
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Anal. Sci. , 05 September 2022
Sec. Biomedical Analysis and Diagnostics
Volume 2 - 2022 | https://doi.org/10.3389/frans.2022.994394
We report on the design of a simple all-DNA circuit with dual functions of signal amplification and signal reporting and its use for detection of human telomerase activity from cancer cells. The system utilizes a catalytic hairpin assembly (CHA) reaction for amplification, which produces split G-quadruplex outputs that assemble to form complete guanine quadruplex structures as reporting modules. As designed, a linear DNA sequence (the target) functions as a catalyst to drive cyclic programmed assembly of two hairpins, producing a DNA duplex with two guanine-rich sequences that assemble to form a complete Gq structure. The formation of the Gq element allows either fluorescence or colorimetric detection of the target. Examples are provided to demonstrate fluorescence detection of cancer cells’ telomerase activities in solution and the first example of a CHA-modulated colorimetric assay for detecting telomerase activities of cancer cells using a simple paper device.
Early diagnosis of human diseases demands simple, cost-effective, highly reliable and ultra-sensitive methods that can detect relevant biomarkers. The use of all-DNA circuits to deliver enzyme-free amplification represents a very attractive option as it removes potential issues with denaturation of protein enzymes, which are required for many DNA amplification systems (Dirks and Pierce, 2004; Yin et al., 2008; Choi et al., 2010; Li et al., 2011; Li et al., 2012a; Allen et al., 2012; Li et al., 2012b; Jiang et al., 2014; Chen et al., 2015; Du et al., 2015; Wu et al., 2015; Kotani and Hughes, 2017). Catalytic hairpin assembly (CHA) offers a particularly simple solution: the system only uses two hairpin-shaped DNA oligonucleotides designed to undergo a chain reaction driven by an input DNA (target) as the catalyst strand (Figure 1) (Yin et al., 2008; Li et al., 2011; Li et al., 2012a; Allen et al., 2012; Li et al., 2012b; Jiang et al., 2014; Chen et al., 2015; Du et al., 2015; Wu et al., 2015). The product of CHA is a DNA duplex formed between the two DNA hairpins. To make the CHA system into a biosensor, a common approach is to use an additional duplex assembly doubly labeled with a fluorophore/quencher pair as the signal output module (Yan et al., 2016). However, the use of the external reporter module not only adds to the cost and design complexity of CHA, but often is inherently associated with a high background signal that significantly reduces the analytical performance of CHA.
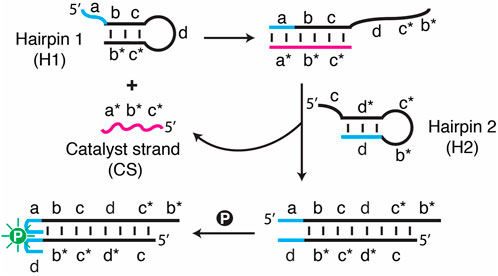
FIGURE 1. Schematic illustration of a catalytic hairpin assembly (CHA) system featuring SGq/PPIX as a fluorescent reporter. All the DNA sequences used for this study are provided in Supplementary Table S1.
Recently, our groups reported on a simplified CHA biosensor that incorporates a split G-quadruplex (SGq) as an internal reporter module for the duplex product (Zhou et al., 2020). Using a 4-way junction as a control element, it was possible to design either turn-on or turn-off sensors based on target-mediated CHA to produce full G-quadruplexes from assembly of the SGq elements, which were detected based on the fluorescence enhancement of protoporphryn IX (PPIX) upon binding to the full G quadruplex. Assembly of G-quadruplexes as fluorescence reporters for telomerase detection has also been reported based on a combined strand-displacement reaction (SDR)-CHA system, and using CHA incorporating a DNAzyme feedback system to improve amplification efficiency (Yan et al., 2016; Zou et al., 2019).
In this work, we evaluate the potential of the SGq-CHA system for ultra-sensitive monitoring of the activity of human telomerase, a proposed diagnostic marker for human cancers, via detection of the telomeric DNA repeats made by the telomerase. To move the assay toward a simple point-of-care sensor, we extend the original design using a fluorescence output to a colorimetric output based on the peroxidase activity of the full Gq (Li et al., 1996; Travascio et al., 1998; Travascio et al., 1999; Travascio et al., 2001; Lee et al., 2004; Xiao et al., 2004; Travascio et al., 2006; Li et al., 2009a; Li et al., 2009b; Li B. et al., 2009; Li et al., 2010a; Li et al., 2010b; Zhu et al., 2010; Zhu et al., 2011). In addition, we demonstrate the translation of the colorimetric assay onto a simple printed paper-based sensor to allow equipment-free detection of telomerase activity (Liu et al., 2016).
DNA strands were purchased from Integrated DNA Technologies (IDT), and are listed in Supplementary Table S1. All DNA oligonucleotides were purified by 10% denaturing (8 M urea) polyacrylamide gel electrophoresis (dPAGE), and their concentrations were determined spectroscopically. Protoporphyrin IX (PPIX) and 3,3′,5,5′-tetramethylbenzidine (TMB) were purchased from Sigma-Aldrich. Other chemicals were of reagent grade and were used without further purification.
Before starting the CHA reaction, the hairpin substrates were separately heated at 90°C for 5 min and slowly cooled down to room temperature (RT, ∼22°C) in Tris buffer (5 mM Tris-HCl, 0.5 mM EDTA, 100 mM NaCl, 20 mM KCl, 10 mM MgCl2, pH 8.0). For a typical CHA reaction, two hairpins (0.5 μM), aliquots of catalyst strand (CS, 0–40 nM) and PPIX (1 μM) were mixed together. After incubating for 1 h at RT, the fluorescence analysis was performed using a Cary Eclipse fluorescence spectrophotometer (Agilent Technologies, USA). The slit widths for excitation and emission were both set at 10.0 nm. The excitation wavelength was set at 410 nm and emission was recorded at 631 nm.
HeLa and MCF7 cells were cultured in D-MEM medium supplemented with 10% fetal calf serum, and were maintained at 37°C in a humidified atmosphere with 5% CO2. MCF10A cells were cultured in D-MEM medium supplemented with 5% (v/v) Horse Serum (Sigma H1138). Cells were collected in the exponential phase of growth, then 5 × 106 cells were dispensed in a 1.5 ml EP tube, washed twice with ice-cold PBS solution, and resuspended in 100 μL of ice-cold CHAPS lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.5% (w/v) CHAPS, 0.1 mM PMSF (freshly prepared in ethanol), 5 μL of RNase Inhibitor (40 U/μL)). The lysate was incubated for 30 min on ice and then centrifuged for 20 min (11,000 g, 4°C). Without disturbing the pellet, the cleared lysate was carefully transferred. The lysate was used immediately or stored at −80°C before use.
Telomerase extracts were diluted to respective numbers of cells using the CHAPS lysis buffer mentioned above. Then, 5 μL of extracts were mixed with dNTPs (1 mM each dNTP) and 0.5 μM primer in Extension Buffer (20 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 63 mM KCl, and 1 mM EGTA). The final volume of the solution was 50 μL. In control experiments, the telomerase extracts were first heated at 90°C for 30 min, and then used to perform the extension experiments as above.
The design of the SGq-CHA system was based on a similar system that was previously published (Zhou et al., 2020). Hairpins H1 and H2 (0.5 μM each), CS (8 nM) and PPIX (1 μM) were added to the above telomerase extension solutions with a final volume of 100 μL. After incubation for 1 h, the fluorescence intensity measurements were carried out. The excitation wavelength was set at 410 nm (λem = 631 nm).
The DNA solutions mixed with 6× loading buffer (TEK buffer, pH 8.0, 50% glycerol, 0.25% bromphenol blue) were analyzed by 10% (or 15% if noted) native polyacrylamide gel electrophoresis. The electrophoresis was conducted in 1× TBE (pH 8.0) at a constant voltage of 110 V for 1 h. The gels were scanned using a Typhoon 9,200 variable mode imager (GE Healthcare) after staining with SYBR Gold.
Paper micro-well plates were first printed using a Xerox ColorQube 8570N solid wax printer.
To immobilize DP on paper for the DNA extension reaction, streptavidin was used to bind the biotinylated DP. Briefly, 5 nmol of DP and 100 μL of 2 mg/ml streptavidin were added to 300 μL of Tris buffer. After incubating at RT for 15 min, free DP was removed by centrifugation through a 30 K membrane (NANOSEP OMEGA, Pall Incorporation) at 5,000 g for 10 min. The as-prepared streptavidin-biotin DNA conjugates were washed twice with Extension Buffer and collected. Then 10 μL of the above solution was added onto each test zone and was incubated overnight. After blocking with 10% BSA for 20 min and washing twice, the obtained bioactive paper was dried at RT and was ready to be used.
In a typical telomerisation experiment, 2 μL of cell extracts, dNTPs (10 mM), and Extension Buffer were added (total volume: 20 μL) to the test zones. The reaction was allowed to proceed at RT for 1 h. For the radioactive assay, 0.5 μL of [α-32P]dGTP was added for the extension reaction; then after being washed three times, the radioactive paper was scanned using a Typhoon 9,200 variable mode imager. For the colorimetric assay with SGq-CHA, H1 (1 μM), H2 (1 μM), CS (10 nM) and hemin (3 μM) was introduced to each well and allowed to react with the extension products for 40 min. Finally, 10 μL of H2O2 and TMB mixture in CDK buffer (26.6 mM citrate, 51.4 mM disodium hydrogen phosphate, 20 mM KCl, pH 5.0) was added and mixed by pipetting to initiate the colorimetric reaction, and the picture was taken using a digital camera.
Among the many possible G-quadruplex outputs, T30695, a 16-nucleotide G-rich DNA molecule with a sequence of 5′-GGGT GGGT GGGT GGGT-3′, exhibits significant peroxidase-like catalytic activity upon binding hemin. In the presence of H2O2, the T30695/hemin complex is capable of catalyzing the H2O2-mediated oxidation of 2,2-azinobis (3-ethylbenzothiazoline)-6-sulfonic acid (ABTS) or 3,3′,5,5′-tetramethylbenzidine (TMB) to produce a color change (Travascio et al., 1998; Travascio et al., 1999; Li et al., 2009a; Li B. et al., 2009; Zhu et al., 2010; Zhu et al., 2011). T30695 is also capable of binding protoporphyrin IX (PPIX) and the resultant DNA-porphyrin complex exhibits significantly enhanced fluorescence over PPIX alone, providing a useful fluorescence output (Li et al., 2010b). Therefore, T30695/PPIX can also act as a fluorescent reporter. Finally, T30695 can also be formed from a “split” configuration to restore the G-quadruplex structure to create a two-probe DNA biosensor (Deng et al., 2008; Kolpashchikov, 2008; Nakayama and Sintim, 2009; Kong et al., 2010; Wang et al., 2011; Ren et al., 2012; Zhou et al., 2012; Zhu et al., 2012; Zhu et al., 2013a; Zhu et al., 2013b; Zhu et al., 2013c; Hou et al., 2013; Jiang et al., 2013; Zhu et al., 2015). We therefore selected T30695 for incorporation into a CHA circuit to allow both fluorimetric and colorimetric detection of telomerase activity.
To generate a fluorimetric assay, the SGq-CHA system consists of three DNA components (Figure 1): DNA hairpin 1 (H1), DNA hairpin 2 (H2) and a catalyst strand (CS, the DNA target), along with PPIX (simplified as “p" in Figure 1) as a fluorescence reporter. H1 and H2 each contain a sequence tag that can assemble into a guanine quadruplex when placed in physical proximity. H1 and H2 are designed in a specific way such that the target CS, which contains sequence elements a*, b* and c*, is complementary to the abc sequence elements of H1 and thus can hybridize with H1 to create the dc*b* tail. The c* element within this tail then initiates the binding to H2 at the c element, followed by hybridization of the d*c*b* elements of H2 onto H1, resulting in the displacement of CS. The newly released CS can then undergo the next cycle of the CHA reaction. Each cycle causes the assembly of a G-quadruplex from the original SGq system, which is capable of binding with PPIX, resulting in a significantly enhanced fluorescence signal.
We initially assessed the performance of the SGq-CHA design via both fluorescence measurements and native polyacrylamide gel electrophoresis (nPAGE). The emission spectra of H1/H2 solutions containing increasing concentrations of CS (Figure 2A) and the fluorescence intensity vs the concentration of CS (Figure 2B) demonstrate the functionality of the system, as the all-DNA system can produce increased fluorescence in response to increasing CS concentrations. The nPAGE data provides direct evidence that CS promotes the formation of the H1/H2 hybrid.
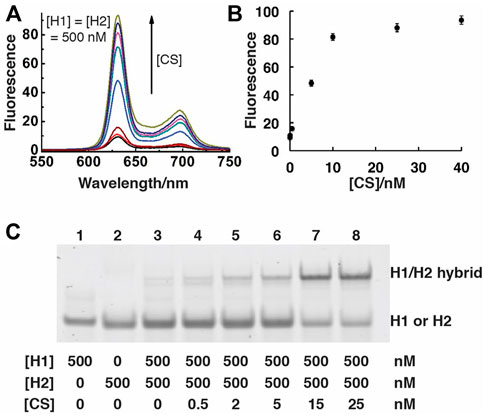
FIGURE 2. Analysis of SGq-CHA reactions. (A) Fluorescence spectra of SGq-CHA-PPIX solutions containing varying concentrations of CS. [CS] = 0, 0.05, 0.5, 5, 8, 10, 25, 40 nM; [H1] = [H2] = 500 nM; [PPIX] = 1 μM. The excitation wavelengths were set at 410 nm, reaction time is 1 h. (B) Plot of fluorescence intensity at 631 nm vs. [CS] from panel (A). (C) 10% native polyacrylamide gel analysis of SGq-CHA reaction solutions containing indicated concentrations of H1, H2 and CS.
Given the excellent performance of the SGq-CHA system, we further investigated its utility for amplified detection. We chose to apply the SGq-CHA system for the detection of human telomerase activity. Telomeres, consisting of tandemly repeated GGGTTA sequences along with several binding proteins, act as the “caps” at the end of linear chromosomes (Harley et al., 1994; Kim et al., 1994; Hou et al., 2001). Human telomerase is a ribonucleoprotein that can synthesize telomeric DNA sequences using its intrinsic RNA template and reverse transcriptase (Morin, 1989; Feng et al., 1995; Cohen et al., 2007; Wang et al., 2012; Wu et al., 2012; Wang et al., 2015). In most healthy human somatic cells, each cell division will shorten the length of telomeres, finally triggering the cellular senescence program and apoptosis when the telomere reaches a critical length (Hastie et al., 1990; Harley et al., 1994; Kim et al., 1994; Hou et al., 2001; Blasco, 2005). However, most malignant human cells are capable of escaping senescence and sustaining infinite proliferation through the activation of telomerase to stabilize their telomere length (Harley et al., 1994; Kim et al., 1994; Shay and Bacchetti, 1997; Hou et al., 2001). Therefore, telomerase has been considered as a potential diagnostic marker for tumors and an attractive target for cancer therapy (Harley et al., 1994; Kim et al., 1994; Shay and Bacchetti, 1997; Hou et al., 2001). Accordingly, there is significant interest in the development of sensitive, simple and cost-effective methods for the analysis of telomerase activity. At present, the telomeric repeat amplification protocol (TRAP) is the “gold standard” method for the determination of telomerase activity (Harley et al., 1994; Kim et al., 1994; Hou et al., 2001). This method, although highly sensitive because telomerase products are amplified by a subsequent polymerase chain reaction (PCR), has inherent drawbacks, including a limited dynamic range, long assay time, use of perishable DNA polymerase, need for a thermocycler and skilled operators, and susceptibility to PCR-derived artifacts (Harley et al., 1994; Kim et al., 1994; Hou et al., 2001).
We investigated the use the proposed SGq-CHA system to replace the current PCR technique with an isothermal amplification strategy for detection of telomerase activity, which can also enable exponential amplification to ensure high sensitivity and avoid the disadvantages of TRAP. As shown in Figure 3A, the basis of such detection is that the primer-telomeric repeats generated in the telomerase reaction can hybridize with CS, thereby “locking” the catalytic ability of CS and suppressing the downstream CHA reaction.
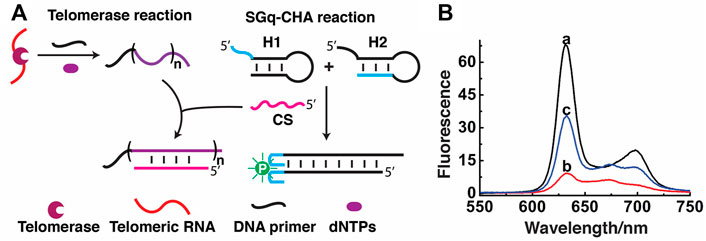
FIGURE 3. Detection of telomerase activity using SGq-CHA reaction. (A) Design principle. (B) Fluorescence emission spectra of reaction solutions containing: curve a: H1/H2/CS/PPIX, curve b: a + telomerase extracts from 2.5 × 105 Hela cells, and curve c: a + heat-treated telomerase extracts from 2.5 × 105 Hela cells. [H1] = [H2] = 500 nM; [CS] = 8 nM; [PPIX] = 1 μM λex = 410 nm.
Fluorescence measurements were first carried out to test the feasibility of the assay. Initially, the fluorescence of the SGq-CHA system with 8 nM CS was high (Figure 3B, curve a). However, the presence of telomerase products greatly inhibited the ability of CS to trigger the CHA reaction, causing a decreased fluorescence signal (Figure 3B, curve b). In a control experiment to show the specificity of the SGq-CHA system for telomerase, a heat-treated telomerase sample was used, which should prevent the process of telomerisation. In this case, the fluorescence of the system remained relatively high (Figure 3B, curve c), owing to a lack of telomere DNA.
We also used nPAGE to detect the amount of the reaction product produced in the presence of different numbers of Hela cells, from which telomerase had been extracted (Figure 4A). With the use of increasing numbers of Hela cells (lane 3 to lane 8), the intensity of the band corresponding to the DNA primer (DP) as well as to the H1/H2 hybrid progressively lessened, indicating that more and more DP was used by the increasing amounts of telomerases from the cells to produce more and more telomeric repeats, accompanied by stronger and stronger suppression of the CHA reaction. As a control, we conducted an additional reaction where heat was applied to denature telomerase (lane 9; note that the same numbers of Hela cells were used for the reactions in lanes 8 and 9). The high intensity of both the DP band and the H1/H2 hybrid band further supports that the presence of telomerase can inhibit the CHA reaction. Moreover, high molecular-weight products were detected by agarose gel electrophoresis (Supplementary Figure S1). Taken together, the nPGAE results indicate that SGq-CHA can specifically detect the telomerase activity from human cancer cells.
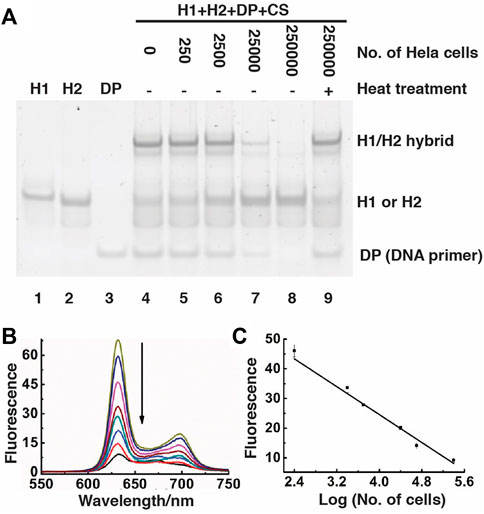
FIGURE 4. Detection of telomerase activity using SGq-CHA reaction. (A) 15% nPAGE analysis of reaction solutions containing telomerase extracted from indicated numbers of Hela cells. DP: DNA primer, which was used at 0.5 μM; [H1] = [H2] = 500 nM; [CS] = 8 nM. (B) Fluorescence spectra of SGq-CHA reactions containing telomerase extracted from 0, 25, 250, 2.5 × 103, 1.0 × 104, 2.5 × 104, 1.0 × 105 and 2.5 × 105 cells. [H1] = [H2] = 500 nM; [CS] = 8 nM λex = 410 nm. (C) Plot of fluorescence intensity at 631 nm vs number of Hela cells in logarithmic scale. λex = 410 nm.
The telomerase/SGq-CHA reactions were further examined using fluorescence spectroscopy (Figure 4B). There existed a linear correlation between the fluorescence intensity at 635 nm and the logarithm of the numbers of Hela cells over the range of 2.5 × 102 to 2.5 × 105 cells (Figure 4C). The detection limit of this method is 10 cells (signal-to-noise of 3) using a 1 h reaction time. Our method shows better or comparable sensitivity to previously reported methods that need long telomerization times or complex preparation procedures (Yang et al., 2011; Wang et al., 2012; Wu et al., 2012; Zhou and Xing, 2012; Liu et al., 2013; Wang et al., 2014; Zhu et al., 2014; Wang et al., 2015; Gao et al., 2016; Yan et al., 2016; Zhang et al., 2017). A list of methods and detection limit of comparable cell-free assays are presented in Supplementary Table S2.
To make the CHA method more convenient for practical use, we also engineered a paper device with color change as the output. As shown in Figure 5, we first used the wax printing technique to produce a 96-microzone paper plate, with the diameter of each test zone being 4 mm. A pre-formed 5′-biotinylated DP-streptavidin conjugate was then printed onto the test zone using inkjet printing. To demonstrate that the proposed strategy allows efficient telomerization on the paper surface, we added cellular extracts and dNTPs on the test zone to initiate the telomerization reaction. In the experiment shown as Figure 5A, we added [α-32P]dGTP in the reaction mixture so that the telomerization product became radioactive. Strong radioactivity was observed for extracts of Hela and MCF7 cells, both of which are known to have high telomerase activity. Much reduced radioactivity was observed in the control experiments with either MCF10A cells (which have low telomerase activity) or a non-binding DNA sequence (named DC) as the primer. These experiments show the high specificity of the paper sensor for the detection of the human telomerase activity from cancer cells.
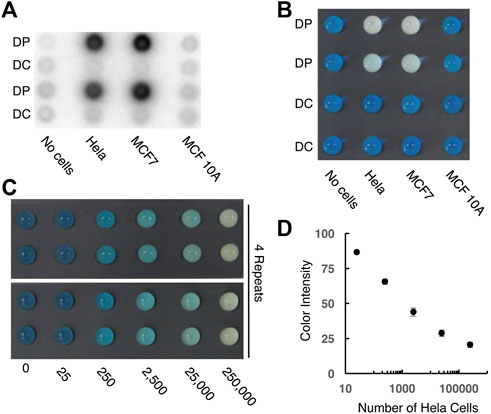
FIGURE 5. Designing paper sensors to detect human telomerase activity. (A) Tracking the telomerization reaction using paper-printed DNA primer and radioactive dNTPs. DP: telomerase reaction with matching DNA primer; DC: telomerase reaction with control DNA primer. 2.5 × 105 cells were used for this experiment. (B) Colorimetric signal output for control and test samples. 2.5 × 105 cells were used for this experiment. (C) Color signal vs number of Hela cells. [H1] = [H2] = 1 μM; [hemin] = 3 μM; [CS] = 10 nM. (D) Plot of color intensity vs number of Hela cells in logarithmic scale.
We then investigated the colorimetric reporting capability of the paper sensor (Figure 5B). In this design, the SGq-CHA reaction produces an intact G-quadruplex, which, upon binding to hemin, can act as a peroxidase to oxidize TMB and produce a blue color. The presence of telomerase leads to the generation of telomeric repeats on the paper surface, which hybridize with CS to supress the SGq-CHA reaction. As shown in Figure 5B, both Hela and MCF7 cells (2.5 × 105 cells) resulted in a decrease in the characteristic blue color seen with the control reactions. Semi-quantitative analysis (Figure 5, panels c and d) indicates that this paper-based method is able to detect as low as 250 cells within 100 min (60 min telomerase reaction followed by 40 min CHA reaction).
We have designed an all-DNA system that can achieve the dual functions of integrated signal-amplification and signal-reporting. It uses three separate DNA molecules to carry out catalytic hairpin assembly (CHA). It differs from the classic CHA design by the incorporation of a half guanine quadruplex (a split guanine quadruplex or sGq) into each of the two hairpins needed for the CHA reaction. This unique feature allows the CHA reaction to produce a complete guanine quadruplex (cGq) as part of the reaction product. The creation of cGq allows for convenient signal output, either via enhanced fluorescence as a result of PPIX-cGq binding, or via the production of a color signal as a result of hemin-cGq binding and subsequent oxidization of TMB. This sGq/cGq design is compatible with many applications that have already been demonstrated for traditional CHA reactions. However, to further expand the utility of CHA as well as illustrate the effectiveness of the sGq/cGq approach, we have applied this method to detect the high human telomerase activity associated with cancer cells in a cell-free assay. For this specific application, a DNA primer is provided to allow the telomerase to produce a repetitive sequence that interferes with the CHA reaction. The method is shown to be capable of detecting telomerase activity from as few as 10 cancer cells using fluorimetric assay, or 250 cancer cells using a simple paper-based colorimetric sensor. We have previously shown that paper sensors printed with DNA-based materials are extremely stable during storage at ambient temperatures and no significant performance reduction was observed after storage for up to 180 days (Liu et al., 2016; Ali et al., 2017; Hsieh et al., 2017). Given these properties, this all-DNA amplification and reporting system has the potential to be developed into a point of care diagnostic test.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YL and JB conceived the project. ZZ and JG designed the CHA system and carried out all experimental work. All authors contributed to the preparation and reviewing of the manuscript.
This work was supported by the Natural Sciences and Engineering Research Council of Canada Discovery Grant program, the Ontario Research Fund and Covalon Technologies Inc.
JB holds the Canada Research Chair in Point-of-Care Diagnostics.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frans.2022.994394/full#supplementary-material
Ali, M. M., Brown, C. L., Jahanshahi-Anbuhi, S., Kannan, B., Li, Y., Filipe, C. D. M., et al. (2017). A printed multicomponent paper sensor for bacterial detection. Sci. Rep. 7, 12335. doi:10.1038/s41598-017-12549-3
Allen, P. B., Arshad, S. A., Li, B., Chen, X., and Ellington, A. D. (2012). DNA circuits as amplifiers for the detection of nucleic acids on a paperfluidic platform. Lab. Chip 12, 2951–2958. doi:10.1039/c2lc40373k
Blasco, M. A. (2005). Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622. doi:10.1038/nrg1656
Chen, J., Zhou, S., and Wen, J. (2015). Concatenated logic circuits based on a three-way DNA junction: A keypad-lock security system with visible readout and an automatic reset function. Angew. Chem. Int. Ed. Engl. 54, 446–450. doi:10.1002/anie.201408334
Choi, H. M. T., Chang, J. Y., Trinh, L. A., Padilla, J. E., Fraser, S. E., and Pierce, N. A. (2010). Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212. doi:10.1038/nbt.1692
Cohen, S. B., Graham, M. E., Lovrecz, G. O., Bache, N., Robinson, P. J., and Reddel, R. R. (2007). Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853. doi:10.1126/science.1138596
Deng, M., Zhang, D., Zhou, Y., and Zhou, X. (2008). Highly effective colorimetric and visual detection of nucleic acids using an asymmetrically split peroxidase DNAzyme. J. Am. Chem. Soc. 130, 13095–13102. doi:10.1021/ja803507d
Dirks, R. M., and Pierce, N. A. (2004). Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U. S. A. 101, 15275–15278. doi:10.1073/pnas.0407024101
Du, Y., Hughes, R. A., Bhadra, S., Jiang, Y. S., Ellington, A. D., and Li, B. (2015). A sweet spot for molecular diagnostics: Coupling isothermal amplification and strand exchange circuits to glucometers. Sci. Rep. 5, 11039. doi:10.1038/srep11039
Feng, J., Funk, W. D., Wang, S. S., Weinrich, S. L., Avilion, A. A., Chiu, C. P., et al. (1995). The RNA component of human telomerase. Science 269, 1236–1241. doi:10.1126/science.7544491
Gao, Y., Xu, J., Li, B., and Jin, Y. (2016). PCR-free and label-free fluorescent detection of telomerase activity at single-cell level based on triple amplification. Biosens. Bioelectron. 81, 415–422. doi:10.1016/j.bios.2016.03.022
Harley, C. B., Kim, N. W., Prowse, K. R., Weinrich, S. L., Hirsch, K. S., West, M. D., et al. (1994). Telomerase, cell immortality, and cancer. Cold Spring Harb. Symp. Quant. Biol. 59, 307–315. doi:10.1101/sqb.1994.059.01.035
Hastie, N. D., Dempster, M., Dunlop, M. G., Thompson, A. M., Green, D. K., and Allshire, R. C. (1990). Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868. doi:10.1038/346866a0
Hou, M., Xu, D., Björkholm, M., and Gruber, A. (2001). Real-time quantitative telomeric repeat amplification protocol assay for the detection of telomerase Activity2. Clin. Chem. 47, 519–524. doi:10.1093/clinchem/47.3.519
Hou, T., Wang, X., Liu, X., Liu, S., Du, Z., and Li, F. (2013). A label-free and colorimetric turn-on assay for coralyne based on coralyne-induced formation of peroxidase-mimicking split DNAzyme. Analyst 138, 4728–4731. doi:10.1039/c3an01024d
Hsieh, P.-Y., Monsur Ali, M., Tram, K., Jahanshahi-Anbuhi, S., Brown, C. L., Brennan, J. D., et al. (2017). RNA protection is effectively achieved by pullulan film formation. Chembiochem 18, 502–505. doi:10.1002/cbic.201600643
Jiang, J., He, Y., Yu, X., Zhao, J., and Cui, H. (2013). A homogeneous hemin/G-quadruplex DNAzyme based turn-on chemiluminescence aptasensor for interferon-gamma detection via in-situ assembly of luminol functionalized gold nanoparticles, deoxyribonucleic acid, interferon-gamma and hemin. Anal. Chim. Acta 791, 60–64. doi:10.1016/j.aca.2013.06.048
Jiang, Y. S., Bhadra, S., Li, B., and Ellington, A. D. (2014). Mismatches improve the performance of strand-displacement nucleic Acid circuits. Angew. Chem. Int. Ed. Engl. 53, 1845–1848. doi:10.1002/anie.201307418
Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D., Ho, P. L., et al. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015. doi:10.1126/science.7605428
Kolpashchikov, D. M. (2008). Split DNA enzyme for visual single nucleotide polymorphism typing. J. Am. Chem. Soc. 130, 2934–2935. doi:10.1021/ja711192e
Kong, D.-M., Wang, N., Guo, X.-X., and Shen, H.-X. (2010). Turn-on” detection of Hg2+ ion using a peroxidase-like split G-quadruplex-hemin DNAzyme. Analyst 135, 545–549. doi:10.1039/b924014d
Kotani, S., and Hughes, W. L. (2017). Multi-arm junctions for dynamic DNA nanotechnology. J. Am. Chem. Soc. 139, 6363–6368. doi:10.1021/jacs.7b00530
Lee, H.-W., Chinnapen, D. J.-F., and Sen, D. (2004). Structure-function investigation of a deoxyribozyme with dual chelatase and peroxidase activities. Pure Appl. Chem. 76, 1537–1545. doi:10.1351/pac200476071537
Li, B., Chen, X., and Ellington, A. D. (2012a). Adapting enzyme-free DNA circuits to the detection of loop-mediated isothermal amplification reactions. Anal. Chem. 84, 8371–8377. doi:10.1021/ac301944v
Li, B., Du, Y., Li, T., and Dong, S. (2009). Investigation of 3, 3’, 5, 5’-tetramethylbenzidine as colorimetric substrate for a peroxidatic DNAzyme. Anal. Chim. Acta 651, 234–240. doi:10.1016/j.aca.2009.09.009
Li, B., Ellington, A. D., and Chen, X. (2011). Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 39, e110. doi:10.1093/nar/gkr504
Li, B., Jiang, Y., Chen, X., and Ellington, A. D. (2012b). Probing spatial organization of DNA strands using enzyme-free hairpin assembly circuits. J. Am. Chem. Soc. 134, 13918–13921. doi:10.1021/ja300984b
Li, T., Dong, S., and Wang, E. (2010a). A lead(II)-driven DNA molecular device for turn-on fluorescence detection of lead(II) ion with high selectivity and sensitivity. J. Am. Chem. Soc. 132, 13156–13157. doi:10.1021/ja105849m
Li, T., Li, B., Wang, E., and Dong, S. (2009a). G-quadruplex-based DNAzyme for sensitive mercury detection with the naked eye. Chem. Commun. 24, 3551–3553. doi:10.1039/b903993g
Li, T., Wang, E., and Dong, S. (2010b). Parallel G-quadruplex-specific fluorescent probe for monitoring DNA structural changes and label-free detection of potassium ion. Anal. Chem. 82, 7576–7580. doi:10.1021/ac1019446
Li, T., Wang, E., and Dong, S. (2009b). Potassium-lead-switched G-quadruplexes: A new class of DNA logic gates. J. Am. Chem. Soc. 131, 15082–15083. doi:10.1021/ja9051075
Li, Y., Geyer, C. R., and Sen, D. (1996). Recognition of anionic porphyrins by DNA aptamers. Biochemistry 35, 6911–6922. doi:10.1021/bi960038h
Liu, J., Lu, C.-Y., Zhou, H., Xu, J.-J., Wang, Z.-H., and Chen, H.-Y. (2013). A dual-functional electrochemical biosensor for the detection of prostate specific antigen and telomerase activity. Chem. Commun. 49, 6602–6604. doi:10.1039/c3cc43532f
Liu, M., Hui, C. Y., Zhang, Q., Gu, J., Kannan, B., Jahanshahi-Anbuhi, S., et al. (2016). Target-Induced and equipment-free DNA amplification with a simple paper device. Angew. Chem. Int. Ed. Engl. 55, 2709–2713. doi:10.1002/anie.201509389
Morin, G. B. (1989). The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59, 521–529. doi:10.1016/0092-8674(89)90035-4
Nakayama, S., and Sintim, H. O. (2009). Colorimetric split G-quadruplex probes for nucleic acid sensing: Improving reconstituted DNAzyme’s catalytic efficiency via probe remodeling. J. Am. Chem. Soc. 131, 10320–10333. doi:10.1021/ja902951b
Ren, J., Wang, J., Wang, J., Luedtke, N. W., and Wang, E. (2012). Contribution of potassium ion and split modes of G-quadruplex to the sensitivity and selectivity of label-free sensor toward DNA detection using fluorescence. Biosens. Bioelectron. 31, 316–322. doi:10.1016/j.bios.2011.10.038
Shay, J. W., and Bacchetti, S. (1997). A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791. doi:10.1016/S0959-8049(97)00062-2
Travascio, P., Bennet, A. J., Wang, D. Y., and Sen, D. (1999). A ribozyme and a catalytic DNA with peroxidase activity: Active sites versus cofactor-binding sites. Chem. Biol. 6, 779–787. doi:10.1016/s1074-5521(99)80125-2
Travascio, P., Li, Y., and Sen, D. (1998). DNA-enhanced peroxidase activity of a DNA-aptamer-hemin complex. Chem. Biol. 5, 505–517. doi:10.1016/s1074-5521(98)90006-0
Travascio, P., Sen, D., and Bennet, A. J. (2006). DNA and RNA enzymes with peroxidase activity — an investigation into the mechanism of action. Can. J. Chem. 84, 613–619. doi:10.1139/v06-057
Travascio, P., Witting, P. K., Mauk, A. G., and Sen, D. (2001). The peroxidase activity of a hemin--DNA oligonucleotide complex: Free radical damage to specific guanine bases of the DNA. J. Am. Chem. Soc. 123, 1337–1348. doi:10.1021/ja0023534
Wang, J., Wu, L., Ren, J., and Qu, X. (2012). Visualizing human telomerase activity with primer-modified Au nanoparticles. Small 8, 259–264. doi:10.1002/smll.201101938
Wang, N., Kong, D.-M., and Shen, H.-X. (2011). Amplification of G-quadruplex DNAzymes using PCR-like temperature cycles for specific nucleic acid and single nucleotide polymorphism detection. Chem. Commun. 47, 1728–1730. doi:10.1039/c0cc04182c
Wang, W.-J., Li, J.-J., Rui, K., Gai, P.-P., Zhang, J.-R., and Zhu, J.-J. (2015). Sensitive electrochemical detection of telomerase activity using spherical nucleic acids gold nanoparticles triggered mimic-hybridization chain reaction enzyme-free dual signal amplification. Anal. Chem. 87, 3019–3026. doi:10.1021/ac504652e
Wu, C., Cansiz, S., Zhang, L., Teng, I.-T., Qiu, L., Li, J., et al. (2015). A nonenzymatic hairpin DNA cascade reaction provides high signal gain of mRNA imaging inside live cells. J. Am. Chem. Soc. 137, 4900–4903. doi:10.1021/jacs.5b00542
Wu, L., Wang, J., Feng, L., Ren, J., Wei, W., and Qu, X. (2012). Label-free ultrasensitive detection of human telomerase activity using porphyrin-functionalized graphene and electrochemiluminescence technique. Adv. Mat. 24, 2447–2452. doi:10.1002/adma.201200412
Wu, L., Wang, J., Ren, J., and Qu, X. (2014). Ultrasensitive telomerase activity detection in circulating tumor cells based on DNA metallization and sharp solid-state electrochemical techniques. Adv. Funct. Mat. 24, 2727–2733. doi:10.1002/adfm.201303818
Xiao, Y., Pavlov, V., Niazov, T., Dishon, A., Kotler, M., and Willner, I. (2004). Catalytic beacons for the detection of DNA and telomerase activity. J. Am. Chem. Soc. 126, 7430–7431. doi:10.1021/ja031875r
Yan, L., Hui, J., Liu, Y., Guo, Y., Liu, L., Ding, L., et al. (2016). A cascade amplification approach for visualization of telomerase activity in living cells. Biosens. Bioelectron. 86, 1017–1023. doi:10.1016/j.bios.2016.07.102
Yang, W., Zhu, X., Liu, Q., Lin, Z., Qiu, B., and Chen, G. (2011). Label-free detection of telomerase activity in HeLa cells using electrochemical impedance spectroscopy. Chem. Commun. 47, 3129–3131. doi:10.1039/c0cc05230b
Yin, P., Choi, H. M. T., Calvert, C. R., and Pierce, N. A. (2008). Programming biomolecular self-assembly pathways. Nature 451, 318–322. doi:10.1038/nature06451
Zhang, X., Lou, X., and Xia, F. (2017). Advances in the detection of telomerase activity using isothermal amplification. Theranostics 7, 1847–1862. doi:10.7150/thno.18930
Zhou, M., Liu, Y., Tu, Y., Tao, G., and Yan, J. (2012). DNAzyme-based turn-on chemiluminescence assays in homogenous media. Biosens. Bioelectron. 35, 489–492. doi:10.1016/j.bios.2012.03.013
Zhou, X., and Xing, D. (2012). Assays for human telomerase activity: Progress and prospects. Chem. Soc. Rev. 41, 4643–4656. doi:10.1039/c2cs35045a
Zhou, Z., Brennan, J. D., and Li, Y. (2020). A multi-component all-DNA biosensing system controlled by a DNAzyme. Angew. Chem. Int. Ed. Engl. 59, 10401–10405. doi:10.1002/anie.202002019
Zhu, J., Li, T., Hu, J., and Wang, E. (2010). A novel dot-blot DNAzyme-linked aptamer assay for protein detection. Anal. Bioanal. Chem. 397, 2923–2927. doi:10.1007/s00216-010-3802-9
Zhu, J., Li, T., Zhang, L., Dong, S., and Wang, E. (2011). G-quadruplex DNAzyme based molecular catalytic beacon for label-free colorimetric logic gates. Biomaterials 32, 7318–7324. doi:10.1016/j.biomaterials.2011.06.040
Zhu, J., Yang, X., Zhang, L., Zhang, L., Lou, B., Dong, S., et al. (2013a). A visible multi-digit DNA keypad lock based on split G-quadruplex DNAzyme and silver microspheres. Chem. Commun. 49, 5459–5461. doi:10.1039/c3cc42028k
Zhu, J., Zhang, L., Dong, S., and Wang, E. (2013b). Four-way junction-driven DNA strand displacement and its application in building majority logic circuit. ACS Nano 7, 10211–10217. doi:10.1021/nn4044854
Zhu, J., Zhang, L., Dong, S., and Wang, E. (2015). How to split a G-quadruplex for DNA detection: New insight into the formation of DNA split G-quadruplex. Chem. Sci. 6, 4822–4827. doi:10.1039/c5sc01287b
Zhu, J., Zhang, L., Li, T., Dong, S., and Wang, E. (2013c). Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mat. 25, 2440–2444. doi:10.1002/adma.201205360
Zhu, J., Zhang, L., and Wang, E. (2012). Measurement of the base number of DNA using a special calliper made of a split G-quadruplex. Chem. Commun. 48, 11990–11992. doi:10.1039/c2cc36693b
Zhu, X., Xu, H., Lin, R., Yang, G., Lin, Z., and Chen, G. (2014). Sensitive and portable detection of telomerase activity in HeLa cells using the personal glucose meter. Chem. Commun. 50, 7897–7899. doi:10.1039/c4cc03553d
Keywords: DNA circuit, guanine quadruplex, telomerase, biosensor, paper-based analytical device
Citation: Zhou Z, Gu J, Brennan JD and Li Y (2022) An all-deoxyribonucleic acid circuit for detection of human telomerase activity in solution and on paper. Front. Anal. Sci. 2:994394. doi: 10.3389/frans.2022.994394
Received: 14 July 2022; Accepted: 11 August 2022;
Published: 05 September 2022.
Edited by:
Ying Liu, Nanjing University, ChinaReviewed by:
Ruo-Can Qian, East China University of Science and Technology, ChinaCopyright © 2022 Zhou, Gu, Brennan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingfu Li, bGl5aW5nQG1jbWFzdGVyLmNh; John D. Brennan, YnJlbm5hbmpAbWNtYXN0ZXIuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.