
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Artif. Intell. , 28 November 2022
Sec. Medicine and Public Health
Volume 5 - 2022 | https://doi.org/10.3389/frai.2022.1046668
This article is a correction to:
DeepCarc: Deep Learning-Powered Carcinogenicity Prediction Using Model-Level Representation
A corrigendum on
DeepCarc: Deep learning-powered carcinogenicity prediction using model-level representation
by Li, T., Tong, W., Roberts, R., Liu, Z., and Thakkar, S. (2021). Front. Artif. Intell. 4:757780. doi: 10.3389/frai.2021.757780
In the published article, there was an error where the compounds were mismatched with the compound's prediction. A correction has been made to the Section “DeepCarc Is Employed to Screen DrugBank and Tox21 Compounds,” Paragraphs 1 and 2. The corrected section appears below:
“The DeepCarc was used as a screening tool for identifying the carcinogenicity potential of the compounds from DrugBank (Figure 5A). The predicted probabilistic values ranging from 0 to 1 were split into 10 intervals with a size of 0.1. Of 9,814 compounds, there were 7,555 (i.e., 7,555/9,814 = 76.98%), 920 (9.37%), 442 (4.50%), 277 (2.82%), 186 (1.90%) compounds with their predicted probabilities belong to the intervals of (0, 0.1), (0.1, 0.2), (0.2, 0.3), (0.3, 0.4), and (0.4, 0.5), respectively, indicating low carcinogenicity concern. In total, 434 compounds (4.42%) were predicted with probabilistic values ≥0.5, indicating compounds with carcinogenicity risk. Of 434 compounds, there were 26 compounds (0.26%) with the predicted probability ≥0.9, indicating high carcinogenicity concern. The predicted probabilistic value of each drug is included in Supplementary Table 4.
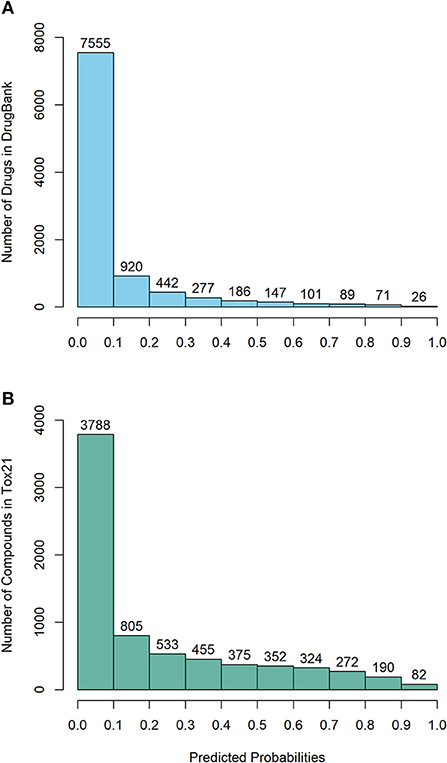
Figure 5. The probability distribution of the DeepCarc prediction of the compounds from (A) DrugBank; (B) Tox21.
The DeepCarc further screened the carcinogenicity potential of the compounds from the Tox21 (Figure 5B). Similarly, the predicted probabilistic values were separated into 10 intervals. Of the 7,176 compounds, there were 3,788 (i.e., 3,788/7,176 = 52.79%), 805 (11.22%), 533 (7.43%), 455 (6.34%), 375 (5.23%) compounds with their predicted probabilities belong to the intervals of (0, 0.1), (0.1, 0.2), (0.2, 0.3), (0.3, 0.4), and (0.4, 0.5), respectively, indicating low carcinogenicity concern. The other 1,220 (17.00%) compounds were predicted with probabilistic values ≥0.5, suggesting the compounds possessed carcinogenicity risk. There were 82 (1.14%) compounds with the predicted probabilistic value ≥0.9, suggesting high carcinogenicity concern (Supplementary Table 5).”
In the published article, there was also an error in Figure 5 as published. The probability distribution is incorrect due to a mismatched compounds' prediction. The corrected Figure 5 appears below.
In the published article, there was an error in Supplementary Tables 4, 5 where the compounds were mismatched with the prediction. These files have now been updated in the original article.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
We would like to express our great appreciation to Sunggun Lee, Dr. Kamel Mansouri, and Dr. Nicole Kleinstreuer from the National Institute of Environmental Health Sciences (NIEHS) for their great contribution to the correction.
The views presented in this article do not necessarily reflect current or future opinions or policies of the US Food and Drug Administration. Any mention of commercial products is for clarification and not intended as an endorsement.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: carcinogenicity, deep learning, QSAR, non-animal models, NCTRlcdb
Citation: Li T, Tong W, Roberts R, Liu Z and Thakkar S (2022) Corrigendum: DeepCarc: Deep learning-powered carcinogenicity prediction using model-level representation. Front. Artif. Intell. 5:1046668. doi: 10.3389/frai.2022.1046668
Received: 16 September 2022; Accepted: 07 October 2022;
Published: 28 November 2022.
Edited and reviewed by: Thomas Hartung, Johns Hopkins University, United States
Copyright © 2022 Li, Tong, Roberts, Liu and Thakkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhichao Liu, emhpY2hhby5saXVAZmRhLmhocy5nb3Y=; Shraddha Thakkar, c2hyYWRkaGEudGhha2thckBmZGEuaGhzLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.