- 1Department of Biochemistry and Molecular Biology, Miller School of Medicine, University of Miami, Miami, FL, United States
- 2Sylvester Comprehensive Cancer Center, Miller School of Medicine, University of Miami, Miami, FL, United States
The formation, inheritance, and removal of DNA methylation in the genome of mammalian cells is directly regulated by two families of enzymes–DNA methyltransferases (DNMTs) and Ten-Eleven Translocation proteins (TETs). DNMTs generate and maintain the inheritance of 5-methylcytosine (5mC), which is the substrate targeted by the TET enzymes for conversion to 5-hydroxymethylcytosine (5hmC) and its downstream oxidized derivatives. The activity of DNMT and TET is dependent on the availability of micronutrients and metabolite co-factors, including essential vitamins, amino acids, and trace metals, highlighting how DNA methylation levels can be directly enhanced, suppressed, or remodeled via metabolic and nutritional perturbations. Dynamic changes in DNA methylation are required during embryonic development, lineage specification, and maintenance of somatic cell function that can be fine-tuned based on the influence of essential micronutrients. As we age, DNA methylation and hydroxymethylation levels drift in patterning, leading to epigenetic dysregulation and genomic instability that underlies the formation and progression of multiple diseases including cancer. Understanding how DNA methylation can be regulated by micronutrients will have important implications for the maintenance of normal tissue function upon aging, and in the prevention and treatment of diseases for improved health and lifespan.
Introduction
DNA methylation in the form of 5-methylcytosine (5mC) can be found in ∼80% of CpGs in the genome and is primarily enriched in heterochromatin, working in positive feedback with silencing histone marks to maintain lineage identity (Lister et al., 2009). 5mC is conventionally known as a repressive epigenetic mark, controlling gene expression and chromatin organization to influence cellular development and differentiation (Guo et al., 2014). During development and in response to environmental stimuli, many 5mC marks are dynamic, including at shores of CpG islands, promoters, enhancers, and across gene bodies of differentially expressed genes (Deaton and Bird, 2011; Nguyen et al., 2022). DNA methyltransferases (DNMTs) catalyze the transfer of a methyl group to the 5’ position of cytosine residues to regulate de novo 5mC formation (DNMT3A and DNMT3B) independently of the cell cycle, or during DNA replication to ensure faithful DNA methylation inheritance (DNMT1) (Okano et al., 1999; Pradhan et al., 1999).
The Ten-eleven translocation (TET1-3) enzymes are considered to have an opposing role to the DNMTs due to their ability to oxidize 5mC to generate 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Tahiliani et al., 2009; Ito et al., 2010; He et al., 2011). TET proteins belong to a superfamily of dioxygenases that utilize Fe2+ and α-ketoglutarate (αKG) as essential cofactors (Lorsbach et al., 2003) [reviewed in (Lu et al., 2015a; Rasmussen and Helin, 2016; Wu and Zhang, 2017; Lio et al., 2020)]. The oxidized methylcytosines generated by TETs can be stable modifications in the genome or transient modifications that promote active or passive DNA demethylation (Tahiliani et al., 2009; Zhang et al., 2010; Bachman et al., 2014). DNMT1 preferentially recognizes hemi-methylated DNA to establish inheritance of methylation at palindromic CpG dinucleotides upon DNA replication (Bashtrykov et al., 2012) however, the inability of DNMT1 to recognize 5hmC causes a passive, cell-cycle dependent loss of DNA methylation (Otani et al., 2013). Unlike 5hmC that does not interfere with RNA or DNA polymerase extension, the iterative oxidation products 5fC and 5caC can slow RNA elongation (Wang L. et al., 2015), cause DNA polymerase pausing (Shibutani et al., 2014), and mimic a T:G mismatch leading to active removal and replacement with an unmethylated cytosine via base excision repair (BER) independently of DNA replication (Cortellino et al., 2011; He et al., 2011).
Essential micronutrients can directly or indirectly influence DNMT and/or TET activity that in turn impact DNA methylation levels (Figure 1). B vitamins can be regarded as pro-methylating micronutrients given their role in one-carbon metabolism that generates both nucleotides for DNA synthesis and the transfer of methyl groups required for the maintenance of DNA methylation (Tong et al., 2009; Lyon et al., 2020). The methyl group used in the formation of 5mC by DNMTs is derived from the coordinated effort of B2, B6, B9, B12, and the essential amino acid methionine. Together, these micronutrients regulate the folate and methionine cycles of one-carbon metabolism to generate S-adenosylmethionine (SAM), the sole methyl donor used by all methyltransferases to methylate proteins, RNA and DNA. TET catalytic activity is also dependent on metabolites and micronutrients whose abundance can be directly regulated by dietary supplementation. More recently it has been shown that vitamin C can act as a direct cofactor of TET enzymes to enhance 5hmC formation (Cimmino et al., 2017; Cimmino et al., 2018) by reducing ferric iron (Fe3+) to ferrous iron (Fe2+). Alterations in iron homeostasis may influence TET activity, as shown in the brain, liver, gut, lymphocytes, and cord blood cells (Jiang et al., 2021; Barks et al., 2022; Gao et al., 2022; Taeubert et al., 2022) and additional micronutrients, such as vitamin A, D, and E, have also been implicated as direct or indirect regulators of DNA methylation (Doig et al., 2013; Hore et al., 2016; Remely et al., 2017). Given that aberrant DNA methylation profiles are associated with aging (Sedivy et al., 2008; Salameh et al., 2020; He et al., 2021; Wilkinson et al., 2021; Lu et al., 2023) and that loss of function mutations in DNMT and TET enzymes are a hallmark of cancer (Delhommeau et al., 2009; Figueroa et al., 2010a; Li et al., 2011; Buscarlet et al., 2017; Ostrander et al., 2020), understanding the connection between DNA methylation maintenance and micronutrient availability may uncover liabilities associated with diseases upon aging (Patel et al., 2023) and reveal novel metabolic interventions that can be implemented to retain faithful DNA methylation patterns and function for disease prevention and longevity.
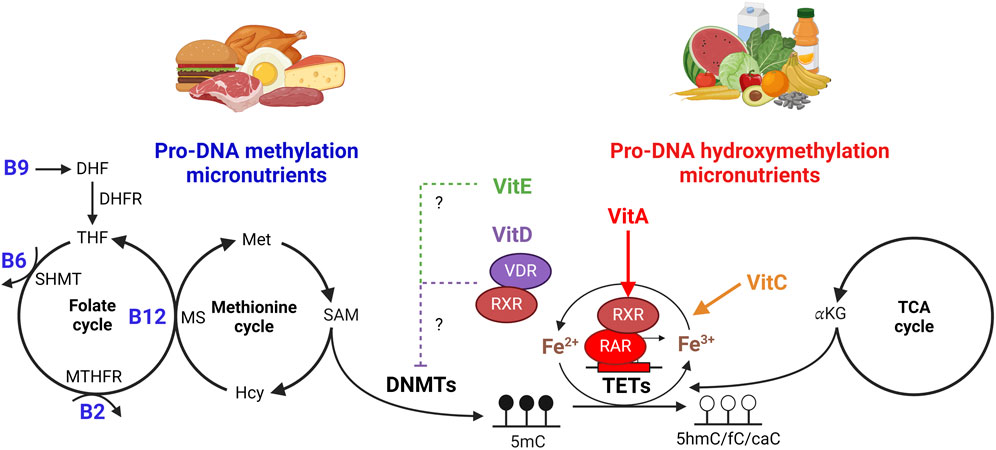
Figure 1. Role of micronutrients on DNA (hydroxy) methylation. One-carbon metabolism is comprised of the folate and methionine cycles that coordinate both nucleotide synthesis and the generation of S-adenosylmethionine (SAM), the sole methyl donor used by DNA methyltransferases to generate 5-methylcytosine (5mC) in the genome. Folate (vitamin B9) is converted to dihydrofolate (DHF) then to tetrahydrofolate (THF) by dihydrofolate reductase (DHFR). Serine hydroxymethyltransferase (SHMT) utilizes vitamin B6 as a cofactor to generate the metabolite 5,10-methyleneTHF, which can then be reduced by methylenetetrahydrofolate reductase (MTHFR) into 5-methyltetrahydrofolate (5-mTHF) using B2 as a cofactor. In conjunction with the methionine cycle, vitamin B12 is a cofactor for methionine synthase (MS), which uses 5-mTHF to transfer a methyl group to homocysteine (Hcy) in the recycling of methionine. Methionine, when converted to S-adenosylmethionine (SAM), provides DNA methyltransferases (DNMTs) with the methyl group needed to generate 5mC. Ten-Eleven Translocation (TET) enzymes hydroxylate 5mC to form 5hmC, 5fC, and/or 5caC using Fe2+ and α-ketoglutarate (αKG) as cofactors. Vitamin C (VitC) enhances TET function by recycling ferric (Fe3+) to ferrous (Fe2+) iron, while αKG is supplied by the tricarboxylic acid (TCA) cycle. Vitamin A (VitA) can directly upregulate TET gene expression via retinoid receptor (RAR/RXR) signaling. Vitamin D (VitD) acts as a ligand for the vitamin D receptor (VDR), which can also form a heterodimer with RXR, and both VitD or vitamin E (VitE) treatment have been shown to inhibit DNMT expression to influence DNA methylation maintenance. Figure created with BioRender.com.
Micronutrient regulation of 5-methylcytosine formation and maintenance (pro-methylating micronutrients)
The role of several B vitamins in the direct regulation of one-carbon metabolism places these essential micronutrients as DNA methylation guardians of the epigenome. One-carbon metabolism is comprised of the folate and methionine cycles that coordinate both pyrimidine synthesis and the generation of S-adenosylmethionine (SAM), the sole methyl donor used by DNA methyltransferases to generate 5mC in the genome. The coordinated regulation of nucleotide synthesis and methylation by one-carbon metabolism ensures that DNA replication is coupled with DNA methylation inheritance (Ducker and Rabinowitz, 2017). A connection between the folate cycle and DNA methylation maintenance has been known for decades. Nutritional vitamin B9 or B12 deficiencies during gestation or polymorphisms in folate cycle enzymes of pregnant women are associated with neural tube defects (NTDs) (Smithells et al., 1976; Prevention of neural tube defects, 1991; Crider et al., 2011). The process of neurulation in embryos requires increased global methylation to promote neural tube fusion (Greene et al., 2011), and NTDs potentially form due to the lack of silencing of Notch1 and Sox2 (Alata Jimenez and Strobl-Mazzulla, 2022). Loss of serine hydroxymethyltransferase 1 (SHMT1) activity, which utilizes B6 as a cofactor, can impair neural tube closure (Beaudin et al., 2011; Beaudin et al., 2012). Moreover, polymorphisms that decrease the activity of methylenetetrahydrofolate reductase (MTHFR) (Prevention of neural tube defects, 1991), which depends on B2 as a cofactor, or methionine synthase (MS) that utilizes B12 as a cofactor (Tong et al., 2009; Guéant et al., 2020), and methionine synthase reductase that regenerates methylated B12 levels from its oxidized form have all been associated with NTDs (Botto and Yang, 2000; Imbard et al., 2013). One-carbon metabolism inhibitors such as the anti-folate methotrexate (MTX) that blocks the activity of dihydrofolate reductase (DHFR) promotes DNA hypomethylation in embryonic tissues and leads to NTDs (Greene et al., 2011; Wang X. et al., 2015). DNA methylation generated by DNMTs is required for closure of the neural tube, therefore as per one-carbon metabolic enzyme gene polymorphisms and B vitamin deficiencies, disruption of DNA methylation caused by Dnmt3b knockout, and DNMT or methylation cycle inhibitors also result in NTDs (Okano et al., 1999; Afman et al., 2005; Dunlevy et al., 2006).
The role of B vitamins in DNA methylation maintenance in other diseases and cell types have also been described. Clinical studies have shown that lower B9 intake leads to DNA hypomethylation in colonic tissue (Pufulete et al., 2003) and an increased risk of colorectal cancer (CRC) (Kim, 2007). A B9-deficient diet in post-menopausal women also causes DNA hypomethylation in lymphocytes (Jacob et al., 1998). Low dose therapy with MTX used in the treatment of rheumatoid arthritis demethylates the FOXP3 locus in developing regulatory T cells, resulting in their enhanced differentiation and suppression of inflammation (Cribbs et al., 2015; Rossetti et al., 2015). Furthermore, higher global DNA methylation in leukocytes is associated with decreased responsiveness to MTX in rheumatoid arthritis patients (Gosselt et al., 2019), indicating the importance of MTX’s hypomethylating activity.
Elevated levels of B9 or B12 supplementation are positively correlated with increased DNA methylation through increased methyl-donor availability. Combined B9 and B12 supplementation leads to increased DNA methylation in leukocytes (Kok DE. et al., 2015; Kok DEG. et al., 2015), and B12 supplementation alone increases DNA methylation in disease-free children (Yadav et al., 2018) and mouse models of depression and meningitis (de Queiroz et al., 2020; Trautmann et al., 2020). Supplementation with B9 increases sperm DNA methylation levels in a mouse model of the MTHFR polymorphism, and supplementation with B2 has also been shown to increase DNA methylation in peripheral leukocytes of healthy individuals or cardiovascular disease patients with an MTHFR polymorphism (Amenyah et al., 2020; Amenyah et al., 2021). Amongst the healthy population, there have been no reports of toxicity for excess B vitamin consumption; however, given that one-carbon metabolism promotes DNA methylation, caution may be warranted for high B vitamin supplementation in disease states driven by DNA hypermethylation phenotypes (Ehrlich, 2019).
Hydroxymethylation formation and DNA methylation removal via micronutrient mediated activation of the TET enzymes (pro-hydroxymethylation micronutrients)
Epigenetic plasticity, including the ability to remodel DNA methylation patterns in the genome, is integral to maintaining stem cell potency, lineage specification, and gene regulation in response to changes in the environment. The TET enzymes (TET1-3) are the only known mammalian DNA demethylases that can trigger the passive or active removal of 5mC in the genome (Tahiliani et al., 2009; Ito et al., 2010; He et al., 2011). TET1 is most highly expressed in embryonic stem cells, primordial germ cells, and neural tissues, whereas TET2 and TET3 are most highly expressed in stem, progenitor, and other differentiated adult cell populations (Lu et al., 2015a; Rasmussen and Helin, 2016; Wu and Zhang, 2017; Lio et al., 2020). While the TET enzymes all exhibit a conserved core C-terminal catalytic domain, they diverge in their N-terminal structure, in which TET2 lacks the CXXC DNA-binding motif found in TET1 and TET3 and therefore relies on protein-binding partners for its recruitment to chromatin (Ko et al., 2013; Rampal et al., 2014). TET activity is primarily associated with gene activation, given the enrichment of 5hmC within enhancers and gene bodies of actively expressed genes (Ficz et al., 2011; Rasmussen and Helin, 2016). The role of TET activity is perceived to oppose the gene silencing role of DNMTs; however, 5mC is an essential requirement for the generation of 5hmC and the iterative oxidized products 5fC and 5caC, thus DNA methylation and hydroxymethylation, as mediated by DNMTs and TETs, must work together to form and remove cytosine methylation in the DNA (Zhang et al., 2016; Gu et al., 2018; Lopez-Moyado et al., 2019; Chao et al., 2022). In addition to the 5mC DNA substrate, TET enzymes require Fe2+ and αKG as obligatory cofactors for their catalytic activity (Tahiliani et al., 2009). Perturbations in Fe2+ homeostasis can influence TET activity, where iron sufficiency is associated with increased TET activity and maintenance of 5hmC formation in cord blood, hepatocytes, enterocytes, and splenocytes (Jiang et al., 2021; Taeubert et al., 2022) whereas deficiencies in iron lead to reduced activity in the cerebellum of rats evidenced by decreased 5hmC levels and neurodevelopmental defects (Barks et al., 2022). However, in settings of excess iron and iron overloading, in which homeostatic responses may block cellular uptake of redox active iron (Conrad et al., 1963), TET enzymatic activity can also become impaired and promote pathogenic T cell expansion and systemic lupus erythematosus pathogenesis (Gao et al., 2022). These findings suggest that at a physiological level, the role of iron homeostasis and the balance between iron stores or redox active labile iron on TET activity requires further clarification.
Based on its role as a cofactor of αKG-dependent dioxygenases (αKGDDs), vitamin C was tested as an enhancer of TET activity. Acting as a targeted antioxidant, vitamin C can directly bind the C-terminal catalytic domain of Tet enzymes, promoting the reduction and recycling of Fe3+ to Fe2+ (Yin et al., 2013; Cimmino et al., 2017). Vitamin C has been shown to increase the levels of 5hmC, 5fC, and 5caC anywhere from 4 to 20-fold in a variety of contexts, including ESCs, fibroblasts and leukemia cells, within as little as 24 h (Wang et al., 2011; Minor et al., 2013; Yin et al., 2013; Cimmino et al., 2017; Mingay et al., 2018; Brabson et al., 2021). Increased 5hmC formation and DNA demethylation upon vitamin C treatment leads to improved reprogramming efficiency during iPSC formation, and enhanced pluripotency of ESC cultures (Hore et al., 2016) by reducing methylation normally gained at CpG islands (CGIs) of pluripotency genes during blastocyst to epiblast transition (Blaschke et al., 2013; Hu et al., 2014). Tissue specific studies of vitamin C treatment in lung, breast, bladder, and kidney cultures have also reported similar increases in 5hmC generation and DNA demethylation (Ge et al., 2018; Sant et al., 2018).
Interestingly, an algal homolog of the TET enzymes uses vitamin C directly instead of αKG as the primary cofactor for the generation of glucosyl-methylation marks in the genome to regulate photosynthesis (Xue et al., 2019). Vitamin C is a structural homolog of αKG, but unlike this TCA cycle intermediate, is an essential micronutrient for humans and therefore imparts a dependence for enhancing TET activity in the context of nutritional availability (Myllyla et al., 1978; Majamaa et al., 1986; Yin et al., 2013). One requirement for increased TET activity may be in the regulation of immune responses. In regulatory T (Treg) cells, vitamin C promotes DNA demethylation of a Foxp3 enhancer, promoting Treg differentiation and immunosuppressive function akin to the role of low-dose MTX in rheumatoid arthritis patients (Yue et al., 2021; Suga et al., 2023). Vitamin C also drives oxidized methylcytosine formation and DNA demethylation within the three enhancers of the Prdm1 locus that are silenced in naive B cells but require activation for plasma cell differentiation (Qi et al., 2020). The effect of vitamin C on lymphocyte differentiation, including Tregs and plasma cells, has been shown to be both Tet2 and Tet3 dependent, which are the most highly expressed of the three TET enzymes in hematopoietic cells (An et al., 2015; Brabson et al., 2023). Furthermore, vitamin C increases Th1-type chemokines and PD-L1 gene expression in a TET2-dependent manner resulting in enhanced sensitivity to anti-PD-1/PD-L1 therapy (Xu et al., 2019).
Deficiencies in vitamin C mimic loss-of-function mutations in TET2, which is the most frequently mutated of the TET enzymes in cancer and primarily in blood cell malignancy causing impaired DNA de-methylation. Aberrant DNA hypermethylation and/or vitamin C deficiency have both been reported in patients with hematological malignancies (Figueroa et al., 2010a; Figueroa et al., 2010b). Oral supplementation with vitamin C can raise plasma levels in patients with hematological malignancies, leading to an overall increase in the 5hmC/5mC ratio in circulating white blood cells (Gillberg et al., 2019) and a reduction in the proportion of hypermethylated loci caused by TET2 mutation (Taira et al., 2023). Normalization of low vitamin C serum levels via supplementation in murine models also leads to a restoration of 5hmC formation in hematopoietic cells that suppresses Tet2-deficient leukemia progression (Agathocleous et al., 2017). These studies provide proof of principle that altering vitamin C supplementation can have a powerful influence on the blood cell DNA methylome.
Recent studies have also implicated vitamin A as a regulator of TET activity and DNA demethylation. Vitamin A comprises multiple active metabolites termed retinoids that play an essential role in growth, development, and differentiation (Gudas, 2013). The active form of vitamin A, retinoic acid (RA), binds to retinoic acid and retinoid X receptors (RAR/RXR) in the nucleus that, in association with other transcriptional coactivators, localize to retinoic acid responsive elements (RAREs) in target genes to activate gene expression (Gudas, 2013; Hore et al., 2016; Coyle et al., 2018). Thymidine DNA glycosylase (TDG) promotes DNA demethylation by forming a complex with acetylated TET2 (Zhang et al., 2017), and RAR/RXR activation upon RA-signaling has been shown to recruit the histone acetyltransferase CBP, TET proteins, and TDG to trigger oxidized methylcytosine formation and DNA demethylation via BER at target gene loci (Leger et al., 2014; Hassan et al., 2017; Hassan et al., 2020). RA-signaling in this manner can initiate TET2-dependent DNA demethylation at the Hic1 locus, which is often hypermethylated in human cancers, such as colon, breast, and brain cancer (Hassan et al., 2020). RA-signaling in breast cancer cells also induces a RARB-TET2 complex, targeted to the promoters of genes involved in cellular differentiation, such as miR-200c (Wu et al., 2017). Defective RARB/TET2 signaling by loss of TET2 and/or deficient miR-200c expression is correlated with RA-resistant breast cancer cell growth and aggressiveness (Wu et al., 2017). RA treatment of human embryonic carcinoma stem cells has also described a TET2-dependent 5mC to 5hmC conversion in the HOXA gene cluster (Bocker et al., 2012). These studies collectively show that RAR, CBP, TET, and TDG enzymes play an interconnected role in the initiation of gene expression in response to RA-signaling. TET expression can also be directly up-regulated by RA-signaling, and alone or in combination with vitamin C, enhances 5hmC formation in ESCs (Hore et al., 2016). A conserved RARE identified in the first intron of the TET2 locus suggests that expression could be mediated via direct RAR binding (Hore et al., 2016). Thus, the ability to directly enhance TET expression and increase the catalytic rate of TET activity via combined treatment with vitamin A and vitamin C suggests that these micronutrients work together to activate gene expression via increased methylcytosine oxidation.
Indirect micronutrient regulation of DNA methylation
Other essential micronutrients have been reported to play an indirect role in the maintenance of DNA methylation via modulation of chromatin states and the regulation of DNA methyltransferase expression. Similar to RAR, the vitamin D receptor (VDR) forms a heterodimer with RXR at vitamin D responsive elements (VDREs) in the DNA (Doig et al., 2013; Krstic et al., 2022; Mirza et al., 2022). The active form of vitamin D, calcitriol [1,25(OH)2D3], when bound to VDR-RXR, triggers the recruitment of co-activators, including steroid receptor and p300/CBP (Fetahu et al., 2014). However, in the absence of its ligand, VDR-RXR is bound by co-repressors such as NCOR1 and SMRT that maintain repressive histone modifications at target loci (Doig et al., 2013; Fetahu et al., 2014). Vitamin D signaling may promote DNA hypomethylation via the downregulated expression of DNMT1 and DNMT3B (Jiao et al., 2019; Lai et al., 2020) in this matter. Likewise, vitamin E can influence the expression of DNMT1; both a decreased expression in liver cells and an increased level in colon cells have been reported (Remely et al., 2017; Zappe et al., 2018). Prostate cancer models treated with vitamin E exhibit decreased DNMT protein expression and reduced CpG methylation at target loci of genes involved in cellular redox homeostasis such as the Nrf2 promoter (Huang et al., 2012a). As oxidative stress is often correlated with increased global levels of 5mC (Garcia-Guede et al., 2020; Goncalves et al., 2021), it is possible that vitamin E can reduce DNA methylation indirectly due to its antioxidant activity (Ryan et al., 2010; Zappe et al., 2018).
Discussion and future directions
The balance of essential micronutrient intake and bioavailability has the potential to significantly alter DNA methylation and hydroxymethylation levels in the body (Figure 2). Research using datasets comprising thousands of samples to track alterations in DNA methylation associated with cellular development, differentiation, aging, and disease progression has led to the identification of DNA methylation biomarkers and epigenetic clocks that accurately estimate chronological age (Hannum et al., 2013; Horvath, 2013; Horvath and Raj, 2018), healthspan and lifespan (Levine et al., 2018; Lu et al., 2019; Lu et al., 2022), or the pace of aging (Belsky et al., 2022) in both humans and animal models. These same DNA methylation signatures are also being used to investigate the impact of lifestyle factors such as diet and nutritional status on disease risk and aging (Quach et al., 2017; Lu et al., 2019; Fitzgerald et al., 2021) that may reveal potential therapeutic interventions to slow aging phenotypes and age-related diseases, including cancer. Plant-based, Mediterranean, and methylation-supportive diets can rejuvenate DNA methylation aging signatures, decreasing chronological age predictions by 1–10 years in as little as 8 weeks of intervention (Gensous et al., 2020; Dwaraka et al., 2023; Fitzgerald et al., 2023). One study (Dwaraka et al., 2023) using blood samples from paired twins reported that a vegan diet compared to an omnivorous diet led to a significant decrease in overall epigenetic age acceleration measured using multiple DNA methylation biomarkers (Levine et al., 2018; Lu et al., 2019; Belsky et al., 2022; Lu et al., 2022) that correlated with fewer hypermethylated loci in the vegan diet, consistent with the notion that vegan diets contribute to a reduced intake and lower serum levels of B12 (Gilsing et al., 2010; Niklewicz et al., 2023) and methionine (McCarty et al., 2009; Sanderson et al., 2019; Allen and Locasale, 2021), two of the major regulators of methyl-donor (SAM) levels. However, no controlled studies have attempted to address how supplementation with specific micronutrients associates with epigenetic clocks and DNA methylation biomarkers of aging or disease.
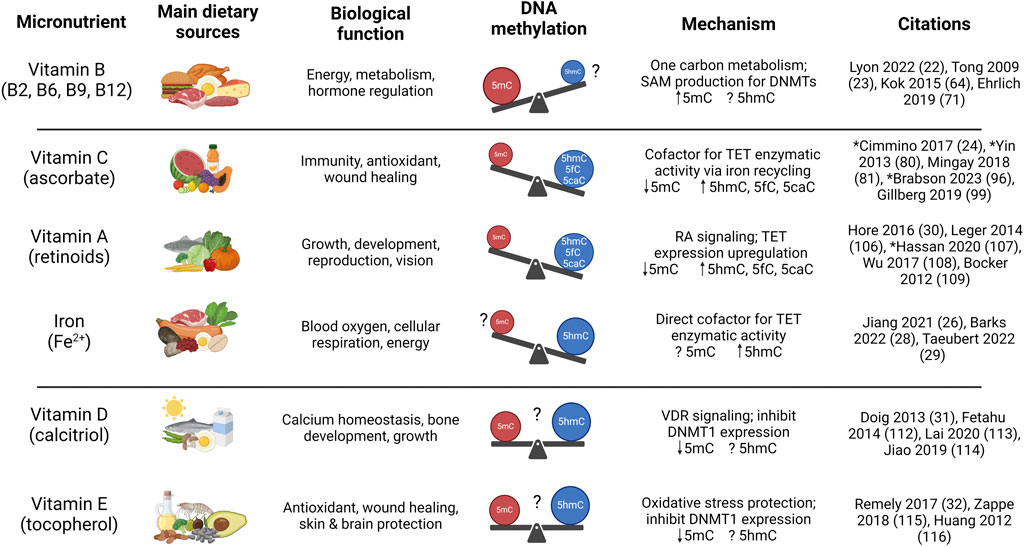
Figure 2. Influence of essential dietary micronutrients on DNA methylation or hydroxymethylation and biological outcome. Summary of biological effects of known micronutrient regulators of DNA methylation and hydroxymethylation. Vitamin A, typically sourced from foods such as carrots, leafy greens, and fish, is essential for development and reproduction, and stimulates TET activity leading to increased 5hmC formation and DNA demethylation. The various B vitamins play a coordinated role in energy production and one-carbon metabolism, are enriched in animal-derived foods, and higher intake can increase SAM production that facilitates DNMT activity and 5mC formation. Vitamin C, a major cellular antioxidant enriched in fruits and vegetables, acts as a cofactor for TET enzymes, driving 5hmC formation and DNA demethylation. Vitamin D is sourced from exposure to sunlight and is enriched in dairy products, whereas vitamin E, sourced from oils and nuts, protects against oxidative stress. Both Vitamin D and E have been shown to inhibit DNMT expression, although the effect on global methylation and hydroxymethylation varies based on cellular context and the mechanism remains unclear. Iron, a trace metal found in legumes, lean meat, and shellfish, is a direct cofactor for TET enzymes and increased supplementation facilitates 5hmC formation. *Studies that quantified micronutrient influence on 5hmC, 5fC, and 5caC levels. Figure created with BioRender.com.
An important caveat to consider when interpreting micronutrient influences on DNA methylation status using existing biomarkers is that the gold standard for measuring global DNA methylation in research and clinical diagnostics has historically relied on CpG hybridization array platforms using bisulfite-treated DNA. Unmethylated cytosines, 5fC, and 5caC, are converted to uracil upon bisulfite treatment, while both 5mC and 5hmC remain protected and measured as cytosine, rendering their contribution to DNA methylation signatures indistinguishable (Huang et al., 2010). DNA immunoprecipitation and base-resolution mapping of 5hmC, 5fC and 5caC in the mammalian genome have shown that most reside in a CpG context, enriched within gene bodies, low density CpG regions (CpG island shores), and at distal regulatory elements such as enhancers, where 5mC and 5hmC can be present in nearly equal representation (Williams et al., 2011; Wu et al., 2011; Huang et al., 2012b; Yu et al., 2012; Lu et al., 2015b; Xia et al., 2015). While the contribution of 5hmC, or the rarer modifications 5fC and 5caC, to the major DNA methylation biomarkers currently remains unknown, recent advances in base resolution mapping using alternate chemical and enzymatic conversion strategies (Huang et al., 2010; Booth et al., 2013; Schutsky et al., 2018) [reviewed in (Berney and McGouran, 2018)] should allow the contribution of nutritional interventions and micronutrient levels on DNMT vs. TET-mediated DNA methylation maintenance, epigenetic clocks, and other predictive disease biomarkers to be determined.
Author contributions
TL: Writing–original draft, Writing–review and editing. PL: Writing–original draft. JP: Writing–original draft. LC: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the NIH (NCI 1R01CA282453-01), Leukemia & Lymphoma Society (7028-22), and Castaways Against Cancer Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afman, L. A., Blom, H. J., Drittij, M. J., Brouns, M. R., and van Straaten, H. W. (2005). Inhibition of transmethylation disturbs neurulation in chick embryos. Dev. Brain Res. 158 (1-2), 59–65. doi:10.1016/j.devbrainres.2005.06.002
Agathocleous, M., Meacham, C. E., Burgess, R. J., Piskounova, E., Zhao, Z., Crane, G. M., et al. (2017). Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549 (7673), 476–481. doi:10.1038/nature23876
Alata Jimenez, N., and Strobl-Mazzulla, P. H. (2022). Folate carrier deficiency drives differential methylation and enhanced cellular potency in the neural plate border. Front. Cell Dev. Biol. 10, 834625. doi:10.3389/fcell.2022.834625
Allen, A. E., and Locasale, J. W. (2021). Metabolomics: insights into plant-based diets. EMBO Mol. Med. 13 (2), e13568. doi:10.15252/emmm.202013568
Amenyah, S. D., McMahon, A., Ward, M., Deane, J., McNulty, H., Hughes, C. F., et al. (2020). Riboflavin supplementation alters global and gene-specific DNA methylation in adults with the MTHFR 677 TT genotype. Biochimie 173, 17–26. doi:10.1016/j.biochi.2020.04.007
Amenyah, S. D., Ward, M., McMahon, A., Deane, J., McNulty, H., Hughes, C., et al. (2021). DNA methylation of hypertension-related genes and effect of riboflavin supplementation in adults stratified by genotype for the MTHFR C677T polymorphism. Int. J. Cardiol. 322, 233–239. doi:10.1016/j.ijcard.2020.09.011
An, J., Gonzalez-Avalos, E., Chawla, A., Jeong, M., Lopez-Moyado, I. F., Li, W., et al. (2015). Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 6, 10071. doi:10.1038/ncomms10071
Bachman, M., Uribe-Lewis, S., Yang, X., Williams, M., Murrell, A., and Balasubramanian, S. (2014). 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6 (12), 1049–1055. doi:10.1038/nchem.2064
Barks, A., Beeson, M. M., Hallstrom, T. C., Georgieff, M. K., and Tran, P. V. (2022). Developmental iron deficiency dysregulates TET activity and DNA hydroxymethylation in the rat Hippocampus and cerebellum. Dev. Neurosci. 44 (2), 80–90. doi:10.1159/000521704
Bashtrykov, P., Jankevicius, G., Smarandache, A., Jurkowska, R. Z., Ragozin, S., and Jeltsch, A. (2012). Specificity of Dnmt1 for methylation of hemimethylated CpG sites resides in its catalytic domain. Chem. Biol. 19 (5), 572–578. doi:10.1016/j.chembiol.2012.03.010
Beaudin, A. E., Abarinov, E. V., Malysheva, O., Perry, C. A., Caudill, M., and Stover, P. J. (2012). Dietary folate, but not choline, modifies neural tube defect risk in Shmt1 knockout mice. Am. J. Clin. Nutr. 95 (1), 109–114. doi:10.3945/ajcn.111.020305
Beaudin, A. E., Abarinov, E. V., Noden, D. M., Perry, C. A., Chu, S., Stabler, S. P., et al. (2011). Shmt1 and de novo thymidylate biosynthesis underlie folate-responsive neural tube defects in mice. Am. J. Clin. Nutr. 93 (4), 789–798. doi:10.3945/ajcn.110.002766
Belsky, D. W., Caspi, A., Corcoran, D. L., Sugden, K., Poulton, R., Arseneault, L., et al. (2022). DunedinPACE, a DNA methylation biomarker of the pace of aging. Elife 11, e73420. doi:10.7554/elife.73420
Berney, M., and McGouran, J. F. (2018). Methods for detection of cytosine and thymine modifications in DNA. Nat. Rev. Chem. 2 (11), 332–348. doi:10.1038/s41570-018-0044-4
Blaschke, K., Ebata, K. T., Karimi, M. M., Zepeda-Martinez, J. A., Goyal, P., Mahapatra, S., et al. (2013). Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500 (7461), 222–226. doi:10.1038/nature12362
Bocker, M. T., Tuorto, F., Raddatz, G., Musch, T., Yang, F. C., Xu, M., et al. (2012). Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat. Commun. 3, 818. doi:10.1038/ncomms1826
Booth, M. J., Ost, T. W., Beraldi, D., Bell, N. M., Branco, M. R., Reik, W., et al. (2013). Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 8 (10), 1841–1851. doi:10.1038/nprot.2013.115
Botto, L. D., and Yang, Q. (2000). 5,10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am. J. Epidemiol. 151 (9), 862–877. doi:10.1093/oxfordjournals.aje.a010290
Brabson, J. P., Leesang, T., Mohammad, S., and Cimmino, L. (2021). Epigenetic regulation of genomic stability by vitamin C. Front. Genet. 12, 675780. doi:10.3389/fgene.2021.675780
Brabson, J. P., Leesang, T., Yap, Y. S., Wang, J., Lam, M. Q., Fang, B., et al. (2023). Oxidized mC modulates synthetic lethality to PARP inhibitors for the treatment of leukemia. Cell Rep. 42 (1), 112027. doi:10.1016/j.celrep.2023.112027
Buscarlet, M., Provost, S., Zada, Y. F., Barhdadi, A., Bourgoin, V., Lepine, G., et al. (2017). DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood 130 (6), 753–762. doi:10.1182/blood-2017-04-777029
Chao, L., Yang, S., Li, H., Long, C., Xi, Q., and Zuo, Y. (2022). Competitive binding of TET1 and DNMT3A/B cooperates the DNA methylation pattern in human embryonic stem cells. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1865 (7), 194861. doi:10.1016/j.bbagrm.2022.194861
Cimmino, L., Dolgalev, I., Wang, Y., Yoshimi, A., Martin, G. H., Wang, J., et al. (2017). Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170 (6), 1079–1095.e20. doi:10.1016/j.cell.2017.07.032
Cimmino, L., Neel, B. G., and Aifantis, I. (2018). Vitamin C in stem cell reprogramming and cancer. Trends Cell Biol. 28 (9), 698–708. doi:10.1016/j.tcb.2018.04.001
Conrad, M. E., Crosby, W. H., and Merrill, B. (1963). Intestinal mucosal mechanisms controlling iron absorption. Blood 22, 406–415. doi:10.1182/blood.v22.4.406.406
Cortellino, S., Xu, J., Sannai, M., Moore, R., Caretti, E., Cigliano, A., et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146 (1), 67–79. doi:10.1016/j.cell.2011.06.020
Coyle, K. M., Dean, C. A., Thomas, M. L., Vidovic, D., Giacomantonio, C. A., Helyer, L., et al. (2018). DNA methylation predicts the response of triple-negative breast cancers to all-trans retinoic acid. Cancers (Basel) 10 (11), 397. doi:10.3390/cancers10110397
Cribbs, A. P., Kennedy, A., Penn, H., Amjadi, P., Green, P., Read, J. E., et al. (2015). Methotrexate restores regulatory T cell function through demethylation of the FoxP3 upstream enhancer in patients with rheumatoid arthritis. Arthritis & Rheumatology 67 (5), 1182–1192. doi:10.1002/art.39031
Crider, K. S., Bailey, L. B., and Berry, R. J. (2011). Folic acid food fortification-its history, effect, concerns, and future directions. Nutrients 3 (3), 370–384. doi:10.3390/nu3030370
Deaton, A. M., and Bird, A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25 (10), 1010–1022. doi:10.1101/gad.2037511
Delhommeau, F., Dupont, S., Valle, V. D., James, C., Trannoy, S., Masse, A., et al. (2009). Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360 (22), 2289–2301. doi:10.1056/nejmoa0810069
de Queiroz, K. B., Cavalcante-Silva, V., Lopes, F. L., Rocha, G. A., D'Almeida, V., and Coimbra, R. S. (2020). Vitamin B(12) is neuroprotective in experimental pneumococcal meningitis through modulation of hippocampal DNA methylation. J. neuroinflammation 17 (1), 96. doi:10.1186/s12974-020-01763-y
Doig, C. L., Singh, P. K., Dhiman, V. K., Thorne, J. L., Battaglia, S., Sobolewski, M., et al. (2013). Recruitment of NCOR1 to VDR target genes is enhanced in prostate cancer cells and associates with altered DNA methylation patterns. Carcinogenesis 34 (2), 248–256. doi:10.1093/carcin/bgs331
Ducker, G. S., and Rabinowitz, J. D. (2017). One-carbon metabolism in health and disease. Cell Metab. 25 (1), 27–42. doi:10.1016/j.cmet.2016.08.009
Dunlevy, L. P., Burren, K. A., Chitty, L. S., Copp, A. J., and Greene, N. D. (2006). Excess methionine suppresses the methylation cycle and inhibits neural tube closure in mouse embryos. FEBS Lett. 580 (11), 2803–2807. doi:10.1016/j.febslet.2006.04.020
Dwaraka, V. B., Aronica, L., Carreras-Gallo, N., Robinson, J. L., Hennings, T., Lin, A., et al. (2023). Unveiling the epigenetic impact of vegan vs. Omnivorous diets on aging: insights from the twins nutrition study (TwiNS). medRxiv.
Ehrlich, M. (2019). DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics 14 (12), 1141–1163. doi:10.1080/15592294.2019.1638701
Fetahu, I. S., Höbaus, J., and Kállay, E. (2014). Vitamin D and the epigenome. Front. Physiol. 5, 164. doi:10.3389/fphys.2014.00164
Ficz, G., Branco, M. R., Seisenberger, S., Santos, F., Krueger, F., Hore, T. A., et al. (2011). Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473 (7347), 398–402. doi:10.1038/nature10008
Figueroa, M. E., Abdel-Wahab, O., Lu, C., Ward, P. S., Patel, J., Shih, A., et al. (2010b). Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18 (6), 553–567. doi:10.1016/j.ccr.2010.11.015
Figueroa, M. E., Lugthart, S., Li, Y., Erpelinck-Verschueren, C., Deng, X., Christos, P. J., et al. (2010a). DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 17 (1), 13–27. doi:10.1016/j.ccr.2009.11.020
Fitzgerald, K. N., Campbell, T., Makarem, S., and Hodges, R. (2023). Potential reversal of biological age in women following an 8-week methylation-supportive diet and lifestyle program: a case series. Aging (Albany NY) 15 (6), 1833–1839. doi:10.18632/aging.204602
Fitzgerald, K. N., Hodges, R., Hanes, D., Stack, E., Cheishvili, D., Szyf, M., et al. (2021). Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging (Albany NY) 13 (7), 9419–9432. doi:10.18632/aging.202913
Gao, X., Song, Y., Wu, J., Lu, S., Min, X., Liu, L., et al. (2022). Iron-dependent epigenetic modulation promotes pathogenic T cell differentiation in lupus. J. Clin. investigation 132 (9), e152345. doi:10.1172/jci152345
Garcia-Guede, A., Vera, O., and Ibanez-de-Caceres, I. (2020). When oxidative stress meets epigenetics: implications in cancer development. Antioxidants (Basel) 9 (6), 468. doi:10.3390/antiox9060468
Ge, G., Peng, D., Xu, Z., Guan, B., Xin, Z., He, Q., et al. (2018). Restoration of 5-hydroxymethylcytosine by ascorbate blocks kidney tumour growth. EMBO Rep. 19 (8), e45401. doi:10.15252/embr.201745401
Gensous, N., Garagnani, P., Santoro, A., Giuliani, C., Ostan, R., Fabbri, C., et al. (2020). One-year Mediterranean diet promotes epigenetic rejuvenation with country- and sex-specific effects: a pilot study from the NU-AGE project. Geroscience 42 (2), 687–701. doi:10.1007/s11357-019-00149-0
Gillberg, L., Orskov, A. D., Nasif, A., Ohtani, H., Madaj, Z., Hansen, J. W., et al. (2019). Oral vitamin C supplementation to patients with myeloid cancer on azacitidine treatment: normalization of plasma vitamin C induces epigenetic changes. Clin. Epigenetics 11 (1), 143. doi:10.1186/s13148-019-0739-5
Gilsing, A. M., Crowe, F. L., Lloyd-Wright, Z., Sanders, T. A., Appleby, P. N., Allen, N. E., et al. (2010). Serum concentrations of vitamin B12 and folate in British male omnivores, vegetarians and vegans: results from a cross-sectional analysis of the EPIC-Oxford cohort study. Eur. J. Clin. Nutr. 64 (9), 933–939. doi:10.1038/ejcn.2010.142
Goncalves, A. C., Alves, R., Baldeiras, I., Marques, B., Oliveiros, B., Pereira, A., et al. (2021). DNA methylation is correlated with oxidative stress in myelodysplastic syndrome-relevance as complementary prognostic biomarkers. Cancers (Basel) 13 (13), 3138. doi:10.3390/cancers13133138
Gosselt, H. R., van Zelst, B. D., de Rotte, M., Hazes, J. M. W., de Jonge, R., and Heil, S. G. (2019). Higher baseline global leukocyte DNA methylation is associated with MTX non-response in early RA patients. Arthritis Res. Ther. 21 (1), 157. doi:10.1186/s13075-019-1936-5
Greene, N. D., Stanier, P., and Moore, G. E. (2011). The emerging role of epigenetic mechanisms in the etiology of neural tube defects. Epigenetics 6 (7), 875–883. doi:10.4161/epi.6.7.16400
Gu, T., Lin, X., Cullen, S. M., Luo, M., Jeong, M., Estecio, M., et al. (2018). DNMT3A and TET1 cooperate to regulate promoter epigenetic landscapes in mouse embryonic stem cells. Genome Biol. 19 (1), 88. doi:10.1186/s13059-018-1464-7
Gudas, L. J. (2013). Retinoids induce stem cell differentiation via epigenetic changes. Seminars Cell & Dev. Biol. 24 (10-12), 701–705. doi:10.1016/j.semcdb.2013.08.002
Guéant, J.-L., Oussalah, A., Zgheib, R., Siblini, Y., Hsu, S. B., and Namour, F. (2020). Genetic, epigenetic and genomic mechanisms of methionine dependency of cancer and tumor-initiating cells: what could we learn from folate and methionine cycles. Biochimie 173, 123–128. doi:10.1016/j.biochi.2020.03.015
Guo, H., Zhu, P., Yan, L., Li, R., Hu, B., Lian, Y., et al. (2014). The DNA methylation landscape of human early embryos. Nature 511 (7511), 606–610. doi:10.1038/nature13544
Hannum, G., Guinney, J., Zhao, L., Zhang, L., Hughes, G., Sadda, S., et al. (2013). Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49 (2), 359–367. doi:10.1016/j.molcel.2012.10.016
Hassan, H. M., Isovic, M., Kolendowski, B., Bauer-Maison, N., Onabote, O., Cecchini, M., et al. (2020). Loss of thymine DNA glycosylase causes dysregulation of bile acid homeostasis and hepatocellular carcinoma. Cell Rep. 31 (1), 107475. doi:10.1016/j.celrep.2020.03.039
Hassan, H. M., Kolendowski, B., Isovic, M., Bose, K., Dranse, H. J., Sampaio, A. V., et al. (2017). Regulation of active DNA demethylation through RAR-mediated recruitment of a TET/TDG complex. Cell Rep. 19 (8), 1685–1697. doi:10.1016/j.celrep.2017.05.007
He, X., Liu, J., Liu, B., and Shi, J. (2021). The use of DNA methylation clock in aging research. Exp. Biol. Med. (Maywood) 246 (4), 436–446. doi:10.1177/1535370220968802
He, Y. F., Li, B. Z., Li, Z., Liu, P., Wang, Y., Tang, Q., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333 (6047), 1303–1307. doi:10.1126/science.1210944
Hore, T. A., von Meyenn, F., Ravichandran, M., Bachman, M., Ficz, G., Oxley, D., et al. (2016). Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naive pluripotency by complementary mechanisms. Proc. Natl. Acad. Sci. U. S. A. 113 (43), 12202–12207. doi:10.1073/pnas.1608679113
Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biol. 14 (10), R115. doi:10.1186/gb-2013-14-10-r115
Horvath, S., and Raj, K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19 (6), 371–384. doi:10.1038/s41576-018-0004-3
Hu, X., Zhang, L., Mao, S. Q., Li, Z., Chen, J., Zhang, R. R., et al. (2014). Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 14 (4), 512–522. doi:10.1016/j.stem.2014.01.001
Huang, Y., Khor, T. O., Shu, L., Saw, C. L., Wu, T. Y., Suh, N., et al. (2012a). A gamma-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J. Nutr. 142 (5), 818–823. doi:10.3945/jn.111.153114
Huang, Y., Pastor, W. A., Shen, Y., Tahiliani, M., Liu, D. R., and Rao, A. (2010). The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One 5 (1), e8888. doi:10.1371/journal.pone.0008888
Huang, Y., Pastor, W. A., Zepeda-Martinez, J. A., and Rao, A. (2012b). The anti-CMS technique for genome-wide mapping of 5-hydroxymethylcytosine. Nat. Protoc. 7 (10), 1897–1908. doi:10.1038/nprot.2012.103
Imbard, A., Benoist, J.-F., and Blom, H. J. (2013). Neural tube defects, folic acid and methylation. Int. J. Environ. Res. Public Health 10 (9), 4352–4389. doi:10.3390/ijerph10094352
Ito, S., D'Alessio, A. C., Taranova, O. V., Hong, K., Sowers, L. C., and Zhang, Y. (2010). Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466 (7310), 1129–1133. doi:10.1038/nature09303
Jacob, R. A., Gretz, D. M., Taylor, P. C., James, S. J., Pogribny, I. P., Miller, B. J., et al. (1998). Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J. Nutr. 128 (7), 1204–1212. doi:10.1093/jn/128.7.1204
Jiang, L., Wang, J., Wang, K., Wang, H., Wu, Q., Yang, C., et al. (2021). RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood 138 (8), 689–705. doi:10.1182/blood.2020008986
Jiao, X., Wang, L., Wei, Z., Liu, B., Liu, X., and Yu, X. (2019). Vitamin D deficiency during pregnancy affects the function of Th1/Th2 cells and methylation of IFN-gamma gene in offspring rats. Immunol. Lett. 212, 98–105. doi:10.1016/j.imlet.2019.06.012
Kim, Y. I. (2007). Folate and colorectal cancer: an evidence-based critical review. Mol. Nutr. food Res. 51 (3), 267–292. doi:10.1002/mnfr.200600191
Ko, M., An, J., Bandukwala, H. S., Chavez, L., Aijo, T., Pastor, W. A., et al. (2013). Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 497 (7447), 122–126. doi:10.1038/nature12052
Kok, D. E., Dhonukshe-Rutten, R. A., Lute, C., Heil, S. G., Uitterlinden, A. G., van der Velde, N., et al. (2015a). The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin. Epigenetics 7, 121. doi:10.1186/s13148-015-0154-5
Kok, D. E. G., Dhonukshe-Rutten, R. A. M., Lute, C., Heil, S. G., Uitterlinden, A. G., van der Velde, N., et al. (2015b). The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin. Epigenetics 7, 121. doi:10.1186/s13148-015-0154-5
Krstic, N., Bishop, N., Curtis, B., Cooper, C., Harvey, N., Lilycrop, K., et al. (2022). Early life vitamin D depletion and mechanical loading determine methylation changes in the RUNX2, RXRA, and osterix promoters in mice. Genes Nutr. 17 (1), 7. doi:10.1186/s12263-022-00711-0
Lai, G. R., Lee, Y. F., Yan, S. J., and Ting, H. J. (2020). Active vitamin D induces gene-specific hypomethylation in prostate cancer cells developing vitamin D resistance. Am. J. Physiology-Cell Physiology 318 (5), C836–C847. doi:10.1152/ajpcell.00522.2019
Leger, H., Smet-Nocca, C., Attmane-Elakeb, A., Morley-Fletcher, S., Benecke, A. G., and Eilebrecht, S. (2014). A TDG/CBP/RARα ternary complex mediates the retinoic acid-dependent expression of DNA methylation-sensitive genes. Genomics Proteomics Bioinforma. 12 (1), 8–18. doi:10.1016/j.gpb.2013.11.001
Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S., et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10 (4), 573–591. doi:10.18632/aging.101414
Li, Z., Cai, X., Cai, C. L., Wang, J., Zhang, W., Petersen, B. E., et al. (2011). Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood 118 (17), 4509–4518. doi:10.1182/blood-2010-12-325241
Lio, C. W. J., Yue, X., Lopez-Moyado, I. F., Tahiliani, M., Aravind, L., and Rao, A. (2020). TET methylcytosine oxidases: new insights from a decade of research. J. Biosci. 45, 21. doi:10.1007/s12038-019-9973-4
Lister, R., Pelizzola, M., Dowen, R. H., Hawkins, R. D., Hon, G., Tonti-Filippini, J., et al. (2009). Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462 (7271), 315–322. doi:10.1038/nature08514
Lopez-Moyado, I. F., Tsagaratou, A., Yuita, H., Seo, H., Delatte, B., Heinz, S., et al. (2019). Paradoxical association of TET loss of function with genome-wide DNA hypomethylation. Proc. Natl. Acad. Sci. U. S. A. 116 (34), 16933–16942. doi:10.1073/pnas.1903059116
Lorsbach, R. B., Moore, J., Mathew, S., Raimondi, S. C., Mukatira, S. T., and Downing, J. R. (2003). TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 17 (3), 637–641. doi:10.1038/sj.leu.2402834
Lu, A. T., Binder, A. M., Zhang, J., Yan, Q., Reiner, A. P., Cox, S. R., et al. (2022). DNA methylation GrimAge version 2. Aging (Albany NY) 14 (23), 9484–9549. doi:10.18632/aging.204434
Lu, A. T., Fei, Z., Haghani, A., Robeck, T. R., Zoller, J. A., Li, C. Z., et al. (2023). Universal DNA methylation age across mammalian tissues. Nat. Aging 3 (9), 1144–1166. doi:10.1038/s43587-023-00462-6
Lu, A. T., Quach, A., Wilson, J. G., Reiner, A. P., Aviv, A., Raj, K., et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11 (2), 303–327. doi:10.18632/aging.101684
Lu, X., Han, D., Zhao, B. S., Song, C. X., Zhang, L. S., Dore, L. C., et al. (2015b). Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 25 (3), 386–389. doi:10.1038/cr.2015.5
Lu, X., Zhao, B. S., and He, C. (2015a). TET family proteins: oxidation activity, interacting molecules, and functions in diseases. Chem. Rev. 115 (6), 2225–2239. doi:10.1021/cr500470n
Lyon, P., Strippoli, V., Fang, B., and Cimmino, L. (2020). B vitamins and one-carbon metabolism: implications in human health and disease. Nutrients 12 (9), 2867. doi:10.3390/nu12092867
Majamaa, K., Gunzler, V., Hanauske-Abel, H. M., Myllyla, R., and Kivirikko, K. I. (1986). Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J. Biol. Chem. 261 (17), 7819–7823. doi:10.1016/s0021-9258(19)57475-0
McCarty, M. F., Barroso-Aranda, J., and Contreras, F. (2009). The low-methionine content of vegan diets may make methionine restriction feasible as a life extension strategy. Med. Hypotheses 72 (2), 125–128. doi:10.1016/j.mehy.2008.07.044
Mingay, M., Chaturvedi, A., Bilenky, M., Cao, Q., Jackson, L., Hui, T., et al. (2018). Vitamin C-induced epigenomic remodelling in IDH1 mutant acute myeloid leukaemia. Leukemia 32 (1), 11–20. doi:10.1038/leu.2017.171
Minor, E. A., Court, B. L., Young, J. I., and Wang, G. (2013). Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 288 (19), 13669–13674. doi:10.1074/jbc.c113.464800
Mirza, I., Mohamed, A., Deen, H., Balaji, S., Elsabbahi, D., Munasser, A., et al. (2022). Obesity-associated vitamin D deficiency correlates with adipose tissue DNA hypomethylation, inflammation, and vascular dysfunction. Int. J. Mol. Sci. 23 (22), 14377. doi:10.3390/ijms232214377
Myllyla, R., Kuutti-Savolainen, E. R., and Kivirikko, K. I. (1978). The role of ascorbate in the prolyl hydroxylase reaction. Biochem. Biophysical Res. Commun. 83 (2), 441–448. doi:10.1016/0006-291x(78)91010-0
Nguyen, T. T., Tran, T. A., Le, N. Q., Pham, D. M., and Ou, Y. Y. (2022). An extensive examination of discovering 5-methylcytosine sites in genome-wide DNA promoters using machine learning based approaches. IEEE/ACM Trans. Comput. Biol. Bioinforma. 19 (1), 87–94. doi:10.1109/tcbb.2021.3082184
Niklewicz, A., Smith, A. D., Smith, A., Holzer, A., Klein, A., McCaddon, A., et al. (2023). The importance of vitamin B(12) for individuals choosing plant-based diets. Eur. J. Nutr. 62 (3), 1551–1559. doi:10.1007/s00394-022-03025-4
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 (3), 247–257. doi:10.1016/s0092-8674(00)81656-6
Ostrander, E. L., Kramer, A. C., Mallaney, C., Celik, H., Koh, W. K., Fairchild, J., et al. (2020). Divergent effects of Dnmt3a and Tet2 mutations on hematopoietic progenitor cell fitness. Stem Cell Rep. 14 (4), 551–560. doi:10.1016/j.stemcr.2020.02.011
Otani, J., Arita, K., Kato, T., Kinoshita, M., Kimura, H., Suetake, I., et al. (2013). Structural basis of the versatile DNA recognition ability of the methyl-CpG binding domain of methyl-CpG binding domain protein 4. J. Biol. Chem. 288 (9), 6351–6362. doi:10.1074/jbc.m112.431098
Patel, P., Selvaraju, V., Babu, J. R., and Geetha, T. (2023). Association of the DNA methylation of obesity-related genes with the dietary nutrient intake in children. Nutrients 15 (13), 2840. doi:10.3390/nu15132840
Pradhan, S., Bacolla, A., Wells, R. D., and Roberts, R. J. (1999). Recombinant human DNA (Cytosine-5) methyltransferase. J. Biol. Chem. 274 (46), 33002–33010. doi:10.1074/jbc.274.46.33002
Prevention of Neural Tube Defects. (1991). Prevention of neural tube defects: results of the medical research council vitamin study. MRC vitamin study research group. Lancet. 338 (8760), 131–137. doi:10.1016/0140-6736(91)90133-A
Pufulete, M., Al-Ghnaniem, R., Leather, A. J., Appleby, P., Gout, S., Terry, C., et al. (2003). Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124 (5), 1240–1248. doi:10.1016/s0016-5085(03)00279-8
Qi, T., Sun, M., Zhang, C., Chen, P., Xiao, C., and Chang, X. (2020). Ascorbic acid promotes plasma cell differentiation through enhancing TET2/3-mediated DNA demethylation. Cell Rep. 33 (9), 108452. doi:10.1016/j.celrep.2020.108452
Quach, A., Levine, M. E., Tanaka, T., Lu, A. T., Chen, B. H., Ferrucci, L., et al. (2017). Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 9 (2), 419–446. doi:10.18632/aging.101168
Rampal, R., Alkalin, A., Madzo, J., Vasanthakumar, A., Pronier, E., Patel, J., et al. (2014). DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 9 (5), 1841–1855. doi:10.1016/j.celrep.2014.11.004
Rasmussen, K. D., and Helin, K. (2016). Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30 (7), 733–750. doi:10.1101/gad.276568.115
Remely, M., Ferk, F., Sterneder, S., Setayesh, T., Kepcija, T., Roth, S., et al. (2017). Vitamin E modifies high-fat diet-induced increase of DNA strand breaks, and changes in expression and DNA methylation of Dnmt1 and MLH1 in C57bl/6J male mice. Nutrients 9 (6), 607. doi:10.3390/nu9060607
Rossetti, M., Spreafico, R., Saidin, S., Chua, C., Moshref, M., Leong, J. Y., et al. (2015). Ex vivo-expanded but not in vitro-induced human regulatory T cells are candidates for cell therapy in autoimmune diseases thanks to stable demethylation of the FOXP3 regulatory T cell-specific demethylated region. J. Immunol. 194 (1), 113–124. doi:10.4049/jimmunol.1401145
Ryan, M. J., Dudash, H. J., Docherty, M., Geronilla, K. B., Baker, B. A., Haff, G. G., et al. (2010). Vitamin E and C supplementation reduces oxidative stress, improves antioxidant enzymes and positive muscle work in chronically loaded muscles of aged rats. Exp. Gerontol. 45 (11), 882–895. doi:10.1016/j.exger.2010.08.002
Salameh, Y., Bejaoui, Y., and El Hajj, N. (2020). DNA methylation biomarkers in aging and age-related diseases. Front. Genet. 11, 171. doi:10.3389/fgene.2020.00171
Sanderson, S. M., Gao, X., Dai, Z., and Locasale, J. W. (2019). Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat. Rev. Cancer 19 (11), 625–637. doi:10.1038/s41568-019-0187-8
Sant, D. W., Camarena, V., Mustafi, S., Li, Y., Wilkes, Z., Van Booven, D., et al. (2018). Ascorbate suppresses VEGF expression in retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 59 (8), 3608–3618. doi:10.1167/iovs.18-24101
Schutsky, E. K., DeNizio, J. E., Hu, P., Liu, M. Y., Nabel, C. S., Fabyanic, E. B., et al. (2018). Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotechnol. 36, 1083–1090. doi:10.1038/nbt.4204
Sedivy, J. M., Banumathy, G., and Adams, P. D. (2008). Aging by epigenetics--a consequence of chromatin damage? Exp. Cell Res. 314 (9), 1909–1917. doi:10.1016/j.yexcr.2008.02.023
Shibutani, T., Ito, S., Toda, M., Kanao, R., Collins, L. B., Shibata, M., et al. (2014). Guanine- 5-carboxylcytosine base pairs mimic mismatches during DNA replication. Sci. Rep. 4, 5220. doi:10.1038/srep05220
Smithells, R. W., Sheppard, S., and Schorah, C. J. (1976). Vitamin dificiencies and neural tube defects. Archives Dis. Child. 51 (12), 944–950. doi:10.1136/adc.51.12.944
Suga, K., Suto, A., Tanaka, S., Sugawara, Y., Kageyama, T., Ishikawa, J., et al. (2023). TAp63, a methotrexate target in CD4+ T cells, suppresses Foxp3 expression and exacerbates autoimmune arthritis. JCI Insight. 8 (10), e164778. doi:10.1172/jci.insight.164778
Taeubert, M. J., de Prado-Bert, P., Geurtsen, M. L., Mancano, G., Vermeulen, M. J., Reiss, I. K. M., et al. (2022). Maternal iron status in early pregnancy and DNA methylation in offspring: an epigenome-wide meta-analysis. Clin. Epigenetics 14 (1), 59. doi:10.1186/s13148-022-01276-w
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y., et al. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324 (5929), 930–935. doi:10.1126/science.1170116
Taira, A., Palin, K., Kuosmanen, A., Valimaki, N., Kuittinen, O., Kuismin, O., et al. (2023). Vitamin C boosts DNA demethylation in TET2 germline mutation carriers. Clin. Epigenetics 15 (1), 7. doi:10.1186/s13148-022-01404-6
Tong, X., Zhao, F., and Thompson, C. B. (2009). The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 19 (1), 32–37. doi:10.1016/j.gde.2009.01.002
Trautmann, C., Bock, A., Urbach, A., Hübner, C. A., and Engmann, O. (2020). Acute vitamin B12 supplementation evokes antidepressant response and alters Ntrk-2. Neuropharmacology 171, 108112. doi:10.1016/j.neuropharm.2020.108112
Wang, L., Zhou, Y., Xu, L., Xiao, R., Lu, X., Chen, L., et al. (2015a). Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex. Nature 523 (7562), 621–625. doi:10.1038/nature14482
Wang, T., Chen, K., Zeng, X., Yang, J., Wu, Y., Shi, X., et al. (2011). The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9 (6), 575–587. doi:10.1016/j.stem.2011.10.005
Wang, X., Guan, Z., Chen, Y., Dong, Y., Niu, Y., Wang, J., et al. (2015b). Genomic DNA hypomethylation is associated with neural tube defects induced by methotrexate inhibition of folate metabolism. PloS one 10 (3), e0121869. doi:10.1371/journal.pone.0121869
Wilkinson, G. S., Adams, D. M., Haghani, A., Lu, A. T., Zoller, J., Breeze, C. E., et al. (2021). DNA methylation predicts age and provides insight into exceptional longevity of bats. Nat. Commun. 12 (1), 1615. doi:10.1038/s41467-021-21900-2
Williams, K., Christensen, J., Pedersen, M. T., Johansen, J. V., Cloos, P. A., Rappsilber, J., et al. (2011). TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473 (7347), 343–348. doi:10.1038/nature10066
Wu, H., D'Alessio, A. C., Ito, S., Wang, Z., Cui, K., Zhao, K., et al. (2011). Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 25 (7), 679–684. doi:10.1101/gad.2036011
Wu, M. J., Kim, M. R., Chen, Y. S., Yang, J. Y., and Chang, C. J. (2017). Retinoic acid directs breast cancer cell state changes through regulation of TET2-PKCζ pathway. Oncogene 36 (22), 3193–3206. doi:10.1038/onc.2016.467
Wu, X., and Zhang, Y. (2017). TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 18 (9), 517–534. doi:10.1038/nrg.2017.33
Xia, B., Han, D., Lu, X., Sun, Z., Zhou, A., Yin, Q., et al. (2015). Bisulfite-free, base-resolution analysis of 5-formylcytosine at the genome scale. Nat. Methods 12 (11), 1047–1050. doi:10.1038/nmeth.3569
Xu, Y. P., Lv, L., Liu, Y., Smith, M. D., Li, W. C., Tan, X. M., et al. (2019). Tumor suppressor TET2 promotes cancer immunity and immunotherapy efficacy. J. Clin. Investigation 129 (10), 4316–4331. doi:10.1172/jci129317
Xue, J. H., Chen, G. D., Hao, F., Chen, H., Fang, Z., Chen, F. F., et al. (2019). A vitamin-C-derived DNA modification catalysed by an algal TET homologue. Nature 569 (7757), 581–585. doi:10.1038/s41586-019-1160-0
Yadav, D. K., Shrestha, S., Lillycrop, K. A., Joglekar, C. V., Pan, H., Holbrook, J. D., et al. (2018). Vitamin B(12) supplementation influences methylation of genes associated with Type 2 diabetes and its intermediate traits. Epigenomics 10 (1), 71–90. doi:10.2217/epi-2017-0102
Yin, R., Mao, S. Q., Zhao, B., Chong, Z., Yang, Y., Zhao, C., et al. (2013). Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 135 (28), 10396–10403. doi:10.1021/ja4028346
Yu, M., Hon, G. C., Szulwach, K. E., Song, C. X., Zhang, L., Kim, A., et al. (2012). Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149 (6), 1368–1380. doi:10.1016/j.cell.2012.04.027
Yue, X., Samaniego-Castruita, D., Gonzalez-Avalos, E., Li, X., Barwick, B. G., and Rao, A. (2021). Whole-genome analysis of TET dioxygenase function in regulatory T cells. EMBO Rep. 22 (8), e52716. doi:10.15252/embr.202152716
Zappe, K., Pointner, A., Switzeny, O. J., Magnet, U., Tomeva, E., Heller, J., et al. (2018). Counteraction of oxidative stress by vitamin E affects epigenetic regulation by increasing global methylation and gene expression of MLH1 and DNMT1 dose dependently in caco-2 cells. Oxidative Med. Cell. Longev. 2018, 1–13. doi:10.1155/2018/3734250
Zhang, H., Zhang, X., Clark, E., Mulcahey, M., Huang, S., and Shi, Y. G. (2010). TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 20 (12), 1390–1393. doi:10.1038/cr.2010.156
Zhang, X., Su, J., Jeong, M., Ko, M., Huang, Y., Park, H. J., et al. (2016). DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 48 (9), 1014–1023. doi:10.1038/ng.3610
Keywords: DNA methylation, Ten-eleven translocation (TET) enzymes, 5-hydroxymethylation, vitamin A, vitamin B, vitamin C
Citation: Leesang T, Lyon P, Pinzone J and Cimmino L (2024) Micronutrient regulation of the DNA methylome. Front. Epigenet. Epigenom. 2:1409355. doi: 10.3389/freae.2024.1409355
Received: 29 March 2024; Accepted: 15 April 2024;
Published: 26 April 2024.
Edited by:
Andre J. van Wijnen, University of Vermont, United StatesReviewed by:
Hu Wang, Hangzhou Normal University, ChinaCopyright © 2024 Leesang, Lyon, Pinzone and Cimmino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luisa Cimmino, bHVpc2EuY2ltbWlub0BtZWQubWlhbWkuZWR1
 Tiffany Leesang
Tiffany Leesang Peter Lyon
Peter Lyon Joey Pinzone1,2
Joey Pinzone1,2 Luisa Cimmino
Luisa Cimmino