- 1Department of Computational Mathematics, Science and Engineering, Michigan State University, East Lansing, MI, United States
- 2Department of Mathematics, Michigan State University, East Lansing, MI, United States
Many and varied methods currently exist for featurization, which is the process of mapping persistence diagrams to Euclidean space, with the goal of maximally preserving structure. However, and to our knowledge, there are presently no methodical comparisons of existing approaches, nor a standardized collection of test data sets. This paper provides a comparative study of several such methods. In particular, we review, evaluate, and compare the stable multi-scale kernel, persistence landscapes, persistence images, the ring of algebraic functions, template functions, and adaptive template systems. Using these approaches for feature extraction, we apply and compare popular machine learning methods on five data sets: MNIST, Shape retrieval of non-rigid 3D Human Models (SHREC14), extracts from the Protein Classification Benchmark Collection (Protein), MPEG7 shape matching, and HAM10000 skin lesion data set. These data sets are commonly used in the above methods for featurization, and we use them to evaluate predictive utility in real-world applications.
1 Introduction
Persistence diagrams are an increasingly useful shape descriptor from Topological Data Analysis. One of their more popular uses to date has been as features for machine learning tasks, with success in several applications to science and engineering. Though many methods and heuristics exist for performing learning with persistence diagrams, evaluating their relative merits is still largely unexplored. Our goal here is to contribute to the comparative analysis of machine learning methods with persistence diagrams.
Starting with topological descriptors of datasets, in the form of persistence diagrams, we provide examples and methodology to create features from these diagrams to be used in machine learning algorithms. We provide the necessary background and mathematical justification for six different methods (in chronological order): the Multi-Scale Kernel, Persistence Landscapes, Persistence Images, Adcock-Carlsson Coordinates, Template Systems, and Adaptive Template Systems. To thoroughly evaluate these methods, we have researched five different data sets and the relevant methods to compute persistence diagrams from them. The datasets, persistence diagrams and code to compute the persistence diagrams is readily available for academic use.
As part of this review, we also provide a user guide for these methods, including comparisons and evaluations across the different types of datasets. After computing the six types of features, we compared the predictive accuracy of a ridge regression, random forest, and support vector machine model to assess the type of featurization that is most useful in predictive models. The code developed for this analysis is available, with some functions developed specifically for use in machine learning applications, and easy-to-use jupyter notebooks showing examples of each function with multiple dataset types.
Of these methods, Persistence Landscapes, Adcock-Carlsson Coordinates, and Template Systems are quite accurate and create features for large datasets quickly. Adaptive Template Systems and Persistence Images took somewhat longer to run, however, the Adaptive Template Systems featurization method did improve accuracy over other methods. The Multi-Scale Kernel was the most computationally intensive, and during our evaluation we did not observe instances of it outperforming other methods.
2 Background
Algebraic topology is the branch of mathematics concerned with the study of shape in abstract spaces. Its main goal is to quantify the presence of features which are invariant under continuous deformations; these include properties like the number of connected components in the space, the existence of holes and whether or not the space is orientable. As an example, Figure 1 shows two spaces: the 2-dimensional sphere on the left, which is the set
The aforementioned properties of shape for these spaces are as follows. Both
The homology of a space is one way in which topologists have formalized measuring the presence of n-dimensional holes in a space (Hatcher, 2002). Indeed, for a space X (e.g., like the sphere or the Möbius band) an integer
where, again, the dimension of
2.1 Persistent Homology
There are several learning tasks where each point in a data set has shape or geometric information relevant to the problem at hand. Indeed, in shape retrieval, database elements are often 3D meshes discretizing physical objects, and the ensuing learning tasks are often related to pose-invariant classification (Pickup et al., 2014). In computational chemistry and drug design, databases of chemical compounds are mined in order to discover new targets with desirable functional properties. In this case, the shape of each molecule (i.e., of the collection of comprising atoms) is closely related to molecular function, and thus shape features can be useful in said data analysis tasks (Bai et al., 2009).
If homology is what topologists use to measure the shape of abstract spaces, then persistent homology is how the shape of a geometric data set can be quantified (Perea, 2019). Persistent homology takes as input an increasing sequence of spaces
Any such sequence is called a filtration. The definition of persistent homology relies on two facts: first, that one can compute homology for each space separately, i.e.,
of vector spaces and induced linear transformations.
The evolution of features in
See section VII.1 of Edelsbrunner and Harer (2010) for more details. In other words,
2.1.1 Filtrations From Point Cloud Data
There are several ways of constructing filtrations from geometric data. Indeed, let X be a set and
where
for
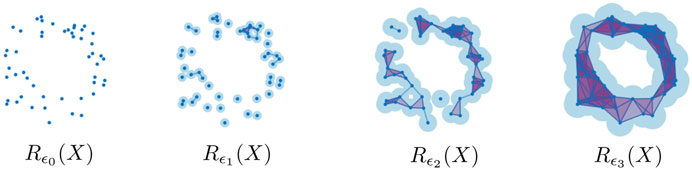
FIGURE 2. The Rips complex on a point cloud
The persistent homology of the Vietoris-Rips filtration, i.e.,
often encode features such as density and curvature, in addition to the presence of holes and voids (Bubenik et al., 2020). Figure 3 shows the Vietoris-Rips persistence diagrams in dimensions
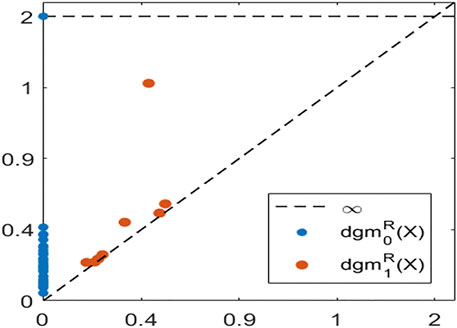
FIGURE 3. The Rips persistence diagrams in dimensions 0 (blue) and 1 (orange), for a point cloud sampled around the unit circle.
2.1.2 Filtrations From Scalar Functions and Image Data
If
define the so called sublevel set filtration of
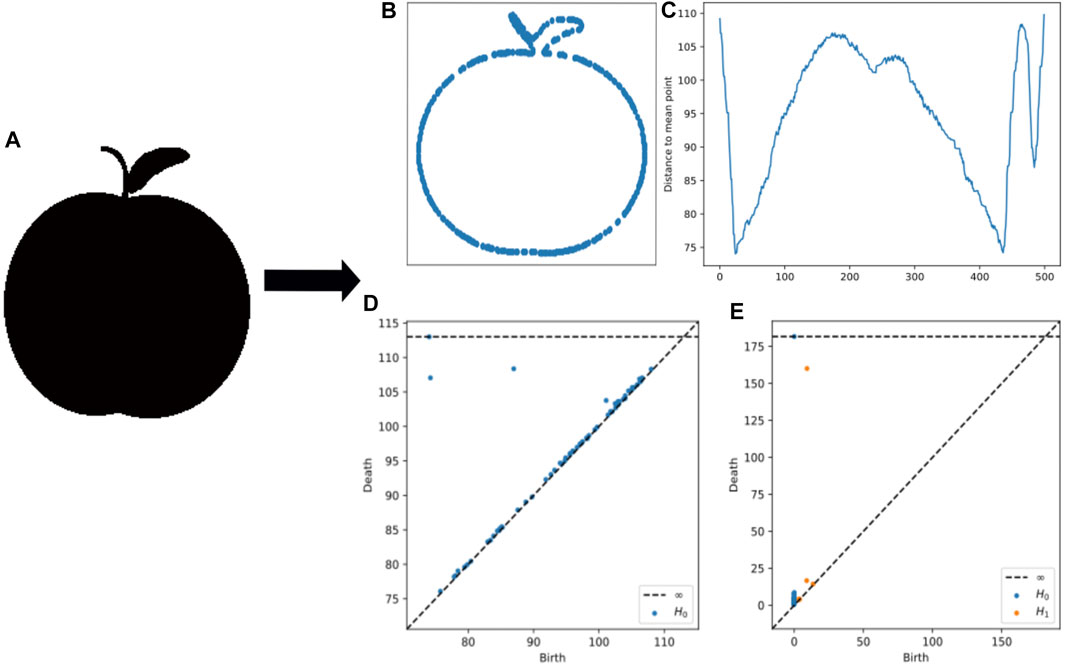
FIGURE 4. (A) An example apple from MPEG7 data. (B) An example image contour used for MPEG7. (C) The distance to mean point calculation used for sublevel set persistence (D) Persistence diagrams from lower star persistence (E) Persistence diagrams from the contour.
Images provide another data modality where sublevel set persistence can be useful. Indeed, an image can be thought of as a function on a grid of pixels; if the image is in grey scale, then we have a single scalar valued function, and if the image is multi-channel (like RGB color representations) then each channel is analyzed independently. The grid yields a triangulated space via a Freudenthal triangulation of the plane, and the values of pixel intensity in each channel can be extended via convex combinations to the faces [see Lemma 1 of Kuhn (1960)]. We will apply this methodology later on to the MNIST hand written digit data base (Figure 5). This approach to computing persistent homology from images is not unique in the literature; other popular methods such as cubical homology (Kaczynski et al., 2004) have been used for this same purpose. This work, however, deals exclusively with simplicial homology as it is the standard approach in many applications.
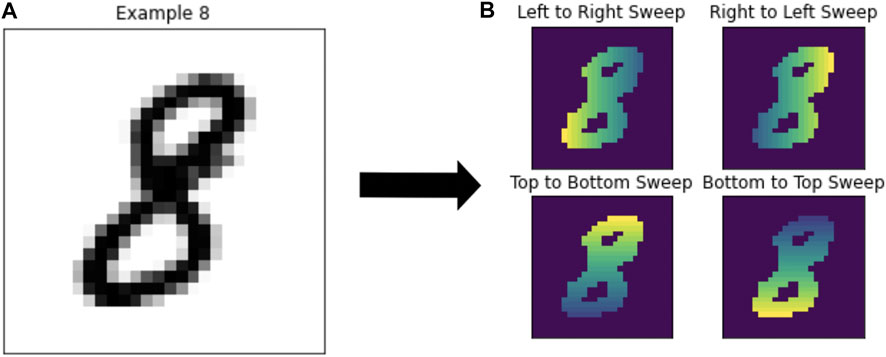
FIGURE 5. (A) Example number 8 from the MNIST dataset. (B) The same number 8 after computing each of the four types of coordinate transforms to compute the persistence diagrams.
2.2 The Space of Persistence Diagrams
Persistence diagrams have shown to be a powerful tool for quantifying shape in geometric data (Carlsson, 2014). Moreover, one of their key properties is their stability with respect to perturbations in the input, which is crucial when dealing with noisy measurements. Indeed, two persistence diagrams D and
If
The bottleneck distance
for every integer
where
In order to develop the mathematical foundations needed for doing machine learning with persistence diagrams, it has been informative to first study the structure of the space they form. Indeed, if
where the infimum runs over all multiset bijections
and that
Doing statistics and machine learning directly on the space of persistence diagrams turns out to be quite difficult. Indeed,
3 Featurization Methods
For each of the methods below, we start with a collection of persistence diagrams. A persistence diagram can be represented in either the birth-death plane or birth-lifetime plane—some methods will require birth-death coordinates and others will require birth-lifetime coordinates. The birth-death plane is the representation pair
3.1 Multi-Scale Kernel
The Multi-Scale Kernel of Reininghaus et al. (2015) defines a Kernel over the space of persistence diagrams, which can then be used in various types of kernel learning methods. In general, a kernel k is by definition a symmetric and positive definite function of two variables. Mathematically, from Reininghaus et al. (2015), given a set X, a function
Reininghaus et al. (2015) propose a multi-scale kernel on
where
where if
The multi-scale kernel is shown to be stable w.r.t the 1-Wasserstein distance by Theorem 2 of Reininghaus et al. (2015), which is a desirable property for classification algorithms. However, by Theorem 3 of Reininghaus et al. (2015), the multi-scale kernel is not stable in the Wasserstein sense for
3.2 Persistence Landscapes
Persistence landscapes are a mapping of persistence diagrams into a function space that is either a Banach space or Hilbert space (Bubenik, 2020). Advantages of persistence landscapes are that they are invertible, stable, parameter-free, and nonlinear. Persistence landscapes can be computed from a persistence diagram as follows.
From Bubenik (2020), for a persistence diagram
and
with kmax as the kth largest element.
The persistence landscape is the sequence of piecewise linear functions,
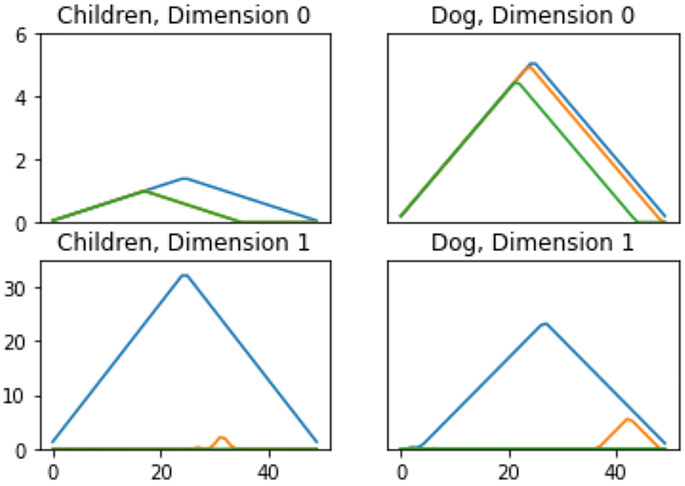
FIGURE 6. Persistence Landscapes from the MPEG7 dataset to show differences in features. Each color corresponds to a different landscape, i.e.,
3.3 Persistence Images
From Adams et al. (2015), persistence images are a mapping sending a persistence diagram to an integrable function, called a persistence surface. Fixing a grid on
More explicitly, let D be a persistence diagram in birth-lifetime coordinates. We take
The persistence surface
with
3.4 Adcock-Carlsson Coordinates: The Ring of Algebraic Functions on Persistence Diagrams
This method is explored by Adcock et al. (2016) where the authors highlight the fact that any persistence diagram with exactly n points can be described by a vector of the form
The inclusions
which provide the basis for studying algebraic functions on the space of persistence diagrams.
With this setting in mind, the main goal of Adcock et al. (2016) is to determine free generating sets for the subalgebra of
Theorem 1The subalgebra of 2-multisymmetric functions invariant under adding points with zero persistence, is freely generated over
for integers
3.5 Template Systems
The goal of this method is to find features for persistence diagrams by finding dense subsets of
The function
This injective featurization allows us to define a template system for
The advantage of working with these template systems is that they can be used to approximate real-valued functions on the space of persistence diagrams as proven by the following theorem [see Theorem 29 Perea et al. (2019)].
Theorem 2Let
for every
where
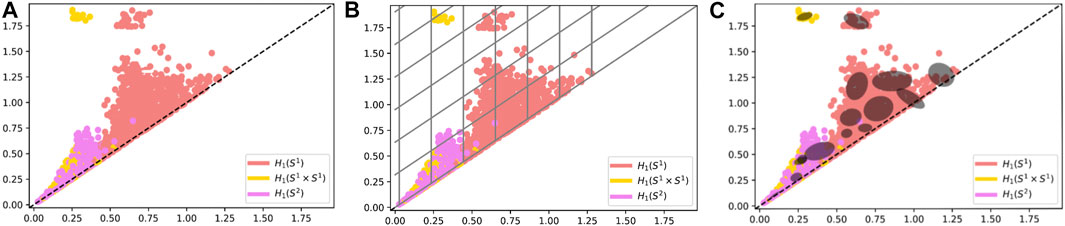
FIGURE 8. (A) Persistence Diagrams (B) Persistence Diagrams with boundaries of the support function for Tent Features (C) Persistence Diagrams with boundaries of the support for Adaptive Templates.
3.6 Adaptive Template Systems
The Adaptive Template Systems methodology of Polanco and Perea (2019a) concerns itself with improving and furthering some of the work presented in Perea et al. (2019). The goal is to produce template systems that are attuned or adaptive to the input data set and the supervised classification problem at hand. One shortcoming of template systems, like tent functions, when applied to Theorem 2 is that without prior knowledge about the compact set
The relationship between template systems and adaptive template systems is demonstrated in Figure 4, showing the adaptive template systems depend on density of data. To do so, given a compact set
Once this family of ellipsoidal domains is computed, we use them to define the following adaptive template functions
3.7 Other Approaches
The featurization methods presented in this section are by no means an exhaustive list of what is available in the literature. Here are some others that the interested reader may find useful:
• The Persistent homology rank functions of Robins and Turner (2016) are similar in spirit to persistent landscapes, in that they provide an injective inclusion of
This is equivalent, for a persistence diagram
where
This approach has shown to be effective in analyzing point processes, and sphere packing patterns.
The Persistent curve (Chung et al., 2018; Giusti et al., 2015) provides another functional summary closely related to persistent rank functions. Specifically, for a persistence diagram
Discretizations of these curves have been useful in computer vision tasks (Chung and Lawson, 2020), as well as in neuroscience applications (Giusti et al., 2015).
Other kernel methods, besides the Multi-Scale kernel of Reininghaus et al. (2015), have appeared in the literature. They correspond to the following choices of kernel function
for parameters
where
Persistence diagrams as features for deep neural networks have also been studied recently. In particular, the PersLay framework of Carrière et al. (2020) leverages the Deep Sets architecture of Zaheer et al. (2017) to implement layers that can process persistence diagrams. Specifically, layers of the form:
where
4 Datasets
The five different datasets considered in this work were chosen from a collection of experiments presented in the literature of topological methods for machine learning. We acknowledge that this selection is inherently bias towards datasets with favorable performance with regards to specific topological methods. Nevertheless, we counterbalance this by applying all the evaluated featurization methods to all the data sets here considered and compare the classification results across all the presented methodologies. This comparative work showcases how the variation between methods results in the need for the user to find suitable combination of featurization methods and parameter tuning to obtain optimal results in a given dataset. As such, readers should view this as a resource for their own analysis, and not as a recommendation for specific techniques.
For all datasets and methods, parameter tuning was done using a grid search method on a subset of data that was not used to report final results, and parameters were chosen based on performance of a ridge regression model, random forest and support-vector machine (SVM) model. It is worth noting a weakness of the analysis in that the same parameters were used in the feature set calculation for all reported models, and run with a single split. This was due to time required for feature calculation.
The ridge regression and random forest classifier were run with default parameters, and the support-vector machine was run using the radial basis function (RBF) with some tuning on the cost parameter (C). The exception is for the Multi-Scale Kernel feature set—we only fit a support-vector machine model. It is important to highlight that results regarding ridge regression with (polynomial and radial basis function) kernel methods are not included in this work as they produce increased computational times while the classification results do not improve significantly compared to the one presented here. Each dataset was sampled for a 10 or 100 trials depending on size, with the exception of the Protein Classification Dataset, which included indices for predefined problems.
Random forest classifiers as presented in Breiman (2001) are used to solve the same classification problems presented for each data set. Parameters such as number of trees in each forest and the size of each tree are chosen based on performance and tuned on the testing set.
4.1 MNIST
The MNIST dataset from LeCun and Cortes (1999) is a database of 70,000 handwritten single digits of numbers zero to nine. An example image from the MNIST database is shown in Figure 7.
The calculation of persistence diagrams for the MNIST dataset is as in Adcock et al. (2016). This method creates a base of 8 different persistence diagrams to use in the application of methods. The persistence diagrams are calculated using a “sweeping” motion in one of four directions: left to right, right to left, top to bottom, or bottom to top, corresponding to the 0-dimensional and 1-dimensional persistence diagrams. To compute this filtration, pixels are converted to a binary response with intensity calculated based on position. This has the effect that depending on the direction of sweep, features will have different birth and death times, providing distinct information for each direction. The number of topological features available for model fitting is dependent on the method. For the Persistent Images, Persistence Landscapes, and Template Systems there are eight features each. The Multi-Scale Kernel produces eight different kernel matrices, and for Adcock-Carlsson Coordinates, 32 different features were computed from these persistence diagrams.
Figure 5 shows the various calculations of persistence diagrams for an example number eight. Both 0-dimensional and 1-dimensional persistence diagrams were used for the MNIST dataset, noting that some observations did not have 1-dimensional persistence diagrams, so these observations were filled with a single diagram of birth-death coordinate of [0,.01].
For the MNIST dataset, a random sample of 1,000 images was used to tune parameters, with 80% used for the training portion, and 20% used for the testing portion. We used the set of 60,000 images corresponding to the training set of MNIST to create our own training and testing sets for model fitting and evaluation. For this set of 60,000, 10 trials were run with an 80% training and 20% testing split to determine model performance.
4.2 SHREC14
We evaluated the SHREC 2014 dataset (Pickup et al., 2014) in the same manner as the authors of Polanco and Perea (2019a). To compute the topological features, the authors of Reininghaus et al. (2015) describe using a heat kernel signature to compute persistence diagrams for both the 0-dimensional and 1-dimensional persistence diagrams. The dataset consists of 15 different labels, corresponding to five different poses for the three classes of male, female, and child figures.
As noted in Polanco and Perea (2019a), parameters in the dataset define different problems due to a different calculation of the heat kernel signature, and for this evaluation we focused on the problem with the highest accuracy as reported in Polanco and Perea (2019a).
For the SHREC14 dataset, a random sample of 90 images (30% of the data) was used to tune the model and determine appropriate model parameters. The remaining 210 observations were split into 80% training and 20% testing for 100 trials to report final model fit. Persistence diagrams for 0-dimensional homology and 1-dimensional homology were computed for this dataset.
Table 1 shows complete results for the SHREC 2014 dataset.
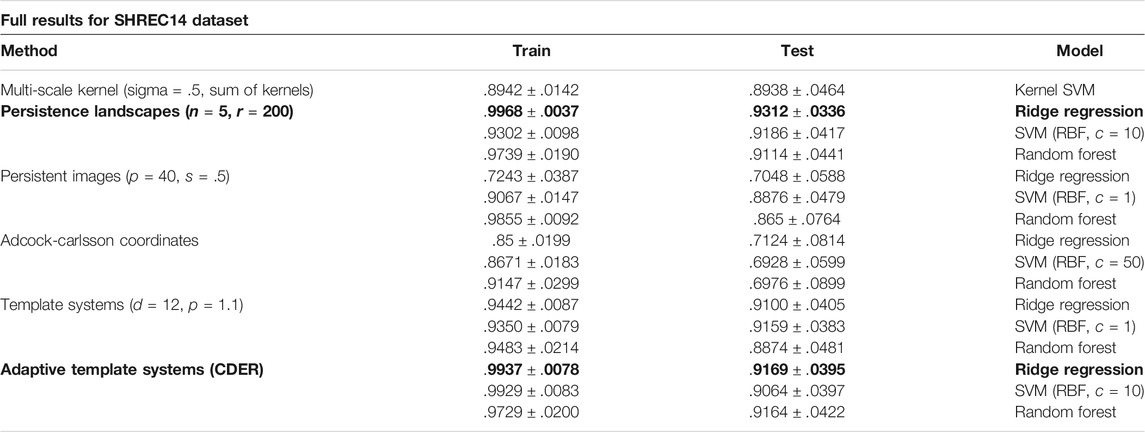
TABLE 1. Results from the Shrec14 Dataset using the average model classification accuracy ± standard deviation over 100 trials.
4.3 Protein Classification
We use the Protein Classification Benchmark dataset PCB00019 Sonego et al. (2007) as another type of data to evaluate the topological methods above. This specific set contains information for 1,357 proteins corresponding to 55 classification problems, and we reported on 54 of the problems using one to tune parameters. The training and testing index were provided, and the mean accuracy was reported for both training and testing sets using these indices. Table 2 shows results from our experiments using the training and testing indices provided in the original dataset.
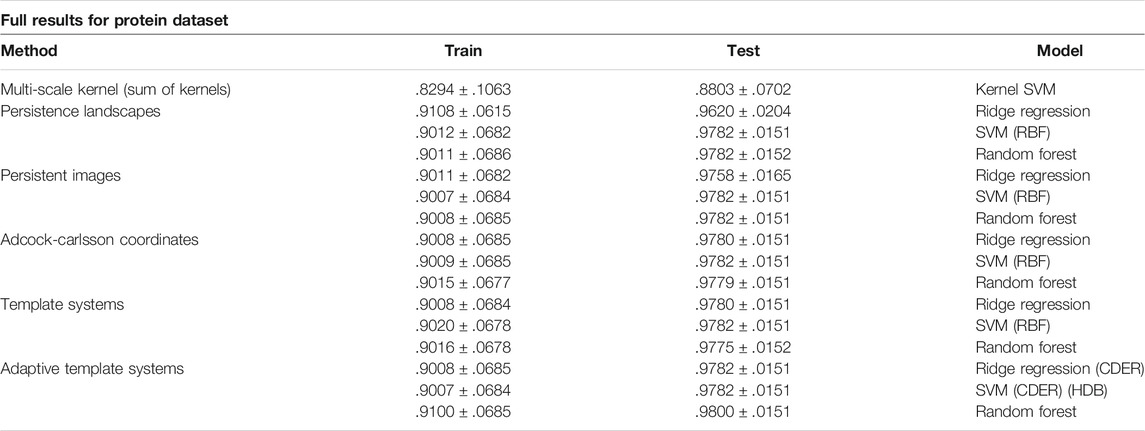
TABLE 2. Results from the Protein Dataset using the average model classification accuracy ± standard deviation over 54 trials corresponding to the predefined indices of the dataset.
Persistence diagrams for this dataset were computed for each protein by considering the 3-D structure [provided in wwPDB consortium (2018)] as a point cloud in
4.4 MPEG7
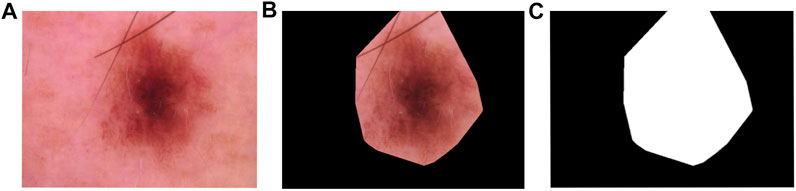
The mpeg-7 dataset from Bai et al. (2009) is a database of object shapes in black and white, with 1,400 shapes in 70 classes. An example from the original dataset is shown in Figure 4 along with the contour as described below.
To compute persistence diagrams, first the image contour is computed by placing observations from the point cloud into a sequence. The distance curve is computed as the distance from the center of the sequence. Sublevel set persistence is taken using the computed distance curves as point cloud data. Persistence diagrams for both 0-dimensional and 1-dimensional homology were computed for this dataset.
We used this dataset for a timing comparison of featurization methods from persistence diagrams. We do not report on the results of this dataset. An example notebook of MPEG7 is provided using only four shapes—apple, children, dog, and bone. This approach is due to the initial difficulty in getting accurate models for the full dataset. Due to the small number of samples (80 total) and lack of repeated sampling, the estimates provided for this dataset are not stable and are not reported.
4.5 HAM10000
The HAM10000 dataset provided by Philipp Tschandl et al. (2018) is a collection of 10,000 images of skin lesions with one 7 potential classifications: Actinic Keratoses and Intraepithelial Carcionma, Basal cell carcinoma, Benign keratosis, Dermatofibroma, Melanocytic nevi, Melanoma, Vascular skin lesions. A total of 18 persistence diagrams for this set were calculated using the methods outlined in Chung et al. (2018), 9 corresponding to the 0-dimensional homology and 9 corresponding to the 1-dimensional homology.
To obtain such diagrams, first a mask is computed by implementing the methodology proposed in Chung et al. (2018). In general terms, this method creates a filtration of binary images obtained from different thresholds to convert the gray scale image into a binary one. Once this binary filtration is obtained, the center most region of the image is computed using the “persistence” of each point in the binary filtration. An example image and this process is shown in Figure 9.
Once the mask is computed it is applied to the original image and then it is converted into three different color models: RGB, HSV , and XYZ. Each color model is split into their corresponding channels, and for each channel we use sublevel set filtration to obtain 0-dimensional and 1-dimensional persistence diagrams. In total, for each image on the data set we obtain 18 persistence diagrams, 9 in homological dimension 0 and 9 for homological dimension 1.
To tune the models, a random sample of 250 images were taken a ridge regression, random forest, and support vector machine model were fit to determine parameters. The remaining 9,750 images were split into an 80% training and 20% testing set to report final results.
To evaluate the HAM10000 dataset, due to the large number of birth and death pairs in each persistence diagram, subsampling of persistent features was required. Each observation in a data set, for example an image, will yield 18 persistence diagrams corresponding to homological features in that observation. In the HAM10000 dataset, there was an average of 5,783 birth-death pairs in each persistence diagram. This was an issue to complete computation for the vectorization methods, even for adaptive templates, so each persistence diagram was subsampled as follows.
The method of subsampling is two steps: Highly persistent features were always included, and a uniform random sampling method (without replacement) was used to sample the remaining points. The threshold for feature lifetime and number of points to sample was determined by using parameters that preserve the distribution of points in each persistence diagram. As a result, features in each persistence diagram with a lifetime of five or more were automatically included, and 5% of the rest of the points were also included. This resulted in sampled persistence diagrams with an average of 290 points each (Table 3).
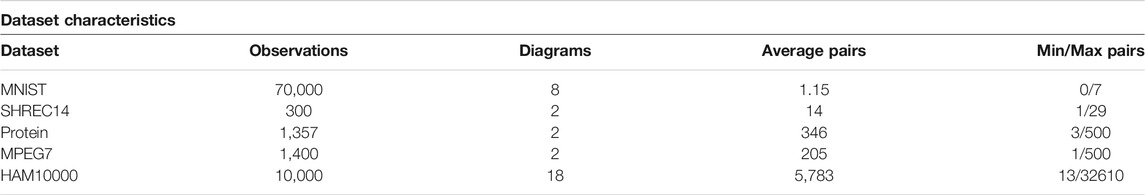
TABLE 3. Characteristics of each dataset. The column headings can be explained as such: Observations—number of observations in the dataset, Diagrams—the number of homological types used to compute persistence diagrams, Average Pairs—the average number of birth/death pairs across the set of persistence diagrams for a single observation in the original dataset, and Min/Max Pairs—the minimum and maximum number of birth/death pairs across the set of persistence diagrams for a single observation in the original dataset.
5 User Guide
5.1 Available Functions
As part of the available code, a function for each method is included. Each function requires two sets of persistence diagrams, a training set and a testing set, and parameters specific to the function. The function returns two feature sets for that method, corresponding to the training and test set respectively. Each function also prints the time in seconds taken at the end of each run. In this section of the user guide each function is described, along with the required parameters for the function.
The Multi-Scale Kernel feature matrix can be computed using the function kernel_features or fast_kernel_features. It is recommended to use fast_kernel_features due to computation time. Both functions require a parameter sigma, denoted as s in the function with a default value of 4. In Reininghaus et al. (2015) this parameter is referred to as the scale parameter. From the closed form distribution of the Multi-Scale Kernel
we note that as sigma, σ, increases the function decreases. Increasing sigma results in a less diffuse kernel matrix, while decreasing sigma results in a more diffuse kernel matrix.
Due to time required for the Multi-Scale Kernel, there are two additional sets of functions that use Numba (Siu Kwan Lam and Seibert, 2015) for significantly faster computation. In the current implementation, these are not able to be combined with multi-core processing (MPI for example), and have a different format than the other functions included. These functions are provided in the github repository for this project, and were used to compute results for the Multi-Scale Kernel for the MNIST dataset.
The Persistence Landscapes features can be computed using the function landscape_features. The Multi-Scale Kernel function, landscape_features requires two parameters: the number of landscapes, n and resolution, r. The number of landscapes parameter, n, controls the number of landscapes used, and the resolution, r, controls the number of samples used to compute the landscapes. The default parameters for n is 5 and r is 100.
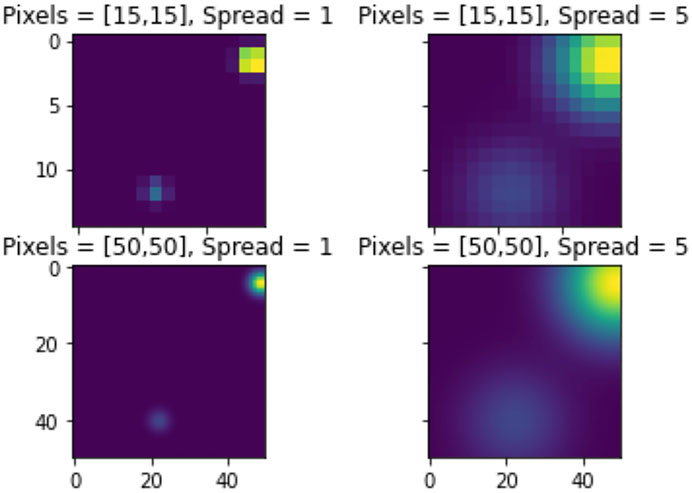
The Persistence Images can be computed using the function persistence_image_features. The persistence_image_features function requires two parameters, pixels and spread. The pixels, p is a pair that defines the number of pixels along each axis. The spread, s, is the standard deviation of the gaussian kernel used to generate the persistence image. It is worth noting that the implementation here uses the gaussian kernel, however, other distributions could be chosen so that s would correspond to parameters specific to the chosen distribution. Additionally, the weighting function is constant for this implementation. Increasing spread increases the variance for each distribution, resulting in larger “hot spots”. Increasing pixels provides a smoother distribution, whereas decreasing pixels yields a less smooth distribution. Note that increasing pixels increases computation time. This is demonstrated in Figure 7 in the methods section.
The Adcock-Carlsson Coordinates features can be computed using the function carlsson_coordinates, does not require any parameters. This function returns four different features for every type of persistence diagram provided. So for datasets that have persistence diagrams corresponding to 0-dimensional and 1-dimensional homology, 8 features are returned for machine learning. The features returned correspond to the four coordinates calculated in Adcock et al. (2016), and are:
The Template Systems features can be computed using the function tent_features, and has a choice of two parameters: d, which defines the number of bins in each axis and padding, which controls the padding around the collection of persistence diagrams. This function returns a training and testing set. This function computes the tent features from Perea et al. (2019).
The Adaptive Template Systems features can be called with the function adaptive_features, and requires the labels for the training set. Users can choose three different types of Adaptive Templates: Gaussian Mixture Modeling (GMM), Cover-Tree Entropy Reduction (CDER), and Hierarchical density-based clustering of applications with noise (HDBSCAN). The parameter d refers to the number of components when using the GMM model type. This would be minimally the number of classes in your data, and ideally represents closer to the number of distributions in the data that correspond to each observation. Details on these methods can be found in Polanco and Perea (2019a), as well as the original references linked in the methods section. During this evaluation, we evaluated adaptive templates using both GMM and CDER methods, but did not formally evaluate HDBSCAN. HDBSCAN was difficult to formally assess as we had difficulty with completion of the algorithm for some datasets. For those datasets we were able to complete, we did not notice an improvement over other adaptive methods.
6 Results
One consideration we must make before analysing the results comes from the computation of Multi-Scale Kernel features. As explained for each dataset in Section 4, more often than not we will compute multiple persistent diagrams per data point in a given data set. Such persistent diagram correspond to 0-dimensional, 1-dimensional, and in some cases 2-dimensional persistent homology (see details in Section 2). To compute Multi-Scale Kernel as given by Eq. 6 we require pairs of persistent diagrams. Since this multi-scale kernel provides a notion of similarity between persistent diagrams (Reininghaus et al., 2015) we require it to be computed between diagrams corresponding to the same dimension homology and method type. For example, the kernel matrix that corresponds to the 0-dimensional homology of a data set is computed using the persistence scale space kernel between two sets of persistence diagrams that represent the 0-dimensional homology. This means that for a dataset that has sets of 0-dimensional homology persistence diagrams and 1-dimensional homology persistence diagrams, two kernel matrices were returned (one per each dimension).
The kernel matrix used in our models is the sum of available kernels, and differs based on the persistence diagrams available for each dataset. While this does improve accuracy significantly over individual kernel matrices, other methods of combining kernel features were not explored in this paper, but is available in Gönen and Alpaydin (2011) for the interested reader. The available parameter, sigma, is consistent across all types of diagrams for our evaluation.
For each of the other methods, Persistence Landscapes, Persistence Images, Adcock-Carlsson Coordinates, Template Systems, and Adaptive Template Systems, each feature matrix was constructed for the relevant set of diagrams, and all topological types were used in fitting the same model.
The datasets used in this analysis were of varying size, both in terms of observations and the size of sets of persistence diagrams. As noted in the descriptions of data, the types of persistence diagrams calculated also differs. A summary of characteristics for each dataset is included in Table 3.
6.1 MNIST
The Multi-Scale Kernel features calculated yielded eight different kernel matrices, and the final kernel matrix was calculated using the unweighted summation of these kernels as in Gönen and Alpaydin (2011). Due to the time needed for computation of the Multi-Scale Kernel, a smaller set of 12,000 observations was used to report final results and a version of the kernel computation using Numba with a gpu target was necessary.
Table 4 shows complete results for the MNIST analysis. Four different methods (highlighted on the table) provided similar results for the MNIST dataset, and we note the SVM model had higher accuracy in each case. This table, and all subsequent results tables, include the method used to construct topological features, training and test accuracy, and model and parameters used for evaluation.
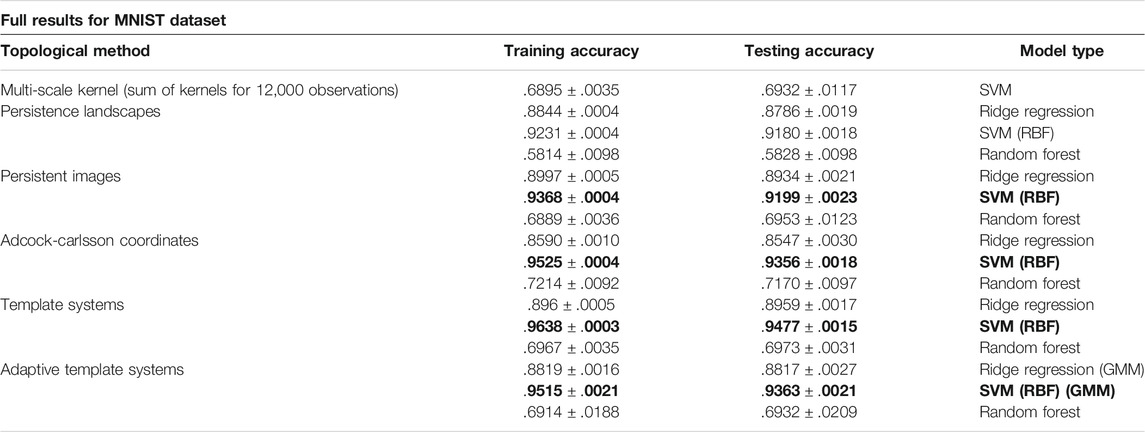
TABLE 4. Results from the MNIST Dataset using the average model classification accuracy ± standard deviation over 10 trials.
6.2 SHREC14
Results are reported in Table 1. Adaptive Template Systems and Persistence Landscapes were the two methods with highest classification accuracy on the test dataset, with Template Systems and the Multi-Scale Kernel performing nearly as well.
6.3 Protein Classification
Nearly all of the topological methods in this paper provided similar classification accuracy for this dataset. We observe the testing accuracy as higher than the training accuracy for this dataset, and the results are similar to those in Polanco and Perea (2019b). The Multi-Scale Kernel though did not perform as well and as shown in Figure 10 is the most computationally intensive. Results are reported in Table 2.
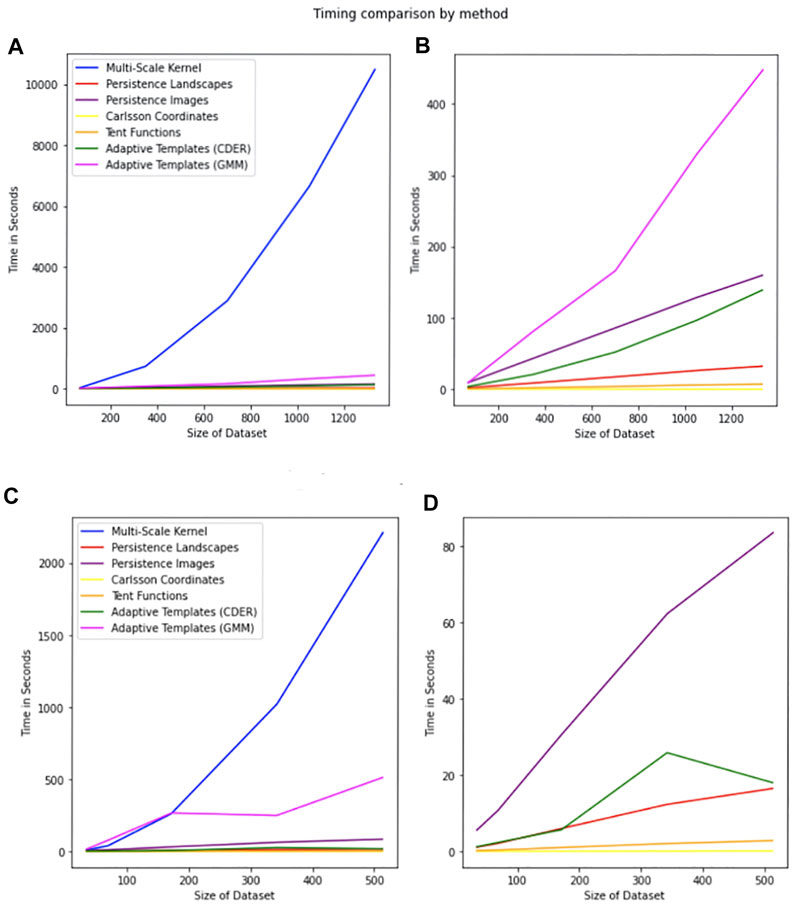
FIGURE 10. Comparison of timing by method. The legend is the same for all plots. The x-axis represents the size of the dataset, and the y-axis represents the time in seconds required for calculation of all of the persistence diagrams associated with the dataset of the given size. (A) Timings for the MPEG7 Dataset including the Multi-Scale Kernel. (B) Timings for the MPEG7 Dataset excluding the Multi-Scale Kernel. (C) Timings for the Protein Dataset including all features. (D) Timings for the Protein Dataset excluding Multi-Scale Kernel and Adaptive Templates (GMM).
6.4 HAM10000
Due to run time for the large number of points in each persistence diagram, even after subsampling, results were not reported for the Multi-Scale Kernel or Adaptive Template Systems.
Results are listed in Table 5. The HAM10000 dataset presented the largest computational challenge during this review, and is a continued area of research.
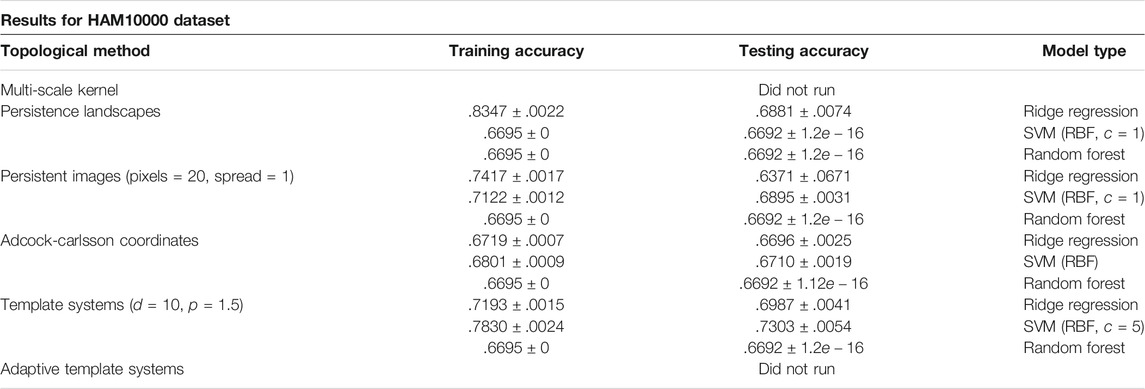
TABLE 5. Results from the HAM10000 Dataset using the average model classification accuracy ± standard deviation over 10 trials.
7 Computation Time of Features
Formal timings were captured for all features for the 0-dimensional persistence diagrams for the MPEG7 and Protein Datasets. A comparison of timings is in Figure 10. The timing reported is for the generation of features from one type of persistence diagram for a dataset of that size. This means when computing a training feature set and testing feature set for multiple types of persistence diagrams, the expected time to generate features can be significantly longer. For example, in the MNIST dataset we compute four different types of persistence diagrams with both 0-dimensional and 1-dimensional homology, giving eight sets of features that can be generated for the sets of persistence diagrams for that dataset. Specific to the multi-scale kernel method, the timing reported is for a symmetric feature matrix that is
Additionally, during the review of these methods, we did not encounter significant issues with model fitting, hence formal timings were not recorded for this portion of the analysis. Conclusions from these timings are addressed in the discussion section.
7.1 Data Availability
The datasets, persistence diagrams (or code to compute the diagrams), and all other associated code for this study can be found in the machine learning methods for persistence diagrams github repository https://github.com/barnesd8/machine_learning_for_persistence.
For each of the five datasets, the following code is available:
• A jupyter notebook that loads and formats the persistence diagrams including images and does a preliminary model fitting on a subset of the data
• A python script that calculates the persistence diagrams from the original dataset - some of these are written using MPI depending on the size of the dataset
• A python script that fits models for random samples of the data to get mean estimates of accuracy for both the training and test dataset
These scripts reference modules included in the github repository, including a persistence methods script that calculates features from persistence diagrams for a training and test dataset. This uses a combination of other available functions and functions written specifically for this paper.
The Template Systems and Adaptive Template Systems methods use functions from https://github.com/lucho8908/adaptive_template_systems, which is the corresponding code repository for Polanco and Perea (2019a). The available methods in our extension include Tent Functions and Adaptive Template Functions (GMM, CDER, and HDBScan methods).
The Adcock-Carlsson Coordinates method is a function developed specifically for this paper, and includes the calculation of the 4 different features used in our analysis. The Persistence Landscape method uses the persistence landscape calculation from the Gudhi Package Dlotko (2021). The Multi-Scale Kernel Method has two included implementations, one is from Nathaniel Saul (2019) and is slower to compute, while the other is a faster implementation that can be used on larger datasets. All of the results in this paper were reported using the implementation written specifically for this paper. The Persistence Images features are also from Nathaniel Saul (2019). Additionally, many functions from Pedregosa et al. (2011) are used throughout.
8 Discussion
Adcock-Carlsson Coordinates, Tent Functions, and Persistence Landscapes scale well, and perform well even for large datasets. It is of note though that parameter choice will affect computation time. This was especially notable in the Template Features (Tent Functions) computation time. As the number of tent functions is increased, the time to compute features also increases. We observed a superlinear increase, however, even with this increase computation time was not a barrier for analysis.
Persistence Images and Adaptive Template Functions do not scale or perform as well, however, do provide good featurizations for accurate models and should be considered depending on the dataset. Specifically, the Adaptive Template Functions was not completed for the full HAM10000 dataset due to computation time.
When using these methods, it should be of note that the Multi-Scale Kernel method is computationally intensive, and does not scale well. Additionally, the accuracy achieved is not better than other methods for the datasets in this paper.
9 Conclusion
This paper reviews six methods for topological feature extraction for use in machine-learning algorithms. Persistence Landscapes, Adcock-Carlsson Coordinates, Template Systems, and Adaptive Template Systems perform consistently with minimal differences between datasets and types of persistence diagrams. These methods are also less expensive in terms of execution time. A main contribution of this paper is the availability of datasets, persistence diagrams, and code for others to use and contribute to the research community.
Author Contributions
JP coordinated the experimental framework and co-wrote the paper with significant contributions to the background sections. LP curated datasets, contributed to the software code and co-wrote the paper with significant contributions to the methods sections. DB curated datasets, ran the experimental results, developed the accompanying software and co-wrote the paper with significant contributions in most areas. All authors reviewed results and the manuscript, with DB approving and reviewing the final version.
Funding
This work was partially supported by the National Science Foundation through grants CCF-2006661, and CAREER award DMS-1943758.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was supported in part by Michigan State University through computational resources provided by the Institute for Cyber-Enabled Research. Special thanks to Elizabeth Munch for providing background information for computing persistence diagrams using a directional transform. We also thank the reviewers for critical feedback that helped improved our review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frai.2021.681174/full#supplementary-material
References
Adams, H., Chepushtanova, S., Emerson, T., Hanson, E., Kirby, M., Motta, F., et al. (2015). Persistence Images: A Stable Vector Representation of Persistent Homology. [Dataset]. http://arxiv.org/abs/1507.06217.
Adcock, A., Carlsson, E., and Carlsson, G. (2016). The Ring of Algebraic Functions on Persistence Bar Codes. Homology, Homotopy Appl. 18, 381–402. doi:10.4310/hha.2016.v18.n1.a21
Bai, X., Yang, X., Latecki, L. J., Liu, W., and Tu, Z. (2009). Learning Context Sensitive Shape Similarity by Graph Transduction. IEEE Trans. Pattern Anal. Mach. Intell. 32, 861–74. doi:10.1109/TPAMI.2009.85
Bendich, P., Marron, J. S., Miller, E., Pieloch, A., and Skwerer, S. (2016). Persistent Homology Analysis of Brain Artery Trees. Ann. Appl. Stat. 10, 198–218. doi:10.1214/15-AOAS886
Blumberg, A. J., Gal, I., Mandell, M. A., and Pancia, M. (2014). Robust Statistics, Hypothesis Testing, and Confidence Intervals for Persistent Homology on Metric Measure Spaces. Found. Comput. Math. 14, 745–789. doi:10.1007/s10208-014-9201-4
Bubenik, P., Hull, M., Patel, D., and Whittle, B. (2020). Persistent Homology Detects Curvature. Inverse Probl. 36, 025008. doi:10.1088/1361-6420/ab4ac0
Bubenik, P. (2020). The Persistence Landscape and Some of its Properties. Topological Data Anal. 15, 77–102. doi:10.1007/978-3-030-43408-3_4
Bubenik, P., and Wagner, A. (2020). Embeddings of Persistence Diagrams into hilbert Spaces. J. Appl. Comput. Topology 4, 339–351. doi:10.1007/s41468-020-00056-w
Campello, R. J. G. B., Moulavi, D., and Sander, J. (2013). “Density-based Clustering Based on Hierarchical Density Estimates,” in Advances in Knowledge Discovery and Data Mining. Editors J. Pei, V. S. Tseng, L. Cao, H. Motoda, and G. Xu (Berlin, Heidelberg: Springer Berlin Heidelberg), 160–172. doi:10.1007/978-3-642-37456-2_14
Carlsson, G. (2014). Topological Pattern Recognition for point Cloud Data. Acta Numerica 23, 289–368. doi:10.1017/s0962492914000051
Carrière, M., Chazal, F., Ike, Y., Lacombe, T., Royer, M., and Umeda, Y. (2020). “Perslay: a Neural Network Layer for Persistence Diagrams and New Graph Topological Signatures,” in International Conference on Artificial Intelligence and Statistics (PMLR), 2786–2796.
Carriere, M., Cuturi, M., and Oudot, S. (2017). “Sliced Wasserstein Kernel for Persistence Diagrams,” in International Conference on Machine Learning PMLR), 664–673.
Chazal, F., De Silva, V., and Oudot, S. (2014). Persistence Stability for Geometric Complexes. Geom Dedicata 173, 193–214. doi:10.1007/s10711-013-9937-z
Chung, Y.-M., and Lawson, A. (2020). Persistence Curves: A Canonical Framework for Summarizing Persistence Diagrams.[Dataset]. http://arxiv.org/abs/1904.07768.
Chung, Y., Hu, C., Lawson, A., and Smyth, C. (2018). “Topological Approaches to Skin Disease Image Analysis,” .in 2018 IEEE International Conference on Big Data (Big Data), 100–105.
Cohen-Steiner, D., Edelsbrunner, H., and Harer, J. (2007). Stability of Persistence Diagrams. Discrete Comput. Geom 37, 103–120. doi:10.1007/s00454-006-1276-5
Dlotko, P. (2021). “Persistence Representations,” in GUDHI User and Reference Manual. 1 edn (GUDHI Editorial Board), 3.4.
Edelsbrunner, H., and Harer, J. (2010). Computational Topology: An Introduction, Vol. 69. Providence, RI: American Mathematical Soc.
Giusti, C., Pastalkova, E., Curto, C., and Itskov, V. (2015). Clique Topology Reveals Intrinsic Geometric Structure in Neural Correlations. Proc. Natl. Acad. Sci. USA 112, 13455–13460. doi:10.1073/pnas.1506407112
Gönen, M., and Alpaydin, E. (2011). Multiple Kernel Learning Algorithms. J. Machine Learn. Res., 2211–2268.
Gromov, M. (2007). Metric Structures for Riemannian and Non-riemannian Spaces. Boston: Springer Science & Business Media.
Kaczynski, T., Mischaikow, K., and Mrozek, M. (2004). Cubical Homology. New York, NY: Springer New York, 39–92. doi:10.1007/0-387-21597-2_2
Kuhn, H. W. (1960). Some Combinatorial Lemmas in Topology. IBM J. Res. Dev. 4, 518–524. doi:10.1147/rd.45.0518
Kusano, G., Hiraoka, Y., and Fukumizu, K. (2016). “Persistence Weighted Gaussian Kernel for Topological Data Analysis,” in International Conference on Machine Learning (PMLR), 2004–2013.
Latschev, J. (2001). Vietoris-rips Complexes of Metric Spaces Near a Closed Riemannian Manifold. Arch. Math. 77, 522–528. doi:10.1007/pl00000526
Le, T., and Yamada, M. (2018). “Persistence fisher Kernel: a Riemannian Manifold Kernel for Persistence Diagrams,” in Proceedings of the 32nd International Conference on Neural Information Processing Systems, 10028–10039.
LeCun, Y., and Cortes, C. (1999). The Mnist Database. [Dataset]. http://yann.lecun.com/exdb/mnist/.
Mileyko, Y., Mukherjee, S., and Harer, J. (2011). Probability Measures on the Space of Persistence Diagrams. Inverse Probl. 27, 124007. doi:10.1088/0266-5611/27/12/124007
Mitra, A., and Virk, Ž. (2021). The Space of Persistence Diagrams on $n$ Points Coarsely Embeds into Hilbert Space. Proc. Amer. Math. Soc. 149, 2693–2703. doi:10.1090/proc/15363
Nathaniel Saul, C. T. (2019). Scikit-tda: Topological Data Analysis for python. doi:10.5281/zenodo.2533369 [Dataset].
Oudot, S. Y. (2015). Persistence Theory: From Quiver Representations to Data Analysis, Vol. 209. Providence, RI: American Mathematical Society Providence.
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011). Scikit-learn: Machine Learning in Python. J. Machine Learn. Res. 12, 2825–2830.
Perea, J. A., Munch, A., and Khasawneh, F. A. (2019). Approximating Continuous Functions on Persistence Diagrams Using Template Functions.CoRR abs/1902.07190
Pickup, D., Sun, X., Rosin, P. L., Martin, R. R., Cheng, Z., Lian, Z., et al. (2014). “SHREC’14 Track: Shape Retrieval of Non-rigid 3d Human Models,” in Proceedings of the 7th Eurographics workshop on 3D Object Retrieval (Eurographics Association), 1–10. EG 3DOR’14
Polanco, L., and Perea, J. A. (2019a). Adaptive template systems: Data-driven feature selection for learning with persistence diagrams. Available at: http://arxiv.org/abs/1910.06741.
Polanco, L., and Perea, J. A. (2019b). “Coordinatizing Data with Lens Spaces and Persistent Cohomology,” in Proceedings of the 31 st Canadian Conference on Computational Geometry (CCCG), 49–57.
Reininghaus, J., Huber, S., Bauer, U., and Kwitt, R. (2015). “A Stable Multi-Scale Kernel for Topological Machine Learning,” in 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 4741–4748.
Reynolds, D. (2009). Gaussian Mixture Models. Boston, MA: Springer US, 659–663. doi:10.1007/978-0-387-73003-5_196
Robins, V., and Turner, K. (2016). Principal Component Analysis of Persistent Homology Rank Functions with Case Studies of Spatial point Patterns, Sphere Packing and Colloids. Physica D: Nonlinear Phenomena 334, 99–117. doi:10.1016/j.physd.2016.03.007
Siu Kwan Lam, A. P., and Seibert, S. (2015). “Numba: a Llvm-Based python Jit Compiler,” in LLVM ’15: Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC, 1–6.
Smith, A., Bendich, P., Harer, J., and Hineman, J. (2017). Supervised Learning of Labeled Pointcloud Differences via Cover-Tree Entropy Reduction. CoRR Abs/1702.07959.
Sonego, P., Pacurar, M., Dhir, S., Kertész-Farkas, A., Kocsor, A., Gáspári, Z., et al. (2007). A Protein Classification Benchmark Collection for Machine Learning. Nucleic Acids Res. 35, D232–D236. doi:10.1093/nar/gkl812
Tralie, C., Saul, N., and Bar-On, R. (2018). Ripser.py: A Lean Persistent Homology Library for python. Joss 3, 925. doi:10.21105/joss.00925
Tschandl, P., Rosendahl, C., and Kittler, H. (2018). The Ham10000 Dataset, a Large Collection of Multi-Source Dermatoscopic Images of Common Pigmented Skin Lesions. Sci. Data 5, 180161. doi:10.1038/sdata.2018.161
Turner, K., Mileyko, Y., Mukherjee, S., and Harer, J. (2014). Fréchet Means for Distributions of Persistence Diagrams. Discrete Comput. Geom 52, 44–70. doi:10.1007/s00454-014-9604-7
Wagner, A. (2019). Nonembeddability of Persistence Diagrams with Wasserstein Metric. arXiv preprint arXiv:1910.13935
wwPDB consortium (2018). Protein Data Bank: the Single Global Archive for 3D Macromolecular Structure Data. Nucleic Acids Res. 47, D520–D528. doi:10.1093/nar/gky949
Keywords: persistent homology, machine learning, topological data analysis, persistence diagrams, barcodes
Citation: Barnes D, Polanco L and Perea JA (2021) A Comparative Study of Machine Learning Methods for Persistence Diagrams. Front. Artif. Intell. 4:681174. doi: 10.3389/frai.2021.681174
Received: 16 March 2021; Accepted: 11 June 2021;
Published: 28 July 2021.
Edited by:
Kathryn Hess, École Polytechnique Fédérale de Lausanne, SwitzerlandReviewed by:
Mathieu Carrière, Institut National de Recherche en Informatique et en Automatique, FranceGregory Henselman-Petrusek, University of Oxford, United Kingdom
Copyright © 2021 Barnes, Polanco and Perea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Danielle Barnes, barnesd8@msu.edu
 Danielle Barnes
Danielle Barnes