- 1Department of Biology, Radford University Carilion, Roanoke, VA, United States
- 2Kapi’olani Medical Center for Women & Children, Hawaii Pacific Health, Honolulu, HI, United States
- 3Department of Ecology & Evolutionary Biology, University of Kansas, Lawrence, KS, United States
Our goal was to resolve phylogenetic relationships among Apis laboriosa, and the Apis dorsata subspecies A. d. dorsata, A. d. binghami, and A. d. breviligula, the last two of which have been proposed as full species by several authors. We carried out a phylogenetic analysis of the giant honey bees using mitochondrial cox1 and cox2 gene sequences analyzed with maximum likelihood methods. We obtained strong support for four clades within A. dorsata in the broad sense: the three subspecies or species mentioned above, and a fourth lineage from south India. However, our analysis did not resolve the phylogenetic relationships among the four lineages. The presence of two genetically distinguishable groups of “A. dorsata” in India parallels the presence there of two cavity-nesting honey bees, A. cerana cerana and A. c. indica (the black hill bees and yellow plains bees, respectively). This suggests that past climatic or geological events may have temporarily isolated Indian populations from populations of the Asian mainland, leading to divergence and possibly speciation of Indian giant and cavity-nesting bees, followed by recolonization of India by eastern Asian forms. Recognition of these distinct lineages is important for conservation planning, so that their individual distributions, ecologies, and migration patterns can be considered, and so that the genetic diversity they represent can be maintained.
Introduction
The giant honey bees have a geographic range centered on south and southeast Asia, extending northwest into Pakistan, eastwards through India, Bangladesh, Nepal, Bhutan, Myanmar, Thailand, southern China, and southeast Asia, and through the islands of Malaysia, Indonesia, and the Philippines (Otis, 1996; Kitnya et al., 2020; Huang et al., 2022; Kitnya et al., 2024; Otis et al., 2024; Voraphab et al., 2024; Warrit et al., 2024). Several earlier writers including Maa (1953) and Ruttner (1988) pointed out diversity among giant honey bee populations based on morphological and morphometric data. In particular they noted that the giant honey bees of the Himalayan region, the Indonesian island of Sulawesi, and the oceanic Philippine islands (i.e., those islands never connected to the Asian mainland) differed from one another and from the more widespread form found elsewhere. Maa divided honey bees into three genera—Micrapis, the dwarf honey bees, Megapis, the giant honey bees, and Apis, the cavity-nesting honey bees—and recognized four giant bee species: Megapis breviligula from the Philippines, M. binghami from Sulawesi and smaller nearby islands, M. laboriosa from high altitude Himalayan regions, and the more widespread M. dorsata. Ruttner, like most subsequent authors, recognized just one genus, Apis, and only one species of giant honey bee, Apis dorsata. He and many subsequent authors (e.g., Engel, 1999, 2002) considered the Himalayan form a subspecies, A. d. laboriosa, but noted that additional information might confirm it as a distinct species.
The taxonomic status of A. laboriosa remained contentious for many years despite numerous studies. Sakagami et al. (1980) made detailed morphological comparisons of A. laboriosa from Nepal and A. dorsata collected from many parts of its range, documenting “distinct and stable differences between them” supporting species status of A. laboriosa. McEvoy and Underwood (1988) reported that they could find no morphological differences between male genitalia (the everted endophallus) of A. laboriosa and A. dorsata, but nonetheless supported species status of A. laboriosa on the basis of other morphological differences, habitat, the presence of two species of braulid parasites (Diptera: Bruaulidae, Megabraula) in nests of A. laboriosa but (apparently) not those of A. dorsata, and genetic differences revealed by allozyme electrophoresis.
However, some authors argued that the characters used to support species status of A. laboriosa—including habitat, color patterns, and morphometric characters—could represent intraspecific variation and adaptation to different habitats, and thus took the conservative position that more data were needed, particularly concerning reproductive isolation of populations occurring in sympatry (e.g., Ruttner, 1988; Engel, 1999). Cao et al. (2012a) carried out morphometric comparisons of A. laboriosa and A. dorsata collected from Yunnan, Guangxi and Hainan provinces in China and again found significant differences between them. Collection sites for the two were in relatively close proximity (on the order of 200-300 km) but not strictly sympatric, and they were found at different elevations (A. laboriosa 1500 m and above, A. dorsata 1300 m and lower, though all but one collection was made at 700 m or lower).
More recently, new distributional records for A. laboriosa (Kitnya et al., 2020) reported A. dorsata and A. laboriosa foraging sympatrically at sites in Arunachal Pradesh, India and in northern Vietnam. Kitnya et al. (2022), found distinct morphological, morphometric, and genetic differences between Indian populations of A. dorsata and A. laboriosa, both in sympatry and in allopatry, providing convincing support for the species status of A. laboriosa.
Until recently the species status of A. d. breviligula and A. d. binghami have received much less attention. Arias and Sheppard (2005) included A. laboriosa, A. dorsata from Thailand and Sri Lanka, and A. binghami in a larger phylogenetic analysis of Apis species using both nuclear (EF-1α intron) and mitochondrial (ND2) sequence data. The giant honey bees were recovered as a monophyletic group and A. laboriosa was consistently recovered as a clade distinct from A. dorsata and A. d. binghami; however, A. dorsata and A. d. binghami were not consistently resolved as separate lineages. Raffiudin and Crozier (2007) used both mitochondrial (cox2, ND2, and the large (16S) ribosomal subunit or rrnL) and nuclear (inositol 1,4,5-triphosphate receptor or itpr) gene sequences in their phylogenetic analysis of Apis taxa, also including the giant honey bees A. dorsata from Sabah, Malaysia, A. d. binghami, and A. laboriosa. Their analyses consistently recovered A. laboriosa as sister to A. dorsata and A. d. binghami. Lo et al. (2010) carried out a more comprehensive coverage of giant honey bees, including A. laboriosa, A. dorsata from Sabah, Malaysia and Palawan Island, the Philippines, A. d. binghami and A. d. breviligula in their phylogenentic analysis of Apis species, using the same set of genes as Raffiudin and Crozier (2007) minus the mitochondrial ND2. Their results strongly supported the species status of A. d. breviligula from the Philippines, though the placement of A. d. binghami remained unresolved.
Kitnya et al. (2024) carried out a taxonomic study of giant honey bees using morphological characters. Their study included A. laboriosa, A. dorsata, and the island lineages A. d. binghami and A. d. breviligula. They found that A. dorsata from mainland Asia differs morphologically from A. d. binghami and A. d. breviligula but concluded that the latter two represent a single morphological species, A. binghami, with two subspecies, A. b. binghami and A. b. breviligula.
In this paper, we accept the species status of A. laboriosa. We use the names A. dorsata dorsata, A. d. breviligula and A. d. binghami for the other distinctive populations of giant honey bees because the species status of the latter two is still subject to investigation. We use the name “A. d. SouthIndia” to refer to a population that appears to be a cryptic unnamed species or subspecies (Smith, 1991; Kitnya et al., 2022). “Apis dorsata in the broad sense” will refer to all giant honey bees excluding A. laboriosa.
The objective of this study is to carry out a phylogenetic analysis for populations of giant honey bees, including representatives from as much of their range as we could obtain, to test whether the lineages within A. dorsata in the broad sense are monophyletic, and to determine relationships among them. Samples include A. laboriosa [Nepal], A. d. dorsata [multiple populations], and the distinctive island populations A. d. binghami [Sulawesi and smaller nearby islands] and A. d. breviligula [the oceanic islands of the Philippines]. We also include the dwarf honey bees, A. florea and A. andreniformis, and the cavity-nesting honey bees A. mellifera and A. cerana as outgroups. We generated partial sequences of the mitochondrial cytochrome c oxidase subunit 1 (cox1) and cytochrome c oxidase subunit 2 (cox2) genes and used Maximum Likelihood methods in MEGA7 to construct phylogenetic trees.
Methods
Field methods
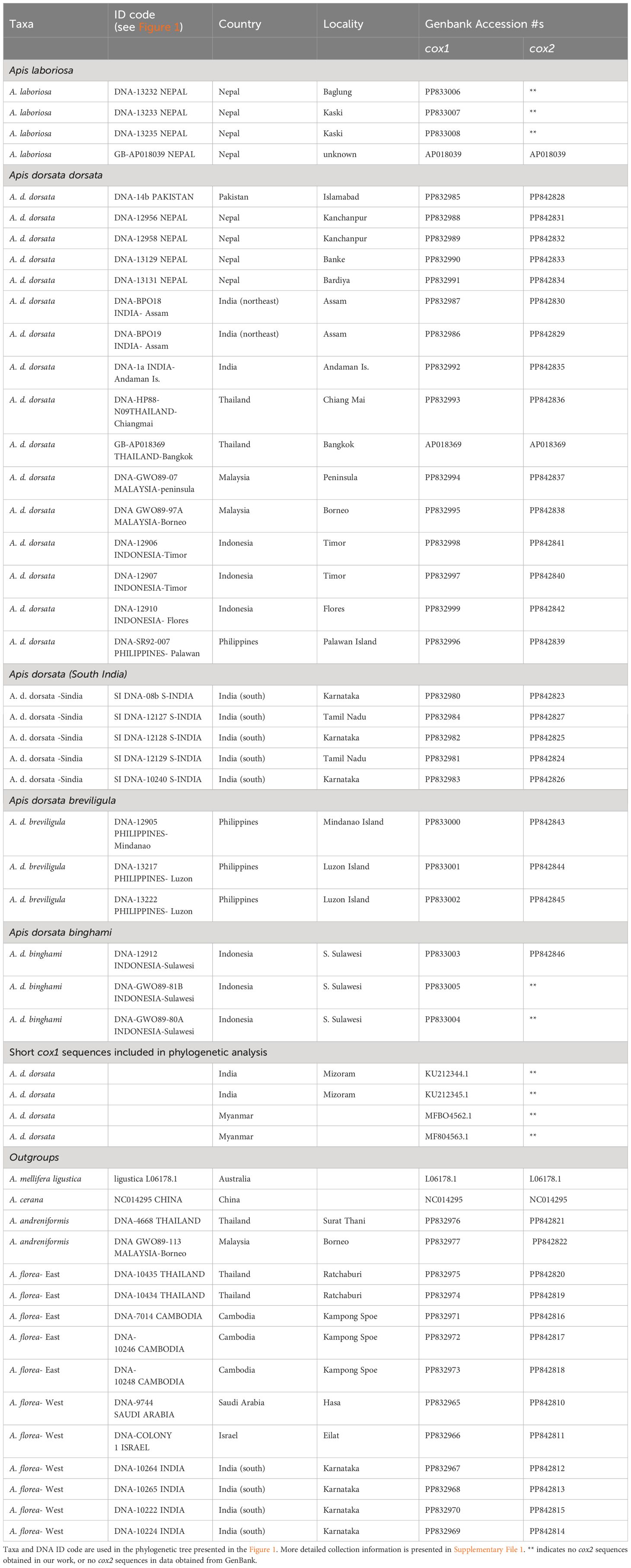
Samples used in this study were collected by multiple researchers from 1989 to 2018 using a variety of collection and preservation techniques. Table 1 gives locality and collection information, and sample IDs corresponding to those used in Figure 1. Most specimens were collected directly from colonies, though some bees were collected while they were foraging. Most specimens are adult worker bees, while a few are pupae collected directly from nests. Individual bees or bee thoraces were preserved in the field in liquid nitrogen (1988–1990) or in 95% ethanol (1991 onwards). Frozen specimens were later stored at −80°C. Ethanol-preserved specimens were stored at 4° to −20°C.
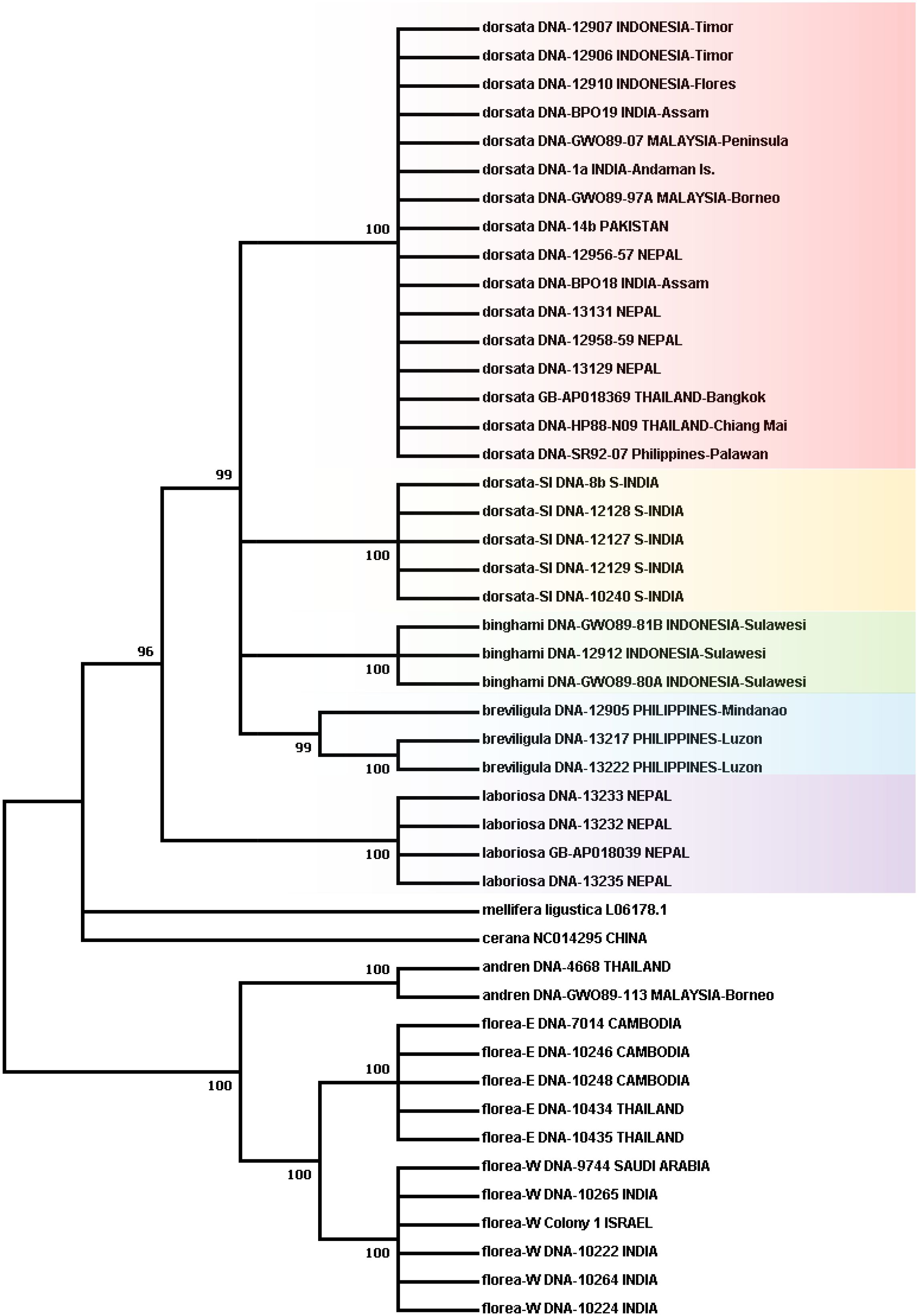
Figure 1 Phylogenetic tree obtained using the Maximum Likelihood method based on the General Time Reversible model. Numbers on branches indicate bootstrap support; partitions with less than 95% support were collapsed. See text for more detailed information on analysis methods.
Laboratory methods
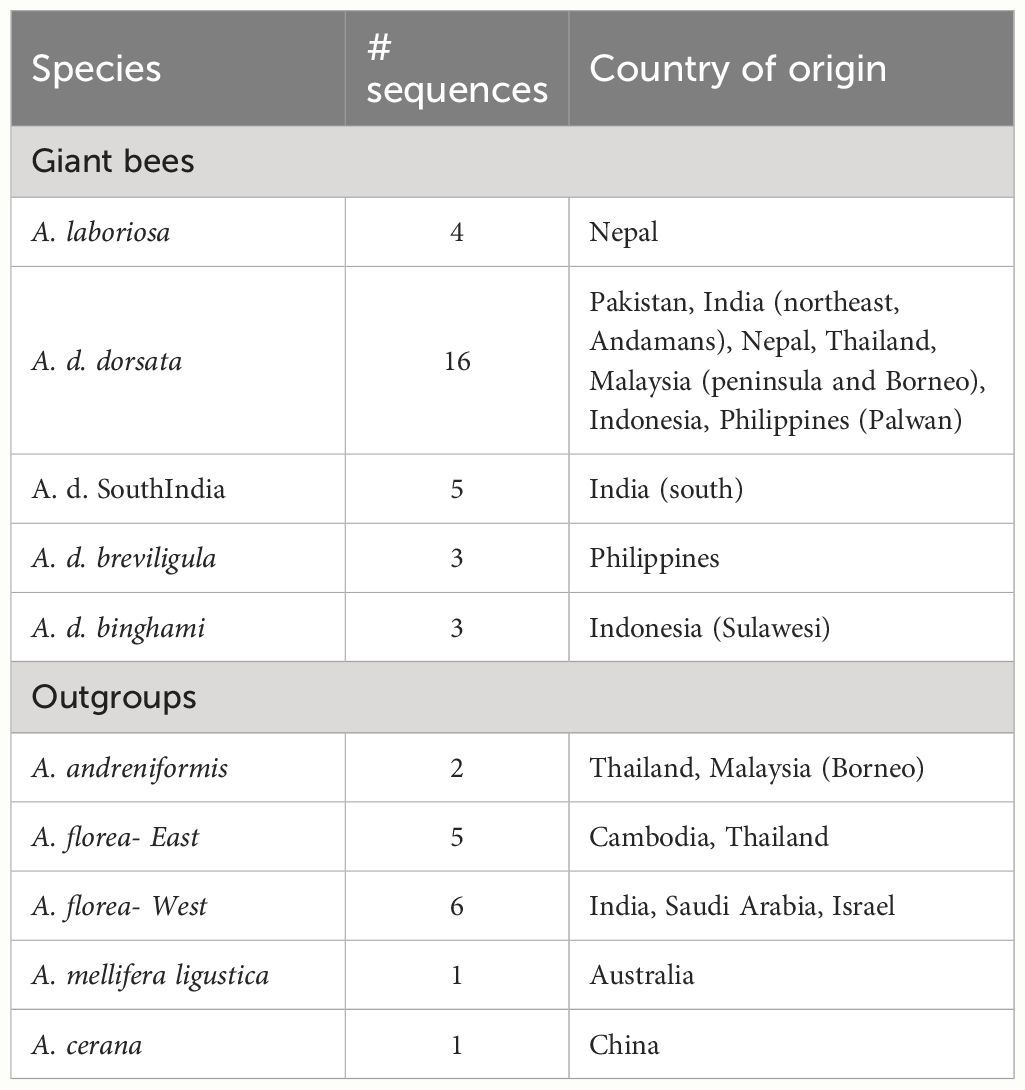
Genomic DNA was extracted from the mitochondrion-rich thoracic flight muscle tissue using DNA spin-columns, primarily the Qiagen DNEasy Blood and Tissue kit (www.Qiagen.com, Ann Arbor, MI USA) and the GenElute Mammalian Genomic DNA Miniprep kit (www.sigmaaldrich.com, St. Louis, MO USA) following the manufacturers’ recommendations. Extracted DNAs were stored at −20°C. Portions of the mitochondrial genome were amplified using the primers shown in Table 2. These sequences included a large portion of cox1, leucine tRNAUUR, a short non-coding sequence, and a portion of cox2. Figure 2 shows the relative position of the primers on the honey bee cox1 to cox2 sequences. Sanger sequencing was carried out at the Idaho State University Molecular Research Core Facility, Pocatello, ID. As only protein-coding sequence was included in the phylogenetic analysis, the tRNA and non-coding sequences were removed after alignment (see below) and the cox1 and cox2 sequences were concatenated. Some of the sequences were also obtained from Genbank (Table 1). The total number of sequences for each taxon and their geographic origins are summarized in Table 3.
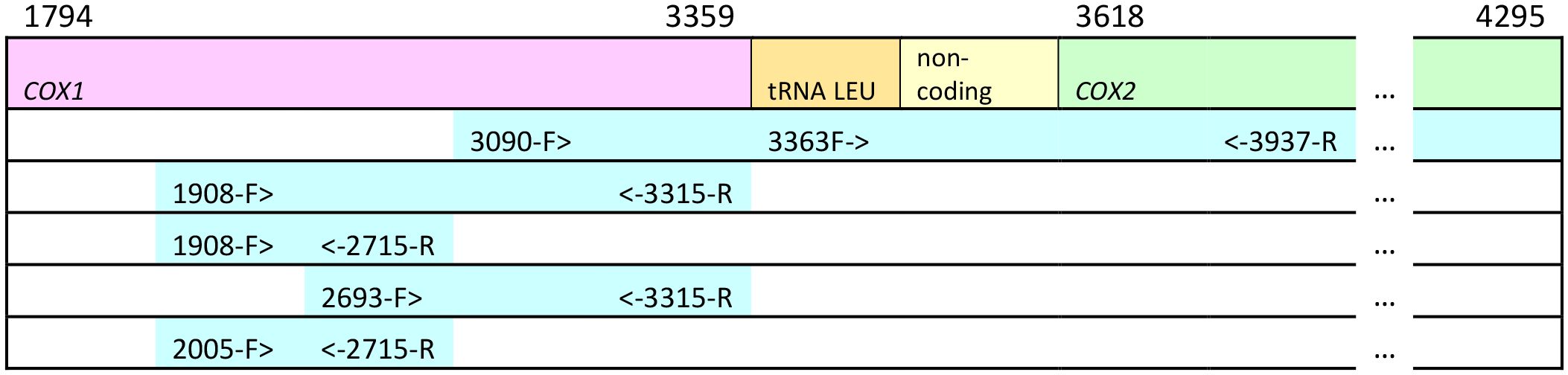
Figure 2 Relative position of primers on the mitochondrial cox1, cox2 and leucine tRNA genes. Numbers above cox1 and cox2 indicate starting and ending position of the genes in the complete mitochondrial genome of Apis mellifera ligustica: numbers in the primer names refer to the position of the 5' end of the primer in the A. m. ligustica mitochondrial genome (Genbank Accession #L06178.1; Crozier and Crozier, 1993). Not drawn to scale.
Phylogenetic analysis
Sequences were aligned manually with cox1 and cox2 sequences from Apis mellifera ligustica (Crozier and Crozier, 1993; Genbank accession L06178) in MEGA7 (Kumar et al., 2016). Sequences were screened for missing bases and correct reading frame by translating DNA sequences to amino acid sequences. In total, 46 sequences were used in the phylogenetic analysis and another four Genbank sequences of A. dorsata dorsata from India (Mizoram) and Myanmar (Table 1) that were too short to include in the phylogenetic analysis were aligned with the larger data set to determine which sequences they matched most closely.
The best model of sequence evolution was selected using MEGA “Model Selection” analysis and the following conditions: maximum likelihood statistical methods, partial deletion of sites with missing data, coverage cutoff of 75%, all codon positions used, moderate branch swapping filter. The model of sequence evolution with the lowest Bayesian Information Criteria (BIC) score was selected for use in the phylogenetic analysis. This model (BIC score 13302.38) was a general time reversible model with non-uniform rates of evolution among sites (gamma distributed) and a fraction of sites seemingly invariable (GTR+G+I).
Phylogenetic trees were constructed using Maximum Likelihood methods in MEGA7 with the following settings: model of evolution gamma distributed with invariant sites (GTR+G+I) with 5 gamma categories, partial deletion of sites with missing data, 75% site coverage cutoff, all codon positions used, maximum likelihood heuristic method Subtree-Pruning-Regrafting-Fast, initial tree generated by Neighbor-Joining, moderate branch swap filter, 3 threads. Support for the branching patterns was evaluated with 1000 bootstrap replicates. Branches with less than 95% bootstrap support were collapsed. A coverage cutoff of 75% was chosen during model choice and tree-building to ensure that inclusion of shorter sequences did not result in elimination of informative data.
Results
Figure 1 presents the phylogenetic tree obtained showing partitions with 95% bootstrap support or better. As has been found in other recent studies, A. laboriosa constitutes a well-supported lineage separate from and sister to all A. dorsata in the broad sense, further supporting its status as a distinct species. Within A. dorsata in the broad sense, we found four distinct lineages: A. d. breviligula from the oceanic Philippine islands, A. d. binghami from the Indonesian island of Sulawesi, A. d. SouthIndia, a genetically distinct population so far known only from southern India, and a more narrowly defined A. dorsata dorsata, represented by our samples from Pakistan, Nepal, northeastern India (Assam and the Andaman Islands), Thailand, Malaysia (Peninsular and Sabah, Borneo), the Philippine island of Palawan, and the Indonesian islands of Timor and Flores. The short sequences from Mizoram, India and Myanmar most closely matched those of the A. dorsata dorsata group and were clearly distinct from the A. d. SouthIndia group (Table 4).
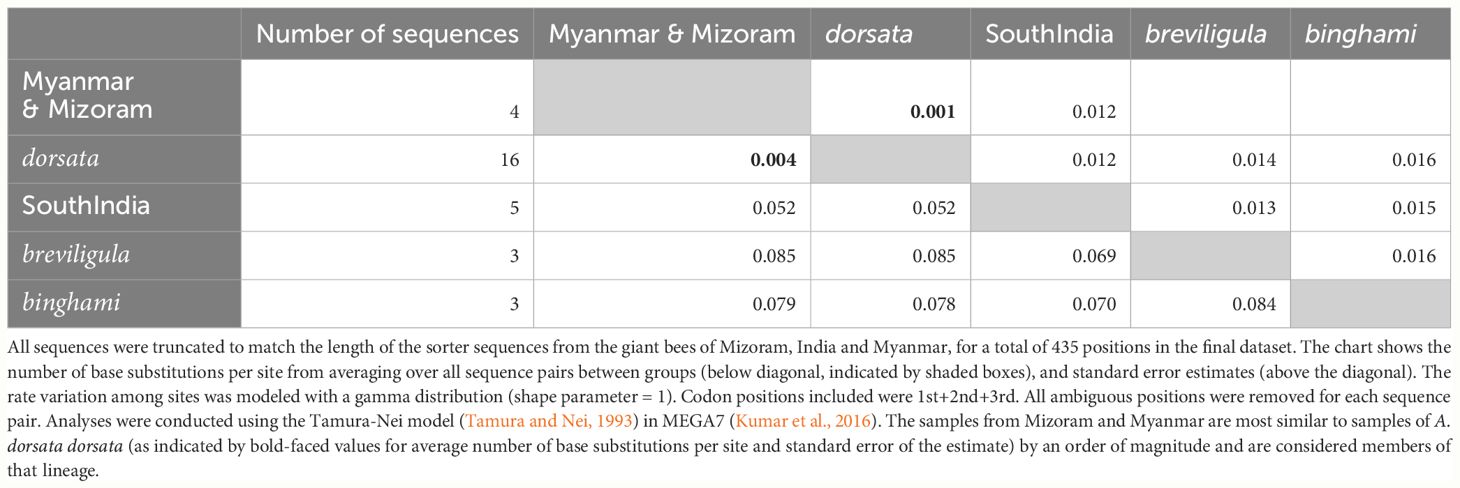
Table 4 Comparison of sequence similarity between samples of A. dorsata from Myanmar and Mizsoram, India (see Table 1) and the giant bee lineages A. dorsata dorsata, A. breviligula (or A. d. breviligula), A. binghami (or A. d. binghami) and a mitochondrially distinct giant bee found in southern India (A. d. SouthIndia).
Unfortunately, although this analysis shows four well-supported lineages within A. dorsata in the broad sense, it does not resolve branching patterns among the four lineages.
Discussion
In this study we support the species status of A. laboriosa and show that Apis dorsata in the broad sense includes four genetically distinguishable lineages: A. dorsata dorsata, A. d. binghami, A. d. breviligula and A. d. SouthIndia, though our data do not resolve branching patterns among the four lineages. Regardless of whether these four lineages merit species status, recognition and continued investigation of these groups are important for the study of honey bee biogeography, for maintenance of existing diversity within the giant honey bees, and even for conservation of the Asian bee fauna.
Honey bee biogeography
Although color, morphometric and morphological differences among A. d. dorsata, A. d. binghami and A. d. breviligula have been reported (e.g., Maa, 1953, but see Kitnya et al., 2024), there are no obvious morphological differences between A. d. SouthIndia and the widespread A. d. dorsata (Kitnya et al., 2024). In their study of A. laboriosa and A. dorsata in India, Kitnya et al. (2022) examined specimens of A. dorsata from Arunachal Pradesh in the extreme northeast of India and Karnataka (specifically Bangalore) in south India. In a dendogram displaying morphometric similarity of the samples, the south India specimens did not form a discrete cluster but were mixed in among the specimens from northeast India. However, in their phylogenetic analysis of the same collections (using a 500 bp fragment of the mitochondrial cox1 gene), the samples from Arunachal Pradesh and south India formed two separate clades with 99% and 100% bootstrap support, respectively. To the best of our knowledge, there is no information on the geographical distributions of A. d. dorsata and A. d. SouthIndia. Although giant honeybees have been collected at points between southern and northern India, at the moment the only way to tell the two apart is by genetic testing.
The distinctive nature of A. d. breviligula and A. d. binghami compared to the more widespread A. d. dorsata has long been recognized (e.g., Maa, 1953; Ruttner, 1988). A. d. breviligula and A. d. binghami are primarily black, with white stripes on metasomal tergites 3,4, and 5 formed by short white hairs, while the metasomal terga 1-3 (and sometimes tergites 4-5)and sterna 1-2 are yellow to brown in A. dorsata dorsata (Kitnya et al., 2024). In addition to differences in coloration, A. dorsata dorsata of mainland Asia differs from the two island taxa based on ocellus size and the spacing of the compound eyes and ocelli (Kitnya et al., 2024). Whether the two island forms constitute separate species remains to be determined. Kitnya et al. (2024) found no morphological basis for separating the two, and considered them a single species, distinct from A. d. dorsata, with two subspecies: A. binghami binghami and A. b. breviligula. Evidence from mitochondrial gene sequences presented here retrieves A. d. binghami and A. d. breviligula as genetically distinct but is insufficient determine species status.
The fact that isolated island populations show traits distinct from those of mainland populations is not surprising. What is more surprising is the presence of a genetically distinct giant honey bee in southern India, along with the more widespread A. dorsata dorsata in northern India. However, a broader view of Indian Apis shows that this pattern has appeared more than once. India is also home to two cavity-nesting bees, the yellow or plains bee, and the hill or black bee (Ruttner, 1988 and references cited therein; Bhatta et al., 2020). According to Engel (2002) the yellow or plains bee corresponds to A. cerana indica Fabricius, 1798 while the black or hill bee corresponds to A. cerana cerana Fabricius, 1793. Genetic evidence collected over the past three decades (e.g., mitochondrial cox1, cox2 and non-coding sequences, Smith, 1991; Smith and Hagen, 1996; mitochondrial and nuclear gene sequences, Lo et al., 2010; and genomic SNPs, Su et al., 2023) support species status of the yellow Indian bee, as proposed by Lo et al., 2010; Smith, 2011, and Su et al., 2023. The dwarf honey bee, Apis florea, also consists of two distinct groups revealed by mitochondrial cox1-cox2 sequences and nuclear SNPs (Smith, 2011; Su et al., 2023, and Figure 1 of this study). These are an eastern lineage including populations from Thailand eastwards, and a western lineage including populations from India westward, including the invasive dwarf honey bee populations in Jordan and Israel, and probably those in east Africa as well. The “switchover” from East to West is apparently in the poorly sampled region from northeastern India through Bangladesh and Myanmar.
Why does India have a distinct variety of cavity-nesting bee, A. cerana indica, along with A. cerana cerana, and a distinct south Indian variety of giant honey bee along with A. dorsata dorsata in northern India? And why does it have a variety of A. florea different from that in eastern Asia? Answering these questions requires (1) information on the ranges of the species and putative species of Apis in India, particularly the distributions of the yellow and black cavity-nesting bees, and A. d. SouthIndia and A. dorsata dorsata, and (2) a phylogeny that resolves the branching patterns of the four lineages within A. dorsata in the broad sense. A robust phylogeny would provide information on the order and timing of diversification events. A time calibrated phylogeny could suggest specific geological and climatic events that could have promoted diversification, and help us determine if the dwarf, giant, and cavity nesting lineages responded to historical events with similar patterns of diversification.
Maintenance of diversity in the giant bees
At least three of the four lineages within A. dorsata in the broad sense exhibit migratory behavior. The vast majority of giant honey bee migration research has been carried out on populations that our study would place in A. d. dorsata (for example, Dyer and Seeley, 1994; Kahono et al., 1999; Neumann et al., 2000; Paar et al., 2000; Itioka et al., 2001; Rattanawannee et al., 2013), which is not surprising, as it is the most widespread. Koeniger and Koeniger (1980) investigated giant honey bee migration in Sri Lanka; though we have not sampled any giant bees from Sri Lanka, we predict that they are part of the A. d. SouthIndia clade, based on the fact that the south Indian plains bee, A. c. indica, is also found in Sri Lanka. At least one set of observations has been made on migration by A. d. binghami in Sulawesi (Nagir et al., 2016). Morse and Laigo (1969, cited in Robinson, 2021) reported that A. d. breviligula in the Philippines does not migrate.
Migration is typically a predictable annual response to seasonal patterns of rainfall and resource availability (e.g., Dyer and Seeley, 1994) or a response to erratically occurring masting events in which forest trees produce a superabundance of blossoms and resources (Itioka et al., 2001). Migrating bees appear to show fidelity to their nesting sites at either end of the migratory route (Neumann et al., 2000; Paar et al., 2000). This alone means that protecting a giant honey bee’s nesting and foraging habitat means protecting more than one location. Migrating colonies of giant honey bees may travel distances that require “rest stops” to forage. Recent work by Robinson, (2012, 2021) has shown that migrating A. d. dorsata in Thailand make use of “traditional” rest stops, where they forage for food and water for variable lengths of time before continuing their journey. These rest stops are likely to be crucial for successful migration. To maintain the genetic diversity represented by linages within A. dorsata in the broad sense, it will be necessary to maintain not only the endpoints of their migratory routes, but quite probably sufficient rest stops along the route too, in conditions that provide the bees with the forage, resting sites and nesting sites they need. This would require tracking migration routes and their timing and noting changes in migration timing or route due to climate change or habitat destruction.
Conservation
Warrit et al. (2024) discuss the challenges facing bee research and bee conservation in Asia, noting, “If we do not know the species present, their distribution and threats, we cannot protect them.” They point to the eusocial bees as “flagship species” for bee conservation measures, as their economic value to humans—through pollination services and honey production—is generally known to the public. In particular, “the honey bee” is likely the only bee most people know, especially in urban populations. Although the giant bees are large, conspicuous, and widespread across the Asian continent, we are still discovering new diversity (at the species or subspecies level) and still lack basic information on the ranges of some lineages such as the south Indian giant bee.
Giant honey bees are not just major pollinators in Asian ecosystems. With their large, conspicuous open-air combs, large aggregations of nests, and migratory behavior, plus the well-publicized harvesting of cliff-side A. laboriosa nests, they are arguably the most charismatic of the Asian social bees. Public support for protection of giant honey bees would also have the effect of protecting habitat for the many other social and solitary Asian bee species.
Our results suggest avenues for additional research, particularly regarding Indian populations. What is the range of the South Indian giant honey bee, and what are its migration patterns? Is the range of the southern Indian cavity-nesting “plains bee” (currently A. cerana indica) congruent with the range of the southern Indian giant honey bee, suggesting similar biogeographic history? Does the south Indian giant honey bee differ in behavior or ecology from A. d. dorsata? And of course, will behavioral and genetic study of giant honey bees from a greater portion of their ranges (e.g., as in Cao et al., 2012b) reveal more diversity?
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the study as no human subjects, other vertebrates, or higher invertebrates were used. This study used preserved insect specimens collected from 1989 to 2018.
Author contributions
CB: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. SZ: Investigation, Methodology, Writing – review & editing. DS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was partially supported by National Science Foundation grants BSR-8918932 to DS and Fred Dyer and USDA-NIFA AFRI 2010-65-104-20533 to O. Rueppell and DS, and by an Undergraduate Research Award from the University of Kansas to SZ. We also benefited from the generosity of many bee-keepers and colleagues who shared specimens with us.
Acknowledgments
We would like to thank the many people who have helped us in the field and by collecting and donating specimens: Ahmed Al-Ghamdi, Nicola Bradbear, Fred Dyer, Steven Goodman, the late Randall Hepburn, Ben Oldroyd, Jurgen Paar, the late Herman Pechhacker, Stephen Petersen, the late Stefan Reyes, Benny Shalmon, Yong-Chao Su, and especially Gard Otis, who helped many bee researchers begin their studies of Asian honey bees. A very large portion of this work was completed by SZ (née Cluff) in partial fulfilment of the requirements for an Honors thesis and Bachelor of Science (Honors) degree in Biological Sciences at the University of Kansas.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frbee.2024.1401851/full#supplementary-material
References
Arias M. C., Sheppard W. S. (2005). Phylogenetic relationships of honeybees (Hymenoptera: Apinae: Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 37, 25–35. doi: 10.1016/j.ympev.2005.02.017
Bhatta C. P., Reddy M. S., Smith D. R. (2020). Scientific note: Varroa jacobsoni and V. destructor on hill and plains strains of Apis cerana in southern India. Apidologie 51, 391–394. doi: 10.1007/s13592-019-00723-7
Cao L.-F., Zheng H.-Q., Chen X., Niu D.-F., Hu F.-L., Hepburn H. R. (2012a). Multivariate morphometric analyses of the giant honeybees, Apis dorsata F. and Apis laboriosa F. in China. J. Apicul. Res. 51, 245–251. doi: 10.3896/IBRA.1.51.3.05
Cao L.-F., Zheng H.-Q., Hu C.-Y., He S.-Y., Kuang H.-O., Hu F.-L. (2012b). Phylogeography of the giant honeybee Apis dorsata (Hymenoptera: Apidae) from China and neighbouring Asian areas. Ann. Entomol. Soc. America 105, 298–304. doi: 10.1603/AN11104
Cornuet J.-M., Garnery L., Solignac M. (1991). Putative origin and function of the intergenic region between COI and COII of Apis mellifera L. Mitochondrial DNA. Genetics 128, 393–403. doi: 10.1093/genetics/128.2.393
Crozier R. H., Crozier Y. C. (1993). The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics 133, 97–117. doi: 10.1093/genetics/133.1.97
Dyer F. C., Seeley T. H. (1994). Colony Migration in the tropical honeybee Apis dorsata F. (Hymenoptera: Apidae). Insectes Sociaux 41, 129–140. doi: 10.1007/BF01240473
Engel M. S. (1999). The taxonomy of recent and fossil honeybees (Hymenoptera: Apidae; Apis). J. Hymenoptera Res. 8, 165–196.
Engel M. S. (2002). The honeybees of India, Hymenoptera: Apidae. J. Bombay Natural History Soc. 99, 3–7.
Hall H. G., Smith D. R. (1991). Distinguishing African and European honeybee matrilines using amplified mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A. 88, 4548–4552. doi: 10.1073/pnas.88.10.4548
Huang M.-J., Hughes A. C., Xu Z.-Y., Miao B.-G., Gao J., Peng Y.-Q. (2022). Mapping the changing distribution of two important pollinating giant honey bees across 21000 years. Glob. Ecol. Conserv. 39, e02282. doi: 10.1016/j.gecco.2022.e02282
Itioka T., Inoue T., Kaliang H., Kato M., Nagamitsu T., Momose K., et al. (2001). Six-year population fluctuation of the giant honeybee Apis dorsata (Hymenoptera: Apidae) in a tropical lowland dipterocarp forest in Sarawak. Ann. Entomol. Soc. America 94, 545–549. doi: 10.1603/0013-8746(2001)094[0545:SYPFOT]2.0.CO;2
Kahono S., Nakamura K., Amir M. (1999). Seasonal migration and colony behavior of the tropical honeybee Apis dorsata F. (Hymenoptera: Apidae). Treubia 31, 283–297. doi: 10.14203/treubia.v31i3.611
Kitnya N., Brockmann B., Otis G. W. (2024). Taxonomic revision and identification keys for the giant honey bees. bioRxiv. doi: 10.1101/2024.04.03.587895
Kitnya N., Otis G. W., Chakravorty J., Smith D. R., Brockmann A. (2022). Apis laboriosa confirmed by morphometric and genetic analyses of giant honeybees (Hymenoptera, Apidae) from sites of sympatry in Arunachal Pradesh, Northeast India. Apidologie 53, 47, 17. doi: 10.1007/s13592-022-00956-z
Kitnya N., Prabhudev M. V., Bhatta C. P., Pham T. H., Nidup T., Megu K., et al. (2020). Geographical distribution of the giant honeybee Apis laboriosa Smith 1871(Hymenoptera, Apidae). Zookeys 951, 67–81. doi: 10.3897/zookeys.951.49855
Koeniger N., Koeniger G. (1980). Observations and experiments on migration and dance communication of Apis dorsata in Sri-Lanka. J. Apicul. Res. 19, 21–34. doi: 10.1080/00218839.1980.11099994
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lo N., Gloag R. S., Anderson D. L., Oldroyd B. P. (2010). A molecular phylogeny of the genus Apis suggests that the giant honeybee of the Philippines, A. breviligula Maa, and the plains honeybee of southern India, A. indica Fabricius, are valid species. Systemat. Entomol. 35, 226–233. doi: 10.1111/j.1365-3113.2009.00504.x
Maa T. C. (1953). An inquiry into the systematics of the tribus Apidini or honeybees (Hymenoptera). Treubia 21, 525–640. doi: 10.14203/treubia.v21i3.2669
McEvoy V. M., Underwood B. A. (1988). Drone and species status of the Himalayan honeybee, Apis laboriosa (Hymenoptera: Apidae). J. Kansas Entomol. Soc. 61, 246–249. https://www.jstor.org/stable/25084995
Morse R. A., Laigo F. M. (1969). Apis dorsata in the Philippines (Laguna, Philippines: Philippine Association of Entomologists).
Nagir M. T., Atmowidi T., Kahono S. (2016). The distribution and nest–site preference of Apis dorsata binghami at Maros Forest, South Sulawesi, Indonesia. J. Insect Biodiversity 4, 1–14. doi: 10.12976/jib/2016.4.23
Neumann P., Koeniger N., Koeniger G., Tingek S., Kryger P., Moritz R. F. A. (2000). Home–site fidelity in migratory honeybees. Nature 406, 474–475. doi: 10.1038/35020193
Otis G. W. (1996). Distributions of recently recognized species of honey bees (Hymenoptera: Apidae; Apis) in Asia. J. Kansas Entomol. Soc. 64 (4) suppl, 311–333. http://www.jstor.org/stable/25085727
Otis G. W., Huang M.-J., Kitnya N., Sheikh U. A. A., Faiz A. H., Phung C. H., et al. (2024). The distribution of Apis laboriosa revisited: range extensions, biogeographic affinities, and species distribution modeling. Front. Bee Sci. 2:1374852. doi: 10.3389/frbee.2024.1374852
Paar J., Oldroyd B. P., Kastberger G. (2000). Giant honeybees return to their nest sites. Nature 406, 475. doi: 10.1038/35020196
Raffiudin R., Crozier R. H. (2007). Phylogenetic analysis of honeybee behavioral evolution. Mol. Phylogenet. Evol. 43, 543–552. doi: 10.1016/j.ympev.2006.10.013
Rattanawannee A., Chanchao C., Lim J., Wongsiri S., Oldroyd B. P. (2013). Genetic structure of a giant honeybee (Apis dorsata) population in northern Thailand: implications for conservation. Insect Conserv. Diversity 6, 38–44. doi: 10.1111/j.1752-4598.2012.00193.x
Robinson W. (2012). Migrating giant honeybees (Apis dorsata) congregate annually at stopover site in Thailand. PloS One 7, e44976. doi: 10.1371/journal.pone.0044976
Robinson W. (2021). Surfing the Sweet Wave: migrating giant honeybees (Hymenoptera: Apidae: Apis dorsata) display spatial and temporal fidelity to annual stopover site in Thailand. J. Insect Sci. 21, 1–12. doi: 10.1093/jisesa/ieab037
Ruttner F. (1988). Biogeography and taxonomy of honeybees (Berlin, Germany: Springer). doi: 10.1007/978-3-642-72649-1
Sakagami S. F., Matsumura T., Ito K. (1980). Apis laboriosa in Himalaya, the little-known world largest honeybee (Hymenoptera, Apidae). Insecta Matsumurana 19, 47–77. http://hdl.handle.net/2115/9801
Smith D. R. (1991). “Mitochondrial DNA and honeybee biogeography,” in Diversity in the genus Apis. Ed. Smith D. R. (Westview Press, Boulder, CO), 131–176.
Smith D. R. (2011). “Asian honeybees and mitochondrial DNA,” in Honeybees of Asia. Eds. Hepburn H. R., Radloff S. E. (Springer Verlag, New York), 69–93. 681 pp.
Smith D. R., Hagen R. H. (1996). The biogeography of Apis cerana as revealed by mitochondrial DNA sequence data. J. Kansas Entomol. Soc. 69, 294–310. https://www.jstor.org/stable/25085726
Su Y.-C., Chiu Y.-F., Warrit N., Otis G. W., Smith D. R. (2023). Phylogeography and species delimitation of the Asian cavity-nesting honeybees. Insect Systemat. Diversity 7, 1–10. doi: 10.1093/isd/ixad015
Tamura K., Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Voraphab I., Chatthanabun N., Nalinrachatakan P., Thanoosing C., Traiyasut P., Kunsete C., et al. (2024). Discovery of the Himalayan giant honey bee, Apis laboriosa, in Thailand: a major range extension. Apidologie 55, 31. doi: 10.1007/s13592-024-01069-5
Keywords: phylogeny, Apis dorsata, Apis laboriosa, cox1 gene, cox2 gene, species discrimination
Citation: Bhatta CP, Zajonz SC and Smith DR (2024) Phylogeography of the giant honey bees based on mitochondrial gene sequences. Front. Bee Sci. 2:1401851. doi: 10.3389/frbee.2024.1401851
Received: 18 March 2024; Accepted: 20 May 2024;
Published: 28 June 2024.
Edited by:
Andrea Galimberti, University of Milano – Bicocca, ItalyReviewed by:
Andrea Ferrari, University of Milan, ItalyPetar Hristov, Bulgarian Academy of Sciences, Bulgaria
Copyright © 2024 Bhatta, Zajonz and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chet P. Bhatta, Y2JoYXR0YUByYWRmb3JkLmVkdQ==; Deborah R. Smith, ZGVic21pdGhAa3UuZWR1
†ORCID: Chet P. Bhatta, orcid.org/0000-0002-2472-6397
Deborah R. Smith, orcid.org/0000-0002-2581-5009
 Chet P. Bhatta
Chet P. Bhatta Sarah C. Zajonz
Sarah C. Zajonz Deborah R. Smith
Deborah R. Smith