- Department of Forensic Science, Sam Houston State University, Huntsville, TX, United States
Introduction: Gas chromatography combined with mass spectrometry (GC/MS) is popular analytical instrumentation for chemical separation and identification. A novel framework for chemical forensics based on the visualization of GC/MS data and transfer learning is proposed.
Methods: To evaluate the framework, 228 GC/MS data collected from two standard cannabis varieties, i.e., hemp and marijuana, were utilized. By processing the raw GC/MS data, analytical features, including retention times, mass-to-charge ratios, intensities, and summed ion mass spectra, were successfully transformed into two types of image representations. The GC/MS data transformed images were fed into a pre-trained convolutional neural network (CNN) to develop intelligent classifiers for the sample classification tasks. The effectiveness of several hyper-parameters for improving classification performance was investigated during transfer learning.
Results: The proposed analytical workflow could classify hemp and marijuana with 97% accuracy. Furthermore, the transfer-learning-based classifiers were established without requiring big data sets and peak alignment.
Discussion: The potential application of the new artificial intelligence (AI)-powered framework for chemical forensics using GC/MS data has been demonstrated. This framework provides unique opportunities for classifying various types of physical evidence using chromatography and mass spectrometry signals.
1 Introduction
Gas chromatography combined with mass spectrometry (GC/MS) is popular analytical instrumentation for chemical separation and identification, including evidence classification tasks in forensics (Gin and Imwinkelried, 2018). The high efficiency of GC/MS in separating components with great sensitivity allows the analysts to perform qualitative and quantitative chemical analyses of physical evidence (Liu et al., 2016). However, the interpretation of GC/MS data typically involves an analyst’s knowledge and highly relies on the analyst’s training, skill, and experience (Baerncopf and Hutches, 2014). Therefore, the turnaround time may be affected in a high-throughput forensic laboratory. Furthermore, the use of GC/MS data to assist evidence classification and association is critical because essential intelligence can be provided to support case investigation (Zadora and Neocleous, 2009). In this scenario, multivariate modeling is constantly adopted to analyze GC/MS data by various approaches (Belmonte-Sánchez et al., 2018; Sales et al., 2019).
Cannabis sativa L. (cannabis) has been known to produce cannabinoids that may have diverse bioactivities (Marsh and Smid, 2021). The Agriculture Improvement Act of 2018 provided a new statutory definition of hemp, which defines the cannabis plant, and any part of it, with a delta-9-tetrahydrocannabinol (THC) concentration of not more than 0.3% on a dry weight basis (United States Congress, 2018]. Therefore, the new law limits the definition of marijuana to include cannabis containing more than 0.3% THC. While hemp is used in manufacturing versatile products, marijuana remains a Schedule I controlled substance under the federal Controlled Substances Act (the United States Drug Enhancement Administration, 2022). To discriminate between hemp and marijuana, validated analytical workflows for cannabis samples have been revised for quantitative analysis of THC and improved by forensic associations and laboratories (McRae and Melanson, 2020; Sgrò et al., 2021; Oregon Environmental Laboratory Accreditation Program, 2022). Headspace sampling provides a solvent-free extraction process that features shorter extraction time, nearly non-destruction, and less sample consumption (Kataoka, 2011; Leghissa et al., 2017). Several laboratories have demonstrated the capability of headspace chemical analysis for cannabis samples (Osman and Caddy, 1985; Ilias et al., 2005; Wiebelhaus et al., 2016). McDaniel and co-workers have developed a heated headspace solid phase microextraction (HHS-SPME)-GC/MS method for marijuana profiling (McDaniel et al., 2018).
Recently, a deep convolutional neural network (CNN) in the framework of artificial intelligence (AI) is emerging as a potential candidate for performing multivariate predictive analysis in analytical chemistry (Lussier et al., 2020; Ayres et al., 2021; Debus et al., 2021). Deep learning has extensive applications in categorizing various kinds of data, such as sounds, speech, and text (Zhang et al., 2021), but particularly gains increasing attention in classifying image tasks (Obaid Kavi et al., 2019). In the structure of a CNN, multiple layers are responsible for identifying features in the images in a hierarchical manner and making inferences on categorical classification (Murata et al., 2018). Though deep CNNs have been praised for extracting relevant features from the images without manual engineering, the downside of this technique is that it fails to perform well when dealing with a small amount of data (Tammina, 2019). As a result, transfer learning is becoming popular for AI development because it is a more efficient learning approach regarding computation, costs, and time than training a CNN from scratch with randomly initialized weights (Khan et al., 2019). The transfer learning approach exploits existing knowledge of a pre-trained CNN, which has been trained to solve a particular classification problem on an extensive data set. The transfer learning process involves fine-tuning the pre-trained model weights to re-train the model for a new task with a smaller data set (Talo et al., 2019). In addition to saving the costs and time for data collection and modeling, the benefits of transfer learning also reflect in achieving higher classification performance (Thenmozhi and Srinivasulu Reddy, 2019). In mass spectrometry, the application of transfer learning is still at its early stage, and a few examples can be found in the retention time predictions (Ju et al., 2021; Osipenko et al., 2021; Yang et al., 2021). Compared to other approaches, for instance, PLS-DA, sparse PLS-DA, Random Forests, the transfer learning approach does not require feature selection, such as the determination of a matrix of abundance values or pretreatment of GC/MS peaks.
Digital image analysis has been investigated to process chromatograms and mass spectra to generate various 2-dimensional (2D) images in recent years. In those data, the retention times in the total ion chromatogram (TIC) and the ion fragments in the mass spectrum can be considered unique chemical profiles of a sample. It has also been reported that the summed ion mass spectrum, derived from adding up the intensities of individual nominal mass overall retention times, is characteristic of different volatile substances (Sigman et al., 2008). Therefore, by taking advantage of data visualization techniques, distinctive patterns can be generated from raw chromatography and mass spectral data, which allow the analysts to conduct a comparative analysis of the samples. For example, Bi et al. created a fingerprint image generation process for GC/MS data to evaluate food flavor quality (Bi et al., 2019). Crutchfield et al. and Zhao et al. employed a pseudo-color image approach to process liquid chromatography and mass spectrometry (LC/MS) data (Crutchfield et al., 2013; Zhao et al., 2021). Du et al. applied a continuous wavelet transform (CWT)-based peak detection algorithm with Mexican Hat wavelet to process mass spectra for improving denoising and peak detection (Du et al., 2006). Hence, it can be expected that the visualization of GC/MS data offers enormous potential for discrimination between different substances, including forensic evidence.
This work aimed to propose a new framework that integrates converting GC/MS data into 2D images and applying transfer learning for solving classification tasks in chemical forensics. Our research question was to explore whether a pre-trained CNN could distinguish the features of the images transformed from a small set of GC/MS data (<250). Additionally, it was hypothesized that, besides retention times, m/z values, and peak intensities, the summed ion mass spectrum could be a viable candidate for generating characteristic patterns in 2D images. To evaluate the feasibility of the framework, the discrimination between hemp and marijuana was chosen in this work. Briefly, HHS-SPME-GC/MS data collected from standard cannabis samples were visualized in 2D image formats using pseudo-color-heat-map-based and scalogram-based strategies. Two classifiers for differentiating hemp samples from marijuana samples were developed by transfer learning. The settings of the hyper-parameters were optimized, and the performance of the classifiers was evaluated through several standard benchmarks. This research work investigated and presented a promising approach that combined GC/MS data visualization and transfer learning as a new framework for solving various forensic classification tasks.
2 Materials and methods
2.1 Cannabis data set, headspace sampling, and instrumentation
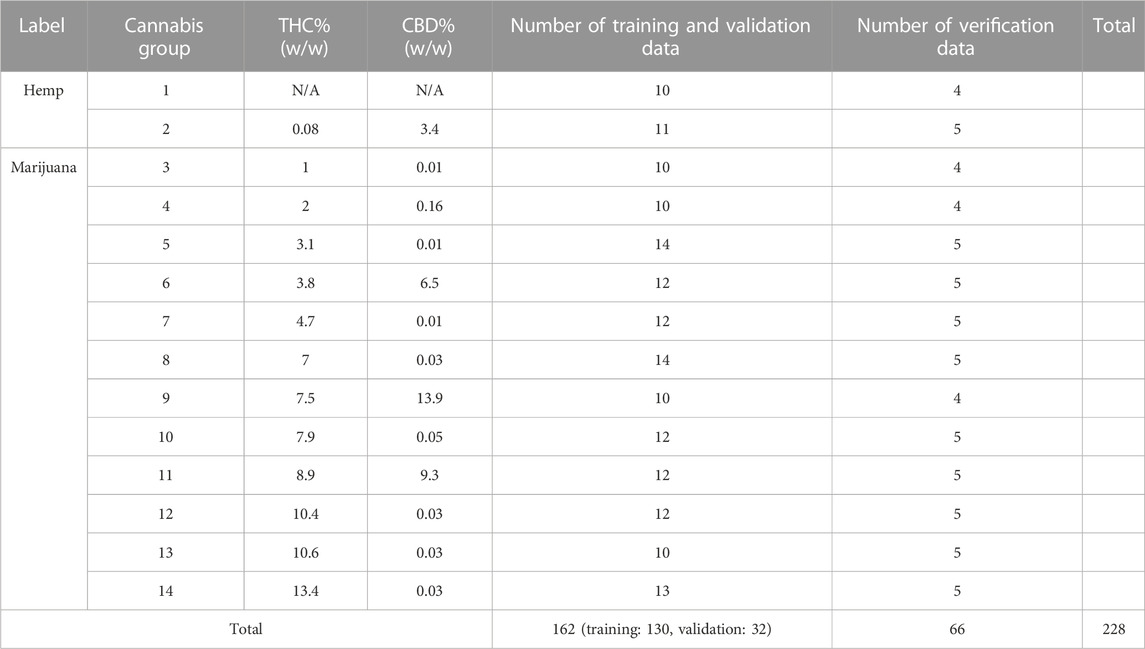
The cannabis data set was built from the data previously collected by McDaniel et al. (McDaniel et al., 2018). As shown in Table 1, the standard reference cannabis samples were provided by the National Institute on Drug Abuse (NIDA). The data set contained 14 different cannabis groups with known levels of THC and cannabidiol (CBD). The cannabis samples comprised mixed dry botanical structures, including buds, leaves, and stems, except groups 1, 3, 4, and 9 had some missing structures, as indicated in Supplementary Table S1. Each group was divided into several sub-portions: individual structure (i.e., buds, leaves, and stems, coded as B, L, S, respectively); a mixture of all structures (coded as W); ground materials from all structures (coded as C). Note that the concentration of THC and CBD were determined by NIDA, the grouping of hemp and marijuana were labeled based on the pre-determined concentration of THC in each variety.
When processing industrial hemp samples, a drying step of the cannabis sample is commonly employed to ensure the sample weight is in fact the dry weight (Health Canada, 2021). In our procedure, the sample provided by NIDA has been dried and packaged in glass bottles. Therefore, the weighting process did not include a drying step. Instead, an aliquot of each cannabis sample, 10 ± 0.1 mg measured by an analytical balance, was transferred into a 20-mL headspace vial for HHS-SPME extraction.
The HHS-SPME extraction parameters were selected from the optimized extraction method reported by Ilias et al. with the implementation of an autosampler (Ilias et al., 2005). All botanical structures were extracted using identical headspace sampling parameters. Briefly, a 23 gauge and 100 μm polydimethylsiloxane (PDMS) SPME fiber obtained from Sigma-Aldrich (St. Louis, MO) was installed onto the autosampler for headspace sampling. The pre-SPME fiber conditioning temperature was 250 °C. The sample pre-SPME incubation temperature was 140°C for 5 min. During HHS-SPME, the fiber was exposed to the headspace of a 20-ml sample vial for 2.5 min. After SPME, the fiber was retracted into a needle and introduced into the GC injection port for thermal desorption. Desorption time was set at 0.5 min. SPME for blank samples, i.e., empty headspace vials, were performed between runs to ensure no contaminants and carryovers on the SPME fiber. An Agilent 7890B and 5975A mass selective detector were used for GC/MS data collection. The GC column for separating cannabinoids was Rxi 35Sil-M3 (15 m × 0.25 mm × 0.25 μm) supplied by Restek (Bellefonte, PA). The initial GC oven temperature was 170 °C (hold 1 min). The temperature program for cannabinoids separation was as follows: 15°C/min to 228 °C (hold 3 min), 10°C/min to 250 °C, 5°C/min to 270 °C (hold 1.4 min). A detailed experimental explanation can be found in a previous report presented by our research group (McDaniel et al., 2018).
In the standard cannabis GC/MS data set, Groups 1 and 2 were labeled as “hemp” class and groups 3 to 14 were labeled as “marijuana” class according to the concentration of THC in the samples. There was a total number of 228 GC/MS data in the entire cannabis data set, which included 30 data in the hemp class and 198 data in the marijuana class. In each class, the GC/MS data were separated into training (total: 162 data) and verification (total: 66 data) subsets for the purposes of transfer learning and confirmation of the training outcomes. In the training data set, 80% (130 data) were used for model training, and 20% (32 data) were used for validation.
The verification data set (66 data) is a separate ground truth data set that was never used in the classifier training and validation process. The verification data were also used to evaluate the efficiency of several hyper-parameters for the proposed classifiers. Specifically, the verification data were obtained by random selection of one object in every sub-portion of the 14 groups, as listed in Supplementary Table S1.
2.2 GC/MS data processing
2.2.1 Data structure construction and signal resampling
The GC/MS data were first exported into NetCDF format using the Agilent ChemStation (Agilent Technologies, Inc., California, United States) to facilitate subsequent data processing in MATLAB (MATLAB 2021a and Bioinformatics Toolbox, MathWorks, Natick, Massachusetts, United States). As a starting point, we constructed a scalar of the vector of retention times (denoted as Time in the following statements) and a two-column matrix named “peaks.” The vector Time has 2,913 elements (i.e., scans) associated with retention times recorded from 0 to 16 min. The “peaks” matrix was representative of the mass spectrum in each scan, where the first column contained m/z values and the second column contained intensity values. It is worth noting that the idea of data matrix construction was to “resample” signals by fitting raw GC/MS data into equal sizes of arrays for all the samples. This task was achieved using the “msppresample” function, where a Gaussian kernel was used to reconstruct the signal. The outcome of resampling comprised recreated m/z values (denoted as MZ) and intensity values (denoted as Y). Consequently, the GC/MS data array consisted of MZ as a column vector of 2,000 elements. The elements were derived from the scanned mass range, initially set from m/z 40 to 450. Therefore, the unified data array enabled the transformation of chemical signals into image presentations using pseudo-color heat maps and scalograms.
2.2.2 GC/MS data selection
The major cannabinoids in cannabis plants include THC, CBD, and cannabinol (CBN). Supplementary Figures S1 shows an example of the total ion chromatogram (TIC) of sample 08059AB1. The THC, CBD, and CBN were detected between 6 and 13 min of retention time. Supplementary Figures S2A–C illustrate the extracted ion profiles (mass spectra) of sample 08059AB1, which represent the three cannabinoids. The primary ion fragments of THC, CBD, and CBN appeared between m/z 210 to 390. Supplementary Table S2 summarizes the retention times and major ions specific to each cannabinoid. Based on those observations, a range of data characteristic of the cannabinoids in vector Time and matrix MZ was selected, as indicated in Supplementary Table S3.
2.2.3 Pseudo-color heat map approach
The first approach for 2D image visualization of GC/MS data proposed in this study was using a pseudo-color heat map. The image displays the intensities for the spectra after a log transformation of the selected range of m/z values at the chosen range of retention times in matrix Y. In a pseudo-color heat map, the x and y-axes depict m/z values and retention times accordingly. Figure 1 shows the pseudo-color heat maps created from a typical hemp and marijuana sample.
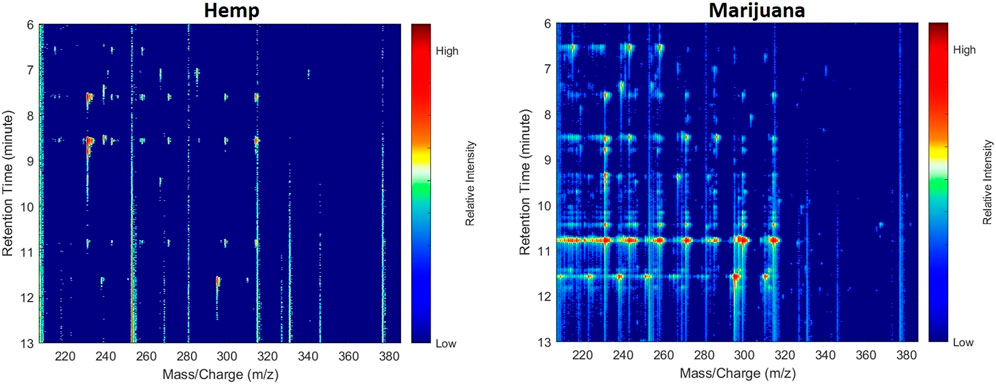
FIGURE 1. Examples of pseudo-color heat maps transformed from cannabis headspace GC/MS data. The axes, labels, and color bars were removed for the transfer learning.
2.2.4 Scalogram approach
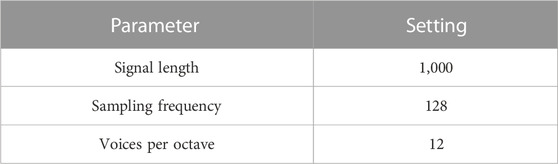
The second proposed 2D image visualization of GC/MS data was to create scalograms based on summed ion mass spectrum. The image was generated through CWT, a technique that has been applied to denoise or detect signals in various research areas, such as bioinformatics (Li et al., 2008; Xie et al., 2009), Raman spectroscopy (Ramos and Ruisánchez, 2005; Cooper et al., 2011; Li et al., 2013), and mass spectrometry (Du et al., 2006; Zheng et al., 2016). The mother wavelet used in this analysis was the Morse wavelet, widely used in processing modulated signals with time-varying amplitude and frequency (Sarraf et al., 2019). In this work, the summed ion mass spectrum was a column vector, denoted sY, by summing up the elements in each row in the extracted region in matrix Y. The summed ion mass spectrum was thereby regarded as modulated-analog signals. More precisely, the vector sY referred to the sum of the intensities of each m/z value over retention times in the selected region. The next step was to compute the vector sY by CWT to produce a wavelet scalogram, with the x and y-axes representing m/z values and the CWT coefficient scale, respectively. The CWT parameters are listed in Table 2. Figure 2 gives examples of scalograms of a hemp and marijuana sample. More information on the application of the Morse wavelet in 2D data processing can be found in research work by Huang et al. (Huang and Yu, 2021) and other related reports (Olhede and Walden, 2002; Lilly and Olhede, 2009; Lilly and Olhede, 2010; Lilly and Olhede, 2012; Lilly, 2016). To be compatible with the pre-trained CNN used in this study, the pseudo-color heat maps and the scalograms were produced in RGB images with an array size of 224 by 224 by 3. Moreover, the axes, labels, and color bars were also removed from the images for the subsequent transfer learning.
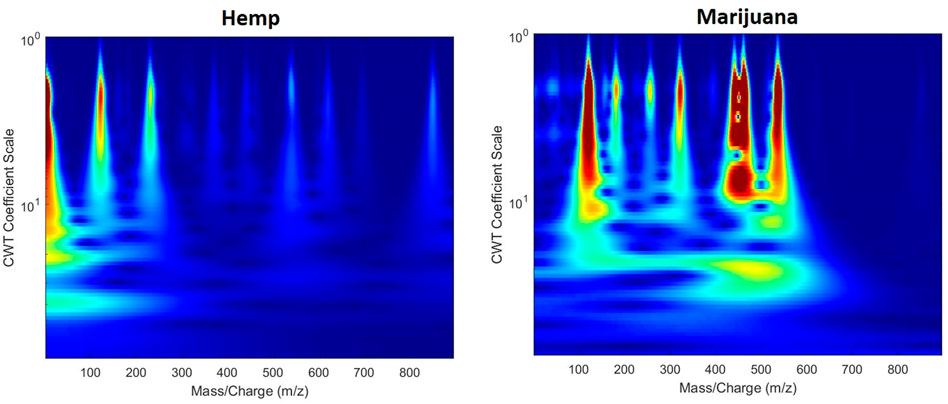
FIGURE 2. Examples of scalograms transformed from cannabis headspace GC/MS data. The axes and labels were removed for the transfer learning.
2.3 Transfer learning
2.3.1 Pre-trained CNN selection
A pre-trained CNN, GoogLeNet (Szegedy et al., 2015), was selected for transfer learning to classify pseudo-color heat maps and scalograms representing standard cannabis samples. This CNN model was originally trained to solve a 1,000-categories classification problem and is well-known for winning the ImageNet Large-Scale Visual Recognition Challenge 2014 (ILSVRC14) with an achievement of a top-5 error rate of 6.67% (Abbasi et al., 2020; Yilmaz and Trocan, 2021). It was hypothesized that GoogleNet can be retrained for the purpose of image analysis of analytical signals using CNN classification algorithm.
2.3.2 Network modification
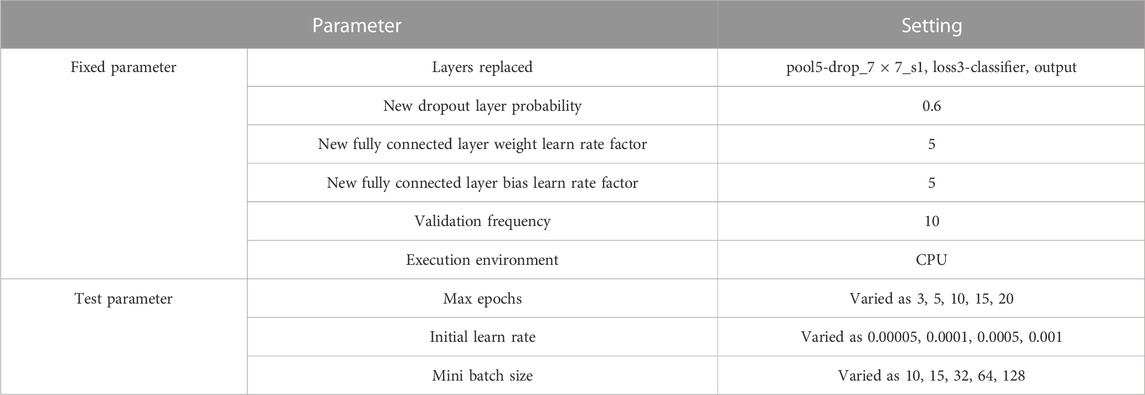
To make the original GoogLeNet adapt to our new data, the initial layers were retained. The final layers, including dropout (pool5-drop_7 × 7_s1), fully connected (loss3-classifier), and classification (output) layers, were replaced with new layers. The goal of employing a new dropout layer was to increase the probability of randomly removing input data from the network to prevent overfitting. A new fully connected layer was revised to the number of filters equaling the number of cannabis classes. Moreover, the learning rate factors for weights and bias at the new fully connected layer were increased to speed up the learning process. Finally, a new classification layer was utilized to specify the output classes investigated in this work.
2.3.3 Classifier training and verification
A stochastic gradient descent with 0.9 momentum was employed as an optimizer to minimize the loss function involved in the iterative process. To optimize the performance of the re-trained GoogLeNet, several hyper-parameters that might affect the classification error or cause overfitting were selected as “test parameters.” Those parameters include maximum epochs, learning rate, and mini batch size, as shown in Table 3. Both classifiers (one for pseudo-color-heat-map-based and another for scalogram-based) were constructed following the above-stated methodology.
2.3.4 Training options setting
The experiments of classifier training were conducted in MATLAB 2021a with 8 GB RAM and Intel(R) Core(TM) i7-5600U @2.60 GHz CPU. During transfer learning, the training data were randomly divided into 80% (130 images) and 20% (32 images) for the training and validation phases. Once the training was completed, the verification data were inputted into the classifiers to evaluate the classifier performance.
2.4 Evaluation of model performance
2.4.1 Performance measures
The effectiveness of the proposed cannabis classification systems was assessed by computing the average classification probability of the images in the verification data set. Five performance measures derived from the confusion matrices of the verification data were calculated. The measures involved accuracy, sensitivity, specificity, precision, and F1 score, computed by the formula in Supplementary Table S4.
2.4.2 Comparison tests
The classification performance of the proposed classifiers was compared with several common machine learning (ML) algorithms, including k-nearest neighbor (KNN), discriminant analysis (DA), Naive Bayes, support vector machine (SVM), and ensemble classification models. The ML and proposed CNN classifiers were developed using the same training data set. To compare with the pseudo-color-heat-map-based classifier, TICs between retention times of 6 and 13 min were chosen to train the ML classifiers. To compare with the scalogram-based classifier, summed ion mass spectra between 6 and 13 min retention times and m/z 210 to 390 were used to train the ML classifiers. Therefore, the data selection for the ML classifiers shared the same data range (retention time and m/z) of the proposed classifiers. The training of the ML classifiers was conducted in MATLAB with the Statistics and Machine Learning Toolbox (The MathWorks Inc, 2021). Once the final classifiers were obtained, bootstrap sampling of the verification data set was utilized to provide a comprehensive and statistical comparison (Harrington, 2006; Lu and Harrington, 2007). The means and uncertainties of the prediction outcome reported in the confusion matrix were computed from the 100 bootstrap samples with a 95% confidence interval. Note that the procedures for multiple ML techniques have been well-developed and detailed in the MATLAB platform. The technical details can be found in the operation manual.
3 Results and discussion
3.1 Optimization of hyper-parameters in transfer learning
To evaluate the effect of hyper-parameters in transfer learning, different settings of hyper-parameters were adjusted to construct initial classifiers. Sixty-six standard cannabis data (9 hemp images and 57 marijuana images) new to the initial classifiers were used to compare their performance using accuracy and prediction probability.
3.1.1 Maximum epochs
To determine the optimal number of epochs for final transfer learning, the training epochs were adjusted at 3, 5, 10, 15, and 20. The learning rate and mini batch size values were set at 0.001 and 10, respectively. As shown in Supplementary Figures S3A,D, both accuracy and average probabilities improved as the number of epochs increased in the pseudo-color heat-map-based classifier. In the scalogram-based classifier, the accuracy reached 0.97 as early as epoch 3 and maintained the same level through epoch 20. However, an increase in average probabilities along with an increment of the number of epochs was observed before epoch 15 both in the pseudo-color-heat-map-based classifier and the scalogram-based classifier. Therefore, the classification performance of the proposed classifiers was improved when the number of epochs increased. The optimal number of epochs for the pseudo-color-heat-map-based and scalogram-based classifiers in our experiments were determined to be 20 and 15, respectively.
3.1.2 Learning rate
Learning rates ranging from 0.00005, 0.0001, 0.0005, and 0.001 with a fixed mini batch size of 0.001 and maximum epochs of 20 and 15 for the pseudo-color-heat-map-based and scalogram-based classifiers were tested. As shown in Supplementary Figures S3B,E, the highest score for accuracy in both image types was obtained when the learning rate was set at 0.001. Further comparison of the average probabilities among all learning rates indicates that the prediction probabilities gradually improved when the value of the learning rate increased. In this study, a higher learning rate decreased the prediction error for the proposed classifiers. Therefore, the optimal learning rate was set to a value of 0.001 for both pseudo-color-heat-map-based and scalogram-based classifiers.
3.1.3 Mini batch size
To evaluate the effects of mini batch size, the initial classifiers were trained with a batch size of 10, 15, 32, 64, and 128. The learning rate was set at 0.001, and the maximum epochs were set at 20 and 15 for the pseudo-color-heat-map-based classifier and the scalogram-based classifier, accordingly. In Supplementary Figures S3C,F, the highest accuracy and average probabilities of both classifiers were observed when the mini batch size was set at 10. Overall, setting a small mini batch size resulted in higher classification performance. Hence, the optimal mini batch size was set at 10 for final transfer learning.
3.2 Deviation of standard cannabis data
During the optimization of the hyper-parameters, two misclassifications were found in all tests for both image transformation approaches. After checking the data, the two misclassified events were produced from samples 07271AS1 and 08171BC1. The former sample was a stem structure. The latter sample was ground cannabis materials from all plant structures. Both samples were within hemp group 1. It was observed that the two samples had relatively higher THC peaks. Comparing the median of peak intensities of THC in all the samples in hemp group 1, the THC peak intensities in 07271AS1 and 08171BC1 exceeded three scaled median absolute deviations away from the median of the THC peak intensities of all group 1 hemp samples. Since cannabis samples are plant materials, cannabinoids could be localized in different plant structures. Because the sampling size in each HHS-SPME-GC/MS test was only 10 mg, the deviations in the THC peak intensities might attribute to the heterogeneity of cannabinoids in the cannabis plants.
3.3 Final training progress
Supplementary Figures S4A,B present the training progress of the final transfer learning by using the optimal hyper-parameter values discussed in the previous section. The elapsed training times were 20 min 41 s for the pseudo-color-heat-map-based and 15 min 26 s for the scalogram-based classifier. There were 260 (pseudo-color-heat-map-based classifier) and 195 (scalogram-based classifier) iterations used for the stochastic gradient descent algorithm to repeatedly evaluate the gradient and update the descent algorithm weights to minimize the loss function for the final classifiers. Every 10 iterations, the classifiers validated the performance using the previously divided 20% of the images in the training data. Supplementary Figures S4A,B display that both image transformation approaches had some variations in the validation accuracy and loss at the beginning of the training due to the GoogLeNet starting to learn the new classification task. By comparing the variations, the oscillation of the validation accuracy and loss were more easily observed in the pseudo-color-heat-map-based approach, indicating an unstable learning process. Therefore, the pseudo-color-heat-map-based classifier required more epochs (11 epochs) and iterations (approximately 138 iterations) to reach 100% validation accuracy and 0% validation loss. In comparison, the scalogram-based approach took 7 epochs and 90 iterations to reach the same training accuracy. Nevertheless, both approaches achieved 100% and 0% for validation accuracy and loss at the end of the training. The training progress result suggests that the GoogLeNet with the proposed hyper-parameter values has successfully developed two new classification systems by transfer learning.
3.4 Comparison of CNN and ML classifier performance
The verification data set was also utilized to compare the performance of the final CNN classifiers and several ML classifiers. Tables 4, 5 shows the confusion matrices obtained from 100 bootstrap sampling of verification data, and Table 6 shows the accuracies and uncertainties at 95% confidence intervals for the CNN and ML classifiers. As shown in Tables 4, 5, the pseudo-color-heat-map-based classifier performs well (57 ± 0) in classifying marijuana samples. The decreased performance in classifying hemp samples was attributed to samples 07271AS1 and 08171BC1, whose THC peak intensities were deviated from the median of the THC peak intensities of all group 1 hemp samples, as discussed previously. A similar prediction outcome was observed for the scalogram-based classifier. When comparing the accuracies of the CNN and ML classifiers, as shown in Table 6, it is found that the pseudo-color-heat-map-based classifier has comparable performance with KNN, DA, and SVM classifiers. In contrast, the scalogram-based and DA classifiers perform better (0.97 ± 0.12 and 0.97 ± 0.1, respectively) than the other ML classifiers.
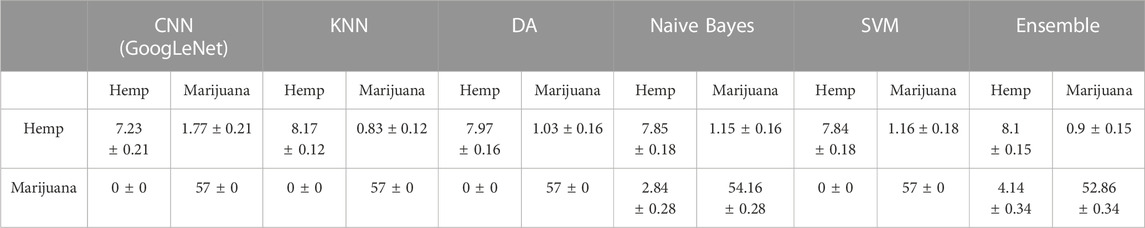
TABLE 4. Comparison of means of prediction outcome and uncertainties at 95% confidence intervals obtained from pseudo-color-heat-map-based classifier and ML classifiers using 100 bootstrap sampling of verification data.
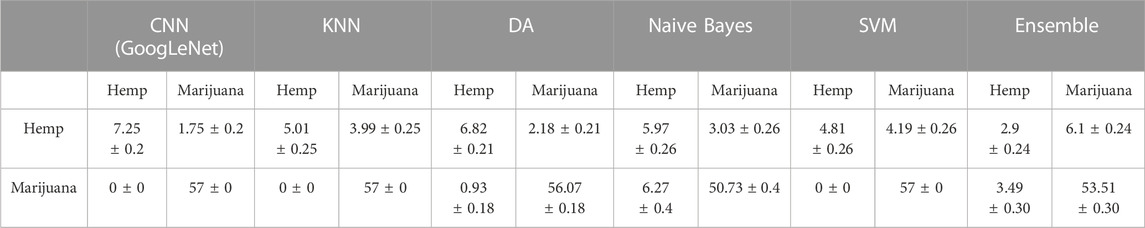
TABLE 5. Comparison of means of prediction outcome and uncertainties at 95% confidence intervals obtained from scalogram-based classifier and ML classifiers using 100 bootstrap sampling of verification data.
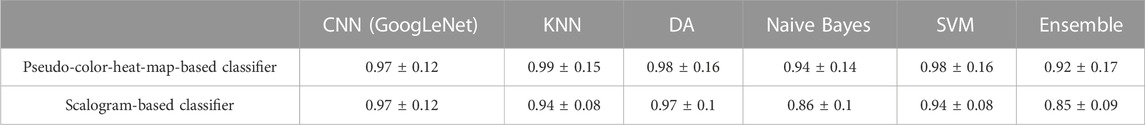
TABLE 6. Comparison of accuracies and uncertainties at 95% confidence intervals for the proposed classifiers and ML classifiers using 100 bootstrap sampling of verification data.
Overall, the results suggest that the pseudo-color-heat-map-based and scalogram-based approaches offered satisfactory capabilities to transform GC/MS data for transfer learning. Compared to the conventional ML methods for GC/MS data classification, the proposed framework can reduce human intervention in feature engineering and eliminate the tedious trial and error process, similar to the work reported by Janiesch et al. (Janiesch et al., 2021).
3.5 Discussion of experimental results
The pre-trained deep learning model, GoogLeNet, was structurally modified and fine-tuned to learn the features of the images transformed from GC/MS data. The experimental results show that both final classifiers obtained 97% accuracy in classifying the verification data into correct cannabis classes using the transfer learning technique. The average prediction probabilities also achieved nearly 100%. This performance not only reveals the successful development of the intelligent classifiers through the proposed framework but also indicates that the framework is feasible for solving GC/MS data classification problems without the need for a big training data set.
Another breakthrough of this study is the visualization of GC/MS data to facilitate transfer learning. The efficiency of the visualization of GC/MS data for cannabis classification has been demonstrated by obtaining high scores in the performance evaluation measures, including sensitivity, specificity, precision, and F1 score. The performance of the final classifiers provided promising results that the pseudo-color heat maps and scalograms can transform the characteristic information of GC/MS data for feature recognition in the transfer learning process. Moreover, the adoption of digital image analysis coupling with CNN transfer learning empowers the classifiers with the capability of translation invariance, meaning to ignore slight changes in the position of the feature in the input image. This type of classifier can effectively eliminate the peak alignment of the same sample analyzed in multiple runs, which is often required in traditional chemometric techniques (Li and Wang, 2019).
To further compare the two strategies for image transformation, Figures 3A,B display the scores of the evaluation measures derived from the hyper-parameter optimization tests. Though both GC/MS data visualization methods achieved good performance, the pseudo-color heat maps approach appeared more difficult than the scalograms approach to attain satisfying classification performance during the tuning process. In other words, the pseudo-color heat maps-based classifier relied more on optimizing critical hyper-parameters than the scalogram-based classifier. It was also found that the pseudo-color heat maps-based classifier encountered a more unstable learning process at the beginning and required more training cycles than the scalogram-based classifier to obtain 100% validation accuracy. Moreover, when using the bootstrap method to compare the classifier performance, the scalogram-based classifier gave better prediction results than the pseudo-color-heat-map-based classifier. These results support our hypothesis that the summed ion mass spectra can provide valuable features when visualized in 2D images. Moreover, it suggested that the GoogLeNet, a pre-trained CNN for image classification, could more readily recognize the chemical features in a scalogram than in a pseudo-color heat map.
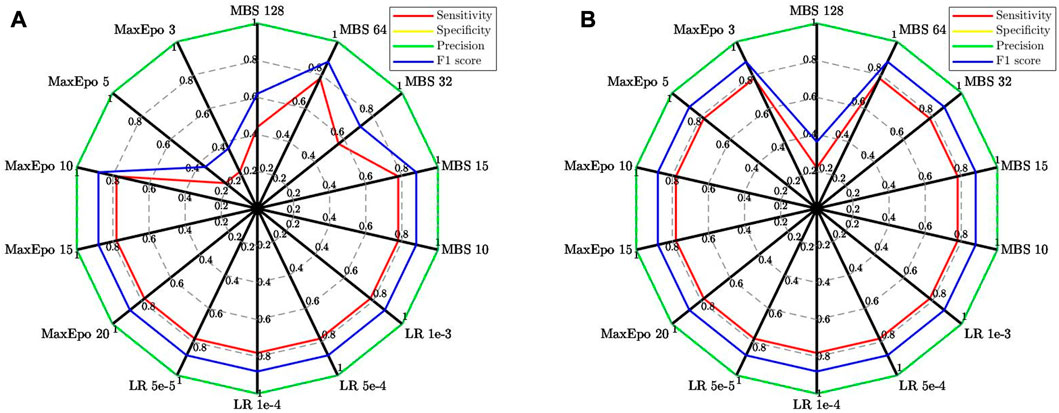
FIGURE 3. Radar charts showing the evaluation measures of the verification data in the hyper-parameter optimization tests. (A) Pseudo-color-heat-map-based classifier; (B) Scalogram-based classifier.
4 Conclusion
We proposed a new framework to successfully demonstrate the construction of AI-powered classification systems for cannabis samples using small data sets. The classifiers were assessed by careful examinations and comparisons of the hyper-parameters. An additional verification data set was prepared and tested to ensure the robustness of the performance checks. The results indicated that the classifiers achieved high performance in several performance measures. In addition, the proposed framework successfully eliminated the pre-processing of GC/MS data required by traditional multivariate modeling approaches. Given the novelty of the proposed framework, the most crucial step was the visualization of GC/MS data by transforming the summed ion mass spectra into scalograms and employing transfer learning of GoogLeNet to recognize the features representing characteristic headspace chemical profiles of the target substance. However, no biological-chemical interpretation of the CNN model may be the limitation of the proposed framework.
Moreover, the probability-based classification outcome given by the proposed framework aids statistical evaluations for unknown samples in some analytical and applied chemical disciplines, such as evidence interpretation in forensics. The workflow was accurate, fast, and relied on minimal manual involvement. The combination of GC/MS data transformation and transfer learning offered a new way for cannabis sample classification. More reliable cannabis sources and attributes can be collected in future work to assess the generalization capacities of this new approach. Due to the heterogeneity of cannabis plants, homogenizing cannabis plant samples and increasing the sample mass for HHS-SPME are highly recommended. In summary, the proposed framework provides a new perspective for chemical forensics in chromatography and mass spectrometry. We envision this new AI-powered classification framework for GC/MS data as a promising tool for other types of forensic evidence in a criminal investigation.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Author contributions
T-YH: Validation, Visualization, Writing–Original Draft. JY: Conceptualization, Writing–Review & Editing, Project administration.
Funding
This work was partly funded by the Ministry of Education, Taiwan. Standard marijuana varieties provided by the National Institute on Drug Abuse (NIDA) Drug Supply Program were appreciated.
Acknowledgments
The authors would like to acknowledge Austin McDaniel for his assistance in performing laboratory experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2023.1125049/full#supplementary-material
References
Abbasi, A. A., Hussain, L., Awan, I. A., Abbasi, I., Majid, A., Nadeem, M. S. A., et al. (2020). Detecting prostate cancer using deep learning convolution neural network with transfer learning approach. Cogn. Neurodynamics 14, 523–533. doi:10.1007/s11571-020-09587-5
Ayres, L. B., Gomez, F. J., Linton, J. R., Silva, M. F., and Garcia, C. D. (2021). Taking the leap between analytical chemistry and artificial intelligence: A tutorial review. Anal. Chim. Acta. 1161, 338403. doi:10.1016/j.aca.2021.338403
Baerncopf, J., and Hutches, K. (2014). A review of modern challenges in fire debris analysis. Forensic Sci. Int. 244, e12–e20. doi:10.1016/j.forsciint.2014.08.006
Belmonte-Sánchez, R., Gherghel, S., Arrebola-Liébanas, J., Romero González, R., Martínez Vidal, J. L., Parkin, I., et al. (2018). Rum classification using fingerprinting analysis of volatile fraction by headspace solid phase microextraction coupled to gas chromatography-mass spectrometry. Talanta 187, 348–356. doi:10.1016/j.talanta.2018.05.025
Bi, K., Zhang, D., Qiu, T., and Huang, Y. (2019). GC-MS fingerprints profiling using machine learning models for food flavor prediction. Processes 8, 23. doi:10.3390/pr8010023
Cooper, G., Kubik, M., and Kubik, K. (2011). Wavelet based Raman spectra comparison. Chemom. Intell. Lab. Syst. 107, 65–68. doi:10.1016/j.chemolab.2011.01.010
Crutchfield, C. A., Olson, M. T., Gourgari, E., Nesterova, M., Stratakis, C. A., and Yergey, A. L. (2013). Comprehensive analysis of LC/MS data using pseudocolor plots. J. Am. Soc. Mass Spectrom. 24, 230–237. doi:10.1007/s13361-012-0524-6
Debus, B., Parastar, H., Harrington, P., and Kirsanov, D. (2021). Deep learning in analytical chemistry. Trac. Trends Anal. Chem. 145, 116459. doi:10.1016/j.trac.2021.116459
Du, P., Kibbe, W. A., and Lin, S. M. (2006). Improved peak detection in mass spectrum by incorporating continuous wavelet transform-based pattern matching. Bioinformatics 22, 2059–2065. doi:10.1093/bioinformatics/btl355
Gin, J., and Imwinkelried, E. J. (2018). Gas chromatography-mass spectrometer (GC/MS): In scientific evidence, even “gold standard” techniques have limitations. SSRN Electron. J. doi:10.2139/ssrn.3245423
Harrington, P. B. (2006). Statistical validation of classification and calibration models using bootstrapped Latin partitions. Trac. Trends Anal. Chem. 25, 1112–1124. doi:10.1016/j.trac.2006.10.010
Health Canada (2021). Industrial hemp technical manual - standard operating procedures for sampling, testing and processing methodology. Available at: https://www.canada.ca/en/health-canada/services/health-concerns/reports-publications/controlled-substances-precursor-chemicals/industrial-hemp-technical-manual-standard-operating-procedures-sampling-testing-processing-methodology.html#a1 (Accessed September 29, 2022).
Huang, T. Y., and Yu, J. C. C. (2021). Development of crime scene intelligence using a hand-held Raman spectrometer and transfer learning. Anal. Chem. 93, 8889–8896. doi:10.1021/acs.analchem.1c01099
Ilias, Y., Rudaz, S., Mathieu, P., Christen, P., and Veuthey, J. L. (2005). Extraction and analysis of different cannabis samples by headspace solid-phase microextraction combined with gas chromatography-mass spectrometry. J. Sep. Sci. 28, 2293–2300. doi:10.1002/jssc.200500130
Janiesch, C., Zschech, P., and Heinrich, K. (2021). Machine learning and deep learning. Electron Mark. 31, 685–695. doi:10.1007/s12525-021-00475-2
Ju, R., Liu, X., Zheng, F., Lu, X., Xu, G., and Lin, X. (2021). Deep neural network pretrained by weighted autoencoders and transfer learning for retention time prediction of small molecules. Anal. Chem. 93, 15651–15658. doi:10.1021/acs.analchem.1c03250
Kataoka, H. (2011). Current developments and future trends in solid-phase microextraction techniques for pharmaceutical and biomedical analyses. Anal. Sci. 27, 893–905. doi:10.2116/analsci.27.893
Khan, S., Islam, N., Jan, Z., Ud Din, I., and Rodrigues, J. J. P. C. (2019). A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 125, 1–6. doi:10.1016/j.patrec.2019.03.022
Leghissa, A., Hildenbrand, Z. L., and Schug, K. A. (2017). A review of methods for the chemical characterization of cannabis natural products. J. Sep. Sci. 41, 398–415. doi:10.1002/jssc.201701003
Li, M., and Wang, X. R. (2019). Peak alignment of gas chromatography–mass spectrometry data with deep learning. J. Chromatogr. A 1604, 460476. doi:10.1016/j.chroma.2019.460476
Li, Z. C., Zhou, X. B., Dai, Z., and Zou, X. Y. (2008). Prediction of protein structural classes by chou’s pseudo amino acid composition: Approached using continuous wavelet transform and principal component analysis. Amino Acids 37, 415–425. doi:10.1007/s00726-008-0170-2
Li, S., Nyagilo, J. O., Dave, D. P., and Gao, J. X. (2013). Continuous wavelet transform based partial least squares regression for quantitative analysis of Raman spectrum. IEEE Trans. NanoBiosci. 12, 214–221. doi:10.1109/tnb.2013.2278288
Lilly, J. M., and Olhede, S. C. (2009). Higher-order properties of analytic wavelets. IEEE Trans. Signal Process 57, 146–160. doi:10.1109/tsp.2008.2007607
Lilly, J. M., and Olhede, S. C. (2010). On the analytic wavelet transform. IEEE Trans. Inf. Theory. 56, 4135–4156. doi:10.1109/tit.2010.2050935
Lilly, J. M., and Olhede, S. C. (2012). Generalized Morse wavelets as a superfamily of analytic wavelets. IEEE Trans. Signal Process 60, 6036–6041. doi:10.1109/tsp.2012.2210890
Lilly, J. M. (2016). jLab: A data analysis package for Matlab, v.1.7.1. Available at: http://www.jmlilly.net/jmlsoft.html (Accessed August 25, 2022).
Liu, C. T., Zhang, M., Yan, P., Liu, H. C., Liu, X. Y., and Zhan, R. T. (2016). Qualitative and quantitative analysis of volatile components of zhengtian pills using gas chromatography mass spectrometry and ultra-high performance liquid chromatography. J. Anal. Methods Chem. 2016, 1206391–1206398. doi:10.1155/2016/1206391
Lu, Y., and Harrington, P. B. (2007). Forensic application of gas chromatography–differential mobility spectrometry with two-way classification of ignitable liquids from fire debris. Anal. Chem. 79, 6752–6759. doi:10.1021/ac0707028
Lussier, F., Thibault, V., Charron, B., Wallace, G. Q., and Masson, J. F. (2020). Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. Trac. Trends Anal. Chem. 124, 115796. doi:10.1016/j.trac.2019.115796
Marsh, D. T., and Smid, S. D. (2021). Cannabis phytochemicals: A review of phytocannabinoid chemistry and bioactivity as neuroprotective agents. Aust. J. Chem. 74, 388–404. doi:10.1071/ch20183
McDaniel, A., Perry, L., Liu, Q., Shih, W. C., and Yu, J. C. C. (2018). Toward the identification of marijuana varieties by headspace chemical forensics. Forensic Chem. 11, 23–31. doi:10.1016/j.forc.2018.08.004
McRae, G., and Melanson, J. E. (2020). Quantitative determination and validation of 17 cannabinoids in cannabis and hemp using liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 412, 7381–7393. doi:10.1007/s00216-020-02862-8
Murata, M., Ariji, Y., Ohashi, Y., Kawai, T., Fukuda, M., Funakoshi, T., et al. (2018). Deep-learning classification using convolutional neural network for evaluation of maxillary sinusitis on panoramic radiography. Oral Radiol. 35, 301–307. doi:10.1007/s11282-018-0363-7
Obaid Kavi, B., Zeebaree Subhi, R. M., and Omar Ahmed, M. (2019). Deep learning models based on image classification: A review. Int. J. Sci. Bus. 4, 75–81.
Olhede, S., and Walden, A. (2002). Generalized Morse wavelets. IEEE Trans. Signal Process. 50, 2661–2670. doi:10.1109/tsp.2002.804066
Oregon Environmental Laboratory Accreditation Program (2022). Protocol for collecting samples of cannabinoids concentrates, extracts and products. ORELAP-SOP-002 Rev3.2.
Osipenko, S., Botashev, K., Nikolaev, E., and Kostyukevich, Y. (2021). Transfer learning for small molecule retention predictions. J. Chromatogr. A 1644, 462119. doi:10.1016/j.chroma.2021.462119
Osman, A., and Caddy, B. (1985). Analysis of cannabis using Tenax-GC. J. Forensic Sci. Soc. 25, 377–384. doi:10.1016/s0015-7368(85)72417-6
Ramos, P. M., and Ruisánchez, I. (2005). Noise and background removal in Raman spectra of ancient pigments using wavelet transform. J. Raman Spectrosc. 36, 848–856. doi:10.1002/jrs.1370
Sales, C., Portolés, T., Johnsen, L., Danielsen, M., and Beltran, J. (2019). Olive oil quality classification and measurement of its organoleptic attributes by untargeted GC–MS and multivariate statistical-based approach. Food Chem. 271, 488–496. doi:10.1016/j.foodchem.2018.07.200
Sarraf, S. Y., Trappen, R., Kumari, S., Bhandari, G., Mottaghi, N., Huang, C. Y., et al. (2019). Application of wavelet analysis on transient reflectivity in ultra-thin films. Opt. Express. 27, 14684–14694. doi:10.1364/oe.27.014684
Sgrò, S., Lavezzi, B., Caprari, C., Polito, M., D’Elia, M., Lago, G., et al. (2021). Delta9-THC determination by the EU official method: Evaluation of measurement uncertainty and compliance assessment of hemp samples. Anal. Bioanal. Chem. 413, 3399–3410. doi:10.1007/s00216-021-03283-x
Sigman, M. E., Williams, M. R., Castelbuono, J. A., Colca, J. G., and Clark, C. D. (2008). Ignitable liquid classification and identification using the summed-ion mass spectrum. Instrum. Sci. Technol. 36, 375–393. doi:10.1080/10739140802151440
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., et al. (2015). “Going deeper with convolutions,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 1–9.
Talo, M., Baloglu, U. B., Yıldırım, Z., and Rajendra Acharya, U. (2019). Application of deep transfer learning for automated brain abnormality classification using MR images. Cognit. Syst. Res. 54, 176–188. doi:10.1016/j.cogsys.2018.12.007
Tammina, S. (2019). Transfer learning using VGG-16 with deep convolutional neural network for classifying images. Int. J. Sci. Res. Publ. (IJSRP) 9, 9420. doi:10.29322/ijsrp.9.10.2019.p9420
The MathWorks, Inc. (2022). Get started with Statistics and machine learning Toolbox. Available at: https://www.mathworks.com/help/stats/getting-started-12.html (Accessed February 22, 2023).
Thenmozhi, K., and Srinivasulu Reddy, U. (2019). Crop pest classification based on deep convolutional neural network and transfer learning. Comput. Electron. Agric. 164, 104906. doi:10.1016/j.compag.2019.104906
United States Congress (2018). H.R.2 - agriculture improvement Act of 2018. Available at: https://www.congress.gov/bill/115th-congress/house-bill/2/text (Accessed September 9, 2022).
United States Drug Enhancement Administration (2022). Drug scheduling. Available at: https://www.dea.gov/drug-information/drug-scheduling (Accessed September 9, 2022).
Wiebelhaus, N., Hamblin, D., Kreitals, N. M., and Almirall, J. R. (2016). Differentiation of marijuana headspace volatiles from other plants and hemp products using capillary microextraction of volatiles (CMV) coupled to gas-chromatography–mass spectrometry (GC–MS). Forensic Chem. 2, 1–8. doi:10.1016/j.forc.2016.08.004
Xie, X. P., Ding, X. H., Wang, H. Q., and Jiang, Y. C. (2009). Continuous wavelet analysis of gene expression signals for cancer classification. J. Biol. Syst. 17, 377–396. doi:10.1142/s0218339009002946
Yang, Q., Ji, H., Fan, X., Zhang, Z., and Lu, H. (2021). Retention time prediction in hydrophilic interaction liquid chromatography with graph neural network and transfer learning. J. Chromatogr. A 1656, 462536. doi:10.1016/j.chroma.2021.462536
Yilmaz, E., and Trocan, M. (2021). A modified version of GoogLeNet for melanoma diagnosis. J. Inf. Telecommun. 5, 395–405. doi:10.1080/24751839.2021.1893495
Zadora, G., and Neocleous, T. (2009). Likelihood ratio model for classification of forensic evidence. Anal. Chim. Acta. 642, 266–278. doi:10.1016/j.aca.2008.12.013
Zhang, X., Wu, F., and Li, Z. (2021). Application of convolutional neural network to traditional data. Expert Syst. Appl. 168, 114185. doi:10.1016/j.eswa.2020.114185
Zhao, F., Huang, S., and Zhang, X. (2021). High sensitivity and specificity feature detection in liquid chromatography–mass spectrometry data: A deep learning framework. Talanta 222, 121580. doi:10.1016/j.talanta.2020.121580
Keywords: transfer learning, convolutional neural network, chemical forensics, pseudo-color heat map, scalogram, gas chromatography-mass spectrometry
Citation: Huang T-Y and Yu JCC (2023) Intelligent framework for cannabis classification using visualization of gas chromatography/mass spectrometry data and transfer learning. Front. Anal. Sci. 3:1125049. doi: 10.3389/frans.2023.1125049
Received: 15 December 2022; Accepted: 03 April 2023;
Published: 17 April 2023.
Edited by:
James J. Harynuk, University of Alberta, CanadaReviewed by:
Véronique Cariou, Agroalimentaire et de l’alimentation de Nantes-Atlantique (Oniris), FranceMartina Foschi, University of L'Aquila, Italy
Copyright © 2023 Huang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jorn Chi Chung Yu, am9ybnl1QHNoc3UuZWR1
 Ting-Yu Huang
Ting-Yu Huang Jorn Chi Chung Yu
Jorn Chi Chung Yu