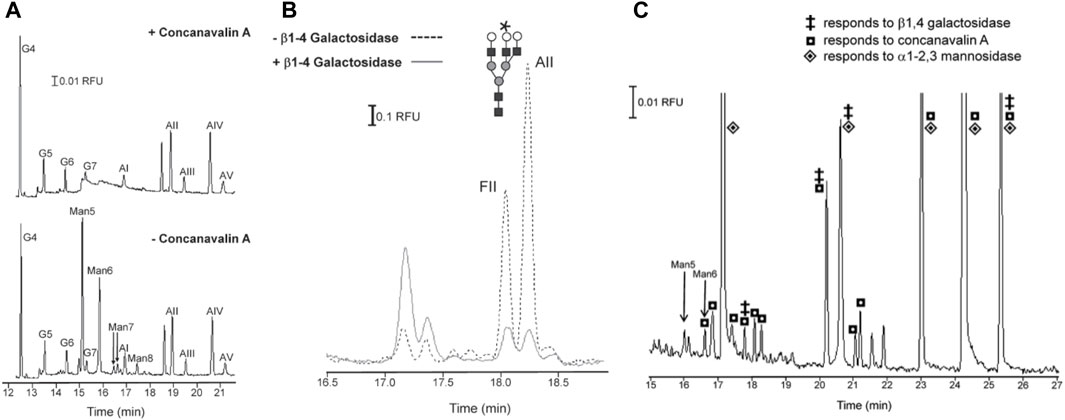
FIGURE 6. Integration of lectin and exoglycosidase treatment of N-glycans with glycan separation by microchip capillary electrophoresis. (A) Separation of amine 8-aminopyrene-1,3,6-trisulfonic acid (APTS)-labeled N-glycan standards derived from two model glycoproteins: Ribonuclease B and 𝛂1-acid glycoprotein. The top trace shows separations of labeled N-glycans after their incubation with the lectin concanavalin A (conA). The bottom trace show separations of glycans without the lectin incubation step. Peaks corresponding to high-mannose type glycans (Man5-8 peaks) are absent in the lower panel as these oligosaccharides were captured during the conA incubation step. (B) Separation of an analogous mixture of APTS-labeled glycan standards. The two traces correspond to separations performed after N-glycans were treated with β1-4 galactosidase (solid line) or without said treatment (dotted line). The abundance of glycan isomers FII and AII decreased after treatment with β1-4 galactosidase, indicating the presence of terminal β1-4 -linked galactose residues. (C) Separation of APTS-labeled N-glycans derived from soluble glycoproteins produced by MCF7 breast cancer cells. Several different separation experiments were performed wherein glycans were treated with conA, β1-4 galactosidase, or 𝛂1-2,3 mannosidase prior to separation. The glycan peaks are annotated with symbols that denote sensitivity to the conA (square), β1-4 galactosidase (double dagger), or 𝛂1-2,3 mannosidase (diamond) treatment. This annotation provides general information about the structure (e.g., presence of terminal β1-4 -linked Gal, 𝛂1-2-linked Man, or 𝛂1- 3-linked Man) and composition (e.g., high mannose) of these oligosaccharides. Reproduced with permission from (S. A. Archer-Hartmann et al., 2011).
Indeed, MCE has shown the most potential as a high-throughput tool for clinical diagnostics and quality control of biological products. Commercial MCE platforms have been used separately and in combination with other microfluidics to aid clinical glycomics assays, including a GlycoHepatoTest (Vanderschaeghe et al., 2010) and detection of glycan biomarkers for ovarian (Zhuang et al., 2011; Mitra et al., 2013) and colorectal (Snyder et al., 2016) cancers. MCE have also been widely used to monitor glycosylation of biotherapeutic glycoproteins. For instance, MCE-SDS was used to monitor the quality of monoclonal antibodies with the same rigor as conventional CE-SDS (Chen et al., 2008). MCE has also facilitated screens for antibody glycans during mammalian cell culture (Primack, Flynn, and Pan, 2011). Hundreds of crude cell culture samples were profiled within a few hours using this method. Kinoshita et al. recently repurposed a DNA analyzer for MCE to profile the glycosylation of multiple FDA-approved mABs (Kinoshita et al., 2021). Their process provides a blueprint for the adoption of MCE for the industrial-scale analysis of biotherapeutics.
4.2 Nanoflow liquid chromatography
Liquid chromatography remains the most widely used technique for separating released glycans and glycoproteins prior to MS analysis. LC methods most often used in glycomics are reverse-phase LC (RP-LC) and hydrophilic interaction liquid chromatography (HILIC). While LC columns with diverse stationary phases have been used, those packed with porous graphitized carbon (PGC) are the most popular. Miniaturized LC has garnered increasing interest in glycoanalysis for enabling the sensitive detection of low-abundant molecules like glycans. This enhanced sensitivity stems from the smaller column dimensions, which accommodate lower volumetric flow rates and thus immensely reduce the sample dilution observed in conventional LC. Miniature LC systems are defined by volumetric flow rate and include microflow (∼100 μL/min), capillary flow (1–15 μL/min), and nanoflow (300 nL/min) LC. Here, we focus on the applications of microflow LC systems or the integration of nanoflow and capillary flow LC into microchip devices for the analysis of glycans.
Most microfluidics used for HPLC are commercial microchips that have been outfitted with nanoscale LC columns. The most popular design reported for glycan analysis features a nanosized enrichment column upstream of the separation column (Niñonuevo et al., 2005; Bynum et al., 2009; Ni, Bones, and Karger, 2013). Porous graphitized carbon (PGC) is the stationary phase of choice for these devices owing to its hydrophilic properties (Niñonuevo et al., 2005; Bynum et al., 2009; Jmeian, Hammad, and Mechref, 2012; Ni, Bones, and Karger, 2013; Yang et al., 2013). Glycan mixtures with varying levels of complexity have been separated by PGC-packed HPLC chips, including mixtures of human milk oligosaccharides (Niñonuevo et al., 2005), O-linked glycans released from mucins (Niñonuevo et al., 2005), and both linkage and positional isomers of N-glycans from human serum proteins (Ni, Bones, and Karger, 2013). A major benefit of microchip LC is that these platforms are easily coupled to MS for on-line analysis as discussed below.
5 Microfluidic platforms for glycan structural analysis
After completing the appropriate sample preparation steps, the structural features of glycans are finally evaluated. Analyses typically probe one or more aspects of glycan structure, including monosaccharide composition, the sequence of sugars, the number and length of branches, positioning of glycosidic linkages, and linkage stereochemistry. Many experiments also seek to quantify the relative abundance of different oligosaccharide structures. The choice of analytical technique(s) depends on the type of glycans under investigation and the level of structural detail required to answer the proposed biological question. Because no one technique can account for every structural feature, most analyses leverage multiple analytical methods to discern different aspects of carbohydrate structure and to increase the confidence of data interpretation. Integrated LC/MS is the preeminent approach for detailed, quantitative analysis of glycans. Carbohydrate binding proteins, like lectins and anti-glycan antibodies, are often employed in studies aiming to identify specific glycan motifs and changes in their expression on soluble or cell surface glycoconjugates. CBP binding assays have, thus, proven essential to clinical glycomics, where a central objective is to define glycosylation patterns, or “glyco-codes”, corresponding to disease onset and progression. Both LC/MS and CBP-based structural analysis have been adapted to microfluidics with the goal of increasing detection sensitivity and the throughput of analysis. This section examines notable examples of such work.
5.1 Microfluidics-assisted glycoprofiling with carbohydrate-binding proteins
A major objective of clinical glycomics is to identify changes in glycosylation that signify concomitant changes in disease. This poses several challenges. First, potential glycan biomarkers often present at trace levels requiring analytical methods with sufficient sensitivity. Second, detection strategies need to accommodate high-throughput and systematic evaluations, criteria that MS and LC methods do not often meet. Carbohydrate-binding proteins, like lectins and anti-glycan antibodies, are attractive alternatives for monitoring the disease-dependent changes in glycosylation. These proteins selectively bind to specific monosaccharide or oligosaccharide moieties, making them exceptional tools for probing glycan composition. Many different CBPs can be arrayed on-chip to perform multiplexed detection of glycan biomarkers and these arrays are, in turn, readily integrated with microfluidics to create high-throughput, low-volume analytical systems.
CBP-functionalized microfluidics have garnered much attention in the field of cancer glycobiology, where they have aided in the identification of glyco-codes unique to cancer type and clinical stage. Glyco-codes could serve as a more accurate diagnostic tool for cancer compared to conventional methods that screen for a single protein biomarker as many of these molecules are elevated in benign disease states or undetectable in early phases of cancer (Hanash, Pitteri, and Faca, 2008). Lectin microarray technology has become a powerful tool for conducting the kind of high-throughput analysis needed to establish glyco-codes. In these platforms, panels of lectins are immobilized on a substrate in array format, allowing for parallel detection of multiple carbohydrate motifs present in a mixture. Glycoconjugates in the sample of interest are fluorescently labeled to visualize their association with various lectin patches. Thus, the presence and relative abundance of a several motifs can be determined and used to define signature glycosylation patterns. The integration of lectin microarrays and microfluidics can further enhance the efficiency and sensitivity of lectin profiling for this purpose. Glyco-codes for several stages of gastric cancer were quickly elucidated by performing glycoprofiling experiments inside of a lectin-functionalized microfluidic (Roy et al., 2014). The device consisted of a PDMS microchannel supported by a glass slide. The latter was derivatized with APTES-glutaraldehyde to covalently bond multiple lectins in an array format. 17 unique lectins were selected to probe for a diverse range of carbohydrate motifs, including known tumor-associated antigens, on glycoproteins extracted from tissue biopsies or blood sera samples. These extracts were taken from patients with gastric cancer, Type B gastritis, Type C gastritis, and from healthy individuals to establish glycan fingerprints for various phases of gastric cancer progression (Figures 7A, B). Following extraction, glycoprotein solutions were fluorescently labeled to visualize the glycan-lectin interactions and then flowed through the microchannel for delivery to the lectin array. The flow conditions reduced both the time and concentration of lectins required for a single experiment by an order of magnitude compared to the analogous macro-scale devices. Because each lectin was immobilized in octuplicate, a single run generated multiple replicates of each binding experiment, improving the accuracy of analysis. In this way, the authors were able to conduct comprehensive and systematic glyco-profiles of gastric disease states across multiple sample types (tissue biopsy and blood serum) in a timely fashion. While no distinctive glyco-codes were detected in serum samples, profiles of tissue biopsies revealed signature glycosylation patterns for chronic gastritis and full-blown gastric cancer with diagnostic and prognostic potential (Figures 7C, D).
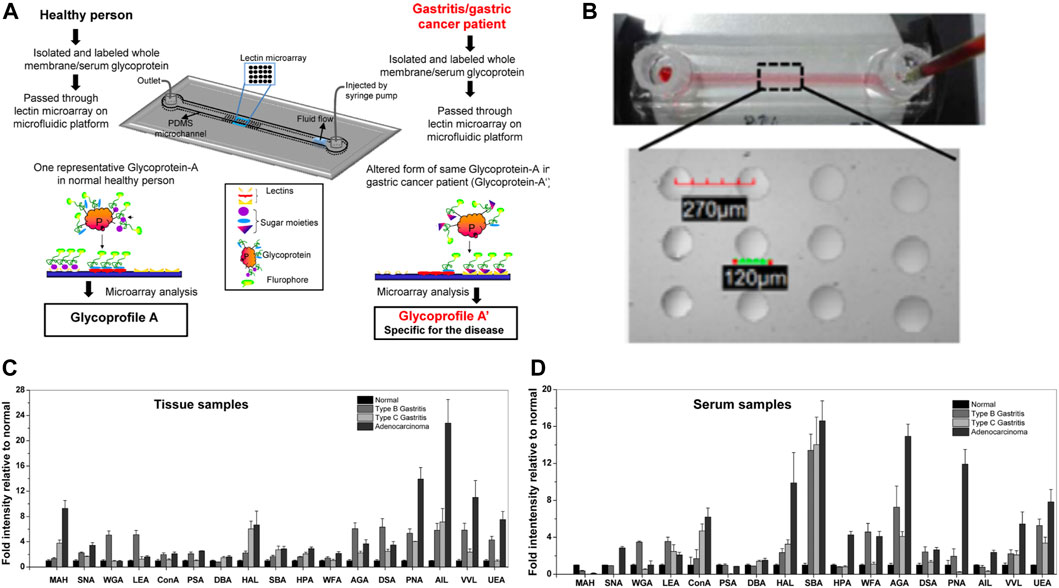
FIGURE 7. Glycoprofiling of gastric disease states using a lectin-functionalized microfluidic. (A) Diagram of the integrated lectin microarray and microfluidic system used to assay the glycan content of tissue biopsies and blood serum samples taken from healthy individuals (normal) or patients with various stages of gastric cancer (type B gastritis, type C gastritis, and adenocarcinoma). The device comprises a microchannel built on top of a glass slide support. The latter is functionalized with an array of 17 different lectins. During analysis glycoproteins were extracted from patient samples, fluorescently labeled, and then passed through the microfluidic channel at optimized flow rates. (B) Image of the physical microfluidic channel (top panel) and the microarray patterned on the glass slide (lower panel). Lectins were immobilized on circular spots of 120 µm diameter and spaced 270 µm apart. Fluorescence plots from lectin binding experiments with (C) tissue-derived glycoproteins and (D) serum-derived glycoproteins isolated from the four patient populations. Large variations in lectin binding were observed between the four clinical groups for the tissue samples but not for the serum samples. Abbreviations: maackia amurensis hemoagglutinin (MAH), elderberry lectin (SNA), wheat germ agglutinin (WGA), Lycopersicon esculentum agglutinin (LEA), concanavalin A (ConA), pisum setivum agglutinin (PSA), dolichos biflorus agglutinin (DBA), helix aspersa lectin (HAL), soybean agglutinin (SBA), helix pometia agglutinin (HPA), westeric floribunda agglutinin (WFA), abrus agglutinin (AGA), datura stramonium agglutinin (DSA), peanut agglutinin (PNA), jacalin (AIL), hair vetch lectin (VVL), ulex europaeus agglutinin (UEA). Reprinted from (Roy et al., 2014) with permission of AIP Publishing.
Antibody-functionalized microfluidics have facilitated glycan-dependent capture and evaluation of circulating tumor cells (CTCs). CTCs are known to predict survival better than other cancer biomarkers (Scher et al., 2015), making them invaluable diagnostic tools for therapeutic decision-making and forecasting therapeutic outcomes. Conventional approaches to CTC capture are consistently limited by the low recovery of this rare cell type, which accounts for less than 0.004% of mononucleated blood cells (Dong et al., 2013). Many of these protocols also rely on the expression of epithelial proteins that are not cancer-specific and leave many relevant CTC populations undetected (Azevedo et al., 2018). Microfluidics allow faster and cheaper isolation of CTCs based on cell-surface protein expression, and the integration of antibodies specific to cancer glycan antigens into these platforms has improved the detection and enrichment of CTCs. Neves et al. demonstrated selective, on-chip enrichment of CTCs over-expressing the sialyl-Tn (STn) cancer glycan antigen (Neves et al., 2019). A multi-chip design was implemented in which CTC isolates from bladder or colorectal cancer patients were first filtered by size on-chip and then fed into a microchip functionalized with an anti-STn antibody to sequester STn-expressing cells from the filtered cell population. The device provided the first glycomic profile of colorectal CTCs. Following a similar concept, a “UriChip” was later developed to enrich and profile exfoliated tumor cells of bladder cancer patients (Carvalho et al., 2020). On-chip immunocapture of circulating bladder cancer cells was performed using antibodies specific to aberrantly glycosylated integrin (Wang et al., 2020). An immunocapture device has also been designed for pancreatic CTCs using antibodies against glycoprotein biomarkers EpCAM and mucin 1 (Thege et al., 2014).
More recently, both lectin and antibody-based glycoprofiling have been paired with microfluidics to study the cell-surface glycome of individual cells. Notable examples include a “lab in a trench” (LiaT) system (Dimov, Kijanka, and Ducrée, 2010) designed to trap and analyze single living cells in an environment that minimizes shear-related changes in cell morphology and physiology (O’Connell et al., 2014). In this case, the cell-surface glycan compositions of trapped lymphoma cells were then assayed by sequential incubation with fluorescently labeled lectins. These single-cell glycomics experiments often employ more complex detection methods to achieve the necessary sensitivity. One group coated quantum dots with lectins (Cao et al., 2012) and phenylboronic acid (Cao et al., 2015) to amplify fluorescence signal whilst also reducing the photobleaching observed with conventional staining techniques. Droplet-based microfluidics integrated with surface-enhanced Raman scattering (SERS), a well-stablished technique for detection of trace analytes, has also been demonstrated (Willner et al., 2018). In this system, 1 cell and the SERS probe functionalized with WGA were encapsulated inside a single droplet. This permitted cell-to-cell and intracellular variability in cell surface sialic acid expression to be monitored in prostate cancer cells. Most recently, bipolar electrode arrays were integrated into a microchannel platform to monitor cell-surface galactosylation of single cells by electrofluorochromic imaging (Tian et al., 2021). Several enzymatic and ligation reactions were required to selectively label galactose with nanoprobes coated with Ferrocene. Labeled cells were captured in the anodic microchannel, where oxidation of ferrocene resulted in the reduction of resazurin to fluorescent resorufin. Fluorescence was captured by confocal microscopy. Overall, these microfluidic experiments could be carried out with significantly fewer cells (∼1000 cells/experiment) relative to bulk and conventional scale techniques.
5.2 Microfluidics coupled to mass spectrometers
Direct hyphenation of mass spectrometry with microfluidics provides several advantages for the structural analysis of glycans. As discussed in the previous sections, microfluidics are excellent platforms for preparing glycans derived from complex biological sources. Fractionation, deglycosylation, and separation of glycan analytes have all been shown to proceed with comparable, if not better, efficiency when conducted in microfluidics (Zhuang et al., 2011; Mai, Sommer, and Hatch, 2012; Mitra et al., 2016; Szigeti et al., 2016). Additionally, mass spectrometers and microdevices generate similar flow rates making it easy to interface the two systems for faster analysis. Lastly, the analysis of carbohydrates by MS is complicated by the low ionization efficiency of glycans. Methods that increase the sensitivity of MS measurements, such as by maximizing analyte concentration and purity, are essential in this regard. Direct coupling of micro-preparatory platforms and MS detection serves this purpose by reducing sample dilution and sample loss commonly observed during the manual transfer of samples between analytical steps.
MS has primarily been coupled to microchip LC systems. The MS instruments used in these platforms often employ gentle ionization techniques, such as matrix assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI), and contain quadrupole or time of flight (TOF) mass analyzers (Alley et al., 2010; Ni, Bones, and Karger, 2013). Coupling microchip LC to MS can reduce the volume of analyte required for rigorous statistical analyses. Reverse-phase micro-chip LC coupled to MS was used to identify a new glycan indicator of late-stage breast cancer from 1 µL of blood serum (Alley et al., 2010). Direct coupling has also facilitated detailed structural analysis of oligosaccharides without the need for extra processing steps, such as exoglycosidase digestion. For instance, interfacing HPLC microchips with negative ion ESI-MS/MS helped resolve the structure of linkage and positional isomers in complex mixtures of N-linked oligosaccharides (Ni, Bones, and Karger, 2013). In this hyphenated format, multiple orthogonal data sets were obtained in a single analysis cycle, including information from the separation of glycans on a PGC-packed nanoLC column, accurate mass analysis via Q-TOF MS, and generation of the MS/MS spectra.
6 Microfluidics for total glycan analysis (μTGA)
Microfluidics capable of combining glycan enrichment, cleavage, separation, and analysis into a single device are highly desired to automate these often manually intensive procedures. Some labs have begun to realize these all-in-one lab-on-chip technologies for potential use in clinical diagnostics, biotherapeutic manufacturing, and biomarker discovery. The following section discusses several illustrative examples of microfluidics designed for total glycan analysis (μTGA).
μTGA platforms have often been designed for the study of protein glycosylation. Bynum and colleagues reported one of the first μTGA platforms, which was designed to characterize N-glycans on recombinant IgG antibodies (Bynum et al., 2009). The device comprised three chips connected in series for enzymatic glycan release, glycan enrichment, and glycan separation. Total analysis time was condensed from a couple of days to a few minutes on-chip, with a turnaround time of 10 min between antibody injection and data collection. Yang and coworkers later integrated glycoprotein capture, glycan derivatization, enzymatic glycan cleavage, and chromatographic separation of glycans into a single device (Yang et al., 2013). Glycoprotein isolation, derivatization, and cleavage steps were performed in a single microchip packed with aldehyde beads for protein immobilization. Immobilized glycoproteins were subjected to a derivatization step before being treated with PNGase F to release N-glycans. Released oligosaccharides were then separated on a second PGC chip before being analyzed offline with MALDI MS. When used by itself, the enrichment chip enabled the detection of 65 glycan structures in human blood serum. Connecting the enrichment chip to the PGC chip greatly enhanced the sensitivity of MS, increasing the number of glycans detected in human blood serum from 65 to 148 structures.
Aside from prototypical flow-based devices, centrifugal microfluidic platforms have proven advantageous as μTGA platforms. Such microfluidics are commonly built in the shape of a compact disc (CD), wherein fluid flow is driven by centrifugal forces. As such, CD-based systems do not require connection to an external pump that pushes fluid through the device. Centrifugation is instead used to trigger fluid movement that results in the separation, mixing, reaction, or detection of molecules (Andersson et al., 2007). Several groups have used CD-based devices to automate the capture, labeling, and separation of glycoproteins for MS analysis (Thuy, Inganäs, and Thorsén, 2011; Thuy and Thorsén, 2013). In these experiments, CDs are subjected to multiple centrifugation cycles to carryout multi-step assays in a single device (Andersson et al., 2007). One such platform was used to automate the analysis of the N-glycoproteome of human serum (Quaranta et al., 2016). The system incorporated several sequential operations, including 1) the capture of biotinylated anti-human transferrin antibody onto streptavidin-coated beads, 2) the selective capture of transferrin from human serum samples, 3) enzymatic release of N-glycans and sialic acid residues, 4) recapture of desialylated glycans on PGC column, and 5) selective elution of glycans for offline MALDI-MS analysis. Each operation was triggered by varying the spinning speed and interval. The platform was shown to be highly selective towards the target protein, transferrin, and enabled diagnosis of chronic alcoholism based on the altered glycosylation of transferrin in the patient’s serum relative to the glycoforms observed in healthy individuals. The CD microfluidic identified alcoholic patients with an accuracy comparable to the sensitivity of traditional clinical tests (74% vs. 79%) but with the added advantages of short duration times, higher levels of automation, fewer reagents, and lower sample volume requirements.
The performance of antibody/lectin assays have also been improved by conducting these assays in μTGA systems. Shang et al. developed and automated antibody-lectin sandwich assays using a multichannel microfluidic (Shang, Zeng, and Zeng, 2016). In these assays, arrays of glycan-specific antibodies or lectins are incubated with complex biological mixtures to capture glycoproteins based on the identity of the core protein or glycan appendages. Captured proteins are then probed with another set of lectins or antibodies to measure the alternate attribute (i.e., the protein or the glycan), and, thus, become sandwiched between antibody/lectin pairs. In this way, both the level of core proteins and glycans are easily measured and compared across different samples. The microfluidic in question contained eight microchannels, each incorporating a microarray of 16 different lectins. Up to eight biological samples were simultaneously pumped through the device resulting in glycan-dependent capture of constituent glycoproteins. The level of glycoprotein associated with each lectin was then measured by antibody binding using a biotinylated antibody and dye-labeled streptavidin (Figure 8A, B). Tissue-dependent changes in the glycosylation of the ovarian cancer biomarker, CA125, were distinguished by this method. Leveraging microfluidics and an antibody lectin sandwich overcame several issues associated with lectin only detection methods. Device dimensions and flow conditions were tailored to make glycan-lectin binding kinetics reaction-limited rather than mass-transported limited, enhancing analytical performance. This culminated in the detection of CA125 at limits far lower than the clinical cutoff for cancer diagnosis. The method was made label-free by using antibody binding, circumventing the need to install fluorescent labels onto the glycoprotein analytes. Furthermore, the non-specific binding of blocking agents and lectins was reduced by rapid on-chip screening of potential blocking agents to identify blockers with minimal noise contribution. Lastly, the sample volume requirements were significantly reduced by integrating the lectin array into a microfluidic.
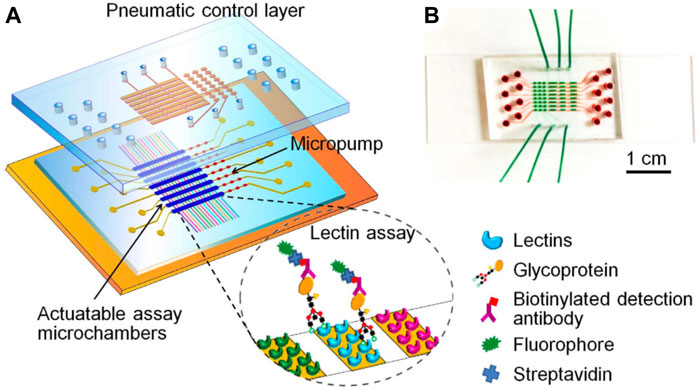
FIGURE 8. Multichannel microfluidic lectin barcode platform. (A) Schematic illustration of the microfluidic system used to monitor the glycosylation of the ovarian cancer biomarker CA125. The device was designed as a two-layer PDMS chip containing eight parallel channels and supported by a glass slide. Each channel is connected to a three-valve pump and actuatable assay chamber. Lectins are arrayed in each channel to capture glycoproteins which are in turn detected by biotinylated antibodies and fluorescently labeled streptavidin. (B) Photo of the physical microfluidic chip filled with red dye in the bottom flow channels and green dye in the pneumatic control channels. Reprinted with permission from (Shang, Zeng, and Zeng, 2016). 13.
7 Summary and concluding remarks
Microfluidics have been successfully integrated at every step of carbohydrate analysis. In every case, miniaturization increased process throughput or detection sensitivity using drastically smaller sample volumes. Most applications of microfluidics have aimed to streamline glycan processing steps conducted upstream to their structural characterization, such as glycan enrichment, cleavage, and separation. Relative to the one pot reactions, microfluidics functionalized with the endoglycosidase PNGaseF reduced the time, costs, and temperature conditions required to efficiently cleave glycans from captured glycoproteins. Miniaturized versions of CE, can now render high-resolution separations of carbohydrates at remarkably high speeds on the order of a few seconds. Microfluidics functionalized with carbohydrate-binding proteins have enabled the purification of rare cells at the population and even single-cell level.
Microfluidics implemented in carbohydrate analysis show the greatest potential for translation into clinical settings, where the ability to process many complex and low-volume samples is highly desired. Global profiling of serum glycans, as opposed to screening for individual glycoprotein biomarkers, is currently being explored as an alternative method for diagnosing cancer with better accuracy and at earlier stages (Hu et al., 2019). Many of these diagnostic tests rely on low-throughput techniques like standard chromatography, electrophoresis, and histochemical staining to screen for glycan biomarkers in blood and urine samples. Replacing macro scale methods with miniaturized systems like MCE could expedite the time to results and lower the burden of testing on patient populations with limited blood volumes, such as pediatric patients. Clinical pathology labs often rely on MS to conduct glycan-based assays which occludes their use in low-resource facilities lacking MS instruments or the expertise to interpret MS data. In these cases, MCE could provide simpler systems for performing diagnostic assays when detailed or de novo structural analyses are not essential. The growing availability of commercial MCE devices could facilitate their adoption in these settings where users are likely to be inexperienced.
Microfluidics also appear well-suited to support the discovery of novel glycan biomarkers and antibody therapeutics. Miniaturized systems have facilitated the efficient capture of low-abundance CTCs from several types of tumors. CTCs are attractive tools for biomarker discovery because their molecular composition mirrors that of the parent tumor. Glycomic profiles of CTCs thus offer a direct way to unearth perturbations in glycan structure or composition unique to specific types and phases of cancer. Unique glycan expression patterns have already been established for several stages of multiple cancers based on the analysis of CTCs isolated with CBP-functionalized microfluidics (Neves et al., 2019; Carvalho et al., 2020). Similar microfluidics may aid the capture and glyco-profiling of other cell-derived materials, like extracellular vesicles, which are known to be rich stores of biomarkers for many chronic diseases but accumulate in low abundance in bodily fluids.
While microfluidics have consistently been shown to streamline challenging analytical procedures, these technologies have been slow to transition into the mainstream. The devices discussed herein were developed for proof-of-concept studies by labs with expertise in micro-analytical systems. Moreover, platforms were microfabricated in-house using custom designs that may be difficult for users with limited knowledge or access to microfabrication facilities to replicate. Broader uptake seems most likely for routine procedures where novice users can employ ready-made platforms requiring little to no customization. Even still, the high cost of manufacturing disposable devices and the lack of standards for device production and application have slowed their translation into non-academic settings (Marques and Szita, 2017; Fernandes, Gernaey, and Krühne, 2018). For many of these prospective users, the benefits of microfluidics may not yet outweigh the hassle of integrating miniaturized systems into tightly regulated work streams.
Within academia, the adoption of microfluidics in glycomics has been narrowly focused on characterizing the N-glycosylation of glycoproteins. We found few examples in which microfluidics were used to investigate other types of glycoconjugates, such as proteoglycans, polysaccharides, or glycolipids. Additionally, most applications have been restricted to the analysis of human or murine N-glycans. This may reflect the dearth of chromatographic and spectral data available in glycan structural databases for interpreting non-mammalian carbohydrate structures (Schäffer and Messner, 2017). As more structural information is gathered for glycans derived from other glycoconjugate classes and biological kingdoms, including bacteria and plants, so too will the feasibility of investigating these structures with micro-technologies. The growing number of bioinformatic tools available to curate glycomic data may assist in interpreting analytical data from lesser-studied organisms (Maass et al., 2007; Hong et al., 2017; Schäffer and Messner, 2017; Birch et al., 2019). The success of PNGaseF microfluidics also points to the potential to develop similar platforms for the removal of glycans from other glycoconjugate classes. Commercial glycosidase specific to O-linked (Fujita et al., 2005; Koutsioulis, Landry, and Guthrie, 2008) and lipid-linked (Ishibashi et al., 2007) oligosaccharides are now available for incorporation into microfluidics. However, unlike PNGaseF, these enzymes exhibit much stricter substrate specificity and may only marginally increase the range of glycans that can be deglycosylated on-chip. Alternatively, microfluidics could also be explored as systems for conducting safer and faster chemical deglycosylation reactions. In this regard, miniaturizing chemical deglycosylation could offer the best first step toward generalizing microfluidics for examining diverse glycoconjugate classes.
Diversifying the device formats could also help broaden the use of microfluidics in carbohydrate analysis. Except for a handful of examples, most platforms discussed in this review use classic microchannel designs operated under continuous, pressure-driven flow. Consequently, the microfluidic technology sampled in glycomics represents a tiny fraction of the device styles, flow modalities, and active detection techniques that have been developed for microanalysis. Exploiting some of these cutting-edge technologies could address key limitations hindering broader use of microfluidics in carbohydrate analysis. For instance, devices made with inexpensive paper or fabric materials could reduce the cost and complexity of device assembly, facilitating their adoption by users lacking the requisite experience or resources for microfabrication. Paper-based MCE systems have already been developed for the analysis of other types of biomolecules (Hasan et al., 2021). Both droplet-based and inertial microfluidics have shown promise for integration with ion-mobility spectrometry (IM-MS), a gas-phase and label-free MS technique that outperforms traditional liquid-phase methods in separation ability and isomer discrimination (Zhang et al., 2021). Thus far, inertial microfluidics-assisted IM-MS has been realized for high-throughput single-cell lipidomics, which suggests that an analogous system for single-cell glycomics could soon follow suit. Droplet-MS platforms have also been developed for the analysis of small molecules, intact proteins (Smith et al., 2013; Kempa et al., 2020), and protein-ligand interactions (D’Amico et al., 2022), all of which provide a blueprint for the creation of similar systems for carbohydrate analysis. Lastly, there is a growing interest in single-cell glycan analysis. Most of the recent reports on microfluidics-assisted glycomics studies were conducted at the single-cell level. Because efficient cell-sorting is integral to these experiments, such applications may benefit from incorporating more active sorting techniques based on acoustic, electric, magnetic, or optical forces. In short, much remains to be explored by glycoscientists in the ever-expanding field of microfluidics.
Author contributions
FP: Conceptualization, Formal Analysis, Investigation, Writing–original draft, Writing–review and editing. JC: Writing–review and editing. SD: Funding acquisition, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation (CMMI-1728049 and MCB-1935370) to SD and a Howard Hughes Medical Institute Gilliam Fellowship (GT11442) to FP and SD.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
APTS, Amine-8-aminopyrene-1,3,6-trisulfonic acid; CBP, Carbohydrate binding protein; CE, Capillary electrophoresis; CTC, Circulating tumor cell; CD, Compact disc; conA, Concanavalin A; EOF, Electroosmotic Flow; ESI, Electrospray ionization; EPO, Erythropoietin; HPLC, High-performance liquid chromatography; HILIC, Hydrophilic interaction liquid chromatography; IMS, Ion-mobility spectrometry; LIF, Laser-induced fluorescence; LC, Liquid chromatography; DDM, n-Dodecyl β-D-maltoside; NMR, Nuclear magnetic resonance; MS, Mass Spectrometry; MALDI, Matrix-assisted laser desorption/ionization; MC, Methyl cellulose; µTAS, Micro-total analysis systems; PNGase F, Peptide-N glycosidase F; PDMS, Polydimethylsiloxane; PMMA, Poly (methyl methacrylate); RPLC, Reverse-phase liquid chromatography; TOF, Time-of-flight.
References
Alley, W. R., Madera, M., Mechref, Y., and Novotny, M. V. (2010). Chip-based reversed-phase liquid chromatography-mass spectrometry of permethylated N-linked glycans: a potential methodology for cancer-biomarker discovery. Anal. Chem. 82 (12), 5095–5106. doi:10.1021/AC100131E
Andersson, P., Jesson, G., Kylberg, G., Ekstrand, G., and Thorsén, G. (2007). Parallel nanoliter microfluidic analysis system. Anal. Chem. 79 (11), 4022–4030. doi:10.1021/AC061692Y
Archer-Hartmann, S. A., Crihfield, C. L., and Holland, L. A. (2011). Online enzymatic sequencing of glycans from Trastuzumab by phospholipid-assisted capillary electrophoresis. Electrophoresis 32 (24), 3491–3498. doi:10.1002/ELPS.201100432
Archer-Hartmann, S. A., Sargent, L. M., Lowry, D. T., and Holland, L. A. (2011). Microscale exoglycosidase processing and lectin capture of glycans with phospholipid assisted capillary electrophoresis separations. Anal. Chem. 83 (7), 2740–2747. doi:10.1021/AC103362R
Ashwell, G., and Morell, A. G. (1974). The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv. Enzym. - Relat. Areas Mol. Biol. 41, 99–128. doi:10.1002/9780470122860.CH3
Azevedo, R., Soares, J., Peixoto, A., Cotton, S., Lima, L., Santos, L. L., et al. (2018). Circulating tumor cells in bladder cancer: emerging technologies and clinical implications foreseeing precision oncology. Urologic Oncol. Seminars Orig. Investigations 36 (5), 221–236. doi:10.1016/J.UROLONC.2018.02.004
Berg, A. van den, and Lammerink, T. S. J. (1998). “Micro total analysis systems: microfluidic aspects, integration concept and applications,” in Microsystem technology in chemistry and life science. Editors A. Manz,, and H. Becker (Berlin, Heidelberg: Springer), 21–49. doi:10.1007/3-540-69544-3_2
Birch, J., Van Calsteren, M. R., Pérez, S., and Svensson, B. (2019). The exopolysaccharide properties and structures database: EPS-DB. Application to bacterial exopolysaccharides. Carbohydr. Polym. 205, 565–570. doi:10.1016/J.CARBPOL.2018.10.063
Bojar, D., Meche, L., Meng, G., Eng, W., Smith, D. F., Cummings, R. D., et al. (2022). A useful guide to lectin binding: machine-learning directed annotation of 57 unique lectin specificities. ACS Chem. Biol. 17 (11), 2993–3012. doi:10.1021/ACSCHEMBIO.1C00689
Burggraf, N., Krattiger, B., de Rooij, N. F., Manz, A., and de Mello, A. J. (1998). Holographic refractive index detector for application in microchip-based separation systems. Analyst 123, 1443–1447. doi:10.1039/A801478G
Burns, M. A., Johnson, B. N., Brahmasandra, S. N., Handique, K., Webster, J. R., Krishnan, M., et al. (1998). An integrated nanoliter DNA analysis device. Science 282 (5388), 484–487. doi:10.1126/SCIENCE.282.5388.484
Bynum, M. A., Yin, H., Felts, K., Lee, Y. M., Monell, C. R., and Killeen, K. (2009). Characterization of IgG N-glycans employing a microfluidic chip that integrates glycan cleavage, sample purification, LC separation, and MS detection. Anal. Chem. 81 (21), 8818–8825. doi:10.1021/AC901326U
Cabezas, J. (1994). The origins of glycobiology. Biochem. Educ. 22 (1), 3–7. doi:10.1016/0307-4412(94)90126-0
Callewaert, N., Contreras, R., Mitnik-Gankin, L., Carey, L., Matsudaira, P., and Ehrlich, D. (2004). Total serum protein N-glycome profiling on a capillary electrophoresis-microfluidics platform. Electrophoresis 25, 3128–3131. doi:10.1002/ELPS.200406020
Cao, J. T., Chen, Z. X., Hao, X. Y., Zhang, P. H., and Zhu, J. J. (2012). Quantum dots-based immunofluorescent microfluidic chip for the analysis of glycan expression at single-cells. Anal. Chem. 84 (22), 10097–10104. doi:10.1021/AC302609Y
Cao, J. T., Zhang, P. H., Liu, Y. M., Abdel-Halim, E. S., and Zhu, J. J. (2015). Versatile microfluidic platform for the assessment of sialic acid expression on cancer cells using quantum dots with phenylboronic acid tags. ACS Appl. Mat. Interfaces 7 (27), 14878–14884. doi:10.1021/ACSAMI.5B03519
Carvalho, S., Abreu, C. M., Ferreira, D., Lima, L., Ferreira, J. A., Santos, L. L., et al. (2020). Phenotypic analysis of urothelial exfoliated cells in bladder cancer via microfluidic immunoassays: sialyl-tn as a novel biomarker in liquid biopsies. Front. Oncol. 10, 1774. doi:10.3389/FONC.2020.01774
Čaval, T., Tian, W., Yang, Z., Clausen, H., and Heck, A. J. R. (2018). Direct quality control of glycoengineered Erythropoietin variants. Nat. Commun. 9, 3342. doi:10.1038/s41467-018-05536-3
Chen, X., Tang, K., Lee, M., and Flynn, G. C. (2008). Microchip assays for screening monoclonal antibody product quality. Electrophoresis 29 (24), 4993–5002. doi:10.1002/ELPS.200800324
D’Amico, C. I., Polasky, D. A., Steyer, D. J., Ruotolo, B. T., and Kennedy, R. T. (2022). Ion mobility-mass spectrometry coupled to droplet microfluidics for rapid protein structure analysis and drug discovery. Anal. Chem. 94 (38), 13084–13091. doi:10.1021/ACS.ANALCHEM.2C02307
Dang, F., Kakehi, K., Cheng, J., Tabata, O., Kurokawa, M., Nakajima, K., et al. (2006). Hybrid dynamic coating with n-dodecyl β-d-Maltoside and methyl cellulose for high-performance carbohydrate analysis on poly(methyl methacrylate) chips. Anal. Chem. 78 (5), 1452–1458. doi:10.1021/AC051702F
Dang, F., Zhang, L., Hagiwara, H., Mishina, Y., and Baba, Y. (2003). Ultrafast analysis of oligosaccharides on microchip with light-emitting diode confocal fluorescence detection. Electrophoresis 24 (4), 714–721. doi:10.1002/ELPS.200390086
Dimov, I. K., Kijanka, G., and Ducrée, J. (2010). Lab-in-a-Trench platform for real-time monitoring of cell surface protein expression. 2010 IEEE 23rd Int. Conf. Micro Electro Mech. Syst. (MEMS), 96–99. doi:10.1109/MEMSYS.2010.5442556
Dong, Y., Skelley, A. M., Merdek, K. D., Sprott, K. M., Jiang, C., Pierceall, W. E., et al. (2013). Microfluidics and circulating tumor cells. J. Mol. Diagnostics 15 (2), 149–157. doi:10.1016/J.JMOLDX.2012.09.004
Fernandes, A. C., Gernaey, K. V., and Krühne, U. (2018). Connecting worlds – a View on microfluidics for a wider application. Biotechnol. Adv. 36 (4), 1341–1366. doi:10.1016/J.BIOTECHADV.2018.05.001
Figeys, D., and Pinto, D. (2000). Lab-on-a-Chip: a revolution in biological and medical sciences. Anal. Chem. 72 (9), 330–335. doi:10.1021/ac002800y
Fujita, K., Oura, F., Nagamine, N., Katayama, T., Hiratake, J., Sakata, K., et al. (2005). Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-Glycan-Specific endo-α-N-acetylgalactosaminidase from bifidobacterium longum. J. Biol. Chem. 280 (45), 37415–37422. doi:10.1074/jbc.M506874200
Fukuda, M., Sasaki, H., Lopez, L., and Fukuda, M. (1989). Survival of recombinant Erythropoietin in the circulation: the role of carbohydrates. Blood 73, 84–89. doi:10.1182/BLOOD.V73.1.84.84
Gaunitz, S., Nagy, G., Pohl, N. L. B., and Novotny, M. V. (2017). Recent advances in the analysis of complex glycoproteins. Anal. Chem. 89, 389–413. doi:10.1021/ACS.ANALCHEM.6B04343
Gooday, G. W. (1990). “The ecology of chitin degradation,” in Advances in microbial ecology (Boston, MA: Springer), 387–430. doi:10.1007/978-1-4684-7612-5_10
Gray, C. J., Migas, L. G., Barran, P. E., Pagel, K., Seeberger, P. H., Eyers, C. E., et al. (2019). Advancing solutions to the carbohydrate sequencing challenge. J. Am. Chem. Soc. 141 (37), 14463–14479. doi:10.1021/JACS.9B06406
Guo, Y., Jia, W., Yang, J., and Zhan, X. (2022). Cancer glycomics offers potential biomarkers and therapeutic targets in the framework of 3P medicine. Front. Endocrinol. 13, 970489. doi:10.3389/FENDO.2022.970489
Hanash, S. M., Pitteri, S. J., and Faca, V. M. (2008). Mining the plasma proteome for cancer biomarkers. Nature 452 (7187), 571–579. doi:10.1038/nature06916
Hasan, M. N., An, R., Akkus, A., Akkaynak, D., Minerick, A. R., Kharangate, C. R., et al. (2021). Dynamic PH and thermal analysis of paper-based microchip electrophoresis. Micromachines 12 (11), 1433. doi:10.3390/MI12111433
Haslam, S. M., Freedberg, D. I., Mulloy, B., Dell, A., Stanley, P., and Prestegard, J. H. (2022). Structural analysis of glycans. Ann. Transl. Med. 2 (2). doi:10.1101/GLYCOBIOLOGY.4E.50
Henry, C. S. (2006). Microchip capillary electrophoresis: an introduction. Methods Mol. Biol. Clift. N.J 339, 1–10. doi:10.1385/1-59745-076-6:1
Hong, P., Sun, H., Sha, L., Pu, Y., Khatri, K., Yu, X., et al. (2017). GlycoDeNovo – an efficient algorithm for accurate de novo glycan topology reconstruction from tandem mass spectra. J. Am. Soc. Mass Spectrom. 28 (11), 2288–2301. doi:10.1007/S13361-017-1760-6
Hsu, C.-H., Hung, S.-C., Wu, C.-Y., and Wong, C.-H. (2011). Toward automated oligosaccharide synthesis. Angew. Chem. Int. Ed. 50 (50), 11872–11923. doi:10.1002/anie.201100125
Hu, M., Lan, Y., Lu, A., Ma, X., and Zhang, L. (2019). Glycan-based biomarkers for diagnosis of cancers and other diseases: past, present, and future. Prog. Mol. Biol. Transl. Sci. 162 (January), 1–24. doi:10.1016/BS.PMBTS.2018.12.002
Ishibashi, Y., Nakasone, T., Kiyohara, M., Horibata, Y., Sakaguchi, K., Hijikata, A., et al. (2007). A novel endoglycoceramidase hydrolyzes oligogalactosylceramides to produce galactooligosaccharides and ceramides. J. Biol. Chem. 282 (15), 11386–11396. doi:10.1074/jbc.M608445200
Jmeian, Y., Hammad, L. A., and Mechref, Y. (2012). Fast and efficient online release of N-glycans from glycoproteins facilitating liquid chromatography-tandem mass spectrometry glycomic profiling. Anal. Chem. 84 (20), 8790–8796. doi:10.1021/AC301855V
Kempa, E. E., Smith, C. A., Li, X., Bellina, B., Richardson, K., Pringle, S., et al. (2020). Coupling droplet microfluidics with mass spectrometry for ultrahigh-throughput analysis of complex mixtures up to and above 30 hz. Anal. Chem. 92 (18), 12605–12612. doi:10.1021/ACS.ANALCHEM.0C02632
Kinoshita, M., Nakajima, K., Yamamoto, S., and Suzuki, S. (2021). High-throughput N-glycan screening method for therapeutic antibodies using a microchip-based DNA analyzer: a promising methodology for monitoring monoclonal antibody N-glycosylation. Anal. Bioanal. Chem. 413 (19), 4727–4738. doi:10.1007/S00216-021-03434-0
Kinoshita, M., and Yamada, K. (2022). Recent advances and trends in sample preparation and chemical modification for glycan analysis. J. Pharm. Biomed. Analysis 207, 114424. doi:10.1016/J.JPBA.2021.114424
Klemm, D., Heublein, B., Fink, H. P., and Bohn, A. (2005). Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 44 (22), 3358–3393. doi:10.1002/ANIE.200460587
Koeller, K. M., and Wong, C. H. (2000). Synthesis of complex carbohydrates and glycoconjugates: enzyme-based and programmable one-pot strategies. Chem. Rev. 100 (12), 4465–4494. doi:10.1021/cr990297n
Koutsioulis, D., Landry, D., and Guthrie, E. P. (2008). Novel endo- -N-acetylgalactosaminidases with broader substrate specificity. Glycobiology 18 (10), 799–805. doi:10.1093/GLYCOB/CWN069
Krenkova, J., Lacher, N. A., and Svec, F. (2009). Multidimensional system enabling deglycosylation of proteins using a capillary reactor with peptide-N-glycosidase F immobilized on a porous polymer monolith and hydrophilic interaction liquid chromatography–mass spectrometry of glycans. J. Chromatogr. A 1216 (15), 3252–3259. doi:10.1016/J.CHROMA.2009.02.036
Krenkova, J., Szekrenyes, A., Keresztessy, Z., Foret, F., and Guttman, A. (2013). Oriented immobilization of peptide-N-glycosidase F on a monolithic support for glycosylation analysis. J. Chromatogr. A 1322, 54–61. doi:10.1016/J.CHROMA.2013.10.087
Laser, D. J., and Santiago, J. G. (2004). A review of micropumps. J. Micromechanics Microengineering 14 (6), R35–R64. doi:10.1088/0960-1317/14/6/R01
Lebrilla, C. B., Liu, J., Widmalm, G., and Prestegard, J. H. (2022). Oligosaccharides and polysaccharides. Essentials Glycobiol. doi:10.1101/GLYCOBIOLOGY.4E.3
Liang, Y., Wu, C., Zhao, Q., Wu, Q., Jiang, B., Weng, Y., et al. (2015). Gold nanoparticles immobilized hydrophilic monoliths with variable functional modification for highly selective enrichment and on-line deglycosylation of glycopeptides. Anal. Chim. Acta 900, 83–89. doi:10.1016/J.ACA.2015.10.024
Lillehoj, E. P., Kato, K., Lu, W., and Kim, K. C. (2013). Cellular and molecular biology of airway mucins. Int. Rev. Cell. Mol. Biol. 303, 139–202. doi:10.1016/B978-0-12-407697-6.00004-0
Maass, K., Ranzinger, R., Geyer, H., von der Lieth, C., and Geyer, R. (2007). “Glyco-peakfinder” – de novo composition analysis of glycoconjugates. Proteomics 7 (24), 4435–4444. doi:10.1002/PMIC.200700253
Mai, J., Sommer, G. J., and Hatch, A. V. (2012). Microfluidic digital isoelectric fractionation for rapid multidimensional glycoprotein analysis. Anal. Chem. 84 (8), 3538–3545. doi:10.1021/AC203076P
Manz, A., Graber, N., and Widmer, H. M. (1990). Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sensors Actuators B Chem. 1 (1–6), 244–248. doi:10.1016/0925-4005(90)80209-I
Marques, M. P., and Szita, N. (2017). Bioprocess microfluidics: applying microfluidic devices for bioprocessing. Curr. Opin. Chem. Eng. 18, 61–68. doi:10.1016/J.COCHE.2017.09.004
Marth, J. D. (2008). A unified vision of the building blocks of life. Nat. Cell. Biol. 10 (9), 1015. doi:10.1038/ncb0908-1015
Matrosovich, M., Herrler, G., and Klenk, H. D. (2015). Sialic acid receptors of viruses. Top. Curr. Chem. 367, 1–28. doi:10.1007/128_2013_466
Mitra, I., Alley, W. R., Goetz, J. A., Vasseur, J. A., Novotny, M. V., and Jacobson, S. C. (2013). Comparative profiling of N-glycans isolated from serum samples of ovarian cancer patients and analyzed by microchip electrophoresis. J. Proteome Res. 12 (10), 4490–4496. doi:10.1021/PR400549E
Mitra, I., Snyder, C. M., Zhou, X., Campos, M. I., Alley, W. R., Novotny, M. V., et al. (2016). Structural characterization of serum N-glycans by methylamidation, fluorescent labeling, and analysis by microchip electrophoresis. Anal. Chem. 88 (18), 8965–8971. doi:10.1021/ACS.ANALCHEM.6B00882
Möckl, L. (2020). The emerging role of the mammalian glycocalyx in functional membrane organization and immune system regulation. Front. Cell. Dev. Biol. 8, 253. doi:10.3389/FCELL.2020.00253
Neves, M., Azevedo, R., Lima, L., Oliveira, M. I., Peixoto, A., Ferreira, D., et al. (2019). Exploring sialyl-tn expression in microfluidic-isolated circulating tumour cells: a novel biomarker and an analytical tool for precision oncology applications. New Biotechnol. 49, 77–87. doi:10.1016/J.NBT.2018.09.004
Nguyen, N.-T., and Wu, Z. (2004). Micromixers—a review. J. Micromechanics Microengineering 15 (2), R1–R16. doi:10.1088/0960-1317/15/2/R01
Ni, W., Bones, J., and Karger, B. L. (2013). In-depth characterization of N-linked oligosaccharides using fluoride-mediated negative ion microfluidic chip LC-MS. Anal. Chem. 85 (6), 3127–3135. doi:10.1021/ac3031898
Nielsen, M. M., and Pedersen, C. M. (2018). Catalytic glycosylations in oligosaccharide synthesis. Chem. Rev. 118 (17), 8285–8358. doi:10.1021/acs.chemrev.8b00144
Niñonuevo, M., An, H., Yin, H., Killeen, K., Grimm, R., Ward, R., et al. (2005). Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis 26 (19), 3641–3649. doi:10.1002/ELPS.200500246
O’Connell, T. M., King, D., Dixit, C. K., O’Connor, B., Walls, D., and Ducrée, J. (2014). Sequential glycan profiling at single cell level with the microfluidic lab-in-a-trench platform: a new era in experimental cell biology. Lab. Chip 14 (18), 3629–3639. doi:10.1039/C4LC00618F
Palm, A. K., and Novotny, M. V. (2005). A monolithic PNGase F enzyme microreactor enabling glycan mass mapping of glycoproteins by mass spectrometry. Rapid Commun. Mass Spectrom. 19 (12), 1730–1738. doi:10.1002/RCM.1979
Panza, M., Pistorio, S. G., Stine, K. J., and Demchenko, A. V. (2018). Automated chemical oligosaccharide synthesis: novel approach to traditional challenges. Chem. Rev. 118 (17), 8105–8150. doi:10.1021/ACS.CHEMREV.8B00051
Pinnock, F., and Daniel, S. (2022). Small tools for sweet challenges: advances in microfluidic technologies for glycan synthesis. Anal. Bioanal. Chem. 414 (18), 5139–5163. doi:10.1007/S00216-022-03948-1
Primack, J., Flynn, G. C., and Pan, H. (2011). A high-throughput microchip-based glycan screening assay for antibody cell culture samples. Electrophoresis 32 (10), 1129–1132. doi:10.1002/ELPS.201000619
Qu, Y., Xia, S., Yuan, H., Wu, Q., Li, M., Zou, L., et al. (2011). Integrated sample pretreatment system for N-linked glycosylation site profiling with combination of hydrophilic interaction chromatography and PNGase F immobilized enzymatic reactor via a strong cation exchange precolumn. Anal. Chem. 83 (19), 7457–7463. doi:10.1021/AC201665E
Quaranta, A., Sroka-Bartnicka, A., Tengstrand, E., and Thorsén, G. (2016). N-glycan profile analysis of transferrin using a microfluidic compact disc and MALDI-MS. Anal. Bioanal. Chem. 408 (17), 4765–4776. doi:10.1007/S00216-016-9570-4
Roy, B., Chattopadhyay, G., Mishra, D., Das, T., Chakraborty, S., and Maiti, T. K. (2014). On-chip lectin microarray for glycoprofiling of different gastritis types and gastric cancer. Biomicrofluidics 8 (3), 034107. doi:10.1063/1.4882778
Rudd, P. M., Karlsson, N. G., Khoo, K.-H., Thaysen-Andersen, M., Wells, L., and Packer, N. H. (2022). Glycomics and glycoproteomics. Essentials Glycobiol. doi:10.1101/GLYCOBIOLOGY.4E.51
Schäffer, C., and Messner, P. (2017). Emerging facets of prokaryotic glycosylation. FEMS Microbiol. Rev. 41 (1), 49–91. doi:10.1093/FEMSRE/FUW036
Scher, H. I., Heller, G., Molina, A., Attard, G., Danila, D. C., Jia, X., et al. (2015). Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 33 (12), 1348–1355. doi:10.1200/JCO.2014.55.3487
Schmid, K. (1950). Preparation and properties of an acid glycoprotein prepared from human plasma. J. Am. Chem. Soc. 72 (6), 2816. doi:10.1021/JA01162A553
Sears, P., and Wong, C.-H. (2001). Toward automated synthesis of oligosaccharides and glycoproteins. Science 291 (5512), 2344–2350. doi:10.1126/SCIENCE.1058899
Seeberger, P. H. (2008). Automated carbohydrate synthesis as platform to address fundamental aspects of glycobiology—current status and future challenges. Carbohydr. Res. 343 (12), 1889–1896. doi:10.1016/J.CARRES.2008.05.023
Shang, Y., Zeng, Y., and Zeng, Y. (2016). Integrated microfluidic lectin barcode platform for high-performance focused glycomic profiling. Sci. Rep. 6 (1), 20297. doi:10.1038/srep20297
Sharon, N., and Lis, H. (1987). A century of lectin research (1888–1988). Trends Biochem. Sci. 12, 488–491. doi:10.1016/0968-0004(87)90236-2
Smith, C. A., Li, X., Mize, T. H., Sharpe, T. D., Graziani, E. I., Abell, C., et al. (2013). Sensitive, high throughput detection of proteins in individual, surfactant-stabilized picoliter droplets using nanoelectrospray ionization mass spectrometry. Anal. Chem. 85 (8), 3812–3816. doi:10.1021/AC400453T
Snyder, C. M., Alley, W. R., Campos, M. I., Svoboda, M., Goetz, J. A., Vasseur, J. A., et al. (2016). Complementary glycomic analyses of sera derived from colorectal cancer patients by MALDI-TOF-MS and microchip electrophoresis. Anal. Chem. 88 (19), 9597–9605. doi:10.1021/ACS.ANALCHEM.6B02310
Sun, S., Yang, M., Kostov, Y., and Rasooly, A. (2010). ELISA-LOC: lab-on-a-chip for enzyme-linked immunodetection. Lab. Chip 10, 2093–2100. doi:10.1039/c003994b
Suzuki, S., Shimotsu, N., Honda, S., Arai, A., and Nakanishi, H. (2001). Rapid analysis of amino sugars by microchip electrophoresis with laser-induced fluorescence detection. Electrophoresis 22 (18). doi:10.1002/1522-2683(200110)22:18<4023::AID-ELPS4023>3.0
Suzuki, Y., Ito, T., Suzuki, T., Holland, R. E., Chambers, T. M., Kiso, M., et al. (2000). Sialic acid species as a determinant of the host range of influenza A viruses. J. Virol. 74 (24), 11825–11831. doi:10.1128/JVI.74.24.11825-11831.2000
Szigeti, M., Bondar, J., Gjerde, D., Keresztessy, Z., Szekrenyes, A., and Guttman, A. (2016). Rapid N-glycan release from glycoproteins using immobilized PNGase F microcolumns. J. Chromatogr. B 1032, 139–143. doi:10.1016/J.JCHROMB.2016.02.006
Temiz, Y., Lovchik, R. D., Kaigala, G. V., and Delamarche, E. (2015). Lab-on-a-Chip devices: how to close and plug the lab? Microelectron. Eng. 132, 156–175. doi:10.1016/J.MEE.2014.10.013
Thaysen-Andersen, M., Packer, N. H., and Schulz, B. L. (2016). Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Mol. Cell. Proteomics 15 (6), 1773–1790. doi:10.1074/MCP.O115.057638
Thege, F. I., Lannin, T. B., Saha, T. N., Tsai, S., Kochman, M. L., Hollingsworth, M. A., et al. (2014). Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab. Chip 14 (10), 1775–1784. doi:10.1039/C4LC00041B
Thuy, T. T., Inganäs, M., and Thorsén, G. (2011). High-throughput profiling of N-linked oligosaccharides in therapeutic antibodies using a microfluidic CD platform and MALDI-MS. Anal. Bioanal. Chem. 399 (4), 1601–1611. doi:10.1007/S00216-010-4469-Y
Thuy, T. T., and Thorsén, G. (2013). Glycosylation profiling of therapeutic antibodies in serum samples using a microfluidic CD platform and MALDI-MS. J. Am. Soc. Mass Spectrom. 24 (7), 1053–1063. doi:10.1007/S13361-013-0623-Z
Tian, Z., Wu, Y., Shao, F., Tang, D., Qin, X., Wang, C., et al. (2021). Electrofluorochromic imaging analysis of glycan expression on living single cell with bipolar electrode arrays. Anal. Chem. 93 (12), 5114–5122. doi:10.1021/ACS.ANALCHEM.0C04785
Vanderschaeghe, D., Szekrényes, Á., Wenz, C., Gassmann, M., Naik, N., Bynum, M., et al. (2010). High-throughput profiling of the serum N-glycome on capillary electrophoresis microfluidics systems: toward clinical implementation of GlycoHepatoTest. Anal. Chem. 82 (17), 7408–7415. doi:10.1021/AC101560A
Varki, A. (2011). Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb. Perspect. Biol. 3 (6), a5462–a5514. doi:10.1101/CSHPERSPECT.A005462
Vilaj, M., Lauc, G., and Trbojević-Akmačić, I. (2021). Evaluation of different PNGase F enzymes in immunoglobulin G and total plasma N-glycans analysis. Glycobiology 31 (1), 2–7. doi:10.1093/glycob/cwaa047
Walsh, G., and Jefferis, R. (2006). Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24 (10), 1241–1252. doi:10.1038/nbt1252
Walther, T., Karamanska, R., Chan, R. W. Y., Chan, M. C. W., Jia, N., Air, G., et al. (2013). Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection. PLOS Pathog. 9 (3), e1003223. doi:10.1371/JOURNAL.PPAT.1003223
Wang, X., Yi, L., Mukhitov, N., Schrell, A. M., Dhumpa, R., and Roper, M. G. (2015). Microfluidics-to-Mass spectrometry: a review of coupling methods and applications. J. Chromatogr. A 1382, 98–116. doi:10.1016/j.chroma.2014.10.039
Wang, Y., Liu, Q., Men, T., Liang, Y., Niu, H., and Wang, J. (2020). A microfluidic system based on the monoclonal antibody BCMab1 specifically captures circulating tumor cells from bladder cancer patients. J. Biomaterials Sci. Polym. Ed. 31 (9), 1199–1210. doi:10.1080/09205063.2020.1748332
Wang, Y. C., Stevens, A. L., and Han, J. (2005). Million-Fold preconcentration of proteins and peptides by nanofluidic filter. Anal. Chem. 77 (14), 4293–4299. doi:10.1021/AC050321Z
Weibel, D. B., Kruithof, M., Potenta, S., Sia, S. K., Lee, A., and Whitesides, G. M. (2005). Torque-actuated valves for microfluidics. Anal. Chem. 77 (15), 4726–4733. doi:10.1021/AC048303P
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442 (7101), 368–373. doi:10.1038/nature05058
Willner, M. R., McMillan, K. S., Graham, D., Vikesland, P. J., and Zagnoni, M. (2018). Surface-enhanced Raman scattering based microfluidics for single-cell analysis. Anal. Chem. 90 (20), 12004–12010. doi:10.1021/ACS.ANALCHEM.8B02636
Wuhrer, M., Koeleman, C. A. M., Deelder, A. M., and Hokke, C. H. (2004). Normal-phase nanoscale liquid chromatography-mass spectrometry of underivatized oligosaccharides at low-femtomole sensitivity. Anal. Chem. 76 (3), 833–838. doi:10.1021/AC034936C
Yamamoto, S., Ueda, M., Kasai, M., Ueda, Y., Kinoshita, M., and Suzuki, S. (2020). A fast and convenient solid phase preparation method for releasing N-glycans from glycoproteins using trypsin- and peptide-N-glycosidase F (PNGase F)-Impregnated polyacrylamide gels fabricated in a pipette tip. J. Pharm. Biomed. Analysis 179, 112995. doi:10.1016/J.JPBA.2019.112995
Yang, S., Clark, D., Liu, Y., Li, S., and Zhang, H. (2017). High-throughput analysis of N-glycans using AutoTip via glycoprotein immobilization. Sci. Rep. 7, 10216. doi:10.1038/s41598-017-10487-8
Yang, S., Li, Y., Shah, P., and Zhang, H. (2013). Glycomic analysis using glycoprotein immobilization for glycan extraction. Anal. Chem. 85 (11), 5555–5561. doi:10.1021/AC400761E
Yang, S., Toghi Eshghi, S., Chiu, H., Devoe, D. L., and Zhang, H. (2013). Glycomic analysis by glycoprotein immobilization for glycan extraction and liquid chromatography on microfluidic chip. Anal. Chem. 85 (21), 10117–10125. doi:10.1021/AC4013013
Yang, Y., Liu, F., Franc, V., Halim, L. A., Schellekens, H., and Heck, A. J. R. (2016). Hybrid mass spectrometry approaches in glycoprotein analysis and their usage in scoring biosimilarity. Nat. Commun. 7, 13397. doi:10.1038/ncomms13397
Zhang, L., Xu, T., Zhang, J., Wong, S. C. C., Ritchie, M., Hou, H. W., et al. (2021). Single cell metabolite detection using inertial microfluidics-assisted ion mobility mass spectrometry. Anal. Chem. 93 (30), 10462–10468. doi:10.1021/ACS.ANALCHEM.1C00106
Zhu, H., Fohlerová, Z., Pekárek, J., Basova, E., and Neužil, P. (2020). Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 153, 112041. doi:10.1016/J.BIOS.2020.112041
Zhuang, Z., Mitra, I., Hussein, A., Novotny, M. V., Mechref, Y., and Jacobson, S. C. (2011). Microchip electrophoresis of N-glycans on serpentine separation channels with asymmetrically tapered turns. Electrophoresis 32 (2), 246–253. doi:10.1002/ELPS.201000461
Keywords: carbohydrate, glycobiology, microfluidics, glycoprotein, bioanalysis, glycan analyses, micro-total analysis systems
Citation: Pinnock F, Carten JD and Daniel S (2024) More small tools for sweet challenges: advances in microfluidic technologies for glycan analysis. Front. Lab. Chip. Technol. 3:1359183. doi: 10.3389/frlct.2024.1359183
Received: 20 December 2023; Accepted: 30 January 2024;
Published: 20 February 2024.
Edited by:
Trieu Nguyen, Honda Research Institute USA Mountain View, United StatesReviewed by:
Feng Shen, Shanghai Jiao Tong University, ChinaSoumya Srivastava, West Virginia University, United States
Copyright © 2024 Pinnock, Carten and Daniel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susan Daniel, c2QzODZAY29ybmVsbC5lZHU=
 Ferra Pinnock
Ferra Pinnock Juliana Debrito Carten
Juliana Debrito Carten Susan Daniel
Susan Daniel