
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Lab Chip Technol., 11 January 2024
Sec. Organ on a Chip
Volume 2 - 2023 | https://doi.org/10.3389/frlct.2023.1337945
This article is part of the Research TopicReviews in Organ on a ChipView all 4 articles
Inflammatory bowel disease (IBD), celiac disease, and other inflammatory conditions of the gastrointestinal tract are highly prevalent in society. Due to the complexity of factors involved, detailed pathogenesis is difficult to determine and thus remains elusive in many cases. The advent of gut-on-chip devices has permitted more accurate modeling of the gut microenvironment with the inclusion of fluid flow and mechanical cues that are true to intestinal architecture and physiology. This review outlines the evolution of gut-on-chip platforms towards greater physiological relevance and elucidates how these devices have been used to model and study intestinal inflammation in humans. In addition, we identify key elements from both technological and disease standpoints that are integral for accurate gut-on-chip models of IBD and celiac disease.
The small and large intestines, which make up a considerable part of the gastrointestinal tract, each have distinct mucosal structures and microenvironments. Both anatomical parts contribute diversely to physiological functions such as digestion, nutrient absorption, microbial interactions, and immune responses. Throughout life, the cellular and molecular composition of the epithelium that lines the mucosa changes in response to evolving functional requirements and environmental exposures (Rodríguez et al., 2015; Elmentaite et al., 2021). Some of these environmental factors, along with diet and microbiota, are thought to contribute to the development of inflammatory bowel disease (IBD), celiac disease and other inflammatory conditions in genetically predisposed individuals (Liu et al., 2015; Huang et al., 2017; Guan, 2019; Krishnareddy, 2019). Accurately modeling the in vivo gut environment, as well as aberrations resulting in inflammation, is therefore crucial for further understanding the pathogenesis of chronic gastrointestinal conditions and for investigating effective treatments.
Conventional methods for recreating the intestinal environment for research purposes comprise immortalized cell lines or primary cell culture in traditional culture wells and Transwell inserts, as well as everted gut sacs (Alam et al., 2012; Yamaura et al., 2016; Altay et al., 2019). While these models have been vital for studying, for example, barrier function and drug metabolism, they do not completely recapitulate the three-dimensional architecture and dynamic microenvironment of the small and large intestines. Although significant developments have been made to incorporate stem cells and organoids, many functional properties and supporting cell populations are still unaccounted for in static cultures (Spence et al., 2011; Gómez and Boudreau, 2021). In terms of inflammatory conditions, conventional methods are largely unable to depict features that are essential for modeling their multifactorial nature, such as the mechanical forces acting on cells and immune and microbe components. These limitations have led to the development of gut-on-chip devices.
In general, organ-on-chip models are microfluidic devices predominantly made of clear, flexible polymers into which several hollow channels or chambers are molded. Specific organs can be mimicked by culturing relevant cell types in the device, which is designed to allow control over parameters such as concentration gradients, mechanical forces, cell interactions, and tissue morphology (Wu et al., 2020). Both single-organ and multi-organ systems have been successfully established, where the latter are typically simpler biologically and prioritize the systemic interactivity between organs. Single-organ organ-on-chips have a greater focus on the detailed function of an organ. For example, by combining biomaterials, microfluidics, and cell culture, gut-on-chip devices enable the recreation of an epithelial barrier of polarized cells with high integrity and functionality. Moreover, incorporation of fluid flow and mechanical deformation supports cell differentiation and facilitates coculture, such as with microbial symbionts, without the overgrowth that is common in static models (Kim et al., 2012). The constantly evolving organ-on-chip technology therefore provides an opportunity for studying the multiple aspects at play in the development and progression of intestinal inflammatory conditions.
The gut lumen is a hollow, tubular passage lined with a protective barrier composed of a diverse range of intestinal epithelial cells (IECs). In the small intestine, the monolayer of IECs forms a crypt-villus axis that affects fluid flow and significantly increases the surface area for the absorption of nutrients (Clevers, 2013). Imaging of the different regions of the small intestine has revealed that villi are taller and more prevalent proximally in the duodenum and jejunum than distally in the ileum (Holmes et al., 1961). Absorptive enterocytes, which largely enable digestive and metabolic functions, make up the majority of the cell population, while several types of secretory cells perform more specialized roles within the epithelium. For instance, goblet cells secrete mucins to produce a physical mucous barrier that prevents large particles from directly contacting the epithelium and enteroendocrine cells produce and release hormones in response to different stimuli (Kim and Khan, 2013; Gribble and Reimann, 2019). Other secretory cells include immune response-mediating tuft cells and Paneth cells, which secrete antimicrobial molecules into the lumen for regulating the gut microbiota (Bevins and Salzman, 2011). In addition, Paneth cells are located adjacent to intestinal stem cells (ISCs) in the intestinal crypts, where they support the rapid renewal of the epithelium (Clevers and Bevins, 2013). Self-renewal and regeneration in response to injury are driven by the production of daughter progenitors by ISCs in the crypts (Clevers, 2013). Subsequently, these progenitor cells migrate towards the luminal surface and undergo multilineage differentiation to generate the different types of IECs.
Longitudinally, the large intestine can also be divided into parts encompassing the cecum, colon, rectum, and anus. In contrast to the small intestine, IECs in the large intestine are arranged into inner crypts which open into a flat luminal surface. Although the cellular compositions are largely similar, cell analyses have revealed that the large intestine houses a greater number of goblet cells and less Paneth cells (Specian and Oliver, 1991; Tanaka et al., 2001). These differences support a shift in function towards the absorption of water, electrolytes, and vitamins, as well as the formation of feces from indigestible and unabsorbed residues. Despite the distinctions between the small and large intestines, IECs collectively establish and maintain both a physical and biochemical barrier, offering protection from harmful pathogens and various other inflammatory stimuli. When the epithelium is intact, mucosal permeability is determined by the dynamic proteins that compose tight junctions, which regulate the paracellular transport of water, ions, and other molecules (Rescigno, 2011; Suzuki, 2012).
IECs also provide an interface for epithelial-microbe-immune interactions to further support a finely tuned system that maintains a delicate balance between the host and its microbial inhabitants. Thus, another important regulator of the gut microenvironment is the microbiota, which mostly comprises non-pathogenic bacteria aiding in metabolism, barrier function, and immune response stimulation (Hillman et al., 2017). Similar to cell composition, microbiota have also been found to have spatial variability throughout the intestines due to differences in flow rate and pH in the lumen (McHardy et al., 2013). Additionally, the distribution of microorganisms is largely dependent on diet, age, and the presence of an oxygen gradient in the mucosa (Rinninella et al., 2019). Luminal microbiota and mucosa-associated microbiota located in the mucus layer can affect the integrity of the gut barrier through activation of signaling pathways in IECs. Conversely, the composition of the microbiota can be modulated by signaling molecules and alternative energy sources provided by IECs. Similar communication occurs between IECs and immune cells located in the lamina propria underlying the IEC layer, which also contains blood vessels, lymphatic vessels, and nerves that further regulate intestinal motility and nutrient transport. IECs mediate the crosstalk between microbiota and immunologically active cells (T cells, B cells, dendritic cells, macrophages) as well as other luminal contents, often resulting in the adjustment of cytokine secretion to maintain intestinal homeostasis.
Furthermore, luminal flow, and peristalsis, which make up the intestinal mechanical microenvironment, are responsible for gut motility and in part inhibit microbial overgrowth. Peristalsis is the result of layers of smooth muscle contracting to propel food through the intestines and as the luminal contents move, shear forces result from the friction against the intestinal wall. These mechanical forces also act on IECs, contributing to normal epithelial differentiation, mucus production, and the establishment of stable symbiosis between the epithelium and resident gut microbiome (Kim et al., 2016; Jeon et al., 2022). In summary, establishing an in vitro intestinal model that more closely resembles the native tissue requires incorporation of the diverse range of IECs and supporting cells, correct epithelial barrier morphology, mechanical forces, and the gut microbiota (Figure 1).
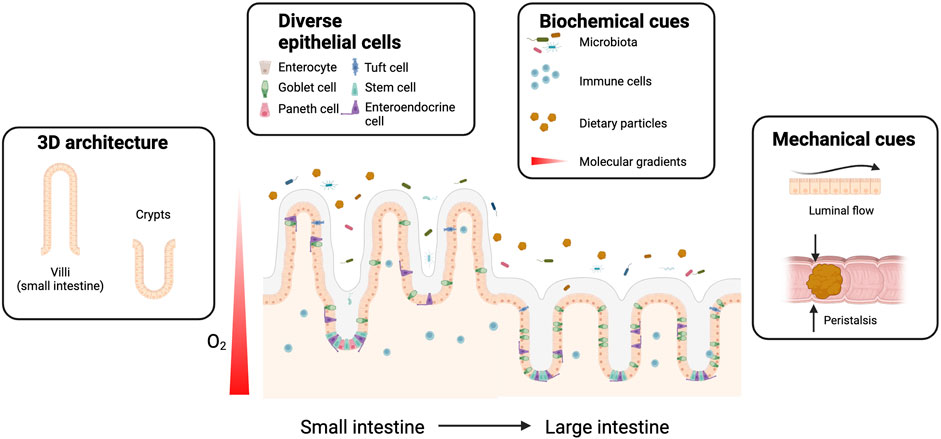
FIGURE 1. Gut-on-chip platforms enable mimicking of the human gut microenvironment. The microenvironment of the human intestines is variable depending on the anatomical region, but several common elements can be pinpointed as integral for normal functioning. These elements include three-dimensional (3D) tissue architecture, epithelial cellular diversity and various biochemical, and mechanical cues. Organ-on-chip technology can facilitate the generation of a more accurate in vitro intestinal model consisting of a combination of these features.
Intestinal diseases like IBD (Crohn’s disease, ulcerative colitis) and celiac disease are characterized by chronic inflammation that affects various regions of the small and large bowels. Although there is some variation between patients, ulcerative colitis typically manifests within the colon, celiac disease in the small intestine, and Crohn’s disease throughout the gastrointestinal tract. Many factors are thought to cause changes in the gut microenvironment and in turn contribute to the development and perpetuation of a dysregulated inflammatory response in these conditions. The proposed mechanisms of IBD suggest an interplay between genetic susceptibility, immune abnormalities, microbiota, and environmental factors (Strober et al., 2007; Lee et al., 2018). The sequence of pathogenetic events is unknown, but include microbial dysbiosis, compromised intestinal epithelium integrity, aberrant immune reactivity against antigens, and increased production of proinflammatory cytokines (Zeissig et al., 2007; Kamada et al., 2013; de Souza and Fiocchi, 2016; Parker et al., 2019). Eventually, a sustained inflammatory state can result in structural damage of the mucosa and increased IEC death coupled with decreased cell turnover (Parker et al., 2019). In celiac disease, small-bowel mucosal inflammation, crypt hyperplasia, and villous atrophy are a result of inappropriate immune responses to certain gluten-derived peptides from wheat, rye, and barley. Repetitive glutamine- and proline-rich regions in gluten resist proteolytic degradation and initiate both adaptive and innate immune responses, eventually leading to mucosal inflammation and IEC destruction (Hausch et al., 2002; Lindfors et al., 2019). Although some of the hallmarks of IBD and celiac disease have been identified, further understanding of the pathogenesis and development of possible novel treatments for these conditions relies on the generation of more accurate in vitro models.
To an extent, the compositions of the mouse small and large intestines are comparable to those of humans, with similar cell types and distributions. Thus, chemically induced, genetically engineered, spontaneous and transgenic murine models have been commonly employed to study different aspects involved in intestinal inflammatory diseases (Prattis and Jurjus, 2015; Mizoguchi et al., 2016; Wirtz et al., 2017). Despite the similarities, the translation of results to humans requires the consideration of significant differences in areas such as microbiota, diet, anatomical structure, and size. Particularly, while mouse models can mimic the phenotypes of inflammatory diseases, they are not as suitable for determining underlying mechanisms and etiology of multifactorial diseases in humans (Jiminez et al., 2015).
The need for more cost-effective, ethically acceptable, and high-throughput research models hasled to the widespread use of cell-based systems. Intestinal barrier permeability and toxicity studies are often performed with colorectal adenocarcinoma-derived Caco-2 cells cultured either on standard well plates or permeable inserts. Caco-2 cells are able to form differentiated and polarized monolayers, exhibiting characteristics similar to intestinal enterocytes (Darling et al., 2020). Though undoubtedly valuable, these static systems fail to model the complexity of the human intestinal tissue because they lack interactions with surrounding cell types and the extracellular matrix (ECM), which are necessary for proper epithelial formation. Compared to the normal human intestine, Caco-2 monolayers can have altered expression of transporters and enzymes affecting overall permeability (Sun et al., 2008). Without incorporation of the mechanical microenvironment, Caco-2 are also unable to adequately portray crypt-villus structures of the small intestine, and coculture with microbiota is difficult due to bacterial overgrowth. The addition of cell culture substrates and additional cell types, such as HT29-MTX cells to increase mucus production, has resulted in the formation of an enhanced epithelial layer. However, the cancerous origin of Caco-2 cells means that possible gene mutations detract from their ability to portray normal epithelium (Bourgine et al., 2012; Hoffmann et al., 2021).
For a more advanced model, intestinal organoids can be derived from primary intestinal crypt stem cells, iPSCs or intact intestinal crypts isolated from biopsy samples (Kasendra et al., 2018; Altay et al., 2019; Yoshida et al., 2020). Unlike Caco-2 cell cultures, organoids represent the stem cell lineage, which further differentiate to encompass the heterogeneous population of the in vivo tissue. The self-organizing capability of intestinal crypt-like units, in combination with the addition of factors such as epidermal growth factor (EGF), Noggin, and R-Spondin-1, allow the crypts to develop into a closed spherical structure (Dignass and Sturm, 2001; Sato and Clevers, 2013). The main drawback of such systems is the inability to access both sides of the epithelium, but they are able to retain correct cellular spatial organization and cellular interactions (Beumer et al., 2020). Biopsy-derived organoids have been successfully cultured as two-dimensional monolayers with exposed apical surfaces. However, expansion is difficult and cannot be done through simple resuspension and passaging (Braverman and Yilmaz, 2018; Thorne et al., 2018). To date, organoids have been valuable in studying human cell mechanisms and pathways despite the lack of functional immune cells and vascularization in most current systems. Other intestinal models include the everted gut sac and Ussing chamber, which are used largely for absorption studies (Thomson et al., 2019). Although highly physiologically relevant, both typically utilize ex vivo tissue, leading to limited viability after approximately 2 h and the speed of absorption is often not comparable to that in vivo.
A majority of gut-on-chip models are composed of polydimethylsiloxane (PDMS) and contain two adjacent hollow channels that are separated by an ECM-coated porous membrane Table 1. Culturing IECs on the upper surface of the porous membrane allows access and manipulation of the conditions in contact with both the apical and basolateral sides of the epithelium. In contrast to conventional static models, the gut-on-chip platform enables continuous exchange of the culture medium, providing adequate oxygenation and nutrients, while also imparting physiological shear stress on cells. For instance, Jalili-Firoozinez et al. fabricated a two-channel PDMS microfluidic device with Caco-2 cells representing the IECs and human intestinal microvascular endothelial cells (HIMECs) representing the vascular endothelium (Figure 2A) (Jalili-Firoozinezhad et al., 2019). Under perfusion flow, Caco-2 cells formed an epithelium of polarized cells connected via tight junctions in the top channel and HIMECs formed a confluent monolayer in the bottom channel (Jalili-Firoozinezhad et al., 2019). In the presence of an oxygen gradient, the microenvironment of the gut-on-chip was also able to support a higher level of microbial diversity when compared to models without oxygen modulation (Jalili-Firoozinezhad et al., 2019). Additionally, IECs isolated from small intestinal biopsy-derived organoids have been cultured in the same two-channel design, resulting in multilineage differentiation, more accurate villus formation, and mucus production (Kasendra et al., 2018).
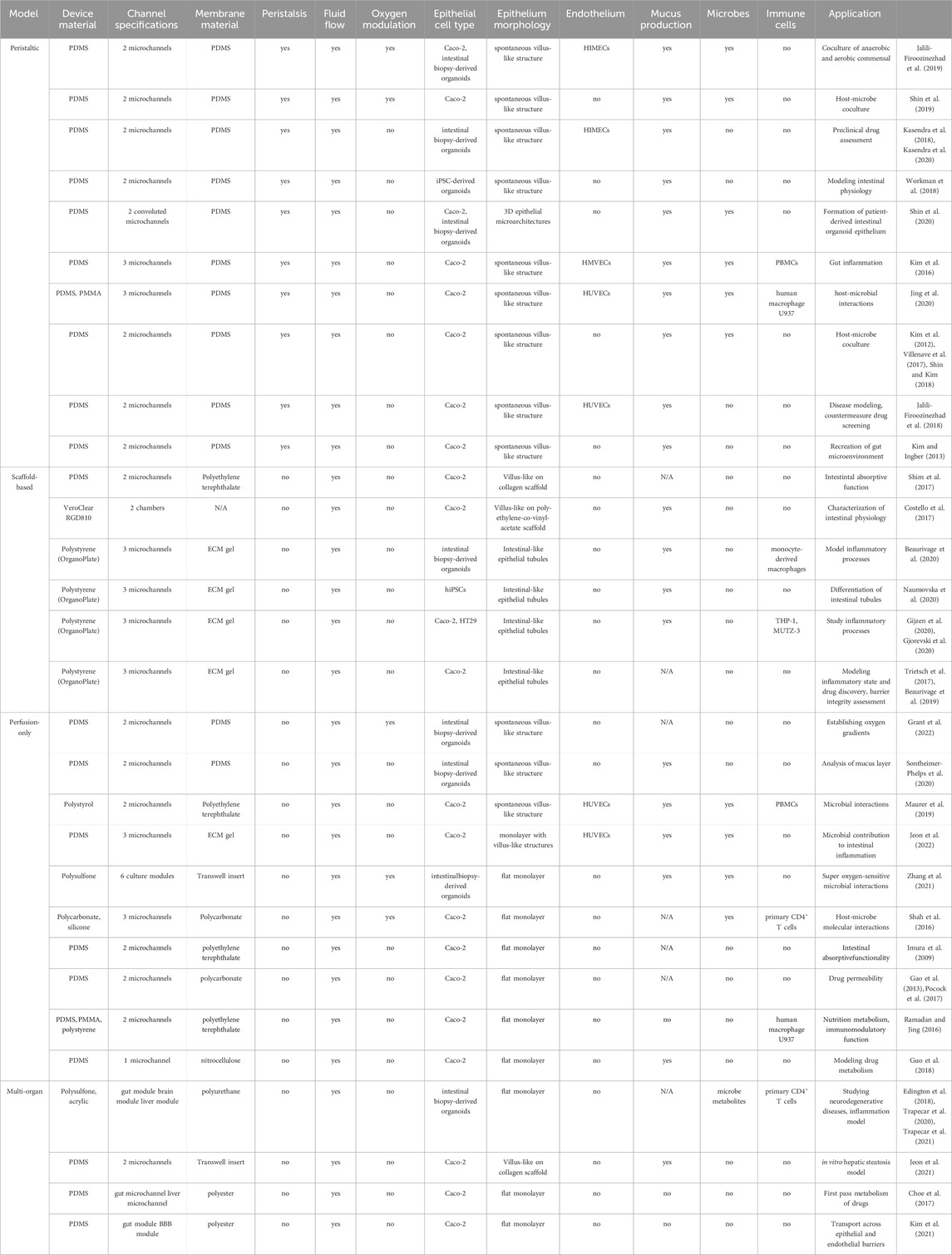
TABLE 1. Features of human gut-on-chip models. PDMS, polydimethylsiloxane; HIMECs, human intestinal microvascular endothelial cells; iPSC, induced pluripotent stem cells; HMVECs, human microvascular endothelial cells; PBMCs, peripheral blood mononuclear cells; PMMA, poly (methyl methacrylate); HUVECs, human umbilical endothelial cells; ECM, extracellular matrix; hiPSC, human induced pluripotent stem cells.
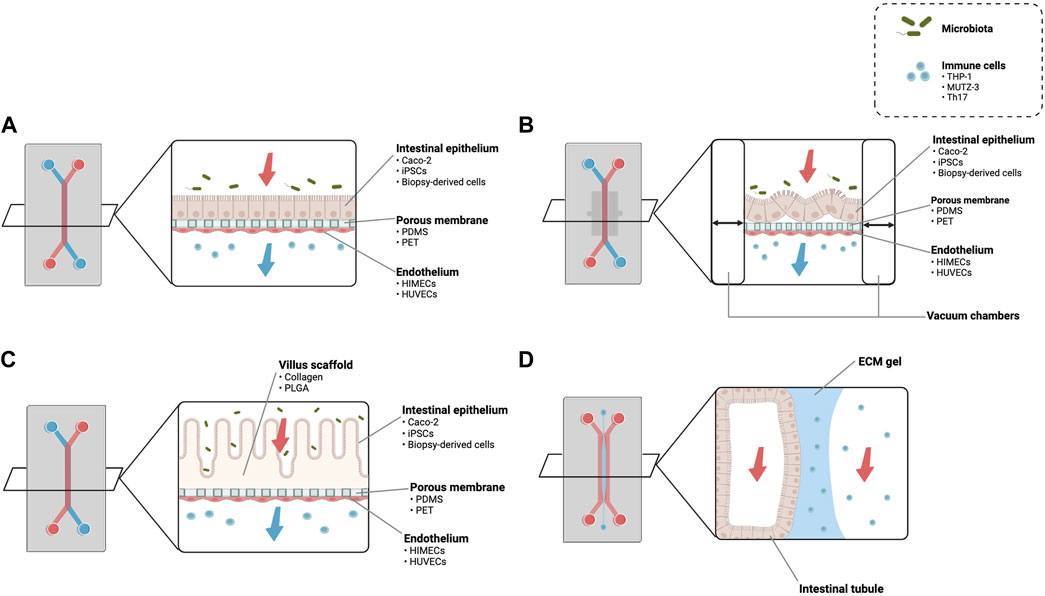
FIGURE 2. Schematic of different types of gut-on-chip devices. (A) A two-channel gut-on-chip with a porous membrane separating microchannels. The upper microfluidic channel contains the intestinal epithelium while the bottom channel can be used to incorporate other cell types, typically the vascular endothelium. Apical and basolateral medium flow enables coculture with factors such as microbiota and immune cells. (B) The application of cyclic suction through the integration of vacuum chambers attached to the porous membrane has been linked to the formation of villus-like structures. (C) Culturing intestinal epithelial cells on engineered villus scaffolds increases the reproducibility of the intestinal three-dimensional architecture. (D) A three-channel gut-on-chip design with an ECM gel in the center microchannel which guides the formation of an intestinal tubule. iPSCs, induced pluripotent stem cells; PDMS, polydimethylsiloxane; PET, polyethylene terephthalate; HIMECs, human intestinal microvascular endothelial cells; HUVECs, human umbilical vein endothelial cells; PLGA, poly (lactic-co-glycolic acid); ECM, extracellular matrix. Images based on (Kim et al., 2012; Shim et al., 2017; Jalili-Firoozinezhad et al., 2019; Beaurivage et al., 2020).
The porous membrane between the channels allows for apical-basal access to the epithelia, but the presence of an artificial membrane could have unwanted effects on permeability and cell morphology. As a solution, Beaurivage et al. utilized a gut-on-chip device (OrganoPlate) where the artificial membrane is replaced by an ECM gel that is patterned so that epithelial cells seeded into the perfusion channel formed a tubule structure (Figure 2D) (Beaurivage et al., 2019). The confluent tubule was defined by tight junction and brush border formation as well as increased expression of glucose and MRP2 transporters, suggesting that the ECM plays an important role in cell differentiation and protein expression (Trietsch et al., 2017).
Although some evidence exists supporting the spontaneous formation of villus-like structures by Caco-2 cells under perfusion flow and mechanical deformation, using a scaffold-based approach (Figure 2C) increases the reproducibility of the small intestinal crypt-villus axis (Sung et al., 2011; Kim and Kim, 2018; Castaño et al., 2019; Creff et al., 2019; Kim and Kim, 2020; Verhulsel et al., 2021). For example, Shim et al. fabricated a collagen scaffold using photolithography on which cultured Caco-2 cells formed intestinal structures resembling human intestinal villi (Shim et al., 2017). The combination of perfusion flow and three-dimensional morphology improved metabolic activity and influenced drug absorption, which were largely attributed to a larger absorptive surface and alterations in the expression of tight junctions and transporter proteins (Shim et al., 2017). The importance of cell-matrix interactions and spatial microstructure has also been noted with similar villus scaffolds made from other materials such as poly-lactic-glycolic acid (PLGA) (Costello et al., 2017; Wang et al., 2017).
Normal gut function also involves the involuntary contraction of smooth muscle underlying the mucosa and submucosa. This process, called peristalsis, has been recently incorporated into gut-on-chip models by attaching the central porous membrane to vacuum chambers (Figure 2B). Compared to static Transwell cultures, application of cyclic strain to cause peristaltic motion of the porous membrane enhanced Caco-2 barrier formation and absorptive functions (Jing et al., 2020). Kim et al. also found that cyclic strain increased the expression of proteins in lipid and carbohydrate metabolism, as well as enzyme activity related to intestinal differentiation (Kim et al., 2012). More advanced gut-on-chip models implement a combination of luminal flow and peristalsis-like movement, supporting the formation of villus architecture and coculture with microbiota and immune cells.
IBD and celiac disease are characterized by chronic inflammation affecting various parts of the GI tract. Existing gut-on-chip models utilize a variety of cell sources and methods of inducing inflammation. In most cases, a healthy model of the gut epithelium is first established, consisting of a polarized monolayer of IECs with tight junction formation. Then, to mimic the loss of barrier integrity, different combinations of inflammatory cytokines or endotoxins are introduced to the microchannels of the device. Several cytokine superfamilies have been identified as playing a key role in IBD pathogenesis, including interleukin (IL), tumor necrosis factor (TNF), and interferon (IFN) families (Neurath, 2014). During IBD-related inflammation, IECs produce and are exposed to these factors which have profound effects on tight junction formation, apoptotic activity, and mucosal healing (Andrews et al., 2018; Friedrich et al., 2019).
By culturing Caco-2 cells in a microenvironment with perfusion flow and peristalsis-like movement, Kim et al. were able to induce the spontaneous formation of in vivo-like intestinal villi with highly polarized cells connected by tight junctions (Kim et al., 2016). Introduction of lipopolysaccharide (LPS) or a combination of nonpathogenic bacteria and immune cells to the Caco-2 villi led to increased production of TNF-α, IL-1β, IL-6, and IL-8 by epithelial cells (Kim et al., 2016). Subsequently, as long as IL-8 was also present, each of these proinflammatory cytokines alone was able to elicit the destruction and shortening of villi and decreased barrier function that is characteristic for IBD patients (Kim et al., 2016). In a separate study with similar chip design by Min et al., proinflammatory cytokines TNF-α and IL-1β successfully impaired the Caco-2 barrier and led to intestinal inflammation (Min et al., 2022). Similarly, a combination of IL-1β, TNF-α and IFN-γ successfully triggered increased cell activation and loss of barrier integrity in Caco-2 tubules formed in the OrganoPlate platform, as supported by increased cytokine release by IECS and a drop in transepithelial electrical resistance (TEER) values (Beaurivage et al., 2019). In all three studies, as wells as a study by Liu et al., anti-inflammatory compounds or probiotic therapies were able to suppress the IEC destruction (Liu et al., 2023).
Some gut-on-chip models have also incorporated other intestinal cell types alongside Caco-2 cells to account for in vivo IEC diversity. In the OrganoPlate, Caco-2 enterocytes and HT29-MTX-E12 goblet cells were co-cultured in one channel and THP-1 and MUTZ-3 immune cells in the other, with the channels being separated by an ECM gel (Gijzen et al., 2020). The epithelial layer exhibited decreased barrier function and inflammation with the introduction of TNF-α and IL-1β (Gijzen et al., 2020). The inflammatory state was prevented with the application of TPCA-1, an anti-inflammatory drug. Gjorevski et al. used a similar experimental setup to study neutrophil infiltration and neutrophil-mediated epithelial damage after triggering inflammation in the Caco-2 tubule using LPS in the presence of THP-1 immune cells (Gjorevski et al., 2020). The addition of pro-inflammatory mediators resulted in a leaky epithelial barrier and the activation of macrophages, which in turn attracted and allowed the migration of neutrophils through the ECM gel to further promote tissue inflammation and damage (Gjorevski et al., 2020). More recently, Jeon et al. also reported similar results using the same device design further enhanced with microelectrode arrays and endothelial cells cultured on the opposite side of the ECM gel (Jeon et al., 2022).
In an effort to improve the in vivo accuracy of the inflammation model, Beaurivage et al. cultured biopsy-derived human intestinal organoids in the OrganoPlate platform and induced IBD hallmarks using LPS and IFN-γ. As a result, the intestinal epithelium displayed an inflammatory state as well as increased cytokine production, attributed to an overrepresentation of cytokine regulation and bacterial response pathways that are commonly aberrant in IBD patients (Beaurivage et al., 2020). They further assessed the interplay between inflammation and the immune system by incorporating monocyte-derived macrophages that, upon cytokine treatment, differentiated into TNF-α and IL-6 secreting M1 inflammatory macrophages (Beaurivage et al., 2020). Shin et al. also created a gut-on-chip model by culturing intestinal organoids derived from patients with Crohn’s disease and ulcerative colitis, showing that the diseased epithelial pathology and protein expression is retained in vitro (Shin et al., 2020). Although biopsy is a somewhat limited and invasive source of tissue, these results hold the promise of patient-specific disease modeling.
Since the causes and effects of IBD-related inflammation are not solely limited to the intestines, Trapecar et al. developed a gut-liver model for studying the effects of short-chain fatty acids (SCFAs) on intestinal inflammation. Again, patient-specific ulcerative colitis organoids were able to recapitulate the diseased condition and the addition of SCFAs led to enrichment of hepatic metabolic pathways and reduction of inflammatory pathways (Trapecar et al., 2020). Addition of regulatory cells (Treg) and T helper 17 (Th17) immune cells resulted in T cell-mediated inflammation and autoimmune hepatitis (Trapecar et al., 2020). Other multiorgan chips with an intestinal component have explored interactions with additional organs such as the brain and kidney (Lee et al., 2021; Trapecar et al., 2021).
Alternatively, dextran sodium sulfate (DSS) has been adapted from murine studies to induce ulcerative colitis-like pathologies in human gut-on-chip models for testing probiotic therapies (Shin and Kim, 2018). Similar to cytokines and LPS, DSS was capable of decreasing barrier integrity, decreasing villus height, and disrupting the mucus layer of a Caco-2 epithelium without cytotoxic effects (Shin and Kim, 2018). The barrier dysfunction contributed to the onset of inflammation by enhancing inflammatory cytokine production and immune cell recruitment, but if the intestinal barrier remained intact, the negative effects of LPS or nonpathogenic bacteria were suppressed. Probiotics were successful in maintaining the intestinal barrier when administered as pretreatment, but not when administered following DSS (Shin and Kim, 2018).
The advancement of gut-on-chip devices with applicable physiological conditions and genetic and environmental factors enables more accurate modeling of IBD and celiac disease compared with standard cell cultures. However, as of yet, there is no singular, established gut-on-chip device that is used to model IBD and none have been developed to model celiac disease. Adapting current microfluidic intestinal inflammation models for more disease-specific purposes requires careful consideration of relevant technological and pathological aspects. Implementation of these features each come with their own limitations (Table 2).
Current state-of-the-art gut-on-chip models strive to incorporate as many physiological features of the intestines as possible, within the confines of engineering limitations and without hindering usability. As previously mentioned, to accurately recreate the mechanical microenvironment, a gut-on-chip device must be equipped with both luminal flow and peristaltic movement. Technologically, this requires perfusion of culture medium using pumps and the application of cyclic strain. Previous studies have sought to achieve a constant flow rate (30–60 μL/h) that imparted a shear stress of 0.02 dyne/cm2, which has been linked to proper cell organization and mucin production (Lindner et al., 2021). As the cells of the intestinal epithelium do not experience constant shear stress in vivo, dynamic flow rates have also been explored, largely with no additional effects on villus formation or protein expression (Shin et al., 2020; Fois et al., 2021). Nevertheless, dynamic flow rates could be capable of representing the transport of particles through the intestines more accurately. To mimic peristalsis, cyclic strain (5%–10% strain, 0.15–0.2 Hz) has typically been implemented by stretching the membrane under the IEC layer using vacuum suction (Kim et al., 2012). In order to study the complex anaerobic and aerobic microbes that are present in the intestines, a gut-on-chip device must also mimic the low oxygen environment of the intestinal lumen and the comparatively well-oxygenated intestinal tissue. To do so, PDMS gut-on-chip devices, due to their gas permeability, have been placed in anaerobic chambers continuously supplied with CO2 (Jalili-Firoozinezhad et al., 2019; Grant et al., 2022). Alternatively, Shah et al. used polycarbonate to segregate the culture area and perfused anoxic medium to control the oxygen concentration (Shah et al., 2016). Due to the small size of microfluidic devices, precisely quantifying properties such as oxygen concentration, pH, and TEER requires integration of different sensing systems. For example, the barrier function of the intestinal epithelium can be monitored with TEER electrodes, or the amount of dissolved oxygen can be measured using optical sensors (Shah et al., 2016; Henry et al., 2017; Lesher-Pérez et al., 2017; Jalili-Firoozinezhad et al., 2018; Jalili-Firoozinezhad et al., 2019; Soucy et al., 2019; Okkelman et al., 2020).
The structural differences between the small and large intestines, as well as regional differences within each, should also be taken into consideration when creating an in vitro model. To date, most gut inflammation models seek to recreate the general crypt-villus architecture of the small intestine, which disregards the distinct regions of the intestines which can have variable responses to inflammation. Already, specific gut-on-chip devices have been constructed using biopsy samples to represent the duodenum and ileum as well as a colon model using human colonic mucosal tissue (Kasendra et al., 2018; Beaurivage et al., 2020; Kasendra et al., 2020; Sontheimer-Phelps et al., 2020). The use of region-specific gut-on-chip devices should be further expanded to study inflammation and disease progression in applicable areas of the GI tract. Similarly, the cellular components of the epithelium must be accurately represented. Caco-2 cells cultured in microfluidic devices have been reported to spontaneously form structures similar to the small intestine, but genomic analysis has revealed lower expression of physiologically relevant genes compared to gut-on-chip cultures of biopsy-derived organoids (Yin et al., 2020). Biopsy samples and iPSC-derived organoids share the same genetic features and develop similar cellular architecture as intestinal epithelium in vivo but with a closed configuration, making them inaccessible from the apical side. A further development for gut-on-chip models would be to use intestinal organoids to generate the epithelial monolayer, or to differentiate and mature iPSCs towards intestinal epithelial cells in 2D (Negoro et al., 2018; Thorne et al., 2018).
Inflammatory conditions of the intestines involve a combination of immune and microbial factors, the coculture of which has been facilitated by microfluidic devices. The multichannel composition of gut-on-chip devices permits the application of different factors to the apical or basolateral surface of the intestinal epithelium to study their effects. In a homeostatic state, most models contain an intact epithelial barrier which separates microbial populations on the apical (lumen) side and immune components on the basal side. PBMCs, THP-1, Th17, and MUTZ-3 immune cells have already been incorporated into gut-on-chip models, specifically into microfluidic channels adjacent to the intestinal epithelium (Shin et al., 2020; Trapecar et al., 2020; Lee et al., 2021; Jeon et al., 2022). An immunocompetent model is especially important with regard to the development of a celiac disease gut-on-chip, where CD4+ T cells have an important role in creating an inflammatory milieu enabling the development of tissue damage (Christophersen et al., 2019; Lindfors et al., 2019). The intestinal microbiota also has several proposed roles in IBD and celiac disease. For example, dysbiosis of the microbiota could alter the permeability of the intestinal barrier, allowing for the translocation of the luminal contents into the lamina propria and subsequently triggering immune responses (Levy et al., 2017). Since the exact role and interplay between immune cells, the intestinal microbiota, and other cell types are largely unknown, further study requires their integration into future gut-on-chip disease models. Unlike most conventional intestinal models, organ-on-chip technology allows for evaluating the relative contributions of the different factors to inflammatory diseases.
Along with immune cells, the lamina propria also contains the intestinal blood supply and lymphatic vasculature. Blood and lymphatic vessels are mainly lined by endothelial cells that can be maladaptive in diseased conditions, playing a role in areas such as cell infiltration, cytokine production, and immunological reactivity (Myrsky et al., 2009; Cromer et al., 2011). In existing two-channel gut-on-chip models it is relatively common to include HUVECs or HIMECs with the result that they form an endothelial monolayer adjacent to the intestinal epithelium. Other cells that are thought to play a role in the progression of mucosal barrier destruction are intraepithelial lymphocytes (IELs) that represent a population of T cells interspersed between IECs. Generally, IELs participate in the protection of the intestinal epithelial layer. However, in celiac disease, these cells are found to be cytotoxic in nature and capable of inducing IEC apoptosis upon the onset of an immune response (Lindfors et al., 2019). The central role of IELs in promoting atrophy of small intestinal villi conveys the need for their representation in an accurate in vitro celiac disease model.
In terms of pathogenesis, ulcerative colitis and Crohn’s disease have different inflammatory mediator profiles, the former being driven by infiltration of Th2 cell-associated cytokines and the latter by Th1 cell-associated cytokines (Nemeth et al., 2017). Some overlap also exists, as both forms of IBD show increased expression of factors such as Th17 cytokine IL-17 and TNF (Atreya et al., 2011; Monteleone et al., 2011). The existence of specific cytokine networks that lead to mucosal inflammation is further supported by the key cytokines linked to the perpetuation of celiac disease, including IFN-γ, IL-15, and IL-21 (Garrote et al., 2008). Thus, especially when using Caco-2 cells, the right combination of triggers is necessary to result in gut-on-chip inflammation that has both structural changes and levels of cytokine release comparable to the specific disease. Utilizing a cell source that retains the disease pathology without the use of an external stimulant could be a way to surpass this challenge.
It is also important to consider whether disease severity can be manipulated within gut-on-chip models, as different forms of IBD and celiac disease present differently in individual patients. Beaurivage et al. compared rapid and prolonged exposure of inflammatory factors to Caco-2 cells, finding that there was no significant effect on cytokine production, TEER values, or E-cadherin localization (Beaurivage et al., 2019). Though there was no change in the severity of the inflammatory state, it was proposed that further optimizing the concentration of cytokines could more accurately depict different disease phenotypes.
Finally, there are currently no standardized methods to trigger inflammation in gut-on-chip models. Delivering inflammatory modulators to apical, basal, or both sides of the intestinal epithelium has resulted in the secretion of different concentrations of pro-inflammatory cytokines. In a study using LPS and IFN-γ as triggers, there was no apparent difference in cytokine levels regardless of whether LPS was applied to the apical or basal compartment, as long as IFN-γ was only applied to the basal side (Beaurivage et al., 2020). When LPS and IFN-γ were simultaneously applied to both sides, there were fluctuations in the secretion of multiple cytokines both apically and basally. In this case, the conclusion was to apply the inflammatory trigger to both sides to promote the complete activation and differentiation of macrophages, but many other studies apply triggers to only one side of the epithelium (Beaurivage et al., 2020). No clear answer exists to how inflammation should be triggered, but the results from different approaches illuminate how even slight variability can have an impact on the overall accuracy of a disease model.
This review outlines the general construction of gut-on-chip devices as well as how they have been adapted to model intestinal inflammation. An ideal device should provide an intact epithelial barrier with mucus production, peristaltic motion, luminal flow, and interactions between IECs and immune and microbial components. Many existing models have been able to incorporate a combination of these features, but limitations still exist as the field is fairly new. Whether these models can be further adapted for modeling multifactorial diseases such as IBD and celiac disease relies on careful consideration of aspects such as inflammatory mediator profiles, cell type, tissue architecture and the application of microbial and immune factors in an in vivo-like manner. Gut-on-chip models are progressively getting closer to mimicking the intestines in both architecture and physiology, providing more opportunities to create accurate models to study complex inflammatory intestinal conditions. While gaining new insights into the pathogenesis and treatment of these diseases can be achieved with a gut-on-chip device, complete modeling most likely requires the integration of multiple different approaches. All in all, organ-on-chip technology provides an excellent platform for accurately modeling intestinal inflammation, with the promise of further development into IBD and celiac disease models.
ST: Writing–original draft, Writing–review and editing. KJ-U: Writing–review and editing. KK: Writing–review and editing. KL: Writing–review and editing. PK: Writing–review and editing. MK: Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Finland-China Network in Food and Health as a pilot of the global program for research and innovation funded by the Ministry of Education and Culture of Finland, the Mary and Georg C. Ehrnrooth Foundation (KJ-U), the Finnish Cultural Foundation (KJ-U), the Paivikki and Sakari Sohlberg Foundation (KJ-U), and the Diabetes Research Foundation (KJ-U).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alam, M. A., Al-Jenoobi, F. I., and Al-mohizea, A. M. (2012). Everted gut sac model as a tool in pharmaceutical research: limitations and applications. J. Pharm. Pharmacol. 64 (3), 326–336. doi:10.1111/j.2042-7158.2011.01391.x
Altay, G., Larrañaga, E., Tosi, S., Barriga, F. M., Batlle, E., Fernández-Majada, V., et al. (2019). Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci. Rep. 9 (1), 10140–10214. doi:10.1038/s41598-019-46497-x
Andrews, C., McLean, M. H., and Durum, S. K. (2018). Cytokine tuning of intestinal epithelial function. Front. Immunol. 9, 1270. doi:10.3389/fimmu.2018.01270
Atreya, R., Zimmer, M., Bartsch, B., Waldner, M. J., Atreya, I., Neumann, H., et al. (2011). Antibodies against tumor necrosis factor (TNF) induce T-cell apoptosis in patients with inflammatory bowel diseases via TNF receptor 2 and intestinal CD14+ macrophages. Gastroenterol. (New York, N.Y. 1943) 141 (6), 2026–2038. doi:10.1053/j.gastro.2011.08.032
Beaurivage, C., Kanapeckaite, A., Loomans, C., Erdmann, K. S., Stallen, J., and Janssen, R. A. J. (2020). Development of a human primary gut-on-a-chip to model inflammatory processes. Sci. Rep. 10 (1), 21475. doi:10.1038/s41598-020-78359-2
Beaurivage, C., Naumovska, E., Chang, Y., Elstak, E., Nicolas, A., Wouters, H., et al. (2019). Development of a gut-on-A-chip model for high throughput disease modeling and drug discovery. Int. J. Mol. Sci. 20 (22), 5661. doi:10.3390/ijms20225661
Beumer, J., Puschhof, J., Bauzá-Martinez, J., Martínez-Silgado, A., Elmentaite, R., James, K. R., et al. (2020). High-Resolution mRNA and secretome atlas of human enteroendocrine cells. Cell 181 (6), 1291–1306. doi:10.1016/j.cell.2020.04.036
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9 (5), 356–368. doi:10.1038/nrmicro2546
Bourgine, J., Billaut-Laden, I., Happillon, M., Lo-Guidice, J. M., Maunoury, V., Imbenotte, M., et al. (2012). Gene expression profiling of systems involved in the metabolism and the disposition of xenobiotics: comparison between human intestinal biopsy samples and colon cell lines. Drug Metabolism Dispos. 40 (4), 694–705. doi:10.1124/dmd.111.042465
Braverman, J., and Yilmaz, Ö. H. (2018). From 3D organoids back to 2D enteroids. Dev. Cell 44 (5), 533–534. doi:10.1016/j.devcel.2018.02.016
Castaño, A. G., García-Díaz, M., Torras, N., Altay, G., Comelles, J., and Martínez, E. (2019). Dynamic photopolymerization produces complex microstructures on hydrogels in a moldless approach to generate a 3D intestinal tissue model. Biofabrication 11 (2), 025007. doi:10.1088/1758-5090/ab0478
Choe, A., Ha, S. K., Choi, I., Choi, N., and Sung, J. H. (2017). Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed. Microdevices 19 (1), 4–11. doi:10.1007/s10544-016-0143-2
Christophersen, A., Lund, E. G., Snir, O., Solà, E., Kanduri, C., Dahal-Koirala, S., et al. (2019). Distinct phenotype of CD4 + T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med. 25 (5), 734–737. doi:10.1038/s41591-019-0403-9
Clevers, H. (2013). The intestinal crypt, A prototype stem cell compartment. Cell 154 (2), 274–284. doi:10.1016/j.cell.2013.07.004
Clevers, H. C., and Bevins, C. L. (2013). Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiology 75 (1), 289–311. doi:10.1146/annurev-physiol-030212-183744
Costello, C. M., Phillipsen, M. B., Hartmanis, L. M., Kwasnica, M. A., Chen, V., Hackam, D., et al. (2017). Microscale Bioreactors for in situ characterization of GI epithelial cell physiology. Sci. Rep. 7 (1), 12515–12610. doi:10.1038/s41598-017-12984-2
Creff, J., Courson, R., Mangeat, T., Foncy, J., Souleille, S., Thibault, C., et al. (2019). Fabrication of 3D scaffolds reproducing intestinal epithelium topography by high-resolution 3D stereolithography. Biomaterials 221, 119404. doi:10.1016/j.biomaterials.2019.119404
Cromer, W. E., Mathis, J. M., Granger, D. N., Chaitanya, G., and Alexander, J. S. (2011). Role of the endothelium in inflammatory bowel diseases. World J. Gastroenterology 17 (4), 578–593. doi:10.3748/wjg.v17.i5.578
Darling, N. J., Mobbs, C. L., González-Hau, A. L., Freer, M., and Przyborski, S. (2020). Bioengineering novel in vitro Co-culture models that represent the human intestinal mucosa with improved caco-2 structure and barrier function. Front. Bioeng. Biotechnol. 8, 992. doi:10.3389/fbioe.2020.00992
de Souza, H. S. P., and Fiocchi, C. (2016). Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterology Hepatology 13 (5), 13–27. doi:10.1038/nrgastro.2015.186
Dignass, A. U., and Sturm, A. (2001). Peptide growth factors in the intestine. Eur. J. Gastroenterology Hepatology 13 (7), 763–770. doi:10.1097/00042737-200107000-00002
Edington, C. D., Chen, W. L. K., Geishecker, E., Kassis, T., Soenksen, L. R., Bhushan, B. M., et al. (2018). Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 8 (1), 4530–4618. doi:10.1038/s41598-018-22749-0
Elmentaite, R., Kumasaka, N., Roberts, K., Fleming, A., Dann, E., King, H. W., et al. (2021). Cells of the human intestinal tract mapped across space and time. Nat. Lond. 597 (7875), 250–255. doi:10.1038/s41586-021-03852-1
Fois, C. A. M., Schindeler, A., Valtchev, P., and Dehghani, F. (2021). Dynamic flow and shear stress as key parameters for intestinal cells morphology and polarization in an organ-on- a-chip model. Biomed. Microdevices 23 (4), 55. doi:10.1007/s10544-021-00591-y
Friedrich, M., Pohin, M., and Powrie, F. (2019). Cytokine networks in the pathophysiology of inflammatory bowel disease. Immun. Camb. Mass.) 50 (4), 992–1006. doi:10.1016/j.immuni.2019.03.017
Gao, D., Liu, H., Lin, J. M., Wang, Y., and Jiang, Y. (2013). Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab a Chip 13 (5), 978–985. doi:10.1039/c2lc41215b
Garrote, J. A., Gómez-González, E., Bernardo, D., Arranz, E., and Chirdo, F. (2008). Celiac disease pathogenesis: the proinflammatory cytokine network. J. Pediatr. Gastroenterology Nutr. 47 (Suppl. 1), S27–S32. doi:10.1097/mpg.0b013e3181818fb9
Gijzen, L., Marescotti, D., Raineri, E., Nicolas, A., Lanz, H. L., Guerrera, D., et al. (2020). An intestine-on-a-chip model of plug-and-play modularity to study inflammatory processes. SLAS Technol. 25 (6), 585–597. doi:10.1177/2472630320924999
Gjorevski, N., Avignon, B., Gérard, R., Cabon, L., Roth, A. B., Bscheider, M., et al. (2020). Neutrophilic infiltration in organ-on-a-chip model of tissue inflammation. Lab a Chip 20 (18), 3365–3374. doi:10.1039/d0lc00417k
Gómez, D. P., and Boudreau, F. (2021). Organoids and their use in modeling gut epithelial cell lineage differentiation and barrier properties during intestinal diseases. Front. Cell Dev. Biol. 9, 732137. doi:10.3389/fcell.2021.732137
Grant, J., Lee, E., Almeida, M., Kim, S., LoGrande, N., Goyal, G., et al. (2022). Establishment of physiologically relevant oxygen gradients in microfluidic organ chips. Lab a Chip 22 (8), 1584–1593. doi:10.1039/d2lc00069e
Gribble, F. M., and Reimann, F. (2019). Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 15 (4), 226–237. doi:10.1038/s41574-019-0168-8
Guan, Q. (2019). A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J. Immunol. Res. 2019, 1–16. doi:10.1155/2019/7247238
Guo, Y., Li, Z., Su, W., Wang, L., Zhu, Y., and Qin, J. (2018). A biomimetic human gut-on-a-chip for modeling drug metabolism in intestine. Artif. Organs 42 (12), 1196–1205. doi:10.1111/aor.13163
Hausch, F., Shan, L., Santiago, N. A., Gray, G. M., and Khosla, C. (2002). Intestinal digestive resistance of immunodominant gliadin peptides. Am. J. Physiology -Gastrointestinal Liver Physiology 283 (4), 996–1003. doi:10.1152/ajpgi.00136.2002
Henry, O. Y. F., Villenave, R., Cronce, M. J., Leineweber, W. D., Benz, M. A., and Ingber, D. E. (2017). Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function. Lab a Chip 17 (13), 2264–2271. doi:10.1039/c7lc00155j
Hillman, E. T., Lu, H., Yao, T., and Nakatsu, C. H. (2017). Microbial ecology along the gastrointestinal tract. Microbes Environ. 32 (4), 300–313. doi:10.1264/jsme2.me17017
Hoffmann, P., Burmester, M., Langeheine, M., Brehm, R., Empl, M. T., Seeger, B., et al. (2021). Caco-2/HT29-MTX co-cultured cells as a model for studying physiological properties and toxin-induced effects on intestinal cells. PLoS One 16, e0257824. doi:10.1371/journal.pone.0257824
Holmes, R., Hourihane, D. O., and Booth, C. C. (1961). The mucosa of the small intestine. Postgrad. Med. J. 37, 717–724. doi:10.1136/pgmj.37.434.717
Huang, H., Fang, M., Jostins, L., Umićević Mirkov, M., Boucher, G., Anderson, C. A., et al. (2017). Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nat. Lond. 547 (7662), 173–178. doi:10.1038/nature22969
Imura, Y., Asano, Y., Sato, K., and Yoshimura, E. (2009). A microfluidic system to evaluate intestinal absorption. Anal. Sci. 25 (12), 1403–1407. doi:10.2116/analsci.25.1403
Jalili-Firoozinezhad, S., Gazzaniga, F. S., Calamari, E. L., Camacho, D. M., Fadel, C. W., Bein, A., et al. (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3 (7), 520–531. doi:10.1038/s41551-019-0397-0
Jalili-Firoozinezhad, S., Prantil-Baun, R., Jiang, A., Potla, R., Mammoto, T., Weaver, J. C., et al. (2018). Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 9 (2), 223–314. doi:10.1038/s41419-018-0304-8
Jeon, J., Lee, S. H., Kim, D., and Sung, J. H. (2021). In vitro hepatic steatosis model based on gut– liver-on a chip. Biotechnol. Prog. 37 (e), e3121. doi:10.1002/btpr.3121
Jeon, M. S., Choi, Y. Y., Mo, S. J., Ha, J. H., Lee, Y. S., Lee, H. U., et al. (2022). Contributions of the microbiome to intestinal inflammation in a gut-on- a chip. Nano Converg. 9 (1), 8. doi:10.1186/s40580-022-00299-6
Jiminez, J. A., Uwiera, T. C., Douglas Inglis, G., and Uwiera, R. R. E. (2015). Animal models to study acute and chronic intestinal inflammation in mammals. Gut Pathog. 7 (29), 29. doi:10.1186/s13099-015-0076-y
Jing, B., Wang, Z. A., Zhang, C., Deng, Q., Wei, J., Luo, Y., et al. (2020). Establishment and application of peristaltic human gut-vessel microsystem for studying host-microbial interaction. Front. Bioeng. Biotechnol. 8, 272. doi:10.3389/fbioe.2020.00272
Kamada, N., Seo, S. U., Chen, G. Y., and Núñez, G. (2013). Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13 (5), 321–335. doi:10.1038/nri3430
Kasendra, M., Luc, R., Yin, J., Manatakis, D. V., Kulkarni, G., Lucchesi, C., et al. (2020). Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. eLife 9, e50135. doi:10.7554/elife.50135
Kasendra, M., Tovaglieri, A., Sontheimer-Phelps, A., Jalili-Firoozinezhad, S., Bein, A., Chalkiadaki, A., et al. (2018). Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 8 (1), 2871–2914. doi:10.1038/s41598-018-21201-7
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab a Chip 12 (12), 2165–2174. doi:10.1039/c2lc40074j
Kim, H. J., and Ingber, D. E. (2013). Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. Camb. 5 (9), 1130–1140. doi:10.1039/c3ib40126j
Kim, H. J., Li, H., Collins, J. J., and Ingber, D. E. (2016). Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. U. S. A. 113 (1), E7–E15. doi:10.1073/pnas.1522193112
Kim, J. J., and Khan, W. I. (2013). Goblet cells and mucins: role in innate defense in enteric infections. Pathogens 2 (1), 55–70. doi:10.3390/pathogens2010055
Kim, M. H., Kim, D., and Sung, J. H. (2021). A Gut-Brain Axis-on-a-Chip for studying transport across epithelial and endothelial barriers. J. Industrial Eng. Chem. 101 (0), 126–134. doi:10.1016/j.jiec.2021.06.021
Kim, W., and Kim, G. (2018). Intestinal villi model with blood capillaries fabricated using collagen-based bioink and dual-cell-printing process. ACS Appl. Mater. Interfaces 10 (48), 41185–41196. doi:10.1021/acsami.8b17410
Kim, W., and Kim, G. H. (2020). An intestinal model with a finger-like villus structure fabricated using a bioprinting process and collagen/SIS-based cell-laden bioink. Theranostics 10 (6), 2495–2508. doi:10.7150/thno.41225
Krishnareddy, S. (2019). The microbiome in celiac disease. Gastroenterology Clin. N. Am. 48 (1), 115–126. doi:10.1016/j.gtc.2018.09.008
Lee, S. H., Kwon, J. E., and Cho, M. L. (2018). Immunological pathogenesis of inflammatory bowel disease. Intestinal Res. 16 (1), 26–42. doi:10.5217/ir.2018.16.1.26
Lee, Y., Kim, M. H., Alves, D. R., Kim, S., Lee, L. P., Sung, J. H., et al. (2021). Gut-kidney Axis on chip for studying effects of antibiotics on risk of hemolytic uremic syndrome by shiga toxin-producing Escherichia coli. Toxins (Basel) 13 (11), 775. doi:10.3390/toxins13110775
Lesher-Pérez, S. C., Kim, G. A., Kuo, C., Leung, B. M., Mong, S., Kojima, T., et al. (2017). Dispersible oxygen microsensors map oxygen gradients in three-dimensional cell cultures. Biomaterials Sci. 5 (10), 2106–2113. doi:10.1039/c7bm00119c
Levy, M., Kolodziejczyk, A. A., Thaiss, C. A., and Elinav, E. (2017). Dysbiosis and the immune system. Nat. Rev. Immunol. 17 (4), 219–232. doi:10.1038/nri.2017.7
Lindfors, K., Ciacci, C., Kurppa, K., Lundin, K. E. A., Makharia, G. K., Mearin, M. L., et al. (2019). Coeliac disease. Nat. Rev. Dis. Prim. 5 (1), 3. doi:10.1038/s41572-018-0054-z
Lindner, M., Laporte, A., Block, S., Elomaa, L., and Weinhart, M. (2021). Physiological shear stress enhances differentiation, mucus-formation and structural 3D organization of intestinal epithelial cells in vitro. Cells (Basel, Switz. 10 (8), 2062. doi:10.3390/cells10082062
Liu, J., Lu, R., Zheng, X., Hou, W., Wu, X., Zhao, H., et al. (2023). Establishment of a gut-on-a-chip device with controllable oxygen gradients to study the contribution of Bifidobacterium bifidum to inflammatory bowel disease. Biomaterials Sci. 11 (7), 2504–2517. doi:10.1039/d2bm01490d
Liu, J. Z., van Sommeren, S., Huang, H., Ng, S. C., Alberts, R., Takahashi, A., et al. (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47 (9), 979–986. doi:10.1038/ng.3359
Maurer, M., Gresnigt, M. S., Last, A., Wollny, T., Berlinghof, F., Pospich, R., et al. (2019). A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 220, 119396. doi:10.1016/j.biomaterials.2019.119396
McHardy, I. H., Goudarzi, M., Tong, M., Ruegger, P. M., Schwager, E., Weger, J. R., et al. (2013). Integrative analysis of the microbiome and metabolome of the human intestinal mucosal surface reveals exquisite inter-relationships. Microbiome 1 (1), 17. doi:10.1186/2049-2618-1-17
Min, S., Than, N., Shin, Y. C., Hu, G., Shin, W., Ambrosini, Y. M., et al. (2022). Live probiotic bacteria administered in a pathomimetic leaky gut chip ameliorate impaired epithelial barrier and mucosal inflammation. Sci. Rep. 12 (1), 22641. doi:10.1038/s41598-022-27300-w
Mizoguchi, A., Takeuchi, T., Himuro, H., Okada, T., and Mizoguchi, E. (2016). Genetically engineered mouse models for studying inflammatory bowel disease. J. Pathology 238 (2), 205–219. doi:10.1002/path.4640
Monteleone, I., Pallone, F., and Monteleone, G. (2011). Th17-related cytokines: new players in the control of chronic intestinal inflammation. BMC Med. 9 (1), 122. doi:10.1186/1741-7015-9-122
Myrsky, E., Caja, S., Simon-Vecsei, Z., Korponay-Szabo, I. R., Nadalutti, C., Collighan, R., et al. (2009). Celiac disease IgA modulates vascular permeability in vitro through the activity of transglutaminase 2 and RhoA. Cell. Mol. Life Sci. 66 (20), 3375–3385. doi:10.1007/s00018-009-0116-1
Naumovska, E., Aalderink, G., Wong Valencia, C., Kosim, K., Nicolas, A., Brown, S., et al. (2020). Direct on-chip differentiation of intestinal tubules from induced pluripotent stem cells. Int. J. Mol. Sci. 21 (14), 4964. doi:10.3390/ijms21144964
Negoro, R., Takayama, K., Kawai, K., Harada, K., Sakurai, F., Hirata, K., et al. (2018). Efficient generation of small intestinal epithelial-like cells from human iPSCs for drug absorption and metabolism studies. Stem Cell Rep. 11 (6), 1539–1550. doi:10.1016/j.stemcr.2018.10.019
Nemeth, Z. H., Bogdanovski, D. A., Barratt-Stopper, P., Paglinco, S. R., Antonioli, L., and Rolandelli, R. H. (2017). Crohn’s disease and ulcerative colitis show unique cytokine profiles. Cureus (Palo Alto, CA) 9 (4), e1177. doi:10.7759/cureus.1177
Neurath, M. F. (2014). Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 14 (5), 329–342. doi:10.1038/nri3661
Okkelman, I. A., Neto, N., Papkovsky, D. B., Monaghan, M. G., and Dmitriev, R. I. (2020). A deeper understanding of intestinal organoid metabolism revealed by combining fluorescence lifetime imaging microscopy (FLIM) and extracellular flux analyses. Redox Biol. 30, 101420. doi:10.1016/j.redox.2019.101420
Parker, A., Vaux, L., Patterson, A. M., Modasia, A., Muraro, D., Fletcher, A. G., et al. (2019). Elevated apoptosis impairs epithelial cell turnover and shortens villi in TNF driven intestinal inflammation. Cell Death Dis. 10 (2), 108. doi:10.1038/s41419-018-1275-5
Pocock, K., Delon, L., Bala, V., Rao, S., Priest, C., Prestidge, C., et al. (2017). Intestine-on-a-Chip microfluidic model for efficient in vitro screening of oral chemotherapeutic uptake. ACS Biomaterials Sci. Eng. 3 (6), 951–959. doi:10.1021/acsbiomaterials.7b00023
Prattis, S., and Jurjus, A. (2015). Spontaneous and transgenic rodent models of inflammatory bowel disease. Laboratory Animal Res. 31 (2), 47–68. doi:10.5625/lar.2015.31.2.47
Ramadan, Q., and Jing, L. (2016). Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed. Microdevices 18 (1), 11–19. doi:10.1007/s10544-016-0035-5
Rescigno, M. (2011). The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol. 32 (6), 256–264. doi:10.1016/j.it.2011.04.003
Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G., Gasbarrini, A., et al. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorg. (Basel) 7 (1), 14. doi:10.3390/microorganisms7010014
Rodríguez, J. M., Murphy, K., Stanton, C., Ross, R. P., Kober, O. I., Juge, N., et al. (2015). The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health and Dis. 26 (1), 1–26050. doi:10.3402/mehd.v26.26050
Sato, T., and Clevers, H. (2013). Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Sci. Am. Assoc. Adv. Sci. 340 (6137), 1190–1194. doi:10.1126/science.1234852
Shah, P., Fritz, J. V., Glaab, E., Desai, M. S., Greenhalgh, K., Frachet, A., et al. (2016). A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 7 (1), 11535. doi:10.1038/ncomms11535
Shim, K. Y., Lee, D., Han, J., Nguyen, N. T., Park, S., and Sung, J. H. (2017). Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed. Microdevices 19 (2), 37–10. doi:10.1007/s10544-017-0179-y
Shin, W., and Kim, H. J. (2018). Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. U. S. A. 115 (45), E10539–E10547. doi:10.1073/pnas.1810819115
Shin, W., Wu, A., Massidda, M. W., Foster, C., Thomas, N., Lee, D. W., et al. (2019). A robust longitudinal Co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front. Bioeng. Biotechnol. 7, 13. doi:10.3389/fbioe.2019.00013
Shin, Y. C., Shin, W., Koh, D., Wu, A., Ambrosini, Y. M., Min, S., et al. (2020). Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines (Basel) 11 (7), 663. doi:10.3390/mi11070663
Sontheimer-Phelps, A., Chou, D. B., Tovaglieri, A., Ferrante, T. C., Duckworth, T., Fadel, C., et al. (2020). Human colon-on-a-chip enables continuous in vitro analysis of colon mucus layer accumulation and physiology. Cell. Mol. Gastroenterology Hepatology 9 (3), 507–526. doi:10.1016/j.jcmgh.2019.11.008
Soucy, J. R., Bindas, A. J., Koppes, A. N., and Koppes, R. A. (2019). Instrumented microphysiological systems for real-time measurement and manipulation of cellular electrochemical processes. iScience 21, 521–548. doi:10.1016/j.isci.2019.10.052
Specian, R. D., and Oliver, M. G. (1991). Functional biology of intestinal goblet cells. Am. J. Physiology - Cell Physiology 260 (2), 183–193. doi:10.1152/ajpcell.1991.260.2.c183
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nat. Lond. 470 (7332), 105–109. doi:10.1038/nature09691
Strober, W., Fuss, I., and Mannon, P. (2007). The fundamental basis of inflammatory bowel disease. J. Clin. Investigation 117 (3), 514–521. doi:10.1172/jci30587
Sun, H., Chow, E. C., Liu, S., Du, Y., and Pang, K. S. (2008). The Caco-2 cell monolayer: usefulness and limitations. Expert Opin. Drug Metabolism Toxicol. 4 (4), 395–411. doi:10.1517/17425255.4.4.395
Sung, J. H., Yu, J., Luo, D., Shuler, M. L., and March, J. C. (2011). Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab. Chip 11 (3), 389–392. doi:10.1039/c0lc00273a
Suzuki, T. (2012). Regulation of intestinal epithelial permeability by tight junctions. Cell. Mol. Life Sci. 70 (4), 631–659. doi:10.1007/s00018-012-1070-x
Tanaka, M., Saito, H., Kusumi, T., Fukuda, S., Shimoyama, T., Sasaki, Y., et al. (2001). Spatial distribution and histogenesis of colorectal Paneth cell metaplasia in idiopathic inflammatory bowel disease. J. Gastroenterology Hepatology 16 (12), 1353–1359. doi:10.1046/j.1440-1746.2001.02629.x
Thomson, A., Smart, K., Somerville, M. S., Lauder, S. N., Appanna, G., Horwood, J., et al. (2019). The Ussing chamber system for measuring intestinal permeability in health and disease. BMC Gastroenterol. 19 (1), 98. doi:10.1186/s12876-019-1002-4
Thorne, C. A., Chen, I. W., Sanman, L. E., Cobb, M. H., Wu, L. F., and Altschuler, S. J. (2018). Enteroid monolayers reveal an autonomous WNT and BMP circuit controlling intestinal epithelial growth and organization. Dev. Cell 44 (5), 624–633.e4. doi:10.1016/j.devcel.2018.01.024
Trapecar, M., Communal, C., Velazquez, J., Maass, C. A., Huang, Y. J., Schneider, K., et al. (2020). Gut-liver physiomimetics reveal paradoxical modulation of IBD- related inflammation by short-chain fatty acids. Cell Syst. 10 (3), 223–239.e9. doi:10.1016/j.cels.2020.02.008
Trapecar, M., Wogram, E., Svoboda, D., Communal, C., Omer, A., Lungjangwa, T., et al. (2021). Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci. Adv. 7 (5), eabd1707. doi:10.1126/sciadv.abd1707
Trietsch, S. J., Naumovska, E., Kurek, D., Setyawati, M. C., Vormann, M. K., Wilschut, K. J., et al. (2017). Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 8 (1), 262–268. doi:10.1038/s41467-017-00259-3
Verhulsel, M., Simon, A., Bernheim-Dennery, M., Gannavarapu, V. R., Gérémie, L., Ferraro, D., et al. (2021). Developing an advanced gut on chip model enabling the study of epithelial cell/fibroblast interactions. Lab a Chip 21 (2), 365–377. doi:10.1039/d0lc00672f
Villenave, R., Wales, S. Q., Hamkins-Indik, T., Papafragkou, E., Weaver, J. C., Ferrante, T. C., et al. (2017). Human gut-on-A-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One 12 (2), e0169412. doi:10.1371/journal.pone.0169412
Wang, Y., Gunasekara, D. B., Reed, M. I., DiSalvo, M., Bultman, S. J., Sims, C. E., et al. (2017). A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55. doi:10.1016/j.biomaterials.2017.03.005
Wirtz, S., Popp, V., Kindermann, M., Gerlach, K., Weigmann, B., Fichtner-Feigl, S., et al. (2017). Chemically induced mouse models of acute and chronic intestinal inflammation. Nat. Protoc. 12 (7), 1295–1309. doi:10.1038/nprot.2017.044
Workman, M. J., Gleeson, J. P., Troisi, E. J., Estrada, H. Q., Kerns, S. J., Hinojosa, C. D., et al. (2018). Enhanced utilization of induced pluripotent stem cell–derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterology Hepatology 5 (4), 669–677.e2. doi:10.1016/j.jcmgh.2017.12.008
Wu, Q., Liu, J., Wang, X., Feng, L., Wu, J., Zhu, X., et al. (2020). Organ-on-a-chip: recent breakthroughs and future prospects. Biomed. Eng. Online 19 (1), 9. doi:10.1186/s12938-020-0752-0
Yamaura, Y., Chapron, B. D., Wang, Z., Himmelfarb, J., and Thummel, K. E. (2016). Functional comparison of human colonic carcinoma cell lines and primary small intestinal epithelial cells for investigations of intestinal drug permeability and first-pass metabolism. Drug Metabolism Dispos. 44 (3), 329–335. doi:10.1124/dmd.115.068429
Yin, S., Ray, G., Kerschner, J. L., Hao, S., Perez, A., Drumm, M. L., et al. (2020). Functional genomics analysis of human colon organoids identifies key transcription factors. Physiol. Genomics 52 (6), 234–244. doi:10.1152/physiolgenomics.00113.2019
Yoshida, S., Miwa, H., Kawachi, T., Kume, S., and Takahashi, K. (2020). Generation of intestinal organoids derived from human pluripotent stem cells for drug testing. Sci. Rep. 10 (1), 5989. doi:10.1038/s41598-020-63151-z
Zeissig, S., Burgel, N., Gunzel, D., Richter, J., Mankertz, J., Wahnschaffe, U., et al. (2007). Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 56 (1), 61–72. doi:10.1136/gut.2006.094375
Keywords: gut-on-a-chip, celiac disease, IBD, tissue engineering, intestinal epithelium
Citation: Taavitsainen S, Juuti-Uusitalo K, Kurppa K, Lindfors K, Kallio P and Kellomäki M (2024) Gut-on-chip devices as intestinal inflammation models and their future for studying multifactorial diseases. Front. Lab. Chip. Technol. 2:1337945. doi: 10.3389/frlct.2023.1337945
Received: 13 November 2023; Accepted: 27 December 2023;
Published: 11 January 2024.
Edited by:
Lydia Sohn, University of California, Berkeley, United StatesReviewed by:
Michael Super, Harvard University, United StatesCopyright © 2024 Taavitsainen, Juuti-Uusitalo, Kurppa, Lindfors, Kallio and Kellomäki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Taavitsainen, c3VzYW5uZS50YWF2aXRzYWluZW5AdHVuaS5maQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.