- 1Diagnostics Wildlife and Molecular Biology Department, SASA, Edinburgh, United Kingdom
- 2Syngenta, Jealott’s Hill International Research Centre, Bracknell, United Kingdom
- 3Acarology Lab, Ecology and Evolutionary Biology Section, School of Biological Sciences, University of Reading, Reading, United Kingdom
Phytoseiidae predatory mites are used as biological control agents in glasshouse environments to control pestiferous mites and insects. The influence of the microbiomes on these mites within mass-reared facilities is largely unknown; this study aims to improve our understanding of the Phytoseiidae microbiome. We used Sanger sequencing and traditional culturing methods to characterise the dominant species within the external and internal bacterial microbiotas of three commercially reared predatory mites: Phytoseiulus persimilis, Typhlodromips (=Amblyseius) swirskii, and Neoseiulus (=Amblyseius) cucumeris. The effects of selected bacterial taxa on the survival, voracity, and oviposition of P. persimilis under lab conditions were also examined. A total of 34 different OTUs were found from three phyla (Actinobacteria, Proteobacteria, and Firmicutes), 12 families, and 22 genera. N. cucumeris had 17 OTUs, compared to 15 from P. persimilis and 12 from T. swirskii. Significant differences were found between internal and external microbiota for each mite species, suggesting functional differences. Principal component analysis (PCA) of bacterial diversity between mite species showed that T. swirskii and N. cucumeris microbiotas were more similar, compared to P. persimilis. This could be due to the characteristics of the environment in which N. cucumeris and T. swirskii are reared, and the use of factitious Astigmata prey. Phytoseiidae microbiotas were found to contain bacterial taxa previously isolated from Astigmata mites. Also, Acaricomes phytoseiuli was isolated from the integument of P. persimilis and T. swirskii; this bacterium has previously been suggested as a pathogen of P. persimilis. Bacteria from gut extracts and body surfaces were cultured and selected taxa were inoculated onto P. persimilis within controlled leaf disc experiments. Of the seven bacterial species tested, Serratia marcescens significantly reduced mite survival, voracity, and oviposition; however, a closely related species, Serratia odorifera, significantly increased mite voracity. Mites showed the highest survival rates and highest mean ages at death in the presence of Elizabethkingia sp. and Staphylococcus kloosii, but these results were not significantly different to the control. The implications of these findings are discussed in the context of benefits to mass rearing and the health of predatory mites in these systems.
1 Introduction
The Phytoseiidae (Acari: Mesostigmata) are a family of fast-moving predatory mites that are found in upper soil layers and on the aerial parts of plants. They feed mainly on microscopic arthropods, particularly other mites, but some species additionally feed on nematodes, fungi, pollens, and plant exudates (Gerson et al., 2003). Phytoseiidae mites have attracted attention due to their use as biological control agents of ubiquitous pests such as spider mites (Gerson et al., 2003; Zhang, 2003), thrips (Chow et al., 2010), and whiteflies (Messelink et al., 2008). They are also able to control many other species of pestiferous mites such as broad mites (van Maanen et al., 2010), cassava green mite, citrus mites, and avocado mites (Vásquez et al., 2023). Phytoseiidae mites continue to attract interest due to their potential role in biological control, especially where alternatives to chemical control are needed (Vásquez et al., 2023). Described Phytoseiidae species total 2,692 within 102 genera and 3 subfamilies (Prasad, 2012); however, less than 10% of species are considered to be well studied in terms of their biological control potential (Kostiainen and Hoy, 1996). Certain species of the Phytoseiidae are well suited for use in biological control as they have high voracity; maintain the prey at low densities; can be efficiently mass-reared, stored, and transported; have fast reproductive and developmental rates; and have a female-biased sex ratio comparable to their prey around 2.5:1 (Hoy, 2011).These attributes allow them to respond to and control a sudden increase in prey density.
The most widely used species come from approximately eight different genera, namely, Neoseiulus, Typhlodromus, Phytoseiulus, Galendromus, Euseius, Amblyseius, Iphiseius, and Typhlodromips. This project focused on three Phytoseiidae species widely used in biological control mainly in glass houses. They are Phytoseiulus persimilis Athias-Henriot, Neoseiulus (=Amblyseius) cucumeris (Oudemans), and Typhlodromips (=Amblyseius) swirskii (Oudemans).
In 1966, P. persimilis became the first commercially available biological control agent (Van Lenteren and Woets, 1988). It is a “type I” specialised predator of Tetranychus species and prefers to complete its life cycle on and around the intense webbing produced by spider mites (McMurtry and Croft, 1997). This mite is an obligate predator that will even cannibalise its own eggs and immatures or exhibit intraguild predation (McMurtry and Croft, 1997; Waltzer et al., 2001; Cakmak et al., 2006; Hoy, 2011). This behaviour can hinder efforts of integrated pest management with multiple Phytoseiidae species and often sees populations of generalist mites become dominant. Additionally, mass rearing of this mite species must be performed in a tri-trophic system, where predators can feed on Tetranychus species reared on beans (Phaseolus vulgaris) or other suitable host plants (Waltzer et al., 2001). Neoseiulus cucumeris and Typhlodromips swirskii are type III generalist predators mainly used for controlling thrips. The potential for mass production of Neoseiulus cucumeris and its use as a biological control agent for Thrips tabaci on greenhouse cucumber was outlined in 1983 (Ramakers, 1983). Contemporarily, it is used to control three species of thrips, Frankliniella occidentalis (Pergande), T. tabaci (Linderman), and Scirtothrips dorsalis (Hood) in greenhouses and open fields in Europe (De Courcy Williams, 2001; Wimmer et al., 2008), Northern Africa (El-Kholy and El-Saiedy, 2009), North America (Van Driesche et al., 2006), and Australia (Manners et al., 2013). Under certain circumstances T. swirskii exhibits superior capacity for biological control of thrips compared to N. cucumeris (Van Houten et al., 2005; Messelink et al., 2006). It is also an effective predator of two species of whitefly Bemisia tabaci (Hoogerbrugge et al., 2005; Fouly et al., 2011) and Trialeurodes vaporariorum (Messelink et al., 2008) and can be used to control outbreaks of these pests on economically important vegetable crops and ornamentals. In mass-rearing facilities, N. cucumeris and T. swirskii can be fed on factitious Astigmata prey mites (Bolckmans and van Houten, 2006; Smytheman, 2011; Midthassel et al., 2013). The predatory mites and Astigmata mites are reared in containers with substrate such as bran or wheat husks, which provides a larger area for prey and predator proliferation, serving as food for the Astigmata preys and allowing dissipation of gasses and metabolic heat.
The importance of Phytoseiidae as effective bio-control agents of numerous economically damaging pests has encouraged research on their biology and ecology, including physiology, reproduction, genetics, behaviour, disease, and applications in biological control. However, little is known about the intrinsic microbiota that exists on Phytoseiidae species. This is especially true for ectosymbionts, which are beneficial symbionts that live on the integument and within the digestive tract of the Phytoseiidae (Sumner-Kalkun et al., 2020). Ectosymbionts provide a link between the mites and their habitat or environment and are vital in influencing the interactions between mite host and the rearing environment of the three species examined in the current study.
In general, for the acari, intrinsic microbiotas have been characterised and analysed in Astigmata, Prostigmata, Mesostigmata, Trombidiiformes, and Oribatida, and the associations provide a number of beneficial roles, similar to those already described from other animal groups. These roles include prolonging the viability of sperm and improving reproduction in Ornithodorus ticks (Perotti and Braig, 2011); providing nutritional benefits in Trichouropoda spp. (Roets et al., 2007; Roets et al., 2011), Tarsonematidae (Bridges and Moser, 1983; Moser, 1985), Astigmata (Smrž et al., 1991; Martinez-Giron et al., 2009; Hubert et al., 2011; Hubert et al., 2012; Erban et al., 2016b; Erban et al., 2016a; Pekas et al., 2017), and Oribatidae (Smrž and Trelová, 1995; Moquin et al., 2012); and conferring pesticide resistance in Tetranychus urticae (Yoon et al., 2010). Examples of morphological adaptations in Tarsonematus mites suggests co-evolution between host and ectosymbionts (Moser, 1985; Roets et al., 2011). Previous studies of microbes associated with specific parts of the Phytoseiidae anatomy showed that microfungi-like organisms and bacteria were commonly found on these mites and bacteria and microsporidia-like organisms were also found in the digestive tract and malphigian tubules of these mites (Sumner-Kalkun et al., 2020).
The most widely studied bacterial associations within Phytoseiidae are from endosymbiotic Rickettsia, Wolbachia, Spiroplasm, and Cardinium (Enigl and Schausberger, 2007; Schütte and Dicke, 2008). Wolbachia are serial endosymbionts of many arthropods and have been found to infect many species and populations of Phytoseiidae mites (Johanowicz and Hoy, 1999). They can have detrimental effects on their host by manipulating genetic systems causing reproductive incompatibility between infected and non-infected individuals, as in Metaseiulus occidentalis (Johanowicz and Hoy, 1998; Hoy and Jeyaprakash, 2005; Hoy and Jeyaprakash, 2008). Wolbachia has been found in seven different species of Phytoseiidae, but its effects on species other than M. occidentalis are only recently elucidated (Schütte and Dicke, 2008). Enigl et al. (2005) attempted to isolate Wolbachia from seven different strains of P. persimilis from the USA, Europe, and Africa, but all were negative. These first attempts originally suggested that Wolbachia did not play a significant role in the biology of this host mite and is probably acquired from prey mite via feeding (Wu and Hoy, 2012b). However, the endosymbiont Cardinium was found to have a closer association to the predatory mite M. occidentalis and is likely the cause of reproductive incompatibility (Wu and Hoy, 2012a).
Mite microbiotas have been shown to be significantly different in terms of diversity and function depending on the life stage of the mite, as in eggs vs. adults of T. urticae (Merlin et al., 2022); however, different host plant environments did not significantly influence diversity and functional aspects of T. urticae and Neoseiulus californicus (McGregor) (Merlin et al., 2022); this suggests that both prey and predatory mites are dependent on their distinct microbiotas for survival.
In one of the few studies of microbes associated with Phytoseiidae that did not focus on endosymbionts or pathogens, examination of the microbes within the gut of M. occidentalis uncovered two species of bacteria that were new to the Phytoseiidae microbiota (Hoy and Jeyaprakash, 2008). One was related to an Enterobacter sp. also found in gypsy moths and the sheep scab mite Psoroptes ovis, and the other to a Sulcia sp. found in the sharpshooter Homalodisca vitripennis (Hemiptera: Cicadellidae). Although their locations were never verified and their effects on the host were never proven, these bacteria were posed to be nutritional symbionts of their host.
In the current study, using Sanger sequencing methods and traditional microbiology approaches, the bacterial diversity of the gut and integument of three economically important Phytoseiid species were characterised and compared. Furthermore, the effects of selected bacterial cultures on the survival, voracity, and oviposition of P. persimilis were examined to identify potential beneficial and pathogenic isolates. This study makes initial contributions towards characterisation of the mass-reared Phytoseiidae bacterial microbiota to understand its diversity and effects on its host species. Particular attention is given to bacterial species that can potentially be manipulated to improve the health of phytoseiid mites within mass-rearing systems or those that can cause disease and reduced performance of the predatory mite.
2 Materials and methods
2.1 Mite rearing, collection of samples, and iso-female lines
Mite specimens of P. persimilis, N. cucumeris, and T. swirskii were obtained from live colonies from “Biological Crop Protection Ltd.” and preserved in 100% ethanol and stored in 1.5-ml microcentrifuge tubes at 2–4°C for subsequent DNA extraction. For N. cucumeris and T. swirskii, the prey diet was bran-based with 1% w/w wheatgerm, a moisture content of ca. 10%. Predator–prey chambers were housed at 23°C ( ± 3) and 65% RH. Starter cultures were renewed every month. The prey used for N. cucumeris was Tyrophagous putrescentiae and the prey used for T. swirskii was Suidasia medanensis. P. persimilis was reared on T. urticae infested dwarf bean plants (var. Prince) grown under glasshouse conditions.
Laboratory stock colonies of prey (Tetranychus urticae) were reared on bean leaves (Phaseolus vulgaris) under a 16:8 light:dark cycle at 20°C ( ± 2). Live batches of P. persimilis were received from Biological Crop Protection Ltd. and used to set up an iso-female colony, 2 weeks before the start of bioassays.
Iso-female colonies of P. persimilis were produced using a single gravid female to reduce genetic variation within experiments. Females were placed singly on 2-cm-diameter leaf disks on cotton wool moistened with sterile distilled deionised water to prevent escape and supplied with 15 adult and 10 juvenile T. urticae, ad libitum. After 3 days of oviposition, eggs were transferred to a fresh bean leaf with all stages of T. urticae and reared to adult. Mated females of around 7–9 days were used in bioassays measuring survivorship, voracity, oviposition rates, and fecundity.
2.2 DNA extraction from mites for microbiota characterisation
Bacteria were isolated in situ from the surface of the mite’s integument and the digestive tract, independently. Bacteria from the integument were obtained via a unique method using chemical disruption of bacterial cells followed by ultrasonic treatment. Mites were placed into 10 μl of molecular-grade H2O in a 1.5-ml microcentrifuge tube and left for 30 min to rehydrate. Twenty mites of each species were used to ensure adequate copy numbers of bacterial DNA. Samples were centrifuged at 13,000 rpm for 2 min at room temperature to ensure full submersion in water. Triton X-100 detergent (0.2 μl) was added to each tube and vortexed for a few seconds. Detergent was used to disrupt bacterial cell membranes and break their attachment on the surface of the mite. Samples were centrifuged at 13,000 rpm for 2 min at room temperature (RT) to ensure submersion in the water-detergent mixture. Samples were sonicated using an ultrasonic bath (Sonicor SC-121TH, Copiague, NY, USA) for 3 min at 50 kHz and RT to dislodge bacteria from the surface of the mite. Samples were returned to the centrifuge at 13,000 rpm for 2 min. Water from the sample was decanted into a fresh 1.5-ml microcentrifuge tube ensuring that the whole mite was not carried over into the new tube. Via this method, it is possible to remove bacteria from the surface of the mite for DNA extraction.
For gut extractions, 20 mites were placed into 20 μl of molecular-grade H2O and rehydrated at room temperature for 1 h. Samples were then washed using 0.2 μl of Triton X-100 and sonicated for 3 min at 50 kHz. Water was then removed from the tube using a pipette ensuring mites remained in the tube. The tube containing the mites was then rinsed three times with 500 μl of molecular-grade H2O. Water was removed from the tube and 10 μl of buffer ATL (Qiagen “DNeasy Blood and Tissue kit”, Hilden, Germany) was added. Mites were macerated inside the tubes using sterile pipette tips and samples were composed of digestive caeca, Malpighian tubules, and anal atria of the mites. Buffer ATL (70 μl) was added to the tube. All mite extracts were subsequently extracted and purified using the “DNeasy Blood and Tissue kit” (QIAGEN, Hilden, Germany).
Extraction controls were also included to account for contamination from kit reagents and plastic ware. No template controls were included to account for contamination in water and PCR reagents. Extraction controls and no-template control were subject to the PCR protocol described below and only samples showing no contamination in no-template controls and extraction controls were taken forward for sequencing. This was checked via electrophoresis gel and absence of amplified DNA in those samples.
2.3 Culturing bacteria from Phytoseiidae extracts
Bacteria were cultured from mite integuments and alimentary tracts. For digestive tracts, mites were collected from live colonies supplied by “Certis Europe BV” and placed into sterile water inside “lysing matrix A” tubes (MP Biomedicals, Santa Ana, USA) containing ¼-inch ceramic spheres. For bacteria cultured from alimentary tracts, a combination of chemical sterilisation and microdissection was necessary. Mites were initially placed at 4°C for 30 min, then surface sterilised in 100% ethanol for 10 s and rinsed in molecular-grade water before being placed onto sterile microscope slides. The edge of a cover slip was pressed against the mite, causing the internal organs to rupture out of the body. In most cases, the digestive caeca, Malpighian tubules, and anal atria were visible due to their darker colour. These organs were then lifted from the slide with a sterile tungsten micropin or microloop and placed into “lysing matrix A” tubes for homogenisation. Samples were homogenised at 5.0 M/s for 6 min for whole mites and 2 min for dissected mite alimentary tracts, in a “FastPrep-24” machine (MP Biomedicals, Santa Ana, USA). Dilution series of each sample were prepared up to dilutions of 10−6 and 100 µl of each was spread onto LB Agar containing cycloheximide to suppress the growth of fungi. Control plates were spread with 100 µl of sterile water to ensure no contamination was present. Bacteria from the integuments of the mites were cultured by allowing mites to walk on LB agar plates for 30 min and then removed carefully from the agar surface. Mites were applied and removed from plates using a sterile brush or micropin.
All plates were incubated for at least 3 days under a 16:8 light/dark cycle at 20–25°C to simulate the conditions of the mite colonies. Colonies were differentiated using morphological differences observed from examination of the colony on the plate, i.e., colour, colony shape, size, and texture. Pure cultures were obtained by streaking single colonies onto plates at least three subsequent times until only one type of colony grew on the plate. Sterile technique was used during preparation of bacterial cultures. Pure cultures were grown in sterile LB broth overnight at 25°C; 1 ml of that broth was added to 1 ml of glycerol (50%) in cryo-vials and placed at −80°C for long-term storage.
2.4 DNA barcoding and clone library preparation of bacterial microbiota
Bacterial DNA in extracted and purified mite samples was amplified using PCR for use in cloning and sequencing. Genomic DNA of bacteria was amplified using universal primers for the 16S ribosomal gene (U16S). Forward primers 27f (5′-AGA GTT TGA TCC TGG CTC AG) and 63f (5′-CAG GCC TAA CAC ATG CAA GTC), in conjunction with 1387r (5′-GGG CGG WGT GTA CAA GGC), were used to amplify 16 rRNA gene sequences for identification of bacterial OTUs. Reactions contained the following reagents:
- 3 µl of DNA sample
- 12.5 µl of Go-taq green Master Mix (Promega, Madison, USA)
- 0.5 µl of each primer (63f and 1387r) or (27f and 1387r)
- 8.5 µl molecular- grade H2O.
Two PCR temperature cycles were used for different DNA extraction samples:
1- Temperature cycle for amplification of bacterial DNA from cultured bacterial colonies:
- 95°C for 5 min
- 35× cycle of 95°C for 30 s; 54°C for 30 s; 72°C for 90 s
- Final extension, 72°C for 6 min.
2- Temperature cycle for amplification of bacterial DNA from culture independent extractions:
- 95°C for 5 min
- 35× cycle of: 95°C for 30 s; ramp to 54°C; 54°C for 30 s; ramp to 72°C; 72°C for 90 s
- Final extension, 72°C for 10 min.
Successful reactions were verified using gel electrophoresis in 1% agarose gels stained using ethidium bromide and visualised in a “Biodoc-it imaging system” (UVP, Upland, USA) benchtop UV trans-illuminator. Positive reactions showed bands at ~1,300 bp. Positive reactions were purified with the “QIAquick PCR purification kit” (Qiagen, Hilden, Germany) and DNA quantities were measured with a “NanoDrop Lite spectrophotometer” (Thermo Scientific, Waltham, USA).
Purified DNA from bands was cloned into competent E. coli cells using the “Strataclone PCR Cloning Kit” (Stratagene, San Diego, USA). Bands were ligated into pSC-A-amp/kan cloning vector and transformed into “Strataclone solopack competent cells” (Stratagene, San Diego, USA). Ligation reactions were performed for 2 h to increase the chance of obtaining positive inserts. After transformation, cells were plated on LB agar containing ampicillin (10 mg/ml) and 2% x-gal. At least 20 white bacterial colonies were picked from each plate and verified for positive inserts using PCR with t3 and t7 primers flanking the insert region. Colonies carrying the correct size insert (~1,300) were cultured overnight in 4 ml of LB broth containing ampicillin (10 mg/ml) at 37°C. Plasmid DNA was recovered from bacterial cells using an “E.Z.N.A Plasmid Mini Kit I-Spin Protocol” (Omega Bio-tek, Georgia, USA). Samples were sent to Macrogen Europe for Sanger sequencing.
Almost entire 16S ribosomal gene DNA strands were obtained by assembling forward and reverse strands for each sample sequence using the “CAP contig assembly program” in “Bioedit” software. Raw sequences were processed in “Geneious” to remove low-quality regions of the sequencing read. Similar or identical sequences from different samples were aligned using “ClustalW multiple alignment”. Sequences were then checked manually for errors or artefacts of sequencing and edited accordingly. The final edited sequences were submitted to BLASTn similarity searches (Altschul et al., 1990) for closest known matches in the GenBank sequence database. Bacterial taxa were identified using “RDP Naive Bayesian rRNA Classifier Version 2.8” (Wang et al., 2007) to complement and check identities recovered using BLASTn. Species were deemed the same as their closest match in the case that genera were not known to be problematic with regard to 16S rDNA identification; sequence similarity was ≥99% and sequences were at least 500 bp in length, but preferably ≥1,300 (Janda and Abbott, 2007).
2.5 Bioassays to examine the influence of Phytoseiidae bacterial microbiota
Five bacterial cultures from the Phytoseiidae microbiota isolated in this study were selected for further study into their effects on P. persimilis in leaf disc bioassays (Table 1).
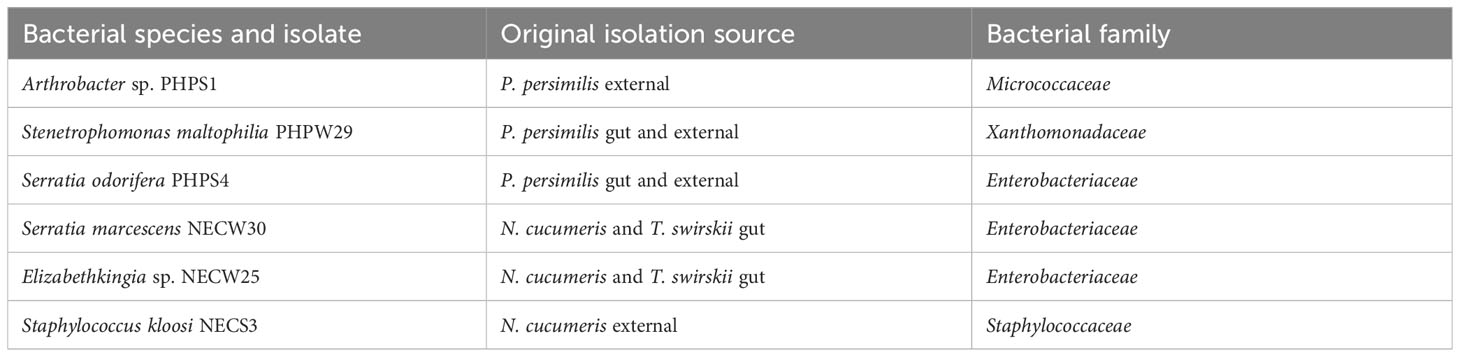
Table 1 Isolation source of each bacterial culture used in leaf-disc bioassays with P. persimilis. [external: external integument].
Individual adult female mites were observed on test arenas treated with one of six bacterial cultures or only sterile water for the control. To set up test arenas, sterile 5-cm3 receptacles were used to contain cotton wool moistened with sterile water on which a 2-cm2 leaf disk cut from primary leaves of Phaseolus vulgaris was placed with the adaxial surface face up. Prior to use in test arenas, leaves were washed in 0.5% sodium hypochlorite for 1 min and rinsed three times with sterile water. Bacterial inoculations were made by removing approximately 0.1 g of bacterial cells from LB agar plates and resuspending them in 1 ml of sterile water. Each leaf arena was then supplied with either an inoculation of one of the six different bacteria in 1 ml of sterile water or the same volume of only sterile water containing no bacteria. Leaf disks were allowed to dry then supplied with prey, 5 adults and 10 juveniles of T. urticae; these were allowed to feed on the leaf matter for at least 30 min before the addition of predatory mites. Female P. persimilis of 7–9 days of age from iso-female colonies were applied singly to each prepared leaf arena supplied with prey. Predatory mite activity was then monitored each day over the next 7 days recording the number of prey eaten (adults and juveniles), the number of eggs laid, and the number of mites that died; any unusual symptoms were also recorded. Prey were replenished every 2 days or as otherwise appropriate so that mites consistently had access to fresh prey mites. Leaf disks were replaced after 4 days of recording to ensure prey had ample leaf matter to feed on, and new leaf disks were subjected to the same method of preparation as outlined above depending on treatment type. Leaf arenas were kept and prepared in a controlled environment of 16:8 light:dark cycle at 20°C (± 2°C) and were kept within a sterile Captair Pyramid (Erlab, Massachusetts, USA) to prevent contamination with microbes from the laboratory environment. Counts and observations were made using a “Leica Zoom 2000” dissecting microscope at 30× magnification in a laminar flow cabinet (Bassaire P5H), and when test arenas were transported, they were protected from external contamination by placing a sterile 5-cm3 plastic container over the top of the rearing receptacle. The following equation was used for calculation of voracity scores:
Counts of the number of predators that died on a particular day were recorded for survivorship analysis. For control mites vs. bacterial culture treatment, voracity scores were analysed using one-way ANOVA and mean voracity was compared using Tukey’s test with p-values adjusted using Bonferroni’s correction. Oviposition rates were analysed using one-way analysis of deviance on generalised linear models fitted to Gaussian error distributions. For analysis of survivorship data, survival curves for each treatment were produced using the Kaplan–Meier formula and models were compared using Chi-squared goodness of fit to show significant differences between groups and ANOVA for model simplification. All statistical analyses were performed in “R” (R core development team, 2012). Libshuff analyses, describing and comparing community structures, were also applied.
3 Results
3.1 Characterisation and comparisons of the Phytoseiidae microbiota
From a total of 170 sequences, 34 different OTUs were found from three phyla (Actinobacteria, Proteobacteria, and Firmicutes), 12 families, and 22 genera. N. cucumeris had the most diverse microbiota with 17 OTUs, compared to 15 from P. persimilis and 12 from T. swirskii. Numbers of OTUs from each species and each isolation location are shown in Table 2. The groups of bacteria making up the Phytoseiidae microbiota were Actinomycetes (15.10%), Alpha-proteobacteria (11.32%), Beta-proteobacteria (26.42%), Gamma-proteobacteria (30.20%), Bacilli (9.43%), and Bacteriodota (7.54%). The predominant OTUs in all phytoseiids were from Proteobacteria making up 68.75% in P. persimilis, 73.91% in N. cucumeris, and 57.14% in T. swirskii. Genera were shared across taxa, being present in all species or on only two of the examined species (Figure 1). Other bacterial isolates were unique to their host and even unique to a specific anatomy. Generally, the integument and gut showed a similar proportion of unique OTUs per mite, apart from in N. cucumeris where the integument made up a higher percentage of unique OTUs compared to the gut (Table 2). The total number of bacteria isolated from the integument of all the Phytoseiidae species comprised 38.24% of the unique bacterial diversity present in the samples, whereas gut isolates comprised 32.35% of the total unique bacterial diversity (Table 2). The Phytoseiidae mites examined here shared a core microbiota of 32.35% of the same OTUs found in at least two species (Supplementary Table 1). The most abundant OTUs from N. cucumeris and T. swirskii were from Serratia sp. and Elizabethkingia sp., whereas the most abundant OTUs from P. persimilis were Stenetrophomonas sp., Serratia sp., and Achromobacter sp. The same OTU was never found in all three mite species. OTUs closely matching Serratia spp. were isolated from all species and body regions (Supplementary Table 1). Culture-dependent and -independent methods of bacterial isolation yielded 26 and 17 OTUs respectively, with some OTUs being detected via both methods (Supplementary Table 1).
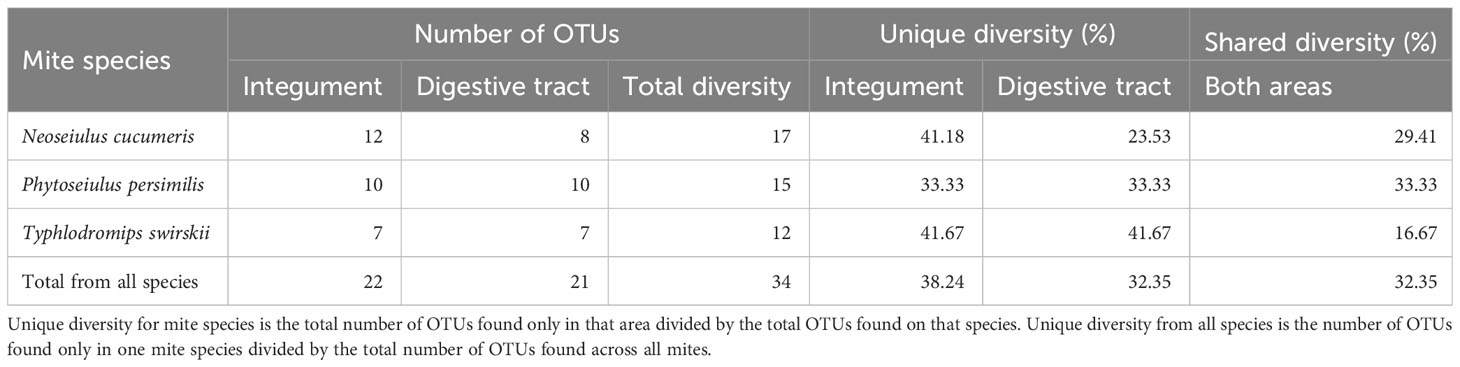
Table 2 A breakdown of total OTUs found in different parts of the anatomy and unique bacterial diversity isolated from those areas.
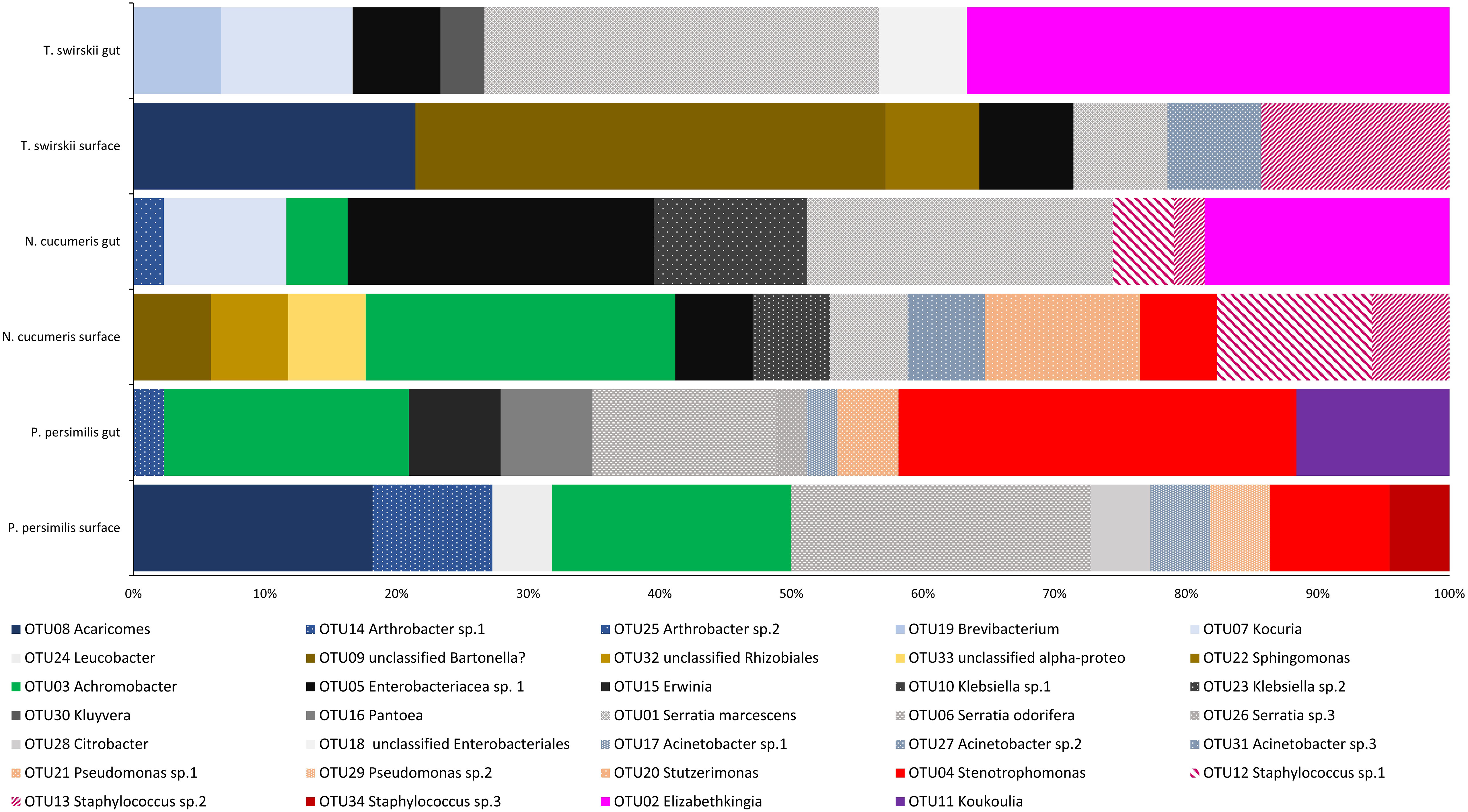
Figure 1 Bacterial diversity (OTUs) across the microbiota of three different species of Phytoseiidae comparing gut and surface-isolated bacteria. OTUs are weighted with relative abundance (number of sequences of each isolate obtained).
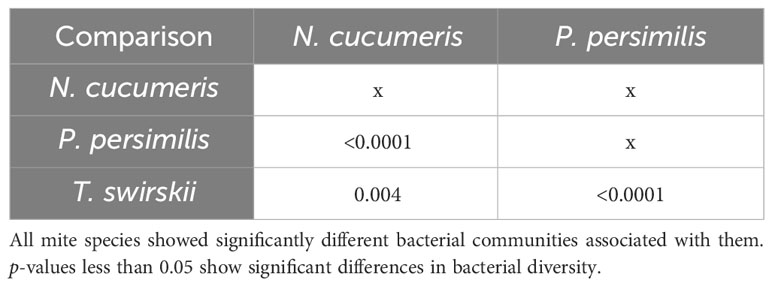
Libshuff analysis used to analyse differences in the total bacterial communities found in mite species showed that diversity was significantly different between species (Table 3). Using different methods of isolation of bacterial DNA enabled characterisation of specific anatomies to assess the differing bacterial diversity associated with these different areas of the mites. The microbiota of the integument and gut differed, showing distinct bacterial communities belonging to each region (Figure 1 and Table 4). Significant differences between the gut microbiota and external microbiota of each species were also found (Table 4). Between-species analyses showed significant differences in all but two cases: the comparison between the gut microbiota (p-value = 0.205) and external microbiota (p-value = 0.1745) of N. cucumeris and T. swirskii.
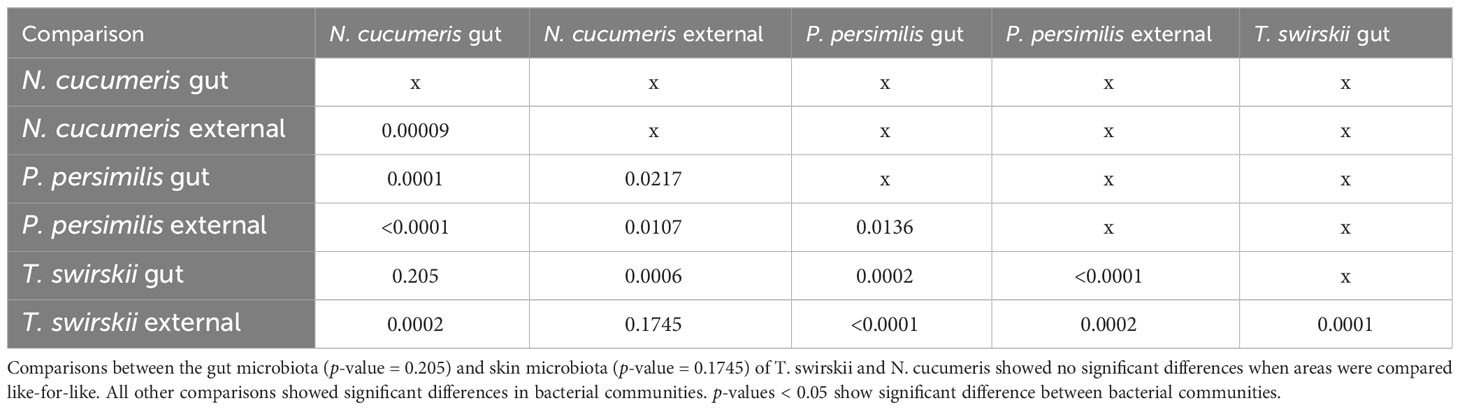
Table 4 Libshuff analysis of bacterial microbiota comparing different species and different anatomies (gut and external microbiota).
The microbiota of the alimentary tract, including the digestive caeca, anal atrium, and Malpighian tubules, was characterised by bacteria from the Enterobacteriaceae and each mite species had at least one genus belonging to this family within their alimentary tract. Comparatively, the microbiota of the integument of each mite species was composed of at least one species of Staphylococcaceae. Bacteria from the genera Achromobacter, Arthrobacter, Pseudomonas, Serratia, Sphingomonas, and Stenetrophomonas were isolated from both the integument and alimentary tracts of mites examined (Figure 1). One OTU isolated from the surface of N. cucumeris and T. swirskii showed high similarity to a Bartonella sp. recently found in Astigmata mites (Supplementary Table 1).
Analysis of bacterial diversity with principal component analysis (PCA) showed which variables best described the variation in the samples. The variables included in this analysis were species and area of isolation, comparing whole microbiotas, external microbiotas and gut microbiotas. Figure 2 shows loading plots of individual OTUs and biplots of different variables represented by arrows.
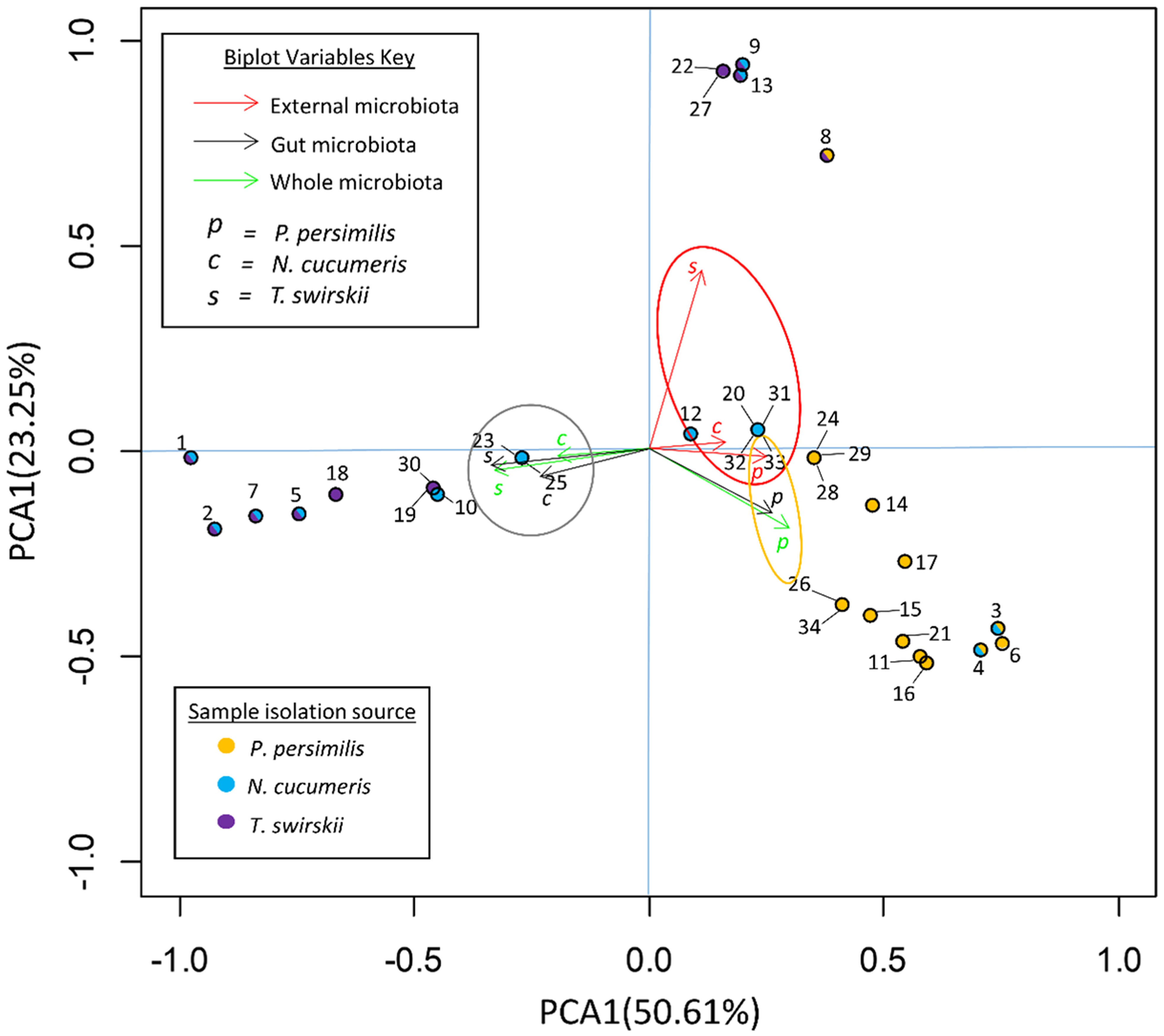
Figure 2 Principal component analysis (PCA) of OTUs in the microbiota of three species of phytoseiid. PC1 explains that 50.61% of the variance in the data set is due to the location of the bacteria (gut/external). PC2 explains that 23.25% is from host species. Arrow biplots represent the effects of different variables on the variance of the data set. Solid circles represent loadings of each OTU within the data set and the colour of the center denotes species from which the OTU was originally isolated.
The variables responsible for the most variation in the data set came from the variation between gut microbiota and external microbiota. External microbiota of each of the three species clustered together and explained much of the variation in the sample as highlighted by the red oval (Figure 2). Most variation between the gut and external microbiota was found in T. swirskii from whom a few rare taxa in the external microbiota caused high variation (Figure 2). Least variation between the gut and external microbiota within a species was found in P. persimilis shown by the clustering of taxa and variables attributed to this species (Figure 2). Comparisons of the taxa in the gut and whole microbiota of T. swirskii and N. cucumeris showed little variation, suggesting similar bacterial communities in these variables (Figure 2).
3.2 Effects of bacterial cultures on survival, oviposition, and voracity of P. persimilis
Most bacterial cultures tested had no significant effects on average voracity of adult female P. persimilis under laboratory conditions (Figure 3). However, the two cultures Serratia odorifera and Serratia marcescens showed significant increase and decrease in voracity, respectively. Voracity scores of predators treated with S. marcescens were significantly lower compared to all other bacterial treatments and the control (p = 0.0000034). Conversely, predatory mites treated with S. odorifera showed a significant increase in average voracity, compared to all other bacterial treatments and the control (p = 0.00012) (Figure 3).
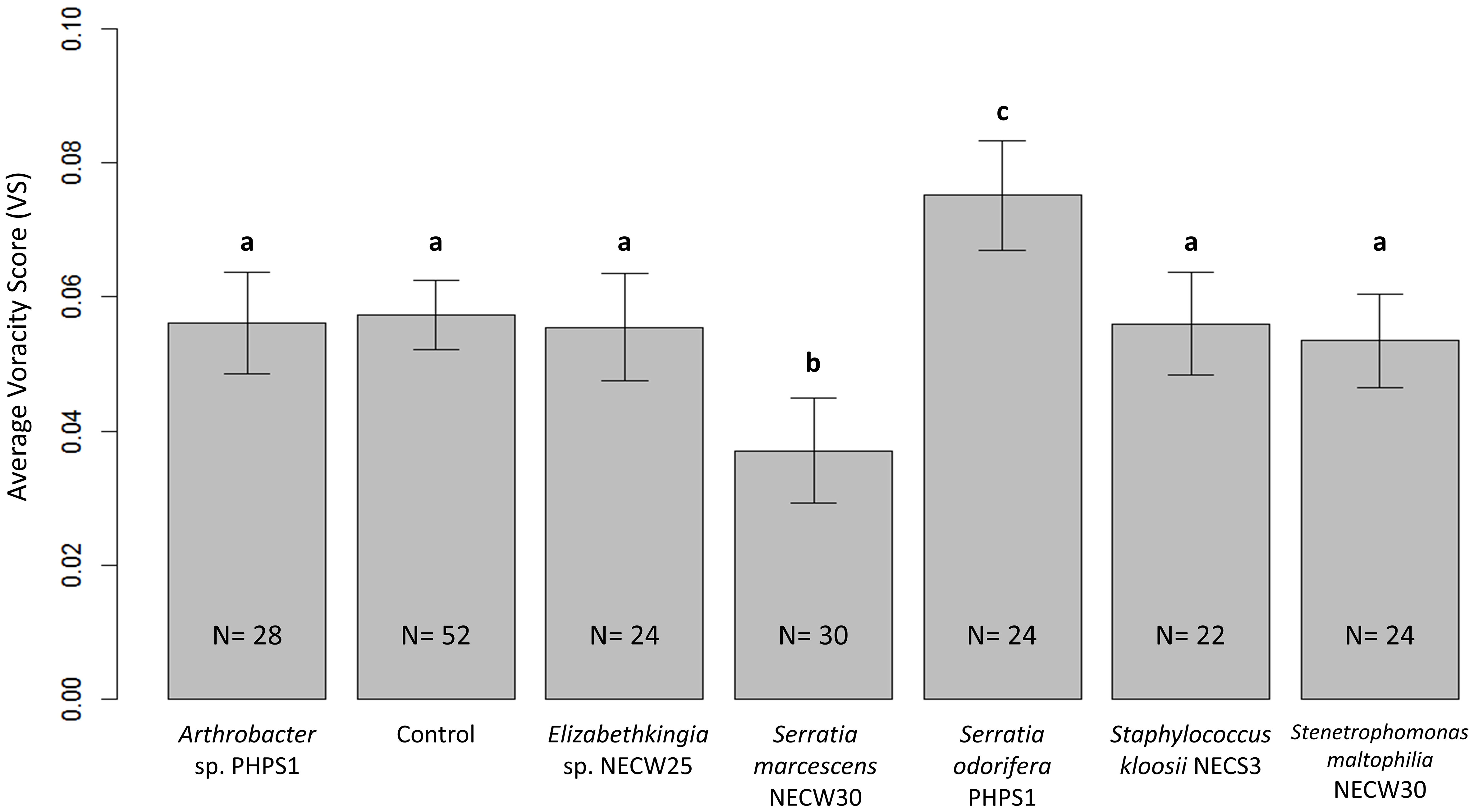
Figure 3 Average voracity scores for P. persimilis females subjected to all bacterial treatments and the control treatment containing no bacterial culture. Error bars are equal to 1 SE of the difference between means; non-overlapping bars show significantly different average voracity for that treatment. Different letters indicate significant differences between average voracity.
Generalised linear models were used to analyse oviposition rates for each bacterial treatment. Using an F-test for goodness-of-fit comparison between the model including the bacterial treatment as an explanatory variable and a model without treatment as an explanatory variable showed that bacterial treatment was a significant factor affecting the rate of oviposition in these laboratory trials on P. persimilis females [F(6,112) = 6.643, p < 0.001]. Therefore, the null hypothesis that bacterial treatments do not affect the oviposition rates of P. persimilis cannot be accepted. To analyse the source of this variation, multiple comparisons of means for each treatment were performed using Tukey contrast fit on the general linear models. These results showed that S. marcescens-treated P. persimilis had significantly lower mean oviposition rates than control mites (p < 0.001) (Figure 4). All other bacterial treatments were not statistically significantly different compared to the control, or when compared with all other treatments except S. marcescens. Mean oviposition rates were greater for every treatment compared to that of S. marcescens (Figure 4) and the median oviposition for mites treated was zero (Figure 4) as most mites in S. marcescens trails died before they had a chance to oviposit. Only one mite from N = 14 exhibited oviposition that was near to the median oviposition of mites from all other treatments.
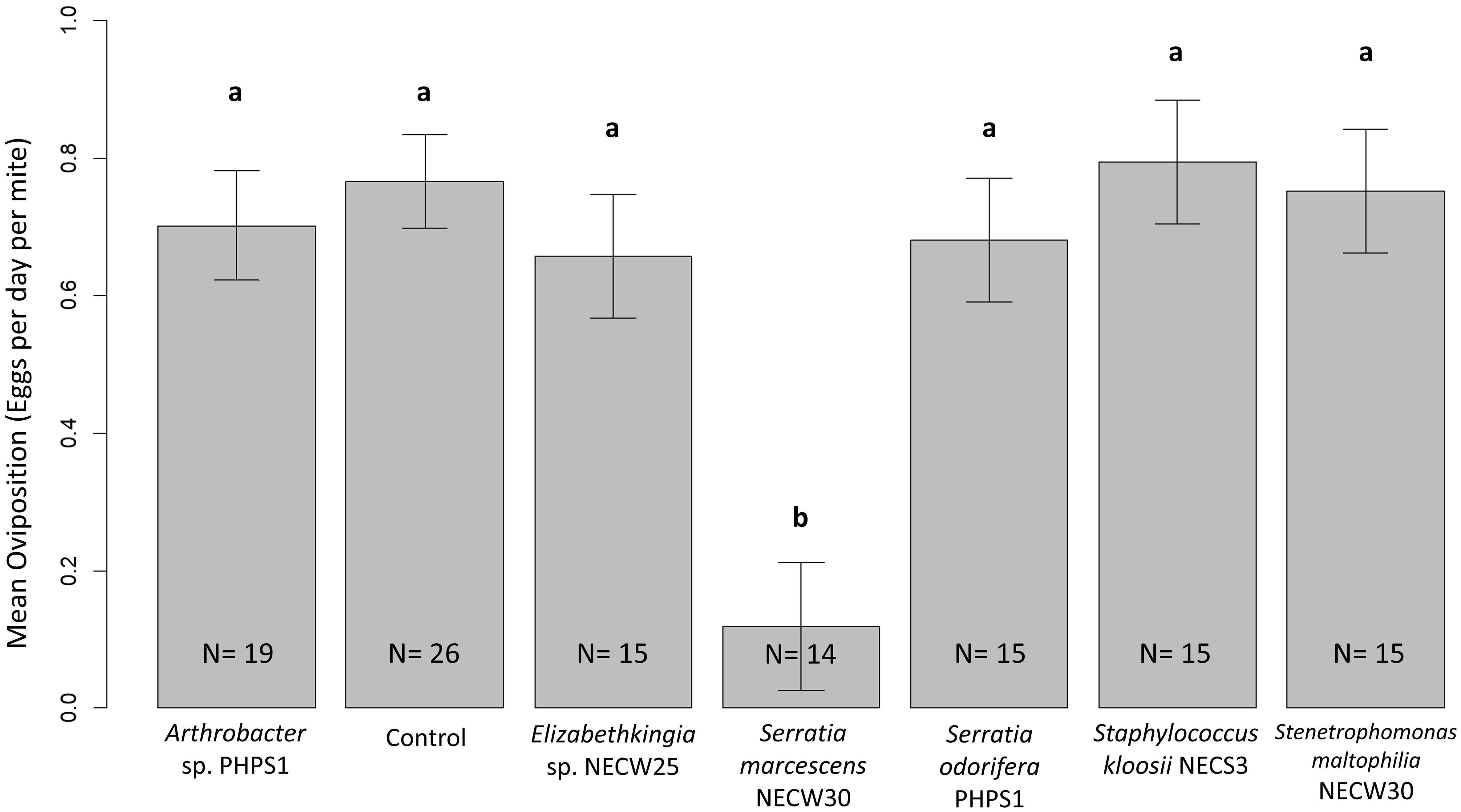
Figure 4 Mean oviposition per day for each treatment. Error bars show one SE of the difference between means. Different letters show significant differences between groups. Only S. marcescens-treated P. persimilis mites showed significantly lowered oviposition rates due to bacterial treatment. All other treatments were statistically similar when using Tukey contrasts performed simultaneously on generalised linear models for hypothesis testing. A p-value of <0.05 showed significant difference between treatments.
Survival analysis (Figure 5) showed significant differences in mean survivorship between bacterial treatments when Kaplan–Meier curves were compared with a Tarone-Ware test [χ2(6, n = 209) = 58.6, p > 0.0001]. Therefore, we cannot accept the null hypothesis that mean survivorship was not different for different bacterial treatments. Comparisons of Kaplan–Meier survival models grouped these data into two statistically different groups. Survival models were simplified to incorporate statistically similar treatments into the same groups and Chi-squared tests were used to compare goodness of fit between groups.
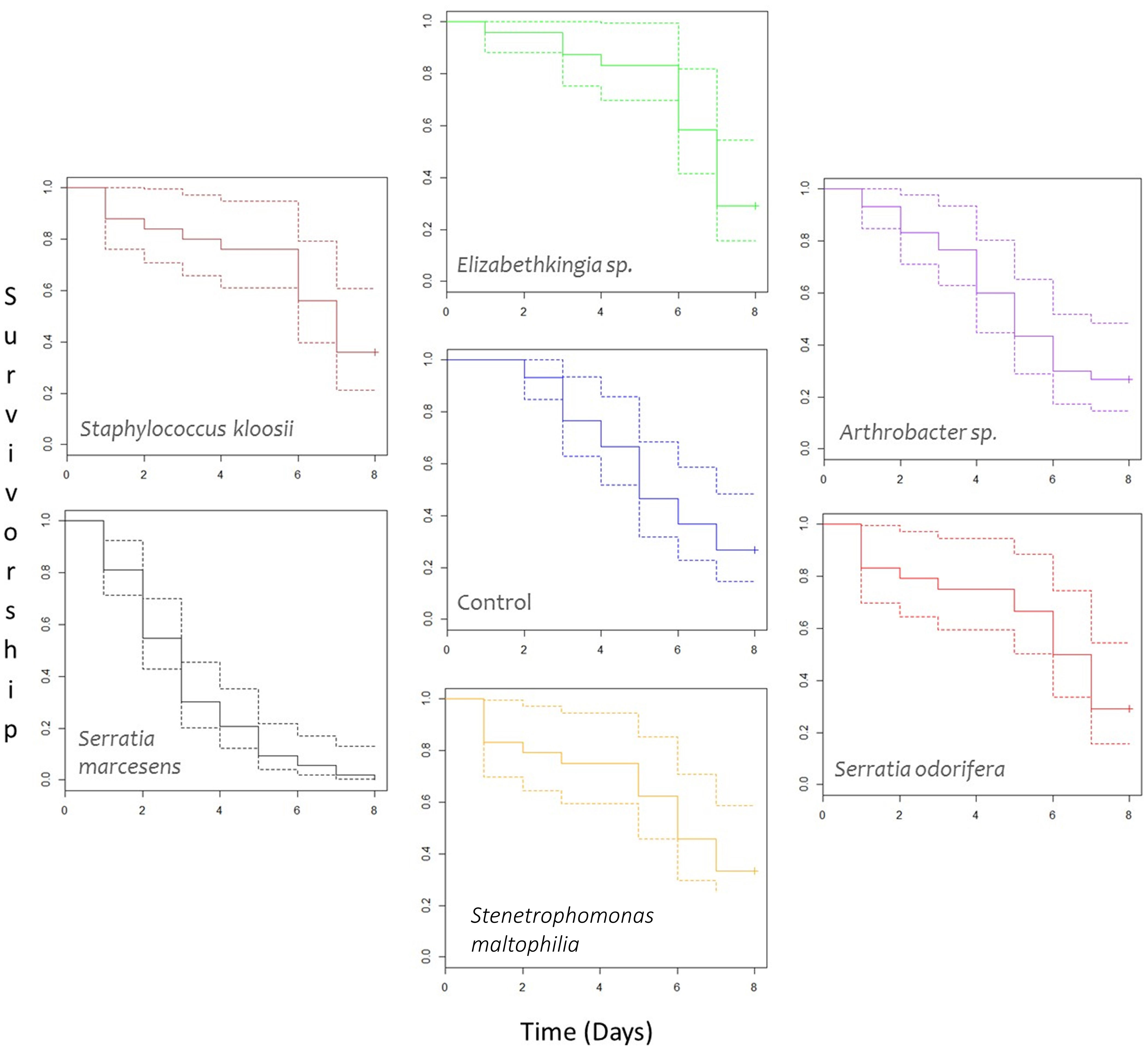
Figure 5 Survivorship curves of female P. persimilis treated with different bacterial cultures over an 8-day period. Dotted lines represent upper and lower confidence intervals, and solid lines represent mean survivorship. Crosses at the end of survivorship curves denote that some individuals involved in the study were still alive after the 8-day period. N for each treatment are as follows: Arthrobacter sp. = 30; Control = 30; Elizabethkingia sp. = 24; Serratia marcescens = 53; Serratia odorifera = 24; Stenetrophomonas maltophilia = 24; and Staphylococcus kloosi = 25.
After model simplification, group A consisted solely of S. marcescens and group B comprised all remaining treatments (Arthrobacter sp., Elizabethkingia sp., S. odorifera, Staphylococcus kloosii, Stenetrophomonas maltophilia, and the control containing no bacteria). P. persimilis treated with an S. marcescens inoculum showed significantly lower mean survival compared to the control and all other treatments [Group A vs. Group B, χ2 (1 d.f., n = 209) = 58.47, p-value > 0.001]. Mites treated with an Elizabethkingia sp. inoculum had the highest mean age at death of 5.647 days; however, this did not prove to be statistically higher than mean age at death for control mites [χ2 (1, n = 54) = 0.86, p = 0.4].
4 Discussion
This comprehensive study examined the Phytoseiidae microbiotas of three economically significant species and the influence of bacterial taxa on the health of P. persimilis, the first organism to be used as a biological control agent. We used molecular and classical microbiology culturing methods to characterise bacteria on the skin and digestive tract of three different Phytoseiidae species. Our study showed that significant differences were found between mite species and gut and skin microbiota were significantly different in diversity, suggesting a functional separation of microbiomes. The Proteobacteria were the most dominant group of bacteria present in all mite species tested making up 68.75% in P. persimilis, 73.91% in N. cucumeris, and 57.14% in T. swirskii. We confirm the status of S. marcescens as a pathogen of P. persimilis and suggest here new beneficial bacterial species.
Similar to the study of microorganisms associated with M. occidentalis (Hoy and Jeyaprakash, 2005), Enterobacter were a large constituent of the microbiota of these three phytoseiids. Enterobacteriales are a common occurrence in the microbiota of Acari and other Arthropods (Dillon and Charnley, 2002; Hubert et al., 2011; Kaltenpoth, 2011; Pekas et al., 2017) and further investigation should focus on understanding their function in reared Phytoseiidae. In the current study, the predominant bacterial group comprising the Phytoseiidae microbiota was Proteobacteria. This supports a previous study where Proteobacteria (particularly gamma-proteobacteria) were the most prevalent in Neoseiulus californicus at >85% of all amplicon sequence variants (ASVs) (Merlin et al., 2022), and a similar trend is also seen in insect gut microbiota where Proteobacteria made up 62.1% of total OTUs from 218 species across 21 different orders (Yun et al., 2014). In the current study, the predominance of Proteobacteria could represent bias in our sampling methods, molecular detection, and culturing (Nemergut et al., 2011). Although non-selective media was used, this could lead to underrepresentation of fastidious microbes that might have significant effects on the mites. Bias in DNA extraction and PCR amplification could also lead to rarer bacterial taxa to be overlooked. It is possible that, because Proteobacteria are also very widely distributed and abundant in the environment (Nemergut et al., 2011), the Phytoseiidae are therefore more likely to acquire them through the environment.
The microbiota of T. swirskii and N. cucumeris guts and integument was not significantly different between these two species, suggesting that the microbial communities of these two species are similar, adding support to the hypothesis that artificial rearing conditions may cause convergence of microbiota of predatory mites reared in this manner. These two species have a similar lifestyle (Type 3 predators) (McMurtry and Croft, 1997) and can persist on similar prey species. Similarities in their microbiotas might also be explained by acquisition of bacterial taxa due to their evolutionary history, as with bees and ants, whereby species sharing similar niches and lifestyles have acquired similar bacterial diversity (Russell et al., 2009; Kaltenpoth, 2011; Martinson et al., 2011). Additionally, the similarities in the bacterial diversity of the guts and external anatomy of these predatory mite species could be a result of the way in which they are reared. N. cucumeris and T. swirskii are both reared on factitious prey mite species in a similar rearing media composed of bran or other dried cereals along with vermiculite or similar substrate (Bolckmans and van Houten, 2006; Smytheman, 2011; Midthassel et al., 2013). Biotic and abiotic factors associated with this environment may influence the bacterial diversity of mites reared in these conditions and possibly cause certain bacterial species to become prevalent within the predator’s microbiota; potentially explaining the similarity in the microbiota of these species. However, previous studies have shown that predator and prey mite species have distinctive microbiota that are not greatly influenced by host plant (Merlin et al., 2022); it is possible that mass-rearing environments where mites are reared on factitious prey mites in synthetic environments can have a bigger impact on mite microbiomes than in more natural rearing systems.
Transmission of microbes from prey to predators (and vice versa) is an interesting phenomenon and may have implications on the behaviour or health of the predator. The microbiota of these two predatory mites contains bacteria from Rhizobiales and Enterobacteriaceae that are similar to bacteria previously isolated from the microbiota of Tyrophagous putrescentiae and Acarus siro (Hubert et al., 2011; Pekas et al., 2017). If different species of astigmatid mites share a similar bacterial diversity, it is likely that bacteria found in the Phytoseiidae was transmitted horizontally to the predator when feeding on its prey Astigmata species in the mass-rearing facility. Rhizobiales are commonly found in the microbiota of Astigmata (Hubert et al., 2011; Pekas et al., 2017). In N. californicus–T. urticae predator–prey systems, some transfer of bacterial taxa between mites was found (Merlin et al., 2022). Enterobacteriaceae in insects have been shown to play an important role in protection of the host and indirectly promote host health by preventing the establishment of pathogenic microbes (Zhang et al., 2021). Citrobacter sp. within the gut of Bactrocera dorsalis conferred improved pesticide resistance due to the activation of phosphatase hydrolase genes (Cheng et al., 2017); in the current study, a Citrobacter sp. was found on the skin surface of P. persimilis (OTU28). Klebsiella oxytoca has been used as a probiotic treatment in the Mediterranean fruit fly (Ceratitis capitata) and significantly increased the sexual competitiveness of male fruit flies and prolonged their survival (Gavriel et al., 2011); similar bacteria were present in the gut and skin of N. cucumeris in the current study (OTU10 and OTU23). We propose similar mechanisms for these bacteria in Phytoseiidae mites and the Enterobacteriaceae may be significant in enabling these mites to survive within the mass-rearing environment. However, further examination of the impact of these bacteria on Phytoseiidae health is necessary.
Within species examined in the current study, there were significant differences between the microbiota of the integument compared to the gut. Gut microbiota may be responsible for aiding in digestion and maintaining gut health, whereas external microbiota may be responsible for preventing the invasion of pathogenic organisms. The unique subsets of bacterial diversity found in the microbiota of the gut and the integument of mites suggest that bacterial communities in these areas carry out specialised functions on distinctive anatomies. Bacteria typically associated with insect gut flora found on the integument of T. swirskii suggest that bacteria can be transferred to the integument of the predator by direct contact with prey or through the environment, as previously suggested (Pekas et al., 2017; Merlin et al., 2022). It has been shown that bacteria can pass into the digestive tract of Phytoseiidae mites via intake of food (Arutunyan, 1985; Wu and Hoy, 2012b; Pekas et al., 2017); thus, it is very likely that the unclassified Enterobacteriaceae within the gut of N. cucumeris originated from its acarid prey source and was transmitted during feeding. To support this, in a previous study, Metaseiulus occidentalis was shown to acquire Wolbachia from their prey species (T. urticae) via feeding and environmental infection but not be a part of M. occidentalis natural microbiota (Wu and Hoy, 2012b). Therefore, mass-reared systems should consider the transfer of microbes from predators to prey and ensure that transfer of microbiome will not cause a detrimental effect on the predator mite’s survivability, or ultimate ability to control its intended pest target. Previous work revealed a range of eukaryotic and prokaryotic microbes associated with the integument and digestive tract of the three Phytoseiidae mite examined in this study (Sumner-Kalkun et al., 2020). This study complements the presence of bacterial taxa within the Phytoseiidae digestive tract as previously seen using fluorescence in situ hybridisation (FISH) and laser confocal microscopy (Sumner-Kalkun et al., 2020).
A study by Pekas et al. (2017) on the microbiome of N. cucumeris also presented similar diversity to that found in the current study. Staphylococcus kloosi was the main constituent of the N. cucumeris microbiome in the Pekas et al. (2017) study and was suggested to make up a large percentage of the mite’s intestinal flora and is possibly passed on from mite to mite via the feces. This is supported by the fact that this bacterium was cultured from the surface of N. cucumeris in our study. Previously, a Kocuria sp. and Brevibacterium spp. also make up a small proportion of the predatory mite microbiome (Pekas et al., 2017). In the current study, we found these bacteria in the Phytoseiidae microbiome, and it is suggested that Kocuria could be an important arthropod pathogen and these bacteria warrant further examination of their role in the Phytoseiidae microbiome and mass-rearing systems.
The pathogenic effects of S. marcescens have previously been observed on many insect species (Poinar and Thomas, 1984) and the Phytoseiidae mite species M. occidentalis (Lighthart et al., 1988). For this reason, it was chosen for this study to examine its effects on the more widely used biological control agent P. persimilis. In laboratory trials, S. marcescens was found to be a strong pathogen of P. persimilis, inducing characteristic pathogenic symptoms and reduced the performance of the mites treated with this bacterium. Mites infected with S. marcescens suffered reduced voracity, a reduction in oviposition, and a significantly lowered survival rate. S. marcescens infection also caused mites to become lethargic and unresponsive to agitation with a fine hairbrush. Moribund mites moved slowly if at all and legs or opisthosoma would often be stuck to the leaf surface. After death, mites would shrivel and legs were often retracted underneath the body. Occasionally, one of the legs would be contracted apparently because it had become stuck to the leaf surface. This provides the first evidence of pathogenicity of S. marcescens against the predatory mite P. persimilis. S. marcescens is likely a generalist pathogen of Phytoseiidae. Compared to the study by Lighthart et al. (1988) in which 50% of mite populations had died out after 4 days of bacterial treatment, populations of P. persimilis examined in this study took less than 3 days to drop below 50%. Further comparisons show concurrence in the ability of S. marcescens to reduce survival rate, and our current study provides new evidence to suggest that S. marcescens can also decrease the fecundity (oviposition) and voracity of Phytoseiidae mites. However, Lighthart et al. (1988) found that infection with S. marcescens did not terminate egg production in already gravid females. As all females used in this study were in the primary stages of egg production after mating, it is possible that S. marcescens were able to prevent the onset of egg development. Additionally, due to the high virulence and subsequent high mortality rate caused by S. marcescens, it is possible that mites died before they had the opportunity to lay any eggs. A previous study showed that S. marcescens could enter the hemocoel of the predatory mirid bug Adelphocoris suturalis and its presence in the predator was influenced by an imbalanced diet whereby only aphids were ingested (Luo et al., 2021). A similar mechanism for transmission and mortality is suggested in the current study for Phytoseiidae–prey, although further study to verify this is necessary. This could be achieved by assessing the titers of selected bacterial taxa in the predator vs. prey using molecular screening such as real-time PCR and FISH as previously demonstrated for the Acari (Hubert et al., 2011; Sumner-Kalkun et al., 2020).
Although Koch’s postulates were not strictly fulfilled in the current study, these data justify the addition of S. marcescens to the list of bacterial pathogens of the Phytoseiidae, making it one of only a few extensively studied bacterial pathogens of P. persimilis (Pukall et al., 2006; Schutte, 2006; Bjørnson, 2008; Schütte et al., 2008). The bacterial culture of S. marcescens used in this study was originally isolated from healthy mass-reared populations of N. cucumeris and was also present in healthy mass-reared populations of T. swirskii. However, the effects of this bacterium on those species are currently unknown. It is possible that pathogenic effects of this bacterium may only be exhibited in mass-rearing systems when conditions are optimal for the growth of this pathogen, or when the pathogen becomes more abundant in the environment. Dosages used in this experiment are likely to be much higher than the number of S. marcescens CFUs present in the natural microbiota, and the host may therefore be able to resist lower levels of infection. In a study of T. urticae and N. californicus microbiomes by Merlin et al. (2022), Serratia were found to be the most abundant ASV on T. urticae eggs but less represented in adult T. urticae and N. californicus. Depending on the identity and role of those Serratia species, it is possible that they represent a form of defense mechanism to prevent egg predation by Phytoseiidae by deterring predators, due to the potential pathogenic nature of some Serratia spp. as shown in the current study. It could also be possible that predatory mites use sensory or olfactory cues from these bacteria to locate eggs.
Future work should examine the effects of S. marcescens on other Phytoseiidae species such as N. cucumeris and T. swirskii, and how it regulates predator–prey interactions. Experiments should also focus on the effects of rearing conditions such as humidity, temperature, and predator/prey density to examine the optimal growth conditions for this bacterium. This information will help in the control of this disease and prevent excessive buildup of this pathogen. Finally, given the pathogenicity of S. marcescens exhibited here to induce pathogenic symptoms in some populations of P. persimilis, it is suggested that S. marcescens should be included in all future experimental designs that require the use of a Phytoseiidae pathogen. Serratia spp. have also been used in other arthropods in para-transgenic strategies for controlling insects such as mosquitoes (Koosha et al., 2019). The use of these bacterial species for controlling pest mite species should be explored further.
This study provides some evidence that the microbiotas of Phytoseiidae contain some beneficial species within its ectosymbionts. Whilst S. marcesens appears to be a pathogen of P. persimilis, its close relative S. odorifera appears to be a beneficial partner. Mites treated with S. odorifera showed significantly higher voracity compared to all other bacterial treatments, including the control. The mechanism by which S. odorifera can improve the voracity of P. persimilis, however, is undetermined and could represent a long-term detriment to the mites, although it did not affect oviposition or survival rates. Speculatively, owing to the finding of Serratia as the most abundant bacterium on T. urticae eggs (Merlin et al., 2022), it is possible that S. odorifera affects the searching behaviour of P. persimilis or could be used for defense against predation by T. urticae. For a biological control agent such as P. persimilis, increasing the rate of consumption of its prey is a desirable trait. If all individuals of a population of predatory mites consume more pests once released into the cropping system, this would greatly improve the time it takes the pest population to be effectively controlled, thus improving the performance of the biological control agent. However further work is necessary to confirm whether S. odorifera can improve the capacity for pest control of a population of P. persimilis on this scale. Future work should focus on the effects of S. odorifera on Phytoseiidae populations on a larger scale such as glasshouse or field trials and on other host species.
Although its effects did not prove to be significant in the current study, Elizabethkingia has been previously shown to improve the health of other arthropods in laboratory populations (Kampfer et al., 2011; Boissière et al., 2012; Ngwa et al., 2013). Given the reported antimicrobial properties of this genus, it could be a promising candidate to improve the health of Phytoseiidae in mass-rearing facilities.
In the current study, bacteria found in N. cucumeris and T. swirskii (OTU09) showed similarity to an uncultured Bartonella sp. first isolated from T. putrescentiae (Erban et al., 2016a). The next closest match to this bacterium according to an NCBI GenBank search (Clark et al., 2016) is only around 95% similar to the nearest Bartonella spp. This OTU could represent a unique species that is associated with Astigmata mites and warrants further study to examine its role in Astigmata and Phytoseiidae. It is likely that this OTU was picked up by the predatory mites from their factitious prey species or rearing environment as this OTU was not found in P. persimilis.
During the current study, Acaricomes phytoseiuli was isolated from the skin surface of populations of P. persimilis and T. swirskii, but only using culture-independent methods. This bacterium was first reported in a population of P. persimilis mites showing symptoms that were detrimental to the health and behaviour of this predatory mite and ultimately decreasing its ability to perform as an efficient biological control agent (Pukall et al., 2006; Schutte, 2006; Schütte et al., 2008). These symptoms include non-responding disorder, shrinking of adult females, reduced voracity, buildup of excretory products in legs and Malpighian tubules, and eventual death (Schütte et al., 2008).
Since 2008, no further information has been published regarding A. phytoseiuli in peer-reviewed journals. This study furthers the knowledge and occurrence of this bacterium. This is the first report of this bacterium in T. swirskii, and its effects on this mite are unknown. All mites examined were from healthy stocks of mass-reared P. persimilis and T. swirskii; whilst some individuals did display white colouration within the Malpighian tubules and anal atrium, no individuals exhibited the shrunken characteristics mentioned as a symptom of disease caused by A. phytoseiuli (Pukall et al., 2006; Schutte, 2006; Schütte et al., 2008) and no complaints of reduced efficacy coincided with the times that these mites were examined. Therefore, the presence of this bacteria within a healthy population of mites is unusual. It is possible that A. phytoseiuli is an opportunist pathogen that requires certain conditions to cause disease. It is also likely that other microorganisms within the Phytoseiidae microflora can prevent A. phytoseiuli from completely invading and producing significant numbers to cause disease.
In previous studies, Stenotrophomonas spp. were considered opportunistic pathogenic bacteria of T. urticae (Zhu et al., 2020). In our study, S. maltophila bacterium was found within the microbiome of P. persimilis, but in bioassays, it did not cause any significant effects to survival, voracity, or oviposition.
From this baseline study, further studies should concentrate on potentially beneficial and pathogenic species and examine their prevalence in Phytoseiidae mites and their prey species. The current study should be used to examine further interaction between Phytoseiidae, their prey and environment to further understand the interactions which influence the Phytoseiidae microbiota and how this microbiome affects the health of these mites. Although the current study did not experimentally verify the exact origin of external and internal bacteria, our results show significant differences in external and internal microbiotas, which justifies the experimental approach and suggests its effectiveness. Highlighting the main constituents of the microbiota of these mite species in rearing systems means we can now develop further experiments to track these species within the mass-rearing systems and we can examine the titers of these important bacteria in predator and prey using advanced molecular techniques such as real-time PCR and FISH. These experiments will advance our knowledge of the prevalence of these bacteria in mass-rearing systems and help us unravel their impacts on the health and performance of Phytoseiidae as biological control agents.
The Phytoseiidae microbiome is a dynamic characteristic of the biology of these mites that must be understood to further understand the biology and health of these mites, especially in mass-rearing environments where the performance of these mites depends on their health status. This study provides a basis for the study of the microbiota of these mites and should be built upon to determine microbiota–Phytoseiidae interactions and the function of the mite microbiome. Based on the results of this study, it is likely that the Phytoseiidae microbiome contains pathogenic and beneficial taxa that could be manipulated to improve mite health and ultimately their performance as biological control agents. The mite microbiome is influenced by the mass-rearing environment and the prey species on which mites are reared; P. persimilis had the most diverse microbiota of the three species examined, possibly because it is reared in an environment that more closely represents its natural habitat. The unique microbiota of gut and integument of these mites suggests a functional difference between these two areas of the body. Further work is needed to understand the mite–microbiome interactions that are significant for their biology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
AP and IB contributed to the design and conception. AP and IB contributed to securing funding for the project. JS-K and AP performed statistical analysis and experimental design. JS-K performed lab work and collected data. JS-K and AP contributed to data storage and curation. JS-K wrote the first draft of the manuscript. AP revised and edited the manuscript. IB dealt with industry permissions to publish. All authors contributed to the article and approved the submitted version.
Funding
This work was funded as part of a BBSRC CASE studentship (BB/H016643/1) at the University of Reading in partnership with Biological Crop Protection Ltd.
Conflict of interest
Author IB was employed by the company Biological Crop Protection Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
The authors declare that this study received funding from Biological Crop Protection as part of a BBSRC Case Studentship. The funder had the following involvement in the study: providing mite samples and expertise regarding the production and maintenance of mite cultures.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frchs.2023.1242716/full#supplementary-material
References
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–105. doi: 10.1016/S0022-2836(05)80360-2
Arutunyan E. S. (1985). Morphologicheskije osobennosti pischevaritelnoho tracta Phytoseiidnvch kleshchej. Structural peculiarities of the digestive tract in phytoseiid mites. (in Russian). Biologicheskii Zhurnal Armenii 38, 590–596.
Bjørnson S. (2008). Natural enemies of mass-reared predatory mites (Family phytoseiidae) used for biological pest control. Exp. Appl. Acarology 46 (1–4), 299–306. doi: 10.1007/s10493-008-9187-1
Boissière A., Tchioffo M. T., Bachar D., Abate L., Marie A., Nsango S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector anopheles Gambiae and interactions with plasmodium falciparum infection. PLoS Pathog. 8, e1002742. doi: 10.1371/journal.ppat.1002742
Bolckmans K., van Houten Y. (2006). Phytoseiid mite rearing methods and methods for biological pest control issued 2006.
Bridges J. R., Moser J. C. (1983). Role of two phoretic mites in transmission of bluestain fungus, Ceratocystis minor. Ecol. Entomology 8, 9–12. doi: 10.1111/j.1365-2311.1983.tb00476.x
Cakmak I., Janssen A., Sabelis M. (2006). Intraguild interactions between the predatory mites neoseiulus californicus and phytoseiulus persimilis. Exp. Appl. Acarology 38, 33–46. doi: 10.1007/s10493-005-6247-7
Cheng D., Guo Z., Riegler M., Xi Z., Liang G., Xu Y. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly bactrocera dorsalis (Hendel). Microbiome 5 (1), 1–125. doi: 10.1186/s40168-017-0236-z
Chow A., Chau A., Heinz K. M. (2010). Compatibility of amblyseius (Typhlodromips) swirskii (Athias-henriot) (Acari: Phytoseiidae) and orius insidiosus (Hemiptera: Anthocoridae) for biological control of frankliniella occidentalis (Thysanoptera: Thripidae) on roses. Biol. Control 53 (2), 188–965. doi: 10.1016/j.biocontrol.2009.12.008
Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W. (2016). GenBank. Nucleic Acids Res. 44 (D1), D67–D72. doi: 10.1093/nar/gkv1276
De Courcy Williams M. (2001). Biological control of thrips on ornamental crops: interactions between the predatory mite Neoseiulus cucumeris (Acari: Phytoseiidae) and western flower thrips, Frankliniella occidentalis (Tysanoptera: Thripidae), on cyclamen. Biocontrol Sci. Technol. 11, 41–55. doi: 10.1080/09583150020029736
Dillon R., Charnley K. (2002). Mutualism between the desert locust schistocerca gregaria and its gut microbiota. Res. Microbiol. 153 (8), 503–595. doi: 10.1016/S0923-2508(02)01361-X
El-Kholy M., El-Saiedy E. (2009). Biological control of thrips tabaci (Lind.) and aphis gossypii (Glover) using different predatory phytoseiid mites and the biocide vertimec on eggplant at behaira governorate. Egypt. Acad. J. Biol. Sci. Entomol 2, 13–22. doi: 10.21608/eajbsa.2009.15425
Enigl M., Schausberger P. (2007). Incidence of the endosymbionts wolbachia, cardinium and spiroplasma in phytoseiid mites and associated prey. Exp. Appl. Acarology 42 (2), 75–855. doi: 10.1007/s10493-007-9080-3
Enigl M., Zchori-Fein E., Schausberger P. (2005). Negative evidence of wolbachia in the predacious mite phytoseiulus persimilis. Exp. Appl. Acarology 36, 249–262. doi: 10.1007/s10493-005-6075-9
Erban T., Klimov P. B., Smrz J., Phillips T. W., Nesvorna M., Kopecky J., et al. (2016a). Populations of stored product mite tyrophagus putrescentiae differ in their bacterial communities. Front. Microbiol. 7 (JUL). doi: 10.3389/fmicb.2016.01046
Erban T., Rybanska D., Harant K., Hortova B., Hubert J. (2016b). Feces derived allergens of Tyrophagus putrescentiae reared on dried dog food and evidence of the strong nutritional interaction between the mite and Bacillus cereus producing protease bacillolysins and exo-chitinases. Front. Physiol. 7, 53. doi: 10.3389/fphys.2016.00053
Fouly A. H., Al-Deghairi M. A., Baky N. F. A. (2011). Biological aspects and life tables of Typhlodromips swirskii (Acari: Phytoseiidae) fed Bemisia tabaci (Hemiptera: Aleyroididae). J. Entomology 8, 52–62. doi: 10.3923/je.2011.52.62
Gavriel S., Jurkevitch E., Gazit Y., Yuval B. (2011). Bacterially enriched diet improves sexual performance of sterile male mediterranean fruit flies. J. Appl. Entomology 135, 564–573. doi: 10.1111/j.1439-0418.2010.01605.x
Gerson U., Smiley R. L., Ochoa R. (2003). Mites (Acari) for pest control (Oxford, UK: Blackwell Science).
Hoogerbrugge H., Calvo J., van Houten Y. M., Bolkmans K. (2005). Biological control of the tobaco whitefly Bemisia tabaci with the predatory mite Amblyseius swirskii in sweet pepper crops. IOBC/WPRS Bull. 28, 119–122.
Hoy M. A. (2011). “The phytoseiidae: effective natural enemies,” in Agricultural Acarology Introduction to Integrated Mite managment (New York, USA: Taylor & Francis), 159–184.
Hoy M. A., Jeyaprakash A. (2005). Microbial diversity in the predatory mite Metaseiulus occidentalis (Acari: Phytoseiidae) and its prey, tetranychus urticae (Acari: Tetranychidae). Biol. Control 32, 427–441. doi: 10.1016/j.biocontrol.2004.12.012
Hoy M. A., Jeyaprakash A. (2008). Symbionts, including pathogens, of the predatory mite metaseiulus occidentalis: current and future analysis. Exp. Appl. Acarology 46, 329–347. doi: 10.1007/s10493-008-9185-3
Hubert J., Kopecký J., Perotti M. A., Nesvorná M., Braig H. R., Ságová-Marečková M., et al. (2011). Detection and identification of species-specific bacteria associated with synanthropic mites. Microbial Ecol. 63, 919–928. doi: 10.1007/s00248-011-9969-6
Hubert J., Nesvorná M., Ságová-Marečková M., Kopecký J. (2012). Shift of bacterial community in synanthropic mite tyrophagus putrescentiae induced by fusarium fungal diet. PLoS One 7 (10). doi: 10.1371/journal.pone.0048429
Janda J.M., Abbott S. L. (2007). 16S RRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45 (9), 2761–2645. doi: 10.1128/JCM.01228-07
Johanowicz D. L., Hoy M. A. (1998). Experimental induction and termination of non-reciprocal reproductive incompatibilities in a parahaploid mite. Entomologia Experimentalis Applicata 87 (1), 51–585. doi: 10.1046/j.1570-7458.1998.00303.x
Johanowicz D. L., Hoy M. A. (1999). Wolbachia infection dynamics in experimental laboratory populations of metaseiulus occidentalis. Entomologia Experimentalis Applicata 93 (3), 259–685. doi: 10.1046/j.1570-7458.1999.00586.x
Kaltenpoth M. (2011). Honeybees and bumblebees share similar bacterial symbionts. Mol. Ecol. 20 (3), 439–440. doi: 10.1111/j.1365-294X.2010.04960.x
Kampfer P., Matthews H., Glaeser S. P., Martin K., Lodders N., Faye I. (2011). Elizabethkingia anophelis sp. Nov., isolated from the midgut of the mosquito anopheles Gambiae. Int. J. Systematic Evolutionary Microbiol. 61, 2670–2675. doi: 10.1099/ijs.0.026393-0
Koosha M., Vatandoost H., Karimian F., Choubdar N., Reza Abai M., Oshaghi M. A. (2019). Effect of serratia AS1 (Enterobacteriaceae: Enterobacteriales) on the fitness of culex pipiens (Diptera: Culicidae) for paratransgenic and RNAi approaches. J. Med. Entomology 56 (2), 553–595. doi: 10.1093/jme/tjy183
Kostiainen T. S., Hoy M. A. (1996). The phytoseiidae as biological control agents of pest mites and insects: A biblography, (1960-1994) (Gainesville: University of Florida, Department of Entomology and Nematology, IFAS).
Lighthart B., Sewall D., Thomas D. R. (1988). Effect of several stress factors on the susceptibility of the predatory mite, Metaseiulus occidentalis (Acari: Phytoseiidae), to the weak bacterial pathogen Serratia marcescens. J. Invertebrate Pathol. 52, 33–42. doi: 10.1016/0022-2011(88)90099-7
Luo J., Cheng Y., Guo L., Wang A., Lu M., Xu L. (2021). Variation of gut microbiota caused by an imbalance diet is detrimental to bugs’ Survival. Sci. Total Environ. 771, 144880. doi: 10.1016/j.scitotenv.2020.144880
Manners A., Dembowski B., Healey M. (2013). Biological control of western flower thrips frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae) in gerberas, chrysanthemums and roses. Aust. J. Entomology 52, 246–258. doi: 10.1111/aen.12020
Martinez-Giron R., Doganci L., Iraola V. (2009). Gregarines in Dermatophagoides spp. (Acari: Pyroglyphidae): light microscopy observation. J. Med. Entomology 46, 367–368. doi: 10.1603/033.046.0223
Martinson V. G., Danforth B. N., Minckley R. L., Rueppell O., Tingek S., Moran N. A. (2011). A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20 (3), 619–285. doi: 10.1111/j.1365-294X.2010.04959.x
McMurtry J. A., Croft B. A. (1997). Life-styles of phytoseiid mites and their roles in biological control. Annu. Rev. Entomology 33, 239–269. doi: 10.1146/annurev.ento.42.1.291
Merlin B. Laís, Moraes G. J., Cônsoli F. L. (2022). The microbiota of a mite prey-predator system on different host plants are characterized by dysbiosis and potential functional redundancy. Microbial Ecol., 85, 1590–1607. doi: 10.1007/s00248-022-02032-6
Messelink G. J., van Maanen R., van Steenpaal S. E. F., Janssen A. (2008). Biological control of thrips and whiteflies by a shared predator: two pests are better than one. Biol. Control 44, 372–379. doi: 10.1016/j.biocontrol.2007.10.017
Messelink G., van Steenpaal S., Ramakers P. (2006). Evaluation of phytoseiid predators for control of western flower thrips on greenhouse cucumber. BioControl 51, 753–768. doi: 10.1007/s10526-006-9013-9
Midthassel A., Leather S. R., Baxter I. H. (2013). Life table parameters and capture success ratio studies of typhlodromips swirskii (Acari: Phytoseiidae) to the factitious prey suidasia medanensis (Acari: Suidasidae). Exp. Appl. Acarology 61 (1), 69–78. doi: 10.1007/s10493-013-9682-x
Moquin S. A., Garcia J. R., Brantley S. L., Takacs-Vesbach U. L., Shepherd C. D. (2012). Bacterial diversity of bryophyte-dominant biological soil crusts and associated mites. J. Arid Environ. 87, 110–117. doi: 10.1016/j.jaridenv.2012.05.004
Moser J. C. (1985). Use of sporothecae by phoretic tarsonemus mites to transport ascospores of coniferous bluestain fungi. Trans. Br. Mycology Soc. 84, 750–753. doi: 10.1016/S0007-1536(85)80138-8
Nemergut D. R., Costello E. K., Hamady M., Lozupone C., Jiang L., Schmidt S. K., et al. (2011). Global patterns in the biogeography of bacterial taxa. Environ. Microbiol. 13 (1), 135–144. doi: 10.1111/j.1462-2920.2010.02315.x
Ngwa C., Glockner V., Abdelmohsen M., Scheuermayer U., Fischer R., Hentschel U., et al. (2013). 16S RRNA gene-based identification of elizabethkingia meningoseptica (Flavobacteriales: Flavobacteriaceae) as a dominant midgut bacterium of the asian malaria vector anopheles stephensi (Diptera: Culicidae) with antimicrobial activities. J. Med. Entomology 50, 405–414. doi: 10.1603/ME12180
Pekas A., Palevsky E., Sumner J. C., Perotti M. A., Nesvorná M., Hubert J. (2017). Comparison of bacterial microbiota of the predatory mite neoseiulus cucumeris (Acari: phytoseiidae) and its factitious prey Tyrophagus putrescentiae (Acari: Acaridae). Sci. Rep. 7. doi: 10.1038/s41598-017-00046-6
Perotti M. A., Braig H. R. (2011). Eukaryotic ectosymbionts of acari. J. Appl. Entomology 135, 514–523. doi: 10.1111/j.1439-0418.2011.01639.x
Poinar J. G., Thomas G. (1984). Laboratory Guide to Insect Pathogens and Parasites (New York, USA: Plenum).
Prasad V. (2012). Checklist of Phytoseiidae of the World (Acari: Mesostigmata) (Michigan, USA: Indira Publishing House).
Pukall R., Schumann P., Schütte R., Gols C., Dicke M. (2006). Acaricomes phytoseiuli gen. Nov., sp. Nov., isolated from the predatory mite phytoseiulus persimilis. Int. J. Systematic Evolutionary Microbiol. 56, 465–469. doi: 10.1099/ijs.0.63930-0
Ramakers P. (1983). Mass production and introduction of amblyseius mckenziei and amblyseius cucumeris. Bull. IOBCWRPS 3, 203–206.
R core development team (2012). R: A language and environment for statistical computing (Vienna, Austria: R foundation for statistical computing).
Roets F., Wingfield M. J., Crous P. W., Dreyer L. L. (2007). Discovery of fungus-mite mutualism in a unique niche. Environ. Entomology 36, 1226–1237. doi: 10.1093/ee/36.5.1226
Roets F., Wingfield M. J., Wingfield B. D., Dreyer L. L. (2011). Mites are the most common vectors of the fungus Gondwanamyces proteae in protea infructescences. Fungal Biol. 115, 343–350. doi: 10.1016/j.funbio.2011.01.005
Russell J. A., Moreau C. S., Goldman-Huertas B., Fujiwara M., Lohman D. J., Pierce N. E. (2009). Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl. Acad. Sci. United States America 106 (50), 21236–21241. doi: 10.1073/pnas.0907926106
Schutte C. (2006). A Novel Bacterial Disease of the Predatory Mite Phytoseiulus Persimilis : Disease Syndrome , Disease Transmission and Pathogen Isolation. (Wageningen University)
Schütte C., Dicke M. (2008). Verified and potential pathogens of predatory mites (Acari: Phytoseiidae). Exp. Appl. Acarology 46 (1–4), 307–328. doi: 10.1007/s10493-008-9188-0
Schütte C., Gols R., Kleespies R. G., Poitevin O., Dicke M. (2008). Novel bacterial pathogen Acaricomes phytoseiuli causes severe disease symptoms and histopathological changes in the predatory mite Phytoseiulus persimilis (Acari, Phytoseiidae). J. Invertebrate Pathol. 98, 127–135. doi: 10.1016/j.jip.2008.03.006
Smrž J., Svobodova J., Catska V. (1991). Synergetic participation of tyrophagus putrescentiae (Schrank) (Acari: Acaridida) and its associated bacteria on the destruction of some soil micromycetes. J. Appl. Entomol 11, 206–210.
Smrž J., Trelová M. (1995). The association of bacteria and some soil mites (Acari: Oribatida and Acaridida). Acta Zoologica Fennica 196, 120–123.
Sumner-Kalkun J. C., Baxter I., Alejandra Perotti M. (2020). Microscopic analysis of the microbiota of three commercial phytoseiidae species (Acari: Mesostigmata). Exp. Appl. Acarology 81 (3), 389–408. doi: 10.1007/s10493-020-00520-3
Van Driesche R. G., Lyon S., Stanek E. J., Xu Bo, Nunn C. (2006). Evaluation of efficacy of neoseiulus cucumeris for control of western flower thrips in spring bedding crops. Biol. Control 36 (2), 203–155. doi: 10.1016/j.biocontrol.2005.08.011
Van Houten Y. M., Ostlie M., Hoogerbrugge H., Bolkmans K. (2005). Biological control of western flower thrips on sweet pepper using the predatory mites Amblyseius cucumeris, Iphiseius degenerans, A. Andersoni and A. Swirskii. IOBC/WPRS Bull. 28, 283–286.
Van Lenteren J. C., Woets J. (1988). Biological and integrated pest control in greenhouses. Annu. Rev. Entomology 33, 239–269. doi: 10.1146/annurev.en.33.010188.001323
van Maanen R., Vila E., Sabelis M. W., Janssen A. (2010). Biological control of broad mites (Polyphagotarsonemus latus) with the generalist predator amblyseius Swirskii. Exp. Appl. Acarology 52 (1), 29–345. doi: 10.1007/s10493-010-9343-2
Vásquez C., Colmenárez Y. C., Greco N., Ramos M. (2023). Current status of phytoseiid mites as biological control agents in latin america and experiences from Argentina using neoseiulus californicus. Neotropical Entomology 52, 240–250. doi: 10.1007/s13744-023-01026-4
Waltzer A., Blumel S., Schausberger P. (2001). Population dynamics of interacting predatory mites, phytoseiulus persimilis and neoseiulus californicus, held on detached bean leaves. Exp. Appl. Acarology 25, 731–743. doi: 10.1023/A:1016332129137
Wang Q., Garrity G., Tiedje J., Cole J. (2007). Naive bayesian classifier for rapid asignment of RRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wimmer D., Hoffman D., Schausberger P. (2008). Prey suitability of western flower thrips, frankliniella occidentalis, and onion thrips, thrips tabaci, for the predatory mite amblyseius swirskii. Biocontrol Sci. Technol. 18, 533–542. doi: 10.1080/09583150802029784
Wu K., Hoy M. A. (2012a). Cardinium is associated with reproductive incompatibility in the predatory mite metaseiulus occidentalis (Acari: phytoseiidae). J. Invertebrate Pathol. 110, 359–365. doi: 10.1016/j.jip.2012.03.027
Wu K., Hoy M. A. (2012b). Extended starvation reduced and eliminated wolbachia, but not cardinium, from metaseiulus occidentalis females (Acari: Phytoseiidae): A need to reassess wolbachia’s status in this predatory mite?E. J. Invertebrate Pathol. 109, 20–26. doi: 10.1016/j.jip.2011.09.005
Yoon C., Indiragandhi P., Anandham R., Cho S., Sa T. M., Kim G. -H. (2010). Bacterial diversity and distribution from the whole mite extracts in acaricide resistant and susceptible populations of twospotted spider mite-Tetranychus urticae (Acari: Tetranychidae). J. Korean Soc. Appl. Biol. Chem. 53, 446–457. doi: 10.3839/jksabc.2010.069
Yun Ji H., Woon Roh S., Whon T. W., Jung Mi Ja, Kim M. S., Park D. S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80 (17), 5254–5264. doi: 10.1128/AEM.01226-14
Zhang Q., Wang S., Zhang X., Zhang K., Liu W., Zhang R., et al. (2021). Enterobacter hormaechei in the intestines of housefly larvae promotes host growth by inhibiting harmful intestinal bacteria. Parasites Vectors 14 (1), 1–15. doi: 10.1186/s13071-021-05053-1
Keywords: microbiome, Phytoseiidae, biocontrol, agriculture, mass rearing, symbionts, microbiota, mite
Citation: Sumner-Kalkun JC, Baxter I and Perotti MA (2023) Bacterial microbiota of three commercially mass-reared predatory mite species (Mesostigmata: Phytoseiidae): pathogenic and beneficial interactions. Front. Arachn. Sci. 2:1242716. doi: 10.3389/frchs.2023.1242716
Received: 19 June 2023; Accepted: 25 August 2023;
Published: 28 September 2023.
Edited by:
Alejandro Cabezas-Cruz, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Elianne Piloto-Sardiñas, National Center for Animal and Plant Health (CENSA), CubaAlejandra Wu Chuang, Agence Nationale de Sécurité Sanitaire de l’Alimentation, de l’Environnement et du Travail (ANSES), France
Copyright © 2023 Sumner-Kalkun, Baxter and Perotti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason C. Sumner-Kalkun, amFzb24uc3VtbmVyLWthbGt1bkBzYXNhLmdvdi5zY290
 Jason C. Sumner-Kalkun
Jason C. Sumner-Kalkun Ian Baxter
Ian Baxter M. Alejandra Perotti
M. Alejandra Perotti