- 1Department of Horticulture, Washington State University, Pullman, WA, United States
- 2Department of Horticultural Sciences, Texas A&M University, College Station, TX, United States
- 3Department of Biochemistry, University of California, Riverside, Riverside, CA, United States
We characterized the effects of crop residue derived biochar on tomato growth, soil microbial diversity, and rhizosphere-level gene expression responses in an organic production system. Shoot fresh biomass and fruit yield were assessed at the end of the growing cycle. The corresponding transcriptomic response of the roots, the soil microbial community profiles, and the active transcripts within the communities were quantified using a metatranscriptomic approach at four different developmental stages of the plant. Biochar treatment did not impact shoot biomass or fruit production; however, metatranscriptome analysis revealed that the gene expression activity of the tomato rhizosphere changes over time in response to the biochar treatment, with a number of bacteria with known benefits to soil health and plant growth displaying increased gene expression (e.g., Rhizobiaceae, Pseudomonadaceae, Micromonosporaceae, Sphingomonadaceae). Streptomycetaceae were expressed at the highest levels in the rhizosphere. Biochar seemed to attenuate the expression of this bacteria by the end of the time course, possibly due to the rise in competition for resources driven by the increased activity of other beneficial microbes. Notably, pathogenic fungi in the soil displayed generally reduced expression in the biochar-amended rhizosphere in comparison with the control. In addition to the assessment of the rhizosphere microbiome, transcriptome analysis and gene ontology analysis of tomato roots revealed functional enrichment of genes associated with nitrogen metabolic processes, regulation of metabolic processes, and production of organic compounds in the biochar treated rhizosphere. Together, these results suggest that biochar amendment enhances gene expression of beneficial soil microbes, and also impacts gene expression in the plant roots, which may in turn lead to improvements in soil and plant health. The results of this study provide foundations and a methodology for using metatranscriptomic approaches to investigate the impacts of biochar or other soil amendments in different crops, varying soil types, and with greater experimental complexity. The findings of such investigations will inform the development of biochar-based soil amendment strategies to enhance soil fertility and crop health in a wide range of production systems.
Introduction
Intensive agricultural practices have negatively impacted soil health over time, thereby decreasing crop yield and quality (Tahat et al., 2020; Tenic et al., 2020; Xu et al., 2023). Thus, establishing sustainable systems that foster efficient production of high-yielding, nutrient-dense crops while simultaneously promoting soil health and fertility is critical. With the goal of reducing potentially harmful chemical inputs to soil (typically in the form of fertilizers and pesticides), additional approaches to promoting fertility beyond the fertilizer regime must be considered.
Fostering a healthy rhizosphere is an important way by which crop health and soil quality can be improved. Comprised of diverse and functionally dynamic bacteria, fungi, and other microbes, the rhizosphere microbiome largely modulates plant health and nutrition, as well as defense signaling and immunity to pathogens (Berendsen et al., 2012; Boparai and Sharma, 2021). Sometimes referred to as the second genome of the plant, the rhizosphere microbiome works in synchrony, often in direct symbiosis, with the associated plant life (Berendsen et al., 2012; Quiza et al., 2015). Therefore, understanding how this microbiome changes under adverse and beneficial conditions in different crops can lend insight into the functional microbial community profiles that are needed to better enhance crop and soil health.
Previous work has demonstrated that microbiome health can be improved through organic soil amendments (Hoitink and Boehm, 1999; Cesarano et al., 2017). In addition to the implementation of an organic fertilizer regime, the utilization of biochar—the product of pyrolyzed organic matter—has arisen as a promising strategy for the enhancement of soil fertility, diversity of beneficial microbes, and long-term sequestration of carbon (Muhammad et al., 2014; Amonette et al., 2016; Tenic et al., 2020; Lehmann et al., 2021; Li et al., 2023; Xu et al., 2023). Biochar is a carbon-rich, highly porous material; these properties facilitate enhanced water, gas, and nutrient exchange, and may result in decreased soil acidification when biochar it is utilized as a soil amendment (Jiang et al., 2014; Dai et al., 2017). While biochar is gaining traction in modern agriculture, different variations of its production have been practiced throughout history, and the use of pyrolyzed organic biomass for the enhancement of soil fertility can be traced back thousands of years (Tenenbaum, 2009; Glaser and Birk, 2012). In efforts to reclaim this millennia-old strategy, and to modernize it to address current global environmental challenges to crop production, biochar-based soil amendment and the consequent impacts on plant growth and yield have been evaluated for a diverse range of crops (Hien et al., 2017; Li and Shangguan, 2018; Velez et al., 2018; Woldetsadik et al., 2018; Faloye et al., 2019; Liu et al., 2019; Nzediegwu et al., 2019; Sánchez-Monedero et al., 2019).
Biochar nutrient properties are influenced by the original feedstock source (i.e., the type of biomass pyrolyzed). While there are numerous feedstocks available, the majority of those used to produce biochar for soil amendment fall into two categories: crop residue-based and wood-based. Biochars derived from wood feedstocks are generally enriched in organic matter and have higher surface area than those derived from crop residues; however, they have inherently lower levels of macronutrients (nitrogen [N], phosphorous [P], and potassium [K]) and display reduced cation exchange capacity (CEC) (Gao and DeLuca, 2018; Tenic et al., 2020). Unlike wood-based biochars, crop residue-derived biochars are rich in N, and have a lower surface area (Xiao et al., 2018; Tenic et al., 2020). Not surprisingly, the nutrient and physical profiles of biochars derived from different feedstock sources interact differently with soil and the rhizosphere to impact plant growth (Sun et al., 2014; Dai et al., 2020; Xu et al., 2023). Because of the elevated nutrient content, crop residue-derived biochars are favored in crop growth trials, but the impact on the microbiome functional activity has yet to be studied in detail. Several recent studies and reviews have indicated that, in addition to enhancing key physical and chemical properties of soil that enhance the growth of certain crops, biochar may also elicit changes in the soil microbiome; this may be due to the increased presence of labile carbon and biochar-bound nutrients that could serve to feed microbial growth (Xu et al., 2016; Li et al., 2020); moreover, the pore structure of biochar could lend protection from predation to certain microbes (Palansooriya et al., 2019; Li et al., 2020).
Understanding of microbial community profiles in diverse soil and rhizosphere environments has been classically understood through metagenomics approaches (White et al., 2017; Boparai and Sharma, 2021), which rely on sequencing of the 16S region of prokaryotes or the ITS region of eukaryotes in the soil. Indeed, metagenomic approaches have revealed the potential for biochar to alter the composition and function of microbial communities in the rhizosphere, with increases in the relative abundance of certain beneficial bacteria (Kolton et al., 2017). While metagenomic analysis provides information on the genetic potential of a microbial community, it does not provide information regarding genes that are actively being expressed and, thus, the level of activity of the microbial community.
In contrast, metatranscriptomic analysis involves sequencing and analyzing the mRNA transcripts present in an environmental sample, which provides information on which genes are being actively expressed within the microbial community (Hurt et al., 2001; Urich et al., 2008). This information allows for the identification of active metabolic pathways at a given time (and in which microbes these pathways are active) and provides insight into the interaction between the plant roots and the rhizosphere microbiome at the gene expression level. Additionally, metatranscriptomic analysis can reveal changes in gene expression of the rhizosphere in response to environmental factors or perturbations, such as the addition of biochar to soil. Despite these benefits, comprehensive gene expression profiling of soil rhizospheres is challenging due to several factors, including the limited amount of RNA present, the propensity of mRNA to degrade, the high content of humic acid (which is often co-extracted with nucleic acids and inhibits enzyme activity and downstream molecular processes required for transcriptome profiling), and low relative abundance of mRNA compared to rRNA (the latter of which accounts for 95%–99% of total RNA, but is not useful for gene expression analysis) (Wang et al., 2009; Mettel et al., 2010; Carvalhais et al., 2013; Bharti et al., 2021).
In this study, we sought to provide foundations for using metatranscriptomics to understand the soil rhizosphere, while expanding upon knowledge of the impacts of biochar amendment on the soil microbiome and plant growth. Using tomato (cv. “Oregon Spring”) as a model, we tested the hypothesis that the transcriptomic responses of tomato roots, as well as soil microbial community profiles and activity, change over time in the presence of biochar in an organic production system. The results of this work provide insight into how biochar impacts crop growth and microbial community profiles in tomatoes. Ultimately, the aim is to utilize metatranscriptome analysis as one of the approaches to be used as an indicator of an effective soil amendment.
Materials and methods
Experimental design
Experiments were conducted using organic-certified biochar provided by Qualterra Inc. (Spokane, WA). A 4 × 30-m study plot at the Eggert Organic Farm at Washington State University (WSU) was utilized for this study. Two days before planting, two evenly spaced rows were dug in the plot to a depth of 20 cm. To one of the rows, biochar was applied at a rate of 2 tons/acre (0.494 kg/m2). The soil was homogenized to incorporate the biochar into the soil for the treatment row, while non-amended organic farm soil was re-incorporated into the control rows. Tomato seeds (Solanum lycopersicum L.) “Oregon Spring” sourced from Territorial Seed Company (Cottage Grove, OR), were started in Sunshine® #1 Natural & Organic potting mix (Sun Gro Horticulture, MA) in the greenhouse. Greenhouse temperatures were maintained at 21.1/18.3°C (day/night) with a 14 h day, supplemented with high-pressure sodium lamps and 10 h night photoperiod. A total of 18, 4-week emergent seedlings with two fully expanded true leaves were transplanted into the organic farm soil for each treatment. Throughout the experiment, all plants in each treatment group were fertilized once a week with 100 mL of dilute (20 mL/L water) organic Alaska Fish Fertilizer 5-1-1 (Pennington Seed Inc., GA).
Time course sampling
The rhizosphere samples were collected from three randomly selected plants for each time point, and flash frozen in liquid nitrogen, at four time points representing key plant developmental stages: 1) 25 days–the emergence of third leaf, 2) 40 days–vegetative growth, pre-flowering, 3) 55 days–initiation of flowering, and 4) 70 days–fruit production (75% of fruit at the red ripe stage). At 70 days, mature fruits were harvested, and above-ground biomass was collected by cutting the plant at the soil level. Fruit biomass and shoot fresh weight were recorded for each treatment group. Differences in the mean biomass and shoot fresh weight were assessed using one-way analysis of variance (ANOVA) using SAS University Edition (SAS Institute Inc., Cary, NC).
Rhizosphere RNA extraction and sequencing
Hexadecyltrimethylammonium bromide (CTAB) extraction buffer was prepared by mixing equal volumes of 10% (wt/vol) CTAB in 0.7 M NaCl with 240 mM potassium phosphate buffer (pH 8.0). RNA precipitation solution was prepared by dissolving 30% (wt/vol) polyethylene glycol 8,000 in a 1.6 M NaCl solution. 0.5 g rhizosphere sample (wet soil + roots) was weighed and transferred to 2 mL screw cap centrifuge tubes containing 0.5 g (0.5 µM) glass. For RNA extraction, 0.5 mL of CTAB extraction buffer and 0.5 mL phenol-chloroform-isoamyl alcohol (25:24:1) (pH 7.15) were added to the soil samples. Samples were then lysed for 3 min using Qiagen TissueLyser-II with machine speed set at 30 1/s. The samples were then centrifuged at 14,000 rpm at 4°C for 10 min. The aqueous phase was then transferred to a fresh 1.5 mL Eppendorf tube and an equal volume of chloroform-isoamyl alcohol (24:1) was added, followed by centrifugation at 14,000 rpm at 4°C for 10 min to remove the residual phenol. The aqueous layer was again transferred to a fresh 1.5 mL Eppendorf tube. For RNA precipitation, 2 volumes of RNA precipitation solution were added to the extracted aqueous layer and incubated at room temperature for 2 h. Samples were then centrifuged at 14,000 rpm at 4°C for 10 min, and the supernatant was gently discarded. The pellets were then washed twice in ice-cold 100% ethanol and air dried prior to resuspension in 50 µL diethyl pyrocarbonate (DEPC)-treated water. Resuspended samples were treated with DNase using an Ambion Turbo DNA-free kit (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s guidelines. RNA was quality checked using agarose gel electrophoresis and quantified using Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States). Ribosomal depletion, library preparation, and sequencing was conducted at the WSU Genomics Core, where RNA was subjected to shearing, size selection, and adapter ligation using the TruSeq Total RNA library with Ribo-zero, during which ribosomal RNA was removed from the sample. The libraries produced were sequenced on an Illumina HiSeq 2,500 as 100-base single-end reads.
Soil metatranscriptome assembly
The fastq files generated from sequencing were preprocessed and assembled using CLC Genomics Workbench (Version 8.0.1) (Qiagen). Reads were trimmed for quality using a limit of 0.01, corresponding to a Phred value of 20; ambiguous nucleotides were trimmed, and terminal nucleotides with low-quality sequences were removed based on the results of the QC quality reports. Reads below length 34 were discarded. To remove host (tomato) reads from putative metatranscriptome reads, individual treatment read sets were mapped to the tomato reference genome (SL3.0, GCA_000188115.3) using a specified length fraction of 0.6 and similarity fraction of 0.9. Reads that mapped to the genome were deemed to be of tomato origin and were thus consolidated for subsequent root transcriptome analysis. Unmapped reads, determined to contain reads of soil microbiome origin, were de novo assembled using the metagenomics module in OmicsBox (version 1.3.11) (BioBam Bioinformatics S.L., Valencia, Spain). This resulted in the production of 43,935 contiguous sequences (contigs). The assembly consensus was exported as a fasta, which was then imported back into CLC for subsequent read mapping steps. For each individual dataset (treatment/time point) the original, non-trimmed reads were mapped back to the metatranscriptome assembly. Default parameters were used, except for the length and similarity fractions, which were set to 0.5 and 0.9, respectively. Mapping resulted in the generation of individual treatment sample reads per contig. The master metatranscriptome was exported as a fasta file for functional annotation and the read counts for each dataset were exported as tab delimited text files for normalization with the Reads Per Kilobase per Million reads (RPKM) method (Supplementary Material S1).
Tomato transcriptome mapping
Reads from each treatment group that mapped to the tomato genome in the original mapping step underwent a second mapping, this time to the reference transcriptome (46,846 contigs) derived from the SL3.0 genome. Read counts for each treatment mapping were exported for normalization with the RPKM method (Supplementary Material S1).
Functional annotation
The master metatranscriptome fasta was functionally annotated in the OmicsBox Transcriptomics Module. Briefly, contig sequences were identified by a blastx alignment against the NCBI “nr” database with an e-value specification of 10.0E-3 to identify the organism from which each contig was derived. Of all the rhizosphere microbiome annotations, 97.6% annotations have an E-value less than 1E-9 (or 10E-10) (see Supplementary File S1). Only 2.3% of the sequencing reads correspond to the annotated contigs with an E-value ranging from 10E-3 to 10E-10. Gene ontology (GO) annotation was assigned using the “Mapping” and “Annotation” features using default parameters to generate a functionally annotated master assembly. InterPro scan was conducted in conjunction with the blastx step, and the results of this scan were merged with GO annotations to provide additional functional information to the contigs. For the differentially expressed (DE) genes originating from the collective microbial transcriptome dataset, only those which displayed a sim mean of 90% were utilized in downstream analyses.
For the tomato transcriptome, annotations corresponding to RNAs derived from the tomato reference genome (SL3.0) were downloaded from GenBank and concatenated with their numeric contig IDs.
Metatranscriptome diversity analysis and heat tree expression maps
Normalized read count data was binned by each taxon identified in the functional annotation step. The sums of normalized reads for each taxon were used to calculate Shannon diversity for the metatranscriptome, and they also served as the input for expression heat tree map generation, which was performed in R using the metacoder package (Foster et al., 2017).
Differential expression analyses
Pairwise differential expression analyses were conducted for the tomato root transcriptome and the soil metatranscriptome in the OmicsBox Transcriptomics Module using NOISeq-sim to compare the treatments at each time point. NOISeq-sim infers significant differential expression without sequencing replicates. NOISeq-sim uses a multinomial distribution to model technical replicate read counts (Tarazona et al., 2011; Tarazona et al., 2013; Tarazona et al., 2015). Default parameters were used, with five simulated replications and a set variability of 0.2 in each replication. Genes with a NOIseq probability of greater than 0.9 and a |log2| fold change expression values greater than 1.0 for at least one treatment and time point was considered to be differentially expressed. We have previously used this approach and these cutoff values to establish thresholds for differential expression (Hewitt et al., 2021).
GO enrichment analyses
GO enrichment analyses were conducted to determine over- and under-represented biological processes among the differentially expressed genes in each treatment/time using the OmicsBox Enrichment Analysis tool, selecting Fisher’s Exact as the test method (Götz et al., 2008). For both the plant transcriptome and the collective microbial transcriptome, the annotated master transcriptome was used as the reference dataset, and the set of genes identified as differentially expressed over time in the treatment group versus the control group was used as the test dataset.
Data availability
The total RNAseq datasets generated during the current study are available in the Short Read Archive (SRA) on the NCBI database. The Bioproject accession number is PRJNA950600 and a temporary link during the review process is provided below: https://dataview.ncbi.nlm.nih.gov/ object/PRJNA950600?reviewer=acnbu6ct527lvh1vas8p3b3m33.
Results and discussion
Effects of biochar treatment on the growth of tomato plants
At the end of the time course, shoot fresh biomass and fruit weight of control and biochar-treated tomatoes grown in organically fertilized soil were assessed (Figure 1). No significant differences were observed (p < 0.05) between treatments for these parameters; control tomato plants had a mean fresh shoot biomass of 1.96 kg, while the biochar treated plants had a mean fresh shoot biomass of 2.07 kg (+5.6%). Furthermore, control plants and plants grown in the biochar-amended soil had nearly identical fruit biomass (5.56 and 5.50 kg, respectively).
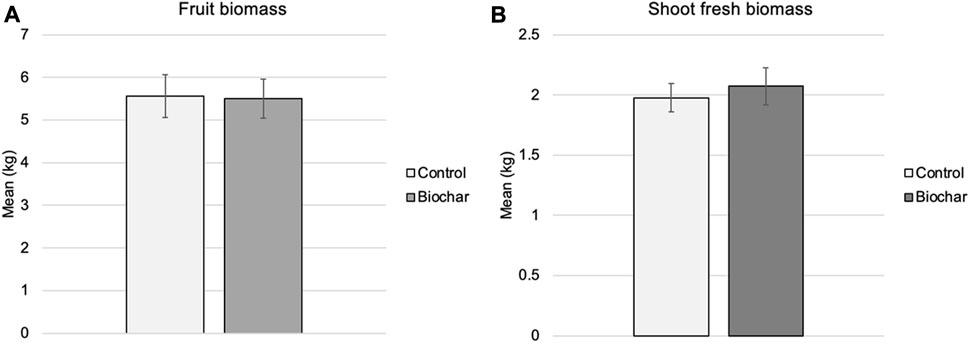
FIGURE 1. Shoot fresh biomass (A) and fruit biomass (B) at time 4 (fruiting, 70 days) of tomatoes grown in biochar-amended organic soil or control soil (no biochar). No significant differences were observed in shoot or fruit biomass (p < 0.05); however mean shoot biomass was higher in the biochar-treated tomato plants.
Many of the soil environments in which biochar has proven beneficial are marginal in nature (Liu et al., 2013; Yu et al., 2019). WSU organic farm represents healthy soils, and it has been demonstrated that the impact of soil amendments is limited in well-balanced soils (Hussain et al., 2017). Therefore, it was not surprising that plant growth and yield were not impacted in this study. Although, the application rate or timing of application of the biochar also needs to be determined. These results lend support to biochar as an avenue for long-term carbon sequestration that does not negatively impact crop production when incorporated into agricultural soils.
Rhizosphere functional activity
A novel aspect of this work is that we approach the characterization of the soil rhizosphere from a metatranscriptomics perspective, rather than metagenomics, to look at functional activity rather than abundance; this is expected to provide a better understanding of not only what microbes are present in the rhizosphere, but also which ones are the most active. Microbial diversity as a function of expressed transcripts pertaining to each taxon identified via functional annotation decreased slightly over time for the control rhizosphere. The biochar-treated rhizosphere displayed lower diversity at the first developmental time point (day 25); however, from day 40–day 70, the biochar-treated rhizosphere displayed higher diversity than that of the control (Figure 2A). In addition to diversity, rhizosphere activity was assessed as a function of the total number of normalized reads corresponding to functionally annotated microbes in each treatment and at each time point (Figures 2B–D). Analysis of rhizosphere bacterial expression (Figure 2B) revealed a quadratic increase in expression in the control throughout the treatment, and lower expression of the bacteria in the biochar-treated rhizosphere overall, particularly by the end of the time course (70 days). Interestingly, we observed that 80% of the rhizosphere transcript reads corresponded to Streptomyces bacteria; thus, to better understand the impact of biochar on the remaining soil bacteria, the full bacterial dataset was analyzed without Streptomyces (Figure 2C). In the latter analysis, the biochar-treated rhizosphere displayed elevated microbial gene expression in comparison with the control from day 25–day 55. By day 70, the treatment and control were approximately the same and had both demonstrated increased overall gene expression over time. With regards to fungal diversity, the main changes were observed at the fruiting stage of the plant (day 70), where biochar treatments resulted in decreased fungal gene expression (Figure 2D). Together these results provide a high-level overview of the impacts of biochar on the overall diversity and expression of the bacterial and fungal communities in the tomato rhizosphere.
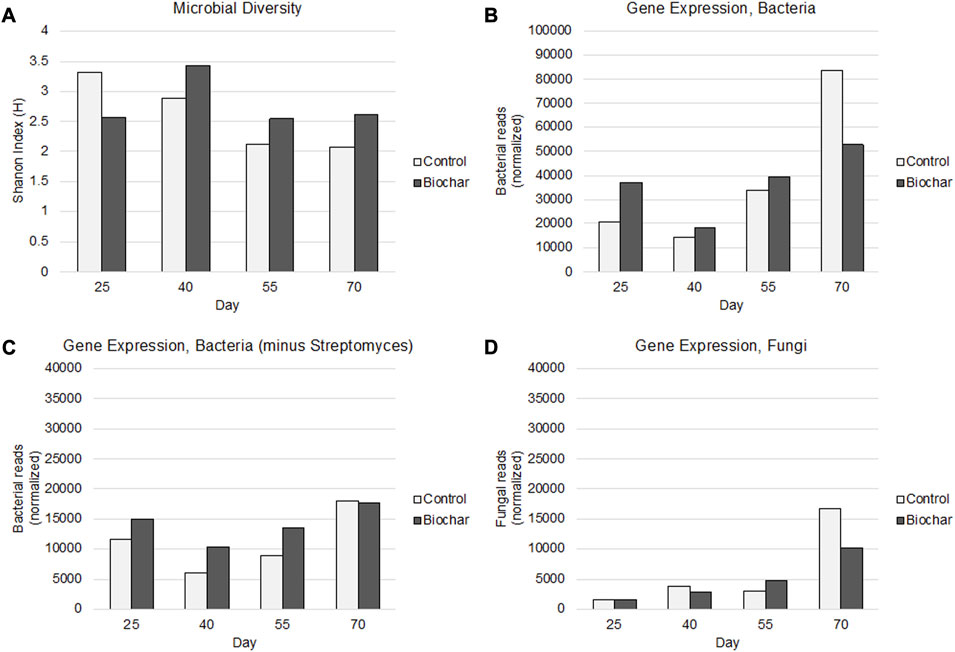
FIGURE 2. Rhizosphere microbial diversity (A); bacterial functional activity (B); bacterial functional activity, minus Streptomyces (which represented an overwhelming majority of the soil microbiome) (C), and fungi functional activity (D) in biochar-treated and control soil regimes. Time points: 1) 25 days–third-leaf stage; 2) 40 days–vegetative stage, pre-flowering; 3) 55 days–flowering stage; 4) 70 days–fruiting stage.
Expression of top microbial taxa
The most highly expressed bacteria in the tomato rhizosphere included the following groups 1) Proteobacteria–Rhizobiaceae, Nitrosomonadaceae, Pseudomonadaceae, Enterobacteriaceae, Comamonadaceae, Sphingomonadaceae, Methylophilaceae, Xanthomonadaceae, Oxalobacteraceae; 2) Actinobacteria–Nocardiaceae, Microbacteriaceae, Micromonosporaceae, Streptomycetaceae; 3) Bacteroidetes–Flavobacteriaceae; and 4) Firmicutes–Paenbacillaceae. Nearly all of these microbes displayed elevated gene activity in response to biochar throughout the time course, namely, Rhizobiaceae, Pseudomonadaceae, Enterobacteriaceae, Sphingomonadaceae, Xanthomonadaceae, Microbacteriaceae, and Micromonosporaceae (Figure 3).
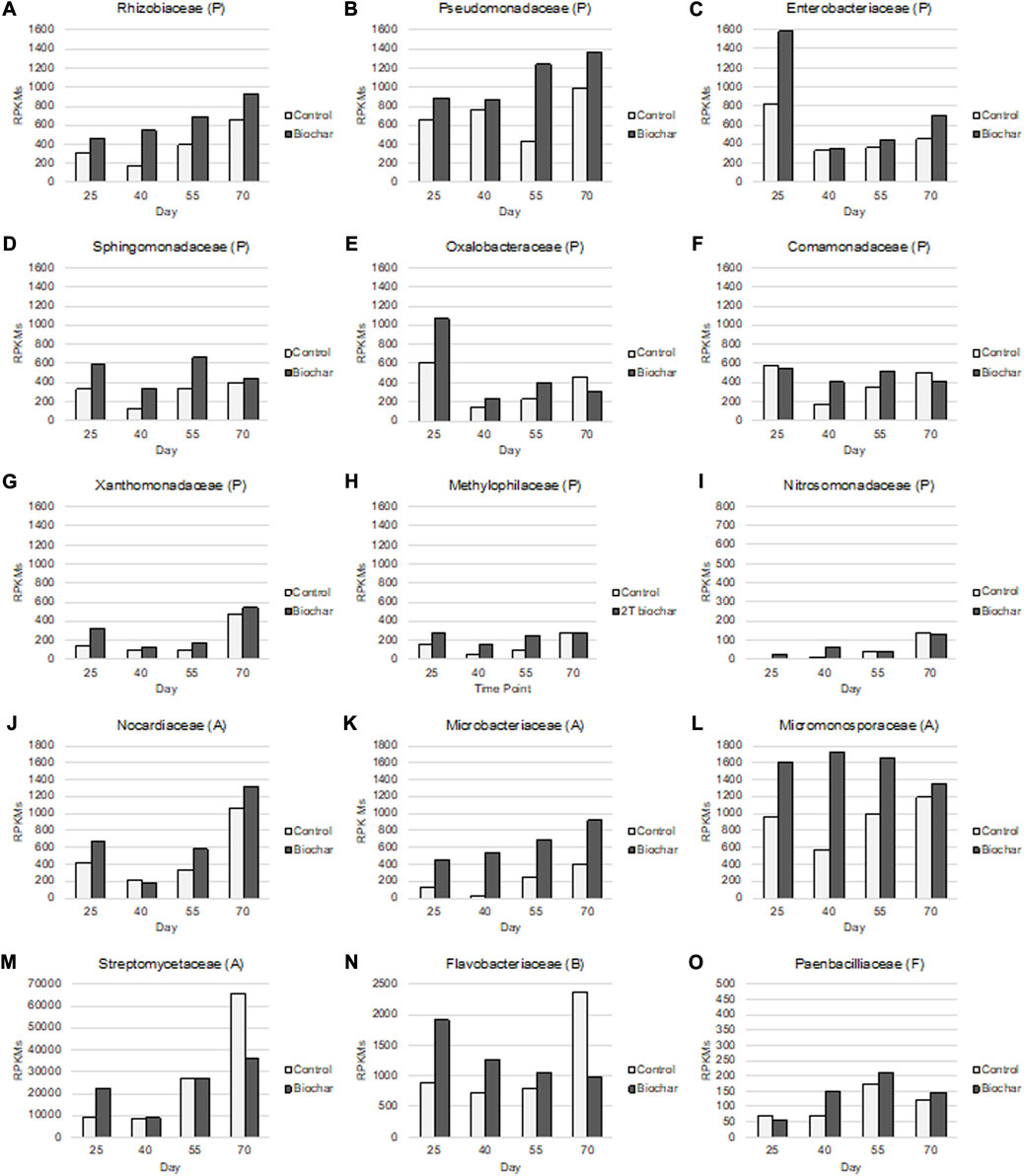
FIGURE 3. Normalized gene expression of the top 15 bacterial families (representing approximately 90% of the gene expression activity in the rhizosphere), identified in the biochar-treated and control tomato rhizospheres throughout four plant developmental stages. Letters in parenthesis indicate the phylum to which each family of bacteria pertains (P, Proteobacteria (A–I); A, Actinomycetes (J–M); B, Bacteroidetes (N); F, Firmicutes (O)). Time points: (1) 25 days—third-leaf stage; (2) 40 days—vegetative stage, pre-flowering; (3) 55 days—flowering stage; (4) 70 days—fruiting stage.
Many of the bacteria that increased in functional activity in the biochar-amended rhizospheres in comparison to the control rhizospheres over time are known to be beneficial to soil health and plant growth, while only a few have potentially detrimental effects. Beneficial features of highly expressed soil bacteria include nitrogen fixation and assimilation (Rhizobiaceae, Enterobacteriaceae, Oxalobacteraceae, Microbacteriaceae); nitrogen cycling (Nitrosomonadaceae, Pseudomonadaceae, Enterobacteriaceae, Comamonadaceae, Oxalobacteraceae, Microbacteriaceae, Paenbacilliaceae); carbon cycling (Pseudomonadaceae, Comamonadaceae, Sphingomonadaceae, Methylophilaceae, Microbacteriaceae, Streptomycetaceae, Flavobacteriaceae); plant growth promotion (Rhizobiaceae, Pseudomonadaceae, Enerobacteriaceae, Comamonadaceae, Sphingomonadaceae, Oxalobacteraceae, Microbacteriaceae, Paenbacilliaceae, Flavobacteriaceae); biodegradation of organic compounds (Pseudomonadaceae, Comamonadaceae, Sphingomonadaceae, Nocardiaceae, Paenbacilliaceae); and antimicrobial, antifungal, disease suppressive, and/or fungistatic properties (Pseudomonadaceae, Oxalobacteriaceae, Xanthomonadaceae, Nocardiaceae, Microbacteriaceae, Micromonosporaceae, Streptomycetaceae, Paenbacillaceae, Flavobacteriaceae) (Table 1).
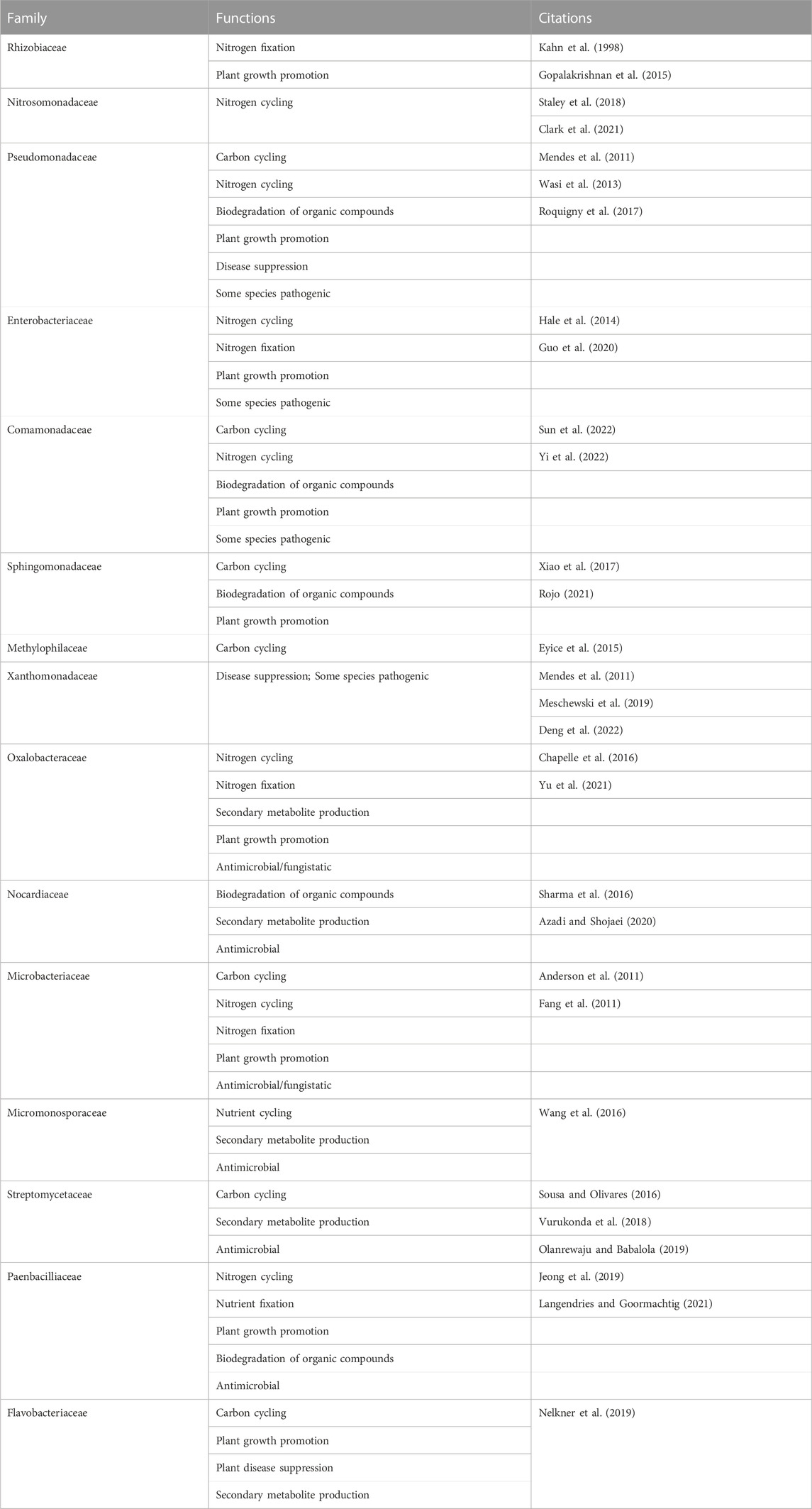
TABLE 1. Soil bacteria with the highest functional activity identified in the biochar-amended tomato rhizosphere and their known functions.
As indicated previously, the microbial taxa most highly represented in all samples pertained to the family Streptomycetaceae (and primarily of the Streptomyces genus). Streptomyces are common soil Actinomycete bacteria. They have been described as important soil microbes due to their production of beneficial bioactive compounds, including enzymes that can enhance soil health and fertility, and have positive impacts on agriculture overall (Olanrewaju and Babalola, 2019). Furthermore, some Streptomyces have been characterized as free-living diazotrophs and are believed to contribute substantial nitrogen to some ecosystems (Dahal et al., 2017). In a recently published study, Streptomyces was reported to be the potential keystone species in an organic system where a shotgun metagenomics study was performed (Khoiri et al., 2021). Interestingly, while the other bacterial families discussed displayed elevated gene expression in response to biochar treatment, Streptomycetaceae displayed initial elevation in gene expression followed by reduced expression by the end of the time course. Reduction in Streptomycetaceae is possibly explained by increased competition for resources that is expected to occur as the other microbial families increase in abundance. Several other recent studies have reported a reduction in Streptomycetaceae bacteria abundance in response to biochar amendment (Yang et al., 2022; Wang et al., 2023).
Similar to the results seen for rhizosphere bacteria, biochar application resulted in changes in the expression of the fungal communities in the soil over time; in particular, it substantially altered the activity of beneficial and detrimental fungi. Of the major fungal taxa that were active in the rhizosphere microbiome, those with benefits to the rhizosphere include decomposers (Mortierella, Spizellomyces, Basidiobolus, and Podospora) and mycorrhizal fungi (Rhizophagus), the latter of which increases surface area for absorption of nutrients through symbiotic relationships with plant roots (Gujre et al., 2021; Chen et al., 2022). Rhizophagus has also been reported to utilize biochar as a medium for growth (Hammer et al., 2014). Expression of beneficial fungi decreased over time, but was generally higher in the biochar-treated rhizosphere than the control, particularly during the initial half of the time course (Figure 4A).
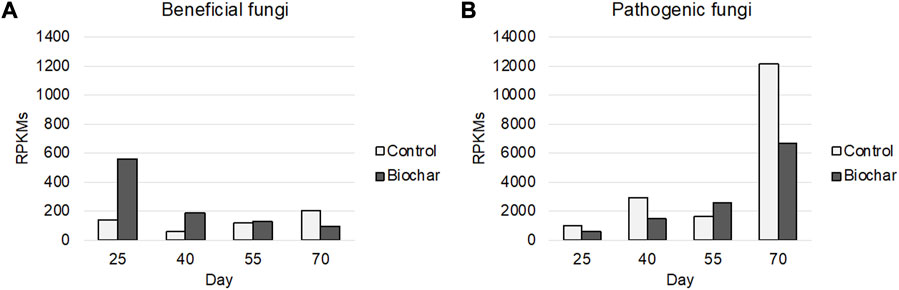
FIGURE 4. Expression of beneficial (A) and pathogenic (B) soil fungi. Beneficial bacteria include (Mortierella, Spizellomyces, Basidiobolus, Podospora, and Rhizophagus). Pathogenic bacteria include (Fusarium, Rhizoctonia, Verticillium, Synchytrium, Colleotrichum, Moniliophthora, Puccinia, Stachybotrys, and Thermothielavioides). Time points: 1) 25 days–third-leaf stage; 2) 40 days–vegetative stage, pre-flowering; 3) 55 days–flowering stage; 4) 70 days–fruiting stage.
Pathogenic fungi present in the rhizosphere include Fusarium, Rhizoctonia, Verticillium, Synchytrium, Colleotrichum, Moniliophthora, Puccinia, Stachybotrys, and Thermothielavioides. Biochar treatment generally decreased the functional activity of pathogenic fungi throughout the developmental time course (Figure 4B).
Previous studies have shown that biochar has the potential to enhance the abundance of beneficial microbes in the soil and reduce the abundance of detrimental microbes, in some cropping systems (Meng et al., 2019; Pathy et al., 2020), thereby contributing to the suppression of diseases caused by soilborne pathogens (Jaiswal et al., 2017). Our study lends important additional insight regarding the impact of biochar on the gene expression activity of microbes in the soil. Together, the observations regarding soil microbial functional activity suggest that biochar presence does indeed influence the temporal expression activity of important soil microbes, with biochar resulting in increased microbial activity and, in many cases, curbing the expression of detrimental microbes.
The porosity of biochar and its chemical properties are thought to be some factors that underly its benefits to soil health (Jaafar et al., 2014; Mousavi et al., 2022). The increased expression of beneficial microbes in the biochar-treated rhizosphere throughout the time course is likely due in part to the increased available carbon, as well as to the presence of increased surface area and improved soil aeration resulting from the porosity of biochar, the latter of which is expected to facilitate enhanced microbial colonization within the biochar pores. Moreover, through direct, and potentially symbiotic, interaction with the tomato roots—which are known to exude different metabolites throughout their development (Jacoby et al., 2020)—the promoted microbial communities are expected to influence soil health, and potentially also plant growth and fruit yield/quality (although the latter effects were not observed within this experimental time frame).
The temporal progression of expression changes of bacteria, down to the genus level, was visualized using heat tree maps (Figure 5). Because Streptomyces were present at significantly higher levels than all other bacteria, their expression was visualized separately to better resolve the expression changes of all other bacteria. This format provides a glimpse of expression changes in both high-level (e.g., phylum, class) and lower-level (family, genus) taxonomic classifications. At the high level, we observed elevated expression for Actinobacteria, Proteobacteria, and Firmicutes in the biochar-treated versus control rhizosphere at all time points; Bacteroidetes displayed elevated expression only at 40 days. At the genus level, highly expressed microbes among the Actinobacteria include 1) nitrogen-fixing, cycling, and/or nitrifying bacteria (e.g., Rhizobium, Klebsiella, Mesorhizobium, Sinorhizobium, Nitrosomonas, Nitrospira, Arthrobacter, and Ensifer), plant growth-promoting bacteria (e.g., Pseudomonas, Paenibacillus, Bacillus, Stenotrophomonas); a number of microbes involved in organic matter degradation, including (e.g., Flavobacterium, Rhodococcus, Mycobacterium, Sphingomonas, Thermoactinomyces, Variovorax, etc.); and bacteria involved in the production of antimicrobial compounds (e.g., Actinoplanes, Plantactinospora, Nonomuraea, Nocardia, and Chromobacterium) (Figure 5). Uncultured Candidatus, many members of which are responsible for plant diseases, displayed reduced abundance at all time points in the biochar-treated rhizosphere than in the control. Streptomyces displayed initial heightened expression in the biochar treated soil, but this expression was curbed over time and was significantly reduced in comparison with the control at the final sampling time point (fruiting stage, 70 days). A similar visualization was performed for fungi (Supplementary Material S1).
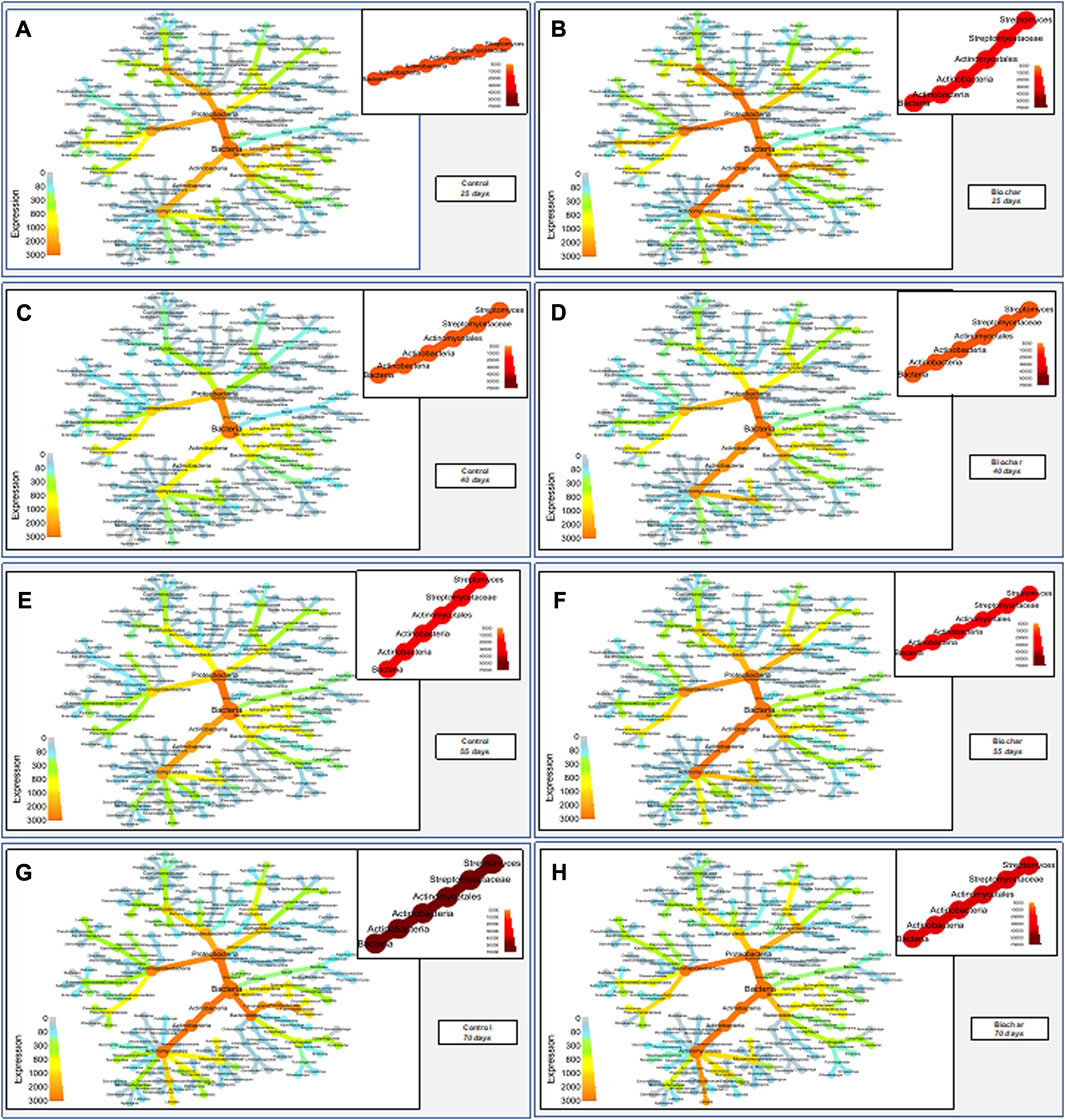
FIGURE 5. Heat tree maps indicating expression of soil bacteria in the control and biochar-treated tomato rhizospheres over time. Streptomyces is shown separately due to their disproportionately high expression in the soil rhizospheres in all treatments/times. Time points: 1) 25 days–third-leaf stage; 2) 40 days–vegetative stage, pre-flowering; 3) 55 days–flowering stage; 4) 70 days–fruiting stage.
Gene ontology enrichment analyses
Bacteria, fungi, and root differentially expressed gene sets were used to conduct GO enrichment analyses for each developmental time point to compare enriched biological processes and molecular functions in the biochar-treated versus the control rhizosphere. For the microbial datasets, due to the high number of enriched ontologies, an adjusted p-value filter of 0.001 was employed to facilitate the identification of the top GOs (Supplementary Materials S3, S4).
For bacteria, several ontologies were highly enriched in the biochar-treated rhizosphere in comparison with the control at all four time points and included nitrogen compound metabolic/biosynthetic processes (GO:1901566, GO:0034641, GO:0044271); biosynthesis of peptides, amides, and other organic substances (GO:0043043, GO:0006518, GO:0044249, GO:0009059, GO:0043604, GO:0043603, GO:1901576); structural molecules (GO:0003735, GO:0005198); and gene expression (GO:0010467). The enrichment of nitrogen compound-associated processes in the biochar-treated rhizosphere in comparison with the control rhizosphere is consistent with the observed increase in the overall activity of nitrogen cycling/fixing bacteria (e.g., J, etc.) observed for this treatment (Table 1; Figure 3). As nitrogen is a major nutrient limiting plant growth (Moreau et al., 2019), the increase in the nitrogen cycling microbes in the biochar-amended rhizosphere is an important finding. In addition to nitrogen, the enriched processes associated with the biosynthesis of organic compounds and structural molecules may result from the secondary compound-producing activity and/or need the direct microbe/substrate or microbe/root interactions.
Among the fungi, there were no ontologies that were significantly enriched throughout the full developmental time course; however, several GOs displayed enrichment from 25–55 days in the biochar-treated vs. control rhizosphere, including organic cyclic compound binding (GO:0097159, GO:1901363, GO:005488), binding of ions and nucleic acids (GO: 0005488, GO: 0043167, GO:0043169, GO:0003676), and oxidoreductase activity (GO: 0016491). Organic cyclic compounds, such as polyaromatic aromatic hydrocarbons (PAH) and those found in lignin and cellulose, are often found in organic matter (PAH found particularly in contaminated soils) and can be difficult to break down (Tomer et al., 2021; Vipotnik et al., 2021). However, some of the soil fungi identified in this study (e.g., Mortierella, Basidiobolus) have the ability to bind to these compounds, break them down via oxidoreductases (e.g., dioxygenases), and use them as a source of carbon and energy (Cerniglia, 1993; Wang et al., 2020; Wang et al., 2021; Sun et al., 2022), a finding consistent with the enrichment results.
In the plant roots, enriched GOs in the biochar-amended tomato rhizosphere versus the control were almost exclusively identified for the final developmental time point (at the fruiting stage). This suggests a lack of major changes in the overall functional activity of the roots during the earlier stages of plant development and may underly the lack of any significant differences in fruit and shoot biomass. At the final sampling stage, however, enriched functions associated with nitrogen metabolism (GO:004721, GO:0034641, GO:0006807, GO:1901566); organic substance metabolism (GO:1901576, GO:0071704); biosynthesis of peptides, amides, and other organic macromolecules (GO:0006412, GO:0006518, GO:0009059, GO:0043043, GO:0043603, GO:0043604, GO:0044249, GO:1901576, GO:0043170, GO:0019538, GO:0009058, GO:0071704); enzyme mediated regulation of biological processes (GO:0065009, GO:0030234, GO:0098772); and gene expression (GO:0010476, GO003723). That these functions are becoming active at the end of time course suggests that there is a delay in the response of the roots to the shift in the soil microbiome. As we did not see any significant differences in shoot biomass or fruit production in this study, these results suggest that perhaps the response to biochar and the shift in microbiome activity do not impact the plant as strongly during the vegetative growth stage but becomes more impactful at the time of fruit production. If the time for microbes to become established, colonize biochar, and interact with the plant roots requires several weeks to months, as appears to be the case for this trial, it is likely that a longer experimental time course would better address the question of how wheat-straw derived biochar impacts tomato growth in an organic soil regime.
Role of biochar in changing the tomato rhizosphere
Based on the results, we propose the following model for biochar’s action in the soil to explain the observations: The specific biochar amendment regime and conditions used in this study result in increased availability of carbon and other nutrients serving as a food source for beneficial microbes in addition to increasing surface area for microbial colonization due to biochar’s porous structure. As the rhizosphere begins to adjust to the new conditions, microbes begin to engage in processes such as nutrient cycling, decomposition of organic matter, and production of plant growth-promoting compounds. However, it may take several weeks, or even months, for the soil microbial community to establish and reach a new equilibrium after biochar amendment. This could be due to factors such as the initial state of the soil and the soil microbiome (e.g., biochar has been shown to elicit greater impacts on marginal soils than in already healthy soils), the amount and type of biochar added (chemical and physical composition varies based on feedstock source and pyrolysis temperature), and the climate and environmental conditions. Once the soil microbiome has adjusted to the new conditions, it can begin to positively influence plant growth by facilitating nutrient uptake, improving soil structure, and providing protection against soilborne diseases. However, the response in the plants may not occur immediately after the changes in the soil microbiome, as is seen in this study; in addition to the aforementioned factors, the plant’s genetic background and level of biochar application may also impact the magnitude and speed of response to biochar. A large number of enriched biological processes and molecular functions in the biochar-treated tomato roots only at the end of the time course, while enriched microbial ontologies were seen throughout the time course, suggests that a longer time course may be necessary in order to observe impacts of biochar on biomass and yield in the context of the given study parameters. The fruiting-stage plants may begin shaping the microbial communities around them by releasing metabolites that can selectively promote the growth of certain microbes while suppressing others (Jacoby et al., 2020). This can help reduce competition from harmful microbes that could affect the quality or yield of the fruit, while further enhancing the activity of beneficial microbes.
Implications of biochar use in organic production systems
Organic farming practices prioritize the preservation of soil health and fertility without heavy reliance on chemical/synthetic inputs. The outcomes of this study indicate that the incorporation of biochar as a soil amendment in organic cropping systems could serve as a valuable approach to positively impact soil health within such systems. The observed upregulation in gene expression activity among beneficial soil microbes associated with essential functions like nitrogen fixation, nutrient cycling, carbon cycling, and plant growth promotion indicates the potential of biochar to improve nutrient availability and cycling in the soil. Additionally, the observed decrease in the expression of pathogenic fungi within the biochar-amended rhizosphere highlights the potential of biochar for suppressing soilborne diseases, offering a natural and sustainable means of disease management. Furthermore, the intriguing observation of reduction in microbial function at the mature fruit stage indicates that biochar might be enabling better communication between the plant and the microbes and reduce the competition for carbon. In future work, it will be of interest to evaluate whether the use of biochar leads to reductions in yield penalties often observed under organic production systems (between 5–34%) (Seufert et al., 2012; Reganold and Wachter, 2016; Kravchenko et al., 2017; Sharpe et al., 2020). By capitalizing on the positive interactions between biochar and the soil microbiome, organic farmers can potentially minimize their dependence on conventional inputs while fostering sustainable soil fertility and crop production.
Conclusion
This work provides a proof-of-principle for using a metatranscriptome approach to understand changes in tomato rhizosphere functional activity in a biochar-amended organic soil regime. While biochar impacts on crop production are variable and depend on a number of factors, in this study, no changes to crop biomass or fruit yield were observed to the single concentration of biochar applied; importantly, however, the soil microbiome displayed the heightened functional activity of a number of beneficial microbes, and reduction in the activity of pathogenic fungi throughout the time course. Enrichment analyses revealed increased nitrogen cycling and breakdown of organic compounds in the soil microbiome throughout the time course, which were linked to increased nitrogen metabolism and primary metabolic processes in the tomato roots at the end of the time course; these findings indicate that, while biochar has a more immediate impact on the soil microbiome, a longer time course is needed to observe a plant-level response. Our findings lay the groundwork for future experiments that will consider additional variables (initial soil health/composition, plant genetic background, biochar type, and biochar rate of application) and measure additional parameters indicative of soil health (e.g., soil CEC, SOM, pH, and available N and C); in doing so, such studies will generate further knowledge that will aid scientists and producers to develop management plans for sustainable crop production and soil fertility.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA950600.
Author contributions
SH assisted with preparation of samples for sequencing, analyzed the metatranscriptome data, and wrote the manuscript. RG developed the protocol for extraction of RNA from the soil rhizosphere and assisted in soil RNA extractions. WT assisted with data analysis and contributed to the interpretation of results. ET conducted the experiment, collected samples, and analyzed tomato biomass data. AD, DI, and ET designed the experiment. AD conceived and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by a WSARE grant GW18-034 to SH. Work in the Dhingra lab was supported in part by the Washington State University Agriculture Center Research Hatch Grant WNP00011 and startup funds from Texas A&M AgriLife Research, Texas A&M University.
Conflict of interest
SH and AD were employed by Qualterra, Inc., at the time that the results of the study were being analyzed. Qualterra, Inc., is a spinout from the Dhingra Research Program at Washington State University. SH serves as a grants specialist and genomics scientist and AD serves as the chief scientific officer. The biochar used in this study was provided as a gift. The funders had no involvement in the collection, analysis, interpretation of data, writing or decision to publish the results.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author AD declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2023.1205583/full#supplementary-material
References
Amonette, J. E., Garcia-Perez, M., Sjoding, D., and Fuchs, M. R. (2016). Biochar from biomass and its potential agronomic and environmental use in Washington: A promising alternative to drawdown carbon from the atmosphere and develop a new industry. Richland, WA (United States): Pacific Northwest National Lab.
Anderson, C. R., Condron, L. M., Clough, T. J., Fiers, M., Stewart, A., Hill, R. A., et al. (2011). Biochar induced soil microbial community change: implications for biogeochemical cycling of carbon, nitrogen and phosphorus. Pedobiol. (Jena) 54, 309–320. doi:10.1016/j.pedobi.2011.07.005
Azadi, D., and Shojaei, H. (2020). Biodegradation of polycyclic aromatic hydrocarbons, phenol and sodium sulfate by Nocardia species isolated and characterized from Iranian ecosystems. Sci. Rep. 10, 21860. doi:10.1038/s41598-020-78821-1
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi:10.1016/j.tplants.2012.04.001
Bharti, M. K., Siddique, R. A., Ranjan, K., Chandra, D., and Singh, N. P. (2021). “Relevance of metatranscriptomics in symbiotic associations between plants and rhizosphere microorganisms,” in Microbial metatranscriptomics belowground (Berlin, Germany: Springer), 59–90.
Boparai, J. K., and Sharma, P. K. (2021). “Metagenomics and metatranscriptomics approaches in understanding and discovering novel molecules in rhizosphere environment,” in Omics science for rhizosphere biology (Berlin, Germany: Springer), 41–52.
Carvalhais, L., Dennis, P., Tyson, G., and Schenk, P. (2013). “Rhizosphere metatranscriptomics: challenges and opportunities,” in Molecular microbial ecology of the rhizosphere, 1 and 2 (New Jersey, United States: Wiley Online Library), 1137–1144.
Cerniglia, C. E. (1993). Biodegradation of polycyclic aromatic hydrocarbons. Curr. Opin. Biotechnol. 4, 331–338. doi:10.1016/0958-1669(93)90104-5
Cesarano, G., De Filippis, F., La Storia, A., Scala, F., and Bonanomi, G. (2017). Organic amendment type and application frequency affect crop yields, soil fertility and microbiome composition. Appl. Soil Ecol. 120, 254–264. doi:10.1016/j.apsoil.2017.08.017
Chapelle, E., Mendes, R., Bakker, P. A. H. M., and Raaijmakers, J. M. (2016). Fungal invasion of the rhizosphere microbiome. ISME J. 10, 265–268. doi:10.1038/ismej.2015.82
Chen, Q., Deng, X., Elzenga, J. T. M., and van Elsas, J. D. (2022). Effect of soil bacterioids on mycorrhizal colonization by rhizophagus irregularis—Interactive effects on maize (Zea mays L) growth under salt stress. Biol. Fertil. Soils 58, 515–525. doi:10.1007/s00374-022-01636-x
Clark, I. M., Hughes, D. J., Fu, Q., Abadie, M., and Hirsch, P. R. (2021). Metagenomic approaches reveal differences in genetic diversity and relative abundance of nitrifying bacteria and archaea in contrasting soils. Sci. Rep. 11, 15905–15909. doi:10.1038/s41598-021-95100-9
Dahal, B., NandaKafle, G., Perkins, L., and Brözel, V. S. (2017). Diversity of free-Living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiol. Res. 195, 31–39. doi:10.1016/j.micres.2016.11.004
Dai, Y., Zheng, H., Jiang, Z., and Xing, B. (2020). Combined effects of biochar properties and soil conditions on plant growth: a meta-analysis. Sci. Total Environ. 713, 136635. doi:10.1016/j.scitotenv.2020.136635
Dai, Z., Zhang, X., Tang, C., Muhammad, N., Wu, J., Brookes, P. C., et al. (2017). Potential role of biochars in decreasing soil acidification-A critical review. Sci. Total Environ. 581, 601–611. doi:10.1016/j.scitotenv.2016.12.169
Deng, X., Zhang, N., Li, Y., Zhu, C., Qu, B., Liu, H., et al. (2022). Bio-organic soil amendment promotes the suppression of Ralstonia solanacearum by inducing changes in the functionality and composition of rhizosphere bacterial communities. New Phytol. 235, 1558–1574. doi:10.1111/nph.18221
Eyice, Ö., Namura, M., Chen, Y., Mead, A., Samavedam, S., and Schäfer, H. (2015). SIP metagenomics identifies uncultivated Methylophilaceae as dimethylsulphide degrading bacteria in soil and lake sediment. ISME J. 9, 2336–2348.
Faloye, O. T., Alatise, M. O., Ajayi, A. E., and Ewulo, B. S. (2019). Effects of biochar and inorganic fertiliser applications on growth, yield and water use efficiency of maize under deficit irrigation. Agric. Water Manag. 217, 165–178. doi:10.1016/j.agwat.2019.02.044
Fang, L. Z., Kun, X. C., Song, Z. C., Qin, X. J., Qiu, H. Y., Qun, D. C., et al. (2011). Fungistatic intensity of agricultural soil against fungal agents and phylogenetic analysis on the actinobacteria involved. Curr. Microbiol. 62, 1152–1159. doi:10.1007/s00284-010-9836-6
Foster, Z. S. L., Sharpton, T. J., and Grünwald, N. J. (2017). Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLOS Comput. Biol. 13, e1005404. doi:10.1371/journal.pcbi.1005404
Gao, S., and DeLuca, T. H. (2018). Wood biochar impacts soil phosphorus dynamics and microbial communities in organically-managed croplands. Soil Biol. biochem. 126, 144–150. doi:10.1016/J.SOILBIO.2018.09.002
Glaser, B., and Birk, J. J. (2012). State of the scientific knowledge on properties and genesis of Anthropogenic Dark Earths in Central Amazonia (terra preta de Índio). Geochim. Cosmochim. Acta 82, 39–51. doi:10.1016/j.gca.2010.11.029
Gopalakrishnan, S., Sathya, A., Vijayabharathi, R., Varshney, R. K., Gowda, C. L. L., and Krishnamurthy, L. (2015). Plant growth promoting rhizobia: challenges and opportunities. 3 Biotech. 5, 355–377. doi:10.1007/s13205-014-0241-x
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi:10.1093/nar/gkn176
Gujre, N., Soni, A., Rangan, L., Tsang, D. C. W., and Mitra, S. (2021). Sustainable improvement of soil health utilizing biochar and arbuscular mycorrhizal fungi: a review. Environ. Pollut. 268, 115549. doi:10.1016/j.envpol.2020.115549
Guo, D.-J., Singh, R. K., Singh, P., Li, D.-P., Sharma, A., Xing, Y.-X., et al. (2020). Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 11, 580081. doi:10.3389/fmicb.2020.580081
Hale, L., Luth, M., Kenney, R., and Crowley, D. (2014). Evaluation of pinewood biochar as a carrier of bacterial strain Enterobacter cloacae UW5 for soil inoculation. Appl. Soil Ecol. 84, 192–199. doi:10.1016/j.apsoil.2014.08.001
Hammer, E. C., Balogh-Brunstad, Z., Jakobsen, I., Olsson, P. A., Stipp, S. L. S., and Rillig, M. C. (2014). A mycorrhizal fungus grows on biochar and captures phosphorus from its surfaces. Soil Biol. biochem. 77, 252–260. doi:10.1016/j.soilbio.2014.06.012
Hewitt, S., Kilian, B., Koepke, T., Abarca, J., Whiting, M., and Dhingra, A. (2021). Transcriptome analysis reveals potential mechanisms for ethylene-inducible pedicel–fruit abscission zone activation in non-climacteric sweet cherry (Prunus avium l). Horticulturae 7, 270. doi:10.3390/HORTICULTURAE7090270
Hien, T. T. T., Shinogi, Y., and Taniguchi, T. (2017). The different expressions of draft cherry tomato growth, yield, quality under bamboo and rice husk biochars application to clay loamy soil. Agric. Sci. 8, 934–948. doi:10.4236/as.2017.89068
Hoitink, H. A. J., and Boehm, M. J. (1999). Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37, 427–446. doi:10.1146/annurev.phyto.37.1.427
Hurt, R. A., Qiu, X., Wu, L., Roh, Y., Palumbo, A. V., Tiedje, J. M., et al. (2001). Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67, 4495–4503. doi:10.1128/aem.67.10.4495-4503.2001
Hussain, M., Farooq, M., Nawaz, A., Al-Sadi, A. M., Solaiman, Z. M., Alghamdi, S. S., et al. (2017). Biochar for crop production: potential benefits and risks. J. Soils Sediments 17, 685–716. doi:10.1007/s11368-016-1360-2
Jaafar, N. M., Clode, P. L., and Abbott, L. K. (2014). Microscopy observations of habitable space in biochar for colonization by fungal hyphae from soil. J. Integr. Agric. 13, 483–490. doi:10.1016/S2095-3119(13)60703-0
Jacoby, R. P., Chen, L., Schwier, M., Koprivova, A., and Kopriva, S. (2020). Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Research 9, F1000. doi:10.12688/f1000research.21796.1
Jaiswal, A. K., Elad, Y., Paudel, I., Graber, E. R., Cytryn, E., and Frenkel, O. (2017). Linking the belowground microbial composition, diversity and activity to soilborne disease suppression and growth promotion of tomato amended with biochar. Sci. Rep. 7, 44382. doi:10.1038/srep44382
Jeong, H., Choi, S.-K., Ryu, C.-M., and Park, S.-H. (2019). Chronicle of a soil bacterium: paenibacillus polymyxa E681 as a tiny guardian of plant and human health. Front. Microbiol. 10, 467. doi:10.3389/fmicb.2019.00467
Jiang, P., Sun, K., Lun, F. M. F., Guo, A. M., Wang, H., Chan, K. C. A., et al. (2014). Methy-pipe: an integrated bioinformatics pipeline for whole genome bisulfite sequencing data analysis. PLoS One 9, e100360. doi:10.1371/journal.pone.0100360
Kahn, M. L., McDermott, T. R., and Udvardi, M. K. (1998). Carbon and nitrogen metabolism in rhizobia. Rhizobiaceae Mol. Biol. Model Plant-Associated Bact. 1998, 461–485.
Khoiri, A. N., Cheevadhanarak, S., Jirakkakul, J., Dulsawat, S., Prommeenate, P., Tachaleat, A., et al. (2021). Comparative metagenomics reveals microbial signatures of sugarcane phyllosphere in organic management. Front. Microbiol. 12, 623799. doi:10.3389/fmicb.2021.623799
Kolton, M., Graber, E. R., Tsehansky, L., Elad, Y., and Cytryn, E. (2017). Biochar-stimulated plant performance is strongly linked to microbial diversity and metabolic potential in the rhizosphere. New Phytol. 213, 1393–1404. doi:10.1111/nph.14253
Kravchenko, A. N., Snapp, S. S., and Robertson, G. P. (2017). Field-scale experiments reveal persistent yield gaps in low-input and organic cropping systems. Proc. Natl. Acad. Sci. U. S. A. 114, 926–931. doi:10.1073/pnas.1612311114
Langendries, S., and Goormachtig, S. (2021). Paenibacillus polymyxa, a Jack of all trades. Environ. Microbiol. 23, 5659–5669. doi:10.1111/1462-2920.15450
Lehmann, J., Cowie, A., Masiello, C., Kammann, C., Woolf, D., Amonette, J., et al. (2021). Biochar in climate change mitigation. Nat. Geosci. 14, 883–892. (in Press. doi:10.1038/s41561-021-00852-8
Li, C., Zhao, C., Zhao, X., Wang, Y., Lv, X., Zhu, X., et al. (2023). Applications of metal-organic frameworks and their derivatives in electrochemical CO2 reduction. Agronomy 13, 113. doi:10.1007/s40820-023-01092-8
Li, S., and Shangguan, Z. (2018). Positive effects of apple branch biochar on wheat yield only appear at a low application rate, regardless of nitrogen and water conditions. J. Soils Sediments 18, 3235–3243. doi:10.1007/s11368-018-1994-3
Li, X., Wang, T., Chang, S. X., Jiang, X., and Song, Y. (2020). Biochar increases soil microbial biomass but has variable effects on microbial diversity: a meta-analysis. Sci. Total Environ. 749, 141593. doi:10.1016/j.scitotenv.2020.141593
Liu, B., Cai, Z., Zhang, Y., Liu, G., Luo, X., and Zheng, H. (2019). Comparison of efficacies of peanut shell biochar and biochar-based compost on two leafy vegetable productivity in an infertile land. Chemosphere 224, 151–161. doi:10.1016/J.CHEMOSPHERE.2019.02.100
Liu, X., Zhang, A., Ji, C., Joseph, S., Bian, R., Li, L., et al. (2013). Effects of arsenic sulfide (As2S2) on B and T lymphoma cell lines and possible underlying mechanisms. Plant Soil 373, 583–588. doi:10.3892/br.2013.119
Mendes, R., Kruijt, M., De Bruijn, I., Dekkers, E., Van Der Voort, M., Schneider, J. H. M., et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Sci. (80-. ) 332, 1097–1100. doi:10.1126/science.1203980
Meng, L., Sun, T., Li, M., Saleem, M., Zhang, Q., and Wang, C. (2019). Soil-applied biochar increases microbial diversity and wheat plant performance under herbicide fomesafen stress. Ecotoxicol. Environ. Saf. 171, 75–83. doi:10.1016/j.ecoenv.2018.12.065
Meschewski, E., Holm, N., Sharma, B. K., Spokas, K., Minalt, N., and Kelly, J. J. (2019). Pyrolysis biochar has negligible effects on soil greenhouse gas production, microbial communities, plant germination, and initial seedling growth. Chemosphere 228, 565–576. doi:10.1016/j.chemosphere.2019.04.031
Mettel, C., Kim, Y., Shrestha, P. M., and Liesack, W. (2010). Extraction of mRNA from soil. Appl. Environ. Microbiol. 76, 5995–6000. doi:10.1128/AEM.03047-09
Moreau, D., Bardgett, R. D., Finlay, R. D., Jones, D. L., and Philippot, L. (2019). A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 33, 540–552. doi:10.1111/1365-2435.13303
Mousavi, S. M., Srivastava, A. K., and Cheraghi, M. (2022). Soil health and crop response of biochar: an updated analysis. Arch. Agron. Soil Sci. 2022, 1–6. doi:10.1080/02701367.2022.2070116
Muhammad, N., Dai, Z., Xiao, K., Meng, J., Brookes, P. C., Liu, X., et al. (2014). Changes in microbial community structure due to biochars generated from different feedstocks and their relationships with soil chemical properties. Geoderma 226–227, 270–278. doi:10.1016/J.GEODERMA.2014.01.023
Nelkner, J., Henke, C., Lin, T. W., Pätzold, W., Hassa, J., Jaenicke, S., et al. (2019). Effect of long-term farming practices on agricultural soil microbiome members represented by metagenomically assembled genomes (MAGs) and their predicted plant-beneficial genes. Genes (Basel). 10, 424. doi:10.3390/genes10060424
Nzediegwu, C., Prasher, S., Elsayed, E., Dhiman, J., Mawof, A., and Patel, R. (2019). Effect of biochar on heavy metal accumulation in potatoes from wastewater irrigation. J. Environ. Manage. 232, 153–164. doi:10.1016/j.jenvman.2018.11.013
Olanrewaju, O. S., and Babalola, O. O. (2019). Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188. doi:10.1007/s00253-018-09577-y
Palansooriya, K. N., Wong, J. T. F., Hashimoto, Y., Huang, L., Rinklebe, J., Chang, S. X., et al. (2019). Response of microbial communities to biochar-amended soils: a critical review. Biochar 1, 3–22. doi:10.1007/s42773-019-00009-2
Pathy, A., Ray, J., and Paramasivan, B. (2020). Biochar amendments and its impact on soil biota for sustainable agriculture. Biochar 2, 287–305. doi:10.1007/s42773-020-00063-1
Quiza, L., St-Arnaud, M., and Yergeau, E. (2015). Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front. Plant Sci. 6, 507. doi:10.3389/fpls.2015.00507
Reganold, J. P., and Wachter, J. M. (2016). Organic agriculture in the twenty-first century. Nat. Plants 2, 15221. doi:10.1038/nplants.2015.221
Rojo, F. (2021). A new global regulator that facilitates the co-metabolization of polyaromatic hydrocarbons and other nutrients in Novosphingobium. Environ. Microbiol. 23, 2875–2877. doi:10.1111/1462-2920.15527
Roquigny, R., Novinscak, A., Biessy, A., and Filion, M. (2017). Pseudomonadaceae: from biocontrol to plant growth promotion. Rhizotrophs Plant Growth Promot. Bioremediation 2017, 39–68.
Sánchez-Monedero, M. A., Cayuela, M. L., Sánchez-García, M., Vandecasteele, B., D’Hose, T., López, G., et al. (2019). Agronomic evaluation of biochar, compost and biochar-blended compost across different cropping systems: perspective from the European project FERTIPLUS. Agronomy 9, 225. doi:10.3390/agronomy9050225
Seufert, V., Ramankutty, N., and Foley, J. (2012). Comparing the yields of organic and conventional agriculture. Nature 485, 229–232. doi:10.1038/nature11069
Sharma, P., Kalita, M. C., and Thakur, D. (2016). Broad spectrum antimicrobial activity of forest-derived soil actinomycete, Nocardia sp. PB-52. Front. Microbiol. 7, 347. doi:10.3389/fmicb.2016.00347
Sharpe, R. M., Gustafson, L., Hewitt, S., Kilian, B., Crabb, J., Hendrickson, C., et al. (2020). Concomitant phytonutrient and transcriptome analysis of mature fruit and leaf tissues of tomato (Solanum lycopersicum L. Cv. Oregon Spring) grown using organic and conventional fertilizer. PLoS One 15, e0227429. doi:10.1371/journal.pone.0227429
Sousa, J. A. de J., and Olivares, F. L. (2016). Plant growth promotion by streptomycetes: ecophysiology, mechanisms and applications. Chem. Biol. Technol. Agric. 3, 24–12. doi:10.1186/s40538-016-0073-5
Staley, C., Breuillin-Sessoms, F., Wang, P., Kaiser, T., Venterea, R. T., and Sadowsky, M. J. (2018). Urea amendment decreases microbial diversity and selects for specific nitrifying strains in eight contrasting agricultural soils. Front. Microbiol. 9, 634. doi:10.3389/fmicb.2018.00634
Sun, Y., Ding, A., Zhao, X., Chang, W., Ren, L., Zhao, Y., et al. (2022). Response of soil microbial communities to petroleum hydrocarbons at a multi-contaminated industrial site in Lanzhou, China. Chemosphere 306, 135559. doi:10.1016/j.chemosphere.2022.135559
Sun, Y., Gao, B., Yao, Y., Fang, J., Zhang, M., Zhou, Y., et al. (2014). Effects of feedstock type, production method, and pyrolysis temperature on biochar and hydrochar properties. Chem. Eng. J. 240, 574–578. doi:10.1016/j.cej.2013.10.081
Tahat, M., Alananbeh, K., Othman, Y., and Leskovar, D. (2020). Soil health and sustainable agriculture. Sustainability 12, 4859. doi:10.3390/su12124859
Tarazona, S., Furió-Tarı, P., Ferrer, A., and Conesa, A. (2013). NOISeq: differential expression in RNA-seq.
Tarazona, S., Furió-Tarí, P., Turrà, D., Pietro, A. D., Nueda, M. J., Ferrer, A., et al. (2015). Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 43, e140. doi:10.1093/nar/gkv711
Tarazona, S., García, F., Ferrer, A., Dopazo, J., and Conesa, A. (2011). NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet. J. 17, 18–19. doi:10.14806/ej.17.b.265
Tenenbaum, D. J. (2009). Biochar: carbon mitigation from the ground up. Environ. Health Perspect. 117, A70–A73. doi:10.1289/ehp.117-a70
Tenic, E., Ghogare, R., and Dhingra, A. (2020). Biochar—a panacea for agriculture or just carbon? Horticulturae 6, 37. doi:10.3390/horticulturae6030037
Tomer, A., Singh, R., Singh, S. K., Dwivedi, S. A., Reddy, C. U., Keloth, M. R. A., et al. (2021). Role of fungi in bioremediation and environmental sustainability. Mycoremediation Environ. Sustain. 3, 187–200.
Urich, T., Lanzén, A., Qi, J., Huson, D. H., Schleper, C., and Schuster, S. C. (2008). Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3, e2527. doi:10.1371/journal.pone.0002527
Velez, T. I., Moonilall, N. I., Reed, S., Jayachandran, K., and Scinto, L. J. (2018). Impact of melaleuca quinquenervia biochar on phaseolus vulgaris growth, soil nutrients, and microbial gas flux. J. Environ. Qual. 47, 1487–1495. doi:10.2134/jeq2017.12.0484
Vipotnik, Z., Michelin, M., and Tavares, T. (2021). Ligninolytic enzymes production during polycyclic aromatic hydrocarbons degradation: effect of soil pH, soil amendments and fungal co-cultivation. Biodegradation 32, 193–215. doi:10.1007/s10532-021-09933-2
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19, 952. doi:10.3390/ijms19040952
Wang, C., Wang, Y., Yan, S., Li, Y., Zhang, P., Ren, P., et al. (2023). Biochar-amended composting of lincomycin fermentation dregs promoted microbial metabolism and reduced antibiotic resistance genes. Bioresour. Technol. 367, 128253. doi:10.1016/j.biortech.2022.128253
Wang, H., Wang, C., Yuan, W., Chen, H., Lu, W., Zhang, H., et al. (2021). The role of phenylalanine hydroxylase in lipogenesis in the oleaginous fungus Mortierella alpina. Microbiology 167, 1062. doi:10.1099/mic.0.001062
Wang, T., Su, D., Wang, X., and He, Z. (2020). Adsorption-degradation of polycyclic aromatic hydrocarbons in soil by immobilized mixed bacteria and its effect on microbial communities. J. Agric. Food Chem. 68, 14907–14916. doi:10.1021/acs.jafc.0c04752
Wang, W., Wang, H., Feng, Y., Wang, L., Xiao, X., Xi, Y., et al. (2016). Consistent responses of the microbial community structure to organic farming along the middle and lower reaches of the Yangtze River. Sci. Rep. 6, 35046–35111. doi:10.1038/srep35046
Wang, Y., Hayatsu, M., and Fujii, T. (2009). Extraction of bacterial RNA from soil: challenges and solutions. Microbes Environ. 2009, 1202170350.
Wasi, S., Tabrez, S., and Ahmad, M. (2013). Use of Pseudomonas spp. for the bioremediation of environmental pollutants: a review. Environ. Monit. Assess. 185, 8147–8155. doi:10.1007/s10661-013-3163-x
White, R. A., Borkum, M. I., Rivas-Ubach, A., Bilbao, A., Wendler, J. P., Colby, S. M., et al. (2017). From data to knowledge: the future of multi-omics data analysis for the rhizosphere. Rhizosphere 3, 222–229. doi:10.1016/j.rhisph.2017.05.001
Woldetsadik, D., Drechsel, P., Marschner, B., Itanna, F., and Gebrekidan, H. (2018). Effect of biochar derived from faecal matter on yield and nutrient content of lettuce (Lactuca sativa) in two contrasting soils. Environ. Syst. Res. 6, 2. doi:10.1186/s40068-017-0082-9
Xiao, X., Chen, B., Chen, Z., Zhu, L., and Schnoor, J. L. (2018). Insight into multiple and multilevel structures of biochars and their potential environmental applications: a critical review. Environ. Sci. Technol. 52, 5027–5047. doi:10.1021/acs.est.7b06487
Xiao, X., Fan, M., Wang, E., Chen, W., and Wei, G. (2017). Interactions of plant growth-promoting rhizobacteria and soil factors in two leguminous plants. Appl. Microbiol. Biotechnol. 101, 8485–8497. doi:10.1007/s00253-017-8550-8
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi:10.1016/j.ejsobi.2016.02.004
Xu, P., Gao, Y., Cui, Z., Wu, B., Yan, B., Wang, Y., et al. (2023). Research progress on effects of biochar on soil environment and crop nutrient absorption and utilization. Sustainability 15, 4861. doi:10.3390/su15064861
Yang, X., Wang, W., Chen, X., Sardans, J., Wang, C., Vancov, T., et al. (2022). Effects of N-enriched biochar on ecosystem greenhouse gas emissions, rice yield, and bacterial community diversity in subtropical rice paddy soils. Eur. J. Soil Biol. 113, 103440. doi:10.1016/j.ejsobi.2022.103440
Yi, M., Zhang, L., Qin, C., Lu, P., Bai, H., Han, X., et al. (2022). Temporal changes of microbial community structure and nitrogen cycling processes during the aerobic degradation of phenanthrene. Chemosphere 286, 131709. doi:10.1016/j.chemosphere.2021.131709
Yu, H., Zou, W., Chen, J., Chen, H., Yu, Z., Huang, J., et al. (2019). Biochar amendment improves crop production in problem soils: a review. J. Environ. Manage. 232, 8–21. doi:10.1016/J.JENVMAN.2018.10.117
Keywords: metatranscriptome analysis, rhizosphere, organic production, plant-soil-microbiome interactions, biochar (BC)
Citation: Hewitt S, Ghogare R, Troxel W, Tenic E, Isaac D and Dhingra A (2023) Metatranscriptomic analysis of tomato rhizospheres reveals insight into plant-microbiome molecular response to biochar-amended organic soil. Front. Anal. Sci. 3:1205583. doi: 10.3389/frans.2023.1205583
Received: 14 April 2023; Accepted: 08 August 2023;
Published: 29 August 2023.
Edited by:
Makusu Tsutsui, Osaka University, JapanReviewed by:
M. Elena Diaz Rubio, The State University of New Jersey, United StatesMaciej Kochanowski, National Veterinary Research Institute (NVRI), Poland
Copyright © 2023 Hewitt, Ghogare, Troxel, Tenic, Isaac and Dhingra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Dhingra, YWRoaW5ncmFAdGFtdS5lZHU=
 Seanna Hewitt
Seanna Hewitt Rishikesh Ghogare
Rishikesh Ghogare William Troxel
William Troxel Elvir Tenic
Elvir Tenic Daylen Isaac1
Daylen Isaac1 Amit Dhingra
Amit Dhingra